Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Zwitterionic polymer with minimal reactivity against PEG antibodies to enhance the therapeutic effects of cytokine-targeting DNA aptamer†
Seojung
Cho
a,
Miyuki
Hori
b,
Ryosuke
Ueki
a,
Yutaro
Saito
 a,
Yukiko
Nagai
a,
Yukiko
Nagai
 a,
Haruka
Iki
a,
Akira
Tsuchiya
a,
Tomohiro
Konno
c,
Kensuke
Owari
b,
Haishun
Piao
b,
Kazunobu
Futami
b and
Shinsuke
Sando
a,
Haruka
Iki
a,
Akira
Tsuchiya
a,
Tomohiro
Konno
c,
Kensuke
Owari
b,
Haishun
Piao
b,
Kazunobu
Futami
b and
Shinsuke
Sando
 *ad
*ad
aDepartment of Chemistry and Biotechnology, Graduate School of Engineering, The University of Tokyo, 7-3-1, Hongo, Bunkyo-ku, Tokyo 113-8656, Japan. E-mail: ssando@chembio.t.u-tokyo.ac.jp
bTAGCyx Biotechnologies Inc. Komaba Open Laboratory, The University of Tokyo, 4-6-1 Komaba, Meguro-ku, Tokyo, 153-0041, Japan
cGraduate School of Pharmaceutical Sciences, Tohoku University, 6-3 Aoba, Aramaki, Aoba-ku, Sendai, 980-8578, Japan
dDepartment of Bioengineering, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan
First published on 4th February 2025
Abstract
Overcoming poor in vivo pharmacokinetics is a critical challenge in developing therapeutic aptamers, and conjugation to poly(ethylene glycol) (PEG) is a well-established technique for aptamers to prolong blood circulation. However, the existence of antibodies that specifically recognize PEG and their adverse effects on in vivo behaviors have been increasingly reported, highlighting the necessity of alternative modification strategies for aptamers. To address this issue, we focused on a zwitterionic polymer, particularly poly(2-methacryloyloxyethyl phosphorylcholine) (PMPC), as a PEG alternative to modify DNA aptamers. We conjugated PMPC to a DNA aptamer targeting IFN-gamma and investigated the properties of the PMPC-conjugated DNA aptamer as a therapeutic agent. PMPC modification did not affect the neutralizing activity of the aptamer. PMPC demonstrated lower reactivity against anti-PEG antibodies than PEG-like aptamer modifiers previously reported to exhibit low reactivity against PEG antibodies. In addition, PMPC extended the blood circulation time of the aptamer as long as or longer than PEG with a similar molecular size. In the LPS-induced inflammation animal model, the survival rate after treatment with the PMPC-aptamer conjugate was significantly superior to that with unmodified aptamer. These results indicate that PMPC has potential as an aptamer or other nucleic acid drug modifier to replace or be compatible with PEG.
Introduction
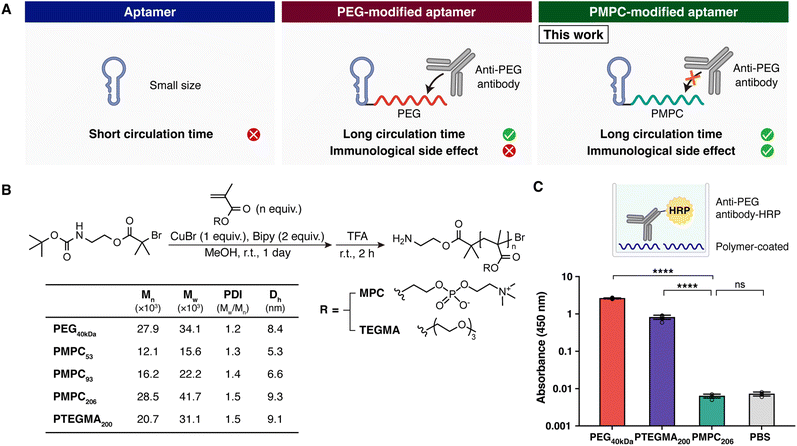
Aptamers, also called chemical antibodies, are highly structured, short, single-stranded oligonucleotides that bind to their targets with high affinity and selectivity.1–3 Because they can be chemically synthesized rather than produced biologically and are resistant to heat denaturation unlike antibodies, researchers have been attempting to develop aptamers for use in targeted therapy, in vitro diagnosis, and in vivo imaging.2Proper modifications are often introduced for nucleic acid-based therapeutics, including aptamers, to tailor their pharmacokinetic properties. Without any modification, aptamers are rapidly eliminated from the in vivo circulation. This is because aptamers are mostly smaller (6–30 kDa) than the glomerular effective size cutoff (30–50 kDa) and are subject to renal filtration.2 Therefore, while several aptamers have been or were in clinical trials so far, most of them were conjugated to polymers to lengthen their circulation times, thereby decreasing the dosing frequency and enhancing their efficacy.3
Poly(ethylene glycol) (PEG) has long been considered to be biocompatible and non-immunogenic, making PEG modification (PEGylation) a primary option for drug delivery.4 However, there is an increasing number of reports that repeated injection of PEG-modified materials can evoke the production of anti-PEG antibodies.5 These anti-PEG antibodies could specifically bind to PEGylated drugs and trigger the immune response.6 In the phase 2b clinical trial of the PEGylated RNA aptamer Pegnivacogin, 3 of 640 patients experienced severe allergic reactions after the first dose, to which a high level of pre-existing anti-PEG antibodies is considered to majorly contribute, resulting in early termination of the trial.7 Due to exposure to various consumer products containing PEG, anti-PEG antibodies were revealed to be prevalent even in patients naive to PEGylated drugs.6 Furthermore, administration of PEGylated pharmaceuticals, including mRNA vaccines for SARS-CoV2, continues to increase, heightening the possibility that more patients possess higher levels of anti-PEG antibodies.8 This emphasizes the necessity for researchers to look for alternatives to PEG for modifying aptamers.
Ozer and colleagues reported that poly(triethylene glycol methyl ether methacrylate) (PTEGMA), a PEG-like brush polymer, conjugated on an aptamer is much less reactive to PEG antibodies compared to the PEGylated aptamer.9 However, it has also been reported that PTEGMA is still recognized by anti-PEG antibodies at a non-negligible level,10 which is reasonable considering the chemical structure of PTEGMA including ethylene glycol groups.
Meanwhile, zwitterionic polymers, known for their antifouling ability achieved by high levels of hydration through electrostatic interactions,11 are an attractive choice for aptamer modification. Poly(2-methacryloyloxyethyl phosphorylcholine) (PMPC) is a bioinspired zwitterionic polymer with a chemical structure similar to that of a biomembrane.12,13 Molecular simulations showed that PMPC exhibits tightly bound and structured hydration via ionic solvation due to the abundance of charges in the side chain, which contributes to the resistance to interactions with other substances,14 thus minimizing the chance of recognition by the immune system.15 Due to their high biocompatibility and non-immunogenicity,16 PMPC-based polymers have been FDA-approved and successfully used as coating materials in medical devices.17 Therefore, PMPC has also been adopted as an alternative to PEG for prolonging the circulation time of nanoparticles and proteins. For instance, Jackson and colleagues used PMPC-based polyplex coronas to deliver siRNA, which surpassed PEG-based polyplexes in terms of circulation half-lives following intravenous delivery and bioactivity in tumors.18 However, the potency of PMPC as an aptamer modifier has not been investigated.
Here in this research, we aim to evaluate the function of the zwitterionic polymer, particularly PMPC, as a DNA aptamer modifier: whether PMPC modification can deal with the problem of the short half-lives of DNA aptamers without impairing the function of DNA aptamers and overcome the immunogenicity issues that arise when modifying aptamers with PEG (Fig. 1A). In the last part of this report, the therapeutic efficacy of a DNA aptamer targeting murine interferon (IFN)-gamma modified with PMPC is studied in an animal model with lipopolysaccharide (LPS)-induced endotoxic shock.
Results and discussion
Synthesis of polymeric aptamer modifiers and evaluation of their reactivity against anti-PEG antibodies
PMPC and PTEGMA used in this study were synthesized by atom transfer radical polymerization using 2-((tert-butoxycarbonyl)amino)ethyl 2-bromo-2-methylpropanoate as a starting compound (Fig. 1B). After the polymerization reaction, the terminal tert-butoxycarbonyl protecting group was deprotected by trifluoroacetic acid (TFA) treatment to afford PMPC and PTEGMA with terminal amino groups. The degree of polymerization and molecular weight profile were determined by 1H NMR and gel permeation chromatography (GPC), respectively.We first investigated the anti-PEG reactivity of the polymers with approximately the same number of repeating units per molecule, PMPC206 and PTEGMA200 (Fig. 1C). Mouse anti-PEG antibodies (1D9-6), selective to the PEG backbone, were used for the assay. PTEGMA did exhibit less binding to anti-PEG antibodies than PEG, as reported. However, it was also reported that anti-PEG antibodies recognizing the ethylene glycol (EG) backbone bind to PEG-like brush polymers when there are more than two EG moieties in repeated units, while poly(diethylene glycol methacrylate) bearing only two EG repeats is not appropriate for in vivo use due to its lower critical solution temperature, above which it becomes hydrophobic and loses its antifouling effect.10,19–21 Consistent with this, PTEGMA, which has 3 EG moieties in each repeating unit, could not completely prevent anti-PEG antibodies from binding, as shown in Fig. 1C. On the other hand, PMPC was less recognized by anti-PEG antibodies than PTEGMA. These results suggest that PMPC, which has the potential to successfully evade the immune system, could be a new polymeric modifier for DNA aptamers.
Synthesis of polymer-aptamer conjugates and evaluation of their reactivity against anti-PEG antibodies
In the present study, an aptamer, TXB0063, that binds to murine IFN-gamma was used (Fig. 2A). TXB0063 is a 62-nucleotide DNA aptamer (19.7 kDa without polymer modification) with the expanded genetic alphabet 7-(2-thienyl)imidazo[4,5-b]pyridine (Ds) for enhanced affinity and a mini-hairpin structure at the 3′-terminus for improved serum stability. TXB0063 binds to IFN-gamma with a KD value of 2.47 nM and inhibits its activity in cellulo,22 but does not function in vivo effectively because of its rapid clearance and requires appropriate modification.Aptamer-polymer conjugates were synthesized by copper-free strain-promoted alkyne–azide cycloaddition (SPAAC, Fig. 2B). PMPC53, 93, 206, PTEGMA200, and PEG40kDa with a terminal amino group, shown in Fig. 1B, were the polymer modifiers used. The dibenzocyclooctyne (DBCO) group was introduced into the polymer terminus (DBCO-polymer, Fig. S1† for typical 1H NMR spectra). TXB0063 was modified with an azide group (TXB0063-azide) through the reaction between an azido-PEG4-N-hydroxysuccinimide ester and an amine group on the thymine residue, shown in red in Fig. 2A. TXB0063-polymer conjugates were prepared using the SPAAC click chemistry between the DBCO-polymer and TXB0063-azide through a gradual freeze-thawing method23 and purified by ion-exchange chromatography (Fig. 2B and S2†).
To characterize the synthesized conjugates, the anti-PEG reactivity of TXB0063-polymer conjugates was examined by the competitive enzyme-linked immunosorbent assay (ELISA) (Fig. 2C). TXB0063-PMPC206 showed a significantly lower reactivity than TXB0063-PTEGMA200 and TXB0063-PEG40kDa. Since TXB0063 without polymer modification did not show non-specific binding to the anti-PEG antibody, the antibody is considered to recognize PTEGMA and PEG, as was the case for the polymer alone (Fig. 1C).
Evaluation of the effect of PMPC modification on the molecular size and in vivo circulation time of aptamers
The conjugates were also analyzed by size exclusion chromatography (SEC) (Fig. 2D). It suggests that modification with PMPC increased the size of the aptamer in a molecular weight-dependent way, with TXB0063-PMPC206 having a comparable size to TXB0063-PEG40kDa.After confirming that TXB0063 with a 3′-mini hairpin structure24 is more stable in serum than single-stranded DNA with no predictable secondary structure and that PMPC modification does not affect the stability of TXB0063 in serum (Fig. S3†), we evaluated the in vivo pharmacokinetic properties. TXB0063 conjugated to PEG40kDa and PMPC53, 93, 206 was intravenously administered (40 nmol kg−1) into rats, and the concentration of aptamers in the blood circulation over time was quantified by qPCR (Fig. 2E). All aptamer-polymer conjugates showed extended half-lives during the distribution phase (α phase t1/2) from 0.09 h for non-modified TXB0063 to 0.18–0.54 h. TXB0063-PMPC conjugates exhibited size-dependent improvement in plasma retention half-lives. Notably, PMPC206 surpassed PEG40kDa, a conventional choice for aptamer modification, improving both the half-life and AUC of TXB0063 better than PEG40kDa.
Considering that PMPC206, determined by GPC and DLS (Fig. 1B), was close in size to PEG40kDa and TXB0063-PMPC206 was shown to be with a similar size to TXB0063-PEG40kDa in SEC (Fig. 2D), this result suggests that PMPC with a comparable molecular size can work correspondingly and be used as a substitute for PEG to prolong the survival of DNA aptamers in blood circulation.
Therefore, we focused on PMPC206 as a modifier capable of enhancing aptamer circulation and reducing the reactivity to anti-PEG antibodies, and proceeded to more detailed in vitro and in vivo evaluation studies of TXB0063-PMPC206.
Evaluation of in cellulo and in vivo function of the PMPC-modified aptamer
We evaluated TXB0063-PMPC206 for its function as an inhibitor for IFN-gamma activity. Initially, we investigated the IFN-gamma inhibitory activity of TXB0063-PMPC206 in cultured cells. IFN-gamma induces the phosphorylation of signal transducer and activator of transcription (STAT1), and TXB0063 can bind to the IFN-gamma aptamer to function as an inhibitor (Fig. 3A).22 In the presence of 0.3–300 nM TXB0063, TXB0063-PMPC206, or TXB0063-PEG40kDa, IFN-gamma (2 ng mL−1, 129 pM as a monomer) was added to the murine fibroblast cell line L929 and incubated at 37 °C for 15 min. STAT1 phosphorylation levels were assessed using flow cytometry after immunostaining (Fig. 3B).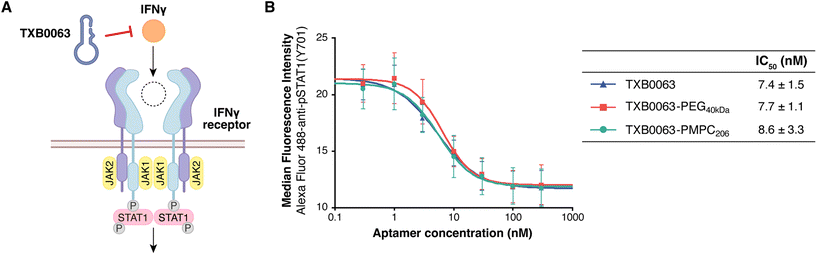 | ||
| Fig. 3 (A) Inhibitory mechanism of TXB0063 in IFN-gamma signaling. (B) Inhibition of STAT1 phosphorylation by TXB0063 and TXB0063-polymer conjugates. L929 cells were treated with IFN-gamma (2 ng mL−1) at 37 °C for 15 minutes in the presence of TXB0063 or TXB0063-polymer conjugates. After being washed with ice-cold PBS, the cells were fixed, permeabilized, and immunostained to measure the STAT1(Tyr701) phosphorylation level by flow cytometry. Plotted curves and the median fluorescence intensities are representative of three independent experiments performed in triplicate. IC50 values are expressed in average ± standard deviation of the values obtained from each experiment. Inhibition experiments using TXB0063-PTEGMA200 and random oligonucleotide control are shown in Fig. S4.† | ||
TXB0063-PMPC206 demonstrated a concentration-dependent neutralizing effect on IFN-gamma, with an IC50 value of 8.6 nM, which is not significantly different from 7.4 nM and 7.7 nM of TXB0063 and TXB0063-PEG40kDa, respectively. These results indicate that PMPC modification does not compromise the aptamer's binding and neutralizing capabilities. Circular dichroism (CD) spectra further support these results. The CD spectrum of TXB0063-PMPC206 shows no notable difference from TXB0063 with no polymer modification (Fig. S5†). These results suggest that PMPC modification does not alter the conformation and inhibitory efficacy of TXB0063.
Finally, we investigated if PMPC modification can realize the therapeutic efficacy of TXB0063 in vivo, by systemic administration. In the previous study, TXB0063 was shown to mitigate bladder inflammation in Hunner-type interstitial cystitis-like model mice, when applied locally to the bladder via intravesical instillation.22 In addition, previous observations on animal models and humans have suggested that neutralization of IFN-gamma could be an effective therapeutic approach to hyperinflammation.25 We, therefore, tested the therapeutic potential of TXB0063 using a lipopolysaccharide (LPS)-induced endotoxic shock model.
IFN-gamma-deficient mice are resistant to endotoxic shock,26,27 and mice treated with anti-IFN-gamma antibodies can better survive LPS-mediated septic shock.28,29 Since TXB0063 is an IFN-gamma binding aptamer, if it functions as intended in vivo with blood circulation extended by PMPC modification, it is expected to neutralize IFN-gamma, alleviate acute inflammatory responses, and improve the survival rate of the LPS-injected mice.
We monitored the survival rate of mice after intravenous administration of PBS, TXB0063, and TXB0063-PMPC199 at a dose of 10 mg kg−1 (aptamer equivalent) injection at 4, 24, and 48 hours after LPS injection (12.5 mg kg−1) (Fig. 4A). During the one-week observation period, only 10% of mice that received PBS control survived longer than 3 days. When treated with TXB0063, the survival rate of mice was no better than the PBS-treated group, with only 10% survival by the 4th day. In contrast, TXB0063-PMPC199 significantly improved survival rates, reaching 50% by the end of the observation week (Fig. 4B). This result clearly shows that PMPC modification significantly enhances the function of cytokine-inhibitory aptamers to achieve therapeutic efficacy in vivo.
Conclusions
In the present study, we report a strategy to improve the pharmacokinetic properties of DNA aptamers by modifying them with the zwitterionic polymer PMPC.DNA aptamers often require polymeric modifications for clinical applications, as their molecular weights are usually below the molecular cut-off for glomerular filtration. PEG has been considered non-immunogenic and commonly used to modify aptamers and various injectable drugs. However, it has been revealed that anti-PEG antibodies are generated and can trigger immune reactions. With the increased sensitivity of assays and shared awareness about the immunogenicity of PEG, it has been increasingly reported that many healthy individuals, even without known exposure to PEGylated drugs, possess anti-PEG antibodies. This is due to daily exposure to PEG-containing products such as food or cosmetics, increasing the probability of possible immune reactions that occur when these antibodies bind to PEGylated aptamers.
PTEGMA, a polymer with three ethylene glycol moieties grafted along its methyl methacrylate backbone, was previously proposed as a candidate to modify aptamers with reduced reactivity against anti-PEG antibodies. However, our investigation showed that PTEGMA cannot fully evade recognition by anti-PEG antibodies due to its PEG-like chemical structure. On the other hand, PMPC, which includes no ethylene glycol backbone structure, was found not to bind to anti-PEG antibodies.
Aside from minimizing the potential for immunological side effects against PEG antibodies, it is notable that PMPC fulfilled its role as a macromolecular modifier to improve the in vivo behavior of DNA aptamers. First, PMPC206 modification did not affect the structure of TXB0063, a DNA aptamer targeting murine IFN-gamma, and retained its neutralizing activity. Second, PMPC206 modification prolonged the half-life of TXB0063, to a level comparable to or slightly superior to PEG40kDa. Finally, prolonged circulation of the PMPC-modified aptamer contributed to its enhanced therapeutic efficacy. While no difference in the survival rate was observed between TXB0063 and PBS control in the LPS-induced endotoxic shock model, the PMPC-modified aptamer significantly improved the survival rate from 10% to 50%. Further investigation on the optimization of the dose and frequency of administration may enhance the efficiency of the therapeutic potential of this aptamer.
These results comprehensively support the potential of PMPC as an aptamer modifier that can replace or be used alongside PEG. It is also expected that our approach of using PMPC as an aptamer modifier may be extended to other aptamer therapeutics, including those previously studied with PEG modifications as well as newly discovered aptamers.
Author contributions
S.S. conceived the project. S.C., M.H., R.U., K.F. and S.S. designed the experiments. S.C., M.H., R.U., Y.S., Y.N., H.I., K.O., H.P., and K.F. carried out experiments and analyzed data, with support from A.T. and T.K. R.U. and S.S. provided resources and supervision. S.C. and S.S. wrote the manuscript, and all authors commented on and approved the manuscript.Ethical approval
All animal experiments were performed with the approval of the Animal Care and Use Committee of the University of Tokyo, in accordance with the guidelines for the care and use of laboratory animals as stated by this institution.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare the following competing financial interests: the authors (S.C., M.H., R.U., Y.S., K.O., H.P., K.F., and S.S.) have filed a patent application (PCT/JP2022/022601). All other authors declare that they have no competing interests.Acknowledgements
This research was partially supported by JSPS KAKENHI [Grant Number JP24KJ0742 (to S. C.)]. We thank Prof. Takai and Dr. Masuda for their support in measuring DLS. This study was financially supported partially by TAGCyx Biotechnologies Inc.References
- C. Chen, S. Zhou, Y. Cai and F. Tang, npj Precis. Oncol., 2017, 1, 37 CrossRef PubMed.
- J. Zhou and J. Rossi, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef CAS PubMed.
- S. Ni, Z. Zhuo, Y. Pan, Y. Yu, F. Li, J. Liu, L. Wang, X. Wu, D. Li, Y. Wan, L. Zhang, Z. Yang, B. T. Zhang, A. Lu and G. Zhang, ACS Appl. Mater. Interfaces, 2021, 13, 9500–9519 CrossRef CAS PubMed.
- Y. Gao, M. Joshi, Z. Zhao and S. Mitragotri, Bioeng. Transl. Med., 2024, 9, e10600 CrossRef CAS PubMed.
- A. Moreno, G. A. Pitoc, N. J. Ganson, J. M. Layzer, M. S. Hershfield, A. F. Tarantal and B. A. Sullenger, Cell Chem. Biol., 2019, 26, 634–644 CrossRef CAS PubMed.
- B. M. Chen, T. L. Cheng and S. R. Roffler, ACS Nano, 2021, 15, 14022–14048 CrossRef CAS PubMed.
- N. J. Ganson, T. J. Povsic, B. A. Sullenger, J. H. Alexander, S. L. Zelenkofske, J. M. Sailstad, C. P. Rusconi and M. S. Hershfield, J. Allergy Clin. Immunol., 2016, 137, 1610–1613 CrossRef CAS PubMed.
- Y. Ju, W. S. Lee, E. H. Pilkington, H. G. Kelly, S. Li, K. J. Selva, K. M. Wragg, K. Subbarao, T. H. O. Nguyen, L. C. Rowntree, L. F. Allen, K. Bond, D. A. Williamson, N. P. Truong, M. Plebanski, K. Kedzierska, S. Mahanty, A. W. Chung, F. Caruso, A. K. Wheatley, J. A. Juno and S. J. Kent, ACS Nano, 2022, 16, 11769–11780 CrossRef CAS PubMed.
- I. Ozer, G. A. Pitoc, J. M. Layzer, A. Moreno, L. B. Olson, K. D. Layzer, A. M. Hucknall, B. A. Sullenger and A. Chilkoti, Adv. Mater., 2022, 34, 2107852 CrossRef CAS PubMed.
- D. Y. Joh, Z. Zimmers, M. Avlani, J. T. Heggestad, H. B. Aydin, N. Ganson, S. Kumar, C. M. Fontes, R. K. Achar, M. S. Hershfield, A. M. Hucknall and A. Chilkoti, Adv. Healthcare Mater., 2019, 8, 1801177 CrossRef PubMed.
- H. Qian, K. Wang, M. Lv, C. Zhao, H. Wang, S. Wen, D. Huang, W. Chen and Y. Zhong, J. Controlled Release, 2022, 343, 492–505 CrossRef CAS PubMed.
- K. Ishihara, Sci. Technol. Adv. Mater., 2022, 23, 498–524 CrossRef CAS PubMed.
- Y. Iwasaki and K. Ishihara, Sci. Technol. Adv. Mater., 2012, 13, 064101 CrossRef CAS PubMed.
- Y. He, J. Hower, S. Chen, M. T. Bernards, Y. Chang and S. Jiang, Langmuir, 2008, 24, 10358–10364 CrossRef CAS PubMed.
- Z. Zeng, S. Chen and Y. Chen, ChemMedChem, 2023, 18, e202300245 CrossRef CAS PubMed.
- H. Ou, T. Cheng, Y. Zhang, J. Liu, Y. Ding, J. Zhen, W. Shen, Y. Xu, W. Yang, P. Niu, J. Liu, Y. An, Y. Liu and L. Shi, Acta Biomater., 2018, 65, 339–348 CrossRef CAS PubMed.
- K. Ishihara, J. Biomed. Mater. Res., Part A, 2019, 107, 933–943 CrossRef CAS PubMed.
- M. A. Jackson, T. A. Werfel, E. J. Curvino, F. Yu, T. E. Kavanaugh, S. M. Sarett, M. D. Dockery, K. V. Kilchrist, A. N. Jackson, T. D. Giorgio and C. L. Duvall, ACS Nano, 2017, 11, 5680–5696 CrossRef CAS PubMed.
- A. Ramírez-Jiménez, C. Alvarez-Lorenzo, A. Concheiro and E. Bucio, Radiat. Phys. Chem., 2014, 99, 53–61 CrossRef.
- J. F. Lutz and A. Hoth, Macromolecules, 2006, 39, 893–896 CrossRef CAS.
- E. Wassel, S. Jiang, Q. Song, S. Vogt, G. Nöll, S. I. Druzhinin and H. Schönherr, Langmuir, 2016, 32, 9360–9370 CrossRef CAS PubMed.
- Y. Akiyama, K. Harada, J. Miyakawa, K. J. Kreder, M. A. O'Donnell, M. Daichi, H. Katoh, M. Hori, K. Owari, K. Futami, S. Ishikawa, T. Ushiku, H. Kume, Y. Homma and Y. Luo, iScience, 2023, 26, 108262 CrossRef CAS PubMed.
- H. Takemoto, K. Miyata, T. Ishii, S. Hattori, S. Osawa, N. Nishiyama and K. Kataoka, Bioconjugate Chem., 2012, 23, 1503–1506 CrossRef CAS PubMed.
- K. I. Matsunaga, M. Kimoto, C. Hanson, M. Sanford, H. A. Young and I. Hirao, Sci. Rep., 2015, 5, 18478 CrossRef CAS PubMed.
- F. De Benedetti, G. Prencipe, C. Bracaglia, E. Marasco and A. A. Grom, Nat. Rev. Rheumatol., 2021, 17, 678–691 CrossRef CAS PubMed.
- S. Nakamura, T. Otani, Y. Ijiri, R. Motoda, M. Kurimoto and K. Orita, J. Immunol., 2000, 164, 3330–3336 CrossRef CAS PubMed.
- B. D. Car, V. M. Eng, B. Schnyder, L. Ozmen, S. Huang, P. Gallay, L. Heumann, M. Aguet and B. Ryffel, J. Exp. Med., 1994, 179, 1437–1444 CrossRef CAS PubMed.
- M. M. U. Zaman, K. Masuda, K. K. Nyati, P. K. Dubey, B. Ripley, K. Wang, J. P. Chalise, M. Higa, H. Hanieh and T. Kishimoto, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 11543–11548 Search PubMed.
- H. Heremans, J. O. Van Damme, C. Dillen, R. Dijkmans and A. Billiau, J. Exp. Med., 1990, 171, 1853–1869 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4bm01541j |
| This journal is © The Royal Society of Chemistry 2025 |