Micro-immobilized enzyme reactors for mass spectrometry proteomics
Zhongjie
Yao
a,
Yilan
Li
 *b and
Wei
Xu
*b and
Wei
Xu
 *a
*a
aSchool of Medical Technology, Beijing Institute of Technology, Beijing 100081, China. E-mail: weixu@bit.edu.cn
bAnalysis & Testing Center, Beijing Institute of Technology, Beijing 102488, China. E-mail: liyilan@bit.edu.cn
First published on 19th June 2025
Abstract
Micro-immobilized enzyme reactors (μ-IMERs) have proven to be superior to traditional enzymatic methods by offering improved enzyme stability, less reagent consumption, and higher reaction efficiency. This review aims to provide an assessment of recent advances in μ-IMER design, fabrication, and proteomic applications. Common immobilization methods including adsorption, covalent binding, affinity binding, and entrapment are discussed, and their respective advantages, challenges and possible future development directions are summarized. The review also covers different μ-IMER designs, including open-tube, packed, monolithic reactors, and membrane-immobilized enzymes, and provides a detailed analysis of their structural architectures and operational performance characteristics. In addition, μ-IMER applications in proteomics are presented, demonstrating their potential for improving sample preparation and analytical workflows. Finally, we summarize the current state of development and the challenges faced, offering new insights into potential future directions.
1. Introduction
Proteomics offers a systemic and dynamic approach for the explanation of the life molecular processes by examining protein expression, post-translational modification, interactions, and function in cells, tissues, and organisms.1,2 Besides enabling the discovery of disease biomarkers and drug targets, proteomics drives basic biological research in the direction of more systematic and precise methods.3,4In modern proteomics, the development of two of the most renowned ionization techniques—electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI)—has significantly advanced the use of mass spectrometry (MS) in the field.5,6 Along with the growing size of protein databases, improvements in mass spectrometry scanning rates and mass precision have rendered mass spectrometers as indispensable equipment for proteomics problem-solving.7,8 Mass analyzers, including quadrupole, ion trap, time-of-flight, orbitrap, and Fourier transform ion cyclotron resonance, provide significant technical support for proteomics investigation.9–11 In 2014, the first near-complete human proteome expression profile was explored, providing a crucial milestone for continued development of our understanding of protein expression and function.5 Over the past few years, improvements in the development of novel search engines have accelerated peptide and protein identification and quantification of MS/MS data, rendering MS more efficient and accurate in proteomics.12–14
Pre-treatment of proteomics samples is a difficult process. Traditional enzymatic digests are performed in solution, where proteins are incubated with free proteases in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 to 1
20 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 enzyme-to-substrate ratios and reaction times of 6 to 12 h.15 During this process, peptide fragments that are produced as a result of the autolysis of the protease are created, which not only add to the sample complexity but also reduce the signal intensity of the target peptide.16,17 This results in low analytical throughput and higher detection limits.
100 enzyme-to-substrate ratios and reaction times of 6 to 12 h.15 During this process, peptide fragments that are produced as a result of the autolysis of the protease are created, which not only add to the sample complexity but also reduce the signal intensity of the target peptide.16,17 This results in low analytical throughput and higher detection limits.
The application of an immobilized enzyme reactor (IMER) in proteomics could be traced back to 1989, when Cobb and Novotny immobilized trypsin in an agarose gel and filled Pyrex tubes with an inner diameter of 30 cm × 1 mm.18 With this method, 50 ng of protein was digestible, and sample usage was reduced by a factor of approximately three orders over solution-based conventional methods. Over the last few years, μ-IMERs—particularly those implemented in microchannels or capillaries—have attracted specific interest, having several benefits over traditional enzymatic reactions performed in solution.19–21 Immobilization of enzymes reduces protease self-degradation, enhancing its storage stability, and reduces the demand for costly sequencing-grade proteases (recombinant trypsin), thereby improving cost-efficiency.22,23 Immobilized enzyme is also easily recyclable, reducing the use of enzyme and obviating the need for reagents such as acetic acid or formic acid to terminate the reaction.24,25 The dense protease distribution across a small volume enhances the rate of the enzymatic reaction, thus reducing the detectable limit of low-abundance samples.26–28 Compared with free enzyme reaction systems, μ-IMERs offer superior control over reaction conditions, reduced inter-batch variability and greater integration—minimizing human error.29,30
This review highlights the recent progress in enzyme immobilization strategies, reactor architectures, and advanced carrier materials, emphasizing their advantages in rapid digestion, high-throughput processing, microfluidic integration, etc. The combination of μ-IMERs and MS was further explored to address challenges in proteomics, including complex sample analysis, structural proteomics, quantitative proteomics, etc. The review also briefly discusses emerging applications in multi-enzyme cascades, automated workflows, and single-cell proteomics, covering the publications over the past 5 years, while also including earlier landmark studies where relevant.
2. Immobilization of enzymes
2.1 Adsorption
Based on this principle, a stable enzyme immobilized μ-IMER with high catalytic performance was thoroughly established, providing a reliable platform for enzymatic reaction. A 5 min rapid enzymatic pretreatment was employed for the sample preparation to examine the glycosylation structure of a monoclonal antibody (mAb). Direct preparation avoided involving steps like alkylation and desalting. The enzymatic digestion provided glycopeptides containing the native sugar structures of the mAb Fc region. This publication has identified eleven principal categories of mAb glycosylation.37
Although both methods are simple to perform, they rely on weak interactions between the enzyme and the support. For example, the enzyme retained only 60% of its activity after storage at 4 °C for 10 days.38,39 Reduced enzymatic activity during storage directly affects digestion efficiency. In addition, enzymes may gradually leach from the support during repeated use, which affects enzyme retention and compromises the overall robustness and reusability of the system, particularly when used in analytical and sensing applications.40 More stable methods such as covalent bonding have therefore been explored.
2.2 Covalent binding
Unlike non-covalent bonds, which rely on weak interactions, covalent bonds are applied for immobilization of enzymes due to their stability and resistance to breakage.41,42 Immobilization via covalent bonding can also enhance enzyme activity by adjusting the distance of spacers between the enzyme and carrier.43 Nouaimi et al., for instance, enhanced the activity of immobilized trypsin on polyester fibers by using spacer molecules such as polyethylene glycol (PEG)-diamine and aldehyde dextran.44 Similarly, Holyavka et al. used high molecular weight chitosan to design longer inter-arms for papain and hence enhance enzyme activity by reducing spatial resistance.45 The right length of inter-arms not only enhances enzyme activity but also enhances enzyme immobilization, allowing more enzyme molecules to bind effectively to the carrier. During covalent immobilization, the use of an enzyme activity inhibitor can block the active site and therefore covalent bonding to the carrier. This protection preserves the catalytic activity of the enzyme and provides long-term stability and efficiency of the reactor. The most commonly used covalent bonding methods are Schiff base or carbodiimide chemistry (Fig. 1a).25 | ||
| Fig. 1 Covalent immobilization for enzyme fixation. (a) Common covalent immobilization methods: Schiff base and carbodiimide reactions. Reprinted with permission from ref. 25. Copyright 2017 Elsevier. (b) Trypsin immobilized on a boronate affinity monolith via Schiff base linkages. Reprinted with permission from ref. 46. Copyright 2022 Elsevier. | ||
For example, Wang et al. used the Schiff base reaction and Michael addition reaction to covalently immobilize trypsin onto a boron affinity monolithic column for mouse proteomics (Fig. 1b).46 Notably, the immobilized enzyme reactor maintained 80% of the initial enzyme activity after 28 days of storage at 4 °C. In another study, Duong et al. employed carbodiimide chemistry to immobilize trypsin and antibodies onto silica particles, achieving a 10 min enzymatic digestion of cytochrome C (Cyt C) at room temperature, which is comparable to 18 h of in-solution digestion at 37 °C.47
2.3 Affinity binding
In surface adsorption and covalent bonding, the relative orientation of the enzyme cannot be controlled, so it is impossible to ensure that the active site has low steric hindrance. However, affinity binding uses molecular recognition to bind the enzyme in a specific direction to ensure the accessibility of the active site. Histidine tag (His tag)-mediated metal ion affinity binding technology is one of the most widely used affinity binding immobilization methods. It achieves directional immobilization by forming a stable coordination between the protease with the His tag and metal ions such as Ni2+ and Co2+.48,49 Since the tag is located primarily at either end of the protein, beyond the region encompassing the active site, the shape and function of the enzyme are not impaired. Also, this strategy is very stable against interference and retains enzyme stability even in cases of high salt levels or in complex matrices. Makrydaki et al. achieved homogeneous and effective modification by directly appending a His tag to β-1,4-galactosyltransferase (β4GalT1) and immobilizing it on a solid-phase carrier to catalyze galactosylation of the immunoglobulin G (IgG), achieving an efficiency of 80.2%–96.3%.50 A novel method of immobilization has been welcomed more recently based on employing a specific nucleic acid chain as a linker: first, a nucleic acid layer is pre-modified over the surface of a solid carrier, followed by deposition of a complementary sequence on the enzyme molecule. On combining, the complementary strands get associated, resulting in ordered and directed immobilization of the enzyme. Fan et al. reduced spatial obstruction of the enzyme active site by building a DNA tetrahedral scaffold on magnetic nanoparticles (MNPs), which allowed rational control of the distribution of tryptic enzymes such that adequate spacing between enzyme molecules was provided.51 Experimental results showed that immobilization system was extremely effective in bovine serum albumin (BSA) enzymatic digestion, achieving 91% sequence coverage within just 2 min.2.4 Entrapment
Entrapment is a method that uses spatial limitation to trap enzymes. In comparison with physical adsorption, entrapment is effective in preventing enzyme loss and increasing enzyme loading. In comparison with covalent immobilization, this process does not require complex chemical reactions, hence preserving the natural conformation of the enzyme and activity. Arad et al. formed a rapid entrapment of pepsin in an elastic hydrogel by optimizing the sodium alginate, calcium carbonate, and formic acid ratios. The hydrogel can be used in microcentrifuge tubes or pipette tips for in situ enzymatic digestion of microsamples.52 Experimental data reveal that the enzymatic digestion efficiency is approximately 120 times higher than that of conventional in-solution digestion, enabling comprehensive peptide profiling of BSA within 1 min (Fig. 2). Increasing the degree of polymerization of the carrier and thickening the supporting matrix to increase the enzyme loading will also increase the steric hindrance between the substrate and the enzyme active site, thereby reducing the reaction efficiency. Therefore, the microenvironment of the carrier needs to be strictly regulated throughout the preparation process to achieve the most effective enzyme immobilization. | ||
| Fig. 2 Pepsin–trypsin immobilization on a microcentrifuge tube or pipette tip for proteomics sample digestion. Reprinted with permission from ref. 52. Copyright 2024 The Authors. | ||
In summary, enzyme immobilization strategies differ in terms of operational complexity, enzyme stability, and suitability for various substrates. While physical adsorption is simple and mild, it suffers from poor stability. Covalent and affinity binding offers stronger retention but requires surface functionalization. Entrapment provides a compromise between structural preservation and loading efficiency. These immobilization methods lay the foundation for subsequent microreactor design, as the effectiveness of enzyme utilization is strongly influenced by the physical and chemical environments provided by the reactor structure.
3. Manufacture of μ-IMERs
The performance of μ-IMERs depends not only on how enzymes are attached, but also on how the reactor supports substrate transport, mechanical stability, and integration with analytical platforms. Different types of μ-IMER configurations were further elaborated.3.1 Open-tubular support
Open-tubular supports refer to capillary-based systems where enzymes are immobilized directly on the inner wall, often forming the functional core of open-tube μ-IMERs. Open-tubular supports, typically implemented using fused silica capillaries, modify the capillary surface charge to facilitate electrostatic adsorption of enzymes.35 However, the inherently low surface area of open-tubular systems limits enzyme loading capacity, making it difficult to process digestion-resistant or complex protein samples. Increasing enzyme layers along the capillary wall improves attachment site availability and enhances substrate diffusion to active sites.To improve immobilization efficiency, the capillary inner wall is often silanized using (3-aminopropyl) triethoxysilane (APTES). The carboxyl groups are further activated by carbodiimide chemistry to directly covalently bind the enzyme, or the enzyme is cross-linked to the capillary wall by glutaraldehyde-mediated imine bond reaction. To extend its service life, it can be stabilized to C–N single bonds using reducing agents. While APTES is widely used, it tends to form non-uniform multilayer coatings.53 These reduce inner wall smoothness and compromise pore structure control, increasing steric hindrance during immobilization and leaving unreacted cross-linkers that may lead to enzyme loss.
(3-Triethoxysilyl) butyraldehyde (TESB) has emerged as an effective alternative. Its aldehyde group forms direct cross-links with enzymes, and the hydroxyl group at the other end can bind to the surface of the substrate to form a uniform coating. Since the aldehyde group directly cross-links with the enzyme, the process does not require chemical modification. This minimizes residual linker effects, ensuring a more uniform coating and reducing potential enzyme loss.54
Among the novel carrier materials explored in recent years, metal–organic frameworks (MOFs) have attracted significant attention for enzyme immobilization due to their structural tunability, chemical stability, and large surface area.
Zhang et al. synthesized a stabilized enzyme microreactor (ChT@ZIF-L-IMER) using a structured encapsulation approach. Chymotrypsin (ChT) was blended with a short DNA strand and immobilized in a microcapillary tube.39 The enzyme was then encapsulated in situ using a zeolitic imidazolate framework (ZIF-L). Negatively charged phosphate groups in the DNA facilitated a superior uniformity and ordered deposition of ZIF-L by entrapment of its precursors. This encapsulation provided a protective shell over the enzyme, significantly enhancing its stability under extreme conditions—such as elevated temperatures (80 °C), high or low pH, and the presence of organic solvents (methanol and acetonitrile)—compared to conventional open-tubular supports.
In addition to improving stability, the ZIF-L encapsulation also enhances the enzyme's substrate affinity. The Michaelis constant (Km) of ChT@ZIF-L-IMER is 0.31 mM, significantly lower than that of the conventional ChT-IMER (2.61 mM), indicating improved substrate accessibility and a more favorable microenvironment provided by the MOF matrix.
Open-tubular supports feature low backpressure and smooth flow paths, enabling rapid digestion with short residence times. However, their inherently low surface-to-volume ratio limits enzyme loading, which may reduce sequence coverage and compromise long-term enzyme stability. These characteristics make them more suitable for fast digestion of simple samples or integration into microfluidic platforms.
3.2 Particulate supports
Particulate-based μ-IMERs enable higher enzyme-loading capacity and shorter substrate diffusion distances, thus improving digestion efficiency.55 These systems typically utilize nano- or micro-sized particles—such as non-magnetic polymer or silica microspheres and MNPs—as the stationary phase within microchannels, similar to liquid chromatography columns. The use of packed particulate supports facilitates dense enzyme distribution and enhances contact between the substrate and catalyst, making them particularly effective for complex protein digestion workflows. Optimizing particle size and packing density further boosts performance: smaller particles (e.g., 20 μm) and higher packing ratios increase the surface area for enzyme attachment, whereas larger particles with lower densities reduce mass transfer resistance and facilitate faster substrate access to active sites.The effectiveness of this approach is demonstrated by existing commercial products. For example, Sigma-Aldrich Inc. has developed silica microspheres with immobilized trypsin (20 μm particle size) that achieve the same digestion efficiency in 15 min as traditional overnight digestion.56 Similarly, Thermo Scientific has successfully implemented fused silica capillaries loaded with immobilized trypsin for high-throughput proteomics analysis of micro-samples (<100 μL) (Fig. 3).57 These technologies highlight the industrial potential of packed μ-IMERs in improving enzyme utilization efficiency and operational flexibility.
 | ||
| Fig. 3 Capillary microreactor with agarose-immobilized trypsin for efficient digestion of trace mammalian cells. Reprinted with permission from ref. 57. Copyright 2020 American Chemical Society. | ||
A key consideration for particulate supports is the effective immobilization of particles for reaction efficiency and stability. Particle mobility or leakage can significantly impair reactor performance. The size of the packing is usually larger than the pore size of the sieve plates. These plates are either installed post-synthesis or formed in situ during fabrication.57,58 In systems using particulate supports within microfluidic chips, particle retention is often achieved by narrowing the channel outlet below the particle size.59
MNPs offer unique advantages as particulate supports. The particulate support can be directly captured by an external magnetic field without using sieves or channel size adjustments, making device manufacturing easier. The magnetic field can also enable easy recovery of particles and precise control of enzyme–substrate reactions by separating the enzyme at a specific time, thereby controlling the reaction progress with accuracy. Bataille et al. developed a magnetic particle microfluidized bed reactor, where an external magnetic field was used to precisely control particle positioning, enhance enzyme–substrate interactions, and accelerate mass transfer. This strategy resulted in improved digestion efficiency and greater sequence coverage.60
In addition to flow-based μ-IMER configurations, MNPs have also been successfully applied to static enzymatic workflows. Dieters-Castator et al. developed a magnetic bead-based cell surface proteomics platform for efficient enzymatic digestion of cell surface proteins under static solution conditions by covalently immobilizing trypsin and lysyl endopeptidase (Lys-C) on the surface of MNPs.61 The system was able to identify up to 900 surface glycoproteins from 25–200 μg of total proteins and effectively retain N-glycosylation site information. This demonstrates the potential of the static MNP system for efficient processing of complex samples.
It will also be helpful in understanding the flow-based and static modes of enzymatic digestion when describing the modes of μ-IMER operation because these will show the similarities and differences between them. The flow-based modes of μ-IMER operation are typically better suited for automated continuous operation and are typically better suited for integration into an LC-MS workflow, whereas the static modes of operation (particularly when MNPs are used) are simpler, offer improved enzyme reusability, and are better suited for small numbers of samples and batch mode operation.
Particulate supports enable high enzyme loading and efficient substrate interaction. While MNPs offer flexible use in both flow and static modes, particle leakage and pressure variability may affect long-term stability, requiring careful reactor design.
3.3 Monolithic supports
Monolithic supports utilize a continuous porous matrix as the immobilization scaffold for the immobilization of enzyme. The system, compared to open-tube reactors, possesses a network structure in three dimensions, hence significantly increasing the surface area available for enzyme immobilization and providing superior enzyme loading.62 Additionally, compared to filled reactors, its innovative through-channel structure reduces the resistance of fluid while not sacrificing the mechanical strength. The structure helps in substrate diffusion and avoids the leakage of the enzyme. Preparation of the reactor is carried out by three significant processes: in situ entrapment of the enzyme through cross-linking of monomers, formation of covalent networks through chemical condensation, and preparation of a porous skeleton through phase separation technology. The water stability, modifiability, and wide pH tolerance of polymer matrices have made them the most sought-after in the field.Non-target molecules, however, can be adsorbed on the channel surface. This is avoidable using hydrophilic surface modification with PEG. Porous materials suffer from the substrate mass transfer limitation problem.63,64 This has been overcome with an advancement in high internal phase emulsion (HIPE) template technology that maximizes mass transfer by the creation of a macroporous scaffold that enables maximum substrate flow and high enzyme loading.65
In recent years, thiol–ene (TE) click chemistry has emerged as a powerful approach for constructing monolithic μ-IMERs on microfluidic platforms, offering high efficiency and precise control over polymerization. Via photo-initiated free radical polymerization this approach allows the in situ generation of porous monoliths in microchannels. The new polymerization mechanism offers strong covalent anchoring to the chip substrate and by varying the thiol to ene molar ratio one can obtain surfaces bearing thiol or alkene groups, which allow various strategies for enzyme coupling.
In contrast to conventional methacrylate-based monoliths, which have to be pretreated by surface silanization, robust composite materials can be prepared directly in unmodified microchannels via thiol–silanol interactions during the polymerization. Using this approach, Procházka et al. assembled a trypsin-immobilized μ-IMER, which digested BSA in 40 s and achieved 73.7% sequence coverage. Notably, the versatility of TE click chemistry has effectively expanded the applicability of μ-IMERs by enabling the efficient immobilization of diverse enzymes—including trypsin, pepsin, and glycosidases—without prior modification.60,66,67
Enzyme reactors based on monolithic carrier immobilization usually show efficient enzymolysis, high sequence coverage and strong enzyme stability. These properties are due to its interconnected porous network structure, which not only increases the enzyme loading capacity but also enhances the mass transfer efficiency of the substrate. The stable covalent connection formed between the enzyme and the carrier further reduces the loss of the enzyme, thereby ensuring its performance stability during repeated use or long-term operation.
3.4 Membrane-based supports
In addition to the aforementioned support types, membrane-based systems have also demonstrated significant advantages in protein digestion. Although these structures sometimes exceed the microscale in physical dimensions, they retain the key characteristics of μ-IMERs, such as flow-through operation and high catalytic efficiency, thereby exhibiting strong practical applicability in real-world scenarios31,68,69 For example, Kjellander et al. immobilized trypsin on anodized alumina membranes to facilitate on-line digestion and real-time peptide analysis in conjunction with mass spectrometry.70 Enzymes are immobilized through covalent or electrostatic binding on nylon membranes, and the ultra-thin (∼100 μm) membrane architecture allows reactions to be carried out in milliseconds to seconds. They are thus best adapted to the restricted digestion under low pressure, yielding precise resolution of structurally heterogeneous protein domains.71Another widely used carrier type is porous ceramic capillary membranes.72,73 These are essentially tubes with an inner diameter of about 1 mm that are produced by extrusion molding. They possess chemical and thermal stability, mechanical strength, and controlled pore size distribution. Such a nature qualifies them for multiphase reaction systems that consist of both aqueous and organic phases. For example, enzyme immobilization catalyzed by carbodiimide, combined with a continuous flow format, enables rapid processing of complex biological samples.73 These carriers are expected to enable microscale reaction control and high-throughput analysis.
Membrane supports usually achieve short enzymatic hydrolysis times due to their high-throughput flow characteristics, making them suitable for rapid online analysis. However, due to the thin stationary phase layer and limited enzyme loading capacity, the sequence coverage will be affected when processing complex substrates. In addition, enzymes lack structural protection in such systems and find it difficult to support long-term stable operation.
4. Multi-enzyme reaction system
The application of μ-IMERs in complex catalytic systems, particularly multi-enzyme cascade reactions, has garnered increasing research interest. Commonly used proteolytic enzymes in the μ-IMER system include trypsin, Lys-C, glutamyl endopeptidase C (Glu-C) and ChT, each with its own specificity. Trypsin specifically hydrolyzes the C-terminus of lysine (K) and arginine (R) residues, but is limited by the uneven distribution of K/R in the target protein. It is often combined with Glu-C, which can cleave glutamic acid (E) and aspartic acid (D), to achieve orthogonal enzymatic cleavage. The cleavage site of Glu-C is greatly affected by the pH value, and appropriate buffer conditions need to be selected according to the substrate. In structurally complex areas, it is difficult for trypsin to achieve efficient enzymatic cleavage at the K position, while Lys-C/trypsin coupling can efficiently cleave K residues by Lys-C, thereby improving the overall enzymatic efficiency. ChT recognizes aromatic amino acids (F, Y, and W), which can supplement the enzymatic blind spots of trypsin in hydrophobic or rigid structural areas and enhance sequence coverage, but it is sensitive to heat and denaturants and needs to be used under mild conditions.To translate these synergistic protease strategies into practical μ-IMER applications, recent developments have focused on co-immobilizing multiple enzymes within a single reactor or arranging multiple μ-IMERs in tandem to enhance digestion efficiency.74,75 Yuan et al. covalently immobilized trypsin and Glu-C in the KIT-6 molecular sieve using glutaraldehyde as a cross-linking agent.76 The three-dimensional pore structure of the KIT-6 material effectively increased the enzyme loading, which was 2 to 3 times higher than that of the traditional method. The orthogonal digestion strategy using immobilized trypsin and Glu-C increased the sequence coverage of BSA from 76.90% (free enzyme) to 91.30%. In addition, Brandtzaeg et al. developed a multi-channel open-tubular support enzyme reactor to co-immobilize trypsin and Lys-C to achieve rapid online digestion of castor bean extracts containing the highly toxic protein ricin. The system does not require reduction or alkylation steps, can achieve efficient digestion in just 5 min, and can accurately identify ricin peptides in complex biological matrices.77
5. μ-IMERs’ proteomics advantage
5.1 Streamlined μ-IMERs for complex samples
In proteomics, the efficiency bottleneck in sample pretreatment is especially evident.78,79 μ-IMERs are gaining popularity as tools to combine sample pretreatment and digestion. This increasing adoption is driven by their efficient digestion capabilities and suitability for automated workflows.80,81 Gilquin's team developed an integrated microfluidic platform that combines cell filtration, protein enrichment, and enzymatic hydrolysis modules, enabling in situ processing of whole blood samples.82 After removing interfering blood cell components using specific adsorption resin, the platform employs immobilized proteases for targeted digestion and is directly connected to a mass spectrometer. Experimental results demonstrate that the system shortens the detection time for low-abundance blood biomarkers—specifically alanine aminotransferase 1 (ALT1)—to just 2 h, while also providing a high level of automation. With a combination of μ-IMERs and cluster hollow fibre membrane interfaces, an integrated plasma proteome sample preparation system was developed by Zhang's group, in which high abundance plasma proteins were initially depleted by an immunoaffinity column, followed by on-line denaturation, reduction, desalting, and tryptic digestion of middle- and low-abundance proteins. Compared to conventional in-solution protocols, not only the sample preparation time could be shortened from 20 h to 20 min, but also the number of identified proteins increased by 1.4 to 2.0 times.83 In addition, to evaluate and mitigate the risk of cross-contamination between samples in the μ-IMER-based workflow, Zhang's group implemented a SILAC-based sequential processing strategy.84 Specifically, heavy- and light-labeled proteins from HepG2 cell lines were processed consecutively using the integrated continuous-flow automated sample treatment (cFAST) device, which combines μ-IMERs with cluster hollow fiber membrane interfaces (cHFMIs). Between the two samples, the system was flushed with organic solvents to remove residual peptides and proteins from the reactor surfaces. After discarding the heavy-labeled fraction, the light-labeled sample was collected and analyzed by nanoLC-MS/MS. The level of heavy peptides detected in the light fraction was used as a measure of carryover. The results demonstrated a minimal peptide carryover of only 5.7% ± 1.8% (n = 3), confirming the system's excellent anti-contamination performance.Furthermore, the complex glycosylation modifications of proteins lead to significant molecular heterogeneity, making traditional processing methods time-consuming, often requiring dozens of hours.85,86 Camperi's team developed an integrated multidimensional separation platform.87 This system combines a reversed-phase chromatography column with an enzymatic column loaded with trypsin/Lys-C, enabling online continuous processing of Fc-fusion proteins. Experimental data showed that this platform achieved over 90% sequence coverage of the target protein, with a missed cleavage rate below 10%. Moreover, it successfully identified three key glycosylation sites.
Monoclonal antibodies (mAbs) have large molecular weight, approximately 150 kDa, the “top-down” analysis strategies find it difficult to achieve high-resolution protein analysis, and the “bottom-up” method requires complex enzymatic hydrolysis of the samples, which makes it difficult to improve the analysis efficiency. Due to such reasons, a “middle-up” strategy has been promoted where the hinge region of mAbs is cleaved using papain to produce fragments of approximately 50 kDa. This strategy takes into account both analytical accuracy and work throughput. For example, Francesca Rinaldi's team developed an innovative technology platform.88 It utilizes high-porosity polymerized high internal phase emulsion (polyHIPE) materials as carriers, with papain stably immobilized onto a three-dimensional porous network through a directional immobilization strategy. The experimental results showed that three well-defined subunits were generated via enzymatic digestion of rituximab (RTX) in situ. After 12 months of storage, the immobilized papain still maintained 66.2% of its initial activity, which provides a stable and efficient detection platform for the quality control of antibody drugs. In addition, Zhang et al. employed a middle-up strategy to enzymatically cleave trastuzumab (an IgG1 monoclonal antibody) into three distinct subunits using papain. Each subunit was subsequently characterized using mobility shift capillary electrophoresis (MCE) and native mass spectrometry (nMS), enabling high-resolution structural characterization of the antibody.89
5.2 μ-IMERs for structural proteomics
In protein structural studies, hydrogen/deuterium exchange mass spectrometry (HDX-MS) is increasingly a trendy technique to investigate dynamic conformational changes of proteins.90 Pepsin is commonly used for HDX-MS analysis, but its poor efficiency at hydrolyzing disulfide-containing regions restricts its spatial resolution.91 To address the problem of low hydrolysis efficiency of pepsin in disulfide-bonded regions, Comamala et al. constructed an integrated pretreatment scheme that in combination with an online electrochemical reduction module can effectively break disulfide bonds (Fig. 4).67 This scheme facilitates the subsequent enzymatic hydrolysis process, and the deglycosylase and pepsin reactors are also configured in series. This method has been successfully used for automated continuous processing of c-Met receptors and enzymatic hydrolysis of hemoglobin, with sequence coverage close to 100%. Although pepsin still reigns supreme over HDX-MS, scientists are attempting other enzymatic hydrolysis approaches. Zheng et al. outlined a novel protocol utilizing nepenthesin II (NepII), immobilized onto a silica support through glutaraldehyde cross-linking.92 NepII produced peptides with an average 1.31 amino acid shorter length and about 2.3 units increased charge over pepsin. This not only enhances the spatial resolution of HDX-MS but also opens up new avenues for its application.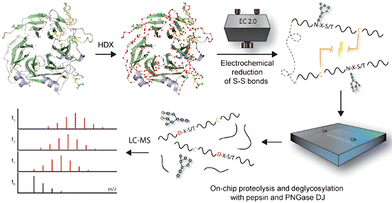 | ||
| Fig. 4 Microfluidic chip-based immobilization of deglycosylase and pepsin for HDX-MS glycoprotein analysis. Reprinted with permission from ref. 67. Copyright 2021 The Authors. | ||
In protein structural studies, precise control of enzymatic cleavage is essential for elucidating highly accessible and flexible regions of proteins. The key is how to minimize the amount of intact protein remaining in the sample while ensuring the generation of sufficient numbers of large peptides. To solve this problem, Dong's group has suggested an electrostatic anchoring technology. In this approach, trypsin is directionally immobilized at the surface of a nylon membrane, and the reaction time is regulated at the millisecond level through the micrometer level thickness of the membrane.71 The reaction proved 92.4% effective in cleaving Cyt C at the 79K site, which was close to the flexible region in the crystal structure. This subsecond time-resolved regulation of enzymatic cleavage therefore holds new promise in the study of dynamic protein conformations.
5.3 μ-IMERs for quantitative proteomics
Quantitative proteomics has limitations inherent in isotope labeling technology. Traditional 18O labeling requires 24 to 48 h to complete, and isotope back-exchange caused by free enzymes can influence quantitative accuracy.93,94 To address these issues, Lee et al. developed an integrated microfluidic platform.95 The platform is advantageous in using immobilized pancreatic enzymes to prevent interference by free enzymes and a multi-channel parallel structure to facilitate simultaneous enzymatic hydrolysis and labeling. This design reduces the whole processing time to just 30 min, which was 48 times quicker than that with conventional methods. The clinical usefulness of the technology was demonstrated by showing screening of lung cancer biomarkers. The platform enables screening of lung cancer biomarkers by merely analyzing serum samples directly. The 76 N-glycopeptides that are frequently encountered and 12 tumor metastasis-associated glycosylation modification sites were successfully identified with this approach. Similarly, Fan et al. developed a DNA tetrahedron-based μ-IMER with immobilized trypsin, achieving rapid digestion of BSA within 2 min, and for the first time, enabling fast and accurate quantification of a human growth hormone reference material after a 4 h immobilized enzyme digestion, demonstrating its promising utility in quantitative proteomics.51Based on the above discussion, the application of μ-IMERs in proteomics is summarized in Table 1.
| Reactor material | Immobilized enzyme | Immobilization strategy | Type of support | Application | Ref. |
|---|---|---|---|---|---|
| Polydimethylsiloxane | Trypsin | Physical adsorption | Open-tubular | Practical rapid enzyme digestion | 33 |
| Nylon | Trypsin and ChT | Electrostatic adsorption | Membrane | Glycoprotein analysis | 69 |
| Fused silica capillary | Trypsin | Electrostatic adsorption | Open-tubular | CE-MS for tear proteomics | 35 |
| Silica particles | Trypsin | Covalent binding | Particulate | Enrichment and enzymolysis of trace level proteins | 47 |
| Polydimethylsiloxane | Trypsin | Covalent binding | Monolithic | Pretreatment-digestion platform | 80 |
| Porous resin | NepII and pepsin | Covalent binding | Particulate | Structural proteomics | 92 |
| Magnetic nanoparticles | Trypsin | Affinity binding | Particulate | Bottom-up proteomic analysis | 51 |
| Molecular sieve | Trypsin and Glu-C | Affinity binding | Monolithic | Large-scale automated digestion | 76 |
| Alginate hydrogel | Trypsin and pepsin | Entrapment | Monolithic | Quantitative proteomics | 52 |
6. Challenges and prospects
Despite the remarkable potential of μ-IMERs in accelerating proteomic workflows, improving enzymatic efficiency, and enabling high-throughput automation, several technical constraints still hinder their broader adoption. These include issues related to system stability, reproducibility, and mechanistic uncertainties in enzyme–substrate interactions under immobilized conditions.This section outlines the primary technical challenges faced during long-term μ-IMER operation, including loss of catalytic activity, degradation of the carrier structure, and accumulation of substrate residues. While flushing protocols and performance monitoring are routinely applied to mitigate the latter two, enzymatic activity retention remains a key concern.
Substantial evidence has demonstrated that diverse μ-IMER systems can maintain acceptable enzymatic activity levels over continuous operation periods ranging from 7 days to 12 months.96–98 However, there are no systematic studies of enzyme molecule leakage. Since the concentration of the leaked enzyme is typically below the detection threshold of MS, the signal of intact enzyme molecules is easily suppressed by competitive ionization from peptides. Although μ-IMER does not require traditional freezing conditions (−20 °C), its storage conditions still have to be stringently controlled. It is generally recommended to store the system in Tris-HCl buffer (pH 8) at 4 °C, and 0.02% sodium azide is included to inhibit microbial growth.99
Notwithstanding the diversity in μ-IMER design, catalytic mechanisms of core enzyme components such as trypsin and pepsin share analogous features. A general evaluation scheme is therefore proposed, using benzoyl-L-arginine ethyl ester (BAEE) as a model substrate. The enzymatic hydrolysis product benzoyl-L-arginine (BA) should be monitored to detect the fluctuation of enzyme activity in various cycles of storage.
Although μ-IMERs show higher catalytic activity than free enzyme systems, the interaction mechanism between immobilized enzymes and substrates is not yet fully understood. Current prediction models of solution enzyme digestion reactions, such as DeepDigest, can reliably mimic enzyme cleavage sites from free enzyme kinetics.100 However, in the μ-IMER system, factors including enzyme molecule steric hindrance, microenvironment shift of the carrier surface, and limiting substrate mass transfer may alter enzyme cleavage specificity. These specificity changes are irrelevant in a single protein sample (e.g., standard proteins). However, in complex biological samples (e.g., mouse liver protein extract), μ-IMERs can produce specific sequence peptides, which do not occur in free enzyme systems, through alterations of the binding mode between the substrate and the enzyme. This influence means that immobilization can introduce heterogeneity to enzyme catalytic activity and hence influence the analysis of enzymatic hydrolysis spectra in complex samples.101
In the last few years, μ-IMERs have increasingly been used in proteomics studies, reducing sample digestion time to minutes from over 10 h. Automated sample pre-treatment platforms have also streamlined workflows by enabling effortless processing from cell preparation to mass spectrometry analysis. By automating, manual intervention is reduced, processing time is decreased, and high-throughput, real-time analysis is enabled. Despite these advancements, a number of constraints remain in developing completely automated systems for handling nanoliter-scale samples with the stability required for single-cell proteomics. The overcoming of these limitations will expand the potential of μ-IMERs as a valuable tool for high-throughput single-cell analysis.
Author contributions
ZJY: conceptualization, investigation, formal analysis, and writing – original draft. YLL: investigation, formal analysis, and writing – review & editing. WX: conceptualization, supervision, formal analysis, and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Data availability
Data availability is not applicable to this article as no new data were created or analyzed in this study.Acknowledgements
This work was supported by NNSFC (22374009).References
- R. Aebersold and M. Mann, Nature, 2003, 422, 198–207 CrossRef CAS PubMed.
- A. I. Nesvizhskii, O. Vitek and R. Aebersold, Nat. Methods, 2007, 4, 787–797 CrossRef CAS PubMed.
- M. Park, H. A. Shin, V. A. Duong, H. Lee and H. Lew, Cells, 2022, 11, 2–16 Search PubMed.
- J. Han, A. Agarwal, J. N. Young, S. Owji, Y. Luu, D. Poplausky and D. Yassky, et al. , Sci. Rep., 2022, 12, 22364–22370 CrossRef CAS PubMed.
- M.-S. Kim, S. M. Pinto, D. Getnet, R. S. Nirujogi, S. S. Manda, R. Chaerkady and A. K. Madugundu, et al. , Nature, 2014, 509, 575–581 CrossRef CAS PubMed.
- M. Wilhelm, J. Schlegl, H. Hahne, A. M. Gholami, M. Lieberenz, M. M. Savitski and E. Ziegler, et al. , Nature, 2014, 509, 582–587 CrossRef CAS PubMed.
- X. Han, A. Aslanian and J. R. Yates 3rd, Curr. Opin. Chem. Biol., 2008, 12, 483–490 CrossRef CAS PubMed.
- Y. Zhang, Q. Cai, Y. Luo, Y. Zhang and H. Li, J. Pharm. Anal., 2023, 13, 63–72 CrossRef PubMed.
- R. M. Searfoss, E. Zahn, Z. Lin and B. A. Garcia, J. Proteome Res., 2025, 24, 1230–1240 CrossRef CAS PubMed.
- Y. He, E. Shishkova, T. M. Peters-Clarke, D. R. Brademan, M. S. Westphall, D. Bergen and J. Huang, et al. , Anal. Chem., 2023, 95, 10655–10663 CrossRef CAS PubMed.
- L. Ramalhete, M. B. Vieira, R. Araujo, E. Vigia, I. Aires, A. Ferreira and C. R. C. Calado, Int. J. Mol. Sci., 2024, 25, 3844–3944 CrossRef PubMed.
- R. Craig and R. C. Beavis, Bioinformatics, 2004, 20, 1466–1467 CrossRef CAS PubMed.
- J. Yuan, R. Zhang, Z. Yang, J. Lee, Y. Liu, J. Tian and X. Qin, et al. , Eur. Urol., 2013, 63, 902–912 CrossRef CAS PubMed.
- A. T. Kong, F. V. Leprevost, D. M. Avtonomov, D. Mellacheruvu and A. I. Nesvizhskii, Nat. Methods, 2017, 14, 513–520 CrossRef CAS PubMed.
- D. López-Ferrer, B. Cañas, J. Vázquez, C. Lodeiro, R. Rial-Otero, I. Moura and J. L. Capelo, TrAC, Trends Anal. Chem., 2006, 25, 996–1005 CrossRef.
- G. Ghafourifar, A. Fleitz and K. C. Waldron, Electrophoresis, 2013, 34, 1804–1811 CrossRef CAS PubMed.
- A. K. Palm and M. V. Novotny, Rapid Commun. Mass Spectrom., 2004, 18, 1374–1382 CrossRef CAS PubMed.
- K. A. Cobb and M. Novotny, Anal. Chem., 1989, 61, 2226–2231 CrossRef CAS PubMed.
- B. Ziaie, A. Baldi, M. Lei, Y. Gu and R. A. Siegel, Adv. Drug Delivery Rev., 2004, 56, 145–172 CrossRef CAS PubMed.
- L. Szekely and A. Guttman, Electrophoresis, 2005, 26, 4590–4604 CrossRef CAS PubMed.
- X. Zhao and H. Bi, Food Chem., 2025, 465, 141868–141878 CrossRef CAS PubMed.
- B. Wang, L. Shangguan, S. Wang, L. Zhang, W. Zhang and F. Liu, J. Chromatogr. A, 2016, 1477, 22–29 CrossRef CAS PubMed.
- L. M. H. Reinders, M. D. Klassen, T. Teutenberg, M. Jaeger and T. C. Schmidt, Anal. Bioanal. Chem., 2021, 413, 7119–7128 CrossRef CAS PubMed.
- Y. Yin, Y. Xiao, G. Lin, Q. Xiao, Z. Lin and Z. Cai, J. Mater. Chem. B, 2015, 3, 2295–2300 RSC.
- K. Meller, M. Szumski and B. Buszewski, Sens. Actuators, B, 2017, 244, 84–106 CrossRef CAS.
- B. Jiang, K. Yang, Q. Zhao, Q. Wu, Z. Liang, L. Zhang and X. Peng, et al. , J. Chromatogr. A, 2012, 1254, 8–13 CrossRef CAS PubMed.
- J. Gan, A. R. Bagheri, N. Aramesh, I. Gul, M. Franco, Y. Q. Almulaiky and M. Bilal, Int. J. Biol. Macromol., 2021, 167, 502–515 CrossRef CAS PubMed.
- C. Garcia-Galan, Á. Berenguer-Murcia, R. Fernandez-Lafuente and R. C. Rodrigues, Adv. Synth. Catal., 2011, 353, 2885–2904 CrossRef CAS.
- Y. Asanomi, H. Yamaguchi, M. Miyazaki and H. Maeda, Molecules, 2011, 16, 6041–6059 CrossRef CAS PubMed.
- L. Niyaz Ahmed and P. T, Chem. Eng. J., 2024, 496, 153858–153873 CrossRef CAS.
- S. B. Sigurdardóttir, J. Lehmann, S. Ovtar, J. C. Grivel, M. D. Negra, A. Kaiser and M. Pinelo, Adv. Synth. Catal., 2018, 360, 2578–2607 CrossRef.
- J. M. Nelson and E. G. Griffin, J. Am. Chem. Soc., 2002, 38, 1109–1115 CrossRef.
- C. Nagy, A. Kecskemeti and A. Gaspar, Anal. Chim. Acta, 2020, 1108, 70–78 CrossRef CAS PubMed.
- M. Liu, Y. Hu, Y. Zhang and H. Lu, Talanta, 2013, 110, 101–107 CrossRef CAS PubMed.
- C. Nagy, R. Szabo and A. Gaspar, Molecules, 2021, 26, 5902–5915 CrossRef CAS PubMed.
- J. Yang, R. Ostafe and M. L. Bruening, Anal. Chem., 2024, 96, 6347–6355 CrossRef CAS PubMed.
- M. Yan, D. Wang, H. Liao, Y. Gong, B. Ji, Y. Liu and X. Tao, et al. , Anal. Chem., 2024, 96, 17300–17309 CrossRef CAS PubMed.
- J. Wan, L. Zhang, B. Yang, B. Jia, J. Yang and X. Su, Chem. Eng. J., 2022, 427, 131976–131983 CrossRef CAS.
- S. Zhang, Y. Gan, H. Wang, X. Qi, P. Su, J. Song and Y. Yang, Anal. Chem., 2024, 96, 9228–9235 CrossRef CAS PubMed.
- A. A. Homaei, R. Sariri, F. Vianello and R. Stevanato, J. Chem. Biol., 2013, 6, 185–205 CrossRef PubMed.
- J. M. Bolivar and F. López-Gallego, Curr. Opin. Green Sustainable Chem., 2020, 25, 100349–100357 CrossRef.
- M. Romero-Fernandez and F. Paradisi, Curr. Opin. Chem. Biol., 2020, 55, 1–8 CrossRef CAS PubMed.
- H. Jian, Y. Wang, Y. Bai, R. Li and R. Gao, Molecules, 2016, 21, 895–908 CrossRef PubMed.
- M. Nouaimi, K. Möschel and H. Bisswanger, Enzyme Microb. Technol., 2001, 29, 567–574 CrossRef CAS.
- M. G. Holyavka, S. S. Goncharova and V. G. Artyukhov, Int. J. Mol. Sci., 2025, 26, 547–570 CrossRef CAS PubMed.
- Y. Wang, X. Zhang, Z.-H. Wei, Y.-J. Jiao, D.-Y. An, Y.-P. Huang and Z.-S. Liu, et al. , J. Chromatogr. A, 2022, 1666, 462848–462858 CrossRef CAS PubMed.
- K. Duong, S. Maleknia, D. Clases, A. Minett, M. P. Padula, P. A. Doble and R. Gonzalez de Vega, Anal. Bioanal. Chem., 2022, 415, 4173–4184 CrossRef PubMed.
- S. Z. Tapdigov, Int. J. Biol. Macromol., 2021, 183, 1676–1696 CrossRef CAS PubMed.
- J. Lee, S. M. Kim, B. W. Jeon, H. W. Hwang, E. G. Poloniataki, J. Kang and S. Lee, et al. , Nat. Chem. Eng., 2024, 1, 354–364 CrossRef.
- E. Makrydaki, R. Donini, A. Krueger, K. Royle, I. Moya Ramirez, D. A. Kuntz and D. R. Rose, et al. , Nat. Chem. Biol., 2024, 20, 732–741 CrossRef CAS PubMed.
- X. Fan, Z. Chu, M. Zhu, Y. Song, Y. Zhao, B. Meng and X. Gong, et al. , Anal. Chem., 2023, 95, 15875–15883 CrossRef CAS PubMed.
- M. Arad, C. Frey, R. Balagtas, R. Hare, K. Ku, D. Jereb and Z. Nestman, et al. , Anal. Chem., 2024, 96, 18880–18889 CrossRef CAS PubMed.
- C. M. Janczak, I. A. C. Calderon, E. Noviana, P. Hadvani, J. R. Lee and C. A. Aspinwall, ACS Appl. Nano Mater., 2019, 2, 1259–1266 CrossRef CAS PubMed.
- K. Ku, C. Frey, M. Arad and G. Ghafourifar, Anal. Methods, 2022, 14, 4053–4063 RSC.
- Y. Cao, L. Wen, F. Svec, T. Tan and Y. Lv, Chem. Eng. J., 2016, 286, 272–281 CrossRef CAS.
- M. R. Robinson, L. A. Vasicek, C. Hoppmann, M. Li, G. Jokhadze and D. S. Spellman, Analyst, 2020, 145, 3148–3156 RSC.
- K. Hata, Y. Izumi, T. Hara, M. Matsumoto and T. Bamba, Anal. Chem., 2020, 92, 2997–3005 CrossRef CAS PubMed.
- H. Salim, R. Pero-Gascon, E. Gimenez and F. Benavente, Anal. Chem., 2022, 94, 6948–6956 CrossRef CAS PubMed.
- A. Kecskemeti and A. Gaspar, Talanta, 2017, 166, 275–283 CrossRef CAS PubMed.
- J. Bataille, A. Viodé, I. Pereiro, J. P. Lafleur, F. Varenne, S. Descroix and F. Becher, et al. , Analyst, 2018, 143, 1077–1086 RSC.
- D. Z. Dieters-Castator, P. Manzanillo, H. Y. Yang, R. V. Modak, M. J. Rardin and B. W. Gibson, J. Proteome Res., 2024, 23, 618–632 CrossRef CAS PubMed.
- Y. Mao, R. Fan, R. Li, X. Ye and U. Kulozik, Electrophoresis, 2021, 42, 2599–2614 CrossRef CAS PubMed.
- K. Yamamoto, K. Morikawa, H. Imanaka, K. Imamura and T. Kitamori, Analyst, 2020, 145, 5801–5807 RSC.
- X. Zhang, Z. Liao, X. Zhang, X. Ruan, H. Gong, X. Wang and W. Zheng, et al. , Sep. Purif. Technol., 2024, 347, 127501–127509 CrossRef CAS.
- Y.-J. Jiao, F.-F. Yuan, P.-R. Fan, Z.-H. Wei, Y.-P. Huang and Z.-S. Liu, Chem. Eng. J., 2021, 407, 127061–127069 CrossRef CAS.
- N. Lu, D. Sticker, A. Kretschmann, N. J. Petersen and J. P. Kutter, Anal. Bioanal. Chem., 2020, 412, 3559–3571 CrossRef CAS PubMed.
- G. Comamala, C. C. Krogh, V. S. Nielsen, J. P. Kutter, J. Voglmeir and K. D. Rand, Anal. Chem., 2021, 93, 16330–16340 CrossRef CAS PubMed.
- Y. Mao and U. Kulozik, Int. Dairy J., 2018, 85, 96–104 CrossRef CAS.
- W. Cao and M. L. Bruening, J. Am. Soc. Mass Spectrom., 2023, 34, 1086–1095 CrossRef CAS PubMed.
- M. Kjellander, E. Billinger, H. Ramachandraiah, M. Boman, S. Bergström Lind and G. Johansson, J. Proteomics, 2018, 172, 165–172 CrossRef CAS PubMed.
- J. Dong, W. Ning, W. Liu and M. L. Bruening, Analyst, 2017, 142, 2578–2586 RSC.
- M. M. Hoog Antink, T. Sewczyk, S. Kroll, P. Árki, S. Beutel, K. Rezwan and M. Maas, Biochem. Eng. J., 2019, 147, 89–99 CrossRef CAS.
- L. Messner, M. H. Antink, T. Guo, M. Maas and S. Beutel, Eng. Life Sci., 2021, 21, 527–538 CrossRef CAS PubMed.
- C. Seidl, A. F. L. Vilela, J. M. Lima, G. M. Leme and C. L. Cardoso, Anal. Chim. Acta, 2019, 1072, 81–86 CrossRef CAS PubMed.
- K. Meller, P. Pomastowski, D. Grzywiński, M. Szumski and B. Buszewski, J. Chromatogr. A, 2016, 1440, 45–54 CrossRef CAS PubMed.
- F.-F. Yuan, P. Wang, X.-J. Han, T.-T. Qin, X. Lu and H.-J. Bai, Sci. Rep., 2024, 14, 15667–15675 CrossRef CAS PubMed.
- O. K. Brandtzaeg, B. T. Røen, S. Enger, E. Lundanes and S. R. Wilson, Anal. Chem., 2017, 89, 8667–8673 CrossRef CAS PubMed.
- Z. Yang and L. Sun, Anal. Methods, 2021, 13, 1214–1225 RSC.
- V. A. Duong and H. Lee, Int. J. Mol. Sci., 2023, 24, 5350–5372 CrossRef CAS PubMed.
- Z.-H. Wei, X. Zhang, X. Zhao, Y.-J. Jiao, Y.-P. Huang and Z.-S. Liu, Talanta, 2021, 224, 121810–121819 CrossRef CAS PubMed.
- B. Wouters, S. A. Currivan, N. Abdulhussain, T. Hankemeier and P. J. Schoenmakers, TrAC, Trends Anal. Chem., 2021, 144, 116419–116427 CrossRef CAS.
- B. Gilquin, M. Cubizolles, R. Den Dulk, F. Revol-Cavalier, M. Alessio, C.-E. Goujon and C. Echampard, et al. , Anal. Chem., 2020, 93, 683–690 CrossRef PubMed.
- Y. Li, H. Yuan, Z. Dai, W. Zhang, X. Zhang, B. Zhao and Z. Liang, et al. , Anal. Chim. Acta, 2021, 1154, 338343–338351 CrossRef CAS PubMed.
- Y. Li, H. Yuan, Z. Dai, W. Zhang, X. Zhang, B. Zhao and Z. Liang, et al. , Anal. Chim. Acta, 2021, 1154, 338343 CrossRef CAS PubMed.
- H. Tang, H. Wang, D. Zhao, M. Cao, Y. Zhu and Y. Li, Anal. Chem., 2022, 94, 5715–5722 CrossRef CAS PubMed.
- H. Yin and J. Zhu, Mass Spectrom. Rev., 2023, 42, 887–917 CrossRef CAS PubMed.
- J. Camperi, S. Dahotre, D. Guillarme and C. Stella, Talanta, 2022, 246, 123519–123526 CrossRef CAS PubMed.
- F. Rinaldi, S. Tengattini, G. Brusotti, G. Tripodo, B. Peters, C. Temporini and G. Massolini, et al. , Front. Mol. Biosci., 2021, 8, 765683–765695 CrossRef CAS PubMed.
- W. Zhang, J. Hong, L. Yang, Z. Xu, Y. Xiang and W. Xu, Chin. Chem. Lett., 2024, 35, 108695–108699 CrossRef CAS.
- J. R. Engen, T. Botzanowski, D. Peterle, F. Georgescauld and T. E. Wales, Anal. Chem., 2020, 93, 567–582 CrossRef PubMed.
- E. I. James, T. A. Murphree, C. Vorauer, J. R. Engen and M. Guttman, Chem. Rev., 2022, 122, 7562–7623 CrossRef CAS PubMed.
- J. Zheng, T. S. Strutzenberg, A. Reich, V. Dharmarajan, B. D. Pascal, G. C. Crynen and S. J. Novick, et al. , Anal. Chem., 2020, 92, 11018–11028 CrossRef CAS PubMed.
- O. T. Schubert, H. L. Rost, B. C. Collins, G. Rosenberger and R. Aebersold, Nat. Protoc., 2017, 12, 1289–1294 CrossRef CAS PubMed.
- F. Calderon-Celis, J. R. Encinar and A. Sanz-Medel, Mass Spectrom. Rev., 2018, 37, 715–737 CrossRef CAS PubMed.
- S. Y. Lee, S. Lee, S. B. Park, K. Y. Kim, J. Hong and D. Kang, J. Chromatogr. B, 2018, 1100–1101, 58–64 CrossRef CAS PubMed.
- S. Tengattini, F. Rinaldi, L. Piubelli, T. Kupfer, B. Peters, T. Bavaro and E. Calleri, et al. , J. Pharm. Biomed. Anal., 2018, 157, 10–19 CrossRef CAS PubMed.
- H. Yuan, L. Zhang and Y. Zhang, J. Chromatogr. A, 2014, 1371, 48–57 CrossRef CAS PubMed.
- S. Moore, S. Hess and J. Jorgenson, J. Chromatogr. A, 2016, 1476, 1–8 CrossRef CAS PubMed.
- H. Lin, C. Zhang, Y. Lin, Y. Chang, J. Crommen, Q. Wang and Z. Jiang, et al. , J. Sep. Sci., 2019, 42, 1980–1989 CrossRef CAS PubMed.
- J. Yang, Z. Gao, X. Ren, J. Sheng, P. Xu, C. Chang and Y. Fu, Anal. Chem., 2021, 93, 6094–6103 CrossRef CAS PubMed.
- Z. Wei, P. Fan, Y. Jiao, Y. Wang, Y. Huang and Z. Liu, Anal. Chim. Acta, 2020, 1102, 1–10 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |



