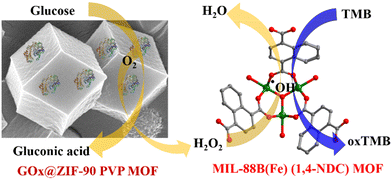
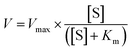Exploring the potential of iron-based metal–organic frameworks as peroxidase nanozymes for glucose detection with various secondary building units†
Sivasankar
Kulandaivel
 a,
Hsin-Tsung
Chen
a,
Hsin-Tsung
Chen
 *b,
Chia-Her
Lin
*b,
Chia-Her
Lin
 *a and
Yi-Chun
Yeh
*a and
Yi-Chun
Yeh
 *a
*a
aDepartment of Chemistry, National Taiwan Normal University, Taipei 11677, Taiwan. E-mail: chiaher@ntnu.edu.tw; yichuny@ntnu.edu.tw
bDepartment of Chemistry, Chung Yuan Christian University, 200 Zhongbei Rd., Zhongli Dist., Taoyuan 320314, Taiwan. E-mail: htchen@cycu.edu.tw
First published on 10th July 2023
Abstract
Finding materials in biosensing that balance enzyme-like reactivity, stability, and affordability is essential for the future. Because of their unique peroxidase properties, including variable pore size, surface area, and Lewis acid active sites, iron-based metal–organic frameworks (MOFs) have evolved as viable possibilities. In this study, we constructed a Fe-MOF and tested its peroxidase-like activity and responsiveness toward H2O2 colorimetric techniques. Using encapsulation, we incorporated glucose oxidase into the ZIF-90 PVP MOF and conducted a sequential reaction with the Fe-MOF to detect glucose. The results showed better peroxidase catalytic activity of the MIL-88B(Fe) (1,4-NDC) MOF and similar secondary building unit (SBU) Fe-MOFs were studied in other peroxidase nanozyme studies. When combined with an enzyme-encapsulating ZIF-90 PVP MOF, they could be sequentially employed for glucose detection purposes. This study highlights the potential of nanozymes as an alternative to natural enzymes, with promising applications in biosensing and beyond.
Introduction
Due to their remarkable specificity and mild reaction conditions, enzymes are essential biocatalysts in various fields, including medicine, biology, and industry.1–4 However, natural enzymes have certain limitations, such as high costs, poor operational stability, and environmental sensitivity, which have hindered their widespread use. Researchers have sought alternative solutions and turned to synthetic nanozymes that exhibit enzyme-like catalytic activity, exceptional stability, and programmable activity.5 Various methods utilizing different nanomaterials, including noble metals/metal–organic frameworks (MOFs)/carbon-based materials, have been created for these artificial mimetic enzymes.6 Compared to natural enzymes, these biomimetic materials, known as nanozymes, offer several advantages, including higher stability, more expansive operational windows, for example, various temperature/pH condition operations, simple synthetic protocols, tunability of catalytic activity, and lower costs.7 Furthermore, the distinct physicochemical features of nanosized materials give nanozymes a plethora of structural and chemical functionalization choices, providing a superior strategy for altering their catalytic activity. Significant biotechnology and nanomaterial research developments have resulted in rational design along with the novel synthesis of nanozymes for biosensor applications in recent years. Given these advantages, the developed nanozymes can potentially overcome the inherent restrictions.8Metal–organic frameworks (MOFs) are crystalline structures that emerge from the self-assembly of organic ligands linked by metal nodes.9–11 MOFs exhibit remarkable properties stemming from their distinctive microstructure, such as an elevated surface area, tunable porosity, and cavities.12 Consequently, they have gained immense popularity and are being utilized in various domains, varying from gas separation/storage to small-molecule sensing and catalysis.13–15 In recent years, significant advancements have been made in using MOFs as catalysts to replicate enzymatic activities, leading to a revolution in catalysis.16 Specifically, Fe, Cu, Mn, Co, Ni, Mo, or Ce-based single or bimetallic MOFs have been used to create peroxidase nanozymes, with Fe-based MOFs and their nanocomposites showing particularly remarkable enzyme-like performances.17–21
Fe-based MOFs have open, active sites that make them suitable for reactant transfer. The Fenton reaction is an advanced oxidation process that involves the redox cycling of Fe(III)/Fe(II) by H2O2,22 to proceed. However, the rate-limiting step in this reaction is the reduction of Fe(III) by H2O2, which ultimately affects the overall efficiency of the reaction. To increase the efficiency of H2O2, utilization and accelerate the redox cycling of Fe(III)/Fe(II), researchers are exploring new strategies to develop more efficient Fenton catalysts. One promising avenue of research involves the use of metal–organic frameworks (MOFs) as catalysts. However, MOF metal nodes are usually fully coordinated by organic linkers, which can reduce the number of Lewis acid sites and make them unavailable for H2O2 activation.23 Despite this limitation, Fe-MOFs have unique secondary building units and open metal sites, which are coordinatively unsaturated (CUS) metal centers, also known as Lewis acid sites. These Fe-MOFs have shown peroxidase nanozyme activity and can be utilized as efficient Fenton catalysts. Some examples of Fe-MOFs that have been explored for their catalytic activity include MIL-88A(Fe), MIL-100(Fe), Fe-MIL-88B-NH2, and MIL-101(Fe).24–31 Ongoing research in this area aims to optimize these catalysts to further improve their efficiency and practical applicability.
