Rational design of polymer-based mRNA delivery systems for cancer treatment
Qianyu
Wan
ab,
Yuji
Sun
a,
Xuanrong
Sun
 *b and
Zhuxian
Zhou
*b and
Zhuxian
Zhou
 *a
*a
aZhejiang Key Laboratory of Smart Biomaterials and Key Laboratory of Biomass Chemical Engineering of Ministry of Education, College of Chemical and Biological Engineering, Zhejiang University, Hangzhou, 310027, China. E-mail: zhouzx@zju.edu.cn
bCollaborative Innovation Center of Yangtze River Delta Region Green Pharmaceuticals and College of Pharmaceutical Science, Zhejiang University of Technology, Hangzhou 310014, China. E-mail: sunxr@zjut.edu.cn
First published on 21st May 2024
Abstract
Messenger RNA (mRNA)-based cancer therapeutics have shown great promise in cancer prevention and treatment, such as mRNA cancer vaccines and protein replacement therapeutics. The key to mRNA cancer therapeutics is to develop safe and effective delivery systems that can efficiently encapsulate mRNA, protect them from degradation, selectively target specific tissues, facilitate cellular uptake and endosomal escape, and ultimately release them into the cytoplasm for protein expression. Polymer-based mRNA delivery systems have received increasing attention for cancer therapy due to their virtues of customizable chemical structures, easy functionalization, and controllable stability, allowing the overcoming of tumor delivery barriers and enhancing therapeutic efficacy. This review introduces the characteristics and applications of mRNA therapeutics, the principles, and classification of polymer-based mRNA delivery systems, and summarizes several advanced mRNA delivery strategies to realize cancer-selective and intracellular delivery, organ-targeted delivery, and tissue-penetrating delivery. The review aims to provide guidance for the design of future polymer-based cancer mRNA delivery systems.
1. Introduction
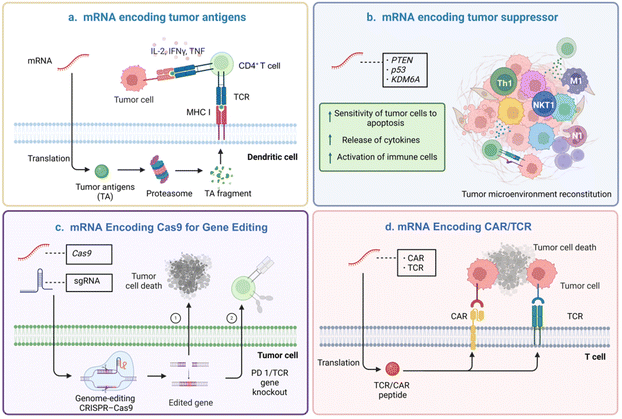
Messenger RNA (mRNA) is a single-stranded ribonucleic acid that is transcribed from a DNA strand, which carries the coding information for protein synthesis and can be further transcribed and processed into functional proteins.1 mRNA is in a special position in the process of information transfer, it expresses the target protein or antigen in the organism according to the central dogma, so that it becomes the “key” to unlock a variety of diseases, thus realizing the purpose of treatment or immunoprophylaxis.2 mRNA therapy has several advantages over traditional therapies. mRNA is safer than DNA drugs as it does not integrate into the host genome.3,4 Secondly, mRNA can produce proteins that are difficult to synthesize in vitro and can be targeted to specific receptors or the circulatory system.5 Moreover, mRNA sequences are highly modifiable and can be rapidly updated and iterated. As a result, mRNA-based cancer therapies have specific applications in cancer prevention and treatment. The common principle of mRNA-based cancer therapies is that mRNA is successfully translated into proteins to inhibit tumor growth or to induce or enhance anti-tumor immune responses. For example, mRNA cancer vaccines can encode specific tumor antigens that activate immune responses. Delivery of essential tumor suppressor mRNA can greatly improve the expression level of tumor suppressor protein and regulate the tumor microenvironment (TME). Encoding Cas 9 with mRNA reduces off-target effects and genotoxicity while encoding chimeric antigen receptor (CAR) or T cell receptor (TAR) with mRNA increases transfection rates and reduces mutation risk (Fig. 1).6–9Nevertheless, the single-stranded structure of the mRNA molecule, which is different from DNA.10–12 Naked mRNA is highly susceptible to destruction by nucleases or hydrolases in blood or body fluids and is rapidly cleared by the kidneys. Unprotected mRNA has been reported to have an extremely short metabolism or systemic half-life of less than 3.8 minutes.13 Second, it is also challenging for mRNA with certain physicochemical characteristics-like being hydrophilic and negatively charged-to get through the cell membrane and enter the cytoplasm. Additionally, mRNA is usually internalized and trapped in acidic endo-lysosomes, where it is degraded by enzymes, restricting its genetic tasks. Therefore, the development of mRNA carriers to get around the challenges above is the key to effective mRNA delivery. Various delivery systems have been developed for mRNA delivery, including viral vectors14,15 and non-viral vectors.16,17 Viral vectors utilize genetic engineering technology to modify viruses as carriers for exogenous gene delivery. Viral vectors mainly consist of retroviruses,18 lentiviruses,19 and adenoviruses,20 which have the advantages of high transfection efficiency and a high expression level of exogenous genes. However, viral vectors have disadvantages of high immunogenicity,21 carcinogenicity risk,22 limited capacity of encapsulated drugs,23 and complicated operation of the preparation process.23 Non-viral mRNA delivery vectors, including lipid-based nanoparticles (NPs),24 polymeric NPs,25 inorganic NPs,26 and biomimetic NPs,27 have been engineered with many favorable properties such as biocompatibility,28 controlled targeting,28,29 low immunogenicity, and high safety.30 However, non-viral mRNA delivery vectors are still limited in gene transfection efficiency compared to the natural viral vectors, holding a great challenge to improve the delivery efficiency.
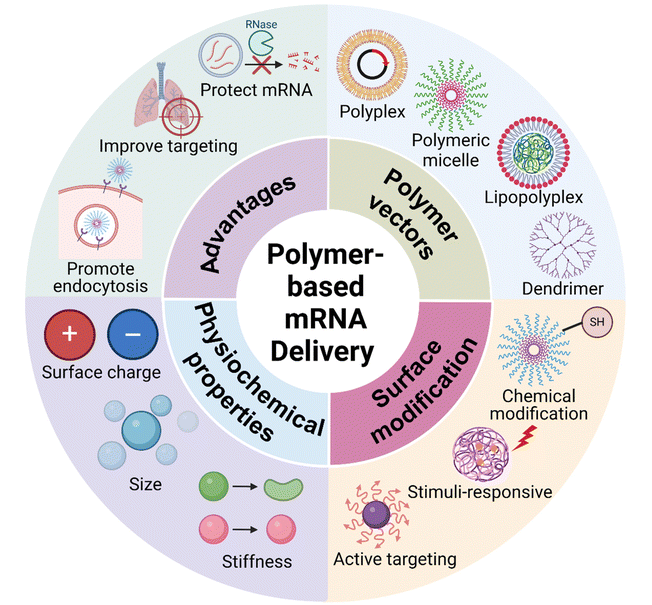
Polymers have been exploited as a major type of non-viral gene delivery carrier, with the merits of simple synthesis, diverse structure, and easy functionalization.31,32 Therefore, they have been widely used for gene delivery of DNA, mRNA, siRNA, etc. (Fig. 2).33–35 Positively charged polymers can electrostatically bind to negatively charged mRNA and form multimeric complexes at physiological pH, protecting the mRNAs from degradation and facilitating intracellular delivery.36,37 In addition, covalent attachment of mRNA to polymers can also be achieved through the use of degradable linkers.38 The systemic delivery of mRNA by polymers involves several steps (Fig. 3): (1) target and accumulate in specific organs and tissues; (2) entry into the cell by endocytosis; (3) endosome escape; (4) intracellular release, and (5) translation into protein.39–41
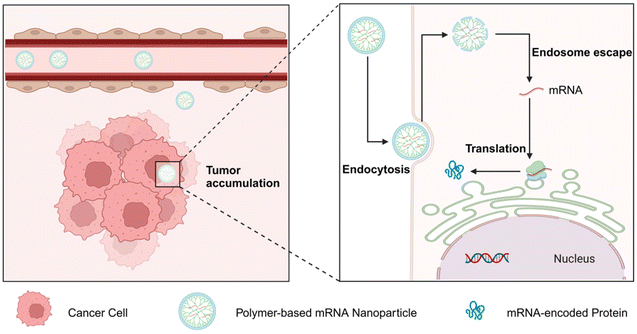 | ||
| Fig. 3 Schematic representation of polymer-based mRNA delivery systems to overcome the systemic biological barriers. | ||
This review summarizes representative non-viral polymeric vectors for mRNA cancer delivery. We highlight the recent advance of polymer-based mRNA delivery systems for enhanced cell entrance and cancer cell-selective delivery, organ-targeted delivery, and tissue-penetrating delivery.
2. Polymer-based nonviral vectors for cancer mRNA delivery
Polymer molecules are usually able to load by electrostatic adsorption with mRNA. An optimal polymer-based mRNA delivery system should: (1) efficiently encapsulate and protect mRNA from nuclease degradation; (2) possess high biocompatibility and safety; (3) promote the penetration and accumulation of specific cells, tissues, and organs; (4) avoid lysosomal degradation in intracellular translocation pathways; (5) enhance the release of mRNA in the cytoplasm to exert the desired effect. In this section, different types of polymer-based delivery of mRNA will be discussed, including polyplexes, lipopolyplexes, etc (Fig. 4, Table 1).| Polymer backbone | Cancer/tissue | Related mRNA | Size (nm) | Zeta potential (mV) | Ref. |
|---|---|---|---|---|---|
| F-PEI | Melanoma | MC38-encoded mRNA | 280.0 | −6.5/−12.3 | 50 |
| β-CD-PEI | — | OVA-encoded mRNA | 234.7 ± 3.5 | 47.3 ± 3.6 | 51 |
| PEG-PEI | L929 fibroblasts (in vitro) | Luciferase (Luc) mRNA | 103.0 ± 7.7 | — | 54 |
| PLGA | p53-deficient hepatocellular carcinoma | p53-encoded mRNA | 105.0 ± 0.1 | −8.1 ± 2.0 | 61 |
| PBAE | Mammary carcinoma | 4-1BB/IL-12-encoded mRNA | 100.0 ± 20.0 | 23.0 ± 2.0 | 65 |
| PBAE | Ovarian carcinoma | IRF5/IKKβ-encoded mRNA | 99.8 ± 24.5 | 3.4 ± 2.2 | 70 |
| APE | — | Luc mRNA | 120.0 ± 30.0 | 5.0 ± 3.0 | 71 |
| PACE | — | Deglycosylation-dependent Renilla Luc/firefly Luc (FLuc)-encoded mRNA | 210.4 | 9.9 | 72 |
| PAsp | — | Cy5-mRNA | 110.0 | 20.0 | 82 |
| PAMAM | — | Cy5-EGFP mRNA | 133.2 | 34.2 | 90 |
| PHTA | Melanoma | mOVA-encoded mRNA | 130.0 ± 6.0 | 25.0 ± 7.0 | 35 |
| Trimannose | Cervical cancer | N1mψ nucleoside modified mRNA | 235.7 ± 4.7 | 46.0 ± 2.1 | 92 |
| Lipid shell-PBAE | Melanoma | OVA-encoded mRNA | 50.0 | — | 93 |
| Lipid shell-dendrimer | Breast carcinoma | Luc mRNA | 138 ± 1.5 | −1.0 ± 0.4 | 94 |
2.1 Polyplex
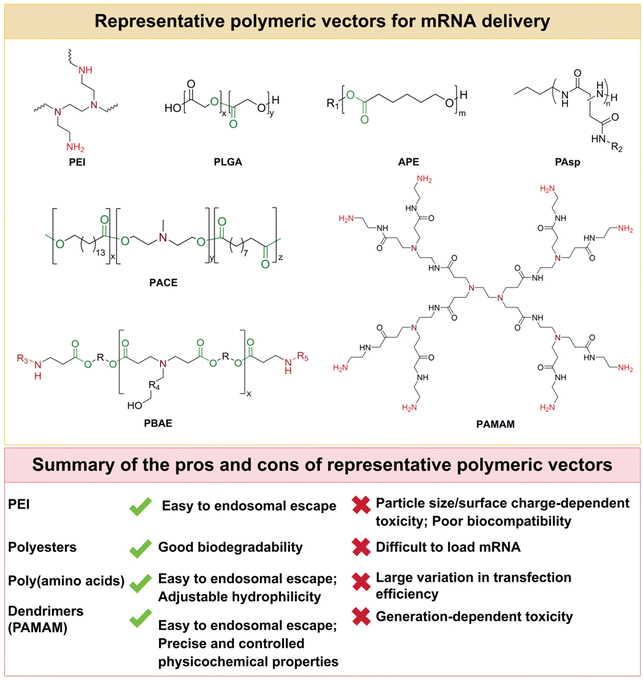
A commonly used vector for delivering nucleic acids is polyplex (i.e. polymeric NP). Polyplex protects nucleic acids from enzymatic degradation and promotes cellular uptake.42 Compared to cationic liposome-based systems, polyplex systems offer a high degree of versatility by controlling molecular weight, structure and composition.43 Commonly used polyplexes as nucleic acid vectors are polyethyleneimine (PEI), polyester, poly(amino acids), etc.The particle size and the surface charge are the main factors affecting the bioactivity and biocompatibility of PEI-based complexes. PEIs with molecular weights between 10 and 30 kDa are severely cytotoxic despite their high mRNA delivery efficiency, whereas PEIs with molecular weights below 1.8 kDa are ineffective, thus limiting their potential for biological applications.48,49 Efforts have been made to improve the delivery efficiency and biocompatibility of PEI. Li et al.50 synthesized fluoroalkane-grafted polyethyleneimine (F-PEI) for mRNA delivery that promoted intracellular delivery of mRNA and activated the Toll-like receptor 4 (TLR4)-mediated signaling pathway. This nanovaccine without additional adjuvants induced DCs maturation, triggered efficient antigen presentation and anti-tumor immune responses, and delayed the growth of established B16-OVA melanomas. Tan et al.51 prepared β-CD-coupled branched PEI (2 kDa) to improve the delivery efficacy of mRNA delivery. This CD-PEI coupler-based mRNA vaccine platform also facilitated the escape of mRNA molecules across the plasma membrane and from endosomes, ensuring high transfection efficiency. Modification of polyethyleneglycol (PEG) to polycations can reduce cytotoxicity and improve the solubility and stability of colloidal complexes.52 PEG fragments like a corona prevent aggregation of the complexes and reduce the adsorption of serum, thus improving the solubility and stability.53 Debus et al.54 first studied the PEG (20 kDa) modified PEI (25 kDa) for mRNA delivery. The results showed PEGylation significantly improved colloidal stability but also reduced transfection efficiency.
