Open Access Article
Open Access ArticleCharacterisation of polymeric nanoparticles for drug delivery
Thomas G. Floyda,
Pratik Gurnani b and
Julia Y. Rho
b and
Julia Y. Rho
 *c
*c
aAdvanced Drug Delivery, Pharmaceutical Sciences, R&D, AstraZeneca, Macclesfield, UK
bUCL School of Pharmacy, University College London, 29-39 Brunswick Square, Bloomsbury, London WC1N 1AX, UK
cDepartment of Chemistry, University College London, 20 Gordon Street, London, WC1H 0AJ, UK. E-mail: j.rho@ucl.ac.uk
First published on 28th February 2025
Abstract
Polymeric nanoparticles represent an innovative approach to drug delivery, particularly for addressing complex diseases like cancer. Their nanoscale dimensions facilitate targeted cellular uptake and effective navigation of biological barriers. With a broad range of polymerisation and functionalisation techniques, these nanoparticles can enable precise drug release, enhanced stability, and improved bioavailability while minimising side effects. Compared to conventional carriers, polymeric nanoparticles offer superior stability and versatility. However, despite these beneficial attributes, challenges remain in understanding their dynamic behaviour and interactions within biological systems. This mini-review aims to highlight key characterisation methods for studying polymeric nanocarriers, explore recent advances, and examine current challenges that must be addressed to optimise their therapeutic potential and advance these promising targeted drug delivery systems.
Introduction
Polymeric nanoparticles, typically ranging from 10 to 1000 nanometers, have garnered significant interest as advanced drug delivery vectors due to their small size, adaptability, and ability to transport and protect therapeutic agents to target sites within the body.1 Their nanoscale dimensions enable cellular uptake and allow these particles to cross biological barriers, including the challenging blood–brain barrier,2,3 which is critical for delivering drugs directly to cells in diseases like cancer.2,4–6A major advantage of polymeric nanoparticles (PNPs) is their chemical versatility, enabling the creation of a virtually limitless range of polymers with tailored properties through various polymerisation methods (e.g., ring-opening, radical, ionic, etc.), functional groups (e.g., sugars, charged moieties, stimuli-responsive chemistries), and controlled chain length and composition.7–18 This versatility supports fine-tuned control over essential delivery parameters, such as release profiles and targeting mechanisms, making polymers ideal for optimised drug delivery.19 Notably, PNPs can encapsulate drugs, improving the stability and solubility of hydrophobic compounds and protecting sensitive molecules—such as peptides, proteins, and small molecules—from premature degradation to ensuring targeted delivery.20–23 Additionally, polymeric nanoparticles can be engineered for controlled release triggered by stimuli like pH, temperature, or enzyme activity, reducing side effects and enhancing therapeutic outcomes.24–30 Importantly, these polymer-based nanomaterials open new avenues for administering drugs previously limited by poor solubility or bioavailability. Additionally, by enhancing drug efficacy at lower doses, polymeric nanoparticles could reduce adverse effects and improve patient outcomes, which is a crucial benefit for administering highly toxic drugs.31–35
In addition to small-molecule drugs, polymers and polymeric nanocarriers have garnered much attention as gene delivery vectors.36–38 Compared to traditional gene delivery systems like liposomes, polymeric nanoparticles offer better stability and tunability, facilitating easier storage and broader accessibility for patients worldwide. Their adaptability and cost-effectiveness—particularly their potential to eliminate the need for cryogenic storage—make them appealing alternatives to protein—or lipid-based carriers,39 which often struggle with stability and scalability issues.
Polymer nanoparticles are capable of self-assembly through a variety of motifs, including hydrophobic effects,40 hydrogen bonding,41–47 crystallisation,48,49 and host–guest interactions.50–52 These self-assembled structures exhibit a diverse array of properties, such as defined sizes, morphologies, charges, surface chemistries, and functional targeting moieties,15,53–55 all of which significantly influence their performance in drug delivery applications.56 The characterisation of these features is crucial for understanding their structure–property relationships and typically involves a collection of complementary techniques, including electron microscopy, light scattering, and fluorescence bioimaging.57,58 These methods can collectively enable precise tuning of nanoparticle properties to optimise therapeutic outcomes.
Despite their exciting potential, characterising polymeric nanoparticles remains a significant hurdle. Advanced techniques are required to study nanoparticle assembly, stability, and behaviour both in vitro and in vivo. These methods are essential for evaluating how nanoparticles interact with cells, traverse biological barriers, and function within complex extracellular environments. Understanding the fate of nanoparticles in circulation, their biodistribution, and their ability to evade the immune system is critical to advancing nanomedicine and achieving targeted delivery.
As polymeric nanoparticles are developed for drug delivery, their performance must be evaluated through in vivo studies, including comprehensive pharmacokinetic and pharmacodynamic analyses. These assessments extend beyond nanoscale characterisation, examining critical parameters such as circulation profiles, biocompatibility, toxicity, and therapeutic efficacy. While this article will focus on the nanoscale characterisation of self-assembled polymeric nanostructures for drug delivery, we acknowledge that studying their in vivo behaviour and translational potential is equally critical to realising their therapeutic promise for patients. Readers are directed to the excellent reviews cited herein for valuable insights and discussions on these topics.59,60
This mini-review highlights key chemical characterisation techniques for studying organic polymer-based nanoparticles (see Fig. 1), emphasising their strengths, limitations, and recent advancements. By discussing how these techniques deepen our understanding of nanoparticle behaviour in biological systems and the current challenges in the field, we aim to advocate for improved characterisation practices within the field. This mini-review aims to provide a valuable resource for newcomers to the field, encouraging them to explore the wide range of methods available for characterising novel polymeric drug delivery systems. It also seeks to inspire those already working in the field to adopt new analytical approaches to deepen understanding and corroborate insights into the complex polymeric delivery systems being developed. Lastly, we hope this summary may attract researchers from other disciplines to join us in building innovative approaches that unlock the full therapeutic potential of polymeric nanomedicines and advance the future of targeted drug delivery.
Characterisation methods
Nuclear magnetic resonance (NMR) spectroscopy
Nuclear Magnetic Resonance (NMR) Spectroscopy is an essential tool in polymer chemistry, providing detailed insights into the conversion and kinetics of polymerisation reactions. While less common methods, such as gas chromatography, can also be used to track volatile vinyl monomers, NMR remains the primary method for monitoring the progression of polymerisation reactions. Regardless of the initiation mechanism, the conversion of monomer to polymer, hence polymerisation, is calculated by tracking the appearance of polymer and the disappearance of monomer peaks.Many self-assembled polymer nanoparticles used for drug delivery are composed of copolymers with distinct block chemistries. These variations drive phase separation into well-defined microdomains, enabling the formation of nanostructures with diverse morphologies. Such nanostructures include spherical and cylindrical micelles, nanofibres, platelets, and vesicles.12,32,61–63 It is hoped that tuning the polymeric morphologies may hold the key to addressing significant challenges in drug delivery, such as controlled release.64
In block copolymer synthesis, the sequential building of blocks relies on maintaining the “living” polymer chain ends, which allows polymerisation to continue when conditions are suitable (e.g., in the presence of additional monomers, initiators, and typically an oxygen-free environment). NMR is particularly valuable for assessing the “livingness” of controlled polymers, as it can monitor polymer growth and confirm the process remains under controlled conditions. By analysing specific signals in 1H NMR spectra, researchers can verify consistent chain growth with low levels of termination or side reactions, ensuring the polymerisation retains the characteristics of a living system.
Moreover, NMR becomes a powerful technique when dyes or drugs are conjugated to the polymer structure. Upon conjugation, new characteristic peaks often emerge in the NMR spectrum, or existing peaks may shift due to the formation of new bonds. By examining these changes, it is possible to confirm the successful conjugation of therapeutic agents or fluorescent dyes, which is essential for applications in targeted drug delivery and imaging. Quantifying the success of conjugation is feasible by comparing expected and observed integration values. However, matching the integration of characteristic peaks does not guarantee that all molecules in the solution are conjugated, as unconjugated species may still be present. Therefore, additional characterisation methods are recommended to corroborate conjugation efficiency, such as HPLC and vide infra.
Two-dimensional NMR techniques, such as COSY, HSQC, and HMBC, offer powerful insights into the local chemical environments near the conjugation site. These techniques can reveal interactions between adjacent atoms and identify specific structural features of the polymer. By mapping out spatial relationships and connectivity, 2D NMR can help confirm that conjugation has occurred at the intended site on the polymer chain, thereby ensuring accurate functionalisation. In the context of drug delivery, these complementary methods are invaluable. 1H NMR enables the confirmation of drug-conjugate linkages, while 13C NMR provides detailed structural information about the polymer backbone. Additionally, quantitative NMR facilitates the precise measurement of drug loading or the density of functional groups. For instance, in PEG–PLGA systems, NMR is frequently employed to validate the successful conjugation of a hydrophobic drug to the hydrophilic polymer, enhancing both solubility and release profiles.
