Open Access Article
Open Access ArticleMicrofluidics in environmental analysis: advancements, challenges, and future prospects for rapid and efficient monitoring
Prakash
Aryal†
a,
Claire
Hefner†
a,
Brandaise
Martinez
a and
Charles S.
Henry
 *abcd
*abcd
aDepartment of Chemistry, Colorado State University, Fort Collins, Colorado 80523, USA. E-mail: chuck.henry@colostate.edu
bDepartment of Chemical and Biological Engineering, Colorado State University, Fort Collins, Colorado 80523, USA
cSchool of Biomedical Engineering, Colorado State University, Fort Collins, Colorado 80523, USA
dMetallurgy and Materials Science Research Institute, Chulalongkorn University, Bangkok 10330, Thailand
First published on 2nd January 2024
Abstract
Microfluidic devices have emerged as advantageous tools for detecting environmental contaminants due to their portability, ease of use, cost-effectiveness, and rapid response capabilities. These devices have wide-ranging applications in environmental monitoring of air, water, and soil matrices, and have also been applied to agricultural monitoring. Although several previous reviews have explored microfluidic devices' utility, this paper presents an up-to-date account of the latest advancements in this field for environmental monitoring, looking back at the past five years. In this review, we discuss devices for prominent contaminants such as heavy metals, pesticides, nutrients, microorganisms, per- and polyfluoroalkyl substances (PFAS), etc. We cover numerous detection methods (electrochemical, colorimetric, fluorescent, etc.) and critically assess the current state of microfluidic devices for environmental monitoring, highlighting both their successes and limitations. Moreover, we propose potential strategies to mitigate these limitations and offer valuable insights into future research and development directions.
1. Introduction
As the dominant species on Earth, humans have altered almost every aspect of the natural world with our activity; however, these alterations have come at a price. Impacts from rapid industrialization, urban expansion, and population growth have caused significant environmental pollution, directly affecting human health and the ecosystem. Studies reviewed by Xu et al. show that environmental pollutants like heavy metals, particulate matter (PM), biogenic toxins, and industrial effluents are associated with adverse health conditions, contributing to about 22% of the global disease burden and 23% of deaths.1–3 Despite significant efforts towards environmental remediation, pollution remains a substantial global problem, particularly in developing regions where large populations are impacted by industrial discharges, poor sanitation, inadequate waste management, compromised water sources, and indoor air pollution from biomass. The lack of quick and cost-effective testing methods exacerbates conditions in resource-constrained areas. Moreover, industrial advancements in developed nations have introduced new pollutants that spread rapidly and surpass natural environmental defenses. Addressing these challenges requires exploring technological interventions that can swiftly and effectively reduce pollutant levels, restoring a safe environment.Conventional methods used for monitoring contaminants in the environment, such as inductively coupled plasma optical emission spectroscopy (ICP-OES), atomic absorption spectroscopy (AAS), fluorescence spectroscopy, ultraviolet-visible (UV-vis) spectroscopy, mass spectrometry (MS), and high-performance liquid chromatography (HPLC), offer high sensitivity and precision even at low analyte concentrations.4–9 However, they have drawbacks like time-consuming processes, expensive equipment, and the need for skilled operators,10 making them impractical for point-of-need testing/monitoring, especially in resource-limited areas. Researchers have focused on developing faster, user-friendly, and environmentally friendly detection technologies to address these limitations.
Microfluidics has emerged as a promising solution because it enables rapid analysis, and automation of multiple chemical processes.11,12 These platforms precisely control microliters of liquid in narrow flow channels using a variety of substrates such as silicon, glass, ceramics, paper, thermoplastics, polydimethylsiloxane (PDMS), and hydrogels.13 In some cases, multiple materials are combined in their fabrication.14–16 The versatility of microfluidic devices are further enhanced by the ability to perform sample pretreatment and preconcentration directly within the device.17
Previous research has extensively explored various microfluidic platforms to monitor and detect contamination in soil,18–21 water,22–24 and air25–28 matrices. Notably, techniques like absorbance-, electrochemical-, fluorescence-, and chemiluminescence-based microfluidic systems have seen significant progress.29–32 Comprehensive reviews have evaluated paper-based microfluidic sensors (μPADs) for environmental monitoring.33–35 Despite assessing microfluidic devices in food-based sensing,23,36 limited reviews regarding microfluidic devices for agricultural monitoring exist.37,38
This review provides an up-to-date overview of the advancements in microfluidic devices for environmental monitoring over the past five years. It spans a variety of environmental domains, including air, water, soil matrices, and agriculture applications for monitoring the most prevalent analytes such as heavy metals, nutrients, pesticides, microorganisms (bacteria, viruses, etc.), and polyfluoroalkyl substances (PFAS). The review will also address the current challenges associated with using microfluidic devices in environmental monitoring, future perspectives, and their commercialization.
2. Detection techniques
Several detection techniques have been explored over the last five years for microfluidics-based environmental quality monitoring systems with fluorescence, electrochemical, and colorimetric detection being the most widely used. Other methods include photoelectrochemical, chemiluminescence, absorbance-based, quartz crystal microbalance, and surface plasmon resonance (SPR). Herein we focus on electrochemical, fluorescence, chemiluminescence, and colorimetric methods which are the most prevalent techniques in recent times.Electrochemical analysis includes various techniques such as conductometry, potentiometry, voltammetry, polarography, amperometry, and coulometry, each dependent on specific electrical characteristics.39 The standard configuration for an electrochemical system involves a three-electrode setup, comprising a working electrode, a counter electrode, and a reference electrode.39–41 Many analytes can be detected based on respective fixed redox potentials on bare electrodes allowing for distinguishable signal generation.39,42 Electrochemical analysis offers low detection limits, capable of extending into the picomole range.43 This represents a significant advantage over widely used techniques such as fluorescence or colorimetric methods, both of which have yet to achieve comparably low limits of detection. In many cases, using electrochemical detection has furthers advantage by using disposable and/or mobile electrochemical systems for detecting heavy metals. A range of electrodes, including screen-printed electrodes, carbon paste electrodes, disposable glass electrodes, and disposable paper electrodes,44–46 as well as portable setups including homemade electronics, portable potentiostat, and smartphone-based systems have also been explored.44–46
Fluorescence-based techniques for environmental detection include the design and synthesis of fluorophores that incorporate recognition elements like ligands, enzymes, aptamers, etc. allowing for effective sensing of environmental analytes (mostly in solution).47,48 These techniques utilize various suitable probes, such as rhodamine, pyrene, anthracene, naphthylamide, aminoquioline, bithiophene, etc., combined with fluorescence detectors to detect a specific analyte of interest.49 A common technique to incorporate fluorescent detection inside microfluidics involves mixing a sample with a particular fluorescent probe for the target analyte, applying it through the device, illuminating it to induce fluorescence, and then measuring the intensity of the emitted light with a photodetector, which directly correlates with analyte concentration. Whereas some techniques pretreat the probes inside the microfluidic channel and florescence is detected when the target analyte is run through the microchannels.
In addition, recent investigations have also emphasized flow-based chemiluminescence (CL) assays involving microfluidic devices.33,50–52 When a molecule undergoes exothermic excitation to reach the singlet excited state, it emits CL characterized by electromagnetic radiation with a distinct wavelength, mainly falling within the visible and near-infrared range. The emitted light's intensity can be correlated with the concentration of the analyte. The flow-based CL substantially reduces sample and reagent consumption, shortens analysis time, and utilizes automated, compact, and highly integrated flow detection systems as compared to the traditional CL assays.
In the past five years, there has been a surge in the development of colorimetric platforms for detecting environmental analytes. Most of the researchers have focused on developing paper-based microfluidic colorimetric sensors as promising approaches for cheap and user-friendly detection. A flow channel, created by incorporating hydrophobic barriers on both sides of the paper, guides the sample flow to a paper pad treated with a colorimetric reagent for detection.53 Colorimetric signal is produced from the reaction of the sample, wicked through the paper pad via capillary action, with the stored colorimetric reagent. Hydrophobic barriers can be implemented into paper substrates to control flow through a variety of techniques including wax printing, plasma treatment, and photolithography.13
Moreover, nanomaterials-based techniques, including those utilizing gold nanoparticles, carbon nanotubes, quantum dots, and graphene, have gained even more popularity in microfluidics for environmental detection.54–56 Their high surface area-to-volume ratio enhances sensitivity, facilitating efficient binding and detecting the analyte of interest.54–57 The tunable size, shape, and surface chemistry enable the design of detection systems tailored to target specific analytes. Functionalization with specific ligands or receptors enhances selectivity and accuracy of detection.58,59
3. Water
The global challenge of accessing safe drinking water is growing, with nearly one-third of the world's population lacking access to clean water.60 Alarming statistics from the World Health Organization (WHO) reveal that approximately 2 billion people rely on contaminated water sources, leading to nearly a million deaths each year.61 Roughly 80% of wastewater worldwide remains untreated, carrying a toxic mix of pollutants, including hazardous industrial effluents and organic waste.62 Water contaminants, such as heavy metals, microbes, PFAS, and pesticides, can permeate through multiple pathways, causing significant harm to human health and ecosystems and resulting in widespread disruptions and negative consequences.63 The situation becomes even more concerning when water sources are not regularly tested for these pollutants. Hence, there is an urgent demand for easier monitoring platforms to address this critical issue.Microfluidics offers a remarkable advantage in detecting water contamination due to its simplicity and versatility in handling water samples.64 Water's natural fluidic properties, such as its low viscosity and surface tension, enable smooth flow and efficient mixing within the microchannels, ensuring precise processing of samples.65 Additionally, microfluidic devices' portability and automation capabilities enable convenient on-site water monitoring, making them valuable in remote or resource-constrained areas where access to safe drinking water is crucial.66,67
3.1 Heavy metals
Heavy metals are naturally occurring metals with densities at least five times greater than water.68 While certain heavy metals such as arsenic (As), cadmium (Cd), and lead (Pb) are naturally present and essential for specific physiological functions within safe limits, exceeding these thresholds can have severe health consequences due to their mutagenic or carcinogenic properties.69–71 Regulatory bodies have established maximum contamination limits (MCL) for heavy metals in drinking water (Table 1) to combat these harmful effects. Heavy metal pollution arises from both natural sources and human activities, with anthropogenic factors playing a substantial role.72 Activities such as industrial waste disposal, mining, and the use of heavy metal-containing pesticides and fertilizers introduce these toxic elements into water sources.73–75 The complex task of heavy metal pollution control is further complicated by their non-biodegradable nature, widespread occurrence, and potential for bioaccumulation as they move up the food chain.76,77 Therefore, active monitoring of heavy metal levels in water is important to safeguard public health. Scheme 1 illustrates the schematic representation of heavy metal detection using microfluidics.| Metals | EPA78 (ppb) | EU79 (ppb) | WHO80 (ppb) | Sources | Health impact |
|---|---|---|---|---|---|
| Nickel | — | 20 | 70 | Forest fires, volcanic eruptions, industrial wastewater, and sewage sludge etc. | Liver toxicity, lung cancer, lung disease, skin disease etc.81–83 |
| Silver | 100 | — | — | Electroplating, smelting, atmospheric deposition etc. | Skin irritation, breathing problems, lung and throat problems, liver, and kidney damage etc.84–86 |
| Zinc | 500 | — | — | Corrosion of pipes under acidic conditions, industrial discharges, Zn metal batteries etc. | Liver toxicity, nausea, vomiting, copper deficiency (with excessive intake of zinc supplements) etc.87,88 |
| Mercury | 2 | 1 | 1 | Mining pollution, volcanic emission, natural deposits, coal combustion, waste combustion treatment etc. | Digestive system, skin rashes, diarrhea, neurological disorders, asthma, cancer, renal failure, acrodynia etc.89–92 |
| Lead | 15 | 10 | 10 | Mining, jewelry, natural deposits, fossil fuel burning, lead batteries, manufacturing process, PVC pipes etc. | Muscular weakness, kidney damage, nervous system impairment, cancer, weight loss, brain damage, hemoprotein, paralysis etc.93–96 |
| Copper | 1300 | 2000 | 2000 | Building construction, electronic products, photovoltaic cells, machinery, tanning, transmission industry etc. | Adreno-cortical hyperactivity, vomiting, liver and kidney damage, lung cancer, alopecia, anemia, Wilson's disease etc.97–100 |
| Chromium | 100 | 50 | 50 | Building construction, electronic products, cement production, tanning, transmission industry, fertilizers, volcano eruption, leather industry etc. | Low blood sugar, diarrhea, skin ulcers, liver and kidney damage, gastrointestinal cancer, teeth abnormalities, lung cancer etc.101–104 |
| Cadmium | 5 | 5 | 3 | Sewage disposable, mining, natural deposits, synthetic rubber, smelting, electroplated parts, tobacco smoking etc. | Hypertension, renal toxicity, cardiovascular issues, DNA damage, kidney disease, pancreatic cancer, and breast cancer etc.105–108 |
| Arsenic | 10 | 10 | 10 | Coal burning, volcanic eruption, sandstorm, metal mining, smelting etc. | Hyper-pigmentation, skin cancer, renal system failure, effect on central nervous system etc.94,109–111 |
Lace et al. created a microfluidic system to detect arsenic in water using leucomalachite green (LMG) dye.112 Arsenic reacts with potassium iodate in an acidic solution, releasing iodine. The iodine then oxidizes leucomalachite green to malachite green, resulting in a green color with a visible absorbance peak at 617 nm. The method was the first integration of LMG in microfluidic device for arsenic detection. The authors also improved upon a colorimetric technique using 1,5-diphenylcarbazide to measure Cr(VI) levels. This optimized approach achieved a very low limit of detection, almost 80% lower than the regulatory limit for chromium in water.113 Moreover, the color complex exhibited long-term stability, making it highly suitable for integration into microfluidic analysis. While these methods are sensitive, their complexity and reliance on absorbance-based design often necessitate using syringe pumps for operation. This requirement adds a level of intricacy that can be challenging to manage in resource-limited environments.
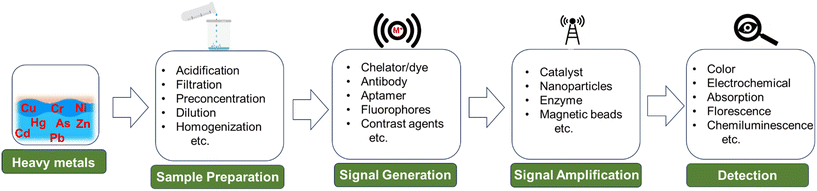
Wang et al. developed a novel sensor that can detect multiple heavy metal ions.114 The device is compact and integrates automatic sample measurement, on-chip reactions, gravitational-magnetic separation, and distance-based readout, making it easy to use and interpret. The chemosensor is made of a 3D-printed PDMS and Norland Optical Adhesive 63 (NOA63) layer. It has three chambers for sample metering, reactions, and separation (Fig. 1A). The chambers are preloaded with deoxyribozymes (DNAzymes), probe-modified magnetic microparticles (MMPs), and polystyrene microparticles (PMPs). When a water sample is added to the first chamber, the particles are reconstituted. The MMPs and PMPs attach to the DNAzyme at its ends, forming a structure called “MMPs-DNAzyme-PMPs”. This structure is stable in the absence of target metal ions. When target metal ions are added, the DNAzyme undergoes cleavage, which separates the MMPs and PMPs. This event disrupts the “MMPs-DNAzyme-PMPs” structure. The PMP trapping distance can then be used to visually quantify the metal ions.
 | ||
| Fig. 1 Examples of microfluidics systems for heavy metals detection in water including (A) a fully integrated, ready-to-use system for heavy metals detection by Wang et al. demonstrating (i) automatic sample metering, (ii) on-chip sequential reaction, and (iii) distance-based readout for visual quantification of multiple heavy ions (reprinted from ref. 114 with permission, copyright 2021, American Chemical Society). (B) Design by Borthakur et al., using paper strip with CuS and NiS nanoparticle-decorated porous-reduced graphene oxide sheets as peroxidase nanozymes (reprinted from ref. 115 with permission, copyright 2021, American Chemical Society). (C) Epitaxial graphene sensor combined with microfluidics by Santangelo et al. (reprinted from ref. 118, copyright 2019, MDPI). (D) 3D SERS chip using all-femtosecond-laser-processing developed by Bai et al., (reprinted from ref. 120 with permission, copyright 2023, John Wiley and Sons). Fabricating 3D microfluidic SERS chips involves three main steps. Initially, a 3D microchannel is created in a glass substrate through femtosecond laser-assisted wet etching (FLAE) (a and b). The second step involves femtosecond laser selective metallization (FLSM) of the Cu–Ag layered thin film within the microchannel (c and d). The final stage encompasses the formation of a 2D periodic metal nanostructure through femtosecond laser-induced periodic surface structure (fs-LIPSS) (e). | ||
Borthakur et al. observed that the presence of Hg(II) ions can inhibit the catalytic activity of nanozymes involved in the oxidation of TMB.115 The sensor is based on a metal sulfide/p-rGO (reduced graphene oxide) nanocomposite that contains nanozymes that are sensitive to Hg(II) ions. When Hg(II) ions are present, they bind to the nanozymes and inhibit their catalytic activity. This results in a decrease in the amount of TMB that is oxidized, which can be visually observed as a change in color (Fig. 1B). The resulting sensor is able to detect Hg(II) ions in the nanomolar concentration range.
Sharifi et al. made a significant advancement by introducing a three-dimensional origami microfluidic paper analytical device (μPAD) in combination with a PVC membrane.116 This novel approach successfully addressed the issues commonly seen in traditional flow-based paper systems, such as the movement of colored products or dye leaching, leading to uneven color distribution in detection zones. By allowing the sample flow to be perpendicular to the surface, the analyte sample could be evenly spread throughout the detection zone. Li et al. also developed a three-dimensional microfluidic paper-based device that utilizes a smartphone and a flat light-emitting diode (LED) lamp to achieve multiplexed colorimetric detection of six metal ions.117 The integration of these components resulted in improved color perception, significantly enhanced sensitivity, and an extended detection range, making it a promising improvement for metal ion analysis.
Santangelo et al. investigated the ability of epitaxial graphene on silicon carbide (4H-SiC) to detect heavy metals, specifically Pb(II) and Cd(II) in water.118 The sensor is a monolayer of epitaxial graphene grown on an on-axis, Si-face 4H-SiC substrate. The graphene layer is grown using a sublimation growth technique (Fig. 1C). The sensor works by monitoring changes in conductivity when Pb and/or Cd ions interact with the graphene surface. The results of the study showed that EG/SiC is a highly sensitive sensor for Pb and Cd, with a detection range of nanomolar to micromolar concentrations.
Gimenez-Gomez et al. developed an innovative technique for automated As(III) determination in waters with an electrochemical sensor integrated into a modular microfluidic system. The system features a gold nanoparticle (AuNP)-modified gold thin-film electrode for highly sensitive As(III) detection using anodic stripping linear sweep voltammetry.119 The microfluidic system facilitates automatic sensor calibration, sample uptake, preconditioning, and the detection of As(III). The sensor demonstrated excellent performance in spike recovery analysis, offering a promising alternative for As(III) quantification.
Ma and coauthors developed a portable microfluidic electrochemical sensing platform that allows rapid detection of hazardous Pb2+. The platform utilizes thermocapillary convection and incorporates a 3D Ag-rGO-f-Ni(OH)2/NF element for signal amplification.44 They chose Pb2+ as a model for detection and proposed a microfluidic electrochemical sensing chip, which can be used with a smartphone-based electrochemical workstation for quick and efficient detection. The system demonstrated an impressive detection limit to ppb levels.
Bai et al. developed a new technique for fabricating 2D periodic metal nanostructures inside 3D glass microfluidic channels.120 The 3D channels are fabricated using femtosecond laser-assisted wet etching. Next, Cu–Ag layered thin films are formed using femtosecond laser direct writing ablation and electroless metal plating. Finally, the Cu–Ag films are nanostructured by irradiation with linearly polarized beams to form periodic surface structures (Fig. 1D). The technique eliminates the need for complicated substrate stacking and bonding procedures or lithography for micro and nanostructuring while developing surface-enhanced Raman spectroscopy (SERS) chips. The concentration of Cd(II) was calculated by monitoring the blue or red shift in the Raman peaks of crystal violet. The resulting SERS microchips exhibit high sensitivity and reproducibility, detecting Cd2+ ions at concentrations as low as 10 ppb.
Huang et al. designed a microfluidic aptamer-based sensor that detects Hg(II) and Pb(II) ions in water.121 The presence of Hg(II) and Pb(II) ions is determined by assessing the alteration in fluorescence intensity of the GO (graphene oxide)/aptamer suspension induced by fluorescence resonance energy transfer (FRET) occurring within the aptamer molecules. The researchers achieved an impressive detection limit in the parts per trillion (ppt) range using the developed sensor.
Conventional microfluidic systems designed for heavy metal detection in water are primarily capable of detecting labile metal ions. However, it's important to note that non-labile metal ions, which are often bound to organic surfaces or form precipitates, can also exist in water, and contribute to overall water toxicity. To accurately measure the total concentration of heavy metals in water, it is essential to convert these non-labile metal ions into their ionic form. This conversion can be achieved by automating an acid breakdown process within the microfluidic platform.
Table 2 summarizes the analytical performance of the microfluidics platforms mentioned above and other notable advancements in the field in recent years for detecting heavy metals in water.
| Work | Method | Metal | Device or material | Detection limit |
|---|---|---|---|---|
| A 3D origami paper-based analytical device combined with PVC membrane116 | Colorimetric | Cu(II) | Paper | 1.7 ppm & 1.9 ppm |
| Three-dimensional microfluidic paper-based device combined with smartphone117 | Colorimetric | Fe(III), Ni(II), Cr(VI), Cu(II), Al(III), Zn(II) | Paper | 0.2, 0.3, 0.1, 0.03, 0.08, and 0.04 ppm respectively |
| Highly selective simultaneous determination of five metal ions122 | Colorimetric | Cu(II), Co(II), Ni(II), Hg(II), and Mn(II) | Paper | 0.32, 0.59, 5.87, 0.20, and 0.11 ppm respectively |
| Chemically functionalized paper-based microfluidic platform for multiplex heavy metal detection123 | Colorimetric | Ni(II), Cr(VI), and Hg(II) | Paper | 0.24, 0.18, and 0.19 ppm |
| Portable smartphone-based PDMS microfluidic kit for the simultaneous colorimetric detection of arsenic and mercury124 | Colorimetric | As(III), and Hg(II) | PDMS | 224 ppb and 3.4 ppb |
| Low-cost and selective identification of Cu(II), Fe(III), and Hg(II) using GQDs-DPA supported amino acids125 | Colorimetric | Cu(II), Fe(III), and Hg(II) | Paper | 0.1 ppm |
| Capillary flow driven microfluidics combined with paper14 | Colorimetric | Ni(II), Cu(II), and Fe(III) | Paper | 2.0, 0.3, and 1.1 ppm |
| Plastic screen-printing126 | Colorimetric | Cr(III) | Polycaprolactone | 15 ppb |
| Triple-Indicator-Based platform127 | Colorimetric | Pb(II), Cr(VI), Ni(II), Cu(II), Fe(III) | Paper | 0.1 μM to 15 μM |
| Dual-gel electromembrane extraction128 | Colorimetric | Cr(III), and Cr(VI) | Paper | 2 ppb and 3 ppb |
| Simultaneous colorimetric detection of metallic salts on paper129 | Colorimetric | Pb(II), Ba(II), Sb(III), Fe(III), Al(III), Zn(II), Mg(II) | Paper | 0.1–0.4 μg |
| (HF-LPME) for highly sensitive detection of hexavalent chromium in water samples130 | Colorimetric | Cr(VI) | Paper | 3 ppb |
| A MEMS-based multi-parameter integrated chip131 | Electrochemical | Cu(II) | MEMS | 2.33 ppb |
| Plug-and-play assembly of paper-based colorimetric and electrochemical devices132 | Electrochemical and colorimetric | Fe(III), Ni(II), Cu(II), Zn(II), Cd(II), Pb(II) | ePAD, and (μPAD) | 0.9 to 10.5 ppb and 0.1 to 0.3 ppm |
| Enhancing the sensitivity of electrochemical sensors by ion concentration polarisation133 | Electrochemical | As(III) | Gold electrodes on glass | 1 ppb and 7 ppb |
| Microfluidic sensor integrated with nanochannel liquid conjunct Ag/AgCl reference electrode134 | Electrochemical | Pb(II) | Glass–silicon–glass | 0.13 ppb |
| Printed paper-based origami electrochemical sensor135 | Electrochemical | Cd(II) and Pb(II) | Paper | 20.39 and 50.80 ppb |
| Thread-based electrodes for interference free detection of As(III)136 | Electrochemical | As(III) | Capillary tube | 0.416 μM |
| Sponge-based microfluidic sensor137 | Potentiometry | Cd2+, and Pb2+ | Polyurethane based sponge | 10 μM, and 1 μM |
| Acidified paper substrates for microfluidic solution sampling integrated with potentiometric sensors138 | Potentiometry | Pb(II) | Paper | N/A |
| Non-equilibrium potentiometric sensors integrated with metal modified paper-based microfluidic solution sampling substrates139 | Potentiometry | Pb(II) | Paper | N/A |
| ZnSe quantum dot-based ion imprinting on paper140 | Fluorescence | Cd(II) and Pb(II) | 3D microfluidic paper chip | 0.245 ppb and 0.335 ppb |
| A three-dimensional pinwheel-shaped paper-based microfluidic analytical device for fluorescence detection141 | Fluorescence | Cu2+, Cd2+, Pb2+, and Hg2 | Paper | 0.007–0.015 ppb |
| A three-dimensional pinwheel-shaped paper-based microfluidic analytical device for fluorescence detection141 | Fluorescence | As(III), Cd(II), Pb(II) | PDMS | 5.03 nM, 41.1 nM, and 4.44 nM, respectively |
| Feedback-controlling digital microfluidic fluorometric sensor142 | Fluorescence | Hg(II) | DMF Chip | 0.7 ppb |
| Nitrogen-doped carbon dots as fluorescence ON–OFF–ON sensor143 | Fluorescence | Cu(II), and Hg(II) | Paper | 6.2 nM and 2.304 nM |
| A suspending-droplet mode paper-based microfluidic platform144 | Colorimetry and fluorescence | Pb(II) | Paper | NA |
| (SERS) chips fabricated by all-femtosecond-laser-processing120 | SERS | Cd(II) | Glass | 10 ppb |
| SERS substrate in microfluidic channel145 | SERS | Hg(II) | Glass and PDMS | 0.1 μM |
| A three-dimensional pinwheel-shaped paper-based microfluidic analytical device146 | Naked eye colorimetric | Pb(II) | Microfluidic particle dam | 0.44 ppb |
| G-quadruplex DNAzyme on microfluidic paper147 | Distance based | Hg(II) | Paper | 0.23 nM |
| Distance-based detection of Ag+ with gold nanoparticles-coated microfluidic paper148 | Distance based | Ag(I) | Paper | 1 ppm |
| Quantitative colorimetric paper analytical devices based on radial distance measurements149 | Distance based | Fe(III), Cu(II), Zn(II) | Paper | 1 ppb, and 2.5 ppb |
| Wireless microfluidic sensor for metal ion detection in water150 | Resonance frequency variation | Pb(II), Cd(II), Mg(II), Ca(II), K(I) | Low temperature co-fired ceramic (LTCC) | 5 μM |
| Cost-effective microabsorbance detection based nanoparticle immobilized microfluidic system151 | Microabsorbance | Pb(II), Cr(VI), Hg(II) | PDMS | 0.5 ppb |
| Graphene-integrated microfluidic devices152 | Differential resistance | Pb(II) | Si-face 4H-SiC | 95 nM |
| Microfluidic detection system based on the leucomalachite green method112 | Spectrometry | As(III) | PMMA | 0.32 ppm |
| DMF diluter-based algal biosensor153 | Microalgal motility measurement | Cu(II), Pb(II) | Digital microfluidic diluter chip | 0.65 μM, and 1.90 μM |
| Thiol-based microfabricated piezoresistive sensors array with ion-selective self-assembled monolayer154 | Resistance measurement | Hg(II) | Piezoresistive microcantilever | 0.75 ppb |
| Aptasensor based on PCB electrodes155 | Interfacial capacitance | Cd(II) | Gold interdigitated electrode (IDE) | 253.16 aM |
3.2 Nutrients
Nutrients (i.e., nitrogen, phosphorus, potassium, etc.) are essential in sustaining the health and balance of our aquatic ecosystems.156 However, excess nutrients in the water from agriculture runoff, wastewater runoff, or industrial discharges can result in severe environmental impacts.156 Eutrophication, which leads to decreased oxygen levels for aquatic life and eventual aquatic death, is one of the severe consequences of nutrient overload. Cyanobacteria in algal blooms can also produce highly toxic compounds hazardous to aquatic life, domestic animals, and humans.156 Therefore, monitoring nutrients in our water systems is critical to understanding overall water quality and sources of pollution. Earlier this year (2023), Li et al. published an extensive review on microfluidic devices for nutrient detection in water, including nitrate, nitrite, ammonia nitrogen, phosphate, and silicate detection devices.157 Given the detailed nature of that publication, we will highlight articles released since the publishing of Li et al.'s review.Catalan-Carrio et al. introduced an ionogel-based (IO) hybrid polymethyl methacrylate (PMMA)-paper handheld device for nitrite and nitrate detection in water. The device introduces the sample to a paper-based microfluidic section where Zn0 is immobilized on the paper surface. Nitrate is then reduced to nitrite upon contact with Zn0. The solution subsequently flows to the IO section of the device, where the Griess reagent reacts with the nitrite in the sample, leading to a measurable color change in the IO. The device exhibited detection limits of 0.47 mg L−1 for nitrite and 2.3 mg L−1 for nitrate. However, it is worth noting that color stabilization may take up to 50 min, which may not be ideal for field-based applications.158
Luy et al. developed a colorimetric-based device for nitrate and phosphate detection underwater. This device, composed of PMMA, comprises two layers with flow channels and inlets to enable simultaneous dual chemistry. Two parallel optical cells were employed for color analysis. The Griess assay was utilized for nitrate detection, while the PMB assay was employed for phosphate detection. The device demonstrated detection limits of 97 nM for nitrate and 15 nM for phosphate. During an eight-day field deployment, the device successfully provided 592 measurements, showcasing its practicality in real-world scenarios.159
Similarly, Zhang et al. presented a colorimetric device for online nitrate detection in surface water samples. Nitrate detection was achieved through UV absorption, with an LED serving as the light source and a photodiode used to measure the resulting signal. To mitigate interference from natural organic matter (NOM), which also absorbs UV light and can hinder accurate nitrate detection, the authors employed a miniaturized capacitive deionization cell to separate nitrate ions from NOM. The device exhibited a limit of detection (LoD) of 0.03 mg L−1 and a limit of quantification (LoQ) of 0.12 mg L−1 for nitrate.160
Salinity, climate, pollution, and other factors can significantly impact the nutrient concentrations in natural water bodies like lakes, rivers, and seas.161 Therefore, low-cost high-frequency testing technologies must be developed to thoroughly understand the presence of nutrients in water systems. In this respect, the high cost of mass production has limited the advancement of microfluidic technology.162 According to Li et al.'s review, most microfluidic nutrition sensors use pumps and valves to regulate fluid flow, which raises the cost and resource demands for operation.157 To accomplish more economical and regular monitoring of water nutrients, less expensive technologies like μPADS should be further explored.
3.3 Pesticides
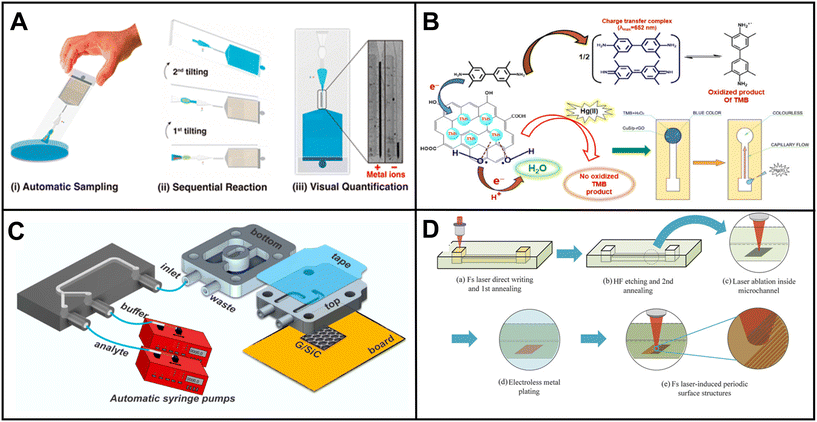
Pesticides play a vital role in modern agriculture by effectively managing pests to meet global food production demands. However, with over three billion kilograms of pesticides consumed worldwide annually, their extensive usage has raised concerns regarding the presence of pesticides and pesticide residues in the environment.163 Exposure to pesticides has been linked to both acute and chronic effects, such as asthma, hormone disruption, cancer, neurological issues, and more. The significant risk to human health necessitates the development of effective detection methods, particularly in water sources susceptible to contamination through agricultural runoff. Pesticides are typically classified based on chemical class or mode of action. Common pesticide classes include organophosphates, carbamates, and organochlorines, with organophosphates making up the most widely applied pesticides in agriculture today.164Over the past five years, research has focused on harnessing the potential of microfluidic devices for pesticide detection in water (Table 3).165,166 Researchers have explored various innovative methods for detection. The commonly employed rapid pesticide detection techniques utilizing microfluidics include enzyme inhibition, immunoassays, molecular imprinting, and other related methods.170 Kim et al. demonstrated a colorimetric paper-based sensor for detecting chlorpyrifos, an organophosphate pesticide. The device relied on a competitive-inhibition reaction involving acetylcholinesterase (AChE), indoxyl acetate chromogenic reagent, and pesticide analyte. Indoxyl acetate transforms into a blue-colored product when acted upon by AChE. However, organophosphate pesticides inhibit AChE, resulting in a less intense colored product. This color change enabled the quantification of the pesticide through image analysis.167 Arduini et al. reported an origami paper-based electrochemical biosensor for the simultaneous detection of pesticides 2,4-D, atrazine, and paraoxon. Their approach also utilized enzymatic inhibition in conjunction with a portable potentiostat. To create the biosensor, they pretreated paper pads with three distinct enzymes: butyrylcholinesterase (BChE), alkaline phosphatase, and tyrosinase. Each enzyme was specifically inhibited by a different class of pesticides: organophosphorus insecticides, phenoxy-acid herbicides, and triazine herbicides. By employing this enzymatic inhibition method, Arduini et al. demonstrated the biosensor's capability to simultaneously detect and differentiate between the three classes of pesticides.168 In addition to enzymatic inhibition, other assay techniques have been developed for pesticide detection in water.
| Work | Method | Pesticide | Device or material | Detection limit |
|---|---|---|---|---|
| Paper based sensor based on competitive-inhibition reaction167 | Colorimetric | Chlorpyrifos | Paper | 8.60 ppm |
| 2D electrode array in paper-based digital microfluidics173 | Colorimetric | Methyl paraoxon | Paper, 2D electrode array, PDMS | 10–20 μM |
| Bioactive microfluidic paper device174 | Colorimetric | Carbaryl and chlorpyrifos | Paper | 0.24 μg L−1, 2.00 μg L−1 |
| Pump-free microfluidic rapid mixer with paper-based channel171 | Colorimetric | Malathion | Paper, transparency film | 10 nmol L−1 (LoQ) |
| Microfluidic paper-based device for type-II pyrethroids detection172 | Colorimetric | Cypermethrin, deltamethrin, cyhalothrin, and fenvalerate | Paper, laminating pouches | 2.50, 1.06, 3.20, and 5.73 μg mL−1 |
| Lipase embedded paper-based device175 | Colorimetric | Chlorpyrifos | Paper | 0.065 mg L−1 |
| Paper-based microfluidic device for carbamate pesticide detection176 | Colorimetric | Carbaryl, carbosulfan, and furathiocarb | Paper | 0.4, 0.24, and 0.46 mg L−1 |
| Three-layered paper-based microfluidic chips coupled to smartphone177 | Colorimetric | Profenofo and methomyl | Paper-based chip | 55 nM and 34 nM |
| Dispersive liquid–liquid microextraction coupled with microfluidic paper-based device178 | Colorimetric | Carbaryl, carbosulfan, chloropyrifos, furathiocarb, malathion, methomyl | Paper | 0.18–0.41 μg L−1 |
| Foldable paper-based device using angle-based readout179 | Colorimetric, angle-based readout | Dimethyl methylphosphonate (DMMP) | Paper | Semi-quantitative |
| Origami multiple paper-based electrochemical biosensor168 | Electrochemical | Paraoxon, 2,4-D, atrazine | Paper, electrode | 2 ppb, 50 ppb, N/A |
| Fe3O4 nanozyme-supported carbon quantum dots and sliver terephthalate MOFs as double catalytic amplification strategy on microfluidic paper-based chip180 | Electrochemical | Parathion-methyl | Paper-chip based | 1.1 × 10−11 mol L−1 |
| Electrochemical microsensor using MIP-based concentrators170 | Electrochemical | Glyphosate | Glass-based chip, PMMA | 247 nM – cal |
| 188 nM – chip | ||||
| Paper-based microfluidic chip using ratiometric fluorescence imaging181 | Fluorescence | 2,4-Dichlorophenoxy acetic acid | Paper | 90 nM |
| SERS-active Au@POM nanostructures in microfluidic device169 | SERS | Paraoxon-methyl | PDMS | 52.1 μg L−1 and 41.4 μg L−1 for Au@PMo and Au@PW chips |
| SERS coupled with microfluidic system for glyphosate in tap water detection182 | SERS | Glyphosate | PFA coils | 40 μg L−1 |
| S,N-doped carbon quantum dots for paper-based chemiluminescence detection of bendiocarb183 | Chemiluminescence | Bendiocarb | Paper | 0.02 μg mL−1 |
Lafuente et al. presented an intriguing method involving the preparation of surface-enhanced Raman spectroscopy (SERS)-active regions within a microfluidic channel to detect the paraoxon-methyl pesticide.169 UV light was utilized to induce the formation of polyoxometalate-decorated gold nanostructures on the microchannel surface (Fig. 2B). The adsorption of the pesticide onto the substrate led to a measurable change in SERS activity.
 | ||
| Fig. 2 Examples of microfluidic devices for pesticide detection in water. (A) Electrochemical chip-based sensor for glyphosate monitoring (adapted from ref. 170 with permission, copyright 2021, American Chemical Society). The figure shows (a) the microfluidic sensor featuring working electrodes with gold electrodeposition and a PMMA flow cell equipped with inlet and outlet ports and (b) cross-sectional view of the microfluidic sensor, highlighting the experimental setup that incorporates MIP-based concentrators. (B) Microfluidic device with SERS-active regions using polyoxometalate-decorated gold nanostructures (Au@POM) (reprinted from ref. 169 with permission, copyright 2020, American Chemical Society). The figure shows (a) the procedure for fabricating PDMS microfluidic chips using three-dimensional (3D) printing assistance and (b) synthesis of Au@POM nanostructures directly driven by UV light within the PDMS channels. (C) Pump-free microfluidic rapid mixer combines transparency film and double-sided adhesive. The detection of an organophosphate pesticide was used to demonstrate the utility of the device (reprinted from ref. 171 with permission, copyright 2020, American Chemical Society). The schematics illustrate the capillary-driven microfluidic mixer. (a) Representation of the assembled mixing device. Detailed depiction of the double-sided adhesives and transparency films for (b) overlapping inlet and (c) side-by-side inlet channels. Layers 1, 3, and 5 are composed of transparency film (light gray), while layers 2 and 4 are constructed with double-sided adhesive (dark gray). (d) A cross-sectional view. (D) μPAD for colorimetric detection of type-II pyrethroids in water. Detection is based on the formation of cyanide from the hydrolysis of type-II pyrethroids (reprinted from ref. 172). | ||
Uka et al. developed an electrochemical-based device for real-time monitoring of glyphosate in water.170 The device incorporated a molecularly imprinted polymer (MIP) concentrator based on solid-phase extraction (SPE) and a glass-based chip with a microelectrode array for glyphosate detection (Fig. 2A). The authors claimed that analysis with the chip could be obtained within minutes. However, the MIP concentration process, involving multiple steps such as equilibrating, pre-washing, washing, loading, cleaning, and eluting, would significantly increase the total assay time.
Detecting pesticides in water using microfluidic sensors presents challenges due to the diverse pesticide classes (herbicides, insecticides, fungicides, etc.). Each class may necessitate distinct detection methods, making a multiplex microfluidic device for simultaneous pesticide class detection desirable. Moreover, environmental factors like geography, farming practices, and seasonal fluctuations contribute to variability in pesticides present in water. Designing adaptable microfluidic devices capable of accommodating these variations is crucial for practical applicability.
Table 3 summarizes the analytical performance of the above-mentioned microfluidics platforms and other notable advancements in the field in recent years for detecting pesticides in water. It includes information on the substrate type, detection method, fabrication approach, and limit of detection (LoD) for each device.
