Emerging lipid–polymer hybrid nanoparticles for genome editing
Mariana
Gameiro
 ,
João F.
Mano
,
João F.
Mano
 * and
Vítor M.
Gaspar
* and
Vítor M.
Gaspar
 *
*
Department of Chemistry, CICECO – Aveiro Institute of Materials, University of Aveiro, Campus Universitário de Santiago, 3810-193, Aveiro, Portugal. E-mail: jmano@ua.pt; vm.gaspar@ua.pt; Tel: +351 234 370 200
First published on 2nd August 2024
Abstract
Genome editing technologies have been key to unlocking new bioengineering strategies as they enable the modification of mammalian cells’ genes in a fully user-programmed mode. Despite major advancements, the development of proficient systems for a safer and more efficient delivery of gene editing machineries into all classes of mammalian cells is still challenging. In this context, new generations of lipid–polymer hybrid nanoparticles are rapidly emerging as potentially valuable alternatives to upgrade mainstream gene delivery toolboxes. Building on this, herein we showcase the most recent advances in designing hybrid nanocarriers for the delivery of genome editing components. Major polymer and lipid features harnessed for optimal CRISPR/Cas9-based gene editing, along with tissue- and cell-targeting strategies are specifically highlighted. Alongside this, key technologies for the formulation of lipid–polymer conjugates are showcased. Such hybrid vehicles, along with the existing chemical toolsets are envisioned to unlock progressively more proficient nonviral platforms for maximizing genome editing efficacy, especially in the most challenging primary cells or tissues.
1. Introduction
Currently available genome editing toolboxes can be leveraged to manipulate the human genome in a permanent mode, ushering a new dawn in biomedical sciences.1–5 Despite the promise of relevant tools, including zinc fingers (ZFNs) and transcription activator-like effector nucleases (TALENs), their multiple drawbacks paved the way for the discovery of the clustered regularly interspaced short palindromic repeat (CRISPR) system.3–10 CRISPR-based editors have marked a transformative breakthrough in the gene editing field, leveraging two main components: a nuclease, such as Cas9, which generates double-stranded breaks (DSBs) in the target DNA locus, directed by a single guide RNA (sgRNA), complementary to the target sequence.2,3,11–17 Such DSBs activate native DNA repair mechanisms, either leading to the knock-in (i.e., insertion), or the knockout (i.e., disruption) of a gene of interest.2 Challenges such as off-target effects and genomic cytotoxicity have prompted the advancement of next-generation CRISPR tools.6,18 In this line, Base Editors (i.e., adenine and cytosine base editors) enable precise base conversions (C-to-T or A-to-G), without introducing DSBs, leveraging a catalytically inactive or a nickase Cas9 nuclease.5 To further achieve even more feasible and safer gene editing interventions, Prime Editors were developed. This tool enables small insertions, deletions and all 12 possible base-to-base conversions, extending the promise to reverse genetic diseases.18 Alongside this, CRISPR interference (CRISPRi) and activation (CRISPRa) have expanded the toolset for gene expression regulation at the transcriptional level and epigenome editing, by fusion with transcriptional repressors or activators, respectively.2 Such technologies have been extensively harnessed for a myriad of applications, including for the treatment of genetic diseases,3,18 improvement of cell-based therapeutics,2in vitro disease modelling,11,12 synthetic biology,12,14–16 drug development,19 molecular sensing19 or cellular imaging.19 Importantly, the world‘s first approved CRISPR-based ex vivo cell therapy – for sickle cell disease and beta thalassemia – in 2023 - represents an unprecedented milestone in the field.20The promise of editing machineries is, however, highly dependent on the efficacy and safety of the nanosized delivery systems designed for their transport, protection, and release at the target site where they can exert their effect, especially for in vivo settings.19,21–25 Notably, contrary to the delivery of classical gene therapies, e.g., with small interfering RNA (siRNA), messenger RNA (mRNA), or antisense oligonucleotides (ASOs), gene editing tools generally have larger cargo sizes that require optimization for an effective packaging into the delivery system and that potentially impact adequate cytosolic trafficking to the nucleus.21,22,26 Moreover, different delivery challenges arise depending on the chosen gene editing format.6 For instance, the large size of the plasmid DNA (pDNA) encoding Cas9 and sgRNA may hinder the encapsulation efficiency, while requiring entry to the nucleus for transcription and translation.27 On the other hand, Cas9 mRNA and sgRNA formats have a shorter half-life and reduced off-target effects, solely requiring to be delivered into the cytoplasm.28 Finally, the Cas9/sgRNA ribonucleoprotein (RNP) format significantly enhances the editing efficiency and specificity, with a rapid action.27 Nevertheless, the potential immunogenicity and large size of Cas9, along with the denaturation risk during formulation of delivery systems can limit the delivery of RNPs.28 Going forward, although viral vectors, i.e., adenoviruses, retroviruses, and adeno-associated viruses, are clinically relevant gene delivery platforms, shortcomings such as adverse immune responses, off-target effects, and limited payload capacity are still to be fully addressed.21,22,27,28 Hence, alternative nonviral nanosized delivery systems have attracted much attention in the field, such as polymeric, lipid, silica, and gold nanoparticles (NPs).6,17,19,22,28,29 Such nanocarriers must proficiently traverse multiple extracellular and intracellular physiological barriers to reach the target cells, being required to possess specific features, including: (i) robust stability in the bloodstream,21,24 (ii) effective cellular internalization,21,27 and (iii) endo/lysosomal escape ability for intracellular release of the genetic payload, which is then required to translocate into the nucleus for genome editing.21,27,30 Moreover, strategies such as surface functionalization, fine-tuning of the physicochemical features of NPs (e.g., size, surface charge, shape), and stimuli-responsiveness have been widely explored to augment the delivery specificity for lower off-target gene editing repercussions.19,24,31–33 From all systems, lipid nanoparticles (LNPs) currently represent the most clinically established class, given their relatively low immunogenicity and high release of the editing machinery in the intracellular milieu.17,28,34–41 Notably, the FDA approval of a siRNA lipid gene therapy (Patisiran™) in 2018 and two mRNA COVID-19 lipid vaccines (BNT162b/COMIRNATY® from Pfizer-BioNTech and mRNA-1273/SPIKEVAX® from ModernaTx), in 2021 and 2022, has prompted major developments. In fact, clinical trials with the first CRISPR-based medicine (NTLA-2001), that is to be systemically administered leveraging lipid carriers, are currently ongoing.17,28,42–45 On the other hand, polymeric nanoparticles have been widely explored for gene therapy, given their exquisite tunability, reproducibility and relatively high transfection capabilities.17,21,22,46–51 Nonetheless, several parameters have hindered their preclinical advancement, particularly the trade-offs between a highly efficient gene transfection associated with a higher cytotoxic profile and immunogenicity concerns.46,47,52–54 In particular, such challenges hinder the translational potential of nonviral-based gene-edited hard-to-transfect cells, such as primary, immune, and stem cells to address clinical needs.55,56 Seeking an alternative to standalone systems, lipid–polymer hybrid nanoparticles (LPHNs) have recently emerged as a novel class of delivery platforms, combining the most valuable features of both lipids and polymers, with the aim to ultimately display an enhanced cargo encapsulation, biological stability, transfection efficacy, and biocompatibility.53,54,57–61
In the last decade, such hybrid multifaceted carriers have been mainly leveraged for the delivery of hydrophobic drugs, cancer therapies and biomedical imaging.53,57,58 Focusing on the gene delivery domain, LPHNs have been widely explored as platforms for transporting and delivering transient nucleic acid therapeutics (e.g., siRNA, mRNA), including in a recently developed COVID-19 mRNA vaccine candidate (SW0123).42,62–74 Considering their unique capabilities, innovative LPHNs have also begun to be explored for the delivery of gene editing machineries, with specific formulations showing highly promising therapeutical outcomes.75–78
Gathering this potential, herein we aim to review emerging lipid–polymer hybrid nanosystems for the delivery of CRISPR/Cas9-based genome editing components to address the current challenges faced by standalone polymer- and lipid-based vehicles. Major polymer and lipid design blueprints and their influence on the transport efficiency, cellular uptake, intracellular cargo release, biocompatibility and overall gene editing performance are discussed in light of recent advancements. Moreover, major targeting moieties exploited for cell- and organ-specific delivery, including surface-attached targeting ligands and spatiotemporal stimuli-responsiveness, are also highlighted in an attempt to open discussion on their relevance in enhancing the specificity of such carriers. As lipid–polymer hybrid systems are still in their infancy, major challenges and potential future directions are critically addressed aiming at fueling discussion and bringing novel insights to expand the potential of such nanovehicles. Hopefully, future developments on these hybrid systems will pave the way for a new generation of nonviral gene editing platforms with increased efficacy, safety and rapid translation to a clinical setting.
2. Lipid–polymer hybrids: design blueprints
The design of emerging lipid–polymer hybrids for gene editing mainly leverages the strategic combination of lipid and polymeric materials that are conventionally explored as standalone components of nanoparticles. To date, a broad scope of polymeric and lipid components has been harnessed to augment: (i) biocompatibility, (ii) biological stability, (iii) genetic payload encapsulation and protection, (iv) transport and targeting specificity, (v) cellular uptake, and (vi) intracellular trafficking (e.g., endo/lysosomal escape) into the cytoplasmatic milieu or nuclear compartment. Considering the diversity of such toolsets, the major polymer and lipid building blocks that have been exploited to date so as to achieve optimal gene editing via CRISPR/Cas9 machinery are discussed in the following sections, along with major approaches for targeted delivery into the desired tissues or cells (Fig. 1).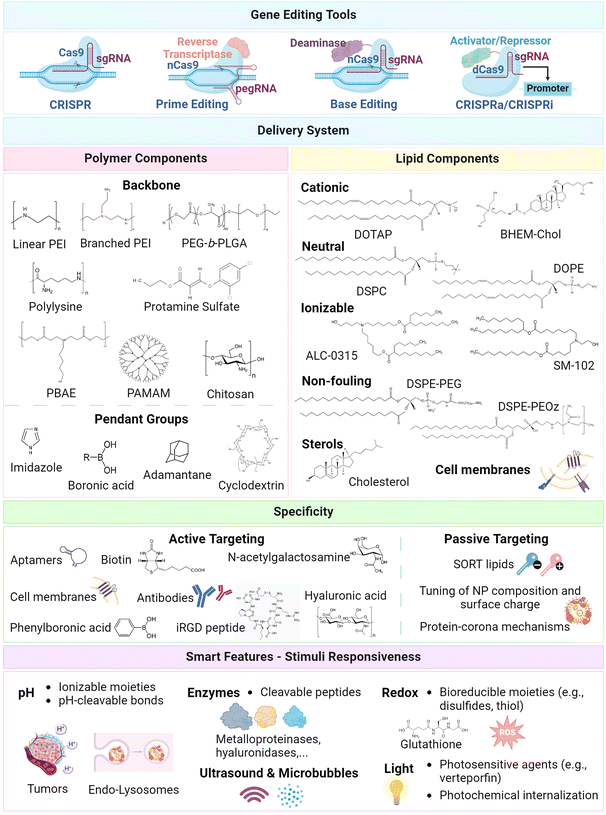 | ||
| Fig. 1 Schematic illustration of major design blueprints harnessed for design of lipid and polymer-based nanoparticles for gene editing. Major gene editing tools include CRISPR, base editing, prime editing, along with CRISPRa and CRISPRi for epigenome editing.2,5,18,79 Polymer components harnessed in delivery systems: (i) backbone: linear and branched PEI (poly(ethylenimine)),80,81 PEG-b-PLGA (poly(ethylene glycol)-block-poly(lactide-co-glycolide)),63,82 polylysine,63,83 PBAE (poly(beta-amino ester)),84,85 PAMAM (poly(amidoamine)),86,87 chitosan,88 protamine sulfate;89–91 pendant groups: imidazole,92,93 boronic acid,86,94 adamantane,87,95,96 cyclodextrin.87,95,97 Lipid components:28,39,42,98,99 cationic lipids DOTAP (1,2-dioleoyl-3-trimethylammonium-propane), BHEM-Chol (N,N-bis(2-hydroxyethyl)-N-methyl-N-(2-cholesteryloxycarbonyl aminoethyl) ammonium bromide); neutral lipids DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine), and DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine); ionizable lipids ALC-0315 ((4-hydroxybutyl)azanediyl)bis(hexane-6,1-diyl)bis(2-hexyldecanoate), SM-102 (heptadecan-9-yl 8-((2-hydroxyethyl)(6-oxo-6-(undecyloxy)hexyl)amino) octanoate); non-fouling components DSPE-PEG (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-poly(ethylene glycol)) lipids, and DSPE-PEOz (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-poly(2-ethyl-2-oxazoline)) lipids; cholesterol; biomimetic cell membranes.100–102 Major moieties harnessed to enhance the specificity of gene editing delivery systems: (i) active targeting approaches with targeting ligands, including aptamers,89,90 antibodies,103,104N-acetylgalactosamine,105 hyaluronic acid,106,107 phenylboronic acid,86 iRGD peptide,76,108 biotin,109 and biomimetic cell membranes;100 (ii) passive approaches relying on the fine-tuning of the nanoparticle composition and surface charge,110,111 SORT lipids,112,113 and protein-corona mechanisms.24,33,112 Major moieties explored for smart vehicles include pH-,83,114 enzyme-,115,116 redox-,48,117 light-,118,119 and ultrasound120-stimuli strategies. Created with Biorender.com. | ||
2.1. Polymer components for optimal CRISPR genome editing
A vast toolset of synthetic and natural polymeric materials has been explored in the gene editing avenue to date. Additionally, new moieties can be further incorporated in the backbone of polymers for augmenting their performance, specifically either to enhance genetic cargo encapsulation (e.g., RNPs versus plasmid/mRNA format), biodegradability, intracellular cargo release, and overall gene editing efficiency.21,27,30,46,49 Several aspects can influence the overall performance of polymer components, such as charge density, chemical and topological structure, molecular weight, degree of polymerization, and the presence of different functional groups. The fine-tuning of such properties is crucial to maximize the applicability of polymeric building blocks, with extensive efforts being put into finding a fine balance between cell-selective features, an enhanced gene transfection efficiency and a low cytotoxicity profile.63 Moreover, the selection criteria for harnessing such polymeric components in hybrid designs may include factors such as the: (i) type of gene editing cargo, (ii) cyto/biocompatibility and (iii) overall genome editing performance.Owing to their cationic nature, PBAEs have been widely harnessed to formulate gene editing polyplexes, bearing amine groups and degradable ester bonds.63,85,128–130 These are widely known for their facile synthesis, commonly via Michael Addition polymerization by reacting amine monomers with diacrylates, enabling researchers to generate combinatorial libraries of PBAEs for structure–activity relationship experiments.49,131 Interestingly, such polycation has been widely explored in lipid–polymer conjugates for mRNA delivery.64,132–134 Moreover, hyperbranched PBAEs have shown an excellent performance for cargo complexation and endosomal escape, owing to their superior amine content.135,136 Also, combinatorial screening of thio-ester hyperbranched PBAEs revealed that the incorporation of thiols dramatically increased the utility of the lead candidate P76 polymer for the delivery of CRISPR-based therapies.84 Nonetheless, as Michael Addition polymerizations may lead to a wide polydispersity, e.g., due to elongation of reaction times, the clinical application of PBAEs can be compromised; thus, the implementation of more controllable methodologies could be beneficial.137
Other systems based on positively charged dendrimers, such as polyamidoamine (PAMAM), and poly(propylene imine) (PPI) constitute well-defined synthetic polymeric materials with low polydispersity and high uniformity, given their synthesis in a stepwise manner with iterative generation of branching structures.22,29,86,87,138,139 In particular, PAMAM dendrimers have been shown to form highly stable polyplexes through electrostatic interactions between their primary amines and negatively charged nucleic acids. Moreover, the tertiary amines of PAMAM additionally aid in the endosomal escape step.63,69 Higher generations of dendrimers show an enhanced gene transfection efficiency, yet at the expense of higher cytotoxic profiles due to the excess cationic charges given the higher density of amine groups.47
Finally, despite not possessing cationic amine groups, the polyester poly(lactic-co-glycolic acid) (PLGA) is widely used in the drug delivery field, being one of the few polymers currently approved by the for human administration by the Food and Drug Administration (FDA), given its biodegradability via hydrolysable ester linkages, structural integrity, biocompatibility, and ease of functionalization.46,52,81,140,141 Nonetheless, in the gene delivery space, its hydrophobic nature and the subsequent inefficient encapsulation of the hydrophilic nucleic acids have hindered its application to formulate standalone PLGA polymeric nanoparticles.63 Conversely, in LPHNs, the combination of PLGA, or the block copolymer poly(ethylene glycol)-block-poly(lactide-co-glycolide) (PEG-b-PLGA) with lipid moieties can promote highly efficient encapsulation, while ensuring more rigid, biocompatible and biodegradable cores, all of which are highly attractive characteristics for the translation of gene editing delivery systems.49,52,63,81,82,142 In addition, other amphiphilic copolymers have been harnessed for the delivery of CRISPR payloads, including the biocompatible, FDA-approved, Pluronic F127, which aids in promoting stable DNA/polycation complexes.143,144
In addition, phenylboronic acid (PBA) can be harnessed as a pendant group to enhance the complexation of nucleic acids, via hydrophobic interactions, having been widely conjugated with low molecular weight PEI.75,86 In essence, these contain boronate groups with high specificity for the vicinal diols groups found in nucleic acids, thus maximizing the polyplex stability.149 Also, PBA moieties have been reported to enhance endosomal escape, by destabilizing the endosome via hydrophobic interactions, and subsequently binding to cytoplasmic adenosine triphosphate (ATP), thus triggering plasmid unpackaging in the cytosol.94
Moreover, adding to electrostatic-based polyplexes, alternative supramolecular chemistries can be explored, namely host–guest chemistries, which have attracted wide attention in drug delivery.47,76 In particular, the biocompatible cyclic oligosaccharides cyclodextrins (CDs) are widely popular host molecules that possess cavities that tightly encapsulate guest molecules, such as Adamantane (Ad), via noncovalent interactions, showing promise in PEI- and PAMAM-based CD–Ad complexes for gene editing.47,87,95,96,138,150 Notably, such chemistry highly contributes to more stable polyplexes.95,96 Moreover, as CD can reduce the charge density, PEI-β-cyclodextrin has been shown to be less cytotoxic, while also mimicking the desirable high transfection efficacy of high-molecular-weight PEI.63,97
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) could efficiently deliver RNPs, emphasizing how advantageous polymeric tunability can be, compared with lipid-based systems.48 Moreover, carboxylated hyperbranched PBAEs promote hydrogen bonding and hydrophobic interplays that enhance polymer–protein interactions, maximizing transfection efficiency.85 Also, anionic polymers such as glutamic acid,151 and poly(propylacrylic acid) (PPAA)152 can stabilize RNPs and potentiate endosome escape of Cas9 nucleases. Moreover, amine-terminated PAMAMs functionalized with PBA pendant groups have achieved unprecedented endosomal escape and cytosolic delivery of CRISPR RNPs, in which PBA could bind with the cationic moieties present in Cas9, via cation–π and nitrogen–boronate interactions.86 Also, guanidinated and fluorinated polymers, either independently or in combination, can promote an enhanced adherence of Cas9, by establishing strong hydrogen bonds and salt bridges between their amides and oxyanions, forming stable nanocomplexes, and promoting fusion with endosome membranes, respectively.93,95 Leveraging the above discussed moieties, universal polymeric-based nanoplatforms have been rationally designed to efficiently condense and deliver all the different formats of CRISPR.92,93,96 In addition, amphiphilic peptide-based materials have been explored for cellular uptake of RNPs, which interact mainly via non-covalent ionic interactions with the cargo.153–157 For generating these materials, solid-phase peptide synthesis is highly valuable as it enables the generation of sequence-defined structures with suitable chemical precision and versatility, such as fluorinated amphiphilic xenopeptides that have shown potent cell internalization of RNPs and endosomal escape.156 Moreover, peptides comprising both cell-penetrating and endosomal leakage domains have been designed to further improve delivery of RNPs in hard-to-transfect human cells.155
1) could efficiently deliver RNPs, emphasizing how advantageous polymeric tunability can be, compared with lipid-based systems.48 Moreover, carboxylated hyperbranched PBAEs promote hydrogen bonding and hydrophobic interplays that enhance polymer–protein interactions, maximizing transfection efficiency.85 Also, anionic polymers such as glutamic acid,151 and poly(propylacrylic acid) (PPAA)152 can stabilize RNPs and potentiate endosome escape of Cas9 nucleases. Moreover, amine-terminated PAMAMs functionalized with PBA pendant groups have achieved unprecedented endosomal escape and cytosolic delivery of CRISPR RNPs, in which PBA could bind with the cationic moieties present in Cas9, via cation–π and nitrogen–boronate interactions.86 Also, guanidinated and fluorinated polymers, either independently or in combination, can promote an enhanced adherence of Cas9, by establishing strong hydrogen bonds and salt bridges between their amides and oxyanions, forming stable nanocomplexes, and promoting fusion with endosome membranes, respectively.93,95 Leveraging the above discussed moieties, universal polymeric-based nanoplatforms have been rationally designed to efficiently condense and deliver all the different formats of CRISPR.92,93,96 In addition, amphiphilic peptide-based materials have been explored for cellular uptake of RNPs, which interact mainly via non-covalent ionic interactions with the cargo.153–157 For generating these materials, solid-phase peptide synthesis is highly valuable as it enables the generation of sequence-defined structures with suitable chemical precision and versatility, such as fluorinated amphiphilic xenopeptides that have shown potent cell internalization of RNPs and endosomal escape.156 Moreover, peptides comprising both cell-penetrating and endosomal leakage domains have been designed to further improve delivery of RNPs in hard-to-transfect human cells.155
2.2. Lipid component features for optimal CRISPR genome editing
In recent years, lipid components have been broadly scrutinized towards the optimization of gene editing delivery systems with an ideal biocompatibility and performance, bringing a new horizon to lipid–polymer hybrid nanocarriers. In this way, a diverse cocktail of amphiphilic lipid molecules has been used, generally containing three domains, e.g., a polar head group, hydrophobic tail, and a linker between the two domains.39 In essence, permanently or ionizable cationic lipids, neutral helper lipids, and cholesterol, along with lipids conjugated with non-fouling components can be explored.28,46,53,98Permanently positively charged cationic lipids, such as 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA), possess a strong cationic quaternary ammonium group.6 Such cationic lipids mainly promote: (i) the complexation of anionic genetic payloads, via electrostatic interactions, forming lipoplexes, and (ii) an enhanced cellular uptake, by electrostatically interacting with the negatively charged cell membranes.27,28,98,158 Also, the cationic N,N-bis(2-hydroxyethyl)-N-methyl-N-(2-cholesteryloxycarbonyl aminoethyl) ammonium bromide (BHEM-Chol) was derived from introducing hydroxyl groups to 3β-[N-(N′,N′-dimethylaminoethane)-carbamoyl]cholesterol (DC-Chol) to improve fusion with cellular membranes.39 Moreover, DOTAP has been shown to increase repulsion between LPHNs, avoiding nanoparticle aggregation, thus enhancing colloidal stability.39,52,64,98,140 Nevertheless, these can lead to an undesired cytotoxicity, e.g., by interacting with negatively charged serum proteins and cell membranes, showing a limited applicability for in vivo settings, thus prompting extensive research on ionizable cationic lipids.22,28,29,39,99 Generally, bearing tertiary amine headgroups with a pKa below the physiological pH 7.0, ionizable lipids: (i) are positively charged at acidic conditions, complexing with gene editing payloads, (ii) become neutral during NP transport in the blood circulation, maintaining physiological stability, and finally (iii) within acidic endosomes, ionizable lipids re-protonate and significantly favor the endosomal escape for intracellular cargo release, by aiding in the transition from a planar bilayer structure in endosomal membranes to a more hexagonal-like structure, thus triggering membrane disruption.24,29,99,140 Also, these have been shown to play key roles as adjuvants for the tolerability and low immunogenicity of LNPs.159,160 Clinically relevant ionizable lipids leverage (6Z,9Z,28Z,31Z)-heptatriaconta-6,9,28,31-tetraen-19-yl 4-(dimethylamino) butanoate (DLin-MC3-DMA; MC3), heptadecan-9-yl 8-((2-hydroxyethyl)(6-oxo-6-(undecyloxy)hexyl)amino) octanoate (Lipid H (SM-102)), and ((4-hydroxybutyl)azanediyl)bis(hexane-6,1-diyl)bis(2-hexyldecanoate) (ALC-0315), included in the FDA-approved Onpattro siRNA drug, and mRNA-1273 and BNT162b COVID-19 vaccines, respectively.28,39,42,99 In contrast to MC3, biodegradable SM-102 and ALC-0315 leverage hydrolysable ester motifs, subsequently showing superior in vivo delivery efficiency and pharmacokinetics.39,161 These have been included in gene editing nanoplatforms, with a myriad of novel ionizable lipids or lipidoids designed so far, including cKK-E12,111,162 LP-01,40 TCL053,160 8-O14B,163 BAMEA-O16B,117 5A2-SC8,37 FTT5,164 and RCB-4–8.165 Notably, research has comprehensively focused on combinatorial libraries of ionizable lipids generally by high-throughput Michael Addition reactions to draw correlations between their chemical structure and activity, as amine headgroups, the length/number and unsaturation of hydrophobic lipid tails and the linkers between these two domains highly influence the final performance.161,163,165–168 For instance, unsaturated and multi-tail ionizable lipids have been correlated with an enhanced endosomal escape.99 Moreover, biodegradable moieties, namely ester, carbonate, or disulfide bonds (see section 2.3.3) are degradable in intracellular environments, which is advantageous for minimizing NP toxicity and accelerating intracellular release of the gene editing machinery.28,39,99,117,167,169 Also, ionizable polymer–lipids, leveraging cationic polymers, such as the lipomer 7C1, have been used for the delivery of siRNA and gene editing machineries in various tissues, including in non-human primates, with negligible toxicity.170–173 Moreover, dendrimer ionizable lipids possessing highly branched tails have also been explored.28,174
To further enhance the stability of lipid formulations for both long-term storage and in vivo circulation, the incorporation of non-cationic charged lipid moieties is also beneficial, namely with phospholipid-based zwitterionic helper lipids, e.g., phosphatidylcholines and phosphatidylethanolamines.27,98,175 As reported in stand-alone LNPs, the degree of phospholipid carbon tail unsaturation and amine head group greatly influenced their performance.158 For instance, the fusogenic 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) contains two unsaturated tails with a conical shape, which promote the adoption of an inverted hexagonal structure at acidic endosomes, destabilizing endosomal membranes and aiding in the endosomal escape step.39,41,158,168,169,176,177 On the other hand, neutral lipids with cylindrical-shaped tails, such as 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), promote highly stable lipid bilayers, significantly stabilizing the structure of nanoparticles, similarly to 1,2-dioctadecanoyl-sn-glycero-3-phosphocholine (DSPC) used in the two lipid COVID-19 mRNA vaccines.39,98 Alternatively, the natural adjuvant lecithin can be harnessed, as it is essentially a mixture of phosphatidylcholines, phosphatidylethanolamines, and phosphatidylserines, endowing the lipid shell with biocompatible neutral and negative charges.178 Zwitterionic amino lipids (ZALs) have also been developed to expand the toolset of available lipid components, combining the properties of zwitterionic and cationic lipids by incorporating a zwitterionic sulfobetaine head group and an amine-rich linker region.179 Moreover, in comparison with the extensively studied cationic ionizable lipids, neutral lipids have not received such attention, as these are not chemically tunable and contain an irreversible zwitterion.175 In this way, recently emerging multi-tailed ionizable phospholipids (iPhos) have been designed by integrating the advantages of ionizable amines with the fusogenic behaviour of phospholipids, ultimately synergistically maximizing the endosomal escape step for augmented gene editing.175
Going further, the naturally occurring cholesterol is widely harnessed in lipid carries, as its hydrophobic moieties promote a high colloidal stability and mechanical rigidity.28,46,98,168 Simultaneously, these aid in the membrane fusion process for efficient cellular uptake and dramatically reduce the amount of potential surface-bound proteins.158,180 In addition, although less explored, optimization of the cholesterol structure has been explored as an alternative approach for augmenting delivery or the endosomal escape step, e.g., with phytosterols (C-24 alkyl cholesterol analogues).180
Moreover, as the systemic delivery of nanocarriers can be particularly challenging owing to the risk of opsonization and rapid clearance in biological fluids, a crucial design step in LPHNs is to ensure a stealth coating to minimize interactions with serum proteins.47,53 As lipid moieties are generally designed to be situated in the outermost part of LPHNs, and subsequently in more contact with the biological environment, these can be strategically conjugated with stabilizing non-fouling materials such as the hydrophilic polymer poly(ethylene glycol) (PEG).22,28,46,47,181 By forming hydrogen bonds with serum water, PEG-lipids (e.g., C18 or C14 lipids-PEG, ceramide-PEG41) create a hydrated shell around the nanoparticle, reducing interactions with biological components and prolonging circulation time for a more efficient gene delivery.46,47 Looking more closely, PEG moieties are usually conjugated with hydrophobic lipid anchors, such as phosphatidylethanolamines.28,46,53,158 The properties of PEGylated lipids can be controlled by tuning both the molar ratio and the length of both the PEG chain and the lipid tail. Notably, intermediately lengthened PEG chains, such as those with a molecular weight of 2000 Da, are more widely used, as these provide a good compromise between an increased half-life in the bloodstream and an efficient gene delivery.158 Moreover, longer fatty acid chains, such as 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE, C18), promote tightly packed and stable hydrophobic PEGylated layers for efficient inhibition of unwanted protein adsorption compared with C14 lipids, e.g., dimyristoyl-glycerol (DMG, C14).158 Nevertheless, denser PEGylated layers can somewhat hinder the endosomal escape or the nanoparticle cellular uptake, for instance, by interfering in the endogenous targeting that may depend on the interaction with specific plasma proteins or receptors on the cell surface, a paradox known as the “PEG dilemma”.158 In addition to such non-fouling behavior, PEG molecules play a key role as colloidal stabilizers by providing steric repulsive forces that minimize particle aggregation during formulation and storage.28,158 This also helps to ensure stable nanoparticle suspensions with a controlled particle size.28,158 Despite such advantages, PEG moieties can be slightly immunogenic, stimulating the development of anti-PEG antibodies and inducing the rapid clearance of nanoparticles – the accelerated blood clearance phenomenon – after multiple administrations of PEGylated NPs.158 Hence, alternative non-fouling components such as poly(2-ethyl-2-oxazoline) (PEOz)-lipid hybrids have been explored, being similarly able to promote negligible interactions with plasma proteins, and potentially showing higher hemocompatibility and less cytotoxicity than PEG-lipid carriers.75,182 Poly(sarcosine) (PSar) has additionally been shown to promote longer in vivo circulation of lipid nanoparticles compared with the PEG coating, constituting an interesting alternative to the more conventionally used PEGylated carriers.183,184
Finally, biomimetic cell membrane coating technologies have emerged as novel strategies to camouflage polymeric nanoparticles owing to the superior properties of such natural lipid-rich structures, e.g. red blood cell membranes, cancer cell membranes, or bacterial vesicles.21,100,102,185 Such structures bring multiple advantages, including inherently circumventing immunogenicity issues and in vivo immune clearance due to the presence of antigen retention.24,91,185
2.3. Targeting features for optimal CRISPR genome editing
Upon systemic delivery of nanosized carriers, these generally have the tendency to accumulate in the liver, spleen, or lung tissues, which can hinder the selective delivery of gene editing apparatus for in vivo treatment of various diseases.27 Moreover, the challenging extracellular barriers that carriers need to traverse to reach the target cell may reduce the gene editing efficiency. Markedly, the translation of gene editing platforms requires the optimization of vehicles towards on-target sites to maximize the efficiency of such tools, while minimizing any unintended off-target gene editing effects in the wrong cells or organs, which can for instance induce harmful mutational outcomes and immunogenic responses.19 In this way, several strategies have been explored in lipid- and polymer-based vehicles, including: (i) surface functionalization approaches, leveraging surface attached-synthetic and naturally derived ligands, along with biomimetic structures, (ii) passive-targeting approaches, which rely on protein corona–NP interactions modulated by the physicochemical properties of NPs, as well as (iii) exogenous and endogenous stimuli-responsive strategies for spatiotemporal control over the delivery.For instance, not only do short cell-penetrating peptides (CPPs) possess an extremely high selectivity for target receptors, but their low molecular weight and inherent tunability also makes these highly attractive targeting building blocks.21 Examples of explored peptides include TAT-NLS89 and T22-NLS90 for combinatorial cell penetrating and nuclear translocation functionalities; the internalizing RGD (iRGD),76,108i.e., a modified derivative of Arg–Gly–Asp peptide, for integrin-iRGD cell-based internalization; and angiopep-2187 to enhance penetration of the blood–brain barrier (BBB). Moreover, the well-known binding affinity of monoclonal antibodies has been harnessed in various nanocarriers and can also increase the circulation time of NPs. For instance, the intercellular adhesion molecule-1 (ICAM-1) antibody has been harnessed for specific binding of tumor cells.104 Moreover, an alternative to typical chemical conjugations, a customizable platform has been developed for antibody-targeted cell-specific delivery, leveraging membrane-anchored lipoproteins – ASSETs – that interact with the antibody crystallizable fragment (Fc) domain.103,188 Looking for lower-cost alternatives, aptamers have emerged as novel “chemical antibodies”, leveraging short single-stranded oligonucleotides that form secondary and tertiary structures with high binding affinity to physiological targets.19,21,27 Also, these can penetrate tumors more effectively compared with antibodies, for instance with AS141189,91,189 and MUC1 aptamers.27,90 Moreover, other ligands have been harnessed for active targeting, including biotin,109 folic acid,190 phenylboronic acid-functionalized lipids,191 galactose,192 and N-acetylgalactosamine,105 for base editing hepatic interventions in non-human primates.
Finally, bio-derived compounds have emerged as promising targeting alternatives to synthetic ligands. Hyaluronic acid has been extensively explored in gene editing nanoplatforms, as it possesses inherent specificity to various surface receptors, including CD4489,106,107,116 – overexpressed in cancer cells – or even lymphatic vessel endothelial hyaluronan receptor 1 (LVE-1).21 Moreover, membrane-coating technologies can be considered as biomimetic functionalization approaches, as these enable inherent tissue and cell targeting properties, most commonly towards cancer cells due to the inherent homologous adhesion property of cancer cells membranes.21,33,193,194
In essence, passive strategies like these have been widely interpreted to be associated with protein corona-based mechanisms, in which serum proteins adsorb on the NP surface, according with the NP composition and surface charge.24,33 Ultimately, these can determine the in vivo interaction of NPs with living cells and tissues, influencing tissue- and cell-targeting. For instance, key corona proteins: (i) ApoE, albumin, (ii) fibrinogen β/γ chain and vitronectin, and (iii) β2-GPI have been correlated with liver, lung, and spleen targeting, respectively.33,110,112 Nonetheless, the fundamental understanding of how protein corona–nanoparticle interactions impact in vivo targeting still requires further research.24,33,112
Also, redox-responsive moieties have been gaining wide attention in the field. In essence, intracellular environments are generally reductive, i.e., contain higher concentrations of glutathione (GSH) reductase, along with cancer cells being generally correlated with higher levels of reactive oxygen species (ROS).27,100,196,201 In this way, disulfide bonds and thiol groups, as well as thioether and diselenide bonds can be harnessed to enhance release of the payload into the cytosol.27 Notably, not only aiming to minimize cytotoxicity, but also to augment the intracellular cargo release, disulfide-containing bioreducible ionizable lipids have been extensively researched in LNPs, enhancing gene editing efficiency.21,27,99,117,163,167 Moreover, a series of bioreducible polymeric-based carriers for the delivery of different formats of CRISPR has been designed with incorporation of disulfide bonds to augment cargo release, including reducible branched ester-amine quadpolymers (rBEAQs),202 bioreducible host–guest supramolecular polyplexes,95,96 and others.48,92,93,203 Also, as enzymes such as matrix metalloproteinases (MMPs) and hyaluronidase (HAse) are generally upregulated in tumors, these have enabled the development of enzyme-responsive gene editing platforms. For instance, MMP-cleavable peptides can be conjugated to PEG moieties, enabling the exposure of polyplexes at tumors for enhanced cellular internalization, while HAses can promote endosomal escape of polyplexes.92,115,116,204,205
On the other hand, exogenous stimuli enable researchers to have high spatiotemporal control over gene delivery nanosystems, offering novel ways to ensure the safety and robustness of in vivo gene editing interventions in a remote manner.21 In this way, new-generation or smart liposome shells have been designed by the incorporation of photothermal agents within liposomes (e.g., IRDye 88CW,206 verteporfin118) to induce destabilization of liposomes upon irradiation, and ultimately enhance endosomal escape and intracellular release of CRISPR payloads in a controllable manner. Moreover, photosensitizers (e.g., chlorin e6,80,122 pheophorbide a119) can be harnessed to generate ROS inside lysosomes upon light stimuli. In this line, the inclusion of ROS-sensitive thioketal moieties in the backbone of polymers has been widely explored, enabling the disassembly of polyplexes and precise control over cargo release to the cytoplasm upon light irradiation.119,122,207 Additionally, ultrasound stimuli can reach deeper tissues in a noninvasive manner as compared with light-based triggers. Essentially, as commonly used in sonodynamic therapy, sonosensitizer molecules (e.g., hematoporphyrin monomethyl ether (HMME)) can be incorporated within liposome shells, as these generate ROS upon ultrasound irradiation, thus triggering intracellular cargo release precisely at targeted locations and minimizing undesired leakages.120 Moreover, ultrasound waves can be harnessed in combination with microbubbles to transiently enhance membrane permeability to enable efficient accumulation of gene editing carriers at the intended tissues.196 Finally, leveraging enzymatic or ROS stimuli, charge-reversal polymers have been exploited in the design of lipid–polymer hybrids which facilitate DNA release upon cellular internalization.67,70,208
3. Technologies for lipid–polymer hybrid formulation
There are several technologies available to formulate nanoplatforms encapsulating genetic payloads. On one hand, lipid-based nanoparticles can be formulated by several methodologies, for instance, ethanol-loading and dilution techniques, thin film hydration, and detergent dialysis.22,27,60 Such techniques have been progressively replaced by rapid-mixing technologies, namely by using microfluidics, in which essentially organic and aqueous phase solutions, containing lipid moieties and the genetic payload, respectively, are introduced in the devices.22 Particularly, chaotic and staggered herringbone mixers have become the gold-standard for LNP assembly, for instance with commercially available mixers, e.g., Nanoassemblr platform (Precision Nanosystems, Vancouver, Canada).27 Such systems enable high control over the mixing process parameters and size of LNPs.22 On the other hand, polymer-based nanoparticles can be generally formulated by methodologies such as nanoprecipitation, impingement jet mixing, emulsification–solvent evaporation, solvent exchange, or microfluidics, although the last is still more prominent in LNPs.31Leveraging such technologies, from an architectural design perspective three major groups of LPHNs have so far been formulated, Fig. 2. On one hand, polymer core–lipid shell nanosystems can be designed based on three main building blocks: (i) an inner polymeric core, condensing the negatively charged cargo (e.g., CRISPR/Cas9 plasmid, Cas9 mRNA/sgRNA, Cas9/sgRNA ribonucleoprotein (RNP)), into a polyplex, along with (ii) an inner biocompatible and protective liposomal shell around the polymeric core, and (iii) an outer lipid shell containing non-fouling components, included so as to minimize the non-specific adsorption of proteins that may affect the efficiency of the delivery.53,75–77
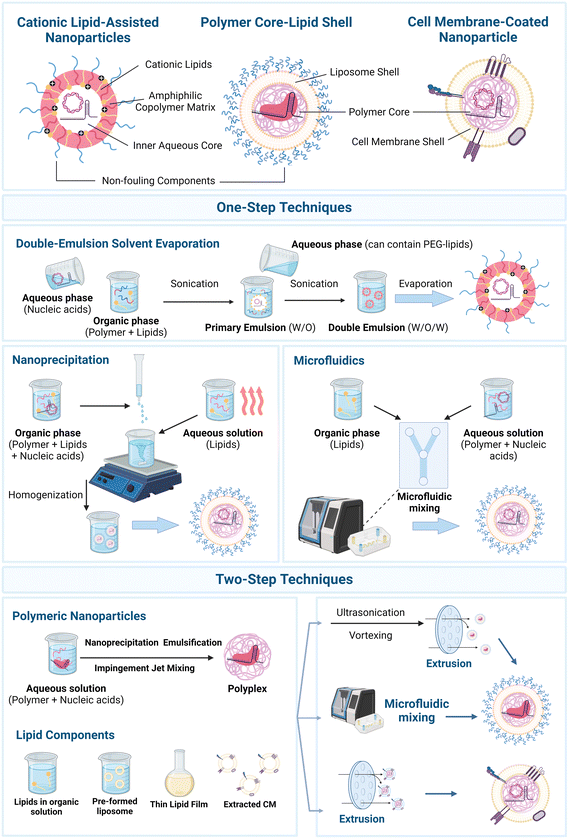 | ||
| Fig. 2 Major groups of lipid–polymer hybrid nanosystems harnessed for CRISPR/Cas9 genome editing applications and formulation technologies. (i) Cationic lipid-assisted nanoparticles (CLANs)78,209,210 are generally constituted by cationic lipids within an amphiphilic copolymer matrix, protecting the gene editing payload, (ii) polymer core–lipid shell hybrid nanoparticles75,76,145 are constituted by an inner polymer core encapsulating the gene editing payload, and further shielded by a liposome shell, while (iii) cell membrane-coated nanoparticles91,100,102 provide a biomimetic camouflage to polyplexes. Major formulation technologies can be divided by (i) one-step techniques, and (ii) two-step techniques: one-step techniques include double-emulsion solvent evaporation,78,82,185 nanoprecipitation,142 and microfluidic mixing.65,73,74 Two-step methodologies encompass the independent formulation of polymer nanoparticles encapsulating the gene payloads (e.g., polyplexes), and subsequent mixing with the lipid components, either by microfluidic mixing62 or extrusion steps.75,76,100,185 Created with Biorender.com. | ||
Moreover, LPHNs named cationic lipid-assisted nanoparticles (CLANs) can be designed by essentially incorporating lipid moieties within an amphiphilic block copolymer to maximize the encapsulation and protection of the genetic payload.82 Also, cell membrane-coating lipid nanoparticles can be formulated, encompassing polyplexes coated by protective biomimetic cell membranes and extracellular vesicles. Such hybrid carriers can be formulated either by: (i) one-step or (ii) two-step methods, Fig. 2.53,57,58,211 In essence, one-step methods enable efficient formulation of particles in a single step, e.g., via emulsification–solvent evaporation, and nanoprecipitation.53,58,211 Emulsification–solvent evaporation can be divided into single or double emulsion methods, in which double emulsion is more widely used for encapsulation of hydrophilic molecules, such as nucleic acids.212 Notably, CLANs are usually formulated by double emulsion solvent evaporation.53,82 In such method, an aqueous solution of nucleic acids is firstly dispersed in an organic solution containing the polymer and lipids, forming a primary water-in-oil (W/O) emulsion. In this step, the cationic lipids tightly self-assemble at the water–oil interface, in which the nucleic acids interact with their hydrophilic headgroups, efficiently encapsulating the genetic cargo into an inner aqueous phase, and avoiding leakages.82 This emulsion is then dispersed in a second aqueous solution, which can contain additional lipids (e.g., PEG-lipids), thus forming a double emulsion water-in-oil-in-water (W/O/W), and the solvent is removed by evaporation, forming hybrid particles.82,213 Despite several advantages, including the low energy input required, this technique still suffers from drawbacks, such as high polydispersity and potential leakages of hydrophilic molecules into the external aqueous phase.53 Aiming to obtain particles with a narrower size distribution, nanoprecipitation brings major advantages, while also enabling automation via microfluidic platforms.53,59,142,211 Nanoprecipitation essentially involves: (i) incubating nucleic acids with the polymer and cationic lipids, in an organic solution, and (ii) adding it drop-wise under vigorous stirring to an aqueous solution containing additional lipids (e.g., PEG-lipids) – previously heated beyond the gel-to-liquid transition temperature of lipids to dissolve these into a dispersed liquid crystalline phase.213 Subsequently, this triggers the precipitation of nanoparticles, and polymer core–lipid shell particles are formed.53,142,211 A few limitations of this technique may involve the potential incomplete mixing of aqueous and organic solutions before precipitation, leading to unevenly small nanoparticles and batch-to-batch variations, thus challenging scale-up processes.60
Going further, two-step methods are based on the independent formulation of: (i) polymeric nanoparticles encapsulating the genetic payloads, e.g., by nanoprecipitation or emulsification–solvent evaporation, and (ii) pre-formed liposomes, or more commonly, dried lipid films.211,213 Finally, both counterparts are mixed, with the lipid shell assembling onto the surface of the polymeric core by electrostatic interactions.53 Looking more closely, the thin film hydration method is widely used to prepare liposome shells, in which the constituent lipids are initially dissolved in organic solvents and further evaporated to yield a dried thin lipid film.27,53,59 Such lipid films are then hydrated with an aqueous solution containing the polymeric nanoparticles, e.g., polyplexes, through vortexing and/or sonication, promoting the encapsulation of the polymeric core within the lipid shell.53 As a final step, to generate monodisperse core–shell particles with a controllable and homogeneous size, membranes with a specific pore size are widely used to extrude LPHNs.53,59 Although two-step methods can be more time-consuming, requiring more resources and being more technically complex than one-step methods, these enable superior control over the separately produced polymeric and lipid components.57 Moreover, to obtain highly homogeneous and monodisperse hybrid nanoparticles, steps of extrusion and/or homogenization are usually performed.53,59,60 Also, physical extrusion is widely exploited to formulate cell membrane-coated polyplexes after initial cell membrane isolation techniques (e.g., freeze-thawing, ultrasonic waves, and homogenization).101 Such extrusion technologies are aided by membranes with a specific pore size that enables a superior size control; nonetheless, optimization for scale-up can be challenging.53,59,60 Lastly, inspired by the widely used microfluidic platforms in LNPs, such cutting-edge and high-throughput technologies can be harnessed to formulate lipid–polymer hybrids.53,62 In essence, such platforms can be used either in: (i) one-step methods, essentially leveraging the principles of nanoprecipitation, or (ii) two-step methods, which can involve the pre-assembly of polymeric nanoparticles encapsulating the genetic payloads, with subsequent mixing with the lipid components within the microfluidic mixers.53 Compared with the previously discussed techniques, microfluidics enables continuous production, allowing for a fine-tuning of formulation process parameters, and high reproducibility, minimizing batch-to-batch variation.22,27,53,59,62 Nonetheless, microfluidic mixing faces several challenges, ranging from clogging issues to the high costs of the equipment.214 Also, most equipment is designed for relatively limited throughputs (mL h−1), compared with clinically relevant production rates (L h−1).27,60,214,215 To address such challenges, the use of parallelized devices can enable high-throughput and reproducible generation of nanoparticles, however always requiring the screening of optimal parameters.214,215 Finally, one relevant aspect to take into consideration for the formulation of LPHNs is the type of gene editing payload intended to be encapsulated. Notably, formulation with CRISPR/Cas9 RNPs is more complex, as organic solvents, acidic/basic conditions, or high temperatures may lead to denaturation and loss of protein activity.28,30,37,216 To counteract this, neutral buffers have been used to preserve the nuclease integrity in microfluidic mixing-based protocols in LNPs, instead of citrate buffer (pH 4.0),37 as well as custom microfluidic devices with 3 inlets to avoid aggregation of RNPs exposed to ethanol.216
4. Advanced lipid–polymer hybrids for CRISPR genome editing machinery
As discussed above, there is a vast toolset of lipid and polymer features currently available, along with formulation technologies that can be exploited to design sophisticated lipid–polymer hybrid vehicles for delivery of genome editing machineries. Notably, emerging LPHNs developed so far have focused on maximizing the packaging, safety profile, and delivery of CRISPR-based genome and epigenome editors. A few synthetic and natural polymers have begun to be harnessed, along with major lipid moieties, Table 1. Moreover, focusing on approaches for cell- and tissue-specific delivery, hybrid carriers have so far been widely designed with surface functionalization moieties, and a few spatiotemporal controllable nanosystems have been developed. Moreover, the vast majority of hybrids have been formulated essentially either by double emulsion or extrusion methodologies. Another aspect to be noted is that, with a few exceptions, the great majority of systems contain a diameter below 200 nm, avoiding rapid clearance from the bloodstream, as well as positive surface charges, enhancing cellular uptake.22,24,28| Hybrid delivery systems composition | Size [nm]; surface charge [mV] | Surface functionalization/targeting | Stimuli-responsivity | In vitro data | In vivo data | Ref. |
|---|---|---|---|---|---|---|
| Abbreviations: Ad, adamantane; Alox12, arachidonate 12-lipoxygenase encoding gene; BAFFR, B-cell activating factor receptor; BAM-TK-TMP, ROS-responsive small-molecule stabilizer for dsCas9 covalently conjugated to oleyl ether-modified poly(ethylene glycol) through a thioketal linker, anchored on the macrophage membrane; BBB, blood–brain barrier; BHEM-Chol, N,N-bis(2-hydroxyethyl)-N-methyl-N-(2-cholesteryloxycarbonyl aminoethyl) ammonium bromide; CD, cyclodextrin; CD40, cluster of differentiation 40; CD80, cluster of differentiation 80; CD86, cluster of differentiation 86; Chol, cholesterol; CLAN, cationic lipid-assisted nanoparticle; CML, chronic myeloid leukemia; cRGD, cyclic arginine–glycine–aspartic acid peptide; DC-chol, 3β-[N-(N′,N′-dimethylaminoethane)-carbamoyl]cholesterol; DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; DOPE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; DOTAP, 1,2-dioleoyl-3-trimethylammonium-propane; DPP-4, dipeptidyl peptidase-4 encoding gene; DSPE-PEG, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-poly(ethylene glycol); DSPE-PEOz, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-poly(2-ethyl-2-oxazoline); eWAT, white adipose tissue; DSS, dextran sulfate sodium; HA, hyaluronic acid; HIF-1α, hypoxia-inducible factor-1 alpha encoding gene; iRGD, internalizing arginine–glycine–aspartic acid peptide; KRAB, Krüppel-associated box repressor; Lcn2, Lipocalin 2 encoding gene; LEX, lexiscan; LHPN, liposome-templated hydrogel nanoparticle; MGMT, (O6-methylguanine-DNA methyltransferase) encoding gene; MHCII, major histocompatibility complex class II; miR-10, microRNA-10b; MTH1, human MuT homolog 1 encoding gene; NE, neutrophil elastase encoding gene; NLRP3, (NLR family pyrin domain containing 3) encoding gene; NP, nanoparticle; Ntn1, netrin-1 encoding gene; OMV, outer membrane vesicle; PBA, phenylboronic acid; PBAE, poly(β-amino ester); PEG, poly(ethylene glycol); PEG-b-PLGA, (poly(ethylene glycol)-block-poly(lactide-co-glycolide)); PEI, polyethyleneimine; PLGA, poly(lactic-co-glycolic acid); PLK1, polo-like kinase 1 encoding gene; PHD2, prolyl hydroxylase domain 2 encoding gene; PTX, paclitaxel; T1D, type-1 diabetes; T2D, type-2 diabetes; TAPP, 5,10,15,20-tetrakis-(4-aminophenyl) porphyrin; TMP, trimethoprim; TMZ, temozolomide; UMMD, ultrasound-mediated microbubble destruction; Ythdf1, YTH N6-methyladenosine RNA binding protein F1 encoding gene. | ||||||
| PEG-b-PLGA; BHEM-Chol | 147.8 nm; 27.6 mV | No | No | Transfection efficiency up to 74.6% in K562 cells after 72 h, in which pCas9/gBCR-ABL-1 achieved a 46.8% knockout efficiency of BCR-ABL fusion gene | Intravenous injection in a CML mouse model resulted in 19.1% and 16.1% knockout of BCR-ABL in the blood and bone marrow, respectively. Mice had significant prolonged survival rate, minimized number of white blood cells and apoptosis of CML cells | 217 |
| PEG-b-PLGA; BHEM-Chol | 129.7 ± 8.1 nm; 18.5 ± 2.8 mV | No | No | Transfection efficiency of 35.9% in primary bone-marrow-derived macrophages, in which pCas9/sgNtn1 resulted in 32.7% knockout of Ntn1 gene | Intravenous injection in T2D mice resulted in >50% downregulation of Ntn1 protein in adipose tissue and reduced inflammatory cytokines, improving glucose tolerance and insulin sensitivity | 210 |
| PEG-b-PLGA/PLGA; BHEM-Chol | 160 ± 6.3 nm; 30.5 ± 3.1 mV | No | No | No in vitro data | Library of 20 CLANs with different PEG density and surface charge was screened for neutrophil targeting | 218 |
| After CLAN optimization, intravenous injection in T2D mice resulted in 19.5% and 27.3% knockout efficiency of NE gene in neutrophils at eWAT and liver, respectively, improving glucose tolerance, insulin sensitivity, and reducing inflammation | ||||||
| PEG-b-PLGA/PLGA; BHEMChol | 151.8 ± 2.4 nm; 12.2 ± 0.7 mV | No | No | pCas9/sgB220 resulted in 29.3% knockout of B220 gene in cultured B cells | Library of CLANs with different PEG density, surface charge and lipid composition was screened for B-cell targeting | 219 |
| Administration of Cas9/sgBAFFR in male DBA/1 mice significantly inhibited rheumatoid arthritis score and ankle diameter, reducing the number of B cells by >50% in lymph nodes | ||||||
| PEG-b-PLGA/PLGA; BHEMChol | 130 nm; 31.5 ± 5.84 mV | No | No | Transfection efficiency of 65.8% in primary bone marrow-derived macrophages | Library of CLANs with different PEG density and surface charge was screened for macrophages targeting | 78 |
| mCas9/sgNLRP3 achieved a dose- and time-dependent NLRP3 gene knockout efficiency of up to 70.2% at 24 h, inhibiting inflammasome activation | Injection in C57BL/6 mice reached 47.1% knockout of NLRP3 gene at 24 h, with low off-target effects. Attenuated systemic and peritoneal inflammation, and improved symptoms of T2D | |||||
| PEG-b-PLGA/PLGA; BHEMChol | 100 nm; 10 mV | No | No | Cellular uptake of >90% in cultured bone-marrow dendritic cells, and transfection efficiency of 26.7% | Model for acute graft rejection was established: back skin of BALB/c mice graft donors was transplanted to C57BL/6 mice recipients | 209 |
| mCas9/gCD40 achieved knockout efficiency of gene CD40 of 25.5% | Intravenous injection into recipient mice after transplantation significantly prolonged graft survival, by decreasing CD40 expression and inhibiting T-cell activation | |||||
| PEG-b-PLGA; BHEMChol | 138 nm; 23 mV | No | No | pCas9/gCD80,86,40/2.5mi efficiently disrupted CD80, CD86, and CD40 (>15%) in bone-marrow dendritic cells, and autoimmune 2.5mi peptide was presented by MHCII on the surfaces of DCs | Intravenous injection of all-in-one nanomedicine in T1D mice significantly reduced incidence of diabetes by >80%, inhibited infiltration, insulitis and inflammation, restoring autoimmune tolerance | 220 |
| Induced tolerogenic phenotype and expansion of Treg and 2.5mi peptide-specific CD4+ T cells | ||||||
| PEG-b-PLGA; OMVs/DOTAP | 180 nm; 26.67 mV | OMVs enabled dendritic cell targeting and immune activity activation | No | OMVs significantly increased internalization (to ∼100%) in primary bone marrow-derived dendritic cells, compared with non-targeted NPs (∼80%) | Tail-based injection in MC38 tumor-bearing mice targeted dendritic cells, triggering CD8+ T-cell-mediated antitumor immunity, and eradicating tumors by 97.72% | 185 |
| pCas9/gYthdf1 resulted in ∼30% reduction of Ythdf1 mRNA level in DC2.4 cells | ||||||
| PLGA; DC-chol/lecithin/DSPE-PEG | 179.6 ± 44.82 nm; −29.6 ± 4.33 mV | cRGD peptide for tumor targeting | Focused ultrasound (FUS) + microbubbles (MBs) for BBB opening | Transfection efficiency of 36.39% in T98G cells after 48 h (>2 times higher than non-targeting NPs) | Intravenous injection in orthotopic T98G glioblastoma-bearing mice inhibited volume of glioblastoma tumor by ∼50% after 42 days, in combination with TMZ, prolonging mouse survival | 142 |
| pCas9/MGMT significantly downregulated MGMT protein expression, re-sensitizing T98G cells to TMZ | ||||||
| DOTAP/Chol/DSPE-PEG; PEI-Ad/PEI-CD | 95 nm; surface charge not reported | In vitro: mHph1 + mHph3 cell-penetration peptides | No | Gene transfection efficiency was 1.3 times higher than with Lipofectamine™ 2000 | Intravenous injection in mice bearing intracranial U87 gliomas inhibited PLK1 protein expression by 60.4% and reduced tumor growth, prolonging survival to 40 days. LEX increased LHNPs accumulation at tumor site | 76 |
| In vivo: iRGD cancer-targeting peptide + LEX to transiently enhance BBB permeability | Delivery of Cas9/minisgPLK1-2 inhibited cell proliferation by 79.3% in U87 cells and 80.2% in GS5 cells | |||||
| Lecithin/Chol/DOGS-NTA-Ni; PEI | 220.2 nm; −5 mV | No | No | Cas9/sgDPP-4 delivery resulted in 67% reduction of DPP-4 mRNA in SNU398 cells | Intravenous injection in T2D mice induced 39% knockout of DPP-4 gene and >50% mRNA expression at the liver, restoring glucose tolerance, insulin resistance, and reducing liver and kidney damage | 178 |
| DSPE-PEOz/lecithin/Chol; PEI-PBA | 118.9 ± 1.8 nm; −31.6 ± 1.80 mV | Binding of PBA to overexpressed sialic acid in tumor cells | pH-responsive PEOz lipids disassembly at acidic pH of ∼6.5 (tumor microenvironment), and pH ∼5.0 (lysosome) + UMMD mediated uptake | 60.13% and 52.31% transfection efficiencies of dCas9-KRAB/sgmiR-10b in 4T1 and MDA-MB-231 cells, respectively | Intravenous injection in 4T1 tumor-bearing mice resulted in significant miR-10b protein repression and reduced number of lung metastatic nodules by ∼84%. UMMD significantly enhanced accumulation at tumor site | 75 |
| DSPE-PEG/Chol/DOPE/DOTAP; protamine/chondroitin sulfate | 156.5 nm; 23.2 mV | No | No | 47.4%, 36.2% and 37.8% transfection efficiencies in A375, PC-3 and MCF-7 cells | Intratumor injection of pCas9/sgPLK-1 into melanoma-bearing mice induced 61.1% downregulation efficiency of PLK-1 protein, reducing >67% of tumor growth | 77 |
| Knockout efficiency of 16.1% in A375 cells (>13 times higher than Lipofectamine™ 2000) | ||||||
| DOPE/DOTAP/Chol/DSPE-PEG-HA; protamine sulfate | 130 nm; 10.4 mV | Hyaluronic acid (HA) for tumor targeting, via HA-CD44 cell surface receptor interaction | No | 80% transfection efficiency of pCas9/sgMTH1 in A549 cells, with 33.1% knockout of MTH1 gene and apoptosis induction rate of 69.4% | Tail vein injection in non-small cell lung cancer-bearing mice induced selective tumor accumulation and low MTH1 protein expression, inhibiting tumor growth by 66.7% and prolonging survival to 65 days | 145 |
| Neoplastic H1299 cell membrane/DSPE-PEG; protamine/Ca2+ | 198 nm; −15 mV | AS1411 aptamers for tumor targeting; neoplastic H1299 cell membrane enabled tumor enrichment and persistent circulation | Light-responsive intracellular cargo release via ROS-generating TAPP photosensitizer | pCas9/sgHIF-1α delivery resulted in 43.3% knockout of HIF-1α gene in H1299 cells | Tail vein injection in H1299 tumor-bearing mice downregulated HIF-1α protein and augmented PTX chemotherapy, causing 38.4% cell apoptosis and reducing primary lung tumors | 91 |
| DOPC/DSPE-PEG-COOH; alginate | 111 nm ± 23; −4.6 ± 3.8 mV | ICAM1 antibody for tumor targeting | No | Delivery of pCas9/sgLcn2 resulted in 80% knockout of Lcn2 gene in MDA-MB-231 and MDA-MB-426 cells | Tail vein injection in MDA-MB-231 tumor-bearing mice induced >81% knockout of Lcn2 gene, suppressing tumor volume by 77% without off-target toxicities | 104 |
| ICAM1 antibody resulted in 2- to 3-fold increase in cell uptake | ||||||
| RAW264.7-derived macrophage membrane; PBAE | 151 nm; ∼−4 mV | Macrophage membrane enabled targeting of inflammatory lesion | ROS-responsive TMP stabilizes dscas9 for conditional activation of editing machinery in inflammatory lesion | ∼40% transfection efficiency in CT26 cells, performing better than Lipofectamine™ 2000 | Intravenous administration in BALB/c mice with DSS-induced colitis induced 14.8% knockout of PHD2 gene (∼4 times higher than without ROS stimuli) with low off-target mutations | 100 |
| Plasmid encoding dsCas9 and sgPHD2 with 29.2% knockout efficiency of PHD2 gene, under H2O2 stimulation | Relieved symptoms of colitis in mice, including loss of body weight and decrease of colon length | |||||
| RAW264.7-derived macrophage membrane; poly(disulfide) (w/cationic diethylenetriamine moieties and guanidyl groups) | −350 nm; ∼−18 mV | Macrophage membrane enabled targeting of inflammatory lesion in liver | Glutathione-triggered poly(disulfide) degradation in intracellular reductive environment | Plasmid encoding Cas editor with synthetic chimeric liver-specific promoter enabled 50% transfection efficiency in HepG2 cells, compared with A549, DU145, SW480 and MCF-7 cells (<5%), at 48 h | Systemic administration in hepatic ischemia-reperfusion injury model mice, successfully knocked-out Alox12 gene (14.3%), alleviating inflammatory responses for treatment of liver damage | 102 |
| Systemic administration in Concanavalin A-induced acute liver injury model mice, disrupted Fas locus (13.2%) and treated liver fibrosis, prolonging survival of mice | ||||||
4.1. PLGA-based lipid–polymer hybrids
Based on the preclinical success of PEG-b-PLGA-based CLANs for systemic siRNA delivery, a series of formulated CLANs has been adapted for efficient delivery of CRISPR/Cas9 genome editing machineries to treat various diseases, Fig. 3 and 4, with negligible cytotoxicities.78,82,209,210,217,218 As previously discussed, the encapsulation of nucleic acids on clinically validated polymers such as the hydrophobic PLGA is highly unsatisfactory, the inclusion of lipid moieties has been demonstrated to increase the encapsulation efficiency, for example from 20.4% to 96.4%.217 Regarding their therapeutical potential, pCas9-loaded CLANs were initially harnessed for the treatment of chronic myeloid leukemia (CML), targeting the BCR-ABL fusion gene – correlated with cell proliferation of myeloid cells and their conversion into CML cells – achieving similar gene editing frequencies to the commercial transfection reagent Lipofectamine™ 2000 (Lipo2000).217 Moreover, such gene disruption in the blood and bone marrow of mice significantly prolonged survival rate up to 100 days, compared with 65 days in the non-treated group.217 Moreover, CLANs have been harnessed for type-2 diabetes (T2D) treatment, by delivering a macrophage-specific promoter-driven pCRISPR/Cas9 (pM330), Fig. 3A.210 Such system promoted an in vitro gene editing efficiency comparable to the widely popular lipid transfection reagent Lipo2000, successfully knocking-out the netrin-1 protein-encoding gene (Ntn1), which is highly involved in T2D disease. Moreover, similarly to the established diabetes drug glyburide, such therapy ameliorated T2D symptoms in vivo, improving glucose tolerance and reducing the inflammatory profile.210 Moreover, as the particle size, surface charge and PEG density influence the drug delivery efficacy, different libraries of CLANs have been screened to modulate the targeting efficiency, based on the modulation of both the surface charge and PEG density.78,218,219 For instance, high surface charges, promoted by the cationic BHEM-Chol, combined with low surface densities of PEG were shown to promote a higher cellular uptake of pCRISPR/Cas9-loaded CLANs to target neutrophils at the epididymal white adipose tissue (eWAT) and the liver of T2D mice, Fig. 3B.218 In the same study, the successful knockout of the neutrophil elastase-encoding gene (NE) further reduced neutrophil infiltration, improving T2D symptoms and increasing anti-inflammatory arginase expression, similarly to the established sivelestat and metformin drugs.218 A similar screening was conducted to maximize the targeting towards B cells, as the dysfunction of such cells often induces autoimmune and inflammatory diseases, Fig. 3C.219 Optimized CLANs had slightly lower PEG surface charges than the previous study in Fig. 3B. In this way, delivery of pCRISPR was able to efficiently disrupt the B220 gene – specifically expressed in B cells – in vitro, and the BAFFR gene – highly important in B-cell survival – in vivo.219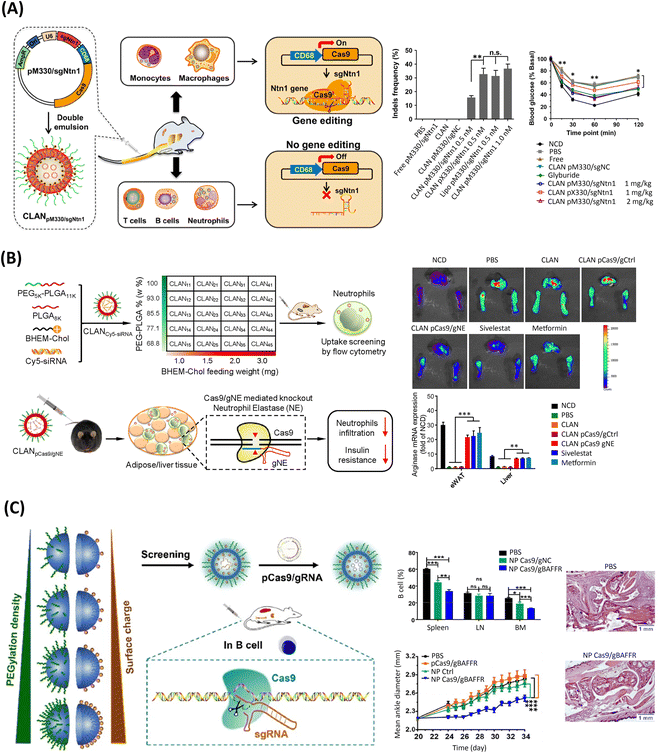 | ||
| Fig. 3 PLGA-based LPHNs for the delivery of genome editing components. (A) Schematics of CLANs for delivery of macrophage-specific promoter-driven Cas9 expression plasmids (pM330) for Ntn1 gene disruption and treatment of T2D. Middle row: gene disruption efficacy in bone-marrow-derived macrophages (BMDMs) based on the indels frequency in the Ntn1 locus taken from T7 Endonuclease I (T7EI) mismatch cleavage assay, after treatment with CLAN(pM330/sgNtn1), CLAN(pX330/sgNtn1), or Lipo(M330/sgNtn1), in vitro. Data shown as the means ± SD (n = 3), **p < 0.01, n.s. p > 0.05. Right row: insulin tolerance test of T2D mice after treatment with glyburide, CLAN(pX330/sgNtn1), or CLAN(pM330/sgNtn1). Data shown as the means ± SD (n = 10), *p < 0.05, **p < 0.01. sgNC: scramble sgRNA (negative control); pX330: plasmid without containing macrophage-specific CD68 promoter; pM330: plasmid containing macrophage-specific CD68 promoter; NCD: healthy control mice. Reprinted with permission from ref. 210. Copyright (2018) American Chemical Society. (B) Schematized screening of CLAN library to encapsulate pCas9/gNE for NE gene knockout and treatment of T2D. Upper right: fluorescence images of the activity of NE protein in eWAT and liver after different treatments in T2D mice. Lower right: mRNA expression of anti-inflammatory arginase in eWAT and liver after CLAN pCas9/gNE treatment. Data shown as the means ± s.d. (n = 3). **p < 0.01, ***p < 0.001. (PBS, unloaded CLAN or CLANpCas9/gCtrl were used as the negative controls, while sivelestat and metformin were used as the positive controls.) Reprinted from ref. 218, Copyright (2018), with permission from Elsevier. (C) Illustration of CLANs screening for delivery of CRISPR-Cas gene editing system for B-cell intervention. Upper middle column: percentage of B cells in the spleen, lymph node, and bone marrow, upon injection of PBS, NP Cas9/gNC, or NP Cas9/gBAFFR in C57BL/6 mice, every 2 days for 12 days. Bars represent the mean ± SEM (n = 4–5 mice per group). Bottom middle column: mean ankle diameter (mm) of mice after treatment with PBS, pCas9/gBAFFR, NP ctrl, or NP Cas9/gBAFFR. Bars represent the mean ± SEM (n = 10 mice per group). Right: histopathological images of the ankle joints of mice, scale bar of 1 mm. Reproduced with permission from Springer Nature, Copyright (2018).219 | ||
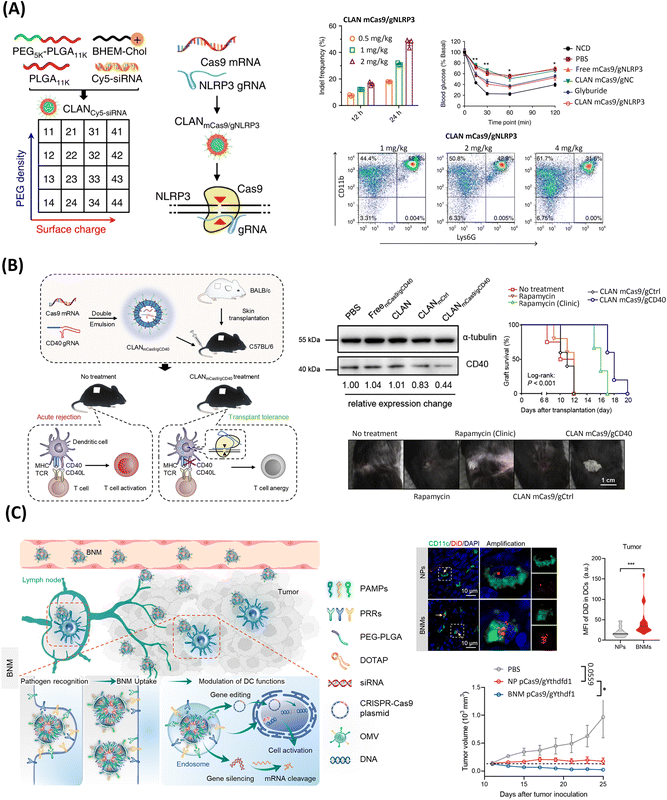 | ||
| Fig. 4 PLGA-based LPHNs for the delivery of genome editing components. (A) Schematics of screening of CLAN for mCas9/gRNA delivery to macrophages for NLRP3 knockout and treatment of inflammation. Middle upper column: gene disruption efficacy in peritoneal macrophages of mice injected with CLAN mCas9/gNLRP3 at 12 or 24 h post-injection, through T7E1 assays of indels introduced at the NLRP3 locus. N = 5 per group. Upper right: insulin tolerance test (ITT) in HFD-induced T2D mice treated with CLAN mCas9/gNLRP3 or other formulations. Two-way ANOVA, *p < 0.05, **p < 0.01. Lower row: fluorescence-activated cell sorting analysis of neutrophil recruitment in the peritoneal cavity of MSU-induced peritonitis, with CD11Bb+ and Ly6G+ labeling. N = 5 per group. Reproduced under the terms of the CC-BY Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/).78 Copyright 2018, The Authors, Published by Springer Nature. (B) Schematized reprogramming of dendritic cells using CLAN mCas9/gCD40 to induce skin graft transplantation tolerance. Upper middle: western blot analysis of CD40 protein expression in dendritic cells after CLAN mCas9/gCD40 injection in mice. Upper right: skin graft survival after intravenous injection of CLAN mCas9/gCD40. Bottom image: photographs of skin graft upon treatment with different conditions, 12 days after transplantation. Reprinted from ref. 209, Copyright (2019), with permission from Elsevier. (C) Illustration of bacterial nanomedicines (BNMs) for pathogen recognition-mediated dendritic cell-specific gene editing for cancer immunotherapy. Upper row: uptake efficiency of NPs and BNMs by dendritic cells in tumors seen at 72 h after injection in MC38 tumor-bearing mice seen in immunofluorescence images (left) (yellow arrows indicate dendritic cells encapsulating NPs or BNMs), and violin plots (right) of DiD gMFl (normalized geometric mean fluorescence intensity) (n = 31 per group). Bottom: antitumor immunity efficiency showed with tumor growth curves of mice after receiving PBS, NP pCas9/gYthdf1, or BNM pCas9/gYthdf1. Reprinted with permission from ref. 185. Copyright (2018) American Chemical Society. | ||
Overall, it successfully downregulated the number of B cells in mice, consequently alleviating major symptoms of the autoimmune disease rheumatoid arthritis and preserving the skeletal structure of the joints.219 This nanoplatform showed promise for treating a wide range of debilitating diseases correlated with B-cell dysfunction. Going further, CLANs were screened for optimal cell internalization of macrophages for the treatment of inflammatory diseases, Fig. 4A.78 In this study, CLANs with a higher surface charge and lower PEG density were better internalized by macrophages, in which increasing the surface charge was shown to be more effective than reducing the PEG density.78 Delivery of mCas9/gNLRP3 significantly disrupted (up to 47.1%) the NLRP3 gene – associated with the progressive release of several proinflammatory cytokines – in mice, ultimately mitigating various NLRP3-dependent inflammatory profiles, including T2D and peritonitis.78 Another remarkable study using the CLAN platform leveraged the encapsulation and delivery of Cas9 mRNA and CD40 gRNA as a strategy to relieve transplant rejection and prolong skin graft survival, Fig. 4B.209 Notably, as traditional immunosuppressants have several immune-adverse effects, gene editing is emerging as a promising safer alternative. As a proof of concept, this successfully minimized the expression of CD40– a costimulatory molecule with a critical role in initiating alloimmune responses – in dendritic cells, thus inhibiting T-cell activation and reducing the skin graft rejection damage in mice.209
As showcased above, CLAN systems have demonstrated remarkable versatility, being applicable to a wide range of diseases and different types of editing cargos. Moreover, the incorporation of clinically validated components in CLANs offers a vast potential for the large-scale manufacturing and clinical translation of such platforms.220 Nonetheless, further inclusion of additional lipid or targeting moieties could be interesting to expand the toolset of CLANs. Notably, the PEG-PLGA block copolymer and DOTAP lipid have been recently harnessed to formulate hybrid bacterial nanomedicines (BNM), in which the addition of bacteria-derived outer membrane vesicles (OMVs) at the nanoparticle surface enabled dendritic cell (DC)-targeted immunotherapy via pCRISPR/Cas9, Fig. 4C.185
The uniqueness of this carrier leveraged a biomimetic targeting strategy that recapitulated the phenomenon of pathogen infection recognition by DCs, found in nature. Looking more closely, nature-derived pathogen-associated molecular patterns (PAMPs) found in OMVs can specifically interact with the pattern recognition receptors (PRR) at the surface of DCs. In this way, such strategy increased the in vitro cellular uptake by ∼2.5 fold compared with non-specific nanoparticles, knocking-out the YTH N6-methyladenosine RNA binding protein F1 encoding-gene (YTHDF1). Ultimately, such therapy successfully activated the DCs, triggering CD8+ T-cell-mediated antitumor immunity, eradicating the tumor growth by 97.72% in mice.185
4.2. PEI-based lipid–polymer hybrids
As shown in Fig. 5, a few PEI-based lipid–polymer hybrids have been explored for gene editing. Notably, one of the earliest studies harnessing the attractive PEI polymer for the establishment of polymer core–lipid shell hybrids was based on liposome-templated hydrogel nanoparticles (LHNPs) for CRISPR/Cas9-based targeted brain tumor therapy, Fig. 5A.76 In particular, LHNPs co-delivered Cas9 protein and a minicircle DNA, instead of a sgRNA, to enhance the efficiency, with a PEI-based hydrogel non-covalently crosslinked via CD–AD host–guest interactions. The most notable aspect of this system relied on the key role played by the soft hydrogel in: (i) promoting a lower cytotoxicity than Lipo2000, and (ii) favoring the maintenance of Cas9 nuclease activity, while simultaneously improving the encapsulation efficiency up to ∼63%, compared with the stand-alone liposome. Markedly, hydrogel-based carriers have been widely applied in the biomedical field, as these contain a high water absorptivity and biocompatibility, thus being highly promising for the delivery of gene editing machineries.29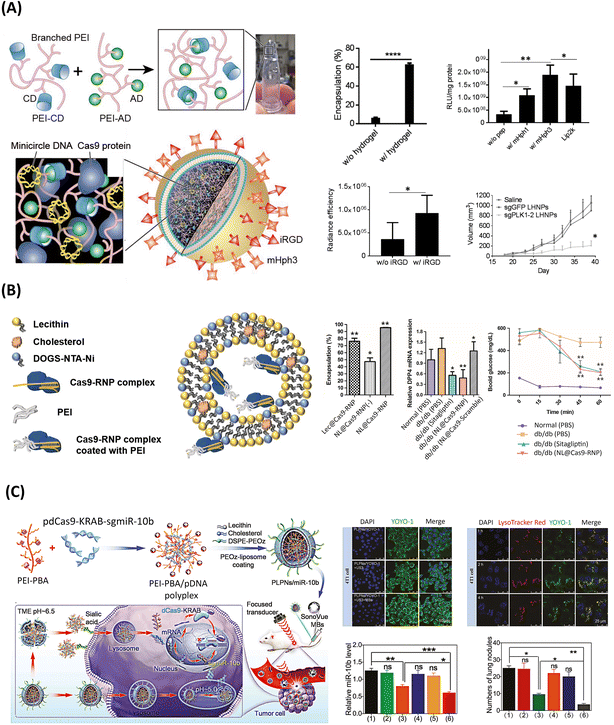 | ||
| Fig. 5 PEI-based LPHNs for the delivery of genome editing components. (A) Schematics of liposome-templated hydrogel nanoparticles (LHNPs) encapsulating Cas9 and minicircle DNA for targeted knock-out of PLK1 gene for cancer therapy. Upper left: Cas9 encapsulation efficiency of LHNPs with and without hydrogel-core. Upper right: in vitro gene delivery efficiency of pGl4.13 (luciferase-encoding plasmid)-loaded LHNPs, testing different ligands, along with Lipo2000, on U87 cells (luciferase signal was detected at 72 h after transfection).* and ** represent p < 0.05 and 0.005, respectively. Lower left: fluorescence intensity-based semi-quantification of LHNPs with and without conjugation of iRGF in flank tumors. Temporal evolution of tumor volumes in U87 tumor-bearing mice after intravenous administration of the treatment (n = 6) (lower right). Reprinted from ref. 76, Copyright (2017), with permission from John Wiley and Sons. (B) Representation of nano-liposomal particles encapsulating Cas9/sgRNA ribonucleoprotein for disruption of DPP-4 gene and T2D treatment. Left column: encapsulation efficiency for different liposomal formulations (Lec@Cas9-RNP: RNP encapsulated in lecithin, NL@Cas9-RNP(−): RNP encapsulated in liposome shell without PEI, NL@Cas9-RNP: RNP encapsulated in liposome shell with PEI). Middle column: expression mRNA levels of DPP4 in extracted liver tissue, after a single administration of the different treatments. Right column: insulin tolerance test results at various times after administration of glucose meal or insulin after treatment with NL@Cas9-RNP or Sitagliptin (n = 3; *p < 0.05). Reproduced under the terms of the CC-BY Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/). Copyright 2019, The Authors, Published by Springer Nature.178 (C) Illustration of pH-responsive PLPNs combined with UMMD, encapsulating CRISPRi-based plasmid, for tumor-specific repression of miR-10b. Upper left: fluorescence images of uptake efficiency in 4T1 cells treated with PLPNs/YOYO-1-pDNA (top), PLPNs/YOYO-1-pDNA plus ultrasound (US) (middle), or PLPNs/YOYO-1-pDNA plus UMMD (bottom). Upper right: fluorescence images of endosomal escape efficiency, based on YOYO-1 (green) labeled pDNA. (Endosomes and lysosomes were stained with LysoTracker Red, and nuclei were stained with DAPI (blue).) Lower left: relative expression levels of miR-10b. Lower right: quantification of tumor nodules in 4T1 tumor-bearing mice, 25 days after repeated injection of PLPNs/miR-10b + UMMD. Data are presented as mean ± SD (n = 5), *p < 0.05, **p < 0.01, ***p < 0.001. (1)–(6) correspond to (1) PBS, (2) PLPNs/negative control (NC), (3) PLPNs/miR-10b, (4) UMMD, (5) PLPNs/NC + UMMD, and (6) PLPNs/miR-10b + UMMD treatment groups. Reprinted from ref. 75, Copyright (2023), with permission from John Wiley and Sons. | ||
Moreover, different peptides, including the surface-conjugated cell penetration peptide iRGD, further enhanced the targeted accumulation of LHNPs at glioma tumors in mice, by specifically binding with αvβ3/αvβ5 integrins and neuropilin-1 at the cancer cell surface.76 The successful knockout of the polo-like kinase 1 (PLK1) gene – frequently overexpressed in cancers – significantly inhibited tumor growth, in which the average tumor volume in the treated group was only 23.5% of that in the control groups receiving saline treatment.76 Moreover, PEI has also been harnessed in lecithin-based liposomal nanocarriers for the delivery of recombinant CRISPR RNPs machinery for treatment of T2D, Fig. 5B.178 Here, PEI moieties mainly served to mitigate the negative repulsive electrostatic interactions between the RNPs and the negative liposome shell, which specifically harbored a negatively charged lipid (DOGS-NTA-Ni) to mediate the binding and encapsulation of Histidine-tagged Cas9.178,221 This system achieved an encapsulation efficiency of ∼95%, and successfully knocked-out the dipeptidyl peptidase-4-encoding gene (DPP-4) in the liver of mice – a gene correlated with the rapid degradation of the glucagon-like peptide-1 hormone, linked with insulin secretion. Such therapy significantly reduced liver damage and restored the glucose tolerance and insulin resistance, similarly to the established drug sitagliptin, which requires multiple doses, in contrast to CRISPR. Moreover, lecithin's natural metabolism in the liver strongly contributed to the in vivo biodistribution of the carrier.178
Going further, simultaneously harnessing physiological endogenous factors and external stimuli can be interesting for developing vehicles with an enhanced delivery specificity, along with spatial and temporal precision.32 As a proof of concept, pH-responsive lipid–polymer hybrids (PLPNs) combined with ultrasound-mediated microbubble destruction (UMMD) were recently developed for the delivery of a CRISPRi-encoding plasmid for targeted tumor therapy, Fig. 5C.75 The non-invasive UMMD has been widely employed in gene delivery systems, as it promotes mechanical perturbations in blood vessels wall and cell membranes, enhancing the permeability of vectors into deep tumor tissues.75,120 Moreover, although CRISPRi does not represent a permanent gene editing tool per se, it may be a safer approach, as it is potentially reversible and does not damage the genome, as it does not involve direct insertion/deletion of sequences.17 Here, the outer lipid shell enhanced the compact packing of the PEI-PBA-based polyplex in an electrostatic manner, by possessing negative charges due to the use of the negatively charged poly(2-ethyl-2-oxazoline) (PEOz)-lipids.75 In essence, upon entry into the acidic tumor microenvironment (pH ∼6.5), or traficking into the lysosomal compartments (pH ∼5.0), the carbonyl group in the side chains of the tertiary amide groups in PEOz easily binds to hydrogen ions in solution, forming numerous hydrogen bonds with other tertiary amide groups within intermolecular or intramolecular PEOz.222–224 This induces the protonation of the amides, which destabilize and disrupt the core–shell structure, facilitating the release of the polyplex for subsequent cellular uptake in a specific and “active” targeting manner. Moreover, the PBA functional groups also aid in the cell internalization, by binding to the commonly overexpressed sialic acid in cancer cells, via reversible boronic acid ester linkages.149,191 Ultimately, combining PLNPs with UMMD resulted in superior tumor enrichment, gene transfection and endosomal escape, effectively silencing miR-19b – highly expressed in metastatic breast tumors – and thus reducing the number of nodules by up to ∼84% in mice.75 One of the novelties of this carrier leveraged the non-fouling PEOz component, which is a promising replacement for PEG. Nonetheless, further comparisons between each would be interesting to critically determine the best performing system, mainly regarding the accelerated blood clearance issue reported in PEGylated particles.
