Synergistic immunotherapy with a calcium-based nanoinducer: evoking pyroptosis and remodeling tumor-associated macrophages for enhanced antitumor immune response†
Fang
Cheng
 ,
Lei
He
,
Jiaqi
Wang
,
Lunhui
Lai
,
Li
Ma
,
Kuiming
Qu
,
Zicheng
Yang
,
Xinyue
Wang
,
Ruyu
Zhao
,
Lixing
Weng
,
Lei
He
,
Jiaqi
Wang
,
Lunhui
Lai
,
Li
Ma
,
Kuiming
Qu
,
Zicheng
Yang
,
Xinyue
Wang
,
Ruyu
Zhao
,
Lixing
Weng
 * and
Lianhui
Wang
* and
Lianhui
Wang
 *
*
State Key Laboratory for Organic Electronics and Information Displays & Jiangsu Key Laboratory for Biosensors, Institute of Advanced Materials (IAM), Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM), Nanjing University of Posts and Telecommunications, Nanjing 210023, P. R. China. E-mail: lxweng@njupt.edu.cn; iamlhwang@njupt.edu.cn
First published on 11th September 2024
Abstract
The challenges posed by low immunogenicity and the immunosuppressive tumor microenvironment (TME) significantly hinder the efficacy of cancer immunotherapy. Pyroptosis, characterized as a pro-inflammatory cell death pathway, emerges as a promising approach to augment immunotherapy by promoting immunogenic cell death (ICD). The predominance of M2 phenotype tumor-associated macrophages (TAMs) in the TME underscores the critical need for TAM reprogramming to mitigate this immunosuppression. Herein, we introduce a calcium-based, intelligent-responsive nanoinducer (CaZCH NPs), designed to concurrently initiate pyroptosis and remodel TAMs, thereby amplifying antitumor immunotherapy effects. Modified with hyaluronic acid, CaZCH NPs can target tumor cells. Once internalized, CaZCH NPs respond to the acidic environment, releasing Ca2+, curcumin and H2O2 to induce mitochondrial Ca2+ overload and oxidation stress, leading to caspase-3/GSDME-mediated cell pyroptosis. Concurrently, O2 produced by CaZCH and pro-inflammatory cytokines from pyroptotic cells work together to shift TAM polarization towards the M1 phenotype, effectively countering TME's immunosuppressive effect. Notably, the synergistic effect of Ca2+-mediated pyroptosis and TAM remodeling demonstrates superior antitumor efficiency in colorectal cancer models. The induced ICD, coupled with M1-type TAMs, effectively enhances immunogenicity and mitigates immunosuppression, promoting dendritic cell maturation and activating CD8+ T cell-dependent systemic antitumor immunity. Our study presents a promising synergistic strategy for achieving highly efficient immunotherapy using a simple calcium-based nanoinducer.
Introduction
Immunotherapy has recently emerged as a revolutionary cancer treatment strategy, promising to suppress and eliminate tumors.1–3 However, its efficacy is limited by low immunogenicity and immunosuppression of the tumor microenvironment (TME).4 Pyroptosis, a pro-inflammatory programmed cell death (PCD), provides a unique opportunity to boost immune response by triggering immunogenic cell death (ICD).5,6 In this process, the gasdermin protein cleaves, releasing the N-terminal fragment that forms pores in the cell membrane, causing cell swelling, membrane rupture, and cellular content release.7–9 Such pyroptotic cells act as an in situ “tumor vaccine”, enhancing immunogenicity and emitting danger-associated molecular patterns (DAMPs) such as calreticulin (CRT) and high mobility group box 1 (HMGB1), which serve as immune adjuvants, recruiting and maturing antigen-presenting cells (APCs) to boost immune activation.10–13 Yet, the immunosuppressive TME remains a significant challenge to pyroptosis’ antitumor efficacy.14Tumor-associated macrophages (TAMs), key players in maintaining the immunosuppressive TME, display two phenotypes: the tumor-suppressing M1 and the tumor-promoting M2.15–20 Tumor metabolism shifts TAMs towards the M2 phenotype,21 but strategies to reprogram TAMs are emerging to combat this issue.22,23 The rapid proliferation of tumor cells significantly contributes to tumor hypoxia, a key factor for inducing M2 polarization. Evidence shows that mitigating TME hypoxia can encourage M2 TAMs to revert to the M1 phenotype, thereby diminishing immunosuppression.24,25 Furthermore, increasing T helper cell (Th1) cytokines, such as interferon-γ (IFN-γ) and lipopolysaccharides (LPS), can also drive TAMs towards the M1 phenotype.26,27 Since pyroptosis releases pro-inflammatory cytokines, it has the potential to reorient TAMs towards the M1 phenotype. Therefore, inducing pyroptosis alongside TAM reprogramming could significantly bolster systemic anti-tumor immunity, offering a novel avenue for cancer therapy.
In this study, a TME-responsive calcium nanoinducer was designed to simultaneously trigger pyroptosis and reprogram TAMs to amplify the antitumor immune response (Scheme 1). Calcium ions, critical for cellular signaling and as immune cell recruiters, can activate T cell receptors and stimulate immune responses when intracellular levels are manipulated.28–31 Furthermore, compared to chemotherapy-induced pyroptosis, the Ca2+-based nanoinducer usually shows reduced side effects and drug resistance.32 Modified with hyaluronic acid (HA), the CaZCH nanoparticles (NPs) internalize into tumor cells through CD44-mediated endocytosis33 and then collapse to release high levels of Ca2+, curcumin (CUR), oxygen (O2), and hydrogen peroxide (H2O2). The combination of Ca2+ and CUR induces mitochondrial Ca2+ overload,34,35 resulting in mitochondrial damage. H2O2 exacerbates intracellular oxidative stress,36 further damaging mitochondria, resulting in cytochrome c (cyt c) release, caspase-3 activation, and gasdermin E (GSDME)-mediated pyroptosis. The concurrent release of O2 and pro-inflammatory cytokines facilitates TAM polarization towards the M1 phenotype, mitigating the TME's immunosuppressive effects. By promoting the maturation of dendritic cells and activating CD8+ T cells, the dual action of Ca2+-induced pyroptosis and TAM reprogramming presents a novel approach to strengthen the immune system's ability to target tumors. This study highlights the synergy between inducing pyroptosis and TAM reprogramming as a potent strategy to invoke a targeted antitumor immune response.