3.4 Microorganisms
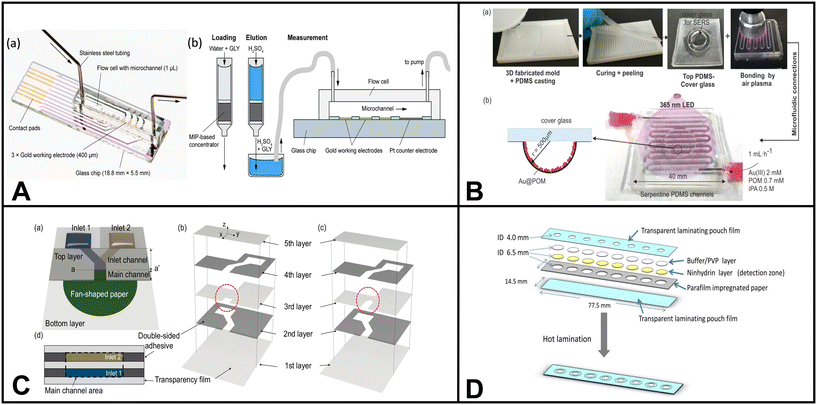
Pathogenic microorganisms are bacteria, viruses, and other microscopic organisms that may cause disease in humans and animals.184 Pathogens can be transmitted through various means, such as airborne particles, bodily fluids, contaminated food, and tainted water sources.184 Water is a vital resource for human consumption and essential activities. Therefore, developing effective detection techniques for monitoring waterborne pathogens becomes paramount in ensuring public health and safety. Traditional methods for detecting microorganisms include surface plasmon resonance (SPR), loop-mediated isothermal amplification (LAMP), polymerase chain reaction (PCR), electrochemistry, spectroscopy, and fluorescence microscopy.23 However, many of these techniques require expensive equipment and are shifting towards integration with microfluidic systems.Altintas et al. demonstrated an electrochemical sensor for bacteria detection, specifically Escherichia coli. The sensor consisted of a biochip with eight Au electrodes integrated into a microfluidic channel formed by PMMA. The detection method employed a sandwich immunoassay approach. Initially, a specific antibody for E. coli was attached to the surface of the sensor chip. Then, E. coli bacteria were introduced into the system, followed by adding a second antibody labeled with horseradish peroxidase (HRP) enzyme, which served as the detection antibody. As the concentration of E. coli increased, the number of HRP-labeled detection antibodies also increased, resulting in a higher measured response when substrate 3,3′5,5′-tetramethylbenzidine (TMB) was added to the system. The authors also incorporated gold nanoparticles into the system to amplify the detection of E. coli, resulting in enhanced sensitivity when quantifying E. coli in tap water samples.185
Chung et al. developed a paper-based microfluidic device for fluorescence detection of norovirus in environmental samples (Fig. 3E).186 The process involved pipetting norovirus containing samples onto a microfluidic paper-based analytical device (μPAD). The norovirus was captured on nitrocellulose paper through electrostatic interactions. Then, fluorescent polystyrene particles conjugated with antibodies specific to norovirus capsid protein were added to the μPAD. The interaction between the antibodies and antigens caused aggregation of the fluorescent particles. The resulting fluorescent aggregates were imaged using a smartphone-based fluorescence microscope and quantified using Image J software.
 | ||
| Fig. 3 Examples of microfluidic devices for microorganism detection in water. (A) Optoelectrical detection platform for bacteria based on plasmonic-assisted electrochemical impedance spectroscopy (PEIS). The microfluidic device consists of 3D gold nano/microislands (NMIs) and graphene nanosheets (adapted from ref. 191 with permission, copyright 2020, American Chemical Society). (B) Microfluidic array chip using PCR-based detection of waterborne bacteria (reprinted from ref. 192). (C) Schematic illustrating the process of continuous bacterial concentration using viscoelastic fluid. (a) Bacteria dispersed in a viscoelastic fluid are introduced randomly at the inlet. (b) Elastic forces cause bacterial cells to concentrate at the center of the microchannel. At the outlet, tightly focused cells are collected at the central outlet (outlet A), while the suspending medium is directed to the side outlets (outlet B). (c) Image depicting the fabricated device employed in this study for bacterial concentration. Microscopic images of (d) the filter zone, (e) the inlet, and (f) the outlet region of the microchannel (reprinted from ref. 189). (D) Colorimetric-based device using immunomagnetic separation to detect E. coli (reprinted from ref. 193). (E) Paper-based device coupled with smartphone technology for fluorescent detection of norovirus in environmental water samples. The figure schematic illustrates (a) the introduction of norovirus solutions and anti-norovirus particle suspension, and (b) the application of Blue LED irradiation to the μPAD from the side (reprinted from ref. 186 with permission, copyright 2019, American Chemical Society, https://pubs.acs.org/doi/10.1021/acsomega.9b00772, further permission related to the material excerpted should be directed to the ACS). | ||
Gowda et al. presented a proof-of-concept microfluidic device for environmental monitoring to detect water bacteria. Droplet digital loop-mediated isothermal amplification (LAMP) was used for bacteria quantification on a centrifugal disk (CD). The authors emphasized that the CD microfluidic platform is also capable of bacterial cell lysis in water samples, DNA extraction, and reagent mixing. These features collectively contribute to simplifying and automating the analysis process. Gowda and coauthors chose enterococcus faecalis as a representative bacterium to validate the device's performance. Their study showcases the potential of this microfluidic system for efficient and automated detection of bacteria in water samples, highlighting its ability to streamline various steps involved in the analysis process while minimizing the need for manual handling by the user.187
Studies have explored preconcentration of bacteria using microfluidic devices, showcasing their potential in addition to detection methods. For instance, Krafft et al. developed a microfluidic device for the concentration and detection of bacteria in drinking water. Using SERS as the detection method, they successfully identified E. coli and P. taiwanensis in tap water samples. The microfluidic device consisted of a nanoporous membrane connected to perpendicular microfluidic channels. The nanoporous membrane served a dual purpose: it acted as a concentration area by electrodriven trapping of silver nanoparticles and bacteria, enriching the target bacteria, and it facilitated SERS-based detection by providing an appropriate surface for enhanced Raman scattering, enabling the identification and quantification of the bacteria of interest.188
Similarly, Choo et al. presented a microfluidic device designed for the continuous concentration of bacterial cells based on viscoelastic non-Newtonian microfluidics (Fig. 3C).189 This technique utilizes fluids containing polymers with both viscous and elastic properties which is the basis for the microfluidic device effectively focusing particles based on size during continuous flow operations. To verify the device's performance, S. aureus was used, resulting in a concentration factor of 20.6-fold.
Microorganisms vary widely in size, shape, and surface properties.190 Designing a sensor that can detect these diverse set of microorganisms in water is a challenge to be addressed. Additionally, maintaining the viability of microorganisms during detection from water sources is necessary for certain applications, like studying environmental microorganisms or identifying live pathogens. Ensuring that the microfluidic environment does not harm or compromise the microorganisms is also a key challenge.
Table 4 provides a comprehensive overview of the analytical performance of microfluidics platforms for detecting microorganisms in water, including the above-mentioned platforms and other notable advancements in the field in recent years.
| Work | Method | Microorganism | Device or material | Detection limit |
|---|---|---|---|---|
| Aptasensor for pathogenic bacteria194 | Colorimetric | E. coli O157:H7, S. typhimurium | Paper | 103 CFU mL−1, 102 CFU mL−1 |
| Microfluidic device with access holes named micro-pupil for colorimetric signal view angle195 | Colorimetric | E. coli | PDMS, glass | 2 CFU/100 mL |
| 3D printed integrated microfluidic chip196 | Colorimetric | SARS-CoV-2, E. coli K12, Enterococcus faecalis, and Salmonella typhimurium | Methacrylate-based resin (3D printed chip) | 100 GE mL−1 and 500 CFU mL−1 |
| Lysis and direct detection of coliforms197 | Colorimetric | E. coli | Fluorinated paper | ∼104 CFU mL−1 |
| Colorimetric lateral flow strips198 | Colorimetric | E. coli | Nitrocellulose | 104 CFU mL−1 |
| Water-based polyurethane acrylate via UV light curing as alternative fabrication method199 | Colorimetric | E. coli BL21 | Paper | 3.7 × 103 CFU mL−1 |
| Microfluidic device with Au biochip185 | Electrochemical | E. coli | PMMA, biochip | 50 CFU mL−1 |
| Nanostructured gold/graphene microfluidic device191 | Plasmonic-assisted electrochemical impedance | E. coli, P. putida, and S. epidermidis | PDMS | ∼20 CFU mL−1 |
| Microfluidic chip and silver nanoparticle-based signal enhancement200 | Impedimetric | E. coli O157:H7 | PDMS, glass, microfluidic chip | 500 CFU mL−1 |
| Electroosmotic flow driven microfluidic device for bacteria isolation using magnetic microbeads201 | Fluorescence | E. coli | PDMS, glass | N/A |
| Ultrasonic nanosieve within microfluidic device202 | Fluorescence | E. coli DH5α | PDMS | 3.25 × 102 CFU mL−1 |
| Digital E. coli counter203 | Fluorescence | E. coli DSM 1103 | PDMS, glass | 100 bead particles/50 μL of volume |
| In liquid-fluorescence in situ hybridization assay on a microfluidic system204 | Fluorescence | E. coli | PDMS, glass | 104 cell per mL in the previous study |
| 102–104 cell per mL could be counted | ||||
| Immunoassay using fluorescence with magnetic nanoparticles for bacteria separation205 | Fluorescence | E. coli and Salmonella enteritidis | N/A | 5 CFU mL−1 and 3 CFU mL−1 |
| Continuous microfluidic concentrator189 | Fluorescence, RT-LAMP | Staphylococcus aureus | PDMS | N/A |
| Smartphone-based paper microfluidic particulometry of norovirus186 | Fluorescence, particulometry | Norovirus | Paper | 1 genome copy per μL (DI water), 10 genome copies per μL (reclaimed wastewater) |
| SERS-based microfluidic device188 | SERS | E. coli DH5α and Pseudomonas taiwanensis VLB120 | PDMS, PCTE (polycarbonate track-etched) membrane, glass slide | N/A |
| Portable pathogen analysis system187 | Colony counting | Enterococcus faecalis | PMMA | N/A |
| Phage-based bioluminescence assay on microfluidic device206 | Luminescence | E. coli | PVDF membrane | 4.1 CFU in 100 mL drinking water |
| Microfluidic array chip192 | PCR | EC H8, Gen bac III, UidA – primers | PDMS, glass, microchip | 71.8 copies (Gen bac III) |
| Bacteroidales, E. coli (target organisms)… | ||||
| Capillary flow dynamics-based sensing207 | Capillary flow based | E. coli K12 (water), Zika virus (blood serum) | Paper | 1 log CFU mL−1, and 20 pg mL−1 |
3.5 PFAS
Per- and polyfluoroalkyl substances (PFAS) are pervasive environmental contaminants with significant global health and ecological implications. Their applications range from textile coatings to components in aqueous film-forming foams (AFFF) to consumer items like food packaging and non-stick appliances.208 Despite their prevalence in air, water, and soil, the majority of PFAS (95%) are released into aquatic environments, making PFAS detection in water crucial.209 While the application of microfluidic devices for PFAS detection remains relatively limited, the few studies that have ventured into this domain have yielded promising outcomes.Cheng et al. introduced an electrochemical-based sensor for detecting perfluorooctanesulfonate (PFOS) using a metal–organic framework (MOF) with a chromium center. This approach relied on the MOF's strong electronic attraction to facilitate the effective capture of PFOS molecules. The captured PFOS was then detected using an interdigitated microelectrode array within a microfluidic channel. The sensor achieved a limit of detection (LoD) of 0.5 mg L−1 and was successfully tested with real groundwater samples spiked with PFOS.210
Breshears et al. developed a paper-based microfluidic device that harnesses competitive molecular interactions during capillary action to detect perfluorooctanoic acid (PFOA). The device operates by exploiting the interactions between PFOA, cellulose fibers, and reagents L-lysine, casein, or albumin. The reagent is preloaded on the paper surface within the device. When the sample solution containing PFOA is added, it interacts with the reagent. This interaction leads to forming a PFOA-reagent complex, reducing surface tension as the complex migrates to the wetting front and slowing down the capillary flow rate on the microfluidic device. It is worth noting that while the assay only provides qualitative yes/no results, its LoD for PFOA is highly impressive, with a LoD of 10 ag μL−1 in DI water and 1 fg uL−1 in processed wastewater.211
In 2022, the EPA updated its interim suggestion for the acceptable level of PFOS to 0.02 parts per trillion (ppt).212 This limit is a stricter compared to the previous recommendation set in 2016,213 further showcasing the negative effects of PFAS on human health and the environment. Creating a single sensor that can accurately detect all these different PFAS structures is tricky because PFAS come in various chain lengths and head groups.214 Although numerous sensors have been created to tell PFOA and PFOS apart, the main objective should be to design a microfluidic platform capable of identifying and quantifying all types of PFAS.
3.6 Other
Other applications of microfluidic devices for water monitoring include the detection of compounds and properties such as pharmaceuticals,24,215 dissolved oxygen content, explosive residues, and pH, among others. For instance, Burtsev et al. developed a microfluidic device to determine insoluble pharmaceuticals in water via extraction and SERS measurements. They demonstrated the device application using ibuprofen and generated a detection limit of 10−8 M.216 Gril et al. presented a fluorescence-based sensor for detecting dissolved oxygen in water. The system is composed of silica capillaries and optical fibers, and upon testing, revealed an accuracy of 0.025 mg L−1 with a response time of less than 60 s.217 Finally, Charles et al. demonstrated a bifurcated 128-microchannel device for environmental monitoring of explosives. The device, constructed of PMMA, used fluorescence-based displacement immunoassay for explosive determination. TNT was used as a representative explosive, and the detection limit was determined as 10 ppt.218 Moradi et al. developed a fluorescence-based pH sensor where the indicator dye 8-hydroxypyrene-1,3,6-trisulfonic acid trisodium salt (HPTS) and test solution are mixed in a microfluidic chip. The resulting fluorescence signal is transduced via optical fibers and measured with a photodiode. A pH range of 2.5 to 9 was tested.2194. Soil
Ensuring sustainable agricultural production and food security relies upon efficient monitoring of soil health for diverse contaminants and nutrients. The traditional approach to soil chemistry monitoring involves the collection and transportation of soil samples to centralized labs for analysis using complex technologies such as gas and liquid chromatography, mass spectrometry, and nuclear magnetic resonance.220–222 These techniques are not suitable for on-site soil monitoring, thus, frequent and timely assessments of soil health is challenging. A promising resolution to bridge the gaps in soil sensing platforms is using microfluidic systems since they offer the potential for automating multiple steps, saving both cost and time.Contrary to water samples, which can be handled quite easily in microfluidics, soil samples need more extensive processing due to their complexity. For example, soil characteristics can vary even within a small sampling area. Additionally, organic debris, minerals, and other impurities can obstruct microfluidic tests and may require a pre-treatment stage. Therefore, to provide precise and trustworthy results in microfluidic applications using soil samples, several crucial sample preparation steps are required. First, the representative soil sample is collected, sieved, and homogenized to remove huge debris, achieve a constant particle size, and ensure homogeneity of the sample. Other treatments could involve processes like wet sieving, centrifugation, or chemical processing. The soil sample may also need to be pre-treated before being separated into its solid and liquid components, frequently using methods like sedimentation or filtering. Finally, the removed liquid phase can then be added to microfluidic systems for additional investigation, which may include different on-chip procedures like mixing, filtering, and detection.
4.1 Heavy metals
Soil stores and circulates essential nutrients, microbes, and minerals that plants require for growth, making them the foundation of nutrition and food security. However, introducing metals into the soil can pose significant threats to this essential function. Specifically, soil areas contaminated with heavy metals can disrupt crucial processes, including microbial activity and the decomposition of organic materials.223 Heavy metals such as Cd, Cu, Pb, etc., when present in the soil, have the potential to severely impede vital functions like nitrogen mineralization, nitrification, and soil respiration.223Table 5 shows the regulatory soil limits for heavy metals. The negative impact of heavy metal contamination on soil health necessitates active and regular monitoring of their levels in the soil. Farmers, land managers, and environmental agencies can gain valuable insights into the potential risks posed by the heavy metal presence and take appropriate actions to mitigate their harmful effects by implementing effective monitoring techniques.| Heavy metal | Maximum concentration in sludge (mg kg−1 or ppm) | Annual pollutant loading rates (kg ha−1 per year) | Annual pollutant loading rates (lb A−1 per year) | Cumulative pollutant loading rates (kg ha−1) | Cumulative pollutant loading rates (lb A−1) |
|---|---|---|---|---|---|
| Arsenic | 75 | 2 | 1.8 | 41 | 36.6 |
| Cadmium | 85 | 1.9 | 1.7 | 39 | 34.8 |
| Chromium | 3000 | 150 | 134 | 3000 | 2679 |
| Copper | 4300 | 75 | 67 | 1500 | 1340 |
| Lead | 420 | 21 | 14 | 420 | 375 |
| Mercury | 840 | 15 | 13.4 | 300 | 268 |
| Molybdenum | 57 | 0.85 | 0.8 | 17 | 15 |
| Nickel | 75 | 0.9 | 0.8 | 18 | 16 |
| Selenium | 100 | 5 | 4 | 100 | 89 |
| Zinc | 7500 | 140 | 125 | 2800 | 2500 |
Sutariya et al., introduced a novel method for the single-step fluorescence detection of As3+, Nd3+, and Br− using a pyrene-linked calix[4]arene compound.225 The sensing probe is incorporated into a cellulose matrix, and the sensing mechanism involves resonance energy transfer. The authors detected Nd3+ in soil matrices. In addition, the authors expanded the utility of calix[4]arene by developing a paper-based probe for chelation-enhanced fluorescence-photoinduced electron transfer (CHEF-PET) fluorescence, which allowed them to detect La3+, Cu2+, and Br−.226 Furthermore, they designed a fluorescence-photoinduced electron transfer probe that effectively detected Mn2+, Cr3+, and F−.227 Arabyarmohammadi et al. focused on the simultaneous immobilization of heavy metals (Cu2+, Pb2+, and Zn2+) in the soil environment using nanoporous biochars derived from pulp and paper. The authors synthesized paper-derived biochar and investigated their mechanisms for effectively immobilizing these heavy metals within contaminated soil.228
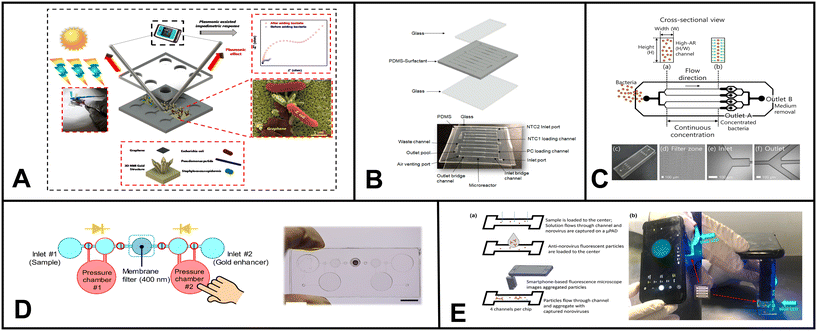
Nashukha et al. developed a new paper-based analytical device (μPAD) for detecting mercury. The μPAD works by generating a tetraiodomercurate(II) ion (HgI42−) from a reaction between mercury and iodide ions.229 The iodine vapor then diffuses through the μPAD, forming a purple-colored tri-iodide starch complex. The intensity of the purple color can be used to quantify the amount of mercury in the sample (Fig. 4C). The μPAD was found to be effective in detecting mercury in both soil and water samples. The results were in good agreement with those obtained using ICP-MS. The μPAD is a simple and portable device that could be used to assess mercury levels in a variety of settings, such as inactive gold mines.
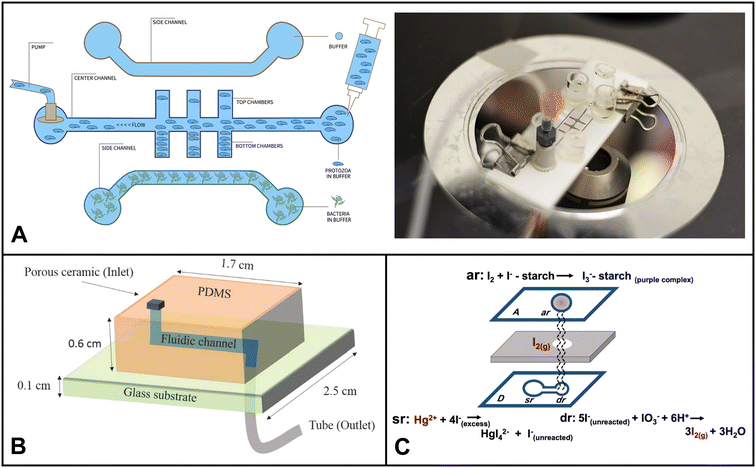 | ||
| Fig. 4 Examples of microfluidics systems for soil monitoring including (A) conceptual design (left) and microfluidic assembly (right) for microfluidic device for the quantitative evaluation of protozoa response to Listeria monocytogenes by Gaines et al. (reprinted from ref. 238, copyright 2019, PLOS). (B) A sensor designed by Böckmann et al. for the extraction of soil solution into a microfluidic chip (reprinted from ref. 234 copyright 2021, MDPI). (C) Equipment-free paper-based device for determination of mercury in contaminated soil by Nashukha et al., (reprinted from ref. 229, copyright 2021, MDPI). | ||
Ding et al. introduced a device that integrated a solid reference electrode with a paper-based microfluidic system for potentiometric ion sensing.230 The device included solid-contact ion-selective electrodes (ISEs) sensitive to metal ions and Cl−. Instead of directly dropping a KCl solution onto the disposable paper substrate (DPS) during analysis, researchers deposited solid KCl on the reference zone beforehand. The researchers found that reference electrodes equipped with paper substrates offering higher KCl levels had shorter equilibration times and higher potential reproducibility. The developed device was then used to directly analyze sludge and sweat samples.
Silva et al. introduced a novel approach for determining heavy metals in complex environmental samples using non-equilibrium potentiometric sensors integrated with metal-modified paper-based substrates.139 The utilization of metal-modified paper substrates helped control the response of lead(II) ion selective electrodes. This non-equilibrium-based potentiometric system accurately determined lead concentrations in challenging environmental samples, showcasing its potential as an effective and practical method for heavy metal analysis.
In addition to the extra sample preparation step, the detection of heavy metals in soil is more challenging than in water due to higher spatial variability and oxidation state of heavy metals. Different reducing and oxidizing microbes present in soil can change the oxidation state of metal ions, which can lead to inaccurate estimations if the detection system is not designed to take this into account. Therefore, it is important to carefully select the sampling sites, collect multiple samples from each site, and use a detection system that is capable of oxidizing or reducing all forms of metal to one oxidation state. The microfluidic chip can be automated to pretreat the sample through an oxidizing/reducing region by incorporating stable reducing or oxidizing reagents in those pretreatment sections.
4.2 Nutrients
Soil is a vital source of essential nutrients crucial for the growth and development of plants. Nitrogen (N), phosphorus (P), and potassium (K), collectively known as NPK, are particularly significant nutrients in the soil. As secondary soil nutrients, calcium, magnesium, and sulfur are also pivotal in supporting overall plant health. Maintaining an optimal concentration of these nutrients ensures optimal plant growth and crop production. Over the past five years, numerous methodologies have emerged for monitoring soil nutrient levels through microfluidics.Using a unique anti-aggregation mechanism, Pinyorospathum et al. developed a colorimetric sensor to determine phosphate ions.231 This sensor is based on using 2-mercaptoethanesulfonate-modified silver nanoplates (MS-AgNPls) and europium ions (Eu3+). The colorimetric response is attributed to the interaction between the negatively charged sulfonate groups on MS-AgNPls and the europium ions, leading to aggregation of the MS-AgNPls and causing a distinct color change. However, in the presence of pre-mixed Eu3+ with PO43−, the color of MS-AgNPls remains unaltered due to the stronger binding affinity of Eu3+ towards PO43−. As a result, PO43− prevents the aggregation of MS-AgNPls by forming stable complexes with Eu3+, resulting in the dispersion of the nanoplates.
Pal et al. developed a low cost and low power colorimetry-based platform to quantify phosphorus and nitrogen in soil and other environmental samples. A PDMS based microfluidic chip was integrated with LED and photodiode to determine the concentration of the nutrients. The authors also developed a mobile app for data transmission and monitoring in short range. The sensor was able to achieve the detection upto an environmentally relevant μM levels. The compact device allows integration with the Internet of Things (IoT) and Bluetooth-based modules for real-time data monitoring, enabling timely interventions and adjustments based on accurate and up-to-the-minute environmental data.232
Dudala et al. developed a microfluidic soil nutrient detection system capable of simultaneously detecting nitrite concentration, pH, and electrical conductivity. The system utilized a multiplexed polydimethylsiloxane (PDMS) device to perform tests for EC and nitrite. The EC measurement employed a conductivity cell containing copper electrodes connected in series with a resistor powered by an oscillating power source. Using an LED and photodiode, a photometric detection method based on the Griess reaction was employed to quantify nitrite levels. Additionally, a trans-impedance amplifier circuit was designed to amplify the output from the photodiode.233
Böckmann and colleagues designed a microfluidic chip with a built-in porous ceramic filter that can extract soil solution on-site with minimal sample requirement.234 The chip had little to no coagulation effect. The device was able to draw out soil water from three different soil types, silt, garden soil, and sand, by using a pump to generate suction through the microfluidic channel (Fig. 5B). The developed system has the potential to be used in both continuous sensing setups and discrete time-point sensing applications in soil (e.g. bacterial monitoring, ecological simulation etc.).
When designing microfluidic technologies for soil nutrient detection, the ultimate objective should be to develop compact, mobile, and versatile applications. However, several challenges need to be addressed for real-world success. First, there is a need to ensure proper soil sample preparation to enable accurate analysis. Second, nutrient levels can vary significantly due to factors like temperature, humidity, topography, and surrounding vegetation. Therefore, the impact of environmental changes on measurement results must be considered. Third, it is vital to make the technology cost-effective and accessible to a broader user base.
4.3 Microorganisms
Soil microbes are fundamental to the food chain, agriculture, and plant growth. Compared to water, soil often has a higher concentration and diversity of microorganisms. Due to its complex structure, accessibility of nutrients, and interactions between organic and inorganic components, the soil environment offers a rich habitat for microorganisms. This diverse group includes bacteria, fungi, viruses, protozoa, and archaea, which are pivotal in regulating vital ecosystem processes. Microorganisms like mycorrhizal fungi, actinobacteria, nitrogen-fixing bacteria, certain archaea, soil-adapted algae, and diverse soil-dwelling animals are more prevalent in soil environments compared to water. They establish symbiotic associations with plants, exhibit decomposition abilities, and facilitate nutrient cycling. Hence, monitoring soil microorganisms is essential for evaluating soil health, nutrient cycling, and the resilience of plants and ecosystems. In the past five years, microfluidics used in soil microbial monitoring predominantly focused on investigating their spatial distribution, functions, and biological processes. Researchers have used these devices to explore critical phenomena, including quorum sensing mechanisms, bacterial chemotaxis, horizontal gene transfer, biofilm streamer formation, and other essential microbial activities.235–237Gaines et al. explored using a microfluidic system to investigate how protozoa (Euglena) respond to chemical cues released by bacterial cells (Listeria monocytogenes).238 They devised a three-channel microfluidic device comprised of a nitrocellulose membrane and glass slides (Fig. 4A). The device facilitates Euglena's suspension's flow through the central channel. The bacterial chemicals served as attractants, diffusing through the membrane and influencing the movement of Euglena. The researchers could make measurable comparisons by analyzing the numbers of Euglena migrating towards each side of the device in the presence of bacterial cells. This innovative microfluidic setup provided valuable insights into the interactions between protozoa and bacteria, shedding light on their chemotactic responses and behavioral changes.
Baranger et al. developed a microfluidic system for monitoring the growth of individual hyphae in confined environments.239 Their microfluidic device enables precise microscopic observations and nutrient perfusion, allowing for both static and dynamic conditions. Through time-lapse microscopy, the researchers simultaneously monitored the growth of multiple Talaromyces helicus and Neurospora crassa hyphae in parallel microchannels.
De Anna et al. employed a microfluidic model system capable of capturing flow disorder and chemical gradients at the pore scale. They investigated the transport and dispersion of the soil-dwelling bacterium Bacillus subtilis in porous media. Through this microfluidic approach, they discovered that chemotaxis significantly influences the bacteria's persistence in low-flow regions of the pore space, leading to a 100% increase in their dispersion coefficient.
Crane et al. conducted a comprehensive investigation to determine the feasibility of using microfluidic quantitative polymerase chain reaction (MFQPCR) as a high-throughput alternative for quantifying functional genes. Their study evaluated 29 established and 12 newly designed primer pairs targeting taxonomic, nitrogen-cycling, and hydrocarbon degradation genes in genomic DNA soil extracts. The researchers tested these primers under three different sets of MFQPCR assay conditions to assess their efficiency, specificity, and sensitivity. The findings revealed that while MFQPCR enables high-throughput quantification of microbial functional genes, it requires meticulous curation of primers to ensure accurate and reliable results.
Microfluidic devices used for monitoring and investigating the biological processes of soil microbes can face limitations such as surface modification and characterization, representing 3D soil structures, and accommodating limited internal volumes.18 First, there is a lack of robust methods to fabricate microfluidic systems that can replicate the intricate surface properties found in natural soils. Second, the use of 2D micromodels in these devices fails to capture the full complexity of 3D porous soil structures. Last, the small internal volumes of microfluidic devices present challenges when it comes to conducting downstream chemical analyses for comprehensive microbial monitoring.
4.4 Pesticides
A study conducted in 2021 by Gunstone et al. found that the extensive use of pesticides in American agriculture poses a grave threat to biodiversity, soil health, and the fight against climate change.240 The study revealed that in 71% of the cases investigated, pesticides caused harm or even death to soil invertebrates, such as earthworms, ants, beetles, and ground-nesting bees.240 Hence, it is critical to monitor pesticide levels in soil matrices.Hondred et al. introduced a method for electrochemical pesticide monitoring using nanoporous gold peel-and-stick biosensors.241 They patterned a 3-electrode system on adhesive polyimide films, creating convenient disposable peel-and-stick tape or wearable sticker sensors. They applied these peel-and-stick sensors to develop a disposable tape pesticide device and a reusable 3D-printed flow cell for detecting organophosphate (paraoxon) in soil samples. In addition, Guselnikova and coauthors introduced a novel and efficient approach for sensitive and selective surface-enhanced Raman scattering (SERS) detection of organophosphorus pesticides.242
Shriver-Lake et al. developed a paper-based electrochemical detection method for chlorate.243 They utilized a paper-based probe impregnated with a vanadium-containing polyoxometalate anion on screen-printed carbon electrodes. Notably, the device exhibited impressive stability, retaining its functionality after eight months of storage.
The efficient extraction of pesticides from soil and their transfer into microfluidic systems is a major challenge. Many pesticide detection methods employ colorimetric enzyme inhibition techniques, but interference from non-targeted pigments in the soil matrix can significantly alter the color of the extracted solution during the extraction process as reported by Jin et al.244 This problem can be minimized by using a more selective extraction solvent or incorporating a cleanup pretreatment step within the microfluidic system.
4.5 Others
Moram et al. presented a novel approach involving Ag/Au nanoparticle-loaded paper-based substrates for versatile surface-enhanced Raman spectroscopy (SERS) detection of explosive molecules, including picric acid, 2,4-dinitrotoluene, and 3-nitro-1,2,4-triazol-5-one.245 Pellegrini and his colleagues have recently introduced an innovative paper sensor technique designed to assess acid volatile sulfides in soil samples on-site.246 Moreover, microfluidics in soil analysis has demonstrated its ability to detect other analytes, such as melamine,247S. chartarum,248 TNT,249,250 pH,233 and conductivity.233As reviewed by Zhu et al., replicating entire soil environments in microfluidic devices is technically impossible.237 Instead, microfluidic soil platforms for active monitoring should aim to mimic specific soil characteristics under defined conditions, such as pore geometries, chemical gradients, and microbe separation. Researchers should clearly define the distinct features of the soil environment when designing microfluidic devices and report results based on soil settings. Table 6 summarizes the different microfluidic platforms for soil monitoring discussed in this section and other notable advancements in the field in recent years.
| Work | Method | Metal | Device or material | Detection limit |
|---|---|---|---|---|
| Paper based device for Hg detection in soil229 | Colorimetric | Hg(II) | Paper | 20 ppm |
| Rapid detection of As(III) in contaminated soil251 | Colorimetric | As(III) | PDMS | 0.71 ppm |
| Investigation of total chromium using uPADs252 | Colorimetric | Total Cr | Paper | 0.25 ppm |
| Paper sensor for acid volatile sulfides in soil246 | Colorimetric | S2− | Paper | <0.1 μmol g−1 |
| Detection of melamine using superhydrophobic preconcentration paper spray ionization mass spectrometry247 | Colorimetric | Melamine | Paper | 1.2 ppt |
| Paper-based colorimetric nanoprobe for the detection of S. chartarum248 | Colorimetric | S. chartarum | Paper | 10 to 100 spores mL−1 |
| Paper-based versatile surface-enhanced Raman spectroscopy substrates for multiple explosives detection245 | Colorimetric | Picric acid, 2,4-dinitrotoluene, and 3-nitro-1,2,4-triazol-5-one | Paper | NA |
| Colorimetric sensor for determination of phosphate ions using anti-aggregation of modified silver nanoplates and europium ions231 | Colorimetric | PO43− | Paper | 0.33 ppm |
| IoT enabled microfluidic colorimetric detection232 | Colorimetric | Nitrite and phosphate | PDMS | 0.021 ppm & 0.095 ppm |
| Paper based device for Hg detection in soil229 | Colorimetric | Hg(II) | Paper | 20 ppm |
| Rapid detection of As(III) in contaminated soil251 | Colorimetric | As(III) | PDMS | 0.71 ppm |
| Investigation of total chromium using uPADs252 | Colorimetric | Total Cr | Paper | 0.25 ppm |
| Paper sensor for acid volatile sulfides in soil246 | Colorimetric | S2− | Paper | <0.1 μmol g−1 |
| Detection of melamine using superhydrophobic preconcentration paper spray ionization mass spectrometry247 | Colorimetric | Melamine | Paper | 1.2 ppt |
| Paper-based colorimetric nanoprobe for the detection of S. chartarum248 | Colorimetric | S. chartarum | Paper | 10 to 100 spores mL−1 |
| Paper-based versatile surface-enhanced Raman spectroscopy substrates for multiple explosives detection245 | Colorimetric | Picric acid, 2,4-dinitrotoluene, and 3-nitro-1,2,4-triazol-5-one | Paper | NA |
| Colorimetric sensor for determination of phosphate ions using anti-aggregation of modified silver nanoplates and europium ions231 | Colorimetric | PO43− | Paper | 0.33 ppm |
| IoT enabled microfluidic colorimetric detection232 | Colorimetric | Nitrite and phosphate | PDMS | 0.021 ppm & 0.095 ppm |
| Nanoporous gold peel-and-stick biosensors created with etching inkjet maskless lithography241 | Electrochemical | Paraoxon | NPGL and metal leaf materials | 0.53 pM |
| Nanoporous gold peel-and-stick biosensors created with etching inkjet maskless lithography241 | Electrochemical | Paraoxon | NPGL and metal leaf materials | 0.53 pM |
| Solid reference electrode integrated with uPADs230 | Potentiometry | K+, Na+ | Paper | 10–4.1±0.1, 10–3.3±0.1 mol dM−3 |
| Solid reference electrode integrated with uPADs230 | Potentiometry | K+, Na+ | Paper | 10–4.1±0.1, 10–3.3±0.1 mol dM−3 |
| Microfluidic soil nutrient detection system233 | Photometric | Nitrite | PDMS | 0.07103 ppm |
| Single step fluorescence recognition using pyrene linked calix[4]arene225 | Fluorescence | As(III), Nd(III) | Paper | 11.53 nM, and 0.65 nM |
| Fluorescent aptasensor for Pb(II) detection253 | Fluorescence | Pb(II) | Paper | 6.1 nM |
| Fluorescence paper based probe for La(III), Cu(II), and Br− (ref. 226) | Fluorescence | La(III), Cu(II) | Paper | 0.88 nM, and 0.19 nM |
| Fluorescent aptasensor for Pb(II) detection253 | Fluorescence | Pb(II) | Paper | 6.1 nM |
| Fluorescence paper based probe for La(III), cu(II), and Br− (ref. 226) | Fluorescence | La(III), Cu(II) | Paper | 0.88 nM, and 0.19 nM |
| Single step fluorescence recognition using pyrene linked calix[4]arene225 | Fluorescence | As(III), Nd(III) | Paper | 11.53 nM, and 0.65 nM |
| MOF-5 coated SERS active gold gratings for the selective detection of organic contaminants in soil242 | SERS | Azo-dye, mycotoxin and pesticide | Gold gratings | Up to 10−14 M |
| MOF-5 coated SERS active gold gratings for the selective detection of organic contaminants in soil242 | SERS | Azo-dye, mycotoxin and pesticide | Gold gratings | Up to 10−14 M |
5. Air
Air pollution is a critical global issue with serious health implications for exposed communities. Prolonged inhalation of gases and particles from human activities can lead to discomfort, allergies, and even cancer.254 The negative effects of these pollutants are influenced by various factors such as their chemical composition, size, shape, exposure duration, and intensity.255 Therefore, active monitoring of air pollutants is essential. In recent years, Lab-on-a-Chip (LoC) devices have emerged as adaptable, rapid, accurate, and cost-effective platforms for detecting and analyzing suspended particles in water-based environments.256,257 However, with gaseous samples, LoC technologies encounter significant constraints, particularly in the analysis of atmospheric aerosols.258 The successful capture and analysis of air particles on microchips pose considerable challenges due to the broad spectrum of sizes, rapid gas diffusion, and complex particle–gas interactions.259 Despite the challenges posed by chemical interactions and inconsistencies in gas-based LoCs, significant strides have been made in using microfluidics for active monitoring of air analytes in past five years.5.1 Heavy metals
Multiple sources release heavy metals into the atmosphere, including industrial emissions, fossil fuel combustion, soil resuspension, waste incineration, and natural processes.260 Exposure to particulate metals poses a significant health risk to diverse populations, especially millions of workers in manufacturing, construction, and transportation industries, who are exposed to these airborne metal particles during their occupational activities.261–263 Inhalation of these metal particulates can lead to a spectrum of health hazards, including immune dysregulation, respiratory carcinogenesis, developmental impairments, hepatic dysfunction, neuropathies etc.264–267 Therefore, it is essential to create simple methods for detecting particle metals, in order to safeguard the public health and environment.Shariati et al. developed an innovative microfluidic paper-based analytical device utilizing gold nanoparticles (AuNPs) functionalized with N,N′-bis(2-hydroxyethyl)dithiooxamide (HEDTO) for the rapid detection of Hg(II) in air and various samples.268 The detection mechanism involves the aggregation of HEDTO–AuNPs with Hg2+ ions in the paper channels, resulting in an immediate color change of the modified AuNPs within the test zone. This observable color change is easily discernible with the naked eye or can be quantified using a digital camera. The device's performance was validated by collecting particulate matter samples on filters and comparing the results obtained through cold vapor atomic absorption spectrometry (CVAAS) and UV-vis spectroscopy.
Sun et al. developed a novel approach for the enhanced on-site detection of trace Ni, Cu, and Fe metals in airborne environments using graphene oxide nanosheets coupled with paper microfluidics.269 Their system comprises GO-nanosheet-coated paper devices, unmanned aerial vehicle multiaxial sampling, and cellphone-based colorimetric detection. The researchers also utilized a Wi-Fi camera, a self-developed app, and a sample pretreatment cartridge to rapidly process and characterize metal in particulate matter (PM) samples within 30 minutes.
Jia et al. presented a study on spatial varying profiling of PM constituents using paper-based microfluidics.270 Their approach involved the integration of mobile aerial sampling, in-flight tethered charging, and smartphone-based colorimetric analysis within a cost-effective paper microfluidic device. The method enabled quantitative profiling of spatiotemporal variations in trace metal components (Fe, Ni, and Mn) at four different geographical locations in Fuzhou, China, over 21 days.
Mettakoonpitak et al. developed a novel Janus electrochemical paper-based analytical device (ePAD) for metal detection in aerosol samples.271 Their study enhanced the Janus ePAD design to include four independent channels and working electrodes, facilitating the simultaneous detection of multiple metals in particulate matter. The device's configuration allows for square-wave anodic stripping voltammetry (SWASV) and square-wave cathodic stripping voltammetry (SWCSV), enabling the simultaneous detection of Pb, Fe, Cd, Cu, and Ni from a single sample at concentrations as low as ppb levels.
Particulate metal toxicity is mostly an occupational health risk in the construction and manufacturing sectors which can release metal particles into the air. Therefore, real-time monitoring systems are essential for assessing occupational exposure to particulate metals and preventing these health problems. Electrochemical sensors are a promising technology for real-time monitoring since they are portable and can be easily integrated with wireless platforms, allowing for remote monitoring. In this regard, microfluidics hold a promising path since it can be integrated into these electrochemical platforms to automate the multiple sample pretreatment and mixing process.
5.2 Organic pollutants
Air pollutants besides heavy metals include volatile organic compounds (VOCs), particulate matter, nitrogen oxides, ozone, and various airborne pathogens.272 Exposure to these pollutants can lead to respiratory problems, cardiovascular diseases, neurological disorders, and even premature death.272 Additionally, they contribute to environmental issues such as smog formation, acid rain, and climate change.272 Accurate and efficient detection methods are crucial to combat the adverse effects of non-metallic air pollutants.Guo et al. developed a microfluidic chip designed to detect gaseous formaldehyde. The chip is based on the 4-aminohydrazine-5-mercapto-1,2,4-triazole (AHMT) method, in which formaldehyde gas reacts with AHMT and specific reagents, leading to the formation of a distinct colored product (Fig. 5D).273 The chip, fabricated using PDMS, contains two reagent reservoirs that effectively mix and direct the reaction into a hydrophobic porous polytetrafluoroethylene (PTFE) membrane within the third chamber. The reaction's resulting color directly correlates to the formaldehyde concentration. Moreover, the microchip showcases excellent selectivity, as demonstrated by minimal interference from acetaldehyde and various VOCs tested against the device.
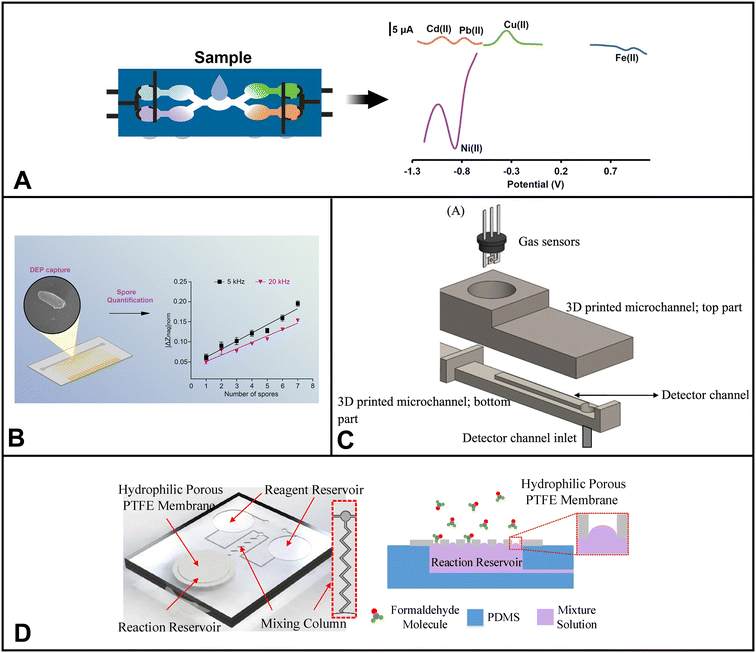 | ||
| Fig. 5 Examples of microfluidic devices for air applications. (A) Electrochemical paper-based device for simultaneous detection of Cd(II), Pb(II), cu(II), Fe(II), and Ni(II) in aerosols (adapted from ref. 271 with permission, copyright 2019, American Chemical Society). (B) Nanoelectrode-activated microwell array for airborne pathogen quantification (reprinted from ref. 277, American Chemical Society, https://pubs.acs.org/doi/10.1021/acsomega.1c04878). (C) Microfluidic gas sensor based on “like dissolves like” (adapted from ref. 282). (D) Microfluidic chip component used in smartphone-based colorimetric sensor for formaldehyde detection (adapted from ref. 273). | ||
Ozhikandathil et al. presented a novel optical method for gaseous ammonia detection based on the reaction of ninhydrin with ammonia. This approach involves integrating ninhydrin into a PDMS polymer matrix, forming a ninhydrin-PDMS composite, which is then integrated into a microfluidic device for ammonia detection. Upon chemisorption of ammonia onto the composite, its optical absorption property undergoes a change, measured using an LED and photoresistor within the device. The resulting purple-colored compound allows for efficient optical detection of ammonia, offering a simple yet effective solution for gas sensing.274
In the pursuit of improved selective detection of VOCs, Ghazi et al. have devised an interesting microfluidic device. Selectivity is significantly enhanced by embedding micro- and nanofeatures within the device and optimizing their characteristics, such as the number, radius, height, and distance between the microfeatures. Furthermore, the optimized microfeatures are coated with graphene oxide, effectively increasing the surface-to-volume ratio of the microchannel. The detection method relies on a MOS sensor, where the interaction between the target analyte and oxygen on the sensing layer results in a change in conductivity. Compared to a non-modified device, the modified microfluidic device exhibits a 64.6% and 120.9% improvement in selectivity with micro-features and nano-microfeatures, respectively.275
Yang et al. have introduced an array-assisted SERS microfluidic chip for gas sensing, using aldehyde gas as a model air pollutant. Their SERS probe is comprised of a composite nanoparticle housing MOF material, Au@Ag nanocubes, and cysteamine (CA), which acts as the gas-capturing agent. A PDMS microfluidic chip with arrays was fabricated, where these nanoparticles were uniformly modified on the side walls of the prisms, serving as the sensing areas. This setup allows for efficient gas detection with enhanced sensitivity, making it an exciting advancement in the field of gas sensing.276
Although some portable LoC platforms for organic pollutant matter have been reported, there is still a lack of real-time analysis devices due to the separation of various technologies involved, especially for airborne PM.28 Micro-electro-mechanical systems and nano-electro-mechanical system (MEMS/NEMS) technologies will be helpful for integrating and continuously monitoring multiple variables related to airborne PM.
5.3 Airborne pathogens
Airborne pathogens represent a specific subset of non-metallic air pollutants, including various infectious microorganisms such as bacteria, viruses, and fungi. These microscopic organisms travel through the air and pose significant health risks to humans and animals alike. Exposure to airborne pathogens can lead to respiratory infections, including influenza, tuberculosis, and other contagious diseases.272 Furthermore, these pathogens can intensify pre-existing health conditions, especially in vulnerable populations. In addition to their impact on human health, airborne pathogens can also harm the environment by affecting ecosystems and wildlife.272 Detecting and monitoring airborne pathogens is essential for preventing disease outbreaks and implementing effective public health measures.Duarte et al. developed a microfluidic device with a nanoelectrode-activated microwell array for capturing and quantifying Sclerotinia sclerotiorum spores, pathogens causing Sclerotinia stem rot in crops (Fig. 6B).277 The device is based on a microfluidic design and contains a nanothick aluminum electrode structure integrated with a picoliter well. This combination enables the effective capture of Sclerotinia sclerotiorum spores using dielectrophoresis, where electric fields are used to manipulate the movement of charged particles (in this case, the spores) toward the wells. Moreover, the device is equipped with an on-chip quantitative detection method by impedimetric sensing. The device achieves over 90% spore trapping rate and high sensitivity, enabling single spore detection, making it valuable for crop disease forecasting.277
Kim et al. presented a paper-based microfluidic chip for directly capturing and quantifying airborne SARS-CoV-2. First, the droplets and aerosolized virus are collected onto the paper microfluidic chip. Then, antibody-conjugated particles are loaded onto the paper channel, which induces antibody–antigen binding and subsequent particle aggregation. A smartphone-based fluorescence microscope is used to quantify the particle aggregation on the paper chip and can be correlated to the presence of airborne SARS-CoV-2. Directly detecting SARS-CoV-2 from the air can be challenging, especially due to the low concentration. However, without using an air sampler, this study directly detects SARS-CoV-2 in the droplets/aerosols form from the air. Additionally, the total capture-to-assay time was less than 30 min, which is impressive when compared to its competitors with several hours.278
Liu et al. demonstrated a microfluidic module for detecting airborne and waterborne pathogens. Liu and coworkers developed three types of chips: one for airborne pathogen enrichment and analysis, one for body fluid sample enrichment and nucleic acid detection, and a third chip for parasite oocysts or cysts enrichment and in situ immunofluorescence detection. For the airborne pathogen chip, the module consists of a microfilter for the concentration of airborne pathogens as well as a LAMP detection zone on a PDMS chip. The microfilter comprised a polycarbonate membrane sandwiched between two pieces of PDMS containing channel features. The module's efficacy was exemplified using Pseudomonas aeruginosa as a representative target.279
Ryu et al. introduced an innovative smartphone-based integrated microsystem designed for the on-site collection and detection of indoor airborne microparticles. The device includes a microfluidic particle trapping chamber and a CMOS photodetector, both efficiently operated using smartphone-based communications. This platform can collect and detect airborne microparticles in less than a minute. The device's capability was demonstrated by differentiating between varying particle densities using representative compounds like Escherichia coli (E. coli), Bacillus subtilis, Micrococcus luteus, and Staphylococcus.280
Despite the progress in airborne pathogen detection using microfluidic devices, there are ongoing areas that need improvement. For instance, many current devices are unable to differentiate between living and deceased pathogens within bioaerosols, leading to inaccuracies in quantification.281 Moreover, many devices contain separate parts for sample collection/concentration and pathogen detection. Integration of the two components is ideal for continuous, automated monitoring, and on-site collection and detection.
Table 7 summarizes the different microfluidic platforms for air monitoring discussed in this section and other notable advancements in the field in the past 5 years.
| Work | Method | Analyte | Device or material | Detection limit |
|---|---|---|---|---|
| Smartphone-based microfluidic colorimetric sensor273 | Colorimetric | Formaldehyde | PDMS with PTFE membrane | 0.01 ppm |
| Membraneless gas-separation microfluidic paper-based device283 | Colorimetric | Hypochlorite | Paper | 6.0 g Cl2 L−1 |
| Microfluidic paper-based analytical device using gold nanoparticles modified with N,N′-bis (2-hydroxyethyl)dithiooxamide284 | Colorimetric | Hg(II) | Paper | 15 nM |
| Graphene oxide nanosheets coupled with paper microfluidics285 | Colorimetric | Fe, Cu, Ni | Paper | 16.6, 5.1, 9.9 ng |
| Paper-based microfluidic device with smartphone-based colorimetric analysis286 | Colorimetric | Fe, Mn, Ni | Paper | 170, 9.2, 81 ng |
| Paper-based microfluidic device for multiplex quantification of metals in PM coupled with smartphone detection287 | Colorimetric | Co, Cu, Fe, Mn, Cr, and Ni | Paper | 8.16, 45.84, 1.86 × 102, 10.08, 1.52 × 102, and 80.40 ng |
| Digital microfluidics288 | Colorimetric | Sulfate, nitrate, and ammonium | Conductive electrodes patterned on an insulating substrate with dielectric layer deposited over electrodes | In droplet: 11 ppm, N/A, 0.26 ppm |
| In air: 0.275 μg m−3, N/A, 6.5 ng m−3 | ||||
| Polymer composite optically integrated lab on chip for detection of ammonia274 | Optical | Ammonia | PDMS – ninhydrin | 2 ppm |
| Gas sensor with embedded cylindrical microfeatures coated with graphene oxide275 | MOS sensor | Methanol, ethanol, propanol, pentanol, hexanal, toluene | VeroClear RGD810 material | N/A |
| Nanoelectrode-activated microwell array277 | Electrochemical | Sclerotinia sclerotiorum | Biopsy punch, salinization | N/A |
| Microfluidic-based olfaction system289 | Metal-oxide semiconductor (MOS sensor) | H2S | PMMA, coatings with metals and polymers | N/A |
| Multiple biosensing system for nerve gases, toxic proteins, and pathogens290 | Electrochemical, LSPR, PCR | Sarin and VX, BTX/A/Hc and ricin, and anthrax | PMMA | N/A |
| Paper microfluidic chip for direct capture and smartphone quantification of SARS-CoV-2 (ref. 278) | Fluorescence | SARS-CoV-2 | Paper | N/A |
| Dye-modified silica–anatase nanoparticles for fluorogenic detection of TATP explosive291 | Fluorescence | Triacetone triperoxide aka: TATP | Eppendorf tube | 93.4 ng L−1 (0.4 nM) |
| Small-volume rotating microfluidic fluorescence chip-integrated sampling system292 | Fluorescence | SARS-CoV-2 | Polytetrafluoroethylene membrane with polycarbonate microchip | 10 copies per μL |
| Gas–liquid microreactor based on the integration of a hydrophobic membrane inside a polymer flat chip293 | Fluorescence | Formaldehyde | PMMA | N/A |
| Coupling of microfluidics and sol–gel chemistry for the detection of toxic gases294 | Fluorescence | Formaldehyde | PDMS, silica–zirconia beads | N/A |
| Array-assisted SERS microfluidic chips276 | SERS | Aldehyde | PDMS | 1 ppb |
| Programmable plasmonic gas microsystem295 | SERS | Formaldehyde, acetaldehyde, acetone, benzaldehyde, cyclohexanone, DNT, indole, benzene, hydrogen sulfide | PDMS | ppb level |
| Microfilter and LAMP chip integrated on a portable device for enrichment and detection296 | LAMP | P. aeruginosa, C. parvum oocysts and G. lamblia cysts | PDMS w polycarbonate membrane | N/A |
| Single photon counting and a real-time spectrometer on microfluidics297 | Single photon counting measurement (SPCM counting) | Microspheres, rice smut spores, aspergillus niger spores, aspergillus charcoal spores | PDMS | N/A |
| Integrated gas–liquid droplet microfluidic platform for airborne targets298 | Droplet-based | Ammonia | PDMS | 500 ppm |
| Hybrid nanophotonic-microfluidic sensor299 | Nanophotonic sensing | DI water and isopropanol | Nanophotonic chip, PDMS with MFCs | 4.46 × 10−6 MRR device, 6.92 × 10−6 MZI device |
| Microfluidic chip to enrich airborne fungal spores directly from gas flow300 | Size-based separation, followed by SEM | Rice false smut spore (crop fungal spore) | PDMS | N/A |
| Smartphone-based microfluidic-biochip system280 | Optoelectronic photodetector | Escherichia coli, Bacillus subtilis, Micrococcus luteus, Staphylococcus | PMMA, PSA layer | 411 CFU mL−1, ∼620, ∼280, ∼400 CFU mL−1 |
6. Agricultural products (crops, fruits, and vegetables)
The ever-changing features of agricultural environments present a challenge when it comes to the in-depth study of fruit crops and vegetables. Fluctuations in nutrient content, texture, water-holding capacity, element mobility, and microbiota affect the growth and development of fruits, crops, and vegetables.301 Therefore, it is essential to monitor these components to ensure effective resource management and optimize agricultural practices.Much like the challenges faced in analyzing soil matrices, one of the challenges in detecting analytes in agri-food systems is laborious sample preparation and the use of heavy instrumentation techniques that can only be used in centralized labs. This presents an issue as the delay caused by transporting samples to labs and the subsequent analysis by professionals may surpass the critical window for farmers to make timely decisions on resource management. Further, there's a risk of crops or plants dying if not promptly treated with the appropriate fertilizers or pesticides. Therefore, real-time monitoring is crucial for analyte detection/monitoring in agri-food systems.
In this context, integration of microfluidics techniques with portable tools such as handheld potentiostats, colorimetric devices paired with smartphone-based color quantification systems, or distance-based detection systems, fluorescence-based microfluidic devices combined with portable fluorometers etc., allows for on-site analysis. These benefits allow farmers to perform real-time analysis, which is crucial for making informed decisions regarding irrigation efficiency, water management, and fertilization.
In the recent half-decade, significant strides have been made in the field of microfluidics to detect different analytes of interest within fruits, crops, and vegetables. Notably, a particular focus has been placed on identifying pesticides in these agricultural products, a concern that is understandable given their increasing use in agriculture. Additionally, considerable efforts have been devoted to identifying other analytes of interest, including heavy metals, bacteria, and the measurement of phenolic compounds found in these products.
Zhao et al. developed a new microfluidic device that uses fluorescent sensors to distinguish between four different types of pesticide residues (carbendazim, diazine, fenvalerate, and pentachloronitrobenzene).302 The device is based on a novel design that combines a microfluidic chip with a spectral recognition system. The results showed that the device could accurately distinguish between all four residues, even at low concentrations. The device could also successfully discriminate between pesticide residues in real cabbage samples.
Yang et al. developed a microfluidic chip integrating enzyme inhibition and a label-free screen-printed carbon electrode (SPCE).303 Three different pesticides were used to test the chip: avermectin, phoxim, and dimethoate. The chip could identify the presence of each pesticide by analyzing the impedance time-sequence spectrum data generated by the enzyme inhibition in 15 minutes. The method's performance was tested in lettuce samples. The detection method showed good stability and specificity, with an identification accuracy of 93%. The method is also less costly and uses less reagents than traditional platforms.
Cao and colleagues developed a system for detecting multiple foodborne bacteria simultaneously using a microfluidic chip with loop-mediated isothermal amplification (LAMP).304 The system utilizes a ten-well microfluidic chip with pre-loaded LAMP primer sets for the quantitative and qualitative detection of Salmonella, Staphylococcus aureus, Escherichia coli O157:H7, and Shigella. The system was successfully applied to detect these bacteria in apples, milk, yogurt, and pork.
Zhang et al. developed a real-time Cd(II) detection system to detect Cd(II) at the sub-femtomolar level in various liquid media.155 The system uses an aptasensor integrated with microfluidic enrichment to improve the sensitivity and specificity of the detection. The system successfully detected Cd(II) in water systems and rice-leaching solutions. Other detected analytes include aflatoxin B1,305 2,4-D,306,307 phenolic compounds,308 and others.309 Besides detecting product analytes, microfluidics has also been used to capture and detect crop airborne disease spores and enrich airborne fungal spores directly from the gas flow.300,310
While on-site quantifications hold immense potential for aiding farmers in agri-food systems, their practical application faces a major limitation that is delaying the scalability. The complex matrices of agricultural products make it difficult to directly integrate microfluidics with them, especially when analyzing contaminant residues. This is because most agricultural products are solid, and microfluidic systems typically require liquid samples. To address this challenge, a sample preparation step is often required to convert solid agricultural products into a liquid form. In recent years, researchers have increasingly recognized the potential of 3D printing platforms to synergize with microfluidics during sample preparation. 3D printing can be used to create custom interfaces that allow agricultural products to be effectively washed and processed within automated microfluidic chips. This streamlining of the entire detection process addresses the difficulty of directly incorporating solid samples into microfluidics. Many current microfluidic systems that use enzymatic inhibition assays require incubation steps, which are impractical for field settings. Portable incubators may be a good option for small-scale projects that need to be moved frequently, while self-heating incubators may be a better option for resource-constrained field settings.
Table 8 provides a comprehensive overview of microfluidic sensors for crops, fruits, and vegetables, including their device design, analytical performance, and applications.
| Work | Product | Method | Analyte | Device or material | Detection limit |
|---|---|---|---|---|---|
| A laser-induced fluorescent detector for pesticide residue302 | Cabbage | Fluorescence | Carbendazim, diazine, fenvalerate and pentachloronitrobenzene | PDMS and glass | <10 ppb |
| Deposition of CdTe quantum dots on microfluidic paper for detection of 2,4-D307 | Bean sprouts | Fluorescence | 2,4-D | Paper | 90 nM |
| uPAD for acetylcholinesterase inhibition assay utilizing organic solvent extraction311 | Head lettuce | Colorimetric | Chlorpyrifos, phoxim, carbaryl, triazophos, carbofuran, and methamidophos | Paper | 0.77, 0.39, 0.25, 1.29, 0.006 and 1.39 ppm |
| Pesticide identification by impedance time-sequence spectrum of enzyme inhibition on multilayer uPAD303 | Lettuce | Electrochemical | Avermectin, phoxim, dimethoate | Paper | NA |
| Printed low-cost microfluidic uPADs for quantitative detection of vitamin C in fruits309 | Pineapples, oranges, guavas, and apples | Colorimetric | Vitamin C | Paper | NA |
| Pump-free microfluidic rapid mixer combined with a paper-based channel312 | Apples, cucumbers, and tomatoes | Colorimetric | Malathion | Paper | 10 nmol L−1 (LoQ) |
| CRISPR-Cas12a based fluorescence assay for organophosphorus pesticides313 | Citrus fruits and cabbage | Fluorescence | Paraoxon, dichlorvos, and demeton | MnO2 Nanosheets | 270, 406, and 218 pg mL−1 |
| C3N4 nanosheets-based oxidase-like 2D fluorescence nanozyme for dual-mode detection of organophosphorus pesticides314 | Green tea, red tea, apple, orange, and cabbage | Fluorescence | Trichlorfon | 2D g-C3N4 nanosheets | 0.083 ng mL−1 |
| Multi-channel fluorescent paper-based microfluidic chip315 | Apple and lettuce | Fluorescence | Pb(II), Hg(II), Cd(II), and As(II) | Paper | 4.20 nM, 1.70 nM, 2.04 nM, and 1.65 nM |
| Visual detection of mixed organophosphorus pesticide using QD-AChE aerogel based microfluidic arrays sensor316 | Apple | Fluorescence | Paraoxon, parathion, dichlorvos and deltamethrin | Aerogel | 0.4 ng mL−1 |
| 3D uPAD-based ratiometric fluorescent sensor317 | Spinach and tomato | Fluorescence | Dichlorvos | Paper | 1.0 μg L−1 |
| Light-shading reaction microfluidic PMMA/paper detection system for detection of cyclamate318 | 10 different foods from Taiwan | Colorimetric | Cyclamate | PMMA/paper | 20 ppm |
| A versatile microfluidic paper chip platform based on MIPs for rapid ratiometric sensing of dual fluorescence signals319 | Cucumber | Fluorescence | 2,4-D | Paper | 0.17 μmol L−1 |
| Fluorescence detection of 2,4-D by ratiometric fluorescence imaging uPAD306 | Soybean sprout | Fluorescence | 2,4-D | Paper | 90 nM |
| A dual-channel and dual-signal microfluidic paper chip320 | Cucumber | Fluorescence | Difenoconazole and mancozeb | Paper | 0.18 and 2 μg mL−1 |
| Real-time Cd2+ detection at sub-femtomolar level by an aptasensor integrated with microfluidic enrichment155 | Rice leaching solution | Capacitive sensing | Cd(II) | Gold IDE | 253.16 aM |
| Nanocolloidal Mn(IV) in a chemiluminescence system for estimating the total phenolic content308 | Pomegranate juices | Chemiluminescence | 8 phenolic compounds | Nanocolloidal MnO2 | 1.3–6.5 ng mL−1 |
| Simultaneous detection of multiple foodborne bacteria by loop-mediated isothermal amplification304 | Apple and pork | Colorimetric and fluorescent | Salmonella, Staphylococcus aureus, Escherichia coli O157:H7, and Shigella | Polycarbonate | 8 × 103 CFU mL−1 |
| Quantum dots-based immunoassay in a recyclable gravity-driven microfluidic chip305 | Corn, rice and milk | Fluorescence | Aflatoxin B1 | PDMS | 0.06 ng mL−1 |
7. Challenges
7.1 Considerations for different detection techniques
When using microfluidic platforms for environmental analysis, researchers must consider a range of factors impacting the reliability and performance of different detection techniques (i.e., fluorescence, electrochemical, colorimetric). For example, in terms of sensitivity and achieving lower detection limits, fluorescence and electrochemical can be more sensitive techniques, followed by UV-vis and colorimetric. Whereas in terms of practicality, colorimetric can be the most user-friendly technique.Fluorescence-based techniques can be susceptible to various issues. For instance, fluorescent molecules are also often unstable over time and in different assay conditions making it challenging to implement in different temperature and humidity conditions inside the microfluidics. The stability of fluorescent molecules can be increased by using stabilizing buffers or chelating agents. Commonly employed materials in microfluidics, like PDMS and glass possess intrinsic fluorescence, making it difficult to distinguish between the blank and sample signals. One way of mitigating the self-fluorescence of materials used is to work with alternate substrates like quartz or acrylic that are not fluorescent or coat the material with a non-fluorescent substance.
Researchers using microfluidic electrochemical detection for environmental analysis also encounter challenges, including limited mass transport, electrode fouling, limited dynamic range, and inconsistencies of performance for disposable electrodes. Moreover, portable potentiostats are typically less accurate, have a smaller voltage range than benchtop potentiostats, and have increased noise levels. The lower accuracy and voltage range of portable potentiostats can make them less suitable for applications that require high precision or a wide range of voltages.
Employing smaller channels and higher flow rates can allow analyte molecules enough time to interact with electrode surfaces to overcome limited mass transport. Electrode fouling can be reduced to a minimum by using foul-resistant materials, such as gold or platinum, or by using coatings that inhibit analyte adherence. In some cases, the limited dynamic range problem could be solved by increasing electrode surface area to extend the observable concentration range.
UV-vis spectroscopy is widely employed for its high sensitivity and ease of detecting contaminants in water.321–323 However, when applied in microfluidic chips, it can suffer from decreased sensitivity due to the shorter optical path length inherent in the miniaturized format.324 Various strategies have been explored to address this challenge, such as incorporating mirrors and modifying channel geometries.325 Another promising approach involves the utilization of LEDs in combination with optical fibers.326,327 LED-based sensors offer the advantage of emitting narrow wavelength bands, negating the need for monochromators. Additionally, these sensors exhibit remarkable resilience to harsh environmental conditions.328
Colorimetric microfluidic sensors encounter sensitivity, integration, sample matrix effects, selectivity, and quantitative analysis challenges. Additionally, environmental robustness, reagent stability, miniaturization constraints, and sensor readout influence their suitability for environmental monitoring. Although many colorimetric platforms have been coupled with smartphone-enabled techniques in the last five years, there is a lack of well-established imaging/color quantification applications available in app stores. To promote further advancements in the field, researchers are encouraged to openly share the codes of their sensing and quantification platforms, facilitating future improvements and innovations.
7.2 Commercialization
While commercialization is often the end goal for microfluidic devices, factors such as scalability, cost-effectiveness, and regulatory considerations introduce challenges that must be addressed. For instance, most scalability concerns arise because there is a need to bridge the gap between proof-of-concept and real-world suitability. This gap is evident in various sensing platforms where researchers often neglect testing the sensors in real-world environmental matrices. It is crucial to acknowledge that the performance/applicability of microfluidic chips tested in controlled lab conditions (i.e. Millipore water/controlled chambers/controlled soil conditions) may not always translate directly to real-world applications.Fabrication costs can also be a real issue. Many Lab-on-a-Chip (LOC) devices rely on external components such as pumps, power sources, on-site calibration tools (microscopes, spectrometers, smartphones), and specialized computer software for data analysis. These additional requirements lower the practicality of miniaturization and contribute to higher costs, deterring widespread adoption. Moreover, environmental assays frequently require off-chip sample pretreatment, adding to the complexity of the device and deterring end-users.
Despite the increasing integration of microfluidics in environmental devices, there is a lack of specific EPA/WHO-recognized standards for microfluidic-based environmental monitoring, which poses a significant obstacle to device development and approval.329 Standardized regulatory guidance documents regarding performance, sample integrity, interference mitigation, and calibration procedures are necessary to ensure accuracy, reliability, and user safety in environmental monitoring applications. In 2022, the Food and Drug Administration's Center for Devices and Radiological Health took a significant step forward in regulating microfluidics-based medical devices by developing a new leakage testing tool. This marks a promising advancement towards establishing standards for the commercialization of microfluidics. However, for comprehensive guidelines, it is crucial that environmental regulatory organizations also actively participate in formulating these guidelines.
To pave the way for widespread commercialization, certain considerations must be prioritized during microfluidic sensor development:330
Overcoming these challenges and emphasizing the considerations listed above are pivotal steps toward realizing the widespread commercialization of microfluidic devices in environmental monitoring.
8. Future perspectives
Despite commercialization problems, the use of microfluidics in academia, business, and the public has expanded thanks to significant improvements in manufacturing methods. Traditional techniques like replica molding with PDMS and wax printing are still valid for research and prototyping. Although commercial wax printers have been discontinued, the utilization of paper-based microfluidics continues to expand due to the emergence of viable alternatives such as thermal transfer printing, laser cutting, screen printing, photolithography, and laminate capillary-driven microfluidics. Lamination-based microfluidics employ layered paper, film, acrylics, and/or glass slides to create robust channels with enhanced performance, eliminating uneven flow and resistance in porous paper microchannels. This approach holds a promising future for improved microfluidic systems. While there are still certain obstacles to commercialization, 3D printing appears promising. Due to 3D printing's intrinsic versatility, it is possible to design microfluidic devices with various geometries and features suited to meet certain applications' needs. The method is notably precise, producing microfluidic devices up to submicron channels. Furthermore, the continual decrease in printer costs has made 3D printing more cost-effective, making it an attractive option for the low-cost prototyping and mass manufacturing of microfluidic devices.The future is promising for advancing microfluidic platforms, particularly in monitoring pesticides, nutrients, and toxins in water and soil matrices. Realizing these possibilities relies on ongoing research, especially in enabling on-site testing and monitoring capabilities. One practical approach would be combining colorimetric tests with advanced technologies, creating user-friendly systems for real-time monitoring. More integration of Internet of Things (IoT) platforms could also transform monitoring efforts.333 This integration would allow volunteers to collect samples and monitor environments, reducing the need for frequent scientist visits.
Another crucial aspect of this progress is smoothly integrating microfluidic devices with sample preparation and detection systems. This combination could speed testing, reduce processing time, and improve safeguards against contamination. While current microfluidic tests often focus on individual analytes or multiplexing for one class of analytes at a time, we must widen our approach and consider various analytes in a single run. This is especially significant in active environmental monitoring, where detecting multiple analytes simultaneously could bring substantial economic and resource benefits.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was funded through a contract (W911QY2120003) awarded by the US Army DEVCOM Center, and the National Science Foundation (Fellow ID: 2022343709) providing financial support for the research.References
- X. Xu, S. Nie, H. Ding and F. F. Hou, Nat. Rev. Nephrol., 2018, 14, 313–324 CrossRef CAS PubMed.
- A. Prüss-Üstün and C. Corvalán, World Health Organization, 2006 Search PubMed.
- A. Laborde, F. Tomasina, F. Bianchi, M.-N. Bruné, I. Buka, P. Comba, L. Corra, L. Cori, C. M. Duffert and R. Harari, Environ. Health Perspect., 2015, 123, 201–209 CrossRef PubMed.
- J. Broekaert, TrAC, Trends Anal. Chem., 1982, 1, 249–253 CrossRef CAS.
- S. Mukherjee, S. Bhattacharyya, K. Ghosh, S. Pal, A. Halder, M. Naseri, M. Mohammadniaei, S. Sarkar, A. Ghosh and Y. Sun, Trends Food Sci. Technol., 2021, 109, 674–689 CrossRef CAS.
- A. Sekar, R. Yadav and N. Basavaraj, New J. Chem., 2021, 45, 2326–2360 RSC.
- J. Murphy and J. P. Riley, Anal. Chim. Acta, 1962, 27, 31–36 CrossRef CAS.
- D. Barceló, E. Eljarrat and M. Petrovic, in Encyclopedia of Analytical Science, ed. P. Worsfold, A. Townshend and C. Poole, Elsevier, 2005, pp. 468–475 Search PubMed.
- A. B. Kanu, J. Chromatogr. A, 2021, 1654, 462444 CrossRef CAS PubMed.
- B. C. Dhar and N. Y. Lee, BioChip J., 2018, 12, 173–183 CrossRef CAS.
- J. C. Jokerst, J. M. Emory and C. S. Henry, Analyst, 2012, 137, 24–34 RSC.
- D. M. Cate, J. A. Adkins, J. Mettakoonpitak and C. S. Henry, Anal. Chem., 2015, 87, 19–41 CrossRef CAS PubMed.
- E. Noviana, T. Ozer, C. S. Carrell, J. S. Link, C. McMahon, I. Jang and C. S. Henry, Chem. Rev., 2021, 121, 11835–11885 CrossRef CAS PubMed.
- P. Aryal, E. Brack, T. Alexander and C. S. Henry, Anal. Chem., 2023, 95, 5820–5827 CrossRef CAS PubMed.
- J.-W. Shangguan, Y. Liu, J.-B. Pan, B.-Y. Xu, J.-J. Xu and H.-Y. Chen, Lab Chip, 2017, 17, 120–127 RSC.
- Y. Lu, B. Lin and J. Qin, Anal. Chem., 2011, 83, 1830–1835 CrossRef CAS PubMed.
- R. B. Channon, M. P. Nguyen, A. G. Scorzelli, E. M. Henry, J. Volckens, D. S. Dandy and C. S. Henry, Lab Chip, 2018, 18, 793–802 RSC.
- X. Zhu, K. Wang, H. Yan, C. Liu, X. Zhu and B. Chen, Environ. Sci. Technol., 2022, 56, 711–731 CrossRef CAS PubMed.
- Y. Cheng, R. M. H. Yang, F. M. Alejandro, F. Li, S. K. Balavandy, L. Wang, M. Breadmore, R. Doyle and R. Naidu, in Smartphone-Based Detection Devices, 2021, pp. 103–128, DOI:10.1016/b978-0-12-823696-3.00010-6.
- M. Ueland, L. Blanes, R. V. Taudte, B. H. Stuart, N. Cole, P. Willis, C. Roux and P. Doble, J. Chromatogr. A, 2016, 1436, 28–33 CrossRef CAS PubMed.
- C. E. Stanley, G. Grossmann, X. C. I. Solvas and A. J. de Mello, Lab Chip, 2016, 16, 228–241 RSC.
- P. K. Rai, M. Islam and A. Gupta, Sens. Actuators, A, 2022, 347, 113926 CrossRef CAS.
- Z. Kotsiri, J. Vidic and A. Vantarakis, J. Environ. Sci., 2022, 111, 367–379 CrossRef PubMed.
- M. E. Barocio, E. Hidalgo-Vázquez, Y. Kim, L. I. Rodas-Zuluaga, W.-N. Chen, D. Barceló, H. N. M. Iqbal, R. Parra-Saldívar and C. Castillo-Zacarías, Case Stud. Chem. Environ. Eng., 2021, 3, 100069 CrossRef CAS.
- F. Pena-Pereira, I. Lavilla, I. de la Calle, V. Romero and C. Bendicho, Anal. Bioanal. Chem., 2023, 415, 4039–4060 CrossRef CAS PubMed.
- M. Khatib and H. Haick, ACS Nano, 2022, 16, 7080–7115 CrossRef CAS PubMed.
- S. Kaaliveetil, J. Yang, S. Alssaidy, Z. Li, Y. H. Cheng, N. H. Menon, C. Chande and S. Basuray, Micromachines, 2022, 13, 1716 CrossRef PubMed.
- S. Ezrre, M. A. Reyna, C. Anguiano, R. L. Avitia and H. Marquez, Biosensors, 2022, 12, 191 CrossRef CAS PubMed.
- F. Schulze, X. Gao, D. Virzonis, S. Damiati, M. R. Schneider and R. Kodzius, Genes, 2017, 8, 244 CrossRef PubMed.
- C.-T. Kung, C.-Y. Hou, Y.-N. Wang and L.-M. Fu, Sens. Actuators, B, 2019, 301, 126855 CrossRef CAS.
- P. K. Rai, M. Islam and A. Gupta, Sens. Actuators, A, 2022, 113926 CrossRef CAS.
- C.-T. Kung, C.-Y. Hou, Y.-N. Wang and L.-M. Fu, Sens. Actuators, B, 2019, 301, 126855 CrossRef CAS.
- D. Zhang, C. Li, D. Ji and Y. Wang, Crit. Rev. Anal. Chem., 2022, 52, 1432–1449 CrossRef CAS PubMed.
- Y. Yang, E. Noviana, M. P. Nguyen, B. J. Geiss, D. S. Dandy and C. S. Henry, Anal. Chem., 2017, 89, 71–91 CrossRef CAS PubMed.
- S. Das, Gagandeep and R. Bhatia, Rev. Anal. Chem., 2022, 41, 112–136 CrossRef.
- K. Kant, M. A. Shahbazi, V. P. Dave, T. A. Ngo, V. A. Chidambara, L. Q. Than, D. D. Bang and A. Wolff, Biotechnol. Adv., 2018, 36, 1003–1024 CrossRef CAS PubMed.
- S. Neethirajan, I. Kobayashi, M. Nakajima, D. Wu, S. Nandagopal and F. Lin, Lab Chip, 2011, 11, 1574–1586 RSC.
- G. Kim, J. Lim and C. Mo, J. Biosyst. Eng., 2016, 41, 116–125 CrossRef.
- S. Li, C. Zhang, S. Wang, Q. Liu, H. Feng, X. Ma and J. Guo, Analyst, 2018, 143, 4230–4246 RSC.
- M. Pohanka and P. Skládal, J. Appl. Biomed., 2008, 6, 57–64 CrossRef CAS.
- G. Zhao, H. Wang and G. Liu, Int. J. Electrochem. Sci., 2017, 12, 8622–8641 CrossRef CAS.
- Y. Lin, D. Gritsenko, S. Feng, Y. C. Teh, X. Lu and J. Xu, Biosens. Bioelectron., 2016, 83, 256–266 CrossRef CAS PubMed.
- J. Baranwal, B. Barse, G. Gatto, G. Broncova and A. Kumar, Chemosensors, 2022, 10, 363 CrossRef CAS.
- S. Ma, W. Zhao, Q. Zhang, K. Zhang, C. Liang, D. Wang, X. Liu and X. Zhan, J. Hazard. Mater., 2023, 448, 130923 CrossRef CAS PubMed.
- M. Zhang, X. Zhang, P. Niu, T. Shen, Y. Yuan, Y. Bai and Z. Wang, Nanotechnol. Precis. Eng., 2021, 4, 013005 CrossRef CAS.
- T. O. Hara and B. Singh, ACS ES&T Water, 2021, 1, 462–478 Search PubMed.
- J. Lian, Q. Xu, Y. Wang and F. Meng, Front. Chem., 2020, 8, 593291 CrossRef CAS PubMed.
- D.-E. Zacharioudaki, I. Fitilis and M. Kotti, Molecules, 2022, 27, 4801 CrossRef CAS PubMed.
- N. De Acha, C. Elosúa, J. M. Corres and F. J. Arregui, Sensors, 2019, 19, 599 CrossRef PubMed.
- Q. Shang, P. Zhang, H. Li, R. Liu and C. Zhang, J. Innovative Opt. Health Sci., 2019, 12, 1950016 CrossRef CAS.
- P. Inpota, D. Nacapricha, P. Sunintaboon, W. Sripumkhai, W. Jeamsaksiri, P. Wilairat and R. Chantiwas, Talanta, 2018, 188, 606–613 CrossRef CAS PubMed.
- H. Shi, S. Jiang, B. Liu, Z. Liu and N. M. Reis, Microchem. J., 2021, 171, 106845 CrossRef CAS.
- G. G. Morbioli, T. Mazzu-Nascimento, A. M. Stockton and E. Carrilho, Anal. Chim. Acta, 2017, 970, 1–22 CrossRef CAS PubMed.
- A. Saifi, R. K. Chaudhary and S. Srivastava, Sens. Lett., 2019, 17, 25–40 CrossRef.
- W. Cui, Z. Ren, Y. Song and C. L. Ren, Sens. Actuators, A, 2022, 113733 CrossRef CAS.
- N. Mohamad Nor, N. H. Ramli, H. Poobalan, K. Qi Tan and K. Abdul Razak, Crit. Rev. Anal. Chem., 2023, 53, 253–288 CrossRef CAS PubMed.
- O. Oktaviani, Jurnal Latihan, 2021, 1, 11–20 Search PubMed.
- E. Priyadarshini and N. Pradhan, Sens. Actuators, B, 2017, 238, 888–902 CrossRef CAS.
- R. Sharma, Mater. Today: Proc., 2023 DOI:10.1016/j.matpr.2023.03.462.
- UNICEF, Geneva: WHO News Release, 2019.
- WHO, World Health Organisation Geneva, 2019.
- P. Weerasekara, Future Food: J. Food Agric. Soc., 2017, 5, 80–81 Search PubMed.
- F. N. Chaudhry and M. Malik, J. Ecosyst. Ecography, 2017, 7, 225–231 Search PubMed.
- J. Saez, R. Catalan-Carrio, R. M. Owens, L. Basabe-Desmonts and F. Benito-Lopez, Anal. Chim. Acta, 2021, 1186, 338392 CrossRef CAS PubMed.
- J. Atencia and D. J. Beebe, Nature, 2005, 437, 648–655 CrossRef CAS PubMed.
- I. E. Araci and P. Brisk, Curr. Opin. Biotechnol., 2014, 25, 60–68 CrossRef CAS PubMed.
- Z. Li, H. Liu, D. Wang, M. Zhang, Y. Yang and T.-L. Ren, TrAC, Trends Anal. Chem., 2022, 116790 Search PubMed.
- M. Al Osman, F. Yang and I. Y. Massey, BioMetals, 2019, 32, 563–573 CrossRef CAS PubMed.
- F. W. Sunderman Jr, Fed. Proc., 1978, 37, 40–46 CAS.
- P. B. Tchounwou, C. G. Yedjou, A. K. Patlolla and D. J. Sutton, Exper. Suppl., 2012, 101, 133–164 Search PubMed.
- M. Balali-Mood, K. Naseri, Z. Tahergorabi, M. R. Khazdair and M. Sadeghi, Front. Pharmacol., 2021, 12, 1–19 Search PubMed.
- Q. Zhang and C. Wang, Water, Air, Soil Pollut., 2020, 231, 1–13 CrossRef.
- E. T. W. Nachana'a Timothy, Chemistry, 2019, 3, 72–82 Search PubMed.
- Z. Li, Z. Ma, T. J. van der Kuijp, Z. Yuan and L. Huang, Sci. Total Environ., 2014, 468, 843–853 CrossRef PubMed.
- C. Zamora-Ledezma, D. Negrete-Bolagay, F. Figueroa, E. Zamora-Ledezma, M. Ni, F. Alexis and V. H. Guerrero, Environ. Technol. Innovation, 2021, 22, 101504 CrossRef CAS.
- N. Verma and M. Singh, BioMetals, 2005, 18, 121–129 CrossRef CAS PubMed.
- M. Jin, H. Yuan, B. Liu, J. Peng, L. Xu and D. Yang, Anal. Methods, 2020, 12, 5747–5766 RSC.
- Usepa, personal communication.
- C. Directive, Off. J. Eur. Communities, 1998, 330, 32–54 Search PubMed.
- WHO, 2022.
- K. Das, S. Das and S. Dhundasi, Indian J. Med. Res., 2008, 128, 412–425 CAS.
- A. Cavani, Toxicology, 2005, 209, 119–121 CrossRef CAS PubMed.
- H. Kitaura, N. Nakao, N. Yoshida and T. Yamada, New Microbiol., 2003, 26, 101–108 CAS.
- P. L. Drake and K. J. Hazelwood, Ann. Occup. Hyg., 2005, 49, 575–585 CAS.
- L. P. Padhye, T. Jasemizad, S. Bolan, O. V. Tsyusko, J. M. Unrine, B. K. Biswal, R. Balasubramanian, Y. Zhang, T. Zhang and J. Zhao, Sci. Total Environ., 2023, 871, 161926 CrossRef CAS PubMed.
- H. T. Ratte, Environ. Toxicol. Chem., 1999, 18, 89–108 CrossRef CAS.
- G. J. Fosmire, Am. J. Clin. Nutr., 1990, 51, 225–227 CrossRef CAS PubMed.
- C. Xu, Y. Zhang, N. Zhang, X. Liu, J. Yi, X. Liu, X. Lu, Q. Ru, H. Lu and X. Peng, Chem. – Asian J., 2020, 15, 3696–3708 CrossRef CAS PubMed.
- H. Satoh, Ind. Health, 2000, 38, 153–164 CrossRef CAS PubMed.
- J. F. Risher and S. N. Amler, Neurotoxicology, 2005, 26, 691–699 CrossRef CAS PubMed.
- L. T. Budnik and L. Casteleyn, Sci. Total Environ., 2019, 654, 720–734 CrossRef CAS PubMed.
- M. C. Houston, J. Clin. Hypertens., 2011, 13, 621–627 CrossRef CAS PubMed.
- A. Ara and J. A. Usmani, Interdiscip. Toxicol., 2015, 8, 55–64 CrossRef PubMed.
- Y. Fan, Y. Jiang, P. Hu, R. Chang, S. Yao, B. Wang, X. Li, Q. Zhou and Y. Qiao, J. Exposure Sci. Environ. Epidemiol., 2016, 26, 464–470 CrossRef CAS PubMed.
- R. A. Goyer, Environ. Health Perspect., 1993, 100, 177–187 CrossRef CAS PubMed.
- G. Lockitch, Clin. Biochem., 1993, 26, 371–381 CrossRef CAS PubMed.
- S. Selvaraj, S. Krishnaswamy, V. Devashya, S. Sethuraman and U. M. Krishnan, Langmuir, 2011, 27, 13374–13382 CrossRef CAS PubMed.
- C. Flemming and J. Trevors, Water, Air, Soil Pollut., 1989, 44, 143–158 CrossRef CAS.
- M. L. Schilsky, Semin. Liver Dis., 1996, 16, 83–95 CrossRef CAS PubMed.
- A. R. G. Georgopoulos, M. J. Yonone-Lioy, R. E. Opiekun and P. J. Lioy, J. Toxicol. Environ. Health, Part B, 2001, 4, 341–394 Search PubMed.
- M. Dotaniya, J. Thakur, V. Meena, D. Jajoria and G. Rathor, Agric. Rev., 2014, 35, 153–157 CrossRef.
- M. Mohanty and H. K. Patra, Rev. Environ. Contam. Toxicol., 2011, 210, 1–34 CAS.
- S. Chowdhury, M. J. Mazumder, O. Al-Attas and T. Husain, Sci. Total Environ., 2016, 569, 476–488 CrossRef PubMed.
- R. Saha, R. Nandi and B. Saha, J. Coord. Chem., 2011, 64, 1782–1806 CrossRef CAS.
- G. Genchi, M. S. Sinicropi, G. Lauria, A. Carocci and A. Catalano, Int. J. Environ. Res. Public Health, 2020, 17, 3782 CrossRef CAS PubMed.
- J.-M. Moulis and F. Thévenod, Occup. Environ. Med., 2010, 23, 763–768 CAS.
- Q. Mahmood, M. Asif, S. Shaheen, M. T. Hayat and S. Ali, in Cadmium toxicity and tolerance in plants, Elsevier, 2019, pp. 141–161 Search PubMed.
- M. Hutton, Ecotoxicol. Environ. Saf., 1983, 7, 9–24 CrossRef CAS PubMed.
- M. F. Hughes, Toxicol. Lett., 2002, 133, 1–16 CrossRef CAS PubMed.
- J. Nriagu, P. Bhattacharya, A. Mukherjee, J. Bundschuh, R. Zevenhoven and R. Loeppert, Trace Met. Other Contam. Environ., 2007, 9, 3–60 CAS.
- S. Shankar and U. Shanker, Sci. World J., 2014, 2014, 304524 Search PubMed.
- A. Lace, D. Ryan, M. Bowkett and J. Cleary, Anal. Methods, 2019, 11, 5431–5438 RSC.
- A. Lace, D. Ryan, M. Bowkett and J. Cleary, Int. J. Environ. Res. Public Health, 2019, 16, 1803 CrossRef CAS PubMed.
- G. Wang, J. Li, S. Wu, T. Jiang and T.-H. Chen, Anal. Chem., 2022, 94, 15925–15929 CrossRef CAS PubMed.
- P. Borthakur, P. K. Boruah and M. R. Das, ACS Sustainable Chem. Eng., 2021, 9, 13245–13255 CrossRef CAS.
- H. Sharifi, J. Tashkhourian and B. Hemmateenejad, Anal. Chim. Acta, 2020, 1126, 114–123 CrossRef CAS PubMed.
- F. Li, Y. Hu, Z. Li, J. Liu, L. Guo and J. He, Anal. Bioanal. Chem., 2019, 411, 6497–6508 CrossRef CAS PubMed.
- M. F. Santangelo, I. Shtepliuk, D. Filippini, D. Puglisi, M. Vagin, R. Yakimova and J. Eriksson, Sensors, 2019, 19, 2393 CrossRef CAS PubMed.
- P. Gimenez-Gomez, A. Baldi, C. Ayora and C. Fernandez-Sanchez, ACS Sens., 2019, 4, 3156–3165 CrossRef CAS PubMed.
- S. Bai, D. Serien, A. Hu and K. Sugioka, Adv. Funct. Mater., 2018, 28, 1706262 CrossRef.
- W.-H. Huang, V.-P. Mai, R.-Y. Wu, K.-L. Yeh and R.-J. Yang, Micromachines, 2021, 12, 1283 CrossRef PubMed.
- P. Kamnoet, W. Aeungmaitrepirom, R. F. Menger and C. S. Henry, Analyst, 2021, 146, 2229–2239 RSC.
- J. P. Devadhasan and J. Kim, Sens. Actuators, B, 2018, 273, 18–24 CrossRef CAS.
- A. Motalebizadeh, H. Bagheri, S. Asiaei, N. Fekrat and A. Afkhami, RSC Adv., 2018, 8, 27091–27100 RSC.
- P. Abdollahiyan, M. Hasanzadeh, F. Seidi and P. Pashazadeh-Panahi, J. Environ. Chem. Eng., 2021, 9, 106197 CrossRef CAS.
- J. Mettakoonpitak, K. Khongsoun, N. Wongwan, S. Kaewbutdee, A. Siripinyanond, A. Kuharuk and C. S. Henry, Sens. Actuators, B, 2021, 331, 129463 CrossRef CAS.
- N. Idros and D. Chu, ACS Sens., 2018, 3, 1756–1764 CrossRef CAS PubMed.
- H. Tabani, F. Dorabadi Zare, W. Alahmad and P. Varanusupakul, Environ. Chem. Lett., 2020, 18, 187–196 CrossRef CAS.
- K. R. Chabaud, J. L. Thomas, M. N. Torres, S. Oliveira and B. R. McCord, Forensic Chem., 2018, 9, 35–41 CrossRef CAS.
- W. Alahmad, N. Tungkijanansin, T. Kaneta and P. Varanusupakul, Talanta, 2018, 190, 78–84 CrossRef CAS PubMed.
- Z. Wu, J. Wang, C. Bian, J. Tong and S. Xia, Micromachines, 2020, 11, 63 CrossRef PubMed.
- H. A. Silva-Neto, T. M. Cardoso, C. J. McMahon, L. F. Sgobbi, C. S. Henry and W. K. Coltro, Analyst, 2021, 146, 3463–3473 RSC.
- V. Subramanian, S. Lee, S. Jena, S. K. Jana, D. Ray, S. J. Kim and P. Thalappil, Sens. Actuators, B, 2020, 304, 127340 CrossRef CAS.
- J. Dai, W. Gao, J. Yin, L. Liang, J. Zou and Q. Jin, Anal. Chim. Acta, 2021, 1164, 338511 CrossRef CAS PubMed.
- P. Madzivhandila, S. Smith, L. Ntuli and K. Land, Fifth Conference on Sensors, MEMS, and Electro-Optic Systems, 2019, p. 11043 Search PubMed.
- J. M. Mohan, K. Amreen, A. Javed, S. K. Dubey and S. Goel, IEEE Trans. NanoBiosci., 2021, 21, 117–124 Search PubMed.
- R. Ding and G. Lisak, Anal. Chim. Acta, 2019, 1091, 103–111 CrossRef CAS PubMed.
- R. Ding, Y. H. Cheong, K. Zhao and G. Lisak, Sens. Actuators, B, 2021, 347, 130567 CrossRef CAS.
- R. Silva, A. Ahamed, Y. H. Cheong, K. Zhao, R. Ding and G. Lisak, Anal. Chim. Acta, 2022, 1197, 339495 CrossRef CAS PubMed.
- J. Zhou, B. Li, A. Qi, Y. Shi, J. Qi, H. Xu and L. Chen, Sens. Actuators, B, 2020, 305, 127462 CrossRef CAS.
- M. Wang, Z. Song, Y. Jiang, X. Zhang, L. Wang, H. Zhao, Y. Cui, F. Gu, Y. Wang and G. Zheng, Anal. Bioanal. Chem., 2021, 413, 3299–3313 CrossRef CAS PubMed.
- Q. Zhang, X. Zhang, X. Zhang, L. Jiang, J. Yin, P. Zhang, S. Han, Y. Wang and G. Zheng, Mar. Pollut. Bull., 2019, 144, 20–27 CrossRef CAS PubMed.
- K. Patir and S. K. Gogoi, Nanoscale Adv., 2019, 1, 592–601 RSC.
- H. Sun, W. Li, Z.-Z. Dong, C. Hu, C.-H. Leung, D.-L. Ma and K. Ren, Biosens. Bioelectron., 2018, 99, 361–367 CrossRef CAS PubMed.
- S. Yan, F. Chu, H. Zhang, Y. Yuan, Y. Huang, A. Liu, S. Wang, W. Li, S. Li and W. Wen, Spectrochim. Acta, Part A, 2019, 220, 117113 CrossRef CAS PubMed.
- G. Wang, L. T. Chu, H. Hartanto, W. B. Utomo, R. A. Pravasta and T.-H. Chen, ACS Sens., 2019, 5, 19–23 CrossRef PubMed.
- C. Wu, G. Gao, K. Zhai, L. Xu and D. Zhang, Food Chem., 2020, 331, 127208 CrossRef CAS PubMed.
- Y. Zhang, Y.-L. Li, S.-H. Cui, C.-Y. Wen, P. Li, J.-F. Yu, S.-M. Tang and J.-B. Zeng, J. Anal. Test., 2021, 5, 11–18 CrossRef.
- J. C. Hofstetter, J. B. Wydallis, G. Neymark, T. H. Reilly III, J. Harrington and C. S. Henry, Analyst, 2018, 143, 3085–3090 RSC.
- Y. Liang, M. Ma, F. Zhang, F. Liu, T. Lu, Z. Liu and Y. Li, ACS Omega, 2021, 6, 9302–9309 CrossRef CAS PubMed.
- N. Mishra, A. Dhwaj, D. Verma and A. Prabhakar, Anal. Chim. Acta, 2022, 1205, 339734 CrossRef CAS PubMed.
- M. Santangelo, I. Shtepliuk, D. Filippini, I. G. Ivanov, R. Yakimova and J. Eriksson, Sens. Actuators, B, 2019, 288, 425–431 CrossRef CAS.
- S. Han, Q. Zhang, X. Zhang, X. Liu, L. Lu, J. Wei, Y. Li, Y. Wang and G. Zheng, Biosens. Bioelectron., 2019, 143, 111597 CrossRef CAS PubMed.
- D. Rotake, A. Darji and N. Kale, in Handbook of Nanomaterials for Sensing Applications, Elsevier, 2021, pp. 139–155 Search PubMed.
- J. Zhang, Y. Zhang, J. Wu, H. Qi, M. Zhao, M. Yi, Z. Li and L. Zheng, Sens. Actuators, B, 2021, 329, 129282 CrossRef CAS.
- A. M. Michalak, E. J. Anderson, D. Beletsky, S. Boland, N. S. Bosch, T. B. Bridgeman, J. D. Chaffin, K. Cho, R. Confesor, I. Daloglu, J. V. Depinto, M. A. Evans, G. L. Fahnenstiel, L. He, J. C. Ho, L. Jenkins, T. H. Johengen, K. C. Kuo, E. Laporte, X. Liu, M. R. McWilliams, M. R. Moore, D. J. Posselt, R. P. Richards, D. Scavia, A. L. Steiner, E. Verhamme, D. M. Wright and M. A. Zagorski, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 6448–6452 CrossRef CAS PubMed.
- Z. Li, H. Liu, D. Wang, M. Zhang, Y. Yang and T.-L. Ren, TrAC, Trends Anal. Chem., 2023, 158, 116790 CrossRef CAS.
- R. Catalan-Carrio, J. Saez, L. A. Fernandez Cuadrado, G. Arana, L. Basabe-Desmonts and F. Benito-Lopez, Anal. Chim. Acta, 2022, 1205, 339753 CrossRef CAS PubMed.
- E. Luy, J. Smith, I. Grundke, C. Sonnichsen, A. Furlong and V. Sieben, Front. Sens., 2022, 3, 1080020 CrossRef.
- H. Zhang, R. Zhao, Y. Yang, Y. Liu and L. Han, Water Qual. Res. J., 2023, 58, 1–13 CrossRef CAS.
- M. Barletta, A. R. Lima and M. F. Costa, Sci. Total Environ., 2019, 651, 1199–1218 CrossRef CAS PubMed.
- J. J. Davis, M. Padalino, A. S. Kaplitz, G. Murray, S. W. Foster, J. Maturano and J. P. Grinias, Anal. Chim. Acta, 2021, 1151, 338230 CrossRef CAS PubMed.
- M. Tudi, H. Li, H. Li, L. Wang, J. Lyu, L. Yang, S. Tong, Q. J. Yu, H. D. Ruan, A. Atabila, D. T. Phung, R. Sadler and D. Connell, Toxics, 2022, 10, 335 CrossRef CAS PubMed.
- M. A. Hassaan and A. El Nemr, Egypt. J. Aquat. Res., 2020, 46, 207–220 CrossRef.
- P. Murugesan, G. Raj and J. A. Moses, Rev. Environ. Sci. Bio/Technol., 2023, 22, 625–652 CrossRef CAS.
- B. Xu, J. Guo, Y. Fu, X. Chen and J. Guo, Electrophoresis, 2020, 41, 821–832 CrossRef CAS PubMed.
- H. J. Kim, Y. Kim, S. J. Park, C. Kwon and H. Noh, BioChip J., 2018, 12, 326–331 CrossRef CAS.
- F. Arduini, S. Cinti, V. Caratelli, L. Amendola, G. Palleschi and D. Moscone, Biosens. Bioelectron., 2019, 126, 346–354 CrossRef CAS PubMed.
- M. Lafuente, I. Pellejero, A. Clemente, M. A. Urbiztondo, R. Mallada, S. Reinoso, M. P. Pina and L. M. Gandia, ACS Appl. Mater. Interfaces, 2020, 12, 36458–36467 CrossRef CAS PubMed.
- B. Uka, J. Kieninger, G. A. Urban and A. Weltin, ACS Sens., 2021, 6, 2738–2746 CrossRef CAS PubMed.
- I. Jang, D. B. Carrao, R. F. Menger, A. R. Moraes de Oliveira and C. S. Henry, ACS Sens., 2020, 5, 2230–2238 CrossRef CAS PubMed.
- S. Pengpumkiat, J. Nammoonnoy, W. Wongsakoonkan, P. Konthonbut and P. Kongtip, Sensors, 2020, 20, 4107 CrossRef CAS PubMed.
- A. T. Jafry, H. Lee, A. P. Tenggara, H. Lim, Y. Moon, S.-H. Kim, Y. Lee, S.-M. Kim, S. Park, D. Byun and J. Lee, Sens. Actuators, B, 2019, 282, 831–837 CrossRef CAS.
- M. D. Fernandez-Ramos, A. L. Ogunneye, N. A. A. Babarinde, M. M. Erenas and L. F. Capitan-Vallvey, Talanta, 2020, 218, 121108 CrossRef CAS PubMed.
- K. Sankar, D. Lenisha, G. Janaki, J. Juliana, R. S. Kumar, M. C. Selvi and G. Srinivasan, Talanta, 2020, 208, 120408 CrossRef CAS PubMed.
- S. Beshana, A. Hussen, S. Leta and T. Kaneta, Bull. Environ. Contam. Toxicol., 2022, 109, 344–351 CrossRef CAS PubMed.
- H. Wu, J. Chen, Y. Yang, W. Yu, Y. Chen, P. Lin and K. Liang, Anal. Bioanal. Chem., 2022, 414, 1759–1772 CrossRef CAS PubMed.
- S. Beshana, A. Hussen, S. Leta and T. Kaneta, Anal. Sci., 2022, 38, 1359–1367 CrossRef CAS PubMed.
- S. Lee, J. Park and J.-K. Park, Sens. Actuators, B, 2018, 273, 322–327 CrossRef CAS.
- S. Li, C. Pang, X. Ma, Y. Zhang, Z. Xu, J. Li, M. Zhang and M. Wang, Microchim. Acta, 2021, 188, 438 CrossRef CAS PubMed.
- Z. Zhang, X. Ma, B. Li, J. Zhao, J. Qi, G. Hao, R. Jianhui and X. Yang, Analyst, 2020, 145, 963–974 RSC.
- G. Emonds-Alt, C. Malherbe, A. Kasemiire, H. T. Avohou, P. Hubert, E. Ziemons, J. M. Monbaliu and G. Eppe, Talanta, 2022, 249, 123640 CrossRef CAS PubMed.
- I. Al Yahyai, H. A. J. Al-Lawati and J. Hassanzadeh, Anal. Methods, 2021, 13, 3461–3470 RSC.
- D. Zhang, H. Bi, B. Liu and L. Qiao, Anal. Chem., 2018, 90, 5512–5520 CrossRef CAS PubMed.
- Z. Altintas, M. Akgun, G. Kokturk and Y. Uludag, Biosens. Bioelectron., 2018, 100, 541–548 CrossRef CAS PubMed.
- S. Chung, L. E. Breshears, S. Perea, C. M. Morrison, W. Q. Betancourt, K. A. Reynolds and J. Y. Yoon, ACS Omega, 2019, 4, 11180–11188 CrossRef CAS PubMed.
- H. N. Gowda, H. Kido, X. Wu, O. Shoval, A. Lee, A. Lorenzana, M. Madou, M. Hoffmann and S. C. Jiang, Sci. Total Environ., 2022, 813, 152556 CrossRef CAS PubMed.
- B. Krafft, A. Tycova, R. D. Urban, C. Dusny and D. Belder, Electrophoresis, 2021, 42, 86–94 CrossRef CAS PubMed.
- S. Choo, H. Lim, T. E. Kim, J. Park, K. B. Park, C. Park, C. S. Lim and J. Nam, Micromachines, 2022, 13, 1093 CrossRef PubMed.
- P. Rajapaksha, A. Elbourne, S. Gangadoo, R. Brown, D. Cozzolino and J. Chapman, Analyst, 2019, 144, 396–411 RSC.
- R. Siavash Moakhar, T. AbdelFatah, A. Sanati, M. Jalali, S. E. Flynn, S. S. Mahshid and S. Mahshid, ACS Appl. Mater. Interfaces, 2020, 12, 23298–23310 CrossRef CAS PubMed.
- L. Gorgannezhad, K. R. Sreejith, J. Zhang, G. Kijanka, M. Christie, H. Stratton and N. T. Nguyen, Micromachines, 2019, 10, 883 CrossRef PubMed.
- Y. Jo, J. Park and J. K. Park, Sensors, 2020, 20, 2267 CrossRef CAS PubMed.
- S. B. Somvanshi, A. M. Ulloa, M. Zhao, Q. Liang, A. K. Barui, A. Lucas, K. M. Jadhav, J. P. Allebach and L. A. Stanciu, Biosens. Bioelectron., 2022, 207, 114214 CrossRef CAS PubMed.
- D. Rodoplu, C. S. Chang, C. Y. Kao and C. H. Hsu, Microchem. J., 2022, 178, 107390 CrossRef CAS.
- K. Yin, X. Ding, Z. Xu, Z. Li, X. Wang, H. Zhao, C. Otis, B. Li and C. Liu, Sens. Actuators, B, 2021, 344, 130242 CrossRef CAS PubMed.
- S. A. Snyder, M. Boban, C. Li, J. S. VanEpps, G. Mehta and A. Tuteja, Lab Chip, 2020, 20, 4413–4419 RSC.
- J. F. Bergua, L. Hu, C. Fuentes-Chust, R. Alvarez-Diduk, A. H. A. Hassan, C. Parolo and A. Merkoci, Lab Chip, 2021, 21, 2417–2426 RSC.
- D. Lin, B. Li, J. Qi, X. Ji, S. Yang, W. Wang and L. Chen, Sens. Actuators, B, 2020, 303, 127213 CrossRef CAS.
- R. Wang, Y. Xu, T. Sors, J. Irudayaraj, W. Ren and R. Wang, Microchim. Acta, 2018, 185, 184 CrossRef PubMed.
- S. Miller, A. A. Weiss, W. R. Heineman and R. K. Banerjee, Sci. Rep., 2019, 9, 14228 CrossRef PubMed.
- B. Ang, R. Habibi, C. Kett, W. H. Chin, J. J. Barr, K. L. Tuck, A. Neild and V. J. Cadarso, Sens. Actuators, B, 2023, 374, 132769 CrossRef CAS.
- S. Rauf, N. Tashkandi, J. I. de Oliveira Filho, C. I. Oviedo-Osornio, M. S. Danish, P. Y. Hong and K. N. Salama, Biosensors, 2022, 12, 34 CrossRef CAS PubMed.
- N. Yamaguchi and S. Goto, Water, Air, Soil Pollut., 2019, 230, 285 CrossRef CAS.
- U. Dogan, E. N. Kasap, F. Sucularli, E. Yildirim, U. Tamer, D. Cetin, Z. Suludere, I. H. Boyaci and N. Ertas, Anal. Methods, 2020, 12, 3788–3796 RSC.
- L. F. Alonzo, T. C. Hinkley, A. Miller, R. Calderon, S. Garing, J. Williford, N. Clute-Reinig, E. Spencer, M. Friend, D. Madan, V. T. T. Dinh, D. Bell, B. H. Weigl, S. R. Nugen, K. P. Nichols and A. M. Le Ny, Lab Chip, 2022, 22, 2155–2164 RSC.
- K. E. Klug, K. A. Reynolds and J. Y. Yoon, Chemistry, 2018, 24, 6025–6029 CrossRef CAS PubMed.
- R. Dhore and G. S. Murthy, Bioresour. Technol., 2021, 341, 125808 CrossRef CAS PubMed.
- R. F. Menger, E. Funk, C. S. Henry and T. Borch, Chem. Eng. J., 2021, 417, 129133 CrossRef CAS PubMed.
- Y. H. Cheng, D. Barpaga, J. A. Soltis, V. Shutthanandan, R. Kargupta, K. S. Han, B. P. McGrail, R. K. Motkuri, S. Basuray and S. Chatterjee, ACS Appl. Mater. Interfaces, 2020, 12, 10503–10514 CrossRef CAS PubMed.
- L. E. Breshears, S. Mata-Robles, Y. Tang, J. C. Baker, K. A. Reynolds and J. Y. Yoon, J. Hazard. Mater., 2023, 446, 130699 CrossRef CAS PubMed.
- USEPA, US Environmental Protection Agency, 2022.