Although the stability and delivery efficiency of PEI-based mRNA vectors can be improved after chemical modification, the clinical application is still hindered by their non-degradability and relatively poor biocompatibility. For example, excessive stability of PEI-mRNA complexes would result in limited release of their loaded mRNAs, thereby affecting their translation efficiency. Therefore, it is important to develop polymeric carriers with good biodegradability and biocompatibility for mRNA delivery. In future studies, when considering the use of PEI as an mRNA delivery vector, its dosage can be rationalized to ensure effective cellular uptake and avoid unnecessary cytotoxicity. In addition, changing the molecular weight of PEI is also a feasible strategy to reduce cytotoxicity. Introducing degradable chemical bonds, such as disulfide bonds, into the PEI structure would provide a pathway for mRNA to be degraded more readily, thereby reducing the potential impact on the organism.55 Meanwhile, modification of the surface of PEI, for example, through chemical modification or physical cross-linking, could enhance its affinity for the target cells, thereby improving the efficiency of mRNA delivery.50,51,56
PLGA is a hydrophobic polyester polymerized from lactic and glycolic acid monomers. The biodegradation rate can be controlled by adjusting the ratio of lactic and glycolic acid and the molecular weights. With this property, PLGA has been widely used as a gene carrier materials for disease treatments, including cancer.59,60 Xiao et al.61 reported the combination therapy of a suppressor p53 mRNA-loaded PLGA NP with an immune checkpoint inhibitor (anti-PD-1) for cancer therapy. This strategy induced p53 reprogramming of the TME by NK and CD8 T cell activation, polarized the predominance of tumor-associated macrophages (TAMs) to an anti-tumor phenotype, and inhibited p53-deficient liver tumors growth and metastasis (Fig. 5).
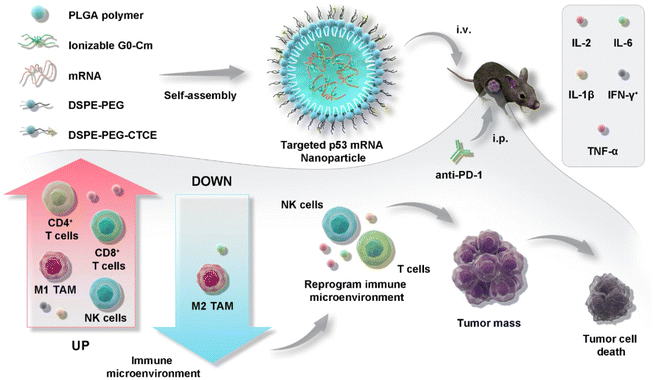 | ||
| Fig. 5 Illustration of PLGA-based nanotherapeutics loaded with p53 mRNA for combination cancer therapy with an anti-PD-1 immune checkpoint inhibitor.61 Copyright 2022, adapted with permission from Springer Nature. | ||
PBAE is a kind of degradable cationic polymer obtained by Michael's addition of primary amine or secondary amine and diacrylate. PBAE can self-assemble in an aqueous solution to form nanoscale discrete structures that enhance the transfection of mRNA.62 The surface charge of PBAE can be altered by changing the side chain and terminal groups to increase the buffering capacity and promote cellular internalization.63,64 PBAE as a vector can be used for reprogramming the TME to enhance cancer immunotherapy. Neshat and colleagues65 used PBAE vectors for the co-delivery of mRNAs encoding immuno-stimulatory cytokine interleukin (IL)-12, the co-stimulatory signaling 2 molecule 4-1BB ligand, and immunostimulatory adjuvants. The flatform induced tumor-associated antigen-presenting cells (tAPCs) and activated cytotoxic T effector cells. As antigen-free cancer immunotherapy, the platform utilizes copolymers to control mRNA release and specifically transfect at TME. This localized treatment dramatically lowers the risk of systemic inflammation and toxicity as compared to prior tumor immunotherapies (Fig. 6). To achieve the anti-tumor effect, TAMs can be converted from the M2 to the M1 phenotype by gene editing,66,67 but it may trigger systemic inflammation due to non-specificity.68,69 To address this challenge, Zhang et al.70 constructed a PBAE-based system carrying in vitro-transcribed mRNA encoded with M1 phenotype transcriptional polarizing factors to reprogram TAMs. The system circumvented the pro-tumor growth and metastatic characteristics of the M2 phenotype and exerted the anti-tumor effects of the M1 phenotype, avoiding systemic toxicity (Fig. 7).
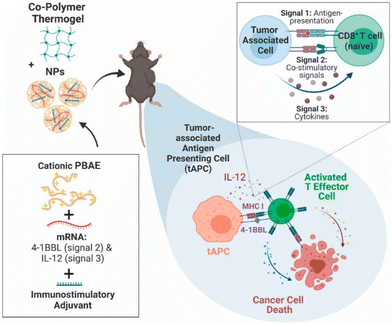 | ||
| Fig. 6 Illustration of PBAE-based delivery system carrying in vitro-transcribed mRNA to reprogram TAMs to an anti-tumor M1 phenotype for cancer immunotherapy.65 Copyright 2023, Elsevier. | ||
 | ||
| Fig. 7 Schematic representation of the anti-tumor effect of macrophages by editing the TAMs gene M2 to M1 phenotype by the PBAE-based mRNA delivery systems.70 Copyright 2019, Springer Nature. | ||
APEs are obtained by ring-opening polymerization of lactones using amino alcohols as initiators. Yan et al.71 synthesized functionalized APE libraries using a thiol-alkene click reaction. To improve the stability of mRNA in serum, the triblock copolymer F127 was added to protect mRNA. The results showed that with an increase in the content of the protective coating, the stability of mRNA was enhanced, but the delivery efficiency was decreased. Therefore, it is necessary to fully balance the relationship between the protective effect and the delivery efficiency when designing the mRNA delivery system.
PACEs are synthesized from three monomers by enzymatic reaction: cationic diol monomer, lactone monomer and diacid monomer. Their roles are to participate in the primary electrostatic interaction with nucleic acid, stabilize the complex through hydrophobic interaction, and form biodegradable ester bonds with the other two monomers.72,73 Endosomal escape is a critical step in the intracellular drug delivery of mRNA.74 To rationally optimize the mRNA transfection efficiency of polymeric material libraries, Jiang et al.72 optimized the mRNA transfection efficiency of PACE material libraries and found that mRNA encapsulation efficiency and endosomal escape are determinants of transfection efficiency rather than uptake.
Polyester has great potential in the field of delivering mRNA due to its good biodegradability and biocompatibility. Polyester with functional groups can deliver mRNA therapeutics specifically into immune cells to induce a strong immune response.75 However, a series of technical challenges need to be overcome to effectively encapsulate mRNA in polyesters. Due to the significant differences between mRNA and polyesters, such as hydrophobicity/hydrophilicity, this leads to repulsion problems when they come into contact with each other. These properties make precise control of polyesters loading on mRNA a complex task. Therefore, specific strategies are needed to address these potential obstacles to ensure that the mRNA can be stabilized and perform its function under appropriate conditions. For example, one potential strategy is to synthesize analogs of specific mRNAs. Such analogs have complementary base pairing with specific mRNA sequences. Such a design not only ensures the structural stability and biological activity of the mRNA, but also enables efficient mRNA loading in polyesters.76
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of PAsp derivatives that can effectively transfect mRNA was around −2.4 at pH 7.3. When the log
P of PAsp derivatives that can effectively transfect mRNA was around −2.4 at pH 7.3. When the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P was greater than −2.4, the mRNA expression was 1000-fold different from that when it was less than −2.4. When the log
P was greater than −2.4, the mRNA expression was 1000-fold different from that when it was less than −2.4. When the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of PAsp derivatives is between −1.8 and −1.3, it can be realized that mRNA reaches the lungs preferentially after systemic administration.
P of PAsp derivatives is between −1.8 and −1.3, it can be realized that mRNA reaches the lungs preferentially after systemic administration.
Lipid-based mRNA delivery systems have been reported to elicit strong cytokine responses upon systemic administration, which may be responsible for adverse reactions leading to autoimmune lesions or systemic inflammatory infections.91 In addition to the types of polymers described in the previous section, several polymeric vectors have been explored to address the problems of lipid-based mRNA delivery systems. Huang et al.35 developed a delivery system for amphiphilic alternating poly(ortho-hydroxy tertiary amine) (PHTA) copolymers encapsulating mRNA encoding ovalbumin (mOVA). The abundant hydroxyl groups in the copolymers' backbone can chelate with metal ions and inhibit the production of reactive oxygen species (ROS) associated with inflammation, which makes this delivery system delivered in vivo without inflammatory side effects. Meanwhile, the vector successfully delivered mRNA cancer vaccines, which triggered strong T-cell-mediated anti-tumor cellular immunity, providing a potential approach for establishing potent mRNA cancer vaccines with a favorable inflammatory safety profile (Fig. 8).
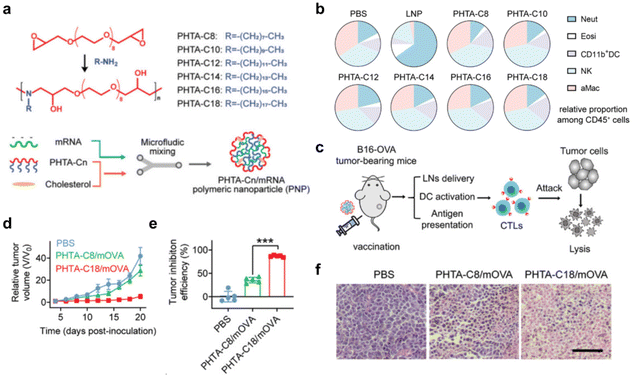 | ||
| Fig. 8 (a) Synthesis of the alternating copolymer PHTA and schematic construction of an integrated polymer vaccine. (b) Percentage of neutrophils (Neut), eosinophils (Eosi), CD11b DCs, NK and alveolar macrophages (aMac) within CD45 cells in lung tissue. (c) Schematic representation of the therapeutic mechanism of this polymeric mRNA cancer vaccine. (d) Mean tumor growth curves of mice receiving the indicated treatments. (e) Tumor suppression efficiency at endpoint compared to PBS group. (f) Hematoxylin and eosin (H&E) stained images of tumor tissues after B16-OVA tumor-bearing mice received PBS, PHTA-C8/mOVA and PHTA-C18/mOVA, respectively. Scale bar: 100 μm.35 Copyright 2022, Wiley. | ||
2.2 Lipopolyplex
Lipopolyplex is a bilayer-structured NP with a polymer-encapsulated mRNA as the core and phospholipid as the shell. When it comes to reducing inflammation in lipid-based delivery systems to improve safety, lipopolyplex can be an effective alternative to lipids as they combine the advantages of increased endocytosis efficiency and reduced cytotoxicity.92,93 Kris Thielemans' group92 studied a lipopolyplex-based mRNA delivery system. The strategy utilizes naturally occurring nucleoside (e.g., N1-methylpseudouridine (N1mψ)) modifications to reduce innate sensing of mRNAs, thereby inhibiting inflammatory cytokine responses. When administered systemically, this lipopolyplex platform demonstrated good hemocompatibility and mostly limited mRNA expression in splenic antigen-presenting cells (APCs). Compared with electroporation and lipid-based delivery systems, immunization with lipopolyplex elicited robust T-cell immunity and showed greater efficacy in modulating tumorigenesis. Persano et al.93 encapsulated mOVA in PBAEs' core, which was packaged in a lipid bilayer shell structure. These core–shell NPs were taken up by DCs through macropinocytosis and effectively stimulated the expression of interferon-β and IL-12 in DCs. In treating lung metastatic B16-OVA melanoma, it significantly reduced tumor nodules by more than 90%. This core–shell structure provides a potential platform for mRNA vaccine delivery. Lipopolyplexes-based mRNA delivery systems have made significant advances in cancer therapy. Moreover, platforms applied to tumor detection and imaging are also particularly important when diagnosing cancer. Therefore, developing mRNA delivery systems that can both image tumors and efficiently produce functional proteins is a challenging task. Xiong et al.94 developed a pH-responsive lipopolyplex-based mRNA delivery system. The system contains PEGylated BODIPY dyes that were used for non-invasive near-infrared (NIR) imaging. The study found that the length of PEG modified in the lipid shell has an effect on mRNA delivery. When PEG length is between 1000–5000 g mol−1, lipopolypexes are protected from aggregation and non-specific cellular uptake. The study also demonstrated a relationship between mRNA expression intensity in vivo and pKa at lipopolyplex. When the pKa is about 6.3, mRNA can usually produce more protein in the liver. The optimal lipopolyplex obtained could successfully mediate mRNA expression in tumors by pH-responsive NIR imaging while illuminating the tumor. The platform is expected to become a suitable method for simultaneous diagnosis and treatment of cancer.3. Strategies for advanced mRNA delivery
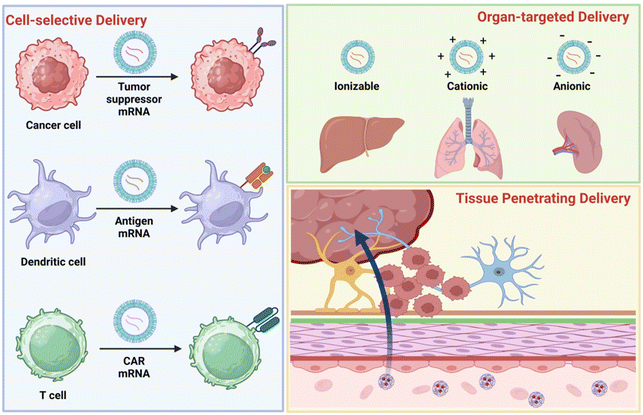
The key to effective mRNA therapy is efficiently delivering mRNA to the target site to produce enough aimed proteins. The off-target of the mRNA will decrease the therapeutic efficacy and increase the toxicity to normal tissues. Therefore, when rationally designing the delivery system to protect mRNA, it is also necessary to consider whether cell-selectivity, organ-selectivity, and penetration of difficult-to-permeate tissues can be achieved during its in vivo delivery. Targeting strategies for polymer-based mRNA delivery systems include passive targeting, endogenous targeting, and active targeting.95 In cell-selective delivery, surface modification is a commonly used method to confer targeting capabilities to delivery systems. In particular, antibodies, peptides, or other molecules (e.g., ligands) are conjugated to polymeric vectors to achieve selective delivery to cancer cells, DCs, and T cells.96 In organ-targeted delivery, the charge of the delivery system plays a key role. Serum proteins recognize NPs with different charges and adsorb them to the surface. This surface-adsorbed protein interacts with homologous receptors in the target organ to facilitate organ-targeted delivery of mRNA. For example, ionizable NPs can be targeted to the liver, cationic NPs to the lungs, and anionic NPs to the spleen. In hard-to-penetrate tissues, the special physicochemical properties of NPs can interact with biological barriers. Because of the interaction, they usually show tissue enrichment and a strong ability to penetrate (Fig. 9). This section will discuss advanced strategies for cell-selective, organ-targeted, and tissue-penetrating polymer-based mRNA delivery systems.3.1 Cancer-selective and intracellular delivery
One of the great challenges in cancer mRNA therapy is the effective targeted delivery of therapeutic genes into cancer cells. Moreover, the mRNA vectors must be able to cross the cell membrane and internalize into the cytoplasm for translation.97 Nanocarriers are modified with tumor-homing ligands to realize cancer-selective delivery and improve the internalization rate.98,99 For example, Chen et al.100 developed a cyclic Arg-Gly-Asp peptide (cRGD)-modified GFP mRNA polymeric micelle. This system utilizes the ability of cRGD to specifically bind to αVβ3 and αVβ5 integrins overexpressed on the membranes of tumor vascular cells and tumor cells. The cRGD-modified polymeric vectors exhibited a 10-fold increase in tumor accumulation and GFP protein expression in the tumor compared to those without the cRGD ligands. The intracellular delivery of the gene vectors can be enhanced by surface chemical modification.APCs process these neoantigens and present them to effector T cells for triggering and activation.101,102 Among them, DCs are the APCs with the strongest antigen-presenting ability, which can take up and process antigens such as tumors or pathogens and deliver them to T cells.102 Fornaguera et al.103 prepared an oligopeptide-end modified PBAEs-based mRNA delivery system. This system utilizes cellular phagocytosis and endocytosis to selectively deliver NPs to splenic DCs. Jordan J. Green's group104 demonstrated that the addition of lipophilic subunits to the PBAE backbone is critical for targeted delivery to DCs. A rational explanation may be that a multitude of innate immune receptors have developed the ability to identify the hydrophobic segment of the molecule. Particles with more hydrophobic surfaces may increase DCs uptake into cells due to interactions with the receptor. Because of this interaction, it allows for cellular uptake and transfection of NPs without PEGylation or targeted ligand modification (Fig. 10).
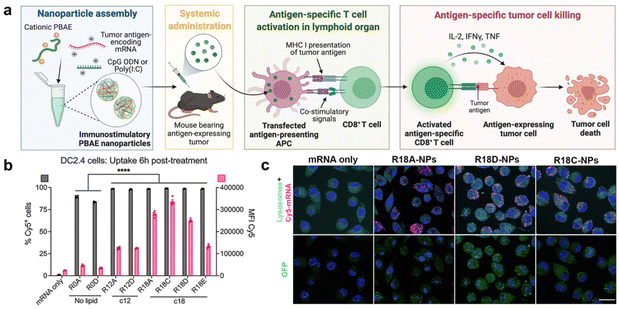 | ||
| Fig. 10 (a) Schematic diagram of a biodegradable PBAE vector system targeted for delivery to splenic DCs. (b) PBAE NPs containing lipophilic side chains exhibit more efficient cellular uptake. (c) In order to observe cellular uptake, NPs' colocalization with endosomes/lysosomes, and GFP transfection, representative pictures of DC2.4 cells labeled with lysosome/endosome dye were taken six hours after treatment with NPs expressing Cy5-labeled GFP-encoding mRNA (scale bars: 20 μm).104 Copyright 2023, National Academy of Sciences. | ||
T cells exert their immune function through lymphatic vessels and blood circulation,105 and their differentiation checkpoints are important for improving cancer immunotherapy.106 However, in the treatment of solid tumors, most T cells cannot effectively enter and activate in tumor tissues.107 Therefore, how to deliver mRNA targeting to T cells and promote their entry into tumors is an urgent problem. To optimize gene delivery for effective regulation of T cells, various cellular immunotherapies have been proposed, such as chimeric antigen receptor T cell (CART) immunotherapy,108 but there are still potential side effects such as neurological toxicity and cytokine storm.109,110 To circumvent the above problems, Paul A. Wender's group111 has developed a new CART-mRNA therapy, namely, charge-altering releasable transporters based on β-amido carbonate (bAC-CARTs). The effects of polymer backbone and side chain spacing on cellular endocytosis have received little attention in previous studies of CARTs. This system used a polymeric backbone to increase the spacing and mobility of lipids, which allowed for better fusion with the cell membrane. The bAC-CARTs convert the initial polycationic CARTs into neutral intermediates, thus enabling lysosomal escape, release, and subsequent translation of the loaded mRNA. In addition, polymer side chain spacing may also affect particle organizing, viscosity, and ion exchange, thus affecting mRNA release. The results showed that the system had 97% spleen targeting. In contrast to previous studies with CARTs, it allows efficient transfection of splenic T cells without targeted ligands and does not cause adverse inflammatory responses.
Notably, due to the sieving effect of biological filtration systems, most NPs are usually engineered to be of the desired size, resulting in a more pronounced accumulation in the corresponding organ.112,113 However, they are not targeted for delivery to the corresponding cell populations. Therefore, in addition to focusing on the biodistribution of NPs in organs, but the fate of NPs in different cells within the organ needs to be given more attention. In conclusion, the precise delivery of mRNA to target cells can undoubtedly significantly enhance drug delivery and effectively reduce potential side effects caused by off-target effects. However, the realization of such a targeting strategy is an extremely complex and challenging process. It involves an in-depth understanding of the structure and function of different cells, as well as the specific mechanism of endocytosis of NPs by the cell membrane.114,115 In addition, multiple intracellular biological barriers such as endosome escape, mRNA stability, and translation efficiency need to be overcome, all of which may affect delivery efficiency and safety.116,117 Therefore, despite the potential advantages of cellular-level targeting, the road to its implementation is still full of unknowns and challenges.
3.2 Organ-targeted mRNA delivery
The systemic delivery of mRNA to tumors remains a challenge that has not yet been fully resolved. Various polymer-based delivery systems have been designed to deliver mRNA to organs. These polymers may contain many of the accessory lipids present in normal lipid NPs for mRNA delivery, and they frequently exhibit comparable properties to lipid systems.118 After systemic delivery, polymeric NPs can cause mRNA expression in different organs. Furthermore, the mechanisms of polymeric NPs targeting to organs are not well defined. In general, variations in formulation composition, such as regulating the internal and/or external charge of the NPs, will affect their expression levels in different organs.119The delivery of circulating NPs to the liver is mediated by adsorption to the surface of soluble apolipoprotein E (ApoE), so that NPs encapsulating mRNAs reach the liver preferentially.120,121 An especially useful “molecular sieve” is the endothelium of the hepatic sinusoids, which allows NPs smaller than 150 nm to freely enter the Disse space where hepatocytes and hepatic stellate cells are located.122,123 In contrast, relatively large NPs are taken up by Kupffer cells surrounding the hepatic sinusoids. However, delivery to the liver through the size of NPs alone is not enough, as the precision of delivery to different cells, tissues, and cancer types in the liver remains to be improved. Using hepatic tumor-specific peptide-modified polymeric vectors is an important strategy to achieve liver-targeted delivery.124,125 Lei et al.126 modified polymer micelles with the hepatocellular carcinoma-specific peptide HCC 167. The HCC 167-modified micelles had more substantial and faster cellular uptake than unmodified micelles. This system demonstrates that HCC 167 can specifically recognize the alkaline phosphatase placental-like 2 receptors on HepG2 cell membranes, forming a high-affinity receptor–ligand complex for liver targeting.
After systemic delivery, the preferential arrival of NPs to the liver can result in excessive homing effects in the liver. Therefore, organ-targeted delivery strategies are particularly important for optimal efficacy and therapeutic precision. Daniel J. Siegwart's group127,128 proposed a selective organ targeting (SORT) strategy. By rationally designing NPs, different ratios of variously charged lipids (called “SORT molecules”) were added to the vector formulation. The results showed that mRNA-loaded vectors with different charges were reliably and precisely delivered to the lungs, spleen, and liver of mice, respectively. Additionally, Zhang et al.129 created a single-component, multifunctional ionizable amphiphilic Janus dendrimer (IAJD)-based mRNA delivery system. The fundamental structure of the hydrophobic region of the IAJD was varied in this by altering the alkyl lengths, and this contributed to a 90.2-fold improvement in the activity of mRNA targeting delivery to the liver, lungs, lymph nodes, and spleen. This result showed that asymmetry in the primary structure of the hydrophobic portion of the vector plays a vital role in organ-target delivery.
Negatively charged polymers and rational modification of polymer side chains enable targeted delivery of mRNA to the spleen.130 However, negatively charged polymers alone are difficult to electrostatically adsorb and successfully encapsulate with similarly negatively charged mRNAs, resulting in inefficient preparation and delivery. To address this problem, strategies of zwitterion and charge reversal have emerged. Liu et al.36 reported a cationic polymer zwitterionic phosphorylation modification strategy. This modification strategy produced a serum-stable amphiphilic structure for polyplexes and introduced hydrophobic alkyl chains to help polyplexes fuse with endosomal membranes. The incorporation of negative phosphate groups into the system effectively enhanced the ability of polyplexes to deliver mRNA targeting the spleen and lymph nodes. Compared with the unmodified cationic polyplexes, the optimal system obtained delivered mRNA with a 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500-fold increase in protein expression (Fig. 11). Shen's group10 presents an esterase-triggered deionization strategy for the design of quaternium lipid-like molecular systems. The system internalizes into cells via macropinocytosis and the clathrin-related pathway. Since the spleen contains appropriate levels of esterase, the system remained stable during circulation and was released in the spleen, allowing for spleen-specific transfection. The system encapsulating mRNA with tumor antigens induced antigen presentation and immune response in APCs and demonstrated effective treatment of melanoma.
500-fold increase in protein expression (Fig. 11). Shen's group10 presents an esterase-triggered deionization strategy for the design of quaternium lipid-like molecular systems. The system internalizes into cells via macropinocytosis and the clathrin-related pathway. Since the spleen contains appropriate levels of esterase, the system remained stable during circulation and was released in the spleen, allowing for spleen-specific transfection. The system encapsulating mRNA with tumor antigens induced antigen presentation and immune response in APCs and demonstrated effective treatment of melanoma.
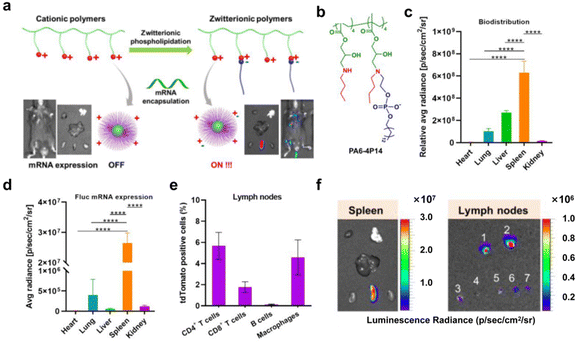 | ||
| Fig. 11 (a) Schematic representation of phospholipidation action converting cationic polymers into zwitterionic mRNA carriers that are selectively expressed in spleen and lymph nodes. (b) Chemical structures of representative ZPP polymers at physiological pH. (c) Biodistribution quantification of representative Cy5-mRNA polyplexes (Cy5-mRNA dose: 0.25 mg kg−1). (d) Representative Cy5-mRNA polyplexes mediate the quantification of fLuc mRNA expression, mostly in the spleen. (e) Quantification of CD4 T cells, CD8 T cells, B cells and macrophages by flow cytometry of lymph node cells 48 hours after intravenous injection. (f) Representative in vitro and in vivo organ images of protein expression after delivery of mRNA polyplexes (Fluc mRNA dose: 0.25 mg kg−1).36 Copyright 2021, adapted with permission from the American Chemical Society. | ||
After systemic delivery, mRNA accumulates mainly in the liver and spleen. To realize the delivery of mRNA to extrahepatic organs, further studies on the potential targeting delivery mechanisms and methodologies are needed. Qiu et al.131 suggested that it is precisely because NPs selectively adsorb specific plasma proteins upon administration that these surface proteins can act as ligands to target organs. The route of administration plays an important role in facilitating the targeting of NPs to organs.132 Therefore, combining particular administration and polymer design will facilitate mRNA organ-targeted delivery. Tang et al.133 reported an innovative design of dual-targeted mRNA nanoformulations based on PBAE and hyaluronic acid. The formulation has ideal stability and efficiently transfects target proteins into lung tissues by inhalation. More importantly, the optimized dual-targeted mRNA-NPs efficiently accumulate in lung tumor cells and inflammatory macrophages, expressing desired proteins like the p53 tumor suppressor, for therapeutic use and effective lung tissue transfection. Similarly, Philip Santangelo's group134 evaluated a PBAE-based NP P76, which efficiently delivered various mRNAs to the lungs of different animals via nebulized inhalation. The dose of this delivery system was greatly reduced compared to previously reported PBAE. The results showed that the delivery system was safe, well tolerated, and had high protein expression after mRNA transfection, providing an idea for inhalable nucleic acid therapy (Fig. 12).
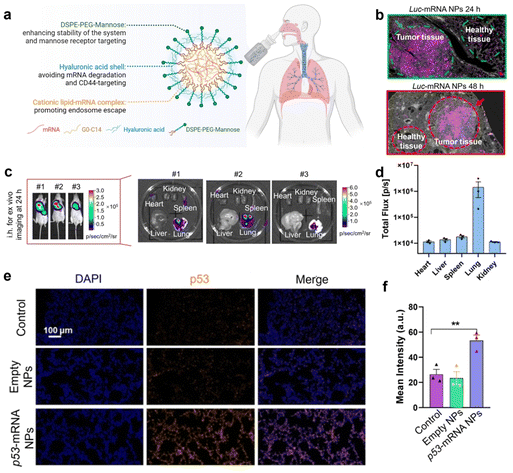 | ||
| Fig. 12 (a) Schematic representation of dual-targeted mRNA nanoformulations based on PBAE and hyaluronic acid. (b) Immunofluorescence staining of lung tissue was performed to assess the location of target protein expression. Luc proteins are indicated in pink. (c) The ex vivo bioluminescence images of different major organs after 24 h of inhalation of the nanoformulations. (d) Quantitative analysis of the luminous flux emitted by different organs. (e) Fluorescence images of mouse lung tissue sections after inhalation of empty nanoformulations or loaded p53-mRNA nanoformulations (nuclei, blue; p53 protein, orange). (f) Quantitative analysis of p53 signaling in lung tissue sections after inhalation of p53-loaded mRNA nanoformulations.133 Copyright 2023, National Academy of Sciences. | ||
3.3 Tissue penetrating mRNA delivery
NPs-based drug delivery systems usually need to take the CAPIR five cascade steps (i.e. Circulation, Accumulation, Penetration, Internalization, Release).135,136 Analysis of the in vivo delivery process of NPs reveals that various biological barriers in organisms often hinder the penetration of NPs. At the same time, the delivery of NPs in solid tumors often suffers from low penetration efficiency. Therefore, further improving the tissue penetration ability of NPs and enhancing their therapeutic efficacy are urgent problems for anti-tumor nanomedicines.The biological barrier of TME is a big obstacle to the penetration of anti-cancer nanomedicine, which mainly occurs in the following situations: (1) vascular abnormality of solid tumors; (2) lymphatic vessel abnormality of solid tumors; and (3) dense extracellular matrix of solid tumors, which greatly impede the penetration of anticancer nanomedicine into the depths of the tumor tissues, and make it difficult for nanomedicine to exert effective anti-cancer effects. The more studied strategies to promote tumor penetration of anti-cancer nanomedicines usually employ regulation of TME and optimization of the physical properties of NPs.137–141 RNA or RNA-related liquid–liquid phase separation methods may be used to provide novel approaches to regulating TME.142 Xing et al.143 introduced cationic polymers into living cells, which were then combined with negatively charged TGF-β1 mRNA, and liquid–liquid phase separation occurred, preventing the translation of TGF-β1 mRNA. As a result, the immunosuppressive ability of tumor cells in TME was reduced, triggering significant anti-tumor responses and improving the efficiency of tumor immunotherapy. In recent years, many advanced strategies to optimize the physicochemical properties of NPs for enhancing tumor penetration have been proposed, such as the design of nano-based delivery systems with mucosal adhesion.144,145 Functional groups, such as amine and sulfydryl groups, can effectively prolong the residence time in tumor-containing organs, thereby promoting the uptake and penetration of NPs into tumor tissues. Kong et al.146 modified the PLGA vector with sulfydryl group to form disulfide bonds with cysteines in bladder tissue mucus glycoproteins, conferring mucosal adhesion properties to this system. This property prolonged the exposure of mRNA encoding the tumor suppressor KDM6A to bladder tumors. The system was internalized into bladder cancer cells via macropinocytosis, effectively inhibiting bladder tumor growth and metastasis.
Transcytosis, being an inherent cellular function, has garnered interest as a possible remedy for the drawbacks related to passive delivery. Transcytosis is a transcellular transport of molecules facilitated by electrostatic adsorption or receptor-mediated endocytosis. It has recently been recognized that the active transport of NPs into solid tumors through transcytosis is an effective way to overcome the biological barriers of the blood vessel wall and tumor extracellular matrix.147–149 Various transcytosis-based nanocarriers have been developed for small-molecule and biomacromolecular drug delivery.150–152 The transcytosis-based strategy may be a promising way for mRNA delivery.
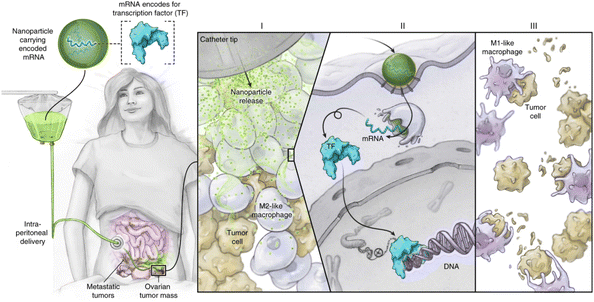
Nanomedicines also need to penetrate various biological barriers in the body during delivery, the most challenging is the brain blood barrier (BBB), as the tight junctions between cells strictly control substance transportation.153 Transcytosis-based gene delivery systems have been developed to overcome BBB. Following the identification of the BBB's increased transport mechanism, a thorough investigation into the potential applications of this route in brain tumor treatment has commenced.154 Liu et al.13 proposed a PEI-poly-L-lysine (PLL)-erythrocyte membrane-based PTEN mRNA delivery system. This system specifically bound the low-density lipoprotein receptor family overexpressed in BBB endothelial and brain tumor cells, utilizing receptor-mediated transcytosis for BBB penetration. The erythrocyte surface was doped with ApoE peptides, which further enhanced BBB penetration and GBM cell targeting. Notably, this system is the first polymer-based mRNA delivery system to overcome the BBB and achieve brain tumor cell targeting (Fig. 13). Kumthekar et al.155 developed a gold NP-based spherical nucleic acid (SNA) delivery system to treat GBM. It was shown that class A scavenger receptors on BBB endothelial cells facilitate the recognition of siRNA oligonucleotide corona in this system. Subsequently, the SNA penetrates the BBB and the tumor via transcytosis. After the SNA enters the cancer cells, the expression of oncogenes is suppressed and triggers cancer cell death. This study marks the first nanomedicine that penetrated the BBB by intravenous injection and entered the Phase 0 clinical study. However, achieving drug delivery across the BBB remains a major challenge, and there are few reports of polymer-delivered mRNAs in this area. The delivery design strategies described above can serve as references for future polymer research. In addition, we can also consider coating the polymer surface with peptides that facilitate BBB penetration or connecting neurotransmitters to act as “guides” when penetrating the BBB.156,157
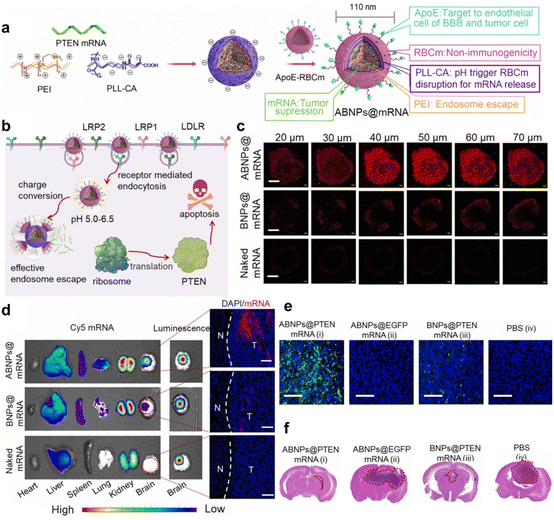 | ||
| Fig. 13 (a) Schematic diagram of PEI-PLL-erythrocyte membrane-encapsulated mRNA for the preparation of functionalized biomimetic NPs. (b) Mechanisms of transfection and antitumor induction by BBB-overexpressed receptor-mediated transcytosis at low pH endosomal escape and mRNA release. (c) Penetration of this polymer mimetic NPs in 3D U87MG tumor spheroids. (d) Biodistribution of major organs 4 h after intravenous injection of polymer-mimicking NPs-mRNA or naked mRNA. (e) TUNEL staining of brain tissue from mice on day 20. (f) H&E staining of brain tissue from mice on day 20.13 Copyright 2023, adapted with permission from Elsevier. | ||
Many biological barriers in the body cause the efficacy of mRNA vaccines to be affected by the site and route of administration.158 Therefore, some strategies administer therapeutic vaccines directly at or near the disease site.146,159 Intranasal delivery is widely used in treating brain and lung tumors due to its specific spatial targeting effect.132,160,161 However, a major problem in developing intranasal mRNA delivery agents is effectively penetrating antigens into the nasal mucosal layer.162 Jeong et al.163 used photochemical internalization (PCI) technology to generate ROS at specific wavelengths of light, which destabilizes endosomal membranes along with cationic polymers. The PCI technology permits polymeric NPs to penetrate the nasal mucosal layer more efficiently and deliver the mRNA to the lungs.