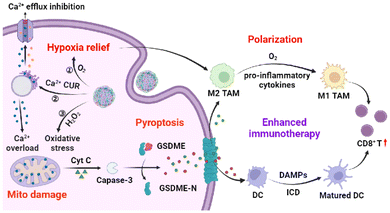 | ||
| Scheme 1 Schematic illustration of TME-responsive CaZCH NPs shows improved immunotherapy via Ca2+-mediated pyroptosis and TAM remodelling. | ||
Experimental
Materials
CaCl2·2H2O (≥99%) and polyvinyl pyrrolidone (PVP, MW ≈ 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) were purchased from Sigma-Aldrich. CUR (98%) and 2-methylimidazole (2-MI, 99%) were purchased from J&K Chemical Ltd. HA (97%) was obtained from Macklin Biochemical Technology Co., Ltd. Zn(NO3)2·6(H2O), NH4OH (25%–28%), H2O2 (30%), ethanol and methanol were purchased from Sinopharm Group Chemical Reagent Co., Ltd. RPMI-1640, DMEM, phosphate buffer solution (PBS) and Hoechst 33342 were purchased from KeyGEN Biotechnology Co., Ltd. Fetal Bovine Serum (FBS) was obtained from Gibco. Hydrogen Peroxide Assay Kit, Calcium Colorimetric Assay Kit, Cell Counting Kit-8, Reactive Oxygen Species Assay Kit, Fluo-4 Calcium Assay Kit, MitoTracker Red CMXRos, ATP Assay Kit, Lactate Dehydrogenase Cytotoxicity Assay Kit, 4% Paraformaldehyde Fix Solution and Triton-X 100 were purchased from Beyotime Biotechnology Co., Ltd. Anti-HMGB1 and anti-CRT were purchased from Sevier Biotechnology Co., Ltd. Anti-GSDME was obtained from Abcam. Goat anti-Rabbit IgG-FITC was obtained from Absin Biotechnology Co., Ltd. APC anti-mouse CD3, FITC anti-mouse CD4, PE anti-mouse CD8a, PE/Cyanine7 anti-mouse CD11c, APC anti-mouse CD80, PE anti-mouse CD86, and TruStain FcX™ (anti-mouse CD16/32) were obtained from BioLegend. All reagents were used as received without any purification, and deionized (DI) water (18.2 MΩ cm−1) was used for solution preparation.
000) were purchased from Sigma-Aldrich. CUR (98%) and 2-methylimidazole (2-MI, 99%) were purchased from J&K Chemical Ltd. HA (97%) was obtained from Macklin Biochemical Technology Co., Ltd. Zn(NO3)2·6(H2O), NH4OH (25%–28%), H2O2 (30%), ethanol and methanol were purchased from Sinopharm Group Chemical Reagent Co., Ltd. RPMI-1640, DMEM, phosphate buffer solution (PBS) and Hoechst 33342 were purchased from KeyGEN Biotechnology Co., Ltd. Fetal Bovine Serum (FBS) was obtained from Gibco. Hydrogen Peroxide Assay Kit, Calcium Colorimetric Assay Kit, Cell Counting Kit-8, Reactive Oxygen Species Assay Kit, Fluo-4 Calcium Assay Kit, MitoTracker Red CMXRos, ATP Assay Kit, Lactate Dehydrogenase Cytotoxicity Assay Kit, 4% Paraformaldehyde Fix Solution and Triton-X 100 were purchased from Beyotime Biotechnology Co., Ltd. Anti-HMGB1 and anti-CRT were purchased from Sevier Biotechnology Co., Ltd. Anti-GSDME was obtained from Abcam. Goat anti-Rabbit IgG-FITC was obtained from Absin Biotechnology Co., Ltd. APC anti-mouse CD3, FITC anti-mouse CD4, PE anti-mouse CD8a, PE/Cyanine7 anti-mouse CD11c, APC anti-mouse CD80, PE anti-mouse CD86, and TruStain FcX™ (anti-mouse CD16/32) were obtained from BioLegend. All reagents were used as received without any purification, and deionized (DI) water (18.2 MΩ cm−1) was used for solution preparation.
Characterization
Scanning electron microscopy (SEM) images were acquired using a Hitachi S-4800 microscope. Transmission electron microscopy (TEM) images were taken using a Hitachi HT7700 microscope. High-angle annular dark-field scanning TEM (HAADF-STEM) images were obtained with an FEI Talos F200X microscope operated at 200 kV. X-ray photoelectron spectroscopy (XPS) was conducted using an X-ray photoelectron spectrometer (Axis Supra). X-ray diffraction (XRD) patterns were tested with a single crystal X-ray diffractometer (Bruker D8 Quest). UV-Vis absorption spectra were recorded using a UV-Vis spectrophotometer (Shimadzu UV-3600). Size distribution and zeta potential were measured by using a dynamic light scattering device (ZetaPALS). Inductively coupled plasma mass spectrometry (ICP-MS) results were recorded using a Nexion 2000 ICP-MS instrument (PerkinElmer). CCK-8 assay was conducted using a microplate reader (SpectraMax M2). Confocal images were acquired using a confocal laser scanning microscope (OLYMPUS, FV1000MPE). Flow cytometry analysis was performed using a NovoCyte flow cytometer, and the data were analyzed using FlowJo software.Synthesis and methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 850g, 15 min), washed thrice with ethanol, and finally redispersed in 10 mL of methanol for further use.
850g, 15 min), washed thrice with ethanol, and finally redispersed in 10 mL of methanol for further use.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 850g, 15 min), washed with methanol, and vacuum-dried into powder. CaO2@ZIF-8/CUR NPs (CaZC NPs) were synthesized using the same method except for the addition of 10 mL of CUR (1 mg mL−1 in methanol) into the reaction solution.
