 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Does the number of cells of individual strains correlate with their contribution to the total substrate turnover within a microbial community?†
Daniel
Buchner
 *,
Johannes
Scheckenbach
,
Philipp R.
Martin
*,
Johannes
Scheckenbach
,
Philipp R.
Martin
 and
Stefan B.
Haderlein
and
Stefan B.
Haderlein

Department of Geosciences, University of Tübingen, Tübingen, Germany. E-mail: daniel.buchner@uni-tuebingen.de
First published on 1st August 2024
Abstract
The contribution of individual bacterial strains within a mixed microbial community to the overall turnover of a specific compound is often assessed using qPCR data quantifying strain-specific 16S rRNA or functional genes. Here we compare the results of a qPCR based approach with those of compound specific stable isotope analysis (CSIA), which relies on strain-specific magnitudes of kinetic isotope fractionation associated with the biotransformation of a compound. To this end, we performed tetrachloroethylene (PCE) transformation experiments using a synthetic binary culture containing two different Desulfitobacterium strains (Desulfitobacterium hafniense strain Y51; εC,PCE = −5.8‰ and Desulfitobacterium dehalogenans strain PCE1; εC,PCE = −19.7‰). Cell abundances were analyzed via qPCR of functional genes and compared to strain-specific PCE turnover derived via carbon isotope fractionation. Repeated spiking of an initially strain Y51 dominated synthetic binary culture with PCE led to a steadily increasing contribution of strain PCE1 to PCE turnover (εC,initial = −5.6 ± 0.6‰ to εC,final = −18.0 ± 0.6‰) which was not or only weakly reflected in the changes of the cell abundances. The CSIA data further indicate that strain-specific PCE turnover varied by more than 75% at similar cell abundances of the two strains. Thus, the CSIA approach provided new and unexpected insights into the evolution of the metabolic activity of the single strains within a synthetic binary culture and indicates that strain-specific substrate turnover appears to be controlled by physiological and enzymatic properties of the strains rather than their cell abundance.
Water impactThe biotransformation potential at contaminated aquifers undergoing bioremediation is often assessed using qPCR data targeting 16S rRNA or functional genes. Yet, data from compound specific isotope analysis demonstrates that the most abundant bacterial strain(s) may not be responsible for the transformation under investigation. These findings need to be considered when assessing the potential for bioremediation of aquifers for drinking water production. |
1. Introduction
The unambiguous identification of a single bacterial strain within a microbial community that metabolizes a specific biogeochemical process is still a major challenge but is a prerequisite to gain insights into bacterial-driven contaminant transformation.1,2 Deciphering the metabolic activity of different key organisms within a microbial community is difficult as (i) several bacterial strains can catalyze the same process and/or (ii) metabolic versatile bacterial strains do not necessarily catalyze the process under investigation, e.g., contaminant transformation. Direct evidence of catalytic activity can be obtained through enzyme-activity assays under specific conditions with respect to pH, temperature, or cofactors.3,4 However, these techniques can be time and cost intensive. Furthermore, the mere detection of a specific enzyme does not provide conclusive evidence of its actual catalytic activity.5 Despite the potential of new (meta-)omics techniques such as metagenomics, metatranscriptomics and metaproteomics,6 evidence for bacterial substrate turnover is often derived from the quantification of 16S rRNA of specific bacteria or of functional genes at the DNA level. This is because this analytical technique is well established and widely available. Such data are used according to the following principles: firstly, an increase in 16S rRNA of specific bacteria or functional gene copy numbers in space and/or time is correlated with a specific process under investigation; secondly, the specific bacterial strain(s) with the highest abundance catalyze the process. However, it is important to note that these data only provide information on the genetic potential of a microbial community but do not give insights into the actual bacterial activity.7,8 Consequently, it is possible to imagine a scenario in which high turnover per cell can compensate for low cell numbers (and vice versa), thereby violating the basic principles. For metabolically flexible bacteria that can also grow on a different substrate, cell numbers and even growth may not be correlated to the compound turnover under investigation at all.Identification (and quantification) of bacterial key players is highly relevant for bioremediation, for instance at sites contaminated with chlorinated ethenes (CEs). Bioremediation is often used as a follow-up treatment after the use of conventional methods (e.g., pump and treat), as the latter have proven ineffective at low residual concentrations of CEs.9–11 These biological remediation approach relies on organohalide-respiring bacteria (OHRB) which use halogenated hydrocarbons like tetrachloroethene (PCE) or trichloroethene (TCE) as terminal electron acceptor to generate growth supporting energy.9,10 OHRB are found in diverse microbial taxa and can be grouped into metabolically versatile (e.g., Desulfuromonas, Desulfitobacterium) or obligate OHRB which are restricted to organohalides (e.g., Dehalococcoides, Dehalobacter).12,13 The enzymes that catalyze the transformation of CEs belong to the group of reductive dehalogenases (RdhA) and are encoded in the genome of OHRB within rdh-gene operons.13,14
Characterization and monitoring of OHRB in microbial (enrichment) cultures and at contaminated sites is routinely carried out by quantitative real-time PCR (qPCR) targeting CE-rdhA-genes or 16S rRNA-genes of OHRB like Desulfitobacterium or Dehalococcoides.15–18 The analyzed gene trends (increase or decrease in time and/or space) are used to estimate the contribution of individual OHRB strains to the complete dichlorination of PCE (e.g., vcrA-gene indicative for the transformation of cis-DCE to VC and ethene19,20). Numerous studies analyzed CE-rdhA- and 16S rRNA-gene abundances of different OHRB from multiple genera.21–25 These studies indicate that typically several OHRB strains of Dehalococcoides, Desulfitobacterium, Dehalobacter, Desulfuromonas coexist within a microbial community and are involved in the complete dehalogenation of PCE to ethene. Furthermore one transformation step (e.g., PCE to TCE is done by Desulfitobacterium, Dehalobacter, Geobacter, and/or Dehalococcoides) was shown to be carried out by several OHRB.10,26 Changes over time in the abundance of rdhA-genes of different OHRB in enrichment cultures also indicated changes in the contribution of the different strains to CE turnover.22,27,28 For obligate OHRB the qPCR data are quite conclusive as growth is directly coupled to organohalide transformation.17,29 Nevertheless there are also potential pitfalls in the determination of the actual substrate turnover of these bacteria, as studies have shown that transformation and growth can be decoupled.30–32 Johnson et al.33 proposed a quorum-sensing-type regulatory mechanism to control maximum cell densities, decoupling the growth of a Dehalococcoides mccartyi strain from dehalogenation. The presence and growth of versatile OHRB cannot be taken as clear evidence for organohalide respiration, as they are not restricted to this process to gain energy. In addition, one needs to consider that strain-specific substrate turnover is not a pure function of the number of cells as also the number of the catalyzing enzymes per cell can change.30 Even further the enzyme activity itself was reported to changes upon environmental factors such as pH, substrate supply or cofactors.34–36 For example, a scenario is conceivable, in which an increase in the cell numbers of a strain is observed, but due to a simultaneous decrease in enzyme activity, the contribution to the turnover of a substance by this strain is actually reduced. Due to these ambiguities, the presented study aims to shed light onto the qPCR derived quantification of functional genes (equivalent to cell numbers) as a proxy for the PCE turnover of a specific strain within a synthetic binary culture.
Next to molecular biological analysis, transformation processes can also be monitored and quantified via compound specific stable isotope analysis (CSIA). This approach relies on typically slightly faster reaction rate constants for molecules containing a light isotope (e.g., 12C or 35Cl) at the reactive position compared to molecules containing a heavy isotope (e.g., 13C or 37Cl). This process, known as kinetic isotope fractionation, results in an enrichment of the heavy isotopes in the residual substrate pool as it undergoes transformation.37,38 The magnitude of isotope fractionation is quantified by the isotope enrichment factor (ε).37,39 Previous studies investigated the effect of different cultivation conditions on the ε values for chlorinated ethenes both in pure cultures and in enrichment cultures in which a single OHRB catalyzed the dehalogenation.30,40 Measured ε values were robust and not affected by repeated chlorinated ethene addition, different electron donor and acceptor concentrations or micronutrient availability.5,30,40 However, the ε value observed in a mixed culture is a lumped parameter comprising the strain-specific enrichment factors and the strains' contributions to substrate turnover.41,42 To this end, the shift of 8‰ in the PCE carbon isotope enrichment factor (εC) in a microbial enrichment culture was correlated with a change in the predominantly dechlorinating OHRB, highlighting the use of CSIA to observe and even quantify changes in the contribution of different transformation processes.43 CSIA is particularly valuable in cases where the substrate is converted to the same products (e.g., PCE to TCE), as this makes it impossible to distinguish between the two transformation pathways based on product formation. The diagnostic power of CSIA is further improved by the analysis of a second isotope system. For chlorinated ethenes the combination of carbon and chlorine CSIA may reveal mass transfer limitation, masking effects as well as changes in the underlying reaction mechanisms of the catalyzing enzymes.44–46
Our study aimed to investigate the reliability of changes in cell abundances (quantification of functional genes via qPCR on DNA level) as proxy for strain-specific substrate turnover during contaminant biotransformation and to gain insights into the development of the metabolic activity of the investigated OHRB. To this end, reductive dechlorination of PCE was investigated within a synthetic binary culture containing Desulfitobacterium dehalogenans strain PCE1 and Desulfitobacterium hafniense strain Y51 (initial PCE1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Y51 ratio of 1
Y51 ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (v/v)). These strains were selected because their isotope enrichment factors for PCE differ considerably (strain PCE1: εC = −19.7‰ and εCl = −6.3‰, strain Y51: εC = −5.8‰ and εCl = −2.2‰),47 allowing the use of isotope fractionation as a proxy for strain-specific PCE turnover. Additionally, cell numbers of the individual strains were tracked quantifying PCE-rdhA-gene abundances of the individual strains (strain PCE1: prdA-gene, strain Y51: pceA-gene; one copy of the respective gene per genome).48–50
5 (v/v)). These strains were selected because their isotope enrichment factors for PCE differ considerably (strain PCE1: εC = −19.7‰ and εCl = −6.3‰, strain Y51: εC = −5.8‰ and εCl = −2.2‰),47 allowing the use of isotope fractionation as a proxy for strain-specific PCE turnover. Additionally, cell numbers of the individual strains were tracked quantifying PCE-rdhA-gene abundances of the individual strains (strain PCE1: prdA-gene, strain Y51: pceA-gene; one copy of the respective gene per genome).48–50
2. Materials and methods
2.1 Chemicals
Chemicals were purchased from Carl Roth (Karlsruhe, Germany), Sigma-Aldrich (Steinheim, Germany) or Merck (Darmstadt, Germany) with the highest available level of purity.2.2 Cultivation of Desulfitobacterium spp. and experimental setups
Desulfitobacterium dehalogenans strain PCE1 (no. 10344) was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). Desulfitobacterium hafniense strain Y51 was kindly provided by Prof. Furukawa of the Department of Food and Bioscience, Beppu University (Japan). Precultures were cultivated in 250 mL serum flasks containing 200 mL liquid medium, 200 μM PCE and 1 mM pyruvate. Serum bottles were placed on a horizontal shaker (140 rpm, 48 h) to dissolve the neat PCE. Serum bottles contained an anoxic, sodium carbonate buffered medium as described by Buchner et al.,30 while the headspace was flushed with N2/CO2 (80%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20%). Precultures were grown on PCE after prior degassing with N2/CO2 (80%
20%). Precultures were grown on PCE after prior degassing with N2/CO2 (80%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20%) for 90 min prior to inoculation to remove the PCE transformation products. Successful removal was screened via GC-MS.
20%) for 90 min prior to inoculation to remove the PCE transformation products. Successful removal was screened via GC-MS.
The re-spike transformation experiments were conducted in 560 mL serum bottles sealed with butyl rubber stoppers. The inoculation was done in an anaerobic glove box (O2 < 1 ppm) by adding 40 mL (8% v/v) of a strain PCE1 preculture to 200 mL of PCE adapted cells of strain Y51. PCE and pyruvate were added pre-dissolved in 250 mL fresh medium as spike solution yielding a final concentration of 200 μM PCE and 1 mM pyruvate. The setup comprised two living-replicates and a cell-free control.
The setup was continuously cultivated within six monitored PCE (re-)spike experiments to investigate development of cell numbers as well as the observable PCE and TCE isotope fractionation. All experiments were conducted with PCE (200 μM) as electron acceptor, except for the 8th spike in which 200 μM TCE were supplied as electron acceptor. After initial PCE transformation PCE was continuously supplied to the culture and selected dehalogenation events were monitored (see Table 1 and Fig. 1). For non-monitored PCE additions, neat PCE was added and dissolved by placing the cultures on a rotary shaker at 140 rpm to achieve a final concentration of 200 μM. Monitored experiments were initiated in the glovebox by adding PCE spike solutions. For the 8th spike, initial TCE transformation was monitored and followed by three additional non-monitored additions of neat TCE (each 200 μM). Each chlorinated ethene addition was accompanied with the addition of 1 mM pyruvate as electron donor. After each complete monitored or non-monitored PCE/TCE transformation, cultures were degassed with N2/CO2 and screened for residual chlorinated ethenes by GC-MS. Experimental flasks were individually maintained including the 7th spike. Thereafter, both living replicates were pooled in a sterilized 1 L glass bottle after monitored transformation experiments and equally divided into sterilized 560 mL serum flasks prior to the following monitored experiment. Cultures were stored in the dark. The control bottle was treated the same way as living replicates, but non-monitored PCE/TCE additions were omitted.
| Experiment | Provided CE (number of transformation event(s)) | Duration in days | Time since last monitored dehalogenation event in days |
|---|---|---|---|
| a Refers to the single microcosms (M) of the individual experiment. | |||
| 1st spike | PCE | <1 | 0 |
| 4th spike | PCE (3) | 1 (M1)a | 22 |
| 2 (M2)a | |||
| 7th spike | PCE (6) | <1 | 36 |
| 8th spike | TCE (1) | 6 (M1)a | 21 |
| 16 (M2)a | |||
| 12th spike | PCE (7) after 4× TCE | <1 | 26 |
| 15th spike | PCE (10) | <1 | 38 |
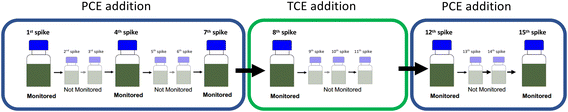 | ||
| Fig. 1 Overview of the (re-)spike experiments of the synthetic binary culture containing Desulfitobacterium dehalogenans strain PCE1 and Desulfitobacterium hafniense strain Y51. | ||
2.3 Sample extraction and analytical methods
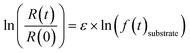 | (1) |
R(t) and R(0) denote the isotope ratios at different sampling points and the initial isotope ratio at the start of the experiment, respectively; f(t)substrate is the remaining fraction of PCE or TCE at sampling point t. Fractions of PCE as well as of the less chlorinated ethenes (TCE and cis-DCE) were calculated based on the timepoint-wise mass balance correction method described by Buchner et al. (2017)55 to account for the cumulative mass removal due to repetitive sampling. Dual isotope slopes (ΛC/Cl) and associated standard errors (SE) were calculated for the individual experiments by regressing measured δ13C and δ37Cl data using the York method.56
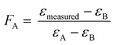 | (2) |
The εmeasured is the measured enrichment factor of the synthetic binary culture and represents the weighted average of strain-specific ε values (εA or εB).37,57 Corresponding errors were calculated by error propagation (eqn (S2)†) In an ideal scenario the values of FA are equal to the fraction of the cell numbers fA derived via qPCR analysis (see eqn (3)).
2.4 Molecular biological analysis
Cell abundances of D. dehalogenans strain PCE1 and D. hafniense strain Y51 were determined using quantitative real-time PCR (qPCR) assays. For strain PCE1, copy numbers of the prdA-gene encoding the PCE transforming PrdA enzyme were quantified.48 For strain Y51, copy numbers of the pceA-gene encoding the PCE transforming PceA enzyme were quantified.49 Based on genome analysis, one copy of the genes is present per genome of the individual strains and measured gene copy numbers therefore equal the cell numbers of strain PCE1 and strain Y51, respectively.48,58 Detailed information on DNA extraction and the quantification of prdA- and pceA-genes are provided in the ESI† as well as the determination of error the qPCR analysis.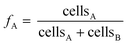 | (3) |
Since measured cell numbers of each microcosm are associated with an error, the error of averaged strain fractions at time point t was calculated via Gaussian error propagation (see eqn (S1)†).
| εC,calculated = (εC,A × fA) + (εC,B × fB) | (4) |
The εC,calculated values represent the weighted average of the strain-specific enrichment factor and the strain-specific contribution. Errors were calculated by error propagation, considering the errors for the strain fractions and the confidence intervals of the pure strain ε values (eqn (S3)†).
3. Results and discussion
3.1 Chlorinated ethene transformation pattern within the synthetic binary culture
Setting up a synthetic binary culture containing Desulfitobacterium dehalogenans strain PCE1 (dechlorination of PCE to TCE) and Desulfitobacterium hafniense strain Y51 (dechlorination of PCE to cis-DCE) we monitored PCE as well as TCE turnover upon continuously PCE and TCE re-spiking (see Table 1 and Fig. 1). To gain insight into strain-specific turnover of PCE and TCE, we conducted carbon (and chlorine) isotope analysis and quantified kinetic isotope fractionation. Furthermore, the cell numbers of each strain were tracked by quantifying functional gene numbers via qPCR. The initial synthetic binary culture containing the strains PCE1 and Y51 at a volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (strainPCE1
5 (strainPCE1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) strainY51) was re-spiked as follows: (i) 1st spike with PCE followed by, (ii) six PCE re-spikes while the 4th and 7th re-spike were monitored, (iii) four spikes of TCE while the 1st TCE spike (equals to the 8th re-spike) was monitored and finally (iv) five PCE spikes while the 12th and 15th re-spike were monitored.
strainY51) was re-spiked as follows: (i) 1st spike with PCE followed by, (ii) six PCE re-spikes while the 4th and 7th re-spike were monitored, (iii) four spikes of TCE while the 1st TCE spike (equals to the 8th re-spike) was monitored and finally (iv) five PCE spikes while the 12th and 15th re-spike were monitored.
For all PCE spikes a complete transformation to TCE and finally to cis-DCE was observed (see Fig. S1–S5†). Single PCE transformation events showed durations of up to two days while TCE transformation in the 8th re-spike was completed after 16 days (see Fig. S6†). As TCE can be only metabolized by strain Y51 the TCE profile allows to draw first conclusions on the activity of this strain within the synthetic binary culture. For comparison, in pure culture experiments with strain Y51 a maximum TCE fractions of 20 to 25% are reported when PCE is provided as initial substrate.47 Thus, high transient TCE fractions such as 65 to 80% (observed in the 7th spike) suggest that strain PCE1 predominantly catalyzed PCE dehalogenation. While low TCE fractions implicate primary catalysis of PCE dehalogenation by strain Y51 as observed in the 1st spike.
3.2 Isotope fractionation as tool to decipher single strain PCE turnover in a synthetic binary culture
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) strainY51 volume ratio of 1
strainY51 volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 a gradual shift from a strain Y51 dominated culture towards a strain PCE1 dominated one was observed. While the initial εC value was −5.6 ± 0.6‰, εC values of −7.3 ± 0.3‰ and −17.4 ± 0.7‰ were observed for the 4th and 7th PCE spike, respectively. The shift was also reflected in εCl and ΛC/Cl values (4th spike εCl = −2.6 ± 0.4‰, ΛC/Cl = 3.0 ± 0.1 and 7th spike εCl = −5.6 ± 0.6‰, ΛC/Cl = 3.13 ± 0.04). After four sequential re-spikes with TCE and therefore a provoked selective stimulation of strain Y51 an εC value of −7.3 ± 0.2‰ was observed for PCE in the 12th spike, while two further PCE additions resulted again in a shift of the εC value to −18.0 ± 0.6‰ in the 15th spike.
5 a gradual shift from a strain Y51 dominated culture towards a strain PCE1 dominated one was observed. While the initial εC value was −5.6 ± 0.6‰, εC values of −7.3 ± 0.3‰ and −17.4 ± 0.7‰ were observed for the 4th and 7th PCE spike, respectively. The shift was also reflected in εCl and ΛC/Cl values (4th spike εCl = −2.6 ± 0.4‰, ΛC/Cl = 3.0 ± 0.1 and 7th spike εCl = −5.6 ± 0.6‰, ΛC/Cl = 3.13 ± 0.04). After four sequential re-spikes with TCE and therefore a provoked selective stimulation of strain Y51 an εC value of −7.3 ± 0.2‰ was observed for PCE in the 12th spike, while two further PCE additions resulted again in a shift of the εC value to −18.0 ± 0.6‰ in the 15th spike.
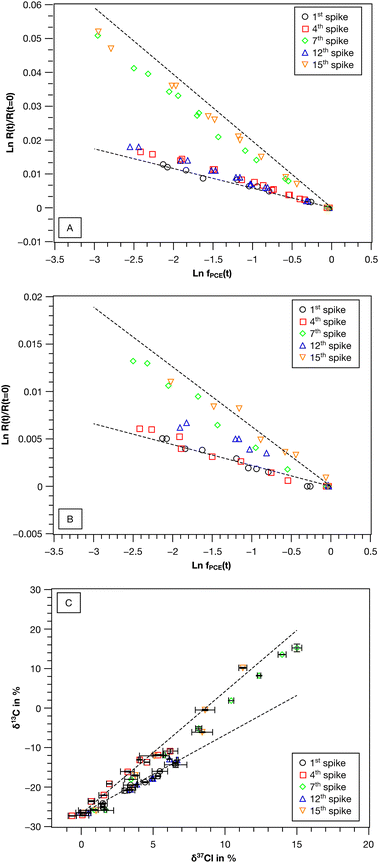 | ||
| Fig. 2 Carbon and chlorine isotope fractionation of PCE in the synthetic binary culture (re-)spike experiment. Shown are double logarithmic Rayleigh plots for (A) carbon and (B) chlorine isotope fractionation and (C) dual isotope plots (δ37Cl vs. δ13C). Error bars represent standard deviation of measurement replicates (n = 2 for δ13C, n = 3 to 5 for δ37Cl). Dashed lines in (A)–(C) represent PCE isotope fractionation of Desulfitobacterium dehalogenans strain PCE1 and Desulfitobacterium hafniense strain Y51 in pure culture after Büsing et al.47 | ||
The observed variability of the εC values in the synthetic binary culture exhibited a clear and reproducible trend regarding the contribution of the two strains to PCE turnover. While in the 1st spike PCE transformation was performed by strain Y51, continuous PCE re-spikes resulted in increasing contributions of strain PCE1 to the overall PCE turnover. The addition of TCE had the potential to reverse this effect, thereby re-establishing a culture in which PCE turnover is once again dominated by strain Y51.
| (Re-)spike experiment | |||
|---|---|---|---|
1st spike (v/v = 1PCE1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5Y51) 5Y51) |
−5.6 ± 0.6 | −2.7 ± 0.2 | 2.1 ± 0.1 |
| 4th spike with PCE | −7.3 ± 0.3 | −2.6 ± 0.4 | 3.0 ± 0.1 |
| 7th spike with PCE | −17.4 ± 0.7 | −5.6 ± 0.6 | 3.1 ± 0.1 |
| 12th spike with PCE (after 4× TCE spikes) | −7.3 ± 0.2 | −3.6 ± 0.8 | 2.1 ± 0.3 |
| 15th spike with PCE | −18.0 ± 0.6 | −5.5 ± 0.9 | 3.3 ±0.3 |
| TCE addition | |||
|---|---|---|---|
| a 95% confidence interval (CI) for the linear regression in the double logarithmic Rayleigh plots. b Standard error (SE) for the York regression calculated after Ojeda et al.56 c Data from Büsing et al.47 d Data from Buchner et al.5 and Buchner et al.30 | |||
| Dsb strain Y51 pured | −8.7 ± 0.2 | −2.7 ± 0.2 | 3.2 ± 0.2 |
| 1st TCE addition (8th spike) | −8.0 ± 0.2 | −2.9 ± 0.1 | 2.8 ± 0.1 |
The ε values (carbon and chlorine) for PCE observed for the synthetic binary culture result from the strain-specific ε values and thus reflect the contributions of the individual strains.41 Therefore, the observed carbon isotope enrichment factors of the synthetic binary culture were used to calculate the strain-specific contribution (F) to the PCE turnover according to eqn (2) (see Table 3). In previous studies in which PCE and TCE isotope fractionation of the two strains was individually investigated in detail, robust strain-specific intrinsic εC and εCl values were observed despite significant changes of the growth conditions.5,30,47 Therefore, it is well justified to assume robust strain-specific ε values within the synthetic binary culture. This assumption is further supported by the observed robust TCE isotope fractionation of strain Y51 in the synthetic binary culture.
| Experiment | Carbon CSIA | qPCR | Deviation fPCE1 and FPCE1e ± error in % | ||
|---|---|---|---|---|---|
| Measured εC ± 95% CI in ‰ | Contribution PCE1 (FPCE1)a ± errorb in % | Average fraction PCE1 (fPCE1) ± errorc in % | Calculated εC ± errord in ‰ | ||
| a Values calculated according to eqn (2). b Values calculated according to eqn (S2).† c Values calculated according to eqn (S1).† d Values calculated according to eqn (S3).† e Deviation was calculated using the formula: dev = (fPCE1 − FPCE1) × 100%. | |||||
| 1st spike | −5.6 ± 0.6 | 0 ± 5 | 18 ± 9 | −8.3 ± 1.8 | 18 ± 10 |
| 4th spike | −7.3 ± 0.3 | 11 ± 3 | 31 ± 22 | −10.1 ± 4.3 | 20 ± 22 |
| 7th spike | −17.4 ± 0.7 | 83 ± 6 | 94 ± 5 | −18.8 ± 1.1 | 11 ± 8 |
| 12th spike (after 4 TCE spikes) | −7.3 ± 0.2 | 11 ± 2 | 92 ± 1 | −18.6 ± 0.5 | 81 ± 2 |
| 15th spike | −18.0 ± 0.6 | 88 ± 5 | 93 ± 4 | −18.9 ± 0.9 | 5 ± 6 |
In the (re-)spike experiments the contribution of strain PCE1 to the overall PCE turnover successively increased during successive PCE addition from no contribution in the 1st PCE spike to 11 ± 3% and 83 ± 6% in 4th re-spike and the 7th re-spike, respectively. A decrease in the contribution to PCE turnover by strain PCE1 was observed after four TCE additions resulting in a contribution of 11 ± 2% in the 12th spike. The following repeated three PCE additions resulted again in an increased PCE turnover of strain PCE1 with a contribution of 88 ± 5% in the 15th re-spike. These results imply that (i) the PCE transformation within the synthetic binary culture develops from a strain Y51 dominated process to a strain PCE1 dominated one and (ii) that this effect can be reversed. Thus, the added chlorinated ethene and the growth history had major impacts on the contribution and subsequently on the currently observed metabolic activities of the two strains. While the PCE turnover of strain PCE1 was enhanced by repeated PCE addition, the PCE turnover of strain Y51 was adversely affected by cultivation in the synthetic binary culture. It is noteworthy that even though TCE can only be used by strain Y51, the strain is unable to use this competitive advantage in order to dominate the synthetic binary culture. On the other hand, repeated TCE addition allows strain Y51 to outcompete strain PCE1 and selectively enhanced the PCE turnover of strain Y51 for the following PCE addition. These results demonstrate that the addition of chlorinated ethene exerts selective pressure on single OHRB strains, which is consistent with previous studies on microbial enrichment cultures and contaminated sites.28,59 Further studies may investigate possible inhibition/competition effects of the two strains to elucidate the factor underlying the increasing dominance of strain PCE1.
3.3 Correlation of PCE-rdhA-gene abundances and strain-specific contribution to PCE turnover
Based on PCE isotope fractionation we demonstrated that PCE turnover of the single strains significantly differed in the synthetic binary culture. In a next step we investigated whether strain-specific PCE turnover is correlated with PCE-rdhA-gene abundances of the strains. To this end PCE/TCE-rdhA-gene copy numbers on DNA level (strain PCE1: prdA and strain Y51: pceA) were quantified at the start of the experiment, the last point with a δ13C value for PCE and after complete transformation of PCE to cis-DCE. Note that measured PCE-rdhA-gene copy numbers correspond to cell numbers of each strain, as there is only one copy of the respective gene per genome.48,58 In the synthetic binary culture, the total bacterial cell numbers are equal to the cell numbers of strain PCE1 plus the cell numbers of strain Y51. Therefore, for the purposes of simplicity, we will only discuss the development of the cell fractions of strain PCE1 (fPCE1) in %.In general, only minor changes in the absolute cell numbers of the two strains and subsequently in the strain fractions were observed within one single PCE transformation event (see Fig. 3). For example, in the 1st PCE spike the cell fraction of strain PCE1 shifted slightly from initially 17% to a final fraction of 12% when PCE was fully transformed to cis-DCE. On the other hand, a significant shift in fPCE1 was observed when comparing the repeated PCE addition events. While the cell fraction of the 1st spike indicates a Y51-dominated culture (fPCE1 = 12%) the cell fraction shifted to be PCE1 dominated (fPCE1 = 93%) in the 7th spike. Thereafter, the synthetic binary culture composition was dominated by strain PCE1, with strain fractions equal to or greater than 90%. Hence, both strains decoupled cell growth from PCE transformation. Similar observations were reported for pure and enrichment cultures of other OHRB when the total cell numbers exceeded 107 cells per mL.29,30,60 Nutrient limitation and accumulation of toxic metabolites can be excluded to limit microbial cell growth in our experiments as with each spike a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 dilution of the cultures with fresh medium and degassing of dehalogenation products was done.
2 dilution of the cultures with fresh medium and degassing of dehalogenation products was done.
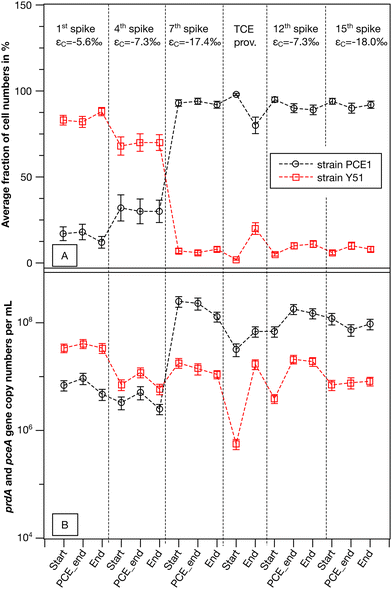 | ||
| Fig. 3 Culture composition and measured carbon isotope enrichment factors for the synthetic binary culture containing Desulfitobacterium dehalogenans strain PCE1 and Desulfitobacterium hafniense strain Y51. (A) Average fraction of the strain PCE1 and Y51 calculated based on total cell numbers in living replicates (n = 2). (B) Averaged cell numbers based on prdA- and pceA-gene copy numbers of living replicates (n = 2). Gene abundances were corrected for dilution due to addition of spike solution. Error bars represent percentage errors calculated via error propagation from qPCR assays. Average strain fractions were calculated according to eqn (3) and corresponding error by Gaussian error propagation (see eqn (S1)†). | ||
Total cell numbers of both strains followed a similar trend over all monitored transformation events (see Fig. 3B). The change in the composition of the synthetic binary culture from strain Y51 to strain PCE1 was due to a smaller decrease in the cell number of strain PCE1 compared to strain Y51 from the 1st to the 4th spike, and a stronger growth of strain PCE1 from the 4th to the 7th spike. The results indicate that strain PCE1 was more adaptable in the synthetic binary culture during repeated PCE spikes compared to strain Y51. Interestingly, strain PCE1 maintained its predominant abundance during TCE addition, although no appropriate electron acceptor for this strain was provided. This may be explained by an efficient generation of metabolic energy by strain PCE1 from fermentation of pyruvate which was added with each chlorinated ethene addition.61
3.4 Comparison of strain-specific PCE turnover: CSIA vs. qPCR
To investigated how strain-specific PCE turnover compares with the measured cell abundances we calculated the contribution of strain PCE1 (FPCE1) according to eqn (2) considering the measured εC values (εC,measured) and the εC values of pure cultures of each strain (see Table 3).The εC,calculated value represents the average of the enrichment factors calculated based on the strain fractions at the beginning and the end of the experiment (see Table S1†). The results show that contribution of strain PCE1 (FPCE1) to PCE turnover in most cases did not correlate with the average PCE1 strain fraction (fPCE1) with deviations ranging from 0 to 82% (Table 3). Based on calculated FPCE1 values, the cell abundance derived PCE turnover of strain PCE1 was overestimated, especially for the initial transformation period, as well as for the 4th and 12th spike. It is notable that at the 1st spike 107 cells per mL of strain PCE1 and 4 × 107 cells per mL of strain Y51 were present corresponding to a fPCE1 values of 18 ± 9%. However, the CSIA data revealed that the transformation of PCE was solely carried out by strain Y51 (FPCE1 = 0 ± 5%). Good correlations were only obtained for 7th and 15th spike where the synthetic binary culture seams to reach a steady state condition and PCE transformation is solely conducted by strain PCE1.
These results demonstrate that strain abundances did not reflect the contribution to PCE turnover of the two strains. Therefore, other physiological factors must have compensated for low cell numbers of strain Y51 at least in the 12th spike. The reductive dehalogenases PrdA (strain PCE1) and PceA (strain Y51) are the catalysts of PCE transformation suggesting that strain-specific dehalogenation contribution was governed by a complex interplay between RdhA de novo synthesis rates, amount of catalytically active RdhA per cell and RhdA specific properties of the strains. Thus, low abundances of strain Y51 were likely compensated by a higher content of active PCE-RdhA per cell and/or faster de novo RdhA synthesis rates compared to strain PCE1. In addition, more beneficial RdhA properties such as high substrate specificity for PCE (i.e., low Michaelis–Menten constant KM) and high enzymatic dehalogenation rates may have contributed to high dehalogenation activities of this strain. The high contribution of strain Y51 in the 12th spike, may be explained by constant upregulation of PceA synthesis during prior TCE additions.30 In contrast, PrdA is not expressed upon exposure to TCE.62 In general, due to previous TCE addition, a higher physiological adaption by strain Y51 compared to strain PCE1 may explain major PCE transformation by strain Y51 despite its lower abundance in the 12th spike. A calculation of the contribution to PCE turnover per cell revealed that the turnover per cell is highly variable. It is notable that the contribution of strain Y51 cells significantly increased (see eqn (S4) and Fig. S7†) after addition of TCE. This further points towards an enhanced metabolic status of strain Y51 cells due to previous TCE additions.
In summary, our data show that the contribution of individual strains to substrate turnover cannot reliably be inferred from the abundance of cell numbers and/or functional genes, at least not for metabolically versatile bacteria. While CE-rdhA-gene and 16S rRNA gene quantification is an essential tool for monitoring and characterizing OHRB in microbial communities, it does not allow for the inference of substrate turnover.
4. Conclusions
Quantifying the turnover of individual bacterial strains is crucial for evaluating and optimizing bioremediation approaches. Well-established techniques include quantification of functional genes encoding key enzymes and CSIA. While functional gene analysis can be used to characterize the general bacterial potential, CSIA can estimate or even quantify in situ biotransformation based on appropriate isotope enrichment factors. As multiple OHRB strains are typically present at contaminated sites, it is important to understand the contribution of each strain to the investigated transformation process. This can be achieved by monitoring the evolution of cell numbers and/or functional gene abundance. Increasing numbers are commonly assumed to indicate the contribution to the transformation process under investigation.In this study, CSIA data revealed that PCE-rdhA-gene analysis of a synthetic binary culture containing two OHRB – the simplest case of a microbial community – the contribution to the substrate turnover of the strains is not necessarily reflected by their relative abundance in terms of cell numbers. Thus, activities of single OHRB strains in microbial communities cannot be reliably assessed by CE-rdhA-gene and/or cell abundances. Our results demonstrate that despite low rdhA-gene abundance, strain-specific RdhA properties and physiological properties may give rise to high dehalogenation turnover.
A combination of CSIA and functional gene analysis in synthetic lab studies can help elucidating strain-specific contaminant turnover. The strain-specific contaminant turnover at a field site can be evaluated by quantifying isotope fractionation and OHRB abundance in laboratory experiments using autochthonous microbial inocula from groundwater and sediment in combination with hydrogeochemical data such as pH and redox sensitive species. Furthermore, the performance of in situ measures such as stimulating microbial activity or bioaugmentation can be better accessed by such a combined approach. Finally, we would like to point out that a comprehensive analysis of the contribution of different microbial strains using CSIA, as done in the presented work, necessitates specific requirements: (i) only two processes are active in the system under investigation, (ii) the enrichment factors of the individual processes must be known, and (iii) exhibit a high degree of variation. Consequently, no standardized procedure can be derived from these findings that describes the use of CSIA to elucidate strain-specific contributions at a field site.
Future studies should therefore attempt to investigate the effect of changing cultivation conditions on isotope fractionation of mixed cultures in continuous chemostat cultures, which allows a more dynamic simulation of changing environmental conditions compared to batch cultures. Under certain settings the combination of CSIA and molecular biological tools quantifying rdhA-gene transcription can then be applied to elucidate the effect of changes in bacterial physiologies on dehalogenation activities and isotope fractionation.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
D. B.: conceptualization, funding acquisition, methodology, supervision, writing – original draft, review & editing. J. S.: conceptualization, methodology, investigation, formal analysis, writing – original draft. P. R. M.: formal analysis, methodology, writing – review & editing. S. B. H: resources, funding acquisition, supervision, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Jana Braun for laboratory assistance. D. hafniense strain Y51 was kindly provided by Prof. Furukawa in the Department of Bioscience and Biotechnology at Kyushu University (Japan).References
- G. K. Druschel and A. Kappler, Geomicrobiology and Microbial Geochemistry, Elements, 2015, 11, 389–394 CrossRef.
- C. M. Kao, H. Y. Liao, C. C. Chien, Y. K. Tseng, P. Tang, C. E. Lin and S. C. Chen, The change of microbial community from chlorinated solvent-contaminated groundwater after biostimulation using the metagenome analysis, J. Hazard. Mater., 2016, 302, 144–150 CrossRef CAS PubMed.
- H. Bisswanger, Enzyme assays, Perspect. Sci., 2014, 1, 41–55 CrossRef.
- D. Turkowsky, N. Jehmlich, G. Diekert, L. Adrian, M. von Bergen and T. Goris, An integrative overview of genomic, transcriptomic and proteomic analyses in organohalide respiration research, FEMS Microbiol. Ecol., 2018, 94, fiy013 CrossRef PubMed.
- D. Buchner, P. R. Martin, J. Scheckenbach, S. Behrens and S. B. Haderlein, Effects of Bacterial Growth Conditions on Carbon and Chlorine Isotope Fractionation Associated with TCE Biotransformation, ACS ES&T Water, 2022, 2, 2510–2518 Search PubMed.
- T. Van Den Bossche, M. O. Arntzen, D. Becher, D. Benndorf, V. G. H. Eijsink, C. Henry, P. D. Jagtap, N. Jehmlich, C. Juste, B. J. Kunath, B. Mesuere, T. Muth, P. B. Pope, J. Seifert, A. Tanca, S. Uzzau, P. Wilmes, R. L. Hettich and J. Armengaud, The Metaproteomics Initiative: a coordinated approach for propelling the functional characterization of microbiomes, Microbiome, 2021, 9, 243 CrossRef PubMed.
- L. A. Achenbach and J. D. Coates, Disparity between Bacterial Phylogeny and Physiology - Comparing 16S rRNA sequences to assess relationships can be a powerful tool, but its limitations need to be considered, ASM news, 2000, 66, 714–715 Search PubMed.
- C. Desai, H. Pathak and D. Madamwar, Advances in molecular and “-omics” technologies to gauge microbial communities and bioremediation at xenobiotic/anthropogen contaminated sites, Bioresour. Technol., 2010, 101, 1558–1569 CrossRef CAS PubMed.
- A. Tiehm, K. R. Schmidt, C. Stoll, A. Müller, S. Lohner, M. Heidinger, F. Wickert and U. Karch, Assessment of natural microbial dechlorination, Ital. J. Eng. Geol. Environ., 2007, 71–77 Search PubMed.
- L. Adrian and F. E. Löffler, Organohalide-Respiring Bacteria, Springer, Berlin Heidelberg, 2016 Search PubMed.
- D. Buchner, M. Schweikhart, S. Behrens, T. Schöndorf, C. Laskov and S. B. Haderlein, Sanierung eines PCE-Schadens in einem makroskopisch oxischen Grundwasserleiter durch Stimulation anaerober dehalogenierender Bakterien, Grundwasser, 2019, 24, 51–63 CrossRef.
- L. A. Hug, F. Maphosa, D. Leys, F. E. Löffler, H. Smidt, E. A. Edwards and L. Adrian, Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases, Philos. Trans. R. Soc., B, 2013, 368, 20120322 CrossRef PubMed.
- F. Maphosa, W. M. de Vos and H. Smidt, Exploiting the ecogenomics toolbox for environmental diagnostics of organohalide-respiring bacteria, Trends Biotechnol., 2010, 28, 308–316 CrossRef CAS PubMed.
- L. A. Hug and E. A. Edwards, Diversity of reductive dehalogenase genes from environmental samples and enrichment cultures identified with degenerate primer PCR screens, Front. Microbiol., 2013, 4, 341 Search PubMed.
- K. M. Ritalahti, B. K. Amos, Y. Sung, Q. Wu, S. S. Koenigsberg and F. E. Loffler, Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains, Appl. Environ. Microbiol., 2006, 72, 2765–2774 CrossRef CAS PubMed.
- H. Van Raemdonck, A. Maes, W. Ossieur, K. Verthe, T. Vercauteren, W. Verstraete and N. Boon, Real time PCR quantification in groundwater of the dehalorespiring Desulfitobacterium dichloroeliminans strain DCA1, J. Microbiol. Methods, 2006, 67, 294–303 CrossRef CAS PubMed.
- P. K. Lee, T. W. Macbeth, K. S. Sorenson, Jr., R. A. Deeb and L. Alvarez-Cohen, Quantifying genes and transcripts to assess the in situ physiology of “Dehalococcoides” spp. in a trichloroethene-contaminated groundwater site, Appl. Environ. Microbiol., 2008, 74, 2728–2739 CrossRef CAS PubMed.
- I. Nijenhuis, M. Nikolausz, A. Koth, T. Felfolfi, H. Weiss, J. Drangmeister, J. Grossmann, M. Kastner and H. H. Richnow, Assessment of the natural attenuation of chlorinated ethenes in an anaerobic contaminated aquifer in the Bitterfeld/Wolfen area using stable isotope techniques, microcosm studies and molecular biomarkers, Chemosphere, 2007, 67(2), 300–311 CrossRef CAS PubMed.
- N. Blazquez-Palli, M. Rosell, J. Varias, M. Bosch, A. Soler, T. Vicent and E. Marco-Urrea, Multi-method assessment of the intrinsic biodegradation potential of an aquifer contaminated with chlorinated ethenes at an industrial area in Barcelona (Spain), Environ. Pollut., 2019, 244, 165–173 CrossRef CAS PubMed.
- B. van der Zaan, F. Hannes, N. Hoekstra, H. Rijnaarts, W. M. de Vos, H. Smidt and J. Gerritse, Correlation of Dehalococcoides 16S rRNA and chloroethene-reductive dehalogenase genes with geochemical conditions in chloroethene-contaminated groundwater, Appl. Environ. Microbiol., 2010, 76, 843–850 CrossRef CAS PubMed.
- N. Yoshida, K. Asahi, Y. Sakakibara, K. Miyake and A. Katayama, Isolation and quantitative detection of tetrachloroethene (PCE)-dechlorinating bacteria in unsaturated subsurface soils contaminated with chloroethenes, J. Biosci. Bioeng., 2007, 104, 91–97 CrossRef CAS PubMed.
- Y. Yang, M. Pesaro, W. Sigler and J. Zeyer, Identification of microorganisms involved in reductive dehalogenation of chlorinated ethenes in an anaerobic microbial community, Water Res., 2005, 39, 3954–3966 CrossRef CAS PubMed.
- K. Rouzeau-Szynalski, J. Maillard and C. Holliger, Frequent concomitant presence of Desulfitobacterium spp. and “Dehalococcoides” spp. in chloroethene-dechlorinating microbial communities, Appl. Microbiol. Biotechnol., 2011, 90, 361–368 CrossRef CAS PubMed.
- D. Ghezzi, M. Filippini, M. Cappelletti, A. Firrincieli, D. Zannoni, A. Gargini and S. Fedi, Molecular characterization of microbial communities in a peat-rich aquifer system contaminated with chlorinated aliphatic compounds, Environ. Sci. Pollut. Res., 2021, 28, 23017–23035 CrossRef CAS PubMed.
- J. Li, A. Hu, S. Bai, X. Yang, Q. Sun, X. Liao and C. P. Yu, Characterization and Performance of Lactate-Feeding Consortia for Reductive Dechlorination of Trichloroethene, Microorganisms, 2021, 9, 751 CrossRef CAS PubMed.
- R. E. Richardson, Genomic insights into organohalide respiration, Curr. Opin. Biotechnol., 2013, 24, 498–505 CrossRef CAS PubMed.
- L. Hermon, J. Hellal, J. Denonfoux, S. Vuilleumier, G. Imfeld, C. Urien, S. Ferreira and C. Joulian, Functional Genes and Bacterial Communities During Organohalide Respiration of Chloroethenes in Microcosms of Multi-Contaminated Groundwater, Front. Microbiol., 2019, 10, 89 CrossRef PubMed.
- C. Lihl, L. M. Douglas, S. Franke, A. Perez-de-Mora, A. H. Meyer, M. Daubmeier, E. A. Edwards, I. Nijenhuis, B. Sherwood Lollar and M. Elsner, Mechanistic Dichotomy in Bacterial Trichloroethene Dechlorination Revealed by Carbon and Chlorine Isotope Effects, Environ. Sci. Technol., 2019, 53, 4245–4254 CrossRef CAS PubMed.
- P. K. Lee, D. R. Johnson, V. F. Holmes, J. He and L. Alvarez-Cohen, Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp, Appl. Environ. Microbiol., 2006, 72, 6161–6168 CrossRef CAS PubMed.
- D. Buchner, S. Behrens, C. Laskov and S. B. Haderlein, Resiliency of Stable Isotope Fractionation (δ(13)C and δ(37)Cl) of Trichloroethene to Bacterial Growth Physiology and Expression of Key Enzymes, Environ. Sci. Technol., 2015, 49, 13230–13237 CrossRef CAS PubMed.
- X. Maymo-Gatell, Y. Chien, J. M. Gossett and S. H. Zinder, Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene, Science, 1997, 276, 1568–1571 CrossRef CAS PubMed.
- D. R. Johnson, A. Nemir, G. L. Andersen, S. H. Zinder and L. Alvarez-Cohen, Transcriptomic microarray analysis of corrinoid responsive genes in Dehalococcoides ethenogenes strain 195, FEMS Microbiol. Lett., 2009, 294, 198–206 CrossRef CAS PubMed.
- D. R. Johnson, E. L. Brodie, A. E. Hubbard, G. L. Andersen, S. H. Zinder and L. Alvarez-Cohen, Temporal Transcriptomic Microarray Analysis of “Dehalococcoides ethenogenes” Strain 195 during the Transition into Stationary Phase, Appl. Environ. Microbiol., 2008, 74, 2864–2872 CrossRef CAS PubMed.
- K. Furukawa, A. Suyama, Y. Tsuboi, T. Futagami and M. Goto, Biochemical and molecular characterization of a tetrachloroethene dechlorinating Desulfitobacterium sp. strain Y51: a review, J. Ind. Microbiol. Biotechnol., 2005, 32, 534–541 CrossRef CAS PubMed.
- J. Yan, B. Simsir, A. T. Farmer, M. Bi, Y. Yang, S. R. Campagna and F. E. Loffler, The corrinoid cofactor of reductive dehalogenases affects dechlorination rates and extents in organohalide-respiring Dehalococcoides mccartyi, ISME J., 2016, 10, 1092–1101 CrossRef CAS PubMed.
- S. T. Lohner and A. M. Spormann, Identification of a reductive tetrachloroethene dehalogenase in Shewanella sediminis, Philos. Trans. R. Soc., B, 2013, 368, 20120326 CrossRef PubMed.
- M. Elsner, Stable isotope fractionation to investigate natural transformation mechanisms of organic contaminants: principles, prospects and limitations, J. Environ. Monit., 2010, 12, 2005–2031 RSC.
- T. B. Hofstetter, R. Bakkour, D. Buchner, H. Eisenmann, A. Fischer, M. Gehre, S. B. Haderlein, P. Höhener, D. Hunkeler, G. Imfeld, M. A. Jochmann, S. Kümmel, P. R. Martin, S. G. Pati, T. C. Schmidt, C. Vogt and M. Elsner, Perspectives of compound-specific isotope analysis of organic contaminants for assessing environmental fate and managing chemical pollution, Nat. Water, 2024, 2, 14–30 CrossRef.
- M. A. Jochmann and T. C. Schmidt, Compound-specific Stable Isotope Analysis, Royal Society of Chemistry, Cambridge, U.K., 2012 Search PubMed.
- K. C. Harding, P. K. Lee, M. Bill, T. E. Buscheck, M. E. Conrad and L. Alvarez-Cohen, Effects of varying growth conditions on stable carbon isotope fractionation of trichloroethene (TCE) by tceA-containing Dehalococcoides mccartyi strains, Environ. Sci. Technol., 2013, 47, 12342–12350 CrossRef CAS PubMed.
- B. M. van Breukelen, Extending the Rayleigh equation to allow competing isotope fractionating pathways to improve quantification of biodegradation, Environ. Sci. Technol., 2007, 41, 4004–4010 CrossRef CAS PubMed.
- B. M. Van Breukelen, Quantifying the degradation and dilution contribution to natural attenuation of contaminants by means of an open system rayleigh equation, Environ. Sci. Technol., 2007, 41, 4980–4985 CrossRef CAS PubMed.
- Y. Dong, E. C. Butler, R. P. Philp and L. R. Krumholz, Impacts of microbial community composition on isotope fractionation during reductive dechlorination of tetrachloroethylene, Biodegradation, 2011, 22, 431–444 CrossRef CAS PubMed.
- S. Cretnik, K. A. Thoreson, A. Bernstein, K. Ebert, D. Buchner, C. Laskov, S. Haderlein, O. Shouakar-Stash, S. Kliegman, K. McNeill and M. Elsner, Reductive dechlorination of TCE by chemical model systems in comparison to dehalogenating bacteria: insights from dual element isotope analysis (13C/12C, 37Cl/35Cl), Environ. Sci. Technol., 2013, 47, 6855–6863 CrossRef CAS PubMed.
- B. Heckel, S. Cretnik, S. Kliegman, O. Shouakar-Stash, K. McNeill and M. Elsner, Reductive Outer-Sphere Single Electron Transfer Is an Exception Rather than the Rule in Natural and Engineered Chlorinated Ethene Dehalogenation, Environ. Sci. Technol., 2017, 51, 9663–9673 CrossRef CAS PubMed.
- J. Renpenning, S. Keller, S. Cretnik, O. Shouakar-Stash, M. Elsner, T. Schubert and I. Nijenhuis, Combined C and Cl isotope effects indicate differences between corrinoids and enzyme (Sulfurospirillum multivorans PceA) in reductive dehalogenation of tetrachloroethene, but not trichloroethene, Environ. Sci. Technol., 2014, 48, 11837–11845 CrossRef CAS PubMed.
- J. Büsing, D. Buchner, S. Behrens and S. B. Haderlein, Deciphering the Variability of Stable Isotope (C, Cl) Fractionation of Tetrachloroethene Biotransformation by Desulfitobacterium strains Carrying Different Reductive Dehalogenases Enzymes, Environ. Sci. Technol., 2020, 54, 1593–1602 CrossRef PubMed.
- T. Kruse, T. Goris, J. Maillard, T. Woyke, U. Lechner, W. de Vos and H. Smidt, Comparative genomics of the genus Desulfitobacterium, FEMS Microbiol. Ecol., 2017, 93, fix135 CrossRef PubMed.
- A. Suyama, M. Yamashita, S. Yoshino and K. Furukawa, Molecular characterization of the PceA reductive dehalogenase of desulfitobacterium sp. strain Y51, J. Bacteriol., 2002, 184, 3419–3425 CrossRef CAS PubMed.
- B. A. van de Pas, J. Gerritse, W. M. de Vos, G. Schraa and A. J. Stams, Two distinct enzyme systems are responsible for tetrachloroethene and chlorophenol reductive dehalogenation in Desulfitobacterium strain PCE1, Arch. Microbiol., 2001, 176, 165–169 CrossRef CAS PubMed.
- A. Bernstein, O. Shouakar-Stash, K. Ebert, C. Laskov, D. Hunkeler, S. Jeannottat, K. Sakaguchi-Soder, J. Laaks, M. A. Jochmann, S. Cretnik, J. Jager, S. B. Haderlein, T. C. Schmidt, R. Aravena and M. Elsner, Compound-specific chlorine isotope analysis: a comparison of gas chromatography/isotope ratio mass spectrometry and gas chromatography/quadrupole mass spectrometry methods in an interlaboratory study, Anal. Chem., 2011, 83, 7624–7634 CrossRef CAS PubMed.
- K. A. Ebert, C. Laskov, M. Elsner and S. B. Haderlein, Calibration bias of experimentally determined chlorine isotope enrichment factors: the need for a two-point calibration in compound-specific chlorine isotope analysis, Rapid Commun. Mass Spectrom., 2017, 31, 68–74 CrossRef CAS PubMed.
- D. Buchner, P. R. Martin, J. Scheckenbach, S. Kummel, F. Gelman and S. B. Haderlein, Expanding the calibration range of compound-specific chlorine isotope analysis by the preparation of a (37) Cl-enriched tetrachloroethylene, Rapid Commun. Mass Spectrom., 2022, 36, e9378 CrossRef CAS PubMed.
- K. M. Scott, X. Lu, C. M. Cavanaugh and J. S. Liu, Optimal methods for estimating kinetic isotope effects from different forms of the Rayleigh distillation equation, Geochim. Cosmochim. Acta, 2004, 68, 433–442 CrossRef CAS.
- D. Buchner, B. Jin, K. Ebert, M. Rolle, M. Elsner and S. B. Haderlein, Experimental Determination of Isotope Enrichment Factors - Bias from Mass Removal by Repetitive Sampling, Environ. Sci. Technol., 2017, 51, 1527–1536 CrossRef CAS PubMed.
- A. S. Ojeda, E. Phillips, S. A. Mancini and B. S. Lollar, Sources of Uncertainty in Biotransformation Mechanistic Interpretations and Remediation Studies using CSIA, Anal. Chem., 2019, 91, 9147–9153 CrossRef CAS PubMed.
- M. Lee, E. Wells, Y. K. Wong, J. Koenig, L. Adrian, H. H. Richnow and M. Manefield, Relative contributions of Dehalobacter and zerovalent iron in the degradation of chlorinated methanes, Environ. Sci. Technol., 2015, 49, 4481–4489 CrossRef CAS PubMed.
- H. Nonaka, G. Keresztes, Y. Shinoda, Y. Ikenaga, M. Abe, K. Naito, K. Inatomi, K. Furukawa, M. Inui and H. Yukawa, Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195, J. Bacteriol., 2006, 188, 2262–2274 CrossRef CAS PubMed.
- A. Perez-de-Mora, A. Lacourt, M. L. McMaster, X. Liang, S. M. Dworatzek and E. A. Edwards, Chlorinated Electron Acceptor Abundance Drives Selection of Dehalococcoides mccartyi (D. mccartyi) Strains in Dechlorinating Enrichment Cultures and Groundwater Environments, Front. Microbiol., 2018, 9, 812 CrossRef PubMed.
- B. G. Rahm, R. M. Morris and R. E. Richardson, Temporal expression of respiratory genes in an enrichment culture containing Dehalococcoides ethenogenes, Appl. Environ. Microbiol., 2006, 72, 5486–5491 CrossRef CAS PubMed.
- J. Gerritse, V. Renard, T. M. Pedro Gomes, P. A. Lawson, M. D. Collins and J. C. Gottschal, Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols, Arch. Microbiol., 1996, 165, 132–140 CrossRef CAS PubMed.
- N. Tsukagoshi, S. Ezaki, T. Uenaka, N. Suzuki and R. Kurane, Isolation and transcriptional analysis of novel tetrachloroethene reductive dehalogenase gene from Desulfitobacterium sp. strain KBC1, Appl. Microbiol. Biotechnol., 2006, 69, 543–553 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ew00511b |
| This journal is © The Royal Society of Chemistry 2024 |
