Modelling biodegradability based on OECD 301D data for the design of mineralising ionic liquids†
Ann-Kathrin
Amsel
 ab,
Suman
Chakravarti
c,
Oliver
Olsson
ab,
Suman
Chakravarti
c,
Oliver
Olsson
 a and
Klaus
Kümmerer
*ab
a and
Klaus
Kümmerer
*ab
aInstitute of Sustainable Chemistry, Leuphana University of Lüneburg, Universitätsallee 1, 21335 Lüneburg, Germany. E-mail: klaus.kuemmerer@leuphana.de
bResearch and Education Hub, International Sustainable Chemistry Collaborative Centre (ISC3), Leuphana University of Lüneburg, Universitätsallee 1, 21335 Lüneburg, Germany
cMultiCASE Inc., 5885 Landerbrook Dr. #210, Mayfield Heights, OH 44124, USA
First published on 22nd May 2024
Abstract
Ionic liquids (ILs) are increasingly used, e.g. as solvents, electrolytes, active pharmaceutical ingredients and herbicides. If ILs enter the environment due to their use or accidental spills at industry sites, they can pollute the environment. To avoid adverse side effects of persistent ILs in the environment, they should be designed to fully mineralise in the environment after they fulfilled their function during application. (Quantitative) structure–biodegradability relationship models ((Q)SBRs) have been successfully applied in the design of benign chemicals. However, (Q)SBR models have not been widely applied to design mineralising ILs. Therefore, in this study we developed five quantitative structure–biodegradability relationship (QSBR) models based on OECD 301D data from the literature and our own in-house biodegradation experiments. These models can potentially be part of a test battery for designing fully mineralising ILs to increase the overall reliability of the biodegradability assessment and reduce uncertainties. Two datasets were formed and randomly divided into a training set with 233 and 321 compounds and a test set with 26 and 36 compounds, respectively. Both classification and regression models were built using molecular fragments with the aim to predict the classification and continuous biodegradation rate, respectively. The internal and external validations produced a R2 of 0.620–0.854 for the regression models and accuracy, true positive rate, and true negative rate were between 62 and 100% for the classification models indicating an adequate performance but also a need for improvement. For the models and the test battery presented in this study, further research is needed to demonstrate their applicability.
1. Introduction
Ionic liquids (ILs) are of interest in various application areas because an IL can be tuned to the desired physical and chemical properties by changing its combination of cations and anions.1 ILs have been examined in application areas such as solvents for cellulose, electrolytes in batteries, solvents for the preparation of perovskite photovoltaics, active pharmaceutical ingredients and herbicides.2–6 Indeed many quaternary ammonium compounds (QACs) like benzylalkyldimethyl ammonium or alkyltrimethylammonium compounds, which are not called ILs in the literature, have commercial applications, e.g. as disinfectants.7 Actually, they should be included in the group of ILs. ILs can be introduced in the environment through environmentally open applications, at the end-of-life of the mentioned products or accidental spills at industry sites. Some ILs have already been detected in surface water, sediments and wastewater effluents.8,9 The environmental impact of ILs is of concern because of their (eco)toxicological effects and persistence.10–14 Many ILs are not biodegradable in the aquatic environment.15There are two categories of ILs: single ILs, which consist of a single, distinct cation and anion, and mixtures of ILs, which contain different cations and anions. For the purpose of this study, the term “ILs” exclusively denote single ILs. In the literature, 16 different cations have been tested for biodegradability ranging from imidazolium, QACs, pyridinium, cholinium, pyrrolidinium, piperidinium, prolinium, piperazinium, phosphonium, morpholinium, quinolinium, 1,4-diazabicyclo(2.2.2)octanium (DABCO), guandinium, sulphonium, thiazolium to triazolium.15 The tested anions were either organic or inorganic. Organic anions were alkylsulphates, α-amino acids, bis(trifluoromethylsulphonyl)amide, carboxylic acids or dicyanamide. Inorganic anions included halides, tetrafluoroborate or hexafluorophosphate.15,16 In total, ready biodegradability data are available for 508 ILs in the literature.15 Of them, 120 ILs have been identified to be more than 60% biodegradable according to ready biodegradability test methods like the OECD 301 series or ISO 14593.15 However, just 34 ILs have reached the pass level for ready biodegradability, which is ≥60% biodegradation within 10 days starting from a degradation level of 10% as defined by the OECD guideline for the 301 series.17–23 Therefore, it is reasonable to consider that IL's design should follow the concept of Benign by Design (BbD).24 Accordingly, ILs should be designed from scratch for full mineralisation in the environment after their intended usage.24 This means ensuring that both the anionic and cationic components of ILs undergo complete mineralisation either during wastewater treatment processes or within the natural environment. This aligns with the goal of creating safe and inherently sustainable chemical compounds, as outlined in the “Chemicals Strategy for Sustainability towards a Toxic-Free Environment” by the European Commission.25
(Quantitative) structure–biodegradability relationship models ((Q)SBR) can be applied to support the design of readily biodegradable and mineralising ILs. (Q)SBR models help make better informed decisions in the design process prior to the synthesis of chemicals, potentially saving time and resources.26–31 Most of the models for biodegradability of non-charged chemicals, e.g. in EPISuite, Vega, MultiCASE and CATALOGIC, use ready biodegradability data measured according to the MITI test (OECD 301C) and predict either a continuous biodegradation rate or a classification into readily biodegradable or not.32–36 Furthermore, the software CATALOGIC offers a model based on OECD 301F data which predicts the biodegradation pathway.34 In fragment-based (Q)SBR models, the modelled relationships between structural fragments and biodegradability, also called alerts, have the advantage that they increase the interpretability of the predictions and help to understand why chemicals are biodegradable or not.37,38 The relationship can give first insights into which structural adjustments might be needed to design a fully mineralising chemical.26,27 ILs differ from non-charged organic compounds as they consist of two charged components, the anion and the cation. Both components have their own biodegradability potential if they are of organic nature. One of the components could be biodegradable, while the other may not. This is not represented in the overall biodegradation rate that is measured in tests according to OECD 301 or ISO 14593 and can lead to false conclusions in biodegradability.39 Inorganic anions or cations lack carbon atoms that could be metabolised by microorganisms and thus do not contribute to the overall biodegradation rate of an IL. Consequently, ILs cannot be treated in modelling approaches similar to uncharged organic compounds and current modelling techniques need to be adapted to accommodate the unique characteristics of ILs.
