Recent advances in sample preparation for analysis of small molecules with surface-assisted laser desorption/ionization time-of-flight mass spectrometry: enrichment methods and applications
Mingyuan
Liu
a,
Tianyu
Hua
a,
Yihan
Zhang
b,
Zifang
Peng
c,
Dan
Yin
a,
Wenfen
Zhang
 bc,
Yanhao
Zhang
bc,
Yanhao
Zhang
 a,
Congcong
Pei
*b and
Shusheng
Zhang
a,
Congcong
Pei
*b and
Shusheng
Zhang
 *bc
*bc
aSchool of Ecology and Environment, Zhengzhou University, Zhengzhou, 450001, PR China
bCollege of Chemistry, Zhengzhou University, Zhengzhou, 450001, PR China. E-mail: ccpei@zzu.edu.cn; zsszz@126.com
cCenter of Advanced Analysis and Gene Sequencing, Zhengzhou University, Zhengzhou, 450001, PR China
First published on 3rd June 2025
Abstract
The detection of small molecules, including metabolites, pollutants, and drugs, is significant for clinical diagnosis, environmental monitoring, and drug analysis. Surface-assisted laser desorption/ionization time-of-flight mass spectrometry (SALDI-TOF MS), which integrates sample preparation and detection in one step, has attracted considerable attention in the analysis of small molecules. In SALDI-TOF MS, sample preparation is a key step to ensure analytical selectivity and sensitivity; however only a few relevant reviews summarize sample preparation. This paper reviews the recent progress of research on targeted and non-targeted enrichment methods for small molecular analytes in complex samples based on the SALDI-TOF MS platform, and discusses their applications in pollutant analysis, in vitro diagnosis, and drug analysis. Finally, the review can potentially inspire researchers to design and select suitable sample preparation methods to solve the challenges of background interference in complex analytical samples, thereby facilitating the development of new platforms for high-throughput and high-sensitivity detection.
1. Introduction
The term ‘small molecules’ is typically employed to denote molecules with a molecular weight of less than 900 Daltons.1 This category encompasses a large number of crucial compounds in the current research area, such as metabolites,2–5 environmental pollutants,6 and pharmaceutical molecules.7 Metabolites are the end products in biological pathways and these bio-molecules have become significant for clinical diagnosis and monitoring.8 Small molecular pollutants, such as potassium perfluorooctanoic sulfonate, 1-hydroxy-pyrene, and p-benzenediamine quinones, are broadly distributed in the environment and threaten the safety of ecological system.9 In addition, small molecular drugs with their advantages of good oral absorption, high stability, and controllable costs have become indispensable therapeutic tools in the treatment of cancer, infectious diseases, and chronic diseases.10 Therefore, the detection of these small molecules is of significance for clinical diagnostics, environmental monitoring, and drug analysis.Among the many analytical techniques, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, with its simple sample preparation, high throughput, and high sensitivity, is widely used to detect small molecular analytes.2,11 However, the organic matrix can hinder the detection of small molecules in complex samples due to poor enrichment ability, non-uniformity of crystallization, and background interference in the low mass range.12,13 As pioneers, Tanaka et al. first proposed that inorganic materials could serve as effective matrices to assist laser desorption/ionization time-of-flight mass spectrometry in detecting macromolecules. Similarly, the inorganic materials could also be ideal matrices in the detection of small molecules due to their stable structure, high light absorption, high electrical conductivity, and high surface area to volume ratio.14,15 Currently, various inorganic materials, including noble metals, metal oxides, metal–organic frameworks, and carbon-based materials, have been successfully developed as effective matrices for detecting small molecules. The new technique has been called surface-assisted laser desorption/ionization time-of-flight mass spectrometry (SALDI-TOF MS).16,17
In the SALDI-TOF MS platform, inorganic materials that have both enrichment and matrix functions are key to the rapid and sensitive detection of small molecules.18 Inorganic matrices, through their surface functionalization modification and porous structure, can directly achieve co-crystallization with analytes and efficiently in situ enrich target analytes in complex samples.19,20 Compared with traditional protein precipitation, liquid–liquid extraction, and solid-phase extraction, sample preparation time for SALDI-TOF MS has been shortened to minutes, eliminating the need for complex operating procedures and equipment, thereby greatly simplifying the enrichment process.21,22 In real samples, the detection of small molecules presents challenges due to low concentrations and strong background interference. Therefore, sample preparation represents a critical step to ensure the sensitivity and accuracy of analysis.23,24 There have been many reviews on the discussion of nanomaterials as LDI-TOF MS matrices, but a systematic investigation of their ability to enrich small molecules in complex samples is still limited.
Herein, this article provides a timely review of the latest studies on the enrichment and detection of small molecules using targeted and untargeted enrichment methods based on SALDI-TOF MS, exploring their applications in pollutant analysis, in vitro diagnosis, and drug analysis. This review highlights the importance of enrichment methods for the detection of small molecules by SALDI-TOF MS, providing new avenues for the development of high-throughput detection methods.
2. Enrichment methods for small molecules in SALDI-TOF MS
Inorganic nanomaterials, as the enrichment carrier and matrix, are at the core of the detection of small molecular analytes in SALDI-TOF MS.25 Initially, small molecular analytes are efficiently enriched on the surface/pore structure of inorganic nanomaterials through physical adsorption,26 chemical bonding,27 and electrostatic interaction,18 significantly increasing the local concentration to overcome the sensitivity limitation for trace substances. After being subjected to laser irradiation, inorganic nanomaterials rapidly absorb more energy due to their excellent photo-responsivity, which helps the enriched analytes desorb and ionize from the surface of the inorganic nanomaterials.28 This synergistic enrichment-desorption/ionization mechanism not only dramatically improves the detection limit,26 but also enables SALDI-TOF MS to directly analyze small molecules with low abundance in complex samples (biological fluids and environmental samples).29 Therefore, the nanomaterials-based enrichment process can not only remove the trade-off bottleneck between interference suppression and sensitivity improvement in the SALDI-TOF MS analysis of small molecules, but also promote the development of mass spectrometry technology in the direction of high throughput, high sensitivity, and low sample consumption.The current enrichment methods can be divided into targeted and untargeted enrichment in order to satisfy a variety of analytical requirements and objectives.
2.1 Targeted enrichment for small molecules in SALDI-TOF MS
Targeted enrichment is an effective method for detecting specific small molecules through specific chemical functional groups,30 metal coordination,31 hydrophobic interaction,32 and electrostatic interaction33 (Table 1). Therefore, inorganic matrices can achieve highly specific trapping of target molecules and significantly improve the detection sensitivity of trace targets.| Enrichment method | Matrix | Small molecule | LOD | Application | Ref. |
|---|---|---|---|---|---|
| Chemical functional groups | 2D boron nanosheets | Glucose, lactose, mannose, and fructose | 1 nM | Detection of lactose in milk samples | 39 |
| GO-VPBA | Adenosine, guanosine, and galactose | Guanosine: 0.63 pmol mL−1 | Urine samples | 40 | |
| BCOFs | cis-Diol compounds | fg mL−1 | Detection of cis-diol compounds in human serum, milk, and Capsicum samples | 41 | |
| Fe3O4@PDA@B-UiO-66 | Glucose | 58.5 nM | Quantitative detection of glucose in complex samples | 13 | |
| UiO-66(NH2)-MUMIPs | Luteolin | 0.5 ng mL−1 | Detection of luteolin and its metabolites | 29 | |
| COFs film | PFOS | 0.5 ng mL−1 | Detection of PFOS in zebrafish, rat kidney and liver tissues | 42 | |
| BNQDs | Bisphenol A | 0.05 nM | Detection of BPA in environmental water | 30 | |
| Metal coordination | AuNPs/ZnO NRs | Glutathione | 150 amol | Detection of GSH in medicine and fruits | 31 |
| IBAs-AuNPs/COF | Insulin | 0.28 μg L−1 | Detection of insulin diabetes and healthy serum samples | 46 | |
| Hydrophobic interaction | 3D monolithic SiO2 | Desipramine and trimipramine | 10 μg mL−1 and 1 ng mL−1 | Detection of hydrophobic antidepressant drugs | 44 |
| Silane monolayer-modified porous silicon | LysoPC | 0.5 ng mL−1 | Detection of LysoPC in plasma | 45 | |
| Electrostatic adsorption | p-AAB/Mxene | PPDQs and DAIs | 10–70 ng mL−1, pg m−3 | Detection of pollutants in beverage and PM2.5 samples | 33 |
| MP-HOFs | Paraquat and chlormequat | 0.001 ng mL−1 and 0.05 ng mL−1 | Detection of pesticides in tap water, river water, and soil | 32 |
The versatility and tunability of chemical functional groups render them amenable to design for selective adsorption of target molecules.18 Chemical functional groups can be tuned to achieve specific recognition of certain small molecular compounds, leading to specific enrichment.34 Specific cis-diol compounds, including sugars, glycoproteins, and nucleosides, have been shown to possess significant physiological functions and pharmacological activities.35 These compounds have been shown to play crucial roles in various biological processes and clinical medicine.36,37 Research has demonstrated that boric acid can form stable cyclic esters with cis-diol groups through covalent interactions. Therefore, a variety of materials with boric acid have been developed for selective enrichment of cis-diol-containing compounds.38 Wang et al.39 prepared two-dimensional boron nanosheets (2DBs) that have a boric acid moiety. Based on the specific affinity between boric acid and cis-diols, 2DBs can be used as an adsorbent material and pristine matrix for the selective enrichment and direct detection of cis-diol compounds by SALDI-TOF MS. The results demonstrated that 2DBs exhibited excellent adsorption capabilities for cis-diol compounds, achieving a detection limit as low as 1 nM for glucose, lactose, mannose, and fructose. Zhang et al.40 prepared a graphene oxide (GO) material that had been functionalized with 4-vinylphenylboronic acid (GO-VPBA). This material was then used as a SALDI-TOF MS matrix for the selective enrichment of small diol-containing bio-molecules (adenosine, guanosine, and galactose). The results showed that the novel GO-VPBA can significantly improve the detection limits (e.g., guanosine: 0.63 pmol mL−1) by about 115-fold and 131-fold compared with conventional GO and 2,5-dihydroxybenzoic acid (DHB), respectively. Covalent organic frameworks (COFs) are a class of novel porous polymers. Their superior enrichment properties are attributed to their larger specific surface area, enhanced stability, and π–π stacking interactions. Hu et al.41 synthesized boronic acid-functionalized COFs (BCOFs). The COFs and boric acid moiety have been proved to enhance the selectivity for capture of cis-diol compounds (Fig. 1a). The enrichment ability of the BCOF matrix was achieved through SALDI-TOF MS. Riboflavin and pyrocatechol were selected as model samples of the cis-diol-containing compound. 4-Nitrophenol was selected as a non-specific compound. The high sensitivity to target analytes in complex samples with a molar ratio of target analyte to non-specific compound of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 was also established. In addition, metal–organic frameworks (MOFs) are porous crystalline materials via coordination bonds. Boric-acid-modified multifunctional Zr-based MOFs (denoted as Fe3O4@PDA@B-UiO-66) were prepared as the adsorbent and matrix for enrichment and analysis of glucose.13 The large specific surface area, abundant boric-acid-active sites, porous structure, and superior UV absorption properties make Fe3O4@PDA@B-UiO-66 an ideal enrichment matrix. The limit of detection (LOD) of glucose on the Fe3O4@PDA@B-UiO-66-based LDI MS platform was 58.5 nM.
100 was also established. In addition, metal–organic frameworks (MOFs) are porous crystalline materials via coordination bonds. Boric-acid-modified multifunctional Zr-based MOFs (denoted as Fe3O4@PDA@B-UiO-66) were prepared as the adsorbent and matrix for enrichment and analysis of glucose.13 The large specific surface area, abundant boric-acid-active sites, porous structure, and superior UV absorption properties make Fe3O4@PDA@B-UiO-66 an ideal enrichment matrix. The limit of detection (LOD) of glucose on the Fe3O4@PDA@B-UiO-66-based LDI MS platform was 58.5 nM.
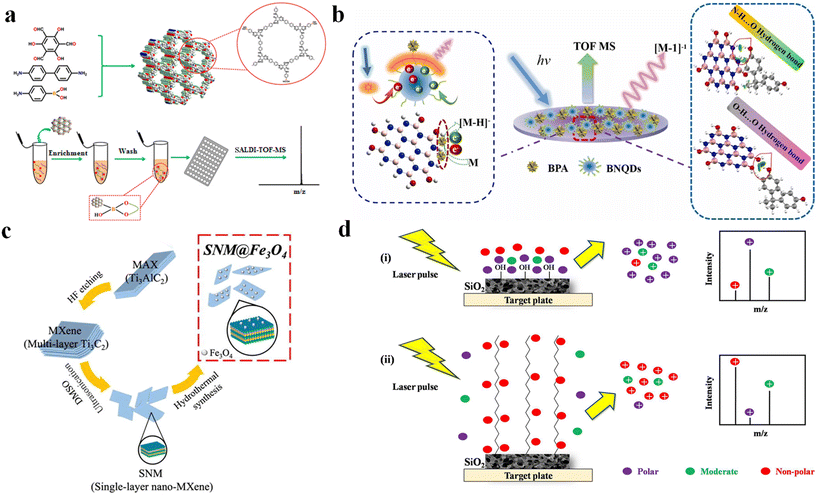 | ||
| Fig. 1 Targeted enrichment of small molecules in SALDI-TOF MS. (a) Synthesis of B-COFs and enrichment of cis-diol compounds from ref. 41; reprinted with permission from American Chemical Society. Copyright © 2019, American Chemical Society. (b) Enrichment of BPA compounds by BNQDs from ref. 30; reprinted with permission from Elsevier. Copyright © 2023 Elsevier. (c) Enrichment and detection of PPD by SNM@Fe3O4 from ref. 43; reprinted with permission from Elsevier. Copyright © 2022 Elsevier. (d) Enrichment of small molecules of different polarities by silica-based monoliths from ref. 44; reprinted with permission from Elsevier. Copyright © 2019 Elsevier. | ||
To effectively improve selectivity, Lu et al.29 combined MOFs with magnetic molecularly imprinted polymers (MIPs) for the selective enrichment and detection of luteolin. The MIPs can achieve efficient and selective enrichment of targeted bio-molecules via a “lock and key” mechanism. The composite of MOFs and MIPs (MUMIPS) was synthesized using luteolin as the template molecule. In the absence of the templated molecule, the composite of the non-molecularly imprinted polymer with M-UiO-66 (NH2) (MUNIPs) was prepared as the control matrix. In the presence of structural analogues, the adsorption capacity of MUMIPS for luteolin (luteolin (Q = 53.8 mg g−1) > myricetin (Q = 25.4 mg g−1)) has a clear advantage over that of MUNIPs. The MUMIPS-SALDI-TOF MS exhibited high selectivity and sensitivity for simultaneously enriching and detecting luteolin.
Due to its moderate bond energy and reversibility, hydrogen bonding exhibits advantages in efficient and selective enrichment. Zhao et al.30 (Fig. 1b) developed a boron nitride quantum dots (BNQDs)-assisted LDI MS platform. The surface of BNQDs is abundant in amino and hydroxyl groups, which selectively enriches bisphenol A (BPA) molecules through hydrogen bonding interactions (e.g., O–H⋯O and N–H⋯O). The BNQDs, as an inorganic matrix, can directly facilitate the quantitative detection of BPA and significantly reduce the background interference in the low-mass region. To overcome the limitations of traditional matrix deposition methods in SALDI-TOF MS imaging, Luo et al.42 developed a novel film prepared from COFs by imprinting the sieved powder of COFs onto the surface of double-sided adhesive tape. The new film comprising COFs demonstrated outstanding reproducibility, with a relative standard deviation (RSD) of 8.37%. In addition, the hydrogen bond interaction between the –OH of COFs and the fluorine of potassium perfluorooctanoic sulfonate (PFOS) contributed to the enrichment of PFOS. This excellent enrichment performance of COFs demonstrated their promising potential for application in PFOS detection at low concentration levels in zebrafish, rat kidney, and liver tissues. Lai et al.14 rapidly prepared magnetic graphene oxide (MGO) probes by combining graphene oxide (GO) with Fe3+ under microwave heating. The MGO can specifically capture aflatoxin B1 (6 min) using π–π interactions. By combining magnetic separation with LDI MS, aflatoxin B1 was rapidly purified and detected (∼10 min). The LOD of MGO-assisted LDI MS was as low as 1 nM, which is a significant improvement in detection efficiency.
Furthermore, physical interactions between the matrix and the target analyte, including electrostatic adsorption, and hydrophilic/hydrophobic and magnetic interactions have also been established. Zhang et al.33 developed p-aminoazobenzene (p-AAB)-modified MXene composites (p-AAB/MXene) to efficiently enrich emerging organic pollutants (p-benzenediamine quinones and diamide insecticides) at low concentrations in the environment via electrostatic adsorption and π–π stacking. SALDI-TOF MS was used to directly detect p-benzenediamine quinones and diamide insecticides in real beverage and PM2.5 samples with LOD of 10–70 ng mL−1. Similarly, Peng et al.43 developed a novel magnetic MXene monolayer nanomaterial (SNM@Fe3O4). The SNM@Fe3O4 was used as a matrix for rapidly detecting four p-phenylenediamine (PPD) antioxidants in SALDI-TOF MS (Fig. 1c). The efficient enrichment of trace targets in water was achieved by hydrogen bonding and van der Waals force interactions between SNM@Fe3O4 and PPD antioxidants. Lv et al.32 developed microporous hydrogen-bonded organic frameworks (MP-HOFs) for the simultaneous enrichment and detection of paraquat (PQ) and chlormequat chloride (CQ) using the SALDI-TOF MS platform. The MP-HOFs, with negatively charged surfaces, were able to specifically capture positively charged PQ and CQ through electrostatic interactions and π–π stacking. After enrichment, the LOD values (0.01 and 0.05 ng mL−1) were 3 and 2 orders of magnitude lower than the LOD values for unenriched PQ and CQ analytes, respectively. Amin et al.44 developed a 3D monolithic SiO2 matrix to achieve selective enrichment of small molecules by modulating its surface hydrophilicity (Fig. 1d). Their results showed that hydrophilic SiO2 has stronger adsorption for short-chain fatty acids (C14), while hydrophobic C18-SiO2 has stronger adsorption for long-chain fatty acids (C20). This observation verified the selectivity via hydrophilicity and hydrophobicity effects based on the length of the fatty acid chains. Similarly, Lavigne et al.45 investigated the effect of silane monolayer-modified porous silicon on the enrichment and detection of lysophosphatidylcholine (LysoPC) in SALDI-TOF MS. The porous silicon surface was modified with short-chain (CH3 short) and long-chain (CH3 long) silane molecules. The selective enrichment of small-molecule LysoPC was achieved by modulating the pore structure (10 nm pore size, 1 μm depth) of porous silicon. The short CH3 monolayer promoted the enrichment of LysoPC inside the pore, which significantly increased the sensitivity of LysoPC detection (LOD = 0.5 ng mL−1).
The metal–ligand interaction has been demonstrated to significantly enhance the enrichment ability and detection sensitivity through specific binding. Dou et al.31 developed a SALDI-TOF MS chip based on a composite of gold nanoparticles (AuNPs) and zinc oxide nanorods (ZnO NRs) for the selective enrichment and detection of glutathione (GSH). The thiol group modified on the surface of AuNPs can selectively capture GSH through Au–S bonds. Furthermore, the increased specific surface area and the vertical alignment of ZnO NRs provided a platform for the uniform loading of AuNPs, thereby enhancing the efficiency for GSH enrichment. Ge et al.46 developed composite aptamer–gold nanoparticle-doped COFs (IBAs-AuNPs/COFs) for the selective enrichment and highly sensitive detection of human insulin. Thiolated insulin aptamers (IBAs) were immobilized on the surface of the AuNPs via Au–S bonds to achieve specific enrichment of insulin. Furthermore, COFs with a large specific surface area (1365 m2 g−1) and microporous structure (2.0 nm) can assist in the enrichment. The sensitivity of the insulin signal remained stable (RSD < 7%) even in the presence of a 50-fold concentration of interferon.
Transition metal dichalcogenides are suitable materials for the sensitive detection of therapeutic drugs using LDI MS. Joh et al.47 enriched dozens of antiepileptic drugs (AEDs) in patient serum using six transition metal dichalcogenides as matrices for LDI-MS. These matrices included molybdenum disulfide, molybdenum diselenide, molybdenum ditelluride, tungsten disulfide, tungsten diselenide, and tungsten ditelluride. These matrices significantly improved the desorption and ionization efficiency of the drugs through enhanced coulombic interactions, with signal intensities increased 10–100-fold compared with conventional organic substrates. The relatively low surface electron density of tellurite-containing transition metal disulfides induces strong coulombic interactions, thereby enhancing the efficiency of laser desorption and ionization. Therefore, the molybdenum ditelluride and tungsten ditelluride matrices are suitable for the sensitive quantification of therapeutic drugs. Earlier, Joh et al.48 developed tungsten disulfide (WS2) nanosheet-based LDI-MS for the quantitative analysis of the immunosuppressive drugs cyclosporine A and tacrolimus in the blood of organ transplant recipients. This substrate avoided low-mass interfering peaks, thereby improving the sensitivity of SALDI-TOF MS by 2–20 times.
2.2 Untargeted enrichment of small molecules in SALDI-TOF MS
Another type of enrichment strategy for SALDI-TOF MS is untargeted enrichment. Untargeted enrichment methods facilitate comprehensive analysis and non-specific adsorption of unknown small molecules in complex samples through the physical adsorption ability of matrix material surfaces (e.g., porous structures and nanoscale crack structures). This enrichment method is mainly used in the field of omics studies (Table 2).| Enrichment method | Matrix | Small molecule | LOD | Application | Ref. |
|---|---|---|---|---|---|
| Porous structures | Mesoporous PdPtAu | Metabolic profiles in human blood | — | Early diagnosis of GC | 52 |
| Au/HPZOMs | Metabolic profiles in human serum | 8.6 pmol | Early diagnosis and surveillance of postmenopausal osteoporosis | 54 | |
| Co3O4/C | Serum metabolic fingerprints | 10–50 μmol L−1 | Detection of SLE activity for pregnant women | 55 | |
| hf-COFs | Serum metabolic fingerprints | — | Early diagnosis of colorectal cancer and GC | 57 | |
| COF-S@AuNPs | Metabolic patterns in serum | pmol level | Triclosan exposure | 51 | |
| Nanoscale rough surface | AuNP arrays | Glucose in cerebrospinal fluid | From 0.2 mM to 1.9 mM | Differentiating early-stage lung cancer patients | 70 |
| Vo-Co3O4 | Serum metabolic fingerprints | 1 × 10−6 M | Screening and prognosis of depression | 18 | |
| Ferric particles | Metabolic patterns in biofluids | — | Disease diagnosis | 59–66 | |
| MO/CMO | Serum metabolic fingerprints | 0.9 μmol L−1 | Early diagnosis of ovarian tumor | 67 | |
| GCM | Serum lipid fingerprints | FA 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ng mL−1; CE 18 10 ng mL−1; CE 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 μg mL−1; TG 51 1 μg mL−1; TG 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 ng mL−1 100 ng mL−1 |
Liver cancer diagnosis | 69 | |
| ZMF flowers | Serum metabolic fingerprints | — | Cardiovascular disease diagnosis | 68 |
The significance of porous structures in sample pretreatment stems from the effect of their unique microscopic morphology.49 These porous materials with pore diameters ranging from 2 to 50 nm offer abundant active sites for capturing target substances in complex samples through highly ordered pore networks and large specific surface areas.50 The pore size of the porous material affects adsorption of molecules of different sizes. Smaller pore sizes are suitable for small molecular analytes while effectively eliminating large molecular interferents through the pore size sieving effect.51 Su et al.52 first designed mesoporous trimetallic alloys and elucidated the relationship between their porous structure and the untargeted metabolic extraction from plasma in SALDI-TOF MS. A nanogap of ≈2 nm was confirmed as the optimized pore size for trapping small molecules and shielding macromolecules. In addition, suitable pore structures can act as hot spots with enhanced electromagnetic fields to desorb and ionize trapped metabolites (Fig. 2a). Their group also designed porous PtAu alloys using a soft-template method.53 The porous structure featuring pores of ∼7 nm can help capture small metabolites. The porous PtAu alloy-assisted LDI MS platform can be used to identify metabolic phenotypes and distinguish diseases in a large cohort of real samples in 30 minutes. Microspheres of AuNP-anchored hierarchically porous ZrO2 (Au/HPZOMs), with pore sizes of 1.6 nm and 2.9 nm, were synthesized by calcination of the hierarchically porous UiO-66-NH2(Zr).54 The porous structures can increase the specific surface area of the matrix and provide additional adsorption sites for analytes. Therefore, the Au/HPZOMs can improve the enrichment effect, charge transfer capability, and enhance desorption/ionization for untargeted metabolites of serum during SALDI-TOF MS analysis. Specifically, the LOD values for arginine, mannitol, and phenylalanine were 8.6, 8.2, and 9.1 pmol, respectively. Wang et al.55 also prepared a hollow Co3O4/C matrix with a pore size of 1–3 nm for the selective adsorption of small molecules in the serum sample of systemic Lupus erythematosus. The enrichment process and detection platform were cost-effective (€0.04 per g) and demonstrated high-throughput (15 min/384 samples). Similarly, porous Co3O4/CuO nanocages derived from MOFs were synthesized.56 The Co3O4/CuO material exhibits a wide distribution of mesopores ranging in size from 5 to 30 nm, which enables the LDI MS platform to quickly adsorb small molecules in 0.03 μL samples and detect them with high sensitivity. Compared with solid materials, porous organic framework materials are widely used in sample pretreatment due to their larger specific surface area and porosity. Yang et al.57 established a hollow flower-shaped COF-assisted LDI MS platform (hf-COFs-LDI-MS) (Fig. 2b). The hf-COFs have a large specific surface area (1439 m2 g−1) and a suitable pore size (3.2 nm), which are beneficial for loading extensive mixtures of small molecular analytes and excluding macromolecular proteins and peptides during the LDI-MS process. 473 high-quality serum metabolic patterns were extracted without any tedious sample pretreatment. The composite material of sulfur-functionalized COF@Au nanoparticles (COF-S@AuNPs), which exhibited an average pore size of around 2.92 nm, were utilized to extract metabolic profiles from mouse serum.51 The LOD values of the COF-S@AuNPs-based LDI MS platform were as low as the ∼pmol level, highlighting the high sensitivity of porous COF-S@AuNPs for small-molecule analysis. Besides the porous structure, the nanogap (3 nm) of the porous ZIF-8 can synergistically afford the size exclusion for large proteins (Fig. 2c). The core–satellite FeOOH@ZIF-8 was designed for fast, sensitive, and selective SALDI-TOF MS analysis of metabolites in serum (circa 1 μL) without any pretreatment.58
 | ||
| Fig. 2 Untargeted enrichment of small molecules in SALDI-TOF MS. (a) Experimental workflow for the extraction of plasma metabolic fingerprints by PdPtAu-assisted LDI MS from ref. 52; reprinted with permission from John Wiley and Sons. Copyright © 2021 Wiley. (b) Establishment of hf-COFs-assisted LDI-MS for high-throughput serum metabolic patterns extraction from ref. 57; reprinted with permission from American Chemical Society. Copyright © 2024, American Chemical Society. (c) Enrichment and LDI MS analysis of metabolites in serum by FeOOH@ZIF-8 from ref. 58; reprinted with permission from John Wiley and Sons. Copyright © 2020 Wiley. (d) Enrichment and detection of small molecule metabolites by MO/CMO from ref. 67; reprinted with permission from John Wiley and Sons. Copyright © 2023 Wiley. | ||
Untargeted enrichment methods rely on the non-specific adsorption of analytes onto the surface of the matrix upon incubation with a real sample. The nanoscale crack structure and nanoscale surface roughness can enrich untargeted metabolites within the crevices, facilitating the in situ separation of proteins in bio-samples to achieve selective SALDI-TOF MS processes for metabolomics. Qian's group synthesized ferric particles with rough nanoscale surfaces to construct ferric particle-enhanced LDI MS (FPELDI MS). The FPELDI MS platform has high throughput, minimal sample consumption (only 0.1–1.5 μL), high speed (∼20 s per sample), and high reproducibility (coefficient of variation <2%) toward extracting metabolic fingerprints from serum,59,60 urine,61 plasma,62 vitreous liquid,63 dried serum spots,18 single-cell samples,64 cerebrospinal fluid,65 and follicular fluid.66 Their innovative work provided the pathway for the subsequent design of a rough nanoscale matrix to support the LDI MS platform for the rapid enrichment and detection of small molecular analytes. Pei et al.67 synthesized an Mn2O3/(Co,Mn)(Co,Mn)2O4 (MO/CMO) composite matrix with rough surfaces using a nanoscale combustion and contraction process (Fig. 2d). Even in the presence of highly concentrated salt (0.5 M KCl or 0.5 M NaCl) and protein (5.00 mg mL−1 bovine serum albumin (BSA)), the signal of the metabolite can be successfully detected with the MO/CMO. In order to further validate the size-selective trapping in MO/CMO, histamine (metabolites; MW < 900 Da) and bovine serum albumin (macromolecules; MW > 900 Da) were selected as representative analytes, and these were then mixed with nanoparticles to form nanoparticle–analyte hybrids. Through calculating the ratio of the signal intensity of carbon on the nanoparticles to that of the background, the elemental mapping analysis of the nanoparticle–analyte hybrids showed a significantly higher molecular-selective trapping rate for histamine than for bovine serum albumin, indicating the ability to enrich untargeted metabolites in the crevices of rough nanoscale surfaces. Vacancy-engineered cobalt oxide,18 with its rough nanoscale surface, was selected as the adsorption agent and matrix and facilitated direct and robust recording of plasma metabolic fingerprints using the SALDI-TOF MS platform. Li et al.68 used a simple method to synthesize a 3D flower-shaped spatially functional nanocage matrix (ZMF flowers) with multiple interspersed lamellae. The 3D flower structure features high-density binding sites, enhancing the adsorption of small molecular analytes and facilitating energy concentration in SALDI-TOF MS, thereby contributing to efficient desorption/ionization of analytes. In complex samples, the ZMF flowers exhibited superior performance. A graphitized carbon matrix (GCM) with rough surface was designed for serum lipidomic profiling in SALDI-TOF MS.69 High-throughput lipid fingerprints of serum could be directly acquired by GCM-assisted LDI MS without extraction or purification. This work has pushed lipidomics in the direction of simplified experimental procedures with reduced errors.
3. Applications
SALDI-TOF MS utilizes inorganic nanomaterials as matrices and integrates sample preparation and analyte detection into one step, significantly improving analytical efficiency and opening a wide range of applications in the analysis of metabolites, pollutants, and drugs.3.1 Metabolic analysis
SALDI-TOF MS is a widely utilized technique in the analysis of metabolites. The abnormal fluctuation of metabolite levels in biofluids may be a sign of disease occurrence and development. Wang et al.70 developed a SALDI-TOF MS analytical platform based on self-assembled gold nanoparticle (AuNP) arrays to enrich metabolites in biofluids (Fig. 3a) successfully. The analytical platform quantified the dynamic changes of glucose levels (from 0.2 mM to 1.9 mM) in cerebrospinal fluid during treatment, thus enabling effective differentiation between patients with brain infections and healthy controls, permitting rapid evaluation of the clinical treatment response. The measurement time is dramatically reduced to 5 minutes, demonstrating excellent speed and sensitivity for practical clinical application.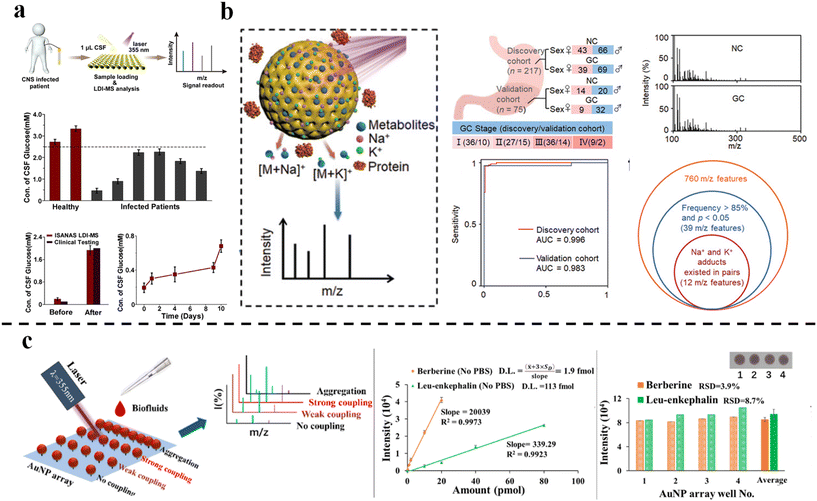 | ||
| Fig. 3 (a) Interfacial self-assembled AuNPs array-assisted LDI-MS analysis of glucose concentrations in cerebrospinal fluid from ref. 70; reprinted with permission from American Chemical Society. Copyright © 2021, American Chemical Society. (b) Extraction of plasma metabolic fingerprints for early diagnosis of GC from ref. 52; reprinted with permission from John Wiley and Sons. Copyright © 2021 Wiley. (c) Sensitive detection and analytical performance of metabolites from biofluids on AuNP arrays from ref. 72; reprinted with permission from American Chemical Society. Copyright © 2022, American Chemical Society. | ||
In metabolomics, untargeted enrichment methods are more widely used to obtain comprehensive data on metabolites. Global analysis of all metabolites in real samples is frequently necessary for clinical diagnosis, identifying potential biomarkers, and studying metabolic pathways. Jiang et al.71 developed a porous silica-assisted LDI MS (PSALDI-MS) technique for enriching metabolites in serum. The average pore size of porous silica particles was 9.86 nm allowing their use to extract metabolic fingerprints from serum. The PSALDI-MS platform demonstrated advantages of high throughput (5 min/96 samples), high sensitivity (LOD = 1 pmol), and reproducibility (with a coefficient of variation (CV) of less than 15%) in the detection of untargeted metabolic fingerprints (UMFs). A cohort, including healthy controls (HC), individuals with chronic hepatitis B (CHB), patients with liver cirrhosis (LC), and patients with hepatocellular carcinoma (HCC), was recruited and 1433 clinical serum samples were provided. Machine learning model trained on the 1433 UMFs classified the samples with accuracy values of 91.2% for HC, 71.4% for CHB, 70.0% for LC, and 95.3% for HCC. Su et al.52 reported an LDI MS technique based on mesoporous PdPtAu alloys, which enriched small-molecule metabolites from blood through the porous structure of 2 nm. Furthermore, the PdPtAu alloys exhibited excellent desorption and ionization abilities for adsorbed metabolites, owing to their preferred charge transfer and enhanced photothermal conversion. The technique was applied to the diagnosis of early gastric cancer (GC), and the sensitivity and specificity of early GC detection reached 92.0% with an area under the curve (AUC) of 0.942, values that were significantly better than those of traditional CEA protein markers (AUC = 0.545) (Fig. 3b). The technique provided a rapid and accurate new method for the early detection of GC. Similarly, Yang et al.57 developed an LDI MS technique based on hollow flower-like covalent organic frameworks (hf-COFs), which achieved the enrichment of metabolites in serum through a mesoporous structure with a pore diameter of 3.2 nm and a large specific surface area of 1439 m2 g−1. Ultimately, they screened 8 specific metabolic markers in 473 clinical samples, establishing an early diagnostic model for colorectal cancer (CRC) and GC, and achieved early differential diagnosis of CRC and GC (AUC = 0.966). Their work provided a new strategy for non-invasive, high-throughput, highly sensitive and highly specific clinical screening for cancer, wherein the excellent enrichment properties of the porous structure enabled efficient enrichment of untargeted metabolic fingerprints in body fluids. Currently, porous PtAu alloys, Au/HPZOMs, hollow Co3O4/C, Co3O4/CuO hollow polyhedral nanocages, and FeOOH@ZIF-8 are designed as the enrichment matrices for SALDI-TOF MS to rapidly extract the UMFs from serum samples, and realize the diagnosis of myocardial infarction, postmenopausal osteoporosis, systemic Lupus erythematosus, epidural-related maternal fever, and gynecological cancers, respectively.
In addition, the surface structures of nanomaterials also enable efficient enrichment of metabolites and are widely used in disease diagnosis and metabolomics analysis. Without complicated sample pretreatment, the FPELDI MS platforms constructed by Qian's group successfully realized the rapid and accurate diagnosis of prostate cancer,60 moyamoya disease,59 and diabetic retinopathy.63 Similarly, the UMFs of epithelial ovarian tumors were obtained using rougher matrix surfaces in MO/CMO-assisted LDI-MS, where high reproducibility and direct detection were achieved without pretreatment. Moreover, machine learning based on UMFs can be used to distinguish malignant ovarian tumors from benign controls with an AUC of 0.987. 7 metabolites related to ovarian tumor progression being screened from UMFs as potential biomarkers. Based on the use of matrices with rougher surfaces and their excellent desorption/ionization capability in untargeted metabolite enrichment, amino-modified polystyrene@Fe3O4 magnetic beads, vacancy-engineered cobalt oxide, and 3D flower-shaped Zr-MOF/Au have been prepared as LDI MS matrices. These matrices were used to extract the UMFs from the biofluids, enabling diagnosis hepatocellular carcinoma, depression, and cardiovascular disease, respectively. The diagnostic accuracy and specificity using UMFs are superior to those of traditional biomarkers.
Although SALDI-TOF MS has the advantage of rapidly enriching metabolites to obtain the UMFs of body fluids, the reproducibility of the matrix is the biggest challenge. Su et al.72 developed a SALDI-TOF MS substrate based on gold nanoparticle (AuNP) arrays placed on a coverslip (Fig. 3c). The arrays were constructed by controlling the coulombic repulsion of positively charged AuNPs. The interparticle arrangement of AuNPs is in the form of covalent bonds with a distance of 16 nm. Compared with self-assembled arrays, the covalently bonded AuNP arrays exhibited superior detection repeatability (RSD < 12%) and were reusable in 10 experiments without significant signal degradation (<15%). In in vitro diagnosis, the AuNP arrays successfully enriched 7 small-molecule metabolites from serum, including dopamine and isopalmitic acid. Combined with machine learning models, the platform achieved perfect classification results, with an AUC of 1.00 in the test cohort, accurately discriminating early-stage lung cancer patients from healthy populations.
3.2 Pollutant analysis
In recent years, SALDI-TOF MS has been extensively used in environmental monitoring, with the aim of detecting various pollutants in the environment, including p-phenylenediamine-quinones (PPDQs), polycyclic aromatic hydrocarbons (PAHs), and pentachlorophenol. Most of these are present in complex samples at low concentrations. Zhang et al.33 developed a new matrix based on p-AAB/MXene to efficiently enrich and detect PPDQs and diamide insecticides (DAIs) in the environment (Fig. 4a). The p-AAB/MXene was successfully applied to the detection of PPDQs and DAIs in beverage and PM2.5 samples with a sensitivity of ng mL−1 (LOD: 10–70 ng mL−1) in beverages and pg m−3 in PM2.5 samples, significantly simplifying the complex pretreatment steps and improving sensitivity. The magnetic single-layer nano-MXene43 (SNM@Fe3O4) was used as the matrix of SALDI-TOF MS to enrich and detect PPD antioxidants in an aqueous environment. The large surface area and the abundant active sites (–O, –F, and –OH) of the SNM@Fe3O4 surface were beneficial for the adsorption of PPD antioxidants. In actual water samples from the north and south of China, N,N′-diphenyl-p-phenylenediamine, N-phenyl-N′-cyclohexyl-p-phenylenediamine, 6PPD, and N,N′-di-2-naphthyl-p-phenylenediamine (DNPD) were successfully detected, with DNPD being the most prevalent (concentration values up to the pg m−3 level). Verification results from LC-MS/MS are highly consistent. However, compared with the LC-MS/MS technique, the SNM@Fe3O4-based LDI MS platform featured results having a cleaner background and obtained at a faster speed, eliminating the time-consuming sample pretreatment process (enrichment occurs in only 4 min). Similarly, Zhang and coworkers73 also developed a 3D-printed target plate (3D-MTP) based on boron, nitrogen, and sulfur-doped MXene quantum dots (BNS-MQDs). The BNS-MQDs-based 3D-MTP can be used both as the substrate for SALDI-TOF MS and also to enrich PPD antioxidants, PPDQs, and PAHs in the environment. It has demonstrated high sensitivity (with a LOD as low as 0.93 ng mL−1), strong anti-interference capability, and high throughput (analyzing a single sample in 30 seconds and 48 samples in 10 minutes) during environmental monitoring. In addition, the three-dimensional spatial distribution of DNPD in zebrafish was visualized by SALDI-TOF MS imaging for the first time, providing an efficient and cost-effective technological platform for environmental toxicology research. | ||
| Fig. 4 (a) Enrichment and detection of pollutants by the matrices of p-AAB and p-AAB/MXene from ref. 33; reprinted with permission from Elsevier. Copyright © 2025 Elsevier. (b) Synthesis of porous TiC ceramic powders for the detection of four environmental pollutants from ref. 75; reprinted with permission from Elsevier. Copyright © 2018 Elsevier. (c) Designed Pt/NiFe-LDH matrix for quantification and identification of polyphenols from ref. 81; reprinted with permission from Elsevier. Copyright © 2024 Elsevier. | ||
Gao et al.74 developed an efficient SALDI-TOF MS method based on 2D TiO2 nanosheets for enriching and detecting toxic small molecules in the environment. The method dramatically simplifies the traditional pretreatment steps. The LOD of malachite green is 10 pg mL−1, and its sensitivity far exceeds the international standard of 2 ng mL−1. It has also been successfully applied to the detection of small molecules in biological fluids and industrial wastewater. Moriwaki et al.75 developed a SALDI-TOF MS platform based on porous titanium carbide (TiC) ceramic powders for adsorption and detection of small molecular contaminants in aqueous environments (Fig. 4b), including pentachlorophenol, PFOS, 2,4,5-T, imidacloprid, sodium dodecyl sulfate-d25, methyl orange, methyl red, and triclosan, which were adsorbed and detected in the negative ion mode of TiC-based LDI-MS. Acetamiprid, benzyldimethyldodecylammonium chloride, methyl red, symetryn, N,N-dimethyl dodecylamine-N-oxide, pyrene, and benzo(a)pyrene were detected in the positive ion mode. The LOD of PFOS by TiC-based LDI-MS was 2 nM, almost on a par with that of solid-phase extraction followed by LC/MS. The method was also employed in the detection of pollutants in municipal wastewater, industrial wastewater, and natural water bodies. This work provided a new strategy for the rapid screening and untargeted analysis of trace pollutants in environmental water samples. Zhao et al.30 developed a nitrogen–sulfur-doped 3D porous carbon (N, S–C) nanosheet as a novel SALDI-TOF MS matrix. The porous structure and large specific surface area of the N, S–C nanosheet effectively enriched environmental pollutants (4-nitrocatechol, 1-hydroxy-pyrene, and nitroguaiacol) in PM2.5. The N, S–C matrices exhibit significantly lower LOD (up to 0.1 μg mL−1) for 1-hydroxypyrene than conventional organic matrices. He et al.76 developed a peptide-modified C18 microcyclone column and coupled it with SALDI-MS, achieving specific enrichment and detection of arsenic(III) ions in environmental waters. The LOD was as low as 0.7 μg L−1. The method has been applied to the detection of arsenic contamination in actual water samples from rivers and ponds.
3.3 Drug analysis
Replacing traditional organic matrices with inorganic nanomaterials in SALDI-TOF MS significantly enhances the detection sensitivity and signal-to-noise ratio for the analysis of small molecule drugs. A core–shell structured MOFs@COFs composite (UiO@TapbTp) was developed by Zheng et al.77 The large surface area and π–π stacking structure effectively enriched three non-steroidal anti-inflammatory drugs (ketoprofen, naproxen, and aspirin) in complex samples. The LOD of UiO@TapbTp-assisted LDI MS was 0.001 mg L−1, which was approximately 1000 times lower than that of conventional methods. The platform was successfully applied to detect all three non-steroidal anti-inflammatory drugs in complex samples (saliva, lake water, and tap water), with recovery rates ranging from 84.8% to 118%, which verified its applicability in practical applications. Dou et al.78 developed a high-density silicon nanopillar and silver nano-island chip as a substrate for SALDI-TOF MS, enabling the sensitive detection of sulfonamides. The heterostructure chip exhibited high detection sensitivity, with a LOD for sulfamethazine as low as 3 amol, making it applicable for the rapid quantification of sulfonamides. Moreover, Amin et al.79 developed a SALDI-TOF MS platform based on carbon black-triiron tetraoxide (CB-Fe3O4) magnetic nanocomposites for the efficient enrichment and detection of drugs, amino acids, and fatty acids. The large specific surface area (115.10 m2 g−1) and abundance of surface functional groups (e.g., C–O, Fe–O) enabled the rapid enrichment of the target analytes, with the adsorption rate of the drug reaching 27.8%–36.1% within 3 min. The LOD was as low as 1–1000 pg mL−1. Furthermore, Sun et al.80 described a method combining pipette-tip solid-phase extraction (PT-SPE) with SALDI-TOF MS for the enrichment and detection of five antidepressants in rat serum. The technique exhibits good linearity (R2 ≥ 0.997), precision (RSD ≤ 7.5%), and low quantification limit (0.05 μg mL−1). In addition, the platform does not require complex pretreatment and can efficiently extract the target drugs from the serum within just 2 minutes.Polyphenols with antioxidant properties are significant in medical and pharmaceutical applications. A 3D-mesoporous graphene (3D-MG) material was designed as an adsorbent and matrix for the detection of polyphenols in biological samples.26 The 3D-MG demonstrated a powerful ability to enrich polyphenols due to π–π stacking interaction and molecular sieving effects. Resveratrol and baicalein were clearly detected in plasma and urine samples following their oral administration. Therefore, the construction of a 3D-MG-based LDI MS platform was of great significance for studying the enrichment methods in complex samples, facilitating the development of new matrices for sample preparation in SALDI-TOF MS. Ding et al.81 developed a dual platform based on a platinum-modified nickel–iron-layered double hydroxide (Pt/NiFe-LDH) for quantification and identification of polyphenols (Fig. 4c). The Pt/NiFe-LDH exhibits peroxidase-like properties, enabling the rapid quantitative detection of polyphenols within 1 min by colorimetric analysis. Furthermore, the Pt/NiFe-LDH can also be used as an adsorbent and enhanced matrix. The Pt/NiFe-LDH-LDI MS platform can accurately identify the polyphenol fingerprints in different teas and directly monitor the oxidation–reduction reaction of phenolic substances. This work provides a reference for the subsequent expansion of dual-platform and multimodal detection methods in the quantification and identification of various substance.
Amin et al.44 developed a SALDI-TOF MS method based on the sequential synthesis of barium ferrite nanoparticles (BaFe2O4 and BaFe12O9) in a droplet microreactor. The nanoparticles produced using the droplet microreactor exhibited relatively small particle sizes and greater ability for photo-responsiveness, demonstrating their superior performance compared to bulk particles in SALDI-TOF MS. The LOD of antihistamine drugs in oral fluid was as low as 1 pg mL−1. This work also provides new ideas and approaches for future designs of multifunctional matrices for SALDI-TOF MS to improve the selectivity and sensitivity of detection methods.
4. Conclusion and outlook
Small molecules, including metabolites, drug compounds, and environmental pollutants, play key roles in the life sciences and environmental research. The dynamic changes of these analytes are directly related to the exploration of disease mechanisms, the evaluation of drug efficacy, and ecological risk assessment. In the detection process, efficient sample preparation techniques (such as separation, enrichment, and purification) for complex samples are the main prerequisites to overcome background interference and achieve accurate quantification, determining directly detection ability and data reliability in the analysis of trace small molecules. SALDI-TOF MS has been widely used in the field of small-molecule analysis due to its high throughput, superior sensitivity, and reproducibility. By regulating the surface physicochemical property of the inorganic matrix, SALDI-TOF MS can simultaneously achieve targeted and untargeted enrichment of trace small molecules in complex samples, greatly simplifying tedious sample pretreatment steps of conventional mass spectrometry (with analysis times reduced to minutes). This review emphasized the recent advances in sample preparation for small molecular analytes in complex samples, and further summarized SALDI-TOF MS approaches for the efficient detection of small molecules in environmental monitoring, metabolite analysis, and drug monitoring.Despite the achievements mentioned above, the challenges of sample preparation in SALDI-TOF MS still require attention. The surface modification and functionalization of inorganic matrices may increase the background interference. The non-uniform dispersion of nanomaterials can exhibit insufficient selective enrichment ability for small molecules, resulting in the omitted detection of low-abundance analytes. These factors will affect the reproducibility and stability of the SALDI-TOF MS approach. Multiple processes could be optimized synergistically: (1) preparation of new matrices to optimize the homogeneity; (2) strict control of temperature and humidity in the crystallization process; (3) optimization of laser energy and calibration of instrumental parameters; (4) multi-point signal averaging and intelligent algorithms for noise reduction; (5) automated sample preparation to reduce operational errors; (6) systematic quality control through QC sampling.
By integrating an “enrichment-detection” analysis platform based on an inorganic matrix, SALDI-TOF MS innovatively integrates sample pretreatment with the detection process. This method not only significantly simplifies the traditionally cumbersome sample-pretreatment process, but also enhances the detection sensitivity and specificity of trace small-molecular analytes in complex samples. It provides support for high-throughput and precise clinical diagnosis, rapid screening of environmental pollutants, and pharmacokinetics studies, extensively promoting the application efficiency of trace analysis in real-time monitoring and large-scale detection scenarios.
Author contributions
Mingyuan Liu: writing – original draft, investigation, formal analysis. Tianyu Hua: investigation. Yihan Zhang: data curation. Zifang Peng: data curation. Dan Yin: data curation. Wenfen Zhang: data curation. Yanhao Zhang: data curation. Congcong Pei: writing – review & editing, supervision, conceptualization. Shusheng Zhang: writing – review & editing, supervision, funding acquisition.Data availability
No primary research results, software or code have been included and no new data were generated or analyzed as part of this review.Conflicts of interest
There are no conflicts to declare in the manuscript.Acknowledgements
We thank the National Natural Science Foundation of China (22476185 and 22276177).References
- H. Zheng, S. Munusamy, S. Zhou, R. Jahani, J. Chen, J. Kong and X. Guan, Small, 2025, 21, 2407184 CrossRef CAS PubMed
.
- W. Shu, M. Zhang, C. Zhang, R. Li, C. Pei, Y. Zeng, L. Zhao, J. Zhao and J. Wan, Adv. Funct. Mater., 2023, 33, 2210267 CrossRef CAS
.
- W. Liang, W. Yan, X. Wang, X. Yan, Q. Hu, W. Zhang, H. Meng, L. Yin, Q. He and C. Ma, Biosens. Bioelectron., 2024, 246, 115903 CrossRef CAS PubMed
.
- Z. Zheng, W. Fu, G. Wang, B. Li, W. Xun, N. Zhou, C. Yang, R. Ren, S. Tang, Y. Du, J. Tan, D. Gu and G. Liao, Meat Sci., 2025, 225, 109820 CrossRef PubMed
.
- M. Etxebeste-Mitxeltorena, H. Flores-Romero, S. Ramos-Inza, E. Masiá, M. Nenchova, J. Montesinos, L. Martinez-Gonzalez, G. Porras, M. Orzáez, M. J. Vicent, C. Gil, E. Area-Gomez, A. J. Garcia-Saez and A. Martinez, J. Med. Chem., 2025, 68, 1179–1194 CrossRef CAS PubMed
.
- H. Zheng, G. Zhang, C. Zhang and S. Zhang, Water Res., 2024, 261, 122015 CrossRef CAS PubMed
.
- R. Shi, P. Pan, R. Lv, C. Ma, E. Wu, R. Guo, Z. Zhao, H. Song, J. Zhou, Y. Liu, G. Xu, T. Hou, Z. Kang and J. Liu, Sci. Adv., 2022, 8, eabl4923 CrossRef CAS PubMed
.
- W. H. Müller, E. De Pauw, J. Far, C. Malherbe and G. Eppe, Prog. Lipid Res., 2021, 83, 101114 CrossRef PubMed
.
- Y. Lu, P. Wang, C. Wang, M. Zhang, X. Cao, C. Chen, C. Wang, C. Xiu, D. Du, H. Cui, X. Li, W. Qin, Y. Zhang, Y. Wang, A. Zhang, M. Yu, R. Mao, S. Song, A. C. Johnson, X. Shao, X. Zhou, T. Wang, R. Liang, C. Su, X. Zheng, S. Zhang, X. Lu, Y. Chen, Y. Zhang, Q. Li, K. Ono, N. C. Stenseth, M. Visbeck and V. Ittekkot, J. Hazard. Mater., 2022, 424, 127570 CrossRef CAS PubMed
.
- G. Ciucci, A. Colliva, R. Vuerich, G. Pompilio and S. Zacchigna, Trends Pharmacol. Sci., 2022, 43, 894–905 CrossRef CAS PubMed
.
- S. A. Iakab, P. Ràfols, M. Tajes, X. Correig-Blanchar and M. García-Altares, ACS Nano, 2020, 14, 6785–6794 CrossRef CAS PubMed
.
- M. Shariatgorji, A. Nilsson, R. J. A. Goodwin, P. Svenningsson, N. Schintu, Z. Banka, L. Kladni, T. Hasko, A. Szabo and P. E. Andren, Anal. Chem., 2012, 84, 7152–7157 CrossRef CAS PubMed
.
- Z. Li, P. Huo, C. Gong, C. Deng and S. Pu, Anal. Bioanal. Chem., 2020, 412, 8083–8092 CrossRef CAS PubMed
.
- Y.-T. Lai, K. Kandasamy and Y.-C. Chen, Anal. Chem., 2021, 93, 7310–7316 CrossRef CAS PubMed
.
- M. Shipkova and H. Valbuena, TrAC, Trends Anal. Chem., 2016, 84, 23–33 CrossRef CAS
.
- Y. Liu, W. Zhang, G. Zhu, T. Pei, G. Guo, X. Wang and Y. Zhao, TrAC, Trends Anal. Chem., 2025, 187, 118206 CrossRef CAS
.
- L. Cao, M. Guler, A. Tagirdzhanov, Y.-Y. Lee, A. Gurevich and H. Mohimani, Nat. Commun., 2021, 12, 3718 CrossRef CAS PubMed
.
- X. Chen, Y. Wang, C. Pei, R. Li, W. Shu, Z. Qi, Y. Zhao, Y. Wang, Y. Lin, L. Zhao, D. Peng and J. Wan, Adv. Mater., 2024, 36, 2312755 CrossRef CAS PubMed
.
- Y. Jin, J. Yan, Z. Cai and Z. Lin, TrAC, Trends Anal. Chem., 2025, 183, 118102 CrossRef CAS
.
- L. Chen, C. Zhou, F. Jiang, L. Zhang and C. Xu, Small Methods, 2024, 8, 2400777 CrossRef CAS PubMed
.
- J. Zhu, S. Pan, H. Chai, P. Zhao, Y. Feng, Z. Cheng, S. Zhang and W. Wang, Small, 2024, 20, 2310700 CrossRef CAS PubMed
.
- F. N. Castañeda, D. L. Prince, S. R. Peirano, S. Giovannoni, R. N. Echevarría, S. Keunchkarian and M. Reta, TrAC, Trends Anal. Chem., 2024, 180, 117924 CrossRef
.
- Y. Zhao, Y. Hou, J. Ji, F. Khan, T. Thundat and D. J. Harrison, Anal. Chem., 2019, 91, 7570–7577 CrossRef CAS PubMed
.
- Y. Dong, M. Ren, J. Tu and Y. Chen, J. Hazard. Mater., 2025, 493, 138410 CrossRef CAS PubMed
.
- Z. Qiu, K. Yang, Z. Huang, H. Zhao, Z. Lin, Q. Kuang and Z. Xie, J. Hazard. Mater., 2025, 492, 138119 CrossRef CAS PubMed
.
- L. Lu, G. Zheng, M. Wang, D. Wang and Z. Xia, Talanta, 2021, 222, 121365 CrossRef CAS PubMed
.
- X.-N. Wang and B. Li, Anal. Chem., 2022, 94, 952–959 CrossRef CAS PubMed
.
- X. Zhang, V. Srot, X. Wu, K. Kern, P. A. van Aken and K. Anggara, Adv. Mater., 2024, 36, 2310817 CrossRef CAS PubMed
.
- L. Lu, Z. Wen, J. Lin, K. Zhang, D. Gao and D. Wang, J. Chromatogr., A, 2022, 1678, 463377 CrossRef CAS PubMed
.
- H. Zhao, X. Zhang, J. Wang and J. Wang, Appl. Surf. Sci., 2023, 623, 157052 CrossRef CAS
.
- S. Dou, J. Du, Q. Zhu, Z. Wang, Y. Wang, Q. Chen and N. Lu, Sens. Actuators, B, 2021, 335, 129709 CrossRef CAS
.
- Y. Lv, X. Qin, K. Hu, F. Ye and S. Zhao, Sens. Actuators, B, 2022, 353, 131132 CrossRef CAS
.
- X. Zhang, X. Ji, Z. Peng, Y. Zhang, Z. Cai and S. Zhang, J. Chromatogr., A, 2025, 1745, 465759 CrossRef PubMed
.
- W. Tan, X. Xu, Y. Lv, W. Lei, K. Hu, F. Ye and S. Zhao, J. Colloid Interface Sci., 2021, 603, 172–181 CrossRef CAS PubMed
.
- D. Li, Y. Chen and Z. Liu, Chem. Soc. Rev., 2015, 44, 8097–8123 RSC
.
- S. Feng, A. Zhang, F. Wu, X. Luo and J. Zhang, J. Chromatogr., A, 2022, 1677, 463281 CrossRef CAS PubMed
.
- H. Li, X. Zhang, L. Zhang, W. Cheng, F. Kong, D. Fan, L. Li and W. Wang, Anal. Chim. Acta, 2017, 985, 91–100 CrossRef CAS PubMed
.
- J. P. Lorand and J. O. Edwards, J. Org. Chem., 1959, 24, 769–774 CrossRef CAS
.
- X. Wang, S. Qin, G. Zheng, W. Wei, F. Li, Y. Luo, J. Tang and K. Zhou, J. Chromatogr., A, 2023, 1705, 464142 CrossRef CAS PubMed
.
- J. Zhang, X. Zheng and Y. Ni, J. Am. Soc. Mass Spectrom., 2015, 26, 1291–1298 CrossRef CAS PubMed
.
- K. Hu, Y. Lv, F. Ye, T. Chen and S. Zhao, Anal. Chem., 2019, 91, 6353–6362 CrossRef CAS PubMed
.
- Y. Luo, S. Ma, J. Zhang, Q. Zhang, Y. Zhang, J. Mao, H. Yuan, G. Ouyang, S. Zhang and W. Zhao, Anal. Chim. Acta, 2024, 1303, 342528 CrossRef CAS PubMed
.
- Z. Peng, W. Zhang, D. Yin, X. Zhang, S. Liu, W. Zhao, J. Mao, Y. Zhang and J. Xie, Chem. Eng. J., 2023, 454, 139978 CrossRef CAS
.
- M. O. Amin, B. D'Cruz and E. Al-Hetlani, Analyst, 2023, 148, 4489–4503 RSC
.
- A. Lavigne, B. Gilquin, T. Géhin, V. Jousseaume, M. Veillerot, Y. Chevolot, M. Phaner-Goutorbe and C. Yeromonahos, ACS Appl. Mater. Interfaces, 2023, 15, 18685–18693 CrossRef CAS PubMed
.
- K. Ge, Y. Peng, Z. Lu, Y. Hu and G. Li, J. Chromatogr., A, 2020, 1615, 460741 CrossRef CAS PubMed
.
- S. Joh, J. Yoo, S. M. Lee, E. Lee, H.-K. Na, J. G. Son, J. Kim, M. S. Jeong, S.-G. Lee and T. G. Lee, ACS Nano, 2024, 18, 17681–17693 CrossRef CAS PubMed
.
- S. Joh, H.-K. Na, J. G. Son, A. Y. Lee, C.-H. Ahn, D.-J. Ji, J.-S. Wi, M. S. Jeong, S.-G. Lee and T. G. Lee, ACS Nano, 2021, 15, 10141–10152 CrossRef CAS PubMed
.
- A. Ali, T. Qin, W. Zhang, S. Zhang, L. He and W. Zhao, Anal. Chim. Acta, 2025, 1355, 343934 CrossRef CAS PubMed
.
- H. Liang, W. Wang, W. Liang, X. Deng, X. Ruan, D. Zhang and Y. Yang, J. Environ. Chem. Eng., 2023, 11, 109126 CrossRef CAS
.
- Y. Jin, J. Chen, W. Xie, J. Zhang, J. Yan, C. Chen, J. Lin, Z. Cai and Z. Lin, ACS Appl. Mater. Interfaces, 2025, 17, 7056–7065 CrossRef CAS PubMed
.
- H. Su, X. Li, L. Huang, J. Cao, M. Zhang, V. Vedarethinam, W. Di, Z. Hu and K. Qian, Adv. Mater., 2021, 33, 2007978 CrossRef CAS PubMed
.
- Z. Zhang, Y. Li, R. Wang, S. Yang, P. Li, K. Zhao, Y. Gu, K. Meng, J. Li, J. Pu, X. Yan, S. Gu, H. Su, X. Kong and K. Qian, Nano Today, 2025, 62, 102702 CrossRef CAS
.
- J. Qi, G. Chen, Z. Deng, Y. Ji, S. An, B. Chen, G. Fan, C. Fang, K. Yang, F. Shi and C. Deng, Anal. Chem., 2025, 97, 345–354 CrossRef CAS PubMed
.
- Y. Wang, W. Shu, S. Lin, J. Wu, M. Jiang, S. Li, C. Liu, R. Li, C. Pei, Y. Ding, J. Wan and W. Di, Small, 2022, 18, e2106412 CrossRef PubMed
.
- H. Zhang, N. Li, F. Shi, F. Yuan, S. Huang, N. Sun and C. Deng, Anal. Chem., 2024, 96, 18824–18833 CrossRef CAS PubMed
.
- C. Yang, H. Yu, W. Li, H. Lin, H. Wu and C. Deng, Anal. Chem., 2024, 96, 6264–6274 CrossRef CAS PubMed
.
- C. Pei, C. Liu, Y. Wang, D. Cheng, R. Li, W. Shu, C. Zhang, W. Hu, A. Jin, Y. Yang and J. Wan, Angew. Chem., Int. Ed., 2020, 59, 10831–10835 CrossRef CAS PubMed
.
- R. Weng, Y. Xu, X. Gao, L. Cao, J. Su, H. Yang, H. Li, C. Ding, J. Pu, M. Zhang, J. Hao, W. Xu, W. Ni, K. Qian and Y. Gu, Adv. Sci., 2025, 12, 2405580 CrossRef CAS PubMed
.
- Y. Su, Z. Peng, Y. Wang, S. Yang, X. Xu, W. Liu, Q. Bao, C. Jiang, K. Qian and X. Fan, Small, 2025, 21, 2411871 CrossRef CAS PubMed
.
- X. Xu, H. Tan, W. Zhang, W. Liu, Y. Chen, J. Zhang, M. Gu, Y. Yang, Q. Chen, Y. Wang, K. Qian and B. Xu, Small Methods, 2025, 2401906 CrossRef PubMed
.
- Y. Zhao, C. Ding, H. Su, A. Wang, A. Tang, H. Zhao, Y. Ma, M. Zhang, W. Liu, R. Wang, Z. Zhang, S. Yang, D. Liang, Y. Huang, K. Qian, L. Huang, Q. Fu and Y. Cao, Adv. Sci., 2025, 12, 2416454 CrossRef CAS PubMed
.
- Y. Wang, S. Li, T. Li, J. Wu, Y. Huang, W. Liu, C. Ding, L. Huang, X. Xu, Y. Wang, S. Gu, K. Liu, K. Qian and X. Sun, Small, 2025, 21, 2412195 CrossRef CAS
.
- J. Cao, Q. J. Yao, J. Wu, X. Chen, L. Huang, W. Liu, K. Qian, J. J. Wan and B. O. Zhou, Cell Metab., 2024, 36, 209–221 CrossRef CAS PubMed
.
- Y. Zhao, R. Boukherroub, L. Liu, H. Li, R.-S. Zhao, Q. Wei, X. Yu and X. Chen, J. Hazard. Mater., 2023, 459, 132336 CrossRef CAS PubMed
.
- J. Wu, C. Liang, X. Wang, Y. Huang, W. Liu, R. Wang, J. Cao, X. Su, T. Yin, X. Wang, Z. Zhang, L. Shen, D. Li, W. Zou, J. Wu, L. Qiu, W. Di, Y. Cao, D. Ji and K. Qian, Adv. Sci., 2023, 10, 2302023 CrossRef CAS PubMed
.
- C. Pei, Y. Wang, Y. Ding, R. Li, W. Shu, Y. Zeng, X. Yin and J. Wan, Adv. Mater., 2023, 35, 2209083 CrossRef CAS PubMed
.
- Z. Li, S. Zhang, Q. Xiao, S. Shui, P. Dong, Y. Jiang, Y. Chen, F. Lan, Y. Peng, B. Ying and Y. Wu, ACS Nano, 2025, 19, 6180–6194 CrossRef CAS PubMed
.
- G. Fan, B. Zhang, J. Wang, N. Wang, S. Qin, W. Zhao and J. Zhang, Talanta, 2024, 268, 125298 CrossRef CAS PubMed
.
- Y. Wang, K. Zhang, T. Tian, W. Shan, L. Qiao and B. Liu, ACS Appl. Mater. Interfaces, 2021, 13, 4886–4893 CrossRef CAS PubMed
.
- X. Jiang, L. Tao, S. Cao, Z. Xu, S. Zheng, H. Zhang, X. Xu, X. Qu, X. Liu, J. Yu, X. Chen, J. Wu and X. Liang, ACS Appl. Mater. Interfaces, 2025, 17, 5893–5908 CrossRef CAS PubMed
.
- Y. Su, X. Lai, K. Guo, X. Wang, S. Chen, K. Liang, K. Pu, Y. Wang, J. Hu, X. Wei, Y. Chen, H. Wang, W. Lin, W. Ni, Y. Lin, J. Zhu and K.-M. Ng, Anal. Chem., 2022, 94, 16910–16918 CrossRef CAS PubMed
.
- Z. Peng, Y. Zhang, Y. Dong, W. Zhao, J. Mao, Q. Zhang, G. Ouyang, S. Zhang and J. Xie, Chem. Eng. J., 2025, 507, 160596 CrossRef CAS
.
- C. Gao, J. Bai, Y. He, Q. Zheng, W. Ma and Z. Lin, ACS Sustainable Chem. Eng., 2019, 7, 18926–18934 CrossRef CAS
.
- H. Moriwaki, T. Otsuka, Y. Kawabe, I. Osaka, A. Miyazato, J. Maruo and O. Yamada, Int. J. Mass Spectrom., 2018, 428, 49–54 CrossRef CAS
.
- H.-Y. He and H.-H. Hsiao, Microchem. J., 2024, 207, 112179 CrossRef CAS
.
- R. Zheng, Y. Yang, C. Yang and Y. Xia, Microchim. Acta, 2021, 188, 179 CrossRef CAS PubMed
.
- S. Dou, J. Lu, Q. Chen, C. Chen and N. Lu, Sens. Actuators, B, 2022, 364, 131846 CrossRef CAS
.
- M. O. Amin, B. D'Cruz, M. Madkour and E. Al-Hetlani, Microchim. Acta, 2019, 186, 503 CrossRef CAS PubMed
.
- Z. Sun, F. Wang, W. Li, R. Ren, P. Zhou, Q. Jia, L. Zhao, D. Chen and L. Zuo, Anal. Bioanal. Chem., 2024, 416, 5013–5023 CrossRef CAS PubMed
.
- C. Ding, Y. Zhu, Z. Huo, S. Yang, Y. Zhou, A. Yiming, W. Chen, S. Liu, K. Qian and L. Huang, Mater. Today Bio, 2024, 26, 101047 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2025 |