In addition to delivering mRNA in vector-encapsulated form, subcutaneous delivery of naked mRNA is considered more efficient.164–166 However, naked mRNA is very susceptible to enzymatic degradation during delivery. In addition, the negative charge of the mRNA makes it difficult to enter the cell as it experiences electrostatic repulsion at the cell membrane. Dissolving microneedles are often used as a transdermal medication delivery platform. The platform is typically made of excipients and water-soluble polymers, allowing mRNA to penetrate the skin's viable layer, break down, and be released.167,168 Koh et al.168 demonstrated for the first time that low molecular weight polyvinylpyrrolidone (PVP) is used for the fabrication of solubilized microneedle RNApatch and maintains sufficient stability of the mRNA, with its transfection efficiency increasing with the depth of delivery. The microneedles, when applied to the E.G7-OVA immunotherapy model, retarded tumor growth more effectively than subcutaneous injection, induced equivalent prophylactic cellular and humoral immunity, and showed higher mean antibody titers.
Although some strategies to optimize penetration have been reported in recent years, their efficacy remains to be improved due to the specificity of TME and the biological barrier in vivo (Table 2). Future studies could focus on the nature of TME and the depth of penetration of NPs across biological barriers. Bioengineering strategies can be considered to optimize the design of polymer-based mRNA delivery systems, enhance penetration of the biological barrier, and improve tissue targeting.
| Advanced delivery | Specific site of action | Backbone type | Targeting strategies | Ref. |
|---|---|---|---|---|
| Cancer-selective and intracellular delivery | U87 cells | cRGD-PEG-PLL-PNIPAM | Receptor specific recognition | 100 |
| APCs | PBAE | Oligopeptide mRNA terminal modification | 103 | |
| Splenic DCs | PBAE | Adjuvant co-delivery | 104 | |
| Splenic T cells | bAC-CART | Chemical modification | 111 | |
| Organ-targeted mRNA delivery | Liver | DMP | Receptor specific recognition | 126 |
| Lungs, spleen, and liver | LNP | SORT strategy | 127 and 128 | |
| Lungs, spleen, and liver | IAJD | Primary structure of the hydrophobic fraction | 129 | |
| Spleen and lymph nodes | ZPP | Charge reversal | 36 | |
| Spleen | Quaternium lipid-like molecule | Charge reversal | 10 | |
| Lung | PBAE-PEG | Administration by inhalation | 133 | |
| Lung | PBAE | Administration by inhalation | 134 | |
| Tissue penetrating mRNA delivery | TME | PEI | Intracellular mRNA phase separation | 143 |
| TME | PLGA | Chemical modification | 146 | |
| BBB | PEI-PLL | Receptor-mediated transcytosis | 13 | |
| BBB | Gold NPs | Receptor-mediated transcytosis | 155 | |
| Nasal cavity mucous membrane layer | PAsp | PCI technology | 163 | |
| Skin barrier | PVP | Dissolvable microneedle delivery platform | 168 |
4. Perspective
Cancer is one of the leading causes of mortality around the globe. Cancer-related fatalities are anticipated to increase to 13.1 million by 2030.169 Despite the great efforts made to overcome cancer, the results so far have been unsatisfactory. Traditional cancer treatments include surgical resection,170 radiotherapy and chemotherapy.171,172 However, existing clinical treatments are unable to effectively deal with metastasis and recurrence of tumors. Treatments that can completely reverse and inhibit the growth of tumors have yet to be developed. Compared with traditional vaccine approaches, mRNA vaccine technology has many advantages:26,173 (1) shorter development cycle. (2) Dual immunization mechanism with inherent immunostimulatory properties and adjuvant action. (3) Higher safety. Most existing cutting-edge research on mRNAs is based on lipid-based delivery systems. Several liposomes have entered clinical trials of mRNA vaccines for the treatment of cancer, such as NCT02410733 and NCT04534205, etc.174–176 Patients currently receiving these therapies do show encouraging clinical efficacy in studies, but many clinical trials of mRNA cancer vaccines are still in the early stages of research. Polymer-based mRNA delivery systems have yet to be utilized on a large scale in clinical treatments. However, the system is characterized by simple synthesis, precise and controllable physicochemical properties, and penetration of biological barriers. Due to these advantages, polymer-based mRNA delivery systems have a non-negligible potential in mRNA cancer therapy.Nevertheless, the application of polymer-based mRNA delivery systems to the treatment of cancer is still full of challenges. Within this field, researchers are continuously working to develop systems that can effectively deliver mRNA to tumor cells. These systems must comprehensively consider the properties of polymeric NPs, including the composition, size, stiffness, and surface charge.112,127,177 These properties may affect the physiological state and the immune system. In addition, due to the tunable and modifiable nature of polymers, the rational design of polymer-based mRNA delivery systems should emphasize precise delivery and efficient biological barrier penetration. Future research on mRNA delivery platforms will continue to focus on improving efficiency and minimizing side effects for highly targeted and safe delivery. In order to achieve effective mRNA delivery, these properties need to be finely regulated. These regulations should ensure that the mRNA can safely penetrate the biological barrier and ultimately reach the inside of the tumor cell to fulfill its therapeutic role. The designing and in-depth understanding of NP properties is critical, as they directly impact mRNA delivery efficiency and safety.
Meanwhile, designing polymeric NPs that can successfully penetrate physiological barriers is essential, but addressing the associated challenges cannot be ignored. For example, the understanding of the interactions between polymeric NPs and complex TMEs remains incomplete.178 Since tumor tissue is a highly complex and heterogeneous microenvironment characterized by hypoxia, abnormal vascular system, dense extracellular matrix, overexpression of immunosuppressive proteins and abnormal metabolic regulation.179 Therefore a deeper understanding of the behavior of polymeric NPs in complex physiological barriers could provide potential strategies to improve their mRNA delivery efficiency.
In cancer treatment, precision and personalized therapies have been important trends in medicine in recent years. This brings more efficient and targeted treatment options for cancer patients.180 With the rapid advancement of genome sequencing technology and the cross-application of big data science, the concepts of precision medicine and personalized treatment are gradually gaining popularity.181,182 The core of precision medicine is the “three right things”: the right patient, the right dose, and the right drug. This means that treatment needs to be patient-specific, using the right dose of the right drug. Through genetic sequencing, it is possible to find the target of a cancer patient's genetic mutation and then use targeted drugs for precise treatment. Several studies have been reported on the use of nanomaterials for precision medicine.183–185 For example, Siemer et al.185 used computational modeling and sequencing techniques to determine the mechanism of cisplatin resistance in head and neck cancer tumor cells. Poly(sarcosine)-based polymeric NPs loaded with cisplatin were constructed for the purpose of overcoming drug resistance. In the future, mRNA delivery technologies can be considered to provide strong support for personalized treatment options. In particular, polymer-based mRNA delivery systems can precisely deliver specific mRNA molecules into tumor cells, thus enabling targeted therapy against specific gene mutations. This treatment modality has the potential to significantly improve therapeutic efficacy and reduce side effects.
In this review, significant advances in polymer-based mRNA delivery systems were elucidated. Past and ongoing studies have demonstrated that polymer-based mRNA delivery systems have great potential for clinical applications. Additionally, advanced delivery strategies are also discussed, which can inform the design of future polymer-based delivery systems to achieve precise targeted delivery. However, to realize the potential for truly effective clinical translation of polymer-based mRNA delivery systems, the delivery challenges mentioned above must be addressed through sustained research.
Author contributions
Qianyu Wan was responsible for writing the manuscript, while corresponding authors. Xuanrong Sun and Zhuxian Zhou were responsible for conceptualizing the ideas, and Yuji Sun, Xuanrong Sun and Zhuxian Zhou all made suggestions. All authors edited and approved the final manuscript.Consent for publication
We have included 9 figures (Fig. 5–8 and 10–13) from previously published literature with required copyright permission from the copyright owners. We have mentioned this in the manuscript with appropriate citations. Fig. 1, 2, 3 and 9 were created with biorender.com.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We thank the National Natural Science Foundation of China (52073249, 52373158 and 22075247), the National Key Research and Development Program of China (2021YFA1201200), and the Zhejiang Provincial Key Research and Development Program (2020C01123).References
- K. Leppek, R. Das and M. Barna, Nat. Rev. Mol. Cell Biol., 2018, 19, 158–174 CrossRef CAS PubMed
.
- R. Balakrishnan, M. Mori, I. Segota, Z. Zhang, R. Aebersold, C. Ludwig and T. Hwa, Science, 2022, 378, eabk2066 CrossRef CAS
.
- Z. Meng, J. O'Keeffe-Ahern, J. Lyu, L. Pierucci, D. Zhou and W. Wang, Biomater. Sci., 2017, 5, 2381–2392 RSC
.
- U. Sahin, K. Karikó and Ö. Türeci, Nat. Rev. Drug Discovery, 2014, 13, 759–780 CrossRef CAS PubMed
.
- X. Han, M.-G. Alameh, K. Butowska, J. J. Knox, K. Lundgreen, M. Ghattas, N. Gong, L. Xue, Y. Xu, M. Lavertu, P. Bates, J. Xu, G. Nie, Y. Zhong, D. Weissman and M. J. Mitchell, Nat. Nanotechnol., 2023, 18, 1105–1114 CrossRef CAS PubMed
.
- A. García, G. Maldonado, J. L. González, Y. Svitkin, D. Cantú, A. García-Carrancá, N. Sonenberg and G. Hernández, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, 2108359118 CrossRef PubMed
.
- R. T. Shroff, P. Chalasani, R. Wei, D. Pennington, G. Quirk, M. V. Schoenle, K. L. Peyton, J. L. Uhrlaub, T. J. Ripperger, M. Jergović, S. Dalgai, A. Wolf, R. Whitmer, H. Hammad, A. Carrier, A. J. Scott, J. Nikolich-Žugich, M. Worobey, R. Sprissler, M. Dake, B. J. LaFleur and D. Bhattacharya, Nat. Med., 2021, 27, 2002–2011 CrossRef CAS PubMed
.
- C. Liu, Q. Shi, X. Huang, S. Koo, N. Kong and W. Tao, Nat. Rev. Cancer, 2023, 23, 526–543 CrossRef CAS
.
- T. Soundara Rajan, A. Gugliandolo, P. Bramanti and E. Mazzon, Int. J. Mol. Sci., 2020, 21, 6514 CrossRef PubMed
.
- R. Zhang, S. Shao, Y. Piao, J. Xiang, X. Wei, Z. Zhang, Z. Zhou, J. Tang, N. Qiu, X. Xu, Y. Liu and Y. Shen, Adv. Mater., 2023, 35, 2303614 CrossRef CAS PubMed
.
- N. Chaudhary, D. Weissman and K. A. Whitehead, Nat. Rev. Drug Discovery, 2021, 20, 817–838 CrossRef CAS PubMed
.
- K. Servick, Science, 2020, 370, 1388–1389 CrossRef CAS PubMed
.
- Y. Liu, D. Zhang, Y. An, Y. Sun, J. Li, M. Zheng, Y. Zou and B. Shi, Nano Today, 2023, 49, 101790 CrossRef CAS
.
- K. Lundstrom, Diseases, 2018, 6, 42 CrossRef PubMed
.
- W. Doerfler, Virus Res., 2021, 302, 198466 CrossRef CAS PubMed
.
- R. Yang, Y. Deng, B. Huang, L. Huang, A. Lin, Y. Li, W. Wang, J. Liu, S. Lu, Z. Zhan, Y. Wang, R. A, W. Wang, P. Niu, L. Zhao, S. Li, X. Ma, L. Zhang, Y. Zhang, W. Yao, X. Liang, J. Zhao, Z. Liu, X. Peng, H. Li and W. Tan, Signal Transduction Targeted Ther., 2021, 6, 213 CrossRef CAS PubMed
.
- M. Kakran, M. Muratani, W. J. Tng, H. Liang, D. B. Trushina, G. B. Sukhorukov, H. H. Ng and M. N. Antipina, J. Mater. Chem. B, 2015, 3, 5842–5848 RSC
.
- J. W. Schott, D. Hoffmann and A. Schambach, Curr. Opin. Pharmacol., 2015, 24, 135–146 CrossRef CAS PubMed
.
- M. C. Milone and U. O'Doherty, Leukemia, 2018, 32, 1529–1541 CrossRef CAS PubMed
.
- M. Watanabe, Y. Nishikawaji, H. Kawakami and K. I. Kosai, Viruses, 2021, 13, 2502 CrossRef CAS PubMed
.
- S. Wu, J. Huang, Z. Zhang, J. Wu, J. Zhang, H. Hu, T. Zhu, J. Zhang, L. Luo, P. Fan, B. Wang, C. Chen, Y. Chen, X. Song, Y. Wang, W. Si, T. Sun, X. Wang, L. Hou and W. Chen, Lancet Infect. Dis., 2021, 21, 1654–1664 CrossRef CAS PubMed
.
- G. N. Nguyen, J. K. Everett, S. Kafle, A. M. Roche, H. E. Raymond, J. Leiby, C. Wood, C.-A. Assenmacher, E. P. Merricks, C. T. Long, H. H. Kazazian, T. C. Nichols, F. D. Bushman and D. E. Sabatino, Nat. Biotechnol., 2021, 39, 47–55 CrossRef CAS PubMed
.
- J. N. Warnock, C. Daigre and M. Al-Rubeai, Methods Mol. Biol., 2011, 737, 1–25 CrossRef CAS PubMed
.
- M. Jeong, Y. Lee, J. Park, H. Jung and H. Lee, Adv. Drug Delivery Rev., 2023, 200, 114990 CrossRef CAS PubMed
.
- A. F. Rodrigues, C. Rebelo, S. Simões, C. Paulo, S. Pinho, V. Francisco and L. Ferreira, Adv. Sci., 2023, 10, 2205475 CrossRef CAS PubMed
.
- S. Chen, X. Huang, Y. Xue, E. Álvarez-Benedicto, Y. Shi, W. Chen, S. Koo, D. J. Siegwart, Y. Dong and W. Tao, Nat. Rev. Methods Primers, 2023, 3, 63 CrossRef CAS
.
- Y. Liang, J. Zhang, C. Xu, J. Wang, W. Han, J. Yang, S. Wu, J. An, J. Liu, Z. Zhang, J. Shi and K. Zhang, ACS Nano, 2023, 17, 15025–15043 CrossRef CAS
.
- J. C. Kaczmarek, A. K. Patel, K. J. Kauffman, O. S. Fenton, M. J. Webber, M. W. Heartlein, F. DeRosa and D. G. Anderson, Angew. Chem., Int. Ed., 2016, 55, 13808–13812 CrossRef CAS PubMed
.
- J. C. Kaczmarek, A. K. Patel, L. H. Rhym, U. C. Palmiero, B. Bhat, M. W. Heartlein, F. DeRosa and D. G. Anderson, Biomaterials, 2021, 275, 120966 CrossRef CAS
.
- M. A. Islam, E. K. G. Reesor, Y. Xu, H. R. Zope, B. R. Zetter and J. Shi, Biomater. Sci., 2015, 3, 1519–1533 RSC
.
- W. Zhang, A. Mehta, Z. Tong, L. Esser and N. H. Voelcker, Adv. Sci., 2021, 8, 2003937 CrossRef CAS PubMed
.
- Y. Zhou, Z. Peng, E. S. Seven and R. M. Leblanc, J. Controlled Release, 2018, 270, 290–303 CrossRef CAS PubMed
.
- C. G. Zamboni, K. L. Kozielski, H. J. Vaughan, M. M. Nakata, J. Kim, L. J. Higgins, M. G. Pomper and J. J. Green, J. Controlled Release, 2017, 263, 18–28 CrossRef CAS PubMed
.
- M. Zoulikha, Q. Xiao, G. F. Boafo, M. A. Sallam, Z. Chen and W. He, Acta Pharm. Sin. B, 2022, 12, 600–620 CrossRef CAS
.
- P. Huang, L. Jiang, H. Pan, L. Ding, B. Zhou, M. Zhao, J. Zou, B. Li, M. Qi, H. Deng, Y. Zhou and X. Chen, Adv. Mater., 2023, 35, 2207471 CrossRef CAS PubMed
.
- S. Liu, X. Wang, X. Yu, Q. Cheng, L. T. Johnson, S. Chatterjee, D. Zhang, S. M. Lee, Y. Sun, T. C. Lin, J. L. Liu and D. J. Siegwart, J. Am. Chem. Soc., 2021, 143, 21321–21330 CrossRef CAS PubMed
.
- A. Jarzębińska, T. Pasewald, J. Lambrecht, O. Mykhaylyk, L. Kümmerling, P. Beck, G. Hasenpusch, C. Rudolph, C. Plank and C. Dohmen, Angew. Chem., Int. Ed., 2016, 55, 9591–9595 CrossRef
.
- W. Yang, L. Mixich, E. Boonstra and H. Cabral, Adv. Healthcare Mater., 2023, 12, 2202688 CrossRef CAS PubMed
.
- Z. Zhou, X. Liu, D. Zhu, Y. Wang, Z. Zhang, X. Zhou, N. Qiu, X. Chen and Y. Shen, Adv. Drug Delivery Rev., 2017, 115, 115–154 CrossRef CAS PubMed
.
- P. Huang, H. Deng, Y. Zhou and X. Chen, Matter, 2022, 5, 1670–1699 CrossRef CAS
.
- E. Rohner, R. Yang, K. S. Foo, A. Goedel and K. R. Chien, Nat. Biotechnol., 2022, 40, 1586–1600 CrossRef CAS PubMed
.
- A. K. Blakney, P. F. McKay, K. Hu, K. Samnuan, N. Jain, A. Brown, A. Thomas, P. Rogers, K. Polra, H. Sallah, J. Yeow, Y. Zhu, M. M. Stevens, A. Geall and R. J. Shattock, J. Controlled Release, 2021, 338, 201–210 CrossRef CAS
.
- S. Uchida, C. Y. J. Lau, M. Oba and K. Miyata, Adv. Drug Delivery Rev., 2023, 199, 114972 CrossRef CAS PubMed
.
- S. Nimesh, Curr. Clin. Pharmacol., 2012, 7, 121–130 CrossRef CAS
.
- J. Cai, Y. Yue, Y. Wang, Z. Jin, F. Jin and C. Wu, J. Controlled Release, 2016, 238, 71–79 CrossRef CAS PubMed
.
- A. Akinc, M. Thomas, A. M. Klibanov and R. Langer, J. Gene Med., 2005, 7, 657–663 CrossRef CAS
.
- G. Chen, B. Zhao, E. F. Ruiz and F. Zhang, Theranostics, 2022, 12, 4081–4109 CrossRef CAS
.
- J. Zhou, J. W. Yockman, S. W. Kim and S. E. Kern, Pharm. Res., 2007, 24, 1079–1087 CrossRef CAS PubMed
.
- R. R. Sawant, S. K. Sriraman, G. Navarro, S. Biswas, R. A. Dalvi and V. P. Torchilin, Biomaterials, 2012, 33, 3942–3951 CrossRef CAS PubMed
.
- J. Li, Y. Wu, J. Xiang, H. Wang, Q. Zhuang, T. Wei, Z. Cao, Q. Gu, Z. Liu and R. Peng, Chem. Eng. J., 2023, 456, 140930 CrossRef CAS
.
- L. Tan, T. Zheng, M. Li, X. Zhong, Y. Tang, M. Qin and X. Sun, Drug Delivery Transl. Res., 2020, 10, 678–689 CrossRef CAS
.
- M. Neu, D. Fischer and T. Kissel, J. Gene Med., 2005, 7, 992–1009 CrossRef CAS PubMed
.
- T. Wu, L. Wang, S. Ding and Y. You, Macromol. Biosci., 2017, 17, 1700114 CrossRef
.
- H. Debus, P. Baumhof, J. Probst and T. Kissel, J. Controlled Release, 2010, 148, 334–343 CrossRef CAS
.
- S. Taranejoo, R. Chandrasekaran, W. Cheng and K. Hourigan, Carbohydr. Polym., 2016, 153, 160–168 CrossRef CAS
.
- J. W. Park, K. H. Bae, C. Kim and T. G. Park, Biomacromolecules, 2011, 12, 457–465 CrossRef CAS
.
- C. Martins, F. Sousa, F. Araújo and B. Sarmento, Adv. Healthcare Mater., 2018, 7, 1701035 CrossRef CAS PubMed
.
- T. Bus, A. Traeger and U. S. Schubert, J. Mater. Chem. B, 2018, 6, 6904–6918 RSC
.
- B. Du, H. Yin, Y. Chen, W. Lin, Y. Wang, D. Zhao, G. Wang, X. He, J. Li, Z. Li, F. Luo, H. Tan and Q. Fu, J. Mater. Chem. B, 2020, 8, 4434–4446 RSC
.
- H. K. Makadia and S. J. Siegel, Polymers, 2011, 3, 1377–1397 CrossRef CAS PubMed
.
- Y. Xiao, J. Chen, H. Zhou, X. Zeng, Z. Ruan, Z. Pu, X. Jiang, A. Matsui, L. Zhu, Z. Amoozgar, D. S. Chen, X. Han, D. G. Duda and J. Shi, Nat. Commun., 2022, 13, 758 CrossRef CAS
.
- H. Guerrero-Cázares, S. Y. Tzeng, N. P. Young, A. O. Abutaleb, A. Quiñones-Hinojosa and J. J. Green, ACS Nano, 2014, 8, 5141–5153 CrossRef
.
- P. Dosta, N. Segovia, A. Cascante, V. Ramos and S. Borrós, Acta Biomater., 2015, 20, 82–93 CrossRef CAS PubMed
.
- K. L. Kozielski, S. Y. Tzeng, B. A. De Mendoza and J. J. Green, ACS Nano, 2014, 8, 3232–3241 CrossRef CAS
.
- S. Y. Neshat, C. H. R. Chan, J. Harris, O. M. Zmily, S. Est-Witte, J. Karlsson, S. R. Shannon, M. Jain, J. C. Doloff, J. J. Green and S. Y. Tzeng, Biomaterials, 2023, 300, 122185 CrossRef CAS PubMed
.
- L. Yang and Y. Zhang, J. Hematol. Oncol., 2017, 10, 58 CrossRef
.
- A. Mantovani, F. Marchesi, A. Malesci, L. Laghi and P. Allavena, Nat. Rev. Clin. Oncol., 2017, 14, 399–416 CrossRef CAS
.
- W. D. Tap, Z. A. Wainberg, S. P. Anthony, P. N. Ibrahim, C. Zhang, J. H. Healey, B. Chmielowski, A. P. Staddon, A. L. Cohn, G. I. Shapiro, V. L. Keedy, A. S. Singh, I. Puzanov, E. L. Kwak, A. J. Wagner, D. D. Von Hoff, G. J. Weiss, R. K. Ramanathan, J. Zhang, G. Habets, Y. Zhang, E. A. Burton, G. Visor, L. Sanftner, P. Severson, H. Nguyen, M. J. Kim, A. Marimuthu, G. Tsang, R. Shellooe, C. Gee, B. L. West, P. Hirth, K. Nolop, M. van de Rijn, H. H. Hsu, C. Peterfy, P. S. Lin, S. Tong-Starksen and G. Bollag, N. Engl. J. Med., 2015, 373, 428–437 CrossRef CAS
.
- N. Butowski, H. Colman, J. F. De Groot, A. M. Omuro, L. Nayak, P. Y. Wen, T. F. Cloughesy, A. Marimuthu, S. Haidar, A. Perry, J. Huse, J. Phillips, B. L. West, K. B. Nolop, H. H. Hsu, K. L. Ligon, A. M. Molinaro and M. Prados, Neuro-Oncology, 2016, 18, 557–564 CrossRef PubMed
.
- F. Zhang, N. N. Parayath, C. I. Ene, S. B. Stephan, A. L. Koehne, M. E. Coon, E. C. Holland and M. T. Stephan, Nat. Commun., 2019, 10, 3974 CrossRef CAS PubMed
.
- Y. Yan, H. Xiong, X. Zhang, Q. Cheng and D. J. Siegwart, Biomacromolecules, 2017, 18, 4307–4315 CrossRef CAS PubMed
.
- Y. Jiang, Q. Lu, Y. Wang, E. Xu, A. Ho, P. Singh, Y. Wang, Z. Jiang, F. Yang, G. T. Tietjen, P. Cresswell and W. M. Saltzman, Nano Lett., 2020, 20, 1117–1123 CrossRef CAS PubMed
.
- M. K. Grun, A. Suberi, K. Shin, T. Lee, V. Gomerdinger, Z. M. Moscato, A. S. Piotrowski-Daspit and W. M. Saltzman, Biomaterials, 2021, 272, 120780 CrossRef CAS PubMed
.
- S. Chatterjee, E. Kon, P. Sharma and D. Peer, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, 2307800120 CrossRef PubMed
.
- W. Chen, Y. Ma, X. Liu and D. Zhu, Explor Target Antitumor Ther., 2022, 3, 117–127 CAS
.
- A. Harguindey, D. W. Domaille, B. D. Fairbanks, J. Wagner, C. N. Bowman and J. N. Cha, Adv. Mater., 2017, 29, 1700743 CrossRef PubMed
.
- H. Xu, Q. Yao, C. Cai, J. Gou, Y. Zhang, H. Zhong and X. Tang, J. Controlled Release, 2015, 199, 84–97 CrossRef CAS PubMed
.
- Y. Meng and J. Wu, ACS Appl. Mater. Interfaces, 2021, 13, 49658–49670 CrossRef CAS PubMed
.
- X. Fan, H. Ren, X. Luo, P. Wang, G. Lv, H. Yuan, H. Li and Y. Yan, J. Biomater. Appl., 2016, 30, 1261–1272 CrossRef CAS PubMed
.
- H. J. Kim, S. Ogura, T. Otabe, R. Kamegawa, M. Sato, K. Kataoka and K. Miyata, ACS Cent. Sci., 2019, 5, 1866–1875 CrossRef CAS PubMed
.
- N. U. Dharmaratne, T. M. M. Jouaneh, M. K. Kiesewetter and R. T. Mathers, Macromolecules, 2018, 51, 8461–8468 CrossRef CAS
.
- J. Yum, B. S. Kim, S. Ogura, R. Kamegawa, M. Naito, Y. Yamasaki, H. J. Kim and K. Miyata, J. Controlled Release, 2022, 342, 148–156 CrossRef CAS PubMed
.
- Z. Zhou, X. Ma, C. J. Murphy, E. Jin, Q. Sun, Y. Shen, E. A. Van Kirk and W. J. Murdoch, Angew. Chem., Int. Ed., 2014, 53, 10949–10955 CrossRef CAS PubMed
.
- J. Yang, K. Wang, Y. Zheng, Y. Piao, J. Wang, J. Tang, Y. Shen and Z. Zhou, Angew. Chem., Int. Ed., 2022, 61, 202202128 CrossRef PubMed
.
- G. Wang, Z. Zhou, Z. Zhao, Q. Li, Y. Wu, S. Yan, Y. Shen and P. Huang, ACS Nano, 2020, 14, 4890–4904 CrossRef CAS PubMed
.
- F. Bi, J. Zhang, Z. Wei, D. Yu, S. Zheng, J. Wang, H. Li, Z. Hua, H. Zhang and G. Yang, Biomacromolecules, 2022, 23, 128–139 CrossRef CAS PubMed
.
- D. Huang and D. Wu, Mater. Sci. Eng., C, 2018, 90, 713–727 CrossRef CAS PubMed
.
- S. Chowdhury, I. Toth and R. J. Stephenson, Biomaterials, 2022, 280, 121303 CrossRef CAS PubMed
.
- J. Chen, D. Zhu, X. Liu and L. Peng, Acc. Mater. Res., 2022, 3, 484–497 CrossRef CAS PubMed
.
- F. Joubert, M. J. Munson, A. Sabirsh, R. M. England, M. Hemmerling, C. Alexander and M. B. Ashford, J. Controlled Release, 2023, 356, 580–594 CrossRef CAS PubMed
.
- S. Ndeupen, Z. Qin, S. Jacobsen, A. Bouteau, H. Estanbouli and B. Z. Igyártó, iScience, 2021, 24, 103479 CrossRef CAS PubMed
.
- K. Van der Jeught, S. De Koker, L. Bialkowski, C. Heirman, P. Tjok Joe, F. Perche, S. Maenhout, S. Bevers, K. Broos, K. Deswarte, V. Malard, H. Hammad, P. Baril, T. Benvegnu, P. A. Jaffrès, S. A. A. Kooijmans, R. Schiffelers, S. Lienenklaus, P. Midoux, C. Pichon, K. Breckpot and K. Thielemans, ACS Nano, 2018, 12, 9815–9829 CrossRef CAS PubMed
.
- S. Persano, M. L. Guevara, Z. Li, J. Mai, M. Ferrari, P. P. Pompa and H. Shen, Biomaterials, 2017, 125, 81–89 CrossRef CAS PubMed
.
- H. Xiong, S. Liu, T. Wei, Q. Cheng and D. J. Siegwart, J. Controlled Release, 2020, 325, 198–205 CrossRef CAS PubMed
.
- S. A. Dilliard and D. J. Siegwart, Nat. Rev. Mater., 2023, 8, 282–300 CrossRef CAS PubMed
.
- Y. Zong, Y. Lin, T. Wei and Q. Cheng, Adv. Mater., 2023, 35, 2303261 CrossRef CAS PubMed
.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC
.
- Y. Hu, X. Liu, M. Ran, T. Yang, T. Li, Y. Wu, Y. Lin, Z. Qian and X. Gao, Mater. Today Nano, 2022, 17, 100151 CrossRef CAS
.
- J. Yoo, C. Park, G. Yi, D. Lee and H. Koo, Cancers, 2019, 11, 640 CrossRef CAS PubMed
.
- Q. Chen, R. Qi, X. Chen, X. Yang, S. Wu, H. Xiao and W. Dong, Mol. Ther., 2017, 25, 92–101 CrossRef CAS PubMed
.
- L. B. Darragh and S. D. Karam, Mol. Carcinog., 2022, 61, 153–164 CrossRef CAS PubMed
.
- S. Jhunjhunwala, C. Hammer and L. Delamarre, Nat. Rev. Cancer, 2021, 21, 298–312 CrossRef CAS PubMed
.
- C. Fornaguera, M. Guerra-Rebollo, M. Ángel Lázaro, C. Castells-Sala, O. Meca-Cortés, V. Ramos-Pérez, A. Cascante, N. Rubio, J. Blanco and S. Borrós, Adv. Healthcare Mater., 2018, 7, 1800335 CrossRef PubMed
.
- E. Ben-Akiva, J. Karlsson, S. Hemmati, H. Yu, S. Y. Tzeng, D. M. Pardoll and J. J. Green, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, 2301606120 CrossRef PubMed
.
- B. V. Kumar, T. J. Connors and D. L. Farber, Immunity, 2018, 48, 202–213 CrossRef CAS PubMed
.
- P. Zhou, H. Shi, H. Huang, X. Sun, S. Yuan, N. M. Chapman, J. P. Connelly, S. A. Lim, J. Saravia, A. Kc, S. M. Pruett-Miller and H. Chi, Nature, 2023, 624, 154–163 CrossRef CAS PubMed
.
- B. L. Cadilha, M. R. Benmebarek, K. Dorman, A. Oner, T. Lorenzini, H. Obeck, M. Vänttinen, M. Di Pilato, J. N. Pruessmann, S. Stoiber, D. Huynh, F. Märkl, M. Seifert, K. Manske, J. Suarez-Gosalvez, Y. Zeng, S. Lesch, C. H. Karches, C. Heise, A. Gottschlich, M. Thomas, C. Marr, J. Zhang, D. Pandey, T. Feuchtinger, M. Subklewe, T. R. Mempel, S. Endres and S. Kobold, Sci. Adv., 2021, 7, eabi5781 CrossRef CAS PubMed
.
- K. M. Maalej, M. Merhi, V. P. Inchakalody, S. Mestiri, M. Alam, C. Maccalli, H. Cherif, S. Uddin, M. Steinhoff, F. M. Marincola and S. Dermime, Mol. Cancer, 2023, 22, 20 CrossRef CAS PubMed
.
- N. G. Holtzman, H. Xie, S. Bentzen, V. Kesari, A. Bukhari, F. El Chaer, F. Lutfi, J. Siglin, E. Hutnick, N. Gahres, K. Ruehle, H. Ahmad, C. Shanholtz, M. H. Kocoglu, A. Z. Badros, J. A. Yared, N. M. Hardy, A. P. Rapoport and S. Dahiya, Neuro-Oncology, 2021, 23, 112–121 CrossRef CAS PubMed
.
- M. L. Schubert, M. Schmitt, L. Wang, C. A. Ramos, K. Jordan, C. Müller-Tidow and P. Dreger, Ann. Oncol., 2021, 32, 34–48 CrossRef CAS PubMed
.
- Z. Li, L. Amaya, R. Pi, S. K. Wang, A. Ranjan, R. M. Waymouth, C. A. Blish, H. Y. Chang and P. A. Wender, Nat. Commun., 2023, 14, 6983 CrossRef CAS PubMed
.
- Y. Hui, X. Yi, F. Hou, D. Wibowo, F. Zhang, D. Zhao, H. Gao and C. X. Zhao, ACS Nano, 2019, 13, 7410–7424 CrossRef CAS PubMed
.
- S. Hamdy, A. Haddadi, R. W. Hung and A. Lavasanifar, Adv. Drug Delivery Rev., 2011, 63, 943–955 CrossRef CAS PubMed
.
- M. Zhang, H. Jiang, L. Wu, H. Lu, H. Bera, X. Zhao, X. Guo, X. Liu, D. Cun and M. Yang, J. Controlled Release, 2022, 352, 422–437 CrossRef CAS PubMed
.
- G. S. Naidu, S. B. Yong, S. Ramishetti, R. Rampado, P. Sharma, A. Ezra, M. Goldsmith, I. Hazan-Halevy, S. Chatterjee, A. Aitha and D. Peer, Adv. Sci., 2023, 10, 2301929 CrossRef CAS PubMed
.
- M. Maugeri, M. Nawaz, A. Papadimitriou, A. Angerfors, A. Camponeschi, M. Na, M. Hölttä, P. Skantze, S. Johansson, M. Sundqvist, J. Lindquist, T. Kjellman, I. L. Mårtensson, T. Jin, P. Sunnerhagen, S. Östman, L. Lindfors and H. Valadi, Nat. Commun., 2019, 10, 4333 CrossRef CAS PubMed
.
- N. Yoshinaga, S. Uchida, A. Dirisala, M. Naito, K. Koji, K. Osada, H. Cabral and K. Kataoka, Adv. Healthcare Mater., 2022, 11, 2102016 CrossRef CAS PubMed
.
- X. Xu and T. Xia, ACS Nanosci. Au, 2023, 3, 192–203 CrossRef CAS PubMed
.
- S. T. LoPresti, M. L. Arral, N. Chaudhary and K. A. Whitehead, J. Controlled Release, 2022, 345, 819–831 CrossRef CAS PubMed
.
- V. Kumar, J. Qin, Y. Jiang, R. G. Duncan, B. Brigham, S. Fishman, J. K. Nair, A. Akinc, S. A. Barros and P. V. Kasperkovitz, Mol. Ther. – Nucleic Acids, 2014, 3, 210 CrossRef
.
- B. L. Mui, Y. K. Tam, M. Jayaraman, S. M. Ansell, X. Du, Y. Y. Tam, P. J. Lin, S. Chen, J. K. Narayanannair, K. G. Rajeev, M. Manoharan, A. Akinc, M. A. Maier, P. Cullis, T. D. Madden and M. J. Hope, Mol. Ther. – Nucleic Acids, 2013, 2, 139 CrossRef PubMed
.
- C. Dobrowolski, K. Paunovska, M. Z. C. Hatit, M. P. Lokugamage and J. E. Dahlman, Adv. Healthcare Mater., 2021, 10, 2002022 CrossRef CAS PubMed
.
- H. Wang, C. A. Thorling, X. Liang, K. R. Bridle, J. E. Grice, Y. Zhu, D. H. G. Crawford, Z. P. Xu, X. Liu and M. S. Roberts, J. Mater. Chem. B, 2015, 3, 939–958 RSC
.
- Y. S. Zhu, K. Tang and J. Lv, Trends Pharmacol. Sci., 2021, 42, 857–869 CrossRef CAS PubMed
.
- P. Lingasamy, A. Tobi, M. Haugas, H. Hunt, P. Paiste, T. Asser, T. Rätsep, V. R. Kotamraju, R. Bjerkvig and T. Teesalu, Biomaterials, 2019, 219, 119373 CrossRef CAS PubMed
.
- S. Lei, X. Chen, Y. Gao, M. Shuai, W. Zhou, J. Li, J. Wu, K. Men and X. Duan, Adv. Funct. Mater., 2022, 32, 2204342 CrossRef CAS
.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Nat. Nanotechnol., 2020, 15, 313–320 CrossRef CAS PubMed
.
- X. Wang, S. Liu, Y. Sun, X. Yu, S. M. Lee, Q. Cheng, T. Wei, J. Gong, J. Robinson, D. Zhang, X. Lian, P. Basak and D. J. Siegwart, Nat. Protoc., 2023, 18, 265–291 CrossRef CAS PubMed
.
- D. Zhang, E. N. Atochina-Vasserman, J. Lu, D. S. Maurya, Q. Xiao, M. Liu, J. Adamson, N. Ona, E. K. Reagan, H. Ni, D. Weissman and V. Percec, J. Am. Chem. Soc., 2022, 144, 4746–4753 CrossRef CAS PubMed
.
- S. T. LoPresti, M. L. Arral, N. Chaudhary and K. A. Whitehead, J. Controlled Release, 2022, 345, 819–831 CrossRef CAS PubMed
.
- M. Qiu, Y. Tang, J. Chen, R. Muriph, Z. Ye, C. Huang, J. Evans, E. P. Henske and Q. Xu, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, 2116271119 CrossRef PubMed
.
- Q. Wan, X. Zhang, D. Zhou, R. Xie, Y. Cai, K. Zhang and X. Sun, J. Nanobiotechnol., 2023, 21, 215 CrossRef CAS PubMed
.
- Z. Tang, X. You, Y. Xiao, W. Chen, Y. Li, X. Huang, H. Liu, F. Xiao, C. Liu, S. Koo, N. Kong and W. Tao, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, 2304966120 CrossRef PubMed
.
- L. Rotolo, D. Vanover, N. C. Bruno, H. E. Peck, C. Zurla, J. Murray, R. K. Noel, L. O'Farrell, M. Araínga, N. Orr-Burks, J. Y. Joo, L. C. S. Chaves, Y. Jung, J. Beyersdorf, S. Gumber, R. Guerrero-Ferreira, S. Cornejo, M. Thoresen, A. K. Olivier, K. M. Kuo, J. C. Gumbart, A. R. Woolums, F. Villinger, E. R. Lafontaine, R. J. Hogan, M. G. Finn and P. J. Santangelo, Nat. Mater., 2023, 22, 369–379 CrossRef CAS PubMed
.
- Q. Sun, X. Sun, X. Ma, Z. Zhou, E. Jin, B. Zhang, Y. Shen, E. A. Van Kirk, W. J. Murdoch, J. R. Lott, T. P. Lodge, M. Radosz and Y. Zhao, Adv. Mater., 2014, 26, 7615–7621 CrossRef CAS PubMed
.
- Q. Sun, Z. Zhou, N. Qiu and Y. Shen, Adv. Mater., 2017, 29, 1606628 CrossRef
.
- L. Li, M. Zhen, H. Wang, Z. Sun, X. Cao, J. Li, S. Liu, Z. Zhao, C. Zhou, C. Wang and C. Bai, Nano Today, 2023, 48, 101702 CrossRef CAS
.
- S. Ma, W. Xu, Y. Fei, D. Li, X. Jia, J. Wang and E. Wang, Adv. Healthcare Mater., 2023, 12, 2301413 CrossRef CAS PubMed
.
- C. Hotz, T. R. Wagenaar, F. Gieseke, D. S. Bangari, M. Callahan, H. Cao, J. Diekmann, M. Diken, C. Grunwitz, A. Hebert, K. Hsu, M. Bernardo, K. Karikó, S. Kreiter, A. N. Kuhn, M. Levit, N. Malkova, S. Masciari, J. Pollard, H. Qu, S. Ryan, A. Selmi, J. Schlereth, K. Singh, F. Sun, B. Tillmann, T. Tolstykh, W. Weber, L. Wicke, S. Witzel, Q. Yu, Y. A. Zhang, G. Zheng, J. Lager, G. J. Nabel, U. Sahin and D. Wiederschain, Sci. Transl. Med., 2021, 13, eabc7804 CrossRef CAS PubMed
.
- K. J. Hassett, J. Higgins, A. Woods, B. Levy, Y. Xia, C. J. Hsiao, E. Acosta, Ö. Almarsson, M. J. Moore and L. A. Brito, J. Controlled Release, 2021, 335, 237–246 CrossRef CAS PubMed
.
- S. Dong, Z. Feng, R. Ma, T. Zhang, J. Jiang, Y. Li, Y. Zhang, S. Li, X. Liu, X. Liu and H. Meng, Nano Lett., 2023, 23, 2137–2147 CrossRef CAS PubMed
.
- S. Mehta and J. Zhang, Nat. Rev. Cancer, 2022, 22, 239–252 CrossRef CAS
.
- Z. Xing, J. Xue, X. Ma, C. Han, Z. Wang, S. Luo, C. Wang, L. Dong and J. Zhang, J. Nanobiotechnol., 2022, 20, 442 CrossRef CAS
.
- C. Mugabe, Y. Matsui, A. I. So, M. E. Gleave, J. H. Baker, A. I. Minchinton, I. Manisali, R. Liggins, D. E. Brooks and H. M. Burt, Clin. Cancer Res., 2011, 17, 2788–2798 CrossRef CAS
.
- Q. Zhang, K. G. Neoh, L. Xu, S. Lu, E. T. Kang, R. Mahendran and E. Chiong, Langmuir, 2014, 30, 6151–6161 CrossRef CAS
.
- N. Kong, R. Zhang, G. Wu, X. Sui, J. Wang, N. Y. Kim, S. Blake, D. De, T. Xie, Y. Cao and W. Tao, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, 2112696119 CrossRef
.
- Q. Zhou, S. Shao, J. Wang, C. Xu, J. Xiang, Y. Piao, Z. Zhou, Q. Yu, J. Tang, X. Liu, Z. Gan, R. Mo, Z. Gu and Y. Shen, Nat. Nanotechnol., 2019, 14, 799–809 CrossRef CAS PubMed
.
- S. Sindhwani, A. M. Syed, J. Ngai, B. R. Kingston, L. Maiorino, J. Rothschild, P. MacMillan, Y. Zhang, N. U. Rajesh, T. Hoang, J. L. Y. Wu, S. Wilhelm, A. Zilman, S. Gadde, A. Sulaiman, B. Ouyang, Z. Lin, L. Wang, M. Egeblad and W. C. W. Chan, Nat. Mater., 2020, 19, 566–575 CrossRef CAS PubMed
.
- Z. Wang, Y. Sun, Y. Shen and Z. Zhou, Adv. NanoBiomed Res., 2023, 2300125 Search PubMed
.
- H. Ren, Q. Hu, J. Yang, X. Zhou, X. Liu, J. Tang, H. Hu, Y. Shen and Z. Zhou, Adv. Healthcare Mater., 2023, 12, 2302210 CrossRef CAS
.
- Y. Qin, G. Wang, L. Chen, Y. Sun, J. Yang, Y. Piao, Y. Shen and Z. Zhou, Adv. Mater., 2023, 2302292 Search PubMed
.
- H. Ren, Q. Hu, Y. Sun, X. Zhou, Y. Zhu, Q. Dong, L. Chen, J. Tang, H. Hu, Y. Shen and Z. Zhou, Biomater. Sci., 2023, 11, 7051–7061 RSC
.
- E. Nance, S. H. Pun, R. Saigal and D. L. Sellers, Nat. Rev. Mater., 2022, 7, 314–331 CrossRef CAS PubMed
.
- L. Wang and S. Wilhelm, Nat. Mater., 2023, 22, 282–283 CrossRef CAS PubMed
.
- P. Kumthekar, C. H. Ko, T. Paunesku, K. Dixit, A. M. Sonabend, O. Bloch, M. Tate, M. Schwartz, L. Zuckerman, R. Lezon, R. V. Lukas, B. Jovanovic, K. McCortney, H. Colman, S. Chen, B. Lai, O. Antipova, J. Deng, L. Li, S. Tommasini-Ghelfi, L. A. Hurley, D. Unruh, N. V. Sharma, M. Kandpal, F. M. Kouri, R. V. Davuluri, D. J. Brat, M. Muzzio, M. Glass, V. Vijayakumar, J. Heidel, F. J. Giles, A. K. Adams, C. D. James, G. E. Woloschak, C. Horbinski and A. H. Stegh, Sci. Transl. Med., 2021, 13, eabb3945 CrossRef CAS PubMed
.
- F. Ma, L. Yang, Z. Sun, J. Chen, X. Rui, Z. Glass and Q. Xu, Sci. Adv., 2020, 6, eabb4429 CrossRef CAS PubMed
.
- C. Fornaguera, M. Lázaro, P. Brugada-Vilà, I. Porcar, I. Morera, M. Guerra-Rebollo, C. Garrido, N. Rubio, J. Blanco, A. Cascante and S. Borrós, Drug Delivery, 2018, 25, 472–483 CrossRef CAS
.
- M. Saxena, S. H. van der Burg, C. J. M. Melief and N. Bhardwaj, Nat. Rev. Cancer, 2021, 21, 360–378 CrossRef CAS
.
- E. Blass and P. A. Ott, Nat. Rev. Clin. Oncol., 2021, 18, 215–229 CrossRef PubMed
.
- L. Tang, R. Zhang, Y. Wang, X. Zhang, Y. Yang, B. Zhao and L. Yang, Acta Biomater., 2023, 155, 521–537 CrossRef CAS PubMed
.
- M. Sheinin, B. Jeong, R. K. Paidi and K. Pahan, Cancers, 2022, 14, 5648 CrossRef CAS PubMed
.
- D. Cahn, M. Amosu, K. Maisel and G. A. Duncan, Nat. Rev.
Bioeng., 2023, 1, 83–84 CrossRef PubMed
.
- H. Jeong, H. Kim, Y. A. Kim, K. S. Kim and K. Na, ACS Appl. Mater. Interfaces, 2023, 15, 56749–56759 CAS
.
- K. K. Phua, K. W. Leong and S. K. Nair, J. Controlled Release, 2013, 166, 227–233 CrossRef CAS PubMed
.
- N. Sultana, A. Magadum, Y. Hadas, J. Kondrat, N. Singh, E. Youssef, D. Calderon, E. Chepurko, N. Dubois, R. J. Hajjar and L. Zangi, Mol. Ther., 2017, 25, 1306–1315 CrossRef CAS PubMed
.
- S. Kreiter, A. Selmi, M. Diken, M. Koslowski, C. M. Britten, C. Huber, O. Türeci and U. Sahin, Cancer Res., 2010, 70, 9031–9040 CrossRef CAS PubMed
.
- S. C. Balmert, C. D. Carey, G. D. Falo, S. K. Sethi, G. Erdos, E. Korkmaz and L. D. Falo, Jr., J. Controlled Release, 2020, 317, 336–346 CrossRef CAS PubMed
.
- K. J. Koh, Y. Liu, S. H. Lim, X. J. Loh, L. Kang, C. Y. Lim and K. K. L. Phua, Sci. Rep., 2018, 8, 11842 CrossRef PubMed
.
- M. J. Thun, J. O. DeLancey, M. M. Center, A. Jemal and E. M. Ward, Carcinogenesis, 2010, 31, 100–110 CrossRef CAS PubMed
.
- K. Washington and F. Rocha, Curr. Treat Options Oncol., 2021, 22, 97 CrossRef PubMed
.
- Y. Pointreau, J. Biau, N. Delaby, J. Thariat and M. Lapeyre, Cancer Radiother., 2022, 26, 199–205 CrossRef CAS PubMed
.
- I. Ali, W. A. Wani, K. Saleem and A. Haque, Adv. Anticancer Agents Med. Chem., 2013, 13, 296–306 CrossRef CAS PubMed
.
- N. Pardi, M. J. Hogan, F. W. Porter and D. Weissman, Nat. Rev. Drug Discovery, 2018, 17, 261–279 CrossRef CAS PubMed
.
- C. L. Lorentzen, J. B. Haanen, Ö. Met and I. M. Svane, Lancet Oncol., 2022, 23, 450–458 CrossRef PubMed
.
- U. Sahin, P. Oehm, E. Derhovanessian, R. A. Jabulowsky, M. Vormehr, M. Gold, D. Maurus, D. Schwarck-Kokarakis, A. N. Kuhn, T. Omokoko, L. M. Kranz, M. Diken, S. Kreiter, H. Haas, S. Attig, R. Rae, K. Cuk, A. Kemmer-Brück, A. Breitkreuz, C. Tolliver, J. Caspar, J. Quinkhardt, L. Hebich, M. Stein, A. Hohberger, I. Vogler, I. Liebig, S. Renken, J. Sikorski, M. Leierer, V. Müller, H. Mitzel-Rink, M. Miederer, C. Huber, S. Grabbe, J. Utikal, A. Pinter, R. Kaufmann, J. C. Hassel, C. Loquai and Ö. Türeci, Nature, 2020, 585, 107–112 CrossRef CAS PubMed
.
- A. J. Barbier, A. Y. Jiang, P. Zhang, R. Wooster and D. G. Anderson, Nat. Biotechnol., 2022, 40, 840–854 CrossRef CAS PubMed
.
- S. Egloff, A. Runser, A. Klymchenko and A. Reisch, Small Methods, 2021, 5, 2000947 CrossRef CAS PubMed
.
- Y. Huang, W. Xiao, S. Ahrari, M. Yu and J. Zheng, Angew. Chem., Int. Ed., 2023, 62, 202308909 CrossRef PubMed
.
- W. Zhen, R. R. Weichselbaum and W. Lin, Adv. Mater., 2023, 35, 2206370 CrossRef CAS PubMed
.
- A. Wahida, L. Buschhorn, S. Fröhling, P. J. Jost, A. Schneeweiss, P. Lichter and R. Kurzrock, Nat. Rev. Cancer, 2023, 23, 43–54 CrossRef CAS PubMed
.
- J. C. Denny and F. S. Collins, Cell, 2021, 184, 1415–1419 CrossRef CAS PubMed
.
- S. M. Schüssler-Fiorenza Rose, K. Contrepois, K. J. Moneghetti, W. Zhou, T. Mishra, S. Mataraso, O. Dagan-Rosenfeld, A. B. Ganz, J. Dunn, D. Hornburg, S. Rego, D. Perelman, S. Ahadi, M. R. Sailani, Y. Zhou, S. R. Leopold, J. Chen, M. Ashland, J. W. Christle, M. Avina, P. Limcaoco, C. Ruiz, M. Tan, A. J. Butte, G. M. Weinstock, G. M. Slavich, E. Sodergren, T. L. McLaughlin, F. Haddad and M. P. Snyder, Nat. Med., 2019, 25, 792–804 CrossRef PubMed
.
- Y. Wang, S. Sun, Z. Zhang and D. Shi, Adv. Mater., 2018, 30, 1705660 CrossRef PubMed
.
- O. Adir, M. Poley, G. Chen, S. Froim, N. Krinsky, J. Shklover, J. Shainsky-Roitman, T. Lammers and A. Schroeder, Adv. Mater., 2020, 32, 1901989 CrossRef CAS PubMed
.
- S. Siemer, T. A. Bauer, P. Scholz, C. Breder, F. Fenaroli, G. Harms, D. Dietrich, J. Dietrich, C. Rosenauer, M. Barz, S. Becker, S. Strieth, C. Reinhardt, T. Fauth, J. Hagemann and R. H. Stauber, ACS Nano, 2021, 15, 18541–18556 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2024 |