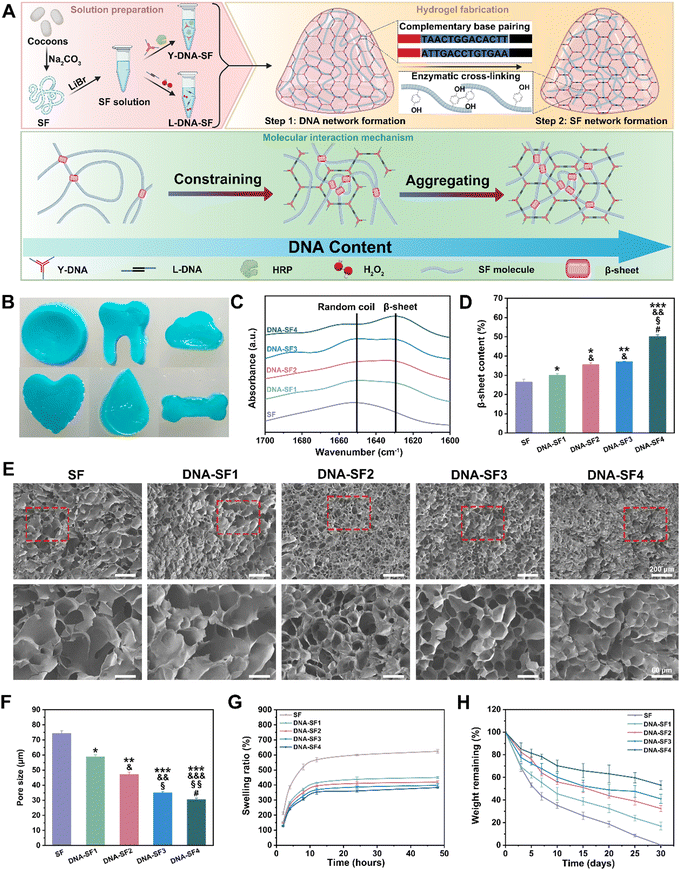
Dual-network DNA–silk fibroin hydrogels with controllable surface rigidity for regulating chondrogenic differentiation†
Ziyang
Zhou‡
abcde,
Peiran
Song‡
abc,
Yan
Wu‡
abc,
Miaomiao
Wang‡
abc,
Congyi
Shen
abcde,
Zhixin
Ma
abcde,
Xiaoxiang
Ren
abc,
Xiuhui
Wang
 abc,
Xiao
Chen
f,
Yan
Hu
f,
Zuhao
Li
abcf,
Qin
Zhang
abc,
Xiao
Chen
f,
Yan
Hu
f,
Zuhao
Li
abcf,
Qin
Zhang
 *abc,
Mengmeng
Li
*abc,
Zhen
Geng
*abc,
Mengmeng
Li
*abc,
Zhen
Geng
 *abc and
Jiacan
Su
*abc and
Jiacan
Su
 *abcf
*abcf
aInstitute of Translational Medicine, Shanghai University, Shanghai, 200444, China. E-mail: sabrina_1985@shu.edu.cn; mengmengli@shu.edu.cn; nanboshan1987@163.com; drsujiacan@163.com
bOrganoid Research Center, Shanghai University, Shanghai, 200444, China
cNational Center for Translational Medicine (Shanghai) SHU Branch, Shanghai University, Shanghai, 200444, China
dSchool of Medicine, Shanghai University, Shanghai, 200444, China
eSchool of Life Sciences, Shanghai University, Shanghai, 200444, China
fDepartment of Orthopedics, Xinhua Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200092, China
First published on 3rd January 2024
Abstract
Osteoarthritis (OA) is a common joint disease known for cartilage degeneration, leading to a substantial burden on individuals and society due to its high disability rate. However, current clinical treatments for cartilage defects remain unsatisfactory due to the unclear mechanisms underlying cartilage regeneration. Tissue engineering hydrogels have emerged as an attractive approach in cartilage repair. Recent research studies have indicated that stem cells can sense the mechanical strength of hydrogels, thereby regulating their differentiation fate. In this study, we present the groundbreaking construction of dual-network DNA–silk fibroin (SF) hydrogels with controllable surface rigidity. The supramolecular networks, formed through DNA base-pairing, induce the development of β-sheet structures by constraining and aggregating SF molecules. Subsequently, SF was cross-linked via horseradish peroxidase (HRP)-mediated enzyme reactions to form the second network. Experimental results demonstrated a positive correlation between the surface rigidity of dual-network DNA–SF hydrogels and the DNA content. Interestingly, it was observed that dual-network DNA–SF hydrogels with moderate surface rigidity exhibited the highest effectiveness in facilitating the migration of bone marrow mesenchymal stem cells (BMSCs) and their chondrogenic differentiation. Transcriptome sequencing further confirmed that dual-network DNA–SF hydrogels primarily enhanced chondrogenic differentiation of BMSCs by upregulating the Wnt and TGF-β signaling pathways while accelerating collagen II synthesis. Furthermore, in vivo studies revealed that dual-network DNA–SF hydrogels with moderate surface rigidity significantly accelerated cartilage regeneration. In summary, the dual-network DNA–SF hydrogels represent a promising and novel therapeutic strategy for cartilage regeneration.
New conceptsIn this work, we have pioneered the combination of DNA with SF to fabricate dual-network DNA–SF hydrogels with tunable surface rigidity. The introduction of a DNA supramolecular network induced a transformation in SF molecules from random coils to β-sheet, allowing for precise control over the surface rigidity of dual-network DNA–SF hydrogels. Notably, hydrogels with moderate surface rigidity were found to be more conducive to chondrogenic differentiation of BMSCs. Transcriptome analysis revealed that these dual-network DNA–SF hydrogels primarily enhanced collagen-containing extracellular matrix synthesis in BMSCs by upregulating Wnt and TGF-β signaling pathways, thereby promoting chondrogenesis. Importantly, hydrogels with moderate surface rigidity significantly facilitated cartilage regeneration in a rat model of articular cartilage defects. In summary, dual-network DNA–SF hydrogels with moderate surface rigidity hold immense potential for effective cartilage repair, offering valuable insights into the design of cartilage repair materials and the construction of cartilage organoids. |
1. Introduction
Osteoarthritis (OA) is a degenerative joint disease characterized by extensive structural alterations within the entire joint, primarily distinguished by cartilage defects.1,2 Cartilage defects result in severe pain for affected patients, sometimes escalating to the risk of amputation.3 With the global aging population and increasing obesity rates, the number of OA patients is expected to surge dramatically in the next decade. This poses a significant burden on both the healthcare system and the socioeconomic aspects of society.4,5 Cartilage, a non-vascularized and non-innervated tissue, is primarily composed of water, collagen II, and proteoglycans.6 It contains only one type of cell, chondrocytes, which exhibit low mitotic activity.7,8 Therefore, the self-repair capability of cartilage is extremely limited. Current treatment strategies for cartilage defects include microfracture, autologous chondrocyte transplantation, and autologous/allogeneic cartilage transplantation.9 However, these widely used clinical treatment strategies still have limitations. For instance, microfracture may lead to cartilage fibrosis, and the autologous chondrocyte transplantation strategy is constrained by the scarcity of available cartilage cells and suboptimal treatment outcomes.10 Moreover, allogeneic/autologous cartilage transplantation also faces challenges such as donor shortage, poor graft integration, and surgical infections.11 Indeed, it is imperative for clinical practice to explore novel strategies to address cartilage defects.Recently, there has been significant progress in using tissue engineering scaffolds to repair cartilage defects.12–15 Particularly, hydrogels have become the most attractive choice in tissue engineering scaffolds.16 Different from other types of scaffolds, hydrogels have garnered extensive attention due to their versatility and structural similarity to the extracellular matrix (ECM). They exhibit remarkable water absorption capacity, biodegradability, biocompatibility, and a 3D porous structure.17–20 Notably, cells can perceive the mechanical properties of biomaterials through mechanotransduction pathways.21 These mechanical properties have a significant impact on regulating cell migration, differentiation, and stimulating the direction of cell secretion related substances.22 For example, Huang et al. prepared a collagen I-based hydrogel with adjustable mechanical properties.23 They demonstrated that hydrogels with adjustable mechanical characteristics could modulate the chondrogenic differentiation of stem cells through Rho-associated kinase-dependent apoptosis. Similarly, Wu et al. engineered polyethylene glycol-lated poly(glycerol sebacate) hydrogels with adjustable mechanical properties by altering the crosslinking degree and composition ratio.24 Their research revealed that stiff hydrogels promoted osteogenic differentiation, hydrogels with moderate mechanical properties favored chondrogenic differentiation, and soft hydrogels facilitated adipogenic differentiation.25–27
Silk fibroin (SF) is a natural macromolecular material renowned for its exceptional mechanical properties, biocompatibility, biodegradability, and processability.28 It has a long history of diverse applications, spanning from ancient silk products to modern essentials in medical domains such as surgical sutures and wound dressings, alongside cosmetic items like facial masks and soaps.29 Presently, an array of scholars have made substantive strides in utilizing SF-based hydrogels for addressing cartilage defects.30–32 However, the synthesis of SF-based hydrogels necessitates intricate degumming and dissolution procedures, which may perturb the native SF protein structure and consequently compromise the mechanical properties of SF-based hydrogels. Deoxyribonucleic acid (DNA) emerges as an exceptional functional constituent of biomaterials, distinguished by its programmable sequence and precisely tailored functionalities.33,34 Within the realm of DNA-based materials, three-dimensional DNA hydrogels manifest outstanding biocompatibility, tunable mechanical characteristics, and suitability for three-dimensional cell printing. These attributes have propelled them into various domains, encompassing tissue engineering, therapeutic interventions, protein engineering, and the arena of soft robotics. DNA-based hydrogels are held in high regard as critical biomedical materials, heralding immense application value and auspicious prospects.35–37 Based on this, we propose the concept of constructing dual-network DNA–SF composite hydrogels. On the one hand, SF can serve as a molecular scaffold for DNA. On the other hand, DNA can reinforce the SF-based hydrogels, endowing them with controllable mechanical properties, thus synergistically contributing to cartilage regeneration and repair.
In this study, we have successfully developed novel dual-network DNA–SF hydrogels with controllable surface rigidity for the first time (Scheme 1). The supramolecular network formed by DNA through base complementary pairing can constrain and aggregate SF molecules, thereby inducing the formation of β-sheet structures. Concurrently, SF molecules form a second network through enzymatic cross-linking mediated by horseradish peroxidase (HRP). The experimental results demonstrate that the surface rigidity of the dual-network DNA–SF hydrogels is positively correlated with the DNA content. Interestingly, dual-network DNA–SF hydrogels with moderate surface rigidity were conducive to bone marrow mesenchymal stem cell (BMSC) migration. Simultaneously, this hydrogel exhibits the optimal promotion in chondrogenic differentiation of BMSCs. Transcriptome sequencing results showed that dual-network DNA–SF hydrogels with moderate surface rigidity up-regulated Wnt and TGF-β signaling pathways, thereby promoting ECM containing collagen expression and inducing chondrogenic differentiation of BMSCs. Finally, in vivo studies demonstrated that the implantation of dual-network DNA–SF hydrogels significantly promoted cartilage regeneration. Therefore, dual-network DNA–SF hydrogels with moderate surface rigidity hold immense potential as a promising approach for treating cartilage defects.
 | ||
| Scheme 1 Schematic illustration of the design of dual network DNA–SF hydrogels with controllable surface rigidity for regulating chondrogenic differentiation to repair cartilage defects. | ||
2. Results and discussion
2.1. Fabrication and characterization of dual-network DNA–SF hydrogels
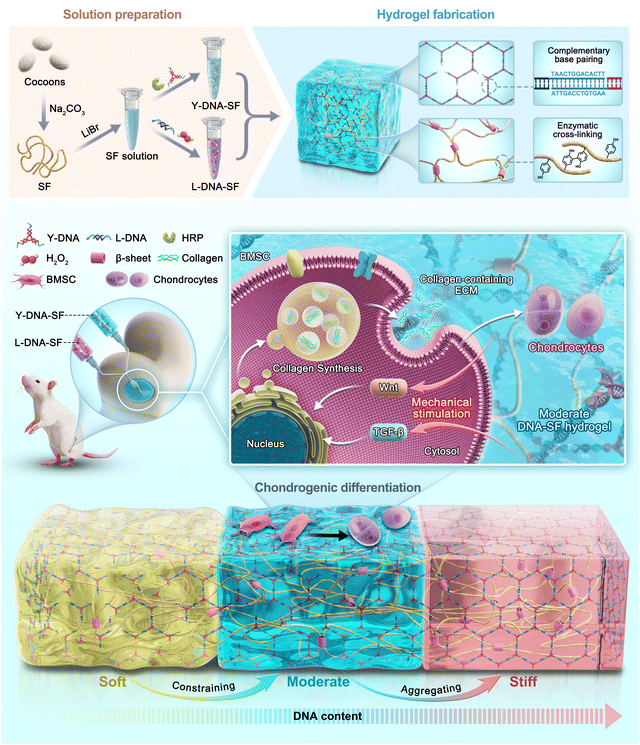
The preparation process of dual-network DNA–SF composite hydrogels is shown in Fig. 1A. The B. mori cocoons were subjected to the process of degumming and dissolution to obtain the SF solution. The Y-shaped DNA modules (Y-DNA)-SF solution was prepared by introducing Y-DNA and HRP into the SF solution, while the L-shaped DNA modules (L-DNA)-SF solution was prepared by combining L-DNA and hydrogen peroxide (H2O2) with the SF solution. Once the two hydrogel precursor solutions were thoroughly mixed, the Y-DNA and L-DNA rapidly engaged in physical cross-linking via base pairing, forming a DNA supramolecular network. Subsequently, SF molecules participated in a secondary network formation through enzymatic cross-linking reaction mediated by HRP/H2O2, resulting in the formation of stable dual-network DNA–SF hydrogels. In theory, the introduction of another macromolecular networks would constrain and aggregate SF molecules, thereby inducing SF molecules to transition into β-sheet conformations.38,39 Therefore, we postulated that an increase in the DNA content within the system would correspondingly increase the β-sheet content within the SF network. To verify this hypothesis, we conducted Fourier-transform infrared (FTIR) tests on all hydrogel groups, plotting the infrared absorption spectra within the range of 1600 to 1700 cm−1 (Fig. 1C and Fig. S1, ESI†). The experimental results unequivocally demonstrated that as the DNA content increased, the absorption peak of the hydrogel at 1650 cm−1 gradually shifted towards 1629 cm−1. This shift signifies that the introduction of the DNA network induced a conformational transition of SF molecules from random coils to β-sheet structures. To quantitatively assess the proportion of the β-sheet content, we employed PeakFit software for Fourier self-deconvolution of the infrared absorption spectra (Fig. 1D and Fig. S2, ESI†). The results indicated that the DNA supramolecular network significantly increased the β-sheet content in SF molecules. Moreover, a higher DNA content corresponded to a greater proportion of β-sheet conformation.We further observed the morphology and structure of the hydrogels. The physical pictures of dual-network DNA–SF composite hydrogels (with blue ink) are shown in Fig. 1B. These hydrogels exhibited not only adaptability to standard circular molds but also a remarkable capacity to conform to complex geometries, including teeth, clouds, hearts, water droplets, and bones. This observation underscored the exceptional adaptability of the dual-network DNA–SF composite hydrogels, rendering them versatile for accommodating various shapes of cartilage defects. To observe the internal structure of the hydrogels, we subjected them to freeze-drying and subsequently examined their cross-sections using a cold field emission scanning electron microscope (SEM) (Fig. 1E). In all groups, the hydrogels exhibited porous architectures. Importantly, the incorporation of DNA had a discernible impact on reducing the pore size within the hydrogels. Quantitative analysis of hydrogel pore sizes, conducted using ImageJ software, confirmed that an increase in the DNA content corresponded to a gradual reduction in the hydrogel pore size, consistent with our earlier hypothesis (Fig. 1F). The porous internal structure of the hydrogels was instrumental in facilitating the transport of vital nutrients and metabolic waste, which were imperative for cellular activities, thereby creating a conducive microenvironment for cellular functions. Subsequently, we investigated the swelling behavior of the hydrogels. Fig. 1G illustrates the swelling behavior of both the SF hydrogel and DNA–SF hydrogels immersed in a phosphate buffered saline (PBS) solution. Notably, the DNA–SF hydrogel exhibited substantially reduced swelling compared to the pure SF hydrogel. This diminished swelling can be primarily attributed to the higher β-sheet content in the dual-network hydrogels. As the DNA content increased, a corresponding decrease in the swelling ratio of the hydrogel was observed. Remarkably, the DNA–SF4 group, possessing the highest DNA content, exhibited a swelling rate of 383.40%. In contrast, the pure SF hydrogel displayed a notably higher swelling rate, reaching 624.16%. These findings underscored the role of DNA in significantly enhancing the stability of the hydrogels. Furthermore, the degradation performance of the hydrogels holds substantial importance in the realm of cartilage repair. Rapid degradation can impede the process of cartilage regeneration. Therefore, we evaluated the degradation behavior of the hydrogels by tracking the remaining mass of the respective hydrogels within enzyme solutions. The degradation profiles are presented in Fig. 1H. After 30 days, the pure SF hydrogel was entirely degraded, while the dual-network DNA–SF hydrogels exhibited relative stability in the enzyme-rich environment under identical conditions. Enzymatic activity predominantly targeted the non-crystalline regions of SF molecules within the hydrogel. This indicated that the dual-network DNA–SF hydrogels possessed a higher proportion of crystalline regions, specifically β-sheet structures, corroborating our earlier FTIR results. This outcome proved the superior structural stability of the DNA–SF hydrogel, with its slower degradation rate presenting favorable conditions for long-term applications in cartilage tissue engineering.
To gain deeper insights into the mechanistic influence of DNA on hydrogels and to mitigate experimental biases arising from variations in the DNA content within the hydrogel system, we prepared and characterized a series of hydrogels-E (hydrogels with equal contents of DNA), namely SF-E, DNA–SF1-E, DNA–SF2-E, DNA–SF3-E, and DNA–SF4-E. These hydrogels were formulated with uniform concentrations of both DNA and SF, achieved by the introduction of non-functional DNA fragments that do not participate in the construction of DNA supramolecular networks within the aforementioned hydrogel system. We conducted FTIR analysis on these hydrogels-E (Fig. S3, ESI†). Experimental results indicated that the β-sheet content of hydrogels-E was comparable to that of the previous hydrogel groups. This suggests that non-functional DNA fragments do not induce the formation of β-sheets in SF molecules. The results demonstrated that the supramolecular network composed of DNA was a critical factor in inducing the formation of β-sheets in SF molecules. Additionally, we performed swelling and degradation experiments on hydrogels-E (Fig. S4, ESI†). The experimental results showed slight variations compared to both pure SF hydrogels and DNA–SF hydrogels, indicating that the impact of DNA fragments on hydrogel properties was subtle, underscoring the primary influence of the DNA supramolecular network.
2.2. Surface rigidity of the dual-network DNA–SF hydrogels
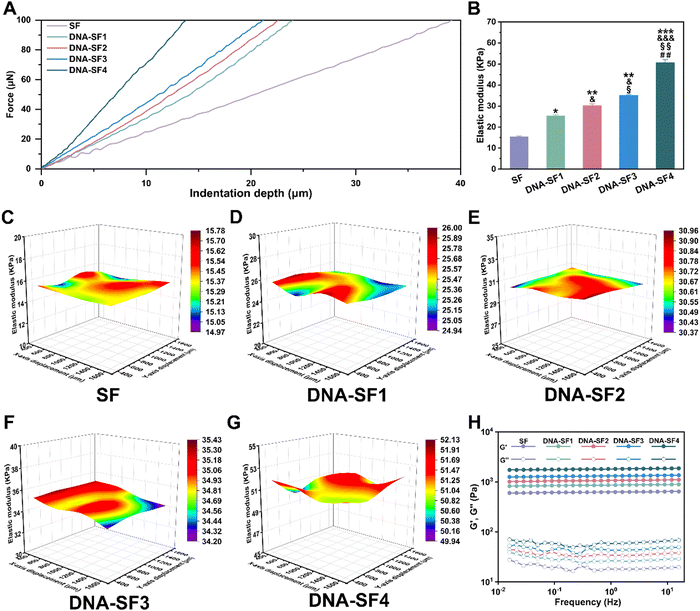
The surface rigidity of SF hydrogels and dual-network DNA–SF hydrogels was meticulously assessed via nanoindentation testing employing a precision nanoindenter. Stress–strain curves and corresponding equivalent elastic moduli (surface rigidity) for SF hydrogels and dual-network DNA–SF hydrogels are illustrated in Fig. 2A and B. These results demonstrated that the strain exhibited by dual-network DNA–SF hydrogels was notably lower than that of SF hydrogels when the applied stress attains 100 μN. Additionally, the surface rigidity of dual-network DNA–SF hydrogels significantly surpassed that of SF hydrogels. Significantly, consonant with variations in the β-sheet content within the hydrogels, a higher DNA content corresponds to an elevated surface rigidity. (The surface rigidity of hydrogels within the low-to-high DNA content groups is 15.41 kPa, 25.28 kPa, 30.21 kPa, 35.02 kPa, and 50.67 kPa, respectively.) We speculated that the DNA network induced the formation of β-sheets in the SF network, thus enhancing the mechanical properties of the dual-network DNA–SF composite hydrogels. To validate this hypothesis, we prepared SF hydrogels with varying β-sheet contents (treated with ethanol for 10, 30, and 60 seconds, respectively) and characterized their surface rigidity (Fig. S5, ESI†). The results revealed that with an increase in the β-sheet content, the surface rigidity of the SF hydrogels also increased. Similarly, we subjected hydrogels-E to surface rigidity testing using a nanoindenter (Fig. S6, ESI†). The results revealed that the incorporation of non-functional DNA fragments had non-significant impact on the surface rigidity of the hydrogels, consistent with our earlier conclusion that the addition of non-functional DNA fragments did not elevate the proportion of β-sheets in the hydrogels. These findings provided further evidence that the direct influence of DNA on the hydrogel was not substantial. Instead, the supramolecular network constituted by DNA effectively constrained and aggregated SF molecules, inducing the formation of β-sheets in SF and thereby enhanced the surface rigidity of the hydrogels. Additionally, to further scrutinize the uniformity of mechanical attributes across SF hydrogels and DNA–SF hydrogels, matrix testing was systematically conducted on each hydrogel group using the nanoindenter. Fig. 2C–F show three-dimensional (3D) distribution maps of surface rigidity obtained through matrix testing across different hydrogel groups. The findings illuminated that within the same hydrogel, the surface rigidity across different regions exhibited remarkable uniformity. Notably, the DNA–SF2 group displayed the narrowest range of surface rigidity, measuring 0.59 kPa. This observation implies that the density of the DNA network within the system induces an ordered arrangement of SF molecules. Intriguingly, with a further increase in DNA network density, the uniformity within the hydrogel diminishes. The DNA–SF4 group exhibited a surface rigidity range of 2.19 kPa. Our conjecture was that the high-density DNA network encourages a higher proportion of β-sheet formation within the SF network, and these β-sheets are attracted and aggregated via hydrophobic interactions, leading to localized structural heterogeneity within the hydrogels.40 Subsequently, we conducted rheological tests on both SF hydrogels and DNA–SF hydrogels. Fig. 2H illustrates the frequency sweep curves for each hydrogel group. All groups exhibited the classical mechanical attributes characteristic of hydrogels. Furthermore, the storage modulus of the hydrogels displayed a direct correlation with the DNA content. This implies that the incorporation of DNA contributes to the enhancement of structural stability within the hydrogels, corroborating the previous findings from the nanoindentation tests. It is noteworthy that a high β-Sheet content in SF molecules may impact the efficiency of the HRP-mediated enzyme crosslinking reaction. Therefore, we conducted time scans using a rheometer on SF hydrogels and DNA–SF hydrogels to explore the time required for the hydrogels to reach a stable state (Fig. S7, ESI†). Experimental results revealed that the introduction of the DNA supramolecular network led to an increase in β-the sheet content in SF, thereby slightly prolonging the time required for the hydrogel to reach a stable state. The higher the DNA content in the hydrogel system, the longer the time required for the hydrogel to reach a stable state. Nevertheless, we posit that this marginal time extension for the hydrogels to reach a stable state has almost negligible the internal repair of cartilage defects (3 months) within the hydrogel system (216 seconds for the SF group, 391 seconds for the DNA–SF4 group). In conclusion, the DNA supramolecular network exerted a significant influence by augmenting the surface rigidity and stability of the dual-network DNA–SF hydrogels.2.3. In vitro proliferation and migration of BMSCs on dual-network DNA–SF composite hydrogels
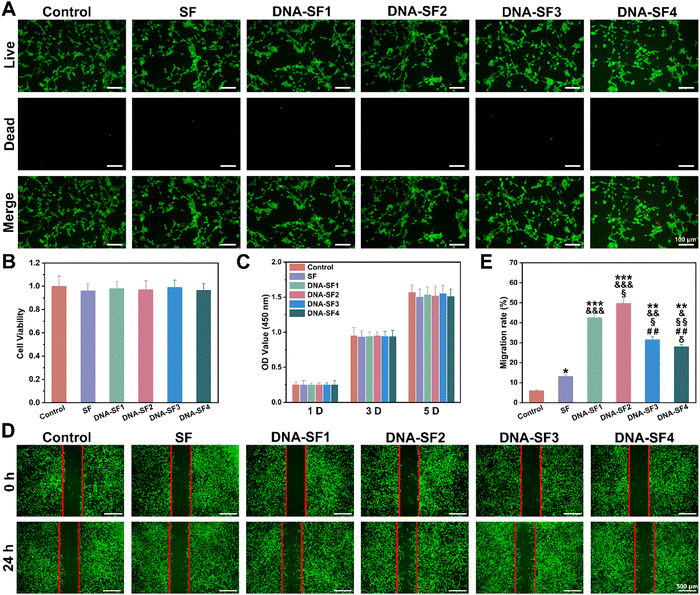
Based on the comprehensive characterization of the physicochemical properties and mechanical performance of the dual-network DNA–SF hydrogels, we conducted an in-depth evaluation of their in vitro cell compatibility through live/dead staining and cell counting kit-8 (CCK-8) analysis. Following a 3-day co-culture of the hydrogels with BMSCs, we performed live/dead staining using the calcein AM/PI double staining kit to assess cell viability (Fig. 3A). Results indicated that both the SF hydrogel and DNA–SF hydrogel group predominantly exhibited viable BMSCs (indicated by green fluorescence) with minimal presence of dead cells (indicated by red fluorescence), consistent with the control group. Quantitative analysis corroborated these findings, as the cell viability of BMSCs in both the SF hydrogel group and the DNA–SF hydrogel group exceeded 96% (Fig. 3B). This suggests that both the SF hydrogel and the DNA–SF hydrogel were non-toxic to cells and exhibited excellent cell compatibility. Additionally, we evaluated the effect of the hydrogels on BMSC proliferation using the CCK-8 assay. The results demonstrated that BMSCs exhibited normal proliferation in both the SF hydrogel and DNA–SF hydrogel groups, with no significant differences compared to the control group (Fig. 3C). This finding reaffirmed the great cell compatibility of both the SF hydrogel and DNA–SF hydrogel, as they did not interfere with proliferation of BMSCs. The migratory behavior of BMSCs assumes paramount importance in cartilage repair. Thus, we examined the impact of the hydrogels on migration behavior of BMSCs by using the scratch assay (Fig. 3D and E). As observed in the images, the presence of the hydrogels provided a supportive substrate for attachment and migration of BMSCs. Furthermore, the DNA–SF2 group exhibited a significantly enhanced BMSC migration rate, reaching 49.76%, compared to other groups. The lower mechanical strength (SF group and DNA–SF1 group) and too small pore size (DNA–SF3 group and DNA–SF3 group) may hinder the migration of BMSCs.2.4. Dual-network DNA–SF composite hydrogels regulated chondrogenic differentiation of BMSCs
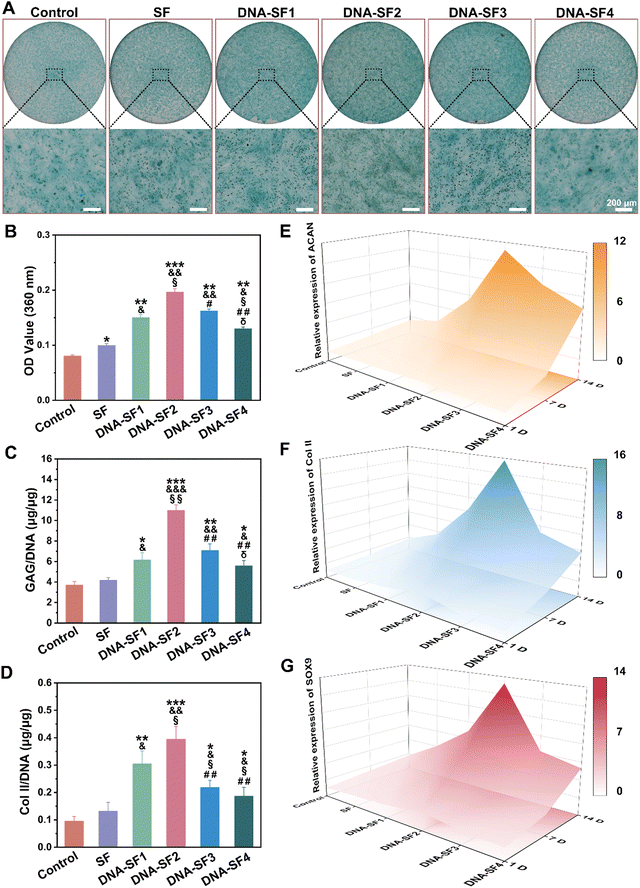
After characterizing the effect of hydrogels on BMSC proliferation and migration, we further investigated their impact on BMSC chondrogenic differentiation. To facilitate the formation of cartilage tissue, we seeded BMSCs on the surfaces of both SF hydrogels and DNA–SF hydrogels for chondrogenic induction. Fig. 4A and B display the alcian blue staining and its quantitative analysis of BMSCs co-cultured with the hydrogels. The staining results demonstrated an initial increase followed by a decrease in positive signals with the rise in the DNA content within the hydrogels, with the DNA–SF2 group exhibiting the most pronounced positive signal. This result proved that, relative to the control group and other hydrogel groups, the DNA–SF2 group provided a more conducive environment for heightened chondrogenic differentiation and the secretion of an increased quantity of proteoglycans by BMSCs. It demonstrated that the DNA–SF2 hydrogel, with moderate DNA content (a surface rigidity of 30.21 kPa) furnished an optimal microenvironment for chondrogenic differentiation. To further scrutinize the impact of various hydrogels on the chondrogenic differentiation of BMSCs, we conducted an analysis of ECM components in the co-cultured and induced cells, focusing on glycosaminoglycans (GAG) and type II collagen (Col II) (Fig. 4C and D). The results mirrored the outcomes of the alcian blue staining, as the main components of chondrocyte staining (GAG and Col II) exhibited an initial increase followed by a subsequent decrease with the augmentation of the DNA content in the hydrogels (the DNA–SF2 group had the highest content). Subsequently, we delved into an in-depth examination of the chondrogenic differentiation of BMSCs on the surfaces of various hydrogel groups through quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Fig. 4E–G and Fig. S8, ESI†). The expression levels of chondrogenic genes (ACAN, Sox9, and Col II) were compared among the groups. After 1, 7 and 14 days of chondrogenic induction, all the dual-network DNA–SF hydrogel groups exhibited significantly elevated expression levels of ACAN, Sox9, and Col II in comparison to SF hydrogels and the control group. Importantly, within the DNA–SF hydrogel groups, an initial surge followed by a subsequent decrease in the expression levels of ACAN, Sox9, and Col II mRNA in the induced cells were observed as the DNA content in the hydrogels increased, with the DNA–SF2 group registering the highest levels.In addition, to eliminate the potential influence of varying DNA content on the hydrogel matrix, we co-cultured BMSCs with hydrogels-E. Fig. S9A and B (ESI†) depict alcian blue staining and its quantitative analysis of BMSCs co-cultured with the hydrogels-E. The experimental results closely resemble those of previous SF hydrogels and DNA–SF hydrogels, with the DNA–SF2-E hydrogel group exhibiting the strongest positive signal. Subsequently, quantitative analyses of ECM chondrogenic differentiation markers in these cells after 7 days of co-culture were performed (Fig. S9C and D, ESI†). Although there were fluctuations in the expression of chondrogenic markers GAG and Col II compared to previous results, the overall trend remained consistent, with the DNA–SF2-E hydrogel group showing the highest expression levels of GAG and Col II. Furthermore, we assessed the expression levels of chondrogenic differentiation marker genes in these cells through qRT-PCR after 1, 7, and 14 days of chondrogenic induction (Fig. S9E–G, ESI†). The DNA–SF2-E hydrogel group consistently exhibited the highest expression levels of chondrogenic differentiation marker genes in co-cultured BMSCs at these three time points. These results indicated that, even after accounting for differences in the DNA content within the hydrogel matrix, the DNA–SF2-E hydrogel, characterized by a moderate surface rigidity, exerted the most favorable effects in promoting BMSC chondrogenic differentiation. This validated that, in this series of hydrogels in our study, the impact of DNA itself on BMSC chondrogenic differentiation is minimal, and the primary regulatory role is played by the surface stiffness of the hydrogel.
2.5. Dynamic analysis of dual-network DNA–SF composite hydrogels
After the preparation of hydrogels, their properties (including mechanical performance and the ability to induce chondrogenic differentiation) are not constant. In SF-based hydrogels, the secondary structure of SF molecules gradually transforms from the α-helix to β-sheet over time. Simultaneously, as the degradation reaction progresses, various properties of SF-based hydrogels also undergo changes. Therefore, SF-based hydrogels are constantly in a state of complex and dynamic evolution. To simulate the dynamic performance changes of dual-network DNA–SF composite hydrogels in vivo, we subjected the prepared SF hydrogels and DNA–SF hydrogels to enzymatic treatment for 7 days and 14 days (based on the previous in vitro degradation experiments), followed by characterizing their surface rigidity and ability to promote chondrogenic differentiation of BMSCs (Fig. S10A and S11A, ESI†).Following 7 days of enzymatic treatment, Fig. S10B and C (ESI†) illustrated the variations in the surface rigidity of SF-7 hydrogels and DNA–SF-7 hydrogels. Compared to D 0, there was a certain degree of increase in surface rigidity for the pure SF hydrogel, DNA–SF1 hydrogel, DNA–SF2 hydrogel, and DNA–SF3 hydrogel. In contrast, the surface rigidity of the DNA–SF4 hydrogel decreased. This indicates an increase in the β-sheet content of SF molecules in the pure SF hydrogel, DNA–SF1 hydrogel, DNA–SF2 hydrogel, and DNA–SF3 hydrogel, while the β-sheet content in the DNA–SF4 hydrogel is already sufficiently high, preventing a further increase. As the DNA–SF4 hydrogel degraded, the surface stiffness decreased. Additionally, BMSCs were co-cultured with SF-7 hydrogels and DNA–SF-7 hydrogels treated with enzyme solution for 7 days. Alcian blue staining results show no significant changes compared to D 0 for SF hydrogels and DNA–SF hydrogels after 7 days of enzyme treatment (Fig. S10D and E, ESI†). Similarly, quantitative detection of GAG and Col II (Fig. S10F and G, ESI†), as well as qRT-PCR analysis (Fig. S10H–J, ESI†), yielded results similar to previous findings. This indicates that after 7 days of enzyme treatment, the DNA–SF2 hydrogel still maintained optimal performance in promoting BMSC chondrogenic differentiation.
Subsequently, we subjected the hydrogels treated with enzyme solution for 14 days to similar characterization. With the progression of the degradation reaction, the surface rigidity of all hydrogel groups exhibited a reduction (Fig. S11B and C, ESI†). Notably, the SF hydrogel experienced the most significant decline, indicating its poorer stability. This outcome corroborated with our previous findings. Alcian blue staining results, as shown in Fig. S11D and E (ESI†), revealed a certain degree of decrease in positive signals for all hydrogel groups compared to D 0 and D 7. This suggests that as the degradation reaction advanced, the hydrogel structure underwent disruption, leading to a decline in the ability to promote BMSC chondrogenic differentiation. It is noteworthy that the positive signals for the DNA–SF2 hydrogel remained the strongest. ECM component detection and qRT-PCR results consistently indicated a reduction in the effectiveness of promoting BMSC chondrogenic differentiation for all hydrogel groups when compared to D 0 and D 7 (Fig. S11F–J, ESI†). Particularly noteworthy is that the DNA–SF2 group hydrogel still outperformed other group hydrogels in its ability to promote BMSC chondrogenic differentiation.
Therefore, the DNA–SF2 hydrogels demonstrated sustained and superior efficacy in promoting BMSC chondrogenic differentiation, and DNA–SF2 hydrogels were selected for further experiments.
2.6. In vivo degradation behavior analysis of DNA–SF hydrogels
The previous dynamic analysis experimentally demonstrated that among all DNA–SF hydrogels, the DNA–SF2 hydrogel with moderate surface rigidity exhibited the most efficient promotion of BMSC chondrogenic differentiation. It is noteworthy that the in vivo degradation behavior of hydrogels was influenced by various factors, including pH, ion concentration, enzymes, mechanical stress, and temperature. To explore the in vivo degradation behavior of DNA–SF hydrogels in rats, an in vivo degradation experiment was conducted by injecting SF hydrogels (left hind legs) and DNA–SF hydrogels (right hind legs) with fluorescent dyes into the joint cavity of rats. The experimental results as shown in Fig. S12 (ESI†) indicat gradual degradation over time for both SF hydrogels and DNA–SF hydrogels, proving their excellent biodegradability. Additionally, at 3 W, the fluorescence signal of the SF hydrogel was extremely weak and nearly disappeared at 4 W. In contrast, the fluorescence signal of the DNA–SF hydrogel remained strong at 4 W. This demonstrates that the stability of the DNA–SF hydrogel was significantly superior to that of the SF hydrogel, facilitating long-term repair of cartilage defects.2.7. The transcriptome sequencing analysis of dual-network DNA–SF hydrogels on BMSCs
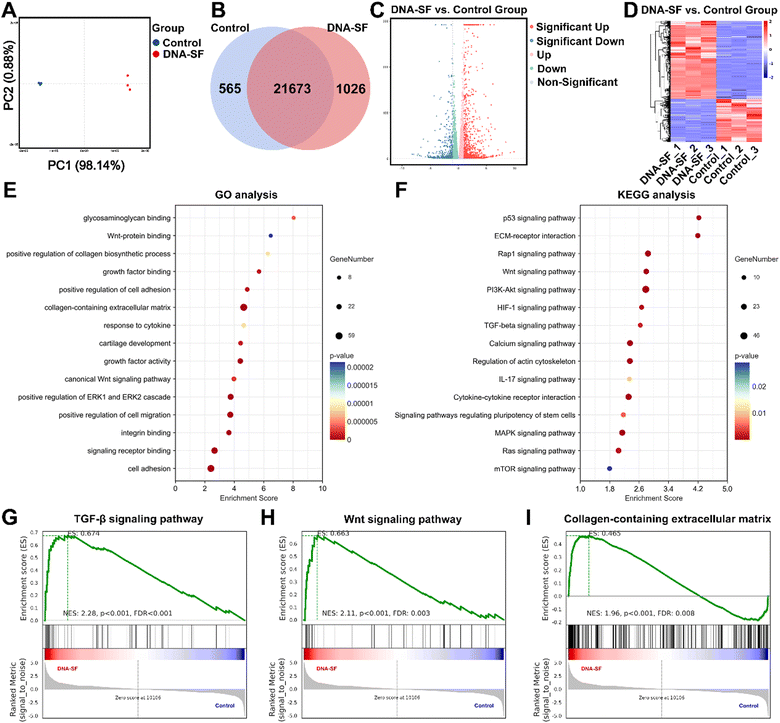
In order to further explore the biological effects of dual-network DNA–SF hydrogels on BMSCs, mRNA-seq analysis was performed. Principal component analysis was performed to compare the similarities and distinctions between the control group and the DNA–SF group (Fig. 5A). The small inter-point distances within each group indicate a high level of reproducibility, whereas the distances between the control group and the DNA–SF group are notably larger. In total, the mRNA-seq analysis revealed 1591 differentially expressed genes (DEGs), consisting of 565 downregulated genes and 1026 upregulated genes (Fig. 5B). Additionally, volcano plots and cluster heatmaps were used to visualize the DEGs within each group, as depicted in Fig. 5C and D, respectively. Gene ontology (GO) analysis was performed to analyze the functions of DEGs at three levels: biological process (BP), cellular component (CC), and molecular function (MF). In comparison to the control group, BMSCs in the DNA–SF group exhibited upregulation of genes associated with cell adhesion and cell migration. Additionally, genes associated with chondroitin sulfate biosynthesis, collagen biosynthetic process, growth factor activity, and cartilage development were upregulated in the DNA–SF group (Fig. 5E). Furthermore, KEGG pathway enrichment analysis was conducted to reveal the functions of DEGs (Fig. 5F). The analysis results were consistent with our previous findings, indicating that the dual-network DNA–SF hydrogel upregulated the mTOR pathway to promote chondrogenic differentiation of BMSCs.41 It is worth noting that the DNA–SF hydrogel upregulated ECM receptor interactions and multiple chondrogenic related signaling pathways. As shown in Fig. 5G–I and Fig. S13 (ESI†), GSEA and pathway gene heatmap results also indicate that compared to the control group, DNA–SF double-network hydrogels could regulate the secretion of collagen-containing ECM by affecting Wnt and TGF-β signaling pathways. The results from the aforementioned GO analysis, KEGG analysis, and GSEA analysis collectively demonstrated that dual-network DNA–SF hydrogels significantly upregulated genes related to adhesion and migration of BMSCs, thereby enhancing the endogenous recruitment and migration efficiency of BMSCs and augmenting the potential for cartilage regeneration. Interestingly, dual-network DNA–SF hydrogels with moderate surface rigidity provide a favorable microenvironment for BMSCs, as they respond to mechanical cues by upregulating various cartilage-promoting signaling pathways, particularly by affecting the Wnt and TGF-β signaling pathways. This led to accelerated synthesis of collagen-containing ECM and promoted chondrogenic differentiation of BMSCs.2.8. In vivo evaluation of dual-network DNA–SF composite hydrogels on BMSCs for cartilage repair
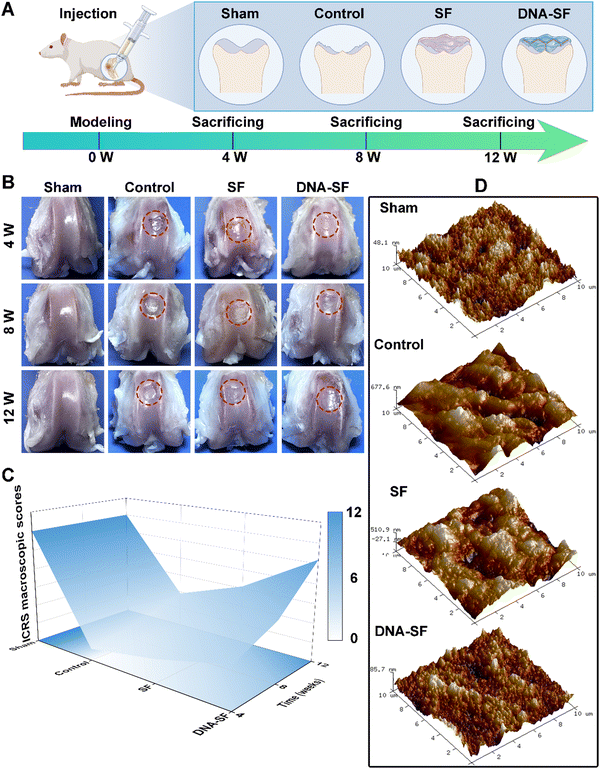
To evaluate the regenerative capacity of dual-network DNA–SF hydrogels on BMSCs in vivo, we employed the rat model of articular cartilage defect (2 × 1.5 mm). As shown in Fig. 6A, the rats were systematically allocated into four distinct groups, namely, the Sham group (representing the positive control), control group (non-treatment), SF group, and DNA–SF group. At 4, 8 and 12 weeks post-surgery, the rats were sacrificed, and tissue samples from the femur, heart, liver, spleen, lung, and kidneys were collected for subsequent experimental analyses.We obtained the distal femoral full-thickness cartilage tissue for gross observation (Fig. 6B). In comparison to the control and SF groups, the cartilage surface within the DNA–SF hydrogel group exhibited enhanced smoothness and continuity, suggesting that the regenerated cartilage closely resembled the characteristics of normal cartilage. Subsequently, we performed repair assessments based on the International Cartilage Repair Society (ICRS) macroscopic assessment (Fig. 6C and Fig. S14, ESI†). The restorative effects observed subsequent to the injection of the DNA–SF hydrogel into the cartilage defect exhibited a significantly superiority over the control and SF groups. In addition, the atomic force microscopy (AFM) images and analysis results are shown in Fig. 6D and Fig. S15 (ESI†). The height information of regenerated cartilage, collected through AFM, reveals significant variations in height among different locations in the control group. In comparison, the SF group exhibited slightly smaller height differences than the control group. In the DNA–SF group, the height differences in regenerated cartilage across various locations approach levels observed in the sham group. This demonstrates that the surfaces of regenerated cartilage in the control and SF groups were relatively rough, while the DNA–SF group displays a smoother surface, resembling the normal cartilage in the sham group.
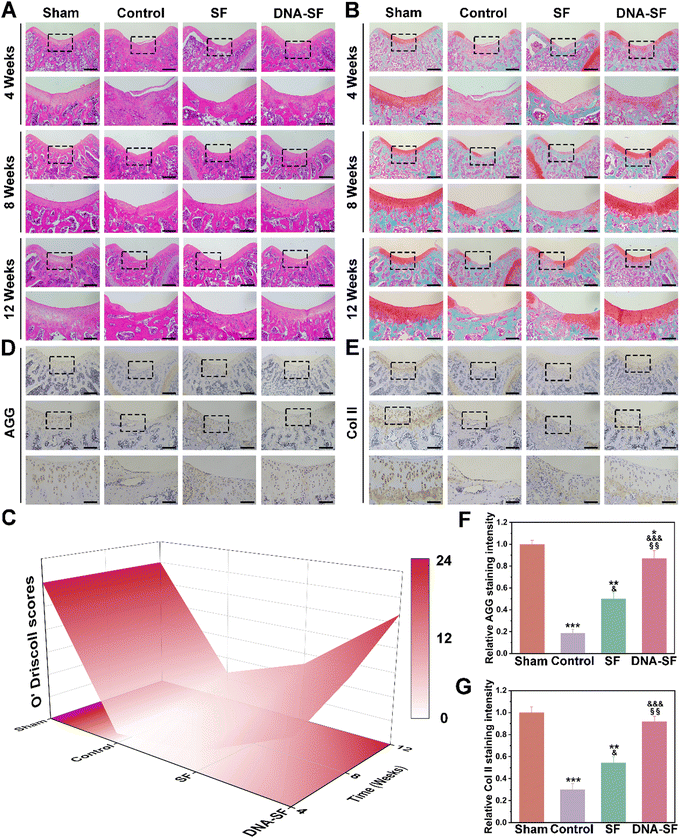
We further analyzed the tissue staining of the regenerated cartilage area, including HE and Safranin O/Fast Green, to evaluate the therapeutic effects (Fig. 7A and B). At 12 weeks post-surgery, a minimal presence of fibrous tissue was observed at the knee groove defect area of control and SF groups. Remarkably, in the DNA–SF hydrogel group, new tissue formation was evident at 8 weeks. Furthermore, at 12 weeks, the surface appeared smooth and resembled the normal tissue. Importantly, the new tissue in the DNA–SF group showed great integration with the surrounding cartilage. Additionally, the average O'Driscoll scores in the DNA–SF hydrogel group closely approached to the Sham group (Fig. 7C and Fig. S16, ESI†).
Immunohistochemical staining was conducted to assess the expression of AGG and Col II within the defect area (Fig. 7D–G). The DNA–SF hydrogel group showed higher expression levels of AGG and Col II compared to the control group and SF hydrogel group. Furthermore, at 12 weeks post-surgery, histological examination of major organs using HE staining proved the great long-term biocompatibility of the DNA–SF hydrogel (Fig. S17, ESI†). No pathological changes were found in the main organs, which further indicates that the hydrogel has good biocompatibility. The liver and kidney function tests also indicated that all parameters in each group of rats were within the normal range (Fig. S18, ESI†). These results indicated that the DNA–SF hydrogels with moderate surface rigidity could promote the secretion of cartilage-related ECM by cells, which was consistent with the transcriptome sequencing results. This further confirms that the DNA–SF hydrogels with moderate surface rigidity possessed excellent ability to promote cartilage regeneration.
Biomaterials play a key role in tissue engineering, and their mechanical properties (include surface rigidity) constitute a significant factor influencing tissue repair.42,43 The mechanical stimulation provided by biomaterials mimics the mechanical environment of developing tissues. Notably, cartilage, a specialized tissue responsible for joint cushioning and support, displays high sensitivity to mechanical stimuli within its growth microenvironment.44 Mechanical properties of biomaterials exert a dual impact. Firstly, they directly influence cellular behaviors. Under optimal mechanical conditions, BMSCs exhibit enhanced adhesion, proliferation, and differentiation. In contrast, unsuitable mechanical properties may disrupt cellular stress equilibrium, thereby affecting cell functions and directing differentiation along unintended pathways.45 Secondly, biomaterial mechanical properties also influence the synthesis of the ECM, which comprises collagen and glycosaminoglycans, the principal constituents providing mechanical support and stability to cartilage tissue.46 Previous research had indicated that biomaterials possessing higher mechanical strength tend to promote osteogenic differentiation of BMSCs, while those with lower strength favor adipogenic differentiation. Only biomaterials characterized by moderate mechanical strength proved conducive to chondrogenic differentiation of BMSCs.24 Thus, to optimize cartilage regeneration effects, the development of biomaterials with tunable mechanical properties becomes imperative.
The SF-based hydrogel, characterized by its natural origin, biocompatibility, bioactivity, and malleability, has emerged as a promising candidate for cartilage tissue engineering. However, the intricate physical and chemical processes involved in hydrogel preparation can disrupt the molecular structure of SF, rendering the pure SF hydrogel inadequate for cartilage repair applications.47 In attempts to bolster the mechanical robustness of SF hydrogels, the introduction of artificially synthesized macromolecules has proven to be most effective.48,49 Nevertheless, the incorporation of such macromolecules may compromise the biodegradability of SF hydrogels. Even for bioinert materials, prolonged in vivo implantation does not favor tissue regeneration and repair.50 Similarly, the incorporation of a secondary network into hydrogels can enhance their mechanical properties. However, many of these double-network hydrogels exhibit suboptimal biocompatibility owing to residual toxic crosslinking agents or photo-initiators.51
To meet the emerging demand as a biocompatible hydrogel with controlled mechanical strength for promoting cartilage repair, we developed novel dual-network DNA–SF hydrogels (Fig. 8). DNA, a ubiquitous biomacromolecule in living organisms, demonstrates outstanding biocompatibility and elicits minimal immune and toxic responses, rendering it highly suitable for repairing cartilage defects. Moreover, DNA can undergo self-assembly and crosslinking via base complementary pairing, effectively circumventing issues related to the toxicity associated with residual toxic crosslinkers or photo-initiators.52 Our findings indicated that the DNA supramolecular network within the hydrogel promoted the transformation of SF molecules into β-sheet structures. Importantly, we could regulate the surface rigidity of the hydrogels by controlling the DNA content. Interestingly, the dual-network DNA–SF hydrogels with moderate surface rigidity were more conducive to migration and chondrogenic differentiation of BMSCs, aligning with previous research results.23 Notably, in this study, the dual-network DNA–SF hydrogels with a 30 kPa modulus were found to be the most conducive to chondrogenic differentiation of BMSCs, in contrast to the previously reported 20 kPa. The divergence in the results may be attributed to the distinct mechanisms underlying the action of DNA–SF and PEG on BMSCs. Furthermore, we used transcriptome sequencing to reveal the mechanisms by which the dual-network DNA–SF hydrogels regulated chondrogenic differentiation of BMSCs. The DNA–SF hydrogels with moderate surface rigidity upregulated multiple chondrogenic-related signaling pathways (mainly the Wnt and TGF-β signaling pathways), promoting the secretion of collagen-containing ECM, thereby inducing chondrogenic differentiation of BMSCs. In contrast, previous studies have found that hydrogels regulate differentiation of BMSCs through the Rap1 signaling pathway.23 Moreover, in vivo experiments also demonstrated that compared to the pure SF hydrogel, the double-network DNA–SF hydrogel exhibits the best cartilage repair effect.
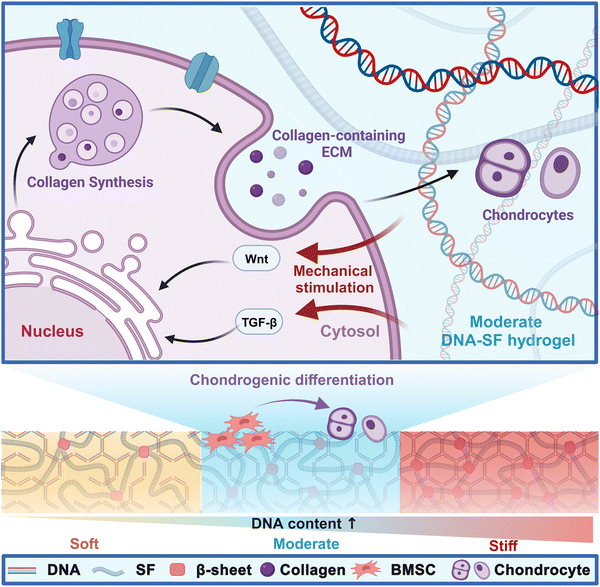 | ||
| Fig. 8 DNA–SF hydrogel regulating chondrogenic differentiation of BMSCs through mechanical stimulation. | ||
This study represents the inaugural utilization of SF in conjunction with DNA to fabricate dual-network hydrogels. It is worth noting that DNA exhibits distinctive programmability. Nevertheless, our investigation in this study primarily focused on the physical properties of DNA, specifically base pairing self-assembly, for the enhancement of SF hydrogels. In subsequent research, we intend to functionalize DNA sequences to explore the environmental responsiveness and drug delivery capabilities of the DNA–SF composite hydrogels. This strategic development may culminate in the construction of cartilage organoids by optimizing the functional attributes of DNA–SF composite hydrogels, potentially ushering in a novel platform for cartilage repair.
3. Conclusions
This study is the pioneering development of novel dual-network DNA–SF hydrogels tailored for cartilage repair applications. Through control of the DNA content within the hydrogels, successful induction of β-sheet structures in SF molecules was achieved, thereby enabling precise modulation of hydrogel surface rigidity. Importantly, we discovered that dual-network DNA–SF hydrogels with moderate surface rigidity provided a conducive microenvironment for chondrogenic differentiation of BMSCs. Mechanistically, these dual-network DNA–SF hydrogels exert mechanical stimuli on BMSCs, orchestrating the regulation of pivotal TGF-β and Wnt signaling pathways. Consequently, this regulation elicits the secretion of collagen-rich ECM and instigates BMSCs toward chondrogenic differentiation. Our findings were further corroborated through animal experiments, conclusively demonstrating the capacity of dual-network DNA–SF hydrogels to significantly expedite cartilage regeneration in a rat model. In conclusion, the dual-network DNA–SF hydrogels present a promising platform for effective cartilage repair.4. Materials and methods
4.1. Materials
B. mori cocoons were purchased from Sericulture Research Institute of Chinese Academy of Agricultural Sciences, China. ssDNA was purchased from Sangon Biotech (Shanghai) Co., Ltd, China. HRP was purchased from Macklin Chemical Reagent Co., Ltd, China. Hydrogen peroxide (H2O2), lithium bromide (LiBr), and sodium carbonate (Na2CO3) were purchased from Aladdin Reagent (Shanghai) Co., Ltd, China.4.2. Preparation of SF solution
The B. mori cocoons were precisely sectioned into small fragments and subsequently introduced into a boiling 0.02 M Na2CO3 solution for a duration of 30 × 30 minutes. This step aimed to facilitate the separation of silk sericin from SF. Following successful separation, the extracted SF underwent a meticulous purification process, drying, and was then solubilized in a 9.3 M LiBr solution, maintained at a constant temperature of 60 °C for a duration of 4 hours. Upon achieving complete dissolution, the resulting SF solution underwent a thorough dialysis procedure, spanning 2 days, with regular changes of deionized water occurring 3–4 times per day. Upon the conclusion of the dialysis process, the SF solution was carefully collected and subjected to centrifugation at 9000 rpm for 20 minutes at a controlled temperature of 4 °C, employing a high-speed centrifuge, to effectively eliminate impurities. Finally, the concentration of the SF solution was accurately determined through a drying method, ultimately yielding a 4% (w/v) SF solution.534.3. Preparation of dual-network DNA–SF hydrogels
The DNA supramolecular networks within the dual-network DNA–SF hydrogels comprise Y-DNA and L-DNA modules. Three single-stranded DNA (ssDNA) molecules, each consisting of 39 nucleotides, undergo self-assembly to form Y-DNA characterized by three 13-nucleotide sticky ends. In addition, two ssDNA molecules, each comprising 44 nucleotides, self-assemble into DNA-linker structures (L-DNA modules) featuring two 13-nucleotide sticky ends (Table S1, ESI†). Importantly, the sticky ends of the Y-DNA exhibit full complementarity with the sticky ends of the L-DNA modules. Consequently, upon further mixing of the Y-DNA and L-DNA, these sticky ends hybridize to engender the desired DNA supramolecular network. The second network within the dual-network DNA–SF hydrogels is formed by SF molecules via an enzymatic cross-linking reaction mediated by HRP and H2O2. To fabricate a series of dual-network DNA–SF hydrogels, Y1, Y2, and Y3 ssDNA were dissolved in the aforementioned SF solution, followed by the addition of HRP (20 U mL−1) to create the Y-DNA–SF solution. Similarly, L1 and L2 ssDNA were dissolved in the aforementioned SF solution, and H2O2 (0.01%) was subsequently added to obtain the L-DNA–SF solution. Finally, the dual-network DNA–SF hydrogels were prepared by blending equal proportions of the Y-DNA–SF solution and L-DNA–SF solution. Based on the DNA content within the hydrogels, they were designated as SF, DNA–SF1, DNA–SF2, DNA–SF3, and DNA–SF4 (Table 1). Additionally, in order to mitigate the influence of varying DNA content within the hydrogels and to explore the mechanisms of interaction between DNA and SF molecules, we introduced non-functional double-stranded DNA fragments (not involved in constructing DNA supramolecular networks) into the aforementioned hydrogel systems, and prepared hydrogels with equivalent DNA and SF contents (SF-E, DNA–SF1-E, DNA–SF2-E, DNA–SF3-E, and DNA–SF4-E).| Group | Y-DNA content (nmol mL−1) | L-DNA content (nmol mL−1) |
|---|---|---|
| SF | 0 | 0 |
| DNA–SF1 | 250 | 375 |
| DNA–SF2 | 500 | 750 |
| DNA–SF3 | 750 | 1125 |
| DNA–SF4 | 1000 | 1500 |
4.4. Characterization of SF and DNA–SF hydrogels
4.5. Cell culture, proliferation and migration
Primary rat BMSCs were cultured in complete medium containing alpha minimal essential medium (α-MEM), 1% penicillin–streptomycin, and 10% fetal bovine serum at 37 °C under 5% CO2. The culture medium was refreshed every 2 days, and BMSCs from the 3rd to the 5th passage were used for subsequent experiments. The pre-hydrogel solution was injected into the cell well plates. After gelation, hydrogels were subjected to UV sterilization for 2 hours. The BMSCs mentioned above were seeded onto SF or DNA–SF hydrogels and cultured for 3 days. Cell viability was assessed by using the calcein AM/PI double staining kit for live/dead staining. Subsequently, BMSCs were co-cultured as described above for 1, 3, and 5 days, followed by exposure to fresh culture medium with CCK-8 for 0.5 h. BMSC proliferation was quantified by measuring the absorbance of the supernatant using a multifunctional microplate reader at 450 nm. Similarly, BMSCs were seeded on SF or DNA–SF hydrogels, and the influence of the hydrogel on BMSC migration was assessed through a scratch assay. The rate of cell migration was analyzed using ImageJ software.4.6. Chondrogenic differentiation
Similarly, SF or DNA–SF hydrogels were prepared in cell well plates, and then BMSCs were seeded onto them. The cells were cultured in chondrogenic induction medium (medium containing 10 ng mL−1 TGF-β3, 1 mM sodium pyruvate, 0.1 μM dexamethasone, 1% insulin-transferrin-selenium (ITS) Premix, and 1 μM ascorbate-2-phosphate), with the medium changed every 2 days. BMSCs were stained with Alcian blue (1%, pH = 2.5). After staining, the cells were washed with 6 M guanidine hydrochloride to elute the dye. The eluted dye solution was then measured at 630 nm using a multifunctional microplate reader. The quantification of GAG and Col II in BMSCs was carried out in accordance with the experimental procedure using the hydroxyproline assay kit and the tissue total GAG content dimethyl methylene blue (DMMB) Colorimetry Kit. Both GAG and COL II contents were subsequently normalized to the total DNA content, which was quantified using the Quant-iT™ PicoGreen® dsDNA Reagent and Kits. The materials were treated in the same way to exclude the influence of material composition on the experimental results. To assess the influence of hydrogels on chondrogenic differentiation of BMSCs, the expression levels of chondrogenic genes (Sox9, ACAN, and Col II) in BMSCs cultured on various hydrogels were measured by using qRT-PCR. The primer sequences for these genes are provided in Table S2 (ESI†).4.7. Transcriptomic analysis (mRNA-seq)
The samples of BMSCs co-cultured with DNA–SF hydrogels were collected and labeled as the DNA–SF group. Total RNA was extracted from each sample using the TRIzol reagent, Construct RNA library, OE Biotech Co., Ltd (China). Transcripts were sequenced. For data analysis, differentially expressed genes (DEGs) were determined using DESeq. p values <0.05, fold change >2 or <0.5 were considered significant. Gene ontology (GO) enrichment, kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment and gene set enrichment analysis (GSEA) were used for in-depth bioinformatics analysis.4.8. In vivo cartilage repair of hydrogels
Male SD rats (6 weeks old, 150 g) were procured from Changzhou Cavens Laboratory Animal Co., Ltd. All animal procedures adhered to the established guidelines and were approved by the Shanghai University Ethics Committee. The rats were categorically assigned to one of the four groups: Sham, control, SF, and DNA–SF. Following general anesthesia achieved through the intraperitoneal injection of pentobarbital sodium at a dosage of 80 mg kg−1, a cartilage defect model was created in the central region of the patellar groove in the SD rats, with a diameter of 2 mm and a depth of 1.5 mm. Subsequently, approximately 5 μL of the hydrogel precursors were injected into the defect site using a syringe. The knee joint capsule and skin were sutured and sterilized. The rats in each group were subjected to standardized care. At 4, 8 and 12 weeks, the rats were euthanized, and specimens of the femurs, heart, liver, spleen, lung, and kidneys were harvested for subsequent assessments.The articular cartilage treatment on the femoral condyle of SD rats was macroscopically observed at 4, 8, and 12 weeks. The femurs from SD rats were immersed in 4% formalin for 48 hours. We utilized atomic force microscopy (AFM) to collect height information on regenerated cartilage 12 weeks post-surgery. Subsequently, the data were analyzed using the Bruker Dimension ICON to assess the surface roughness of the regenerated cartilage. The femoral condyle condyles were subjected to a 6-week decalcification process, followed by paraffin embedding at the appropriate position. Subsequently, 10 μm thick sections were obtained through microtome slicing, after ethanol dehydration in a graded manner. These sections were subjected to staining with Safranin O/Fast Green and Hematoxylin and Eosin (H&E). Immunohistochemistry was employed to assess the expression of aggrecan (AGG) and collagen II within the defect area. After 12 weeks, major organs (heart, liver, spleen, lung, kidneys, and blood) were collected and subjected to pathological staining and liver and kidney function tests, including alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TBIL), direct bilirubin (DBIL), albumin (ALB), alkaline phosphatase (ALP), γ-glutamyl transferase (γ-GT), total bile acid (TBA), urea (UREA), creatinine (CREA), and uric acid (UA), to evaluate the biocompatibility of the hydrogels.
4.9. Statistical analysis
All experimental data were obtained from three or more independent experiments and are presented as mean ± SD. The t-test was used to determine the statistical significance of the difference between groups, for multiple group comparisons, one-way analysis of variance (ANOVA) was used, and a p value < 0.05 was considered statistically significant.Abbreviations
| OA | Osteoarthritis |
| DNA | Deoxyribonucleic acid |
| SF | Silk fibroin |
| HRP | Horseradish peroxidase |
| ECM | Extracellular matrix |
| BMSCs | Bone marrow mesenchymal stem cells |
| Y-DNA | Y-shaped DNA modules |
| L-DNA | L-shaped DNA modules |
| FTIR | Fourier-transform infrared |
| SEM | Scanning electron microscope |
| PBS | Phosphate buffered saline |
| Hydrogels-E | Hydrogel with equal content of DNA |
| 3D | Three-dimensional |
| CCK-8 | Cell Counting Kit-8 |
| GAG | Glycosaminoglycans |
| Col II | Type II collagen |
| AGG | Aggrecan |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| ICRS | International cartilage repair society |
| AFM | Atomic force microscopy |
| H2O2 | Hydrogen peroxide |
| LiBr | Lithium bromide |
| Na2CO3 | Sodium carbonate |
| α-MEM | Alpha minimal essential medium |
| ITS | Insulin-transferrin-selenium |
| DMMB | Dimethyl methylene blue |
| mRNA-seq | Transcriptomic analysis |
| DEGs | Differentially expressed genes |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| GSEA | Gene set enrichment analysis |
| H&E | Hematoxylin and eosin |
| ALT | Alanine aminotransferase |
| AST | Aspartate aminotransferase |
| TBIL | Total bilirubin |
| DBIL | Direct bilirubin |
| ALB | Albumin |
| ALP | Alkaline phosphatase |
| γ-GT | γ-glutamyl transferase |
| TBA | Total bile acid |
| UREA | Urea |
| CREA | Creatinine |
| UA | Uric acid |
Data availability
Data supporting the findings of this study can be obtained from the corresponding author upon request.Author contributions
Z. Z., Q. Z., M. L., Z. G., and J. S. conceived the experiments. S. W., H. Z., and S. W. performed the experiments, characterization and manuscript editing. C. S., Z. M., X. R., X. W., X. C., Y. H. and Z. L. contributed to the discussion of the results. Q. Z. and M. L. contributed to the writing – review & editing. Z. G. and J. S. provided the concept, supervision, writing – review & editing and financial support. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the Integrated Project of Major Research Plan of National Natural Science Foundation of China (92249303), Key Project of the National Natural Science Foundation of China (82230071), National Natural Science Foundation of China (32101084) and Science and Technology Commission of Shanghai Municipality (22ZR1424900).References
- K. H. Collins, K. L. Lenz, E. N. Pollitt, D. Ferguson, I. Hutson, L. E. Springer, A. K. Oestreich, R. Tang, Y. R. Choi, G. A. Meyer, S. L. Teitelbaum, C. T. N. Pham, C. A. Harris and F. Guilak, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2021096118 CrossRef CAS.
- L. Zheng, Z. Zhang, P. Sheng and A. Mobasheri, Ageing Res. Rev., 2021, 66, 101249 CrossRef CAS PubMed.
- Z. Peng, H. Sun, V. Bunpetch, Y. Koh, Y. Wen, D. Wu and H. Ouyang, Biomaterials, 2021, 268, 120555 CrossRef CAS.
- D. J. Hunter and S. Bierma-Zeinstra, Lancet, 2019, 393, 1745–1759 CrossRef CAS.
- A. Singh, D. D'Amico, P. A. Andreux, A. M. Fouassier, W. Blanco-Bose, M. Evans, P. Aebischer, J. Auwerx and C. Rinsch, Cell Rep. Med., 2022, 3, 100633 CrossRef.
- P. Onnerfjord, A. Khabut, F. P. Reinholt, O. Svensson and D. Heinegard, J. Biol. Chem., 2012, 287, 18913–18924 CrossRef PubMed.
- C. A. Vilela, C. Correia, J. M. Oliveira, R. A. Sousa, J. Espregueira-Mendes and R. L. Reis, ACS Biomater. Sci. Eng., 2015, 1, 726–739 CrossRef PubMed.
- W. Zhang, H. Ouyang, C. R. Dass and J. Xu, Bone Res., 2016, 4, 15040 CrossRef PubMed.
- X. Zhao, D. A. Hu, D. Wu, F. He, H. Wang, L. Huang, D. Shi, Q. Liu, N. Ni, M. Pakvasa, Y. Zhang, K. Fu, K. H. Qin, A. J. Li, O. Hagag, E. J. Wang, M. Sabharwal, W. Wagstaff, R. R. Reid, M. J. Lee, J. M. Wolf, M. El Dafrawy, K. Hynes, J. Strelzow, S. H. Ho, T. C. He and A. Athiviraham, Front. Bioeng. Biotechnol., 2021, 9, 603444 CrossRef PubMed.
- A. Galperin, R. A. Oldinski, S. J. Florczyk, J. D. Bryers, M. Zhang and B. D. Ratner, Adv. Healthcare Mater., 2013, 2, 872–883 CrossRef.
- Y. P. Singh, J. C. Moses, N. Bhardwaj and B. B. Mandal, J. Mater. Chem. B, 2018, 6, 5499–5529 RSC.
- T. Su, M. Zhang, Q. Zeng, W. Pan, Y. Huang, Y. Qian, W. Dong, X. Qi and J. Shen, Bioact. Mater., 2021, 6, 579–588 Search PubMed.
- W. Chen, Y. Xu, Y. Li, L. Jia, X. Mo, G. Jiang and G. Zhou, Chem. Eng. J., 2020, 382, 122986 CrossRef.
- J. Wei, D. T. Baptista-Hon, Z. Wang, G. Li, T. Herrler, C. Dai, K. Liu, B. Yu, X. Chen, M. Yang, D. Han, Y. Gao, R. L. Huang, L. Guo, K. Zhang and Q. Li, Cell Rep. Med., 2023, 4, 101156 CrossRef.
- W. C. Ross, H. McWilliam, Z. Liu, J. Wang, F. Han, R. A. Black, J. Wu, X. Luo, B. Li and W. Shu, Biomater. Transl., 2023, 4, 104–114 Search PubMed.
- H. Li, P. Yang, J. Hwang, P. Pageni, A. W. Decho and C. Tang, Biomater. Transl., 2022, 3, 162–171 Search PubMed.
- N. A. Peppas, J. Z. Hilt, A. Khademhosseini and R. Langer, Adv. Mater., 2006, 18, 1345–1360 CrossRef.
- W. Chen, Z. Zhou, D. Chen, Y. Li, Q. Zhang and J. Su, Gels, 2021, 7 Search PubMed.
- G. P. Zhang, Z. L. Xie, J. Jiang, Y. T. Zhao, K. Lei, Z. L. Lin, S. L. Chen, T. H. Su, L. Tan, S. Peng, J. Wang, C. Liu and M. Kuang, Cell Rep. Med., 2023, 4, 101128 CrossRef.
- P. Maturavongsadit, W. Wu, J. Fan, I. B. Roninson, T. Cui and Q. Wang, Biomater. Transl., 2022, 3, 152–161 Search PubMed.
- J. S. Park, J. S. Chu, A. D. Tsou, R. Diop, Z. Tang, A. Wang and S. Li, Biomaterials, 2011, 32, 3921–3930 CrossRef.
- A. Elosegui-Artola, A. Gupta, A. J. Najibi, B. R. Seo, R. Garry, C. M. Tringides, I. de Lazaro, M. Darnell, W. Gu, Q. Zhou, D. A. Weitz, L. Mahadevan and D. J. Mooney, Nat. Mater., 2023, 22, 117–127 CrossRef PubMed.
- D. Huang, Y. Li, Z. Ma, H. Lin, X. Zhu, Y. Xiao and X. Zhang, Sci. Adv., 2023, 9, eade9497 CrossRef PubMed.
- Z. Wu, Z. Yang, D. Sha, Y. Ma, B. Y. S. Kim, W. Jiang, Y. Yuan and C. Liu, Chem. Eng. J., 2022, 434 Search PubMed.
- E. Schuh, S. Hofmann, K. S. Stok, H. Notbohm, R. Muller and N. Rotter, J. Tissue Eng. Regen. Med., 2012, 6, e31–e42 CrossRef.
- K. Lee, Y. Chen, X. Li, N. Kawazoe, Y. Yang and G. Chen, J. Mater. Sci. Technol., 2021, 63, 1–8 CrossRef.
- L. Fu, L. Li, Q. Bian, B. Xue, J. Jin, J. Li, Y. Cao, Q. Jiang and H. Li, Nature, 2023, 618, 740–747 CrossRef PubMed.
- Z. Zhou, J. Cui, S. Wu, Z. Geng and J. Su, Theranostics, 2022, 12, 5103–5124 CrossRef CAS PubMed.
- C. Holland, K. Numata, J. Rnjak-Kovacina and F. P. Seib, Adv. Healthcare Mater., 2019, 8, e1800465 CrossRef PubMed.
- Z. N. Mao, X. W. Bi, C. A. Wu, Y. F. Zheng, X. Shu, S. J. Wu, J. Guan and R. O. Ritchie, Adv. Healthcare Mater., 2022, 12, 2201588 CrossRef.
- Z. C. Cao, H. M. Wang, J. L. Chen, Y. A. Zhang, Q. Y. Mo, P. Zhang, M. Y. Wang, H. Y. Liu, X. Y. Bao, Y. Z. Sun, W. Zhang and Q. Q. Yao, Bioact. Mater., 2023, 20, 221–242 CAS.
- A. Mahajan, A. Singh, D. Datta and D. S. Katti, ACS Appl. Mater. Interfaces, 2022, 14, 7531–7550 CrossRef CAS.
- J. Tang, J. Ou, C. Zhu, C. Yao and D. Yang, Adv. Funct. Mater., 2021, 32 Search PubMed.
- F. Mo, K. Jiang, D. Zhao, Y. Wang, J. Song and W. Tan, Adv Drug Delivery Rev., 2021, 168, 79–98 CrossRef CAS.
- Y. Shao, H. Jia, T. Cao and D. Liu, Acc. Chem. Res., 2017, 50, 659–668 CrossRef CAS.
- M. Shin, J. H. Ryu, J. P. Park, K. Kim, J. W. Yang and H. Lee, Adv. Funct. Mater., 2015, 25, 1270–1278 CrossRef CAS.
- S. Iqbal, F. Ahmed and H. Xiong, Chem. Eng. J., 2021, 420 Search PubMed.
- A. Ibanez-Fonseca, D. Orbanic, F. J. Arias, M. Alonso, D. I. Zeugolis and J. C. Rodriguez-Cabello, Small, 2020, 16, e2001244 CrossRef.
- Y. Zhao, Z. S. Zhu, J. Guan and S. J. Wu, Acta Biomater., 2021, 125, 57–71 CrossRef CAS PubMed.
- J. K. Sahoo, J. Choi, O. Hasturk, I. Laubach, M. L. Descoteaux, S. Mosurkal, B. Wang, N. Zhang and D. L. Kaplan, Biomater. Sci., 2020, 8, 4176–4185 RSC.
- S. Wu, H. Zhang, S. Wang, J. Sun, Y. Hu, H. Liu, J. Liu, X. Chen, F. Zhou, L. Bai, X. Wang and J. Su, Mater. Horiz., 2023, 10, 3507–3522 RSC.
- O. Chaudhuri, L. Gu, D. Klumpers, M. Darnell, S. A. Bencherif, J. C. Weaver, N. Huebsch, H.-P. Lee, E. Lippens, G. N. Duda and D. J. Mooney, Nat. Mater., 2015, 15, 326–334 CrossRef.
- O. Chaudhuri, J. Cooper-White, P. A. Janmey, D. J. Mooney and V. B. Shenoy, Nature, 2020, 584, 535–546 CrossRef CAS.
- M. J. Zuscik, M. J. Hilton, X. Zhang, D. Chen and R. J. O'Keefe, J. Clin. Invest., 2008, 118, 429–438 CrossRef CAS.
- C. Liu, R. Abedian, R. Meister, C. Haasper, C. Hurschler, C. Krettek, G. von Lewinski and M. Jagodzinski, Biomaterials, 2012, 33, 1052–1064 CrossRef CAS.
- X. Chen, J. Li, E. Wang, Q. Zhao, Z. Kong and X. Yuan, J. Biomed. Mater. Res. A, 2015, 103, 3886–3895 CrossRef CAS PubMed.
- D. Su, M. Yao, J. Liu, Y. Zhong, X. Chen and Z. Shao, ACS Appl. Mater. Interfaces, 2017, 9, 17489–17498 CrossRef CAS PubMed.
- K. Wang, Q. Ma, Y.-M. Zhang, G.-T. Han, C.-X. Qu and S.-D. Wang, Cellulose, 2019, 27, 1845–1852 CrossRef.
- K. Luo, Y. Yang and Z. Shao, Adv. Funct. Mater., 2016, 26, 872–880 CrossRef CAS.
- Z. Peng, C. Xie, S. Jin, J. Hu, X. Yao, J. Ye, X. Zhang, J. X. Lim, B. Wu, H. Wu, R. Liang, Y. Wen, J. Huang, X. Zou and H. Ouyang, Biomaterials, 2023, 301, 122234 CrossRef CAS.
- W. Xiao, X. Qu, J. Li, L. Chen, Y. Tan, K. Li, B. Li and X. Liao, Eur. Polym. J., 2019, 118, 382–392 CrossRef CAS.
- Y. Hu and J. Y. Ying, Mater. Today, 2023, 63, 188–209 CrossRef CAS.
- R. Sheng, J. Chen, H. Wang, Y. Luo, J. Liu, Z. Chen, Q. Mo, J. Chi, C. Ling, X. Tan, Q. Yao and W. Zhang, Adv. Healthcare Mater., 2022, 11, e2200602 CrossRef.
- X. Hu, K. Shmelev, L. Sun, E. S. Gil, S. H. Park, P. Cebe and D. L. Kaplan, Biomacromolecules, 2011, 12, 1686–1696 CrossRef CAS PubMed.
- W. Jiang, X. Xiang, M. Song, J. Shen, Z. Shi, W. Huang and H. Liu, Mater. Today Bio, 2022, 17, 100485 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3mh01581e |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2024 |