Open Access Article
Open Access ArticleBacterial outer membrane vesicles and their functionalization as vehicles for bioimaging, diagnosis and therapy
Kaikai
Xue
,
Lu
Wang
 * and
Jinyao
Liu
* and
Jinyao
Liu
 *
*
Shanghai Key Laboratory for Nucleic Acid Chemistry and Nanomedicine, Institute of Molecular Medicine, State Key Laboratory of Oncogenes and Related Genes, Shanghai Cancer Institute, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai 200127, China. E-mail: jyliu@sjtu.edu.cn; luwang@shsmu.edu.cn
First published on 2nd August 2022
Abstract
Bacterial outer membrane vesicles (OMVs) are spherical nanostructures spontaneously released from Gram-negative bacteria. Natural OMVs that are abundant in lipopolysaccharides, phospholipids, proteins, and nucleic acids have diverse biological functions including extracellular communication, transferring contents to host cells, and modulating immune responses, which are involved in multiple processes in human health. In addition to their native characteristics, functionalization of OMVs has sparked substantial biomedical applications due to their advantages of nanoscale size and the ability to carry a broad variety of payloads. In this review, we summarize the bioactivities of OMVs, including biogenesis, immunogenicity, and interactions with host cells, followed by a discussion on the strategies of utilizing functionalized OMVs as delivery vehicles for bioimaging, diagnosis, and therapy. Particularly, the applications of engineered OMVs as therapeutics to treat inflammatory bowel diseases and several types of cancer and also as vaccines to prevent infections have been elaborated. We also highlight the challenges and future perspectives of using OMVs for different biomedical applications. This work would inspire new thoughts on the development of innovative OMV-based biomedical nanomaterials through synthetic bioengineering and physicochemical modification.
1. Introduction
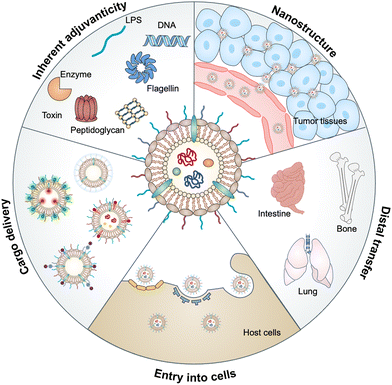
Bacterial outer membrane vesicles (OMVs), which are extracellular vesicles discharged from all types of Gram-negative bacteria during growth, have been studied for decades due to their roles in cell-to-cell communication, biofilm formation, pathogenesis and stress responses.1,2 OMVs were first identified in the envelopes of Vibrio cholera3 and Neisseria meningitidis4 during their normal growth. Subsequently, the detection of OMVs in the cerebrospinal fluid of patients with meningococcal infection5 has revealed that OMVs may be associated with bacterial pathogenesis, which has attracted great attention to the functionalization of these vesicles. In the past decade, natural OMVs derived from both pathogenic and commensal bacteria have been verified to orchestrate the mucosal immune system and even distal tissues to broadly affect the therapeutic responses of human diseases, including tumors, gastrointestinal disorders, and metabolic syndrome.6 OMVs derived from bacterial outer membranes are spherical lipid bilayer nanostructures with a size ranging from approximately 20 to 250 nm.7 Notably, the structure of OMVs allows them to carry cargo molecules either on the surface or inside of OMVs via genetic engineering or chemical modification. Proteomic and biochemical analyses have disclosed that OMVs contain substantial biological substrates from parent bacteria, such as periplasmic and membrane-bound proteins, lipopolysaccharides (LPS), toxins, peptidoglycans (PG), DNA, RNA and enzymes.8 Based on the interaction of these bacterial pathogen-associated molecular patterns (PAMPs) with host pattern recognition receptors (PRRs), OMVs possess inherent adjuvanticity to provoke immune responses.9 Thus, OMVs have been developed for a variety of biomedical applications in drug delivery, diagnostics, cancer immunotherapy, vaccines, and others. In this review, we aim to provide a comprehensive understanding of the biological characteristics of OMVs and then introduce the interactions between OMVs and host cells, followed by highlighting the advances of OMVs in biomedical applications (Fig. 1).2. Biogenesis of OMVs
Compared to Gram-positive species, Gram-negative bacteria possess a lipopolysaccharide-rich outer membrane located outside the thin PG layer.10 The formation of OMVs is a highly spontaneous and conserved process that requires the outward budding and fission of an outer membrane lacking membrane–peptidoglycan bonds.11 In principle, OMVs can be obtained in a liquid bacterial culture without the addition of any foreign molecules. However, the spontaneous release of OMVs is insufficient, resulting in low yields.12 Thus, methods capable of increasing OMV production are needed for practical biological applications. Although the information about the molecular mechanisms of their dynamic production and shedding is limited, crosslinks bridging the outer membrane to PG, the lipid composition dictating membrane fluidity, and the accumulation of misfolded molecules may play essential roles in the biogenesis of OMVs.13In fact, these factors have demonstrated to be able to regulate the production of OMVs, depending on the species of bacteria. Several genetic mutations involved in crosslinks between the outer membrane and PG, such as the Tol–Pal system, outer membrane protein A (OmpA), and lipoprotein (Lpp), are closely associated with enhanced OMV production.14,15 For example, the Tol–Pal system, composed of YbgC, TolQ, TolA, TolR, TolB, Pal and YbgF proteins, exhibits a non-covalent interaction with the PG and is important for outer membrane invagination during OMV biogenesis in Escherichia coli.16 Mutations in each of the seven Tol–Pal components can substantially increase OMV production. Meanwhile, lipoprotein diacylglyceryl transferase (Lgt) is required for lipidation and maturation of Lpp by catalyzing the acylation of Lpp.17 The mutation of Lgt leads to lack of Lpp, which increases the cytoplasmic membrane fluidity and vesicle formation of E. coli.18 Furthermore, the removal of LPS with detergents, such as EDTA, is a traditional method to increase the vesicle release and has been widely used for scale up.19 Vesiculation levels also display temperature-dependent alteration, which is presumably modulated by membrane lipid dynamics.20 Additionally, bacteria exposed to stress factors, such as antibiotics, ultraviolet radiation and oxidative stress, tend to increase bulging of the outer membrane to remove the misfolded proteins in periplasm and envelope.21–23 Similarly, the lack of nutrients, including metal ions, lysine and hemin, can also cause an overproduction of OMVs to obtain carbon or iron from the surrounding environment via carrying more degradative enzymes or transferrin-binding proteins, respectively.24 Strikingly, in addition to yields, the cargo of OMVs can also be regulated by environments. For example, under iron-limiting growth conditions, OMVs produced by Helicobacter pylori reduce the expressions of virulence factors, such as VacA, whereas they increase the expressions of proteolytic enzymes.25 The polysaccharide portion of Porphyromonas gingivalis allows gingivalase to be preferentially packaged into OMVs and selectively excludes the outer membrane proteins RagA and RagB from OMVs.26,27 Collectively, different approaches have been utilized to increase the yields of OMVs, basing on the requirement for a given component or application and a specific bacterial strain.
