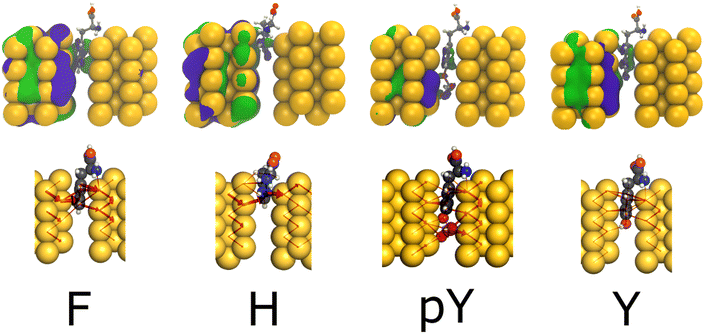Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Detection and distinction of amino acids and post-translational modifications with gold nanojunctions†
Rodrigo G.
Amorim
 *a,
Filipe C. D. A.
Lima
*a,
Filipe C. D. A.
Lima
 b,
Fábio Arthur
Leão de Souza
b,
Fábio Arthur
Leão de Souza
 c,
Wanderlã L.
Scopel
c,
Wanderlã L.
Scopel
 d,
Jariyanee
Prasongkit
d,
Jariyanee
Prasongkit
 e and
Ralph H.
Scheicher
e and
Ralph H.
Scheicher
 *f
*f
aDepartamento de Física, ICEx, Universidade Federal Fluminense - UFF, Volta Redonda, RJ, Brazil. E-mail: rgamorim@id.uff.br
bFederal Institute of Education, Science and Techonology of São Paulo, Matão, SP, Brazil
cInstituto Federal de Educação, Ciência e Tecnologia do Espírito Santo - IFES, Ibatiba, ES, Brazil. E-mail: fabio.souza@ifes.edu.br
dDepartamento de Física, Universidade Federal do Espirito Santo-UFES, Vitória, ES, Brazil. E-mail: wanderla.scopel@ufes.br
eDivision of Physics, Faculty of Science, Nakhon Phanom University, Nakhon Phanom 48000, Thailand. E-mail: jariyanee.prasongkit@npu.ac.th
fDivision of Materials Theory, Department of Physics and Astronomy, Uppsala University, Sweden. E-mail: ralph.scheicher@physics.uu.se
First published on 17th January 2025
Abstract
Amino acids are fundamental building blocks of proteins, playing critical roles in medical diagnostics, environmental monitoring, and biomarker identification. The development of nanoscale electronic sensors capable of single-amino-acid recognition has gained significant attention due to their potential for label-free, real-time detection. In this study, we investigate the electronic transport properties of amino acids in two gold-based nanodevices with distinct architectures: a gold nanojunction and a gold-capacitor system. Using density functional theory (DFT) in combination with nonequilibrium Green's function (NEGF) calculations, we explore the sensing mechanism and conductance variations induced by different amino acids, including select phosphorylated variants. Each device was assigned a reference amino acid, F (M), for a capacitor (nanojunction) to differentiate its conductance from other molecules. Our results reveal distinct conductance that enables amino acid classification based on their electronic signatures, demonstrating that molecular discrimination is primarily governed by conductance variations as a function of the binding energy differences. The nanojunction exhibited remarkable differentiation for the amino acids S, pS, Y, and pY, rendering it especially proficient in distinguishing between structurally analogous molecules. These findings highlight the strong potential of gold-based nanodevices for precise amino acid detection, paving the way for advancements in biosensing technologies, molecular diagnostics, and biomedical applications.
1. Introduction
Amino acids, the building blocks of proteins, are crucial in a variety of fields, such as medical diagnosis, environmental monitoring, and food safety.1 Recent research has demonstrated the potential of amino acid recognition-based electronic nanodevices for biosensing and bioelectronics applications.1–3The field of single-molecule protein sequencing is rapidly advancing with the development of new nanodevices.2–6 Various different techniques6 have been used to explore this issue, for example: recognition tunneling, biological nanopores, single-molecule fluorosequencing, mass spectroscopy, anti-body detection and solid-state biosensors. Some notable examples include the study by Reed et al.,4 which demonstrated a dynamic approach using dye-labeled N-terminal amino acid recognizers, aminopeptidases to discriminate single amino acid substitutions, and post-translational modifications with integrated semiconductor chips.
Brinkerhoff et al.5 presented a nanopore-based proteomics tool capable of sequencing and identifying proteins and single amino acid substitutions, accurately discriminating different variants with average accuracy of 87% and the ability to “rewind” the reading. Aksimentiev et al.7,8 proposed a computational method called the steric exclusion model (SEM) of nanopore conductance to predict ionic currents produced by different sequences. This approach combines the computational efficiency of a finite element solver with the atomic precision of a nanopore conductance map, carried out by molecular dynamics calculations. Ohshiro et al.9 presented a novel method utilizing electron tunneling (ET) currents to detect post-translational modifications in single peptides. Their study successfully identified 12 distinct amino acids and phosphotyrosine sites. Additionally, the authors demonstrated the capability to partially sequence peptides derived from an epidermal growth factor receptor substrate, distinguishing between a peptide and its phosphorylated variant. Similarly, Zhao et al.10 introduced recognition tunneling (RT) as an innovative approach for identifying individual peptides. The RT method involves confining molecules between two electrodes coated with recognition molecules and subsequently measuring the ET current across the junction. The research findings showcase RT's capacity to discriminate between various binding motifs, as well as identify D and L enantiomers, a methylated amino acid, isobaric isomers, and short peptides. These outcomes suggest the possibility of achieving direct electronic sequencing of single proteins by sequential measurement of products from processive exopeptidase digestion, or by employing a molecular motor to guide proteins through a nanopore-integrated tunnel junction. Both studies underscore the potential of ET-based methods in the identification and characterization of individual amino acids and peptides. The research by Zhao et al.10 further encompasses the ability to differentiate between diverse binding motifs. These advancements in single-protein analysis techniques provide valuable insights for biomarker identification and the realization of real-time diagnostic capabilities.
The identification of individual peptides and amino acids using ET currents and RT has been presented with notable progress. In this context, computational modeling is a key ingredient for a deeper understanding of the sensing mechanism at the single-molecule level. As an example, diamondoid-functionalized gold electrodes have been employed to sense and sequence DNA11–13 showing a high-resolution sensitivity. The investigation of 2D materials with nanopores for identifying single biomolecules was extensively studied using different materials.14–17 In addition, there are some advances in nanomaterials and bioelectronics enhancing amino acid sensing through high-performance sensors and data-driven approaches.18
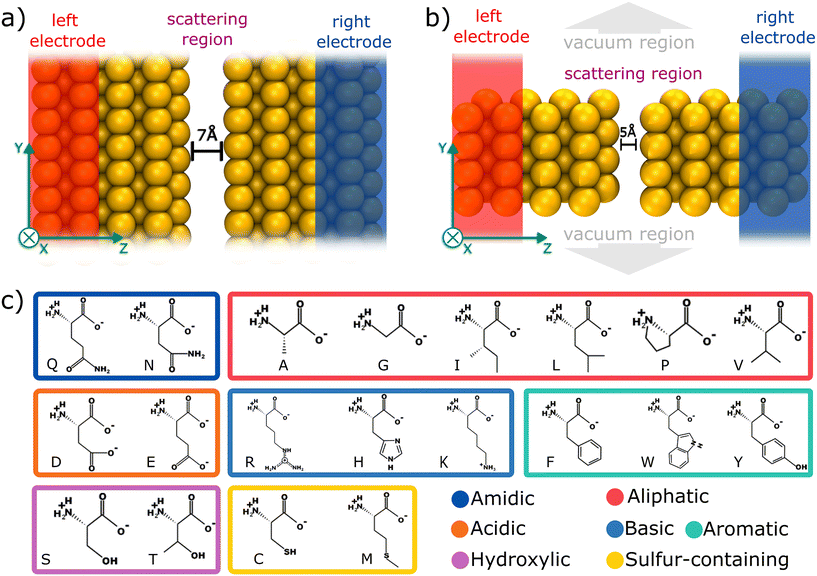
Here, we have employed computational modeling based on density functional theory (DFT), in combination with quantum electronic transport calculations, to investigate the electric sensing mechanism for amino acid identification. For this purpose, we have explored two different devices based on gold electrodes: (i) a gold infinite capacitor with a 7.0 Å gap; and (ii) a gold nanojunction constituting a 5.0 Å gap. These devices were inspired by the experimental configuration suggested by Ohshiro et al.,9 in which the amino acids translocate vertically through the junction. For each nanodevice, 20 individual amino acids and three post-translational modifications were evaluated. By considering the electronic transport properties and the strength of interaction of the molecules with the device, one can discriminate groups of amino acids with similar conductance features. For individual molecule electrical discrimination, we considered one reference amino acid to calculate the relative sensitivity. Our investigation demonstrates that the 5.0 Å gap nanojunction device is more effective in terms of amino acid discrimination than the 7.0 Å gap infinite capacitor due to the higher signal and sensitivity of the former.
2. Methodology
Electronic structure calculations were performed using DFT as implemented in the SIESTA code.19,20 Norm-conserving Troullier–Martin pseudopotentials21 and the Perdew–Burke–Ernzerhof generalized gradient approximation (PBE–GGA)22 for exchange–correlation potential was used, with a double-zeta polarized (DZP) basis set.23 Here, van der Waals corrections are not considered because the molecule-device distances are significantly less than 3.4 Å,24 making the interaction predominantly governed by strong electrode-molecule coupling rather than long-range dispersion forces.Real-space and k-point meshes were set to a cutoff energy of 200 Ry and (3 × 1 × 2) k-points in Monkhorst–Pack scheme,25 respectively. Structural relaxations were carried out with a convergence threshold for all atomic component forces as 0.01 eV Å−1. Given the confined space inside the junction and the resulting constriction in the degrees of freedom, multiple relaxation attempts with different initial orientations consistently converged to similar local minima, indicating that the structural configuration is primarily dictated by the strong interactions within the nanojunction.
For the electronic transport calculations, the non-equilibrium Green's functions (NEGF) method combined with DFT was employed as implemented in the Transiesta code.26 Schematic representations of the two nano-devices employed in this study are depicted in Fig. 1a and b. Electronic transport is simulated to occur along the z-direction. For each system, four gold layers of atoms are used as electrodes, where they play the role of the source and drain, respectively. To numerically solve the large tridiagonal matrix eigenvalue problem, we consider the Green's function:
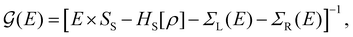 | (1) |
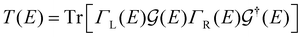 | (2) |
The coupling matrices are defined as 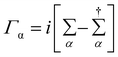 , with α ≡ {L, R}.
, with α ≡ {L, R}. 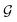 (
(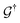 ) is the retarded (advanced) Green's function. Zero-bias calculations are performed in which the chemical potential of both electrodes are at the same level, leading to equal chemical potentials (μL = μR), where the electrical current is zero. Thus, our transmission calculations can be regarded as predicting the conductance of the device in the limit of small bias voltages. Further details on the theory and its numerical implementation can be found elsewhere.27,28
) is the retarded (advanced) Green's function. Zero-bias calculations are performed in which the chemical potential of both electrodes are at the same level, leading to equal chemical potentials (μL = μR), where the electrical current is zero. Thus, our transmission calculations can be regarded as predicting the conductance of the device in the limit of small bias voltages. Further details on the theory and its numerical implementation can be found elsewhere.27,28
The local current (LC) consists of the transmission projections between two neighboring sites, N and M, and it is possible to write:
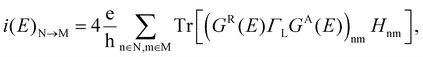 | (3) |
3. Results & discussions
To elucidate the mechanism underlying the detection of individual amino acids, we propose two distinct gold based devices, as depicted in Fig. 1a and b. Primarily, an infinite capacitor with a tailored 7.0 Å gap was computationally designed, as illustrated in Fig. 1a. Its scattering region possesses periodicity in the xy directions. Subsequently, we conceived a nanojunction featuring a 5.0 Å gap (Fig. 1b), in contrast with the infinite capacitor device (non-periodic in xy plane). According to Fyta et al.,11 the bare capacitor conductance tends to be negligible for gaps that are at least 5.0 Å wide, thus in both devices, the detection/identification mechanism is given by electron tunneling mediated by the amino acid residing the nanojuction.Fig. 1c depicts each of the 20 amino acids (AA) investigated, namely: alanine (ALA, A), arginine (ARG, R), asparagine (ASN, N), aspartic acid (ASP, D), cysteine (CYS, C), glutamine (GLN, Q), glutamic acid (GLU, E), glycine (GLY, G), histidine (HIS, H), isoleucine (ILE, I), leucine (LEU, L), lysine (LYS, K), methionine (MET, M), phenylalanine (PHE, F), proline (PRO, P), serine (SER, S), threonine (THR, T), tryptophan (TRP, W), tyrosine (TYR, Y), and valine (VAL, V). Additionally, we investigated the phosphorylated post-translational modifications of three amino acids: pY, pT, and pS. Hereafter, we will use a single-letter representation to refer to each amino acid throughout the text. Each of the 23 amino acids was positioned within the two nanogaps and a full structural relaxation was performed to obtain the most stable configuration.
The interaction between the amino acids and the two nano-devices was evaluated by calculating the binding energy, as reported in previous studies.17 Additionally, the conductance was determined in the limit of zero bias voltage, where it follows the relationship 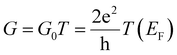 , with
, with 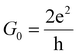 standing for the quantum of conductance and T(EF) representing the transmission at the Fermi energy (EF). The distinction among the target molecules can be achieved by comparing two quantities: (i) conductance of the combined system (nanogap/amino acid); and (ii) the absolute binding energy (Eb)16 of each of them with the device. The latter one can be directly connected to the dwell time of the target molecule inside the nanojuction.
standing for the quantum of conductance and T(EF) representing the transmission at the Fermi energy (EF). The distinction among the target molecules can be achieved by comparing two quantities: (i) conductance of the combined system (nanogap/amino acid); and (ii) the absolute binding energy (Eb)16 of each of them with the device. The latter one can be directly connected to the dwell time of the target molecule inside the nanojuction.
In Fig. 2a, the binding energy is plotted against the conductance for the infinite capacitor device. Notably, amino acid groups are identified, and some of them can be uniquely detected. For instance, M and pT exhibit similar conductance values around 0.20G0, but the Eb for M is approximately four times smaller, which should enable a distinction via the dwell time. Similarly, the group consisting of E, D, I, and L shows alike conductance values, while their binding energies have a deviation of 0.25 eV. For R and pY amino acids, a quite similar binding energy is noted with different conductance of about 0.25G0. Amino acids W, F, and Y display comparable conductance values as well as binding energies, posing difficulty to their distinction. The group comprising P, T, Q, pS, and C exhibits comparable binding energies, albeit with slightly different conductance values. Interestingly, the device seems to be capable of distinguishing, via conductance difference, the amino acids S, T, and Y from their modified counterparts pS, pT, and pY, respectively.
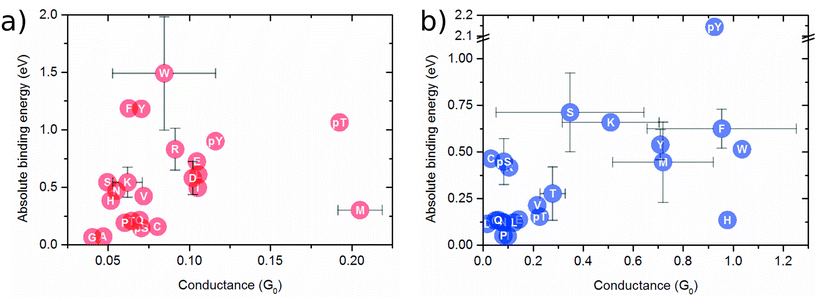 | ||
| Fig. 2 Binding energy as a function of the conductance (at Fermi level) for (a) capacitor of 7.0 Å nanogap; (b) 5.0 Å nanojunction. | ||
Fig. 2b shows the binding energy as a function of the conductance for the nanojunction device. The amino acids and the three mutants presenting a larger conductance range compared to the infinite capacitor device. S and K display distinct conductance. Conversely, the molecules pY, F, W, and H exhibit quite similar conductance values while pY, F/W and H possess different binding energies. Further, the groupings consisting of: (i) C, Ps and R; (ii) Q, E, L, N, D, P; and (iii) V, pT, T display similar conductance and binding values, thereby allowing for the distinction of each one group only.
Aiming to discriminate each amino acid, we quantified the differences in the conductance/resistance (sensitivity) of the proposed devices toward each target molecule, where a specific amino acid is chosen as reference, namely, M for the nanojunction and F for the infinite capacitor device, respectively. The sensitivity is defined as follows:
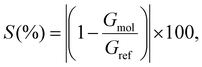 | (4) |
 ratio, observing that F is the optimal choice for the 7.0 Å infinite capacitor, while M is the optimal choice for the 5.0 Å nanojunction.
ratio, observing that F is the optimal choice for the 7.0 Å infinite capacitor, while M is the optimal choice for the 5.0 Å nanojunction.
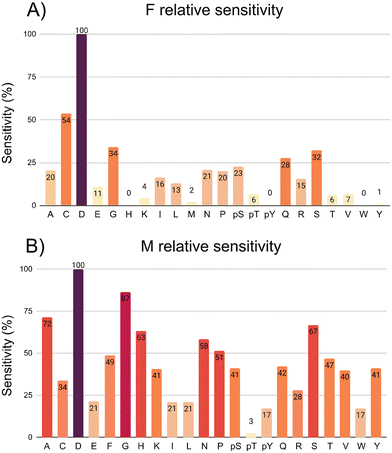
For the 7.0 Å infinite capacitor (Fig. 3a), the amino acid D stands out with an exceptional level of distinguishability, showcasing the infinite capacitor's high-resolution capabilities. This finding suggests that the device is particularly adept at recognizing and isolating D within the complex matrix of amino acids. Further analysis reveals varying degrees of sensitivity across different amino acids. C, G, Q, and S exhibit a robust sensitivity range, spanning from roughly 30% up to 50%. In the moderate sensitivity range, amino acids A, E, I, L, N, P, pS, and R present values fluctuating between 11% and 23%. While not as pronounced as the high-sensitivity group, this range signifies a considerable ability of the infinite capacitor to detect these amino acids with a satisfactory level of accuracy. Amino acids pT, T, K, and V fall into the low sensitivity category, displaying values ranging between 4% and 7%. Notably, H, pY, W, and Y are undetected by the 7.0 Å device. An interesting point is that both S and pS emerge as distinguishable entities. This indicates a specificity in the infinite capacitor's ability to differentiate among these similar amino acids, showcasing a level of precision in its discriminatory power.
 | ||
| Fig. 3 Graphical representation depicting the sensitivity (%) of the following devices: (a) 7.0 Å infinite capacitor; (b) 5.0 Å nanojunction. | ||
The 5.0 Å nanojunction excels in distinguishing various amino acids compared to the infinite capacitor, providing valuable insights into its sensitivity across a diverse set. In both cases, amino acid D stands out with exceptional distinguishability, underscoring its high-resolution capabilities. G exceeds an impressive threshold of 85%, showcasing heightened responsiveness. In the range of 49% up to 70%, A, H, S, N, P, and F demonstrate substantial sensitivity, indicating a considerable ability to detect and distinguish these molecules with a high level of accuracy. The molecules C, K, pS, Q, R, V, and Y fall into sensitivity range of the 30% up to 40%, still showcasing discerning capability. In the 10% up to 20% sensitivity range, E, I, L, pY, and W demonstrate lower levels. AA pT remains undetected by the 5.0 Å nanojunction, suggesting a limitation in its recognition capabilities for this specific marker. Both Y and pY emerge as distinguishable amino acids, emphasizing its ability to differentiate between phosphorylation. Similarly, S and pS showcase distinctive signals, indicating notable specificity in the nanojunction's discriminatory potential against the 7.0 Å infinite capacitor.
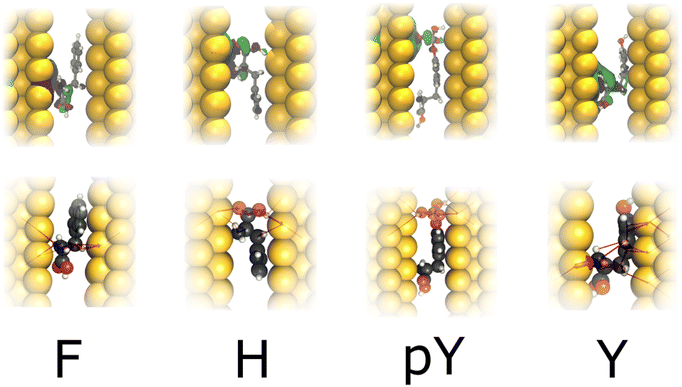
Fig. 4 depicts the wavefunction (WF) and local current (LC) for F, H, pY, and Y in the infinite capacitor device (7.0 Å). We note a WF with higher degree of localization in the molecules. Here, considering H and F, we verify that the WF and also LC are localized in the backbone, which could explain their similar conductance (≤0.025G0). While Y presents LC and WF spreading by both the backbone and the radical, whereas pY shows this characteristic majority in the phosphorylated group, which contributes to additional conducting channels, as can be seen in Fig. 2a.
Fig. 5 provides the WF and LC profiles for the amino acids F, H, pY, and Y within the 5.0 Å nanojunction device. In general, the upper panel exhibits the WF localized on the left electrode and the molecule part appearing between electrodes. The conductance value hierarchy, at Fermi energy, follows the order (pY > Y and F ∼ H). The H, F, and Y exhibit a pentagon ring, a phenyl group, and a phenol group between the electrodes, respectively. These observations are in agreement with the LC, where it passes through the molecules to reach the right side by tunneling. Likewise in the capacitor device, for pY and Y, the WF and LC are more localized at the phosphorylated group, leading to a higher conductance for the former than the latter one (≥0.20G0). Thus, regarding the pY and Y, the nanojunction is more sensitive to the presence of a phosphorylated group than the infinite capacitor device.
Through the utilization of our two gold-electrodes devices tailored for single amino acid electronic detection, we conducted a comparative analysis between the infinite capacitor and nanojunction. Remarkably, the nanojunction exhibited significantly higher resolution than the infinite capacitor. Regarding the nanojunctions device and using the M molecule as a reference, we achieved exceptional sensitivity resolution. Additionally, the higher conductance of the nanojunction detector suggests a possible attenuation of the signal-to-noise ratio, as supported by the wavefunction and local current results. These compelling findings underscore the nanojunction device as a promising frontrunner for the precise electric detection of single amino acids, positioning it as a potential breakthrough in the field.
It is important to note that a real experimental apparatus contains an aqueous solution, leading to thermal fluctuations. Consequently, to incorporate the effects of temperature and water, an enhanced model employing molecular dynamics and QM/MM methodologies should be feasible. Prior research indicates that the inclusion of an aqueous solvent is probable to maintain the transmission signature of the most stable configuration.30 This work posits that the equilibrium configuration, demonstrating optimal coupling between the device and the target molecule, will dominate all other possible arrangements when averaged across the total transmission curve. Consequently, we have concentrated solely on this configuration.
4. Conclusions
In summary, using density functional theory (DFT) and the nonequilibrium Green's function (NEGF) method, we have investigated two distinct gold-based nanodevices for single amino acid detection. Our analysis demonstrates that amino acids can be grouped based on conductance and binding energy values, revealing distinct electronic signatures that enable molecular discrimination. We selected amino acid F as a reference to calculate the relative sensitivity for the infinite capacitor and M for the nanojunction. In the case of the former, only the amino acids A, C, D, E, G, I, L, N, P, pS, Q, R, and S exhibited relative sensitivity levels exceeding 11%. However, the nanojunction exhibited a broader range of conductance variations, a sixfold increase in signal magnitude, and superior sensitivity and selectivity compared to the capacitor system. Notably, the nanojunction displayed exceptional differentiation for amino acids S, pS, Y, and pY, making it particularly effective in distinguishing between structurally similar molecules. Our results indicate that electronic transport properties provide a robust mechanism for amino acid identification, with the nanojunction outperforming the infinite capacitor in both sensitivity and noise reduction. These findings suggest that gold-based nanojunctions hold strong potential for real-time biomarker detection, biosensing applications, and future integration into nanotechnology-driven diagnostic platforms. Further studies could explore the impact of external gating and environmental conditions to optimize device performance for practical applications.Data availability
This study was carried out using publicly available data from RCSB PDB at https://www.rcsb.org/.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge financial support from the Brazilian agencies CAPES (001), FAPERJ, CNPq, INCT-Materials Informatics, FAPES (TO - 1043/2022) and computational resources of Sci-Com/ES and CENAPAD-SP as well as resources provided by the National Academic Infrastructure for Supercomputing in Sweden (NAISS), partially funded by the Swedish Research Council through grant agreement no. 2022-06725.References
- T. Li, X.-M. Lu, M.-R. Zhang, K. Hu and Z. Li, Bioact. Mater., 2022, 11, 268–282 Search PubMed.
- M. D. Ventra and M. Taniguchi, Nat. Nanotechnol., 2016, 11, 117–126 CrossRef PubMed.
- S. J. Heerema and C. Dekker, Nat. Nanotechnol., 2016, 11, 127–136 CrossRef PubMed.
- B. D. Reed, M. J. Meyer, V. Abramzon, O. Ad, O. Ad, P. Adcock, F. R. Ahmad, G. Alppay, J. A. Ball, J. Beach, D. Belhachemi, A. Bellofiore, M. Bellos, J. F. Beltrán, A. Betts, M. W. Bhuiya, K. Blacklock, R. Boer, D. Boisvert, N. D. Brault, A. Buxbaum, S. Caprio, C. Choi, T. D. Christian, R. Clancy, J. Clark, T. Connolly, K. F. Croce, R. Cullen, M. Davey, J. Davidson, M. M. Elshenawy, M. Ferrigno, D. Frier, S. Gudipati, S. Hamill, Z. He, S. Hosali, H. Huang, L. Huang, A. Kabiri, G. Kriger, B. Lathrop, A. Li, P. Lim, S. Liu, F. Luo, C. Lv, X. Ma, E. McCormack, M. Millham, R. Nani, M. Pandey, J. Parillo, G. Patel, D. H. Pike, K. Preston, A. Pichard-Kostuch, K. Rearick, T. Rearick, M. Ribezzi-Crivellari, G. Schmid, J. Schultz, X. Shi, B. Singh, N. Srivastava, S. F. Stewman, T. Thurston, T. R. Thurston, P. Trioli, J. Tullman, X. Wang, Y.-C. Wang, E. A. G. Webster, Z. Zhang, J. Zuniga, S. S. Patel, A. D. Griffiths, A. M. van Oijen, M. McKenna, M. D. Dyer and J. M. Rothberg, Science, 2022, 378, 186–192 CrossRef CAS PubMed.
- H. Brinkerhoff, A. S. W. Kang, J. Liu, A. Aksimentiev and C. Dekker, Science, 2021, 374, 1509–1513 CrossRef CAS PubMed.
- J. A. Alfaro, P. Bohländer, M. Dai, M. Filius, C. J. Howard, X. F. van Kooten, S. Ohayon, A. Pomorski, S. Schmid, A. Aksimentiev, E. V. Anslyn, G. Bedran, C. Cao, M. Chinappi, E. Coyaud, C. Dekker, G. Dittmar, N. Drachman, R. Eelkema, D. Goodlett, S. Hentz, U. Kalathiya, N. L. Kelleher, R. T. Kelly, Z. Kelman, S. H. Kim, B. Kuster, D. Rodriguez-Larrea, S. Lindsay, G. Maglia, E. M. Marcotte, J. P. Marino, C. Masselon, M. Mayer, P. Samaras, K. Sarthak, L. Sepiashvili, D. Stein, M. Wanunu, M. Wilhelm, P. Yin, A. Meller and C. Joo, Nat. Methods, 2021, 18, 604–617 CrossRef CAS PubMed.
- J. Wilson, K. Sarthak, W. Si, L. Gao and A. Aksimentiev, ACS Sens., 2019, 4, 634–644 CrossRef CAS PubMed.
- J. Yoo, D. Winogradoff and A. Aksimentiev, Curr. Opin. Struct. Biol., 2020, 64, 88–96 CrossRef CAS PubMed.
- T. Ohshiro, M. Tsutsui, K. Yokota, M. Furuhashi, M. Taniguchi and T. Kawai, Nat. Nanotechnol., 2014, 9, 835–840 CrossRef CAS.
- Y. Zhao, B. Ashcroft, P. Zhang, H. Liu, S. Sen, W. Song, J. Im, B. Gyarfas, S. Manna, S. Biswas, C. Borges and S. Lindsay, Nat. Nanotechnol., 2014, 9, 466–473 CrossRef CAS.
- G. Sivaraman, R. G. Amorim, R. H. Scheicher and M. Fyta, Nanoscale, 2016, 8, 10105–10112 RSC.
- G. Sivaraman, R. G. Amorim, R. H. Scheicher and M. Fyta, Nanotechnology, 2016, 27, 414002 CrossRef.
- G. Sivaraman, R. G. Amorim, R. H. Scheicher and M. Fyta, RSC Adv., 2017, 7, 43064–43072 RSC.
- R. L. Kumawat, M. K. Jena and B. Pathak, J. Phys. Chem. C, 2020, 124, 27194–27202 CrossRef CAS.
- S. Mittal, S. Manna and B. Pathak, ACS Appl. Mater. Interfaces, 2022, 14, 51645–51655 Search PubMed.
- F. A. de Souza, G. Sivaraman, M. Fyta, R. H. Scheicher, W. L. Scopel and R. G. Amorim, Nanoscale, 2020, 12, 18289–18295 RSC.
- F. A. L. de Souza, R. G. Amorim and R. H. Scheicher, et al. , Phys. Chem. Chem. Phys., 2019, 21, 24884–24890 Search PubMed.
- S. Mittal, M. K. Jena and B. Pathak, Chem. – Eur. J., 2023, 29, e202301667 CrossRef CAS.
- P. Hohenberg and W. Kohn, Phys. Rev. B, 1964, 136, 864 CrossRef.
- J. M. Soler, E. Artacho, J. D. Gale, A. García, J. Junquera, P. Ordejón and D. Sánchez-Portal, J. Phys.: Condens. Matter, 2002, 14, 2745 CrossRef CAS.
- D. Hamann, M. Schlüter and C. Chiang, Phys. Rev. Lett., 1979, 43, 1494 CrossRef CAS.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865 CrossRef CAS.
- J. Junquera, Ó. Paz, D. Sánchez-Portal and E. Artacho, Phys. Rev. B: Condens. Matter Mater. Phys., 2001, 64, 235111 Search PubMed.
- S. Grimme, A. Hansen, J. G. Brandenburg and C. Bannwarth, Chem. Rev., 2016, 116, 5105–5154 CrossRef CAS PubMed.
- H. J. Monkhorst and J. D. Pack, Phys. Rev. B, 1976, 13, 5188–5192 CrossRef.
- M. Brandbyge, J. Mozos, P. Ordejón, J. Taylor and K. Stokbro, Phys. Rev. B: Condens. Matter Mater. Phys., 2002, 65, 165401 CrossRef.
- S. Datta, Quantum transport: Atom to Transistor, Cambridge University Press, 2005 Search PubMed.
- A. Rocha, V. García-Suárez, S. Bailey, C. Lambert, J. Ferrer and S. Sanvito, arXiv preprint cond-mat/0510083, 2005.
- M. Paulsson and M. Brandbyge, Phys. Rev. B: Condens. Matter Mater. Phys., 2007, 76, 115117 CrossRef.
- J. Prasongkit, E. de Freitas Martins, F. A. De Souza, W. L. Scopel, R. G. Amorim, V. Amornkitbamrung, A. R. Rocha and R. H. Scheicher, J. Phys. Chem. C, 2018, 122, 7094–7099 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr03359k |
| This journal is © The Royal Society of Chemistry 2025 |