Open Access Article
Open Access ArticleSingle-cell droplet microfluidics for biomedical applications
Dan
Liu
,
Meilin
Sun
,
Jinwei
Zhang
 ,
Rui
Hu
,
Wenzhu
Fu
,
Tingting
Xuanyuan
and
Wenming
Liu
,
Rui
Hu
,
Wenzhu
Fu
,
Tingting
Xuanyuan
and
Wenming
Liu
 *
*
Departments of Biomedical Engineering and Pathology, School of Basic Medical Science, Central South University, Changsha, Hunan 410013, China. E-mail: liuwenming0229@csu.edu.cn
First published on 9th April 2022
Abstract
Single-cell manipulation and analysis is critical to the study of many fundamental biological processes and uncovering cellular heterogeneity, and presents the potential for extremely valuable applications in biomedical fields, including neuroscience, regenerative therapy, early diagnosis, and drug screening. The use of microfluidic technologies in single-cell manipulation and analysis is one of the most promising approaches and enables the creation of innovative conditions that are impractical or impossible to achieve using conventional methods. Herein, an overview of the technological development of single-cell droplet microfluidics is presented. The significant advantages of microfluidic droplet technology, the dynamic parameters affecting droplet production, and the geometric structures of microfluidic devices are emphasized. Furthermore, the progress to date in passive and active droplet generation methods based on microfluidics and various microfluidic tools for the production of single-cell droplets and hydrogel microspheres are summarized. Their key features, achievements, and limitations associated with single-cell droplet and hydrogel formation are discussed. The recent popularized applications of single-cell droplet microfluidics in biomedicine involving small-molecule detection, protein analysis, and drug screening and genetic analysis of single cells are explored too. Finally, the challenges that must be overcome to enable future applications in single-cell droplet microfluidics are highlighted.
1. Introduction
Cells with diverse structures and functions interact to form complex organisms.1 Because of the diversity of their functions, cells in living organisms have obvious heterogeneity in terms of, e.g., cell size, morphology, growth rate, and biomolecule expression.2,3 These differences often play a crucial role in the growth, differentiation, and development of living organisms. The size of an individual cell is very small (approximately 1–15 μm) and single-cell secretion occurs at very low levels, meaning the detection of single-cell-secreted molecules requires a high degree of sensitivity.4 Conventional methods for cell analysis often rely on the use of cell populations. Although this can ensure a sufficient content of detection substance, it can obscure data signals produced by small numbers of abnormal cells, resulting in the loss of important information.5,6 Single-cell analysis not only overcomes the problem of loss of small-sample abnormal cell data in cell population analysis, but can also reveal differences between cells more accurately. As such, it has become an important emerging field in biological and biomedical research. The analysis of cells at the single-cell level is an effective way for detecting cell heterogeneity and differentiation, and also aids in the understanding of cell behavior and metabolic activities to enable a comprehensive analysis of the behavior of tissues, organs, and even the entire organism.7Single-cell analysis holds great promise for understanding cell diversity and discovering abnormal cells formed during disease development. Single-cell technologies have been applied in many biomedical fields, including neuroscience,8 stem cell biology,9 early diagnosis of diseases,10,11 and drug screening.12–14 High-throughput single-cell analysis is needed to accurately describe and ultimately clarify the underlying causes of the heterogeneity of cells in organisms. Classic methods of single-cell analysis include flow cytometry,15 atomic force microscopy,16 optical17 and magnetic tweezing,18 and single-molecule fluorescence spectroscopy,19etc. However, each of these methods has limitations: for example, although conventional flow cytometry is automated and enables high-throughput, high-efficiency cell sorting in single-cell analysis, it requires the use of expensive instruments and complex cell surface markers, and has long sample pretreatment times, reduced cell viability, and a risk of sample contamination.20 Furthermore, other methods allow accurate manipulation of single cells, but suffer more or less from high cost, complex operation, and low detection flux problems.
Because of advantages such as low reagent consumption, high detection sensitivity, and large-scale subsystem integration, microfluidics has become a widely used technology in the life sciences and has proved to be a powerful tool for efficient single-cell analysis. The fundamental capability of microfluidic technology in terms of single-cell analysis is the manipulation of single cells to enable, for instance, single-cell capture. Microfluidic single-cell capture technologies implement a wide variety of capture strategies, including microwells,21 micropatterns,22 microdroplets,23 and microtraps.24 Microwells for single-cell capture have the advantages of simple operation and fabrication and allow for the formation of highly uniform single-cell arrays. However, microwell approaches often require that the reagents be distributed across the entire array, making it necessary to achieve instantaneous deployment of liquid flows and thus hindering the dynamic monitoring of rapid cell responses. Although micropatterns for single-cell capture share some of the advantages of microwells, they require complex pretreatment, which increases the operation time and cost. Microtraps can be used to quickly and conveniently produce high-throughput, high-density single-cell arrays; however, cells are generally squeezed by the liquid flow and chip structure during the capture process, which can potentially damage them. Droplet microfluidics is a promising novel technology for studying the generation, manipulation, and application of microdroplets with sizes ranging from several to hundreds of microns. Unlike the other methods described above, microdroplet technology can encapsulate single cells in monodisperse droplets to enable high-throughput analysis and precise control of the local extracellular environment. Microfluidic droplet technology also has high degree of repeatability, low consumption of samples and reagents, fast reaction speeds, and easily attained precise controllability. Single-cell droplet microfluidics has been widely used in a number of biological experiments, including cell culture,25,26 polymerase chain reaction (PCR),27 protease activity detection,28 and cell arrangement and sorting,29,30 and has become an important platform for high-throughput single-cell manipulation and analysis in biomedicine.
Previously, droplet microfluidics for single-cell manipulation and analysis has been reviewed concerning certain aspects of its progress, including single-cell encapsulation,31 single-cell omics,32–35 and single-cell microgels for diagnostics and therapeutics.36 Besides, several review papers on microfluidic technology for single-cell analysis37 and droplet microfluidics for biomedical applications38,39 have emerged too. However, there are few reviews on single-cell droplet microfluidics for biomedical applications with emphasis on recent advancements. Over the past six years in particular, there has been considerable progress of single-cell droplet microfluidics in single-cell encapsulation,40 operation,41 and analysis,42,43 which has extremely enhanced the potential for developing innovative analytical microsystems for biomedical purposes in the future.
Herein, we overview the recent studies and advances in microfluidic single-cell droplet technology for use in biomedicine (Scheme 1). Our review begins with an introduction of the important advantages of microfluidic droplet technology, the dynamic parameters affecting droplet production, and the geometric structure of microfluidic devices. We also discuss current microfluidics-based passive and active droplet generation methods. We then systematically summarize techniques for the preparation of single-cell droplets and hydrogel microspheres using microfluidic droplet technology. Representative cases are introduced in terms of the pivotal features, defects, and technical challenges of each single-cell droplet/hydrogel production method. Next, we discuss recent applications of single-cell droplet microfluidics in the field of biomedicine, including small-molecule detection, protein analysis, and drug screening and genetic analysis of single cells. Finally, we conclude with a discussion of the development of microfluidic single-cell droplet technologies for biomedical applications and highlight the technical challenges to the widespread application of single-cell droplet microfluidics as well as perspectives relating to the future development of microfluidic single-cell droplet systems.
 | ||
| Scheme 1 Overview of the recent advances in single-cell droplet microfluidics for biomedical utilization. | ||
2. Microfluidic droplet technology
Microfluidic droplet technology refers to technology used to manipulate nanoliter-to-picoliter (nL-to-pL) liquids within microfluidic chips. The basic process of droplet generation involves the production of a continuous-phase liquid flow in a microchannel through an external injection pump, followed by the division of the dispersed-phase liquid into a series of microdroplets by providing shear force in the process of flow. In general, there are three types of formed droplets, which are oil-in-water, water-in-oil, and water-in-water types. Droplet diameter can be regulated at the micron level by modifying the technical parameters (e.g., flow velocity, fluid viscosity, and microchannel geometry), allowing the droplet volume to be controllable according to the experimental requirements.442.1 Important advantages
Microfluidic droplet technology is an important branch of microfluidic technology. Through a combination of microfluidic and droplet technologies, fluids can be operated under mild conditions, while avoiding cross-contamination between samples. This technology has become a reliable scientific research platform, especially for chemical and biological analysis. Microfluidic droplet technology has a number of outstanding advantages: (1) it reduces sample and reagent consumption for analyzing a single cell. The volume of a produced droplet containing a single cell is within the nL-to-pL range, which is remarkably less than the operating volume using the conventional single-cell macro-manipulation (typically microliter). (2) The monodispersity of microdroplets reduces cross-contamination (especially for cells, macromolecules, and most water-soluble small molecules) between themselves. Increasing the spacing between droplets can also extremely reduce the exchange of materials between droplets.45 The independence of each microdroplet provides a relatively stable culture and reaction condition for single-cell analysis. (3) It promotes a rapid detection. When an individual cell is encapsulated in a nL-to-pL droplet for secretion determination, the molecules released by the cell accumulate around it, causing the concentration of cell secretion to increase rapidly to the detection limit, thereby reducing the time required for analysis. (4) The high throughput of microfluidic droplet technology makes it suitable for carrying out single-cell analysis. For an instance, by encapsulating the single-cell DNA templates and primers together within a microdroplet, large numbers of reaction units can be generated per unit time to enable high-throughput single-cell DNA amplification.462.2 Dimensionless numbers
The primary process applied in droplet generation is the application of a force sufficient to disturb the interfacial tension between the dispersed and continuous phases to make them unstable, causing the dispersed phase to break through the shackles of the interfacial tension and enter the continuous phase to form dispersed droplets. In this process, shear force, gravity, surface tension, and inertial and viscous forces act together, with interfacial tension and viscous force constituting the dominant forces.47 Dimensionless parameters such as the Reynolds number, Weber number, bond number, capillary number, viscosity ratio, and flow rate ratio are used to characterize the respective effects.The Reynolds number reflects the features of fluid flow in the microfluidic channel and represents the ratio of inertial to viscous force.48 Generally speaking, there are two states of fluid flow: laminar (low Reynolds number <1) or turbulent (high Reynolds number >1000).49 In microscale channels, the Reynolds number is typically less than one and, therefore, the inertial force is negligible relative to the viscous force and the flow follows typical laminar. Turbulence is dominated by inertial forces, which produce random eddies, vortices, and other chaotic fluctuations. It is worth noting that the mixing of fluids during chemical reactions in a microfluidic system is primarily controlled by diffusion. Although the Reynolds number can effectively characterize the dynamics of single-phase flow in microfluidic channels, it is not often used to describe the process of droplet generation. The bond number, a dimensionless parameter used to investigate the action of gravity in multiphase flow, characterizes the ratio of gravitational force to interfacial tension. In microscale flows, the bond number is generally on the order of 10−3. This means that the interface force is dominant and the influence of gravity can be ignored.50 The droplet size and formation rate can be adjusted based on the bond number by modifying its impact factors such as the density and interfacial tension of the fluid. The Weber number is the ratio of inertial force to interfacial tension. As the inertial force of the fluid in a microfluidic channel is negligible (typically <1),51 the interface effect is very important for the formation of droplets. In microscale flow, viscous force and interfacial tension play important roles in the process of droplet formation, making the relative effect of viscous force on interfacial tension, as expressed by capillary number, a key parameter.52 The capillary number is usually between 10−3 and 10. At small capillary number, the interfacial tension is dominant, the interfacial surface area is minimum, and spherical droplets are formed. At large capillary number, by contrast, the viscous force plays a dominant role, causing the droplet interfaces to gradually deform and the originally spherically symmetrical droplets to transition into non-spherical droplets.47 The regulation of the capillary number and the Weber number can influence the droplet formation state, such as dripping and jetting states in co-flowing liquid streams.36
2.3 Device geometry
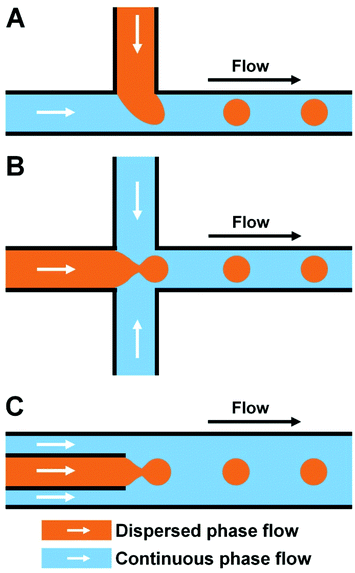
The most important technical requirement of microfluidic droplet technology is the stable formation of droplets. Currently, a number of chips with diverse structures are used to achieve robustness, controllability, and high-throughput generation of different droplets: commonly, these can be divided into T-junction, flow-focusing, and coaxial focusing structures.2.4 Droplet generation
Depending on whether an external force is needed, droplet generation typically includes two types, that is, passive and active generation of microdroplets, as shown in Table 1.58,63–76 Passive methods require no external force, and have simple structure of the microfluidic chip. The formation of droplets is controlled mainly by modifying the microchannel structure and two-phase velocity ratio. Active methods produce droplets through external force, and generally have complex device geometries. For more information on the specific principles of droplet generation, the reader is referred to previous reviews.77–79 A description with recent research reports is presented here.| Droplet generation | External energy | Mechanism | Device geometry | Type of droplet generation | Ref. |
|---|---|---|---|---|---|
| Passive | No | Fluid power | T-junction | Random | 58 |
| No | Fluid power | Flow focusing | Random | 63 | |
| No | Fluid power | Co-flow | Random | 64 | |
| Active | Electrical | DC and AC electric fields | Flow focusing | Tunable | 65 |
| Co-planar electrodes | T-junction | On-demand | 70 | ||
| Magnetic | Non-uniform magnetic field | T-junction | Tunable | 66 | |
| Square wave magnetic field | Flow focusing | Tunable | 71 | ||
| Centrifugal | Rotation-induced centrifugal force | T-junction | Tunable | 72 | |
| Tabletop mini-centrifuge | Co-flow | On-demand | 73 | ||
| Optical | Infrared laser | Flow focusing | On-demand | 68 | |
| Thermal | Temperature | Flow focusing | Tunable | 74 | |
| Pulse heating | T-junction | On-demand | 75 | ||
| Acoustic | Surface acoustic wave | T-junction | On-demand | 69 | |
| Mechanical | Mechanical vibration | Co-flow | Tunable | 67 | |
| Pneumatic valve | Flow focusing | Tunable | 76 |
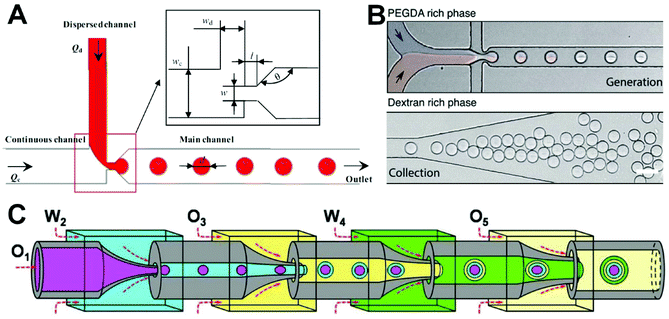 | ||
| Fig. 2 Passive microfluidic droplet generation. (A) Monodisperse microdroplet production in a T-junction microfluidic device with a constriction microchannel. Reproduced from ref. 58 with permission from Springer Nature, copyright 2018. (B) Water-in-water microdroplet production in a flow-focusing microfluidic device. Reproduced from ref. 63 with permission from the Royal Society of Chemistry, copyright 2020. (C) Microdroplet generation of Trojan-horse-like microcapsules with capsule-in-capsule structures for diverse programmed sequential release using a glass-capillary microfluidic device based on coaxial focusing principle. Reproduced from ref. 64 under the terms of the Creative Commons Attribution License. Copyright 2018, the Authors, published by John Wiley and Sons. | ||
Most microdroplets are oil–water or water–oil droplets and generally require the use of organic reagents and surfactants, which limits their wider application in biological and cellular research. Therefore, it would be more appropriate to prepare water-in-water microdroplets without oil and surfactants. Mazutis's group proposed a droplet preparation approach using an aqueous two-phase system comprising dextran and acrylate-modified polyethylene glycol (Fig. 2B).63 Their method was used to encapsulate Gram-negative and -positive bacterial cells and to test the efficiency of the genome amplification reaction. This system has obvious advantages in terms of good biocompatibility, flexibility, and high throughput, and has potential uses in a variety of chemical and biological applications such as microcarrier preparation and tissue engineering. However, the strategy for encapsulating of most cells under passive droplet generation depends on the use of cells with a Poisson spatial distribution and, as a result, the number of microdroplets containing individual cells will be very small. Therefore, the post-separation to obtain droplets containing single cells is necessary.
Recently, multi-compartment microcapsules have been widely used in biomedical fields such as drug delivery, combined cancer therapy, tissue regeneration, and restriction enzyme reactions because they allow multiple components to be encapsulated separately. Mou et al. reported a method on the basis of a coaxial microfluidic device to prepare uniform Trojan-horse-like stimuli-responsive microcapsules (Fig. 2C).64 Specifically, uniform O1/W2/O3/W4/O5 microcapsules are prepared using the four-emulsion method. Each microcapsule contains two stimulation-responsive hydrogel shells that can release the contents of the capsule compartment via shell decomposition or fracturing. The prepared microcapsules can be used as candidate materials for controlled release and microreactions. In addition, Qin's group designed a flow-focusing microfluidic device, which forms a stable water–water interface through spontaneous phase separation of dextran and polyethylene glycol from core and shell flows, and then forms multi-compartment microcapsules under the action of oil phase shear.83
There has been some recent progress in developing active droplet generation methods in which various forces, including electrical, magnetic, mechanical, optical, and acoustic forces, are used to manipulate the microfluid to produce droplets. Because of their rapid response speeds and good compatibility with microchannel structures, electric fields are widely used in the generation of droplets. Yin et al. demonstrated a method for accurately forming oil-in-water droplets with controllable sizes in microchannels without surface treatment through the application of DC- and AC-based electric fields (Fig. 3A).65 They found that the sizes of droplets formed under the control of a square-wave electric field followed an inverse linear relationship with the field frequency and could be changed by adjusting the voltage and frequency of the field. The primary advantages of electric field methods are their fast responses, ability to accurately control droplet formation, and elimination of the need for surface treatment required in conventional PDMS microchannels, which greatly reduces the hydrodynamic characteristic requirements and solves the problem of wettability of materials in the process of droplet formation. Furthermore, Samlali et al. developed a hybrid microfluidic platform based on a co-planar electrode component to precisely control the droplet generation for single-cell isolation.70 Unfortunately, this system can only achieve the operation of a dozen droplets/single cells and its throughput needs to improve.
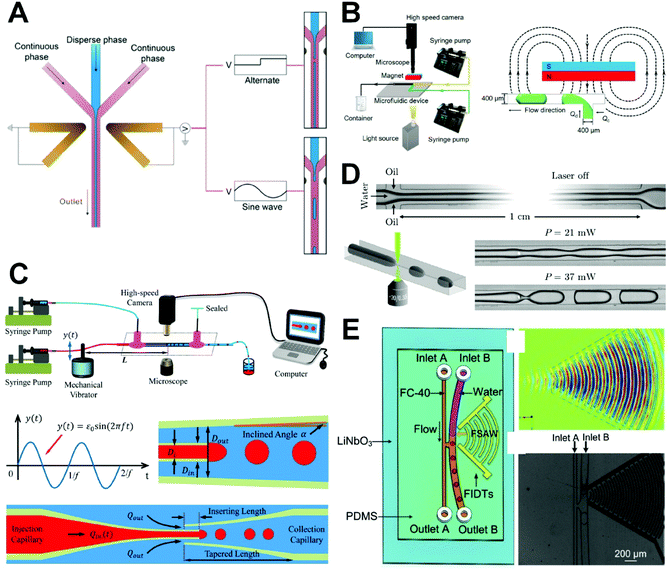 | ||
| Fig. 3 Active microfluidic droplet generation. (A) Microdroplet production using DC and AC electric fields in a flow-focusing microfluidic device. Reproduced from ref. 65 with permission from American Chemical Society, copyright 2019. (B) Ferrofluid droplet formation using magnetic field in a T-junction microfluidic device. Reproduced from ref. 66 with permission from Elsevier, copyright 2018. (C) Microdroplet production in a coaxial focusing microfluidic device with mechanical vibration. Reproduced from ref. 67 with permission from Springer Nature, copyright 2016. (D) Droplet generation using a focused laser in a flow-focusing microfluidic device. Reproduced from ref. 84 with permission from American Physical Society, copyright 2015. (E) Microdroplet formation induced by focused surface acoustic waves in a T-junction microfluidic device. Reproduced from ref. 69 with permission from Elsevier, copyright 2019. | ||
The use of magnetic fields to generate droplets has the advantages of non-heat generation, operational simplicity, and avoidance of contact with samples. In addition, environmental parameters such as temperature, ion concentration, and pH have little effect on magnetic fields and magnetic forces, which enables new approaches to microfluidic droplet technology. Zhang et al. proposed a method for generating droplets using a magnetic field in a T-junction microfluidic device (Fig. 3B)66 in which a permanent magnet was placed on one side of the junction to form a non-uniform magnetic field. They explored the influence of the two-phase flow rate and magnetic flux density on droplet size, demonstrating that it increases with the magnetic flux density and decreases as the two-phase flow ratio and capillary number increase. The size of ferrofluid droplets can be adjusted too by changing the square wave magnetic field in a flow-focusing device.71 The precise manipulation of ferromagnetic droplets using magnetic force presents new prospects for a wide range of applications in the fields of biology, medicine, and precision instrument control.
Mechanical vibration for droplet production can be achieved by disturbing a dispersed-phase microtubule using a mechanical vibrator. Whereas most active droplet generation methods require integration of additional components such as electrodes into the chip, mechanical vibration relies on the use of a chip-exterior vibrator to disturb the microtubule, enabling simple and convenient operation. Zhu et al. quantitatively described the generation of droplets in coaxial focusing microfluidic channels affected by mechanical vibration by measuring the internal fluid flow fluctuations caused by the vibration (Fig. 3C).67 They found that the droplet frequency was synchronized with the vibrational frequency and that the size of the droplets could be modulated over a large range. Because the dependence of droplet size on microchannel geometry is very weak under mechanical droplet generation, this approach provides an effective method for overcoming the longstanding constriction of droplet size by microchannel geometry. Recently, pneumatic manipulation was demonstrated by Qin's group to be a pretty good way for controllable generation of water-in-water droplets.76 The switch cycle of pneumatic valve acting on the dispersed phase and the flow rates of the dispersed and continuous phases impacted precisely on the size of microdroplets in their integrated microfluidic system, presenting high stability, controllability, and flexibility.
Droplets can also be generated on demand on the basis of optical manipulation using a focused laser. The light forces spatially the deformation of the liquid interface by local Marangoni stresses and causes the thread-to-droplet transition as reported by Delville's group (Fig. 3D).84 Recently, Wang et al. studied the control of droplet generation using a focused infrared laser with a wavelength of 1550 nm.68 They assessed the characteristics of laser-controlled droplets produced under different flow rates, beam powers, and spot positions and compared the conditions required with and without laser control. This work provides a comprehensive resource for understanding infrared laser-controlled droplet generation and for promoting its application in microfluidic droplet technology.
Surface acoustic waves are widely used in on-demand droplet generation because they are high energy, can be confined to propagate along the substrate surface, allow for efficient fluid–solid coupling, and are clean and pollution-free. Jin et al. reported a method of microdroplet formation based on the driving of microfluid by a focused surface acoustic wave and studied the associated mechanism and process of droplet formation (Fig. 3E).69 They observed that size of the droplets was mainly determined by the frequency of the focusing surface acoustic waves, driving voltage, and velocity of the dispersed phase. Their method overcomes the limitations imposed by microchannel structure and capillary number in the general microfluidic droplet technology and has a high degree of uniformity along with a reduced system response time.
Additionally, there are still other active droplet generation methods depending on centrifugal and thermal effects. The centrifugal force induced by the rotation of microfluidic device72 or the centrifugation of glass capillaries using a tabletop mini-centrifuge73 assists the droplet formation in the T-junction or co-flowing channel, respectively. Moreover, the liquid–vapor phase transformation induced by electro-heating and ice pack-aided cooling can help to produce microdroplets with relatively small size,74 and the microwave heating can lead to the change of Laplace pressure at the interface between two fluids and then enable droplet generation on demand.75
In developing microfluidic droplet approaches for cell study, manufacturing convenience, droplet production rate, and biocompatibility are all key parameters that must be evaluated. In view of the limited number of studies on active droplet production methods, however, it is unclear which method is most suitable for cell encapsulation. None of the active methods listed in Table 1 satisfy these conditions optimally. For example, the biocompatibility of lasers and high-voltage electric fields is poor, the formation of centrifugal force-driven droplets is generally harmful to cells, and acoustics, magnetism, and other external driving methods involve either relatively slow generation rates or complex chip integration. Passive methods have disadvantages in terms of low single-cell encapsulation rates and the inability to generate droplets on demand. However, these shortcomings only reflect the current development situation and do not represent inherent limitations, as the use of microfluidic droplet technology for single-cell encapsulation remains underdeveloped. Thus, the application of microfluidic droplet technology in single-cell research has broad research prospects.
3. Single-cell encapsulation
Single-cell analysis has become a popular and indispensable strategy for the improved exploration of the unknown features and dynamics of cells, which vary broadly by type and state (i.e., cell heterogeneity), and for a deeper understanding of the behaviors of tissues, organs, and even entire organisms. Microfluidic droplet technology can be used to generate microdroplets with high throughput, making it possible to conduct large-scale parallel research on single cells. Although the encapsulation rate of single cells is relatively low using a classic microfluidic droplet method, especially based on passive droplet generation, the total number and throughput of single cells are much higher than that using other microfluidic single-cell manipulation methods like microwells and micropatterns.4 Moreover, microdroplets can encapsulate individual cells within nanoliter droplets, which can further reduce the dilution/cross-contamination/adsorption of substances, improve the exchange efficiencies of oxygen, nutrients, and metabolites, maintain cell activity and biological functions, and facilitate cell investigation at the single-cell level. Finally, microfluidic droplet technology enables a flexible single-cell operation and allows the integration of different cell research processes including cell encapsulation, culture, separation/sorting, and detection in a single device. In short, the emergence of single-cell encapsulation based on microfluidic droplet technology provides a new approach for single-cell analysis.3.1 Single-cell encapsulation in droplets
Single-cell encapsulation in droplets is mainly achieved using fluid dynamics and surface tension manipulation. The droplet size is determined by the viscosity of the continuous phase, interfacial tension, microchannel size, and flow rate of the dispersed and continuous phases, with droplet diameters ranging from tens to hundreds of microns. Single-cell microdroplets have high flux and small size, characteristics that make them suitable for high-throughput single-cell analysis. However, the encapsulation of single cells is generally random, especially when based on the passive droplet generation methods which are commonly used for single-cell analysis. The control of single-cell encapsulation needs to be strengthened. To improve encapsulation efficiency for practical application, random encapsulation is usually carried out, followed by further screening according to the process requirements. In addition, the channel structure is tailored to ensure optimal arrangement and encapsulation of cells.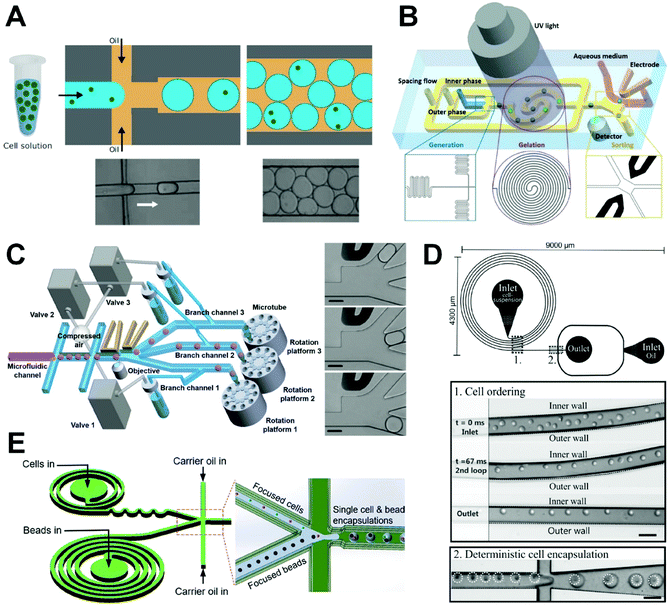 | ||
| Fig. 4 Single-cell encapsulation in droplets. (A) Single-cell microdroplet generation in a flow-focusing microfluidic device. The quantity of cells per microdroplet follows the Poisson distribution. Reproduced from ref. 86 with permission from John Wiley and Sons, copyright 2017. (B) A microfluidic system combined with a sorting electrode for purifying single-cell encapsulation. Reproduced from ref. 88 with permission from John Wiley and Sons, copyright 2019. (C) On-demand droplet collection system based on a combination of droplet microfluidic technology and fluorescence-activated sorting technology to collect required microdroplets continuously. Reproduced from ref. 25 with permission from John Wiley and Sons, copyright 2020. (D) A droplet microfluidic system with curved microchannel structure for generation of microdroplets to achieve significant improvement in single-cell capture efficiency through inertial sequencing caused by Dean force. Reproduced from ref. 91 with permission from the Royal Society of Chemistry, copyright 2012. (E) A microfluidic system combining spiral and serpentine channels to focus cells and microbeads for high-efficiency single-cell pairing. Reproduced from ref. 92 with permission from American Chemical Society, copyright 2019. | ||
In a random encapsulation method, the number of cells per droplet depends on the initial concentration of the cell suspension and the droplet volume but will follow a highly random distribution in which the proportion of single-cell droplets is low. Thus, the degree to which the number of cells encapsulated within an individual droplet can be controlled must be further strengthened. To improve the efficiency of single-cell encapsulation, some researchers have adopted a method of random encapsulation followed by sorting. For example, Navi et al. proposed a method for creating water-in-water droplets using a flow-focused microfluidic device.87 In their device, ferrofluid is integrated into an all-water droplet microfluidic system to enable the diamagnetic separation of single-cell encapsulated and empty droplets. The primary advantage of their platform with respect to the existing platforms is good biocompatibility, with no harm being rendered to the cells, and a chip that is simple and easy to operate. By using an antimagnetic force to manipulate the cell-encapsulating droplets, the droplets do not need to be magnetized and the cells therefore do not come into direct contact with the magnetic material, enabling an unlabeled cell operation with a separation purity of 100%. This simple and biocompatible microfluidic platform is quite applicable in single-cell analysis. Further, Nan et al. introduced a microfluidic system to produce and sort single-cell microencapsulations (Fig. 4B),88 through which they were able to achieve high-throughput preparation of microdroplets and microgels in microfluidic devices. Importantly, an on-chip sorting electrode was used to complete the selection of single-cell microencapsulations while transferring them to the culture medium. At a cell density of 3.05 × 106 cell per mL and a droplet diameter of 50 μm, they achieved a single-cell encapsulation rate of 16%. After sorting, the single-cell encapsulation rate increased to more than 80%. This method can be applied to integrate droplet gelation, single-cell-laden microgel sorting, and transference to the medium to enable high-throughput analysis at the single-cell level in comprehensively assessing cell heterogeneity. More recently, Shum's group developed an on-demand droplet collection system based on a combination of microfluidic droplet and fluorescence-activated sorting technology, that could collect required droplets continuously and quantitatively into microtubules without significant sample loss or microfluidic interruption (Fig. 4C).25 Specifically, their approach allows each branch channel to be alternately sorted, distributed, and collected following droplet generation to achieve the continuous collection of single-cell droplets. The encapsulated single-cell droplets are then effectively captured on a culture plate, and further cultured to study cell behavior and fully analyze the specificity of individual cells. Through the use of fluorescence-activated droplets sorting and valve-driven distribution, this method can screen single cells at a frequency of more than 200 Hz with an individual cell encapsulation efficiency and recovery rate of 98.5% and 99%, respectively. This approach is much more efficient than conventional manual cell selection and laser-capture microdissection technology and has broad applicational prospects in single-cell-level analysis tasks, such as cytokine detection in immunology, mass spectrometry in proteomics, and whole-genome sequencing of the transcriptome.
Kemna et al. were the first to use Dean flow to form single-cell arrangements, followed by single-cell encapsulation (Fig. 4D).91 Dean flow can induce the cells to gather within balanced positionings to achieve desired spatial arrangements, ensure that the cell flow cycle is consistent with the droplet generation frequency, and improve the encapsulation efficiency to 77%. Their method uses the microstructure to control the regular arrangement of cells and then complete the encapsulation of single cells, which effectively improves the ratio of single-cell droplets. However, this requires not only control of the formation of Dean flow but also the synchronization of cell and droplet frequencies, which is difficult to achieve.
He's group combined spiral and serpentine channels to efficiently focus cells and microbeads. They used their approach to encapsulate both barcoded microbeads and human mouse cells and then characterized the cell heterogeneity via sequencing (Fig. 4E).92 Using this method, they achieved 300% and 40% increases in cell utilization rate relative to the conventional Drop-seq device and the device applied only for focusing microbeads,93 respectively. The results supported an enhanced operation efficiency of their microfluidic chip. This chip design has great potential for achieving efficient single-cell expression profiles.
3.2 Single-cell encapsulation in hydrogels
In addition to simulating a three-dimensional (3D) microenvironment to support the activities and functions of cells, the encapsulation of single cells in microscale hydrogels can act as a good tool for conducting independent cell manipulation and monitoring, which is critical for exploring single-cell activities in an in vivo-like microenvironment. Unlike single-cell encapsulated droplets, microgels can form 3D network structures with morphologies that can be controlled by adjusting the concentrations of gel monomers or polymers. This ensures the supply of oxygen, nutrients, and growth factors and the timely discharge of cell metabolic waste, which makes it possible to culture cells in gel microspheres for periods of time. Through the application of appropriate chemical or physical methods, the uncured gel in a droplet can be activated to transform into a gel microsphere. Chemical gelation can be accomplished via light-induced cross-linking,88 redox-induced polymerization94 or intermolecular chemical cross-linking reactions.95 Although hydrogels prepared by chemical gelation have greater stabilities, longer durabilities, and better mechanical properties, the use of chemical cross-linking agents may lead to cytotoxicity and the low specificity of some chemical reactions may lead to unexpected interactions with proteins and cells.96,97 Physical gelation can be achieved via thermally mediated sol–gel transition98 or physical cross-linking reaction between ions.99 In thermally produced gelation processes, the droplets containing the polymer are in a liquid state when the temperature is higher than the gelation temperature, and reducing the temperature to below the gelation temperature will cause the linear molecules of the polymer to polymerize and form a gel network structure, inducing the droplets to solidify into gel microspheres. Another gelation method is physical cross-linking based on electrostatic interactions. One common physical cross-linking gelation method is the conversion of sodium alginate to calcium alginate gel via the chelation of calcium ions and carboxylic acid groups in the glucose unit of sodium alginate.100 Although the physical gelation process can be carried out under mild conditions, the physical hydrogel network will generally have a low mechanical strength and poor stability within the tissue.97 Thus, the physical method is more suitable for encapsulating cells and has better biocompatibility than the chemical cross-linking method. Two types of materials—synthetic and natural polymers—are used to produce hydrogel microspheres for encapsulating single cells.In general, a synthetic polymer polymerized by ultraviolet light is applied to form gel microspheres for encapsulating cells. A cell suspension containing a photoinitiator, cross-linking agent, and monomer is used as the dispersed phase. After the microdroplets have been produced in the microfluidic chip, the photoinitiator, under irradiation by ultraviolet light, promotes the cross-linking reaction of the monomer under the action of a cross-linking agent to produce gelated microspheres. Kamperman et al. used polyethylene glycol diacrylate as a raw material to form microdroplets encapsulated with pluripotent human mesenchymal stem cells or bovine chondrocytes in a microfluidic flow-focusing device at an encapsulating speed of 1 kHz (Fig. 5A).101 The droplets were photocrosslinked through an external ultraviolet light source in the downstream channel to prepare the cell-loading microgels and maintain high cell activity. Single-cell microgels (>90% purity) sorted by flow cytometry were then mixed with different biocompatible materials such as polyethylene glycol diacrylate and alginate to create various modular bioinks, which can be used for 3D printing to rebuild the cellular microenvironment at the single-cell resolution. However, ultraviolet irradiation has some toxic effects on cells. There are two potential ways for addressing this problem: (1) the exposure time to ultraviolet irradiation can be reduced or the ultraviolet wavelength can be adjusted; (2) new polymerization methods can be developed to avoid cell damage.
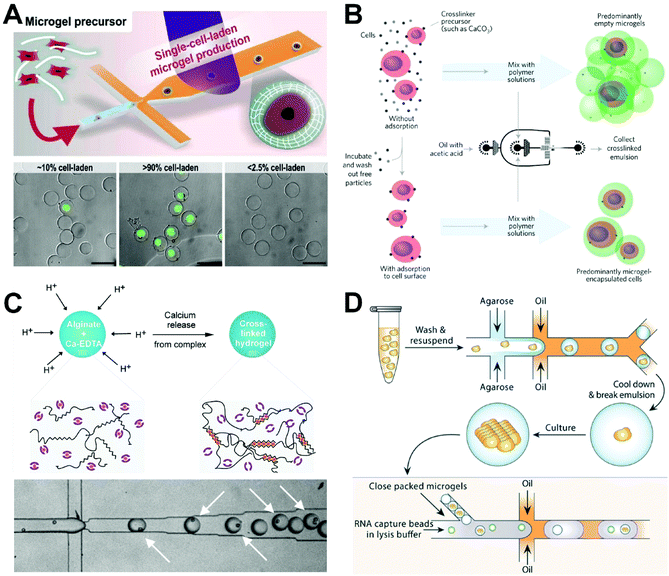 | ||
| Fig. 5 Single-cell encapsulation in hydrogels. (A) Production of single cell-laden microgels using polyethylene glycol diacrylate to maintain high cell activity. Reproduced from ref. 101 with permission from John Wiley and Sons, copyright 2017. (B) Deterministic encapsulation of single cells in thin alginate layer by an internal gelation method. Reproduced from ref. 111 with permission from Springer Nature, copyright 2017. (C) Microfluidic fabrication of cell-laden microgels with uniform structures. Reproduced from ref. 104 with permission from John Wiley and Sons, copyright 2015. (D) Colony-containing agarose microgels produced by encapsulating single yeast cells into agarose microdroplets, retrieving microgels, and then culturing for colony formation within the microgels. Reproduced from ref. 114 with permission from the Royal Society of Chemistry, copyright 2019. | ||
Alginate is a water-soluble linear macromolecule polysaccharide compound extracted from brown algae that is formed from β-D-manuronic and α-L-guluronic acid units in different proportions.108 The carboxyl group in a guluronic acid unit can react with divalent cations such as calcium, barium, and magnesium, resulting in rapid gel formation.100 Because sodium alginate is non-toxic and has a mild gelation mode and good biocompatibility, it is often used to generate gel microspheres for encapsulating cells and can be applied in cell biology, tissue engineering, drug screening, and other biological fields. In general, there are two methods for controlling the gelation process of sodium alginate: external gelation, in which the cross-linking agent diffuses outside of the droplets; and internal gelation, in which the cross-linking agent is loaded into the aqueous phase and triggered by stimulation.
Using external gelation, Liao et al. encapsulated cells in biocompatible sodium alginate droplets and enveloped them in oil droplets to form double emulsions based on a double flow-focusing regime.109 Oil-diffused Ca2+ enters into the sodium alginate to form microgels. Although this gelation method is helpful in maintaining high cell viability, complete gelation of a microgel cannot be achieved within short time frames because of the difficulty of calcium diffusion. To solve the problem of incomplete gelation, researchers have proposed a droplet-fusing method to better prepare single-cell microgels. A new method for the preparation of single-cell microgels based on microfluidic droplet technology was reported by Liu et al.110 They used a cell suspension containing 2% sodium alginate or 1% CaCl2 solution as the dispersed phase and soybean oil as the continuous phase to produce pairs of distinct droplets. The two droplets met and fused downstream and Ca2+ gelated the sodium alginate to form gel microspheres with encapsulated cells. However, because the cross-linking occurred before the calcium ions were uniformly distributed, the homogeneity of the microgel was reduced. In addition, fusion typically led to an increase in the volume of the final cross-linked alginate microgel.
Mooney's group developed an internal gelation method for preparing single-cell microgels using a flow-focusing microfluidic device (Fig. 5B).111 Specifically, cells are suspended in a solution of calcium carbonate nanoparticles, which are adsorbed onto the cell surfaces. After washing of the excess nanoparticles, the cell suspension is mixed with a sodium alginate solution as the dispersed phase to form water-in-oil droplets. Because of the presence of acetic acid in the oil phase, H+ diffusion releases calcium, which mediates the gelation of sodium alginate to form microspheres. Although this method can significantly improve the efficiency of single-cell encapsulation without additional downstream treatment, only the droplets containing cells will be cross-linked and the structure of the resulting gel microspheres will be uneven. To solve the problem of uneven gel microsphere structure, Utech et al. proposed a method to prepare monodisperse alginate microgels with uniform structures using microfluidic droplet technology (Fig. 5C).104 The use of a water-soluble calcium complex as a cross-linking precursor allows for the homogeneous distribution of calcium ions within the generated alginate droplets. Specifically, following encapsulation of single mesenchymal stem cells in droplets, acetic acid is added to the continuous phase to dissociate the complexes and release calcium ions, which in turn react with sodium alginate to form alginate microgels with a uniform structure.
Agarose is a natural linear polysaccharide extracted from the cell walls of red algae that is composed of β-D-galactose and α-3,6-galactoside. It is a typical natural polymer that is initiated by temperature in a solution at 37 °C and then cross-linked into a gel at 20 °C.112 Agarose—in particular low-melting-point agarose—is suitable for cell encapsulation experiments due to its good biocompatibility and mild gelation conditions.113 For example, Liu et al. proposed isogenic colony sequencing as a general method for the high-throughput analysis of gene expression in cells (Fig. 5D).114 In their process, single yeast cells are encapsulated with low-melting-point agarose to form agarose droplets which are collected into a 50 mL centrifuge tube placed on ice to form agarose microgels. The agarose microgels are resuspended in a suitable medium for overnight culturing to form single-yeast-cell colony microgels, which provide sufficient RNA for colony deep sequencing and reduce the errors arising in individual single-cell gene expression profiles.
Gelatin is a mixture of amino acids such as glycine, proline, and hydroxyproline that forms through the degradation of collagen in connective tissues such as skin, bone, and muscle tendon.112 Gelatin also forms as a network structure through temperature-induced polymerization. Li et al. used gelatin as a hydrogel material to encapsulate microalgae cells (E. gracilis and C. reinhardtii) for single-cell culture and screening.106 Based on the flow cytometry-assisted sorting, the cultured monoclonal populations of microalgae with rapid biomass productivity or high lipid productivity can be selected for multiple screening. In particular, thermally responsive gelatin can maintain a liquid culture of cells in the range of 20–27 °C, with gelation to form microgels occurring at 4 °C for sorting and degelation occurring for effective cell recovery when the temperature is raised to 35 °C. Although the overall process does not harm the cells, the complex gelatinization mechanism has certain disadvantages, including a long gelatinization time in which it generally takes several hours to form a moderately hard gel, that lead to low experimental efficiency. To overcome this shortcoming, researchers have applied chemical modification to gelatin. Nan et al. used methacrylic gelatin, an optically cross-linked gelatin, that can be rapidly gelatinized by ultraviolet irradiation. Their on-chip gel technology was seamlessly combined with microfluidic droplet technology to achieve high-throughput preparation of gel microspheres.88
To better simulate the physiological environment of cells in vivo, researchers have used multi-composed materials (e.g., gelatin and collagen) to prepare single-cell microgels. The mechanical properties of collagen are poor when it is used alone as a scaffold, but the addition of gelatin improves the mechanical properties of the gel by supporting cell adhesion while retaining the regulatory properties of the collagen. The material properties (stiffness, pore diameter, cross-linking degree, and expansion ratio) of a microgel can be controlled by changing the composition of the droplets and the cross-linking time.115 Encapsulated cell viability is highly dependent on cross-linking time, with longer cross-linking times leading to reduced cell viability. Approximately 70% of the cells in a microgel with a cross-linking time of 8 min were able to survive for one week, indicating that the formation of gel beads does not damage cells and can potentially be used in future single-cell analysis. In addition, single cells in microgels are mostly off-center. To preventing cell escape and realize the special manipulation purpose (e.g., long-term cell culture), Kamperman et al. presented a microfluidic droplet method for centering single cells in microgels being composed of hyaluronic acid and dextran by delaying enzymatic cross-linking.95 This single-cell centering achieved a constant prevention (over 28 d) of cell escape from microgels, which greatly improved the reliability of long-term culture of single cells.
4. Biomedical applications of single-cell droplets and hydrogels
Single-cell heterogeneity has been highlighted in various biomedical fields including physiology, oncology, neurobiology, clinical diagnosis and therapy. The methodological developments of previous microfluidic strategies have achieved dynamic single-cell manipulation, monitoring, and analysis.116–119 Because of its high fluxes, controllable size, and use of independent components, microfluidic droplet technology has attracted a significant amount of attention as a tool for cell-based high-throughput analysis in biomedical applications such as proteolytic activity detection,79,120,121 cytokine secretion,76,122 and gene sequencing.123 Here, we review single-cell droplet applications—including small-molecule detection, protein analysis, drug screening analysis, and genetic analysis—that have been introduced in biomedical fields over the last six years.4.1 Small-molecule detection of single cells
The detection of small molecules in single-cell analysis is a necessary strategy for obtaining a better understanding of living organisms, exploring cell heterogeneity, and the early diagnosis of diseases. Intercellular small molecular substances, such as lactate and reactive oxygen species, are widely involved in the signaling pathways of cells and play an important role in many physiological and pathological processes. For example, one of the salient features of cancer is high glucose metabolism and lactate production. Cancer cells convert glucose into lactate through aerobic glycolysis, which plays an important role in cellular signaling activation, proliferation, and disease progression. Single-cell droplet microfluidics has recently been applied to detect these small molecules. Mongersun et al. proposed a droplet microfluidic method in which passive force is used to place droplet-encapsulated cells into arrays. This method can quickly and quantitatively determine the release of lactate from many single cells.124 Through the conversion of lactate into fluorescent products using a commercial lactate kit, the release rate of lactate in a single cell is determined by the increase of droplet fluorescence. Based on this approach, they developed a method to determine the intercellular differences in lactate release in K562 leukemia and U87 glioblastoma cells and under the condition of chemically inhibited lactate efflux, which provided a new tool for the study of highly heterogeneous cell populations. The fluorescence detection of lactate in single circulating tumor cells (CTC) from metastatic patients was demonstrated too by Scoles’ group using a microfluidic droplet device with T-junction channel in the same year.125 They achieved the detection of 10 tumor cells from 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 blood cells. This work provides a proof-of-concept demonstration of single CTC identification involving metabolism.
000 blood cells. This work provides a proof-of-concept demonstration of single CTC identification involving metabolism.
More recently, Xu's group developed a surface-enhanced Raman scattering (SERS)-microfluidic droplet platform to enable the label-free simultaneous analysis of multiplexed metabolites at the single-cell level via a versatile magnetic SERS substrate composed of silver nanoparticle-decorated Fe3O4 magnetic microspheres (Fig. 6A).126 They achieved label-free, nondestructive, simultaneous determination of three single-cell metabolites: pyruvate, adenosine triphosphate, and lactate. Their metal-magnetic composite substrate is useful in achieving the efficient adsorption of single-cell metabolites and the fast separation from complex matrices, and has high SERS sensitivity making the SERS-microdroplet platform a powerful tool for exploring single-cell heterogeneity at the metabolic level. Single-cell droplets can also be sorted based on small-molecule dynamics. Zielke et al. developed a droplet microfluidic device to separate cancer cell subpopulations based on glycolysis, an approach that does not require the use of markers or active sorting components.127 Their technology is used to control the secretion of protons in the process of glycolysis in breast cancer cells under specific surfactant environments to decrease the pH values of the droplets. As the droplet pH value decreases, the interfacial tension of droplets increases, enabling the separation of cells at different glycolytic levels. Because cancer cells are exposed to different oxygen and nutrient environments, they differ in terms of their glycolysis reactions. This method can be used to separate cancer cells from normal cells based on selected metabolic differences and to identify cells with similar physical characteristics.
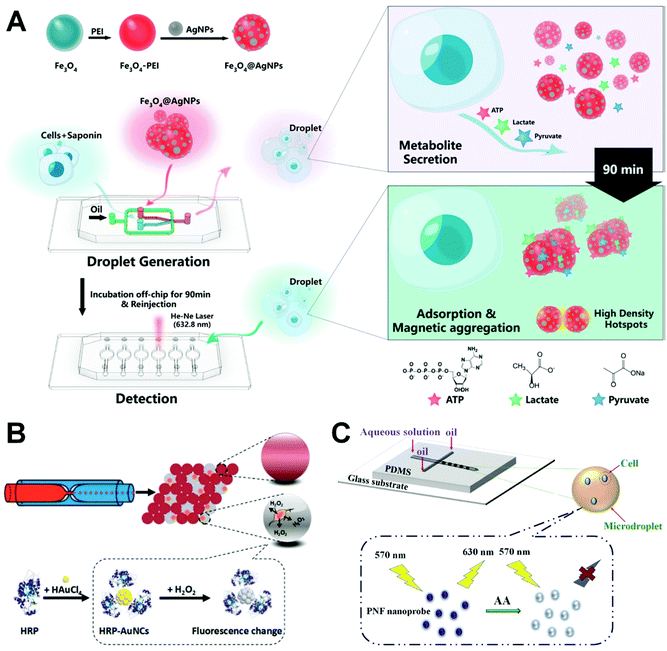 | ||
| Fig. 6 Applications of single-cell droplet microfluidics for small-molecule detection. (A) A SERS-microfluidic droplet system for single-cell encapsulation and simultaneous detection of three metabolites produced by single cells. Reproduced from ref. 126 with permission from American Chemical Society, copyright 2019. (B) Detecting hydrogen peroxide in the single-cell droplets in combination with horseradish peroxidase-modified gold nanoclusters. Reproduced from ref. 130 with permission from American Chemical Society, copyright 2018. (C) Rapid detection and imaging ascorbic acid in single cells using a droplet microfluidic platform. Reproduced from ref. 131 with permission from Elsevier, copyright 2020. | ||
Intracellular reactive oxygen species, such as superoxide anion128 and hydrogen peroxide (H2O2),129 participate in the regulation of the immune system, cell growth and migration, metabolic regulation, important biological substance synthesis, and other physiological processes. Liu's group developed a highly sensitive droplet microfluidic method for the quantitative determination of H2O2 secreted by different types of single cells combined with Au nanoclusters (Fig. 6B).130 H2O2 secreted by a single cell can induce significant fluorescence changes in horseradish peroxidise-modified Au nanoclusters. Their results showed that fast-growing cancer cells can produce secretions with higher H2O2 content than normal cell lines, which provides a useful tool for studying the intercellular differences in H2O2 secretion at the single-cell level.
Ascorbic acid (AA), also known as vitamin C, is a strong antioxidant and a cofactor in enzymatic reactions, and can prevent free radical-induced diseases, especially Parkinson's disease and cancers. AA can efficiently scavenge toxic free radicals and other reactive oxygen species that are associated with several forms of tissue damage and disease, making it useful to develop facile, rapid and simple analytical methods for the highly selective and reliable measurement of AA. Recently, Alizadeh et al. reported a droplet microfluidic approach for the intracellular imaging of AA in living cells (Fig. 6C).131 By isolating small microdroplets, they were able to rapidly detect and image AA in single cells. Their sensing system is based on a non-oxidation reduction strategy in which a fluorescent polymer nanocomposite is applied. Their technique provides a robust method and an ideal platform for studying single cells and a promising novel tool for molecular conducting biology assays in clinical assessment.
4.2 Protein analysis of single cells
Protein is a biological macromolecule with a specific function in living cells that is directly related to the specificity of cell behavior and metabolism. The proteins expressed in different cells have obvious heterogeneities, and there are many types of protein within a single cell. Although there is a high abundance of protein types, the number of proteins with important functions is very low. Thus, the quantitative analysis of specific proteins based on single cells is very important in revealing the heterogeneity of cells in a population to improve the early diagnosis of diseases (such as cancer) caused by a single cell or a small group of cells. Flow cytometry, the classic single-cell protein analysis method, can detect the content of specific functional proteins by immunolabelling a variety of functional proteins.132 However, dynamic detection and analysis of specific proteins secreted by single cells based on the conventional flow cytometry is still challenging due to the low abundance of secreted proteins, lack of sufficient sensitivity and requirement of cell fixing. Microfluidic droplet technology has broad applicational prospects in single-cell protein analysis because of its small size, high detection sensitivity, and high throughput. In the following, we discuss the quantitative analysis of proteins such as enzymes and cytokines secreted by single cells to express the heterogeneity of cell populations.In the tumor metastasis process, malignant tumor cells break through a surrounding physiological barrier and migrate. Tumor and immune cells have different migration abilities in vivo. Secreted products such as matrix metalloproteinases (MMPs) represent promising drug targets and biomarker candidates and provide valuable information for evaluating cancer metastasis and, based on an analysis of their activity, allow for the screening of new drugs to regulate inflammatory responses and tumorigenesis.133–135 Yu et al. reported a microfluidic approach for the detection of MMP9 (a gelatinase subgroup of MMPs) enzyme activity in individual tumor cell droplets using a flow-focusing capillary microfluidic device (Fig. 7A).121 MMP9 secreted by cells is able to cleave the peptide modified by a pair of fluorophore/quenching molecules (FITC and DABCYL). This can be used to measure the activity of MMP9 enzyme in microdroplets in terms of fluorescence intensity, enabling the quantitative analysis of the proteolysis activity of MMP9 secreted by a single cancer cell with a high degree of reproducibility to evaluate the invasive function of cancer cells. The ultra-high sensitivity of this method in the detection of cellular aggressiveness makes it applicable in high-throughput screening for cell diagnosis and cancer research. Because of the fluidity of carrier oil, however, existing droplet-based single cell matrix metalloproteinase (MMP) analysis methods are rarely able to continuously track the proteolysis content of droplets. To improve this, Mu's group described the use of a thermosetting oil in the real-time monitoring of MMP analysis of droplets through a process in which droplets are fixed by converting them into solids following formation (Fig. 7B).135 This method can enable the real-time monitoring of single-cell proteolytic activity without compromising the flexibility of the droplet microfluidics, allowing for the calculation of a reaction rate according to a real-time fluorescence curve. The obvious cellular heterogeneities in MMP activity revealed by this process indicate its potential for use in other types of cell-based high-resolution dynamic information analysis.
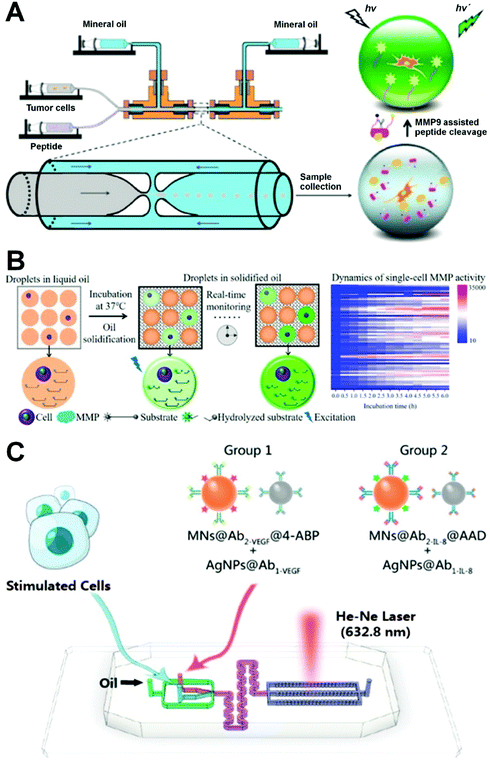 | ||
| Fig. 7 Applications of single-cell droplet microfluidics for protein analysis. (A) A glass-capillary microfluidic device for single-cell encapsulation and detection of the enzymatic activities of MMP9 secreted by single cells in microdroplets. Reproduced from ref. 121 with permission from American Chemical Society, copyright 2017. (B) Dynamic detection of single-cell MMP activity in a droplet array immobilized by thermosetting oil. Reproduced from ref. 135 with permission from American Chemical Society, copyright 2021. (C) A droplet microfluidic platform for single-cell encapsulation and detection of VEGF and IL-8 secreted by single cells. Reproduced from ref. 138 with permission from American Chemical Society, copyright 2019. | ||
Cytokines secreted by mammalian cells (e.g., immune cells, endothelial cells, and cancer cells) play a key role in infection, immune response, inflammation, and disease development.136 In particular, the increased expression of cytokines is related to tumor vascularization and growth, which in turn can generate new tumor blood vessels and amplify existing ones.137 Hsu et al. established a microfluidic droplet system for single-cell multiplexed secretomic analysis with high sensitivity.94 A generated droplet contained an individual cell and a hydrogel microparticle synthesized by multiple antibody-modified poly(N-isopropylacrylamide), allowing the simultaneous detection of multiple secretions of the single cancer cell including interleukin-6, interleukin-8 (IL-8), and monocyte chemoattractant protein-1. The system can also accomplish the determination of significant heterogeneity for cell secretions from 6000 cells within 60 min. Furthermore, Xu's group established a SERS-droplet microfluidic platform for the rapid, ultra-sensitive, and simultaneous detection of cytokines including vascular endothelial growth factor (VEGF) and IL-8 secreted by individual cancer cells (Fig. 7C).138 Their results revealed that cell-to-cell interactions can promote cancer cell angiogenesis by up-regulating VEGF and IL-8. The high sensitivity of this method arises from the amplification effect of metal plasma enhancement and magnetic field-induced aggregation, which allow for the formation of high-throughput water-in-oil droplets containing single cells and four immune particles. Furthermore, cell sorting can be performed according to different excretion capacities to explore the intercellular heterogeneity of cytokine release from single cells. This technique provides a new approach to understanding the biological role of various cytokines in tumor vascularization and aggressive tumor growth. Recently, Wimmers et al. presented a microfluidic single-cell droplet system for the immunofluorescence detection of type I interferon (IFN) production in human plasmacytoid dendritic cells (pDCs).139 Type I IFN is a key cytokine of immunity associated with infection and cancer. Their results supported that type I IFN production is restricted to a small amount of individually stimulated pDCs, and indicated stochastic differences in pDC-driven type I IFN production. In addition, Yuan et al. used microfluidic devices to quickly encapsulate single natural killer NK-92 MI cells and their K562 target cells into microdroplets to improve the ability of the single cells to secrete IFN-γ.140 Droplet encapsulation can prevent secretions from spreading to adjacent cells and significantly reduce the false positive and negative rates obtained under non-microfluidic droplet approaches.
4.3 Drug screening analysis of single cells
Because of the heterogeneity among cells, different cells have different responses to certain drugs. Although drug screening using traditional miniaturized microtiter plates is well established and simple to execute, drug screening for single cells still faces some technical obstacles, including the uncontrolled evaporation of the distributed liquid in an open environment.141 Conventional miniaturized microtiter plates can only statically culture cells, which limits their applicability because this approach cannot simulate the natural extracellular microenvironment. By contrast, microfluidic devices can be used perform 3D cell culturing through continuous infusion, giving them the ability to simulate the microenvironment associated with cell physiology in vivo along with the advantages of low sample consumption and cost and high throughput. Brouzes et al. proposed a drop-based microfluidic device that can achieve the cytotoxic effect of a drug library on U937 cells.142 In their approach, they first label the drug concentration library with unique fluorescence and combine it with droplets containing single cells. The combined droplets are incubated for 24 h and then injected into another device to be fused with droplets containing cellular-reactive fluorescent dyes and then briefly incubated on the chip. Finally, fluorescence analysis is performed to reveal the heterogeneity of the single cells. Because of its accurate cell encapsulation, high-throughput droplet generation and operation, and the potential for use in combined screening, the device represents an ideal platform for single-cell drug screening. Recently, Scheler et al. developed a droplet-based digital minimum inhibitory concentration screen that serves as a practical analytical platform for quantifying the single-cell distribution of phenotypic responses to antibiotics and allows for the measuring of the inoculum effect with high accuracy (Fig. 8A).143 Based on results obtained through the treatment of beta-lactamase-carrying Escherichia coli with cefotaxime, they found that antibiotic efficacy is determined by the amount of antibiotic used per bacterial colony forming unit rather than the absolute antibiotic concentration.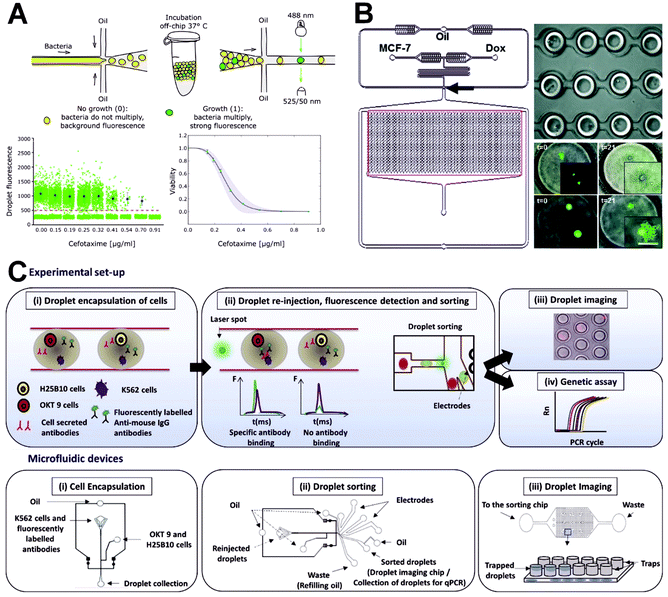 | ||
| Fig. 8 Applications of single-cell droplet microfluidics for drug screening analysis. (A) A droplet-based digital minimum inhibitory concentration screen platform for quantifying the single-cell distribution of phenotypic responses to antibiotics. Reproduced from ref. 143 under the terms of the Creative Commons Attribution License. Copyright 2020, the Authors, published by Springer Nature. (B) An integrated droplet microfluidic array platform for analyzing the uptake and cytotoxicity of doxorubicin in wild-type and doxorubicin-resistant breast cancer cells. Reproduced from ref. 144 with permission from the Royal Society of Chemistry, copyright 2015. (C) A high-throughput droplet microfluidic system for antibody binding detection and antibody screening by using a dual-color normalized fluorescence readout. Reproduced from ref. 146 under the terms of the Creative Commons Attribution License. Copyright 2018, the Authors, published by Elsevier. | ||
Unlike flow cytometry analysis, single-cell drug screening tests are carried out using microdroplet approaches in which the selected compounds are added to isolated individual cells rather than to a cell population. This allows for interactions between cells to be avoided and, because different cells have different sensitivities to drugs, makes it possible to isolate drug-resistant cells from groups of cells for further analysis, potentially providing for the development of new drugs for the early treatment of cancer. For example, Sarkar et al. used an integrated microfluidic droplet array platform to analyze the uptake and cytotoxicity of doxorubicin in wild-type and doxorubicin-resistant breast cancer cells (Fig. 8B).144 They found that drug-sensitive cells were more prone than drug-resistant cells to dying in the presence or absence of doxorubicin (Dox). They also observed polyphasic uptake in individual drug-sensitive cells, whereas the drug-resistant cells demonstrated low uptake and retention. Bithi et al. proposed a pipette-based microfluidic cell separation technique for generating microdroplet arrays.145 Their platform can be used for single-cell drug analysis of small numbers of tumor cells or tumor cell clusters within a small sample volume. They used the platform to carry out single-cell analysis of the response of tumor cells to doxorubicin and found that, although the drug uptakes of individual tumor cells differed, the occurrence of apoptosis was determined by the accumulation of the key intracellular concentration of doxorubicin. Their method provides an ideal platform for understanding tumor cell–drug interactions.
Monoclonal antibodies are one of the largest classes of biopharmaceuticals used clinically against cancer, autoimmune and inflammatory diseases, and other clinical conditions. Technologies to screen antibodies for the binding of cell-surface receptors face certain limitations. Conventional hybridoma screens typically do not allow for the assaying of more than a few thousand clones, are cost intensive, and take several weeks to complete. Furthermore, microtiter-plate-based formats are dependent on cell proliferation to obtain sufficient amounts of antibodies for screening. Thus, it remains a challenge to screen antibodies for the binding of cell-surface receptors (the most important class of drug targets) or for binding to target cells rather than purified proteins. To address this, Shembekar et al. established a high-throughput droplet microfluidic system employing a dual-color normalized fluorescence readout to detect antibody binding (Fig. 8C).146 Their platform can obtain quantitative data on target cell recognition, using as little as 33 fg of IgG per assay. Starting with an excess of hybridoma cells releasing non-specific antibodies, individual clones secreting specific binders can be enriched 220-fold following the sorting of 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 clones in a single experiment, demonstrating the advantages of the system in screening antibodies for the specific binding of cancer target cells in a high-throughput single-cell format. Their results point the way toward new therapeutic antibody discoveries, particularly given the fact that the single-cell approach is also applicable to primary human plasma cells. Eyer et al. also proposed a microfluidic droplet strategy for detection of antibody secretion by individual cells from mice immunized with tetanus toxoid.147 Single cells were encapsulated in microdroplets using a flow-focusing device and then analyzed in a droplet array chip using a fluorescence relocation-based immunoassay. This microfluidic platform allows quantitative, sensitive, and simultaneous analysis of antibody secretion rate, specificity, and affinity for tetanus toxoid. Furthermore, Gérard et al. presented a high-throughput screening of antibodies from millions of non-immortalized cells using a integrated microfluidic droplet device with a sorting electrode component.148 The screening of different antibodies against tetanus toxoid, glucose-6-phosphate isomerase, and tetraspanin-8 is mainly based on fluorescence-assisted single-cell bioassay and sorting, paired VH–VL sequencing of antibodies using in-droplet single-cell barcoded reverse transcription, and bioinformatics analysis. The microfluidic system enabled recovery of ∼450–900 antibody sequences from ∼2200 antibody-secreting cells in each run, which can facilitate the potential antibody identification for therapeutic purposes.
000 clones in a single experiment, demonstrating the advantages of the system in screening antibodies for the specific binding of cancer target cells in a high-throughput single-cell format. Their results point the way toward new therapeutic antibody discoveries, particularly given the fact that the single-cell approach is also applicable to primary human plasma cells. Eyer et al. also proposed a microfluidic droplet strategy for detection of antibody secretion by individual cells from mice immunized with tetanus toxoid.147 Single cells were encapsulated in microdroplets using a flow-focusing device and then analyzed in a droplet array chip using a fluorescence relocation-based immunoassay. This microfluidic platform allows quantitative, sensitive, and simultaneous analysis of antibody secretion rate, specificity, and affinity for tetanus toxoid. Furthermore, Gérard et al. presented a high-throughput screening of antibodies from millions of non-immortalized cells using a integrated microfluidic droplet device with a sorting electrode component.148 The screening of different antibodies against tetanus toxoid, glucose-6-phosphate isomerase, and tetraspanin-8 is mainly based on fluorescence-assisted single-cell bioassay and sorting, paired VH–VL sequencing of antibodies using in-droplet single-cell barcoded reverse transcription, and bioinformatics analysis. The microfluidic system enabled recovery of ∼450–900 antibody sequences from ∼2200 antibody-secreting cells in each run, which can facilitate the potential antibody identification for therapeutic purposes.
4.4 Genetic analysis of single cells
The genetic differences between cells are pretty significant in development, differentiation, signal transduction, and disease, and it is important to develop analytical methods for single-cell gene profiling to explore these life processes. Beyond the separation of single cells, droplet microfluidics is become a promising tool for the separation of single-cell genome, making it an ideal technology for high-throughput research of single-cell genomics for cell biology and clinical medicine.Lan et al. described a method for ultra-high-throughput single-cell genome sequencing using microfluidic droplet technology (Fig. 9A).149 In their approach, individual cells are encapsulated in microgels, which can permeate molecules such as enzymes, detergents, and small molecules with hydraulic diameters smaller than their pore sizes, but can also spatially capture large molecules such as genomic DNA. The use of microgels facilitates the cleaning of encapsulated cells to perform necessary steps such as cell lysis and genome processing while maintaining the separation of individual genomes. Using their approach, they easily carried out a series of operations like cell lysis and genome processing on the encapsulated cells, while maintaining partitions between individual genomes. Through its combination of microgel and microfluidic droplet technology, their method can process 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells in a short time and achieve ultra-high-throughput single-cell genome sequencing. However, their encapsulating process follows a Poisson distribution resulting in a low encapsulation rate. Furthermore, Moon et al. developed a new droplet microfluidic platform, in which high-concentration microbeads containing oligonucleotide barcodes are spontaneously arranged in spiral channels under the effect of inertia, and then coencapsulated with individual cells in droplets for single-cell sequencing (Fig. 9B).93 Through the deterministic encapsulation of inertially ordered beads, their platform significantly improves throughput and reduces barcode errors, significantly reducing intermittent errors in multi-bead packaging relative to the nondeterministic Drop-seq system. Guo et al. developed a rapid, PCR-free single-cell miRNA assay to amplify the target miRNA signal using a DNA hybridization chain reaction in a droplet microfluidic platform with a photomultiplier (PMT) sensor (Fig. 9C),150 in which single cells and lysates are encapsulated in droplets and target miRNA released by individual cells interacts with DNA amplifiers to trigger the hybridization reaction and produce fluorescence signals. The target sequence is cycled without the use of PCR thermal cycling to significantly amplify the fluorescence signal. Their method effectively converts laboratory desktop PCR analysis into real-time droplet analysis using post-reaction fluorescence for reading to achieve ultra-high-throughput single-cell sequencing (300–500 cells per minute) for rapid biomedical identification. In addition, Segaliny et al. reported a single-cell droplet microfluidic method for identifying T cells expressing engineered T cell receptor (TCR) by dynamic monitoring of TCR T cell activation, reverse-transcription PCR, and sequencing of TCR chains at a single-cell level.151
000 cells in a short time and achieve ultra-high-throughput single-cell genome sequencing. However, their encapsulating process follows a Poisson distribution resulting in a low encapsulation rate. Furthermore, Moon et al. developed a new droplet microfluidic platform, in which high-concentration microbeads containing oligonucleotide barcodes are spontaneously arranged in spiral channels under the effect of inertia, and then coencapsulated with individual cells in droplets for single-cell sequencing (Fig. 9B).93 Through the deterministic encapsulation of inertially ordered beads, their platform significantly improves throughput and reduces barcode errors, significantly reducing intermittent errors in multi-bead packaging relative to the nondeterministic Drop-seq system. Guo et al. developed a rapid, PCR-free single-cell miRNA assay to amplify the target miRNA signal using a DNA hybridization chain reaction in a droplet microfluidic platform with a photomultiplier (PMT) sensor (Fig. 9C),150 in which single cells and lysates are encapsulated in droplets and target miRNA released by individual cells interacts with DNA amplifiers to trigger the hybridization reaction and produce fluorescence signals. The target sequence is cycled without the use of PCR thermal cycling to significantly amplify the fluorescence signal. Their method effectively converts laboratory desktop PCR analysis into real-time droplet analysis using post-reaction fluorescence for reading to achieve ultra-high-throughput single-cell sequencing (300–500 cells per minute) for rapid biomedical identification. In addition, Segaliny et al. reported a single-cell droplet microfluidic method for identifying T cells expressing engineered T cell receptor (TCR) by dynamic monitoring of TCR T cell activation, reverse-transcription PCR, and sequencing of TCR chains at a single-cell level.151
 | ||
| Fig. 9 Applications of single-cell droplet microfluidics for genetic analysis. (A) Ultra-high throughput single-cell genome sequencing using droplet microfluidics. Reproduced from ref. 149 with permission from Springer Nature, copyright 2017. (B) A droplet microfluidic device containing a bead-ordering unit for high throughput single-cell RNA-sequencing. Reproduced from ref. 93 with permission from the Royal Society of Chemistry, copyright 2018. (C) A microfluidics-based online PMT analysis system for single-cell encapsulation and single-cell miRNA detection and screening. Reproduced from ref. 150 with permission from the Royal Society of Chemistry, copyright 2018. (D) Droplet-based single-cell detection of 16S rRNA for pathogen identification and antimicrobial susceptibility testing from clinical urine samples. Reproduced from ref. 153 under the terms of the Creative Commons Attribution License. Copyright 2021, the Authors, published by John Wiley and Sons. | ||
More recently, single-cell droplet microfluidics-based genetic analysis has increasingly been applied by research scholars for the application in clinical diagnosis. Rivello et al. described a general method for investigating metabolism of circulating stromal cells associated with metastatic disease to reveal the prognosis of prostate cancer using a droplet-based microfluidic chip.152 They performed in-droplet extracellular pH measurement of single cells from metastatic prostate cancer patients for detecting and separating highly metabolically active cells (HMCs). Single-cell mRNA-sequencing analysis of HMCs was then conducted, and the results showed that approximately 87% were circulating stromal cells with high metabolism. Kaplan–Meier analysis reveals that those patients with over 5 HMCs have a considerably poorer survival probability than patients with fewer than 5 HMCs. Therefore, HMCs could be an indicator of poor prognosis in prostate cancer. The single-cell droplet microfluidic method is potentially useful for prostate cancer diagnosis. Additionally, Wang's group established a single-cell-droplet-based microfluidic system for pathogen identification and antimicrobial susceptibility testing from clinical urine specimens (Fig. 9D).153 Fluorescent hybridization probes were used to detect 16S Ribosomal RNA (16S rRNA) from single bacteria in microdroplets, allowing molecular discrimination of uropathogenic bacteria from clinical urine specimens within 16 min and quantitative indication of the susceptibility of bacteria to the antibiotic. The microfluidic system achieved a brilliant performance for urinary pathogen analysis within a short turnaround time (<30 min). The diversity genetic analysis of single cells from clinical samples using droplet-based microfluidics is quite valuable for accelerating the development of clinical diagnosis/treatment and precision medicine.
5. Conclusions and future outlook
In this review, we introduced and discussed recent advances with regard to single-cell droplet microfluidics for biomedical applications. Microfluidic droplet technology was presented in terms of important advantages, dimensionless numbers, device geometry, and droplet generation. A variety of recent microfluidic droplet manipulation methods used for single-cell encapsulation in droplets based on a random or controlled mode, as well as in hydrogels using synthetic or natural polymers, were described and discussed. Finally, we summarized the important applications of single-cell droplet microfluidics in small-molecule detection, protein analysis, drug screening, and genetic analysis.Although some progress has been made in single-cell manipulation and analysis using microfluidic droplet technology, there remain bottlenecks. First, microfluidic droplet technology has a low single-cell encapsulation rate, and the detection tools used for single-cell analysis are still quantitatively limited and have relatively low sensitivities. The development of simpler, cheaper, and more integrated droplet microfluidic devices will significantly improve single-cell analysis. Furthermore, the gel materials used to encapsulate single cells are functionally limited, and it is necessary to develop multi-composite/functional gel materials for culturing to enable the effective recreation of the 3D microenvironment of cells in vivo for the high-throughput analysis of single cells. Finally, the complexity of the cell itself and the fact that the current trends in single-cell analysis involve multimodal characterization encourage the development of methods for the synchronous detection of more types of intercellular molecules. These challenges will promote the further development of droplet microfluidics for operational and detection purposes and in multi-component single-cell analysis, providing a scientific basis for further biomedical research on single cells and leading to more accurate and comprehensive understanding of the functions and regulatory mechanisms of the molecules involved in biological processes.
Author contributions
Dan Liu: conceptualization, formal analysis, investigation, writing – original draft, writing – review and editing. Meilin Sun: conceptualization, formal analysis, investigation, writing – review and editing. Jinwei Zhang: formal analysis, investigation. Rui Hu: formal analysis. Wenzhu Fu: formal analysis. Tingting Xuanyuan: formal analysis. Wenming Liu: conceptualization, supervision, investigation, funding acquisition, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 32171450, 31971328, 31470971, 31100726), the Fundamental Research Funds for the Central Universities of Central South University (202045001, 2021zzts0930), and the Department of Education of Hunan Province (21B0008).References
- X. Xu, J. Wang, L. Wu, J. Guo, Y. Song, T. Tian, W. Wang, Z. Zhu and C. Yang, Small, 2020, 16, 1903905 CrossRef CAS PubMed
.
- H. Sun, Z. Miao, X. Zhang, U. Chan, S. Su, S. Guo, C. K. H. Wong, X. Xu and C. Deng, J. Biol. Chem., 2018, 293, 8315–8329 CrossRef CAS PubMed
.
- J. P. Junker and A. van Oudenaarden, Cell, 2014, 157, 8–11 CrossRef CAS PubMed
.
- S. Lindström and H. Andersson-Svahn, Lab Chip, 2010, 10, 3363–3372 RSC
.
- S. J. Altschuler and L. F. Wu, Cell, 2010, 141, 559–563 CrossRef CAS PubMed
.
- F. S. O. Fritzsch, C. Dusny, O. Frick and A. Schmid, Annu. Rev. Chem. Biomol. Eng., 2012, 3, 129–155 CrossRef CAS PubMed
.
- D. Gao, F. Jin, M. Zhou and Y. Jiang, Analyst, 2019, 144, 766–781 RSC
.
- C. Mayer, C. Hafemeister, R. C. Bandler, R. Machold, R. B. Brito, X. Jaglin, K. Allaway, A. Butler, G. Fishell and R. Satija, Nature, 2018, 555, 457–462 CrossRef CAS PubMed
.
- W. Fan, X. Chen, Y. Ge, Y. Jin, Q. Jin and J. Zhao, Biosens. Bioelectron., 2019, 145, 111730 CrossRef CAS PubMed
.
- M. Pellegrino, A. Sciambi, S. Treusch, R. Durruthy-Durruthy, K. Gokhale, J. Jacob, T. X. Chen, J. A. Geis, W. Oldham, J. Matthews, H. Kantarjian, P. A. Futreal, K. Patel, K. W. Jones, K. Takahashi and D. J. Eastburn, Genome Res., 2018, 28, 1345–1352 CrossRef CAS PubMed
.
- D. Sun, F. Cao, L. Cong, W. Xu, Q. Chen, W. Shi and S. Xu, Lab Chip, 2019, 19, 335–342 RSC
.
- D. Lin, P. Li, J. Feng, Z. Lin, X. Chen, N. Yang, L. Wang and D. Liu, Small, 2020, 16, 1901001 CrossRef CAS PubMed
.
- L. Pang, J. Ding, Y. Ge, J. Fan and S. Fan, Anal. Chem., 2019, 91, 8318–8325 CrossRef CAS PubMed
.
- J. R. Heath, A. Ribas and P. S. Mischel, Nat. Rev. Drug Discovery, 2016, 15, 204–216 CrossRef CAS PubMed
.
- D. Holmes, D. Pettigrew, C. H. Reccius, J. D. Gwyer, C. van Berkel, J. Holloway, D. E. Davies and H. Morgan, Lab Chip, 2009, 9, 2881–2889 RSC
.
- Y. Shi, M. Cai, L. Zhou and H. Wang, Semin. Cell Dev. Biol., 2018, 73, 31–44 CrossRef CAS PubMed
.
- X. Wang, X. Gou, S. Chen, X. Yan and D. Sun, J. Micromech. Microeng., 2013, 23, 075006 CrossRef CAS
.
- S. M. Le, R. C. Liu, C. T. Lim and J. Yan, Methods, 2016, 94, 13–18 CrossRef CAS PubMed
.
- P. Tinnefeld and M. Sauer, Angew. Chem., Int. Ed., 2005, 44, 2642–2671 CrossRef CAS PubMed
.
- L. Liu, D. Chen, J. Wang and J. Chen, Cells, 2020, 9, 1271 CrossRef CAS PubMed
.
- L. Huang, Y. Chen, Y. Chen and H. Wu, Anal. Chem., 2015, 87, 12169–12176 CrossRef CAS PubMed
.
- C. Wu, X. Zhu, T. Man, P. Chung, M. A. Teitell and P. Y. Chiou, Lab Chip, 2018, 18, 3074–3078 RSC
.
- X. An, P. Zuo and B. Ye, Talanta, 2020, 209, 120571 CrossRef CAS PubMed
.
- M. Sauzade and E. Brouzes, Lab Chip, 2017, 17, 2186–2192 RSC
.
- L. Nan, M. Y. A. Lai, M. Y. H. Tang, Y. K. Chan, L. L. M. Poon and H. C. Shum, Small, 2020, 16, 1902889 CrossRef CAS PubMed
.
- L. Zhang, K. Chen, H. Zhang, B. Pang, C. H. Choi, A. S. Mao, H. Liao, S. Utech, D. J. Mooney, H. Wang and D. A. Weitz, Small, 2018, 14, 1702955 CrossRef PubMed
.
- S. C. Kim, I. C. Clark, P. Shahi and A. R. Abate, Anal. Chem., 2018, 90, 1273–1279 CrossRef CAS PubMed
.
- E. X. Ng, M. A. Miller, T. Y. Jing and C. H. Chen, Biosens. Bioelectron., 2016, 81, 408–414 CrossRef CAS PubMed
.
- K. Perez-Toralla, A. Olivera-Torres, M. A. Rose, A. M. Esfahani, K. Reddy, R. Yang and S. A. Morin, Adv. Sci., 2020, 7, 2000769 CrossRef CAS PubMed
.
- P. Zhang and A. R. Abate, Adv. Mater., 2020, 32, 2005346 CrossRef PubMed
.
- D. J. Collins, A. Neild, A. deMello, A. Q. Liu and Y. Ai, Lab Chip, 2015, 15, 3439–3459 RSC
.
- N. Wen, Z. Zhao, B. Fan, D. Chen, D. Men, J. Wang and J. Chen, Molecules, 2016, 21, 881 CrossRef PubMed
.
- K. Matuła, F. Rivello and W. T. S. Huck, Adv. Biosyst., 2020, 4, 1900188 CrossRef PubMed
.
- Y. Ding, P. D. Howes and A. J. deMello, Anal. Chem., 2020, 92, 132–149 CrossRef CAS PubMed
.
- L. R. Shang, Y. Cheng and Y. J. Zhao, Chem. Rev., 2017, 117, 7964–8040 CrossRef CAS PubMed
.
- R. Dubay, J. N. Urban and E. M. Darling, Adv. Funct. Mater., 2021, 31, 2009946 CrossRef CAS
.
- J. D. Sun, L. Gao, L. P. Wang and X. L. Sun, Talanta, 2021, 234, 122671 CrossRef CAS PubMed
.
- L. Amirifar, M. Besanjideh, R. Nasiri, A. Shamloo, F. Nasrollahi, N. R. de Barros, E. Davoodi, A. Erdem, M. Mahmoodi, V. Hosseini, H. Montazerian, J. Jahangiry, M. A. Darabi, R. Haghniaz, M. R. Dokmeci, N. Annabi, S. Ahadian and A. Khademhosseini, Biofabrication, 2022, 14, 022001 CrossRef PubMed
.
- H. Feng, T. Zheng, M. Li, J. Wu, H. Ji, J. Zhang, W. Zhao and J. Guo, Electrophoresis, 2019, 40, 1580–1590 CrossRef CAS PubMed
.
- S. S. Terekhov, I. V. Smirnov, A. V. Stepanova, T. V. Bobik, Y. A. Mokrushina, N. A. Ponomarenko, A. A. Belogurov, M. P. Rubtsova, O. V. Kartseva, M. O. Gomzikova, A. A. Moskovtsev, A. S. Bukatin, M. V. Dubina, E. S. Kostryukova, V. V. Babenko, M. T. Vakhitova, A. I. Manolov, M. V. Malakhova, M. A. Kornienko, A. V. Tyakht, A. A. Vanyushkina, E. N. Ilina, P. Masson, A. G. Gabibov and S. Altman, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 2550–2555 CrossRef CAS PubMed
.
- A. I. Segaliny, G. Li, L. Kong, C. Ren, X. Chen, J. K. Wang, D. Baltimore, G. Wu and W. Zhao, Lab Chip, 2018, 18, 3733–3749 RSC
.
- K. Grosselin, A. Durand, J. Marsolier, A. Poitou, E. Marangoni, F. Nemati, A. Dahmani, S. Lameiras, F. Reyal, O. Frenoy, Y. Pousse, M. Reichen, A. Woolfe, C. Brenan, A. D. Griffiths, C. Vallot and A. Gérard, Nat. Genet., 2019, 51, 1060–1066 CrossRef CAS PubMed
.
- A. M. Kaushik, K. Hsieh and T. H. Wang, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2018, 10, e1522 Search PubMed
.
- T. P. Lagus and J. F. Edd, J. Phys. D: Appl. Phys., 2013, 46, 114005 CrossRef
.
- P. Gruner, B. Riechers, B. Semin, J. Lim, A. Johnston, K. Short and J. C. Baret, Nat. Commun., 2016, 7, 10392 CrossRef CAS PubMed
.
- X. Li, D. Zhang, W. Ruan, W. Liu, K. Yin, T. Tian, Y. Bi, Q. Ruan, Y. Zhao, Z. Zhu and C. Yang, Anal. Chem., 2019, 91, 13611–13619 CrossRef CAS PubMed
.
- E. Y. Basova and F. Foret, Analyst, 2015, 140, 22–38 RSC
.
- D. Di Carlo, Lab Chip, 2009, 9, 3038–3046 RSC
.
- T. Trantidou, M. S. Friddin, A. Salehi-Reyhani, O. Ces and Y. Elani, Lab Chip, 2018, 18, 2488–2509 RSC
.
- H. Gu, M. H. G. Duits and F. Mugele, Int. J. Mol. Sci., 2011, 12, 2572–2597 CrossRef CAS PubMed
.
- A. S. Utada, A. Fernandez-Nieves, H. A. Stone and D. A. Weitz, Phys. Rev. Lett., 2007, 99, 094502 CrossRef PubMed
.
- J. K. Nunes, S. S. H. Tsai, J. Wan and H. A. Stone, J. Phys. D: Appl. Phys., 2013, 46, 114002 CrossRef PubMed
.
- T. Thorsen, R. W. Roberts, F. H. Arnold and S. R. Quake, Phys. Rev. Lett., 2001, 86, 4163–4166 CrossRef CAS PubMed
.
- Y. Ding, X. Casadevall i Solvas and A. deMello, Analyst, 2015, 140, 414–421 RSC
.
- M. Rhee, P. Liu, R. J. Meagher, Y. K. Light and A. K. Singh, Biomicrofluidics, 2014, 8, 034112 CrossRef PubMed
.
- W. Han and X. Chen, Microgravity Sci. Technol., 2019, 31, 855–864 CrossRef
.
- R. A. Letteri, C. F. S. Chalarca, Y. Bai, R. C. Hayward and T. Emrick, Adv. Mater., 2017, 29, 1702921 CrossRef PubMed
.
- Z. Liu, J. Zhao, Y. Pang and X. Wang, Microfluid. Nanofluid., 2018, 22, 124 CrossRef
.
- K. Vijayakumar, S. Gulati, A. J. deMello and J. B. Edel, Chem. Sci., 2010, 1, 447–452 RSC
.
- S. L. Anna, N. Bontoux and H. A. Stone, Appl. Phys. Lett., 2003, 82, 364–366 CrossRef CAS
.
- R. Dreyfus, P. Tabeling and H. Willaime, Phys. Rev. Lett., 2003, 90, 144505 CrossRef PubMed
.
- C. Cramer, P. Fischer and E. J. Windhab, Chem. Eng. Sci., 2004, 59, 3045–3058 CrossRef CAS
.
- G. Leonaviciene, K. Leonavicius, R. Meskys and L. Mazutis, Lab Chip, 2020, 20, 4052–4062 RSC
.
- C. Mou, W. Wang, Z. Li, X. Ju, R. Xie, N. Deng, J. Wei, Z. Liu and L. Chu, Adv. Sci., 2018, 5, 1700960 CrossRef PubMed
.
- S. Yin, Y. Huang, T. N. Wong, W. H. Chong and K. T. Ooi, J. Phys. Chem. C, 2019, 123, 25643–25650 CrossRef CAS
.
- Q. Zhang, H. Li, C. Zhu, T. Fu, Y. Ma and H. Z. Li, Colloids Surf., A, 2018, 537, 572–579 CrossRef CAS
.
- P. Zhu, X. Tang and L. Wang, Microfluid. Nanofluid., 2016, 20, 47 CrossRef
.
- Z. Wang, R. Chen, X. Zhu, Q. Liao, D. Ye, B. Zhang, X. He and W. Li, AIP Adv., 2018, 8, 015302 CrossRef
.
- S. Jin, X. Wei, Z. Liu, J. Ren, Z. Jiang, R. Abe and Z. Yu, Sens. Actuators, B, 2019, 291, 1–8 CrossRef CAS
.
- K. Samlali, F. Ahmadi, A. B. V. Quach, G. Soffer and S. C. C. Shih, Small, 2020, 16, 2002400 CrossRef CAS PubMed
.
- Q. Yan, S. Xuan, X. Ruan, J. Wu and X. Gong, Microfluid. Nanofluid., 2015, 19, 1377–1384 CrossRef
.
- Y. Ren and W. W. F. Leung, Micromachines, 2016, 7, 17 CrossRef PubMed
.
- M. Morita, H. Yamashita, M. Hayakawa, H. Onoe and M. Takinoue, J. Visualized Exp., 2016, 108, e53860 Search PubMed
.
- K. Wang, L. Xie, Y. Lu and G. Luo, Lab Chip, 2013, 13, 73–76 RSC
.
- W. Cui, G. Yesiloz and C. L. Ren, Anal. Chem., 2021, 93, 1266–1270 CrossRef CAS PubMed
.
- H. Liu, H. Wang, W. Wei, H. Liu, L. Jiang and J. Qin, Small, 2018, 14, 1801095 CrossRef PubMed
.
- Z. Z. Chong, S. H. Tan, A. M. Gañán-Calvo, S. B. Tor, N. H. Loh and N. T. Nguyen, Lab Chip, 2016, 16, 35–58 RSC
.
- P. Zhu and L. Wang, Lab Chip, 2017, 17, 34–75 RSC
.
- C. H. Chen, A. Sarkar, Y. A. Song, M. A. Miller, S. J. Kim, L. G. Griffith, D. A. Lauffenburger and J. Han, J. Am. Chem. Soc., 2011, 133, 10368–10371 CrossRef CAS PubMed
.
- I. Gal, R. Edri, N. Noor, M. Rotenberg, M. Namestnikov, I. Cabilly, A. Shapira and T. Dvir, Small, 2020, 16, 1904806 CrossRef CAS PubMed
.
- C. W. Beh, Y. L. Fu, C. R. Weiss, C. Hu, A. Arepally, H. Q. Mao, T. H. Wang and D. L. Kraitchman, Lab Chip, 2020, 20, 3591–3600 RSC
.
- R. Seemann, M. Brinkmann, T. Pfohl and S. Herminghaus, Rep. Prog. Phys., 2012, 75, 016601 CrossRef PubMed
.
- H. Wang, H. Liu, F. He, W. Chen, X. Zhang, M. Zhao, L. Wang and J. Qin, Adv. Mater. Technol., 2020, 5, 2000045 CrossRef CAS
.
- M. R. de Saint Vincent, H. Chraïbi and J. P. Delville, Phys. Rev. Appl., 2015, 4, 044005 CrossRef
.
- S. Köster, F. E. Angilè, H. Duan, J. J. Agresti, A. Wintner, C. Schmitz, A. C. Rowat, C. A. Merten, D. Pisignano, A. D. Griffiths and D. A. Weitz, Lab Chip, 2008, 8, 1110–1115 RSC
.
- P. K. P. Rajeswari, H. N. Joensson and H. Andersson-Svahn, Electrophoresis, 2017, 38, 305–310 CrossRef PubMed
.
- M. Navi, N. Abbasi, M. Jeyhani, V. Gnyawali and S. S. H. Tsai, Lab Chip, 2018, 18, 3361–3370 RSC
.
- L. Nan, Z. Yang, H. Lyu, K. Y. Y. Lau and H. C. Shum, Adv. Biosyst., 2019, 3, 1900076 CrossRef PubMed
.
- X. Zhang, X. Wei, X. Men, Z. Jiang, W. Ye, M. Chen, T. Yang, Z. Xu and J. Wang, Anal. Chem., 2020, 92, 6604–6612 CrossRef CAS PubMed
.
- J. F. Edd, D. Di Carlo, K. J. Humphry, S. Köster, D. Irimia, D. A. Weitz and M. Toner, Lab Chip, 2008, 8, 1262–1264 RSC
.
- E. W. M. Kemna, R. M. Schoeman, F. Wolbers, I. Vermes, D. A. Weitz and A. van den Berg, Lab Chip, 2012, 12, 2881–2887 RSC
.
- L. Li, P. Wu, Z. Luo, L. Wang, W. Ding, T. Wu, J. Chen, J. He, Y. He, H. Wang, Y. Chen, G. Li, Z. Li and L. He, ACS Sens., 2019, 4, 1299–1305 CrossRef CAS PubMed
.
- H. S. Moon, K. Je, J. W. Min, D. Park, K. Y. Han, S. H. Shin, W. Y. Park, C. E. Yoo and S. H. Kim, Lab Chip, 2018, 18, 775–784 RSC
.
- M. N. Hsu, S. C. Wei, S. Guo, D. T. Phan, Y. Zhang and C. H. Chen, Small, 2018, 14, 1802918 CrossRef PubMed
.
- T. Kamperman, S. Henke, C. W. Visser, M. Karperien and J. Leijten, Small, 2017, 13, 1603711 CrossRef PubMed
.
- Y. Jiang, J. Chen, C. Deng, E. J. Suuronen and Z. Zhong, Biomaterials, 2014, 35, 4969–4985 CrossRef CAS PubMed
.
- W. Jiang, M. Li, Z. Chen and K. W. Leong, Lab Chip, 2016, 16, 4482–4506 RSC
.
- A. Khater, O. Abdelrehim, M. Mohammadi, M. Azarmanesh, M. Jonmaleki, R. Salahandish, A. Mohamad and A. Sanati-Nezhad, Lab Chip, 2020, 20, 2175–2187 RSC
.
- Y. Sun, B. Cai, X. Y. Wei, Z. X. Wang, L. Rao, Q. F. Meng, Q. Q. Liao, W. Liu, S. S. Guo and X. Z. Zhao, Electrophoresis, 2019, 40, 961–968 CrossRef CAS PubMed
.
- Ý. A. Mørch, I. Donati, B. L. Strand and G. Skjåk-Bræk, Biomacromolecules, 2006, 7, 1471–1480 CrossRef PubMed
.
- T. Kamperman, S. Henke, A. van den Berg, S. R. Shin, A. Tamayol, A. Khademhosseini, M. Karperien and J. Leijten, Adv. Healthcare Mater., 2017, 6, 1600913 CrossRef PubMed
.
- K. J. Park, K. G. Lee, S. Seok, B. G. Choi, M. K. Lee, T. J. Park, J. Y. Park, D. H. Kim and S. J. Lee, Lab Chip, 2014, 14, 1873–1879 RSC
.
- C. An, W. Liu, Y. Zhang, B. Pang, H. Liu, Y. Zhang, H. Zhang, L. Zhang, H. Liao, C. Ren and H. Wang, Acta Biomater., 2020, 111, 181–196 CrossRef CAS PubMed
.
- S. Utech, R. Prodanovic, A. S. Mao, R. Ostafe, D. J. Mooney and D. A. Weitz, Adv. Healthcare Mater., 2015, 4, 1628–1633 CrossRef CAS PubMed
.
- H. Zhang, G. Jenkins, Y. Zou, Z. Zhu and C. Yang, Anal. Chem., 2012, 84, 3599–3606 CrossRef CAS PubMed
.
- M. Li, M. van Zee, C. T. Riche, B. Tofig, S. D. Gallaher, S. S. Merchant, R. Damoiseaux, K. Goda and D. Di Carlo, Small, 2018, 14, 1803315 CrossRef PubMed
.
- M. Yamada, A. Hori, S. Sugaya, Y. Yajima, R. Utoh, M. Yamato and M. Seki, Lab Chip, 2015, 15, 3941–3951 RSC
.
- L. W. Chan, Y. Jin and P. W. S. Heng, Int. J. Pharm., 2002, 242, 255–258 CrossRef CAS PubMed
.
- Q. Liao, S. Zhao, B. Cai, R. He, L. Rao, Y. Wu, S. Guo, Q. Liu, W. Liu and X. Zhao, Sens. Actuators, A, 2018, 279, 313–320 CrossRef CAS
.
- K. Liu, Y. Deng, N. Zhang, S. Li, H. Ding, F. Guo, W. Liu, S. Guo and X. Zhao, Microfluid. Nanofluid., 2012, 13, 761–767 CrossRef CAS
.
- A. S. Mao, J. W. Shin, S. Utech, H. Wang, O. Uzun, W. Li, M. Cooper, Y. Hu, L. Zhang, D. A. Weitz and D. J. Mooney, Nat. Mater., 2017, 16, 236–243 CrossRef CAS PubMed
.
- D. Velasco, E. Tumarkin and E. Kumacheva, Small, 2012, 8, 1633–1642 CrossRef CAS PubMed
.
- C. Mulas, A. C. Hodgson, T. N. Kohler, C. C. Agley, P. Humphreys, H. Kleine-Brüggeney, F. Hollfelder, A. Smith and K. J. Chalut, Lab Chip, 2020, 20, 2580–2591 RSC
.
- L. Liu, C. K. Dalal, B. M. Heineike and A. R. Abate, Lab Chip, 2019, 19, 1838–1849 RSC
.
- S. Ma, M. Natoli, X. Liu, M. P. Neubauer, F. M. Watt, A. Fery and W. T. S. Huck, J. Mater. Chem. B, 2013, 1, 5128–5136 RSC
.
- C. H. Lin, Y. H. Hsiao, H. C. Chang, C. F. Yeh, C. K. He, E. M. Salm, C. C. Chen, I. M. Chiu and C. H. Hsu, Lab Chip, 2015, 15, 2928–2938 RSC
.
- Y. Sun, W. Song, X. Sun and S. Zhang, ACS Appl. Mater. Interfaces, 2018, 10, 31054–31060 CrossRef CAS PubMed
.
- L. Li, H. Wang, L. Huang, S. A. Michael, W. Huang and H. Wu, Anal. Chem., 2019, 91, 15908–15914 CrossRef CAS PubMed
.
- C. K. He, Y. W. Chen, S. H. Wang and C. H. Hsu, Lab Chip, 2019, 19, 1370–1377 RSC
.
- T. Y. Jing, R. Ramji, M. E. Warkiani, J. Han, C. T. Lim and C. H. Chen, Biosens. Bioelectron., 2015, 66, 19–23 CrossRef CAS PubMed
.
- Z. Yu, L. Zhou, T. Zhang, R. Shen, C. Li, X. Fang, G. Griffiths and J. Liu, ACS Sens., 2017, 2, 626–634 CrossRef CAS PubMed
.
- V. Chokkalingam, J. Tel, F. Wimmers, X. Liu, S. Semenov, J. Thiele, C. G. Figdor and W. T. S. Huck, Lab Chip, 2013, 13, 4740–4744 RSC
.
- I. C. Clark, C. L. Delley, C. Sun, R. Thakur, S. L. Stott, S. Thaploo, Z. R. Li, F. J. Quintana and A. R. Abate, Anal. Chem., 2020, 92, 14616–14623 CrossRef CAS PubMed
.
- A. Mongersun, I. Smeenk, G. Pratx, P. Asuri and P. Abbyad, Anal. Chem., 2016, 88, 3257–3263 CrossRef CAS PubMed
.
- F. Del Ben, M. Turetta, G. Celetti, A. Piruska, M. Bulfoni, D. Cesselli, W. T. S. Huck and G. Scoles, Angew. Chem., Int. Ed., 2016, 55, 8581–8584 CrossRef CAS PubMed
.
- D. Sun, F. Cao, Y. Tian, A. Li, W. Xu, Q. Chen, W. Shi and S. Xu, Anal. Chem., 2019, 91, 15484–15490 CrossRef CAS PubMed
.
- C. Zielke, C. W. Pan, A. J. G. Ramirez, C. Feit, C. Dobson, C. Davidson, B. Sandel and P. Abbyad, Anal. Chem., 2020, 92, 6949–6957 CrossRef CAS PubMed
.
- L. Qu, D. Li, L. Qin, J. Mu, J. S. Fossey and Y. Long, Anal. Chem., 2013, 85, 9549–9555 CrossRef CAS PubMed
.
- Z. Chen, Q. Li, Q. Sun, H. Chen, X. Wang, N. Li, M. Yin, Y. Xie, H. Li and B. Tang, Anal. Chem., 2012, 84, 4687–4694 CrossRef CAS PubMed
.
- R. Shen, P. Liu, Y. Zhang, Z. Yu, X. Chen, L. Zhou, B. Nie, A. Zaczek, J. Chen and J. Liu, Anal. Chem., 2018, 90, 4478–4484 CrossRef CAS PubMed
.
- N. Alizadeh, F. Ghasemi, A. Salimi, R. Hallaj, F. Fathi and F. Soleimani, Dyes Pigm., 2020, 173, 107875 CrossRef CAS
.
- C. Pitzler, G. Wirtz, L. Vojcic, S. Hiltl, A. Böker, R. Martinez and U. Schwaneberg, Chem. Biol., 2014, 21, 1733–1742 CrossRef CAS PubMed
.
- M. Pytliak, V. Vargová and V. Mechírová, Onkologie, 2012, 35, 49–53 CrossRef PubMed
.
- H. Huang, Sensors, 2018, 18, 3249 CrossRef PubMed
.
- W. Wu, S. Zhang, T. Zhang and Y. Mu, ACS Appl. Mater. Interfaces, 2021, 13, 6081–6090 CrossRef CAS PubMed
.
- G. Dranoff, Nat. Rev. Cancer, 2004, 4, 11–22 CrossRef CAS PubMed
.
- C. Fischbach, H. J. Kong, S. X. Hsiong, M. B. Evangelista, W. Yuen and D. J. Mooney, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 399–404 CrossRef CAS PubMed
.
- D. Sun, F. Cao, W. Xu, Q. Chen, W. Shi and S. Xu, Anal. Chem., 2019, 91, 2551–2558 CrossRef CAS PubMed
.
- F. Wimmers, N. Subedi, N. van Buuringen, D. Heister, J. Vivié, I. Beeren-Reinieren, R. Woestenenk, H. Dolstra, A. Piruska, J. F. M. Jacobs, A. van Oudenaarden, C. G. Figdor, W. T. S. Huck, I. J. M. de Vries and J. Tel, Nat. Commun., 2018, 9, 3317 CrossRef PubMed
.
- Y. Yuan, J. Brouchon, J. M. Calvo-Calle, J. Xia, L. Sun, X. Zhang, K. L. Clayton, F. Ye, D. A. Weitz and J. A. Heyman, Lab Chip, 2020, 20, 1513–1520 RSC
.
- J. Zhai, S. H. Yi, Y. W. Jia, P. I. Mak and R. P. Martins, TrAC, Trends Anal. Chem., 2019, 117, 231–241 CrossRef CAS
.
- E. Brouzes, M. Medkova, N. Savenelli, D. Marran, M. Twardowski, J. B. Hutchison, J. M. Rothberg, D. R. Link, N. Perrimon and M. L. Samuels, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14195–14200 CrossRef CAS PubMed
.
- O. Scheler, K. Makuch, P. R. Debski, M. Horka, A. Ruszczak, N. Pacocha, K. Sozański, O. P. Smolander, W. Postek and P. Garstecki, Sci. Rep., 2020, 10, 3282 CrossRef CAS PubMed
.
- S. Sarkar, N. Cohen, P. Sabhachandani and T. Konry, Lab Chip, 2015, 15, 4441–4450 RSC
.
- S. S. Bithi and S. A. Vanapalli, Sci. Rep., 2017, 7, 41707 CrossRef CAS PubMed
.
- N. Shembekar, H. Hu, D. Eustace and C. A. Merten, Cell Rep., 2018, 22, 2206–2215 CrossRef CAS PubMed
.
- K. Eyer, R. C. L. Doineau, C. E. Castrillon, L. Briseño-Roa, V. Menrath, G. Mottet, P. England, A. Godina, E. Brient-Litzler, C. Nizak, A. Jensen, A. D. Griffiths, J. Bibette, P. Bruhns and J. Baudry, Nat. Biotechnol., 2017, 35, 977–982 CrossRef CAS PubMed
.
- A. Gérard, A. Woolfe, G. Mottet, M. Reichen, C. Castrillon, V. Menrath, S. Ellouze, A. Poitou, R. Doineau, L. Briseno-Roa, P. Canales-Herrerias, P. Mary, G. Rose, C. Ortega, M. Delincé, S. Essono, B. Jia, B. Iannascoli, O. Richard-Le Goff, R. Kumar, S. N. Stewart, Y. Pousse, B. Q. Shen, K. Grosselin, B. Saudemont, A. Sautel-Caillé, A. Godina, S. McNamara, K. Eyer, G. A. Millot, J. Baudry, P. England, C. Nizak, A. Jensen, A. D. Griffiths, P. Bruhns and C. Brenan, Nat. Biotechnol., 2020, 38, 715–721 CrossRef PubMed
.
- F. Lan, B. Demaree, N. Ahmed and A. R. Abate, Nat. Biotechnol., 2017, 35, 640–646 CrossRef CAS PubMed
.
- S. Guo, W. N. Lin, Y. W. Hu, G. Y. Sun, D. T. Phan and C. H. Chen, Lab Chip, 2018, 18, 1914–1920 RSC
.
- A. I. Segaliny, G. D. Li, L. S. Kong, C. Ren, X. M. Chen, J. K. Wang, D. Baltimore, G. K. Wu and W. A. Zhao, Lab Chip, 2018, 18, 3733–3749 RSC
.
- F. Rivello, K. Matula, A. Piruska, M. Smits, N. Mehra and W. T. S. Huck, Sci. Adv., 2020, 6, eaaz3849 CrossRef CAS PubMed
.
- A. M. Kaushik, K. Hsieh, K. E. Mach, S. Lewis, C. M. Puleo, K. C. Carroll, J. C. Liao and T. H. Wang, Adv. Sci., 2021, 8, 2003419 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2022 |