Barycki et al. developed AquaBoxIL to predict the environmental distribution of an IL between water, sediment and organic matter.40 The models for biodegradability in AquaBoxIL were the first and the only ones reported for ILs until now to the best of our knowledge. They were based on 77 ILs with OECD 310 (CO2 headspace test) data. The training set included 52 ILs and the test set 25 ILs. 2D and 3D molecular descriptors were used for model building.40 A classification tree assigns the query IL either to the readily biodegradable or not readily biodegradable class. Depending on the classification result a linear regression quantitative structure–biodegradability relationship (QSBR) model for persistent ILs (training set consisted only of ILs that are biodegradable by ≤60%) or one for readily biodegradable ILs (training set consisted only of ILs that are biodegradable by ≥60%) is applied to predict the percentage of biodegradability for the query IL.40 AquaBoxIL was mainly built to predict the environmental distribution. Since the applied molecular descriptors are not always easy to interpret regarding structure–biodegradability relationships (SBRs), decisions on specific structural changes in the design for improved biodegradability are not straightforward.41 Fragment-based models comprising structural alerts are needed for making better-informed decisions in the structural design of ILs.30,42
Therefore, this study presents newly developed fragment-based QSBR models based on a newly compiled OECD 301D dataset which comply with the OECD principles for validating (quantitative) structure–activity relationship ((Q)SAR) models.43,44 We utilised data derived from OECD 301D, recognised as the most rigorous method within the OECD 301 series because readily biodegradable compounds according to this test will be completely biodegradable in surface water. Furthermore, this choice was made to ensure the comprehensive representation of various common ILs, such as imidazolium, pyridinium, QACs, and cholinium ILs, which have been extensively tested through OECD 301D, within our training dataset.15 In total, five fragment-based QSBR models were developed using MultiCASE's FlexFilters platform45 with regard to the ease of interpretation and deriving SBRs to support design decisions. Ordinary least squares (OLS) and logistic regression (LR) were used as modelling approaches to support prediction outcomes of continuous biodegradation rate and classification in biodegradable and non-biodegradable ILs, respectively. Additionally, an in silico test battery as part of the workflow proposed by Lorenz et al. for designing fully mineralising ILs was developed to discuss the possible applications of the models.30
2. Materials and methods
2.1 Experimental ready biodegradability data according to OECD 301D
From the Institute of Sustainable Chemistry (INSC) at Leuphana University (Prof. Kümmerer's working group), a dataset based on the in-house OECD 301D biodegradation experiment was provided. The OECD 301D guideline determines the ready biodegradability under aerobic conditions in water.23 The dataset included 105 ILs (total 116 data points with measured biodegradation rates), 4 organic anions combined with an inorganic cation (6 data points) and 79 non-charged organic compounds (101 data points), which were structurally related to the ILs. These data include information on whether the test substance is readily biodegradable or not and whether the test was valid. With the help of these data, the biodegradability of the individual compounds was evaluated. The in-house OECD 301D test was described in previous studies.17,22 The same OECD 301D test protocol and the same inoculum source were used to generate the data. A test compound is considered to be readily biodegradable if it was degraded by ≥60% within a 10-day window starting after 10% degradation was reached.23As per the protocol the biodegradation results were valid if the following conditions are met:
(1) the degradation rates in the duplicates of the test suspension did not differ by >20% after 28 d,
(2) the compound did not inhibit the degradation of the reference compound (sodium acetate) in the toxicity control (sodium acetate must be degraded ≥25% within 14 d based on its share of the total theoretical oxygen demand (ThOD),
(3) the oxygen concentration in the test vessels must not be <0.5 mg L−1,
(4) sodium acetate was degraded by ≥60% within 14 d in the positive control,
(5) the oxygen consumption is ≤1.5 mg L−1 after 28 d in the blank.
2.2 Compiling the training and test sets
Two datasets, set_IL and set_ILNI, were compiled to examine the influence of the larger dataset set_ILNI on model performance compared to the smaller dataset set_IL.46 Two data sources were used to compile OECD 301D data, (a) data from the INSC in-house biodegradation experiments (section 2.1) and (b) literature data based on OECD 301D as compiled in Amsel et al.15 The following criteria had to be met by the literature data of each IL: (i) tests lasted 28 d, (ii) the allowed concentration of the compound and inoculum of the allowed source was used, and (iii) mineralisation as the ratio between the biochemical oxygen demand (BOD) and the ThOD or chemical oxygen demand (COD) was measured. Sometimes none or just a few validation principles were reported for the data in the literature. Nevertheless, the data were used to expand the dataset. To increase set_ILNI the study on benzalkonium chloride of Sütterlin et al. was added.47 The raw data were available and the applied OECD 301D method was similar to the one at INSC.The literature data and the INSC data were combined. Duplicates were combined into one IL by calculating the mean biodegradation rate. The set_IL is just composed of ILs. For the ILs measured at the INSC, stereochemistry was included in the structures. However, the models were not able to consider stereochemistry in their predictions. The set_ILNI contained ILs, anions, and non-charged compounds. ILs differing in stereochemistry were considered as duplicates.
Without considering the structures, both set_IL and set_ILNI were randomly divided into a training and test set. For the test set 10% of the compounds in set_IL and set_ILNI were used as suggested.48 The train_set_IL contained 233 ILs and the test_set_IL 26 ILs. The set_ILNI was randomly divided into the train_set_ILNI of 321 compounds and the test_set_ILNI of 36 compounds. The train_set_ILNI contained 73 non-ionic compounds, four anions and 244 ILs. The test_set_ILNI contained six non-ionic compounds and 30 ILs.
To characterise the training and test sets, the biodegradability data of the ILs, anions and non-ionic compounds were classified (red: 0–19%, amber: 20–59%, green: ≥60%). The classification is based on the OECD guidelines.23,49 No or minimal biodegradability equals 0–19% degradation. Inherently biodegradable are compounds that degrade by 20–59% in ready biodegradability tests like OECD 301D.49 Compounds classified as ≥60% are possibly readily biodegradable. If ≥60% of ThOD was removed within 10 days starting from a degradation level of 10% ThOD, compounds are readily biodegradable.23
2.3 Model building
Predictive QSBR models were built using the MultiCASE's FlexFilters platform.45 The OECD principles for validating (Q)SAR models were followed to increase models’ reliability and ensure that the models can be used for REACH registration.43 The principles are as follows: (1) a defined endpoint, (2) an unambiguous algorithm, (3) a defined domain of applicability, (4) appropriate measures of goodness-of–fit, robustness and predictivity, and (5) a mechanistic interpretation, if possible.44 Model building was essentially done in three steps: (i) fragmentation of the training compounds, (ii) selection of the most representative fragments (privileged fragments/substructures) that explain the variation of biodegradability of the training set chemicals, and (iii) building a regression model (OLS and LR for continuous regression and classification models, respectively) using these privileged substructures as descriptors. The details of the fragmentation and selection of the most representative fragments are described in the ESI.†Two types of fragment descriptors were used: (i) fragments based on extended connectivity fingerprint (ECFP) type circular fragments50 and (ii) element of a special continuous valued fingerprint containing 600 elements developed by Chakravarti.51 The variable selection using L1 regularisation/Lasso regression was needed to limit the number of unique fragments obtained from the training set chemicals to prevent overfitting.37,52–54 In the variable selection those structural fragments were picked that were relevant to the biodegradability potential of the training chemicals.
For the model IL_FP_cont 600 elements of the fingerprints were considered as descriptors. However, after the variable selection step using L1 regularisation/Lasso regression, only 61 elements were found to be relevant to biodegradability potential (Table S2†). For the models IL_Al_cont, IL_Al_class, ILNI_Al_cont and ILNI_Al_class 70, 29, 130 and 60 fragments, respectively, were found to be relevant (Tables S3, S4, S5 and S6†). These fragments are also called alerts.
Several modelling approaches are available, e.g. multiple linear regression, partial least squares, artificial neural network, random forest, support vector machine and many more, which all have their strengths and weaknesses.55 In this study, simple and well-known OLS and LR modelling in conjugation with fragment descriptors were used (second OECD principle for validating (Q)SAR models). Both approaches, OLS and LR, were chosen since they differ in the prediction outcome (continuous biodegradability rate and classification, respectively). Furthermore, they are easy to interpret due to the linear relationship between descriptors and the target property. On this basis, five models were developed to compare the different modelling approaches and training sets. Models using alerts as descriptors, OLS or LR, were built for both training sets, train_set_IL and train_set_ILNI. Additionally, OLS and elements of fingerprint as descriptors were used for a model based on train_set_IL (Table 1). The constructed regression models were then used for ready biodegradability prediction of new ILs. The continuous regression models’ endpoint was ready biodegradability potential based on OECD 301D (ranging between 0 and 100%) (IL_FP_cont, IL_Al_cont, ILNI_Al_cont, Table 1). The classification models (IL_Al_class, ILNI_Al_class, Table 1) produced a probability value (ranging between 0.0 and 1.0), which can be separated in two classes (readily biodegradable and not readily biodegradable based on OECD 301D) by applying a threshold (usually 0.5) (first OECD principle for validating (Q)SAR models).
| Model 1 IL_FP_cont | Model 2 IL_Al_cont | Model 3 IL_Al_class | Model 4 ILNI_Al_cont | Model 5 ILNI_Al_class | |
|---|---|---|---|---|---|
| Training set number of chemicals | 233 Ionic liquids | 321 Ionic liquids and non-ionic compounds | |||
| Test set number of chemicals | 26 Ionic liquids | 36 Ionic liquids and non-ionic compounds | |||
| Number of descriptors/alerts | 61 | 70 | 29 | 130 | 60 |
| Techniques | Fingerprints, OLS | Alerts, OLS | Alerts, LR, | Alerts, OLS | Alerts, LR, |
| Prediction outcome | Continuous rate in % of the ThOD | Continuous rate in % of the ThOD | Classification in biodegradable or not | Continuous rate in % of the ThOD | Classification in biodegradable or not |
All models have in common that they were built using rigorously identified fragment-based activity privileged substructures, providing easy interpretability and a mechanistic interpretation to enable better-informed decisions in the design of readily biodegradable ILs. These fragments are annotated with the quantitative relationship with biodegradability (regression coefficients). While predicting a test chemical, these fragments are identified in the test chemical and therefore the mechanistic explanations for the predictions can be constructed (fifth OECD principle for validating (Q)SAR models). Hence, a non-biodegradable fragment could be replaced by a better biodegradable one to increase the biodegradability of the whole IL. Both approaches, ordinary and logistic regression were chosen since they have an advantage over the other at different steps in the design process. A classification is appropriate for design decisions at the beginning of the process as they can be used to separate ILs into biodegradable and non-biodegradable ones. The classification provides first insights into which ILs should be focused on to develop readily biodegradable ILs. In contrast, after the first classification step continuous biodegradability rates help to answer questions like which IL of the biodegradable ones is the best biodegradable IL or which IL is the best candidate for further structural adjustments when all ILs are not biodegradable.
2.4 Model validation
The validation of QSBRs was divided into internal and external validation as proposed by OECD to assess the goodness-of-fit and the predictivity, respectively (details are described in the ESI†).44 The typical performance measures accuracy, sensitivity (true positive rate, TPR), specificity (true negative rate, TNR) and area under the curve (AUC) were evaluated for classification models (Table S7†), since they help to understand the model's performance in predicting both classes, biodegradable and non-biodegradable ILs.44,48 For the OLS models the commonly used squared correlation coefficient R2 was evaluated which ranges from 0.0 to 1.0 (Table S7†).44,48For the development of new (Q)SBR models, it is important to define the domain of applicability (AD) of the models to prevent potentially unreliable results for query chemicals with very different chemistry. The AD is a “theoretical region in chemical space” and depends on the chemicals in the training set and the descriptors used to model the endpoint.56 In general, the AD informs about to which chemical structures the models can be applied.57 Clustering was performed using the “R” package rtsne to visualise and study the chemical space defined by the ILs.58 The two-dimensional (2D) t-distributed stochastic neighbour embedding (t-SNE) methodology was applied.59 The 600-element continuous-valued fingerprints of the ILs (section 2.3) were used for clustering.
2.5 Developing an in silico test battery for designing fully mineralising ILs
The ECHA recommends applying all available independent and valid models for one endpoint to increase the overall reliability of the prediction.43 Independent models means that the models differ in descriptors, structural alerts or training sets.43 Therefore, an in silico test battery was developed to structure the application of the newly developed models in the design process of mineralising ILs. As outlined in the workflow for the benign design of newly or redesigned chemicals using in silico tools in a study by Lorenz et al. an in silico test battery supports the identification of the most promising molecules regarding improved environmental biodegradability.30 The workflow by Lorenz et al. started with a pool of molecules that were generated by one of the BbD approaches, which are the targeted or non-targeted de novo and targeted or non-targeted redesign.30 This pool was the starting point for the development of the in silico test battery in this study, which aims to limit the pool of molecules to the most promising ones regarding environmental biodegradability by combining different models for this property and guide their application.3. Results and discussion
3.1 Training and test sets used for modelling
Applying the criteria defined in section 2.2 to the literature data, 25 studies out of 31 containing OECD 301D data were appropriate for the dataset. From the literature in total 231 data points were collected for 201 ILs (dataset, Table 2). 7 of 25 studies contained data measured in the in-house OECD 301D biodegradation experiment at INSC. These 7 studies reported 77 data points for 75 ILs. For set_IL 192 ILs from the literature were combined with 75 ILs measured in the in-house OECD 301D biodegradation experiment at INSC (Table 2). After removing the duplicates, set_IL contained 259 ILs that differed in the organic cation and the side chains attached to it and were combined with organic or inorganic anions. For set_ILNI 196 ILs from the literature were used after removing the stereoisomers from the dataset. These data were combined with 90 ILs, four organic anions combined with an inorganic cation, 79 non-ionic compounds from the INSC in-house OECD 301D data leading to a total number of 357 compounds (Table 2).| Dataset | set_IL | set_ILNI | ||
|---|---|---|---|---|
| Literature data | ILs (data points) | 201 (231) of them measured at INSC: 75 (77) | 192 (222) of them measured at INSC: 67 (69) | 196 (231) of them measured at INSC: 70 (77) |
| Anions (data points) | 0 | 0 | 0 | |
| Non-charged compounds (data points) | 0 | 0 | 0 | |
| INSC in-house OECD 301D data | ILs (data points) | 105 (116) | 75 (79) | 90 (116) |
| Anions (data points) | 4 (6) | 0 | 4 (6) | |
| Non-charged compounds (data points) | 79 (101) | 0 | 79 (101) | |
| Number after removing duplicates | ILs | 294 | 259 | 274 |
| Anions | 4 | 0 | 4 | |
| Non-charged compounds | 79 | 0 | 79 | |
| Total number of compounds | 377 | 259 | 357 | |
| Characteristics | Stereoisomers, ILs, anions, non-charged compounds | Stereoisomers, just ILs | No stereoisomers, ILs, anions, non-charged compounds |
The ILs in set_IL and set_ILNI were the only ones for which OECD 301D data were available that complied with the criteria defined for the literature data in section 2.2. Using OECD 301D data ensured the inclusion of many common ILs, like imidazolium, pyridinium, QACs and cholinium ILs.15 More than 50% of the compounds in set_IL and set_ILNI were measured in the same laboratory at the INSC using the same OECD 301D test protocol and the same inoculum source. Data from the INSC were used for 139 of 259 ILs in set_IL and for 240 of 357 compounds in set_ILNI. The set_IL and set_ILNI were randomly divided into a training set of 233 and 321 compounds and a test set with 26 and 36 compounds, respectively.
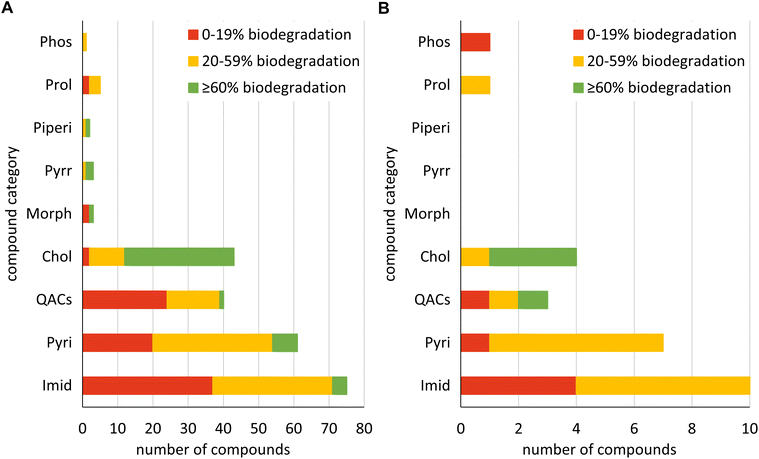
The number of compounds per compound category and biodegradability in the train_set_IL and the test_set_IL are shown in Fig. 1. The prevalent cations in the train_set_IL were imidazolium (75 ILs), pyridinium (61 ILs), QACs (40 ILs) and cholinium (43 ILs) (Fig. 1A). Just a few morpholinium (3 ILs), pyrrolidinium (3 ILs), piperidinium (2 ILs), prolinium (5 ILs) and phosphonium (1IL) ILs were in train_set_IL (Fig. 1A). Most of the ILs in the test_set_IL were imidazolium (10 ILs), pyridinium (7 ILs), QACs (3 ILs) and cholinium (4 ILs). Additionally, one prolinium IL and one phosphonium IL were included in the test_set_IL (Fig. 1B).
In each category, most of the compounds in train_set_IL and test_set_IL were not equally distributed over the biodegradability classes. Just for QACs in the test_set_IL there is an equal number of compounds per class. Since set_IL was randomly divided into a training set and a test set, the distribution of ILs over biodegradability classes and compound category was not influenced. Without considering the compound category, the compounds were not equally distributed over the biodegradability classes as well. In the train_set_IL, 87, 99 and 47 ILs can be assigned to the biodegradability classes 0–19%, 20–59% and ≥60%, respectively. In the train_set_IL, the number of biodegradable ILs is less than that for non-biodegradable and slightly biodegradable ILs. In the test_set_IL seven ILs were biodegradable by 0–19%, 15 ILs by 20–59% and four ILs by ≥60% showing a higher number for slightly biodegradable ILs than for biodegradable and non-biodegradable ILs.
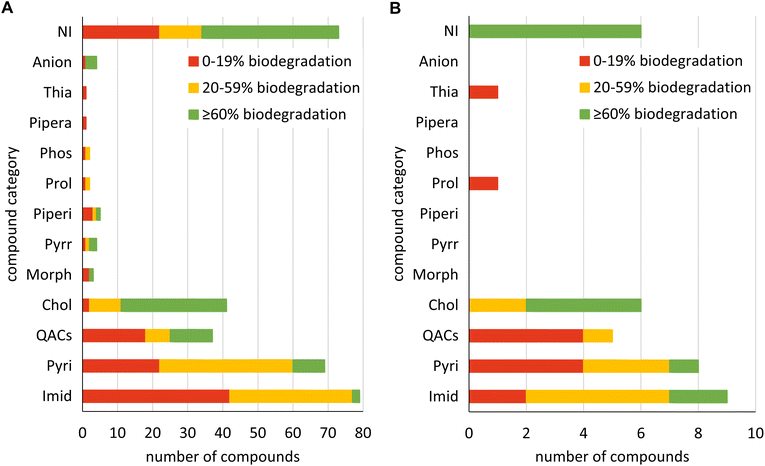
Compared to set_IL, set_ILNI comprised additional compounds to examine the influence of the larger dataset set_ILNI on model performance. Therefore, one piperazinium and one thiazolium IL, as well as four anions (organic anion in combination with inorganic cation) and 79 non-ionic compounds that are structurally related to the ILs were used for set_ILNI. Similar to train_set_IL and test_set_IL most of the ILs belong to the categories of imidazolium (79 ILs), pyridinium (69 ILs), QACs (37 ILs) and cholinium ILs (41 ILs) (Fig. 2). Additionally, the test_set_ILNI contained one prolinium and one thiazolium IL. The non-ionic compounds were not divided into different categories to show the ratio between ILs and non-ionic compounds in Fig. 2. Regarding the biodegradability of the compounds in both subsets, in each category, the compounds in train_set_ILNI and test_set_ILNI were not equally distributed over the biodegradability classes. Without considering the categories, the compounds are nearly equally distributed over the biodegradability classes. In the train_set_ILNI of a total of 321 compounds, 117 compounds were biodegradable by 0–19%, 105 by 20–59% and 99 by ≥60%. In the test_set_ILNI of a total of 36 compounds 12 compounds were biodegradable by 0–19%, 11 by 20–59% and 13 by ≥60%.
Fig. 1 and 2 show that the number of ILs in the presented datasets is not equally distributed among the compound categories. It was shown that more valid biodegradation data based on OECD 301D is needed for morpholinium, pyrrolidinium, piperidinium, prolinium, phosphonium, piperazinium, thiazolium, guandinium, DABCO, quinolinium, sulphonium and triazolium ILs.15
3.2 Biodegradability models for ILs and validation results
The five models were developed with respect to the OECD principles for validating (Q)SAR models (Table S8†). The endpoint was biodegradability according to OECD 301D, and the models predict a continuous value as the percentage of ThOD (models IL_FP_cont, IL_Al_cont, ILNI_Al_cont) or a classification in biodegradable or non-biodegradable ILs (models IL_Al_class, ILNI_Al_class).The results of internal and external validation (section 2.4) are summarised in Table 3. The model IL_AL_class had better accuracy in the training (98%) and test set (96%) compared to model ILNI_Al_class (92% in the training set and 81% in the test set) meaning it classified the ILs more correctly into biodegradable and non-biodegradable. Furthermore, the model IL_AL_class showed better sensitivity and specificity in both the training set (91% and 100% respectively) and test set (75% and 100% respectively). Therefore, IL_AL_class also assigned the ILs more correctly to single classes, biodegradable and non-biodegradable, than ILNI_Al_class. Both models have in common that they had a better specificity than sensitivity meaning they were better in predicting non-biodegradable compounds correctly. The AUC value of the test set was larger for the model ILNI_Al_class (0.90) than for the model IL_AL_class (0.82) (Table 3). In contrast, the AUC in the training set for IL_AL_class was 0.99 and therefore larger than that for ILNI_Al_class (AUC of 0.97 in the training set). Since the AUC is higher than 0.5 the models are able to discriminate between biodegradable and non-biodegradable ILs.
| Model 1 IL_FP_cont | Model 2 IL_Al_cont | Model 3 IL_Al_class | Model 4 ILNI_Al_cont | Model 5 ILNI_Al_class | ||
|---|---|---|---|---|---|---|
| Internal validation | Accuracy | — | — | 98% | — | 92% |
| TPR | — | — | 91% | — | 80% | |
| TNR | — | — | 100% | — | 96% | |
| AUC | — | — | 0.99 | — | 0.97 | |
| R 2 | 0.814 | 0.843 | — | 0.788 | — | |
| External validation | Accuracy | — | — | 96% | — | 81% |
| TPR | — | — | 75% | — | 62% | |
| TNR | — | — | 100% | — | 91% | |
| AUC | — | — | 0.82 | — | 0.90 | |
| R 2 | 0.854 | 0.687 | — | 0.620 | — | |
Of all OLS models, IL_Al_cont had the best goodness-of-fit (R2 of 0.843 for the training set). The model ILNI_Al_cont had the worst goodness-of-fit (R2 of 0.788 for the training set) and the worst predictivity (R2 of 0.620 for the test set) (Table 3). The model IL_FP_cont showed a R2 of 0.854 for the test set. Compared to model IL_Al_cont (R2 of 0.687 for the test set) and ILNI_Al_cont (R2 of 0.620 for the test set) model IL_FP_cont had therefore the best predictivity of continuous values for biodegradability. The plots for comparison of the predicted vs. experimental biodegradation rates for the continuous regression models are visualised in Fig. S1–S6.† The validation results for the models ILNI_Al_cont and ILNI_Al_class were worse than those for the models IL_Al_class and IL_Al_cont. The results showed that the train_set_ILNI compared to train_set_IL did not increase the performance. Overfitting of the models is not very likely. On the one hand, the results indicate that the performance metrics of the internal and external validation are not very different. On the other hand, the number of descriptors is lower than the training data points and was limited to the relevant ones for the biodegradability potential using L1 regularisation/Lasso regression.
In order to compare the performance of the newly developed models with models for biodegradability from the literature for charged and non-charged compounds the performance measures and the training sets have to be considered. To assess whether a model's algorithm is better than the other, the same training and test set and performance measures have to be used.60 Since the used training sets in this study were not used in other studies, it is not possible to assess whether the models from the literature are better or worse performing. The classification model in the AquaBoxIL showed an accuracy, sensitivity and specificity for the test set of 96%, 94% and 100%, respectively.40R2 was 0.726 for the model for persistent ILs and 0.881 for the model for readily biodegradable ILs.40 The accuracy, sensitivity, specificity and R2 related to the test set of the classification model and the two linear regression models are larger than those of the models presented here in this study. However, the training set of the model in Barycki et al.40 contained 52 ILs and is therefore smaller than the training sets used in this study (233 and 321 ILs). The smaller the number of ILs, the less skewed the distribution of readily biodegradable and not readily biodegradable ILs, and the structural similarity of biodegradable and non-biodegradable ILs could have positively influenced the performance.61 This cannot be proven since the experimental biodegradation data used for AquaBoxIL and information on the structures in the training and test set were missing.40
The performance of previous models for non-charged compounds was between 69 and 92% for accuracy, sensitivity, and specificity and between 0.7 and 0.9 for R2.32–35,42,62–64 The performance is similar to the newly developed models IL_FP_cont, IL_Al_class and ILNI_Al_class in this study. However, Table 3 shows that models IL_Al_cont and ILNI_Al_cont are not in the range of 0.7–0.9 for R2 of models described in the literature. Nevertheless, the combination of fragment-based descriptors with OLS and LR for model building resulted in adequate models for predicting the biodegradability of ILs. To improve the models’ performance, an increase of the size of the datasets while covering a wider variety of structural classes might be considered.
3.3 Clustering of the training sets
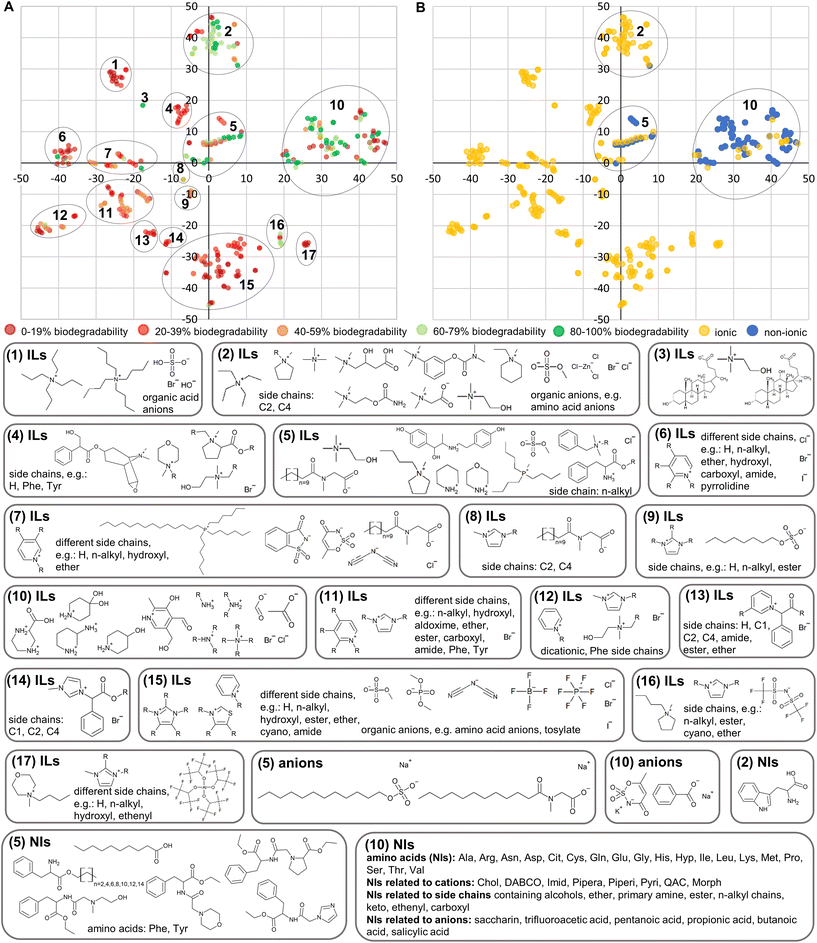
Fig. 1A and 2A provide first insights into the AD since they show which cations were included in the training sets. The models can only be applied to ILs incorporating these cations. However, for every new query IL a check is needed whether it falls within the AD or not since the side chains attached to the cation and the anion have an influence on this, too. Examples for anions, cations and side chains attached to the cation are visualised for train_set_IL in Fig. 3 and for train_set_ILNI in Fig. 4. The tSNE coordinates are available in Tables S9 and S10† for every compound. In the t-SNE plots in Fig. 3 and 4 compounds with different structural fragments are located away from each other, while similar compounds are located close to each other. The 600-element continuous-valued fingerprints were able to separate the compounds regarding their structural fragments and their biodegradability as clusters of the same colour show (Fig. 3 and 4). Imidazolium, pyridinium ILs, and QACs are mainly non-biodegradable as can been seen from the structural fragments shown for each cluster. Most of the biodegradable ILs belong to the group of cholinium ILs. Some α-amino acids, which are just available in train_set_ILNI and highlighted as non-ionic compounds in Fig. 4B, are biodegradable as well.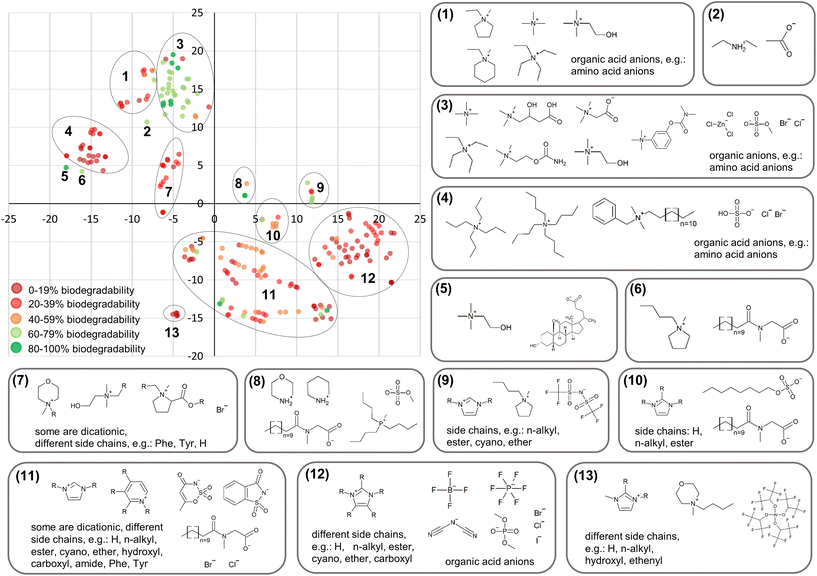 | ||
| Fig. 3 Structural fragments of compounds in train_set_IL and t-SNE plot highlighted for the biodegradability. Phenylalanine (Phe) and tyrosine (Tyr). | ||
ILs were not available for every combination of cations, anions and side chains. Predictions for ILs that are structurally not related to the training set compounds would be based on extrapolations and could be possibly unreliable.37 Therefore, the model's performance (Table 3) is only provided within the AD.
Most of the ILs available in the training sets are imidazolium, pyridinium, QACs and cholinium ILs. Only a few morpholinium, pyrrolidinium, piperidinium, prolinium, phosphonium, piperazinium and thiazolium ILs were included. Therefore, the AD is broader for imidazolium, pyridinium, QACs and cholinium ILs than for the under-represented ILs. The models cannot predict the biodegradability of ILs that are not represented in the training sets and therefore not structurally related to the training set compounds, e.g. DABCO, guandinium, quinolinium, sulphonium and triazolium, and of mixtures of ILs. Therefore, there is a need for more experimental data based on OECD 301D for morpholinium, pyrrolidinium, piperidinium, prolinium, phosphonium, piperazinium, thiazolium, guandinium, DABCO, quinolinium, sulphonium and triazolium ILs to enlarge the AD.
The ADs of the models for biodegradability prediction in the AquaBoxIL were visualised in a William's and Insubria plot.40 Both plots identify response outliers and chemicals that are outside the AD due to their structure. The plots differ in their applicability. The William's plot can be used for chemicals for which experimental data are available, while the Insubria plot is used for chemicals without experimental data.48,65 However, it was not mentioned, which structural features the training and test set contained. Therefore, it is not possible to compare the AD of AquaBoxIL and the models presented in this study.
3.4 Application of models for designing environmentally mineralising ILs
The five models were developed to support the design of biodegradable ILs. Since they differ in the training set or descriptors or structural alerts, they can be considered as independent from each other. Therefore, according to ECHA43 all five models should be applied to increase the reliability of the overall biodegradability assessment of ILs. Hence, this section explores how these models could be applied in an in silico test battery (Fig. 5) that is part of the workflow for the benign design of chemicals presented in Lorenz et al.30 The test battery starts with a pool of ILs either in the de novo or redesign of chemicals (Fig. 5). Just statistical QSBR models are included since these are the only ones available for ILs and the endpoint ready biodegradability according to OECD 301D.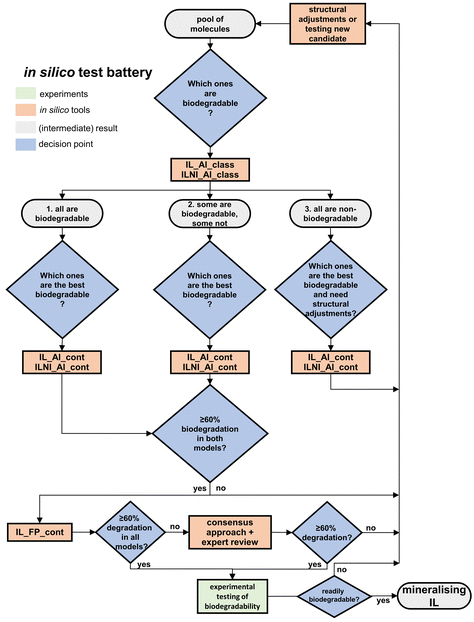 | ||
| Fig. 5 Possible applications of models in an in silico test battery for designing fully mineralising ILs. | ||
The advantage of biodegradability alerts was demonstrated in the rational redesign of atenolol and metoprolol.26,27 Accordingly, Fig. 5 proposes to apply IL_Al_class and ILNI_Al_class at first to gain insights from two independent models and two different sets of alerts (one for each model). The models facilitate to separate the pool of new or redesigned ILs into biodegradable and non-biodegradable ILs (Fig. 5). The model ILNI_Al_class performs not as well as IL_Al_class, but contains more alerts compared to IL_Al_class (60 compared to 29). Therefore, ILNI_Al_class helps to understand why some ILs are biodegradable and others are not. In this step three different outcomes are possible: 1. all ILs in the pool of newly developed or redesigned ILs are biodegradable, 2. some are biodegradable and some are not, and 3. no IL is biodegradable.
However, after the classification, a continuous biodegradation rate is needed to decide which IL in the class of biodegradable ILs (outcomes 1 and 2) or non-biodegradable ILs (outcome 3) is the best candidate to change structural fragments and to design a biodegradable IL. The approach of first using a classification model and then a model predicting a continuous value was demonstrated to be useful for prioritising chemicals in chemical safety assessment regarding their carcinogenicity.66 Therefore, this study suggests the combination of a classification and a continuous biodegradation rate in Fig. 5 to support the prioritisation. In this respect, both models IL_Al_cont and ILNI_Al_cont are suitable since they generate a continuous biodegradation rate and their performance is adequate (Table 3). If in outcomes 1 and 2 the predicted biodegradation rate is ≤60% structural adjustments are needed and the workflow would start from the beginning with the pool of molecules. The models’ alerts and the identified SBRs in Amsel et al. could support to identify which structural changes are needed.16 In particular, the 130 alerts in model ILNI_Al_cont, the model with the most alerts, give insights into SBRs and could help to design a fully mineralising IL.
The model IL_FP_cont should be applied to confirm the predictions of IL_Al_cont and ILNI_Al_cont since it generates continuous rates with the best performance in the test set compared to IL_Al_cont and ILNI_Al_cont (Table 3). If all three models indicate that the IL is biodegradable by ≥60%, its ready biodegradability should be tested in a laboratory experiment (Fig. 5). The pass level for ready biodegradability in OECD 301D is ≥60% removal of ThOD within 10 days starting from a degradation level of 10%.23 If the three models do not agree with their outcome, a consensus approach or an expert review as proposed by the workflow for the benign design of chemicals in Lorenz et al. might be helpful to increase the confidence of the assessment and reduce uncertainties.30 If the outcome indicates that the IL is not biodegradable, structural changes can be made to possibly increase the biodegradability. The redesigned IL would be included in the pool of molecules and its biodegradability be predicted. If the consensus approach or expert review confirms a biodegradation rate of ≥60%, the IL should be tested in the laboratory for ready biodegradability. A mineralising IL was designed if it is readily biodegradable in experimental testing. Non-biodegradable ILs that differ in structural fragments with training set ILs might be tested in the laboratory as well. The new data of biodegradable and non-biodegradable ILs could be included in the training sets and possibly improve the models’ performance.
3.5 Evaluation of the developed models for biodegradability of ILs
Since around 80% of the wastewater is not treated worldwide, ILs can be introduced into the environment via wastewater or leakages.67 The endpoint ready biodegradability according to OECD 301D was chosen since it is the most stringent method of the OECD 301 series and many of the common ILs were tested according to this test method.15 Therefore, this study developed models for the endpoint ready biodegradability according to OECD 301D of ILs using literature data and INSC in-house OECD 301D data addressing this topic for the first time.Both datasets, set_IL and set_ILNI are unique since more than 50% of the compounds were measured in the same laboratory at the INSC using the same OECD 301D test protocol, validation criteria and similar inoculum leading to increased data quality compared to the literature data for which different inoculum sources, concentrations and microorganism diversities were used and not all validation criteria were reported.
For every individual model, the training set defined the AD as it determined the representative fragment descriptors and the alerts (section 3.3). The models might not cover important SBRs that are relevant for biodegradability predictions of a query compound. Hence, the models are not able to make reliable predictions for a query IL that differs in too many fragments from training set compounds.37,68 Therefore, just within the model's AD reliable predictions can be made according to the model's performance, and extrapolations in predictions are avoided.
As the validation results showed, the QSBR models successfully predicted the biodegradability of common ILs, like imidazolium, pyridinium, QACs and cholinium ILs (section 3.2). The models can be applied in a test battery to design environmentally readily mineralising ILs. Uncertainties regarding the biodegradability of a newly designed IL are addressed after the in silico design process by testing the biodegradability in the laboratory. Hence, QSBR models are versatile tools for planning of experiments and selecting the most promising candidates.
4. Conclusion
Previous biodegradation models for ILs focused on modelling the environmental distribution of ILs between water, sediment and organic matter. These models used OECD 310 (CO2 headspace test) literature data of 77 ILs. In our study, we used 294 ILs’ biodegradability data (OECD 301D, ready biodegradability) for five fragment-based QSBR models using the MultiCASE's FlexFilters platform. Well-known and easily interpretable modelling approaches were applied, OLS and LR, to build models with two different outcomes, a continuous biodegradation rate and a classification model, respectively. The models successfully predicted the biodegradability of common ILs, like imidazolium, pyridinium, QACs and cholinium ILs. Additionally, the models were developed in agreement with the OECD principles for the validation to increase their reliability and their acceptance for regulatory purposes. Thus, this application showed that OECD principles can be implemented in biodegradation prediction models of ILs, even for the most stringent method of the OECD 301 series, OECD 301D. The internal and external validation results were adequate to predict the biodegradability of ILs. The train_set_ILNI did not increase the model's performance compared to train_set_IL even though it contained more ILs. Furthermore, the reasonably good prediction performance suggests an application of the models in a test battery for the design of environmentally mineralising ILs to increase the overall reliability of the assessment of newly developed or redesigned ILs. The test battery supports the candidate selection for synthesis and testing while saving time. In the test battery different models for environmental biodegradability according to OECD 301D were applied. The in silico test battery as part of the workflow for the benign design of newly developed or redesigned chemicals using in silico tools was successfully demonstrated. The test battery will help practitioners to understand when which model could be applied in the assessment of biodegradability to limit the pool of newly developed or redesigned ILs to the mineralising ones. However, best practice examples are needed to demonstrate the applicability of the models and the test battery and the ease of interpretation of the alerts. Better performance could be possibly achieved by increasing the size of the datasets while covering a wider variety of structural classes. Biodegradability is a central endpoint for a benign IL regarding its end-of-life. Bioaccumulation and (eco)toxicity have to be examined as well for which (Q)SAR models should be developed if not yet available to support the design of benign ILs.Author contributions
Ann-Kathrin Amsel: conceptualisation, methodology, data curation, formal analysis, investigation, visualisation, writing – original draft, writing – review & editing, and validation; Suman Chakravarti: conceptualisation, methodology, software, data curation, formal analysis, investigation, supervision, resources, visualisation, writing – original draft, writing – review & editing, and validation; Oliver Olsson: conceptualisation, data curation, investigation, visualisation, writing – original draft, writing – review & editing, and validation; Klaus Kümmerer: conceptualisation, methodology, investigation, supervision, resources, writing – original draft, writing – review & editing, and validation.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
A.-K. A. and K. K. would like to thank the German Federal Ministry for the Environment, Nature Conservation, Nuclear Safety and Consumer Protection (BMUV) and the German Umweltbundesamt (UBA) for their support with the International Sustainable Chemistry Collaborative Centre (ISC3) activities.References
- N. V. Plechkova and K. R. Seddon, Chem. Soc. Rev., 2008, 37, 123–150 RSC.
- M. Watanabe, M. L. Thomas, S. Zhang, K. Ueno, T. Yasuda and K. Dokko, Chem. Rev., 2017, 117, 7190–7239 CrossRef CAS PubMed.
- K. S. Egorova, E. G. Gordeev and V. P. Ananikov, Chem. Rev., 2017, 117, 7132–7189 CrossRef CAS PubMed.
- J. Zhang, J. Wu, J. Yu, X. Zhang, J. He and J. Zhang, Mater. Chem. Front., 2017, 1, 1273–1290 RSC.
- F. Wang, D. Duan, M. Singh, C. M. Sutter-Fella, H. Lin, L. Li, P. Naumov and H. Hu, Energy Environ. Mater., 2023, 6, e12435 CrossRef CAS.
- W. Wilms, M. Woźniak-Karczewska, A. Syguda, M. Niemczak, Ł. Ławniczak, J. Pernak, R. D. Rogers and Ł. Chrzanowski, J. Agric. Food Chem., 2020, 68, 10456–10488 CrossRef CAS PubMed.
- C. Zhang, F. Cui, G. Zeng, M. Jiang, Z. Yang, Z. Yu, M. Zhu and L. Shen, Sci. Total Environ., 2015, 518–519, 352–362 CrossRef CAS PubMed.
- S. Brand, M. P. Schlüsener, D. Albrecht, U. Kunkel, C. Strobel, T. Grummt and T. A. Ternes, Water Res., 2018, 136, 207–219 CrossRef CAS PubMed.
- S. G. Pati and W. A. Arnold, Environ. Sci.: Processes Impacts, 2020, 22, 430–441 RSC.
- M. Amde, J.-F. Liu and L. Pang, Environ. Sci. Technol., 2015, 12611–12627 CrossRef CAS PubMed.
- S. P. F. Costa, A. M. O. Azevedo, P. C. A. G. Pinto and M. L. M. F. S. Saraiva, ChemSusChem, 2017, 10, 2321–2347 CrossRef CAS PubMed.
- P. G. Jessop, Faraday Discuss., 2018, 206, 587–601 RSC.
- T. P. T. Pham, C.-W. Cho and Y.-S. Yun, Water Res., 2010, 44, 352–372 CrossRef CAS PubMed.
- E. M. Siedlecka, M. Czerwicka, J. Neumann, P. Stepnowski, J. Fernández and J. Thöming, in Ionic liquids: Theory, properties, new approaches, ed. A. Kokorin, InTech, Rijeka, Croatia, 2011, pp. 701–722 Search PubMed.
- A.-K. Amsel, O. Olsson and K. Kümmerer, Chemosphere, 2022, 299, 134385 CrossRef CAS PubMed.
- A.-K. Amsel, O. Olsson and K. Kümmerer, Green Chem., 2023, 9226–9250 RSC.
- A. Haiß, A. Jordan, J. Westphal, E. Logunova, N. Gathergood and K. Kümmerer, Green Chem., 2016, 18, 4361–4373 RSC.
- J. R. Harjani, R. D. Singer, M. T. Garcia and P. J. Scammells, Green Chem., 2008, 10, 436–438 RSC.
- X.-D. Hou, Q.-P. Liu, T. J. Smith, N. Li and M.-H. Zong, PLoS One, 2013, 8, e59145 CrossRef CAS PubMed.
- S. Morrissey, B. Pegot, D. Coleman, M. T. Garcia, D. Ferguson, B. Quilty and N. Gathergood, Green Chem., 2009, 11, 475–483 RSC.
- B. Peric, J. Sierra, E. Martí, R. Cruañas, M. A. Garau, J. Arning, U. Bottin-Weber and S. Stolte, J. Hazard. Mater., 2013, 261, 99–105 CrossRef CAS PubMed.
- M. Suk, A. Haiß, J. Westphal, A. Jordan, A. Kellett, I. V. Kapitanov, Y. Karpichev, N. Gathergood and K. Kümmerer, Green Chem., 2020, 22, 4498–4508 RSC.
- OECD, OECD guideline for testing of chemicals. Ready biodegradability, 1992 Search PubMed.
- K. Kümmerer, Green Chem., 2007, 9, 899 RSC.
- European Commission, Chemicals Strategy for Sustainability. Towards a Toxic-Free Environment,Brussels, 2020 Search PubMed.
- T. Rastogi, C. Leder and K. Kümmerer, Chemosphere, 2014, 111, 493–499 CrossRef CAS PubMed.
- T. Rastogi, C. Leder and K. Kümmerer, RSC Adv., 2015, 5, 27–32 RSC.
- T. Rastogi, C. Leder and K. Kümmerer, Environ. Sci. Technol., 2015, 49, 11756–11763 CrossRef CAS PubMed.
- C. Leder, M. Suk, S. Lorenz, T. Rastogi, C. Peifer, M. Kietzmann, D. Jonas, M. Buck, A. Pahl and K. Kümmerer, ACS Sustainable Chem. Eng., 2021, 9, 9358–9368 CrossRef CAS.
- S. Lorenz, A.-K. Amsel, N. Puhlmann, M. Reich, O. Olsson and K. Kümmerer, ACS Sustainable Chem. Eng., 2021, 9, 12461–12475 CrossRef CAS.
- J. van Dijk, H. Flerlage, S. Beijer, J. C. Slootweg and A. P. van Wezel, Chemosphere, 2022, 296, 134050 CrossRef CAS PubMed.
- R. S. Boethling, D. G. Lynch and G. C. Thom, Environ. Toxicol. Chem., 2003, 22, 837–844 CrossRef CAS PubMed.
- A. Lombardo, F. Pizzo, E. Benfenati, A. Manganaro, T. Ferrari and G. Gini, Chemosphere, 2014, 108, 10–16 CrossRef CAS PubMed.
- S. Dimitrov, T. Pavlov, N. Dimitrova, D. Georgieva, D. Nedelcheva, A. Kesova, R. Vasilev and O. Mekenyan, SAR QSAR Environ. Res., 2011, 22, 719–755 CrossRef CAS PubMed.
- J. Jaworska, S. Dimitrov, N. Nikolova and O. Mekenyan, SAR QSAR Environ. Res., 2002, 13, 307–323 CrossRef CAS PubMed.
- G. Klopman and M. Tu, Environ. Toxicol. Chem., 1997, 16, 1829–1835 CAS.
- P. Gramatica, Int. J. Quantum Struct. Prop. Relatsh., 2020, 5, 61–97 Search PubMed.
- G. J. Myatt, L. D. Beilke and K. P. Cross, in Comprehensive Medicinal Chemistry III, Elsevier, 2017, pp. 156–176 Search PubMed.
- S. Stolte, S. Steudte, A. Igartua and P. Stepnowski, Curr. Org. Chem., 2011, 15, 1946–1973 CrossRef CAS.
- M. Barycki, A. Sosnowska and T. Puzyn, Green Chem., 2018, 20, 3359–3370 RSC.
- D. T. Stanton, J. Chem. Inf. Comput. Sci., 2003, 43, 1423–1433 CrossRef CAS PubMed.
- A. Sedykh and G. Klopman, SAR QSAR Environ. Res., 2007, 18, 693–709 CrossRef CAS PubMed.
- ECHA, Practical guide. How to use and report (Q)SARs, 2016 Search PubMed.
- OECD, Guidance Document on the Validation of (Quantitative) Structure-Activity Relationships [(Q)SAR] Models, 2007 Search PubMed.
- S. K. Chakravarti and S. R. M. Alla, in QSAR in Safety Evaluation and Risk Assessment, ed. H. Hong, Elsevier, 2023, pp. 219–234 Search PubMed.
- A.-K. Amsel, O. Olsson and K. Kümmerer, Ready biodegradability data of ionic liquids, OECD 301D (Closed Bottle Test), 2024, V1, PubData, Leuphana University Lüneburg, 2024, available at: DOI:10.48548/pubdata-151, accessed 21 February 2024.
- H. Sütterlin, R. Alexy, A. Coker and K. Kümmerer, Chemosphere, 2008, 72, 479–484 CrossRef PubMed.
- P. Gramatica, in Computational Toxicology. Methods in Molecular Biology, ed. B. Reisfeld and A. Mayeno, Humana Press, Totowa, NJ, vol 930, 2013, pp. 499–526 Search PubMed.
- OECD, Revised Introduction to the OECD Guidelines for Testing of Chemicals, Section 3. Part 1: Principles and Strategies related to the Testing of Degradation of Organic Chemicals, OECD, 2006 Search PubMed.
- D. Rogers and M. Hahn, J. Chem. Inf. Model., 2010, 50, 742–754 CrossRef CAS PubMed.
- S. K. Chakravarti, ACS Omega, 2018, 3, 2825–2836 CrossRef CAS PubMed.
- D. M. Hawkins, J. Chem. Inf. Comput. Sci., 2004, 44, 1–12 CrossRef CAS PubMed.
- J. Friedman, T. Hastie and R. Tibshirani, J. Stat. Softw., 2010, 33, 1–22 Search PubMed.
- R. Tibshirani, J. Bien, J. Friedman, T. Hastie, N. Simon, J. Taylor and R. J. Tibshirani, J. R. Stat. Soc. B, 2012, 74, 245–266 CrossRef PubMed.
- L. C. Yee and Y. C. Wei, in Statistical Modelling of Molecular Descriptors in QSAR/QSPR, ed. M. Dehmer, K. Varmuza and D. Bonchev, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, 2012, pp. 1–31 Search PubMed.
- P. Gramatica, QSAR Comb. Sci., 2007, 26, 694–701 CrossRef CAS.
- T. I. Netzeva, A. Worth, T. Aldenberg, R. Benigni, M. T. D. Cronin, P. Gramatica, J. S. Jaworska, S. Kahn, G. Klopman, C. A. Marchant, G. Myatt, N. Nikolova-Jeliazkova, G. Y. Patlewicz, R. Perkins, D. Roberts, T. Schultz, D. W. Stanton, J. J. M. van de Sandt, W. Tong, G. Veith and C. Yang, ATLA, 2005, 33, 155–173 CAS.
- J. H. Krijthe, Rtsne: T-Distributed Stochastic Neighbor Embedding using a Barnes-Hut Implementation, 2015, available at: https://github.com/jkrijthe/Rtsne, accessed 2 February 2024 Search PubMed.
- L. van der Maaten and G. Hinton, J. Mach. Learn. Res., 2008, 9, 2579–2605 Search PubMed.
- S. Koutsoukos, F. Philippi, F. Malaret and T. Welton, Chem. Sci., 2021, 12, 6820–6843 RSC.
- A. Tropsha, Mol. Inf., 2010, 29, 476–488 CrossRef CAS PubMed.
- R. S. Boethling, D. G. Lynch, J. S. Jaworska, J. L. Tunkel, G. C. Thom and S. Webb, Environ. Toxicol. Chem., 2004, 23, 911–920 CrossRef CAS PubMed.
- S. Dimitrov, G. Dimitrova, T. Pavlov, N. Dimitrova, G. Patlewicz, J. Niemela and O. Mekenyan, J. Chem. Inf. Model., 2005, 45, 839–849 CrossRef CAS PubMed.
- J. Tunkel, P. H. Howard, R. S. Boethling, W. Stiteler and H. Loonen, Environ. Toxicol. Chem., 2000, 19, 2478–2485 CrossRef CAS.
- S. Brandmaier, W. Peijnenburg, M. K. Durjava, B. Kolar, P. Gramatica, E. Papa, B. Bhhatarai, S. Kovarich, S. Cassani, P. P. Roy, M. Rahmberg, T. Öberg, N. Jeliazkova, L. Golsteijn, M. Comber, L. Charochkina, S. Novotarskyi, I. Sushko, A. Abdelaziz, E. D'Onofrio, P. Kunwar, F. Ruggiu and I. V. Tetko, ATLA, 2014, 42, 13–24 CAS.
- C. Toma, A. Manganaro, G. Raitano, M. Marzo, D. Gadaleta, D. Baderna, A. Roncaglioni, N. Kramer and E. Benfenati, Molecules, 2021, 26, 127 CrossRef CAS PubMed.
- United Nations World Water Assessment Programme, The United Nations World Water Development Report 2017. Wasterwater: The untapped resource, UNESCO, Paris, 2017, vol. 2017 Search PubMed.
- J. S. Jaworska, R. S. Boethling and P. H. Howard, Environ. Toxicol. Chem., 2003, 22, 1710–1723 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4gc00889h |
| This journal is © The Royal Society of Chemistry 2024 |