 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Single-visible-light performed STORM imaging with activatable photoswitches†
Fanghui Li‡
a,
Mengqi Li‡a,
Yiqi Shia,
Xinyun Biana,
Ning Lva,
Shaomeng Guoa,
Ying Wanga,
Weijun Zhao *ab and
Wei-Hong Zhu
*ab and
Wei-Hong Zhu *ab
*ab
aKey Laboratory for Advanced Materials and Joint International Research Laboratory of Precision Chemistry and Molecular Engineering, Feringa Nobel Prize Scientist Joint Research Center, Shanghai Key Laboratory of Functional Materials Chemistry, Institute of Fine Chemicals, East China University of Science & Technology, Shanghai 200237, China. E-mail: whzhu@ecust.edu.cn; zhwj@ecust.edu.cn
bCenter of Photosensitive Chemicals Engineering, East China University of Science and Technology, Shanghai, 200237, China
First published on 10th July 2025
Abstract
Stochastic optical reconstruction microscopy (STORM) overcomes the diffraction limit of optical imaging, facilitating high-resolution visualization of cellular substructures at the nanoscale. Essential to this technique is the development of fluorescent photoswitches. However, existing photoswitches typically rely on sophisticated dual-beam systems that involve harmful UV-light and lack specific recognition of biomolecules. Here we develop unique intracellular biomolecule-activatable photoswitches tailored for single-visible-light performed STORM imaging. Upon incorporating intramolecular proton transfer (IPT) units into the photochromic diarylethene, the all-visible-light driven photoswitches are established with excellent photoresponsive efficiency, high brightness and fluorescence ON-to-OFF contrast ratio, guaranteeing STORM imaging using a single-visible-light (488 nm) by regulating the activation, excitation and deactivation processes. Furthermore, we functionalized the IPT units with biomolecular recognition motifs, creating photoswitches capable of sensing the expression levels of intracellular biomolecules (like glutathione (GSH) or β-galactosidase (β-Gal)) with super-resolution. Our objective is to engineer single-visible-light driven, biomolecule-activatable photoswitches, which will significantly streamline the STORM technique and expand the applicability of super-resolution imaging for the precise mapping of intracellular substructures.
Introduction
The stochastic optical reconstruction microscopy (STORM) technique overcomes the classical optical diffraction limit,1,2 emerging as a powerful super-resolution imaging tool for visualizing biological structures with sub-diffraction spatial resolution.3,4 Essential to this technique is the selection of fluorescent photoswitches, whose properties largely determine the quality of STORM images.5,6 While existing photoswitch systems have achieved remarkable progress, some limitations persist. For example, reversible photoswitches convert between fluorescent (on) and dark (off) states by two light sources/lasers, which typically involve harmful UV-light,7–9 supplemental additives (e.g., oxygen scavenging systems and thiols) are essential for conventional photoswitches (e.g., Cy3B, Alexa Fluor 647) to improve photoresponsive performance and photostability,10–12 and the lack of biomolecule-specific responsive photoswitches limits the capability to visualize intracellular biomolecules with high precision.13,14 Accordingly, it is desirable to develop novel photoswitches to break through these limitations and broaden the application scope of super-resolution imaging in biological research.Previous work on STORM systems has produced simpler (single laser wavelength performed STORM) and easier (without additives) protocols,15–17 typically involving spontaneous blinking dyes and photochromic dyes as well as quantum dots. Spontaneous blinking dyes18,19 (e.g., cyanines, rhodamine derivatives) spontaneously switch between fluorescent and dark states without additional activation of light, but they operate in specific toxic and non-physiological imaging buffers; photochromic dyes, such as turn-on mode fluorescent SDAE,20 can use the “hot-band” phenomenon to enable single-visible-light control of activation, excitation and deactivation of fluorescence without additional buffers.21 However, due to the extremely low molar extinction coefficients of hot-band absorption, high laser intensities are required for super-resolution imaging; quantum dots22,23 have emerged as attractive alternatives through their intrinsic reversible blinking behavior for STORM. Nevertheless, fundamental challenges persist regarding their unclear photoswitching mechanisms and difficulties in functionalization modification.
Here, we present novel fluorescent photoswitches for single-visible-light driven STORM imaging by incorporating the intramolecular proton transfer (IPT) units of 2-(2-hydroxyphenyl)-benzothiazole (HBT)24,25 into photochromic diarylethene (DAE) (Fig. 1a, right). DAE systems,26–29 which feature reversible photoinduced fluorescence regulation between the open and closed forms without additional reagents, have attracted increasing attention in super-resolution imaging.30–35 Intramolecular proton transfer (IPT) is an important feature of hydrogen-bonded systems (e.g., salicylidene Schiff base and derivatives). The equilibrium between two tautomers OH (proton bonded to oxygen) and NH (proton bonded to nitrogen) determines their optical behavior, with the NH form exhibiting a redshifted absorption band compared to the OH form.36–38 Leveraging the distinctive IPT effect, the absorption of the open form is efficiently extended into the visible region, overlapping with that of the closed form, realizing a fully visible-light driven photoresponsive system. Furthermore, a strongly electron-donating morpholine unit was attached to HBT to realize the red-shift of the absorption peak of the open form and the enhancement of the molar extinction coefficient within the visible region. Importantly, the incorporation of the morpholine unit simultaneously improves the fluorescence quantum yield and the visible-light photoresponsive performance of the photoswitch, thus enabling a single 488 nm light performed STORM imaging and greatly simplifying the STORM technique. Finally, we developed two intracellular biomolecule-activatable photoswitches, DH-Mor-GSH and DH-Mor-β-Gal (Fig. 1a, left), by integrating specific recognition motifs to modulate the IPT switching effect. These rationally designed photoswitches exhibit specific responsiveness toward glutathione (GSH) or β-galactosidase (β-Gal), enabling high-resolution visualization of endogenous biomolecules (GSH or β-Gal) (Fig. 1b). Given these activatable photoswitches, we employed the STORM single-molecule counting method to analyze the expression levels of GSH and β-Gal in various cancer cells, thus offering a convenient and intuitive approach for the semiquantitative analysis of intracellular biomolecules. In summary, we offer single-visible-light driven and biomolecule-activatable photoswitches for broadening STORM super-resolution imaging for visualizing intracellular substructures.
Results and discussion
All-visible-light driven photoswitches via the IPT effect
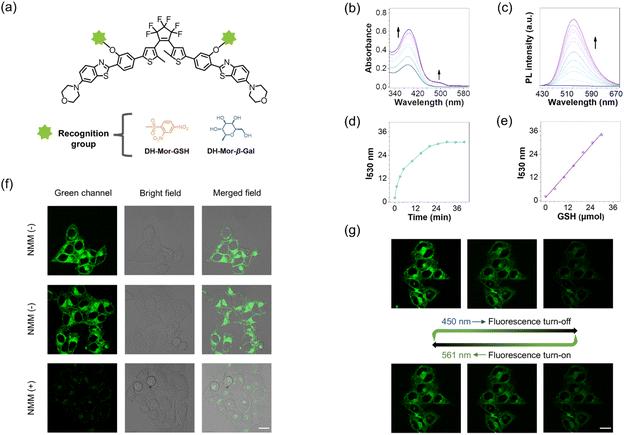
Since the progress in developing photochromic DAE dyes for STORM imaging is hindered by the requirement for harmful UV-light to induce photocyclization,39–41 we rationally designed a series of all-visible-light modulated fluorescent photoswitches, namely DH, DH-OMe and DH-Mor (synthetic methods are described in the ESI†). These novel photoswitches feature the fluorophore HBT which has an IPT effect, and methoxy and morpholine units with varying electron-donating ability to modulate their photoresponsive and fluorescent performance (Fig. 2a).As anticipated, the open forms of the three designed photoswitches (DH, DH-OMe and DH-Mor) exhibited an additional absorption band in the visible-light region (Fig. 2b). Compared to DH and DH-OMe, DH-Mor with a stronger electron-donating morpholine group showed a more red-shifted absorption band and a significantly higher molar extinction coefficient in the visible-light range (400–500 nm), reaching 0.8 × 104 M−1 cm−1 at 450 nm. Extending π-conjugation in DAEs is a common strategy to red-shift the absorption band into the visible region,42–44 but often leads to a significant reduction in photoresponsive efficiency. With the introduction of the IPT strategy, the three designed photoswitches maintain excellent photoresponsive behavior upon all-visible-light irradiation and show excellent fatigue resistance (Fig. 2c and S1–S3†). The photocyclization quantum yields (Φo–c) at 450 nm were measured to be 36% for DH-Mor, accompanied by an excellent photocyclization conversion ratio as high as 92%. Desirable photoswitching efficiencies were also achieved for both DH and DH-OMe upon visible light irradiation at 450 nm (Table 1). Notably, the photocyclization quantum yields measured at 450 nm were lower than those obtained under 365 nm irradiation. This decrease is attributed to enhanced competitive photocycloreversion reactions at the longer wavelength. Surprisingly, DH-Mor exhibited both the highest photocyclization quantum yield and conversion ratio under 450 nm light irradiation, with a photocyclization rate approximately 47 times higher than that of the DH-OMe (Fig. S7†). In summary, the rational design of IPT-based DAE realizes desirable all-visible-light driven photoresponsive performance, and DH-Mor with the strong electron-donating morpholine group shows the highest photoresponsive efficiency.
| Compounda | Photocyclization | Photocycloreversion | ||||
|---|---|---|---|---|---|---|
| λirrb [nm] | CRo–cc [%] | Φo–cc [%] | λirrb [nm] | CRc–od [%] | Φc–od [%] | |
| a Solvent for DHs: DMSO.b λirr represents the wavelength of light for irradiation.c CRo–c and Φo–c represent the conversion ratio and quantum yield of photocyclization, respectively.d CRc–o and Φc–o represent the conversion ratio and quantum yield of photocycloreversion, respectively. | ||||||
| DH | 365 | 93 | 37 | 600 | >99 | 7.6 |
| 450 | 62 | 27 | ||||
| DH-OMe | 365 | 98 | 40 | 600 | >99 | 5.5 |
| 450 | 69 | 29 | ||||
| DH-Mor | 365 | >99 | 42 | 600 | >99 | 2.3 |
| 450 | 92 | 36 | ||||
Efficient fluorescence modulation via a morpholine moiety
In addition to photoresponsive performance, the fluorescence characteristics, such as fluorescence brightness and the ability to modulate between fluorescence ON and OFF states, are critical factors for fluorescent photoswitches.45,46 Notably, the open form of DH-Mor exhibited brightest green fluorescence, with a fluorescence quantum yield (ΦF) of 52%, approximately four times higher than those of DH and DH-OMe. Upon dual visible-light (450 and 561 nm) irradiation, the green fluorescence of DH-Mor was gradually and reversibly switched, with a fluorescence ON-to-OFF contrast ratio of 100.3, significantly higher than those of DH (13.6) and DH-OMe (19.5) (Fig. 2d and e). DH-Mor is a classical turn-off mode fluorescent molecule: the open form is fluorescent, while the closed form remains in a dark state. This efficient fluorescence conversion can be attributed to the intermolecular energy transfer process between the fluorescent IPT unit and the photochromic DAE core (Fig. 2f). Overall, through donor molecular engineering, the fluorescence photoswitch DH-Mor exhibited both the highest brightness and fluorescence ON-to-OFF contrast ratio, making it highly promising for STORM imaging.Biomolecule-activated fluorescent photoswitches
Glutathione (GSH) and β-galactosidase (β-Gal)47–51 are vital biomolecules in living organisms, crucial for cellular physiological processes. However, the nano-level visualization of intracellular GSH or β-Gal remains challenging due to the limited resolution of traditional microscopes and the lack of responsive photoswitches.52–54 Given the high-performance photoswitch DH-Mor, we constructed activatable photoswitches DH-Mor-GSH and DH-Mor-β-Gal by incorporating GSH or β-Gal recognition motifs into the phenolic hydroxyl reactive oxygen site within the IPT motif (Fig. 3a). Prior to cellular experiments, we also validated the performance of DH-Mor under physiologically relevant aqueous conditions (PBS) to ensure intracellular functionality. As shown in Fig. S8,† DH-Mor maintains all-visible-light driven photoresponsive performance and good fatigue resistance in aqueous media. As illustrated in Fig. 3b–d, both the absorbance and fluorescence of DH-Mor-GSH rapidly responded to GSH, reaching equilibrium within 30 min at physiological temperatures (37 °C). The fluorescence intensity of DH-Mor-GSH had a positive linear relationship to GSH concentration (0–30 μM) (Fig. 3e), indicating the potential for quantitative detection of endogenous GSH. Notably, DH-Mor-GSH retained excellent all-visible-light modulated photoresponsive performance when reacting with GSH (Fig. S9†). Similarly, DH-Mor-β-Gal demonstrated a promising activatable response towards β-Gal in vitro (Fig. S11†).To validate the effectiveness of DH-Mor-GSH in detecting GSH and the photoresponsive properties within the cells, intracellular confocal imaging experiments were first conducted. The photoswitches showed low cytotoxicity in living cells (Fig. S12†), making them suitable for intracellular imaging. Upon treatment with DH-Mor-GSH, HeLa cells exhibited intense fluorescence (Fig. 3f), in contrast with the very weak fluorescence in cells pretreated with NEM (a well-known thiol scavenger), confirming the specific responsiveness of DH-Mor-GSH to endogenous GSH. Meanwhile, the reversible modulation of intracellular fluorescence with alternating irradiation at 450 and 561 nm demonstrated excellent photoresponsive properties within the cells (Fig. 3g). The β-Gal activatable photoswitch DH-Mor-β-Gal also exhibited outstanding recognition ability for endogenous β-Gal (Fig. S13 and S14†).
Single-visible-light performed STORM imaging
Conventional STORM technology typically requires two light sources to reversibly convert photoswitches between fluorescence ON and OFF states. Our photoswitch DH-Mor has an effective overlap of the absorption spectra of its open and closed forms in the visible range, making it highly capable of modulating fluorescence with a single light source.Here we propose a mechanism illustration for STORM imaging using a single 488 nm laser (spectral adaptation to DH-Mor) (Fig. 4a): (1) exciting the probe's fluorescence, (2) switching off the fluorescence by inducing cyclization to its fluorescence OFF state, (3) activating the photoswitch to its fluorescence ON state via cycloreversion. To validate this mechanism, we conducted confocal imaging using a 488 nm laser on DH-Mor labeled micelles (formed from PSt38k-b-PEO11k copolymer self-assembly). Initially, the fluorescence of the open form of DH-Mor molecules was excited by a 488 nm laser, the micelles exhibit intense fluorescence, which slightly decreases upon irradiation and eventually reaches a photostationary state. Initial fluorescence (100% open form) decreased to 92% at the PSS state (Fig. S15†). This imaging process clearly reflects the dynamic fluorescence switching of DH-Mor driven by a single 488 nm laser. In detail, the 488 nm laser excites each DH-Mor molecule to emit photons and enter the fluorescence OFF state, while simultaneously reactivating it to return to the fluorescence ON state, enabling reversible switching between fluorescence ON and OFF states. By employing a single 488 nm laser, we observed distinct fluorescent blinking behavior within the micelles. The STORM imaging of DH-Mor labeled micelles significantly enhances image resolution with a full-width at half-maximum (FWHM) of 47 nm, a ninefold reduction compared to conventional widefield fluorescence imaging (Fig. S16†). Moreover, the spatial resolution was analyzed using Fourier ring correlation (FRC),55–57 a method recently often used to determine the resolution of localization-based super-resolution fluorescence imaging. The resolution of the localizations was determined to be about 63 nm (Fig. S17†).
We further conducted STORM imaging using intracellular biomolecule-activatable photoswitches, DH-Mor-GSH and DH-Mor-β-Gal, with a single laser beam. Upon intracellular GSH activation, we performed STORM imaging using a single 488 nm laser. The super-resolution imaging exhibited a significant improvement in spatial resolution compared to conventional wide-field and confocal microscopy. As shown in Fig. 4b and S18,† traditional imaging approaches demonstrated limited capacity to resolve the nanoscale spatial distribution of intracellular GSH due to their inherent diffraction-limited resolution constraints. In contrast, STORM imaging successfully resolved intracellular GSH localization with a measured overall spatial resolution of 69 nm (Fig. S20†) as measured by FRC. Notably, STORM image exhibited a clear distinction between two localized points separated by a distance as small as 47 nm (Fig. 4c). The STORM image exhibited an average photon count (636 per switch) and average localization precision (29 per switch) (Fig. 4d and e), enabling the localization and visualization of endogenous GSH with high precision. Super-resolution imaging has clearly revealed the distribution characteristics of GSH within HeLa cells. GSH is predominantly enriched in the cytoplasm, where it participates in a variety of critical biochemical reactions, sustaining the normal physiological activities of the cell.58 Upon activation by β-Gal, the activatable photoswitch DH-Mor-β-Gal also realized a single-light performed STORM imaging (Fig. S19†).
STORM single-molecule counting mode for analyzing the biomolecules' expression levels
STORM technology provides precise localization of single-molecules59,60 when adopting our activatable photoswitch capable of specific biomolecule recognition; it becomes an ideal tool for directly analyzing intracellular biomolecule expression levels by quantifying the number of localized molecules in STORM images. As reported, β-Gal expression levels varied across different cancer cells and were typically evaluated using two methods: confocal imaging (based on fluorescence intensity) and western blotting (based on protein bands gray-scale analysis) (Fig. 5a). Using the activatable photoswitch DH-Mor-β-Gal, STORM imaging can clearly demonstrate the distribution of intracellular β-Gal in all three types of cancer cells (Fig. 5b). The counting mode of STORM imaging can provide the localization number per μm2, ranked from highest to lowest as follows: OVCAR3, HepG2, and HeLa cells (Fig. 5c), consistent with the trends observed in confocal imaging (Fig. S21†) and western blotting experiments (Fig. 5d), thereby validating the accuracy and convenience of the STORM-based counting method. Moreover, using the STORM single-molecule counting mode, the differentiated expression levels of GSH in HeLa, HepG2, and OVCAR3 cells can be evaluated by activatable probe DH-Mor-GSH (Fig. S22 and S23†), showing the GSH expression levels ranked from highest to lowest as follows: HeLa cells, HepG2 and OVCAR3. These results suggest that the STORM-based single-molecule counting method offers a simpler, more intuitive approach to semiquantitative analysis of intracellular biomolecules, providing a powerful alternative with improved resolution.Conclusions
In summary, we have successfully developed novel biomolecule-activatable photoswitches, specifically driven by single-visible-light, eliminating the need for harmful UV-light and supplemental additives. Diverging from conventional methodologies that achieve all-visible-light-regulated diarylethene through extending π-conjugation and utilizing the “hot-band” phenomena, our innovative strategy is to ingeniously integrate morpholine and IPT units into the photoswitch DAE. On the one hand, introducing IPT units generates a strong emerged absorption band in the visible-light region, under 450 nm irradiation, while DH-Mor exhibits a high photocyclization quantum yield (36%) and conversion ratio (92%). On the other hand, the well-chosen morpholine groups endow DH-Mor with a high fluorescence quantum yield (52%) and an enhanced fluorescence ON-to-OFF contrast ratio, ensuring effective fluorescence modulation. These two design strategies synergistically contribute to single-visible-light performed STORM imaging, along with the following advantages: favorable cell permeability and biocompatibility, as well as the elimination of supplemental additives during STORM data acquisition. Furthermore, recognition units for vital biomolecules, such as GSH and β-Gal, are incorporated to switch the IPT effect, yielding unique biomolecule-activatable photoswitches that enable localization of intracellular GSH and β-Gal with high precision. Given these single-visible-light driven and activatable photoswitches, we can visualize expression levels of GSH and β-Gal in various cancer cells using STORM single-molecule counting methods, offering a convenient and precise approach for the semiquantitative analysis of intracellular biomolecules. This innovative design strategy not only simplifies the STORM imaging process but also expands the application scope of super-resolution imaging in biological research.Data availability
All relevant data is presented in the paper and ESI.†Author contributions
W. Zhu and W. Zhao supervised the study and directed the scientific research, and prepared the manuscript. F. L., Y. S., N. L., Y. W. and S. G. carried out the design, synthesis, and optical determination. F. L. and M. L. did the data analysis and wrote the manuscript. F. L. and X. B. performed the bioimaging experiments and analyzed the data.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Research and Development Program of China (2021YFA0910000); Basic Science Center of National Natural Science Foundation (T2488302); National Natural Science Foundation of China (9235630033, 22338006, 22105069); Science and Technology Commission of Shanghai Municipality (grant no. 24DX1400200); and Innovation Program of Shanghai Municipal Education Commission (2023FGS01). The authors thank the staff members of the Integrated Laser Microscopy System at the National Facility for Protein Science in Shanghai (NFPS), Shanghai Advanced Research Institute, Chinese Academy of Sciences, China for data collection and analysis. The authors are grateful to Lili Wang from Nikon Precision (Shanghai) Co., Ltd for useful assistance on the NIKON N-STORM.Notes and references
- M. Lelek, M. T. Gyparaki, G. Beliu, F. Schueder, J. Griffié, S. Manley, R. Jungmann, M. Sauer, M. Lakadamyali and C. Zimmer, Nat. Rev. Methods Primers, 2021, 1, 39–66 CrossRef CAS PubMed.
- K. Xu, G. Zhong and X. Zhuang, Science, 2013, 339, 452–456 CrossRef CAS PubMed.
- X. Zhu, Q. Chen, H. Zhao, Q. Yang, Goudappagouda, M. Gelléri, S. Ritz, D. Ng, K. Koynov, S. H. Parekh, V. K. Chetty, B. K. Thakur, C. Cremer, K. Landfester, K. Müllen, M. Terenzio, M. Bonn, A. Narita and X. Liu, J. Am. Chem. Soc., 2024, 146, 5195–5203 CrossRef CAS PubMed.
- D. Englert, E.-M. Burger, F. Grün, M. S. Verma, J. Lackner, M. Lampe, B. Bühler, J. Schokolowski, G. U. Nienhaus, A. Jäschke and M. Sunbul, Nat. Commun., 2023, 14, 3879 CrossRef CAS PubMed.
- G. T. Dempsey, J. C. Vaughan, K. H. Chen, M. Bates and X. Zhuang, Nat. Methods, 2011, 8, 1027–1036 CrossRef CAS.
- S. Samanta, W. Gong, W. Li, A. Sharma and J. S. Kim, Coord. Chem. Rev., 2019, 380, 17–34 CrossRef CAS.
- F. M. Jradi and L. D. Lavis, ACS Chem. Biol., 2019, 14, 1077–1090 CrossRef CAS PubMed.
- Y. Yang, Y. Ma, J. F. Berengut, L. K. Lee, R. D. Tilley, K. Gaus and J. J. Gooding, Nat. Photonics, 2024, 18, 713–720 CrossRef CAS.
- C. S. Wijesooriya, J. A. Peterson, P. Shrestha, E. J. Gehrmann, A. H. Winter and E. A. Smith, Angew. Chem., Int. Ed., 2018, 57, 12685–12689 CrossRef CAS PubMed.
- B. Wang, M. Xiong, J. Susanto, X. Li, W. Y. Leung and K. Xu, Angew. Chem., Int. Ed., 2022, 61, e202113612 CrossRef CAS PubMed.
- S. van de Linde, A. Löschberger, T. Klein, M. Heidbreder, S. Wolter, M. Heilemann and M. Sauer, Nat. Protoc., 2011, 6, 991–1009 CrossRef CAS PubMed.
- X. Zhang, D. Guan, Y. Liu, J. Liu, K. Sun, S. Chen, Y. Zhang, B. Zhao, T. Zhai, Y. Zhang, F. Li and Q. Liu, Angew. Chem., Int. Ed., 2022, 61, e202211767 CrossRef CAS PubMed.
- X. Qi, Z. Li, J. Zhang, H. Li, G. Zhang, M. Li, B. Li, Y. Fu, M. Cai, H. Wang, T. Tong and J. Gao, iScience, 2023, 26, 106360 CrossRef CAS PubMed.
- H. Liu, Z. Ye, Y. Deng, J. Yuan, L. Wei and L. Xiao, Trends Anal. Chem., 2023, 169, 117359 CrossRef CAS.
- W.-L. Gong, J. Yan, L.-X. Zhao, C. Li, Z.-L. Huang, B. Z. Tang and M.-Q. Zhu, Photochem. Photobiol. Sci., 2016, 15, 1433–1441 CrossRef CAS.
- W. Chi, D. Tan, Q. Qiao, Z. Xu and X. Liu, Angew. Chem., Int. Ed., 2023, 62, e202306061 CrossRef CAS.
- S. Zong, X. Jiang, Z. Wang, C. Chen, J. Lu, L. Wang, D. Zhu and Y. Cui, Nanoscale, 2016, 8, 19110–19119 RSC.
- P. Werther, K. Yserentant, F. Braun, N. Kaltwasser, C. Popp, M. Baalmann, D.-P. Herten and R. Wombacher, Angew. Chem., Int. Ed., 2020, 59, 804–810 CrossRef CAS PubMed.
- J.-Z. Lai, C.-Y. Lin, S.-J. Chen, Y.-M. Cheng, M. Abe, T.-C. Lin and F.-C. Chien, Angew. Chem., Int. Ed., 2024, 63, e202404942 CrossRef CAS.
- K. Uno, H. Niikura, M. Morimoto, Y. Ishibashi, H. Miyasaka and M. Irie, J. Am. Chem. Soc., 2011, 133, 13558–13564 CrossRef CAS PubMed.
- Y. Arai, S. Ito, H. Fujita, Y. Yoneda, T. Kaji, S. Takei, R. Kashihara, M. Morimoto, M. Irie and H. Miyasaka, Chem. Commun., 2017, 53, 4066–4069 RSC.
- J. A. Erstling, J. A. Hinckley, N. Bag, J. Hersh, G. B. Feuer, R. Lee, H. F. Malarkey, F. Yu, K. Ma, B. A. Baird and U. B. Wiesner, Adv. Mater., 2021, 33, 2006829 CrossRef CAS.
- P. Hoyer, T. Staudt, J. Engelhardt and S. W. Hell, Nano Lett., 2011, 11, 245–250 CrossRef CAS PubMed.
- M. Ikegami and T. Arai, J. Chem. Soc., Perkin Trans. 2, 2002, 1296–1301 RSC.
- P. Majumdar and J. Zhao, J. Phys. Chem. B, 2014, 119, 2384–2394 CrossRef PubMed.
- M. Li and W.-H. Zhu, Acc. Chem. Res., 2022, 55, 3136–3149 CrossRef CAS PubMed.
- T. Mei, D. Xiang and D. Mayer, Natl. Sci. Rev., 2017, 4, 666–667 CrossRef CAS.
- G. Naren, W. Larsson, C. Benitez-Martin, S. Li, E. Perez-Inestrosa, B. Albinsson and J. Andreasson, Chem. Sci., 2021, 12, 7073–7078 RSC.
- Z. Xu, Q. T. Liu, X. Wang, Q. Liu, D. Hean, K. C. Chou and M. O. Wolf, Chem. Sci., 2020, 11, 2729–2734 RSC.
- C. Li, K. Xiong, Y. Chen, C. Fan, Y. L. Wang, H. Ye and M. Q. Zhu, ACS Appl. Mater. Interfaces, 2020, 12, 27651–27662 CrossRef CAS.
- J. X. Liu, B. Xin, C. Li, N. H. Xie, W. L. Gong, Z. L. Huang and M. Q. Zhu, ACS Appl. Mater. Interfaces, 2017, 9, 10338–10343 CrossRef CAS PubMed.
- Z. Qiang, K. M. Shebek, M. Irie and M. Wang, ACS Macro Lett., 2018, 7, 1432–1437 CrossRef CAS PubMed.
- D. Kim, K. Jeong, J. E. Kwon, H. Park, S. Lee, S. Kim and S. Y. Park, Nat. Commun., 2019, 10, 3089–3099 CrossRef PubMed.
- O. Nevskyi, D. Sysoiev, A. Oppermann, T. Huhn and D. Wöll, Angew. Chem., Int. Ed., 2016, 55, 12698–12702 CrossRef CAS PubMed.
- B. Roubinet, M. Weber, H. Shojaei, M. Bates, M. L. Bossi, V. N. Belov, M. Irie and S. W. Hell, J. Am. Chem. Soc., 2017, 139, 6611–6620 CrossRef CAS PubMed.
- C. T. Cao, L. Li, C. Cao and J. Liu, J. Phys. Org. Chem., 2020, 34, 4164–4173 CrossRef.
- H. Xi, Z. Zhang, W. Zhang, M. Li, C. Lian, Q. Luo, H. Tian and W. H. Zhu, J. Am. Chem. Soc., 2019, 141, 18467–18474 CrossRef CAS PubMed.
- M. Ziółek, J. Kubicki, A. Maciejewski, R. Naskrȩcki and A. Grabowska, J. Chem. Phys., 2006, 124, 124518–124528 CrossRef PubMed.
- D. Bléger and S. Hecht, Angew. Chem., Int. Ed., 2015, 54, 11338–11349 CrossRef.
- Z. Zhang, W. Wang, M. O'Hagan, J. Dai, J. Zhang and H. Tian, Angew. Chem., Int. Ed., 2022, 61, 05758–05782 Search PubMed.
- Z. Zhang, W. Wang, P. Jin, J. Xue, L. Sun, J. Huang, J. Zhang and H. Tian, Nat. Commun., 2019, 10, 4232 CrossRef.
- S. Tang, F. Song, M. Lu, K. Han and X. Peng, Sci. China: Chem., 2019, 62, 451–459 CrossRef CAS.
- D. G. Patel, T. B. Mitchell, S. D. Myers, D. A. Carter and F. A. Novak, J. Org. Chem., 2020, 85, 2646–2653 CrossRef CAS PubMed.
- Z. Li, Y. Dai, Z. Lu, Y. Pei, H. Chen, L. Zhang, Y. Duan and H. Guo, Chem. Commun., 2019, 55, 13430–13433 RSC.
- L. Wang, M. S. Frei, A. Salim and K. Johnsson, J. Am. Chem. Soc., 2019, 141, 2770–2781 CrossRef CAS PubMed.
- Z. Liu, J. Liu, X. Wang, F. Mi, D. Wang and C. Wu, Bioconjugate Chem., 2020, 31, 1857–1872 CrossRef CAS PubMed.
- S. Emmert, G. Quargnali, S. Thallmair and P. Rivera-Fuentes, Nat. Chem. Biol., 2023, 15, 1415–1421 CrossRef CAS PubMed.
- X. Jiang, J. Chen, A. Bajić, C. Zhang, X. Song, S. L. Carroll, Z.-L. Cai, M. Tang, M. Xue, N. Cheng, C. P. Schaaf, F. Li, K. R. MacKenzie, A. C. M. Ferreon, F. Xia, M. C. Wang, M. Maletić-Savatić and J. Wang, Nat. Commun., 2017, 8, 16087 CrossRef CAS PubMed.
- Q. Yu, L. Zhang, M. Jiang, L. Xiao, Y. Xiang, R. Wang, Z. Liu, R. Zhou, M. Yang, C. Li, M. Liu, X. Zhou and S. Chen, Angew. Chem., Int. Ed., 2023, 62, 13137–13146 Search PubMed.
- M. Mwafulirwa, K. Abdalla, W. Bian, H. Wei, L. Xu, W. Yu, Z. Hui, Q. Yang and X. Sun, Smart Mol., 2024, 2, e20240044 CrossRef CAS PubMed.
- K. Gu, W. Qiu, Z. Guo, C. Yan, S. Zhu, D. Yao, P. Shi, H. Tian and W.-H. Zhu, Chem. Sci., 2019, 10, 398–405 RSC.
- X. Chai, H.-H. Han, A. C. Sedgwick, N. Li, Y. Zang, T. D. James, J. Zhang, X.-L. Hu, Y. Yu, Y. Li, Y. Wang, J. Li, X.-P. He and H. Tian, J. Am. Chem. Soc., 2020, 142, 18005–18013 CrossRef CAS.
- A. Morozumi, M. Kamiya, S.-n. Uno, K. Umezawa, R. Kojima, T. Yoshihara, S. Tobita and Y. Urano, J. Am. Chem. Soc., 2020, 142, 9625–9633 CAS.
- E. A. Halabi, Z. Thiel, N. Trapp, D. Pinotsi and P. Rivera-Fuentes, J. Am. Chem. Soc., 2017, 139, 13200–13207 CrossRef CAS PubMed.
- W. Zhao, X. Huang, J. Yang, L. Qu, G. Qiu, Y. Zhao, X. Wang, D. Su, X. Ding, H. Mao, Y. Jiu, Y. Hu, J. Tan, S. Zhao, L. Pan, L. Chen and H. Li, Light: Sci. Appl., 2023, 12, 298 CrossRef CAS PubMed.
- R. P. J. Nieuwenhuizen, K. A. Lidke, M. Bates, D. L. Puig, D. Grünwald, S. Stallinga and B. Rieger, Nat. Methods, 2013, 10, 557–562 CrossRef CAS PubMed.
- W. Wang, W. Yang, Z. Zhang, J. Dai, Y. Xu and J. Zhang, Chem. Sci., 2024, 15, 5539–5547 RSC.
- Y. Wang, K. Huang, T. Wang, L. Liu, F. Yu, W. Sun, W. Yao, H. Xiong, X. Liu, H. Jiang and X. Wang, Small, 2024, 20, 2310300 CrossRef CAS PubMed.
- L. Schermelleh, A. Ferrand, T. Huser, C. Eggeling, M. Sauer, O. Biehlmaier and G. P. C. Drummen, Nat. Cell Biol., 2019, 21, 72–84 CrossRef CAS PubMed.
- K. Kikuchi, L. D. Adair, J. Lin, E. J. New and A. Kaur, Angew. Chem., Int. Ed., 2022, 62, 04745–04770 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5sc03224e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:
