Open Access Article
Open Access ArticleIn silico exploration of acetic acid driven multicomponent synthesis: design, characterization, and antioxidant evaluation of spiroacridines and spiroquinolines†
Subham G.
Patel
 ab,
Dipti B.
Upadhyay
a,
Nirajkumar V.
Shah
b,
Mehul P.
Parmar
a,
Paras J.
Patel
ab,
Dipti B.
Upadhyay
a,
Nirajkumar V.
Shah
b,
Mehul P.
Parmar
a,
Paras J.
Patel
 a,
Apoorva
Malik
c,
Rakesh K.
Sharma
a,
Apoorva
Malik
c,
Rakesh K.
Sharma
 c and
Hitendra M.
Patel
c and
Hitendra M.
Patel
 *a
*a
aDepartment of Chemistry, Sardar Patel University, Vallabh Vidyanagar-388120, Gujarat, India. E-mail: hm_patel@spuvvn.edu
bJ & J College of Science, Nadiad-387001, Kheda, Gujarat, India
cSustainable Materials and Catalysts Research Laboratory (SMCRL), Department of Chemistry, Indian Institute of Technology, Jodhpur, India
First published on 20th March 2024
Abstract
In this study, a highly efficient green synthetic protocol was developed for the synthesis of spiroacridines and spiroquinolines via a multicomponent reaction of 3,4-methylenedioxyaniline, isatin and cyclic 1,3-diones in glacial acetic acid. This metal-free multicomponent approach offers mild reaction conditions, shorter reaction time and easy product purification without column chromatography. Frontier molecular orbitals (FMOs), various quantum chemical descriptors (QCDs) and molecular electrostatic potential (MEP) surfaces were computed using density functional theory (DFT) with a B3LYP/6-311G+(d,p) basis set. All newly synthesized compounds were screened for their antioxidant properties. The compounds 4a and 4v are the most potent ABTS and DPPH radical scavengers to the standard ascorbic acid. In silico molecular docking studies were conducted to determine the binding affinities of the most potent 4a and 4v with targeted antioxidant protein, LD-carboxypeptidase, which show a strong binding affinity of −10.5 kcal mol−1 and −10.7 kcal mol−1, respectively. The most potent antioxidants 4a and 4v were evaluated for in silico drug-likeness and ADME prediction.
Sustainability spotlightWe synthesized a novel spiroacridines and spiroquinolines using one pot multicomponent reactions of 3,4-methylenedioxyaniline, isatin and cyclic 1,3-diones in presence of glacial acetic acid. This reactions protocol covers broad sustainable approaches such as metal-free, mild reaction conditions, shorter reaction time and easy product purification without using column chromatography. The claimed derivatives provides a promising antioxidants property and drug likeness properties. |
1. Introduction
Reactive oxygen species (ROS) play an integral role in the development of many major illnesses, including cancer, diabetes, arteriosclerosis, heart disease, and cataracts.1 The detrimental effects of free radicals that may cause biological damage are referred to as oxidative stress. Free radicals have a pathogenic role in the development of a wide range of chronic degenerative illnesses in humans, including cancer and autoimmune, inflammatory, and cardiovascular neurodegenerative disorders.2,3 Free radicals are chemical entities that possess an unpaired electron. By grasping an electron from a neighbouring biomolecule, these free radicals initiate a chain reaction in the human body, causing them to behave abnormally in living systems. Antioxidants are compounds that donate electrons to free radicals in order to transform them into non-harmful substances. In a nutshell, antioxidants protect the body from oxidative stress by suppressing the oxidation process.4 As a result, the necessity to produce antioxidant drugs has grown in recent years.In present times, the use of sustainable synthetic protocols has received much more attention for the construction of novel heterocycles.5 For the construction of various heterocycles of biological interest, multicomponent reactions are most commonly used to form multiple C–C and C–N bonds without the isolation of an intermediate. MCRs are more vulnerable than classical multistep synthesis owing to their higher reaction efficiency and straightforward operation.6,7 New spiroacridines and spiroquinolines were made using this multicomponent synthetic approach. It has encouraged the synthetic community to seek out more environmentally friendly syntheses and reactions in order to speed up preparation while using less energy and effort.1,8
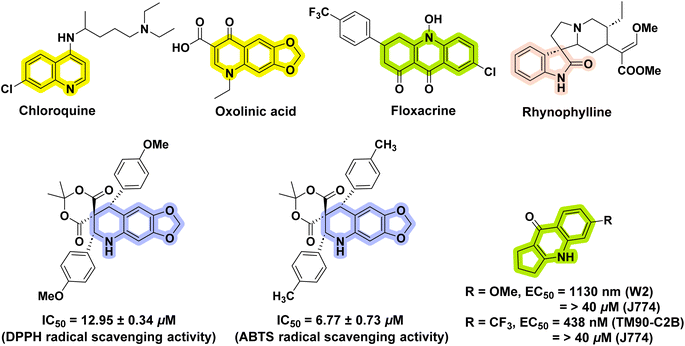
Nitrogen-containing heterocycles are significant in medicinal chemistry and drug design, as evidenced by their preponderance in marketed drugs.9 Spiroindolines have grabbed the interest of researchers due to their privileged structural moiety and occurrence in a wide range of bioactive natural products and synthesised drugs.10 Acridines and quinolines are interesting moieties in heterocyclic chemistry that attract the most attention to chemical modifications and are promising chemical entities both synthetically and medicinally due to their diverse pharmacological applications such as anticancer,11,12 SRC kinase inhibitory,13,14 antioxidant,15,16 and antiallergic activity.17Fig. 1 depicts several acridine,18 quinoline,19,20 spiroindoline21 and 1,3-benzodioxole22 conjugates with promising biological applications. The antioxidant activity of the spiroacridine and spiroquinoline moieties has received little attention. We were inspired to create acridine and quinoline-containing spiroacridines and spiroquinolines as a result of this.
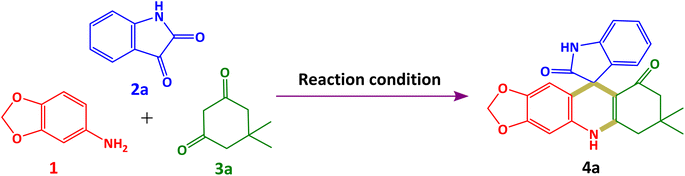
In continuation to our efforts towards the construction of novel spiro molecules,16,23–26 herein, we explored a metal-free multicomponent synthesis1,27–29 of spiroacridines and spiroquinolines via a one-pot reaction of 3,4-methylenedioxyaniline, isatin and cyclic 1,3-diones in the presence of glacial acetic acid, which is vital for sustainability (Scheme 1). To acquire theoretical insights, the frontier molecular orbital (FMO), the molecular electrostatic potential (MEP), and several quantum chemical descriptors (QCDs) were computed using the DFT using the B3LYP/6-311G+(d,p) basis set. All the newly synthesised spiro molecules were screened for their antioxidant properties.
 | ||
| Scheme 1 Multicomponent synthesis of 4a from 3,4-methylenedioxyaniline 1, isatins 2a–h, and cyclic 1,3-diones 3a–c. | ||
2. Results and discussion
2.1. Chemistry
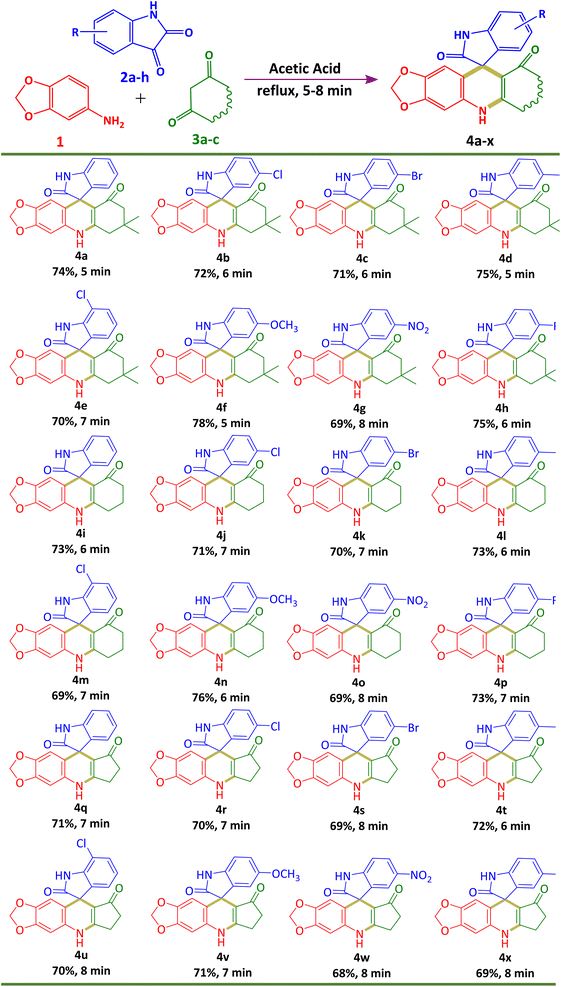
In this protocol, we selected 3,4-methylenedioxyaniline 1 (1 mmol), isatin 2a (1 mmol), and dimedone 3a (1 mmol) as a model reaction in order to achieve our target spiro heterocyclic moiety 7′,7′-dimethyl-7′,8′-dihydro-5′H-spiro[indoline-3,10′-[1,3]dioxolo[4,5-b]acridine]-2,9′(6′H)-dione 4a under catalyst-free conditions via multicomponent synthesis. In order to check the effect of temperature and solvents, we optimized our model reaction by using a variety of solvents at different temperatures. In the initial phase, we initiated our model reaction employing environmentally friendly solvents, water and ethanol, under room temperature conditions (Table 1, entries 1 and 2). However, no discernible transformation of isatin 2a into our target compound 4a was observed. In pursuit of our desired molecule 4a, we executed our model reaction by elevating the temperature from room temperature to reflux. Subsequently, our model reaction was conducted utilizing ethanol at its reflux temperature (Table 1, entry 3). During this experiment, we noted the production occurring within the reaction medium at its reflux temperature. Thin-layer chromatography analysis indicated a full transformation of 2a into 4a, accompanied by the emergence of a novel fluorescence spot on the TLC plate. After that, we performed our model reaction using different organic solvents such as methanol, ethyl L-lactate, acetic acid, DMF and acetonitrile (Table 1, entries 4–8). The best outcomes were achieved when glacial acetic acid was used (Table 1, entry 6). In the presence of glacial acetic acid, we observed rapid transformation of 2a into 4a within just 5 minutes under reflux conditions. Therefore, we selected it as the best optimum condition for the synthesis of our target moiety 7′,7′-dimethyl-7′,8′-dihydro-5′H-spiro[indoline-3,10′-[1,3]dioxolo[4,5-b]acridine]-2,9′(6′H)-dione 4a. It is noteworthy that acetic acid is the sole solvent containing acidic hydrogen among all solvents, playing a dual role as both the catalyst and solvent. This dual functionality enhances reaction efficiency and reduces overall reaction time.| Entry | Solventb | Temperature | Time | Conversion relative to isatinc |
|---|---|---|---|---|
| a Reaction conditions: 1 mmol 3,4-methylenedioxyaniline 1, 1 mmol isatin 2a and 1 mmol dimedone 3a, reflux. b 3 mL solvent. c Observed from TLC analysis. | ||||
| 1 | Water | rt | 6 h | NR |
| 2 | Ethanol | rt | 6 h | NR |
| 3 | Ethanol | Reflux | 45 min | 100% |
| 4 | Methanol | Reflux | 45 min | 100% |
| 5 | Ethyl L-lactate | Reflux | 90 min | 100% |
| 6 | Acetic acid | Reflux | 5 min | 100% |
| 7 | DMF | Reflux | 4 h | Incomplete |
| 8 | Acetonitrile | Reflux | 60 min | 100% |
Using this best optimum condition, we systematically explored the substrate scope for synthesizing spiroacridines 4a–p and spiroquinolines 4q–x. This exploration involved utilizing 3,4-methylenedioxyaniline 1 (1 mmol), various substituted isatins 2a–h (1 mmol), and diverse cyclic 1,3-diones 3a–c (1 mmol) in the presence of glacial acetic acid at reflux temperature. In the presence of various substituted isatins 2a–h and various cyclic 1,3-diones, all reactions proceeded smoothly and furnished final moieties 4a–x with good isolated yield ranging from 68–78% within the stipulated time range. The results are summarized in Table 2. Fortunately, we discovered the solidification of the final product following the reaction transformation in each set of studies. The primary investigation for the confirmation of the desired product is carried out by the thin layer chromatography method using 70% ethyl acetate and 30% n-hexane as the mobile phase. The structures of all newly synthesized compounds are characterized by 1H NMR, 13C NMR and HRMS spectral analysis.
| a Reaction conditions: 1 mmol 3,4-methylenedioxyaniline 1, 1 mmol isatins 2a–h and 1 mmol cyclic 1,3-diones 3a–c, 3 mL glacial acetic acid, reflux. |
|---|
|
2.2. Density functional theory calculations
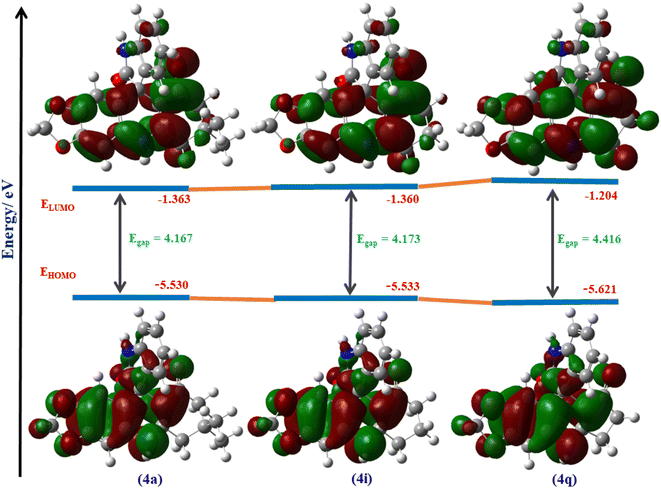 | ||
| Fig. 2 The HOMO–LUMO energy diagram of molecules 4a, 4i and 4j calculated at DFT/B3LYP/6-311G+(d,p) in the gaseous phase. | ||
HOMO and LUMO energies are also used to illustrate various quantum chemical descriptors (QCDs) such as ionisation potential (IP), electron affinity (EA), chemical hardness (η) and softness (σ), electronegativity (χ), chemical potential (μ), electrophilicity index (ω), nucleophilicity index (εγ) and maximum charge transfer index (ΔNmax). These QCDs were computed using the B3LYP/6-311G+(d,p) basis set, according to Koopman's theorem.30 These QCDs play an essential role in emphasising the chemical behaviour and reactivity of the target molecule. The computed QCDs of spiro molecules 4a–x are listed in Table 3.
| Molecule | E HOMO (eV) | E LUMO (eV) | E g (eV) | IP (eV) | EA (eV) | η (eV) | σ (eV) | χ (eV) | μ (eV) | ω (eV) | ε γ (eV) | N (eV) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a DFT/B3LYP/6-311G+(d,p) was used in a gaseous state. | ||||||||||||
| 4a | −5.530 | −1.363 | 4.167 | 5.530 | 1.363 | 2.083 | 0.240 | 3.447 | −3.447 | 2.851 | −7.181 | 1.654 |
| 4b | −5.645 | −1.487 | 4.158 | 5.645 | 1.487 | 2.079 | 0.240 | 3.566 | −3.566 | 3.058 | −7.413 | 1.715 |
| 4c | −5.649 | −1.484 | 4.165 | 5.649 | 1.484 | 2.082 | 0.240 | 3.567 | −3.567 | 3.054 | −7.427 | 1.713 |
| 4d | −5.506 | −1.340 | 4.166 | 5.506 | 1.340 | 2.083 | 0.240 | 3.423 | −3.423 | 2.812 | −7.129 | 1.643 |
| 4e | −5.641 | −1.473 | 4.168 | 5.641 | 1.473 | 2.084 | 0.240 | 3.557 | −3.557 | 3.036 | −7.413 | 1.707 |
| 4f | −5.429 | −1.285 | 4.144 | 5.429 | 1.285 | 2.072 | 0.241 | 3.357 | −3.357 | 2.719 | −6.956 | 1.620 |
| 4g | −5.831 | −2.547 | 3.284 | 5.831 | 2.547 | 1.642 | 0.305 | 4.189 | −4.189 | 5.343 | −6.878 | 2.551 |
| 4h | −5.627 | −1.460 | 4.167 | 5.627 | 1.460 | 2.084 | 0.240 | 3.544 | −3.544 | 3.013 | −7.384 | 1.701 |
| 4i | −5.533 | −1.360 | 4.173 | 5.533 | 1.360 | 2.086 | 0.240 | 3.446 | −3.446 | 2.847 | −7.191 | 1.652 |
| 4j | −5.651 | −1.480 | 4.171 | 5.651 | 1.480 | 2.085 | 0.240 | 3.565 | −3.565 | 3.047 | −7.435 | 1.710 |
| 4k | −5.656 | −1.485 | 4.170 | 5.656 | 1.485 | 2.085 | 0.240 | 3.571 | −3.571 | 3.057 | −7.446 | 1.712 |
| 4l | −5.509 | −1.336 | 4.173 | 5.509 | 1.336 | 2.086 | 0.240 | 3.422 | −3.422 | 2.807 | −7.140 | 1.640 |
| 4m | −5.646 | −1.472 | 4.174 | 5.646 | 1.472 | 2.087 | 0.240 | 3.559 | −3.559 | 3.035 | −7.428 | 1.706 |
| 4n | −5.462 | −1.309 | 4.152 | 5.462 | 1.309 | 2.076 | 0.241 | 3.385 | −3.385 | 2.760 | −7.029 | 1.631 |
| 4o | −5.842 | −2.543 | 3.299 | 5.842 | 2.543 | 1.649 | 0.303 | 4.192 | −4.192 | 5.328 | −6.915 | 2.542 |
| 4p | −5.634 | −1.460 | 4.173 | 5.634 | 1.460 | 2.087 | 0.240 | 3.547 | −3.547 | 3.014 | −7.401 | 1.700 |
| 4q | −5.621 | −1.204 | 4.416 | 5.621 | 1.204 | 2.208 | 0.226 | 3.413 | −3.413 | 2.637 | −7.536 | 1.545 |
| 4r | −5.736 | −1.322 | 4.413 | 5.736 | 1.322 | 2.207 | 0.227 | 3.529 | −3.529 | 2.822 | −7.787 | 1.599 |
| 4s | −5.701 | −1.306 | 4.394 | 5.701 | 1.306 | 2.197 | 0.228 | 3.504 | −3.504 | 2.793 | −7.698 | 1.595 |
| 4t | −5.596 | −1.181 | 4.415 | 5.596 | 1.181 | 2.208 | 0.226 | 3.388 | −3.388 | 2.600 | −7.480 | 1.535 |
| 4u | −5.732 | −1.153 | 4.579 | 5.732 | 1.153 | 2.289 | 0.218 | 3.442 | −3.442 | 2.588 | −7.881 | 1.504 |
| 4v | −5.568 | −1.153 | 4.415 | 5.568 | 1.153 | 2.207 | 0.227 | 3.360 | −3.360 | 2.558 | −7.418 | 1.522 |
| 4w | −5.920 | −2.614 | 3.306 | 5.920 | 2.614 | 1.653 | 0.302 | 4.267 | −4.267 | 5.508 | −7.053 | 2.582 |
| 4x | −5.720 | −1.306 | 4.415 | 5.720 | 1.306 | 2.207 | 0.227 | 3.513 | −3.513 | 2.795 | −7.755 | 1.591 |
2.3. In vitro antioxident activity
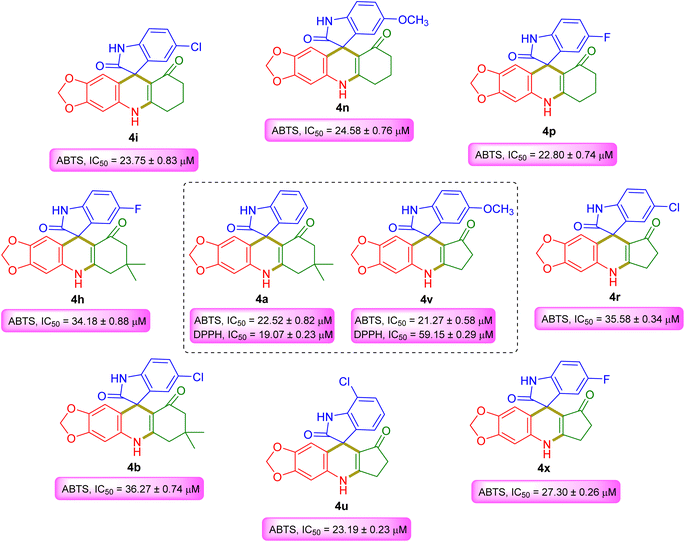
To identify antioxidant properties of all newly synthesized spiroacridines 4a–p and spiroquinolines 4q–x, in vitro 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) and 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging activity was tested. As a standard, antioxidant ascorbic acid was used. For ABTS and DPPH radical scavenging assay, it exhibits 41.84 ± 0.25 μM and 90.10 ± 0.70 μM IC50 values, respectively. Table 4 displays the antioxidant activities of 4a–x. All spiro compounds show good radical scavenging activity. Among them 4a (ABTS, IC50 = 22.52 ± 0.82 μM; DPPH, IC50 = 19.07 ± 0.23 μM) and 4v (ABTS, IC50 = 21.27 ± 0.58 μM; DPPH, IC50 = 59.15 ± 0.29 μM) are the most potent radical scavengers against both ABTS & DPPH radicals which display excellent scavenging activity to standard ascorbic acid (Fig. 4). Meanwhile, all spiro compounds 4a–x are good ABTS radical scavengers. Spiroacridines 4b (ABTS, IC50 = 36.27 ± 0.74 μM), 4h (ABTS, IC50 = 34.18 ± 0.88 μM), 4i (ABTS, IC50 = 23.75 ± 0.83 μM), 4n (ABTS, IC50 = 24.58 ± 0.76 μM), and 4p (ABTS, IC50 = 22.80 ± 0.74 μM) and spiroquinolines 4r (ABTS, IC50 = 35.85 ± 0.34 μM), 4u (ABTS, IC50 = 23.19 ± 0.23 μM), and 4x (ABTS, IC50 = 27.30 ± 0.26 μM) show excellent ABTS radical scavenging activity.| Compound name | ABTS radical scavenging activity | DPPH radical scavenging activity |
|---|---|---|
| IC50 ± SEMa (μM) | IC50 ± SEMa (μM) | |
| a SEM (standard error mean). b Standard for ABTS and DPPH radical scavenging activity. Bold values show the lowest IC50s. | ||
| 4a | 22.52 ± 0.82 | 19.07 ± 0.23 |
| 4b | 36.27 ± 0.74 | 156.64 ± 0.49 |
| 4c | 66.48 ± 0.19 | 768.70 ± 0.27 |
| 4d | 66.92 ± 0.53 | 686.08 ± 0.72 |
| 4e | 56.39 ± 0.42 | 695.75 ± 0.64 |
| 4f | 76.40 ± 0.31 | 253.01 ± 0.52 |
| 4g | 48.12 ± 0.36 | 318.36 ± 0.37 |
| 4h | 34.18 ± 0.88 | 179.63 ± 0.48 |
| 4i | 23.75 ± 0.83 | 245.18 ± 0.23 |
| 4j | 57.70 ± 0.19 | 897.43 ± 0.18 |
| 4k | 46.31 ± 0.26 | 427.61 ± 0.28 |
| 4l | 48.81 ± 0.38 | 364.29 ± 0.45 |
| 4m | 48.97 ± 0.85 | 942.74 ± 0.34 |
| 4n | 24.58 ± 0.76 | 262.80 ± 0.72 |
| 4o | 42.89 ± 0.38 | 503.96 ± 0.16 |
| 4p | 22.80 ± 0.74 | 901.44 ± 0.83 |
| 4q | 42.75 ± 0.59 | 526.25 ± 0.44 |
| 4r | 35.58 ± 0.34 | 1088.76 ± 0.58 |
| 4s | 47.96 ± 0.19 | 924.27 ± 0.32 |
| 4t | 46.96 ± 0.42 | 119.50 ± 0.34 |
| 4u | 23.19 ± 0.23 | 904.15 ± 0.46 |
| 4v | 21.27 ± 0.58 | 59.15 ± 0.29 |
| 4w | 112.09 ± 0.66 | 726.31 ± 0.41 |
| 4x | 27.30 ± 0.26 | 231.09 ± 0.17 |
| Ascorbic acidb | 41.84 ± 0.25 | 90.10 ± 0.70 |
2.4. In silico docking study
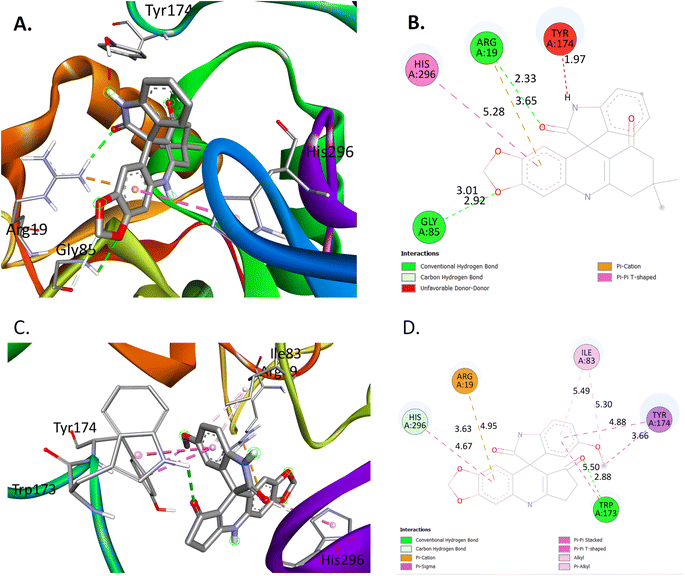
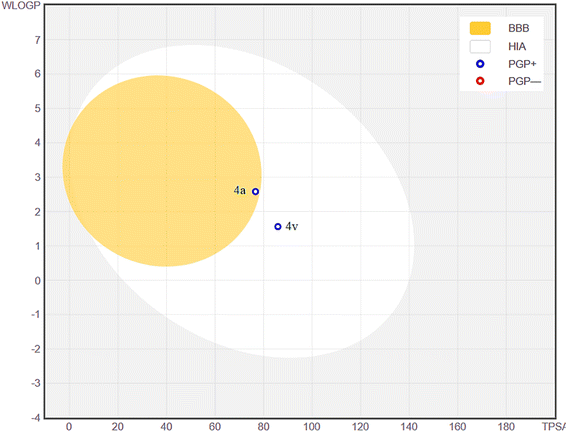
Following the observation of outstanding antioxidant activity exhibited by the potent spiroacridine 4a and spiroquinoline 4v, a subsequent docking study was conducted. Gram-positive Listeria monocytogenes is a widespread intracellular pathogen which is responsible for numerous outbreaks of foodborne illness.31 So, for the docking study, we have chosen the Listeria monocytogenes targeted antioxidant protein in the LD-carboxypeptidase domain. According to the docking analysis, 4a and 4v exhibit proficient binding to the receptor's active sites, specifically site-1 and site-3, with respective binding affinities of −10.5 kcal mol−1 and −10.7 kcal mol−1, which are subsequently better than −5.4 kcal mol−1, exhibited by the standard drug (ascorbic acid) employed in the antioxidant activity investigation (Fig. 5). Among the receptor's two active sites, 4a interacts with the active site-1 via conventional H-bonds, π-cation type of interaction, carbon–hydrogen bonds, π–π T-shapes, and unfavorable donor–donor type of interaction. The H-atom attached to the indole ring of spiroacridine 4a shows unfavorable donor–donor type interaction with TYR A:174 (1.94 Å). The carbonyl oxygen of the indole ring and an oxygen atom of 1,3-benzodioxole show conventional H-bonding with ARG A:19 (2.33 Å) and GLY A:85 (2.92 Å), respectively. An aryl ring of 1,3-benzodioxol of 4a interacts with ARG A:19 (3.65 Å) and HIS A:296 (5.28 Å) through π-cation and π–π T-shaped types of interactions, respectively. Furthermore, the methylene group of 1,3-bonzodioxol of 4a interacts with GLY A:85 (3.01 Å) through a carbon–hydrogen bond.4v interacts with the receptor at active site-3 through conventional H-bonding, π–π stacked type of bond, carbon–hydrogen bonds, π–π T-shapes, and π-cation, alkyl, π-sigma, and π-alkyl types of bonds. Carbonyl oxygen atoms of the quinoline ring and indole ring of 4v interact with TRP A:173 (2.88 Å) and HIS A:296 (4.67 Å) through conventional H-bonds and π–π stacked type of bond, respectively. An aryl ring of indole of 4a interacts with ILE A:83 (5.49 Å), TYR A:174 (4.88 Å) and TRP A:173 (5.50 Å) through π-alkyl, π–π stacked and π–π T-shaped interactions, respectively. The methoxy group attached to the aryl ring of 4a interacts with ILE A:83 (5.30 Å) and TYR A:174 (4.88 Å) through alkyl and π-sigma types of bonds, respectively. Furthermore, an aryl ring of 1,3-benzodioxol interacts with HIS A:296 (4.67 Å) and ARG A:19 (4.95 Å) through π–π T-shaped and π-cation types of bonds. These interactions make the binding of 4a and 4v strong enough in the binding sites of the Listeria monocytogenes targeted antioxidant protein of the LD-carboxypeptidase domain. So, 4a and 4v inhibit radical growth and show a good minimum inhibitory concentration value.
2.5. In silico ADME prediction
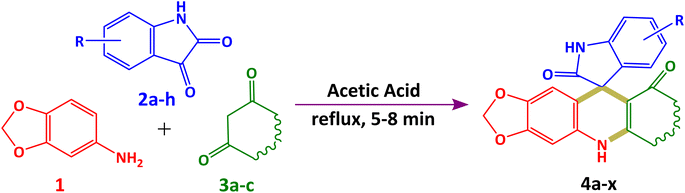
Prediction of absorption, distribution, metabolism, and excretion (ADME) properties before experimental trials is an essential aspect of the drug discovery and development process. The most potent antioxidants 4a and 4v were evaluated for in silico drug-likeness and ADME prediction. Several filters such as the Lipinski filter, Ghose filter, Veber filter, Egan filter and Muegge filter help predict drug-likeness based on their physicochemical properties. It was observed that both 4a and 4v obeyed all necessary rules and had no violation of such rules, proving them to be important drug candidate. The calculated physicochemical properties of potent compounds are summarized in Table S1 (mentioned in the ESI†).The calculated bioavailability score was found to be 0.55 for 4a and 4v. BOILED-Egg delineation was used to predict high gastrointestinal absorption (HIA) and blood–brain barrier (BBB) permeability of 4a and 4v. BOILED-Egg delineation of these 4a and 4v is illustrated in Fig. 6 “The white region is the physicochemical space of molecules with the highest probability of being absorbed by the gastrointestinal tract, and the yellow region (yolk) is the physicochemical space of molecules with the highest probability to permeate to the brain”.324v shows high gastrointestinal absorbance, and it has no blood–brain permeability, while 4a shows blood–brain permeability. Blue dots for 4a and 4v denoted that they are P-gp substrates. From the SVM model, 4a was found to be a CYP1A2 inhibitor except for 4v, whereas both 4a and 4v were found to be inhibitors of CYP2C19, CYP2C9, CYP2D6 and CYP3A4 which can perform metabolic biotransformation. Furthermore, there were no PAINS and Brenk filter alerts of toxicity found for 4a and 4v.
3. Experimental
3.1. General
All chemicals used in this synthesis are commercially available and used without further purification. Melting points were obtained by the open capillary tube method and are uncorrected. High-resolution mass spectra (HRMS) were acquired using an Agilent technologies model G6564 QTOF. Nuclear magnetic resonance spectra (1H NMR & 13C NMR) were obtained using a Bruker 500 MHz WB FT-NMR spectrometer having proton noise decoupling mode with a standard 5 mm probe using DMSO-d6 solution. Abbreviations used for the 1H NMR signal are as follows: s = singlet, d = doublet, t = triplet, q = quartet, qu = quintet, dd = double doublet, td = triple doublet, and m = multiplet. The chemical shifts are reported in parts per million and coupling constants (J) are provided in Hertz.3.2. General procedure for the synthesis of spiroacridines 4a–p and spiroquinolines 4q–x
In a 50 mL round bottom flask, a mixture of 3,4-methylenedioxy aniline 1 (1 mmol), substituted isatins 2a–h (1 mmol) and cyclic 1,3-diones 3a–c (1 mmol) in 3 mL glacial acetic acid was refluxed for 5–8 min. On completion of the reaction (TLC), 5 mL of distilled water was added to the reaction mixture and stirred at room temperature for complete precipitation of the product. The furnished solid mass was filtered off, dried and then thoroughly washed again with 5 mL 5% ethyl acetate solution in n-hexane to afford the desired products 4a–x.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.86 (s, 1H, OC
aHbO), 5.86 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.50–2.36 (m, 2H, CH2), 2.08 (d, J = 16 Hz, 1H, COC
bHaO), 2.50–2.36 (m, 2H, CH2), 2.08 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHb), 1.92 (d, J = 16 Hz, 1H, COC
aHb), 1.92 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHa), 1.04 (s, 3H, CH3), 0.99 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 190.7, 179.7, 150.9, 145.6, 142.3, 140.0, 138.9, 128.7, 126.0, 121.7, 120.8, 114.6, 107.8, 104.2, 102.5, 100.1, 95.8, 50.2, 49.0, 39.5, 31.1, 27.4, 25.7; HRMS (ESI-MS) m/z calcd for C23H21N2O4 [M + H]+: 389.1501, found: 389.1518.
bHa), 1.04 (s, 3H, CH3), 0.99 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 190.7, 179.7, 150.9, 145.6, 142.3, 140.0, 138.9, 128.7, 126.0, 121.7, 120.8, 114.6, 107.8, 104.2, 102.5, 100.1, 95.8, 50.2, 49.0, 39.5, 31.1, 27.4, 25.7; HRMS (ESI-MS) m/z calcd for C23H21N2O4 [M + H]+: 389.1501, found: 389.1518.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.88 (s, 1H, OC
aHbO), 5.88 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.46 (d, J = 4.5 Hz, 2H, CH2), 2.07 (d, J = 16 Hz, 1H, COC
bHaO), 2.46 (d, J = 4.5 Hz, 2H, CH2), 2.07 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHb), 1.98 (d, J = 16 Hz, 1H, COC
aHb), 1.98 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHa), 1.04 (s, 3H, CH3), 1.01 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 190.9, 179.3, 151.3, 145.9, 142.5, 140.6, 139.1, 128.7, 126.0, 124.2, 121.6, 113.7, 109.3, 104.0, 101.9, 100.2, 96.0, 50.5, 48.9, 39.4, 31.2, 27.0, 26.1; HRMS (ESI-MS) m/z calcd for C23H19ClN2O4 [M]+: 422.1033, found: 422.1038.
bHa), 1.04 (s, 3H, CH3), 1.01 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 190.9, 179.3, 151.3, 145.9, 142.5, 140.6, 139.1, 128.7, 126.0, 124.2, 121.6, 113.7, 109.3, 104.0, 101.9, 100.2, 96.0, 50.5, 48.9, 39.4, 31.2, 27.0, 26.1; HRMS (ESI-MS) m/z calcd for C23H19ClN2O4 [M]+: 422.1033, found: 422.1038.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.88 (s, 1H, OC
aHbO), 5.88 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.46 (s, 2H, CH2), 2.07 (d, J = 16 Hz, 1H, COC
bHaO), 2.46 (s, 2H, CH2), 2.07 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHb), 1.99 (d, J = 16 Hz, 1H, COC
aHb), 1.99 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHa), 1.04 (s, 3H, CH3), 1.01 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 190.9, 179.2, 151.3, 145.9, 142.5, 141.0, 139.5, 128.8, 128.6, 124.3, 111.9, 109.9, 104.0, 101.9, 100.2, 96.0, 78.0, 48.9, 39.4, 31.1, 27.9, 27.0, 26.1; HRMS (ESI-MS) m/z calcd for C23H20BrN2O4 [M + H]+: 467.0606, found: 467.0602.
bHa), 1.04 (s, 3H, CH3), 1.01 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 190.9, 179.2, 151.3, 145.9, 142.5, 141.0, 139.5, 128.8, 128.6, 124.3, 111.9, 109.9, 104.0, 101.9, 100.2, 96.0, 78.0, 48.9, 39.4, 31.1, 27.9, 27.0, 26.1; HRMS (ESI-MS) m/z calcd for C23H20BrN2O4 [M + H]+: 467.0606, found: 467.0602.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.86 (s, 1H, OC
aHbO), 5.86 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.11 (s, 2H, CH2), 1.96–1.90 (m, 2H, CH2), 1.29 (s, 3H, ArCH3), 1.04 (s, 3H, CH3), 1.00 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 190.9, 179.3, 151.4, 146.0, 142.6, 141.1, 139.6, 128.9, 128.7, 124.4, 113.7, 112.0, 110.0, 104.1, 102.0, 100.3, 96.1, 49.0, 39.5, 31.2, 28.0, 27.6, 27.1, 26.2; HRMS (ESI-MS) m/z calcd for C24H23N2O4 [M + H]+: 403.1658, found: 403.1654.
bHaO), 2.11 (s, 2H, CH2), 1.96–1.90 (m, 2H, CH2), 1.29 (s, 3H, ArCH3), 1.04 (s, 3H, CH3), 1.00 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 190.9, 179.3, 151.4, 146.0, 142.6, 141.1, 139.6, 128.9, 128.7, 124.4, 113.7, 112.0, 110.0, 104.1, 102.0, 100.3, 96.1, 49.0, 39.5, 31.2, 28.0, 27.6, 27.1, 26.2; HRMS (ESI-MS) m/z calcd for C24H23N2O4 [M + H]+: 403.1658, found: 403.1654.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.87 (s, 1H, OC
aHbO), 5.87 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.45 (m, 2H, CH2), 2.09 (d, J = 16 Hz, 1H, COC
bHaO), 2.45 (m, 2H, CH2), 2.09 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHb), 1.93 (d, J = 16 Hz, 1H, COC
aHb), 1.93 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHa), 1.04 (s, 3H, CH3), 0.98 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 192.3, 181.3, 152.5, 147.2, 143.9, 141.5, 140.5, 130.2, 127.6, 123.3, 122.0, 116.2, 109.4, 105.8, 104.1, 101.7, 97.4, 51.8, 50.6, 41.1, 32.6, 29.0, 27.3; HRMS (ESI-MS) m/z calcd for C23H19ClN2O4 [M]+: 422.1033, found: 422.1038.
bHa), 1.04 (s, 3H, CH3), 0.98 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 192.3, 181.3, 152.5, 147.2, 143.9, 141.5, 140.5, 130.2, 127.6, 123.3, 122.0, 116.2, 109.4, 105.8, 104.1, 101.7, 97.4, 51.8, 50.6, 41.1, 32.6, 29.0, 27.3; HRMS (ESI-MS) m/z calcd for C23H19ClN2O4 [M]+: 422.1033, found: 422.1038.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.87 (s, 1H, OC
aHbO), 5.87 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 3.58 (s, 3H, OCH3), 2.08 (d, J = 16 Hz, 1H, COC
bHaO), 3.58 (s, 3H, OCH3), 2.08 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHb), 1.94 (d, J = 16 Hz, 1H, COC
aHb), 1.94 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHa), 1.91 (s, 2H, CH2), 1.04 (s, 3H, CH3), 1.00 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 192.0, 180.5, 152.5, 147.1, 143.6, 141.8, 140.3, 129.8, 127.1, 125.4, 122.8, 114.8, 110.5, 105.2, 103.1, 101.4, 97.2, 51.7, 50.1, 40.6, 32.3, 28.2, 27.3, 21.1; HRMS (ESI-MS) m/z calcd for C24H20N2O4Na [M + Na] + [−H2O]: 423.1321, found: 423.1113.
bHa), 1.91 (s, 2H, CH2), 1.04 (s, 3H, CH3), 1.00 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 192.0, 180.5, 152.5, 147.1, 143.6, 141.8, 140.3, 129.8, 127.1, 125.4, 122.8, 114.8, 110.5, 105.2, 103.1, 101.4, 97.2, 51.7, 50.1, 40.6, 32.3, 28.2, 27.3, 21.1; HRMS (ESI-MS) m/z calcd for C24H20N2O4Na [M + Na] + [−H2O]: 423.1321, found: 423.1113.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.89 (s, 1H, OC
aHbO), 5.89 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.07 (d, J = 16 Hz, 1H, COC
bHaO), 2.07 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHb), 1.99 (d, J = 16 Hz, 1H, COC
aHb), 1.99 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHa), 1.90 (s, 2H, CH2), 1.04 (s, 3H, CH3), 1.03 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 191.4, 179.7, 151.8, 146.4, 143.0, 141.6, 140.0, 129.3, 129.2, 124.9, 114.2, 112.4, 110.4, 104.5, 102.5, 100.7, 96.5, 49.4, 39.9, 31.6, 28.4, 27.5, 26.6; HRMS (ESI-MS) m/z calcd for C23H19N3O6 [M]+: 433.1274, found: 433.1240.
bHa), 1.90 (s, 2H, CH2), 1.04 (s, 3H, CH3), 1.03 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 191.4, 179.7, 151.8, 146.4, 143.0, 141.6, 140.0, 129.3, 129.2, 124.9, 114.2, 112.4, 110.4, 104.5, 102.5, 100.7, 96.5, 49.4, 39.9, 31.6, 28.4, 27.5, 26.6; HRMS (ESI-MS) m/z calcd for C23H19N3O6 [M]+: 433.1274, found: 433.1240.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.87 (s, 1H, OC
aHbO), 5.87 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.44 (m, 2H, CH2), 2.07 (d, J = 16 Hz, 1H, COC
bHaO), 2.44 (m, 2H, CH2), 2.07 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHb), 1.96 (d, J = 16 Hz, 1H, COC
aHb), 1.96 (d, J = 16 Hz, 1H, COC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHa), 1.04 (s, 3H, CH3), 1.00 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm):192.5, 181.1, 159.3, 157.5, 154.4, 147.2, 143.7, 141.9 (d, J = 25 Hz), 137.7, 130.0, 115.2, 113.5 (d, J = 90 Hz), 110.8 (d, J = 95 Hz), 109.7 (d, J = 30 Hz), 105.4, 104.8, 101.5, 97.3, 62.3, 52.2, 36.8, 27.6, 25.8, 21.3; HRMS (ESI-MS) m/z calcd for C23H20FN2O4 [M + H]+: 407.1407, found: 407.1393.
bHa), 1.04 (s, 3H, CH3), 1.00 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm):192.5, 181.1, 159.3, 157.5, 154.4, 147.2, 143.7, 141.9 (d, J = 25 Hz), 137.7, 130.0, 115.2, 113.5 (d, J = 90 Hz), 110.8 (d, J = 95 Hz), 109.7 (d, J = 30 Hz), 105.4, 104.8, 101.5, 97.3, 62.3, 52.2, 36.8, 27.6, 25.8, 21.3; HRMS (ESI-MS) m/z calcd for C23H20FN2O4 [M + H]+: 407.1407, found: 407.1393.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.85 (s, 1H, OC
aHbO), 5.85 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.57 (t, J = 6 Hz, 2H, CH2), 2.05–2.18 (m, 2H, CH2), 1.83–1.92 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 192.6, 181.4, 154.4, 147.2, 143.8, 141.5, 140.6, 130.1, 127.6, 123.5, 121.9, 116.2, 109.4, 105.5, 101.7, 97.5, 97.2, 51.9, 37.1, 27.8, 21.5; HRMS (ESI-MS) m/z calcd for C21H16N2O4 [M]+: 360.1110, found: 360.1110.
bHaO), 2.57 (t, J = 6 Hz, 2H, CH2), 2.05–2.18 (m, 2H, CH2), 1.83–1.92 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 192.6, 181.4, 154.4, 147.2, 143.8, 141.5, 140.6, 130.1, 127.6, 123.5, 121.9, 116.2, 109.4, 105.5, 101.7, 97.5, 97.2, 51.9, 37.1, 27.8, 21.5; HRMS (ESI-MS) m/z calcd for C21H16N2O4 [M]+: 360.1110, found: 360.1110.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.87 (s, 1H, OC
aHbO), 5.87 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 3.77 (qu, J = 12 Hz, 6 Hz, 2H, CH2), 2.58 (dd, J = 11 Hz, 5 Hz, 2H, CH2), 2.16–2.12 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 179.5, 153.2, 145.9, 142.4, 140.7, 139.0, 128.5, 125.9, 124.2, 121.8, 113.6, 109.3, 103.9, 103.3, 100.2, 95.9, 60.9, 50.5, 35.4, 26.1, 24.3, 19.9; HRMS (ESI-MS) m/z calcd for C21H15ClN2O4 [M]+: 394.0720, found: 394.0734.
bHaO), 3.77 (qu, J = 12 Hz, 6 Hz, 2H, CH2), 2.58 (dd, J = 11 Hz, 5 Hz, 2H, CH2), 2.16–2.12 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 179.5, 153.2, 145.9, 142.4, 140.7, 139.0, 128.5, 125.9, 124.2, 121.8, 113.6, 109.3, 103.9, 103.3, 100.2, 95.9, 60.9, 50.5, 35.4, 26.1, 24.3, 19.9; HRMS (ESI-MS) m/z calcd for C21H15ClN2O4 [M]+: 394.0720, found: 394.0734.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.88 (s, 1H, OC
aHbO), 5.88 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 3.78 (qu, J = 6 Hz, 2.5 Hz, 1H, 2H, CH2), 2.59 (dd, J = 12 Hz, 5.5 Hz, 2H, CH2), 2.17–2.12 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 191.2, 179.3, 153.2, 145.9, 142.4, 141.2, 139.4, 128.8, 128.5, 124.5, 113.7, 112.0, 109.8, 104.0, 103.3, 100.2, 95.9, 50.5, 35.4, 26.1, 24.3, 19.9; HRMS (ESI-MS) m/z calcd for C21H16BrN2O4 [M + H]+: 439.0293, found: 439.0289.
bHaO), 3.78 (qu, J = 6 Hz, 2.5 Hz, 1H, 2H, CH2), 2.59 (dd, J = 12 Hz, 5.5 Hz, 2H, CH2), 2.17–2.12 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 191.2, 179.3, 153.2, 145.9, 142.4, 141.2, 139.4, 128.8, 128.5, 124.5, 113.7, 112.0, 109.8, 104.0, 103.3, 100.2, 95.9, 50.5, 35.4, 26.1, 24.3, 19.9; HRMS (ESI-MS) m/z calcd for C21H16BrN2O4 [M + H]+: 439.0293, found: 439.0289.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.88 (s, 1H, OC
aHbO), 5.88 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.59 (t, J = 6.5 Hz, 2H, CH2), 2.21–2.07 (m, 2H, CH2), 1.94–1.84 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 179.6, 153.3, 145.9, 142.5, 140.8, 139.0, 128.6, 126.0, 124.3, 121.8, 113.7, 109.3, 104.0, 103.3, 100.2, 96.0, 61.0, 50.6, 35.4, 26.2, 24.3; HRMS (ESI-MS) m/z calcd for C21H15ClN2O4 [M + H]+: 395.0799, found: 395.0796.
bHaO), 2.59 (t, J = 6.5 Hz, 2H, CH2), 2.21–2.07 (m, 2H, CH2), 1.94–1.84 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 179.6, 153.3, 145.9, 142.5, 140.8, 139.0, 128.6, 126.0, 124.3, 121.8, 113.7, 109.3, 104.0, 103.3, 100.2, 96.0, 61.0, 50.6, 35.4, 26.2, 24.3; HRMS (ESI-MS) m/z calcd for C21H15ClN2O4 [M + H]+: 395.0799, found: 395.0796.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.86 (s, 1H, OC
aHbO), 5.86 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 3.59 (s, 1H, OCH3), 2.57 (t, J = 5.5 Hz, 2H, CH2), 2.15–2.07 (m, 2H, CH2), 1.91–1.88 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 192.8, 180.9, 154.8, 147.4, 144.0, 142.8, 141.0, 130.4, 130.1, 126.1, 115.2, 113.6, 111.4, 110.1, 105.6, 104.9, 101.8, 97.5, 62.5, 52.1, 37.0, 25.9, 21.5; HRMS (ESI-MS) m/z calcd for C22H18N2O5 [M]+: 390.1216, found: 390.1179.
bHaO), 3.59 (s, 1H, OCH3), 2.57 (t, J = 5.5 Hz, 2H, CH2), 2.15–2.07 (m, 2H, CH2), 1.91–1.88 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 192.8, 180.9, 154.8, 147.4, 144.0, 142.8, 141.0, 130.4, 130.1, 126.1, 115.2, 113.6, 111.4, 110.1, 105.6, 104.9, 101.8, 97.5, 62.5, 52.1, 37.0, 25.9, 21.5; HRMS (ESI-MS) m/z calcd for C22H18N2O5 [M]+: 390.1216, found: 390.1179.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.88 (s, 1H, OC
aHbO), 5.88 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.67–2.57 (m, 2H, CH2), 2.18–2.09 (m, 2H, CH2), 1.90–1.88 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 193.0, 181.7, 155.2, 148.5, 147.7, 144.2, 142.6, 141.0, 130.2, 125.4, 118.6, 114.5, 109.6, 105.5, 104.6, 101.9, 97.7, 51.9, 36.8, 27.7, 21.4; HRMS (ESI-MS) m/z calcd for C21H16N3O6 [M + H]+: 406.1039, found: 406.1037.
bHaO), 2.67–2.57 (m, 2H, CH2), 2.18–2.09 (m, 2H, CH2), 1.90–1.88 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 193.0, 181.7, 155.2, 148.5, 147.7, 144.2, 142.6, 141.0, 130.2, 125.4, 118.6, 114.5, 109.6, 105.5, 104.6, 101.9, 97.7, 51.9, 36.8, 27.7, 21.4; HRMS (ESI-MS) m/z calcd for C21H16N3O6 [M + H]+: 406.1039, found: 406.1037.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.87 (s, 1H, OC
aHbO), 5.87 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 3.77 (qu, J = 12 Hz, 6 Hz, 2H, CH2), 2.57 (t, J = 6 Hz, 2H, CH2), 2.15–2.08 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 192.6, 181.3, 159.5, 157.7, 154.6, 147.4, 143.9, 142.1 (d, J = 30 Hz), 137.9, 130.1, 115.4, 113.7 (d, J = 95 Hz), 111.0 (d, J = 100 Hz), 109.9 (d, J = 35 Hz), 105.6, 105.0, 101.7, 97.5, 52.4, 37.0, 26.0, 21.5; HRMS (ESI-MS) m/z calcd for C21H16FN2O4 [M + H]+: 379.1094, found: 379.1090.
bHaO), 3.77 (qu, J = 12 Hz, 6 Hz, 2H, CH2), 2.57 (t, J = 6 Hz, 2H, CH2), 2.15–2.08 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 192.6, 181.3, 159.5, 157.7, 154.6, 147.4, 143.9, 142.1 (d, J = 30 Hz), 137.9, 130.1, 115.4, 113.7 (d, J = 95 Hz), 111.0 (d, J = 100 Hz), 109.9 (d, J = 35 Hz), 105.6, 105.0, 101.7, 97.5, 52.4, 37.0, 26.0, 21.5; HRMS (ESI-MS) m/z calcd for C21H16FN2O4 [M + H]+: 379.1094, found: 379.1090.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.94 (s, 1H, ArH), 5.92 (s, 1H, OC
aHbO), 5.94 (s, 1H, ArH), 5.92 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.73 (dd, J = 10.5 Hz, 4.5 Hz, 2H, CH2), 2.23 (t, J = 5 Hz, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 199.0, 179.5, 166.3, 147.8, 144.3, 140.9, 131.8, 128.3, 126.2, 124.5, 115.1, 111.3, 108.5, 106.5, 102.0, 98.1, 51.6, 33.3, 26.0; HRMS (ESI-MS) m/z calcd for C20H14ClN2O4 [M + H]+: 381.0642, found: 381.0641.
bHaO), 2.73 (dd, J = 10.5 Hz, 4.5 Hz, 2H, CH2), 2.23 (t, J = 5 Hz, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 199.0, 179.5, 166.3, 147.8, 144.3, 140.9, 131.8, 128.3, 126.2, 124.5, 115.1, 111.3, 108.5, 106.5, 102.0, 98.1, 51.6, 33.3, 26.0; HRMS (ESI-MS) m/z calcd for C20H14ClN2O4 [M + H]+: 381.0642, found: 381.0641.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.94 (s, 1H, ArH), 5.92 (s, 1H, OC
aHbO), 5.94 (s, 1H, ArH), 5.92 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.72 (t, J = 8.5 Hz, 2H, CH2), 2.23 (t, J = 5 Hz, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 197.5, 177.8, 164.8, 146.2, 142.7, 139.7, 138.8, 130.2, 129.5, 125.6, 113.5, 112.4, 110.3, 106.9, 104.9, 100.4, 96.5, 49.9, 31.7, 23.1; HRMS (ESI-MS) m/z calcd for C20H14BrN2O4 [M + H]+: 425.0136, found: 425.0133.
bHaO), 2.72 (t, J = 8.5 Hz, 2H, CH2), 2.23 (t, J = 5 Hz, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 197.5, 177.8, 164.8, 146.2, 142.7, 139.7, 138.8, 130.2, 129.5, 125.6, 113.5, 112.4, 110.3, 106.9, 104.9, 100.4, 96.5, 49.9, 31.7, 23.1; HRMS (ESI-MS) m/z calcd for C20H14BrN2O4 [M + H]+: 425.0136, found: 425.0133.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.90 (s, 1H, OC
aHbO), 5.90 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 5.88 (s, 1H, ArH), 2.71 (dd, J = 10 Hz, 4.5 Hz, 2H, CH2), 2.22–2.20 (m, 2H, CH2), 2.14 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 199.0, 179.8, 166.1, 147.5, 144.1, 139.3, 138.5, 131.7, 131.2, 128.6, 125.0, 116.1, 109.5, 109.2, 106.6, 101.9, 97.4, 51.4, 33.4, 24.6, 21.1; HRMS (ESI-MS) m/z calcd for C21H17N2O4 [M + H]+: 361.1188, found: 361.1189.
bHaO), 5.88 (s, 1H, ArH), 2.71 (dd, J = 10 Hz, 4.5 Hz, 2H, CH2), 2.22–2.20 (m, 2H, CH2), 2.14 (s, 3H, CH3); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 199.0, 179.8, 166.1, 147.5, 144.1, 139.3, 138.5, 131.7, 131.2, 128.6, 125.0, 116.1, 109.5, 109.2, 106.6, 101.9, 97.4, 51.4, 33.4, 24.6, 21.1; HRMS (ESI-MS) m/z calcd for C21H17N2O4 [M + H]+: 361.1188, found: 361.1189.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.94 (s, 1H, ArH), 5.91 (s, 1H, OC
aHbO), 5.94 (s, 1H, ArH), 5.91 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.73 (t, J = 5 Hz, 2H, CH2), 2.43–2.21 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm):199.0, 179.6, 166.2, 147.8, 144.3, 139.7, 131.7, 128.4, 123.7, 123.2, 115.2, 114.0, 108.7, 106.5, 102.0, 98.1, 79.6, 52.2, 33.3, 24.7; HRMS (ESI-MS) m/z calcd for C20H14ClN2O4 [M + H]+: 381.0642, found: 381.0677.
bHaO), 2.73 (t, J = 5 Hz, 2H, CH2), 2.43–2.21 (m, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm):199.0, 179.6, 166.2, 147.8, 144.3, 139.7, 131.7, 128.4, 123.7, 123.2, 115.2, 114.0, 108.7, 106.5, 102.0, 98.1, 79.6, 52.2, 33.3, 24.7; HRMS (ESI-MS) m/z calcd for C20H14ClN2O4 [M + H]+: 381.0642, found: 381.0677.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.91 (s, 1H, OC
aHbO), 5.91 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 5.90 (s, 1H, ArH), 3.61 (s, 3H, OCH3), 2.74–2.70 (m, 2H, CH2), 2.22 (t, J = 5 Hz, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 199.0, 179.7, 166.2, 155.6, 147.5, 144.1, 139.5, 135.2, 131.7, 115.9, 112.8, 111.4, 110.1, 109.0, 106.5, 101.9, 98.0, 55.8, 51.8, 33.4, 24.6; HRMS (ESI-MS) m/z calcd for C21H20N3O5 [M + NH4]+: 394.1403, found: 394.1426.
bHaO), 5.90 (s, 1H, ArH), 3.61 (s, 3H, OCH3), 2.74–2.70 (m, 2H, CH2), 2.22 (t, J = 5 Hz, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 199.0, 179.7, 166.2, 155.6, 147.5, 144.1, 139.5, 135.2, 131.7, 115.9, 112.8, 111.4, 110.1, 109.0, 106.5, 101.9, 98.0, 55.8, 51.8, 33.4, 24.6; HRMS (ESI-MS) m/z calcd for C21H20N3O5 [M + NH4]+: 394.1403, found: 394.1426.
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) aHbO), 5.94 (s, 1H, ArH), 5.91 (s, 1H, OC
aHbO), 5.94 (s, 1H, ArH), 5.91 (s, 1H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) bHaO), 2.71 (t, J = 7.5 Hz, 2H, CH2), 2.3 (t, J = 5 Hz, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 198.3, 179.0, 165.5, 154.9, 146.8, 143.4, 138.8, 134.5, 131.0, 115.2, 112.1, 110.7, 109.4, 108.3, 105.8, 101.2, 97.3, 55.1, 32.7, 23.9; HRMS (ESI-MS) m/z calcd for C20H14N3O6 [M + H]+: 392.0883, found: 392.0882.
bHaO), 2.71 (t, J = 7.5 Hz, 2H, CH2), 2.3 (t, J = 5 Hz, 2H, CH2); 13C-NMR (125 MHz, DMSO-d6) (δ, ppm): 198.3, 179.0, 165.5, 154.9, 146.8, 143.4, 138.8, 134.5, 131.0, 115.2, 112.1, 110.7, 109.4, 108.3, 105.8, 101.2, 97.3, 55.1, 32.7, 23.9; HRMS (ESI-MS) m/z calcd for C20H14N3O6 [M + H]+: 392.0883, found: 392.0882.
3.3. Computational details
The Gaussian 16 software package33 was utilized for computational investigations and all DFT calculations were performed using the B3LYP method with a 6-311G+(d,p) basis set in the ground state. The chemical reactivity and stability of the molecule were investigated using Becke's three parameter (B3LYP) diffusion functionals at the 6-311G+(d,p) basis set. The frontier molecular orbital (FMO) and molecule electrostatic potential (MEP) surface analyses were accomplished by the DFT/B3LYP method using a 6-311G+(d,p) basis set. The orbital energy gap (Egap) and other quantum chemical descriptors (QCDs) were calculated from frontier molecular orbitals (FMOs) with the same conceptual method and by using well-known equations. Gauss view 6.1.1 (ref. 34) was used for geometry optimization and data visualization.3.4. ABTS radical scavenging assay
The ABTS free radical cation scavenging activity of compounds was determined by a standard procedure. First, a 7 mM concentrated solution of ABTS was prepared and then a 2.45 mM concentrated solution of potassium persulfate was added to the ABTS solution. After adding potassium persulfate to the ABTS solution, the solution was kept in a dark place for 12–16 h at room temperature. The sample solutions of the test compound were prepared in absolute alcohol ranging from 0.01 mg mL−1 to 1 mg mL−1. Then the test sample was added to the ABTS solution and incubated for 30 min at 37 °C. The absorbance was measured at a wavelength of 734 nm, and the procedure was repeated for ascorbic acid as a reference standard. The % inhibition of radical scavenging activity was determined using the given formula.3.5. DPPH radical scavenging assay
The free radical scavenging activity of DPPH was tested using a standard procedure. 0.3 mM DPPH concentrated solution in ethanol was incubated for 30 min at 37 °C with various test samples. The absorption of the sample was taken at 517 nm. The same procedure was followed for ascorbic acid as for the standard. The % inhibition of radical scavenging activity was determined using the given formula.3.6. In silico molecular docking
3.7. In silico ADMET prediction
In silico ADMET prediction of the most potent antioxidants 4a and 4q were calculated using web tool SwissADME (http://www.swissadme.ch/).364. Conclusion
In conclusion, we have developed a green synthetic approach for the synthesis of spiroacridines and spiroquinolines via a multicomponent reaction of 3,4-methylenedioxyaniline, isatin and cyclic 1,3-diones in glacial acetic acid. The key features of this protocol are the metal-free multicomponent approach, mild reaction conditions, faster reaction time, moderate to good yield and easy product purification without column chromatography. FMOs, various QCDs and MEP of all spiro molecules were obtained using DFT calculations using the B3LYP method with a 6-311G+(d,p) basis set. Furthermore, the antioxidant properties of all newly synthesized compounds were evaluated. According to in silico molecular docking studies of the most potent 4a and 4v with the targeted antioxidant protein, LD-carboxypeptidase, they show a strong binding affinity of −10.5 kcal mol−1 and −10.7 kcal mol−1, respectively. In addition, 4a and 4v are the most potent radical scavengers as they possess good drug-likeness as well as in silico ADME prediction.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
SGP, PJP, DBU and HMP are grateful to the Department of Chemistry, Sardar Patel University, for providing computational & lab facilities. SGP is grateful to the UGC, New Delhi, for a UGC-JRF (NTA Ref. No. 201610157514; dated: 01.04.2021). MPP and DBU are thankful to the Knowledge Consortium of Gujarat for the SHODH fellowship (Student Reference No. 2021016434 & 2021016413, respectively).References
- P. J. Patel, S. G. Patel, D. B. Upadhyay, L. Ravi, A. Dhanasekaran and H. M. Patel, RSC Adv., 2023, 13, 24466–24473 RSC.
- A. Lozynskyi, V. Zasidko, D. Atamanyuk, D. Kaminskyy, H. Derkach, O. Karpenko, V. Ogurtsov, R. Kutsyk and R. Lesyk, Mol. Diversity, 2017, 21, 427–436 CrossRef CAS PubMed.
- E. N. Bentz, A. B. Pomilio and R. M. J. C. Lobayan, Comput. Theor. Chem., 2017, 1110, 14–24 CrossRef CAS.
- U. Asmat, K. Abad and K. Ismail, Saudi Pharm. J., 2016, 24, 547–553 CrossRef PubMed.
- A. Sarkar, S. Santra, S. K. Kundu, A. Hajra, G. V. Zyryanov, O. N. Chupakhin, V. N. Charushin and A. Majee, Green Chem., 2016, 18, 4475–4525 RSC.
- D. M. Patel and H. M. Patel, ACS Sustain. Chem. Eng., 2019, 7, 18667–18676 CrossRef CAS.
- B. H. Rotstein, S. Zaretsky, V. Rai and A. K. Yudin, Chem. Rev., 2014, 114, 8323–8359 CrossRef CAS PubMed.
- D. M. Patel, R. M. Vala, M. G. Sharma, D. P. Rajani and H. M. Patel, ChemistrySelect, 2019, 4, 1031–1041 CrossRef CAS.
- E. Vitaku, D. T. Smith and J. T. Njardarson, J. Med. Chem., 2014, 57, 10257–10274 CrossRef CAS PubMed.
- M. M. M. Lobe and S. M. N. Efange, R. Soc. Open Sci., 2020, 7, 191316 CrossRef CAS PubMed.
- S. S. Duarte, D. K. F. Silva, T. M. H. Lisboa, R. G. Gouveia, R. C. Ferreira, R. O. de Moura, J. M. da Silva, E. D. A. Lima, S. Rodrigues-Mascarenhas and P. M. da Silva, Anticancer Res., 2020, 40, 5049–5057 CrossRef CAS PubMed.
- R. K. Bhaskarachar, V. G. Revanasiddappa, S. Hegde, J. P. Balakrishna and S. Y. Reddy, Med. Chem. Res., 2015, 24, 3516–3528 CrossRef CAS.
- P. Varakumar, K. Rajagopal, B. Aparna, K. Raman, G. Byran, C. M. Gonçalves Lima, S. Rashid, M. H. Nafady, T. B. Emran and S. Wybraniec, Molecules, 2022, 28, 193 CrossRef PubMed.
- M. Ramadan, Y. A. M. M. Elshaier, A. A. Aly, M. Abdel-Aziz, H. M. Fathy, A. B. Brown, J. R. Pridgen, K. N. Dalby and T. S. Kaoud, Bioorg. Chem., 2021, 116, 105344 CrossRef CAS PubMed.
- S. S. Duarte, D. K. F. Silva, T. M. H. Lisboa, R. G. Gouveia, C. C. N. de Andrade, V. M. de Sousa, R. C. Ferreira, R. O. de Moura, J. N. S. Gomes and P. M. da Silva, Pharmacol. Rep., 2022, 74, 545–554 CrossRef CAS PubMed.
- P. J. Patel, D. M. Patel, R. M. Vala, S. G. Patel, D. B. Upadhyay, Y. Pannerselvam and H. M. Patel, ACS Omega, 2022, 8, 444–456 CrossRef PubMed.
- I. R. Ager, A. C. Barnes, G. W. Danswan, P. W. Hairsine, D. P. Kay, P. D. Kennewell, S. S. Matharu, P. Miller and P. Robson, J. Med. Chem., 1988, 31, 1098–1115 CrossRef CAS PubMed.
- W. Raether, B. Enders, J. Hofmann, U. Schwannecke, H. Seidenath, H. Hänel and M. Uphoff, Parasitol. Res., 1989, 75, 619–626 CrossRef CAS PubMed.
- P. J. Patel, D. M. Patel, R. M. Vala, S. G. Patel, D. B. Upadhyay, Y. Pannerselvam and H. M. Patel, ACS Omega, 2023, 8, 444–456 CrossRef CAS PubMed.
- R. M. Cross, J. R. Maignan, T. S. Mutka, L. Luong, J. Sargent, D. E. Kyle and R. Manetsch, J. Med. Chem., 2011, 54, 4399–4426 CrossRef CAS PubMed.
- N. Kausar, A. A. Masum, M. M. Islam and A. R. Das, Mol. Diversity, 2017, 21, 325–337 CrossRef CAS PubMed.
- D. Song, X. Cao, J. Wang and S. Ke, Bioorg. Med. Chem. Lett., 2020, 30, 126826 CrossRef CAS PubMed.
- D. M. Patel, H. J. Patel, J. M. Padrón and H. M. Patel, RSC Adv., 2020, 10, 19600–19609 RSC.
- D. B. Upadhyay, R. M. Vala, S. G. Patel, P. J. Patel, C. Chi and H. M. Patel, J. Mol. Struct., 2023, 1273, 134305 CrossRef CAS.
- S. G. Patel, P. J. Patel, D. B. Upadhyay, A. Puerta, A. Malik, N. K. Kandukuri, R. K. Sharma, J. M. Padrón and H. M. Patel, J. Mol. Struct., 2023, 1292, 136174 CrossRef CAS.
- D. M. Patel and H. M. Patel, ACS Sustain. Chem. Eng., 2019, 7, 18667–18676 CrossRef CAS.
- S. G. Patel, R. M. Vala, P. J. Patel, D. B. Upadhyay, V. Ramkumar, R. L. Gardas and H. M. Patel, RSC Adv., 2022, 12, 18806–18820 RSC.
- S. G. Patel, A. González-Bakker, R. M. Vala, P. J. Patel, A. Puerta, A. Malik, R. K. Sharma, J. M. Padrón and H. M. Patel, RSC Adv., 2022, 12, 30404–30415 RSC.
- P. J. Patel, R. M. Vala, S. G. Patel, D. B. Upadhyay, D. P. Rajani, F. Damiri, M. Berrada and H. M. Patel, J. Mol. Struct., 2023, 1285, 135467 CrossRef CAS.
- I. Tankov, R. Yankova, S. Genieva, M. Mitkova and D. Stratiev, J. Mol. Struct., 2017, 1139, 400–406 CrossRef CAS.
- J. M. Farber and P. I. Peterkin, Microbiol. Rev., 1991, 55, 476–511 CrossRef CAS PubMed.
- A. Daina and V. Zoete, ChemMedChem, 2016, 11, 1117–1121 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson and H. Nakatsuji, Gaussian 16, Wallingford CT, 2016, vol. 421 Search PubMed.
- T. A. Keith and J. M. Millam, gausview 6.1.1, Semichem Inc., 2016 Search PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS PubMed.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4su00024b |
| This journal is © The Royal Society of Chemistry 2024 |