Engineering considerations in the design of tissue specific bioink for 3D bioprinting applications†
Shivi
Tripathi
ab,
Madhusmita
Dash
c,
Ruchira
Chakraborty
d,
Harri Junaedi
Lukman
 e,
Prasoon
Kumar
e,
Prasoon
Kumar
 *d,
Shabir
Hassan
fg,
Hassan
Mehboob
e,
Harpreet
Singh
h and
Himansu Sekhar
Nanda
*d,
Shabir
Hassan
fg,
Hassan
Mehboob
e,
Harpreet
Singh
h and
Himansu Sekhar
Nanda
 *abi
*abi
aBiomaterials and Biomanufacturing Laboratory, Discipline of Mechanical Engineering, PDPM Indian Institute of Information Technology Design and Manufacturing, Jabalpur 482005, MP, India. E-mail: himansu@iiitdmj.ac.in
bInternational Centre for Sustainable and Net Zero Technologies, PDPM-Indian Institute of Information Technology Design and Manufacturing Jabalpur, Madhya Pradesh 482005, India
cSchool of Minerals, Metallurgical and Materials Engineering, Indian Institute of Technology Bhubaneswar, Argul, Khordha, Odisha 752050, India
dBiodesign and Medical Device Laboratory, Department of Biotechnology and Medical Engineering, National Institute of Technology, Rourkela, 769008, Odisha, India. E-mail: kumarprasoon@nitrkl.ac.in
eDepartment of Engineering and Management, College of Engineering, Prince Sultan University, Riyadh 12435, Saudi Arabia
fDepartment of Biological Sciences, Khalifa University, Abu Dhabi, United Arab Emirates
gBiotechnology Centre (BTC), Khalifa University, Abu Dhabi, United Arab Emirates
hDr B R Ambedkar National Institute of Technology Jalandhar, Grand Trunk Road, Barnala Amritsar Bypass Rd, Jalandhar, Punjab 14401111, India
iTerasaki Institute for Biomedical Innovation, 21100 Erwin, St Los Angeles, CA 91367, USA
First published on 23rd October 2024
Abstract
Over eight million surgical procedures are conducted annually in the United Stats to address organ failure or tissue losses. In response to this pressing need, recent medical advancements have significantly improved patient outcomes, primarily through innovative reconstructive surgeries utilizing tissue grafting techniques. Despite tremendous efforts, repairing damaged tissues remains a major clinical challenge for bioengineers and clinicians. 3D bioprinting is an additive manufacturing technique that holds significant promise for creating intricately detailed constructs of tissues, thereby bridging the gap between engineered and actual tissue constructs. In contrast to non-biological printing, 3D bioprinting introduces added intricacies, including considerations for material selection, cell types, growth, and differentiation factors. However, technical challenges arise, particularly concerning the delicate nature of living cells in bioink for tissue construction and limited knowledge about the cell fate processes in such a complex biomechanical environment. A bioink must have appropriate viscoelastic and rheological properties to mimic the native tissue microenvironment and attain desired biomechanical properties. Hence, the properties of bioink play a vital role in the success of 3D bioprinted substitutes. This review comprehensively delves into the scientific aspects of tissue-centric or tissue-specific bioinks and sheds light on the current challenges of the translation of bioinks and bioprinting.
1. Introduction
3D bioprinting is cutting-edge technology in tissue engineering and regenerative medicine that enables the construction of 3D tissue structures with specific shapes and patterns.1 This technology can create skin, muscle, cartilage, bone, nerve tissues, etc. The market size for 3D bioprinting was $2.13 billion in 2022. It is expected to grow to $8.3 billion by 2030, with a compound annual growth rate (CAGR) of 18.51%.2 The bioprinting industry witnesses a yearly increase in the number of companies, including newcomers like Bico (CELLINK) and Poietis, as well as established players like GE Healthcare, venturing into the field. The market demand is driven by the increasing need for tissue and organ transplantation due to the rise in chronic diseases, which can damage tissues and organs, reduce quality of life, and resist treatment.3 While organ transplantation can effectively address organ dysfunction, the availability of suitable donor organs is a considerable constraint. For example, a new name is added to the organ transplant waiting list every 15 minutes in the United States.4 However, only one-third of patients on this list receive matched organs from donors. The stagnant supply of organs, meeting only 10% of global demand, highlights the urgent need for alternatives. In response, 3D bioprinting is advancing rapidly due to the demand for personalized treatments and alternatives to animal testing. While innovations currently position bioprinted tissues for medical research and drug development applications, their clinical adoption still faces issues due to bioprinter incapabilities, bioink concerns, and ethical and legal barriers.5Among all the challenges, addressing bioink concerns, such as mechanical properties and biocompatibility, is crucial for fabricating functional 3D constructs that mimic actual tissues. Bioink is a blend of biomaterials, biomolecules, and living cells used as the input for a bioprinter to create solid structures that closely match the specific geometry of targeted tissues or organs.6 Hydrogels, commonly used as bioinks in 3D bioprinting, replicate the extracellular matrix (ECM) to support cells and create tissue-like structures.6,7 For example, a blend of alginate, gelatin, and hyaluronic acid is used to create cartilage-specific bioinks that replicate the cartilage ECM and promote chondrocyte differentiation and growth, producing accurate and detailed cartilage replicas.8 Similarly, bone-specific bioinks are also being produced by amalgamating collagen, hydroxyapatite, and glycosaminoglycans.9 These specialized bioinks are designed to mimic the composition of bone tissue, providing a biomimetic environment that supports the growth and maturation of bone cells. These bioinks, combining collagen for support, hydroxyapatite for mineralization, and glycosaminoglycans for cell signalling, create a specialized microenvironment that enhances osteoblast activity and enables the precise printing of complex bone structures.
Despite their benefits, biomaterials used in bioinks for 3D-printed tissue constructs intended for implantation can provoke immune responses by activating the immune cells. For example, hyaluronic acid activates cells through CD44, and when used in hydrogel form, it can induce foreign body reactions within four to eight weeks after transplantation.10 In certain cases, polyethylene glycol (PEG) triggers a significant immune reaction, resulting in the production of anti-PEG IgM and IgG antibodies, as well as the formation of memory T cells.11 Likewise, polyglycolic acid (PGA) can trigger inflammatory responses from xenogeneic sources.12 As a result, these materials should be approached with caution when used as bioink for bioprinting tissue intended for transplantation.
Tissue-centric or tissue-specific bioinks solve this issue of immune reactions by allowing the incorporation of decellularized extracellular matrix (dECM) from specific tissues in the bioink for 3D bioprinting applications.13 dECM is created by removing the cells from the ECM of the tissue or organ while preserving the native composition and architecture of the ECM.14 dECMs can promote cell proliferation and differentiation while preserving the distinct biochemical and mechanical characteristics of original tissues. dECMs from diverse tissues, such as the heart, liver, and lung, are being utilized by researchers to formulate bioinks that can be used to print functional tissue structures.15 These bioinks could create complex tissue designs with high accuracy and fidelity by facilitating cell proliferation and differentiation. Kim et al.16 created a liver-derived dECM bioink to enhance the biomimicry of 3D liver tissue models. The incorporation of dECM into the bioink offers a liver-specific microenvironment for hepatocytes that enhance the function of 3D liver tissue models, with a 4.3-times increase in albumin secretion, a 2.5-times increase in urea secretion, and a 2.0-times increase in cytochrome enzyme activity compared to the tissue models without dECM. It also showed 1.8 times better drug responsiveness than the tissue model without dECM, highlighting its potential for developing functional in vitro liver models for drug toxicity testing. Similarly, Chae et al.17 employed 3D cell-printing technology to create biomimetic tendon and ligament tissue constructs using stem cells and tissue-specific bioinks. The tissue-specific bioinks enabled prolonged cell culture with high cell viability and enhanced tendon formation, leading to better alignment of cells and structure with the progress of time. Apart from tissue specific cues, the ideal bioink should offer excellent printability, mechanical stability, insolubility in cell culture media, appropriate biodegradability, non-toxicity, non-immunogenicity, and support for cellular adhesion.18 Therefore, it is crucial to simultaneously consider multiple factors for the successful transition of bioinks and bioprinting from research to clinical use. For example, Jia et al.19 developed a set of 30 alginate solutions to study the effects of fundamental material properties, density, and viscosity of alginate solutions on their printability. This investigation aimed to identify an appropriate range of alginate material properties for bioprinting applications. However, the high expense of bioinks and the time-intensive nature of printing trials have stimulated increasing interest in computational simulations within this area. In response to these challenges, the existing literature highlights various strategies to overcome current limitations in 3D bioprinting for tissue engineering, demonstrating continued efforts to advance the field. However, the reliance on generic bioinks hinders the clinical application of this technology. These generic bioinks lack tissue-specific cues, provide inadequate cell support, suffer from poor vascularization, and have mismatched mechanical properties, all affecting the quality of the printed tissue.20
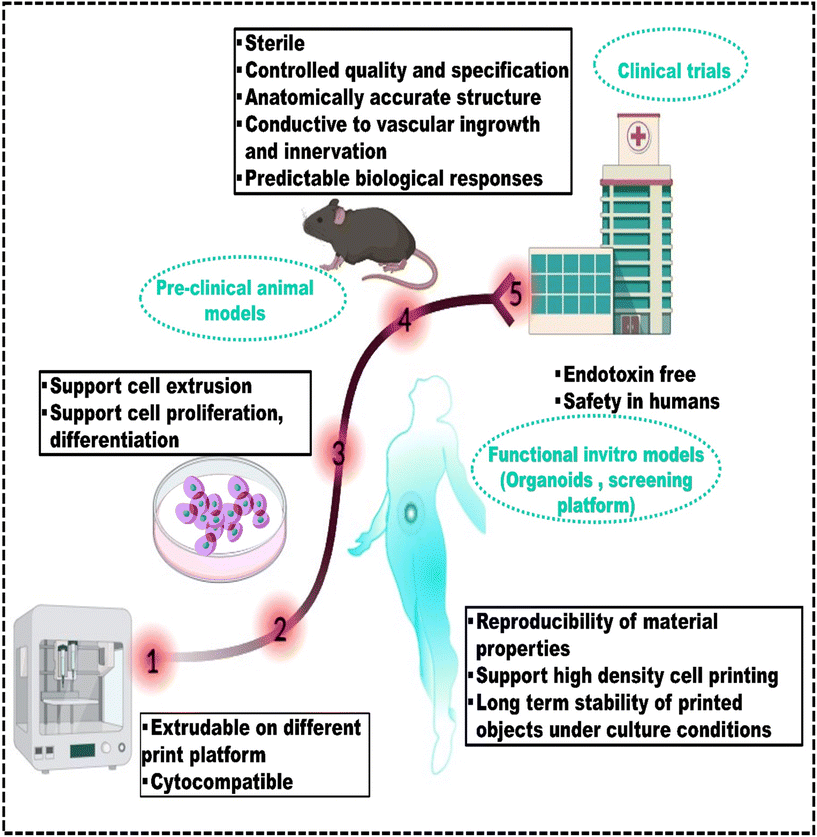
Therefore, this review fills this gap of underexplored challenges in the existing literature by providing recent strategies used to create robust bioinks suitable for 3D bioprinting of bone, skin, cartilage, and liver tissues. It provides an overview of additive manufacturing techniques and the different considerations for developing bioinks tailored to specific methods, along with polymer-based bioinks, essential for understanding 3D bioprinting. The article covers tissue-specific bioinks for mimicking target tissues, computational methods, machine learning for predicting bioink properties, and discusses the challenges in advancing 3D bioprinting for healthcare applications. It also provides a future outlook on these developments.6,9,14,15,19–28Fig. 1 shows the entire biofabrication process, from MRI scanning to bioink formulation to 3D bioprinting.29 This process forms the core framework for all applications discussed in this review and highlights its importance in bioprinting.30
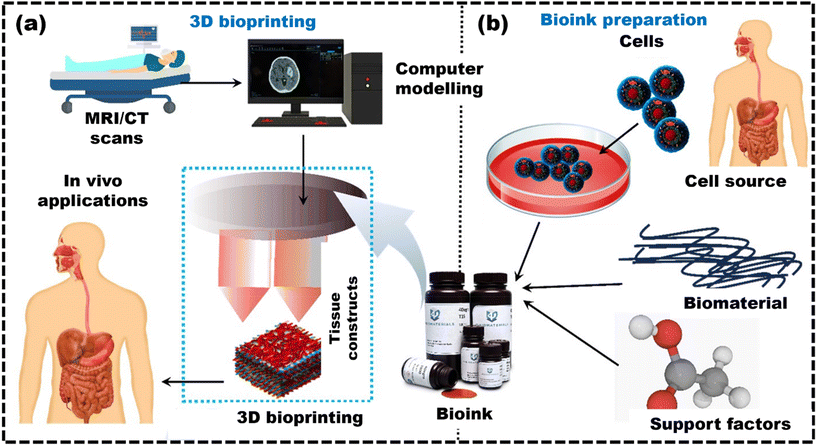 | ||
| Fig. 1 The diagram shows the overall process of 3D bioprinting, highlighting the key phases: (a) schematic shows the complex processing of data, reproduced from Ripley et al.29 with permission from John Wiley and Sons, copyright (2016); (b) bioink preparation for 3D bioprinting and its applications, reproduced from Jose et al.30 with permission from Elsevier, copyright (2024). | ||
2. Additive manufacturing techniques for bioprinting
Additive manufacturing (AM) offers extensive customization for printing structural components for medical applications based on patient-specific data. AM has shown its potential for a number of activities like medical device prototyping, surgical planning, tissue or organ printing, and so on.31 The automated production of versatile 3D human tissues and organs could simplify organ transplantation and provide the solution for pre-clinical drug and biological testing.32 Various methods have been devised to create 3D models from metals, ceramics, polymers, etc. The strategies and capabilities of AM vary depending on the materials employed, cross-linking mechanisms, and material deposition processes. Every procedure upholds a specific scope of control over model design, mechanical properties, and biodegradation.33 3D bioprinting, a subtype of AM, uses biological materials such as cells, biomaterials, and growth factors to create functional 3D living tissues and organ constructs.34 Commonly used 3D bioprinting methods include extrusion-based printing, vat photopolymerization, and jetting-based techniques. Jetting-based bioprinting can be categorized into various types: laser-based printing, inkjet printing, microvalve bioprinting, and electrohydrodynamic printing. The desired bioinks and the specific properties of each printing method are discussed in subsequent sections.2.1 Extrusion-based bioprinting
Extrusion-based bioprinting is increasingly popular because it is accessible, cost-effective, and does not use potentially harmful energy sources like lasers, making it an affordable method for printing a variety of bioinks with high cell viability.35 A key advantage of this bioprinting technique is the flexibility of bioinks, which can incorporate various hydrogels, such as collagen and ECM proteins, allowing the replication of actual tissues. Additionally, enhancing these biomaterials with growth factors or peptides can boost cell adhesion, proliferation, and differentiation, facilitating the creation of custom-designed tissues.36 For fabricating tissue constructs, it involves a dispensing head and an automated three-axis robotic system.37 The syringe with a nozzle filled with bioink is deposited on a stage either by a dispensing head that moves along three axes or by moving the stage while the head remains fixed.38 Inside the syringe, cells experience different velocity gradients based on their positions from the center to the edge. The highest velocity gradient and maximum shear stress occur near the nozzle wall, where cells close to the wall move more slowly than those in the center, resulting in more significant cell deformation.39 Shear stress, a mechanical force that deforms a material along a plane parallel to the stress direction, is regarded as the primary cause of cell damage or death in extrusion-based bioprinting. Theoretically, cell damage increases exponentially with the rising shear stresses, starting at zero in the absence of stress and reaching up to 100% under high-stress conditions.39 However, different cell types vary in their sensitivity and response to shear stress, leading to variations in cell viability. Despite having cell viability issues, the versatility of the bioink used is one of the primary advantages of this bioprinting technique. Zhao et al.40 utilized extrusion-based 3D printing technology to create chitosan ducts for soft tissue engineering applications. The preparation process can be refined using various acidic solutions, such as formic acid, acetic acid, glycolic acid, and lactic acid, to manufacture chitosan ducts. The optimal solvent for chitosan was 30 wt% glycolic acid solution.37 The Young's modulus, tensile strength, and fracture strain values for chitosan ducts were 12.38 ± 1.19 MPa, 10.98 ± 0.61 MPa, and 146.03 ± 15.05%, respectively, similar to the soft tissue properties. These characteristics make chitosan ducts suitable for soft-tissue restoration.41 However, like other biological structures, chitosan ducts need an appropriate microenvironment to support cell adhesion, proliferation, and differentiation. Cell-laden micro-scaffold-based bioinks can tackle these challenges by offering a more physiologically relevant microenvironment for cells to further support tissue regeneration.Tan et al.42 utilized a micropipette extrusion-based 3D bioprinting method to fabricate living multicellular tissues using cell-laden micro-scaffold-based bioinks. The bioink consists of poly(lactic-co-glycolic acid) (PLGA) porous microspheres with a thin encapsulation of agarose–collagen (AC) hydrogel. L929 mouse fibroblasts were cultured to assess the biocompatibility of the printed construct. The bioprinted construct demonstrated excellent biocompatibility, with more than 90% cell viability maintained over the 2 days, 7 days, and 14 days of cell culture. Furthermore, compared to the AC hydrogel, the mechanical strength of the construct was significantly improved, exceeding it by more than 100 times. However, single-component bioinks typically lack essential traits like cell-specific bioactivity and optimal rheological properties. This limitation results in a narrow bio-fabrication window, restricting the applicability of extrusion bioprinting in tissue engineering and regenerative medicine. Selecting specific additional polymers, micro-, or nanoparticles for existing single-component bioinks enables the design of multicomponent bioinks. Xin et al.43 investigated the hydrogel microparticle dissipation process during printing, analyzing both external resistance from the apparatus and the internal physicochemical properties of the hydrogel microparticles through experimental and computational methods. These offer a broader bio-fabrication window, enabling enhanced functionality and mechanical stability to mimic targeted physiological tissues.44 Overall, 3D extrusion bioprinting is poised to make significant advancements with novel bioinks and improved printing techniques, paving the way for promising medical applications.
2.2 Vat photopolymerization-based bioprinting
Vat photopolymerization (VP) has been widely used in tissue engineering and regenerative medicine due to its superior spatiotemporal control over bioresin.45 This capability enables the fabrication of intricate 3D hierarchical structures that closely resemble human tissues and organs.46 VP bioprinting technology uses the curing properties of photocurable materials by directing a light beam onto the precursor solution, allowing for on-demand material curing. This method enables the creation of various 3D structures by controlling where and how long the light beam is projected.47 Among them, stereolithography (SLA), digital light projection (DLP), and volumetric bioprinting technology are commonly used VP techniques. SLA technology uses photosensitive liquid resin as bioink, which solidifies upon exposure to ultraviolet light.48 A computer-controlled laser traces each layer, solidifying the resin, and the process is repeated layer-by-layer to create 3D constructs. DLP uses a UV lamp or laser with a digital mirror device to cure resin layer-by-layer, forming 3D constructs. Unlike SLA, which uses a moving laser, DLP employs a dynamic mask for light exposure.49 Both methods demonstrate the versatility of VP techniques, allowing for the accurate creation of complex shapes in various applications. For instance, Guillaume et al.50 developed photo-cross-linkable poly(trimethylene carbonate) (PTMC) resins containing 20% and 40% hydroxyapatite (HA) nanoparticles and employed SLA technology to fabricate scaffolds with precise macro-architecture. The results indicated that rabbits implanted with PTMA-MA scaffolds containing 40% nano-HAP showed the most significant degree of vascularization at the lesion site, effectively promoting the formation of new bone tissue. Likewise, Ma et al.51 employed the DLP technique of VP to fabricate a gelatin methacrylate/methacrylated hyaluronic acid hydrogel liver model, matching the stiffness of actual liver tissue. The hepatic model provided a 3D environment for hiPSC-derived hepatic cells co-cultured with supporting cells (human umbilical vein endothelial cells) in a hepatic lobule-like structure. It enabled in vitro maturation and helped maintain the functional integrity of hiPSC-derived hepatic cells within a biomimetic setting. Although extensive studies have reported on VP-based bioprinting of human tissues such as the liver, skin, and bone, achieving the complete fabrication of transplantable tissues remains elusive. Human organs have complex structures composed of various cell types and materials. Therefore, utilizing multiple materials is essential to replicate human tissue in VP bioprinting accurately.52 However, the vat storage of bioresin in VP poses more challenges for multimaterial printing than extrusion and inkjet bioprinting. The bioresin used in VP-based bioprinting must have low viscosity, as higher viscosity can cause the sedimentation of encapsulated cells. Furthermore, bioresins for VP need to be transparent to enable light penetration and initiate photopolymerization.47 However, there is a limited availability of highly transparent biomaterials suitable for bioresins.53 Therefore, more research is needed to address these challenges and create universal bioresin toolboxes that effectively manage viscosity while allowing for cell encapsulation.2.3 Jetting-based bioprinting
The 3D bioprinted stromal structures adhered to the host tissue, indicating successful tissue integration potential. This highlights the potential of LAB 3D bioprinting for creating functional tissue constructs that integrate with native tissues and contribute to their regeneration.64 Similarly, Keriquel et al.54 demonstrated the utilization of LAB technology for on-site bone regeneration by precisely printing specific biological components (nHA collagen material) and mesenchymal stromal cells (MSCs) within a defect in the calvaria (upper part of the skull) of mice (Fig. 2a). Different cell geometries, with distinctive cellular repartitions (such as disc and ring), were also tested to show their impact on bone regeneration. The results showed that one month after printing, the nHA-collagen and nHA-collagen + MSC cells printed in a ring shape showed only marginal tissue reconstruction, mainly at the periphery of the defect. In contrast, the nHA-collagen + MSC cells printed in a disk shape showed significant new bone formation, well distributed throughout the defect. Hence, LAB proves effective at printing mammalian cells with minimal impact on their viability and functionality, presenting a promising avenue for in situ bioprinting of the tissues. However, a key challenge is developing functional blood vessels to improve the integration and effectiveness of synthetic bone grafts. Kédanet al.65 employed LAB to directly print endothelial cells within a mouse calvaria bone defect. The defect was pre-filled with collagen-containing vascular endothelial growth factor (VEGF) and mesenchymal stem cells (MSCs). This approach aims to establish pre-vascularization with a well-defined architecture and promoting in vivo bone regeneration. The influence of different cellular arrangements (disc, crossed, and random) on bone regeneration (br) and vascular organization (vr) was examined in vivo. After two months, vr and br showed statistically significant improvements in the ‘disc’ pattern (vr: +203.6%, br: +294.1%) and ‘crossed circle’ pattern (vr: +355%, br: +602.1%) compared to the ‘random seeding’ conditions.
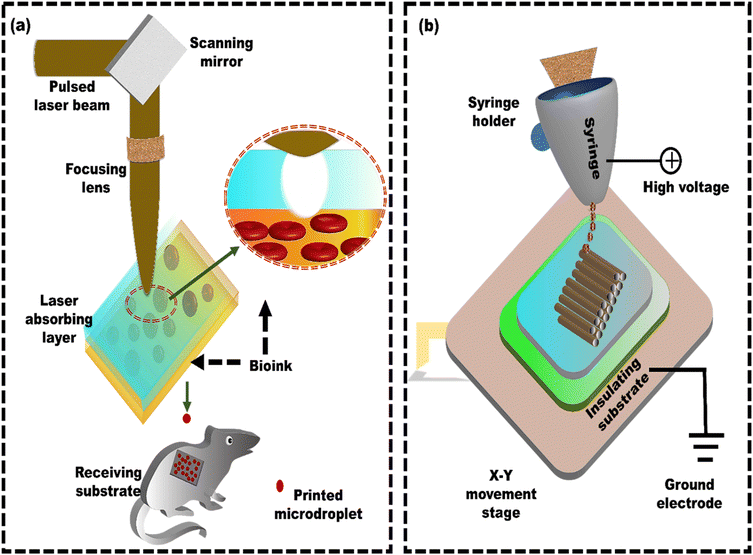 | ||
| Fig. 2 Schematic of laser-assisted bioprinting (LAB) and electrohydrodynamic printing: (a) a standard LAB setup consists of a pulsed laser beam, a focusing system, and a ribbon that generates microdroplets, reproduced from Keriquel et al.54 with permission from Elsevier, copyright (2017), and (b) electrohydrodynamic printing using a coaxial nozzle for creating microscale constructs with embedded cells, reproduced from Liang et al.94 with permission from Elsevier, copyright (2018). | ||
In addition to its use in calvaria bone defect repair, LAB technology is gaining attention in treating pancreatic ductal adenocarcinoma (PDAC). Hakobyan et al.66 used LAB to create 3D pancreatic cell spheroid arrays and monitored their phenotypic changes over time through image analysis and phenotypic characterization for PDAC. The results showed that the 3D model of spheroids containing various combinations of acinar and ductal exocrine pancreas cells was suitable for studying the early stages of PDAC development. As LAB enables precise control over cell placement, shape, and production speed, this high-throughput spheroid array model holds significant promise as a substitute for standard 2D cultures or more intricate and time-consuming 3D methods.67–69
Despite having various advantages like precision and resolution enabling the creation of intricate structures, LAB also faces several technical challenges. A support bath or environment is often needed to print complex 3D structures to maintain their shape and prevent deformations because the viscosities accommodated in the LAB are often too low to construct a stable structure without support.64 Scaling from the laboratory benchtop to factory production poses another challenge. The current printing speeds of LAB technology need to be faster to efficiently produce large-scale bioproducts.64,70 Regarding commercial availability, laser-based bioprinters are relatively expensive and require significant expertise. Thus, more investigations and advancements are required to tackle these technological obstacles to increase the use of LAB in both academic and industrial settings.
Shear stress in inkjet bioprinting primarily develops during two key phases, which include the ejection of cell-laden droplets from the nozzle and their landing on the substrate. Extended exposure to mechanical shear stress can affect the phenotype and viability of biological cells, potentially leading to cell lysis.75 Inkjet bioprinting exposes cells to significantly higher shear stress levels, reaching up to 500 Pa, but only for about 100 μs.75 The response to shear stress is strongly influenced by the type of cell, the shear stress's magnitude, and the exposure time. Increasing the concentration of cells in the DOD inkjet bioprinting system causes the droplets to jet out at a slower impact velocity. This slower droplet speed helps protect the cells by reducing splashing, improving the overall quality of the printed material. Additionally, it is essential to limit the printing time for each layer to prevent the droplets from evaporating too much, which helps maintain cell viability.76 For example, Ng et al.77 used a thermal inkjet system to dispense sub-nanoliter cell-laden droplets, identifying two critical factors, droplet impact velocity and droplet volume, which significantly affected the viability and proliferation of printed cells. The results demonstrated that under the optimal conditions, with a concentration of 4 million cells per mL and a printing time of less than 2 minutes, the cells showed elongation by day 1 and proliferated efficiently, covering almost 90% of the surface area by day 7. Therefore, it is crucial to optimize the magnitude of shear stress and the exposure time to enhance the benefits of inkjet bioprinting.
Still, the resolution is an issue in microvalve bioprinting that needs attention because microvalves offer lower resolution (∼150 μm) compared to inkjet printing (∼20 μm).84 Additionally, the impingement shear stress is comparable to the nozzle wall shear stress. It may even exceed it in some instances. Therefore, minimizing impingement shear stress is essential in microvalve bioprinting to reduce process-induced cell death. Nasehi et al.80 showed that the distance between the nozzle and the platform influenced the amplitude of impingement shear stress. Consequently, this important factor should be addressed by adjusting the nozzle-to-platform distance to optimize cell viability in the case of microvalve bioprinting. Overall, this technique is better suited for co-printing larger volumes of biomaterials, such as cells, genes, hydrogels like collagen and alginate, and growth factors, which require a larger nozzle size.
EHD bioprinting typically involves minimizing the nozzle-to-collector distance to a few millimeters. This modification ensures a singular material flow, facilitating stable and controlled deposition of minute fibers.87 EHD offers several advantages over other bioprinting techniques, including its ability to print various biomaterials (hydrogels, biopolymers, and living cells), creating functional tissues and organs that closely mimic the actual tissues. It also offers a precision of approximately 50 nm high-resolution and creates heterogeneous, gradient structures.88 These structures are achieved by controlling the electric field and the flow rate of the bioink, which can be used to create structures with varied cell densities, stiffness, and porosity.89 These heterogeneous and gradient structures are essential for creating tissues with complex functions, such as muscles and organs, which require different cell types and environments to function correctly.90
It simplifies transferring micro/nano-architectures from photomasks to biologically relevant materials, demonstrating flexibility in producing tiny 3D architectures.91 Similarly, in contrast to light-based micro/nanoscale 3D printing techniques like projection micro-stereolithography and two-photon polymerization, EHD bioprinting is a cost-effective method.92
Cell printing is extensively utilized in biomedical fields because it creates living tissue constructs with precise control over the cell arrangements. However, producing cell-laden 3D structures with high cell viability and high resolution is still difficult. EDH bioprinting is increasingly recognized as a method to address the challenge of achieving high resolution and cell viability in 3D constructs.93 Liang et al.94 developed a coaxial nozzle-assisted EHD cell printing strategy to fabricate 3D cell-laden constructs (Fig. 2b). The results demonstrated that the proposed strategy could print 3D hydrogel structures with uniform filament dimensions (around 80 μm) and consistent cell distribution, with more than 90% cell viability. Still, the fabrication of complex, heterogeneous 3D living constructs with multiple cell types and diverse hydrogel compositions remains challenging. Altun et al.95 used the EHD strategy to fabricate a bacterial cellulose/polycaprolactone (BC/PCL) composite scaffold for tissue engineering applications. The BC/PCL composite scaffold fibers were spaced approximately 100 μm apart, and the depth of the fibers was 4.7 ± 1.03 μm. The BC/PCL composite scaffold demonstrated over 155% cell viability compared to the PCL scaffold, indicating its strong potential for tissue engineering applications with improved bioactivity. Despite their advantages, most scaffolds in the literature feature a single pore size or porosity. However, native biological tissues, like cartilage and skin, have a layered architecture with zone-specific pore sizes and mechanical properties. As a result, functionally graded materials are becoming increasingly popular for addressing these challenges by incorporating zones with different pore sizes and mechanical properties. Vijayavenkataraman et al.96 used the EHD technique to develop PCL-based functionally graded scaffolds (FGS) with various gradient patterns, such as radial, sectional, diagonal, concentric, and axial directions for interfacial tissue engineering, and in vitro cancer metastasis. The FGS patterns were tested for mechanical properties under tensile loading and compared with simulated results. The computational analysis aligns with experimental observations in all cases. Mechanical testing confirmed that all the FGS had a mean yield stress from 6 MPa to 8 MPa, whereas the mean yield strain was between 3% and 9%. He et al.97 explored the potential of utilizing solution-based EHD 3D printing to produce microscale scaffolds made of PCL, including multi-walled carbon nanotubes (MWCNTs) (Fig. 3a). The solution-based strategy aimed to overcome the challenges of uniformly incorporating functional or bioactive nanomaterials into printed microfibers in viscous melted polymers. When the printing process used 8 wt% PEO and 5 wt% PCL concentrations, 3D fibrous structures with fibers measuring 10 μm in diameter, like the size of living cells, were developed (Fig. 3b and c). The results revealed that although MWCNTs have a detrimental effect on cell attachment, the developed PEO–PCL–MWCNT scaffolds promoted cell alignment (Fig. 3d). Cells adhered and aligned along the microfibers of the PEO–PCL–MWCNT scaffolds (Fig. 3e). Similarly, Choe et al.98 employed electrohydrodynamic jetting (EHDJ) to fabricate a PCL/cellulose composite scaffold consisting of coil-shaped struts and controllable macro/micropores. This coil-shaped framework demonstrated substantial cell migration of 79 ± 1.7%, whereas the control (PCL coil-shaped scaffold) showed a lower cell movement of 51 ± 2.4%. The uniform distribution of cellulose within coil-shaped PCL/cellulose composite scaffolds directly contributes to their high cell proliferation rates, indicating their potential application in hard tissue regeneration.
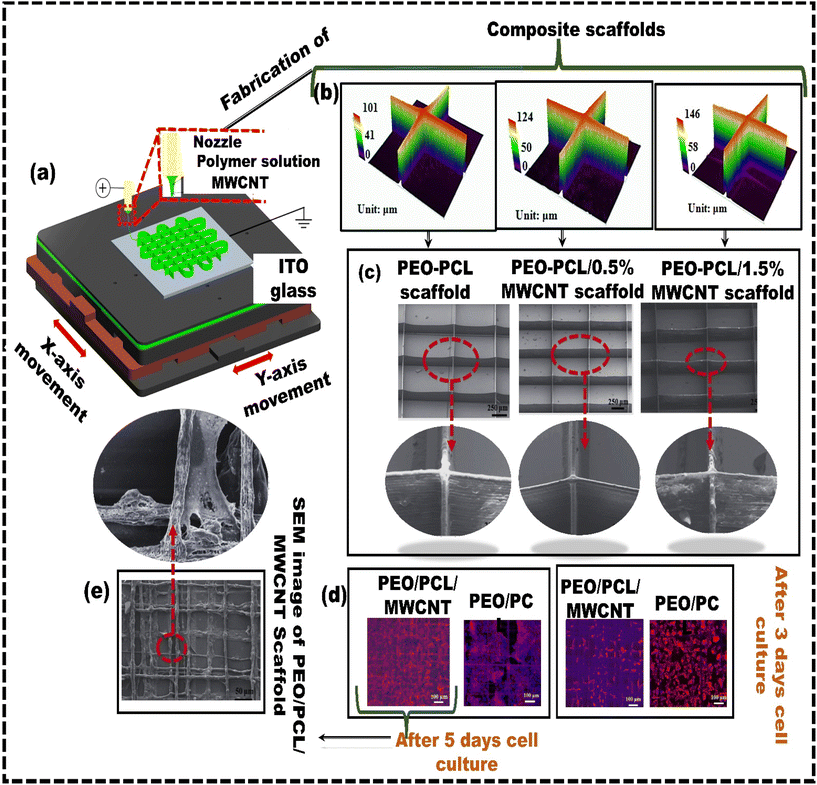 | ||
| Fig. 3 Solution-based electrohydrodynamic 3D printing: (a) schematic representing the electrohydrodynamic method used for preparing microscale PEO–PCL–MWCNT composite scaffolds; (b) profiles of 3D composite scaffolds with varying MWCNT content; (c) SEM images; (d) fluorescent microscopy images showing cellular structures on PEO–PCL–MWCNT and PEO–PCL scaffolds; (e) SEM images depicting cell morphology on PEO–PCL–MWCNT scaffolds following a five-day cell culture period, reproduced from Jiankang et al.97 with permission from IOP Publishing, Ltd, copyright (2017). | ||
Apart from several advantages of EDH, specific challenges hinder the broader implementation of EHD bioprinting, and addressing these challenges is crucial to unlocking its full potential. The key issues include the limited height of 3D micro/nanofibrous structures, reaching approximately 5 mm due to variations in electrical force during stacking.99 Strategies such as dynamic voltage adjustments and using paper substrates have been explored to overcome these challenges. While EHD bioprinting offers high resolution, precisely depositing tiny fibers for intricate microstructures and efficiently fabricating large tissue scaffolds with EHD bioprinting are significant challenges. These issues have been addressed using electrostatic forces to control filament paths, achieving printing speeds of up to 0.5 m s−1 horizontally and 0.4 m s−1 vertically.100 This electrostatic deflection technique shows great promise for ultrafast printing. It significantly enhances the scalability of EHD bioprinting, especially for large and complex tissue scaffolds.101 Despite these advancements, several challenges persist, such as biomaterial scarcity, particularly for high-resolution EHD bioprinting.102 This emphasizes the need for innovative, material-independent solutions to broaden the application of EHD in biomedical engineering.
Overall, the detailed overview of 3D bioprinting methods covers every aspect, from materials to capabilities and applications in tissue engineering. The most widely used 3D bioprinting technologies adapted for fabricating tissue constructs and their resolution, materials, and possible applications are summarized in Table 1.
| Printing technique | Resolution | Material | Application |
|---|---|---|---|
| Extrusion based | 150–600 μm (ref. 103) | Hydrogels, thermoplastics | Cartilage,104 adipose,105 a cellular scaffold,106 cell-free scaffold107 |
| Laser based | 20–100 μm (ref. 108 and 109) | Hydrogels, ceramics, etc. | Bone,55 vascular,110 adipose,111 organoid,112 cornea,63 skin68 |
| Electrohydrodynamic | 50 nm (ref. 113 and 114) | PCL, chitosan, GelMA | Bone,115 blood vessel,116 tendon/ligament, corneal stroma,117 cartilage,118 cardiac patch119 |
| Inkjet bioprinting | <50–75 μm (ref. 120) | Hydrogel-based materials such as alginate, calcium chloride and, acrylated PEG, etc. | Skin (dermo-epidermal skin constructs),121 DNA microarray,71 pharmaceutical development71 |
3. Considerations for developing bioinks for 3D bioprinting
Bioinks play a fundamental role in 3D bioprinting, serving as the foundation for the process and ensuring the desired printability and structural stability of the printed structure.122 This stability is crucial for the success of the bioprinting process, as it directly impacts the capability to create intricate tissue structures with exceptional resolution and reproducibility.123 Wettability, rheological properties, crosslinking methods, and immunogenicity are interconnected factors significantly impacting bioprinting success. Wettability affects bioink adhesion to the print bed; a smaller contact angle improves stability during layer deposition, which is essential for vertical fidelity and preventing displacement.124–127 Rheological properties, such as viscosity and shear-thinning behaviour, influence the flow of bioink during printing, balancing the stabilization of encapsulated cells with ease of extrusion to ensure cell viability and high-resolution prints.128 Crosslinking methods like thermal, ionic, enzymatic, and photo-crosslinking create stable structures, with the choice affecting both mechanical strength and cytocompatibility. Lastly, minimizing immunogenicity is crucial for long-term success, as materials should reduce immune responses to avoid inflammation and promote biodegradability and biocompatibility for cell growth and function.129 In extrusion-based bioprinting, the choice of bioink and its properties is vital for achieving structural stability and maintaining shape fidelity during printing. While, in inkjet bioprinting, where cells are suspended in low-viscosity fluids, the impact of bioink properties on stability may be less pronounced due to inherent characteristics of the fluid. Therefore, by optimizing bioink formulations tailored to the specific bioprinting method, it is possible to enhance the stability of the printed constructs, leading to improved cell viability, tissue regeneration, and functional outcomes. Besides the properties of the bioink, for translation purposes, bioinks must be sterile, free from endotoxins, and not induce pyrogenicity.130 So, it is vital to consider these bioink aspects and the challenges that must be addressed for successful commercialization and clinical adoption.1313.1 Designing bioink for extrusion-based 3D bioprinting
Choosing a suitable bioink material for extrusion bioprinting requires establishing a thorough performance index to assess its suitability.126 Bioinks must satisfy different performance requirements in extrusion bioprinting at each stage—before, during, and after extrusion.103 In the pre-extrusion stage, key characteristics of bioinks include viscosity, cell distribution, and biocompatibility, while during extrusion, shear-thinning is crucial as it helps reduce shear stress.132 Shear-thinning relies on the reversible breaking of crosslinked bonds within hydrogel materials, primarily formed by non-covalent interactions. When subjected to high shear forces, these non-covalent bonds break, causing a reduction in the bioink's viscosity.133 Once the shear forces are removed, the bonds re-form through molecular non-covalent forces, increasing viscosity and preserving the bioink's shape stability.134 The minimal force needed for the bioink to begin flowing is known as yield stress, and too much of it can harm cells and 3D printing apparatus.135 Static viscosity and yield stress provide an initial basis for evaluating a material's suitability for bioink preparation in extrusion-based printing. A material is not suitable for extrusion bioprinting if its viscosity barely changes with shear force or the yield stress is too high.44In the second stage, known as the shear-thinning phase following extrusion, the viscosity of the bioink decreases with increasing shear rate. This enables continuous extrusion, the creation of uniform filaments, and enhances cell survival during the process.136 But there is a threshold below which viscosity reduction causes the bioink to extrude as a droplet instead of a homogeneous filament. The third stage is the post-print recovery stage. In this stage, the post-print curing capability of the bioink is assessed by examining the relationship between viscosity and time, which is linked to the stability of size and cell dispersity.137 For example, slow viscosity recovery may result in the sedimentation of cells.125 Additionally, temperature is also critical in extrusion-based bioprinting in managing bioink rheology and crosslinking. Biomaterials like gelatin and collagen, which are temperature-sensitive, shift from liquid to gel at around 37 °C, allowing for smooth extrusion and proper solidification.138 Hence, crosslinking methods such as ionic, thermal, or photochemical require precise temperature control to ensure optimal printability and adequate bonding after extrusion. Among various cross-linking materials, thermally crosslinked hydrogels are becoming increasingly popular because they enable flexible crosslinking through adjustments to the temperature and duration.139 For instance, Moncal et al.140 designed a novel thermally-controlled extrusion unit for the bioprinting of collagen/Pluronic composite bioinks. The rat bone marrow-derived stem cells (rBMSCs) were incorporated into the composite bioink, and their viability and proliferation were assessed over a week via in vitro cell culture. The formulated bioink showed promising performance for bioprinted cells. The cells could attach, spread, and increase within the collagen fibers. Due to its capability to regulate the alignment of collagen fibers, this composite bioink holds potential promise for future applications in cartilage,141 skin,142 or corneal143 tissue engineering. Additional research is necessary to create extrudable biomaterial bioinks that possess suitable mechanical properties and bioactivity, specifically adapted to the requirements of different organs and tissue elasticity.
3.2 Designing bioink for jetting-based 3D bioprinting
Surface tension, inertia, and viscosity are the main forces influencing the jetting phenomena in bioprinting.144 The relative importance of these forces in basic jetting phenomena can be evaluated using Reynolds and Weber dimensionless numbers.126 Droplet dispensing is effective only at a specific ratio of these forces. The Reynolds number (Re) indicates the ratio of inertial forces to viscous forces, as shown below in eqn (1).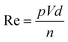 | (1) |
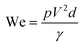 | (2) |
The Ohnesorge number (Oh) effectively summarizes the physical properties of the bioink and the characteristic length scale as follows in eqn (3).
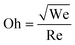 | (3) |
These properties are sometimes represented as the inverse of the Ohnesorge number (Z = Oh−1) or as the Laplace number (La = Oh−2). The value of the Ohnesorge number is independent of the driving conditions, i.e., fluid velocity.148 When the Ohnesorge number exceeds 1 (Oh > 1), viscous forces dominate and obstruct the separation of droplets from the nozzle.149 When the Ohnesorge number is too low (Oh < 0.1), the viscous forces are unable to stabilize the breaking jet, causing it to break apart into several unwanted satellite droplets.150 Satellite droplets have much less momentum than a single large droplet, making them more prone to being altered by air currents between the droplet and the medium. This results in uncertainty in their placement.151 Hence, designing an effective bioink for jetting-based bioprinting necessitates thoroughly understanding the abovementioned physical factors. Optimizing these parameters is crucial for improving the performance of jetting-based technologies and thereby advancing the field of tissue engineering.
3.3 Designing bioink for VAT photopolymerization-based 3D bioprinting
In VP, several important factors impact the quality and performance of the printed constructs, with viscosity playing a crucial role.152 It affects multiple elements of the printing process and the final product, including the polymerization rate, oxygen inhibition, cure depth, green strength, and the forces applied to the construct during the peeling process. Most importantly, viscosity affects the resin's printability by controlling the recoating process between layers. As mentioned earlier, each printed layer requires a new resin coating before photopolymerization can continue. When the resin flows more slowly, this step takes longer, increasing the time between layers and extending the print duration.153 Resins with high viscosity can cause bubbles or result in an incomplete recoating. Therefore, high-viscosity polymers should be avoided, and low-viscosity resins are preferred to improve the efficiency of the recoating process.154 Viscosities appropriate for VP typically vary from 1 Pa s to 10 Pa s, depending on the molecular weight.155 The resin's viscosity affects the cure rate by influencing monomer mobility. Generally, lower viscosity enhances monomer mobility, allowing reactive functionalities to interact more quickly and speeding up cure rates.156 Accelerating the conversion from liquid resin to solid construction leads to reduced overall construction times. Higher viscosity increases cross-linking near the surface, improving the green strength of the construct.157 However, it also raises capillary forces during layer separation, negatively impacting low-modulus hydrogels used in tissue engineering. Therefore, it is crucial to understand how viscosity influences the printing process when developing new bioresins for VP.3.4 Sterilization techniques
Sterilization eradicates all types of microbial life, including bacteria, yeast, and viruses.158 According to guidelines like ISO 11737, medical equipment must be sterilized to guarantee that no pathogens are present on it.158 Thus, while using 3D bioprinted constructs in clinical settings, it's essential to consider practical measures that lower the risk of implantation-related infections. The commonly used sterilization methods include chemical and physical methods.159 Irradiation, autoclaving, steam treatment, plasma, and syringe filtration are examples of physical sterilizing techniques, whereas using peracetic acid (PAA), ethanol, and ethylene oxide (EtO) are the chemical methods.160 For example, microorganisms are made inactive in EtO through the alkylation of nucleic acid groups, including hydroxyl, carboxyl, phenolic, and amino groups. As a result, the microbes suffer damage or die, which stops their ability to replicate and metabolize. Rizwan et al.161 studied the impact of different sterilizing techniques like autoclaving, EtO treatment, and gamma (γ) irradiation on GelMA-based bioinks. The findings suggested that sterilization using EtO reduced the viability of the encapsulated cells. Both autoclaved GelMA samples and samples treated with EtO showed a notable decrease in compressive modulus. γ irradiation compromised the printability of GelMA, likely due to its detrimental effect on the sol–gel transition. Overall, autoclaving showed no cytotoxic effects and did not affect printability, indicating its suitability for GelMA-based bioinks. Similarly, Hodder et al.162 examined the impact of different sterilization methods on alginate/methyl cellulose (alg/MC) bioinks containing bovine primary chondrocytes for 3D bioprinting. The investigation suggested exposing the bioink to γ irradiation at a dose of 25 kGy notably decreased the stability and viscosity of alginate/methyl cellulose (alg/MC) after bioprinting compared to other sterilization methods. This indicates that γ irradiation might not be appropriate for 3D bioprinting using alg/MC bioinks. To date, bioinks that contain cells cannot be sterilized without causing any damage to the cells. Therefore, new sterilization methods are needed to prevent cytotoxic effects, material deterioration, discoloration, and embrittlement. These methods should preserve the mechanical properties of materials used in bioink and be less damaging to cells and other biological factors. This challenge introduces a new research area with the potential for significant advancements in translational research. Hence, future studies in 3D bioprinting should explore the impact of sterilization on rheology, printability, and the mechanical and biological properties of the printed constructs.For successful clinical application of bioprinted structures, it is crucial to maintain their structural integrity, ensure sufficient mechanical strength, and guarantee long-term viability. This necessitates developing bioinks with specific properties and investigating sterilization methods that minimize damage to the bioprinted materials. Hence, there is a need to standardize the properties of bioink using specific criteria and tools for measuring performance at different stages of development, as shown in Fig. 4.163
 | ||
| Fig. 4 Navigating the bioinks’ translational path to the clinic: standardizing properties through performance criteria, reproduced from Gu et al.163 with permission from Wiley-VCH GmbH, copyright (2021). | ||
4. Synthetic and natural polymer-based bioinks
Polymers are commonly used as bioink materials in 3D bioprinting because their diverse and attractive features complement the 3D printing process. Natural polymers, such as alginate, collagen, gelatin, and hyaluronic acid (HA), have gained popularity due to inherent biocompatibility and similarity to human tissue ECM.164 Collagen is a significant component of the ECM and has been used in bioinks to regenerate cartilage and skin.165 HA is a major component of synovial fluid and is widely used in bioinks for cartilage and bone regeneration.166 Gelatin, derived from collagen, has been used as a bioink for cell encapsulation and the regeneration of various tissues, including bone, cartilage, and skin.167 In recent developments of natural polymer bioinks, the integration of growth factors and cell signaling molecules has become a notable trend for promoting tissue regeneration.168 Simultaneously, the application of 3D printing techniques is utilized to construct hierarchical structures and gradients within the printed tissues.Similarly, synthetic polymers such as polycaprolactone (PCL) and polyethylene glycol (PEG) are commonly used as bioinks due to their well-demonstrated biocompatibility and tunable mechanical properties.169 PCL has excellent mechanical properties and can be processed into various structures, making it an ideal material for bone tissue engineering.170 Due to the high water solubility, biocompatibility, and ease of modification of PEG, it can also be widely used as a bioink.171 It can also be modified to include specific functional groups to enhance cell adhesion and proliferation.172,173 The polymeric bioinks should align with the 3D printing process and create a biocompatible environment that closely mimics actual tissue. Ideally, these bioinks should degrade in a controlled manner without producing any harmful by-products.174 Both synthetic and natural polymers have advantages and disadvantages, and the choice of polymer depends on the specific application and desired properties of the printed tissue or organ. However, most polymeric bioinks initiate gelation after extrusion, typically requiring high polymer fraction solutions (>5 wt%) for sufficient printing viscosity.39 These dense matrices can impede in vivo matrix remodelling and vascularization, making them not suitable for tissue engineering. Moreover, high polymer fractions in cell-encapsulating bioinks can restrict cell activities, limiting their use in cell-laden structures. Rutz et al.175 introduced a single bioink method to generate extrudable, gel-phase bioinks from various synthetic and natural materials (Fig. 5a). While gels typically behave as solids and do not flow, careful control of chemical cross-linking enables gel bioinks to be extruded through fine nozzles (200 μm) while maintaining their integrity (Fig. 5b). Bioinks were printed layer-by-layer to create well-defined, self-supporting structures where each layer remained stable without collapsing (Fig. 5c). PEGX-fibrinogen and PEGX-gelatin were successfully co-printed, demonstrating the ability to spatially organize multiple ECM types within a single 3D construct (Fig. 5d). Cell viability in PEGX-gelatin and PEGX-fibrinogen bioinks was qualitatively assessed using the live/dead assay, confirming the method's ability to support human dermal fibroblast (HDF) and human umbilical vein endothelial cell (HUVEC) viability after the printing process (Fig. 5e). PEGX-PEG and PEGX-gelatin were co-printed into a single construct, where HDFs selectively adhered to the gelatin struts, showing the ability to direct cell adhesion to specific areas of the scaffold (Fig. 5f).176,177
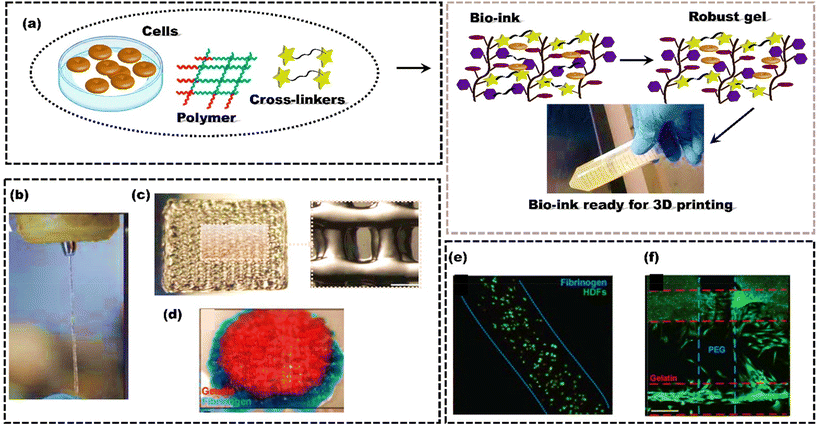 | ||
| Fig. 5 Schematic representing the formulation of bioink to form high-strength scaffolds; (a) Polymer, cells and cross-linkers are mixed to form robust gel through cross-linking; (b) extrusion through a 200 μm tip; (c) 15 × 15 mm2 printed, 4 layers; (d) PEGX-gelatin (red) and PEGX-fibrinogen (blue) co-printed cylinder, 15 mm diameter; (e) cell viability using a live/dead assay at one-day post-printing using 3 w/v% fibrinogen at a 0.2 PEG ratio, scale bars 200 μm; (f) PEGX-PEG and PEGX-gelatin co-printed structure seeded with HDFs (green, Calcein AM). Cells preferentially adhere to PEGX-gelatin, reproduced from Rutz et al.175 with permission from Wiley-VCH, copyright (2015). | ||
Polymeric bioinks derived from supramolecular assemblies are increasingly used in biomedical applications, especially for injectable systems delivering therapeutics and cells.178 Heterogeneity impacts drug release profiles and cellular interactions, emphasizing the importance of biochemical and biophysical signal scales, while controlling microstructural evolution in bioinks enhances methods for introducing structural heterogeneity. Rodell et al.179 modified hyaluronic acid (HA) with β-cyclodextrin (β-CD) or adamantane (Ad) to create CD–HA and Ad–HA, which formed supramolecular assemblies through host–guest interactions. These mechanisms resulted in pore diameters spanning three orders of magnitude and void fractions of up to 93.3 ± 2.4%. The findings enhance the understanding of polymer assembly and microstructure in supramolecular hydrogels, facilitating the development of structurally complex systems. However, network relaxations after printing prevent covalent cross-linking from producing highly durable structures. Employing dual cross-linking can mitigate this issue, resulting in improved stability and durability of the printed constructs. Ouyang et al.180 used a dual crosslinking strategy to stabilize printed HA filaments with supramolecular bonds immediately after extrusion. This initial stabilization ensured short-term stability, further strengthened by covalent crosslinking. The printed structures stayed stable for up to a month during incubation. On the other hand, HA-based hydrogel inks that used only single crosslinking, either supramolecular interactions or covalent cross-linking, lost their filament structure or became unstable after just a few layers. The dual-crosslinking approach eliminates the need for additional support materials like gelatin or alginate. Likewise, Hong et al.181 developed a tough hydrogel composed of PEG and sodium alginate, suitable for cell encapsulation. The product hydrogel of covalently cross-linked PEG and ionically cross-linked alginate possesses a high fracture toughness of 1500 J m−2 (more than the articular cartilage's toughness value182). High cell viability, ranging from 86.0 ± 3.8% to 75.5 ± 11.6%, was observed consistently throughout the 7 day culture period. The resulting hydrogel promotes the long-term viability of cell culture by facilitating nutrient diffusion and waste transport.
Ying et al.183 developed a bioink by combining poly(ethylene oxide) (PEO) and gelatin methacryloyl (GelMA) in an aqueous two-phase emulsion, resulting in bioprinted structures with well-connected pores. The stiffness of the GelMA–PEO hydrogel increased from 0.9 ± 0.3 to 1.4 ± 0.1 kPa as the PEO concentration increased from 0.5% to 1.6%. With a constant PEO concentration, increasing the GelMA-to-PEO ratio from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 4
1 to 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 raised the Young's modulus from 2.3 ± 0.4 to 9.8 ± 1.6 kPa. Live/dead assays with different mammalian cells (HepG2, HUVECs, NIH/3T3 fibroblasts) showed that the emulsion bioink and resulting porous hydrogels enhanced cell growth and spreading compared to the non-porous hydrogels. Raphael et al.184 described a new methodology for 3D bioprinting of cell-laden constructs with high geometrical definition, mechanical stability, and cell viability. The idea revolves around using a new commercial self-assembling peptide-based hydrogel. This hydrogel forms a nanofibrous bioink through the self-assembly of synthetic peptides when subjected to ionic strengths like those found in the human body. Optimizing the printing parameters enables the system to print both soft and stiff bioinks without compromising the viability of encapsulated cells. This 3D printing method allowed for stacking individual droplets of soft matter bioink to develop larger structures. It was also used to print branched alginate microvasculature.185 Similar to peptide-based hydrogels, Pluronic also has good printing properties due to shear thinning behaviour and good shear recovery, which are crucial for accurate 3D extrusion printing.186 Though Pluronic has demonstrated promising in vivo outcomes as a material for cartilage tissue engineering, in vitro studies have demonstrated that long-term cell viability is low in the presence of more than 5% Pluronic.187 Unfortunately, significant concentrations of Pluronic are needed to display gelation and the necessary rheological behaviour for extrusion printing. Therefore, the “nanostructuring” method was employed by Müller et al.188 with the aim of enhancing the long-term survival of encapsulated cells while maintaining superior printing qualities. Diacrylate Pluronic F127 (PF127-DA) was mixed with regular Pluronic F127 for 3D bioprinting of bovine chondrocytes. After printing, photocrosslinking stabilized PF127-DA, and washing out PF127 created a nanostructured network. This nanostructuring improved the long-term viability of bioprinted chondrocytes to 86%, compared to 62% for pure acrylated Pluronic hydrogel.
1 raised the Young's modulus from 2.3 ± 0.4 to 9.8 ± 1.6 kPa. Live/dead assays with different mammalian cells (HepG2, HUVECs, NIH/3T3 fibroblasts) showed that the emulsion bioink and resulting porous hydrogels enhanced cell growth and spreading compared to the non-porous hydrogels. Raphael et al.184 described a new methodology for 3D bioprinting of cell-laden constructs with high geometrical definition, mechanical stability, and cell viability. The idea revolves around using a new commercial self-assembling peptide-based hydrogel. This hydrogel forms a nanofibrous bioink through the self-assembly of synthetic peptides when subjected to ionic strengths like those found in the human body. Optimizing the printing parameters enables the system to print both soft and stiff bioinks without compromising the viability of encapsulated cells. This 3D printing method allowed for stacking individual droplets of soft matter bioink to develop larger structures. It was also used to print branched alginate microvasculature.185 Similar to peptide-based hydrogels, Pluronic also has good printing properties due to shear thinning behaviour and good shear recovery, which are crucial for accurate 3D extrusion printing.186 Though Pluronic has demonstrated promising in vivo outcomes as a material for cartilage tissue engineering, in vitro studies have demonstrated that long-term cell viability is low in the presence of more than 5% Pluronic.187 Unfortunately, significant concentrations of Pluronic are needed to display gelation and the necessary rheological behaviour for extrusion printing. Therefore, the “nanostructuring” method was employed by Müller et al.188 with the aim of enhancing the long-term survival of encapsulated cells while maintaining superior printing qualities. Diacrylate Pluronic F127 (PF127-DA) was mixed with regular Pluronic F127 for 3D bioprinting of bovine chondrocytes. After printing, photocrosslinking stabilized PF127-DA, and washing out PF127 created a nanostructured network. This nanostructuring improved the long-term viability of bioprinted chondrocytes to 86%, compared to 62% for pure acrylated Pluronic hydrogel.
Overall, the studies mentioned above highlight the benefits of composite bioinks formed by blending natural and synthetic polymers or different synthetic polymers. This approach leverages their complementary properties to overcome individual advantages and disadvantages.189 Still, there is a need to include tissue-specific cues to replicate the native ECM and biomimetic environment to support cell adhesion, proliferation, and differentiation.
5. Tissue-specific bioinks
More recently, the quest for a suitable bioink to create a conducive microenvironment for cellular activities has been emphasized. This is because conventional bioinks struggle to replicate or inherit the intrinsic cues present in a native ECM.190 Therefore, bioinks must be designed to mimic the specific properties of the target tissue, such as biocompatibility, mechanical strength, and appropriate degradation rates to replicate or inherit the intrinsic cues present in a native ECM. For example, bioinks created for skin tissue printing must imitate the unique traits of skin, such as stretchability and flexibility. Conversely, bioinks used for bone tissue printing must replicate the ability of the bone to endure mechanical stress. The ongoing progress in biomaterials science is anticipated to further refine the development of tissue-specific bioinks for 3D bioprinting.5.1 Bioinks for skin tissue engineering
Skin, the largest organ of the human body, has a multidimensional architecture with three distinct layers: the epidermis, dermis, and hypodermis.191 The epidermis acts as a barrier against pathogens and UV radiation. The dermis provides structural support, contains nerve endings for sensation, and maintains the vascular network for thermoregulation. The hypodermis is crucial for temperature regulation, serving as an insulating layer. The adipose tissue within it traps heat and helps maintain the core temperature of the body by minimizing heat loss to the environment. The interaction of these layers and their microstructures ensures the essential function of skin tissue.192 The protective function of the skin is crucial for preventing injuries and maintaining integrity. However, skin injuries or burns and damage significantly impact global healthcare spending, accounting for over half of annual expenditures.193 This emphasizes the importance of understanding and addressing the function of skin and vulnerabilities to prevent and treat skin-related health problems effectively. Skin tissue engineering offers a promising solution for treating skin injuries and disorders, using grafts from the patient or donors.194 However, traditional methods like skin grafts and flaps have limitations such as donor site morbidity and availability issues. The limitations of traditional methods in creating precise microarchitecture and replicating native skin can be overcome by 3D bioprinting.195 However, one of the main challenges in bioprinting skin substitutes is accurately depositing layers to replicate the complex structure of skin tissue.196 Thus, achieving success in this field necessitates selecting an appropriate bioink with biomaterials, cell sources, and the growth factors that support cell proliferation, differentiation, and tissue regeneration.197 The bioprinted skin grafts must replicate the composition and properties of native skin layers to encourage de novo tissue formation with functional characteristics.198 Recently, dECM has paved the way for innovative therapeutic products in skin tissue engineering and advanced biomimetic in vitro systems for skin modeling. It offers significant potential and has shown promising results for wound healing and in vitro skin modeling. dECM derived from the porcine dermis can be used to create a printable bioink containing human dermal fibroblasts. Jang et al.199 created a skin-like structure using a bioink containing skin-derived dECM, keratinocytes, and fibroblasts with 3D bioprinting technology for surgeries involving skin transplantation and extensive burns.The 3D-printed skin showed rapid re-epithelialization and superior tissue regeneration in histological and immunohistochemical analyses of the animal experiment.
Likewise, Lee et al.200 developed a soft tissue-specific bioink using decellularized porcine skin powder (PSP) (Fig. 6a). The study showed that higher PSP concentrations increased the viscosity, making the bioink suitable for 3D printing (Fig. 6b). Cell viability was approximately 75%, and there was notable preservation of elastin, suggesting that PSP-inks could be a viable option for skin dermis bioink in 3D bioprinting (Fig. 6c).
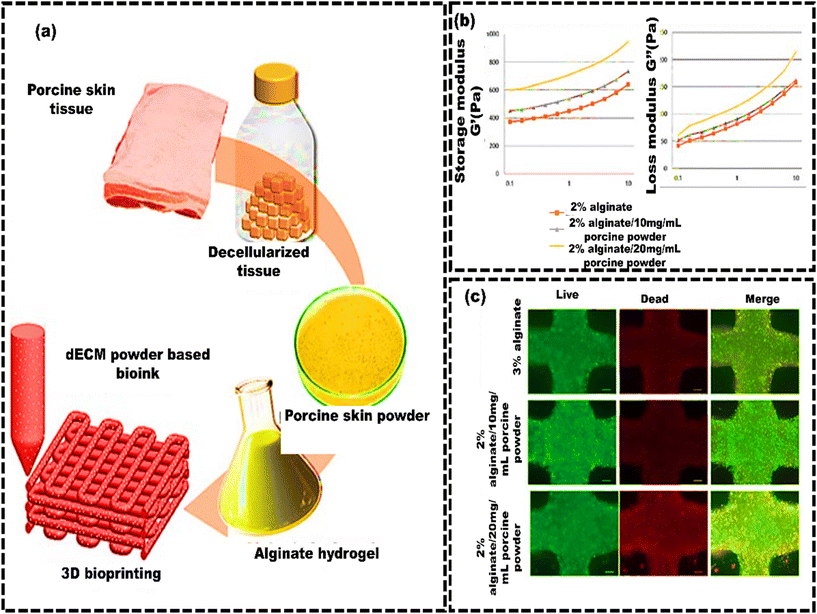 | ||
| Fig. 6 Skin decellularized bioink for soft tissue engineering. (a) Schematic of the manufacturing process of porcine skin powder (PSP)-based inks, which involves decellularized porcine skin tissue for bioprinting; (b) mechanical properties of PSP-inks: logarithmic plots of storage modulus G′ and loss modulus G′′ against frequency (0.1–10 Hz at 24 °C); (c) cellular metabolic activity and viability assessment of printed cells in PSP inks using fluorescence imaging of live/dead staining. Scale bars represent 200 μm, reproduced from Lee et al.205 with permission from MDPI, copyright (2020). | ||
Ramakrishnan et al.201 developed a multicomponent bioink system comprising gelatin, alginate, dECM, and fibrinogen for 3D printing skin tissues with dermal and epidermal histology. This bioink exhibited excellent shape fidelity, shear-thinning properties, and muscular mechanical strength. Histological studies revealed collagen-I and CK14 markers, indicating maintained cellular phenotype and functionality. The cells expressed skin-specific genes and secreted proteins, with regulated ECM deposition supporting effective tissue regeneration, making it suitable for wound healing applications. Despite these advantages, some challenges still need to be addressed that are quite complex, such as fabricating skin constructs with features like hair follicles, sweat glands, and sebaceous glands.202 Achieving the accurate colour and texture to imitate natural skin can be challenging.203 Despite challenges, skin-specific bioinks hold immense potential in skin tissue engineering, offering a promising path for advanced skin substitutes in wound healing, cosmetics, and disease modeling.204
5.2 Bioinks for bone tissue engineering
The available methods for restoring bone defects, like allografts and autografts, face several challenges.206 These include limited donor tissue supply, the risk of complications, transplant rejection, and biocontamination.207 Bone tissue engineering has gained traction as a promising approach involving replacing bone tissue with a 3D scaffold containing cells.208,209 3D scaffolds are designed to mimic bone ECM and can also contain bioactive factors to stimulate bone regeneration.210 Numerous fabrication methods for bionic 3D scaffolds have been developed, such as phase separation,211 electrospinning,212 and salt leaching, etc.213 These methods create porous matrices with controlled surface morphology and adjustable processing variables. However, they may occasionally result in less controlled structures. In contrast, 3D bioprinting uses bioink to create scaffolds with complex geometries, evenly distributing cells and releasing signaling factors in an orderly manner.214 Establishing a complex 3D structure with a personalized and enriched ECM composition in bone tissue regeneration is a fundamental initial step.215 This requires cytocompatible and osteoinductive bioink for successfully regenerating functional bone tissue.216 Synthetic self-assembling peptides, with a nanofibrous structure resembling the native ECM, are an excellent component of bioinks. Amorphous magnesium phosphates (AMPs) are synthetic self-assembling peptides that demonstrate enhanced resorption efficacy while maintaining elevated biocompatibility, osteoinductive properties, and mitigated inflammatory response.217 Dubey et al.218 developed a bioink made of ECM hydrogel and AMP encapsulated with dental pulp stem cells (DPSCs) for 3D bioprinting of craniomaxillofacial bone tissue (Fig. 7a and b). The ECM/AMP bioink showed improved osteogenic differentiation compared to the ECM bioink without requiring chemical inducers (Fig. 7c). This bioink enables the fabrication of 3D constructs with good shape fidelity and osteogenic potential, suggesting that AMP can trigger osteogenic differentiation in encapsulated DPSCs. Compared to the ECM group, the ECM/AMP formulation had 1.7 times higher bone volume at 4 weeks and 1.4 times higher at 8 weeks. Bone density in ECM/AMP increased significantly from 4 to 8 weeks (Fig. 7d). This demonstrates that AMP in the bioink significantly enhanced bone formation, supporting its potential for in situ bioprinting. Similarly, the concept of nanoengineered ionic covalent entanglement (NICE) for 3D bone bioprinting can also be used to further enhance the osteogenic differentiation of the stem cells without the need for any osteogenic medium. The NICE bioinks provide precise control over printability, mechanical properties, and degradation, allowing customized 3D biofabrication of resilient, cellularized structures. Chimene D et al.219 developed a NICE bioink to address the issues of developing a bioink compatible with cells and featuring excellent printability to repair craniomaxillofacial bone defects. The study showed that the osteoinductive bioink stimulated the production of osteo-related mineralized ECM by encapsulating hMSCs without the need for growth factors. These NICE bioinks offer a precise method to reconstruct bone structures from CT scans of actual patients. They serve as a customizable and user-friendly alternative to autografts, providing surgeons with more options for bone surgery. Although these reconstructed structures using NICE bioink provide anatomical resemblance, additional bone-specific cues are needed to imitate the complex environment of native bone tissue more effectively. Lee et al.220 developed a process to generate bioactive alginate-based bioink with dECM obtained from porcine bone. The bone derived dECM was modified through the methacrylate (Ma) reaction, referred to as Ma-dECM, to avoid the issue of low printability of the dECM. Ma-dECM was incorporated into alginate to create a composite bioink. The printability and cell viability were then assessed using human adipose-derived stem cells. The developed alginate-based cell-laden structures significantly affected the ability of human adipose-derived stem cells to differentiate into bone-forming cells when incorporated into the bioink. The composite bioink showed high in situ cell viability of more than 90%, suggesting that the polysaccharide-based biomaterial composite bioink had potential for bone tissue engineering applications. Kim et al.221 proposed a novel bioink comprising bioceramic (β-TCP), fibrillated collagen and cells to create a 3D porous cell-laden composite structure. The composite structures demonstrated good mechanical stability and substantial cell viability (∼90%) in both preosteoblasts (MC3T3-E1) and human adipose stem cells (hASCs). Furthermore, compared to the pure collagen bioink (control), the composite structure showed a considerable increase in osteogenic activity. Moreover, the hASC-loaded composite structure exhibited elevated levels of osteogenic gene expression even without an osteogenic medium, indicating that β-TCP enhanced the osteogenic differentiation of hASCs.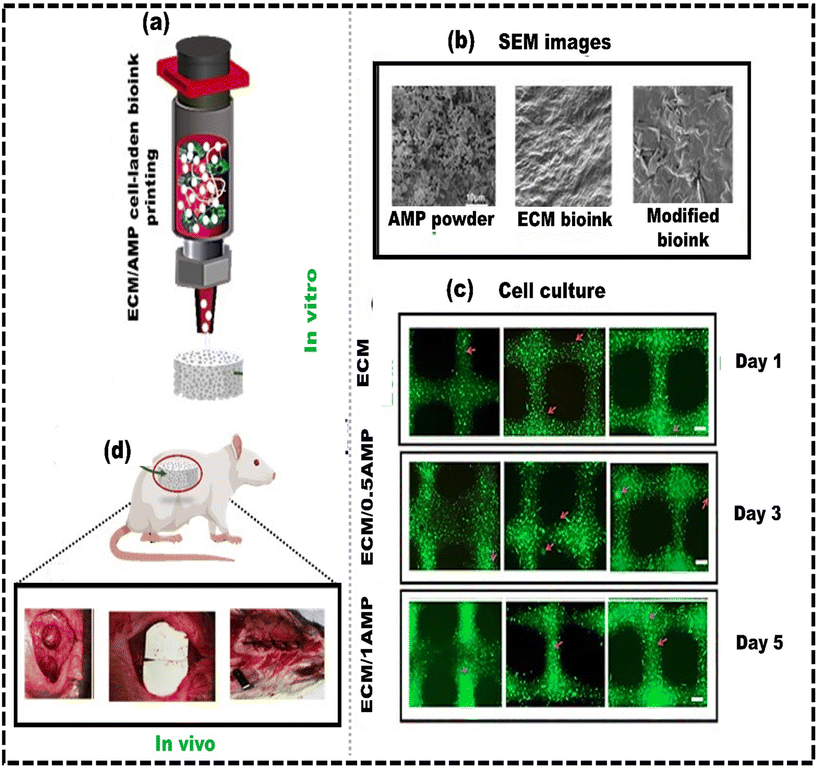 | ||
| Fig. 7 Development of a bone-specific bioink for 3D constructs with high shape fidelity and osteogenic potential: (a) 3D bioprinting of ECM/AMP cell-laden bioink; (b) SEM images of AMP powder, ECM bioink, and AMP modified bioink; (c) Calcein AM (green) and PI (red) staining assay for live and dead cell analysis showing the elongated morphology in AMP modified constructs; (d) image showing the defect after wound cleaning, implantation and suture performed in rats, reproduced from Dubey et al.218 with permission from American Chemical Society, copyright (2020). | ||
Overall, reconstructing defective bone tissue is complex, but bioprinting offers significant benefits for creating biomimetic implants. Significant progress has been made in selecting bioink components and formulating hydrogel-based bioinks containing cells and dECM. These bioinks promise to improve scaffold compatibility and mechanical properties, which are essential for successfully fabricating tissue-engineered constructs. However, bionic scaffolds do not yet possess the same level of intelligence as actual tissues.
Therefore, improving bioink formulations for bone tissue engineering requires the carefully selection and combination of materials replicating the native ECM, optimizing mechanical properties, and ensuring high cell viability and proliferation. The key strategies include incorporating a decellularized bone matrix and using hydrogels with tunable properties to enhance interactions between printed tissues, cells, and their microenvironment. Ongoing research and development on optimization in these areas is essential for advancing 3D bioprinting in bone tissue engineering applications.
5.3 Bioinks for cartilage tissue engineering
Articular cartilage is smooth white tissue with the primary role of offering a seamless, lubricated surface for movement and enabling the transfer of loads with minimal friction.222 It consists of a compact ECM with a limited number of highly specialized cells known as chondrocytes. The ECM mainly consists of water, collagen, and proteoglycans, with smaller amounts of non-collagenous proteins and glycoproteins. These components collectively retain water, which is crucial for preserving the distinctive mechanical properties of ECM. Articular cartilage lacks blood vessels, nerves, or lymphatic channels, which makes it different from other tissues. Due to this, cartilage tissue exhibits a limited capacity for self-repair to the damage caused by congenital conditions, trauma, cancer, or age-related diseases.223 Traditional cartilage repair methods (e.g., grafts, microfracture, and autologous chondrocyte implantation) have limitations like donor-site issues and tissue integration. Despite various advancements in repairing cartilage defects, the restricted spatial complexity of tissue-engineered implants, particularly in terms of cell types, materials, and active factors, has hindered the overall success of engineered cartilage.The utilization of bioprinting technology in cartilage tissue engineering holds the potential to tackle cartilage defects and regeneration methods.224 Selecting a bioink with a suitable composition and mechanical properties is essential for developing adequate cartilage substitutes. Antich et al.225 developed a composite bioink with HA and alginate, co-printed with polylactic acid (PLA), using 3D bioprinting. The hydrogel in bioink forms cross-linked gels in the presence of calcium as a crosslinker, creating a hybrid construct for AC regeneration. The HA-based bioink and 3D hybrid construct exhibited compression and shear modulus values like those in healthy human articular cartilage. Live/dead staining showed over 85% cell viability before and after printing, demonstrating effective cell preservation due to the shear-thinning properties of bioink. Genomic stability of chondrocytes, assessed by G banding, revealed no karyotype changes, confirming a typical diploid karyotype and stable chromosomal integrity post-bioprinting. However, using chemical cross-linkers can result in adverse effects like cell toxicity, structural alterations in polymers, and increased costs, emphasizing the importance of crosslinker-free alternatives.226 Singh et al.227 created a bioink without crosslinkers, which is ideal for extrusion and promotes cartilage repair. This ink combines B. mori and P. ricini silk, initiating self-gelling. Gelatin is then added to form a viscoelastic ink suitable for bioprinting. Porcine chondrocytes were encapsulated in this bioink and evaluated for printability, fidelity, biological function, and compatibility in vitro and in vivo. Optimal concentrations for cell-laden bioprinting were 5%–9% gelatin and 1%–2% silk fibroin. The bioink had the lowest viscosity between 25 and 35 °C, exhibiting gel-like properties at 4–25 °C and 35–45 °C and more liquid-like behaviour at 25–35 °C. Calcein-AM staining confirmed chondrocyte viability with a spherical morphology. In vivo testing by implanting the construct fabricated from that bioink under mouse skin showed cellular distribution within the construct, supporting its use in cartilage tissue engineering.
Rathan et al.228 developed a hybrid construct for treating focal articular cartilage defects using a 3D-printed PCL network and cartilage-ECM (cECM)-functionalized bioink and evaluated its mechanical properties and biocompatibility. The biofabricated constructs demonstrated mechanical properties that closely mimicked the native articular cartilage. The bioinks are 3D printable, support MSC viability post-printing, and promote robust chondrogenesis in vitro. Enhanced chondrogenesis in cECM-functionalized bioinks is linked to a pathway resembling natural cartilage development, as seen through increased RUNX2 expression and calcium deposition in vitro. However, single-layer scaffolds cannot replicate the depth-dependent zonal structure of native cartilage, which is crucial for articular cartilage regeneration. Kang et al.229 developed a trilayer scaffold using poly(ethylene glycol)-diacrylate-co-6-aminocaproic acid copolymers (bioink) to support the continuous differentiation of donor cells into cartilage tissue and to stimulate bone formation by recruiting endogenous cells. The trilayer scaffold consists of a biomineralized bottom layer mimicking the calcium phosphate (CaP)-rich bone microenvironment, a cryogel middle layer with anisotropic pore architecture, and a hydrogel top layer. When the trilayer scaffold was implanted in vivo, these scaffolds promoted osteochondral tissue development with a cartilage surface rich in lubricin.
Despite these accomplishments, enhancing the mechanical properties of engineered cartilage constructs is crucial because joints experience tension, shear, and compression loads during daily activities.230 So, the artificial cartilage replacements must be strong enough for appropriate joint reconstruction and capable enough to restore their properties after being exposed to changing external factors such as pH, temperature, shear stress, etc. Therefore, the research communities are more attracted to self-healing hydrogels for the formulation of bioinks to restore damaged cartilage tissues.231 Overall, the cartilage-specific bioinks for fabricating the cartilage tissue equivalents are currently in the initial stages of preclinical trials. For example, over the past five years, only a few clinical trials evaluating cartilage tissue equivalents have been registered on ClinicalTrials.gov.232 Therefore, more development is needed in bioinks designed explicitly for cartilage tissue engineering, which will likely increase the preclinical and clinical trials on cartilage-specific bioinks.
5.4 Bioinks for liver tissue engineering
The liver is pivotal in facilitating over 500 distinct biochemical processes like metabolic activities, bile synthesis, detoxification, coagulation, immunological functions, thermogenesis, the regulation of water and electrolyte levels, and so on.233 The liver houses several types of resident cells, including hepatocytes, hepatic stellate cells, liver sinusoid endothelial cells, Kupffer cells, and biliary epithelial cells. Liver conditions like acute liver failure (ALF),234 chronic liver condition (CLD),235 fibrosis, viral hepatitis,236 and cancer are causing higher mortality. Hence, the global burden of liver disease is rising, leading to around 2 million deaths annually and resulting in substantial economic costs and decreased quality of life.237 Essential surgeries like hepatectomy and liver transplants are crucial. For patients with severe liver dysfunction, transplantation is the only effective treatment.However, due to limited donors, immune rejection, and surgical complications, liver transplants face challenges in clinical practice.238 Liver tissue engineering holds promise in addressing the aforementioned challenges, and 3D bioprinting is an innovative approach to improve these procedures.239 Bioinks for liver tissue engineering are formulated using readily available biomaterials like gelatin, collagen, alginate, etc.240 However, these bioinks cannot replicate the ideal biological environment for liver tissue regeneration because they lack liver-specific biochemical components. When liver-derived material serves as a bioink, achieving uniform printing often involves enhancing it with poly(ethylene glycol) diacrylate (PEGDA) and UV cross-linking. However, UV light or radicals from the photoinitiator can unexpectedly affect cell behaviour, potentially causing toxicity in the bioink. Lee et al.241 developed a liver-dECM bioink for 3D cell printing and compared its performance with widely available commercial collagen bioink for different biomedical applications (Fig. 8a and b). The liver dECM bioink effectively preserved the essential ECM components of the liver. It demonstrated excellent printability, with minimal cell death observed during printing, indicating its non-toxic nature. The study evaluated stem cell differentiation and HepG2 cell function in the liver dECM bioink. The results showed that the liver dECM bioink promoted stem cell differentiation and improved HepG2 cell function compared to a commercial collagen bioink. However, the pepsin digestion process, employed to solubilize decellularized tissue for these conventional dECM bioink preparations, degrades the native structure and mechanical strength of the tissue.242 Furthermore, various researchers have reported that the process of digestion may denature specific biochemical constituents of dECM, such as growth factors and proteins.243 As a result, the main obstacles to the application of conventional dECM bioink in 3D bioprinting are its weak mechanical properties, inconsistent physical properties, slow crosslinking speed, and reduced biochemical components. Therefore, improving the 3D printability and mechanical properties of conventional dECM bioink is gaining tremendous attention. In this regard, Kim et al.244 created a dECM bioink with improved 3D printability and mechanical properties using dECM micro-particles (approximately 13.4 μm) prepared from decellularized porcine liver via freeze-milling. These dECM microparticles were incorporated into a gelatin mixture to develop a novel bioink named dECM powder-based bioink (dECM pBioink) (Fig. 9a). The conventional dECM bioink and the dECM pBioink both exhibited a micro-fibril network, with the conventional dECM bioink showing a more compact fibril structure than the dECM pBioink (Fig. 9b). The results demonstrated significant improvement in its printability and mechanical properties. In contrast to conventional dECM bioink, it exhibited a maximum increase in elastic modulus by 9.17 times. Micro-patterns containing viable cells were successfully fabricated with 93% cell viability (Fig. 9c and d). Moreover, the dECM pBioink exhibited better performance in layer stacking for 3D printing than the conventional bioink, which had difficulty in maintaining its shape. These advancements have led to the creation of clinically relevant liver scaffold constructs that are decellularized and recellularized with intact vascular linings, showing promising applications in regenerative medicine. However, despite these advancements, the immunogenicity of decellularized liver scaffolds made from dECM bioink is a concern due to potential antigenic molecules remaining even after cellular removal, which can trigger immune responses upon implantation, leading to scaffold rejection and failure of engineered liver tissue. To address this concern, it is essential to repopulate the parenchymal area within decellularized scaffolds with functional hepatocytes. This approach aims to create a biocompatible environment that integrates well with host tissue, reducing the risk of immune rejection. While primary hepatocytes have consistently maintained their properties, their function has initially demonstrated capabilities such as temporarily stabilizing serum ammonia levels. However, further research is needed to optimize the functionality of repopulated hepatocytes within decellularized scaffolds for achieving long-term, stable liver-specific functions.
 | ||
| Fig. 8 Diagram depicting (a) the formulation of liver dECM bioink and (b) the utilization of liver dECM bioink in 3D cell printing for liver tissue engineering, reproduced from Lee et al.241 with permission from American Chemical Society, copyright (2017). | ||
 | ||
| Fig. 9 Development of liver derived powder based dECM bioink for enhanced printability and mechanical properties: (a) porcine liver tissue is decellularized, powdered, and incorporated into a gelatin mixture to create dECM powder-based bioink (dECM pBioink) for 3D bioprinting; (b) SEM pictures of crosslinked bioinks. The presence of loaded dECM micro-particles is highlighted by white arrows (scale bar: 2 μm). (c) The degradation of conventional dECM bioink and dECM pBioink (1, 2, and 3 wt%); (d) fluorescence images depicting cell viability on day 0 and day 7, reproduced from Kim et al.244 with permission from IOP Publishing, Ltd, copyright (2020). | ||
Overall, the bioinks derived from decellularized tissues for skin, bone, cartilage, and liver tissue engineering create a biomimetic microenvironment closely resembling the native ECM of the target tissue. By incorporating tissue-specific biochemical cues and preserving the native ECM composition and structure, dECM bioinks support cell adhesion, proliferation, and differentiation. This leads to the formation of complex and heterogeneous tissue constructs that mimic the cellular organization and microenvironment of native tissues. However, controlling compositional changes during decellularization is challenging, leading to batch-to-batch variations due to the differences in tissue composition.243 Due to variations in the amounts of ECM proteins and cues present in each bioink formulation, these differences influence cell behaviour in bioprinting applications.245 To address this concern, one potential solution is to decellularize tissues in larger quantities and then blend them for use in dECM-based bioink formulations. Further investigation is required to overcome these challenges because dECM-based bioinks exhibit considerable potential as viable options for in situ bioprinting in various applications, owing to their superior biochemical properties.246Table 2 presents several kinds of bioinks that are currently employed in the construction of different tissue constructs.
| Bioink | Printing technique | Tissue | Reference |
|---|---|---|---|
| Cryo bioink | Extrusion based | Vasculature | El Assal et al.247 |
| Magnetic iron oxide nanoparticles | Hybrid nano-printing system | Vasculature networks | Yildirim Ö et al.248 |
| AgNPs in hydrogels | Extrusion-based printing | Cyborg organs/cartilage | Hassan et al.249 |
| Au NPs with thiol-modified hyaluronic acid and gelatin (AuNP-sECMs) | Extrusion-based printing | Vascular | Skardal et al.250 |
| Poly(vinyl alcohol) (PVA) | Extrusion-based 3D printing | Artificial cartilage re-constructs | Meng et al.251 |
| ECM/AMP hydrogel containing 2% octapeptide FEFEFKFK | Microvalve bioprinting | Cranio-maxillofacial bone tissue | Dubey et al.252 |
| Graphene–polyurethane composite | Conventional bio-printer | Neural tissue constructs | Huang et al.253 |
| Matrigel with alginate | Pneumatic dispensing system enabled | Human endothelial progenitor cells (EPCs) laden constructs | Poldervaart et al.254 |
| Ru/SPS with dECM | DLP bioprinting system and extrusion-based printing | Various tissues | Kim et al.255 |
| dECM-based bioink | Bio-architect 3D bioprinter | 3D primary ovarian cell-laden | Zheng et al.256 |
| GelMA with liver dECM. | DLP (digital light processing) | Liver tissue | Mao et al.257 |
| Silk-dECM construct + TGF-β encapsulated | Extrusion-based bioprinting system | Cartilage tissue | Zhang et al.258 |
| Hyaluronic acid methacrylate (HAMA) | Extrusion-based bioprinting system | 3D-printed islet organoid | Wang et al.259 |
| Hyaluronic acid (hyaluronan, HA) | Extruder and microvalve-based print | Cartilage tissue constructs | Hauptstein et al.260 |
| fibrinogen with PEG or a PEG–gelatin mixture | Thermal inkjet printing technology | Micro-vasculature networks | Cui et al.261 |
| Fibrinogen | Laser-based 3D bioprinters | 3D assembly of multi-cellular arrays | Gruene et al.111 |
| Fibrin and alginate | Inkjet bioprinter | Cartilage tissue | Nakamura et al.262 |
| Gelatin–alginate composite | Extrusion-based bioprinter | Cell-laden aortic valve conduits | Duan et al.263 |
| Collagen | Laser-based 3D bioprinters | 3D skin tissue | Koch et al.264 |
6. Computational methods for predicting bioink properties
The thrust for high-performance bioink in 3D bioprinting opens up new possibilities for designing functional tissue constructs replicating the complex geometries and properties of native tissues.265 Extensive research offers experimental support for the notion that different geometric characteristics and tunable material attributes of bioinks can greatly influence cell viability and appropriate development of the target tissue architecture. For example, in the case of drop-on-demand 3D bioprinting, loss of cell viability is impacted by shear stress.266 Cell viability is a crucial parameter of cell behaviour that can be influenced by the injection process and droplet impact during DoD printing. During the impact process, cell damage is mainly caused by von Mises stress and surface area strain.267Shear stress is a critical factor to consider for cell survival during the injection process.268 Furthermore, shear stress has a significant impact on cell biology. For example, moderate shear stress has been shown to promote stem cell differentiation,269 whereas excessive shear stress can damage the cell membrane, leading to cell rupture.270 Hence, optimizing shear stress is essential for improving cell viability and ensuring that cellular behaviours, such as proliferation and differentiation, occur correctly. However, to mitigate the issue of shear stress to some extent, microgels are emerging as a star player as a new bioink in 3D bioprinting.271
Microgels are water-based particles with micrometer-scale diameters organized similarly to hydrogels, often through dense packing or jamming.272 The unique rheological characteristics of a microgel can safeguard encapsulated cells from shear forces during the printing process.273 Due to their distinctive dynamic structure, excellent biocompatibility, and adjustable mechanical properties, microgels hold substantial potential in tissue engineering.274 However, sometimes cells areencapsulated in an asymmetrical position within the microgel, resulting in partial encapsulation and possible cell escape during gelation and subsequent culturing.275 Furthermore, incomplete encapsulation or off-center cells in a microgel expose cells unevenly to biochemical and biomechanical stimuli, which can negate the intended functions of the microgel, such as immunoprotection.276 Therefore, there is a need to overcome these challenges of shear stress during irregular cell distributions and limited cell density for the successful implementation of this 3D bioprinting in regenerative medicine and for effectively addressing organ shortages.58 Computational simulation has emerged as a valuable resource in this optimization process, effectively reducing the need for extensive experimental trials and the associated costs.277 Numerous research studies have utilized simulation and modeling in conjunction with experimental methods to improve and predict the mechanical, biological, and rheological behaviour of bioinks.278,279 One example was the coaxial printing of sodium alginate along with the crosslinking agent calcium chloride by Sun et al.280 to fabricate vascular scaffolds. Finite element simulation helped analyze bioink flow behaviour and the extrusion process, thereby optimizing the printing parameters to control cross-linking to obtain the desired cellular distribution and proliferation within the vascular scaffold. Additionally, by predicting how the crosslinking process affects the mechanical environment within the bio-printed scaffold, finite element modelling simulation enables the optimization of printing parameters, leading to improved cell viability and differentiation. Likewise, using computational fluid dynamics (CFD), a powerful tool that simulates material behaviour via parametric modifications to rheological parameters, can reduce the requirement for thorough testing and iterations. Göhl et al.281 utilized a CFD simulation tool (IPS IBOFlow) to investigate the effect of different printing parameters on a bioprinting process. IPS IBOFlow imitates the final structure of printed material and the deposition of bioink by utilizing a unique surface tension model and a viscoelastic rheology model. The ability of the simulation tool to replicate 3D bioprinting using ink with diverse viscoelastic properties indicated its potential applicability across various viscoelastic solutions and hydrogels. Choe et al.282 developed a 3D solid mechanics model to investigate the effect of shear stress on the hydrogel scaffold. First, the simulation examined the distribution of stress and deformation of composite scaffolds made from materials used for osteochondral tissue regeneration. Assessments were conducted on four PCL/polyethylene glycol diacrylate (PEGDA) and four PCL/GelMA interface scaffolds (PCL alone, 1 PCL:1 PEGDA, 2 PCL:1 PEGDA, and 3 PCL:1 PEGDA). Second, the simulation examined the distribution of stress and the deformation of several different biphasic interface scaffolds. Shear stresses were applied to these bioink scaffolds with varying moduli (high and low). The scaffolds made up of only PCL demonstrated the uniform distribution of von Mises stress across the interface and exhibited the lowest average von Mises stress values (PCL/PEGDA: 103.31 kPa; PCL/GelMA: 103.42 kPa) (Fig. 10a). Conversely, interface scaffolds with mechanical interlocking designs experienced an elevated von Mises stress at the interface, primarily due to stress concentration within the hydrogel struts forming the interface layer (Fig. 10b). von Mises stress values were highest for PCL/GelMA and PCL/PEGDA interface scaffolds (139.19 kPa and 137.78 kPa, respectively) out of all interface scaffolds with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 interlocking patterns. The 1
1 interlocking patterns. The 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PCL/GelMA and 1
1 PCL/GelMA and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PCL/PEGDA scaffolds with mechanically interlocking interfaces exhibited a notable increase in an average von Mises stress (PCL/PEGDA: 33%; PCL/GelMA: 35%) compared to their respective PCL scaffolds. In all PCL/GelMA and PCL/PEGDA scaffolds, the cartilage layer showed the highest displacement, while the bone layer exhibited the lowest displacement (Fig. 10c and d).
1 PCL/PEGDA scaffolds with mechanically interlocking interfaces exhibited a notable increase in an average von Mises stress (PCL/PEGDA: 33%; PCL/GelMA: 35%) compared to their respective PCL scaffolds. In all PCL/GelMA and PCL/PEGDA scaffolds, the cartilage layer showed the highest displacement, while the bone layer exhibited the lowest displacement (Fig. 10c and d).
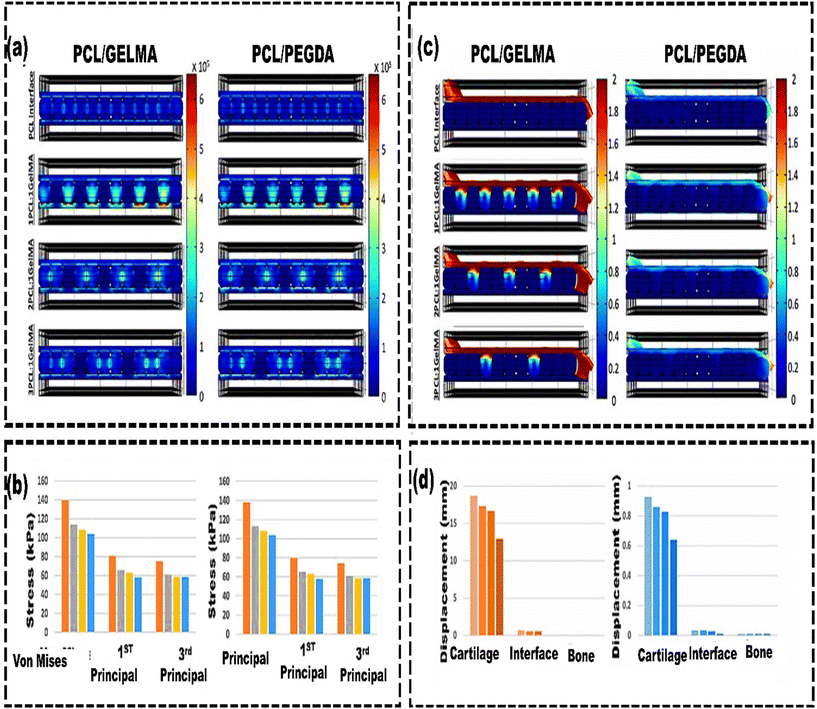 | ||
| Fig. 10 Stress and displacement distribution in various hydrogel scaffolds: (a) von Mises stress contours for PCL/GelMA and PCL/PEGDA interface scaffolds. Stress is uniform in PCL-only scaffolds. The greatest stress is at the ends of GelMA struts. (b) Comparing stresses in PCL/GelMA and PCL/PEGDA interface scaffolds. Scaffolds with mechanical interlocking patterns show higher interface stresses than PCL-only scaffolds. 1 PCL:1 GelMA and 1 PCL:1 PEGDA scaffolds have the highest von Mises and principal stresses among the four types. (c) Comparison of displacement contours: PCL/GelMA vs. PCL/PEGDA interface scaffolds. GelMA scaffolds with mechanical interlocking exhibit higher interface displacement than PEGDA, while PCL-only scaffolds show minimal displacement upon shear. (d) Comparing vertical displacement along the y-axis in the cartilage, interface, and bone layers of PCL/GelMA scaffolds. Mechanical interlocking patterns increase displacement compared to PCL-only scaffolds. 1 PCL:1 GelMA scaffolds show the highest displacements. PCL/GelMA scaffolds exhibit about ten times more displacement in cartilage and interface layers than PCL/PEGDA scaffolds, reproduced from Choe et al.282 with permission from IOP Publishing Ltd, copyright (2022). | ||
Cells subjected to mild shear stress (<5 kPa) showed excellent cell viability (up to 96%), while cells subjected to higher shear pressures (5–10 and >10 kPa) had decreased cellular vitality (91% and 76%, respectively).283 Similarly, CFD can also predict the effect of the temperature of the CELLINK Bioink (commercial bioink) on the velocity and shear stress to optimize the cell viability of the bioprinted constructs. In most bioinks, there are specific functionalities, and their properties can vary significantly with changes in temperature. Therefore, Gómez-Blancoet al.284 used CFD analysis to investigate bioink behaviour at different temperatures (15, 25, and 37 °C) and three conical tip geometries (20, 22, and 25G) as inlet parameters. The study examined the effects of these variables on shear stress, pressure, and velocity in CELLINK Bioink. The simulation results showed that pressure, velocity, and shear stress combinations were almost constant. Using this bioink at 37 °C is recommended to not only reduce shear stress but also enhance the volumetric flow. Increased volumetric flow results in higher bioprinting speeds, which in turn reduces the time for which the cells are subjected to pressure. Moreover, the shear stress predicted by simulations suggested that cell viability remained adequate at all temperatures as long as the values of shear stress were below 5 kPa. Consequently, computational modelling can effectively reduce the need for trial-and-error in 3D printing while improving the results of tissue engineering. So far, most of the studies reported in the literature are focused on optimizing the properties of bioink, probably because 3D bioprinted constructs for tissue engineering depend on the behaviour of the bioink.285 However, there are a few limitations that hinder accurate computational simulation. For example, simulations often assume conditions such as using cell-free bioinks and no interaction between the printing substrate and filaments in extrusion-based printing. These assumptions can lead to discrepancies between the simulated model and the actual 3D-printed construct.286 Despite being in the early stages, utilizing computational modelling integrated with experimental data could mitigate processing errors or facilitate real-time error management.
7. Machine learning to accelerate bioink development
Recent developments in bioprinting methods, biofabrication strategies, and innovations in materials and chemistries have significantly improved the replication of physiologically relevant tissue structures.5 This advancement facilitates incorporating multiple cell types and diverse material properties within a single structure. For bioprinting to succeed, an ideal bioink must enable efficient printing, preserve cell viability, and create an environment that supports cell proliferation and differentiation and mimics natural cellular behaviour after printing.287 The material requirements for pre- and post-printing can be drastically different. Many bioinks demonstrate excellent pre-printing properties but have insufficient post-printing performance, mainly when providing the environment for effective cell and tissue development.288 For example, structures printed with sodium alginate, Pluronic F-127, or gelatin demonstrate strong structural integrity and maintain high shape fidelity, creating filaments.289 However, sodium alginate and Pluronic F-127 do not contain cell binding sites for attachment unless they undergo further modification. On the other hand, gelatin includes binding sites for cell attachment; however, it turns into a liquid solution at 37 °C. This requires a modification to transform it into a more stable, permanent form. In situations where post-printing crosslinking is needed for lasting shape stability and mechanical strength, highly crosslinked hydrogels can reduce oxygen and nutrient diffusion, compromising the survival of encapsulated cells. Traditional computational simulations are helpful in optimizing processes, reducing the need for extensive experiments, and lowering costs. However, they often rely on oversimplified assumptions, which may not fully represent the complexity of biological materials.290 Additionally, modelling theories have limited utility because only a few can accurately predict material properties, and those that do are generally applicable only to single-component inks within a narrow concentration range.291Analytical techniques such as machine learning, a subset of artificial intelligence, and multiple regression can be used to identify and predict the complex relationships among ink formulations, ink properties, and printability.292 For example, Lee et al.293 identified a 3D-printable bioink through machine learning. The first step was selecting suitable rheological characteristics of biomaterials and then modifying them through ink composition design. Multiple regression was utilized to confirm the machine learning results and predict printability according to the composition of collagen and fibrin in the bioink. Eqn (4) presents the prediction algorithm that facilitated achieving a high storage modulus (G′) and low yield stress (τy) through multiple regression, resulting in a printable ink with high shape fidelity
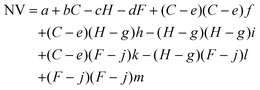 | (4) |
ML provides a systematic approach for identifying uncontrollable factors, improving the reliability of the bioprinting process, and optimizing biomaterials, bioinks, and process parameters. Various studies have shown the potential to customize bioprinted constructs and control their cell culture responses as expected.298 However, the limited size and quality of datasets for ML pose significant challenges. The costly and labor-intensive nature of data collection means that only a small dataset of adequate quality is currently available for ML applications in bioprinting, particularly for material optimization and cell performance analysis.299 Although ML is still in its infancy, integrating ML for biomaterial ink optimization provides significant advantages, making multi-component biomaterial ink potentially suitable for tissue engineering applications.300
8. Challenges and future outlook
3D bioprinting enables the accurate placement of cells and materials within constructs, making it well-suited for complex tissue engineering applications.61 However, despite considerable advancements, several unresolved issues must be addressed before 3D-printed tissues and organs can be effectively used in clinical settings. Important considerations in 3D bioprinting include minimizing cell damage, developing optimal bioinks, ensuring the quality of 3D-printed constructs, hybrid bioprinting, and navigating various regulatory aspects. These factors highlight the necessity for further advancements in the field, which are discussed in detail below.8.1. Printing-induced cell damage
The difficulty of accurately placing cells and constructing intricate structures needs to be resolved before the use of 3D bioprinted tissues in regenerative medicine.283 To date, there is no single technique used to address this issue. For instance, extrusion-based 3D printing requires a small nozzle diameter to achieve maximum cell resolution. However, it significantly increases shear stress on the printed cells and reduces cell viability.301 Although laser-based technology avoids the extrusion process and focuses on the 2D patterning of cells, its high cost and inability to construct complex 3D architectures have hindered its wider adoption. Similarly, the applications of droplet technologies to tissue creation are restricted to cartilage structures, fibro-cartilage interfaces, and bone tissues. High cell survivability and sub-100 μm droplet widths are among the benefits of inkjet bioprinters.302 Nevertheless, they also have drawbacks, such as clogging, restricted biopolymer compatibility, and very low cell density.Although many studies have investigated DOD bioprinting, most have concentrated on the droplet formation process (printability) while giving minimal attention to the post-impact viability of the printed cells.58 According to a study, shear stress during printing is not as harmful as droplet impact or substrate stiffness. There is still a need to fully understand how the droplet formation process affects the viability of cells after printing. The adverse effects of droplet impact on the substrate surface can be lessened by lowering the pressure in the printing chamber to 0.3 atm.267 Under this reduced atmospheric pressure, the released droplet spreads gently on the substrate without causing any splashing.303
8.2 Bioink development
Advancements in bioprinting are restricted by the limited availability of suitable bioinks, which need to satisfy rigorous standards for both printability and biocompatible cross-linking methods.304 The interaction between cells and their surrounding environment is vital for replicating cell–biomaterial dynamics. However, challenges such as inadequate tissue formation and limited cell–biomaterial connections remain significant concerns in bioprinted cell-encapsulated structures.305 These polymeric materials do not adequately mimic the intricate structure of natural ECMs, which is crucial for directing tissue development and replicating important cell-to-cell interactions.306 This review emphasized the importance of tissue specificity in preserving essential cellular functions and phenotypes by utilizing cells and ECMs derived from specific tissues and organs, providing detailed insights. While a dECM is a promising biomaterial, incomplete removal of cellular remnants can lead to potential pro-inflammatory responses.306 One potential solution is to recreate the unique ECM niche by arranging specific biomaterials in designated regions. This approach focuses on developing a biomimetic microenvironment that better replicates the actual conditions required for specific cell types.307 To measure its effectiveness, cytometry can be employed to analyze the physical and chemical characteristics of the suspended cells, providing insights into their health within these bioinks. By evaluating cell viability (live/dead assay using propidium iodide and Calcein-AM),308 apoptosis (by Annexin V and 7-AAD detection, prevalent in case of apoptosis),309 proliferation (Ki-67 and BrdU in actively dividing cells)310 differentiation before and after printing.311 Cytometry can ensure that the cells within the bioink remain healthy, leading to more successful and reliable bio-printed tissues and organs. This technique enables researchers to optimize bioink formulations and refine printing processes, minimizing cell stress and death.312 Cytometry also enables the development of tailored bioinks that meet the specific needs of different cell types, which is essential for creating complex, multicellular structures that closely mimic natural tissues. For instance, a 3D-printed annular ring-like scaffold was printed with a cellular mixture of human cardiac AC16 cardiomyocytes, fibroblasts, and microvascular endothelial cells.312 Moreover, integrating cytometry into the bioprinting workflow enhances the consistency and reproducibility of bio-printed products, making it easier to scale up production and meet regulatory standards.308 This is especially important for bioprinting to advance toward clinical applications, where maintaining cell health is critical for the functionality of bio-printed tissues.8.3 3D bioprinted constructs
The absence of adequate vascularization in the designed structures is among the most pertinent concerns.313 Cartilage, skin, and bladder grafts have been successfully employed in clinical practice because they can rely on the blood vessels of the host to meet their oxygen and nutrient requirements. However, vascularization takes a longer time for larger tissue grafts. The delay in delivering sufficient nutrients to cells within the large 3D scaffolds poses challenges in tissue engineering, as it leads to limitations in mass transport. This issue arises because of the restricted diffusion of oxygen and nutrients from the scaffold surface to deeper layers, which can result in cell death after a few hundred microns.314 Hence, the huge 3D printed structures get damaged without a perfusable circulatory system, which restricts the dimensions of these constructs that are clinically relevant. Therefore, several techniques, such as the creation of a vascular network by using the sacrificial template approach, have been developed to combat these vascularization concerns. For instance, Tocchio et al.315 proposed a flexible sacrificial molding technique for the creation of a robust vascular network in large, porous scaffolds. Highly resilient thermoplastic sacrificial templates made of poly(vinyl alcohol) were used to create these embedded fluidic systems. One major benefit of this technology is that it only requires filling the space surrounding the template and cross-linking the matrix to generate the vascularized scaffold. Another distinctive feature is the ability to create a perfusable matrix with regulated porosity and adjustable fluidic characteristics to create a scaffold that closely mimics the biological environment. Due to this, it may be possible to produce large-scale vascularized scaffolds, paving the way for a wide range of clinical uses.Additionally, when trying to replicate a tissue or cellular and extracellular components of an organ very precisely, the resulting tissue construct often does not function exactly like the native tissues. Therefore, innovative design approaches may be necessary to prioritize both the function and fabrication efficiency of the construct. This could involve 3D-printed constructs that efficiently support tissue function, even if they do not precisely mimic the original size or shape of the tissue. For example, constructs like sheets or patches could assist in liver or pancreas function.316 This method can significantly enhance biomanufacturing efficiency and decrease production expenses. However, it may require identifying suitable implantation sites, promoting connection with existing blood vessels, and ensuring sustained long-term integration and function. Likewise, utilizing automated designs in personalized biomanufacturing can regulate control mechanisms for predictable outcomes. 3D imaging and modelling software enable precise computational procedures, producing customized structures that align with the anatomy of a patient. This method assigns predefined hatching patterns, cell types, and materials to specific areas of the bioprinted structure using a 3D model derived from medical imaging (e.g., CT or MRI).317 A comprehensive understanding of biomaterial design, the successful integration of engineered tissue constructs with surrounding host tissue, and the biological processes involved in tissue healing and repair are crucial for meeting patient needs and advancing the clinical application of bioprinted product tissue engineering solutions.
8.4 Need for hybrid bioprinting
Despite various reports on bioprinted cell-encapsulated constructs, the primary concerns still need to improve tissue formation and limited cell–biomaterial interactions.59 Surface topography significantly affects cellular behaviour. Microscale features can impact cell shape and movement, while nanoscale textures can influence the differentiation and proliferation of stem cells.318 When it comes to producing these nanoscale structures, which are essential for controlling cellular differentiation, the bioprinting method has its limitations. To overcome these limitations, hybrid systems that combine bioprinting and electrospinning can be employed to create complex constructs with detailed nanoscale structures while accurately depositing highly viable cells in designated areas.319 For example, Graham et al.302 presented a highly accurate cell printing technique with a 1 nL droplet resolution. High-resolution 3D constructs, including a branching cell junction, a slanted junction, and a bone–cartilage interface, were printed with a resolution smaller than 200 μm. The printed constructs were cultured for a period of 35 days, demonstrating more than 90% cell viability. The study demonstrates that ovine mesenchymal stem cells can differentiate into cartilage cells and create cartilage-like structures rich in type II collagen, suggesting their promise for cartilage regeneration. Still, more improvement in scientific and technological platforms is needed to use 3D bioprinting in a hybrid manner with other printing techniques for fabricating bone, skin, liver, and cartilage constructs.320 These technologies serve as reference points for assessing the current state of bioprinting with tissue-specific bioinks. Manufacturing speed is crucial for achieving large-scale tissue and organ fabrication. Combining microvalve and extrusion-based bioprinting methods enables precise drop-on-demand cell placement while continuously extruding hydrogel filaments, allowing for the creation of large, heterogeneous 3D cell-laden structures at high throughput.788.5 Regulatory challenges
Despite the evolution of diverse manufacturing methods, the FDA currently evaluates conventionally manufactured products and 3D-printed healthcare devices using the same guidelines. However, some guidelines are specifically for 3D bioprinting, like the “21st Century Cures Act”.321 This act outlines regenerative medicine treatments that could qualify for the “regenerative medicine advanced therapy (RMAT)” designation. These treatments include tissue-engineering products, human cells and tissue products, combined therapies or products, gene therapies, and cell therapies that permanently affect human cells and tissues, often through genetic changes.322 Hence, to shape the future of tissue-specific bioink in 3D bioprinting, the FDA has provided extensive guidance to 3D printer manufacturers. Still, numerous regulatory challenges remain relevant for functional 3D-bioprinted constructs, including bioactive materials and living cells. For example, bioprinted constructs with living cells and bioactive materials pose unique regulatory challenges due to their intricate mechanisms and unknown long-term effects. Defining their potency is complex due to the multiple active components. Even minor manufacturing changes can lead to significant and unpredictable alterations in product characteristics. Hence, it is important to outline how existing laws and regulations governing device manufacturing will apply to emerging creators, like academic institutions and medical facilities producing personalized 3D-printed devices.323 Apart from this, no established standards exist for 3D bioprinting technology, materials, or the overall process. While there are some guidelines for additive manufacturing technologies (ISO/DIS 17296-1)323 and 3D printer manufacturers, there is a critical need for increased production guidelines and standardization for tissue specific bioink in 3D bioprinting.Along with regulatory constraints, establishing standards for bioprinting materials like standardized cell-expansion basal media would streamline product development.324 This standardization would also expedite clinical application and enable manufacturers to refine the processes for specific constructs, reducing the resource needs for base materials and technical processes. Logistics also become intricate when producing custom tissues or organs for individual patients. Patient-care facilities are unlikely to have 3D printing or the capacity for producing medical-grade materials. The existing cold storage, shipping methods, etc., were designed many years ago and did not meet the precise requirements for transporting living biological products.325 A novel logistic system that guarantees the necessary communication, data collection, stability, and documented compliance needs to be developed for emerging bioinks.
Overall, the roadmap of 3D bioprinting illustrates its hierarchical structure (Fig. 11).326 It starts with nanoscale molecules and biomacromolecules, advances to microscale cell manipulation, and progresses to the assembly of centimeter-scale tissue pieces. Ultimately, these tissues can be combined into decimeter-scale organs designed to replicate the essential functions of natural organs for potential implantation.
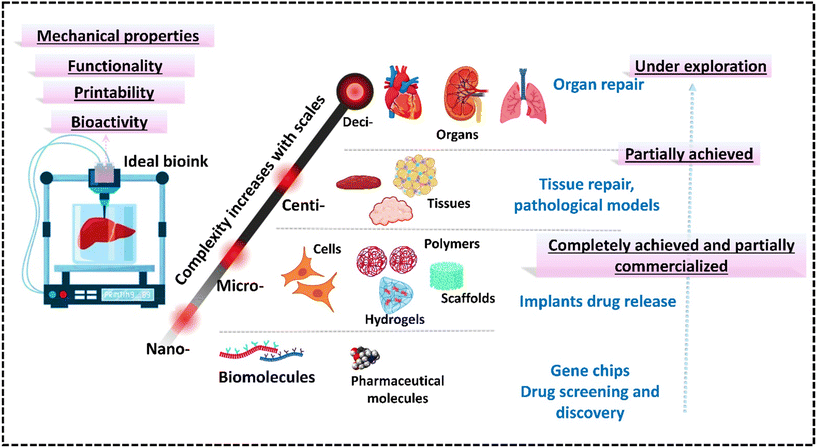 | ||
| Fig. 11 Properties of an ideal bioink and the roadmap for bioprinting. The items in the lower stages are components that contribute to the stages above them, and the complexity of assembly increases as the scale becomes larger. The right side shows typical bioprinting applications for each stage and their levels of completion, reproduced from Compaan et al.326 with permission from American Chemical Society, copyright (2017). | ||
9. Conclusion
Bioinks have attracted worldwide attention in tissue engineering because of their capability to create biomimetic, complex, and functional tissue constructs. This review explores existing and emerging bioinks and bioprinting methods. It covers up-to-date developments in fabricating tissue constructs that closely mimic native tissues, aiming to address the organ shortage crisis through advancements in 3D bioprinting. Polymeric materials are widely used as bioinks in 3D bioprinting due to their diverse properties, effectively supporting the printing process. However, these materials are insufficient for fully replicating the native ECM and biomimetic environment needed for optimal cell adhesion, proliferation, and differentiation. To enhance the performance of 3D bioprinting, the review emphasizes developing tissue-specific bioinks that incorporate cues for bone, skin, cartilage, and liver tissue engineering. Furthermore, computational modeling can substantially reduce the time and cost of experimental trial-and-error optimizing a bioink formulation. The properties of bioink before, during, and after gelation are vital for its printability, affecting shape fidelity, structural resolution, and cell survival. By utilizing computational modeling, these properties can be effectively optimized. Machine learning is also documented as a method to enhance the reliability of the bioprinting process and to optimize biomaterials, bioinks, and process parameters. This review also discussed the complex challenges of utilizing 3D bioprinted tissues in regenerative medicine, particularly the critical issue of nutrient and oxygen diffusion, which can lead to cell death. It emphasizes the importance of developing vascular networks to improve nutrient delivery and the precision needed in placing cells within complex structures. Additionally, cytometry is recognized as an effective method for monitoring cell health in bioinks, decreasing cell mortality, and boosting the viability of bioprinted structures. Moreover, it discusses the regulatory hurdles associated with the multifaceted nature of bioprinted materials and the lack of established standards in the field. Therefore, with the rise of personalized medicine, it is necessary to develop bioinks that incorporate bioactive factors specific to each patient. Additionally, new regulatory frameworks may be necessary to address the challenges of bioprinting and other advanced medical products. ‘Process-based’ frameworks could be more beneficial than traditional ‘product-based’ approaches, as they are better suited to handle product variability and customization. Hence, further research is required to overcome the current constraints of tissue-specific bioinks and develop personalized bioinks that can effectively support the growth, differentiation, and function of cells tailored to individual patients.Author contributions
All the authors listed have made a substantial, direct, and intellectual contribution to the work and approved its publication. More details on the specific contributions from each author are available in the ESI† under CRediT contributor roles.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
The authors declare no conflicts of interest in the publication of this article.Acknowledgements
This work was supported by start-up research grants (SRG/2019/001504 and SRG/2021/000859) from the Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India. The authors acknowledge the support from Faculty Initiation Grant and International Centre for Sustainable and Net Zero Technologies, PDPM-IIITDM Jabalpur and teaching assistantship fromMinistry of Education (MOE), Government of India. The authors would like to thank Prince Sultan University for their support.References
- Z. Xie, M. Gao, A. O. Lobo and T. J. Webster, Polymers, 2020, 12, 1717 CrossRef PubMed.
- S. Vijayavenkataraman, J. 3D Print. Med., 2023, 7, 3DP8 CrossRef.
- B. Giri, S. Dey, T. Das, M. Sarkar, J. Banerjee and S. K. Dash, Biomed. Pharmacother., 2018, 107, 306–328 CrossRef PubMed.
- G. M. Abouna, Transplant. Proc., 2008, 40, 34–38 CrossRef PubMed.
- S. Tripathi, S. S. Mandal, S. Bauri and P. Maiti, MedComm, 2023, 4, e194 CrossRef PubMed.
- P. Ramiah, L. C. du Toit, Y. E. Choonara, P. P. D. Kondiah and V. Pillay, Front. Mater., 2020, 7, 1–13 CrossRef.
- W. Fang, M. Yang, L. Wang, W. Li, M. Liu, Y. Jin, Y. Wang, R. Yang, Y. Wang, K. Zhang and Q. Fu, Int. J. Bioprint., 2023, 9, 759 CrossRef PubMed.
- J. Huang, J. Xiong, D. Wang, J. Zhang, L. Yang, S. Sun and Y. Liang, Gels, 2021, 7, 144 CrossRef CAS PubMed.
- N. Li, R. Guo and Z. J. Zhang, Front. Bioeng. Biotechnol., 2021, 9, 630488 CrossRef PubMed.
- M. Y. Kwon, C. Wang, J. H. Galarraga, E. Puré, L. Han and J. A. Burdick, Biomaterials, 2019, 222, 119451 CrossRef CAS PubMed.
- H. Wang, Y. Wang, C. Yuan, X. Xu, W. Zhou, Y. Huang, H. Lu, Y. Zheng, G. Luo, J. Shang and M. Sui, npj Vaccines, 2023, 8, 169 CrossRef CAS PubMed.
- K. Ceonzo, A. Gaynor, L. Shaffer, K. Kojima, C. A. Vacanti and G. L. Stahl, Tissue Eng., 2006, 12, 301–308 CrossRef CAS PubMed.
- W. Park, G. Gao and D.-W. Cho, Int. J. Mol. Sci., 2021, 22, 7837 CrossRef CAS.
- X. Zhang, X. Chen, H. Hong, R. Hu, J. Liu and C. Liu, Bioact. Mater., 2022, 10, 15–31 Search PubMed.
- M. Zhe, X. Wu, P. Yu, J. Xu, M. Liu, G. Yang, Z. Xiang, F. Xing and U. Ritz, Materials, 2023, 16, 3197 CrossRef PubMed.
- M. K. Kim, W. Jeong, S. Jeon and H. W. Kang, Front. Bioeng. Biotechnol., 2023, 11 DOI:10.3389/fbioe.2023.1305023.
- S. Chae, Y.-J. Choi and D.-W. Cho, Biofabrication, 2022, 14, 025013 CrossRef PubMed.
- J. Kim, Bioengineering, 2023, 10, 457 CrossRef PubMed.
- J. Jia, D. J. Richards, S. Pollard, Y. Tan, J. Rodriguez, R. P. Visconti, T. C. Trusk, M. J. Yost, H. Yao, R. R. Markwald and Y. Mei, Acta Biomater., 2014, 10, 4323–4331 CrossRef PubMed.
- Y. Zhao, Y. Li, S. Mao, W. Sun and R. Yao, Biofabrication, 2015, 7, 045002 CrossRef PubMed.
- Z. Xie, M. Gao, A. O. Lobo and T. J. Webster, Polymers, 2020, 12, 1717 CrossRef PubMed.
- A. S. Theus, L. Ning, B. Hwang, C. Gil, S. Chen, A. Wombwell, R. Mehta and V. Serpooshan, Polymers, 2020, 12, 1–19 CrossRef.
- Z. Wang, W. Kapadia, C. Li, F. Lin, R. F. Pereira, P. L. Granja, B. Sarmento and W. Cui, J. Controlled Release, 2021, 329, 237–256 CrossRef.
- A. M. Shabbirahmed, R. Sekar, L. A. Gomez, M. R. Sekhar, S. P. Hiruthyaswamy, N. Basavegowda and P. Somu, Biomimetics, 2023, 8, 16 CrossRef.
- J. R. Fuchs, B. A. Nasseri and J. P. Vacanti, Ann. Thorac. Surg., 2001, 72, 577–591 CrossRef PubMed.
- S. J. Forbes and N. Rosenthal, Nat. Med., 2014, 20, 857–869 CrossRef CAS PubMed.
- E. Jacques and E. J. Suuronen, Clin. Transl. Sci., 2020, 13, 440–450 CrossRef PubMed.
- M. Hospodiuk, M. Dey, D. Sosnoski and I. T. Ozbolat, Biotechnol. Adv., 2017, 35, 217–239 CrossRef PubMed.
- B. Ripley, D. Levin, T. Kelil, J. L. Hermsen, S. Kim, J. H. Maki and G. J. Wilson, J. Magn. Reson. Imaging, 2017, 45, 635–645 CrossRef PubMed.
- J. Jose, A. Peter, K. Y. Thajudeen, M. Gomes Pereira, V. P. Athira, S. G. Bhat and H. Michel, Results Eng., 2024, 22, 102060 CrossRef.
- J. Li, C. Wu, P. K. Chu and M. Gelinsky, Mater. Sci. Eng., R, 2020, 140, 100543 CrossRef.
- M. P. Sekar, H. Budharaju, A. Zennifer, S. Sethuraman, N. Vermeulen, D. Sundaramurthi and D. M. Kalaskar, J. Tissue Eng., 2021, 12, 204173142110276 Search PubMed.
- V. S. Telang, R. Pemmada, V. Thomas, S. Ramakrishna, P. Tandon and H. S. Nanda, Curr. Opin. Biomed. Eng., 2021, 17, 100264 CrossRef.
- S. V. Murphy and A. Atala, Nat. Biotechnol., 2014, 32, 773–785 CrossRef PubMed.
- A. Rossi, T. Pescara, A. M. Gambelli, F. Gaggia, A. Asthana, Q. Perrier, G. Basta, M. Moretti, N. Senin, F. Rossi, G. Orlando and R. Calafiore, Front. Bioeng. Biotechnol., 2024, 12 DOI:10.3389/fbioe.2024.1393641.
- N. Guan, Z. Liu, Y. Zhao, Q. Li and Y. Wang, Drug Delivery, 2020, 27, 1438–1451 CrossRef PubMed.
- M. A. A. Ansari, A. A. Golebiowska, M. Dash, P. Kumar, P. K. Jain, S. P. Nukavarapu, S. Ramakrishna and H. S. Nanda, Biomater. Sci., 2022, 10, 2789–2816 RSC.
- M. Kuss and B. Duan, in Biofabrication and 3D Tissue Modeling, The Royal Society of Chemistry, 2019, pp. 22–48 Search PubMed.
- S. Boularaoui, G. Al Hussein, K. A. Khan, N. Christoforou and C. Stefanini, Bioprinting, 2020, 20, e00093 CrossRef.
- C. Q. Zhao, W. G. Liu, Z. Y. Xu, J. G. Li, T. T. Huang, Y. J. Lu, H. G. Huang and J. X. Lin, Carbohydr. Polym., 2020, 236, 116058 CrossRef CAS PubMed.
- H. S. Kim, S. G. Kumbar and S. P. Nukavarapu, Curr. Opin. Biomed. Eng., 2021, 17, 100260 CrossRef CAS.
- Y. J. Tan, X. Tan, W. Y. Yeong and S. B. Tor, Sci. Rep., 2016, 6, 1–13 CrossRef.
- S. Xin, K. A. Deo, J. Dai, N. K. R. Pandian, D. Chimene, R. M. Moebius, A. Jain, A. Han, A. K. Gaharwar and D. L. Alge, Sci. Adv., 2021, 7, eabk3087 CrossRef CAS PubMed.
- X. Cui, J. Li, Y. Hartanto, M. Durham, J. Tang, H. Zhang, G. Hooper, K. Lim and T. Woodfield, Adv. Healthcare Mater., 2020, 9, 1901648 CrossRef CAS PubMed.
- P. Chansoria, R. Rizzo, D. Rütsche, H. Liu, P. Delrot and M. Zenobi-Wong, Chem. Rev., 2024, 124, 8787–8822 CrossRef CAS PubMed.
- C. Bai, X. Yang, D. Hu, P. Jiang and X. Wang, in Vat Photopolymerization Additive Manufacturing, Elsevier, 2024, pp. 439–494 Search PubMed.
- Y. Li, X. Zhang, X. Zhang, Y. Zhang and D. Hou, Polymers, 2023, 15, 3940 CrossRef CAS PubMed.
- J. Shao, Y. Liu, Y. Li, Y. Wang, R. Li, L. Yao, Y. Chen, Y. Fei, J. Wang and A. Zhao, Mater. Today Commun., 2024, 40, 109645 CrossRef CAS.
- C. E. Garciamendez-Mijares, F. J. Aguilar, P. Hernandez, X. Kuang, M. Gonzalez, V. Ortiz, R. A. Riesgo, D. S. R. Ruiz, V. A. M. Rivera, J. C. Rodriguez, F. L. Mestre, P. C. Castillo, A. Perez, L. M. Cruz, K. S. Lim and Y. S. Zhang, Appl. Phys. Rev., 2024, 11 DOI:10.1063/5.0187558.
- O. Guillaume, M. A. Geven, D. W. Grijpma, T. T. Tang, L. Qin, Y. X. Lai, H. Yuan, R. G. Richards and D. Eglin, Polym. Adv. Technol., 2017, 28, 1219–1225 CrossRef CAS.
- X. Ma, X. Qu, W. Zhu, Y.-S. Li, S. Yuan, H. Zhang, J. Liu, P. Wang, C. S. E. Lai, F. Zanella, G.-S. Feng, F. Sheikh, S. Chien and S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2206–2211 CrossRef CAS PubMed.
- U. Shaukat, E. Rossegger and S. Schlögl, Polymers, 2022, 14, 2449 CrossRef.
- W. L. Ng, J. M. Lee, M. Zhou, Y.-W. Chen, K.-X. A. Lee, W. Y. Yeong and Y.-F. Shen, Biofabrication, 2020, 12, 022001 CrossRef PubMed.
- V. Keriquel, H. Oliveira, M. Rémy, S. Ziane, S. Delmond, B. Rousseau, S. Rey, S. Catros, J. Amédée, F. Guillemot and J. C. Fricain, Sci. Rep., 2017, 7, 1–10 CrossRef.
- V. Keriquel, H. Oliveira, M. Rémy, S. Ziane, S. Delmond, B. Rousseau, S. Rey, S. Catros, J. Amédée, F. Guillemot and J.-C. Fricain, Sci. Rep., 2017, 7, 1778 CrossRef PubMed.
- M. Alonzo, S. AnilKumar, B. Roman, N. Tasnim and B. Joddar, Transl. Res., 2019, 211, 64–83 CrossRef.
- T. Salih, M. Caputo and M. T. Ghorbel, Biomolecules, 2024, 14, 861 CrossRef PubMed.
- H.-Q. Xu, J.-C. Liu, Z.-Y. Zhang and C.-X. Xu, Mil. Med. Res., 2022, 9, 70 Search PubMed.
- G. Huang, Y. Zhao, D. Chen, L. Wei, Z. Hu, J. Li, X. Zhou, B. Yang and Z. Chen, Biomater. Sci., 2024, 12, 1425–1448 RSC.
- L. Koch, M. Gruene, C. Unger and B. Chichkov, Curr. Pharm. Biotechnol., 2013, 14, 91–97 Search PubMed.
- D. Song, Y. Xu, S. Liu, L. Wen and X. Wang, Polymers, 2021, 13, 1–32 Search PubMed.
- P. S. Gungor-Ozkerim, I. Inci, Y. S. Zhang, A. Khademhosseini and M. R. Dokmeci, Biomater. Sci., 2018, 6, 915–946 RSC.
- A. Sorkio, L. Koch, L. Koivusalo, A. Deiwick, S. Miettinen, B. Chichkov and H. Skottman, Biomaterials, 2018, 171, 57–71 CrossRef.
- C. Dou, V. Perez, J. Qu, A. Tsin, B. Xu and J. Li, ChemBioEng Rev., 2021, 8, 517–534 CrossRef.
- O. Kédan, D. Hakobyan, M. Rémy, S. Ziane, N. Dusserre, J. C. Fricain, S. Delmond, N. B. Thébaud and R. Devillard, Biofabrication, 2019, 11, 045002 CrossRef.
- D. Hakobyan, C. Médina, N. Dusserre, M. L. Stachowicz, C. Handschin, J. C. Fricain, J. Guillermet-Guibert and H. Oliveira, Biofabrication, 2020, 12, 035001 CrossRef.
- B. Guillotin, A. Souquet, S. Catros, M. Duocastella, B. Pippenger, S. Bellance, R. Bareille, M. Rémy, L. Bordenave, J. Amédée and F. Guillemot, Biomaterials, 2010, 31, 7250–7256 CrossRef PubMed.
- S. Michael, H. Sorg, C. T. Peck, L. Koch, A. Deiwick, B. Chichkov, P. M. Vogt and K. Reimers, PLoS One, 2013, 8, e57741 CrossRef.
- S. Catros, J. C. Fricain, B. Guillotin, B. Pippenger, R. Bareille, M. Remy, E. Lebraud, B. Desbat, J. Amédée and F. Guillemot, Biofabrication, 2011, 3, 025001 CrossRef PubMed.
- S. Ruiz-Alonso, I. Villate-Beitia, I. Gallego, M. Lafuente-Merchan, G. Puras, L. Saenz-del-Burgo and J. L. Pedraz, Pharmaceutics, 2021, 13, 308 CrossRef PubMed.
- X. Li, B. Liu, B. Pei, J. Chen, D. Zhou, J. Peng, X. Zhang, W. Jia and T. Xu, Chem. Rev., 2020, 120, 10793–10833 CrossRef PubMed.
- W. L. Ng, H. Xi, V. Shkolnikov, G. L. Goh, R. Suntornnond and W. Y. Yeong, Int. J. Bioprint., 2021, 8, 424 CrossRef.
- V. V. Nayak, V. Sanjairaj, R. K. Behera, J. E. Smay, N. Gupta, P. G. Coelho and L. Witek, J. Biomed. Mater. Res., Part B, 2024, 112, e35402 CrossRef PubMed.
- D. Kang, J. A. Park, W. Kim, S. Kim, H. Lee, W. Kim, J. Yoo and S. Jung, Adv. Sci., 2021, 8, 2004990 CrossRef PubMed.
- W. L. Ng and V. Shkolnikov, Int. J. Bioprint., 2024, 2135 CrossRef.
- I. Angelopoulos, M. C. Allenby, M. Lim and M. Zamorano, Biotechnol. Bioeng., 2020, 117, 272–284 CrossRef PubMed.
- W. L. Ng, X. Huang, V. Shkolnikov, G. L. Goh, R. Suntornnond and W. Y. Yeong, Int. J. Bioprint., 2022, 8, 1–17 Search PubMed.
- W. L. Ng, J. M. Lee, W. Y. Yeong and M. Win Naing, Biomater. Sci., 2017, 5, 632–647 RSC.
- J. Dudman, A. M. Ferreira, P. Gentile, X. Wang and K. Dalgarno, Cells, 2021, 10, 3329 CrossRef PubMed.
- R. Nasehi, S. Aveic and H. Fischer, Int. J. Bioprint., 2023, 9, 743 CrossRef PubMed.
- N. Okubo, A. J. Qureshi, K. Dalgarno, K. L. Goh and S. Derebail, Bioprinting, 2019, 13, e00043 CrossRef.
- H. Lee, S. H. Kim, J. S. Lee, Y. J. Lee, O. J. Lee, O. Ajiteru, M. T. Sultan, S. W. Lee and C. H. Park, Adv. Healthcare Mater., 2023, 12, 2202664 CrossRef PubMed.
- L. Horváth, Y. Umehara, C. Jud, F. Blank, A. Petri-Fink and B. Rothen-Rutishauser, Sci. Rep., 2015, 5, 7974 CrossRef PubMed.
- S. Rahmani Dabbagh, M. Rezapour Sarabi, M. T. Birtek, N. Mustafaoglu, Y. S. Zhang and S. Tasoglu, Aggregate, 2023, 4, e197 CrossRef.
- X. Zhu, Q. Xu, H. Li, M. Liu, Z. Li, K. Yang, J. Zhao, L. Qian, Z. Peng, G. Zhang, J. Yang, F. Wang, D. Li and H. Lan, Adv. Mater., 2019, 31, 1902479 CrossRef PubMed.
- Z. Gu, J. Fu, H. Lin and Y. He, Asian J. Pharm. Sci., 2020, 15, 529–557 CrossRef PubMed.
- J. He, B. Zhang, Z. Li, M. Mao, J. Li, K. Han and D. Li, Biofabrication, 2020, 12, 042002 CrossRef CAS PubMed.
- T. G. Papaioannou, D. Manolesou, E. Dimakakos, G. Tsoucalas, M. Vavuranakis and D. Tousoulis, Acta Cardiol. Sin., 2019, 35, 284–289 Search PubMed.
- M. Kuzucu, G. Vera, M. Beaumont, S. Fischer, P. Wei, V. P. Shastri and A. Forget, ACS Biomater. Sci. Eng., 2021, 7, 2192–2197 CrossRef CAS PubMed.
- L. Gasperini, D. Maniglio, A. Motta and C. Migliaresi, Tissue Eng., Part C, 2015, 21, 123–132 CrossRef CAS.
- Q. Gao, P. Zhao, R. Zhou, P. Wang, J. Fu and Y. He, Bio-Des. Manuf., 2019, 2, 1–9 CrossRef CAS.
- B. Brenken, E. Barocio, A. Favaloro, V. Kunc and R. B. Pipes, Addit. Manuf., 2018, 21, 1–16 CAS.
- J. Vajda, M. Milojević, U. Maver and B. Vihar, Biomedicines, 2021, 9, 1–27 CrossRef PubMed.
- H. Liang, J. He, J. Chang, B. Zhang and D. Li, Int. J. Bioprint., 2018, 4, 1–8 CAS.
- E. Altun, N. Ekren, S. E. Kuruca and O. Gunduz, Mater. Lett., 2019, 234, 163–167 CrossRef.
- S. Vijayavenkataraman, S. Zhang, W. F. Lu and J. Y. H. Fuh, J. Mater. Res., 2018, 33, 1999–2011 CrossRef.
- J. He, F. Xu, R. Dong, B. Guo and D. Li, Biofabrication, 2017, 9, 015007 CrossRef PubMed.
- Y. E. Choe and G. H. Kim, Virtual Phys. Prototyp., 2020, 15, 403–416 CrossRef.
- Z. Qiu, H. Zhu, Y. Wang, A. Kasimu, D. Li and J. He, Bio-Des. Manuf., 2023, 6, 136–149 CrossRef.
- N. Mkhize and H. Bhaskaran, Small Sci., 2022, 2, 2100073 CrossRef.
- A. Reizabal, B. Tandon, S. Lanceros-Méndez and P. D. Dalton, Small, 2023, 19, 2205255 CrossRef PubMed.
- M. Mao, H. Liang, J. He, A. Kasimu, Y. Zhang, L. Wang, X. Li and D. Li, Int. J. Bioprint., 1970, 7, 362 CrossRef PubMed.
- G. Gillispie, P. Prim, J. Copus, J. Fisher, A. G. Mikos, J. J. Yoo, A. Atala and S. J. Lee, Biofabrication, 2020, 12, 022003 CrossRef PubMed.
- M. Petretta, G. Desando, B. Grigolo and L. Roseti, Materials, 2021, 14, 3528 CrossRef.
- L. Tytgat, L. Van Damme, M. Ortega Arevalo, H. Declercq, H. Thienpont, H. Otteveare, P. Blondeel, P. Dubruel and S. Van Vlierberghe, Int. J. Biol. Macromol., 2019, 140, 929–938 CrossRef.
- Y. Wang, X. Yuan, B. Yao, S. Zhu, P. Zhu and S. Huang, Bioact. Mater., 2022, 17, 178–194 Search PubMed.
- N. Khoshnood and A. Zamanian, Bioprinting, 2020, 19, e00088 CrossRef.
- S. Agarwal, S. Saha, V. K. Balla, A. Pal, A. Barui and S. Bodhak, Front. Mech. Eng., 2020, 6, 589171 CrossRef.
- R. D. Ventura, Med. Lasers, 2021, 10, 76–81 CrossRef.
- M. Gruene, M. Pflaum, A. Deiwick, L. Koch, S. Schlie, C. Unger, M. Wilhelmi, A. Haverich and B. N. Chichkov, Biofabrication, 2011, 3, 015005 CrossRef.
- M. Gruene, M. Pflaum, C. Hess, S. Diamantouros, S. Schlie, A. Deiwick, L. Koch, M. Wilhelmi, S. Jockenhoevel, A. Haverich and B. Chichkov, Tissue Eng., Part C, 2011, 17, 973–982 CrossRef PubMed.
- R. Gaebel, N. Ma, J. Liu, J. Guan, L. Koch, C. Klopsch, M. Gruene, A. Toelk, W. Wang, P. Mark, F. Wang, B. Chichkov, W. Li and G. Steinhoff, Biomaterials, 2011, 32, 9218–9230 CrossRef PubMed.
- C. Cong, X. Li, W. Xiao, J. Li, M. Jin, S. H. Kim and P. Zhang, Nanotechnol. Rev., 2022, 11, 3305–3334 CrossRef.
- Y. Wu, Acta Biomater., 2021, 128, 21–41 CrossRef.
- S. H. Ahn, H. J. Lee and G. H. Kim, Biomacromolecules, 2011, 12, 4256–4263 CrossRef.
- T. Jungst, I. Pennings, M. Schmitz, A. J. W. P. Rosenberg, J. Groll and D. Gawlitta, Adv. Funct. Mater., 2019, 29, 1905987 CrossRef.
- B. Kong, Y. Chen, R. Liu, X. Liu, C. Liu, Z. Shao, L. Xiong, X. Liu, W. Sun and S. Mi, Nat. Commun., 2020, 11, 1435 CrossRef.
- J. L. Li, Y. L. Cai, Y. L. Guo, J. Y. H. Fuh, J. Sun, G. S. Hong, R. N. Lam, Y. S. Wong, W. Wang, B. Y. Tay and E. S. Thian, J. Biomed. Mater. Res., Part B, 2014, 102, 651–658 CrossRef PubMed.
- M. Mao, J. He, Z. Li, K. Han and D. Li, Acta Biomater., 2020, 101, 141–151 CrossRef.
- Y. Liu, T. Huang, N. A. Yap, K. Lim and L. A. Ju, Bioact. Mater., 2024, 42, 328–344 CAS.
- M. Abellan Lopez, L. Hutter, E. Pagin, M. Vélier, J. Véran, L. Giraudo, C. Dumoulin, L. Arnaud, N. Macagno, R. Appay, L. Daniel, B. Guillet, L. Balasse, H. Caso, D. Casanova, B. Bertrand, F. Dignat, L. Hermant, H. Riesterer, F. Guillemot, F. Sabatier and J. Magalon, Front. Bioeng. Biotechnol., 2023, 11, 1217655 CrossRef.
- S. A. Oluwole, W. D. Weldu, K. Jayaraman, K. A. Barnard and C. Agatemor, 2024Article number: 4c00537, ACS Appl. Bio Mater., 2024, 4c00537, DOI:10.1021/acsabm.4c00537.
- R. Byrne, A. Carrico, M. Lettieri, A. K. Rajan, R. J. Forster and L. R. Cumba, Mater. Today Bio, 2024, 28, 101185 CrossRef PubMed.
- S. Naghieh and X. Chen, J. Pharm. Anal., 2021, 11, 564–579 CrossRef PubMed.
- A. Schwab, R. Levato, M. D'Este, S. Piluso, D. Eglin and J. Malda, Chem. Rev., 2020, 120, 11028–11055 CrossRef PubMed.
- W. L. Ng and V. Shkolnikov, Bio-Des. Manuf., 2024, 7, 771–799 CrossRef.
- T. D. Ngo, A. Kashani, G. Imbalzano, K. T. Q. Nguyen and D. Hui, Composites, Part B, 2018, 143, 172–196 CrossRef.
- A. Habib, R. Sarah, S. Tuladhar, B. Khoda and S. M. Limon, Bioprinting, 2024, 38, e00332 CrossRef.
- J. Park and D. Kim, Adv. Healthcare Mater., 2024, 2304496, DOI:10.1002/adhm.202304496.
- H. Liu, Y. Gong, K. Zhang, S. Ke, Y. Wang, J. Wang and H. Wang, Gels, 2023, 9, 195 CrossRef PubMed.
- D. Gogoi, M. Kumar and J. Singh, Ann. 3D Print. Med., 2024, 15, 100159 CrossRef.
- W. Liu, M. A. Heinrich, Y. Zhou, A. Akpek, N. Hu, X. Liu, X. Guan, Z. Zhong, X. Jin, A. Khademhosseini and Y. S. Zhang, Adv. Healthcare Mater., 2017, 6, 1601451 CrossRef PubMed.
- S. H. Moon, T. Y. Park, H. J. Cha and Y. J. Yang, Mater. Today Bio, 2024, 25, 100973 CrossRef CAS PubMed.
- M. A. Habib and B. Khoda, J. Manuf. Process., 2022, 76, 708–718 CrossRef.
- D. Venkata Krishna and M. Ravi Sankar, Eng. Regen., 2022, 76, 708–718 Search PubMed.
- X. B. Chen, A. Fazel Anvari-Yazdi, X. Duan, A. Zimmerling, R. Gharraei, N. K. Sharma, S. Sweilem and L. Ning, Bioact. Mater., 2023, 28, 511–536 CAS.
- S. Tavakoli, N. Krishnan, H. Mokhtari, O. P. Oommen and O. P. Varghese, Adv. Funct. Mater., 2024, 34, 2307040 CrossRef.
- X. Li, F. Zheng, X. Wang, X. Geng, S. Zhao, H. Liu, D. Dou, Y. Leng, L. Wang and Y. Fan, Int. J. Bioprint., 2022, 9, 649 CrossRef.
- Z. Ahmad, S. Salman, S. A. Khan, A. Amin, Z. U. Rahman, Y. O. Al-Ghamdi, K. Akhtar, E. M. Bakhsh and S. B. Khan, Gels, 2022, 8, 167 CrossRef.
- K. K. Moncal, V. Ozbolat, P. Datta, D. N. Heo and I. T. Ozbolat, J. Mater. Sci. Mater. Med., 2019, 30, 55 CrossRef.
- F. You, B. F. Eames and X. Chen, Int. J. Mol. Sci., 2017, 18, 1597 CrossRef PubMed.
- M. Hovakimyan, R. F. Guthoff and O. Stachs, J. Ophthalmol., 2012, 2012, 1–12 CrossRef PubMed.
- Y. Yu, K. K. Moncal, J. Li, W. Peng, I. Rivero, J. A. Martin and I. T. Ozbolat, Sci. Rep., 2016, 6, 28714 CrossRef PubMed.
- R. Sridharan, A. R. Cameron, D. J. Kelly, C. J. Kearney and F. J. O'Brien, Mater. Today, 2015, 18, 313–325 CrossRef.
- G. H. McKinley and M. Renardy, Phys. Fluids, 2011, 23, 127101 CrossRef.
- B. Derby, Annu. Rev. Mater. Res., 2010, 40, 395–414 CrossRef.
- T. C. de Goede, N. Laan, K. G. de Bruin and D. Bonn, Langmuir, 2018, 34, 5163–5168 CrossRef.
- J. Eggers and E. Villermaux, Rep. Prog. Phys., 2008, 71, 036601 CrossRef.
- D. Jang, D. Kim and J. Moon, Langmuir, 2009, 25, 2629–2635 CrossRef.
- H. Wee, C. R. Anthony and O. A. Basaran, Phys. Rev. Fluids., 2022, 7, L112001 CrossRef.
- A. Goswami and Y. Hardalupas, J. Fluid Mech., 2023, 961, A17 CrossRef.
- Z. Lu, W. Gao, F. Liu, J. Cui, S. Feng, C. Liang, Y. Guo, Z. Wang, Z. Mao and B. Zhang, Addit. Manuf., 2024, 94, 104443 Search PubMed.
- S. K. Paral, D.-Z. Lin, Y.-L. Cheng, S.-C. Lin and J.-Y. Jeng, Polymers, 2023, 15, 2716 CrossRef PubMed.
- T. Aziz, F. Haq, A. Farid, L. Cheng, L. F. Chuah, A. Bokhari, M. Mubashir, D. Y. Y. Tang and P. L. Show, Carbon Lett., 2024, 34, 477–494 CrossRef.
- R. H. Bean, G. Nayyar, M. K. Brown, J. Wen, Y. Fu, K. I. Winey, C. B. Williams and T. E. Long, Addit. Manuf., 2024, 92, 104391 Search PubMed.
- W. Li, L. S. Mille, J. A. Robledo, T. Uribe, V. Huerta and Y. S. Zhang, 2020Volume: 9Article number: 2000156, Adv. Healthcare Mater., 2020, 9, 2000156 CrossRef.
- M. Shah, A. Ullah, K. Azher, A. U. Rehman, W. Juan, N. Aktürk, C. S. Tüfekci and M. U. Salamci, RSC Adv., 2023, 13, 1456–1496 RSC.
- C. D. O'Connell, C. Onofrillo, S. Duchi, X. Li, Y. Zhang, P. Tian, L. Lu, A. Trengove, A. Quigley, S. Gambhir, A. Khansari, T. Mladenovska, A. O'Connor, C. Di Bella, P. F. Choong and G. G. Wallace, Biofabrication, 2019, 11, 035003 CrossRef.
- A. Machado, I. Pereira, F. Costa, A. Brandão, J. E. Pereira, A. C. Maurício, J. D. Santos, I. Amaro, R. Falacho, R. Coelho, N. Cruz and M. Gama, Clin. Oral Investig., 2023, 27, 979–994 CrossRef PubMed.
- A. Bernhardt, M. Wehrl, B. Paul, T. Hochmuth, M. Schumacher, K. Schütz and M. Gelinsky, PLoS One, 2015, 10, e0129205 CrossRef.
- M. Rizwan, S. W. Chan, P. A. Comeau, T. L. Willett and E. K. F. Yim, Biomed. Mater., 2020, 15, 065017 CrossRef PubMed.
- E. Hodder, S. Duin, D. Kilian, T. Ahlfeld, J. Seidel, C. Nachtigall, P. Bush, D. Covill, M. Gelinsky and A. Lode, J. Mater. Sci. Mater. Med., 2019, 30, 10 CrossRef.
- Y. Gu, A. Forget and V. P. Shastri, Adv. Sci., 2022, 9, 2103469 CrossRef.
- M. Gomez-Florit, A. Pardo, R. M. A. Domingues, A. L. Graça, P. S. Babo, R. L. Reis and M. E. Gomes, Molecules, 2020, 25, 5858 CrossRef.
- E. O. Osidak, V. I. Kozhukhov, M. S. Osidak and S. P. Domogatsky, Int. J. Bioprint., 2024, 6, 270 CrossRef PubMed.
- Y. S. Kim and F. Guilak, Int. J. Mol. Sci., 2022, 23, 8662 CrossRef PubMed.
- I. Lukin, I. Erezuma, L. Maeso, J. Zarate, M. F. Desimone, T. H. Al-Tel, A. Dolatshahi-Pirouz and G. Orive, Pharmaceutics, 2022, 14, 1–19 CrossRef PubMed.
- A. Fatimi, O. V. Okoro, D. Podstawczyk, J. Siminska-Stanny and A. Shavandi, Gels, 2022, 8, 179 CrossRef PubMed.
- S. Vanaei, M. S. Parizi, S. Vanaei, F. Salemizadehparizi and H. R. Vanaei, Eng. Regen., 2021, 2, 1–18 Search PubMed.
- C. I. Idumah, Int. J. Polym. Mater. Polym. Biomater., 2024, 1–30 Search PubMed.
- T. Insperger and G. Stépán, Appl. Math. Sci., 2011, 178, 93–149 Search PubMed.
- F. Liu and X. Wang, Polymers, 2020, 12, 1765 CrossRef PubMed.
- M. Gharibshahian, M. Salehi, N. Beheshtizadeh, M. Kamalabadi-Farahani, A. Atashi, M.-S. Nourbakhsh and M. Alizadeh, Front. Bioeng. Biotechnol., 2023, 11, 1168504 CrossRef PubMed.
- A. Schwab, R. Levato, M. D'Este, S. Piluso, D. Eglin and J. Malda, Chem. Rev., 2020, 120, 11028–11055 CrossRef PubMed.
- A. L. Rutz, K. E. Hyland, A. E. Jakus, W. R. Burghardt and R. N. Shah, Adv. Mater., 2015, 27, 1607–1614 CrossRef PubMed.
- B. A. G. de Melo, Y. A. Jodat, E. M. Cruz, J. C. Benincasa, S. R. Shin and M. A. Porcionatto, Acta Biomater., 2020, 117, 60–76 CrossRef PubMed.
- H. Rastin, R. T. Ormsby, G. J. Atkins and D. Losic, ACS Appl. Bio Mater., 2020, 3, 1815–1826 CrossRef PubMed.
- D. Petta, U. D'Amora, L. Ambrosio, D. W. Grijpma, D. Eglin and M. D'Este, Biofabrication, 2020, 12, 032001 CrossRef PubMed.
- C. B. Rodell, C. B. Highley, M. H. Chen, N. N. Dusaj, C. Wang, L. Han and J. A. Burdick, Soft Matter, 2016, 12, 7839–7847 RSC.
- L. Ouyang, C. B. Highley, C. B. Rodell, W. Sun and J. A. Burdick, ACS Biomater. Sci. Eng., 2016, 2, 1743–1751 CrossRef PubMed.
- S. Hong, D. Sycks, H. F. Chan, S. Lin, G. P. Lopez, F. Guilak, K. W. Leong and X. Zhao, Adv. Mater., 2015, 27, 4035–4040 CrossRef PubMed.
- N. K. Simha, C. S. Carlson and J. L. Lewis, J. Mater. Sci. Mater. Med., 2004, 15, 631–639 CrossRef PubMed.
- G. Ying, N. Jiang, S. Maharjan, Y. Yin, R. Chai, X. Cao, J. Yang, A. K. Miri, S. Hassan and Y. S. Zhang, Adv. Mater., 2018, 30, 1805460 CrossRef PubMed.
- B. Raphael, T. Khalil, V. L. Workman, A. Smith, C. P. Brown, C. Streuli, A. Saiani and M. Domingos, Mater. Lett., 2017, 190, 103–106 CrossRef.
- K. Pataky, T. Braschler, A. Negro, P. Renaud, M. P. Lutolf and J. Brugger, Adv. Mater., 2012, 24, 391–396 CrossRef PubMed.
- P. A. Amorim, M. A. d’Ávila, R. Anand, P. Moldenaers, P. Van Puyvelde and V. Bloemen, Bioprinting, 2021, 22, e00129 CrossRef.
- N. Cui, C.-Y. Dai, X. Mao, X. Lv, Y. Gu, E.-S. Lee, H.-B. Jiang and Y. Sun, Gels, 2022, 8, 360 CrossRef PubMed.
- M. Müller, J. Becher, M. Schnabelrauch and M. Zenobi-Wong, Biofabrication, 2015, 7, 35006 CrossRef PubMed.
- P. Ramiah, L. C. du Toit, Y. E. Choonara, P. P. D. Kondiah and V. Pillay, Front. Mater., 2020, 7, 00076 CrossRef.
- B. S. Kim, H. Kim, G. Gao, J. Jang and D.-W. Cho, Biofabrication, 2017, 9, 034104 CrossRef PubMed.
- S. P. Tarassoli, Z. M. Jessop, A. Al-Sabah, N. Gao, S. Whitaker, S. Doak and I. S. Whitaker, J. Plast. Reconstr. Aesthet. Surg., 2018, 71, 615–623 CrossRef PubMed.
- G. K. Menon and A. M. Kligman, Skin Pharmacol. Physiol., 2009, 22, 178–189 CrossRef CAS PubMed.
- N. Bhardwaj, D. Chouhan and B. B. Mandal, Curr. Pharm. Des., 2017, 23, 3455–3482 CrossRef CAS PubMed.
- F. P. W. Mamsen, C. H. Kiilerich, J. Hesselfeldt-Nielsen, I. Saltvig, C. L.-N. Remvig, H. Trøstrup and V.-J. Schmidt, J. Pers. Med., 2022, 12, 2067 CrossRef PubMed.
- T. Hodgkinson and A. Bayat, Arch. Dermatol. Res., 2011, 303, 301–315 CrossRef CAS.
- M. Sheikholeslam, M. E. E. Wright, M. G. Jeschke and S. Amini-Nik, Adv. Healthcare Mater., 2018, 7, 1700897 CrossRef PubMed.
- K. Vig, A. Chaudhari, S. Tripathi, S. Dixit, R. Sahu, S. Pillai, V. A. Dennis and S. R. Singh, Int. J. Mol. Sci., 2017, 18, 789 CrossRef PubMed.
- B. S. Kim, Y. W. Kwon, J. S. Kong, G. T. Park, G. Gao, W. Han, M. B. Kim, H. Lee, J. H. Kim and D. W. Cho, Biomaterials, 2018, 168, 38–53 CrossRef.
- K.-S. Jang, S.-J. Park, J.-J. Choi, H.-N. Kim, K.-M. Shim, M.-J. Kim, I.-H. Jang, S.-W. Jin, S.-S. Kang, S.-E. Kim and S.-H. Moon, Materials, 2021, 14, 5177 CrossRef.
- S. J. Lee, J. H. Lee, J. Park, W. D. Kim and S. A. Park, Materials, 2020, 13, 3522 CrossRef.
- R. Ramakrishnan, N. Kasoju, R. Raju, R. Geevarghese, A. Gauthaman and A. Bhatt, Carbohydr. Polym. Technol. Appl., 2022, 3, 100184 Search PubMed.
- W. L. Ng, S. Wang, W. Y. Yeong and M. W. Naing, Trends Biotechnol., 2016, 34, 689–699 CrossRef PubMed.
- R. D. Abbott and D. L. Kaplan, Trends Biotechnol., 2015, 33, 401–407 CrossRef PubMed.
- J. Zhang, S. Yun, A. Karami, B. Jing, A. Zannettino, Y. Du and H. Zhang, Bioprinting, 2020, 19, e00089 CrossRef.
- S. J. Lee, J. H. Lee, J. Park, W. D. Kim and S. A. Park, Materials, 2020, 13, 1–9 Search PubMed.
- S. van de Vijfeijken, T. Münker, R. Spijker, L. Karssemakers, W. Vandertop, A. Becking, D. Ubbink, A. Becking, L. Dubois, L. Karssemakers, D. Milstein, S. van de Vijfeijken, P. Depauw, F. Hoefnagels, W. Vandertop, C. Kleverlaan, T. Münker, T. Maal, E. Nout, M. Riool and S. Zaat, World Neurosurg., 2018, 117, 443–452 CrossRef PubMed.
- L. Vidal, C. Kampleitner, M. Á. Brennan, A. Hoornaert and P. Layrolle, Front. Bioeng. Biotechnol., 2020, 8, 00061 CrossRef PubMed.
- H. Qu, H. Fu, Z. Han and Y. Sun, RSC Adv., 2019, 9, 26252–26262 RSC.
- B. Dalisson, B. Charbonnier, A. Aoude, M. Gilardino, E. Harvey, N. Makhoul and J. Barralet, Acta Biomater., 2021, 136, 37–55 CrossRef CAS PubMed.
- P. Baldwin, D. J. Li, D. A. Auston, H. S. Mir, R. S. Yoon and K. J. Koval, J. Orthop. Traumatol., 2019, 33, 203–213 Search PubMed.
- E. Capuana, F. Lopresti, F. Carfì Pavia, V. Brucato and V. La Carrubba, Polymers, 2021, 13, 2041 CrossRef CAS PubMed.
- S. Anjum, F. Rahman, P. Pandey, D. K. Arya, M. Alam, P. S. Rajinikanth and Q. Ao, Int. J. Mol. Sci., 2022, 23, 9206 CrossRef CAS PubMed.
- D. Y. Kwon, J. Y. Park, B. Y. Lee and M. S. Kim, Polymers, 2020, 12, 2210 CrossRef CAS PubMed.
- M. Kamaraj, G. Sreevani, G. Prabusankar and S. N. Rath, Mater. Sci. Eng., C, 2021, 131, 112478 CrossRef CAS.
- C. Li and W. Cui, Eng. Regen., 2021, 2, 195–205 Search PubMed.
- Q. Yan, H. Dong, J. Su, J. Han, B. Song, Q. Wei and Y. Shi, Engineering, 2018, 4, 729–742 CrossRef CAS.
- M. Nabiyouni, T. Brückner, H. Zhou, U. Gbureck and S. B. Bhaduri, Acta Biomater., 2018, 66, 23–43 CrossRef CAS PubMed.
- N. Dubey, J. A. Ferreira, J. Malda, S. B. Bhaduri and M. C. Bottino, ACS Appl. Mater. Interfaces, 2020, 12, 23752–23763 CrossRef.
- D. Chimene, L. Miller, L. M. Cross, M. K. Jaiswal, I. Singh and A. K. Gaharwar, ACS Appl. Mater. Interfaces, 2020, 12, 15976–15988 CrossRef PubMed.
- J. Lee, J. Hong, W. J. Kim and G. H. Kim, Carbohydr. Polym., 2020, 250, 116914 CrossRef PubMed.
- W. Kim and G. Kim, Biofabrication, 2019, 0–31 Search PubMed.
- A. J. Sophia Fox, A. Bedi and S. A. Rodeo, Sports Health, 2009, 1, 461–468 CrossRef.
- T. Stampoultzis, P. Karami and D. P. Pioletti, Curr. Res. Transl. Med., 2021, 69, 103299 Search PubMed.
- X. Yang, Z. Lu, H. Wu, W. Li, L. Zheng and J. Zhao, Mater. Sci. Eng., C, 2018, 83, 195–201 CrossRef.
- C. Antich, J. de Vicente, G. Jiménez, C. Chocarro, E. Carrillo, E. Montañez, P. Gálvez-Martín and J. A. Marchal, Acta Biomater., 2020, 106, 114–123 CrossRef PubMed.
- L. Roseti, C. Cavallo, G. Desando, V. Parisi, M. Petretta, I. Bartolotti and B. Grigolo, Materials, 2018, 11, 1749 CrossRef PubMed.
- Y. P. Singh, A. Bandyopadhyay and B. B. Mandal, ACS Appl. Mater. Interfaces, 2019, 11, 33684–33696 CrossRef.
- S. Rathan, L. Dejob, R. Schipani, B. Haffner, M. E. Möbius and D. J. Kelly, Adv. Healthcare Mater., 2019, 8, 1–11 Search PubMed.
- H. Kang, Y. Zeng and S. Varghese, Acta Biomater., 2018, 78, 365–377 CrossRef PubMed.
- C. J. Little, N. K. Bawolin and X. Chen, Tissue Eng., Part B, 2011, 17, 213–227 CrossRef PubMed.
- V. K. A Devi, R. Shyam, A. Palaniappan, A. K. Jaiswal, T.-H. Oh and A. J. Nathanael, Polymers, 2021, 13, 3782 CrossRef PubMed.
- Z. Zhang and L. Schon, Cartilage, 2022, 13, 194760352210930 Search PubMed.
- Y. Cen, G. Lou, J. Qi, M. Zheng and Y. Liu, Cell Commun. Signaling, 2023, 21, 214 CrossRef PubMed.
- Y.-M. Wang, K. Li, X.-G. Dou, H. Bai, X.-P. Zhao, X. Ma, L.-J. Li, Z.-S. Chen and Y.-C. Huang, in Acute Exacerbation of Chronic Hepatitis B, Springer Netherlands, Dordrecht, 2019, pp. 273–370 Search PubMed.
- P. A. Cortesi, S. Conti, L. Scalone, A. Jaffe, A. Ciaccio, S. Okolicsanyi, M. Rota, L. Fabris, M. Colledan, S. Fagiuoli, L. S. Belli, G. Cesana, M. Strazzabosco and L. G. Mantovani, Liver Int., 2020, 40, 2630–2642 CrossRef PubMed.
- H. B. El-Serag, Gastroenterology, 2012, 142, 1264–1273 CrossRef PubMed.
- D. (Danielle) Huang, S. B. Gibeley, C. Xu, Y. Xiao, O. Celik, H. N. Ginsberg and K. W. Leong, Adv. Funct. Mater., 2020, 30, 1909553 CrossRef PubMed.
- N. Badwei, World J. Transplant., 2024, 14, 96637 Search PubMed.
- Q. Wang, Y. Feng, A. Wang, Y. Hu, Y. Cao, J. Zheng, Y. Le and J. Liu, iLIVER, 2024, 3, 100080 CrossRef.
- C. Benwood, J. Chrenek, R. L. Kirsch, N. Z. Masri, H. Richards, K. Teetzen and S. M. Willerth, Bioengineering, 2021, 8, 1–19 CrossRef PubMed.
- H. Lee, W. Han, H. Kim, D. H. Ha, J. Jang, B. S. Kim and D. W. Cho, Biomacromolecules, 2017, 18, 1229–1237 CrossRef CAS PubMed.
- F. Al-Hakim Khalak, F. García-Villén, S. Ruiz-Alonso, J. L. Pedraz and L. Saenz-del-Burgo, Int. J. Mol. Sci., 2022, 23, 12930 CrossRef CAS PubMed.
- M. K. Kim, W. Jeong and H.-W. Kang, J. Funct. Biomater., 2023, 14, 417 CrossRef CAS PubMed.
- M. K. Kim, W. Jeong, S. M. Lee, J. B. Kim, S. Jin and H. W. Kang, Biofabrication, 2020, 12, 025003 CrossRef CAS PubMed.
- A. Abaci and M. Guvendiren, Adv. Healthcare Mater., 2020, 9, 2000734 CrossRef CAS.
- N. Ashammakhi, S. Ahadian, C. Xu, H. Montazerian, H. Ko, R. Nasiri, N. Barros and A. Khademhosseini, Mater. Today Bio, 2019, 1, 100008 CrossRef CAS PubMed.
- R. El Assal, S. Guven, U. A. Gurkan, I. Gozen, H. Shafiee, S. Dalbeyler, N. Abdalla, G. Thomas, W. Fuld, B. M. W. Illigens, J. Estanislau, J. Khoory, R. Kaufman, C. Zylberberg, N. Lindeman, Q. Wen, I. Ghiran and U. Demirci, Adv. Mater., 2014, 26, 5815–5822 CrossRef CAS PubMed.
- Ö. Yildirim and A. Arslan-Yildiz, Biomater. Sci., 2022, 10, 6707–6717 RSC.
- R. U. Hassan, S. M. Khalil, S. A. Khan, S. Ali, J. Moon, D.-H. Cho and D. Byun, Polymers, 2022, 14, 4373 CrossRef PubMed.
- A. Skardal, J. Aleman, S. Forsythe, S. Rajan, S. Murphy, M. Devarasetty, N. Pourhabibi Zarandi, G. Nzou, R. Wicks, H. Sadri-Ardekani, C. Bishop, S. Soker, A. Hall, T. Shupe and A. Atala, Biofabrication, 2020, 12, 025017 CrossRef PubMed.
- Y. Meng, J. Cao, Y. Chen, Y. Yu and L. Ye, J. Mater. Chem. B, 2020, 8, 677–690 RSC.
- N. Dubey, J. A. Ferreira, J. Malda, S. B. Bhaduri and M. C. Bottino, ACS Appl. Mater. Interfaces, 2020, 12, 23752–23763 CrossRef PubMed.
- C.-T. Huang, L. Kumar Shrestha, K. Ariga and S. Hsu, J. Mater. Chem. B, 2017, 5, 8854–8864 RSC.
- M. T. Poldervaart, H. Gremmels, K. van Deventer, J. O. Fledderus, F. C. Öner, M. C. Verhaar, W. J. A. Dhert and J. Alblas, J. Controlled Release, 2014, 184, 58–66 CrossRef PubMed.
- H. Kim, B. Kang, X. Cui, S. Lee, K. Lee, D. Cho, W. Hwang, T. B. F. Woodfield, K. S. Lim and J. Jang, Adv. Funct. Mater., 2021, 31, 2011252 CrossRef.
- J. Zheng, Y. Liu, C. Hou, Z. Li, S. Yang, X. Liang, L. Zhou, J. Guo, J. Zhang and X. Huang, Int. J. Bioprint., 2022, 8, 597 CrossRef PubMed.
- Q. Mao, Y. Wang, Y. Li, S. Juengpanich, W. Li, M. Chen, J. Yin, J. Fu and X. Cai, Mater. Sci. Eng., C, 2020, 109, 110625 CrossRef PubMed.
- X. Zhang, Y. Liu, C. Luo, C. Zhai, Z. Li, Y. Zhang, T. Yuan, S. Dong, J. Zhang and W. Fan, Mater. Sci. Eng., C, 2021, 118, 111388 CrossRef PubMed.
- D. Wang, Y. Guo, J. Zhu, F. Liu, Y. Xue, Y. Huang, B. Zhu, D. Wu, H. Pan, T. Gong, Y. Lu, Y. Yang and Z. Wang, Acta Biomater., 2023, 165, 86–101 CrossRef PubMed.
- J. Hauptstein, T. Böck, M. Bartolf-Kopp, L. Forster, P. Stahlhut, A. Nadernezhad, G. Blahetek, A. Zernecke-Madsen, R. Detsch, T. Jüngst, J. Groll, J. Teßmar and T. Blunk, Adv. Healthcare Mater., 2020, 9, 2000737 CrossRef.
- X. Cui and T. Boland, Biomaterials, 2009, 30, 6221–6227 CrossRef PubMed.
- M. Nakamura, S. Iwanaga, C. Henmi, K. Arai and Y. Nishiyama, Biofabrication, 2010, 2, 014110 CrossRef PubMed.
- B. Duan, L. A. Hockaday, K. H. Kang and J. T. Butcher, J. Biomed. Mater. Res., Part A, 2013, 101A, 1255–1264 CrossRef PubMed.
- L. Koch, A. Deiwick, S. Schlie, S. Michael, M. Gruene, V. Coger, D. Zychlinski, A. Schambach, K. Reimers, P. M. Vogt and B. Chichkov, Biotechnol. Bioeng., 2012, 109, 1855–1863 CrossRef PubMed.
- S. Ramesh, O. L. A. Harrysson, P. K. Rao, A. Tamayol, D. R. Cormier, Y. Zhang and I. V. Rivero, Bioprinting, 2021, 21, e00116 CrossRef.
- J. Shi, B. Wu, S. Li, J. Song, B. Song and W. F. Lu, Biomed. Phys. Eng. Express, 2018, 4, 045028 CrossRef.
- P. He, Y. Liu and R. Qiao, Microfluid. Nanofluid., 2015, 18, 569–585 CrossRef.
- K. H. K. Wong, J. M. Chan, R. D. Kamm and J. Tien, Annu. Rev. Biomed. Eng., 2012, 14, 205–230 CrossRef PubMed.
- K. M. Kim, Y. J. Choi, J.-H. Hwang, A. R. Kim, H. J. Cho, E. S. Hwang, J. Y. Park, S.-H. Lee and J.-H. Hong, PLoS One, 2014, 9, e92427 CrossRef PubMed.
- Y. Koo and G. Kim, Biofabrication, 2016, 8, 025010 CrossRef PubMed.
- R. Levato, T. Jungst, R. G. Scheuring, T. Blunk, J. Groll and J. Malda, Adv. Mater., 2020, 32, 1906423 CrossRef PubMed.
- C. Du, W. Huang and Y. Lei, Int. J. Bioprint., 2023, 9, 753 CrossRef PubMed.
- M. Sreepadmanabh, A. B. Arun and T. Bhattacharjee, Biophys. Rev., 2024, 5, 021304 CrossRef PubMed.
- K. Wang, Z. Wang, H. Hu and C. Gao, Supramol. Mater., 2022, 1, 100006 Search PubMed.
- T. Kamperman, S. Henke, C. W. Visser, M. Karperien and J. Leijten, Small, 2017, 13, 1603711 CrossRef PubMed.
- L. S. Moreira Teixeira, J. Feijen, C. A. van Blitterswijk, P. J. Dijkstra and M. Karperien, Biomaterials, 2012, 33, 1281–1290 CrossRef PubMed.
- X. Lin, X. Li and X. Lin, Molecules, 2020, 25, 1375 CrossRef PubMed.
- A. Neagu, J. 3D Print. Med., 2017, 1, 103–121 CrossRef.
- R. Naranjo-Alcazar, S. Bendix, T. Groth and G. Gallego Ferrer, Gels, 2023, 9, 230 CrossRef PubMed.
- J. Sun, Y. Gong, M. Xu, H. Chen, H. Shao and R. Zhou, Micromachines, 2024, 15, 463 CrossRef PubMed.
- J. Göhl, K. Markstedt, A. Mark, K. Håkansson, P. Gatenholm and F. Edelvik, Biofabrication, 2018, 10, 034105 CrossRef PubMed.
- R. Choe, E. Devoy, B. Kuzemchak, M. Sherry, E. Jabari, J. D. Packer and J. P. Fisher, Biofabrication, 2022, 14, 025015 CrossRef PubMed.
- A. Blaeser, D. F. Duarte Campos, U. Puster, W. Richtering, M. M. Stevens and H. Fischer, Adv. Healthcare Mater., 2016, 5, 326–333 CrossRef PubMed.
- J. C. Gómez-Blanco, E. Mancha-Sánchez, A. C. Marcos, M. Matamoros, A. Díaz-Parralejo and J. B. Pagador, Processes, 2020, 8, 865 CrossRef.
- M. Nooranidoost, D. Izbassarov, S. Tasoglu and M. Muradoglu, Phys. Fluids, 2019, 31, 081901 CrossRef.
- J. C. Gómez-Blanco, J. B. Pagador, V. P. Galván-Chacón, L. F. Sánchez-Peralta, M. Matamoros, A. Marcos and F. M. Sánchez-Margallo, Int. J. Bioprint., 2023, 9, 730 CrossRef PubMed.
- G. Decante, J. B. Costa, J. Silva-Correia, M. N. Collins, R. L. Reis and J. M. Oliveira, Biofabrication, 2021, 13, 032001 CrossRef PubMed.
- J. Karvinen and M. Kellomäki, Bioprinting, 2023, 32, e00274 CrossRef.
- T. A. Mappa, C.-M. Liu, C.-C. Tseng, M. Ruslin, J.-H. Cheng, W.-C. Lan, B.-H. Huang, Y.-C. Cho, C.-C. Hsieh, H.-H. Kuo, C.-H. Tsou and Y.-K. Shen, Polymers, 2023, 15, 3223 CrossRef PubMed.
- S. Freeman, S. Calabro, R. Williams, S. Jin and K. Ye, Front. Bioeng. Biotechnol., 2022, 10 DOI:10.3389/fbioe.2022.913579.
- C. Bailey, S. Stoyanov, H. Lu, T. Tilford, C. Yin and N. Strusevich, in Nanopackaging, Springer International Publishing, Cham, 2018, pp. 45–82 Search PubMed.
- C. Yu and J. Jiang, Int. J. Bioprint., 2020, 6, 253 CrossRef PubMed.
- J. Lee, S. J. Oh, S. H. An, W.-D. Kim and S.-H. Kim, Biofabrication, 2020, 12, 035018 CrossRef PubMed.
- S. Tian, R. Stevens, B. McInnes and N. Lewinski, Micromachines, 2021, 12, 780 CrossRef PubMed.
- H. Zhang and S. K. Moon, ACS Appl. Mater. Interfaces, 2021, 13, 53323–53345 CrossRef PubMed.
- P. I. Frazier and J. Wang, 2016, Bayesian Optimization for Materials Design, pp. 45–75 Search PubMed.
- A. Hashemi, M. Ezati, I. Zumberg, T. Vicar, L. Chmelikova, V. Cmiel and V. Provaznik, Mater. Today Commun., 2024, 40, 109777 CrossRef.
- J. Sun, K. Yao, J. An, L. Jing, K. Huang and D. Huang, Int. J. Bioprint., 2024, 9, 717 CrossRef PubMed.
- L. Jin, X. Zhai, K. Wang, K. Zhang, D. Wu, A. Nazir, J. Jiang and W.-H. Liao, Mater. Des., 2024, 244, 113086 CrossRef.
- K. S. Kaswan, J. S. Dhatterwal, R. Batra, B. Balusamy and E. Gangadevi, in Computational Intelligence in Bioprinting, Wiley, 2024, pp. 303–321 Search PubMed.
- R. Chang, J. Nam and W. Sun, Tissue Eng., Part A, 2008, 14, 41–48 Search PubMed.
- A. D. Graham, S. N. Olof, M. J. Burke, J. P. K. Armstrong, E. A. Mikhailova, J. G. Nicholson, S. J. Box, F. G. Szele, A. W. Perriman and H. Bayley, Sci. Rep., 2017, 7, 7004 CrossRef PubMed.
- Y. Yonemoto, K. Tashiro, K. Shimizu and T. Kunugi, Sci. Rep., 2022, 12, 5093 CrossRef PubMed.
- J. Adhikari, A. Roy, A. Das, M. Ghosh, S. Thomas, A. Sinha, J. Kim and P. Saha, Macromol. Biosci., 2021, 21, e2000179 CrossRef PubMed.
- H. Mao, L. Yang, H. Zhu, L. Wu, P. Ji, J. Yang and Z. Gu, Prog. Nat. Sci.: Mater. Int., 2020, 30, 618–634 CrossRef.
- Z. Chen, C. Du, S. Liu, J. Liu, Y. Yang, L. Dong, W. Zhao, W. Huang and Y. Lei, Giant, 2024, 19, 100323 CrossRef.
- A. A. Golebiowska, J. T. Intravaia, V. M. Sathe, S. G. Kumbar and S. P. Nukavarapu, Bioact. Mater., 2024, 32, 98–123 Search PubMed.
- D. E. Godar, C. Gurunathan and I. Ilev, Photochem. Photobiol., 2019, 95, 581–586 CrossRef PubMed.
- M. de Villiers and L. H. Du Plessis, Biomed. Mater., 2023, 18, 045031 CrossRef PubMed.
- X. Sun and P. D. Kaufman, Chromosoma, 2018, 127, 175–186 CrossRef CAS PubMed.
- B. Parisi, M. Sünnen, R. Chippalkatti and D. K. Abankwa, STAR Protoc., 2023, 4, 102637 CrossRef CAS PubMed.
- M. Alonzo, R. El Khoury, N. Nagiah, V. Thakur, M. Chattopadhyay and B. Joddar, ACS Appl. Mater. Interfaces, 2022, 14, 21800–21813 CrossRef CAS PubMed.
- K. Zheng, M. Chai, B. Luo, K. Cheng, Z. Wang, N. Li and X. Shi, Smart Mater. Med., 2024, 5, 183–195 CrossRef.
- D. P. Forrestal, T. J. Klein and M. A. Woodruff, Biotechnol. Bioeng., 2017, 114, 1129–1139 CrossRef CAS PubMed.
- A. Tocchio, M. Tamplenizza, F. Martello, I. Gerges, E. Rossi, S. Argentiere, S. Rodighiero, W. Zhao, P. Milani and C. Lenardi, Biomaterials, 2015, 45, 124–131 CrossRef CAS PubMed.
- Y. Kim, Y. W. Kim, S. B. Lee, K. Kang, S. Yoon, D. Choi, S.-H. Park and J. Jeong, Biomaterials, 2021, 274, 120899 CrossRef CAS PubMed.
- C. Kengla, E. Renteria, C. Wivell, A. Atala, J. J. Yoo and S. J. Lee, 3D Print. Addit. Manuf., 2017, 4, 239–247 CrossRef.
- L. Xiao, Y. Sun, L. Liao and X. Su, J. Mater. Chem. B, 2023, 11, 2550–2567 RSC.
- K. Loukelis, Z. A. Helal, A. G. Mikos and M. Chatzinikolaidou, Gels, 2023, 9, 103 CrossRef CAS PubMed.
- S. V. Murphy, P. De Coppi and A. Atala, Nat. Biomed. Eng., 2019, 4, 370–380 CrossRef PubMed.
- S. Bonamici, H.R.34–21st Century Cures Act, 2016 Search PubMed.
- V. Mansouri, N. Beheshtizadeh, M. Gharibshahian, L. Sabouri, M. Varzandeh and N. Rezaei, Biomed. Pharmacother., 2021, 141, 111875 CrossRef PubMed.
- Guidance for Industry and Food and Drug Administration Staff, Technical Considerations for Additive Manufactured Medical Devices.
- J. G. Hunsberger, S. Goel, J. Allickson and A. Atala, Curr. Stem Cell Rep., 2017, 3, 77–82 CrossRef.
- R. Genç, Tohoku J. Exp. Med., 2008, 216, 287–296 CrossRef PubMed.
- A. M. Compaan, K. Christensen and Y. Huang, ACS Biomater. Sci. Eng., 2017, 3, 1519–1526 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm01192a |
| This journal is © The Royal Society of Chemistry 2025 |