4.3. Protamine-based lipid–polymer hybrids
A few protamine-based hybrid vehicles have also shown promise, Fig. 6. For instance, a protamine core–lipid shell was designed for the delivery of a pCRISPR/Cas9-mediated tumor therapy.77 The inclusion of the anionic chondroitin sulfate within the core was critical for maximizing the electrostatic interactions between the plasmid and the positively charged protamine, forming a highly condensed anionic ternary core that is highly stable and minimizing the size of the cargo.77 Further coating with the biocompatible cationic lipid shell facilitated cell internalization, achieving an in vitro transfection efficiency of about 47%. | ||
| Fig. 6 Protamine and alginate-based LPHNs for the delivery of genome editing components. (A) Schematic illustration of a multifunctional protamine-based carrier encapsulating CRISPR/Cas9 plasmids for disruption of MTH1 gene and targeted non-small cell lung cancer (NSCLC) therapy. Top: PS@HA-Lip/pMTH1-induced in vitro cytotoxicity analyzed by the inhibition rate of A549 cells growth after treatment with different formulations. Bottom: in vivo targeting effect of PS@HA-Lip/pMTH1 seen as the biodistribution of YOYO-1 labeled vectors in normal lung and tumor site in NSCLC-bearing mice. P-Lip (non-specific liposomal NP), PS@P-Lip (non-specific hybrid NP), HA-Lip (HA-targeted liposomal NP), PS@HA-Lip (HA-targeted hybrid NP). Reprinted from ref. 145, Copyright (2022), with permission from Elsevier. (B) Schematics of a biomimetic protamine-based system with controllable laser irradiation for the delivery of pCRISPR-Cas9/sgRNA for targeted tumor cell reprogramming. Top: fluorescence images of endosomal escape after transfection of YOYO-1-labeled pCas9 and of GFP-tagged-Cas9-based fluorescence distribution, in H1299 cells, with and without laser irradiation. Bottom left: flow cytometry analysis of the GFP-tag expression in H1299 cells after different treatments. Bottom right: number of metastatic nodules. Data are presented as mean ± SD (n = 5). P values were calculated by one-way ANOVA (***p > 0.001, ****p < 0.0001). NP + L (non-specific particle with light), CM-NP + L (cell membrane-targeted nanoparticle with light), ATCM-NP + L (cell membrane and aptamer-targeted nanoparticle with light). Reprinted from ref. 91, Copyright (2023), with permission from Elsevier. (C) Schematic illustration of an alginate-based tumor-targeted nanolipogel system (tNLG) encapsulating pCRISPR/Cas9 for knock-out of the Lcn2 gene and treatment of triple-negative breast cancer (TNBC). Top left: protein expression of Lcn2 in TNBC cells before and after Lcn2 knockout measured by IF staining. Top right: images of excised TNBC tumors from mice treated with PBS, tNLG-SCR, nNLG-Lcn2KO, or tNLG-Lcn2KO under a 28-day treatment regimen. N = 5 per group. (Scale bar: 1 cm.) Bottom left: in vitro relative cell migration in MDA-MB-231 cells with Lcn2 CRISPR knockout. Bottom right: in vivo genome editing efficiency of tNLG-Lcn2KO and other treatments, determined by qRT-PCR. (**p < 0.01, ***p < 0.001). Reprinted from ref. 104, Copyright (2019), with permission from PNAS. | ||
Moreover, the successful knockout of the PLK1 gene resulted in significant inhibition of melanoma growth (∼67%) in mice, achieving a better performance than Lipo2000. In addition, this system was also more mechanically stable than Lipo2000, as its diameter did not vary with different serum concentrations in the medium.77 Despite such promising results, the inclusion of targeting ligands may perhaps result in an enhanced tumor delivery specificity. Bearing this in mind, the incorporation of hyaluronic acid (HA) at the surface of a protamine sulfate-based hybrid was attempted to enhance the tumor-selective accumulation, Fig. 6A.145 An relatively high in vitro transfection efficiency (∼80%) was achieved, attributed to the inclusion of protamine and HA moieties. Moreover, the knockout of the Human MuT homolog 1 (MTH1) gene – correlated with tumorigenesis – further induced growth inhibition of ∼67% of non-small cell lung cancer in mice.145 Despite the inclusion of HA, the in vivo tumor inhibition was identical to the previously mentioned non-targeting study. Perhaps to legitimately achieve robust performances, the combination of targeting ligands with stimuli-responsive moieties may be a better strategy. Notably, a light-controlled biomimetic hybrid consisting of pCRISPR/Cas9 targeting the cancer metastasis-related hypoxia-inducible factor-1 alpha-encoding gene (HIF-1α) was developed for augmented tumor therapy, Fig. 6B.91 The inclusion of calcium ions in the protamine core greatly enhanced the permeability of the nuclear pore complex, while the neoplastic H1299 cell membrane (CM) coating camouflage and AS1411 aptamers enhanced the tumor enrichment and induced a negative surface charge.91 In particular, CMs have attracted wide attention in drug delivery, as these possess inherent homotypic recognition and immune escaping functions, while promoting tissue targeting with greater safety and fewer side effects, given their natural origins.24,91 On the other hand, remote light stimulus is also widely applied in tumor therapy, owing to its desirable safety profile and spatiotemporal precision.21,27 In essence, after cell targeting and internalization, upon on-demand laser irradiation (i.e., 660 nm) the surface-embedded 5,10,15,20-tetrakis-(4-aminophenyl) porphyrin (TAPP) photosensitizer induced the formation of excessive ROS, which in turn enhanced the endocytic membrane permeability and CM leakage, ultimately maximizing lysosomal escape and cargo intracellular release.91 In this way, successful knockout of the HIF-1α gene promoted cell reprogramming and enhanced cell sensitivity to the established chemotherapeutical agent paclitaxel (PTX). The synergistic effects of CRISPR and PTX efficiently inhibited tumor metastasis in the lungs of mice.91 Not only does this study demonstrate: (i) the advantages of exogenous stimuli for the augmentation of gene editing, but also, (ii) the advantages of harnessing gene editing tools for combinatorial treatments with already established therapeutics.
4.4. Alginate-based lipid–polymer hybrids
Finally, based on alginate, one intriguing and unique LPHN has been developed for breast cancer therapy, Fig. 6C.104 Essentially, an alginate hydrogel network-based nanoliposome was formulated for pCRISPR/Cas9 delivery, with the inclusion of ICAM1 antibody for high-affinity tumor targeting.104 On one hand, owing to the non-cationic nature of both the hydrogel core and the liposomal shell, the system showed very low cytotoxicity, and could efficiently entrap the CRISPR toolset within the polysaccharide hydrogel network without relying on electrostatic interactions.104 On the other hand, the deformable hydrogel core promoted low particle elasticity (with an elastic modulus of 1.3 MPa), which is considerably different from the conventionally more rigid solid lipid and polymer stand-alone nanoparticles (with elastic moduli ranging from 0.76 to 1.2 GPa).104 In turn, this low elasticity induced an augmented and selective extravasation of the tumor endothelial barrier compared with normal endothelial barriers. Moreover, upon cellular uptake, the low stiffness also induced direct delivery of the genetic payload into the cytosol via a receptor-mediated membrane fusion pathway, avoiding endosomal entrapment.104 This represents another remarkable advantage in comparison with conventional nanocarriers, which face several challenges regarding endosome entrapment. Finally, a highly efficient knockout (>81%) of the Lipocalin 2 gene (Lcn2) – a known breast cancer oncogene – significantly attenuated tumor volume by 77% in mice.104 This demonstrated the known advantages of including low-elastic hydrogel-based cores and non-cationic liposomal shells in LPHNs to bypass the challenges faced by conventional nanoparticles.5. Outlook and future directions
Cutting-edge genome-editing technologies are now beginning to show their potential for being applied in a wide spectrum of biomedical applications and becoming more streamlined. However, the fine-tuning of the nanoparticle physicochemical properties, along with the inclusion of organ- or cell-targeting building blocks, or stimuli-responsive moieties for cargo release/delivery control, is still highly required for designing platforms that are more efficient, selective and safer for gene editing. Among the library of available delivery platforms, CLANs mainly stand out for their simplicity of design, versatility and clinical translation prospective, while polymer core–lipid shell hybrids show multiple advantages, including intrinsic liposomal biocompatibility, along with the independent design of the core and shell counterparts, enabling the meticulous assembly of innovative carriers.The adoption of optimal design blueprints harnessed in stand-alone vehicles could bring outstanding innovation for the design of novel lipid–polymer conjugates. Notably, exploring more in depth the versatile polymer chemistry in future research could bring many advantageous features, either to enhance the biodegradability, endosomal escape, or complexation of the cargo, as widely explored in conventional carriers. Moreover, for maximum control over the polyplex properties, customizable in situ polymerization of synthetic polymer chains could enable truly personalized hybrids.17,47,48,203 Moving forward to the lipid components, as only a few hybrids have included all major primary lipid components, future research could greatly benefit from advancing lipid formulations, as broadly encouraged by LNPs; for instance, either by incorporating emerging and safer lipid derivatives, or tuning lipid chemical moieties in a combinatorial manner with polymeric components. In particular, as permanent cationic lipids can show in vivo cytotoxicity, ionizable cationic lipids are envisioned to unlock game-changer hybrids, as these have shown superior performances and currently represent the most exploited components in LPNs for the delivery of CRISPR editors.28,37,46,99,117,225 In addition to enhancing the circulation stability owing to their neutral charge at physiological pH, these could augment the endosomal escape step by protonating the acidic endosomes, working in harmony with polymer chains to achieve unprecedented dual performances.24,99,131,158,159,161 The overarching question remains whether such dual approaches could legitimately promote much faster and exceptional gene transfection and editing efficiencies, as compared with conventional ionizable lipids or polymers. Inspired by the ionizable lipids ALC-0315 and SM-102 found in the two approved COVID-19 lipid vaccines, respectively, the recent hybrid core–shell COVID-19 mRNA vaccine also contains an ionizable lipid; however, neither the specific nature of the ionizable lipid nor the kind of polymer harnessed have yet been publicly disclosed.62,99,158 Going further, despite their recognized biocompatibility, standalone mRNA lipid nanoparticles have been shown to trigger adverse immune responses in patients suffering, for example, from COVID-19, potentially caused by the ionizable components.184,226,227 Nonetheless, a more comprehensive understanding of how lipid nanoparticles and each lipid moiety impact the immune system is still lacking.184 Moreover, given the immunogenicity challenges of PEG in drug delivery, a strategic direction to follow may be to explore in more depth stealth coating alternatives for LPHNs, as PEOz and cell membranes have already shown promise, for instance.75,91 Nevertheless, the exact mechanism and impact of anti-PEG antibodies is still unclear, namely regarding COVID-19 mRNA LNP vaccines.228 Moreover, as in vivo gene editing therapies are envisioned to be ideally administered in a single dose, to what extent could these suffer from the same immunogenicity issues as the ones reported with repeated vaccine administrations? In particular, harnessing biomimetic cell membrane coatings for increasingly superior and biocompatible lipid–polymer hybrids seems strikingly promising for future research. As a proof of concept, such structures have been combined with PBAE-based polyplexes and lipid-anchored ROS-responsive components,100 or even with poly(disulfide)-based polyplexes102 to augment gene editing targeting and specificity.
Going forward, surface functionalization strategies so far appear to be key components in hybrid carriers to maximize the targeting efficiency of in vivo interventions. Nonetheless, these can however contribute to more complex designs and cytotoxicity, in the case of synthetic ligands, currently being no existing examples of clinically approved nanoparticles with such targeting moieties.33 Hence, exploring more prospective clinical passive-targeting approaches in hybrid carriers could be a tremendously interesting route to take, especially for extrahepatic in vivo gene editing, for instance with SORT lipids.33 Would the integration between lipids and polymers alter protein corona-based targeting mechanisms seen in LNPs, and how could these be tuned to further enhance tissue and cell selectivity? Also, as spatially and temporally stimuli-responsive strategies have not been widely explored in LPHNs so far, future research could greatly benefit from these, as shown in a multitude of interesting polymeric and lipid vehicles. In particular, designing multi-responsive vehicles in cascade-like behaviors, i.e., with combinatorial intelligent liposomal and polymeric building blocks, could potentially maximize the targeting specificity and gene editing outcome, paving the way for truly ingenious innovative hybrids.116,122 Moreover, as in vivo studies have mainly focused on the systemic delivery of in vivo interventions, different approaches could be extremely interesting to consider in further research, such as nebulized or inhalable formulations for pulmonary gene editing.84,229,230 Moreover, the fine-tuning of the physicochemical properties of nanoparticles for targeted delivery upon systemic administration can be complex, particularly when considering hybrid systems such as LPHNs. Also, precise spatiotemporal control over the delivery of gene editing machineries can be critical to clinically ensure safe and efficient interventions. In this way, there is an emergence of injectable hydrogels for minimally invasive localized therapies with a proficient nanoparticle accumulation in target tissues.231–234 Notably, the vast majority of the so far approved gene therapies have focused on local delivery.22 Given the outstanding mechanical properties and ease of surface functionalization that lipid–polymer nanoparticles can possess, these could be highly promising building blocks to formulate novel injectable colloidal supramolecular hydrogels for in vivo genes for localized delivery of gene-editing cargo.229,231,232,234 Also, in order to further interrogate in more depth how superior lipid–polymer hybrids can be, further research on hard-to-transfect cells, such as adipose stem cells (ASCs) would be particularly interesting.55 Could lipid–polymer conjugates surpass conventional delivery approaches and promote maximum transfection efficiencies to expedite the development of stem cell-based therapies? Also, as nonviral approaches have begun to being tackled for the delivery of next-generation larger sized editing tools, e.g., Base and Prime Editors, lipid–polymer hybrids could be particularly interesting as a way to ensure a superior encapsulation and protection of such larger payloads. Going forward, another key aspect to consider is the scalability and reproducibility needed to expand the clinical potential of emerging hybrids. Leading-edge microfluidic mixing technologies are envisioned to aid in the establishment of formulations with robust size controllability, reproducibility, and even desired tissue tropism outcomes, as broadly demonstrated in LPNs, along with advanced lipid–polymer hybrids for delivery of mRNA and siRNA transient gene therapies.17,27,28,165,235 Also, lipid–polymer hybrids are envisioned to tackle the challenges broadly faced by lipid and polymer-based nanoparticles regarding long-term stability for storage. Nevertheless, lyophilization methods could be explored in hybrids to further maximize their storage stability and their potential for clinical translation, especially in the more unexplored cell membrane-coated nanoparticles.22 Moreover, the refinement of humanized in vitro 3D disease models such as organoids and organ-on-chips is expected to accelerate the accurate validation and translation of these vehicles, as these have been progressively adopted in the preclinical stages of newly developed cell and gene therapies.22,236 Finally, from a regulatory perspective, the approval of LPHNs is more challenging than conventional carriers, as there are no currently existing specific regulatory guidelines for the clinical development of LPHNs.237 Moreover, as part of the FDA guidelines for the approval of genome editing-based therapeutics, delivery vehicles should be as simple as possible.17 Hence, as hybrid carriers combine macromolecules with different natures, these are inherently more complex, which may compromise their translational clinical potential and regulatory approval. Hence, appropriate methodologies for quality control assays should be adopted to accurately facilitate the in vitro and in vivo characterization of lipid–polymer hybrids. Current research lacks, for instance, key validation experiments drawing comparisons between hybrids and stand-alone lipid or polymeric nanoparticles, with regards to features such as biocompatibility and gene transfection performance. Such analysis could facilitate the comprehensive assessment of the distinct features of lipid–polymer hybrids, and potentially accelerate clinical translation. Also, as stand-alone polymeric nanoparticles have faced many hurdles regarding the workflows for their screening, the adoption of high-throughput techniques more widely explored for lipid carriers could be highly beneficial for LPHNs; for instance, (i) the use of Design of Experiments (DoE) for the optimization of the formulation parameters, and/or (ii) DNA barcoding for multiplex assessment of their in vivo performance and biodistribution.17,47,65,171,216,225,235,238 As the rational design of novel nanocarriers usually requires extensive and laborious formulation scans, the adoption of in silico screening could be a cost-effective alternative to minimize the extensive trial-and-error methods and the economic burdens during research stages.22 Moreover, the rise of cutting-edge artificial intelligence (AI)-based models and machine learning algorithms is envisioned to accelerate the research of polymeric and lipid excipients, tailored to specific delivery systems and target cells, with significant advances in this direction being expected in the upcoming years.
In conclusion, as genome-editing tools continue to evolve into authentic programmable machineries for precision medicine, there is a continuous need for the refinement of clinically safe and efficient delivery systems. Emerging lipid–polymer hybrids are widely promising for unlocking a new generation of nonviral genome editing drugs and bringing unparallel advancements to the field.
Data availability
No new data were generated or analyzed in this review. All the primary data supporting this review are available within the articles cited in the manuscript.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was developed within the scope of the project CICECO-Aveiro Institute of Materials, UIDB/50011/2020 DOI: 10.54499/UIDB/50011/2020), UIDP/50011/2020 (DOI: 10.54499/UIDP/50011/2020), and LA/P/0006/2020 (DOI: 10.54499/LA/P/0006/2020), financed by national funds through the FCT/MEC (PIDDAC). The funding of the European Research Council for the project REBORN (ERC-2019-ADG-883370) is acknowledged. This work was also funded by EdiGenT (Grant ID: 101070903) project on the framework of the European Union's Horizon Europe research and innovation programme.References
- A. Pickar-Oliver and C. A. Gersbach, The next generation of CRISPR–Cas technologies and applications, Nat. Rev. Mol. Cell Biol., 2019, 20, 490–507 CrossRef CAS PubMed.
- C. J. Bashor, I. B. Hilton, H. Bandukwala, D. M. Smith and O. Veiseh, Engineering the next generation of cell-based therapeutics, Nat. Rev. Drug Discovery, 2022, 21, 655–675 CrossRef CAS PubMed.
- M. Chavez, X. Chen, P. B. Finn and L. S. Qi, Advances in CRISPR therapeutics, Nat. Rev. Nephrol., 2023, 19, 9–22 CrossRef CAS PubMed.
- J. Y. Wang and J. A. Doudna, CRISPR technology: A decade of genome editing is only the beginning, Science, 2024, 379, eadd8643 CrossRef PubMed.
- A. V. Anzalone, L. W. Koblan and D. R. Liu, Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors, Nat. Biotechnol., 2020, 38, 824–844 CrossRef CAS PubMed.
- B. D. Kevadiya, F. Islam and P. Deol, et al., Delivery of gene editing therapeutics, Nanomedicine, 2023, 54, 102711 CrossRef CAS PubMed.
- L. Cong, F. A. Ran and D. Cox, et al., Multiplex Genome Engineering Using CRISPR/Cas Systems, Science, 2013, 339, 819–823 CrossRef CAS PubMed.
- M. Adli, The CRISPR tool kit for genome editing and beyond, Nat. Commun., 2018, 9, 1911 CrossRef PubMed.
- T. Li, Y. Yang and H. Qi, et al., CRISPR/Cas9 therapeutics: progress and prospects, Signal Transduction Targeted Ther., 2023, 8, 36 CrossRef PubMed.
- J. Li, J. J. Røise, M. He, R. Das and N. Murthy, Non-viral strategies for delivering genome editing enzymes, Adv. Drug Delivery Rev., 2021, 168, 99–117 CrossRef CAS PubMed.
- M. H. Geurts and H. Clevers, CRISPR engineering in organoids for gene repair and disease modelling, Nat. Rev. Bioeng., 2023, 1, 32–45 CrossRef.
- C. Trentesaux, T. Yamada, O. D. Klein and W. A. Lim, Harnessing synthetic biology to engineer organoids and tissues, Cell Stem Cell, 2023, 30, 10–19 CrossRef CAS PubMed.
