Recent microfluidic advances in submicron to nanoparticle manipulation and separation
Samith
Hettiarachchi
 a,
Haotian
Cha
a,
Lingxi
Ouyang
a,
Amith
Mudugamuwa
a,
Haotian
Cha
a,
Lingxi
Ouyang
a,
Amith
Mudugamuwa
 b,
Hongjie
An
b,
Hongjie
An
 a,
Gregor
Kijanka
a,
Navid
Kashaninejad
a,
Gregor
Kijanka
a,
Navid
Kashaninejad
 a,
Nam-Trung
Nguyen
a,
Nam-Trung
Nguyen
 *a and
Jun
Zhang
*a and
Jun
Zhang
 *a
*a
aQueensland Micro- and Nanotechnology Centre, Griffith University, Nathan, Queensland 4111, Australia. E-mail: nam-trung.nguyen@griffith.edu.au; jun.zhang@griffith.edu.au
bUniversity of Moratuwa, Katubedda 10400, Sri Lanka
First published on 1st November 2022
Abstract
Manipulation and separation of submicron and nanoparticles are indispensable in many chemical, biological, medical, and environmental applications. Conventional technologies such as ultracentrifugation, ultrafiltration, size exclusion chromatography, precipitation and immunoaffinity capture are limited by high cost, low resolution, low purity or the risk of damage to biological particles. Microfluidics can accurately control fluid flow in channels with dimensions of tens of micrometres. Rapid microfluidics advancement has enabled precise sorting and isolating of nanoparticles with better resolution and efficiency than conventional technologies. This paper comprehensively studies the latest progress in microfluidic technology for submicron and nanoparticle manipulation. We first summarise the principles of the traditional techniques for manipulating nanoparticles. Following the classification of microfluidic techniques as active, passive, and hybrid approaches, we elaborate on the physics, device design, working mechanism and applications of each technique. We also compare the merits and demerits of different microfluidic techniques and benchmark them with conventional technologies. Concurrently, we summarise seven standard post-separation detection techniques for nanoparticles. Finally, we discuss current challenges and future perspectives on microfluidic technology for nanoparticle manipulation and separation.
1 Introduction
Submicron (0.10–1.0 μm) and nanoparticles (1.0–100 nm) such as extracellular vesicles (EVs),1 bacteria,2 viruses,3 metal,4 carbon,5 and polymer nanoparticles,6etc. have broad applications in disease diagnostics, drug delivery and material synthesis. The capability to manipulate and separate these tiny particles for isolating and enriching a specific population with uniform properties (e.g., size, shape, and charge) is critical for these applications. For example, exosomes, a subset of EVs released from cells, have been recognised as an essential biomarker for diagnosing various diseases and monitoring therapy efficiency.7 Exosome-containing biological fluids carry other EVs, such as microvesicles (MVs), apoptotic bodies and macromolecules (e.g., proteins, proteases and nuclease), which may interfere with the analysis of exosomes. Therefore, efficient isolation and purification of exosomes from biological liquids have become increasingly important.8 Moreover, the size fractionation of nanoparticles from the synthesised nanoparticles of broad size distribution is essential for practical applications because the functions and toxicities of nanoparticles are size-dependent.9–11Many technologies have been developed for separating and concentrating nanoparticles, including ultracentrifugation,12,13 ultrafiltration,14 size exclusion chromatography,15 precipitation,16 and immunoaffinity capture.17 Ultracentrifugation is the most common method for separating and purifying nanoparticles. The particles deposit on the bottom of the tube by centrifugal force induced by an ultra-high rotational speed. Furthermore, ultrafiltration and size exclusion chromatography use nanostructures on the membranes or columns to separate particles based on their size. In addition, the precipitation technique allows particles to aggregate by modifying the surface chemistry of nanoparticles with a solvent. Immunoaffinity capture utilises immobilised antibodies to selectively bind target particles through specific antigen interactions. These conventional techniques have high separation efficiency, ease of use, high yield and good reproducibility.7 However, they suffer from several notable limitations, such as longer processing time, membrane clogging, high cost, low purity, or risk of damaging biological particles.18 Therefore, extensive research efforts have been carried out to overcome these bottlenecks.
Microfluidics, the science of manipulating fluids in channels with dimensions of tens of micrometres, has emerged as a promising technology platform with many applications in chemistry, biology, medicine and physical science.19 Benefiting from the precise control of fluid flow in microchannels, microfluidic technology offers an unprecedented resolution for manipulating and separating micro and nanoparticles.20 Microfluidic techniques are broadly categorised as active and passive methods based on the origin of manipulating forces.21 Active techniques use an external force field such as acoustic,22 electric,23 magnetic,24 or optical25 to precisely control particles. In contrast, the passive methods do not use any physical fields other than the intrinsic hydrodynamics and channel geometry. Examples of passive methods are inertial microfluidics,26 deterministic lateral displacement,27 microfluidic filtration,28 pinched flow fractionation,29 and viscoelastic microfluidics.30
Both active and passive methods have merits and demerits in particle manipulation. Active methods precisely control the particles with varying external forces in real time.31 However, since they require a longer time to be exposed to the force field, the flow rate is generally slow, resulting in a relatively low throughput. In contrast, passive microfluidic devices for particle manipulation are simple, easy to operate and provide a relatively high throughput. Nevertheless, the controllability and accuracy are generally lower than active microfluidic techniques.32 A recent trend is integrating multiple active and passive methods to overcome the limitations of each technique.33 To date, microfluidic techniques have been employed for manipulating, focusing and isolating various biological, metal and polymer nanoparticles for disease diagnostics, drug delivery, therapeutics, material synthesis, etc.34–36
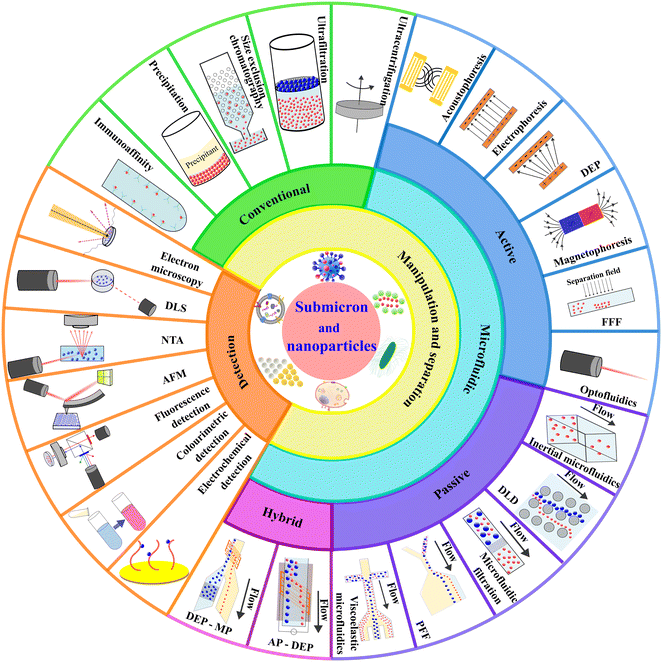
Although a few recent literature review articles summarised the development of microfluidic technology for isolating specific bio nanoparticles such as EVs and exosomes,7,20,37–39 this review paper aims to study the topic more comprehensively from a fundamental technological aspect to cover broad particle types and to provide an update on the latest technology developments. This paper first summarises the conventional techniques for the manipulation and separation of nanoparticles. Then, we elaborate on the current development of microfluidic technology based on the classification of active, passive, and hybrid microfluidics. We explain the physics of each manipulating force, the device design, working mechanism and applications. We also compare and discuss the advantages and limitations of the reported microfluidic technologies and compare them with conventional technologies. Concurrently, we summarise the seven most typical detection techniques for nanoparticles. Fig. 1 shows an overview of this paper, covering the scope of nanoparticle manipulation, separation, and detection. Lastly, we discuss the current challenges in the field and perspectives on future developments of microfluidic technology on nanoparticle separation.
2 Conventional techniques
Many technologies have been developed to manipulate and separate nanoparticles. Instead of exhaustively listing all these technologies, we focus on the five most common technologies for nanoparticle separation: ultracentrifugation, ultrafiltration, size exclusion chromatography, precipitation, and immunoaffinity capture. Following this, we discuss their primary working principle and compare the technological features such as manipulating particle size and type, separation performance, throughput, etc., Table 1.| Technique | Principle | Particle size | Particle types | Separation efficiency | Throughput | Merits | Demerits | Ref. | |
|---|---|---|---|---|---|---|---|---|---|
| Conventional techniques | UC | Centrifugal force by rotation | 50–200 nm | Exosomes, polymersomes, gold nanoparticles | 23–70% | 106–6 × 108 cells per h | High throughput | Low yield, time-consuming, need for specialised equipment | 12, 44, 48, 74 |
| UF | Membrane pore size | 50–250 nm | Exosomes, natural organic matters | 70–82% | 0.5–1.6 ml min−1 | High separation efficiency, simple operation | Membrane clogging, possibility of damage due to shear stress | 14, 54, 55 | |
| SEC | Different elution speeds in a porous medium | 50–200 nm | Exosomes, vesicles, gold nanoparticles, quantum dots | 80–90% | 25–250 μl min−1 | Shorter processing time, small sample volume, good reproducibility | Need for specialised equipment, possible adsorption to the stationary phase | 53, 56, 58–60 | |
| Precipitation | Converting a solution into a solid | 50–150 nm | EVs, viruses | 28–85% | 0.35–8 μl min−1 | Less expensive, can process large sample volume, high throughput, easy integration with others | Time-consuming, co-precipitation of non-target particles, potential interference with the biological functions | 16, 55, 66 | |
| Immunoaffinity capture | Capturing by tagging antibodies | 30–200 nm | Exosomes, EVs | — | 4–160 μl min−1 | High specificity and purity | Low yield, time-consuming, high cost | 17, 58 | |
| Active microfluidic techniques | AP | Acoustic radiation force | 100–900 nm | MVs, bacteria | >90% | 4–80 μl min−1 | High separation efficiency, contactless, highly controllable, biocompatible, wide versatility | Complex fabrication, low throughput, induced thermal energy | 78, 92, 103 |
| Electrophoresis | Electrostatic force | 3.5–150 nm | Exosomes, gold nanoparticles | 65–98% | 2–40 μl min−1 | High separation efficiency, highly controllable | Induce thermal energy, electrochemical reaction | 4, 115, 120, 123 | |
| DEP | Interaction of electrical polarisation and a non-uniform electrical field | 50–490 nm | Exosomes, viruses, bacteria, polystyrene particles | 77–83% | 3–16 μl min−1 | High separation efficiency, controllable, minimum particle damage, high throughput | Require a strong electric field, joule heating, electrochemical reaction | 79, 80, 83, 138 | |
| MP | Magnetic force | 5–200 nm | EVs, bacteria | 80–90% | 0.3–1000 μl min−1 | High throughput, contactless, low cost, non-contact, no heat generation | Time-consuming and labour-intensive sample preparation, exposure to the paramagnetic medium may affect the integrity of biomolecules | 2, 147, 151 | |
| FFF | Perpendicular force field to the flow | 5–250 nm | Exosomes, gold nanoparticles, nanocolloids | 90% | 45–400 μl min−1 | High separation efficiency, high throughput, versatility by using various force fields | Longer setting up time, the limitation of the external force fields | 162, 166, 170 | |
| Optofluidics | Optical radiation scattering and gradient force | 50–200 nm | Gold nanoparticles, silver nanoparticles, yeast cells | — | 100–300 particles per min | High separation efficiency, precise controllability | Low throughput, induce thermal energy affects biocompatibility | 172, 176, 177 | |
| Passive microfluidic techniques | Inertial microfluidics | Inertial lift and Dean drag forces | 0.2–2 μm | EVs, polystyrene particles | 15–97% | 80–100 μl min−1 | High throughput, simple, good biocompatible | Inertial forces are weak, low separation resolution | 180, 202 |
| DLD | Particles displace differentially in tilted pillar array | 51 nm–1.5 μm | EVs, polystyrene particles | 50–98.5% | 0.1–0.2 nl min−1 | Simple, easy to operate, high separation resolution | Low throughput, channel clogging, high fabrication cost | 185, 217, 254 | |
| Microfluidic filtration | Nano membrane in microfluidics | 30–200 nm | Exosomes, viruses | 73–89% | 40–500 μl min−1 | High separation efficiency, simple, easy to operate | Membrane clogging, possible damage due to shear stress | 188, 220, 222, 224 | |
| PFF | Pinching particles with hydrodynamic flows | 30 nm–2 μm | EVs, viruses, polystyrene particles | 82–90% | 0.1–2.8 μl min−1 | Simple design, easy fabrication | Low throughput and efficiency, significant sample dilution by secondary flow | 189, 190, 233 | |
| Viscoelastic microfluidics | Elastic force due to imbalance of normal stresses in a viscoelastic fluid | 49 nm–2 μm | EVs, polystyrene particles | 55–95% | 90–1300 μl min−1 | Simple, higher separation resolution, low-cost biocompatible | Low throughput, potential contamination | 191, 249, 250 | |
| Hybrid microfluidic techniques | DEP–MP | Connection of DEP and MP in a serial manner | 500 nm | Bacteria | 95.6–99.74% | 2.5 × 107 cells per h | High separation efficiency, high throughput, sort multiple particles | Complex fabrication, time consumption | 251 |
| AP–DEP | Force coupling of AP and DEP | 300–500 nm | Exosomes, polystyrene particles | 81–95% | 0.75–3 μl min−1 | High separation efficiency, high precision | Induce thermal energy | 252, 253 |
2.1 Ultracentrifugation
Ultracentrifugation (UC) is predominantly used to separate particles based on particle size, shape, and density.40 Particles deposit due to the centrifugal force induced by ultra-high rotational speed. UC generally uses a rotational speed of as high as 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–200
000–200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g to separate nanoparticles such as exosomes.41 Differential centrifugation and density gradient ultracentrifugation (DGUC) are two main modes of UC.42 In differential centrifugation, multiple fractions within a mixture can be separated using differential centrifugation speeds.13 The larger and denser particles form a sediment at a relatively lower rotational speed and are isolated from the mixture. A higher centrifugation speed is subsequently applied and isolates smaller and lighter particles from the supernatant, Fig. 2(A).43 In DGUC, solutions with different densities are loaded into the centrifugal tube to create a density gradient medium.44 During the ultracentrifugation, particles move through the medium and are retained where their densities match the surrounding medium due to the balance between centrifugal and buoyancy forces. Particles with various densities can be separated by differential fraction collection.45 UC has been widely employed and is considered the gold standard for particle separation.46,47 However, UC suffers from low yield, long processing time, inconsistent outcomes, the necessity for a large sample volume, high-cost equipment, and skilled labour requirement.48–50 Moreover, biological particles may be damaged by forces induced by high rotational speed, which diminishes downstream analysis.51
000g to separate nanoparticles such as exosomes.41 Differential centrifugation and density gradient ultracentrifugation (DGUC) are two main modes of UC.42 In differential centrifugation, multiple fractions within a mixture can be separated using differential centrifugation speeds.13 The larger and denser particles form a sediment at a relatively lower rotational speed and are isolated from the mixture. A higher centrifugation speed is subsequently applied and isolates smaller and lighter particles from the supernatant, Fig. 2(A).43 In DGUC, solutions with different densities are loaded into the centrifugal tube to create a density gradient medium.44 During the ultracentrifugation, particles move through the medium and are retained where their densities match the surrounding medium due to the balance between centrifugal and buoyancy forces. Particles with various densities can be separated by differential fraction collection.45 UC has been widely employed and is considered the gold standard for particle separation.46,47 However, UC suffers from low yield, long processing time, inconsistent outcomes, the necessity for a large sample volume, high-cost equipment, and skilled labour requirement.48–50 Moreover, biological particles may be damaged by forces induced by high rotational speed, which diminishes downstream analysis.51
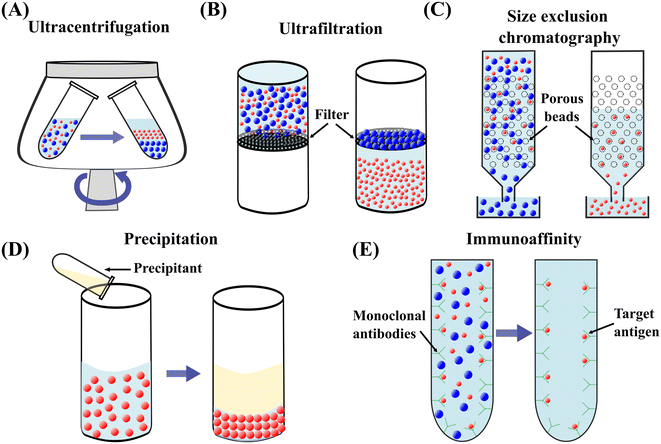 | ||
| Fig. 2 Schematic diagrams of conventional techniques for submicron and nanoparticle separation. (A) Ultracentrifugation.13 (B) Ultrafiltration.53 (C) Size exclusion chromatography.59 (D) Precipitation.61 (E) Immunoaffinity capture.17 | ||
2.2 Ultrafiltration
Ultrafiltration (UF) utilises porous membranes to separate particles based on the membrane pore sizes.52 The particles are infiltrated through the membrane by applying pressure or centrifugation.53,54 Particles larger than the cut-off size are trapped on the membrane while smaller particles can pass through it, Fig. 2(B).36 UF is a faster alternative to UC and provides a higher yield with increasing isolation efficiency.55 However, particles experience a relatively high stress during the passage through the membrane pores, potentially damaging the morphology or functionality of particles. The large particles trapped in the membrane may cause clogging. Membrane clogging prevents the passage of desired particles from crossing the membrane and lowers the throughput and recovery rate.14 Moreover, the co-existence of particles of similar size reduces the purity of target particles.382.3 Size exclusion chromatography
Size exclusion chromatography (SEC) is a size-based particle isolation technique that uses a stationary phase of porous beads with defined pore size. Particles smaller than the pores of porous beads can travel through them, whereas larger particles cannot penetrate through and can only move between the porous beads, Fig. 2(C). The travelling time for smaller particles is longer than the larger particles, resulting in a later elution.56 This technique is easy to use and low cost and has shown a high reproducibility in isolating nanoparticles with low volume.57,58 However, SEC needs specialised equipment, and purity may be compromised due to the coexistence of particles of similar sizes.59 Irreversible adsorption of nanoparticles to the stationary phase is another drawback of this technique.15 Recent approaches combining SEC and UF have significantly enhanced the purity of isolated exosomes.602.4 Precipitation
Precipitation converts particles in solution into a solid phase by transforming the substance into its insoluble form or oversaturating the solution.61 Polymer-induced precipitation is employed to isolate biological particles such as exosomes and EVs. The water-soluble polymer such as polyethylene glycol is added to the sample and incubated for a period of time, Fig. 2(D). The precipitated EVs or exosomes form sediment in the form of a pellet after low-speed centrifugation or filtration.53 Precipitation is also used to synthesise non-biological nanoparticles.62,63 For example, ionic metals are converted to an insoluble form by a chemical reaction of a soluble metal and a reagent. Subsequently, the formed metal particles can be removed by filtration.64 Precipitation-based isolation is low-cost as it does not need special equipment requirements.16,53,65 This technique can process large sample volumes and be easily integrated with other separation methods.39 However, the precipitation reagents may interfere with the biological characteristics of particles.66,67 Co-precipitation of non-target particles can cause the contamination of target particles, such as co-precipitation of protein and polymeric substances during exosome precipitation using polymer solutions.72.5 Immunoaffinity capture
Immunoaffinity capture is widely employed to isolate biological particles from a mixture based on the specific interaction of an antibody and an antigen.17 The surfaces of many bioparticles comprise antigens that can be bound exclusively by particular antibodies.68 The antibodies can be immobilised on the surface of a solid phase, such as magnetic beads, plates, and chromatography matrices. Due to the specific binding between the antibody and antigen, the target bioparticles are immobilised on the solid phase. In contrast, unbound, non-specific particles are freely dispersed in the medium and can be eluted, Fig. 2(E).69 Therefore, target bioparticles can be selectively isolated and concentrated.70 For the isolation of exosomes, the most commonly utilised biomarkers are tetraspanins such as CD9, CD63, CD81, CD82, and CD151.71–73 Immunoaffinity-based capturing methods provide a high separation specificity. Still, this method is challenged by relatively low yields, limited accessibility of particular antibodies, and potential damage to the biological particles.38,743 Active microfluidic techniques
Active microfluidic technologies utilise external force fields to manipulate and separate particles on top of the intrinsic hydrodynamic forces.21 Acoustic, electric, magnetic, and optical force fields are the most commonly used in active microfluidics.75 Due to their versatility and high controllability, active microfluidic technologies have been tailored to process many submicron and nanoparticles, including exosomes,76,77 bacteria,2,78,79 viruses,80 platelets,81 polystyrene and gold nanoparticles,4,82,83etc. In the following, we will explain the physics of each manipulation force, the current status of the research, the working mechanism of the reported devices, and their application to the manipulation of submicron and nanoparticles.3.1 Acoustophoresis
Acoustophoresis (AP) is defined as the movement of particles by sound waves.84 In this technique, ultrasound waves exert acoustic radiation forces on particles, and particles migrate toward the acoustic pressure nodes or antinodes based on the relative compressibility of particles and fluids. The strength of the acoustic radiation force on a particle depends on the particle size, density, and compressibility.85 The magnitude of the acoustic radiation force (FR) acting on a spherical particle with a diameter of d can be expressed as:86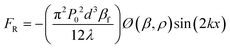 | (1) |
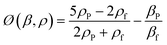 | (2) |
Particle manipulation by an acoustic field has been widely known as acoustic tweezers.88 Surface acoustic waves (SAWs)89 and bulk acoustic waves (BAWs)90 are the two main platforms for microparticle manipulation.91–93 SAWs are mechanical waves travelling along a substrate surface, and they are generally induced by interdigitated transducers (IDTs) with different electrode shapes such as focused94 and straight95 on the surface of piezoelectrics.96 SAWs can further be divided into standing surface acoustic waves (SSAWs) and travelling surface acoustic waves (TSAWs).97,98 BAWs are the compressional waves propagating through a bulk material.90 With an acoustic resonator, particles suspended in a medium can be effectively manipulated by BAW.88,99 Acoustophoresis is simple, robust, contactless, and biocompatible for particle manipulation. This technique has been successfully applied for the separation and purification of submicron and nanoparticles such as platelets,81 MVs,100 lipid particles,101 bacteria,78,102 and polystyrene nanoparticles.82
Lee et al.103 reported on an acoustic nanofilter system to separate MVs from biological samples using the SSAW principle. In their device, the two sheath flows pinched the sample to the middle of the microchannel at the inlet. Next, acoustic radiation force pulls larger MVs to the pressure nodes near two sidewalls, whereas the smaller MVs remained at the central flow due to the weak acoustic force, Fig. 3(A). The device isolated exosomes with diameters less than 200 nm from cell culture media and erythrocyte-derived vesicles from blood samples with a recovery rate of more than 80%. Meanwhile, using the same principle with tilted IDTs, a fractionation of nanoparticles of varying sizes could also be achieved.11 110 nm and 220 nm polystyrene particles were separated from 900 nm and 600 nm ones. The recovery rates of 110 nm and 220 nm particles were 90.6% and 85.6%, and the depletion ratios of 900 nm and 600 nm particles were 96.6% and 80.4%, respectively. Furthermore, the same group successfully isolated exosomes from undiluted blood with a purity of 98% by integrating two sequential tilted angle SSAW modules.100 In this platform, the first module removed the larger blood components, such as red blood cells (RBCs), white blood cells (WBCs), and platelets. The second module in the downstream isolated exosomes from the remaining EVs. In addition to symmetric acoustic transducers where interdigitated electrodes are patterned symmetrically on the two sides of microchannels, asymmetric acoustic waves have also been reported. Collins et al.82 developed a focused TSAW device that manipulates micro and nanoparticles using an asymmetric focused acoustic wave, Fig. 3(B). Different focus points for separating nanoparticles of 100 nm, 300 nm, and 500 nm diameters were observed regardless of their initial starting position.
