Open Access Article
Open Access ArticleGeneration of nanobodies with conformational specificity for tau oligomers that recognize tau aggregates from human Alzheimer's disease samples†
Nikki
McArthur
 a,
Jay D.
Squire
a,
Jay D.
Squire
 a,
Ogechukwu J.
Onyeachonam
a,
Nemil N.
Bhatt
bc,
Cynthia
Jerez
bc,
Abigail L.
Holberton
a,
Ogechukwu J.
Onyeachonam
a,
Nemil N.
Bhatt
bc,
Cynthia
Jerez
bc,
Abigail L.
Holberton
 d,
Peter M.
Tessier
efgh,
Levi B.
Wood
dij,
Rakez
Kayed
bc and
Ravi S.
Kane
d,
Peter M.
Tessier
efgh,
Levi B.
Wood
dij,
Rakez
Kayed
bc and
Ravi S.
Kane
 *aij
*aij
aSchool of Chemical & Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia 30332, USA. E-mail: ravi.kane@chbe.gatech.edu
bMitchell Center for Neurodegenerative Disease, University of Texas Medical Branch, Galveston, Texas 77555, USA
cDepartment of Neurology, University of Texas Medical Branch, Galveston, Texas 77555, USA
dGeorge W. Woodruff School of Mechanical Engineering, Georgia Institute of Technology, Atlanta, Georgia 30332, USA
eDepartment of Chemical Engineering, University of Michigan, North Campus Research Complex, 2800 Plymouth Road, Ann Arbor, MI 48109, USA
fBiointerfaces Institute, University of Michigan, Ann Arbor, MI 48109, USA
gDepartment of Pharmaceutical Sciences, University of Michigan, Ann Arbor, MI 48109, USA
hDepartment of Biomedical Engineering, University of Michigan, Ann Arbor, MI 48109, USA
iWallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology, Atlanta, Georgia 30332, USA
jParker H. Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, Georgia 30332, USA
First published on 22nd October 2024
Abstract
Tauopathies are neurodegenerative diseases that involve tau misfolding and aggregation in the brain. These diseases, including Alzheimer's disease (AD), are some of the least understood and most difficult to treat neurodegenerative disorders. Antibodies and antibody fragments that target tau oligomers, which are especially toxic forms of tau, are promising options for immunotherapies and diagnostic tools for tauopathies. In this study, we have developed conformational, tau oligomer-specific nanobodies, or single-domain antibodies. We demonstrate that these nanobodies, OT2.4 and OT2.6, are highly specific for tau oligomers relative to tau monomers and fibrils. We used epitope mapping to verify that these nanobodies bind to discontinuous epitopes on tau and to support the idea that they interact with a conformation present in the oligomeric, and not monomeric or fibrillar, forms of tau. We show that these nanobodies interact with tau oligomers in brain samples from AD patients and from healthy older adults with primary age-related tauopathy. Our results demonstrate the potential of these nanobodies as tau oligomer-specific binding reagents and future tauopathy therapeutics and diagnostics.
Introduction
Neurodegenerative diseases that involve the misfolding and deposition of aggregates of the protein tau, termed tauopathies, are among the most common neurodegenerative diseases and include Alzheimer's disease (AD) and other less widespread disorders such as progressive supranuclear palsy, Pick's disease, and corticobasal degeneration.1 These neurodegenerative diseases disproportionately affect older adults, so as the size and life expectancy of the human population increase, so will the impact tauopathies have on society. Despite the growing burden of tauopathies, there are few options for safe and effective disease-modifying treatments for them.2 In addition to the lack of effective therapies for tauopathies, there is a need for better diagnostics and research tools to study these diseases. Conformational antibodies that target aggregates of the amyloidogenic proteins involved in neurodegenerative diseases, namely tau, amyloid-β, and α-synuclein, are promising candidates for treatments to slow the progression of neurodegenerative diseases and are useful as reagents to understand the aggregation of amyloid proteins and the role of aggregation in disease progression.3–7In AD, the aggregation of both tau and amyloid-β proteins plays a role in the progression of the disease. Studies show that amyloid-β oligomerization precedes and may promote tau oligomerization.8,9 In AD patients, amyloid-β aggregation into amyloid plaques is followed by elevated levels of hyperphosphorylated tau and tau aggregation.8,9 Tau pathology is more closely related to cognitive decline and clinical symptoms of AD than amyloid-β pathology, suggesting that tau pathology is likely responsible for neurodegeneration.8,9 Lecanemab is a monoclonal antibody that binds specifically to large, soluble amyloid-β protofibrils and is approved by the FDA for the treatment of AD.10 Treatment with lecanemab reduced brain amyloid levels and cognitive decline in patients with early AD.10 The success of lecanemab motivates the further study of amyloid oligomers and protofibrils as targets for drug development.
Tau belongs to the family of microtubule-associated proteins and natively exists as an unfolded monomer bound to microtubules.11 Tau pathology is closely related to the progression and symptoms of AD.11 In AD, tau becomes hyperphosphorylated and undergoes other posttranslational modifications leading to its dissociation from microtubules, misfolding, and aggregation into oligomers and larger fibrils, the primary component of neurofibrillary tangles (NFTs).12 Many studies have indicated that soluble tau oligomers, and not fibrils, are the most toxic form of tau.12–17 Prefibrillar soluble aggregates or oligomers and protofibrils of tau, not NFTs, correlate with cognitive deficits in transgenic mouse models of AD and in humans.14,15 Additionally, small, soluble oligomers, rather than fibrils, are prion-like in nature, exhibit seeding behavior, and are responsible for the spread of tau pathology throughout the brain.15,16 Finally, oligomers, not fibrils, propagate or induce toxic effects in vivo.17 For example, when administered to wild-type mice, tau oligomers, not monomers or fibrils, induce synaptic damage, mitochondrial dysfunction, and cognitive deficits.17 This evidence suggests that tau oligomers are an attractive target for passive immunotherapy.
Many groups have developed antibodies that interact with tau oligomers, but few antibodies are available that conformationally and specifically target tau oligomers.18–21 Castillo-Carranza et al. report the development of a tau oligomer-specific monoclonal antibody (TOMA), via the immunization of mice with tau oligomers formed spontaneously from recombinant tau monomers.4 They found that a single injection of TOMA in P301L tau (JNPL3) mice, a mouse model of tauopathy, or in htau mice (over-expressing human tau) along with the administration of brain-derived tau oligomers, conferred protection against the accumulation of tau oligomers and locomotor and memory deficits.4,5 TOC1, a monoclonal antibody that binds dimeric and larger tau oligomers but not fibrils, was developed by Patterson et al. by immunization of mice with cross-linked tau dimers and oligomers.22 When used to stain human tissue from an AD patient, TOC1 colocalizes with Tau pS422, an early marker of tau pathology, and not with NFT markers.22 Tai et al. describe the development of APNmAb005, a monoclonal antibody that preferentially binds early-stage oligomers over tau monomers and late-stage oligomers through the immunization of mice with tau aggregates encapsulated in artificial vesicles.23 In a tauopathy mouse model, rTg4510, treatment with APNmAb005 partially rescued neuronal loss.23 Ongoing phase I clinical trials with a humanized APNmAb005 began in May 2022. The specificity and binding sites of these antibodies, other tau oligomer-targeting antibodies, and tau-targeting antibodies can be found in ESI Table 1.†
The success of these approaches emphasizes the diagnostic and therapeutic value of antibodies targeting tau oligomers. In this work, we have taken a different approach focusing on nanobodies or single-domain antibodies. Specifically, we have generated conformational, tau oligomer-specific nanobodies through the screening of a synthetic yeast surface display nanobody library against recombinant tau oligomers. Nanobodies are single-domain antibody fragments derived from heavy-chain camelid antibodies. They exhibit high binding affinity and selectivity towards a target antigen, high thermal stability, and can be expressed in bacterial and yeast cells.24 Because of their single-domain nature, they are easily engineered in a multivalent format to enhance avidity. Their small size and extended CDR3 loop allow them to bind concave or less accessible epitopes on antigens.25
While a few tau-targeting nanobodies have been developed,26–35 to our knowledge, there are no reports of conformational tau oligomer-specific nanobodies. A few nanobodies specific for oligomers of the small amyloidogenic peptide amyloid-β have been identified;36–38 however, no conformational nanobodies specific for oligomers of α-synuclein, another large amyloidogenic protein, have been developed either. The development of amyloid oligomer-specific nanobodies is thus a relatively unexplored field.
Here, we establish a method to develop conformational nanobodies that bind specifically to tau oligomers over monomers or fibrils and identify and characterize two of these tau oligomer-specific nanobodies. We show that our nanobodies bind to nonoverlapping epitopes on tau, which are exposed and properly folded only in the oligomeric conformation. Our nanobodies bind to tau oligomers of various sizes present in the brains of older adults and AD patients. Because of their smaller size and the shape of their paratope, these single-domain antibodies likely interact with tau oligomers at three-dimensional epitopes different from the previously reported tau oligomer-specific monoclonal antibodies. These nanobodies could be developed into disease-modifying treatments for AD and other tauopathies and are valuable reagents for the identification and characterization of tau oligomers.
Materials and methods
Expression and purification of tau monomers and oligomers
The gene encoding 2N4R human tau with mutations F8W, C291A, and C322A was synthesized and cloned into the pET-28b plasmid without a His-tag from Gene Universal Inc. (Newark, DE). This mutant 2N4R tau was expressed and purified as described previously.26 The tau-containing plasmid was transformed into BL21(DE3) E. coli (New England Biolabs) according to the manufacturer's protocol. Four 5 mL cultures of LB media with 50 μg mL−1 kanamycin were inoculated with the transformed E. coli. The culture was incubated overnight at 37 °C with rotation. The next day, the four 5 mL cultures were used to inoculate 1 L of LB media with 50 μg mL−1 kanamycin. The culture was grown until it reached an OD600 value between 0.6 and 0.8 at 37 °C and 225 rpm. 0.9 mM IPTG was added to the culture to induce protein expression, and the culture was incubated with rotation at 225 rpm overnight at 22 °C.The next day, the cells were pelleted by centrifugation at 7000g for 7 min and resuspended in 20 mL of lysis buffer (20 mM Tris, 100 mM NaCl, 1 mM EDTA, 1 mM PMSF, protease inhibitor tablet [Sigma-Aldrich]). The cell slurry was sonicated eight times at 25% amplitude with 40 s on and 60 s off pulses, and centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min was used to separate the cell debris from the lysate. The lysate was boiled in a water bath at 100 °C for 30 min, and then the boiled lysate was centrifuged at 15
000g for 10 min was used to separate the cell debris from the lysate. The lysate was boiled in a water bath at 100 °C for 30 min, and then the boiled lysate was centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. The pellet containing precipitated, denatured proteins was discarded, and tau was purified from the supernatant using immobilized metal affinity chromatography (IMAC). Although there is no His-tag on this tau construct, it still interacts with IMAC resin in the absence of imidazole. 2.5 mL of HisPur Ni-NTA resin (Thermo Fisher Scientific) was mixed with the tau-containing supernatant for 1 h at 4 °C. The resin slurry was then loaded into a gravity flow column (G-Biosciences), and the resin was washed with 20 mL of IMAC equilibration buffer (20 mM Tris HCl, 500 mM NaCl, pH 8.0). Tau was eluted in 8 mL of imidazole-containing elution buffer (20 mM Tris HCl, 500 mM NaCl, 100 mM imidazole, pH 8.0) and then concentrated with a 10 kDa MWCO centrifugal filter (MilliporeSigma). To separate tau monomers from tau oligomers that form spontaneously during the expression and purification process, size exclusion chromatography (SEC) with a Superdex 200 Increase 10/300 GL column (Cytiva) in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) was conducted. Tau monomer and oligomer concentrations were determined by the bicinchoninic acid (BCA) assay (Thermo Scientific) and protein purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
000g for 10 min. The pellet containing precipitated, denatured proteins was discarded, and tau was purified from the supernatant using immobilized metal affinity chromatography (IMAC). Although there is no His-tag on this tau construct, it still interacts with IMAC resin in the absence of imidazole. 2.5 mL of HisPur Ni-NTA resin (Thermo Fisher Scientific) was mixed with the tau-containing supernatant for 1 h at 4 °C. The resin slurry was then loaded into a gravity flow column (G-Biosciences), and the resin was washed with 20 mL of IMAC equilibration buffer (20 mM Tris HCl, 500 mM NaCl, pH 8.0). Tau was eluted in 8 mL of imidazole-containing elution buffer (20 mM Tris HCl, 500 mM NaCl, 100 mM imidazole, pH 8.0) and then concentrated with a 10 kDa MWCO centrifugal filter (MilliporeSigma). To separate tau monomers from tau oligomers that form spontaneously during the expression and purification process, size exclusion chromatography (SEC) with a Superdex 200 Increase 10/300 GL column (Cytiva) in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) was conducted. Tau monomer and oligomer concentrations were determined by the bicinchoninic acid (BCA) assay (Thermo Scientific) and protein purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Tau biotinylation
EZ-Link Sulfo-NHS-LC-Biotin (Thermo Scientific) was used to biotinylate tau monomers and oligomers according to the manufacturer's protocol. The biotinylated protein was buffer exchanged into TBS (20 mM Tris HCl, 100 mM NaCl, pH 7.4) with a 10 kDa MWCO centrifugal filter (MilliporeSigma) to remove excess biotin reagent. The extent of biotinylation was measured by characterizing mixtures of streptavidin (Rockland Immunochemicals) and biotinylated tau monomers or oligomers with SDS-PAGE at 4 °C.Tau fibril formation
To form tau fibrils, 5 μM tau monomers, 2.5 μM heparin, and PBS were mixed in a final volume of 1 mL. This mixture was incubated at 37 °C at 250 rpm for 3–4 days. To remove leftover tau monomers, the tau fibrils were buffer exchanged with a 100 kDa MWCO centrifugal filter (MilliporeSigma) into HEPES buffer (20 mM HEPES, 100 mM NaCl, 1 mM EDTA, pH 7.4), and the fibril concentration was determined by the BCA assay (Thermo Scientific).Dynamic light scattering
100 μL of tau monomers, oligomers, or fibrils at 70 μg mL−1 were added to a ZEN0040 cuvette (Malvern) and loaded into a Zetasizer Nano ZS instrument (Malvern). Zetasizer software (Malvern) was used to record 5 size measurements per sample at 25 °C using refractive index and absorption parameters for a protein sample in PBS with a 173° backscatter measurement angle.SDS-PAGE
1 μg of protein was diluted in Nu-PAGE lithium dodecyl sulfate (LDS) sample buffer (Invitrogen). Protein samples and PageRuler Plus Prestained Protein Ladder (Thermo Scientific) were loaded onto a 4–12% Bis-Tris gel (Invitrogen) and the gel was run for 50 min at 120 V. Gels that were not transferred onto a membrane for western blots were rinsed with water, stained with Imperial Protein Stain (Thermo Scientific), destained in water, and imaged using the ChemiDoc MP imaging system (Bio-Rad).Tau immunoblot characterization
Dot blots and western blots with Tau5 and T2239 antibodies were used to characterize tau monomers, oligomers, fibrils, and biotinylated tau monomers and oligomers. Dot blots were conducted by spotting 0.1 μg of tau protein or BSA in 1.3 μL PBS onto a 0.2 μm nitrocellulose membrane in triplicate. The membrane was allowed to dry for 30 min and then blocked with 5% BSA in TBST (20 mM Tris, 150 mM NaCl, 0.1% Tween-20, pH 7.4) for 1 h at room temperature. The membrane was next incubated with Tau5 primary antibody (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) in 5% BSA in TBST for 2 h at room temperature. The membrane was washed with TBST three times for 5 min and incubated with goat anti-mouse IgG Alexa Fluor 647 (Invitrogen, 1
5000) in 5% BSA in TBST for 2 h at room temperature. The membrane was washed with TBST three times for 5 min and incubated with goat anti-mouse IgG Alexa Fluor 647 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) secondary antibody in 5% BSA in TBST for 1 h at room temperature. The membrane was washed with TBST three times for 5 min and imaged with the ChemiDoc MP imaging system (Bio-Rad). With the same membrane, primary and secondary antibody incubations were repeated with T22 (1
2000) secondary antibody in 5% BSA in TBST for 1 h at room temperature. The membrane was washed with TBST three times for 5 min and imaged with the ChemiDoc MP imaging system (Bio-Rad). With the same membrane, primary and secondary antibody incubations were repeated with T22 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) and donkey anti-rabbit IgG Alexa Fluor 488 (Invitrogen, 1
250) and donkey anti-rabbit IgG Alexa Fluor 488 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000), respectively, and the membrane was imaged with the ChemiDoc MP imaging system (Bio-Rad).
2000), respectively, and the membrane was imaged with the ChemiDoc MP imaging system (Bio-Rad).
Western blots were conducted with tau monomers, oligomers, and biotinylated tau monomers and oligomers. 1 μg of protein was run on SDS-PAGE as described and proteins were transferred to a 0.2 μm nitrocellulose membrane using a Trans-Blot Turbo Mini 0.2 μm Nitrocellulose Transfer Pack (Bio-Rad) and a Trans-Blot Turbo Transfer System (Bio-Rad). The membrane was rinsed with TBST and then, to confirm protein transfer, was stained with Ponceau S solution (Rockland Immunochemicals) for 15 min. The membrane was rinsed with water and then imaged with the ChemiDoc MP imaging system (Bio-Rad). The membrane was washed with TBST three times for 5 min to remove the Ponceau S solution and then blocked with 5% BSA in TBST for 1 h at room temperature. Antibody incubations and membrane imaging were conducted as described for dot blots.
Yeast cell culture
Yeast cells expressing the synthetic nanobody library40 were grown overnight in tryptophan deficient SD-Trp media (3.8 g L−1 yeast synthetic drop-out medium supplements without tryptophan, 6.7 g L−1 yeast nitrogen base, 20 g L−1 glucose, 100 U mL−1 penicillin–streptomycin, pH 6.0) at 30 °C, 225 rpm, and 107 cells per mL. Nanobody expression on the surface of the cells was induced by culturing the yeast in SG-Trp media (3.8 g L−1 yeast synthetic drop-out medium supplements without tryptophan, 6.7 g L−1 yeast nitrogen base, 20 g L−1 galactose, 100 U mL−1 penicillin–streptomycin, pH 6.0) overnight at 25 °C and 250 rpm.Magnetic-activated cell sorting
Magnetic-activated cell sorting (MACS) was performed as described previously.26 Two rounds of MACS were conducted to select tau oligomer-binding nanobodies. In each round, negative selections were followed by a positive selection against tau oligomer-coated magnetic Dynabeads Biotin Binder beads (Invitrogen).Dynabeads were prepared following the manufacturer's protocol. For MACS Round 1, to prepare oligomer-coated beads for positive selection, 107 beads were first washed twice using a magnet and PBS with 0.1% BSA and then incubated with 12.5 μg of biotinylated tau oligomers in 500 μL of 0.1% BSA in PBS overnight at 4 °C with rotation. 107 beads for negative sorts containing no tau protein were washed and incubated in 0.1% BSA in PBS overnight at 4 °C with rotation. The following day, the beads were washed with 0.1% BSA in PBS and resuspended in 100 μL of 0.1% BSA in PBS. 1010 nanobody-expressing library cells were mixed with the unlabeled Dynabeads and incubated for 1 h at room temperature with rotation. Unbound yeast cells were then collected with a magnet, and this negative selection step against unlabeled beads was repeated once. After the negative selections, the remaining yeast cells were incubated with tau oligomer-coated beads for 1 h at room temperature with rotation. Unbound cells were discarded, and the beads were washed 5 times with 1 mL of 0.1% BSA in PBS using a magnetic tube rack. The beads were transferred to 5 mL of SD-Trp media and yeast cells bound to the beads were grown for 48 h at 30 °C and 250 rpm.
During MACS Round 2, two sets of selections starting with 109 nanobody-expressing MACS Round 1 yeast cells were performed in parallel. Negative selections against unlabeled Dynabeads were first performed as described with both sets of yeast. After negative selections against the unlabeled Dynabeads, one set of yeast underwent a negative selection against 106 Dynabeads coated with 1.25 μg of biotinylated tau monomers. Finally, a positive selection with both sets of yeast against tau oligomer-coated beads was performed as described. Oligomer-coated beads for the positive selection were prepared by mixing 106 Dynabeads with 1.25 μg of biotinylated tau oligomers in 50 μL of 0.1% BSA in PBS overnight at 4 °C with rotation.
Fluorescence-activated cell sorting
Two rounds of fluorescence-activated cell sorting (FACS) were conducted with yeast from each of the two MACS Round 2 sorts—one without a negative selection against tau monomers and one with a negative selection against tau monomers. Induced yeast cells from the previous MACS or FACS rounds were labeled for FACS as described previously.26 107 yeast cells were labeled with 10 nM biotinylated tau oligomers and with an anti-HA tag rabbit antibody (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) to check for nanobody expression in 100 μL of 0.1% BSA in PBS for 20 min at room temperature with rotation. Yeast cells were washed with 0.1% BSA in PBS and then incubated with streptavidin R-phycoerythrin conjugate (Invitrogen, 1
200) to check for nanobody expression in 100 μL of 0.1% BSA in PBS for 20 min at room temperature with rotation. Yeast cells were washed with 0.1% BSA in PBS and then incubated with streptavidin R-phycoerythrin conjugate (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) and donkey anti-rabbit IgG Alexa Fluor 488 (Invitrogen, 1
250) and donkey anti-rabbit IgG Alexa Fluor 488 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) in 100 μL of 0.1% BSA for 10 min on ice. Cells were washed with 0.1% BSA in PBS and sorted on a BD FACSAria Fusion cytometer. Selected yeast cells were added to 5 mL of SD-Trp media and cultured for 48 h.
500) in 100 μL of 0.1% BSA for 10 min on ice. Cells were washed with 0.1% BSA in PBS and sorted on a BD FACSAria Fusion cytometer. Selected yeast cells were added to 5 mL of SD-Trp media and cultured for 48 h.
Individual nanobody selection
Yeast cells from each of the four FACS rounds were plated on SD-Trp agar plates (SD-Trp media supplemented with 15 g L−1 agar) and grown for 48 h at 30 °C. 15 colonies from each round of sorting were randomly selected and used to inoculate 5 mL SD-Trp cultures. The cultures were allowed to grow for 24–48 h, and then plasmid DNA was extracted from the yeast using a Zymoprep Yeast Plasmid Miniprep II kit (Zymo Research). The extracted plasmids were transformed into NEB 5-alpha Competent E. coli cells (New England Biolabs) according to the manufacturer's protocol. The transformed E. coli was grown in 5 mL of LB media with 100 μg mL−1 ampicillin overnight at 37 °C with rotation. Plasmid DNA was extracted from the E. coli using the E.Z.N.A. Plasmid Mini Kit I (Omega Bio-tek) and sequenced by MCLAB (San Francisco, CA).Flow cytometry
107 nanobody-expressing yeast cells were labeled with biotinylated tau oligomers or biotinylated tau monomers and with an anti-HA tag rabbit antibody (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) in 100 μL of 0.1% BSA in PBS for 20 min at room temperature with rotation. Yeast cells were washed once with 0.1% BSA in PBS and incubated in 100 μL of 0.1% BSA for 10 min on ice with streptavidin R-phycoerythrin conjugate (Invitrogen, 1
200) in 100 μL of 0.1% BSA in PBS for 20 min at room temperature with rotation. Yeast cells were washed once with 0.1% BSA in PBS and incubated in 100 μL of 0.1% BSA for 10 min on ice with streptavidin R-phycoerythrin conjugate (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) and donkey anti-rabbit IgG Alexa Fluor 488 (Invitrogen, 1
250) and donkey anti-rabbit IgG Alexa Fluor 488 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500). Cells were washed with 0.1% BSA in PBS and tau binding was evaluated on a CytoFLEX S flow cytometer (Beckman Coulter). Data analysis was performed using the CytExpert software (Beckman Coulter).
500). Cells were washed with 0.1% BSA in PBS and tau binding was evaluated on a CytoFLEX S flow cytometer (Beckman Coulter). Data analysis was performed using the CytExpert software (Beckman Coulter).
Expression and purification of bivalent OT2.4-Fc, OT2.6-Fc, and MT3.1-Fc
Genes encoding bivalent OT2.4 fused to a rabbit IgG Fc and bivalent OT2.6 fused to a mouse IgG Fc (used for epitope mapping analysis and dot blotting) and bivalent OT2.4 or bivalent OT2.6 fused to a human IgG Fc (used for immunoblot analysis) were synthesized and cloned into the TGEX-HC plasmid (Antibody Design Labs) from Gene Universal (Newark, DE). These plasmids and a TGEX-HC plasmid for the expression of bivalent MT3.1 fused to a human IgG Fc were transfected into Expi293F cells (Thermo Fisher Scientific) using the ExpiFectamine 293 Transfection Kit (Thermo Fisher Scientific) and protocol. The Fc fusion proteins were expressed for six days in the Expi293F cells and purified as previously described.26 Cell cultures were centrifuged at 5000g for 10 min to pellet out the Expi293F cells, and the supernatant was loaded onto a 1 mL HiTrap MabSelect SuRe column (GE). Fc fusion proteins were purified according to the manufacturer's protocol. Protein elution was conducted with a 20 column volume linear gradient of 0.1 M sodium citrate, pH 3.5, and the purified Fc fusion proteins were dialyzed into PBS. Protein concentrations were determined using the BCA assay (Thermo Scientific) and purity was confirmed by SDS-PAGE.OT2.4 and OT2.6 western blot characterization
Western blots with 1 μg of tau monomers and oligomers were conducted as described above to probe binding of OT2.4 and OT2.6 to tau monomers and oligomers. 50 nM bivalent OT2.4-Fc or bivalent OT2.6-Fc were used as primary antibodies and goat anti-human IgG Alexa Fluor 488 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) was used as the secondary antibody.
2000) was used as the secondary antibody.
OT2.4 and OT2.6 dot blot characterization
Two-fold serial dilutions of tau monomers, oligomers, or fibrils starting at 256 ng μL−1 in 1 μL of PBS were spotted onto a 0.2 μm nitrocellulose membrane in triplicate. Dot blots were probed as described above with 50 nM bivalent OT2.4-Fc or bivalent OT2.6-Fc. Goat anti-human IgG Alexa Fluor 647 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) was used as the secondary antibody. Significance was determined by Brown-Forsythe and Welch ANOVA tests between binding to 256 ng of each antigen. Statistical analysis was carried out using Prism 10 (GraphPad).
2000) was used as the secondary antibody. Significance was determined by Brown-Forsythe and Welch ANOVA tests between binding to 256 ng of each antigen. Statistical analysis was carried out using Prism 10 (GraphPad).
For EC50 calculations, 0.1 μg of tau oligomer 1, oligomer 2, or oligomer 3 were spotted onto 0.2 μm nitrocellulose membranes in triplicate. Oligomer 1 and oligomer 2 were separate preparations of tau oligomers produced with the method described above. Oligomer 3 was produced with a different protocol as described previously.41,42 Dot blots were probed as described above with three-fold serial dilutions of bivalent OT2.4 fused to a rabbit Fc or bivalent OT2.6-Fc. Donkey anti-rabbit IgG Alexa Fluor 488 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) or goat anti-human IgG Alexa Fluor 647 (Invitrogen, 1
2000) or goat anti-human IgG Alexa Fluor 647 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) were used as secondary antibodies for probing OT2.4 or OT2.6 binding, respectively. Binding data for each combination of tau oligomer antigen and nanobody were fit to a binding isotherm using global nonlinear least squares regression.43 Maximum absorbance values for each repeat were used to normalize the data. A single EC50 value for each antigen and nanobody combination was determined as a fitted parameter across all three repeats.
2000) were used as secondary antibodies for probing OT2.4 or OT2.6 binding, respectively. Binding data for each combination of tau oligomer antigen and nanobody were fit to a binding isotherm using global nonlinear least squares regression.43 Maximum absorbance values for each repeat were used to normalize the data. A single EC50 value for each antigen and nanobody combination was determined as a fitted parameter across all three repeats.
Bis-ANS assay
200 nM tau oligomers were incubated with various concentrations of bivalent OT2.4-Fc, bivalent OT2.6-Fc, or a control Fc fusion nanobody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, and 1
4, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 molar ratios of tau oligomers
8 molar ratios of tau oligomers![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nanobody) for 90 minutes. These mixtures or tau oligomers with no nanobody were then used to seed freshly prepared 20 μM tau monomers. Samples were incubated for 48 hours with shaking at 300 rpm. Bis-ANS assays were performed using a BioTek Synergy H1 Multimode Reader. In brief, 50 μL of each incubated sample was added to a clear-bottom 96-well plate at a final concentration of 2 μM protein, with 40 μM bis-ANS. Bis-ANS fluorescence was measured at 490 nm following excitation at 380 nm. Each sample was tested in triplicate, and data were analyzed using GraphPad Prism. Significance was determined by two-way ANOVA.
nanobody) for 90 minutes. These mixtures or tau oligomers with no nanobody were then used to seed freshly prepared 20 μM tau monomers. Samples were incubated for 48 hours with shaking at 300 rpm. Bis-ANS assays were performed using a BioTek Synergy H1 Multimode Reader. In brief, 50 μL of each incubated sample was added to a clear-bottom 96-well plate at a final concentration of 2 μM protein, with 40 μM bis-ANS. Bis-ANS fluorescence was measured at 490 nm following excitation at 380 nm. Each sample was tested in triplicate, and data were analyzed using GraphPad Prism. Significance was determined by two-way ANOVA.
Binding epitope analysis
Epitope mapping with OT2.4 and OT2.6 was performed using a membrane containing overlapping 15 amino acid peptides scanning the full length 2N4R human tau sequence. The peptides were synthesized on a PepSpots cellulose membrane (JPT Peptide Technologies, Berlin, Germany). Peptides overlapped by 11 amino acids. The peptide membrane was rinsed with methanol for 5 min and washed three times with TBST (50 mM Tris, 137 mM NaCl, 2.7 mM KCl, 0.05% Tween-20, pH 8.0) for 3 min. The membrane was blocked with 5% BSA in TBST at 4 °C overnight and then incubated with 1 μg mL−1 bivalent OT2.4 fused to a rabbit Fc (12.8 nM) in 5% BSA in TBST at 4 °C overnight. The membrane was washed three times with TBST for 5 min and incubated with HRP-conjugated donkey anti-rabbit IgG (Cytiva, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) in 5% BSA in TBST for 1 h at room temperature. The membrane was washed three times with TBST for 5 min and then incubated with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) for 30 s and imaged using the ChemiDoc MP imaging system (Bio-Rad).
5000) in 5% BSA in TBST for 1 h at room temperature. The membrane was washed three times with TBST for 5 min and then incubated with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) for 30 s and imaged using the ChemiDoc MP imaging system (Bio-Rad).
The membrane was also incubated with 1 μg mL−1 bivalent OT2.6 fused to a mouse Fc (12.5 nM) in 5% BSA in TBST at 4 °C overnight. The membrane was washed three times with TBST for 5 min and incubated with a goat anti-mouse IgG Alexa Fluor 546 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) in 5% BSA in TBST for 1 h at room temperature. The membrane was washed three times with TBST for 5 min and imaged using the ChemiDoc MP imaging system (Bio-Rad).
2000) in 5% BSA in TBST for 1 h at room temperature. The membrane was washed three times with TBST for 5 min and imaged using the ChemiDoc MP imaging system (Bio-Rad).
Western blotting of human tissue lysates
Tissues were collected post-mortem from patients who had provided prior informed consent and cryopreserved at the Emory University Goizueta Alzheimer's Disease Research Center in accordance with guidelines approved by the Emory University Institutional Review Board. Postmortem brain tissue from human subjects with AD (n = 5) and control subjects (n = 5), obtained from the Goizueta Alzheimer's Disease Center at Emory University, were lysed in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). Sections of approximately 100 mg were cut from frozen tissues and placed into 300 μL ice cold PBS containing cOmplete™, Mini Protease Inhibitor Cocktail (Roche, 1 tablet per 5 mL). Tissues were lysed using a TissueLyser II (Qiagen) and one 5 mm stainless steel bead (Qiagen) per sample for 4 min at 30 Hz. The stainless steel beads were removed, and the lysates were centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 4 °C for 10 min. The pellet was discarded, and a BCA assay (Thermo Scientific) was performed with the supernatant to determine the total protein concentration. The tissue lysate was aliquoted and stored at −80 °C.
000 rpm and 4 °C for 10 min. The pellet was discarded, and a BCA assay (Thermo Scientific) was performed with the supernatant to determine the total protein concentration. The tissue lysate was aliquoted and stored at −80 °C.
Frozen lysates were thawed on ice and 15 μg of total protein was diluted in Laemmli sample buffer (Bio-Rad) and run on two identical western blots as described above. First, 5 nM bivalent OT2.4-Fc (membrane 1) or 5 nM bivalent OT2.6-Fc (membrane 2) primary antibodies and goat anti-human IgG Alexa Fluor 488 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) secondary antibodies were used to probe OT2.4 and OT2.6 binding to the tissue samples. Both membranes were then incubated with 3 nM Phospho-Tau (Ser202, Thr205) monoclonal antibody (AT8) (Invitrogen) primary antibody, washed with TBST (20 mM Tris, 150 mM NaCl, 0.1% Tween-20, pH 7.4), and incubated with goat anti-mouse IgG Alexa Fluor 647 (Invitrogen, 1
2000) secondary antibodies were used to probe OT2.4 and OT2.6 binding to the tissue samples. Both membranes were then incubated with 3 nM Phospho-Tau (Ser202, Thr205) monoclonal antibody (AT8) (Invitrogen) primary antibody, washed with TBST (20 mM Tris, 150 mM NaCl, 0.1% Tween-20, pH 7.4), and incubated with goat anti-mouse IgG Alexa Fluor 647 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) secondary antibody. Membrane 2 was incubated with T22 (1
2000) secondary antibody. Membrane 2 was incubated with T22 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) primary antibody, washed with TBST, and incubated with HRP-conjugated donkey anti-rabbit IgG (Cytiva, 1
250) primary antibody, washed with TBST, and incubated with HRP-conjugated donkey anti-rabbit IgG (Cytiva, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) to confirm the presence of tau oligomers. Next, antibodies were stripped from both membranes using Restore Western Blot Stripping Buffer (Thermo Scientific) for 30 min at 37 °C with shaking. The membranes were washed three times for 5 min with TBST, blocked with 5% BSA in TBST for 1 h at room temperature, incubated with GAPDH primary antibody (GeneTex, 1
5000) to confirm the presence of tau oligomers. Next, antibodies were stripped from both membranes using Restore Western Blot Stripping Buffer (Thermo Scientific) for 30 min at 37 °C with shaking. The membranes were washed three times for 5 min with TBST, blocked with 5% BSA in TBST for 1 h at room temperature, incubated with GAPDH primary antibody (GeneTex, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), washed with TBST, and incubated with goat anti-mouse IgG Alexa Fluor 647 (Invitrogen, 1
1000), washed with TBST, and incubated with goat anti-mouse IgG Alexa Fluor 647 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000) secondary antibody. All images were taken using the ChemiDoc MP imaging system (Bio-Rad).
2000) secondary antibody. All images were taken using the ChemiDoc MP imaging system (Bio-Rad).
Results and discussion
Identification of tau oligomer-specific nanobodies
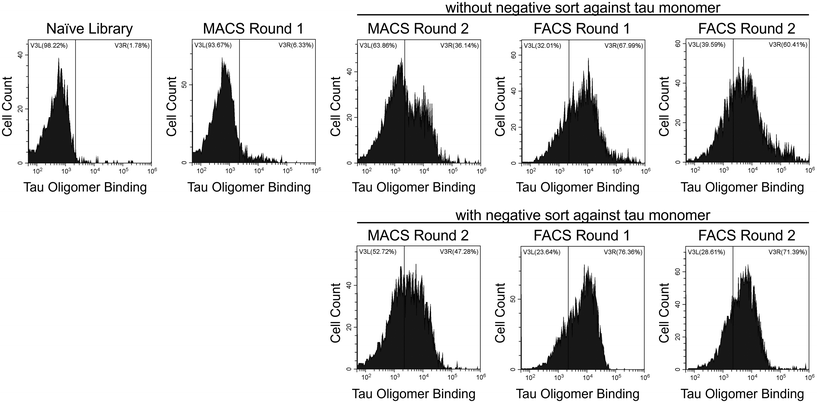
To identify tau oligomer-specific nanobodies, we screened a synthetic yeast surface display nanobody library against recombinant tau oligomers. The nanobody library was created by McMahon et al. and designed to recapitulate the natural amino acid diversity seen in the CDRs of camelid heavy-chain antibodies.40 Our recombinant oligomers form spontaneously during the expression and purification of the 2N4R isoform of human tau in E. coli. We separated oligomeric tau from monomeric tau using SEC (ESI Fig. 1†) and characterized their sizes using dynamic light scattering (DLS; ESI Fig. 2†). Additionally, we characterized our tau proteins via dot blot (ESI Fig. 3a and b†) and western blot (ESI Fig. 3c–f†). We verified the conformation of our tau oligomers using a tau oligomer-specific polyclonal antibody, T22, in these immunoblots (ESI Fig. 3b and f†).39We performed MACS and FACS to select our nanobodies. During MACS Round 2, two sets of screens were performed in parallel. The first included a positive selection against tau oligomers, and the second included a negative selection against tau monomers and a positive selection against tau oligomers. FACS screens after MACS Round 2 were conducted under identical conditions on the yeast from the MACS screen both without and with the negative selection against tau monomers. After each round of sorting, yeast cells were labeled with tau oligomers (Fig. 1) and monomers (data not shown) to monitor the enrichment of binding to tau oligomers and the level of undesired binding to tau monomers. Over the course of the screens, binding to tau oligomers was greatly enriched.
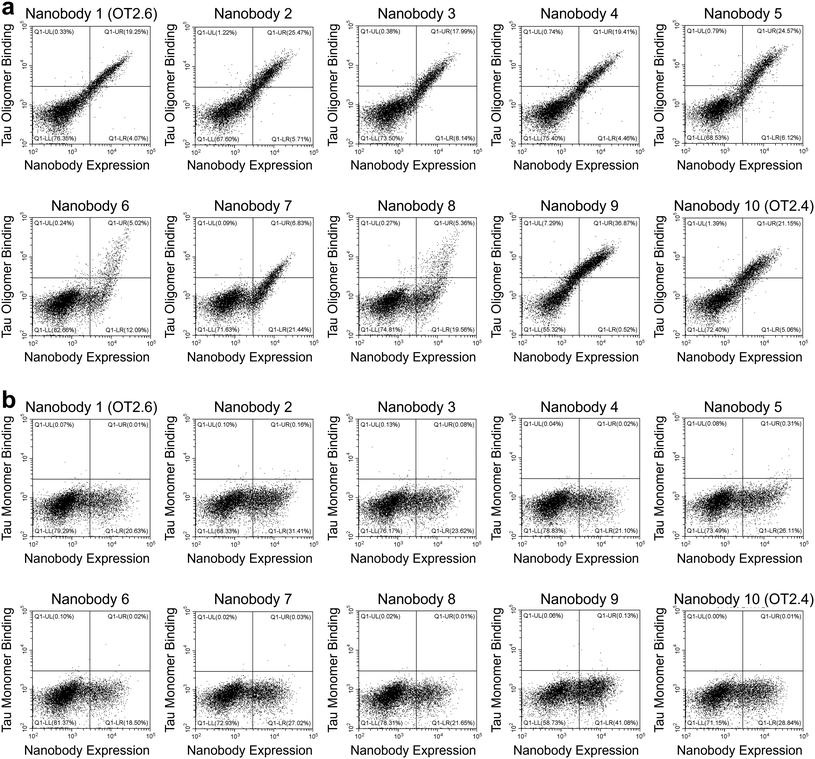
After FACS Round 2, 15 nanobodies from each of the four FACS screens were selected and sequenced. From the 60 nanobodies sequenced, ten unique tau oligomer-binding nanobodies were identified (Table 1, and ESI Table 2†). Yeast cells expressing these nanobodies were labeled with a range of concentrations of tau oligomers (1 nM–10 nM) and of tau monomers (10 nM–1000 nM) and the extent of binding was assessed with flow cytometry (Fig. 2a and b). All ten nanobodies showed binding to tau oligomers and much less binding to tau monomers. Two of the nanobodies, OT2.4 and OT2.6, appeared to be either stronger binders to tau oligomers or more specific binders to tau oligomers relative to tau monomers compared to the other selected nanobodies. These nanobodies were chosen for further characterization.
| Nanobody | CDR1 | CDR2 | CDR3 |
|---|---|---|---|
| 1 (OT2.6) | GRTFRYNAM | ELVAAITVRTGSTYY | AVDRDYLVRYSQLYREYGY |
| 2 | GITFRSYAM | EFVAAITSGGASTYY | AARRPYKPYDY |
| 3 | GRTFYRYTM | ELVAAISFRAGRTYY | AADQYLSADYDY |
| 4 | GSIFRANAM | ELVAAITTGSRTYY | ARRALYLPQRINYSDAMDY |
| 5 | GYTFGRNTM | EFVAAITQSGGNTYY | NARLRPPYGWKYGY |
| 6 | GFTFGGANVM | ELVAAITYGGGSTYY | AARSYRYWTQILYDY |
| 7 | GRTFTSYTM | ELVAAITDRGGRTYY | NTVWGYHGGDEVDH |
| 8 | GRTFVWNAM | ELVAAITYRGASTYY | NARKYVTLKYDY |
| 9 | GRTFGRNAM | ELVAAITTGGSTNY | AATRWRKWYYY |
| 10 (OT2.4) | GIISNNNAM | EFVAAISTSGGSTYY | NRRVVERYWRGYWYREDGY |
Characterization of nanobodies confirms specificity to tau oligomers
We designed, expressed, and purified bivalent versions of the nanobodies OT2.4 and OT2.6 fused to a human IgG Fc (ESI Fig. 4†). These Fc fusion constructs were used to confirm and further examine the specificity of the nanobodies to tau oligomers over tau monomers and fibrils using western blots and dot blots. Western blots with recombinant tau monomers and tau oligomers show binding of bivalent OT2.4-Fc and bivalent OT2.6-Fc to the oligomers and not monomers (Fig. 3), verifying the specificity seen in flow cytometry experiments. Next, we probed the binding of bivalent OT2.4-Fc, bivalent OT2.6-Fc, and bivalent MT3.1-Fc to serial dilutions of tau monomers, oligomers, and fibrils on dot blots (Fig. 4). MT3.1 is a pan-tau nanobody developed by our laboratory that binds to tau monomers, oligomers, and fibrils.26 These dot blots showed high levels of specificity for bivalent OT2.4-Fc and bivalent OT2.6-Fc to tau oligomers over both monomers and fibrils. In contrast, bivalent MT3.1-Fc preferentially binds tau fibrils, and to some extent, tau monomers, over tau oligomers. Bivalent OT2.4-Fc, in particular, was extremely selective towards tau oligomers relative to monomers and fibrils.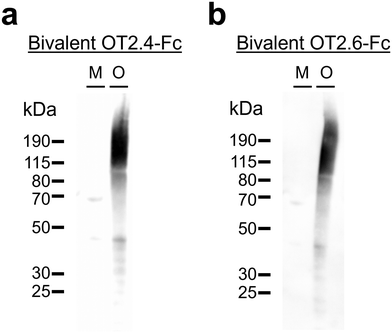 | ||
| Fig. 3 Western blotting with OT2.4 and OT2.6. (a and b) Specific binding of (a) bivalent OT2.4-Fc and (b) bivalent OT2.6-Fc was evaluated with western blots containing recombinant tau monomers (M) and recombinant tau oligomers (O). Ponceau S staining of these membranes is shown in ESI Fig. 8a and b, respectively, and unprocessed images of these membranes are shown in ESI Fig. 9a and b,† respectively. | ||
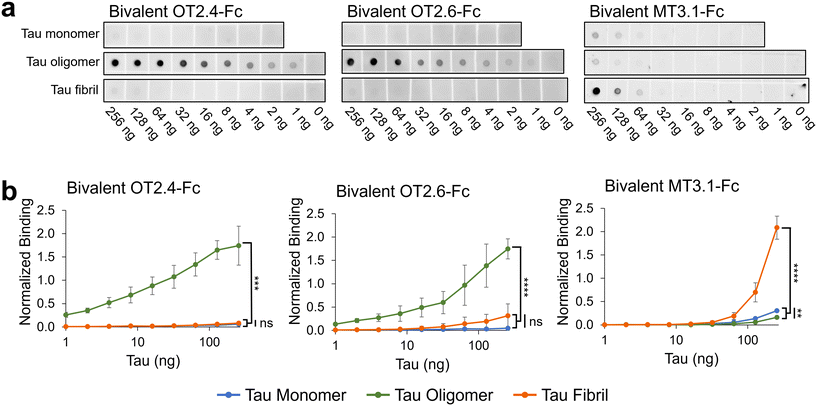 | ||
| Fig. 4 Analysis of OT2.4 and OT2.6 specificity towards tau oligomers. (a) Dot blots containing serial dilutions of tau monomers, tau oligomers, and tau fibrils were probed with bivalent OT2.4-Fc, bivalent OT2.6-Fc, and bivalent MT3.1-Fc. Shown are representative images from six repeats from two independent assays. Unprocessed images of these membranes are shown in ESI Fig. 10 a, c, and e.† Images of membranes from a second independent repeat are shown in ESI Fig. 10 b, d, and f.† (b) Binding of bivalent OT2.4-Fc, bivalent OT2.6-Fc, and bivalent MT3.1-Fc to tau monomers (blue), tau oligomers (green), and tau fibrils (orange) on dot blots was quantified and plotted. Data points are averages from six repeats from two independent assays and error bars indicate standard deviation. ****P < 0.0001, ***P < 0.001, **P < 0.01, ns: not significant, determined by Brown-Forsythe and Welch ANOVA tests. | ||
Since tau oligomers are known to be highly heterogeneous, we confirmed binding of bivalent OT2.4-Fc and bivalent OT2.6-Fc to tau oligomers from different sources. These tau oligomer samples were created in two different laboratories with different preparation methods. Both nanobodies bound to all three tau oligomer samples. Bivalent OT2.4-Fc bound with EC50s of 27 nM, 19 nM, and 160 nM to oligomer preparations 1, 2, and 3, respectively (ESI Fig. 5†). Bivalent OT2.6-Fc bound with EC50s of 1.0, 0.53, and 240 nM to oligomer preparations 1, 2, and 3, respectively (ESI Fig. 5†). Through bis-ANS fluorescence spectroscopy, we also showed that OT2.4 and OT2.6 inhibited the oligomer-seeded aggregation of tau monomers in a dose-dependent manner (ESI Fig. 6†).
Epitope mapping reveals that nanobodies interact with tau oligomers via multiple non-overlapping epitopes
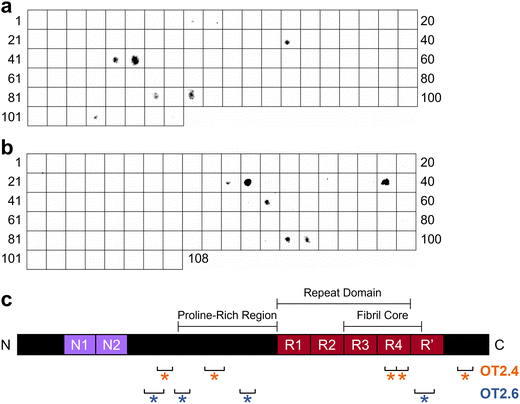
To determine the epitopes at which OT2.4 and OT2.6 bind to tau, we performed epitope mapping. To conduct this mapping, we used a membrane containing 15-mer peptides spanning the full length of the 2N4R tau isoform with 11 amino acid overlaps. Incubation of the membrane with bivalent OT2.4-Fc and bivalent OT2.6-Fc revealed that both nanobodies bind to multiple discontinuous epitopes on tau (Fig. 5). OT2.4 bound to peptides containing residues 133–147, 177–195, 343–367, and 413–427 (Fig. 5a and c). These four epitopes lie within the projection domain, proline-rich domain, R4 repeat of the repeat domain and microtubule binding domain, and C-terminal end of tau, respectively. The epitope containing residues 343–367 is buried within the fibril core of tau when it assembles into tau fibrils, while the other three epitopes are within the disordered fuzzy coat of the fibrils.44 OT2.6 bound to peptides containing residues 121–139, 149–163, 209–223, and 373–391 (Fig. 5b and c). The first of these epitopes is within the projection domain, the second and third are within the proline-rich domain, and the fourth is within the R′ repeat in the microtubule binding domain. The first three epitopes lie in the fuzzy coat of a tau fibril while the last epitope is partially within the fibril core.44 The third OT2.6 epitope, residues 209–223, overlap with the binding epitope of the conformational tau oligomer-specific antibody TOC1 (209–224).22,45 All of the binding epitopes of both nanobodies are present in all six isoforms of tau.Discontinuous, or conformational, epitopes are epitopes that contain multiple groups of residues essential to antibody binding that are separated in the antigen's primary sequence but brought together in spatial proximity when the antigen is properly folded.46 Based on our immunoblot specificity analysis and the location of these epitopes, we believe that they are only accessible and properly folded in oligomeric tau, not monomeric or fibrillar tau. From these results, we conclude that OT2.4 and OT2.6 are conformational, tau oligomer-specific nanobodies.
OT2.4 and OT2.6 recognize tau oligomers from human Alzheimer's disease and healthy older adult brain tissues
Next, we investigated the ability of OT2.4 and OT2.6 to recognize tau oligomers present in brain lysate samples from AD patients and healthy older adults. Tau oligomers are found in the brains of humans with AD and can be detected at early stages of the disease.14,39,47 Additionally, tau oligomers are present in the brain tissue and serum of older adults without neurodegenerative diseases and appear during the aging process.48,49 Kolarova et al. reported that in healthy older adults, tau oligomers are present in serum and tau oligomer levels correlate with aging.48 In a study by Maeda et al., tau oligomers were detected in the brains of older adults with Braak stage I neuropathology, a stage before the onset of clinical symptoms of AD and the presence of NFTs in the frontal cortex.49 They found no significant difference between tau oligomer levels in Braak I, II, and V stage frontal cortex samples.49 They also observed tau oligomers at low levels in Braak stage 0 samples, but these levels were significantly lower than those in Braak I, II, and I samples.49We conducted western blots with brain samples from five individuals with AD (ages 77 to 87, Braak stage VI) and from five control individuals with primary age-related tauopathy (PART) (ages 70 to >89, Braak stages I, II, or III) (ESI Table 3†). We detected tau oligomers with bivalent OT2.4-Fc (Fig. 6a) and bivalent OT2.6-Fc (Fig. 6b) in all of the AD tissues and non-AD (control) tissues. We note that the detection of tau oligomers in tissues of older adults without AD is expected and consistent with the studies described above. Staining of these tissues with phospho-tau antibody AT8 (Fig. 6c and d) overlapped with bivalent OT2.4-Fc and bivalent OT2.6-Fc staining at 150 kDa and higher molecular weight bands. To confirm that the bands stained by bivalent OT2.4-Fc and bivalent OT2.6-Fc are tau oligomers, we probed binding with T22, an antibody that has previously been shown to bind to tau oligomers present in AD brain tissue,39 and saw an overlap of staining at bands at 50 kDa and 150 kDa (ESI Fig. 7†). These results indicate that nanobodies OT2.4 and OT2.6 may be used to assess tau oligomer levels in the brain for early AD diagnosis as increased levels of tau oligomers may represent an early sign of neurodegeneration and can be used as a presymptomatic marker of AD.
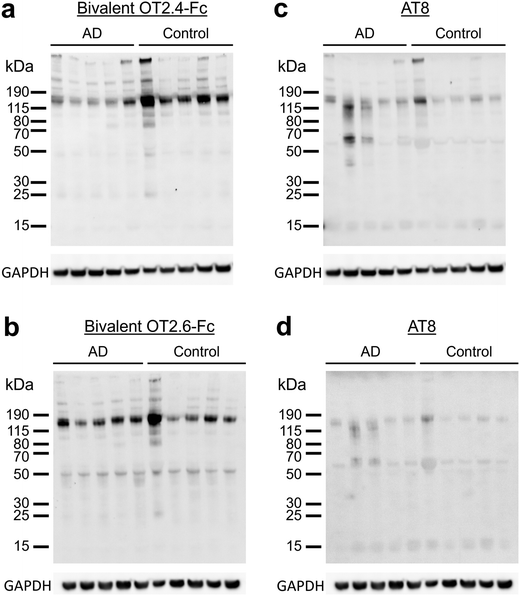 | ||
| Fig. 6 Binding of OT2.4 and OT2.6 to tau oligomers from human brain samples. (a and b) Lysates of human brain tissue samples from five patients with AD and five controls were run on western blots and binding with (a) bivalent OT2.4-Fc and (b) bivalent OT2.6-Fc was assessed. Ponceau S staining of these membranes is shown in ESI Fig. 8c and d, respectively, and unprocessed images of these membranes are shown in ESI Fig. 11a and d,† respectively. (c and d) These membranes were also stained with phospho-tau antibody AT8. Unprocessed images of these membranes are shown in ESI Fig. 11b and e,† respectively. Unprocessed images of GAPDH staining are shown in ESI Fig. 11c and f.† | ||
Conclusions
There is a need for more effective disease-modifying treatments, diagnostics, and research tools for tauopathies like AD. Conformational antibodies that target tau oligomers, suspected to be the most toxic form of tau present in AD, are promising candidates for passive immunotherapy and diagnostic reagents. Nanobodies, in particular, are exciting options for tau oligomer-specific binders as they can target concave or hidden epitopes inaccessible to traditional IgG antibodies. Therefore, a conformational tau oligomer-specific nanobody may interact with tau oligomers differently from the few available tau oligomer-specific antibodies. Other benefits of nanobodies include their single-domain nature which makes them inexpensive to produce in yeast or bacterial cells and easy to link together in multivalent or Fc fusion formats. Nanobodies are not likely to be cell permeable and will not be able to target intracellular tau oligomers if they are unmodified, but methods including conjugation to cell-penetrating peptides or intracellular expression can be used to deliver them intracellularly.50 Extracellular nanobodies can also target tau oligomers that have escaped neurons to the extracellular space and prevent the cell-to-cell spread of tau oligomers.In this work, we report the development and characterization of the first conformational, tau oligomer-specific nanobodies, OT2.4 and OT2.6. These nanobodies were identified through the screening of a yeast surface display nanobody library40 against recombinant tau oligomers. We have demonstrated the specificity of these nanobodies towards tau oligomers over monomers and fibrils with western blot and dot blot analysis. With epitope mapping, we have confirmed that OT2.4 and OT2.6 bind to discontinuous epitopes on tau, making them conformational binders. Finally, we have shown that OT2.4 and OT2.6 recognize tau oligomers of different sizes present in the brains of AD patients and healthy older adults with primary age-related tauopathy (PART).
Future work will be conducted to study if and how these nanobodies interact with tau present in the brains of younger, healthy adults and towards their development into viable therapeutic or diagnostic tools for AD and other tauopathies. Overall, this work demonstrates significant progress in developing conformational, amyloid oligomer-specific antibodies and antibody fragments that have great potential in the diagnosis and treatment of neurodegenerative diseases.
Author contributions
NM: conceptualization, investigation, methodology, project administration, visualization, writing – original draft, and writing – review and editing; JDS: investigation; OJO: investigation; NNB: investigation, methodology, visualization, and writing – review and editing; CJ: investigation; ALH: investigation and methodology; PMT: funding acquisition and writing – review and editing; LBW: funding acquisition, methodology, resources, and writing – review and editing; RK: funding acquisition and resources; RSK: conceptualization, funding acquisition, project administration, supervision, writing – original draft, and writing – review and editing.Data availability
The data required to support the conclusions of the study have been included in the manuscript and ESI.† Sequences for the identified nanobodies have been included in ESI Table 2.†Conflicts of interest
PMT is a member of the scientific advisory boards for Nabla Bio, Aureka Biotechnologies, and Dualitas Therapeutics. The other authors have no conflicts of interest to declare.Acknowledgements
The authors acknowledge funding from the National Institutes of Health (RF1AG059723 to RSK and PMT, and R35GM136300 to PMT). This material is based upon work (by NM) supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-2039655. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. The synthetic nanobody yeast surface display library was a gift from the Kruse Lab at Harvard University. Postmortem brain tissue samples from human subjects were obtained from the Goizueta Alzheimer's Disease Center at Emory University (P30 AG066511). LBW acknowledges funding from the George W. Woodruff School of Mechanical Engineering at Georgia Institute of Technology.References
- Y. Zhang, K. M. Wu, L. Yang, Q. Dong and J. T. Yu, Tauopathies: New Perspectives and Challenges, Mol. Neurodegener., 2022, 17(1), 1–29, DOI:10.1186/S13024-022-00533-Z.
- C. Song, J. Shi, P. Zhang, Y. Zhang, J. Xu, L. Zhao, R. Zhang, H. Wang and H. Chen, Immunotherapy for Alzheimer's Disease: Targeting β-Amyloid and Beyond, Transl. Neurodegener., 2022, 11(1), 1–17, DOI:10.1186/S40035-022-00292-3.
- X. Guo, L. Yan, D. Zhang and Y. Zhao, Passive Immunotherapy for Alzheimer's Disease, Ageing Res. Rev., 2024, 94, 102192, DOI:10.1016/J.ARR.2024.102192.
- D. L. Castillo-Carranza, U. Sengupta, M. J. Guerrero-Muñoz, C. A. Lasagna-Reeves, J. E. Gerson, G. Singh, D. M. Estes, A. D. T. Barrett, K. T. Dineley, G. R. Jackson and R. Kayed, Passive Immunization with Tau Oligomer Monoclonal Antibody Reverses Tauopathy Phenotypes without Affecting Hyperphosphorylated Neurofibrillary Tangles, J. Neurosci., 2014, 34(12), 4260, DOI:10.1523/JNEUROSCI.3192-13.2014.
- D. L. Castillo-Carranza, J. E. Gerson, U. Sengupta, M. J. Guerrero-Muñoz, C. A. Lasagna-Reeves and R. Kayed, Specific Targeting of Tau Oligomers in Htau Mice Prevents Cognitive Impairment and Tau Toxicity Following Injection with Brain-Derived Tau Oligomeric Seeds, J. Alzheimer’s Dis., 2014, 40(S1), S97–S111, DOI:10.3233/JAD-132477.
- P. He, P. Schulz and M. R. Sierks, A Conformation-Specific Antibody against Oligomeric β-Amyloid Restores Neuronal Integrity in a Mouse Model of Alzheimer's Disease, J. Biol. Chem., 2021, 296, 100241, DOI:10.1074/jbc.RA120.015327.
- M. X. Henderson, D. J. Covell, C. H. Y. Chung, R. M. Pitkin, R. M. Sandler, S. C. Decker, D. M. Riddle, B. Zhang, R. J. Gathagan, M. J. James, J. Q. Trojanowski, K. R. Brunden, V. M. Y. Lee and K. C. Luk, Characterization of Novel Conformation-Selective α-Synuclein Antibodies as Potential Immunotherapeutic Agents for Parkinson's Disease, Neurobiol. Dis., 2020, 136, 104712, DOI:10.1016/J.NBD.2019.104712.
- G. Gallardo and D. M. Holtzman, Amyloid-β and Tau at the Crossroads of Alzheimer's Disease, in Advances in Experimental Medicine and Biology, Springer, 2019, vol. 1184, pp. 187–203. DOI:10.1007/978-981-32-9358-8_16.
- A. Roda, G. Serra-Mir, L. Montoliu-Gaya, L. Tiessler and S. Villegas, Amyloid-Beta Peptide and Tau Protein Crosstalk in Alzheimer's Disease, Neural Regener. Res., 2022, 17(8), 1666–1674, DOI:10.4103/1673-5374.332127.
- Y. Zhang, H. Chen, R. Li, K. Sterling and W. Song, Amyloid β-Based Therapy for Alzheimer's Disease: Challenges, Successes and Future, Signal Transduction Targeted Ther., 2023, 8(1), 1–26, DOI:10.1038/s41392-023-01484-7.
- Y. Guo, S. Li, L.-H. Zeng and J. Tan, Tau-Targeting Therapy in Alzheimer’s Disease: Critical Advances and Future Opportunities, Ageing Neurodegener. Dis., 2022, 2(3), 11, DOI:10.20517/and.2022.16.
- E. E. Congdon, C. Ji, A. M. Tetlow, Y. Jiang and E. M. Sigurdsson, Tau-Targeting Therapies for Alzheimer Disease: Current Status and Future Directions, Nat. Rev. Neurol., 2023, 19(12), 715–736, DOI:10.1038/s41582-023-00883-2.
- G. Niewiadomska, W. Niewiadomski, M. Steczkowska and A. Gasiorowska, Tau Oligomers Neurotoxicity, Life, 2021, 11(1), 1–28, DOI:10.3390/LIFE11010028.
- M. D. C. Cárdenas-Aguayo, L. Gómez-Virgilio, S. DeRosa and M. A. Meraz-Ríos, The Role of Tau Oligomers in the Onset of Alzheimer's Disease Neuropathology, in ACS Chemical Neuroscience. American Chemical Society, 2014, pp. 1178–1191. DOI:10.1021/cn500148z.
- S. S. Shafiei, M. J. Guerrero-Muñoz and D. L. Castillo-Carranza, Tau Oligomers: Cytotoxicity, Propagation, and Mitochondrial Damage, Front. Aging Neurosci., 2017, 9 DOI:10.3389/FNAGI.2017.00083.
- G. Ghag, N. Bhatt, D. V. Cantu, M. J. Guerrero-Munoz, A. Ellsworth, U. Sengupta and R. Kayed, Soluble Tau Aggregates, Not Large Fibrils, Are the Toxic Species That Display Seeding and Cross-Seeding Behavior, Protein Sci., 2018, 27(11), 1901–1909, DOI:10.1002/PRO.3499.
- J. E. Gerson, D. L. Castillo-Carranza and R. Kayed, Advances in Therapeutics for Neurodegenerative Tauopathies: Moving toward the Specific Targeting of the Most Toxic Tau Species, ACS Chem. Neurosci., 2014, 5(9), 752–769, DOI:10.1021/CN500143N.
- G. A. Jicha, R. Bowser, I. G. Kazam and P. Davies, Alz-50 and MC-1, a New Monoclonal Antibody Raised to Paired Helical Filaments, Recognize Conformational Epitopes on Recombinant Tau, J. Neurosci. Res., 1997, 48, 128–132, DOI:10.1002/(SICI)1097-4547(19970415)48:2<128::AID-JNR5>3.0.CO;2-E.
- R. Kayed, I. Canto, L. Breydo, S. Rasool, T. Lukacsovich, J. Wu, R. Albay, A. Pensalfini, S. Yeung, E. Head, J. L. Marsh and C. Glabe, Conformation Dependent Monoclonal Antibodies Distinguish Different Replicating Strains or Conformers of Prefibrillar Aβ Oligomers, Mol. Neurodegener., 2010, 5(1), 1–10, DOI:10.1186/1750-1326-5-57.
- B. Combs, C. Hamel and N. M. Kanaan, Pathological Conformations Involving the Amino Terminus of Tau Occur Early in Alzheimer's Disease and Are Differentially Detected by Monoclonal Antibodies, Neurobiol. Dis., 2016, 94, 18–31, DOI:10.1016/J.NBD.2016.05.016.
- R. Abskharon, P. M. Seidler, M. R. Sawaya, D. Cascio, T. P. Yang, S. Philipp, C. K. Williams, K. L. Newell, B. Ghetti, M. A. DeTure, D. W. Dickson, H. V. Vinters, P. L. Felgner, R. Nakajima, C. G. Glabe and D. S. Eisenberg, Crystal Structure of a Conformational Antibody That Binds Tau Oligomers and Inhibits Pathological Seeding by Extracts from Donors with Alzheimer's Disease, J. Biol. Chem., 2020, 295(31), 10662, DOI:10.1074/JBC.RA120.013638.
- K. R. Patterson, C. Remmers, Y. Fu, S. Brooker, N. M. Kanaan, L. Vana, S. Ward, J. F. Reyes, K. Philibert, M. J. Glucksman and L. I. Binder, Characterization of Prefibrillar Tau Oligomers in Vitro and in Alzheimer Disease, J. Biol. Chem., 2011, 286(26), 23063–23076, DOI:10.1074/jbc.M111.237974.
- H.-C. Tai, H.-T. Ma, S.-C. Huang, M.-F. Wu, C.-L. Wu, Y.-T. Lai, Z.-L. Li, R. Margolin, A. J. Intorcia, G. E. Serrano, T. G. Beach, M. Nallani, B. Navia, M.-K. Jang and C.-Y. Tai, The Tau Oligomer Antibody APNmAb005 Detects Early-Stage Pathological Tau Enriched at Synapses and Rescues Neuronal Loss in Long-Term Treatments. DOI:10.1101/2022.06.24.497452.
- S. Steeland, R. E. Vandenbroucke and C. Libert, Nanobodies as Therapeutics: Big Opportunities for Small Antibodies, Drug Discovery Today, 2016, 21(7), 1076–1113, DOI:10.1016/J.DRUDIS.2016.04.003.
- G. Bao, M. Tang, J. Zhao and X. Zhu, Nanobody: A Promising Toolkit for Molecular Imaging and Disease Therapy, EJNMMI Res., 2021, 11(1), 1–13, DOI:10.1186/S13550-021-00750-5.
- N. McArthur, B. Kang, F. G. Rivera Moctezuma, A. T. Shaikh, K. Loeffler, N. N. Bhatt, M. Kidd, J. M. Zupancic, A. A. Desai, N. Djeddar, A. Bryksin, P. M. Tessier, R. Kayed, L. B. Wood and R. S. Kane, Development of a Pan-Tau Multivalent Nanobody That Binds Tau Aggregation Motifs and Recognizes Pathological Tau Aggregates, Biotechnol. Prog., 2024, e3463, DOI:10.1002/BTPR.3463.
- J. M. Zupancic, M. D. Smith, H. Trzeciakiewicz, M. E. Skinner, S. P. Ferris, E. K. Makowski, M. J. Lucas, N. McArthur, R. S. Kane, H. L. Paulson and P. M. Tessier, Quantitative Flow Cytometric Selection of Tau Conformational Nanobodies Specific for Pathological Aggregates, Front. Immunol., 2023, 14, 1164080, DOI:10.3389/FIMMU.2023.1164080.
- A. A. Desai, J. M. Zupancic, H. Trzeciakiewicz, J. E. Gerson, K. N. DuBois, M. E. Skinner, L. M. Sharkey, N. McArthur, S. P. Ferris, N. N. Bhatt, E. K. Makowski, M. D. Smith, H. Chen, J. Huang, C. Jerez, R. S. Kane, N. M. Kanaan, H. L. Paulson and P. M. Tessier, Flow Cytometric Isolation of Drug-like Conformational Antibodies Specific for Amyloid Fibrils, BioRxiv, 2023, 2023.07.04.547698. DOI:10.1101/2023.07.04.547698.
- C. Danis, E. Dupré, O. Zejneli, R. Caillierez, A. Arrial, S. Bégard, J. Mortelecque, S. Eddarkaoui, A. Loyens, F.-X. Cantrelle, X. Hanoulle, J.-C. Rain, M. Colin, L. Buée and I. Landrieu, Inhibition of Tau Seeding by Targeting Tau Nucleation Core within Neurons with a Single Domain Antibody Fragment, Mol. Ther., 2022, 30(4), 1484–1499, DOI:10.1016/j.ymthe.2022.01.009.
- E. Dupré, C. Danis, A. Arrial, X. Hanoulle, M. Homa, F.-X. Cantrelle, H. Merzougui, M. Colin, J.-C. Rain, L. Bueé and I. Landrieu, Single Domain Antibody Fragments as New Tools for the Detection of Neuronal Tau Protein in Cells and in Mice Studies, ACS Chem. Neurosci., 2019, 10, 3997–4006, DOI:10.1021/acschemneuro.9b00217.
- T. Li, M. Vandesquille, F. Koukouli, C. Dudeffant, I. Youssef, P. Lenormand, C. Ganneau, U. Maskos, C. Czech, F. Grueninger, C. Duyckaerts, M. Dhenain, S. Bay, B. Delatour and P. Lafaye, Camelid Single-Domain Antibodies: A Versatile Tool for in Vivo Imaging of Extracellular and Intracellular Brain Targets, J. Controlled Release, 2016, 243, 1–10, DOI:10.1016/J.JCONREL.2016.09.019.
- R. Abskharon, H. Pan, M. R. Sawaya, P. M. Seidler, E. J. Olivares, Y. Chen, K. A. Murray, J. Zhang, C. Lantz, M. Bentzel, D. R. Boyer, D. Cascio, B. A. Nguyen, K. Hou, X. Cheng, E. Pardon, C. K. Williams, A. L. Nana, H. V. Vinters, S. Spina, L. T. Grinberg, W. W. Seeley, J. Steyaert, C. G. Glabe, R. R. Ogorzalek Loo, J. A. Loo and D. S. Eisenberg, Structure-Based Design of Nanobodies That Inhibit Seeding of Alzheimer’s Patient-Extracted Tau Fibrils, Proc. Natl. Acad. Sci. U. S. A., 2023, 120(41), e2300258120, DOI:10.1073/pnas.2300258120.
- Y. Jiang, Y. Lin, S. Krishnaswamy, R. Pan, Q. Wu, L. A. Sandusky-Beltran, M. Liu, M. H. Kuo, X. P. Kong, E. E. Congdon and E. M. Sigurdsson, Single-Domain Antibody-Based Noninvasive in Vivo Imaging of α-Synuclein or Tau Pathology, Sci. Adv., 2023, 9(19), eadf3775, DOI:10.1126/SCIADV.ADF3775.
- E. E. Congdon, R. Pan, Y. Jiang, L. A. Sandusky-Beltran, A. Dodge, Y. Lin, M. Liu, M.-H. Kuo, X.-P. Kong and E. M. Sigurdsson, Single Domain Antibodies Targeting Pathological Tau Protein: Influence of Four IgG Subclasses on Efficacy and Toxicity, EBioMedicine, 2022, 84, 104249, DOI:10.1016/j.ebiom.2022.104249.
- N. De Leiris, P. Perret, C. Lombardi, B. Gözel, S. Chierici, P. Millet, M. Debiossat, S. Bacot, B. B. Tournier, P. Chames, J. L. Lenormand, C. Ghezzi, D. Fagret and M. Moulin, A Single-Domain Antibody for the Detection of Pathological Tau Protein in the Early Stages of Oligomerization, J. Transl. Med., 2024, 22(1), 1–14, DOI:10.1186/S12967-024-04987-1.
- P. Lafaye, I. Achour, P. England, C. Duyckaerts and F. Rougeon, Single-Domain Antibodies Recognize Selectively Small Oligomeric Forms of Amyloid β, Prevent Aβ-Induced Neurotoxicity and Inhibit Fibril Formation, Mol. Immunol., 2009, 46(4), 695–704, DOI:10.1016/J.MOLIMM.2008.09.008.
- A. Bigi, L. Napolitano, D. M. Vadukul, F. Chiti, C. Cecchi, F. A. Aprile and R. Cascella, A Single-Domain Antibody Detects and Neutralises Toxic Aβ42 Oligomers in the Alzheimer's Disease CSF, Alzheimers Res. Ther., 2024, 16(1), 1–19, DOI:10.1186/S13195-023-01361-Z.
- S. Kasturirangan, L. Li, S. Emadi, S. Boddapati, P. Schulz and M. R. Sierks, Nanobody Specific for Oligomeric Beta-Amyloid Stabilizes Nontoxic Form, Neurobiol. Aging, 2012, 33(7), 1320–1328, DOI:10.1016/J.NEUROBIOLAGING.2010.09.020.
- C. A. Lasagna-Reeves, D. L. Castillo-Carranza, U. Sengupta, J. Sarmiento, J. Troncoso, G. R. Jackson and R. Kayed, Identification of Oligomers at Early Stages of Tau Aggregation in Alzheimer's Disease, FASEB J., 2012, 26(5), 1946–1959, DOI:10.1096/fj.11-199851.
- C. McMahon, A. S. Baier, R. Pascolutti, M. Wegrecki, S. Zheng, J. X. Ong, S. C. Erlandson, D. Hilger, S. G. F. Rasmussen, A. M. Ring, A. Manglik and A. C. Kruse, Yeast Surface Display Platform for Rapid Discovery of Conformationally Selective Nanobodies, Nat. Struct. Mol. Biol., 2018, 25(3), 289–296, DOI:10.1038/s41594-018-0028-6.
- U. Sengupta, M. Carretero-Murillo and R. Kayed, Preparation and Characterization of Tau Oligomer Strains, in Methods in Molecular Biology, Humana Press Inc., 2018, vol. 1779, pp. 113–146. DOI:10.1007/978-1-4939-7816-8_9.
- C. A. Lasagna-Reeves, D. L. Castillo-Carranza, M. J. Guerrero-Muñoz, G. R. Jackson and R. Kayed, Preparation and Characterization of Neurotoxic Tau Oligomers, Biochemistry, 2010, 49(47), 10039–10041, DOI:10.1021/bi1016233.
- N. Gera, M. Hussain and B. M. Rao, Protein Selection Using Yeast Surface Display. Methods, Academic Press, 2013, pp. 15–26. DOI:10.1016/j.ymeth.2012.03.014.
- A. W. P. Fitzpatrick, B. Falcon, S. He, A. G. Murzin, G. Murshudov, H. J. Garringer, R. A. Crowther, B. Ghetti, M. Goedert and S. H. W. Scheres, Cryo-EM Structures of Tau Filaments from Alzheimer's Disease, Nature, 2017, 547(7662), 185–190, DOI:10.1038/nature23002.
- S. M. Ward, D. S. Himmelstein, J. K. Lancia, Y. Fu, K. R. Patterson and L. I. Binder, TOC1: Characterization of a Selective Oligomeric Tau Antibody, J. Alzheimer's Dis., 2013, 37(3), 593, DOI:10.3233/JAD-131235.
- P. Sun, H. Ju, Z. Liu, Q. Ning, J. Zhang, X. Zhao, Y. Huang, Z. Ma and Y. Li, Bioinformatics Resources and Tools for Conformational B-Cell Epitope Prediction, Comput. Math Methods Med., 2013, 2013, 946346, DOI:10.1155/2013/943636.
- M. A. Meraz-Ríos, K. I. Lira-De León, V. Campos-Peña, M. A. De Anda-Hernández and R. Mena-López, Tau Oligomers and Aggregation in Alzheimer's Disease, J. Neurochem., 2010, 112(6), 1353–1367, DOI:10.1111/J.1471-4159.2009.06511.X.
- M. Kolarova, U. Sengupta, A. Bartos, J. Ricny and R. Kayed, Tau Oligomers in Sera of Patients with Alzheimer's Disease and Aged Controls, J. Alzheimer's Dis., 2017, 58(2), 471–478, DOI:10.3233/JAD-170048.
- S. Maeda, N. Sahara, Y. Saito, S. Murayama, A. Ikai and A. Takashima, Increased Levels of Granular Tau Oligomers: An Early Sign of Brain Aging and Alzheimer's Disease, Neurosci. Res., 2006, 54(3), 197–201, DOI:10.1016/J.NEURES.2005.11.009.
- D. Schumacher, J. Helma, A. F. L. Schneider, H. Leonhardt and C. P. R. Hackenberger, Nanobodies: Chemical Functionalization Strategies and Intracellular Applications, Angew. Chem., Int. Ed., 2018, 57(9), 2314–2333, DOI:10.1002/ANIE.201708459.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm00707g |
| This journal is © The Royal Society of Chemistry 2024 |