3. Mechanisms of OMVs entry into host cells
Mucosal tissues, including the gastrointestinal tract, respiratory tract, and reproductive tract, represent the major barrier between the host and the external environment. Particularly, the mucosal epithelial layer, forming a contiguous interface, is the first line of the barrier between the luminal contents and the host.28 A number of studies have reported that OMVs released from various commensal or pathogenic bacteria in the lumina either maintain or disrupt host homeostasis by direct entry into mucosal epithelial cells.29 In addition, OMVs can also cross the mucosal barrier through paracellular and transepithelial pathways to interact with myeloid cell subsets and further modulate immune responses. The entry of OMVs into different types of host cells can occur via multiple endocytic routes (Fig. 2), including micropinocytosis, lipid raft-mediated uptake, clathrin- and caveolin-mediated endocytosis, and PRR recognition.30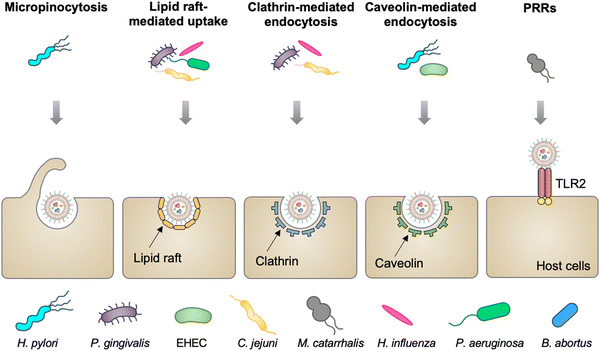 | ||
| Fig. 2 Schematic diagram of different routes of OMVs to enter host cells, including micropinocytosis, lipid raft-mediated uptake, clathrin- and caveolin-mediated endocytosis, and extracellular PRRs. | ||
Vesicles released from Pseudomonas aeruginosa,31P. gingivalis,32Haemophilus influenza,33 and Campylobacter jejuni34 enter gastric and airway epithelial cells through cholesterol-dependent fusion with lipid rafts, which is the most frequently reported route. While OMVs released by enterohemorrhagic E. coli (EHEC),35H. pylori,36 and Brucella abortus37 are internalized by epithelial cells or macrophage via clathrin-dependent, cholesterol-independent endocytosis. Vanaja et al. have provided the evidence that adaptor protein 2 (AP2) is probably involved in clathrin-mediated endocytosis, in which macrophage treated with EHEC-derived OMVs after the knockdown of AP2 display decreased internalization and alleviated inflammatory responses.38 It is noted that the size and composition of OMVs appear to affect the route of the cell entry. For example, smaller OMVs (20–100 nm in diameter) from H. pylori preferentially enter epithelial cells via clathrin-mediated endocytosis, whereas larger OMVs (90–450 nm in diameter) enter these cells via micropinocytosis.39 For the effect of composition, LPS with an intact O antigen guides E. coli-derived OMVs to enter HeLa cells via lipid raft-mediated endocytosis, yet the deficiency of the O antigen causes clathrin-mediated endocytosis.40 Moreover, Moraxella catarrhalis produces OMVs that interact with Toll-like receptor 2 (TLR2) and subsequently enter alveolar epithelial cells to induce the chronic obstructive pulmonary disease.41 It is indicated that extracellular PRRs participate in modulating the entry of OMVs into host cells, although the mechanism remains unclear. Together, the capacity of OMVs to enter cells enables them to serve as delivery vehicles to transfer cargo into eukaryotic cells for different applications.
4. Immunogenicity of OMVs
Since OMVs contain multiple MAMPs, including LPS, lipoproteins, PG, flagellin, DNA, and RNA, it is presumable that the recognition and uptake of OMVs can engage with the host PRRs to trigger innate and adaptive immune responses.42,43 There are several types of PRRs, such as Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), NOD-like receptors (NLRs), and cytosolic sensors of DNA, which recognize individual PAMPs.44 Given that the content and composition of OMVs depends on bacterial species, PRR signaling pathways induced by OMVs are different.LPS, a large glycolipid molecule and composed of the lipid A, core oligosaccharide, and O antigen, is one of the most potent and classical PAMP.45 The LPS binding protein delivers LPS from OMVs to CD14 on cell surfaces, where it is recognized by the TLR4–MD2 complex.46–48 Generally, OMVs produced by E. coli, P. aeruginosa, and N. meningitidis contain LPS and can effectively drive TLR4 dimerization, which recruits downstream adaptor molecules to mount an inflammatory response.49–51 OMVs derived from P. aeruginosa initiate the TLR adaptor myeloid differentiation primary response protein 88 (MyD88)-dependent nuclear factor-kappa B (NF-κB) signaling pathway,52 whereas OMVs released from H. pylori activate the mitogen-activated protein kinase (MAPK) and activator protein 1 (AP-1) signaling pathway.53 Specifically, upon uptake by macrophages, OMVs discharged from P. aeruginosa, H. pylori, Neisseria gonorrhoeae, and EHEC can activate inflammasome signaling, which causes the LPS-dependent secretion of inflammatory cytokines, mitochondrial apoptosis, and eventual cellular pyroptosis.54–56 Unlike pathogenic bacteria, Bacteroides fragilis, as a symbiotic strain, releases the capsular polysaccharide A (PSA) in OMVs to induce anti-inflammatory effects through TLR2 on the surface of dendritic cells (DCs), depending on the growth arrest and DNA-damage-inducible protein Gadd45α.57 Occasionally, multiple PRRs are simultaneously involved in the recognition of OMVs. For example, M. catarrhalis, one of the major respiratory tract pathogens in humans, indirectly releases OMV containing MID and unmethylated CpG-DNA motifs to activate host B cells via both TLR2 and TLR9.58 Additionally, H7 flagellin of OMVs from EHEC O157 induces the interleukin 8 (IL-8) production via both TLR4 and TLR5 signaling in intestinal epithelial cells.59
During the uptake of OMVs derived from H. pylori, P. aeruginosa, and N. gonorrhoeae by gastric epithelial cells, bacterial PG fragments are specifically recognized by nucleotide-binding oligomerization domain (NOD) proteins NOD1 and NOD2 in the cytosol to activate NF-κB.42,60,61 Intriguingly, certain components are deficient in OMVs compared to parent bacteria and hence allow them to activate different signaling pathways than their parent bacteria. For example, TLR13 recognizing bacterial ribosomal RNA is involved in bacteria-mediated innate responses but not in OMVs, owing to the absence of ribosomal RNA.62 Generally, OMV-mediated PRR signal transduction can also induce host epithelial cells to produce antimicrobial peptides, thereby promoting the clearance of pathogenic bacteria.30 Furthermore, protein glycosylation is an emerging field to study bacterial virulence and pathogenicity. It has been reported that the N-linked protein glycosylation of C. jejuni, as the most abundant post-translation modification of proteins, plays an important role in intestinal adhesion, colonization and invasion.63 OMVs generated by C. jejuni have been identified with 16 glycoproteins via proteomic analysis, which may be involved in the mechanism of OMV–host cell interaction.34 In short, the diversified PRRs are mainly responsible for OMVs to launch innate immune responses and shape adaptive immune responses to specific pathogens.
