Using micro pillar array columns (μPAC) for the analysis of permethylated glycans†
Byeong Gwan
Cho
 ,
Peilin
Jiang
,
Mona
Goli
,
Sakshi
Gautam
and
Yehia
Mechref
,
Peilin
Jiang
,
Mona
Goli
,
Sakshi
Gautam
and
Yehia
Mechref
 *
*
Department of Chemistry and Biochemistry, Texas Tech University, Lubbock, TX 79409-1061, USA. E-mail: yehia.mechref@ttu.edu; Fax: +1 806-742-1289; Tel: +1 806-742-3059
First published on 9th June 2021
Abstract
Glycosylation is a complex and common post-translational modification of proteins. To study glycosylation, liquid chromatography-mass spectrometry (LC-MS) is often used to profile and structurally characterize the glycans in biological systems. While bed packed reverse phase columns are frequently utilized for the separation of permethylated glycans, the use of newly commercialized micro array pillar nanoLC columns (μPAC) have not been demonstrated previously. Owing to its advantages such as low back pressure, reproducibility, and durability, we have investigated the viability of the μPAC for the analysis of permethylated glycans. In this work, we demonstrate the online purification ability of μPAC trapping column compared against PepMap trapping column. We also found that the 50 cm μPAC can be used for the analysis of both permethylated N- and O-glycans. The use of 50 cm μPAC was compared against the previous method. The use of 200 cm μPAC was also investigated for the permethylated glycan analysis. 200 cm μPAC demonstrated efficient separation of oligomannose glycan isomers as well as other complex glycans.
Introduction
Glycans have been investigated extensively due to their roles in various biological processes, including cell signaling,1,2 cell–cell interaction,3,4 immune response,4,5 and protein stability.6,7 Roles in numerous vital biological processes make glycosylation a crucial post-translational modification.8,9 Various branched and isomeric linkage forms introduce the complexity and structural heterogeneity in glycan molecules,10,11 which renders glycomics a challenging avenue.8,12 Alterations in the expression of glycans and their isomers have been observed in various diseases including Alzheimer's disease,13–15 cardiovascular diseases,16,17 immune disorders,18,19 and cancers.18,20,21 Thus, glycans and their isomers are considered important candidates for disease biomarker studies.22–24 Most recently, glycans in coronavirus spike proteins have been extensively investigated both for their role in facilitating viral membrane protein interaction with the host cell membrane, as well as to develop targeted therapies against the virus.25,26For decades, mass spectrometry (MS) has been predominantly employed for profound glycan structural studies.8 Liquid chromatography in conjunction with MS (LC-MS) is the most popular technique. It combines efficient glycan separation obtained from LC with the highly sensitive and abundant structural information from MS.27–29 Several LC separation techniques have been used for glycans, such as hydrophilic interaction liquid chromatography (HILIC),30,31 porous graphitized carbon (PGC) chromatography,27–29 and reverse-phase liquid chromatography (RPLC).15,32 The choice of LC technique typically depends on the type of chemical derivatization used for the glycans.
Several derivatization strategies have been used to chemically modify the glycans to improve sensitivity and enhance stability.33 Permethylation is one such derivatization method, where hydrogens on the hydroxyl groups within the glycan molecule are converted to methyl groups using iodomethane. This chemical modification renders stability to glycan molecules by preventing sialic acid loss and fucose rearrangement, especially with MS analysis. Additionally, it enhances the ionization capabilities of glycans in the gas phase, thus improving sensitivity.28 Permethylation also enhances the hydrophobicity of the molecules, thereby making them ideal for RPLC-based separation.33
Packed C18 columns are the most widely used column for RPLC-based separation of glycans. These columns comprise octadecyl chains (C18) bonded to silica as stationary phase.27 Although these columns have been commonly used for the separation of permethylated as well as reducing end labeled glycans, they have demonstrated limited glycan isomeric separation efficiencies27 which prompted the use of PGC columns to facilitate efficient isomeric separation of permethylated glycans.34,35
Besides bed packed C18 and PGC columns, the recent commercialization of a microchip-based chromatography column has demonstrated its use in proteomics. Its reproducibility and chromatographic performance have been well-documented by Muller et al.36 and Stadlmann et al.37 The absence of a packed bed configuration in the column provides uniformity which in turn eliminates peak dispersion resulting from Eddy diffusion,38,39 while maintaining uniformly placed micro-pillars coated with C18 to facilitate RPLC. This micropillar array column (μPAC, Pharmafluidics) also benefits from its bi-directional flow and its resistance to clogging due to its lack of particles or frits. Because of this, μPAC nanoLC columns can operate at a much higher flowrate enabled by the low back pressure of the column, which can be useful to perform high-throughput analyses.
While it has been established for use in bottom-up proteomics experiments, its utilization has not been shown in glycan analysis. In this work, the use of both 50 and 200 cm μPAC nanoLC columns for the analysis of both permethylated N- and O-glycan analysis is demonstrated. Also, the performance of the μPAC is compared to our previously established permethylated glycan LC-MS methodology.29 The ability to use a 200 cm μPAC nanoLC column to attain an efficient isomeric glycan separation is also shown. Our results show that μPAC nanoLC columns can be utilized in glycan analysis and offer an alternative to traditional C18 bed packed columns.
Experimental
Materials
Two model glycoproteins, bovine ribonuclease B (RNase B) and bovine fetuin as well as pooled human blood serum (HBS) were purchased from Sigma Aldrich (St Louis, MO). Other chemicals acquired from Sigma Aldrich included ammonium bicarbonate (ABC), sodium hydroxide beads (20–40 mesh, 97%), dimethyl sulfoxide (DMSO, 99.9%), iodomethane, trifluoracetic acid, and borane-ammonia complex (97% purity). Glycan standards were purchased from Chemily Glycoscience (Atlanta, GA). HPLC-grade solvents including acetonitrile (MeCN), methanol, formic acid, and HPLC water were obtained from Fisher Scientific (Pittsburgh, PA). Dextran MW 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 was also purchased from Fisher Scientific (Pittsburgh, PA). Intact monoclonal antibody mass check standard (mouse IgG1 protein) was acquired from Waters (Milford, MA). PNGase F (glycerol-free, 500
000 was also purchased from Fisher Scientific (Pittsburgh, PA). Intact monoclonal antibody mass check standard (mouse IgG1 protein) was acquired from Waters (Milford, MA). PNGase F (glycerol-free, 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units ml−1) was purchased from New England Biolabs (Ipswich, MA). Empty micro spin columns and activated charcoal micro spin columns were obtained from Harvard Apparatus (Cambridge, MA).
000 units ml−1) was purchased from New England Biolabs (Ipswich, MA). Empty micro spin columns and activated charcoal micro spin columns were obtained from Harvard Apparatus (Cambridge, MA).
N-Glycan release
N-Glycans were released from model glycoproteins and HBS as previously reported.40 Briefly, 20 μg of fetuin, RNase B, and IgG1 were dissolved in 50 mM ABC buffer and denatured at 90 °C water bath for 30 min. For HBS, 10 μL of HBS was diluted to 100 μL with 50 mM ABC buffer then incubated at 90 °C water bath for 30 min. After cooling down the sample to room temperature, 1 μL of PNGase F stock solution (500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units mL−1) was added to each sample and incubated at 37 °C for 18 hours. After PNGase F digestion, de-N-glycosylated proteins were removed by adding 900 μL of ice-cold ethanol and keeping it at −20 °C for 1 hour for precipitation. By centrifugation, the supernatant was collected and then dried.
000 units mL−1) was added to each sample and incubated at 37 °C for 18 hours. After PNGase F digestion, de-N-glycosylated proteins were removed by adding 900 μL of ice-cold ethanol and keeping it at −20 °C for 1 hour for precipitation. By centrifugation, the supernatant was collected and then dried.
Reduction and permethylation
Dried released N-glycans from 20 μg glycoproteins were reduced by adding 10 μL of 10 μg μL−1 borane-ammonia complex and incubated at 60 °C for 1 hour. A washing step with methanol was repeated 3–4 times to remove excess borane. A solid-phase permethylation method was applied to reduced N-glycans as previously reported.41 Briefly, reduced glycan samples were resuspended in 30 μL DMSO and 1.2 μL water. DMSO-soaked sodium hydroxide beads were packed in empty micro spin columns and washed with 200 μL DMSO by centrifuging the column at 1800 rpm for 2 min. After adding 20 μL iodomethane to the sample solution, samples were loaded to the prepared sodium hydroxide columns and incubated at room temperature for 25 min. Then, another 20 μL iodomethane was added to those samples and incubated for an additional 15 min. After this, permethylated glycans were eluted by centrifuging the columns at 1800 rpm for 2 min. Another 30 μL ACN was added and centrifuged to elute permethylated glycans off the column. The final elution was dried overnight in a speed vacuum and resuspended in 20% ACN and 0.1% formic acid, ready for LC-MS analysis.Dextran ladder preparation
The dextran ladder was prepared by first performing partial acid hydrolysis of dextran using trifluoroacetic acid (TFA) as described by Ashwood et al.42 Partially hydrolyzed dextran was then extracted using activated charcoal micro spin columns as previously described by Benktander et al.43 Extracted partially hydrolyzed dextran was then reduced and permethylated as described above.LC-MS analysis
The separation was achieved with an Ultimate 3000 nanoLC system (Thermo Scientific, Sunnyvale, CA). A bed packed column, a C18 Acclaim PepMap column (75 μm i.d. × 15 cm, 2 μm particle sizes, 100 Å pore sizes, Thermo Scientific, San Jose, CA), was used in conjunction with an online trapping column, a C18 Acclaim PepMap 100 trapping column (75 μm i.d. × 2 cm, 3 μm particle sizes, 100 Å pore sizes, Thermo Scientific, San Jose, CA). Loading was performed with 0.1% formic acid in 98% water and 2% acetonitrile at a flowrate of 3 μL min−1. The mobile phase A was 98% water, 2% acetonitrile, and 0.1% formic acid and B was 100% acetonitrile and 0.1% formic acid. Separation was achieved with a nano pump flowrate of 0.35 μL min−1 with a gradient elution of 20% mobile phase B over 10 min, then increased to 42% B (10–11 min), 42% to 55% B (11–48 min), 55% to 90% B (48–49 min), 90% B (49–54 min), 90% to 20% B (54–55 min), then 20% B (55–60 min) as previously described.40For μPAC, a C18 trapping column (5 μm pillar diameter, 2.5 μm inter pillar distance, 18 μm pillar height,10 mm bed length, 100–200 Å pore sizes, PharmaFluidics) was used for online purification with the same loading solution as the bed packed trapping column a with flowrate of either 3 or 10 μL min−1. For 50 cm μPAC (5 μm pillar diameter, 2.5 μm inter pillar distance, 18 μm pillar height, 50 cm bed length, 100–200 Å pore sizes, PharmaFluidics), three different gradients were used. One used the gradient from the bed packed column described previously, with 0.1% formic acid in 100% acetonitrile as a mobile phase B. The other two gradients, with 0.1% formic acid in 80% acetonitrile and 20% water for N-glycan analysis, began with 40% B for the first 4 min, then increased to 70% B (4–64 min), 70% to 97.5% B (64–69 min), 97.5% B (69–89 min), 97.5% to 40% B (89–94 min) then, 40% B (94–114 min). For O-glycan analysis, the gradient began with 25% B for the first 9 min, then increased to 70% B (9–99 min), 70% to 97.5% B (99–104 min), 97.5% B (104–114 min), 97.5% to 40% B (114–119 min), then 25% B (119–129 min).
For the 200 cm μPAC (5 μm pillar diameter, 2.5 μm inter pillar distance, 18 μm pillar height, 200 cm bed length, 100–200 Å pore sizes, PharmaFluidics), two different gradients were used based on the mobile phase B; 0.1% formic acid in 100% acetonitrile and 0.1% formic acid in 80% acetonitrile. For 0.1% formic acid in 100% acetonitrile, the gradient started at 20% B for the first 4 min. Then it was increased to 42% in 5 min, to 55% in 130 min, and to 95% in 5 min. It was kept at 95% for 25 min then decreased to 20% in 5 min and kept constant for 16 min. For 0.1% formic acid in 80% acetonitrile, the gradient started at 40% B for the first 4 min. Then it was increased to 64% in 240 min and to 97.5% in 5 min. It was kept at 97.5% for 34 min then decreased to 40% in 5 min and kept constant for 34 min.
For the analysis of permethylated glycans from model glycoproteins, an Exactive (Thermo Scientific, Sunnyvale CA) mass spectrometer was used. It was operated in positive mode and set to a resolution of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and a scan range of 700–2000 m/z. ESI voltage was 1.6 kV, and the capillary temperature was 300 °C. For permethylated glycans from human blood serum, an LTQ Orbitrap Velos instrument was used with spray voltage of 1.6 kV. Mass resolution was set at 60
000 and a scan range of 700–2000 m/z. ESI voltage was 1.6 kV, and the capillary temperature was 300 °C. For permethylated glycans from human blood serum, an LTQ Orbitrap Velos instrument was used with spray voltage of 1.6 kV. Mass resolution was set at 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 with data dependent acquisition (DDA) for MS2 scans where the top 8 precursor ions were subjected for collision induced dissociation (CID). A normalized collision energy of 35% with 3 m/z isolation window was selected. The experimental scheme is summarized in Fig. S1†
000 with data dependent acquisition (DDA) for MS2 scans where the top 8 precursor ions were subjected for collision induced dissociation (CID). A normalized collision energy of 35% with 3 m/z isolation window was selected. The experimental scheme is summarized in Fig. S1†
Data processing
Chromatograms and mass spectra were made with Xcalibur 4.3 software. The same software was used to analyze LC-MS/MS data. A mass tolerance of 10 ppm was used for EICs. Glycoworkbench was utilized for depiction of glycan structures. Tables were made with Microsoft Excel. Bar graphs and X–Y plots were made with GraphPad Prism 9.Results and discussion
Evaluation of μPAC trapping column for permethylated glycans
The use of trapping columns for permethylated glycan online enrichment on the liquid chromatography unit was previously performed by Desantos-Garcia et al.,44 where online purification was demonstrated to be more sensitive than either liquid–liquid extraction or offline C18 solid phase extraction protocols. Trapping capabilities of permethylated glycans by μPAC trapping columns were assessed similarly to evaluate the optimal trapping conditions. Unlike the bed packed trapping columns, μPAC offers a unique bi-directional flow mode as well as the ability to enrich at a much higher flowrate up to 20 μL min−1 which allows faster enrichment, reducing the analysis time. Moreover, the bi-directional flow mode permits loading the analytes in one direction but eluting them in the other direction, unlike the bed packed columns where the analytes are loaded in the same direction as the elution. This can be advantageous because eluting the analytes in the same direction as the loading may prompt the trap columns to affect the separation. Eluting the analytes in the opposite direction to the loading eliminates this possibility, since the different loading directions minimize the time, the analytes spend on the trapping column with organic solvents. Due to these functionalities, different flow rates and enrichment times were compared against the bed packed C18 trapping column using permethylated N-glycans derived from human blood serum with a bed packed C18 separation column. As shown in Fig. S2a,† a μPAC trapping column with reverse elution at 4 minutes, 10 μL min−1 loading flow rate, demonstrated comparable or higher abundances compared to the benchmark method (bed packed trapping column) among the ten various glycan structures while, other methods showed diminishing abundances except for the case of Man9. Reverse elution at 4 min with a 10 μL min−1 loading flow rate was chosen for the evaluation from among other methods since it was the optimized method provided by Pharmafluidics for proteomics experiments. Injection to injection variability were also measured by plotting % relative standard deviation (%RSD) against the same methods with the same glycans shown in Fig. S2b.† Reverse elution at 4 minutes, 10 μL min−1 loading flow rate and forward elution at 10 minutes, 10 μL min−1 loading flow rate showed less than 15% %RSD for all ten glycans, while other methods including the benchmark method showed higher than 15% %RSD for at least one glycan composition (Fig. S2b†). Because reverse elution at 4 minutes, 10 μL min−1 loading flow rate showed higher intensity with consistency across the ten glycans, this method was chosen for other experiments.Dextran ladder
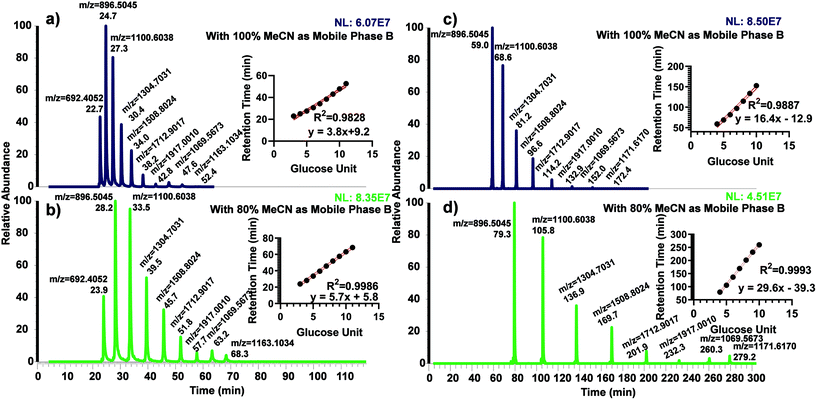
The use of dextran ladder was first demonstrated by Guile et al.45 to normalize the retention time by the elution order of glucose units (GU). We recently investigated the use of GU to predict and normalize the retention time of permethylated glycans using permethylated dextrin on both C18 and porous graphitized carbon columns.40 Permethylated partially hydrolyzed dextran was separated using both 50 cm and 200 cm μPAC columns to examine the possibility of using the glucose unit index (GUI) (Fig. 1). For both columns, two different organic mobile phases were explored; 100% acetonitrile and 80% acetonitrile, both with 0.1% formic acid as an acid modifier. The former was used to directly compare against the benchmark method (Bed packed C18, 15 cm) while the latter was investigated since it was utilized in several proteomics experiments.36,46,47 Interestingly, using the 50 cm μPAC with 100% acetonitrile (Fig. 1a) demonstrated less linearity than with 80% acetonitrile (Fig. 1b) as shown by R2 (0.9986 vs. 0.9828). The 200 cm μPAC also indicated a similar pattern, as the use of 80% acetonitrile as mobile phase B (Fig. 1d) resulted in more linearity than 100% acetonitrile (Fig. 1c), R2 (0.9993 vs. 0.9887). It is important to note that GU 11 for 200 cm column was excluded in the retention time vs. GU plot due to the elution during the washing step of the programmed gradient. This result is contrary to the recent work by Gautam et al.40 where permethylated GU plots from a C18 bed packed column were non-linear and therefore 3rd order polynomial fitting was used instead of linear. However, another recent work by Kurz et al.48 also demonstrated a linear relationship between retention time and GU using 80% MeCN as the organic mobile phase. A linear plot of GU allows the prediction of glycan elution by a simple linear equation rather than polynomial equations.Permethylated N-glycan analysis using 50 cm μPAC
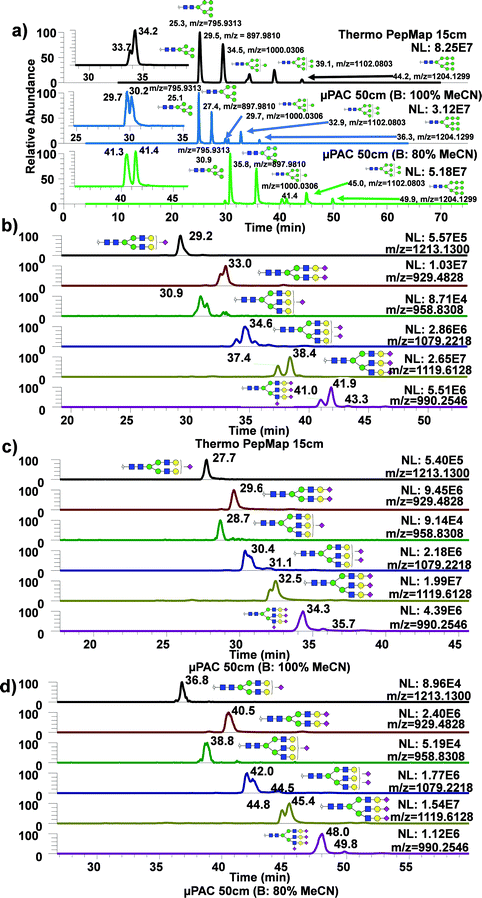
To assess the use of 50 cm μPAC for analysis of permethylated N-glycans, glycans derived from standard glycoproteins were analyzed with the same gradient as for the dextran ladder, using either 100% or 80% MeCN with 0.1% formic acid as an acid modifier. These two approaches were then directly compared with a routine glycomics method from our group using a C18 bed packed column (75 μm, 2 μm, 15 cm).49Fig. 2a demonstrated extracted ion chromatograms (EICs) of permethylated oligomannose glycans derived from ribonuclease B using three different techniques, with a table describing the performance of each technique using three glycans: Man5, 6, and 9. Man7 and Man8 were excluded from this list due to the observation of partial isomeric separation, as demonstrated by the insets showing Man7 partial isomeric separation from each technique. As shown in the Table S1,† the use of 50 cm μPAC column generally exhibited higher theoretical plate (N) value for high mannose type glycans and sialylated glycans. The use of 80% MeCN as mobile phase B with 50 cm μPAC also showed some improvement over the use of 100% MeCN as mobile phase B in terms of theoretical plate numbers. The same experiment was conducted using acidic glycans derived from bovine fetuin as shown in Fig. 2b–d. Similarly, several glycans with three antennas and multiple sialic acids showed partial isomeric separations, though this was observed more frequently with bed packed column (Fig. 2b) compared to the 50 cm μPAC (Fig. 2c and d). This experiment demonstrated the viability of using 50 cm μPAC as a separation column for the analysis of permethylated glycans, even though not much isomeric separation was observed. Also, 50 cm μPAC has generally outperformed 15 cm PepMap column significantly in terms of theoretical plate numbers although there were some cases where PepMap column performed better than the μPAC. In terms of FWHM, the results shown in this work largely agree with the previous work on peptides by Stadlmann et al.37 which demonstrated general improvement in FWHM from μPAC compared to the PepMap column.An advantage of using 50 cm μPAC is the ability to adjust column flowrate, which can be used up to 2 μL min−1, whereas the flowrate of the bed packed column used in this work is limited by the backpressure. The use of a higher flowrate decreases the LC run time, facilitating higher throughput analysis. Fig. 3a are the EICs of permethylated N-glycans derived from human blood serum with 50 cm μPAC at 0.3 μL min−1 while Fig. 3b are the EICs of the same glycans with the same column at 0.6 μL min−1. As expected, there was a noticeable shift in retention time due to the higher flowrate. Although operating at a higher flowrate also showed a visible gain in peak sharpness, there was no improvement in isomeric separation. While there are advantages of performing separation at a higher flowrate, the disadvantage was the loss of sensitivity as shown in Fig. S3† where the substantial loss of signal was observed for complex glycans. Interestingly, however, Man5 and 6 showed a slight gain in sensitivity.
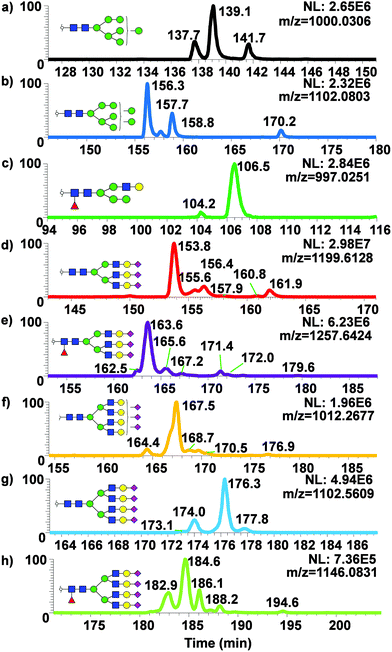
Isomeric separation of permethylated N-glycans using 200 cm μPAC
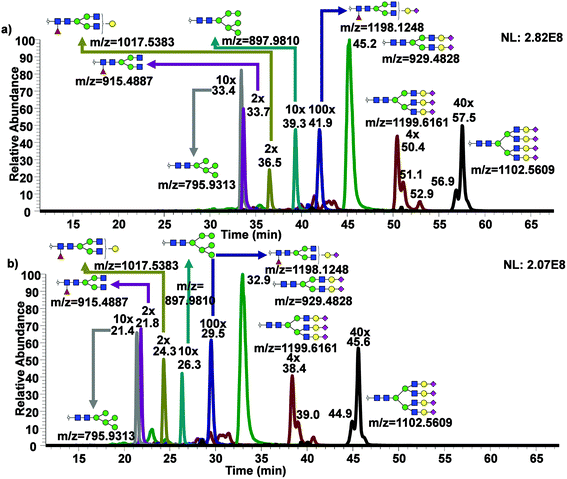
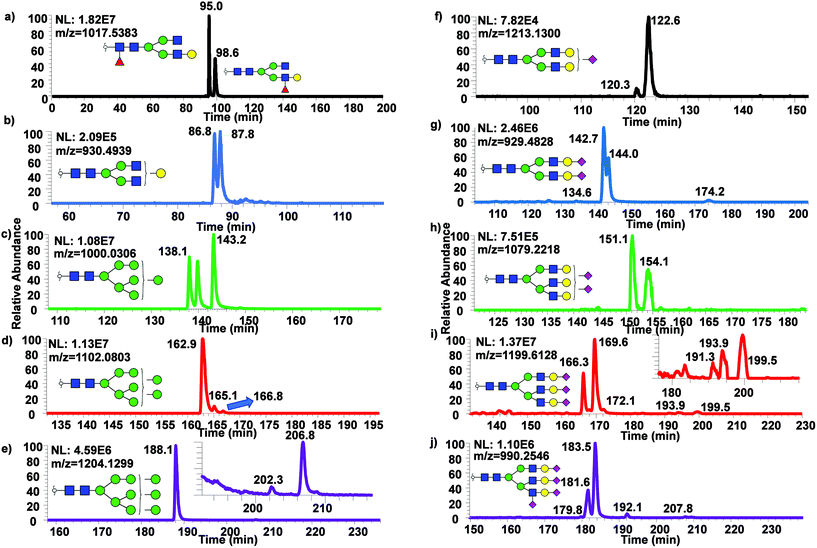
While only partial isomeric separation was observed with 50 cm μPAC, an attempt to achieve better isomeric separation using 200 cm μPAC was performed. Similar experiments were performed as previously described for the 50 cm μPAC, where permethylated N-glycans derived from model glycoproteins as well as glycan standards were analyzed. EICs of permethylated N-glycans were shown in Fig. 4a (GlcNAc4Man3Gal1deoxyHex1 isomer standards), Fig. 4b (Mouse Immunoglobin G), Fig. 4c–e (Ribonuclease B), and Fig. 4f–j (Bovine Fetuin) using 200 cm μPAC. There was a baseline resolved separation of GlcNAc4Man3Gal1deoxyHex1 fucose positional isomers (Fig. 4a), with branched fucosylated isomer eluting later than core fucosylated isomer. Isomeric separation of non-sialylated complex glycan (biantennary with terminal galactose structure) derived from mouse immunoglobin G was also separated (Fig. 4b). However, Glc4Man3Gal1Fuc1 (Fig. S4d†) only displayed shouldering between the two isomers of terminal galactose linked to different branches. Isomeric separation of oligomannose glycans was distinct, especially in the cases of Man7, 8 and 9 shown in Fig. 4c–e. Extensive isomeric separation of permethylated N-glycans were observed for complex sialylated triantennary N-glycans derived from bovine fetuin (Fig. 4f–j). However, only partial separation of biantennary disialylated glycans was achieved (Fig. 4g) using this method although previous PGC-based separation of permethylated glycans have demonstrated baseline resolution separation of biantennary disialylated glycan isomers.34 This result suggested that more work is needed to achieve baseline separation of permethylated glycan isomers using 200 cm μPAC, since the previous work by Zhou et al.34 has demonstrated baseline separation of biantennary disialylated glycan isomers using PGC at high temperature.To examine the broad spectrum of glycan isomers, permethylated glycans derived from human blood serum were analyzed using 200 cm μPAC. Isomeric separation of glycans were observed especially in the cases of oligomannose structures such as Man7 (Fig. 5a) and 8 (Fig. 5b) at 0.3 μL min−1. Interestingly, 4 peaks of permethylated Man 8 were displayed. Although MS2 spectra for these peaks were examined to determine the structures (Fig. S5†), and to investigate whether Man8 isomers previously determined by Prien et al.50 would match the isomers found in this study, no diagnostic ions were found in the spectra. Even though the existence of these Man8 isomers can be confirmed by both MS1 and MS2 spectra, precise structural determination of these isomers was not achieved. Unlike the EICs shown with a 50 cm μPAC, the 200 cm μPAC was able to show extensive isomeric separation of complex N-glycans with multiple terminal sialic acids as depicted in Fig. 5d–h. This suggests that 200 cm μPAC is capable of separating permethylated glycan isomers, although improvements to peak resolution are needed as well as the determination of complex glycan isomers including sialic acid linkage isomers.
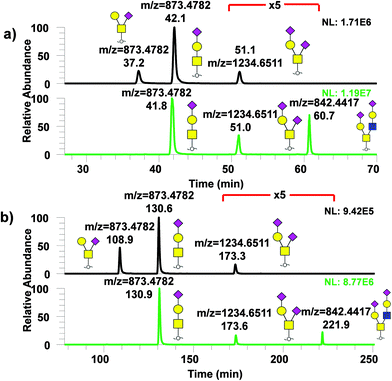
Analysis of permethylated O-glycans using μPAC columns
Similarly, analysis of permethylated O-glycans derived from model glycoproteins such as bovine fetuin and κ-casein with μPAC columns was performed as shown in Fig. 6 with (a) showing the chromatogram for permethylated O-glycans from κ-casein (top) and from fetuin (bottom) using 50 cm μPAC and (b) using 200 cm μPAC. In both cases, two isomers, GalNAc1Gal1NeuAc1 and GalNAc1(Gal1)NeuAc1, were baseline resolved while 200 cm μPAC provided substantially more separation than the 50 cm μPAC, as expected. However, it must be noted that even though 200 cm μPAC demonstrated much more separation than 50 cm, the run time was substantially higher which reduced the throughput. The first O-glycan eluted, GalNAc1(Gal1)NeuAc1 from 200 cm μPAC, took more than 100 minutes to elute. This was due to the column's void volume (10 μL) which caused a significant amount of time for the analyte to pass through with a flowrate of 0.3 μL min−1. This perhaps could be addressed by increasing the flowrate in the beginning of the run, then lowering to the intended flowrate as previously demonstrated by Muller et al.36 with proteomics experiments.Conclusion
In this work, we demonstrated the use of μPAC to perform glycomics analysis for the first time. Since μPAC is a RPLC column, glycans were permethylated in order to facilitate the separation of both N- and O-glycans. The use of μPAC trapping column was accessed to determine the optimal condition for online purification of the permethylated glycans. Using both 50 and 200 cm μPAC, dextran ladders were established with linear correlation which was contrary to the previous work by Gautam et al.40 Then, using the established mobile phases and the gradients, N- and O-glycans were derived from various model glycoproteins as well as human blood serum as a biological matrix. Our results showed that 50 cm μPAC was able to perform glycomics analysis through permethylation. The results from 50 cm μPAC were compared against the conventional bed packed column and found that 50 cm μPAC outperformed bed packed column in terms of column performance. We also tested the analysis at a higher flowrate to examine higher throughput. However, a decrease in signal intensity was observed which was an expected result. Although 50 cm μPAC was not able to separate isomers, 200 cm μPAC was able to resolve some permethylated glycan isomers. However, both columns have a large internal volume relative to the optimal flow rate. To perform high-throughput analysis, increasing the flow rate at the beginning of the run to reduce void time as well as the washing step would be a viable solution.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by grants from the National Institutes of Health, NIH (1R01GM112490, 1R01GM130091).References
- P. Stanley and T. Okajima, in Curr Top Dev Biol, Elsevier, 2010, vol. 92, pp. 131–164 Search PubMed.
- J. H. Dewald, F. Colomb, M. Bobowski-Gerard, S. Groux-Degroote and P. Delannoy, Cells, 2016, 5, 43 CrossRef PubMed.
- K. Ohtsubo and J. D. Marth, Cell, 2006, 126, 855–867 CrossRef CAS PubMed.
- P. M. Rudd, M. R. Wormald, R. L. Stanfield, M. Huang, N. Mattsson, J. A. Speir, J. A. DiGennaro, J. S. Fetrow, R. A. Dwek and I. A. Wilson, J. Mol. Biol., 1999, 293, 351–366 CrossRef CAS PubMed.
- M. Sperandio, C. A. Gleissner and K. Ley, Immunol. Rev., 2009, 230, 97–113 CrossRef CAS PubMed.
- H. S. Lee, Y. Qi and W. Im, Sci. Rep., 2015, 5, 8926 CrossRef PubMed.
- D. Shental-Bechor and Y. Levy, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 8256–8261 CrossRef CAS PubMed.
- X. Dong, Y. Huang, B. G. Cho, J. Zhong, S. Gautam, W. Peng, S. D. Williamson, A. Banazadeh, K. Y. Torres-Ulloa and Y. Mechref, Electrophoresis, 2018, 39, 3063–3081 CrossRef CAS PubMed.
- C.-H. Wong, J. Org. Chem., 2005, 70, 4219–4225 CrossRef CAS.
- F. Higel, A. Seidl, F. Sörgel and W. Friess, Eur. J. Pharm. Biopharm., 2016, 100, 94–100 CrossRef CAS PubMed.
- H. J. An, J. W. Froehlich and C. B. Lebrilla, Curr. Opin. Chem. Biol., 2009, 13, 421–426 CrossRef CAS PubMed.
- R. D. Cummings and J. M. Pierce, Chem. Biol., 2014, 21, 1–15 CrossRef CAS PubMed.
- M. Frenkel-Pinter, M. D. Shmueli, C. Raz, M. Yanku, S. Zilberzwige, E. Gazit and D. Segal, Sci. Adv., 2017, 3, e1601576 CrossRef PubMed.
- Q. Zhang, C. Ma, L.-S. Chin and L. Li, Sci. Adv., 2020, 6, eabc5802 CrossRef CAS PubMed.
- B. G. Cho, L. Veillon and Y. Mechref, J. Proteome Res., 2019, 18, 3770–3779 CrossRef CAS PubMed.
- C. Menni, I. Gudelj, E. Macdonald-Dunlop, M. Mangino, J. Zierer, E. Bešić, P. K. Joshi, I. Trbojević-Akmačić, P. J. Chowienczyk and T. D. Spector, Circ. Res., 2018, 122, 1555–1564 CrossRef CAS PubMed.
- D. Marques-da-Silva, R. Francisco, D. Webster, V. dos Reis Ferreira, J. Jaeken and T. Pulinilkunnil, J. Inherited Metab. Dis., 2017, 40, 657–672 CrossRef CAS PubMed.
- C. Reily, T. J. Stewart, M. B. Renfrow and J. Novak, Nat. Rev. Nephrol., 2019, 1 Search PubMed.
- J. Ugonotti, S. Chatterjee and M. Thaysen-Andersen, Mol. Aspects Med., 2020, 100882 Search PubMed.
- S. S. Pinho and C. A. Reis, Nat. Rev. Cancer, 2015, 15, 540–555 CrossRef CAS PubMed.
- W. Peng, S. Zhou, P. Mirzaei and Y. Mechref, Sci. Rep., 2019, 9, 1–13 Search PubMed.
- D. Thomas, A. K. Rathinavel and P. Radhakrishnan, Biochim. Biophys. Acta, Rev. Cancer, 2020, 188464 Search PubMed.
- Q. Li, M. J. Kailemia, A. A. Merleev, G. Xu, D. Serie, L. M. Danan, F. G. Haj, E. Maverakis and C. B. Lebrilla, Anal. Chem., 2019, 91, 5433–5445 CrossRef CAS PubMed.
- P. Regan, P. L. McClean, T. Smyth and M. Doherty, Medicines, 2019, 6, 92 CrossRef CAS PubMed.
- Y. Watanabe, Z. T. Berndsen, J. Raghwani, G. E. Seabright, J. D. Allen, O. G. Pybus, J. S. McLellan, I. A. Wilson, T. A. Bowden and A. B. Ward, Nat. Commun., 2020, 11, 1–10 Search PubMed.
- M. Sanda, L. Morrison and R. Goldman, Anal. Chem., 2020, 93, 2003–2009 CrossRef PubMed.
- L. Veillon, Y. Huang, W. Peng, X. Dong, B. G. Cho and Y. Mechref, Electrophoresis, 2017, 38, 2100–2114 CrossRef CAS PubMed.
- S. Zhou, X. Dong, L. Veillon, Y. Huang and Y. Mechref, Anal. Bioanal. Chem., 2017, 409, 453–466 CrossRef CAS PubMed.
- S. Zhou, Y. Hu and Y. Mechref, Electrophoresis, 2016, 37, 1506–1513 CrossRef CAS PubMed.
- T. Klarić and I. Gudelj, in High-Throughput Glycomics and Glycoproteomics, Springer, 2017, pp. 207–216 Search PubMed.
- A. Messina, A. Palmigiano, F. Esposito, A. Fiumara, A. Bordugo, R. Barone, L. Sturiale, J. Jaeken and D. Garozzo, Glycoconjugate J., 2020, 1–11 CAS.
- S. Zhou, K. M. Wooding and Y. Mechref, in High-Throughput Glycomics and Glycoproteomics, Springer, 2017, pp. 83–96 Search PubMed.
- J. Zhou, C. Liu, D. Si, B. Jia, L. Zhong and Y. Yin, Anal. Chim. Acta, 2017, 972, 62–72 CrossRef CAS PubMed.
- S. Zhou, Y. Huang, X. Dong, W. Peng, L. Veillon, D. A. S. Kitagawa, A. J. A. Aquino and Y. Mechref, Anal. Chem., 2017, 89, 6590–6597 CrossRef CAS PubMed.
- Y. Huang, S. Zhou, J. Zhu, D. M. Lubman and Y. Mechref, Electrophoresis, 2017, 38, 2160–2167 CrossRef CAS PubMed.
- J. B. Muller, P. E. Geyer, A. R. Colaco, P. V. Treit, M. T. Strauss, M. Oroshi, S. Doll, S. Virreira Winter, J. M. Bader, N. Kohler, F. Theis, A. Santos and M. Mann, Nature, 2020, 582, 592–596 CrossRef PubMed.
- J. Stadlmann, O. Hudecz, G. Krssakova, C. Ctortecka, G. Van Raemdonck, J. Op De Beeck, G. Desmet, J. M. Penninger, P. Jacobs and K. Mechtler, Anal. Chem., 2019, 91, 14203–14207 CrossRef CAS PubMed.
- P. Gzil, N. Vervoort, G. Baron and G. Desmet, Anal. Chem., 2003, 75, 6244–6250 CrossRef CAS PubMed.
- W. D. Malsche, H. Gardeniers and G. Desmet, Anal. Chem., 2008, 80, 5391–5400 CrossRef PubMed.
- S. Gautam, W. Peng, B. G. Cho, Y. Huang, A. Banazadeh, A. Yu, X. Dong and Y. Mechref, Analyst, 2020, 145, 6656–6667 RSC.
- P. Kang, Y. Mechref and M. V. Novotny, Rapid Commun. Mass Spectrom., 2008, 22, 721–734 CrossRef CAS PubMed.
- C. Ashwood, B. Pratt, B. X. MacLean, R. L. Gundry and N. H. Packer, Analyst, 2019, 144, 3601–3612 RSC.
- J. D. Benktander, S. T. Gizaw, S. Gaunitz and M. V. Novotny, J. Am. Soc. Mass Spectrom., 2018, 29, 1125–1137 CrossRef CAS PubMed.
- J. L. Desantos-Garcia, S. I. Khalil, A. Hussein, Y. Hu and Y. Mechref, Electrophoresis, 2011, 32, 3516–3525 CrossRef CAS PubMed.
- G. R. Guile, P. M. Rudd, D. R. Wing, S. B. Prime and R. A. Dwek, Anal. Bioanal. Chem., 1996, 240, 210–226 CAS.
- T. Beyer, F. Klose, A. Kuret, F. Hoffmann, R. Lukowski, M. Ueffing and K. Boldt, J. Proteomics, 2021, 231, 103947 CrossRef CAS PubMed.
- C. Huber, I. Stamm, W. Ziebuhr, G. Marincola, M. Bischoff, B. Strommenger, G. Jaschkowitz, T. Marciniak, C. Cuny, W. Witte, J. Doellinger, C. Schaudinn, A. Thurmer, L. Epping, T. Semmler, A. Lubke-Becker, L. H. Wieler and B. Walther, Sci. Rep., 2020, 10, 14787 CrossRef CAS PubMed.
- S. Kurz, M. O. Sheikh, S. Lu, L. Wells and M. Tiemeyer, Mol. Cell. Proteomics, 2021, 20, 100045 CrossRef CAS PubMed.
- S. Zhou, K. M. Wooding and Y. Mechref, Methods Mol. Biol., 2017, 1503, 83–96 CrossRef CAS PubMed.
- J. M. Prien, D. J. Ashline, A. J. Lapadula, H. Zhang and V. N. Reinhold, J. Am. Soc. Mass Spectrom., 2009, 20, 539–556 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an00643f |
| This journal is © The Royal Society of Chemistry 2021 |