Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
CRISPR for companion diagnostics in low-resource settings†
Xu Qian*a,
Qiang Xua,
Christopher J. Lyonbc and
Tony Y. Hu *bc
*bc
aDepartment of Clinical Laboratory, Zhejiang Cancer Hospital, Hangzhou Institute of Medicine (HIM), Chinese Academy of Sciences, Hangzhou 310022, China. E-mail: Qian_michelle2014@163.com
bCenter for Cellular and Molecular Diagnostics, Tulane University School of Medicine, 1430 Tulane Ave, New Orleans, LA 70112, USA. E-mail: tonyhu@tulane.edu
cDepartment of Biochemistry and Molecular Biology, Tulane University School of Medicine, 1430 Tulane Ave, New Orleans, LA 70112, USA
First published on 5th September 2024
Abstract
New point-of-care tests (POCTs), which are especially useful in low-resource settings, are needed to expand screening capacity for diseases that cause significant mortality: tuberculosis, multiple cancers, and emerging infectious diseases. Recently, clustered regularly interspaced short palindromic repeats (CRISPR)-based diagnostic (CRISPR-Dx) assays have emerged as powerful and versatile alternatives to traditional nucleic acid tests, revealing a strong potential to meet this need for new POCTs. In this review, we discuss CRISPR-Dx assay techniques that have been or could be applied to develop POCTs, including techniques for sample processing, target amplification, multiplex assay design, and signal readout. This review also describes current and potential applications for POCTs in disease diagnosis and includes future opportunities and challenges for such tests. These tests need to advance beyond initial assay development efforts to broadly meet criteria for use in low-resource settings.
1 Introduction
Rapid and accurate clinical tests are essential for effective patient care in multiple disease conditions, as they can provide critical information for disease diagnosis, treatment planning, and prognosis. Such tests can also play essential roles in efforts to contain various infectious diseases, as was demonstrated during the COVID-19 pandemic, when multiple tests for SARS-CoV-2 were rapidly developed and employed in screening efforts intended to reduce virus transmission. But access to sensitive and accurate diagnostic tests is often limited in many parts of the world regarded as low-resource settings. These tests often require expensive equipment and well-trained personnel and are thus frequently restricted to central laboratories that have the resources and infrastructure required to perform them. Self-contained point-of-care tests (POCTs), which do not require extensive training or additional resources to perform, have the potential to extend the use of critical diagnostic/prognostic assays into low-resource settings to improve healthcare disparities and patient outcomes and inform disease control strategies for future outbreaks.Ideal POCTs should adhere to the ASSURED criteria (affordable, sensitive, specific, user-friendly, rapid, equipment-free, delivered),1 and it has recently been proposed that these should be expanded to the REASSURED criteria, including real-time connectivity, ease of specimen collection, and environmental friendliness.2 Sensitivity and specificity should largely be maintained in REASSURED assays for low-resource settings, but performance may drop slightly compared with that of central laboratory tests to allow for accessibility and affordability in these settings. Tremendous progress has been made in the development and adoption of assays that meet ASSURED criteria; these assays include diagnostic tests for HIV-1, malaria, and syphilis.2 But major challenges remain for many other diseases, particularly for those requiring nucleic acid (NA)-based assays, which may need careful refinement to ensure acceptable performance after exposure to a variety of conditions during assay shipping, storage, and use.
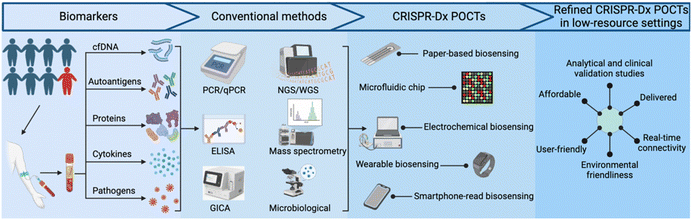
The rapid expansion of available genomic data over the past two decades serves as an excellent resource for new NA tests designed to identify specific microbial pathogens, predict drug susceptibility or disease progression, or provide other useful clinical information. Such NA tests can have excellent diagnostic or prognostic value, but require highly-trained personnel, careful sample preparation, and costly equipment and/or reagents that limit their utility. Adopting various isothermal amplification approaches can alleviate the need for thermocyclers, required by conventional PCR assays, but these alternative approaches often have their own distinct limitations. Clustered regularly interspaced short palindromic repeats (CRISPR)-based diagnostic (CRISPR-Dx) assays, such as SHERLOCK,3 DETECTR,4 and HOLMES,5 have emerged as powerful and versatile tools that complement traditional NA assays for sensitive detection of specific NA sequences in complex samples. Other target types, including proteins and small molecules, can also be detected by CRISPR-based methods that employ aptamers or proteins that recognize these targets. CRISPR-Dx assays thus exhibit robust potential for adaptation to POCTs, as has been reported in studies that have used paper-based,6 microfluidic chip,7,8 electrochemical,9 wearable10 and smartphone-read biosensor11 approaches that can be refined for use in low-resource settings (Fig. 1).7,8,12–16 Importantly, similar to the development of clinical assays, the development of refined CRISPR-Dx POCTs requires analytical and clinical validation. These tests should also meet REASSURED criteria.
This review describes how the inherent features of CRISPR-based assays align with the needs of POCTs and highlights progress made on CRISPR-Dx applications for infectious disease. We also discuss new applications of CRISPR-based assays in oncology, including their broad potential for cancer prevention, screening, and chronic disease management.
Finally, we discuss barriers remaining for the broad adoption of CRISPR-based applications, their utility in low-resource settings, and the future outlook for these assays.
2 CRISPR-Dx assays
2.1 Mechanism of CRISPR/Cas-based diagnosis
CRISPR/CRISPR-associated protein (CRISPR/Cas) complexes function as adaptive immune system factors in bacteria and archaea by targeting foreign NAs for destruction.17 Each CRISPR/Cas complex employs a short (17 to 20 nucleotide) CRISPR RNA sequence (crRNA) transcribed from a CRISPR array to recognize and directly cleave previously encountered pathogen-specific NA target sequences (Fig. 2), which may or may not contain a conserved protospacer adjacent motif (PAMs) at its 3′ end, depending upon the CRISPR/Cas system. The potential of these Cas/crRNA complexes to cleave a diverse array of NA target sequences was rapidly recognized and CRISPR/Cas systems have since become the gold-standard approach for genome editing. After binding their recognition sequence, some Cas protein types exhibit both target-specific (cis), and non-specific collateral (trans) cleavage activities. Such trans-cleavage activities can indiscriminately cleave single-stranded DNA (e.g., Cas12 (ref. 4 and 18) or RNA (e.g., Cas13 (ref. 19)), depending on the Cas effector, and have thus been employed as an efficient means of cleaving a labelled reporter probe or other substrates that can induce a detectable assay signal.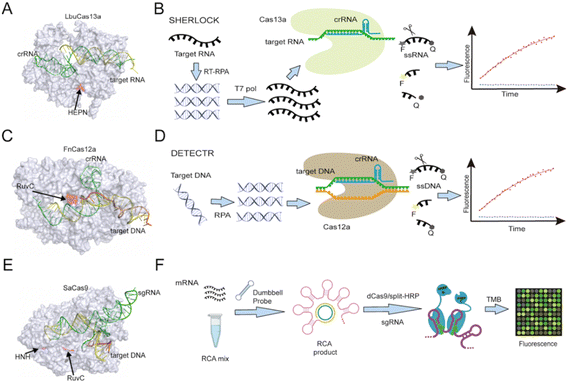 | ||
| Fig. 2 (A) Structure of Cas13a (PDB ID: 5XWP) protein complexed with crRNA and bound to a target NA sequence recognized by the crRNA sequence. (B) Cas13-based SHERLOCK assay workflow scheme. (C) Structure of Cas12a (PDB ID: 6GTG) protein with crRNA and a target NA sequence. (D) Cas12a-based DETECTR assay workflow scheme. (E) Cas9 (PDB ID: 5AXW) with crRNA and a target NA sequence. (F) A dCas9-based platform utilizes split horseradish peroxidase for detection of microRNA. RuvC domain catalytic sites are marked in Cas12a and Cas9 protein, as is the HEPN domain catalytic site in Cas13a and the HNH domain catalytic site in Cas9. RCA: rolling circle amplification; TMB: 3,3′,5,5′-tetramethylbenzidine. | ||
The most commonly used Cas systems for CRISPR-Dx assays are Cas13, Cas12, Cas9, and Cas14. These are all class 2 CRISPR–Cas systems that employ a single Cas effector protein. Within class 2, Cas9 is classified as type II; Cas12 and Cas14, as type V; and Cas13, as type VI. These Cas systems have distinct features, including different crRNA structures, PAM sequences, target specificities, and cleavage activities, which give them specific utility for applications that detect various NA targets.
The SHERLOCK (specific high-sensitivity enzymatic reporter unLOCKing) platform was the first reported assay based on Cas13 collateral RNase activity (Fig. 2B).3,21,22 This assay amplifies its targeted DNA or RNA of interest by recombinase polymerase amplification (RPA) or reverse transcription (RT) plus RPA. It then employs T7 RNA polymerase to transcribe its amplicons into RNA that can be recognized by a target-specific Cas13/crRNA complex to trigger Cas13-mediated trans-cleavage of a reporter RNA with both a fluorophore and a quencher. The fluorescent signal produced by this process can be monitored in real time, enabling attomolar detection of a target RNA, which is comparable to the detection achieved by digital droplet PCR (ddPCR) or quantitative PCR (qPCR). SHERLOCK was tested for its ability to detect Zika and Dengue viruses in clinical isolates, genotype bacteria, and detect single-nucleotide polymorphisms (SNPs). In the latter, it could detect low-frequency cancer-associated mutations (EGFR L858R and BRAF V600E) present in cell-free DNA (cfDNA) fragments spiked into mock patient samples. Several modifications have been made to the original sample processing, target amplification, and signal readout methods of the SHERLOCK platform to adapt it for new applications and make the platform more accessible to its end users, some of these modifications are discussed in later sections.
Efforts have also been made to modify Cas13 activity, including a recent study that demonstrated that Cas13 collateral cleavage activity could be enhanced by tethering its RNA binding domain adjacent to the active site responsible for these trans-cleavage events. These engineered Cas13 variants can exhibit catalytic efficiencies for reporter cleavage that are two- to three-fold higher than wildtype Cas13.23 This study suggests the potential for protein engineering to further improve detection sensitivities beyond what's currently achievable with Cas13-based assays and suggests that similar optimization efforts could possibly also improve the detection sensitivities of assays based on other Cas protein systems.
3 CRISPR/Cas-based applications and their potential for adaptation to POCTs
The development of CRISPR-Dx assays typically follows a standard workflow. First, using synthetic or mock samples, the researcher tests signal amplification and readout methods to evaluate the assay's sensitivity and specificity for its biomarker target and decides whether a different approach, or a target preamplification step, is necessary to achieve desired performance. Next, isolation procedures employed to isolate target material from clinical specimens (if used) are revised to improve target detection and simplify the assay workflow, as necessary. Finally, successful assays can then be adapted for incorporation into multiplex assays or point-of-care devices. We discuss each of these steps in the following sections.3.1 Preamplification methods
CRISPR-based assays are often employed to detect low abundance targets since Cas/crRNA complexes can exhibit high affinity for their target sequences and their cleavage activity can often be used for signal amplification. However, slow enzyme kinetics and background signal may still require an assay to employ a target NAA step to improve the LOD for scarce NA targets or to decrease the reaction time required to produce a detectable signal for a chosen assay readout method. NAA-based CRISPR assays often report LODs in the attomolar range while LODs reported for non-NAA CRISPR assays typically fall in the femtomolar range. Isothermal NAA methods are frequently selected for this step, since they do not require a thermocycler and often function effectively over a relatively broad temperature range, making them more suitable for use in low-resource settings. RPA and LAMP are the most popular choices, but some assays have also employed RCA,49,50 strand-displacement amplification (SDA),51,52 exponential amplification reaction (EXPAR),53,54 and NA sequence-based amplification (NASBA).55CRISPR-based assays that combine an NAA step and CRISPR reaction in one tube are highly desirable for low-resource settings and can simplify sample handling procedures, reducing the risk for operator error and for cross-contamination. This combined approach, however, can be challenging as it requires simultaneous optimization of the NAA and CRISPR reactions for target sensitivity and specificity, and these reactions can have distinct preferences for reagent composition, buffer system and reaction temperature. For example, LAMP prefers higher temperature (55–70 °C) than tolerated by standard Cas proteins, thus integrating LAMP and CRISPR into one tube assays can requires the use of thermostable Cas protein variants such as AapCas12b56 One-pot reactions can employ either sequential57,58 or parallel24,59 NAA and CRISPR reactions. Sequential one-pot reactions are usually achieved by separating the CRISPR reagents from the NAA reaction until after its completion and then using a physical force, from centrifugation or shaking, to mix these reagents without opening the tube, although interesting alternatives have also been used, namely, a photo-controlled CRISPR–Cas12a reaction60,61 and a dynamic aqueous multiphase reaction.62 One-pot assays that employ parallel NAA and CRISPR reactions can have simpler workflows but must be designed so that temperature, reagent concentrations, and reagent compositions are optimal for both reactions so that CRISPR activity cleaves NAA amplicons at a rate that does not markedly reduce the sensitivity or kinetics of the assay. One approach to address this is to slow the cis-cleavage rate through the use of suboptimal PAMs, although this can reduce assay sensitivity.63 A recent study proposed that amplicon cleavage in one-pot assays can be delayed by allowing Cas–crRNA complexes to form during the NAA reaction, instead of adding preformed complex.64 This approach exploited the slow association kinetics between Cas12a and its crRNA to reduce competition between the NAA and cleavage reactions by delaying cleavage of the target amplicon and was reported to significantly improve the detection limit. This new assay was also compared with a collection of NAAT–CRISPR–Cas one-pot diagnostics assays that increase sensitivity by mitigating the competition between amplification and detection (for detailed data please go to Table S1†).64
Detection of RNA targets in one-pot assays is further complicated by the need to integrate a reverse transcription step. RNA–cDNA heteroduplexes that remain after reverse transcription can also slow NAA initiation to decrease sensitivity, and Feng et al. have reported that adding RNase H to eliminate these RNA–cDNA hybrids can improve the sensitivity of RNA detection.65
3.2 Signal readout
Many different approaches have been used to couple reporter systems to Cas protein activity to enable sensitive detection or quantification of NA targets or other molecules from which NAs can be generated or released as indirect signals. Most of these assays generate optical or electrical signals. Each of these signal transduction methods has strengths and limitations, and the choice of method may depend on whether the assay will be used as a laboratory test or a POCT.Cas9-based assays that employ Cas9, dCas9, or nCas9 can also employ fluorescence for signal output. SYBR Green I fluorescence, induced upon the binding of this dye to dsDNA, has been used for real-time monitoring of amplified dsDNA products generated in nCas9 assays, including those produced by CAS-EXPAR30 and an nCas9-mediated strand displacement amplification approach.31 Cas9 assays can also utilize the Cas9-mediated cleavage of ssDNA reporters containing both fluorophore and quencher molecules to produce a fluorescent signal that is proportional to the assay target sequence. But this approach requires that these reporters be hybridized to the complimentary sequence region of a denatured target amplicon, which results in cleavage of both the reporter and its recognition sequence and thus does not allow for signal amplification, requiring that a sequence-specific reporter be designed and synthesized for each assay target.70
Bioluminescence and chemiluminescence reporter systems have also been incorporated into CRISPR assays to permit sensitive detection of assay target molecules. For example, one group developed an assay in which the small and large domains of a luciferase reporter protein were fused to dCas9 proteins. The fused products were then used to generate dCas9/crRNA complexes with different target specificities so that their dual binding to a dsDNA target produced a bioluminescent luciferase signal.25,71 An RT-RPA SARS-CoV-2 assay using this system was reported to detect SARS-CoV-2 viral RNA at ∼200 copies per μL within ∼20 min.71 Chemiluminescence has also been used to enhance the signal output of CRISPR-Dx assays. One group developed a portable CRISPR/Cas13a chip assay (PECL-CRISPR) in which Cas13a recognition of a target miRNA induced the cleavage of an NA construct to generate primer for an EXPAR amplification reaction, and used enhanced chemiluminescent (ECL) signal produced upon intercalation of the “light-switch” dye [Ru(phen)2dppz]2+ into the resulting amplicons as the assay readout.72 Another group developed a NAA-free Cas12 assay with an ECL readout to detect HPV-16 virus in undiluted human blood samples at an reported LOD of 0.48 picomolar. This group used an approach where target recognition cleaved a ferrocene-tagged thiolated ssDNA quencher to release it from the readout electrode and permit ECL signal production from L-methionine-stabilized gold nanoclusters conjugated to this sensor.73 Similarly, another group used a CuS nanoparticle (NP) with a biotinylated ssDNA tether to suppress a Cu2+ ion-catalyzed luminol–H2O2 reaction.74 In this assay, the recognition of a target miRNA by a complementary oligonucleotide permitted its ligation to serve as the substrate for an RCA reaction that produced a Cas12 target whose recognition induced the cleavage of the ssDNA tether of the CuS NP. Subsequent addition of AgNO3 then initiated a cation exchange reaction to induce the Cu2+ ion-catalyzed luminol–H2O2 reaction. Bioluminescence and chemiluminescence assays do not require incident light, unlike fluorescence assays that require specific excitation spectra, but instead require other reagents that have the potential to interfere with the reactions that lead to the production of the assay signal.
Real-time and endpoint fluorescent signals can be monitored by instruments, such as plate readers, fluorescent imagers, or spectrophotometers, commonly found in well-equipped research labs or clinical laboratories. Compact devices can also be used to excite the cleaved reporter molecules, producing a fluorescent signal that can then be detected by eye, by a smartphone camera, or by an integrated device. Such devices facilitate POC testing, and handheld readers or integrated systems (e.g., MASTR Pouch75) have been developed for this purpose.
The SHERLOCKv2 assay69 employs an LFA format in which ssRNA reporters with FAM and biotin tags are cleaved by Cas13a/crRNA complexes bound to the target RNA, while intact reporters are captured by streptavidin conjugated to the first line on the LFA strip and detected by gold nanoparticle (AuNP)-conjugated antibodies specific for FAM. Cleavage of the reporter prevents these (AuNP)-conjugated antibodies from binding and allows them to migrate to the second line where they are captured by protein A. Nanoparticle binding produces a colorimetric signal at these two strip regions, and the presence and relative abundance of the assay target is determined by the loss of colorimetric signal at the test line as it shifts to the control line. This LFA AuNP-signal approach has been applied to detect various targets, including SARS-CoV-2,57,76–78 HPV,79 and cytomegalovirus (CMV) and BK polyomavirus (BKV).80 A similar AuNP loss-of-signal readout approach has also been used to detect target-dependent Cas12 cleavage of an ssDNA reporter in proportion to target amplicon abundance.81,82 Finally, commercial pregnancy test strips have been employed to detect HPV and SARS-CoV-2, using an approach where target recognition by Cas12a cleaves a dsDNA oligonucleotide sequence that tethers a human chorionic gonadotropin (hCG) protein reporter to an NP that otherwise prevents its transit across, and detection by, the LFA test strip.11,83
AuNP loss-of-signal readout approaches are best suited to detect high abundance targets, since, with low abundance targets, it can be difficult to detect minor changes in colorimetric signals by eye or with LFA readout devices. Nonetheless, many different approaches have been employed as CRISPR-assay LFA readouts, including ones that produce positive colorimetric signal proportional to DNA target concentration. For example, one such approach used streptavidin to capture Cas9/crRNA complexes bound to biotinylated assay amplicons at the test line, where they were detected by their hybridization with AuNPs conjugated with horseradish peroxidase and an ssDNA complementary to an exposed crRNA hairpin region, while excess AuNPs were bound to an oligo containing the same recognition sequence that was immobilized at the control line.84,85 In both these LFAs, AuNP binding was detected by the colorimetric conversion of the peroxidase substrate 3,3′-diaminobenzidine to produce a colorimetric signal detectable by eye. Similarly, the binding of two dCas9/crRNA complexes to adjacent regions on a target sequence has been used to induce the dimerization of Cas9 fusion proteins containing the N- or the C-terminal domains of HRP to catalyze the oxidation of 3,3-5,5-tetramethylbenzidine (TMB) and produce a colorimetric signal that corresponds to target concentration.26
Several CRISPR assays have also used AuNPs to produce and/or regulate their assay signal readouts. For example, one assay used Cas9 cleavage activity and a nicking enzyme to produce target ssDNA for subsequent RCA. Then, it detected the colorimetric signal produced when AuNPs conjugated with ssDNA complementary to the target sequence bound these sequential amplicons to induce their aggregation and enhance their scatter effect.86 Another Cas12-based assay used a dsDNA reporter with an ssDNA overhang. This reporter connects AuNPs of different size (20 and 60 nanometer) at each of its termini and has a fluorophore placed immediately before the start of its ssDNA overhang region. Upon target recognition, colorimetric and fluorophore-induced signals can be produced.87 In this approach, energy transfer from the smaller of the two AuNPs could stimulate a fluorescent signal from the fluorophore. But this signal was quenched by the larger adjacent AuNP in the intact reporter molecule until its release by Cas12 trans-cleavage activity acting on the ssDNA reporter region following target detection, which also shifted the colorimetric signal produced by this reporter.
Cas/crRNA complex binding and cleavage activity can also be coupled to many different colorimetric reactions to produce assay readout signals suitable for target detection or quantification. For example, one group developed a Cas9-mediated SDA assay, where recognition of a concatenated target amplicon sequence by the ssDNA overhang region of a dsDNA oligonucleotide conjugated to a magnetic nanoparticle displaces the dsDNA region to release an ssDNA strand conjugated to glucose oxidase.51 The assay signal is then read by removing the magnetic beads and analysing the conversion of a colorimetric substrate by the activity of the glucose oxidase displaced from these beads.
3.3 NAA-free methods
Standard LFA and fluorescence-based CRISPR assays that do not employ NA preamplification steps may be limited to picomolar sensitivities due to the slow Cas enzyme kinetics and by background noise derived from nonspecific probe degradation or incomplete quenching systems that employ fluorophore-quencher probes for signal readouts. However, adding an NAA step increases the assay run time and the risk for non-specific amplification or cross-contamination. Combining isothermal amplification and CRISPR into one-pot assays can reduce cross-contamination, but this requires simultaneous optimization of both reactions, which increases their difficulty. Researchers have therefore pursued many strategies to develop NAA-free assays, as discussed in recent reviews.12,113,114One promising approach is to design a feedback circuit where signal production is coupled to Cas activity, as this can transform a linear Cas cleavage response into an exponential signal. One example of this approach is the CRISPR–Cas-only amplification network (CONAN).115 In this platform, recognition of the target DNA activates transducer 2, comprised of Cas12a and a dsDNA probe, and the resulting Cas12a trans-cleavage activity liberates a caged crRNA that can then target transducer 2, creating a positive feedback circuit that generates an exponentially increasing fluorescent signal. This approach may be promising for POCTs since it employs a single enzyme, requires one 37 °C reaction step, and can reportedly achieve attomolar detection sensitivity. Autocatalysis-driven feedback amplification strategies have also been reported for Cas13-based assays for RNA targets that use hairpin RNA116 or a three-stranded RNA probe117 to mediate signal amplification.
Another popular approach to overcome the Cas kinetics limitation is to employ picoliter or femtoliter droplets or a microchamber to reduce the reaction volume and thus effectively increase the reactant concentration.118–122 Such assays can provide greater sensitivity and quantitative results for targeted molecules, but setup of their spatially confined platforms requires additional equipment and training. To make such assays suitable for low-resource settings, researchers must integrate the reaction setup and signal readout into a user-friendly microfluidic system. Other methods, such as protein engineering for faster kinetics23 and using multiple Cas/crRNA complexes to detect different regions of the target,58,123,124 have also been explored for NAA-free assays.
As discussed in 2.1.5, there is also controversy regarding LODs attained by some assays that do not use preamplification steps, as some reported LODs were deemed to be physically impossible.47 NAA-free assays are frequently reported at the early stages of development. However, future assays, especially those intended for use in low-resource settings, need to report their adherence to the REASSURED criteria and have their performance verified against a reference assay. For example, a a one-step NAA-free Cas12a-aptamer fluorescent detection strategy for prostate-specific antigen (PSA) was compared to enzyme-linked immunosorbent assay (ELISA), surface plasmon resonance, ECL, SERS, electrochemistry, and radioimmunoassay approaches to evaluate their relative performance, procedure times and complexity and sample pre-treatment and equipment requirements.125 The study reported that the LODs and working ranges for this assay and ELISA were 0.16 ng ml−1 and 0.35–5 ng ml−1 versus 0.19 ng ml−1 and 0.31–20 ng ml−1, respectively.
3.4 Sample processing
Diagnostic assays can analyse a variety of specimen types (e.g., nasopharyngeal swabs, blood, saliva, and urine), which must often be processed prior to their analysis. Traditional NA extraction approaches can be cumbersome and time-consuming, and they can also introduce a contamination risk. Some CRISPR applications do not require purified NA samples, however, and simple NA release protocols, such as the widely adopted heating unextracted diagnostic samples to obliterate nucleases (HUDSON) method21,126—which uses heat and chemicals to inactivate sample nucleases and lyse viral particles—have been developed to streamline sample processing for these assays. As an alternative, other studies have used proteinase K to degrade sample proteins and enhance the release of NA targets.127 Such streamlined processing methods are not appropriate for all assays and targets, since NA purification and concentration steps are often needed to sensitively detect low-concentration NA targets in complex samples. NA extraction methods that use magnetic beads56 or membranes128 as affinity matrices for NA capture have been developed as rapid NA isolation and concentration methods suitable for use with, or integration into, CRISPR assays, including assays where the NA extraction and purification steps are integrated into microfluidic devices.129–1313.5 Multiplex CRISPR assays
Multiplexed CRISPR-Dx assays could have substantial value for clinical applications, since it is often necessary to simultaneously monitor several biomarkers in a sample to obtain accurate information for diagnosis and treatment decisions or to screen for multiple target pathogens in epidemiologic surveillance studies, but these assays are subject to technical challenges. Multiple Cas/crRNA complexes can be used in one assay to recognize different targets, but sensitive detection methods often employ Cas trans-cleavage activity for signal amplification, which can limit the discrimination of signals arising from different markers in a sample. SHERLOCKv2 has attempted to address this issue by employing four Cas proteins (PsmCas13b, LwaCas13a, Cca-Cas13b, and AsCas12a) that have different preferences for NA targets (ssDNA or dsDNA) and trans-cleavage sequences (poly AU, UC, AC, or GA),69,132 although this approach is limited by the number of Cas proteins with desired target and substrate preferences and thus is not scalable. Massively multiplexed parallel detection can, however, be achieved by using an array of crRNAs with a single Cas protein type in a microfluidic-based labelled nanodroplet assay via the combinatorial arrayed reactions for multiplexed evaluation of nucleic acids (CARMEN) platform, which can test more than 4500 crRNA-target pairs on a single array.133 This assay causes nanodroplet pairs containing CRISPR detection reagents and amplified targets to fuse in a microwell array, detects the specific fluorescent dyes added to each distinct Cas/crRNA and amplification reaction to identify the nanodroplet pairs present in each well, and reads the fluorescent signal produced by Cas13a trans-cleavage of the reporter in each well. This assay was used to detect and distinguish 169 human viruses, including SARS-CoV-2 variants, in eight samples. A subsequent study developed a revised assay, which detected a panel of 21 respiratory viruses including SARS-CoV-2, other coronaviruses, and individual influenza H and N subtypes, and also found that this approach could identify an array of HIV mutations associated with drug resistance.134 Other multiplex assays using spatially separated reactions have also been designed to detect HPV,67 miRNAs,70,91 foodborne pathogens,135 SARS-CoV-2,136 and other targets.3.6 Microfluidic-based CRISPR assays and wearable devices
Most if not all CRISPR-Dx procedures, including steps of NA extraction, amplification, Cas reaction, signal transduction, and readout, can be integrated into custom microfluidic devices, but this has been recently reviewed elsewhere7,8,137 and will not be discussed in detail here. Notably, however, a few CRISPR-Dx assays have been fully integrated into wearable devices.138,139 One group developed a face mask with a lyophilized CRISPR sensor, designed to detect SARS-CoV-2 in respiratory droplets, consisting of a push-button controlled fluid reservoir; an aerosol collection membrane; a sample processing area containing spatially separated lyophilized lysis, RT-RPA, and CRISPR reagents; and an LFA signal readout sensor.139 In this device, pressing the button on the fluid reservoir caused captured material to flow through the lyophilized reagents to initiate a room-temperature RT-RPA CRISPR reaction that could be read within 90 min and had a SARS-CoV-2 detection limit comparable to that of qPCR. Another group designed a wearable CRISPR microneedle patch that could directly interact with the interstitial fluid of the skin to allow extended (10 day) online detection and monitoring of Epstein–Barr virus (EBV) DNA and changes in cfDNA associated with tissue injury accompanying sepsis and donor kidney rejection.138 The patch detected changes in the electric current in the microelectrode upon interaction of a microneedle dCas9/crRNA-activated graphene interface with its NA target. These wearable approaches may have significant potential for rapid, real-time monitoring of patients at risk for specific disease conditions and could possibly assess the effectiveness of therapeutic approaches initiated to treat these conditions.4 Application of CRISPR-based molecular diagnostics for infectious diseases
4.1 COVID-19
CRISPR-Dx assays have the potential to become POCTs that provide rapid and accurate results for on-site screening and diagnosis.140 One successful example is the use of CRISPR in the detection of SARS-CoV-2 viral RNA (Fig. 3) during the COVID-19 pandemic, as has been reviewed elsewhere.141,142 One group employed the Cas13-based SHERLOCK system to detect NAA of SARS-CoV-2 viral gene targets (spike, nucleoprotein [N], and replicase polyprotein 1ab) on a lateral-flow readout system with an internal control to detect ribonuclease contamination events that could produce false-negative results.143 This LFA had an approximate 1 hour sample-to-result time, a reported LOD of 42 copies per reaction, a diagnostic sensitivity of 97%, and a specificity of 100% upon visual inspection of its colorimetric signal. A subsequent NAA-free fluorescent CRISPR Cas13a assay, read by a mobile phone microscope, reported an LOD of 100 copies per μL after 30 min,144 but this result was questioned due to its unlikely kinetics and high senstivity.47 Similarly, another group developed a RT-LAMP and CRISPR Cas12a-based DETECTR assay targeting the SARS-CoV-2 nucleoprotein E and N genes, which had a 45 min sample-to-result time and an LOD of 10 copies per μL.76 Initial tests used nasal swab samples, but CRISPR/Cas12a-based assays work not only for nasal swab samples145 but also for longitudinal plasma samples146 and for multiple systemic tissues,147 having a versatility that improves diagnosis. A saliva test was also developed; this smartphone-read CRISPR/Cas12a-based test for SARS-CoV-2 subsequently demonstrated a 15 min sample-to-result time and an LOD of 0.38 copies per μL, lower than the LOD of the RT-PCR reference assay.148 Subsequent CRISPR assays for SARS-CoV-2 were also developed using crRNAs that targeted signature mutations of the alpha, beta, delta, and omicron SARS-CoV-2 variants.149–151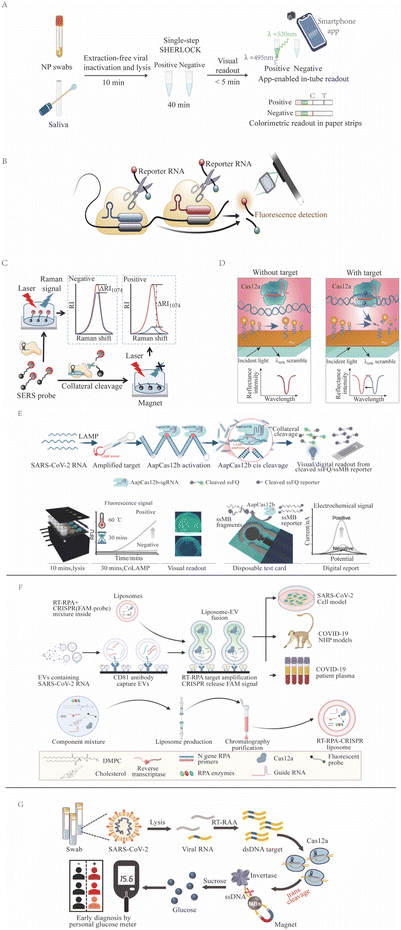 | ||
| Fig. 3 CRISPR-based SARS-CoV-2 detection methods. (A) SHERLOCK employs Cas13a target recognition to cleave a reporter, detecting signal from unextracted diagnostic samples (adapted from ref. 126 with permission). (B) An NNA-free Cas13-based assay employs multiple CAS13/crRNA complexes with different specificities to enhance target signal (adapted from ref. 144 with permission). (C) SERS-CRISPR assays use Cas12a recognition of a DNA target to cleave a quenched reporter and yield SERS signal (adapted from ref. 152 with permission). (D) A Cas12a assay where target recognition is used to cleave a modified ssDNA reporter bound to a sensor to induce a surface plasmon resonance signal (adapted from ref. 94 with permission). (E) CoLAMP assays employ AapCas12a recognition of a LAMP-amplified target to induce trans cleavage of quenched reporters, which can produce fluorescent or electrochemical assay readouts (adapted from ref. 153 with permission). (F) A liposome-fusion RT-RPA CRISPR assay that detects SARS-CoV-2 RNA-positive extracellular vesicles in plasma after their affinity capture on an assay well surface (adapted from ref. 154 with permission). (G) A Cas12a assay employs target recognition to cleave a ssDNA tether linking invertase to a magnetic bead. After magnetic bead removal, invertase-mediated conversion of sucrose to glucose can be read as an assay signal with a personal glucose meter (adapted from ref. 106 with permission). | ||
Several one-pot SARS-CoV-2 CRISPR-Dx assays have also been developed as POCTs that have also demonstrated lower limits of detection than the RT-PCR reference assays. For example, the RT-free one-pot enzyme-catalyzed RCA-assisted CRISPR/FnCas12a detector (OPERATOR) assay155 integrated ligation-mediated RCA and FnCas12a reactions with a fluorescent LFA readout to achieve a 30 min sample-to-result time with an reported LOD of 0.081 copies per μL. The CRISPR-based one-pot loop-mediated isothermal amplification (CoLAMP) assay reported slightly lower sensitivity (0.5 copies per μL with a 40 min sample-to-result time), but balanced the LAMP and CRISPR reactions to deplete the assay target and reduce the risk for subsequent aerosol contamination.156
4.2 Tuberculosis
Several CRISPR assays have been developed to diagnose tuberculosis (TB), which remains a leading cause of death worldwide but is significantly underdiagnosed by current screening approaches and assays.157 Most individuals who develop Mycobacterium tuberculosis infections after exposure to TB develop an asymptomatic latent TB infection that can progress to active pulmonary TB or extrapulmonary TB disease at any point in their lifetime,158 and current screening methods have limitations, making it difficult to identify and treat new TB cases (Table 1). In 2023, the WHO recommended that TB household contacts, HIV-infected individuals, and other high-risk groups be screened for TB by rapid TB diagnostics (e.g., Xpert MTB/RIF and Xpert Ultra).159 But these molecular diagnostic tests are expensive and difficult to implement in low-resource settings. Rapid, sensitive, and user-friendly POCTs are thus urgently needed to improve TB diagnosis and identify drug- resistant TB cases. CRISPR-based molecular diagnostics integrated into POCT platforms have the potential to meet this demand (Fig. 4), and several methods have been developed to date that have sample-to-result times of 2 hours or less and limits of detection that are similar to, or lower than, the Xpert reference assay (Table S1†). The sensitivity estimates of these assays range from 79% to 97.2% and the specificity estimates, from 95.2% to 100%, approaching or exceeding those obtained with Xpert. One clinical study is evaluating a CRISPR-based test using sputum or bronchoalveolar lavage fluid from individuals with suspected pulmonary TB (NCT04074369). CRISPR-based TB diagnostics can also be applied to analyse serum and plasma samples, with at least one study reporting that testing of such samples can effectively diagnose extrapulmonary TB, paucibacillary TB, and paediatric TB cases with and without HIV-1 coinfection, which are challenging cases to diagnose by the current reference methods.160 Notably, serum levels of M. tuberculosis cfDNA decreased after treatment initiation in this study, suggesting the CRISPR-Dx assays that detect M. tuberculosis cfDNA levels in serum could not only increase the coverage of TB screening efforts, but also permit rapid evaluation of treatment efficacy.| Diagnostic methods | Advantages | Limitations | Ref |
|---|---|---|---|
| Abbreviations: AFB: microscopic examination for acid-fast mycobacteria bacilli; IGRA: interferon-gamma release assay; WGS: whole genome sequencing; NGS: next generation sequencing; LAMP: loop-mediated isothermal amplification; CRISPR: clustered regularly interspaced short palindromic repeats; BCG: bacillus Calmette–Guérin; cfDNA: cell-free DNA; POCT: point-of-care testing. | |||
| Mtb culture and drug susceptibility testing | • Gold standard | • Time consuming: 10–21 days | 161 |
| • High specificity >99% | |||
| AFB staining | • Rapid, practical | • Low sensitivity | 162 |
| • Sputum | • Unable to differentiate different strains | ||
| Tuberculin skin test | • Low cost | • Cannot distinguish latent and active TB | 163 |
| • Widely used, practical | • Low sensitivity in an immune-compromised individual | ||
| • Influenced by BCG exposure or other atypical mycobacteria | |||
| • 2–3 days | |||
| IGRA | • Blood | • High cost | 164 |
| • No BCG influence | • 2–3 days | ||
| PCR: line probe assay | • Multidrug resistance | • High cost | 165, 166 |
| • Sputum | • Low sensitivity in paucibacillary patients and non-respiratory samples | ||
| WGS or NGS | • Drug resistance | • High cost | 167 |
| • 3 days | |||
| • Need well-trained experts | |||
| LAMP: nucleic acid amplification test | • High specificity and sensitivity | • Low sensitivity in paucibacillary patients | 168, 169 |
| • Rapid, practical | |||
| • POCT | |||
| CRISPR-based detection | • Rapid, practical | • Need further validation and POCT development | 160 |
| • POCT | |||
| • Blood cfDNA | |||
| • Available for EPTB and immune-compromised individuals | |||
| • High sensitivity | |||
| • Low cost | |||
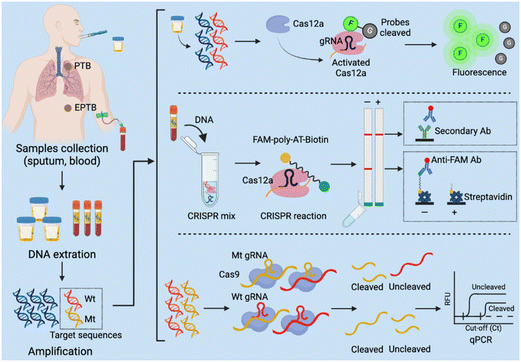 | ||
| Fig. 4 CRISPR-based diagnostic (CRISPR-Dx) assays for Mycobacterium tuberculosis detection. PTB: pulmonary tuberculosis; EPTB: extrapulmonary tuberculosis. | ||
Rapid molecular diagnostics are also needed to improve screening efforts to detect and treat the increasing number of drug-resistant and multidrug-resistant TB cases,157 and CRISPR diagnostics could play a significant role in meeting this need. For example, a recent study described a Cas9/crRNA-assisted quantitative real-time PCR (CARP) assay designed to directly detect SNPs at two sites in the rifampicin-resistance area that are mutated in 60% to 86% of rifampicin-resistant TB cases.170 Another group used a multiplex Cas9-based CRISPR reaction to improve the analysis of 52 candidate genes associated with resistance to first- or second-line anti-TB drugs. This group used crRNAs specific for these genes to cleave the sequences into fragments appropriately sized for an efficient next-generation sequencing analysis to improve the sequencing depth of these target sequences.171
4.3 Other infectious diseases
To diagnose other infectious diseases, groups have developed CRISPR assays such as LAMP-Cas12a172 and recombinase-aided amplification (RAA)-Cas13a assays173 that detect HBV DNA in clinical samples and a multiplex RPA-Cas12a/Cas13a assay that simultaneously detects the F3L and B6R genes of monkeypox virus via Cas12a and Cas13a cleavage of differently labelled ssDNA and ssRNA reporters.174 Multiplexed and portable CRISPR assays have also been developed for SARS-CoV-2, dengue virus, Zika virus, and influenza A and have streamlined workflows that render them suitable for POCTs, and laboratory-based CARMEN microwell assays can perform 1000 tests per chip and detect 169 human viruses.1334.4 CRISPR-Dx assays for low-resource settings
POCT CRISPR-Dx assays intended for use in remote or low-resource settings may have requirements not shared by similar tests designed for use in well-equipped clinical laboratories. Assays intended for low-resource settings are normally optimized to reduce material expenses; storage, equipment, and operator requirements; and may also employ decentralized reporting strategies (i.e., connect with a portable smart diagnostic device or smartphone) to facilitate telehealth and epidemiologic efforts. Adapting clinical laboratory CRISPR-Dx assays for use as POCTs in low-resource settings usually involves trade-offs between assay performance and cost, portability, or other considerations that can affect their overall utility. For example, POCTs intended for use in low-resource settings usually employ lyophilized reagents to eliminate or minimize cold-chain requirements during transport and storage, but this comes at the cost of some loss of reagent activity and assay sensitivity. Thus, such trade-offs may produce POCTs that exhibit lower sensitivity and specificity than laboratory-based tests but still retain significant diagnostic value, particularly in the absence of a practical alternative tests. For example, the Cas13-based streamlined highlighting of infections to navigate epidemics, version 2 (SHINEv.2) SARS-CoV-2 assay was designed for use outside centralized laboratories, as it had minimal sample processing and no instrument requirements but a reported LOD of 200 copies per μL.175 This assay was also reported to achieve a 90.5% sensitivity 100% specificity when used on unextracted nasopharyngeal swab samples, comparable to rapid antigen tests used to diagnose SARS-CoV-2 infection.Cost can influence choices for all steps in a POCT workflow, from sample preparation to signal readout. These choices include the selection of assay reagents and the materials and fabrication processes for disposable assay cartridges and their portable readers. One study estimated that per sample costs of several CRISPR- and PCR-based diagnostics intended for use in low-resource settings range from US$7 to $20, although it was not clear if the estimate included laboratory instrumentation and infrastructure costs.176 Several groups have attempted to develop inexpensive CRISPR-Dx POCTs. For example, one study described LAMP/CRISPR HPV assay that employed a gold leaf electrode to obtain a sensitivity of 104 copies per test and cost ∼$2.30 per test.177,178
The minimally instrumented SHERLOCK (miSHERLOCK)128 POC saliva assay for SARS-CoV-2 and its mutations was reported to have a LOD of 1240 copies per mL and cost $15 per test, although this could be reduced to $11 per test by reusing its electronics and heaters. The major assay expense (∼$9) derived from its use of commercial enzymes (RPA and reverse transcriptase) which should be reduced at scale. Another SARS-CoV-2 assay, the Cas9-based FnCas9 editor linked uniform detection assay (FELUDA), had a reported $7 per test cost,179 but used PCR for target amplification and thus required a thermocycler.
Incorporating CRISPR assays into microfluidic devices made using inexpensive materials and manufacturing methods can substantially reduce assay costs. For example, one study has reported that a compact, fully-integrated thermoplastic cartridge can function as an automated device to perform a CRISPR-based SARS-CoV-2 assay,129 and most assay costs derived from the reagents ($7.84) rather than this cartridge ($0.30). However, this assay was still read by a relatively large and expensive portable device, which could limit its utility in low-resource settings. Several groups have attempted to address similar instrumentation issues by using inexpensive portable devices, including smartphone cameras or smartphone-coupled devices, as assay readers. For example, one group developed a finger-actuated microfluidic biosensor with a one-pot multiplex NAA/CRISPR assay for seven common foodborne pathogens where the assay's fluorescent signal was captured and analysed with a smartphone device and app.135 This assay had a reported LOD of <500 CFU mL−1 with a 1 hour sample-to-result time, a microfluidic chip per-test cost of <$4, and an assay device cost of $150, excluding the smartphone itself. Microfluidic/electrochemical paper-based analytical devices are also of interest because paper is inexpensive, portable, and disposable,180,181 although high-quality paper may be costly in low-resource settings.
Table 2 summarizes efforts to develop assay workflows suitable for use in low-resource settings.175 However, there are currently no commercial CRISPR-Dx POCTs approved for use in low-resource settings. SHERLOCK- and DETECTR-based assays for SARS-CoV-2 were authorized by the FDA for emergency use, as was the Cas9-based FELUDA179 in India, but these assays are for certified laboratories only.
| Objective | Effort | Representative studies |
|---|---|---|
| Abbreviations: NA: nucleic acid; PES: polyethersulfone, NAA: nucleic acid amplification. | ||
| Simplified sample processing | Heat-mediated nuclease inactivation and NA release | Myhrvold21 Arizti-Sanz126 |
| Magnetic bead-assisted NA purification | Joung56 | |
| PES membrane-mediated NA concentration | de Puig128 | |
| Integrated microfluidic sample processing | Chen,129 Wu 2021 (ref. 130) | |
| Simplified assay workflow | One-pot amplification and detection | Ali57 Li24 Ding59 Chen60 Feng65 |
| NAA-free assay approach | Shi115 Yang23 | |
| Simplified signal readout | Lateral flow assays | Gootenberg69 Mukama81 Tang11 |
| Colorimetric assays | Qiu26 Gong51 Hu84 Wang85 | |
| Fluorescence assays with minimal instrumentation | de Puig128 Ning148 Wang182 | |
| Electrochemical assays | Zeng183 Han184 | |
| Improved temperature stability | Lyophilized reagents | de Puig128 Nguyen139 Arizti-Sanz175 |
| Affordability | Reduced instrument cost | de Puig128 Wang182 |
| Reduced consumable expenses | Zamani185 Chen129 | |
5 CRISPR-based diagnostics for early detection of cancer
Early cancer detection leads to early intervention and improves survival rates, as seen with successful screening strategies for cervical, breast, and colorectal cancers. However, there were still 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 958
958![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 310 new cancer cases and 609
310 new cancer cases and 609![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 820 cancer-related deaths in the United States in 2023,186 and there are expected to be 28.4 million new cancer cases worldwide in 2040.187 Further, about 50% of cancers are diagnosed at a late stage where the odds of long-term survival are substantially worse.188 It is therefore crucial to implement cancer prevention and early detection strategies, including those that can effectively serve low-resource settings.
820 cancer-related deaths in the United States in 2023,186 and there are expected to be 28.4 million new cancer cases worldwide in 2040.187 Further, about 50% of cancers are diagnosed at a late stage where the odds of long-term survival are substantially worse.188 It is therefore crucial to implement cancer prevention and early detection strategies, including those that can effectively serve low-resource settings.
CRISPR-Dx POCTs have shown promise for infectious disease diagnosis and have great potential for cancer, as they could help identify high-risk populations to facilitate cancer prevention and screening efforts in low-resource settings. For example, at-home HPV self-sampling could be a future option for cervical cancer screening, but this approach still requires that samples be sent to a lab. This shortcoming could be addressed by the development of at-home tests for early cancer detection, similar to those developed during the COVID-19 pandemic.
Rapid turnaround time is not as essential for cancer diagnosis as it is for infectious disease diagnosis but developing accurate and low-cost assays with minimal training and equipment requirements will be necessary to permit early cancer screening efforts in large populations in low-resource settings. We discuss some potential CRISPR application scenarios below.