850g, 15 min), washed with methanol, and vacuum-dried into powder. CaO2@ZIF-8/CUR NPs (CaZC NPs) were synthesized using the same method except for the addition of 10 mL of CUR (1 mg mL−1 in methanol) into the reaction solution.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 under stirring overnight. The product was collected by centrifugation (7000g, 15 min) and washed with DI water three times. CaO2@ZIF-8-HA NPs (CaZH NPs) were prepared using the same method, except for that CaZ NPs were used instead of CaZC NPs.
10 under stirring overnight. The product was collected by centrifugation (7000g, 15 min) and washed with DI water three times. CaO2@ZIF-8-HA NPs (CaZH NPs) were prepared using the same method, except for that CaZ NPs were used instead of CaZC NPs.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min, and the resulting supernatant was utilized to determine the concentration of Ca2+ using a calcium colorimetric assay kit following the manufacturer's instructions. Additionally, the intracellular Ca2+ concentration was assessed by staining the cells with Fluo-4 AM and visualizing with CLSM in parallel.
000g for 5 min, and the resulting supernatant was utilized to determine the concentration of Ca2+ using a calcium colorimetric assay kit following the manufacturer's instructions. Additionally, the intracellular Ca2+ concentration was assessed by staining the cells with Fluo-4 AM and visualizing with CLSM in parallel.
Results and discussion
Synthesis and pH-responsive decomposition of CaZCH NPs
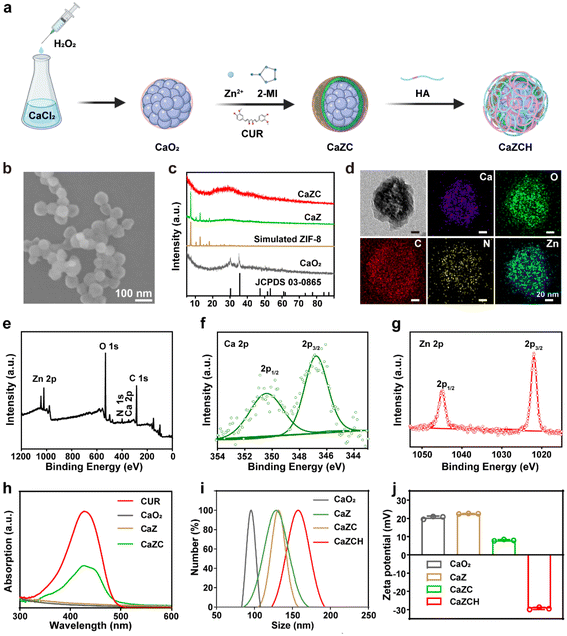
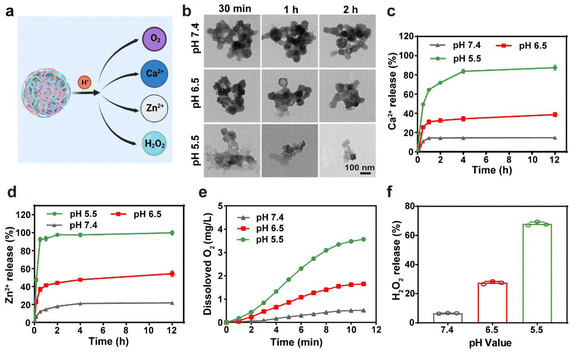
Fig. 1a presents the synthesis of CaZCH NPs, starting with CaO2 NPs prepared from CaCl2 and·H2O2.38 These NPs were encapsulated with ZIF-8 and CUR via a one-pot method to produce CaZC NPs, which were further modified with HA to improve tumor targeting, resulting in CaZCH NPs. The SEM image reveals CaZC NPs as spherical and uniform, averaging 100 nm in diameter (Fig. 1b and Fig. S1†), slightly larger than the initial 90 nm CaO2 NPs (Fig. S2†). The XRD pattern of CaZ NPs exhibits the characteristic peaks of CaO2 and ZIF-8, verifying the successful coating of CaO2 NPs with ZIF-8 (Fig. 1c). The broad peaks observed in the CaZC NPs’ XRD pattern suggest CUR's impact on NPs’ crystallinity. The TEM image and elemental mappings demonstrate uniform elemental distribution in CaZC NPs (Fig. 1d), while XPS further confirms the presence of Ca, O, N, C, and Zn (Fig. 1e–g and Fig. S3†). The absorption band of CUR at 427 nm in the UV-vis spectra of CaZC NPs signifies the successful incorporation of CUR (Fig. 1h). HA modification altered the zeta potential from positive (+7.9 ± 0.54 mV) to negative (−29.5 ± 1.0 mV) and increased the hydrodynamic diameter from 129 nm to 158 nm, indicating an effective surface modification (Fig. 1i–j). The stability of CaZCH NPs was assessed, and the nearly constant hydrodynamic diameter and PDI demonstrate the excellent stability of the synthesized NPs (Fig. S4†). Biocompatibility tests by red blood cell hemolysis analysis showed minimal hemolytic activity, affirming CaZCH NPs’ excellent biocompatibility (Fig. S5†).The pH-sensitive ZIF-8 outer layer of CaZCH NPs allows for their response to the acidic TME, facilitating the rapid release of Ca2+, Zn2+, O2, and H2O2 (Fig. 2a). TEM analysis of CaZCH NPs dispersed in phosphate-buffered saline (PBS) at varying pH levels showed structural stability in pH 7.4 PBS for 2 h, confirming stability under physiological conditions. Nevertheless, decomposition began at pH 6.5, with substantial breakdown at pH 5.5 within 30 min, evidencing pH-sensitive decomposition (Fig. 2b). ICP-MS further highlighted a pronounced ion release of 87.6% for Ca2+ and 98.6% for Zn2+ at pH 5.5 over 12 h, in stark contrast to minimal release at pH 7.4 (14.8% and 11.9%, respectively), aligning with the TEM observations (Fig. 2c and d). The breakdown of the ZIF-8 outer layer triggers CaO2 NPs to react with the acidic TME, generating O2 and H2O2. Oxygen levels measured by a portable dissolved oxygen meter, increased with decreasing pH, reaching up to 3.6 mg L−1 at pH 5.5 within 10 min (Fig. 2e). Similarly, H2O2 production also showed a pH-dependent increase from 6.6% to 67.0% as the pH shifted from 7.4 to 5.5 (Fig. 2f). These results emphasize the pH-responsive decomposition and ion release capabilities of CaZCH NPs.
