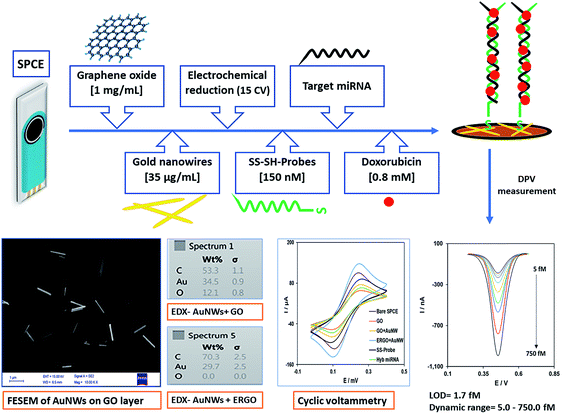
Open Access Article
Open Access ArticleEarly detection of Alzheimer's disease using a biosensor based on electrochemically-reduced graphene oxide and gold nanowires for the quantification of serum microRNA-137
Mostafa Azimzadehabc,
Navid Nasirizadeh†
 *d,
Mahdi Rahaie†
*d,
Mahdi Rahaie† *c and
Hossein Naderi-Manesh†*e
*c and
Hossein Naderi-Manesh†*e
aStem Cell Biology Research Center, Yazd Reproductive Sciences Institute, Shahid Sadoughi University of Medical Sciences, PO Box: 89195-999, Yazd, Iran
bDepartment of Advanced Medical Sciences and Technologies, School of Paramedicine, Shahid Sadoughi University of Medical Sciences, Yazd, 8916188635, Iran
cDepartment of Life Science Engineering, Faculty of New Sciences and Technologies, University of Tehran, Tehran, Iran 1439957131. E-mail: mrahaie@ut.ac.ir; Fax: +98 21 88497324; Tel: +98 21 86093408
dDepartment of Textile and Polymer Engineering, Yazd Branch, Islamic Azad University, Yazd, Iran 8916871967. E-mail: nasirizadeh@iauyazd.ac.ir; Fax: +98 35 38211109; Tel: +98 35 38211109
eDepartment of Nanobiotechnology/Biophysics, Faculty of Biological Sciences, Tarbiat Modares University, Tehran, Iran 14115-154. E-mail: naderman@modares.ac.ir; Fax: +98 21 82884420; Tel: +98 21 82884420
First published on 8th December 2017
Abstract
Alzheimer's disease (AD) is the most common form of dementia, and its early detection using reliable molecular biomarkers is believed to be best approach to controlling and even curing it. Herein, we develop an ultrasensitive electrochemical nanobiosensor to quantify serum miR-137 as a validated biomarker of AD. Electrochemically-reduced graphene oxide (ERGO) and gold nanowires (AuNWs) are used to modify the surface of a screen-printed carbon electrode (SPCE) with the application of an intercalated label, doxorubicin (Dox). The fabrication steps are analysed via field emission scanning electron microscopy (FE-SEM) and energy dispersive spectroscopy (EDS), as well as two reliable electrochemical methods, cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS). The results show that the linear range of the nanobiosensor is from 5.0 to 750.0 fM and its limit of detection is 1.7 fM. In addition, the nanobiosensor shows a great performance in specificity experiments and is able to discriminate between target oligos versus non-specific oligos (one-base mismatch target; three-base mismatch target, non-specific miR-21 and miR-155) very well. The evaluation of the sensing mechanism of the developed nanobiosensor in actual human serum reveals its potential clinical applications for the early detection of AD in the future.
1. Introduction
In recent decades, the field of biosensors has been growing, and the application of nanotechnology has developed as one of the biggest opportunities to achieve higher sensitivity for nanobiosensors.1,2 Among the different types of nanobiosensors, those based on electrochemical methods are still the most attractive due to their advantages of cost-effectiveness, easy production and easy use.3–5 There are many publications, patents and commercial products related to electrochemical nanobiosensors and their application for the detection of different types of biomarkers for different purposes.1,6 Undoubtedly, the early and fast detection of disease are the most important steps in curing them, in which electrochemical nanobiosensors have become fundamentally useful.1,7,8MicroRNAs (miRNAs) are a hot topic today, especially circulating miRNAs which are known as modern and reliable biomarkers for disease detection.9–11 They are short oligonucleotides that can specifically bind to their target mRNA in order to decrease or inhibit expression of the target gene and consequently production of the protein.9,12 Since 2007, electrochemical nanobiosensors for miRNA quantification has been rapidly developing.7,13–17
MicroRNA-137 (miR-137) (23 nucleotides) is known as a reliable non-invasive plasma biomarker for Alzheimer's disease (AD) and has been proven reliable for the detection of AD.18–22 AD is a neurodegenerative disease and is the most common type of dementia, which can cause different mental disabilities ranging from memory-loss to death.23–26 AD affects over 47 million worldwide which is expected to increase to more than 131 million in 2050. Nevertheless, there is still no commercial treatment available for this disease.20,27,28 Therefore, the early detection of AD is the most important step to prevent its progress; however, the conventional methods for AD detection, including medical imaging techniques, suffer from several problems and bias.29,30
Currently, there are some biosensors/nanobiosensors available for the quantification of different AD biomarkers using methods other than electrochemical methods,31 including fluorescent methods,19,32 surface plasmon resonance (SPR),33 localized surface plasmon resonance (LSPR),34 chemiluminescence35 and optical methods.36,37
Additionally, there are numerous publications regarding electrochemical biosensors/nanobiosensors for AD detection using different types of nanoparticles,38–43 where most of them used biomarkers other than miRNAs such as protease BACE1,44 apolipoprotein E,45,46 amyloid beta peptide,47,48 tau protein49 and acetylcholine.50
Also, there are a limited number of recent publications on electrochemical biosensors/nanobiosensors utilising miRNA for the detection of AD41 such as miR-34a;51–53 miR-16, -15a and -660 (ref. 54); miR-15a55 and miR-1306.56 However, thus far, to the best of our knowledge, there are no reports on electrochemical methods for the quantification of miR-137 for AD early detection and the present study is the first.
In this study, the application of gold nanowires decorated on the surface of electrochemically-reduced graphene oxide is used to modify a screen-printed carbon electrode to enhance the sensitivity of the proposed electrochemical nanobiosensor.
Graphene has been attracting much interest since its introduction in 2004 compared to other carbon-based materials, which is mainly due to its special fascinating properties especially in the biosensing field.57–59 The large surface area (its 2D structure constitutes the absolute maximum surface area to volume ratio in layered materials), superior conductivity, rapid heterogeneous electron transfer, and very high mechanical strength are the most important characteristics of graphene-based nanomaterials, which make them highly attractive nano/biomaterials for use in electrochemical biosensors to enhance their sensitivity.7,57,60–62 Furthermore, a derivative of graphene, namely graphene oxide (GO), possesses oxygen-containing groups which enhance the adsorption and desorption of molecules/nanomaterials to be used to improve sensitivity.57,58,63,64 In recent years, electrochemically reduced graphene oxide (ERGO) has been produced by the electrochemical reduction of graphene oxide, which is an efficient method to enhance its conductivity and electron transfer rate and can be easily applied to the graphene oxide layer on the electrode surface.65–67 It has been shown that the conductivity of ERGO is much higher than that of GO due to the elimination of oxygen-containing groups although some groups cannot be removed by electrochemical reduction such as carboxylic groups which can still be used for the attachment of molecules and materials.60
Gold nanowires (AuNWs) have been used in electrochemical nanobiosensors to enhance their sensitivity via different approaches, which takes advantage of the wonderful characteristics of gold nanoparticles with high aspect ratios.68–70
To date, to the best of our knowledge, the combination of AuNWs and ERGO has not been reported. Although, different forms of nanocomposites containing reduced graphene and gold nanoparticles have been used in electrochemical nanobiosensors.7,11,71–73 Based on our literature review, doxorubicin (Dox) has not been used as an electrochemical label in miRNA biosensors. We use it as an electrochemical label based on two important specifications. First, the mechanism of Dox and DNA interaction is proven to be an intercalation interaction, which means that Dox molecules prefer to be inserted between double stranded oligonucleotides (intercalation mechanism)74–76 which is helpful in the recognition of hybridized miRNA–DNA probe oligonucleotides over the single-stranded DNA probe. Second, the electroactive nature of the Dox molecule, which has also been proven in previous studies, makes it the perfect candidate as an electrochemical label in DNA/miRNA hybridization biosensors since its reduction peak can be easily measured in DPV experiments.74,77,78
2. Experimental
2.1. Chemicals and oligos
Ebedoxo (doxorubicin hydrochloride) (2.0 mg mL−1) as purchased from Ebewe (Ebewe Pharma Co., Austria). Bare CTAB modified gold nanowires (AuNWs) in distilled water with excess CTAB capping agent (35.0 μg mL−1) (A14-2000-CTAB-5) with a diameter of 75.0 nm and length of 2000.0 nm (aspect ratio of about 27) were purchased from Nanopartz Company (Nanopartz Inc. USA). Graphene oxide (GO) (1.0 mg mL−1), dispersion in H2O was purchased from Sigma-Aldrich Co. (USA). All other materials were analytical grade purchased from either Merck or Aldrich (upon availability) and used as received without further purification. Oligonucleotides were purchased from Generay Biotech. Co. (Shanghai, China) in HPLC purified-lyophilized powder form. miRNAs were ordered as miRNA-mimics oligos (thymine (T) was replaced by uracil (U)) for convenience. All miRNAs sequences were taken from the miRBase database (http://www.mirbase.org), and miRNA-137 (5′-TTATTGCTTAAGAATACGCGTAG-3′) was chosen as the target miRNA, and miRNA-21 was chosen as unspecific sequences in the selectivity experiments. One and three nucleotide replacements were made in target miR-137 to obtain the single- and three-base mismatched targets oligos, respectively. All solutions were prepared based on previous protocols.72.2. Electrochemical system, electrodes and modifications
An Autolab potentiostat/galvanostat model PGSTAT 101 (EcoChemic, Utrecht, Netherlands) and NOVA software (version 2.1) were used as the electrochemical system. A screen-printed carbon electrode (SPCE) (Model DRP-C110) (Dropsens Co., Spain) was used in which the working electrode (4.0 mm diameter) was made of carbon, the counter electrode was made of platinum and the reference electrode was made of silver.The working electrode of the SPE (carbon) was gently washed with 50% ethanol solution and then double-distilled sterilized water and dried with nitrogen stream before use. A 4.0 μL drop of graphene oxide solution (1.0 mg mL−1 in H2O) was applied on the working electrode which was kept in an isolated container until it completely dried. Afterwards, a 4.0 μL drop of gold nanowires (35.0 μg mL−1 in H2O, CTAB modified) was applied on the GO-modified working electrode for the self-assembly decoration of the positively-charged (CTAB modified) gold nanowires on the GO layer which does have oxygen-containing functional groups, and was kept in an isolated high-humidity container until the drop completely dried. Then, the electrochemical reduction of the GO was done based on a previously reported protocol67 by performing 15 continuous cyclic voltammetry cycles between 0.0 and −1.4 V in 50.0 μL, 0.1 M phosphate buffer at pH 6.5. Next, a 3.5 μL drop of ssDNA thiolated probe in immobilization solution (150.0 nM) was applied on the modified electrode which was also kept level in a high-humid environment for 100 minutes for the self-assembly attachment of the thiolated probes on the gold nanowire surfaces. Subsequently, the electrode was washed with sterile double distilled water to remove the unattached and disoriented probes. Then, the electrode was immersed in MCH solution for 5 min to fill the unattached area. Finally, the electrode was rinsed with an ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water solution (80
water solution (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 v/v) and then distilled water.
20 v/v) and then distilled water.
2.3. miRNA hybridization and electrochemical measurement
Once the modified electrode (named nanobiosensor) was ready, it was subjected to a 2.5 μL drop of the complementary miRNA sequence for 120 min which was prepared by dissolving different concentrations of the target oligonucleotide in the hybridization solution, and then the electrode was rinsed with washing solution. Next, a drop of homogenized Dox solution (0.8 mM final Dox concentration in 0.1 mM phosphate buffer solution (pH 7.0)) was applied to the surface of the working electrode for 15 min which was then rinsed twice with sterile double distilled water. The reduction signals of the accumulated Dox on the double strand hybridized oligonucleotides were measured via differential pulse voltammetry (DPV) at an amplitude of 25 mV, modulation time 0.05 s, and step potential of 50 mV in 1.0 M phosphate buffer solution (pH 7.0). The differential pulse voltammograms were subjected to moving average baseline correction with a peak width of 0.01.2.4. Characterization of electrode modification steps
Field emission scanning electron microscopy (FE-SEM) imaging and energy dispersive spectroscopy (EDS) analysis were performed using a Zeiss Sigma 500 VP FESEM instrument with an integrated EDS Oxford Instrument microanalysis detector to verify the exact decoration and composition of the nanomaterials on the surface of the electrode.Additionally, after each preparation step cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) experiments were performed on the modified electrode in [Fe(CN)6]3−/4− solution based on a previously reported protocol.5
2.5. Specificity and real sample study
The selectivity of the biosensor was evaluated by testing in different mixed solutions of specific (target miR-137) and non-specific oligonucleotides (one- and three-base mismatch oligos) and completely non-specific miRNAs (miR-155 and miR-21). The rest of the experiment was the same as that the previous sections and the DPV currents were used to construct a bar chart. All experiments were performed in four replicates.In addition to the specificity assessment, a real sample environment (human serum) was used to evaluate the performance of the nanobiosensor in real clinical samples with biological and chemical moieties as interferences. For this, isolated human serum was used as the real sample environment to dissolve synthetic miRNA to simulate the actual serum of an Alzheimer's patient. To assess the clinical application of the proposed nanobiosensor, three different concentrations of miR-137 (20.0, 100.0, and 500.0 fM) were prepared in the serum of a healthy normal person (33 year old man) and then the prepared modified electrode was applied to quantify the concentrations of miRNA using a similar protocol to that in the synthetic environment experiments. These experiments were performed in four replicates. All experiments were performed according to the ethical principles and national guidelines for conducting medical research on human tissue and organs in Iran, developed by the Research Ethics Committee of the Ministry of Health and Medical Education, Iran, and was approved by the Faculty of New Sciences and Technologies Bioethics Committee of the University of Tehran. In addition, informed consent was obtained from the involved serum donor.
3. Results and discussion
A brief schematic overview of the stages of the nanobiosensor preparation, concentrations of materials and characterization are presented in Scheme 1, as was described previously in the Materials and methods section.3.1. Characterization of the modified electrode
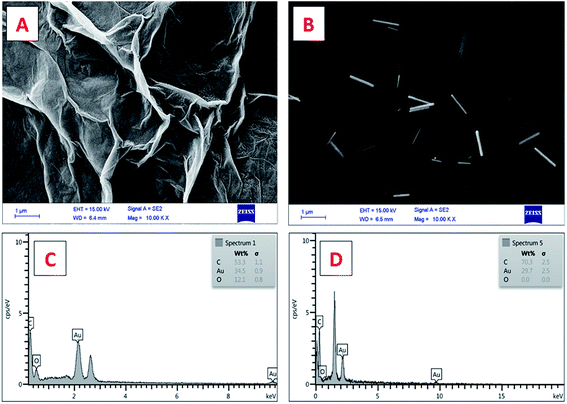
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×) of the working electrode surface modified with GO (Fig. 1A) and ERGO + AuNWs (Fig. 1B) was used to confirm the applied modifications. As it can be seen in Fig. 1A, a layer of GO covered the electrode surface and in Fig. 1B, the AuNWs are homogenously distributed and decorated with different alignments and depth on the GO surface. In addition, the size, wire-form morphology and shape of the gold nanowires are exhibited in Fig. 1B.
000×) of the working electrode surface modified with GO (Fig. 1A) and ERGO + AuNWs (Fig. 1B) was used to confirm the applied modifications. As it can be seen in Fig. 1A, a layer of GO covered the electrode surface and in Fig. 1B, the AuNWs are homogenously distributed and decorated with different alignments and depth on the GO surface. In addition, the size, wire-form morphology and shape of the gold nanowires are exhibited in Fig. 1B.
Furthermore, the chemical composition of the surface of the modified electrode was confirmed by EDX analysis, which is one of the important characterization methods for evaluating the surface of electrodes in electrochemical experiments. The EDX spectra of the GO AuNWs modified electrode before (Fig. 1C) and after electrochemical reduction of GO to make the ERGO + AuNWs modified electrode (Fig. 1D) are presented. The spectrum in Fig. 1C clearly depicts the presence of carbon and oxygen which originate from the graphene oxide sheet and gold element corresponding to the AuNWs. In contrast, in Fig. 1D, only carbon and gold elements are present on the surface of the electrode. From the comparison of the two spectra, the percentage of oxygen element dramatically decreased due to the electrochemical reduction of graphene oxide and elimination of the oxygen containing functional groups.
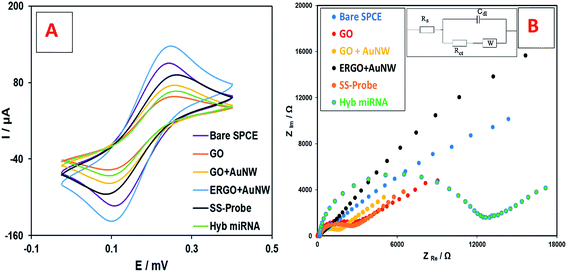 | ||
| Fig. 2 Electrochemical analysis of the different stages of nanobiosensor preparation using (A) CV, and (B) EIS analysis in [Fe(CN)6]3−/4− solution. | ||
Fig. 2B shows the Nyquist plots from the EIS study and the electrical equivalent circuit. It can be seen that the Rct of the curve of the bare SPCE electrode is low (1860 Ω) which increased by the addition GO to the SPCE surface (3329 Ω) due to its insulating nature. However, the addition of AuNWs increased the electron transfer ability and therefore the Rct decreased (1944 Ω). After electrochemical reduction, the Rct decreased even lower than the bare SPCE (1335 Ω). This is mainly due to the reduction of graphene oxide, which turned it to a very conductive material. Afterwards, the addition of SS-probes and the target miRNA increased the Rct to 2449 Ω and 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 813 Ω, respectively. The trend of Rct changes during the preparation of the nanobiosensor is consistent with the results of the CV study, which was explained earlier. Therefore, both the CV and EIS studies explain the step-by-step modifications of the electrode surface in the fabrication of the nanobiosensor.
813 Ω, respectively. The trend of Rct changes during the preparation of the nanobiosensor is consistent with the results of the CV study, which was explained earlier. Therefore, both the CV and EIS studies explain the step-by-step modifications of the electrode surface in the fabrication of the nanobiosensor.
3.2. Specificity of the nanobiosensor
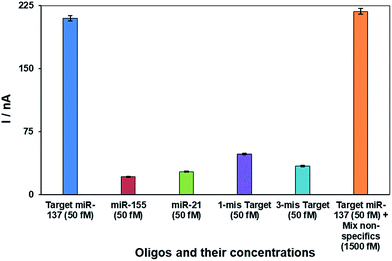
The selectivity of the nanobiosensor for the target miRNA (miR-137) over non-specific oligos (one-base mismatch target; three-base mismatch target, non-specifics miR-21 and miR-155) is shown in Fig. 3. The error bars are from four replicates of each experiment. The chart clearly depicts that the proposed nanobiosensor successfully distinguishes between 50.0 fM of target miR-137 and non-specific oligos. In addition, a mixture solution of non-specific miRNAs (containing 1500.0 fM of miR-21 and miR-155) with a 30-fold higher concentration of target miRNA (50.0 fM of miR-137) was tested by the nanobiosensor and the results show the high selectivity of the nanobiosensor even at higher concentrations of non-specific oligos over the target oligo. These results are attributed to the intercalation mechanism of doxorubicin,76,81 which has a greater affinity to double-stranded DNA oligos than single-stranded oligos. Therefore, the nanobiosensor has the ability to distinguish between the target miRNA, which forms a complete double strand with the SS-probe, and non-specific oligos which are unable to form a complete double-stranded structure.3.3. Analytical performance of the nanobiosensor
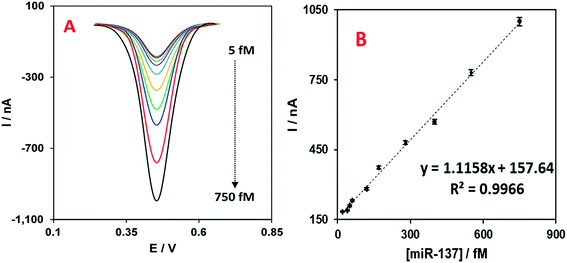
The ideal analytical performance of designed detection/quantification systems is to detect low concentrations and wide linear ranges of target molecule, as well as exhibiting acceptable reproducibility. To check the performance quality, we assessed the developed miR-137 electrochemical nanobiosensor in different concentrations of the target oligo to determine the linear range of concentrations it can detect. The DPV linear range concentration results are shown in Fig. 4A with 10 different concentrations (5.0; 25.0; 50.0; 60.0; 120; 170.0; 280.0; 400.0; 550.0 and 750.0 fM of miR-137). In addition, the calibration plot of the nanobiosensor for these concentrations versus their DPV peak currents are shown in Fig. 4B and in addition to the trend line and regression coefficient. The detection limit was calculated to be 1.7 fM based on the slope of the trendline of the calibration plot and the method explained before.5 The error bars in Fig. 4B were calculated based on four replicates for each experiment.The reproducibility of the nanobiosensor was tested by performing 14 replicate experiments with the nanobiosensor detecting 50.0 fM of the target miR-137. From the results, the mean DPV peak current was 210.0 ± 5.2 nA which is about 2.5% of the relative standard deviation (RSD). This low RSD represents the good reproducibility of the nanobiosensor and the functionality of the fabrication method for future potential applications.
The stability of the nanobiosensor or its storage ability was assessed for different time periods. The fabricated nanobiosensor was stored in 0.1 M PBS pH 7.0 solution inside the refrigerator (4 °C) for 5 to 20 days and then hybridized with 50.0 fM of target miR-137 and analysed with the same protocol reported earlier in Section 2.3. DNA hybridization and electrochemical measurement. The reduction peak current of Dox on the hybridized SS-probe-target miRNA was compared to that for the freshly fabricated nanobiosensor and the percentage of the performance is reported together with the RSD value for 4 replicate experiments in Table 1.
As it can be seen in the Table 1, the performance of the biosensor after 20 days of storage in a refrigerator just decreased to 19.6 percent, which is acceptable for a nanobiosensor composed of nanomaterials and SSDNA probes. This result indicates that the storage ability of the fabricated electrochemical nanobiosensor is another good characteristic for its commercial application in clinics.
| Duration of storage (days) | Average response of the nanobiosensor (%) | RSD (%) |
|---|---|---|
| 0 | 100 | 3.6 |
| 5 | 96.4 | 4.5 |
| 10 | 91.9 | 4.3 |
| 15 | 86.2 | 3.9 |
| 20 | 80.4 | 4.8 |
Table 2 presents a comparison of our results with previously published papers regarding the electrochemical early detection of Alzheimer's miRNAs. As it can be seen in this table, the proposed nanobiosensor shows a better linear range and detection limit in comparison with the others.51–55 Additionally, the combination of ERGO and AuNWs has not been used before in the references listed in Table 2.
| miRNA | Electrode/method | Nanomaterials | Linear range | LOD | Ref. |
|---|---|---|---|---|---|
| miR-34a | PGE/EIS | GO | 0.1–10.0 mg mL−1 | 1.9 mg mL−1 | 51 |
| miR-34a | PGE/DPV | GO | 5.0–35.0 μg mL−1 | 7.5 μg mL−1 | 52 |
| miR-34a | SPE (graphite)/DPV | CNF | 25.0–100.0 μg mL−1 | 10.9 μg mL−1 | 53 |
| miR-16 | MUX-SPE16/DPV | Magnetic beads | 5.0–100.0 μg mL−1 | 1.4 pmol μL−1 | 54 |
| miR-15a | MUX-SPE16/LSV | Magnetic beads | 0.5–3.0 μg mL−1 | 11.4 fmol μL−1 | 55 |
| miR-1306 | Ni/EIS | Graphene | 0.1 pM to 1.0 nM | 0.8 fM | 56 |
| miR-137 | SPCE/DPV | ERGO + AuNWs | 5.0–750.0 fM | 1.7 fM | This work |
The higher sensitivity of our biosensor could be explained by combined application of SPCE electrodes, ERGO, AuNWs and Dox. Previous studies reported some advantages of SPE electrodes including low production cost, high reproducibility and reliability, and also portability of SPE.82,83 Furthermore, the application of ERGO84,85 and AuNWs86,87 has been reported to work well in enhancing the sensitivity of nanobiosensors, which is mainly due to their high electron transport rate in electrochemical studies,88–90 as was explained herein. This is also concluded from comparing our results to three previous publications on electrochemical nanobiosensor for oligonucleotides. The results of the present study are relatively better than our previous report which used graphene oxide and gold nanorods on a glassy carbon electrode for the detection of serum miR-155,7 and also much better sensitivity than other miRNA electrochemical systems without the application of nanomaterials with a detection limit of 13.5 pM.5 The improved sensitivity in the present research might originate from the fact that ERGO was used instead of the GO which possesses better electrical conductivity, and also the application of gold nanowires instead of gold nanorods which are better media for guided electron transfer. Moreover, the sensitivity of the present nanobiosensor is also better than the study by Aghili and colleagues, in which they applied a combination of exfoliated graphene oxide and gold nano-urchins resulting in a sensitivity of 13.0 fM for DNA detection.91 Finally, Hajihosseini and colleagues applied graphene oxide and gold nanoparticles for DNA detection and they achieved the detection limit of 27.0 pM of for bacterial DNA.92
In addition to application of nanomaterials, the use of an appropriate electrochemical intercalating label, such as doxorubicin, can result in sensitivity enhancement in electrochemical nanobiosensors.74,76,93 Dox could produce reduction signals with very low noise. Therefore, the selectivity and sensitivity of our nanobiosensor were enhanced by using Dox as an electrochemical label. Moreover, the low reduction potential of Dox is an advantage in real sample studies, which enables the nanobiosensor to selectively recognize the target miRNA among other electroactive biological molecules and moieties available in the human serum involved in signalling at higher potentials and thus interfere with the signalling.
3.4. Assessment in spiked serum sample
The developed nanobiosensor was also tested in the human serum environment to assess the functionality of its sensing mechanism for the detection of the circulating biomarker of Alzheimer's disease, miR-137, in real clinical samples. Based on the standard addition method, the isolated human serum from a healthy donor was tested for miR-137 initial concentration which showed zero concentration of target miR-137 and then, three known concentrations of target miRNA were added to the isolated human serum and the nanobiosensor was used again to quantify them. The results of the relative recovery percentage of the three concentrations are listed in Table 3, as well as the relative standard deviation of four replicates of each experiment. As it can be concluded from Table 3, the RSD values are good and acceptable and the recovery percentages are close to 100 percent which is a sign of the great detection ability of the biosensor in a real sample environment without being affected by the other molecules and moieties in human serum. In conclusion, the proposed nanobiosensor can be considered as a new and reliable method with high sensitivity, selectivity and stable performance in real clinical sample evaluations for the early detection of the Alzheimer's disease.| Added miR-137 (fM) | Detected miR-137 (fM) | Relative recovery (%) (4 replicates) | Relative standard deviation (%) |
|---|---|---|---|
| 20.0 | 20.06 | 100.3 | 3.82 |
| 100.0 | 98.21 | 98.2 | 3.18 |
| 500.0 | 498.01 | 99.6 | 2.90 |
4. Conclusions
There has been great development in the area of miRNA electrochemical nanobiosensing in the past decade since the first report on this topic in 2007. In this field, the main aim of scientists is to develop sensitive, selective and applicable detection methods and make diagnosis and prognosis easier for clinical technicians and medical doctors. The present study aimed to improve the diagnosis of Alzheimer's disease in its early stages using an easier, cheaper and more accurate approach than the current methods. The application of nanomaterials such as electrochemical reduced graphene oxide and gold nanowires, together with the application of screen-printed electrodes and also an electrochemical label with an intercalating mechanism enabled the proposed nanobiosensor to achieve very good sensitivity as low as 1.7 fM and also a broad linear range for the quantification of miR-137 as a confirmed and reliable biomarker for the early detection of Alzheimer's disease. The accuracy and reliability of the nanobiosensor fabrication steps were verified through different highly valued techniques and also the functionality of the nanobiosensor was demonstrated in a real sample of human plasma containing miR-137. Due to its advantages over the previous publications in this area, the proposed nanobiosensor has potential future clinical application in the early detection of Alzheimer's disease even before main clinical symptoms appear, which can stop the disease progress in its earliest stages.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are sincerely grateful to the University of Tehran, Tarbiat Modares University and Islamic Azad University of Yazd, for their supports.Notes and references
- S. N. Topkaya, M. Azimzadeh and M. Ozsoz, Electrochemical Biosensors for Cancer Biomarkers Detection: Recent Advances and Challenges, Electroanalysis, 2016, 28, 1402–1419 CrossRef CAS
.
- C. I. Justino, T. A. Rocha-Santos, S. Cardoso and A. C. Duarte, Strategies for enhancing the analytical performance of nanomaterial-based sensors, TrAC, Trends Anal. Chem., 2013, 47, 27–36 CrossRef CAS
.
- L. Syedmoradi, M. Daneshpour, M. Alvandipour, F. A. Gomez, H. Hajghassem and K. Omidfar, Point of care testing: the impact of nanotechnology, Biosens. Bioelectron., 2017, 87, 373–387 CrossRef CAS PubMed
.
- S. Shojaei, N. Nasirizadeh, M. Entezam, M. Koosha and M. Azimzadeh, An Electrochemical Nanosensor Based on Molecularly Imprinted Polymer (MIP) for Detection of Gallic Acid in Fruit Juices, Food Analytical Methods, 2016, 1–11 Search PubMed
.
- M. Azimzadeh, M. Rahaie, N. Nasirizadeh and H. Naderi-Manesh, Application of Oracet Blue in a novel and sensitive electrochemical biosensor for the detection of microRNA, Anal. Methods, 2015, 7(22), 9495–9503 RSC
.
- M. Daneshpour, K. Omidfar and H. Ghanbarian, A novel electrochemical nanobiosensor for the ultrasensitive and specific detection of femtomolar-level gastric cancer biomarker miRNA-106a, Beilstein J. Nanotechnol., 2016, 7(1), 2023–2036 CrossRef CAS PubMed
.
- M. Azimzadeh, M. Rahaie, N. Nasirizadeh, K. Ashtari and H. Naderi-Manesh, An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer, Biosens. Bioelectron., 2016, 77, 99–106 CrossRef CAS PubMed
.
- Q. Liang, H. Liu, T. Zhang, Y. Jiang, H. Xing and A.-h. Zhang, Metabolomics-based screening of salivary biomarkers for early diagnosis of Alzheimer's disease, RSC Adv., 2015, 5(116), 96074–96079 RSC
.
- J. Li, S. Tan, R. Kooger, C. Zhang and Y. Zhang, MicroRNAs as novel biological targets for detection and regulation, Chem. Soc. Rev., 2014, 43(2), 506–517 RSC
.
- H. H. Cheng, H. S. Yi, Y. Kim, E. M. Kroh, J. W. Chien, K. D. Eaton, M. T. Goodman, J. F. Tait, M. Tewari and C. C. Pritchard, Plasma processing conditions substantially influence circulating microRNA biomarker levels, PLoS One, 2013, 8(6), e64795 CAS
.
- M. Azimzadeh, M. Rahaie, N. Nasirizadeh, M. Daneshpour and H. Naderi-Manesh, Electrochemical miRNA Biosensors: The Benefits of Nanotechnology, Nanomed. Res. J., 2017, 1(3), 158–171 Search PubMed
.
- V. Ambros, The functions of animal microRNAs, Nature, 2004, 431(7006), 350–355 CrossRef CAS PubMed
.
- L. Wang, Q. Xiong, F. Xiao and H. Duan, 2D nanomaterials based electrochemical biosensors for cancer diagnosis, Biosens. Bioelectron., 2017, 89, 136–151 CrossRef CAS PubMed
.
- S. N. Topkaya, M. Azimzadeh and M. Ozsoz, Electrochemical Biosensors for Cancer Biomarkers Detection: Recent Advances and Challenges, Electroanalysis, 2016, 28, 1402–1419 CrossRef CAS
.
- M. Keshavarz, M. Behpour and H.-A. Rafiee-pour, Recent trends in electrochemical microRNA biosensors for early detection of cancer, RSC Adv., 2015, 5(45), 35651–35660 RSC
.
- P. Miao, Y. Tang and J. Yin, MicroRNA detection based on analyte triggered nanoparticle localization on a tetrahedral DNA modified electrode followed by hybridization chain reaction dual amplification, Chem. Commun., 2015, 51(86), 15629–15632 RSC
.
- P. Miao, Y. Tang, B. Wang, C. Jiang, L. Gao, B. Bo and J. Wang, Nuclease assisted target recycling and spherical nucleic acids gold nanoparticles recruitment for ultrasensitive detection of microRNA, Electrochim. Acta, 2016, 190, 396–401 CrossRef CAS
.
- V. Viswambharan, I. Thanseem, M. M. Vasu, S. A. Poovathinal and A. Anitha, miRNAs as biomarkers of neurodegenerative disorders, Biomarkers, 2017, 11(2), 151–167 CrossRef CAS PubMed
.
- S. Delkhahi, M. Rahaie and F. Rahimi, Design and Fabrication a Gold Nanoparticle-DNA Based Nanobiosensor for Detection of microRNA Involved in Alzheimer's Disease, J. Fluoresc., 2016, 1–8 Search PubMed
.
- H. Z. Y. Wu, K. L. Ong, K. Seeher, N. J. Armstrong, A. Thalamuthu, H. Brodaty, P. Sachdev and K. Mather, Circulating microRNAs as biomarkers of Alzheimer's disease: a systematic review, J. Alzheimer's Dis., 2016, 49(3), 755–766 CrossRef CAS PubMed
.
- H. Geekiyanage and C. Chan, MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid β, novel targets in sporadic Alzheimer's disease, J. Neurosci., 2011, 31(41), 14820–14830 CrossRef CAS PubMed
.
- S. Kumar and P. H. Reddy, Are circulating microRNAs peripheral biomarkers for Alzheimer's disease?, Biochim. Biophys. Acta, Mol. Basis Dis., 2016, 1862(9), 1617–1627 CrossRef CAS PubMed
.
- M. T. Heneka, D. T. Golenbock and E. Latz, Innate immunity in Alzheimer's disease, Nat. Immunol., 2015, 16(3), 229–236 CrossRef CAS PubMed
.
- C. Reitz and R. Mayeux, Alzheimer disease: epidemiology, diagnostic criteria, risk factors and biomarkers, Biochem. Pharmacol., 2014, 88(4), 640–651 CrossRef CAS PubMed
.
- J. M. Hill and W. J. Lukiw, microRNA (miRNA)-mediated pathogenetic signaling in Alzheimer's disease (AD), Neurochem. Res., 2016, 41(1–2), 96–100 CrossRef CAS PubMed
.
- Z. Liu, A. Zhang, H. Sun, Y. Han, L. Kong and X. Wang, Two decades of new drug discovery and development for Alzheimer's disease, RSC Adv., 2017, 7(10), 6046–6058 RSC
.
- B. Imtiaz, A.-M. Tolppanen, M. Kivipelto and H. Soininen, Future directions in Alzheimer's disease from risk factors to prevention, Biochem. Pharmacol., 2014, 88(4), 661–670 CrossRef CAS PubMed
.
- M. Prince, A. Comas-Herrera, M. Knapp, M. Guerchet and M. Karagiannidou, World Alzheimer report 2016: improving healthcare for people living with dementia: coverage, quality and costs now and in the future, 2016 Search PubMed
.
- S. Teipel, A. Drzezga, M. J. Grothe, H. Barthel, G. Chételat, N. Schuff, P. Skudlarski, E. Cavedo, G. B. Frisoni and W. Hoffmann, Multimodal imaging in Alzheimer's disease: validity and usefulness for early detection, Lancet Neurol., 2015, 14(10), 1037–1053 CrossRef PubMed
.
- C. Laske, H. R. Sohrabi, S. M. Frost, K. López-de-Ipiña, P. Garrard, M. Buscema, J. Dauwels, S. R. Soekadar, S. Mueller and C. Linnemann, Innovative diagnostic tools for early detection of Alzheimer's disease, Alzheimer's Dementia, 2015, 11(5), 561–578 CrossRef PubMed
.
- S. Laurent, M. R. Ejtehadi, M. Rezaei, P. G. Kehoe and M. Mahmoudi, Interdisciplinary challenges and promising theranostic effects of nanoscience in Alzheimer's disease, RSC Adv., 2012, 2(12), 5008–5033 RSC
.
- L. Lai, C. Zhao, X. Li, X. Liu, H. Jiang, M. Selke and X. Wang, Fluorescent gold nanoclusters for in vivo target imaging of Alzheimer's disease, RSC Adv., 2016, 6(36), 30081–30088 RSC
.
- K. Hegnerová, M. Bockova, H. Vaisocherová, Z. Krištofiková, J. Říčný, D. Řípová and J. Homola, Surface plasmon resonance biosensors for detection of Alzheimer disease biomarker, Sens. Actuators, B, 2009, 139(1), 69–73 CrossRef
.
- A. J. Haes, W. P. Hall, L. Chang, W. L. Klein and R. P. Van Duyne, A Localized Surface Plasmon Resonance Biosensor: First Steps toward an Assay for Alzheimer's, Molecules, 2004, 5, 7–9 Search PubMed
.
- B. Li and Z. Zhang, Chemiluminescence flow biosensor for determination of total D-amino acid in serum with immobilized reagents, Sens. Actuators, B, 2000, 69(1), 70–74 CrossRef CAS
.
- A. J. Haes, L. Chang, W. L. Klein and R. P. Van Duyne, Detection of a biomarker for Alzheimer's disease from synthetic and clinical samples using a nanoscale optical biosensor, J. Am. Chem. Soc., 2005, 127(7), 2264–2271 CrossRef CAS PubMed
.
- R.-a. Doong, P.-S. Lee and K. Anitha, Simultaneous determination of biomarkers for Alzheimer's disease using sol–gel-derived optical array biosensor, Biosens. Bioelectron., 2010, 25(11), 2464–2469 CrossRef CAS PubMed
.
- D. Brambilla, B. Le Droumaguet, J. Nicolas, S. H. Hashemi, L.-P. Wu, S. M. Moghimi, P. Couvreur and K. Andrieux, Nanotechnologies for Alzheimer's disease: diagnosis, therapy, and safety issues, Nanomedicine, 2011, 7(5), 521–540 CrossRef CAS PubMed
.
- H. V. Ganesh, A. M. Chow and K. Kerman, Recent advances in biosensors for neurodegenerative disease detection, TrAC, Trends Anal. Chem., 2016, 79, 363–370 CrossRef CAS
.
- A. Kaushik, R. D. Jayant, S. Tiwari, A. Vashist and M. Nair, Nano-biosensors to detect beta-amyloid for Alzheimer's disease management, Biosens. Bioelectron., 2016, 80, 273–287 CrossRef CAS PubMed
.
- S. Scarano, S. Lisi, C. Ravelet, E. Peyrin and M. Minunni, Detecting Alzheimer's disease biomarkers: from antibodies to new bio-mimetic receptors and their application to established and emerging bioanalytical platforms-A critical review, Anal. Chim. Acta, 2016, 940, 21–37 CrossRef CAS PubMed
.
- A. Veloso and K. Kerman, Advances in electrochemical detection for study of neurodegenerative disorders, Anal. Bioanal. Chem., 2013, 405(17), 5725–5741 CrossRef CAS PubMed
.
- Y. Wang, Y. Zhang, C. Hou and M. Liu, Magnetic Fe3O4@MOFs decorated graphene nanocomposites as novel electrochemical sensor for ultrasensitive detection of dopamine, RSC Adv., 2015, 5(119), 98260–98268 RSC
.
- F. Qu, M. Yang and A. Rasooly, Dual Signal Amplification Electrochemical Biosensor for Monitoring the Activity and Inhibition of the Alzheimer's Related Protease β-Secretase, Anal. Chem., 2016, 88(21), 10559–10565 CrossRef CAS PubMed
.
- L. Wu, H. Ji, H. Sun, C. Ding, J. Ren and X. Qu, Label-free ratiometric electrochemical detection of the mutated apolipoprotein E gene associated with Alzheimer's disease, Chem. Commun., 2016, 52(81), 12080–12083 RSC
.
- A. Mars, W. Argoubi, S. B. Aoun and N. Raouafi, Induced conformational change on ferrocenyl-terminated alkyls and their application as transducers for label-free immunosensing of Alzheimer's disease biomarker, RSC Adv., 2016, 6(3), 2414–2421 RSC
.
- P. Carneiro, J. Loureiro, C. Delerue-Matos, S. Morais and M. do Carmo Pereira, Alzheimer's disease: development of a sensitive label-free electrochemical immunosensor for detection of amyloid beta peptide, Sens. Actuators, B, 2017, 239, 157–165 CrossRef CAS
.
- J. Gonçalves, L. Lima, M. Moraes and S. Ribeiro, Immunosensor for diagnosis of Alzheimer disease using amyloid-β 1–40 peptide and silk fibroin thin films, Mater. Sci. Eng., C, 2016, 68, 338–342 CrossRef PubMed
.
- S. X. Wang, D. Acha, A. J. Shah, F. Hills, I. Roitt, A. Demosthenous and R. H. Bayford, Detection of the tau protein in human serum by a sensitive four-electrode electrochemical biosensor, Biosens. Bioelectron., 2017, 92, 482–488 CrossRef CAS PubMed
.
- F. T. Moreira, M. G. F. Sale and M. Di Lorenzo, Towards timely Alzheimer diagnosis: a self-powered amperometric biosensor for the neurotransmitter acetylcholine, Biosens. Bioelectron., 2017, 87, 607–614 CrossRef CAS PubMed
.
- G. Congur, E. Eksin and A. Erdem, Impedimetric detection of microRNA at graphene oxide modified sensors, Electrochim. Acta, 2015, 172, 20–27 CrossRef CAS
.
- D. Isin, E. Eksin and A. Erdem, Graphene oxide modified single-use electrodes and their application for voltammetric miRNA analysis, Mater. Sci. Eng., C, 2017, 75, 1242–1249 CrossRef CAS PubMed
.
- A. Erdem, E. Eksin and G. Congur, Indicator-free electrochemical biosensor for microRNA detection based on carbon nanofibers modified screen printed electrodes, J. Electroanal. Chem., 2015, 755, 167–173 CrossRef CAS
.
- A. Erdem and G. Congur, Label-free voltammetric detection of MicroRNAs at multi-channel screen printed array of electrodes comparison to graphite sensors, Talanta, 2014, 118, 7–13 CrossRef CAS PubMed
.
- A. Erdem, G. Congur and E. Eksin, Multi channel screen printed array of electrodes for enzyme-linked voltammetric detection of microRNAs, Sens. Actuators, B, 2013, 188, 1089–1095 CrossRef CAS
.
- D. H. Seo, S. Pineda, J. Fang, Y. Gozukara, S. Yick, A. Bendavid, S. K. H. Lam, A. T. Murdock, A. B. Murphy and Z. J. Han, Single-step ambient-air synthesis of graphene from renewable precursors as electrochemical genosensor, Nat. Commun., 2017, 8, 14217 CrossRef CAS PubMed
.
- M. Pumera, Graphene in biosensing, Mater. Today, 2011, 14(7), 308–315 CrossRef CAS
.
- M. S. Artiles, C. S. Rout and T. S. Fisher, Graphene-based hybrid materials and devices for biosensing, Adv. Drug Delivery Rev., 2011, 63(14), 1352–1360 CrossRef CAS PubMed
.
- W. Yang, K. R. Ratinac, S. P. Ringer, P. Thordarson, J. J. Gooding and F. Braet, Carbon nanomaterials in biosensors: should you use nanotubes or graphene?, Angew. Chem., Int. Ed., 2010, 49(12), 2114–2138 CrossRef CAS PubMed
.
- M. Pumera, Graphene-based nanomaterials and their electrochemistry, Chem. Soc. Rev., 2010, 39(11), 4146–4157 RSC
.
- T. Gan and S. Hu, Electrochemical sensors based on graphene materials, Microchim. Acta, 2011, 175(1–2), 1 CrossRef CAS
.
- D. Chen, L. Tang and J. Li, Graphene-based materials in electrochemistry, Chem. Soc. Rev., 2010, 39(8), 3157–3180 RSC
.
- D. Chen, H. Feng and J. Li, Graphene oxide: preparation, functionalization, and electrochemical applications, Chem. Rev., 2012, 112(11), 6027–6053 CrossRef CAS PubMed
.
- M. Veerapandian, M.-H. Lee, K. Krishnamoorthy and K. Yun, Synthesis, characterization and electrochemical properties of functionalized graphene oxide, Carbon, 2012, 50(11), 4228–4238 CrossRef CAS
.
- M. A. Raj and S. A. John, Fabrication of electrochemically reduced graphene oxide films on glassy carbon electrode by self-assembly method and their electrocatalytic application, J. Phys. Chem. C, 2013, 117(8), 4326–4335 CAS
.
- X. Xu, D. Huang, K. Cao, M. Wang, S. M. Zakeeruddin and M. Grätzel, Electrochemically reduced graphene oxide multilayer films as efficient counter electrode for dye-sensitized solar cells, Sci. Rep., 2013, 3, 1489 CrossRef PubMed
.
- S. Kurbanoglu, L. Rivas, S. A. Ozkan and A. Merkoçi, Electrochemically reduced graphene and iridium oxide nanoparticles for inhibition-based angiotensin-converting enzyme inhibitor detection, Biosens. Bioelectron., 2017, 88, 122–129 CrossRef CAS PubMed
.
- E. Ogabiela, S. B. Adeloju, J. Cui, Y. Wu and W. Chen, A novel ultrasensitive phosphate amperometric nanobiosensor based on the integration of pyruvate oxidase with highly ordered gold nanowires array, Biosens. Bioelectron., 2015, 71, 278–285 CrossRef CAS PubMed
.
- E. Spain, A. McCooey, K. Joyce, T. E. Keyes and R. J. Forster, Gold nanowires and nanotubes for high sensitivity detection of pathogen DNA, Sens. Actuators, B, 2015, 215, 159–165 CrossRef CAS
.
- E. T. Carlen and v. d. A. Berg, Nanowire electrochemical sensors: can we live without labels?, Lab Chip, 2007, 7, 19–23 RSC
.
- S. Liu, J. Yan, G. He, D. Zhong, J. Chen, L. Shi, X. Zhou and H. Jiang, Layer-by-layer assembled multilayer films of reduced graphene oxide/gold nanoparticles for the electrochemical detection of dopamine, J. Electroanal. Chem., 2012, 672, 40–44 CrossRef CAS
.
- F.-Y. Kong, M.-T. Xu, J.-J. Xu and H.-Y. Chen, A novel lable-free electrochemical immunosensor for carcinoembryonic antigen based on gold nanoparticles–thionine–reduced graphene oxide nanocomposite film modified glassy carbon electrode, Talanta, 2011, 85(5), 2620–2625 CrossRef CAS PubMed
.
- J. Li, H. Feng, J. Li, Y. Feng, Y. Zhang, J. Jiang and D. Qian, Fabrication of gold nanoparticles-decorated reduced graphene oxide as a high performance electrochemical sensing platform for the detection of toxicant Sudan I, Electrochim. Acta, 2015, 167, 226–236 CrossRef CAS
.
- H. C. Yau, H. L. Chan and M. Yang, Electrochemical properties of DNA-intercalating doxorubicin and methylene blue on n-hexadecyl mercaptan-doped 5′-thiol-labeled DNA-modified gold electrodes, Biosens. Bioelectron., 2003, 18(7), 873–879 CrossRef CAS PubMed
.
- F. Zunino, A. Di Marco, A. Zaccara and R. A. Gambetta, The interaction of daunorubicin and doxorubicin with DNA and chromatin, Biochim. Biophys. Acta, Nucleic Acids Protein Synth., 1980, 607(2), 206–214 CrossRef CAS
.
- D. Agudelo, P. Bourassa, G. Bérubé and H.-A. Tajmir-Riahi, Intercalation of antitumor drug doxorubicin and its analogue by DNA duplex: structural features and biological implications, Int. J. Biol. Macromol., 2014, 66, 144–150 CrossRef CAS PubMed
.
- B. P. Ting, J. Zhang, Z. Gao and J. Y. Ying, A DNA biosensor based on the detection of doxorubicin-conjugated Ag nanoparticle labels using solid-state voltammetry, Biosens. Bioelectron., 2009, 25(2), 282–287 CrossRef CAS PubMed
.
- Y. Hahn and H. Y. Lee, Electrochemical behavior and square wave voltammetric determination of doxorubicin hydrochloride, Arch. Pharmacal Res., 2004, 27(1), 31–34 CrossRef CAS
.
- P. Velasquez, D. Leinen, J. Pascual, J. R. Ramos-Barrado, P. Grez, H. Gomez, R. Schrebler, R. Del Río and R. Cordova, A chemical, morphological, and electrochemical (XPS, SEM/EDX, CV, and EIS) analysis of electrochemically modified electrode surfaces of natural chalcopyrite (CuFeS2) and pyrite (FeS2) in alkaline solutions, J. Phys. Chem. B, 2005, 109(11), 4977–4988 CrossRef CAS PubMed
.
- N. Wang, M. Lin, H. Dai and H. Ma, Functionalized gold nanoparticles/reduced graphene oxide nanocomposites for ultrasensitive electrochemical sensing of mercury ions based on thymine–mercury–thymine structure, Biosens. Bioelectron., 2016, 79, 320–326 CrossRef CAS PubMed
.
- M. R. Smith, B. Chako, G. Benavides, M. Johnson, Y.-M. G. Kang, D. Jones and V. Darley-Usmar, 453-Effects of Doxorubicin on Cellular Bioenergetics and Metabolism in Platelets, Free Radicals Biol. Med., 2016, 100, S187 CrossRef
.
- O. D. Renedo, M. Alonso-Lomillo and M. A. Martinez, Recent developments in the field of screen-printed electrodes and their related applications, Talanta, 2007, 73(2), 202–219 CrossRef CAS PubMed
.
- S. Jampasa, W. Wonsawat, N. Rodthongkum, W. Siangproh, P. Yanatatsaneejit, T. Vilaivan and O. Chailapakul, Electrochemical detection of human papillomavirus DNA type 16 using a pyrrolidinyl peptide nucleic acid probe immobilized on screen-printed carbon electrodes, Biosens. Bioelectron., 2014, 54, 428–434 CrossRef CAS PubMed
.
- M. Zhou, Y. Zhai and S. Dong, Electrochemical sensing and biosensing platform based on chemically reduced graphene oxide, Anal. Chem., 2009, 81(14), 5603–5613 CrossRef CAS PubMed
.
- B. Li, G. Pan, N. D. Avent, R. B. Lowry, T. E. Madgett and P. L. Waines, Graphene electrode modified with electrochemically reduced graphene oxide for label-free DNA detection, Biosens. Bioelectron., 2015, 72, 313–319 CrossRef CAS PubMed
.
- A. K. Wanekaya, W. Chen, N. V. Myung and A. Mulchandani, Nanowire-based electrochemical biosensors, Electroanalysis, 2006, 18(6), 533–550 CrossRef CAS
.
- U. Yogeswaran and S.-M. Chen, A review on the electrochemical sensors and biosensors composed of nanowires as sensing material, Sensors, 2008, 8(1), 290–313 CrossRef CAS PubMed
.
- Y. Zhang, W. Chu, A. D. Foroushani, H. Wang, D. Li, J. Liu, C. J. Barrow, X. Wang and W. Yang, New gold nanostructures for sensor applications: a review, Materials, 2014, 7(7), 5169–5201 CrossRef PubMed
.
- K. Dawson, J. r. Strutwolf, K. P. Rodgers, G. g. Herzog, D. W. Arrigan, A. J. Quinn and A. O'Riordan, Single nanoskived nanowires for electrochemical applications, Anal. Chem., 2011, 83(14), 5535–5540 CrossRef CAS PubMed
.
- S. Pud, A. Kisner, M. Heggen, D. Belaineh, R. Temirov, U. Simon, A. Offenhäusser, Y. Mourzina and S. Vitusevich, Features of transport in ultrathin gold nanowire structures, Small, 2013, 9(6), 846–852 CrossRef CAS PubMed
.
- Z. Aghili, N. Nasirizadeh, A. Divsalar, S. Shoeibi and P. Yaghmaei, A Nanobiosensor Composed of Exfoliated Graphene Oxide and Gold Nano-Urchins, for Detection of GMO Products, Biosens. Bioelectron., 2017, 95, 72–80 CrossRef CAS PubMed
.
- S. Hajihosseini, N. Nasirizadeh, M. S. Hejazi and P. Yaghmaei, A sensitive DNA biosensor fabricated from gold nanoparticles and graphene oxide on a glassy carbon electrode, Mater. Sci. Eng., C, 2016, 61, 506–515 CrossRef CAS PubMed
.
- C. Pérez-Arnaiz, N. Busto, J. M. Leal and B. a. García, New insights into the mechanism of the DNA/doxorubicin interaction, J. Phys. Chem. B, 2014, 118(5), 1288–1295 CrossRef PubMed
.
Footnote |
| † Authors with equal contribution. |
| This journal is © The Royal Society of Chemistry 2017 |