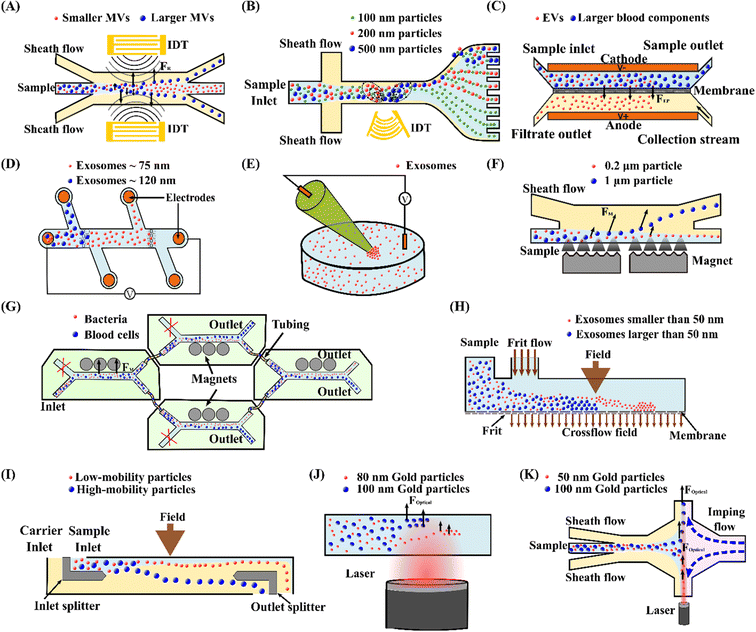 | ||
| Fig. 3 Active microfluidic techniques for manipulation, focus, and separation of submicron and nanoparticles. (A) Separation of MVs by acoustophoresis.103 (B) Focused TSAW device for separating 100 nm, 200 nm and 500 nm polystyrene particles.82 (C) Isolation EVs from whole blood by DC electrophoresis.115 (D) Simultaneous capture and separation of different subpopulations of exosomes by DC insulator-based dielectrophoresis.137 (E) Entrapment of exosomes by a low voltage nanopipette DEP device.76 (F) Separation of 0.2 μm and 1 μm fluorescence polystyrene particles by magnetophoresis.146 (G) Separation of bacteria by integrating multiple magnetophoresis devices.2 (H) Size fractionation of exosomes by field-flow fractionation.166 (I) SPLITT device for improved fractionation of cells/particles.168 (J) Trapping of 80 nm and 100 nm gold particles by optofluidics.176 (K) Optofluidic sorting of gold nanoparticles of 50 nm and 100 nm diameters.172 | ||
In contrast to SAW, the BAW technique has rarely been studied for manipulating submicron and nanoparticles.104,105 Ohlsson et al.78,106 successfully utilised the BAW technique to separate and enrich bacteria from the blood. In their device, bacteria were first isolated from RBCs by acoustic force and then enriched using acoustic trapping. Due to the operational frequency range of BAW, separating nanoparticles is challenging for this technique.
In summary, acoustophoresis has been used in a wide range of applications in biomedical research, such as diagnostics and therapeutics.104,107 In general, it possesses the advantages of being contactless, biocompatible, highly controllable, and versatile in submicron and nanoparticle manipulation. However, the processing throughput is relatively low, the fabrication process for these devices is complex, and the induced thermal energy may deteriorate the performance of the devices. Moreover, since biological fluids contain components of similar size and acoustic characteristics, acoustic separation of these components may be challenging.108
3.2 Electrophoresis
Electrophoresis is the relative motion of charged particles over fluid in an electric field.109 Due to the electrostatic force, charged particles are drawn towards the oppositely charged electrode, whereas the field does not affect uncharged particles. The electrophoretic force (FEP) acting on a spherical particle with a radius a can be expressed as:110| FEP = 6πζpεmaE | (3) |
Mogi et al.77 developed a microfluidic device capable of damage-less handling of exosomes using an ion-depletion zone. A direct current (DC) voltage of 100 V was applied between two microchannels to create an ion depletion region through a Nafion membrane. This region acts as a barrier against any charged particles. Since exosomes are negatively charged, they are pushed away by the ion depletion zone. Using this technique, the team achieved an exosome yield of 98% with less damage than the conventional techniques. In addition, a combination of DC electrophoresis and a membrane filter was proposed to isolate EVs from whole blood.115 A DC voltage of 10 V was applied over two electrodes with a separation distance of 1.5 cm, Fig. 3(C). A porous monolithic membrane with pore sizes of less than 500 nm allows EVs to pass through the membrane between two electrodes. Furthermore, a similar technique has been used to separate EVs from proteins in the blood plasma.120 The membrane pore size was 30 nm so that proteins could pass through the membrane, but the EVs were retained. This work recovered 65% of EVs, and 83.6% of the protein was removed within 30 minutes. Gold nanoparticles are used in biomedical imaging and diagnostics, but their cytotoxicity depends on particle size.121 Thus, it is crucial to isolate monodisperse metallic nanoparticles.122 Using commercially available CE equipment in an evaporative light scattering detector (ELSD), Bouri et al.4,123 separated gold nanoparticles of 3.5 nm, 6.5 nm, and 10.5 nm.
In summary, electrophoresis is simple in design and controllable but requires trained users and tedious operation steps.36 Moreover, direct contact between sample solution and electrodes and potentially excessive heat generation due to the high voltages may also affect biological components such as exosomes.124
3.3 Dielectrophoresis
Dielectrophoresis (DEP) is the movement of dielectric particles when subjected to a non-uniform electric field.125 The force exerted on the particle depends on the dielectric properties of the particles and the suspending medium.126,127 The actuating electric field can be alternating current (AC) or DC.128 For a homogeneous spherical particle, the DEP force (FDEP) is expressed as:129| FDEP = 2πa3εm Re[K(ω)]∇|Erms|2 | (4) |
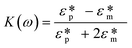 | (5) |
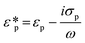 | (6) |
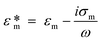 | (7) |
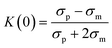 | (8) |
Depending on the CM factor, DEP is categorised as positive or negative DEP. If the permittivity or the conductivity of the particle is greater than that of the medium (K(ω) > 0), it is called positive DEP, and particles move towards the maximum electric field. Conversely, if the polarisation of particles is lower than the medium (K(ω) < 0), it is the negative DEP force that migrates particles towards the minima of the electric field.125,132,133 Both the positive and negative DEP forces can be used to separate submicron and nanoparticles83 and biomolecules, such as viruses,80 bacteria,79,134 and exosomes.135,136
Zhao et al.83 developed a novel DC-DEP microfluidic device for continuous separation of nanoparticles with similar sizes but different electric properties (i.e. 140 nm polystyrene and 150 nm magnetic particles). The non-uniform electric field was generated using a pair of asymmetric orifices in their device. Particles experienced distinct positive and negative DEP forces due to different electrical conductivities and were separated into various outlets. Apart from the electrical properties, DC-DEP could also separate submicron particles by size. Ayala-Mar et al.137 developed a DC insulator-based DEP (DC-iDEP) device to capture and separate two different subpopulations of exosomes by size, Fig. 3(D). The device has two different channel sections filled with oval-shaped insulating posts to trap the exosomes of diverse populations. The first post array has a gap space of 15 μm to trap exosomes with a mean diameter of 113.23 ± 10.34 nm, while the second array with 10 μm gap spaces was capable of trapping the exosomes with a mean diameter of 72.86 ± 8.71 nm. Successful trapping was achieved with an electric potential difference of 2000 V. After the trapping, electric stimulation was removed to release the exosomes, and a DC voltage of 200 V was applied to side channels to direct the released exosomes to the respective reservoirs by electroosmosis. In addition, an alternative approach was introduced by Shi et al.76 by applying a significantly lower DC voltage (10 V cm−1) across a glass nanopipette, Fig. 3(E). The device is capable of rapid entrapment of nanoparticles and exosomes. When a DC voltage was applied across the pipette, the particles suspended in the medium were attracted towards the pipette tip due to the balance between dielectrophoretic, electroosmotic, and electrophoretic forces. Exosomes from undiluted plasma, serum and saliva were isolated separately from four parallelly connected pipettes with a negative electrical polarity.135
Meanwhile, an AC electrokinetic (ACE) microarray device was developed to isolate various drug delivery nanoparticles with diameters between 100 nm and 200 nm from blood plasma.35 A positive DEP force trapped particles at the electrode edges with a high electric field in the device. A washing buffer subsequently eluted the non-trapped particles, and the particles held by DEP force could be released and collected by removing the AC electric field. Due to the distinct dielectric properties, the recovery of each type of drug delivery nanoparticle required a different AC electrical strength. Gel-filled nanoliposomes, hollow silica shells, empty nanoliposomes, and solid polymer nanoparticles were isolated at an electric voltage of 12 Vpp, 8 Vpp, 18 Vpp, and 15 Vpp, respectively. Later, the same group successfully applied the ACE microarray chip to isolate glioblastoma exosomes from 30–50 μl undiluted human plasma in less than 30 minutes.138 Another study proposes utilising DEP to rapidly detect the dengue virus (DENV).80 The microfluidic device consists of a guiding electrode and a capturing electrode. First, mouse anti-flavivirus monoclonal antibody (4G2) coated beads were infused into the device and captured on the electrode. Then the DENV-labelled fluorescent probe was injected to interact with 4G2-coated beads, so that the virus could be detected based on the fluorescence intensity. In addition, a periodically controlled positive DEP chip is developed to continuously isolate rare pathogenic bacteria from the blood.79 The device is divided into three main sections: focusing, separation, and collection. The sample pinches towards the sidewall by a sheath flow in the focusing region. The bacteria experience a positive DEP force, but no net DEP force is exerted in WBCs in the separating region. The trajectory of bacteria is separated from that of WBCs. Finally, bacteria and WBCs are collected at distinct outlets in the collecting region. This device recovered 82.1% E. coli, 83.3% S. aureus, and 77.1% P. aeruginosa from WBCs.
The DEP technique is promising for manipulating biological particles with adequate controllability, easy operation, high efficiency, and minimum particle damage.128,139 Furthermore, DEP is label-free and suitable for low-volume sample processing. However, DEP has challenges, such as creating non-uniform electric fields at the nanoscale to achieve precise, flexible, and large-scale manipulation of nanomaterials.140 Moreover, microfluidic devices may suffer from the adhesion of particles and chemical reactions on the surfaces of electrodes, resulting in a reduction in the recovery yield. In addition, a strong electric field is needed to manipulate nanoparticles. However, an excessive electric field may affect the viability and functionality of bioparticles.79
3.4 Magnetophoresis
Magnetophoresis (MP) refers to the movement of particles relative to the fluid under an external magnetic field.141 The magnetic field can either be generated by a permanent magnet or an electromagnet. The magnetic force (FM) acting on a particle can be expressed as:142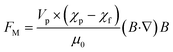 | (9) |
Zeng et al.146 developed a magnetofluidic device to separate 0.2 μm-polystyrene particles from 1 μm particles using negative magnetophoresis. At the inlet region, sample flow is focused along the side of a magnetic pole array by a co-flowing sheath flow. Large (1 μm) particles experienced a more significant negative magnetophoretic force in the separation channel and were repelled further away than the small (0.2 μm) particles, Fig. 3(F). In order to generate an ultrahigh magnetic intensity and gradient for separating nanoscale particles, high-permeability alloys, on-chip integrated magnetic micro-pole arrays, and a strong external NdFeB permanent magnet were incorporated into the microfluidic device. Later, they modified the device to enhance the sample throughput by taking advantage of the separation space between both sidewalls.147 In the modified device, the sheath flow symmetrically focused the sample flow along both sidewalls at the inlet. The high-permeability alloys, on-chip magnetic micro-pole arrays and permanent NdFeB magnets have been symmetrically patterned on both sides of the separation channel so that symmetric ultrahigh magnetic fields repelled nanoparticles toward the channel centre. They applied the device to separate small extracellular vesicles (sEVs) with dimensions 30–200 nm from the cell culture supernatant, and a recovery rate of 85.8% and purity of 80.45% were achieved. Using a similar ferrohydrodynamic technique, Liu et al.151 successfully separated 30–150 nm exosome-like particles from a biological sample with a recovery rate of 94.3% and a purity of 87.9%. This work used a quadrupole configuration of four permanent magnets to provide both high magnetic flux density and gradient.
Furthermore, positive magnetophoresis has been used to isolate target bioparticles by magnetic labelling. Separating bacteria from blood cells is vital for rapidly diagnosing bloodstream-related infections.152 Lee et al.2 developed a microfluidic system to separate E. coli from blood using bis-Zn-DPA modified magnetic nanoparticles, which can bind to both Gram-positive and Gram-negative bacteria. 88% E. coli were removed in the first cycle, and complete removal was achieved after the second cycle. Later the team enhanced the performance of the microfluidic platform by integrating multiple devices in series and parallel so that the separation could be completed in a single pass, Fig. 3(G). In addition, Oh et al.150 proposed a novel magnetisable micropipette tip to separate E. coli by positive magnetophoresis. The microtip contains a nickel mesh with rectangular micropores, and a permanent magnet could instantaneously magnetise it. Bacteria labelled with magnetic nanoparticles were attracted and attached to the mesh. After washing off the non-targets and demagnetising the tip, the E. coli were recovered at a 90.5% recovery rate.
Magnetophoresis is advantageous in non-contact, high throughput isolation, low cost, has selective controllability, and minimal heat generation.153–155 Despite the advantages, sample preparation for magnetophoresis is time-consuming and labour-intensive. The separation efficiency depends on the properties, such as the load capacity of the magnetic beads.144 Furthermore, the accumulation of magnetic nanoparticles and prolonged exposure to the paramagnetic medium may also affect the integrity of biomolecules.156
3.5 Field-flow fractionation
Field-flow fractionation (FFF) is a chromatography-like technique that uses an external force field perpendicular to the main flow to fractionate particles, Fig. 3(H).157 The external force can be hydraulic, thermal, electric, magnetic, or gravitational, and they can displace particles laterally to the side walls.158 The migration speed of the particles is affected by the physical parameters of the particle, the applied force field and the parabolic flow profile.159 Giddings first invented this technique in 1966.160 FFF partially overlaps with the scope of other active or passive techniques. FFF has been successfully utilised to isolate and characterise biological and non-biological sub-micro and nanoparticles.161–165The FFF technique has been used to separate EVs based on their hydrodynamic diameter. Kang et al.166 developed a flow FFF to fractionate exosomes according to their hydrodynamic diameter, Fig. 3(H). The team created an asymmetrical fluid flow as the external field and accumulated particles at the bottom wall. When an external field was applied, the small particles were eluted earlier than the larger ones, resulting in the size-based separation of particles. Their device isolated exosomes larger than 50 nm and smaller than 50 nm, respectively. A similar study fractioned urine exosomes ranging from 28.2 ± 4.3 nm to 136.7 ± 27.9 nm in diameter. The results indicated that the exosome fractions obtained from urine samples of healthy donors differed from those of prostate cancer.167
In addition, the same principle was used to separate a gold nanoparticle mixture by Calzolai et al.162 A cross-flow field separated 5 nm, 15 nm, and 45 nm gold nanoparticles from the nanoparticle mixture. Subsequently, a novel split-flow lateral-transport thin (SPLITT) technique was introduced by Giddings to improve cell/particle fractionation, Fig. 3(I).168 In this approach, splitters were added to the inlet and outlet to direct the particles to different outlets while an external force field acted perpendicular to the flow direction. The inlet splitter prevents the carrier stream from being in contact with the sample until they are under the influence of the external force field. The acting force on the particles transferred them into the carrier medium. The outlet splitters divided the stream into two outlets, consequently enabling the collection of separate sample fractions. De Momi et al.169,170 implemented this technique to fractionate colloids and particles in lake water, which facilitated the understanding of trace pollutant fate and behaviour in the environment. They were able to fractionate nanoparticles less than 10 nm to 250 nm.
Although FFF is advantageous in separating particles that differ in size, it can also sort particles differentiated by their mass, thermal, electrical, and magnetic characteristics since various physical forces can be employed. However, FFF has challenges such as a relatively long processing time for setting up and analysis, the inability to distinguish between differently shaped particles with the same mean hydrodynamic diameter or mass, and requiring optimisation at different external fields.161
3.6 Optofluidics
Holding and moving microscopic and sub-microscopic objects like atoms, nanoparticles and droplets using a focused laser beam is called optical tweezers, which Ashkin discovered in 1970.171 The optical force that acts on a particle is the sum of the radiation scattering force and gradient force. The scattering force pushes the particle along the direction of the light, and the gradient force pulls the particles towards the highest intensity gradient. The induced optical force depends on the particle size, shape, and reflective index. Optical tweezers have been widely used in chemistry, physics, biology, and medicine.172–175Nan et al.176 trapped 150 nm silver nanoparticles and sorted 80 nm and 100 nm gold nanoparticles with the synchronisation of an optical phase gradient and fluid drag forces, Fig. 3(J). When the particle reaches the trapping plane, the optical line can tightly confine the nanoparticle in the perpendicular direction of the flow. Then a strong phase gradient creates a lateral force that balances the fluid drag force and traps the particle. Due to the size-dependent nature of optical and fluid drag forces, the 80 nm and 100 nm particles were trapped in an optical line with a distance of ∼5 μm. Meanwhile, Wu. et al.172 reported the separation of gold nanoparticles with different diameters (50 nm/100 nm and 100 nm/200 nm) in a continuous manner, Fig. 3(K). In this device, two sheath flows confined the sample to a narrow region, and a counter flow created a stagnation point from the opposite side. The larger particles were pushed to the buffer stream, whereas the smaller particles remained within the sample flow under a laser beam so that the particles could be collected at two different outlets. Two-line ‘Y’-shaped optical tweezers patterned in a microfluidic channel were developed to sort yeast cells of various sizes by tuning optical intensities and wavelengths.177 The force acting on larger and smaller cells varies by the two-line tweezers in a way that the particles can be sorted into two different outlets. The optimal sorting was achieved when one line laser power was kept at 35 mW, while the second line laser power was altered from 100 to 190 mW.
Since light has better directionality and can be controlled precisely, it provides excellent efficiency in separating submicron and nanoparticles. Even though optofluidics can easily be integrated within a microfluidic device, heat generation during the operation limits applicability to biological particles due to biocompatibility concerns.178 In addition, the throughput of optofluidics is relatively low since the optical force is generally weaker than other forces.
4 Passive microfluidic techniques
Passive microfluidic techniques utilise intrinsic hydrodynamic forces and channel geometry for particle manipulation. The reported passive microfluidic technologies for submicron and nanoparticle separation include inertial microfluidics,179–182 deterministic lateral displacement,183–185 microfluidic filtration,186–188 pinched flow fractionation,189,190 and viscoelastic microfluidics.191–1934.1 Inertial microfluidics
Inertial microfluidics is a passive microfluidic technology that employs hydrodynamic forces to manipulate particles at a high inertia flow. In contrast to conventional microfluidics, where fluid inertia is negligible and flow is at the Stokes flow region (Re ≪ 1), the flow inertia is significant (1 < Re < 100) and cannot be neglected for inertial microfluidics. The finite fluid inertia causes two intriguing effects: inertial migration and secondary flow. Inertial migration is a phenomenon where randomly dispersed particles migrate to a particular cross-sectional equilibrium position in a straight channel due to a hydrodynamic force, called the inertial lift force. This inertial lift force has two major components: shear gradient lift force and wall lift force. Shear gradient lift force arises due to the interaction of a particle with the background parabolic profile of the fluid velocity, and it tends to move the particle towards the channel walls. The wall lift force occurs due to the flow field interaction between particles and their nearby walls, repelling the particles away from the wall.194 Based on the assumption that the particle is much smaller than the channel dimension, the net inertial lift force (FL) can be expressed as:195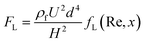 | (10) |
The secondary flow is usually induced in a curved channel or a channel with disturbance obstacles, leveraging from the fluid momentum mismatch in the centre and near-wall region within the curvature or obstacle. The counterbalance of secondary flow drag (FD) and inertial force (FL) determines the final focusing positions and patterns of particles, Fig. 4(A). If FL ≫ FD, particles will focus at the equilibrium positions due to the dominant inertial lift force. If FL and FD are in the same order, the original equilibrium positions will be modified due to the counterbalance of inertial lift force and secondary flow. If FL ≪ FD, the secondary flow becomes dominant, and no focusing will be observed since the particles remain in the mixing streamlines.194,201
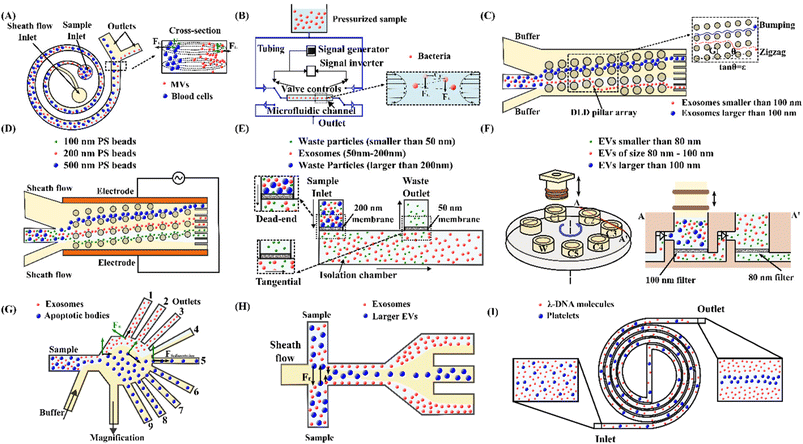 | ||
| Fig. 4 Passive microfluidic techniques for manipulation, focus, and separation of submicron and nanoparticles. (A) High-resolution Dean flow fractionation of MVs in a spiral microchannel.202 (B) Oscillatory microfluidic device for focusing bacteria.179 (C) Separating exosomes based on their sizes using a DLD microfluidic device.183 (D) Tuning of 100 nm, 200 nm and 500 nm polystyrene bead trajectories using an electrokinetic DLD microfluidic device.216 (E) Integrated double-filtration microfluidic device for isolation of exosomes.187 (F) Size-selective filtration of EVs with a movable plunger and rotational chip.224 (G) Microfluidic PFF for separation of exosomes and apoptotic bodies.189 (H) Isolation of exosomes from EVs by microfluidic viscoelastic flows.193 (I) A double spiral microfluidic device for sorting of λ-DNA and blood platelets by viscoelastic microfluidics.191 | ||
In a straight channel, inertial focusing of submicron to nanoparticles is challenging because the inertial lift force is too weak on the tiny particles. According to eqn (10), the inertial lift force is approximately 10−15–10−13 N on 1 μm particles, and the efficient focusing of 1 μm particles requires an unpractical channel length of 1 m to 1 km. To overcome the channel length limitation, Mutlu et al.179 proposed an oscillatory microfluidic device with a straight channel, Fig. 4(B). In this device, the flow direction is altered at a high frequency so that particles move back and forth, enabling an infinite axial movement within a fixed channel length. As flow velocity is symmetrical along the flow axis, the directionality of the inertial lift force acting on the particles is preserved despite switching of the flow direction. This technique demonstrates a successful inertial focusing of synthetic particles as small as 500 nm and a submicron bacterium, Staphylococcus aureus.
In a curved channel, the secondary flow is prone to retain the submicron/nanoparticles within the rotating streamlines because of a tiny inertial lift force. To confine the rotating streams of the secondary flow and suppress its mixing effect, shrunk asymmetric serpentine channels with smaller cross sections (width = 10–20 μm, height = 5–10 μm) were used to focus the submicron (200 nm, 600 nm, 920 nm and 2 μm) polystyrene spheres.180 2 μm and 920 nm particles were concentrated near the channel wall, while the 200 nm and 600 nm particles remained evenly distributed. Although inertial focusing of particles below 1 μm is challenging in a spiral channel, separating these particles was achieved using size-dependent movement trajectories of particles within a cross-sectional rotating stream.182,202 Tay et al.202 developed a high-resolution Dean flow fractionation (HiDFF) technique using a spiral channel to purify sub-micrometre particles. The device comprises two inlets separately for injecting the sheath flow and the sample. The transport distance varies with particle size during the Dean vortices-induced migration, Fig. 4(A). As a proof of concept, fractionation of drug-loaded particles for enhanced drug release and anti-tumour effects and isolation of circulating EVs from whole blood using HiDFF were conducted. Recently, this technique was tailored to simultaneously separate exosomes, MVs and platelets from diluted whole blood for vascular risk profiling in type 2 diabetes mellitus (T2DM).181 The team achieved a separation efficiency of 15 ± 3.8% for nanovesicles, which is three times better than UC.
The inertial microfluidic technology has the advantage of simplicity, high throughput, ease of operation, and good biocompatibility.203–208 However, it is challenging to manipulate submicron and nanoscale particles because the inertial life forces are relatively weak and secondary flow is dominant. Moreover, this method lacks the capacity to further size-fractionated particles within submicron scales.
4.2 Deterministic lateral displacement
Deterministic lateral displacement (DLD) is a hydrodynamic-based microfluidic technique that uses tilted pillars to create unique flow streamlines and displace particles in predetermined paths according to their size.209 Particles either follow a zigzag or bumping mode depending on their size, Fig. 4(C). The cut-off between these two modes is the critical diameter (Dc). Particles smaller than the critical diameter follow the initial streamline, defined as zigzag mode, whereas larger particles bump onto pillars and displace laterally to another streamline, termed as the bumping mode.210 An empirical formula was derived for Dc by Davis et al.211| Dc = 1.4Gε0.48 | (11) |
Wunsch et al.183 developed a nano-DLD technology with a pillar gap of 235 nm to analyse, sort, and collect exosomes based on their sizes, Fig. 4(C). The nano-DLD array displaces larger exosomes (∼100 nm) to the right side of the channel, whereas the smaller exosomes follow a zigzag or partial pumping mode and flow out of the array into the left outlet. However, the single array nano-DLD system has severe limitations in processing throughput. Therefore, the same group developed an integrated nano-DLD chip with 1024 parallel arrays interconnected by a fluidic bus network to enrich EVs from serum and urine.184 Using pillar gaps of 225 nm, the team achieved an EV yield of approximately 50% for both serum and urine. In addition to a high-pressure source, electroosmotic flow (EOF) was employed to drive fluids and particles into a nanopillar chip.185 The design consists of four reservoirs to create EOF with electrical potentials. The flow velocity and direction can be easily controlled by adjusting the electric field applied to the reservoirs. The continuous fractionation of EVs was achieved in this device, where small vesicles of 50–400 nm and larger aggregates of more than 400 nm were collected in different reservoirs.
The critical diameter (Dc) is generally fixed for a DLD device with specific microstructure dimensions. To address this limitation, Gillams et al. demonstrated tuning of particle trajectories and deflected particles smaller than the Dc by incorporating an AC electric field orthogonal to the fluid flow in a DLD device, Fig. 4(D).216 In the AC electric field, particles deflect due to nDEP force, making particles switch from zigzag to bumping mode. Without the electric field, polystyrene particles of 100 nm, 200 nm and 500 nm suspended in high conductivity electrolyte KCl showed a zigzag trajectory because they are significantly below the Dc (1–1.5 μm) of the device. In contrast, by applying 250 VPP at 100 kHz frequency, 500 nm, 200 nm and 100 nm particles were deflected by 900 μm, 540 μm and 100 μm, respectively. Meanwhile, Zeming et al.217 proposed a real-time Dc modulation using an electrostatic force. Various salt ionic concentrations created different thicknesses of the electric double layer, modulating the electrostatic force interactions between the nanoparticle and the pillar walls. In a low ionic concentration, particles experience an increasing lateral displacement, whereas the lateral displacement was lower for identical particles at a high ionic concentration. The team designed a DLD device with a pore size of 2 μm and a gradient space of pillar array, and the corresponding critical diameters ranged from 350 nm to 1000 nm in steps of 50 nm. The device could separate 51 nm and 190 nm fluorescent polystyrene beads in ultrapure DI water and 1000 nm and 1500 nm particles in 25 mM NaCl solution. In addition, to circular pillars, the same group studied the effects of pillar and particle shape and implemented an I-shaped pillar array for bacteria separation.212 The team evaluated the performance of their devices using four species of bacteria. The experimental results indicated that the I-shaped design was the most effective for separating spherical and non-spherical particles.
The DLD technique can separate various particles with high resolution due to its simplicity, easy operation, and high precision. However, the limitations of DLD include low throughput, vulnerability to channel clogging, bulky setup and high fabrication cost.210,218,219
4.3 Microfluidic filtration
Microfluidic filtration employs porous membranes to separate particles by miniaturising ultrafiltration in a microfluidic platform. Particles smaller than the pore sizes pass through the membranes, while larger particles are trapped. There are two types of filtration strategies: dead-end and tangential flow. The sample moves perpendicularly to the membrane surface in the dead-end filtration. In contrast, the particles migrate parallelly along the membrane surface in the tangential flow filtration. This approach has successfully separated exosomes,220 EVs,120,221 viruses,222 and nanoparticles.223A tangential flow filtration device was developed to isolate and purify exosomes.186 A nanoporous polycarbonate track etched (PCTE) membrane with a uniform pore size of 100 nm was stacked by two polymethyl methacrylate (PMMA) layers. Two identical serpentine channels were embedded in the top and bottom PMMA layers. The diluted plasma was injected into the top PMMA layer. The membrane trapped the exosomes while proteins and other small particles were washed out. The trapped exosomes were eluted by injecting DI water through a separate inlet. In a similar study, an enhanced microfluidic device was developed to isolate exosomes by combining dead-end and tangential flow filtrations, Fig. 4(E).187 The sample was injected through the first 200 nm pore membrane as dead-end filtration to eliminate large debris. Next, a second 50 nm pore membrane trapped exosomes through a tangential flow strategy. A similar device was proposed by Liang et al.188 to isolate EVs from urine, using a 200 nm and 30 nm pore membrane, respectively. The device retained and enriched exosomes with a 30–200 nm diameter at a 74.2% recovery rate. Another size-based exosome total isolation chip (ExoTIC) with a series of filter membranes of 200 nm, 100 nm, 80 nm and 50 nm pore sizes was developed to isolate EVs in specific size ranges.220 Clinical samples passed through nanoporous membranes to enrich and purify intact EVs, but free nucleic acids, proteins, lipids and other small fragments were flushed out. This modular device successfully isolated EVs from clinical samples, including plasma, urine, and lavage, providing a 1000-fold yield compared to UC. A similar microfluidic device consisting of a vertically mobile plunger and a rotationally mobile chip was implemented for isolating EVs of multiple sizes, Fig. 4(F).224 The rotational chip has five filtration chambers with nanopore filters with pore diameters of 200 nm, 100 nm, 50 nm and 30 nm, respectively. These chambers are sequentially connected via check valves that allow unidirectional flow when the plunger moves downward to push the fluids. Different chamber connections can separate EVs with various size intervals, and separated EVs can be collected from the respective chambers. Using Pluronic coating on the chamber surfaces to prevent nonspecific adsorption of EVs, the team obtained a high EV recovery rate of 89% from blood plasma.
Furthermore, Wang et al.222 developed a disposable, pump-free, size exclusion-based filter microchip that can be used to separate viruses from unprocessed whole blood. An HIV-spiked whole blood sample was introduced to the microchip by pipetting. The filter membrane retained the blood components larger than the pore size, and a phosphate-buffered saline (PBS) solution was pipetted to push the filtered virus to the outlet. The team was able to separate HIV with a recovery rate ranging from 74.2 ± 7.3% to 84.3 ± 4.7% using a 1 μm pore size filter and 73.1 ± 8.3% to 82.5 ± 4.1% using a 2 μm pore size filter.
Microfluidic filtration is a simple, label-free, and flexible method.225 The cut-off size can be easily controlled by membrane pore size. However, like ultrafiltration, the trapped particles can accumulate on the membrane, causing clogging and fouling issues.226 Furthermore, biological particles may be exposed to shear stresses when squeezed through filter pores. The shear stress may negatively impact the biological integrity of filtered particles.227
4.4 Pinched flow fractionation
Pinched flow fractionation (PFF) employs pinching particles with hydrodynamic flows and separating them according to their size. PFF devices consist of two inlets for sample and secondary flows, a contraction that works as the pinched section, an expansion section and multiple outlets. A secondary flow continuously pinches all particles suspended in a fluid toward the sidewall.228 Smaller particles stay closer to the wall compared to the larger particles. In the expansion section, the lateral difference of differently-sized particles is amplified due to the expanding fluid streamlines, enabling size-based separation of various particles via multiple outlets.229 PFF has been employed for the isolation of synthetic and biological nanoparticles.189,190,230–232Controlling the distribution of the fluid streamline is critical for the sorting of particles in PFF. A PFF device with a magnification channel was reported for non-invasive size-based EV separation.189 In the device, a magnification channel draws partial flow before the expansion section, which helps to alter the streamline separation and vesicle allocation to the outlets, Fig. 4(G). This work defined a magnification ratio as the flow rate ratio in the magnification channel to the total flow rate. The introduction of the magnification channel achieved increased efficiency at minimum sample dilution. A magnification ratio of 75% allowed the authors to observe exosomes at outlets 1 to 3 and apoptotic bodies at outlets 5 to 9. Exosomes with cup-shaped morphology of 30–100 nm were collected at outlet 2, whereas aggregates with sizes of 500 to 2000 nm were gathered at outlet 8. Meanwhile, Hamacher et al.233 developed a microfluidic chip based on PFF to separate spermatozoa from virus-spiked semen. With the optimised flow rate ratios (total flow/sample flow) of 24–32 and fluid removal fraction (percentage of flow to waste outlet) of 2.7%, they recovered 86 ± 6% of spermatozoa and removed 84 ± 4% of cowpea chlorotic mottle virus (CCMV). This technology may help reduce the spread of disease in the context of artificial insemination in the veterinary industry.
Furthermore, another study employed a polydimethylsiloxane (PDMS) membrane microvalve at one outlet to tune the effluent position of the particles.190 The microfluidic device comprised outlet 1 with the microvalve and four other valveless outlets. Flow rate tuning at the corresponding outlet was achieved by controlling the microvalve using positive pneumatic pressure. When the microvalve was partially closed, the flow rate at outlet 1 was reduced, allowing the larger particles to shift away from outlet 1, while the effluent position of smaller particles remained unchanged. Initially, fluorescent polystyrene beads with diameters of 0.5 μm and 0.86 μm both exited through outlet 1 without actuation of the microvalve. But after applying 18.0 kPa to the microvalve, 0.5 μm and 0.86 μm particles were separated successfully, with a purity of 90% at outlet 1 for 0.5 μm particles and 82% at outlet 2 for 0.86 μm particles.
PFF is excellent for isolating particles of various sizes due to its simple design and convenient fabrication processes. However, this technique provides low throughput and low efficiency compared to the other hydrodynamic-based separation techniques.234,235 The substantial sample dilution is another significant drawback of this technique.
4.5 Viscoelastic microfluidics
Viscoelastic microfluidics manipulates particles using elastic effects of a non-Newtonian medium.236 An elastic force arises due to the imbalance of normal stresses in viscoelastic fluid flow. Both the first normal stress difference (N1 = τxx − τyy) and the second normal stress difference (N2 = τyy − τzz) contribute to the elastic force. τxx, τyy and τzz are the normal stresses in streamwise, transverse, and vorticity directions, respectively.237–239 In a diluted viscoelastic solution, N2 is negligible as its magnitude is significantly lower than N1.240 Therefore, the elastic lift force (FE) is proportional to the variation of N1 over the particle size and can be expressed as:241FE = CeLd3∇N1 = −2CeLd3ηpλ∇![[small gamma, Greek, dot above]](https://www.rsc.org/images/entities/i_char_e0a2.gif) 2 2 | (12) |
![[small gamma, Greek, dot above]](https://www.rsc.org/images/entities/i_char_e0a2.gif) is the average shear rate. The combination of the inertial and elastic effects determines the focusing regions of particles in the microchannel.242–244 In viscoelastic fluids, particles migrate to the centre of the channel under the elasto-inertial effect, and the migration speed is proportional to particle size. This size-dependent lateral migration speed creates a lateral gap between various particles. These phenomena have been used in a wide range of particle manipulation and separation applications.191,192,245–247
is the average shear rate. The combination of the inertial and elastic effects determines the focusing regions of particles in the microchannel.242–244 In viscoelastic fluids, particles migrate to the centre of the channel under the elasto-inertial effect, and the migration speed is proportional to particle size. This size-dependent lateral migration speed creates a lateral gap between various particles. These phenomena have been used in a wide range of particle manipulation and separation applications.191,192,245–247
Kim et al.192 firstly discovered that submicron colloidal particles (500, 200 and 100 nm in diameter) and macromolecules (λ-DNA and T4-DNA with radii of gyration of ∼0.69 and 1.5 μm, respectively) could be focused decently in a viscoelastic fluid flow. The discovery indicated the potential of viscoelastic microfluidics for submicron particle manipulation. Since then, this technique has attracted increasing attention for submicron particle focusing and separation.193,246,248–250 For instance, Liu et al.193 proposed a viscoelastic microfluidic device for the size-based separation of exosomes from larger EVs. The device consists of a straight main channel, two inlets for the sample and sheath flow, and three outlets, Fig. 4(H). Both sample and sheath flow contain a low concentration of polyethylene oxide (PEO), so that the elastic lift force exerted on the nanoparticles controls their lateral migration. Larger EVs migrate faster to the centre of the microchannel and are collected from the middle outlets, whereas exosomes moving very slowly are collected at the side outlets. Later, the team employed a modified viscoelastic medium by dissolving λ-DNA in Tris–borate–EDTA (TBE) buffer as the central sample flow and a Newtonian TBE buffer for the sheath flow. They applied the modified viscoelastic microfluidic device to fractionate EV subpopulations, i.e., exosomes, MVs, and apoptotic bodies.246 Nam et al.248 reported a sheathless viscoelastic microfluidic device to isolate submicrometer platelet-derived microparticles (PDMPs) from whole blood. Since the diameters of RBCs and platelets are significantly higher than the diameter of PDMPs, they can be concentrated at the channel centre and filtered out from the middle outlet. In contrast, PDMPs are uniformly distributed and partially extracted from the side outlets.
Apart from straight channels, spiral and wavy channels have also been explored for viscoelastic manipulation of submicron and nanoparticles. A double spiral microchannel has been developed to manipulate synthetic nanoparticles and λ-DNA molecules, Fig. 4(I).191 In this geometry, the smaller particles (49, 100, 500 and 1000 nm in diameter) were focused along the channel centre. In contrast, the larger particles (2000 nm in diameter) focused off the centre near the two side walls. The device demonstrated high-quality sorting of a binary mixture of nanoparticles. Both polystyrene particles (100/2000 nm) and λ-DNA/blood platelet mixtures were sorted with separation efficiencies above 95%. Recently, a spiral channel device was applied to separate U87 glioblastoma cell-derived small and medium size EVs. In this work, medium EVs (mEVs) could be focused, but sEVs were too small to be focused and randomly dispersed.250 They recovered 55% of sEVs with 6% contamination of mEVs and 80% of mEVs with 22% contamination of sEVs after one round of recycling.
Furthermore, a wavy microchannel was developed for submicron particle focusing and exosome sorting using viscoelastic fluids.249 Besides the elastic lift force, the wavy channel induces an additional Dean flow, facilitating effective focusing of particles towards the channel centre. Large particles (300, 500 and 1000 nm) were effectively focused, whereas small particles (100 nm) were kept widely distributed at the end of the channel. After applying a sheath flow to pinch all particles along the sidewalls at the inlet, the device was used to sort exosomes from large EVs based on the size-dependent migration speed.
Viscoelastic microfluidics is a promising technique for manipulating submicron particles. This technique is label-free, simple, and biocompatible.49 It has a much higher separation resolution (∼O(100 nm)) than inertial microfluidics (∼O(1 μm)), since the elastic lift force is at least one order of magnitude higher than that of the inertial lift force for submicron particles.193 In addition, viscoelastic microfluidics uses relatively larger channel space than DLD and microfiltration, lessening channel blockage issues and fabrication costs. One limitation is the need for specialised viscoelastic medium preparation. The elasticity enhancers may contaminate biological samples, and the enhanced viscosity may require a higher pressure drop to drive the sample.238 Moreover, the throughput is still low compared to the conventional techniques. Finally, manipulating nanoscale particles is still a major challenge and has been less explored.191
5 Hybrid microfluidic techniques
Hybrid microfluidic techniques integrate two or more techniques discussed above in a serially connected or physically coupled manner.33 Hybrid microfluidics can combine the advantages of each method and potentially overcome the limitations of individual ones through the synergy of inherent characteristics. Therefore, superior performance and more versatility are expected. However, due to the complexity of the integration, physical coupling and device design, very few studies have been reported on manipulating submicron and nanoparticles using hybrid microfluidic techniques.5.1 DEP–MP
Integrating DEP and magnetophoresis in a single microfluidic device achieved high-purity sorting of multiple bacterial targets.251 The integrated dielectrophoretic–magnetic activated cell sorter (iDMACS) consists of a dielectrophoretic separation module and a serial magnetic separation module, Fig. 5(A). Three distinct bacterial clones of an E. coli strain were labelled with DEP, magnetic and no tags. In the DEP separation module, the bacteria strain labelled with DEP tags was efficiently deflected from other bacteria and eluted from the upper outlet. In contrast, the magnetic tagged and non-tagged strains were retained in the fluid streamline and entered into the downstream magnetic separation module. In the magnetic separation module, the external magnetic field activated the titanium/nickel structure and captured the magnetically tagged bacteria, whereas non-tagged bacteria continuously passed through. After the entire sample was processed, the magnetic tagged bacteria strain was retrieved by removing the external magnetic field. Target bacteria strains with DEP and magnetic tags achieved a separation purity of 98.6% and 95.6%, respectively, without cross-contamination between targeted strains. The waste fraction consisted mainly of non-tagged cells (99.74%), and the de-labelling of tagged bacteria during the separation process probably contributed to the slight contamination.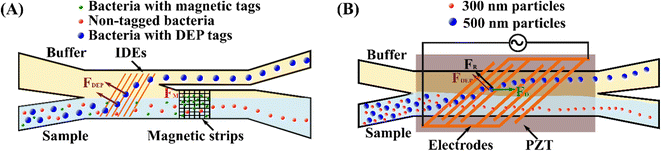 | ||
| Fig. 5 Hybrid microfluidic techniques for sorting submicron and nanoparticles. (A) Sorting multiple bacterial targets by serially combining DEP and MP in an integrated DEP–MP cell sorter.251 (B) A vDLD system for sorting sub-micrometre particles by coupling acoustic and DEP forces.252 | ||
5.2 AP–DEP
A virtual DLD (vDLD) microfluidic device was developed by integrating acoustic and DEP forces, Fig. 5(B).252 An array of IDTs on a piezoelectric lithium niobate substrate can generate both acoustic and electric force fields. Acoustic forces allow for isolating particles based on mechanical properties such as compressibility and density, whereas DEP can sort particles based on electrical properties such as permittivity. Either of these forces can be employed for the separation of particles. By tuning the strength of the dominant force, larger particles are captured in the force field and deflected laterally, while smaller particles are not affected. The device demonstrated the separation of 500 nm particles from 300 nm particles with an efficiency of 87%. Later, the vDLD device was modified to sort exosomes and MVs, taking advantage of combined acoustic and DEP forces with tilted-angle IDTs.253 In this device, the cut-off size of particles for separation (or critical diameter) was determined by coupling acoustic radiation, DEP and fluid drag forces. The concurrent acoustic and DEP forces significantly reduced the critical diameter while doubling the lateral displacement of particles compared to the previous version. The combined acoustic/electric approach achieved exosome isolation of more than 95% purity and 81% recovery.6 Nanoparticle detection techniques after separation
Due to the vast utilisation of nanoparticles, more precise and reproducible characterisation techniques are also required after separation. Various nanoparticle detection techniques have been developed based on their inherent physical and chemical properties. Here, we mainly discuss the seven most common methods: electron microscopy, dynamic light scattering, nanoparticle tracking analysis, atomic force microscopy, fluorescence analysis, colourimetric detection, and electrochemical detection.6.1 Electron microscopy
Compared with the traditional optical microscopy, electron microscopy (EM) uses electrons as the source of illuminating radiation instead of visible light to obtain sample images, Fig. 6(A). The shorter wavelength of the electron beam overcomes visible light's resolution limitation, which renders higher resolution up to 1–5 nm. This technique has emerged as a powerful tool for characterising a wide range of biological and non-biological particles.255 Typically, according to the different imaging mechanisms, EM can be divided into two main types: transmission electron microscopy (TEM) and scanning electron microscopy (SEM). TEM uses a high-energy electron beam transmitted through the specimen to obtain a projection image. Meanwhile, the electron diffraction pattern gained by transmitted electrons resulted in Bragg scattering which provides crystal structure information of the samples. Therefore, TEM would be an ideal technology for metal or crystal particle identification and analysis. Under specific sample preparation processes, TEM can also be utilised to analyse nanoparticle size and shape, providing the most accurate estimations of nanoparticle homogeneity for applications such as cell biology.256 In contrast, SEM works by scanning the surface of an object with a focused beam of electrons and images the sample by collecting electrons reflected from or knocked off the sample surface, Fig. 6(A).257 As a result, SEM can acquire clear 3D morphology with a depth of field and information about the sample's surface and composition.258–260 In addition, SEM can operate in transmission mode, known as transmission scanning electron microscopy (TSEM). It has shown immense potential in the accurate measurement of dimensions of nanoparticles.261 Although the extremely high resolution of TEM and SEM advances the characterisation of particles, the complicated sample preparation procedures (dehydration, conductive treatment, thinning) and harsh testing requirements (high vacuum, high-energy electron beam) make it complicated to get in situ results, and may also cause damage to the subtle samples. Approaches like cryo-electron microscopy have been implemented to avoid the effects of the aforementioned drawbacks. EM is not an excellent choice for real-time and non-invasive particle measurement.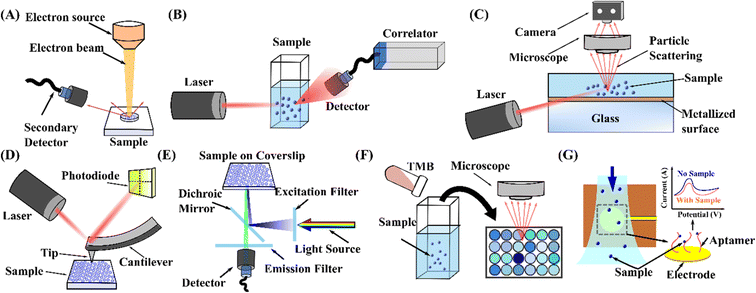 | ||
| Fig. 6 Schematic representation of the working principle of nanoparticle measurement and detection techniques. (A) Electron microscopy.260 (B) DLS.265 (C) NTA.267 (D) AFM.279 (E) Fluorescence detection.286 (F) Colourimetric detection.294 (G) Electrochemical detection.304 | ||
6.2 Dynamic light scattering
Dynamic light scattering (DLS) is commonly used to measure the size of nanoparticles suspended in a liquid with a laser beam passing through the bulk sample. A photodetector is usually positioned at an angle with the incident light to collect the scattered light intensity from the particles, Fig. 6(B). The fluctuation of the scattered light due to the Brownian motion of particles correlates with their velocity. An autocorrelator estimates the diffusion coefficient and the hydrodynamic radius using the Stokes–Einstein relationship.262 DLS is a non-invasive, highly reproducible, time-efficient, and low labour-intensity technique for characterising nanoparticles.263 However, due to the difficulty of extracting specific particle size information from the autocorrelation function, DLS cannot distinguish between polydispersed particles of a similar size range and becomes less reliable when measuring samples with broad particle size distribution.264 In addition, as the obtained decay rate of the correlation from the initial state is an average value, the distribution data acquired from DLS may not be reliable. The particle aggregation may also result in more significant measurement errors.265 Nonetheless, the high sensitivity of DLS can be utilised for disease diagnosis, such as rapid screening of virus–antibody binding by detecting the formation of virus–antibody aggregates.2666.3 Nanoparticle tracking analysis
Nanoparticle tracking analysis (NTA) utilises light scattering to acquire the size, distribution and concentration of nanoparticles in a dispersed medium.264 In contrast to DLS, NTA technology is equipped with a charge-coupled device (CCD) or complementary metal oxide semiconductor (CMOS) sensor camera to enable the activity processing of particles, estimating their speed due to Brownian motion, Fig. 6(C).267 Therefore, the hydrodynamic diameter of a particle can be evaluated by the Stokes–Einstein equation.268 NTA provides information on the size distribution and concentrations of monodisperse and polydisperse particle samples and can also be employed to differentiate single nanoparticles based on their reflective index.269,270 Furthermore, NTA offers greater flexibility and robustness in characterising biological nanoparticles and importantly, it is not biased towards larger nanoparticles or aggregates compared to other measurement techniques.271–273 Nevertheless, NTA is time-consuming and requires expert operational skills to conduct the measurements.269 Moreover, a large particle in the solution, such as debris or impurities, will scatter at high intensity and introduce significant errors in the analysis. Furthermore, since the measuring chip of the fluid channel only contains a small volume, repetitive measurements should be carried out to obtain reliable results.6.4 Atomic force microscopy
Atomic force microscopy (AFM) provides high spatial resolution in nanoparticle measurements.274 A tiny sharp tip connected to an ultra-small force-sensitive cantilever is used as the probe for x–y grid scanning the samples by direct contact with the sample surface. A laser beam detects and measures the deflection of the cantilever in AFM, Fig. 6(D).267 Therefore, AFM can simultaneously measure the size distribution and mechanical properties such as friction, energy dissipation, tension, pressure, and reversible and irreversible deformation.275–277 Since the operating procedure does not require specific sample preparation and measurement environment, AFM can also characterise biological samples in liquids, such as those found under physiological conditions.278 Due to these superior capabilities, AFM has the potential to be used as a diagnostic tool for cancer. For instance, the changes in ultrastructure and mechanical properties within tumour tissues and cells can be employed as a marker for clinical diagnostics.279 However, AFM measurement of soft matter shows errors in height due to tip pressing and lateral size due to tip-broadening effects. To overcome this drawback, a tapping mode has been utilised to measure soft matter.280 A constant amplified oscillation was applied to the cantilever during the tapping mode. The piezo signal varies when the tip comes close to the surface, enabling the morphology scanning of samples such as proteins and exosomes. Meanwhile, a novel image reconstruction algorithm has also been developed for high-speed AFM known as localisation AFM (LAFM), allowing the extraction of high-resolution information in the study of single biomolecules beyond the tip radius resolution limit.281 Moreover, the scanning measurement mechanism allows AFM to obtain accurate size distribution information of the polydisperse system, which is not applicable in DLS.282 Nevertheless, bubbles or droplets of similar size and shape in the sample can interfere with particle classification. Therefore, combining the characterisation results with other techniques is encouraged for better results.6.5 Fluorescence detection
Fluorescence imaging technology is adapted for detecting nanoparticles in microfluidics due to its accuracy and sensitivity.227 Fluorescence is the emission of light from a substance that has absorbed light or other electromagnetic radiation. Its intensity, wavelength and luminescence time depend on fluorophore material properties. Fluorescent molecules are used to tag biological particles to facilitate detection in microfluidic channels with the aid of fluorescence emission parameters.138,283 Fluorescence imaging has shown good accuracy and high sensitivity in exosome detection. Exosomes labelled with fluorescent dyes can be quantified using standard plate readers by measuring the fluorescence intensities at excitation and emission wavelengths, Fig. 6(E).284 In addition, immunofluorescently stained exosomes can be characterised and quantified using an inverted fluorescent microscope.285 This technique provides the highest spatial resolution for disease diagnosis at the microscopic level.286 Due to its unique advantages, it has been employed for various chemical and biological applications.287,288 However, prolonged exposure to fluorescent light can result in loss of intensity, affecting the accuracy of the detection results.2896.6 Colourimetric detection
Colourimetric detection allows direct visualisation of samples to detect nanoparticles. This method works by comparing or measuring the colour depth of chromogenic substances. 3,3′,5,5′-Tetramethylbenzidine (TMB) is widely used as a chromogenic substance for exosome detection due to its high sensitivity and high colour purity.290,291 The oxidation of TMB provides the ability to obtain naked eye read-outs by converting a colourless solution to blue colour.292 The absorbance change is directly proportional to the concentration of target particles so that captured particles can be quantitively determined, Fig. 6(F).293 Recently, microfluidic technology has been combined with colourimetric detection to visualise exosomes and EVs.221,294 Furthermore, this detection technique enables cost-effective and straightforward monitoring of heavy metal ions associated with environmental pollution and health-related risks.295 Nevertheless, it is less sensitive and requires higher sample concentration.296,297 Moreover, colourimetric detection exhibits constraints in determining individual components from a mixture.2986.7 Electrochemical detection
The electrochemical detection technique detects electrical signal changes between electrodes by a target-induced chemical reaction.299 Due to chemical or biochemical reactions, electrical properties around electrodes can vary, and changes in the electrical signal, such as current, can be detected. With the maturity of micromachining technologies, the integrated microelectrodes implemented on microfluidic platforms have enabled the detection of biomolecules with high sensitivity.300–302 For instance, electrodes modified with a CD63-specific aptamer have been developed to capture exosomes and emit an electrochemical signal for quantification.8,303,304 Electrochemical detection has many advantages, such as reduced reagent requirements, high sensitivity, low cost, and rapid detection.305–307 Besides, it is independent of the optical path length and sample turbidity.308 However, the inability to detect non-electroactive nanoparticles and the need for solutions with strong acidic or oxidant characters prevent colourimetric detection from being a universal detection technique.3097 Discussion and perspectives
This paper studied the conventional and microfluidic technologies for the manipulation and separation of sub-micron and nanoparticles. We discussed the principle of five standard traditional techniques: ultracentrifugation, ultrafiltration, size exclusion chromatography, precipitation and immunoaffinity capture. We subsequently classified the microfluidic technologies as active, passive and hybrid technologies based on the sources of the main manipulating forces. We elaborated on the physics of manipulating forces and the working mechanism of reported devices and discussed the features, limitations and applications of these techniques. Table 1 summarises and compares the principles, applicable particle sizes, particle types, separation efficiencies and throughput, as well as the merits and demerits of both conventional and microfluidic techniques. The conventional techniques generally possess the advantages of time efficiency, high yield, ease of use and good reproducibility, but they are limited by the high cost and low purity. Microfluidic technologies, which can manipulate fluid flow and particles precisely in a microscale space, are promising alternatives to provide complementary capacity for enhanced separation resolution. Although significant progress has been achieved in microfluidic technology with a broad range of applications, there is still a considerable gap to be filled to enable microfluidic technology to be as mature as conventional technologies.The major challenge associated with the effective manipulation and separation of submicron and nanoparticles is the minuscule size of the particles. Both the active and passive manipulating forces are proportional to a higher order of particles size (F ∼ an; n > 1, e.g., n = 3 for dielectrophoresis, magnetophoresis, and viscoelastic forces) than the fluidic drag force (F ∼ a), and the size reduction of particles toward the nanoscale sharply decreases the manipulating forces. Moreover, the manipulating forces drop much faster than the drag force when reducing the particle size. In contrast, Brownian motion becomes more significant for nanoparticles, negatively impacting the effective manipulation of nanoparticles. To address these issues, possible solutions may arise from the following perspectives: (i) the amplification of the overall size of target nanoparticles; (ii) the enlargement of local physical strength and gradient; and (iii) the combination of multiple manipulating forces. One way to amplify the overall size of target nanoparticles is by binding the nanoparticles specifically with larger microscale beads. Target nanoparticles can be tagged to microbeads of distinct sizes. Subsequently, the manipulating force will be scaled up as on microparticles, and the size difference of nanoparticles in the nanoscale can be translated into the difference in microscale, significantly lessening the technical challenges of separation. A successful example was reported by Sarkar et al.,310 who used microbeads of different sizes to selectively bind multiple proteins and separate these microbeads successfully using an inertial microfluidic device. Therefore, the various proteins can be divided in a continuous manner. Another microscale approach is to induce the self-aggregation of target nanoparticles using methods such as precipitation. This method also results in a larger cluster of nanoparticles, but the co-precipitation of non-target particles should be avoided.
Enlarging the local physical strength and its gradient is another feasible method to amplify manipulating forces, because manipulating forces are based on electric and magnetic fields and depend not only on the strength of these physical fields but also on their gradients, as shown in eqn (4) and (9). Apart from applying a high electrical or magnetic field, well-defined microscale structures (e.g., insulating microposts,132 microelectrodes,134,135 and magnetic micropole arrays146) with microscale nonuniformity close to the manipulation region can induce high physical gradients. Therefore, the resulting forces are sufficient to manipulate nanoscale particles. Furthermore, combining multiple manipulating forces is also a strongly feasible approach. Although a single manipulating force is not strong enough or not sensitive enough to distinguish nanoparticle populations, adding up multiple forces on a mixture of nanoparticles could enhance the manipulation capacity. The design of multiple physical coupling is challenging, and only one successful work has been reported thus far. Ai's group has developed a novel microfluidic device combining acoustic and DEP forces to sort exosomes and MVs.253 In their work, coupling acoustic with DEP forces resulted in a much larger net force to counterbalance the mainstream drag force, enabling an effective manipulation of nanoparticles with a significantly reduced size. Besides, a more significant lateral net force allowed an enhanced deflection of particles, magnifying the lateral displacement of particles. The combined acoustic/electric technology improved exosome isolation in purity and recovery. A precise design of each manipulating physical force and how they are coupled is critical to ensure the successful coupling of multiple physics. More efforts are needed in the future to explore different coupling schemes of various forces. Another way to combine various force fields is to connect several manipulating sections consecutively. In each section, a manipulating force field dominates the isolation of particles based on the local selection criterium. Multistage processing, passing through several consecutive stages, is expected to promote separation purity and enhance the versatility of devices in the fractionation of complex samples. For consecutive connection, the adjacent stages are linked by the fluid flow, and the flow rate of the outlet upstream must match the inlet of the second stage. The flow speed needs to be tuned to meet the operating flow range for upstream and downstream stages. One way is to modify the channel cross-sectional area at different sections to optimise the linear flow speed. The other way is to exhaust partial fluids upstream or supplement additional fluid to lower or enhance the downstream flow speed. In addition, a pumping unit between the connective units can adjust the flow speed for the second stage. In this case, a large reservoir may collect and store the outlet sample from the first stage.33
For passive microfluidic technologies, the channels or gap spaces were generally scaled down to submicron and even nanoscale to adapt to the nanoparticles.180,183 However, minuscule fluidic channels and gap spaces inevitably bring about a very high fluidic resistance, and thus, a powerful pumping system is needed to drive the sample into the device. Moreover, the high bonding strength of device layers is required to prevent delamination, and a more robust assembly connecting tubing to the microfluidic device input ports is also necessary. Furthermore, a deformation of the microchannel may become more significant under high pressure, which may impair the functionality of the device. These all add to the technical challenges of microfluidic technology for processing nanoparticles. Therefore, device materials, fabrication, assembling, and pumping are critical in designing microfluidics for nanoscale particles.
The miniaturisation of conventional techniques into a microfluidic platform can reduce reagent consumption and sample volume. Due to the significant advantages of precisely controlling fluid flow in microfluidics, controlling fluid motion and the procedure of multiple fluids to filtrate multiple components becomes feasible. The miniaturisation of ultrafiltration successfully formed microfluidic filtration by embedding membranes into microfluidic channels.311 The nanoparticles can be controlled to move at different orientations toward the membranes and at multiple stages to achieve the versatility of the devices. More recent work reported the miniaturisation of size exclusion chromatography (SEC) in microfluidics.312 Benefiting from the precise control of fluid flow in microfluidics, the authors successfully regulated the hydraulic resistances in a T-junction channel to address the significant challenge of on-chip sample injection. Combining precipitation with microfluidics is also promising but not fully explored. Microfluidic devices can potentially be utilised to control the mixing of precipitation agents and samples. In addition, microfluidic devices can continuously remove the precipitated target particles from the suspension without requiring centrifugation or filtration.
Microfluidics can miniaturise conventional techniques to improve the separation resolution. It also has the benefit of handling samples of small volumes. However, a larger sample volume is required for some applications, such as isolation of exosomes for therapeutic purposes313 or treatment of the environment and industry nanoparticles.314,315 The throughput of microfluidic technologies needs to be scaled up to reach or even exceed the level of conventional technologies in fulfilling these tasks. Parallelisation of microfluidic channels is a possible way forward, and careful design and optimisation are necessitated to ensure uniformity between multiple channels. For passive microfluidic techniques, the devices work only by channel geometry or hydrodynamic forces. For the microchannels with one inlet, scaling up the throughput can be readily achieved by patterning multiple functional channels along the horizontal, radial, and vertical directions.316–319 However, more efforts should be made to the microchannels with two or more inlets to ensure fluid flow consistency between parallel channels. Moreover, upscaling the design for active microfluidic devices is more challenging. Active microfluidic techniques use external acoustic, electric, magnetic and optical force fields to manipulate particles. Consistency in fluid flow and external force fields between multiple parallel channels demands a delicate design. In addition to an elevated throughput, a parallelisation of channels can also reduce the overall fluidic resistance since the net channel cross-sectional area is amplified by the number of parallel channels and the fluidic resistance is reversed to the channel cross-sectional area.
In conclusion, microfluidic technologies have distinct advantages and benefits compared to conventional techniques regarding separation resolution and purity. Although significant progress has been achieved in microfluidic submicron and nanoparticle manipulation, it is still in its early development stage. A broad field region is still not explored and exploited in the aspects of the fundamental mechanism, device design and applications. Besides the manipulation and separation, integrating other functional units, such as mixing, synthesis, counting, detection, and analysis, into one microfluidic platform is an excellent prospect for novel automated and portable solutions. To elevate microfluidic technologies to the maturity of conventional methods, further significant efforts are urgently needed to tackle the current limitations on throughput, manipulating accuracy, separation resolution, device material, and connection strength.
Lists of abbreviations
| EVs | Extracellular vesicles |
| MVs | Microvesicles |
| DEP | Dielectrophoresis |
| FFF | Field flow fractionation |
| DLD | Deterministic lateral displacement |
| PFF | Pinched flow fractionation |
| AP–DEP | Acoustic–dielectrophoresis |
| DEP–MP | Dielectrophoretic mangenetophoresis |
| AFM | Atomic force microscopy |
| NTA | Nanoparticle tracking analysis |
| DLS | Dynamic light scattering |
| UC | Ultracentrifugation |
| DGUC | Density gradient ultracentrifugation |
| UF | Ultrafiltration |
| SEC | Size exclusion chromatography |
| AP | Acoustophoresis |
| SAW | Surface acoustic wave |
| BAW | Bulk acoustic wave |
| IDT | Interdigitated transducer |
| SSAW | Standing surface acoustic wave |
| TSAW | Travelling surface acoustic wave |
| RBC | Red blood cell |
| WBC | White blood cell |
| CP | Capillary electrophoresis |
| GE | Gel electrophoresis |
| DC | Direct current |
| ELDS | Evaporative light scattering detector |
| AC | Alternating current |
| CM | Clausius–Mossotti |
| DC-iDEP | Direct current insulator-based dielectrophoresis |
| ACE | Alternating current electrokinetics |
| DENV | Dengue virus |
| 4G2 | Mouse anti-flavivirus monoclonal antibody |
| MP | Magnetophoresis |
| SPLITT | Split-flow lateral-transport thin |
| HiDFF | High-resolution Dean flow fractionation |
| T2DM | Type 2 diabetes mellitus |
| EOF | Electroosmotic flow |
| PCTE | Polycarbonate track etched |
| PMMA | Polymethyl methacrylate |
| ExoTIC | Exosome total isolation chip |
| PBS | Phosphate-buffered saline |
| CCMV | Cowpea chlorotic mottle virus |
| PDMS | Polydimethylsiloxane |
| PEO | Polyethylene oxide |
| TBE | Tris–borate–EDTA |
| PDMP | Platelet-derived microparticles |
| sEV | Small extracellular vesicle |
| mEV | Medium extracellular vesicle |
| iDMACS | Integrated dielectrophoretic–magnetic activated cell sorter |
| vDLD | Virtual deterministic lateral displacement |
| EM | Electron microscopy |
| TEM | Transmission electron microscopy |
| SEM | Scanning electron microscopy |
| 3D | Three dimensional |
| TSEM | Transmission scanning electron microscopy |
| CCD | Charge-coupled device |
| CMOS | Complementary metal oxide semiconductor |
| LAFM | Localisation atomic force microscopy |
| TMB | 3,3′,5,5′-Tetramethylbenzidine |
Author contributions
J. Z. and N. T. N. conceived the idea and designed and supervised the project. S. H. conducted the literature study, drew the figures, and wrote the manuscript. J. Z. contributed to the discussion and perspectives. H. C. and A. M. contributed to drawing the figs. L. O. contributed to the measurement and detection techniques. J. Z., N. T. N., H. A., G. K., and N. K. revised and modified the manuscript. All the authors provided critical feedback and read and approved the manuscript.Conflicts of interest
The authors have declared no conflict of interest.Acknowledgements
The authors acknowledge the support from the Australian Research Council (ARC) Discovery Project (Grant No. DP180100055), ARC DECRA fellowship (Grant No. DE210100692) and ARC Future Fellowships (FT180100361).References
- C. M. Abreu, B. Costa-Silva, R. L. Reis, S. C. Kundu and D. Caballero, Lab Chip, 2022, 22, 1093–1125 RSC.
- J.-J. Lee, K. J. Jeong, M. Hashimoto, A. H. Kwon, A. Rwei, S. A. Shankarappa, J. H. Tsui and D. S. Kohane, Nano Lett., 2014, 14, 1–5 CrossRef PubMed.
- T. Hamacher, J. T. W. Berendsen, J. E. van Dongen, R. M. van der Hee, J. J. L. M. Cornelissen, M. L. W. J. Broekhuijse and L. I. Segerink, Lab Chip, 2021, 21, 4477–4486 RSC.
- M. Bouri, R. Salghi, M. Algarra, M. Zougagh and A. Ríos, RSC Adv., 2015, 5, 16672–16677 RSC.
- D. Maiti, X. Tong, X. Mou and K. Yang, Front. Pharmacol., 2019, 9, 1401 CrossRef PubMed.
- Y. Xie, J. Rufo, R. Zhong, J. Rich, P. Li, K. W. Leong and T. J. Huang, ACS Nano, 2020, 14, 16220–16240 CrossRef PubMed.
- S. Z. Shirejini and F. Inci, Biotechnol. Adv., 2022, 54, 107814 CrossRef CAS PubMed.
- X. Ma, Y. Hao and L. Liu, Int. J. Nanomed., 2021, 16, 7575–7608 CrossRef CAS PubMed.
- W. Jiang, B. Y. S. Kim, J. T. Rutka and W. C. W. Chan, Nat. Nanotechnol., 2008, 3, 145–150 CrossRef.
- A. Albanese, P. S. Tang and W. C. W. Chan, Annu. Rev. Biomed. Eng., 2012, 14, 1–16 CrossRef.
- M. Wu, Z. Mao, K. Chen, H. Bachman, Y. Chen, J. Rufo, L. Ren, P. Li, L. Wang and T. J. Huang, Adv. Funct. Mater., 2017, 27, 1606039 CrossRef.
- P. Arosio, T. Müller, L. Mahadevan and T. P. J. Knowles, Nano Lett., 2014, 14, 2365–2371 CrossRef.
- M. A. Livshits, E. Khomyakova, E. G. Evtushenko, V. N. Lazarev, N. A. Kulemin, S. E. Semina, E. V. Generozov and V. M. Govorun, Sci. Rep., 2015, 5, 17319 CrossRef.
- J. Lohwacharin and S. Takizawa, J. Membr. Sci., 2009, 326, 354–362 CrossRef CAS.
- G.-T. Wei and F.-K. Liu, J. Chromatogr. A, 1999, 836, 253–260 CrossRef.
- K. Y. Chung, J. M. Quek, S. H. Neo and H. P. Too, J. Mol. Diagn., 2020, 22, 610–618 CrossRef.
- D. Brambilla, L. Sola, A. M. Ferretti, E. Chiodi, N. Zarovni, D. Fortunato, M. Criscuoli, V. Dolo, I. Giusti, V. Murdica, K. Kluszczyńska, L. Czernek, M. Düchler, R. Vago and M. Chiari, Anal. Chem., 2021, 93, 5476–5483 CrossRef PubMed.
- A. Liga, A. D. B. Vliegenthart, W. Oosthuyzen, J. W. Dear and M. Kersaudy-Kerhoas, Lab Chip, 2015, 15, 2388–2394 RSC.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef PubMed.
- J. C. Contreras-Naranjo, H.-J. Wu and V. M. Ugaz, Lab Chip, 2017, 17, 3558–3577 RSC.
- M. Sivaramakrishnan, R. Kothandan, D. K. Govindarajan, Y. Meganathan and K. Kandaswamy, Curr. Opin. Biomed. Eng., 2020, 13, 60–68 CrossRef.
- S. Zhao, M. Wu, S. Yang, Y. Wu, Y. Gu, C. Chen, J. Ye, Z. Xie, Z. Tian, H. Bachman, P.-H. Huang, J. Xia, P. Zhang, H. Zhang and T. J. Huang, Lab Chip, 2020, 20, 1298–1308 RSC.
- M. Dimaki, M. H. Olsen, N. Rozlosnik and W. E. Svendsen, Micromachines, 2022, 13, 866 CrossRef PubMed.
- S. Khizar, H. Ben Halima, N. M. Ahmad, N. Zine, A. Errachid and A. Elaissari, Electrophoresis, 2020, 41, 1206–1224 CrossRef PubMed.
- H. Zhao, L. K. Chin, Y. Shi, P. Y. Liu, Y. Zhang, H. Cai, E. P. H. Yap, W. Ser and A.-Q. Liu, Sens. Actuators, B, 2021, 331, 129428 CrossRef.
- Q. Zhao, D. Yuan, J. Zhang and W. Li, Micromachines, 2020, 11, 461 CrossRef.
- M. Al-Fandi, M. Al-Rousan, M. A. K. Jaradat and L. Al-Ebbini, Robot. Comput.-Integr. Manuf., 2011, 27, 237–244 CrossRef.
- Y. Cheng, Y. Wang, Z. Ma, W. Wang and X. Ye, Lab Chip, 2016, 16, 4517–4526 RSC.
- A. L. Vig and A. Kristensen, Appl. Phys. Lett., 2008, 93, 203507 CrossRef.
- G. Romeo, G. D'Avino, F. Greco, P. A. Netti and P. L. Maffettone, Lab Chip, 2013, 13, 2802–2807 RSC.
- J. Zhou, P. Mukherjee, H. Gao, Q. Luan and I. Papautsky, APL Bioeng., 2019, 3, 041504 CrossRef.
- S. Yan, J. Zhang, C. Pan, D. Yuan, G. Alici, H. Du, Y. Zhu and W. Li, J. Micromech. Microeng., 2015, 25, 084010 CrossRef.
- H. Cha, H. Fallahi, Y. Dai, D. Yuan, H. An, N.-T. Nguyen and J. Zhang, Lab Chip, 2022, 22, 423–444 RSC.
- S. Lin, Z. Yu, D. Chen, Z. Wang, J. Miao, Q. Li, D. Zhang, J. Song and D. Cui, Small, 2020, 16, 1903916 CrossRef PubMed.
- S. Ibsen, A. Sonnenberg, C. Schutt, R. Mukthavaram, Y. Yeh, I. Ortac, S. Manouchehri, S. Kesari, S. Esener and M. J. Heller, Small, 2015, 11, 5088–5096 CrossRef PubMed.
- T. Salafi, K. K. Zeming and Y. Zhang, Lab Chip, 2016, 17, 11–33 RSC.
- M. Tayebi, Y. Zhou, P. Tripathi, R. Chandramohanadas and Y. Ai, Anal. Chem., 2020, 92, 10733–10742 CrossRef PubMed.
- D. Yang, W. Zhang, H. Zhang, F. Zhang, L. Chen, L. Ma, L. M. Larcher, S. Chen, N. Liu, Q. Zhao, P. H. L. Tran, C. Chen, R. N. Veedu and T. Wang, Theranostics, 2020, 10, 3684–3707 CrossRef.
- J. Chen, P. Li, T. Zhang, Z. Xu, X. Huang, R. Wang and L. Du, Front. Bioeng. Biotechnol., 2022, 9, 811971 CrossRef PubMed.
- F. Gao, F. Jiao, C. Xia, Y. Zhao, W. Ying, Y. Xie, X. Guan, M. Tao, Y. Zhang, W. Qin and X. Qian, Chem. Sci., 2019, 10, 1579–1588 RSC.
- S. Cai, B. Luo, P. Jiang, X. Zhou, F. Lan, Q. Yi and Y. Wu, Nanoscale, 2018, 10, 14280–14289 RSC.
- L. Ding, X. Yang, Z. Gao, C. Y. Effah, X. Zhang, Y. Wu and L. Qu, Small, 2021, 17, 2007174 CrossRef.
- J. D. Robertson, L. Rizzello, M. Avila-Olias, J. Gaitzsch, C. Contini, M. S. Magoń, S. A. Renshaw and G. Battaglia, Sci. Rep., 2016, 6, 27494 CrossRef PubMed.
- X. Sun, S. M. Tabakman, W.-S. Seo, L. Zhang, G. Zhang, S. Sherlock, L. Bai and H. Dai, Angew. Chem., Int. Ed., 2009, 48, 939–942 CrossRef.
- W. Wang, J. Luo and S. Wang, Adv. Healthcare Mater., 2018, 7, 1800484 CrossRef.
- H. Shao, H. Im, C. M. Castro, X. Breakefield, R. Weissleder and H. Lee, Chem. Rev., 2018, 118, 1917–1950 CrossRef CAS PubMed.
- C. Gardiner, D. D. Vizio, S. Sahoo, C. Théry, K. W. Witwer, M. Wauben and A. F. Hill, J. Extracell. Vesicles, 2016, 5, 32945 CrossRef.
- G. K. Patel, M. A. Khan, H. Zubair, S. K. Srivastava, M. Khushman, S. Singh and A. P. Singh, Sci. Rep., 2019, 9, 5335 CrossRef.
- J. Wang, P. Ma, D. H. Kim, B.-F. Liu and U. Demirci, Nano Today, 2021, 37, 101066 CrossRef CAS PubMed.
- C. Chen, J. Skog, C.-H. Hsu, R. T. Lessard, L. Balaj, T. Wurdinger, B. S. Carter, X. O. Breakefield, M. Toner and D. Irimia, Lab Chip, 2010, 10, 505–511 RSC.
- S.-C. Guo, S.-C. Tao and H. Dawn, J. Extracell. Vesicles, 2018, 7, 1508271 CrossRef PubMed.
- S. Benfer, P. Árki and G. Tomandl, Adv. Eng. Mater., 2004, 6, 495–500 CrossRef.
- F. A. W. Coumans, A. R. Brisson, E. I. Buzas, F. Dignat-George, E. E. E. Drees, S. El-Andaloussi, C. Emanueli, A. Gasecka, A. Hendrix, A. F. Hill, R. Lacroix, Y. Lee, T. G. van Leeuwen, N. Mackman, I. Mäger, J. P. Nolan, E. van der Pol, D. M. Pegtel, S. Sahoo, P. R. M. Siljander, G. Sturk, O. de Wever and R. Nieuwland, Circ. Res., 2017, 120, 1632–1648 CrossRef.
- X. Xiang, F. Guan, F. Jiao, H. Li, W. Zhang, Y. Zhang and W. Qin, Anal. Methods, 2021, 13, 1591–1600 RSC.
- R. J. Lobb, M. Becker, S. Wen Wen, C. S. F. Wong, A. P. Wiegmans, A. Leimgruber and A. Möller, J. Extracell. Vesicles, 2015, 4, 27031 CrossRef PubMed.
- L. Pitkänen and A. M. Striegel, TrAC, Trends Anal. Chem., 2016, 80, 311–320 CrossRef.
- S. Süβ, C. Metzger, C. Damm, D. Segets and W. Peukert, Powder Technol., 2018, 339, 264–272 CrossRef.
- R. Stranska, L. Gysbrechts, J. Wouters, P. Vermeersch, K. Bloch, D. Dierickx, G. Andrei and R. Snoeck, J. Transl. Med., 2018, 16, 1 CrossRef CAS.
- C.-S. Hong, S. Funk, L. Muller, M. Boyiadzis and T. L. Whiteside, J. Extracell. Vesicles, 2016, 5, 29289 CrossRef.
- S. L. Shu, Y. Yang, C. L. Allen, E. Hurley, K. H. Tung, H. Minderman, Y. Wu and M. S. Ernstoff, J. Extracell. Vesicles, 2019, 9, 1692401 CrossRef.
- G. Sharma, A. Kumar, S. Sharma, Mu. Naushad, R. Prakash Dwivedi, Z. A. ALOthman and G. T. Mola, J. King Saud Univ., Sci., 2019, 31, 257–269 CrossRef.
- G. Sharma, V. K. Gupta, S. Agarwal, A. Kumar, S. Thakur and D. Pathania, J. Mol. Liq., 2016, 219, 1137–1143 CrossRef CAS.
- P. Y. Reyes, J. A. Espinoza, M. E. Treviño, H. Saade and R. G. López, J. Nanomater., 2010, 2010, e948941 Search PubMed.
- Y. Dahman, in Nanotechnology and Functional Materials for Engineers, ed. Y. Dahman, Elsevier, 2017, pp. 121–144 Search PubMed.
- K. P. De Sousa, I. Rossi, M. Abdullahi, M. I. Ramirez, D. Stratton and J. M. Inal, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, e1835 Search PubMed.
- L. Paolini, A. Zendrini, G. D. Noto, S. Busatto, E. Lottini, A. Radeghieri, A. Dossi, A. Caneschi, D. Ricotta and P. Bergese, Sci. Rep., 2016, 6, 23550 CrossRef.
- Y. K. Yoo, J. Lee, H. Kim, K. S. Hwang, D. S. Yoon and J. H. Lee, Micromachines, 2018, 9, 634 CrossRef PubMed.
- C. F. Ruivo, B. Adem, M. Silva and S. A. Melo, Cancer Res., 2017, 77, 6480–6488 CrossRef.
- M. L. Yarmush, A. M. Weiss, K. P. Antonsen, D. J. Odde and D. M. Yarmush, Biotechnol. Adv., 1992, 10, 413–446 CrossRef.
- D. D. Taylor and S. Shah, Methods, 2015, 87, 3–10 CrossRef CAS PubMed.
- C.-L. Shih, K.-Y. Chong, S.-C. Hsu, H.-J. Chien, C.-T. Ma, J. W.-C. Chang, C.-J. Yu and C.-C. Chiou, New Biotechnol., 2016, 33, 116–122 CrossRef CAS PubMed.
- J. Kowal, G. Arras, M. Colombo, M. Jouve, J. P. Morath, B. Primdal-Bengtson, F. Dingli, D. Loew, M. Tkach and C. Théry, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E968–E977 CrossRef CAS.
- Z. Andreu and M. Yáñez-Mó, Front. Immunol., 2014, 5, 442 Search PubMed.
- B. J. Tauro, D. W. Greening, R. A. Mathias, H. Ji, S. Mathivanan, A. M. Scott and R. J. Simpson, Methods, 2012, 56, 293–304 CrossRef CAS PubMed.
- P. Sajeesh and A. K. Sen, Microfluid. Nanofluid., 2014, 17, 1–52 CrossRef.
- L. Shi, A. Rana and L. Esfandiari, Sci. Rep., 2018, 8, 6751 CrossRef.
- K. Mogi, K. Hayashida and T. Yamamoto, Anal. Sci., 2018, 34, 875–880 CrossRef CAS PubMed.
- P. Ohlsson, K. Petersson, P. Augustsson and T. Laurell, Sci. Rep., 2018, 8, 9156 CrossRef PubMed.
- T. Yoon, H. S. Moon, J. W. Song, K. A. Hyun and H. I. Jung, Cytometry, Part A, 2019, 95, 1135–1144 CrossRef.
- E. Iswardy, T.-C. Tsai, I.-F. Cheng, T.-C. Ho, G. C. Perng and H.-C. Chang, Biosens. Bioelectron., 2017, 95, 174–180 CrossRef.
- Y. Chen, M. Wu, L. Ren, J. Liu, P. H. Whitley, L. Wang and T. J. Huang, Lab Chip, 2016, 16, 3466–3472 RSC.
- D. J. Collins, Z. Ma, J. Han and Y. Ai, Lab Chip, 2016, 17, 91–103 RSC.
- K. Zhao and D. Li, Sens. Actuators, B, 2017, 250, 274–284 CrossRef CAS.
- A. Lenshof and T. Laurell, in Encyclopedia of Nanotechnology, ed. B. Bhushan, Springer Netherlands, Dordrecht, 2014, pp. 1–6 Search PubMed.
- S. Mohanty, I. S. M. Khalil and S. Misra, Proc. R. Soc. A, 2020, 476, 20200621 CrossRef PubMed.
- Y. Xie, H. Bachman and T. J. Huang, TrAC, Trends Anal. Chem., 2019, 117, 280–290 CrossRef CAS PubMed.
- A. Fornell, K. Cushing, J. Nilsson and M. Tenje, Appl. Phys. Lett., 2018, 112, 063701 CrossRef.
- L. Meng, F. Cai, F. Li, W. Zhou, L. Niu and H. Zheng, J. Phys. D: Appl. Phys., 2019, 52, 273001 CrossRef CAS.
- K. Länge, B. E. Rapp and M. Rapp, Anal. Bioanal. Chem., 2008, 391, 1509–1519 CrossRef.
- I. Leibacher, P. Reichert and J. Dual, Lab Chip, 2015, 15, 2896–2905 RSC.
- Z. Chen, X. Liu, M. Kojima, Q. Huang and T. Arai, Appl. Sci., 2020, 10, 1260 CrossRef CAS.
- D. J. Collins, Z. Ma and Y. Ai, Anal. Chem., 2016, 88, 5513–5522 CrossRef CAS PubMed.
- L. Johansson, J. Enlund, S. Johansson, I. Katardjiev and V. Yantchev, Biomed. Microdevices, 2012, 14, 279–289 CrossRef PubMed.
- G. Destgeer, J. H. Jung, J. Park, H. Ahmed, K. Park, R. Ahmad and H. J. Sung, RSC Adv., 2017, 7, 22524–22530 RSC.
- J. Shi, H. Huang, Z. Stratton, Y. Huang and T. J. Huang, Lab Chip, 2009, 9, 3354–3359 RSC.
- W. Connacher, N. Zhang, A. Huang, J. Mei, S. Zhang, T. Gopesh and J. Friend, Lab Chip, 2018, 18, 1952–1996 RSC.
- R. Weser, A. Winkler, M. Weihnacht, S. Menzel and H. Schmidt, Ultrasonics, 2020, 106, 106160 CrossRef CAS.
- G. Destgeer, B. H. Ha, J. Park, J. H. Jung, A. Alazzam and H. J. Sung, Phys. Procedia, 2015, 70, 34–37 CrossRef.
- Y. Yang, L. Zhang, K. Jin, M. He, W. Wei, X. Chen, Q. Yang, Y. Wang, W. Pang, X. Ren and X. Duan, Sci. Adv., 2022, 8, eabn8440 CrossRef.
- M. Wu, Y. Ouyang, Z. Wang, R. Zhang, P.-H. Huang, C. Chen, H. Li, P. Li, D. Quinn, M. Dao, S. Suresh, Y. Sadovsky and T. J. Huang, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 10584–10589 CrossRef.
- F. Petersson, A. Nilsson, C. Holm, H. Jönsson and T. Laurell, Lab Chip, 2005, 5, 20–22 RSC.
- Y. Ai, C. K. Sanders and B. L. Marrone, Anal. Chem., 2013, 85, 9126–9134 CrossRef PubMed.
- K. Lee, H. Shao, R. Weissleder and H. Lee, ACS Nano, 2015, 9, 2321–2327 CrossRef CAS PubMed.
- M. Wu, A. Ozcelik, J. Rufo, Z. Wang, R. Fang and T. Jun Huang, Microsyst. Nanoeng., 2019, 5, 32 CrossRef.
- M. Antfolk, P. B. Muller, P. Augustsson, H. Bruus and T. Laurell, Lab Chip, 2014, 14, 2791–2799 RSC.
- P. Ohlsson, M. Evander, K. Petersson, L. Mellhammar, A. Lehmusvuori, U. Karhunen, M. Soikkeli, T. Seppä, E. Tuunainen, A. Spangar, P. von Lode, K. Rantakokko-Jalava, G. Otto, S. Scheding, T. Soukka, S. Wittfooth and T. Laurell, Anal. Chem., 2016, 88, 9403–9411 CrossRef CAS PubMed.
- Y. Li, S. Cai, H. Shen, Y. Chen, Z. Ge and W. Yang, Biomicrofluidics, 2022, 16, 031502 CrossRef CAS.
- Y. Gao, M. Wu, Y. Lin and J. Xu, Micromachines, 2020, 11, E921 CrossRef.
- K. D. Dorfman, in Encyclopedia of Microfluidics and Nanofluidics, ed. D. Li, Springer, New York, NY, 2015, pp. 926–934 Search PubMed.
- D. Li, in Interface Science and Technology, Elsevier, 2004, vol. 2, pp. 542–616 Search PubMed.
- X. Ou, P. Chen, X. Huang, S. Li and B.-F. Liu, J. Sep. Sci., 2020, 43, 258–270 CrossRef CAS PubMed.
- M. Dawod, N. E. Arvin and R. T. Kennedy, Analyst, 2017, 142, 1847–1866 RSC.
- K. D. Dorfman, S. B. King, D. W. Olson, J. D. P. Thomas and D. R. Tree, Chem. Rev., 2013, 113, 2584–2667 CrossRef CAS PubMed.
- E. O. Adekanmbi and S. K. Srivastava, Lab Chip, 2016, 16, 2148–2167 RSC.
- R. T. Davies, J. Kim, S. C. Jang, E.-J. Choi, Y. S. Gho and J. Park, Lab Chip, 2012, 12, 5202–5210 RSC.
- S. Marczak, K. Richards, Z. Ramshani, E. Smith, S. Senapati, R. Hill, D. B. Go and H.-C. Chang, Electrophoresis, 2018, 39, 2029–2038 CrossRef CAS PubMed.
- B. Franze and C. Engelhard, Anal. Chem., 2014, 86, 5713–5720 CrossRef CAS.
- C. Adelantado, N. Rodríguez-Fariñas, R. C. Rodríguez Martín-Doimeadios, M. Zougagh and Á. Ríos, Anal. Chim. Acta, 2016, 923, 82–88 CrossRef CAS PubMed.
- D.-T. Phan, S. A. M. Shaegh, C. Yang and N.-T. Nguyen, Sens. Actuators, B, 2016, 222, 735–740 CrossRef CAS.
- S. Cho, W. Jo, Y. Heo, J. Y. Kang, R. Kwak and J. Park, Sens. Actuators, B, 2016, 233, 289–297 CrossRef CAS.
- Y. Pan, S. Neuss, A. Leifert, M. Fischler, F. Wen, U. Simon, G. Schmid, W. Brandau and W. Jahnen-Dechent, Small, 2007, 3, 1941–1949 CrossRef CAS PubMed.
- B. Kowalczyk, I. Lagzi and B. A. Grzybowski, Curr. Opin. Colloid Interface Sci., 2011, 16, 135–148 CrossRef CAS.
- M. Bouri, R. Salghi, M. Zougagh and A. Ríos, Anal. Chem., 2013, 85, 4858–4862 CrossRef CAS PubMed.
- S. Hassanpour Tamrin, A. Sanati Nezhad and A. Sen, ACS Nano, 2021, 15, 17047–17079 CrossRef CAS.
- S. Grimnes and Ø. G. Martinsen, in Bioimpedance and Bioelectricity Basics, ed. S. Grimnes and Ø. G. Martinsen, Academic Press, Oxford, 3rd edn, 2015, pp. 179–254 Search PubMed.
- R. Pethig, Biomicrofluidics, 2010, 4, 022811 CrossRef PubMed.
- T. Ghomian and J. Hihath, IEEE Trans. Biomed. Eng., 2022, 1–15 Search PubMed.
- C. Qian, H. Huang, L. Chen, X. Li, Z. Ge, T. Chen, Z. Yang and L. Sun, Int. J. Mol. Sci., 2014, 15, 18281–18309 CrossRef CAS PubMed.
- A. Alazzam, B. Mathew and F. Alhammadi, J. Sep. Sci., 2017, 40, 1193–1200 CrossRef CAS PubMed.
- R. Pethig, in Dielectrophoresis, John Wiley & Sons, Ltd, 2017, pp. 119–144 Search PubMed.
- E. A. Kwizera, M. Sun, A. M. White, J. Li and X. He, ACS Biomater. Sci. Eng., 2021, 7, 2043–2063 CrossRef CAS.
- H. Song, J. M. Rosano, Y. Wang, C. J. Garson, B. Prabhakarpandian, K. Pant, G. J. Klarmann, A. Perantoni, L. M. Alvarez and E. Lai, Lab Chip, 2015, 15, 1320–1328 RSC.
- H. Zhang, H. Chang and P. Neuzil, Micromachines, 2019, 10, 423 CrossRef PubMed.
- U. Kim, J. Qian, S. A. Kenrick, P. S. Daugherty and H. T. Soh, Anal. Chem., 2008, 80, 8656–8661 CrossRef CAS PubMed.
- L. Shi, D. Kuhnell, V. J. Borra, S. M. Langevin, T. Nakamura and L. Esfandiari, Lab Chip, 2019, 19, 3726–3734 RSC.
- S. Ayala-Mar, R. C. Gallo-Villanueva and J. González-Valdez, Mater. Today: Proc., 2019, 13, 332–340 CAS.
- S. Ayala-Mar, V. H. Perez-Gonzalez, M. A. Mata-Gómez, R. C. Gallo-Villanueva and J. González-Valdez, Anal. Chem., 2019, 91, 14975–14982 CrossRef CAS PubMed.
- S. D. Ibsen, J. Wright, J. M. Lewis, S. Kim, S.-Y. Ko, J. Ong, S. Manouchehri, A. Vyas, J. Akers, C. C. Chen, B. S. Carter, S. C. Esener and M. J. Heller, ACS Nano, 2017, 11, 6641–6651 CrossRef CAS PubMed.
- P. Modarres and M. Tabrizian, Sens. Actuators, B, 2017, 252, 391–408 CrossRef CAS.
- L. Liu, K. Chen, N. Xiang and Z. Ni, Electrophoresis, 2019, 40, 873–889 CrossRef CAS.
- S. Yaman, M. Anil-Inevi, E. Ozcivici and H. C. Tekin, Front. Bioeng. Biotechnol., 2018, 6, 192 CrossRef.
- N.-T. Nguyen, Microfluid. Nanofluid., 2012, 12, 1–16 CrossRef.
- M. D. Tarn, N. Hirota, A. Iles and N. Pamme, Sci. Technol. Adv. Mater., 2009, 10, 014611 CrossRef PubMed.
- D. Robert, N. Pamme, H. Conjeaud, F. Gazeau, A. Iles and C. Wilhelm, Lab Chip, 2011, 11, 1902–1910 RSC.
- M. D. Krebs, R. M. Erb, B. B. Yellen, B. Samanta, A. Bajaj, V. M. Rotello and E. Alsberg, Nano Lett., 2009, 9, 1812–1817 CrossRef PubMed.
- L. Zeng, X. Chen, J. Du, Z. Yu, R. Zhang, Y. Zhang and H. Yang, Nanoscale, 2021, 13, 4029–4037 RSC.
- L. Zeng, S. Hu, X. Chen, P. Zhang, G. Gu, Y. Wang, H. Zhang, Y. Zhang and H. Yang, Lab Chip, 2022, 22, 2476–2488 RSC.
- C. Park, J. Lee, Y. Kim, J. Kim, J. Lee and S. Park, J. Microbiol. Methods, 2017, 132, 128–133 CrossRef CAS PubMed.
- Z. M. Wang, R. G. Wu, Z. P. Wang and R. V. Ramanujan, Sci. Rep., 2016, 6, 26945 CrossRef CAS.
- S. Oh, S. H. Jung, H. Seo, M.-K. Min, B. Kim, Y. K. Hahn, J. H. Kang and S. Choi, Sens. Actuators, B, 2018, 272, 324–330 CrossRef CAS.
- Y. Liu, W. Zhao, R. Cheng, M. Logun, M. D. M. Zayas-Viera, L. Karumbaiah and L. Mao, Lab Chip, 2020, 20, 3187–3201 RSC.
- A. J. Mach, O. B. Adeyiga and D. D. Carlo, Lab Chip, 2013, 13, 1011–1026 RSC.
- S. S. Leong, Z. Ahmad, S. C. Low, J. Camacho, J. Faraudo and J. Lim, Langmuir, 2020, 36, 8033–8055 CrossRef PubMed.
- F. Alnaimat, S. Dagher, B. Mathew, A. Hilal-Alnqbi and S. Khashan, Chem. Rec., 2018, 18, 1596–1612 CrossRef PubMed.
- P. Su, C. Ren, Y. Fu, J. Guo, J. Guo and Q. Yuan, Sens. Actuators, A, 2021, 332, 113180 CrossRef.
- A. Munaz, M. J. A. Shiddiky and N.-T. Nguyen, Biomicrofluidics, 2018, 12, 031501 CrossRef PubMed.
- P. Skupin-Mrugalska, P. A. Elvang and M. Brandl, Int. J. Mol. Sci., 2021, 22, 10456 CrossRef PubMed.
- H. Kato, in Characterization of Nanoparticles, ed. V.-D. Hodoroaba, W. E. S. Unger and A. G. Shard, Elsevier, 2020, pp. 249–264 Search PubMed.
- A. Lenshof and T. Laurell, Chem. Soc. Rev., 2010, 39, 1203–1217 RSC.
- J. C. Giddings, Sep. Sci., 2006, 123–125 Search PubMed.
- C. Contado, Anal. Bioanal. Chem., 2017, 409, 2501–2518 CrossRef CAS PubMed.
- L. Calzolai, D. Gilliland, C. P. Garcìa and F. Rossi, J. Chromatogr. A, 2011, 1218, 4234–4239 CrossRef CAS PubMed.
- H. Zhang, D. Freitas, H. S. Kim, K. Fabijanic, Z. Li, H. Chen, M. T. Mark, H. Molina, A. B. Martin, L. Bojmar, J. Fang, S. Rampersaud, A. Hoshino, I. Matei, C. M. Kenific, M. Nakajima, A. P. Mutvei, P. Sansone, W. Buehring, H. Wang, J. P. Jimenez, L. Cohen-Gould, N. Paknejad, M. Brendel, K. Manova-Todorova, A. Magalhães, J. A. Ferreira, H. Osório, A. M. Silva, A. Massey, J. R. Cubillos-Ruiz, G. Galletti, P. Giannakakou, A. M. Cuervo, J. Blenis, R. Schwartz, M. S. Brady, H. Peinado, J. Bromberg, H. Matsui, C. A. Reis and D. Lyden, Nat. Cell Biol., 2018, 20, 332–343 CrossRef CAS PubMed.
- J. Gigault, H. El Hadri, S. Reynaud, E. Deniau and B. Grassl, Anal. Bioanal. Chem., 2017, 409, 6761–6769 CrossRef CAS PubMed.
- B. Roda, A. Zattoni, P. Reschiglian, M. H. Moon, M. Mirasoli, E. Michelini and A. Roda, Anal. Chim. Acta, 2009, 635, 132–143 CrossRef CAS PubMed.
- D. Kang, S. Oh, S.-M. Ahn, B.-H. Lee and M. H. Moon, J. Proteome Res., 2008, 7, 3475–3480 CrossRef CAS.
- J. S. Yang, J. C. Lee, S. K. Byeon, K. H. Rha and M. H. Moon, Anal. Chem., 2017, 89, 2488–2496 CrossRef CAS PubMed.
- J. C. Giddings, Sep. Sci. Technol., 1985, 20, 749–768 CrossRef CAS.
- A. De Momi and J. R. Lead, Sci. Total Environ., 2008, 405, 317–323 CrossRef PubMed.
- A. De Momi and J. R. Lead, Environ. Sci. Technol., 2006, 40, 6738–6743 CrossRef.
- A. Ashkin, Phys. Rev. Lett., 1970, 24, 156–159 CrossRef.
- W. Wu, X. Zhu, Y. Zuo, L. Liang, S. Zhang, X. Zhang and Y. Yang, ACS Photonics, 2016, 3, 2497–2504 CrossRef.
- I. Fernandez-Cuesta, A. Llobera and M. Ramos-Payán, Anal. Chim. Acta, 2022, 1192, 339307 CrossRef.
- B. Landenberger, H. Höfemann, S. Wadle and A. Rohrbach, Lab Chip, 2012, 12, 3177–3183 RSC.
- P. Paiè, T. Zandrini, R. M. Vázquez, R. Osellame and F. Bragheri, Micromachines, 2018, 9, 200 CrossRef.
- F. Nan and Z. Yan, Nano Lett., 2018, 18, 7400–7406 CrossRef.
- B. Ma, B. Yao, F. Peng, S. Yan, M. Lei and R. Rupp, J. Opt., 2012, 14, 105702 CrossRef.
- H. Zhang and K.-K. Liu, J. R. Soc., Interface, 2008, 5, 671–690 CrossRef PubMed.
- B. R. Mutlu, J. F. Edd and M. Toner, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 7682–7687 CrossRef.
- L. Wang and D. S. Dandy, Adv. Sci., 2017, 4, 1700153 CrossRef.
- H. M. Tay, S. Y. Leong, X. Xu, F. Kong, M. Upadya, R. Dalan, C. Y. Tay, M. Dao, S. Suresh and H. W. Hou, Lab Chip, 2021, 21, 2511–2523 RSC.
- M. R. Condina, B. A. Dilmetz, S. R. Bazaz, J. Meneses, M. E. Warkiani and P. Hoffmann, Lab Chip, 2019, 19, 1961–1970 RSC.
- B. H. Wunsch, J. T. Smith, S. M. Gifford, C. Wang, M. Brink, R. L. Bruce, R. H. Austin, G. Stolovitzky and Y. Astier, Nat. Nanotechnol., 2016, 11, 936–940 CrossRef.
- J. T. Smith, B. H. Wunsch, N. Dogra, M. E. Ahsen, K. Lee, K. K. Yadav, R. Weil, M. A. Pereira, J. V. Patel, E. A. Duch, J. M. Papalia, M. F. Lofaro, M. Gupta, A. K. Tewari, C. Cordon-Cardo, G. Stolovitzky and S. M. Gifford, Lab Chip, 2018, 18, 3913–3925 RSC.
- Y. Hattori, T. Shimada, T. Yasui, N. Kaji and Y. Baba, Anal. Chem., 2019, 91, 6514–6521 CrossRef PubMed.
- Z. Han, C. Peng, J. Yi, D. Zhang, X. Xiang, X. Peng, B. Su, B. Liu, Y. Shen and L. Qiao, Sens. Actuators, B, 2021, 333, 129563 CrossRef.
- F. Inci, Langmuir, 2022, 38, 1897–1909 CrossRef PubMed.
- L.-G. Liang, M.-Q. Kong, S. Zhou, Y.-F. Sheng, P. Wang, T. Yu, F. Inci, W. P. Kuo, L.-J. Li, U. Demirci and S. Wang, Sci. Rep., 2017, 7, 46224 CrossRef.
- S. Shin, D. Han, M. C. Park, J. Y. Mun, J. Choi, H. Chun, S. Kim and J. W. Hong, Sci. Rep., 2017, 7, 9907 CrossRef PubMed.
- Y. Sai, M. Yamada, M. Yasuda and M. Seki, J. Chromatogr. A, 2006, 1127, 214–220 CrossRef.
- C. Liu, B. Ding, C. Xue, Y. Tian, G. Hu and J. Sun, Anal. Chem., 2016, 88, 12547–12553 CrossRef.
- J. Y. Kim, S. W. Ahn, S. S. Lee and J. M. Kim, Lab Chip, 2012, 12, 2807–2814 RSC.
- C. Liu, J. Guo, F. Tian, N. Yang, F. Yan, Y. Ding, J. Wei, G. Hu, G. Nie and J. Sun, ACS Nano, 2017, 11, 6968–6976 CrossRef PubMed.
- J. Zhang, S. Yan, D. Yuan, G. Alici, N.-T. Nguyen, M. E. Warkiani and W. Li, Lab Chip, 2015, 16, 10–34 RSC.
- D. D. Carlo, Lab Chip, 2009, 9, 3038–3046 RSC.
- R. Nasiri, A. Shamloo, S. Ahadian, L. Amirifar, J. Akbari, M. J. Goudie, K. Lee, N. Ashammakhi, M. R. Dokmeci, D. Di Carlo and A. Khademhosseini, Small, 2020, 16, 2000171 CrossRef.
- W. Tang, S. Zhu, D. Jiang, L. Zhu, J. Yang and N. Xiang, Lab Chip, 2020, 20, 3485–3502 RSC.
- X. Lu, J. J. M. Chow, S. H. Koo, T. Y. Tan, B. Jiang and Y. Ai, Anal. Chem., 2020, 92, 15579–15586 CrossRef.
- M. Li, M. van Zee, K. Goda and D. D. Carlo, Lab Chip, 2018, 18, 2575–2582 RSC.
- M. E. Warkiani, A. K. P. Tay, B. L. Khoo, X. Xiaofeng, J. Han and C. T. Lim, Lab Chip, 2015, 15, 1101–1109 RSC.
- D. Di Carlo, D. Irimia, R. G. Tompkins and M. Toner, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 18892–18897 CrossRef PubMed.
- H. M. Tay, S. Kharel, R. Dalan, Z. J. Chen, K. K. Tan, B. O. Boehm, S. C. J. Loo and H. W. Hou, NPG Asia Mater., 2017, 9, e434–e434 CrossRef.
- D. Huang, J. Man, D. Jiang, J. Zhao and N. Xiang, Electrophoresis, 2020, 41, 2166–2187 CrossRef CAS PubMed.
- G.-Y. Kim, J.-I. Han and J.-K. Park, BioChip J., 2018, 12, 257–267 CrossRef CAS.
- Y. Gou, Y. Jia, P. Wang and C. Sun, Sensors, 2018, 18, 1762 CrossRef PubMed.
- H. Amini, W. Lee and D. D. Carlo, Lab Chip, 2014, 14, 2739–2761 RSC.
- Y. Deng, M. Kizer, M. Rada, J. Sage, X. Wang, D.-J. Cheon and A. J. Chung, Nano Lett., 2018, 18, 2705–2710 CrossRef PubMed.
- A. J. Mach and D. Di Carlo, Biotechnol. Bioeng., 2010, 107, 302–311 CrossRef PubMed.
- D. W. Inglis, J. A. Davis, R. H. Austin and J. C. Sturm, Lab Chip, 2006, 6, 655–658 RSC.
- A. Hochstetter, R. Vernekar, R. H. Austin, H. Becker, J. P. Beech, D. A. Fedosov, G. Gompper, S.-C. Kim, J. T. Smith, G. Stolovitzky, J. O. Tegenfeldt, B. H. Wunsch, K. K. Zeming, T. Krüger and D. W. Inglis, ACS Nano, 2020, 14, 10784–10795 CrossRef CAS PubMed.
- J. A. Davis, Doctor of Philosophy, Princeton University, 2008 Search PubMed.
- S. Ranjan, K. K. Zeming, R. Jureen, D. Fisher and Y. Zhang, Lab Chip, 2014, 14, 4250–4262 RSC.
- L. R. Huang, E. C. Cox, R. H. Austin and J. C. Sturm, Science, 2004, 304, 987–990 CrossRef CAS PubMed.
- E. Pariset, C. Parent, Y. Fouillet, B. François, N. Verplanck, F. Revol-Cavalier, A. Thuaire and V. Agache, Sci. Rep., 2018, 8, 17762 CrossRef CAS PubMed.
- K. K. Zeming, T. Salafi, C.-H. Chen and Y. Zhang, Sci. Rep., 2016, 6, 22934 CrossRef CAS PubMed.
- R. J. Gillams, V. Calero, R. Fernandez-Mateo and H. Morgan, Lab Chip, 2022, 22, 3869–3876 RSC.
- K. K. Zeming, N. V. Thakor, Y. Zhang and C.-H. Chen, Lab Chip, 2015, 16, 75–85 RSC.
- T. Salafi, Y. Zhang and Y. Zhang, Nano-Micro Lett., 2019, 11, 77 CrossRef PubMed.
- J. McGrath, M. Jimenez and H. Bridle, Lab Chip, 2014, 14, 4139–4158 RSC.
- F. Liu, O. Vermesh, V. Mani, T. J. Ge, S. J. Madsen, A. Sabour, E.-C. Hsu, G. Gowrishankar, M. Kanada, J. V. Jokerst, R. G. Sierra, E. Chang, K. Lau, K. Sridhar, A. Bermudez, S. J. Pitteri, T. Stoyanova, R. Sinclair, V. S. Nair, S. S. Gambhir and U. Demirci, ACS Nano, 2017, 11, 10712–10723 CrossRef PubMed.
- H.-K. Woo, V. Sunkara, J. Park, T.-H. Kim, J.-R. Han, C.-J. Kim, H.-I. Choi, Y.-K. Kim and Y.-K. Cho, ACS Nano, 2017, 11, 1360–1370 CrossRef PubMed.
- S. Wang, D. Sarenac, M. H. Chen, S.-H. Huang, F. F. Giguel, D. R. Kuritzkes and U. Demirci, Int. J. Nanomed., 2012, 7, 5019–5028 Search PubMed.
- H. Bolze, J. Riewe, H. Bunjes, A. Dietzel and T. P. Burg, Chem. Eng. Technol., 2021, 44, 457–464 CrossRef.
- I. Seder, H. Moon, S. J. Kang, S. Shin, W. J. Rhee and S.-J. Kim, Lab Chip, 2022, 22, 3699–3707 RSC.
- N. Debnath and M. Sadrzadeh, J. Indian Inst. Sci., 2018, 98, 137–157 CrossRef.
- Y. Yoon, S. Kim, J. Lee, J. Choi, R.-K. Kim, S.-J. Lee, O. Sul and S.-B. Lee, Sci. Rep., 2016, 6, 26531 CrossRef CAS PubMed.
- S. Lin, Z. Yu, D. Chen, Z. Wang, J. Miao, Q. Li, D. Zhang, J. Song and D. Cui, Small, 2020, 16, 1903916 CrossRef CAS PubMed.
- M. Yamada, M. Nakashima and M. Seki, Anal. Chem., 2004, 76, 5465–5471 CrossRef CAS PubMed.
- T. Morijiri, S. Sunahiro, M. Senaha, M. Yamada and M. Seki, Microfluid. Nanofluid., 2011, 11, 105–110 CrossRef.
- H. W. Nho, N. Yang, J. Song, J. S. Park and T. H. Yoon, Sens. Actuators, B, 2017, 249, 131–141 CrossRef CAS.
- S. Wang, Z. Liu, S. Wu, H. Sun, W. Zeng, J. Wei, Z. Fan, Z. Sui, L. Liu and X. Pan, Electrophoresis, 2021, 42, 2223–2229 CrossRef CAS PubMed.
- A. Vig Larsen, L. Poulsen, H. Birgens, M. Dufva and A. Kristensen, Lab Chip, 2008, 8, 818–821 RSC.
- T. Hamacher, J. T. W. Berendsen, J. E. van Dongen, R. M. van der Hee, J. J. L. M. Cornelissen, M. L. W. J. Broekhuijse and L. I. Segerink, Lab Chip, 2021, 21, 4477–4486 RSC.
- N. Nivedita and I. Papautsky, Biomicrofluidics, 2013, 7, 054101 CrossRef PubMed.
- A. Jain and J. D. Posner, Anal. Chem., 2008, 80, 1641–1648 CrossRef CAS PubMed.
- D. Stoecklein and D. Di Carlo, Anal. Chem., 2019, 91, 296–314 CrossRef CAS PubMed.
- N. Xiang, X. Zhang, Q. Dai, J. Cheng, K. Chen and Z. Ni, Lab Chip, 2016, 16, 2626–2635 RSC.
- J. Zhou and I. Papautsky, Microsyst. Nanoeng., 2020, 6, 1–24 CrossRef PubMed.
- Y. Zhou, Z. Ma and Y. Ai, Lab Chip, 2020, 20, 568–581 RSC.
- K. W. Seo, H. J. Byeon, H. K. Huh and S. J. Lee, RSC Adv., 2013, 4, 3512–3520 RSC.
- A. M. Leshansky, A. Bransky, N. Korin and U. Dinnar, Phys. Rev. Lett., 2007, 98, 234501 CrossRef PubMed.
- S. Yang, J. Y. Kim, S. J. Lee, S. S. Lee and J. M. Kim, Lab Chip, 2011, 11, 266–273 RSC.
- G. Li, G. H. McKinley and A. M. Ardekani, J. Fluid Mech., 2015, 785, 486–505 CrossRef.
- J. Nam, H. Lim, D. Kim, H. Jung and S. Shin, Lab Chip, 2012, 12, 1347–1354 RSC.
- D. Yuan, J. Zhang, R. Sluyter, Q. Zhao, S. Yan, G. Alici and W. Li, Lab Chip, 2016, 16, 3919–3928 RSC.
- C. Liu, J. Zhao, F. Tian, J. Chang, W. Zhang and J. Sun, J. Am. Chem. Soc., 2019, 141, 3817–3821 CrossRef CAS PubMed.
- M. A. Faridi, H. Ramachandraiah, I. Banerjee, S. Ardabili, S. Zelenin and A. Russom, J. Nanobiotechnol., 2017, 15, 3 CrossRef PubMed.
- J. Nam, J. Yoon, H. Jee, W. S. Jang and C. S. Lim, Adv. Mater. Technol., 2020, 5, 2000612 CrossRef CAS.
- Y. Zhou, Z. Ma, M. Tayebi and Y. Ai, Anal. Chem., 2019, 91, 4577–4584 CrossRef CAS PubMed.
- F. Shiri, H. Feng, K. E. Petersen, H. Sant, G. T. Bardi, L. A. Schroeder, M. L. Merchant, B. K. Gale and J. L. Hood, Sci. Rep., 2022, 12, 6146 CrossRef CAS PubMed.
- U. Kim and H. T. Soh, Lab Chip, 2009, 9, 2313–2318 RSC.
- D. J. Collins, T. Alan and A. Neild, Lab Chip, 2014, 14, 1595–1603 RSC.
- M. Tayebi, D. Yang, D. J. Collins and Y. Ai, Nano Lett., 2021, 21, 6835–6842 CrossRef CAS PubMed.
- S. M. Santana, M. A. Antonyak, R. A. Cerione and B. J. Kirby, Biomed. Microdevices, 2014, 16, 869–877 CrossRef CAS PubMed.
- C. Théry, K. W. Witwer, E. Aikawa, M. J. Alcaraz, J. D. Anderson, R. Andriantsitohaina, A. Antoniou, T. Arab, F. Archer, G. K. Atkin-Smith, D. C. Ayre, J.-M. Bach, D. Bachurski, H. Baharvand, L. Balaj, S. Baldacchino, N. N. Bauer, A. A. Baxter, M. Bebawy, C. Beckham, A. Bedina Zavec, A. Benmoussa, A. C. Berardi, P. Bergese, E. Bielska, C. Blenkiron, S. Bobis-Wozowicz, E. Boilard, W. Boireau, A. Bongiovanni, F. E. Borràs, S. Bosch, C. M. Boulanger, X. Breakefield, A. M. Breglio, M. Á. Brennan, D. R. Brigstock, A. Brisson, M. L. Broekman, J. F. Bromberg, P. Bryl-Górecka, S. Buch, A. H. Buck, D. Burger, S. Busatto, D. Buschmann, B. Bussolati, E. I. Buzás, J. B. Byrd, G. Camussi, D. R. Carter, S. Caruso, L. W. Chamley, Y.-T. Chang, C. Chen, S. Chen, L. Cheng, A. R. Chin, A. Clayton, S. P. Clerici, A. Cocks, E. Cocucci, R. J. Coffey, A. Cordeiro-da-Silva, Y. Couch, F. A. Coumans, B. Coyle, R. Crescitelli, M. F. Criado, C. D'Souza-Schorey, S. Das, A. Datta Chaudhuri, P. de Candia, E. F. De Santana, O. De Wever, H. A. del Portillo, T. Demaret, S. Deville, A. Devitt, B. Dhondt, D. Di Vizio, L. C. Dieterich, V. Dolo, A. P. D. Rubio, M. Dominici, M. R. Dourado, T. A. Driedonks, F. V. Duarte, H. M. Duncan, R. M. Eichenberger, K. Ekström, S. E. L. Andaloussi, C. Elie-Caille, U. Erdbrügger, J. M. Falcón-Pérez, F. Fatima, J. E. Fish, M. Flores-Bellver, A. Försönits, A. Frelet-Barrand, F. Fricke, G. Fuhrmann, S. Gabrielsson, A. Gámez-Valero, C. Gardiner, K. Gärtner, R. Gaudin, Y. S. Gho, B. Giebel, C. Gilbert, M. Gimona, I. Giusti, D. C. Goberdhan, A. Görgens, S. M. Gorski, D. W. Greening, J. C. Gross, A. Gualerzi, G. N. Gupta, D. Gustafson, A. Handberg, R. A. Haraszti, P. Harrison, H. Hegyesi, A. Hendrix, A. F. Hill, F. H. Hochberg, K. F. Hoffmann, B. Holder, H. Holthofer, B. Hosseinkhani, G. Hu, Y. Huang, V. Huber, S. Hunt, A. G.-E. Ibrahim, T. Ikezu, J. M. Inal, M. Isin, A. Ivanova, H. K. Jackson, S. Jacobsen, S. M. Jay, M. Jayachandran, G. Jenster, L. Jiang, S. M. Johnson, J. C. Jones, A. Jong, T. Jovanovic-Talisman, S. Jung, R. Kalluri, S. Kano, S. Kaur, Y. Kawamura, E. T. Keller, D. Khamari, E. Khomyakova, A. Khvorova, P. Kierulf, K. P. Kim, T. Kislinger, M. Klingeborn, D. J. Klinke, M. Kornek, M. M. Kosanović, Á. F. Kovács, E.-M. Krämer-Albers, S. Krasemann, M. Krause, I. V. Kurochkin, G. D. Kusuma, S. Kuypers, S. Laitinen, S. M. Langevin, L. R. Languino, J. Lannigan, C. Lässer, L. C. Laurent, G. Lavieu, E. Lázaro-Ibáñez, S. Le Lay, M.-S. Lee, Y. X. F. Lee, D. S. Lemos, M. Lenassi, A. Leszczynska, I. T. Li, K. Liao, S. F. Libregts, E. Ligeti, R. Lim, S. K. Lim, A. Linē, K. Linnemannstöns, A. Llorente, C. A. Lombard, M. J. Lorenowicz, Á. M. Lörincz, J. Lötvall, J. Lovett, M. C. Lowry, X. Loyer, Q. Lu, B. Lukomska, T. R. Lunavat, S. L. Maas, H. Malhi, A. Marcilla, J. Mariani, J. Mariscal, E. S. Martens-Uzunova, L. Martin-Jaular, M. C. Martinez, V. R. Martins, M. Mathieu, S. Mathivanan, M. Maugeri, L. K. McGinnis, M. J. McVey, D. G. Meckes, K. L. Meehan, I. Mertens, V. R. Minciacchi, A. Möller, M. Møller Jørgensen, A. Morales-Kastresana, J. Morhayim, F. Mullier, M. Muraca, L. Musante, V. Mussack, D. C. Muth, K. H. Myburgh, T. Najrana, M. Nawaz, I. Nazarenko, P. Nejsum, C. Neri, T. Neri, R. Nieuwland, L. Nimrichter, J. P. Nolan, E. N. Nolte-'t Hoen, N. N. Hooten, L. O'Driscoll, T. O'Grady, A. O'Loghlen, T. Ochiya, M. Olivier, A. Ortiz, L. A. Ortiz, X. Osteikoetxea, O. Østergaard, M. Ostrowski, J. Park, D. M. Pegtel, H. Peinado, F. Perut, M. W. Pfaffl, D. G. Phinney, B. C. Pieters, R. C. Pink, D. S. Pisetsky, E. P. von Strandmann, I. Polakovicova, I. K. Poon, B. H. Powell, I. Prada, L. Pulliam, P. Quesenberry, A. Radeghieri, R. L. Raffai, S. Raimondo, J. Rak, M. I. Ramirez, G. Raposo, M. S. Rayyan, N. Regev-Rudzki, F. L. Ricklefs, P. D. Robbins, D. D. Roberts, S. C. Rodrigues, E. Rohde, S. Rome, K. M. Rouschop, A. Rughetti, A. E. Russell, P. Saá, S. Sahoo, E. Salas-Huenuleo, C. Sánchez, J. A. Saugstad, M. J. Saul, R. M. Schiffelers, R. Schneider, T. H. Schøyen, A. Scott, E. Shahaj, S. Sharma, O. Shatnyeva, F. Shekari, G. V. Shelke, A. K. Shetty, K. Shiba, P. R.-M. Siljander, A. M. Silva, A. Skowronek, O. L. Snyder, R. P. Soares, B. W. Sódar, C. Soekmadji, J. Sotillo, P. D. Stahl, W. Stoorvogel, S. L. Stott, E. F. Strasser, S. Swift, H. Tahara, M. Tewari, K. Timms, S. Tiwari, R. Tixeira, M. Tkach, W. S. Toh, R. Tomasini, A. C. Torrecilhas, J. P. Tosar, V. Toxavidis, L. Urbanelli, P. Vader, B. W. van Balkom, S. G. van der Grein, J. Van Deun, M. J. van Herwijnen, K. Van Keuren-Jensen, G. van Niel, M. E. van Royen, A. J. van Wijnen, M. H. Vasconcelos, I. J. Vechetti, T. D. Veit, L. J. Vella, É. Velot, F. J. Verweij, B. Vestad, J. L. Viñas, T. Visnovitz, K. V. Vukman, J. Wahlgren, D. C. Watson, M. H. Wauben, A. Weaver, J. P. Webber, V. Weber, A. M. Wehman, D. J. Weiss, J. A. Welsh, S. Wendt, A. M. Wheelock, Z. Wiener, L. Witte, J. Wolfram, A. Xagorari, P. Xander, J. Xu, X. Yan, M. Yáñez-Mó, H. Yin, Y. Yuana, V. Zappulli, J. Zarubova, V. Žėkas, J. Zhang, Z. Zhao, L. Zheng, A. R. Zheutlin, A. M. Zickler, P. Zimmermann, A. M. Zivkovic, D. Zocco and E. K. Zuba-Surma, J. Extracell. Vesicles, 2018, 7, 1535750 CrossRef.
- M. Winey, J. B. Meehl, E. T. O'Toole and T. H. Giddings, Mol. Biol. Cell, 2014, 25, 319–323 CrossRef.
- K. Akhtar, S. A. Khan, S. B. Khan and A. M. Asiri, in Handbook of Materials Characterization, ed. S. K. Sharma, Springer International Publishing, Cham, 2018, pp. 113–145 Search PubMed.
- Y.-S. Chen, C. Chen, C. P.-K. Lai and G.-B. Lee, Sens. Actuators, B, 2022, 358, 131473 CrossRef CAS.
- B. Lee, S. Yoon, J. W. Lee, Y. Kim, J. Chang, J. Yun, J. C. Ro, J.-S. Lee and J. H. Lee, ACS Nano, 2020, 14, 17125–17133 CrossRef CAS PubMed.
- A. E. Vladár and V.-D. Hodoroaba, in Characterization of Nanoparticles, ed. V.-D. Hodoroaba, W. E. S. Unger and A. G. Shard, Elsevier, 2020, pp. 7–27 Search PubMed.
- E. Buhr, N. Senftleben, T. Klein, D. Bergmann, D. Gnieser, C. G. Frase and H. Bosse, Meas. Sci. Technol., 2009, 20, 084025 CrossRef.
- P. M. Carvalho, M. R. Felício, N. C. Santos, S. Gonçalves and M. M. Domingues, Front. Chem., 2018, 6, 237 CrossRef.
- J. Lim, S. P. Yeap, H. X. Che and S. C. Low, Nanoscale Res. Lett., 2013, 8, 381 CrossRef PubMed.
- F. Caputo, J. Clogston, L. Calzolai, M. Rösslein and A. Prina-Mello, J. Controlled Release, 2019, 299, 31–43 CrossRef.
- A. P. Ramos, in Nanocharacterization Techniques, ed. A. L. Da Róz, M. Ferreira, F. de Lima Leite and O. N. Oliveira, William Andrew Publishing, 2017, pp. 99–110 Search PubMed.
- Y. H. Lai, S. Koo, S. H. Oh, E. A. Driskell and J. D. Driskell, Anal. Methods, 2015, 7, 7249–7255 RSC.
- S. Mourdikoudis, R. M. Pallares and N. T. K. Thanh, Nanoscale, 2018, 10, 12871–12934 RSC.
- P. Hole, K. Sillence, C. Hannell, C. M. Maguire, M. Roesslein, G. Suarez, S. Capracotta, Z. Magdolenova, L. Horev-Azaria, A. Dybowska, L. Cooke, A. Haase, S. Contal, S. Manø, A. Vennemann, J.-J. Sauvain, K. C. Staunton, S. Anguissola, A. Luch, M. Dusinska, R. Korenstein, A. C. Gutleb, M. Wiemann, A. Prina-Mello, M. Riediker and P. Wick, J. Nanopart. Res., 2013, 15, 2101 CrossRef.
- V. Filipe, A. Hawe and W. Jiskoot, Pharm. Res., 2010, 27, 796–810 CrossRef CAS.
- E. van der Pol, F. A. W. Coumans, A. Sturk, R. Nieuwland and T. G. van Leeuwen, Nano Lett., 2014, 14, 6195–6201 CrossRef CAS.
- E. van der Pol, F. Coumans, Z. Varga, M. Krumrey and R. Nieuwland, J. Thromb. Haemostasis, 2013, 11, 36–45 CrossRef.
- R. A. Dragovic, C. Gardiner, A. S. Brooks, D. S. Tannetta, D. J. P. Ferguson, P. Hole, B. Carr, C. W. G. Redman, A. L. Harris, P. J. Dobson, P. Harrison and I. L. Sargent, Nanomed.: Nanotechnol., Biol. Med., 2011, 7, 780–788 CrossRef CAS PubMed.
- D. Bachurski, M. Schuldner, P.-H. Nguyen, A. Malz, K. S. Reiners, P. C. Grenzi, F. Babatz, A. C. Schauss, H. P. Hansen, M. Hallek and E. Pogge von Strandmann, J. Extracell. Vesicles, 2019, 8, 1596016 CrossRef CAS PubMed.
- A. Rao, M. Schoenenberger, E. Gnecco, T. Glatzel, E. Meyer, D. Brändlin and L. Scandella, J. Phys.: Conf. Ser., 2007, 61, 971–976 CrossRef.
- Y. Brill-Karniely, Front. Bioeng. Biotechnol., 2020, 8, 605153 CrossRef.
- A. Calò, D. Reguera, G. Oncins, M.-A. Persuy, G. Sanz, S. Lobasso, A. Corcelli, E. Pajot-Augy and G. Gomila, Nanoscale, 2014, 6, 2275–2285 RSC.
- M. Krieg, G. Fläschner, D. Alsteens, B. M. Gaub, W. H. Roos, G. J. L. Wuite, H. E. Gaub, C. Gerber, Y. F. Dufrêne and D. J. Müller, Nat. Rev. Phys., 2019, 1, 41–57 CrossRef.
- Y. F. Dufrêne, Nat. Rev. Microbiol., 2008, 6, 674–680 CrossRef PubMed.
- X. Deng, F. Xiong, X. Li, B. Xiang, Z. Li, X. Wu, C. Guo, X. Li, Y. Li, G. Li, W. Xiong and Z. Zeng, J. Nanobiotechnol., 2018, 16, 102 CrossRef CAS PubMed.
- M. Li, N. Xi and L. Liu, Nanoscale, 2021, 13, 8358–8375 RSC.
- G. R. Heath, E. Kots, J. L. Robertson, S. Lansky, G. Khelashvili, H. Weinstein and S. Scheuring, Nature, 2021, 594, 385–390 CrossRef CAS PubMed.
- C. M. Hoo, N. Starostin, P. West and M. L. Mecartney, J. Nanopart. Res., 2008, 10, 89–96 CrossRef CAS.
- Z. Zhao, Y. Yang, Y. Zeng and M. He, Lab Chip, 2016, 16, 489–496 RSC.
- S. S. Kanwar, C. J. Dunlay, D. M. Simeone and S. Nagrath, Lab Chip, 2014, 14, 1891–1900 RSC.
- S. Fang, H. Tian, X. Li, D. Jin, X. Li, J. Kong, C. Yang, X. Yang, Y. Lu, Y. Luo, B. Lin, W. Niu and T. Liu, PLoS One, 2017, 12, e0175050 CrossRef PubMed.
- X. Han, K. Xu, O. Taratula and K. Farsad, Nanoscale, 2019, 11, 799–819 RSC.
- A. Zuber, M. Purdey, E. Schartner, C. Forbes, B. van der Hoek, D. Giles, A. Abell, T. Monro and H. Ebendorff-Heidepriem, Sens. Actuators, B, 2016, 227, 117–127 CrossRef CAS.
- M.-A. D'Hallewin, L. Bezdetnaya and F. Guillemin, Eur. Urol., 2002, 42, 417–425 CrossRef.
- A.-L. Robson, P. C. Dastoor, J. Flynn, W. Palmer, A. Martin, D. W. Smith, A. Woldu and S. Hua, Front. Pharmacol., 2018, 9, 80 CrossRef PubMed.
- Q.-L. Wang, W.-X. Huang, P.-J. Zhang, L. Chen, C.-K. Lio, H. Zhou, L.-S. Qing and P. Luo, Microchim. Acta, 2019, 187, 61 CrossRef PubMed.
- F. He, H. Liu, X. Guo, B.-C. Yin and B.-C. Ye, Anal. Chem., 2017, 89, 12968–12975 CrossRef CAS PubMed.
- H. Di, Z. Mi, Y. Sun, X. Liu, X. Liu, A. Li, Y. Jiang, H. Gao, P. Rong and D. Liu, Theranostics, 2020, 10, 9303–9314 CrossRef.
- Z. Chen, S.-B. Cheng, P. Cao, Q.-F. Qiu, Y. Chen, M. Xie, Y. Xu and W.-H. Huang, Biosens. Bioelectron., 2018, 122, 211–216 CrossRef PubMed.
- R. Vaidyanathan, M. Naghibosadat, S. Rauf, D. Korbie, L. G. Carrascosa, M. J. A. Shiddiky and M. Trau, Anal. Chem., 2014, 86, 11125–11132 CrossRef PubMed.
- E. Priyadarshini and N. Pradhan, Sens. Actuators, B, 2017, 238, 888–902 CrossRef.
- J. Xu and S. Wu, in Nano-Inspired Biosensors for Protein Assay with Clinical Applications, ed. G. Li, Elsevier, 2019, pp. 91–111 Search PubMed.
- M. G. Saad, H. Beyenal and W.-J. Dong, Biosensors, 2021, 11, 518 CrossRef PubMed.
- M. J. Kangas, R. M. Burks, J. Atwater, R. M. Lukowicz, P. Williams and A. E. Holmes, Crit. Rev. Anal. Chem., 2017, 47, 138–153 CrossRef CAS PubMed.
- Z. Li and M. Zhu, Chem. Commun., 2020, 56, 14541–14552 RSC.
- S. Kwakye, V. N. Goral and A. J. Baeumner, Biosens. Bioelectron., 2006, 21, 2217–2223 CrossRef CAS.
- M. Safavieh, M. U. Ahmed, M. Tolba and M. Zourob, Biosens. Bioelectron., 2012, 31, 523–528 CrossRef CAS PubMed.
- T. Chen, C. Huang, Y. Wang and J. Wu, Chin. Chem. Lett., 2022, 33, 1180–1192 CrossRef CAS.
- Y.-G. Zhou, R. M. Mohamadi, M. Poudineh, L. Kermanshah, S. Ahmed, T. S. Safaei, J. Stojcic, R. K. Nam, E. H. Sargent and S. O. Kelley, Small, 2016, 12, 727–732 CrossRef CAS.
- Q. Zhou, A. Rahimian, K. Son, D.-S. Shin, T. Patel and A. Revzin, Methods, 2016, 97, 88–93 CrossRef CAS.
- B. J. Sanghavi, J. A. Moore, J. L. Chávez, J. A. Hagen, N. Kelley-Loughnane, C.-F. Chou and N. S. Swami, Biosens. Bioelectron., 2016, 78, 244–252 CrossRef CAS PubMed.
- Y. Wang, H. Xu, J. Luo, J. Liu, L. Wang, Y. Fan, S. Yan, Y. Yang and X. Cai, Biosens. Bioelectron., 2016, 83, 319–326 CrossRef CAS PubMed.
- S. R. Shin, T. Kilic, Y. S. Zhang, H. Avci, N. Hu, D. Kim, C. Branco, J. Aleman, S. Massa, A. Silvestri, J. Kang, A. Desalvo, M. A. Hussaini, S.-K. Chae, A. Polini, N. Bhise, M. A. Hussain, H. Lee, M. R. Dokmeci and A. Khademhosseini, Adv. Sci., 2017, 4, 1600522 CrossRef PubMed.
- T. H. Fang, N. Ramalingam, D. Xian-Dui, T. S. Ngin, Z. Xianting, A. T. Lai Kuan, E. Y. Peng Huat and G. Hai-Qing, Biosens. Bioelectron., 2009, 24, 2131–2136 CrossRef CAS PubMed.
- D. Martín-Yerga, Biosensors, 2019, 9, 47 CrossRef PubMed.
- A. Sarkar, H. W. Hou, A. E. Mahan, J. Han and G. Alter, Sci. Rep., 2016, 6, 23589 CrossRef CAS PubMed.
- I. Bouhid de Aguiar and K. Schroën, Membranes, 2020, 10, 316 CrossRef PubMed.
- S. Y. Leong, H. B. Ong, H. M. Tay, F. Kong, M. Upadya, L. Gong, M. Dao, R. Dalan and H. W. Hou, Small, 2022, 18, 2104470 CrossRef CAS PubMed.
- Y. Zhang, M. Chopp, Y. Meng, M. Katakowski, H. Xin, A. Mahmood and Y. Xiong, J. Neurosurg., 2015, 122, 856–867 Search PubMed.
- S. Dubascoux, F. Von Der Kammer, I. Le Hécho, M. P. Gautier and G. Lespes, J. Chromatogr. A, 2008, 1206, 160–165 CrossRef CAS PubMed.
- C. Liu, H. Gong, W. Liu, B. Lu and L. Ye, Ind. Eng. Chem. Res., 2019, 58, 4695–4703 CrossRef CAS.
- S. C. Hur, H. T. K. Tse and D. D. Carlo, Lab Chip, 2010, 10, 274–280 RSC.
- H. Jeon, T. Kwon, J. Yoon and J. Han, Lab Chip, 2022, 22, 272–285 RSC.
- N. Xiang, R. Zhang, Y. Han and Z. Ni, Anal. Chem., 2019, 91, 5461–5468 CrossRef PubMed.
- H. Ren, Z. Zhu, N. Xiang, H. Wang, T. Zheng, H. An, N.-T. Nguyen and J. Zhang, Sens. Actuators, B, 2021, 337, 129758 CrossRef.
| This journal is © The Royal Society of Chemistry 2023 |