- A. Cordner, V. Y. De La Rosa, L. A. Schaider, R. A. Rudel, L. Richter and P. Brown, J. Exposure Sci. Environ. Epidemiol., 2019, 29, 157–171 CrossRef PubMed.
- R. F. Menger, E. Funk, C. S. Henry and T. Borch, Chem. Eng. J., 2021, 417, 129133 CrossRef CAS PubMed.
- A. L. Campana, S. L. Florez, M. J. Noguera, O. P. Fuentes, P. Ruiz Puentes, J. C. Cruz and J. F. Osma, Biosensors, 2019, 9, 41 CrossRef CAS PubMed.
- V. Burtsev, M. Erzina, O. Guselnikova, E. Miliutina, Y. Kalachyova, V. Svorcik and O. Lyutakov, Analyst, 2021, 146, 3686–3696 RSC.
- D. Gril and D. Donlagic, Sensors, 2023, 23, 4984 CrossRef CAS PubMed.
- P. T. Charles, V. Wadhwa, A. Kouyate, K. J. Mesa-Donado, A. A. Adams, J. R. Deschamps and A. W. Kusterbeck, Sensors, 2018, 18, 1568 CrossRef PubMed.
- V. Moradi, M. Akbari and P. Wild, Sens. Actuators, A, 2019, 297, 111507 CrossRef CAS.
- D. Gaurav, A. K. Malik and P. Rai, Crit. Rev. Anal. Chem., 2007, 37, 227–268 CrossRef CAS.
- L. Heighton, W. F. Schmidt, C. P. Rice and R. L. Siefert, 2008.
- V. Kumar, N. Upadhay, A. Wasit, S. Singh and P. Kaur, Curr. World Environ., 2013, 8, 313 CrossRef.
- H. Hattori, Curr. World Environ., 1992, 38, 93–100 CAS.
- U. S. D. O. Agriculture, 2000.
- P. G. Sutariya, H. Soni, S. A. Gandhi and A. Pandya, New J. Chem., 2019, 43, 737–747 RSC.
- P. G. Sutariya, H. Soni, S. A. Gandhi and A. Pandya, J. Lumin., 2019, 212, 171–179 CrossRef CAS.
- P. G. Sutariya, H. Soni, S. A. Gandhi and A. Pandya, J. Lumin., 2019, 208, 6–17 CrossRef CAS.
- H. Arabyarmohammadi, A. K. Darban, M. Abdollahy and B. Ayati, J. Environ. Health Sci. Eng., 2018, 16, 109–119 CrossRef CAS PubMed.
- H. L. Nashukha, J. Sitanurak, H. Sulistyarti, D. Nacapricha and K. Uraisin, Molecules, 2021, 26, 2004 CrossRef CAS PubMed.
- R. Ding, M. Fiedoruk-Pogrebniak, M. Pokrzywnicka, R. Koncki, J. Bobacka and G. Lisak, Sens. Actuators, B, 2020, 323, 128680 CrossRef CAS.
- C. Pinyorospathum, P. Rattanarat, S. Chaiyo, W. Siangproh and O. Chailapakul, Sens. Actuators, B, 2019, 290, 226–232 CrossRef CAS.
- A. Pal, S. K. Dubey and S. Goel, Comput. Electron. Agric., 2022, 195, 106856 CrossRef.
- S. Dudala, S. K. Dubey and S. Goel, IEEE Sens. J., 2020, 20, 4504–4511 CAS.
- S. Böckmann, I. Titov and M. Gerken, AgriEngineering, 2021, 3, 783–796 CrossRef.
- Y. Bai, M. Gao, L. Wen, C. He, Y. Chen, C. Liu, X. Fu and S. Huang, Biotechnol. J., 2018, 13, 1700170 CrossRef PubMed.
- C. Roggo, C. Picioreanu, X. Richard, C. Mazza, H. Van Lintel and J. R. Van Der Meer, Environ. Microbiol., 2018, 20, 241–258 CrossRef CAS PubMed.
- X. Zhu, K. Wang, H. Yan, C. Liu, X. Zhu and B. Chen, Environ. Sci. Technol., 2022, 56, 711–731 CrossRef CAS PubMed.
- A. Gaines, M. Ludovice, J. Xu, M. Zanghi, R. J. Meinersmann, M. Berrang, W. Daley and D. Britton, PLoS One, 2019, 14, e0222484 CrossRef CAS PubMed.
- C. Baranger, A. Fayeulle and A. Le Goff, R. Soc. Open Sci., 2020, 7, 191535 CrossRef CAS PubMed.
- T. Gunstone, T. Cornelisse, K. Klein, A. Dubey and N. Donley, Front. Environ. Sci., 2021, 9, 122 CrossRef.
- J. A. Hondred, Z. T. Johnson and J. C. Claussen, J. Mater. Chem. C, 2020, 8, 11376–11388 RSC.
- O. Guselnikova, P. Postnikov, R. Elashnikov, E. Miliutina, V. Svorcik and O. Lyutakov, Anal. Chim. Acta, 2019, 1068, 70–79 CrossRef CAS PubMed.
- L. C. Shriver-Lake, D. Zabetakis, W. J. Dressick, D. A. Stenger and S. A. Trammell, Sensors, 2018, 18, 328 CrossRef PubMed.
- S. Jin, Z. Xu, J. Chen, X. Liang, Y. Wu and X. Qian, Anal. Chim. Acta, 2004, 523, 117–123 CrossRef CAS.
- S. S. B. Moram, C. Byram, S. N. Shibu, B. M. Chilukamarri and V. R. Soma, ACS Omega, 2018, 3, 8190–8201 CrossRef CAS PubMed.
- E. Pellegrini, M. Contin, L. Vittori Antisari, G. Vianello, C. Ferronato and M. De Nobili, Environ. Toxicol. Chem., 2018, 37, 3025–3031 CrossRef CAS PubMed.
- P. Basuri, A. Baidya and T. Pradeep, Anal. Chem., 2019, 91, 7118–7124 CrossRef CAS PubMed.
- G. A. Suaifan and M. Zourob, Microchim. Acta, 2019, 186, 1–11 CrossRef CAS PubMed.
- M.-G. Lee, H.-W. Yoo, S. H. Lim and G.-R. Yi, Korean J. Chem. Eng., 2020, 37, 2171–2178 CrossRef CAS.
- P. T. Charles, V. Wadhwa, A. Kouyate, K. J. Mesa-Donado, A. A. Adams, J. R. Deschamps and A. W. Kusterbeck, Sensors, 2018, 18, 1568 CrossRef PubMed.
- M. F. Siddiqui, S. Kim, H. Jeon, T. Kim, C. Joo and S. Park, Sensors, 2018, 18, 777 CrossRef PubMed.
- A. Muhammed, A. Hussen, M. Redi and T. Kaneta, Anal. Sci., 2021, 37, 585–592 CrossRef CAS PubMed.
- Z. Suo, R. Liang, R. Liu, M. Wei, B. He, L. Jiang, X. Sun and H. Jin, Anal. Chim. Acta, 2023, 1239, 340714 CrossRef CAS PubMed.
- M. Kampa and E. Castanas, Environ. Pollut., 2008, 151, 362–367 CrossRef CAS PubMed.
- M. Steenhof, N. A. Janssen, M. Strak, G. Hoek, I. Gosens, I. S. Mudway, F. J. Kelly, R. M. Harrison, R. H. Pieters and F. R. Cassee, Inhalation Toxicol., 2014, 26, 141–165 CrossRef CAS PubMed.
- W.-T. Liu and C. Lay, Water Sci. Technol.: Water Supply, 2007, 7, 165–172 CAS.
- J. Gardeniers and A. Van den Berg, Anal. Bioanal. Chem., 2004, 378, 1700–1703 CrossRef CAS PubMed.
- A. R. Metcalf, S. Narayan and C. S. Dutcher, Aerosol Sci. Technol., 2018, 52, 310–329 CrossRef CAS.
- S. Kaaliveetil, J. Yang, S. Alssaidy, Z. Li, Y.-H. Cheng, N. H. Menon, C. Chande and S. Basuray, Micromachines, 2022, 13, 1716 CrossRef PubMed.
- I. B. Koki, A. S. Bayero, A. Umar and S. Yusuf, Afr. J. Pure Appl. Chem., 2015, 9, 204–210 CrossRef CAS.
- R. Chen, H. Yin, I. S. Cole, S. Shen, X. Zhou, Y. Wang and S. Tang, Chemosphere, 2020, 259, 127452 CrossRef CAS PubMed.
- V. Søyseth, H. L. Johnsen and J. Kongerud, Curr. Opin. Pulm. Med., 2013, 19, 158–162 CrossRef PubMed.
- P. C. Zeidler-Erdely, A. Erdely and J. M. Antonini, J. Immunotoxicol., 2012, 9, 411–425 CrossRef PubMed.
- S. Kumari, M. K. Jain and S. P. Elumalai, J. Health Pollut., 2021, 11, 210305 CrossRef PubMed.
- Y. M. Coyle, A. T. Minahjuddin, L. S. Hynan and J. D. Minna, J. Thorac. Oncol., 2006, 1, 654–661 Search PubMed.
- M. Franklin, P. Koutrakis and J. Schwartz, Epidemiology, 2008, 19, 680 CrossRef PubMed.
- P. Wild, E. Bourgkard and C. Paris, Cancer Epidemiol., 2009, 139–167 CAS.
- S. Shariati and G. Khayatian, New J. Chem., 2020, 44, 18662–18667 RSC.
- H. Sun, Y. Jia, H. Dong and L. Fan, Microsyst. Nanoeng., 2019, 5, 4 CrossRef PubMed.
- Y. Jia, W. Wu, J. Zheng, Z. Ni and H. Sun, Biomicrofluidics, 2019, 13, 054103 CrossRef PubMed.
- J. Mettakoonpitak, J. Volckens and C. S. Henry, Anal. Chem., 2019, 92, 1439–1446 CrossRef PubMed.
- I. Manisalidis, E. Stavropoulou, A. Stavropoulos and E. Bezirtzoglou, Front. Public Health, 2020, 8, 14 CrossRef PubMed.
- X. L. Guo, Y. Chen, H. L. Jiang, X. B. Qiu and D. L. Yu, Sensors, 2018, 18, 3141 CrossRef PubMed.
- J. Ozhikandathil, S. Badilescu and M. Packirisamy, J. Electrochem. Soc., 2018, 165, B3078–B3083 CrossRef CAS.
- M. Ghazi, S. Janfaza, H. Tahmooressi, N. Tasnim and M. Hoorfar, J. Hazard. Mater., 2022, 424, 127566 CrossRef CAS PubMed.
- K. Yang, S. Zong, Y. Zhang, Z. Qian, Y. Liu, K. Zhu, L. Li, N. Li, Z. Wang and Y. Cui, ACS Appl. Mater. Interfaces, 2020, 12, 1395–1403 CrossRef CAS PubMed.
- P. A. Duarte, L. Menze, L. Shoute, J. Zeng, O. Savchenko, J. Lyu and J. Chen, ACS Omega, 2022, 7, 459–468 CrossRef CAS PubMed.
- S. Kim, P. Akarapipad, B. T. Nguyen, L. E. Breshears, K. Sosnowski, J. Baker, J. L. Uhrlaub, J. Nikolich-Zugich and J. Y. Yoon, Biosens. Bioelectron., 2022, 200, 113912 CrossRef CAS PubMed.
- Q. Liu, X. Zhang, Y. Yao, W. Jing, S. Liu and G. Sui, Sens. Actuators, B, 2018, 258, 1138–1145 CrossRef CAS.
- B. Ryu, J. Chen, K. Kurabayashi, X. Liang and Y. Park, Analyst, 2020, 145, 6283–6290 RSC.
- J. Wang, L. Yang, H. Wang and L. Wang, Micromachines, 2022, 13, 1576 CrossRef PubMed.
- M. Paknahad, C. McIntosh and M. Hoorfar, Sci. Rep., 2019, 9, 161 CrossRef PubMed.
- J. Sitanurak, N. Wangdi, T. Sonsa-Ard, S. Teerasong, T. Amornsakchai and D. Nacapricha, Talanta, 2018, 187, 91–98 CrossRef CAS PubMed.
- S. Shariati and G. Khayatian, New J. Chem., 2020, 44, 18662–18667 RSC.
- H. Sun, Y. Jia, H. Dong and L. Fan, Microsyst. Nanoeng., 2019, 5, 4 CrossRef PubMed.
- Y. Jia, W. Wu, J. Zheng, Z. Ni and H. Sun, Biomicrofluidics, 2019, 13, 054103 CrossRef PubMed.
- H. Sun, Y. Jia, H. Dong, L. Fan and J. Zheng, Anal. Chim. Acta, 2018, 1044, 110–118 CrossRef CAS PubMed.
- S. Huang, J. Connolly, A. Khlystov and R. B. Fair, Sensors, 2020, 20, 1281 CrossRef CAS PubMed.
- M. M. Montazeri, N. De Vries, A. D. Afantchao, A. O'Brien, P. Kadota and M. Hoorfar, IEEE Sens. J., 2018, 18, 7772–7778 CAS.
- M. Saito, N. Uchida, S. Furutani, M. Murahashi, W. Espulgar, N. Nagatani, H. Nagai, Y. Inoue, T. Ikeuchi, S. Kondo, H. Uzawa, Y. Seto and E. Tamiya, Microsyst. Nanoeng., 2018, 4, 17083 CrossRef CAS.
- S. Lapcinska, A. Revilla-Cuesta, I. Abajo-Cuadrado, J. V. Cuevas, M. Avella, P. Arsenyan and T. Torroba, Mater. Chem. Front., 2021, 5, 8097–8107 RSC.
- H. Xiong, X. Ye, Y. Li, J. Qi, X. Fang and J. Kong, Anal. Chem., 2021, 93, 4270–4276 CrossRef CAS PubMed.
- D. Măriuţa, L. Baldas, S. Colin, S. L. Calvé, J. G. Korvink and J. J. Brandner, Int. J. Chem. Eng. Appl., 2020, 11, 23–28 Search PubMed.
- L. Mugherli, A. Lety-Stefanska, N. Landreau, R. F. Tomasi and C. N. Baroud, Lab Chip, 2020, 20, 236–243 RSC.
- K. Yang, C. Zhang, K. Zhu, Z. Qian, Z. Yang, L. Wu, S. Zong, Y. Cui and Z. Wang, ACS Nano, 2022, 16, 19335–19345 CrossRef CAS PubMed.
- X. J. Liu, S. W. Hu, B. Y. Xu, G. Zhao, X. Li, F. W. Xie, J. J. Xu and H. Y. Chen, Talanta, 2018, 182, 202–209 CrossRef CAS PubMed.
- N. Yang, T. Li, S. Dong, S. Zhang, Y. Jia, H. Mao, Z. Zhang, F. Zhang, X. Pan, X. Zhang and Z. Dong, Lab Chip, 2022, 22, 4995–5007 RSC.
- P. Tirandazi and C. H. Hidrovo, Sens. Actuators, B, 2018, 267, 279–293 CrossRef CAS.
- A. Kuzin, V. Chernyshev, V. Kovalyuk, P. An, A. Golikov, R. Ozhegov, D. Gorin, N. Gippius and G. Goltsman, Opt. Lett., 2022, 47, 2358–2361 CrossRef CAS PubMed.
- P. Wang, S. Yuan, N. Yang, A. Wang, A. Fordjour and S. Chen, Aerosol Air Qual. Res., 2020, 20, 72–79 CrossRef.
- V. Kamat, L. Burton, V. Venkadesh, K. Jayachandran and S. Bhansali, ECS Sens. Plus, 2023, 2, 043201 CrossRef.
- S. Zhao, J. Lei, D. Huo, C. Hou, P. Yang, J. Huang and X. Luo, Anal. Methods, 2018, 10, 5507–5515 RSC.
- N. Yang, X. Zhou, D. Yu, S. Jiao, X. Han, S. Zhang, H. Yin and H. Mao, J. Food Process Eng., 2020, 43, e13544 CrossRef CAS.
- Y. Cao, C. Ye, C. Zhang, G. Zhang, H. Hu, Z. Zhang, H. Fang, J. Zheng and H. Liu, Food Control, 2022, 134, 108694 CrossRef CAS.
- X. Xiang, Q. Ye, Y. Shang, F. Li, B. Zhou, Y. Shao, C. Wang, J. Zhang, L. Xue and M. Chen, Biosens. Bioelectron., 2021, 190, 113394 CrossRef CAS PubMed.
- Z. Zhang, X. Ma, B. Li, J. Zhao, J. Qi, G. Hao, R. Jianhui and X. Yang, Analyst, 2020, 145, 963–974 RSC.
- Z. Zhang, X. Ma, M. Jia, B. Li, J. Rong and X. Yang, Analyst, 2019, 144, 1282–1291 RSC.
- B. Al Mughairy, H. A. Al-Lawati and F. O. Suliman, Sens. Actuators, B, 2018, 277, 517–525 CrossRef CAS.
- U. Andayani, M. I. Sari and A. Sabarudin, IOP Conf. Ser.: Mater. Sci. Eng., 2019, 546, 032028 Search PubMed.
- X. Zhang, F. Bian, Y. Wang, L. Hu, N. Yang and H. Mao, Foods, 2022, 11, 3462 CrossRef CAS PubMed.
- L. Jin, Z. Hao, Q. Zheng, H. Chen, L. Zhu, C. Wang, X. Liu and C. Lu, Anal. Chim. Acta, 2020, 1100, 215–224 CrossRef CAS PubMed.
- I. Jang, D. B. Carrão, R. F. Menger, A. R. Moraes de Oliveira and C. S. Henry, ACS Sens., 2020, 5, 2230–2238 CrossRef CAS PubMed.
- R. Fu, Y. Wang, Y. Liu, H. Liu, Q. Zhao, Y. Zhang, C. Wang, Z. Li, B. Jiao and Y. He, Food Chem., 2022, 387, 132919 CrossRef CAS PubMed.
- Y. Shen, X. Gao, H. Chen, Y. Wei, H. Yang and Y. Gu, J. Hazard. Mater., 2023, 451, 131171 CrossRef CAS PubMed.
- M. Yuan, C. Li, Y. Zheng, H. Cao, T. Ye, X. Wu, L. Hao, F. Yin, J. Yu and F. Xu, Talanta, 2023, 125112 Search PubMed.
- T. Hu, J. Xu, Y. Ye, Y. Han, X. Li, Z. Wang, D. Sun, Y. Zhou and Z. Ni, Biosens. Bioelectron., 2019, 136, 112–117 CrossRef CAS PubMed.
- X. Tong, G. Cai, L. Xie, T. Wang, Y. Zhu, Y. Peng, C. Tong, S. Shi and Y. Guo, Biosens. Bioelectron., 2023, 222, 114981 CrossRef CAS PubMed.
- C.-C. Liu, C.-H. Ko, L.-M. Fu and Y.-L. Jhou, Food Chem., 2023, 400, 134063 CrossRef CAS PubMed.
- G. Hao, Z. Zhang, X. Ma, R. Zhang, X. Qin, H. Sun, X. Yang and J. Rong, Microchem. J., 2020, 157, 105050 CrossRef CAS.
- G. Hao, H. Tian, Z. Zhang, X. Qin, T. Yang, L. Yuan and X. Yang, Microchem. J., 2023, 190, 108674 CrossRef CAS.
- A. Ahmed, A. Singh, B. Padha, A. K. Sundramoorthy, A. Tomar and S. Arya, Chemosphere, 2022, 303, 135208 CrossRef CAS PubMed.
- L. N. Sibal and M. P. B. Espino, Int. J. Environ. Anal. Chem., 2018, 98, 536–554 CrossRef CAS.
- S. Kulkarni, S. Dhokpande and J. Kaware, Int. J. Adv. Eng. Res. Sci., 2015, 2, 35–38 Search PubMed.
- P. J. Viskari and J. P. Landers, Electrophoresis, 2006, 27, 1797–1810 CrossRef CAS PubMed.
- F. Pena-Pereira, I. Costas-Mora, V. Romero, I. Lavilla and C. Bendicho, TrAC, Trends Anal. Chem., 2011, 30, 1637–1648 CrossRef CAS.
- A. Llobera, R. Wilke and S. Büttgenbach, Lab Chip, 2004, 4, 24–27 RSC.
- P. Yeh, N. Yeh, C.-H. Lee and T.-J. Ding, Renewable Sustainable Energy Rev., 2017, 75, 461–468 CrossRef CAS.
- A. Lace and J. Cleary, Chemosensors, 2021, 9, 60 CrossRef CAS.
- R. Natu, L. Herbertson, G. Sena, K. Strachan and S. Guha, Micromachines, 2023, 14, 1293 CrossRef PubMed.
- M. Tebrake, Science Behind the Strip: Key Considerations for Scaling Microfluidic Devices, https://multimedia.3m.com/mws/media/1751429O/3m-medical-materials-technologies-key-considerations-for-scaling-microfluidic-devices.pdf.
- B. H. Weigl, R. L. Bardell and C. Cabrera, Handbook of Biosensors and Biochips, 2008 Search PubMed.
- B. K. Gale, A. R. Jafek, C. J. Lambert, B. L. Goenner, H. Moghimifam, U. C. Nze and S. K. Kamarapu, Inventions, 2018, 3, 60 CrossRef.
- K. Rose, S. Eldridge and L. Chapin, The internet society (ISOC), 2015, vol. 80, pp. 1–50 Search PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |