Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Long-wavelength fluorescent boronate probes for the detection and intracellular imaging of peroxynitrite†
Adam C.
Sedgwick
 *a,
Hai-Hao
Han
b,
Jordan E.
Gardiner
*a,
Hai-Hao
Han
b,
Jordan E.
Gardiner
 a,
Steven D.
Bull
a,
Steven D.
Bull
 *a,
Xiao-Peng
He
*a,
Xiao-Peng
He
 *b and
Tony D.
James
*b and
Tony D.
James
 *a
*a
aDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk; s.d.bull@bath.ac.uk
bKey Laboratory for Advanced Materials & Feringa Nobel Prize Scientist Joint Research Center, East China University of Science and Technology, 130 Meilong Rd., Shanghai 200237, P. R. China. E-mail: xphe@ecust.edu.cn
First published on 8th November 2017
Abstract
Two boronate fluorescent probes have been developed for the detection of peroxynitrite (TCFB1 and TCFB2). TCFB1 was shown to have a low sensitvity towards peroxynitrite and have a poor solubility in aqueous solution whereas TCFB2 demonstrated high sensitivity towards peroxynitrite and mitochondria localisation with the ability to detect exogenous and endogenous peroxynitrite in live cells (Hep-G2, RAW 264.7, HeLa and A459).
Peroxynitrite (ONOO−) is a highly reactive nitrogen species that is formed via the diffusion controlled reaction between superoxide (−O2) and nitric oxide (NO).1,2 ONOO− acts as a signalling molecule in vivo for a number of pathways.1,3 However, ONOO− is more commonly known for its deleterious properties, causing irreversible damage to a range of biological targets such as lipids, proteins and DNA.4 Therefore, ONOO− has been implicated as a key pathogenic factor for a number of diseases, which include inflammation, cancer, ischemia-reperfusion and neurodegenerative diseases.5–7 In biological systems, ONOO− is difficult to measure due to it being short-lived with a half-life ∼10–20 ms.1 Therefore, the development of powerful tools for the detection of ONOO− is of significant interest.
With our research, we are particularly interested in the development of small molecule fluorescent probes for the detection of biologically relevant analytes in vivo owing to their high sensitivity, selectivity and high spatial and temporal resolution. In the past few years, a number of ONOO− fluorescent probes have been developed for imaging in live cells and mice.8–13 However, despite significant progress in this area of research, there is a lack of long-wavelength ONOO− fluorescent probes. The development of long wavelength/near infrared (NIR) probes is of particular interest because longer excitation/emission wavelengths allows deeper tissue penetration and minimalises background auto-fluorescence from proteins and photodamage to the biological samples.14,15
In the literature, Sikora et al. reported that the reaction rates of ONOO− with aromatic boronates are 200 times faster than hypochlorous acid (HOCl/ClO−) and a million times faster than hydrogen peroxide (H2O2).16 Therefore, a number of boronate fluorescent probes have been recently developed for the detection of ONOO−.8,17,18
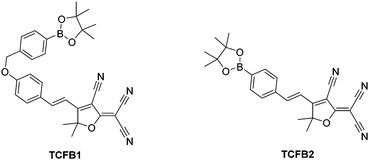
2-Dicyanomethylene-3-cyano-4,5,5-trimethyl-2,5-dihydrofuran (TCF)-based fluorophores have an internal charge transfer (ICT) donor-π-acceptor (D-π-A) structure with long emission wavelengths. As a result, TCF fluorophores have been used in many applications such as non-linear optic chromophores and molecular probes.19–25 With this research, we developed two boronate TCF-based fluorescent probes for the detection of ONOO− (TCFB1 and TCFB2). The TCF fluorophore unit was synthesised in one step using the reaction of 3-hydroxy-3-methyl-2-butanone, malonitrile and NaOEt in EtOH. With the TCF unit in hand, the (D-π-A) systems TCFB1 and TCFB2 were isolated in high yield using microwave reaction conditions.26 The microwave irradiation of a mixture of piperidine (Cat.), TCF and 4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)benzaldehyde in EtOH followed by filtration led to the isolation of the desired TCFB2. For the synthesis of TCFB1, microwave irradiation of a mixture of piperidine (Cat.), TCF and 4-hydroxybenzaldehyde in EtOH followed by filtration led to the isolation of the intermediate TCF-OH. This was subsequently alkylated with 2-(4-(bromomethyl)phenyl)-4,4,5,5-tetramethyl-1,3,2-dioxaborolane using K2CO3 and NaI in MeCN to afford TCFB1 in a reasonable yield (47%).
We initially evaluated the UV-Vis (Fig. S2, ESI†) and fluorescence behaviour (Fig. 1 and Fig. S3, ESI†) of TCFB1, in pH 8.0 buffer solution (20% DMSO). DMSO was required to improve the aqueous solubility of TCFB1. Under these conditions, TCFB1 produced an up to 6.5-fold fluorescence “turn on” in the presence of ONOO− (0–100 μM). (Schemes S1, S2 and Fig. S1, ESI†) However, in comparison to our previously reported ESIPT probe, TCFB1 was less sensitive towards ONOO− despite a larger “turn on” response.8
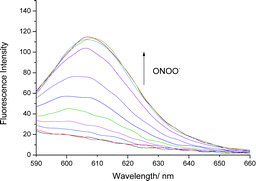 | ||
| Fig. 1 Fluorescence spectra of TCFB1 (10 μM) with addition of ONOO− (0–100 μM) in PBS buffer solution, 20% DMSO, pH 8.00 at 25 °C. λex = 560 nm. Slit widths ex = 10 nm and em = 15 nm. | ||
Subsequently, we evaluated the selectivity of TCFB1 towards other ROS (Fig. S4, S5 and S11, ESI†). TCFB1 demonstrated an excellent selectivity for ONOO−, which permitted the evaluation of its ability to detect exogenous and endogenous ONOO− in live cells. Unfortunately, due to its poor aqueous solubility, large amounts of precipitate with TCFB1 was observed (data not shown).
Therefore, we turned our attention towards the evaluation of the UV-Vis and fluorescence properties of TCFB2, which has previously been reported for the detection of ClO−.20 As previously reported for other aryl boronate fluorescent probes,27,28TCFB2 was found to be initially non-fluorescent with no UV absorption beyond ∼525 nm (Fig. S6, ESI†). The addition of ONOO− to TCFB2 resulted in the appearance of a large emission peak at 606 nm (Fig. 2 and Fig. S7, ESI†). This was accompanied by a colorimetric response (yellow to pink) and the appearance of a large UV absorption peak at ∼590 nm. TCFB2 demonstrated high sensitivity and rapid reaction (Fig. S8, ESI†) with ONOO− and was able to detect very low concentrations (0–10 μM). As predicted, both ClO− and H2O2 also resulted in a fluorescence response (Fig. S9, S10 and S12, ESI†), however, larger concentrations and reaction times were required. These observations clearly, demonstrated the greater reactivity of the boronate towards ONOO−.
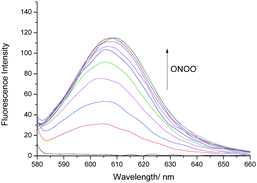 | ||
| Fig. 2 Fluorescence spectra of TCFB2 (10 μM) with addition of ONOO− (0–10 μM) in PBS buffer solution, 20% DMSO, pH 8.00 at 25 °C. λex = 560 nm. Slit widths ex = 10 nm and em = 15 nm. | ||
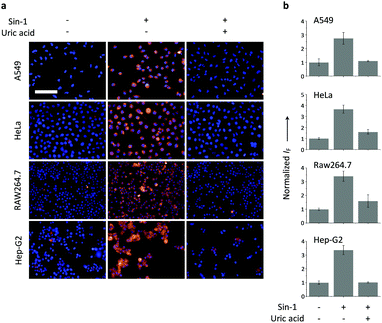
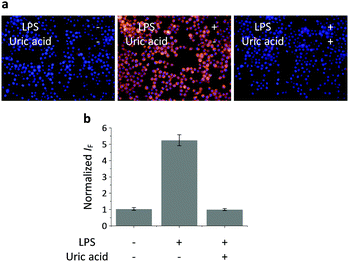
Having determined the selectivity of TCFB2, we evaluated its ability to image endogenous and exogenous ONOO− in live cells. TCFB2 was evaluated in a number of different cell lines (Hep-G2: human hepatoma, HeLa: human cervical cancer, RAW 264.7: mouse macrophage and A549 cells: human lung cancer), which were incubated with TCFB2 (10 μM) for 30 minutes and washed with PBS buffer solution three times. As shown in Fig. 3, TCFB2 demonstrated a clear “turn on” response with the addition of SIN-1 (ONOO− donor). No “turn on” response was observed when the cells were pre-treated with the ONOO− scavenger uric acid. TCFB2 also provided a clear “turn on” response for the detection of stimulated ONOO−. RAW 264.7 cells were used in which ONOO− was stimulated using lipopolysaccharide (LPS).29 This led to the activation of the TCFB2 fluorescence intracellularly (Fig. 4). In contrast, no “turn on” response was observed in the presence of uric acid indicating the selectivity for ONOO− in cells. A cell proliferation assay showed that the compound was not toxic towards all the cell lines used with concentrations well above that used for imaging (Fig. S13, ESI†).
