Open Access Article
Open Access ArticleIntegration of transcriptomics and proteomics data for understanding the mechanisms of positive effects of carbon-based nanomaterials on plant tolerance to salt stress†
Sajedeh Rezaei
Cherati
and
Mariya V.
Khodakovskaya
 *
*
Department of Biology, University of Arkansas at Little Rock, Little Rock, Arkansas 72204, USA. E-mail: mvkhodakovsk@ualr.edu
First published on 18th June 2025
Abstract
Carbon-based nanomaterials (CBNs) can regulate plant responses to environmental stresses. Understanding of the biological mechanisms underlying these positive effects is limited. Integrating transcriptomics, proteomics, and metabolomics data—known as multi-omics data integration—is a powerful strategy for uncovering the molecular mechanisms underlying the effects of CBNs on a plant's molecular level, providing detailed insights into their biological impacts. Here, we combined transcriptomic (RNA-Seq) and proteomics (Tandem MS) data to understand mechanisms of improvement of tolerance to salt stress in tomato plants exposed to CBNs (carbon nanotubes (CNTs) and graphene). At the proteome level, exposure to CNTs resulted in complete restoration of the expression of 358 proteins and partial restoration of the expression of 697 proteins in tomato seedlings exposed to salt stress. Similarly, exposure to graphene resulted in the complete restoration of 587 proteins and the partial restoration of 644 proteins affected by salt stress. In the integrative analysis of transcriptomics and proteomics data 86 upregulated and 58 downregulated features showed the same expression trend (restoration expression towards normal level) at both “omics” levels in NaCl-stressed seedlings exposed to CBNs. Our data indicated that elevated salt tolerance of CBN-treated tomato plants can be associated with the activation of MAPK and inositol signaling pathways, enhancing the ROS clearance, stimulation of hormonal and sugar metabolisms, regulation of water uptake through work of aquaporins, regulation of the production of heat-shock proteins, and promotion of the production of secondary metabolites with defense functions.
Environmental significanceUnderstanding the impact of CBNs, such as CNTs and graphene on plants under salt stress is essential for evaluating their environmental significance. These nanomaterials have emerged as promising tools for enhancing plant resilience to environmental stressors, including salinity, which remains a significant challenge for global agriculture. CBNs have been shown to improve plant stress tolerance by modulating key physiological and molecular pathways, such as water transport, reactive oxygen species (ROS) scavenging, and hormone signaling. To gain deeper insight into their mode of action, an integrative “omics” approach—combining transcriptomics and proteomics—offers a powerful strategy to analyze CBN-induced molecular changes in stressed plants. This approach enables the identification of conserved stress-responsive genes, proteins, and metabolites, providing a comprehensive understanding of the regulatory networks influenced by CBNs. Such findings are critical for optimizing the use of nanomaterials in sustainable agriculture while assessing their broader ecological impact. |
Introduction
Over the past few decades, engineered nanomaterials (ENMs) have emerged as key innovations in modern agricultural practices.1 Their applications primarily focus on nanopesticides, nanoherbicides, nanofertilizers, nanosensors, nanocarriers, and soil supplements.2–7 Among these, CBNs have been identified as highly effective nano-regulators of seed germination,8 plant growth,9 and plant resilience to environmental stress.10,11 Understanding the precise biological mechanisms underlying the beneficial effects of CBNs is essential for their future commercialization in plant agriculture.12 However, elucidating these mechanisms remains challenging due to the nanoscale size of ENMs and the inherent difficulties in visualizing and quantifying nanoparticle uptake in plant cells.13 Nevertheless, investigating the molecular-level effects of CBNs such as their influence on gene expression, protein synthesis, and metabolite production offers valuable insights into their mode of action. Recent studies have identified several genes affected by CBN exposure, particularly those involved in water channel regulation. For instance, aquaporin-related genes exhibited significant upregulation in tomato plants14 and salt-treated broccoli plants exposed to CNTs.15 This suggests a potential link between enhanced water uptake, aquaporin activation, and improved growth in CBN-treated crops.14,15 Transcriptome-wide analyses have further revealed that CBN-induced gene expression changes are highly complex, simultaneously activating and suppressing various genes involved in key signaling pathways.11 Specifically, CNTs and graphene have been shown to partially or fully restore the expression of multiple genes negatively impacted by salt and water-deficit stresses in agricultural crops such as tomatoes, sorghum, and rice.11 These genes are associated with abscisic acid (ABA), inositol triphosphate (InsP3), and mitogen-activated protein kinase (MAPK) stress signaling pathways.11 Such findings highlight the importance of integrating multi-omics approaches in the study of plant-nanoparticle interactions. Given that nanomaterials influence the entire multi-omics network, analyzing the effects of CBNs on gene expression must be complemented by comprehensive assessments at the proteomic and metabolomic levels. Proteomic analyses can provide deeper mechanistic insights by examining the functional translation of genes and their impact on plant physiology.16 Several reports have been published about using the proteomics approach to clarify the molecular base of the plant response to the application of different nanomaterials. Recently, Li et al. examined the impact of molybdenum (Mo)-based nanofertilizer and copper (Cu)-based nanopesticide on wheat using physiological measurements, metal uptake analysis, and targeted proteomics.17 Mo exposure, particularly through roots, led to significant upregulation of 16 proteins across 11 metabolic pathways, showing a dose-dependent response affecting physiological measurements. Cu exposure resulted in tissue-specific effects, notably downregulation of 18 proteins involved in 11 metabolic pathways, in leaf tissues, emphasizing the plants' rapid response to Cu-induced stress. These findings elucidate plant responses to tested nanomaterials, providing valuable information for crop nutrient management practices.17Data collected from metabolomics studies can not only further explain the biological mechanism of nanomaterials interacting with crops but also provide valuable information for risk assessment of nanomaterials potentially reaching the food chain. Previously, McGehee et al. demonstrated that using CNTs as plant growth regulators can significantly affect the total metabolome of tomato fruits.18 Particularly, in biosynthesis of secondary metabolites including alkaloid and flavanol biosynthetic pathways were affected in fruits by CNTs added to the hydroponics system.18 This discovery paved the way for a biotechnological application involving CNTs: utilizing them as cost-effective yet powerful activators to boost the production of pharmacologically active plant alkaloids in easily reproducible cell cultures of medicinal plants.18 In another study, the total plant metabolome of CNT-exposed plants was investigated by Liquid Chromatography and Mass Spectrometry (LC-MS) with the goal of understanding if any potentially toxic tomato metabolites are overproduced in CNT-contaminated tomato organs.19 This risk-assessment study demonstrated that using CNTs to regulate plant growth did not lead to the enhanced synthesis of potentially toxic tomato metabolites such as tomatine.19
It can be concluded that “mono-omics” investigations serve as powerful tools for the detailed characterization of specific aspects of plant-nanomaterial interactions. However, the “mono-omics” approach fails to provide insights into the cascading effects from one “omics” level to the next.20 Recently, the integrated analysis of “multi-omics” data has emerged as a promising strategy for mechanistic studies. This approach enables a comprehensive examination from various angles, enhancing our understanding of molecular functions and the underlying mechanisms at play in biological systems.20 Additionally, the combined analysis of “multi-omics” data produced by different methodologies could significantly aid in bridging the gap between fundamental research and real-world agricultural applications.21,22 However, the integration of data from different “omics” levels for the characterization of the positive effects of CBN in planta and their phytotoxicity in high doses remains relatively limited at this time.23–26 One report employed multiple “omics” analyses to investigate the impact of CNTs on Solanum nigrum L. growth under cadmium and arsenic stresses.27 Application of 500 mg kg−1 CNTs notably enhanced S. nigrum growth, particularly in root tissues, leading to significant increases in shoot length, root length, and fresh biomass. Transcriptomic analysis revealed that CNTs upregulated advantageous biological processes and reprogrammed metabolism related to the defense system, leading to the accumulation of 4-hydroxyphenylpyruvic acid (amino acid), 4-hydroxycinnamic acid (xenobiotic), and (S)-abscisic acid (lipid). Integration of transcriptomic with following metabolomic analyses identified key pathways affected by CNTs, highlighting their potential application in soil remediation.27 In another study, Li et al. investigated the underlying molecular mechanisms of graphene-based nanomaterials (GBNs) towards their phytotoxicity in edible crops using a multi-omics approach.28 GBNs were injected into pepper plant stems at varying concentrations, leading to the regulation of plant defense mechanisms by reducing calcium content, intercellular CO2 concentration, transpiration rate, and stomatal conductance. Nontargeted proteomics and metabolomics analyses revealed the downregulation of carbohydrate metabolism and upregulation of amino acid metabolism as the main mechanisms underlying observed phytotoxicity and defense mechanisms in GBN-treated plants.28 Chen et al. studied phytotoxicity effects and molecular mechanisms induced by graphene in alfalfa (Medicago sativa L.) by integrating transcriptomic and metabolomics analysis.29 This study examined how alfalfa leaves respond physiologically to graphene stress using metabolome and transcriptome analyses with two contrasting genotypes: tolerant and sensitive. It was noticed that graphene disrupted antioxidant defense systems and photosynthesis, with metabolomic analysis revealing changes in amino acids, flavonoids, organic acids, and sugars. Transcriptomic analysis identified core graphene-responsive genes in alfalfa, with the susceptible genotype showing greater disturbance by graphene stress at both transcriptional and metabolic levels.29
In this work, we aimed to get a deeper insight into the possible effects of CBNs on plants' stress tolerance by integration of transcriptomics and proteomics datasets generated from tomato seedlings exposed to NaCl in the presence of two types of CBNs (CNTs and graphene) (Fig. 1). Integrative analysis of the tomato transcriptome and proteome uncovered a shared CBNs-imposed phenomenon of restoration of the expression of transcripts and corresponding proteins that were negatively impacted by salt stress. This approach provided valuable insights into the regulatory networks and signaling mechanisms within plants, significantly enhancing tolerance in CBN-exposed plants to environmental challenges such as salt stress.
Materials and methods
Plant materials, CBNs, and salt treatment
Tomato (cv. Micro-Tom) seeds were purchased from Reimer Seed Co. Inc., MD, USA. After being washed with 70% ethanol for 2 minutes, rinsed with double-distilled water, submerged in a 50% bleach solution, and then vortexed for 30 minutes, tomato seeds were sterilized. After sterilization, seedlings underwent ten rounds of sterile water rinsing. Commercially available CBNs were used for the study. Cheap Tubes (Brattleboro, VT) provided the multiwalled CNTs (MWCNT-COOH, OD 13–18 nm; length 1–12 μm, referred to as “CNTs” for the purposes of this paper) and the graphene nanoplatelets (3 layers; lateral dimension 1–2 μm). As described by Lahiani et al., endotoxins present in the CBNs were removed by autoclaving the solutions three times at 121 °C and 15 lb in−2 pressure for 20 minutes.30Sterilized tomato seeds were placed on either Murashige and Skoog (MS) medium, MS medium supplemented by 100 mM of NaCl, MS medium supplemented by 100 mM NaCl + 100 μg ml−1 CNTs, MS medium supplemented by 100 mM NaCl + 100 μg ml−1 graphene. In a growth chamber, seeds were incubated for 21 days at 24 °C with 12-hour photoperiods and 105 μmol s−1 m−2 of light intensity.
Protein extraction and total proteome analysis
Total proteins were extracted from frozen leaf tissues of 21-day-old tomato seedlings, following the protocol for resolubilization of TCA-precipitated plant proteins for 2-D electrophoresis.31 Fresh leaf tissue was ground into fine powder in liquid nitrogen. Then, ice-cold TCA containing 1% v/v β-mercaptoethanol, 5 ml g−1 tissue was added to the powder. Mixtures were transferred into conical tubes and kept in ice for 5 min. Samples were run in a 4 °C centrifuge at 1000 × g for 5 min and supernatant was discarded. This step was repeated until a clear supernatant was achieved. The pellet was washed with ice-cold acetone twice. The tubes were inverted on a clean piece of absorbent paper to drain the acetone. Following that, phenol containing 0.5% w/v DTT, 2 ml g−1 tissue was added to tubes and mixed thoroughly. Then, tubes were kept at room temperature for 10 min, followed by a centrifuge at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C, and the supernatant was transferred to a new tube. This step was repeated three times, and all the supernatants were assembled. Subsequently, five volumes of cold methanol containing 0.1 M ammonium acetate were added to the collected phenol. The solution was mixed thoroughly and kept at −20 °C freezer for at least 20 min to precipitate the protein. Then it was centrifuged at 10
000 × g at 4 °C, and the supernatant was transferred to a new tube. This step was repeated three times, and all the supernatants were assembled. Subsequently, five volumes of cold methanol containing 0.1 M ammonium acetate were added to the collected phenol. The solution was mixed thoroughly and kept at −20 °C freezer for at least 20 min to precipitate the protein. Then it was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C for 10 min, and the supernatant was discarded carefully. 1 ml ice-cold methanol was added to wash the pellet, followed by centrifuging at 10
000 × g at 4 °C for 10 min, and the supernatant was discarded carefully. 1 ml ice-cold methanol was added to wash the pellet, followed by centrifuging at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C for 5 min, and the supernatant was discarded carefully. This step was repeated three times to remove the residual ammonium acetate and phenol. Protein pellets were kept in double distilled water in a −80 °C freezer for the next steps.
000 × g at 4 °C for 5 min, and the supernatant was discarded carefully. This step was repeated three times to remove the residual ammonium acetate and phenol. Protein pellets were kept in double distilled water in a −80 °C freezer for the next steps.
Total protein quantification was determined by the Thermo Scientific™ Coomassie Plus™ Kit.
Protein samples were sent to Novogene Co., Ltd. (Beijing, China). The iTRAQ (isobaric tag for relative and absolute quantification) technique with initial proteomics data analysis was performed in Novogene Co., Ltd. Provided data set included samples quality control, protein enzymolysis and desalination, isotope labeling, tandem mass spectrometry, and bioinformatic data analysis.
Validation of proteomics analysis in tomato plants
For bioinformatics analysis and proteomics data validation, the western blotting technique has been applied to identify and quantify the presence of two different proteins in the 21-day-old tomato seedlings' protein lysate. Plant tissues were ground in liquid nitrogen and were transferred to sample tubes. 1 ml of RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Triton X-100, 0.1% SDS) was added to each tube, mixed thoroughly, and then centrifuged at 4 °C and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min. Supernatants were collected, and protein concentrations were determined. 20 μl of protein sample was loaded into SDS-PAGE wells with running buffer (25 mM Tris base, 190 mM glycine, 0.1% SDS), including 2 μl of precision plus as molecular weight marker. After gel running, proteins were transferred from gel to PVDF membrane by preparing a transfer sandwich and running in an electrophoresis device at 100v for 1 h inside the cold transfer buffer (25 mM Tris base, 190 mM glycine, 10% methanol). After protein transfer, the gel was gently removed from the transfer sandwich and stained overnight by Coomassie Blue staining to help ensure that proteins were completely transferred from the gel to the membrane. The membrane was kept in blocking buffer (TBST buffer: 20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Tween 20, 5% dry milk) overnight at 4 °C. Antibody was purchased from Agrisera (FBA | Fructose-bisphosphate aldolase 1, 1
000 rpm for 20 min. Supernatants were collected, and protein concentrations were determined. 20 μl of protein sample was loaded into SDS-PAGE wells with running buffer (25 mM Tris base, 190 mM glycine, 0.1% SDS), including 2 μl of precision plus as molecular weight marker. After gel running, proteins were transferred from gel to PVDF membrane by preparing a transfer sandwich and running in an electrophoresis device at 100v for 1 h inside the cold transfer buffer (25 mM Tris base, 190 mM glycine, 10% methanol). After protein transfer, the gel was gently removed from the transfer sandwich and stained overnight by Coomassie Blue staining to help ensure that proteins were completely transferred from the gel to the membrane. The membrane was kept in blocking buffer (TBST buffer: 20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Tween 20, 5% dry milk) overnight at 4 °C. Antibody was purchased from Agrisera (FBA | Fructose-bisphosphate aldolase 1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4000, AS164093). Antibody was diluted to a working concentration in 1× TBST with 5% dry milk. Membranes were incubated in primary antibody solution for 2 h at room temperature. Then, blots were washed 3 times for 10 min in 1× TBST at room temperature with gentle rocking. A secondary antibody was purchased from ABCAM (AB6721 Goat pAb to RbIgG (HRP)). At room temperature, the membrane was incubated with the appropriate-diluted secondary antibody in 1× TBST for 1 hour. The membrane was rewashed three times at room temperature in 1× TBST with gentle rocking. In the next step, chemiluminescent substrate (Thermo Scientific SuperSignal® West Pico Substrate) with the recommended concentration according to the manufacturer, was applied to the membrane and incubated for 5 min at room temperature. The membrane was transferred to the Autoradiography Cassette (Fisher Scientific, FBCA 57) for X-ray imaging in the dark room.
4000, AS164093). Antibody was diluted to a working concentration in 1× TBST with 5% dry milk. Membranes were incubated in primary antibody solution for 2 h at room temperature. Then, blots were washed 3 times for 10 min in 1× TBST at room temperature with gentle rocking. A secondary antibody was purchased from ABCAM (AB6721 Goat pAb to RbIgG (HRP)). At room temperature, the membrane was incubated with the appropriate-diluted secondary antibody in 1× TBST for 1 hour. The membrane was rewashed three times at room temperature in 1× TBST with gentle rocking. In the next step, chemiluminescent substrate (Thermo Scientific SuperSignal® West Pico Substrate) with the recommended concentration according to the manufacturer, was applied to the membrane and incubated for 5 min at room temperature. The membrane was transferred to the Autoradiography Cassette (Fisher Scientific, FBCA 57) for X-ray imaging in the dark room.
Additionally, to western blotting analysis, we utilize real-time PCR analysis for validation of proteomics data. RNA was isolated from tomato plants. According to the manufacturer protocol, cDNA was generated from 1 μg of total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen, USA) with a dT20 oligonucleotide as a primer. cDNA samples were diluted and used for real-time quantitative PCR analysis with SsoAdvanced Universal SYBR® Green Supermix (BIO-RAD, USA) in a CFX Opus 96 Real-time PCR system (Bio-Rad, USA). Eukaryotic translation initiation factor 3 subunit J and RNA-binding protein 8A-like were amplified as tomato genes. To amplify eukaryotic translation initiation factor 3 subunit J, 5′-GGTGGTGATGACAAGACCCT-3′ (forward) and 5′-AACGGTCGCAGCTTATGAGA-3′ (reverse) primers were used. Similarly, for amplifying RNA-binding protein 8A-like, 5′-AGGAGGCTGTGGATTTCGAG-3′ (forward) and 5′-CGCCACCTGTAATGGCTGAT-3′ primers were used. The housekeeping gene (18 s) was amplified for all samples using primers: 5′-AGGCCGCGGAAGTTTGAGGC-3′ and 5′-ATCAGTGTAGCGCGCGTGGG-3′. Three independent biological replicates were used in the analysis. For each biological replica, three technical replicas were run. The real-time PCR data were analyzed by the “comparative count” method to obtain the relative mRNA expression of each tissue, as described in the CFX Opus 96 manual (Bio-Rad).
Integration of transcriptomics and proteomics data
We used raw data obtained from previous RNA-Seq analysis11 and new proteomics raw files to integrate the two level of transcriptomics and proteomics. The data was analyzed at UAMS bioinformatic lab using R packages of MixOmics and MOGSA. MixOmics supervised analysis was employed to discern essential features from integrating multi-omics data. The MixOmics R package offers a diverse array of multivariate methods designed to develop and validate various biological systems, facilitating more profound insights into omics analysis. Multivariate techniques are particularly well-suited for handling large omics datasets where the number of variables, such as genes and proteins, far exceeds the number of samples. These methods possess advantageous characteristics, including dimensionality reduction through the use of instrumental variables (components) that amalgamate all variables. These components are then leveraged to generate informative graphical representations, enhancing comprehension of the relationships and correlation structures among the integrated datasets. Subsequently, multi-omics-gene-set analysis (MOGSA) was employed to integrate gene and protein expression from identical sample sets. This method utilizes a low-dimensional representation of the most variant-correlated features across different data types and standardizes the features to a standard scale.Results and discussion
Justification for selection of experimental conditions
The experimental conditions used in the proteomics study including plant type and age, growth medium, and cultivation practices were identical to those employed in our previous transcriptomics project.11 A NaCl concentration of 100 mM was selected for both proteomics and transcriptomics studies based on prior detailed salt stress experiments,10 which demonstrated that this concentration induces a consistent and measurable level of salt stress under controlled laboratory conditions. This allowed for a robust molecular-level analysis of how CBNs influence plant responses under well-defined stress parameters. Importantly, the use of 100 mM NaCl is not limited to artificial laboratory scenarios. Field studies have shown that soil salinity levels can reach or exceed this threshold, particularly in arid and semi-arid regions where natural salt accumulation and irrigation practices contribute to elevated salinity.32 For example, salinity at or above 100 mM NaCl has been reported to severely impair crop performance—rice often fails to reach maturity, and wheat shows significant yield reduction under such conditions.33 Thus, the use of 100 mM NaCl in our experimental design not only enables the simulation of acute stress responses but also accurately reflects salinity levels encountered in real-world agricultural environments.Tandem mass tag (TMT) quantitative proteomics analysis
In the quantitative proteomics analysis of tomato seedlings exposed to salt stress in presence and absence of two types of CBNs (CNTs, graphene), 336![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 341 spectra were generated using label-free analysis in protein samples extracted from tomato seedlings. Proteome Discoverer 2.2 software was used to do further filtration on the search results to improve the quality of analysis results and reduce the false-positive rate. A total of 105
341 spectra were generated using label-free analysis in protein samples extracted from tomato seedlings. Proteome Discoverer 2.2 software was used to do further filtration on the search results to improve the quality of analysis results and reduce the false-positive rate. A total of 105![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 560 Peptide Spectrum Matches (PSMs) with confidence levels greater than 99% have been selected. From the number of PSMs, 50
560 Peptide Spectrum Matches (PSMs) with confidence levels greater than 99% have been selected. From the number of PSMs, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 542 peptides were identified, and 6501 proteins that contain at least one unique peptide are confident. From the confident proteins, 6478 total proteins can be quantified in all samples with FDR ≤ 0.01 (Fig. S1†). Following protein quantification and identification, functional annotation on the identified proteins was performed to understand the functional characteristics of different proteins. The general function databases that provide annotations include GO (Gene Ontology), KEGG (Kyoto Encyclopedia of Genes and Genomes), COG (Cluster of Orthologous Groups of Proteins), and IPR (InterPro classifications of proteins). In GO function annotation, most proteins in the biological process category were identified in the oxidation–reduction process. In the category of the cellular component, most of the identified proteins belonged to the integral component of the membrane. In the molecular function category, protein binding included the majority of identified proteins (Fig. S2A†). KEGG function annotation shows the number of proteins in different categories of pathways (Fig. S2B†). COG is categorized by the systematic evolutionary relationship of encoded proteins based on a complete sequence genome of bacteria, algae, and eukaryotes. COG can annotate a specific protein sequence through matching. Each cluster of COG is composed of orthologous sequences; thus, the sequence's function can be inferred. COG database can be divided into 26 classes according to functions. Fig. S2C† shows the distribution of the identified proteins into those classes. IPR was used to recognize protein domains and functional sites. To annotate domains more comprehensively, IPR integrates some commonly used domain databases, including Pfam, ProDom, SMART, and other domain databases. Pattern structures and features are used for domain annotation of proteins with unknown functions. Fig. S2D† illustrates the IPR annotated domains and their associated number of identified proteins. After the protein is synthesized in the ribosome, it is transported to a specific organelle by a protein sorting signal, and part of the protein is secreted out of the cell or left in the cytoplasm. Only when it is transported to the correct location can it participate in various life activities of the cell. Understanding the subcellular localization information of proteins is critical for understanding biological mechanisms. The annotation of subcellular location information is shown in Fig. S2E.† Principal component analysis (PCA) was performed to demonstrate the significant difference in total protein expression between experimental groups and the degree of variability between samples within the groups (Fig. 2). PCA analysis showed that the proteome of untreated seedlings (control group) differs significantly from that of all treated seedlings. Moreover, the proteome of NaCl-treated seedlings (NaCl group) displayed distinct differences from the proteomes of seedlings treated with CBNs (CNTs and graphene groups). These findings confirm that both salt stress and the application of CBNs exert substantial effects on the proteome of tomato plants. Additionally, incorporating CBNs produces a notable impact on seedlings exposed to NaCl, highlighting the potential for CBNs to mitigate stress-related proteomic changes (Fig. 2).
542 peptides were identified, and 6501 proteins that contain at least one unique peptide are confident. From the confident proteins, 6478 total proteins can be quantified in all samples with FDR ≤ 0.01 (Fig. S1†). Following protein quantification and identification, functional annotation on the identified proteins was performed to understand the functional characteristics of different proteins. The general function databases that provide annotations include GO (Gene Ontology), KEGG (Kyoto Encyclopedia of Genes and Genomes), COG (Cluster of Orthologous Groups of Proteins), and IPR (InterPro classifications of proteins). In GO function annotation, most proteins in the biological process category were identified in the oxidation–reduction process. In the category of the cellular component, most of the identified proteins belonged to the integral component of the membrane. In the molecular function category, protein binding included the majority of identified proteins (Fig. S2A†). KEGG function annotation shows the number of proteins in different categories of pathways (Fig. S2B†). COG is categorized by the systematic evolutionary relationship of encoded proteins based on a complete sequence genome of bacteria, algae, and eukaryotes. COG can annotate a specific protein sequence through matching. Each cluster of COG is composed of orthologous sequences; thus, the sequence's function can be inferred. COG database can be divided into 26 classes according to functions. Fig. S2C† shows the distribution of the identified proteins into those classes. IPR was used to recognize protein domains and functional sites. To annotate domains more comprehensively, IPR integrates some commonly used domain databases, including Pfam, ProDom, SMART, and other domain databases. Pattern structures and features are used for domain annotation of proteins with unknown functions. Fig. S2D† illustrates the IPR annotated domains and their associated number of identified proteins. After the protein is synthesized in the ribosome, it is transported to a specific organelle by a protein sorting signal, and part of the protein is secreted out of the cell or left in the cytoplasm. Only when it is transported to the correct location can it participate in various life activities of the cell. Understanding the subcellular localization information of proteins is critical for understanding biological mechanisms. The annotation of subcellular location information is shown in Fig. S2E.† Principal component analysis (PCA) was performed to demonstrate the significant difference in total protein expression between experimental groups and the degree of variability between samples within the groups (Fig. 2). PCA analysis showed that the proteome of untreated seedlings (control group) differs significantly from that of all treated seedlings. Moreover, the proteome of NaCl-treated seedlings (NaCl group) displayed distinct differences from the proteomes of seedlings treated with CBNs (CNTs and graphene groups). These findings confirm that both salt stress and the application of CBNs exert substantial effects on the proteome of tomato plants. Additionally, incorporating CBNs produces a notable impact on seedlings exposed to NaCl, highlighting the potential for CBNs to mitigate stress-related proteomic changes (Fig. 2).
Protein differential analysis was used to determine the significance of the difference and the relative quantification values of each protein in the paired groups. Based on the threshold for screening the differentially expressed proteins (DEPs) and analysis of volcano plots (|log2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FC| ≥ 1.2, P-value ≤ 0.05), there were 998 and 860 up and downregulated DEPs in samples treated with salt compared to control. Similarly, the total number of 1135 and 742 upregulated and downregulated DEPs have been found in seedlings treated with salt in the presence of CNTs compared to control, respectively. On the other hand, in samples treated with salt in the presence of graphene, 1879 and 1076 proteins were up and downregulated, respectively (Fig. S3,†Table 1). Statistical analysis of subcellular localization indicated that 3024 differential proteins were localized in all treated groups. From this amount, 651 proteins were localized in the cytoplasm 178 (20.58% of all DEPs in salt-treated samples compared to control) in the salt group, 196 proteins (22.79% of all DEPs in the salt-treated in the presence of CNTs compared to the control), and 277 proteins (21.32% of all DEPs in the salt-treated in the presence of graphene compared to the control). Additionally, a total of 433 proteins were predicted to be localized in the chloroplast, and 349 proteins were predicted to be localized in the nucleus in all treatment groups. These results indicate that DEPs localized in subcellular regions can form a complex regulatory network for tomato seedlings treated with CBNs and subjected to salt stress (Fig. S4†).
FC| ≥ 1.2, P-value ≤ 0.05), there were 998 and 860 up and downregulated DEPs in samples treated with salt compared to control. Similarly, the total number of 1135 and 742 upregulated and downregulated DEPs have been found in seedlings treated with salt in the presence of CNTs compared to control, respectively. On the other hand, in samples treated with salt in the presence of graphene, 1879 and 1076 proteins were up and downregulated, respectively (Fig. S3,†Table 1). Statistical analysis of subcellular localization indicated that 3024 differential proteins were localized in all treated groups. From this amount, 651 proteins were localized in the cytoplasm 178 (20.58% of all DEPs in salt-treated samples compared to control) in the salt group, 196 proteins (22.79% of all DEPs in the salt-treated in the presence of CNTs compared to the control), and 277 proteins (21.32% of all DEPs in the salt-treated in the presence of graphene compared to the control). Additionally, a total of 433 proteins were predicted to be localized in the chloroplast, and 349 proteins were predicted to be localized in the nucleus in all treatment groups. These results indicate that DEPs localized in subcellular regions can form a complex regulatory network for tomato seedlings treated with CBNs and subjected to salt stress (Fig. S4†).
| Additions to the growth medium | Total number of unique quantified proteins (FDR ≤ 0.01) | Total number of differentially expressed proteins compared to control (|log2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FC| ≥ 1.2, P-value ≤ 0.05) FC| ≥ 1.2, P-value ≤ 0.05) |
Number of proteins that fully or partially restored the level of expression as a result of the introduction of CBNs to medium supplemented with NaCl | ||
|---|---|---|---|---|---|
| NaCl (100 mM) | 6478 | 1858 | Up: 998 | NA | |
| Down: 860 | |||||
| NaCl + CNTs | 1877 | Up: 1135 | 1055 | Up: 450 | |
| Down: 742 | Down: 605 | ||||
| NaCl + graphene | 2955 | Up: 1879 | 1231 | Up: 486 | |
| Down: 1076 | Down: 745 | ||||
| Control (no supplements) | NA | NA | |||
Previously, we discovered a new property of CBNs: the ability to fully or partially restore the expression of a large number of plant genes negatively affected by environmental stresses (salt and drought).11 In the proteomic analysis of tomato seedlings, the experimental conditions were replicated from the earlier transcriptomics study.11 We aimed to determine if the observed phenomenon of expression restoration at the transcriptomic level would also manifest at the proteomic level. To comprehend the effects of CBNs on protein restoration levels, we scrutinized the DEP results based on the differentially expressed genes (DEG) analysis.11 In total, 1858 proteins exhibited differential regulation in tomato seedlings following exposure to 100 mM NaCl. Of these, 53.7% displayed upregulation, while 46.3% were downregulated compared to the control levels (Table 1). Among these proteins, a subset showed either full or partial restoration towards control levels, achieved through the upregulation of proteins suppressed by NaCl or downregulation of proteins activated by NaCl. Introducing CNTs to the saline medium in the presence of CNTs (NaCl + CNTs) positively influenced 1055 proteins, restoring 450 proteins through upregulation and 605 proteins through downregulation towards the control level. Similarly, applying graphene led to the full or partial restoration of 1231 tomato proteins affected by NaCl, with 486 proteins by upregulation and 745 proteins by downregulation towards the control level (Tables 1 and S1–S8†). We validated the proteomics data by examining the expression of selected proteins through Western blotting. Fig. S5† illustrates the expression patterns of FBA (Fructose-bisphosphate aldolase 1) protein in control seedlings and those treated with NaCl, NaCl + CNTs, and NaCl + graphene. Furthermore, in addition to Western blotting, we confirmed protein expression by selecting associated genes and employing RT-PCR. Fig. S6† depicts the amplification of tomato EIF3J and RBM8A genes via RT-PCR in control seedlings and those subjected to all treatments. The RT-PCR results corroborated the trends observed in the proteomics data, indicating the suppression of EIF3J and RBM8A gene-associated proteins by NaCl and the restoration of gene expression upon applying CNTs and graphene (Fig. S6†).
GO functional enrichment analysis of different groups of NaCl, NaCl + CNTs, and NaCl + graphene compared to the control group indicated that in biological process category, metabolic process, single organism process, and organonitrogen compound metabolic process have been enriched in NaCl, NaCl + CNTs, and NaCl + graphene comparing to control, respectively (Fig. S7†). Similarly, in the cellular component category, membrane protein complex, protein complex, and cell part were enriched in the NaCl, NaCl + CNTs, and NaCl + graphene samples compared to control, respectively. Furthermore, in the molecular function category, catalytic activity has been enriched the most in the NaCl-exposed samples compared to the control. In contrast, ion binding has been enriched the most in NaCl + CNTs and NaCl + graphene samples compared to the control samples (Fig. S7A–C†). KEGG enrichment analysis indicated significantly enriched pathways in DEPs compared to the total identified proteins. The analysis revealed that DEPs in the groups of seedlings exposed to salt stress were significantly enriched in metabolic pathways and biosynthesis of secondary metabolites. However, in salt-treated samples in the presence of CNTs, DEPs were highly enriched in metabolic pathways and carbon metabolism. In the presence of graphene, DEPs were enriched in ribosome and protein processing in the endoplasmic reticulum (Fig. S8A–C†). IPR domain enrichment analysis revealed that DEPs in the NaCl-exposed seedlings were significantly enriched in the “thioredoxin-like fold”, “haem peroxidase, plant/fungal/bacterial”, and “glutathione S-transferase, N-terminal”. Similarly, in the salt stress treatment in the presence of CNTs, “thioredoxin-like fold”, “haem peroxidase, plant/fungal/bacterial”, and “alpha crystallin/Hsp20” domain were highly enriched. In the presence of graphene, “thioredoxin-like fold”, “thioredoxin domain”, and “chlorophyll A–B binding protein” were enriched (Fig. S9A–C†).
Integration of transcriptomics and proteomics datasets
An integrated analysis of the proteome and transcriptome11 of tomato seedlings treated with NaCl (100 mM) in the presence and absence of CBNs was conducted to determine the correlation between the two datasets generated in the same experimental conditions. In this approach, we investigated the correlation between previously published RNA-Seq data11 and proteome analysis presented here (Fig. 3). In total, 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 958 transcripts and 6478 proteins were identified and quantified across the transcriptome11 and proteome data sets, respectively. 6055 features were shared between the two “omics” levels based on the Ensembl and UniProt IDs. We investigated how much correlation was between transcript and protein expression within the samples. From the total 6055 common features based on Ensembl ID for genes and proteins, there were positive correlation values of 0.36, 0.27, and 0.39 for the seedling exposed to NaCl compared to control, seedlings exposed to NaCl + CNTs compared to control, and seedlings exposed to NaCl + graphene compared to control, respectively (Fig. 3). The majority of differentially expressed genes and proteins had similar expression patterns. The log2 fold change for protein expression is shown on the y-axis, while the gene expression log2 fold change is on the x-axis (Fig. 3). The significant features from each dataset are plotted on the four outside quadrants of the correlation plot, and the middle quadrants show the non-significant features (Fig. 3).
958 transcripts and 6478 proteins were identified and quantified across the transcriptome11 and proteome data sets, respectively. 6055 features were shared between the two “omics” levels based on the Ensembl and UniProt IDs. We investigated how much correlation was between transcript and protein expression within the samples. From the total 6055 common features based on Ensembl ID for genes and proteins, there were positive correlation values of 0.36, 0.27, and 0.39 for the seedling exposed to NaCl compared to control, seedlings exposed to NaCl + CNTs compared to control, and seedlings exposed to NaCl + graphene compared to control, respectively (Fig. 3). The majority of differentially expressed genes and proteins had similar expression patterns. The log2 fold change for protein expression is shown on the y-axis, while the gene expression log2 fold change is on the x-axis (Fig. 3). The significant features from each dataset are plotted on the four outside quadrants of the correlation plot, and the middle quadrants show the non-significant features (Fig. 3).
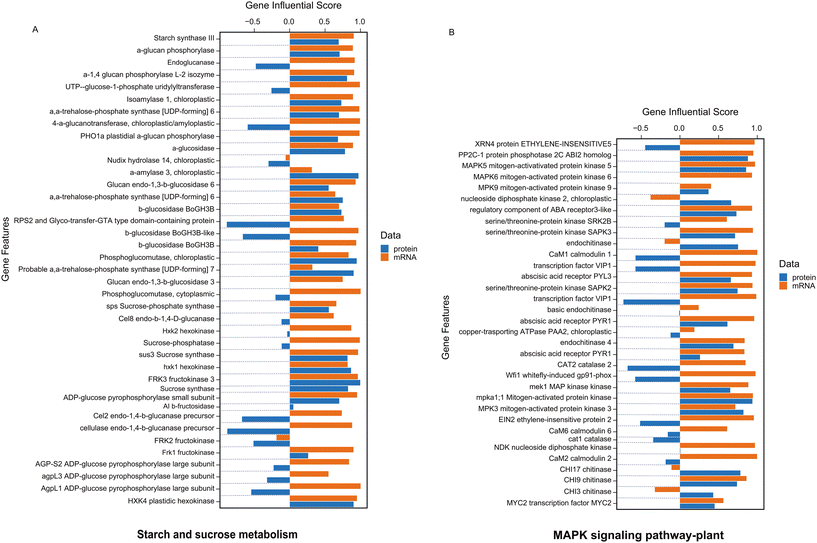
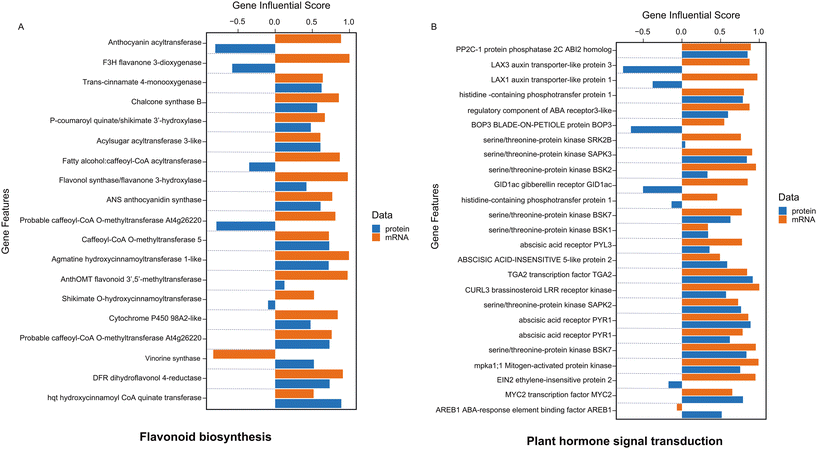
In our analysis, we utilized the DIABLO method to integrate identical biological samples across each dataset. DIABLO is a supervised approach within the MixOmics N-integrative framework, allowing users to combine multiple datasets while elucidating their association with a categorical outcome variable (Fig. 4A).34Fig. 4A demonstrates the top 50 most significant features (P-value <0.05) that are common between RNAseq and proteomics datasets. The integrated gene score is then computed using MOGSA package based on the most informative features within each data type (Fig. 4B).35 MOGSA heatmap shows the Gene Set Score (GSS) for significantly regulated molecular pathways associated with features exhibiting the same expression trends in both transcriptomics and proteomics datasets (Fig. 4B). Using MOGSA, we analyzed the characteristics of each level of “multi-omics” regulation to determine which dataset had the greatest impact in identifying significant pathways. We noticed that the most CBN-affected molecular pathways associated with common features between the two datasets included the MAPK signaling pathway, starch and sucrose metabolism, flavonoid biosynthesis, plant hormone signal transduction, stilbenoid, diarylheptanoid, and gingerol biosynthesis, as well as the biosynthesis of unsaturated fatty acids (Fig. 5, 6, and S10†). Results correlated with previously performed analysis of the total metabolome of tomato organs collected from plants grown in presence of CNTs.18,19 Thus, the strong impact of CNTs on metabolic pathways related to a few branches of secondary metabolism and stress signal transduction including the MAPK signaling pathway was clearly demonstrated at the metabolomics level. This study revealed a notable upregulation of several metabolites derived from flavonoid biosynthetic pathways in tomato fruits exposed to CNTs.18
Identification of conserved features across transcriptomic and proteomic datasets
Plants possess an array of adaptive mechanisms that allow them to tolerate environmental stresses, including salinity, which can significantly affect their growth and development.36 It was discovered that CBNs can positively influence the entire transcriptome of plants under environmental stress by restoration of gene expression negatively affected by stresses such as salt and drought.11 By this effect, CBNs have been able to mitigate the adverse effects of environmental stresses, improving the overall phenotype of the plants exposed to stress.11 Abiotic stress can cause significant fluctuations in protein expression levels within plant cells.37 However, changes in protein accumulation may not always directly correspond to alterations in gene expression, as protein synthesis and regulation are influenced by various factors such as protein targeting, translocation, and post-translational modification, all of which are impacted by stress conditions.37 Exposure to abiotic stress often results in regulating groups of functionally related proteins, including those associated with metabolism, storage, and protein synthesis, even if they are not directly involved in defense mechanisms.37 Here, we found that the effect of restoration of expression in plants induced by CBNs introduced to the salty environment previously documented at the transcriptome level,11 can also be observed at the proteome level. Thus, among all the identified DEPs affected by salt, 56.8%, and 66.2% were fully or partially restored by applying CNTs and graphene, respectively. Of the annotated 1231 proteins (both upregulated and downregulated) that fully and partially restored their expression levels after the addition of graphene (Tables S1–S4†), 38.6% were related to stress-responsive biological processes, while 35.3% of the annotated 1055 proteins (upregulated and downregulated) fully and partially restored their expression levels after the addition of CNTs (Tables S5–S8†) belonged to stress-responsive biological processes. Nonetheless, proteins responsive to abiotic stress can be classified into six main groups based on their functions:1 Osmoprotectant regulators oversee the distribution of osmolyte molecules like sucrose synthase or sugar transporter;2 Reactive Oxygen Species (ROS) scavengers act as the primary defense against oxidative stress, involving peroxidases, superoxide dismutase, or catalase;3 ion transporter proteins manage the flux of ions like H+, Na+, Cl−, or K+, influencing cytosolic pH and transmembrane electrical potential. These proteins may include Na+/H+ antiporters and plasma membrane H+-ATPase;4 water channel proteins, such as aquaporins, are essential for adjusting water content in plant cells;5 Molecular chaperones facilitate protein folding by binding to newly synthesized glycoproteins, with most heat shock proteins falling under this category;6 proteolysis-related proteins regulate the degradation of misfolded, unassembled, or mutated proteins in cells, such as ubiquitins.37 Referring to this classification, several proteins from stress-responsive groups demonstrated a trend in restoring expression affected by salt stress upon CBNs treatment. For instance, proteins associated with osmoprotectant regulators, like sucrose synthase, sucrose transferase, proline ligase, and proline deoxygenase, exhibited full or partial restoration of expression in response to graphene in a saline medium (Tables S1–S4†). This finding aligns with previous transcriptomic analyses, which indicated the restoration of expression of related genes involved in osmoprotectants, such as polyol transporter, a sugar symporter.11 Under salt stress, plants primarily accumulate osmoprotectants to maintain cell turgor pressure through osmoregulation and protect cellular components by reducing ionic toxicity.38 Moreover, these osmoprotective regulators enhance the plant's antioxidative defense system by scavenging harmful ROS and preserving essential antioxidative enzymes.39Furthermore, our results indicated that CBNs may be able to regulate the expression of ROS scavengers. Both peroxidase glutathione and superoxide dismutase exhibited complete or partial restoration of expression in response to CNTs and graphene (Tables S1 and S3–S5†). Additionally, peroxidase-regulating genes also showed restored expression in the gene expression analysis.11 Superoxide dismutase proteins, as a type of antioxidant enzyme, play a crucial role in protecting plants from the harmful effects of ROS, thereby significantly influencing plant growth, development, and their responses to abiotic stress.37 For instance, in a study conducted by Rahman et al., superoxide dismutase was notably induced in perennial ryegrass plant tissue in response to combined heat and drought stress, highlighting the importance of these antioxidants in mitigating ROS damage.40 Moreover, ion transporters, such as plasma membrane antiporters, have demonstrated restored expression following the application of CBNs (Tables S3 and S6†). Salinity-induced disruptions in ion equilibrium, including Na+, H+, K+, and Cl−, within cells can be rectified by these transporters. Crucial subcellular components like the plasma membrane and vacuoles play significant roles in maintaining ion balance.37 For instance, a study by Xu et al. illustrated that transgenic Arabidopsis plants overexpressing H+-ATPase exhibited enhanced salinity tolerance compared to wild-type plants.41 This heightened tolerance was attributed to the decreased Na+ content in transgenic Arabidopsis plants under salt stress, indicating the involvement of plasma membrane ATPase in facilitating Na+ efflux and thereby conferring salt resistance.41 Our transcriptomic analysis further underscored the consistent complete restoration of calcium-transporting ATPase plasma membrane in response to graphene application.11 Furthermore, aquaporins such as TIP 3;2, TIP 1;2, aquaporin PIP-type, and PIP2 bind domain-containing proteins have exhibited full or partial expression restoration upon applying both CNTs and graphene (Tables S1–S6 and S8†). This observation correlates with our previous findings indicating the CBN-treated upregulation of various aquaporin genes in diverse plant tissues, including crop seeds,8 tomato leaves,42 and seedlings.10 The consistency between aquaporin expression levels at both transcriptomic and proteomic levels underscores the relevance of our results.11 Molecular chaperones, which aid in the properly folding and assembly of secretory proteins, are typically located in the endoplasmic reticulum, where newly synthesized proteins undergo folding.37 Our proteomics analysis indicates that molecular chaperones may assist plants by regulating the expression of heat shock proteins following the application of CBNs in response to salinity (Tables S7 and S8†). Specifically, mitochondrial small heat shock proteins have partially restored expression levels towards the control (Tables S4 and S8†). Our transcriptomic analysis further revealed that DNAJ gene-encoded proteins associated with heat shock proteins also restored expression towards the control level in the presence of graphene and CNTs.11 Heat shock proteins are significant regulators of plants' abiotic stress response and growth.43 For instance, a study by Escobar et al. analyzed the function of three mitochondrial small heat shock proteins (M-SHSPs) in Arabidopsis knock-down lines lacking function of M-SHSPs genes during plant growth. The triple knock-down plants exhibited the unusual phenotype and alterations in proteins primarily involved in photosynthesis and oxidative defense compared to control plants.44
Furthermore, ubiquitin, a group of proteins, plays a crucial role in the protein breakdown mechanism. As abiotic stress leads to the accumulation of misfolded and unfolded proteins, protein breakdown and recycling become essential features of the plant's response to environmental stress.37 We observed full restoration of expression of small ubiquitin-related modifiers in response to salinity following the application of graphene (Table S1†). Similarly, the E3 ubiquitin-protein ligase gene exhibited a similar trend in response to salt stress after the application of graphene.11 Studies have demonstrated that rice contains numerous RING finger ligases, such as the salt-induced ring protein family (SIRP), which are implicated in managing salt stress. Specifically, three proteins are recognized as negative regulators of responses to salt stress. Thus, in Arabidopsis, the overexpression of OsSIRP1 gene leads to decreased salinity tolerance during seed germination and root growth due to the breakdown of undefined proteins.45
Several proteins, including the protein kinase domain-containing protein, mitogen-activated protein kinase related to MAPK signaling, Inositol-tetrakisphosphate 1-kinase associated with inositol metabolism, gamma-aminobutyrate transaminase 1, and mitochondrial proteins involved in carbon metabolism and the TCA cycle (Tables S1–S8†). These proteins exhibited full or partial restoration of their expression in salt-stressed seedlings treated with CNTs or graphene, mirroring the trends observed at the transcriptomic level.
However, 23 DEPs matched their corresponding DEGs but showed opposite trends of dysregulation (Table S10†). Similarly, in seedlings treated with salt in the presence of CNTs, 3074 DEGs, and 1877 DEPs were identified, with 25 DEPs matching their corresponding DEGs and showing the same dysregulation trend (Table S11†). In contrast, 12 DEPs had opposite dysregulation trends compared to their corresponding DEGs (Table S12†). Subsequently, in seedlings treated with salt in the presence of graphene, out of 3151 DEGs and 2955 DEPs, 32 DEPs matched their corresponding DEGs. They exhibited the same trend of dysregulation (Table S13†), while 11 DEGs had corresponding DEPs with opposite dysregulation trends (Table S14†).
The correlation between the transcriptome and proteome levels is visualized in Fig. 3. In each graph, the upper right quadrant represents genes and proteins upregulated at both levels (red dots). Similarly, the lower left quadrant shows genes and proteins consistently downregulated at both levels (dark blue dots) (Fig. 3). According to the correlation analysis, most DEG-associated DEPs showed no dysregulation (yellow, gray, and purple dots). This observation was consistent with the pathway analysis of selected MAPK signaling (Fig. 7 and 8). Notably, not all genes expressed at the transcriptomics level led to the expression of corresponding proteins at the proteomics level. This has been demonstrated in Fig. 7 and 8 for MAPK signaling pathway. The KEGG analysis of pathway were done separately solely based on the transcriptomics (Fig. 7) and proteomics (Fig. 8) level. This analysis demonstrates the differences of the expression of the genes in transcription level and translation level in the pathway. As it correlated with the integration analysis, the number of expressed proteins in translation level is less than in transcription level, showing the link between the two levels. It is also demonstrated that in the transcription level, dysregulation of expressed genes had been more affected by the application of CBNs in the salty environment. The number of up and downregulated genes at the transcription level is greater than at the translation level (Fig. 7 and 8).
Integration analysis also facilitated the generation of a list of pathways most significantly regulated in both gene and protein datasets (Fig. 4B). From this list, we selectively analyzed the gene influential score of pathways including starch and sucrose metabolism, MAPK signaling, flavonoid biosynthesis, plant hormone signal transduction, stilbenoid, diarylheptanoid and gingerol biosynthesis, and the biosynthesis of unsaturated fatty acids (Fig. 5, 6, and S10†). These pathways have been shown to have the most consistent influential score in both level of protein and transcripts (Fig. 5, 6, and S10†).
Starch and sucrose are key molecules in plant responses to abiotic stresses, such as drought, high salinity, extreme temperatures, and water deficit. Plants remobilize starch to provide energy and carbon during abiotic stress.46 The released sugars and other metabolites support plant growth by: functioning as osmoprotectants, functioning as compatible solutes, and mitigating the negative effects of stress.46,47 Stress can affect starch metabolism in plants in a number of ways, depending on the type of stress, its intensity, and duration. For example, BAM1(β-amylases) and AMY3 (amylase 3) activity increases in leaves under osmotic stress, which increases starch degradation and the biosynthesis of sugar and proline in response to water stress.48,49 Moreover, sucrose synthase (SuSy) is a glycosyl transferase enzyme that plays a key role in sugar metabolism, primarily in sink tissues. Salt stress increases SPS (sucrose phosphate synthase) activity resulting in an increase in sucrose content.47,50Fig. 5A shows that a number of genes and associated proteins including amylase, sucrose synthase has positive correlation in both level of transcripts and protein. The MAPK (mitogen activated protein kinase) signaling pathway is involved in plants' responses to abiotic stress, such as wounding, high and low temperatures, high salinity, UV radiation, ozone, ROS, drought, and heavy metals.51 MAPK cascades are implicated in ABA (abscisic acid) signaling, and ABA induces protein kinases of the SnRK family to mediate a number of its responses. The MAPK signaling pathway is also stimulated by growth factors, and pathogen-associated molecular patterns.52 MAPK signaling also alters levels of phytohormones, including jasmonic acid (JA) and ethylene, reshaping the transcriptome and thus the proteome in preparation to defend against attack.53 The result of our analysis has also demonstrated the importance of MAPK cascades in activating ABA receptors, MAPK5, MAPK6, and MAPK9 (Fig. 5B). Following abiotic stress in plants, the biosynthesis and accumulation of flavonoids contribute to scavenging reactive oxygen species (ROS) in various plant cell organelles, thereby reducing oxidative damage in plant cells.54 Flavonoids are secondary metabolites known for their antioxidant capabilities, effectively scavenging reactive oxygen species (ROS) and preventing their formation.55 A study reported that a transgenic Arabidopsis line UGT76E11, which overproduces flavonoids, demonstrated increased antioxidant capacity, lower ROS levels, and improved resistance to NaCl and mannitol stress.56 Our analysis showed that enzymes of flavonoid biosynthesis such as chalcone synthase and shikimate transferase were regulated in both levels of transcriptome and proteome (Fig. 6A). Chalcone synthase is a key modulator in shikimate pathway which leads to the production and accumulation of flavonoids in response to stress.57 Plant hormones are signaling compounds that regulate growth, development, and environmental stress responses.58 Gibberellin (GA) and abscisic acid (ABA) are major plant hormones involved response to abiotic stress.59 GA stimulates cell elongation and division, which regulates plant growth and stress responses.60 GA signaling also integrates information from other hormone signaling pathways, such as ethylene and ABA.61 ABA interacts with the jasmonic acid (JA) and salicylic acid (SA) signaling pathways to channel resources into mitigating the effects of abiotic stresses.62Fig. 6B shows activation and regulation of several GA and ABA receptors in our analysis. In plants, the biosynthesis pathway for stilbenoids, diarylheptanoids, and gingerol takes place in the endoplasmic reticulum. This pathway involves coumaroyl-CoA sourced either directly from phenylpropanoid biosynthesis or derived from cinnamoyl-CoA.63 Recent studies of transcriptomics analysis demonstrated that biosynthesis pathway for stilbenoids, diarylheptanoids, and gingerol have significantly enriched in Morella cerifera seedlings in response to alkali stress and leaf transcriptomic response mediated by cold stress in two maize inbred lines.64,65 Fig. S10A† shows that regulating of biosynthesis pathway for stilbenoids, diarylheptanoids, and gingerol not only affect the mRNA level but also will continue to regulate the level of proteomics in our experiments. Plants use unsaturated fatty acids (UFAs) to store energy.66 These UFAs are used to produce aliphatic compounds, such as membrane glycerolipids, TAG (triacylglycerol), cutin/suberin, jasmonates, and nitroalkenes (NO2-FAs). These products, as well as the UFAs themselves, play a key role in plant defense against abiotic and biotic stresses.66 At low temperatures, the degree of unsaturation of fatty acids increases through complex biosynthesis pathways. Cold acclimation increases the ratio of unsaturated to saturated fatty acids.67 in our integrative analysis, several genes such as acyl CoA thioesterase, long chain enoyl CoA reductase which participate in TAG regulation has been regulated at both gene and protein level (Fig. S10B†). Overall, through integration transcriptomics and proteomics data sets, we identified the features that exhibited the same trend to restoration expression in response to CBNs application and can be linked to mechanisms of such effect with high level of confidence.
Integrating multiple “omics” approaches not only can enhance our understanding of the beneficial biological effects of nanomaterials but also can play a critical role in advancing comprehensive risk assessment of use of nanomaterials in agriculture. The application of CBNs in agricultural environments may lead to certain ecotoxicological concerns that merit detailed examination. One of the primary risks is leaching, as these nanomaterials can migrate from treated soils into nearby water bodies, potentially affecting aquatic ecosystems.68,69 Documented chemical stability and persistence of CBNs in the environment mean they may accumulate over time, leading to long-term exposure risks that are not yet fully understood. Furthermore, CNTs and graphene may interact with soil microbiota, which play crucial roles in nutrient cycling, organic matter decomposition, and plant health. Studies have shown that these nanomaterials can potentially alter microbial community composition and enzymatic activity, potentially disrupting these essential functions.70,71 Our group demonstrated that the use of CNTs for nanofertilization at very modest doses did not impact the overall diversity or richness of soil microbial communities. However, it did lead to changes in the relative abundance of specific bacterial groups. While the presence of CNTs caused shifts in microbial community composition, the dominant operational taxonomic units (OTUs) remained consistent across treatments.9 These findings underscore that although CNTs and graphene hold great promise for agricultural applications; their use must be carefully balanced against potential environmental risks. Comprehensive ecotoxicological evaluations are essential to fully assess and mitigate these risks. To achieve this, the application of “multi-omics” approaches is critically important. These tools enable a more holistic understanding of how nanomaterials impact biological systems at multiple levels, from changes in microbial community structure to alterations in gene expression, protein activity, and metabolic function. By integrating data across these “omics” layers, researchers can gain deeper insight into the subtle and potentially cumulative effects of nanomaterial exposure, which may not be evident from single-method studies alone. This systems-level perspective is essential for accurately predicting ecological consequences and guiding the responsible use of nanotechnologies in agriculture.
Conclusion
The generated data demonstrated that integrating multiple “omics” levels is a powerful strategy for uncovering the biological mechanisms underlying nanomaterial interactions with plants. By applying bioinformatics to correlate transcriptomic and proteomic data, we identified key molecular components responsible for the positive effects of CBNs on plant tolerance to environmental stress. Using a tomato model exposed to salinity stress, with and without CBN treatment, we identified genes and proteins that exhibited consistent expression trends across both “omics” levels. This integrative approach enabled us to refine and validate a set of conserved molecular features and associated pathways that likely play a crucial role in CBN-enhanced salt stress tolerance. Specifically, reactive oxygen species (ROS) scavengers, heat shock proteins, aquaporins, ubiquitins, protein kinases linked to MAPK and inositol signaling, and hormone receptors emerged as key conserved elements. Overall, this multi-omics methodology provided valuable insights into the regulatory networks connecting transcriptomic and proteomic responses in CBN-treated plants. Furthermore, it allowed us to fully validate mechanistic hypotheses initially proposed based on transcriptomic data alone, reinforcing the importance of multi-omics integration in plant nanobiotechnology research.Data availability
The part of the data has been included in the ESI.† Transcriptomics data used for the integrative analysis were published early (https://doi.org/10.1021/acsabm.1c00108). Both data sets (transcriptions and proteomics) were deposited in public repositories. The transcriptomics data set was deposited in NIH BioProject Database (https://www.ncbi.nlm.nih.gov/bioproject/1270704) under PRJNA1270704. The proteomics data set was deposited in the PRIDE Proteomics Database under the project number PXD065030.Author contributions
Sajedeh Rezaei Cherati: investigation, methodology, writing original draft, visualization, review and editing. Mariya Khodakovskaya: conceptualization, methodology, supervision, writing, review and editing.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors express their gratitude to Novogene, Co., Ltd. for conducting the proteomic analysis of tomato samples. They also extend their appreciation to Dr. Stephanie D. Byrum and her team at the Department of Biochemistry and Molecular Biology, University of Arkansas for Medical Sciences, for their assistance in integrating transcriptomic and proteomic data. The authors thank Kari Vinzant for help with the improvement of the several Figures. The foundational infrastructure for this research was initially established with funding from USDA-NIFA (ARFI 2020-04096 – Award to MVK).References
- A. Husen, Engineered Nanomaterials for Sustainable Agricultural Production, Soil Improvement and Stress Management, Academic Press, 2022, p. 548 Search PubMed.
- S. R. Djiwanti and S. Kaushik, Nanopesticide: Future Application of Nanomaterials in Plant Protection. In: Plant Nanobionics, Approaches in Nanoparticles, Biosynthesis, and Toxicity, ed. R. Prasad, Springer International Publishing, Cham, 2019, vol. 2, pp. 255–298, Available from: DOI:10.1007/978-3-030-16379-2_10.
- J. Sangeetha, R. Hospet, D. Thangadurai, C. O. Adetunji, S. Islam and N. Pujari, et al., Nanopesticides, Nanoherbicides, and Nanofertilizers: The Greener Aspects of Agrochemical Synthesis Using Nanotools and Nanoprocesses Toward Sustainable Agriculture, In: Handbook of Nanomaterials and Nanocomposites for Energy and Environmental Applications, ed. O. V. Kharissova, L. M. Torres-Martínez and B. I. Kharisov, Springer International Publishing, Cham, 2021, pp. 1663–1677, Available from: DOI:10.1007/978-3-030-36268-3_44.
- T. Thirugnanasambandan, Advances of Engineered Nanofertilizers for Modern Agriculture. In: Plant-Microbes-Engineered Nano-particles (PM-ENPs) Nexus in Agro-Ecosystems: Understanding the Interaction of Plant, Microbes and Engineered Nano-particles (ENPS), ed. P. Singh, R. Singh, P. Verma, R. Bhadouria, A. Kumar and M. Kaushik, Springer International Publishing, Cham, 2021, pp. 131–152, Available from: DOI:10.1007/978-3-030-66956-0_9.
- R. Bhagat, A. P. Ingle and H. Chen, 5 - Nanosensors and nanobiosensors for sustainable agriculture. In: Nanotechnology in Agriculture and Agroecosystems, ed. A. P. Ingle, Elsevier, 2023, pp. 93–112, (Micro and Nano Technologies), Available from: https://www.sciencedirect.com/science/article/pii/B9780323994460000143 Search PubMed.
- X. Xin, J. D. Judy, B. B. Sumerlin and Z. He, Nano-enabled agriculture: from nanoparticles to smart nanodelivery systems, Environ. Chem., 2020, 17(6), 413–425 CrossRef CAS , Available from: https://www.publish.csiro.au/en/EN19254.
- S. S. Salem and A. Husen, Effect of engineered nanomaterials on soil microbiomes and their association with crop growth and production. In: Engineered Nanomaterials for Sustainable Agricultural Production, Soil Improvement and Stress Management, ed. A. Husen, Academic Press, ch. 14, 2023, pp. 311–336, (Plant Biology, sustainability and climate change), Available from: https://www.sciencedirect.com/science/article/pii/B9780323919333000106 Search PubMed.
- M. H. Lahiani, E. Dervishi, J. Chen, Z. Nima, A. Gaume and A. S. Biris, et al., Impact of Carbon Nanotube Exposure to Seeds of Valuable Crops, ACS Appl. Mater. Interfaces, 2013, 5(16), 7965–7973 CrossRef CAS PubMed.
- M. V. Khodakovskaya, B. S. Kim, J. N. Kim, M. Alimohammadi, E. Dervishi and T. Mustafa, et al., Carbon Nanotubes as Plant Growth Regulators: Effects on Tomato Growth, Reproductive System, and Soil Microbial Community, Small, 2013, 9(1), 115–123 CrossRef CAS PubMed , Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/smll.201201225.
- K. Pandey, M. H. Lahiani, V. K. Hicks, M. K. Hudson, M. J. Green and M. Khodakovskaya, Effects of carbon-based nanomaterials on seed germination, biomass accumulation and salt stress response of bioenergy crops, PLoS One, 2018, 13(8), e0202274 CrossRef PubMed.
- S. Rezaei Cherati, S. Shanmugam, K. Pandey and M. V. Khodakovskaya, Whole-Transcriptome Responses to Environmental Stresses in Agricultural Crops Treated with Carbon-Based Nanomaterials, ACS Appl. Bio Mater., 2021, 4(5), 4292–4301, DOI:10.1021/acsabm.1c00108.
- R. Javed, M. Bilal, J. S. Ali, S. Khan and M. Cheema, Nanotechnology: A Tool for the Development of Sustainable Agroindustry, In: Agricultural and Environmental Nanotechnology: Novel Technologies and their Ecological Impact, ed. F. Fernandez-Luqueno and J. K. Patra, Springer Nature, Singapore, 2023, pp. 317–339, Available from: DOI:10.1007/978-981-19-5454-2_11.
- A. A. Sembada and I. W. Lenggoro, Transport of Nanoparticles into Plants and Their Detection Methods, Nanomaterials, 2024, 14(2), 131 CrossRef CAS PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10819755/.
- H. Villagarcia, E. Dervishi, K. de Silva, A. S. Biris and M. V. Khodakovskaya, Surface chemistry of carbon nanotubes impacts the growth and expression of water channel protein in tomato plants, Small, 2012, 8(15), 2328–2334 CrossRef CAS PubMed.
- M. C. Martínez-Ballesta, L. Zapata, N. Chalbi and M. Carvajal, Multiwalled carbon nanotubes enter broccoli cells enhancing growth and water uptake of plants exposed to salinity, J. Nanobiotechnol., 2016, 14(1), 42, DOI:10.1186/s12951-016-0199-4.
- S. Majumdar and A. A. Keller, Omics to address the opportunities and challenges of nanotechnology in agriculture, Crit. Rev. Environ. Sci. Technol., 2021, 51(22), 2595–2636, DOI:10.1080/10643389.2020.1785264.
- W. Li and A. A. Keller, Assessing the Impacts of Cu and Mo Engineered Nanomaterials on Crop Plant Growth Using a Targeted Proteomics Approach, ACS Agric. Sci. Technol., 2024, 4(1), 103–117, DOI:10.1021/acsagscitech.3c00431.
- D. L. McGehee, M. H. Lahiani, F. Irin, M. J. Green and M. V. Khodakovskaya, Multiwalled Carbon Nanotubes Dramatically Affect the Fruit Metabolome of Exposed Tomato Plants, ACS Appl. Mater. Interfaces, 2017, 9(38), 32430–32435, DOI:10.1021/acsami.7b10511.
- S. Rezaei Cherati, M. Anas, S. Liu, S. Shanmugam, K. Pandey and S. Angtuaco, et al., Comprehensive Risk Assessment of Carbon Nanotubes Used for Agricultural Applications, ACS Nano, 2022, 16(8), 12061–12072, DOI:10.1021/acsnano.2c02201.
- Y. Li, L. Ma, D. Wu and G. Chen, Advances in bulk and single-cell multi-omics approaches for systems biology and precision medicine, Briefings Bioinf., 2021, 22(5), bbab024, DOI:10.1093/bib/bbab024.
- T. Stuart and R. Satija, Integrative single-cell analysis, Nat. Rev. Genet., 2019, 20(5), 257–272 CrossRef CAS PubMed , Available from: https://www.nature.com/articles/s41576-019-0093-7.
- D. Leng, L. Zheng, Y. Wen, Y. Zhang, L. Wu and J. Wang, et al., A benchmark study of deep learning-based multi-omics data fusion methods for cancer, Genome Biol., 2022, 23(1), 171, DOI:10.1186/s13059-022-02739-2.
- H. Chen, Metal based nanoparticles in agricultural system: behavior, transport, and interaction with plants, Chem. Speciation Bioavailability, 2018, 30(1), 123–134 CrossRef CAS.
- S. Majumdar, L. Pagano, J. A. Wohlschlegel, M. Villani, A. Zappettini and J. C. White, et al., Proteomic, gene and metabolite characterization reveal the uptake and toxicity mechanisms of cadmium sulfide quantum dots in soybean plants, Environ. Sci.:Nano, 2019, 6(10), 3010–3026 RSC , Available from: https://rsc.66557.net/en/content/articlelanding/2019/en/c9en00599d.
- H. Salehi, A. Chehregani, L. Lucini, A. Majd and M. Gholami, Morphological, proteomic and metabolomic insight into the effect of cerium dioxide nanoparticles to Phaseolus vulgaris L. under soil or foliar application, Sci. Total Environ., 2018, 616–617, 1540–1551 CrossRef CAS PubMed.
- L. Zhao, H. Zhang, J. Wang, L. Tian, F. Li and S. Liu, et al., C60 Fullerols Enhance Copper Toxicity and Alter the Leaf Metabolite and Protein Profile in Cucumber, Environ. Sci. Technol., 2019, 53(4), 2171–2180 CrossRef CAS PubMed.
- X. Chen, J. Wang, R. Wang, D. Zhang, S. Chu and X. Yang, et al., Insights into growth-promoting effect of nanomaterials: Using transcriptomics and metabolomics to reveal the molecular mechanisms of MWCNTs in enhancing hyperaccumulator under heavy metal(loid)s stress, J. Hazard. Mater., 2022, 439, 129640 CrossRef CAS PubMed , Available from: https://www.sciencedirect.com/science/article/pii/S0304389422014339.
- X. Li, S. Sun, S. Guo and X. Hu, Identifying the Phytotoxicity and Defense Mechanisms Associated with Graphene-Based Nanomaterials by Integrating Multiomics and Regular Analysis, Environ. Sci. Technol., 2021, 55(14), 9938–9948, DOI:10.1021/acs.est.0c08493.
- Z. Chen, Z. Guo, J. Niu, N. Xu, X. Sui and H. A. Kareem, et al., Phytotoxic effect and molecular mechanism induced by graphene towards alfalfa (Medicago sativa L.) by integrating transcriptomic and metabolomics analysis, Chemosphere, 2022, 290, 133368 CrossRef CAS PubMed , Available from: https://www.sciencedirect.com/science/article/pii/S004565352103842X.
- M. H. Lahiani, K. Gokulan, K. Williams, M. V. Khodakovskaya and S. Khare, Graphene and carbon nanotubes activate different cell surface receptors on macrophages before and after deactivation of endotoxins, J. Appl. Toxicol., 2017, 37(11), 1305–1316 CrossRef CAS PubMed.
- E. Zhang, X. Chen and X. Liang, Resolubilization of TCA precipitated plant proteins for 2-D electrophoresis, Electrophoresis, 2011, 32(6–7), 696–698 CrossRef CAS PubMed , Available from: https://onlinelibrary.wiley.com/doi/10.1002/elps.201000557.
- S. Hussain, M. Shaukat, M. Ashraf, C. Zhu, Q. Jin and J. Zhang, Salinity Stress in Arid and Semi-Arid Climates: Effects and Management in Field Crops, Climate Change and Agriculture, IntechOpen, 2019, Available from: DOI:10.5772/intechopen.87982.
- H. M. Alkharabsheh, M. F. Seleiman, O. A. Hewedy, M. L. Battaglia, R. S. Jalal and B. A. Alhammad, et al., Field Crop Responses and Management Strategies to Mitigate Soil Salinity in Modern Agriculture: A Review, Agronomy, 2021, 11(11), 2299 CrossRef CAS , Available from: https://www.mdpi.com/2073-4395/11/11/2299.
- F. Rohart, B. Gautier, A. Singh and K. A. L. Cao, mixOmics: An R package for ‘omics feature selection and multiple data integration, PLoS Comput. Biol., 2017, 13(11), e1005752 CrossRef PubMed , Available from: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005752.
- C. Meng, A. Basunia, B. Peters, A. M. Gholami, B. Kuster and A. C. Culhane, MOGSA: Integrative Single Sample Gene-set Analysis of Multiple Omics Data, Mol. Cell. Proteomics, 2019, 18(8 suppl 1), S153–S168 CrossRef CAS PubMed.
- T. Balasubramaniam, G. Shen, N. Esmaeili and H. Zhang, Plants' Response Mechanisms to Salinity Stress, Plants, 2023, 12(12), 2253 CrossRef CAS PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10300796/.
- Z. Hossain, M. Z. Nouri and S. Komatsu, Plant cell organelle proteomics in response to abiotic stress, J. Proteome Res., 2012, 11(1), 37–48 CrossRef CAS PubMed.
- P. Singh, K. K. Choudhary, N. Chaudhary, S. Gupta, M. Sahu and B. Tejaswini, et al., Salt stress resilience in plants mediated through osmolyte accumulation and its crosstalk mechanism with phytohormones, Front. Plant Sci., 2022, 13, 1006617 CrossRef PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9552866/.
- N. Tuteja, Mechanisms of High Salinity Tolerance in Plants, In: Methods in Enzymology, ed. D. Häussinger and H. Sies, Academic Press, ch. 24, 2007, vol. 428, pp. 419–438, (Osmosensing and Osmosignaling), Available from: https://www.sciencedirect.com/science/article/pii/S0076687907280243 Search PubMed.
- M. A. Rahman, J. H. Woo, Y. Song, S. H. Lee, M. M. Hasan and M. A. K. Azad, et al., Heat Shock Proteins and Antioxidant Genes Involved in Heat Combined with Drought Stress Responses in Perennial Rye Grass, Life, 2022, 12(9), 1426 CrossRef CAS PubMed , Available from: https://www.mdpi.com/2075-1729/12/9/1426.
- Z. Xu, P. Marowa, H. Liu, H. Du, C. Zhang and Y. Li, Genome-Wide Identification and Analysis of P-Type Plasma Membrane H+-ATPase Sub-Gene Family in Sunflower and the Role of HHA4 and HHA11 in the Development of Salt Stress Resistance, Genes, 2020, 11(4), 361 CrossRef CAS PubMed , Available from: https://www.mdpi.com/2073-4425/11/4/361.
- M. V. Khodakovskaya, K. de Silva, D. A. Nedosekin, E. Dervishi, A. S. Biris and E. V. Shashkov, et al., Complex genetic, photothermal, and photoacoustic analysis of nanoparticle-plant interactions, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(3), 1028–1033 CrossRef CAS PubMed.
- C. J. Park and Y. S. Seo, Heat Shock Proteins: A Review of the Molecular Chaperones for Plant Immunity, Plant Pathol. J., 2015, 31(4), 323–333 CrossRef CAS PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4677741/.
- M. R. Escobar, I. Feussner and E. M. Valle, Mitochondrial Small Heat Shock Proteins Are Essential for Normal Growth of Arabidopsis thaliana, Plant Pathol. J., 2021, 12, 600426 Search PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7902927/.
- R. Al-Saharin, H. Hellmann and S. Mooney, Plant E3 Ligases and Their Role in Abiotic Stress Response, Cells., 2022, 11(5), 890 CrossRef CAS PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8909703/.
- M. Thalmann and D. Santelia, Starch as a determinant of plant fitness under abiotic stress, New Phytol., 2017, 214(3), 943–951 CrossRef CAS PubMed , Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/nph.14491.
- C. Li, Y. Li, P. Chu, Z. Hao-hao, Z. Wei and Y. Cheng, et al., Effects of salt stress on sucrose metabolism and growth in Chinese rose (Rosa chinensis), Biotechnol. Biotechnol. Equip., 2022, 36(1), 706–716, DOI:10.1080/13102818.2022.2116356.
- C. Ribeiro, M. Stitt and C. T. Hotta, How Stress Affects Your Budget—Stress Impacts on Starch Metabolism, Front. Plant Sci., 2022, 13 Search PubMed , Available from: https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2022.774060/full.
- Y. Du, Q. Zhao, L. Chen, X. Yao, W. Zhang and B. Zhang, et al., Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings, Plant Physiol. Biochem., 2020, 146, 1–12 CrossRef CAS PubMed , Available from: https://www.sciencedirect.com/science/article/pii/S0981942819304541.
- O. Stein and D. Granot, An Overview of Sucrose Synthases in Plants, Front. Plant Sci., 2019, 10, 95 CrossRef PubMed.
- G. Taj, P. Agarwal, M. Grant and A. Kumar, MAPK machinery in plants, Plant Signaling Behav., 2010, 5(11), 1370–1378 CrossRef CAS PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3115236/.
- A. Danquah, A. de Zelicourt, J. Colcombet and H. Hirt, The role of ABA and MAPK signaling pathways in plant abiotic stress responses, Biotechnol. Adv., 2014, 32(1), 40–52 CrossRef CAS PubMed.
- C. Hettenhausen, M. C. Schuman and J. Wu, MAPK signaling – a key element in plant defense response to insects, Insect Sci., 2015, 22(2), 157–164 CrossRef CAS PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5295641/.
- A. Shomali, S. Das, N. Arif, M. Sarraf, N. Zahra and V. Yadav, et al., Diverse Physiological Roles of Flavonoids in Plant Environmental Stress Responses and Tolerance, Plants, 2022, 11(22), 3158 CrossRef CAS PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9699315/.
- M. Di Ferdinando, C. Brunetti, A. Fini and M. Tattini, Flavonoids as Antioxidants in Plants Under Abiotic Stresses, In: Abiotic Stress Responses in Plants: Metabolism, Productivity and Sustainability, ed. P. Ahmad and M. N. V. Prasad, Springer, New York, NY, 2012, pp. 159–179, DOI:10.1007/978-1-4614-0634-1_9.
- Q. Li, H. M. Yu, X. F. Meng, J. S. Lin, Y. J. Li and B. K. Hou, Ectopic expression of glycosyltransferase UGT76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis, Plant Biol., 2018, 20(1), 10–19 CrossRef CAS PubMed.
- S. M. Nabavi, D. Šamec, M. Tomczyk, L. Milella, D. Russo and S. Habtemariam, et al., Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering, Biotechnol. Adv., 2020, 38, 107316 CrossRef CAS PubMed.
- R. Waadt, C. A. Seller, P. K. Hsu, Y. Takahashi, S. Munemasa and J. I. Schroeder, Plant hormone regulation of abiotic stress responses, Nat. Rev. Mol. Cell Biol., 2022, 23(10), 680–694 CrossRef CAS PubMed , Available from: https://www.nature.com/articles/s41580-022-00479-6.
- K. Shu, W. Zhou, F. Chen, X. Luo and W. Yang, Abscisic Acid and Gibberellins Antagonistically Mediate Plant Development and Abiotic Stress Responses, Front. Plant Sci., 2018, 9 Search PubMed , Available from: https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2018.00416/full.
- F. N. Ritonga, D. Zhou, Y. Zhang, R. Song, C. Li and J. Li, et al., The Roles of Gibberellins in Regulating Leaf Development, Plants, 2023, 12(6), 1243 CrossRef CAS PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10051486/.
- E. H. Colebrook, S. G. Thomas, A. L. Phillips and P. Hedden, The role of gibberellin signalling in plant responses to abiotic stress. Davies SA, Dow JAT, Lukowiak K, editors, J. Exp. Biol., 2014, 217(1), 67–75, DOI:10.1242/jeb.089938.
- Y. S. Ku, M. Sintaha, M. Y. Cheung and H. M. Lam, Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses, Int. J. Mol. Sci., 2018, 19(10), 3206 CrossRef PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6214094/.
- PubChem. Stilbenoid, Diarylheptanoid, and Gingerol Biosynthesis. [cited 2024 May 21]. Available from: https://pubchem.ncbi.nlm.nih.gov/pathway/PathBank:SMP0063461.
- Y. Jiao, R. J. Xie and H. M. Jia, Identification of Potential Pathways of Morella cerifera Seedlings in Response to Alkali Stress via Transcriptomic Analysis, Plants, 2022, 11(8), 1053 CrossRef CAS PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9026155/.
- T. Yu, J. Zhang, J. Cao, Q. Cai, X. Li and Y. Sun, et al., Leaf transcriptomic response mediated by cold stress in two maize inbred lines with contrasting tolerance levels, Genomics, 2021, 113(2), 782–794 CrossRef CAS PubMed , Available from: https://www.sciencedirect.com/science/article/pii/S0888754321000446.
- M. He and N. Z. Ding, Plant Unsaturated Fatty Acids: Multiple Roles in Stress Response, Front. Plant Sci., 2020, 11, 562785 CrossRef PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7500430/.
- G. Zheng, B. Tian, F. Zhang, F. Tao and W. Li, Plant adaptation to frequent alterations between high and low temperatures: remodeling of membrane lipids and maintenance of unsaturation levels, Plant, Cell Environ., 2011, 34(9), 1431–1442 CrossRef CAS PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3980542/.
- P. C. Ray, H. Yu and P. P. Fu, Toxicity and environmental risks of nanomaterials: challenges and future needs, J. Environ. Sci. Health, Part C:Environ. Carcinog. Ecotoxicol. Rev., 2009, 27(1), 1–35 CrossRef CAS PubMed.
- A. Freixa, V. Acuña, J. Sanchís, M. Farré, D. Barceló and S. Sabater, Ecotoxicological effects of carbon based nanomaterials in aquatic organisms, Sci. Total Environ., 2018, 619–620, 328–337 CrossRef CAS PubMed.
- A. Mukherjee, S. Majumdar, A. D. Servin, L. Pagano, O. P. Dhankher and J. C. White, Carbon Nanomaterials in Agriculture: A Critical Review, Front. Plant Sci., 2016, 7, 172 Search PubMed , Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4762280/.
- H. Qian, M. Ke, Q. Qu, X. Li, B. Du and T. Lu, et al., Ecological Effects of Single-Walled Carbon Nanotubes on Soil Microbial Communities and Soil Fertility, Bull. Environ. Contam. Toxicol., 2018, 101(4), 536–542 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Statistical results of quantitative proteomics analysis; function annotation description of GO, KEGG, COG, IPR, and subcellular localization; volcano plots of differently expressed proteins; subcellular localization of the differently expressed proteins; analysis of quantification of FBA protein using western blot technique; relative transcript abundance of tomato EIF3J and RBM8A genes; GO enrichment analysis of differently expressed proteins in tomato seedlings; KEGG pathway enrichment analysis of differently expressed proteins in tomato seedlings; IPR domain enrichment analysis of differently expressed proteins in tomato seedlings; gene influential score of selected pathways; annotated proteins that fully or partially restored their expression in the presence of CBNs in tomato seedlings; integrated genes and proteins with matched and unmatched dysregulation in tomato seedlings treated with salt, seedlings treated with salt in the presence of CNTs, and seedlings treated with salt in the presence of graphene compared with control (PDF). See DOI: https://doi.org/10.1039/d5en00327j |
| This journal is © The Royal Society of Chemistry 2025 |