In vitro antitumor efficacy of CaZCH NPs
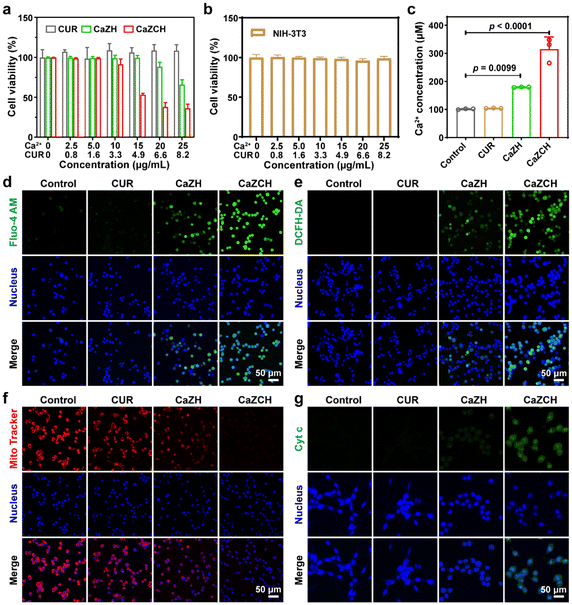
The successful synthesis, biocompatibility, and pH-responsive behavior of CaZCH NPs led us to evaluate their in vitro therapeutic effect. Colorectal cancer (CRC) CT26 cells incubated with these NPs showed increased mean fluorescence intensity (MFI), indicating the maximum cellular uptake at 8 h, as analysed by flow cytometry (FCM) (Fig. S6†). This enhanced fluorescence was attributed to the CUR fluorescence being initially quenched within the NPs and restored upon decomposition, confirming the pH-responsive decomposition of CaZCH NPs within cancer cells. Furthermore, cytotoxicity assessments via cell-counting kit-8 (CCK-8) assays revealed significant inhibition of CT26 cells, demonstrating a robust antitumor activity of the NPs (Fig. 3a). Conversely, NIH-3T3 cell viability remained above 90% (Fig. 3b), confirming the excellent biocompatibility of the NPs. This selective cytotoxicity is attributed to the HA-modified NPs’ ability to target CD44 receptors,39 enhancing their tumor-targeting capability and facilitating ion and drug release in an acidic environment. To verify HA-induced targeting ability, CLSM was employed to analyse the internalization of the NPs. Significantly higher fluorescence intensities were observed in the CaZCH NP group compared to the CaZC NP group at various time points, demonstrating the enhanced targeting ability attributed to HA modification (Fig. S7†).Further investigations into the therapeutic mechanisms revealed that treatment with CaZH and CaZCH NPs resulted in a 1.8-fold and 3.2-fold increase in Ca2+ levels, respectively, indicating Ca2+ overload (Fig. 3c). This finding was corroborated by CLSM using Fluo-4 AM as a probe, which showed significant green fluorescence enhancement in cells treated with CaZH and CaZCH, signalling elevated intracellular Ca2+ levels (Fig. 3d). The more pronounced fluorescence in CaZCH-treated cells is attributed to combined Ca2+ and CUR effects, aligning with calcium assay results. Furthermore, H2O2 generation, a key reactive oxygen species (ROS) source, was monitored using 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA), revealing enhanced green fluorescence in both CaZH and CaZCH groups due to H2O2-induced ROS (Fig. 3e). Notably, the CaZCH group showed higher fluorescence than the CaZH group, attributed to amplified oxidative stress by CUR induced Ca2+ overload. CUR can enhance Ca2+ overload by not only increasing Ca2+ release but also inhibiting Ca2+ efflux out of cells.40 This Ca2+ overload could further amplify intracellular oxidative stress.41
Mitochondria, recognized as the powerhouses of mammalian cells, are crucial for maintaining cellular homeostasis and stress response.42,43 Ca2+ overload and oxidation stress often result in mitochondrial dysfunction.44 Mitochondrial integrity was assessed using MitoTracker Deep Red FM. The CaZCH group exhibited the least red fluorescence, indicating substantial mitochondrial damage from combined Ca2+ and oxidative stress (Fig. 3f). Cyt c, a crucial protein in the apoptotic pathway, resides in the intermembrane space of mitochondria and can be released from these organelles upon mitochondrial damage.45,46 Thus, the levels of cyt c in the cytoplasm were evaluated using immunofluorescence (IF) staining. A 10.1-fold increase in green fluorescence in the CaZCH group compared to the control group indicates effective cyt c release following the CaZCH NP treatment (Fig. 3g and Fig. S8†).
CaZCH NP induced pyroptosis and TAM remodelling
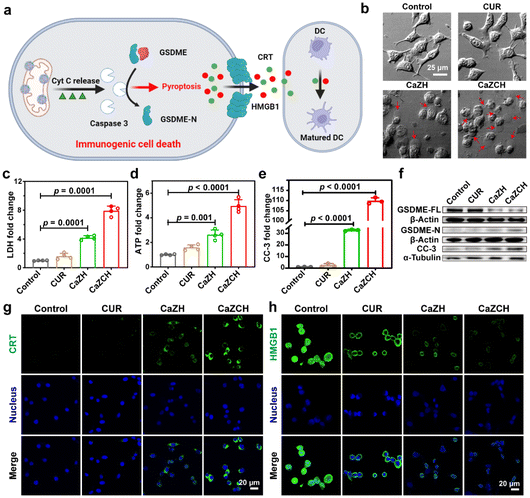
Despite cyt c typically initiating apoptosis, recent research indicates that cells expressing GSDME can undergo a transition from apoptosis to pyroptosis.47,48 Given the high expression of GSDME in the CRC CT26 cell line, we investigated the potential of CaZCH NPs to induce GSDME-mediated pyroptosis and ICD (Fig. 4a). Optical microscopy of treated cells revealed significant swelling and bubbling (indicated by red arrows) in the CaZCH group, suggesting pyroptosis induction, while the control and CUR-treated groups showed few pyroptotic changes (Fig. 4b). Subsequently, several crucial markers of pyroptosis were assessed. Given that pyroptosis-induced membrane rupture leads to the release of cellular contents, the levels of released LDH and ATP in the supernatant were measured. The results revealed an 8.0-fold increase in LDH release and a 4.9-fold increase in ATP release in CaZCH-treated cells compared to the control group (Fig. 4c and d). Caspase-3, a trigger of pyroptosis, cleaves GSDME-FL into GSDME-N. To confirm GSDME-mediated cell pyroptosis, the levels of caspase-3, GSDME-FL, and GSDME-N were analysed. FCM showed a 110-fold increase in CC-3 in the CaZCH group compared to the control group (Fig. 4e and Fig. S9†). Western blot analysis showed a significant decrease in GSDME-FL, along with a concurrent increase in GSDME-N and CC-3 in the CaZCH group (Fig. 4f and Fig. S10†).Pyroptosis, as a pro-inflammatory type of PCD, can rapidly release DAMPs and TAAs, thereby triggering ICD and bolstering antitumor immunity.10–13 In line with this, CRT and HMGB1, the key ICD markers, were evaluated via IF imaging. The CaZCH-treated group displayed marked CRT expression and HMGB1 release (Fig. 4g and h), with relative fluorescence intensities indicating a 28.7-fold increase in CRT and a 2.8-fold increase in HMGB1 compared to the controls (Fig. S11 and S12†), confirming effective ICD induction. These results confirm that CaZCH NPs trigger GSDME-mediated pyroptosis and subsequent ICD.
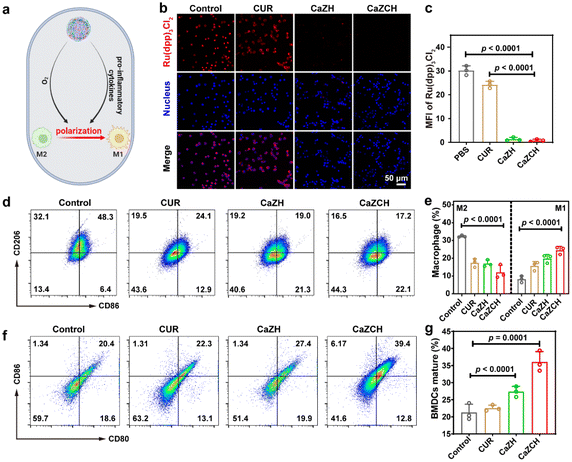
While pyroptosis-induced ICD can substantially enhance the immunogenicity of tumor cells, the immunosuppressive TME mediated by M2-type TAMs remains a significant obstacle to immunotherapeutic efficiency. Given the elevated pro-inflammatory cytokines from pyroptosis and O2 production from CaZCH decomposition, we hypothesized that these factors could remodel TAMs, reversing the immunosuppressive TME (Fig. 5a). To test this, the alleviation of intracellular hypoxia was evaluated using Ru(dpp)3Cl2 as an O2 indicator, whose fluorescence could be quenched by O2.49 Compared to the strong red fluorescence in the control group, fluorescence intensity significantly decreased in tumor cells post CaZCH treatment, indicating the effective hypoxia alleviation (Fig. 5b and c). Furthermore, to examine TAM repolarization, we induced M2-type TAMs in RAW 264.7 macrophages with IL-4 and assessed CD86 and CD206 expressions via FCM to determine M1 and M2 phenotypes. In the CaZCH group, M2-type TAMs (CD206) significantly dropped from 32.1% to 16.5%, while M1-type TAMs (CD86) increased from 6.4% to 22.1% relative to the control, indicating a successful reversal of the immunosuppressive TME through TAM repolarization (Fig. 5d and e).
To evaluate immune activation following CaZCH-mediated ICD and TAM remodeling, BMDCs were co-cultured with the CT26 cell supernatant after various treatments. We measured the expression of costimulatory molecules (CD80 and CD86) to determine DC maturation in vitro. Fig. 5f and g show that mature DCs in the CaZCH group reached 39.4%, significantly exceeding that of all other groups, indicating CaZCH's potent antitumor immune activation. This evidence underscores CaZCH's role in sparking adaptive immune responses via pyroptosis and TAM remodelling.
Antitumor mechanism of CaZCH NPs by transcriptome analyses
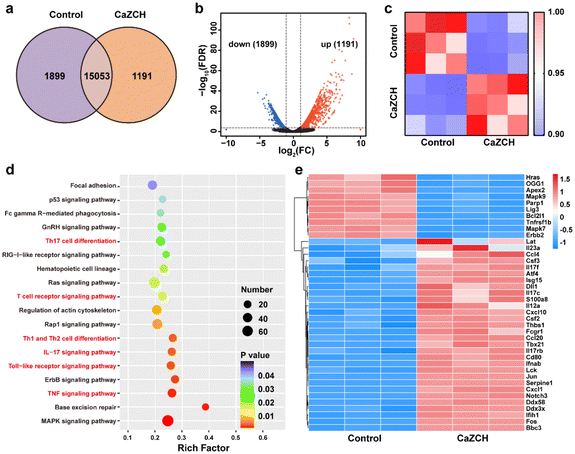
Transcriptome analysis of CT26 cells, with and without the CaZCH NP treatment, was performed to uncover the synergistic antitumor mechanisms involved. Fig. 6a and b illustrate a marked difference in the gene expression between control and CaZCH-treated cells, with 1191 genes upregulated and 1899 genes downregulated. Pearson correlation analysis was conducted on gene expression data from three independent replicates. The results confirmed CaZCH's significant impact on the gene expression and the high consistency across samples (Fig. 6c).Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was conducted on the differentially expressed genes (DEGs) to elucidate the major pathways involved in the process. Fig. 6d illustrates that the predominantly affected signaling pathways include the MAPK pathway, base excision repair, and various inflammatory pathways (such as the TNF signaling pathway and the IL-17 signaling pathway) and immune pathways (such as the Toll-like receptor signaling pathway, T cell receptor signaling pathway, Th1 and Th12 cell differentiation, and Th17 cell differentiation). The gene heatmap indicated 30 upregulated genes and 10 downregulated genes (Fig. 6e).
