Open Access Article
Open Access ArticleBiosynthesis of multifunctional transformable peptides for downregulation of PD-L1†
Yufei
Di
ab,
Zhiwen
Yang
 ab,
Gang
Song
ab,
Gang
Song
 ab,
Qi
Shen
ab,
Qi
Shen
 a,
Haotian
Bai
a,
Haotian
Bai
 a,
Yiming
Huang
a,
Yiming
Huang
 a,
Fengting
Lv
a,
Fengting
Lv
 *a and
Shu
Wang
*a and
Shu
Wang
 *ab
*ab
aBeijing National Laboratory for Molecular Sciences, Key Laboratory of Organic Solids, Institute of Chemistry, Chinese Academy of Sciences, Beijing 100190, China. E-mail: wangshu@iccas.ac.cn; lvft@iccas.ac.cn
bCollege of Chemistry, University of Chinese Academy of Sciences, Beijing 100049, China
First published on 3rd September 2024
Abstract
Here, we present a biosynthesized material M1 for immune checkpoint blocking therapy. M1 could realize a morphological transformation from globular to fibrous in situ in the presence of cathepsin B (CtsB) after entering tumor cells. The GO203 peptides of M1 are exposed, which could bind to mucin 1 (MUC1) to suppress the homodimerization process of MUC1, thereby downregulating PD-L1 expression.
In recent years, significant advancements have been made in the field of cancer immunotherapy, which has made it a prominent treatment modality alongside surgery, chemotherapy, and radiotherapy.1,2 In contrast to traditional cancer treatments, immunotherapy does not directly target tumor cells, but instead activates the body's immune system to recognize and fight tumor cells.3,4 Chimeric antigen receptor T-cell immunotherapy (CAR-T)5–7 and immune checkpoint blocking (ICB)8–11 have achieved outstanding clinical efficacy, providing a new treatment modality for many types of cancer by reactivating T cells to exert their anti-tumor efficacy. The most common ICB therapies block the programmed cell death 1 (PD1)/PD1 ligand 1 (PD-L1) pathway12–16 using antibodies, peptides, small molecule drugs, and DNA.17,18 Nevertheless, the clinical response of the ICB inhibitors is only 10–30%,19 accompanying severe side effects, owing to non-targeting and unresponsiveness of the inhibitors. Therefore, it is of great significance to explore and develop cancer immunotherapy with high efficiency and low side effects.20,21
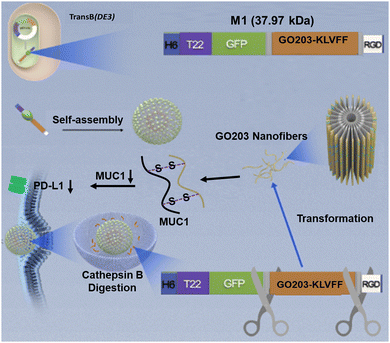
Researchers are dedicated to the study of downregulating the expression of PD-L1 in cancer cells, strengthening the effect of antitumor immunity.22 Peptides, small interfering RNAs (siRNA), and small molecule inhibitors can down-regulate PD-L1 expression levels to block the activation of the PD1/PD-L1 pathway,23–25 which could reactivate T cells to exert their anti-tumor efficacy, inhibiting tumor proliferation. For instance, GO203 peptide could target mucin 1 (MUC1) in multiple tumor models, suppressing PD-L1 expression levels.26,27 However, the current strategy was only able to maintain a single sequence, which was non-responsive and could not work specifically in the tumor area.28,29 Very recently, we reported a biosynthetic peptide-based material (P1) that induced tumor cell apoptosis in vivo by decaging the functional peptide moieties with the morphology transformation of this material.30 Here, we introduced a functional peptide (GO203) to construct peptide-based material M1 by replacing the BAK moiety of P1, inspired by the morphology transformation of nanoparticles in situ.31–34 This enzyme catalytic transformable recombinant protein M1 (H6-T22-GFP-GO203-KLVFF-RGD) integrates multiple functions, consisting of a targeting motif (T22 and RGD),35,36 imaging segment (GFP), tailoring motif (GFLG), functional region (GO203), β-sheet peptide KLVFF and delivery segment (RGD). M1 possesses the ability of morphological transformation from globular to fibrous in situ for downregulating the expression of PD-L1 (Fig. 1). Meanwhile, the control peptide M2 without enzyme-cleavable site GFLG was biosynthesized with the identical method. Peptide M3, the enzyme digestion product of M1, was prepared by standard solid-phase peptide synthesis. Due to the amphiphilic characteristics, M1 was able to self-assemble into nanoparticles. The presence of the CtsB enzyme cleavage site and intermolecular hydrogen bonding allow the nanoparticle structure to transform into nanofibers within the cell, thus releasing GO203 peptide which could bind to MUC1 with an active CQC site to inhibit the homodimerization process of MUC1.37,38 M1 could down-regulate PD-L1 expression in tumor cells thereby activating anti-tumor immunity. Compared to other strategies, the biosynthesized peptide-based material has not only good biocompatibility, but also tumor-targeted morphology transformation capability to downregulate PD-L1 expression.
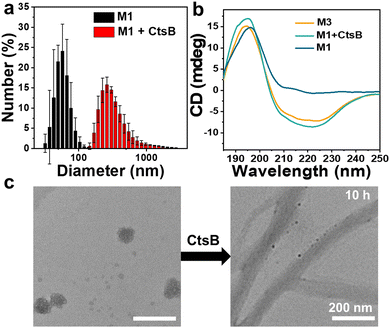
The recombinant protein His-T22-GFP-GO203-KLVFF-RGD was successfully produced in Escherichia coli Trans B (DE3), purified by Ni-NTA affinity chromatography to purify the 6× His-tagged protein products and detected as a single protein species with the expected molecular mass of 37.97 kDa (Fig. S2a, ESI†). The purified peptide M1 exhibited the characteristic fluorescence of GFP with an emission maximum at 520 nm, as depicted in Fig. S1 (ESI†). The successful expression of M1 by bacteria was verified. Meanwhile, the SDS-PAGE assessment of the control peptide M2 was performed and shown in Fig. S2b (ESI†). The molecular transformation of M1 incubated with CtsB was investigated with high-performance liquid chromatography mass spectrometry (LC-MS). A new peak corresponding to M3 in LC-MS as illustrated in Fig. S3 (ESI†) verified that M1 was cleaved by CtsB at the designed digestion site GFLG. Then the enzyme catalytic transformation behavior of the M1 assemblies was examined by transmission electron microscopy (TEM) and circular dichroism (CD). TEM of the purified extracts revealed that the spherical form of M1 was about 60 nm in diameter (Fig. 2c), which was in accordance with the dynamic light scattering (DLS) result of 61.3 ± 9.2 nm (Fig. 2a). There was no spherical to fibrous morphology change of M1 observed after treatment with CtsB for 5 h (Fig. S4, ESI†), while obvious nanofibers were observed with the incubation time up to 10 h, which proved that the morphology of M1 could transform from nanoparticles to nanofibers. On the contrary, the control peptide-based material M2 without CtsB cleavable sites kept the spherical morphology even after the treatment of CtsB for 10 h, which is shown in Fig. S5 (ESI†). The morphology change of M1 was further confirmed by dynamic light scattering measurement, as presented in Fig. 2a, and an obvious positive shift of the particle size of M1 was observed upon the addition of CtsB for 10 h. To investigate the chemical driving force of nanofiber formation, the secondary structure was explored by circular dichroism. After incubation with CtsB for 10 h, the M1 assemblies showed a positive CD signal at 195 nm and negative signals at 220 nm, which confirm the typical β-sheet fibrous structure.39 In contrast, M1 showed a positive band around 195 nm in the absence of CtsB and exhibited a predominant random structure (Fig. 2b). Furthermore, to demonstrate that GO230 could form a complex with MUC1 in cells, MUC1 and GO230 were customized and cocultured. The sulfur crosslinking product of them could be tested by HRMS (Fig. S6, ESI†), proving the successful formation of the GO203-MUC1 fragment complex, which effectively prevents the dimerization of MUC1.
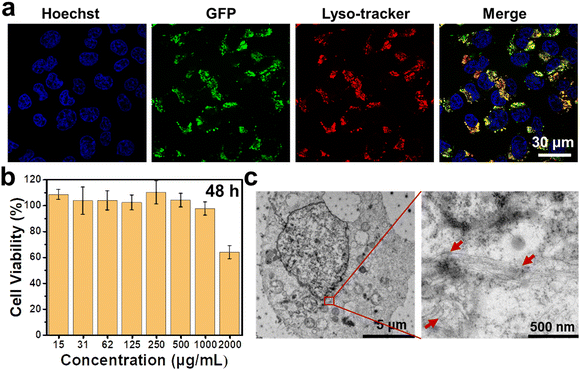
Before investigating the morphological transformation of M1 in living cells, internalization behaviour and biosafety of this peptide were firstly evaluated. For A375 cells treated with 1 mg mL−1 M1, their fluorescence enhanced with increasing the incubation time and reached a plateau at 12 h, then maintained the intensity until 24 h, as shown in Fig. S7 (ESI†). It was suggested that efficient cellular uptake of M1 by the cells occurred, which may be ascribed to its superior hydrophilicity. To evaluate the biosafety of M1, A375 cells were incubated with different concentrations of M1. As illustrated in Fig. 3b, the cells showed high survival rates at concentrations up to 1 mg mL−1, which verified its superior biocompatibility. Given that CtsB is mainly distributed in lysosomes,40 efficient CtsB-catalytic transformation requires lysosomal cellular endocytosis.
To probe the location of M1 in cells, fluorescent co-localization of M1 and lysotracker was performed. As expected, M1 with intensive green fluorescence from GFP shared obvious overlapped fluorescence with lysotracker, which was labelled with red fluorescence (Fig. 3a). These results demonstrated the efficient cellular endocytosis ability and lysosomal location of M1, which satisfy the requirement for CtsB-catalyzed transformation.
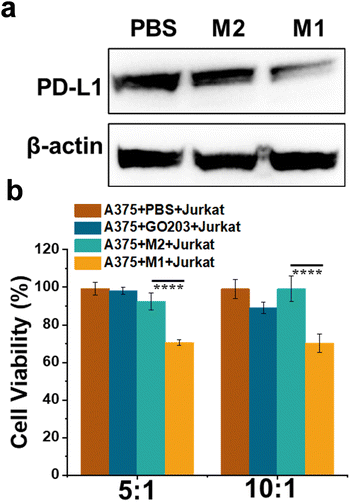
Bio-TEM was employed to study the intracellular morphology transformation of M1. After M1 entered the A375 cells, fibrous structures were observed by TEM, as presented in Fig. 3c, while there was no similar structure found for M2-treated cells or PBS-treated cells (Fig. S8, ESI†). The introduction of the enzyme cleavable peptide GFLG was critical for intracellular transformation of M1 to fibrous structures, which was consistent with the phenomenon in solution. As shown in Fig. 4a, both M1 and M2 could reduce the PD-L1 expression level of A375 cells, and M1 displayed a greater degree than M2, indicating that more GO203 moieties were exposed in the presence of CtsB enzyme to bind MUC1, therefore, leading to better suppression of the PD-L1 expression levels (Fig. S9, ESI†). The cellular immunity could recognize and lyse tumor cells, and the PD-1/PD-L1 interaction has been reported to affect T cell functions.41 We tested our designed peptide M1 for cellular immunity with Jurkat T cells. A375 cells were pretreated with PBS, the single-sequence GO203, M2 and M1 to influence the expression of PD-L1, and subsequently, these A375 cells were cocultured with Jurkat T cells. As illustrated in Fig. 4b, Jurkat T cells killed A375 cells treated with M1 more effectively than those treated with M2 or PBS, which suggested that the designed peptide M1 could downregulate the expression of PD-L1, inducing effective cellular immunity. Moreover, the immunotherapeutic effect of self-assembled GO203 is better than that of single-sequence.
In summary, we have established a biosynthesized protein-based material (M1) with intracellular morphology transformation ability that can effectively downregulate the PD-L1 expression of cancer cells. These findings identify that self-assembly peptide-based materials with morphology transformation ability contribute better immunotherapeutic performance than single sequence and nanostructured GO203, which reduces to some extent the off-target and side effects confronted for clinical immunotherapy. It is believed that better therapeutic performance and more advanced specific biochemical applications will be presented with further functionalization and integration with other therapeutic modalities.
We are grateful for financial support from the National Natural Science Foundation of China (No. 22074150, 22307119, U23A20109, and 22020102005) and Beijing Natural Science Foundation (Z220025, 2222042).
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- J. Prieto, I. Melero and B. Sangro, Nat. Rev. Gastroenterol. Hepatol., 2015, 12, 681–700 CrossRef CAS PubMed.
- A. Nazha and M. A. Sekeres, Annu. Rev. Med., 2017, 68, 127–137 CrossRef CAS PubMed.
- Daniel S. Chen and I. Mellman, Immunity, 2013, 39, 1–10 CrossRef CAS PubMed.
- B. Bao, Q. Yuan, Q. Feng, L. Li, M. Q. Li and Y. Tang, CCS Chem., 2024, 6, 693–708 CrossRef CAS.
- R. C. Larson and M. V. Maus, Nat. Rev. Cancer, 2021, 21, 145–161 CrossRef CAS PubMed.
- A. Rodriguez-Garcia, R. C. Lynn, M. Poussin, M. A. Eiva, L. C. Shaw, R. S. O’Connor, N. G. Minutolo, V. Casado-Medrano, G. Lopez, T. Matsuyama and D. J. Powell, Nat. Commun., 2021, 12, 877 CrossRef CAS PubMed.
- L. C. Wang, A. Lo, J. Scholler, J. Sun, R. S. Majumdar, V. Kapoor, M. Antzis, C. E. Cotner, L. A. Johnson, A. C. Durham, C. C. Solomides, C. H. June, E. Puré and S. M. Albelda, Cancer Immunol. Res., 2014, 2, 154–166 CrossRef CAS PubMed.
- P. Sharma and J. P. Allison, Science, 2015, 348, 56–61 CrossRef CAS PubMed.
- Y. Yang, J. Xu, Y. Sun, L. Mo, B. Liu, X. Pan, Z. Liu and W. Tan, J. Am. Chem. Soc., 2021, 143, 8391–8401 CrossRef CAS PubMed.
- C. Wang, J. Wang, X. Zhang, S. Yu, D. Wen, Q. Hu, Y. Ye, H. Bomba, X. Hu, Z. Liu, G. Dotti and Z. Gu, Sci. Transl. Med., 2018, 10, eaan3682 CrossRef PubMed.
- L. Barrueto, F. Caminero, L. Cash, C. Makris, P. Lamichhane and R. R. Deshmukh, Transl. Oncol., 2020, 13, 100738 CrossRef PubMed.
- A. Mantovani, F. Marchesi, A. Malesci, L. Laghi and P. Allavena, Nat. Rev. Clin. Oncol., 2017, 14, 399–416 CrossRef CAS PubMed.
- S. Rafiq, O. O. Yeku, H. J. Jackson, T. J. Purdon, D. G. van Leeuwen, D. J. Drakes, M. Song, M. M. Miele, Z. Li, P. Wang, S. Yan, J. Xiang, X. Ma, V. E. Seshan, R. C. Hendrickson, C. Liu and R. J. Brentjens, Nat. Biotechnol., 2018, 36, 847–856 CrossRef CAS PubMed.
- J. Pan, X. Li, B. Shao, F. Xu, X. Huang, X. Guo and S. Zhou, Adv. Mater., 2022, 34, 2106307 CrossRef CAS PubMed.
- Z. Xiao, Z. Su, S. Han, J. Huang, L. Lin and X. Shuai, Sci. Adv., 2020, 6, eaay7785 CrossRef CAS PubMed.
- Y. Chao and Z. Liu, Nat. Rev. Bioeng., 2023, 1, 125–138 CrossRef.
- A. Ribas and J. D. Wolchok, Science, 2018, 359, 1350–1355 CrossRef CAS PubMed.
- Y. Zhai, J. Wang, T. Lang, Y. Kong, R. Rong, Y. Cai, W. Ran, F. Xiong, C. Zheng, Y. Wang, Y. Yu, H. H. Zhu, P. Zhang and Y. Li, Nat. Nanotechnol., 2021, 16, 1271–1280 CrossRef CAS PubMed.
- J. E. Rosenberg, J. Hoffman-Censits, T. Powles, M. S. van der Heijden, A. V. Balar, A. Necchi, N. Dawson, P. H. O'Donnell, A. Balmanoukian, Y. Loriot, S. Srinivas, M. M. Retz, P. Grivas, R. W. Joseph, M. D. Galsky, M. T. Fleming, D. P. Petrylak, J. L. Perez-Gracia, H. A. Burris, D. Castellano, C. Canil, J. Bellmunt, D. Bajorin, D. Nickles, R. Bourgon, G. M. Frampton, N. Cui, S. Mariathasan, O. Abidoye, G. D. Fine and R. Dreicer, Lancet, 2016, 387, 1909–1920 CrossRef CAS PubMed.
- E. Zhang, Y. Chen, F. Lv, Z. Zuo, F. He, Y. Li, Y. Huang, Y. Li and S. Wang, CCS Chem., 2023, 6, 641–651 CrossRef.
- P. Sharma and J. P. Allison, Cell, 2015, 161, 205–214 CrossRef CAS PubMed.
- H. Li, C.-W. Li, X. Li, Q. Ding, L. Guo, S. Liu, C. Liu, C.-C. Lai, J.-M. Hsu, Q. Dong, W. Xia, J. L. Hsu, H. Yamaguchi, Y. Du, Y.-J. Lai, X. Sun, P. B. Koller, Q. Ye and M.-C. Hung, Gastroenterology, 2019, 156, 1849–1861 CrossRef CAS PubMed.
- S. Yang, M. K. Shim, W. J. Kim, J. Choi, G.-H. Nam, J. Kim, J. Kim, Y. Moon, H. Y. Kim, J. Park, Y. Park, I.-S. Kim, J. H. Ryu and K. Kim, Biomaterials, 2021, 272, 120791 CrossRef CAS PubMed.
- X. Guan, L. Sun, Y. Shen, F. Jin, X. Bo, C. Zhu, X. Han, X. Li, Y. Chen, H. Xu and W. Yue, Nat. Commun., 2022, 13, 2834 CrossRef CAS PubMed.
- M. Mamuti, Y. Wang, Y.-D. Zhao, J.-Q. Wang, J. Wang, Y.-L. Fan, W.-Y. Xiao, D.-Y. Hou, J. Yang, R. Zheng, H.-W. An and H. Wang, Adv. Mater., 2022, 34, 2109432 CrossRef CAS PubMed.
- T. Maeda, M. Hiraki, C. Jin, H. Rajabi, A. Tagde, M. Alam, A. Bouillez, X. Hu, Y. Suzuki, M. Miyo, T. Hata, K. Hinohara and D. Kufe, Cancer Res., 2018, 78, 205–215 CrossRef CAS PubMed.
- A. Bouillez, D. Adeegbe, C. Jin, X. Hu, A. Tagde, M. Alam, H. Rajabi, K.-K. Wong and D. Kufe, OncoImmunology, 2017, 6, e1338998 CrossRef PubMed.
- J. Dai, J.-J. Hu, X. Dong, B. Chen, X. Dong, R. Liu, F. Xia and X. Lou, Angew. Chem., Int. Ed., 2022, 61, e202117798 CrossRef CAS PubMed.
- H. Yao, J. Lan, C. Li, H. Shi, J.-P. Brosseau, H. Wang, H. Lu, C. Fang, Y. Zhang, L. Liang, X. Zhou, C. Wang, Y. Xue, Y. Cui and J. Xu, Nat. Biomed. Eng., 2019, 3, 306–317 CrossRef CAS PubMed.
- Y. Di, Q. Shen, Z. Yang, T. Fang, Y. Liu, Y. Liu, G. Song, Q. Luo, F. Wang, X. Yan, H. Bai, Y. Huang, F. Lv and S. Wang, Small, 2023, 19, 2303035 CrossRef CAS PubMed.
- H. He, S. Liu, D. Wu and B. Xu, Angew. Chem., Int. Ed., 2020, 59, 16445–16450 CrossRef CAS PubMed.
- Y. Di, E. Zhang, Z. Yang, Q. Shen, X. Fu, G. Song, C. Zhu, H. Bai, Y. Huang, F. Lv, L. Liu and S. Wang, Angew. Chem., Int. Ed., 2022, 61, e202116457 CrossRef CAS PubMed.
- X. Liu, W. Zhan, G. Gao, Q. Jiang, X. Zhang, H. Zhang, X. Sun, W. Han, F.-G. Wu and G. Liang, J. Am. Chem. Soc., 2023, 145, 7918–7930 CrossRef CAS PubMed.
- D.-B. Cheng, D. Wang, Y.-J. Gao, L. Wang, Z.-Y. Qiao and H. Wang, J. Am. Chem. Soc., 2019, 141, 4406–4411 CrossRef CAS PubMed.
- U. Unzueta, M. V. Cespedes, N. Ferrer-Miralles, I. Casanova, J. Cedano, J. L. Corchero, J. Domingo-Espin, A. Villaverde, R. Mangues and E. Vazquez, Int. J. Nanomed., 2012, 7, 4533–4544 CAS.
- L. Sanchez-Garcia, N. Serna, P. Alamo, R. Sala, M. V. Cespedes, M. Roldan, A. Sanchez-Chardi, U. Unzueta, I. Casanova, R. Mangues, E. Vazquez and A. Villaverde, J. Controlled Release, 2018, 274, 81–92 CrossRef CAS PubMed.
- D. Raina, M. Kosugi, R. Ahmad, G. Panchamoorthy, H. Rajabi, M. Alam, T. Shimamura, G. I. Shapiro, J. Supko, S. Kharbanda and D. Kufe, Mol. Cancer Ther., 2011, 10, 806–816 CrossRef CAS PubMed.
- M. Hasegawa, R. K. Sinha, M. Kumar, M. Alam, L. Yin, D. Raina, A. Kharbanda, G. Panchamoorthy, D. Gupta, H. Singh, S. Kharbanda and D. Kufe, Clin. Cancer Res., 2015, 21, 2338–2347 CrossRef CAS PubMed.
- P.-P. Yang, Q. Luo, G.-B. Qi, Y.-J. Gao, B.-N. Li, J.-P. Zhang, L. Wang and H. Wang, Adv. Mater., 2017, 29, 1605869 CrossRef PubMed.
- X. Ai, C. J. H. Ho, J. Aw, A. B. E. Attia, J. Mu, Y. Wang, X. Wang, Y. Wang, X. Liu, H. Chen, M. Gao, X. Chen, E. K. L. Yeow, G. Liu, M. Olivo and B. Xing, Nat. Commun., 2016, 7, 10432 CrossRef CAS PubMed.
- J. Bu, A. Nair, M. Iida, W.-J. Jeong, M. J. Poellmann, K. Mudd, L. J. Kubiatowicz, E. W. Liu, D. L. Wheeler and S. Hong, Nano Lett., 2020, 20, 4901–4909 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc03146f |
| This journal is © The Royal Society of Chemistry 2024 |