In addition, further recyclability remains a challenge, despite their success in peroxidase nanozyme and glucose oxidase cascade reactions. Various techniques have been explored to preserve enzyme stability and activity using porous materials, including enzyme immobilization, encapsulation, pore infiltration, and surface-bound methods to address this challenge.32–36 Encapsulation, also called enzyme@MOF, involves synthesizing a MOF in the presence of an enzyme, resulting in a biocomposite where the enzyme is enclosed within MOF crystals.32 Promising biocompatible synthesis conditions have been developed at room temperature using H2O, and noteworthy MOFs include ZIF-8 and ZIF-90. The enzyme@ZIF-8 and enzyme@ZIF-90 MOFs have been studied and demonstrate advantages in terms of reusability and stability.
This study indicates the better peroxidase catalytic activity of the MIL-88B(Fe) (1,4-NDC) MOF and similar secondary building unit (SBU) Fe-MOFs were studied in other peroxidase nanozyme studies. First, glucose oxidase was immobilized into the ZIF-90 PVP MOF using the encapsulation method for detection. This two-step process involved GOx@ZIF-90 PVP MOF converting glucose into gluconic acid and H2O2. Then, the solution was introduced into the Fe-MOF peroxidase reaction, where H2O2 oxidized the TMB substrate via Fe-MOF catalytic reaction (Scheme 1).
The advantages of encapsulating enzymes into the ZIF-90 PVP MOF and utilizing peroxidase nanozyme (Fe-MOF) for further analysis are evident in their stability and reusability. Thus, this study highlights the potential of nanozymes as an alternative to natural enzymes, with promising applications in biosensing and beyond.
Experimental
Chemicals and materials
Unless otherwise specified, all compounds were obtained from commercial suppliers and were not further purified.Synthesis of MIL-88B(Fe) (1,4-NDC) MOFs
1.5 mmol of FeSO4·7H2O and 1.0 mmol of 1,4-NDC were placed in a 25 mL Teflon-lined autoclave with 8 mL of N,N-dimethylformamide (DMF), 1 mL of methanol, and 1 mL of ddH2O. After adding a few drops of concentrated sulfuric acid (H2SO4), the reactor was heated in an electric oven at 80 °C for 24 hours. After the reaction, the mixture was allowed to cool to ambient temperature, and centrifugation was used to obtain a solid product. The product was washed with DMF, ddH2O, and methanol and then dried for 12 hours in a vacuum oven at 60 °C. The powder that resulted was saved in a sample vial for future use.Synthesis of the GOx@ZIF-90-PVP MOF composite
The enzyme@ZIF-90-PVP MOF was synthesized following a modified version of the literature procedure.37 First, a solution was prepared by dissolving 46 mg of 2-imidazolecarboxaldehyde (2-ICA) and 5 mg of PVP (MW = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) in 2 mL of deionized water at 65 °C and named solution A. Once the starting materials were dissolved, the solution was cooled to 42 °C. Then 2 mg of GOx in 0.2 mL of deionized water was added to Solution A and stirred. Then, 0.3 mL of a 0.62 M zinc nitrate hexahydrate solution was added to the mixture while stirring at room temperature. After mixing, the reaction mixture quickly became milky. After stirring for about 10 minutes, the product was recovered by centrifuging at 14
000) in 2 mL of deionized water at 65 °C and named solution A. Once the starting materials were dissolved, the solution was cooled to 42 °C. Then 2 mg of GOx in 0.2 mL of deionized water was added to Solution A and stirred. Then, 0.3 mL of a 0.62 M zinc nitrate hexahydrate solution was added to the mixture while stirring at room temperature. After mixing, the reaction mixture quickly became milky. After stirring for about 10 minutes, the product was recovered by centrifuging at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 minutes and rinsed thrice with ddH2O. At last, the enzyme@MOF composite was freeze-dried for 24 hours under liquid nitrogen.
000 rpm for 3 minutes and rinsed thrice with ddH2O. At last, the enzyme@MOF composite was freeze-dried for 24 hours under liquid nitrogen.
Peroxidase-like activity of MIL-88B(Fe) (1,4-NDC) MOFs
The following procedure was used to detect H2O2 and determine the peroxidase nanozyme activity: first, a reaction mixture was prepared in a 1.6 mL microcentrifuge tube by sequentially adding 200 μL of 1 mg mL−1 MIL-88B(Fe) (1,4-NDC) MOF in 10 mM NaAc–HAc (pH 3.6) buffer, 200 μL of 1 mM TMB, and 200 μL of H2O2 solution at different concentrations. The mixture was then incubated at 37 °C for 5 minutes, followed by centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 minutes. The resulting solution was then analyzed using UV-vis spectroscopy at ambient temperatures over 300 to 700 nm. For quantitative analysis, the absorbance at 652 nm was measured.
000 rpm for 2 minutes. The resulting solution was then analyzed using UV-vis spectroscopy at ambient temperatures over 300 to 700 nm. For quantitative analysis, the absorbance at 652 nm was measured.
The nanozyme activity was validated by measuring the reaction product using UV-vis absorbance at 652 nm, utilizing the extinction coefficient of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 M−1 cm−1. Additionally, the peroxidase reaction kinetics were calculated using the Michaelis–Menten equation.
300 M−1 cm−1. Additionally, the peroxidase reaction kinetics were calculated using the Michaelis–Menten equation.
Detection of glucose using GOx@ZIF-90-PVP MOF
Sequential experiments were conducted to investigate the GOx@ZIF-90-PVP MOF composite's catalytic performance. First, 50 μL of glucose with varying concentrations was added to 50 μL of the GOx@ZIF-90-PVP MOF composite, followed by incubation at room temperature for 10 min. Next, the reaction solution was centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 min to remove the GOx@ZIF-90-PVP MOF composite. Next, the resulting 100 μL of the solution was added to a reaction mixture containing 200 μL of 1 mg mL−1 MIL-88B(Fe) (1,4-NDC) MOF in 10 mM NaAc–HAc (pH 3.6) buffer, 100 μL of 10 mM NaAc–HAc (pH 3.6) buffer, and 100 μL of 1 mM TMB. The reaction was conducted for 5 minutes, and the absorbance at 652 nm was recorded in a UV spectrum.
000 rpm for 3 min to remove the GOx@ZIF-90-PVP MOF composite. Next, the resulting 100 μL of the solution was added to a reaction mixture containing 200 μL of 1 mg mL−1 MIL-88B(Fe) (1,4-NDC) MOF in 10 mM NaAc–HAc (pH 3.6) buffer, 100 μL of 10 mM NaAc–HAc (pH 3.6) buffer, and 100 μL of 1 mM TMB. The reaction was conducted for 5 minutes, and the absorbance at 652 nm was recorded in a UV spectrum.
Characterization
The MOFs were characterized using various techniques to investigate their crystal structure, morphology, elemental composition, surface area, and pore volume. Powder X-ray diffraction (PXRD) using CuKα radiation (λ = 1.54178) on a Bruker D8 Advance Eco instrument was employed to determine the crystal structure of the MOFs. Field emission scanning electron microscopy (FE-SEM) was utilized to study the morphology of the MOF samples, and elemental analysis was conducted using FE-SEM coupled with energy dispersive X-ray spectroscopy (EDS) on a JEOL JSM-7600F instrument with an Oxford X-Max 80 detector. The BET surface area and pore volume were calculated using N2 sorption isotherms and evaluated on a Micromeritics ASAP 2020 surface area and pore size analyzer at 77 K under liquid nitrogen. The non-linear density functional theory (NL-DFT) model determined the pore size distribution. Before testing, the samples were degassed for 12 hours under high vacuum at 120 °C. X-ray photoelectron spectroscopy (XPS) was performed using a system from Thermofisher Scientific, Waltham, MA, USA, to determine the elemental composition of Fe-MOF. UV-vis spectra were collected using a BioTek Synergy HT microplate reader in a transparent 96-vial plate.Computational methods
All density functional theory (DFT) calculations with spin polarization were performed by using the DMol3 package. The exchange–correlation functional in the Kohn–Sham equations used the generalized gradient approximation (GGA) in the form of the Perdew–Burke–Ernzerhof (PBE). The DFT semi-core pseudopotential (DSPP) and double-numerical plus polarization (DNP) function were applied to describe the core-electrons and basis set treatment. The interaction energy was calculated by the equation of Eint = EA/MOF − (EA + EMOF), where EA and EMOF are the calculated total energies of the isolated TMA molecule and MOF. EA/MOF is the total energy of the TMB molecule in the MOF; wherein a more negative value denotes a stronger interaction between the TMB molecule and MOF.Results and discussion
Structural characterization of MIL-88B(Fe) (1,4-NDC) MOFs
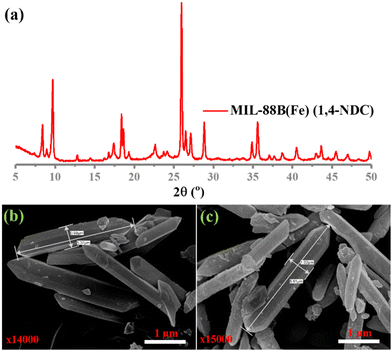
In this study, MIL-88B(Fe) (1,4-NDC), MIL-100(Fe), MIL-88B(Fe) (1,4-BDC), and Fe-1,4-NDC MOF were synthesized using procedures reported in the literature.38–40 However, the MIL-88B(Fe) (1,4-NDC) MOF was synthesized using a solvothermal method with a specific combination of dimethyl formamide (DMF), methanol, and water. DMF played a crucial role in controlling the rate of deprotonation and nucleation during the synthesis of MIL-88B(Fe) (1,4-NDC) MOF, and an appropriate amount was added to the reaction mixture. The aqueous solution's high dispersity and stability are crucial for the reliable application of MOF-based peroxidase mimics. The Fe-based MIL-88B(Fe) (1,4-NDC) MOF is highly compatible with aqueous environments due to its hydrophobic bridging ligands, which make it a suitable candidate for such applications. MOFs such as MIL-100(Fe) and MIL-88B(Fe) (1,4-BDC) MOFs possess peroxidase-like activity due to their porous structure exposing many Fe(III) and Fe(II) activity centers on the surface.29,41 At the solid or liquid interface, these activity centers accelerate heterogeneous Fenton-like reactions.42 In aqueous acidic solutions, the water or hydroxide molecules bridging the SBUs can terminate and become unsaturated open iron sites. Fe(III) can be reduced to Fe(II) by substituting non-bridging ligands with the Lewis base H2O2, which can then react with H2O2 to generate hydroxyl radicals.23 Solid MOF structures with Fe-based reaction sites can limit the rapid diffusion of iron ions and radicals in the surrounding environment during Fe(III)/Fe(II) conversion cycles, improving the catalytic activity.42Characterization techniques, including SEM imaging, PXRD analysis, and nitrogen adsorption studies, were used to study the resulting MOF. The PXRD patterns of both the synthesized and incubated in an acidic buffer of MIL-88B(Fe) (1,4-NDC) MOFs are presented in Fig. 1(a) and Fig. S1–S3 (ESI†), indicating that the synthesized MOF matched the MOF incubated in the acidic buffer condition MIL-88B-Fe structure. Our results showed that the synthesized and incubated in the acidic buffer of MIL-88B(Fe) (1,4-NDC) MOF particles had a rod-like morphology with an average length of 1 μm and width of 5.37 μm and were highly crystalline (Fig. 1(b) and (c)). These results indicate that the MOFs are very stable under acidic conditions. The MIL-100(Fe), MIL-88B(Fe) (1,4-BDC), MIL-88B(Fe) (1,4-NDC), and Fe-1,4-NDC MOF structure, particle morphology, PRXD, N2 sorption isotherm, and pore size distribution are shown in ESI,† Fig. S4–S14.
The N2 adsorption–desorption isotherm (Fig. S13, ESI†) revealed that the MIL-88B(Fe) (1,4-NDC) MOF has a surface area of less than 22 m2 g−1, indicating a non-porous structure. In comparison, the BET-specific surface areas of MIL-88B(Fe) (1,4-BDC) MOF, MIL-100(Fe), and Fe-1,4-NDC MOF were found to be 66, 1693, and 4 m2 g−1. Additionally, the surface area and the pore size distribution shown in Fig. S11–S14 (ESI†) indicated that the MOFs had arranged the order of porous to non-porous structure, MIL-100(Fe) > MIL-88B(Fe) (1,4-BDC) > Fe-MIL-88B (1,4-NDC) ≥ Fe-1,4-NDC, which could be advantageous for catalytic reactions. The MIL-88B(Fe) (1,4-NDC) activated MOF shows non-porous properties in N2 sorption and PXRD shows the closed form of the MIL-88B(Fe) structure. The dried form of MIL-88B(Fe) (1,4-NDC) has pores that are likely comparable to or smaller than the diameters of nitrogen gas (3.8 Å), implying that the gas may be unable to permeate the cavity. PXRD characterization was conducted for the as-synthesized MOF, after immersion in acidic buffer solution and the MOF was dried using acidic buffer solution (Fig. S15, ESI†). The as-synthesized and dried MOF shows a closed form and in solution shows an open form of MIL-88B(Fe) (1,4-NDC). These findings provide a comprehensive understanding of the structure and characteristics of Fe MIL-88B and highlight its potential applications in catalysis. Therefore, comparing different Fe-MOFs with their unique secondary building units can determine their peroxidase-like activity.
To determine the elemental composition and surface chemical state of MIL-88B(Fe) (1,4-NDC) MOF, XPS analysis was conducted. The XPS survey spectrum in Fig. 2(a) confirmed the presence of Fe, C, and O elements in the MIL-88B(Fe) (1,4-NDC) MOF. The C 1s XPS spectra (Fig. 2(b)) were deconvoluted into three peaks at 284.7, 286.3, and 288.7 eV, representing the C–C, C–O–C, and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds, respectively. Similarly, the O 1s XPS spectrum (Fig. 2(c)) was fitted into three peaks at 530.1, 531.9, and 533.7 eV, which were assigned to Fe–O–Fe, C–O–Fe, and HO– bonds, respectively. After curve fitting, the peaks of Fe 2p1/2 and Fe 2p3/2 for MIL-88B(Fe) (1,4-NDC) MOF (Fig. 2(d)) were observed at ca. 710.8, 713.5, 723.7, and 726.2 eV, which are assignable to Fe(III),43,44 and the additional peak at 709.4 eV was observed, which is assigned to Fe(II).45 The XPS quantitative analysis was performed to determine Fe(II) and Fe(III) atomic percentage (%), Fe 2p3/2 (Fe2+: 10.03, Fe3+: 29.3, sat: 26.27) and Fe 2p1/2 (Fe2+: 10.2, Fe3+: 11.96, sat: 12.23) demonstrate that the Fe in MIL-88B(Fe) (1,4-NDC) MOF is predominately Fe(III). These results provide valuable information on the surface chemistry of the MIL-88B(Fe) (1,4-NDC) MOF and confirm its composition for further potential applications.
O bonds, respectively. Similarly, the O 1s XPS spectrum (Fig. 2(c)) was fitted into three peaks at 530.1, 531.9, and 533.7 eV, which were assigned to Fe–O–Fe, C–O–Fe, and HO– bonds, respectively. After curve fitting, the peaks of Fe 2p1/2 and Fe 2p3/2 for MIL-88B(Fe) (1,4-NDC) MOF (Fig. 2(d)) were observed at ca. 710.8, 713.5, 723.7, and 726.2 eV, which are assignable to Fe(III),43,44 and the additional peak at 709.4 eV was observed, which is assigned to Fe(II).45 The XPS quantitative analysis was performed to determine Fe(II) and Fe(III) atomic percentage (%), Fe 2p3/2 (Fe2+: 10.03, Fe3+: 29.3, sat: 26.27) and Fe 2p1/2 (Fe2+: 10.2, Fe3+: 11.96, sat: 12.23) demonstrate that the Fe in MIL-88B(Fe) (1,4-NDC) MOF is predominately Fe(III). These results provide valuable information on the surface chemistry of the MIL-88B(Fe) (1,4-NDC) MOF and confirm its composition for further potential applications.
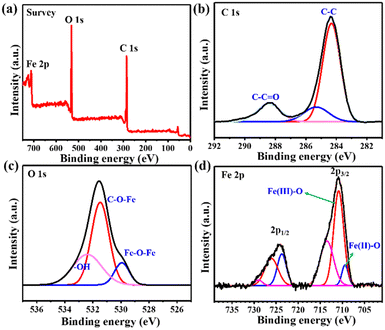 | ||
| Fig. 2 (a) The XPS spectra of MIL-88B(Fe) (1,4-NDC) MOF. (b)–(d) XPS spectrum of C 1s, O 1s, and Fe 2p. | ||
Peroxidase-like catalytic activity of MIL-88B(Fe) (1,4-NDC) MOFs
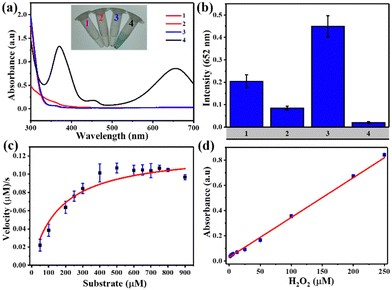
TMB was used as a mimic peroxidase substrate to assess the catalytic efficacy of Fe-MOFs. Fig. 3(a) demonstrates that the MIL-88B(Fe) (1,4-NDC) MOF added to the H2O2/TMB system led to significant absorption at 652 nm, while TMB solution with H2O2 or MIL-88B(Fe) (1,4-NDC) MOF did not produce noticeable absorption at 652 nm. We evaluated the peroxidase activity of four different iron-based metal–organic frameworks (MOFs) – MIL-88B(Fe) (1,4-NDC), MIL-100(Fe), MIL-88B(Fe) (1,4-BDC), and Fe-1,4-NDC – as peroxidase-like nanozymes. Our results show that MIL-88B(Fe) (1,4-NDC) exhibited better peroxidase activity among the four MOFs, with a reaction time of only 5 minutes (Fig. 3(b)). Therefore, we conclude that MIL-88B(Fe) (1,4-NDC) has significant potential as a peroxidase-like nanozyme.The catalytic activity of MIL-88B(Fe) (1,4-NDC) MOF on TMB and H2O2 substrates was assessed, considering the impact of temperature and pH. The optimal conditions of the MIL-88B(Fe) (1,4-NDC) MOF were 28 °C, pH 3.6, and a reaction time of 5 minutes (Fig. S16–S18, ESI†). Moreover, the MOF demonstrated excellent catalytic activity with over 90% relative activity achieved in less than 5 minutes. However, for consistency in data evaluation, the total reaction time of 5 minutes was deemed appropriate. As a result, subsequent studies were conducted under these conditions, at room temperature (28 °C) and pH 3.6.
Michaelis–Menten kinetic curves of MIL-88B(Fe) (1,4-NDC) with a constant TMB concentration and a varying H2O2 concentration are presented in Fig. 3(c). Using two apparent kinetic parameters, where Km is 160 μM and Vmax is 12.5 × 10−8 M s−1 for MIL-88B(Fe) (1,4-NDC) MOF, the values reveal that this MOF had a lower Km value for H2O2 indicating stronger bond between the nanozyme and the substrate are better than other nanozymes, and HRP enzyme.24,46–48 Using the MIL-88B(Fe) (1,4-NDC) MOF nanozyme instead of HRP may result in better H2O2 and glucose detection sensitivity. Meanwhile, as the H2O2 concentration increased, the absorbance at 652 nm increased linearly, and the LOD value was determined to be 2.07 μM (3σ/S) (R2 = 0.996) (Fig. 3(d)).
We also investigated the zeta potential of the Fe-MOFs incubated in an acidic buffer with a pH of 3.6. Our findings showed that MIL-100(Fe) had a negative surface charge with a zeta potential of −2.4 mV, while MIL-88B(Fe) (1,4-BDC) and MIL-88B(Fe) (1,4-NDC) MOF had positive surface charges with zeta potentials of 22.25 mV and 8.65 mV, respectively (Fig. S19, ESI†). In addition, we observed that MIL-100(Fe) had higher catalytic activity compared to MIL-88B(Fe) (1,4-BDC), possibly because the negatively charged MIL-100(Fe) MOF can easily interact with cationic TMB.49 However, when comparing the catalytic activity of MIL-100(Fe) and MIL-88B(Fe) (1,4-NDC) MOF, we expected MIL-100(Fe) to have higher activity because of its negative surface charge. Surprisingly, our results showed that MIL-88B(Fe) (1,4-NDC) had higher activity. We hypothesize that the strong electrostatic interaction and π–π interaction between the 1,4-naphthalene dicarboxylic acid ligand in MIL-88B(Fe) (1,4-NDC) and the TMB molecule contributed to its enhanced catalytic activity. The DFT calculations (Fig. S20, ESI†) were carried out and revealed that the interaction (−73.59 kJ mol−1) between MIL-88B(Fe) (1,4-NDC) and TMB molecule is stronger than that (−64.24 kJ mol−1) in MIL-100(Fe) and TMB molecule. Overall, our findings demonstrate the potential of MIL-88B(Fe) (1,4-NDC) as a highly efficient peroxidase-like nanozyme, which could have significant implications for various biosensor applications. To illustrate this non-porous Fe-MOF peroxidase-like catalytic activity, we further conducted the sequential reaction method to evaluate its applicability.
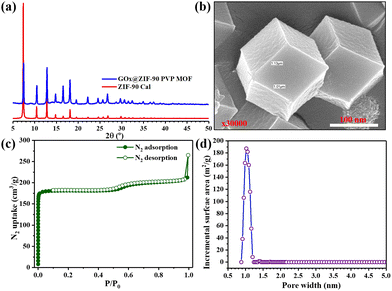
GOx@ZIF-90 PVP MOF characterization
The imidazole-2-carboxaldehyde solution was heated to 65 °C with the addition of PVP for five minutes to dissolve the starting materials quickly. After cooling to room temperature, GOx and metal solutions were added, and the MOF nanoparticles were effectively stabilized. This stabilization technique enabled the precise localization of GOx within the ZIF-90-PVP crystal. The immobilization process was conducted gently and was eco-friendly through co-precipitation in an aqueous solution, preserving the enzymatic activity of GOx even after encapsulation within the ZIF-90-PVP structure. Notably, the XRD patterns of the nanocomposites GOx@ZIF-90 PVP were almost indistinguishable from that of the ZIF-90 calculated pattern, suggesting that the incorporation of GOx had minimal impact on the phase of the ZIF-90 hosts Fig. 4(a). The FE-SEM was used to analyze the morphology of the GOx@ZIF-90 PVP-based nanocomposites, as depicted in Fig. 4(b), revealing a uniform size morphology. To assess its porosity, we measured the nitrogen adsorption–desorption isotherms of GOx@ZIF-90 PVP at 77 K, as depicted in Fig. 4(c). At low relative pressure, the adsorption of GOx@ZIF-90 PVP reached saturation. Our calculations indicate that the BET surface area of GOx@ZIF-90 PVP is 762 m2 g−1, and the pore size distribution curve (Fig. 4(d)) with a maximum of 11 Å matches more precisely with prior literature.37GOx@ZIF-90 PVP MOF and MIL-88B(Fe) (1,4-NDC) MOF catalytic activity
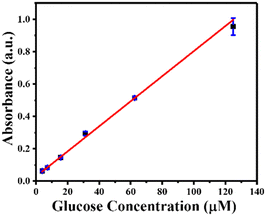
Using GOx@ZIF-90-PVP as a reusable catalyst, MIL-88B(Fe) (1,4-NDC) MOF as a peroxidase nanozyme, and TMB as a chromogenic substrate, glucose was detected with high catalytic efficiency. In just 10 minutes, GOx@ZIF-90-PVP converted glucose to gluconic acid. In addition, it generated H2O2 in the reaction solution, which was then used as a substrate for MIL-88B(Fe) (1,4-NDC) MOF to oxidize TMB, producing TMBox that could be detected at 652 nm. The ZIF-90 PVP MOF failed to convert glucose into gluconic acid without GOx (Fig. S21, ESI†). The absorbance at 652 nm was measured following a total reaction time of 15 minute at room temperature, (i) first step: GO@ZIF-90-PVP composite containing glucose (2.5–125 μM) 10 minutes reaction, (ii) second step: MIL-88B(Fe) (1,4-NDC) MOF and TMB (1 mM), exhibiting excellent linearity between the absorbance and glucose concentration within the range of 2.5–125 μM (R2 = 0.998), with a low detection limit of 2.23 μM (Fig. 5).Moreover, to ensure reproducibility, the peroxidase-like catalytic activity of MIL-88B(Fe) (1,4-NDC) MOF was detected using a sequential reaction method for glucose detection across multiple batches (Fig. S22, ESI†). It's worth noting that in the first step, GOx@ZIF-90-PVP MOF was centrifuged, washed, and then reintroduced into the reaction system. However, due to the small sample size in the reaction, the PXRD analysis revealed partial crystallinity of MIL-88B(Fe) (1,4-NDC) MOF after the sixth cycle (Fig. S23, ESI†). As seen in the XRD data, the reaction involving TMB oxidation and subsequent entrapment or attachment of some of the oxidized TMB molecules into the pores or surface of the MIL-88B(Fe) MOF leads to a decrease in crystallinity. This decrease in crystallinity may affect the flexibility of the MIL-88B(Fe) MOF, which has previously been reported to demonstrate a breathing effect in the literature. The presence of trapped or surface-attached oxidized TMB molecules alters the long-range order of crystallinity, causing the MOF's crystallinity to diminish across several cycles.
Conclusions
In conclusion, our study highlights the potential of different Fe-based metal–organic frameworks (MOFs) as efficient peroxidase-mimetic nanozymes. We observed that MIL-88B(Fe) (1,4-NDC) demonstrated the highest catalytic activity among the four MOFs tested. Its hydrophobic bridging ligands make it highly compatible with aqueous environments, and its unique secondary building units and surface charges could influence its peroxidase-mimicking activity. Specifically, the MIL-88B(Fe) (1,4-NDC) MOF exhibited higher activity than expected due to strong electrostatic and π–π interactions. Additionally, our findings suggest that the MOF encapsulation method for enzyme immobilization offers reusability and stability advantages. Moreover, these findings have significant implications for various biotechnology and environmental monitoring applications and highlight the potential of nanozymes as a promising alternative to natural enzymes.Author contributions
Sivasankar Kulandaivel: investigation, methodology conceptualization, data treatment, and writing–original draft; Hsin-Tsung Chen: investigation, and writing–reviewing and editing; Chia-Her Lin: conceptualization, supervision, and writing–reviewing and editing; Yi-Chun Yeh: conceptualization, supervision, project organization, funding acquisition, and writing–reviewing and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The Ministry of Science and Technology of Taiwan supported this research through projects 110-2113-M-003-009- and 110-2113-M-003-011-MY3. Dr Kulandaivel Sivasankar thanks the Taiwan Ministry of Science and Technology for the postdoctoral fellowship 111-2811-M-003-512-. Furthermore, the Taiwan Ministry of Education (MOE) gratefully acknowledges financial support from the National Taiwan Normal University (NTNU) within the Higher Education Sprout Project framework. The authors would like to express their gratitude to Ms. Natalie Kiatsupaibul for her valuable experimental contributions.References
- E. L. Bell, W. Finnigan, S. P. France, A. P. Green, M. A. Hayes, L. J. Hepworth, S. L. Lovelock, H. Niikura, S. Osuna, E. Romero, K. S. Ryan, N. J. Turner and S. L. Flitsch, Nat. Rev. Methods Primers, 2021, 1, 46 CrossRef CAS.
- J. L. Porter, R. A. Rusli and D. L. Ollis, ChemBioChem, 2016, 17, 197–203 CrossRef CAS PubMed.
- J. P. Adams, M. J. B. Brown, A. Diaz-Rodriguez, R. C. Lloyd and G. D. Roiban, Adv. Synth. Catal., 2019, 361, 2421–2432 CAS.
- C.-W. Chang, Y.-H. Lin, C.-H. Tsai, S. Kulandaivel and Y.-C. Yeh, Anal. Chim. Acta, 2022, 1202, 339641 CrossRef CAS.
- Y. Lin, J. Ren and X. Qu, Acc. Chem. Res., 2014, 47, 1097–1105 CrossRef CAS PubMed.
- Y. Ai, Z. N. Hu, X. Liang, H. B. Sun, H. Xin and Q. Liang, Adv. Funct. Mater., 2022, 32, 2110432 CrossRef CAS.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- J. Wu, X. Wang, Q. Wang, Z. Lou, S. Li, Y. Zhu, L. Qin and H. Wei, Chem. Soc. Rev., 2019, 48, 1004–1076 RSC.
- S.-H. Lo, C.-H. Chien, Y.-L. Lai, C.-C. Yang, J. J. Lee, D. S. Raja and C.-H. Lin, J. Mater. Chem. A, 2013, 1, 324–329 RSC.
- S.-H. Lo, L. Feng, K. Tan, Z. Huang, S. Yuan, K.-Y. Wang, B.-H. Li, W.-L. Liu, G. S. Day, S. Tao, C.-C. Yang, T.-T. Luo, C.-H. Lin, S.-L. Wang, S. J. L. Billinge, K.-L. Lu, Y. J. Chabal, X. Zou and H.-C. Zhou, Nat. Chem., 2020, 12, 90–97 CrossRef PubMed.
- M. Ding, X. Cai and H.-L. Jiang, Chem. Sci., 2019, 10, 10209–10230 RSC.
- D. Liu, D. Zou, H. Zhu and J. Zhang, Small, 2018, 14, 1801454 CrossRef PubMed.
- P. Kumar, A. Deep and K.-H. Kim, TrAC, Trends Anal. Chem., 2015, 73, 39–53 CrossRef CAS.
- H. Li, L. Li, R.-B. Lin, W. Zhou, Z. Zhang, S. Xiang and B. Chen, EnergyChem, 2019, 1, 100006 CrossRef.
- Y. Zhao, H. Zeng, X.-W. Zhu, W. Lu and D. Li, Chem. Soc. Rev., 2021, 50, 4484–4513 RSC.
- X. Huang, S. Zhang, Y. Tang, X. Zhang, Y. Bai and H. Pang, Coord. Chem. Rev., 2021, 449, 214216 CrossRef CAS.
- F. Wang, L. Chen, D. Liu, W. Ma, P. Dramou and H. He, TrAC, Trends Anal. Chem., 2020, 133, 116080 CrossRef CAS.
- L. Mei, S. Zhu, Y. Liu, W. Yin, Z. Gu and Y. Zhao, Chem. Eng. J., 2021, 418, 129431 CrossRef CAS.
- X. Ren, D. Chen, Y. Wang, H. Li, Y. Zhang, H. Chen, X. Li and M. Huo, J. Nanobiotechnol., 2022, 20, 92 CrossRef CAS PubMed.
- S. Kulandaivel, C. H. Lin and Y. C. Yeh, Chem. Commun., 2022, 58, 569–572 RSC.
- L. Zheng, F. Wang, C. Jiang, S. Ye, J. Tong, P. Dramou and H. He, Coord. Chem. Rev., 2022, 471, 214760 CrossRef CAS.