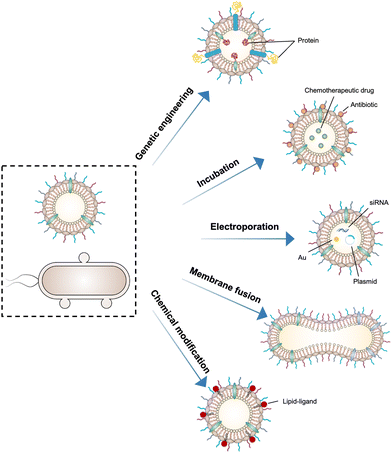
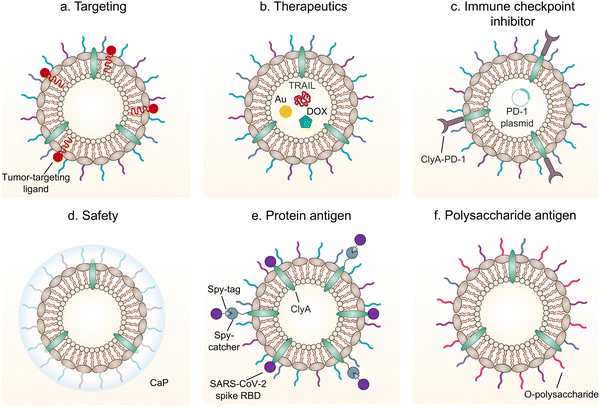
5. Functionalized OMVs as delivery vehicles
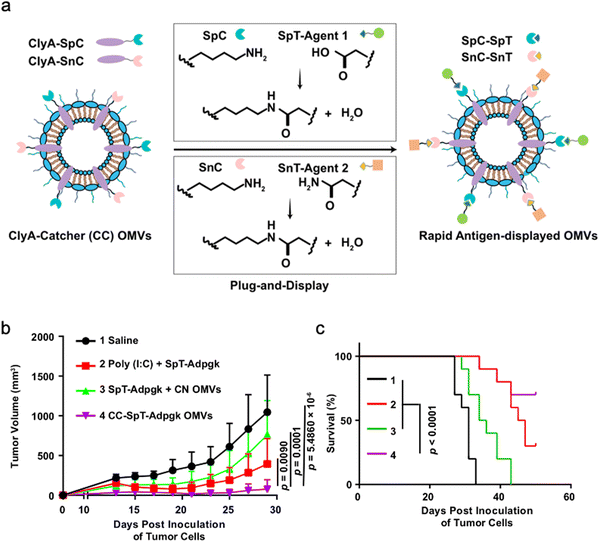
As OMVs are resistant to degradation by various enzymes, they can be transported over long distances and can reach target sites. Meanwhile, by virtue of rapid detection and internalization by host cells, OMVs have been extensively developed as delivery vehicles to transport substances for different biomedical applications.64 To date, a couple of methods have been used for loading various substances into OMVs (Fig. 3), such as genetic engineering, incubation, electroporation, membrane fusion and chemical modification.First, genetic engineering is the most widely used approach to load desired proteins to either the exterior or interior of OMVs by transforming plasmids into parent bacteria. In this way, sophisticated anchor points are essential for protein location. For example, SlyB, sorted into the periplasmic side of the bacterial outer membrane by its N-terminus lipid moiety, is a native and conserved Lpp in Gram-negative bacteria.65 Nanoluciferase (Nluc) is a small, highly stable, and ATP-independent bioluminescent protein, relying on its substrate to produce extremely robust luminescence.66 It can be loaded within OMVs by co-expression with the SlyB protein in the parent bacteria. To enhance the targeting ability of OMVs, the SpyCatcher/Tag or SnoopCatcher/Tag system has been applied to display the functional cargo on the vesicle surface.67,68 Kim et al. have suggested that a single chain Fv antibody fragment can be successfully displayed on the OMV surface by creating a chimeric ClyA fusion protein.69 Cheng et al. have also established a flexible “Plug-and-Display” platform in which antigen-tag can spontaneously bind to the protein catcher expressed in fusion with ClyA on the OMV membrane to simultaneously display multiple tumor antigens (Fig. 4).70
Furthermore, it seems that incubating OMVs or parent cells with cargo to obtain drug-loaded OMVs represents a simple loading strategy. In previous studies, OMVs have been directly incubated with chemotherapeutic drugs or NIR dyes, followed by the removal of the free cargo through ultracentrifugation to carry payloads.71,72 The cargo can be alternatively incubated with the parent bacteria during the growth phase. In this case, substances can be engulfed by bacteria and sorted into OMVs through shedding, which is often applied for antibiotic loading, such as ceftriaxone, amikacin, azithromycin, ampicillin, levofloxacin, ciprofloxacin, and norfloxacin.73 Interestingly, direct incubation with OMVs is incapable of loading antibiotics, which is probably ascribed to the drug efflux mechanism of live bacteria.74 Electroporation is a nucleic acid delivery technique that employs short high-voltage pulses to form transient pores in biological membranes for the entry of substances. This technique recently has been used to promote the loading of siRNA and gold nanoparticles into OMVs without disrupting membrane integrity.75 It is well-known that eukaryotic cell fusion relying on the structure of lipid bilayers is a general process, which induces changes in the cellular phenotype or function. Similarly, the aggregation and fusion of different types of OMVs induced by electrolytes or low pH can introduce exogenous antigens and generate new functions.76 Sonication and physical extrusion have been frequently reported as efficient methods to prepare fused OMVs.77 However, this method is occasionally suffering from membrane disruption, leading to disastrous leakage of cytoplasmic contents in OMVs.
Additionally, abundant lipids and proteins on the OMV surface provide opportunities to anchor cargo molecules through chemical modifications including covalent binding and non-covalent binding. Indeed, due to the similar composition of DSPE to phospholipids of OMV membranes, the RGD peptide, as a targeting ligand, can be readily inserted into OMV membranes using DSPE as a lipid head.71 In another study, Qing et al. have utilized calcium phosphate (CaP) to encapsulate OMVs, in which the acidic residues on the OMV surface offer the “nucleation sites” to chelate calcium ions.78 Furthermore, different approaches can be cooperated to decorate OMVs for introducing multiple functions. For instance, OMVs derived from E. coli can be conjugated with maleimide (Mal) groups on the surface by a coupling reaction between the amines associated with membrane proteins of OMVs and NHS ester in Mal-PEG4-NHS.79 Upon NIR irradiation, Mal groups on OMVs can form stable thioether bonds with the released tumor antigens in the tumor site, which promotes antigen uptake by DCs. Simultaneously, Mal functionalized OMVs are interiorly loaded with an inhibitor of indoleamine 2,3-dioxygenase (IDO), which is an immune suppressive checkpoint, via electroporation to enhance the efficacy of immune modulation. Taken together, various loading strategies have been explored to load cargo molecules into OMVs, which paves the way for their applications in a variety of biomedical applications.
6. Application of engineered OMVs for bioimaging and diagnosis
Bioimaging techniques play an important role in guiding the early diagnosis and treatment of diseases.80 Exogenous bioimaging probes, which provide a visual signal with an optical, a magnetic or a nuclear method, can be designed and anchored onto OMVs (Fig. 5). Benefiting from this characteristic, it is feasible to examine OMV-mediated communication mechanisms between bacteria and hosts.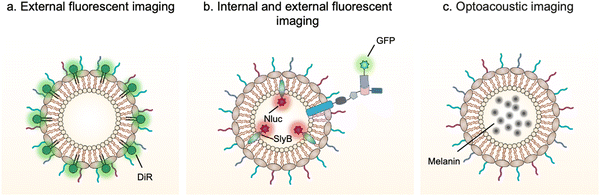 | ||
| Fig. 5 Schematic illustration of the application of engineered OMVs for bioimaging and diagnosis via (a and b) fluorescence or (c) optoacoustic approach. | ||
For instance, DiR iodide is a lipophilic fluorescent dye and widely used to label membranes. By labeling OMVs with DiR, Liu et al. have demonstrated that OMVs secreted from Akkermansia muciniphila can enter and accumulate in bone tissues to generate protective effects via augmenting osteogenic activity and inhibiting osteoclast formation.81 However, lipophilic fluorescent dyes via non-covalent binding are unstable and can be rapidly quenched to lost fluorescence emission. It is necessary to design and anchor bioimaging probes inside OMVs, which weaken fluorescence quenching and reduce leakage during systemic circulation. Based on this point, Chen et al. have developed a modular platform for biosensing applications by simultaneously modifying the interior and exterior of OMVs.65 Leveraging the principle that SlyB is localized at the inner side of the outer membranes, Nluc can be loaded into OMVs through fusion with SlyB. Luminescence signals emitted from engineered OMVs containing SlyB–Nluc are highly visible both in vitro and in vivo. It is worth noting that a trivalent Scaf3 scaffold is sandwiched between the ice nucleation protein (INP) anchor and the antibody-binding Z-domain for the modification of additional functional proteins, such as green fluorescent protein (GFP). These fluorescent OMVs are stabilized and extremely appropriate to explore the dysbiosis of commensal bacteria in digestive and airway systems as the lung commensal bacteria belonging to the genera Bacteroides and Prevotella have been reported to promote pulmonary fibrosis through their OMVs.82
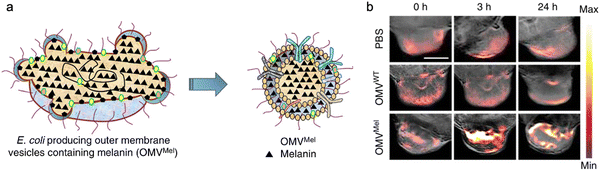
In addition, bacterial vesicles can also be engineered for optoacoustic imaging (Fig. 6). Melanin, naturally synthesized in many organisms, is suitable for optoacoustic imaging given their broad optical absorption.83Via overexpressing tyrosinase in E. coli, a key enzyme in melanin synthesis, melanin can be naturally packaged into OMVs, which generate an enhanced multi-spectral optoacoustic tomography signal and produce local heating upon irradiation.84 After systemic administration, the engineered OMVs can accumulate in the tumor tissue of mice for both imaging and photothermal therapy. In line with this, polydopamine nanoparticles generated by oxidative polymerization of dopamine is a melanin-like material, which can be encapsulated into the OMV–cancer cell hybrid membrane for tumor-targeted photoacoustic imaging and photothermal therapy.85 In general, these achievements indicate the potential of OMVs in bioimaging and disease diagnosis.
7. Application of OMVs for disease treatment
Given the inherent immunoregulatory properties and the ability to deliver exogenous antigens and therapeutic molecules,86 either naturally produced or engineered bacterial OMVs have been intensely applied as therapeutics for treating different diseases, such as inflammatory bowel disease (IBD) and tumors. OMVs have also been modified and used as vaccines to prevent bacterial and viral infections. In the following subsections, applications of OMVs for disease treatment are highlighted and discussed (Fig. 7).7.1 IBD
IBD, including Crohn's disease (CD) and ulcerative colitis (UC), is characterized by chronic or recurring inflammation of intestines.87 The breakdown of intestinal homeostasis maintained by the mucosal barrier and mucosal immunity is the key event in the pathogenesis of IBD.88 Recently, substantial evidences have demonstrated the strong relationship between the gut microbiota and intestinal homeostasis.89 OMVs derived from commensal bacteria are able to maintain intestinal homeostasis by interactions with intestinal epithelia and the mucosal immune system. For example, through releasing PSA-encapsulated OMVs, B. fragilis can be sensed by DCs through TLR2, thereby inducing the differentiation of IL-10-producing regulatory T cells (Tregs).57,86 It is well-known that Tregs, which are differentiated from CD4+ T cells, mainly participate in alleviated IBD by restraining inflammatory responses.90 Treatment with OMVs from B. fragilis protects mice from intestinal inflammation and experimental colitis.91 In a similar fashion, OMVs derived from Bacteroides thetaiotaomicron significantly stimulate the IL-10 expression by colonic DCs in healthy individuals, but not in patients with CD or UC. The ability of B. thetaiotaomicron-derived OMVs to regulate immune responses enables them to be considered as prospective candidates for IBD therapy.92A. muciniphila, a promising probiotic in the gut, plays an important role in the treatment of metabolic diseases. OMVs collected from A. muciniphila elevate the tight junction expression in epithelial cells and reduce the inflammation to increase the integrity of the intestinal barrier, which may rely on the amuc_1100, an outer membrane protein of A. muciniphila.93 As expected, the oral administration of A. muciniphila-derived OMVs has shown protective effects in dextran sodium sulfate (DSS)-induced colitis.94 In addition, compared with free antibiotics, OMV-carried antibiotics exhibit excellent intracellular delivery due to the advantages of OMVs in targeting and biocompatibility, thereby allowing the removal of intracellular bacteria.95 Undisputedly, OMV-carried antibiotics are promising for fighting against the growing threat of antibiotic resistance in infectious diseases.7.2 Tumors
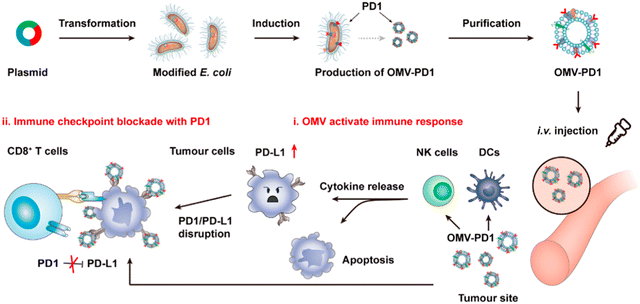
OMVs possess the ability to accumulate in the tumor tissue through the enhanced permeability and retention (EPR) effect. Together with the innate immunogenicity, OMVs can be developed as a promising delivery platform to enhance the efficacy of immunotherapy. Melanoma is a malignant tumor locating in the basal layer of the epidermis and the dermis and characterized with rapid progression, fast relapse, and high metastasis.96 As nanoscale OMVs are skin-permeable and can pass through the stratum corneum, they are appropriate drug delivery vehicles for melanoma therapy. Gao et al. have constructed the E. coli expressing TNF-related apoptosis inducing ligand (TRAIL) protein and modified the derived OMVs with the αvβ3 integrin peptide, targeting ligand and indocyanine green for the targeted therapy of melanoma. The resulting multifunctional OMVs can induce transdermal photo-TRAIL-programmed treatment in skin melanoma to enhance antitumor performance.72 In addition, Tang et al. have modified attenuated Salmonella derived OMVs with the tumor-targeting ligand Arg–Gly–Asp (RGD) peptide to improve tumor-targeting ability. The modified OMVs have been further used to coat chemotherapy drug-loaded polymeric micelles to greatly improve the synergistic therapeutic efficacy of cancer immunotherapy. The systemic injection of OMVs can not only elicit effectively protective immunity to prevent the occurrence of melanoma, but also significantly inhibit the growth and metastasis of tumor and prolong the survival rate of tumor-bearing mice.71Chen et al. have successfully synthesized the complex of gold nanoparticles and E. coli-derived OMVs. They have observed that the combination of this complex with radiotherapy significantly increases the intracellular reactive oxygen species (ROS) level, resulting in effective radio-sensitization and immunoactivation as well as successfully inhibited tumor growth in glioblastoma-bearing mice.97 Other studies have shown that mixing attenuated Klebsiella pneumoniae-derived OMVs with chemotherapeutic DOX can induce an appropriate immune response, which effectively fight against lung cancer without significant toxicity in vivo.98 Kim et al. have demonstrated the potential of bacterial OMVs to treat various cancers. The systemic administration of OMVs obtained from E. coli W3110 can accumulate in the tumor tissue and subsequently induce the interferon-γ (IFN-γ)-dependent antitumor effect. It is noted that despite OMVs are thought to initiate strong IFN-γ-mediated antitumor responses, IFN-γ paradoxically upregulates immunosuppressive factors, especially PD-L1 in the TME, which hamper the T-cell function and restrict the effectiveness of immunotherapeutics.99 Based on this fact, Li et al. have genetically engineered attenuated E. coli to express the ectodomain of PD-1 on the surface (Fig. 8). OMVs collected from this bioengineered strain can bind with PD-L1 on the tumor cell surface and lead to the retarded tumor growth in the B16 or CT26 mouse model, which are attributed to the cooperation between immune stimulation and checkpoint inhibition.100 Furthermore, Pan et al. have also extracted OMVs from E. coli which is modified with the tumor-targeting peptide Lyp1 and subsequently introduced the PD-1 plasmid into OMVs by electroporation. In this situation, the presence of Lyp1 guides the entry of these vesicles into tumor cells and subsequent release of the PD-1 plasmid to the nucleus. PD-1 overexpressed on the surface of tumor cells effectively binds to PD-L1, thereby blocking the PD-1/PD-L1 pathway and preventing immune escape. Simultaneously, OMVs recruit cytotoxic T lymphocytes and natural killer cells and induce the secretion of INF-γ to enhance antitumor immune responses.101 The immunosuppressive tumor microenvironment (TME) mediates immune tolerance and evasion in solid tumors, which reduces the efficacy of immunotherapy. By coating the OMV surface with a CaP shell, Qing et al. have developed a potent OMVbased strategy for reprogramming the immunosuppressive TME and enhancing cancer immunotherapy. The pHsensitive CaP shell can prevent the clearance of OMVs and decrease the toxicity of OMVs, facilitating the neutralization of acidic TME and the polarization of macrophages into the M1 type.78 Although it has been extensively reported that OMVs play a crucial role in the treatment of different tumors, the fundamental understanding of the interface between OMVs and tumor cells or the immune system needs to be further investigated for applications in treating diverse tumors.
7.3 Vaccines
OMVs are mainly composed of bacterial outer membrane components, which inherently carry key antigenic substrates that are required to induce a protective immune response. Different from live bacteria, inanimate OMVs are safe and moderately reactogenic and have been proposed as a vaccine strategy against bacterial pathogens shortly after their discovery. With the advances in genetic engineering and nanotechnology, OMVs with favorable safety, cost-effectiveness, and affordability have been developed as an ideal vaccine candidate to prevent epidemic diseases.102Meningococcal disease, caused by N. meningitidis, induces the infection and inflammation in both the brain and bloodstream. OMVs derived from the group B meningococci, well-known as Cuban vaccine, are the first OMV-based vaccine. Furthermore, they are also the first effective vaccine against the meningococcal disease worldwide, showing long-acting and high titers of bactericidal antibodies.103 With the advance of genetic engineering, the European Medicines Agency (EMA) and the US Food and Drug Administration (FDA) have approved a multi-component type B meningococcal vaccine 4CMenB for the human use.104 4CMenB is a multi-component vaccine prepared by displaying three highly immunogenic antigens of fHbp, NadA, and NHBA on the surface of OMVs.105 The combination of recombinant proteins and group B meningococci derived OMVs can trigger a strong bactericidal immune response against various serotype B isolates in adults, adolescents, and infants.104 Although other OMV-based vaccines have not entered the clinical stage yet, prominent progress has been achieved in the past few years. For example, Shigella disease, also known as bacillary dysentery, is caused by Shigella sonnei from food, which leads to bacterial spreading and invasion in the intestinal epithelia and results in diarrhea.106 Considering that the S. sonnei O-antigen polysaccharide is effective in stimulating the production of protective antibodies, Tian et al. have developed the O-antigen based OMV vaccine by expressing the O-antigen in recombinant Salmonella via the LPS synthesis pathway.107 Inspired by this work, vaccines basing on polysaccharide antigen-loaded bacterial OMVs have received increasing attention. In addition, OMVs can also be encapsulated in polyanhydride nanoparticles by a solvent displacement method using a copolymer of poly (methyl vinyl ether-co-maleic anhydride). Immunization with OMVs encapsulated in these nanoparticles by the nasal or oral route protects mice from traumatic S. sonnei infection.108
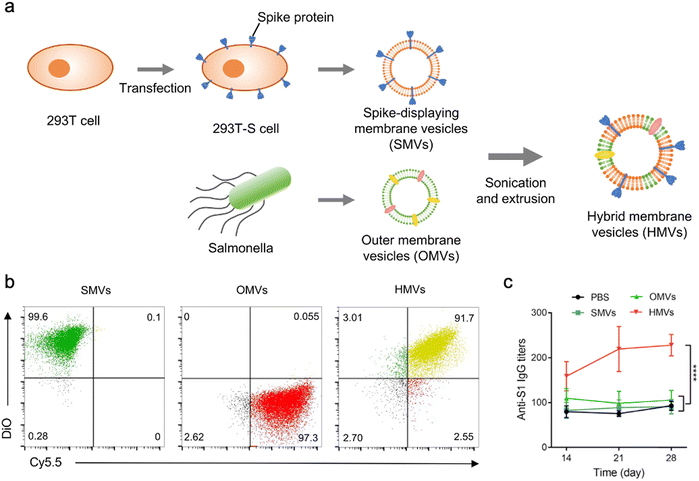
In addition to bacterial infections, genetic engineering techniques and biomimetic nanotechnology allow OMVs to be manipulated to resist viral infections. For instance, the receptor binding domain (RBD) in the SARS-CoV-2 spike protein, which leads to viral invasion via specifically binding to angiotensin-converting enzyme 2 (ACE2) in host cells, is considered as an appealing subunit vaccine target. Zhang et al. have obtained envelop virus-mimetic hybrid membrane-derived vesicles (HMVs) by fusing the eukaryotic membrane vesicle overexpressing spike protein with OMVs. The HMVs present an intact antigen structure and enable to boost antiviral immune responses (Fig. 9).109 Furthermore, Yang et al. have directly achieved the outward exposure of the fusion protein of ClyA–RBD on bacterial membranes and subsequently the presentation of the RBD on the vesicles using a high-pressure homogenization technology.110 Following the subcutaneous injection, these RBD-displaying OMVs can be accumulated in lymph nodes and elicit SARS-CoV-2-specific cellular and humoral immune responses in mice, which suggests the potential of using OMVs as a vaccine development platform.
8. Outlook
In recent years, the explorations of functionalized OMVs and their applications in biomedicine have increased continuously. OMVs possess a variety of unique properties, including a nanoscale structure, immunogenicity, and the ability to carry cargo molecules, which make them acquire high potential for bioimaging, diagnosis, and therapy. However, a few challenges remain before OMVs are ready for broad clinical applications.First, the mechanism of OMV generation has not been fully understood. Previous studies on OMVs have mainly focused on specific molecules related to their functions, which is beneficial for exploring potential biological applications. The explanation for the mechanism of OMV formation is primarily concentrated on several molecules, such as the Tol–Pal complex, mDAP, Lpp, and OmpA, whereas understanding the process of OMV generation can promote the development of artificial approaches for the directional modification of OMVs. To address this challenge, interdisciplinary collaboration is necessary for studying the mechanism of OMV generation. Second, isolation methods of OMVs are relatively limited. Currently, methods capable of separating OMVs can be mainly classified into two categories.111 One is size-based separation, such as ultracentrifugation or size exclusion chromatography, which usually requires a large volume of samples and is time-consuming and susceptible to interference from other substrates with a similar size.112 The other approach is based on the specific binding of OMVs to other substates, such as immunomagnetic beads and polymer-based precipitation. Meanwhile, this separation method is not universal and may affect downstream analysis due to impurities introduced during the separation process. Therefore, it is pivotal to reveal the specific and ubiquitous markers for different OMVs through multi-omics data analysis. New methods that can efficiently isolate OMVs without affecting their native bioactivity are also an urgent need. Third, the presence of abundant endotoxin and virulence factors in OMVs, especially LPS, largely restricts their clinical translation. Despite the genetic engineering technology has been used to develop endotoxin-reduced OMVs, it is needed to further reduce endotoxin levels before clinical trials.113 To this end, more comprehensive investigation of virulence factors on OMVs is required to ensure safety. Lastly, compared to the complexity of genetic engineering, chemical modification has been widely employed to functionalize exosomes, but not OMVs. The utilization of covalent conjugation or supramolecular interactions to modify OMVs may facilitate the generation of functionalized OMV-based nanomaterials and expand a broad range of biomedical applications. We anticipate that this work would inspire the innovation of next-generation nanomedicines for disease diagnosis, prevention and treatment.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Key Basic Research Program (SQ2021YFA090162), the National Natural Science Foundation of China (21875135), and the Innovative Research Team of High-Level Local Universities in Shanghai (SHSMU-ZDCX20210900).References
- C. Schwechheimer and M. J. Kuehn, Nat. Rev. Immunol., 2015, 13, 605–619 CAS.
- M. D. Balhuizen, E. J. A. Veldhuizen and H. P. Haagsman, Front. Microbiol., 2021, 12, 629090 CrossRef PubMed.
- S. N. Chatterjee and J. Das, J. Gen. Microbiol., 1967, 49, 1–11 CrossRef CAS PubMed.
- I. W. Devoe and J. E. Gilchrist, J. Exp. Med., 1973, 138, 1156–1167 CrossRef CAS PubMed.
- I. W. DeVoe and J. E. Gilchrist, J. Exp. Med., 1975, 141, 297–305 CrossRef CAS PubMed.
- D. N. Villageliu and D. R. Samuelson, Front. Microbiol., 2022, 13, 828704 CrossRef PubMed.
- Y. Tashiro, A. Inagaki, M. Shimizu, S. Ichikawa, N. Takaya, T. Nakajima-Kambe, H. Uchiyama and N. Nomura, Biosci., Biotechnol., Biochem., 2011, 75, 605–607 CrossRef CAS PubMed.
- A. T. Jan, Front. Microbiol., 2017, 8, 1053 CrossRef PubMed.
- F. Mancini, O. Rossi, F. Necchi and F. Micoli, Int. J. Mol. Sci., 2020, 21, 4416 CrossRef CAS PubMed.
- T. J. Silhavy, D. Kahne and S. Walker, Cold Spring Harb. Perspect. Biol., 2010, 2, a000414 Search PubMed.
- M. Toyofuku, N. Nomura and L. Eberl, Nat. Rev. Microbiol., 2019, 17, 13–24 CrossRef CAS PubMed.
- Z. Zhu, F. Antenucci, K. R. Villumsen and A. M. Bojesen, mBio, 2021, 12, e0170721 CrossRef PubMed.
- E. D. Avila-Calderón, M. D. S. Ruiz-Palma, M. G. Aguilera-Arreola, N. Velázquez-Guadarrama, E. A. Ruiz, Z. Gomez-Lunar, S. Witonsky and A. Contreras-Rodríguez, Front. Microbiol., 2021, 12, 557902 CrossRef PubMed.
- Y. Ojima, T. Sawabe, K. Konami and M. Azuma, Biotechnol. Bioeng., 2020, 117, 701–709 CrossRef CAS PubMed.
- F. Berlanda Scorza, A. M. Colucci, L. Maggiore, S. Sanzone, O. Rossi, I. Ferlenghi, I. Pesce, M. Caboni, N. Norais, V. Di Cioccio, A. Saul and C. Gerke, PLoS One, 2012, 7, e35616 CrossRef PubMed.
- H. Hirakawa, K. Suzue and H. Tomita, Vaccines, 2022, 10, 422 CrossRef CAS PubMed.
- S. Payen, D. Roy, A. Boa, M. Okura, J. P. Auger, M. Segura and M. Gottschalk, Microorganisms, 2021, 9, 2386 CrossRef CAS PubMed.
- A. Mychack and A. Janakiraman, J. Bacteriol., 2021, 203, e00640 CrossRef CAS PubMed.
- R. Acevedo, S. Fernández, C. Zayas, A. Acosta, M. E. Sarmiento, V. A. Ferro, E. Rosenqvist, C. Campa, D. Cardoso, L. Garcia and J. L. Perez, Front. Immunol., 2014, 5, 121 Search PubMed.
- P. Briaud, A. Frey, E. C. Marino, R. A. Bastock, R. E. Zielinski, R. E. Wiemels, R. A. Keogh, E. R. Murphy, L. N. Shaw and R. K. Carroll, mSphere, 2021, 6, e0067621 CrossRef PubMed.
- M. Hadadi-Fishani, S. Najar-Peerayeh, S. D. Siadat, M. Sekhavati and A. M. Mobarez, Iran. J. Microbiol., 2021, 13, 824–831 Search PubMed.
- V. Zarantonello, T. P. Silva, N. P. Noyma, J. P. Gamalier, M. M. Mello, M. M. Marinho and R. C. N. Melo, Front. Microbiol., 2018, 9, 272 CrossRef PubMed.
- M. J. H. Gerritzen, R. H. W. Maas, J. van den Ijssel, L. van Keulen, D. E. Martens, R. H. Wijffels and M. Stork, Microb. Cell Factories, 2018, 17, 157 CrossRef CAS PubMed.
- C. W. Choi, E. C. Park, S. H. Yun, S. Y. Lee, Y. G. Lee, Y. Hong, K. R. Park, S. H. Kim, G. H. Kim and S. I. Kim, J. Proteome Res., 2014, 13, 4298–4309 CrossRef CAS PubMed.
- J. I. Keenan and R. A. Allardyce, Eur. J. Gastroenterol. Hepatol., 2000, 12, 1267–1273 CrossRef CAS PubMed.
- P. D. Veith, Y. Y. Chen, D. G. Gorasia, D. Chen, M. D. Glew, N. M. O'Brien-Simpson, J. D. Cecil, J. A. Holden and E. C. Reynolds, J. Proteome Res., 2014, 13, 2420–2432 CrossRef CAS PubMed.
- M. F. Haurat, J. Aduse-Opoku, M. Rangarajan, L. Dorobantu, M. R. Gray, M. A. Curtis and M. F. Feldman, J. Biol. Chem., 2011, 286, 1269–1276 CrossRef CAS PubMed.
- E. Izquierdo, J. Rodriguez-Coira, M. I. Delgado-Dolset, C. Gomez-Casado, D. Barber and M. M. Escribese, J. Invest. Allergol. Clin. Immunol., 2022, 32, 81–96 CrossRef CAS PubMed.
- D. Werling and T. W. Jungi, Vet. Immunol. Immunopathol., 2003, 91, 1–12 CrossRef CAS PubMed.
- M. Kaparakis-Liaskos and R. L. Ferrero, Nat. Rev. Immunol., 2015, 15, 375–387 CrossRef CAS PubMed.
- S. J. Bauman and M. J. Kuehn, BMC Microbiol., 2009, 9, 26 CrossRef PubMed.
- N. Furuta, K. Tsuda, H. Omori, T. Yoshimori, F. Yoshimura and A. Amano, Infect. Immun., 2009, 77, 4187–4196 CrossRef CAS PubMed.
- S. W. Sharpe, M. J. Kuehn and K. M. Mason, Infect. Immun., 2011, 79, 4361–4369 CrossRef CAS PubMed.
- A. Elmi, E. Watson, P. Sandu, O. Gundogdu, D. C. Mills, N. F. Inglis, E. Manson, L. Imrie, M. Bajaj-Elliott, B. W. Wren, D. G. Smith and N. Dorrell, Infect. Immun., 2012, 80, 4089–4098 CrossRef CAS PubMed.
- M. Bielaszewska, C. Rüter, L. Kunsmann, L. Greune, A. Bauwens, W. Zhang, T. Kuczius, K. S. Kim, A. Mellmann, M. A. Schmidt and H. Karch, PLoS Pathog., 2013, 9, e1003797 CrossRef PubMed.
- A. Olofsson, L. Nygård Skalman, I. Obi, R. Lundmark and A. Arnqvist, mBio, 2014, 5, e00979 CrossRef PubMed.
- J. J. Lee, D. G. Kim, D. H. Kim, H. L. Simborio, W. Min, H. J. Lee, M. Her, S. C. Jung, M. Watarai and S. Kim, J. Biol. Chem., 2013, 288, 28049–28057 CrossRef CAS PubMed.
- R. Pascolutti, V. Algisi, A. Conte, A. Raimondi, M. Pasham, S. Upadhyayula, R. Gaudin, T. Maritzen, E. Barbieri, G. Caldieri, C. Tordonato, S. Confalonieri, S. Freddi, M. G. Malabarba, E. Maspero, S. Polo, C. Tacchetti, V. Haucke, T. Kirchhausen, P. P. Di Fiore and S. Sigismund, Cell Rep., 2019, 27, 3049–3061.e3046 CrossRef CAS PubMed.
- L. Turner, N. J. Bitto, D. L. Steer, C. Lo, K. D'Costa, G. Ramm, M. Shambrook, A. F. Hill, R. L. Ferrero and M. Kaparakis-Liaskos, Front. Immunol., 2018, 9, 1466 CrossRef PubMed.
- E. J. O'Donoghue, N. Sirisaengtaksin, D. F. Browning, E. Bielska, M. Hadis, F. Fernandez-Trillo, L. Alderwick, S. Jabbari and A. M. Krachler, PLoS Pathog., 2017, 13, e1006760 CrossRef PubMed.
- V. Schaar, S. P. de Vries, M. L. Perez Vidakovics, H. J. Bootsma, L. Larsson, P. W. Hermans, A. Bjartell, M. Mörgelin and K. Riesbeck, Cell. Microbiol., 2011, 13, 432–449 CrossRef CAS PubMed.
- M. Kaparakis, L. Turnbull, L. Carneiro, S. Firth, H. A. Coleman, H. C. Parkington, L. Le Bourhis, A. Karrar, J. Viala, J. Mak, M. L. Hutton, J. K. Davies, P. J. Crack, P. J. Hertzog, D. J. Philpott, S. E. Girardin, C. B. Whitchurch and R. L. Ferrero, Cell. Microbiol., 2010, 12, 372–385 CrossRef CAS PubMed.
- M. J. Kuehn and N. C. Kesty, Genes Dev., 2005, 19, 2645–2655 CrossRef CAS PubMed.
- H. Kumar, T. Kawai and S. Akira, Biochem. Biophys. Res. Commun., 2009, 388, 621–625 CrossRef CAS PubMed.
- R. Di Guida, A. Casillo, J. M. Tomás, S. Merino and M. M. Corsaro, Int. J. Mol. Sci., 2022, 23, 1204 CrossRef CAS PubMed.
- L. Gu, R. Meng, Y. Tang, K. Zhao, F. Liang, R. Zhang, Q. Xue, F. Chen, X. Xiao, H. Wang, H. Wang, T. R. Billiar and B. Lu, Shock, 2019, 51, 256–265 CrossRef CAS PubMed.
- D. M. Pilla, J. A. Hagar, A. K. Haldar, A. K. Mason, D. Degrandi, K. Pfeffer, R. K. Ernst, M. Yamamoto, E. A. Miao and J. Coers, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6046–6051 CrossRef CAS PubMed.
- J. C. Santos, M. S. Dick, B. Lagrange, D. Degrandi, K. Pfeffer, M. Yamamoto, E. Meunier, P. Pelczar, T. Henry and P. Broz, EMBO J., 2018, 37, e98089 CrossRef PubMed.
- T. N. Ellis, S. A. Leiman and M. J. Kuehn, Infect. Immun., 2010, 78, 3822–3831 CrossRef CAS PubMed.
- A. Zariri, J. Beskers, B. van de Waterbeemd, H. J. Hamstra, T. H. Bindels, E. van Riet, J. P. van Putten and P. van der Ley, Infect. Immun., 2016, 84, 3024–3033 CrossRef CAS PubMed.
- M. R. Mirlashari and T. Lyberg, Med. Sci. Monit., 2003, 9, Br316–Br324 Search PubMed.
- K. Zhao, X. Deng, C. He, B. Yue and M. Wu, Infect. Immun., 2013, 81, 4509–4518 CrossRef CAS PubMed.
- C. C. Allison, T. A. Kufer, E. Kremmer, M. Kaparakis and R. L. Ferrero, J. Immunol., 2009, 183, 8099–8109 CrossRef CAS PubMed.
- P. Deo, S. H. Chow, M. L. Han, M. Speir, C. Huang, R. B. Schittenhelm, S. Dhital, J. Emery, J. Li, B. T. Kile, J. E. Vince, K. E. Lawlor and T. Naderer, Nat. Microbiol., 2020, 5, 1418–1427 CrossRef CAS PubMed.
- J. D. Cecil, N. M. O'Brien-Simpson, J. C. Lenzo, J. A. Holden, W. Singleton, A. Perez-Gonzalez, A. Mansell and E. C. Reynolds, Front. Immunol., 2017, 8, 1017 CrossRef PubMed.
- J. Yang, I. Hwang, E. Lee, S. J. Shin, E. J. Lee, J. H. Rhee and J. W. Yu, Front. Immunol., 2020, 11, 581165 CrossRef CAS PubMed.
- Y. Shen, M. L. Giardino Torchia, G. W. Lawson, C. L. Karp, J. D. Ashwell and S. K. Mazmanian, Cell Host Microbe, 2012, 12, 509–520 CrossRef CAS PubMed.
- M. L. Vidakovics, J. Jendholm, M. Mörgelin, A. Månsson, C. Larsson, L. O. Cardell and K. Riesbeck, PLoS Pathog., 2010, 6, e1000724 CrossRef PubMed.
- M. Bielaszewska, M. Marejková, A. Bauwens, L. Kunsmann-Prokscha, A. Mellmann and H. Karch, Int. J. Med. Microbiol., 2018, 308, 882–889 CrossRef CAS PubMed.
- M. A. Cañas, M. J. Fábrega, R. Giménez, J. Badia and L. Baldomà, Front. Microbiol., 2018, 9, 498 CrossRef PubMed.
- B. Thay, A. Damm, T. A. Kufer, S. N. Wai and J. Oscarsson, Infect. Immun., 2014, 82, 4034–4046 CrossRef PubMed.
- A. Hidmark, A. von Saint Paul and A. H. Dalpke, J. Immunol., 2012, 189, 2717–2721 CrossRef CAS.
- S. Abouelhadid, S. J. North, P. Hitchen, P. Vohra, C. Chintoan-Uta, M. Stevens, A. Dell, J. Cuccui and B. W. Wren, mBio, 2019, 10, e00297 CrossRef CAS PubMed.
- J. Nicolás-Ávila, J. M. Adrover and A. Hidalgo, Immunity, 2017, 46, 15–28 CrossRef PubMed.
- Q. Chen, S. Rozovsky and W. Chen, Chem. Commun., 2017, 53, 7569–7572 RSC.
- S. Yu, Z. Li, J. Li, S. Zhao, S. Wu, H. Liu, X. Bi, D. Li, J. Dong, S. Duan and B. D. Hammock, Sens. Actuators B Chem., 2021, 336, 129717 CrossRef CAS PubMed.
- H. B. van den Berg van Saparoea, D. Houben, M. I. de Jonge, W. S. P. Jong and J. Luirink, Appl. Environ. Microbiol., 2018, 84, e02567 CrossRef PubMed.
- H. B. van den Berg van Saparoea, D. Houben, C. Kuijl, J. Luirink and W. S. P. Jong, Front. Microbiol., 2020, 11, 890 CrossRef PubMed.
- J. Y. Kim, A. M. Doody, D. J. Chen, G. H. Cremona, M. L. Shuler, D. Putnam and M. P. DeLisa, J. Mol. Biol., 2008, 380, 51–66 CrossRef CAS PubMed.
- K. Cheng, R. Zhao, Y. Li, Y. Qi, Y. Wang, Y. Zhang, H. Qin, Y. Qin, L. Chen, C. Li, J. Liang, Y. Li, J. Xu, X. Han, G. J. Anderson, J. Shi, L. Ren, X. Zhao and G. Nie, Nat. Commun., 2021, 12, 2041 CrossRef CAS PubMed.
- Q. Chen, H. Bai, W. Wu, G. Huang, Y. Li, M. Wu, G. Tang and Y. Ping, Nano Lett., 2020, 20, 11–21 CrossRef CAS PubMed.
- L. H. Peng, M. Z. Wang, Y. Chu, L. Zhang, J. Niu, H. T. Shao, T. J. Yuan, Z. H. Jiang, J. Q. Gao and X. H. Ning, Sci. Adv., 2020, 6, eaba2735 CrossRef CAS PubMed.
- W. Huang, Q. Zhang, W. Li, M. Yuan, J. Zhou, L. Hua, Y. Chen, C. Ye and Y. Ma, J. Controlled Release, 2020, 317, 1–22 CrossRef CAS PubMed.
- S. Fazal and R. Lee, Pharmaceutics, 2021, 13, 1430 CrossRef CAS PubMed.
- V. Gujrati, S. Kim, S. H. Kim, J. J. Min, H. E. Choy, S. C. Kim and S. Jon, ACS Nano, 2014, 8, 1525–1537 CrossRef CAS PubMed.
- Y. M. D. Gnopo, A. Misra, H. L. Hsu, M. P. DeLisa, S. Daniel and D. Putnam, J. Colloid Interface Sci., 2020, 578, 522–532 CrossRef CAS PubMed.
- R. Roberts, G. Moreno, D. Bottero, M. E. Gaillard, M. Fingermann, A. Graieb, M. Rumbo and D. Hozbor, Vaccine, 2008, 26, 4639–4646 CrossRef CAS PubMed.
- S. Qing, C. Lyu, L. Zhu, C. Pan, S. Wang, F. Li, J. Wang, H. Yue, X. Gao, R. Jia, W. Wei and G. Ma, Adv. Mater., 2020, 32, e2002085 CrossRef PubMed.
- Y. Li, K. Zhang, Y. Wu, Y. Yue, K. Cheng, Q. Feng, X. Ma, J. Liang, N. Ma, G. Liu, G. Nie, L. Ren and X. Zhao, Small, 2022, 18, e2107461 CrossRef PubMed.
- H. Gu, H. Tang, P. Xiong and Z. Zhou, Nanomaterials, 2019, 9, 130 CrossRef PubMed.
- J. H. Liu, C. Y. Chen, Z. Z. Liu, Z. W. Luo, S. S. Rao, L. Jin, T. F. Wan, T. Yue, Y. J. Tan, H. Yin, F. Yang, F. Y. Huang, J. Guo, Y. Y. Wang, K. Xia, J. Cao, Z. X. Wang, C. G. Hong, M. J. Luo, X. K. Hu, Y. W. Liu, W. Du, J. Luo, Y. Hu, Y. Zhang, J. Huang, H. M. Li, B. Wu, H. M. Liu, T. H. Chen, Y. X. Qian, Y. Y. Li, S. K. Feng, Y. Chen, L. Y. Qi, R. Xu, S. Y. Tang and H. Xie, Adv. Sci., 2021, 8, 2004831 CrossRef CAS PubMed.
- D. Yang, X. Chen, J. Wang, Q. Lou, Y. Lou, L. Li, H. Wang, J. Chen, M. Wu, X. Song and Y. Qian, Immunity, 2019, 50, 692–706.e697 CrossRef CAS PubMed.
- D. L. Longo, R. Stefania, S. Aime and A. Oraevsky, Int. J. Mol. Sci., 2017, 18, 1719 CrossRef PubMed.
- V. Gujrati, J. Prakash, J. Malekzadeh-Najafabadi, A. Stiel, U. Klemm, G. Mettenleiter, M. Aichler, A. Walch and V. Ntziachristos, Nat. Commun., 2019, 10, 1114 CrossRef PubMed.
- D. Wang, C. Liu, S. You, K. Zhang, M. Li, Y. Cao, C. Wang, H. Dong and X. Zhang, ACS Appl. Mater. Interfaces, 2020, 12, 41138–41147 CrossRef CAS PubMed.
- Y. J. Yu, X. H. Wang and G. C. Fan, Acta Pharmacol. Sin., 2018, 39, 514–533 CrossRef CAS PubMed.
- G. Bouma and W. Strober, Nat. Rev. Immunol., 2003, 3, 521–533 CrossRef CAS PubMed.
- C. C. Bain and A. M. Mowat, Immunol. Rev., 2014, 260, 102–117 CrossRef CAS PubMed.
- J. L. Round and S. K. Mazmanian, Nat. Rev. Immunol., 2009, 9, 313–323 CrossRef CAS PubMed.
- J. B. Yan, M. M. Luo, Z. Y. Chen and B. H. He, J. Immunol. Res., 2020, 2020, 8813558 Search PubMed.
- S. Ahmadi Badi, S. H. Khatami, S. H. Irani and S. D. Siadat, Cell J., 2019, 21, 57–61 Search PubMed.
- L. Durant, R. Stentz, A. Noble, J. Brooks, N. Gicheva, D. Reddi, M. J. O'Connor, L. Hoyles, A. L. McCartney, R. Man, E. T. Pring, S. Dilke, P. Hendy, J. P. Segal, D. N. F. Lim, R. Misra, A. L. Hart, N. Arebi, S. R. Carding and S. C. Knight, Microbiome, 2020, 8, 88 CrossRef CAS PubMed.
- L. Wang, L. Tang, Y. Feng, S. Zhao, M. Han, C. Zhang, G. Yuan, J. Zhu, S. Cao, Q. Wu, L. Li and Z. Zhang, Gut, 2020, 69, 1988–1997 CrossRef CAS PubMed.
- C. S. Kang, M. Ban, E. J. Choi, H. G. Moon, J. S. Jeon, D. K. Kim, S. K. Park, S. G. Jeon, T. Y. Roh, S. J. Myung, Y. S. Gho, J. G. Kim and Y. K. Kim, PLoS One, 2013, 8, e76520 CrossRef CAS PubMed.
- W. Huang, L. Meng, Y. Chen, Z. Dong and Q. Peng, Acta Biomater., 2022, 140, 102–115 CrossRef CAS PubMed.
- Y. R. Woo, S. H. Cho, J. D. Lee and H. S. Kim, Int. J. Mol. Sci., 2022, 23, 1813 CrossRef CAS PubMed.
- M. H. Chen, T. Y. Liu, Y. C. Chen and M. H. Chen, Nanomaterials, 2021, 11, 1661 CrossRef CAS PubMed.
- K. Kuerban, X. Gao, H. Zhang, J. Liu, M. Dong, L. Wu, R. Ye, M. Feng and L. Ye, Acta Pharm. Sin. B, 2020, 10, 1534–1548 CrossRef CAS PubMed.
- O. Y. Kim, H. T. Park, N. T. H. Dinh, S. J. Choi, J. Lee, J. H. Kim, S. W. Lee and Y. S. Gho, Nat. Commun., 2017, 8, 626 CrossRef PubMed.
- Y. Li, R. Zhao, K. Cheng, K. Zhang, Y. Wang, Y. Zhang, Y. Li, G. Liu, J. Xu, J. Xu, G. J. Anderson, J. Shi, L. Ren, X. Zhao and G. Nie, ACS Nano, 2020, 14, 16698–16711 CrossRef CAS PubMed.
- J. Pan, X. Li, B. Shao, F. Xu, X. Huang, X. Guo and S. Zhou, Adv. Mater., 2022, 34, e2106307 CrossRef PubMed.
- Z. Kis, R. Shattock, N. Shah and C. Kontoravdi, J. Biotechnol., 2019, 14, e1800376 Search PubMed.
- G. V. Sierra, H. C. Campa, N. M. Varcacel, I. L. Garcia, P. L. Izquierdo, P. F. Sotolongo, G. V. Casanueva, C. O. Rico, C. R. Rodriguez and M. H. Terry, NIPH Ann., 1991, 14, 195–207 CAS ; discussion 208-110.
- D. Serruto, M. J. Bottomley, S. Ram, M. M. Giuliani and R. Rappuoli, Vaccine, 2012, 30(Suppl. 2), B87–B97 CrossRef CAS PubMed.
- M. Pizza, V. Scarlato, V. Masignani, M. M. Giuliani, B. Aricò, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi and R. Rappuoli, Science, 2000, 287, 1816–1820 CrossRef CAS PubMed.
- K. L. Kotloff, Curr. Opin. Pediatr., 2022, 34, 147–155 CrossRef CAS PubMed.
- H. Tian, B. Li, T. Xu, H. Yu, J. Chen, H. Yu, S. Li, L. Zeng, X. Huang and Q. Liu, Appl. Environ. Microbiol., 2021, 87, e0096821 CrossRef PubMed.
- A. I. Camacho, J. M. Irache, J. de Souza, S. Sánchez-Gómez and C. Gamazo, Vaccine, 2013, 31, 3288–3294 CrossRef CAS PubMed.
- M. Zhang, L. Wang, J. Liu and Y. Pang, iScience, 2022, 25, 104490 CrossRef CAS PubMed.
- Z. Yang, L. Hua, M. Yang, S. Q. Liu, J. Shen, W. Li, Q. Long, H. Bai, X. Yang, Z. Ren, X. Zheng, W. Sun, C. Ye, D. Li, P. Zheng, J. He, Y. Chen, W. Huang, X. Peng and Y. Ma, Nano Lett., 2021, 21, 5920–5930 CrossRef CAS PubMed.
- J. Klimentová and J. Stulík, Microbiol. Res., 2015, 170, 1–9 CrossRef PubMed.
- L. L. Yu, J. Zhu, J. X. Liu, F. Jiang, W. K. Ni, L. S. Qu, R. Z. Ni, C. H. Lu and M. B. Xiao, BioMed Res. Int., 2018, 2018, 3634563 Search PubMed.
- H. C. Watkins, C. G. Rappazzo, J. S. Higgins, X. Sun, N. Brock, A. Chau, A. Misra, J. P. B. Cannizzo, M. R. King, T. R. Maines, C. A. Leifer, G. R. Whittaker, M. P. DeLisa and D. Putnam, Mol. Ther., 2017, 25, 989–1002 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |