Open Access Article
Open Access ArticleCell-specific spatial profiling of targeted protein expression to characterize the impact of intracortical microelectrode implantation on neuronal health†
Lindsey N.
Druschel
ab,
Niveda M.
Kasthuri
ab,
Sydney S.
Song
ab,
Jaime J.
Wang
 ab,
Allison
Hess-Dunning
ab,
E. Ricky
Chan
c and
Jeffrey R.
Capadona
ab,
Allison
Hess-Dunning
ab,
E. Ricky
Chan
c and
Jeffrey R.
Capadona
 *ab
*ab
aDepartment of Biomedical Engineering, Case Western Reserve University, Cleveland, OH 44106, USA. E-mail: jrc35@case.edu
bAdvanced Platform Technology Center, Louis Stokes Cleveland Veterans Affairs Medical Center, Cleveland, OH 44106, USA
cCleveland Institute for Computational Biology, Case Western Reserve University, Cleveland, OH 44106, USA
First published on 23rd October 2024
Abstract
Intracortical microelectrode arrays (MEAs) can record neuronal activity and advance brain-computer interface (BCI) devices. Implantation of the invasive MEA kills local neurons, which has been documented using immunohistochemistry (IHC). Neuronal nuclear protein (NeuN), a protein that lines the nuclei of exclusively neuronal cells, has been used as a marker for neuronal health and survival for decades in neuroscience and neural engineering. NeuN staining is often used to describe the neuronal response to intracortical microelectrode array (MEA) implantation. However, IHC is semiquantitative, relying on intensity readings rather than directly counting expressed proteins. To supplement previous IHC studies, we evaluated the expression of proteins representing different aspects of neuronal structure or function: microtubule-associated protein 2 (MAP2), neurofilament light (NfL), synaptophysin (SYP), myelin basic protein (MBP), and oligodendrocyte transcription factor 2 (OLIG2) following a neural injury caused by intracortical MEA implantation. Together, these five proteins evaluate the cytoskeletal structure, neurotransmitter release, and myelination of neurons. To fully evaluate neuronal health in NeuN-positive (NeuN+) regions, we only quantified protein expression in NeuN+ regions, making this the first-ever cell-specific spatial profiling evaluation of targeted proteins by multiplex immunochemistry following MEA implantation. We performed our protein quantification along with NeuN IHC to compare the results of the two techniques directly. We found that NeuN immunohistochemical analysis does not show the same trends as MAP2, NfL, SYP, MBP, and OLIG2 expression. Further, we found that all five quantified proteins show a decreased expression pattern that aligns more with historic intracortical MEA recording performance.
Introduction
Implanted intracortical neural electrodes, such as microelectrode arrays (MEAs), can help characterize the underlying pathophysiology of various diseases or allow for interfacing between human patients and external systems.1–3 MEAs implanted directly into the cortex can record neurons with single-unit resolution, making them extremely valuable for brain-computer interfacing (BCI) applications.4–7 BCIs are devices that allow for communication between the brain and an external device, including prosthetics or wheelchairs, to aid patients with loss of motor function.8,9 However, challenges in MEA reliability restrict the long-term clinical implementation of BCIs with MEA input. MEA recording quality is highly variable and generally decreases with time.10–15 MEA failures weeks to months post-implantation are mostly attributed to the tissue's biological response to the implant.16MEAs are implanted in the cortex of the brain and, like many neural injuries, result in the activation of microglia and macrophages, the formation of a glial scar, the dieback of local neurons, and demyelination.15,17 Microglia are macrophage cells localized to the brain and are immediately activated by MEA implantation.18 Astrocytes are activated by microglia signaling and will migrate to the MEA surface, forming a tight glial scar that encapsulates the MEA and separates it from the neurons.19 Immune cells such as macrophages and T-cells will be recruited from the periphery to the MEA implant site.20,21 Collectively, the neuroinflammatory response to implanted MEAs contributes to the breakdown of electrode material, increased impedance between the neurons and the microelectrode contacts, dieback of local neurons, production of an impermeable glial scar, and therefore reduction in the electrode recording quality.15,22 Many different mechanisms have been investigated as major contributors to chronic MEA failure, including BBB permeability, material degradation, glial scar impedance, innate immune activation, and neuronal death.23–27 The complexity of device failure and the tissue response make it difficult to develop a therapeutic that adequately stabilizes recording quality.
Upon implantation, mechanical stress will rupture the cell bodies or the axons of the neurons in the path of the MEA.28 The initial injury of MEA insertion is considered the primary injury.29,30 Neurons injured during the primary stage will release damage-associated molecular pattern molecules (DAMPs), activating local microglia and heightening the acute inflammatory response.31,32 Chronic neuroinflammatory responses to the MEAs result in a secondary neuronal injury. M1 microglia and A1 astrocytes will secrete pro-inflammatory factors that cause immune cell recruitment and proliferation.33,34 Reactive oxygen species (ROS) are highly reactive free radicals that are produced by inflammatory processes following inflammation. ROS are known to contribute to aging and neurodegeneration in injured states.35 ROS will react with DNA, RNA, proteins, and lipid cell membranes in the neural environment, damaging local cells.35,36 The recording window required for an MEA to obtain single unit action potentials is approximately 50–150 μm from the implant,37 and many neurons within this region are killed post-implantation.38
The current standard for neuronal evaluation following MEA implantation is immunohistochemistry. The complex mechanisms of neuronal dieback are most commonly evaluated by immunohistochemistry using neuronal nuclear protein (NeuN).1,13–15,27,39–47 NeuN is a protein abundant in neuronal nuclei that binds to the nuclear membrane of mature neurons. Though the function of NeuN is not fully understood, it is hypothesized that NeuN is a transcription factor that regulates the general neuronal phenotype.48 NeuN is present exclusively in neurons but is not detected in some neuronal cell types, such as Purkinje cells in the cerebral cortex and mitral cells in the olfactory bulb.49,50 Bedell et al. and Nguyen et al. have shown that NeuN expression returns to a healthy baseline level 50–100 μm from the MEA implant site.23,42 Essentially, NeuN expression initially declines and then replenishes with time post-MEA implantation. However, MEA neuronal recording performance decreases with time post-implantation.10,51 The stark difference between trends in neuronal recordings and NeuN expression could be interpreted to suggest that neuronal health is not a main contributor to chronic MEA failure. A more holistic understanding of the changes in targeted protein expression within neurons following MEA implantation is necessary to drive forward therapeutics to improve recording performance.
Our lab recently conducted a proteomic study of inflammation and neuronal health following MEA implantation in the motor and sensory cortices, evaluating the expression of several key proteins for neuronal and oligodendrocyte health and functionality using the NanoString Digital Spatial Profiler (DSP).52 The NanoString DSP quantifies the proteins found in individual regions of interest (ROIs) using UV capture with <1-micron resolution, allowing for customizable and precise calculations of protein expression. Proteomic evaluation within 180 μm of the implant site revealed that several key proteins, including neurofilament light (NfL) and myelin basic protein (MBP), exhibit more than a 50% decrease in implanted animals compared to controls. The proteomic deficits did not include cell-specific or distance-specific capture of proteins. We are expanding on our previous proteomic analysis by adding segmentation to our multiplex immunochemistry collection process. In previous papers, we utilized much larger panels, which could be called proteomics. Here, due to the focus on a subset of the proteins examined in that panel, we will more accurately refer to this subset of the larger proteomics as multiplex immunochemistry. The present study is the first to segment multiplex immunochemistry quantification based on factors like distance from the MEA implant or cell type. Utilizing the DSP's functionality of ROI-specific collection, we split collection regions based on both distance from the implant (90 μm rings), as well as by cell type using NeuN immunohistochemistry (IHC). Essentially, we collected proteins only within NeuN+ regions in both implanted and naïve control samples. The additional segmentation methods will allow us to isolate neuronal proteins and quantify the damage based on distance from the MEA.
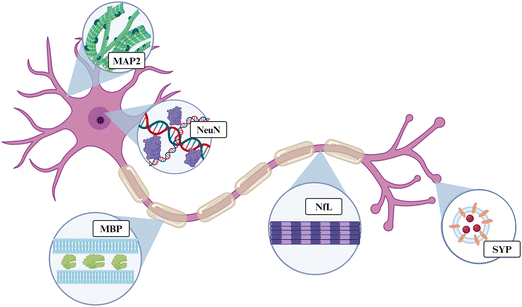
Here, we evaluated the expression of five proteins associated with neuronal and oligodendrocyte health: microtubule-associated protein 2 (MAP2), neurofilament light (NfL), synaptophysin (SYP), myelin basic protein (MBP), and oligodendrocyte transcription factor 2 (OLIG2). Taken together, this group of proteins can give an indication of a neuron's structure and ability to properly conduct an action potential or release neurotransmitters. MAP2, NfL, SYP, MBP, or OLIG2 are found in nearly all major regions of neurons: the dendrites, nucleus, cytoplasm, axon, and axon terminal (Fig. 1). MAP2 is a cytoskeletal protein abundant within neuronal dendrites that increases microtubule bundling and polymerization.53 Downregulation of MAP2 could lead to increased microtubule collapse or depolymerization in the dendrites, reducing connections between neurons, and leading to neuronal hypoexcitability.54 NfL is a cytoskeletal protein that makes up most of the proteins within an axon. NfL is an intermediate filament or cluster of fibers containing rod-like domains that comprise the cytoskeleton. NfL is also present in the cytoplasm of the cell body, but it is not as abundant in that region.55,56 SYP is a protein that forms the walls of synaptic vesicles that store and release neurotransmitters in the synaptic terminals. SYP is used as a marker for synaptic density.57,58 Neurotransmitter release is crucial to the function of a neuron because it allows for the excitation of the post-synaptic neuron in the neural circuit. Therefore, SYP downregulation causes neuronal hypoexcitability.59 MBP is the second-most abundant protein in oligodendrocytes, making up 30% of the total proteins. Oligodendrocytes are the myelin cells of the central nervous system and are made up of proteolipid membranes. MBP binds to the lipid membranes and helps to form the structure of the myelin.60,61 Downregulation of MBP is likely indicative of demyelination of local neurons. OLIG2 is a transcription factor expressed highly in oligodendrocytes and oligodendrocyte precursor cells, while some expression has also been observed in astrocytes.62 OLIG2 is considered an important protein for myelin healing after disease or injury.63
In addition to the 5-protein panel, we performed immunohistochemical analysis of NeuN, consistent with previous MEA studies.13,40,42,46 The direct comparison of NeuN immunohistochemical analysis to quantifying other neuronal health proteins could advance our understanding of neuronal health following MEA implantation. Under the assumption that NeuN represents neuronal health surrounding the MEA, other proteins involved in neuronal functionality would be expected to show similar trends. Evaluating additional proteins could also provide a more insightful understanding of neurodegenerative mechanisms and the extent of neuronal damage. We can call upon a vast collection of MEA recording literature evaluating chronic device performance,10,16 and neuronal proteins that align with MEA recording trends could be further evaluated as markers that indicate recording capabilities. Finally, adding distance and cell segmentation, we present new innovative techniques for quantifying protein expression following MEA implantation.
Methods
Animal preparation
Mice were housed at Case Western Reserve University and cared for in compliance with an IACUC-approved protocol. The mice used for this study were male C57BL/6J mice. For the experimental groups, mice underwent intracortical MEA implantation surgery once they reached 8 weeks of age. Surgical procedures were conducted according to established laboratory protocols approved by the Case Western Reserve University IACUC.64,65 For each mouse in the experimental groups, four non-functional probes were implanted into the motor and sensory cortices on both the left and right sides of the brain, with coordinates 1.5 mm lateral and 1.0 mm anterior and posterior to the bregma. Mice were sacrificed at 4 or 8 weeks following MEA implantation (4 mice per group, each with 4 implants). The naïve control mice were sacrificed without any craniotomy or implant (4 mice).Mice were sacrificed via cardiac perfusion. Animals were anesthetized and treated with ketamine and xylazine intraperitoneal injections. While the heart was still beating, incisions were made in the chest to expose the heart, and a butterfly needle attached to a peristaltic pump was inserted into the left ventricle. An incision was made in the right atrium to relieve the pressure on the heart and allow the outflow of blood and PBS. First, 30 ml of 1× phosphate-buffered saline (PBS) was circulated through the bloodstream. Next, 30 ml of 30% sucrose in 1× PBS was pumped through the mouse to cryoprotect the brain. Brains were extracted, flash frozen on dry ice suspended in optimum cutting temperature compound (OCT, Sakura Finetek USA, Item Number: 25608-930), and kept at −80° until cryo-slicing.
Microelectrode probes
The probes used in this study were non-functional silicon probes in the shape of single-shank, Michigan-style electrodes. Probes were 2 mm long, 150 μm wide at the widest section of the shank, and 15 μm thick. The Pancrazio Laboratory at the University of Texas Dallas generously provided these probes.Analysis for spatially resolved protein expression
The full protocol for spatial protein expression analysis was provided by NanoString Technologies (https://university.nanostring.com/geomx-dsp-manual-slide-preparation-user-manual) using the FFPE protocol with amendments for fresh frozen tissue. The main steps are illustrated in Fig. 2. This protocol is described in depth in previous proteomic analyses.52 Isolated brains were cryo-sliced into five μm sections along the length of the cortex. These slices were fixed in 10% neutral buffered formalin (NBF) overnight at 4 °C. Slides were washed thrice for five minutes each in 1× tris-buffered saline with 0.1% tween 20 detergent (TBST). Following fixation and washing, slides were placed in 1× citrate buffer and underwent antigen retrieval in the TintoRetriever Pressure Cooker (Bio SB, Item Number: BSB 7008) on high temperature and pressure settings. Slides were then left at room temperature for 25 minutes and washed once again with 1× TBST. Following this, slides were stained with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 anti-NeuN (Alexa Fluor® 532 GA-5, Item Number: NBP2-33184AF532) and 1
10 anti-NeuN (Alexa Fluor® 532 GA-5, Item Number: NBP2-33184AF532) and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 anti-GFAP antibodies (Alexa Fluor® 647 EPR12763, Item Number: ab190565), which were used to identify the implant site.
40 anti-GFAP antibodies (Alexa Fluor® 647 EPR12763, Item Number: ab190565), which were used to identify the implant site.
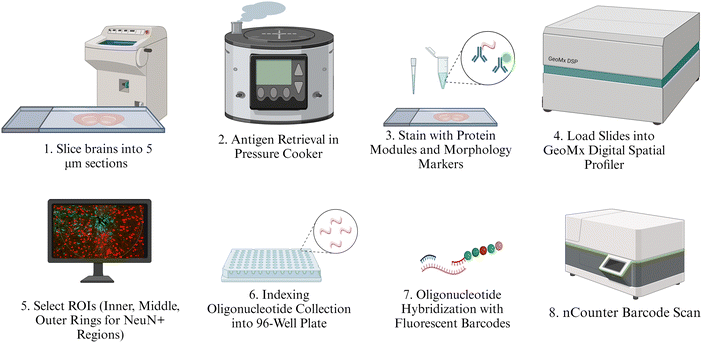 | ||
| Fig. 2 Illustration of protein quantification methods. Intermediate steps such as washes or fixation are not illustrated but are described at length in the provided protocol. | ||
The protein panels purchased for this experiment include the Neural Cell Profiling Core (25 proteins, Item Number: 121300120), which was paired with the Glial Cell Subtyping Module (10 proteins, Item Number: 121300125), and Autophagy Module (10 proteins, Item Number: 121300124), which were analyzed previously. Our former proteomics analysis used the same protein panels but focused largely on immune cell activation and potential immunotherapy targets.52 For this study, we only included the expression of a subset of proteins that we thought would best represent the structure, health, and function of neurons and oligodendrocytes. The five proteins used in this study are: MAP2, NfL, SYP, MBP, and OLIG2. The slides were stained with custom antibodies that are each associated with a unique fluorescent identifier, or “barcode.” Each barcode uniquely corresponds to a protein that was quantified in this study. After staining with morphological and NanoString-specific panel antibodies, slides were incubated in a humidity chamber overnight at 4 °C.
The following day, slides were washed in 1× TBST three times for 10 minutes each, and then fixed with 10% NBF in a humidity chamber for 30 minutes at room temperature. Next, the slides were washed twice with 1× TBST for five minutes each. Slides were then placed in a humidity chamber and stained with SYTO-13 (NanoString Technologies, Item Number:121300303) as a nuclear stain to better visualize the implant site. The slides were allowed to incubate for 15 minutes at room temperature, after which they were washed with 1× TBST two times. Finally, slides were loaded into the NanoString GeoMx Digital Spatial Profiler to image the implant site and select a region of interest (ROI), which was segmented into NeuN+ and GFAP+ regions. While NeuN and GFAP were both used to identify the implant sites, only NeuN was used to identify collection sites. Images were exported to FIJI, where custom segmentation was created to segment each NeuN+ region into three concentric rings 0–90 μm (inner), 90–180 μm (middle), and 180–270 μm (outer) from the implant site (Fig. 3). These rings will hereafter be referred to as “inner,” “middle,” and “outer” rings, respectively. The 270 μm collection region was selected to fit into the maximum ROI radius of the GeoMx DSP, which is 300 μm. The 90 μm interval was chosen for consistency with transcriptomic studies to be reported in future publications. A minimum area of tissue for the collection of at least 20 cells for transcriptomic analysis in the area closest to the MEAs. To later be able to correlate the protein expression with transcriptomics, we binned at 90 μm. Segmented images were imported back into the GeoMx and assigned separate areas of interest from which to collect protein counts. The GeoMx shined a UV light on NeuN+ area in each ring, cleaving the barcodes from the antibodies they were bound to. Barcodes were collected in a 96-well plate. GeoMx Hybridization Codes (NanoString Technologies, Item Number: 121300401) were added to the rows in the 96-well plate to allow for pooling each column and streamlining the 96-well plate into a 12-strip tube. Following the pooling of the samples, the 12-strip tube was loaded into the NanoString nCounter MAX/FLEX System, which transferred samples into a clear cartridge and scanned the barcodes, generating the counts of each barcode, and thus each protein, present within each selected NeuN+ ring.
Protein expression statistics
The raw counts of each measured protein were exported from the NanoString nCounter MAX/FLEX system and analyzed for differential expression in MATLAB. A custom MATLAB script normalized the protein counts to three housekeeping proteins included in the protein panels: histone H3, S6, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The MATLAB script also calculated the log2(fold change) (“log2FC”) for each protein, which is the log of the average of the experimental group protein expression divided by the average of the control group protein expression. The control group, or baseline, for this study is always naïve controls. A positive log2FC indicates that the ratio of experimental to control expression is greater than 1, which means the given protein is found in higher concentration in the experimental group. We denote higher experimental expression as “upregulation.” Likewise, a negative log2FC value is “downregulation” of the experimental group compared to naïve controls. The experimental group is always one of the implanted groups, either 4- or 8-weeks post-implantation. The MATLAB script also generated the p-values and the adjusted p-values (padj) for each protein. The p-values were adjusted using the Benjamini–Hochberg correction to filter out random significance. The R packages “tidyverse” and “readxl” were used to generate probability density functions (PDFs) for each protein using a kernel density estimation.66,67 A kernel density estimation is used to PDFs visualize the likelihood of a normalized protein count appearing at a specific value. PDFs are used to show the distribution, such as normal, bimodal, or skewed, of a protein within a specific comparison.68 GraphPad Prism 10 (GraphPad Software, Boston, Massachusetts) was used to generate volcano plots and heat maps for data visualization. Within the volcano plots, all proteins on the left side of the x-axis (negative values) are downregulated in the experimental group, and all proteins on the right side of the x-axis (positive values) are upregulated in the experimental group. The volcano plots show the significance cutoff of padj < 0.05.Analysis of neuronal nuclear counts using NeuN
Images from the ROI selection step of the experimental pipeline was exported from the GeoMx. The exact inner, middle, and outer rings used in the protein expression analysis were used for the NeuN nuclear count analysis (Fig. 3). The NeuN stain was used to manually count NeuN+ cells in each ring separately for the experimental groups. Cell counts were uploaded to an Excel (Microsoft, Redmond, Washington) spreadsheet for statistical analysis. The NeuN+ cell count for each ring was normalized by the area of the individual ring. This gave us an area density of neuronal nuclei per square millimeter for each 90 μm ring of the experimental groups. The control groups did not consist of rings, as there was no implant, but instead, an equivalent area to the combined inner, middle, and outer rings (270 μm radius) was collected at the same coordinates as the implants in the experimental groups. The average nuclear density in the naïve control mice region is referred to as the control baseline. An unpaired, two-tailed t-test was performed on densities with a significance cutoff of p < 0.05. Data from the neuronal nuclear counts was visualized using GraphPad Prism 10.Results
Analysis of neuronal and oligodendrocyte health using multiplex protein panel
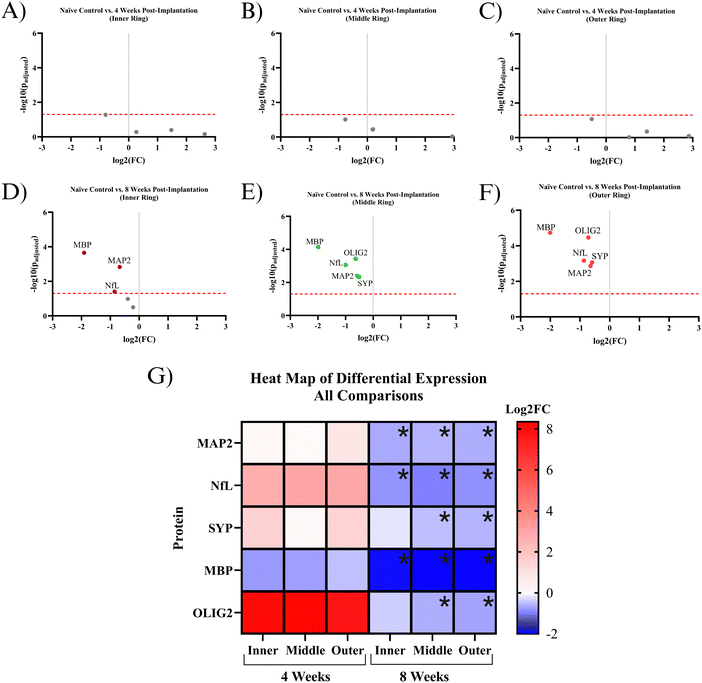
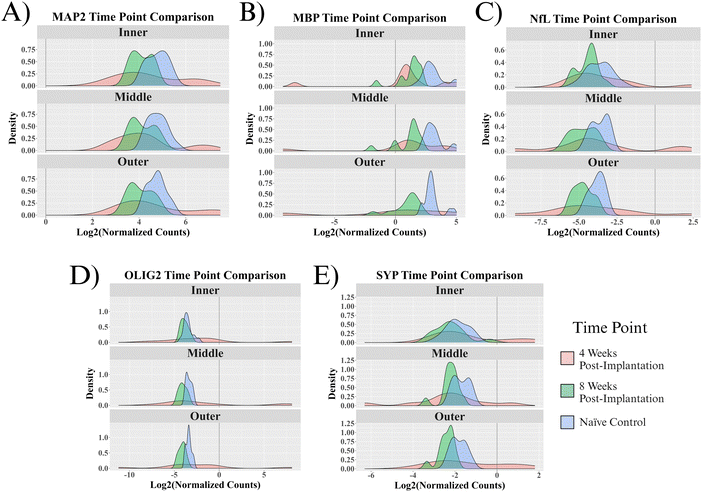
Five proteins were quantified in NeuN+ regions using the NanoString GeoMx Digital Spatial Profiler: MAP2, NfL, SYP, MBP, and OLIG2. Six comparisons were made for protein expression: a combination of the two time points (4- and 8 weeks post-implantation) and three 90 μm concentric rings (inner, middle, outer) extending from the implant site. All comparisons have a baseline of healthy, unimplanted naïve control mice, meaning that the reported results represent changes from healthy tissue.At 4 weeks post-MEA implantation, no proteins were found to be significantly differentially expressed. MAP2, NfL, SYP, and OLIG2 show insignificant upregulation in the implanted mice compared to naïve controls. OLIG2 exhibited a relatively high average log2FC value of ∼8.0. Though insignificant, MBP was the only downregulated protein, with a log2FC of ∼−0.8 (Fig. 4A–C and G). The protein expression 4 weeks post-implantation is highly variable, which resulted in a lack of significance at a threshold of padj < 0.05. Fig. 5 shows the probability density functions of all 5 measured proteins, illustrating the protein expression distribution within each comparison.
By 8 weeks post-implantation, MAP2, NfL, and MBP show significant downregulation in all three rings extending to 270 μm from the MEA-tissue interface. The average log2FC of MAP2, MBP, and NfL between the three rings were ∼−0.6, −2.0, and −0.9, respectively. In the inner ring, OLIG2 and SYP show insignificant downregulation with a log2FC of ∼−0.4 and −0.2, respectively (Fig. 4D–F and G). In the middle and outer rings, OLIG2 and SYP are significantly downregulated with log2FC values ranging from ∼−0.6 to −0.7 and −0.5 to −0.6, respectively.
Immunohistochemistry using NeuN as a marker for neuronal nuclei
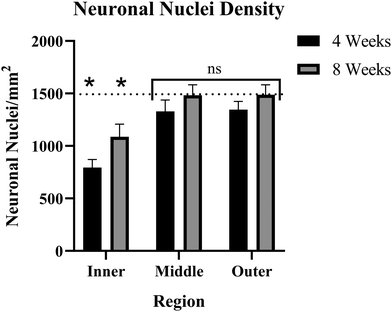
The number of NeuN+ cells was counted on a per-region basis. The naïve control tissue was evaluated to create a baseline for comparison to implanted tissue. Here, baseline NeuN+ cell density was determined to be ∼1534 neuronal nuclei per square millimeter (NN mm−2). Both 4-weeks and 8-weeks post-implantation groups showed significant decreases in nuclear density within the inner ring (0–90 μm). The 4-week post-implantation inner ring averaged ∼860 NN mm−2, while the 8-week post-implantation inner ring nuclear count was ∼1124 NN mm−2. Both inner ring comparisons had a p < 0.05, in the 4-week comparison p = 0.00006 and in the 8-week comparison p = 0.0202. The middle rings (90–180 μm) in the 4-week and 8-week post-implantation groups showed no significant deficits in neuronal nuclear density, averaging ∼1370 and ∼1540 NN mm−2, respectively. In the 4-week middle ring comparison, p = 0.2786, and in the 8-week middle ring comparison, p = 0.9681. Like the middle rings, the outer rings (180–270 μm) showed no significant neuronal deficits, with the 4-week group average of ∼1386 NN mm−2 (p = 0.2710) and the 8-week group average of ∼1528 NN mm−2 (p = 0.9667) (Fig. 6). When comparing the 4-week group and the 8-week groups, the inner rings were not significantly different from one another (p = 0.1193), and neither were the middle or outer rings (p = 0.2487 and 0.2581, respectively). When comparing rings within 4- and 8-week time points, for both groups, the inner rings were significantly different from both the middle and outer rings (p < 0.02 in all cases), but the middle and outer rings were not significantly different than one another (p > 0.9 in both cases).Consistent with a previous study from our lab,69 both the middle and outer rings show reduced nuclear count in the 4-week post-implantation group compared to baseline, though the effect is not statistically significant (Fig. 6). To determine what sample size would be required to reject the null hypothesis at a significance level of p < 0.05, we ran a power analysis. Our power analysis revealed that in the 4-week implanted vs. naïve control middle ring comparison, a sample size of ∼100 per group would be required to yield a p < 0.05. The 4-week outer ring comparison would require ∼96 samples per group to yield a p < 0.05. A distinction between the current and prior study was that the binning intervals used here were 90 μm while previous studies have used 50 μm intervals.
Discussion
Analysis of neuronal and oligodendrocyte health using multiplex immunochemistry protein panel
In the present study, we aimed to conduct a thorough evaluation of proteins that would provide insights into the effect of MEA implantation on recording performance as a function of neuronal density and integrity. We evaluated proteins that make up the dendrites (MAP2), axons (NfL), synaptic terminals (SYP), and myelination (MBP, OLIG2) of neurons in the motor cortex. Four weeks post-MEA implantation, MAP2, NfL, SYP, and OLIG2 are all upregulated (insignificantly) compared to a healthy baseline. These results are insignificant because there is extremely high variance between implant sites (Fig. 6). NfL and MAP2 upregulation in implanted mice could be due to axonal swelling and microtubule aggregation that occurs during neural disease or injury.70,71 Additionally, the upregulation of MAP2 and NfL along with SYP could indicate the neurons attempting to extend and remodel following injury. Neuroplasticity, or the remodeling of neuronal circuitry within the brain, occurs in healthy tissue but can ramp up following neural injury.72,73 Neuroplasticity consists of dendrites and axons remodeling and extending to rearrange the local neural networks. The dendritic expansion and axonal sprouting result in synaptogenesis, or the formation of new synapses. Successful remodeling would likely be indicated by the proteins returning to a healthy baseline value by 8 weeks post-implantation. Our results show that this is not the case, as by 8 weeks post-MEA implantation, all the proteins are downregulated in at least two rings (Fig. 4).In the 8 w post-implantation mice, all five proteins show downregulation in at least two of the three measured NeuN+ rings (Fig. 4D–F and G). Highly variable protein expression at 4 weeks post-implantation seems to converge to the same outcome by 8 weeks post-implantation. Fig. 5 shows that the ranges of normalized protein values are much smaller for the 8-week post-implantation mice. The 8-week post-MEA implantation mice protein values show nearly normal distributions, typically similar in shape to naïve control distributions but shifted left toward lower values. The two neuronal cytoskeletal proteins, MAP2 and NfL, show downregulation in all three rings of the 8-week post-implantation mice (Fig. 4D–F and G). NfL and MAP2 are known to be ischemia-sensitive and are used as markers of damage following ischemic stroke.74,75 Neuronal cytoskeletal degeneration is linked to the transition between reversible and irreversible tissue damage,74,75 meaning the NfL and MAP2 deficits caused by MEA implantation suggest damage that may never heal. Dendritic degeneration, as shown by the downregulation of MAP2, can hinder neuronal firing and is believed to contribute to neuronal atrophy.54,76,77 NfL release from axons has been linked to cognitive decline and neurodegeneration.78 Synaptophysin is a protein abundant in synaptic vesicles that does not have a clearly defined function.79 However, decreased synaptophysin has been associated with neurodegenerative disease and neuronal dysfunction.80 Taken together, MAP2, NfL, and SYP represent damage to three neuronal regions that span the length of the cell and can drastically impact neuronal functionality (Fig. 1).
OLIG2 is a protein that regulates the transcription of myelin-associated proteins. Along with their main function of insulating portions of the neuronal axons, oligodendrocytes have also been reported to have neuroprotective effects on injured neurons.81 Olig2 and MBP mRNA are upregulated up to six weeks post-MEA implantation.82 Four weeks post-implantation, OLIG2 has an average log2FC of ∼8. The high protein average log2FC shows that the OLIG2 protein is being successfully upregulated in some mice but not others, which is why it is not significantly upregulated (Fig. 6D). However, this effect does not last until 8 weeks post-implantation, which is where we see a shift towards downregulation of OLIG2 (Fig. 4D–F and G). OLIG2 is downregulated in all three NeuN+ regions but is only significantly downregulated in the middle and outer rings. In traumatic brain injury, OLIG2 is upregulated immediately after injury and remains upregulated for up to 3 months, aiding in the remyelination of the tissue.83,84 In the case of IME implantation, we see the opposite effect with OLIG2 downregulation at 8 weeks post-implantation. OLIG2 downregulation could represent that the continued presence of the implant in the tissue is hindering pathways for myelin healing.
MBP is a structural protein found in the myelin sheaths of oligodendrocytes. Loss of MBP can trigger myelin breakdown or loss of compact myelin.85,86 Demyelination can change axon morphology, causing the axons to swell or deform. Additionally, demyelination affects protein transport through the axon and disrupts the sodium, potassium, and calcium channels that enable healthy action potentials.87 Demyelination can have drastic impacts on neural function, as shown by myelin-associated diseases such as multiple sclerosis (MS). Demyelination in the motor cortex is associated with symptoms such as muscle spasticity, fatigue, and tremors.88–90 In a previous study, our lab was the first to perform a large-scale quantification of protein expression surrounding the MEA implant,52 and the present study expands on prior work by introducing cell segmentation for protein collection. We used the NanoString GeoMx Digital Spatial Profiler to image the MEA implant sites, select all NeuN+ regions, and collect only from those regions within each 90 μm bin. ROI selection was performed using 2D images of 5 μm-thick brain tissue slices (Fig. 3). Therefore, every pixel that detected a neuronal nucleus resulted in all proteins within the 5 μm thickness being collected, generally considered a limitation of proteomic technology that preserves the tissue in situ. However, the 5 μm thick slices allow for targeting the neuronal nuclei while also collecting from the adjacent regions, including the cell body, dendrites, axons, and synaptic terminals. Neurons form highly interwoven circuits, and one can use this colocalization in protein label collection. Here, we collected in NeuN+ regions specifically due to the protein's significance within our field, but other labs could utilize proteins that are not localized to neuronal nuclei, such as MAP2, NfL, or beta-tubulin (or a combination of several markers) to segment for neurons. Future studies could also use cell segmentation to evaluate other cell types that contribute to MEA failure, including astrocytes, microglia, or macrophages.
Immunohistochemistry using NeuN as a marker for neuronal nuclei
The nuclear count using NeuN immunohistochemistry demonstrated that only the inner rings, within 90 μm of the implant site, of both the 4-week and 8-week post-implantation groups have neuronal damage or loss from the MEA implantation. The middle and outer rings for both time points showed no significant deficits compared to naïve controls (Fig. 6). Our power analysis, which resulted in a required sample size of ∼100 for both 4 weeks middle and outer rings, confirmed that in both comparisons, NeuN neuronal nuclear counts were not sufficient to indicate damage within a reasonable sample size. However, the multiplex protein panel of MAP2, MBP, NfL, OLIG2, and SYP suggested a larger region of neurodegeneration over an extended time course. Together, our combined histological analysis suggests that certain elements of the neuron show decreased protein expression only near the implant site (decreased NeuN expression in the nucleus), while others have deficits that do not manifest until the chronic time points over the broader 270 μm region of interest explored here (MAP2, NfL, SYP, MBP and OLIG2 downregulation).IHC and the cell-specific protein expression assay performed in this study represent two methods evaluating the same molecules: proteins, but with very different approaches. IHC is quicker, easier, and can generate images mapping the exact locations of each protein. IHC is extremely useful for proof-of-concept experiments or applications where knowing the location is vital. Other cell-specific proteomic methods, such as FACS-sorted mass spectrometry, can give the advantage of quantifying thousands of proteins at once, but typically are performed in bulk on isolated samples. The isolated samples cannot be reconstructed to determine the exact location of that protein in the brain.91,92 The NanoString DSP provides exact expression data within specified locations, depending on the user's ROIs. The advantages of the DSP methodology are preserving the sample in situ and the customizability of the ROIs. However, it is limited in the number of proteins it can quantify based on available panels. Each method has its own pros and cons, and researchers should explore different options to determine what best fits the needs of their experiment.
Implications for recording and neuronal health post-MEA implantation
Single-unit recordings from intracortical MEAs have been shown to decline in quality with time.93 Optimizing MEA performance is crucial to clinical applications due to the invasiveness of the surgical implant procedure and the persisting need for BCIs to perform basic tasks. Traditional analyses of the MEA biological response have utilized immunohistochemistry to visualize and provide a semiquantitative assessment of the progression of both neuroinflammation and neurodegeneration.15,94 Based largely on histological reports, the general understanding in the field of neural interfacing is that the neuroinflammatory response to intracortical MEA implantation is a major contributor to the decline in signal quality.16,20,95,96 However, there is a disconnect between the documented timeline of neuronal “dieback” adjacent intracortical MEAs69,97 the decline in recording performance,93 and the required proximity of neuronal nuclei to obtain single-unit recordings.98 The density of neurons around the implant site has been described as the most accurate barometer for chronic electrode performance.38 Promoting neuronal growth and survival has been shown to improve cortical recordings, meaning that neuronal health is at least a partial contributor to chronic failure.NeuN has historically been used to denote neuronal loss or “dieback”.38 However, injured, or damaged neurons can also temporarily stop expressing NeuN, while NeuN has also been shown to lose immunoreactivity while still being present in injured or damaged tissue.48 NeuN IHC results can also vary between MEA studies. Some NeuN IHC results show a general trend of an initial decrease and steady increase toward a healthy baseline over time (Fig. 6),13,42 while others show variations from the standard trend, specifically a decline in NeuN over chronic time points.1,97
The purpose of adding NeuN staining to this study was to determine if, within the same tissue, neuronal nuclei counts obtained from NeuN immunohistochemistry and multiplex immunochemistry protein quantification of other neuronal proteins would yield the same conclusion concerning neuronal health. Our findings suggest that they do not. Neurons may still have NeuN proteins present in their nuclei while exhibiting major defects in structure and functionality. To be useful to an organism, neurons need to be able to conduct bioelectric signals, which would be impeded by diminished MAP2 and NfL for maintaining the structure of the neuron and its connections, or SYP, a major structural protein in synaptic vesicles that allows the exocytosis of neurotransmitters. MBP and OLIG2 may not be expressed in neurons directly, but their loss would limit the conduction of action potentials down the length of the axon.87
Our goal is to better understand the correlation, if any, between the histological response to MEA implantation and the time course of signal degradation. In the current study, NeuN expression increases from 4 weeks to 8 weeks post-implantation, while recording performance typically declines over the same interval – suggesting NeuN to be an unreliable marker for MEA recording performance (Fig. 6). MAP2, NfL, SYP, MBP, and OLIG2 are not significantly differentially expressed at 4 weeks after MEA implantation but are significantly downregulated in protein expression at 8 weeks post-implantation. The expression of the 5 proteins quantified in this study more closely correlates with documented trends in decreased MEA performance – suggesting that a combination of MAP2, NfL, SYP, MBP, and OLIG2 could represent more appropriate histological markers than NeuN alone to predict MEA performance. Finally, strategies to prevent the downregulation of MAP2, NfL, OLIG2, and SYP or approaches to increase the expression of MAP2, NfL, OLIG2, and SYP may prove valuable in improving the chronic recording performance of intracortical MEAs.
Ultimately, a major goal of our lab and others investigating the neuroinflammatory response to MEAs is to improve chronic recording performance due to the improved quality of life MEAs and BCIs could offer patients. However, neuronal health impacts more than just recording performance in live human patients. MEA implantation in the motor cortex of healthy rats causes fine motor deficits,99 showing that the device causes enough damage to noticeably affect brain function. MEAs have been applied beyond the motor cortex, to the greater frontal cortex, such as the sensory cortex or Broca's area, where neuronal damage could affect a person's senses or speech.100,101 Preserving neurons that cannot be regenerated is crucial to the safety of MEAs in clinical applications.
Conclusions
This study intended to utilize cell-specific, spatially resolved multiplex protein analysis techniques to perform a comprehensive analysis of neuronal health following MEA implantation. We found that MAP2, NfL, SYP, MBP, and OLIG2 significantly decreased expression at 8 weeks post-implantation but not 4 weeks post-implantation. We directly compared the multiplex protein panel results to the field standard, NeuN immunohistochemistry, to assess the utility of histological protocols to predict or correlate with the recording performance of intracortical MEAs. The expression profiles for MAP2, NfL, SYP, MBP, and OLIG2 more closely correlate with documented trends in decreased MEA performance – suggesting that a combination of MAP2, NfL, SYP, MBP, or OLIG2 could represent more appropriate histological markers than NeuN to predict MEA performance.Author contributions
LD and NK contributed to the conception and design of the work. LD, NK, SS, and JW contributed to the methodology, software analysis, validation, formal analysis, investigation, and data curation. ERC guided the statistical analysis of the work. LD wrote the original draft. LD, NK, JW, and JRC prepared figures. All authors edited the final manuscript. JRC provided the funding and resources to conduct the study.Disclosures
The contents do not represent the views of the U.S. Department of Veterans Affairs, the National Institutes of Health, or the United States Government.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was supported in part by Merit Review Award I01RX002611 (Capadona) and Senior Research Career Scientist Award # I02RX003077 (Capadona) from the United States (US) Department of Veterans Affairs Rehabilitation Research and Development Service. Additionally, this work was also supported in part by the National Institute of Health, National Institute of Neurological Disorders and Stroke R01NS110823 (Capadona/Pancrazio) and R01NS131502 (Ware/Pancrazio/Capadona), and the National Institute for Biomedical Imaging and Bioengineering, T32EB004314, (Capadona/Kirsch). Finally, partial funding was provided from discretionary funding from the Donnell Institute Professorship endowment (Capadona) and the Case School of Engineering Research Incentive Program (Capadona).References
- S. M. Wellman, et al., Aberrant accumulation of age- and disease-associated factors following neural probe implantation in a mouse model of Alzheimer’s disease, J. Neural Eng., 2023, 20(4), 046044 CrossRef PubMed.
- C. Fan, et al., Plug-and-Play Stability for Intracortical Brain-Computer Interfaces: A One-Year Demonstration of Seamless Brain-to-Text Communication, Adv. Neural Inf. Process. Syst., 2023, 36, 42258–42270 Search PubMed.
- A. Benazzouz, et al., Intraoperative microrecordings of the subthalamic nucleus in Parkinson's disease, Mov. Disord, 2002, 17(Suppl 3), S145–S149 CrossRef PubMed.
- A. B. Rapeaux and T. G. Constandinou, Implantable brain machine interfaces: first-in-human studies, technology challenges and trends, Curr. Opin. Biotechnol., 2021, 72, 102–111 CrossRef CAS PubMed.
- A. M. Badakva, N. V. Miller and L. N. Zobova, [Artificial Feedback for Invasive Brain-Computer Interfaces], Fiziol. Chel., 2016, 42(1), 128–136 CAS.
- J. J. Shih, D. J. Krusienski and J. R. Wolpaw, Brain-computer interfaces in medicine, Mayo Clin. Proc., 2012, 87(3), 268–279 CrossRef PubMed.
- J. M. Carmena, et al., Learning to control a brain-machine interface for reaching and grasping by primates, PLoS Biol., 2003, 1(2), E42 CrossRef PubMed.
- S. Saha, et al., Progress in Brain Computer Interface: Challenges and Opportunities, Front. Syst. Neurosci., 2021, 15, 578875 CrossRef PubMed.
- J. Tang, et al., Towards BCI-actuated smart wheelchair system, Biomed. Eng. Online, 2018, 17(1), 111 CrossRef PubMed.
- J. O. Usoro, et al., Intracortical Microelectrode Array Unit Yield under Chronic Conditions: A Comparative Evaluation, Micromachines, 2021, 12(8), 972 CrossRef PubMed.
- G. C. McConnell, et al., Implanted neural electrodes cause chronic, local inflammation that is correlated with local neurodegeneration, J. Neural Eng., 2009, 6(5), 056003 CrossRef PubMed.
- S. Debnath, et al., Long-term stability of neural signals from microwire arrays implanted in common marmoset motor cortex and striatum, Biomed. Phys. Eng. Express, 2018, 4(5), 055025 CrossRef PubMed.
- R. Biran, D. C. Martin and P. A. Tresco, Neuronal cell loss accompanies the brain tissue response to chronically implanted silicon microelectrode arrays, Exp. Neurol., 2005, 195(1), 115–126 CrossRef CAS PubMed.
- T. D. Kozai, et al., Comprehensive chronic laminar single-unit, multi-unit, and local field potential recording performance with planar single shank electrode arrays, J. Neurosci. Methods, 2015, 242, 15–40 CrossRef PubMed.
- M. Jorfi, et al., Progress towards biocompatible intracortical microelectrodes for neural interfacing applications, J. Neural Eng., 2015, 12(1), 011001 CrossRef PubMed.
- J. C. Barrese, et al., Failure mode analysis of silicon-based intracortical microelectrode arrays in non-human primates, J. Neural Eng., 2013, 10(6), 066014 CrossRef PubMed.
- K. Chen, et al., In vivo spatiotemporal patterns of oligodendrocyte and myelin damage at the neural electrode interface, Biomaterials, 2021, 268, 120526 CrossRef CAS PubMed.
- A. Sharon, et al., Inflammatory Foreign Body Response Induced by Neuro-Implants in Rat Cortices Depleted of Resident Microglia by a CSF1R Inhibitor and Its Implications, Front. Neurosci., 2021, 15, 646914 CrossRef PubMed.
- J. W. Salatino, et al., Glial responses to implanted electrodes in the brain, Nat. Biomed. Eng., 2017, 1(11), 862–877 CrossRef CAS PubMed.
- T. Saxena, et al., The impact of chronic blood-brain barrier breach on intracortical electrode function, Biomaterials, 2013, 34(20), 4703–4713 CrossRef CAS PubMed.
- M. Ravikumar, et al., The Roles of Blood-derived Macrophages and Resident Microglia in the Neuroinflammatory Response to Implanted Intracortical Microelectrodes, Biomaterials, 2014, 35(28), 8049–8064 CrossRef CAS PubMed.
- A. Campbell and C. Wu, Chronically Implanted Intracranial Electrodes: Tissue Reaction and Electrical Changes, Micromachines, 2018, 9(9), 430 CrossRef PubMed.
- H. W. Bedell, et al., Targeting CD14 on blood derived cells improves intracortical microelectrode performance, Biomaterials, 2018, 163, 163–173 CrossRef CAS PubMed.
- E. Azemi, C. F. Lagenaur and X. T. Cui, The surface immobilization of the neural adhesion molecule L1 on neural probes and its effect on neuronal density and gliosis at the probe/tissue interface, Biomaterials, 2011, 32(3), 681–692 CrossRef CAS PubMed.
- G. F. Hoeferlin, et al., Antioxidant Dimethyl Fumarate Temporarily but Not Chronically Improves Intracortical Microelectrode Performance, Micromachines, 2023, 14(10), 1902 CrossRef PubMed.
- A. Prasad, et al., Abiotic-biotic characterization of Pt/Ir microelectrode arrays in chronic implants, Front. Neuroeng., 2014, 7, 2 CAS.
- J. C. Barrese, J. Aceros and J. P. Donoghue, Scanning electron microscopy of chronically implanted intracortical microelectrode arrays in non-human primates, J. Neural Eng., 2016, 13(2), 026003 CrossRef PubMed.
- E. Fernandez, et al., Acute human brain responses to intracortical microelectrode arrays: challenges and future prospects, Front. Neuroeng., 2014, 7, 24 Search PubMed.
- Z. Gao, et al., Restoring After Central Nervous System Injuries: Neural Mechanisms and Translational Applications of Motor Recovery, Neurosci. Bull., 2022, 38(12), 1569–1587 CrossRef PubMed.
- B. Yang, et al., Strategies and prospects of effective neural circuits reconstruction after spinal cord injury, Cell Death Dis., 2020, 11(6), 82–89 Search PubMed.
- J. K. Hermann and J. R. Capadona, Understanding the Role of Innate Immunity in the Response to Intracortical Microelectrodes, Crit. Rev. Biomed. Eng., 2018, 46(4), 341–367 CrossRef PubMed.
- A. Picca, et al., Extracellular Vesicles and Damage-Associated Molecular Patterns: A Pandora’s Box in Health and Disease, Front. Immunol., 2020, 11, 601740 CrossRef CAS PubMed.
- S. A. Liddelow, et al., Neurotoxic reactive astrocytes are induced by activated microglia, Nature, 2017, 541(7638), 481–487 CrossRef CAS PubMed.
- S. Guo, H. Wang and Y. Yin, Microglia Polarization From M1 to M2 in Neurodegenerative Diseases. Frontiers in Aging, Neuroscience, 2022, 14, 815347 CAS.
- E. S. Ereifej, et al., Implantation of Neural Probes in the Brain Elicits Oxidative Stress, Front. Bioeng. Biotechnol., 2018, 6, 9 CrossRef PubMed.
- Y. Che, et al., Oxidative damage to RNA but not DNA in the hippocampus of patients with major mental illness, J. Psychiatry Neurosci., 2010, 35(5), 296–302 CrossRef PubMed.
- G. Buzsaki, Large-scale recording of neuronal ensembles, Nat. Neurosci., 2004, 7(5), 446–451 CrossRef CAS PubMed.
- V. S. Polikov, P. A. Tresco and W. M. Reichert, Response of brain tissue to chronically implanted neural electrodes, J. Neurosci. Methods, 2005, 148(1), 1–18 CrossRef PubMed.
- Z. Liu, et al., Beneficial effects of gfap/vimentin reactive astrocytes for axonal remodeling and motor behavioral recovery in mice after stroke, Glia, 2014, 62(12), 2022–2033 CrossRef PubMed.
- T. D. Kozai, et al., Chronic tissue response to carboxymethyl cellulose based dissolvable insertion needle for ultra-small neural probes, Biomaterials, 2014, 35(34), 9255–9268 CrossRef CAS PubMed.
- M. E. Urdaneta, et al., Layer-dependent stability of intracortical recordings and neuronal cell loss, Front. Neurosci., 2023, 17, 1096097 CrossRef PubMed.
- J. K. Nguyen, et al., Mechanically-compliant intracortical implants reduce the neuroinflammatory response, J. Neural Eng., 2014, 11(5), 056014 CrossRef PubMed.
- J. R. Abbott, et al., Planar amorphous silicon carbide microelectrode arrays for chronic recording in rat motor cortex, Biomaterials, 2024, 308, 122543 CrossRef CAS PubMed.
- J. Gaire, et al., The role of inflammation on the functionality of intracortical microelectrodes, J. Neural Eng., 2018, 15(6), 066027 CrossRef PubMed.
- Q. Whitsitt, et al., Spatial transcriptomics at the brain-electrode interface in rat motor cortex and the relationship to recording quality, J. Neural Eng., 2024, 21, 046033 CrossRef PubMed.
- H. W. Bedell, et al., Understanding the Effects of Both CD14-Mediated Innate Immunity and Device/Tissue Mechanical Mismatch in the Neuroinflammatory Response to Intracortical Microelectrodes, Front. Neurosci., 2018, 12, 772 CrossRef PubMed.
- P. R. Patel, et al., Utah array characterization and histological analysis of a multi-year implant in non-human primate motor and sensory cortices, J. Neural Eng., 2023, 20(1), 014001 CrossRef PubMed.
- V. V. Gusel'nikova and D. E. Korzhevskiy, NeuN As a Neuronal Nuclear Antigen and Neuron Differentiation Marker, Acta Nat., 2015, 7(2), 42–47 CrossRef.
- H. K. Wolf, et al., NeuN: a useful neuronal marker for diagnostic histopathology, J. Histochem. Cytochem., 1996, 44(10), 1167–1171 CrossRef CAS PubMed.
- R. J. Mullen, C. R. Buck and A. M. Smith, NeuN, a neuronal specific nuclear protein in vertebrates, Development, 1992, 116(1), 201–211 CrossRef CAS PubMed.
- X. Liu, et al., Evaluation of the stability of intracortical microelectrode arrays, IEEE Trans. Neural Syst. Rehabil. Eng., 2006, 14(1), 91–100 Search PubMed.
- S. Song, et al., Comprehensive proteomic analysis of the differential expression of 62 proteins following intracortical microelectrode implantation, Sci. Rep., 2024, 14, 17596 CrossRef CAS PubMed.
- K. Nishida, et al., Effects of three microtubule-associated proteins (MAP2, MAP4, and Tau) on microtubules' physical properties and neurite morphology, Sci. Rep., 2023, 13(1), 8870 CrossRef CAS PubMed.
- R. A. DeGiosio, et al., More than a marker: potential pathogenic functions of MAP2, Front. Mol. Neurosci., 2022, 15, 974890 CrossRef CAS PubMed.
- H. Herrmann and U. Aebi, Intermediate Filaments: Structure and Assembly, Cold Spring Harbor Perspect. Biol., 2016, 5(1), 3–11 Search PubMed.
- M. T. Sainio, et al., Neurofilament Light Regulates Axon Caliber, Synaptic Activity, and Organelle Trafficking in Cultured Human Motor Neurons, Front. Cell Dev. Biol., 2021, 9, 820105 CrossRef PubMed.
- E. A. van Vliet, et al., Decreased expression of synaptic vesicle protein 2A, the binding site for levetiracetam, during epileptogenesis and chronic epilepsy, Epilepsia, 2009, 50(3), 422–433 CrossRef CAS PubMed.
- X. M. Wu, et al., Effect of connexin 36 blockers on the neuronal cytoskeleton and synaptic plasticity in kainic acid-kindled rats, Transl. Neurosci., 2015, 6(1), 252–258 CrossRef CAS PubMed.
- M. E. Serrano, et al., Imaging Synaptic Density: The Next Holy Grail of Neuroscience?, Front. Neurosci., 2022, 16, 796129 CrossRef PubMed.
- G. Harauz, V. Ladizhansky and J. M. Boggs, Structural polymorphism and multifunctionality of myelin basic protein, Biochemistry, 2009, 48(34), 8094–8104 CrossRef CAS PubMed.
- V. Martinsen and P. Kursula, Multiple sclerosis and myelin basic protein: insights into protein disorder and disease, Amino Acids, 2022, 54(1), 99–109 CrossRef CAS PubMed.
- H. Wang, et al., Region-specific distribution of Olig2-expressing astrocytes in adult mouse brain and spinal cord, Mol. Brain, 2021, 14(1), 36 CrossRef CAS PubMed.
- K. Zhang, et al., The Oligodendrocyte Transcription Factor 2 OLIG2 regulates transcriptional repression during myelinogenesis in rodents, Nat. Commun., 2022, 13(1), 1423 CrossRef CAS PubMed.
- S. Song, et al., Neuroinflammatory Gene Expression Analysis Reveals Pathways of Interest as Potential Targets to Improve the Recording Performance of Intracortical Microelectrodes, Cells, 2022, 11(15), 2348 CrossRef CAS PubMed.
- H. W. Bedell, et al., Differential expression of genes involved in the acute innate immune response to intracortical microelectrodes, Acta Biomater., 2020, 102, 205–219 CrossRef CAS PubMed.
- Hadley Wickham, J.B., readxl: Read Excel Files. 2023.
- H. Wickham, M. Averick, J. Bryan, W. Chang, L. McGowan, R. François, G. Grolemund, A. Hayes, L. Henry, J. Hester, M. Kuhn, T. L. Pedersen, E. Miller, S. M. Bache, K. Müller, J. Ooms, D. Robinson, D. P. Seidel, V. Spinu, K. Takahashi, D. Vaughan, C. Wilke, K. Woo and H. Yutani, Welcome to the Tidyverse, J. Open Source Software, 2019, 4(43), 1686–1692 CrossRef.
- G. Shu, et al., Performance assessment of kernel density clustering for gene expression profile data, Comp. Funct. Genomics, 2003, 4(3), 287–299 CrossRef CAS PubMed.
- K. A. Potter, et al., The effect of resveratrol on neurodegeneration and blood brain barrier stability surrounding intracortical microelectrodes, Biomaterials, 2013, 34(29), 7001–7015 CrossRef CAS PubMed.
- J. Wang and Z. He, NAD and axon degeneration: from the Wlds gene to neurochemistry, Cell Adhes. Migr., 2009, 3(1), 77–87 CrossRef PubMed.
- Y. Yong, et al., Axonal spheroids in neurodegeneration, Mol. Cell. Neurosci., 2021, 117, 103679 CrossRef CAS PubMed.
- V. Zotey, et al., Adaptive Neuroplasticity in Brain Injury Recovery: Strategies and Insights, Cureus, 2023, 15(9), e45873 Search PubMed.
- R. J. Nudo, Recovery after brain injury: mechanisms and principles, Front. Hum. Neurosci., 2013, 7, 887 Search PubMed.
- D. Dewar and D. A. Dawson, Changes of cytoskeletal protein immunostaining in myelinated fibre tracts after focal cerebral ischaemia in the rat, Acta Neuropathol., 1997, 93(1), 71–77 CrossRef CAS PubMed.
- N. Ogata, et al., Degradation of neurofilament protein in cerebral ischemia, J. Neurosurg., 1989, 70(1), 103–107 CAS.
- C. Kirch and L. L. Gollo, Single-neuron dynamical effects of dendritic pruning implicated in aging and neurodegeneration: towards a measure of neuronal reserve, Sci. Rep., 2021, 11(1), 1309 CrossRef CAS PubMed.
- L. C. Pettigrew, et al., Microtubular proteolysis in focal cerebral ischemia, J. Cereb. Blood Flow Metab., 1996, 16(6), 1189–1202 CrossRef CAS PubMed.
- S. Narayanan, et al., Neurofilament light: a narrative review on biomarker utility, Fac. Rev., 2021, 10, 46 CAS.
- J. W. Ahn, et al., Serum neurofilament light chain levels are correlated with the infarct volume in patients with acute ischemic stroke, Medicine, 2022, 101(39), e30849 CrossRef CAS PubMed.
- T. C. Wheeler, et al., Regulation of synaptophysin degradation by mammalian homologues of seven in absentia, J. Biol. Chem., 2002, 277(12), 10273–10282 CrossRef CAS PubMed.
- K. Chen, F. Cambi and T. D. Y. Kozai, Pro-myelinating clemastine administration improves recording performance of chronically implanted microelectrodes and nearby neuronal health, Biomaterials, 2023, 301, 122210 CrossRef CAS PubMed.
- C. H. Thompson, et al., Spatiotemporal patterns of gene expression around implanted silicon electrode arrays, J. Neural Eng., 2021, 18(4), 045005 CrossRef PubMed.
- R. G. Mira, M. Lira and W. Cerpa, Traumatic Brain Injury: Mechanisms of Glial Response, Front. Physiol., 2021, 12, 740939 CrossRef PubMed.
- H. Konishi, S. Koizumi and H. Kiyama, Phagocytic astrocytes: Emerging from the shadows of microglia, Glia, 2022, 70(6), 1009–1026 CrossRef PubMed.
- M. Meschkat, et al., White matter integrity in mice requires continuous myelin synthesis at the inner tongue, Nat. Commun., 2022, 13(1), 1163 CrossRef CAS PubMed.
- M. T. Weil, et al., Loss of Myelin Basic Protein Function Triggers Myelin Breakdown in Models of Demyelinating Diseases, Cell Rep., 2016, 16(2), 314–322 CrossRef CAS PubMed.
- J. S. Coggan, et al., Physiological Dynamics in Demyelinating Diseases: Unraveling Complex Relationships through Computer Modeling, Int. J. Mol. Sci., 2015, 16(9), 21215–21236 CrossRef CAS PubMed.
- G. Leodori, et al., Neural bases of motor fatigue in multiple sclerosis: A multimodal approach using neuromuscular assessment and TMS-EEG, Neurobiol. Dis., 2023, 180, 106073 CrossRef PubMed.
- K. Makhoul, et al., Tremor in Multiple Sclerosis-An Overview and Future Perspectives, Brain Sci., 2020, 10(10), 722 CrossRef PubMed.
- H. Ford, Clinical presentation and diagnosis of multiple sclerosis, Clin. Med., 2020, 20(4), 380–383 CrossRef PubMed.
- E. Maes, et al., FACS-Based Proteomics Enables Profiling of Proteins in Rare Cell Populations, Int. J. Mol. Sci., 2020, 21(18), 6557 CrossRef CAS PubMed.
- S. Al-Amrani, et al., Proteomics: Concepts and applications in human medicine, World J. Biol. Chem., 2021, 12(5), 57–69 CrossRef PubMed.
- J. Usoro, et al., On the definition of ‘chronic’ for intracortical microelectrode array applications, Micromachines, 2021, 12(8), 972 CrossRef PubMed.
- D. H. Szarowski, et al., Brain responses to micro-machined silicon devices, Brain Res., 2003, 983(1–2), 23–35 CrossRef CAS PubMed.
- L. Karumbaiah, et al., Relationship between intracortical electrode design and chronic recording function, Biomaterials, 2013, 34(33), 8061–8074 CrossRef CAS PubMed.
- T. D. Y. Kozai, et al., Brain Tissue Responses to Neural Implants Impact Signal Sensitivity and Intervention Strategies, ACS Chem. Neurosci., 2015, 6(1), 48–67 CrossRef CAS PubMed.
- K. A. Potter, et al., Stab injury and device implantation within the brain results in inversely multiphasic neuroinflammatory and neurodegenerative responses, J. Neural Eng., 2012, 9(4), 046020 CrossRef PubMed.
- G. Buzsaki, C. A. Anastassiou and C. Koch, The origin of extracellular fields and currents-EEG, ECoG, LFP and spikes, Nat. Rev. Neurosci., 2012, 13(6), 407–420 CrossRef CAS PubMed.
- M. Goss-Varley, et al., Microelectrode implantation in motor cortex causes fine motor deficit: Implications on potential considerations to Brain Computer Interfacing and Human Augmentation, Sci. Rep., 2017, 7(1), 15254 CrossRef PubMed.
- T. J. Smith, Behavioral Paradigm for the Evaluation of Stimulation-Evoked Somatosensory Perception Thresholds in Rats, Front. Neurosci., 2023, 17, 1202258 CrossRef PubMed.
- F. R. Willett, et al., A high-performance speech neuroprosthesis, Nature, 2023, 620(7976), 1031–1036 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb01628a |
| This journal is © The Royal Society of Chemistry 2024 |