- Y. Zhu, R. Zhu, Y. Xi, J. Zhu, G. Zhu and H. He, Appl. Catal., B, 2019, 255, 117739 CrossRef CAS.
- D. Wu, J. Jiang, N. Tian, M. Wang, J. Huang, D. Yu, M. Wu, H. Ni and P. Ye, RSC Adv., 2021, 11, 32383–32393 RSC.
- W. Xu, L. Jiao, H. Yan, Y. Wu, L. Chen, W. Gu, D. Du, Y. Lin and C. Zhu, ACS Appl. Mater. Interfaces, 2019, 11, 22096–22101 CrossRef CAS PubMed.
- J. Hassanzadeh, H. A. J. Al Lawati and N. Bagheri, Biosens. Bioelectron., 2022, 207, 114184 CrossRef CAS.
- A. Yuan, Y. Lu, X. Zhang, Q. Chen and Y. Huang, J. Mater. Chem. B, 2020, 8, 9295–9303 RSC.
- L. Ai, L. Li, C. Zhang, J. Fu and J. Jiang, Chem. – Eur. J., 2013, 19, 15105–15108 CrossRef CAS PubMed.
- Y. L. Liu, X. J. Zhao, X. X. Yang and Y. F. Li, Analyst, 2013, 138, 4526–4531 RSC.
- J.-W. Zhang, H.-T. Zhang, Z.-Y. Du, X. Wang, S.-H. Yu and H.-L. Jiang, Chem. Commun., 2014, 50, 1092–1094 RSC.
- J. Xu, J. Peng, X. Wang and X. Hou, ACS Sustainable Chem. Eng., 2022, 10, 9315–9324 CrossRef CAS.
- X. Liang, Y. Chen, K. Wen, H. Han and Q. Li, J. Mater. Chem. B, 2021, 9, 6811–6817 RSC.
- X. Lian, Y. Fang, E. Joseph, Q. Wang, J. Li, S. Banerjee, C. Lollar, X. Wang and H.-C. Zhou, Chem. Soc. Rev., 2017, 46, 3386–3401 RSC.
- J. Mehta, N. Bhardwaj, S. K. Bhardwaj, K.-H. Kim and A. Deep, Coord. Chem. Rev., 2016, 322, 30–40 CrossRef CAS.
- S. S. Nadar, L. Vaidya and V. K. Rathod, Int. J. Biol. Macromol., 2020, 149, 861–876 CrossRef CAS PubMed.
- H. Xia, N. Li, X. Zhong and Y. Jiang, Front. Bioeng. Biotechnol., 2020, 8, 695 CrossRef PubMed.
- X. Wang, P. C. Lan and S. Ma, ACS Cent. Sci., 2020, 6, 1497–1506 CrossRef CAS.
- F.-K. Shieh, S.-C. Wang, C.-I. Yen, C.-C. Wu, S. Dutta, L.-Y. Chou, J. V. Morabito, P. Hu, M.-H. Hsu and K. C. W. Wu, J. Am. Chem. Soc., 2015, 137, 4276–4279 CrossRef CAS.
- P. B. So, H.-T. Chen and C.-H. Lin, Microporous Mesoporous Mater., 2020, 309, 110495 CrossRef CAS.
- P. Horcajada, F. Salles, S. Wuttke, T. Devic, D. Heurtaux, G. Maurin, A. Vimont, M. Daturi, O. David, E. Magnier, N. Stock, Y. Filinchuk, D. Popov, C. Riekel, G. Férey and C. Serre, J. Am. Chem. Soc., 2011, 133, 17839–17847 CrossRef CAS PubMed.
- J.-J. Chen, Y.-T. Chen, D. S. Raja, Y.-H. Kang, P.-C. Tseng and C.-H. Lin, Materials, 2015, 8, 5336–5347 CrossRef CAS.
- M. Fu, B. Chai, J. Yan, C. Wang, G. Fan, G. Song and F. Xu, Appl. Phys. A: Mater. Sci. Process., 2021, 127, 928 CrossRef CAS.
- Q. Jiang, Y. Xiao, A. N. Hong, Z. Gao, Y. Shen, Q. Fan, P. Feng and W. Zhong, ACS Appl. Mater. Interfaces, 2022, 14, 41800–41808 CrossRef CAS.
- X. Zheng, L. Zhang, Z. Fan, Y. Cao, L. Shen, C. Au and L. Jiang, Chem. Eng. J., 2019, 374, 793–801 CrossRef CAS.
- H. Lv, H. Zhao, T. Cao, L. Qian, Y. Wang and G. Zhao, J. Mol. Catal. A: Chem., 2015, 400, 81–89 CrossRef CAS.
- A. P. Grosvenor, B. A. Kobe, M. C. Biesinger and N. S. McIntyre, Surf. Interface Anal., 2004, 36, 1564–1574 CrossRef CAS.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- Q. Wang, L. Zhang, C. Shang, Z. Zhang and S. Dong, Chem. Commun., 2016, 52, 5410–5413 RSC.
- L. Su, W. Qin, H. Zhang, Z. U. Rahman, C. Ren, S. Ma and X. Chen, Biosens. Bioelectron., 2015, 63, 384–391 CrossRef CAS PubMed.
- S.-H. Huo and X.-P. Yan, J. Mater. Chem., 2012, 22, 7449–7455 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb00981e |
| This journal is © The Royal Society of Chemistry 2023 |