Diffusion-ordered (DOSY) NMR has long been a powerful technique for determining the molecular weight of polymers. Its potential was first demonstrated in 1989 by von Meerwall, who showed that pulsed field gradient NMR (PFG NMR) was sufficiently sensitive to detect differences in molecular size and, hence, the molecular weight of polymers.65 Subsequent work by Johnson and colleagues expanded this approach, enabling the determination of molecular weight distributions of polymers using constrained regularisation algorithms in diffusion-ordered NMRs.66 With the rapid expansion of polymer chemistry, significant efforts were devoted to developing accurate methods for determining molecular weights across a diverse range of polymers. These advancements addressed the challenges posed by varying solution properties, which depended on the choice of polymer and solvent systems. Accurate molecular weight determination became critical for understanding polymer behaviour, optimising synthesis, and tailoring properties for specific applications.67–69
Recent advances in flow chemistry and the development of compact benchtop NMR systems have significantly revitalised the utility of NMR for polymer characterisation.70 Notably, the work by the Leibfarth,71 Warren,72 and Junkers groups has demonstrated increasingly efficient,73 solvent-independent,74 and universal approaches to molecular weight determination.75 With the growing incorporation of AI in data collection, processing, and molecular weight prediction, these high-throughput methodologies are poised to become even more critical, enabling rapid input and feedback loops for polymer development. In a recent article, Gormley discusses the intricate interplay of complex interactions that govern the effectiveness of drug delivery systems.76 The article underscores the interconnected nature of key factors such as polymer properties, their behaviour in diverse environments (in vitro and in vivo), and the mechanisms driving drug release. Referred to as the “curse of dimensionality” in drug delivery, the author points out how interdependent parameters like crystallinity, water absorption/swelling, hydrolysis, and erosion, rely on a broad range of polymer/particles attributes (e.g., molecular weight, size, morphology, porosity) and external conditions (e.g., pH, temperature, osmolarity, location). This highlights the substantial challenges involved in designing efficient and reliable polymer-based drug delivery systems. However, the author expresses optimism about the potential of machine learning to address these critical issues.
From this, two key parameters to highlight are zeta potential and porosity. Zeta potential, which measures the electrophoretic mobility of particles, is a crucial indicator of nanoparticle surface charge as it affects colloidal stability, aggregation tendencies, and interactions with biological membranes. A high absolute zeta potential (either positive or negative) generally enhances stability by preventing aggregation, while moderate values may be beneficial for controlled interactions with cells and biomolecules. Zeta potential can be measured using instruments such as electrophoretic light scattering (ELS), phase analysis light scattering (PALS), streaming potential instruments, electrokinetic analysers (EKA), and nanoparticle tracking analysis (NTA). It's important to consider that zeta potential is strongly influenced by pH and salt concentration. As nanoparticles are introduced into biologically representative media like blood and plasma, changes in the media, including pH, ions, and proteins (which form the nanoparticle's protein corona), must be considered carefully.
Porosity in nanoparticles plays a crucial role in drug delivery by enhancing drug encapsulation efficiency and enabling controlled or sustained drug release. Additionally, porosity affects density, influencing sedimentation behaviour, diffusion, and transport within biological systems. Together, these physicochemical properties are key to optimising polymeric nanoparticles for targeted, efficient drug delivery. High porosity increases drug loading capacity, improving efficiency and bioavailability. It allows for sustained, targeted release, reducing side effects and improving patient outcomes. Moreover, the porous structure facilitates better drug diffusion and responsiveness to environmental triggers, making porous nanoparticles ideal for precision medicine. Porosity can be measured using techniques such as Brunauer–Emmett–Teller (BET) analysis (for surface area and pore size), mercury intrusion porosimetry (MIP) for pore volume, and electron microscopy (TEM/SEM) for direct imaging. SAXS provides nanostructural details, while TGA assesses porosity through weight loss analysis. Advanced methods like NMR and X-ray tomography offer 3D imaging of porous structures, all helping to optimise drug loading and controlled release in drug delivery applications.
This level of detail is crucial for applications requiring precise polymer architectures, as even small deviations in conjugation sites can alter the material's properties and performance in drug delivery systems.
Size exclusion chromatography (SEC)
Size Exclusion Chromatography (SEC) is a foundational technique in polymer chemistry, providing critical insights into polymer conversion and the evolution of molecular weight. By tracking changes in molecular weight over time, SEC can help monitor the kinetics of polymerisations, offering valuable information about the progress and control of the reaction. SEC also provides data on the molecular weight distribution, which is important for assessing the uniformity of polymer chains. A narrow symmetrical molecular weight distribution indicates a controlled polymerisation process, where chains start growing at the same time (initiation stage) and grow at a similar rate (propagation stage). In contrast, a broader or asymmetrical distribution may suggest uncontrolled polymerisation and multiple side reactions, leading to variations in chain length.A double molecular weight peak in the SEC profile can indicate chain–chain coupling, where two polymer chains have joined to form a larger macromolecule. This phenomenon may occur more often during certain polymerisation processes or under specific conditions, such as a high concentration of active polymer chains. Importantly, the degree of chain–chain coupling can influence the uniformity of polymer chains, thereby affecting the properties of the final polymer (e.g. solubility and self-assembly behaviour).
SEC is particularly useful in monitoring chain extension during block copolymerisation, which is essential for designing self-assembling polymers. As mentioned above, the difference in block chemistries is often used in polymer nanoparticle synthesis to generate core–shell nanoparticles. The most common example is a block copolymer with hydrophilic and hydrophobic domains, which self-assembles into nanoparticles. The hydrophobic chains are assembled in the core away from the water, and the hydrophilic chains stabilise the nanoparticles, forming the corona.
Detectors compatible with SEC, such as RI, UV, fluorescence, and viscometry, can be used to assess polymer characteristics. If the polymer contains a UV-active group, such as a conjugated dye or a functional group, it can be monitored as it separates by size. For example, in reversible addition–fragmentation chain-transfer (RAFT) polymerisation, the trithiocarbonate group absorbs at 309 nm and can be detected by coupling SEC with UV detection. This allows for the selective monitoring of polymers with UV-active groups, such as dyes or drugs, which are attached via conjugation chemistry. By confirming the presence and distribution of these groups, researchers can verify successful functionalisation and optimise the system for applications requiring precise drug or dye loading.
Due to the increasing complexity of polymeric nanomaterials for drug delivery, molecular weight determination is more complex than for linear polymers. Often, multi-detector SEC (MD-SEC) is required to accurately determine the molecular weight, requiring a concentration detector, such as RI or UV, a light-scattering detector and a viscometer. Working in tandem, these detectors are able to accurately determine the molecular weight of the analyte and other parameters such as concentration, incremental refractive index, size, branching, or, in the case of polymer–drug conjugates, drug loading.77 or polymer–drug conjugates, understanding the release rate of the active pharmaceutical ingredient (API) from the nanoparticle is crucial to understanding its efficacy. Whilst SEC is not commonly applied to determine the rate of API release, it can be used to monitor the release rate by studying the appearance of a peak associated with the API, likely at higher retention volumes, or in the case of APIs with unique UV absorbance from the nanoparticle, monitoring the decrease in UV signal for the nanoparticle.78
A further application of SEC to polymeric nanomaterials is to understand their physical stability in aqueous media. SEC is able to reveal the presence of aggregates in a sample, indicated by peaks at lower retention volumes. This is further confirmed by light scattering detectors, where due to the intensity of scattered light being proportional to the size to the power of six, aggregates have a much greater intensity compared to their corresponding intensity determined by a concentration detector, such as RI. Carrying out a complimentary SEC analysis in an organic solvent can be beneficial in confirming if the aggregation is due to non-covalent interactions or caused by covalent coupling.79
In drug delivery, GPC is crucial for determining a polymer's molecular weight (Mw) and polydispersity index (PDI), ensuring consistency from batch to batch. It also monitors molecular weight changes during degradation, providing insights into the polymer's stability. For example, in hydrogel-based drug carriers, GPC identified an optimal Mw range that enabled sustained drug release over several weeks, aligning with therapeutic objectives.
Mass spectrometry (MS)
Mass spectrometry (MS) is a powerful technique for characterising both homopolymers and copolymers, providing valuable insights into their molecular structure. Matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) MS has been commonplace for the characterisation of homopolymers, such as polyethene glycol. MALDI-TOF is particularly effective for analysing polymers with well-defined end-group functionalisation. This technique allows the identification of the polymer's molecular weight and the detection of end groups, providing crucial information on the degree of polymerisation and ensuring the desired functionalisation at the chain ends.Due to the number of exponential fragment patterns, characterising copolymers using MS is more challenging. Haddleton and coworkers showed that MALDI, in conjunction with laser-induced dissociation (LID) fragmentation techniques (MALDI-LID-ToF/ToF), could be used to isolate and fragment statistical and diblock polyacrylate copolymers.80 Additionally, methods that use two-dimensional or Kendrick mass defect plots to process the mass spectra of polymers have been incredibly powerful.81–83 Volmer and coworkers were able to map the genealogical links in mixtures of lignin depolymerisation products using two-dimensional mass defect matrix plots. These tools have the potential to be extended to other classes of polymers (e.g. copolymers or terpolymers).83
Since then, double-resonance experiments, using electron capture dissociation (ECD) and Fourier transform ion cyclotron resonance (FT-ICR) tandem mass spectrometry, have been able to characterise acrylamide homo- and block copolymers.83 More recently, polymer MS using two-dimensional methodologies has also shown that the precursors that produce independent fragments can be separated via modulation frequency. Furthermore, these techniques can be employed to understand and map the specific sequences of random copolymers, gaining insights into how monomers are distributed along a polymer backbone.84 These advances allow us to analyse increasingly complex and dispersed polymer samples that have previously not been able to be characterised through MS due to the lack of effective separation through chromatographic or isolation techniques.85 In the future, with advances in computation and machine learning, these methods could become more commonplace to differentiate between various copolymer compositions, providing insights into their structural heterogeneity, sequence, and molecular weight distribution.
High-performance liquid chromatography (HPLC)
High-Performance Liquid Chromatography (HPLC), though widely underutilised by the polymer field, is a highly effective technique for polymer characterisation. By optimising solvent composition and selecting appropriate stationary phases, HPLC enables the separation of polymers based on their polarity. This approach is crucial for evaluating the purity, molecular distribution, and structural integrity of polymer products, providing valuable insights into their composition and performance. Notably, Hawker and coworkers utilised the polarity-based separation method (flash chromatography) to successfully isolate polymers with specific degrees of polymerisation (DP).86 Though this process may seem laborious and time-consuming, it represents a significant milestone in polymer synthesis by enabling the isolation of individual polymer chain lengths. This advancement is crucial because it allows for the production of polymers with specific, well-defined molecular weights. In applications where uniformity is essential—such as in drug delivery systems—the ability to isolate distinct polymer chain lengths becomes particularly important. Achieving increasingly uniform or even monodisperse polymers can improve consistency, efficacy, and safety, which are critical factors for meeting regulatory standards and ensuring optimal performance in various biomedical applications.HPLC is also useful for tracking changes in copolymer composition. By monitoring the polymer's retention time (and thus changes in polarity), shifts in composition during polymerisation can be observed. For example, a study by Perrier and Peltier used HPLC to monitor the composition of copolymers (statistical versus block), providing detailed insights into how polymer composition and comonomer distribution vary within a specific polymer chain.87
Another important application of HPLC is its ability to monitor the conjugation of polymers with dyes, drugs, or peptides, a critical process in drug delivery and diagnostic imaging. With built-in UV and fluorescence detectors, HPLC offers valuable tracking of the conjugation process, providing quantitative insights into both reactants and products before and after conjugation, see Fig. 2.88 By analysing the UV absorbance or fluorescence emission signals, researchers can assess the efficiency of the conjugation, as well as the progress of the reaction. This allows for precise control over the functionalisation process, ensuring that the desired loading, composition, and overall quality of the conjugates are achieved for optimal performance in their intended application.
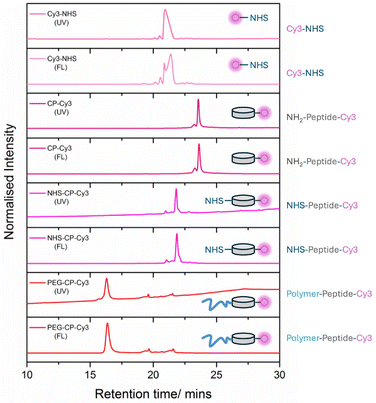 | ||
| Fig. 2 High-performance liquid chromatography (HPLC) of peptide-polymer-dye conjugate. UV absorption at 280 nm (tryptophan absorption to follow the peptide), fluorescence emission measured at 570 nm upon excitation at 555 nm (to follow the Cyanine 3 dye). Adapted from Rho et al., reproduced with permission from Wiley.88 | ||
As mentioned above, understanding the release of drugs from polymer–drug conjugates is a crucial parameter to understand. Not only can we determine the rate of release in biologically relevant media, but we can also determine the effectiveness of a stimuli-responsive linker. As with SEC, HPLC can be utilised to quantify the release of drugs from the polymeric nanoparticle. Suppose the drug is chemically conjugated to the drug. In that case, this can be straightforward, with the release media containing the nanomaterial and released API directly injected into the HPLC and the peak area calculated. However, for drugs physically encapsulated within a polymeric micelle, this proves more challenging. HPLC will disrupt the encapsulation and will not be able to distinguish between released and encapsulated API. Methods for separation are discussed in detail in the review published by Ghezzi et al.89 However, the most common procedure involves the employment of dialysis to separate the free API from the micelle. The filtrate is then analysed, and the concentration of the released drug is determined. For both systems, once the API peak area has been determined, it can be plotted versus time to understand the release profile of the drug from the nanoparticle.
Microscopy
Transmission electron microscopy (TEM)
Microscopy has significantly advanced nanoparticle characterisation, providing valuable insights into their nanoscopic structures in both solution and cellular environments. Transmission Electron Microscopy (TEM) is particularly useful for visualising nanoparticles due to its high resolution (down to a few nanometers). However, organic polymeric nanoparticles can be challenging to image due to their low electron density, which can reduce contrast and clarity compared to inorganic nanoparticles. Despite this, TEM remains essential for understanding the morphology and behaviour of polymeric nanoparticles. The use of contrast agents or stains, such as phosphotungstic acid (PTA), uranyl acetate, osmium tetroxide (OsO4), and ruthenium tetroxide (RuO4), can be used to enhance imaging clarity. However, these stains require careful use and handling due to factors like pH sensitivity and inherent toxicity, which must be managed to ensure accurate and reliable imaging.A key feature of polymer nanoparticles designed as drug carriers is the reversibility of their self-assembled structures, which are formed through non-covalent interactions. This dynamic property allows the nanoparticles to maintain stability during circulation and drug delivery while also enabling disassembly at the target site to release the therapeutic cargo. These metastable nanostructures require sufficient stability for administration through the bloodstream and into targeted cells but must also dynamically disassemble to release drugs at therapeutic levels without causing significant toxicity or instability during transport. Advanced TEM techniques, such as liquid-cell TEM90 and cryo-TEM,91,92 are critical for studying these systems under more representative solution-phase or intracellular conditions. These approaches provide invaluable insights into the stability, behaviour, and drug release mechanisms of polymeric nanoparticles, furthering their development as effective drug delivery vectors.
Key contributions to the field include the work of Granick and coworkers, who demonstrated the visualisation of individual macromolecules like polystyrene sulfonate and poly(ethylene oxide) in aqueous solutions using graphene-based liquid-cell TEM. This approach achieved nanometer-resolution imaging without metal-ion labelling.93 It minimises electron-induced damage and background scattering, enabling detailed analyses of polymer sizes, conformational fluctuations, and adsorption–desorption events with standard TEM instruments.
A powerful example of pushing the resolution of these techniques for self-assembled polymeric nanostructures includes work by Manners and coworkers using high-resolution cryo-TEM. In this work, the corona and crystalline core of nanofibres in vitrified solution were directly observed, revealing a 2D pseudo-hexagonal symmetry in the core's packing and a detailed molecular model of polymeric nanofibres. These findings provide unprecedented insight into the structural order of nanofibres with crystalline cores, with implications for analogous systems, such as π-conjugated block copolymer nanofibers. Moreover, open-source Python libraries have endeavoured to simplify image analysis for TEM and in situ TEM.94
Recent studies by Wu et al. expanded the capabilities of polymer electron microscopy by highlighting advances in TEM instrumentation, including focused ion beams, liquid-phase sample holders, monochromated sources, and direct electron detectors. These reduce radiation sensitivity constraints and promise transformative impacts for imaging soft materials.95
Scanning electron microscopy (SEM)
Another invaluable electron microscopy (EM) technique for polymer nanoparticle characterisation is Scanning Electron Microscopy (SEM). While both SEM and TEM are effective for determining particle size, they excel in different areas. TEM is ideal for internal structural analysis, providing detailed insights into nanoparticle morphology, composition, and assembly. In contrast, SEM focuses on surface morphology and particle aggregates, making it particularly useful for studying coatings and overall topology. The choice between TEM and SEM depends on the specific characterisation goals and required resolution. TEM offers sub-nanometer resolution, while SEM typically resolves to tens of nanometers. However, TEM is more sensitive to beam-induced degradation, particularly at higher voltages, making SEM a more robust option for delicate polymer samples.96When analysing polymeric nanoparticles, two critical factors must be considered: the potential for electron beam-induced damage or alteration of the sample, which can affect imaging accuracy, and the difference between experimental conditions and the biological environment, which raises questions about how well in vitro studies replicate in vivo behaviour. Addressing these challenges, along with advancements in imaging techniques and experimental design, will be crucial for a deeper understanding of how self-assembled polymers behave in living cells and dynamic biological systems.
Atomic force microscopy (AFM)
Atomic Force Microscopy (AFM) complements many of the EM techniques by providing additional insights into the surface morphology, mechanical properties, and three-dimensional topology of polymeric nanostructures. While EM excels in revealing the internal crystalline structure and molecular organisation, AFM offers high-resolution imaging of the nanofibre surface in real space and under conditions closer to ambient. As with living systems, synthetic supramolecular systems can also ‘grow’ in a ‘living’ manner. In situ methods to visualise the growth process have been achieved using AFM,97 TEM,98,99 and iSCAT.100 Going back to the examples of polymeric nanoparticles assembled through crystallisation, Manners and coworkers were able to use AFM to study the growth of nanofibers on a silicon surface.97 The paper also revealed unidirectional growth in certain orientations, where seed alignment restricted growth to one terminus. These insights provide a deeper understanding of BCP nanofiber growth and its potential for controlled nanoparticle fabrication.In related work by O'Reilly, Dove, and coworkers, uniform two-dimensional platelets formed through crystallization-driven self-assembly were effectively characterised using TEM, AFM, and STED, revealing their multilayered structure.101 These multilayered platelets offer significant potential for designing polymeric systems with tailored chemistries, enabling precise control over drug release profiles.102 In other related areas, the field of artificial cells has advanced significantly, particularly through the use of amphiphilic block copolymers to form vesicular bilayers, often referred to as polymersomes.103–108 Structurally analogous to liposomes but composed of polymers, polymersomes provide enhanced stability and tunability, making them ideal candidates for drug delivery and other biomedical applications.104
All these examples offer valuable insights into the assembly and disassembly processes of polymeric nanoparticles. Gaining a deeper understanding of their dynamic behaviour is crucial for optimising drug delivery systems, ensuring efficient transport, and achieving controlled release from polymer-based nanocarriers.
Fluorescence imaging
Fluorescence imaging is crucial for characterising polymer nanoparticles as it can provide real-time, non-invasive insights into their behaviour and interactions in both solution, in vitro and in vivo. It enables the tracking of fluorescently labelled nanoparticles in biological systems, elucidating processes such as cellular uptake, biodistribution, and drug release mechanisms.109 Furthermore, the ability to label specific components of nanoparticles allows for a detailed investigation of core–shell architectures and their dynamic behaviour in complex biological systems.Fluorescent polymeric nanoparticles (NPs) have proven valuable for in vivo applications such as tracking cell fate and tumour development. For example, AIE-based NPs were used to label cancerous cells, enabling their imaging in mice over 21 days, while Rhodamine B-based NPs monitored stem cell implantation in mouse brains.110 Biocompatibility and low toxicity are critical for these applications, with studies showing minimal cytotoxicity for dye-loaded NPs at relevant concentrations. Encapsulation of dyes further reduces toxicity, and biodegradable polymers like PLGA or PCL make these NPs suitable for biomedical use. Emerging applications now leverage dye-loaded NPs not only as labels but also as multifunctional fluorescent probes, paving the way for theranostics and other advanced biomedical technologies. See previous sections on the characterisation of polymer-dye conjugates.
However, the resolution of fluorescence imaging is limited by the diffraction limit of light, approximately 200–250 nm, which can obscure finer structural details at the nanoscale. Recent advancements, such as super-resolution techniques like STED, PALM, and STORM, have extended fluorescence imaging's capabilities to achieve resolutions down to 20–30 nm, enabling more precise characterisation of polymer nanoparticles.111 These innovations bridge the gap between nanoscale resolution provided by electron microscopy and the dynamic, functional insights offered by fluorescence imaging, making it an indispensable tool for nanoparticle research.
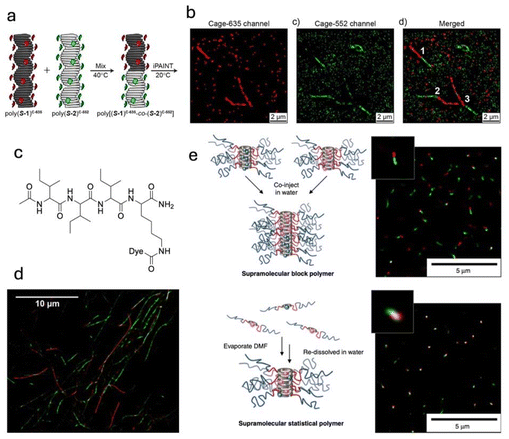
Early work by Albertazzi and Meijer showed that homogeneous exchange could be observed in the BTA-based supramolecular polymers using super-resolution imaging (Fig. 3a and b).112 This could be attributed to disordered domains within the ordered hydrogen-bonded structure, where weaker monomer interactions facilitate the exchange process.113,114 The STORM technique revealed that this exchange occurs uniformly along the polymer backbone, with no evidence of fragmentation, fusion, or polymerization–depolymerization at the chain ends, suggesting a need to reassess the dynamic behaviour of supramolecular fibres.112,115 Cox et al. combined AFM and STORM to investigate the self-assembly process of peptide surfactants, revealing the formation of helical fibres ranging from 5 to 10 μm (Fig. 3c and d). Using two-colour STORM, they were able to study the dynamic self-assembly process of these peptides after mixing. Building on this work, Perrier and coworkers demonstrated that dual-labeling strategies with STORM could also visualise the formation of supramolecular block-like structures, where two individual metastable peptide-polymer nanotubes fused together (Fig. 3e).
Similar studies using other super-resolution techniques, such as stimulated emission-depletion (STED) microscopy, have demonstrated the versatility of non-covalent Alexa-488 labelling for imaging cationic self-assembling peptides and peptide-functionalised gold nanoparticles.116 This approach provides a simple and effective method for electrostatically labelling cationic peptide nanostructures, enabling real-time, in situ imaging of dynamic processes under physiological conditions.
The success of many super-resolution methodologies hinges on striking a balance between fluorescently labelling biological molecules within cells and the synthetic nanostructures of interest (i.e., your drug-loaded nanoparticle). Albertazzi and colleagues developed a simple, versatile workflow using DNA-PAINT to standardise and enhance the quantification of biological molecule density and distribution on synthetic substrates and cell membranes.117–119 This approach improves accuracy, sensitivity, and precision, as demonstrated by the quantification of docking strands on sensor surfaces and PD1 and EGFR receptors on cellular models while effectively filtering out non-specific interactions and artefacts.117
Building on this work, the group also designed nanoparticles functionalised with targeting ligands to selectively deliver therapeutics to cancer cells by exploiting overexpressed receptors. However, moderate receptor expression in healthy cells presents challenges with off-tumour toxicity. To address this, they synthesised a panel of aptamer-functionalised silica-supported lipid bilayers (SSLB) to investigate the interplay of valency, aptamer affinity, and epidermal growth factor receptor (EGFR) density on targeting specificity and selectivity.120 The study revealed that combining high-affinity aptamers with low-valency SSLBs enhanced selectivity for high-density EGFR cells while minimising accumulation in non-tumour tissues, marking a step forward in the rational design of cancer nanotherapeutics.
Recent advancements in the field highlight the powerful combination of techniques like correlative light and electron microscopy (CLEM) to study nanoparticle (NP) behaviour. By integrating STORM and TEM, this method enables precise localisation and quantification of NPs within cellular compartments, providing crucial insights into endo–lysosomal trafficking and the impact of treatments, such as chloroquine, on endosomal escape and therapeutic efficacy.121
Light scattering
Light scattering methods are indispensable for characterising polymer nanoparticles, especially in drug delivery, where understanding their size, morphology, and assembly is critical. Techniques like dynamic light scattering (DLS) and static light scattering (SLS) allow researchers to measure particle size, distribution, and molecular weight in solution, offering a non-invasive way to study these aggregates in conditions mimicking physiological environments.Such analyses go beyond simply determining size and shape, providing insights into the arrangement of polymer chains within the nanoparticle, a critical factor for drug encapsulation and release. While resolving the exact atomic positions within assemblies is currently beyond reach, advancements in scattering methods are progressively enhancing our ability to probe these intricate structures with greater precision. Importantly, light scattering complements other techniques by enabling scalable and reproducible measurements essential for translating polymer nanoparticles from research to therapeutic applications.
A tutorial review by O'Reilly and coworkers serves as an excellent resource for the practical analysis of self-assembled polymer nanoparticles in solution.122 It highlights the use of scattering and microscopic techniques to characterise these materials, offering detailed guidance on evaluating their size, morphology, and molecular arrangement. This review bridges the gap between synthetic advances and reliable analytical methodologies, providing valuable insights for researchers aiming to design precise and functional polymer-based nanostructures. This progress mirrors the natural precision found in biological nanostructures like enzymes and viruses, which serve as inspiration for synthetic analogues. By leveraging light scattering alongside microscopic methods, researchers can achieve a detailed understanding of the self-assembly and functional performance of polymer nanoparticles, ultimately advancing their design for drug delivery and other biomedical applications.
A crucial parameter that must be understood for self-assembled nanomaterials is their critical micelle concentration (CMC). The CMC defines the concentration at which unimers self-assemble to form particles. Upon administration of self-assembled particles in vivo, they will undergo a dilution effect, which could cause the particles to disassemble if the concentration decreases below the CMC.123 DLS can be utilised to determine this parameter by measuring the scattering intensity as a function of concentration. Particle formation results in a strong increase in scattering intensity, with the inflection point indicating the CMC.124 Several other techniques are available to determine CMC, such as conductivity or surface tension measurements or the encapsulation of a hydrophobic fluorophore.125
Small-angle X-ray scattering (SAXS) has proven to be an indispensable tool for monitoring polymerization-induced self-assembly (PISA), offering real-time insights into nanostructure evolution during synthesis.126–129 Extensive work by Armes and coworkers highlights the use of SAXS to characterise morphological transitions—such as from spheres to worms to vesicles—while providing detailed metrics like particle size, aggregation number, and inter-chain separation. This method also unveils critical formation mechanisms, such as inward growth in vesicle membranes, enhancing the precision of in situ characterisation and deepening our understanding of dynamic self-assembly processes.
Small-angle neutron scattering (SANS) is a powerful technique for studying the structural parameters of self-assembled systems, such as size, morphology, and aggregation number, under varying environmental conditions like pH, temperature, or solvent.130 By modelling data with appropriate form factors (e.g., Gaussian coil, comb, or hairy cylinder), SANS provides detailed insights into the architecture of conjugated polymer systems, distinguishing between non-assembled and self-assembled species. This approach enables accurate characterisation of supramolecular structures, advancing understanding of their dynamic behaviours in different environments.
Asymmetric flow field-flow fractionation (AF4) has emerged as an exciting characterisation technique for polymeric nanoparticles, offering unique insights into their size, composition, and behaviour under dynamic conditions. By employing multiple detectors, such as refractive index (RI), multi-angle light scattering (MALS), UV, and fluorescence, AF4 provides a comprehensive analysis of self-assembled structures. The technique uses a gentle cross-flow and diffusion to separate polymers, proteins, and nanostructures based on size, enabling the characterisation of self-assembled systems held together by dynamic non-covalent interactions, which are commonly found in polymeric nanoparticles used for drug delivery.
Kariuki et al. used AF4 in combination with small-angle neutron scattering (SANS) to analyse cyclic peptide-polymer conjugates, revealing the delicate balance between hydrophobic and hydrophilic forces in nanotube stability and elongation.131 Similarly, Louie and coworkers demonstrated the power of AF4 with fluorescence detection (AF4-FLD) in studying drug release profiles from PLGA nanoparticles, showing its superiority over dialysis by resolving temperature-dependent behaviours and enabling size-resolved release profiles.132 However, AF4's advantages come with caveats. Significant method optimisation is required, posing a barrier to entry for less-experienced users, and the resulting separation methods may only partially replicate realistic cellular conditions. Furthermore, even with gentle cross-flow, there is a risk of disassembly, particularly in concentration-dependent systems prone to dynamic changes upon dilution. Despite these challenges, AF4 remains a versatile and invaluable technique for advancing our understanding of polymer nanoparticles in drug delivery.
Several of the methods above allow us to obtain particle size distributions. We can use nanoparticle image analysis, which involves acquiring images using TEM, SEM, and AFM to study their size, shape, and distribution. Image processing methods such as contrast enhancement, noise reduction, thresholding, and edge detection help in segmentation and analysis. Quantitative measurements include size distribution, shape analysis, and aggregation detection. Software tools like ImageJ, MATLAB, and Python (OpenCV) are commonly used for automated processing. Alternatively, solution-phase particle analysis methods, like many of the scattering techniques (DLS, SLS, SAXS, SANS), may not necessarily provide particle distribution analyses but important allow us to calculate the average molecular weights and particle sizes in solution. One of the most powerful methods is AF4, as this method will enable us to obtain size and distribution data. However, as mentioned above, the is a highly specialised instrument, not as commonly used or found in most labs, and often requires significant method development before discernible data is obtained.
In summary, materials characterisation plays a crucial role in drug delivery applications by ensuring the proper design and functionality of nanoparticles. Techniques such as electron microscopy, spectroscopy, and porosity measurements allow researchers to assess key properties like size, surface morphology, drug loading capacity, and release profiles. The importance of a diverse range of characterisation methods lies in building a deeper understanding of nanoparticle behaviour, with the power of complementary data helping to validate findings and provide a stronger, more comprehensive picture. Accurate characterisation enables the optimisation of nanoparticle formulations for enhanced stability, biocompatibility, and controlled drug release. By understanding the material's behaviour at the molecular and nanoscale level, researchers can design more effective and targeted drug delivery systems.
Process analytical technologies for polymer nanoparticle drug delivery system manufacture
The characterisation techniques laid out above highlight critical examples to fully assess the properties of polymer nanoparticle drug delivery systems during the discovery research and development phase. However, analytical tools are also heavily integrated with the manufacturing process of modern pharmaceuticals, typically integrated as process analytical technologies (PATs).133 The goal of PATs is to promote real-time analysis of the critical quality attributes (CQAs) and relate this to the critical process parameters (CPPs) which control said CQAs.134 In principle, this can be integrated with quality-by-design risk matrices where analytical feedback loops can adjust the manufacturing process based on the reported quality attributes at each stage of the manufacturing process to ensure the CQA threshold levels are met for batch release.135PATs can be integrated in several different ways depending on the amount of time it takes to receive feedback on the manufacturing process. In the context of polymeric nanomedicines, most of the characterisation techniques described above can be integrated as PATs during various phases of manufacture, polymer synthesis, drug conjugation or the drug formulation stage either online (within the manufacturing path), offline or at-line. Light scattering and spectroscopic techniques (e.g. UV-Vis, fluorescence, Raman) can be easily integrated online with direct feedback of particle size and chemical fingerprints in real-time.136,137 In contrast, benchtop techniques such as SEC, NMR, mass spectrometry, imaging techniques, and AF4 can, in principle, be integrated at-line. At the same time, SAXS and SANS, which commonly require specialist facilities, would be fully offline.
As there are no approved polymer nanoparticle drug delivery systems, polymer characterisation PAT approaches would likely be inspired by polymer-conjugate therapies (e.g. PegIntron and Plegridy). On the other hand, nanoparticle characterisation approaches could utilise technologies from approved Adeno-associated virus (AAV) gene therapies (e.g. Luxturna)138 or the approved lipid nanoparticle drugs and vaccines (e.g. Doxil, Onpattro, Comirnaty and SpikeVax).139 Understanding how these characterisation tools may be implemented and integrated with state-of-the-art digital manufacturing tools such as digital twins and building on underlying polymer-self assembly theory is critical for the translation of these new nanomedicines to the clinic.
Current challenges in polymer nanoparticle delivery
Polymer-based nanoparticles hold significant promise as drug delivery vehicles, but several challenges must be addressed to realise their full potential. A key issue is understanding how the properties of polymeric nanoparticles evolve as they circulate through the body. Their size, shape, surface charge, and the presence of targeting moieties can change due to interactions with biological components such as proteins, lipids, and enzymes. These transformations critically influence systemic delivery, biodistribution, circulation stability, and eventual clearance, as highlighted by a review from Mitchell, Peppas and Langer.7 Identifying the final properties of nanoparticles upon reaching their cellular target remains a significant gap in current knowledge. However, it is worth noting that the article also includes some therapies that are FDA-approved nanomedicines, and there are also many promising new therapies that utilise nanoparticles that are currently in the clinic.9,140The pharmaceutical industry's infrastructure is predominantly tailored for small-molecule drugs and conventional delivery systems. Transitioning to polymer-based drug delivery vectors would necessitate significant investment in new technologies, regulatory frameworks, and manufacturing processes. This structural shift presents logistical and economic barriers, delaying the adoption of these advanced systems. Polymer-based nanoparticles often require intricate designs to achieve precise targeting, controlled release, and stability in physiological conditions. However, this increased complexity comes at a higher cost. A critical question remains: do the improvements in efficacy and efficiency justify the added expense of developing and producing these therapies? Balancing innovation with affordability is essential for making these systems scalable and accessible.
Despite their sophisticated designs, polymer nanoparticles face significant challenges in drug delivery efficiency. Some nanoparticles are so stable that, even after successful cellular internalisation, they fail to effectively release their drug cargo, undermining the system's therapeutic efficacy. Conversely, overly unstable nanoparticles may disassemble before reaching their target, losing the benefits of their nanoscale morphology and protective architecture. Premature disassembly compromises their ability to shield the drug or biologic from degradation, reducing their therapeutic potential.
Addressing these challenges requires a multidisciplinary approach, integrating advances in material science/characterisation, pharmacology, and systems biology. By improving our understanding of nanoparticle transformations, rethinking pharmaceutical infrastructure, and balancing complexity with cost, polymer-based drug delivery systems could overcome inefficiencies and unlock their potential as next-generation therapeutics.
Conclusions: future approaches
To unlock the full potential of polymer-based nanoparticles as drug delivery vehicles, a deeper understanding of their self-assembly and disassembly processes within living cells is crucial. Advanced characterisation techniques capable of monitoring these nanoparticles in situ are necessary to correlate their dynamic behaviour with drug delivery outcomes directly. Such insights will provide a clearer picture of how nanoparticles interact with complex cellular environments and release their therapeutic payloads effectively.The challenge lies in the multivariable nature of biological systems. Parameters such as temperature, pH, concentration, molecular crowding, and dynamic cellular processes continually shift, influencing nanoparticle behaviour in ways that are not yet fully understood. Unlike traditional structure–activity relationships, these multivariant systems require sophisticated analytical methods to untangle the interplay between these factors. For instance, changes in ionic composition resulting from gene regulation could alter the local microenvironment of the nanoparticle, shifting the kinetics of disassembly and ultimately impacting therapeutic efficacy.
The future of polymer nanoparticle drug delivery depends on our ability to model and predict these complex interactions. Multidisciplinary approaches combining advanced imaging, machine learning, and systems biology will be key to uncovering these relationships. By moving beyond traditional frameworks and embracing the complexity of cellular environments, researchers can develop more effective, reliable, and personalised drug delivery systems, paving the way for a new era of precision medicine.
Author contributions
All authors contributed to the preparation, review and editing of this manuscript.Data availability
No new data were generated or analysed in this mini-review article. All data referenced are publicly available through the repositories listed in the respective studies, with access details provided in the References section.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Prof. Sébastien Perrier for inspiring them to explore beyond their immediate fields in pursuit of impactful discoveries.References
- M. E. Napier and J. M. DeSimone, Polym. Rev., 2007, 47, 321–327 CrossRef CAS.
- W. Zhang, A. Mehta, Z. Tong, L. Esser and N. H. Voelcker, Adv. Sci., 2021, 8, 2003937 CrossRef CAS PubMed.
- S. Ohta, E. Kikuchi, A. Ishijima, T. Azuma, I. Sakuma and T. Ito, Sci. Rep., 2020, 10, 18220 CrossRef CAS PubMed.
- F. Alexis, E. M. Pridgen, R. Langer and O. C. Farokhzad, in Drug Delivery, ed. M. Schäfer-Korting, Springer Berlin Heidelberg, 2010, pp. 55–86 Search PubMed.
- S. Wang, K. Cheng, K. Chen, C. Xu, P. Ma, G. Dang, Y. Yang, Q. Lei, H. Huang, Y. Yu, Y. Fang, Q. Tang, N. Jiang, H. Miao, F. Liu, X. Zhao and N. Li, Nano Today, 2022, 45, 101512 CrossRef CAS.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed.
- A. B. Cook and S. Perrier, Adv. Funct. Mater., 2019, 30, 1901001 CrossRef.
- A. C. Anselmo and S. Mitragotri, Bioeng. Transl. Med., 2019, 4, e10143 CrossRef PubMed.
- M. C. Arno, J. D. Simpson, L. D. Blackman, R. P. Brannigan, K. J. Thurecht and A. P. Dove, Biomater. Sci., 2023, 11, 908–915 RSC.
- Y. Bae, S. Fukushima, A. Harada and K. Kataoka, Angew. Chem., 2003, 115, 4788–4791 CrossRef.
- K. Bunyamin, E. Lars, T. D. Hien, S. B. Johan, B. Cyrille and P. D. Thomas, Polym. Chem., 2013, 5, 350–355 Search PubMed.
- W. Chen, S. Ji, X. Qian, Y. Zhang, C. Li, W. Wu, F. Wang and X. Jiang, Polym. Chem., 2017, 8, 2105–2114 RSC.
- R. Dong, Y. Zhou, X. Huang, X. Zhu, Y. Lu and J. Shen, Adv. Mater., 2015, 27, 498–526 CrossRef CAS PubMed.
- P. Gurnani and S. Perrier, Prog. Polym. Sci., 2020, 102, 101209 CrossRef CAS.
- S. Shukla, F. J. Eber, A. S. Nagarajan, N. A. DiFranco, N. Schmidt, A. M. Wen, S. Eiben, R. M. Twyman, C. Wege and N. F. Steinmetz, Adv. Healthcare Mater., 2015, 4, 874–882 CrossRef CAS PubMed.
- C. Weizhi, J. Shilu, Q. Xiaoping, Z. Yajun, L. Cheng, W. Wei, W. Fei and J. Xiqun, Polym. Chem., 2017, 8, 2105–2114 RSC.
- J. Yang, X. Yu, J. I. Song, Q. Song, S. C. L. Hall, G. Yu and S. Perrier, Angew. Chem., Int. Ed., 2022, 61, e202115208 CrossRef CAS PubMed.
- R. Langer, Science, 1990, 249, 1527–1533 CrossRef CAS PubMed.
- R. M. Broyer, G. N. Grover and H. D. Maynard, Chem. Commun., 2011, 47, 2212–2226 RSC.
- C. F. N. João, K. G. Michelle, A. R. Daniel, O. F. Alexander, S. John, P. Andrew, B. Caroline, J. S. Christopher and C. Vijay, Chem. Commun., 2019, 55, 7671–7674 RSC.
- Y. Krishnan, H. A. Rees, C. P. Rossitto, S.-E. Kim, H.-H. K. Hung, E. H. Frank, B. D. Olsen, D. R. Liu, P. T. Hammond and A. J. Grodzinsky, Biomaterials, 2018, 183, 218–233 CrossRef CAS PubMed.
- M. B. Rebecca, N. G. Gregory and D. M. Heather, Chem. Commun., 2011, 47, 2212–2226 RSC.
- I. Cobo, M. Li, B. S. Sumerlin and S. Perrier, Nat. Mater., 2015, 14, 143–159 CrossRef CAS PubMed.
- P. Cotanda, D. B. Wright, M. Tyler and R. K. O'Reilly, J. Polym. Sci., Part A: Polym. Chem., 2013, 51, 3333–3338 CrossRef CAS.
- G. Vancoillie, W. L. A. Brooks, M. A. Mees, B. S. Sumerlin and R. Hoogenboom, Polym. Chem., 2016, 7, 6725–6734 RSC.
- M. Hartlieb, S. Catrouillet, A. Kuroki, C. Sanchez-Cano, R. Peltier and S. Perrier, Chem. Sci., 2019, 10, 5476–5483 RSC.
- S. K. Rajendrakumar, K. Cherukula, H. J. Park, S. Uthaman, Y. Y. Jeong, B.-I. Lee and I.-K. Park, J. Controlled Release, 2018, 276, 72–83 CrossRef CAS PubMed.
- D. Roy, J. N. Cambre and B. S. Sumerlin, Prog. Polym. Sci., 2010, 35, 278–301 CrossRef CAS.
- D. Roy, J. N. Cambre and B. S. Sumerlin, Chem. Commun., 2009, 2106–2108 RSC.
- M. Amit, S. Yuran, E. Gazit, M. Reches and N. Ashkenasy, Adv. Mater., 2018, 30, 1707083 CrossRef PubMed.
- M. Dergham, S. Lin and J. Geng, Angew. Chem., Int. Ed., 2022, 61, e202114267 CrossRef CAS PubMed.
- S. C. Larnaudie, J. C. Brendel, I. Romero-Canelón, C. Sanchez-Cano, S. Catrouillet, J. Sanchis, J. P. C. Coverdale, J.-I. Song, A. Habtemariam, P. J. Sadler, K. A. Jolliffe and S. Perrier, Biomacromolecules, 2017, 19, 239–247 CrossRef PubMed.
- F. Richter, P. Mapfumo, L. Martin, J. I. Solomun, F. Hausig, J. J. Frietsch, T. Ernst, S. Hoeppener, J. C. Brendel and A. Traeger, J. Nanobiotechnol., 2021, 19, 70 CrossRef CAS PubMed.
- C. Zhu and J. Nicolas, Biomacromolecules, 2022, 23, 3043–3080 CrossRef CAS PubMed.
- R. Kumar, N. Le, F. Oviedo, M. E. Brown and T. M. Reineke, JACS Au, 2022, 2, 428–442 CrossRef CAS PubMed.
- R. Kumar, C. F. Santa Chalarca, M. R. Bockman, C. V. Bruggen, C. J. Grimme, R. J. Dalal, M. G. Hanson, J. K. Hexum and T. M. Reineke, Chem. Rev., 2021, 121, 11527–11652 CrossRef CAS PubMed.
- S. T. G. Street, J. Chrenek, R. L. Harniman, K. Letwin, J. M. Mantell, U. Borucu, S. M. Willerth and I. Manners, J. Am. Chem. Soc., 2022, 144, 19799–19812 CrossRef CAS PubMed.
- C. Zylberberg, K. Gaskill, S. Pasley and S. Matosevic, Gene Ther., 2017, 24, 441–452 CrossRef CAS PubMed.
- N. J. W. Penfold, J. Yeow, C. Boyer and S. P. Armes, ACS Macro Lett., 2019, 8, 1029–1054 CrossRef CAS PubMed.
- L. Brunsveld, B. J. B. Folmer, E. W. Meijer and R. P. Sijbesma, Chem. Rev., 2001, 101, 4071–4098 CrossRef CAS PubMed.
- M. K. Henk, P. S. Rint and E. W. Meijer, Eur. J. Org. Chem., 2004, 2553–2555, DOI:10.1002/ejoc.200300752.
- T. F. A. De Greef, M. M. J. Smulders, M. Wolffs, A. P. H. J. Schenning, R. P. Sijbesma and E. W. Meijer, Chem. Rev., 2009, 109, 5687–5754 CrossRef CAS PubMed.
- M. R. Ghadiri, J. R. Granja, R. A. Milligan, D. E. McRee and N. Khazanovich, Nature, 1993, 366, 324–327 CrossRef CAS PubMed.
- G. J. t. C. Mattijs and G. B. Hans, Macromol. Chem. Phys., 2007, 208, 1437–1446 CrossRef.
- I. W. Hamley, Angew. Chem., Int. Ed., 2014, 53, 6866–6881 CrossRef CAS PubMed.
- J. Y. Rho and S. b. Perrier, ACS Macro Lett., 2021, 10, 258–271 CrossRef CAS PubMed.
- M. Inam, G. Cambridge, A. Pitto-Barry, Z. P. L. Laker, N. R. Wilson, R. T. Mathers, A. P. Dove and R. K. O'Reilly, Chem. Sci., 2017, 8, 4223–4230 RSC.
- L. MacFarlane, C. Zhao, J. Cai, H. Qiu and I. Manners, Chem. Sci., 2021, 12, 4661–4682 RSC.
- J. Zhang, H. Sun and P. X. Ma, ACS Nano, 2010, 4, 1049–1059 CrossRef CAS PubMed.
- C. Stoffelen and J. Huskens, Small, 2016, 12, 96–119 CrossRef CAS PubMed.
- Q.-D. Hu, G.-P. Tang and P. K. Chu, Acc. Chem. Res., 2014, 47, 2017–2025 CrossRef CAS PubMed.
- C. A. Stevens, K. Kaur and H.-A. Klok, Adv. Drug Delivery Rev., 2021, 174, 447–460 CrossRef CAS PubMed.
- P. Gurnani, A. B. Cook, R. A. E. Richardson and S. Perrier, Polym. Chem., 2019, 10, 1452–1459 RSC.
- P. Gurnani, C. Sanchez-Cano, H. Xandri-Monje, J. Zhang, S. H. Ellacott, E. D. H. Mansfield, M. Hartlieb, R. Dallmann and S. Perrier, Small, 2022, 18, 2203070 CrossRef CAS PubMed.
- R. X. Zhang, T. Ahmed, L. Y. Li, J. Li, A. Z. Abbasi and X. Y. Wu, Nanoscale, 2017, 9, 1334–1355 RSC.
- J. Tang, J. M. Li, G. Li, H. T. Zhang, L. Wang, D. Li and J. S. Ding, Int. J. Nanomed., 2017, 12, 6687–6704 CrossRef PubMed.
- P. Das and N. R. Jana, ACS Appl. Polym. Mater., 2021, 3, 4791–4811 CrossRef CAS.
- R. Ferrari, M. Sponchioni, M. Morbidelli and D. Moscatelli, Nanoscale, 2018, 10, 22701–22719 RSC.
- N. Kamaly, Z. Xiao, P. M. Valencia, A. F. Radovic-Moreno and O. C. Farokhzad, Chem. Soc. Rev., 2012, 41, 2971–3010 RSC.
- N. M. Amanda and D. H. Jeffrey, Acc. Chem. Res., 2017, 50, 714–722 CrossRef PubMed.
- Y. Mai and A. Eisenberg, Chem. Soc. Rev., 2012, 41, 5969–5985 RSC.
- H.-A. Klok and S. Lecommandoux, Adv. Mater., 2001, 13, 1217–1229 CrossRef CAS.
- A. Rösler, G. W. M. Vandermeulen and H.-A. Klok, Adv. Drug Delivery Rev., 2012, 64, 270–279 CrossRef.
- E. D. von Meerwall, J. Magn. Reson., 1982, 50, 409–416 CAS.
- A. Chen, D. Wu and C. S. Johnson Jr., J. Am. Chem. Soc., 1995, 117, 7965–7970 CrossRef CAS.
- N. Cherifi, A. Khoukh, A. Benaboura and L. Billon, Polym. Chem., 2016, 7, 5249–5257 RSC.
- C. Chamignon, D. Duret, M.-T. Charreyre and A. Favier, Macromol. Chem. Phys., 2016, 217, 2286–2293 CrossRef CAS.
- K. Ute, R. Nagao and K. Watanabe, in NMR Methods for Characterization of Synthetic and Natural Polymers, ed. R. Zhang, T. Miyoshi and P. Sun, The Royal Society of Chemistry, 2019, ch. 5, pp. 80–100 Search PubMed.
- O. Tooley, W. Pointer, R. Radmall, M. Hall, T. Swift, J. Town, C. Aydogan, T. Junkers, P. Wilson, D. Lester and D. Haddleton, ACS Polym. Au, 2024, 4, 311–319 CrossRef CAS PubMed.
- M. Reis, F. Gusev, N. G. Taylor, S. H. Chung, M. D. Verber, Y. Z. Lee, O. Isayev and F. A. Leibfarth, J. Am. Chem. Soc., 2021, 143, 17677–17689 CrossRef CAS PubMed.
- S. T. Knox, S. Parkinson, R. Stone and N. J. Warren, Polym. Chem., 2019, 10, 4774–4778 RSC.
- P.-J. Voorter, M. Wagner, C. Rosenauer, J. Dai, P. Subramanian, A. McKay, N. R. Cameron, J. J. Michels and T. Junkers, Polym. Chem., 2023, 14, 5140–5146 RSC.
- P. J. Voorter, A. McKay, J. Dai, O. Paravagna, N. R. Cameron and T. Junkers, Angew. Chem., Int. Ed., 2021, 61, e202114536 CrossRef PubMed.
- I. W. F. Silva, A. McKay, A. Sokolova and T. Junkers, Polym. Chem., 2024, 15, 1303–1309 RSC.
- A. J. Gormley, J. Controlled Release, 2024, 373, 23–30 CrossRef CAS PubMed.
- A. M. Striegel, Anal. Bioanal. Chem., 2008, 390, 303–305 CrossRef CAS PubMed.
- N. Akhtar, M. B. Ashford, L. Beer, A. Bowes, T. Bristow, A. Broo, D. Buttar, S. Coombes, R. Cross, E. Eriksson, J.-B. Guilbaud, S. W. Holman, L. P. Hughes, M. Jackman, M. J. Lawrence, J. Lee, W. Li, R. Linke, N. Mahmoudi, M. McCormick, B. MacMillan, B. Newling, M. Ngeny, C. Patterson, A. Poulton, A. Ray, N. Sanderson, S. Sonzini, Y. Tang, K. E. Treacher, D. Whittaker and S. Wren, J. Pharm. Sci., 2023, 112, 844–858 CrossRef CAS PubMed.
- S. Sonzini, F. Caputo, D. Mehn, L. Calzolai, S. E. F. Borgos, A. Hyldbakk, K. Treacher, W. Li, M. Jackman, N. Mahmoudi, M. J. Lawrence, C. Patterson, D. Owen, M. Ashford and N. Akhtar, Int. J. Pharm., 2023, 637, 122905 CrossRef CAS PubMed.
- J. S. Town, G. R. Jones and D. M. Haddleton, Polym. Chem., 2018, 9, 4631–4641 RSC.
- K. Ishitsuka, T. Kakiuchi, H. Sato and T. N. J. Fouquet, Rapid Commun. Mass Spectrom., 2020, 34, e8584 CrossRef CAS PubMed.
- E. Kendrick, Anal. Chem., 1963, 35, 2146–2154 CrossRef CAS.
- T. E. Morgan, S. H. Ellacott, C. A. Wootton, M. P. Barrow, A. W. T. Bristow, S. Perrier and P. B. O'Connor, Anal. Chem., 2018, 90, 11710–11715 CrossRef CAS PubMed.
- T. E. Morgan, T. G. Floyd, B. P. Marzullo, C. A. Wootton, M. P. Barrow, A. W. T. Bristow, S. Perrier and P. B. O'Connor, Polym. Chem., 2022, 13, 4162–4169 RSC.
- T. E. Morgan, C. A. Wootton, B. Marzullo, J. Paris, A. Kerr, S. H. Ellacott, M. A. van Agthoven, M. P. Barrow, A. W. T. Bristow, S. Perrier and P. B. O'Connor, J. Am. Soc. Mass Spectrom., 2021, 32, 2153–2161 CrossRef CAS PubMed.
- J. Lawrence, S.-H. Lee, A. Abdilla, M. D. Nothling, J. M. Ren, A. S. Knight, C. Fleischmann, Y. Li, A. S. Abrams, B. V. K. J. Schmidt, M. C. Hawker, L. A. Connal, A. J. McGrath, P. G. Clark, W. R. Gutekunst and C. J. Hawker, J. Am. Chem. Soc., 2016, 138, 6306–6310 CrossRef CAS PubMed.
- R. Peltier, A. Bialek, A. Kuroki, C. Bray, L. Martin and S. Perrier, Polym. Chem., 2018, 9, 5511–5520 RSC.
- J. Y. Rho, J. C. Brendel, L. R. MacFarlane, E. D. H. Mansfield, R. Peltier, S. Rogers, M. Hartlieb and S. Perrier, Adv. Funct. Mater., 2018, 28, 1704569 CrossRef.
- M. Ghezzi, S. Pescina, C. Padula, P. Santi, E. Del Favero, L. Cantù and S. Nicoli, J. Controlled Release, 2021, 332, 312–336 CrossRef CAS PubMed.
- W. Gibson, J. T. Mulvey, S. Das, S. Selmani, J. G. Merham, A. M. Rakowski, E. Schwartz, A. I. Hochbaum, Z. Guan, J. R. Green and J. P. Patterson, ACS Nano, 2024, 18, 11898–11909 CrossRef CAS PubMed.
- J. Kuntsche, J. C. Horst and H. Bunjes, Int. J. Pharm., 2011, 417, 120–137 CrossRef CAS PubMed.
- P. Renz, M. Kokkinopoulou, K. Landfester and I. Lieberwirth, Macromol. Chem. Phys., 2016, 217, 1879–1885 CrossRef CAS.
- K. H. Nagamanasa, H. Wang and S. Granick, Adv. Mater., 2017, 29, 1703555 CrossRef PubMed.
- G. Ing, A. Stewart, G. Battaglia and L. Ruiz-Perez, PLoS One, 2023, 18, e0285691 CrossRef CAS PubMed.
- H. Wu, H. Sun, R. A. J. F. Oerlemans, S. Li, J. Shao, J. Wang, R. R. M. Joosten, X. Lou, Y. Luo, H. Zheng, L. K. E. A. Abdelmohsen, H. H. P. Garza, J. C. M. van Hest and H. Friedrich, Adv. Mater., 2024, 2402987 CrossRef CAS PubMed.
- C. I. C. Crucho and M. T. Barros, Mater. Sci. Eng., C, 2017, 80, 771–784 CrossRef CAS PubMed.
- R. L. Harniman, S. Pearce and I. Manners, J. Am. Chem. Soc., 2022, 144, 951–962 CrossRef CAS PubMed.
- M. A. Touve, C. A. Figg, D. B. Wright, C. Park, J. Cantlon, B. S. Sumerlin and N. C. Gianneschi, ACS Cent. Sci., 2018, 4, 543–547 CrossRef CAS PubMed.
- G. M. Scheutz, M. A. Touve, A. S. Carlini, J. B. Garrison, K. Gnanasekaran, B. S. Sumerlin and N. C. Gianneschi, Matter, 2021, 4, 722–736 CrossRef CAS.
- Y. Guo, T. Xia, V. Walter, Y. Xie, J. Y. Rho, L. Xiao, R. O. O’Reilly and M. Wallace, ChemRxiv, 2024 DOI:10.26434/chemrxiv-2024-d1gd3.
- T. Xia, Z. Tong, Y. Xie, M. C. Arno, S. Lei, L. Xiao, J. Y. Rho, C. T. J. Ferguson, I. Manners, A. P. Dove and R. K. O'Reilly, J. Am. Chem. Soc., 2023, 145, 25274–25282 CrossRef CAS PubMed.
- M. Khan, T. R. Guimarães, R. P. Kuchel, G. Moad, S. Perrier and P. B. Zetterlund, Angew. Chem., Int. Ed., 2021, 60, 23281–23288 CrossRef CAS PubMed.
- K. T. Kim, J. J. L. M. Cornelissen, R. J. M. Nolte and J. C. M. van Hest, Adv. Mater., 2009, 21, 2787–2791 CrossRef CAS.
- R. J. R. W. Peters, M. Marguet, S. Marais, M. W. Fraaije, J. C. M. v. Hest and S. Lecommandoux, Angew. Chem., Int. Ed., 2014, 53, 146–150 CrossRef CAS PubMed.
- D. D. Lane, F. Y. Su, D. Y. Chiu, S. Srinivasan, J. T. Wilson, D. M. Ratner, P. S. Stayton and A. J. Convertine, Polym. Chem., 2014, 6, 1255–1266 RSC.
- S. Varlas, R. Keogh, Y. Xie, S. L. Horswell, J. C. Foster and R. K. O'Reilly, J. Am. Chem. Soc., 2019, 141, 20234–20248 CrossRef CAS PubMed.
- H. Che and J. C. M. van Hest, ChemNanoMat, 2019, 5, 1092–1109 CrossRef CAS.
- C. G. Palivan, L. Heuberger, J. Gaitzsch, B. Voit, D. Appelhans, B. B. Fernandes, G. Battaglia, J. Du, L. Abdelmohsen, J. C. M. van Hest, J. Hu, S. Liu, Z. Zhong, H. Sun, A. Mutschler and S. Lecommandoux, Biomacromolecules, 2024, 25, 5454–5467 CrossRef CAS PubMed.
- A. Reisch and A. S. Klymchenko, Small, 2016, 12, 1968–1992 CrossRef CAS PubMed.
- K. Li, W. Qin, D. Ding, N. Tomczak, J. Geng, R. Liu, J. Liu, X. Zhang, H. Liu, B. Liu and B. Z. Tang, Sci. Rep., 2013, 3, 1150 CrossRef PubMed.
- W. Li, G. S. Kaminski Schierle, B. Lei, Y. Liu and C. F. Kaminski, Chem. Rev., 2022, 122, 12495–12543 CrossRef CAS PubMed.
- L. Albertazzi, D. van der Zwaag, C. M. A. Leenders, R. Fitzner, R. W. van der Hofstad and E. W. Meijer, Science, 2014, 344, 491–495 CrossRef CAS PubMed.
- L. Albertazzi, D. v. d. Zwaag, C. M. A. Leenders, R. Fitzner, R. W. v. d. Hofstad and E. W. Meijer, Science, 2014, 344, 491–495 CrossRef CAS PubMed.
- M. H. Bakker, C. C. Lee, E. W. Meijer, P. Y. W. Dankers and L. Albertazzi, ACS Nano, 2016, 10, 1845–1852 CrossRef CAS PubMed.
- R. M. P. da Silva, D. van der Zwaag, L. Albertazzi, S. S. Lee, E. W. Meijer and S. I. Stupp, Nat. Commun., 2016, 7, 11561 CrossRef CAS PubMed.
- M. Kumar, J. Son, R. H. Huang, D. Sementa, M. Lee, S. O'Brien and R. V. Ulijn, ACS Nano, 2020, 14, 15056–15063 CrossRef CAS PubMed.
- R. Riera, E. Archontakis, G. Cremers, T. de Greef, P. Zijlstra and L. Albertazzi, ACS Sens., 2023, 8, 80–93 CrossRef CAS PubMed.
- M. M. E. Tholen, B. J. H. M. Rosier, R. T. Vermathen, C. A. N. Sewnath, C. Storm, L. Woythe, C. Izquierdo-Lozano, R. Riera, M. van Egmond, M. Merkx and L. Albertazzi, ACS Nano, 2023, 17, 11665–11678 CrossRef CAS PubMed.
- M. M. E. Tholen, R. P. Tas, Y. Wang and L. Albertazzi, Chem. Commun., 2023, 59, 8332–8342 RSC.
- L. Woythe, D. Porciani, T. Harzing, S. van Veen, D. H. Burke and L. Albertazzi, J. Controlled Release, 2023, 355, 228–237 CrossRef CAS PubMed.
- T. Andrian, Y. Muela, L. Delgado, L. Albertazzi and S. Pujals, Nanoscale, 2023, 15, 14615–14627 RSC.
- J. P. Patterson, M. P. Robin, C. Chassenieux, O. Colombani and R. K. O'Reilly, Chem. Soc. Rev., 2014, 43, 2412–2425 RSC.
- S. C. Owen, D. P. Y. Chan and M. S. Shoichet, Nano Today, 2012, 7, 53–65 CrossRef CAS.
- Q. Yu, R. M. England, A. Gunnarsson, R. Luxenhofer, K. Treacher and M. B. Ashford, Macromolecules, 2022, 55, 401–412 CrossRef CAS.
- R. Wu, M. Tian, C. Shu, C. Zhou and W. Guan, Soft Matter, 2022, 18, 8920–8930 RSC.
- E. E. Brotherton, F. L. Hatton, A. A. Cockram, M. J. Derry, A. Czajka, E. J. Cornel, P. D. Topham, O. O. Mykhaylyk and S. P. Armes, J. Am. Chem. Soc., 2019, 141, 13664–13675 CrossRef CAS PubMed.
- M. J. Derry, L. A. Fielding, N. J. Warren, C. J. Mable, A. J. Smith, O. O. Mykhaylyk and S. P. Armes, Chem. Sci., 2016, 7, 5078–5090 RSC.
- R. Takahashi, S. Miwa, F. H. Sobotta, J. H. Lee, S. Fujii, N. Ohta, J. C. Brendel and K. Sakurai, Polym. Chem., 2020, 11, 1514–1524 RSC.
- S. K. Filippov, J. M. Franklin, P. V. Konarev, P. Chytil, T. Etrych, A. Bogomolova, M. Dyakonova, C. M. Papadakis, A. Radulescu, K. Ulbrich, P. Stepanek and D. I. Svergun, Biomacromolecules, 2013, 14, 4061–4070 CrossRef CAS PubMed.
- E. D. H. Mansfield, M. Hartlieb, S. Catrouillet, J. Y. Rho, S. C. Larnaudie, S. E. Rogers, J. Sanchis, J. C. Brendel and S. Perrier, Soft Matter, 2018, 14, 6320–6326 RSC.
- M. Kariuki, J. Y. Rho, S. C. L. Hall and S. Perrier, Macromolecules, 2023, 56, 6618–6632 CrossRef CAS PubMed.
- H. Wu, A. Homawoo, S. Shariati, C. E. Astete, D. F. Rodrigues, C. M. Sabliov, E. H. Fini and S. M. Louie, RSC Pharm., 2024, 1, 994–1007 RSC.
- G. Gerzon, Y. Sheng and M. Kirkitadze, J. Pharm. Biomed. Anal., 2022, 207, 114379 CrossRef CAS PubMed.
- N. Aguiam, L. I. F. Moura, M. Oliveira, H. Florindo and J. A. Lopes, in Artificial Intelligence for Drug Product Lifecycle Applications, ed. A. Pais, C. Vitorino, S. Nunes and T. Cova, Academic Press, 2025, pp. 169–203 Search PubMed.
- S. Cunha, C. P. Costa, J. N. Moreira, J. M. Sousa Lobo and A. C. Silva, Nanomed.: Nanotechnol. Biol. Med., 2020, 28, 102206 CrossRef CAS PubMed.
- X. Feng, G. Huang, J. Qiu, L. Peng, K. Luo, D. Liu and P. Han, Opt. Commun., 2023, 531, 129225 CrossRef CAS.
- G. Gkogkos, L. Panariello, E. Grammenou, M. A. Cornwell, A. Afrashtehpour, A. J. MacRobert, I. P. Parkin and A. Gavriilidis, Chem. Eng. J., 2023, 467, 143350 CrossRef CAS.
- T. Dobrowsky, D. Gianni, J. Pieracci and J. Suh, Curr. Opin. Biomed. Eng., 2021, 20, 100353 CrossRef.
- Z. Kis, R. Shattock, N. Shah and C. Kontoravdi, Biotechnol. J., 2019, 14, 1800376 CrossRef CAS PubMed.
- A. C. Anselmo and S. Mitragotri, Bioeng. Transl. Med., 2021, 6, e10246 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |