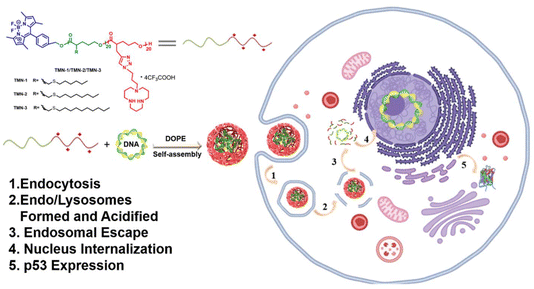
Functionalized polyesters based on valerolactones and [12]aneN3 as effective non-viral gene vectors in HepG2 cells†
Xi
Zhang
a,
De-Zhong
Xu
a,
Zi-Han
Zhang
a,
Zhi-Xuan
Ren
a,
Jia-Rao
Wu
a,
Yun-Jie
Xu
a,
Zhong-Lin
Lu
 *a,
Rui
Liu
*a and
Yang
Liu
*a,
Rui
Liu
*a and
Yang
Liu
 *b
*b
aKey Laboratory of Radiopharmaceutics, Ministry of Education; College of Chemistry, Beijing Normal University, Beijing 100875, P. R. China. E-mail: luzl@bnu.edu.cn; rliu@bnu.edu.cn
bChina National Institute for Food and Drug Control, Institute of Chemical Drug Control, HuaTuo Road 29, Beijing, 100050, China. E-mail: yangliu@nifdc.org.cn
First published on 27th July 2024
Abstract
Polymer vectors hold great promise for application in gene therapy. However, some challenges such as low transfection efficiency, high toxicity, and insufficient cell selectivity still need to be addressed. Here, we proposed a simple and efficient approach to synthesize polyester vectors via a ring-opening polymerization (ROP) process. Three distinct block copolymers, TMN-1/2/3, were prepared by using valerolactones with alkyl sulfide chains (6, 8, and 10 carbon side chains, respectively) and propargyl valerolactone as monomers and BODIPY derivatives as fluorescent units and polymerization initiators, and further modification through a click reaction with 3-azidopropyl-[12]aneN3 as positive units. The three block copolymers integrate multiple functionalities, including imaging capability, degradability, and DNA condensation ability. Their DNA binding ability and gene transfection efficiency were systematically studied and showed good structure–activity relationship. Among them, TMN-3 with a 10 carbon alkyl sulfide chain was the most effective in DNA binding ability and gene transfection efficiency, which was 18.9 times higher than that of 25 kDa PEI in the HepG2 cell line. The efficient cellular uptake, lysosomal escape, and nuclear entry of the TMN-3-DOPE/pDNA polyplex resulted in its excellent performance. Moreover, it was demonstrated that TMN-3-DOPE/pDNA exhibited significant anticancer effects upon internalization into HepG2 cells. The results indicated that TMN-3 has the potential to serve as an effective non-viral gene vector for tumor treatment.
1. Introduction
Over the past few decades, the field of gene therapy has progressed significantly, transitioning from preclinical investigations to clinical research and expanding its scope from monogenic recessive disorders to complex diseases like cancer.1–6 Gene therapy holds immense potential for fundamentally altering the landscape of cancer treatment and has brought new hope to change the status quo of cancer treatment. However, the success of gene therapy critically relies on the development of safe and effective gene vectors. Nucleic acids face challenges such as electrostatic repulsion from cell membranes and rapid degradation in physiological environments.7–10 As a result, the design of efficient gene vectors is pivotal for achieving successful gene therapy outcomes. Benefiting from their natural ability to infect cells, viral vectors have always been the “main force” in clinical trials of gene therapy, but non-viral gene vectors are steadily gaining prominence as a safer alternative to viral vectors due to their low immunogenicity and ease of mass production.11–13Cationic polymers have great potential for clinical transformation in numerous non-viral gene vectors, among which degradable polymers compensate for the high toxicity defects of cationic polymers and have attracted extensive attention.14–16 The development of biodegradable polymer vectors has reached remarkable achievements, particularly in cancer treatment as a nucleic acid delivery tool.17–20 However, challenges persist, including low transfection efficiency and low expression selectivity; moreover the understanding of each step in the transfection process is still incomplete.21–23
Mao and Wang groups have demonstrated that biodegradable vectors possess the ability to selectively deliver and express nucleic acids within specific cells by targeting the intracellular environment.24–26 Meanwhile, Waymouth and Dong groups have discovered that vectors that are responsive to acids and esterases can enhance nucleic acid delivery.27–30 Hence, it is imperative to design a nucleic acid delivery vector that is capable of achieving passive targeting and spatiotemporal control through the tumor microenvironment.31–33 Furthermore, the low transfection efficiency of non-viral vectors may be attributed to issues such as cell uptake, lysosomal escape, and reversible release of nucleic acids. Therefore, introducing fluorescent groups into vectors to target gene delivery is beneficial for us to deepen our understanding of the transfection process and design delivery vectors with higher transfection efficiency.34–36 Nonetheless, the development of a multifunctional vector that integrates stimulation response, fluorescence imaging, and cell selectivity for nucleic acid expression remains a tough challenge in tumor therapy.
To address the aforementioned challenges, we devised a novel approach wherein BODIPY was employed as fluorescent units and polymerization initiators and valerolactone derivatives served as monomers, enabling the formation of degradable polyester scaffolds through a ROP reaction. Additionally, by utilizing a click reaction, we successfully introduced a cationic unit [12]aneN3. These efforts yielded a range of novel block copolymers, designated as TMN-1/2/3, exhibiting diverse alkyl side chain lengths. [12]aneN3 acted as a positively charged unit for enhancing cell uptake and condensation of nucleic acids, while BODIPY was employed to confer the vector with visualizing capabilities during gene transfection (Scheme 1). The backbone of the vector was designed with ester bonds to ensure its degradability under tumor microenvironmental stimuli. In a comprehensive analysis, we assessed the DNA-binding capacity and gene transfection efficiency of these vectors in combination with the helper lipid DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), shedding light on their structure–activity relationship. Remarkably, TMN-3-DOPE demonstrated cell-selective, highly efficient nucleic acid expression in HepG2 cells, boasting a striking transfection efficiency to be 18.9 times that of PEI. We elucidated its selective behavior through investigations into cellular uptake, lysosomal escape, and nuclear tracing. More importantly, in vitro anti-cancer experiments showed that TMN-3-DOPE is universal for the transfection of different tumor suppressor genes. Collectively, these findings establish TMN-3-DOPE as an exceptional multifunctional non-viral gene carrier, demonstrating its immense potential in gene therapy.
2. Results and discussion
2.1 Synthesis and characterization
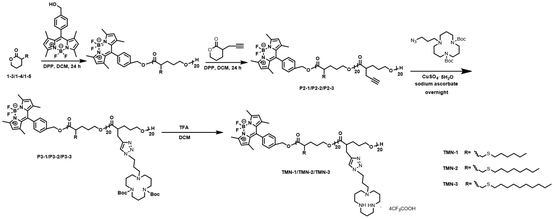
The synthesis route of polyesters TMN-1/2/3 is depicted in Scheme 2. Initially, valerolactones with varying lengths of hydrophobic alkyl sulfide chains underwent ROP reactions under the influence of fluorescent groups. Subsequently, propargyl valerolactone was introduced for block copolymerization. The resulting active site acetylene triple bonds were further connected to macrocyclic polyamine [12]aneN3via a click reaction. Finally, the target products TMN-1/2/3 are obtained by removing Boc using trifluoroacetic acid.The reaction process and chemical structures were confirmed through 1H NMR analysis. Taking the synthesis process of polymer TMN-2 as an example, the conversion rate of compounds 1–4 was determined by 1H NMR (Fig. S1†). Before polymerization, the chemical shift of proton Ha was observed at 4.3 ppm; it shifted towards a higher field after polymerization. Conversions of the polymerization reactions were evaluated by the resonance intensities of proton Ha/(the resonance intensities of Ha + the resonance intensities of Ha*). The disappearance of the characteristic peak at 4.3 ppm of proton Ha from the monomer indicated the successful purification of polymer P3-2 (Fig. S2†). The changes of 1H NMR shown in Fig. 1 illustrated the results of the click reaction and N-Boc deprotection reaction. The complete reaction of polymer P3-2 was evidenced by the disappearance of acetylenic proton H19 (at 2.02 ppm) and the appearance of triazole proton H20 (at 7.32 ppm). Furthermore, the successful removal of the Boc group was indicated by the disappearance of proton H27 (at 1.41 ppm).
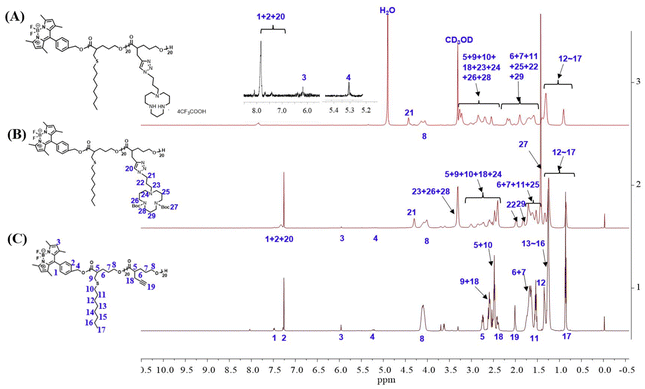 | ||
| Fig. 1 1H NMR spectra of (A) product TMN-2, (B) product P4-2 from the click reaction, and (C) block polyester P3-2 after the subsequent deprotection. | ||
Additionally, the degree of polymerization and molecular weight of the polymer were determined based on the ratio of protons H2 (at 7.28 ppm), H17 (at 0.85 ppm), and H21 (at 4.42 ppm). By integrating the areas in the 1H NMR spectra, the degrees of polymerization of the block polymer were determined to be both 20. This demonstrated that the block polymers were prepared with controllable reactions and well-defined structures through the ROP reaction. The 1H NMR spectra and GPC data of all other target polyesters can be found in Table S1 and Fig. S3.†
2.2 Photophysical properties
Over the past few decades, BODIPY has gradually evolved from a single fluorescent molecule to a functional component in a wide range of applications in biomedicine.37 Considering the influence of initiator steric hindrance on the ring-opening of valerolactone, we designed and synthesized an easily available BODIPY derivative as the initiator of the ROP reaction. This modification also endowed the polyester with fluorescence properties. The UV-Vis and fluorescence spectra of TMN-1/2/3 were systematically studied in various solvents. As shown in Fig. S4,† the polymers TMN-1/2/3 present sharp narrow peaks in the UV-Vis spectrum, which are consistent with the spectral characteristics of the BODIPY group.38 The three polymers exhibit similar optical properties, and the change of solvent polarity has little effect on the absorption. The maximum absorption peak is around 508 nm, corresponding to the π–π* transition of the large conjugated system in the molecule. Using light with a maximum excitation wavelength of 508 nm, the fluorescence emission spectra of polymers TMN-1/2/3 in different solvents were studied (Fig. S5†). The maximum emission peaks of the three polymers were around 523 nm, which did not change significantly in different solvents, but their intensities were weakened obviously in water, which can be attributed to the aggregation induced quenching effect, since the structure of the fluorescent unit BODIPY is relatively rigid and the fluorescence was weakened by π–π stacking in the aggregated state.2.3 DNA condensation/release
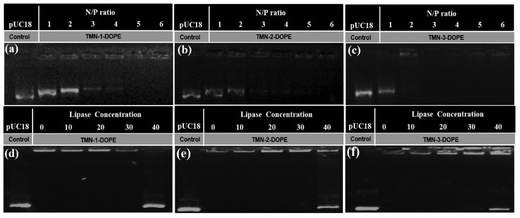
The presence of phosphate groups in the nucleic acid makes it negatively charged, which generates electrostatic repulsion with cell membranes. To enable cellular uptake, non-viral gene vectors must “wrap” nucleic acids to create a positively charged surface. DOPE is commonly used as a helper lipid to promote the transformation of lamellar liposomes into a hexagonal structure, which is conducive to the improvement of transfection efficiency. Agarose gel electrophoresis was used to investigate the condensation ability of the three polyesters combined with DOPE on the pUC18 plasmid. As shown in Fig. 2, the three polyesters can completely condense pUC18 at an N/P ratio of 3–4. With the increase of the length of the hydrophobic alkyl side chain, the condensation ability to the nucleic acid was enhanced. TMN-3-DOPE showed the strongest binding capacity, probably because longer carbon chains were favoured to wrap pUC18. Considering that DOPE itself does not condense DNA,39 these results indicated that these polyesters have a strong ability to bind DNA. Compared with the reported literature, these polyesters exhibited a good condensation effect.40,41In order to ensure that nucleic acids smoothly enter the nucleus for transcription or translation in the cytoplasm, it is well known that gene carriers often need to have the ability to reversibly release nucleic acids after undergoing endocytosis and protecting them from lysosomal escape. As shown in Fig. 2d–f, the carriers were able to accomplish the reversible release of nucleic acids under the influence of lipase. This suggests that the large amount of esterase within the tumor cells may trigger the hydrolysis of ester bonds on the polymeric backbone, leading to its decomposition and subsequent release of nucleic acids. To further verify this conclusion, degradation of polyesters was studied by the GPC method. As depicted in Fig. S6,† the molecular weight of TMN-3 decreased after incubation with lipase for 24 hours. Additionally, the polydispersity index (PDI) exhibited an increase, further corroborating the degradation of the polyester structure.
2.4 SEM and DLS
The size and geometric shape of nanoparticles play a pivotal role in governing their flow dynamics, edge properties, and adhesion characteristics within the bloodstream, as well as their circulating half-life and accumulation sites in the body.42 To comprehend these essential characteristics, scanning electron microscopy (SEM) and dynamic light scattering (DLS) experiments were conducted. Notably, as the length of the hydrophobic alkyl chain increased, so did the size of the nanoparticles, reaching a scale of approximately 200 nm (Fig. S7†). The nanoparticles exhibited relatively large surface area-to-volume ratios and surface energies, leading them to reside in a non-thermodynamically favorable state at room temperature and standard pressure.43 Therefore, their stability in PBS using TMN-3-DOPE as a representative was investigated. The size of the nanoparticles fluctuated in the range of 4 nm to 8 nm within 6 days, indicating that the formed polyplexes were very stable (Fig. S8†). The three polyesters formed uniform spherical nanoparticles after complexing with the pUC18 plasmid (Fig. 3). Spherical nanoparticles have a fast cell internalization rate, indicating that these three polyplexes were able to be effectively taken up by cells. In addition, the sizes of the above three polyplexes were around 150 nm, which is an ideal size to have an EPR effect (100–200 nm).42 In addition, under the action of lipase, the nanoparticles were observed to gradually dissociate and increase in size (Fig. S9†). In summary, the aforementioned three polyesters were able to effectively coalesce nucleic acids, safeguarding them against degradation, ultimately yielding uniform spherical complexes of suitable dimensions, and they showed a lipase response to reversely release the nucleic acid. These attributes positioned them as promising non-viral gene vectors for further research.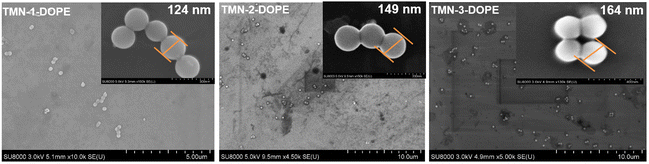 | ||
| Fig. 3 SEM images of pUC18 DNA complexed with polymers TMN-1/2/3-DOPE in 20 mM Tris-HCl buffer (pH = 7.4). | ||
2.5 Cytotoxicity and cellular transfection
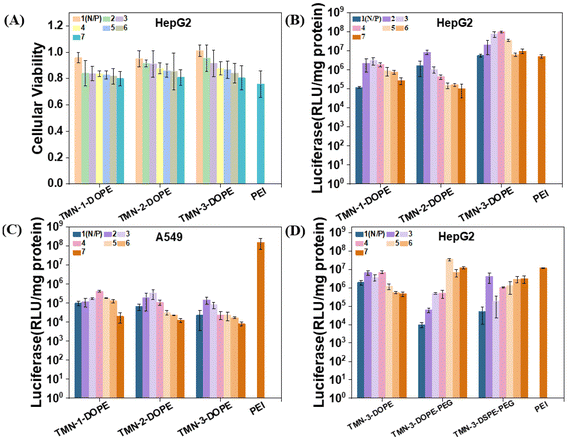
The persistent issue of high cytotoxicity stemming from the non-degradability of polymer gene vectors has garnered significant attention. The development of stimuli-responsive and biodegradable polymers stands out as a promising avenue to mitigate this challenge. In order to better clarify the biocompatibility advantages of the degradable polymer gene vectors synthesized in this work, MTT experiments were conducted to investigate the toxicity of TMN-1/2/3-DOPE@pLuci in HepG2, A549, HeLa, and PC-3 cells (all the cell lines used were obtained from the Cell Resource Center of Peking Union Medical College). The viability of these cells demonstrated a gradual decline with increasing N/P ratio, which is attributed to the disruption of cell membrane stability caused by the positive charge on the surface of nanoparticles.44 Notably, even at an N/P ratio of 7, HepG2 cell viability remained at approximately 80% (Fig. 4A). In A549, HeLa and PC-3 cells, the cell viability of the polyplexes can still reach about 70% at a high N/P ratio (Fig. S10†). The low toxicity of these polyplexes is primarily attributed to two key factors: (1) the main chain of the polymer is constructed by multiple ester bonds. In the low pH microenvironment of cancer cells, ester bonds were stimulated and broken, transformed into smaller fragments, and metabolized; (2) the presence of a triazole unit disperses the positive charge density of [12]aneN3, thereby reducing cytotoxicity.45 Consequently, these polyplexes demonstrated safety merit and potential for further investigation into their transfection ability.The transfection efficiencies of TMN-1/2/3-DOPE were quantitatively evaluated in four cell lines of A549, HeLa, HepG2, and PC-3 through the luciferase expression experiment, using the commercial transfection reagent 25 kDa branched PEI as the control.46–48 Across the four cell lines, a trend in transfection efficiency with increasing N/P ratio was observed, showing an initial rise followed by a decrease, attributable to heightened cytotoxicity at higher N/P ratios (Fig. S11†). The transfection efficiencies of TMN-1/2/3-DOPE in A549, HeLa, and PC-3 cells were all less than 0.1 times that of PEI. However, a contrasting effect was observed in HepG2 cells (Fig. 4B). Specifically, the transfection efficiency of TMN-2-DOPE and TMN-3-DOPE in HepG2 cells exceeded that of PEI by 1.6 times and 18.9 times, respectively (Table S2†). These differences may be attributed to two factors: (1) reduced cytotoxicity of the polyplexes in HepG2 cells and (2) excessive expression of hepatic triglyceride lipase in HepG2 cells that promoted the hydrolysis of the ester bond,49,50 so that the complex dissociates rapidly and DNA is efficiently released, thereby increasing gene transfection efficiency. TMN-3-DOPE surpasses other polymers in transfection efficiency due to the pivotal influence of its hydrophobic alkyl chain length. An elongation of this chain promotes heightened polymer flexibility and fluid dynamics, thereby enhancing its capacity to adapt to and fuse with cellular membranes. This improved membrane adaptability and fusion process facilitates more efficient cellular internalization, or uptake, of the transfection agent. Furthermore, the extended chain length also fortifies polymer stability, acting as a protective barrier against premature intracellular degradation. As a result, there is an augmentation in the ability of the vector to evade lysosomal degradation, fostering effective cytoplasmic release of DNA.51–53 Remarkably, the transfection efficiency of TMN-3-DOPE in HepG2 reached a high level of approximately 108 RLU per mg protein, whereas in A549 cells, it only reached 105 RLU per mg protein, indicating specific nucleic acid expression in HepG2 cells in a cell-selective manner (Fig. 4C). Regarding the specific selectivity of TMN-3-DOPE, the ability of cell uptake and lysosomal escape was studied in detail lately.
While the transfection efficiency of TMN-3-DOPE decreased significantly in 10% serum, TMN-3-DOPE-PEG in HepG2 still exhibited 2.9 times the efficiency of PEI (Fig. 4D). Conversely, TMN-3-DSPE-PEG showed the poorest transfection efficiency, reaching only 0.3 times that of PEI, suggesting that the type of helper lipid could influence the transfection effect (Table S3†).
To visually assess the gene transfection efficiency of the three vectors, their performances in delivering red FL protein (pRFP) into HepG2, A549, HeLa, and PC-3 cells were qualitatively investigated by using confocal laser microscopy (CLSM). As depicted in Fig. 5, TMN-1/2/3-DOPE@pRFP showed more red fluorescent spots in HepG2 cells, while demonstrating lower red fluorescence densities in the other three cell lines, which were consistent with luciferase expression, further indicating that the type of nucleic acid delivery did not affect the specific selectivity of TMN-1/2/3-DOPE for HepG2 cells. In conclusion, TMN-3-DOPE exhibited low cytotoxicity, high transfection efficiency, and specific selectivity for various cell lines, which make it an ideal polymer gene vector. Furthermore, the antiserum property of TMN-3-DOPE-PEG suggests its potential for clinical applications.
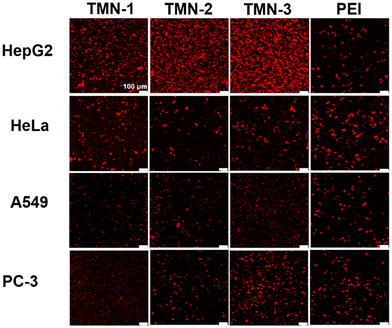 | ||
| Fig. 5 Confocal microscopy images of RFP expression for TMN-1/2/3-DOPE in HepG2, HeLa, A549, and PC-3 cells. [pRFP] = 10 μg mL−1, scale bar: 100 μm. | ||
2.6 Cell uptake
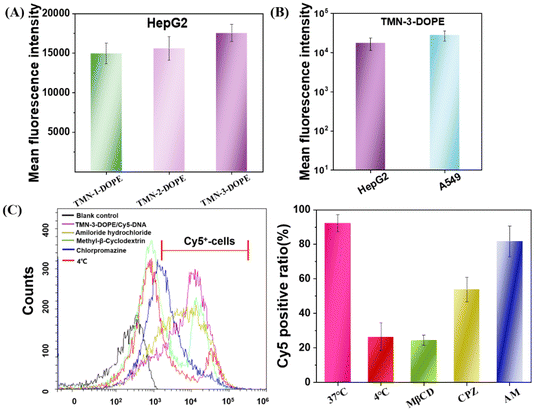
The first obstacle for gene vectors is traversing the cell membrane, which directly impacts the speed and quantity of nucleic acid uptake, thereby influencing gene expression. Therefore, the factors that affect the transfection efficiency were investigated from the perspective of cell uptake. As depicted in Fig. 6A, TMN-3-DOPE exhibited the highest Cy5-DNA uptake in HepG2 cells at 2 hours after post-administration, followed by TMN-2-DOPE, with TMN-1-DOPE showing the least uptake. The above results indicated that the higher transfection efficiency of TMN-3-DOPE in HepG2 cells may be due to its higher cellular uptake rate. Notably, a greater uptake rate of the TMN-3-DOPE@Cy5-DNA complex in A549 cells was observed, with the average fluorescence intensity reaching 2.79 × 104 (Fig. 6B). However, it showed lower transfection, which was not related to the inability of the complex to undergo uptake.Nanoparticles enter cells through various endocytic pathways, and different endocytic pathways exhibit different intracellular transport pathways and subcellular localization. Therefore, a comprehensive understanding of the endocytic mechanism is the key to tracking of the fate of nanoparticles after internalization into tumor cells, which can further elucidate the reasons for the different therapeutic effects of intracellular cargoes.54 Owing to the high transfection efficiency of TMN-3-DOPE@Cy5-DNA, an extensive investigation into its endocytic pathway in HepG2 was conducted by utilizing cell flow cytometry. As illustrated in Fig. 6C, at a low temperature (4 °C), the number of Cy5-positive cells decreased, indicating that the endocytosis of TMN-3-DOPE@Cy5-DNA into cells is an energy-dependent process. After pretreatment of cells with methyl-β-cyclodextrin (CvME endocytosis inhibitor), chlorpromazine (CME endocytosis inhibitor), and amiloride hydrochloride (macropinocytosis inhibitor), the number of Cy5 positive cells respectively decreased to 24%, 53%, and 81%. These outcomes implied that the polyplexes primarily entered cells mainly through caveolae-mediated endocytosis (CvME) and clathrin-mediated endocytosis (CME), with CvME being the dominant route. These experimental results confirmed that the better transfection efficiency of TMN-3-DOPE may be due to the fact that the uptake pathway of CvME is non-acidic and non-degradable, maintaining the integrity of the nucleic acid and avoiding its degradation.55
2.7 Spatiotemporal bio-imaging of the delivery process and endo/lysosomal escape
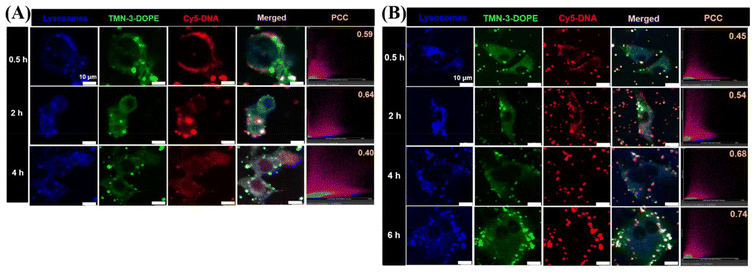
Lysosomal escape is another bottleneck for effective delivery of therapeutic agents at the subcellular level.56–58 For nanocarriers, timely release of the “cargo” from lysosomes is critical to avoid cargo degradation and inactivation. In previous studies, it was found that TMN-3-DOPE entered cells via clathrin-mediated endocytosis, which led to the entry of polyplexes into lysosomes. Therefore, CLSM was used to investigate whether TMN-3-DOPE@Cy5-DNA could escape quickly after entering the lysosomes of HepG2 cells (Fig. 7A). At 0.5 h, TMN-3-DOPE@Cy5-DNA polyplexes only surrounded the cell membrane of HepG2 cells. When the administration time reached 2 h, the polyplexes gradually entered the cytoplasm, and the white areas (blue, green, and red overlapping colors) increased, indicating that TMN-3-DOPE@Cy5-DNA entered the lysosome. At 4 h, it was evident that the overlap of Cy5-DNA with blue lysosomes and the green dots decreased. At this point, DNA has achieved lysosomal escape, avoiding the risk of degradation. The co-localization coefficient between Cy5-DNA and lysosomes provided a more intuitive representation of lysosomal escape in time and space. As time went on, Pearson's co-localization coefficient changed as follows: 0.59/0.5 h, 0.64/2 h, and 0.40/4 h.In order to investigate the reason why TMN-3-DOPE has specific selectivity for HepG2 cells, the lysosomal escape ability of TMN-3-DOPE in A549 cells was studied (Fig. 7B). As time went on, the overlap area between Cy5-DNA and blue lysosomes gradually increased in A549 cells, with DNA failing to escape even after 6 hours. The changes in Pearson's co-localization coefficient were: 0.45/0.5 h, 0.54/2 h, 0.68/4 h, and 0.74/6 h.
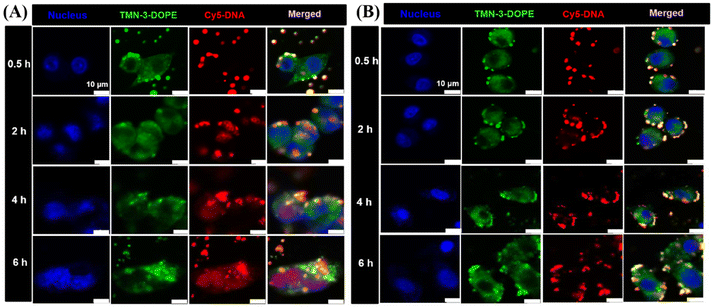
To further verify the results of the lysosomal escape experiments, the nuclear delivery of TMN-3-DOPE@Cy5-DNA in HepG2 and A549 cells was studied. As shown in Fig. 8A, after 0.5 hours of incubation in HepG2 cells, the polyplexes were mainly attached to the cell membrane. At 2 h, purple dots (red and blue superimposed) were seen, indicating that there is little Cy5-DNA entering the nucleus. At 4 h and 6 h, it can be seen that the entire nucleus has turned purple, indicating that most of the Cy5-DNA has entered the nucleus. In A549 cells, the incubation time was increased from 0.5 h to 6 h, and no purple dots were seen, indicating that Cy5-DNA failed to enter the nucleus, which again confirmed the result that the polyplexes could not achieve lysosomal escape in A549 cells (Fig. 8B). In summary, TMN-3-DOPE displayed specific selectivity for HepG2 due to its unique ability to achieve lysosomal escape solely in HepG2 cells. In contrast, other cell lines trapped the polyplexes in lysosomes, preventing their translocation to the nucleus for transcription and resulting in low transfection efficiency.
The 3D tumor model is closer to clinical trials than the gene expression in 2D cells, and this system has a stronger ability to predict clinical efficacy.59 The tissue penetration ability of TMN-3-DOPE@Cy5-DNA was explored by using a HeLa sphere model. Notably, red fluorescence of Cy5-DNA was observable at 120 μm within the sphere, demonstrating excellent tumor penetration ability and clinical application potential for TMN-3-DOPE (Fig. S12†).
In conclusion, it was found that TMN-3-DOPE has specific selectivity in HepG2 cells. This finding highlighted the importance of lysosomal escape for effective gene delivery, which is of great significance for improving transfection efficiency and improving clinical outcomes of gene therapy.
2.8 RFP transfection in vivo
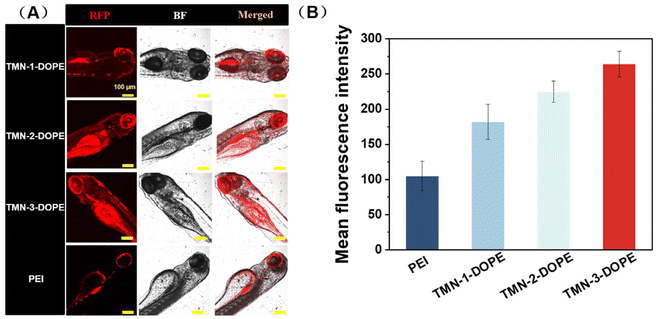
Given the low toxicity and remarkable transfection performance of TMN-3-DOPE at the cellular level, a comparative study was conducted to evaluate the transfection efficiency in zebrafish, by using TMN-1/2-DOPE and PEI as positive controls. As shown in Fig. 9, TMN-3-DOPE@pRFP exhibited the strongest red fluorescence in zebrafish. Through quantitative analysis, it was found that the fluorescence intensity of TMN-1/2/3-DOPE was 1.73, 2.14, and 2.5 times higher than that of PEI. It indicated that the polyester vectors reported in this paper can successfully overcome the obstacles in the physiological environment and effectively achieve gene transfection in zebrafish. It is noteworthy that these vectors facilitated RFP expression predominantly in the eyes, which warrants further investigation into their potential for targeted gene delivery in vivo.2.9 In vitro antitumor efficacy
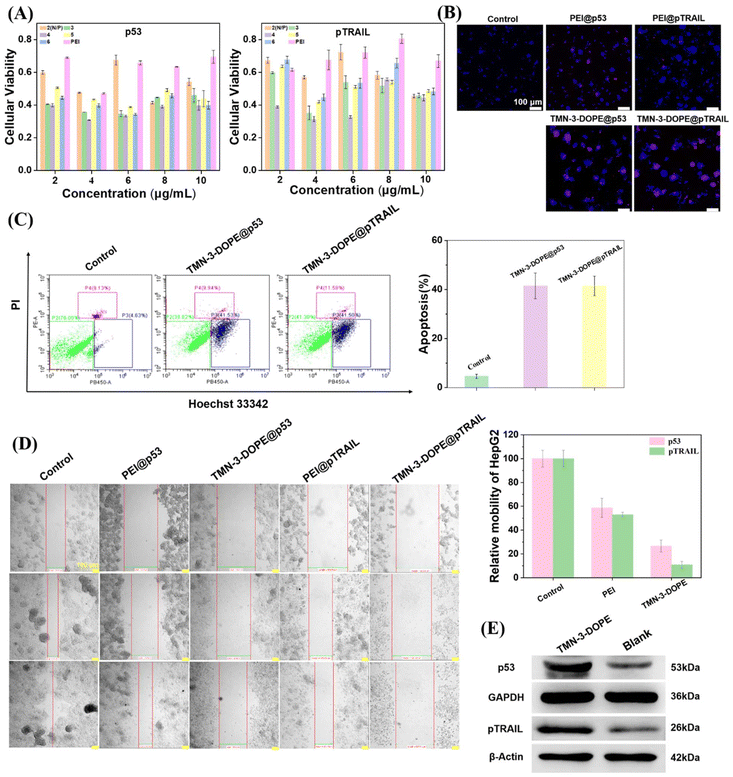
To maximize the practical application of TMN-3-DOPE, we explored its efficacy in targeting HepG2 cells by delivering tumor suppressor genes p53 and pTRAIL. Firstly, we evaluated the impact of different concentrations of the two tumor suppressor genes carried by TMN-3-DOPE on the survival rate of HepG2 cells using MTT assays. Notably, as depicted in Fig. 10A, at an N/P ratio of 4 and a mass concentration of 4 μg mL−1, both tumor suppressor genes exhibited significant apoptotic effects, with cell survival rates plummeting to only 30% (TMN-3-DOPE@p53) and 31% (TMN-3-DOPE@pTRAIL). These observations were further corroborated through cell cloning and scratch experiments. The proliferation ability of HepG2 cells markedly decreased, yielding a limited number of clones (Fig. S13†). Qualitative and quantitative analysis further substantiated the potent effects of TMN-3-DOPE carrying two different tumor suppressor genes on inducing apoptosis or necrosis of HepG2 cells by live–death staining (Fig. 10B) and cell flow cytometry (Fig. 10C). Both TMN-3-DOPE@p53 and TMN-3-DOPE@pTRAIL demonstrated the capacity to induce high levels of apoptosis in HepG2 cells, with the number of apoptotic cells reaching 41.5%. Moreover, the invasion and migration abilities of HepG2 cells were notably impaired following treatment, with migration rates of 26.6% for TMN-3-DOPE@p53 and 10.8% for TMN-3-DOPE@pTRAIL, as depicted in Fig. 10D. In addition, western blotting experiments also showed that the vector TMN-3-DOPE was able to effectively transfect HepG2 cells to produce the p53 protein and the pTRAIL protein (Fig. 10E).3. Conclusions
In summary, three functional polyester-based vectors (TMN-1/2/3) have been synthesized by using valerolactones with varying lengths of hydrophobic alkyl chains and propargyl valerolactone as monomers, employing step-wise ring-opening polymerizations and click reactions. TMN-1/2/3-DOPE were able to effectively aggregate nucleic acids, shielding them from degradation while possessing the capability for reversible nucleic acid release. Moreover, TMN-1/2/3-DOPE proved to condense pUC18 plasmids into uniform spherical nanoparticles of approximately 150 nm, facilitating cellular uptake and enhancing the EPR effect. Biocompatibility assessments via MTT assays confirmed the compatibility of these polyester-based carriers, particularly evident in HepG2 cells with an N/P ratio of 7 resulting in an 80% cell survival rate. Notably, luciferase expression experiments demonstrated the specific selectivity of TMN-3-DOPE, exhibiting significantly higher transfection efficiency in HepG2 cells, reaching 18.9 times that of 25 kDa PEI. The superior transfection efficiency of TMN-3-DOPE in HepG2 cells was elucidated through cellular uptake, lysosomal escape, and nuclear tracking experiments. Specifically, TMN-3-DOPE's longer alkyl chains promoted enhanced membrane fusion and increased cellular uptake rates, while its rapid lysosomal escape in HepG2 cells facilitated shifting of DNA into the nucleus. In vitro anti-cancer experiments underscored the universal transfection capability of TMN-3-DOPE for various tumor suppressor genes in HepG2 cells, demonstrating effective anti-cancer activity regardless of the delivered gene.Author contributions
Xi Zhang: synthesis, property measurement, data collection and analysis, writing – original draft preparation; De-Zhong Xu: property measurement, data collection and analysis; Zi-Han Zhang: property measurement, data collection and analysis; Zhi-Xuan Ren: synthesis, property measurement; Jia-Rao Wu: property measurement, data collection; Yun-Jie Xu: property measurement, data collection; Rui Liu: visualization, investigation, supervision; Zhong-Lin Lu: conceptualization, investigation, writing – reviewing and editing; Yang Liu: conceptualization, investigation, writing – reviewing.Data availability
The data are available in the ESI† and on request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Beijing Municipal Natural Science Foundation, China (2232011 to Z-LL) and the National Natural Science Foundation of China (21801020 to RL).References
- L. Naldini, Nature, 2015, 526, 351–360 CrossRef CAS PubMed.
- C. E. Dunbar, K. A. High, J. K. Joung, D. B. Kohn, K. Ozawa and M. Sadelain, Science, 2018, 359, 175 CrossRef CAS PubMed.
- F. Arabi, V. Mansouri and N. Ahmadbeigi, Biomed. Pharmacother., 2022, 153, 113324 CrossRef CAS PubMed.
- C.-C. Ma, Z.-L. Wang, T. Xu, Z.-Y. He and Y.-Q. Wei, Biotechnol. Adv., 2020, 40, 107502 CrossRef CAS PubMed.
- M. E. Gore and M. K. Collins, Eur. J. Cancer, 1994, 30, 1047–1049 CrossRef PubMed.
- L. Tortajada, C. Felip-León and M. J. Vicent, Polym. Chem., 2022, 13, 718–735 RSC.
- D. Ibraheem, A. Elaissari and H. Fessi, Int. J. Pharm., 2014, 459, 70–83 CrossRef CAS PubMed.
- Y. Zhang, C. Sun, C. Wang, K. E. Jankovic and Y. Dong, Chem. Rev., 2021, 121, 12181–12277 CrossRef CAS PubMed.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Nat. Rev. Genet., 2014, 15, 541–555 CrossRef CAS PubMed.
- J. Kaiser, Science, 2020, 367, 131–131 CrossRef CAS PubMed.
- D. W. Pack, A. S. Hoffman, S. Pun and P. S. Stayton, Nat. Rev. Drug Discovery, 2005, 4, 581–593 CrossRef CAS PubMed.
- D. Wang, P. W. L. Tai and G. Gao, Nat. Rev. Drug Discovery, 2019, 18, 358–378 CrossRef CAS PubMed.
- R. Mohammadinejad, A. Dehshahri, V. S. Madamsetty, M. Zahmatkeshan, S. Tavakol, P. Makvandi, D. Khorsandi, A. Pardakhty, M. Ashrafizadeh, E. G. Afshar and A. Zarrabi, J. Controlled Release, 2020, 325, 249–275 CrossRef CAS PubMed.
- R. Kumar, C. F. Santa Chalarca, M. R. Bockman, C. Van Bruggen, C. J. Grimme, R. J. Dalal, M. G. Hanson, J. K. Hexum and T. M. Reineke, Chem. Rev., 2021, 121, 11527–11652 CrossRef CAS PubMed.
- N. Kamaly, B. Yameen, J. Wu and O. C. Farokhzad, Chem. Rev., 2016, 116, 2602–2663 CrossRef CAS PubMed.
- J. Hao, P. Kos, K. Zhou, J. B. Miller, L. Xue, Y. Yan, H. Xiong, S. Elkassih and D. J. Siegwart, J. Am. Chem. Soc., 2015, 137, 9206–9209 CrossRef CAS PubMed.
- Y. Li, R. Tian, J. Xu, Y. Zou, T. Wang and J. Liu, Polym. Chem., 2024, 15, 1908–1931 RSC.
- Q. Wan, Y. Sun, X. Sun and Z. Zhou, Polym. Chem., 2024, 15, 2437–2456 RSC.
- Y. Zhang, M. Zhang, H. Song, Q. Dai and C. Liu, Small Methods, 2024, 2400278 CrossRef PubMed.
- W. Yang, L. Mixich, E. Boonstra and H. Cabral, Adv. Healthcare Mater., 2023, 12, 2202688 CrossRef CAS PubMed.
- J. Chen, K. Wang, J. Wu, H. Tian and X. Chen, Bioconjugate Chem., 2019, 30, 338–349 CrossRef CAS PubMed.
- N. Veiga, M. Goldsmith, D. Rosenblum, N. Dammes, R. Kedmi, S. Ramishetti and D. Peer, Nat. Commun., 2018, 9, 4493 CrossRef PubMed.
- F. Tang, Q. Wang, Y.-N. Gao, Y.-S. Zhang, Y.-X. Liang, Z.-L. Lu, R. Liu and A.-X. Ding, Bioconjugate Chem., 2022, 33, 929–937 CrossRef CAS PubMed.
- X. Yang, J. Chang, Y. Jiang, Q. Xu, M. Wang and L. Mao, CCS Chem., 2021, 3, 780–790 CrossRef CAS.
- Z. Chen, Z. Wang and Z. Gu, Acc. Chem. Res., 2019, 52, 1255–1264 CrossRef CAS PubMed.
- L. Li, Z. Yang and X. Chen, Acc. Chem. Res., 2020, 53, 2044–2054 CrossRef CAS PubMed.
- K. A. Hajj, J. R. Melamed, N. Chaudhary, N. G. Lamson, R. L. Ball, S. S. Yerneni and K. A. Whitehead, Nano Lett., 2020, 20, 5167–5175 CrossRef CAS PubMed.
- X. Zhang, B. Lin, X. Luo, W. Zhao, J. Jiang, C. Zhang, M. Gao, X. Chen and Y. Dong, Appl. Mater. Interfaces, 2017, 9, 25481–25487 CrossRef CAS PubMed.
- M. L. Ibba, G. Ciccone, C. L. Esposito, S. Catuogno and P. H. Giangrande, Adv. Drug Delivery Rev., 2021, 177, 113930 CrossRef CAS PubMed.
- C. J. McKinlay, J. R. Vargas, T. R. Blake, J. W. Hardy, M. Kanada, C. H. Contag, P. A. Wender and R. M. Waymouth, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 448–456 CrossRef PubMed.
- H. Ding, P. Tan, S. Fu, X. Tian, H. Zhang, X. Ma, Z. Gu and K. Luo, J. Controlled Release, 2022, 348, 206–238 CrossRef CAS PubMed.
- S. A. Dilliard and D. J. Siegwart, Nat. Rev. Mater., 2023, 8, 282–300 CrossRef CAS PubMed.
- J. Tian, L. Ding, H.-J. Xu, Z. Shen, H. Ju, L. Jia, L. Bao and J.-S. Yu, J. Am. Chem. Soc., 2013, 135, 18850–18858 CrossRef CAS PubMed.
- M. Durymanov and J. Reineke, Front. Pharmacol., 2018, 9, 971 CrossRef PubMed.
- A. B. Hill, M. Chen, C. K. Chen, B. A. Pfeifer and C. H. Jones, Trends Biotechnol., 2016, 34, 91–105 CrossRef CAS PubMed.
- I. M. S. Degors, C. Wang, Z. U. Rehman and I. S. Zuhorn, Acc. Chem. Res., 2019, 52, 1750–1760 CrossRef CAS PubMed.
- H. B. Cheng, X. Cao, S. Zhang, K. Zhang, Y. Cheng, J. Wang, J. Zhao, L. Zhou, X. J. Liang and J. Yoon, Adv. Mater., 2022, e2207546 Search PubMed.
- S. Bose, A. H. Ngo and L. H. Do, J. Am. Chem. Soc., 2017, 139, 8792–8795 CrossRef CAS PubMed.
- M. Morille, C. Passirani, A. Vonarbourg, A. Clavreul and J. P. Benoit, Biomaterials, 2008, 29, 3477–3496 CrossRef CAS PubMed.
- L. Song, A. X. Ding, K. X. Zhang, B. Gong, Z. L. Lu and L. He, Org. Biomol. Chem., 2017, 15, 6567–6574 RSC.
- X. Y. Liu, J. B. Yang, T. T. Duan, C. Y. Wu, Q. Tang, Z. L. Lu, L. He and W. Sun, Bioorg. Chem., 2021, 116, 105299 CrossRef CAS PubMed.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed.
- H. T. Phan and A. J. Haes, J. Phys. Chem. C, 2019, 123, 16495–16507 CrossRef CAS PubMed.
- H. Lv, S. Zhang, B. Wang, S. Cui and J. Yan, J. Controlled Release, 2006, 114, 100–109 CrossRef CAS PubMed.
- J.-Y. Liu, X.-Y. Liu, R. Liu, F. Tang, J.-B. Yang, Q. Tang, Z.-L. Lu, H.-J. Qiao and L. He, Bioorg. Chem., 2021, 113, 104983 CrossRef CAS PubMed.
- X. Liu, J. Xiang, D. Zhu, L. Jiang, Z. Zhou, J. Tang, X. Liu, Y. Huang and Y. Shen, Adv. Mater., 2016, 28, 1743–1752 CrossRef CAS PubMed.
- Z. Zhang, N. Qiu, S. Wu, X. Liu, Z. Zhou, J. Tang, Y. Liu, R. Zhou and Y. Shen, Adv. Mater., 2021, 33, 2102219 CrossRef CAS PubMed.
- Q. Jiang, Y. Nie, X. Chen, Y. He, D. Yue and Z. Gu, Adv. Funct. Mater., 2017, 27, 1701571 CrossRef.
- C. Chai, M. Rivkin, L. Berkovits, A. Simerzin, E. Zorde-Khvalevsky, N. Rosenberg, S. Klein, D. Yaish, R. Durst, S. Shpitzen, S. Udi, J. Tam, J. Heeren, A. Worthmann, C. Schramm, J. Kluwe, R. Ravid, E. Hornstein, H. Giladi and E. Galun, Gastroenterology, 2017, 153, 1404–1415 CrossRef CAS PubMed.
- K. T. Ong, M. T. Mashek, S. Y. Bu, A. S. Greenberg and D. G. Mashek, Hepatology, 2011, 53, 116–126 CrossRef CAS PubMed.
- Z. Y. Ong, C. Yang, W. Cheng, Z. X. Voo, W. Chin, J. L. Hedrick and Y. Y. Yang, Acta Biomater., 2017, 54, 201–211 CrossRef CAS PubMed.
- L. Zhu, J. M. Simpson, X. Xu, H. He, D. Zhang and L. Yin, Appl. Mater. Interfaces, 2017, 9, 23476–23486 CrossRef CAS PubMed.
- H. Liu, X. Zhao, Y. Bai, H. Xie, Z. Yang, Y. Wang, Y. Chen, Y. Luo, M. Ma, W. Lu, J. Ma, T. Yang, B. Jin and P. Ding, Mater. Des., 2022, 219, 110758 CrossRef CAS.
- X. Wang, Y. Qiu, M. Wang, C. Zhang, T. Zhang, H. Zhou, W. Zhao, W. Zhao, G. Xia and R. Shao, Int. J. Nanomed., 2020, 15, 9447–9467 CrossRef CAS PubMed.
- K. L. Douglas, Biotechnol. Prog., 2008, 24, 871–883 CrossRef CAS PubMed.
- R. A. Petros and J. M. DeSimone, Nat. Rev. Drug Discovery, 2010, 9, 615–627 CrossRef CAS PubMed.
- D. Pei and M. Buyanova, Bioconjugate Chem., 2019, 30, 273–283 CrossRef CAS PubMed.
- A. K. Varkouhi, M. Scholte, G. Storm and H. J. Haisma, J. Controlled Release, 2011, 151, 220–228 CrossRef CAS PubMed.
- J. Friedrich, C. Seidel, R. Ebner and L. A. Kunz-Schughart, Nat. Protoc., 2009, 4, 309–324 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4py00610k |
| This journal is © The Royal Society of Chemistry 2024 |