Open Access Article
Open Access ArticleDrug-delivery and biological activity in colorectal cancer of a supramolecular porous material assembled from heptameric chromium-copper-adenine entities†
Sandra
Mena-Gutiérrez
 a,
Ekain
Maiza-Razkin
a,
Jon
Pascual-Colino
a,
Ekain
Maiza-Razkin
a,
Jon
Pascual-Colino
 ab,
Marcos J.
Araúzo-Bravo
ab,
Marcos J.
Araúzo-Bravo
 cde,
Garikoitz
Beobide
ab,
Oscar
Castillo
cde,
Garikoitz
Beobide
ab,
Oscar
Castillo
 *ab,
Ainara
Castellanos-Rubio
cfg,
Daniela
Gerovska
*ab,
Ainara
Castellanos-Rubio
cfg,
Daniela
Gerovska
 d,
Antonio
Luque
d,
Antonio
Luque
 *ab and
Sonia
Pérez-Yáñez
*ab and
Sonia
Pérez-Yáñez
 ab
ab
aDepartamento de Química Orgánica e Inorgánica, Facultad de Ciencia y Tecnología, Universidad del País Vasco/Euskal Herriko Unibertsitatea, UPV/EHU, Apartado 644, E-48080 Bilbao, Spain. E-mail: oscar.castillo@ehu.eus; antonio.luque@ehu.eus
bBCMaterials, Basque Center for Materials, Applications and Nanostructures, UPV/EHU Science Park, E-48940 Leioa, Spain
cIKERBASQUE, Basque Foundation for Science, E-48011, Bilbao, Spain
dComputational Biology and Systems Biomedicine Research Group, Biogipuzkoa Health Research Institute, Donostia, Spain
eDepartment of Cell Biology and Histology, Faculty of Medicine and Nursing, University of Basque Country (UPV/EHU), Spain
fBiobizkaia Research Institute, E-480903 Barakaldo, Bizkaia, Spain
gDepartamento de Genetica, Antropologia Fisica y Fisiologia Animal, UPV-EHU, E-48940 Leioa, Bizkaia, Spain
First published on 27th September 2024
Abstract
The therapeutic application of drugs often faces challenges due to non-specific distribution, inadequate dosification and degradation, which limits their efficacy. Two primary strategies are employed to overcome these issues: the use of derivatives of the active substances and incorporation of those into porous materials. The latter, involving materials such as zeolites, metal–organic frameworks (MOFs), and hydrogels, has shown promising results in protecting the active ingredients from degradation and enabling a controlled release. This study focuses on supramolecular metal–organic frameworks (SMOFs), which leverage supramolecular interactions for enhanced pore size control. [Cu6Cr(μ-adeninato-κN3:κN9)6(μ3-OH)6(μ-OH2)6](SO4)1.5·nH2O (Cu6Cr) was chosen for its flexible porous structure, water-stability, and paramagnetic properties. Magnetic sustentation studies showed that this compound was able to capture several drug molecules: 5-fluorouracil (5-FU), 5-aminosalicylic acid (5-ASA), 4-aminosalicylic acid (4-ASA) and theophylline (THEO). Their release follows a pseudo-first-order kinetics with desorption half-lives ranging from 2.2 to 4.7 hours. In this sense, a novel approach is proposed using bulkier raffinose and cholesterol as pore-blocking molecules. Cholesterol exhibited the best performance as a blocking molecule increasing the desorption half-life up to 8.2 hours. Cytotoxicity and RNA-seq transcriptomic assays carried out on human colorectal cancer cell cultures showed, on one hand, that the Cu6Cr porous material exhibits a proliferative effect, probably coming from the over-expression of MIR1248 and SUMO2 genes, and on the other hand, that there is a delay in the emergence of the cytotoxicity of 5-FU as expected for a slower release.
1. Introduction
The use of drugs to treat diseases remains an area requiring ongoing development and improvement, as their distribution often lacks specificity, which hinders reaching the therapeutic target fully and limits their practical application.1 Despite demonstrating efficacy in in vitro experiments, certain active ingredients may encounter degradation or distribution issues when translated into real treatments.2 Two main strategies have been proposed to address this challenge. The first option involves using derivatives of the active substance, which can facilitate its release upon degradation in the body.3 However, this approach is limited due to the unique development required for each active substance, variable chemical conditions within the body, or the involvement of symbiont bacteria. Therefore, an extensively researched second option involves incorporating the active substance into a porous material,4 which not only prevents its degradation but also enables a controlled release.Adsorbent materials employed for this purpose include zeolites,5,6 MOFs,7,8 and hydrogels,9 which also have diverse applications beyond drug delivery7,10,11 and have been the subject of extensive research over the last few decades. In this study, we focus on SMOFs, a variant of MOFs where supramolecular interactions replace covalent bonds connecting adjacent clusters.12 The utilization of these materials is increasingly successful due to the regularity and flexibility of their pore size, offering better control over both adsorption and controlled drug release.13 SMOFs provide some properties that are not so common for MOFs such as structural flexibility, adaptability to the guest molecules and reversible solubilization/recrystallization under mild conditions. These features can also have an impact in many areas, including photocatalytic production of H2 as reported by Tian et al.14 Supramolecular interactions also play a key role in creating supramolecular nanovalves by functionalizing the external surface of a particle with linear molecules that are supramolecularly bound to cyclodextrin or cyclodextrin-like molecules as gate keepers.15 A clear example of the latter is the stimuli-responsive supramolecular delivery systems (SDSs) based on pillararenes for tumor therapy.16
This paper employs a previously reported material with formula [Cu6Cr(μ-adeninato-κN3:κN9)6(μ3-OH)6(μ-OH2)6](SO4)1.5·nH2O17 (Cu6Cr) which is a porous, water-stable, and insoluble compound exhibiting paramagnetic behaviour at room temperature due to its metal centers. The room temperature paramagnetism of this compound allows us to quantify drug adsorption and release using a technique also developed by our research group.13,18,19 This technique employs a magnetic field to attract the paramagnetic particles to the pole of an electromagnet. The critical magnetic field value, at which the particles are detached from the electromagnet, correlates with the amount of adsorbate within the material. It is worth mentioning that in one of our previous works where we used compounds related to Cu6Cr, they showed a significant ability to capture small molecules, including various drugs, and to deliver them, which underscored the potential of these types of compounds.13 However, the release of the drug molecules was still too fast for these materials to constitute an efficient drug delivery system. Therefore, this study will provide a new approach to slow down the drug release rates. To illustrate this approach, the drugs shown in Scheme 1 have been selected. 5-ASA is utilized in managing inflammatory bowel diseases such as ulcerative colitis and Crohn's disease.20 4-ASA is a drug for the treatment of tuberculosis.21 5-FU is an antimetabolite medication primarily employed in the treatment of colorectal cancer.22 THEO is used as a bronchodilator in the treatment of asthma and chronic obstructive pulmonary disease (COPD).23
Herein, we found that the Cu6Cr compound presents good adsorption capacities for these drug molecules but a kinetic release profile that needs to be slowed down to get a more controlled and less abrupt drug delivery. The release kinetics of a specific molecule depends on its shape and size but also on the complex interactions taking place between the molecule and the carrier and between the molecules themselves.24,25 Therefore, achieving the desired release kinetics remains a challenge in many cases. Several strategies have been developed to slow down the release kinetics,26 including coating particles with degradable polymers,27 chemically modifying pore openings to create stimuli-sensitive porous cages,28 or using swellable hydrogels.29 Our study employs a novel approach in which a second molecule is employed in addition to the loaded drug molecule to block the particle's external pore aperture and delay the release of the drug, an approach previously employed for other types of molecules.30 The key to the success of this approach is the careful selection of this second molecule, as it needs to be small enough to interact with the particle's external aperture of the pores but big enough to prevent it from getting deep into the pore channels. In addition, it should present a chemical affinity for the surface of the porous material. All in all, raffinose (RAFF), a hydrophilic and soluble compound, and cholesterol (CHOL), a hydrophobic and highly insoluble lipid, are tested for this role of the goalkeeper of the particle's external pore aperture (Scheme 2). Additionally, the biocompatibility of Cu6Cr porous material is tested using cytotoxicity and RNA-seq transcriptomics assays carried out on human colorectal cancer cell cultures. It is worth noting that the underlying cellular mechanisms that are behind the effect this class of materials (MOFs and SMOFs) produces on the cells are still rarely explored31 and using transcriptomic analysis allows us to monitor the drug effects by interrogating all the genes of the genome.
2. Experimental section
2.1. Chemicals
The following reagents were used for the preparation of the Cu6Cr compound: copper(II) sulfate pentahydrate (Sigma-Aldrich, ≥98%), chromium(III) potassium sulfate dodecahydrate (Sigma-Aldrich, ≥98%), adenine (Sigma-Aldrich, >99%), methanol (Labkem, AGR), triethylamine (Sigma-Aldrich, ≥99%) and sulphuric acid (Labkem, 25% v/v). The used drugs were as follows: 5-FU, (TCI, >99%), 5-ASA (Sigma-Aldrich, ≥99%), 4-ASA (Sigma-Aldrich, 99%), and THEO (Sigma-Aldrich, ≥98%). Molecules used as calibration standards were D-(+)-glucose (Sigma-Aldrich, >99.5%), benzyl alcohol (Sigma-Aldrich, ≥99%) and isopropanol (Labkem, >99.9%). The used pore-blocking molecules were D-(+)-raffinose pentahydrate (Sigma-Aldrich, ≥99%) and cholesterol (Sigma-Aldrich, ≥99%).2.2. Synthesis of compound Cu6Cr: [Cu6Cr(μ-adeninato-κN3:κN9)6(μ3-OH)6(μ-OH2)6](SO4)1.5·17H2O
The synthesis of the compound Cu6Cr was carried out using a previously reported procedure changing the chromium salt with chromium(III) potassium sulfate dodecahydrate.17 A methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water mixture (20 mL, 1:1) of adenine (0.108 g, 0.8 mmol), heated to 70 °C, was added to a solution containing Cu(SO4)·5H2O (0.200 g, 0.8 mmol) and CrK(SO4)2·12H2O (0.100 g, 0.2 mmol) in 20 mL of deionized water. The resulting green suspension, pH 3.0, was dissolved through acidification with sulphuric acid until obtaining a clear green solution (pH ∼1.5). After that, the solution was basified (pH ∼ 9.2) by adding triethylamine, and a green suspension was obtained. Triethylamine was employed to achieve the desired pH and also acted as a modulator to obtain the final compound structure. After 2–3 days, the suspension recrystallized as green needle-shaped crystals (yield: 70–75% based on copper).