Upregulated genes such as IL17f, IL17rb, IL17c, and IL12a, associated with inflammatory processes, are likely responsible for pyroptosis-induced inflammation. The upregulation of macrophage-related genes such as Csf2, Csf3, and Tbx21 was consistent with the TAM reprogramming induced by CaZCH. Moreover, the upregulation of immune genes (Ccl10, Ccl20, Ccl40, Cd80, and Fcgr1) suggests CaZCH's role in immune activation. Downregulated genes mainly related to the MAPK pathway and base excision repair imply CaZCH's effect on cell proliferation and damage repair mechanisms of tumor cells. Collectively, these results indicate that CaZCH activates pathways associated with inflammation, immunity, and macrophage function while affecting cell growth and repair, underscoring its potential to suppress tumors and enhance immune responses.
In vivo antitumor ability and immune activation by CaZCH NPs
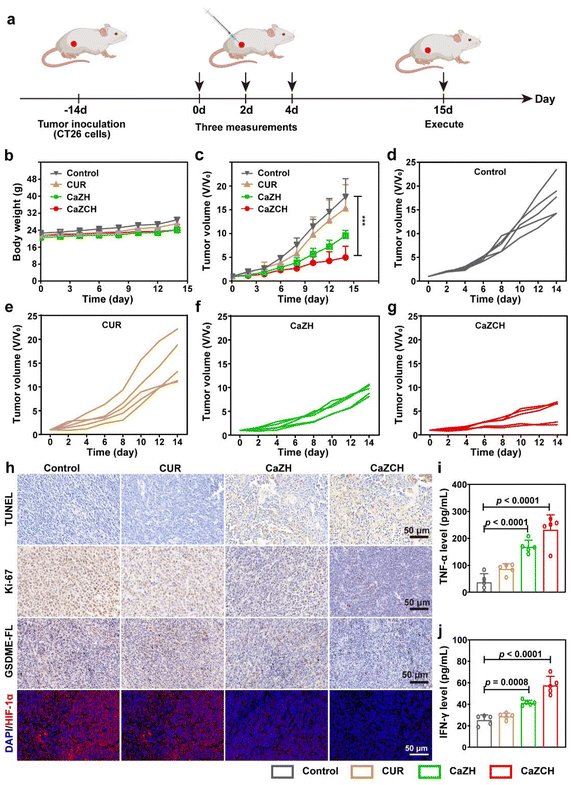
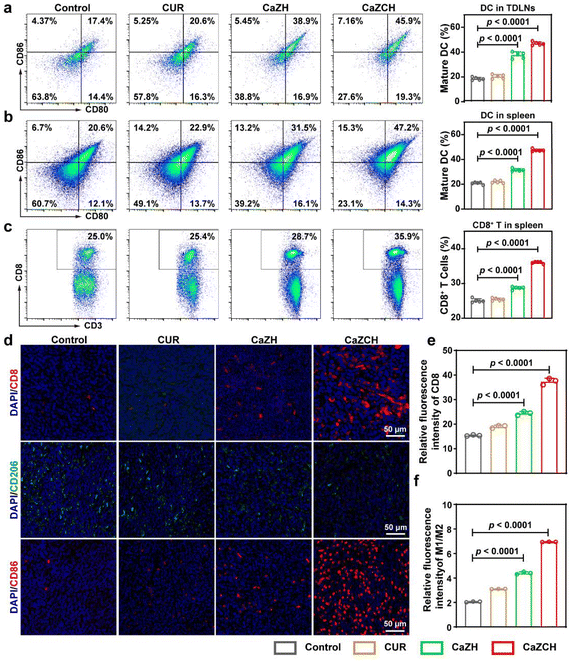
Driven by the notable antitumor capabilities and immune-stimulating effects of CaZCH NPs observed in vitro, we conducted a comprehensive assessment of their therapeutic potential in vivo using a subcutaneous CRC mouse model (Fig. 7a). Throughout the experiment, all mice consistently gained weight, indicating the minimal side effects of CaZCH NPs, as shown in Fig. 7b. H&E staining of tumors and vital organs, detailed in Fig. S13,† confirmed the efficacy of the CaZCH treatment; tumor sections from treated mice showed significant nuclear condensation and reduced area, signifying effective tumor cell eradication. Importantly, there was no evidence of morphological changes in major organs, attesting to the high biocompatibility of CaZCH NPs. Notably, the CaZCH group exhibited substantial tumor suppression, in contrast to the rapid tumor growth observed in both control and CUR groups, highlighting the superior antitumor performance of CaZCH NPs (Fig. 7c–g). Immunohistochemical analysis, including TUNEL staining, identified pronounced apoptosis/necrosis in CaZCH-treated tumors, aligning with Ki-67 staining results that indicated a marked decrease in tumor proliferation (Fig. 7h). The significant reduction of GSDME-FL in the CaZCH group, along with increased levels of TNF-α and IFN-γ measured by ELISA, validated the induction of pyroptosis in vivo (Fig. 7i and j). Additionally, the notable decrease in the hypoxia-inducible factor 1-alpha (HIF-1α) expression in the CaZCH group corroborated in vitro findings of hypoxia mitigation.To determine the in vivo antitumor immune response elicited by CaZCH, tumor-draining lymph nodes (TDLNs), spleens, and tumors were analyzed immunologically. Single-cell suspensions from TDLNs and spleens were stained with fluorochrome-conjugated anti-mouse antibodies for FCM analysis. Fig. 8a shows that CaZCH significantly enhanced DC maturation in TDLN, compared to the control group, with mature DC levels being 2.6-fold higher than in controls. Additionally, the CaZCH group exhibited a notable increase in mature DCs and CD8+ T cells in the spleen, surpassing other groups (Fig. 8b and c). To assess the infiltration of CD8+ T cells into the tumor, fluorescence staining of tumor tissues was performed. As shown in Fig. 8d and e, the most pronounced infiltration of the CD8+ T cell was observed in the CaZCH-treated tumors. To evaluate tumor-infiltrating TAMs, the tumor tissues were stained with CD206 and CD86 to analyze the proportion of M1 and M2 type TAMs.
Quantitative analysis revealed the significantly increased proportion of M1/M2 in the CaZCH group, manifesting the successful remodeling of TAMs in vivo (Fig. 8f). These results highlight the capacity of CaZCH to evoke a robust tumor immune response through pyroptosis and TAM reprogramming.
Conclusions
In conclusion, we have developed an intelligent-responsive calcium-based nanoinducer to harness the synergistic effects of Ca2+-mediated pyroptosis and TAM reprogramming. The synthesized CaZCH NPs degrade under the acidic TME, resulting in the rapid release of Ca2+, CUR, H2O2 and O2. This degradation leads to mitochondrial damage, facilitated by Ca2+ and CUR-induced mitochondrial Ca2+ overload, alongside H2O2-induced oxidative stress, ultimately resulting in cyt c release, caspase-3 activation, and subsequent GSDME-mediated pyroptosis. Additionally, the O2 generated by CaZCH NPs and pro-inflammatory cytokines released from pyroptotic cells collaboratively polarize TAMs toward the M1 phenotype, alleviating the immunosuppressive TME. Overall, CaZCH NPs not only induce immunogenic pyroptosis but also successfully remodel the immunosuppressive TME, providing an efficient strategy to elicit heightened antitumor immune responses. Thus, this collaborative approach involving Ca2+-mediated pyroptosis and TAM reprogramming represents a promising therapeutic avenue through CD8+ T cell-dependent systemic antitumor immunity.Author contributions
Conceptualization: F. C., L. W. and L. W.; methodology: L. H., J. W., L. L. and K. Q.; formal analysis: F. C., L. H., J. W. and L. L.; investigation: L. H., J. W., L. L., and L. M.; visualization: F. C., L. H., J. W. and L. L.; resources: F. C., L. W. and L. W.; writing—original draft preparation: F. C. and L. L.; writing—review and editing: F. C., X. W., L. M. and R. Z.; supervision: F. C., L. W. and L. W.; and funding acquisition: F. C., L. W. and L. W. All authors have read and agreed to the published version of the manuscript.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (22371127, 21902079, 62235008, and 62288102), the Natural Science Foundation of Jiangsu Province-Major Project (BK20212012), the Project of State Key Laboratory of Organic Electronics and Information Displays, Nanjing University of Posts and Telecommunications (GZR2022010028).References
- C. G. Drake, E. J. Lipson and J. R. Brahmer, Nat. Rev. Clin. Oncol., 2014, 11, 24–37 CrossRef CAS.
- Y. Zhang and Z. Zhang, Cell. Mol. Immunol., 2020, 17, 807–821 CrossRef CAS PubMed.
- L. Kraehenbuehl, C.-H. Weng, S. Eghbali, J. D. Wolchok and T. Merghoub, Nat. Rev. Clin. Oncol., 2021, 19, 37–50 CrossRef PubMed.
- D. Ciardiello, P. P. Vitiello, C. Cardone, G. Martini, T. Troiani, E. Martinelli and F. Ciardiello, Cancer Treat. Rev., 2019, 76, 22–32 CrossRef CAS.
- D. Wu, S. Wang, G. Yu and X. Chen, Angew. Chem., Int. Ed., 2021, 60, 8018–8034 CrossRef CAS PubMed.
- W. Xiao, K. Qu, W. Zhang, L. Lai, L. He, F. Cheng and L. Wang, Small Sci., 2024, 4, 2300164 CrossRef CAS.
- X. Liu, S. Xia, Z. Zhang, H. Wu and J. Lieberman, Nat. Rev. Drug Discovery, 2021, 20, 384–405 CrossRef CAS PubMed.
- F. Shao, Nat. Rev. Immunol., 2021, 21, 620–621 CrossRef CAS PubMed.
- J. Ding, K. Wang, W. Liu, Y. She, Q. Sun, J. Shi, H. Sun, D. C. Wang and F. Shao, Nature, 2016, 535, 111–116 CrossRef CAS PubMed.
- M. Li, J. Kim, H. Rha, S. Son, M. S. Levine, Y. Xu, J. L. Sessler and J. S. Kim, J. Am. Chem. Soc., 2023, 145, 6007–6023 CrossRef CAS PubMed.
- L. Li, M. Jiang, L. Qi, Y. Wu, D. Song, J. Gan, Y. Li and Y. Bai, Cancer Sci., 2021, 112, 3979–3994 CrossRef CAS PubMed.
- Z. Li, F. Mo, Y. Wang, W. Li, Y. Chen, J. Liu, T. J. Chen-Mayfield and Q. Hu, Nat. Commun., 2022, 13, 6321 CrossRef CAS PubMed.
- Q. Wang, Y. Wang, J. Ding, C. Wang, X. Zhou, W. Gao, H. Huang, F. Shao and Z. Liu, Nature, 2020, 579, 421–426 CrossRef CAS PubMed.
- Z. Zhang, Y. Zhou, S. Zhao, L. Ding, B. Chen and Y. Chen, Adv. Sci., 2022, 9, 2203583 CrossRef CAS.
- Y. Yin, B. Liu, Y. Cao, S. Yao, Y. Liu, G. Jin, Y. Qin, Y. Chen, K. Cui, L. Zhou, Z. Bian, B. Fei, S. Huang and Z. Huang, Adv. Sci., 2022, 9, 2102620 CrossRef CAS PubMed.
- D. G. DeNardo and B. Ruffell, Nat. Rev. Immunol., 2019, 19, 369–382 CrossRef CAS PubMed.
- M. De Palma and C. E. Lewis, Cancer Cell, 2013, 23, 277–286 CrossRef CAS PubMed.
- X. Zhou, X. Liu and L. Huang, Adv. Funct. Mater., 2021, 31, 2006220 CrossRef CAS PubMed.
- A. Mantovani, F. Marchesi, A. Malesci, L. Laghi and P. Allavena, Nat. Rev. Clin. Oncol., 2017, 14, 399–416 CrossRef CAS PubMed.
- A. Mantovani, P. Allavena, F. Marchesi and C. Garlanda, Nat. Rev. Drug Discovery, 2022, 21, 799–820 CrossRef CAS PubMed.
- X. Shi, B. Xiao, X. Xu, Y. Pan, J. Xiang, S. Shao, Z. Zhou, F. Huang, J. Gao, N. K. H. Slater, Y. Shen and J. Tang, Adv. Funct. Mater., 2023, 34, 2310364 CrossRef.
- D. M. Mosser and J. P. Edwards, Nat. Rev. Immunol., 2008, 8, 958–969 CrossRef CAS.
- S. Zanganeh, G. Hutter, R. Spitler, O. Lenkov, M. Mahmoudi, A. Shaw, J. S. Pajarinen, H. Nejadnik, S. Goodman, M. Moseley, L. M. Coussens and H. E. Daldrup-Link, Nat. Nanotechnol., 2016, 11, 986–994 CrossRef CAS.
- R. Wang, M. Qiu, L. Zhang, M. Sui, L. Xiao, Q. Yu, C. Ye, S. Chen and X. Zhou, Adv. Mater., 2023, 35, 2306748 CrossRef CAS.
- E. B. Rankin and A. J. Giaccia, Science, 2016, 352, 175–180 CrossRef CAS.
- Q. Yang, N. Guo, Y. Zhou, J. Chen, Q. Wei and M. Han, Acta Pharm. Sin. B, 2020, 10, 2156–2170 CrossRef CAS.
- Y. Zheng, Y. Han, Q. Sun and Z. Li, Exploration, 2022, 2, 20210166 CrossRef CAS PubMed.
- S. Marchi, C. Giorgi, L. Galluzzi and P. Pinton, Mol. Cell, 2020, 78, 1055–1069 CrossRef CAS PubMed.
- J. I. E. Bruce and A. D. James, Cancers, 2020, 12, 2351 CrossRef CAS PubMed.
- X. Shi, Y. Bi, W. Yang, X. Guo, Y. Jiang, C. Wan, L. Li, Y. Bai, J. Guo, Y. Wang, X. Chen, B. Wu, H. Sun, W. Liu, J. Wang and C. Xu, Nature, 2012, 493, 111–115 CrossRef CAS PubMed.
- M. Trebak and J.-P. Kinet, Nat. Rev. Immunol., 2019, 19, 154–169 CrossRef CAS PubMed.
- Y. Liu, Y. Wang, S. Song and H. Zhang, Chem. Sci., 2021, 12, 12234–12247 RSC.
- G. Abatangelo, V. Vindigni, G. Avruscio, L. Pandis and P. Brun, Cells, 2020, 9, 1743 CrossRef CAS PubMed.
- P. Zheng, B. Ding, G. Zhu, C Li and J. Lin, Angew. Chem., 2022, 134, 202204904 CrossRef.
- Y. Feng, J. Wang, J. Cao, F. Cao and X. Chen, Exploration, 2024, 1, 20230019 CrossRef.
- Y. Wang, S. Li, X. Zhang, F. Jiang, Q. Guo, P. Jiang and Y. Liu, Small, 2024, 20, 2400254 CrossRef CAS.
- S. Shen, M. Mamat, S. Zhang, J. Cao, Z. D. Hood, L. Figueroa-Cosme and Y. Xia, Small, 2019, 15, 1902118 CrossRef.
- S. Shen, M. Mamat, S. Zhang, J. Cao, Z. D. Hood, L. Figueroa-Cosme and Y. Xia, Small, 2019, 15, e1902118 CrossRef.
- W. J. Xu, J. M. Qian, G. H. Hou, T. B. Wang, J. L. Wang, Y. P. Wang, L. J. Yang, X. K. Cui and A. L. Suo, Adv. Funct. Mater., 2022, 32, 2205013 CrossRef CAS.
- P. Zheng, B. Ding, R. Shi, Z. Jiang, W. Xu, G. Li, J. Ding and X. Chen, Adv. Mater., 2021, 33, 2007426 CrossRef CAS PubMed.
- B. Liu, Y. Bian, S. Liang, M. Yuan, S. Dong, F. He, S. Gai, P. Yang, Z. Cheng and J. Lin, ACS Nano, 2022, 16, 617–630 CrossRef CAS.
- J. B. Spinelli and M. C. Haigis, Nat. Cell Biol., 2018, 20, 745–754 CrossRef CAS PubMed.
- V. Eisner, M. Picard and G. Hajnoczky, Nat. Cell Biol., 2018, 20, 755–765 CrossRef CAS.
- Y. Li, S. Zhou, H. Song, T. Yu, X. Zheng and Q. Chu, Biomaterials, 2021, 277, 121080 CrossRef CAS PubMed.
- C. Garrido, L. Galluzzi, M. Brunet, P. E. Puig, C. Didelot and G. Kroemer, Cell Death Differ., 2006, 13, 1423–1433 CrossRef CAS.
- C. Wang, F. Yu, X. Liu, S. Chen, R. Wu, R. Zhao, F. Hu and H. Yuan, Adv. Healthcare Mater., 2019, 8, 1900501 CrossRef PubMed.
- Y. Wang, W. Gao, X. Shi, J. Ding, W. Liu, H. He, K. Wang and F. Shao, Nature, 2017, 547, 99–103 CrossRef CAS PubMed.
- Z. Zhang, Y. Zhang, S. Xia, Q. Kong, S. Li, X. Liu, C. Junqueira, K. F. Meza-Sosa, T. M. Y. Mok, J. Ansara, S. Sengupta, Y. Yao, H. Wu and J. Lieberman, Nature, 2020, 579, 415–420 CrossRef CAS PubMed.
- M. Huo, P. Liu, L. Zhang, C. Wei, L. Wang, Y. Chen and J. Shi, Adv. Funct. Mater., 2021, 31, 2010196 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr01497a |
| This journal is © The Royal Society of Chemistry 2024 |