- X. Yan, X. Liu, C. Zhao and G. Q. Chen, Applications of synthetic biology in medical and pharmaceutical fields, Signal Transduction Targeted Ther., 2023, 8, 199 CrossRef CAS PubMed.
- T. Kitada, B. DiAndreth, B. Teague and R. Weiss, Programming gene and engineered-cell therapies with synthetic biology, Science, 2018, 359(6376), eaad1067 CrossRef PubMed.
- N. Shakiba, R. D. Jones, R. Weiss and D. Del Vecchio, Context-aware synthetic biology by controller design: Engineering the mammalian cell, Cell Syst., 2021, 12, 561–592 CrossRef CAS PubMed.
- C. A. DeForest, B. E. Kirkpatrick and K. S. Anseth, Engineering native biological complexity from the inside–out and outside–in, Nat. Chem. Eng., 2024, 1, 2–5 CrossRef PubMed.
- V. Madigan, F. Zhang and J. E. Dahlman, Drug delivery systems for CRISPR-based genome editors, Nat. Rev. Drug Discovery, 2023, 22, 875–894 CrossRef CAS PubMed.
- Z. Zhao, P. Shang, P. Mohanraju and N. Geijsen, Prime editing: advances and therapeutic applications, Trends Biotechnol., 2023, 41, 1000–1012 CrossRef CAS PubMed.
- R. Chowdhry, S. Z. Lu, S. Lee, S. Godhulayyagari, S. B. Ebrahimi and D. Samanta, Enhancing CRISPR/Cas systems with nanotechnology, Trends Biotechnol., 2023, 41, 1549–1564 CrossRef CAS PubMed.
- C. Sheridan, The world's first CRISPR therapy is approved: who will receive it?, Nat. Biotechnol., 2024, 42, 3–4 CrossRef CAS PubMed.
- C. Chen, W. Zhong and S. Du, et al., Intelligent nanotherapeutic strategies for the delivery of CRISPR system, Acta Pharm. Sin. B, 2023, 13, 2510–2543 CrossRef CAS PubMed.
- B. B. Mendes, J. Conniot and A. Avital, et al., Nanodelivery of nucleic acids, Nat. Rev. Methods Primers, 2022, 2, 24 CrossRef CAS PubMed.
- Z. Glass, M. Lee, Y. Li and Q. Xu, Engineering the Delivery System for CRISPR-Based Genome Editing, Trends Biotechnol., 2018, 36, 173–185 CrossRef CAS PubMed.
- S. A. Dilliard and D. J. Siegwart, Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs, Nat. Rev. Mater., 2023, 8, 282–300 CrossRef CAS PubMed.
- T. C. Roberts, R. Langer and M. J. A. Wood, Advances in oligonucleotide drug delivery, Nat. Rev. Drug Discovery, 2020, 19, 673–694 CrossRef CAS PubMed.
- X. Pan, H. Veroniaina and N. Su, et al., Applications and developments of gene therapy drug delivery systems for genetic diseases, Asian J. Pharm. Sci., 2021, 16, 687–703 CrossRef PubMed.
- D. K. Sahel, L. K. Vora and A. Saraswat, et al., CRISPR/Cas9 Genome Editing for Tissue-Specific In Vivo Targeting: Nanomaterials and Translational Perspective, Adv. Sci., 2023, 10, 2207512 CrossRef CAS PubMed.
- P. Kazemian, S. Y. Yu, S. B. Thomson, A. Birkenshaw, B. R. Leavitt and C. J. D. Ross, Lipid-Nanoparticle-Based Delivery of CRISPR/Cas9 Genome-Editing Components, Mol. Pharmaceutics, 2022, 19, 1669–1686 CrossRef CAS PubMed.
- S. Wei Hu, T. Ding, H. Tang, H. Guo, W. Cui and Y. Shu, Nanobiomaterial vectors for improving gene editing and gene therapy, Mater. Today, 2023, 66, 114–136 CrossRef.
- Y. Lin, E. Wagner and U. Lächelt, Non-viral delivery of the CRISPR/Cas system: DNA versus RNA versus RNP, Biomater. Sci., 2022, 10, 1166–1192 RSC.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Engineering precision nanoparticles for drug delivery, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed.
- P. Ma, Q. Wang, X. Luo, L. Mao, Z. Wang and E. Ye, et al., Recent advances in stimuli-responsive polymeric carriers for controllable CRISPR/Cas9 gene editing system delivery, Biomater. Sci., 2023, 11, 5078–5094 RSC.
- L. Fu, Y. Zhang, R. A. Farokhzad, B. B. Mendes, J. Conde and J. Shi, ‘Passive’ nanoparticles for organ-selective systemic delivery: design, mechanism and perspective, Chem. Soc. Rev., 2023, 52, 7579–7601 RSC.
- J. Yan, D. D. Kang and Y. Dong, Harnessing lipid nanoparticles for efficient CRISPR delivery, Biomater. Sci., 2021, 9, 6001–6011 RSC.
- S. Zhen and X. Li, Liposomal delivery of CRISPR/Cas9, Cancer Gene Ther., 2020, 27, 515–527 CrossRef CAS PubMed.
- X. Yin, R. Harmancey, D. D. McPherson, H. Kim and S. L. Huang, Liposome-Based Carriers for CRISPR Genome Editing, Int. J. Mol. Sci., 2023, 24, 12844 CrossRef CAS PubMed.
- T. Wei, Q. Cheng, Y. L. Min, E. N. Olson and D. J. Siegwart, Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing, Nat. Commun., 2020, 11, 1 CrossRef PubMed.
- H. Onuma, Y. Sato and H. Harashima, Lipid nanoparticle-based ribonucleoprotein delivery for in vivo genome editing, J. Controlled Release, 2023, 355, 406–416 CrossRef CAS PubMed.
- X. Hou, T. Zaks, R. Langer and Y. Dong, Lipid nanoparticles for mRNA delivery, Nat. Rev. Mater., 2021, 6, 1078–1094 CrossRef CAS PubMed.
- J. D. Finn, A. R. Smith, M. C. Patel, L. Shaw, M. R. Youniss and J. van Heteren, et al., A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing, Cell Rep., 2018, 22, 2227–2235 CrossRef CAS PubMed.
- J. Pil Han, M. Kim, B. Seok Choi, J. Hyeon Lee, G. Seong Lee and M. Jeong, et al., In vivo delivery of CRISPR-Cas9 using lipid nanoparticles enables antithrombin gene editing for sustainable hemophilia A and B therapy, Sci. Adv., 2022, 8, eabj6901 CrossRef PubMed.
- X. Huang, N. Kong, X. Zhang, Y. Cao, R. Langer and W. Tao, The landscape of mRNA nanomedicine, Nat. Med., 2022, 28, 2273–2287 CrossRef CAS PubMed.
- D. Adams, A. Gonzalez-Duarte, W. D. O'Riordan, C. C. Yang, M. Ueda and A. V. Kristen, et al., Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis, N. Engl. J. Med., 2018, 379, 11–21 CrossRef CAS PubMed.
- M. D. Shin, S. Shukla, Y. H. Chung, V. Beiss, S. K. Chan and O. A. Ortega-Rivera, et al., COVID-19 vaccine development and a potential nanomaterial path forward, Nat. Nanotechnol., 2020, 15, 646–655 CrossRef CAS PubMed.
- J. D. Gillmore, E. Gane, J. Taubel, J. Kao, M. Fontana and M. L. Maitland, et al., CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis, N. Engl. J. Med., 2021, 385, 493–502 CrossRef CAS PubMed.
- B. Ashok, N. A. Peppas and M. E. Wechsler, Lipid- and polymer-based nanoparticle systems for the delivery of CRISPR/Cas9, J. Drug Delivery Sci. Technol., 2021, 65, 102728 CrossRef CAS PubMed.
- R. Kumar, C. F. Santa Chalarca, M. R. Bockman, C. V. Bruggen, C. J. Grimme and R. J. Dalal, et al., Polymeric Delivery of Therapeutic Nucleic Acids, Chem. Rev., 2021, 121, 11527–11652 CrossRef CAS PubMed.
- G. Chen, A. A. Abdeen, Y. Wang, P. K. Shahi, S. Robertson and R. Xie, et al., A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing, Nat. Nanotechnol., 2019, 14, 974–980 CrossRef CAS PubMed.
- W. Yang, L. Mixich, E. Boonstra and H. Cabral, Polymer-Based mRNA Delivery Strategies for Advanced Therapies, Adv. Healthcare Mater., 2023, 12, 2202688 CrossRef CAS PubMed.
- Z. Tan, Y. Jiang, M. S. Ganewatta, R. Kumar, A. Keith and K. Twaroski, et al., Block Polymer Micelles Enable CRISPR/Cas9 Ribonucleoprotein Delivery: Physicochemical Properties Affect Packaging Mechanisms and Gene Editing Efficiency, Macromolecules, 2019, 52, 8197–8206 CrossRef CAS.
- B. Winkeljann, D. C. Keul and O. M. Merkel, Engineering poly- and micelleplexes for nucleic acid delivery – A reflection on their endosomal escape, J. Controlled Release, 2023, 353, 518–534 CrossRef CAS PubMed.
- S. Beg, W. H. Almalki, F. Khatoon, K. S. Alharbi, S. Alghamdi and M. H. Akhter, et al., Lipid/polymer-based nanocomplexes in nucleic acid delivery as cancer vaccines, Drug Discovery Today, 2021, 26, 1891–1903 CrossRef CAS PubMed.
- V. Dave, K. Tak, A. Sohgaura, A. Gupta, V. Sadhu and K. R. Reddy, Lipid-polymer hybrid nanoparticles: Synthesis strategies and biomedical applications, J. Microbiol. Methods, 2019, 160, 130–142 CrossRef CAS PubMed.
- K. R. Gajbhiye, R. Salve, M. Narwade, A. Sheikh, P. Kesharwani and V. Gajbhiye, Lipid polymer hybrid nanoparticles: a custom-tailored next-generation approach for cancer therapeutics, Mol. Cancer, 2023, 22, 160 CrossRef PubMed.
- Y. Xue, Y. Zhang, Y. Zhong, S. Du, X. Hou and W. Li, et al., LNP-RNA-engineered adipose stem cells for accelerated diabetic wound healing, Nat. Commun., 2024, 15, 1 Search PubMed.
- X. Han, Z. Liu, M. C. Jo, K. Zhang, Y. Li and Z. Zeng, et al., CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation, Sci. Adv., 2015, 1, 7 Search PubMed.
- B. Mandal, H. Bhattacharjee, N. Mittal, H. Sah, P. Balabathula and L. A. Thoma, et al., Core-shell-type lipid-polymer hybrid nanoparticles as a drug delivery platform, Nanomedicine, 2013, 9, 474–491 CrossRef CAS PubMed.
- K. Hadinoto, A. Sundaresan and W. S. Cheow, Lipid-polymer hybrid nanoparticles as a new generation therapeutic delivery platform: A review, Eur. J. Pharm. Biopharm., 2013, 85, 427–443 CrossRef CAS PubMed.
- L. Khalili, G. Dehghan, N. Sheibani and A. Khataee, Smart active-targeting of lipid-polymer hybrid nanoparticles for therapeutic applications: Recent advances and challenges, Int. J. Biol. Macromol., 2022, 213, 166–194 CrossRef CAS PubMed.
- M. Mehta, T. A. Bui, X. Yang, Y. Aksoy, E. M. Goldys and W. Deng, Lipid-Based Nanoparticles for Drug/Gene Delivery: An Overview of the Production Techniques and Difficulties Encountered in Their Industrial Development, ACS Mater. Au, 2023, 3, 600–619 CrossRef CAS PubMed.
- R. Das, P. Kanjilal, J. Medeiros and S. Thayumanavan, What's Next after Lipid Nanoparticles? A Perspective on Enablers of Nucleic Acid Therapeutics, Bioconjugate Chem., 2022, 33, 1996–2007 Search PubMed.
- R. Yang, Y. Deng, B. Huang, L. Huang, A. Lin and Y. Li, et al., A core-shell structured COVID-19 mRNA vaccine with favorable biodistribution pattern and promising immunity, Signal Transduction Targeted Ther., 2021, 6, 1 CrossRef PubMed.
- P. Huang, H. Deng, Y. Zhou and X. Chen, The roles of polymers in mRNA delivery, Matter, 2022, 5, 1670–1699 CrossRef CAS.
- Y. Cao, Z. He, Q. Chen, X. He, L. Su and W. Yu, et al., Helper-Polymer Based Five-Element Nanoparticles (FNPs) for Lung-Specific mRNA Delivery with Long-Term Stability after Lyophilization, Nano Lett., 2022, 22, 6580–6589 CrossRef CAS PubMed.
- J. C. Kaczmarek, K. J. Kauffman, O. S. Fenton, K. Sadtler, A. K. Patel and M. W. Heartlein, et al., Optimization of a Degradable Polymer-Lipid Nanoparticle for Potent Systemic Delivery of mRNA to the Lung Endothelium and Immune Cells, Nano Lett., 2018, 18, 6449–6454 CrossRef CAS PubMed.
- C. Zhang, J. Chen, Y. Song, J. Luo, P. Jin and X. Wang, et al., Ultrasound-Enhanced Reactive Oxygen Species Responsive Charge-Reversal Polymeric Nanocarriers for Efficient Pancreatic Cancer Gene Delivery, ACS Appl. Mater. Interfaces, 2022, 14, 2587–2596 CrossRef CAS PubMed.
- X. Liu, J. Xiang, D. Zhu, L. Jiang, Z. Zhou and J. Tang, et al., Fusogenic Reactive Oxygen Species Triggered Charge-Reversal Vector for Effective Gene Delivery, Adv. Mater., 2016, 28, 1743–1752 CrossRef CAS PubMed.
- T. Yu, W. Nie, Z. Hong, Y. He, J. Chen and X. Mi, et al., Synergy of Immunostimulatory Genetherapy with Immune Checkpoint Blockade Motivates Immune Response to Eliminate Cancer, Adv. Funct. Mater., 2021, 31, 22 Search PubMed.
- K. W. Huang, F. F. Hsu, J. T. Qiu, G. J. Chern, Y. A. Lee and C. C. Chang, et al., Highly efficient and tumor-selective nanoparticles for dual-targeted immunogene therapy against cancer, Sci. Adv., 2020, 15, eaax5032 CrossRef PubMed.
- G. Wang, D. Zhu, Z. Zhou, Y. Piao, J. Tang and Y. Shen, Glutathione-Specific and Intracellularly Labile Polymeric Nanocarrier for Efficient and Safe Cancer Gene Delivery, ACS Appl. Mater. Interfaces, 2020, 12, 14825–14838 CrossRef CAS PubMed.
- F. Perche, R. Clemençon, K. Schulze, T. Ebensen, C. A. Guzmán and C. Pichon, Neutral Lipopolyplexes for In Vivo Delivery of Conventional and Replicative RNA Vaccine, Mol. Ther.–Nucleic Acids, 2019, 17, 767–775 CrossRef CAS PubMed.
- S. Persano, M. L. Guevara, Z. Li, J. Mai, M. Ferrari and P. P. Pompa, et al., Lipopolyplex potentiates anti-tumor immunity of mRNA-based vaccination, Biomaterials, 2017, 125, 81–89 CrossRef CAS PubMed.
- W. Wei, J. Sun, X. Y. Guo, X. Chen, R. Wang and C. Qiu, et al., Microfluidic-Based Holonomic Constraints of siRNA in the Kernel of Lipid/Polymer Hybrid Nanoassemblies for Improving Stable and Safe in Vivo Delivery, ACS Appl. Mater. Interfaces, 2020, 12, 14839–14854 CrossRef CAS PubMed.
- W. Zhao, C. Zhang, B. Li, X. Zhang, X. Luo and C. Zeng, et al., Lipid Polymer Hybrid Nanomaterials for mRNA Delivery, Cell. Mol. Bioeng., 2018, 11, 397–406 CrossRef CAS PubMed.
- Y. Li, P. Wu, M. Zhu, M. Liang, L. Zhang and Y. Zong, et al., High-Performance Delivery of a CRISPR Interference System via Lipid-Polymer Hybrid Nanoparticles Combined with Ultrasound-Mediated Microbubble Destruction for Tumor-Specific Gene Repression, Adv. Healthc. Mater., 2023, 12, 10 Search PubMed.
- Z. Chen, F. Liu, Y. Chen, J. Liu, X. Wang and A. T. Chen, et al., Targeted Delivery of CRISPR/Cas9-Mediated Cancer Gene Therapy via Liposome-Templated Hydrogel Nanoparticles, Adv. Funct. Mater., 2017, 27, 46 Search PubMed.
- L. Zhang, P. Wang, Q. Feng, N. Wang, Z. Chen and Y. Huang, et al., Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy, NPG Asia Mater., 2017, 9, 10 Search PubMed.
- C. Xu, Z. Lu, Y. Luo, Y. Liu, Z. Cao and S. Shen, et al., Targeting of NLRP3 inflammasome with gene editing for the amelioration of inflammatory diseases, Nat. Commun., 2018, 9, 1 CrossRef PubMed.
- H. A. Rees and D. R. Liu, Base editing: precision chemistry on the genome and transcriptome of living cells, Nat. Rev. Genet., 2018, 19, 770–788 CrossRef CAS PubMed.
- N. Wang, C. Liu, Y. Li, D. Huang, X. Wu and X. Kou, et al., A cooperative nano-CRISPR scaffold potentiates immunotherapy via activation of tumour-intrinsic pyroptosis, Nat. Commun., 2023, 14, 1 Search PubMed.
- X. Zhang, H. Jin, X. Huang, B. Chaurasiya, D. Dong and T. P. Shanley, et al., Robust genome editing in adult vascular endothelium by nanoparticle delivery of CRISPR-Cas9 plasmid DNA, Cell Rep., 2022, 38, 1 CAS.
- C. F. Xu, S. Iqbal, S. Shen, Y. L. Luo, X. Yang and J. Wang, Development of “CLAN” Nanomedicine for Nucleic Acid Therapeutics, Small, 2019, 15, 1900055 CrossRef PubMed.
- Q. Liu, J. Cai, Y. Zheng, Y. Tan, Y. Wang and Z. Zhang, et al., NanoRNP Overcomes Tumor Heterogeneity in Cancer Treatment, Nano Lett., 2019, 19, 7662–7672 CrossRef CAS PubMed.
- L. Rotolo, D. Vanover, N. C. Bruno, H. E. Peck, C. Zurla and J. Murray, et al., Species-agnostic polymeric formulations for inhalable messenger RNA delivery to the lung, Nat. Mater., 2023, 22, 369–379 CrossRef CAS PubMed.
- Y. Rui, D. R. Wilson, J. Choi, M. Varanasi, K. Sanders and J. Karlsson, et al., Carboxylated branched poly(β-amino ester) nanoparticles enable robust cytosolic protein delivery and CRISPR-Cas9 gene editing, Sci. Adv., 2019, 5, eaay3255 CrossRef CAS PubMed.
- C. Liu, T. Wan, H. Wang, S. Zhang, Y. Ping and Y. Cheng, A boronic acid-rich dendrimer with robust and unprecedented efficiency for cytosolic protein delivery and CRISPR-Cas9 gene editing, Sci. Adv., 2019, 5, eaaw8922 CrossRef CAS PubMed.
- P. Yang, S. J. Chou, J. Li, W. Hui, W. Liu and N. Sun, et al., Supramolecular nanosubstrate-mediated delivery system enables CRISPR-Cas9 knockin of hemoglobin beta gene for hemoglobinopathies, Sci. Adv., 2020, 6, eabb7107 CrossRef CAS PubMed.
- J. Qiao, W. Sun, S. Lin, R. Jin, L. Ma and Y. Liu, Cytosolic delivery of CRISPR/Cas9 ribonucleoproteins for genome editing using chitosan-coated red fluorescent protein, Chem. Commun., 2019, 55, 4707–4710 RSC.
- X. Y. He, X. H. Ren, Y. Peng, J. P. Zhang, S. L. Ai and B. Y. Liu, et al., Aptamer/Peptide-Functionalized Genome-Editing System for Effective Immune Restoration through Reversal of PD-L1-Mediated Cancer Immunosuppression, Adv. Mater., 2020, 32, 17 Search PubMed.
- X. H. Ren, C. Xu, L. L. Li, Y. Zuo, D. Han and X. Y. He, et al., A targeting delivery system for effective genome editing in leukemia cells to reverse malignancy, J. Controlled Release, 2022, 343, 645–656 CrossRef CAS PubMed.
- L. Qiao, M. Gao, X. Yi, H. Peng, R. Zhang and W. Yao, et al., Biomimetic gene editing system for precise tumor cell reprogramming and augmented tumor therapy, J. Controlled Release, 2023, 356, 663–677 CrossRef CAS PubMed.
- G. Chen, B. Ma, Y. Wang and S. Gong, A Universal GSH-Responsive Nanoplatform for the Delivery of DNA, mRNA, and Cas9/sgRNA Ribonucleoprotein, ACS Appl. Mater. Interfaces, 2018, 10, 18515–18523 CrossRef CAS PubMed.
- J. Guo, T. Wan, B. Li, Q. Pan, H. Xin and Y. Qiu, et al., Rational Design of Poly(disulfide)s as a Universal Platform for Delivery of CRISPR-Cas9 Machineries toward Therapeutic Genome Editing, ACS Cent. Sci., 2021, 7, 990–1000 CrossRef CAS PubMed.
- N. Yoshinaga, J. K. Zhou, C. Xu, C. H. Quek, Y. Zhu and D. Tang, et al., Phenylboronic Acid-Functionalized Polyplexes Tailored to Oral CRISPR Delivery, Nano Lett., 2023, 23, 757–764 CrossRef CAS PubMed.
- T. Wan, Y. Chen, Q. Pan, X. Xu, Y. Kang and X. Gao, et al., Genome editing of mutant KRAS through supramolecular polymer-mediated delivery of Cas9 ribonucleoprotein for colorectal cancer therapy, J. Controlled Release, 2020, 322, 236–247 CrossRef CAS PubMed.
- Y. Wang, B. Ma, A. A. Abdeen, G. Chen, R. Xie and K. Saha, et al., Versatile Redox-Responsive Polyplexes for the Delivery of Plasmid DNA, Messenger RNA, and CRISPR-Cas9 Genome-Editing Machinery, ACS Appl. Mater. Interfaces, 2018, 10, 31915–31927 CrossRef CAS PubMed.
- Z. Zhang, T. Wan, Y. Chen, Y. Chen, H. Sun and T. Cao, et al., Cationic Polymer-Mediated CRISPR/Cas9 Plasmid Delivery for Genome Editing, Macromol. Rapid Commun., 2019, 40, 5 Search PubMed.
- Y. Zhang, C. Sun, C. Wang, K. E. Jankovic and Y. Dong, Lipids and Lipid Derivatives for RNA Delivery, Chem. Rev., 2021, 121, 12181–12277 CrossRef CAS PubMed.
- X. Han, H. Zhang, K. Butowska, K. L. Swingle, M. G. Alameh and D. Weissman, et al., An ionizable lipid toolbox for RNA delivery, Nat. Commun., 2021, 12, 7233 CrossRef CAS PubMed.
- X. Yan, Q. Pan, H. Xin, Y. Chen and Y. Ping, Genome-editing prodrug: Targeted delivery and conditional stabilization of CRISPR-Cas9 for precision therapy of inflammatory disease, Sci. Adv., 2021, 7, eabj0624 CrossRef CAS PubMed.
- H. Liu, Y. Y. Su, X. C. Jiang and J. Q. Gao, Cell membrane-coated nanoparticles: a novel multifunctional biomimetic drug delivery system, Drug Delivery Transl. Res., 2023, 13, 716–737 CrossRef CAS PubMed.
- X. Xu, H. Tang, J. Guo, H. Xin and Y. Ping, A dual-specific CRISPR-Cas nanosystem for precision therapeutic editing of liver disorders, Signal Transduction Targeted Ther., 2022, 7, 269 CrossRef PubMed.
- D. Rosenblum, A. Gutkin, R. Kedmi, S. Ramishetti, N. Veiga and A. M. Jacobi, et al., CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy, Sci. Adv., 2020, 6, eabc9450 CrossRef CAS PubMed.
- P. Guo, J. Yang, J. Huang, D. T. Auguste and M. A. Moses, Therapeutic genome editing of triple-negative breast tumors using a noncationic and deformable nanolipogel, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 18295–18303 CrossRef CAS PubMed.
- L. N. Kasiewicz, S. Biswas, A. Beach, H. Ren, C. Dutta and A. M. Mazzola, et al., GalNAc-Lipid nanoparticles enable non-LDLR dependent hepatic delivery of a CRISPR base editing therapy, Nat. Commun., 2023, 14, 1 Search PubMed.
- C. Yang, Y. Fu, C. Huang, D. Hu, K. Zhou and Y. Hao, et al., Chlorin e6 and CRISPR-Cas9 dual-loading system with deep penetration for a synergistic tumoral photodynamic-immunotherapy, Biomaterials, 2020, 255, 120194 CrossRef CAS PubMed.
- K. Ma, W. Li, G. Zhu, S. Sun, H. Chi and Y. Yin, et al., Functionalized PDA/DEX-PEI@HA nanoparticles combined with sleeping-beauty transposons for multistage targeted delivery of CRISPR/Cas9 gene, Biomed. Pharmacother., 2021, 142, 112061 CrossRef CAS PubMed.
- S. Deng, X. Li and S. Liu, et al., Codelivery of CRISPR-Cas9 and Chlorin E6 for Spatially Controlled Tumor-Specific Gene Editing with Synergistic Drug Effects, Sci. Adv., 2020, 6, eabb4005 CrossRef CAS PubMed.
- B. Y. Liu, X. Y. He, C. Xu, L. Xu, S. L. Ai and S. X. Cheng, et al., A Dual-Targeting Delivery System for Effective Genome Editing and in Situ Detecting Related Protein Expression in Edited Cells, Biomacromolecules, 2018, 19, 2957–2968 CrossRef CAS PubMed.
- M. Qiu, Y. Tang, J. Chen, R. Muriph, Z. Ye and C. Huang, et al., Lung-selective mRNA delivery of synthetic lipid nanoparticles for the treatment of pulmonary lymphangioleiomyomatosis, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2116271119 CrossRef CAS PubMed.
- K. Paunovska, A. J. Da Silva Sanchez, C. D. Sago, Z. Gan, M. P. Lokugamage and F. Z. Islam, et al., Nanoparticles Containing Oxidized Cholesterol Deliver mRNA to the Liver Microenvironment at Clinically Relevant Doses, Adv. Mater., 2019, 31, 14 Search PubMed.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing, Nat. Nanotechnol., 2020, 15, 313–320 CrossRef CAS PubMed.
- T. Wei, Y. Sun, Q. Cheng, S. Chatterjee, Z. Traylor and L. T. Johnson, et al., Lung SORT LNPs enable precise homology-directed repair mediated CRISPR/Cas genome correction in cystic fibrosis models, Nat. Commun., 2023, 14, 1 Search PubMed.
- D. Sun, Z. Sun, H. Jiang, A. M. Vaidya, R. Xin and N. R. Ayat, et al., Synthesis and Evaluation of pH-Sensitive Multifunctional Lipids for Efficient Delivery of CRISPR/Cas9 in Gene Editing, Bioconjugate Chem., 2019, 30, 667–678 CrossRef CAS PubMed.
- C. Liu, N. Wang, R. Luo, L. Li, W. Yang and X. Wang, et al., A programmable hierarchical-responsive nanoCRISPR elicits robust activation of endogenous target to treat cancer, Theranostics, 2021, 11, 9833–9846 CrossRef CAS PubMed.
- J. Yang, Z. Li, M. Shen, Y. Wang, L. Wang and J. Li, et al., Programmable Unlocking Nano-Matryoshka-CRISPR Precisely Reverses Immunosuppression to Unleash Cascade Amplified Adaptive Immune Response, Adv. Sci., 2021, 8, 13 Search PubMed.
- J. Liu, J. Chang, Y. Jiang, X. Meng, T. Sun and L. Mao, et al., Fast and Efficient CRISPR/Cas9 Genome Editing In Vivo Enabled by Bioreducible Lipid and Messenger RNA Nanoparticles, Adv. Mater., 2019, 31, 33 CrossRef.
- Y. A. Aksoy, B. Yang, W. Chen, T. Hung, R. P. Kuchel and N. W. Zammit, et al., Spatial and Temporal Control of CRISPR-Cas9-Mediated Gene Editing Delivered via a Light-Triggered Liposome System, ACS Appl. Mater. Interfaces, 2020, 12, 52433–52444 CrossRef CAS PubMed.
- Y. Xing, J. Yang, Y. Wang, C. Wang, Z. Pan and F. L. Liu, et al., Remodeling Tumor Immunogenicity with Dual-Activatable Binary CRISPR Nanomedicine for Cancer Immunotherapy, ACS Nano, 2023, 17, 5713–5726 CrossRef CAS PubMed.
- H. Yin, L. Sun, Y. Pu, J. Yu, W. Feng and C. Dong, et al., Ultrasound-Controlled CRISPR/Cas9 System Augments Sonodynamic Therapy of Hepatocellular Carcinoma, ACS Cent. Sci., 2021, 7, 2049–2062 CrossRef CAS PubMed.
- H. Lu, Q. Zhang, S. He, S. Liu, Z. Xie and X. Li, et al., Reduction-Sensitive Fluorinated-Pt(IV) Universal Transfection Nanoplatform Facilitating CT45-Targeted CRISPR/dCas9 Activation for Synergistic and Individualized Treatment of Ovarian Cancer, Small, 2021, 17, 41 Search PubMed.
- J. Yang, L. Bai, M. Shen, X. Gou, Z. Xiang and S. Ma, et al., A Multiple Stimuli-Responsive NanoCRISPR Overcomes Tumor Redox Heterogeneity to Augment Photodynamic Therapy, ACS Nano, 2023, 17, 11414–11426 CrossRef CAS PubMed.
- J. D. Ziebarth and Y. Wang, Understanding the protonation behavior of linear polyethylenimine in solutions through Monte Carlo simulations, Biomacromolecules, 2010, 11, 29–38 CrossRef CAS PubMed.
- A. Zakeri, M. A. J. Kouhbanani and N. Beheshtkhoo, et al., Polyethylenimine-based nanocarriers in co-delivery of drug and gene: a developing horizon, Nano Rev. Exp., 2018, 9, 1488497 CrossRef PubMed.
- J. Yang, L. Bai, M. Shen, X. Gou, Z. Xiang and S. Ma, et al., A Multiple Stimuli-Responsive NanoCRISPR Overcomes Tumor Redox Heterogeneity to Augment Photodynamic Therapy, ACS Nano, 2023, 17, 11414–11426 CrossRef CAS PubMed.
- H. X. Wang, Z. Song, Y. H. Lao, X. Xu, J. Gong and D. Cheng, et al., Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4903–4908 CrossRef CAS PubMed.
- M. K. Grun, A. Suberi, K. Shin, T. Lee, V. Gomerdinger and Z. M. Moscato, et al., PEGylation of poly(amine-co-ester) polyplexes for tunable gene delivery, Biomaterials, 2021, 272, 120780 CrossRef CAS PubMed.
- R. El-Kharrag, K. E. Berckmueller, R. Madhu, M. Cui, G. Campoy and H. M. Mack, et al., Efficient polymer nanoparticle-mediated delivery of gene editing reagents into human hematopoietic stem and progenitor cells, Mol. Ther., 2022, 30, 2186–2198 CrossRef CAS PubMed.
- G. Niu, Z. Jin, C. Zhang, D. He, X. Gao and C. Zou, et al., An effective vaginal gel to deliver CRISPR/Cas9 system encapsulated in poly (β-amino ester) nanoparticles for vaginal gene therapy, EBioMedicine, 2020, 58, 102897 CrossRef PubMed.
- T. Rodgers, N. Muzzio, C. Watson and G. Romero, Stabilization of poly (β-amino ester) nanoparticles for the efficient intracellular delivery of piggybac transposon, Bioengineering, 2021, 8, 1–14 CrossRef PubMed.
- Y. Xiao, Z. Tang, X. Huang, W. Chen, J. Zhou and H. Liu, et al., Emerging mRNA technologies: delivery strategies and biomedical applications, Chem. Soc. Rev., 2022, 51, 3828–3845 RSC.
- Y. Rui, D. R. Wilson, S. Y. Tzeng, H. M. Yamagata, D. Sudhakar and M. Conge, et al., High-throughput and high-content bioassay enables tuning of polyester nanoparticles for cellular uptake, endosomal escape, and systemic in vivo delivery of mRNA, Sci. Adv., 2022, 8, eabk2855 CrossRef CAS PubMed.
- M. P. Lokugamage, D. Vanover, J. Beyersdorf, M. Z. C. Hatit, L. Rotolo and E. S. Echeverri, et al., Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs, Nat. Biomed. Eng., 2021, 5, 1059–1068 CrossRef CAS PubMed.
- J. C. Kaczmarek, A. K. Patel, L. H. Rhym, U. C. Palmiero, B. Bhat and M. W. Heartlein, et al., Systemic delivery of mRNA and DNA to the lung using polymer-lipid nanoparticles, Biomaterials, 2021, 275, 120966 CrossRef CAS PubMed.
- X. Gao, Z. Jin, X. Tan, C. Zhang, C. Zou and W. Zhang, et al., Hyperbranched poly(β-amino ester) based polyplex nanopaticles for delivery of CRISPR/Cas9 system and treatment of HPV infection associated cervical cancer, J. Controlled Release, 2020, 321, 654–668 CrossRef CAS PubMed.
- J. O'Keeffe Ahern, I. Lara-Sáez, D. Zhou, R. Murillas, J. Bonafont and Á. Mencía, et al., Non-viral delivery of CRISPR–Cas9 complexes for targeted gene editing via a polymer delivery system, Gene Ther., 2022, 29, 157–170 CrossRef PubMed.
- Y. Liu, Y. Li, D. Keskin and L. Shi, Poly(β-Amino Esters): Synthesis, Formulations, and Their Biomedical Applications, Adv. Healthc. Mater., 2019, 8, 1801359 CrossRef PubMed.
- T. Wan, Q. Pan and Y. Ping, Microneedle-assisted genome editing: A transdermal strategy of targeting NLRP3 by CRISPR-Cas9 for synergistic therapy of inflammatory skin disorders, Sci. Adv., 2021, 7, eabe2888 CrossRef CAS PubMed.
- C. Dufès, I. F. Uchegbu and A. G. Schätzlein, Dendrimers in gene delivery, Adv. Drug Delivery Rev., 2005, 57, 2177–2202 CrossRef PubMed.
- A. Wahane, A. Waghmode, A. Kapphahn, K. Dhuri, A. Gupta and R. Bahal, Role of lipid-based and polymer-based non-viral vectors in nucleic acid delivery for next-generation gene therapy, Molecules, 2020, 25, 2866 CrossRef CAS PubMed.
- L. J. Cruz, T. van Dijk, O. Vepris, T. M. W. Y. Li, T. Schomann and F. Baldazzi, et al., PLGA-Nanoparticles for Intracellular Delivery of the CRISPR-Complex to Elevate Fetal Globin Expression in Erythroid Cells, Biomaterials, 2021, 268, 120580 CrossRef CAS PubMed.
- Q. Yang, Y. Zhou, J. Chen, N. Huang, Z. Wang and Y. Cheng, Gene therapy for drug-resistant glioblastoma via lipid-polymer hybrid nanoparticles combined with focused ultrasound, Int. J. Nanomed., 2021, 16, 185–199 CrossRef PubMed.
- C. Zhang, H. Ren, G. Liu, J. Li, X. Wang and Y. Zhang, Effective Genome Editing Using CRISPR-Cas9 Nanoflowers, Adv. Healthc. Mater., 2022, 11, 10 Search PubMed.
- Y. H. Lao, M. Li, M. A. Gao, D. Shao, C. W. Chi and D. Huang, et al., HPV Oncogene Manipulation Using Nonvirally Delivered CRISPR/Cas9 or Natronobacterium gregoryi Argonaute, Adv. Sci., 2018, 5, 7 Search PubMed.
- Y. Wang, Y. Tang, X. m. Zhao, G. Huang, J. h. Gong and S. d. Yang, et al., A multifunctional non-viral vector for the delivery of MTH1-targeted CRISPR/Cas9 system for non-small cell lung cancer therapy, Acta Biomater., 2022, 153, 481–493 CrossRef CAS PubMed.
- I. Ruseska, K. Fresacher, C. Petschacher and A. Zimmer, Use of protamine in nanopharmaceuticals—a review, Nanomaterials, 2021, 11, 1508 CrossRef CAS PubMed.
- D. Tang, Y. Yan, Y. Li, Y. Li, J. Tian and L. Yang, et al., Targeting DAD1 gene with CRISPR-Cas9 system transmucosally delivered by fluorinated polylysine nanoparticles for bladder cancer intravesical gene therapy, Theranostics, 2023, 14, 203–219 CrossRef PubMed.
- M. Chiper, N. Tounsi, R. Kole, A. Kichler and G. Zuber, Self-aggregating 1.8 kDa polyethylenimines with dissolution switch at endosomal acidic pH are delivery carriers for plasmid DNA, mRNA, siRNA and exon-skipping oligonucleotides, J. Controlled Release, 2017, 246, 60–70 CrossRef CAS PubMed.
- T. Lan and Q. Guo, Phenylboronic acid-decorated polymeric nanomaterials for advanced bio-application, Nanotechnol. Rev., 2019, 8, 548–561 CAS.
- J. Wankar, N. G. Kotla, S. Gera, S. Rasala, A. Pandit and Y. A. Rochev, Recent Advances in Host–Guest Self-Assembled Cyclodextrin Carriers: Implications for Responsive Drug Delivery and Biomedical Engineering, Adv. Funct. Mater., 2020, 30, 1909049 CrossRef CAS.
- D. N. Nguyen, T. L. Roth, P. J. Li, P. A. Chen, R. Apathy and M. R. Mamedov, et al., Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency, Nat. Biotechnol., 2020, 38, 44–49 CrossRef CAS PubMed.
- B. C. Evans, R. B. Fletcher, K. V. Kilchrist, E. A. Dailing, A. J. Mukalel and J. M. Colazo, et al., An anionic, endosome-escaping polymer to potentiate intracellular delivery of cationic peptides, biomacromolecules, and nanoparticles, Nat. Commun., 2019, 10, 1 CrossRef CAS PubMed.
- Y. Lin, U. Wilk and J. Pöhmerer, et al., Folate Receptor-Mediated Delivery of Cas9 RNP for Enhanced Immune Checkpoint Disruption in Cancer Cells, Small, 2023, 19, 2205318 CrossRef CAS PubMed.
- T. Del’Guidice, J. P. Lepetit-Stoffaes and L. J. Bordeleau, et al., Membrane permeabilizing amphiphilic peptide delivers recombinant transcription factor and CRISPR-Cas9/Cpf1 ribonucleoproteins in hard-to-modify cells, PLoS One, 2018, 13, e0195558 CrossRef PubMed.
- S. Krishnamurthy, C. Wohlford-Lenane and S. Kandimalla, et al., Engineered amphiphilic peptides enable delivery of proteins and CRISPR-associated nucleases to airway epithelia, Nat. Commun., 2019, 10, 4906 CrossRef PubMed.
- Y. Lin, X. Luo and T. Burghardt, et al., Chemical Evolution of Amphiphilic Xenopeptides for Potentiated Cas9 Ribonucleoprotein Delivery, J. Am. Chem. Soc., 2023, 145, 15171–15179 CrossRef CAS PubMed.
- D. V. Foss, J. J. Muldoon and D. N. Nguyen, et al., Peptide-mediated delivery of CRISPR enzymes for the efficient editing of primary human lymphocytes, Nat. Biomed. Eng., 2023, 7, 647–660 CrossRef CAS PubMed.
- C. Hald Albertsen, J. A. Kulkarni, D. Witzigmann, M. Lind, K. Petersson and J. B. Simonsen, The role of lipid components in lipid nanoparticles for vaccines and gene therapy, Adv. Drug Delivery Rev., 2022, 188, 114416 CrossRef CAS PubMed.
- B. Li, A. Y. Jiang, I. Raji, C. Atyeo, T. M. Raimondo and A. G. R. Gordon, et al., Enhancing the immunogenicity of lipid-nanoparticle mRNA vaccines by adjuvanting the ionizable lipid and the mRNA, Nat. Biomed. Eng., 2023 DOI:10.1038/s41551-023-01082-6.
- E. Kenjo, H. Hozumi, Y. Makita, K. A. Iwabuchi, N. Fujimoto and S. Matsumoto, et al., Low immunogenicity of LNP allows repeated administrations of CRISPR-Cas9 mRNA into skeletal muscle in mice, Nat. Commun., 2021, 12, 1 CrossRef PubMed.
- A. M. Jörgensen, R. Wibel and A. Bernkop-Schnürch, Biodegradable Cationic and Ionizable Cationic Lipids: A Roadmap for Safer Pharmaceutical Excipients, Small, 2023, 19, e2206968 CrossRef PubMed.
- H. Yin, C. Q. Song, S. Suresh, Q. Wu, S. Walsh and L. H. Rhym, et al., Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing, Nat. Biotechnol., 2017, 35, 1179–1187 CrossRef CAS PubMed.
- M. Wang, J. A. Zuris, F. Meng, H. Rees, S. Sun and P. Deng, et al., Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2868–2873 CrossRef CAS PubMed.
- X. Zhang, W. Zhao, G. N. Nguyen, C. Zhang, C. Zeng and J. Yan, et al., Functionalized lipid-like nanoparticles for in vivo mRNA delivery and base editing, Sci. Adv., 2020, 6, eabc2315 CrossRef CAS PubMed.
- B. Li, R. S. Manan, S. Q. Liang, A. Gordon, A. Jiang and A. Varley, et al., Combinatorial design of nanoparticles for pulmonary mRNA delivery and genome editing, Nat. Biotechnol., 2023, 41, 1410–1415 CrossRef CAS PubMed.
- Y. Li, T. Yang, Y. Yu, N. Shi, L. Yang and Z. Glass, et al., Combinatorial library of chalcogen-containing lipidoids for intracellular delivery of genome-editing proteins, Biomaterials, 2018, 178, 652–662 CrossRef CAS PubMed.
- Y. Li, J. Bolinger, Y. Yu, Z. Glass, N. Shi and L. Yang, et al., Intracellular delivery and biodistribution study of CRISPR/Cas9 ribonucleoprotein loaded bioreducible lipidoid nanoparticles, Biomater. Sci., 2019, 7, 596–606 RSC.
- Y. Li, A. C. Li and Q. Xu, Intracellular Delivery of His-Tagged Genome-Editing Proteins Enabled by Nitrilotriacetic Acid–Containing Lipidoid Nanoparticles, Adv. Healthc. Mater., 2019, 8, 6 Search PubMed.
- M. Qiu, Z. Glass, J. Chen, M. Haas, X. Jin and X. Zhao, et al., Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2020401118 CrossRef CAS PubMed.
- C. D. Sago, M. P. Lokugamage, K. Paunovska, D. A. Vanover, C. M. Monaco and N. N. Shah, et al., High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E9944–E9952 CrossRef CAS PubMed.
- C. D. Sago, M. P. Lokugamage, F. Z. Islam, B. R. Krupczak, M. Sato and J. E. Dahlman, Nanoparticles That Deliver RNA to Bone Marrow Identified by in Vivo Directed Evolution, J. Am. Chem. Soc., 2018, 140, 17095–17105 CrossRef CAS PubMed.
- J. E. Dahlman, C. Barnes, O. F. Khan, A. Thiriot, S. Jhunjunwala and T. E. Shaw, et al., In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight, Nat. Nanotechnol., 2014, 9, 648–655 CrossRef CAS PubMed.
- O. F. Khan, P. S. Kowalski, J. C. Doloff, J. K. Tsosie, V. Bakthavatchalu and C. B. Winn, et al., Endothelial siRNA delivery in nonhuman primates using ionizable low-molecular weight polymeric nanoparticles, Sci. Adv., 2018, 4, eaar8409 CrossRef PubMed.
- L. Farbiak, Q. Cheng, T. Wei, E. Álvarez-Benedicto, L. T. Johnson and S. Lee, et al., All-In-One Dendrimer-Based Lipid Nanoparticles Enable Precise HDR-Mediated Gene Editing In Vivo, Adv. Mater., 2021, 33, 30 CrossRef PubMed.
- S. Liu, Q. Cheng, T. Wei, X. Yu, L. T. Johnson and L. Farbiak, et al., Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing, Nat. Mater., 2021, 20, 701–710 CrossRef CAS PubMed.
- A. Mackensen, J. B. A. G. Haanen, C. Koenecke, W. Alsdorf, E. Wagner-Drouet and P. Borchmann, et al., CLDN6-specific CAR-T cells plus amplifying RNA vaccine in relapsed or refractory solid tumors: the phase 1 BNT211–01 trial, Nat. Med., 2023, 29, 2844–2853 CrossRef CAS PubMed.
- L. Miao, J. Lin, Y. Huang, L. Li, D. Delcassian and Y. Ge, et al., Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver, Nat. Commun., 2020, 11, 1 CrossRef PubMed.
- E. Y. Cho, J. Y. Ryu, H. A. R. Lee, S. H. Hong, H. S. Park and K. S. Hong, et al., Lecithin nano-liposomal particle as a CRISPR/Cas9 complex delivery system for treating type 2 diabetes, J. Nanobiotechnol., 2019, 17, 1 CrossRef PubMed.
- J. B. Miller, S. Zhang, P. Kos, H. Xiong, K. Zhou and S. S. Perelman, et al., Non–Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co–Delivery of Cas9 mRNA and sgRNA, Angew. Chem., 2017, 129, 1079–1083 CrossRef.
- S. Patel, N. Ashwanikumar, E. Robinson, Y. Xia, C. Mihai and J. P. Griffith, et al., Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA, Nat. Commun., 2020, 11, 1 CrossRef PubMed.
- M. Gautam, A. Jozic, G. L. N. Su, M. Herrera-Barrera, A. Curtis and S. Arrizabalaga, et al., Lipid nanoparticles with PEG-variant surface modifications mediate genome editing in the mouse retina, Nat. Commun., 2023, 14, 1 Search PubMed.
- T. Lorson, M. M. Lübtow, E. Wegener, M. S. Haider, S. Borova and D. Nahm, et al., Poly(2-oxazoline)s based biomaterials: A comprehensive and critical update, Biomaterials, 2018, 178, 204–280 CrossRef CAS PubMed.
- S. S. Nogueira, A. Schlegel and K. Maxeiner, et al., Polysarcosine-Functionalized Lipid Nanoparticles for Therapeutic mRNA Delivery, ACS Appl. Nano Mater., 2020, 3, 10634–10645 CrossRef CAS.
- Y. Lee, M. Jeong, J. Park, H. Jung and H. Lee, Immunogenicity of lipid nanoparticles and its impact on the efficacy of mRNA vaccines and therapeutics, Exp. Mol. Med., 2023, 55, 2085–2096 CrossRef CAS PubMed.
- M. Li, H. Zhou, N. Wu, W. Deng, W. Dong and X. Sun, et al., Pathogen Recognition-Driven Dendritic Cell-Specific Gene Silencing and Editing, Nano Lett., 2023, 23, 2733–2742 CrossRef CAS PubMed.
- I. Menon, M. Zaroudi, Y. Zhang, E. Aisenbrey and L. Hui, Fabrication of active targeting lipid nanoparticles: Challenges and perspectives, Mater. Today Adv., 2022, 16, 100299 CrossRef CAS.
- W. Ruan, M. Jiao, S. Xu, M. Ismail, X. Xie and Y. An, et al., Brain-targeted CRISPR/Cas9 nanomedicine for effective glioblastoma therapy, J. Controlled Release, 2022, 351, 739–751 CrossRef CAS PubMed.
- R. Kedmi, N. Veiga, S. Ramishetti, M. Goldsmith, D. Rosenblum and N. Dammes, et al., A modular platform for targeted RNAi therapeutics, Nat. Nanotechnol., 2018, 13, 214–219 CrossRef CAS PubMed.
- X. He, Q. Long, Z. Zeng, L. Yang, Y. Tang and X. Feng, Simple and Efficient Targeted Intracellular Protein Delivery with Self-Assembled Nanovehicles for Effective Cancer Therapy, Adv. Funct. Mater., 2019, 29, 50 Search PubMed.
- Q. Li, X. Lv, C. Tang and C. Yin, Co-delivery of doxorubicin and CRISPR/Cas9 or RNAi-expressing plasmid by chitosan-based nanoparticle for cancer therapy, Carbohydr. Polym., 2022, 287, 119315 CrossRef CAS PubMed.
- Q. Tang, J. Liu, Y. Jiang, M. Zhang, L. Mao and M. Wang, Cell-Selective Messenger RNA Delivery and CRISPR/Cas9 Genome Editing by Modulating the Interface of Phenylboronic Acid-Derived Lipid Nanoparticles and Cellular Surface Sialic Acid, ACS Appl. Mater. Interfaces, 2019, 11, 46585–46590 CrossRef CAS PubMed.
- W. Sun, J. Wang, Q. Hu, X. Zhou, A. Khademhosseini and Z. Gu, CRISPR-Cas12a delivery by DNA-mediated bioresponsive editing for cholesterol regulation, Sci. Adv., 2020, 6, eaba2983 CrossRef CAS PubMed.
- H. Liu, Q. Zhang, S. Wang, W. Weng, Y. Jing and J. Su, Bacterial extracellular vesicles as bioactive nanocarriers for drug delivery: Advances and perspectives, Bioact. Mater., 2022, 14, 169–181 CAS.
- R. Li, Y. He, S. Zhang, J. Qin and J. Wang, Cell membrane-based nanoparticles: a new biomimetic platform for tumor diagnosis and treatment, Acta Pharm. Sin. B, 2018, 8, 14–22 CrossRef PubMed.
- S. T. LoPresti, M. L. Arral, N. Chaudhary and K. A. Whitehead, The replacement of helper lipids with charged alternatives in lipid nanoparticles facilitates targeted mRNA delivery to the spleen and lungs, J. Controlled Release, 2022, 345, 819–831 CrossRef CAS PubMed.
- T. Fang, X. Cao, M. Ibnat and G. Chen, Stimuli-responsive nanoformulations for CRISPR-Cas9 genome editing, J. Nanobiotechnol., 2022, 20, 354 CrossRef CAS PubMed.
- R. Xie, Y. Wang and S. Gong, External stimuli-responsive nanoparticles for spatially and temporally controlled delivery of CRISPR-Cas genome editors, Biomater. Sci., 2021, 9, 6012–6022 RSC.
- D. K. Sahel, S. G. Goswami, R. Jatyan, A. Kumari, A. Mittal and S. Ramalingam, et al., Lipopolymeric Nanocarrier Enables Effective Delivery of CRISPR/Cas9 Expressing Plasmid, Macromol. Rapid Commun., 2023, 44, 14 CrossRef PubMed.
- C. S. Wang, C. H. Chang, T. Y. Tzeng, A. M. Y. Lin and Y. L. Lo, Gene-editing by CRISPR-Cas9 in combination with anthracycline therapy via tumor microenvironment-switchable, EGFR-targeted, and nucleus-directed nanoparticles for head and neck cancer suppression, Nanoscale Horiz., 2021, 6, 729–743 RSC.
- K. Tu, H. Deng, L. Kong, Y. Wang, T. Yang and Q. Hu, et al., Reshaping Tumor Immune Microenvironment through Acidity-Responsive Nanoparticles Featured with CRISPR/Cas9-Mediated Programmed Death-Ligand 1 Attenuation and Chemotherapeutics-Induced Immunogenic Cell Death, ACS Appl. Mater. Interfaces, 2020, 12, 16018–16030 CrossRef CAS PubMed.
- Z. Zhang, Q. Wang, Q. Liu, Y. Zheng, C. Zheng and K. Yi, et al., Dual-Locking Nanoparticles Disrupt the PD-1/PD-L1 Pathway for Efficient Cancer Immunotherapy, Adv. Mater., 2019, 31, 51 Search PubMed.
- Y. Rui, D. R. Wilson, K. Sanders and J. J. Green, Reducible Branched Ester-Amine Quadpolymers (rBEAQs) Codelivering Plasmid DNA and RNA Oligonucleotides Enable CRISPR/Cas9 Genome Editing, ACS Appl. Mater. Interfaces, 2019, 11, 10472–10480 CrossRef CAS PubMed.
- Y. Xing, J. Yang, C. Wang, Z. Kang, Z. Pan and J. Tang, et al., Time-programmed activation of CD47 disruption and immunogenic cell death with Cas9 ribonucleoprotein nanocapsule for improved cancer immunotherapy, Chem. Eng. J., 2023, 474, 145796 CrossRef CAS.
- R. Xie, X. Wang, Y. Wang, M. Ye, Y. Zhao and B. S. Yandell, et al., pH-Responsive Polymer Nanoparticles for Efficient Delivery of Cas9 Ribonucleoprotein With or Without Donor DNA, Adv. Mater., 2022, 34, 23 Search PubMed.
- N. Wang, C. Liu, Z. Lu, W. Yang, L. Li and S. Gong, et al., Multistage Sensitive NanoCRISPR Enable Efficient Intracellular Disruption of Immune Checkpoints for Robust Innate and Adaptive Immune Coactivation, Adv. Funct. Mater., 2020, 30, 45 Search PubMed.
- J. Fan, Y. Liu, L. Liu, Y. Huang, X. Li and W. Huang, A Multifunction Lipid-Based CRISPR-Cas13a Genetic Circuit Delivery System for Bladder Cancer Gene Therapy, ACS Synth. Biol., 2020, 9, 343–355 CrossRef CAS PubMed.
- Y. Lyu, S. He, J. Li, Y. Jiang, H. Sun and Y. Miao, et al., A Photolabile Semiconducting Polymer Nanotransducer for Near–Infrared Regulation of CRISPR/Cas9 Gene Editing, Angew. Chem., 2019, 131, 18365–18369 CrossRef.
- N. Qiu, J. Gao, Q. Liu, J. Wang and Y. Shen, Enzyme-Responsive Charge-Reversal Polymer-Mediated Effective Gene Therapy for Intraperitoneal Tumors, Biomacromolecules, 2018, 19, 2308–2319 CrossRef CAS PubMed.
- Y. Zhang, S. Shen, G. Zhao, C. F. Xu, H. B. Zhang and Y. L. Luo, et al., In situ repurposing of dendritic cells with CRISPR/Cas9-based nanomedicine to induce transplant tolerance, Biomaterials, 2019, 217, 119302 CrossRef CAS PubMed.
- Y. L. Luo, C. F. Xu, H. J. Li, Z. T. Cao, J. Liu and J. L. Wang, et al., Macrophage-Specific in Vivo Gene Editing Using Cationic Lipid-Assisted Polymeric Nanoparticles, ACS Nano, 2018, 12, 994–1005 CrossRef CAS PubMed.
- F. Persano, G. Gigli and S. Leporatti, Lipid-polymer hybrid nanoparticles in cancer therapy: current overview and future directions, Nano Express, 2021, 2, 012006 CrossRef.
- M. Iqbal, N. Zafar, H. Fessi and A. Elaissari, Double emulsion solvent evaporation techniques used for drug encapsulation, Int. J. Pharm., 2015, 496, 173–190 CrossRef CAS PubMed.
- A. Mukherjee, A. K. Waters, P. Kalyan, A. S. Achrol, S. Kesari and V. M. Yenugonda, Lipid-polymer hybrid nanoparticles as a nextgeneration drug delivery platform: State of the art, emerging technologies, and perspectives, Int. J. Nanomed., 2019, 14, 1937–1952 CrossRef CAS PubMed.
- H. Zhang, J. Yang, R. Sun, S. Han, Z. Yang and L. Teng, Microfluidics for nano-drug delivery systems: From fundamentals to industrialization, Acta Pharm. Sin. B, 2023, 13, 3277–3299 CrossRef CAS PubMed.
- S. J. Shepherd, C. C. Warzecha and S. Yadavali, et al., Scalable mRNA and siRNA Lipid Nanoparticle Production Using a Parallelized Microfluidic Device, Nano Lett., 2021, 21, 5671–5680 CrossRef CAS PubMed.
- Y. Suzuki, H. Onuma, R. Sato, Y. Sato, A. Hashiba and M. Maeki, et al., Lipid nanoparticles loaded with ribonucleoprotein–oligonucleotide complexes synthesized using a microfluidic device exhibit robust genome editing and hepatitis B virus inhibition, J. Controlled Release, 2021, 330, 61–71 CrossRef CAS PubMed.
- Y. Liu, G. Zhao, C. F. Xu, Y. L. Luo, Z. D. Lu and J. Wang, Systemic delivery of CRISPR/Cas9 with PEG-PLGA nanoparticles for chronic myeloid leukemia targeted therapy, Biomater. Sci., 2018, 6, 1592–1603 RSC.
- Y. Liu, Z. T. Cao, C. F. Xu, Z. D. Lu, Y. L. Luo and J. Wang, Optimization of lipid-assisted nanoparticle for disturbing neutrophils-related inflammation, Biomaterials, 2018, 172, 92–104 CrossRef CAS PubMed.
- M. Li, Y. N. Fan, Z. Y. Chen, Y. L. Luo, Y. C. Wang and Z. X. Lian, et al., Optimized nanoparticle-mediated delivery of CRISPR-Cas9 system for B cell intervention, Nano Res., 2018, 11, 6270–6282 CrossRef CAS.
- C. F. Xu, J. Wang, Y. L. Luo, L. F. Liang, Y. J. Gan and J. Liu, et al., An all-in-one nanomedicine consisting of CRISPR-Cas9 and an autoantigen peptide for restoring specific immune tolerance, ACS Appl. Mater. Interfaces, 2020, 12, 48259–48271 CrossRef PubMed.
- J. Y. Ryu, E. J. Won, H. A. R. Lee, J. H. Kim, E. Hui and H. P. Kim, et al., Ultrasound-activated particles as CRISPR/Cas9 delivery system for androgenic alopecia therapy, Biomaterials, 2020, 232, 119736 CrossRef CAS PubMed.
- X. Wang, Y. Zhao, M. Yan, X. Liang, N. Zhao and T. Lu, iRGD mediated pH-responsive mesoporous silica enhances drug accumulation in tumors, Eur. J. Pharm. Sci., 2024, 195, 106725 CrossRef CAS PubMed.
- Y. Zhao, Y. Zhou and D. Wang, et al., PH-responsive polymeric micelles based on poly(2-ethyl-2-oxazoline)-poly(d,l-lactide) for tumor-targeting and controlled delivery of doxorubicin and P-glycoprotein inhibitor, Acta Biomater., 2015, 17, 182–192 CrossRef CAS PubMed.
- G. H. Hsiue, C. H. Wang, C. L. Lo, C. H. Wang, J. P. Li and J. L. Yang, Environmental-sensitive micelles based on poly(2-ethyl-2-oxazoline)-b-poly(l-lactide) diblock copolymer for application in drug delivery, Int. J. Pharm., 2006, 317, 69–75 CrossRef CAS PubMed.
- K. Paunovska, C. J. Gil, M. P. Lokugamage, C. D. Sago, M. Sato and G. N. Lando, et al., Analyzing 2000 in vivo drug delivery data points reveals cholesterol structure impacts nanoparticle delivery, ACS Nano, 2018, 12, 8341–8349 CrossRef CAS PubMed.
- S. P. Chen and A. K. Blakney, Immune response to the components of lipid nanoparticles for ribonucleic acid therapeutics, Curr. Opin. Biotechnol, 2024, 85, 103049 CrossRef CAS PubMed.
- A. Alesci, M. Gitto and M. Kotańska, et al., Immunogenicity, effectiveness, safety and psychological impact of COVID-19 mRNA vaccines, Hum. Immunol., 2022, 83, 755–767 CrossRef CAS PubMed.
- Y. Ju, J. M. Carreño, V. Simon, K. Dawson, F. Krammer and S. J. Kent, Impact of anti-PEG antibodies induced by SARS-CoV-2 mRNA vaccines, Nat. Rev. Immunol., 2023, 23, 135–136 CrossRef CAS PubMed.
- M. P. Lokugamage, D. Vanover, J. Beyersdorf, M. Z. C. Hatit, L. Rotolo and E. S. Echeverri, et al., Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs, Nat. Biomed. Eng., 2021, 5, 1059–1068 CrossRef CAS PubMed.
- A. K. Patel, J. C. Kaczmarek, S. Bose, K. J. Kauffman, F. Mir and M. W. Heartlein, et al., Inhaled Nanoformulated mRNA Polyplexes for Protein Production in Lung Epithelium, Adv. Mater., 2019, 31, 8 Search PubMed.
- R. Zhong, S. Talebian, B. B. Mendes, G. Wallace, R. Langer and J. Conde, et al., Hydrogels for RNA delivery, Nat. Mater., 2023, 22, 818–831 CrossRef CAS PubMed.
- L. Gonçalves, P. Lavrador, A. J. R. Amaral, L. P. Ferreira, V. M. Gaspar and J. F. Mano, Double-Interlinked Colloidal Gels for Programable Fabrication of Supraparticle Architectures, Adv. Funct. Mater., 2023, 33, 45 CrossRef.
- B. Freitas, P. Lavrador, R. J. Almeida, V. M. Gaspar and J. F. Mano, Self-Assembled Bioactive Colloidal Gels as Injectable Multiparticle Shedding Platforms, ACS Appl. Mater. Interfaces, 2020, 12, 31282–31291 CrossRef CAS PubMed.
- S. Correa, A. K. Grosskopf, J. H. Klich, H. Lopez Hernandez and E. A. Appel, Injectable liposome-based supramolecular hydrogels for the programmable release of multiple protein drugs, Matter, 2022, 5, 1816–1838 CrossRef CAS PubMed.
- D. M. Strelkova Petersen, N. Chaudhary, M. L. Arral, R. M. Weiss and K. A. Whitehead, The mixing method used to formulate lipid nanoparticles affects mRNA delivery efficacy and organ tropism, Eur. J. Pharm. Biopharm., 2023, 192, 126–135 CrossRef CAS PubMed.
- A. Loewa, J. J. Feng and S. Hedtrich, Human disease models in drug development, Nat. Rev. Bioeng., 2023, 1, 545–559 CrossRef PubMed.
- S. Shah, P. Famta, R. S. Raghuvanshi, S. B. Singh and S. Srivastava, Lipid polymer hybrid nanocarriers: Insights into synthesis aspects, characterization, release mechanisms, surface functionalization and potential implications, Colloids Interface Sci. Commun., 2022, 46, 100570 CrossRef CAS.
- Y. Qin, A. A. Walters, N. Rouatbi, J. T. W. Wang, H. M. Abdel-Bar and K. T. Al-Jamal, Evaluation of a DoE based approach for comprehensive modelling of the effect of lipid nanoparticle composition on nucleic acid delivery, Biomaterials, 2023, 299, 122158 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |
