Open Access Article
Open Access ArticleGelatin methacryloyl granular hydrogel scaffolds for skin wound healing†
Arian Jaberi‡
 a,
Pejman Ghelich‡b,
Mohamadmahdi Samandarib,
Sina Kheirabadi
a,
Pejman Ghelich‡b,
Mohamadmahdi Samandarib,
Sina Kheirabadi a,
Zaman Ataie
a,
Zaman Ataie a,
Alexander Kedzierskic,
Alireza Hassani Najafabadi
a,
Alexander Kedzierskic,
Alireza Hassani Najafabadi bd,
Ali Tamayol
bd,
Ali Tamayol *b and
Amir Sheikhi
*b and
Amir Sheikhi *acefg
*acefg
aDepartment of Chemical Engineering, The Pennsylvania State University, University Park, PA 16802, USA. E-mail: sheikhi@psu.edu
bDepartment of Biomedical Engineering, University of Connecticut, Farmington, CT 06030, USA. E-mail: atamayol@uchc.edu
cDepartment of Biomedical Engineering, The Pennsylvania State University, University Park, PA 16802, USA
dTerasaki Institute for Biomedical Innovation, Los Angeles, CA 90024, USA
eHuck Institutes of the Life Sciences, The Pennsylvania State University, University Park, PA 16802, USA
fDepartment of Chemistry, The Pennsylvania State University, University Park, PA 16802, USA
gDepartment of Neurosurgery, College of Medicine, The Pennsylvania State University, Hershey, PA 17033, USA
First published on 15th April 2025
Abstract
The pore size and structure of hydrogel scaffolds play a key role in regulating host-scaffold interactions. Incorporating macropores within bulk hydrogels may increase cell ingrowth and modulate scaffold-induced inflammation. To this end, granular hydrogel scaffolds (GHS) have been developed via assembling hydrogel microparticles (microgels). GHS have interconnected cell-scale pores, tailored by microgel size, which are readily accessible to cells. Although bulk gelatin methacryloyl (GelMA) hydrogel scaffolds have frequently been used for tissue regeneration, the efficacy of GelMA GHS in wound healing remains unknown. Here, GelMA GHS are fabricated using microfluidic-generated near-uniform microgels to study the effect of macropores on macrophage behavior in vitro, followed by assessing wound healing in a murine model of full thickness skin injury. Compared with the bulk hydrogel counterpart, macrophages interfaced with GHS secrete less interferon gamma (IFN-γ) and more insulin-like growth factor 1 (IGF-1), which show a transition to pro-healing activities. In addition, GelMA GHS improve the quality of wound healing via increasing the thickness of granulation tissue and downregulating inflammatory markers without affecting the wound closure rate. This work is a step forward in engineering GelMA scaffolds with tailored porosity for wound care.
Wound healing occurs in a cascade of biological processes, including hemostasis, inflammation, proliferation (granulation and contraction), and maturation (remodeling).1,2 Multiple strategies, such as cell therapy,3 applying electrical or mechanical stimuli,4,5 dressings,6,7 and cytokine and growth factor (GF) delivery8 have been investigated to enhance the quality of wound healing.9 Biomaterials, specifically hydrogels providing a supportive temporary structure for tissue regeneration, have emerged as platforms to facilitate the processes pertaining to wound healing.10–12 Biocompatible hydrogels made up of crosslinked hydrophilic polymer networks, such as gelatin methacryloyl (GelMA), are capable of delivering biologics to promote tissue regeneration;13,14 however, their tissue integration is hampered by physical restrictions associated with nanoscale pores, which must undergo degradation and/or remodeling to allow cell infiltration, often causing excessive inflammation.13,14
Macroporous scaffolds overcome some of the barriers of bulk hydrogel scaffolds. Several studies demonstrated the benefits of GelMA scaffold macropores in promoting cell ingrowth and tissue regeneration.15–17 Granular hydrogel scaffolds (GHS), formed by interlinking packed (jammed) hydrogel microparticles (microgels), have microscale pores that are interconnected and enable rapid cell infiltration, tissue integration, and immunomodulation.13,18 GHS may be fabricated using varying microgel sizes to provide a range of cell-scale void spaces that promote cell infiltration and metabolite transport without requiring biodegradation.19–21 Cell infiltration within the interconnected void spaces enables the secretion and transport of GFs and cytokines that enhance cell signalling and downregulate pro-inflammatory responses to scaffolds.20 While GelMA bulk scaffolds have benefits in improving wound healing,22 the immune response and the quality of granulation tissue following healing with GelMA GHS have not yet been investigated. Here, we aim to investigate the pro-healing potential of GelMA GHS via in vitro macrophage function assessment, followed by in vivo implantation in a full-thickness murine wound model.
GelMA microgels are fabricated in two sizes, as shown in Fig. 1A and B, to assemble GHS with varying pore size distributions. The schematic (Fig. 1A) presents three biomaterial groups studied in this work, including bulk GelMA hydrogel (bulk) that is known to have nanoscale pores in its hydrated state,23–25 and GHS comprising large microgels (GHS-L) or small microgels (GHS-S), representing scaffolds with macroscale pores. Fig. 1Ai shows the GelMA chemical structure, and Fig. 1Aii schematically presents the inter- and intra-microgel covalent bond formation of GelMA polymer in GHS. Fig. 1B shows representative optical images of photocrosslinked microgels, which are near-uniform in size. Fig. 1C shows that the average diameter of photocrosslinked microgels is 83 ± 4 and 34 ± 4 μm for the large and small microgels, respectively.
The physically crosslinked microgels are packed and photocrosslinked using ultraviolet (UV) light to form GHS with interconnected void spaces, which are analyzed via fluorescence microscopy in Fig. 1D. The fluorescent dye occupies the void spaces of GHS without penetrating the microgels as a result of a high molecular weight. Fig. 1Di and Dii present the orthographic view of GHS-L and GHS-S, respectively. Fig. 1Diii and Div show the fluorescence images of void spaces among the microgels, and Fig. 1Dv and Dvi present the detected void area using a MATLAB code. The detected area was used to calculate the equivalent diameter of representative circles with the same area as the void spaces, showing that GHS-L had larger pores than GHS-S. Fig. 1E and F present the GHS void fraction and median equivalent pore diameter, respectively. The void fraction of GelMA GHS-L and GHS-S is 24 ± 1 and 25 ± 2% v/v, respectively. Additionally, the median equivalent pore diameter of GelMA GHS-L and GHS-S is 16 ± 2 and 9 ± 2 μm, respectively.
The mechanical properties of scaffolds were analyzed via compression tests and rheology. Fig. 1G shows the compressive stress–strain curves of a bulk scaffold, GHS-L, and GHS-S. At a given compressive strain, the compressive stress of GHS is smaller than the bulk counterpart as a result of macropores. The compressive modulus of scaffolds is calculated based on the slope of the compressive stress versus compressive strain in the linear region. Fig. 1H presents the average compressive modulus of the bulk scaffold, GHS-L, and GHS-S. The GHS-L has the lowest compressive modulus compared with the GHS-S and bulk GelMA scaffolds, which is attributed to the larger void space and weakened connections among the GelMA microgels per unit volume. The lower number density of microgel contact points and larger void space in GHS compared with the crosslinked polymeric network in the nanoporous (bulk) scaffold result in the difference in compressive modulus.13
Fig. 1I presents the scanning electron microscopy (SEM) images of dehydrated GHS-L and GHS-S, showing microgel-microgel connections. The dynamic moduli of scaffolds are shown in Fig. S1.† Fig. S1A† shows a schematic for the frequency sweep test. Fig. S1B† presents examples of scaffold dynamic moduli versus angular frequency at a constant, low oscillatory strain. The average storage modulus (G′) of GHS-L, GHS-S, and bulk scaffolds at the angular frequency of 1 rad s−1 and strain of 0.1% is 6400 ± 960, 6500 ± 1600, and 6700 ± 1800 Pa, respectively. The storage modulus of GHS is not significantly different than that of the bulk scaffold, whereas the compressive modulus of bulk scaffold is much higher than the GHS. In addition, the average loss modulus (G′′) of GHS-L, GHS-S, and bulk scaffold at the same angular frequency and strain is 1060 ± 270, 390 ± 70, and 530 ± 160 Pa, respectively, which are again not significantly different. Accordingly, the GHS and bulk scaffold behave similarly under oscillatory shear; however, the bulk scaffold has a significantly higher compressive modulus.
The GHS toxicity is assessed by interfacing them with NIH/3T3 murine fibroblast cells in vitro. Cells are seeded on top of GHS and allowed to penetrate, as schematically presented in Fig. 2A, followed by culturing for 5 days and conducting the live/dead (Fig. 2B) and PrestoBlue assays. Fig. 2B presents fluorescence images of live and dead cells in GHS-L, GHS-S, and the bulk hydrogel counterpart on days 1 and 5 of culture. Live cells are stained green with calcein acetoxymethyl (AM), while dead cell nuclei are stained red with Bobo-3 iodide. Dominant green areas indicate that the majority of cells are alive, and the red area show that some cells are dead. Fig. 2C shows the effect of microgel size on cell migration 3 days after seeding. Cell immediate penetration on day 0 (4 h after seeding) and migration length on day 3 are analyzed via measuring the distance of green CellTracker-stained cells from the surface of scaffolds. Qualitatively, the results show that the highest immediate penetration and long-term migration occur in GHS-L. Fig. 2D presents the cell viability quantification on days 1 and 5 after seeding, which show that the biomaterial and fabrication process do not induce any toxicity as the cell viability is ∼97 ± 3% on day 1 and ∼96 ± 2% on day 5. Interestingly, the PrestoBlue assay indicates a higher metabolic activity for the cells cultured in the GHS-L compared with those cultured in the GHS-S and bulk scaffolds (Fig. 2E). The metabolic activity increases ∼3.5, ∼2.6, and ∼1.8 fold on day 5 of culture for GHS-L, GHS-S, and bulk, respectively, compared with day 1. This may be a result of GHS-L larger pores, which enhance cell migration inside the scaffolds without the need for network degradation, providing the cells with more space to proliferate. Fig. 2F shows the average cell migration length in the scaffolds, topically seeded with the cells. The average cell immediate penetration length for GHS-L and GHS-S is 236 ± 28 and 93 ± 18 μm, respectively, 4 h after seeding. Additionally, the cell migration length increases to 308 ± 17 and 100 ± 18 μm for GHS-L and GHS-S, respectively, 3 days after seeding. Results suggest that the bulk scaffold does not support cell migration as a result of nanoscale pores. Additionally, in 3 days, cells migrate approximately 71 ± 29 μm in GHS-L and 7 ± 3 μm in GHS-S. Thus, GHS-L facilitate cell migration and proliferation significantly more than the other study groups.
The immune response to the GHS is evaluated in vitro using bone marrow-derived macrophages (BMDM). The isolated BMDM are cultured and activated overnight with lipopolysaccharide (LPS) to create an inflammatory environment in vitro, as shown in Fig. 2G. The culture media are then harvested after 2 days of cell culture with the scaffolds, and flow cytometry and an enzyme-linked immunosorbent assay (ELISA) are used to measure phenotype changes and the concentration of key secretomes, respectively. Fig. S2† shows the polarization of pro-inflammatory macrophages toward anti-inflammatory phenotypes, identified using flow cytometry. The results show the ratio of CD38+ cells (M1) is 78 ± 6% for GHS-L, compared with approximately 95% in the other study groups. Additionally, the average ratio of CD206+ cells (M2) is 22 ± 2% for GHS-L, while it remains around 3% in the other study groups. Accordingly, GHS-L macropores can modulate the inflammatory function of macrophages and direct them toward anti-inflammatory phenotypes. Fig. 2H–K show the ELISA analysis of interferon-gamma (IFN-γ), vascular endothelial growth factor (VEGF), insulin-like growth factor 1 (IGF-1), and interleukin 10 (IL-10) secretion, respectively, in the harvested media of control (no scaffold), bulk scaffold, GHS-L, and GHS-S. Fig. 2H shows a decrease in IFN-γ secretion in GHS-L and GHS-S compared with the control (no treatment). Additionally, GHS-L yields a lower IFN-γ secretion compared with its bulk counterpart. Fig. 2I and J show an increase in immunomodulatory GFs, VEGF and IGF-1, in GHS-L and GHS-S compared with the control. Interestingly, the secretion of IGF-1 in both GHS is significantly higher than that in the bulk scaffold counterpart. Fig. 2K shows that there are no significant differences in the secretion of IL-10 cytokine among the study groups. These results indicate the immunomodulatory capabilities of porous GHS-L and GHS-S, stimulating the secretion of anti-inflammatory markers and GFs by the immune cells, which are in accordance with other types of GHS.20,26
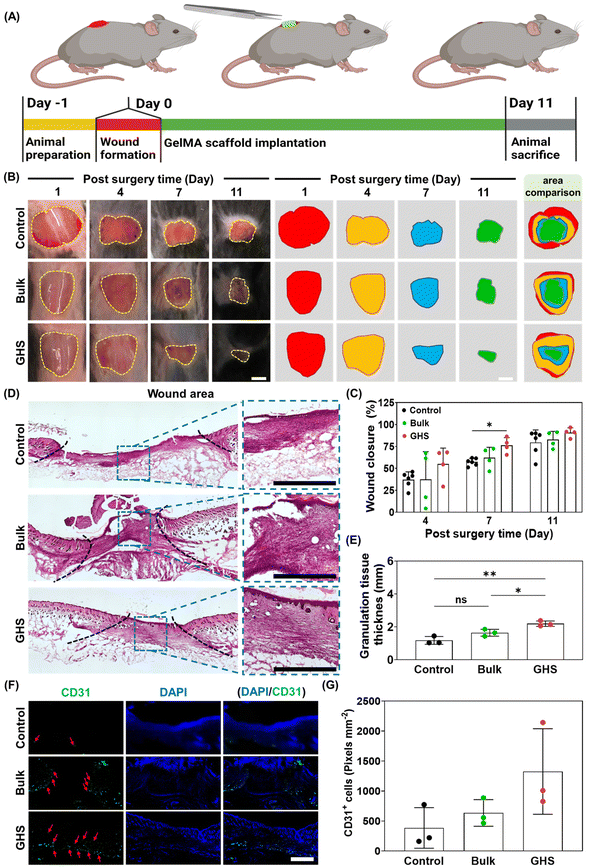
According to the in vitro results, specifically comparing the cell migration length, metabolic activity, and in vitro immune response in GHS-L and GHS-S, we select GHS-L (called GHS for simplicity through the rest of this work), as they provide a more suitable microenvironment for cell migration, which is beneficial for wound healing.21 The cascade of cell behavior pertaining to the healing of skin wounds may be improved by mechanical (e.g., an external substrate) and chemical stimuli (e.g., cytokines and GFs).27 Wound healing requires a complex interplay of immune response (i.e., pro-inflammation and anti-inflammation),12,27 cell migration,21 and tissue regeneration.26,28 Accordingly, an animal study is conducted using a full-thickness murine wound model. The wound is made using an ∼1 cm circular biopsy punch,29,30 as schematically shown in Fig. 3A. After forming the wound on the dorsum, a prefabricated scaffold is implanted as a treatment and secured with a Tegaderm, covering the wound area. Wound closure is monitored for 11 days after surgery. The splint-free wound model has previously been shown to enable wound closure of untreated wounds with an initial diameter of 6 mm within 8 days.31 Fig. 3B presents the wound closure area on days 1, 4, 7, and 11 after surgery for the control (no treatment), bulk scaffold, and GHS. The results show an improvement in the healing rate of all the study groups in 11 days. A similar wound closure rate is observed on day 11 for all the study groups, possibly as a result of the healthy condition of wild-type animals in which wounds close gradually through natural regeneration mechanisms (Fig. 3C). Wound closure mechanism in the wild-type animal model, while different from humans, results from re-epithelialization and granulation tissue formation, which are also involved in human wound healing.31,32
Wound closure is one of the many factors reflecting the effectiveness of a treatment for wound healing. It has been reported that excessive wound contraction leads to scarring and poor-quality wound healing.33 We further investigate wound healing quality through histological analyses. Hematoxylin and eosin (H&E) staining and Masson's trichrome combined with immunofluorescence staining for cluster of differentiation 31 (CD31), an endothelial cell marker demonstrating the infiltration of endothelial cells, F4/80, a macrophage cell marker, and CD80, a pro-inflammatory cell marker, are used to assess the granulation tissue and collagen deposition, as well as endothelial cell, macrophage, and pro-inflammatory cell infiltration in different treatment groups, respectively. Fig. 3D shows the H&E-stained images of harvested tissue for the control, bulk, and GHS. The wounded area is shown, and the granulation tissue thickness is analyzed by measuring the thickness of regenerated tissue. Additionally, Fig. S3A† presents the schematic of skin wound and normal tissue, as well as an H&E image of normal skin tissue, showing the epidermis and dermis layers of tissue prior to the wound formation. Fig. 3E presents the granulation tissue thickness (also see Fig. S3B and S3C†) of control, bulk, and GHS. Additionally, the dermis thickness of normal skin (without wound) is analyzed (Fig. S3D†). The granulation tissue thickness using the GHS treatment increases ∼1.8 and ∼1.3fold compared with the control and bulk scaffold, respectively. As shown in Fig. 3D and E, the granulation tissue thickness is significantly higher in GHS compared with the bulk hydrogel scaffold counterpart and no treatment, which may be attributed to a higher tissue integration with the porous GHS. Consistent with our results, GelMA porous scaffolds enabled tissue regeneration after volumetric muscle loss,34 and PEG-based GHS triggered tissue regeneration.26
Angiogenesis throughout the granulation tissue plays a significant role in the wound healing process.35 Accordingly, the infiltrated endothelial cells are stained with the CD31 marker. Fig. 3F shows the fluorescence images of CD31 stained cells in green and DAPI (cell nuclei) in blue for the control, bulk, and GHS. Qualitatively, the area marked in green is higher in GHS and bulk compared with the control. Fig. 3G presents the CD31 positive cell quantification for the study groups. The average of CD31 positive cells (total positive pixels) normalized with the tissue area is 384 ± 338, 634 ± 222, and 1325 ± 713 pixel per mm2 for the control, bulk, and GHS, respectively. While no significant difference in CD31 positive cells is obtained among the study groups, an increase in the average of endothelial cell number in GHS and bulk scaffold compared with the control is noticed. This may be attributed to a significantly higher level of VEGF secretion in GHS and bulk scaffold compared with the control (Fig. 2I). Overall, the results are consistent with our previous findings that introducing porosity in biodegradable scaffolds increases cell infiltration, enables vascularization, and remodels the scaffolds toward tissue regeneration.34
Additionally, to assess collagen deposition and inflammation within the wound bed, respectively, tissue sections are stained with Masson's trichrome (Fig. S4A†) and immunofluorescence-stained with F4/80, a well-established marker for mouse macrophage populations, and CD80, a marker for inflammation (Fig. S4C†).36 The average of collagen deposition area within the wound bed is 9.9 ± 6.7, 17.2 ± 5.0, and 19.5 ± 6.0 mm2 for the control, bulk, and GHS, respectively (Fig. S4B†). While no significant difference in the area of collagen deposition is obtained among the study groups, an increase in the average collagen deposition in GHS and bulk scaffolds compared with the control is noticed. The low number of F4/80 positive and CD80 positive signals shows that GelMA scaffolds do not induce inflammation within the wound bed (Fig. S4D and S4E†).
Together, the results imply that GelMA GHS enhance skin wound healing quality by promoting cell infiltration, metabolic activity, and partial pro-healing immune responses. Additionally, GelMA GHS improve granulation tissue thickness compared with the bulk GelMA scaffold counterparts. These findings highlight the potential of GelMA GHS as effective scaffolds for wound healing, which may warrant further biomaterial modifications to promote vascularization and immune modulation.
Experimental section/methods
GelMA synthesis
GelMA was synthesized according to an established protocol.24,37 Briefly, a 10% w/v solution of gelatin (type A from porcine skin, gel strength ∼300 g Bloom, MilliporeSigma, MA, USA) in Dulbecco's phosphate-buffered saline (DPBS, MilliporeSigma, MA, USA) was reacted with 8% v/v (final concentration) methacrylic anhydride (MilliporeSigma, MA, USA) for 2 h at 50 °C. The reaction was ended by diluting the solution two times using DPBS. The solution was dialyzed against 40 °C Milli-Q water (resistivity = 18.2 MΩ cm, Millipore Corporation, MA, USA) for 10 days. On day 10, the solution was diluted two times the initial volume using Milli-Q water. Afterward, the GelMA was filtered (vacuum filters, 0.2 μm, VWR, PA, USA), frozen at −80 °C for 24 h, and lyophilized for further use. The degree of substitution of GelMA was around 67 ± 2%, measured according to an established protocol using proton nuclear magnetic resonance (1H NMR, 400 MHz Bruker NEO, MA, USA) spectroscopy at the NMR facilities of Penn State (Fig. S5†).Microfluidic device fabrication
Step emulsification microfluidic devices were fabricated as previously described.37,38 Briefly, soft lithography was used for fabricating high-throughput step emulsification microfluidic devices in the nanofabrication facilities of the Materials Research Institute (MRI) at Penn State. Additionally, Sylgard 184 polydimethylsiloxane (PDMS, Dow Corning, MI, USA) with the base![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) crosslinker weight ratio of 10
crosslinker weight ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were mixed and poured over nanofabricated wafers and cured at 70 °C for 1 h. After air plasma treatment (Plasma Cleaner, Harrick Plasma, NY, USA), the cured PDMS devices were bonded to microscope glass slides (VWR, PA, USA) in an oven at 70 °C for 1 h.
1 were mixed and poured over nanofabricated wafers and cured at 70 °C for 1 h. After air plasma treatment (Plasma Cleaner, Harrick Plasma, NY, USA), the cured PDMS devices were bonded to microscope glass slides (VWR, PA, USA) in an oven at 70 °C for 1 h.
GelMA microgel fabrication
GelMA microgel formation was described in our previous protocols.37 In summary, the lyophilized GelMA was dissolved in a solution of lithium phenyl-2,4,6-trimethylbenzoylphosphinate (0.1% w/v, LAP, Allevi, MA, USA) in DPBS to prepare a GelMA solution with a final concentration of 7% w/v. The GelMA solution was used as an aqueous phase, and a solution of 2% v/v surfactant (Pico-Surf, Sphere Fluidics, Cambridge, UK) in Novec 7500 (3M, MN, USA) was used as an oil phase. The solutions were injected into step emulsification microfluidic devices with a step size of 8 μm or 27 μm to produce GelMA-in-oil emulsions, resulting in small (∼34 ± 4 μm) or large (∼83 ± 4 μm) GelMA droplets, respectively. The GelMA droplets were stored at 2 °C overnight, yielding physically crosslinked GelMA microgels.GelMA bulk scaffold fabrication
A GelMA solution (7% w/v) was prepared by dissolving the lyophilized GelMA in a LAP solution (0.1% w/v) in DPBS. GelMA bulk hydrogel scaffolds were fabricated by transferring the solution to acrylic molds (diameter = 10 mm and height = 3 mm), followed by physically crosslinking GelMA using ice for 10 min and chemically photocrosslinking for 30 s using the UV light (wavelength = 395–405 nm, intensity = 15 mW cm−2).GelMA GHS fabrication
GelMA microgels were washed with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio of a solution including 20% v/v 1H,1H,2H,2H-perfluoro-1-octanol (PFO, Alfa Aesar, MA, USA) in Novec 7500 and centrifuged at 300g for 15 s to remove the oil phase. The GelMA microgels were suspended in the LAP solution (0.1% w/v of LAP in DPBS) and were packed via centrifugation at 3000g for 15 s. The packed microgels were transferred to acrylic molds (diameter = 10 mm and height = 3 mm) and photocrosslinked via UV light (395–405 nm) exposure for 30 s at the intensity of 15 mW cm−2.
1 volume ratio of a solution including 20% v/v 1H,1H,2H,2H-perfluoro-1-octanol (PFO, Alfa Aesar, MA, USA) in Novec 7500 and centrifuged at 300g for 15 s to remove the oil phase. The GelMA microgels were suspended in the LAP solution (0.1% w/v of LAP in DPBS) and were packed via centrifugation at 3000g for 15 s. The packed microgels were transferred to acrylic molds (diameter = 10 mm and height = 3 mm) and photocrosslinked via UV light (395–405 nm) exposure for 30 s at the intensity of 15 mW cm−2.
GelMA GHS pore characterization
The photocrosslinked GelMA GHS were incubated with a high molecular weight fluorescein isothiocyanate–dextran (FITC–dextran) solution (Mw ∼2 MDa, 15 μM in Milli-Q water, MilliporeSigma, MA, USA) for 10 min, to fill the void spaces among the microgels with the fluorescence dye. The GelMA GHS were imaged and analyzed using a Leica DMi8 fluorescence microscope (THUNDER Imager 3D Cell Culture, Leica Microsystems, Germany). The Z-stack images were captured over a depth of around 100 μm within each sample, and the void fraction was analyzed by adjusting the threshold using the built-in microscope software (Leica Application Suite X, LAS X, 5.0.3 Life Science Microscope Software Platform, Germany). In addition, the median pore equivalent diameter was quantified using a custom-written MATLAB code (MATLAB, version 2020b), as previously described.19,37Compression test
The GelMA GHS and bulk scaffolds were fabricated in cylindrical acrylic molds (8 mm diameter and 3 mm height). The scaffolds were incubated in DPBS at ambient temperature for about 1 h before conducting a compression test. The compression test was performed using an Instron 5542 mechanical tester (Universal Testing Systems, Instron, MA, USA) at 1 mm min−1 strain rate. The compressive modulus was calculated based on the ratio of stress over strain at the linear range of low strain (∼0.5–0.15 mm mm−1).Rheological test
The GelMA GHS and bulk scaffolds were fabricated in cylindrical acrylic molds (diameter = 8 mm and height = 3 mm). Oscillatory frequency sweep was performed at frequency of 10−1 to 102 rad s−1 and 0.1% strain using a DHR-2 rheometer (TA instrument, DE, USA), equipped with the sandblasted parallel plates geometry with a diameter of 8 mm at 25 °C. The storage (G′) and loss (G′′) moduli at 1 rad s−1 and 0.1% oscillatory strain were used for comparing the rheological properties of scaffolds.Macrophage harvest, culture, flow cytometry, and ELISA assessments
BMDM were harvested as previously described.39 Bone marrow was flushed from the femur and tibia bones of C57BL/6 mice (8–10-week-old, female) with 10 mL complete culture media, containing (i) Dulbecco's modified Eagle's medium (DMEM, Gibco, MA, USA), (ii) 10% v/v fetal bovine serum (FBS, Gibco, MA, USA), and (iii) 1% v/v penicillin–streptomycin (PS, Gibco, MA, USA), followed by straining with 70 μm cell strainer (Corning, NJ, USA). Isolated BMDM were plated at 1 × 107 cells per 10 cm2 culture dish (Corning, NJ, USA) in the Roswell Park Memorial Institute-1640 medium (RPMI-1640, Gibco, IL, USA), containing 10% v/v FBS (Gibco, MA, USA), 55 μM β-mercaptoethanol (Gibco, MA, USA), 5 ng mL−1 macrophage colony-stimulating factor (MCSF, Thermofisher, IL, USA), and 100 U mL−1 PS (Gibco, MA, USA). Half of the culture media was replaced with fresh media on days 3 and 5. After 8 days, BMDM were scraped and plated at 1 × 106 cells per well in 12-well cell culture plates (Fisher scientific, IL, USA). BMDM cells were activated with 100 ng mL−1 lipopolysaccharide (LPS, E. coli 0111:B4, InvivoGen, CA, USA). After 24 h, LPS was removed and BMDM were washed three times with PBS. For the study groups, GHS and bulk scaffolds were placed over the cultured cells, while no scaffold was used for the control group. After 48 h of culture, supernatants were collected, and the secretion of different cytokines and GFs by BMDM were measured via ELISA (BioLegend, CA, USA) per manufacturer protocols. BMDMs were then rinsed twice with fluorescent activated cell sorting (FACS) buffer (1% bovine serum albumin (BSA, MilliporeSigma, MA, USA) in PBS), incubated with anti-CD16/32 (Invitrogen, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, CA, USA) at room temperature, and then stained on ice with fluorophore-labeled antibodies against CD38 (BioLegend, 1
20, CA, USA) at room temperature, and then stained on ice with fluorophore-labeled antibodies against CD38 (BioLegend, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, CA, USA), CD206 (BioLegend, 1
100, CA, USA), CD206 (BioLegend, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, CA, USA), and e45o fixable viability dye (eBioscience, 1
100, CA, USA), and e45o fixable viability dye (eBioscience, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, CA, USA). Cells were fixed, then washed twice with FACS buffer and analyzed by flow cytometry (Bio-Rad, CA, USA).40,41 The experiments were performed with n ≥ 4 for each group.
1000, CA, USA). Cells were fixed, then washed twice with FACS buffer and analyzed by flow cytometry (Bio-Rad, CA, USA).40,41 The experiments were performed with n ≥ 4 for each group.
Cell viability and metabolic activity analyses
The viability and metabolic activity of NIH/3T3 murine fibroblast cells (ATCC, VA, USA), cultured on GelMA GHS or bulk hydrogel scaffolds were studied. The cells were cultured to reach up to 90% confluency, trypsinized using a trypsin-ethylenediaminetetraacetic acid (EDTA) solution (0.25%, Gibco, MA, USA), and resuspended in complete culture media (cell concentration = 1 × 106 cells per mL). Disc-shaped scaffolds (diameter = 8 mm, height = 3 mm) were placed in 48-well cell culture plates (CELLSTAR, Greiner Bio-One, Austria), followed by the topical seeding of a 10 μL cell suspension. Then, the scaffolds were incubated at 37 °C in a humidified incubator (Eppendorf C170i, Germany) with 5% v/v CO2 for 30 min. Subsequently, 500 μL of culture media was added to each well, followed by placing the culture dish in an incubator under 5% v/v CO2 at 37 °C.Cell viability was measured using live/dead cell imaging kit (Invitrogen, MA, USA) on days 1 and 5 after seeding (n = 3) in accordance with the manufacturer protocol. Live cells were stained in green using calcein-AM (1 mL, 1 μM), and dead cells were stained in red using BOBO-3 iodide (1 μL, 5 μM). The samples were incubated at 25 °C for 30 min, rinsed with DPBS, and imaged using the Leica DMi8 fluorescence microscope (Leica Microsystems, Germany).
The metabolic activity of cells was analyzed using the PrestoBlue cell viability assay (10% v/v in DMEM, Invitrogen, MA, USA) (n = 5) on days 1, 3, and 5 post-seeding. For each scaffold, 1 mL of PrestoBlue solution (10% v/v) was incubated at 37 °C for 3 h, followed by collecting 100 μL of supernatant and transferring to 96-well cell culture plates (CELLSTAR, Greiner Bio-One, Austria). The fluorescence intensity was recorded (excitation wavelength = 530 nm and emission wavelength = 590 nm) using a Tecan Infinite M Plex microplate reader (Männedorf, Switzerland).
In vitro cell migration assay
To assess cell migration, NIH/3T3 cells were labeled with green CellTracker™ 5-chloromethylfluorescein diacetate (CMFDA, Invitrogen, MA, USA) dye, followed by topically seeding on the scaffolds. Prior to cell seeding, the scaffolds were incubated in 1% v/v PS in DPBS for 3 h in a 24-well cell culture plate (non-treated, CELLSTAR, Greiner Bio-One, Austria) at ambient temperature. Then, the DPBS was removed, and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 stained cells in 20 μL of complete culture media were seeded on each scaffold, followed by incubation at ambient temperature for 30 min to allow cell adhesion. Later, complete culture media were added, and the scaffolds were incubated at 37 °C under a 5% v/v CO2 atmosphere for 72 h. Finally, samples were sectioned using a razor blade (VWR, PA, USA) and imaged using the Leica DMi8 fluorescence microscope (Leica Microsystems, Germany). Image analysis was conducted using the ImageJ software (FIJI, version 1.53t, NIH, MD, USA).42
000 stained cells in 20 μL of complete culture media were seeded on each scaffold, followed by incubation at ambient temperature for 30 min to allow cell adhesion. Later, complete culture media were added, and the scaffolds were incubated at 37 °C under a 5% v/v CO2 atmosphere for 72 h. Finally, samples were sectioned using a razor blade (VWR, PA, USA) and imaged using the Leica DMi8 fluorescence microscope (Leica Microsystems, Germany). Image analysis was conducted using the ImageJ software (FIJI, version 1.53t, NIH, MD, USA).42
In vivo wound healing
Animal studies were performed at UCONN Health according to protocol (AP-200055-0723), approved by Institutional Animal Care and Use Committee (IACUC). Fourteen 8-week-old C57BL/6 female mice (The Jackson Laboratory, CT, USA) were acclimated for at least one week. The animals were shaved one day before surgery and received ketoprofen (5 mL kg−1, Zoetis, NJ, USA) as a pain killer on the day of surgery and the day after. Mice were anesthetized using 2% isoflurane in an oxygen carrier, followed by wound formation using a 1 cm circular biopsy punch.33,43 At least 4 animals per study group, including control (no treatment), bulk hydrogel scaffold, and GHS, were used. Subsequently, the wound site and scaffolds were covered by a Tegaderm transparent film dressing (3M, MA, USA). The wound closure was monitored on days 4, 7, and 11 after surgery. On day 11, the animals were euthanized using CO2 (3 L min−1), and their skin tissue was harvested and fixed in paraformaldehyde (PFA, 4% v/v, Thermo Scientific Chemicals, USA) overnight.Histological analyses: H&E, Masson's trichrome staining, and immunohistochemistry (IHC)
Fixed tissues were rehydrated in a sucrose solution (30% w/v, MilliporeSigma, MA, USA) overnight, followed by embedding in an optimal cutting temperature (OCT, Fisher scientific, IL, USA) solution and storing at −80 °C for 1 h. Then, samples were sectioned (thickness = 20 μm) using a Leica cryostat system (Leica Biosystems, Germany), and sections were temporarily mounted on plastic slides using a water droplet and cryofilm type 2C (Section-Lab Co., Japan). The sections were mounted permanently on positively charged glass slides (Fisher scientific, IL, USA) using a UV-curable adhesive (Norland Products, Inc., NJ, USA). To make sure the extra adhesive was removed, the sections were vertically stored in a fridge overnight. Samples were exposed to UV light (wavelength = 254 nm) with the power of 1 W (Stratalinker 1800, Stratagene, CA, USA) for 5 min, followed by soaking in DPBS for 10 min.Sectioned samples were stained with H&E according to a standard protocol.44 Briefly, samples were soaked in 95% v/v ethanol, followed by rinsing in deionized (DI) water. Samples were then incubated in hematoxylin (Leica Biosystems, IL, USA) for 2 min and rinsed with DI water, followed by incubation for 30 s in the bluing agent (Leica Biosystems, IL, USA) and rinsing with DI water. Later, samples were incubated in ethanol 95% v/v for 30 s, followed by 3 min incubation in eosin (Leica Biosystems, IL, USA). Stained sections were drained and washed with 100% v/v ethanol three times, and finally washed with toluene (Fisher scientific, IL, USA). A cover glass was mounted over the stained tissues using Permount Mounting Medium (Fisher scientific, IL, USA). Stained samples were imaged using the brightfield channel of a Zeiss Axio Observer microscope (Zeiss, Germany).
Preserved samples were stained with CD31, an endothelial marker. Briefly, samples were rinsed three times with DPBS, followed by antigen retrieval using citrate buffer (pH = 6, MilliporeSigma, MA, USA) at 60 °C overnight. Samples were blocked with normal goat serum (2.5% w/v, MilliporeSigma, MA, USA) for 30 min. The sections were incubated with rat anti-CD31 (ab56299, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, Abcam, CA, USA) overnight at 4 °C. The sectioned samples were rinsed three times with DPBS and incubated with goat anti-rat immunoglobulin G (IgG) Alexa Fluor 488 (A-11006, 1
100, Abcam, CA, USA) overnight at 4 °C. The sectioned samples were rinsed three times with DPBS and incubated with goat anti-rat immunoglobulin G (IgG) Alexa Fluor 488 (A-11006, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Thermo Fisher Scientific, IL, USA) secondary antibody at room temperature for 1 h, followed by rinsing three times with DPBS and mounting with a cover glass using glycerin (50% v/v, Fisher scientific, USA) with DAPI (DAPI
1000, Thermo Fisher Scientific, IL, USA) secondary antibody at room temperature for 1 h, followed by rinsing three times with DPBS and mounting with a cover glass using glycerin (50% v/v, Fisher scientific, USA) with DAPI (DAPI![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerin = 1
glycerin = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, Invitrogen, IL, USA). CD31-stained samples were imaged using the Zeiss Axio Observer fluorescence microscope (Carl Zeiss Inc, Germany).
100, Invitrogen, IL, USA). CD31-stained samples were imaged using the Zeiss Axio Observer fluorescence microscope (Carl Zeiss Inc, Germany).
Masson's trichrome stain kit (connective tissue stain, ab150686, Abcam, CA, USA) was used according to a standard protocol. Briefly, samples were hydrated in distilled water for 165 min. Bouin's fluid was preheated to 56–64 °C in a water bath under a fume hood, and samples were immersed in the fluid for 60 min. The slides were then rinsed under tap water until becoming clear, followed by rinsing in distilled water. Afterward, samples were stained with Weigert's iron hematoxylin for 5 min, then rinsed under running tap water for 2 min. Biebrich scarlet/acid fuchsin solution was applied for 15 min, followed by another rinse in distilled water. The phosphomolybdic/phosphotungstic acid solution was used for differentiating the samples for 15 min, followed by applying the aniline blue solution for 10 min and a rinse in distilled water. Acetic acid solution (1%) was applied to the samples for 5 min. Samples were dehydrated quickly through two changes of 95% v/v ethanol, followed by two changes of 100% v/v ethanol. They were then cleared in xylene, and a cover glass was mounted over the stained tissues using Permount Mounting Medium (Fisher scientific, IL, USA). Stained samples were imaged using the brightfield channel of a Keyence microscope (BZ-X800, Keyence Corporation, Japan).
Immunofluorescent staining was conducted on skin tissue samples following the treatment described in the H&E section. Preserved samples were stained with FITC anti-mouse F4/80 antibody (123107, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, BioLegend, CA, USA), a well-established marker for detecting macrophages in mice, and PE anti-mouse CD80 antibody (104707, 1
200, BioLegend, CA, USA), a well-established marker for detecting macrophages in mice, and PE anti-mouse CD80 antibody (104707, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, BioLegend, CA, USA), a subtype associated with pro-inflammatory responses.36,40 The CD80 marker served as an indicator of inflammation in the treated areas. Briefly, tissue sections were first rehydrated in PBS for 5 min, followed by incubation with 10% normal goat serum for 30 min in a humid chamber. Samples were incubated overnight at 4 °C with anti-mouse F4/80, CD80, and DAPI to visualize macrophages, cells with pro-inflammatory markers, and cell nuclei, respectively. Imaging was performed using a Keyence microscope (BZ-X800), and widefield fluorescent images were captured; green is F4/80+ cells, red is CD80+ cells, and blue is DAPI. Image analysis was conducted via quantifying the number of stained pixels within a region of interest (0.5 mm × 0.5 mm) in the wound bed using the ImageJ software (FIJI, version 1.53t, NIH, MD, USA).42
200, BioLegend, CA, USA), a subtype associated with pro-inflammatory responses.36,40 The CD80 marker served as an indicator of inflammation in the treated areas. Briefly, tissue sections were first rehydrated in PBS for 5 min, followed by incubation with 10% normal goat serum for 30 min in a humid chamber. Samples were incubated overnight at 4 °C with anti-mouse F4/80, CD80, and DAPI to visualize macrophages, cells with pro-inflammatory markers, and cell nuclei, respectively. Imaging was performed using a Keyence microscope (BZ-X800), and widefield fluorescent images were captured; green is F4/80+ cells, red is CD80+ cells, and blue is DAPI. Image analysis was conducted via quantifying the number of stained pixels within a region of interest (0.5 mm × 0.5 mm) in the wound bed using the ImageJ software (FIJI, version 1.53t, NIH, MD, USA).42
Statistical analyses
GraphPad Prism software (version 9.4.0 or 10.4.0) was used for the statistical analyses. The unpaired two-tailed t-test was used to compare the differences between two groups. Additionally, the one-way analysis of variance (ANOVA) was used for more than two groups and two-way ANOVA was used for the wound closure rate analysis, followed by the Tukey's multiple comparison test. The G*Power software (version 3.1.9.6) was used to calculate the number of mice (n = 4) based on at least 80% power, 0.05 probability of type I error, and 1.4 effect size.45 For the wound closure analysis, the control group included six animals. Additionally, for immunostaining, we were unable to analyze one of the scaffolds, thus for an 80% power and n = 3, the effect size was 2.1. Statistical significance was considered when p-values were lower than 0.05 (*p < 0.05), 0.01 (**p < 0.01), 0.001 (***p < 0.001), and 0.0001 (****p < 0.0001).Conclusions
In conclusion, in vitro and in vivo studies were conducted to assess the capability of GelMA GHS for skin wound healing. Cell proliferation and migration in the GHS with a larger pore diameter (GHS-L) were significantly higher than GHS-S and the bulk hydrogel counterpart. We speculate that this was partly because of higher accessible surface area to volume ratio and improved nutrient access throughout the scaffolds. The analysis of macrophage secretomes when interfaced with GelMA bulk scaffold and GHS showed that GHS decreased the IFN-γ secretion from macrophages and increased IGF-1 secretion compared with the control (no scaffold) and bulk counterpart. Additionally, GelMA scaffolds increased VEGF secretion from macrophages compared with the control. Based on the in vitro outcome, the hydrogel scaffold with a median pore size comparable to cell sizes (GHS-L) was selected for the skin wound healing, which promoted wound healing quality by increasing the granulation tissue thickness while yielding a similar wound closure rate to the control. This study shows the importance of macropores in GHS-based skin wound healing and lays the foundation of engineering this biomaterial platform for regenerative engineering. Future studies may investigate the performance of these scaffolds in animal models with wound healing processes more closely resembling those in humans.Author contributions
Arian Jaberi: experimental design, data curation, formal analysis, methodology, data processing, and writing – original draft. Pejman Ghelich: data curation, formal analysis, methodology, data processing, and writing – review and editing. Mohamadmahdi Samandari: data curation, formal analysis, methodology, and writing – review and editing. Sina Kheirabadi, Zaman Ataie, Alexander Kedzierski, and Alireza Hassani Najafabadi: data curation, data processing, and writing – review and editing. Ali Tamayol and Amir Sheikhi: conceptualization, methodology, experimental design, writing – review & editing, and funding. All authors who have made substantial contributions to the work reported in the manuscript are mentioned in this statement.Data availability
The data supporting this article have been included as part of the manuscript and ESI.† Raw data are available from the corresponding authors upon reasonable request.Conflicts of interest
M. Samandari and A. Tamayol are co-founders of InPrint Bio. A. Tamayol is also a co-founder of 3D PenBone. Other authors declare no competing conflict of interest.Acknowledgements
This research was partially supported by the National Heart, Lung, And Blood Institute (NHLBI) of the National Institutes of Health (NIH) under award number R01HL167939 (A. S.). Support from the Dorothy Foehr Huck and J. Lloyd Huck Early Career Chair and The Pennsylvania State University is acknowledged (A. S.). The financial support from the National Institutes of Health (R01GM126831, R01AR073822, and R01AR077132) and the University of Connecticut is gratefully acknowledged (A. T.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to thank S. Zavari for help with fluorescence microscopy. Parts of Fig. 1–3, S3, graphical abstract, and cover image were created using BioRender.com.References
- G. C. Gurtner, S. Werner, Y. Barrandon and M. T. Longaker, Nature, 2008, 453, 314–321 CrossRef CAS PubMed.
- M. Rodrigues, N. Kosaric, C. A. Bonham and G. C. Gurtner, Physiol. Rev., 2019, 99, 665–706 CrossRef CAS PubMed.
- E. Coalson, E. Bishop, W. Liu, Y. Feng, M. Spezia, B. Liu, Y. Shen, D. Wu, S. Du, A. J. Li, Z. Ye, L. Zhao, D. Cao, A. Li, O. Hagag, A. Deng, W. Liu, M. Li, R. C. Haydon, L. Shi, A. Athiviraham, M. J. Lee, J. M. Wolf, G. A. Ameer, T. C. He and R. R. Reid, Genes Dis., 2019, 6, 342–358 CrossRef PubMed.
- M. Verdes, K. Mace, L. Margetts and S. Cartmell, Curr. Opin. Biotechnol., 2022, 75, 102710 CrossRef CAS PubMed.
- L. A. Barnes, C. D. Marshall, T. Leavitt, M. S. Hu, A. L. Moore, J. G. Gonzalez, M. T. Longaker and G. C. Gurtner, Adv. Wound Care, 2018, 7, 47–56 CrossRef PubMed.
- R. Dong and B. Guo, Nano Today, 2021, 41, 101290 CrossRef CAS.
- J. Boateng and O. Catanzano, J. Pharm. Sci., 2015, 104, 3653–3680 CrossRef CAS PubMed.
- Y. Niu, Q. Li, Y. Ding, L. Dong and C. Wang, Adv. Drug Delivery Rev., 2019, 146, 190–208 CrossRef CAS PubMed.
- A. B. Shodeinde, A. C. Murphy, H. F. Oldenkamp, A. S. Potdar, C. M. Ludolph and N. A. Peppas, Adv. Funct. Mater., 2020, 30, 1909556 CrossRef CAS PubMed.
- A. K. Gaharwar, I. Singh and A. Khademhosseini, Nat. Rev. Mater., 2020, 5, 686–705 CrossRef CAS.
- D. Chouhan, N. Dey, N. Bhardwaj and B. B. Mandal, Biomaterials, 2019, 216, 119267 CrossRef CAS PubMed.
- M. Kharaziha, A. Baidya and N. Annabi, Adv. Mater., 2021, 33, 2100176 CrossRef CAS PubMed.
- A. C. Daly, L. Riley, T. Segura and J. A. Burdick, Nat. Rev. Mater., 2019, 5, 20–43 CrossRef PubMed.
- A. E. Widener, A. Roberts and E. A. Phelps, Adv. Healthcare Mater., 2023, 2303005 Search PubMed.
- Y. Endo, M. Samandari, M. Karvar, A. Mostafavi, J. Quint, C. Rinoldi, I. K. Yazdi, W. Swieszkowski, J. Mauney, S. Agarwal, A. Tamayol and I. Sinha, Biomaterials, 2023, 296, 122058 CrossRef CAS PubMed.
- W. Li, Z. Su, Y. Hu, L. Meng, F. Zhu, B. Xie, J. Wan and Q. Wu, Int. J. Biol. Macromol., 2023, 241, 124102 CrossRef CAS PubMed.
- F. Fu, L. Shang, Z. Chen, Y. Yu and Y. Zhao, Sci. Rob., 2018, 3, eaar8580 CrossRef PubMed.
- Q. Feng, D. Li, Q. Li, X. Cao and H. Dong, Bioact. Mater., 2022, 9, 105–119 CAS.
- A. Sheikhi, J. de Rutte, R. Haghniaz, O. Akouissi, A. Sohrabi, D. di Carlo and A. Khademhosseini, Biomaterials, 2019, 192, 560–568 CrossRef CAS PubMed.
- Y. Liu, A. Suarez-Arnedo, L. Riley, T. Miley, J. Xia and T. Segura, Adv. Healthcare Mater., 2023, 12, 2300823 CrossRef CAS PubMed.
- D. R. Griffin, W. M. Weaver, P. O. Scumpia, D. Di Carlo and T. Segura, Nat. Mater., 2015, 14, 737–744 CrossRef CAS PubMed.
- Y. Piao, H. You, T. Xu, H. P. Bei, I. Z. Piwko, Y. Y. Kwan and X. Zhao, Eng. Regener., 2021, 2, 47–56 Search PubMed.
- C. F. Guimarães, L. Gasperini, A. P. Marques and R. L. Reis, Nat. Rev. Mater., 2020, 5, 351–370 CrossRef.
- Z. Ataie, S. Horchler, A. Jaberi, S. V. Koduru, J. C. El-Mallah, M. Sun, S. Kheirabadi, A. Kedzierski, A. Risbud, A. R. A. E. Silva, D. J. Ravnic and A. Sheikhi, Small, 2023, 22, 2307928 Search PubMed.
- A. Jaberi, A. Kedzierski, S. Kheirabadi, Y. Tagay, Z. Ataie, S. Zavari, M. Naghashnejad, O. Waldron, D. Adhikari, G. Lester, C. Gallagher, A. Borhan, D. Ravnic, E. Tabdanov and A. Sheikhi, Adv. Healthcare Mater., 2024, 13, 2402489 CrossRef CAS PubMed.
- Y. Liu, A. Suarez-Arnedo, E. L. P. Caston, L. Riley, M. Schneider and T. Segura, Adv. Mater., 2023, 35, 2304049 CrossRef CAS PubMed.
- O. A. Peña and P. Martin, Nat. Rev. Mol. Cell Biol., 2024, 25, 599–616 CrossRef PubMed.
- J. P. Quint, M. Samandari, L. Abbasi, E. Mollocana, C. Rinoldi, A. Mostafavi and A. Tamayol, Nanoscale, 2022, 14, 797–814 RSC.
- M. Yampolsky, I. Bachelet and Y. Fuchs, Nat. Protoc., 2024, 19, 184–206 CrossRef CAS PubMed.
- G. Chen, F. Wang, X. Zhang, Y. Shang and Y. Zhao, Sci. Adv., 2023, 9, eadg3478 CrossRef CAS PubMed.
- R. D. Galiano, J. Michaels V, M. Dobryansky, J. P. Levine and G. C. Gurtner, Wound Repair Regener., 2004, 12, 485–492 CrossRef PubMed.
- A. Grada, J. Mervis and V. Falanga, J. Invest. Dermatol., 2018, 138, 2095–2105 CrossRef CAS PubMed.
- K. Nuutila, M. Samandari, Y. Endo, Y. Zhang, J. Quint, T. A. Schmidt, A. Tamayol and I. Sinha, Bioact. Mater., 2022, 8, 296–308 CAS.
- A. Mostafavi, M. Samandari, M. Karvar, M. Ghovvati, Y. Endo, I. Sinha, N. Annabi and A. Tamayol, Appl. Phys. Rev., 2021, 8, 041415 CAS.
- M. G. Tonnesen, X. Feng and R. A. F. Clark, J. Invest. Dermatol. Symp. Proc., 2000, 5, 40–46 CrossRef CAS PubMed.
- M. Mahmoodi, M. A. Darabi, N. Mohaghegh, A. Erdem, A. Ahari, R. Abbasgholizadeh, M. Tavafoghi, P. Mir Hashemian, V. Hosseini, J. Iqbal, R. Haghniaz, H. Montazerian, J. Jahangiry, F. Nasrolahi, A. Mirjafari, E. Pagan, M. Akbari, H. Bae, J. V. John, H. Heidari, A. Khademhosseini and A. Hassani Najafabadi, Adv. Funct. Mater., 2024, 34, 2315040 CrossRef CAS PubMed.
- Z. Ataie, A. Jaberi, S. Kheirabadi, A. Risbud and A. Sheikhi, J. Visualized Exp., 2022, e64829 Search PubMed.
- J. M. de Rutte, J. D. Koh and D. Di Carlo, Adv. Funct. Mater., 2019, 29, 1900071 CrossRef.
- A. H. Najafabadi, Z. I. N. Abadi, M. E. Aikins, K. E. Foulds, M. M. Donaldson, W. Yuan, E. B. Okeke, J. Nam, Y. Xu, P. Weerappuli, T. Hetrick, D. Adams, P. A. Lester, A. M. Salazar, D. H. Barouch, A. Schwendeman, R. A. Seder and J. J. Moon, J. Controlled Release, 2021, 337, 168–178 CrossRef CAS PubMed.
- N. Mohaghegh, A. Ahari, C. Buttles, S. Davani, H. Hoang, Q. Huang, Y. Huang, B. Hosseinpour, R. Abbasgholizadeh, A. L. Cottingham, N. Farhadi, M. Akbari, H. Kang, A. Khademhosseini, V. Jucaud, R. M. Pearson and A. Hassani Najafabadi, ACS Nano, 2024, 18, 27764–27781 CrossRef CAS PubMed.
- A. Hassani Najafabadi, J. Zhang, M. E. Aikins, Z. I. Najaf Abadi, F. Liao, Y. Qin, E. B. Okeke, L. M. Scheetz, J. Nam, Y. Xu, D. Adams, P. Lester, T. Hetrick, A. Schwendeman, M. S. Wicha, A. E. Chang, Q. Li and J. J. Moon, Nano Lett., 2020, 20, 7783–7792 CrossRef CAS PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- R. Yu, H. Chen, J. He, Z. Zhang, J. Zhou, Q. Zheng, Z. Fu, C. Lu, Z. Lin, F. Caruso and X. Zhang, Adv. Mater., 2024, 36, 2307680 CrossRef CAS PubMed.
- A. T. Feldman and D. Wolfe, in Histopathology, 2014, pp. 31–43 Search PubMed.
- J. Charan and N. Kantharia, J. Pharmacol. Pharmacother., 2013, 4, 303–306 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Proton nuclear magnetic resonance (1H NMR) spectroscopy, GHS drying and SEM imaging, and Fig. S1–S5. See DOI: https://doi.org/10.1039/d4bm01062k |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2025 |