water mixture (20 mL, 1:1) of adenine (0.108 g, 0.8 mmol), heated to 70 °C, was added to a solution containing Cu(SO4)·5H2O (0.200 g, 0.8 mmol) and CrK(SO4)2·12H2O (0.100 g, 0.2 mmol) in 20 mL of deionized water. The resulting green suspension, pH 3.0, was dissolved through acidification with sulphuric acid until obtaining a clear green solution (pH ∼1.5). After that, the solution was basified (pH ∼ 9.2) by adding triethylamine, and a green suspension was obtained. Triethylamine was employed to achieve the desired pH and also acted as a modulator to obtain the final compound structure. After 2–3 days, the suspension recrystallized as green needle-shaped crystals (yield: 70–75% based on copper).
2.3. Methods
The purity of the bulk samples was checked by powder X-ray diffraction (PXRD) and Fourier-transform infrared (FTIR) spectroscopy. The crystals undergo a partial dehydration when extracted from the mother liquor; therefore, the PXRD experiments were performed over samples introduced in a Lindemann capillary filled with the synthesis mother liquor. A Rigaku Smartlab automatic diffractometer with a capillary fixation head was employed and the diffraction data were collected in continuous rotation in the range 3°< 2θ< 65°. Routine PXRD measurements were performed using a Phillips X’PERT diffractometer (equipped with Cu–Kα radiation, λ = 1.5418 Å) over the range 5 < 2θ < 70° with a step size of 0.02°, a variable automatic divergence slit, and an acquisition time of 2.5 s per step at 293 K (Fig. S1, ESI†). Magnetic sustentation adsorption quantification was performed using a dipole electromagnet (Newport Pagnell England Electromagnet Type C sourced by a Hewlett-Packard 6655A system DC power supply) to determine the critical magnetic field at which the particles are detached from the electromagnet pole for the unloaded and loaded samples.13–16 Each measurement was repeated 5 times to provide the corresponding associated error. The data to obtain the calibration curve were determined by 1H-NMR spectroscopy using a Bruker AVANCE 500 (one-bay; 500 MHz) spectrometer at 293 K after duplicating the adsorption procedure using D2O instead of water and using an internal standard.2.4. Cytotoxicity and RNA-seq transcriptomic analysis
To test the cytotoxicity, the human colorectal cancer line HCT116 (ATCC–CCL247) was employed. Cells were seeded at 3000 cells per well in 96-well plates and incubated at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and antibiotics. After overnight incubation, the compounds were added to the cells and the cells were collected for crystal violet staining at 12, 24, 48, 72, 84 and 108 h incubation time points. Subsequently, the incubated cells were fixed with paraformaldehyde and stained with 0.1% crystal violet. The staining was washed 3 times; 10% acetic acid was added, and plates were incubated by shaking for 20 min. Absorbance was measured at 590 nm and relative proliferation was calculated. The following concentrations of the compounds were used: 2.00 μg/100 μL of compound Cu6Cr and 0.77 μg/100 μL of 5-FU. For the 5-FU@Cu6Cr, RAFF@5-FU@Cu6Cr, and CHOL@5-FU@Cu6Cr components, 2.77 μg/100 μL was used to ensure equal concentration and obtain comparative results.For RNA sequencing studies, 30.000 cells were seeded in 12 well plates, compounds were added to the cells after overnight incubation and cells were grown for 72 h. Three subsequent cell passages were used for this experiment. After incubation, RNA was extracted from the cultures using the NucleoSpin RNA kit (Macherey Nagel) and submitted for RNA sequencing analyses to the Genomic Platform of the Biobizkaia Health Research Institute (Barakaldo, Spain).
RNA-sequencing (RNA-seq) transcriptomics data analysis was conducted as previously described.32 We used HISAT233 to align the RNA-seq reads to the human reference genome hg38 and to calculate the Fragments Per Kilobase of transcript per Million fragments mapped (FPKMs) and Cufflinks34 to annotate them. We merged the transcriptomics data into a single text file and used it in the downstream analysis using in-house functions developed in MATLAB (MathWorks).35 We equalized the data and stabilized them through the log2 transform of the data plus one – to avoid the undefined values of log2 of zero; calculated the average values for each of the two groups: NT and Cu6Cr of three biological replicates each; selected the differentially expressed genes (DEGs) whose absolute difference in mean values between the two groups was less than the selection threshold θDEG = 1 of the fold change (FC) in the log2 scale; and selected the statistically significant DEGs using an unpaired Student's t test with equal variances and with a significance threshold of αDEG = 0.05. DEG sets were subjected to gene ontology (GO) enrichment analysis (https://geneontology.org). The functional GO enrichment of the DEGs was calculated using the hypergeometric test over the mid-p-values with a significance threshold of αGO = 0.05.
3. Results and discussion
3.1. Crystal structure
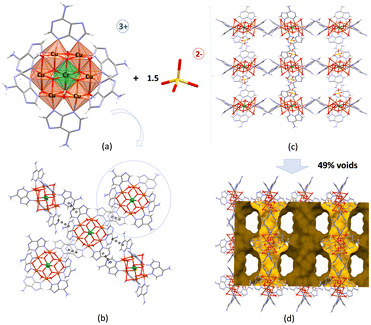
The compound Cu6Cr crystallizes in the monoclinic C2/c space group and contains rigid wheel-shaped [Cu6Cr(μ-adeninato)6(μ3-OH)6(μ-OH2)6]3+ cations consisting of a central [Cr(OH)6]3− core connected to six outer copper(II) metal centers through μ3-hydroxido bridges in a radial and planar arrangement (Fig. 1a).17 The external copper atoms are connected by water molecules and bidentate μ-κN3:κN9 adeninato ligands.13,36 In the crystal structure, each heptameric cation is surrounded by four other ones and held together by adeninato–adeninato offset face-to-face π–π stacking interactions (Fig. 1b). The sulphate counterions are anchored to each cationic entity by means of hydrogen bonds involving the water and hydroxido coordination sites as donors (Fig. 1c). This supramolecular assembling leads to a three-dimensional network of interconnected pores (Table S1 and Fig. S2, ESI†) with a total volume of 4509 Å3 which represents ca. 50% of the volume of the crystallographic unit cell (Fig. 1d). The walls of these pores are essentially lined by the exocyclic amino group and the non-coordinated N sites of the adeninato ligands together with the oxygen atoms of the sulphate anion. The pores of the as-prepared compound are filled by the crystallization water molecules.3.2. Drug sorption and delivery capacity
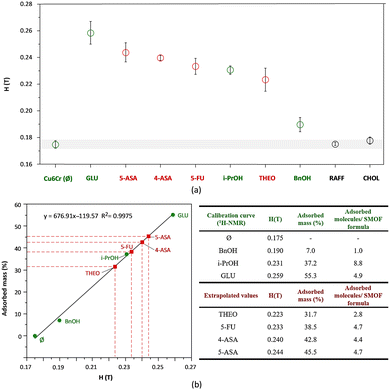
The viability of Cu6Cr as a controlled drug delivery system has been tested by incorporating various pharmaceutically active molecules such as the antitumour 5-FU, the antibiotics 4-ASA and 5-ASA, and the bronchodilator THEO. The drug loadings were carried out from an aqueous solution and monitored by a magnetic sustentantion technique that enables to monitor in-solution sorption processes.13,18 This method allowed the amount of guest molecules incorporated in the cavities of Cu6Cr to be determined using its intrinsic room temperature paramagnetism. The procedure is based on the determination of the critical magnetic field (H) required to keep the particles of this material attached to the poles of a magnet, which correlates with the loaded mass percentage. The greater the deviation from the critical magnetic value of the pristine material, the greater the mass of the trapped guest molecules. Unlike classical quantification methods (1H-NMR, ultraviolet-visible (UV-vis) spectroscopy or chromatography), this technique enables the in situ analysis of sorption values and also tests the adsorption of not completely soluble analytes, as many drugs are, in aqueous media.The magnetic sustentation method requires a calibration curve for the adsorbent, which usually implies determining the adsorption of the calibration standards by any other technique, 1H-NMR in this work.
The calibration samples were prepared using three soluble molecules: benzyl alcohol (BnOH), propan-2-ol (i-PrOH), and glucose (GLU) in addition to the data corresponding to the pristine Cu6Cr. The procedure consisted of placing 50 mg of Cu6Cr and 50 mg of the adsorptive molecules in 2 mL of water (deionized water for the magnetic sustentation measurements and deuterated water for the 1H-NMR data acquisition) and shaking at a rotatory mixer the resulting mixture for one day at 30 °C. Magnetic sustentation measurements were performed in deionized water to determine the critical magnetic field for the adsorbate@Cu6Cr particles. Simultaneously, the amount of loaded adsorbate in terms of mass percentage was calculated from the 1H-NMR spectra of the initial and final deuterated aqueous solution resulting from the adsorption process. To do this, 1H-NMR data acquisition was performed with 1 mL of deuterated water and 100 μL of a 5% solution of tert-butanol (t-BuOH) (Fig. S4–S6, ESI†) was added as an internal standard. Representing the data of the adsorbed mass percentage against the critical magnetic field, a linear relationship must be observed, as shown in Fig. 2.
After the calibration line was obtained, the loaded mass percentage for the rest of the molecules, drugs included, was calculated from the interpolation of their critical magnetic field value on this line.
The obtained adsorption values for each drug molecule seem to correlate with the size (Fig. S11, ESI†) of their aromatic system. Those containing a single ring have higher adsorption mass percentages (38–45%) under these conditions than THEO (32%), which contains fused hexagonal and pentagonal rings (Scheme 1).
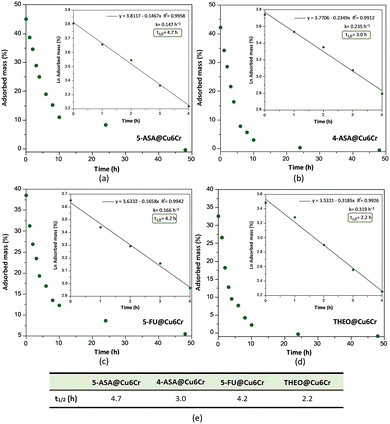
In addition to determining the adsorption capacity of Cu6Cr with respect to these drug molecules, desorption kinetic studies were performed for all of them in water at 35 °C (Fig. 3). In this sense, the critical magnetic field was measured every hour during the first 4 h, then measurements were taken every two hours until the tenth hour and finally at 24 and 48 h. After each measurement, the aqueous medium was replaced with fresh deionized water. The time-dependent desorption curves seem to follow a pseudo-first-order kinetic, at least during the first stages of desorption (the first 4 hours). The kinetic constant and the derived desorption half-live for each drug molecule are shown in the insets of Fig. 3. The desorption half-lives range between 2.2 and 4.7 h but no clear correlation with the molecular size is observed in this case.
3.3. Slowing the desorption kinetics
The above-obtained desorption kinetics could be still too fast for some potential applications related to the controlled release of the drug molecules. Therefore, at this point, we started looking for a method that could slow down the intrinsic desorption rate of these drug@Cu6Cr samples. Although there are different strategies that can be used to modify the desorption process, in this work, we aimed to use a new approach that can be described, using an analogy, as “tap in the bottle”. The goal of this approach is to identify molecules that are small enough to interact with the outer particle apertures of the pores, but large enough to prevent their diffusion into the inner particle pore system (Fig. 4). The magnetic sustentation technique is very helpful in this task, because the molecules that are more likely to act as a cap should present a critical magnetic field which is just above the value for the pristine porous material. Of all the possible candidate molecules, those with a biocompatible nature are the most interesting ones. Raffinose (RAFF), a natural trisaccharide, and cholesterol (CHOL) provided the most promising results for this purpose. | ||
| Fig. 4 (a) Schematic representation of the capture and release processes without a capping molecule and (b) using cholesterol as a release controller of the “tap in a bottle” approach. | ||
These two molecules (with van der Waals dimensions of 440 and 420 Å3 for raffinose and cholesterol, respectively) are relatively large (Fig. S12, ESI†) to diffuse through the pore system of the material (with pore apertures of 4.6 and 6.3 Å of diameter). Therefore, they will only be adsorbed at the near-surface level, which explains the only slight increase in the critical magnetic field. Despite their similarities in size, raffinose and cholesterol differ chemically. Raffinose is a highly hydrophilic molecule that is highly soluble in water, while cholesterol is hydrophobic and relatively insoluble in water. This different chemical nature will probably affect their discharge from the blocked pore entrance where they are stacked. Of all the adsorbed drugs, we have used 5-FU for this study because of its wide use as an antitumour agent. The experimental procedure to accomplish this “tap in a bottle” approach consisted of two stages. In the first stage, the porous matrix was loaded with 5-FU at 30 °C for 24 hours, as previously explained (5-FU@Cu6Cr). In the second stage, the supernatant solution was removed, and 2 mL of a 0.04 M aqueous solution of raffinose or 2 mL of a saturated solution of cholesterol were added and stirred for 1 h at 30 °C. The critical magnetic field was measured at the end of both stages not observing any significant change between both values. The latter indicates that 5-FU has not been released during the second stage.
The desorption kinetics were measured for the resulting samples (RAFF@5-FU@Cu6Cr and CHOL@5-FU@Cu6Cr), observing a delay in the release kinetic (Fig. 5). In the case of the raffinose-blocked sample, the t1/2 value slightly increases from 4.2 h to 5.2 h. However, the cholesterol-blocked sample shows a qualitatively different behaviour. There is a 1 h induction time during which there is no significant release of the captured mass. Later, the previously explained pseudo-first order desorption kinetic is followed with a t1/2 value of 7.2 h. This behaviour is attributed to the presence of a layer of cholesterol covering the external surface of the particles, because of its hydrophobicity, which would be responsible for the observed induction time. This coating is added to the “tap in a bottle” effect that would be responsible for the observed smaller desorption kinetic constant. Consequently, the time required for the release of half of the 5-FU drug content is the sum of the induction time (1 h) and the desorption kinetic t1/2 (7.2 h), a total of 8.2 h (Fig. 6).
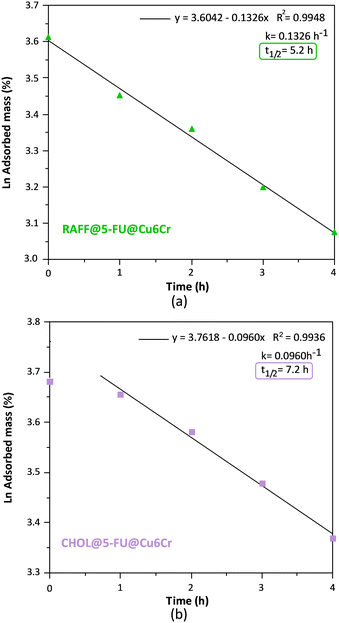 | ||
| Fig. 6 Plots of the fit to the first-order kinetics considering the first 4 h of the desorption process of the samples (a) RAFF@5-FU@Cu6Cr and (b) CHOL@5-FU@Cu6Cr. | ||
3.4. Cytotoxicity and RNA-seq transcriptomic analysis
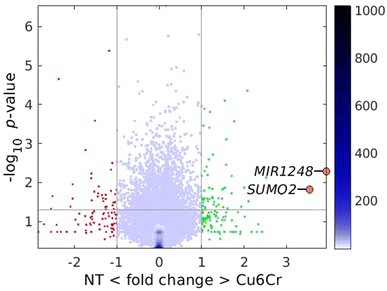
The porous substrate can present biological activity by itself and, prior to any drug delivery study on a cell culture, it is necessary to ensure which is the contribution of the substrate. For this purpose, cytotoxicity and transcriptomic (Fig. S13 and S14, ESI†) studies have been carried out on cultured cells exposed to the porous substrate (2 μg/100 μL). Interestingly, the porous substrate shows a positive proliferative effect on the growth of the human colorectal cancer line HCT116 (ATCC–CCL247), despite the copper and chromium content. A similar proliferative effect has also been reported in the literature for analogous copper(II) only heptameric entities based on the 7-azaindole ligand instead of adenine.37 As this behaviour is very interesting, it was decided to conduct whole genome transcriptomic studies. The comparison between the NT (non-treated) cells and the cells cultured in the presence of Cu6Cr does not show major differences (Fig. 7). However, a careful analysis of the more overexpressed genes: MIR1248 (“microRNA 1248”; FC = 3.9; p = 0.005) and SUMO2 (“small ubiquitin-like modifier 2”; FC = 3.6; p = 0.016) can provide some light on the reason for the greater proliferation rate observed in the Cu6Cr cultured cells. MIR1248 is a member of the microRNAs (miRNAs) which are short (20–24 nt) non-coding RNAs that are involved in the post-transcriptional regulation of gene expression. MIR1248 has been identified as having the potential to be used as a novel therapeutic target for wound healing in diabetes mellitus patients by restoring the wound-healing ability of diabetics human adipose-derived stem cells.38 SUMO2 gene codifies a ubiquitin-like protein that can be covalently attached to proteins as a monomer or as a lysine-linked polymer. This post-translational modification on lysine residues of proteins plays a crucial role in a number of cellular processes such as nuclear transport, DNA replication and repair, mitosis and signal transduction.39 Therefore, both overexpressed genes can have a role in the observed increased proliferation rate of the colon cancer-human cells.Finally, we conducted a study analysing the effect of the slower release of the cancer treatment drug 5-FU on cell cultivars. Prior to this study, the 5-FU@Cu6Cr, RAFF@5-FU@Cu6Cr, and CHOL@5-FU@Cu6Cr samples were purified from any remaining particles of the highly insoluble 5-FU. The room temperature paramagnetism of the SMOF enabled us to purify the sample by using the magnetic sustentation phenomenon to retain the 5-FU@Cu6Cr particles and remove the excess 5-FU present as diamagnetic particles. Pure samples of Cu6Cr and 5-FU were also tested for comparison. Cytotoxicity tests were conducted on all the aforementioned compounds. The results indicate (Fig. 8) a delay in the appearance of the cytotoxic effect, following the same order observed during desorption kinetic studies. The effect with the cytotoxic 5-FU drug alone is seen at 24 h, with the relative growth lying below the basal cell culture, but if you load it on the porous substrate (5-FU@Cu6Cr), this effect is delayed till 72 h. Blocking the external pores with raffinose (RAFF@5-FU@Cu6Cr) does not make a great change. The effect is only observable till 72 h, although the decrease in the cell growth with respect to the basal cell culture is not so notorious as with 5-FU alone. Finally, when using cholesterol (CHOL@5-FU@Cu6Cr), the cytotoxic effect only becomes evident after 84 h.
4. Conclusions
This work clearly illustrates the capacities of the supramolecular porous materials as drug delivery platforms with controllable release kinetics. Usually, these materials are not mechanically strong enough to withstand the removal of the solvent molecules located inside their pores, but they can be chemically very robust, as in the case of the Cu6Cr heptameric compound. This porous material retains its porosity while in water behaving as an efficient adsorbent material. The great amount of drug molecules and the more conventional molecules employed to generate the calibration line for the magnetic sustentation technique is evidence of this fact. Another feature common within the supramolecular porous materials, especially notorious in this material in which the crystal structure is sustained by π-stacking interactions, is their flexible nature. Since a previous work, we already knew that this compound can reversibly transit from a collapsed, non-porous and almost amorphous state when it is removed from the mother liquid to the crystalline, porous, open structure when exposed again to a humidity-saturated atmosphere or immersed in water. However, in this work, it has become clear that this structural flexibility also allows the crystal structure to change upon the incorporation of different molecules, especially the bigger ones, into its pores (as inferred from their powder X-ray diffraction patterns).This latter feature is extremely interesting because the flexibility of the supramolecular structure also means that this material can also accommodate, to some extent, molecules that are slightly greater than the pore aperture. These molecules are probably going to be trapped at the outer part of the channel system because of the distortion their presence generates on the supramolecular structure and the presence of narrower windows in the pore system that hinders the diffusion of these molecules. All in all, this class of relatively big molecules with chemical affinity towards the porous material can be employed to cap or deter the release of the molecules that have gone deeper into the channel system. The latter can also be understood as a means to slow the delivery of the adsorbate molecules which can be very interesting when translating it to the field of drug delivery systems. Furthermore, the above-detailed features that these bigger/capping molecules should have imply that their adsorbed amount necessarily must be very small. Fortunately, the magnetic sustentation technique provides an easy way to detect them: candidate molecules are those whose adsorption amounts are so low that their critical magnetic field is just slightly above the uncertainty range of the neat porous material. In this case, to test this hypothesis and to ensure the biocompatibility of the developed drug delivery system, two biomolecules were selected: raffinose trisaccharide and cholesterol. Raffinose is very soluble in water while cholesterol has a very limited solubility. The best capping molecule, as shown by the 5-FU drug release kinetic studies, is cholesterol that combines a relatively big size and a great tendency to interact with the outer pores of the porous material because of its dislike towards water. It is worth mentioning that the biocompatibility of the so-generated 5-FU drug delivery system does not seem to be negatively affected by the presence of copper and chromium as components of the porous material. Instead of that, a proliferative effect has been observed on HCT116 colon cancer cell line cytotoxicity studies which seems to be corroborated by the overexpression of MIR1248 and SUMO2 genes.
In the near future, we aim to extrapolate this approach of controlling the release of drug molecules by means of external capping molecules for some other flexible supramolecular porous materials but also for more rigid MOF analogues to check the impact the structural flexibility has on the viability of this approach. If these future works corroborate that this new approach for controlling the desorption rate is broadly applicable to different porous materials, drugs and blocking molecules, it could imply a significant breakthrough in the drug delivery technologies.
Author contributions
The manuscript was written through the contributions of all authors that gave approval to the final version. S. M.-G. performed the experiments and with J. P.-C. prepared the manuscript and the supplementary information. E. M.-R. performed and analysed the magnetic sustentation experiments. O. C., A. L., G. B, and S. P.-Y. designed the experiments and contributed to formal analysis. A. C. R., D. G. and M. J. A.-B. performed and analysed the biological assays. O. C. and A. L. contributed to funding acquisition, project administration, supervision, and writing – review and editing.Data availability
Supporting data associated with this work area are included in the ESI.† The transcriptomic data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO)40 and are accessible through GEO Series accession number GSE271345 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE271345).Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support from the Basque Government (IT1722-22 and ELKARTEK KK-2022/00032), the Spanish Ministry of Science and Innovation (TED2021-129810B-C22 financed by MCIN/AEI/10.13039/501100011033 and by the European Union NextGenerationEU/PRTR, and PID2022-138968NB-C22 project funded by MCIN/AEI/10.13039/501100011033) and the University of the Basque Country, UPV/EHU (EHU-N23/51). S. M.-G. thanks the Basque Government and European Union NextGeneration EU (Investigo Program PIFINVE22/47). Technical and human support provided by SGIker (UPV/EHU, MICINN, GV/EJ, ESF) is also acknowledged.Notes and references
- D. Sun, W. Gao, H. Hu and S. Zhou, Acta Pharm. Sin. B, 2022, 12, 3049–3062 CrossRef.
- L. K. Vora, A. D. Gholap, K. Jetha, R. R. S. Thakur, H. K. Solanki and V. P. Chavda, Pharmaceutics, 2023, 15, 1916 CrossRef.
- M. Al Sharabati, R. Sabouni and G. A. Husseini, Nanomaterials, 2022, 12, 277 CrossRef PubMed.
- P. Gao, Y. Chen, W. Pan, N. Li, Z. Liu and B. Tang, Angew. Chem., Int. Ed., 2021, 60, 2–16 CrossRef.
- E. Pérez-Botella, S. Valencia and F. Rey, Chem. Rev., 2022, 122, 17647–17695 CrossRef.
- G. T. M. Kadja, N. T. U. Culsum and R. M. Putri, Results Chem., 2023, 5, 100910 CrossRef.
- S. He, L. Wu, X. Li, H. Sun, T. Xiong, J. Liu, C. Huang, H. Xu, H. Sun, W. Chen, R. Gref and J. Zhang, Acta Pharm. Sin. B, 2021, 11, 2362–2395 CrossRef CAS.
- S. Naghdi, M. M. Shahrestani, M. Zendehbad, H. Djahaniani, H. Kazemian and D. Eder, J. Hazard. Mater., 2023, 442, 130127 CrossRef CAS.
- Z. Sun, C. Song, C. Wang, Y. Hu and J. Wu, Mol. Pharm., 2020, 17, 373–391 CAS.
- B. Maranescu and A. Visa, Int. J. Mol. Sci., 2022, 23, 4458 CrossRef CAS PubMed.
- M. Servatan, P. Zarrintaj, G. Mahmodi, S. J. Kim, M. R. Ganjali, M. R. Saeb and M. Mozafari, Drug Discov. Today, 2020, 25, 642–656 CrossRef CAS PubMed.
- S. Pérez-Yáñez, G. Beobide, O. Castillo, J. Cepeda and A. Luque, Supramolecular metal-organic frameworks based on metal-nucleobase entities, Nova Science Publishers, Inc., Bilbao, 2017 Search PubMed.
- S. Mena-Gutiérrez, J. Pascual-Colino, G. Beobide, O. Castillo, A. Castellanos-Rubio, A. Luque, E. Maiza-Razkin, J. Mentxaka and S. Pérez-Yáñez, Inorg. Chem., 2023, 62, 18496–18509 CrossRef PubMed.
- J. Tian, Z. Y. Xu, D. W. Zhang, H. Wang, S. H. Xie, D. W. Xu, Y. H. Ren, H. Wang, Y. Liu and Z. T. Li, Nat. Commun., 2016, 7, 11580 CrossRef CAS PubMed.
- Z. Li, N. Song and Y. W. Yang, Matter, 2019, 1, 345–368 CrossRef.
- X. Li, M. Shen, J. Yang, L. Liu and Y. W. Yang, Adv. Mater., 2024, 36, 2313317 CrossRef CAS PubMed.
- R. Pérez-Aguirre, B. Artetxe, G. Beobide, O. Castillo, I. de Pedro, A. Luque, S. Pérez-Yáñez and S. Wuttke, Cell Rep. Phys. Sci., 2021, 2, 100421 CrossRef.
- N. Barroso, J. Andreo, G. Beobide, O. Castillo, A. Luque, S. Pérez-Yáñez and S. Wuttke, Commun. Chem., 2023, 6, 1–9 CrossRef.
- J. Pascual-Colino, R. Pérez-Aguirre, G. Beobide, O. Castillo, I. de Pedro, A. Luque, S. Mena-Gutiérrez and S. Pérez-Yáñez, Inorg. Chem. Front., 2023, 10, 2250–2261 RSC.
- A. Murray, T. M. Nguyen, C. E. Parker, B. G. Feagan and J. K. MacDonald, Cochrane Database Syst. Rev., 2020, 8, CD000544 Search PubMed.
- S. Cherukuvada, G. Bolla, K. Sikligar and A. Nangia, Cryst. Growth Des., 2013, 13, 1551–1557 CrossRef CAS.
- S. Vodenkova, T. Buchler, K. Cervena, V. Veskrnova, P. Vodicka and V. Vymetalkova, Pharmacol. Ther., 2020, 206, 107447 CrossRef CAS PubMed.
- P. M. Boylan, M. Abdalla, B. Bissell, M. A. Malesker, M. Santibañez and Z. Smith, Pharmacotherapy, 2023, 43, 963–990 CrossRef.
- X. W. Yan, S. Tarasi, S. J. Wang, K. Rostamizadeh, M. L. Hu, A. Morsali, A. Ramazani, R. Tarasi and Y. Ahmadi, Arabian J. Chem., 2023, 16, 104887 CrossRef CAS.
- M. Ding, W. Liu and R. Gref, Adv. Drug Deliv. Rev., 2022, 190, 114496 CrossRef CAS.
- M. Moharramnejad, A. Ehsani, M. Shahi, S. Gharanli, H. Saremi, R. E. Malekshah, Z. S. Basmenj, S. Salmani and M. Mohammadi, J. Drug Delivery Sci. Technol., 2023, 81, 104285 CrossRef CAS.
- X. Chen, Y. Zhuang, N. Rampal, R. Hewitt, G. Divitini, C. A. O’Keefe, X. Liu, D. J. Whitaker, J. W. Wills, R. Jugdaohsingh, J. J. Powell, H. Yu, C. P. Grey, O. A. Scherman and D. Fairen-Jimenez, J. Am. Chem. Soc., 2021, 143, 13557–13572 CrossRef CAS PubMed.
- Z. Zhou, M. Vázquez-González and I. Willner, Chem. Soc. Rev., 2021, 50, 4541–4563 RSC.
- Y. Liu, J. Huang, S. Li, Z. Li, C. Chen, G. Qu, K. Chen, Y. Teng, R. Ma, X. Wu and J. Ren, Biomater. Sci., 2023, 12, 837–862 RSC.
- N. Barroso, S. Dutta, J. Andreo, G. Beobide, O. Castillo, A. Luque, S. Pérez-Yáñez and S. Wuttke, J. Mater. Chem. A, 2023, 11, 21300–21311 RSC.
- D. Chen, B. Li, L. Jiang, Y. Li, Y. Yang, Z. Luo and J. Wang, ACS Appl. Bio Mater., 2020, 3, 4081–4094 CrossRef CAS PubMed.
- D. Gerovska and M. J. Araúzo-Bravo, Int. J. Mol. Sci., 2023, 24, 2736 CrossRef CAS.
- M. Pertea, D. Kim, G. M. Pertea, J. T. Leek and S. L. Salzberg, Nat. Protoc., 2016, 11, 1650–1667 CrossRef CAS PubMed.
- C. Trapnell, A. Roberts, L. Goff, G. Pertea, D. Kim, D. R. Kelley, H. Pimentel, S. L. Salzberg, J. L. Rinn and L. Pachter, Nat. Protoc., 2012, 7, 562–578 CrossRef CAS PubMed.
- D. Gerovska, G. Larrinaga, J. D. Solano-Iturria, J. Márquez, P. G. Gallastegi, A. M. Khatib, G. Poschmann, K. Stühler, M. Armesto, C. H. Lawrie, I. Badiola and M. J. Araúzo-Bravo, Cancers, 2020, 12, 2380 CrossRef CAS.
- J. Pascual-Colino, G. Beobide, O. Castillo, P. Lodewyckx, A. Luque, S. Pérez-Yáñez, P. Román and L. F. Velasco, J. Inorg. Biochem., 2020, 202, 110865 CrossRef CAS PubMed.
- J. A. Przyojski, N. N. Myers, H. D. Arman, A. Prosvirin, K. R. Dunbar, M. Natarajan, M. Krishnan, S. Mohan and J. A. Walmsley, J. Inorg. Biochem., 2013, 127, 175–181 CrossRef CAS PubMed.
- S. Xiao, D. Zhang, Z. Liu, W. Jin, G. Huang, Z. Wei, D. Wang and C. Deng, Aging, 2020, 12, 6947–6965 CrossRef PubMed.
- P. Richard, S. Feng and J. L. Manley, Genes Dev., 2013, 27, 2227–2232 CrossRef PubMed.
- R. Edgar, M. Domrachev and A. E. Lash, Nucleic Acids Res., 2002, 30, 207–210 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb01521e |
| This journal is © The Royal Society of Chemistry 2024 |