Open Access Article
Open Access ArticleAdvancements in nanosensors for detecting pathogens in healthcare environments†
Antonia
Bruno
 a,
Farida
Tripodi
a,
Alice
Armanni
a,
Farida
Tripodi
a,
Alice
Armanni
 a,
Linda
Barbieri
a,
Linda
Barbieri
 a,
Alessandro
Colombo
a,
Sara
Fumagalli
a,
Hind
Moukham
a,
Alessandro
Colombo
a,
Sara
Fumagalli
a,
Hind
Moukham
 a,
Giulia
Tomaino
a,
Ekaterina
Kukushkina
b,
Roberto
Lorenzi
a,
Giulia
Tomaino
a,
Ekaterina
Kukushkina
b,
Roberto
Lorenzi
 b,
Letizia
Marchesi
c,
Angelo
Monguzzi
bd,
Alberto
Paleari
b,
Letizia
Marchesi
c,
Angelo
Monguzzi
bd,
Alberto
Paleari
 b,
Alessandra
Ronchi
b,
Valeria
Secchi
b,
Laura
Sironi
cd and
Miriam
Colombo
b,
Alessandra
Ronchi
b,
Valeria
Secchi
b,
Laura
Sironi
cd and
Miriam
Colombo
 *ad
*ad
aDepartment of Biotechnology and Biosciences, University of Milano-Bicocca, Milan, Italy. E-mail: miriam.colombo@unimib.it
bDepartment of Materials Science, University of Milano-Bicocca, Milan, Italy
cDepartment of Physics, University of Milano-Bicocca, Milan, Italy
dNanomedicine Center NANOMIB, University of Milano-Bicocca, Milan, Italy
First published on 9th September 2024
Abstract
Hospitals serve as critical environments for the management of diverse medical conditions, ranging from routine illnesses to life-threatening emergencies. However, alongside providing healthcare services, hospitals represent reservoirs for the transmission of microbial pathogens. Understanding the distribution and dynamics of pathogens within hospital settings is crucial for effective infection control and prevention strategies. Concurrently, the integration of cutting-edge technologies for the early detection and monitoring of target bacteria stands as a pivotal strategy in this battle against nosocomial infections. This critical review aims to provide a systematic insight into the main threatening microbes in hospitals and the detection of pathogens in different environments, ranging from intensive care units to general wards, including hospital entrances, bathrooms, high-touch surfaces, patient bed rails, medical equipment, and floors, which are often contaminated. We discuss recent scientific and technological advances in pathogen detection by exploring innovative methods that leverage nanotechnology to improve biosensing effectiveness and selectivity. This review is divided into sections focusing on various types of hospital environments, classes of mostly represented pathogens and kinds of available nanobiosensors. We include two comprehensive tables summarizing bacterial contamination in hospital wards and the materials and substrates associated with the nanobiosensors that have been developed. Eventually, we highlight the open challenges and perspectives in nanotechnology-based healthcare-environment monitoring and remediation as a promising solution to counteract pathogen emergence and spread.
Environmental significanceThe COVID-19 pandemic has confronted us with a deeply altered human and environmental scenario that affects both psychological and physical health. In particular, an exponential increase in attention to and care for the cleanliness of habitable spaces and public facilities has been a common experience. This has created a new mentality that requires high environmental control, especially in places and surfaces where pathogens are more likely to accumulate and thrive. This issue is particularly relevant in hospital environments and various hospital wards and departments. In this review, we focused on the various types of pathogens that are most frequently encountered and the most innovative methods based on the recent advances in nanotechnology that are used for their detection and identification, with the aim of making healthcare environments increasingly safe and habitable. |
1. Introduction
According to the global report on infection prevention and control by the World Health Organization (WHO) in 2022, the frequency of health care-associated infection (HAI) varies between countries and according to economic conditions, ranging from 3.2% in the US to 6.5–8.9% in the UAE and up to 9–11% in Southeast Asia and Eastern Mediterranean region (https://iris.who.int/bitstream/handle/10665/354489/9789240051164-eng.pdf?sequence=1). The consequences of HAIs can range from a prolonged hospital stay to long-term complications and disability and premature deaths. These are often associated with the occurrence of sepsis, which has a global incidence of 15.4 cases per 1000 in adult patients and an incredibly high 112.9 cases per 1000 among neonates (https://iris.who.int/bitstream/handle/10665/354489/9789240051164-eng.pdf?sequence=1).Despite great efforts to prevent these hospital-associated infections, there is still a great urgency to implement procedures to limit the risk factors determining these infections. In this context, the development of new technologies to detect pathogens in a fast, cheap and easy-to-use way would be of great help in limiting nosocomial infections.
2. Pathogenic scenario across the main hospital wards
Hospitals are dynamic ecosystems in which patients with diverse medical conditions receive care, ranging from routine treatments to critical interventions. However, amidst the provision of healthcare services, hospitals also harbor a complex interplay of microbial pathogens. Here, we address the hospital microbial landscape as a climax of selective pressure.To do this, we started with a Web of Science-based search, according to the following query: “(hospital ward bacteria detection) OR (hospital care bacteria detection)”. The research was limited to the title and abstract, and it was restricted by publication year (2015/01/01–2024/04/11). We obtained 327 records. Most of the hits (n = 186) are in the intensive care unit, but many records are also focused on surgical contexts (n = 19) and to a lesser extent on general wards (n = 6). Nevertheless, most of the retrieved papers are centered on human-derived samples rather than environmental samples, necessitating further exploration of the literature (ESI S1†).
Tracing their trajectory from intensive care units (ICUs) to general wards, the journey of pathogens across different hospital wards is influenced by selective pressures. The increase in selective pressure, driven by factors such as antimicrobial usage, invasive procedures, and compromised host immunity, fosters the evolution and dissemination of multidrug-resistant organisms (MDROs). As MDROs traverse across hospital settings, they encounter diverse microbial communities and environmental conditions, further shaping their evolutionary trajectories.
2.1 Neonatal intensive care unit (NICU)
There are approximately 6500 newborn deaths every day, amounting to 47% of all child deaths under the age of 5 years (WHO). The neonatal intensive care unit (NICU) plays a crucial role in hospitals by providing intensive care to newborns who face significant health challenges right from birth. National point prevalence surveys conducted in NICUs in the United States in 1999 and in Europe in 2011 revealed that HAIs affected 11.2% of neonates in the US and 10.7% in Europe, respectively.1,2 The most common infections in the NICU fall into two categories: infections that are acquired during the labor and birth process, and hospital-acquired infections that babies contract when they are patients in the NICU. In fact, bacterial contamination was identified on various surfaces, including neonatal incubators, suction tips, ventilators, stethoscopes, door handles, weighing scales, mother beds, laryngoscopes, telephones, and ultrafiltrate bags. These pathogens can survive for varying durations, further complicating the efforts to maintain a sterile environment.3 An additional challenge in preventing infections in the NICU comes from colonized healthcare workers and patients, who can act as sources of pathogens. Contributory factors to the spread of infections include poor hand hygiene, overcrowding, understaffing, inadequate training of staff, and insufficient disinfection or fumigation practices.4 In NICUs, contamination can result from various significant bacterial and fungal pathogens, presenting substantial health risks. Notably, Gram-negative bacteria have been identified as the leading cause of outbreaks, representing 54% with 21 out of 39 reported incidents.5 Key pathogens include Staphylococcus aureus, species of Klebsiella, Escherichia coli, Pseudomonas, Acinetobacter, and Enterococcus species. A systematic study about pathogen concentrations in NICU is still lacking.2.2 Intensive care unit (ICU)
Intensive care units treat patients with severe or life-threatening illnesses and injuries. Immunocompromised patients account for an increasing proportion of the typical ICU cases,6 making them more vulnerable to HAIs from pathogens. The frequent use of antimicrobials and the high cleaning practices, added to the clinical conditions of patients, create a unique microbiota characterized by common bacteria, opportunistic pathogens,7 and MDROs.8 This makes treatment more difficult9 and contributes to substantial mortality and morbidity. ICUs are the hospital wards with the highest number of nosocomial infections, regarding lower respiratory tract infections, urinary tract infections and bloodstream infections in order of decreasing incidence rates.8,10,11 Infections can be caused by the use of invasive devices12 and by the transmission of pathogens from contaminated surfaces and the gloves of healthcare personnel.13 In the ICUs, there is nearly the same likelihood for sanitary workers to have contamination on their hands or gloves after touching the environment in a room where a patient harboring pathogens resides because there is after directly touching the colonized patient and their surroundings.13 This situation starts a circular process: the patients, the gloves or the environment could be the starting point of the infectious transmission.14 Different surfaces are contaminated by several microorganisms, such as S. aureus, coagulase-negative staphylococci, Pseudomonas aeruginosa, Acinetobacter baumannii, Corynebacterium spp., and Bacillus spp.2.3 General surgical ward
Patients admitted to general surgical wards are those who have undergone surgical procedures or require postoperative care for their surgical conditions. In addition to the general medical ward, it caters to different patient populations and medical needs. However, patients in general surgical wards typically have shorter lengths of stay following surgical procedures although this may vary depending on the complexity of the surgery and postoperative recovery. This means a high heterogeneity in the composition of patients undergoing different treatments. Bacterial pathogens in these wards can contribute to surgical site infections (SSIs). Thus, prior to surgical procedures, patients in general surgical wards may receive prophylactic antibiotics to reduce the risk of surgical site infections. These antibiotics are typically administered shortly before surgery and discontinued within 24 hours postoperatively, adhering to antimicrobial stewardship principles. The most threatening infections are due to multidrug-resistant E. coli.15E. coli is a ubiquitous bacterium implicated in SSIs, urinary tract infections, and bloodstream infections in general surgical wards. These bacteria can contaminate surgical sites through fecal–oral transmission or cross-contamination from the gastrointestinal tract.16 Notably, Cassini et al.15 demonstrated that a large proportion of the burden of antibiotic resistance bacteria was due to health-care-associated bloodstream, respiratory tract, or surgical site infections, and that more than half of health-care-associated infections are considered preventable. The mean bacterial colony count on surfaces in surgical wards was reported at 48.8 CFU cm−2, indicating a potential source of contamination.172.4 General medical ward
The general medical ward provides acute medical services for adults of all ages across a wide variety of specialties (such as gastroenterology, endocrinology, respiratory medicine, rheumatology, and cardiology) but does not necessitate specialized treatment or monitoring found in ICUs or specialty wards.Due to the diverse range of medical conditions and patients admitted, and to the dynamicity of this environment, a high heterogeneity of pathogens can be encountered: S. aureus, Streptococcus pneumoniae, Klebsiella pneumoniae, E. coli, and P. aeruginosa are some examples.18 Common areas show shared bacteria according to exposure (e.g., high touch and high foot traffic sites, such as patient bed rails, medical equipment, and floor). Moreover, more than one study18,19 demonstrated that the detection rate of bacteria in general wards is higher than that in intensive care units, supporting the evidence that different cleaning procedures can affect bacteria proliferation. Noteworthy, methicillin-resistant S. aureus (MRSA), but not vanB-positive vancomycin-resistant enterococci (VRE), was also detected at a high rate in a newly opened hospital. Bacterial load plateaued at a significantly higher level in common areas than in inpatient rooms (p < 0.001, common area median: 2.44 × 105 CFU per swab [2.42 × 105]; inpatient area median [IQR]: 1.10 × 105 CFU per swab [2.17 × 105]).20
Understanding the microbial landscape of general medical wards is essential for patient outcomes and healthcare-associated infection rates.
2.5 Main entrance
The main entrances of healthcare facilities are environments characterized by the largest flux of people, including patients, visitors and staff. Hospital lobby areas and bathrooms are characterized by the presence of viruses,21 fungi,22 and bacteria, such as Citrobacter freundii, Stenotrophomonas maltophilia,23 and S. aureus,24 as reported in Table 1. Because S. aureus is a skin commensal,25 it is found in the most crowded hospital areas and surfaces.26 The main sources of bacteria in this environment are people and the exchange between indoor and outdoor air. Moreover, floor cleaning and maintenance activities contribute to increased bacteria and fungi levels (the average levels of bacteria and fungi were 7.2 × 102 CFU m−3 and 7.7 × 10 CFU m−3, respectively).27 Despite the number of people, the major transmission of HAIs occurs in areas frequented by the patients and the medical staff, so areas with visitors, such as the main entrances, are less contaminated by pathogens or opportunistic bacteria.26| Hospital wards | Surfaces | Microorganisms | Ref. |
|---|---|---|---|
| ICU | Electrocardiography leads | Enterococcus (VRE) | 29 |
| Coagulase-negative staphylococci | 30 | ||
| P. aeruginosa | |||
| A. baumannii (CRAB) | 31 | ||
| Enterobacteriaceae spp. | 32 | ||
| Cardiac monitor | A. baumannii | 33 | |
| Blood pressure cuffs | S. aureus (MRSA) | 34 | |
| A. baumannii (CRAB) | 31 | ||
| C. difficile | 35 | ||
| Enterobacteriaceae spp | 32 | ||
| Ventilator (e.g., buttons, circuits) | S. aureus | 36 | |
| P. aeruginosa | |||
| Suction system | S. aureus | 36 | |
| P. aeruginosa | |||
| S. maltophilia | 37 | ||
| Medical charts | Coagulase-negative staphylococci | 8 | |
| A. baumannii | |||
| K. pneumoniae | |||
| Portable radiography equipment | S. aureus (MRSA) | 38 | |
| Enterococcus (VRE) | |||
| A. baumannii | |||
| K. pneumoniae | |||
| P. aeruginosa | |||
| S. maltophilia | |||
| Ultrasound machine | S. aureus (MRSA, MSSA) | 39 | |
| Coagulase-negative staphylococci | 40 | ||
| P. aeruginosa | |||
| A. baumannii | |||
| Corynebacterium spp. | |||
| Bacillus spp. | |||
| Bed | A. baumannii | 41 | |
| S. aureus (MRSA) | 42 | ||
| E. faecium (VRE) | 9 | ||
| Enterobacteriaceae spp. | 32 | ||
| Stethoscopes | S. aureus | 43 | |
| A. baumannii | |||
| Personnel's uniforms and hands | A. baumannii | 44 | |
| A. baumannii (CRAB) | 31 | ||
| Enterococcus (VRE) | 13 | ||
| Telephone/cell phones | A. baumannii | 45 | |
| A. baumannii (CRAB) | 31 | ||
| Coagulase-negative staphylococci | 46 | ||
| S. aureus | |||
| Non-fermenting Gram-negative bacteria | |||
| Computer (keyboards and/or mouse) | Coagulase-negative staphylococci | 47 | |
| Non-fermenting Gram-negative bacteria | |||
| S. aureus (MRSA) | 33 | ||
| Television | A. baumannii | 33 | |
| S. aureus (MRSA) | |||
| Sink | Klebsiella spp. | 48 | |
| A. baumannii | 33 | ||
| S. aureus (MRSA) | 14 | ||
| K. oxytoca | 48 | ||
| K. pneumoniae | |||
| E. cloacae | |||
| E. asburiae | |||
| C. freundii | |||
| E. coli | |||
| Pantoea spp. | |||
| S. marcescens | |||
| P. aeruginosa | 49 | ||
| Ultrafiltrate bag | P. aeruginosa | 49 | |
| Floor | S. aureus (MRSA) | 33 | |
| Sanitary equipment and toilet | E. coli | 50 | |
| K. pneumoniae | |||
| P. aeruginosa | |||
| S. aureus | |||
| Chair | A. baumannii | 33 | |
| Patient's table | A. baumannii | 33 | |
| S. aureus (MRSA) | 14 | ||
| Door handle or push plate | S. aureus (MRSA) | 14 | |
| A. baumannii (CRAB) | 31 | ||
| NICU | Neonatal incubator | S. marcescens | 51 |
| 52 | |||
| P. aeruginosa | 3 | ||
| Klebsiella spp. | |||
| E. coli | |||
| Enterococcus spp. | |||
| E. faecium | 53 | ||
| S. aureus | |||
| Weighing machine | Klebsiella spp. | 3 | |
| E. coli | |||
| Enterococcus spp. | |||
| Laryngoscope | E. coli | 3 | |
| S. aureus | |||
| Ventilator (e.g., buttons, circuits) | S. marcescens | 51 | |
| E. coli | 3 | ||
| Suction system | E. coli | 3 | |
| P. aeruginosa | |||
| Klebsiella spp. | |||
| A. baumannii | |||
| Enterococcus spp. | |||
| Bed | E. coli | 3 | |
| P. aeruginosa | |||
| Klebsiella spp. | |||
| A. baumannii | |||
| Stethoscopes | S. aureus | 3 | |
| Enterococcus spp. | |||
| Personnel's uniforms and hands | S. marcescens | 51 | |
| Telephone/cell phones | A. baumannii | 3 | |
| Enterococcus spp. | |||
| Sink | S. marcescens | 51 | |
| P. aeruginosa | 54 | ||
| Ultrafiltrate bag | Klebsiella spp. | 3 | |
| Door handle or push plate | S. aureus | 3 | |
| Enterococcus spp. | |||
| General surgical ward | Bed rails | Bacillus cereus group, Enterococcus (faecalis and faecium), M. luteus, Staphylococcus spp., Streptococcus spp. | 55 |
| Keyboard | Bacillus cereus group, M. luteus, Staphylococcus spp. | 55 | |
| Simulation manikin | Bacillus cereus group, M. luteus, Staphylococcus spp. | 55 | |
| Table | Bacillus cereus group, E. faecalis, M. luteus, Staphylococcus spp., Streptococcus spp. | 55 | |
| Workstations-on-wheels | Bacillus cereus group, E. faecalis, M. luteus, Staphylococcus spp., Streptococcus spp. | 55 | |
| Faucet, basin, and drain of sinks | E. coli | 56 | |
| Aeromonas spp. | |||
| S. aureus | |||
| S. epidermidis | |||
| Bed, taps, door handles | S. aureus | 57 | |
| Pseudomonas spp. | |||
| Enterobacteria | |||
| E. faecalis | |||
| Floor, walls, equipment, instruments, operation tables, sink, light switch, chairs, beds, patient cloths, door/locker handlers, trolley, stretchers, sinks/faucets, intravenous stands, and oxygen cylinder | S. aureus | 17 | |
| Klebsiella spp. | |||
| General medical ward | Door handles/knobs | S. aureus | 18 |
| E. coli | |||
| Citrobacter spp. | |||
| K. pneumoniae | |||
| P. aeruginosa | |||
| S. pneumoniae | |||
| Proteus vulgaris | |||
| Bacillus spp. | |||
| Acinetobacter spp. | |||
| Coagulase negative | |||
| Staphylococcus | |||
| Enterobacter spp. | |||
| Enterococcus spp. | |||
| Micrococcus spp. | |||
| Diphtheroids | |||
| Bedrail, bedroom floor and toilet flush | S. aureus | 20 | |
| Enterococcus spp. | |||
| Hospital bed units | P. aeruginosa | 19 | |
| E. cloacae | |||
| Medical instruments | K. pneumoniae | 19 | |
| Water taps, thermos bottles, treatment carts, and dishcloths | P. aeruginosa | 19 | |
| E. cloacae | |||
| K. pneumoniae | |||
| Main entrance | Sink | C. freundii | 24 |
| S. maltophilia | 23 | ||
| Door handle or push plate | S. aureus (MSSA) | 24 |
2.6 Environmental sampling challenges in hospital wards
There is a growing body of evidence indicating that hospital surface environments contribute significantly to the dissemination of pathogens. However, the optimal methods for sampling these surfaces remain unclear, and there is a lack of standardized guidelines or legislation to direct these practices. Currently, there is no legal mandate requiring hospitals to conduct routine environmental monitoring of surface contamination. Hospitals that opt to conduct sampling often rely on in-house protocols or guidelines adapted from the food and pharmaceutical industries. Comprehensive, evidence-based guidelines specific to hospital surface sampling are notably absent, and there are limited studies on the efficacy of various sampling methods under the diverse conditions present in hospital environments. Recent reviews58,59 have detailed the methodologies for sample collection, highlighting the advantages and disadvantages of various approaches. Specifically, it has been noted that both elution-dependent methods (such as pre-moistened swabs, sponges, and wipes) and elution-independent methods (such as replicate organism detection and counting plates, 3M Petrifilm™ plates, and dipslides) require the presence of moisture and neutralizers during sampling to enhance recovery rates.Most of the studies reviewed were conducted in laboratory settings rather than in actual hospital environments, where numerous variables can influence sampling recoveries. Previous studies do not provide a comprehensive understanding of the hospital surface microbiome mainly due to the scarcity of studies examining the general environment outside of outbreak situations, the tendency to focus on specific organisms or pathogens, and the variability in sampling methods, result analyses, and units of measurement (e.g., few studies report results in colony-forming units per square centimeter). This variability complicates cross-study comparisons.
Thus, integrating surface sampling methodologies into the workflow of hospital environments presents several challenges. One significant hurdle is the lack of standardized protocols, as mentioned before, which necessitates the development of customized guidelines that can be consistently applied across various hospital settings. This customization is time-consuming and requires substantial training of staff to ensure accurate and reliable sampling.
Moreover, the hospital environment is characterized by a high degree of variability, including differences in surface materials, cleaning practices, and the presence of diverse microbial communities. These factors can influence the effectiveness of sampling methods, making it difficult to develop a one-size-fits-all approach. Additionally, the integration of routine sampling into the daily operations of a hospital requires coordination across multiple departments and disciplines, potentially disrupting clinical workflows and patient care activities.
Finally, the implementation of routine environmental monitoring must be supported by robust data management and reporting systems to track and analyze trends over time. This requires the integration of new technologies and software, which can be a complex and resource-intensive process. Overcoming these challenges is essential to ensure that environmental monitoring effectively contributes to infection control and patient safety in hospital environments. Finally, the awareness that hospitals are fully fledged ecosystems, representing one of the most peculiar built environments with its microbiome: beyond pathogens, a large community of microorganisms, many harmless and some even potentially beneficial, lives in hospitals. These microbial communities could form a kind of “immune system”, decreasing opportunistic pathogen accumulation and persistence in hospitals.60
3. The most concerning pathogens in the hospital environment
Six species of pathogens are recognized as particularly threatening due to their potential MDR mechanisms and pathogenicity. These are called ESKAPEE pathogens due to their ability to “escape” from commonly used antibiotics due to their increasing multi-drug resistance. They include Enterococcus faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, Enterobacter spp., and E. coli;61,62 along with Clostridioides difficile, these are the most common bacteria causing nosocomial infections.63 A short description and pathogenicity of these bacteria are presented below.3.1 Enterococcus faecium
Enterococci, particularly Enterococcus faecium, have emerged as significant causative agents of infections in humans.60 They are known to be associated with hospital-acquired infections and are linked to a high rate of mortality.61 In fact, studies in population genetics and genomics have revealed the existence of two separate subpopulations within the species E. faecium. The first group is primarily found as harmless inhabitants of the gastrointestinal tract, rarely causing clinical infections. However, the second group consists of hospital-associated E. faecium strains.62 In addition, E. faecium can resist antibiotics and environmental stressors.63 Therefore, the continual use of antibiotics in the hospital environment has significantly contributed to the evolution of E. faecium into a highly adept pathogen within hospital environments. It is important to highlight that a significant majority of modern E. faecium isolates exhibit strong resistance to ampicillin, and a considerable number of these isolates, varying by geographic location, show resistance to glycopeptides.63 The rapid rise in hospital-acquired infections from E. faecium, coupled with limited treatment options, is due to the bacterium's rising resistance to antibiotics and the prevalent challenge of biofilm-associated infections.64–683.2 Staphylococcus aureus
S. aureus is a cocci-shaped Gram-positive bacterium that tends to cluster in “grape-like” bunches. It takes the name “aureus” from the Latin word “gold” due to the golden colonies observed in culture medium.69S. aureus inhabits the environment, and it is part of the human microbiota; however, when entered into the bloodstream or internal tissue, it is responsible for a wide variety of clinical manifestations25,70 such as ocular and skin infections, endocarditis, central nervous system infections, and pneumonia.71S. aureus pathogenic strains include vancomycin-resistant S. aureus (VRSA) group and MRSA group63 that survive on surfaces from 6 h on stainless steel to 3 years on polyethylene.35 In high-income countries, such as those in the European Union and European Economic Area (EU/EEA), MRSA is one of the three most impactful antibiotic-resistant microorganisms, together with E. coli and carbapenem-resistant P. aeruginosa. They are commonly acquired in healthcare settings and determine 70% of the burden of AMR in terms of disability and premature mortality, such as disability-adjusted life-years72 (Global Report on Infection Prevention and Control, 2022). As presented in Table 1, S. aureus has a wide distribution in different sites and wards of hospitals, and this has brought the draft of the guidelines for MRSA in 52.5% of countries in 2021. The globally medium proportion of MRSA was 24.9% (interquartile range (IQR) 11.4–42.7) in 2020, and it was 15.5% in EU/EEA countries in 2019.72,73 The percentage of MRSA isolates with resistance found in North America, Europe and Northeast Asian countries varies from less than 5% in the Scandinavian region to 60% in the U.S.A and China in 2022.74 Moreover, patients infected with MRSA infections have an essential increase in post-infection length of stay, septic shock and mortality compared with those with methicillin-susceptible S. aureus (MSSA) infections, where the risk for discharge to long-term care is more than doubled.723.3 Klebsiella pneumoniae
K. pneumoniae is a Gram-negative bacterium of the Enterobacteriaceae family that can be symbiotic with its host by colonizing intestinal mucosa, skin and nasopharynx. However, it can also be a pathogen in humans, often causing nosocomial infections, such as urinary tract infections, blood infections and pneumonia. For other bacteria, the excessive use and misuse of antibacterial agents has led to an increase in resistance to antibiotics and the emergence of carbapenem-resistant (CRKP) and MDR K. pneumoniae strains.75 Antibiotic resistance of this bacterium is further enhanced by biofilm formation, which can protect the pathogen from the host immune responses and can decrease the antibiotic effects, thus making clinical management of K. pneumoniae infection more complicated.763.4 Acinetobacter baumannii
A. baumannii is a Gram-negative bacterium that causes HAIs, especially affecting patients in intensive care units. It is responsible for hospital-acquired bloodstream infections and pneumonia, and it is particularly prone to cause outbreaks owing to its ability to survive prolonged periods on dry surfaces under unfavorable environmental conditions and to acquire antibiotic resistance.77 During an outbreak, A. baumannii can be found on linen, furniture, and sinks, as well as on medical equipment, such as ventilator tubing. It can also form biofilms on both non-living and biological surfaces, increasing its resistance to antibiotic agents and yielding medical device-related infections. It can be transmitted through air droplets or the skin of infected patients; however, the most common way of transmission is through the hands of healthcare workers.78 Consequently, A. baumannii carbapenem-resistant (CRAB) bacterium is considered one of the priority pathogens for WHO.793.5 Pseudomonas aeruginosa
P. aeruginosa is a Gram-negative bacterium of the family Pseudomonadaceae present in multiple ecological niches, such as soil and aquatic environments, and plant and animal tissues, due to its metabolic versatility.80 It is also an opportunistic pathogen that causes acute or chronic infections in immunocompromised individuals, such as patients with cystic fibrosis and cancer, as well as in patients in the intensive care unit. It can colonize medical equipment and the hospital environment, leading to HAIs, such as pneumonia, urinary tract infections and bloodstream infections.81 Strikingly, P. aeruginosa can exist in both planktonic form and biofilm, which are especially dangerous because they can infect medical devices but are also particularly harmful for patients with cystic fibrosis who frequently succumb to a chronic infection of the lungs.82 In fact, P. aeruginosa infections are extremely difficult to treat due to antibiotic resistance.83 For these reasons, P. aeruginosa is listed in the critical category of the WHO's priority list of bacterial pathogens.793.6 Enterobacter spp.
Enterobacter comprises a group of common Gram-negative bacteria characterized by rod-shaped, facultative anaerobic properties. Flagella in Enterobacter species serve multiple functions, including adhesion, biofilm formation, protein export, and motility. Each species within the genus produces unique endotoxins, and as Gram-negative bacteria, they possess a lipopolysaccharide capsule that aids in evading phagocytosis and triggering inflammatory responses. Among these bacteria, certain strains can cause opportunistic infections in individuals with compromised immune systems, particularly those in hospital settings or those undergoing mechanical ventilation. Infections most commonly affect the urinary and respiratory tracts. Over the past three and a half decades, Enterobacter aerogenes (now known as Klebsiella aerogenes) and Enterobacter cloacae have emerged as significant threats in neonatal wards and intensive care units, especially among mechanically ventilated patients.84 Around 2010, E. cloacae became more prevalent than E. aerogenes as the most frequently isolated species. It is important to note that within the E. cloacae complex, other clinically relevant members exist, often posing challenges for accurate species identification using standard tests.85 MDR Enterobacter species are increasingly causing infections acquired in hospital settings. Prior to 2005, almost all Enterobacter strains were susceptible to carbapenems, but carbapenem resistance has now been reported in all WHO health regions.863.7 Escherichia coli
E. coli is a common Gram-negative bacterium of the family Enterobacteriaceae naturally present as a commensal of the intestinal tract of humans and other animals, with an important role in digestion. However, several pathotypes also exist, which cause infections, such as enteric/diarrhoeal disease, urinary tract infections and sepsis/meningitis, leading to two million deaths annually.87,88 The intestinal E. coli strains can be divided into six well-described categories: enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC) and diffusely adherent E. coli (DAEC).88 Human infection can be acquired through contaminated food/water or via direct contact with an infected person, while in neonates, E. coli infections often occur through the maternal genital tract.89 Clinically, E. coli infections are commonly treated with ciprofloxacin, levofloxacin, fosfomycin and fluoroquinolones; however, resistance to multiple antibiotics has been reported, making E. coli one of the more dangerous pathogens. In 2019, E. coli infections were responsible for more than 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths associated with antimicrobial resistance in Europe.90
000 deaths associated with antimicrobial resistance in Europe.90
3.8 Clostridioides difficile (previously known as Clostridium difficile)
C. difficile is a rod-shaped Gram-positive anaerobic spore-forming bacterium.91 Due to its difficulty in isolation and slow growth, it was given the name “Bacillus difficilis” from the Latin of difficult, changed to C. difficile in the 1970s.92 It is part of animal and human gut microbiota93 but becomes pathogenic in C. difficile infections (CDI). CDIs are caused primarily by clostridial toxin A (TcdA) and/or toxin B (TcdB) and are the leading cause of hospital-acquired diarrhea and colitis.94 These diseases are often related to antibiotic treatments because they unbalance the gut microbiota composition; in this situation, C. difficile has the opportunity to multiply and produce its toxins.92 Moreover, it can survive on various materials, from 15 minutes on dry surfaces to 6 hours on wet items, while its spores exhibit high resistance and can be found on floors and equipment for up to 5 months.35 Due to its pathogenicity, the involvement with antibiotic treatments and the multidrug-resistance of some strains,95 23.7% of countries in the world have guidelines for C. difficile.96As clearly appears, different surfaces can harbor different bacterial species, and different wards constitute peculiar environments. This is due to various factors; for instance, cleaning procedures, surface material, temperature, relative humidity, and initial titer can affect the range of survival times of bacteria. Moreover, survival is species specific and can vary largely among different microorganisms. However, it is worth mentioning that extensive studies have been conducted on selected species, such as E. coli and A. baumannii, but the survivability of many species remains widely unexplored. One reason for the lack of specific data is the unavailability of cellular model systems to study the respective pathogens. Many clinically relevant bacteria remain infectious on inanimate surfaces and can survive for months on dry surfaces. In vitro studies can provide initial indications to assess the risk of transmission of a particular pathogen by fomites; however, the conditions presented in various experimental studies often do not resemble real-life scenarios (e.g., large inoculums and small surface areas) and therefore require careful interpretation. Moreover, the fraction of pathogens transferred depends on multiple factors, including species and surface material. The efficiency of the transfer of a pathogen between fomite and skin is a critical parameter for modeling its potential for transmission and implementing effective hygiene measures while avoiding unnecessary ones.
Thus, there is a significant gap in knowledge regarding this specific issue, as research is often fragmented into individual, non-standardized studies.97
Fig. 1 and Table 1 present the most common areas (wards and surfaces) where the most concerning pathogens have been detected.
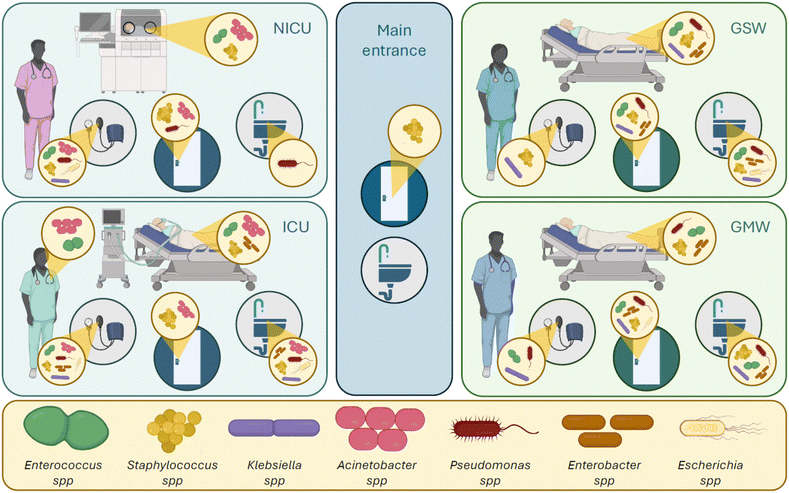 | ||
| Fig. 1 Most concerning pathogens in different hospital wards and surfaces. Starting from Table 1, the image shows various surfaces in different hospital departments contaminated with ESKAPEE pathogens and other congeneric pathogen species. The surfaces are divided into personnel uniforms and/or hands, represented by the nurse image; beds and/or bed rails and sheets, represented by the hospital bed and the neonatal incubator in NICU; medical equipment, represented by the sphygmomanometer; room furniture and/or door handles, represented by the door; and bathroom, represented by the sink. The hospital wards are ICU – intensive care unit; NICU – neonatal intensive care unit; main entrance; GSW – general surgical ward; GMW – general medical ward. At the bottom is the bacterial genera legend the image is made using Biorender. | ||
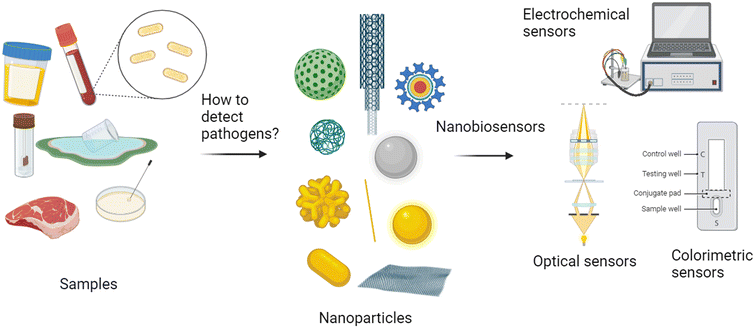
4. Innovative methods for pathogen detection exploiting nanotechnology
In conventional laboratories, colony counting culture, polymerase chain reaction (PCR), Gram staining, and analytical profile index (API) systems are some of the most widely used techniques for identifying the presence of pathogens in clinical samples.98–100 These strategies are often analytically concrete in terms of selectivity and reliability. However, these methods are expensive, time-consuming, and require qualified staff to interpret the results when the experiment is completed.101–103With the increasing attention to point-of-care (POC) testing, there is a need for rapid and reliable diagnostic tools that can be used directly at the patient's bedside or in community settings. Rapid detection platforms fulfill this need by providing quick results for on-the-spot decision-making.104 Biosensors, with their unique features, play an important role in meeting the demand for POC tests. They provide rapid and real-time results, are portable and cost-effective, and generally exhibit high sensitivity and selectivity, providing quick, accurate, and user-friendly diagnostic solutions that are ideal for decentralized healthcare settings.105,106
A biosensor is a device with a biological sensing component integrated into or closely connected to a transducer. The ability to miniaturize the transduction element and the absence of an economical production method are frequently the primary obstacles to developing POC and sensing devices.107
Nanotechnologies have become highly valuable in the field of biosensing due to their versatility and exceptional properties.108,109 Specifically, characteristics of nanomaterials, such as their high reactive capacity, high adsorption, quantum size effects, and surface-to-volume ratio compared to their bulk form, are essential for developing biosensing methods.110 Furthermore, as nanomaterials can be easily tailored in terms of size and shape, it is possible to modify or immobilize their surfaces with various biological species through covalent or non-covalent bonding, improving the biosensing characteristics in terms of high sensitivity, selectivity, and quick response to the analytes in the sample.111–113
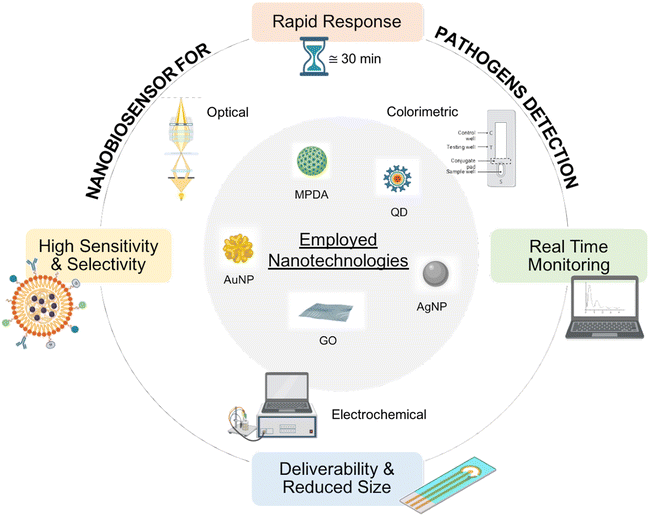
POC biosensors typically leverage one or more of the following six approaches to signal transduction: I) optical, II) electrochemical, III) mechanical, IV) magnetic, V) thermometric, and VI) microgravimetric. Electrochemical and optical techniques are the most frequently used and sensitive ones for chemo- and biosensors.114 Among the optical techniques, the most appealing for the production of point-of-care devices is colorimetric because it does not require sophisticated instruments and qualified personnel.115,116
4.1 Two fundamental parameters: sensitivity and selectivity towards pathogenic subtypes
To enable efficient risk assessment critical for healthcare, food safety analysis, and environmental monitoring, effective pathogen diagnostic methods have to be rapid, ultrasensitive, specific and affordable to be applied in low-resource settings.117 When designing biosensors, sensitivity is a crucial parameter to consider because it is the ability to quantify the analyte within a wide range of concentrations. Another equally important aspect is the sensitivity of the device, expressed as the limit of detection (LoD) of the colony forming units (CFU) per mL or the ng mL−1 of the analyte to be detected. Ideally, the higher the sensitivity, the lower the minimum concentration of the analyte that can be detected.In this respect, the use of nanotechnologies plays a significant role in improving the sensing capability of the detection of biosensors. Various nanomaterials, such as gold nanoparticles (AuNPs), quantum dots (QDs), carbon nanotubes (CNT), metal nanoclusters (MNCs), and up-conversion nanoparticles (UCNPs), are integrated into biosensors to enhance the sensitivity and stability of these devices by amplifying their signal and expanding detection limits.118 Some examples of nanotechnology-based biosensors are provided in the next paragraph. These nanomaterials can be manipulated to contain functionality for specific molecular recognition. Several recognition elements have been explored so far, including enzymes, antibodies, nucleic acids, aptamers, and cells. Their selective interaction with a particular analyte determines the effectiveness of the biosensor. Thus, the selectivity of the biosensor towards a certain target or strain changes depending on the recognition element used on the biosensor. To obtain excellent detection results, the development of bacterial recognition elements with higher efficiency and specificity is significantly needed.119
Antibodies used as recognition elements provide high specificity due to their unique antigen–antibody interactions but are often associated with high costs and lack of reproducibility. Moreover, the instability of protein-based recognition elements presents challenges in maintaining long-term sensor performance.120 A less expensive alternative is represented by aptamers, short, single-stranded DNA or RNA molecules (20–100 nucleotides in length) with defined structures that can specifically bind to a wide range of targets via three-dimensional structures. However, aptamers are prone to degradation, and assays using a single aptamer as a recognition element are less specific.121 Combinations of antibiotics and aptamers as dual recognition elements can be used to increase selectivity. For example, Shen et al. developed the broad-spectrum glycopeptide antibiotic vancomycin (Van) and aptamer-based dual-recognition CD nanoprobe combined with quantum dots to detect S. aureus via ratiometric fluorescence.122 The detection time of the method is 30 min with a LoD of 1.0 CFU mL−1.119 Other examples are presented in Table 2. Overall, by carefully choosing the optimal combination of nanoparticles and recognition elements, one can design highly selective and sensitive nanotechnology-based biosensors. Nevertheless, further advancements are required to reduce their limitations and enhance their efficiency.
| Pathogen | Transduction method | Nanotechnology | Recognition element | Assay time | Linear range | LOD | Sample | Ref. |
|---|---|---|---|---|---|---|---|---|
| Abbreviations: Au: gold; Cit-Ag NPs: citrate capped silver nanoparticles; NCND: nitrogen-doped carbon nanodots; GO: graphene oxide; MNPs: magnetic nanoparticles; AuNPs: gold nanoparticles; Ag: silver; AgNPs: silver nanoparticles; AuNCs: gold nanoclusters; CDs: carbon dots; Van: vancomycin; BCDs: blue carbon dots; Tb: terbium; MOF: metal–organic framework; COF-BA: boronic acid-functionalized covalent-organic framework; @: conjugation; Fe3O4/Au-PEI NPs: polyethyleneimine coated magnetic gold nanoparticles; CO NPs: cupric oxide nanoparticles; ICA: indole-5-carboxylic acid; MPDA: mesoporous polydopamine; PtNPs: platinum nanoparticles; UCNPs: up-conversion nanoparticles; CQDs: carbon quantum dots; NFs: nanofibers; CNPs: carbon nanoparticles; QDs: quantum dots; ALP: alkaline phosphatase. | ||||||||
| Acinetobacter baumannii | Electrochemical | Au-electrode with beta cyclodextrin | DNA probe | 105 min | 0.3 nM to 0.24 μM | 0.14 nM | Food | 123 |
| Cit-Ag NPs | DNA probe | 2 min after DNA hybridization | 1 μM to 1 ZM | 1 ZM (LLOQ) | Bacteria DNA | 124 | ||
| Optical | Au nanoprisms with Fe(III) siderophore | LSPR-based whole-cell sensing with aptamer-based molecular recognition motifs | 3h | 4 × 102 to 4 × 106 CFU mL−1 | 80 cell per mL | Bacterial culture | 125 | |
| NCND/GO | Aptamer ssDNA | 20 min | 2 × 103 to 4.5 × 107 CFU mL−1 | 3 × 102 CFU mL−1 | Bacterial culture, urine sample | 126 | ||
| MNPs | Tail fiber proteins | 10 min | 1 × 104 to 1 × 105 cells per mL | 4.48 × 104 cell per mL | FBS | 127 | ||
| Clostridioides difficile | Electrochemical | AuNPs | Antibody | — | 1 pg mL−1 to 100 pg mL−1 of toxin | 0.61 pg mL−1; 0.60 pg mL−1 of toxin | Stool sample | 128 |
| Enterococcus faecalis | Electrochemical | Au crystals | DNA aptamer, toluidine blue (TB) as DNA hybridization indicator | 6 h (immobilization time); 5 min (TB binding time) | 10−17 to 10−10 M | 4.7 × 10−20 M | Urine and stool sample | 129 |
| Cysteine-modified AuNPs | Clavanin A peptide | — | 101 to 104 CFU mL−1 | 10 CFU mL−1 | Bacterial culture | 130 | ||
| Colorimetric | Quercetin-mediated AgNPs | LAMP amplification DNA | — | 10 to 105 CFU mL−1 | 10 CFU mL−1 | Food | 131 | |
| Escherichia coli | Optical | AuNCs | Cu reduction | <30.0 min | 103 to 106 CFU mL−1 | 89 CFU mL−1 | Bacterial culture | 132 |
| AuNPs plus CeO2NPs | Aptamer and azithromycin | <30.0 min | 10 to 1.5 × 105 CFU mL−1 | 1.04 CFU mL−1 | Food | 133 | ||
| AgNCs plus MNPs | RNA-cleaving DNAzyme probe | — | 102 to 107 CFU mL−1 | 60 CFU mL−1 | Food, tap water | 134 | ||
| MNPs | Fluorescent proteins | <30.0 min | — | 108 CFU mL−1 | Bacterial culture | 135 | ||
| CDs | Receptors (boronic acid, polymixin and Van) | >60.0 min | — | OD600 = 1.0 | Tap water | 136 | ||
| BCDs | Cu quenching and reduction | — | 103 to 107 CFU mL−1 | 1.5 × 102 CFU mL−1 | Food | 137 | ||
| Tb-MOF | Antibody | 5.0 min | 1.3 × 102 to 1.3 × 108 CFU mL−1 | 3 CFU mL−1 | Food | 138 | ||
| COF-BA Au@Ag nanoparticles | Magnetic IgG@Fe3O4 nanoparticles | 30 min | 10 to 103 CFU mL−1 | 10 CFU mL−1 | Bacterial culture | 139 | ||
| Colorimetric | Fe3O4/Au-PEI NPs | Antibody | 60 min | 10 to 107 CFU mL−1 | 0.52 CFU mL−1 | Clinical sample | 140 | |
| Electrochemical | SiO2-NPs | Polyclonal antibodies | 30 min | 8 × 104 to 8 × 106 CFU mL−1 | 2 × 103 CFU mL−1 | Bacterial culture | 141 | |
| CO NPs | Cu reduction | 60 min | 103 to 107 CFU mL−1 | 2 CFU mL−1 | Food and water | 83 | ||
| Klebsiella pneumoniae | Colorimetric | AuNPs | Aptamer | 1 min | 102–108 CFU mL−1 | 3.4 × 103 CFU mL−1 | Bacterial culture, clinical sample, urine | 142 |
| Optical | COF-BA Au@Ag nanoparticles | Magnetic IgG@Fe3O4 nanoparticles | 30 min | 10 to 103 CFU mL−1 | 10 CFU mL−1 | Bacterial culture | 139 | |
| Electrochemical | GO–ICA hybrid film | ssDNA aptamer | — | 10−6 to 10−10 M | 3 × 10−11 M | — | 143 | |
| Pseudomonas aeruginosa | Electrochemical | AuNPs | ssDNA aptamers | 10 min | 60 to 6 × 107 CFU mL−1 | 60 CFU mL−1 | Bacterial culture | 144 |
| CO NPs | Cu reduction | 60 min | 103 to 107 CFU mL−1 | 1.6 × 104 CFU mL−1 | Food and water | 83 | ||
| Colorimetric | MNPs and gold | Specific protease substrate peptide | 1 min | 45 to 4.5 × 107 CFU mL−1 | 102 CFU mL−1 | Clinical sample | 145 | |
| Optical | COF-BA Au@Ag NPs | Magnetic IgG@Fe3O4 nanoparticles | 30 min | 10 to 103 CFU mL−1 | 10 CFU mL−1 | Bacterial culture | 139 | |
| Staphylococcus aureus | Electrochemical | MPDA/MnO2 | SA31 aptamer | — | 5 to 107 CFU mL−1 | 3 CFU mL−1 | Food | 146 |
| PtNPs@Van | Aptamer-coated magnetic CuFe2O4 nanoprobes | — | 5 to 104 CFU mL−1 | 1 CFU mL−1 | Bacterial culture, clinical sample, food | 147 | ||
| Fe3O4@Au NPs | Van and aptamer | Within 50 min | 10 to 107 cells per mL | 3 CFU mL−1 | Bacterial culture | 148 | ||
| AuNP | ssDNA aptamer | — | 6.2 × 102 to 6.2 × 105 CFU mL−1 | 3 CFU mL−1; 2.51 fg μL−1 for genomic DNA | Bacterial culture | 149 | ||
| Optical | UCNPs | Aptamer | — | 50 to 106 CFU mL−1 | 25 CFU mL−1 | Food | 150 | |
| COF-BA Au@Ag NPs | Magnetic IgG@Fe3O4 nanoparticles | 30 min | 10 to 103 CFU mL−1 | 10 CFU mL−1 | Bacterial culture | 139 | ||
| MNPs | Fluorescent proteins | <30.0 min | — | 108 CFU mL−1 | Bacterial culture | 135 | ||
| CDs | Antibody | — | 1 to 2 × 102 CFU mL−1 | 1 CFU mL−1 | Food | 151 | ||
| MOF | Bacteriophages | — | 40 to 4 × 108 CFU mL−1 | 31 CFU mL−1 | Food | 152 | ||
| CQDs plus NFs | Aptamer | 10 to 108 CFU mL−1 | 10 CFU mL−1 | Food | 190 | |||
| CNPs plus QDs | Van and aptamer-based dual-recognition CD nanoprobe | <30.0 min | 10 to 106 CFU mL−1 | 1 CFU mL−1 | Food | 122 | ||
| Au nanodisk | ssDNA aptamer | 120 s | 103–108 CFU mL−1 | 103 CFU mL−1 | Bacterial culture, food | 163 | ||
| Colorimetric | ALP-labeled Fe3O4 and Au NPs | Aptamer | Within 60 min | 10 to 106 CFU mL−1 | 2.4 CFU mL−1; 50 CFU mL−1 (naked eye) | Bacterial culture | 191 | |
| Au NPs | S. aureus protein A gene | 10–15 min | 5 to 40 ng μL−1 | 8.73 ng μL−1 | Stool and urine samples | 192 | ||
Some recent nanotechnology-based biosensors for the detection of the ESKAPEE pathogen are presented in Table 2 with a focus on the sensitivity of the device, the recognition element used to increase the selectivity and the reaction time. Particularly, in the context of pathogen diagnostics critical for healthcare, food safety analysis, and environmental monitoring, the development of a biosensor with rapidity and high accuracy is essential. Compared to traditional diagnostic methods, which are characterized by long reaction times, nanotechnology-based biosensors can be tailored to achieve faster reaction times.
4.2 Nanosensing methods for pathogen detection
This subsection briefly discusses the fundamental principles of signal transduction that exploit nanomaterials, with a focus on electrochemical, optical, and colorimetric methods for the detection of the pathogens presented above (Fig. 2).C. difficile is the leading cause of hospital-acquired diarrhea (see Subsection 3.8). The two major toxins, TcdA and TcdB, have been studied intensively since their initial recognition as major C. difficile virulence factors,156 and they are related to the microorganism infection.157 Thus, Zhu et al. developed a sandwich-type electrochemical impedance immunosensor based on single-domain antibody-conjugated AuNPs applied to amplify the detection signal. In these biosensors, the primary antibody is immobilized on a gold electrode; then, a solution of monodispersed AuNPs conjugated with the secondary antibody is added. When proteins are adsorbed to the electrode surface, they form an inert electron transfer blocking layer and hence increase electron transfer resistance.128
Silica nanoparticles (SNPs) have recently emerged as one of the most up-to-date biocompatible materials because they have strong surface properties, high stability, chemical inertness, and facile functionalization.158 To specifically identify and bind E. coli, Mathelié-Guinlet et al. created an electrochemical biosensor that employs SNPs modified with specific polyclonal antibodies. The gold electrode is initially coated with a polyelectrolyte multilayer to enable the electrostatic immobilization of a layer of NPs functionalized with specific polyclonal antibodies. The transducer without nanoparticles recognizes bacteria although quantification is difficult due to random oscillations in many parameters detected by cyclic voltammetry. However, with the presence of nanoparticles in the biosensor, bacteria are consistently and reliably detected over the measured range.141
Gram-negative bacteria, such as P. aeruginosa, K. pneumoniae, E. coli, and A. baumannii, can tolerate an environment with a large amount of Cu. After internalization, the enzyme cupric reductase starts reducing Cu2+ to Cu+.159 Recently, a biosensor has been developed that exploits the mechanism of copper homeostasis of Gram-negative bacteria associated with the intrinsic oxidase-like activity of cupric oxide nanoparticle (CuONP). In the presence of gram-negative bacteria, reduced Cu+ catalyzes the oxidation of o-phenylenediamine (OPD) to form 2,3-diaminophenazine (oxOPD), which has a fluorescence emission at 573 nm under excitation at 423 nm.54
Nazari-Vanani et al. developed an innovative electrochemical biosensor designed via the electrodeposition of a new gold nanostructure of ice crystals-like as the sensing substrate combined with toluidine blue as the DNA hybridization indicator. Here, a particular thiolated ssDNA was stabilized on the transducer superficies, and the hybridization of the DNA was assessed by differential pulse voltammetry129 (Fig. 3).
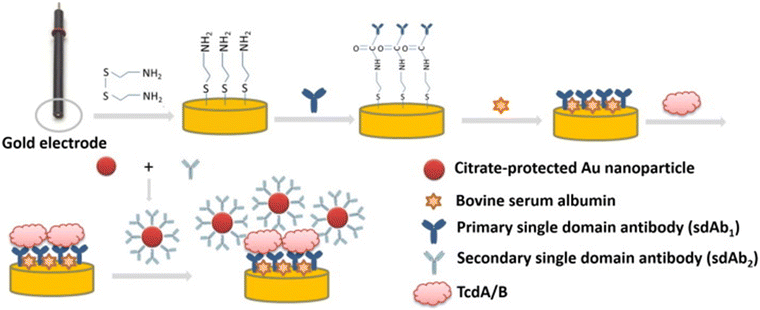 | ||
| Fig. 3 Sandwich-type electrochemical immunosensor for the detection of TcdA and TcdB. Toxins are bonded onto the electrode through antigen–antibody interaction; then, secondary antibody-coated AuNPs are introduced onto the electrode surface as an amplifying probe to optimize the immunosensing performance. Reprinted from Bioelectrochemistry, Zanzan Zhu, Lianfa Shi, Hanping Feng, H. Susan Zhou, “Single domain antibody-coated gold nanoparticles as enhancers for Clostridioides difficile toxin detection by applying electrochemical impedance immunosensors”,128 copyright 2015, with permission from Elsevier. | ||
Localized surface plasmon resonance is an optical phenomenon that occurs when a dielectric surrounds a group of electrons in a metal. The extremely intense and highly confined electromagnetic fields of the LSPR provide a very sensitive probe for detecting small changes in the environment around the nanostructures, which is particularly attractive for sensor applications.162 LSPR-based aptasensors have become important biological and chemical sensing tools for detecting target analytes in real time without labeling agents. Khateb et al. used hole-mask colloidal lithography to fabricate arrays of gold nanodisks on a glass substrate. These nanodisks are then functionalized with DNA aptamers to capture S. aureus within the local optical field of the metal nanostructure, providing a signal readout with a total analysis time of 120 seconds.163
Peptides can act as reducing and stabilizing agents to synthesize AuNPs, thereby regulating the size, morphology, hydrophobicity, and surface charge of the nanoparticles.164 Yu et al. used a one-pot method to synthesize peptide-functionalized gold nanoparticles (P_AuNPs). These P_AuNPs are made with different positive/negative charges and hydrophilic/hydrophobic characteristics tailored for bacterial recognition. The interaction between P_AuNPs and bacteria produces a response in the LSPR spectrum that acts as a bacterial fingerprint. The antibiotic-resistant and antibiotic-susceptible strains of ESKAPEE pathogens are then identified using machine-learning algorithms based on the bacterial fingerprint supplied by the plasmonic nanosensor. The study shows that the surface chemistry of AuNPs changes their plasmonic capabilities, allowing for the fabrication of highly sensitive biosensors for bacterial identification.121
FRET is a non-radiative phenomenon with the energy transferred from an excited donor fluorophore to an acceptor fluorophore through intermolecular dipole–dipole coupling.165 Nanoparticles, such as semiconductor quantum dots (QDs), graphene quantum dots (GQDs), and upconversion nanoparticles (UCNPs), have gained significant attention as photo-stable fluorescence probes and potential donors in FRET. Furthermore, nanoparticles with relatively large sizes have unique electrical properties that account for their quenching capacity. AuNPs and graphene oxide (GO) are two examples of effective fluorescence quenchers in FRET assays.166 Nevertheless, Bahari et al. employed carbon dots (CD) as the donor species and graphene oxide as the acceptor in the FRET process. When the modified ortho-phenylenediamines carbon dot (o-CD) with aptamer (o-CD-ssDNA) binds to the GO surface, the fluorescence of o-CD is efficiently quenched. The aptamer (ssDNA) acts as a biorecognition element and specifically binds with A. baumannii. This binding permits the release of the o-CD-ssDNA from GO, leading to the recovery of the fluorescence signal of o-CD.126
Wang et al. defined a SERS-based sensor for the simultaneous detection of S. aureus, E. coli, P. aeruginosa, and K. pneumoniae. SERS spectra exhibit fingerprint-like patterns that are highly specific to the molecular composition of the analyte, allowing the detection of different bacteria even in complex samples. In particular, the biosensor proposed exploits Au@Ag nanoparticles coated with polydopamine and functionalized with boronic acid as a SERS tag. The nanoprobe forms a sandwich structure by combining the magnetic separation element (IgG@Fe3O4) with the Raman amplification element (SERS tag). The use of IgG-functionalized magnetic nanoparticles enables more efficient capture and isolation of bacteria.139,167,168
Zheng et al. established a method for producing highly photoluminescent AuAg nanoclusters by employing silver ions (Ag+) as linkers to connect the Au-thiol motifs, resulting in Au/Ag–thiol motifs on thiolated Au nanoclusters. This approach significantly increased the photoluminescence in the AuAg bimetallic nanoclusters. When photoluminescent AuAg nanoclusters are exposed to the bacterium A. baumannii, their fluorescence is selectively quenched169 (Fig. 4).
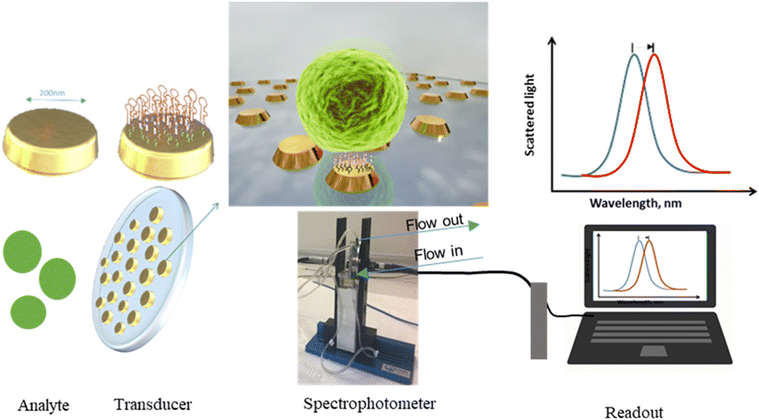 | ||
| Fig. 4 Schematic representation of a label-free optical nanobiosensor via aptamer recognition of Staphylococcus aureus. Arrays of plasmonic gold nanodisks with disk diameters of either 100 or 200 nm are employed to enable refractometric detection of the bacteria. The LSPR spectra shift is measured using a compact custom-built fiber spectrometer and showed a maximum absorption peak at 650 nm for the 100 nm disks and 990 nm for the 200 nm disks. Reprinted with permission from Khateb, Heba et al.163 Development of a label-free LSPR-Apta sensor for Staphylococcus aureus detection, ACS Applied Bio Materials, vol. 3 and 5, 2020, 3066–3077. Copyright 2020, American Chemical Society. | ||
Gold nanoparticles have outstanding optical features because of their distinct size and shape-dependent interactions with light. The color of AuNPs can be controlled by modulating their size, shape, or the surrounding media. Smaller nanoparticles are red, while larger ones are blue.172 Madkour et al. developed a sensor based on the optical properties of gold nanoparticles by synthesizing AuNPs of 10 nm and functionalizing them with a thiolated oligonucleotide probe. The detecting element is designed using protein A sequence data obtained from the gene bank. This protein is specifically tailored to bind to the target DNA sequence of the S. aureus–SPA gene, an important virulence factor of S. aureus. Gene detection is based on a color change that can be observed with the naked eye and quantitatively measured using a UV-vis spectrophotometer. The nanosensor exhibits a stable red color in the presence of the target DNA, while the color changes to blue in the absence of the target. This colorimetric assay allows for the rapid detection of DNA samples without DNA amplification.173 The same process is exploited by Tondro et al., who conjugated gold nanoparticles with a thiolated oligonucleotide probe from a partial sequence of the 16S ribosomal RNA gene of Enterococcus faecalis. In this case, the aggregation is induced by the addition of an acid solution, but when the target sequence is present, it hybridizes with the sequence conjugated on the surface of AuNPs, preventing aggregation.174
Sivakumar et al. exploited the characteristic properties of silver for the selective detection of E. faecium and A. baumannii. In this sensor, nanoparticles are not directly synthesized: the formation of nanoparticles occurs instead under specific conditions to produce a signal that could be detected by the naked eye. The recognition element is the loop-mediated isothermal amplification (LAMP) amplicons of the target DNA. The interaction between the LAMP amplicons, Ag+ ions, and quercetin, a polyphenolic flavonoid with high reducing potential, leads to the formation of silver nanoparticles (AgNPs), which display an intense brown color.131
A lateral flow assay (LFA) is a diagnostic technique used to detect target analytes rapidly and easily in various samples. In an LFA, the sample containing the target analyte is applied to a test strip made up of several components, such as a sample application pad, conjugate pad, nitrocellulose membrane, and an adsorption pad. The nitrocellulose membrane usually contains test and control lines that immobilize specific biorecognition molecules, such as antibodies. When the sample flows through the strip via capillary action, the target analyte interacts with the labeled molecules and moves along the strip. If the target analyte is present, it binds to the capture molecules on the test line, producing a visible signal that indicates a positive result.175
A way to detect P. aeruginosa was developed by Alhogail et al. They created a biosensor on paper by applying a special protease substrate. This substrate was designed to allow the LasA protease from P. aeruginosa to access and break it down easily. The substrate is a peptide linked to magnetic nanoparticles (MNPs) at one end. At the other end, a cysteine allowed the peptide–MNP complex to form a self-assembled monolayer on a gold sensor surface. When P. aeruginosa protease cleaves the peptide, the MNP portion is detached, revealing the golden color of the sensor145 (Fig. 5).
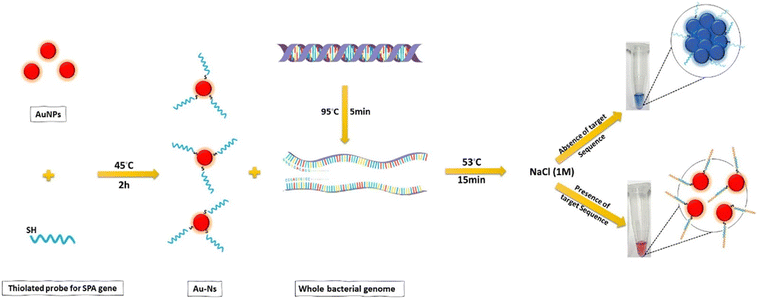 | ||
| Fig. 5 Colorimetric detection strategy for the Staphylococcal protein A (SPA) gene based on gold nanosensors (Au-Ns). AuNP surface is functionalized with a specific thiolated probe. S. aureus genome is denatured by heating at 95 °C for 5 min, then mixed with 50 μL Au-Ns and incubated for 15 minutes. Finally, 5 μl of NaCl solution was added to the mixture, and the result was evaluated by the naked eye.167 | ||
5. Future perspectives suggested by promising approaches based on nanosensors
The recent COVID-19 global pandemic has further confirmed the need to develop miniaturized diagnostic devices that can be run at home by a non-technical operator, thereby supporting the healthcare system in containing infections and allowing for immediate clinical decision making. Even the World Health Organization has stressed the importance of creating such devices by summarizing the required characteristics for an ideal POC with the acronym ASSURED: affordable, sensitive, specific, user-friendly, robust and rapid, equipment-free, and deliverable.176–178 Although traditional methods for detecting pathogens exhibit adequate sensitivity, their speed and flexibility fall short of effectively addressing a significant outbreak. POC analyses emerge as the sole practical approach to handle such situations rapidly, with minimal operator demands on a large scale.179In this review, we focused our attention on the integration of nanotechnology and pathogen detection devices considering that the unique physicochemical properties of nanoparticles have led to the development of all the nanobiosensors listed above. The application of nanotechnologies to produce POC has several advantages, such as increased sensitivity, miniaturization, multiplexing capabilities, improved specificity, enhanced imaging and visualization, and paves the way to personalized medicine.
Electrochemical nanobiosensors are considered the most widespread class of sensors for tracking bacteria. They can offer high sensitivity because they can detect target molecules even at very low concentrations. Moreover, they can be designed to be very selective towards specific biomolecules such as the nanosensor developed by Sohouli et al., which can detect even small amounts of S. aureus in human serum samples. These nanosensors can also be miniaturized, allowing for portable and on-site detection systems for risk evaluation. Even if significant progress has been made in the development of such devices, there are still some issues to solve, such as improving their biocompatibility through surface modification and coating. Furthermore, most electrochemical biochips are currently engineered for single use; thus, future research should focus on making detectors reusable in sensor design.117,155
Optical biosensors have attracted considerable interest as substitutes for conventional methods due to their rapid, straightforward, and accurate outcomes. The integration of nanotechnology has introduced numerous strategies to upgrade conventional optical biosensors into smart, advanced, and efficient optical biosensing platforms.180 Compared with traditional strategies, nanomaterials used in optical biosensors have excellent chemical, physical, and optical properties, which make them suitable for obtaining low detection limits and high sensitivity for identifying viruses and bacteria, depending on their color change or fluorescence conversion.181 Although the use of biosensors employing nanomaterials has shown promising results in laboratory settings, several challenges must still be overcome to implement them effectively in practical applications. At present, optical and photoelectrochemical biosensors primarily rely on precious metals, such as gold, platinum, and silver, for construction. However, their limited availability and high expenses hinder the commercial availability of these sensors. Hence, it has become crucial to explore alternative nanomaterials sourced abundantly and at lower costs to promote the widespread use of these sensors.182
The colorimetric detection technique is one of those interesting optical methods that provides a straightforward and easily interpretable output by producing visible color changes in response to the presence of the target analyte. These nanosensors can also be designed and fabricated to be portable, enabling on-site testing without sophisticated instrumentation, and they can detect pathogenic bacteria in a very short period.161 Compared with fluorescence sensors, colorimetric ones have the advantages of simplicity, fast response, and visualization detection. Although they have been applied to differentiate many chemicals and bioanalytes, there are few reports on colorimetric sensors for identifying bacteria.183 This could be because the interpretation of the color changes associated with the results may introduce variability, impacting reproducibility. Moreover, this kind of sensor is usually solely qualitative and cannot indicate the amount of the pathogen present in the tested sample.176
Focusing on nanobiosensors applied to healthcare-associated infections, the examples reported above (Table 2) were mostly tested on food and biological samples, such as serum, stool, and urine samples. To the best of our knowledge, only a few nanobiosensors have been applied to detect pathogens in personnel, equipment, devices, hospital surfaces and environments, and patient support accessories, which are the main mediated factors responsible for the spread of nosocomial infection pathogens. One strategy to minimize the risk of patient-to-patient transmission of pathogens from other contaminated items is to perform regular sanitary controls of all inanimate surfaces and to implement quick methods for determining cleanliness and hygiene.184,185 Nanotechnologies have the potential to contribute to the implementation of these new rapid, reliable, and sensitive sensors that can improve the traditional detection systems applied in hospitals. Now, this family of nanosensors remains a goal to reach. We can speculate that to develop such a sensor, it is crucial to address challenges related to standardization, regulatory approval, and ethical considerations. Moreover, further studies are required to ensure the accuracy and reliability of the nanobiosensors, which must be validated in real hospital settings.
Other challenges that can hinder the implementation of nanobiosensors in monitoring pathogens on hospital surfaces include diverse microbe-rich background matrices found in environmental samples and low pathogen concentrations. To overcome the latter issue, different strategies have been developed to enrich pathogen concentration besides being applied so far mainly to agriculture, food and bioaerosol monitoring.186,187 These methods can be divided into material-based enrichment methods, electric-based enrichment methods and bio-organism-based enrichment methods. Among the material-based enrichment strategies, noble metal ion characteristics have been exploited to build up nanoparticle aggregation-based enrichment methods where the aggregation of the nanoparticles on the target analytes leads to a change in the color of the solution in a proportional manner.188,189 Recently, to improve the stability, sensitivity and specificity of these nanoparticle-based enrichment assays, research has focused on the development of systems based on the cooperation between DNA/RNA chains and nanoparticles. These strategies are yet to be tested in hospital settings with real environmental samples.
Nevertheless, with the increasing demand for POC devices, nanotechnologies and their properties can be a real resource for designing a new generation of efficient yet easy-to-use pathogen detection and identification assays for clinical environment surveillance. To move toward this new concept, a change in mentality is likely to come with the advent of new technologies not only in the diagnosis and treatment of human diseases but also in the more general management of the healthcare system (Fig. 6).
Abbreviations
| AgNPs | Silver nanoparticles |
| AuNPs | Gold nanoparticles |
| CDI | Clostridioides difficile infections |
| CFU | Colony forming unit |
| CNT | Carbon nanotube |
| CRAB | Carbapenem-resistant Acinetobacter baumannii |
| CRKP | Carbapenem-resistant Klebsiella pneumoniae |
| CuONPs | Cupric oxide nanoparticles |
| DAEC | Diffusely adherent Escherichia coli |
| EU/EEA | European Union and European Economic Area |
| EPEC | Enteropathogenic Escherichia coli |
| EHEC | Enterohemorrhagic Escherichia coli |
| ETEC | Enterotoxigenic Escherichia coli |
| EAEC | Enteroaggregative Escherichia coli |
| EIEC | Enteroinvasive Escherichia coli |
| ESKAPEE | Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter species, Escherichia coli |
| FRET | Fluorescence resonance energy transfer |
| GO | Graphene oxide |
| GQDs | Graphene quantum dots |
| HAI | Healthcare-associated infection |
| ICA | Indole-5-carboxylic acid |
| ICU | Intensive care unit |
| LAMP | Loop-mediated isothermal amplification |
| LFA | Lateral flow assay |
| LoD | Limit of detection |
| LSPR | Localized surface plasmon resonance |
| MDR | Multiple drug resistance |
| MDRO | Multidrug-resistant organism |
| MNCs | Metal nanoclusters |
| MNPs | Magnetic nanoparticles |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| MSSA | Methicillin-susceptible Staphylococcus aureus |
| NCNOs | Nitrogen-doped carbon nano-onions |
| NICU | Neonatal intensive care unit |
| o-CD | Ortho-phenylenediamines carbon dot |
| P_AuNPs | Peptide-functionalized gold nanoparticles |
| POC | Point of care |
| PCR | Polymerase chain reaction |
| QDs | Quantum dots |
| SERS | Surface-enhanced Raman spectroscopy |
| SNPs | Silica nanoparticles |
| SPA | Staphylococcal protein A |
| SSI | Surgical site infection |
| TcdA | Toxin Clostridioides difficile A |
| TcdB | Toxin Clostridioides difficile B |
| UCNPs | Upconversion nanoparticles |
| Van | Vancomycin |
| VRE | Vancomycin-resistant Enterococcus |
| VRSA | Vancomycin-resistant Staphylococcus aureus |
| WHO | World Health Organization |
Data availability
No primary research results, software or code has been included and no new data were generated or analysed as part of this review.Author contributions
Miriam Colombo: conceptualization, methodology, resources, writing – review & editing, supervision, project administration, funding acquisition. Antonia Bruno: conceptualization, methodology, resources, writing – original draft, supervision. Farida Tripodi: conceptualization, methodology, resources, writing – original draft, project administration, funding acquisition. Alice Armanni, Linda Barbieri, Alessandro Colombo, Giulia Tomaino: methodology, resources, writing – original draft, visualization. Sara Fumagalli, Hind Moukham: methodology, resources, writing – original draft. Ekaterina Kukushkina, Roberto Lorenzi, Letizia Marchesi, Alberto Paleari, Alessandra Ronchi, Valeria Secchi: writing – review & editing. Laura Sironi, Angelo Monguzzi: conceptualization, supervision, project administration, funding acquisition. All authors read and approved the submitted version.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the National Plan for NRRP Complementary Investments (PNC, established with the decree-law 6 May 2021, no. 59, converted by law no. 101 of 2021) in the call for the funding of research initiatives for technologies and innovative trajectories in the health and care sectors (directorial decree no. 931 of 06-06-2022) – project no. PNC0000003 – AdvaNced Technologies for Human-centrEd Medicine (project acronym: ANTHEM). This work reflects only the authors' views and opinions, neither the Ministry for University and Research nor the European Commission can be considered responsible for them.Notes and references
- W. Zingg, S. Hopkins, A. Gayet-Ageron, A. Holmes, M. Sharland, C. Suetens and ECDC PPS study group, Health-care-associated infections in neonates, children, and adolescents: an analysis of paediatric data from the European Centre for Disease Prevention and Control point-prevalence survey, Lancet Infect. Dis., 2017, 17, 381–389 CrossRef PubMed.
- A. H. Sohn, D. O. Garrett, R. L. Sinkowitz-Cochran, L. A. Grohskopf, G. L. Levine, B. H. Stover, J. D. Siegel, W. R. Jarvis and Pediatric Prevention Network, Prevalence of nosocomial infections in neonatal intensive care unit patients: Results from the first national point-prevalence survey, J. Pediatr., 2001, 139, 821–827 CrossRef CAS PubMed.
- D. R. Bhatta, S. Hosuru Subramanya, D. Hamal, R. Shrestha, E. Gauchan, S. Basnet, N. Nayak and S. Gokhale, Bacterial contamination of neonatal intensive care units: How safe are the neonates?, Antimicrob. Resist. Infect. Control, 2021, 10, 26 CrossRef PubMed.
- J. Johnson, I. C. Akinboyo and J. K. Schaffzin, Infection Prevention in the Neonatal Intensive Care Unit, Clin. Perinatol., 2021, 48, 413–429 CrossRef PubMed.
- J. Johnson and C. Quach, Outbreaks in the Neonatal Intensive Care Unit: A Review of the Literature, Curr. Opin. Infect. Dis., 2017, 30, 395–403 CrossRef PubMed.
- L. Kreitmann, J. Helms, I. Martin-Loeches, J. Salluh, G. Poulakou, F. Pène and S. Nseir, ICU-acquired infections in immunocompromised patients, Intensive Care Med., 2024, 50, 332–349 CrossRef PubMed.
- A. Cresti, P. Baratta, F. De Sensi, E. Aloia, B. Sposato and U. Limbruno, Clinical Features and Mortality Rate of Infective Endocarditis in Intensive Care Unit: A Large-Scale Study and Literature Review, Anatolian J. Cardiol., 2024, 28, 44–54 Search PubMed.
- J. Teng, S. Imani, A. Zhou, Y. Zhao, L. Du, S. Deng, J. Li and Q. Wang, Combatting resistance: Understanding multi-drug resistant pathogens in intensive care units, Biomed. Pharmacother., 2023, 167, 115564 CrossRef CAS PubMed.
- K. Vickery, A. Deva, A. Jacombs, J. Allan, P. Valente and I. B. Gosbell, Presence of biofilm containing viable multiresistant organisms despite terminal cleaning on clinical surfaces in an intensive care unit, J. Hosp. Infect., 2012, 80, 52–55 CrossRef CAS PubMed.
- Healthcare-associated infections acquired in intensive care units – Annual Epidemiological Report for 2020, https://www.ecdc.europa.eu/en/publications-data/healthcare-associated-infections-acquired-intensive-care-units-annual, (accessed 18 March 2024).
- Y. Li, C. Liu, W. Xiao, T. Song and S. Wang, Incidence, Risk Factors, and Outcomes of Ventilator-Associated Pneumonia in Traumatic Brain Injury: A Meta-analysis, Neurocrit. Care, 2020, 32, 272–285 CrossRef CAS PubMed.
- S. Edwardson and C. Cairns, Nosocomial infections in the ICU, Anaesthesiol. Intensivmed., 2019, 20, 14–18 Search PubMed.
- M. K. Hayden, D. W. Blom, E. A. Lyle, C. G. Moore and R. A. Weinstein, Risk of Hand or Glove Contamination After Contact With Patients Colonized With Vancomycin-Resistant Enterococcus or the Colonized Patients' Environment, Infect. Control Hosp. Epidemiol., 2008, 29, 149–154 CrossRef PubMed.
- S. J. Dancer, Importance of the environment in meticillin-resistant Staphylococcus aureus acquisition: the case for hospital cleaning, Lancet Infect. Dis., 2008, 8, 101–113 CrossRef PubMed.
- A. Cassini, L. D. Högberg, D. Plachouras, A. Quattrocchi, A. Hoxha, G. S. Simonsen, M. Colomb-Cotinat, M. E. Kretzschmar, B. Devleesschauwer, M. Cecchini, D. A. Ouakrim, T. C. Oliveira, M. J. Struelens, C. Suetens, D. L. Monnet, R. Strauss, K. Mertens, T. Struyf, B. Catry, K. Latour, I. N. Ivanov, E. G. Dobreva, A. T. Andraševic, S. Soprek, A. Budimir, N. Paphitou, H. Žemlicková, S. S. Olsen, U. W. Sönksen, P. Märtin, M. Ivanova, O. Lyytikäinen, J. Jalava, B. Coignard, T. Eckmanns, M. A. Sin, S. Haller, G. L. Daikos, A. Gikas, S. Tsiodras, F. Kontopidou, Á. Tóth, Á. Hajdu, Ó. Guólaugsson, K. G. Kristinsson, S. Murchan, K. Burns, P. Pezzotti, C. Gagliotti, U. Dumpis, A. Liuimiene, M. Perrin, M. A. Borg, S. C. de Greeff, J. C. Monen, M. B. Koek, P. Elstrøm, D. Zabicka, A. Deptula, W. Hryniewicz, M. Caniça, P. J. Nogueira, P. A. Fernandes, V. Manageiro, G. A. Popescu, R. I. Serban, E. Schréterová, S. Litvová, M. Štefkovicová, J. Kolman, I. Klavs, A. Korošec, B. Aracil, A. Asensio, M. Pérez-Vázquez, H. Billström, S. Larsson, J. S. Reilly, A. Johnson and S. Hopkins, Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis, Lancet Infect. Dis., 2019, 19, 56–66 CrossRef PubMed.
- E. Righi, N. T. Mutters, X. Guirao, M. D. del Toro, C. Eckmann, A. W. Friedrich, M. Giannella, J. Kluytmans, E. Presterl, E. Christaki, E. L. A. Cross, A. Visentin, G. Sganga, C. Tsioutis and E. Tacconelli, ESCMID/EUCIC clinical practice guidelines on perioperative antibiotic prophylaxis in patients colonized by multidrug-resistant Gram-negative bacteria before surgery, Clin. Microbiol. Infect., 2023, 29, 463–479 CrossRef CAS PubMed.
- H. Getachew, A. Derbie and D. Mekonnen, Surfaces and Air Bacteriology of Selected Wards at a Referral Hospital, Northwest Ethiopia: A Cross-Sectional Study, Int. J. Microbiol., 2018, 2018, e6413179 Search PubMed.
- B. K. Sharma, B. P. Sharma, A. Kunwar, N. Basnet, P. D. Magar and S. Adhikari, Prevalence of Extended Spectrum β-Lactamase Producers (ESBLs) with antibiotic resistance pattern of Gram negative pathogenic bacteria isolated from door handles in hospitals of Pokhara, Western Nepal, J. Genet. Eng. Biotechnol., 2023, 21, 139 CrossRef PubMed.
- H.-P. Wang, H.-J. Zhang, J. Liu, Q. Dong, S. Duan, J.-Q. Ge, Z.-H. Wang and Z. Zhang, Antimicrobial resistance of 3 types of gram-negative bacteria isolated from hospital surfaces and the hands of health care workers, Am. J. Infect. Control, 2017, 45, e143–e147 CrossRef CAS PubMed.
- A. Ashokan, J. M. Choo, S. L. Taylor, D. Lagana, D. R. Shaw, M. S. Warner, S. L. Wesselingh and G. B. Rogers, Environmental dynamics of hospital microbiome upon transfer from a major hospital to a new facility, J. Infect., 2021, 83, 637–643 CrossRef PubMed.
- J. A. Otter, J. Zhou, J. R. Price, L. Reeves, N. Zhu, P. Randell, S. Sriskandan, W. S. Barclay and A. H. Holmes, SARS-CoV-2 surface and air contamination in an acute healthcare setting during the first and second pandemic waves, J. Hosp. Infect., 2023, 132, 36–45 CrossRef CAS PubMed.
- D. G. Falvey and A. J. Streifel, Ten-year air sample analysis of Aspergillus prevalence in a university hospital, J. Hosp. Infect., 2007, 67, 35–41 CrossRef CAS PubMed.
- M. Rose, D. Landman and J. Quale, Are community environmental surfaces near hospitals reservoirs for gram-negative nosocomial pathogens?, Am. J. Infect. Control, 2014, 42, 346–348 CrossRef PubMed.
- A. L. Casey, D. Adams, T. J. Karpanen, P. A. Lambert, B. D. Cookson, P. Nightingale, L. Miruszenko, R. Shillam, P. Christian and T. S. J. Elliott, Role of copper in reducing hospital environment contamination, J. Hosp. Infect., 2010, 74, 72–77 CrossRef CAS PubMed.
- B. P. Howden, S. G. Giulieri, T. Wong Fok Lung, S. L. Baines, L. K. Sharkey, J. Y. H. Lee, A. Hachani, I. R. Monk and T. P. Stinear, Staphylococcus aureus host interactions and adaptation, Nat. Rev. Microbiol., 2023, 21, 380–395 CrossRef CAS PubMed.
- R. Ragusa, G. Giorgianni, G. Faro, A. Lazzara, M. A. Bellia and M. Marranzano, Are Visitors Dangerous Carriers of Pathogens in The Hospital? an Observational Study in an University Hospital in Sicily, Hosp. Top., 2019, 97, 80–86 CrossRef PubMed.
- D.-U. Park, J.-K. Yeom, W. J. Lee and K.-M. Lee, Assessment of the Levels of Airborne Bacteria, Gram-Negative Bacteria, and Fungi in Hospital Lobbies, Int. J. Environ. Res. Public Health, 2013, 10, 541–555 CrossRef PubMed.
- V. Russotto, A. Cortegiani, S. M. Raineri and A. Giarratano, Bacterial contamination of inanimate surfaces and equipment in the intensive care unit, J. Intensive Care, 2015, 3, 54 CrossRef PubMed.
- P. S. Falk, J. Winnike, C. Woodmansee, M. Desai and C. G. Mayhall, Outbreak of Vancomycin-Resistant Enterococci in a Burn Unit, Infect. Control Hosp. Epidemiol., 2000, 21, 575–582 CrossRef CAS PubMed.
- T. Lestari, S. Ryll and A. Kramer, Microbial contamination of manually reprocessed, ready to use ECG lead wire in intensive care units, GMS Hyg. Infect. Control, 2013, 8, Doc07 Search PubMed.
- Y. Jiang, Y. Ding, Y. Wei, C. Jian, J. Liu and Z. Zeng, Carbapenem-resistant Acinetobacter baumannii: A challenge in the intensive care unit, Front. Microbiol., 2022, 13, 1045206 CrossRef PubMed.
- G. Aygün, O. Demirkiran, T. Utku, B. Mete, S. Ürkmez, M. Yılmaz, H. Yaşar, Y. Dikmen and R. Öztürk, Environmental contamination during a carbapenem-resistant Acinetobacter baumannii outbreak in an intensive care unit, J. Hosp. Infect., 2002, 52, 259–262 CrossRef PubMed.
- F. A. Manian, S. Griesenauer, D. Senkel, J. M. Setzer, S. A. Doll, A. M. Perry and M. Wiechens, Isolation of Acinetobacter baumannii Complex and Methicillin-Resistant Staphylococcus aureus from Hospital Rooms Following Terminal Cleaning and Disinfection: Can We Do Better?, Infect. Control Hosp. Epidemiol., 2011, 32, 667–672 CrossRef PubMed.
- M. Matsuo, S. Oie and H. Furukawa, Contamination of blood pressure cuffs by methicillin-resistant Staphylococcus aureus and preventive measures, Ir. J. Med. Sci., 2013, 182, 707–709 CrossRef CAS PubMed.
- A. Jabłońska-Trypuć, M. Makuła, M. Włodarczyk-Makuła, E. Wołejko, U. Wydro, L. Serra-Majem and J. Wiater, Inanimate Surfaces as a Source of Hospital Infections Caused by Fungi, Bacteria and Viruses with Particular Emphasis on SARS-CoV-2, Int. J. Environ. Res. Public Health, 2022, 19, 8121 CrossRef PubMed.
- Y.-S. Sui, G.-H. Wan, Y.-W. Chen, H.-L. Ku, L.-P. Li, C.-H. Liu and H.-S. Mau, Effectiveness of Bacterial Disinfectants on Surfaces of Mechanical Ventilator Systems, Respir. Care, 2012, 57, 250–256 CrossRef PubMed.
- M. Guy, P. Vanhems, C. Dananché, M. Perraud, A. Regard, M. Hulin, O. Dauwalder, X. Bertrand, J. Crozon-Clauzel, B. Floccard, L. Argaud, P. Cassier and T. Bénet, Outbreak of pulmonary Pseudomonas aeruginosa and Stenotrophomonas maltophilia infections related to contaminated bronchoscope suction valves, Lyon, France, 2014, Eurosurveillance, 2016, 21, 30286 CrossRef PubMed.
- P. D. Levin, O. Shatz, S. Sviri, D. Moriah, A. Or-Barbash, C. L. Sprung, A. E. Moses and C. Block, Contamination of Portable Radiograph Equipment With Resistant Bacteria in the ICU, Chest, 2009, 136, 426–432 CrossRef PubMed.
- H. Shokoohi, P. Armstrong and R. Tansek, Emergency department ultrasound probe infection control: challenges and solutions, Open Access Emerg. Med., 2015, 7, 1–9 Search PubMed.
- H. Koibuchi, K. Kotani and N. Taniguchi, Ultrasound probes as a possible vector of bacterial transmission, Med. Ultrasound, 2013, 15, 41–44 Search PubMed.
- M. Catalano, L. S. Quelle, P. E. Jeric, A. Di Martino and S. M. Maimone, Survival of Acinetobacter baumannii on bed rails during an outbreak and during sporadic cases, J. Hosp. Infect., 1999, 42, 27–35 CrossRef CAS PubMed.
- A. P. R. Wilson, D. Smyth, G. Moore, J. Singleton, R. Jackson, V. Gant, A. Jeanes, S. Shaw, E. James, B. Cooper, G. Kafatos, B. Cookson, M. Singer and G. Bellingan, The impact of enhanced cleaning within the intensive care unit on contamination of the near-patient environment with hospital pathogens: a randomized crossover study in critical care units in two hospitals, Crit. Care Med., 2011, 39, 651–658 CrossRef PubMed.
- A. M. Whittington, G. Whitlow, D. Hewson, C. Thomas and S. J. Brett, Bacterial contamination of stethoscopes on the intensive care unit, Anaesthesia, 2009, 64, 620–624 CrossRef CAS PubMed.
- L. S. Munoz-Price, K. L. Arheart, J. P. Mills, T. Cleary, D. DePascale, A. Jimenez, Y. Fajardo-Aquino, G. Coro, D. J. Birnbach and D. A. Lubarsky, Associations between bacterial contamination of health care workers' hands and contamination of white coats and scrubs, Am. J. Infect. Control, 2012, 40, e245–e248 CrossRef PubMed.
- A. Borer, J. Gilad, R. Smolyakov, S. Eskira, N. Peled, N. Porat, E. Hyam, R. Trefler, K. Riesenberg and F. Schlaeffer, Cell Phones and Acinetobacter Transmission, Emerging Infect. Dis., 2005, 11, 1160–1161 CrossRef PubMed.
- F. Ulger, S. Esen, A. Dilek, K. Yanik, M. Gunaydin and H. Leblebicioglu, Are we aware how contaminated our mobile phones with nosocomial pathogens?, Ann. Clin. Microbiol. Antimicrob., 2009, 8, 7 CrossRef PubMed.
- W. A. Rutala, M. S. White, M. F. Gergen and D. J. Weber, Bacterial Contamination of Keyboards: Efficacy and Functional Impact of Disinfectants, Infect. Control Hosp. Epidemiol., 2006, 27, 372–377 CrossRef PubMed.
- D. Roux, B. Aubier, H. Cochard, R. Quentin and N. van der Mee-Marquet, Contaminated sinks in intensive care units: an underestimated source of extended-spectrum beta-lactamase-producing Enterobacteriaceae in the patient environment, J. Hosp. Infect., 2013, 85, 106–111 CrossRef CAS PubMed.
- F. Salm, M. Deja, P. Gastmeier, A. Kola, S. Hansen, M. Behnke, D. Gruhl and R. Leistner, Prolonged outbreak of clonal MDR Pseudomonas aeruginosa on an intensive care unit: contaminated sinks and contamination of ultra-filtrate bags as possible route of transmission?, Antimicrob. Resist. Infect. Control, 2016, 5, 53 CrossRef PubMed.
- M. Robakowska, M. Bronk, A. Tyrańska-Fobke, D. Ślęzak, J. Kraszewski and Ł. Balwicki, Patient Safety Related to Microbiological Contamination of the Environment of a Multi-Profile Clinical Hospital, Int. J. Environ. Res. Public Health, 2021, 18, 3844 CrossRef PubMed.
- M. L. Cristina, M. Sartini and A. M. Spagnolo, Serratia marcescens Infections in Neonatal Intensive Care Units (NICUs), Int. J. Environ. Res. Public Health, 2019, 16, 610 CrossRef CAS PubMed.
- Á. Morillo, V. González, J. Aguayo, C. Carreño, M. J. Torres, D. Jarana, M. J. Artacho, F. Jiménez, M. Conde and J. Aznar, A six-month Serratia marcescens outbreak in a Neonatal Intensive Care Unit, Enferm. Infecc. Microbiol. Clin., 2016, 34, 645–651 CrossRef PubMed.
- I. Lange, B. Edel, K. Dawczynski, H. Proquitté, M. W. Pletz, F. Kipp and C. Stein, Influence of the Incubator as Direct Patient Environment on Bacterial Colonization of Neonates, Microorganisms, 2021, 9, 2533 CrossRef PubMed.
- S. Deng, Y. Tu, L. Fu, J. Liu and L. Jia, A label-free biosensor for selective detection of Gram-negative bacteria based on the oxidase-like activity of cupric oxide nanoparticles, Microchim. Acta, 2022, 189, 471 CrossRef CAS PubMed.
- C. Jinadatha, T. Navarathna, J. Negron-Diaz, G. Ghamande, B. A. Corona, A. Adrianza, J. D. Coppin, H. Choi and P. Chatterjee, Understanding the significance of microbiota recovered from health care surfaces, Am. J. Infect. Control, 2024, 52, 220–224 CrossRef CAS PubMed.
- M. Aleem, A. R. Azeem, S. Rahmatullah, S. Vohra, S. Nasir and S. Andleeb, Prevalence of Bacteria and Antimicrobial Resistance Genes in Hospital Water and Surfaces, Cureus, 2021, 13, e18738 Search PubMed.
- V. La Fauci, C. Genovese, A. Facciolà, M. A. R. Palamara and R. Squeri, Five-year microbiological monitoring of wards and operating theatres in southern Italy, J. Prev. Med. Hyg., 2017, 58, E166–E172 CAS.
- S. Rawlinson, L. Ciric and E. Cloutman-Green, How to carry out microbiological sampling of healthcare environment surfaces? A review of current evidence, J. Hosp. Infect., 2019, 103, 363–374 CrossRef CAS PubMed.
- J. Chai, T. Donnelly, T. Wong and E. Bryce, Environmental sampling of hospital surfaces: Assessing methodological quality, Can. J. Infect. Control, 2018, 33, 138–145 Search PubMed.
- A. Bruno, S. Fumagalli, G. Ghisleni and M. Labra, The Microbiome of the Built Environment: The Nexus for Urban Regeneration for the Cities of Tomorrow, Microorganisms, 2022, 10, 2311 CrossRef PubMed.
- M. S. Mulani, E. E. Kamble, S. N. Kumkar, M. S. Tawre and K. R. Pardesi, Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review, Front. Microbiol., 2019, 10 DOI:10.3389/fmicb.2019.00539.
- A. Rajput, Y. Seif, K. S. Choudhary, C. Dalldorf, S. Poudel, J. M. Monk and B. O. Palsson, Pangenome Analytics Reveal Two-Component Systems as Conserved Targets in ESKAPEE Pathogens, mSystems, 2021, 6(1) DOI:10.1128/msystems.00981-20.
- R. H. Katzenberger, A. Rösel and R.-P. Vonberg, Bacterial survival on inanimate surfaces: a field study, BMC Res. Notes, 2021, 14, 97 CrossRef CAS PubMed.
- X. Zhou, R. J. L. Willems, A. W. Friedrich, J. W. A. Rossen and E. Bathoorn, Enterococcus faecium: from microbiological insights to practical recommendations for infection control and diagnostics, Antimicrob. Resist. Infect. Control, 2020, 9, 130 CrossRef PubMed.
- S. Brinkwirth, O. Ayobami, T. Eckmanns and R. Markwart, Hospital-acquired infections caused by enterococci: a systematic review and meta-analysis, WHO European Region, 1 January 2010 to 4 February 2020, Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull., 2021, vol. 26, p. 2001628 Search PubMed.
- R. J. L. Willems, J. Top, M. van Santen, D. A. Robinson, T. M. Coque, F. Baquero, H. Grundmann and M. J. M. Bonten, Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex, Emerging Infect. Dis., 2005, 11, 821–828 CrossRef CAS.
- V. Cattoir and J.-C. Giard, Antibiotic resistance in Enterococcus faecium clinical isolates, Expert Rev. Anti-infect. Ther., 2014, 12, 239–248 CrossRef CAS PubMed.
- F. L. Paganelli, R. J. Willems, P. Jansen, A. Hendrickx, X. Zhang, M. J. Bonten and H. L. Leavis, Enterococcus faecium biofilm formation: identification of major autolysin AtlAEfm, associated Acm surface localization, and AtlAEfm-independent extracellular DNA Release, mBio, 2013, 4(2), e00154 CrossRef PubMed.
- S. Yousaf Kazmi, The etymology of microbial nomenclature and the diseases these cause in a historical perspective, Saudi J. Biol. Sci., 2022, 29, 103454 CrossRef CAS PubMed.
- F. D. Lowy, Staphylococcus aureus Infections, N. Engl. J. Med., 1998, 339, 520–532 CrossRef CAS PubMed.
- D. S. Ondusko and D. Nolt, Staphylococcus aureus, Pediatr. Rev., 2018, 39, 287–298 CrossRef PubMed.
- W. H. Organization, Global antimicrobial resistance and use surveillance system (GLASS) report: 2021, World Health Organization, 2021 Search PubMed.
- Antimicrobial resistance in the EU/EEA (EARS-Net) – Annual Epidemiological Report for 2020, https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-eueea-ears-net-annual-epidemiological-report-2020, (accessed 14 March 2024).
- C. J. L. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. R. Aguilar, A. Gray, C. Han, C. Bisignano, P. Rao, E. Wool, S. C. Johnson, A. J. Browne, M. G. Chipeta, F. Fell, S. Hackett, G. Haines-Woodhouse, B. H. K. Hamadani, E. A. P. Kumaran, B. McManigal, S. Achalapong, R. Agarwal, S. Akech, S. Albertson, J. Amuasi, J. Andrews, A. Aravkin, E. Ashley, F.-X. Babin, F. Bailey, S. Baker, B. Basnyat, A. Bekker, R. Bender, J. A. Berkley, A. Bethou, J. Bielicki, S. Boonkasidecha, J. Bukosia, C. Carvalheiro, C. Castañeda-Orjuela, V. Chansamouth, S. Chaurasia, S. Chiurchiù, F. Chowdhury, R. C. Donatien, A. J. Cook, B. Cooper, T. R. Cressey, E. Criollo-Mora, M. Cunningham, S. Darboe, N. P. J. Day, M. D. Luca, K. Dokova, A. Dramowski, S. J. Dunachie, T. D. Bich, T. Eckmanns, D. Eibach, A. Emami, N. Feasey, N. Fisher-Pearson, K. Forrest, C. Garcia, D. Garrett, P. Gastmeier, A. Z. Giref, R. C. Greer, V. Gupta, S. Haller, A. Haselbeck, S. I. Hay, M. Holm, S. Hopkins, Y. Hsia, K. C. Iregbu, J. Jacobs, D. Jarovsky, F. Javanmardi, A. W. J. Jenney, M. Khorana, S. Khusuwan, N. Kissoon, E. Kobeissi, T. Kostyanev, F. Krapp, R. Krumkamp, A. Kumar, H. H. Kyu, C. Lim, K. Lim, D. Limmathurotsakul, M. J. Loftus, M. Lunn, J. Ma, A. Manoharan, F. Marks, J. May, M. Mayxay, N. Mturi, T. Munera-Huertas, P. Musicha, L. A. Musila, M. M. Mussi-Pinhata, R. N. Naidu, T. Nakamura, R. Nanavati, S. Nangia, P. Newton, C. Ngoun, A. Novotney, D. Nwakanma, C. W. Obiero, T. J. Ochoa, A. Olivas-Martinez, P. Olliaro, E. Ooko, E. Ortiz-Brizuela, P. Ounchanum, G. D. Pak, J. L. Paredes, A. Y. Peleg, C. Perrone, T. Phe, K. Phommasone, N. Plakkal, A. Ponce-de-Leon, M. Raad, T. Ramdin, S. Rattanavong, A. Riddell, T. Roberts, J. V. Robotham, A. Roca, V. D. Rosenthal, K. E. Rudd, N. Russell, H. S. Sader, W. Saengchan, J. Schnall, J. A. G. Scott, S. Seekaew, M. Sharland, M. Shivamallappa, J. Sifuentes-Osornio, A. J. Simpson, N. Steenkeste, A. J. Stewardson, T. Stoeva, N. Tasak, A. Thaiprakong, G. Thwaites, C. Tigoi, C. Turner, P. Turner, H. R. van Doorn, S. Velaphi, A. Vongpradith, M. Vongsouvath, H. Vu, T. Walsh, J. L. Walson, S. Waner, T. Wangrangsimakul, P. Wannapinij, T. Wozniak, T. E. M. W. Y. Sharma, K. C. Yu, P. Zheng, B. Sartorius, A. D. Lopez, A. Stergachis, C. Moore, C. Dolecek and M. Naghavi, Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis, Lancet, 2022, 399, 629–655 CrossRef CAS PubMed.
- J. Y. Lin, Z. C. Zhu, J. Zhu, L. Chen and H. Du, Antibiotic heteroresistance in Klebsiella pneumoniae: Definition, detection methods, mechanisms, and combination therapy, Microbiol. Res., 2024, 283, 127701 CrossRef CAS PubMed.
- L. Li, X. Gao, M. Li, Y. Liu, J. Ma, X. Wang, Z. Yu, W. Cheng, W. Zhang, H. Sun, X. Song and Z. Wang, Relationship between biofilm formation and antibiotic resistance of Klebsiella pneumoniae and updates on antibiofilm therapeutic strategies, Front. Cell. Infect. Microbiol., 2024, 14, 1324895 CrossRef CAS PubMed.
- J. Garnacho-Montero and J.-F. Timsit, Managing Acinetobacter baumannii infections, Curr. Opin. Infect. Dis., 2019, 32, 69–76 CrossRef PubMed.
- M. Nguyen and S. G. Joshi, Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: a scientific review, J. Appl. Microbiol., 2021, 131, 2715–2738 CrossRef CAS PubMed.
- E. Tacconelli, E. Carrara, A. Savoldi, S. Harbarth, M. Mendelson, D. L. Monnet, C. Pulcini, G. Kahlmeter, J. Kluytmans, Y. Carmeli, M. Ouellette, K. Outterson, J. Patel, M. Cavaleri, E. M. Cox, C. R. Houchens, M. L. Grayson, P. Hansen, N. Singh, U. Theuretzbacher, N. Magrini and WHO Pathogens Priority List Working Group, Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis, Lancet Infect. Dis., 2018, 18, 318–327 CrossRef PubMed.
- J. Botelho, F. Grosso and L. Peixe, Antibiotic resistance in Pseudomonas aeruginosa – Mechanisms, epidemiology and evolution, Drug Resistance Updates, 2019, 44, 100640 CrossRef PubMed.
- M. W. Azam and A. U. Khan, Updates on the pathogenicity status of Pseudomonas aeruginosa, Drug Discovery Today, 2019, 24, 350–359 CrossRef PubMed.
- L. R. Mulcahy, V. M. Isabella and K. Lewis, Pseudomonas aeruginosa Biofilms in Disease, Microb. Ecol., 2014, 68, 1–12 CrossRef CAS PubMed.
- S. Qin, W. Xiao, C. Zhou, Q. Pu, X. Deng, L. Lan, H. Liang, X. Song and M. Wu, Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics, Signal Transduction Targeted Ther., 2022, 7, 1–27 CrossRef PubMed.
- A. Davin-Regli and J.-M. Pagès, Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment, Front. Microbiol., 2015, 6, 392 Search PubMed.
- M. L. Mezzatesta, F. Gona and S. Stefani, Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance, Future Microbiol., 2012, 7, 887–902 CrossRef CAS PubMed.
- D. M. P. De Oliveira, B. M. Forde, T. J. Kidd, P. N. A. Harris, M. A. Schembri, S. A. Beatson, D. L. Paterson and M. J. Walker, Antimicrobial Resistance in ESKAPE Pathogens, Clin. Microbiol. Rev., 2020, 33(3), e00181-19 CrossRef PubMed.
- M. K. Abban, E. A. Ayerakwa, L. Mosi and A. Isawumi, The burden of hospital acquired infections and antimicrobial resistance, Heliyon, 2023, 9(10), e20561 CrossRef PubMed.
- J. B. Kaper, J. P. Nataro and H. L. T. Mobley, Pathogenic Escherichia coli, Nat. Rev. Microbiol., 2004, 2, 123–140 CrossRef CAS PubMed.
- S. Makvana and L. R. Krilov, Escherichia coli Infections, Pediatr. Rev., 2015, 36, 167–170 CrossRef PubMed.
- T. Mestrovic, G. R. Aguilar, L. R. Swetschinski, K. S. Ikuta, A. P. Gray, N. D. Weaver, C. Han, E. E. Wool, A. G. Hayoon, S. I. Hay, C. Dolecek, B. Sartorius, C. J. L. Murray, I. Y. Addo, B. O. Ahinkorah, A. Ahmed, M. A. Aldeyab, K. Allel, R. Ancuceanu, A. E. Anyasodor, M. Ausloos, F. Barra, A. S. Bhagavathula, D. Bhandari, S. Bhaskar, N. Cruz-Martins, A. Dastiridou, K. Dokova, E. Dubljanin, O. C. Durojaiye, A. F. Fagbamigbe, S. Ferrero, P. A. Gaal, V. B. Gupta, V. K. Gupta, V. K. Gupta, C. Herteliu, S. Hussain, I. M. Ilic, M. D. Ilic, E. Jamshidi, T. Joo, A. Karch, A. Kisa, S. Kisa, T. Kostyanev, H. H. Kyu, J. Lám, G. Lopes, A. G. Mathioudakis, A.-F. A. Mentis, I. M. Michalek, M. A. Moni, C. E. Moore, F. Mulita, I. Negoi, R. I. Negoi, T. Palicz, A. Pana, J. Perdigão, I.-R. Petcu, N. Rabiee, D. L. Rawaf, S. Rawaf, M. Z. Shakhmardanov, A. Sheikh, L. M. L. R. Silva, V. Y. Skryabin, A. A. Skryabina, B. Socea, A. Stergachis, T. Z. Stoeva, C. D. Sumi, A. Thiyagarajan, M. R. Tovani-Palone, M. Yesiltepe, S. B. Zaman and M. Naghavi, The burden of bacterial antimicrobial resistance in the WHO European region in 2019: a cross-country systematic analysis, Lancet Public Health, 2022, 7, e897–e913 CrossRef PubMed.
- W. J. Bradshaw, A. K. Roberts, C. C. Shone and K. R. Acharya, The structure of the S-layer of Clostridium difficile, J. Cell Commun. Signaling, 2018, 12, 319–331 CrossRef PubMed.
- M. Kazanowski, S. Smolarek, F. Kinnarney and Z. Grzebieniak, Clostridium difficile: epidemiology, diagnostic and therapeutic possibilities—a systematic review, Tech. Coloproctol., 2014, 18, 223–232 CrossRef CAS PubMed.
- W. K. Smits, D. Lyras, D. B. Lacy, M. H. Wilcox and E. J. Kuijper, Clostridium difficile infection, Nat. Rev. Dis. Primers, 2016, 2, 1–20 Search PubMed.
- M. Kachrimanidou and N. Malisiovas, Clostridium difficile Infection: A Comprehensive Review, Crit. Rev. Microbiol., 2011, 37, 178–187 CrossRef CAS PubMed.
- M. Sebaihia, B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeño-Tárraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell and J. Parkhill, The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome, Nat. Genet., 2006, 38, 779–786 CrossRef PubMed.
- E. Tartari, S. Tomczyk, D. Pires, B. Zayed, A. P. Coutinho Rehse, P. Kariyo, V. Stempliuk, W. Zingg, D. Pittet and B. Allegranzi, Implementation of the infection prevention and control core components at the national level: a global situational analysis, J. Hosp. Infect., 2021, 108, 94–103 CrossRef CAS PubMed.
- J. E. Wißmann, L. Kirchhoff, Y. Brüggemann, D. Todt, J. Steinmann and E. Steinmann, Persistence of Pathogens on Inanimate Surfaces: A Narrative Review, Microorganisms, 2021, 9, 343 CrossRef PubMed.
- N. A. Binte, M. Salleh, Y. Tanaka, L. Sutarlie and X. Su, Detecting bacterial infections in wounds: a review of biosensors and wearable sensors in comparison with conventional laboratory methods, Analyst, 2022, 147, 1756–1776 RSC.
- J. W.-F. Law, N.-S. Ab Mutalib, K.-G. Chan and L.-H. Lee, Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations, Front. Microbiol., 2015, 5 DOI:10.3389/fmicb.2014.00770.
- F. Yeni, S. Acar, Ö. G. Polat, Y. Soyer and H. Alpas, Rapid and standardized methods for detection of foodborne pathogens and mycotoxins on fresh produce, Food Control, 2014, 40, 359–367 CrossRef CAS.
- P. Rajapaksha, A. Elbourne, S. Gangadoo, R. Brown, D. Cozzolino and J. Chapman, A review of methods for the detection of pathogenic microorganisms, Analyst, 2019, 144, 396–411 RSC.
- M. Bühlmann, K. Bögli-Stuber, S. Droz and K. Mühlemann, Rapid Screening for Carriage of Methicillin-Resistant Staphylococcus aureus by PCR and Associated Costs, J. Clin. Microbiol., 2008, 46, 2151–2154 CrossRef PubMed.
- M. Hoyos-Nogués, F. J. Gil and C. Mas-Moruno, Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies, Molecules, 2018, 23, 1683 CrossRef PubMed.
- N. Jiang, R. Ahmed, M. Damayantharan, B. Ünal, H. Butt and A. K. Yetisen, Lateral and Vertical Flow Assays for Point-of-Care Diagnostics, Adv. Healthcare Mater., 2019, 8(14) DOI:10.1002/adhm.201900244.
- Y. Manmana, T. Kubo and K. Otsuka, Recent developments of point-of-care (POC) testing platform for biomolecules, TrAC, Trends Anal. Chem., 2021, 135, 116160 CrossRef CAS.
- J. R. Choi, Development of Point-of-Care Biosensors for COVID-19, Front. Chem., 2020, 8, 517 CrossRef CAS PubMed.
- D. Grieshaber, R. MacKenzie, J. Vörös and E. Reimhult, Electrochemical Biosensors – Sensor Principles and Architectures, Sensors, 2008, 8, 1400–1458 CrossRef CAS PubMed.
- A. Mokhtarzadeh, R. Eivazzadeh-Keihan, P. Pashazadeh, M. Hejazi, N. Gharaatifar, M. Hasanzadeh, B. Baradaran and M. de la Guardia, Nanomaterial-based biosensors for detection of pathogenic virus, TrAC, Trends Anal. Chem., 2017, 97, 445–457 CrossRef CAS PubMed.
- E. Cesewski and B. N. Johnson, Electrochemical biosensors for pathogen detection, Biosens. Bioelectron., 2020, 159, 112214 CrossRef CAS PubMed.
- M. Srivastava, N. Srivastava, P. K. Mishra and B. D. Malhotra, Prospects of nanomaterials-enabled biosensors for COVID-19 detection, Sci. Total Environ., 2021, 754, 142363 CrossRef CAS PubMed.
- L. Sutarlie, S. Y. Ow and X. Su, Nanomaterials-based biosensors for detection of microorganisms and microbial toxins, Biotechnol. J., 2016, 12(4) DOI:10.1002/biot.201500459.
- Z. Farka, T. Juřík, D. Kovář, L. Trnková and P. Skládal, Nanoparticle-Based Immunochemical Biosensors and Assays: Recent Advances and Challenges, Chem. Rev., 2017, 117, 9973–10042 CrossRef CAS PubMed.
- G. Maduraiveeran, M. Sasidharan and V. Ganesan, Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications, Biosens. Bioelectron., 2018, 103, 113–129 CrossRef CAS PubMed.
- C. Dincer, R. Bruch, E. Costa-Rama, M. T. Fernández-Abedul, A. Merkoçi, A. Manz, G. A. Urban and F. Güder, Disposable Sensors in Diagnostics, Food, and Environmental Monitoring, Adv. Mater., 2019, 31, 1806739 CrossRef PubMed.
- L. Rodríguez-Lorenzo, R. de la Rica, R. A. Álvarez-Puebla, L. M. Liz-Marzán and M. M. Stevens, Plasmonic nanosensors with inverse sensitivity by means of enzyme-guided crystal growth, Nat. Mater., 2012, 11, 604–607 CrossRef PubMed.
- R. de la Rica and M. M. Stevens, Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye, Nat. Nanotechnol., 2012, 7, 821–824 CrossRef CAS PubMed.
- I. Bobrinetskiy, M. Radovic, F. Rizzotto, P. Vizzini, S. Jaric, Z. Pavlovic, V. Radonic, M. V. Nikolic and J. Vidic, Advances in Nanomaterials-Based Electrochemical Biosensors for Foodborne Pathogen Detection, Nanomaterials, 2021, 11, 2700 CrossRef CAS PubMed.
- X. Lin, M. Zhao, T. Peng, P. Zhang, R. Shen and Y. Jia, Detection and discrimination of pathogenic bacteria with nanomaterials-based optical biosensors: A review, Food Chem., 2023, 426, 136578 CrossRef CAS PubMed.
- G. Wu, H. Qiu, X. Liu, P. Luo, Y. Wu and Y. Shen, Nanomaterials-based fluorescent assays for pathogenic bacteria in food-related matrices, Trends Food Sci. Technol., 2023, 142, 104214 CrossRef CAS.
- A. P. F. Turner, Biosensors: sense and sensibility, Chem. Soc. Rev., 2013, 42, 3184–3196 RSC.
- T. Yu, Y. Fu, J. He, J. Zhang and Y. Xianyu, Identification of Antibiotic Resistance in ESKAPE Pathogens through Plasmonic Nanosensors and Machine Learning, ACS Nano, 2023, 17, 4551–4563 CrossRef CAS PubMed.
- Y. Shen, T. Wu, Y. Zhang, N. Ling, L. Zheng, S.-L. Zhang, Y. Sun, X. Wang and Y. Ye, Engineering of a Dual-Recognition Ratiometric Fluorescent Nanosensor with a Remarkably Large Stokes Shift for Accurate Tracking of Pathogenic Bacteria at the Single-Cell Level, Anal. Chem., 2020, 92, 13396–13404 CrossRef CAS PubMed.
- Y. Wang, H. He, H. Liu, C. Feng and Z. Yao, An Electrochemical DNA-Hybridization Assay for Acinetobacter baumannii Detection, Int. J. Electrochem. Sci., 2018, 13, 1051–1061 CrossRef CAS.
- F. Bahavarnia, P. Pashazadeh-Panahi, M. Hasanzadeh and N. Razmi, DNA based biosensing of Acinetobacter baumannii using nanoparticles aggregation method, Heliyon, 2020, 6, e04474 CrossRef PubMed.
- J. Hu, M. Ghosh, M. J. Miller and P. W. Bohn, Whole-cell biosensing by siderophore-based molecular recognition and localized surface plasmon resonance, Anal. Methods, 2019, 11, 296–302 RSC.
- D. Bahari, B. Babamiri, A. Salimi and H. Salimizand, Ratiometric fluorescence resonance energy transfer aptasensor for highly sensitive and selective detection of Acinetobacter baumannii bacteria in urine sample using carbon dots as optical nanoprobes, Talanta, 2021, 221, 121619 CrossRef CAS PubMed.
- Y.-L. Bai, M. Shahed-Al-Mahmud, K. Selvaprakash, N.-T. Lin and Y.-C. Chen, Tail Fiber Protein-Immobilized Magnetic Nanoparticle-Based Affinity Approaches for Detection of Acinetobacter baumannii, Anal. Chem., 2019, 91, 10335–10342 CrossRef CAS PubMed.
- Z. Zhu, L. Shi, H. Feng and H. Susan Zhou, Single domain antibody coated gold nanoparticles as enhancer for Clostridium difficile toxin detection by electrochemical impedance immunosensors, Bioelectrochemistry, 2015, 101, 153–158 CrossRef CAS PubMed.
- R. Nazari-Vanani, N. Sattarahmady, H. Yadegari and H. Heli, A novel and ultrasensitive electrochemical DNA biosensor based on an ice crystals-like gold nanostructure for the detection of Enterococcus faecalis gene sequence, Colloids Surf., B, 2018, 166, 245–253 CrossRef CAS PubMed.
- J. L. de Miranda, M. D. L. Oliveira, I. S. Oliveira, I. A. M. Frias, O. L. Franco and C. A. S. Andrade, A simple nanostructured biosensor based on clavanin A antimicrobial peptide for gram-negative bacteria detection, Biochem. Eng. J., 2017, 124, 108–114 CrossRef CAS.
- R. Sivakumar, S. Y. Park and N. Y. Lee, Quercetin-Mediated Silver Nanoparticle Formation for the Colorimetric Detection of Infectious Pathogens Coupled with Loop-Mediated Isothermal Amplification, ACS Sens., 2023, 8, 1422–1430 CrossRef CAS PubMed.
- R. Yan, Z. Shou, J. Chen, H. Wu, Y. Zhao, L. Qiu, P. Jiang, X.-Z. Mou, J. Wang and Y.-Q. Li, On–Off–On Gold Nanocluster-Based Fluorescent Probe for Rapid Escherichia coli Differentiation, Detection and Bactericide Screening, ACS Sustainable Chem. Eng., 2018, 6, 4504–4509 CrossRef CAS.
- S. Ghayyem and F. Faridbod, Detection of pathogenic bacteria in milk and whey samples using a fluorescence resonance energy transfer aptasensor based on cerium oxide nanoparticles, Anal. Methods, 2022, 14, 813–819 RSC.
- L. Zheng, P. Qi and D. Zhang, DNA-templated fluorescent silver nanoclusters for sensitive detection of pathogenic bacteria based on MNP-DNAzyme-AChE complex, Sens. Actuators, B, 2018, 276, 42–47 CrossRef CAS.
- C. Zong, L. Fang, F. Song, A. Wang and Y. Wan, Fluorescent fingerprint bacteria by multi-channel magnetic fluorescent nanosensor, Sens. Actuators, B, 2019, 289, 234–241 CrossRef CAS.
- L. Zheng, P. Qi and D. Zhang, Identification of bacteria by a fluorescence sensor array based on three kinds of receptors functionalized carbon dots, Sens. Actuators, B, 2019, 286, 206–213 CrossRef CAS.
- L. Fu, Q. Chen and L. Jia, Carbon dots and gold nanoclusters assisted construction of a ratiometric fluorescent biosensor for detection of Gram-negative bacteria, Food Chem., 2022, 374, 131750 CrossRef CAS PubMed.
- A. Gupta, M. Garg, S. Singh, A. Deep and A. L. Sharma, Highly Sensitive Optical Detection of Escherichia coli Using Terbium-Based Metal-Organic Framework, ACS Appl. Mater. Interfaces, 2020, 12, 48198–48205 CrossRef CAS PubMed.
- Y. Wang, Q. Li, R. Zhang, K. Tang, C. Ding and S. Yu, SERS-based immunocapture and detection of pathogenic bacteria using a boronic acid-functionalized polydopamine-coated Au@Ag nanoprobe, Microchim. Acta, 2020, 187, 290 CrossRef CAS PubMed.
- H. Ilhan, B. Guven, U. Dogan, H. Torul, S. Evran, D. Çetin, Z. Suludere, N. Saglam, İ. H. Boyaci and U. Tamer, The coupling of immunomagnetic enrichment of bacteria with paper-based platform, Talanta, 2019, 201, 245–252 CrossRef CAS PubMed.
- M. Mathelié-Guinlet, T. Cohen-Bouhacina, I. Gammoudi, A. Martin, L. Béven, M.-H. Delville and C. Grauby-Heywang, Silica nanoparticles-assisted electrochemical biosensor for the rapid, sensitive and specific detection of Escherichia coli, Sens. Actuators, B, 2019, 292, 314–320 CrossRef.
- A. Deb, M. Gogoi, T. K. Mandal, S. Sinha and P. S. G. Pattader, Specific Instantaneous Detection of Klebsiella pneumoniae for UTI Diagnosis with a Plasmonic Gold Nanoparticle Conjugated Aptasensor, ACS Appl. Bio Mater., 2023, 6, 3309–3318 CrossRef CAS PubMed.
- Z. Zhang, H.-W. Yu, G.-C. Wan, J.-H. Jiang, N. Wang, Z.-Y. Liu, D. Chang and H.-Z. Pan, A Label-Free Electrochemical Biosensor Based on a Reduced Graphene Oxide and Indole-5-Carboxylic Acid Nanocomposite for the Detection of Klebsiella pneumoniae, J. AOAC Int., 2017, 100, 548–552 CrossRef CAS PubMed.
- R. Das, A. Dhiman, A. Kapil, V. Bansal and T. K. Sharma, Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme, Anal. Bioanal. Chem., 2019, 411, 1229–1238 CrossRef CAS PubMed.
- S. Alhogail, G. A. R. Y. Suaifan, F. J. Bikker, W. E. Kaman, K. Weber, D. Cialla-May, J. Popp and M. M. Zourob, Rapid Colorimetric Detection of Pseudomonas aeruginosa in Clinical Isolates Using a Magnetic Nanoparticle Biosensor, ACS Omega, 2019, 4, 21684–21688 CrossRef CAS PubMed.
- A. Cui, Y. Hou, J. Zhang, X. Mu, H. Wang, Y. Sun, H. Xu and G. Shan, Dual-mode sensing platform based on aptamer-tunable catalytic activity of mesoporous polydopamine/MnO2 nanozymes for detecting S. aureus, Sens. Actuators, B, 2023, 393, 134218 CrossRef CAS.
- J. Li, H. Jiang, X. Rao, Z. Liu, H. Zhu and Y. Xu, Point-of-Care Testing of Pathogenic Bacteria at the Single-Colony Level via Gas Pressure Readout Using Aptamer-Coated Magnetic CuFe2O4 and Vancomycin-Capped Platinum Nanoparticles, Anal. Chem., 2019, 91, 1494–1500 CrossRef CAS PubMed.
- Y. Pang, N. Wan, L. Shi, C. Wang, Z. Sun, R. Xiao and S. Wang, Dual-recognition surface-enhanced Raman scattering(SERS)biosensor for pathogenic bacteria detection by using vancomycin-SERS tags and aptamer-Fe3O4@Au, Anal. Chim. Acta, 2019, 1077, 288–296 CrossRef CAS PubMed.
- L. Huang, N. Yuan, W. Guo, Y. Zhang and W. Zhang, An electrochemical biosensor for the highly sensitive detection of Staphylococcus aureus based on SRCA-CRISPR/Cas12a, Talanta, 2023, 252, 123821 CrossRef CAS PubMed.
- S. Wu, N. Duan, Z. Shi, C. Fang and Z. Wang, Simultaneous Aptasensor for Multiplex Pathogenic Bacteria Detection Based on Multicolor Upconversion Nanoparticles Labels, Anal. Chem., 2014, 86, 3100–3107 CrossRef CAS PubMed.
- L. Yang, W. Deng, C. Cheng, Y. Tan, Q. Xie and S. Yao, Fluorescent Immunoassay for the Detection of Pathogenic Bacteria at the Single-Cell Level Using Carbon Dots-Encapsulated Breakable Organosilica Nanocapsule as Labels, ACS Appl. Mater. Interfaces, 2018, 10, 3441–3448 CrossRef CAS PubMed.
- N. Bhardwaj, S. K. Bhardwaj, J. Mehta, K.-H. Kim and A. Deep, MOF–Bacteriophage Biosensor for Highly Sensitive and Specific Detection of Staphylococcus aureus, ACS Appl. Mater. Interfaces, 2017, 9, 33589–33598 CrossRef CAS PubMed.
- M. Sharafeldin and J. J. Davis, Point of Care Sensors for Infectious Pathogens, Anal. Chem., 2021, 93, 184–197 CrossRef CAS PubMed.
- T. Sakata, Signal transduction interfaces for field-effect transistor-based biosensors, Commun. Chem., 2024, 7, 1–14 CrossRef PubMed.
- E. Sohouli, M. Ghalkhani, T. Zargar, Y. Joseph, M. Rahimi-Nasrabadi, F. Ahmadi, M. E. Plonska-Brzezinska and H. Ehrlich, A new electrochemical aptasensor based on gold/nitrogen-doped carbon nano-onions for the detection of Staphylococcus aureus, Electrochim. Acta, 2022, 403, 139633 CrossRef CAS.
- D. E. Voth and J. D. Ballard, Clostridium difficile Toxins: Mechanism of Action and Role in Disease, Clin. Microbiol. Rev., 2005, 18, 247–263 CrossRef CAS PubMed.
- N. Bagdasarian, K. Rao and P. N. Malani, Diagnosis and Treatment of Clostridium difficile in Adults: A Systematic Review, JAMA, J. Am. Med. Assoc., 2015, 313, 398 CrossRef PubMed.
- R. Abedi-Firoozjah, H. Ebdali, M. Soltani, P. Abdolahi-Fard, M. Heydari, E. Assadpour, M. Azizi-Lalabadi, F. Zhang and S. M. Jafari, Nanomaterial-based sensors for the detection of pathogens and microbial toxins in the food industry; a review on recent progress, Coord. Chem. Rev., 2024, 500, 215545 CrossRef CAS.
- C. Rensing and G. Grass, Escherichia coli mechanisms of copper homeostasis in a changing environment, FEMS Microbiol. Rev., 2003, 27, 197–213 CrossRef CAS PubMed.
- G. Zanchetta, R. Lanfranco, F. Giavazzi, T. Bellini and M. Buscaglia, Emerging applications of label-free optical biosensors, Nanophotonics, 2017, 6, 627–645 CAS.
- A. Ahangari, P. Mahmoodi and A. Mohammadzadeh, Advanced nano biosensors for rapid detection of zoonotic bacteria, Biotechnol. Bioeng., 2023, 120, 41–56 CrossRef CAS PubMed.
- B. Sepúlveda, P. C. Angelomé, L. M. Lechuga and L. M. Liz-Marzán, LSPR-based nanobiosensors, Nano Today, 2009, 4, 244–251 CrossRef.
- H. Khateb, G. Klös, R. L. Meyer and D. S. Sutherland, Development of a Label-Free LSPR-Apta Sensor for Staphylococcus aureus Detection, ACS Appl. Bio Mater., 2020, 3, 3066–3077 CrossRef CAS PubMed.
- X. Liu, Q. Zhang, W. Knoll, B. Liedberg and Y. Wang, Rational Design of Functional Peptide–Gold Hybrid Nanomaterials for Molecular Interactions, Adv. Mater., 2020, 32, 2000866 CrossRef CAS PubMed.
- R. M. Clegg, Fluorescence resonance energy transfer, Curr. Opin. Biotechnol., 1995, 6, 103–110 CrossRef CAS PubMed.
- J. Shi, F. Tian, J. Lyu and M. Yang, Nanoparticle based fluorescence resonance energy transfer (FRET) for biosensing applications, J. Mater. Chem. B, 2015, 3, 6989–7005 RSC.
- K. C. Bantz, A. F. Meyer, N. J. Wittenberg, H. Im, Ö. Kurtuluş, S. Hoon Lee, N. C. Lindquist, S.-H. Oh and C. L. Haynes, Recent progress in SERS biosensing, Phys. Chem. Chem. Phys., 2011, 13, 11551–11567 RSC.
- R. A. Tripp, R. A. Dluhy and Y. Zhao, Novel nanostructures for SERS biosensing, Nano Today, 2008, 3, 31–37 CrossRef CAS.
- Y. Zheng, X. Wang and H. Jiang, Label-free detection of Acinetobacter baumannii through the induced fluorescence quenching of thiolated AuAg nanoclusters, Sens. Actuators, B, 2018, 277, 388–393 CrossRef CAS.
- Z. Chen, Z. Zhang, J. Qi, J. You, J. Ma and L. Chen, Colorimetric detection of heavy metal ions with various chromogenic materials: Strategies and applications, J. Hazard. Mater., 2023, 441, 129889 CrossRef CAS PubMed.
- S. Paterson and R. de la Rica, Solution-based nanosensors for in-field detection with the naked eye, Analyst, 2015, 140, 3308–3317 RSC.
- X. Huang and M. A. El-Sayed, Gold nanoparticles: Optical properties and implementations in cancer diagnosis and photothermal therapy, J. Adv. Res., 2010, 1, 13–28 CrossRef.
- E. Madkour, A. Abou Zeid, S. Abdel Ghany, F. M. Alshehrei, D. El-Ghareeb and M. Abdel-Hakeem, Sensitive and selective colorimetric detection of Staphylococcus aureus-SPA gene by engineered gold nanosensor, Saudi J. Biol. Sci., 2023, 30, 103559 CrossRef CAS PubMed.
- G. H. Tondro, R. Dehdari Vais and N. Sattarahmady, An optical genosensor for Enterococcus faecalis using conjugated gold nanoparticles-rRNA oligonucleotide, Sens. Actuators, B, 2018, 263, 36–42 CrossRef CAS.
- Z. Wang, J. Zhao, X. Xu, L. Guo, L. Xu, M. Sun, S. Hu, H. Kuang, C. Xu and A. Li, An Overview for the Nanoparticles-Based Quantitative Lateral Flow Assay, Small Methods, 2022, 6, 2101143 CrossRef CAS PubMed.
- O. Pashchenko, T. Shelby, T. Banerjee and S. Santra, A Comparison of Optical, Electrochemical, Magnetic, and Colorimetric Point-of-Care Biosensors for Infectious Disease Diagnosis, ACS Infect. Dis., 2018, 4, 1162–1178 CrossRef CAS PubMed.
- J. Deng, S. Zhao, Y. Liu, C. Liu and J. Sun, Nanosensors for Diagnosis of Infectious Diseases, ACS Appl. Bio Mater., 2021, 4, 3863–3879 CrossRef CAS PubMed.
- C. Jin, Z. Wu, J. H. Molinski, J. Zhou, Y. Ren and J. X. J. Zhang, Plasmonic nanosensors for point-of-care biomarker detection, Mater. Today Bio, 2022, 14, 100263 CrossRef CAS PubMed.
- N. M. Noah and P. M. Ndangili, Current Trends of Nanobiosensors for Point-of-Care Diagnostics, J. Anal. Methods Chem., 2019, 2019, 2179718 Search PubMed.
- S. Yadav, A. Parihar, M. A. Sadique, P. Ranjan, N. Kumar, A. Singhal and R. Khan, Emerging Point-of-Care Optical Biosensing Technologies for Diagnostics of Microbial Infections, ACS Appl. Opt. Mater., 2023, 1, 1245–1262 CrossRef CAS.
- X. Lin, M. Zhao, T. Peng, P. Zhang, R. Shen and Y. Jia, Detection and discrimination of pathogenic bacteria with nanomaterials-based optical biosensors: A review, Food Chem., 2023, 426, 136578 CrossRef CAS PubMed.
- M. Ahmad, M. Hasan, N. Tarannum, M. Hasan and S. Ahmed, Recent advances in optical and photoelectrochemical nanobiosensor technology for cancer biomarker detection, Biosens. Bioelectron.: X, 2023, 14, 100375 CAS.
- B. Li, X. Li, Y. Dong, B. Wang, D. Li, Y. Shi and Y. Wu, Colorimetric Sensor Array Based on Gold Nanoparticles with Diverse Surface Charges for Microorganisms Identification, Anal. Chem., 2017, 89, 10639–10643 CrossRef CAS PubMed.
- S. Fijan, S. Š. Turk and U. Rozman, Comparison of methods for detection of four common nosocomial pathogens on hospital textiles, Slovenian Journal of Public Health, 2014, 53, 17–25 Search PubMed.
- R. Akbari, M. F. Bafghi and H. Fazeli, Nosocomial Infections Pathogens Isolated from Hospital Personnel, Hospital Environment and Devices, J. Med. Bacteriol., 2018, 7, 22–30 Search PubMed.
- H. R. Kim, S. An and J. Hwang, Aerosol-to-Hydrosol Sampling and Simultaneous Enrichment of Airborne Bacteria For Rapid Biosensing, ACS Sens., 2020, 5, 2763–2771 CrossRef CAS PubMed.
- Y. Lu, Q. Yang and J. Wu, Recent advances in biosensor-integrated enrichment methods for preconcentrating and detecting the low-abundant analytes in agriculture and food samples, TrAC, Trends Anal. Chem., 2020, 128, 115914 CrossRef CAS.
- J. V. Rohit and S. K. Kailasa, Simple and selective detection of pendimethalin herbicide in water and food samples based on the aggregation of ractopamine-dithiocarbamate functionalized gold nanoparticles, Sens. Actuators, B, 2017, 245, 541–550 CrossRef CAS.
- X. Liu, Z. Wu, Q. Zhang, W. Zhao, C. Zong and H. Gai, Single Gold Nanoparticle-Based Colorimetric Detection of Picomolar Mercury Ion with Dark-Field Microscopy, Anal. Chem., 2016, 88, 2119–2124 CrossRef CAS PubMed.
- A. B. Pebdeni, M. Hosseini and A. Barkhordari, Smart fluorescence aptasensor using nanofiber functionalized with carbon quantum dot for specific detection of pathogenic bacteria in the wound, Talanta, 2022, 246, 123454 CrossRef CAS PubMed.
- Y. Liu, X. Wang, X. Shi, M. Sun, L. Wang, Z. Hu, F. Liu, Q. Liu, P. Wang, J. Li and C. Zhao, A colorimetric sensor for Staphylococcus aureus detection based on controlled click chemical-induced aggregation of gold nanoparticles and immunomagnetic separation, Microchim. Acta, 2022, 189, 104 CrossRef CAS PubMed.
- R. Shahbazi, M. Salouti, B. Amini, A. Jalilvand, E. Naderlou, A. Amini and A. Shams, Highly selective and sensitive detection of Staphylococcus aureus with gold nanoparticle-based core-shell nano biosensor, Mol. Cell. Probes, 2018, 41, 8–13 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary file 1. See DOI: https://doi.org/10.1039/d4en00381k |
| This journal is © The Royal Society of Chemistry 2024 |