Recent developments of polymeric delivery systems in gene therapeutics
Yijia
Li
ab,
Ruizhen
Tian
ab,
Jiayun
Xu
b,
Yingping
Zou
 a,
Tingting
Wang
*b and
Junqiu
Liu
a,
Tingting
Wang
*b and
Junqiu
Liu
 *b
*b
aState Key Laboratory of Powder Metallurgy, College of Chemistry and Chemical Engineering, Central South University, Changsha 410083, China
bCollege of Material, Chemistry and Chemical Engineering, Key Laboratory of Organosilicon Chemistry and Material Technology, Ministry of Education, Key Laboratory of Organosilicon Material Technology of Zhejiang Province, Hangzhou Normal University, Hangzhou 311121, China. E-mail: wangtt@hznu.edu.cn; junqiuliu@jlu.edu.cn
First published on 24th April 2024
Abstract
Gene therapy has achieved remarkable results in treating diseases by transmitting exogenous functional genes to target cells, inducing gene silencing, gene expression, and gene editing. However, nucleic acids are susceptible to degradation by nucleic acid endonucleases and clearance by the immune system. Therefore, safe and efficient gene delivery vectors are needed to facilitate gene therapy's clinical implementation further. Although viral vectors exhibit high transfection efficiency, their safety issues limit their further application. Despite their promising clinical performance, lipid nanoparticles suffer from poor stability and difficult storage. Polymer cationic vectors (PCVs) with low cost, low immunogenicity, and tunability have attracted the attention of scientists. This article reviews the main strategies to improve the gene transfection efficiency of polymeric delivery systems in recent years and their applications in certain diseases. However, there are still challenges in gene delivery with polymer vectors. Hence, the paper concludes with an outlook on the future development of PCVs, which will hopefully help in the design of PCVs.
1. Introduction
Gene therapy has shown unlimited potential for treating both hereditary and acquired diseases.1 According to Emergency Research's market research, internationally relevant gene therapy companies are expected to generate approximately $6.6 billion in revenue by 2027.2 The treatment of relevant diseases is achieved by introducing exogenous nucleic acids into target cells, inducing gene silencing (inhibition of pathological protein production),3 gene expression (promotion of therapeutic protein expression),4 and gene editing (modification of dysfunctional genes).5 However, multiple intra- and extracellular barriers must be overcome for effective gene transfection.6 DNA and RNA are a group of large negatively charged biological macromolecules that are rapidly cleared by macrophages and the reticuloendothelial system, have a short half-life in the organism, are easily degraded by endonucleases, and have strong electrostatic repulsive effects with the cellular membrane, and are refractory to escape from endosomes as well as to enter the nucleus.7–10 Therefore, exploiting safe and efficient gene delivery vectors is one of the most potent strategies to overcome multiple biological barriers during gene delivery.Delivery vectors are mainly divided into two categories: viral vectors and non-viral vectors. Approximately 70% of clinical gene therapy trials use viral vectors, such as retroviruses, lentiviruses, and adenoviruses.11 Due to their specific infectious properties, viral vectors typically exhibit excellent transfection efficiency in vivo. However, viral vectors pose clinical safety problems, such as their inherent high immunogenicity and the risk of integrating their own sequences into the host genome. Additionally, viral vectors have certain limitations for therapeutic use, such as low gene loading, delivery of only small-sized genes, complex preparation processes, and the inability to repeat administration.11 By contrast, non-viral vectors, especially lipid nanoparticles (LNPs) and cationic polymers, have been favored by scientists due to their advantages, such as high gene loading efficiency, excellent safety profile, and simplicity of preparation. The low toxicity, safety, and translational capacity of LNPs vectors have made it possible to use in clinical applications, such as the COVID-19 mRNA vaccine.12 Nevertheless, LNPs still suffer from poor stability, difficulty in storage,13 and liver tropism.14 Polymer-based gene delivery systems present less clinical performance than lipids, but they have lower enzymatic degradability, higher stability, lower cost, and more accessible physical properties to manipulate. Moreover, polymer vectors allow the assembly of different nanostructures under aqueous conditions, with lyophilization, long-term storage capabilities, and unique pharmacokinetics.15 Polymer nanoparticles have different chemical and physical properties and can be synthesized, modified, and tailored according to different applications. However, polymer vectors still remain challenging in gene delivery.
Cationic groups in polymeric cationic vectors (PCVs) can form stable nanocomplexes with phosphate groups in nucleic acids through electrostatic/hydrophobic interactions, with sizes ranging from tens to hundreds of nanometers.1 Adequate considerations are necessary in designing efficient PCVs. The first is the influence of the PCVs’ surface charge. Upon an increase in positive charge density, PCVs enhances the capacity to compress nucleic acids but increases the difficulty of releasing nucleic acids inside the cell.16 Higher positive charge densities also lead to significant cytotoxicity and adsorption of serum proteins, causing the aggregation of nanoparticles to be cleared by the reticuloendothelial system.17,18 In contrast, an overly low positive charge density is detrimental to cellular uptake of the nanocomplexes, leading to a reduced gene transfection efficiency. Therefore, exploring the optimal positive charge density is crucial for efficient gene delivery. Secondly, the effect of polymer topology, hydrophobicity of chain segments, and size and shape of the nanocomplexes should be considered. The density and molecular weight of the hydrophilic chain segments have a remarkable effect on the stability of the nanoparticles, and excessive hydrophilicity is unfavorable for efficient DNA cohesion and stabilization.1 Nanoparticles smaller than 50 nm are normally cleared rapidly by the kidneys, whereas nanoparticles larger than 300 nm might elicit an immune response in vivo.19 In addition, the effects of endosomes/lysosomes need to be considered. The acidic environment within endosomes/lysosomes, along with various nucleic acid endonucleases, results in the degradation of nanocomplexes and naked nucleic acids.20 For endosomal/lysosomal escape, scientists have proposed two main pathways, “proton sponge effect” and “membrane fusion”.21 In the case of DNA, how to enter the nucleus more efficiently for transcription needs to be considered.22 Although RNA functions in the cytoplasm, it is essential to consider how to release it quickly and efficiently. Regarding the in vivo organ tropic properties of PCVs, how to make the nanocomplexes more productive in target cells or organs dramatically affects the final therapeutic effectiveness. In recent years, scientists have developed a wide range of PCVs with excellent performance through unremitting efforts. This review is mainly to outline the main strategies to improve the gene transfection efficiency of PCVs recently and their applications in certain diseases, then conclude with an outlook on the future development of PCVs, which will hopefully shed some light on the development of this field (Fig. 1).
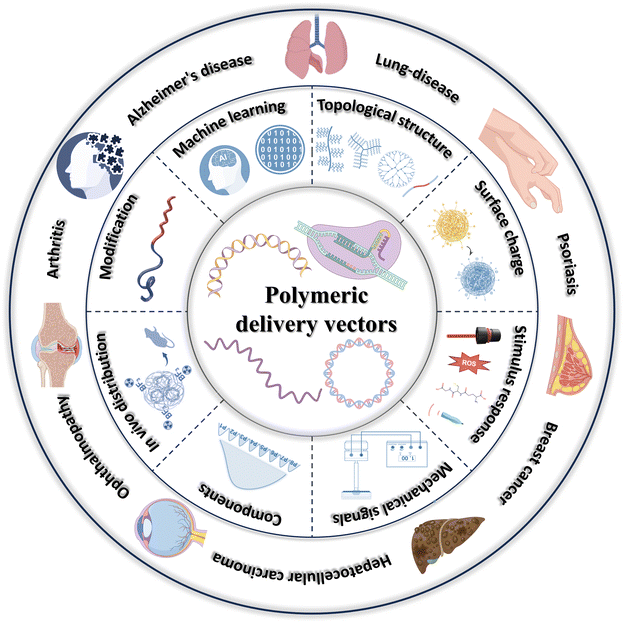 | ||
| Fig. 1 A brief overview of main strategies to improve the gene transfection efficiency of PCVs recently and their applications in certain diseases. | ||
2. Strategies to improve gene transfection efficiency
Numerous cationic polymers have been employed in the exploitation of gene delivery vectors, such as dextran, chitosan, polylysine (PLL), polyarginine (PLR), polyamide-amine (PAMAM) dendrimers, polyethyleneimine (PEI) poly (β-amino esters) (PBAEs), and others. However, some problems still constrain the efficiency of gene transfection, such as stability, cytotoxicity, targeting, cellular uptake, and endosomal escape. Therefore, effective strategies to address these challenges are required to be developed, and this section summarizes some feasible strategies, including modulation of surface charge, alteration of topological structure, modification of polymers, introduction of stimulus-responsive units, incorporation of machine learning, utilization of mechanistic perturbations, assessment of in vivo distribution, probing the effect of distribution of different molecular weight fractions, and combined use of multiple means.2.1 Modulation of surface charge
High doses of cationic carriers disrupt cell membranes23 and may induce immunogenicity and inflammation,24,25 resulting in severe cytotoxicity. Shielding the charge of cationic polymers with hydrophilic or anionic polymers is an effective means of reducing cytotoxicity.26 By adjusting the ratio between anionic and cationic polymers, cytotoxic effects can be mitigated with little loss of transfection efficiency.27The surface charge can also be decreased by removing the cationic portion of the polymer. Huang et al. designed and synthesized a cationic triblock copolymer, cRGD – PEG – PAsp (MEA) PAsp (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N-DETA), which consists of a cRGD-capped poly(ethylene glycol) block (cRGD-PEG), a poly(aspartic acid) block with a sulfhydryl side group [PAsp(MEA)]and a poly(aspartic acid) block with a sulfhydryl side group grafted with the cationic divinylenetriamine (DETA) through a pH-sensitive imine bond [PAsp(C
N-DETA), which consists of a cRGD-capped poly(ethylene glycol) block (cRGD-PEG), a poly(aspartic acid) block with a sulfhydryl side group [PAsp(MEA)]and a poly(aspartic acid) block with a sulfhydryl side group grafted with the cationic divinylenetriamine (DETA) through a pH-sensitive imine bond [PAsp(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N-DETA)] (Fig. 2a).28 After complexing siRNA with the above copolymers, the sulfhydryl groups of the intermediate layer were oxidized to disulfide bonds, thus crosslinking the complex. Then, a noncationic nano-delivery vector was constructed by breaking the imine bond and removing the cationic DETA side chain by dialysis at Ph = 5.0. The vector could encapsulate siRNA efficiently and resist protein adsorption in serum, and the modification of cRGD gave the vector suitable cancer cell targeting properties. In addition, the disulfide bond in the vector would break under the environment of high intracellular glutathione (GSH) pairs, which prompted the release of siRNA and achieved tumor-targeted gene silencing in vivo.
N-DETA)] (Fig. 2a).28 After complexing siRNA with the above copolymers, the sulfhydryl groups of the intermediate layer were oxidized to disulfide bonds, thus crosslinking the complex. Then, a noncationic nano-delivery vector was constructed by breaking the imine bond and removing the cationic DETA side chain by dialysis at Ph = 5.0. The vector could encapsulate siRNA efficiently and resist protein adsorption in serum, and the modification of cRGD gave the vector suitable cancer cell targeting properties. In addition, the disulfide bond in the vector would break under the environment of high intracellular glutathione (GSH) pairs, which prompted the release of siRNA and achieved tumor-targeted gene silencing in vivo.
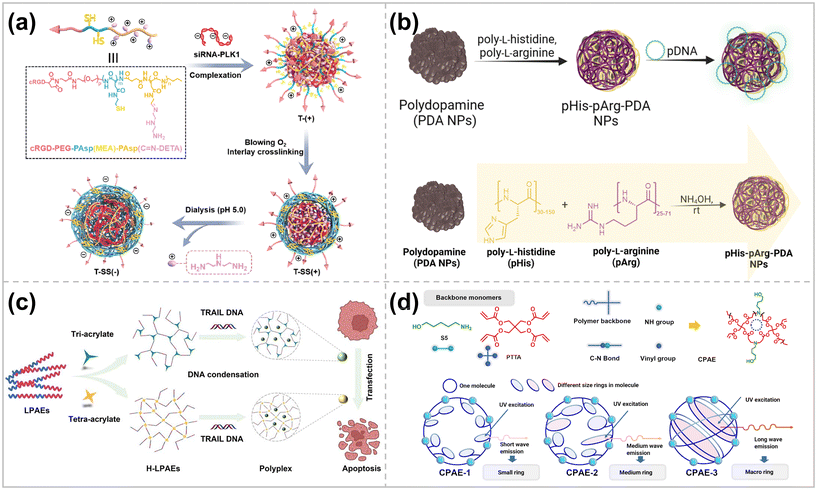 | ||
| Fig. 2 (a) Preparation and intracellular fate of cation-free disulfide bond-crosslinked polymer-siRNA nanocapsule T-SS(−) ref. 28. Copyright 2023, Elsevier. (b) Schematic representation of the synthesis, pDNA binding and gene delivery using pHis-pArg-PDA NPs ref. 34. Copyright 2023, Royal Society of Chemistry. (c) H-LPAEs condenses TRAIL DNA to formulate nano-sized polyplexes and induced tumor cell apoptosis for various cancer cells ref. 42. Copyright 2023, Springer Nature. (d) Structure of the monomer 5-amino-1-pentanol (S5) and pentaerythritol tetra acrylate (PTTA) used for CPAE synthesis and schematic illustration of the relationship between different cyclic structures of CPAEs ref. 2. Copyright 2023, American Chemical Society. | ||
Biopolymer nanoparticles are excellent candidates for gene carriers due to their favorable physicochemical properties, biodegradability, and biocompatibility.29 Their size, surface charge, and functional group tunability enable them to modulate in vivo circulation time, cellular uptake, and nucleic acid adsorption behaviors.30 Biopolymerized polydopamine nanoparticles (PDA NPs) are regarded as one of the typical platforms for drug delivery.
However, PDA NPs are inherently negatively charged, therefore, not conducive to efficient nucleic acid loading. By modifying PEI on the surface of PDA NPs, efficient delivery of genes was achieved, but at the same time, the biotoxicity of the delivery system was increased.31 In nature, encapsulation and protection of DNA is usually achieved by protamine enriched with cationic arginine residues.32 Inspired by this, PDA NPs modified with arginine-rich compounds proved to be beneficial for cellular uptake,33 but the low buffering capacity of poly-L-arginine (pArg) still hinders intracellular gene release and its endosomal escape. To overcome this challenge, Franck et al. modified PDA NPs with a mixture of poly-L-arginine and poly-L-histidine (pHis) for efficient delivery of pDNA (Fig. 2b).34 The presence of a sizeable ionizable imidazole fraction of poly-L-histidine promotes proton buffering inside the internal endocytic body, which in turn leads to their rupture through the proton sponge effect.35 By adjusting the ratio of pHis to pArg and the ratio of NPs to pDNA, comparable transfection efficiencies of Lipofectamine could be monitored by utilizing self-reported EGFP-pDNA. In addition, using commercially available mixtures of pArg and pHis dramatically reduces the cost of the delivery system.
2.2 Alteration of topological structure
It is promising to utilize different topologies of polymers to develop efficient polymeric gene delivery vectors. For example, more than 2500 linear PBAEs (LPBAEs) have been synthesized so far.36,37 Although the results of gene delivery tests of LPBAEs are encouraging, the linear nature of these polymers limits the potential for optimizing the structures. In 2016, Wang et al. constructed highly branched PBAEs (HPBAEs) by a facile A2 + B3 + C2 Michael addition strategy.38 The results showed that the transition from linear to branched structures introduced a large number of functional end-groups, enhanced the interaction with DNA, and optimized the assembly behavior of the polymers. Thus, the gene transfection performance of HPBAEs was significantly higher than LPBAEs.38,39 This discovery has made it more straightforward to scientists that the polymer vectors’ macromolecular structure significantly impacts their transfection efficiency. Therefore, scientists have further developed various topological structures of polymers, including linear, branched, dendritic, star, and ring structures, to explore the effects of different topologies on gene transfection efficiency.40,41Zhao et al. exploited a linear oligomer combinatorial branching strategy to create highly branched linear poly(β-amino esters) (H-LPBAEs). This unique strategy enabled a more homogeneous distribution of linear chain segments and branched units of H-LPAEs with molecular weights ranging from 11.8 kg mol−1 to 19.0 kg mol−1 (Fig. 2c).42 The optimized H-LPBAEs exhibited enhanced DNA condensation ability, and the positively charged H-LPBAE/DNA complexes demonstrated a high level of gene transfection efficiency in different cell lines. In SW1353 and BMSC cells, the optimized H-LPBAEs achieved 58.0% and 33.4% gene transfection efficiencies, 2.5 and 2.0 times higher than the commercially available gene transfection reagent Lipofectamine 3000, respectively. Teo et al. synthesized hetero-armed star polymer nanoparticles consisting of two main components, poly(dimethylamino ethyl methacrylate) (PDMAEMA) and poly[oligo poly(ethylene glycol)methyl ether methacrylate] (POEGMA), with cores connected by a cysteamine-based crosslinker.43 The positively charged polyelectrolyte PDMAEMA can be rapidly combined with anionic siRNA through electrostatic interaction. POEGMA is a brush polymer that reduces the positive charge of star nanoparticles, increases their hydrophilicity, diminishes cytotoxicity, and improves serum resistance. Compared to linear DMAEMA polymers, star polymers offer higher siRNA transfection efficiency, lower cytotoxicity, and more functionality due to multiple polymer arms radiating outward from a central core. These arms have the potential to be designed to have stimuli-responsive or tumor-targeting elements.44
Cyclopolymers have attracted significant attention due to their unique 3D structures and properties. Previous studies have shown that polymers with cyclized structures typically have better biocompatibility and DNA encapsulation than polymers with linear or branched structures.39,45 Nanoparticles formed by cyclic polymers and genes are more compact and smaller than linear polymers. Li et al. used a cyclization stage control strategy to construct 3D cyclic poly(β-amino esters) (CPBAEs) with different ring sizes and ring topologies by regulating the cyclization tendency at various stages of step-growth polymerization (SGP) (Fig. 2d).2 The other ring shapes and end-group distributions of the individual CPBAEs were verified using fluorescence spectroscopy and 2D-NMR. Macrocyclic PAE (MCPBAE) and its complexes had significantly enhanced DNA condensation, cellular uptake, and DNA protection compared to other CPBAEs and HPBAEs, which promoted the expression of transfected genes. The best-performing MCPAE-C, -G, and -M exhibited higher transfection efficiencies than the best commercially available reagents, including Lipo 3000, jetPEI, and Xfect.2 In addition, MCPBAE with optimized end-groups could efficiently deliver CRISPR plasmids encoding both Staphylococcus aureus Cas9 nuclease and dual-guide sg RNA for in vitro gene editing.
2.3 Introduction of stimulus-responsive units
The smart stimulus-responsive delivery system responds to intrinsic stimuli from the TME, such as acidic pH, high redox, and overexpressed enzymes, enabling efficient gene delivery to the tumor location.18Polycarboxyl betaine, poly(N-{N′-[N′′-(2-carboxyethyl)-2-aminoethyl]-2-aminoethyl}glutamide) [PGlu(DET-Car)], where the ethylenediamine portion has two different acid dissociation constants. The PGlu (DET-Car) coated nano-particles show a neutral net charge at pH 7.4, transforming into a cation in acidic environments.46 Shen et al. constructed polymeric complex micelles from PGlu (DET-Car) (PGDC) conjugated branched PEIs for efficient pDNA delivery.47 The PGDC shells show a neutral charge under normal physiological conditions, resulting in prolonged blood circulation time. At pH 6.5, the PGDC shell becomes slightly cationized after enrichment in tumors through the leaky vasculature system, enhancing its retention effect at the tumor location and the efficiency of cellular internalization. The endosomal/lysosomal environment (pH 5.5) increases the PGDC shell layer's cationic charge, facilitating endosomal escape and enabling efficient gene transfection (Fig. 3a).
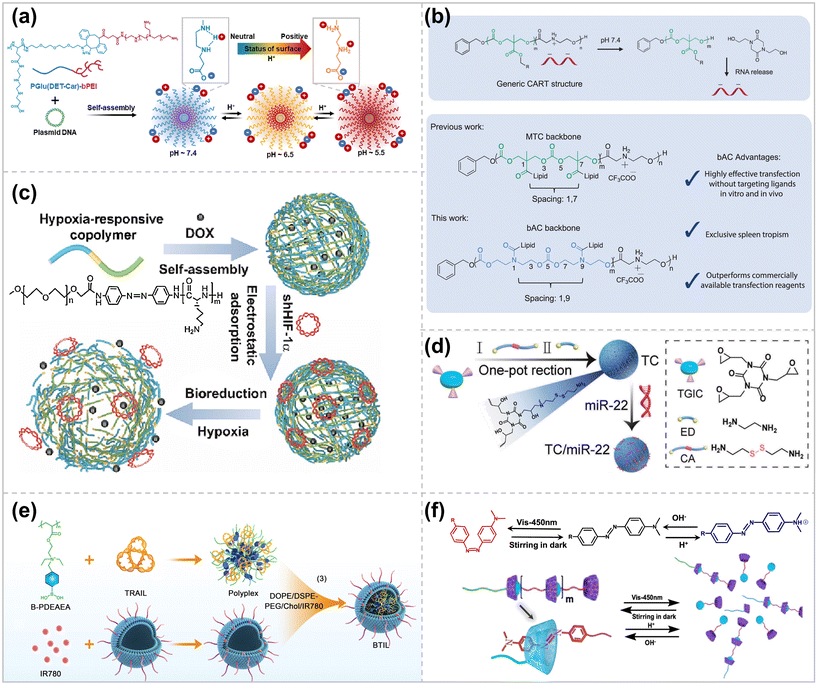 | ||
| Fig. 3 Schematic design of stimulus-responsive polymer vector. (a) Stepwise pH-triggered charge conversion of PGlu(DET-Car)-coated polyplex micelle (PGDC PM) for the effective gene delivery ref. 47. Copyright 2023, Elsevier. (b) CART/mRNA complexes and their pH-driven nitrogen-to-oxygen acyl shift triggering charge cancellation and mRNA release and bAC CARTs possess a polymeric backbone with distinct lipid spacing, leading to improved T-cell delivery ref. 57. Copyright 2023, Springer Nature. (c) The micelles were self-assembled from the copolymer composed of mPEG (blue segment) and PLL (green segment) with AZO (yellow segment) as a hypoxia-responsive bridge ref. 59. Copyright 2023, Elsevier. (d) Schematic illustration of the preparation process of reduction-responsive cationic polymer TC ref. 63. Copyright 2023, Elsevier. (e) Diagram for the ultrasound-enhanced ROS responsive charge-reversal polymeric nanocarriers for pancreatic cancer gene delivery ref. 65. Copyright 2023, American Chemical Society. (f) Schematic illustration of the reversible polymerization/depolymerization behavior of a cationic SBC triggered by various environmental stimuli ref. 71. Copyright 2023, American Chemical Society. | ||
The tertiary amine groups make it feasible to become more hydrophobic upon deprotonation or more cationic upon protonation, modulating the polymer's hydrophobicity and cationic charge density by altering the pH. Xie et al. employed this pH-responsive amphiphilic polymer, m PEG-PC7A, to simultaneously load Cas9 RNP and single-stranded oligodeoxynucleotides (ss ODN) via electrostatic and hydrophobic interactions.48 Intramuscular injection of the complex could effectively restore the muscle strength of the Duchenne muscular dystrophy mouse model.
The use of charge-changed releasable transporters (CARTs) allows for the efficient delivery of polyanionic cargos, including mRNA, plasmid DNA, circRNA, and combinations thereof.49–52 CARTs prepared by organocatalytic ring-opening polymerization (OROP) are block oligomers consisting of an initiator, one or more lipid blocks, and a polycationic block. At pH < 5.5, CART cationic blocks can bind polyanions by electrostatic interaction. At physiological pH (∼7.4), the CART delivery system undergoes an oxygen-to-nitrogen (O-to-N) acyl transfer, which allows for an irreversible conversion to a neutral lactam, contributing to the release of the polyanion (Fig. 3b).50 Using a combinatorial lipid-screening strategy, McKinlay et al. prepared CARTs that bind both oleoyl and nonenyl lipids (named Oleyl-Nonene amino CARTs), which showed significantly improved transfection efficiency (up to ∼80%) in Jurkat cells in T-lymphocyte cell line.53 ONA CARTs have been successfully used in the clinical development of prophylactic (COVID) and therapeutic (cancer and metastatic disease) vaccines54,55 and the generation of CAR-NK cells. However, the transfection efficiency of ONA CARTs on primary T cells is low, only 10%–20%. To address this problem, their group used fingolimod as an initiator of CARTs, a small molecule targeting sphingosine-1 phosphate receptor (S1P1) on T and B lymphocytes,56 which showed better transfection ability of Jurkat cells in vitro. However, transfection of T cells was not improved in the mouse model. Therefore, they developed a CART delivery system with a β-amino carbonate (bAC) skeleton and synthesized a library of 24 bAC CARTs with different lipid compositions using an organocatalytic ring-opening oligomerization reaction (Fig. 3b).57 Among them, the most potent bAC CARTs showed up to 70% primary T cell transfection in vitro, and, more importantly, in vivo delivery of the optimal bAC CARTs produced up to 97% splenic tropism and transfected 8% of primary splenic T cells that did not contain T cell-targeting ligands. This spleen tropism of bAC CARTs contributes to biomedical applications such as T-cell immunotherapy while minimizing off-target effects.
2.4 Utilization of mechanistic stimuli
In living organisms, chemical and mechanical signals continuously influence and coordinate the activities of cellular life, such as motility, proliferation, and phenotypic transition events.73 Endocytosis and transport processes are similarly regulated by interactions between the cell membrane and the underlying cytoskeleton.74 Different mechanical stimuli can improve gene transfer by modulating specific steps in the transmission pathway, including internalization, cytoplasmic transport, and nuclear entry.75 Mechanical loads applied to cells cause specific cellular responses, such as mechanical regulation of membrane transport76 and cytoskeletal remodeling.77,78 In this context, cyclic stretching,79 shear stress,80 and vibrational loading81 may improve the transfection efficiency of non-viral gene delivery vectors by enhancing their uptake and intracellular transport. Ponti et al. propose a method to enhance intracellular delivery and improve transfection efficiency by using mechanical stimulation to initiate cells. A short-lived high-frequency vibrational load (t = 5 min, f = 1000 Hz) applied vertically to the plane of the cell culture plate induces abrupt and extensive plasma membrane growth, triggering the clathrin-mediated endocytosis pathway that makes it easier for the cells to internalize the polymer gene vector, leading to an increase in the transfection efficiency from 10-fold to 100-fold.81 This process is safe for the cells, which recover after 1 hour through plasma membrane remodeling.2.5 Modification of polymers
Rational modification of polymers can enable polymeric vectors to possess high transfection efficiency while maintaining excellent biocompatibility. For example, polyethyleneimine (PEI) is the most widely studied polycationic vector with different molecular weights (0.2–800 kDa), and linear and branched structures.82 Due to its high transfection efficiency in cells, the branched PEI 25 kDa is generally regarded as the “gold standard” of polycationic non-viral vectors.83 However, the cytotoxicity of PEI increases as its molecular weight rises, giving an antagonistic relationship between high transfection efficiency and low cytotoxicity. Modifications such as polyethylene glycol (PEG), lipid, or fluoride have been introduced into PEI polymers to address this critical issue and reduce the biotoxicity of the delivery system.84 PEG-modified PEIs can shield the positively charged surface of the complexes by forming a hydrophilic corona, which reduces damage to the cell membrane. Due to the amphiphilicity of lipids, lipid-modified PEI makes PCC more stable and has better biocompatibility.85 Perfluoroalkyl-containing polymers exhibit favorable serum resistance due to the hydrophobicity and lipophobicity of fluorocarbon chains.86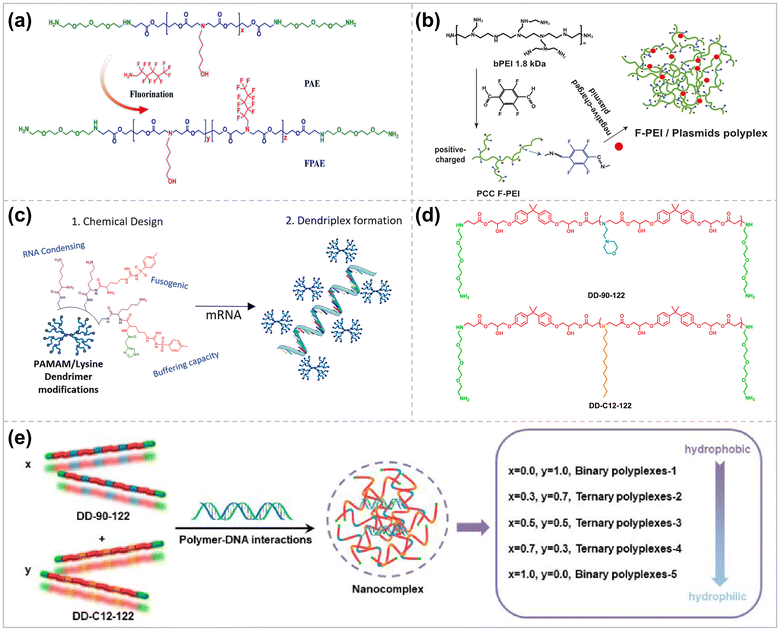 | ||
| Fig. 4 Rationally modified polymer vectors for higher delivery efficiencies. (a) Synthesis strategy of FPAE ref. 93. Copyright 2023, Elsevier. (b) Schematic illustration of PCC F-PEI chemical synthesis route and mechanism of action in vivo ref. 94. Copyright 2023, Elsevier. (c) Dendritic polymer PAMAM modified with amino acids ref. 102. Copyright 2023, Elsevier. (d) Hydrophilic and hydrophobic LPAEs. (e) Modular manipulation of the amphiphilicity of the LPAE/DNA polyplexes for efficient DNA delivery ref. 116. Copyright 2023, American Chemical Society. | ||
Amino acid-modified polymer delivery systems have dramatically increase in biocompatibility and transfection efficiency. This is attributed to the fact that amino acid modification reduces the charge density of the cationic polymer carrier and increases the interaction with nucleic acids while improving cellular uptake efficiency and endosomal escape. Polyamide-amines grafted with histidine and arginine (second-generation PAM) can have higher endosomal escape and higher transfection efficiency than unmodified PAM due to the proton buffering and protein transduction peptide effects of histidine and arginine.100 Partial modification of the PAMAM dendrimer with tosyl arginine allowed the dendrimer to increase its transfection efficiency while minimizing its cytotoxicity.101 Joubert et al. precisely introduced p-toluenesulfonyl arginine and imidazole groups on the lysine-carrying PAMAM, which effectively improved its fusion properties, buffering capacity, and facilitated endosomal escape (Fig. 4c).102 The arginine-rich polymer, which can effectively form pores in the cell membrane and translocate into them, avoids endo/lysosomal trapping, resulting in a significant increase in the efficiency of cellular internalization.103 By screening for structure–activity relationships, the polymer with a side chain containing two arginine residues and a flexible hexanoic acid linker (PTn-R2-C6) showed higher transfection efficiency and lower cytotoxicity than the gold standard transfection reagent (PEI 25K).
L-PBAEs are a class of polymers with promising biocompatibility and biodegradability.104 Due to the presence of basic amine groups, L-PBAEs are able to condense nucleic acids in the form of nanocomplexes, making them a strong candidate for gene vectors. Their transfection efficiency and selectivity for different cell types can also be tuned by synthetically modifying the PBAE backbone or the polymer ends.105 For instance, due to the excellent biocompatibility of peptides, Borrós’ group developed a library of linear oligopeptide-modified PBAEs (OM-PBAEs) with terminal oligopeptides as the cationic portion.106,107 Among them, peptides such as lysine, arginine, and histidine are susceptible to protonation at intestinal luminal pH, which is more conducive to endosomal escape owing to the “proton sponge effect”. Hence, these OM-PBAE polymers show promising applications such as protection of mRNA for vaccine purposes,108 encapsulation of interfering RNA for cardiovascular disease therapy,109 incorporation of various gene editing systems (CRISPR/Cas) for the treatment of rare monogenic diseases,110 preparation of hydrogels for localized siRNA therapy for breast cancer or microneedles loaded with immune-modulating components.106
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 cast ratio, which significantly improved the stability of the complex and enhanced the transfection efficiency of DNA.113 Liu et al. introduced undecylamine (C11) into branched poly(β-amino ester), which enhanced DNA's condensation, delivery, and transfection efficiency.114 However, this chemical modification approac makes the chemical composition of the resulting polymers complex and uncontrollable, and most importantly, this strategy is time-consuming and complicated to operate. Inspired by the modularity strategy, Li et al. fabricated β-cyclodextrin-based cationic modules and adamantyl-functionalized guanidinium-based polymer modules via host–guest interaction to form a micellar delivery system for the efficient delivery of pDNA.115 Shi et al. then synthesized hydrophilic LPAE and hydrophobic LPAE separately, which were then co-condensed with DNA to form a ternary complex. By adjusting the proportion of hydrophilic and hydrophobic LPAEs, the hydrophilicity, size, ζ-potential, and other physicochemical properties of the ternary complexes could be altered, contributing to the regulation of gene transfection (Fig. 4d and e).116
3 cast ratio, which significantly improved the stability of the complex and enhanced the transfection efficiency of DNA.113 Liu et al. introduced undecylamine (C11) into branched poly(β-amino ester), which enhanced DNA's condensation, delivery, and transfection efficiency.114 However, this chemical modification approac makes the chemical composition of the resulting polymers complex and uncontrollable, and most importantly, this strategy is time-consuming and complicated to operate. Inspired by the modularity strategy, Li et al. fabricated β-cyclodextrin-based cationic modules and adamantyl-functionalized guanidinium-based polymer modules via host–guest interaction to form a micellar delivery system for the efficient delivery of pDNA.115 Shi et al. then synthesized hydrophilic LPAE and hydrophobic LPAE separately, which were then co-condensed with DNA to form a ternary complex. By adjusting the proportion of hydrophilic and hydrophobic LPAEs, the hydrophilicity, size, ζ-potential, and other physicochemical properties of the ternary complexes could be altered, contributing to the regulation of gene transfection (Fig. 4d and e).116
2.6 Incorporation of machine learning
Machine-learning based techniques permit more efficient experiment design by obtaining optimal results with fewer experiments.126 High-throughput computer simulation evaluation methods for biomaterials can accelerate delivery device optimization and development by exploring large chemical design spaces more quickly and cost-effectively. Gong et al. apply state-of-the-art machine learning algorithms to a dataset of synthetic biodegradable PBAEs, which have excellent therapeutic gene delivery capabilities in vitro and in vivo.127 Polymer properties are the input part of this dataset, and polymer nanoparticle transfection performance and nanoparticle toxicity in different cells are the output part of this dataset. These data were used to train and evaluate several state-of-the-art machine learning algorithms to predict transfection and the ability to understand structure–function relationships (Fig. 5a). By developing a coding scheme to vectorize the structure of PBAE polymers in a machine-readable format, it was proved that the Random Forest model could predict in vitro DNA transfection based on the chemical structure of PBAE polymers in a cell line-dependent manner. The PBAE gene delivery system prepared based on this model was tested for transfection in RAW 264.7 macrophages and Hep3B hepatocellular carcinoma cells, and the results showed Spearman's R correlation between experimental/tested transfection significance of 0.57 and 0.66, respectively.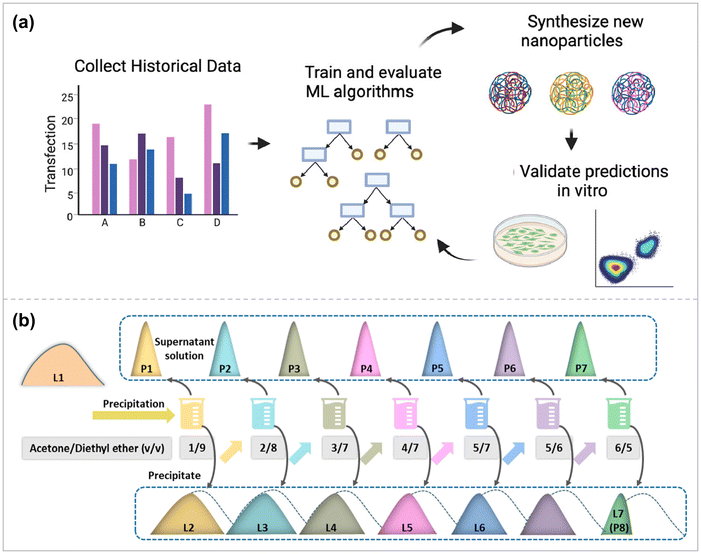 | ||
| Fig. 5 (a) A novel computational pipeline to encode PBAE nanoparticles with chemical descriptors and demonstrate utility in a de novo experimental context ref. 127. Copyright 2022, Elsevier. (b) Schematic illustration of the LPAE polymerization and fractionation processes ref. 132. Copyright 2023, American Chemical Society. | ||
Gaussian process (GP) modeling has succeeded dramatically due to its non-parametric nature, enabling fast black-box optimization of very few data points. It is the best tool for hyperparametric tuning in machine-learning experiments.128 In order to investigate whether oppositely charged hybrid micelles maintain high transfection efficiency while having excellent cytocompatibility, Leer et al. synthesized two sets of oppositely charged double-block terpolymers.128 The two sets of copolymers contain the same hydrophobic poly(n-butyl acrylate) (PnBA) and hydrophilic 4-acryloylmethane (NAM). The charge difference lies in the fact that the cationic copolymer is modified with 3-guanidinoacrylamide (GPAm), and the anionic copolymer is modified with 2-carboxyethylacrylamide (CEAm), which is negatively charged under neutral conditions. Variations in copolymer composition and the form of different ratios between the blended polymers allow for a myriad of possibilities for micellar structures, and optimizing such systems requires a large number of experiments ranging from chemical synthesis to biological studies. A machine learning approach was therefore applied to determine the optimal GPAm/CEAm ratio, resulting in higher transfection efficiency and cell survival with less resource overhead. After two runs, the optimal ratio to overcome the toxicity-efficiency dilemma was determined. Applying machine learning to polymer chemistry holds great potential to efficiently process many possible combinations for probing robust gene vectors.
2.7 Probing the effect of distribution of different molecular weight fractions
Unlike viruses and lipids with defined chemical structures, synthetic polymeric vectors (e.g., PAE, PEI, PLL, DMAEMA) are mixtures of different molecular weight (MW) components, which perform their respective functions and play a synergistic role in the process of gene delivery.129,130 Polymer components with high molecular weight can act as a framework to stabilize the polymer structure and protect the core DNA.129 In contrast, high molecular weight components cannot effectively shield the DNA charge due to their sizeable steric hindrance, whereas mobile small molecular weight components can sufficiently shield the negative residue charge on the phosphate group, thus assisting the DNA in further coiling and folding.131 In addition, other molecular weight components may facilitate cellular uptake, endosomal escape, and nuclear pore entry of the polymer vector.Therefore, to improve the efficiency of gene transfection, it is necessary to determine the optimal combination ratio of different polymer components in polymer vectors, i.e., polymer component distribution (PCD). Li et al. used PBAE as the research object to investigate the effects of different properties of different components of polymers (high MW and low MW components) and the synergistic effect on gene delivery after combining them in appropriate ratios (Fig. 5b).132 The relationship between PCD and gene transfer performance was investigated using an artificial intelligence (AI) analysis. Guided by this analysis, a series of highly efficient polymeric vectors superior to the current commercial reagents jetPEI and Lipo3000 were developed, among which the transfection efficiency of the PAE – B1-based complex in U251 cells was about 1.5 times that of Lipo3000 and 2.0 times that of jetPEI.
2.8 Combined use of multiple means
Improvement of polymeric vectors by a single means is always limited for gene transfection efficiency, and the combined use of multiple means will have a greater chance of facilitating gene transfection, reducing the dosage of drugs, and driving clinical transformation. Karlsson et al. designed reductively cleavable PBAEs (XbNPs) containing photocrosslinked polymers to cross-link and load encapsulated siRNAs.133 The disulfide bridge in XbNPs is conducive to the intracellular release of siRNA under weak acid conditions. Photocrosslinking improves the stability of XbNPs, shields the surface charge, and reduces the adsorption of serum proteins. Under high serum conditions, XbNPs showed better siRNA-mediated knockdown effects in various glioblastoma cell lines and melanoma cells than uncrosslinked preparations.To achieve stable co-encapsulation and efficient delivery of siRNA and hydrophilic chemicals for anti-inflammatory treatment of ulcerative colitis (UC), Xu et al. synthesized poly(ethylene glycol)-b-poly(trimethylene carbonate-co-dithiolocyclic trimethylene carbonate)-b-polyethyleneimine (PEG-P(TMC-DTC))-PEI, which self-assembled to form polymer vesicles with PEG in the outer shell and PEI facing an inner hydrophilic core.134 The disulfide heterocyclic pentane ring suspended in the P(TMC-DTC) block can be oxidized to a disulfide bond while enhancing colloidal stability. The macrophage-targeting peptide TKPR was further modified at the PEG end to make it macrophage-targeting. Electrostatic interactions between PEI and siTNF – αsiRNA/DSP promoted efficient cargo encapsulation. Upon targeted internalization into macrophages, the disulfide bond is fractured by GSH reduction, which accelerates the dissociation of polymer vesicles and the release of cargoes to achieve the anti-inflammatory effect of synergistic anti-UC. Li et al. developed novel pH-responsive lipid polymer nanoparticles (PLPNs) combined with Ultrasound-mediated microbubble destruction (UMMD) technology to achieve optimal CRISPRi systemic delivery, leading to efficient target gene inhibition135 (Fig. 6a). UMMD induces mechanical perturbation of vascular walls and cell membranes, thereby enhancing the permeability of malignant tissues to DNA carriers.136 PLPNs have a core–shell structure in which the CRISPR plasmid DNA (pDNA) with PBA functionalized low molecular weight polyethyleneimine PEI1.8 k (PEI-PBA) condensed to nucleate. The PEIPBA/pDNA complex was further encapsulated in pH-activated “detachable” PEOz lipids. The PEOz lipid shell gives PLPNs a negatively charged surface with good serum resistance and effectively reduces immune clearance. Upon entry into the tumor tissue, the outer shell of PLPNs breaks down in the acidic microenvironment, releasing the PEI-PBA/pDNA complex, which is capable of enhancing the internalization of the complex by cancer cells through the binding of salivary acid (SA). PEI in the complex triggers endosomal rupture through a “proton sponge” effect, releasing pDNA into the cytoplasm. In contrast, some of the undecomposed PLPNs enter the cell via endocytosis, and in an acidic (pH 5.0) environment, the outer shell is almost completely decomposed, which triggers the release of pDNA. In addition, in vivo studies demonstrated that UMMD administration significantly increased the tumor enrichment of PLPNs and improved gene repression efficiency. Zhang et al. constructed a peptide containing nuclear localization signals (NLS) and a Tefilin-modified disulfide-bonded crosslinked polymer (bPEI-SSPEG-T) for efficient delivery of interleukin-10 (IL-10) pDNA.137 First, NLS formed a complex with pDNA by electrostatic interaction to enhance the nucleation efficiency. The NLS/DNA complexes were then combined with bPEI-SS-PEG-T to construct bPEI-SS-PEG-T/NLS/DNA NPs with a particle size of 168.4 nm and a zeta potential of +10.9 mV (Fig. 6b). bPEI-SS-PEG-T is responsive to intracellular glutathione, is biodegradable, and Tefilin peptides interacted with the Fc receptor and neuropilin-1 receptor on macrophages enhanced macrophage-mediated phagocytosis and the nano complexes exhibited better biocompatibility, higher cellular uptake, and nucleation efficiency, leading to better IL-10 plasmid transfection.
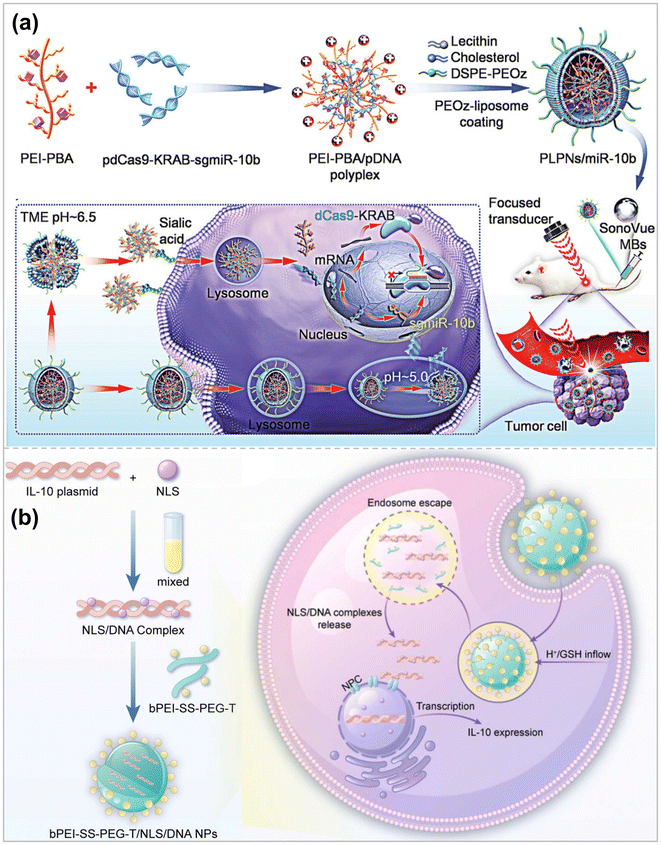 | ||
| Fig. 6 (a) Schematic illustration of the preparation of pH-responsive PLPNs and the enhanced cellular uptake and tumor accumulation by the combination of UMMD and PLPNs ref. 135. Copyright 2023, Wiley-VCH. (b) Illustration of formation of bPEI-SS-PEG-T/NLS/DNA NPs and the intracellular fate of bPEI-SS-PEG-T/NLS/DNA NPs ref. 137. Copyright 2023, Elsevier. | ||
3. Biological application
3.1 Cancer treatment
Breast cancer is one of the most common types of cancer, and its incidence is increasing every year, with more than 3 million new breast cancer cases expected to be diagnosed by 2040.138 Surgery and radiation are the first-line treatments for localized or early-stage breast cancer, and chemotherapy is the treatment of choice for metastatic or advanced disease.139 Although breast cancer is one of the solid tumors sensitive to chemotherapy, the emergence of drug resistance can render chemotherapy ineffective. Especially in triple-negative breast cancer (TNBC), the recurrence rate increases, and long-term survival decreases after conventional treatment. In breast cancer research, scientists have identified various therapeutic targets to inhibit tumor metastasis and recurrence through effective gene delivery and silencing responsive targets.140 Li et al. selected metastatic breast cancer as an in vivo gene inhibition model for the CRISPRi system.135 MicroRNA-10b (miR-10b) is highly expressed in metastatic breast tumors and becomes a therapeutic target for anti-metastasis. Thus, sgRNAs targeting miR-10b were developed. Subsequently, PLPNs/miR-10b were constructed and combined with the UMMD to introduce a powerful gene suppression toolbox into tumor cells and tissues to inhibit tumor metastasis efficiently. Zhang et al. chose antisense miR-21 oligonucleotide (anti21) as the therapeutic sequence for cancer cells via fluoropolymer-coated DNA nanocrystals. Silencing miR-21 can inhibit the secretion of vascular endothelial growth factor (VEGF) secretion, inhibiting tumor angiogenesis.95 The fluorinated polymeric coating was also doped with quantum dots with NIR-IIb (1500–1700 nm) fluorescence and NIR photothermal properties. In the humanized breast cancer model, after anti21 was effectively delivered to breast cancer cells through circulation (Fig. 7a), under NIR light irradiation, anti21 could be released in the cytoplasm on demand, achieving high transfection efficiency, effective silencing of target miRNAs, and inhibition of angiogenesis for tumor ablation (Fig. 7b). In addition, non-invasive volumetric imaging of the gene vector delivery process can be performed under NIR-IIb emission.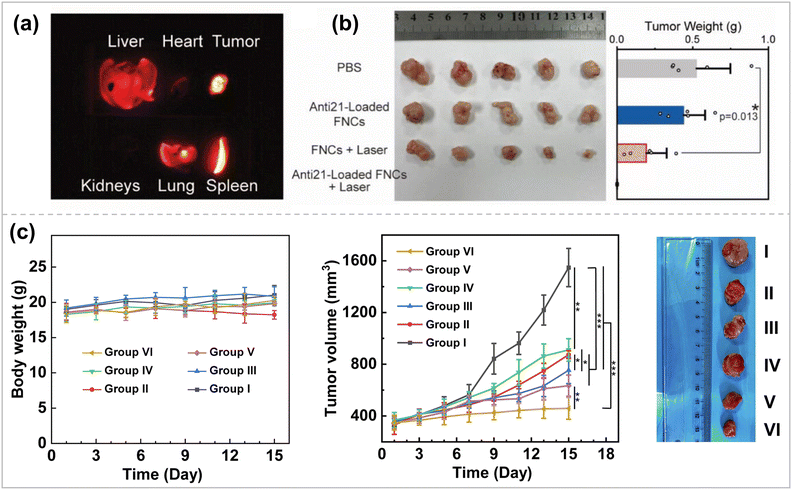 | ||
| Fig. 7 (a) NIR-IIb imaging. (b) Image (left) and weights (right) of tumors after excision from each group ref. 95. Copyright 2023, Wiley-VCH. (c) Body weights, relative tumor volume, and representative tumor pictures of mice during treatment in different groups. I, PBS; II, free DOX; III, G5-G3-D; IV, CBAA-G5/G3-Man/YTHDF1 siRNA; V, free DOX + CBAA-G5/G3-Man/YTHDF1 siRNA; and VI, G5-G3-D + CBAA-G5/G3-Man/YTHDF1 siRNA ref. 143. Copyright 2023, Elsevier. | ||
The efficacy of tumor therapy will be further enhanced when co-delivering multiple therapeutic and adjuvant mRNAs. Neshat et al. utilized biodegradable, lipophilic PBAE for co-delivery of mRNA structures encoding Signal 2 co-stimulatory molecules (4-1BBL) and Signal 3 immunostimulatory cytokines (IL-12), as well as nucleic acid-based immunomodulatory adjuvants (CpG).141 The nanocarriers were further encapsulated with a thermosensitive hydrogel, enhancing their retention time at the tumor site. This triblock thermosensitive copolymer poly(lactic acid–ethanolic acid)-poly(ethylene glycol)-poly (lactic acid–ethanolic acid) (PLGA-PEG-PLGA) was liquid and injectable at 4 °C but formed a gel structure at 37 °C, thereby confining the nanocarriers to the tumor injection site. In vivo results showed that PBAE NPs loaded with 4-1BBL, IL12, and CpG adjuvants effectively reduced E0771 mammary tumor load in a mouse model when combined with an immune checkpoint blocker (anti-PD1) while prolonging the animal's life cycle and avoiding distal effects.
Exploiting effective nanomedicines to counteract tumor immunogenicity and immunosuppression is essential to improve the efficacy of immunotherapy. Liu et al. PBA-modified ε-polylysine targeted nanoplatforms for co-delivery of P-gp siRNA, Bcl-2 siRNA, and Adriamycin to treat multidrug-resistant breast cancer.142 The complex has promising targeting ability and ATP-responsive cytoplasmic nucleic acid release properties with sound inhibitory effects on invasion, proliferation, and clone formation of MCF-7/ADR cells. The abundant groups on the surface of the PAMAM and internal cavities make it easy to confer dendritic macromolecule targeting and better loading of cargoes for tumor therapeutic applications. Song et al. developed two nanomodules based on 5th-generation polyamide-amine dendrimers (G5)/3rd-generation polyamide-amine dendrimers (G3) core–shell structured dendrimers (CSTD) for chemoimmunotherapy in an in situ breast tumor model.143 Acetylated G5/G3 CSTDs wrapped around DOX constituted the first nanomodule that induced immunogenic cell death (ICD) in cancer cells. Carboxybetaine acrylamide (CBAA)-G5/G3-mannose (Man)/YTHDF1 siRNA complexes served as another nanomodule to stimulate the maturation of DCs by inhibiting YTHDF1 expression in DCs. Enhanced T-cell-mediated tumor immunotherapy through tumor ICD effects and genetic engineering of DCs to stimulate DCs maturation led to in vivo enhanced chemoimmunotherapy in orthotopic mouse breast cancer models (Fig. 7c).
Hepatocellular carcinoma (HCC) accounts for 90% of all liver cancers, and its 5-year survival rate is only 10%, posing a severe threat to human health.144 Although surgery or liver transplantation can cure some patients, the majority of patients present with invasive hepatocellular carcinoma, which can only be treated locally, whereas chemotherapy and radiotherapy are off-target toxic to healthy hepatocytes.145 Gene therapy has shown improved targeting and safety. Vaughan et al. used PBAE NPs to deliver a CpG-free plasmid with mutant herpes simplex virus type 1 sr39 thymidine kinase (sr39) DNA to human HCC cells.146 Transfection of sr39 allows the prodrug ganciclovir to kill cancer cells and accumulate 9-(4-18F-fluoro-3-hydroxymethylbutyl) guanine (18F-FHBG) for in vivo imaging. Targeting was achieved using the CpG-free human alpha-fetoprotein (AFP) promoter (CpGf-AFP-sr39). It was reached only in AFP-producing HCC cells, enabling selective transfection of bit HCC xenografts. CpGf AFP-sr39NP treatment resulted in a 62% reduction in tumor volume, and expression of the therapeutic gene was detectable using PET.
3.2 Psoriasis treatment
Psoriasis is a chronic inflammatory skin disease manifested by increased pro-inflammatory cytokine tumor necrosis factor α (TNFα) expression.147 Suzuki et al. developed a siRNA delivery system based on hybrid polymeric lipid nanoparticles (PLNs) in combination with photochemical internalization (PCI) to optimize the endosomal escape of TNF α siRNA, intending to employ the system as a topical agent to treat psoriasis.148 The PCI technique disrupts endosomal membranes and facilitates the release of the cargo in the cytoplasm after activation of the photosensitizer by light irradiation. The PLNs consisted of 2.0% Compritol & 888 ATO (lipid), 1.5% Porloxacin 188, and 0.1% cationic polymer poly(allylamine) hydrochloride, with an average size of 142 nm and a zeta potential of +25 mV. It can efficiently encapsulate the photosensitizer TPPS2a and complex siRNA. The system exhibited excellent biocompatibility together with a high cellular uptake rate. In an in vitro delivery assay in a porcine skin model, PLNs could deliver siRNA and TPPS2a into the skin. In an imiquimod cream-induced hairless mouse model of psoriasis, PLNTPPS2a-TNFα siRNA, in combination with PCI, significantly reduced TNF α levels, resulting in redness and scaling of mouse skin. The results suggest that gene therapy combined with PCI has potential in the local treatment of psoriasis.3.3 Alzheimer's disease treatment
It has been long since brain-derived neurotrophic factor (BDNF) was applied to treating neurodegenerative diseases. However, the therapy still suffers from certain exogenous limitations, such as the production of side effects like neuropathic pain and seizures.149In addition, there are no positive clinical trial results for BDNF-based gene therapy. Therefore, a new method of delivering BDNF is necessary, and Li et al. has designed and synthesized two PBAEs to deliver mRNAs directly to the brain and spinal cord via ventricular pumps.150 In order to obtain mRNAs with higher and more stable transcription levels, Linear Design, an AI algorithm for the secondary structure of mRNAs, was applied to modify the mRNA sequences. Meanwhile, to avoid over-activation of neurons, the 3′ untranslated region was further modified with neuron-specific miRNA targeting sequences. In this way, off-target delivery to neurons was achieved to avoid seizures. The mRNA is then commonly expressed in astrocytes (Fig. 8a), releasing BDNF to support neurons, thereby improving memory in amyloid precursor protein/progerin 1 double transgenic AD model mice.
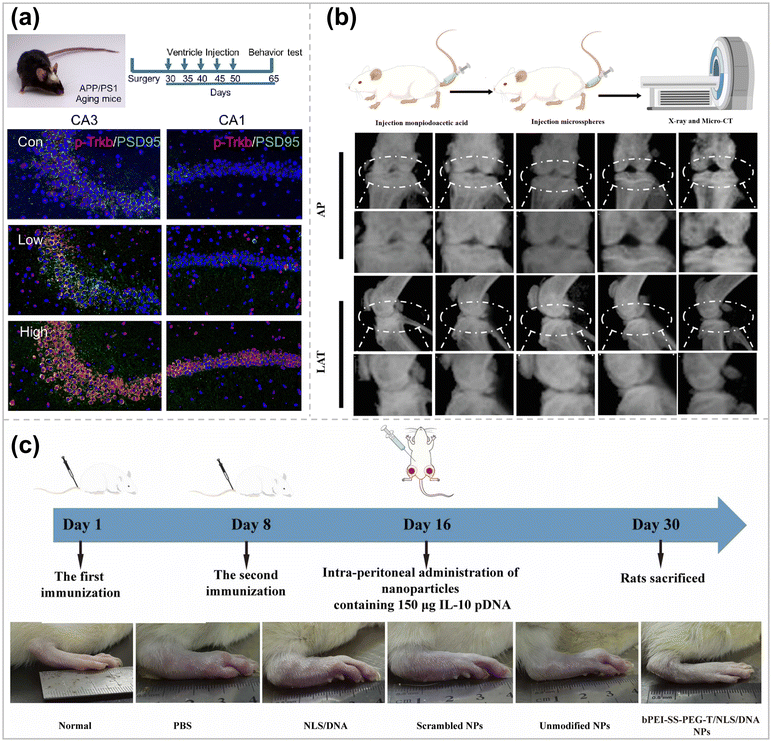 | ||
| Fig. 8 (a) Ventricle catheter system model in AD mice and treatment schedules and immunostaining of brain slices (p-Trkb in red and PSD 95 in green). Scale bar = 100 μm ref. 150. Copyright 2023, Elsevier. (b) Overview of animal experiments and X-ray images in anterior–posterior (AP) and lateral (LAT) view of knee joints ref. 154. Copyright 2023, Wiley-VCH (c) Schematic of CIA rats and treatment with intra-peritoneal injection and photographs of representative paws from the rats treated with different formulations ref. 137. Copyright 2024, Elsevier. | ||
3.4 Arthritis treatment
Continued damage to articular cartilage causes cartilage degeneration, leading to joint dysfunction and disability in patients.151 Compared with normal cartilage, the expression of hypoxia-inducible factor 2α (HIF-2α) is significantly increased in articular chondrocytes of osteoarthritis (OA) patients, which induces calcification as well as apoptosis of hypertrophic chondrocytes around the perichondral bone, leading to cartilage matrix degradation.152 Constructing a suitable gene vector to silence the HIF2α gene in articular chondrocytes will be the key to alleviating articular cartilage degeneration and treating OA. However, articular cartilage itself has no blood vessels, nerve tissue, and lymphatic distribution, and the extracellular matrix of chondrocytes is dense (pore size of about 60 nm) and has a negative electrical barrier, making it difficult to deliver drugs/genes to chondrocytes.153 Therefore, it is necessary to ensure the effective retention of gene carriers in the joint cavity while enhancing their enrichment around chondrocytes.Based on hexachlorocyclotriphosphazene as the core, Chen et al. synthesized a phosphorus-containing dendrimer derivative (G1-NC5·HCl) with piperidine on the surface by an iterative modification method, which can be effectively combined with HIF2α siRNA (G1-NC5·HC@siRNA).154 The cationic nanocarriers achieved efficient penetration into the cartilage damage site (pH = 6.6) and effective anchoring to the cartilage matrix (pH = 7.6). G1-NC5·HC@siRNA was loaded into hydrogel microspheres by ionic bond coordination and microfluidics to enhance the residence time of G1-NC5·HC@siRNA in the articular cavity and maintains its stable and sustained release in the articular cavity. After the hydrogel microspheres were injected into a rat model of traumatic osteoarthritis in a minimally invasive manner, the expression of MMP-13 protein was down-regulated by silencing the HIF2α gene, which inhibited the degeneration of cartilage tissues and the formation of osteoblasts, promoted cartilage regeneration, and then delayed the development of osteoarthritis (Fig. 8b). This system provides a new design idea and strategy for constructing matrix delivery drug nanocarriers.
Rheumatoid arthritis (RA) is a common chronic inflammatory disease that is currently not ideally treated due to the buffer barrier effect of the RA microenvironment.155 Interleukin-10 (IL-10) can regulate the rheumatoid arthritis microenvironment through metabolic reprogramming of macrophages.156 However, the short half-life, low targeting efficiency, and poor tissue penetration of IL-10 have led to its challenging clinical application. Zhang et al. combined bPEI-SS-PEG-modified with Tefilin with NLS/DNA complexes via classical interactions to form a macrophage-targeted IL-10 pDNA delivery system.137 After intraperitoneal injection, gene carriers can be efficiently taken up by peritoneal macrophages and subsequently carried to the site of inflammation by macrophages, thus favoring gene carrier aggregation at the site of RA inflammation. Expressed IL-10 can reduce the production of pro-inflammatory cytokines (TNF-α, IL-1β) by changing the proportion of M1-type and M2-type macrophages, thereby alleviating inflammatory symptoms (Fig. 8c). Compared with current therapeutic approaches, this macrophage-carrying gene delivery system has the advantages of excellent targeting and minimal side effects, which provides ideas for gene therapy of similar inflammatory diseases.
3.5 Lung-disease treatment
Multiple lung diseases, including influenza, cystic fibrosis, SARS-CoV-2, and asthma, make the lung a direct target for RNA therapeutic and preventive drugs.Aerosolized inhalation drug delivery is an approach to deliver RNA cargo directly to lung tissue, which allows for direct delivery of drugs to lesions, reducing the risk of side effects on healthy organs and tissues, but at the same time, must overcome physiological barriers such as respiratory mucosa, mucociliary clearance, and cell phagocytosis.157 Blanchard, E. L. et al. employed HPBAE to achieve lung nebulization delivery of mRNA-expressed CRISPR-Cas13a and guide RNA10 (Fig. 9a).158 The therapeutic effects of Cas13a expression and guide RNA delivery on influenza and SARS-CoV-2 were evaluated in experimental models of mice and hamsters, respectively. Based on this, Rotolo et al. designed and synthesized 166 polymers using hDD90-118 PBAE as a precursor, which share some common features, including (1) diacrylate electrophilic backbone, (2) multifunctional amines, amino-alcohols, and aminothiols as linear or branched nucleophilic components and (3) amino-terminal.159 Functional screening of luciferase reporter genes led to the discovery of candidate P76, a poly-β-amino-thioester (PBATE), which efficiently delivers mRNAs to mice, hamsters, ferrets, cows, and macaques by aerosolization and shows minimal toxicity. The thiols in P76 enable it to efficiently encode short crRNAs and long mRNAs, which were found to be highly effective in the SARS-In the SARS-CoV-2 model of attack on golden gophers, P76 delivered Cas13a mRNA at a dose 4-fold lower than previously reported for PBAE, comparable to the gold standard efficacy of systemic neutralizing antibody therapy (Fig. 9b). Fang et al. attached two antisense oligonucleotides (ASOs) via covalent bonding to a bottle-brush polymer backbone with 30 PEG10k side chains (pacDNA).160 pacDNA has a higher density of PEG than conventional PEGylated drugs, with a hydrodynamic radius of 30 nm and a slight negative charge, characteristics which make pacDNA an ideal mucus-permeable vehicle and thus uniformly distributed throughout the mouse lung airways and parenchyma. In addition, the high density of PEG of pacDNA did not affect ASO hybridization to complementary target sequences. In an NCI-H358 in situ non-small cell lung cancer mouse model, inhalation of pacDNA targeting wild-type KRAS mRNA suppressed KRAS expression and inhibited lung tumor growth at a much lower dose than intravenous pacDNA injection. Zhang et al. designed an inhalable nano-formulation to deliver messenger RNA for MMP13 (mMMP13) and keratinocyte growth factor (KGF) into fibrotic lung tissue for removing large amounts of deposited extracellular matrix (ECM) from the lungs and re-epithelializing ruptured alveoli, thereby reversing the pulmonary fibrosis established in a bleomycin-induced mouse model.123 The nano-formulation was assembled sequentially from ribosomal protein-concentrated mMMP13 cores, bifunctional peptide-modified crowns, and KGF with a PEGylated shielding shell. When inhaled through a nebulizer, microdroplets carrying the nano-formulation were deposited in the alveoli and subsequently penetrated the fibrotic foci, leading to the shedding of the outer KGFs due to the triggering of matrix metalloproteinase 2 (MMP2). After the RGD-grafted cores were revealed, the nano-formulation could specifically target integrin-rich cells for intracellular delivery of mMMP13. The bifunctional peptide used in this nano-formulation significantly enhanced targeting and cellular uptake efficiency. Zhuang et al. developed a hybrid nanoparticle combining dexamethasone-coupled polyethyleneimine (DP) with cell membranes (CM) from LA-4 lung epithelial cells.125 The aim was to improve the delivery efficiency of pDNA to the lungs. The CM in the hybrid nanoparticles interacted with the plasma membrane of the target cells, promoting cellular uptake of pDNA. The highest transfection efficiency to LA-4 cells was observed with DP/CM/pDNA at a mass ratio 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Zhang et al. achieved the preparation of dendritic vesicle nanoparticles by injecting ionizable amphiphilic Janus dendritic macromolecules (IAJD) and mRNA in acetate buffer and were able to deliver mRNA to the pulmonary system efficiently.161
1. Zhang et al. achieved the preparation of dendritic vesicle nanoparticles by injecting ionizable amphiphilic Janus dendritic macromolecules (IAJD) and mRNA in acetate buffer and were able to deliver mRNA to the pulmonary system efficiently.161
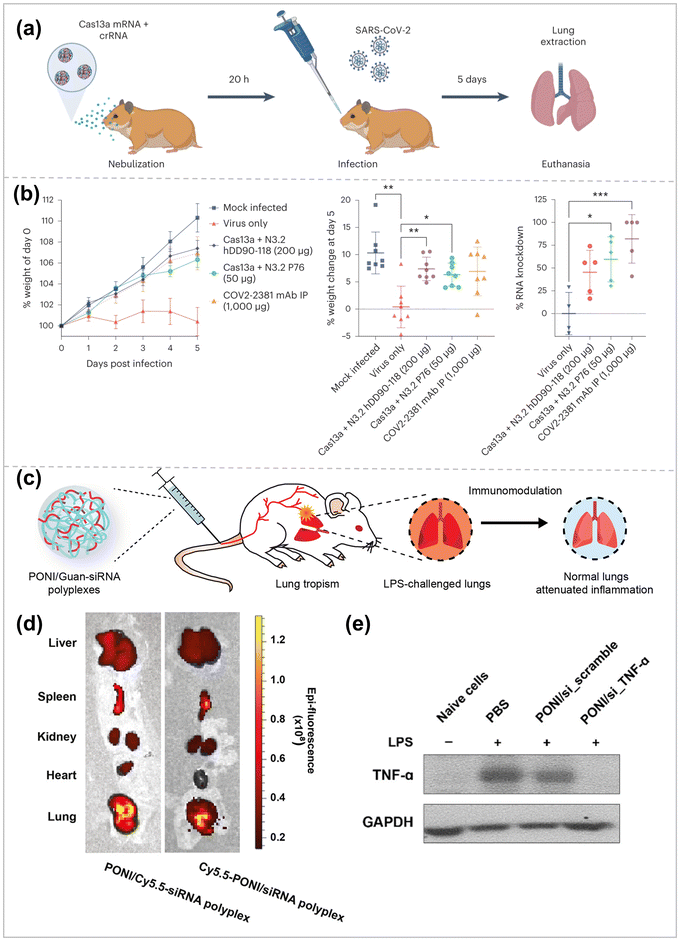 | ||
| Fig. 9 (a) Hamsters were treated with either nebulized P76 or hDD90-118-formulated Cas13 mRNA and crRNA. 20 h later, the hamsters were intranasally inoculated with 103 PFU of WA-1 SARS-CoV-2. The hamsters were euthanized on day 5 and the lungs were extracted and processed for viral load quantification. (b) Percent normalized hamster weight over time, per-cent hamster weight at day 5 post infection and per-cent knockdown of SARS-CoV-2 RNA in the lung at day 5 post infection ref. 158. Copyright 2023, Springer Nature. (c) Systemically administered polyplexes distribute to lung tissue, with preferential localization to inflamed lungs. (d) Representative in vivo imaging system (IVIS) organ images of polyplexes labeled with Cy5.5-PONI-Guan (left) or Cy5.5-siRNA (right), showing colocalization and lung accumulation following systemic administration in LPS-challenged mice. (e) Western blot analysis of TNF-α in RAW 264.7 cells that were untreated or treated with the indicated siRNA for 48 h ref. 174. Copyright 2023, American Chemical Society. | ||
Achieving mucus transport and cell membrane penetration can enhance siRNA delivery to lung tissues. Ge et al. developed guanidinylated and fluorinated bifunctional helical peptides for lung delivery of siRNA targeting tumor necrosis factor-α (TNF-α).162 The guanidinium-based structural domains and α-helices are structurally similar to those of natural cell-penetrating peptides, which allows the peptides to have strong cell membrane penetration. The fluorocarbon chain segment inhibits the adsorption of mucin glycoproteins on the surface of the complex, enhancing the peptide's mucus permeability by 240-fold. When TNF-αsiRNA was delivered intratracheally, peptide P7F7 induced 96% knockdown at 200 μg kg−1 siRNA and exerted a significant anti-inflammatory effect in acute lung injury.
Amine-containing end-capped polymers have been shown to facilitate targeting the lungs, as well as increased mRNA expression.163–165 Poly(amine-co-ester) (PACE) is a class of biodegradable polymers. For certain specific polymer compositions, PACE forms complexes with mRNA through electrostatic interactions between the mildly cationic polymer and the negatively charged nucleic acid phosphate backbone and hydrophobic interactions between the polymer chain segments (PACE-mRNA). The amino-terminated PACE polymer can promote the escape of mRNA from endosomes to the cytoplasm, thereby improving the transfection efficiency.166 PEG-stabilized PACE-mRNA complexes can further improve mRNA delivery in vivo.167 Suberi et al. further optimized protein expression after inhalation delivery to the respiratory tract by screening a library of delivery vectors with different modifications of amino-terminal groups and PEG content.168 The mucus layer includes highly cross-linked mucin chains, water, and other gel-like components, forming a natural barrier.169 At the same time, the highly negative charge of the lung mucus layer makes simple cationic polymer-carriers unable to pass through the mucus effectively. The development of amphiphilic polymers is conducive to the stability and penetration of the delivery carrier mucus while reducing its biological toxicity. Adams et al. prepared a new type of high molecular weight, brush-like spermine-based polyacrylamide and then copolymerized it with decyl acrylamide to increase the hydrophobicity of the polymer (Sp AA-co-DAA).170 When the ratio of cationic and hydrophobic monomer subunits was 43/57, Sp AA-co-DAA showed better lung cell uptake than lipofectamine and 10 times higher than PEI complex. Compared with commercially available transfection reagents, Sp AA-co-DAA was more effective in gene silencing G protein-coupled receptor protease-activated receptor 2 (PAR2) in fibrotic human lung explants. The biomimetic lipid shell can also help the polymer carrier achieve mucus penetration and cell internalization. LNPs composed of poly PLGA core and DPPC lipid shell (hNPs) could effectively transport siRNA to the lungs.171 The siRNA targeting NFκB was encapsulated in hNPs, and effective NFκB gene silencing was achieved in LPS-stimulated human bronchial epithelial cells.
Intravenous administration is a direct method to reach the treatment site. Still, it is usually limited to organs outside the reticuloendothelial system (RES), such as the liver, spleen, and kidneys, rendering the drug more off-target effects and cytotoxicity in treating inflammation in the lungs.172,173 Therefore, Jeon et al. delivered siRNAs to the site of inflammation in the lungs by systemic drug delivery using guanidinium-functionalized poly(oxanorbornene)imide (PONI-Guan) polymers as a gene carrier to knock down tumor necrosis factor α (TNF-α) in lung cells for attenuating the lung inflammation (Fig. 9c).174 Specifically, PONI-Guan homopolymers self-assembled with siRNA via electrostatic interactions to form discrete (∑170 nm) and cationic surface-charged nanocomplexes. These complexes transport siRNA (0.14–0.28 mg kg−1) directly into the cytosol via a membrane fusion-like uptake process, achieving efficient (>70%) GFP reporter gene knockdown in macrophages (mice peritoneal macrophages (raw264.7)). Intravenous injection of the PONI – Guan/siRNA complex in LPS-stimulated mice resulted in a 3-fold increase in lung localization (Fig. 9d), showing an internal tendency for inflamed lung tissue while efficiently knocking down serum TNF-α (>80%) (Fig. 9e). The system efficiently delivers siRNA to the cytoplasm, administering the drug at significantly lower levels than contemporary clinical studies while reducing the risk of off-target effects.
3.6 Others
Delivery of nucleic acids to the retina remains a major challenge due to the eye's unique anatomy and various barriers. Tan et al. developed a core–shell delivery platform for targeted delivery to the retina, with an inner core consisting of amino acid-functionalized dendrimers and nuclear localization signals for DNA complexation, nuclear translocation, and efficient transfection.175 The inner core was encapsulated in a pH-sensitive lipid bilayer. Hyaluronic acid (HA)-1,2-dioleoyl phosphatidylethanolamine (DOPE) is used as the outermost shell layer for retinal cell targeting. The environment in the vitreous does not affect the movement of negatively charged vectors and can diffuse into the retina, enter retinal cells via the CD44-mediated internalization pathway, and finally conclude into the nucleus via NLS translocation for efficient transfection. Guo et al. designed copolymers (PACDs) consisting of hydrophilic PEG blocks, siRNA-binding blocks, and pH-responsive blocks.176 The PACDs and siRNA can be simply mixed and self-assembled into nano-sized polymeric micelles. Due to the pH-responsiveness, PACD/siRNA complexes exhibit excellent endosomal escape efficiency and gene silencing efficiency both in vitro and in vivo. The hydrophilic shell, small size, and slight positive charge of the PACD/siRNA complexes enable them to effectively circumvent the intraocular clearance mechanism after a single injection, rapidly distribute in all retina layers, and remain in the retina for more than a week. In addition, the PACD/siVEGFA complex inhibited angiogenesis in a mouse model of oxygen-induced retinopathy without causing any inflammatory or histopathological changes. This study has implications for the development of efficient ocular gene delivery systems.The yes-associated protein (YAP) is a crucial driver of epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) resistance. Inhibition of YAP expression is a potential therapeutic option for treating non-small cell lung cancer (NSCLC). Huang et al. proposed a nanococktail therapeutic strategy utilizing a stimuli-responsive blocked dendritic polymer, Ppa-conjugate polyOEGMA-blockpoly[Dendron(G3)-Ppa] (Polymer), to simultaneously co-deliver YAP siRNA, a photosensitizer (Ppa) and EGFR – TKI gefitinib (Gef) 3 therapeutic units to achieve targeted drug/gene/photodynamic therapy.177 The formed complexes were efficiently internalized into Gef-resistant NSCLC cells and successfully escaped from late endosomes/lysosomes. The disulfide bonds in the complex break in response to the intracellular reducing environment, efficiently releasing Gef and YAP – siRNA. Released Gef inhibits the EGFR signaling pathway, while YAP – siRNA blocks the Gef resistance bypass pathway. Photodynamic therapy (PDT) elevated the intracellular ROS level to promote apoptosis in tumor cells. In addition, inhibition of glycolysis also promoted apoptosis by down-regulating HIF – 1 α expression, which significantly alleviated PDT-induced hypoxia.
4. Conclusions and perspectives
In recent years, gene therapy has made significant progress both in basic research and clinical fields due to the extensive development of efficient delivery systems. To date, a variety of polymers, such as derivatives based on PEI, polyesters, polyamino acids, and dendrimers, have been used as gene delivery vectors. Scientists have adopted various strategies to improve transfection efficiency in vitro and in vivo, modifying and designing polymer structures at the molecular level, tuning the complexes at the nanoparticle level, and combining artificial intelligence and machine learning to improve the efficiency of gene delivery. The polymeric gene vectors have achieved excellent therapeutic effects in some disease models, providing a basis for future clinical translation.Based on the broad prospect of gene therapy, we look forward to the future development of high-performance gene vectors. (1) Since most of the current studies focus on the transfection efficiency of cells, the research on the transfection of polymer vectors still needs to be completed. Therefore, further clarification of the relationship between the composition and structure of the vectors and the various stages in the transfection process will help to design high-performance polymer gene vectors. (2) The PDI of polymers is extensive, and polymers with different molecular weights play different roles in the delivery process. Therefore, preparing polymers with narrower molecular weight distributions will help to clarify the relationship between polymer structure and function. (3) The development of novel bio-responsive and photo-/thermo-/ultrasound/magneto-responsive delivery vectors to spatiotemporally control the release of genes will contribute to the on-demand delivery of drugs and precise treatment of diseases. A thorough understanding of the cellular internalization pathways of polymeric nanocarriers and their fates in cells will contribute to the optimization of polymeric vectors and bring out the full function of genes in the cell. (4) Prime Editing (PE) is a gene editing technology that enables arbitrary base substitutions, DNA fragment insertion, and deletion.178 The development of polymer vectors suitable for PE delivery will promote the further development of gene therapy. (5) Animal experiments always have deviations in predicting human results due to significant differences between species. Therefore, it is necessary to develop more appropriate and standardized disease models that can better represent physiological conditions, to provide highly relevant data for developing human gene drugs. (6) Utilizing high-throughput evaluation methods to establish a more comprehensive and unified evaluation system will help integrate and analyze data to design high-performance polymer vectors. Scientists have now developed and tested the performances of a large number of polymer vectors, which is a vast database. Therefore, an effective combination of computer-aided learning strategies, using data-driven analysis of the relationship between polymer structure and performances to make accurate predictions of next-generation polymers, would significantly save time and resources.
The development of efficient and safe polymeric gene delivery vectors requires collaborative efforts among scientists from multiple fields, such as molecular biology, polymer chemistry, physical chemistry, biomedicine, and computer artificial intelligence. It is foreseeable that shortly, more and more polymer nanomaterials can be used as effective gene nanocarriers, which can be validated in clinical experiments and applied to the effective treatment of various diseases.
Author contributions
Y. L., R. T. and J. L. were involved in conceptualization and project administration. Y. L., R. T., J. X., T. W. and Y. Z. were involved in visualization. Y. L. and R. T. were involved in writing the original draft. All authors were involved in editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the National Natural Science Foundation of China (No. 22201058, 22161142015, and 22275046) and the National Key R&D Program of China (Grant No. 2020YFA0908500).References
- C. Wang, C. Pan, H. Yong, F. Wang, T. Bo, Y. Zhao, B. Ma, W. He and M. Li, J. Nanobiotechnol., 2023, 21, 272 CrossRef PubMed.
- Y. Li, X. Wang, Z. He, M. Johnson, A. Sigen, I. Lara-Sáez, J. Lyu and W. Wang, J. Am. Chem. Soc., 2023, 145, 17187–17200 CrossRef CAS PubMed.
- I. Lostalé-Seijo and J. Montenegro, Nat. Rev. Chem., 2018, 2, 258–277 CrossRef.
- C. E. Dunbar, K. A. High, J. K. Joung, D. B. Kohn, K. Ozawa and M. Sadelain, Science, 2018, 359, eaan4672 CrossRef PubMed.
- A. V. Anzalone, P. B. Randolph, J. R. Davis, A. A. Sousa, L. W. Koblan, J. M. Levy, P. J. Chen, C. Wilson, G. A. Newby, A. Raguram and D. R. Liu, Nature, 2019, 576, 149–157 CrossRef CAS PubMed.
- P. Huang, H. Deng, Y. Zhou and X. Chen, Matter, 2022, 5, 1670–1699 CrossRef CAS.
- D. W. Pack, A. S. Hoffman, S. Pun and P. S. Stayton, Nat. Rev. Drug Discovery, 2005, 4, 581–593 CrossRef CAS PubMed.
- M. A. Mintzer and E. E. Simanek, Chem. Rev., 2009, 109, 259–302 CrossRef CAS PubMed.
- J. Gilleron, W. Querbes, A. Zeigerer, A. Borodovsky, G. Marsico, U. Schubert, K. Manygoats, S. Seifert, C. Andree, M. Stöter, H. Epstein-Barash, L. Zhang, V. Koteliansky, K. Fitzgerald, E. Fava, M. Bickle, Y. Kalaidzidis, A. Akinc, M. Maier and M. Zerial, Nat. Biotechnol., 2013, 31, 638–646 CrossRef CAS PubMed.
- R. Kumar, C. F. S. Chalarca, M. R. Bockman, C. V. Bruggen, C. J. Grimme, R. J. Dalal, M. G. Hanson, J. K. Hexum and T. M. Reineke, Chem. Rev., 2021, 121, 11527–11652 CrossRef CAS PubMed.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Nat. Rev. Genet., 2014, 15, 541–555 CrossRef CAS PubMed.
- K. A. Hajj and K. A. Whitehead, Nat. Rev. Mater., 2017, 2, 1–17 Search PubMed.
- D. Zhang, E. N. Atochina-Vasserman, D. S. Maurya, N. Huang, Q. Xiao, N. Ona, M. Liu, H. Shahnawaz, H. Ni, K. Kim, M. M. Billingsley, D. J. Pochan, M. J. Mitchell, D. Weissman and V. Percec, J. Am. Chem. Soc., 2021, 143, 12315–12327 CrossRef CAS PubMed.
- M. Qiu, Z. Glass, J. Chen, M. Haas, X. Jin, X. Zhao, X. Rui, Z. Ye, Y. Li, F. Zhang and Q. Xu, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2020401118 CrossRef CAS PubMed.
- H. Guerrero-Cázares, S. Y. Tzeng, N. P. Young, A. O. Abutaleb, A. Quiñones-Hinojosa and J. J. Green, ACS Nano, 2014, 8, 5141–5153 CrossRef PubMed.
- A. A. Eltoukhy, D. J. Siegwart, C. A. Alabi, J. S. Rajan, R. Langer and D. G. Anderson, Biomaterials, 2012, 33, 3594–3603 CrossRef CAS PubMed.
- A. I. S. van den Berg, C.-O. Yun, R. M. Schiffelers and W. E. Hennink, J. Controlled Release, 2021, 331, 121–141 CrossRef CAS PubMed.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed.
- M. D. Giron-Gonzalez, R. Salto-Gonzalez, F. J. Lopez-Jaramillo, A. Salinas-Castillo, A. B. Jodar-Reyes, M. Ortega-Muñoz, F. Hernandez-Mateo and F. Santoyo-Gonzalez, Bioconjugate Chem., 2016, 27, 549–561 CrossRef CAS PubMed.
- Q. Cheng, T. Wei, Y. Jia, L. Farbiak, K. Zhou, S. Zhang, Y. Wei, H. Zhu and D. J. Siegwart, Adv. Mater., 2018, 30, 1805308 CrossRef PubMed.
- X. Li, C. O. T. Qizi, A. M. Khamis, C. Zhang and Z. Su, Pharm. Res., 2022, 39, 1065–1083 CrossRef CAS PubMed.
- X. Guo and L. Huang, Acc. Chem. Res., 2012, 45, 971–979 CrossRef CAS PubMed.
- M. Roursgaard, K. B. Knudsen, H. Northeved, M. Persson, T. Christensen, P. E. K. Kumar, A. Permin, T. L. Andresen, T. Gjetting, J. Lykkesfeldt, L. K. Vesterdal, S. Loft and P. Møller, Toxicol. in Vitro, 2016, 36, 164–171 CrossRef CAS PubMed.
- K. B. Knudsen, H. Northeved, P. Kumar, A. Permin, T. Gjetting, T. L. Andresen, S. Larsen, K. M. Wegener, J. Lykkesfeldt, K. Jantzen, S. Loft, P. Møller and M. Roursgaard, Nanomedicine, 2015, 11, 467–477 CrossRef CAS PubMed.
- J. E. Zuckerman and M. E. Davis, Nat. Rev. Drug Discovery, 2015, 14, 843–856 CrossRef CAS PubMed.
- P.-F. Cui, L.-Y. Qi, Y. Wang, R.-Y. Yu, Y.-J. He, L. Xing and H.-L. Jiang, J. Controlled Release, 2019, 303, 253–262 CrossRef CAS PubMed.
- F. Richter, K. Leer, L. Martin, P. Mapfumo, J. I. Solomun, M. T. Kuchenbrod, S. Hoeppener, J. C. Brendel and A. Traeger, J. Nanobiotechnol., 2021, 19, 292 CrossRef CAS PubMed.
- X. Huang, J. Li, G. Li, B. Ni, Z. Liang, H. Chen, C. Xu, J. Zhou, J. Huang and S. Deng, Acta Biomater., 2023, 161, 226–237 CrossRef CAS PubMed.
- D. Putnam, Nat. Mater., 2006, 5, 439–451 CrossRef CAS PubMed.
- N. S. K. Gowthaman, H. N. Lim, T. R. Sreeraj, A. Amalraj and S. Gopi, in Biopolymers and Their Industrial Applications: From Plant, Animal, and Marine Sources, to Functional Products, Elsevier, 2021, pp. 351–372 Search PubMed.
- P. Zhang, Q. Xu, J. Du and Y. Wang, RSC Adv., 2018, 8, 34596–34602 RSC.
- J. DeRouchey, B. Hoover and D. C. Rau, Biochemistry, 2013, 52, 3000–3009 CrossRef CAS PubMed.
- M. A. Crosio, M. A. Via, C. I. Cámara, A. Mangiarotti, M. G. D. Pópolo and N. Wilke, Biomolecules, 2019, 9, 625 CrossRef CAS PubMed.
- C. O. Franck, A. B. Popov, I. Ahmed, R. E. Hewitt, L. Franslau, P. Tyagi and L. Fruk, Nanoscale Horiz., 2023, 8, 1588–1594 RSC.
- T. Bus, A. Traeger and U. S. Schubert, J. Mater. Chem. B, 2018, 6, 6904–6918 RSC.
- D. M. Lynn and R. Langer, J. Am. Chem. Soc., 2000, 122, 10761–10768 CrossRef CAS.
- D. G. Anderson, A. Akinc, N. Hossain and R. Langer, Mol. Ther., 2005, 11, 426–434 CrossRef CAS PubMed.
- D. Zhou, L. Cutlar, Y. Gao, W. Wang, J. O'Keeffe-Ahern, S. McMahon, B. Duarte, F. Larcher, B. J. Rodriguez, U. Greiser and W. Wang, Sci. Adv., 2016, 2, e1600102 CrossRef PubMed.
- B. Newland, Y. Zheng, Y. Jin, M. Abu-Rub, H. Cao, W. Wang and A. Pandit, J. Am. Chem. Soc., 2012, 134, 4782–4789 CrossRef CAS PubMed.
- Y. Li, X. Wang, Z. He, M. Johnson, A. Sigen, I. Lara-Sáez, J. Lyu and W. Wang, J. Am. Chem. Soc., 2023, 145, 17187–17200 CrossRef CAS PubMed.
- C. Wang, W. He, F. Wang, H. Yong, T. Bo, D. Yao, Y. Zhao, C. Pan, Q. Cao, S. Zhang and M. Li, J. Nanobiotechnol., 2024, 22, 40 CrossRef CAS PubMed.
- Y. Zhao, T. Bo, C. Wang, D. Yao, C. Pan, W. Xu, H. Zhou, M. Li and S. Zhang, J. Nanobiotechnol., 2023, 21, 394 CrossRef CAS PubMed.
- J. Teo, M. McCarroll, C. Boyer and P. A. Phillips, Biomacromolecules, 2016, 17, 2337–2351 CrossRef CAS PubMed.
- J. M. Ren, T. G. McKenzie, Q. Fu, E. H. H. Wong, J. Xu, Z. An, S. Shanmugam, T. P. Davis, C. Boyer and G. G. Qiao, Chem. Rev., 2016, 116, 6743–6836 CrossRef CAS PubMed.
- C. Wang, X. Huang, L. Sun, Q. Li, Z. Li, H. Yong, D. Che, C. Yan, S. Geng, W. Wang and D. Zhou, Chem. Commun., 2022, 58, 2136–2139 RSC.
- A.-H. Ranneh, H. Takemoto, S. Sakuma, A. Awaad, T. Nomoto, Y. Mochida, M. Matsui, K. Tomoda, M. Naito and N. Nishiyama, Angew. Chem., Int. Ed., 2018, 57, 5057–5061 CrossRef CAS PubMed.
- X. Shen, A. Dirisala, M. Toyoda, Y. Xiao, H. Guo, Y. Honda, T. Nomoto, H. Takemoto, Y. Miura and N. Nishiyama, J. Controlled Release, 2023, 360, 928–939 CrossRef CAS PubMed.
- R. Xie, X. Wang, Y. Wang, M. Ye, Y. Zhao, B. S. Yandell and S. Gong, Adv. Mater., 2022, 34, 2110618 CrossRef CAS PubMed.
- R. Chen, S. K. Wang, J. A. Belk, L. Amaya, Z. Li, A. Cardenas, B. T. Abe, C.-K. Chen, P. A. Wender and H. Y. Chang, Nat. Biotechnol., 2023, 41, 262–272 CrossRef CAS PubMed.
- T. R. Blake, W. C. Ho, C. R. Turlington, X. Zang, M. A. Huttner, P. A. Wender and R. M. Waymouth, Chem. Sci., 2020, 11, 2951–2966 RSC.
- N. N. Parayath, S. B. Stephan, A. L. Koehne, P. S. Nelson and M. T. Stephan, Nat. Commun., 2020, 11, 6080 CrossRef CAS PubMed.
- C. J. McKinlay, J. R. Vargas, T. R. Blake, J. W. Hardy, M. Kanada, C. H. Contag, P. A. Wender and R. M. Waymouth, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E448–E456 CrossRef CAS PubMed.
- C. J. McKinlay, N. L. Benner, O. A. Haabeth, R. M. Waymouth and P. A. Wender, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E5859–E5866 CrossRef PubMed.
- L. Amaya, L. Grigoryan, Z. Li, A. Lee, P. A. Wender, B. Pulendran and H. Y. Chang, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2302191120 CrossRef CAS PubMed.
- O. A. W. Haabeth, J. J. K. Lohmeyer, A. Sallets, T. R. Blake, I. Sagiv-Barfi, D. K. Czerwinski, B. McCarthy, A. E. Powell, P. A. Wender, R. M. Waymouth and R. Levy, ACS Cent. Sci., 2021, 7, 1191–1204 CrossRef CAS PubMed.
- A. J. Wilk, N. L.-B. Weidenbacher, R. Vergara, O. A. W. Haabeth, R. Levy, R. M. Waymouth, P. A. Wender and C. A. Blish, Blood Adv., 2020, 4, 4244–4255 CrossRef CAS PubMed.
- Z. Li, L. Amaya, R. Pi, S. K. Wang, A. Ranjan, R. M. Waymouth, C. A. Blish, H. Y. Chang and P. A. Wender, Nat. Commun., 2023, 14, 6983 CrossRef CAS PubMed.
- M. Akman, D. C. Belisario, I. C. Salaroglio, J. Kopecka, M. Donadelli, E. De Smaele and C. Riganti, J. Exp. Clin. Cancer Res., 2021, 40, 28 CrossRef CAS PubMed.
- X. Liu, J. Sun, J. Gu, L. Weng, X. Wang, L. Zhu, Q. Luo and Z. Chen, Chem. Eng. J., 2023, 470, 144271 CrossRef CAS.
- R. Cheng, F. Feng, F. Meng, C. Deng, J. Feijen and Z. Zhong, J. Controlled Release, 2011, 152, 2–12 CrossRef CAS PubMed.
- G. Wang, D. Zhu, Z. Zhou, Y. Piao, J. Tang and Y. Shen, ACS Appl. Mater. Interfaces, 2020, 12, 14825–14838 CrossRef CAS PubMed.
- J.-J. Nie, B. Qiao, S. Duan, C. Xu, B. Chen, W. Hao, B. Yu, Y. Li, J. Du and F.-J. Xu, Adv. Mater., 2018, 30, 1801570 CrossRef PubMed.
- D. Chen, C. Lei, W. Liu, M. Shao, M. Sun, J. Guo, J. Cao, J.-J. Nie, P. Luo, Y. Luo, B. Yu, R. Wang, S. Duan and F.-J. Xu, Bioact. Mater., 2023, 28, 376–385 CAS.
- W. Tao and Z. He, Asian J. Pharm., 2018, 13, 101–112 Search PubMed.
- C. Zhang, J. Chen, Y. Song, J. Luo, P. Jin, X. Wang, L. Xin, F. Qiu, J. Yao, G. Wang and P. Huang, ACS Appl. Mater. Interfaces, 2022, 14, 2587–2596 CrossRef CAS PubMed.
- N. Ma, Y. Li, H. Xu, Z. Wang and X. Zhang, J. Am. Chem. Soc., 2010, 132, 442–443 CrossRef CAS PubMed.
- B. Xianyu, S. Pan, S. Gao, H. Xu and T. Li, Small, 2023, 2306225 CrossRef PubMed.
- J. Wang, D. Li, Y. Fan, M. Shi, Y. Yang, L. Wang, Y. Peng, M. Shen and X. Shi, Nanoscale, 2019, 11, 22343–22350 RSC.
- J. Kim, Y. M. Lee, H. Kim, D. Park, J. Kim and W. J. Kim, Biomaterials, 2016, 75, 102–111 CrossRef CAS PubMed.
- J. Liu, G. Li, H. Guo, C. Ni, Y. Gao, X. Cao, J. Xia, X. Shi and R. Guo, ACS Appl. Mater. Interfaces, 2023, 15, 12809–12821 CrossRef CAS PubMed.
- W. Yasen, B. Li, A. Aini, Z. Li, Y. Su, L. Zhou, D. Guo, Q. Qian, D. Chen, X. Zhu and R. Dong, ACS Appl. Mater. Interfaces, 2023, 15, 41817–41827 CrossRef CAS PubMed.
- G. Kuang, H. Lu, S. He, H. Xiong, J. Yu, Q. Zhang and Y. Huang, Adv. Healthcare Mater., 2021, 10, 2100938 CrossRef CAS PubMed.
- D. E. Jaalouk and J. Lammerding, Nat. Rev. Mol. Cell Biol., 2009, 10, 63–73 CrossRef CAS PubMed.
- D. A. Fletcher and R. D. Mullins, Nature, 2010, 463, 485–492 CrossRef CAS PubMed.
- V. Graceffa, J. Genet. Eng. Biotechnol., 2021, 19, 90 CrossRef PubMed.
- J. G. Joseph and A. P. Liu, Adv. Biosyst., 2020, 4, 1900278 CrossRef PubMed.
- S.-M. Lim, J. P. Trzeciakowski, H. Sreenivasappa, L. J. Dangott and A. Trache, Integr. Biol., 2012, 4, 615–627 CrossRef CAS PubMed.
- K. H. Vining and D. J. Mooney, Nat. Rev. Mol. Cell Biol., 2017, 18, 728–742 CrossRef CAS PubMed.
- R. C. Geiger, W. Taylor, M. R. Glucksberg and D. A. Dean, Gene Ther., 2006, 13, 725–731 CrossRef CAS PubMed.
- K. M. Takeda, Y. Yamasaki, A. Dirisala, S. Ikeda, T. A. Tockary, K. Toh, K. Osada and K. Kataoka, Biomaterials, 2017, 126, 31–38 CrossRef CAS PubMed.
- F. Ponti, N. Bono, L. Russo, P. Bigini, D. Mantovani and G. Candiani, J. Nanobiotechnol., 2022, 20, 363 CrossRef CAS PubMed.
- M. Kubczak, S. Michlewska, M. Karimov, A. Ewe, S. Noske, A. Aigner, M. Bryszewska and M. Ionov, Int. J. Pharm., 2022, 614, 121468 CrossRef CAS PubMed.
- B. Dube, A. Pandey, G. Joshi, R. Mulherkar and K. Sawant, Int. J. Nanomed., 2018, 13, 83–85 CrossRef CAS PubMed.
- L. Liu, Z.-M. Zong, Q. Liu, S.-S. Jiang, Q. Zhang, L.-Q. Cen, J. Gao, X.-G. Gao, J.-D. Huang, Y. Liu and H. Yao, Biomaterials, 2018, 184, 20–30 CrossRef CAS PubMed.
- X. Yu, S. Liu, Q. Cheng, T. Wei, S. Lee, D. Zhang and D. J. Siegwart, Adv. Healthcare Mater., 2020, 9, 1901487 CrossRef CAS PubMed.
- Z. Yuan, X. Guo, M. Wei, Y. Xu, Z. Fang, Y. Feng and W.-E. Yuan, NPG Asia Mater., 2020, 12, 1–11 CrossRef.
- M. Wang and Y. Cheng, Biomaterials, 2014, 35, 6603–6613 CrossRef CAS PubMed.
- H. Liu, Y. Wang, M. Wang, J. Xiao and Y. Cheng, Biomaterials, 2014, 35, 5407–5413 CrossRef CAS PubMed.
- X. Cai, R. Jin, J. Wang, D. Yue, Q. Jiang, Y. Wu and Z. Gu, ACS Appl. Mater. Interfaces, 2016, 8, 5821–5832 CrossRef CAS PubMed.
- M. E. Johnson, J. Shon, B. M. Guan, J. P. Patterson, N. J. Oldenhuis, A. C. Eldredge, N. C. Gianneschi and Z. Guan, Bioconjugate Chem., 2016, 27, 1784–1788 CrossRef CAS PubMed.
- J.-H. Gong, Y. Wang, L. Xing, P.-F. Cui, J.-B. Qiao, Y.-J. He and H.-L. Jiang, Int. J. Pharm., 2018, 535, 180–193 CrossRef CAS PubMed.
- E. Tan, J. Lv, J. Hu, W. Shen, H. Wang and Y. Cheng, J. Mater. Chem. B, 2018, 6, 7230–7238 RSC.
- Y. Deng, J. Zhang, X. Sun, L. Li, M. Zhou, S. Liu, F. Chen, C. Pan, Z. Yu, M. Li, W. Zhong and M. Zeng, J. Controlled Release, 2023, 363, 597–605 CrossRef CAS PubMed.
- Y. Jin, W. Yu, W. Zhang, C. Wang, Y. Liu, W.-E. Yuan and Y. Feng, J. Colloid Interface Sci., 2023, 648, 287–298 CrossRef CAS PubMed.
- P. Zhang, R. Guo, H. Zhang, W. Yang and Y. Tian, Adv. Sci., 2023, 10, 2304633 CrossRef CAS PubMed.
- J. Yang, Q. Zhang, H. Chang and Y. Cheng, Chem. Rev., 2015, 115, 5274–5300 CrossRef CAS PubMed.
- L. Jakubcová, M. Vozárová, J. Hollý, K. Tomčíková, M. Fogelová, K. Polčicová, F. Kostolanský, E. Fodor and E. Varečková, J. Gen. Virol., 2019, 100, 1282–1292 CrossRef PubMed.
- K. Shigeta, S. Kawakami, Y. Higuchi, T. Okuda, H. Yagi, F. Yamashita and M. Hashida, J. Controlled Release, 2007, 118, 262–270 CrossRef CAS PubMed.
- F. Wang, K. Hu and Y. Cheng, Acta Biomater., 2016, 29, 94–102 CrossRef CAS PubMed.
- Y. Lee, J. Lee, M. Kim, G. Kim, J. S. Choi and M. Lee, J. Controlled Release, 2021, 330, 907–919 CrossRef CAS PubMed.
- H. Fang, Z. Guo, L. Lin, J. Chen, P. Sun, J. Wu, C. Xu, H. Tian and X. Chen, J. Am. Chem. Soc., 2018, 140, 11992–12000 CrossRef CAS PubMed.
- F. Joubert, M. J. Munson, A. Sabirsh, R. M. England, M. Hemmerling, C. Alexander and M. B. Ashford, J. Controlled Release, 2023, 356, 580–594 CrossRef CAS PubMed.
- Z. Kang, Q. Liu, Z. Zhang, Y. Zheng, C. Wang, Z. Pan, Q. Li, Y. Liu and L. Shi, Adv. Healthcare Mater., 2022, 11, 2200371 CrossRef CAS PubMed.
- D. M. Lynn and R. Langer, J. Am. Chem. Soc., 2000, 122, 10761–10768 CrossRef CAS.
- R. A. Cordeiro, A. Serra, J. F. J. Coelho and H. Faneca, J. Controlled Release, 2019, 310, 155–187 CrossRef CAS PubMed.
- P. Dosta, N. Segovia, A. Cascante, V. Ramos and S. Borrós, Acta Biomater., 2015, 20, 82–93 CrossRef CAS PubMed.
- C. Fornaguera, M. Guerra-Rebollo, M. Á. Lázaro, C. Castells-Sala, O. Meca-Cortés, V. Ramos-Pérez, A. Cascante, N. Rubio, J. Blanco and S. Borrós, Adv. Healthcare Mater., 2018, 7, 1800335 CrossRef PubMed.
- C. Fornaguera, M. Guerra-Rebollo, M. Á. Lázaro, C. Castells-Sala, O. Meca-Cortés, V. Ramos-Pérez, A. Cascante, N. Rubio, J. Blanco and S. Borrós, Adv. Healthcare Mater., 2018, 7, e1800335 CrossRef PubMed.
- P. Dosta, I. Tamargo, V. Ramos, S. Kumar, D. W. Kang, S. Borrós and H. Jo, Adv. Healthcare Mater., 2021, 10, 2001894 CrossRef CAS PubMed.
- M. Parés, C. Fornaguera, F. Vila-Julià, S. Oh, S. H. Y. Fan, Y. K. Tam, N. Comes, F. Vidal, R. Martí, S. Borrós and J. Barquinero, Hum. Gene Ther., 2021, 32, 1210–1223 CrossRef PubMed.
- S. Yonezawa, H. Koide and T. Asai, Adv. Drug Delivery Rev., 2020, 154–155, 64–78 CrossRef CAS PubMed.
- Y. Weng, C. Li, T. Yang, B. Hu, M. Zhang, S. Guo, H. Xiao, X.-J. Liang and Y. Huang, Biotechnol. Adv., 2020, 40, 107534 CrossRef CAS PubMed.
- A. A. Eltoukhy, D. Chen, C. A. Alabi, R. Langer and D. G. Anderson, Adv. Mater., 2013, 25, 1487–1493 CrossRef CAS PubMed.
- S. Liu, Z. Sun, D. Zhou and T. Guo, J. Mater. Chem. B, 2017, 5, 5307–5310 RSC.
- Y. Wen, H. Bai, J. Zhu, X. Song, G. Tang and J. Li, Sci. Adv., 2020, 6, eabc2148 CrossRef CAS PubMed.
- J. Shi, Y. Zhang, B. Ma, H. Yong, D. Che, C. Pan, W. He, D. Zhou and M. Li, ACS Appl. Mater. Interfaces, 2023, 15, 42130–42138 CrossRef CAS PubMed.
- J. H. Ryu, G. J. Lee, Y.-R. V. Shih, T. Kim and S. Varghese, Curr. Med. Chem., 2019, 26, 6797–6816 CrossRef CAS PubMed.
- C. Liu, T. Wan, H. Wang, S. Zhang, Y. Ping and Y. Cheng, Sci. Adv., 2019, 5, eaaw8922 CrossRef CAS PubMed.
- J. Yang, Q. Zhang, H. Chang and Y. Cheng, Chem. Rev., 2015, 115, 5274–5300 CrossRef CAS PubMed.
- Y. Zou, X. Sun, Q. Yang, M. Zheng, O. Shimoni, W. Ruan, Y. Wang, D. Zhang, J. Yin, X. Huang, W. Tao, J. B. Park, X.-J. Liang, K. W. Leong and B. Shi, Sci. Adv., 2022, 8, eabm8011 CrossRef CAS PubMed.
- R. Sharma, K. Liaw, A. Sharma, A. Jimenez, M. Chang, S. Salazar, I. Amlani, S. Kannan and R. M. Kannan, J. Controlled Release, 2021, 337, 179–192 CrossRef CAS PubMed.
- F. Perrone, E. F. Craparo, M. Cemazar, U. Kamensek, S. E. Drago, B. Dapas, B. Scaggiante, F. Zanconati, D. Bonazza, M. Grassi, N. Truong, G. Pozzato, R. Farra, G. Cavallaro and G. Grassi, J. Controlled Release, 2021, 330, 1132–1151 CrossRef CAS PubMed.
- R. Zhang, W. Jing, C. Chen, S. Zhang, M. Abdalla, P. Sun, G. Wang, W. You, Z. Yang, J. Zhang, C. Tang, W. Du, Y. Liu, X. Li, J. Liu, X. You, H. Hu, L. Cai, F. Xu, B. Dong, M. Liu, B. Qiang, Y. Sun, G. Yu, J. Wu, K. Zhao and X. Jiang, Adv. Mater., 2022, 34, 2107506 CrossRef CAS PubMed.
- S. Han, Y. Lee and M. Lee, J. Controlled Release, 2021, 338, 22–32 CrossRef CAS PubMed.
- C. Zhuang, C. Piao, M. Kang, J. Oh and M. Lee, Biomater. Sci., 2023, 11, 3354–3364 RSC.
- A. Hasanzadeh, M. R. Hamblin, J. Kiani, H. Noori, J. M. Hardie, M. Karimi and H. Shafiee, Nano Today, 2022, 47, 101665 CrossRef CAS PubMed.
- D. Gong, E. Ben-Akiva, A. Singh, H. Yamagata, S. Est-Witte, J. K. Shade, N. A. Trayanova and J. J. Green, Acta Biomater., 2022, 154, 349–358 CrossRef CAS PubMed.
- K. Leer, L. S. Reichel, J. Kimmig, F. Richter, S. Hoeppener, J. C. Brendel, S. Zechel, U. S. Schubert and A. Traeger, Small, 2024, 2306116 CrossRef CAS PubMed.
- T.-M. Ketola, M. Hanzlíková, A. Urtti, H. Lemmetyinen, M. Yliperttula and E. Vuorimaa, J. Phys. Chem. B, 2011, 115, 1895–1902 CrossRef CAS PubMed.
- I. Lostalé-Seijo and J. Montenegro, Nat. Rev. Chem., 2018, 2, 258–277 CrossRef.
- W. T. Godbey, K. K. Wu and A. G. Mikos, J. Controlled Release, 1999, 60, 149–160 CrossRef CAS PubMed.
- Y. Li, Z. He, A. Sigen, X. Wang, Z. Li, M. Johnson, R. Foley, I. L. Sáez, J. Lyu and W. Wang, ACS Appl. Mater. Interfaces, 2023, 15, 36667–36675 CrossRef CAS PubMed.
- J. Karlsson, S. Y. Tzeng, S. Hemmati, K. M. Luly, O. Choi, Y. Rui, D. R. Wilson, K. L. Kozielski, A. Quiñones-Hinojosa and J. J. Green, Adv. Funct. Mater., 2021, 31, 2009768 CrossRef CAS PubMed.
- X. Xu, W. Yang, Q. Liang, Y. Shi, W. Zhang, X. Wang, F. Meng, Z. Zhong and L. Yin, Nano Res., 2019, 12, 659–667 CrossRef CAS.
- Y. Li, P. Wu, M. Zhu, M. Liang, L. Zhang, Y. Zong and M. Wan, Adv. Healthcare Mater., 2023, 12, 2203082 CrossRef CAS PubMed.
- S. Hernot and A. L. Klibanov, Adv. Drug Delivery Rev., 2008, 60, 1153–1166 CrossRef CAS PubMed.
- X. Zhang, Y. Liu, W. Liu, L. Chen, M. Jin, Z. Gao and W. Huang, Nano Today, 2024, 54, 102068 CrossRef CAS.
- S. Lei, R. Zheng, S. Zhang, S. Wang, R. Chen, K. Sun, H. Zeng, J. Zhou and W. Wei, Cancer Commun., 2021, 41, 1183–1194 CrossRef PubMed.
- C. Denkert, C. Liedtke, A. Tutt and G. von Minckwitz, Lancet, 2017, 389, 2430–2442 CrossRef CAS PubMed.
- H. M. Aliabadi, R. Bahadur, E. Bousoik, R. Hall, A. Barbarino, B. Thapa, M. Coyle, P. Mahdipoor and H. Uludağ, Acta Biomater., 2020, 102, 351–366 CrossRef CAS PubMed.
- S. Y. Neshat, C. H. R. Chan, J. Harris, O. M. Zmily, S. Est-Witte, J. Karlsson, S. R. Shannon, M. Jain, J. C. Doloff, J. J. Green and S. Y. Tzeng, Biomaterials, 2023, 300, 122185 CrossRef CAS PubMed.
- H. Liu, D. Ma, J. Chen, L. Ye, Y. Li, Y. Xie, X. Zhao, H. Zou, X. Chen, J. Pu and P. Liu, Nano Res., 2022, 15, 6306–6314 CrossRef CAS.
- C. Song, M. Zhan, Z. Ouyang, Y. Yao, Y. Gao, M. Shen and X. Shi, J. Controlled Release, 2023, 358, 601–611 CrossRef CAS PubMed.
- P. Golabi, S. Fazel, M. Otgonsuren, M. Sayiner, C. T. Locklear and Z. M. Younossi, Medicine, 2017, 96, e5904 CrossRef PubMed.
- J. D. Yang, P. Hainaut, G. J. Gores, A. Amadou, A. Plymoth and L. R. Roberts, Nat. Rev. Gastroenterol. Hepatol., 2019, 16, 589–604 CrossRef PubMed.
- H. J. Vaughan, C. G. Zamboni, L. F. Hassan, N. P. Radant, D. Jacob, R. C. Mease, I. Minn, S. Y. Tzeng, K. L. Gabrielson, P. Bhardwaj, X. Guo, D. Francisco, M. G. Pomper and J. J. Green, Sci. Adv., 2022, 8, eabo6406 CrossRef CAS PubMed.
- H. Nemati, M.-H. Ghahramani, R. Faridi-Majidi, B. Izadi, G. Bahrami, S.-H. Madani and G. Tavoosidana, J. Controlled Release, 2017, 268, 259–268 CrossRef CAS PubMed.
- I. L. Suzuki, M. M. de Araujo, V. S. Bagnato and M. V. L. B. Bentley, J. Controlled Release, 2021, 338, 316–329 CrossRef CAS PubMed.
- M. E. Jönhagen, A. Nordberg, K. Amberla, L. Bäckman, T. Ebendal, B. Meyerson, L. Olson, Å. Seiger, M. Shigeta, E. Theodorsson, M. Viitanen, B. Winblad and L.-O. Wahlund, Dementia Geriatr. Cognit. Disord., 1998, 9, 246–257 CrossRef PubMed.
- H. Li, Y. Cao, J. Ye, Z. Yang, Q. Chen, X. Liu, B. Zhang, J. Qiao, Q. Tang, H. Yang, J. Li, Z. Shi and Y. Mao, Chem. Eng. J., 2023, 466, 143152 CrossRef CAS.
- S. Camarero-Espinosa, B. Rothen-Rutishauser, E. J. Foster and C. Weder, Biomater. Sci., 2016, 4, 734–767 RSC.
- Z.-J. Zhang, Y.-K. Hou, M.-W. Chen, X.-Z. Yu, S.-Y. Chen, Y.-R. Yue, X.-T. Guo, J.-X. Chen and Q. Zhou, J. Nanobiotechnol., 2023, 21, 18 CrossRef CAS PubMed.
- E. Ren, H. Chen, Z. Qin, S. Guan, L. Jiang, X. Pang, Y. He, Y. Zhang, X. Gao, C. Chu, L. Zheng and G. Liu, Adv. Healthcare Mater., 2022, 11, 2101715 CrossRef CAS PubMed.
- L. Chen, J. Zhang, J. Wang, J. Lin, X. Luo and W. Cui, Adv. Funct. Mater., 2023, 33, 2305635 CrossRef CAS.
- N. Feng and F. Guo, J. Controlled Release, 2020, 325, 380–393 CrossRef CAS PubMed.
- W. K. E. Ip, N. Hoshi, D. S. Shouval, S. Snapper and R. Medzhitov, Science, 2017, 356, 513–519 CrossRef CAS PubMed.
- M. Zoulikha, Q. Xiao, G. F. Boafo, M. A. Sallam, Z. Chen and W. He, Acta Pharm. Sin. B, 2022, 12, 600–620 CrossRef CAS PubMed.
- E. L. Blanchard, D. Vanover, S. S. Bawage, P. M. Tiwari, L. Rotolo, J. Beyersdorf, H. E. Peck, N. C. Bruno, R. Hincapie, F. Michel, J. Murray, H. Sadhwani, B. Vanderheyden, M. G. Finn, M. A. Brinton, E. R. Lafontaine, R. J. Hogan, C. Zurla and P. J. Santangelo, Nat. Biotechnol., 2021, 39, 717–726 CrossRef CAS PubMed.
- L. Rotolo, D. Vanover, N. C. Bruno, H. E. Peck, C. Zurla, J. Murray, R. K. Noel, L. O'Farrell, M. Araínga, N. Orr-Burks, J. Y. Joo, L. C. S. Chaves, Y. Jung, J. Beyersdorf, S. Gumber, R. Guerrero-Ferreira, S. Cornejo, M. Thoresen, A. K. Olivier, K. M. Kuo, J. C. Gumbart, A. R. Woolums, F. Villinger, E. R. Lafontaine, R. J. Hogan, M. G. Finn and P. J. Santangelo, Nat. Mater., 2023, 22, 369–379 CrossRef CAS PubMed.
- Y. Fang, J. Cai, M. Ren, T. Zhong, D. Wang and K. Zhang, ACS Nano, 2023, 18, 592–599 CrossRef PubMed.
- D. Zhang, E. N. Atochina-Vasserman, D. S. Maurya, M. Liu, Q. Xiao, J. Lu, G. Lauri, N. Ona, E. K. Reagan, H. Ni, D. Weissman and V. Percec, J. Am. Chem. Soc., 2021, 143, 17975–17982 CrossRef CAS PubMed.
- C. Ge, J. Yang, S. Duan, Y. Liu, F. Meng and L. Yin, Nano Lett., 2020, 20, 1738–1746 CrossRef CAS PubMed.
- A. Jarzębińska, T. Pasewald, J. Lambrecht, O. Mykhaylyk, L. Kümmerling, P. Beck, G. Hasenpusch, C. Rudolph, C. Plank and C. Dohmen, Angew. Chem., Int. Ed., 2016, 55, 9591–9595 CrossRef PubMed.
- P. S. Kowalski, U. C. Palmiero, Y. Huang, A. Rudra, R. Langer and D. G. Anderson, Adv. Mater., 2018, 30, 1801151 CrossRef PubMed.
- L. Rotolo, D. Vanover, N. C. Bruno, H. E. Peck, C. Zurla, J. Murray, R. K. Noel, L. O'Farrell, M. Araínga, N. Orr-Burks, J. Y. Joo, L. C. S. Chaves, Y. Jung, J. Beyersdorf, S. Gumber, R. Guerrero-Ferreira, S. Cornejo, M. Thoresen, A. K. Olivier, K. M. Kuo, J. C. Gumbart, A. R. Woolums, F. Villinger, E. R. Lafontaine, R. J. Hogan, M. G. Finn and P. J. Santangelo, Nat. Mater., 2023, 22, 369–379 CrossRef CAS PubMed.
- Y. Jiang, Q. Lu, Y. Wang, E. Xu, A. Ho, P. Singh, Y. Wang, Z. Jiang, F. Yang, G. T. Tietjen, P. Cresswell and W. M. Saltzman, Nano Lett., 2020, 20, 1117–1123 CrossRef CAS PubMed.
- M. K. Grun, A. Suberi, K. Shin, T. Lee, V. Gomerdinger, Z. M. Moscato, A. S. Piotrowski-Daspit and W. M. Saltzman, Biomaterials, 2021, 272, 120780 CrossRef CAS PubMed.
- A. Suberi, M. K. Grun, T. Mao, B. Israelow, M. Reschke, J. Grundler, L. Akhtar, T. Lee, K. Shin, A. S. Piotrowski-Daspit, R. J. Homer, A. Iwasaki, H.-W. Suh and W. M. Saltzman, Sci. Transl. Med., 2023, 15, eabq0603 CrossRef CAS PubMed.
- M. García-Díaz, D. Birch, F. Wan and H. M. Nielsen, Adv. Drug Delivery Rev., 2018, 124, 107–124 CrossRef PubMed.
- F. Adams, C. M. Zimmermann, D. Baldassi, T. M. Pehl, P. Weingarten, I. Kachel, M. Kränzlein, D. C. Jürgens, P. Braubach, I. Alexopoulos, M. Wygrecka and O. M. Merkel, Small, 2023, 2308775 CrossRef PubMed.
- G. Conte, G. Costabile, D. Baldassi, V. Rondelli, R. Bassi, D. Colombo, G. Linardos, E. V. Fiscarelli, R. Sorrentino, A. Miro, F. Quaglia, P. Brocca, I. d'Angelo, O. M. Merkel and F. Ungaro, ACS Appl. Mater. Interfaces, 2022, 14, 7565–7578 CrossRef CAS PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Nat. Rev. Drug Discovery, 2009, 8, 129–138 CrossRef CAS PubMed.
- A. Wittrup, A. Ai, X. Liu, P. Hamar, R. Trifonova, K. Charisse, M. Manoharan, T. Kirchhausen and J. Lieberman, Nat. Biotechnol., 2015, 33, 870–876 CrossRef CAS PubMed.
- T. Jeon, D. C. Luther, R. Goswami, C. Bell, H. Nagaraj, Y. A. Cicek, R. Huang, J. A. Mas-Rosario, J. L. Elia, J. Im, Y.-W. Lee, Y. Liu, F. Scaletti, M. E. Farkas, J. Mager and V. M. Rotello, ACS Nano, 2023, 17, 4315–4326 CrossRef CAS PubMed.
- G. Tan, D. Liu, R. Zhu, H. Pan, J. Li and W. Pan, Acta Biomater., 2021, 134, 605–620 CrossRef CAS PubMed.
- S. Guo, C. Li, C. Wang, X. Cao, X. Liu, X.-J. Liang, Y. Huang and Y. Weng, Acta Pharm. Sin. B, 2023, 14, 781–794 CrossRef PubMed.
- J. Huang, C. Zhuang, J. Chen, X. Chen, X. Li, T. Zhang, B. Wang, Q. Feng, X. Zheng, M. Gong, Q. Gong, K. Xiao, K. Luo and W. Li, Adv. Mater., 2022, 34, 2201516 CrossRef CAS PubMed.
- A. V. Anzalone, P. B. Randolph, J. R. Davis, A. A. Sousa, L. W. Koblan, J. M. Levy, P. J. Chen, C. Wilson, G. A. Newby, A. Raguram and D. R. Liu, Nature, 2019, 576, 149–157 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |
