Open Access Article
Open Access ArticleMultiplexed no-wash cellular imaging using BenzoTag, an evolved self-labeling protein†
Bryan J.
Lampkin‡
 *,
Benjamin J.
Goldberg
*,
Benjamin J.
Goldberg
 and
Joshua A.
Kritzer
and
Joshua A.
Kritzer
 *
*
Department of Chemistry, Tufts University, Medford, MA 02155, USA. E-mail: lampkinb@uic.edu; joshua.kritzer@tufts.edu
First published on 9th October 2024
Abstract
Self-labeling proteins are powerful tools for exploring biology as they enable the precise cellular localization of a synthetic molecule, often a fluorescent dye. HaloTag7 is the most popular self-labeling protein due to its broad utility, its bio-orthogonality, and the simplicity of its chloroalkane ligand. However, reaction rates of HaloTag7 with different chloroalkane-containing substrates are highly variable and rates are only very fast for rhodamine-based dyes. This is a major limitation for the HaloTag system because fast labeling rates are critical for live-cell assays. Here, we use yeast surface display to produce a HaloTag variant, BenzoTag, with improved performance with a fluorogenic benzothiadiazole dye. Molecular evolution improved conjugation kinetics and increased the signal from the dye–protein complex, allowing for robust, no-wash fluorescence labeling in live cells. The new BenzoTag–benzothiadiazole system has improved performance compared to the best existing HaloTag7–silicon rhodamine system, including saturation of intracellular enzyme in under 100 seconds and robust labeling at dye concentrations as low as 7 nM. The BenzoTag system was also found to be sufficiently orthogonal to the HaloTag7–silicon rhodamine system to enable multiplexed no-wash labeling in live cells. The BenzoTag system will be immediately useful for a large variety of cell-based assays monitoring biological processes and drug action in real time.
Introduction
Biology relies heavily on monitoring fluorescently tagged proteins in living systems.1,2 Recent advancements in both synthetic dye chemistry and protein engineering have produced chemo-genetic constructs for superior cellular imaging. Specifically, self-labeling enzymes such as HaloTag7 allow organic dyes, which are often brighter and more photostable than fluorescent proteins, to be introduced to cellular environments with chemical and genetic precision.3–9 HaloTag7, a modified alkyl dehalogenase, reacts with a linear chloroalkane ligand which can be appended to any molecule of interest, most commonly fluorescent dyes (Fig. 1a) but also a wide variety of substrates including biomolecules. It has been used in a wide array of cell-based and live-animal assays that have revealed new insights into protein–protein interactions, drug action at the cell surface, cell-surface receptor recycling, protein homeostasis, targeted protein degradation, and cell penetration of peptide and oligonucleotide drugs, among other areas.10–16 HaloTag7 has also been reacted with metal-chelating ligands for catalysis,17,18 DNA nanostructures,19 photoactivatable substrates for light-induced release of reactive metabolites,20 and bifunctional substrates for chemically induced protein dimerization.21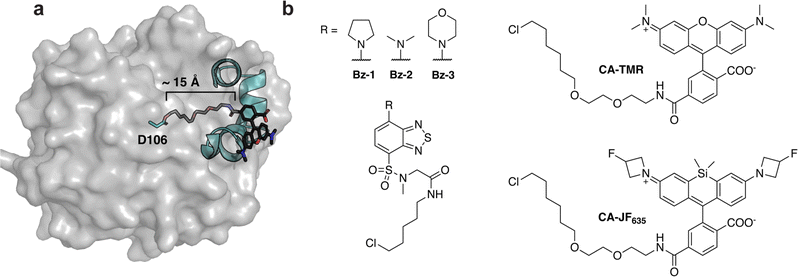 | ||
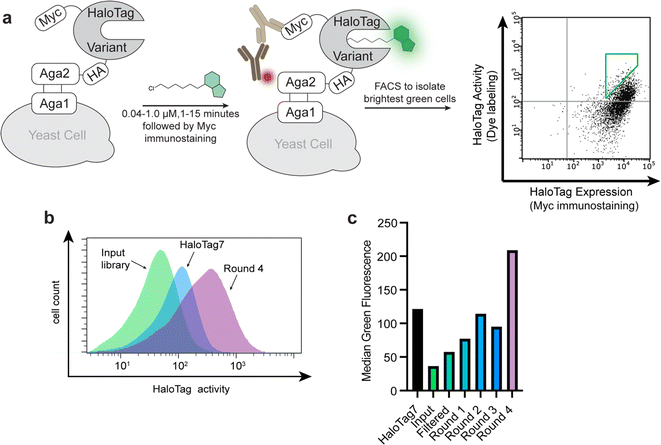
| Fig. 1 HaloTag7 and dye-containing substrates. (a) Crystal structure of HaloTag7 covalently reacted with substrate chloroalkane-tetramethylrhodamine (CA-TMR, PDB: 6Y7A).22 The model highlights how the catalytic residue, D106, is at the bottom of a ∼15 Å hydrophobic channel. This model also highlights how interactions between the rhodamine dye and the surface helices of HaloTag7 drive binding, illustrating why non-rhodamine substrates have slower kinetics. (b) Structures of fluorogenic dyes Bz-1, Bz-2, and Bz-3, and structures of the widely used HaloTag7 ligands CA-TMR and CA-JF635. | ||
HaloTag7 has proven to be very versatile given its genetic encodability and substrate modularity. However, its reaction rate with chloroalkane-tagged substrates is highly variable and substrate-specific.22 Specifically, HaloTag7 reacts with chloroalkane-tetramethylrhodamine (CA-TMR, Fig. 1) with second-order rate constants greater than 107 M−1 s−1 but this rate is much slower with non-rhodamine dyes. For example, the chloroalkane conjugate of AlexaFluor488, a synthetic derivative of fluorescein that still bears a xanthene core, has a reaction rate of 2.5 × 104 M−1 s−1, which is 1000-fold slower than that of CA-TMR.22 When non-xanthene dyes are used, this rate further plummets below 103 M−1 s−1.22–25 HaloTag7's preference for CA-TMR can be explained by the fact that CA-TMR was the substrate in the original HaloTag7 engineering efforts – HaloTag is no exception to the maxim “you get what you screen for.”26,27 In cellular assays, slow reaction kinetics results in sluggish or incomplete HaloTag7 labeling28 and/or the need for higher concentrations of dye, which leads to high background fluorescence.11
HaloTag7's substrate bias has been accommodated by using rhodamines and other xanthene-based dyes that range in spectral properties.29–33 Fluorogenic HaloTag systems, which use dyes that become fluorescent upon conjugation with HaloTag7, are especially attractive as they increase sensitivity and eliminate the need for washout steps, allowing for real-time monitoring of biochemical processes.34–36 However, many fluorogenic HaloTag7 ligands deviate from the rhodamine scaffold and thus suffer from slow reaction rates.23–25,37,38 Rhodamine-based fluorogenic dyes such as CA-JF635 (Fig. 1b) have been developed, and their conjugation kinetics are faster than non-rhodamine dyes but they do not approach the super-fast kinetics of CA-TMR.11,22,39,40 Given the utility of HaloTag7, we sought to broaden its substrate scope to enable more rapid kinetics with a larger variety of substrates. We were especially interested in improving HaloTag7's performance with benzothiadiazoles, a class of fluorogenic dyes that are non-fluorescent in aqueous solution and fluorescent in non-polar environments including the HaloTag7 active site channel as originally demonstrated by Liu, Zhang and coworkers.23,41 Based on that earlier work, we recently developed benzothiadiazole Bz-1 (Fig. 1b) as a cell-penetrant fluorogenic dye with very low background in mammalian cells.25 When conjugated to HaloTag7, Bz-1 had spectral properties that align with GFP and AlexaFluor488 allowing for the use of common blue lasers and blue/green filter sets.25,42,43 Further, Bz-1's small size, high photostability, large Stokes shift of 70 nm which limits self-absorption, and ease of derivatization renders it a nearly ideal dye for turn-on fluorescence labeling in cells. Despite these favorable properties, the reaction rate of HaloTag7 conjugation to Bz-1 was slower than the rates of many commonly used HaloTag7 substrates.25 Thus, we sought to improve the fluorogenic system by developing HaloTag7 variants with improved reaction kinetics with Bz-1.
Some recent efforts have sought to alter HaloTag7 to improve its performance with non-rhodamine substrates. Liang, Ward, and coworkers screened a library of 73 recombinantly expressed and purified single-mutant HaloTag variants for improved activity of a catalytic metal center and, in a separate report, a similar library was screened for improved fluorogenic and labeling properties of a styrylpyridium dye.17,44 Frei, Johnsson, and coworkers engineered HaloTag to modulate the fluorescent lifetimes of fluorogenic rhodamine dyes to enable multiplexed fluorescence lifetime imaging.45 They employed a HaloTag7 library generated by site-saturation mutagenesis of 10 pre-selected residues followed by screening bacterial lysates. While both strategies produced improved HaloTag7 variants for their given application, the screening throughput was limited. In this work, we developed a molecular evolution system for HaloTag that can screen 107 to 108 variants for optimal properties including faster conjugation kinetics. The system was applied to produce an optimized HaloTag7 variant with improved kinetics with Bz-1. We then demonstrated the application of the new self-labeling enzyme, BenzoTag, in rapid, wash-free intracellular labeling in live mammalian cells. The new BenzoTag·Bz-1 system performs comparably to the state-of-the-art HaloTag7·JF635 system but with faster in-cell labeling kinetics. BenzoTag·Bz-1 also shows sufficient orthogonality to that system to enable multiplexed, wash-free fluorescent labeling in live cells.
Results
Evolving HaloTag7 using yeast surface display
We sought to employ a molecular evolution method that would enable higher throughput than previous methods. We adapted yeast surface display for this purpose,46–50 which enabled us to screen libraries using fluorescence-activated cell sorting and to include epitope tags to allow independent measurements of protein activity and expression level (Fig. 2a).51 HaloTag7 was incorporated into a yeast display construct and activity of HaloTag7 on the yeast surface was verified by treating yeast with CA-TMR. Robust CA-TMR signal was observed for yeast cells expressing HaloTag7 but not cells expressing the catalytically inactive D106A mutant (ESI Fig. S1a). CA-TMR signal was linearly correlated with immunostaining signals detecting the HA or Myc tags, demonstrating independent measurements of labeling activity and expression levels (Fig. 2a, ESI Fig. S1b†). This was important to avoid bias towards high-expressing variants in subsequent screens. We used error-prone PCR to generate four sub-libraries with 2 to 6 mutations per variant (ESI Tables S3 and S4†). After verifying that each sub-library retained some activity (ESI Fig. S2†) they were pooled to yield an input library of 2.5 × 108 variants. We filtered the input library to remove catalytically dead variants by treating a pool of over 4 × 109 yeast with excess chloroalkane-biotin and then isolating biotinylated yeast using magnetic streptavidin beads. This pre-screen produced a filtered input library of functional HaloTag variants exceeding 5 × 107 unique members.To validate the HaloTag yeast display system, we screened the HaloTag variant library against Bz-1, a benzothiadiazole dye that we recently developed as a fluorogenic HaloTag ligand (Fig. 1b).25 We subjected the filtered library to iterative rounds of screening using fluorescence-activated cell sorting with substrate Bz-1. In each round, we isolated the top 0.5% of cells with high green fluorescence relative to expression level (Fig. 2a). Stringency was increased after each round by decreasing the concentration of Bz-1 and decreasing the incubation time, with round 4 applying 40 nM Bz-1 for one minute (ESI Table S5†). After four rounds of screening, there was a clear increase in fluorescence of the sorted variants when treated with Bz-1 compared to HaloTag7 (Fig. 2b and c).
HaloTag mutations increase reaction rate with Bz-1 and alter the spectral properties of the complex
We sequenced 110 colonies from rounds 3 and 4. There were few duplicate sequences, but numerous enriched mutations were observed. 15 HaloTag variants that included the highly enriched mutations were identified for further analysis. Notably, most enriched mutations were at residues that were not altered in prior HaloTag engineering efforts (ESI Table S2, see ESI Discussion†)5 with the exception of V245A, which was identified in an earlier HaloTag7 evolution screen with a non-rhodamine, styrylpyridium dye.44 After comparing the activities of these 15 variants on the surface of yeast (ESI Fig. S4, S5 and Table S6†) we selected six of the best-performing variants for recombinant expression and purification (variants 1–6, Table 1). All six variants demonstrated faster Bz-1 labeling kinetics than HaloTag7 (5- to 27-fold, Fig. 3a and b, ESI Fig. S6, S7 and Table S9†). The identified mutations also modulated the fluorescence properties of the enzyme·Bz-1 complex (Fig. 3c). For example, all six variants had increased fluorescence emission intensity compared to HaloTag7, with variant 1 producing the largest enhancement (14% greater than HaloTag7). Further, all mutants bearing the V245A mutation produced a slightly red-shifted emission maximum (Table 1). Examination of a crystal structure of the related dye Bz-2 conjugated with HaloTag7 suggested that all six variants have mutations that alter the environment near the dye's benzothiadiazole core and/or donor amine group (Fig. 3d).23,44 To explore whether these mutations altered interactions with Bz-1's benzothiadiazole core or its donor amine, we compared the kinetics for variants 5 and 6 reacting with Bz-1, Bz-2, and Bz-3 which have pyrrolidine, dimethylamine, and morpholine as their amine donors, respectively.25 The rates of Bz-2 and Bz-3 reacting with variants 5 and 6 were approximately 10-fold slower than Bz-1 but were approximately 10-fold faster than their rates with HaloTag7 (ESI Fig. S8†). These results implied that the newly evolved HaloTag variants specifically recognize both the benzothiadiazole core and the pyrrolidine donor group of Bz-1.| Variant | Mutations | Second-order rate constant (×103 M−1 s−1) | Fold change in fluorescence relative to HaloTag7 | Emission maximum (nm) |
|---|---|---|---|---|
| HaloTag7 | — | 1.5 ± 0.17 | — | 532 |
| Variant 1 | F144S|L246F | 9.3 ± 2.0 | 1.14 | 532 |
| Variant 2 | V245A | 11.0 ± 0.9 | 1.08 | 536 |
| Variant 3 | N195S|V245A|E251K|S291P | 8.3 ± 1.0 | 1.11 | 536 |
| Variant 4 | L161W|V245A | 10.3 ± 1.4 | 1.08 | 536 |
| Variant 5 | I211V|V245A | 41.3 ± 1.6 | 1.07 | 536 |
| Variant 6 | F144L|L221S|V245A | 33.4 ± 2.4 | 1.09 | 536 |
| Variant 7 | F144L|L221S|V245A|L246F | 15.6 ± 1.4 | 1.13 | 534 |
| Variant 8 | F144L|I211V|L221S|V245A | 68.8 ± 8.2 | 1.14 | 536 |
| Variant 9 | F144L|I211V|L221S|V245A|L246F | 44.6 ± 4.1 | 1.15 | 532 |
| Variant 10 (BenzoTag) | F144L|I211V|V245A | 95.4 ± 9.2 | 1.16 | 536 |
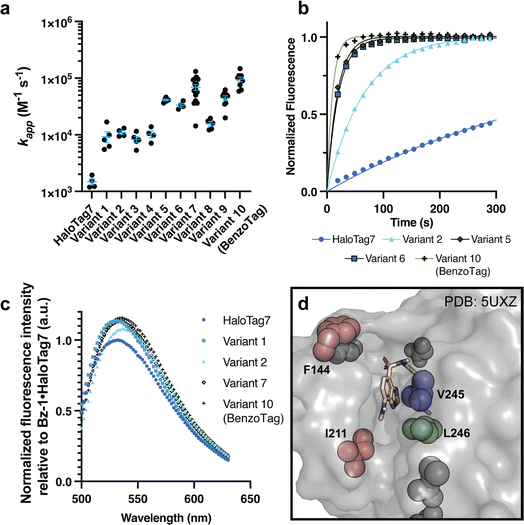 | ||
| Fig. 3 Characterization of recombinantly expressed HaloTag variants with improved fluorogenic properties. (a) Summary of second-order rate constants measured for Bz-1 conjugation to HaloTag7 and variants 1–10. See ESI† for experimental details and representative data. (b) Representative raw kinetic data for 0.25 μM Bz-1 reacting with 1.0 μM HaloTag7 or selected variant. (c) Representative emission spectra of 2.5 μM Bz-1 when conjugated to 5.0 μM HaloTag7 or selected variant after one hour of incubation, normalized to the maximum fluorescence intensity of the HaloTag7·Bz-1 complex. (d) Crystal structure of Bz-2 with HaloTag7 (PDB: 5UXZ)23 showing the locations of key mutations observed in variants with improved fluorogenic properties. | ||
We next generated variants 7–10 that combined different mutations from the six selected variants. We observed that combining mutations V245A, F144L, and L211V (variants 8–10) led to faster rates compared to variants with only two of these mutations (variants 5–7). We also observed that variants with the adjacent mutations V245A and L246F showed an overall decrease in reaction rate (variants 7 and 9, compared to 6 and 8). Lastly, L221S appeared to be a spectator mutation co-isolated with more beneficial mutations in variant 6, as it is located distal to the active site channel and it decreases overall activity (variant 10 compared to 8).
Bz-1 rapidly labels BenzoTag in live cells with low background
We selected variant 10, which we named BenzoTag, for testing in live mammalian cells. BenzoTag was cloned as a fusion to Histone 2B (H2B) to localize it to the nucleus and the fusion was transiently transfected into U-2 OS cells.45 Cells expressing BenzoTag or HaloTag7 were treated for 10 minutes with concentrations of Bz-1 between 7 and 1000 nM (Fig. 4a). Even at the highest concentration tested, Bz-1 showed minimal background fluorescence in non-expressing cells. BenzoTag dramatically outperformed HaloTag7 in live cell labeling with Bz-1, enabling robust fluorescence at 10- to 20-fold lower concentration of Bz-1. Saturation of the turn-on signal was not observed for HaloTag7-expressing cells even at 1000 nM Bz-1, while turn-on signal saturated for BenzoTag at 250 nM. Between 30 and 250 nM, BenzoTag-expressing cells had 5- to 8-fold higher fluorescence over background compared to HaloTag7-expressing cells. BenzoTag·Bz-1 labeling was also very sensitive – after labeling for 10 minutes with only 7 nM Bz-1, BenzoTag-expressing cells showed greater than 200-fold signal over background (Fig. 4a). These results highlight that the intrinsic properties of the Bz-1 dye, including high cell permeability and very low background fluorescence,25 synergize with the increased reaction rate to allow robust fluorescence detection at very low dye concentrations.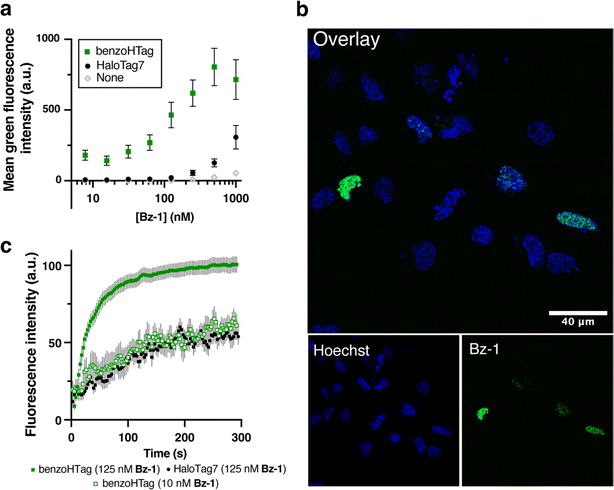 | ||
Fig. 4 Comparing BenzoTag and HaloTag7 in no-wash, live-cell labeling experiments. (a) Labeling with varied concentrations of Bz-1 for 10 minutes in non-transfected U-2 OS cells and cells transfected with H2B–BenzoTag or H2B–HaloTag7 (transfection efficiency was roughly 20%, ESI Fig. S10†). Data are shown as the mean and standard error of six biological replicates with 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells analyzed per experiment. (b) Confocal microscopy imaging of H2B–BenzoTag-expressing U-2 OS cells treated with 10 nM Bz-1. Cells were co-stained with Hoechst to highlight the nucleus. (c) Increase of Bz-1 fluorescence over time when added to U-2 OS cells that were transiently transfected with H2B–BenzoTag or H2B–HaloTag7. Fluorescence intensities of at least 10 transfected cells for each condition were measured over the course of five minutes with the average background fluorescence of 10 non-transfected cells subtracted. See ESI† for details. 000 cells analyzed per experiment. (b) Confocal microscopy imaging of H2B–BenzoTag-expressing U-2 OS cells treated with 10 nM Bz-1. Cells were co-stained with Hoechst to highlight the nucleus. (c) Increase of Bz-1 fluorescence over time when added to U-2 OS cells that were transiently transfected with H2B–BenzoTag or H2B–HaloTag7. Fluorescence intensities of at least 10 transfected cells for each condition were measured over the course of five minutes with the average background fluorescence of 10 non-transfected cells subtracted. See ESI† for details. | ||
We next evaluated the performance of the BenzoTag·Bz-1 system in no-wash, live cell fluorescence microscopy. U-2 OS cells transfected with the H2B–BenzoTag fusion were treated with 125 nM or 10 nM Bz-1 and imaged without exchanging media. Robust nuclear labeling was observed in BenzoTag-expressing cells while non-expressing cells within the same image had no observable background fluorescence (Fig. 4b, ESI Fig. S13†). We captured movies of Bz-1-treated cells (see ESI Video†) and quantified the appearance of fluorescence over time. In BenzoTag-expressing cells treated with 125 nM Bz-1, fluorescence approached saturation within 60 seconds, and BenzoTag-expressing cells saturated with twice the fluorescence signal compared to HaloTag7-expressing cells (Fig. 4c, ESI Fig. S14†). Notably, BenzoTag-expressing cells treated with only 10 nM Bz-1 also labeled within seconds and showed in-cell labeling kinetics similar to HaloTag7-expressing cells treated with 125 nM Bz-1. By contrast, no signal could be detected for HaloTag7-expressing cells when treated with only 10 nM Bz-1. This experiment also highlights the photostability of Bz-1, as it was under continuous irradiation for the five-minute video acquisition time without a decrease in signal. Indeed, superior photostability of benzothiadiazole dyes has been characterized up to 30 minutes of continuous irradiation without loss of signal.52
BenzoTag and HaloTag7 can be used for simultaneous, multiplexed labeling in live cells
Given that BenzoTag recognizes multiple parts of Bz-1, we wondered whether the BenzoTag system had evolved away from HaloTag7's large preference for rhodamine-based substrates. To test this, we measured the kinetics of recombinantly purified BenzoTag with CA-TMR and its fluorogenic silicon rhodamine analog, CA-JF635. CA-TMR reacted with BenzoTag with a second-order rate constant of 2.1 × 104 M−1 s−1 and CA-JF635 reacted with a rate of 1.1 × 102 M−1 s−1, which represent 900- and 9000-fold rate decreases, respectively, relative to their rates with HaloTag7 (Fig. 5a, ESI Fig. S9 and Table S10†). Overall, our kinetic data indicated that Bz-1 reacts 65-fold faster with BenzoTag than with HaloTag7, while CA-JF635 reacts 9000-fold faster with HaloTag7 than with BenzoTag (see ESI Discussion†). These results suggested that BenzoTag·Bz-1 and HaloTag7·CA-JF635 might be orthogonal enough for multiplexed labeling in cells. We then compared dye fluorescence in cells expressing either BenzoTag or HaloTag7 localized to the nucleus. We observed that CA-JF635 preferentially labeled cells expressing HaloTag7 with very little labeling in cells expressing BenzoTag, and Bz-1 preferentially labeled cells expressing BenzoTag with very little labeling in cells expressing HaloTag7 (Fig. 5b, ESI Fig. S11†).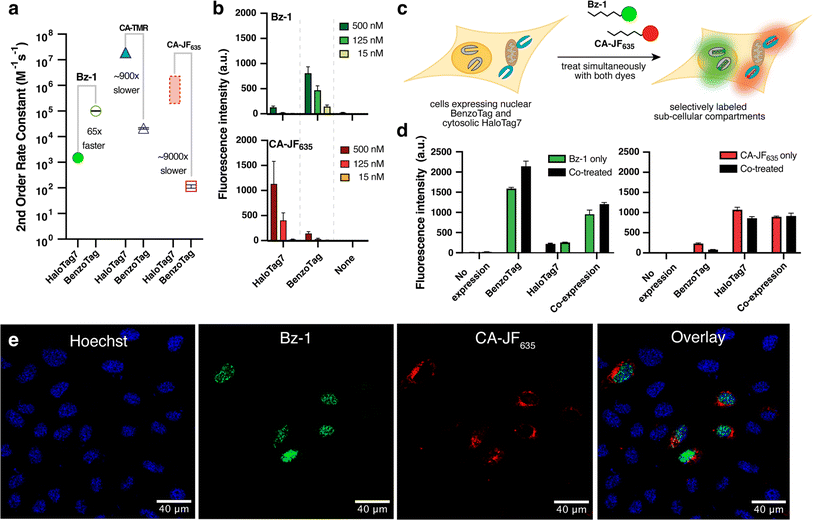 | ||
Fig. 5 Multiplexed labeling using the BenzoTag and HaloTag7 systems. (a) Second-order rate constants for Bz1, CA-TMR, and CA-JF635 with recombinantly expressed and purified HaloTag7 and BenzoTag. The rate of CA-TMR with HaloTag7 was obtained and reported by Johnsson and coworkers.22 The rate of CA-JF635 has not been reported, but rates of analogous Si rhodamines were reported in the range of 105–106 M−1 s−1.11,22,29,39 See ESI† for more details. (b) Comparison of Bz-1 and CA-JF635 labeling for 10 minutes in live U-2 OS cells transiently expressing H2B–BenzoTag or H2B–HaloTag7. Data are shown as the mean and standard error of biological triplicates with 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells analyzed per replicate. (c) Schematic of multiplexed labeling experiments. U-2 OS cells were transiently transfected with H2B–BenzoTag (nuclear) and Tomm20–HaloTag7 (cytosolic, outer mitochondrial membrane fusion) and treated simultaneously with Bz-1 and CA-JF635. (d) Flow cytometry data in orthogonal labeling experiments with 50 nM Bz-1 and 125 nM CA-JF635 for 60 minutes. U-2 OS cells expressing either Tomm20–HaloTag7, H2B–BenzoTag, or both were treated with either dye or co-treated with both dyes. The 15% most fluorescent cells of the 10 000 cells analyzed per replicate. (c) Schematic of multiplexed labeling experiments. U-2 OS cells were transiently transfected with H2B–BenzoTag (nuclear) and Tomm20–HaloTag7 (cytosolic, outer mitochondrial membrane fusion) and treated simultaneously with Bz-1 and CA-JF635. (d) Flow cytometry data in orthogonal labeling experiments with 50 nM Bz-1 and 125 nM CA-JF635 for 60 minutes. U-2 OS cells expressing either Tomm20–HaloTag7, H2B–BenzoTag, or both were treated with either dye or co-treated with both dyes. The 15% most fluorescent cells of the 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events read were analyzed in each experiment because transient transfection efficiencies were roughly 20% (Fig. S9†). Data are shown as the mean and standard error of biological triplicates. (e) Confocal microscopy images of U-2 OS cells transiently transfected with both H2B–BenzoTag and Tomm20–HaloTag7. Cells were stained with nuclear Hoechst dye, washed, and then treated with 50 nM Bz-1 and 125 nM CA-JF635 for 60 minutes. No washing was performed prior to imaging. See Fig. S17 and S18† for corresponding bright-field image and for zoom-ins of individual cells. 000 events read were analyzed in each experiment because transient transfection efficiencies were roughly 20% (Fig. S9†). Data are shown as the mean and standard error of biological triplicates. (e) Confocal microscopy images of U-2 OS cells transiently transfected with both H2B–BenzoTag and Tomm20–HaloTag7. Cells were stained with nuclear Hoechst dye, washed, and then treated with 50 nM Bz-1 and 125 nM CA-JF635 for 60 minutes. No washing was performed prior to imaging. See Fig. S17 and S18† for corresponding bright-field image and for zoom-ins of individual cells. | ||
Encouraged by these results, we evaluated the ability to multiplex the BenzoTag·Bz-1 and HaloTag7·CA-JF635 systems in wash-free, live-cell labeling experiments. Bz-1, CA-JF635 or both dyes were added to U-2 OS cells transiently expressing BenzoTag localized to the nucleus, HaloTag7 localized to the outer mitochondrial membrane, or both. Using 125 nM dye and analyzing cell populations by flow cytometry after 10 minutes of incubation, we observed that Bz-1 predominately labeled BenzoTag while CA-JF635 predominantly labeled HaloTag7 (ESI Fig. S15b†). We further analyzed cells co-transfected with both constructs using confocal fluorescence microscopy, treating the cells with both dyes and imaging directly with no washes (Fig. 5c, ESI Fig. S15c†). When cells were treated with 125 nM of each dye for 10 minutes, we observed robust labeling by Bz-1 but no observable labeling by CA-JF635. Extending the incubation to 60 minutes revealed CA-JF635 labeling at the mitochondria, with Bz-1 labeling at both the nucleus and mitochondria (ESI Fig. S15c†). These results are consistent Bz-1 having faster cell penetration compared to CA-JF635, which exposed mitochondrially localized HaloTag7 to higher concentrations of Bz-1 during the 60 minute incubation. We found that co-treating cells with 50 nM Bz-1 and 125 nM CA-JF635 for 60 minutes resulted in robust, orthogonal labeling (Fig. 5d). Bz-1 fluorescence was entirely localized to the nucleus and CA-JF635 fluorescence was entirely localized to the mitochondria, indicating robust orthogonality between the BenzoTag·Bz-1 and HaloTag7·CA-JF635 systems under these optimized conditions (Fig. 5e). Colocalization analysis confirmed that there was no association between Bz-1 signal and CA-JF635, and positive association between Bz-1 and nuclear Hoechst staining (ESI Fig. S18†). Quantification by flow cytometry under these conditions confirmed orthogonal labeling between the systems (Fig. 5d).
Discussion
Self-labeling enzymes like HaloTag7 have become a mainstay in chemical biology and biomedical research. HaloTag7 is used for applications with a large variety of chloroalkane-tagged compounds, but the enzyme's properties, especially its reaction kinetics, depend greatly on the nature of the substrate attached to the chloroalkane. Some prior work sought to modify HaloTag7 to improve the labeling rates with non-rhodamine substrates. An early example includes the mutation of negatively charged residues around the entrance to the active site channel of HaloTag to promote faster conjugation of chloroalkane-tagged oligonucleotides.19 Most recently, several other groups have modified HaloTag7 to better accompany new substrates in a semi-rational approach using techniques that screen 102–104 variants at a time.17,44,45,53 In this work, we developed a yeast display system capable of screening 107–108 HaloTag7 variants at a time. We anticipate this system will greatly accelerate the development of HaloTag variants that work better with non-rhodamine substrates. Indeed, in this initial application, with a single round of diversification, the larger screening capability enabled the discovery of multiple cooperative mutations at unexpected positions; this result would have been highly unlikely using prior methods.44,45 The yeast display format allows for a variety of positive and negative selections, rapid follow-up assays, and built-in controls for expression level. We anticipate that the ability to rapidly evolve specific HaloTag7 variants for a given chloroalkane-tagged substrate will produce customizable enzyme·substrate pairs to enable applications across the chemical and biological sciences including drug development, photophysics, and catalysis. Further, it was recently demonstrated that the key H272F mutation that converted the delahogenase DhaA into a self-labeling enzyme is translatable across the dehalogenase family.27 This suggests that additional dehalogenases can be immediately incorporated into the yeast display pipeline to capitalize on the large structural diversity afforded by this class of enzymes.In this first application, we provide ample evidence that yeast display produced HaloTag variants with improved conjugation kinetics and brighter fluorescent complexes. The optimized variant, BenzoTag, had a 63-fold enhancement in reaction rate with Bz-1 compared to HaloTag7. Our prior work optimized the fluorescent properties of the benzothiadiazole dye by replacing the dimethylamine donor in Bz-2 with a pyrrolidine in Bz-1, which improved the fluorescent quantum yield of the dye when conjugated to HaloTag7 by 50% while decreasing the background in cells.25 Thus, combining dye engineering and protein engineering, we improved the original HaloTag7·Bz-2 system by 1.5-fold in terms of brightness and by 300-fold in terms of reaction rate.23,25 We expect this roadmap – first optimizing the substrate for ideal functional properties, then optimizing the self-labeling enzyme for faster labeling – will enable additional systems to be developed with improved functionality compared to HaloTag7.
Prior to this work, the most well-developed fluorogenic substrate for HaloTag7 was a silicon rhodamine (SiR) dye, originally reported by Johnsson and colleagues39 and later optimized to CA-JF635 by Lavis and colleagues.29,40,54 We compared the performance of BenzoTag·Bz-1 and HaloTag7·CA-JF635 in live-cell labeling using both flow cytometry and fluorescence microscopy (Fig. 5). The JF635 dye was reported to have a very high extinction coefficient when measured in ethanol, but this value was significantly lower when the chloroalkane–dye conjugate CA-JF635 was bound to HaloTag7 in aqueous solution.40 Thus, despite the approximately 20-fold larger extinction coefficient of JF635 and similar quantum yields of fluorescence (0.56 vs. 0.58), HaloTag7·CA-JF635 only has 5.4-fold higher fluorescent brightness than BenzoHTag·Bz-1 in experiments with recombinant protein (ESI Table S11†). Further, in cell-based experiments the increase in fluorescence signal from CA-JF635 in HaloTag7-expressing compartments compared to background cellular labeling was reported at only 9.2-fold, likely due to background fluorescence.40 These observations explain why Bz-1, with its much lower extinction coefficient, nonetheless had higher signal-over-background in cell-based labeling experiments (ESI Fig. S11 and S19†). Under its best-performing conditions using flow cytometry (125 nM dye and 10 minutes of incubation), we observed that HaloTag7·CA-JF635 had 366-fold signal-over-background (comparing HaloTag7-expressing cells to non-expressing cells). By contrast, BenzoTag·Bz-1 had 3000-fold signal-over-background under its best-performing conditions, which required only 7 nM dye and only 10 minutes of incubation. Notably, concentrations exceeding 62 nM JF635 for 60 minute incubation times resulted in larger non-specific fluorescence in non-expressing cells when compared to the shorter, 10 minute incubation (ESI Fig. S11 and S12†). These results suggest that brighter dyes can produce larger raw fluorescence signals and are more well-suited for single-molecule applications, but fluorogenic dyes with moderate brightness and lower background can be superior for many other cell-based applications.
We also observed differences in the kinetics and saturation of in-cell labeling for the two systems. As described above, the signal-over-background of the two systems were comparable in cells, but signal saturation was achieved much faster and at lower dye concentrations in the BenzoTag·Bz-1 system (Fig. 5b, ESI Fig. S11 and S12†). CA-JF635 is typically used in live-cell imaging experiments at concentrations of 100 nM to 500 nM, and sometimes exceeding 1 μM, with labeling times of 30 minutes or more.29–31,39,40,54 Similar conditions were required for robust cellular labeling of HaloTag7·CA-JF635 in our experiments. By contrast, robust wash-free labeling was observed using only 10 nM Bz-1 with 18-fold signal-over-background as measured by confocal microscopy (ESI Fig. S19†), and nucleus-localized BenzoTag was saturated by 125 nM Bz-1 in under 100 seconds (Fig. 4c). We interpret these results as indicating superior cell permeability for the smaller, uncharged Bz-1 compared to the larger, zwitterionic CA-JF635 (Fig. 1b). We directly observed that Bz-1 internalizes within seconds, but CA-JF635 requires up to 60 minutes for comparable signal-over-background (ESI Fig. S11 and S12†). This interpretation is further supported by the observation that the percentage of cells labeled after 10 minutes was not dependent on concentration of Bz-1 but was highly dependent on concentration of CA-JF635 (ESI Fig. S11c†). The rapid labeling of the BenzoTag·Bz-1 system suggests unique applications for monitoring fast cellular processes, such as endosomal recycling.11 The no-wash nature of this system will also enable measurements of biological processes in real-time including protein folding in the cell, endocytosis and exocytosis rates, protein and organellar degradation processes, and endosomal escape of biologic therapeutics.13,55–60 These applications will be accelerated by the production of stable BenzoTag-expressing cell lines.
There are several implementations of multiplexed fluorescent labeling using two different self-labeling proteins, like HaloTag7 and SNAP-tag,29,45,61 and even a few reports of multiplexed no-wash fluorescent labeling.62 However, HaloTag7 is often preferred over SNAP-tag because SNAP-tag has slower reaction rates, its ligands have higher nonspecific interactions in the cell, and its complexes have weaker photophysical properties.7,22,29,63 Therefore, it would be advantageous to use multiple HaloTag-derived self-labeling proteins in a wash-free multiplexed labeling experiments using orthogonal chloroalkane substrates. We found that BenzoTag·Bz-1 and HaloTag7·CA-JF635 support multiplexed no-wash labeling experiments in live cells (Fig. 5e). Given the ability to perform positive and negative selections using yeast display, we anticipate that additional HaloTag7 variants can be evolved for improved orthogonality and for specificity to other, spectrally orthogonal fluorogenic dyes that will allow multiplexing using three or more colors. Moreover, this strategy could be interfaced with recent advances in protein/peptide tags64–66 and fluorescence lifetime imaging45,67 to offer even more degrees of multidimensional multiplexing.
Conclusion
We have introduced the commonly used self-labeling protein HaloTag7 into a yeast display system for directed evolution of improved variants. This display platform can produce novel systems for in-cell chemistry, including biosensing and biocatalysis, that were previously inaccessible.4,5 We used this system to develop BenzoTag, an evolved HaloTag7 triple mutant that has improved conjugation kinetics to a fluorogenic benzothiadiazole dye, Bz-1. The BenzoTag·Bz-1 system enables robust intracellular labeling of live cells at concentrations as low as 7 nM, in seconds and without washes. The BenzoTag·Bz-1 system exhibits similar in vitro kinetics (within an order of magnitude) and larger signal-over-background than the previously reported HaloTag·CA-JF635 system but has faster and more sensitive in-cell labeling. The fast in-cell labeling rate will be especially useful for real-time monitoring of biological processes, especially intracellular processes, with fast time scales.11,59,68 Finally, the BenzoTag·Bz-1 system was found to be orthogonal to the HaloTag7·CA-JF635 system, allowing for simultaneous application of both systems for wash-free multiplexed imaging in live cells.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
B. L. and J. A. K. designed the project. B. L. did all experiments with assistance from B. G. in bacterial culture, yeast culture, and protein expression and purification. B. L. and B. G. compiled methods and data, and B. L. and J. A. K. analyzed the data and wrote the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by NIH 5F32GM139371 to B. J. L. and NIH GM148407 to J. A. K. The authors would like to thank Prof. James Van Deventer for his thoughtful insights and for providing the RJY100 yeast strain, Dr Luke Lavis for providing CA-JF635 samples, and Dr Matt Lindley and the TAMIC core imaging facility for the confocal microscopy facilities, which is supported by the NIH infrastructure grant NIH S10 OD021624.References
- E. C. Greenwald, S. Mehta and J. Zhang, Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks, Chem. Rev., 2018, 118(24), 11707–11794, DOI:10.1021/acs.chemrev.8b00333.
- M. Wang, Y. Da and Y. Tian, Fluorescent proteins and genetically encoded biosensors, Chem. Soc. Rev., 2023, 52(4), 1189–1214, 10.1039/D2CS00419D.
- G. V. Los, L. P. Encell, M. G. McDougall, D. D. Hartzell, N. Karassina, C. Zimprich, M. G. Wood, R. Learish, R. F. Ohana and M. Urh, et al., HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis, ACS Chem. Biol., 2008, 3(6), 373–382, DOI:10.1021/cb800025k.
- C. G. England, H. Luo and W. Cai, HaloTag Technology: A Versatile Platform for Biomedical Applications, Bioconjugate Chem., 2015, 26(6), 975–986, DOI:10.1021/acs.bioconjchem.5b00191.
- A. Cook, F. Walterspiel and C. Deo, HaloTag-Based Reporters for Fluorescence Imaging and Biosensing, ChemBioChem, 2023, 24(12), e202300022, DOI:10.1002/cbic.202300022.
- V. Liss, B. Barlag, M. Nietschke and M. Hensel, Self-labelling enzymes as universal tags for fluorescence microscopy, super-resolution microscopy and electron microscopy, Sci. Rep., 2015, 5(1), 17740, DOI:10.1038/srep17740.
- R. S. Erdmann, S. W. Baguley, J. H. Richens, R. F. Wissner, Z. Xi, E. S. Allgeyer, S. Zhong, A. D. Thompson, N. Lowe and R. Butler, et al., Labeling Strategies Matter for Super-Resolution Microscopy: A Comparison between HaloTags and SNAP-tags, Cell Chem. Biol., 2019, 26(4), 584–592.e586, DOI:10.1016/j.chembiol.2019.01.003.
- C. Deo, A. S. Abdelfattah, H. K. Bhargava, A. J. Berro, N. Falco, H. Farrants, B. Moeyaert, M. Chupanova, L. D. Lavis and E. R. Schreiter, The HaloTag as a general scaffold for far-red tunable chemigenetic indicators, Nat. Chem. Biol., 2021, 17(6), 718–723, DOI:10.1038/s41589-021-00775-w.
- L. Hellweg, A. Edenhofer, L. Barck, M.-C. Huppertz, M. S. Frei, M. Tarnawski, A. Bergner, B. Koch, K. Johnsson and J. Hiblot, A general method for the development of multicolor biosensors with large dynamic ranges, Nat. Chem. Biol., 2023, 19(9), 1147–1157, DOI:10.1038/s41589-023-01350-1.
- B. C. Shields, E. Kahuno, C. Kim, P. F. Apostolides, J. Brown, S. Lindo, B. D. Mensh, J. T. Dudman, L. D. Lavis and M. R. Tadross, Deconstructing behavioral neuropharmacology with cellular specificity, Science, 2017, 356(6333), eaaj2161, DOI:10.1126/science.aaj2161.
- C. T. H. Jonker, C. Deo, P. J. Zager, A. N. Tkachuk, A. M. Weinstein, E. Rodriguez-Boulan, L. D. Lavis and R. Schreiner, Accurate measurement of fast endocytic recycling kinetics in real time, J. Cell Sci., 2020, 133(2), jcs231225, DOI:10.1242/jcs.231225.
- T. Machleidt, C. C. Woodroofe, M. K. Schwinn, J. Méndez, M. B. Robers, K. Zimmerman, P. Otto, D. L. Daniels, T. A. Kirkland and K. V. Wood, NanoBRET—A Novel BRET Platform for the Analysis of Protein–Protein Interactions, ACS Chem. Biol., 2015, 10(8), 1797–1804, DOI:10.1021/acschembio.5b00143.
- D. L. Buckley, K. Raina, N. Darricarrere, J. Hines, J. L. Gustafson, I. E. Smith, A. H. Miah, J. D. Harling and C. M. Crews, HaloPROTACS: Use of Small Molecule PROTACs to Induce Degradation of HaloTag Fusion Proteins, ACS Chem. Biol., 2015, 10(8), 1831–1837, DOI:10.1021/acschembio.5b00442 , from NLM..
- L. Peraro, K. L. Deprey, M. K. Moser, Z. Zou, H. L. Ball, B. Levine and J. A. Kritzer, Cell Penetration Profiling Using the Chloroalkane Penetration Assay, J. Am. Chem. Soc., 2018, 140(36), 11360–11369, DOI:10.1021/jacs.8b06144.
- K. Deprey, N. Batistatou, M. F. Debets, J. Godfrey, K. B. VanderWall, R. R. Miles, L. Shehaj, J. Guo, A. Andreucci and P. Kandasamy, et al., Quantitative Measurement of Cytosolic and Nuclear Penetration of Oligonucleotide Therapeutics, ACS Chem. Biol., 2022, 17(2), 348–360, DOI:10.1021/acschembio.1c00830.
- K. H. Jung, S. F. Kim, Y. Liu and X. Zhang, A Fluorogenic AggTag Method Based on Halo- and SNAP-Tags to Simultaneously Detect Aggregation of Two Proteins in Live Cells, ChemBioChem, 2019, 20(8), 1078–1087, DOI:10.1002/cbic.201800782.
- S. Fischer, T. R. Ward and A. D. Liang, Engineering a Metathesis-Catalyzing Artificial Metalloenzyme Based on HaloTag, ACS Catal., 2021, 11(10), 6343–6347, DOI:10.1021/acscatal.1c01470.
- P. K. Mishra, N. Sharma, H. Kim, C. Lee and H.-W. Rhee, GEN-Click: Genetically Encodable Click Reactions for Spatially Restricted Metabolite Labeling, ACS Cent. Sci., 2023, 9(8), 1650–1657, DOI:10.1021/acscentsci.3c00511.
- K. J. Koßmann, C. Ziegler, A. Angelin, R. Meyer, M. Skoupi, K. S. Rabe and C. M. Niemeyer, A Rationally Designed Connector for Assembly of Protein-Functionalized DNA Nanostructures, ChemBioChem, 2016, 17(12), 1102–1106, DOI:10.1002/cbic.201600039.
- H.-Y. Lin, J. A. Haegele, M. T. Disare, Q. Lin and Y. Aye, A Generalizable Platform for Interrogating Target- and Signal-Specific Consequences of Electrophilic Modifications in Redox-Dependent Cell Signaling, J. Am. Chem. Soc., 2015, 137(19), 6232–6244, DOI:10.1021/ja5132648.
- E. R. Ballister, C. Aonbangkhen, A. M. Mayo, M. A. Lampson and D. M. Chenoweth, Localized light-induced protein dimerization in living cells using a photocaged dimerizer, Nat. Commun., 2014, 5, 5475, DOI:10.1038/ncomms6475 , from NLM..
- J. Wilhelm, S. Kühn, M. Tarnawski, G. Gotthard, J. Tünnermann, T. Tänzer, J. Karpenko, N. Mertes, L. Xue and U. Uhrig, et al., Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag, Biochemistry, 2021, 60(33), 2560–2575, DOI:10.1021/acs.biochem.1c00258.
- Y. Liu, K. Miao, N. P. Dunham, H. Liu, M. Fares, A. K. Boal, X. Li and X. Zhang, The Cation−π Interaction Enables a Halo-Tag Fluorogenic Probe for Fast No-Wash Live Cell Imaging and Gel-Free Protein Quantification, Biochemistry, 2017, 56(11), 1585–1595, DOI:10.1021/acs.biochem.7b00056.
- S. A. Clark, V. Singh, D. Vega Mendoza, W. Margolin and E. T. Kool, Light-Up “Channel Dyes” for Haloalkane-Based Protein Labeling in Vitro and in Bacterial Cells, Bioconjugate Chem., 2016, 27(12), 2839–2843, DOI:10.1021/acs.bioconjchem.6b00613.
- B. J. Lampkin and J. A. Kritzer, Engineered fluorogenic HaloTag ligands for turn-on labelling in live cells, Chem. Commun., 2024, 60(2), 200–203, 10.1039/D3CC05536A.
- L. P. Encell, R. Friedman Ohana, K. Zimmerman, P. Otto, G. Vidugiris, M. G. Wood, G. V. Los, M. G. McDougall, C. Zimprich and N. Karassina, et al., Development of a dehalogenase-based protein fusion tag capable of rapid, selective and covalent attachment to customizable ligands, Curr. Chem. Genomics, 2012, 6, 55–71, DOI:10.2174/1875397301206010055 , from NLM..
- S. M. Marques, M. Slanska, K. Chmelova, R. Chaloupkova, M. Marek, S. Clark, J. Damborsky, E. T. Kool, D. Bednar and Z. Prokop, Mechanism-Based Strategy for Optimizing HaloTag Protein Labeling, JACS Au, 2022, 2(6), 1324–1337, DOI:10.1021/jacsau.2c00002.
- J. V. Thevathasan, M. Kahnwald, K. Cieśliński, P. Hoess, S. K. Peneti, M. Reitberger, D. Heid, K. C. Kasuba, S. J. Hoerner and Y. Li, et al., Nuclear pores as versatile reference standards for quantitative superresolution microscopy, Nat. Methods, 2019, 16(10), 1045–1053, DOI:10.1038/s41592-019-0574-9.
- J. B. Grimm, A. N. Tkachuk, L. Xie, H. Choi, B. Mohar, N. Falco, K. Schaefer, R. Patel, Q. Zheng and Z. Liu, et al., A general method to optimize and functionalize red-shifted rhodamine dyes, Nat. Methods, 2020, 17(8), 815–821, DOI:10.1038/s41592-020-0909-6.
- N. Lardon, L. Wang, A. Tschanz, P. Hoess, M. Tran, E. D'Este, J. Ries and K. Johnsson, Systematic Tuning of Rhodamine Spirocyclization for Super-resolution Microscopy, J. Am. Chem. Soc., 2021, 143(36), 14592–14600, DOI:10.1021/jacs.1c05004.
- L. Wang, M. Tran, E. D'Este, J. Roberti, B. Koch, L. Xue and K. Johnsson, A general strategy to develop cell permeable and fluorogenic probes for multicolour nanoscopy, Nat. Chem., 2020, 12(2), 165–172, DOI:10.1038/s41557-019-0371-1.
- Q. Zheng, A. X. Ayala, I. Chung, A. V. Weigel, A. Ranjan, N. Falco, J. B. Grimm, A. N. Tkachuk, C. Wu and J. Lippincott-Schwartz, et al., Rational Design of Fluorogenic and Spontaneously Blinking Labels for Super-Resolution Imaging, ACS Cent. Sci., 2019, 5(9), 1602–1613, DOI:10.1021/acscentsci.9b00676.
- D. Si, Q. Li, Y. Bao, J. Zhang and L. Wang, Fluorogenic and Cell-Permeable Rhodamine Dyes for High-Contrast Live-Cell Protein Labeling in Bioimaging and Biosensing, Angew. Chem., Int. Ed., 2023, 62(45), e202307641, DOI:10.1002/anie.202307641.
- E. Prifti, L. Reymond, M. Umebayashi, R. Hovius, H. Riezman and K. Johnsson, A Fluorogenic Probe for SNAP-Tagged Plasma Membrane Proteins Based on the Solvatochromic Molecule Nile Red, ACS Chem. Biol., 2014, 9(3), 606–612, DOI:10.1021/cb400819c.
- C. Li, A. G. Tebo and A. Gautier, Fluorogenic Labeling Strategies for Biological Imaging, Int. J. Mol. Sci., 2017, 18(7), 1473, DOI:10.3390/ijms18071473 , from NLM.
- Y. Chen, H. Jiang, T. Hao, N. Zhang, M. Li, X. Wang, X. Wang, W. Wei and J. Zhao, Fluorogenic Reactions in Chemical Biology: Seeing Chemistry in Cells, Chem. Biomed. Imaging, 2023, 1(7), 590–619, DOI:10.1021/cbmi.3c00029.
- S. P. J. T. Bachollet, Y. Shpinov, F. Broch, H. Benaissa, A. Gautier, N. Pietrancosta, J.-M. Mallet and B. Dumat, An expanded palette of fluorogenic HaloTag probes with enhanced contrast for targeted cellular imaging, Org. Biomol. Chem., 2022, 20(17), 3619–3628, 10.1039/D1OB02394B.
- S. P. J. T. Bachollet, C. Addi, N. Pietrancosta, J.-M. Mallet and B. Dumat, Fluorogenic Protein Probes with Red and Near-Infrared Emission for Genetically Targeted Imaging, Chem.–Eur. J., 2020, 26(63), 14467–14473, DOI:10.1002/chem.202002911.
- G. Lukinavičius, K. Umezawa, N. Olivier, A. Honigmann, G. Yang, T. Plass, V. Mueller, L. Reymond, I. R. Corrêa Jr and Z.-G. Luo, et al., A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins, Nat. Chem., 2013, 5(2), 132–139, DOI:10.1038/nchem.1546.
- J. B. Grimm, A. K. Muthusamy, Y. Liang, T. A. Brown, W. C. Lemon, R. Patel, R. Lu, J. J. Macklin, P. J. Keller and N. Ji, et al., A general method to fine-tune fluorophores for live-cell and in vivo imaging, Nat. Methods, 2017, 14(10), 987–994, DOI:10.1038/nmeth.4403 , from NLM..
- B. A. D. Neto, J. R. Correa and J. Spencer, Fluorescent Benzothiadiazole Derivatives as Fluorescence Imaging Dyes: A Decade of New Generation Probes, Chem.–Eur. J., 2022, 28(4), e202103262, DOI:10.1002/chem.202103262.
- D. Ruhlandt, M. Andresen, N. Jensen, I. Gregor, S. Jakobs, J. Enderlein and A. I. Chizhik, Absolute quantum yield measurements of fluorescent proteins using a plasmonic nanocavity, Commun. Biol., 2020, 3(1), 627, DOI:10.1038/s42003-020-01316-2 , from NLM..
- L. D. Lavis and R. T. Raines, Bright Ideas for Chemical Biology, ACS Chem. Biol., 2008, 3(3), 142–155, DOI:10.1021/cb700248m.
- C. Miró-Vinyals, A. Stein, S. Fischer, T. R. Ward and A. Deliz Liang, HaloTag Engineering for Enhanced Fluorogenicity and Kinetics with a Styrylpyridium Dye, ChemBioChem, 2021, 22(24), 3398–3401, DOI:10.1002/cbic.202100424.
- M. S. Frei, M. Tarnawski, M. J. Roberti, B. Koch, J. Hiblot and K. Johnsson, Engineered HaloTag variants for fluorescence lifetime multiplexing, Nat. Methods, 2021, 19, 65–70, DOI:10.1038/s41592-021-01341-x.
- D. W. Colby, B. A. Kellogg, C. P. Graff, Y. A. Yeung, J. S. Swers and K. D. Wittrup, Engineering antibody affinity by yeast surface display, Methods Enzymol., 2004, 388, 348–358, DOI:10.1016/s0076-6879(04)88027-3 , from NLM..
- I. Chen, B. M. Dorr and D. R. Liu, A general strategy for the evolution of bond-forming enzymes using yeast display, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(28), 11399–11404, DOI:10.1073/pnas.1101046108.
- J. A. Van Deventer and K. D. Wittrup, Yeast Surface Display for Antibody Isolation: Library Construction, Library Screening, and Affinity Maturation, Humana Press, 2014, pp. 151–181 Search PubMed.
- T. C. Branon, J. A. Bosch, A. D. Sanchez, N. D. Udeshi, T. Svinkina, S. A. Carr, J. L. Feldman, N. Perrimon and A. Y. Ting, Efficient proximity labeling in living cells and organisms with TurboID, Nat. Biotechnol., 2018, 36(9), 880–887, DOI:10.1038/nbt.4201 , from NLM..
- M.-A. Plamont, E. Billon-Denis, S. Maurin, C. Gauron, F. M. Pimenta, C. G. Specht, J. Shi, J. Quérard, B. Pan and J. Rossignol, et al., Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(3), 497–502, DOI:10.1073/pnas.1513094113.
- G. Chao, W. L. Lau, B. J. Hackel, S. L. Sazinsky, S. M. Lippow and K. D. Wittrup, Isolating and engineering human antibodies using yeast surface display, Nat. Protoc., 2006, 1(2), 755–768, DOI:10.1038/nprot.2006.94 , from NLM..
- A. M. Thooft, K. Cassaidy and B. A. VanVeller, Small Push–Pull Fluorophore for Turn-on Fluorescence, J. Org. Chem., 2017, 82(17), 8842–8847, DOI:10.1021/acs.joc.7b00939.
- M.-G. Kang, H. Lee, B. H. Kim, Y. Dunbayev, J. K. Seo, C. Lee and H.-W. Rhee, Structure-guided synthesis of a protein-based fluorescent sensor for alkyl halides, Chem. Commun., 2017, 53(66), 9226–9229, 10.1039/C7CC03714G.
- J. B. Grimm, B. P. English, J. Chen, J. P. Slaughter, Z. Zhang, A. Revyakin, R. Patel, J. J. Macklin, D. Normanno and R. H. Singer, et al., A general method to improve fluorophores for live-cell and single-molecule microscopy, Nat. Methods, 2015, 12(3), 244–250, DOI:10.1038/nmeth.3256.
- A. J. Samelson, E. Bolin, S. M. Costello, A. K. Sharma, E. P. O'Brien and S. Marqusee, Kinetic and structural comparison of a protein's cotranslational folding and refolding pathways, Sci. Adv., 2018, 4(5), eaas9098, DOI:10.1126/sciadv.aas9098.
- N. Luo, A. Yan and Z. Yang, Measuring Exocytosis Rate Using Corrected Fluorescence Recovery After Photoconversion, Traffic, 2016, 17(5), 554–564, DOI:10.1111/tra.12380 , from NLM..
- Y. Liu, D. Zhang, Y. Qu, F. Tang, H. Wang, A. Ding and L. Li, Advances in Small-Molecule Fluorescent pH Probes for Monitoring Mitophagy, Chem. Biomed. Imaging, 2024, 2(2), 81–97, DOI:10.1021/cbmi.3c00070.
- H. Koga, M. Martinez-Vicente, F. Macian, V. V. Verkhusha and A. M. Cuervo, A photoconvertible fluorescent reporter to track chaperone-mediated autophagy, Nat. Commun., 2011, 2(1), 386, DOI:10.1038/ncomms1393.
- A. Sahni, Z. Qian and D. Pei, Cell-Penetrating Peptides Escape the Endosome by Inducing Vesicle Budding and Collapse, ACS Chem. Biol., 2020, 15(9), 2485–2492, DOI:10.1021/acschembio.0c00478.
- P. Lönn, A. D. Kacsinta, X. S. Cui, A. S. Hamil, M. Kaulich, K. Gogoi and S. F. Dowdy, Enhancing Endosomal Escape for Intracellular Delivery of Macromolecular Biologic Therapeutics, Sci. Rep., 2016, 6, 32301, DOI:10.1038/srep32301 , from NLM..
- F. Bottanelli, E. B. Kromann, E. S. Allgeyer, R. S. Erdmann, S. Wood Baguley, G. Sirinakis, A. Schepartz, D. Baddeley, D. K. Toomre and J. E. Rothman, et al., Two-colour live-cell nanoscale imaging of intracellular targets, Nat. Commun., 2016, 7(1), 10778, DOI:10.1038/ncomms10778.
- A. Martin and P. Rivera-Fuentes, A general strategy to develop fluorogenic polymethine dyes for bioimaging, Nat. Chem., 2024, 16(1), 28–35, DOI:10.1038/s41557-023-01367-y.
- D. M. Presman, D. A. Ball, V. Paakinaho, J. B. Grimm, L. D. Lavis, T. S. Karpova and G. L. Hager, Quantifying transcription factor binding dynamics at the single-molecule level in live cells, Methods, 2017, 123, 76–88, DOI:10.1016/j.ymeth.2017.03.014 , from NLM..
- A. Gautier, Fluorescence-Activating and Absorption-Shifting Tags for Advanced Imaging and Biosensing, Acc. Chem. Res., 2022, 55(21), 3125–3135, DOI:10.1021/acs.accounts.2c00098.
- L. Xue, I. A. Karpenko, J. Hiblot and K. Johnsson, Imaging and manipulating proteins in live cells through covalent labeling, Nat. Chem. Biol., 2015, 11(12), 917–923, DOI:10.1038/nchembio.1959.
- A. Suyama, K. L. Devlin, M. Macias-Contreras, J. K. Doh, U. Shinde and K. E. Beatty, Orthogonal Versatile Interacting Peptide Tags for Imaging Cellular Proteins, Biochemistry, 2023, 62(11), 1735–1743, DOI:10.1021/acs.biochem.2c00712.
- M. S. Frei, B. Koch, J. Hiblot and K. Johnsson, Live-Cell Fluorescence Lifetime Multiplexing Using Synthetic Fluorescent Probes, ACS Chem. Biol., 2022, 17(6), 1321–1327, DOI:10.1021/acschembio.2c00041.
- T. Komatsu, K. Johnsson, H. Okuno, H. Bito, T. Inoue, T. Nagano and Y. Urano, Real-Time Measurements of Protein Dynamics Using Fluorescence Activation-Coupled Protein Labeling Method, J. Am. Chem. Soc., 2011, 133(17), 6745–6751, DOI:10.1021/ja200225m.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc05090h |
| ‡ Current address: Department of Pharmaceutical Sciences, University of Illinois Chicago, Chicago IL 60612. |
| This journal is © The Royal Society of Chemistry 2024 |