Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Cell-free riboswitches
Takeshi
Tabuchi
 and
Yohei
Yokobayashi
and
Yohei
Yokobayashi
 *
*
Nucleic Acid Chemistry and Engineering Unit, Okinawa Institute of Science and Technology Graduate University, Onna, Okinawa 904-0495, Japan. E-mail: yohei.yokobayashi@oist.jp
First published on 4th August 2021
Abstract
The emerging community of cell-free synthetic biology aspires to build complex biochemical and genetic systems with functions that mimic or even exceed those in living cells. To achieve such functions, cell-free systems must be able to sense and respond to the complex chemical signals within and outside the system. Cell-free riboswitches can detect chemical signals via RNA–ligand interaction and respond by regulating protein synthesis in cell-free protein synthesis systems. In this article, we review synthetic cell-free riboswitches that function in both prokaryotic and eukaryotic cell-free systems reported to date to provide a current perspective on the state of cell-free riboswitch technologies and their limitations.
1. Introduction
Riboswitches are gene switches composed of ribonucleic acid (RNA). They have been found in diverse species of bacteria and in few eukaryotes, and they are used to sense and respond to a variety of metabolites such as coenzymes, nucleobases, ions, and amino acids.1–5 A canonical bacterial riboswitch consists of an aptamer domain and an expression platform that are located in the 5′ untranslated region (UTR) of an mRNA. The aptamer domain directly and specifically binds a small molecule metabolite while the expression platform mediates a local structural change of the mRNA upon aptamer–ligand binding. The structural change results in upregulation or downregulation of expression of the protein(s) encoded in the same mRNA.1,2,4–10 It has been suggested that these riboswitches may be remnants of the ancient RNA world.8,11The possibility of chemical regulation of gene expression without direct involvement of proteins (e.g., transcription factors) has inspired many researchers to develop synthetic riboswitches that respond to non-natural molecules using RNA aptamers selected in vitro. While many synthetic riboswitches mimicking natural mechanisms have been designed in bacteria, synthetic riboswitches that function in other cells and organisms, and riboswitches based on mechanisms not found in nature have also been developed.9,12–19
Bottom-up or cell-free synthetic biology is emerging as a frontier in synthetic biology whose goal is to design and build biochemical or genetic networks (cell-free systems) that display complex functions without using living cells.20–22 Building such cell-free systems can improve our understanding of the design principles of biological systems. Alternatively, with fewer experimental constraints compared to living cells, researchers may be able to build cell-free systems with functions or properties that are difficult or impossible using in vivo systems, for example, production of toxic secondary metabolites or hybrid systems with nonbiological components. Cell-free protein synthesis (CFPS) systems based on cell extracts or a reconstituted translational apparatus play key roles in these efforts. Synthetic riboswitches that can function in these cell-free platforms are attractive tools for interfacing the cell-free systems with diverse chemical signals.
Cell-free riboswitches, however, have not received as much attention as the synthetic riboswitches that function in living cells. To our knowledge, no comprehensive reviews focusing on cell-free riboswitches have been published. Consequently, we surveyed the literature on synthetic cell-free riboswitches that function in CFPS systems to provide a current perspective on the various types of cell-free riboswitches designed and their emerging applications. The cell-free riboswitches discussed in this review are summarized in Tables 1 and 2.
| Mechanism | Riboswitch | Ligand | ORF | On/off ratio (ligand conc.) | CFPS* | Year | Ref. |
|---|---|---|---|---|---|---|---|
| *CFPS (cell-free protein synthesis) systems. PURE: PURE systems.a PURESYSTEM classic II (Post-Genome Institute).b PURESYSTEM custom (Post-Genome Institute) without RF1.c Modified Shimizu's PURE system with T7 RNAP replaced by T3 RNAP.d PURExpress (New England Biolabs).e PUREfrex 1.0 (Gene Frontier). CE: cell extracts.f Commercial S30 cell extract from E. coli B strain SL119 (Promega).g In-house prepared E. coli cell extract from Rosetta2 (DE3).h In-house prepared E. coli cell extract from BL21 Star (DE3). AC: artificial cells.i Water-in-oil emulsions.j Lipid vesicles/liposomes. cGMP: cyclic guanosine monophosphate, TMR: tetramethylrhodamine, TPP: thiamine pyrophosphate, αHL: α-hemolysin, C23DO: catechol-2,3-dioxygenase, GFP: green fluorescent protein (sf-: super-folder, -uv: ultraviolet), LacZ: β-galactosidase, Luc: firefly luciferase, PLC: phospholipase C, RNAP: RNA polymerase, YPet: yellow fluorescent protein for energy transfer. | |||||||
| Aptazyme-driven RBS release | duplex5-Theo | Theophylline | Luc | ON ∼9 × (2 mM) | PUREa | 2007 | 36 |
| LacZ | ON ∼30 × (2 mM) | ||||||
| duplex5-cGMP | cGMP | Luc | ON ∼10 × (10 mM) | PUREa | 2007 | 36 | |
| DelC-M3 | TPP | GFP | ON ∼48 × (0.3 mM) | PUREc | 2012 | 39 | |
| Theo/HHR | Theophylline | GFP | ON ∼15 × (2 mM) | PUREc | 2012 | 39 | |
| Nonsense suppression | AST4m | Theophylline | Luc | ON 11.6 × (1 mM) | PUREb | 2008 | 41 and 115 |
| RBS sequestration | pLac-thiM#2 | TPP | GFPuv | ON 6 × (1 mM) | CEf | 2009 | 42 |
| mCherry | ON 14 × (1 mM) | ||||||
| Luc | ON 20 × (1 mM) | ||||||
| pLac-tenA#59 | TPP | Luc | ON 6 × (1 mM) | CEf | 2009 | 42 | |
| pT7-theo | Theophylline | YPet | ON 6 × (500 μM) | PUREd + ACij | 2011 | 43 and 107 | |
| pTac-theo | Theophylline | YPet | ON 8 × (500 μM) | CEf + ACij | 2011 | 43 | |
| JF001A | Theophylline | αHL | ON ∼10 × (1.5 mM) | PUREd + ACj | 2014 | 108 | |
| pTac-C | Theophylline | Luc | ON ∼65 × (2 mM) | CEf | 2014 | 44 | |
| TMR-10 | TMR | Luc | ON ∼16.5 × (30 μM) | CEf | 2014 | 46 | |
| Dopa-5 | Dopamine | Luc | ON 2 × (1 mM) | CEf | 2014 | 46 | |
| T4-2 | Thyroxine | Luc | ON 2.4 × (150 μM) | CEf | 2014 | 46 | |
| Theo-αHL | Theophylline | αHL | N/D | CEg + ACj | 2017 | 109 | |
| H2 | Histamine | mCherry | ON 30.7 × (5 mM) | PUREe + ACj | 2019 | 45 | |
| αHL | N/D | ||||||
| PLC | N/D | ||||||
| Theo-GFP-MG | Theophylline | sfGFP | ON ∼12 × (10 mM) | CEh, PUREd | 2021 | 31 | |
| Transcription termination | pTac-ade/ydhL | Adenine | YPet | ON 1.7 × (1 μM) | CEf + ACij | 2011 | 43 |
| FRR/CrcB | Fluoride (F−) | sfGFP | ON ∼20 × (3.5 mM) | CEg | 2020 | 48 | |
| C23DO | N/D | ||||||
| Mechanism | Riboswitch | Ligand | ORF | On/off ratio (ligand conc.) | CFPS* | Year | Ref. |
|---|---|---|---|---|---|---|---|
| *CFPS (cell-free protein synthesis) systems. WGE: wheat germ extracts.a Promega.b WEPRO1240 (CellFree Sciences). RRL: rabbit reticulocyte lysate system (Promega). cGMP: cyclic guanosine monophosphate, FMN: flavin mononucleotide, H33258: Hoechst dye 33258, nDNA: nano-sized ssDNA or pentadeoxyribonucleotide, TMR: tetramethylrhodamine, SRB: sulphorhodamine B, CAT: chloramphenicol acetyltransferase, eGFP: enhanced green fluorescent protein, Luc: firefly luciferase, NLuc: NanoLuc, RSETA: ORF of the undigested cloning site from pRSET-A plasmid (Invitrogen), YPet: yellow fluorescent protein for energy transfer. | |||||||
| Ribosome blocking | tob3-RSETA | Tobramycin | RSETA | OFF ∼7 × (60 μM) | WGEa | 1998 | 76 |
| H2-RSETA | H33258 | RSETA | OFF ∼12 × (80 μM) | WGEa | 1998 | 76 | |
| (Th)3 | Theophylline | CAT | OFF ∼8 × (1 mM) | WGEa | 2002 | 77 | |
| B3 | Biotin | CAT | OFF ∼9 × (1 mM) | WGEa, RRL | 2002 | 77 | |
| th+0-U7 | Theophylline | YPet | OFF 5.9 × (1 mM) | WGEb | 2018 | 78 | |
| NLuc | OFF 7.2 × (1 mM) | ||||||
| TMR+0-U7 | TMR | NLuc | OFF 5.1 × (500 μM) | WGEb | 2018 | 78 | |
| Aptazyme-regulated translation | mRNA6-Theo | Theophylline | Luc | ON ∼50 × (500 μM) | WGEb | 2009 | 87 |
| mRNA6-cGMP | cGMP | Luc | ON ∼10 × (750 μM) | WGEb | 2009 | 87 | |
| IRES-modulated translation | theo5 | Theophylline | Luc | ON 9.6 × (1 mM) | WGEb | 2011 | 88 and 116 |
| eGFP | ON 5.1 × (1 mM) | ||||||
| FMN4 | FMN | Luc | ON 7.5 × (300 μM) | WGEb | 2011 | 88 | |
| tc7 | Tetracycline | Luc | ON 29 × (300 μM) | WGEb | 2011 | 88 | |
| eGFP | ON 35 × (300 μM) | ||||||
| sr4 | SRB | Luc | ON 4.2 × (300 μM) | WGEb | 2011 | 88 | |
| theoN5 | Theophylline | NLuc | OFF 5.8 × (1 mM) | WGEb | 2012 | 89 | |
| tc-N5 | Tetracycline | NLuc | OFF 4.9 × (300 μM) | WGEb | 2012 | 89 | |
| FMN-N5 | FMN | NLuc | OFF 4.7 × (300 μM) | WGEb | 2012 | 89 | |
| theoA3-rS | Theophylline | Luc | OFF 14 × (1 mM) | WGEb | 2017 | 90 | |
| nDNA-547-N4 | nDNA | NLuc | ON 21 × (300 μM) | WGEb | 2020 | 114 | |
| Ribosomal shunting | theoS1 | Theophylline | Luc | ON 14.4 × (1 mM) | WGEb | 2013 | 96 and 117 |
| tmrS1 | TMR | Luc | ON 5.4 × (333 μM) | WGEb | 2013 | 96 | |
| Nonsense suppression | theo(th1)-MS(4) | Theophylline | Luc | ON 7.8 × (1 mM) | WGEb | 2015 | 97 and 115 |
| tc(th1)-MS(4) | Tetracycline | Luc | ON 81 × (100 μM) | WGEb | 2015 | 97 and 115 | |
| FMN(th1)-MS(4) | FMN | Luc | ON ∼4 × (30 μM) | WGEb | 2015 | 97 and 115 | |
| 3′ CITE-modulated translation | 5SL-BYm2-theo | Theophylline | YPet | ON 7.7 × (1 mM) | WGEb | 2017 | 99 |
| NLuc | ON 7.3 × (1 mM) | ||||||
| 5SL-BYm2-TMR | TMR | NLuc | ON 5.8 × (100 μM) | WGEb | 2017 | 99 | |
2. Cell-free systems for riboswitch research
Synthetic riboswitches have been designed and studied in both prokaryotic and eukaryotic CFPS systems. Prokaryotic CFPS systems that have been used for cell-free riboswitches include conventional Escherichia coli lysate-based systems (S30 extracts),23–26 and reconstituted in vitro translation systems based on the PURE (stands for Protein synthesis Using Recombinant Elements) system originally developed by Shimizu et al.27,28 S30 extracts are relatively inexpensive and simple to produce, can be scaled up, and exhibit high protein yields.26,29,30 The extracts also contain numerous nonessential cellular components and may reflect the intracellular environment more accurately. PURE systems, on the other hand, consist of purified, mostly recombinant, components from E. coli, therefore, are much more expensive and less scalable. However, they offer a flexible, well-defined, and reproducible platform that allows more freedom to control biochemical parameters if desired. The reconstituted systems also contain low amounts of ribonucleases which can significantly influence riboswitch performance.31Although a number of eukaryotic CFPS systems are currently available, nearly all of the eukaryotic cell-free riboswitches have been studied using wheat germ extract (WGE). In dry state, wheat germ embryos naturally contain all the components required for translation, ready to start protein synthesis as germination begins.32,33 The Endo group developed a stable WGE system with high translation efficiency after removing endogenous translation inhibitory components that limited the life span of the conventional WGEs.34 The current WGE systems offer the highest translation efficiency among eukaryotic CFPS systems35 and can produce high quality proteins in folded state.32 The complexity of translation regulation mechanisms in eukaryotes compared to those in prokaryotes presents both unique challenges and opportunities in designing eukaryotic cell-free riboswitches.
3. Prokaryotic cell-free riboswitches
3.1. Aptazyme-based riboswitches
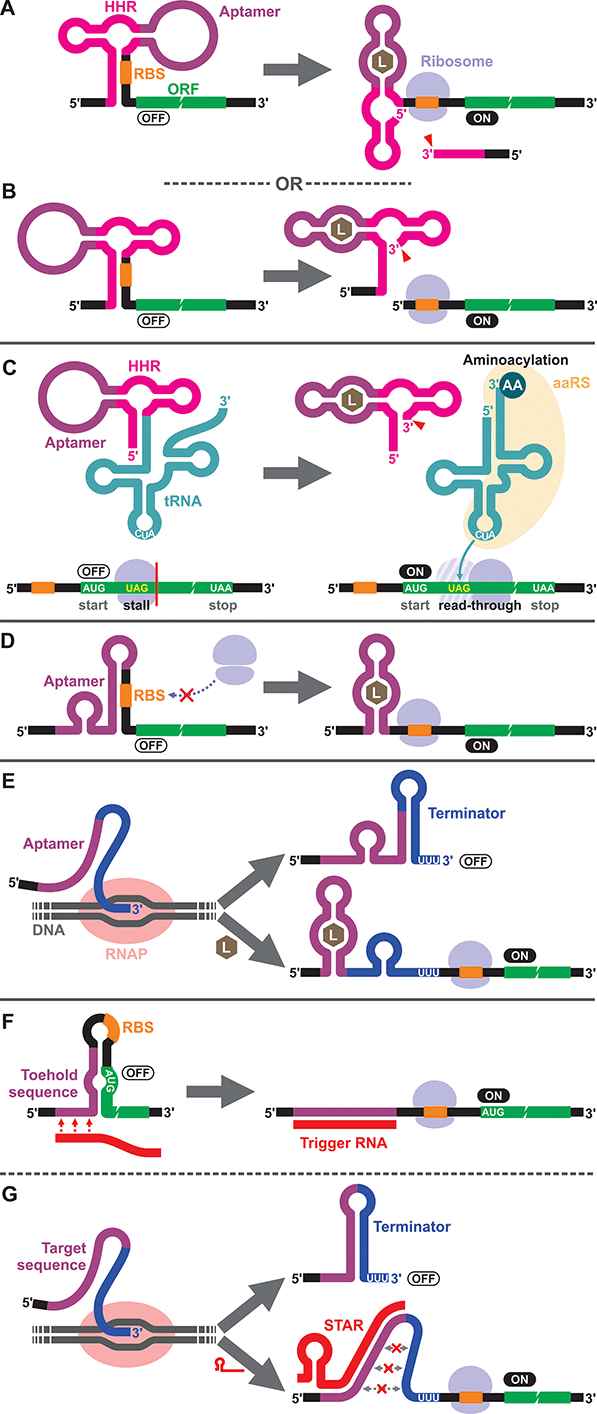
To our knowledge, the first synthetic cell-free riboswitch in a prokaryotic CFPS system was reported by Ogawa and Maeda in 2007 using the PURE system.36 In this work, the researchers adapted the theophylline-activated hammerhead ribozyme (aptazyme) reported by the Breaker group37,38 so that the ribosome binding site (RBS) of the mRNA was sequestered by a complementary sequence upstream of the ribozyme (anti-RBS). Self-cleavage of the ribozyme in the presence of theophylline releases the RBS, allowing protein translation to be activated (Fig. 1A). They also constructed a riboswitch that responds to cyclic guanosine monophosphate (cGMP) based on the same strategy.36 The same theophylline riboswitch and another riboswitch based on a thiamine pyrophosphate (TPP)-activated aptazyme12,13 (Fig. 1B) were further analyzed by Kobori et al. in a modified PURE system for the purpose of optimizing riboswitch performance based on kinetic modeling.39 It should also be noted that synthetic riboswitches developed based on the same design strategy have been shown to function in E. coli by Ogawa and Maeda,40 and the Hartig group.12,13 In another study, Ogawa and Maeda tethered the theophylline-responsive aptazyme to the 5′ end of a nonsense-suppressor tRNA (sup-tRNA) to regulate translation read-through of a gene that contains an amber stop codon in a customized PURE system (Fig. 1C).41 In this strategy, the aptazyme activity mimics the canonical 5′ terminus cleavage mediated by RNase P in vivo which is absent in the PURE system.3.2. Translationally regulated riboswitches
The majority of prokaryotic cell-free riboswitches operate by regulating the translation efficiency of the associated mRNA in response to aptamer–ligand binding. While the aptazyme-based riboswitches achieve translational regulation by irreversible self-cleavage that physically separates the RBS from the anti-RBS sequence, a class of natural riboswitches regulate the local structure near the RBS through a structural change triggered by aptamer–ligand interaction (Fig. 1D). A series of cell-free riboswitches have been designed inspired by these natural prokaryotic riboswitches (Table 1). Muranaka et al. used an E. coli S30 extract system to characterize several TPP-responsive synthetic riboswitches that were originally engineered in E. coli, and observed comparable riboswitch performance in the cell-free system.42 Similarly, a number of cell-free riboswitches of this type were originally engineered in E. coli and simply adapted to or further optimized in cell-free systems.43,44 In contrast, some cell-free riboswitches have been directly engineered in prokaryotic cell-free systems.45,463.3. Transcriptionally regulated riboswitches
Another major class of natural riboswitches regulate premature transcription termination upstream of the start codon by modulating the transcription terminator structure upon aptamer–ligand binding (Fig. 1E). Although transcriptionally regulated riboswitches represent the most common riboswitch class in bacteria,47 there are only two reported cell-free riboswitches of this type, both based on naturally occurring sequences. Martini and Mansy studied the adenine-responsive riboswitch associated with the ydhL gene of Bacillus subtillis using an E. coli S30 extract, and observed modest activation (1.7-fold) of gene expression in the presence of adenine.43 More recently, Thavarajah et al. used the fluoride-responsive riboswitch that activates expression of the efflux pump CrcB in B. cereus in lyophilized E. coli cell-free extract in their effort to develop a biosensor for fluoride detection in water.48 Both systems used E. coli RNA polymerase rather than T7 RNA polymerase for cell-free transcription which is reasonable considering the low termination efficiency of T7 RNA polymerase at canonical bacterial transcription terminators.49,50 The scarcity of engineered riboswitches that are transcriptionally regulated also extends to E. coli and other bacteria, with only a handful of such riboswitches reported.51–56 This could partly be due to the fact that Gram-negative bacteria, including E. coli, seem to prefer translationally regulated riboswitches, whereas Gram-positive bacteria favor transcriptionally regulated ones.5,7,57,58 Genes of the Gram-positive bacteria are more frequently organized in larger polycistronic operons which are regulated more efficiently by transcriptionally regulated riboswitches.7,573.4. DNA/RNA-responsive cell-free switches
We would like to briefly point out the existence of related cell-free switches/sensors that sequence-specifically respond to nucleic acids (DNA/RNA) in cell-free systems. These switches modulate transcription or translation through mechanisms similar to those of the natural bacterial riboswitches except that they respond to specific DNA or RNA sequences through Watson–Crick base pairing. Earlier examples come from Aoyama and coworkers who employed a molecular beacon like structure to regulate cell-free translation in response to short oligo DNA or RNA.59,60 More recent and sophisticated sensors called toehold switches61 also regulate cell-free translation in response to specific RNA sequences (Fig. 1F). Small transcription activating RNAs (STARs)62 exploit RNA–RNA hybridization to regulate premature transcription termination in cell-free systems (Fig. 1G). These RNA/DNA-responsive cell-free switches have been used to prototype genetic circuits63–66 and to develop biosensors for detecting RNAs from viruses,67,68 bacteria,69 and a variety of RNA markers.69,704. Eukaryotic cell-free riboswitches
4.1. Ribosome blocking
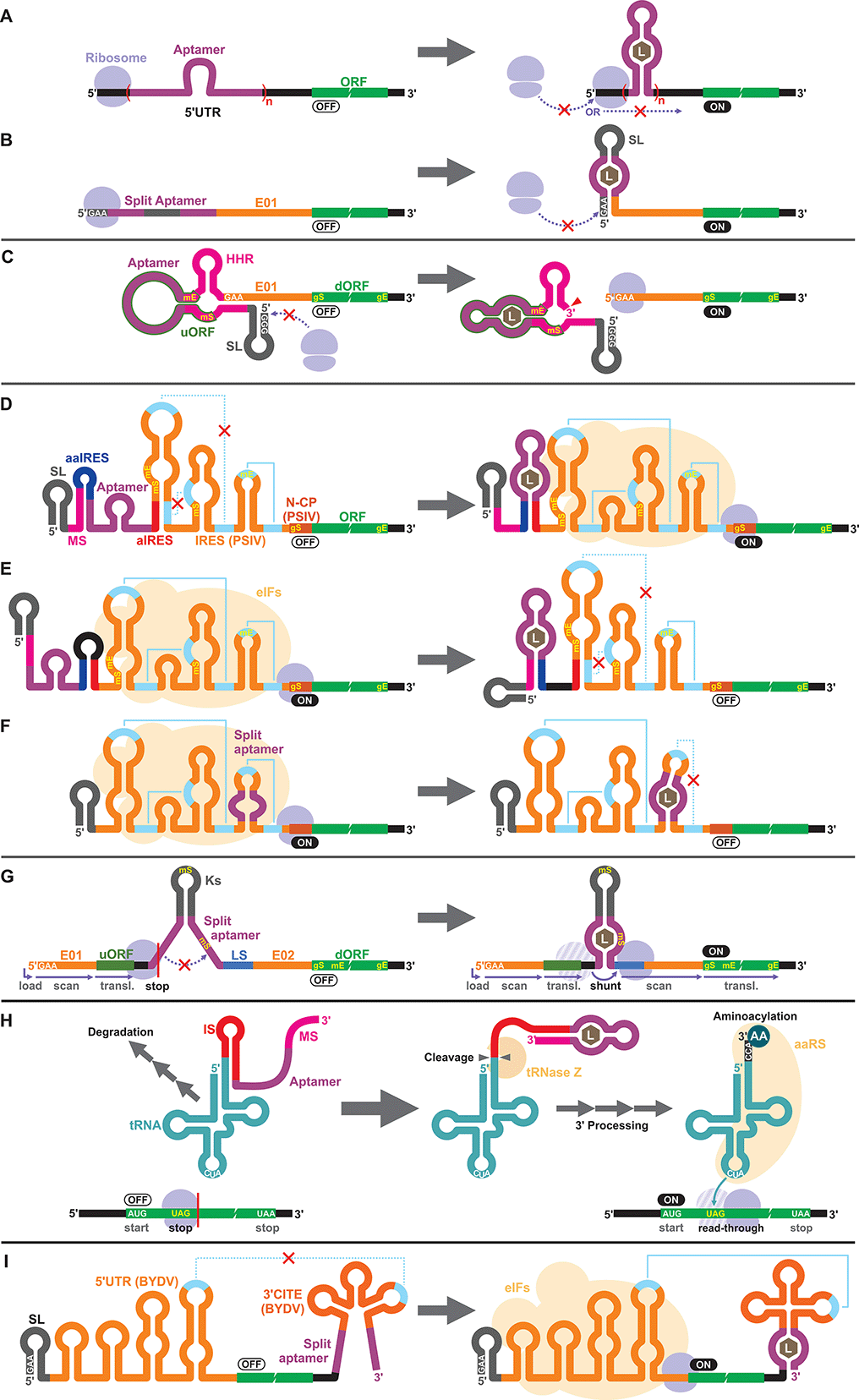
Apart from the TPP riboswitches that modulate pre-mRNA splicing in fungi and plants,6,71–73 there are no other known natural riboswitches in eukaryotes whose posttranscriptional regulatory mechanisms are very different from those in prokaryotes. Consequently, synthetic cell-free riboswitches that function in eukaryotic CFPS systems have been designed to operate via non-natural regulatory mechanisms.Interestingly, earlier reports on eukaryotic cell-free riboswitches predated the discovery of bacterial riboswitches. Inspired by the observation that a stable RNA structure at the 5′ terminus of a eukaryotic mRNA can interfere with ribosome loading or mRNA scanning resulting in lower gene expression,74,75 Werstuck and Green inserted an aptamer that binds tobramycin or Hoechst dye 33258 in the 5′ UTR of an mRNA. Addition of the aptamer ligand suppressed translation in WGE presumably due to the stabilization of the aptamer structure upon ligand binding.76 Similarly, Harvey et al. controlled the synthesis of chloramphenicol acetyltransferase (CAT) in WGE and rabbit reticulocyte lysate by inserting multiple copies of theophylline or biotin aptamers in the 5′ UTR (Fig. 2A).77 More recently, Ogawa et al. designed split aptamers that induce mRNA stabilization at the 5′ UTR to block ribosome loading in WGE (Fig. 2B).78 It is worth noting that similar riboswitch designs have been shown to function in yeast79–82 and in mammalian cells.76
4.2. Aptazyme-based riboswitches
Aptazymes are frequently used to regulate gene expression living eukaryotic cells.18,83,84 Ogawa, who reported the first prokaryotic cell-free riboswitches based on an aptazyme also designed the first aptazyme-based cell-free riboswitches in a eukaryotic cell-free system. Incidentally, Ogawa and coworkers have designed the majority of the synthetic riboswitches that function in eukaryotic cell-free systems, specifically, WGE (Table 2). To engineer their aptazyme-based riboswitches, Ogawa exploited a property of the WGE described by Endo and coworkers.85,86 WGE can efficiently initiate translation from an uncapped mRNA that contains 5′ GAA trinucleotide followed by an enhancer sequence (E01). Ogawa inserted a theophylline-activated aptazyme directly upstream of the GAA trinucleotide such that the 5′ GAA is exposed upon ribozyme cleavage. Without cleavage, the lack of 5′ GAA, the presence of the relatively stable aptazyme structure, and the presence of a short upstream open reading frame (uORF) embedded within the aptazyme sequence repress reporter gene expression (Fig. 2C).874.3. IRES
Next, Ogawa and coworkers shifted their attention to internal ribosomal entry sites (IRES). An IRES is a viral RNA element that recruits the translation initiation complex in the absence of the 5′ cap structure. Specifically, Ogawa identified the minimum functional sequence of the IRES from Plautia stali intestine virus (PSIV), and rationally fused an RNA aptamer to disrupt or induce critical IRES structural elements in the presence of the ligand. For example, Ogawa engineered an ON-switch that activates gene expression in the presence of theophylline following a rational design strategy. First, an 8-nt anti-IRES (aIRES) sequence was introduced upstream of the IRES to disrupt a critical pseudoknot structure, successfully repressing gene expression (OFF). Then, an anti-anti-IRES (aaIRES) and the theophylline aptamer was added further upstream to restore gene expression (ON) by sequestering the aIRES. Finally, a modulator sequence (MS) was appended to the 5′ end of the mRNA interfering with the aaIRES and the aptamer to suppress gene expression (OFF) (Fig. 2D). Ogawa systematically optimized the stability of the MS-aaIRES/aptamer interaction to engineer riboswitches that shift the IRES from the OFF to the ON structure in the presence of theophylline and other ligands.88 Ogawa later designed OFF-switches by rearranging the riboswitch elements (Fig. 2E)89 demonstrating the flexibility of this design strategy. More recently, Ogawa et al. reported a simpler OFF-switch design in which an aptamer was embedded within the IRES structure (Fig. 2F).904.4. Other mechanisms
Ogawa has developed additional cell-free riboswitches in WGE based on other regulatory mechanisms. He exploited the ribosome shunting mechanism used by some viruses to bypass mRNA scanning91–95 by incorporating two halves of a split aptamer separated by an intervening sequence. Aptamer–ligand binding brings the two regions to close proximity to induce ribosome shunting, resulting in efficient gene expression (Fig. 2G).96In another design, Ogawa and Tabuchi engineered riboswitches that control readthrough of premature translation termination codons (nonsense amber mutations) using sup-tRNAs in WGE97 through a mechanism reminiscent of their aptazyme-based riboswitch that operates in a prokaryotic cell-free system (Fig. 1C).41 In the eukaryotic riboswitch, the aptamer was fused to the 3′ end of the sup-tRNA via an inhibitory sequence (IS) that interferes with the natural 3′ processing of the tRNA (cleavage by tRNase Z and addition of CCA by tRNA nucleotidyltransferases). Aptamer–ligand interaction exposes the 3′ terminus of the tRNA allowing its rapid maturation followed by suppression of the amber codon (Fig. 2H).97 The same design was later adapted by Ogawa et al. to develop sup-tRNA switches triggered by complementary DNA oligonucleotides in WGE.98
More recently, Ogawa and colleagues manipulated a viral translation initiation mechanism mediated by the 3′ cap-independent translation element (3′ CITE) from barley yellow dwarf virus (BYDV). The viral RNA element located in the 3′ UTR activates translation by forming a kissing loop interaction with another element located in the 5′ UTR, mimicking the canonical 5′ cap-3′ poly(A) interaction mediated by eukaryotic translation initiation factors (eIFs). Ogawa and colleagues inserted a split aptamer within the 3′ CITE to enhance the kissing loop interaction upon aptamer–ligand binding (Fig. 2I).99
5. Applications of cell-free riboswitches
5.1. Biosensors
Cell-free riboswitches possess several potential advantages as a versatile platform for biosensing. First, natural and laboratory evolved RNA aptamers have been reported for a plethora of compounds. Therefore, in principle, one can expect to be able to develop riboswitches for a variety of analytes. While such laboratory evolved aptamers can potentially work in cellular systems, very few aptamers have been used to engineer riboswitches in vivo. Various biological factors such as intracellular environment, or cell permeability, stability, or toxicity of the ligand can hinder the development of cellular riboswitches using these aptamers, while cell-free riboswitches can circumvent some of these constraints.Second, riboswitches can trigger expression of any gene(s), broadening the options for the detection method. Additionally, signal amplification inherent to translation (multiple proteins translated per mRNA molecule) and provided by some reporter genes (enzymes such as luciferase or β-galactosidase) can improve sensitivity of the biosensor. There are some challenges, however, including low stability of CFPS systems and sensitivity to contaminants.
Performance indicators as biosensors such as detection limit, dose–response, and signal-to-noise ratio of some cell-free riboswitches have been analyzed.36,41,42,46,88,97,100 However, most of those cell-free riboswitches have never been put to test by measuring more practically relevant samples. A recent notable exception was reported by Thavarajah et al. who engineered a cell-free fluoride riboswitch for field application as a biosensor.48 They implemented the riboswitch in a lyophilized E. coli extract format previously optimized for toehold switches63,67–69,101 and used catechol 2,3-dioxygenase (C23DO) as the reporter gene that provides a visual colorimetric output suitable for field use. The practical potential as a biosensor was demonstrated by on-site detection of fluoride levels as low as 50 μM (∼1 ppm) in real-world water samples.48
5.2. Artificial cells
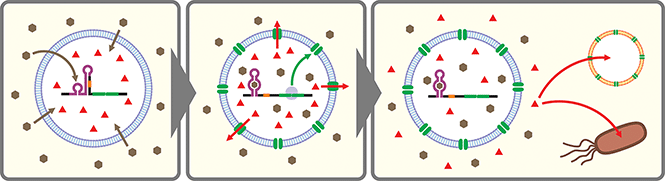
Artificial cells are emerging as a new frontier in synthetic biology.102–104 In many of these artificial cells a CFPS system is encapsulated in a cell-sized compartment along with DNA encoding various genes.105,106 Naturally, researchers started building artificial cells that respond to chemical signals by controlling gene expression. Along with canonical transcription factor-based switches well established in bacterial systems (e.g., LacI–IPTG, AraC–arabinose, LuxR–AHL), some synthetic riboswitches have also been introduced in different types of artificial cells such as water-in-oil emulsions,43,107 and lipid vesicles.43,45,107–109 In a recent report, a riboswitch-regulated DNA was trapped inside silk fibroin-based microcapsules to demonstrate protein expression in the presence of theophylline. However, the riboswitch function was not demonstrated in the protein compartment.100Theophylline riboswitches have been used in lipid vesicle-based artificial cells to mediate chemical signaling between artificial cells and bacteria. In this work, Lentini et al. encapsulated IPTG inside liposomes along with DNA encoding an α-hemolysin gene controlled by a theophylline-activated riboswitch.108 Addition of theophylline triggers expression of α-hemolysin which forms nanometer sized pores on the membrane of the artificial cells. The pores allow IPTG to diffuse out of the artificial cells and to induce GFP expression in co-cultured E. coli cells. Similarly, Adamala et al. constructed riboswitch-controlled artificial cells that release doxycycline to achieve communication between different artificial cells (Fig. 3).109 However, in both examples, performance of the theophylline-responsive riboswitches was not directly characterized in the artificial cells.
More recently, Dwidar et al. applied Systematic Evolution of Ligands by EXponential enrichment (SELEX)110,111 to discover a novel aptamer that recognizes histamine, and they designed and optimized histamine-responsive cell-free riboswitches in the PURE system.45 They thoroughly characterized the riboswitch in liposome-based artificial cells using fluorescence microscopy and flow cytometry to confirm the robust and dynamic response to histamine. Finally, artificial cells that express α-hemolysin or phospholipase C under the control of their riboswitch were constructed to demonstrate either molecular cargo release or self-destruction triggered by histamine.
5.3. Mechanistic understanding of riboswitches
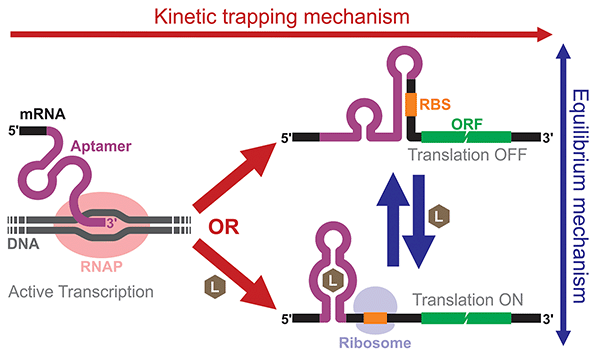
Building and analyzing cell-free riboswitches can lead to fundamental insights into the regulatory mechanisms and the design principles of natural and synthetic riboswitches. An advantage of working with cell-free riboswitches is the presence of fewer experimental constraints compared to working with riboswitches in living cells. For example, researchers can precisely control various parameters such as ligand concentration and the timing of ligand addition. Cell-free riboswitches can also circumvent other biological constraints such as ligand toxicity, ligand permeability, and metabolic burden.An elegant example by Mishler and Gallivan illustrates the advantage of using a cell-free system to study riboswitch mechanisms.44 The researchers used an E. coli S30 extract to study their theophylline-activated riboswitches that they previously engineered in bacteria. By adding theophylline during or after transcription, they unambiguously showed that theophylline must be available during transcription for their riboswitches to function, demonstrating that the kinetic trapping mechanism plays a more dominant role than the thermodynamic equilibrium mechanism for the activation of their riboswitch (Fig. 4). They also showed that the high concentration of theophylline needed to activate the riboswitches was due to this kinetic trapping mechanism and not due to the low permeability of theophylline through the cell membrane as was previously suggested.112,113
Independent reports from Kobori et al.39 and Chushak et al.31 illustrate how kinetic modeling of riboswitch mechanisms based on experimental data of cell-free riboswitches can provide not only insights into the riboswitch mechanisms, but also hints for optimizing riboswitch performance. Espah Borujeni et al. built a more detailed model considering additional kinetic and biophysical parameters such as co-transcriptional folding and free energy of ribosome binding.46 Importantly, they used the model to automate riboswitch design using arbitrary aptamers, and they experimentally validated the designed riboswitches in either E. coli cells or cell-free S30 extracts. While the designed cell-free riboswitches showed mixed performance, the ability of such a model-driven design to generate functional riboswitches is an important achievement.
6. Conclusions and future directions
As reviewed in this article, while a steady number of cell-free riboswitches have been reported over the last two decades, cell-free riboswitches have attracted relatively little attention compared to cellular riboswitches. This trend may change in the near future with the growing interest in cell-free synthetic biology and its applications. With fewer experimental constraints compared to living cells, cell-free systems can allow more precise and extensive control over the genetic circuits and metabolic pathways built by the researchers. Such cell-free systems, much like their cell-based counterparts, will need gene switches to sense and respond to a variety of chemical signals. As discussed above, riboswitches have a number of favorable characteristics as cell-free gene switches.It should be acknowledged, however, that cell-free riboswitches still need more efforts to prove that they are valuable tools for cell-free synthetic biology. For example, OFF-switches are conspicuously missing in the prokaryotic cell-free systems (Table 1). While several cell-free riboswitches show high ON/OFF ratios greater than 30, many others display more modest responses. Nonetheless, the variety of ligands that cell-free riboswitches have been engineered to detect are quite broad compared to cell-based riboswitches. Although theophylline has been, by far, the most popular trigger molecule, cell-free riboswitches that respond to biotin,77 tetramethylrhodamine (TMR),46,78,96 flavin mononcleotide (FMN),88,89,97 cGMP,36,87 TPP,39,42 fluoride,48 histamine,45 dopamine,46 thyroxine,46 sulphorhodamine B,88 Hoechst dye 33258,76 tobramycin,76 tetracycline,88,97 and pentadeoxyribonucleotides114 have been designed (Tables 1 and 2). With the exception of histamine, however, these cell-free riboswitches have been constructed using known aptamers from natural riboswitches or discovered through SELEX intended for other applications. Additional aptamer–ligand combinations adapted to cell-free riboswitches will lead to better understanding of what makes an aptamer–ligand pair more suitable (or not) for cell-free riboswitches.
Finally, more efficient strategies for designing cell-free riboswitches are highly desirable. While many bacterial riboswitches have been shown to function similarly in prokaryotic cell-free systems, optimization of riboswitches in bacteria is not always possible because many desirable ligands for cell-free riboswitches may be toxic to the cells, impermeable through the cell wall or membrane, or unstable inside living cells. Most cell-free riboswitches not derived from bacterial analogs have been designed by trial-and-error or computational modeling, which can be laborious or unreliable. Since most successful design strategies for synthetic riboswitches in living cells involve some form of high-throughput screening or selection, a similar strategy for cell-free riboswitches should greatly facilitate the design process and improve performance. Availability of cell-free riboswitches that can interface cell-free systems with diverse chemical signals should accelerate development of innovative cell-free systems with complex functions and practical applications.
Conflicts of interest
There are no conflicts to declare.References
- W. C. Winkler and R. R. Breaker, Annu. Rev. Microbiol., 2005, 59, 487–517 CrossRef CAS PubMed
.
- A. Nahvi, N. Sudarsan, M. S. Ebert, X. Zou, K. L. Brown and R. R. Breaker, Chem. Biol., 2002, 9, 1043–1049 CrossRef CAS PubMed
.
- P. J. McCown, K. A. Corbino, S. Stav, M. E. Sherlock and R. R. Breaker, RNA, 2017, 23, 995–1011 CrossRef CAS
.
- T. S. Lotz and B. Suess, Microbiol. Spectr., 2018, 6, RWR-0025 Search PubMed
.
- N. Pavlova, D. Kaloudas and R. Penchovsky, Gene, 2019, 708, 38–48 CrossRef CAS
.
- N. Sudarsan, J. E. Barrick and R. R. Breaker, RNA, 2003, 9, 644–647 CrossRef CAS
.
- E. Nudler and A. S. Mironov, Trends Biochem. Sci., 2004, 29, 11–17 CrossRef CAS
.
- A. G. Vitreschak, D. A. Rodionov, A. A. Mironov and M. S. Gelfand, Trends Genet., 2004, 20, 44–50 CrossRef CAS
.
- B. J. Tucker and R. R. Breaker, Curr. Opin. Struct. Biol., 2005, 15, 342–348 CrossRef CAS PubMed
.
- A. D. Garst, A. L. Edwards and R. T. Batey, Cold Spring Harb. Perspect. Biol., 2011, 3, a003533 Search PubMed
.
- R. R. Breaker, Cold Spring Harb. Perspect. Biol., 2012, 4, a003566 Search PubMed
.
- M. Wieland, A. Benz, B. Klauser and J. S. Hartig, Angew. Chem., Int. Ed., 2009, 48, 2715–2718 CrossRef CAS PubMed
.
- M. Wieland and J. S. Hartig, Angew. Chem., Int. Ed., 2008, 47, 2604–2607 CrossRef CAS PubMed
.
- B. Suess and J. E. Weigand, RNA Biol., 2008, 5, 24–29 CrossRef CAS PubMed
.
- Y. Nomura, L. Zhou, A. Miu and Y. Yokobayashi, ACS Synth. Biol., 2013, 2, 684–689 CrossRef CAS
.
- W. G. Scott, L. H. Horan and M. Martick, Prog. Mol. Biol. Transl. Sci., 2013, 120, 1–23 Search PubMed
.
- P. Ketzer, J. K. Kaufmann, S. Engelhardt, S. Bossow, C. von Kalle, J. S. Hartig, G. Ungerechts and D. M. Nettelbeck, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E554–562 CrossRef CAS
.
- K. Mustafina, K. Fukunaga and Y. Yokobayashi, ACS Synth. Biol., 2020, 9, 19–25 CrossRef CAS
.
- A. Ender, M. Etzel, S. Hammer, S. Findeiß, P. Stadler and M. Mörl, Nucleic Acids Res., 2021, 49, 1784–1800 CrossRef CAS PubMed
.
- J. A. Rollin, T. K. Tam and Y. H. P. Zhang, Green Chem., 2013, 15, 1708–1719 RSC
.
- Y. Lu, Synth. Syst. Biotechnol., 2017, 2, 23–27 CrossRef
.
- N. Laohakunakorn, L. Grasemann, B. Lavickova, G. Michielin, A. Shahein, Z. Swank and S. J. Maerkl, Front. Bioeng. Biotechnol., 2020, 8, 213 CrossRef PubMed
.
-
J. M. Pratt, in Transcription and Translation: A Practical Approach, ed. B. D. Hames and S. J. Higgins, IRL Press, Oxford, 1984, pp. 179–209 Search PubMed
.
- D. E. Nevin and J. M. Pratt, FEBS Lett., 1991, 291, 259–263 CrossRef CAS PubMed
.
- J. R. Swartz, M. C. Jewett and K. A. Woodrow, Methods Mol. Biol., 2004, 267, 169–182 CAS
.
- N. Krinsky, M. Kaduri, J. Shainsky-Roitman, M. Goldfeder, E. Ivanir, I. Benhar, Y. Shoham and A. Schroeder, PLoS One, 2016, 11, e0165137 CrossRef
.
- Y. Shimizu, A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa and T. Ueda, Nat. Biotechnol., 2001, 19, 751–755 CrossRef CAS
.
- Y. Shimizu, T. Kanamori and T. Ueda, Methods, 2005, 36, 299–304 CrossRef CAS PubMed
.
- J. F. Zawada, G. Yin, A. R. Steiner, J. Yang, A. Naresh, S. M. Roy, D. S. Gold, H. G. Heinsohn and C. J. Murray, Biotechnol. Bioeng., 2011, 108, 1570–1578 CrossRef CAS PubMed
.
- J. G. Perez, J. C. Stark and M. C. Jewett, Cold Spring Harb. Perspect. Biol., 2016, 8, a023853 CrossRef
.
- Y. Chushak, S. Harbaugh, K. Zimlich, B. Alfred, J. Chavez and N. Kelley-Loughnane, RNA Biol., 2021 DOI:10.1080/15476286.2020.1868149
.
- Y. Endo and T. Sawasaki, Curr. Opin. Biotechnol, 2006, 17, 373–380 CrossRef CAS PubMed
.
- E. H. Morita, T. Sawasaki, R. Tanaka, Y. Endo and T. Kohno, Protein Sci., 2003, 12, 1216–1221 CrossRef CAS PubMed
.
- K. Madin, T. Sawasaki, T. Ogasawara and Y. Endo, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 559–564 CrossRef CAS PubMed
.
- M. Harbers, FEBS Lett., 2014, 588, 2762–2773 CrossRef CAS PubMed
.
- A. Ogawa and M. Maeda, Bioorg. Med. Chem. Lett., 2007, 17, 3156–3160 CrossRef CAS PubMed
.
- G. A. Soukup and R. R. Breaker, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 3584–3589 CrossRef CAS PubMed
.
- G. A. Soukup, G. A. Emilsson and R. R. Breaker, J. Mol. Biol., 2000, 298, 623–632 CrossRef CAS PubMed
.
- S. Kobori, N. Ichihashi, Y. Kazuta, T. Matsuura and T. Yomo, RNA, 2012, 18, 1458–1465 CrossRef CAS
.
- A. Ogawa and M. Maeda, ChemBioChem, 2008, 9, 206–209 CrossRef CAS PubMed
.
- A. Ogawa and M. Maeda, ChemBioChem, 2008, 9, 2204–2208 CrossRef CAS PubMed
.
- N. Muranaka, V. Sharma, Y. Nomura and Y. Yokobayashi, Anal. Lett., 2009, 42, 108–122 CrossRef CAS
.
- L. Martini and S. S. Mansy, Chem. Commun., 2011, 47, 10734–10736 RSC
.
- D. M. Mishler and J. P. Gallivan, Nucleic Acids Res., 2014, 42, 6753–6761 CrossRef CAS PubMed
.
- M. Dwidar, Y. Seike, S. Kobori, C. Whitaker, T. Matsuura and Y. Yokobayashi, J. Am. Chem. Soc., 2019, 141, 11103–11114 CrossRef CAS PubMed
.
- A. Espah Borujeni, D. M. Mishler, J. Wang, W. Huso and H. M. Salis, Nucleic Acids Res., 2016, 44, 1–13 CrossRef
.
- J. E. Barrick and R. R. Breaker, Genome Biol., 2007, 8, R239 CrossRef
.
- W. Thavarajah, A. D. Silverman, M. S. Verosloff, N. Kelley-Loughnane, M. C. Jewett and J. B. Lucks, ACS Synth. Biol., 2020, 9, 10–18 CrossRef CAS
.
- S. T. Jeng, J. F. Gardner and R. I. Gumport, J. Biol. Chem., 1990, 265, 3823–3830 CrossRef CAS
.
- S. T. Jeng, J. F. Gardner and R. I. Gumport, J. Biol. Chem., 1992, 267, 19306–19312 CrossRef CAS PubMed
.
- P. Ceres, J. J. Trausch and R. T. Batey, Nucleic Acids Res., 2013, 41, 10449–10461 CrossRef CAS
.
- P. Ceres, A. D. Garst, J. G. Marcano-Velazquez and R. T. Batey, ACS Synth. Biol., 2013, 2, 463–472 CrossRef CAS PubMed
.
- M. Wachsmuth, S. Findeiß, N. Weissheimer, P. F. Stadler and M. Mörl, Nucleic Acids Res., 2013, 41, 2541–2551 CrossRef CAS PubMed
.
- M. Wachsmuth, G. Domin, R. Lorenz, R. Serfling, S. Findeiß, P. F. Stadler and M. Mörl, RNA Biol., 2015, 12, 221–231 CrossRef PubMed
.
- G. Domin, S. Findeiß, M. Wachsmuth, S. Will, P. F. Stadler and M. Mörl, Nucleic Acids Res., 2017, 45, 4108–4119 CAS
.
- C. Günzel, F. Kühnl, K. Arnold, S. Findeiß, C. E. Weinberg, P. F. Stadler and M. Mörl, RNA Biol., 2020, 18, 457–467 CrossRef PubMed
.
- W. C. Winkler and R. R. Breaker, ChemBioChem, 2003, 4, 1024–1032 CrossRef CAS PubMed
.
- J. K. Soukup and G. A. Soukup, Curr. Opin. Struct. Biol., 2004, 14, 344–349 CrossRef CAS PubMed
.
- S. Sando, A. Narita, K. Abe and Y. Aoyama, J. Am. Chem. Soc., 2005, 127, 5300–5301 CrossRef CAS PubMed
.
- A. Narita, K. Ogawa, S. Sando and Y. Aoyama, Angew. Chem., Int. Ed., 2006, 45, 2879–2883 CrossRef CAS
.
- A. A. Green, P. A. Silver, J. J. Collins and P. Yin, Cell, 2014, 159, 925–939 CrossRef CAS PubMed
.
- J. Chappell, M. K. Takahashi and J. B. Lucks, Nat. Chem. Biol., 2015, 11, 214–220 CrossRef CAS PubMed
.
- K. Pardee, A. A. Green, T. Ferrante, D. E. Cameron, A. DaleyKeyser, P. Yin and J. J. Collins, Cell, 2014, 159, 940–954 CrossRef CAS PubMed
.
- M. K. Takahashi, J. Chappell, C. A. Hayes, Z. Z. Sun, J. Kim, V. Singhal, K. J. Spring, S. Al-Khabouri, C. P. Fall, V. Noireaux, R. M. Murray and J. B. Lucks, ACS Synth. Biol., 2015, 4, 503–515 CrossRef CAS PubMed
.
- F. X. Lehr, M. Hanst, M. Vogel, J. Kremer, H. U. Goringer, B. Suess and H. Koeppl, ACS Synth. Biol., 2019, 8, 2163–2173 CrossRef CAS PubMed
.
- D. Jeong, M. Klocke, S. Agarwal, J. Kim, S. Choi, E. Franco and J. Kim, Methods Protoc., 2019, 2, 39 CrossRef CAS PubMed
.
- K. Pardee, A. A. Green, M. K. Takahashi, D. Braff, G. Lambert, J. W. Lee, T. Ferrante, D. Ma, N. Donghia, M. Fan, N. M. Daringer, I. Bosch, D. M. Dudley, D. H. O'Connor, L. Gehrke and J. J. Collins, Cell, 2016, 165, 1255–1266 CrossRef CAS PubMed
.
- D. Ma, L. Shen, K. Wu, C. W. Diehnelt and A. A. Green, Synth. Biol., 2018, 3, ysy018 CrossRef CAS PubMed
.
- M. K. Takahashi, X. Tan, A. J. Dy, D. Braff, R. T. Akana, Y. Furuta, N. Donghia, A. Ananthakrishnan and J. J. Collins, Nat. Commun., 2018, 9, 3347 CrossRef PubMed
.
- T. H. T. Chau and E. Y. Lee, Clin. Chim. Acta, 2020, 510, 619–624 CrossRef CAS PubMed
.
- T. Kubodera, M. Watanabe, K. Yoshiuchi, N. Yamashita, A. Nishimura, S. Nakai, K. Gomi and H. Hanamoto, FEBS Lett., 2003, 555, 516–520 CrossRef CAS PubMed
.
- M. T. Cheah, A. Wachter, N. Sudarsan and R. R. Breaker, Nature, 2007, 447, 497–500 CrossRef CAS PubMed
.
- A. Wachter, RNA Biol., 2010, 7, 67–76 CrossRef CAS PubMed
.
- M. R. Vega Laso, D. Zhu, F. Sagliocco, A. J. Brown, M. F. Tuite and J. E. McCarthy, J. Biol. Chem., 1993, 268, 6453–6462 CrossRef CAS
.
- J. R. Babendure, J. L. Babendure, J. H. Ding and R. Y. Tsien, RNA, 2006, 12, 851–861 CrossRef CAS PubMed
.
- G. Werstuck and M. R. Green, Science, 1998, 282, 296–298 CrossRef CAS PubMed
.
- I. Harvey, P. Garneau and J. Pelletier, RNA, 2002, 8, 452–463 CrossRef CAS PubMed
.
- A. Ogawa, Y. Murashige and H. Takahashi, Bioorg. Med. Chem. Lett., 2018, 28, 2353–2357 CrossRef CAS PubMed
.
- D. Grate and C. Wilson, Bioorg. Med. Chem., 2001, 9, 2565–2570 CrossRef CAS
.
- S. Hanson, K. Berthelot, B. Fink, J. E. McCarthy and B. Suess, Mol. Microbiol., 2003, 49, 1627–1637 CrossRef CAS PubMed
.
- B. Suess, S. Hanson, C. Berens, B. Fink, R. Schroeder and W. Hillen, Nucleic Acids Res., 2003, 31, 1853–1858 CrossRef CAS PubMed
.
- J. E. Weigand, M. Sanchez, E. B. Gunnesch, S. Zeiher, R. Schroeder and B. Suess, RNA, 2008, 14, 89–97 CrossRef CAS PubMed
.
- Y. Yokobayashi, Curr. Opin. Chem. Biol., 2019, 52, 72–78 CrossRef CAS PubMed
.
- N. Shanidze, F. Lenkeit, J. S. Hartig and D. Funck, Plant Physiol., 2020, 182, 123–135 CrossRef CAS PubMed
.
- T. Sawasaki, T. Ogasawara, R. Morishita and Y. Endo, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 14652–14657 CrossRef CAS PubMed
.
- N. Kamura, T. Sawasaki, Y. Kasahara, K. Takai and Y. Endo, Bioorg. Med. Chem. Lett., 2005, 15, 5402–5406 CrossRef CAS PubMed
.
- A. Ogawa, ChemBioChem, 2009, 10, 2465–2468 CrossRef CAS PubMed
.
- A. Ogawa, RNA, 2011, 17, 478–488 CrossRef CAS PubMed
.
- A. Ogawa, Bioorg. Med. Chem. Lett., 2012, 22, 1639–1642 CrossRef CAS PubMed
.
- A. Ogawa, H. Masuoka and T. Ota, ACS Synth. Biol., 2017, 6, 1656–1662 CrossRef CAS PubMed
.
- J. Fütterer, Z. Kiss-László and T. Hohn, Cell, 1993, 73, 789–802 CrossRef
.
- W. Schmidt-Puchta, D. Dominguez, D. Lewetag and T. Hohn, Nucleic Acids Res., 1997, 25, 2854–2860 CrossRef CAS PubMed
.
- D. I. Dominguez, L. A. Ryabova, M. M. Pooggin, W. Schmidt-Puchta, J. Futterer and T. Hohn, J. Biol. Chem., 1998, 273, 3669–3678 CrossRef CAS PubMed
.
- L. A. Ryabova and T. Hohn, Genes Dev., 2000, 14, 817–829 CrossRef CAS
.
- M. M. Pooggin, L. A. Ryabova, X. He, J. Futterer and T. Hohn, RNA, 2006, 12, 841–850 CrossRef CAS PubMed
.
- A. Ogawa, ChemBioChem, 2013, 14, 1539–1543 CrossRef CAS PubMed
.
- A. Ogawa and J. Tabuchi, Org. Biomol. Chem., 2015, 13, 6681–6685 RSC
.
- A. Ogawa, J. Tabuchi, Y. Doi and M. Takamatsu, Bioorg. Med. Chem. Lett., 2016, 26, 3658–3661 CrossRef CAS PubMed
.
- A. Ogawa, Y. Murashige, J. Tabuchi and T. Omatsu, Mol. BioSyst., 2017, 13, 314–319 RSC
.
- I. Drachuk, S. Harbaugh, J. L. Chavez and N. Kelley-Loughnane, ACS Appl. Mater. Interfaces, 2020, 12, 48329–48339 CrossRef CAS PubMed
.
- M. T. Smith, S. D. Berkheimer, C. J. Werner and B. C. Bundy, Biotechniques, 2014, 56, 186–193 CrossRef CAS PubMed
.
- C. Xu, S. Hu and X. Chen, Mater. Today, 2016, 19, 516–532 CrossRef CAS PubMed
.
- B. C. Buddingh and J. C. M. van Hest, Acc. Chem. Res., 2017, 50, 769–777 CrossRef CAS PubMed
.
- L. Damiano and P. Stano, Front. Bioeng. Biotechnol., 2020, 8, 953 CrossRef PubMed
.
- T. Sunami, T. Matsuura, H. Suzuki and T. Yomo, Methods Mol. Biol., 2010, 607, 243–256 CrossRef CAS PubMed
.
- A. C. Spencer, P. Torre and S. S. Mansy, J. Visualized Exp., 2013, 80, e51304 Search PubMed
.
- L. Martini and S. S. Mansy, Methods Mol. Biol., 2014, 1111, 153–164 CrossRef CAS PubMed
.
- R. Lentini, S. P. Santero, F. Chizzolini, D. Cecchi, J. Fontana, M. Marchioretto, C. Del Bianco, J. L. Terrell, A. C. Spencer, L. Martini, M. Forlin, M. Assfalg, M. Dalla Serra, W. E. Bentley and S. S. Mansy, Nat. Commun., 2014, 5, 4012 CrossRef CAS PubMed
.
- K. P. Adamala, D. A. Martin-Alarcon, K. R. Guthrie-Honea and E. S. Boyden, Nat. Chem., 2017, 9, 431–439 CrossRef CAS PubMed
.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed
.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed
.
- S. K. Desai and J. P. Gallivan, J. Am. Chem. Soc., 2004, 126, 13247–13254 CrossRef CAS PubMed
.
- T. S. Bayer and C. D. Smolke, Nat. Biotechnol., 2005, 23, 337–343 CrossRef CAS PubMed
.
- A. Ogawa and Y. Itoh, ACS Synth. Biol., 2020, 9, 2648–2655 CrossRef CAS PubMed
.
- A. Ogawa, Anal. Sci., 2021, 37, 407–414 CrossRef CAS PubMed
.
- A. Ogawa, Methods Mol. Biol., 2014, 1111, 165–181 CrossRef CAS PubMed
.
- A. Ogawa, Methods Enzymol., 2015, 550, 109–128 CAS
.
| This journal is © The Royal Society of Chemistry 2021 |