Open Access Article
Open Access ArticleA holistic approach to understanding biochemical degradation of human tissues using high resolution MALDI MS†
Jerika Hoabc,
Naomi L. Stock c,
Vaughn Mangal
c,
Vaughn Mangal d,
Shari L. Forbes
d,
Shari L. Forbes a and
Theresa E. Stotesbury
a and
Theresa E. Stotesbury *b
*b
aDepartment of Chemistry and Biochemistry, University of Windsor, 401 Sunset Ave, Windsor, ON N9B 3P4, Canada. E-mail: hojerik@uwindsor.ca
bFaculty of Science, Forensic Science, Ontario Tech University, 2000 Simcoe St. N, Oshawa, ON L1G 0C5, Canada.. E-mail: theresa.stotesbury@ontariotechu.ca
cWater Quality Center, Trent University, 1600 W Bank Dr, Peterborough, ON K9L 0G2, Canada
dDepartment of Chemistry, Brock University, 1812 Sir Isaac Brock Way, St. Catharines, ON L2S 3A1, Canada
First published on 18th July 2025
Abstract
Estimating the postmortem interval (PMI) is crucial in medico-legal death investigations, but existing methods are highly influenced by environmental and physiological factors. This study proposes a novel biochemical strategy for PMI estimation using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry (MALDI MS) to detect molecular changes in decomposing human soft tissues. Samples from the quadriceps and shoulder were collected from 13 human donors at the Research in Experimental and Social Thanatology (REST) facility in Quebec, Canada, spanning accumulated degree days (ADD) ranging from 9.75 to 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 455.86. Tissues (14 μm) were cryosectioned, mounted on carbon tape, dried under nitrogen gas, and coated with a 2,5-dihydroxybenzoic acid matrix solution. Hematoxylin and eosin staining provided histological context for the sampled tissues. MALDI MS data were acquired using a Bruker SolariX XR Fourier-transform Ion Cyclotron Resonance MS. Van Krevelen diagrams and principal component analysis revealed decomposition-linked trends, with shifts in biochemical profiles over time. N/C and N/O ratios increased, reflecting protein and peptide degradation and microbial activity, while O/C ratios declined, likely due to the loss of oxygen-rich compounds. Mixed-effects models showed moderate associations (R2 marginal = 0.06–0.49) between elemental ratios and ADD, with conditional R2 values up to 0.55. Spearman's rank correlation identified compounds significantly associated with ADD. Six candidate biomarkers were identified, with variable model fit (R2 marginal = 0.003–0.80) and unresolved donor-level effects. This study demonstrates the novel application of MALDI MS for PMI biomarker discovery and contributes one of the largest datasets in forensic decomposition chemistry research.
455.86. Tissues (14 μm) were cryosectioned, mounted on carbon tape, dried under nitrogen gas, and coated with a 2,5-dihydroxybenzoic acid matrix solution. Hematoxylin and eosin staining provided histological context for the sampled tissues. MALDI MS data were acquired using a Bruker SolariX XR Fourier-transform Ion Cyclotron Resonance MS. Van Krevelen diagrams and principal component analysis revealed decomposition-linked trends, with shifts in biochemical profiles over time. N/C and N/O ratios increased, reflecting protein and peptide degradation and microbial activity, while O/C ratios declined, likely due to the loss of oxygen-rich compounds. Mixed-effects models showed moderate associations (R2 marginal = 0.06–0.49) between elemental ratios and ADD, with conditional R2 values up to 0.55. Spearman's rank correlation identified compounds significantly associated with ADD. Six candidate biomarkers were identified, with variable model fit (R2 marginal = 0.003–0.80) and unresolved donor-level effects. This study demonstrates the novel application of MALDI MS for PMI biomarker discovery and contributes one of the largest datasets in forensic decomposition chemistry research.
1. Introduction
Accurate determination of postmortem interval (PMI) is crucial in medico-legal death investigations, aiding in the estimation of death timelines and legal proceedings.1,2 The challenge of PMI estimation becomes especially pronounced in complex forensic cases, where remains have been exposed to dynamic and/or extreme outdoor environmental conditions. Forensic taphonomy, the study of postmortem processes, plays a pivotal role in uncovering the effects of decomposition on the human body.3 PMI estimation is also challenging when human remains are discovered weeks to years after death, a timeframe during which traditional methods become increasingly unreliable.4–6Traditional methods of PMI estimation typically rely on observable postmortem changes such as rigor mortis (stiffening of muscles), livor mortis (settling of blood), and algor mortis (cooling of the body).7–9 While these physiological markers are useful in the early postmortem stages, their reliability declines as time progresses, particularly when exposed to extrinsic factors, including temperature, humidity, and the body's surroundings, which can introduce significant variability and uncertainty in PMI estimations.4,10,11 The use of entomology to estimate PMI, based on patterns of colonization and larval development, is another widely applied method; however, it too is highly dependent on environmental factors and may be unreliable in cases of delayed insect access or extreme climates.12,13 Remains in extreme temperature environments, whether high heat (>30 °C) or freezing temperatures (<0 °C), cause decomposition at rates that differ significantly from those in temperate conditions.14–17 For instance, in Canada, decomposition may slow dramatically during subzero winter temperatures and then accelerate rapidly as temperatures rise in the spring.16 In such cases, traditional PMI indicators become less reliable, often leading forensic pathologists to provide broader PMI estimates from weeks to months, as opposed to hours or days. This variability has led to increasing interest in molecular-based forensic approaches, particularly those which apply ‘omics’ techniques to analyze biochemical changes in human remains.
Recent advances in metabolomics, proteomics, and lipidomics offer a more detailed understanding of the molecular changes that occur during decomposition.18–22 Focusing on small-molecule metabolites, metabolomics provides insights into the cellular degradation and microbial activity.22 In tandem, proteomics, investigates protein degradation and modifications such as proteolysis and deamidation.21 Lipidomics has revealed shifts in lipid profiles in decomposing tissues, which may also correlate with decomposition stages.6,23,24 Throughout decomposition, lipids undergo various transformations, including hydrolysis, where enzymatic activity breaks down the phospholipids and triglycerides into free fatty acids and glycerol; oxidation, which leads to the degradation of polyunsaturated fatty acids into aldehydes, ketones, and generation of volatile organic compounds (VOCs); and hydrogenation, where microbial activity reduces unsaturated fatty acids to saturated fatty acids.6,23–26 Other potential lipids transformations include dehydration, esterification, decarboxylation, and polymerization.27 Given the complexity of these molecular changes, mass spectrometry-based techniques have emerged as powerful tools for high-resolution detection, quantification, and characterization of proteins, lipids, and metabolites associated with decomposition.
Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry (MALDI MS) has become an increasingly popular tool in forensic science, with applications spanning from toxicology to tissue analysis.28–31 One of its most significant forensic contributions has been in the analysis of latent fingerprints, where MALDI MS enables both chemical imaging and compound-specific identification within fingerprint residues.32,33 This work demonstrates how MALDI MS, when paired with high-resolution mass analyzers, offers both exceptional mass accuracy and spatial resolution for the forensic analysis of complex samples.33–35 This technique has the added advantage of requiring minimal sample preparation and manipulation, allowing for the direct analysis of complex biological tissues without extensive preprocessing steps that might otherwise damage or alter a fragile, and often trace forensic sample.33 MALDI MS can analyze lipids, proteins, peptides, and small molecules, making it an ideal tool for studying fragile evidence like degraded human remains.33,36
Several other chromatographic techniques paired with MS have been used in PMI estimation studies, including gas chromatography-mass spectrometry (GC-MS),37,38 liquid chromatography-mass spectrometry (LC-MS),11,39,40 and multidimensional gas chromatography.41–48 While other MS studies have identified lipid degradation products, fatty acids, and peptide fragments as promising PMI-related biomarkers,6,23,26,49,50 the application of MALDI MS to PMI estimation presents an opportunity to fill an important gap in forensic taphonomy.
This study investigates the novel application of MALDI MS for PMI biomarker discovery in degrading human soft tissue, combining high-resolution mass spectrometry and multivariate statistical modelling. To aid in interpreting these complex changes, Van Krevelen (VK) diagrams are used.51–54 By plotting the hydrogen-to-carbon (H/C) and oxygen-to-carbon (O/C) ratios of organic compounds, VK plots help infer chemical and biochemical reactions that drive decomposition such as oxidation, hydrolysis, and hydrogenation. Beyond the standard VK plots, heteroatom-centered variations can further provide insights into nucleic acid degradation and microbial processes, offering a broad understanding of biochemical transformations during decomposition.51
To identify candidate biomarkers and characterize temporal trends, this study applied Spearman's rank correlation, principal component analysis (PCA), linear regression, and mixed-effects modelling. Spearman correlation assessed monotonic relationships between compound intensity and accumulated degree days (ADD), while PCA was applied to explore variation in molecular composition. Linear regression evaluated time-dependent patterns in elemental ratios. Mixed-effects models were used to assess the relationship between elemental ratios and ADD, incorporating donor as a random effect to account for inter-donor variability. The same modeling approach was applied to biomarker intensity to evaluate its association with ADD. Together, these analyses offer a comprehensive strategy for capturing biochemical changes and identifying candidate molecular markers of PMI in human soft tissue.
2. Experimental
2.1 Donor information
The REST (Research in Experimental and Social Thanatology) facility is the first Canadian human taphonomic facility that is dedicated to research and training related to human decomposition in a northern continental climate.10 The bodies of thirteen human donors were donated through the Body Donation Program at the Université du Québec à Trois-Rivières (UQTR). Human ethics approval for sample collection from the donors was approved by UQTR with certification numbers CER-19-261-07.10 and SCELERA 22-06. The sampling period began in May 2023 and was completed in April 2024. Sampling was reduced in the fall months and paused in the winter months due to extensive snow coverage.The selection of donors consisted of two females and eleven males, ranging from 54 to 86 years old at the time of death. More donor information including date and time of death, arrival at anatomy lab, arrival at REST, age, sex, weight, height, BMI, and cause of death can be found in Table S1.† The bodies of all donors were kept refrigerated (4 °C) at the morgue within the UQTR Anatomy Laboratory for approximately 24 hours prior to being placed at the REST facility. To maintain consistency with other projects, the donor ID number remained the same as assigned by the REST facility.
2.2 Sample collection
Soft tissue samples from four donors (Donors 22 to 25) were collected from the earliest day of deposition at REST. Samples from the remaining nine donors were taken at time points starting from the study's commencement date. Soft tissue samples were collected using an 8 mm biopsy punch (Integra Miltex, USA) or scalpel (Fisher Scientific, ON, Canada) (approximately 1 cm × 1 cm sample) from the upper arm and/or quadriceps on each donor. For donors with minimal soft tissues, biopsies were taken from regions where soft tissue remained, which were generally obtained from the lower leg. Biopsy samples were stored in 1.5 mL microcentrifuge tubes (Fisherbrand, ON, Canada) at −20 °C until analysis. Accelerated progression of decomposition was observed at the site of sampling locations.2.3 Calculation of accumulated degree days (ADD)
In this study, the maximum and minimum temperature were collected from the onsite weather station at the REST facility,4 and any missing data were retrieved from the closest Environment Canada and Climate Change weather station. Accumulated Degree Days (ADD) were calculated from this data where the average daily temperatures were summed together from the date of body placement at REST until the final day of sampling for this study (Fig. 1). All negative temperature values were treated at zero values when calculating ADD as per standard protocols.4 The maximum temperature measured during this period was 34.8 °C and the minimum temperature measured was −21.6 °C.2.4 Tissue sectioning and staining
Tissue sections were created using a cryostat microtome (Leica Microsystems CM 1900, UK), with the chamber temperature and specimen head set to −22 °C. As optimal cutting temperature (OCT) medium (Sakura Finetek USA, CA, USA) contains polyethylene glycol (PEG), a known MS ion suppressor, a minimal amount was used to mount the sample cores to the cryostat chuck. Cross sections were sectioned at 14 μm and lift transferred to a MALDI plate 384 well ground-steel target plate, purchased from Bruker Corp. (Billerica, MA, USA), using a paint brush. Double sided carbon conductive tape (Electron Microscopy Sciences, PA, USA) was used to adhere the tissue slices to the MALDI plate.55,56 The MALDI plate was placed into a beaker with a line of high purity nitrogen gas (Linde Canada, Peterborough ON) and allowed to dry for 10 minutes prior to matrix application. Additional 8 μm tissue sections were collected in duplicate on positively charged microscope slides (Leica Microsystems, Germany) and stained using traditional haematoxylin and eosin staining methods.2.5 Matrix application
A matrix solution of 5 mg mL−1 2,5 dihydroxybenzoic acid (DHB) (Sigma Aldrich, Oakville, ON, Canada) was prepared in 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 vol/vol (v/v) acetonitrile (ACN) (Sigma Aldrich, Oakville, ON, Canada): high-purity water (18.2 MΩ cm2 resistivity) with 0.1% (v/v) trifluoroacetic acid (TFA) (Sigma Aldrich, ON, Canada). TFA was also used as an external lock mass. To apply the matrix to the surface of the MALDI plate, a MedPro Compressor Nebulizer (AMG Medical Inc., QC, Canada) was used to create an aerosol of matrix (particle size 0.5 μm to 5 μm).57 The MALDI plate and nebulizer chamber were placed in a container and approximately 6 mL of matrix solution was placed in the nebulizer chamber and the nebulizer was run for 5 minutes (Note. S1 and Fig. S1†). The matrix solution was allowed to settle onto the plate within the container for 15 minutes before the MALDI plate was removed and dried under the high-purity nitrogen for another 10 minutes.
2 vol/vol (v/v) acetonitrile (ACN) (Sigma Aldrich, Oakville, ON, Canada): high-purity water (18.2 MΩ cm2 resistivity) with 0.1% (v/v) trifluoroacetic acid (TFA) (Sigma Aldrich, ON, Canada). TFA was also used as an external lock mass. To apply the matrix to the surface of the MALDI plate, a MedPro Compressor Nebulizer (AMG Medical Inc., QC, Canada) was used to create an aerosol of matrix (particle size 0.5 μm to 5 μm).57 The MALDI plate and nebulizer chamber were placed in a container and approximately 6 mL of matrix solution was placed in the nebulizer chamber and the nebulizer was run for 5 minutes (Note. S1 and Fig. S1†). The matrix solution was allowed to settle onto the plate within the container for 15 minutes before the MALDI plate was removed and dried under the high-purity nitrogen for another 10 minutes.
2.6 MALDI MS
All MALDI mass spectra were acquired with a Bruker SolariX XR FTICRMS (Billerica, MA, USA) located at the Water Quality Centre at Trent University and equipped with a 7 T superconducting magnet and SmartBeam laser optics. An external calibration was performed prior to analysis using electrospray ionization (ESI) in negative ionization mode and sodium trifluoroacetic acid (NaTFA, 0.1 mg mL−1 in methanol). MALDI data was collected in negative ion mode using the Bruker ftms Control software (version 2.1.0), over a mass range of m/z 53.7600 to 2200.0000. During the acquisition, the SmartBeam laser operated at 1000 Hz with laser focus set to medium and laser power set to 65%. Analysis consisted of 200 laser shots per scan and 75 scans per sample, over a 1000 μm wide range in each well with a grid increment of 15 μm and an offset of 1 μm. Other parameters were as follows: plate offset voltage, −100.0 V; deflector plate voltage, −200.0 V; 4 million data points per scan and free ion decay (FID) 1.0486 s. Peak alignment by external lock mass calibration to the internal standard TFA was completed prior to extracting mass peak lists. Matrix-only and matrix-plus-tape blanks were also analyzed to account for background signals and potential contamination.2.7 Data analysis
Mass peak lists were exported from Bruker Compass DataAnalysis Software (version 5.2). Peaks with a signal-to-noise ratio below 25 were excluded to minimize low-abundance peaks. The processed datasets were imported into the UltraMassExplorer (UME) database for blank subtraction and preliminary formula assignment, using a mass error tolerance of ±0.5 ppm. UME is an open-access web-based platform that supports high-resolution formula prediction, data visualization, and exploratory mass spectrometric analysis.58 Although UME was originally developed for the analysis of dissolved organic matter, its capabilities for high-resolution formula assignment and visualization make it suitable for application to biological samples in this study. In cases where multiple molecular formulae matched the same m/z value, the formula with the lowest mass error (ppm) was selected as the most probable candidate. The resulting peak tables were exported as CSV files for further analysis in RStudio (version 2024.12.0 + 467).The following analyses were conducted in RStudio, using cor.plot() to visualize relationships between variables.59 Van Krevelen diagrams were constructed to visualize changes in elemental composition over time based on the predicted molecular formula provided by the UME. The general regions for each compound class on the Van Krevelen diagrams (e.g., carbohydrates, lignin, amino sugars) were delineated based on established elemental ratio boundaries reported in prior literature.51 The total number of points within each compound class region was summed, and principal component analysis (PCA) was performed. To account for variation in point counts across samples and donors, the data were normalized prior to analysis.