5.1 Gene mutations
Early-stage cancer diagnosis remains a major challenge for cancer prevention, partly because cancer screening diagnostics have low specificity and sensitivity for protein biomarkers. NA biomarkers (e.g., circulating tumor DNAs [ctDNAs] and miRNAs) have demonstrated potential as biomarkers for targeted therapy, immune-checkpoint inhibitor treatment, and early cancer diagnosis, but their low concentrations can lead to high false-negative rates when analysed by current detection methods. CRISPR-based assays (Table 3) can enhance sensitive detection of NA targets and potentially distinguish alternate SNPs, and at least one group has employed restriction enzymes to provide target specificity. This excision-amplification-synchronous Cas12a-targeted checkout (EasyCatch) assay approach integrates a restriction enzyme to cleave the wild-type site during RPA-mediated amplification of the NA target sequence, allowing preferential amplification of rare mutant alleles against a high wild-type allele background (0.001% mutant) to permit rapid (<1 h) and sensitive detection of these mutations in cancer samples.189 Notably, this approach was used to detect two EGFR mutations (e19del and L858R) and several FLT3 mutations (D835Y/H/V/F) associated with resistance to tyrosine kinase inhibitors, indicating its potential to inform cancer treatment decisions. Cas9- and Cas12a-based biosensing can successfully detect EGFR T790M (93.9% sensitivity and 100% specificity) and L858R mutations in plasma and tissues from patients with lung cancer.155,190 Given the ability of CRISPR-based assays to detect SNPs and other mutations associated with specific cancers, cancer subtypes, and drug resistance phenotypes, there is significant potential for the development of CRISPR-based POCTs for common cancer-associated NA biomarkers.| Cas effector | Target | NAA and/or enrichment | Specimens | Cancer | LOD | Sensitivity/specificity | Ref |
|---|---|---|---|---|---|---|---|
| Abbreviations: RT-RAA: reverse transcriptase-recombinase aided amplification; PCR: polymerase chain reaction; ICP: ion concentration polarization; NSCLC: non-small cell lung cancer; HNSCC: head and neck squamous cell carcinoma; AML: acute myeloid leukemia; cfDNA: cell-free DNA; RPA: recombinase polymerase amplification; RE-RPA: restriction enzyme integrated RPA. | |||||||
| Cas9 | PCA3 and KLK3 | RT-RAA | Urine and peripheral blood | Prostate cancer | 500 and 50 fg μL−1 LNCaP cell RNA | — | 191 |
| Cas9 | EGFR mutants | PCR | Blood | Lung cancer | 10−3–10−4 DNA dilution | — | 192 |
| Cas9 | EGFR T790M mutant | PCR | Plasma | NSCLC | <10 copies per mL plasma | 93.9%/100% | 190 |
| Cas9 | EGFR L858R mutant | ICP | Synthetic samples | — | 10−3 target DNA dilution | — | 193 |
| LbCas12a | EGFR L858R mutant | Isothermal amplification | Lung cancer tissues | NSCLC | 0.3 copies per μL (0.498 aM) in mock multiplex cfDNA samples | — | 122 |
| LbCas12a | TP53 R273 mutations | PCR | Cell lines HNSCC biopsies | HNSCC | 3–6% target DNA dilution | — | 194 |
| Cas12a | FLT3 D835 mutations | RE-RPA | Blood | AML | 10−5 target DNA dilution | — | 189 |
| Cas13a | EGFR L858R | RPA | Mock cfDNA samples | — | 10−3 target DNA dilution | — | 3 |
| BRAF V600E | |||||||
| Cas13, Cas12a, and Csm6 | EGFR L858R | Synthetic and liquid biopsy cfDNA samples | NSCLC | — | — | 69 | |
| EGFR exon 19 deletion | |||||||
| EGFR T790M | |||||||
| APC:c.1262G>A | |||||||
5.2 Oncogenic viral infections
Persistent HPV infections can increase the risk for several cancers, including cervical cancer and head and neck cancers.195,196 According to The Global Cancer Observatory (GLOBOCAN) 2020, the incidence and mortality of cervical cancer remain high in multiple countries.197,198 And the WHO now recommends using assays that detect HPV DNA as a primary test in cervical cancer screening and treatment approaches.195 Streamlined and inexpensive HPV POCTs are thus required to achieve the WHO's screening and treatment targets and to help eliminate cervical cancer as a public health problem.Several NAA-based and NAA-free CRISPR-based assays have been reported as potential means to detect HPV subtypes in clinical specimens (Table 4). LODs reported for the NAA-based (RPA, PCR, and RAA) CRISPR assays tended to cluster near 1 attomolar, although other reported LODs corresponding to 1 copy/reaction values for their assay volumes (0.03 to 0.26 attomolar) or substantially higher values (240–664 attomolar). Studies that provided results for non-NAA controls reported LODs in the low picomolar range (3–50 picomolar), with a few in the nanomolar range (0.5–10 nanomolar).
| Effector Cas protein | HPV subtypes | LOD | Reported assay time | NAA method | Readout | Sensitivity specificity | Clinical sample | Ref |
|---|---|---|---|---|---|---|---|---|
| Reported assay times: only one study specifically reported the assay sample-to-answer time (*). Abbreviations: Cas: CRISPR-associated protein; HPV: human papillomavirus; PCR: polymerase chain reaction; HRCA: hyperbranched rolling circle amplification; dCas9: deactivated Cas9; HCR: hybridization chain reaction; LbCas12a/LbaCas12a: Lachnospiraceae bacterium ND2006 Cas12a; RPA: recombinase polymerase amplification; LAMP: loop-mediated-isothermal amplification; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; LFA: lateral flow assay; LtCas12a: ≤80% sequence similarity with LbCas12a and AsCas12a (Acidaminococcus sp. Cas12a), 1296 amino acids, MW: 152.7 kDa; RAA: recombinase-aided amplification; PB19: Parvovirus B19; HBV: hepatitis B virus; ICP-MS: inductively coupled plasma mass spectrometry; ECL: electrochemiluminescence; LbuCas13a: Leptotrichia buccalis Cas13a. | ||||||||
| Cas9 | HPV16,18 | — | 3–4 h | PCR | Fluorescence | — | Cervical mucus exfoliated cells | 196 |
| Cas9 | HPV16,18 | — | 3 h | PCR | Fluorescence | — | Cervical mucus exfoliated cells | 199 |
| Cas9 | HPV16, 18, 33, 35, 45, 51, 52, 56, 58, 59 | — | 2 h | PCR | Fluorescence | — | Cervical mucus exfoliated cells | 200 |
| Cas9 | HPV16, 18, 33, 35, 45, 51, 52, 56, 58, 59 | — | 2–3 h | PCR | Fluorescence | — | Cervical mucus exfoliated cells | 201 |
| Cas9 | HPV16, 18, 6, 11, 33, 35, 40,45, 51 | — | 90 min | HRCA | Visual, fluorescence | — | Cervical mucus exfoliated cells | 202 |
| dCas9 | HPV16,18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66,68, 73 | — | 30 min | HCR | Fluorescence | — | Cervical sample | 203 |
| LbCas12a | HPV16, 18 | No-RPA: 10 pm | 1 h | RPA | Fluorescence | — | Anal swab | 4 |
| RPA: 1 aM | ||||||||
| LbCas12a | HPV16 | 280 aM | 3 h | RPA | Fluorescence (using LFA) | — | Plasma | 79 |
| HPV18 | 240 aM | |||||||
| LbaCas12a | HPV16 | 10 copies (0.83 aM) | 1 h | RPA | Fluorescence | — | Plasma, cervical swab | 62 |
| HPV18 | 100 copies (8.3 aM) | |||||||
| Cas12a | HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68 | 100 copies (13 aM) to 500 copies (66 aM) | 35 min | RPA | Fluorescence | — | Cytological scrape | 204 |
| LbCas12a | HPV16 | No sensor: 100 pM | — | RPA | Fluorescence, electrochemical | — | Vaginal swab | 205 |
| Sensor: 1 pM | ||||||||
| LbCas12a | HPV16,18 | No-PCR: 50 pM | — | PCR | Fluorescence, LFA | 94.7%/100% | Anal swab | 66 |
| PCR: 1 copy per rxn (0.1 aM) | ||||||||
| Cas12a | HPV16,18 and HIV | 104 copies (664 aM) | — | LAMP | Electrochemical | 100%/89% | Cervical swab | 177 |
| Cas12a | HPV16 | No RPA: 3 pM | — | RPA | Chemiluminescent, visual image | 88.9%/100% | Clinical samples | 206 |
| RPA: 1 copy per rxn (0.03 aM) | ||||||||
| LbCas12a | HPV6, 11, 16, 18, 31, 33, 45, 52, 58 | 0.26 aM | 40 min | RPA | Fluorescence | 97.8%/ | Cervical swab | 67 |
| 98.1% | ||||||||
| LbCas12a | HPV16 | No RPA: 17 pM | — | RPA | Chemiluminescent, visual image | 87.5%/100% | Clinical samples | 207 |
| RPA: 1 copy per rxn (0.09 aM) | ||||||||
| LbCas12a | HPV16 | No RPA: 50 pM | — | RPA | Colorimetric signal on the PTS | — | Vaginal or urethral discharge | 11 |
| RPA: 2 copy per uL (3.3 aM) | ||||||||
| SARS-CoV-2 | ||||||||
| LbCas12a | HPV16,18 | No RPA: 20 pM | 30 min | RPA | Fluorescence | 92.3%/100% | Cervical cell specimen | 208 |
| RPA: 1 aM | ||||||||
| LbCas12a | HPV16,18 | No RPA: 0.5 nM | 30 min* | RPA | Fluorescence, (microchip+LFA) | — | Cervical swab sample | 209 |
| RPA: 1 aM | ||||||||
| LbaCas12a | HPV16,18 | No-RPA: 10 nM | — | RPA | Fluorescence | — | Cervical brush specimen | 210 |
| RPA: 2 aM | ||||||||
| Cas12a | HPV16,18 | 1 aM | 80 min | RPA | Fluorescence | 100%/93.8% | Clinical samples | 211 |
| LtCas12a | HPV16,18 | 30 copies (2 aM) | — | RAA | Fluorescence, LFA | 90% /96.2% (16) | Sanitary napkin blood samples | 212 |
| 92.3% (18) | ||||||||
| Cas12a | HPV16,18 | 1 copy per μL (2 aM) | — | RAA | Fluorescence | 97.1%/100% | Cervical epithelial tissue | 213 |
| Cas13a | ||||||||
| LbCas12a | HPV16 | 50 pM | — | No NAA | Electrochemical | — | No | 88 |
| PB19 | ||||||||
| LbaCas12a | HPV16 | 0.1 pM | — | No NAA | Electrochemical | — | No | 214 |
| LbaCas12a | HPV16 | 3.2 pM | 50 min | No NAA | Electrochemical | — | HPV-spiked serum | 215 |
| Cas12a | HPV16 | 1.6 pM | — | No NAA | Photocurrent | — | No | 216 |
| Cas12a | HPV16 | 1.2 pM | — | No NAA | Photocurrent | — | Cervical brush samples | 183 |
| Cas12a | HPV16 | 1 pM | — | No NAA | Photocurrent | — | HPV-spiked serum | 217 |
| Cas12a | HPV16,18,52 | 0.22 pM | — | No NAA | ICP-MS signal | — | Cervical swab sample | 218 |
| Cas12a | HPV16 | 42.9 pM | 30 min | No NAA | Lateral flow biosensor | — | No | 219 |
| HPV18 | ||||||||
| 0.21 pM | ||||||||
| Cas12a | HPV16 | 3.2 fM | — | No NAA | ECL | — | No | 220 |
| Cas12a | HPV16 | 8.9 fM | 100 min | No NAA | ECL | — | HPV-spiked serum | 221 |
| dCas9 | HPV 16 | — | — | No NAA | Raman spectroscopy | — | No | 222 |
| Cas12a | HPV16,18 and HBV | 1 aM | 20 min | No NAA | Raman spectroscopy | — | No | 223 |
| LbaCas12a | HPV18 | DNA 10 aM | 30 min | No NAA | Fluorescence (digital droplet) | 100%/100% | Cervical epithelial cells | 224 |
| LbuCas13a | SARS-CoV-2 | RNA 100 aM | ||||||
Most NAA-free CRISPR assays reported LODs in the low picomolar range (0.1–50 picomolar), consistent with the non-NAA controls of the NAA assays. Lower LODs were reported for assays employing electrochemoluminescent (ECL, 3.2–8.9 femtomolar), digital droplet (10–100 attomolar) and Raman spectrometry (1 attomolar) readouts, but these results appear feasible given the high sensitivity of each of these approaches.
Multiple NAA approaches have been used for the HPV assays. For example, one group developed a CRISPR-associated hyperbranched RCA technique (CART) assay that used two Cas9/crRNA complexes, one specific for the L1 region of HPV16 and the other, for the L1 region of HPV18. These complexes excised the intervening DNA region, which was then ligated to generate a circular DNA target for an isothermal RCA reaction; the amplified target was then detected by agarose gel electrophoresis or a fluorescent signal produced upon dye intercalation.205 Another group subsequently developed a CRISPR-typing PCR (ctPCR) HPV assay that employed two Cas9/crRNA complexes recognizing conserved regions in the HPV genome. These complexes excised an NA target region, which was then ligated to linkers containing universal primer sequences to allow multiplex amplification of the corresponding region of multiple HPV strains for subsequent analysis.201 Several variants of this assay approach have been developed (ctPCR 2.0, 3.0 and 4.0).196,200,202 But neither the CART nor the ctPCR assays were further adapted to formats suitable for POCTs. Cas9 may also not be the best candidate for a CRISPR-based HPV diagnostic due to its potential for off-target cleavage events.225 A Cas12a-based DETECTR assay, however, could specifically and rapidly detect HPV DNA with attomolar sensitivity.4 CRISPR/Cas12a trans-cleavage activity can also be used to degrade synthetic assay probes that produce a variety of signals (fluorescent,62 electrochemical,205 and colorimetrõic79) detectable on LFA or microfluidic chips through visual inspection or by a handheld device. For example, one group employed a Cas12a-based assay to detect HPV DNA targets in diluted plasma samples (without DNA isolation) using a visual LFA readout, which detected positive HPV16 and HPV18 signals in 93% and 30% of the plasma samples from a small cohort of patients with cervical cancer.79
Several studies have also used NAA-free CRISPR assays to detect HPV (Table 4), but while these assays can be faster, less expensive, and more streamlined than amplification-based assays, they also often require strategies to increase signal production or detection from low-concentration targets. For example, one group developed a portable photoelectrochemical (PEC) assay for HPV16.183 In this assay, target-induced Cas12a trans cleavage depleted a catalytic G-quadruplex, alleviating its inhibition of a photoactivated screen-printed electrode, which induced a photocurrent signal that was detectable by a smartphone device. Similarly, another group developed an electrochemical biosensor, in which target-mediated Cas12a trans-cleavage activity was employed to cleave a methylene blue-labelled ssDNA reporter from the assay biosensor to induce an electrochemical signal that had a 50 picomolar LOD.88 A third group developed a polydisperse droplet digital CRISPR/Cas (pddCas)-based HPV assay that used a standard vortex mixer to generate picoliter droplets, and reported LODs of 10 to 100 attomolar for DNA and RNA targets.224 Droplet digital CRISPR/Cas (pddCas)-based HPV assay that used a standard vortex mixer to generate picoliter droplets, and this assay had an LOD of 10 to 100 attomolar.224
Clinical laboratory-based PCR tests for EBV are used for diagnosis of nasopharyngeal carcinoma and other EBV-related diseases.226 EBV-based screening for nasopharyngeal carcinoma is recommended in high-risk regions.226 EBV-based screening for nasopharyngeal carcinoma is recommended in high-risk regions.156 One prospective study in Hong Kong227 analysed plasma samples from over 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 participants by real-time PCR and participants with EBV-positive were retested after 4 weeks. This study determined that 34 of the 309 participants with persistent EBV-positive results later received nasopharyngeal carcinoma diagnoses, while only one individual with EBV-negative results developed nasopharyngeal carcinoma within a year.
000 participants by real-time PCR and participants with EBV-positive were retested after 4 weeks. This study determined that 34 of the 309 participants with persistent EBV-positive results later received nasopharyngeal carcinoma diagnoses, while only one individual with EBV-negative results developed nasopharyngeal carcinoma within a year.
Several groups have employed various approaches to reduce the equipment demands for such screening efforts. One group recently developed a Cas12a-based colorimetric assay that permits visual detection of EBV-positive serum samples analysed under nonlaboratory conditions. This assay employed Cas12a trans-cleavage activity to degrade an ssDNA region of an oligonucleotide used to cross-link AuNPs that were spiked into RPA reactions of diagnostic serum samples. This degradation reduced the centrifugal precipitation of disrupted AuNP aggregates and produced a colorimetric signal proportional to the amount of AuNPs retained in suspension.228 A second group developed an NAA-free microfluidic Cas12a-based digital droplet assay to detect EBV in serum samples.121 A third group described a proof-of-concept real-time, wearable Cas9 assay patch, designed to allow NAA-free detection of EBV cfDNA in interstitial fluid.138 There is also an ongoing clinical trial to evaluate the performance of a Cas12a-based assay for EBV DNA detection in nasopharyngeal brushing and plasma samples (NCT05447169).
5.3 miRNAs
Small (19 to 25 nucleotide) noncoding miRNAs have also revealed promise as blood-based biomarkers in cancer screening applications.229 For example, results from a large, randomized trial have indicated that low-dose computed tomography and serum miRNA results can predict lung cancer risk.230 Several miRNAs (miR-155, miR-197, and miR-182) have potential for early detection of lung cancer, and ultrasensitive electrochemical biosensors have been established, as reviewed by Shaterabadi et al.231 Several groups have also established biosensors for CRISPR-based assays to detect miRNA and to increase specificity and provide platforms for POCT applications, as summarized in a recent review.232 For example, one group used an array of femtoliter chambers for NAA-free detection of an RNA target by Cas13a cleavage of a quenched fluorescent reporter in a digital droplet assay, achieving femtomolar limits of detection for multiple SARS-CoV-2 RNA targets.233 This assay should be adaptable for the detection of miRNA biomarker targets; however, it was read by a plate reader and may not work for low-resource settings or a POCT format. This assay approach also remains substantially less sensitive than CRISPR assays employing amplification steps, such as an amplification-based CRISPR assay using a Cas12a-based chemiluminescence biosensor, which detected miRNA-21 at an LOD of 16 attomolar (ref. 74) and may be more readily adapted to a POCT. Differential miRNA expression signatures may have greater diagnostic value than the evaluation of individual miRNAs, and biosensors that permits multiplex detection of several distinct miRNAs have been developed. For example, one group developed an NAA-free CRISPR/Cas13a-based microfluidic assay for eight miRNAs, including two miRNAs dysregulated in paediatric medulloblastoma.91 None of assays described above are ready for use as clinical applications or POCTs, as all of them require additional refinement to stabilize their reagents, followed by analytical and clinical validation studies, before they are ready for adoption as diagnostic tests.6 Limitations and perspectives for future development
New CRISPR-based POCTs for infectious disease and cancer screening, diagnosis, or disease management should be user-friendly, employ minimally invasive or non-invasive diagnostic specimens, and provide rapid and clinically valid results to guide healthcare decisions. CRISPR assays using well-established POCT methods to provide user-friendly readouts (e.g., LFA or personal glucose monitor results) may be best suited for self-monitoring applications (perhaps for individuals with a suspected disease condition) or for clinical decisions by healthcare personnel in low-resource settings. But no single approach is universally appropriate, as different assay designs may have distinct advantages and disadvantages for different specimens or types of NA biomarkers. Assay suitability can be influenced by sample processing and analysis workflows, readout formats, sensitivity and specificity, data interpretation and reporting demands, and sample-to-result times. Therefore, new CRISPR-based POCTs for specific diseases and infections must take these all considerations into account during the initial assay design stage.Many CRISPR assays use a previous or parallel target amplification reaction to enhance or permit the detection of low-concentration NA targets in clinical specimens. CRISPR assays that amplify and analyse targets in parallel are generally more suitable for use in POCTs than those that require a target preamplification procedure, unless the amplification and analysis reactions can be integrated into a single device to avoid sample manipulation, which can lead to assay workflow errors and inaccurate results. Integrating these two reactions into the same device or well can require careful optimization of the sample processing workflow, assay reagents, and reaction conditions to maximize sensitivity and accuracy of the final POCT. However, even with such careful optimization, assays that employ single reactions for target amplification and CRISPR detection usually have decreased sensitivity, and thus there is frequently a trade-off between sensitivity and simplicity when designing CRISPR assays for POCTs.
Although CRISPR assays without a target amplification step often lack satisfactory sensitivity, these assays can sometimes have acceptable sensitivity when used to detect more abundant NA targets or when coupled with ultrasensitive readout approaches, such as SERS. But even when sensitive readout approaches are applied to detect low concentration targets, their expense and support requirements usually preclude their use in low-resource settings or in POCTs. It may be possible to improve the detection sensitivity of NAA-free assays by identifying, selecting, or engineering new Cas proteins with enhanced binding affinity and cis- or trans-cleavage activities, but this would require significant effort, and the activity and sensitivity increases that can be achieved through these approaches are not clear.
CRISPR-assays that accurately quantify an NA target may offer significant clinical value by providing a means to estimate the target's abundance in a sample from a patient or an infectious agent, but accurate measurements typically require the use of standard curves or laboratory equipment, which are not feasible for low-resource settings or for POCTs. For example, several groups have developed CRISPR-based droplet assays to quantify NA targets,119,121,224 but analysis of these assays requires the use of specialized instruments and costly consumables.
New software for improving the selection of conserved genome sequence regions, amplification primers, and crRNA sequences suitable for use in NA-based diagnostics could streamline assay development. Recent research indicates that a machine learning-based system can automate these processes to significantly improve assay design; one such method required only 2 hours to design CRISPR/Cas13a-based diagnostics for 1933 virus species that infect vertebrate hosts.234 Specifically, this method searched the target virus genomes, identifying and scoring potential amplicon regions by their predicted amplification potential and the relative activity of optimal amplification primer sets designed to provide high coverage of sequence diversity occurring at their recognition sites. This and similar approaches could greatly improve the selection of NA targets.
Robust analytical validation studies that include synthetic samples (e.g., target material spiked into a diagnostic specimen obtained from a heathy individual) or real clinical samples are critical to identify assays that have the best potential for success prior to performing clinical validation studies. Such analytical validation studies, however, are often complicated by the lack of accepted internal standards for POCTs or in vitro diagnostics. Subsequent clinical validation studies should also analyse samples obtained from patients and controls that reflect the diagnostic complications encountered in the target population, have sufficient power to provide reliable estimates of diagnostic sensitivity and specificity, and include a direct comparison to a reference assay. Most studies describing the development of new CRISPR-based diagnostic assays analyse samples from small case–control studies or from synthetic samples, neither of which may accurately reflect the confounding factors present in target patient populations, and thus have a reduced chance to successfully translate into a clinical application.
Single-visit screening and diagnosis approaches that use noninvasive sample types are preferred to counteract the trend toward more complex diagnostic tests requiring high-throughput sequencing approaches, which can limit testing capacity and coverage of affected patient populations.235 New developments allowing for highly sensitive and specific detection of NA biomarkers in minimally invasive or noninvasive liquid biopsy specimens using one-step, quantitative, and multiplexed CRISPR-Dx POCTs236 have the potential to meet this demand for simpler and more broadly available diagnostics.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
X. Q. and T. Y. H. conceived and designed this review. X. Q. and Q. X. drafted the manuscript. X. Q., C. J. L. and T. Y. H. revised the manuscript. All authors approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
X. Q. was supported by the National Natural Science Foundation of China (82272406), the Zhejiang Leading Innovation and Entrepreneurship Team (2022R01006), and the National Key R&D Program of China (2019YFA0905800). T. Y. H. was supported by the National Institute of Allergy and Infectious Diseases (R01AI144168, R01AI173021, R01AI174964, R01AI175618, R01AI177986, and R01AI179714) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01HD090927 and R01HD103511).Notes and references
- D. Mabey, R. W. Peeling, A. Ustianowski and M. D. Perkins, Nat. Rev. Microbiol., 2004, 2, 231–240 CrossRef CAS PubMed.
- K. J. Land, D. I. Boeras, X. S. Chen, A. R. Ramsay and R. W. Peeling, Nat. Microbiol., 2019, 4, 46–54 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer, C. A. Freije, C. Myhrvold, R. P. Bhattacharyya, J. Livny, A. Regev, E. V. Koonin, D. T. Hung, P. C. Sabeti, J. J. Collins and F. Zhang, Science, 2017, 356, 438–442 CrossRef CAS PubMed.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436–439 CrossRef CAS PubMed.
- S. Y. Li, Q. X. Cheng, J. M. Wang, X. Y. Li, Z. L. Zhang, S. Gao, R. B. Cao, G. P. Zhao and J. Wang, Cell Discovery, 2018, 4, 20 CrossRef PubMed.
- A. Sen, M. Masetty, S. Weerakoon, C. Morris, J. S. Yadav, S. Apewokin, J. Trannguyen, M. Broom and A. Priye, Biosens. Bioelectron., 2024, 257, 116292 CrossRef CAS.
- Y. Chen, S. Qian, X. Yu, J. Wu and J. Xu, Trends Biotechnol., 2023, 41, 557–574 CrossRef CAS PubMed.
- A. S. Avaro and J. G. Santiago, Lab Chip, 2023, 23, 938–963 RSC.
- D. Wachholz Junior and L. T. Kubota, Talanta, 2024, 278, 126467 CrossRef CAS.
- S. Chandra and T. Hu, ACS Nano, 2022, 16, 9985–9993 CrossRef CAS.
- Y. Tang, L. Qi, Y. Liu, L. Guo, R. Zhao, M. Yang, Y. Du and B. Li, Angew. Chem., Int. Ed., 2022, 61, e202115907 CrossRef CAS PubMed.
- Z. Huang, C. J. Lyon, J. Wang, S. Lu and T. Y. Hu, Adv. Sci., 2023, e2301697, DOI:10.1002/advs.202301697.
- Z. Huang, C. J. Lyon and T. Y. Hu, Nat. Rev. Bioeng., 2023, 1, 230–231 CrossRef PubMed.
- J. E. van Dongen, J. T. W. Berendsen, R. D. M. Steenbergen, R. M. F. Wolthuis, J. C. T. Eijkel and L. I. Segerink, Biosens. Bioelectron., 2020, 166, 112445 CrossRef CAS PubMed.
- D. J. Brogan and O. S. Akbari, Biochemistry, 2022, 62, 3488–3492 CrossRef PubMed.
- U. Ganbaatar and C. Liu, Front. Cell. Infect. Microbiol., 2021, 11, 663949 CrossRef CAS PubMed.
- R. Barrangou, Curr. Opin. Immunol., 2015, 32, 36–41 CrossRef CAS PubMed.
- S. Y. Li, Q. X. Cheng, J. K. Liu, X. Q. Nie, G. P. Zhao and J. Wang, Cell Res., 2018, 28, 491–493 CrossRef CAS PubMed.
- O. O. Abudayyeh, J. S. Gootenberg, S. Konermann, J. Joung, I. M. Slaymaker, D. B. Cox, S. Shmakov, K. S. Makarova, E. Semenova, L. Minakhin, K. Severinov, A. Regev, E. S. Lander, E. V. Koonin and F. Zhang, Science, 2016, 353, aaf5573 CrossRef.
- A. East-Seletsky, M. R. O'Connell, S. C. Knight, D. Burstein, J. H. Cate, R. Tjian and J. A. Doudna, Nature, 2016, 538, 270–273 CrossRef CAS PubMed.
- C. Myhrvold, C. A. Freije, J. S. Gootenberg, O. O. Abudayyeh, H. C. Metsky, A. F. Durbin, M. J. Kellner, A. L. Tan, L. M. Paul, L. A. Parham, K. F. Garcia, K. G. Barnes, B. Chak, A. Mondini, M. L. Nogueira, S. Isern, S. F. Michael, I. Lorenzana, N. L. Yozwiak, B. L. MacInnis, I. Bosch, L. Gehrke, F. Zhang and P. C. Sabeti, Science, 2018, 360, 444–448 CrossRef CAS PubMed.
- M. J. Kellner, J. G. Koob, J. S. Gootenberg, O. O. Abudayyeh and F. Zhang, Nat. Protoc., 2019, 14, 2986–3012 CrossRef CAS PubMed.
- J. Yang, Y. Song, X. Deng, J. A. Vanegas, Z. You, Y. Zhang, Z. Weng, L. Avery, K. D. Dieckhaus, A. Peddi, Y. Gao, Y. Zhang and X. Gao, Nat. Chem. Biol., 2023, 19, 45–54 CrossRef CAS.
- L. Li, S. Li, N. Wu, J. Wu, G. Wang, G. Zhao and J. Wang, ACS Synth. Biol., 2019, 8, 2228–2237 CrossRef CAS PubMed.
- Y. Zhang, L. Qian, W. Wei, Y. Wang, B. Wang, P. Lin, W. Liu, L. Xu, X. Li, D. Liu, S. Cheng, J. Li, Y. Ye, H. Li, X. Zhang, Y. Dong, X. Zhao, C. Liu, H. M. Zhang, Q. Ouyang and C. Lou, ACS Synth. Biol., 2017, 6, 211–216 CrossRef CAS PubMed.
- X. Y. Qiu, L. Y. Zhu, C. S. Zhu, J. X. Ma, T. Hou, X. M. Wu, S. S. Xie, L. Min, D. A. Tan, D. Y. Zhang and L. Zhu, ACS Synth. Biol., 2018, 7, 807–813 CrossRef CAS PubMed.
- W. Yang, L. Restrepo-Perez, M. Bengtson, S. J. Heerema, A. Birnie, J. van der Torre and C. Dekker, Nano Lett., 2018, 18, 6469–6474 CrossRef CAS PubMed.
- R. Hajian, S. Balderston, T. Tran, T. deBoer, J. Etienne, M. Sandhu, N. A. Wauford, J. Y. Chung, J. Nokes, M. Athaiya, J. Paredes, R. Peytavi, B. Goldsmith, N. Murthy, I. M. Conboy and K. Aran, Nat. Biomed. Eng., 2019, 3, 427–437 CrossRef CAS PubMed.
- B. Koo, D. E. Kim, J. Kweon, C. E. Jin, S. H. Kim, Y. Kim and Y. Shin, Sens. Actuators, B, 2018, 273, 316–321 CrossRef CAS PubMed.
- M. Huang, X. Zhou, H. Wang and D. Xing, Anal. Chem., 2018, 90, 2193–2200 CrossRef CAS PubMed.
- T. Wang, Y. Liu, H. H. Sun, B. C. Yin and B. C. Ye, Angew. Chem., Int. Ed., 2019, 58, 5382–5386 CrossRef CAS PubMed.
- L. B. Harrington, D. Burstein, J. S. Chen, D. Paez-Espino, E. Ma, I. P. Witte, J. C. Cofsky, N. C. Kyrpides, J. F. Banfield and J. A. Doudna, Science, 2018, 362, 839–842 CrossRef CAS PubMed.
- Y. Wei, Z. Yang, C. Zong, B. Wang, X. Ge, X. Tan, X. Liu, Z. Tao, P. Wang, C. Ma, Y. Wan and J. Li, Angew. Chem., Int. Ed., 2021, 60, 24241–24247 CrossRef CAS PubMed.
- K. Yoshimi, K. Takeshita, S. Yamayoshi, S. Shibumura, Y. Yamauchi, M. Yamamoto, H. Yotsuyanagi, Y. Kawaoka and T. Mashimo, iScience, 2022, 25, 103830 CrossRef CAS PubMed.
- Z. Yu, J. Xu and Q. She, Int. J. Mol. Sci., 2023, 24, 2857 CrossRef CAS PubMed.
- A. Santiago-Frangos, L. N. Hall, A. Nemudraia, A. Nemudryi, P. Krishna, T. Wiegand, R. A. Wilkinson, D. T. Snyder, J. F. Hedges, C. Cicha, H. H. Lee, A. Graham, M. A. Jutila, M. P. Taylor and B. Wiedenheft, Cell Rep. Med., 2021, 2, 100319 CrossRef CAS PubMed.
- J. A. Steens, Y. Zhu, D. W. Taylor, J. P. K. Bravo, S. H. P. Prinsen, C. D. Schoen, B. J. F. Keijser, M. Ossendrijver, L. M. Hofstra, S. J. J. Brouns, A. Shinkai, J. van der Oost and R. H. J. Staals, Nat. Commun., 2021, 12, 5033 CrossRef CAS PubMed.
- S. Sridhara, H. N. Goswami, C. Whyms, J. H. Dennis and H. Li, Nat. Commun., 2021, 12, 5653 CrossRef CAS.
- S. Gruschow, C. S. Adamson and M. F. White, Nucleic Acids Res., 2021, 49, 13122–13134 CrossRef CAS PubMed.
- I. M. Slaymaker, P. Mesa, M. J. Kellner, S. Kannan, E. Brignole, J. Koob, P. R. Feliciano, S. Stella, O. O. Abudayyeh, J. S. Gootenberg, J. Strecker, G. Montoya and F. Zhang, Cell Rep., 2019, 26, 3741–3751.e3745 CrossRef CAS PubMed.
- I. M. Slaymaker, P. Mesa, M. J. Kellner, S. Kannan, E. Brignole, J. Koob, P. R. Feliciano, S. Stella, O. O. Abudayyeh, J. S. Gootenberg, J. Strecker, G. Montoya and F. Zhang, Cell Rep., 2021, 34, 108865 CrossRef CAS PubMed.
- E. M. J. S. Chen, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2021, 371, 436–439 CrossRef PubMed.
- A. Ramachandran and J. G. Santiago, Anal. Chem., 2021, 93, 7456–7464 CrossRef CAS PubMed.
- E. A. Nalefski, N. Patel, P. J. Y. Leung, Z. Islam, R. M. Kooistra, I. Parikh, E. Marion, G. J. Knott, J. A. Doudna, A. M. Le Ny and D. Madan, iScience, 2021, 24, 102996 CrossRef CAS PubMed.
- H. Lv, J. Wang, J. Zhang, Y. Chen, L. Yin, D. Jin, D. Gu, H. Zhao, Y. Xu and J. Wang, Front. Microbiol., 2021, 12, 766464 CrossRef PubMed.
- D. A. Huyke, A. Ramachandran, V. I. Bashkirov, E. K. Kotseroglou, T. Kotseroglou and J. G. Santiago, Anal. Chem., 2022, 94, 9826–9834 CrossRef CAS PubMed.
- J. G. Santiago, QRB Discov., 2022, 3, e9 CrossRef PubMed.
- W. Feng, H. Zhang and X. C. Le, Anal. Chem., 2023, 95, 206–217 CrossRef CAS PubMed.
- R. Wang, X. Zhao, X. Chen, X. Qiu, G. Qing, H. Zhang, L. Zhang, X. Hu, Z. He, D. Zhong, Y. Wang and Y. Luo, Anal. Chem., 2020, 92, 2176–2185 CrossRef CAS PubMed.
- M. Huang, R. Huang, H. Yue, Y. Shan and D. Xing, Sens. Actuators, B, 2020, 325, 128799 CrossRef CAS.
- S. Gong, S. Zhang, X. Wang, J. Li, W. Pan, N. Li and B. Tang, Anal. Chem., 2021, 93, 15216–15223 CrossRef CAS PubMed.
- W. Wang, J. Liu, L. A. Wu, C. N. Ko, X. Wang, C. Lin, J. Liu, L. Ling and J. Wang, Anal. Chim. Acta, 2022, 1195, 339479 CrossRef CAS PubMed.
- S. Zhou, L. Deng, J. Dong, P. Lu, N. Qi, Z. Huang, M. Yang, D. Huo and C. Hou, Microchim. Acta, 2023, 190, 113 CrossRef CAS PubMed.
- H. Yu, Q. Pu, Z. Weng, X. Zhou, J. Li, Y. Yang, W. Luo, Y. Guo, H. Chen, D. Wang and G. Xie, Biosens. Bioelectron., 2021, 194, 113625 CrossRef CAS PubMed.
- K. Pardee, A. A. Green, M. K. Takahashi, D. Braff, G. Lambert, J. W. Lee, T. Ferrante, D. Ma, N. Donghia, M. Fan, N. M. Daringer, I. Bosch, D. M. Dudley, D. H. O'Connor, L. Gehrke and J. J. Collins, Cell, 2016, 165, 1255–1266 CrossRef CAS PubMed.
- J. Joung, A. Ladha, M. Saito, N. G. Kim, A. E. Woolley, M. Segel, R. P. J. Barretto, A. Ranu, R. K. Macrae, G. Faure, E. I. Ioannidi, R. N. Krajeski, R. Bruneau, M. W. Huang, X. G. Yu, J. Z. Li, B. D. Walker, D. T. Hung, A. L. Greninger, K. R. Jerome, J. S. Gootenberg, O. O. Abudayyeh and F. Zhang, N. Engl. J. Med., 2020, 383, 1492–1494 CrossRef CAS PubMed.
- Z. Ali, R. Aman, A. Mahas, G. S. Rao, M. Tehseen, T. Marsic, R. Salunke, A. K. Subudhi, S. M. Hala, S. M. Hamdan, A. Pain, F. S. Alofi, A. Alsomali, A. M. Hashem, A. Khogeer, N. A. M. Almontashiri, M. Abedalthagafi, N. Hassan and M. M. Mahfouz, Virus Res., 2020, 288, 198129 CrossRef CAS PubMed.
- Y. Wang, Y. Ke, W. Liu, Y. Sun and X. Ding, ACS Sens., 2020, 5, 1427–1435 CrossRef CAS PubMed.
- X. Ding, K. Yin, Z. Li, R. V. Lalla, E. Ballesteros, M. M. Sfeir and C. Liu, Nat. Commun., 2020, 11, 4711 CrossRef CAS PubMed.
- Y. Chen, X. Xu, J. Wang, Y. Zhang, W. Zeng, Y. Liu and X. Zhang, Anal. Chem., 2022, 94, 9724–9731 CrossRef CAS PubMed.
- M. Hu, Z. Qiu, Z. Bi, T. Tian, Y. Jiang and X. Zhou, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2202034119 CrossRef CAS PubMed.
- K. Yin, X. Ding, Z. Li, H. Zhao, K. Cooper and C. Liu, Anal. Chem., 2020, 92, 8561–8568 CrossRef CAS PubMed.
- S. Lu, X. Tong, Y. Han, K. Zhang, Y. Zhang, Q. Chen, J. Duan, X. Lei, M. Huang, Y. Qiu, D. Y. Zhang, X. Zhou, Y. Zhang and H. Yin, Nat. Biomed. Eng., 2022, 6, 286–297 CrossRef CAS PubMed.
- J. M. Lesinski, T. Moragues, P. Mathur, Y. Shen, C. Paganini, L. Bezinge, B. Verberckmoes, B. Van Eenooghe, S. Stavrakis, A. J. deMello and D. A. Richards, Anal. Chem., 2024, 96, 10443–10450 CrossRef CAS PubMed.
- W. Feng, H. Peng, J. Xu, Y. Liu, K. Pabbaraju, G. Tipples, M. A. Joyce, H. A. Saffran, D. L. Tyrrell, S. Babiuk, H. Zhang and X. C. Le, Anal. Chem., 2021, 93, 12808–12816 CrossRef CAS PubMed.
- T. Li, R. Hu, J. Xia, Z. Xu, D. Chen, J. Xi, B. F. Liu, J. Zhu, Y. Li, Y. Yang and M. Liu, Biosens. Bioelectron., 2021, 187, 113292 CrossRef CAS PubMed.
- Z. Xu, D. Chen, T. Li, J. Yan, J. Zhu, T. He, R. Hu, Y. Li, Y. Yang and M. Liu, Nat. Commun., 2022, 13, 6480 CrossRef CAS PubMed.
- Y. Li, T. Li, B. F. Liu, R. Hu, J. Zhu, T. He, X. Zhou, C. Li, Y. Yang and M. Liu, Chem. Commun., 2020, 56, 12526–12529 RSC.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Science, 2018, 360, 439–444 CrossRef CAS PubMed.
- M. J. Osborn, A. Bhardwaj, S. P. Bingea, F. Knipping, C. J. Feser, C. J. Lees, D. P. Collins, C. J. Steer, B. R. Blazar and J. Tolar, Bioengineering, 2021, 8, 23 CrossRef CAS PubMed.
- H. J. van der Veer, E. A. van Aalen, C. M. S. Michielsen, E. T. L. Hanckmann, J. Deckers, M. van Borren, J. Flipse, A. J. M. Loonen, J. P. H. Schoeber and M. Merkx, ACS Cent. Sci., 2023, 9, 657–667 CrossRef CAS PubMed.
- T. Zhou, R. Huang, M. Huang, J. Shen, Y. Shan and D. Xing, Adv. Sci., 2020, 7, 1903661 CrossRef CAS PubMed.
- P. F. Liu, K. R. Zhao, Z. J. Liu, L. Wang, S. Y. Ye and G. X. Liang, Biosens. Bioelectron., 2021, 176, 112954 CrossRef CAS PubMed.
- Y. Zhou, S. Xie, B. Liu, C. Wang, Y. Huang, X. Zhang and S. Zhang, Anal. Chem., 2023, 95, 3332–3339 CrossRef CAS PubMed.
- J. Wei, W. Wang, Q. Yu, M. Zhang, F. Xue, B. Fan, T. Zhang, Y. Gao, J. Li, X. Meng and B. Pang, Sens. Actuators, B, 2023, 390, 133950 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C. Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS PubMed.
- D. Xiong, W. Dai, J. Gong, G. Li, N. Liu, W. Wu, J. Pan, C. Chen, Y. Jiao, H. Deng, J. Ye, X. Zhang, H. Huang, Q. Li, L. Xue, X. Zhang and G. Tang, PLoS Biol., 2020, 18, e3000978 CrossRef CAS PubMed.
- L. T. Nguyen, B. M. Smith and P. K. Jain, Nat. Commun., 2020, 11, 4906 CrossRef CAS PubMed.
- J. H. Tsou, Q. Leng and F. Jiang, Transl. Oncol., 2019, 12, 1566–1573 CrossRef PubMed.
- M. M. Kaminski, M. A. Alcantar, I. T. Lape, R. Greensmith, A. C. Huske, J. A. Valeri, F. M. Marty, V. Klambt, J. Azzi, E. Akalin, L. V. Riella and J. J. Collins, Nat. Biomed. Eng., 2020, 4, 601–609 CrossRef CAS PubMed.
- O. Mukama, J. Wu, Z. Li, Q. Liang, Z. Yi, X. Lu, Y. Liu, Y. Liu, M. Hussain, G. G. Makafe, J. Liu, N. Xu and L. Zeng, Biosens. Bioelectron., 2020, 159, 112143 CrossRef CAS PubMed.
- T. Yuan, O. Mukama, Z. Li, W. Chen, Y. Zhang, J. de Dieu Habimana, Y. Zhang, R. Zeng, C. Nie, Z. He and L. Zeng, Analyst, 2020, 145, 6388–6394 RSC.
- P. Shen, Z. Si, D. Huang, Z. Xu, Z. Wang, M. Fang and Z. Xu, Analyst, 2023, 148, 2573–2581 RSC.
- M. Hu, C. Yuan, T. Tian, X. Wang, J. Sun, E. Xiong and X. Zhou, J. Am. Chem. Soc., 2020, 142, 7506–7513 CrossRef CAS PubMed.
- X. Wang, E. Xiong, T. Tian, M. Cheng, W. Lin, H. Wang, G. Zhang, J. Sun and X. Zhou, ACS Nano, 2020, 14, 2497–2508 CrossRef CAS PubMed.
- W. Chang, W. Liu, Y. Liu, F. Zhan, H. Chen, H. Lei and Y. Liu, Microchim. Acta, 2019, 186, 243 CrossRef PubMed.
- J. H. Choi, J. Lim, M. Shin, S. H. Paek and J. W. Choi, Nano Lett., 2021, 21, 693–699 CrossRef CAS PubMed.
- Y. Dai, R. A. Somoza, L. Wang, J. F. Welter, Y. Li, A. I. Caplan and C. C. Liu, Angew. Chem., Int. Ed., 2019, 58, 17399–17405 CrossRef CAS.
- W. Xu, T. Jin, Y. Dai and C. C. Liu, Biosens. Bioelectron., 2020, 155, 112100 CrossRef CAS PubMed.
- R. Bruch, J. Baaske, C. Chatelle, M. Meirich, S. Madlener, W. Weber, C. Dincer and G. A. Urban, Adv. Mater., 2019, 31, e1905311 CrossRef PubMed.
- R. Bruch, M. Johnston, A. Kling, T. Mattmuller, J. Baaske, S. Partel, S. Madlener, W. Weber, G. A. Urban and C. Dincer, Biosens. Bioelectron., 2021, 177, 112887 CrossRef CAS PubMed.
- D. Najjar, J. Rainbow, S. Sharma Timilsina, P. Jolly, H. de Puig, M. Yafia, N. Durr, H. Sallum, G. Alter, J. Z. Li, X. G. Yu, D. R. Walt, J. A. Paradiso, P. Estrela, J. J. Collins and D. E. Ingber, Nat. Biomed. Eng., 2022, 6, 968–978 CrossRef CAS PubMed.
- F. Zheng, Z. Chen, J. Li, R. Wu, B. Zhang, G. Nie, Z. Xie and H. Zhang, Adv. Sci., 2022, 9, e2105231 CrossRef PubMed.
- Z. Chen, J. Li, T. Li, T. Fan, C. Meng, C. Li, J. Kang, L. Chai, Y. Hao, Y. Tang, O. A. Al-Hartomy, S. Wageh, A. G. Al-Sehemi, Z. Luo, J. Yu, Y. Shao, D. Li, S. Feng, W. J. Liu, Y. He, X. Ma, Z. Xie and H. Zhang, Natl. Sci. Rev., 2022, 9, nwac104 CrossRef CAS.
- Y. Chen, Z. Chen, T. Li, M. Qiu, J. Zhang, Y. Wang, W. Yuan, A. H. Ho, O. Al-Hartomy, S. Wageh, A. G. Al-Sehemi, X. Shi, J. Li, Z. Xie, L. Xuejin and H. Zhang, ACS Nano, 2023, 17, 12903–12914 CrossRef CAS PubMed.
- H. Kim, S. Lee, H. W. Seo, B. Kang, J. Moon, K. G. Lee, D. Yong, H. Kang, J. Jung, E. K. Lim, J. Jeong, H. G. Park, C. M. Ryu and T. Kang, ACS Nano, 2020, 14, 17241–17253 CrossRef CAS PubMed.
- S. Balderston, J. J. Taulbee, E. Celaya, K. Fung, A. Jiao, K. Smith, R. Hajian, G. Gasiunas, S. Kutanovas, D. Kim, J. Parkinson, K. Dickerson, J. J. Ripoll, R. Peytavi, H. W. Lu, F. Barron, B. R. Goldsmith, P. G. Collins, I. M. Conboy, V. Siksnys and K. Aran, Nat. Biomed. Eng., 2021, 5, 713–725 CrossRef CAS PubMed.
- H. Li, J. Yang, G. Wu, Z. Weng, Y. Song, Y. Zhang, J. A. Vanegas, L. Avery, Z. Gao, H. Sun, Y. Chen, K. D. Dieckhaus, X. Gao and Y. Zhang, Angew. Chem., Int. Ed., 2022, 61, e202203826 CrossRef CAS PubMed.
- H. Yu, H. Zhang, J. Li, Z. Zhao, M. Deng, Z. Ren, Z. Li, C. Xue, M. G. Li and Z. Chen, ACS Sens., 2022, 7, 3923–3932 CrossRef CAS PubMed.
- N. E. Weckman, N. Ermann, R. Gutierrez, K. Chen, J. Graham, R. Tivony, A. Heron and U. F. Keyser, ACS Sens., 2019, 4, 2065–2072 CrossRef CAS PubMed.
- R. Nouri, Y. Jiang, X. L. Lian and W. Guan, ACS Sens., 2020, 5, 1273–1280 CrossRef CAS PubMed.
- L. Liu, Z. Xu, K. Awayda, S. J. Dollery, M. Bao, J. Fan, D. Cormier, M. O'Connell, G. J. Tobin and K. Du, Adv. Mater. Technol., 2022, 7, 2101550 CrossRef CAS PubMed.
- M. A. English, L. R. Soenksen, R. V. Gayet, H. de Puig, N. M. Angenent-Mari, A. S. Mao, P. Q. Nguyen and J. J. Collins, Science, 2019, 365, 780–785 CrossRef CAS PubMed.
- R. V. Gayet, H. de Puig, M. A. English, L. R. Soenksen, P. Q. Nguyen, A. S. Mao, N. M. Angenent-Mari and J. J. Collins, Nat. Protoc., 2020, 15, 3030–3063 CrossRef CAS.
- D. Huang, D. Ni, M. Fang, Z. Shi and Z. Xu, Anal. Chem., 2021, 93, 16965–16973 CrossRef CAS PubMed.
- D. Huang, Z. Shi, J. Qian, K. Bi, M. Fang and Z. Xu, Biotechnol. Bioeng., 2021, 118, 1587–1596 CAS.
- S. Gong, J. Li, W. Pan, N. Li and B. Tang, Anal. Chem., 2021, 93, 10719–10726 CrossRef CAS PubMed.
- R. Liu, Y. He, T. Lan and J. Zhang, Analyst, 2021, 146, 3114–3120 RSC.
- J. Park, H. Han, J. H. Jeung, H. Jang, C. Park and J. K. Ahn, Biosens. Bioelectron.: X, 2022, 12, 100283 CAS.
- C. Zhou, D. Huang, Z. Wang, P. Shen, P. Wang and Z. Xu, J. Food Sci., 2022, 87, 4137–4147 CrossRef CAS PubMed.
- Z. Jia, Z. Li and C. Liu, Sens. Actuators, B, 2023, 390, 133994 CrossRef CAS PubMed.
- Z. Li, N. Uno, X. Ding, L. Avery, D. Banach and C. Liu, ACS Nano, 2023, 17, 3966–3975 CrossRef CAS PubMed.
- H. Li, Y. Xie, F. Chen, H. Bai, L. Xiu, X. Zhou, X. Guo, Q. Hu and K. Yin, Chem. Soc. Rev., 2023, 52, 361–382 RSC.
- Y. Li, Y. Liu, X. Tang, J. Qiao, J. Kou, S. Man, L. Zhu and L. Ma, ACS Sens., 2023, 8, 4420–4441 CrossRef CAS.
- K. Shi, S. Xie, R. Tian, S. Wang, Q. Lu, D. Gao, C. Lei, H. Zhu and Z. Nie, Sci. Adv., 2021, 7, eabc7802 CrossRef CAS PubMed.
- H. Zeng, P. Zhang, X. Jiang, C. Duan, Y. Yu, Q. Wu and X. Yang, Anal. Chim. Acta, 2022, 1217, 340009 CrossRef CAS PubMed.
- Z. Feng, Y. Xue, Y. Yun, Z. Liu and J. Zhang, Sens. Diagn., 2023, 2, 632–639 RSC.
- X. Ding, K. Yin, Z. Li, M. M. Sfeir and C. Liu, Biosens. Bioelectron., 2021, 184, 113218 CrossRef CAS.
- H. Shinoda, Y. Taguchi, R. Nakagawa, A. Makino, S. Okazaki, M. Nakano, Y. Muramoto, C. Takahashi, I. Takahashi, J. Ando, T. Noda, O. Nureki, H. Nishimasu and R. Watanabe, Commun. Biol., 2021, 4, 476 CrossRef CAS PubMed.
- T. Tian, B. Shu, Y. Jiang, M. Ye, L. Liu, Z. Guo, Z. Han, Z. Wang and X. Zhou, ACS Nano, 2021, 15, 1167–1178 CrossRef CAS PubMed.
- H. Yue, B. Shu, T. Tian, E. Xiong, M. Huang, D. Zhu, J. Sun, Q. Liu, S. Wang, Y. Li and X. Zhou, Nano Lett., 2021, 21, 4643–4653 CrossRef CAS PubMed.
- C. Zhang, Z. Cai, Z. Zhou, M. Li, W. Hong, W. Zhou, D. Yu, P. Wei, J. He, Y. Wang, C. Huang, X. Wang and J. Wu, Biosens. Bioelectron., 2023, 222, 114956 CrossRef CAS PubMed.
- P. Fozouni, S. Son, M. Díaz de León Derby, G. J. Knott, C. N. Gray, M. V. D'Ambrosio, C. Zhao, N. A. Switz, G. R. Kumar, S. I. Stephens, D. Boehm, C. L. Tsou, J. Shu, A. Bhuiya, M. Armstrong, A. R. Harris, P. Y. Chen, J. M. Osterloh, A. Meyer-Franke, B. Joehnk, K. Walcott, A. Sil, C. Langelier, K. S. Pollard, E. D. Crawford, A. S. Puschnik, M. Phelps, A. Kistler, J. L. DeRisi, J. A. Doudna, D. A. Fletcher and M. Ott, Cell, 2021, 184, 323–333.e329 CrossRef CAS PubMed.
- Y. Sha, R. Huang, M. Huang, H. Yue, Y. Shan, J. Hu and D. Xing, Chem. Commun., 2021, 57, 247–250 RSC.
- H. Fan, S. H. Luo, Y. Zhu, J. Shi, F. Yin and J. Li, Chem. Commun., 2023, 59, 13151–13154 RSC.
- J. Arizti-Sanz, C. A. Freije, A. C. Stanton, B. A. Petros, C. K. Boehm, S. Siddiqui, B. M. Shaw, G. Adams, T. F. Kosoko-Thoroddsen, M. E. Kemball, J. N. Uwanibe, F. V. Ajogbasile, P. E. Eromon, R. Gross, L. Wronka, K. Caviness, L. E. Hensley, N. H. Bergman, B. L. MacInnis, C. T. Happi, J. E. Lemieux, P. C. Sabeti and C. Myhrvold, Nat. Commun., 2020, 11, 5921 CrossRef CAS PubMed.
- A. Bhatt, Z. Fatima, M. Ruwali, C. S. Misra, S. S. Rangu, D. Rath, A. Rattan and S. Hameed, J. Appl. Microbiol., 2022, 133, 410–421 CrossRef CAS PubMed.
- H. de Puig, R. A. Lee, D. Najjar, X. Tan, L. R. Soeknsen, N. M. Angenent-Mari, N. M. Donghia, N. E. Weckman, A. Ory, C. F. Ng, P. Q. Nguyen, A. S. Mao, T. C. Ferrante, G. Lansberry, H. Sallum, J. Niemi and J. J. Collins, Sci. Adv., 2021, 7, eabh2944 CrossRef PubMed.
- F. E. Chen, P. W. Lee, A. Y. Trick, J. S. Park, L. Chen, K. Shah, H. Mostafa, K. C. Carroll, K. Hsieh and T. H. Wang, Biosens. Bioelectron., 2021, 190, 113390 CrossRef CAS PubMed.
- H. Wu, S. Qian, C. Peng, X. Wang, T. Wang, X. Zhong, Y. Chen, Q. Yang, J. Xu and J. Wu, ACS Sens., 2021, 6, 4048–4056 CrossRef CAS PubMed.
- S. Xu, X. Wang, C. Wu, X. Zhu, X. Deng, Y. Wu, M. Liu, X. Huang, L. Wu and H. Huang, Biosens. Bioelectron., 2024, 247, 115925 CrossRef CAS PubMed.
- M. Patchsung, A. Homchan, K. Aphicho, S. Suraritdechachai, T. Wanitchanon, A. Pattama, K. Sappakhaw, P. Meesawat, T. Wongsatit, A. Athipanyasilp, K. Jantarug, N. Athipanyasilp, J. Buahom, S. Visanpattanasin, N. Niljianskul, P. Chaiyen, R. Tinikul, N. Wichukchinda, S. Mahasirimongkol, R. Sirijatuphat, N. Angkasekwinai, M. A. Crone, P. S. Freemont, J. Joung, A. Ladha, O. Abudayyeh, J. Gootenberg, F. Zhang, C. Chewapreecha, S. Chanarat, N. Horthongkham, D. Pakotiprapha and C. Uttamapinant, CRISPR J., 2023, 6, 99–115 CrossRef CAS PubMed.
- C. M. Ackerman, C. Myhrvold, S. G. Thakku, C. A. Freije, H. C. Metsky, D. K. Yang, S. H. Ye, C. K. Boehm, T. F. Kosoko-Thoroddsen, J. Kehe, T. G. Nguyen, A. Carter, A. Kulesa, J. R. Barnes, V. G. Dugan, D. T. Hung, P. C. Blainey and P. C. Sabeti, Nature, 2020, 582, 277–282 CrossRef CAS PubMed.
- N. L. Welch, M. Zhu, C. Hua, J. Weller, M. E. Mirhashemi, T. G. Nguyen, S. Mantena, M. R. Bauer, B. M. Shaw, C. M. Ackerman, S. G. Thakku, M. W. Tse, J. Kehe, M. M. Uwera, J. S. Eversley, D. A. Bielwaski, G. McGrath, J. Braidt, J. Johnson, F. Cerrato, G. K. Moreno, L. A. Krasilnikova, B. A. Petros, G. L. Gionet, E. King, R. C. Huard, S. K. Jalbert, M. L. Cleary, N. A. Fitzgerald, S. B. Gabriel, G. R. Gallagher, S. C. Smole, L. C. Madoff, C. M. Brown, M. W. Keller, M. M. Wilson, M. K. Kirby, J. R. Barnes, D. J. Park, K. J. Siddle, C. T. Happi, D. T. Hung, M. Springer, B. L. MacInnis, J. E. Lemieux, E. Rosenberg, J. A. Branda, P. C. Blainey, P. C. Sabeti and C. Myhrvold, Nat. Med., 2022, 28, 1083–1094 CrossRef CAS PubMed.
- G. Xing, Y. Shang, X. Wang, H. Lin, S. Chen, Q. Pu and L. Lin, Biosens. Bioelectron., 2023, 220, 114885 CrossRef CAS PubMed.
- K. Yin, X. Ding, Z. Li, M. M. Sfeir, E. Ballesteros and C. Liu, Lab Chip, 2021, 21, 2730–2737 RSC.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, Nature, 2014, 507, 181–189 CrossRef CAS PubMed.
- B. Yang, J. Kong and X. Fang, Nat. Commun., 2022, 13, 3999 CrossRef CAS PubMed.
- P. Q. Nguyen, L. R. Soenksen, N. M. Donghia, N. M. Angenent-Mari, H. de Puig, A. Huang, R. Lee, S. Slomovic, T. Galbersanini, G. Lansberry, H. M. Sallum, E. M. Zhao, J. B. Niemi and J. J. Collins, Nat. Biotechnol., 2021, 39, 1366–1374 CrossRef CAS PubMed.
- H. Chen, X. Zhou, M. Wang and L. Ren, J. Funct. Biomater., 2023, 14, 14 Search PubMed.
- R. Nouri, Z. Tang, M. Dong, T. Liu, A. Kshirsagar and W. Guan, Biosens. Bioelectron., 2021, 178, 113012 CrossRef CAS PubMed.
- S. Perveen, A. Negi, V. Gopalakrishnan, S. Panda, V. Sharma and R. Sharma, Clin. Chim. Acta, 2023, 538, 139–156 CrossRef CAS PubMed.
- M. Patchsung, K. Jantarug, A. Pattama, K. Aphicho, S. Suraritdechachai, P. Meesawat, K. Sappakhaw, N. Leelahakorn, T. Ruenkam, T. Wongsatit, N. Athipanyasilp, B. Eiamthong, B. Lakkanasirorat, T. Phoodokmai, N. Niljianskul, D. Pakotiprapha, S. Chanarat, A. Homchan, R. Tinikul, P. Kamutira, K. Phiwkaow, S. Soithongcharoen, C. Kantiwiriyawanitch, V. Pongsupasa, D. Trisrivirat, J. Jaroensuk, T. Wongnate, S. Maenpuen, P. Chaiyen, S. Kamnerdnakta, J. Swangsri, S. Chuthapisith, Y. Sirivatanauksorn, C. Chaimayo, R. Sutthent, W. Kantakamalakul, J. Joung, A. Ladha, X. Jin, J. S. Gootenberg, O. O. Abudayyeh, F. Zhang, N. Horthongkham and C. Uttamapinant, Nat. Biomed. Eng., 2020, 4, 1140–1149 CrossRef CAS.
- P. Fozouni, S. Son, M. Diaz de Leon Derby, G. J. Knott, C. N. Gray, M. V. D'Ambrosio, C. Zhao, N. A. Switz, G. R. Kumar, S. I. Stephens, D. Boehm, C. L. Tsou, J. Shu, A. Bhuiya, M. Armstrong, A. R. Harris, P. Y. Chen, J. M. Osterloh, A. Meyer-Franke, B. Joehnk, K. Walcott, A. Sil, C. Langelier, K. S. Pollard, E. D. Crawford, A. S. Puschnik, M. Phelps, A. Kistler, J. L. DeRisi, J. A. Doudna, D. A. Fletcher and M. Ott, Cell, 2021, 184, 323–333 CrossRef CAS PubMed , e329.
- Z. Huang, D. Tian, Y. Liu, Z. Lin, C. J. Lyon, W. Lai, D. Fusco, A. Drouin, X. Yin, T. Hu and B. Ning, Biosens. Bioelectron., 2020, 164, 112316 CrossRef CAS PubMed.
- Z. Huang, B. Ning, H. S. Yang, B. M. Youngquist, A. Niu, C. J. Lyon, B. J. Beddingfield, A. C. Fears, C. H. Monk, A. E. Murrell, S. J. Bilton, J. P. Linhuber, E. B. Norton, M. L. Dietrich, J. Yee, W. Lai, J. W. Scott, X.-M. Yin, J. Rappaport, J. E. Robinson, N. S. Saba, C. J. Roy, K. J. Zwezdaryk, Z. Zhao and T. Y. Hu, J. Clin. Invest., 2021, 131, e146031 CrossRef CAS PubMed.
- Z. Huang, L. Zhang, C. J. Lyon, B. Ning, B. M. Youngquist, A. Niu, B. J. Beddingfield, N. J. Maness, N. S. Saba, C.-Z. Li, C. J. Roy and T. Y. Hu, Emerging Microbes Infect., 2022, 11, 629–638 CrossRef CAS.
- B. Ning, T. Yu, S. Zhang, Z. Huang, D. Tian, Z. Lin, A. Niu, N. Golden, K. Hensley, B. Threeton, C. J. Lyon, X. M. Yin, C. J. Roy, N. S. Saba, J. Rappaport, Q. Wei and T. Y. Hu, Sci. Adv., 2021, 7, eabe3703 CrossRef CAS PubMed.
- Y. Liang, H. Lin, L. Zou, X. Deng and S. Tang, Biosens. Bioelectron., 2022, 205, 114098 CrossRef CAS PubMed.
- Y. Liang, L. Zou, H. Lin, B. Li, J. Zhao, H. Wang, J. Sun, J. Chen, Y. Mo, X. Yang, X. Deng and S. Tang, ACS Synth. Biol., 2022, 11, 1811–1823 CrossRef CAS PubMed.
- B. Ning, B. M. Youngquist, D. D. Li, C. J. Lyon, A. Zelazny, N. J. Maness, D. Tian and T. Y. Hu, Cells Rep. Methods, 2022, 2, 100173 CrossRef CAS PubMed.
- J. Liang, P. Teng, W. Xiao, G. He, Q. Song, Y. Zhang, B. Peng, G. Li, L. Hu, D. Cao and Y. Tang, J. Nanobiotechnol., 2021, 19, 273 CrossRef CAS PubMed.
- Y. Cao, X. Lu, H. Lin, A. F. Rodriguez Serrano, G. C. Y. Lui and I. M. Hsing, Biosens. Bioelectron., 2023, 236, 115402 CrossRef CAS PubMed.
- B. Ning, Z. Huang, B. M. Youngquist, J. W. Scott, A. Niu, C. M. Bojanowski, K. J. Zwezdaryk, N. S. Saba, J. Fan, X. M. Yin, J. Cao, C. J. Lyon, C. Z. Li, C. J. Roy and T. Y. Hu, Nat. Nanotechnol., 2021, 16, 1039–1044 CrossRef CAS PubMed.
- Z. Zhu, Y. Guo, C. Wang, Z. Yang, R. Li, Z. Zeng, H. Li, D. Zhang and L. Yang, Biosens. Bioelectron., 2023, 228, 115179 CrossRef CAS PubMed.
- W. K. J. Lam, A. D. King, J. A. Miller, Z. Liu, K. J. Yu, M. L. K. Chua, B. B. Y. Ma, M. Y. Chen, B. A. Pinsky, P. J. Lou, J. K. S. Woo, W. L. Hsu, J. Simon, D. L. Doolan, T. Waterboer, E. P. Hui, H. Li, R. K. Tsang, K. C. W. Wong, J. P. Goh, A. C. Vlantis, Q. Y. Ai, L. M. Wong, V. Abdullah, J. C. Lin, C. J. Chen, R. M. Pfeiffer, Q. T. Le, A. W. M. Lee, M. Ji, S. Cao, J. Ma, A. T. C. Chan, K. C. A. Chan and A. Hildesheim, J. Natl. Cancer Inst., 2023, 115, 355–364 CrossRef CAS PubMed.
- World Health Organization, https://www.who.int/teams/global-tuberculosis-programme/tb-reports, 2022, Acessed on 2023.11.20.
- X. Qian, D. T. Nguyen, J. Lyu, A. E. Albers, X. Bi and E. A. Graviss, Emerging Microbes Infect., 2018, 7, 102 Search PubMed.
- World Health Organization, WHO standard: universal access to rapid tuberculosis diagnostics, https://www.who.int/publications/i/item/9789240071315, 2023, Acessed on 2023.11.20.
- Z. Huang, S. M. LaCourse, A. W. Kay, J. Stern, J. N. Escudero, B. M. Youngquist, W. Zheng, D. Vambe, M. Dlamini, G. Mtetwa, L. M. Cranmer, I. Njuguna, D. C. Wamalwa, E. Maleche-Obimbo, D. G. Catanzaro, C. J. Lyon, G. John-Stewart, A. DiNardo, A. M. Mandalakas, B. Ning and T. Y. Hu, Lancet Microbe, 2022, 3, e482–e492 CrossRef CAS.
- F. Ardito, B. Posteraro, M. Sanguinetti, S. Zanetti and G. Fadda, J. Clin. Microbiol., 2001, 39, 4440–4444 CrossRef CAS PubMed.
- J. C. Ngabonziza, W. Ssengooba, F. Mutua, G. Torrea, A. Dushime, M. Gasana, E. Andre, S. Uwamungu, A. U. Nyaruhirira, D. Mwaengo and C. M. Muvunyi, BMC Infect. Dis., 2016, 16, 660 CrossRef.
- M. Farhat, C. Greenaway, M. Pai and D. Menzies, Int. J. Tuberc. Lung Dis., 2006, 10, 1192–1204 CAS.
- T. K. Venkatappa, R. Punnoose, D. J. Katz, M. P. Higgins, N. Banaei, E. A. Graviss, R. W. Belknap, C. S. Ho and C. Tuberculosis Epidemiologic Studies, J. Clin. Microbiol., 2019, 57 DOI:10.1128/JCM.00985-19.
- E. MacLean, M. Kohli, S. F. Weber, A. Suresh, S. G. Schumacher, C. M. Denkinger and M. Pai, J. Clin. Microbiol., 2020, 58, e01582–01519 CrossRef CAS PubMed.
- A. Dhana, Y. Hamada, A. P. Kengne, A. D. Kerkhoff, M. X. Rangaka, T. Kredo, A. Baddeley, C. Miller, S. Singh, Y. Hanifa, A. D. Grant, K. Fielding, D. Affolabi, C. S. Merle, A. P. Wachinou, C. Yoon, A. Cattamanchi, C. J. Hoffmann, N. Martinson, E. T. Mbu, M. S. Sander, T. T. Balcha, S. Skogmar, B. W. P. Reeve, G. Theron, G. Ndlangalavu, S. Modi, J. Cavanaugh, S. Swindells, R. E. Chaisson, F. Ahmad Khan, A. A. Howard, R. Wood, S. S. Thit, M. M. Kyi, J. Hanson, P. K. Drain, A. E. Shapiro, T. Kufa, G. Churchyard, D. T. Nguyen, E. A. Graviss, S. Bjerrum, I. S. Johansen, J. K. Gersh, D. J. Horne, S. M. LaCourse, H. A. A. Al-Darraji, A. Kamarulzaman, R. R. Kempker, N. Tukvadze, D. A. Barr, G. Meintjes and G. Maartens, Lancet Infect. Dis., 2022, 22, 507–518 CrossRef PubMed.
- N. A. Makhado, E. Matabane, M. Faccin, C. Pincon, A. Jouet, F. Boutachkourt, L. Goeminne, C. Gaudin, G. Maphalala, P. Beckert, S. Niemann, J. C. Delvenne, M. Delmee, L. Razwiedani, M. Nchabeleng, P. Supply, B. C. de Jong and E. Andre, Lancet Infect. Dis., 2018, 18, 1350–1359 CrossRef PubMed.
- X. Yang, X. Chen, J. Huang, Y. Chen, W. Zheng, W. Chen, H. Chen, S. Lei and S. Li, ACS Infect. Dis., 2023, 9, 762–772 CrossRef CAS PubMed.
- L. Yan, H. Xiao and Q. Zhang, Tuberculosis, 2016, 96, 75–86 CrossRef CAS PubMed.
- L. Augustin and N. Agarwal, Gene, 2023, 857, 147173 CrossRef CAS PubMed.
- T. T. B. Tram, V. T. N. Ha, L. P. T. Trieu, P. M. Ashton, E. D. Crawford, D. D. A. Thu, N. L. Quang, G. E. Thwaites, T. M. Walker, C. Anscombe and N. T. T. Thuong, J. Clin. Microbiol., 2023, 61, e0163422 CrossRef PubMed.
- R. Ding, J. Long, M. Yuan, X. Zheng, Y. Shen, Y. Jin, H. Yang, H. Li, S. Chen and G. Duan, Int. J. Mol. Sci., 2021, 22, 4842 CrossRef CAS PubMed.
- Y. Tian, Z. Fan, L. Xu, Y. Cao, S. Chen, Z. Pan, Y. Gao, H. Li, S. Zheng, Y. Ma, Z. Duan, X. Zhang and F. Ren, Emerging Microbes Infect., 2023, 12, e2177088 CrossRef PubMed.
- T. Jiang, G. Li, R. Liu, J. Zhou, N. Gao and J. Shen, J. Med. Virol., 2023, 95, e28905 CrossRef CAS PubMed.
- J. Arizti-Sanz, A. Bradley, Y. B. Zhang, C. K. Boehm, C. A. Freije, M. E. Grunberg, T. F. Kosoko-Thoroddsen, N. L. Welch, P. P. Pillai, S. Mantena, G. Kim, J. N. Uwanibe, O. G. John, P. E. Eromon, G. Kocher, R. Gross, J. S. Lee, L. E. Hensley, B. L. MacInnis, J. Johnson, M. Springer, C. T. Happi, P. C. Sabeti and C. Myhrvold, Nat. Biomed. Eng., 2022, 6, 932–943 CrossRef CAS PubMed.
- S. Gulati, S. Maiti and D. Chakraborty, Trends Genet., 2021, 37, 776–779 CrossRef CAS PubMed.
- M. Zamani, J. M. Robson, A. Fan, M. S. Bono, Jr., A. L. Furst and C. M. Klapperich, ACS Cent. Sci., 2021, 7, 963–972 CrossRef CAS PubMed.
- M. Zamani, C. M. Klapperich and A. L. Furst, Lab Chip, 2023, 23, 1410–1419 RSC.
- M. Azhar, R. Phutela, M. Kumar, A. H. Ansari, R. Rauthan, S. Gulati, N. Sharma, D. Sinha, S. Sharma, S. Singh, S. Acharya, S. Sarkar, D. Paul, P. Kathpalia, M. Aich, P. Sehgal, G. Ranjan, R. C. Bhoyar, K. Singhal, H. Lad, P. K. Patra, G. Makharia, G. R. Chandak, B. Pesala, D. Chakraborty and S. Maiti, Biosens. Bioelectron., 2021, 183, 113207 CrossRef CAS PubMed.
- S. R. Benjamin, F. de Lima, V. A. D. Nascimento, G. M. de Andrade and R. B. Oriá, Biosensors, 2023, 13, 689 CrossRef CAS PubMed.
- N. Sritong, M. Sala de Medeiros, L. A. Basing and J. C. Linnes, Lab Chip, 2023, 23, 888–912 RSC.
- Y. Wang, H. Chen, K. Lin, Y. Han, Z. Gu, H. Wei, K. Mu, D. Wang, L. Liu, R. Jin, R. Song, Z. Rong and S. Wang, Nat. Commun., 2024, 15, 3279 CrossRef CAS PubMed.
- R. Zeng, H. Gong, Y. Li, Y. Li, W. Lin, D. Tang and D. Knopp, Anal. Chem., 2022, 94, 7442–7448 CrossRef CAS PubMed.
- C. Han, W. Li, Q. Li, W. Xing, H. Luo, H. Ji, X. Fang, Z. Luo and L. Zhang, Biosens. Bioelectron., 2022, 200, 113922 CrossRef CAS PubMed.
- M. Zamani, A. L. Furst and C. M. Klapperich, Acc. Chem. Res., 2021, 54, 3772–3779 CrossRef CAS PubMed.
- R. L. Siegel, K. D. Miller, N. S. Wagle and A. Jemal, Ca-Cancer J. Clin., 2023, 73, 17–48 CrossRef PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- D. Crosby, S. Bhatia, K. M. Brindle, L. M. Coussens, C. Dive, M. Emberton, S. Esener, R. C. Fitzgerald, S. S. Gambhir, P. Kuhn, T. R. Rebbeck and S. Balasubramanian, Science, 2022, 375, eaay9040 CrossRef CAS PubMed.
- Y. Liu, Y. Chen, L. Dang, Y. Liu, S. Huang, S. Wu, P. Ma, H. Jiang, Y. Li, Y. Pan, Y. Wei, X. Ma, M. Liu, Q. Ji, T. Chi, X. Huang, X. Wang and F. Zhou, Mol. Cancer, 2021, 20, 157 CrossRef CAS PubMed.
- B. Kim, Y. Kim, S. Shin, S. T. Lee, J. Y. Cho and K. A. Lee, Cancer Cell Int., 2022, 22, 82 CrossRef CAS PubMed.
- W. Chen, S. Wu, G. Li, X. Duan, X. Sun, S. Li, Y. Zhao, D. Gu, G. Zeng and H. Liu, Biosens. Bioelectron., 2023, 220, 114854 CrossRef CAS PubMed.
- L. Wang, J. Jiang, X. Li, K. Li, R. He, J. Li, L. Duan, W. Luo, Z. Hu and D. Luo, Acta Biochim. Biophys. Sin., 2020, 52, 1316–1324 CrossRef CAS PubMed.
- S. Lee, S. Hong, J. Park, Y. Koh, H. Lee, J. Yang, S. W. Seo and S. J. Kim, Anal. Chem., 2023, 95, 5045–5052 CrossRef CAS PubMed.
- K. A. V. Kohabir, L. O. Nooi, A. Brink, R. H. Brakenhoff, E. A. Sistermans and R. M. F. Wolthuis, CRISPR J., 2023, 6, 127–139 CrossRef CAS PubMed.
- World Health Organization, https://www.who.int/publications/i/item/9789240030824, 2021, Accessed on 2023.11.20.
- Q. Wang, B. Zhang, X. Xu, F. Long and J. Wang, Sci. Rep., 2018, 8, 14126 CrossRef PubMed.
- D. Singh, J. Vignat, V. Lorenzoni, M. Eslahi, O. Ginsburg, B. Lauby-Secretan, M. Arbyn, P. Basu, F. Bray and S. Vaccarella, Lancet Global Health, 2023, 11, e197–e206 CrossRef CAS PubMed.
- M. Cao, H. Li, D. Sun, S. He, Y. Yu, J. Li, H. Chen, J. Shi, J. Ren, N. Li and W. Chen, Cancer Lett., 2021, 506, 120–127 CrossRef CAS PubMed.
- B. Zhang, Q. Wang, X. Xu, Q. Xia, F. Long, W. Li, Y. Shui, X. Xia and J. Wang, Anal. Bioanal. Chem., 2018, 410, 2889–2900 CrossRef CAS PubMed.
- B. Zhang, Q. Xia, Q. Wang, X. Xia and J. Wang, Anal. Biochem., 2018, 561–562, 37–46 CrossRef CAS PubMed.
- J. Gao, L. Wu, D. Yang, W. Gong and J. Wang, J. Mol. Diagn., 2021, 23, 46–60 CrossRef CAS PubMed.
- B. Zhang, M. Li, Y. Wei, J. Wang, Y. Wang, P. Shi, H. Tang and Z. Song, Anal. Biochem., 2022, 658, 114940 CrossRef CAS PubMed.
- X. Xu, T. Luo, J. Gao, N. Lin, W. Li, X. Xia and J. Wang, CRISPR J., 2020, 3, 487–502 CrossRef CAS PubMed.
- J. Gong, G. Zhang, W. Wang, L. Liang, Q. Li, M. Liu, L. Xue and G. Tang, Sci. Rep., 2021, 11, 12800 CrossRef CAS PubMed.
- Z. Li, X. Ding, K. Yin, Z. Xu, K. Cooper and C. Liu, Biosens. Bioelectron., 2021, 192, 113498 CrossRef CAS PubMed.
- X. Ke, Y. Ou, Y. Lin and T. Hu, Biosens. Bioelectron., 2022, 212, 114428 CrossRef CAS PubMed.
- T. Hu, X. Ke, Y. Ou and Y. Lin, Anal. Chem., 2022, 94, 8506–8513 CrossRef CAS PubMed.
- Y. Zhao, D. Chen, Z. Xu, T. Li, J. Zhu, R. Hu, G. Xu, Y. Li, Y. Yang and M. Liu, Anal. Chem., 2023, 95, 3476–3485 CrossRef CAS PubMed.
- H. Zhou, Z. Xu, L. He, Z. Wang, T. Zhang, T. Hu, F. Huang, D. Chen, Y. Li, Y. Yang and X. Huang, Anal. Chem., 2023, 95, 3379–3389 CrossRef CAS PubMed.
- Y. H. Roh, C. Y. Lee, S. Lee, H. Kim, A. Ly, C. M. Castro, J. Cheon, J. H. Lee and H. Lee, Adv. Sci., 2023, 10, e2206872 CrossRef PubMed.
- J. Han, J. Shin, E. S. Lee, B. S. Cha, S. Kim, Y. Jang, S. Kim and K. S. Park, Biosens. Bioelectron., 2023, 232, 115323 CrossRef CAS PubMed.
- Y. Chen, L. Meng, B. Lang, L. Li, J. Liu, Y. Wang, Z. Huang, X. Tian, R. Tian and Z. Hu, Cells Rep. Methods, 2023, 3, 100444 CrossRef CAS PubMed.
- X. Zheng, Y. Li, M. Yuan, Y. Shen, S. Chen and G. Duan, Anal. Methods, 2022, 14, 5065–5075 RSC.
- J. Su, Y. Ke, N. Maboyi, X. Zhi, S. Yan, F. Li, B. Zhao, X. Jia, S. Song and X. Ding, Small Methods, 2021, 5, e2100935 CrossRef PubMed.
- J. Liu, Q. Wan, R. Zeng and D. Tang, Anal. Methods, 2021, 13, 3227–3232 RSC.
- H. Gong, Y. Wu, R. Zeng, Y. Zeng, X. Liu and D. Tang, Chem. Commun., 2021, 57, 8977–8980 RSC.
- Y. Li, R. Zeng, W. Wang, J. Xu, H. Gong, L. Li, M. Li and D. Tang, ACS Sens., 2022, 7, 1593–1601 CrossRef CAS PubMed.
- X. Zhan, J. Zhou, Y. Jiang, P. An, B. Luo, F. Lan, B. Ying and Y. Wu, Biosens. Bioelectron., 2023, 229, 115229 CrossRef CAS PubMed.
- L. Li, H. Tian, G. Wang, S. Ren, T. Ma, Y. Wang, S. Ge, Y. Zhang and J. Yu, Biosens. Bioelectron., 2023, 228, 115224 CrossRef CAS PubMed.
- L. Li, S. Yu, J. Wu and H. Ju, Anal. Chem., 2023, 95, 7396–7402 CrossRef CAS PubMed.
- L. Yu, Y. Peng, M. Sheng, Q. Wang, J. Huang and X. Yang, ACS Sens., 2023, 8, 2852–2858 CrossRef CAS PubMed.
- A. Su, Y. Liu, X. Cao, J. Zhao, W. Xu, C. Liang, P. Li and S. Xu, Anal. Chem., 2023, 95, 5927–5936 CrossRef CAS PubMed.
- J. H. Choi, M. Shin, L. Yang, B. Conley, J. Yoon, S. N. Lee, K. B. Lee and J. W. Choi, ACS Nano, 2021, 15, 13475–13485 CrossRef CAS PubMed.
- Y. Xue, X. Luo, W. Xu, K. Wang, M. Wu, L. Chen, G. Yang, K. Ma, M. Yao, Q. Zhou, Q. Lv, X. Li, J. Zhou and J. Wang, Anal. Chem., 2023, 95, 966–975 CAS.
- C. Guo, X. Ma, F. Gao and Y. Guo, Front. Bioeng. Biotechnol., 2023, 11, 1143157 CrossRef PubMed.
- J. Zhu, R. Fang, Z. Pan and X. Qian, BMC cancer, 2022, 22, 716 CrossRef CAS PubMed.
- K. C. A. Chan, J. K. S. Woo, A. King, B. C. Y. Zee, W. K. J. Lam, S. L. Chan, S. W. I. Chu, C. Mak, I. O. L. Tse, S. Y. M. Leung, G. Chan, E. P. Hui, B. B. Y. Ma, R. W. K. Chiu, S. F. Leung, A. C. van Hasselt, A. T. C. Chan and Y. M. D. Lo, N. Engl. J. Med., 2017, 377, 513–522 CrossRef CAS PubMed.
- Z. R. Bi, M. L. Hu, Y. Z. Jiang, E. H. Xiong, B. W. Shu, S. Q. Li, H. W. Chen, X. H. Chen and X. M. Zhou, J. Anal. Test., 2022, 6, 353–364 CrossRef PubMed.
- T. Kim and C. M. Croce, Exp. Mol. Med., 2023, 55, 1314–1321 CrossRef CAS PubMed.
- U. Pastorino, M. Boeri, S. Sestini, F. Sabia, G. Milanese, M. Silva, P. Suatoni, C. Verri, A. Cantarutti, N. Sverzellati, G. Corrao, A. Marchiano and G. Sozzi, Ann. Oncol., 2022, 33, 395–405 CrossRef CAS PubMed.
- D. Shaterabadi, M. Zamani Sani, F. Rahdan, M. Taghizadeh, M. Rafiee, N. Dorosti, A. Dianatinasab, M. Taheri-Anganeh, P. Asadi, S. H. Khatami and A. Movahedpour, Clin. Chim. Acta, 2024, 552, 117676 CrossRef CAS PubMed.
- Z. Weng, Z. You, J. Yang, N. Mohammad, M. Lin, Q. Wei, X. Gao and Y. Zhang, Angew. Chem., Int. Ed., 2023, 62, e202214987 CrossRef CAS PubMed.
- Y. Xu, C. Wang, G. Liu, X. Zhao, Q. Qian, S. Li and X. Mi, Biosens. Bioelectron., 2022, 217, 114671 CrossRef CAS PubMed.
- H. C. Metsky, N. L. Welch, P. P. Pillai, N. J. Haradhvala, L. Rumker, S. Mantena, Y. B. Zhang, D. K. Yang, C. M. Ackerman, J. Weller, P. C. Blainey, C. Myhrvold, M. Mitzenmacher and P. C. Sabeti, Nat. Biotechnol., 2022, 40, 1123–1131 CrossRef CAS PubMed.
- S. Hersey, S. Keller, J. Mathews, L. King, A. Bandukwala, F. Berisha, M. Birchler, J. Bower, V. Clausen, J. Duarte, F. Garofolo, S. Hopper, S. Kar, O. Mabrouk, J. C. Marshall, K. McGuire, M. Naughton, Y. Saito, I. Schuhmann, G. Sperinde, P. Teixeira, A. Vitaliti, Y. M. Wang, R. Wnek, Y. Zhang, S. Spitz, V. Decman, S. Eck, J. Estevam, P. Goihberg, E. G. Alcaide, C. Gonneau, M. N. Hedrick, G. Hopkins, F. Junker, S. Nuti, U. Sommer, N. Standifer, C. Stevens, E. Stevens, C. Hendricks, M. Wadhwa, A. Torri, M. Ma, S. Harris, S. Kumar, M. A. Partridge, T. Caiazzo, S. Chilewski, I. Cludts, K. Coble, B. Gorovits, C. Grimaldi, G. Jordan, J. Kamerud, B. Leary, M. Liang, H. Lim, A. Mayer, E. O'Connor, N. Palackal, J. Poetzl, S. Prior, M. R. Abhari, N. Savoie, C. Soo, M. Ware, B. Wu, Y. Xu, T. Y. Yang and J. Zoghbi, Bioanalysis, 2021, 14, 627–692 CrossRef PubMed.
- Y. Shen, K. Hu, M. Yuan, G. Duan, Y. Guo and S. Chen, J. Appl. Microbiol., 2023, 134, lxad035 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00 340c |
| This journal is © The Royal Society of Chemistry 2024 |