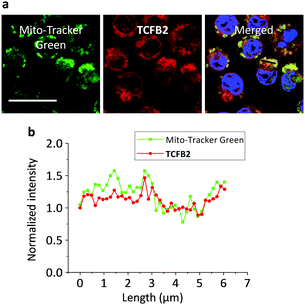
The production of superoxide occurs mainly through the mitochondrial electron transport pathway;30 therefore the mitochondria are the main source of ONOO− in macrophages. Commercial Mito-tracker Green was used to localise in the mitochondrial compartments of RAW 264.7. We then used TCFB2 to investigate the subcellular distribution of ONOO−. The results indicated that the fluorescence of the probe co-localised with that of the tracker resulting in a Pearson coefficient of 0.84 (Fig. 5). We have also carried out an additional lysosome co-localisation assay, and the result showed that the probe did not co-localise well with lysosome (Pearson's correlation = 0.38) (Fig. S14, ESI†). This suggests that ONOO− was produced at the mitochondria.
In conclusion, we have developed two long-wavelength reaction based fluorescent probes for the detection of ONOO−. Unfortunately, TCFB1 had a low solubility in aqueous solution, which led to the observation of precipitates in cell imaging experiments. A glycosylation strategy31,32 to improve the water solubility of the insoluble TCFB1 is currently underway in our laboratories. However, TCFB2 displayed selective and sensitive “turn on” with the addition of ONOO−. The large fluorescence response observed for TCFB2 facilitated its use in cell imaging experiments. Therefore, TCFB2 was able to detect exogenous and endogenous ONOO− with a large fluorescence “turn on” over a range of cell lines (Hep-G2, RAW 264.7, HeLa and A459). Mitochondrial localisation of TCFB2 was observed by co-localisation with Mito-Tracker Green. Overall, these results demonstrate that TCFB2 is a useful tool to understand the role of ONOO− in biological systems and could lead to systems capable of disease diagnosis.
We would like to thank the EPSRC and the University of Bath for funding. A. C. S. and J. E. G. thank the EPSRC for studentships. T. D. J. wishes to thank the Royal Society for a Wolfson Research Merit Award. NMR characterisation facilities were provided through the Chemical Characterisation and Analysis Facility (CCAF) at the University of Bath (http://www.bath.ac.uk/ccaf). The EPSRC UK National Mass Spectrometry Facility at Swansea University is thanked for analyses. X.-P. He thanks the National Natural Science Foundation of China (21722801 and 21572058), the Science and Technology Commission of Shanghai Municipality (15540723800), the Fundamental Research Funds for the Central Universities (222201717003) and the Shanghai Rising-Star Program (16QA1401400) (to X.-P. He) for financial support. The Catalysis And Sensing for our Environment (CASE) network is thanked for research exchange opportunities. T. D. J. thanks ECUST for a guest professorship. Professors Jia Li and Yi Zang are thanked for their helpful advice on the cellular experiments. All data supporting this study are provided as supplementary information accompanying this paper.
Conflicts of interest
There are no conflicts to declare.Notes and references
- P. Pacher, J. S. Beckman and L. Liaudet, Physiol. Rev., 2007, 87, 315 CrossRef CAS PubMed.
- J. S. Beckman and W. H. Koppenol, Am. J. Physiol.: Cell Physiol., 1996, 271, C1424 CAS.
- A. Weidinger and A. V. Kozlov, Biomolecules, 2015, 5, 472 CrossRef CAS PubMed.
- P. Ascenzi, A. di Masi, C. Sciorati and E. Clementi, BioFactors, 2010, 36, 264 CrossRef CAS PubMed.
- H. Ischiropoulos and J. S. Beckman, J. Clin. Invest., 2003, 111, 163 CrossRef CAS PubMed.
- P. Sarchielli, F. Galli, A. Floridi and V. Gallai, Amino Acids, 2003, 25, 427 CrossRef CAS PubMed.
- D. A. Wink, Y. Vodovotz, J. Laval, F. Laval, M. W. Dewhirst and J. B. Mitchell, Carcinogenesis, 1998, 19, 711 CrossRef CAS PubMed.
- A. C. Sedgwick, X. L. Sun, G. Kim, J. Yoon, S. D. Bull and T. D. James, Chem. Commun., 2016, 52, 12350 RSC.
- Z. N. Sun, H. L. Wang, F. Q. Liu, Y. Chen, P. K. H. Tam and D. Yang, Org. Lett., 2009, 11, 1887 CrossRef CAS PubMed.
- X. Li, R.-R. Tao, L.-J. Hong, J. Cheng, Q. Jiang, Y.-M. Lu, M.-H. Liao, W.-F. Ye, N.-N. Lu, F. Han, Y.-Z. Hu and Y.-H. Hu, J. Am. Chem. Soc., 2015, 137, 12296 CrossRef CAS PubMed.
- F. B. A. Yu, P. Li, G. Y. Li, G. J. Zhao, T. S. Chu and K. L. Han, J. Am. Chem. Soc., 2011, 133, 11030 CrossRef CAS PubMed.
- F. B. Yu, P. Li, B. S. Wang and K. L. Han, J. Am. Chem. Soc., 2013, 135, 7674 CrossRef CAS PubMed.
- D. Cheng, Y. Pan, L. Wang, Z. B. Zeng, L. Yuan, X. B. Zhang and Y. T. Chang, J. Am. Chem. Soc., 2017, 139, 285 CrossRef CAS PubMed.
- L. Yuan, W. Y. Lin, K. B. Zheng, L. W. He and W. M. Huang, Chem. Soc. Rev., 2013, 42, 622 RSC.
- R. Weissleder, Nat. Biotechnol., 2001, 19, 316 CrossRef CAS PubMed.
- A. Sikora, J. Zielonka, M. Lopez, J. Joseph and B. Kalyanaraman, Free Radical Biol. Med., 2009, 47, 1401 CrossRef CAS PubMed.
- X. Sun, Q. Xu, G. Kim, S. E. Flower, J. P. Lowe, J. Yoon, J. S. Fossey, X. Qian, S. D. Bull and T. D. James, Chem. Sci., 2014, 5, 3368 RSC.
- S. Palanisamy, P. Y. Wu, S. C. Wu, Y. J. Chen, S. C. Tzou, C. H. Wang, C. Y. Chen and Y. M. Wang, Biosens. Bioelectron., 2017, 91, 849 CrossRef CAS PubMed.
- Y. H. Yang, J. L. Liu, H. Y. Xiao, Z. Zhen and S. H. Bo, Dyes Pigm., 2017, 139, 239 CrossRef CAS.
- W. Shu, L. G. Yan, Z. K. Wang, J. Liu, S. Zhang, C. Y. Liu and B. C. Zhu, Sens. Actuators, B, 2015, 221, 1130 CrossRef CAS.
- Y. J. Wang, Y. Shi, Z. Y. Wang, Z. F. Zhu, X. Y. Zhao, H. Nie, J. Qian, A. J. Qin, J. Z. Sun and B. Z. Tang, Chem. – Eur. J., 2016, 22, 9784 CrossRef CAS PubMed.
- Y. R. Wang, L. Feng, L. Xu, Y. Li, D. D. Wang, J. Hou, K. Zhou, Q. Jin, G. B. Ge, J. N. Cui and L. Yang, Chem. Commun., 2016, 52, 6064 RSC.
- B. C. Zhu, H. Kan, J. K. Liu, H. G. Liu, Q. Wei and B. Du, Biosens. Bioelectron., 2014, 52, 298 CrossRef CAS PubMed.
- C. Y. Li, M. Li, Y. Li, Z. S. Shi, Z. J. Li, X. B. Wang, J. Sun, J. W. Sun, D. M. Zhang and Z. C. Cui, J. Mater. Chem. C, 2016, 4, 8392 RSC.
- T. Yu, G. X. Yin, P. Yin, Y. Zeng, H. T. Li, Y. Y. Zhang and S. Z. Yao, RSC Adv., 2017, 7, 24822 RSC.
- M. Ipuy, C. Billon, G. Micouin, J. Samarut, C. Andraud and Y. Bretonniere, Org. Biomol. Chem., 2014, 12, 3641 CAS.
- E. W. Miller, A. E. Albers, A. Pralle, E. Y. Isacoff and C. J. Chang, J. Am. Chem. Soc., 2005, 127, 16652 CrossRef CAS PubMed.
- B. C. Dickinson, C. Huynh and C. J. Chang, J. Am. Chem. Soc., 2010, 132, 5906 CrossRef CAS PubMed.
- A. Vazquez-Torres, J. Jones-Carson and E. Balish, Infect. Immun., 1996, 64, 3127 CAS.
- M. D. Brand, C. Affourtit, T. C. Esteves, K. Green, A. J. Lambert, S. Miwa, J. L. Pakay and N. Parker, Free Radical Biol. Med., 2004, 37, 755 CrossRef CAS PubMed.
- X.-P. He, Y. Zang, T. D. James, J. Li, G.-R. Chen and J. Xie, Chem. Commun., 2017, 53, 82 RSC.
- J. Zhang, Y. Fu, H.-H. Han, Y. Zang, J. Li, X.-P. He, B. L. Feringa and H. Tian, Nat. Commun., 2017, 8, 987 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7cc07845e |
| This journal is © The Royal Society of Chemistry 2017 |