Average ratio values for each elemental ratio (e.g., H/C, O/C) were determined for each donor and linear regression analysis was initially used to explore gross temporal trends. To account for repeated measures and inter-donor variability, mixed-effects models were applied to elemental ratios, with donor included as a random intercept. Model performance was evaluated using marginal R2 (variance explained by fixed effects) and conditional R2 (variance explained by both fixed and random effects). No additional donor covariates (e.g., age, sex, BMI) were incorporated into the models.
To further assess temporal trends, Spearman's rank correlation was performed between compound intensities and ADD. Only compounds detected in ≥7 samples and with Spearman p-values ≤ 0.05 were retained for analysis.60 Compounds meeting this criterion were cross-referenced against the Human Metabolome Database (HMDB) and Metabolomics Workbench for tentative biological identification. Compounds without matches in these databases were subsequently compared against PubChem for additional tentative identification based on predicted molecular formula and m/z provided from the UME. Mixed-effects models were applied to the tentatively identified compounds to determine correlations between the observed intensity for each compound and ADD with donor included as a random effect.
3. Results and discussion
3.1 Physical and histological changes
Observations on the stage of decomposition were recorded on each sampling day using field notes. During the sampling period, Donors 2, 6, 9–13, 16, and 17 were all in the advanced or skeletonized stages of decomposition. Since minimal external changes were observed in these donors, the following description focuses on donors classified as “fresh” (Donors 22–25) at the beginning of the study. However, data from all donors were included in the biochemical and statistical analyses.The fresh stage of decomposition is characterized by minimal skin discoloration, the onset of insect activity, and limited bloating.4,9 As ADD progressed for Donors 22–25, the production of gases in the abdomen and extremities caused visible bloating. Skin discoloration intensified, with marbling patterns developing across all four donors, consistent with the breakdown of hemoglobin and subsequent reactions with decomposition gases.61 Notably, the head, neck, and shoulders decayed more rapidly than other regions, aligning with prior literature.4,17,62 As decomposition is not a discrete process, the sampling sites showed more rapid decay than the remainder of the body. This can be attributed to the nature of biopsy sampling itself as it adds more open locations for insect and microbial activity to thrive.63,64 It also exposes the remaining internal soft tissues to environmental elements.
The advanced stage of decomposition can persist for an extended period, depending on environmental conditions such as temperature and humidity.65–67 Eventually, the remains reached the skeletonized/dry remains stage, where most or all soft tissues have degraded, leaving only bones and dried remnants.
Fig. 2 shows images of histological stains of early to mid postmortem time points for Donor 23, revealing a progressive loss of connective structure within adipose tissue, resulting in tissue morphology that becomes increasingly difficult to distinguish. At these initial stages, the epidermal and dermal layers of the skin were indistinguishable, possibly due to the interval (∼24–96 hours) between death and deposition. This observation was also reported by Wei (2020), who documented histological changes in human skin up to 32 days postmortem, with changes in the morphology of the epidermis and dermis occurring within 24 to 72 hours after death.68
Between 18.18 ADD and 62.03 ADD, histological alterations became more pronounced, with a noticeable decline in structural integrity (Fig. 2B). These tissue-level changes reflect the early biochemical breakdown of structural proteins such as collagen and elastin, consistent with proteolysis driven by autolysis and microbial enzyme activity.68 Furthermore, observations between 18.18 ADD and 104.07 ADD indicate a gradual compression of adipose cells over time, likely resulting from enzymatic degradation and tissue breakdown (Fig. 2C). This corresponds with lipid hydrolysis and oxidation.24
Finally, a lack of distinct muscle tissue was observed in Donors 22–25 at ADD values of 164.89, 203.76, 127.99, and 172.35, respectively. This loss may be attributed to increased insect activity during these sampling periods. The remaining tissue was structurally consistent with skin in gross appearance but lacked identifiable cellular features histologically and was molecularly indistinguishable (Fig. 2D).
The following MALDI MS analysis was used to confirm and expand upon these biochemical patterns, linking histological degradation with changes in compound composition over time.
3.2 Compound class distribution analysis
MALDI mass spectra were acquired over the range of m/z 53.7600 to 2200.0000, with an average of approximately 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 peaks. Only peaks with a signal-to-noise ratio greater than 25 were retained for analysis. Peaks corresponding to common matrix components, including DHB and TFA, were consistently observed across all samples.
000 peaks. Only peaks with a signal-to-noise ratio greater than 25 were retained for analysis. Peaks corresponding to common matrix components, including DHB and TFA, were consistently observed across all samples.
Van Krevelen (VK) diagrams reveal temporal shifts in compound class composition associated with biochemical decomposition processes (Fig. 3A–F). In early PMI, higher hydrogen-to-carbon (H/C) and oxygen-to-carbon (O/C) ratios are more prevalent, showcasing the contributions of carbohydrates and amino-sugars as seen in Fig. 3(A–C). These hydrogen- and oxygen-rich molecules degrade early, but decomposition by-products from larger carbohydrates and proteins contribute to a steady increase in the carbohydrate and amino-sugar region over time.25,69 This trend supports the idea of compound turnover, in which early-degrading molecules are continuously replaced by decomposition by-products, rather than simple depletion during the early stages of decomposition. Furthermore, there is an emergence of condensed aromatic-like and lignin-like compounds indicating oxidative transformation.26 Later time points, Fig. 3(D–F), show a decrease in the overall number of compounds over time reflecting the progressive degradation and volatilization of compounds.
 | ||
| Fig. 3 Van Krevelen diagrams of Donor 23 at different time points. Different compound categories are represented in different colours. Shown areas are an approximation based on previous compound classification of organic matter studies.51,54 These include lipids (low O/C, high H/C), carbohydrates (high O/C, high H/C), proteins/peptides and amino sugars (moderate O/C and H/C), and condensed aromatics (low H/C, variable O/C). Point colour corresponds to relative signal intensity as shown on the right, with darker blue representing lower and red representing higher intensities. (A) PMI 2/18.18 ADD (B) PMI 6/104.07 ADD (C) PMI 29/609.26 ADD (D) PMI 104/1966.70 ADD (E) PMI 138/2328.78 ADD (F) PMI 299/2506.31 ADD. | ||
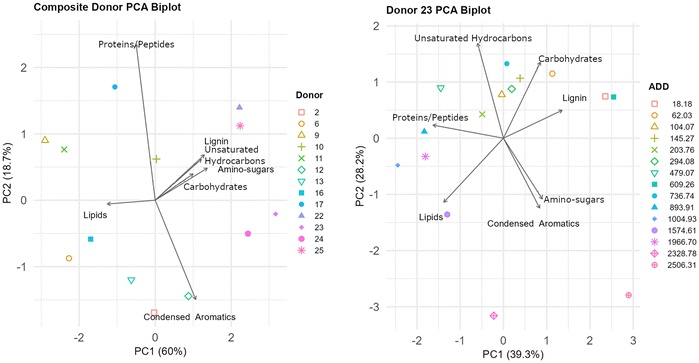
To explore how different compound categories contribute to the chemical profile of each donor, a principal component analysis (PCA) biplot is presented in Fig. 4 (left). The number of compounds within each chemical category (e.g., lipids, amino sugars, unsaturated hydrocarbons) was summed for each sample (Table S2†), and the resulting compound class distribution was used as the input for PCA. The first principal component (PC1) accounts for 60% of the variance and appears to broadly separate early and late-stage donors, while the second component (PC2) explains 18.7% variance. The direction and magnitude of the arrows reflect the influence of each compound class on sample distribution. Fresh donors (Donors 22 to 25) cluster on the right side of the plot, suggesting they are more similar to each other than to the more decomposed donors. This grouping aligns with the positioning of amino-sugars, carbohydrates, and unsaturated hydrocarbons, which are compounds commonly present during the earlier stages of decomposition.8 In contrast, lipids are oriented closer to the more decomposed samples, consistent with their chemical stability and persistence.23,24 The protein/peptide vector contributes minimally to the separation, potentially due to limited detection in the analyzed m/z range.
In the PCA biplot of Donor 23 (Fig. 4, right), samples show a noticeable shift in distribution across PC1 (39.9%), which may reflect changes in compound class composition over time. For this donor, PC1 and PC2 account for 39.9% and 28.2% of the variance, respectively, capturing a substantial portion of the overall variation. Early ADD values (18.18 to 203.76) are positioned near carbohydrates, and unsaturated hydrocarbons, suggesting a predominance of oxygen-containing compounds during the initial stages of decomposition. As ADD increases, samples progressively shift toward the lower left quadrant, aligning with lipid and condensed aromatic vectors. This transition is consistent with the accumulation of degradation-resistant compounds and transformation products as decomposition advances.23 The spatial arrangement of Donor 23 time points parallels the broader compound class trends observed in the composite donor plot, confirming the temporal evolution of biochemical profiles within a single individual.
3.3 Heteroatom trends in elemental ratios
Temporal changes in elemental composition across the decomposition interval revealed distinct patterns for hydrogen, oxygen, nitrogen, sulfur, and phosphorus relative to carbon and oxygen (Fig. 5, 6, Table 1; Fig. S3 and Table S3†). These heteroatom-linked ratios reflect macromolecular degradation processes including hydrolysis, oxidation, and microbial transformation, and were influenced by both postmortem interval and individual donor characteristics. | ||
| Fig. 5 Elemental ratios (H/C, O/C, N/C, N/O, P/C, P/O, S/C, and S/O) plotted as a function of PMI for Donors 22 to 25. Bars represent the average ratio values calculated for each ADD(PMI). | ||
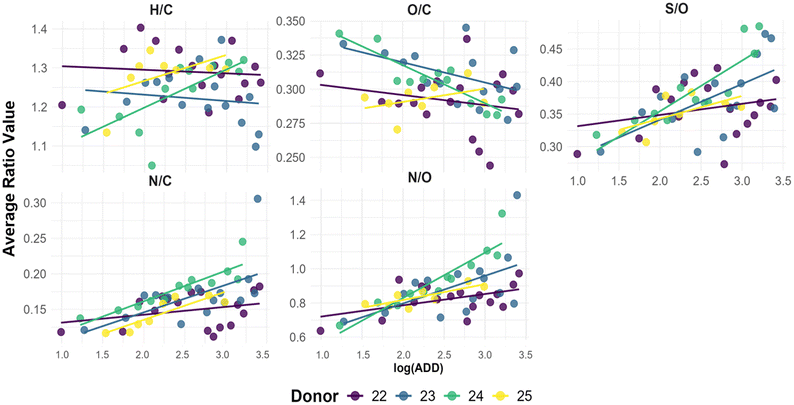 | ||
| Fig. 6 Scatter plots of average elemental ratios (H/C, O/C, S/O, N/C, N/O) as a function of log(ADD), with linear regression trend lines showing correlation patterns for Donor 22 to 25. | ||
| Ratio | Est. | Std. error | CI low | CI high | R2 marg. | R2 con. |
|---|---|---|---|---|---|---|
| H/C | 0.033 | 0.016 | 0.002 | 0.064 | 0.061 | 0.230 |
| O/C | −0.016 | 0.004 | −0.023 | −0.009 | 0.215 | 0.340 |
| N/O | 0.152 | 0.019 | 0.114 | 0.190 | 0.449 | 0.489 |
| N/C | 0.034 | 0.004 | 0.026 | 0.042 | 0.490 | 0.547 |
| S/O | 0.035 | 0.008 | 0.018 | 0.052 | 0.199 | 0.307 |
The hydrogen-to-carbon (H/C) ratio showed an initial increase in early PMI samples, attributed to the breakdown of lipids and fatty acids and the release of hydrogen-rich by-products through hydrolysis.26 As decomposition advanced, a subsequent decline in H/C was observed, likely reflecting further degradation or volatilization of these compounds. Despite limited visual changes at later stages, shifts in H/C indicate ongoing biochemical activity. Mixed-effects modelling revealed low marginal R2 (0.0613), suggesting that most of the variation was driven by donor-specific characteristics (conditional R2 = 0.2304), potentially linked to differences in tissue composition or lipid content.
The oxygen-to-carbon (O/C) ratio exhibited a consistent downward trend over time, reflecting the progressive loss of oxygen-rich biomolecules such as carbohydrates and peptides. These are prone to hydrolysis, forming volatile short-chain acids and alcohols that leach into the environment.70 Donors 22–24 demonstrated a more pronounced decrease, while Donor 25 remained relatively stable. Older donors, including 9, 13, 16, and 17, maintained low O/C ratios, consistent with tissue dominated by lipids and long-chain hydrocarbons. Mixed modelling confirmed a statistically significant negative trend (estimate = –0.0157, marginal R2 = 0.2155), with improved fit when donor was included as a random effect (conditional R2 = 0.3396).
The nitrogen-to-carbon (N/C) and nitrogen-to-oxygen (N/O) ratios increased over time, indicative of protein and amino acid degradation. Donor 23, for example, showed rising N/C and N/O values between 18.18 and 203.76 ADD, corresponding with external signs of decomposition such as bloating, purging, and insect activity. Subsequent declines in these ratios may be related to nitrogen loss through leaching or volatilization, including ammonia release. Notably, the N/O ratio showed the strongest positive relationship with ADD (estimate = 0.1522, marginal R2 = 0.4494), while N/C had the highest conditional R2 (0.5468), reinforcing the importance of accounting for donor-level variation in nitrogen profiles. Temporal spikes in these ratios, such as those observed in Donor 23 between 203.76 and 294.08 ADD, may also reflect environmental influences like rainfall.
The sulfur-to-carbon (S/C) and sulfur-to-oxygen (S/O) ratios exhibited moderate increases over time, particularly in donors at earlier decomposition stages. These trends are consistent with the release and transformation of sulfur-containing amino acids (e.g., cysteine, methionine) and the formation of sulfur-based volatile organic compounds (VOCs) such as hydrogen sulfide and dimethyl disulfide.8,49,71 In later stages, or during rain events (e.g., Donor 22 between 612.80 and 1183.68 ADD), these ratios decreased, likely due to environmental leaching.72 The S/O ratio demonstrated moderate explanatory power in mixed models (marginal R2 = 0.1997; conditional R2 = 0.3073), again highlighting inter-donor variability.
Phosphorus-to-carbon (P/C) and phosphorus-to-oxygen (P/O) ratios were comparatively low across all samples, reflecting the limited abundance of phosphorus relative to other heteroatoms. Still, P/C increased gradually over time, likely due to the degradation of phosphorus-rich compounds such as nucleic acids and phospholipids. These findings align with simultaneous degradation pathways affecting proteins, lipids, and nucleic acids, and suggest that phosphorus dynamics may act as a marker of biochemical progression.
To quantify these patterns, mixed-effects models were constructed using ADD as a fixed effect and donor as a random intercept term (Table 1).
The modelling framework allowed the separation of time-driven effects from inter-donor variability. Ratios like N/C and N/O showed strong associations with ADD, while others (e.g., H/C and S/O) were more sensitive to donor-specific influences. The inclusion of donor improved overall model fit, underscoring the need to account for factors such as age, body composition, and microbiome in postmortem chemistry.
3.4 Biomarker identification
Spearman's rank correlation was performed to determine compounds of interest across the dataset. This non-parametric measure helps determine the strength and direction of association between compound abundance and ADD. Only compounds with p ≤ 0.05 and presence in at least seven samples were retained for further interpretation, ensuring statistical robustness.In Fig. 7, each point represents a unique compound, coloured by Spearman's rho indicating the correlation between observed compound intensity and ADD. Green tones indicate a positive correlation with ADD, while purple tones reflect a negative correlation. Condensed aromatics appear to be positively correlated, suggesting the progressive oxidation and condensation of biomolecules into more structurally stable forms. The lipid region has both positive and negative correlations with ADD which shows variation within this region.
 | ||
| Fig. 7 Van Krevelen diagram containing compounds significantly correlated with ADD across all donors and time points (Spearman p ≤ 0.05, compounds present in ≥7 samples). | ||
All compounds were tentatively identified based on accurate mass (m/z) and predicted molecular formula. Compounds were categorized into three groups based on their likely origin: biological compounds included those with known associations to human, microbial, or bacterial processes; environmental compounds were attributed to external environmental sources; and chemical compounds encompassed all remaining identifiable species in PubChem that lacked an explicit connection to human decomposition (Table 2). These identifications are based on high-resolution molecular formula assignment however, each annotation remains tentative. Given the absence of MS/MS data or reference standards, biomarker annotations should be interpreted as provisional, reflecting molecular formulas that may correspond to multiple structural isomers or isobars.
| Observed m/z | Chemical formula | Tentative identification | Mass Accuracy (ppm) | Spearman's rho (ρ) | p value |
|---|---|---|---|---|---|
| Biological | |||||
| 93.00944 | C4H2N2O | Isocyanato acrylonitrile | 0.04 | −0.7857 | 0.0279 |
| 199.09760 | C10H16O4 | Decenedioic acid * | 0.09 | 0.6723 | 0.0473 |
| 217.00772 | C10H6N2O2S | 3-Thiocyanato-1H-indole-6-carboxylic acid | −0.01 | 0.5330 | 0.0088 |
| 242.01620 | C6H13NO5S2 | S-(3-Sulfopropyl)cysteine | −0.16 | 0.7063 | 0.0133 |
| 255.23300 | C16H32O2 | Palmitic acid * | 0.18 | 0.6446 | 0.0174 |
| 264.00098 | C8H3N5O6 | 5-[(6-Oxido-2,4-dioxo-1H-pyrimidin-5-yl)imino]-2,6-dioxopyrimidin-4-olate | −0.29 | 0.4968 | 0.0135 |
| 281.24862 | C18H34O2 | Oleic acid * | 0.06 | 0.4995 | 0.0031 |
| 299.25913 | C18H36O3 | 2-Hydroxyoctadecanoic acid * | −0.13 | 0.5730 | 0.0407 |
| 301.01118 | C14H10N2O2S2 | (2-Carbamimidoyl-1-benzothiophen-6-yl) thiophene-2-carboxylate | 0.29 | 0.7186 | 0.0446 |
| 326.01888 | C9H13NO10S | Pyridinium oxalate oxalic acid dihydrate | 0.43 | 0.6322 | 0.0498 |
| 328.18781 | C15H27N3O5 | Thr-Ile-Pro * | 0.05 | −0.7711 | 0.0251 |
| 411.20240 | C21H32O8 | ST 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2;O8 * 2;O8 * |
−0.10 | 0.8073 | 0.0154 |
| 455.26819 | C23H41N2O5P | Cyclohexanamine;[[4-(decanoylamino)phenyl]-hydroxymethyl]phosphonic acid | 0.34 | −0.6621 | 0.0265 |
| Environmental | |||||
| 209.11830 | C12H18O3 | Jasmonic acid | −0.09 | 0.5024 | 0.0172 |
| 228.00059 | C5H11NO5S2 | S-(2-Sulfoethyl)-L-cysteine | 0.01 | 0.2912 | 0.0295 |
| Chemical | |||||
| 126.00185 | C5H5NOS | Pyrithione | −0.46 | 0.5645 | 0.0227 |
| 205.00369 | C4H6N4O4S | 4,6-Diamino-2-hydroxy-5-pyrimidinesulfonic acid | −0.05 | 0.6925 | 0.0182 |
| 205.00769 | C9H6N2O2S | 5-[(Z)-4-Pyridinylmethylidene]-1,3-thiazolane-2,4-dione | −0.16 | 0.4626 | 0.0347 |
| 205.00943 | C7H11O3PS | (3-Propan-2-ylthiophen-2-yl) dihydrogen phosphite | 0.26 | 0.4133 | 0.0168 |
| 206.01576 | C5H10N3O2PS | Ligand 4272 | −0.47 | 0.4332 | 0.0149 |
| 228.00013 | C7H8N3O2PS | Azinphos-methyl | −0.34 | 0.4765 | 0.0456 |
| 228.00104 | C5H3N5O6 | N-(3,5-Dinitropyridin-2-yl)nitramide | −0.07 | 0.6602 | 3.93 × 10–5 |
| 228.00191 | C6H7N5OS2 | 5-{[(5-Methyl-1,2,4-oxadiazol-3-yl)methyl]sulfanyl}-1,3,4-thiadiazol-2-amine | −0.07 | 0.5526 | 0.0141 |
| 229.00769 | C11H6N2O2S | 2-(1,1-Dioxo-1-benzothiophen-3-ylidene)propanedinitrile | −0.14 | 0.7890 | 0.0008 |
| 229.00927 | C9H11O3PS | Fenamiphos | −0.46 | 0.4731 | 0.0261 |
| 229.01014 | C7H6N2O7 | 1,3-Dinitrosalicylic acid | −0.37 | 0.7499 | 4.36 × 10–6 |
| 230.01667 | C5H5N5O6 | Methyl (3,5-dinitro-1H-1,2,4-triazol-1-yl)acetate | −0.16 | 0.6619 | 0.0011 |
| 230.01750 | C6H9N5OS2 | 2-Methyl-4-[5-(methylsulfanyl)-1,3,4-thiadiazol-2-yl]-1,2,4-triazolidin-3-one | −0.33 | 0.5288 | 0.0137 |
| 231.02498 | C9H13O3PS | Phosphorothioic acid, O-ethyl S-(4-methylphenyl) ester | −0.20 | 0.5798 | 0.0074 |
| 233.03897 | C11H10N2O2S | 2-(Acetoacetamido)benzothiazole | −0.22 | 0.8627 | 0.0124 |
| 240.00187 | C7H7N5OS2 | 2-[(7-Sulfanylidene-3,6-dihydroimidazo[4,5-d]pyridazin-4-yl)sulfanyl]acetamide | −0.23 | 0.7804 | 0.0001 |
| 241.00933 | C10H11O3PS | 3-Dimethoxyphosphoryl-1-benzothiophene | −0.19 | 0.6527 | 2.86 × 10–5 |
| 242.01287 | C9H9NO5S | Oxine sulfate | 0.01 | 0.6258 | 0.0018 |
| 242.01666 | C6H5N5O6 | Diamino trinitrobenzene | −0.19 | 0.4537 | 0.0445 |
| 252.00100 | C7H3N5O6 | 3,5,7-Trinitro-1H-indazole | −0.22 | 0.5673 | 0.0006 |
| 252.00190 | C8H7N5OS2 | 2-(Pyrimidin-2-ylsulfanyl)-N-(1,3,4-thiadiazol-2-yl)acetamide | −0.10 | 0.6632 | 0.0010 |
| 253.01105 | C10H10N2O2S2 | N-(2-Aminophenyl)thiophene-2-sulfonamide | −0.17 | 0.8675 | 0.0053 |
| 254.01680 | C7H5N5O6 | Nitroso azoxy dinitrotoluene | 0.37 | 0.6090 | 0.0003 |
| 255.02334 | C13H8N2O2S | 6-Nitro-2-phenylbenzothiazole | −0.13 | 0.9550 | 0.0008 |
| 264.00193 | C9H7N5OS2 | N-(3-Cyanothiophen-2-yl)-2-(1H-1,2,4-triazol-3-ylsulfanyl)acetamide | 0.02 | 0.5041 | 0.0073 |
| 265.00697 | C6H10N4O4S2 | 1,3-Benzenedisulfonic acid, 1,3-dihydrazide | −0.38 | 0.6041 | 0.0490 |
| 265.01106 | C11H10N2O2S2 | N-(3-(Methylcarbamoyl)thiophen-2-yl)thiophene-2-carboxamide | −0.13 | 0.7785 | 0.0229 |
| 265.01183 | C8H11O8P | 2-Phosphanylbutane-1,2,3,4-tetracarboxylic acid | −0.18 | 0.8729 | 0.0103 |
| 277.00784 | C15H6N2O2S | 2,7-Dicyanoxanthone | 0.42 | 0.6445 | 0.0237 |
| 278.01628 | C9H13NO5S2 | 6-Isopropylsulfonyl-2-pyridyl methanesulfonate | 0.15 | 0.6810 | 0.0434 |
| 279.02260 | C7H12N4O4S2 | tert-Butyl N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)carbamate | −0.43 | 0.6786 | 0.0108 |
| 302.01844 | C9H10N3O7P | [5-(4-Azanyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-furan-2-yl]methyl dihydrogen phosphate | 0.26 | 0.5800 | 0.0147 |
| 303.02345 | C17H8N2O2S | 2,3-Dihydro-1H-indeno[2,1-b]quinoxaline-2,3-dione | 0.26 | 0.5404 | 0.0169 |
| 305.03913 | C17H10N2O2S | 2-(4-Phenylthiazol-2-yl)isoindoline-1,3-dione | 0.35 | 0.6662 | 0.0048 |
| 307.05475 | C17H12N2O2S | 2-[(E)-2-(3-Nitrophenyl)vinyl]quinoline | 0.25 | 0.7748 | 0.0408 |
| 325.01053 | C8H14N4O4S3 | Methyl N-[[(1-methylsulfanylethenylamino)oxycarbonylamino]sulfanylcarbamoyloxy]ethanimidothioate | 0.27 | 0.8929 | 0.0123 |
| 325.01108 | C16H10N2O2S2 | 2,2′-(1,3)Thiazolo(5,4-d)(1,3)thiazole-2,5-diyldiphenol | −0.04 | 0.9190 | 0.0034 |
| 326.01621 | C13H13NO5S2 | Methyl 3-{4-[(methylsulfonyl)amino]phenoxy}-2-thiophenecarboxylate | −0.09 | 0.4419 | 0.0395 |
| 348.00403 | C12H15NO5S3 | Sulopenem | 0.20 | 0.6154 | 0.0373 |
| 353.03846 | C13H14N4O4S2 | N-Carbamimidoyl-4-[(phenylsulfonyl)amino]benzenesulfonamide | 0.25 | −0.8539 | 0.0070 |
| 421.00872 | C17H15N2O5PS2 | Phosphonic acid, [[(benzo[b]thien-2-ylsulfonyl)amino]methyl]-,mono[4-(cyanomethyl)phenyl] ester | −0.01 | −0.6754 | 0.0459 |
| 455.26843 | C21H44O8S | Heptapropylene glycol | 0.04 | −0.7857 | 0.0480 |
Of these, six compounds matched entries in the Human Metabolome Database (HMDB) or Metabolomics Workbench and are marked with an asterisk (*) in Table 2. These reference-based matches suggest a likely endogenous or microbiome-derived human origin, supporting their relevance as decomposition biomarkers. Only the *-marked compounds were selected for mixed-effects modelling, which assessed the effect of ADD over time while accounting for inter-individual variability.
Among the tentatively identified compounds, several lipid-related molecules demonstrated significant positive correlations with time, supporting their role in the later stages of decomposition. Palmitic acid (C16H32O2, m/z 255.23300) and oleic acid (C18H34O2, m/z 281.24862) are well-established fatty acids associated with cellular membranes and adipose tissue.6 The detection of 2-hydroxyoctadecanoic acid (C18H36O3, m/z 199.09760), a hydroxylated fatty acid, further supports ongoing lipid transformation processes, possibly through microbial oxidation or enzymatic hydrolysis.24 All three compounds exhibited a positive correlation with ADD and their persistence over time highlights the relative chemical stability of lipids during decomposition, consistent with previous studies suggesting that lipids are among the most resistant biomolecular classes in decaying soft tissue.6,26,70
However, mixed-effects modelling of these compounds (Table 3) revealed limitations in estimating donor-level variance. While 2-hydroxyoctadecanoic acid yielded a strong conditional R2 (0.9923)—indicating robust donor-driven variation— the other five compounds failed to compute conditional R2 values. This is likely due to the limited presence of these compounds within multiple donors.
| Tentative identification | Est. | Std. error | CI low | CI high | R2 marg. | R2 con. |
|---|---|---|---|---|---|---|
| Decenedioic acid | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 961 961![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 223 223 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 244 244![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 836 836 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 561 561![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 424 424 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 361 361![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 022 022 |
0.540 | N/A |
| Palmitic acid | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 098 098![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 777 777 |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 931 931![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 148 148 |
−5 × 107 | 1.34 × 108 | 0.029 | N/A |
| Oleic acid | 2.41 × 108 | 4.55 × 108 | −6.5 × 108 | 1.13 × 109 | 0.012 | N/A |
| 2-Hydroxyoctadecanoic acid | 7.61 × 109 | 3.08 × 109 | 1.58 × 109 | 1.36 × 1010 | 0.188 | 0.992 |
| Thr-Ile-Pro | 2.86 × 10–6 | 1.91 × 10–5 | −3.45 × 10–5 | 4.02 × 10–5 | 0.003 | N/A |
ST 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2;O8 2;O8 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 856 856![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 540 540 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077 077![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 186 186 |
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 785 785![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 331 331 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 927 927![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 749 749 |
0.796 | N/A |
In contrast, the tentatively assigned tripeptide Thr-Ile-Pro (C15H27N3O5, m/z 328.18781) exhibited a strong negative correlation with ADD (p = −0.7711). This decline suggests progressive proteolysis and peptide degradation, a hallmark of early decomposition stages.73 The decrease in peptide intensity over time reflects the breakdown of structural and functional proteins, as well as the activity of both endogenous enzymes and microbial proteases.74 The low marginal R2 (0.0032) in mixed effects modelling implies a weak overall trend and points towards its specificity to early-stage decomposition.
Another biologically relevant compound tentatively identified, decenedioic acid (C10H16O4, m/z 328.18781), showed a positive association with ADD (ρ = 0.6723) and a moderate model fit (marginal R2 = 0.5403). This dicarboxylic acid may be a by-product of oxidative lipid degradation and microbial metabolism, marking its potential utility as a mid-to-late decomposition marker.69
Together, these findings demonstrate potential biochemical signatures associated with decomposition progression. Lipids and their derivatives generally persist or accumulate, while peptides decline over time, consistent with known biochemical pathways of postmortem soft tissue breakdown.6,23,24
3.5 Limitations and future considerations
As with many studies in taphonomy and PMI estimation, several limitations and challenges must be acknowledged when interpreting these findings. Inter-donor variability is known to influence results, highlighting the need for a larger dataset. Although this study includes 13 human donors—making it one of the largest biochemical datasets in high-resolution taphonomic research—this variability still underscores the need for even broader sampling across diverse biological and environmental backgrounds. Tissue type and sampling location also play an important role in shaping decomposition chemistry. Biochemical profiles may differ significantly between tissue type and anatomical regions. This introduces complexity in interpreting whole-body trends and highlights the value of future studies incorporating layer-specific or region-specific profiling to refine these findings. Additionally, while the current analysis was limited to soft tissue, incorporating other forensically relevant materials such as DNA, RNA, surrounding soil, and associated microbial communities could offer a more comprehensive and integrated perspective on postmortem changes.From a modelling perspective, although several potential compounds were significantly correlated to ADD, the conditional R2 could not be calculated for most. Similar challenges were observed in the modelling of the elemental ratios where low marginal R2 values indicated that much of the variability could not be explained by ADD alone. These findings suggest that elemental trends may also be heavily influenced by individual donor factors or other unmeasured variables, warranting further exploration in larger and more diverse cohorts.
All compound annotations were derived solely from accurate mass and predicted molecular formulae. These annotations are intended to illustrate biologically relevant examples that correspond to the proposed molecular formula. Given the high potential for isomeric overlap, the presence of isobars with identical formulas, and the limitations of current spectral databases, these assignments remain tentative, and the possibility of false positives are likely.
Despite these challenges, the findings presented here offer compelling insights into the chemical evolution of decomposing soft tissue and lay the groundwork for future predictive modelling. With continued expansion of the dataset and refinement of compound-specific and elemental markers, the development of a robust, high-resolution PMI estimation tool remains a feasible and promising direction for future forensic taphonomic applications.
4. Conclusions
Applying MALDI MS to decomposing human soft tissue marks a significant advancement in forensic taphonomy. This study demonstrates that high-resolution mass spectrometry can detect elemental and compound-level changes that correlate strongly with postmortem interval (PMI). Increases in N/C and N/O, reflected progressive protein and peptide degradation and microbial activity, while declines in O/C, suggested the loss of oxygen-rich volatile compounds. Sulfur-based ratios (S/C, S/O) exhibited stage-specific increases consistent with amino acid degradation and microbial by-products, while phosphorus-based ratios revealed subtle trends linked to nucleic acid and phospholipid breakdown. Linear mixed-effects modelling identified N/O and N/C as being strongly associated with ADD, with donor-level random effects improving model fit and highlighting inter-donor variability. Decomposition was shown to be highly donor-dependant, influenced by tissue composition, environmental exposure, and microbial dynamics.With data from 13 human donors spanning a wide range of PMIs, this study contributes one of the largest high-resolution biochemical datasets in forensic decomposition chemistry research. Integrating MALDI MS with elemental profiling provides a sensitive, compound-level lens through which decomposition can be monitored and quantified. As tissue- and environment-specific sampling improves, the path toward robust, chemically driven PMI models becomes increasingly feasible.
Author contributions
Jerika Ho: formal analysis, investigation, methodology, visualization, writing – original draft. Naomi L. Stock: methodology, writing – review & editing. Vaughn Mangal: formal analysis, writing – reviewing & editing. Shari Forbes: conceptualization, funding acquisition, project administration, supervision, writing – reviewing & editing. Theresa Stotesbury: conceptualization, funding acquisition, project administration, supervision, methodology, writing – reviewing & editing.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the ESI.† Additional data collected from human participants, are not available for confidentiality reasons.Acknowledgements
This research was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) through a Discovery Horizons Grant (DH-2022-00198). The authors would like to thank Colin Elliott, Lesley Hewett, Irina Badell, Dr Darshil Patel, and Dr Agathe Ribéreau-Gayon for their invaluable support in sampling and fieldwork. Thank you to Dr Mark Jermy for his advice with the matrix sprayer. We are also grateful to the Canadian Foundation for Innovation and Ontario Research Fund for funding the Bruker SolariX XR MALDI FT-ICR-MS in the Water Quality Centre at Trent University. Special thanks are extended to the members of the Forensic Chemistry and Materials Laboratory at Ontario Tech University for insights and feedback on writing and data visualization.References
- M. A. Iqbal, K. D. Nizio, M. Ueland and S. L. Forbes, TrAC, Trends Anal. Chem., 2017, 91, 112–124 CrossRef CAS.
- M. A. Iqbal, M. Ueland and S. L. Forbes, Aust. J. Forensic Sci., 2020, 52, 107–123 CrossRef.
- J. T. Pokines and S. A. Symes, Manual of Forensic Taphonomy, CRC Press, 1st edn, 2013 Search PubMed.
- M. S. Megyesi, S. P. Nawrocki and N. H. Haskell, J. Forensic Sci., 2005, 50, 1–9 CrossRef.
- S. J. Marhoff-Beard, S. L. Forbes and H. Green, Forensic Sci. Int., 2018, 291, 158–166 CrossRef PubMed.
- M. Ueland, S. Collins, L. Maestrini, S. L. Forbes and S. Luong, Forensic Chem., 2021, 24, 100335 CrossRef CAS.
- A. Mathur and Y. K. Agrawal, Aust. J. Forensic Sci., 2011, 43, 275–285 CrossRef.
- M. L. Goff, Exp. Appl. Acarol., 2009, 49, 21–36 CrossRef PubMed.
- J. A. Payne, Ecology, 1965, 46, 592–602 CrossRef.
- E. L. Pecsi, G. Bronchti, F. Crispino and S. L. Forbes, Forensic Sci. Int., 2020, 2, 287–292 Search PubMed.
- A. N. D. Nolan, R. J. Mead, G. Maker, S. Bringans and S. J. Speers, Forensic Sci. Int., 2019, 303, 109957 CrossRef CAS PubMed.
- J. Amendt, C. S. Richards, C. P. Campobasso, R. Zehner and M. J. R. Hall, Forensic Sci., Med., Pathol., 2011, 7, 379–392 CrossRef CAS PubMed.
- S. E. Donovan, M. J. R. Hall, B. D. Turner and C. B. Moncrieff, Med. Vet. Entomol., 2006, 20, 106–114 CrossRef CAS PubMed.
- J. K. Suckling, M. K. Spradley and K. Godde, J. Forensic Sci., 2016, 61, 19–25 CrossRef PubMed.
- L. Iancu, A. Bonicelli and N. Procopio, Front. Microbiol., 2024, 15, 1392716 CrossRef PubMed.
- A. Ribéreau-Gayon, D. O. Carter and S. Forbes, J. Forensic Sci., 2023, 68, 536–548 CrossRef PubMed.
- A. Ribéreau-Gayon, C. Rando, R. M. Morgan and D. O. Carter, Sci. Justice, 2018, 58, 167–176 CrossRef PubMed.
- N. Procopio and A. Bonicelli, Proteomics, 2024, 24, 1–47 CrossRef PubMed.
- A. Bonicelli, H. L. Mickleburgh, A. Chighine, E. Locci, D. J. Wescott and N. Procopio, eLife, 2022, e83658 CrossRef CAS PubMed.
- B. K. Pesko, S. Weidt, M. McLaughlin, D. J. Wescott, H. Torrance, K. Burgess and R. Burchmore, OMICS, 2020, 24, 649–659 CrossRef CAS PubMed.
- R. E. Keane, R. J. Tidy, G. J. Parker, J. P. A. Gummer and C. Priddis, WIREs Forensic Sci., 2024, 1–16 Search PubMed.
- L. J. Ward, S. Kling, G. Engvall, C. Söderberg, F. C. Kugelberg, H. Green and A. Elmsjö, iScience, 2024, 27 DOI:10.1016/j.isci.2024.109794.
- S. Collins, B. Stuart and M. Ueland, Forensic Sci. Int., 2023, 343, 111547 CrossRef CAS PubMed.
- S. Collins, L. Maestrini, M. Ueland and B. Stuart, Talanta Open, 2022, 5, 100100 CrossRef.
- A. N. Nolan, R. J. Mead, G. Maker and S. J. Speers, Aust. J. Forensic Sci., 2020, 52, 477–488 CrossRef.
- S. L. Forbes, B. H. Stuart and B. B. Dent, Forensic Sci. Int., 2005, 154, 24–34 CrossRef PubMed.
- D. Carter and M. Tibbett, Soil Analysis in Forensic Taphonomy, CRC Pre, Boca Raton, 2008 Search PubMed.
- M. Vandenbosch, S. P. Nauta, A. Svirkova, M. Poeze, R. M. A. A. Heeren, T. P. Siegel, E. Cuypers and M. Marchetti-Deschmann, Anal. Bioanal. Chem., 2021, 413, 2683–2694 CrossRef CAS PubMed.
- S. Francese, Aust. J. Forensic Sci., 2019, 51, 623–635 CrossRef.
- K. Kennedy, L. Cole, M. Witt, M. Sealey and S. Francese, Molecules, 2022, 27, 2039 CrossRef CAS PubMed.
- K. Kennedy, C. Gannicliffe, L. M. Cole, M. Sealey and S. Francese, Sci. Justice, 2022, 62, 602–609 CrossRef PubMed.
- K. Yeh, W. S. Burr, N. L. Stock and T. Stotesbury, Forensic Chem., 2020, 20, 100274 CrossRef CAS.
- P. J. Hart, S. Francese, E. Claude, M. N. Woodroofe and M. R. Clench, Anal. Bioanal. Chem., 2011, 401, 115–125 CrossRef CAS PubMed.
- P. M. Vaysse, R. M. A. Heeren, T. Porta and B. Balluff, Analyst, 2017, 142, 2690–2712 RSC.
- A. R. Buchberger, K. DeLaney, J. Johnson and L. Li, Anal. Chem., 2018, 90, 240–265 CrossRef CAS PubMed.
- R. J. A. Goodwin, J. C. Dungworth, S. R. Cobb and A. R. Pitt, Proteomics, 2008, 8, 3801–3808 CrossRef CAS PubMed.
- M. Statheropoulos, C. Spiliopoulou and A. Agapiou, Forensic Sci. Int., 2005, 153, 147–155 CrossRef CAS PubMed.
- M. Statheropoulos, A. Agapiou, E. Zorba, K. Mikedi, S. Karma, G. C. Pallis, C. Eliopoulos and C. Spiliopoulou, Forensic Sci. Int., 2011, 210, 154–163 CrossRef CAS PubMed.
- J. I. Muñoz, E. Costas, M. S. Rodríguez-Calvo, J. M. Suárez-Peñaranda, M. López-Rivadulla and L. Concheiro, Hum. Exp. Toxicol., 2006, 25, 279–281 CrossRef PubMed.
- L. Brockbals, S. Garrett-Rickman, S. Fu, M. Ueland, D. McNevin and M. P. Padula, Anal. Bioanal. Chem., 2023, 415, 5487–5498 CrossRef CAS PubMed.
- K. A. Perrault, T. Rai, B. H. Stuart and S. L. Forbes, Anal. Methods, 2015, 7, 690–698 RSC.
- J. Dekeirsschieter, P. H. Stefanuto, C. Brasseur, E. Haubruge and J. F. Focant, PLoS One, 2012, 7, e39005 CrossRef CAS PubMed.
- K. A. Perrault, K. D. Nizio and S. L. Forbes, Chromatographia, 2015, 78, 1057–1070 CrossRef CAS.
- S. Stadler, P. H. Stefanuto, M. Brokl, S. L. Forbes and J. F. Focant, Anal. Chem., 2013, 85, 998–1005 CrossRef CAS PubMed.
- C. Brasseur, J. Dekeirsschieter, E. M. J. Schotsmans, S. de Koning, A. S. Wilson, E. Haubruge and J. F. Focant, J. Chromatogr. A, 2012, 1255, 163–170 CrossRef CAS PubMed.
- A. Agapiou, E. Zorba, K. Mikedi, L. McGregor, C. Spiliopoulou and M. Statheropoulos, Anal. Chim. Acta, 2015, 883, 99–108 CrossRef CAS PubMed.
- L. Irish, S. R. Rennie, G. M. Parkes and A. Williams, Sci. Justice, 2019, 59, 503–515 CrossRef CAS PubMed.
- J. Ho, D. Patel, W. S. Burr, C. Samson and S. L. Forbes, Forensic Chem., 2024, 38, 100561 CrossRef CAS.
- M. Statheropoulos, A. Agapiou, C. Spiliopoulou, G. C. Pallis and E. Sianos, Sci. Total Environ., 2007, 385, 221–227 CrossRef CAS PubMed.
- M. A. Sacco, F. Cordasco, C. Scalise, P. Ricci and I. Aquila, Diagnostics, 2022, 12, 1490 CrossRef CAS PubMed.
- A. Rivas-Ubach, Y. Liu, T. S. Bianchi, N. Tolić, C. Jansson and L. Paša-Tolić, Anal. Chem., 2018, 90, 6152–6160 CrossRef CAS PubMed.
- H. J. Smith, M. Tigges, J. D'Andrilli, A. Parker, B. Bothner and C. M. Foreman, Limnol. Oceanogr. Lett., 2018, 3, 225–235 CrossRef CAS PubMed.
- V. Mangal, B. R. Stenzler, A. J. Poulain and C. Guéguen, Environ. Sci. Technol., 2019, 53, 157–165 CrossRef CAS PubMed.
- J. R. Laszakovits and A. A. MacKay, J. Am. Soc. Mass Spectrom., 2022, 33, 198–202 CrossRef CAS PubMed.
- R. J. A. Goodwin, A. Nilsson, D. Borg, P. R. R. Langridge-Smith, D. J. Harrison, C. L. Mackay, S. L. Iverson and P. E. Andrén, J. Proteomics, 2012, 75, 4912–4920 CrossRef CAS PubMed.
- P. Chaurand, J. L. Norris, D. S. Cornett, J. A. Mobley and R. M. Caprioli, J. Proteome Res., 2006, 5, 2889–2900 CrossRef CAS PubMed.
- A. M. Inc., Nebulizer User Instruction Manual – Model 705-470.
- T. Leefmann, S. Frickenhaus and B. P. Koch, Rapid Commun. Mass Spectrom., 2019, 33, 193–202 CrossRef CAS PubMed.
- D. Makowski, M. S. Ben-Shachar, I. Patil and D. Lüdecke, Journal of Open Source Software, 2020, 5, 2306 CrossRef.
- V. Mangal, W. Y. Lam, H. Huang, E. J. S. Emilson, R. W. Mackereth, C. P. J. Mitchell, E. J. S. Emilson and R. W. Mackereth, Biogeochemistry, 2022, 1, 127–144 CrossRef.
- J. W. Brooks, Vet. Pathol., 2016, 53, 929–940 CrossRef CAS PubMed.
- G. R. Dabbs, M. Connor and J. A. Bytheway, J. Forensic Sci., 2016, 61, 445–451 CrossRef PubMed.
- A. Sutherland, J. Myburgh, M. Steyn and P. J. Becker, Forensic Sci. Int., 2013, 231, 257–262 CrossRef CAS PubMed.
- C. P. Campobasso, G. Di Vella and F. Introna, Forensic Sci. Int., 2001, 120, 18–27 CrossRef CAS PubMed.
- D. O. Carter, D. Yellowlees and M. Tibbett, Appl. Soil Ecol., 2008, 40, 129–137 CrossRef.
- A. Galloway, W. H. Birkby, A. M. Jones, T. E. Henry and B. O. Parks, J. Forensic Sci., 1989, 34, 12680J CrossRef.
- A. Deo, S. L. Forbes, B. H. Stuart and M. Ueland, Aust. J. Forensic Sci., 2020, 52, 654–664 CrossRef.
- W. Wei, Q. Michu, D. Wenjuan, W. Jianrong, H. Zhibing, Y. Ming, J. Bo and L. Xia, Sci. Rep., 2020, 10, 1–7 CrossRef PubMed.
- V. A. Boumba, K. S. Ziavrou and T. Vougiouklakis, Forensic Sci. Int., 2008, 174, 133–151 CrossRef CAS PubMed.
- S. Collins, B. Stuart and M. Ueland, Aust. J. Forensic Sci., 2020, 52, 428–438 CrossRef.
- P. Armstrong, K. D. Nizio, K. A. Perrault and S. L. Forbes, Heliyon, 2016, 2 DOI:10.1016/j.heliyon.2016.e00070.
- D. Clases, M. Ueland, R. G. de Vega, P. Doble and D. Pröfrock, Talanta, 2021, 221, 121424 CrossRef CAS PubMed.
- A. Di Meo, D. Sohaei, I. Batruch, P. Alexandrou, I. Prassas and E. P. Diamandis, J. Proteome Res., 2021, 20, 444–452 CrossRef CAS PubMed.
- E. Rosier, E. Cuypers, M. Dekens, R. Verplaetse, W. Develter, W. Van De Voorde, D. Maes and J. Tytgat, Anal. Bioanal. Chem., 2014, 406, 3611–3619 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5an00588d |
| This journal is © The Royal Society of Chemistry 2025 |