Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A bodipy based hydroxylamine sensor†‡
Adam C.
Sedgwick
 a,
Robert S. L.
Chapman
a,
Robert S. L.
Chapman
 a,
Jordan E.
Gardiner
a,
Jordan E.
Gardiner
 a,
Lucy R.
Peacock
a,
Gyoungmi
Kim
b,
Juyoung
Yoon
a,
Lucy R.
Peacock
a,
Gyoungmi
Kim
b,
Juyoung
Yoon
 *b,
Steven D.
Bull
*b,
Steven D.
Bull
 *a and
Tony D.
James
*a and
Tony D.
James
 *a
*a
aDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk; s.d.bull@bath.ac.uk
bDepartment of Chemistry and Nano Science, Ewha Womans University, Seoul 120-750, Korea. E-mail: jyoon@ewha.ac.kr
First published on 24th August 2017
Abstract
With this research, we have developed a bodipy based system as the first “turn-on” fluorescence system for the detection hydroxylamine.
Hydroxylamine, (HA) NH2OH, is an oxygenated form of ammonia, which is widely used in industrial and pharmaceutical processes.1 HA is widely known as a nitric oxide (NO) donor and participates in a wide range of biological processes.2–4 In cellular metabolism, HA is an intermediate in the conversion L-arginine to nitric oxide (NO).5,6 This process involves the hydrolysis of oxime arginine to L-citrulline and HA. HA is then converted by catalase to NO and superoxide (−O2) in the presence of hydrogen peroxide.7 It is also believed that HA can be converted to NO by the non-enzymatic attack by −O2. Therefore, in inflammatory cells a reaction between HA and −O2 could take place to generate NO.3,8 For example, HA has shown to be a vasodilator in a dose-dependent manner in the blood vessels of rats’ kidneys.9 HA has also shown to be an inhibitor for the release of insulin and HA has been shown to activate K+ channels.3,10
Despite HA being a product of metabolism and a NO donor, it is moderately toxic to humans, animals and plants. HA toxicity occurs at concentrations that are substantially greater than the normal physiological concentrations. Exposure to HA has been shown to be hemotoxic, mutagenic whilst also being an enzyme and virus inhibitor.11
Given these important biological aspects of HA we set out to develop a fluorescence probe for the detection of HA. To our knowledge, there are currently no HA fluorescence probes. HA is currently detected using: HPLC, GC, potentiometric, polarographic, biamperometric and electrochemical methods.12–19
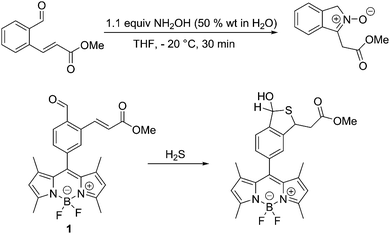
Previously, we have developed a synthetic route to nitrones by reacting an aromatic aldehyde containing an ortho α,β-unsaturated ester with NH2OH (50% wt in H2O) in THF at −20 °C to produce isoindole nitrones in high yields,20Scheme 1. Therefore, we set out to integrate this reaction with a fluorophore for the detection of HA. Luckily, an appropriate system (probe 1) had previously been prepared for the detection of hydrogen sulphide (H2S)21,22Scheme 1.
The synthesis of probe 1, is given in the ESI.†
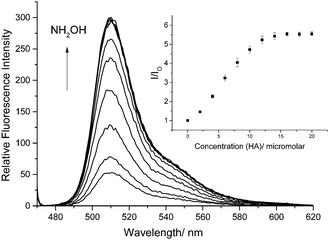
We also confirmed that probe 1 reacts as predicted with hydroxylamines as shown by the 1H NMR, 13C NMR and MS (ESI†). More importantly, when HA is added to a 0.5 μM solution of probe 1 in a 1% DMSO PBS Buffer solution at pH 7.4 the fluorescence is significantly enhanced as the concentration of HA is increased (Fig. 1).
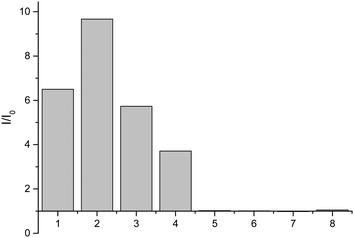
Having shown that probe 1 works with HA we set out to investigate the selectivity amongst several hydroxylamines: N-(benzyl)hydroxylamine, N-(propargyl)hydroxylamine, N-(tert-butyl)hydroxylamine, O-(benzyl)hydroxylamine, and N-(phenyl)hydroxylamine (Fig. 2).
From these results, it is clear that the system works for simple primary hydroxylamines (1) hydroxylamine, (2) N-(methyl)hydroxylamine (3) N-(benzyl)hydroxylamine, (4) N-(propargyl)hydroxylamine, however, larger alkyl groups or aromatic hydroxylamines such as (5) N-(tert-butyl)hydroxylamine and (7) N-(phenyl)hydroxylamine do not work. It is evident that the system also requires the hydroxyl of the hydroxylamine (6) O-(benzyl)hydroxylamine. These observations are consistent with the proposed mechanism of the reaction (Scheme 2). Where, a nucleophillic nitrogen23 and exchangeable OH are required to facilitate condensation of hydroxylamines with probe 1.
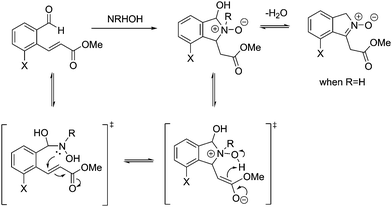 | ||
| Scheme 2 Mechanism for the reaction of probe 1 with hydroxylamines (adapted from ref. 20). | ||
We have carried out a more extensive screen amongst relevant biological species (ESI†), and as expected only H2S (50 μM) produced a fluorescence response.
We then attempted to detect hydroxylamines in a cellular environment using probe 1. HeLa cells were incubated HA (0–150 μM) for 30 minutes and washed with DPBS then 1 μM of probe 1 was added and left for 30 minutes, fluorescence images were then acquired by confocal microscopy. From these fluorescence images it is clear that probe 1 was fluorescent in HeLa cells even without added HA (ESI,† Fig. S3). The strong background fluorescence is attributed to cellular thiols including endogenous H2S present in the HeLa cells.
In conclusion, we have developed a fluorescence turn-on probe for the detection of hydroxylamines. The system works well in vitro but with the in vivo cellular experiments endogenous thiols react to turn on probe 1. We are currently working to develop a system with enhanced selectivity for hydroxylamines over thiols. However, it is interesting to note that the system using N-(propargyl)hydroxylamine could be used as fluorescent “click” tag.
We would like to thank the EPSRC and the University of Bath for funding. ACS, JEG, and LRP thank the EPSRC for studentships. RSLC thanks the EPSRC Doctoral Training Centre in Sustainable Chemical Technologies: EP/G03768X/1 for a studentship. JY thanks to the support from the National Research Foundation of Korea (NRF), which was funded by the Korea government (MSIP) (No. 2012R1A3A2048814). Catherine Lyall is thanked for help in obtaining NMR spectra using NMR characterisation facilities provided through the Chemical Characterisation and Analysis Facility (CCAF) at the University of Bath (http://www.bath.ac.uk/ccaf). The EPSRC UK National Mass Spectrometry Facility at Swansea University is thanked for analyses.
Conflicts of interest
There are no conflicts to declare.Notes and references
- M. Kumasaki, Y. Fujimoto and T. Ando, J. Loss Prev. Process Ind., 2003, 16, 507–512 CrossRef.
- P. Saransaari and S. S. Oja, Neuroscience, 1999, 89, 1103–1111 CrossRef CAS PubMed.
- P. G. Wang, M. Xian, X. P. Tang, X. J. Wu, Z. Wen, T. W. Cai and A. J. Janczuk, Chem. Rev., 2002, 102, 1091–1134 CrossRef CAS PubMed.
- M. S. Ryba, W. Gordon-Krajcer, M. Walski, M. Chalimoniuk and S. J. Chrapusta, Brain Res., 1999, 850, 225–233 CrossRef CAS PubMed.
- G. H. Tang, L. Y. Wu and R. Wang, FASEB J., 2003, 17, A1050–A1050 Search PubMed.
- E. G. Demaster, L. Raij, S. L. Archer and E. K. Weir, Biochem. Biophys. Res. Commun., 1989, 163, 527–533 CrossRef CAS PubMed.
- M. Klink, A. Swierzko and Z. Sulowska, APMIS, 2001, 109, 493–499 CrossRef CAS PubMed.
- P. Vetrovsky, J. C. Stoclet and G. Entlicher, Int. J. Biochem. Cell Biol., 1996, 28, 1311–1318 CrossRef CAS PubMed.
- P. K. Moore, L. Burrows and R. Bhardwaj, J. Pharm. Pharmacol., 1989, 41, 426–429 CrossRef CAS PubMed.
- M. H. Antoine, R. Ouedraogo, J. Sergooris, M. Hermann, A. Herchuelz and P. Lebrun, Eur. J. Pharmacol., 1996, 313, 229–235 CrossRef CAS PubMed.
- P. Gross, Crit. Rev. Toxicol., 1985, 14, 87–99 CrossRef CAS PubMed.
- W. D. Korte, J. Chromatogr., 1992, 603, 145–150 CrossRef CAS.
- A. Afkhami, T. Madrakian and A. Maleki, Anal. Sci., 2006, 22, 329–331 CrossRef CAS PubMed.
- Y. Seike, R. Fukumori, Y. Senga, H. Oka, K. Fujinaga and M. Okumura, Anal. Sci., 2004, 20, 139–142 CrossRef CAS PubMed.
- R. Christova, M. Ivanova and M. Novkirishka, Anal. Chim. Acta, 1976, 85, 301–307 CrossRef.
- D. R. Canterford, Anal. Chim. Acta, 1978, 98, 205–214 CrossRef CAS.
- C. Zhao and J. F. Song, Anal. Chim. Acta, 2001, 434, 261–267 CrossRef CAS.
- Y. Wang, L. Wang, H. H. Chen, X. Y. Hu and S. Q. Ma, ACS Appl. Mater. Interfaces, 2016, 8, 18173–18181 CAS.
- C. H. Zhang, G. F. Wang, M. Liu, Y. H. Feng, Z. D. Zhang and B. Fang, Electrochim. Acta, 2010, 55, 2835–2840 CrossRef CAS.
- L. R. Peacock, R. S. L. Chapman, A. C. Sedgwick, M. F. Mahon, D. Amans and S. D. Bull, Org. Lett., 2015, 17, 994–997 CrossRef CAS PubMed.
- Y. Qian, J. Karpus, O. Kabil, S. Y. Zhang, H. L. Zhu, R. Banerjee, J. Zhao and C. He, Nat. Commun., 2011, 2, 7 Search PubMed.
- X. Li, S. Zhang, J. Cao, N. Xie, T. Liu, B. Yang, Q. J. He and Y. Z. Hu, Chem. Commun., 2013, 49, 8656–8658 RSC.
- F. Brotzel, Y. C. Chu and H. Mayr, J. Org. Chem., 2007, 72, 3679–3688 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7cc05872a |
| ‡ All data created during this research are openly available from the University of Bath data archive at http://doi.org/10.15125/BATH-00381. |
| This journal is © The Royal Society of Chemistry 2017 |