Open Access Article
Open Access ArticleNanoparticle-based biosensors for virus detection in food systems: from farm to fork
Riann Martin Sarza
 ab,
Laura Sutarlie
ab,
Laura Sutarlie
 b,
Sam Fong Yau Li
b,
Sam Fong Yau Li
 *ac and
Xiaodi Su
*ac and
Xiaodi Su
 *ab
*ab
aDepartment of Chemistry, Faculty of Science, National University of Singapore, Block S8, Level 3, 3 Science Drive 3, 117543, Singapore. E-mail: chmlifys@nus.edu.sg; xd-su@imre.a-star.edu.sg
bInstitute of Materials Research and Engineering (IMRE), Agency for Science, Technology, and Research (A*STAR), 2 Fusionopolis Way, Innovis #08-03, 138634, Singapore
cNUS Environmental Research Institute, National University of Singapore, T-Lab Building, 5A Engineering Drive 1, 117411, Singapore
First published on 10th July 2025
Abstract
Viral contamination in food systems poses significant risks to public health, agricultural productivity, and economic stability due to the persistence of viruses across key stages in the food system. This review examines the expanding role of nanoparticle-based biosensors for detecting these viral threats. A key novelty of this work is its system-wide perspective, which, unlike most reviews that focus on a single virus category, connects viral threats and technological solutions across the entire food chain, from production to consumption. It encompasses critical foodborne viruses like norovirus, major livestock viruses such as avian influenza virus, and economically important crop-affecting viruses such as maize chlorotic mottle virus. The review begins by first outlining the major viral challenges with the food system, providing a holistic context for detection needs. Following this, an overview of key nanoparticles and viral analytes central to biosensor design is presented. The core of this work is the critical assessment of nanobiosensor innovations for four major foodborne viruses, key livestock viruses, and multiple crop viruses, evaluating the performance and practical limitations of each technology. Finally, the review addresses the overarching challenges and future perspectives crucial for translating these technologies from the lab to the field. We provide a detailed analysis of biological hurdles like non-culturability, as well as logistical barriers including the food matrix effect, and manufacturing scalability. Promising future directions, such as multiplexing and AI integration, are also explored. While most developments remain at the proof-of-concept stage, this review concludes that nanoparticle-enhanced biosensors show clear potential for becoming a needed tool to strengthen food system resilience.
1. Introduction
Food systems include all the actions and activities related to producing and consuming food, from farm to fork, performed at local to global levels with diverse stakeholders. Their efficiency impacts food security, nutrition, public health, and economic stability. However, modern food systems face significant threats, including climate change, extreme weather, supply chain disruptions, and labor shortages, which undermine food security. Among many food contamination issues, pathogens, such as bacteria, fungi, and viruses, cause foodborne illnesses and disrupt supply chains.1 While food safety research has primarily focused on bacterial contamination, the detection and study of viral contamination present unique challenges.Viruses, ranging from 20 to 300 nanometers, cause diseases in plants, animals, and humans, impacting food safety and economic stability. Fig. 1 illustrates common viruses associated with the major stages or components of food systems, namely production, processing, distribution, consumption and waste management. At the production level, for example, they cause staggering agricultural losses, with viral infections responsible for USD 30 billion in annual crop losses and single outbreaks of livestock diseases like avian influenza costing over USD 2.5 billion.2 At the consumer level, they are responsible for an estimated 30% of all foodborne diseases.3 This pervasive risk is amplified by two key viral characteristics: they can spread through multiple vectors (e.g., contaminated water, soil, infrastructure, and infected handlers), and they can withstand food-processing conditions that typically inactivate bacteria.4–6
Addressing these multi-stage threats requires rapid and robust detection. However, conventional methods such as viral culture, PCR, and immunoassays, while valuable, are often too slow, expensive, or lab-bound to be effective for on-site monitoring at a farm, processing plant, or port of entry. Furthermore, techniques like PCR may detect non-viable viral particles, complicating immediate risk assessment.7 This creates a demand for new technologies that are not only sensitive but also portable and field-ready.
Biosensors offer a promising alternative for virus detection in food systems, using optical, electrochemical, and piezoelectric mechanisms to identify contaminants such as norovirus, hepatitis A virus, and rotavirus.8 They also detect viruses affecting crops and livestock.9–12 Compared with conventional methods, biosensors provide rapid results, cost efficiency, and portability, making them suitable for field use.13 However, challenges remain, such as achieving high sensitivity and specificity for low viral loads in complex food matrices. Furthermore, biosensors must also be portable and robust for real-world deployment.14
Nanoparticle-based biosensors, or nanobiosensors, are emerging as a powerful solution to this challenge, offering the potential for rapid, cost-effective, and on-site virus detection. Owing to their unique physicochemical properties, the addition of nanoparticles to traditional biosensors has greatly improved sensitivity, selectivity, and detection limits. While existing reviews provide excellent coverage of these technologies for consumer-level foodborne viruses (e.g., Zhu et al., 2023),15 a holistic assessment connecting threats and technological solutions across the entire food system, spanning production, processing, and consumption, has yet to be presented.
This review systematically analyzes nanoparticle-based biosensors for viral threats across all critical stages of the food system: production (livestock and crops), processing, and consumption. We first establish a foundation by outlining major viral challenges at each stage, followed by an overview of key nanoparticles and viral analytes central to biosensor design. The core of the review critically assesses nanobiosensor innovations for detecting four major foodborne viruses, three key livestock viruses, and several crop viruses, evaluating their performance and limitations. Finally, we discuss translational challenges and future perspectives, including multiplexing and AI integration, that are crucial for strengthening global food system resilience against viral threats.
2. Major viruses concerning food systems
Viruses in food systems pose threats to public health, trade, and economic stability. As seen in Fig. 1, viruses can enter any part of the food system, affecting various commodities. Table 1 outlines different food commodities, their vulnerable stages in the food system, the viruses affecting them, detectable samples, and associated risks. Although some commodities, like livestock, are linked to specific food system stages, their vulnerability extends across production, processing, distribution, consumption, and waste management, emphasizing the need for holistic detection and mitigation strategies.| Food commodity | Vulnerable stage(s) in the food system | Virus species | Biological structure of the viruses | Detectable samples | Risks | Ref. |
|---|---|---|---|---|---|---|
| Livestock | ● Production | Foot-and-mouth disease virus (FMDV) | RNA virus, spherical, ∼25 nm | Saliva, milk, and excreta of infected livestock | Highly contagious, causing economic losses in agriculture and trade restrictions | 16 |
| ● Processing | ||||||
| ● Distribution and retail | ||||||
| ● Production | African swine fever virus (ASFV) | DNA virus, icosahedral, ∼200 nm | Contaminated meat and waste products | High mortality in swine populations, major economic impacts on pig farming | 17 | |
| ● Processing | ||||||
| ● Waste management | ||||||
| ● Production | Avian influenza virus (AIV) | RNA virus, pleomorphic, ∼80–120 nm | Infected poultry, eggs, and feathers | High pathogenicity, zoonotic risks, and global economic losses | 18 | |
| ● Processing | ||||||
| ● Distribution and retail | ||||||
| ● Consumption | ||||||
| Crop | ● Production | Maize chlorotic mottle virus (MCMV) | RNA virus, spherical, ∼40 nm | Infected maize plants | Causes stunting and necrosis in maize, reducing yields | 19 |
| ● Waste management | ||||||
| ● Production | Tomato spotted wilt virus (TSWV) | RNA virus, spherical, ∼80–110 nm | Infected vegetables and fruits | Reduces crop production; impacts availability and quality | 18 | |
| ● Waste management | ||||||
| ● Production | Sugarcane mosaic virus (SCMV) | RNA virus, spherical, ∼700–750 nm | Infected sugarcane | Reduces sugar yield and stalk productivity | 20 | |
| ● Waste management | ||||||
| Ready-to-eat food | ● Processing | Norovirus | RNA virus, spherical, ∼27 nm | ∼10−2 to 103 copies per mL in food and water | Acute gastroenteritis, highly infectious, outbreaks in communal settings | 21 |
| ● Distribution and retail | ||||||
| ● Consumption | ||||||
| ● Processing | Hepatitis A virus (HAV) | RNA virus, spherical, ∼27–32 nm | 10−4 to 103 copies per mL in contaminated produce | Liver infections, outbreaks from poor sanitation | 22 | |
| ● Consumption | ||||||
| Shellfish | ● Production | Norovirus | RNA virus, spherical, ∼27–38 nm | Bivalves harvested from polluted waters | Gastroenteritis, risks from consuming raw or undercooked shellfish | 23 |
| ● Processing | ||||||
| ● Consumption | ||||||
| ● Production | Hepatitis E virus (HEV) | RNA virus, spherical, ∼32–34 nm | Sewage-contaminated aquatic systems | Liver infections, severe for pregnant women and immunocompromised individuals | 24 | |
| ● Consumption | ||||||
| Beverages | ● Processing | Rotavirus | RNA virus, spherical, ∼70 nm | Unpasteurized water and juices | Severe diarrhea in children, risk increases with poor sanitation | 25 |
| ● Consumption | ||||||
| ● Processing | Hepatitis A virus (HAV) | RNA virus, spherical, ∼27–32 nm | Bottled water from unsafe sources | Liver infection, waterborne outbreaks | 26 | |
| ● Consumption |
During production, both livestock and crop viruses have profound implications for food security and international trade. Diseases such as foot-and-mouth disease (FMD) and highly pathogenic avian influenza (HPAI) have caused severe disruptions, with FMD alone estimated to result in global economic losses ranging from $6.5 to $21 billion due to reduced productivity, herd disruptions, market restrictions, and control measures9,27 Beyond economic losses, HPAI also raises public health concerns through its zoonotic potential, further compounding its impact on consumer confidence and the global poultry market. The resulting waste from culling operations and contaminated products also places significant strain on waste management systems.27,28
In crop systems, viruses such as tobacco mosaic virus (TMV) and wheat streak mosaic virus (WSMV) also harm production. TMV infects over 125 plant species, causing 65–75% yield losses in regions like Vietnam.29 WSMV reduces yields by up to 50%, worsening to 96% with co-infections like Triticum mosaic virus (TriMV).30 Highly resilient viruses like cucumber mosaic virus (CMV) infect over 800 species, complicating outbreak control and contributing to food shortages.31 These disruptions impact downstream food availability and increase supply chain losses.
Foodborne viruses such as norovirus (NoV) and hepatitis A virus (HAV) pose direct consumer risks, contaminating shellfish, fresh produce, and prepared foods.6,8 These viruses withstand freezing, acidity, and cooking processes that typically inactivate bacteria.5 NoV is highly infectious at doses as low as 20 particles, with contamination levels ranging from 10−2 to 103 copies per mL, while HAV in produce and shellfish ranges from 10−4 to 103 copies per mL, making detection difficult.4,5
Contamination risks arise throughout the food system, often originating in the earlier stages. For example, irrigation with fecal-contaminated water contributes to NoV and HAV outbreaks in fresh produce like strawberries, raspberries, and lettuce.32 Shellfish, soft berries, and salad vegetables are highly susceptible due to direct environmental exposure. These upstream vulnerabilities lead to widespread illness, particularly in low-immunity populations. Poor sanitation and infected food handlers during packaging further spread contamination, demonstrating how production issues impact consumer safety.33
As food moves along the food system, ready-to-eat products such as salads, sandwiches, and bakery items become increasingly vulnerable to contamination during handling and processing. Equipment surfaces, aerosols, and infected food handlers may facilitate NoV transmission.34 NoV persists on surfaces, easily spreading through contact. Deli meats and cold desserts also risk recontamination, emphasizing the need for rigorous monitoring and handling protocols. These vulnerabilities across production and processing stages amplify risks for consumers.35
The interconnectedness of production and consumption is evident in how viral risks propagate through food systems. HPAI reduces poultry availability while eroding consumer confidence, affecting demand and trade. Similarly, foodborne viruses like NoV and HAV originate in production and handling but severely impact consumers. These examples illustrate why robust detection and mitigation strategies must address risks across all stages.
Mitigating the systemic risks detailed above requires detection tools that can be deployed at critical control points, i.e., on the farm, in the processing plant, and at the port of entry. Traditional nucleic acid-based methods, while sensitive, are laboratory-bound and cannot provide the immediate data needed for such on-site risk management.7 This highlights the urgent need for new technologies. Biosensors have emerged as a promising solution, offering the potential for the rapid, portable, and cost-effective detection necessary to build a truly resilient food system.15,36
3. Nanoparticles in biosensor applications
The choice of nanoparticle (NP) for a biosensor is dictated by the target virus, food matrix, and detection method requirements. NPs serve as critical components by enabling signal transduction, signal amplification, or target isolation. Their properties, which depend on size, shape, and composition, allow for addressing specific viral detection challenges across food systems. These materials can be categorized into three groups: inorganic, organic, and hybrid nanoparticles.Inorganic nanoparticles are central to most biosensors, with material selection determined by the specific characteristics of the food sample. For optical detection, noble metals like gold (AuNPs) and silver (AgNPs) are widely used for their strong localized plasmon resonance (LSPR) signals in colorimetric assays. AgNPs exhibit sharper LSPR bands and greater sensitivity, while AuNPs offer superior biocompatibility. While effective in clear liquids, their optical signal can be obscured by the turbidity of food homogenates, limiting their performance in complex sample matrices.37,38 In such cases, fluorescent nanoparticles are often preferred. Quantum dots (QDs) and upconversion nanoparticles (UCNPs) enable fluorescence-based detection in food matrices but address autofluorescence differently. QDs provide tunable emission spectra, allowing partial interference mitigation through spectral optimization.39–41 UCNPs, however, eliminate autofluorescence from chlorophyll, riboflavin, and other food components entirely via near-infrared-to-visible photon upconversion, ensuring background-free signals even in complex samples like dairy or plant extracts.39 For electrochemical applications, which are inherently robust against sample color and turbidity, materials are chosen for their conductivity and catalytic activity. Carbon-based materials like graphene and carbon nanotubes offer exceptional conductivity for highly sensitive signal transduction.42 Platinum (PtNPs) and various metal oxides serve as powerful catalysts that enhance the electrochemical reaction, amplifying the signal to detect low viral loads.43,44 To achieve even greater performance, bimetallic nanoparticles (BMNPs) create synergistic effects. By combining metals like Au, Ag, and Pt, BMNPs can enhance both catalytic and optical properties simultaneously, enabling ultrasensitive detection through techniques like surface-enhanced Raman scattering (SERS) for clear liquids or high-efficiency electrocatalysis for opaque food samples.45–47 Separate from signal generation, magnetic nanoparticles (MNPs) play a unique and critical role in sample preparation, a mandatory step for most food matrices. Their superparamagnetism allows for efficient immunomagnetic separation to capture and isolate viruses from complex and viscous samples like milk, meat slurries, or wastewater.48 This crucial pre-analytical step purifies the sample by removing inhibitors that would otherwise interfere with detection, significantly improving the reliability and sensitivity of any subsequent measurement.49
Organic and hybrid nanoparticles offer advanced functionalities focused on stability and specific interactions within challenging food environments. Organic nanoparticles, including polymer-based structures, play a crucial role due to their biocompatibility and versatility. Synthesized through methods such as emulsion polymerization or nanoprecipitation, their size, shape, and surface chemistry can be precisely controlled. This allows for straightforward functionalization with biomolecules like antibodies or aptamers for highly specific virus detection. This tunability is especially valuable for food applications, as surfaces can be engineered to be “bio-inert” to repel the non-specific adhesion of proteins and fats from a food matrix that would otherwise cause false signals.50,51 Furthermore, advanced designs can incorporate stimulus-responsive polymers that react to specific environmental cues, for example, triggering signal release only in the low pH of spoiled milk or fermented products, thereby enhancing context-specific detection.52,53
Hybrid nanoparticles, such as metal–organic frameworks (MOFs), integrate organic and inorganic components into highly porous crystalline structures that excel at combining selective capture with robust protection. These materials offer an exceptionally high surface area and diverse functionalization options, with metal nodes providing structural stability and catalytic activity, while the organic linkers enable selective molecular interactions.54 For food biosensing, this is a powerful combination. The porous structure can act as a molecular cage, physically shielding fragile biorecognition elements like antibodies from degradative proteases found in samples like meat or dairy.12,55 At the same time, their tunable pore sizes can be engineered to act as a selective filter, allowing small target viruses to enter while blocking larger, interfering particles like cell debris, making MOFs a highly promising platform for robust detection in unprocessed food samples.56,57
The single greatest hurdle for any food-based biosensor is the “matrix effect”. Food samples are complex mixtures of proteins, fats, carbohydrates, and salts that can interfere with sensor performance.58,59 For example, the high ionic strength in shellfish homogenates can disrupt the electrostatic balance of unmodified AuNPs, causing them to aggregate and produce a false signal.60,61 Similarly, proteins and fats can non-specifically adsorb to the sensor surface, blocking access for the target virus.62 A primary strategy to overcome this is surface functionalization, often by coating the nanoparticle with a “stealth” layer of an inert polymer like polyethylene glycol (PEG). This coating prevents aggregation and minimizes non-specific binding, ensuring the sensor responds only to the target virus.63,64
Ultimately, the thoughtful selection and combination of these nanoparticles are what enable the development of biosensors that are not just sensitive in the lab, but robust and reliable in the complex and demanding context of the food system. The high surface-area-to-volume ratio allows for dense immobilization of capture probes, while the unique optical, electrical, and magnetic properties form the basis for diverse and powerful detection strategies.65,66
4. Viral analytes: what to target—nucleic acids, proteins or viral particles?
In biosensor design, selecting an appropriate viral target according to the virus type, the intended application, and the complexity of the food matrix is the first step. The choice between targeting nucleic acids, proteins, or whole particles fundamentally affects the sensitivity, specificity, and, most importantly, the relevance of the result for assessing food safety risks.Nucleic acids, widely employed in PCR-based assays, provide high specificity by targeting unique viral DNA or RNA sequences, allowing for precise identification and quantification at concentrations as low as 8 copies per mL.67 However, a significant limitation in food safety is that nucleic acid detection does not indicate viral viability. PCR often amplifies genetic material from non-infectious viruses that may persist in a food product, complicating risk assessment and outbreak control since viral presence does not always imply infection risk.4,68 Moreover, such methods require extensive sample processing, which increases assay time and contamination risk, thereby reducing their field applicability.
Viral proteins, particularly the structural capsid and envelope components involved in host recognition, represent an alternative biosensor target. Protein detection is more closely linked to viral viability, as these components are typically present in intact particles. This is especially relevant in the context of food processing, where many non-enveloped viruses like norovirus and hepatitis A possess robust capsids that allow them to retain structural integrity, and thus infectivity, despite exposure to common preservation methods like mild heating, acidification (e.g., in dressings or marinades), and freezing that would typically inactivate bacteria.4,5 Although protein-based assays may reduce false positives compared with nucleic acid detection, they cannot confirm infectivity outright, as the presence of free, non-infectious viral proteins can limit this correlation.15
For developing sensors against highly pathogenic or unculturable viruses, virus-like particles (VLPs), which are self-assembled capsid proteins without genetic material, offer a non-infectious yet structurally relevant platform. For instance, norovirus VLPs mimic native capsids, enhancing biosensor specificity without requiring live virus samples.69 A key advantage of using proteins or VLPs as targets is the ability to simulate natural virus–host interactions, using affinity ligands like antibodies or aptamers that bind to specific conformational shapes on the viral surface.
This focus on the capsid proteins leads to the ultimate goal for risk assessment: detecting the intact, potentially infectious virion itself. While no method can perfectly confirm infectivity outside of cell culture, assays that target capsid proteins on a whole particle provide a much stronger indication of risk than those detecting isolated genetic fragments. Both nucleic acid and protein-based approaches contribute essential capabilities. PCR methods offer exceptional sensitivity, often detecting 1–10 viral particles, while protein-based assays (e.g., antibody-based tests) typically detect 10–100 infectious units and depend heavily on antibody quality.70
This distinction in capabilities means that detection strategies are tailored to fit specific needs, from regulatory enforcement to on-site screening. For regulatory purposes and clinical diagnostics, the ultra-sensitive quantification of PCR is prioritized. In contrast, for on-site use, stakeholders like manufacturers and retailers prefer portable, rapid tests such as lateral flow assays, which provide actionable results within minutes even if they require higher viral titers for detection. The future of viral detection in food systems will likely involve a combination of these approaches: rapid on-site screening to identify potential contamination, followed by laboratory-based molecular methods for confirmation.
5. Nanobiosensors for viruses in food systems
Integrating biosensors into food monitoring systems can greatly enhance food safety by enabling real-time virus detection and supporting timely interventions.7,13,15 This section focuses on biosensors, specifically nanosensors, designed for detecting viruses in two, out of the five, critical segments of the food system: consumption and production. These segments are emphasized as they represent the primary entry points for viruses into the food system.13,15 On the consumption side, four foodborne viruses, namely norovirus (NoV), hepatitis A virus (HAV), hepatitis E virus (HEV), and rotavirus, will be examined. Biosensors targeting foodborne viruses help mitigate health and safety risks to the consumer. On the production end, three major viruses affecting livestock, namely the foot-and-mouth disease virus (FMDV), avian influenza virus (AIV), and African swine fever virus (ASFV), and several viruses affecting crops, like maize chlorotic mottle virus (MCMV), are explored. These biosensors aid in early detection, improving productivity and agricultural yields. Table 2 provides a summary of the biosensors discussed and highlights their performance in detecting their respective target viruses.| Virus | Detection method/principle | Target | Nanoparticle | Reaction time | Performance | Ref. |
|---|---|---|---|---|---|---|
| Nanosensors for foodborne viruses | ||||||
| Norovirus | Impedimetric | Intact virus/viral capsid | WS2 decorated with peptide-functionalized AuNPs | 1st incubation: 60 min | LOD: 2.37 copies per mL (water), 6.21 copies per mL (oyster) | 71 |
| 2nd incubation: 15 min | ||||||
| AuNP–polyaniline nanocomposite | Not indicated | LOD: 60 ag mL−1 | 72 | |||
| Fluorescence | Intact virus/viral capsid | AuNP and carbon QD mixture | Not indicated | LOD: 80.3 pg mL−1 | 73 | |
| Fe3O4 MNPs and liposomes | 20 min | LOD: 136 copies per mL | 74 | |||
| Colorimetric | Intact virus/viral capsid | AuNP | Not indicated | LOD: 10 pfu mL−1 | 75 | |
| Colorimetric and electrochemical | Intact virus/viral capsid | CuO2@COF-NH2 nanocomposite | Colorimetric: 4.5 h | LOD: 0.125 copies per mL (colorimetric), 0.152 copies per mL (electrochemical) | 76 | |
| Electrochemical: 2 h | ||||||
| V2O5 nanoparticle-encapsulated liposomes | 20 min | LOD: 0.34 pg mL−1 (colorimetric), 4.1 fg mL−1 (electrochemical) | 77 | |||
| Linear range: 1 pg mL−1 to 100 ng mL−1 (colorimetric), 10 fg mL−1 to 10 pg mL−1 | ||||||
| Electrochemical | Intact virus/viral capsid | Au@BP@Ti3C2-MXene and Au@ZnFe2O4@COF | 30 min | LOD: 136 copies per mL | 78 | |
| Hepatitis A virus | Fluorescence | DNA | CdTe QD with carboxy group capping | 35 min | LOD: 13 pM | 79 |
| Percent recovery: 93.1–102.8% | ||||||
| Immunoassay/ELIME | Intact virus/viral capsid | Fe3O4@poly(dopamine) MNPs | 10 min | LOD: 1 × 10−11 IU mL−1 | 80 | |
| Resonance light scattering | MIP nanoprobe with DMA and MIL-101 MOF nanoparticles | 20 min | LOD: 0.1 pM | 56 | ||
| Hepatitis E virus | Fluorescence | DNA | QD-MNP nanocomposite | 30 min | LOD: 1.2 fg mL−1 | 81 |
| Electrochemical | DNA | MNP (for separation and concentration) | Not indicated | LOD: 0.81 fg mL−1 | 82 | |
| Pt–Co3O4 hollow cage (sensing material) | ||||||
| Colorimetric and electrochemical | DNA | AuNP-labelled reporter probes and MNP-labelled capture probes | Not indicated | LOD: 10 pM (visual), 1 fM (electrochemical) | 83 | |
| Rotavirus | SERS-lateral flow | Viral capsid | Au@Ag core–shell NPs | 20 min | LOD: 8 pg mL−1 | 84 |
| LSPR | Viral capsid | Triangular Au nanopillars | Not indicated | LOD: ∼250 virions in 2 μL solution (∼126 PFU mL−1) | 85 | |
| Electrochemical | RNA | Gold nanoflowers MXene and polypyrrole nanocomposite | 120 min | LOD: ∼0.8 aM | 86 | |
| Detection range: 10−18 to 10−7 | ||||||
| SERS | RNA | AuNPs deposited on Blu-ray DVD surface | 60 min | LOD: 10 ng μL−1 | 87 | |
| Reproducibility: 94% | ||||||
| AgNPs deposited on paper | 60 min | LOD: 20 nM (malachite green model system); unclear for actual rotavirus | 88 | |||
| Nanosensors for livestock viruses | ||||||
| Foot-and-mouth disease virus | Immunoassay | Intact virus/viral capsid | Antibody conjugated-AuNPs | 60 min | LOD: 10−4 sample dilution | 89 |
| Nano-PCR | RNA | AuNP | Not indicated | Efficiency: 94.5% | 90 | |
| Specificity: 100% towards FMDV | ||||||
| LAMP | RNA | AuNP and graphene oxide nanocomposite | 60 min | Minimized non-specific amplification | 91 | |
| Sensitivity: 100-fold increase | ||||||
| Reaction time: reduction by 7 min | ||||||
| Electrochemical and fluorescence | Intact virus/viral capsid | Antibody conjugated-MNPs | 2.5 h | LOD: log (5.9) to log (6.7) copies per mL (fluorescent), log (6.1) to log (6.9) copies per mL (electrochemical) | 92 | |
| Avian influenza virus | LSPR | DNA | Hollow spike-like AuNP | 10 min | LOD: 1 pM | 93 |
| SERS | DNA | Ag nanorods | Not indicated | LOD: 31 aM (H7 gene), 44 am (N9 gene) | 94 | |
| SERS-lateral flow | Intact virus/viral capsid | ATP functionalized AuAg@Ag 3-layer nanoparticle | 20 min | LOD: 0.0018 HA units | 95 | |
| Lateral flow | Intact virus/viral capsid | Highly chromatic red silica nanoparticles | 15 min | LOD: 0.08 pg mL−1 | 96 | |
| African swine fever virus | Rolling circle amplification | DNA | AuNP | 3 h | LOD: 3.56 fmol | 97 |
| Lateral flow | Viral genome | AuNP | 2 min | LOD: 200 copies of viral genome | 98 | |
| DNA | AuNP | 30 min | LOD: 100 copies per μL | 12 | ||
| Specificity: 100% towards ASFV | ||||||
| Fluorescence | Intact virus/viral capsid | Eu-labelled polystyrene NP | 15 min | Detection limit: 90 pg per test | 99 | |
| Affinity chromatography | Intact virus/viral capsid | MNPs | 30 min | LOD: 19.8 pM | 100 | |
| Reaction time: 30 min at room temperature | ||||||
| Nanosensors for crop viruses | ||||||
| Rice tungro bacilliform virus and rice tungro spherical virus | Electrochemical | Intact viruses | AuNP | Not indicated | LOD: 1.4 × 10−5 mg mL−1 | 101 |
| Sugarcane mosaic virus and sugarcane streak mosaic virus | Lateral flow | Intact virus/viral capsid | Cysteamine-capped AuNP | Incubation time: 1–2 h | LOD: 10−12 | 102 |
| Reaction time: 5 min | Detection sensitivity: 10−6 to 10−9 | |||||
| Maize chlorotic mottle virus | Colorimetric | DNA | AuNP | 90 min | LOD: 30 pg μL−1 | 103 |
| Lateral flow | Intact virus/viral capsid | AuNP | ∼40 min | LOD: 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600-fold g mL−1 dilution of crude extract 600-fold g mL−1 dilution of crude extract |
104 | |
| Lateral flow | DNA or RNA | AuNP | ∼20–30 min | LOD: 2.5 copies (coat protein gene), 0.96 pg (total RNA extract) | 105 | |
| Begomovirus | Electrochemical | DNA | Carbon nanotube and copper nanoparticle nanocomposite | ∼2 min | LOD: 0.01 ng μL−1 | 106 |
| Quantification limit: 0.04 ng nL−1 | ||||||
| Colorimetric | DNA | AuNP | 20 min | LOD: 500 ag μL−1 | 107 | |
| Tomato yellow leaf curl virus | Colorimetric | DNA | AuNP | 20 min | LOD: 1 copy per μL | 108 |
| LSPR | DNA | AuNP | 15 min | LOD: 5 ng μL−1 | 11 | |
| Potato virus X | Lateral flow | Intact virus/viral capsid | AuNP | 10 min | LOD: 17 pg mL−1 | 10 |
| Lateral flow | Intact virus/viral capsid | AuNP and MNP | 25 min | LOD: 0.25 ng mL−1 | 109 | |
| Potato virus Y | Vertical flow | Intact virus/viral capsid | AuNP | 3–4 min | LOD: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution 000 dilution |
110 |
| Relative error: 0.27–2.5% | ||||||
| Banana bunchy top virus | Immunoassay | Intact virus/viral capsid | AuNP | 1.5 h | LOD: 10−12 dilution of crude extract | 111 |
| Citrus tristeza virus | Electrochemical | RNA | AuNP | 65 min | LOD: 100 nM | 112 |
| Assay time: 65 min | ||||||
| Bean typical mosaic virus | Lateral flow | RNA | AuNP | 25 min | LOD: 10−5 dilution of the crude extract | 113 |
5.1. Nanobiosensors for foodborne viruses
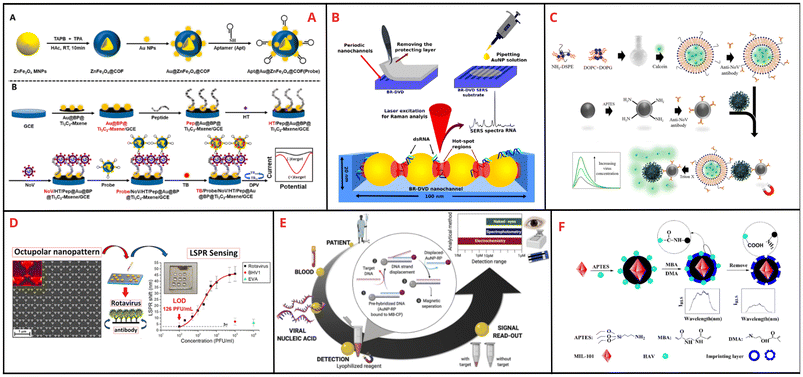
The emergence of foodborne diseases continues to be a significant public health concern globally, necessitating the development of advanced monitoring systems to ensure food safety. Recent advancements in biosensor technology, particularly nanoparticle-based biosensors, have shown considerable promise in addressing this challenge. These nanobiosensors enable the rapid and sensitive detection of viral contaminants in food, providing critical support in preventing outbreaks.7 The following section delves into the recent developments in nanoparticle-based biosensors specifically designed for detecting the four most prevalent foodborne viruses: norovirus, hepatitis A virus, hepatitis E virus, and rotavirus. These are considered as major viral threats due to their high global disease burden, low infectious doses, and remarkable persistence during food processing and handling.13,15 Fig. 2 shows some examples of these biosensors developed to detect foodborne viruses.Several nanosensors have been developed for NoV detection, utilizing various types of nanoparticle and detection mechanism. For example, impedimetric nanosensors, such as the WS2@AuNPs sensor synthesized by Baek et al., use WS2 nanoflowers decorated with AuNPs functionalized with peptides specific to the NoV. When NoV binds to the nanomaterial, it elicits a change in the impedance of the biosensor. Notably, this study demonstrated success not only in buffer but also in deliberately infected oyster samples, though the sensitivity was lower (a higher LOD of 6.21 copies per mL) than in buffer (LOD of 2.37 copies per mL), highlighting the real-world impact of the food matrix. To address the challenge of creating a stable and well-oriented bioreceptor layer.71 Nasrin et al. developed an impedimetric biosensor using a conductive polyaniline–gold (Au–PAni) nanocomposite. They employed a highly specific streptavidin–biotin interaction to immobilize the antibody, which ensures a consistent and robust sensor surface with minimal signal variance. This advanced bioconjugation strategy yielded a very low detection limit of 60 ag mL−1. While this sophisticated surface chemistry provides excellent stability and sensitivity, the multi-step fabrication process adds complexity and cost compared with simpler, direct immobilization techniques, which could be a factor in its potential for mass production.72
Fluorescence nanosensors have also been explored for NoV detection. For instance, Alzahrani et al. synthesized a mixture of AuNPs and carbon quantum dots (CQD) to create a fluorescence-based nanosensor that achieved an LOD of 80.3 pg mL−1. While this was ten times more sensitive than commercial kits, the validation was performed in human serum, leaving its performance against food-specific inhibitors untested.73 Colorimetric and electrochemical nanosensors have also been developed. Alhadrami et al. devised a simple and clever immunoassay using lactoferrin-functionalized cotton swabs and antibody-conjugated AuNPs, allowing for visual detection on the surfaces of foods like cucumber and lettuce. This approach is excellent for rapid, low-cost screening but provides a qualitative “yes or no” result rather than precise quantification.75 Another method employs Fe3O4 magnetic nanoparticles to capture NoV, which is then detected by the release of fluorophores from signal-amplifying liposomes (Fig. 2C). This amplification strategy enabled a low limit of detection of 136 copies per mL, a sensitivity that approaches the range of RT-qPCR, while the final detection step is completed in only 20 minutes, a significant reduction in time compared with the hours required for conventional molecular analysis. However, the use of both magnetic separation and liposome amplification creates a more complex, multi-step assay that may be better suited for a lab setting than for rapid field testing.74
Additionally, a dual-mode, CuO2@COF-NH2-based nanosensor enables detection of NoV through both electrochemical and colorimetric assays. This nanocomposite acts as a peroxidase mimic for colorimetric detection while its electrochemical properties generate a measurable signal. The dual-mode functionality provides valuable internal validation, but the stability of nanozymes like CuO2 in diverse food pH conditions requires further investigation.76 Liu et al. synthesized a highly advanced nanocomposite of Au@BP@Ti3C2-MXene and magnetic Au@ZnFe2O4@COF (Fig. 2A), yielding an LOD of 0.003 copies per mL, the lowest reported sensitivity for NoV detection. While the sensitivity is groundbreaking, being higher than that of PCR, the sensor relies on a sophisticated, multi-component nanomaterial that would be challenging to synthesize consistently and cost-effectively on a commercial scale.78 Ganganboina et al. developed another dual-mode biosensor that utilizes V2O5 nanoparticle-encapsulated, antibody-conjugated liposomes for NoV detection. The V2O5 nanoparticles act as both peroxidase mimics and electrochemical redox indicators. When NoV binds to the liposomes, the liposome structure is disrupted, releasing the V2O5 nanoparticles. This disruption triggers a visible color change for colorimetric detection and generates redox signals for electrochemical assays. This is another innovative example of signal amplification, but like other liposome-based systems, practical application would depend on ensuring the long-term stability and preventing premature leakage of the liposomes during storage.77
Although the abovementioned studies claim high specificity for NoV and report very low LODs (i.e., ag mL−1 or 10−3 copies per mL concentrations), several issues remain unaddressed. Firstly, most methods have only been tested on NoV particle solutions rather than actual food samples, raising questions about their applicability with real sample matrices. Secondly, these studies do not indicate whether these biosensors can detect multiple NoV strains, which would be necessary for field testing since various strains have caused different reported outbreaks. Third, given the absence of standardized guidelines for NoV detection thresholds, the practical utility of biosensors with exceptionally low detection limits remains uncertain.116 Finally, most studies focused on laboratory-based assays, with minimal research on field-deployable biosensors. Field testing is essential for promptly investigating and managing foodborne disease outbreaks once and as they occur.33 Overall, while NoV biosensors demonstrate promising sensitivity and specificity in lab settings, addressing these challenges will be crucial to advancing their field applicability.
Several nanosensors have been developed to detect HAV. A fluorescence nanosensor by Xie et al. used a CdTe quantum dot (QD)-based probe to detect HAV DNA via a FRET mechanism, achieving a limit of detection (LOD) of 13 pM. While the sensor showed excellent recovery rates (93.1–102.8%), its validation in spiked tap water does not fully address the challenges of more complex food matrices like shellfish, which are known to concentrate HAV.79 In a different approach, Micheli et al. developed an enzyme-linked immunomagnetic electrochemical assay (ELIME) using magnetic nanobeads coated with polydopamine (MNP-pDA) as the capture platform. This polydopamine layer provides a versatile and robust surface for immobilizing the capture antibody. The entire “sandwich” complex (MNP-pDA–Antibody–HAV–Enzyme-labeled-antibody) is then magnetically captured on a screen-printed electrode for electrochemical detection. The major advantage of this ELIME format is that it combines magnetic pre-concentration with a sensitive enzymatic signal, and the sensor was successfully applied to detect HAV in tap water. However, the system requires multiple incubation and washing steps, making it more suitable for batch processing in a lab than for rapid, on-the-spot field testing.80 Additionally, a molecularly imprinted polymer (MIP) nanoprobe was designed by Luo et al., using dimethylaminoethyl methacrylate (DMA) and MIL-101 MOF nanoparticles (Fig. 2F). This probe allows for selective virus capture through pH-induced swelling and employs resonance light scattering (RLS) to improve detection sensitivity. This antibody-free approach is promising for reducing costs, though creating consistent viral imprints with MIPs can be a fabrication challenge.56
For HEV, a variety of nanosensors utilize different mechanisms. Ganganboina et al. created a dual-modality sensor using hollow magnetic-fluorescent nanoparticles to isolate and detect HEV DNA with an LOD of 1.2 fg mL−1. The synthesis of such multifunctional nanoparticles, however, is a complex multi-step process.81 In another study, an electrochemical sensor using Pt–Co3O4 hollow-cage nanoparticles achieved an exceptional LOD of 0.81 fg mL−1 for HEV antigen detection.82 The high sensitivity is notable, but the reliance on platinum may impact the sensor's cost-effectiveness for widespread use. Another innovative HEV nanosensor uses a nucleic acid displacement approach. In this setup, AuNP-labeled nucleic acid reporter probes are pre-hybridized on capture probes attached to MNPs (Fig. 2E). When the target DNA is present, these reporter probes are displaced by stronger binding of the target DNA to the capture probe. Detection occurs colorimetrically, spectrometrically, or electrochemically, with LODs of 10 pM for visual and spectrometric methods, and 1 fM for electrochemical detection.83
Overall, the biosensors developed for HAV and HEV show high sensitivity and specificity through diverse detection mechanisms. They provide essential tools for rapid hepatitis virus detection, especially when conventional food processing falls short. However, significant challenges remain, including the need to validate these sensors against the various genotypes of each virus and to demonstrate their reliability in real, high-risk food products beyond simplified lab samples.
Several nanosensors have been developed to detect rotavirus. Y. Zhang et al. developed a portable immunochromatographic assay (ICA) using Au@Ag core–shell NPs that serve as surface-enhanced Raman scattering (SERS)-based labeling agents. The signal from these core–shell NPs is significantly stronger than traditional colloidal AuNPs, enhancing the sensitivity. The surface of the nanoparticles was functionalized with anti-rotavirus monoclonal antibodies, and this device showed an LOD of 8 pg mL−1, 10× lower than naked-eye observation. While it demonstrated high specificity for rotavirus, the authors noted that the device's fabrication process is complex, and the core–shell NPs used exhibit relatively low stability.84
Rippa and colleagues designed a plasmonic sensor array using octupolar triangular Au nanopillars (Fig. 2D), claiming enhanced plasmonic properties. The nanopillars, functionalized with antibodies specific to the rotavirus capsid, create strong LSPR fields that enhance light–matter interaction and sensitivity, enabling the detection of approximately 126 PFU mL−1. While the sensor was specific for rotavirus, it was only tested against two other viruses, and its synthesis process, involving electron beam lithography, is costly and complex.85
Ketabi et al. developed an electrochemical nanosensor for rotavirus RNA detection using a nanocomposite of hierarchical flower-like Au nanostructures (HFGNs), MXene, and polypyrrole as a signal amplification tag. Immobilized on a glassy carbon electrode with antisense ssDNA sequences, it allowed highly efficient RNA hybridization detection. The nanosensor likewise demonstrated high specificity against mismatched sequences, remained stable for 24 days, and successfully detected rotavirus in clinical samples. Its successful application in clinical samples is a significant step, but its performance against potential inhibitors in food or environmental water samples, which differ from clinical matrices, was not evaluated.86
In a different approach, Biswas et al. fabricated a SERS-based nanosensor by immobilizing citrate-reduced AuNPs on the surface of a Blu-ray digital versatile disc (DVD) as an SERS substrate (Fig. 2B). This method cleverly utilizes the disc's inherent nanochannels to trap the nanoparticles and enhance the Raman signal, detecting rotavirus RNA concentrations as low as 10 ng µL−1 with 94% reproducibility. This is a highly innovative use of a commodity material, though the study focused on reproducibility and did not examine selectivity against other types of RNA.87 The same research group later developed an even more accessible sensor using AgNPs on common printing paper as the SERS substrate. This paper-based SERS sensor is remarkably inexpensive (approx. USD 0.06 per substrate), making it a strong candidate for low-resource settings. However, while the paper reports detection of rotavirus samples at a 1% concentration in clinical stool, a precise limit of detection (LOD) in standard units was not established, making direct performance comparison with other methods difficult.88
While the reported nanosensors for rotavirus show great promise in terms of sensitivity and innovative design, significant translational hurdles remain. The primary challenge is moving from detecting RNA in buffer or clinical samples to reliably detecting intact virus particles in diverse and complex food matrices, such as leafy greens or prepared salads. Future work should focus not only on improving the stability and simplifying the fabrication of these platforms but also on validating their performance with real-world food samples to demonstrate their practical value in preventing foodborne outbreaks.
5.2. Nanobiosensors for livestock viruses
The food safety challenges described previously extend beyond the consumer level, with viral threats also originating at the production stage. In this context, livestock viruses present unique risks to both animal health and global food security. At the production level of the food system, viral diseases in livestock represent a critical threat with widespread consequences. These infections cause direct and massive economic losses through reduced productivity, animal mortality, and costly control measures. Crucially, disease outbreaks trigger severe international trade restrictions, disrupting supply chains and impacting food prices and availability globally.9,119 Furthermore, many of these viruses pose a direct threat to public health. Zoonotic pathogens, for instance, can spread from infected animals to humans through contact or the consumption of contaminated products, blurring the line between animal and public health and undermining consumer confidence.120 To illustrate the diverse challenges in this domain, this section focuses on three livestock viruses selected for their profound global impact: foot-and-mouth disease virus (FMDV) for its trade-disrupting potential, avian influenza virus (AIV) for its high-profile zoonotic risk, and African swine fever virus (ASFV) for its devastating mortality and lack of a vaccine. The following sections will critically assess the nanobiosensor technologies being developed to provide the rapid, on-farm detection necessary to manage these significant threats.Jain et al. developed a sensor by conjugating FMDV-specific antibodies to AuNPs, utilizing the nanoparticles’ LSPR properties to enhance sensitivity and specificity. This biosensor achieved an LOD of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 serum dilution, demonstrating high specificity when tested against other viruses. While this simple dot-blot assay is well-suited for low-cost screening, it provides a qualitative result and its performance on non-serum samples like saliva or milk, which is important for surveillance, was not demonstrated.89
000 serum dilution, demonstrating high specificity when tested against other viruses. While this simple dot-blot assay is well-suited for low-cost screening, it provides a qualitative result and its performance on non-serum samples like saliva or milk, which is important for surveillance, was not demonstrated.89
Hamdy and colleagues used AuNPs to improve PCR specificity and efficiency by conjugating FMDV-specific primers to AuNP surfaces. This directly enhanced the gold-standard method; when tested on the same 31 clinical isolates, their AuNP-enhanced rRT-PCR consistently lowered Ct values by 3 to 3.5 cycles compared with conventional rRT-PCR. This increase in efficiency resulted in a tenfold improvement in the analytical limit of detection, achieving an LOD of 1 RNA copy versus 10 RNA copies for the traditional method. While innovative, this approach only enhances a laboratory-based method and still relies on a thermal cycler, limiting its use for true pen-side diagnostics where rapid, equipment-free answers are most needed.90
Meanwhile, a recent study used a nanocomposite of AuNPs and graphene oxide in a modified LAMP assay for FMDV detection (Fig. 3D). This nanocomposite inhibits undesired amplification and reduces non-specific signals by adsorbing single-stranded DNA (ssDNA) through π–π interactions and electrostatic effects, preventing mismatched hybridizations and stabilizing primers before target pairing. The combined large surface area and synergistic interaction of AuNPs and graphene oxide enhance signal quality. The use of isothermal LAMP is a significant step towards field-friendliness, but the study did not compare its sensitivity against the seven different FMDV serotypes, which is a critical consideration for a globally applicable diagnostic.91
Lastly, Hwang et al. created a dual-mode immunoassay that combines fluorescence and electrochemical detection for FMDV. This sandwich assay uses MNPs functionalized with FMDV-specific antibodies along with a secondary antibody conjugated with β-glucosidase. When the virus binds, the secondary antibody attaches, activating β-glucosidase to produce both resorufin and glucose, detectable via fluorescence and electrochemical signals, respectively. The assay achieved LODs which are 1–2 orders of magnitude lower than the LODs of conventional PCR-based methods. Additionally, this approach provides four-fold greater sensitivity compared with commercially available lateral flow assay kits, highlighting its potential for enhanced diagnostics. This dual-mode approach provides excellent sensitivity and a valuable internal cross-check for results. However, its reliance on two separate reading instruments (a fluorometer and a glucometer) adds a layer of complexity and cost for field deployment in low-resource settings.92
Collectively, these biosensors offer effective and highly sensitive approaches for FMDV detection. However, practical application faces significant hurdles. A critical challenge for future development will be creating platforms capable of differentiating infected from vaccinated animals (a DIVA-compliant test), as well as identifying specific viral serotypes, which is crucial for epidemiological tracking and targeted vaccine deployment. Furthermore, simplifying protocols for robust use outside of laboratory conditions remains a primary goal for FMDV surveillance.
Several biosensors using various nanoparticles have been developed for AIV detection. One approach involved a hollow spike-like Au nanoparticle (hAuSN) functionalized with a three-way junction (3WJ) multifunctional DNA aptamer (Fig. 3C). The unique spike-like structure of the hAuSN enhances the LSPR effect and signal sensitivity, enabling the detection of hemagglutinin (HA) of AIV at 1 pM in both PBS buffer and chicken serum. However, as the authors noted, comprehensive data on the sensor's specificity against other common poultry respiratory viruses or different AIV subtypes were not provided.93
Another study used a silver nanorod (AgNR) array as an SERS substrate to detect AIV. AgNRs are ideal for SERS due to their ability to create highly localized electromagnetic fields at their sharp tips, significantly enhancing Raman signal sensitivity. The array utilizes Exo III-assisted cycling amplification with three ssDNAs—capture, replacement, and probe—to amplify the target DNA strand. The amplified product interacts with the AgNR array, which SERS detects, achieving LODs of 31 aM and 44 aM for the H7 and N9 genes, respectively. While this demonstrates exceptional analytical sensitivity, the oblique angle deposition method used to create the AgNR array is a specialized laboratory technique, and ensuring the large-scale, low-cost production of uniform SERS substrates remains a key challenge for commercialization.94
Xiao et al. developed a three-layer nanoparticle composed of alternating Ag and Au layers, with the Au layer functionalized with ATP. The synergistic electromagnetic enhancement from both metals and the protective outer silver layer resulted in a highly sensitive and reproducible signal. After conjugating the nanoparticle with AIV antibodies, it served as the reporter molecule in a SERS-based lateral flow assay. This device achieved an LOD of 0.0018 HA units, providing a sensitivity three orders of magnitude better than conventional assays and comparable to PCR-based methods. The performance is outstanding, but the synthesis of these complex, multi-layer core–shell nanoparticles is intricate and could present challenges for ensuring the high batch-to-batch consistency required for a commercial diagnostic test.95
Su et al. developed an immunochromatographic assay using highly chromatic red silica nanoparticles (SiNPs), enhanced by multiple layers of dye deposition via layer-by-layer electrostatic self-assembly (Fig. 3A). The high chromaticity and stability of these SiNPs, achieved through dense polyelectrolyte films, amplify visual signal intensity and ensure robustness across a wide pH range. Conjugated with AIV-specific antibodies, these SiNPs enable precise antigen binding and sensitive detection. Compared with traditional AuNPs, the red SiNPs provide superior optical performance, allowing both qualitative and quantitative virus detection with greater sensitivity and specificity. While visually impressive and stable, the layer-by-layer assembly process is inherently more time-consuming than one-step nanoparticle synthesis methods, which could impact the overall cost and speed of manufacturing.96
The biosensors developed for AIV detection showcase substantial progress in sensitivity. However, the primary remaining hurdle for practical food system application is the development of multiplexed platforms that can simultaneously detect and differentiate between common and emerging subtypes (e.g., H5, H7, H9) in a single, rapid test. This is critical for effective surveillance and guiding specific response strategies. Furthermore, improving the cost-efficiency and ruggedness of these technologies to withstand the demanding environmental conditions of poultry farms and wild bird monitoring programs is essential for their widespread adoption.
Several nanoparticle-based biosensors have been developed for ASFV detection, employing various amplification and signal enhancement strategies. One of the recent methods combines isothermal rolling circle amplification (RCA) with AuNPs for ASFV detection (Fig. 3B). RCA uses a circular probe to synthesize long tandem repeats of the target DNA, and the interaction of these products with DNA-conjugated AuNPs results in a visible color change. This assay demonstrated an LOD of 3.56 fmol in serum samples. While the colorimetric readout is ideal for low-cost screening, the authors noted that specificity against other common swine viruses was not tested, which is a critical step for validation.97
Meanwhile, a CRISPR-based assay using the Cas12a enzyme in tandem with AuNPs was developed for ASFV detection. Cas12a, a stable and cost-effective DNA endonuclease, was coupled with an LFA to enhance detection. This assay reached an LOD of 200 copies of the viral genome, matching the sensitivity of qPCR, and was shown to be highly specific with no cross-reactivity to other swine viruses. The integration of CRISPR provides an exceptional layer of specificity, making this a very promising approach for an instrument-free field test, though its performance on diverse sample types beyond blood, such as oral fluids or environmental swabs, would need further evaluation.98
Another AuNP-based method combined lateral flow assays with recombinase polymerase amplification (RPA), an isothermal technique that amplifies DNA and RNA at low temperatures (37–42 °C). In this setup, RPA amplifies the ASFV target gene, and the amplicons are visually detected (appearance of a red color) on a test strip via AuNPs. The low-temperature requirement of RPA is a significant advantage for use in settings without reliable electricity, but the sensitivity may be lower than methods incorporating additional signal amplification.12
Aira et al. developed a lateral flow assay using europium-labeled polystyrene nanoparticles as fluorescent reporters for detecting an ASFV-specific antigen. These nanoparticles provide intense, stable fluorescence, significantly enhancing the sensitivity compared with traditional AuNPs. This assay achieved a detection limit of 90 pg per test, outperforming commercial assays by a factor of 16, and detected more positive cases in blood samples than existing kits. While highly effective, this LFA requires a portable fluorescence reader to quantify the results, which adds an instrument cost and a logistical step compared with a purely visual-readout assay.99
Lastly, a portable affinity chromatography setup using magnetic nanoclusters was introduced for ASFV detection. This method employs a hybridization chain reaction (HCR), an enzyme-free polymerization technique that forms a long DNA strand around an MNP upon target recognition, which is then isolated via affinity chromatography. The enzyme-free nature of HCR increases the robustness of the assay, but the chromatography setup, while portable, is more complex to operate than a simple dipstick-style test.100
These diverse nanobiosensor approaches showcase significant potential for ASFV detection. However, since ‘test-and-cull’ is the primary control strategy, any diagnostic tool must be exceptionally reliable to prevent the unnecessary slaughter of healthy animals. Future work must focus on simplifying protocols for use by non-specialists, ensuring robust performance with diverse and challenging sample types from the farm environment (e.g., oral fluids, feces, contaminated feed), and validating these assays across the many different ASFV genotypes circulating globally.
Livestock virus nanobiosensors have advanced significantly, with lateral flow and portable formats now enabling rapid, on-site detection of key pathogens like FMDV, AIV, and ASFV. These technologies demonstrate strong specificity and sensitivity in laboratory settings, but their practical application is often limited by the complexity of real-world samples such as saliva, milk, or environmental swabs. Further efforts are needed to ensure these sensors perform reliably in diverse sample types and under field conditions, supporting timely disease management and outbreak prevention.
5.3. Nanobiosensors for crop viruses
In addition to animal health, plant health is the other foundational pillar of the food system's production stage. Viral diseases in crops are responsible for billions of dollars in annual yield losses, directly impacting food security and agricultural economies worldwide. Unlike for foodborne and livestock viruses, the literature on nanoparticle-based biosensors for crop viruses is more sparse, with most phytopathogens having only a few available studies. This is reflected in some of the foundational publication dates cited in this section. The reasons for this are twofold: the sheer biological diversity of plant pathogens, which requires highly specific biosensor designs for each virus–crop pair, and the fact that research has historically prioritized bacterial and fungal diseases. Therefore, to provide a representative overview of the current landscape, this section reviews biosensors developed for viruses that pose a significant economic threat to major global food staples, including rice, sugarcane, maize, potato, and key fruit crops. Below are representative crop viruses and the corresponding nanoparticle-based biosensors developed for their detection. Selected schematic illustrations are shown in Fig. 4.Rice tungro disease, caused by a complex of rice tungro bacilliform virus (RTBV) and rice tungro spherical virus (RTSV), is one of the most destructive diseases of rice in Asia. Its pathogenesis involves insect-vectored transmission by the green leafhopper, leading to severe stunting and reduced tillering, which can cause 100% yield loss in susceptible rice varieties, directly threatening food security in rice-dependent regions. Uda et al. developed an electrochemical sensor by conjugating antibodies specific to both viruses onto an AuNP–polypyrrole nanocomposite deposited on a screen-printed carbon electrode (SPCE). The system achieved an LOD of 1.4 × 10−5 mg mL−1 and demonstrated selective detection despite antibody cross-reactivity. While this is a pioneering effort for this virus complex, the use of polyclonal antibodies can lead to batch-to-batch variability, and the sensor's performance was not validated using crude extracts from infected rice leaves, which is a critical test for field applicability.101
For sugarcane mosaic virus (SCMV) and sugarcane streak mosaic virus (SCSMV), which can reduce both stalk yield and sucrose content by up to 50%, impacting the global sugar and biofuel industries, Thangavelu et al. designed a dual analyte lateral flow assay for the two viruses. They used cysteamine-capped AuNPs and the cationic surface improved antibody conjugation and thus the assay sensitivity, achieving an LOD of 10−12 and a detection range of 10−6 to 10−9, which outperforms the standard citrate–AuNP LFAs. This simple LFA is ideal for on-field screening by farmers or mill operators, but its specificity against other viruses common in sugarcane was not reported, which could be a concern for accurate diagnosis, due to the potential cross reactivity of the antibodies.102
Maize chlorotic mottle virus (MCMV) mainly causes a mild mosaic appearance, severe stunting, and leaf necrosis in maize plants. Its pathogenesis is particularly problematic as it can be transmitted by vectors, seeds, and mechanical means, facilitating rapid, widespread epidemics of maize lethal necrosis when in co-infection with other viruses. One method to detect the virus was developed by Liu et al., with a colorimetric assay using ssDNA probes and AuNPs that aggregate and change color in the presence of the target viral gene. This instrument-free approach is promising for low-resource settings; however, the study was based on amplified RT-PCR products, and its direct application to crude plant extracts, which contain interfering polyphenols, would be challenging.103 On the other hand, Zhang et al. produced a Dot-ELISA and an AuNP-based LFA. Notably, their Dot-ELISA, which detected MCMV in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 240 dilution, was demonstrated to be 12.1 times more sensitive than a conventional RT-PCR assay run on the same samples. The LFA component is particularly valuable as a rapid (<10 min) point-of-care tool, although the highly sensitive Dot-ELISA portion still requires laboratory equipment and several hours to complete, a common trade-off for ELISA-based formats.104 The MCMV sensor developed by Lei et al. uses a Cas12a-LFA system (Fig. 4D) and established an LOD of 2.5 copies of the coat protein gene. This represents exceptional sensitivity for a field-deployable assay, reaching a level of single-digit copy number detection that is typically only achievable with laboratory-based RT-qPCR. The high sensitivity of this CRISPR-based system is a major advantage for early warning in seed certification programs, but the reliance on enzymes like Cas12a requires careful handling to maintain activity in variable field temperatures.105
240 dilution, was demonstrated to be 12.1 times more sensitive than a conventional RT-PCR assay run on the same samples. The LFA component is particularly valuable as a rapid (<10 min) point-of-care tool, although the highly sensitive Dot-ELISA portion still requires laboratory equipment and several hours to complete, a common trade-off for ELISA-based formats.104 The MCMV sensor developed by Lei et al. uses a Cas12a-LFA system (Fig. 4D) and established an LOD of 2.5 copies of the coat protein gene. This represents exceptional sensitivity for a field-deployable assay, reaching a level of single-digit copy number detection that is typically only achievable with laboratory-based RT-qPCR. The high sensitivity of this CRISPR-based system is a major advantage for early warning in seed certification programs, but the reliance on enzymes like Cas12a requires careful handling to maintain activity in variable field temperatures.105
For begomoviruses, a group of plant viruses responsible for diseases in several essential crops like cotton and tomato, which are transmitted by whiteflies, making their spread rapid and difficult to control, an electrochemical biosensor was developed using a composite of multi-walled carbon nanotubes and copper nanoparticles (CuNP) (Fig. 4C). The nanocomposite was said to improve the detection performance by providing an increased surface area and higher conductivity. An LOD of 0.01 ng μL−1 was determined for the sensor. However, the authors noted the sensor had a limited usable life of 8 weeks due to nanocomposite oxidation, a key consideration for practical deployment.106 In a more recent study, Lavanya and Arun developed a sensor that uses the same principle as that of Liu et al., where dsDNA formation in the presence of the target gene interacts with AuNPs differently than the ssDNA probe. Their biosensor has an LOD of 500 ag μL−1. This exceptional sensitivity is noteworthy, but the assay's robustness against different plant matrices was not explored.107
Aside from the sensors that allow for the detection of begomoviruses, biosensors have also been developed specific to the tomato yellow leaf curl virus (TYLCV), an example of a begomovirus. Wang and Yang fabricated an RPA-LFA system that uses standard AuNP reporter molecules for detecting the virus at 1 copy per μL concentration (Fig. 4B). The authors make a direct claim of superior performance, stating that this detection limit is more sensitive than both traditional PCR and real-time qPCR, while completing the entire process in only 20 minutes at a constant low temperature. The isothermal RPA step makes this suitable for on-site application, but like RT-PCR, it does not distinguish between active and remnant infections.108 Meanwhile, Razmi and colleagues developed a biosensor system similar to the previously mentioned method by Liu et al. (2015).11,103 Without further amplification, this sensor can detect the virus at concentrations as low as 5 ng μL−1. While this method avoids the need for enzymes, its sensitivity is significantly lower, making it better suited for symptomatic, high-titer infections.11
Potato virus X (PVX) and potato virus Y (PVY) are two viruses that mainly infect potatoes. These viruses often occur in co-infections, leading to synergistic disease complexes that can cause up to 90% yield loss in susceptible potato cultivars. Panferov et al. developed a modified LFA to detect PVX where AuNPs were allowed to grow further after the standard LFA procedure by adding HAuCl4 and H2O2, which improved the assay's sensitivity.10 In contrast, Razo et al. developed an LFA using both AuNP and MNP. The addition of the MNP serves to increase the analyte concentration and the visibility of the test lines (Fig. 4E), achieving a 32× more sensitive LOD.109 In detecting PVY, Verma et al. developed a vertical flow lateral assay using standard AuNPs, which overcomes limitations of traditional LFA like the hook effect. These examples show clever engineering to improve the classic LFA format, but validation is often performed using purified virus; performance with tuber or leaf sap, which can contain enzymatic inhibitors and debris, is the true test of field utility.110
Another devastating disease, banana bunchy top virus (BBTV), mainly affects bananas and is considered one of its most severe pathogens, capable of causing 100% crop loss in a plantation. Its pathogenesis involves aphid transmission, leading to “bunchy top” symptoms where new leaves are stunted and bunched together, halting fruit production entirely and making early detection critical for a farm's survival. One nanoparticle-based biosensor was developed using a Dot-ELISA coupled with LFA. The sensor can detect the virus in up to 10−12 dilutions of the crude extract. While the sensitivity is exceptionally high, the Dot-ELISA component, like other ELISA-based methods, still requires multiple washing and incubation steps in a lab setting, limiting its use for immediate on-site diagnosis by farmers.111
Citrus tristeza virus (CTV) mainly infects citrus trees, resulting in stem pitting, vein corking, and reduced fruit quality. Historically, its pathogenesis has led to “quick decline” epidemics responsible for the loss of hundreds of millions of trees grafted on susceptible rootstock, posing a constant threat to the global citrus industry. An electrochemical biosensor was developed wherein AuNPs and DNA probes were deposited on the surface of the electrode (Fig. 4A), allowing for the detection of the virus at an LOD of 100 nM with an assay time of 65 min. This method offers a good balance of speed and sensitivity for the lab-based screening of nursery stock, but its specificity against other citrus viruses that might be present in field samples was not reported.112
Lastly, an RPA-LFA biosensor was developed to detect the bean common mosaic virus (BCMV), a virus that affects all types of bean crop, causing significant yield losses. Its seed-borne nature is a major aspect of its pathogenesis, as it allows the virus to be introduced into new fields via contaminated seed lots, making seed certification a crucial control point. The RPA-LFA biosensor was established to have an LOD of 10−5 dilution of the crude extract, performing considerably better than the standard PCR method. The ability to use crude extracts is a major advantage for on-field testing, and the isothermal RPA component enhances its portability. However, the study did not report on whether the assay could differentiate between the various and often co-infecting strains of BCMV, which can be important for resistance breeding programs.113
The development of nanoparticle-based biosensors for crop viruses, while less mature than for animal or human pathogens, demonstrates clear potential. The prevalent use of LFAs and simple colorimetric assays reflects the agricultural sector's need for rapid, low-cost, on-site diagnostics that can be used directly by growers or inspectors. However, a major and recurring challenge is the lack of robust, commercially available biorecognition elements for the vast diversity of plant viruses, which often forces researchers to rely on lab-grade reagents. Future progress will likely depend on creating platform technologies that can be easily adapted for different plant pathogens and integrating these sensors with smartphone-based imaging for data mapping, enabling precision agriculture and early outbreak warnings.
Earlier examples point to a clear pattern of technological trade-offs across the diverse viral threats facing the food system—from production to consumption. The widespread development of lateral flow assays for livestock and crop viruses highlights the urgent demand for rapid, low-cost, and equipment-free diagnostic tools that can be deployed directly on farms or in the field. These platforms prioritize speed and usability, enabling timely interventions to prevent the spread of highly contagious diseases. In contrast, the advancement of electrochemical and molecular biosensors for detecting foodborne pathogens such as norovirus reflects a different set of priorities: achieving ultra-low detection limits to identify trace contamination in processed foods and safeguard public health. Together, these trends illustrate a fundamental divide in biosensor design, driven by the distinct risk-management needs at different stages of the food system.
A second recurring theme is the dual role of nanoparticles as either signal amplifiers or sample preparation tools. While materials like AuNPs, QDs, and various nanozymes are primarily used to enhance the visibility or electrical readout of a signal, magnetic nanoparticles (MNPs) repeatedly appear as a crucial component for overcoming the “matrix effect”. Their ability to isolate and concentrate target viruses from complex sample backgrounds like oyster tissue, meat products, or plant sap is often the key step that enables a sensor to function reliably outside of pristine laboratory buffers. This highlights that for food system applications, the elegance of the detection chemistry is often secondary to the robustness of the sample preparation.
Despite these innovations, this review also reveals a significant and persistent translational gap. The vast majority of studies validate their sensors using purified virus or spiked samples, with testing on naturally contaminated, real-world samples being a rarity. Furthermore, critical parameters such as specificity against closely related viral strains, long-term stability under non-ideal storage conditions, and the potential for scalable, low-cost manufacturing are often unaddressed. While the scientific potential of nanobiosensors is undeniable, these overarching challenges must be confronted to move these technologies from the laboratory bench to the farm, the factory, and the kitchen.
6. Challenges and future perspectives
As demonstrated in the preceding sections, a remarkable diversity of nanoparticle-based biosensors has been developed to target viral threats across the food system. However, the journey from a proof-of-concept with the desired low LOD in the lab, to a robust, reliable tool adaptable on a farm or in a factory has been limited due to several key biological, technical, and logistical challenges. This section addresses these challenges and explores the future perspectives that hold promise for overcoming them.A major biological challenge is that many food-system viruses, such as norovirus, are non-culturable,7,114 which severely limits research and the validation of new biosensors. Unlike culturable bacteria and fungi,36,125 these viruses require reliance on less-than-perfect surrogates. For example, biosensor development for norovirus often depends on using virus-like particles (VLPs) or utilizing surrogate viruses with similar properties, such as murine norovirus (MNV) or feline calicivirus (FCV), for controlled laboratory testing. While these are invaluable tools, genetic differences can affect detection accuracy, and they do not fully replicate the behavior of the human pathogen in food matrices.7,126 Another significant hurdle is the high mutation rate of many RNA viruses. This rapid genetic variability, as seen with SARS-CoV-2 variants, can reduce the binding efficiency of highly specific antibodies or nucleic acid probes, potentially rendering a sensor obsolete as new strains emerge.127 To solve this problem, one strategy is to design biosensors that target parts of the virus that rarely change, like stable proteins or essential genes. Another approach is to use broader recognition molecules, like host-cell receptors that viruses naturally bind to, which can often detect multiple strains in one go.128
A persistent theme throughout this review is the gap between performance in a controlled buffer and in a real-world food matrix, or the robustness of the sensor performance over different analytical conditions. Most studies report impressive LODs using spiked samples, but the “matrix effect”—interference from fats, proteins, salts, and enzymes in samples like milk, meat, or plant sap—can drastically reduce sensitivity and cause false signals. For example, as highlighted in a WS2NF/AuNP-based electrochemical impedance spectroscopic sensor by Baek et al., the LOD for norovirus was nearly three times higher in oyster samples than in buffer, demonstrating the profound impact of the matrix. Overcoming this requires not just advanced nanoparticle chemistry to reduce the non-specific matrix component binding, as in this case, but also robust and integrated sample preparation that can largely remove the interference. Furthermore, the performance of these sensors must be standardized to be commercially viable. Unlike bacterial and fungal detection, which follow established protocols from organizations such as the Codex Alimentarius, virus detection guidelines in food are inconsistent, complicating result comparisons. Efforts toward standardization, such as the ISO/TS 15216:2013 standard for detecting hepatitis A and norovirus in food matrices, are crucial first steps. However, as multi-laboratory validation studies have shown, further refinement is needed to improve inter-laboratory reliability.129 Finally, questions of manufacturability and economic viability must be addressed. Many of the highest-performing sensors described herein rely on sophisticated nanomaterials requiring complex and/or low-yield fabrication methods. The transition to scalable manufacturing techniques that ensure high batch-to-batch consistency at a low cost is a non-trivial engineering challenge. As for the economic viability, many biosensors (e.g. SERS, electrochemical nanosensors, etc.) can be more cost-effective than standard methods, like PCR and ELISA, due to needing less expensive reagents (e.g. antibodies, enzymes, etc.) and equipment. On the other hand, the target cost for a disposable on-site biosensor must still be low enough to be affordable in the context of what the food products being tested are and which part of the food value chain the test is being performed on.
While these challenges are significant, several promising research directions are emerging to address them directly. A critical goal is to better distinguish infectious from non-infectious viral particles. Nanobiosensors that target viral capsid integrity or host-specific interactions, such as the peptide–aptamer sandwich approach by Liu et al. that recognizes the assembled NoV capsid, offer a promising solution by being less likely to detect harmless viral fragments.78
The development of multiplexed biosensors is also a critical focus. While multiplexing has been widely explored for bacterial detection,130 its application to viral detection in food remains limited, largely due to the challenges of developing sets of highly specific, non-cross-reacting recognition elements for the vast structural diversity of viruses. To address such issue, viral receptors can be additional affinity ligands being used in combination with antibodies and DNA aptamers etc. to achieve sophisticated assay designs for multiplexing.128 Crucially, it could also differentiate between bacterial and viral pathogens that cause clinically indistinguishable symptoms like gastroenteritis, thereby enabling more targeted public health responses and preventing the unnecessary use of antibiotics.13 While examples of this exist for respiratory viruses like SARS-CoV-2,127 transferring this success to the complex matrices and diverse viral targets in the food system is a significant and important future direction.
Smartphone-based biosensors are emerging as powerful tools for real-time virus detection, leveraging the fact that 69% of the global population now owns a smartphone, to create accessible, field-friendly solutions.131 The core of this integration is using the device's camera and processing power to quantify signals from colorimetric or fluorescent biosensor assays, effectively replacing bulky laboratory readers.14 While this technology is advancing rapidly, its application has been uneven. For clinical viruses like HIV, integrated point-of-care systems are becoming highly sophisticated.132 However, in food safety, the primary focus of smartphone-based biosensors has been on bacterial pathogens, as comprehensively reviewed by Yang et al.14 This reveals a significant gap: robust, smartphone-integrated platforms for viruses of specific concern to the food system remain remarkably scarce. Therefore, adapting these advanced, automated smartphone platforms for the specific viral targets of the food system should be a key priority for future development.
Finally, the integration of artificial intelligence (AI) has created vast potential for enhancing biosensor applications, particularly in viral detection within food systems. AI, through machine learning (ML), offers tools for analyzing complex biosensor data with high precision and speed. Specifically, AI can optimize biosensors by improving signal processing, feature extraction, and noise reduction, enabling the detection of viruses at lower concentrations with greater accuracy.133,134 For instance, ML algorithms can distinguish subtle variations in sensor signals that indicate viral contamination, even in the presence of background noise, a task that traditional methods often struggle to achieve.135–137 Moreover, the integration of AI with biosensors allows real-time, multifaceted data analysis, enabling dynamic adaptability to different food matrices and contamination scenarios. In food systems, AI-driven biosensors can also predict contamination trends, flag anomalies, and facilitate predictive maintenance of food safety protocols.138 The combination of nanosensors with AI can detect viral genetic material or protein markers in food at sensitivities comparable to laboratory PCR techniques but with faster, on-site results.135,139 The resulting AI-powered platforms can outpace conventional methods, enhancing early warning systems and reducing foodborne virus outbreaks. Furthermore, such systems support continuous learning, allowing biosensors to improve their detection capabilities over time by adapting to newly emerging viral strains.133,134,138
This review has provided a system-wide analysis of nanoparticle-based biosensors for detecting viral threats to public health, agricultural productivity, and economic stability. By examining their application from production-level threats like FMDV and crop viruses to consumer-level pathogens like norovirus, a clear pattern emerges: a trade-off exists between the rapid, field-deployable assays needed for on-farm screening and the ultra-sensitive platforms required for verifying the safety of processed foods. The practical adoption of these promising technologies is consistently limited by the food matrix effect, manufacturing scalability, and the need for standardized validation. Future advancements in multiplexed platforms, smartphone-integrated readers, and AI-driven analysis will be essential to overcome these hurdles. The continued development of these integrated systems is critical for establishing nanobiosensors as reliable, efficient, and accessible tools capable of strengthening food system resilience worldwide.
Author contributions
Riann Martin Sarza: conceptualization, writing – original draft, writing – review & editing. Laura Sutarlie: conceptualization, writing – review & editing, funding acquisition, resources. Sam Fong Yau Li: conceptualization, writing – review & editing, supervision. Xiaodi Su: conceptualization, writing – review & editing, funding acquisition, resources, supervision.Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results, software or code have been included, and no new data were generated or analysed as part of this review.Acknowledgements
This project was supported by The National Medical Research Council (NMRC) and National Centre for Infectious Diseases (NCID), Singapore [PREPARE-CS1-2022-003 and PREPARE-OC-ETM-Dx-2023-010]. RM Sarza also acknowledges the assistance of Alexcia Caacbay and Adriana Bombase in creating the figures and proofreading the final version of this article.References
- J. Fanzo, A. L. Bellows, M. L. Spiker, A. L. Thorne-Lyman and M. W. Bloem, Am. J. Clin. Nutr., 2021, 113, 7–16 CrossRef CAS PubMed.
- FAIRR, Industry Reinfect: Avian Flu, https://www.fairr.org/resources/reports/industry-reinfected-avian-flu.
- WHO, Daily-adjusted life years (DALYs), The Global Health Observatory, https://www.who.int/data/gho/indicator-metadata-registry/imr-details/158.
- A. Bosch, E. Gkogka, F. S. Le Guyader, F. Loisy-Hamon, A. Lee, L. van Lieshout, B. Marthi, M. Myrmel, A. Sansom, A. C. Schultz, A. Winkler, S. Zuber and T. Phister, Int. J. Food Microbiol., 2018, 285, 110–128, DOI:10.1016/j.ijfoodmicro.2018.06.001.
- N. Nasheri, J. Harlow, A. Chen, N. Corneau and S. Bidawid, Food Environ. Virol., 2021, 13, 107–116 CrossRef CAS PubMed.
- G. Di Cola, A. C. Fantilli, M. B. Pisano and V. E. Ré, Int. J. Food Microbiol., 2021, 338, 108986, DOI:10.1016/j.ijfoodmicro.2020.108986.
- S. Mukherjee, P. Strakova, L. Richtera, V. Adam and A. Ashrafi, Advanced Biosensors for Virus Detection: Smart Diagnostics to Combat SARS-CoV-2, Elsevier, 2022, pp. 391–405 Search PubMed.
- C. Hennechart-Collette, O. Dehan, M. Laurentie, A. Fraisse, S. Martin-Latil and S. Perelle, Int. J. Food Microbiol., 2021, 337, 108931, DOI:10.1016/j.ijfoodmicro.2020.108931.
- T. Chanchaidechachai, H. Saatkamp, C. Inchaisri and H. Hogeveen, Front. Vet. Sci., 2022, 9, 904630, DOI:10.3389/fvets.2022.904630.
- V. G. Panferov, I. V. Safenkova, A. V. Zherdev and B. B. Dzantiev, Microchim. Acta, 2018, 185, 506, DOI:10.1007/s00604-018-3052-7.
- A. Razmi, A. Golestanipour, M. Nikkhah, A. Bagheri, M. Shamsbakhsh and S. Malekzadeh-Shafaroudi, J. Virol. Methods, 2019, 267, 1–7 CrossRef CAS PubMed.
- Z. Wang, W. Yu, R. Xie, S. Yang and A. Chen, Anal. Bioanal. Chem., 2021, 413, 4665–4672, DOI:10.1007/s00216-021-03408-2.
- X. Wang, Y. Luo, K. Huang and N. Cheng, Adv. Agrochem, 2022, 3–6, DOI:10.1016/j.aac.2022.08.002.
- T. Yang, Z. Luo, T. Bewal, L. Li, Y. Xu, S. M. Jafari and X. Lin, Food Chem., 2022, 394, 133534, DOI:10.1016/j.foodchem.2022.133534.
- X. Zhu, T. Y. Kim, S. M. Kim, K. Luo and M. C. Lim, J. Agric. Food Chem., 2023, 71, 15942–15953, DOI:10.1021/acs.jafc.3c05166.
- S. Chakraborty, N. Kumar, K. Dhama, A. K. Verma, R. Tiwari, A. Kumar, S. Kapoor and V. Singh, Advances in Animal and Veterinary Sciences, 2013, 2(2S), 1–18 CrossRef.
- Y. Revilla, D. Pérez-Núñez and J. A. Richt, Advances in Virus Research, Academic Press Inc., 2018, vol. 100, pp. 41–74 Search PubMed.
- E. Spackman, in Animal Influenza Virus, ed. E. Spackman, Humana Press, 3rd edn, 2020, p. 83 Search PubMed.
- P. Signoret, in Encyclopedia of Virology, ed. B.W. J. Mahy and M. H. V. Van Regenmortel, 3rd edn, 2008 Search PubMed.
- G. Lu, Z. Wang, F. Xu, Y. B. Pan, M. P. Grisham and L. Xu, Microorganisms, 2021, 9, 1984, DOI:10.3390/microorganisms9091984.
- V. Morton, J. Jean, J. Farber and K. Mattison, Appl. Environ. Microbiol., 2009, 75, 4641–4643 CrossRef CAS PubMed.
- K. Nemes, S. Persson and M. Simonsson, Viruses, 2023, 15, 1725, DOI:10.3390/v15081725.
- M. Koopmans and E. Duizer, Int. J. Food Microbiol., 2004, 90, 23–41 CrossRef PubMed.
- E. H. Teshale and D. J. Hu, World J. Hepatol., 2011, 3, 285–291 CrossRef PubMed.
- B. S. Coulson, Curr. Opin. Virol., 2015, 15, 90–96, DOI:10.1016/j.coviro.2015.08.012.
- G. R. Takuissu, S. Kenmoe, J. T. Ebogo-Belobo, C. Kengne-Ndé, D. S. Mbaga, A. Bowo-Ngandji, J. L. Ndzie Ondigui, R. Kenfack-Momo, S. Tchatchouang, J. Kenfack-Zanguim, R. L. Fogang, E. Z. Menkem, G. I. Kame-Ngasse, J. N. Magoudjou-Pekam, C. Veneri, P. Mancini, G. B. Ferraro, M. Iaconelli, L. Orlandi, C. Del Giudice, E. Suffredini and G. La Rosa, Int. J. Environ. Res. Public Health, 2023, 20, 1054, DOI:10.3390/ijerph20021054.
- R. A. Farahat, S. H. Khan, A. A. Rabaan and J. A. Al-Tawfiq, New Microbes New Infect., 2023, 53, 101122, DOI:10.1016/j.nmni.2023.101122.
- A. Blagodatski, K. Trutneva, O. Glazova, O. Mityaeva, L. Shevkova, E. Kegeles, N. Onyanov, K. Fede, A. Maznina, E. Khavina, S. J. Yeo, H. Park and P. Volchkov, Pathogens, 2021, 10, 630, DOI:10.3390/pathogens10050630.
- N.Đ Khoa, T. V. Xạ and L. T. Hào, Physiol. Mol. Plant Pathol., 2017, 100, 57–66 CrossRef CAS.
- E. Byamukama, S. N. Wegulo, S. Tatineni, G. L. Hein, R. A. Graybosch, P. S. Baenziger and R. French, Plant Dis., 2014, 98, 127–133 CrossRef CAS PubMed.
- P. Caciagli, Enc. Virol., 2008, 282–290 Search PubMed.
- C. P. Gerba and W. Q. Betancourt, Pathogens, 2019, 8, 107, DOI:10.3390/pathogens8030107.
- D. Dewey-Mattia, K. Manikonda, A. Hall, M. Wise and S. Crowe, Surveillance for Foodborne Disease Outbreaks – United States, 2009–2015, 2018 Search PubMed.
- J. Mans, Viruses, 2019, 11, 341, DOI:10.3390/v11040341.
- M. Rönnqvist, E. Aho, A. Mikkelä, J. Ranta, P. Tuominen, M. Rättö and L. Maunula, Appl. Environ. Microbiol., 2014, 80, 5403–5410 CrossRef PubMed.
- L. Singh and V. S. Sharanagat, Nutr. Food Sci., 2024, 54, 207–237, DOI:10.1108/NFS-01-2023-0012.
- N. Ibrahim, N. D. Jamaluddin, L. L. Tan and N. Y. M. Yusof, Sensors, 2021, 21, 5114, DOI:10.3390/s21155114.
- W. Wang, Y. You and S. Gunasekaran, Compr. Rev. Food Sci. Food Saf., 2021, 20, 5829–5855 CrossRef CAS PubMed.
- R. Peltomaa, E. Benito-Peña, H. H. Gorris and M. C. Moreno-Bondi, Analyst, 2021, 146, 13–32, 10.1039/d0an01883j.
- S. M. A. Färkkilä, M. Mortimer, R. Jaaniso, A. Kahru, V. Kiisk, A. Kikas, J. Kozlova, I. Kurvet, U. Mäeorg, M. Otsus and K. Kasemets, Nanomaterials, 2024, 14, 10, DOI:10.3390/nano14010010.
- Y. Zou, Y. Shi, T. Wang, S. Ji, X. Zhang, T. Shen, X. Huang, J. Xiao, M. A. Farag, J. Shi and X. Zou, Compr. Rev. Food Sci. Food Saf., 2024, 23, e13339, DOI:10.1111/1541-4337.13339.
- A. El Moutaouakil, S. Poovathy, M. Belmoubarik and W. K. Peng, in Review: Graphene-based biosensor for Viral Detection, 2020 Search PubMed.
- Z. Alhalili, Molecules, 2023, 28, 3086, DOI:10.3390/molecules28073086.
- M. A. R. Khan, M. S. Al Mamun and M. H. Ara, Microchem. J., 2021, 171, 106840, DOI:10.1016/j.microc.2021.106840.
- R. Stephanie, M. W. Kim, S. H. Kim, J. K. Kim, C. Y. Park and T. J. Park, TrAC, Trends Anal. Chem., 2021, 135, 116159, DOI:10.1016/j.trac.2020.116159.
- N. Duan, T. Yao, C. Li, Z. Wang and S. Wu, Talanta, 2022, 245, 123445, DOI:10.1016/j.talanta.2022.123445.
- F. Wu, H. Wang, J. Lv, X. Shi, L. Wu and X. Niu, Biosens. Bioelectron., 2023, 236, 115417, DOI:10.1016/j.bios.2023.115417.
- W. Li, F. Xiao, X. Bai and H. Xu, Chem. Eng. J., 2023, 465, 142816, DOI:10.1016/j.cej.2023.142816.
- X. Yu, T. Zhong, Y. Zhang, X. Zhao, Y. Xiao, L. Wang, X. Liu and X. Zhang, J. Agric. Food Chem., 2022, 70, 46–62, DOI:10.1021/acs.jafc.1c03675.
- V. Ayerdurai, M. Cieplak and W. Kutner, TrAC, Trends Anal. Chem., 2023, 158, 116830, DOI:10.1016/j.trac.2022.116830.
- P. Ray, R. Chakraborty, O. Banik, E. Banoth and P. Kumar, ACS Omega, 2023, 8, 3606–3629, DOI:10.1021/acsomega.2c05983.
- Z. Yilmaz-Sercinoglu, C. İlke Kuru and F. Ulucan-Karnak, Sensing Tools and Techniques for COVID-19: Developments and Challenges in Analysis and Detection of Coronavirus, Elsevier, 2022, pp. 57–82 Search PubMed.
- Y. Jeong, J. Kim, J. Lee, S. Seo, S. Roh, G. Lee, B. G. Choi, N. H. Bae, J. Jung, T. Kang, K. G. Lee and E. K. Lim, Mater. Horiz., 2025, 12, 451–457, 10.1039/d4mh01062k.
- S. M. Sheta and S. M. El-Sheikh, Anal. Biochem., 2022, 648, 114680, DOI:10.1016/j.ab.2022.114680.
- O. Prakash, D. Verma and P. C. Singh, J. Mater. Chem. B, 2024, 12, 10198–10214, 10.1039/d4tb01556h.
- L. Luo, F. Zhang, C. Chen and C. Cai, Microchim. Acta, 2020, 187, 140, DOI:10.1007/s00604-020-4122-1.
- Z. Zhang, Y. Zhang, H. Jayan, S. Gao, R. Zhou, N. Yosri, X. Zou and Z. Guo, Food Chem., 2024, 448, 139051, DOI:10.1016/j.foodchem.2024.139051.
- S. Nath, Sustainable Food Technol., 2024, 2, 976–992, 10.1039/d4fb00094c.
- J. Zhang, X. Zhang, Y. Zhang, X. Yang, L. Guo, C. Man, Y. Jiang, W. Zhang and X. Zhang, npj Sci. Food, 2025, 9, 84, DOI:10.1038/s41538-025-00444-5.
- M. A. Elaguech, Y. Yin, Y. Wang, B. Shao, C. Tlili and D. Wang, Sens. Diagn., 2023, 2, 1612–1622 RSC.
- M. S. Rashed, E. A. Abdelkarim, T. Elsamahy, M. Sobhy, H. S. El-Mesery and A. Salem, Food Chem.: X, 2025, 26, 102336, DOI:10.1016/j.fochx.2025.102336.
- S. Giberti, S. Dutta, S. Corni, M. Frasconi and G. Brancolini, Nanoscale, 2025, 17, 4389–4399, 10.1039/d4nr04199b.
- Y. Wang, J. E. Q. Quinsaat, T. Ono, M. Maeki, M. Tokeshi, T. Isono, K. Tajima, T. Satoh, S. Sato, Y. Miura and T. Yamamoto, Nat. Commun., 2020, 11, 6089, DOI:10.1038/s41467-020-19947-8.
- W. A. A. Rosero, A. B. Barbezan, C. D. de Souza and M. E. C. M. Rostelato, Pharmaceutics, 2024, 16, 255, DOI:10.3390/pharmaceutics16020255.
- H. Kumar, K. Kuča, S. K. Bhatia, K. Saini, A. Kaushal, R. Verma, T. C. Bhalla and D. Kumar, Sensors, 2020, 20, 1966, DOI:10.3390/s20071966.
- A. Mokhtarzadeh, R. Eivazzadeh-Keihan, P. Pashazadeh, M. Hejazi, N. Gharaatifar, M. Hasanzadeh, B. Baradaran and M. de la Guardia, TrAC, Trends Anal. Chem., 2017, 97, 445–457, DOI:10.1016/j.trac.2017.10.005.
- Y. S. Chung, N. J. Lee, S. H. Woo, J. M. Kim, H. M. Kim, H. J. Jo, Y. E. Park and M. G. Han, Sci. Rep., 2021, 11, 14817, DOI:10.1038/s41598-021-94196-3.
- M. Eshaghi Gorji, M. T. H. Tan, M. Y. Zhao and D. Li, Pathogens, 2021, 10, 846, DOI:10.3390/pathogens10070846.
- Y. Zheng, X. Song, Z. Fredj, S. Bian and M. Sawan, Anal. Chim. Acta, 2023, 1244, 340860, DOI:10.1016/j.aca.2023.340860.
- A. A. Bergwerff and S. B. Debast, Foods, 2021, 10, 832, DOI:10.3390/foods10040832.
- S. H. Baek, C. Y. Park, T. P. Nguyen, M. W. Kim, J. P. Park, C. Choi, S. Y. Kim, S. K. Kailasa and T. J. Park, Food Control, 2020, 114, 107225, DOI:10.1016/j.foodcont.2020.107225.
- F. Nasrin, I. M. Khoris, A. D. Chowdhury, J. Boonyakida and E. Y. Park, Sens. Actuators, B, 2022, 369, 132390, DOI:10.1016/j.snb.2022.132390.
- A. Alzahrani, T. Alsulami, A. M. Salamatullah and S. R. Ahmed, J. Biol. Eng., 2023, 17, 33, DOI:10.1186/s13036-023-00351-x.
- A. D. Chowdhury, S. Sharmin, F. Nasrin, M. Yamazaki, F. Abe, T. Suzuki, E. Y. Park and E. Y. Park, ACS Appl. Bio Mater., 2020, 3, 3560–3568 CrossRef CAS PubMed.
- H. A. Alhadrami, S. Al-Amer, Y. Aloraij, F. Alhamlan, R. Chinnappan, K. M. Abu-Salah, S. Almatrrouk and M. M. Zourob, ACS Omega, 2020, 5, 12162–12165 CrossRef CAS PubMed.
- G. Ning, Q. Duan, H. Liang, H. Liu, M. Zhou, C. Chen, C. Zhang, H. Zhao and C. P. Li, Food Sci. Hum. Wellness, 2024, 13, 920–931 CrossRef CAS.
- A. B. Ganganboina, A. D. Chowdhury, I. M. Khoris, F. Nasrin, K. Takemura, T. Hara, F. Abe, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2020, 157, 112169, DOI:10.1016/j.bios.2020.112169.
- H. Liu, S. Ma, G. Ning, R. Zhang, H. Liang, F. Liu, L. Xiao, L. Guo, Y. Zhang, C. P. Li and H. Zhao, Talanta, 2023, 258, 124433, DOI:10.1016/j.talanta.2023.124433.
- J. Xie, P. Zhou, M. Wen, J. Bai, F. Yang and C. Duan, Anal. Methods, 2019, 11, 3350–3357 RSC.
- L. Micheli, A. Fasoli, A. Attar, D. T. Donia, M. Divizia, A. Amine, G. Palleschi, P. A. Salazar Carballo and D. Moscone, Talanta, 2021, 234, 122672, DOI:10.1016/j.talanta.2021.122672.
- A. B. Ganganboina, A. D. Chowdhury, I. M. Khoris, R. Doong, T. C. Li, T. Hara, F. Abe, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2020, 170, 112680, DOI:10.1016/j.bios.2020.112680.
- A. B. Ganganboina, I. M. Khoris, A. D. Chowdhury, T. C. Li and E. Y. Park, ACS Appl. Mater. Interfaces, 2020, 12, 50212–50221, DOI:10.1021/acsami.0c13247.
- T. Ngamdee, L. S. Yin, S. Vongpunsawad, Y. Poovorawan, W. Surareungchai and B. Lertanantawong, Anal. Chim. Acta, 2020, 1134, 10–17 CrossRef CAS PubMed.
- Y. Zhang, G. Wu, J. Wei, Y. Ding, Y. Wei, Q. Liu and H. Chen, Microchim. Acta, 2021, 188, 3, DOI:10.1007/s00604-020-04670-4.
- M. Rippa, R. Castagna, S. Brandi, G. Fusco, M. Monini, D. Chen, J. Zhou, J. Zyss and L. Petti, ACS Appl. Nano Mater., 2020, 3, 4837–4844 CrossRef CAS.
- K. Ketabi, H. Soleimanjahi, A. Teimoori, B. Hatamluyi, M. Rezayi and Z. Meshkat, Microchim. Acta, 2023, 190, 293, DOI:10.1007/s00604-023-05871-3.
- S. Biswas, Y. D. Devi, D. Sarma, N. D. Namsa and P. Nath, J. Biophotonics, 2022, 15, e202200138, DOI:10.1002/jbio.202200138.
- S. Biswas, Y. D. Devi, D. Sarma, D. Hatiboruah, N. Chamuah, N. D. Namsa and P. Nath, Spectrochim. Acta, Part A, 2023, 295, 122610, DOI:10.1016/j.saa.2023.122610.
- B. Jain, U. Lambe, A. Tewari, S. K. Kadian and M. Prasad, VirusDisease, 2018, 29, 192–198 CrossRef PubMed.
- M. E. Hamdy, M. Del Carlo, H. A. Hussein, T. A. Salah, A. H. El-Deeb, M. M. Emara, G. Pezzoni and D. Compagnone, J. Nanobiotechnol., 2018, 16, 48, DOI:10.1186/s12951-018-0374-x.
- J. W. Kim, K. W. Park, M. Kim, K. K. Lee and C. S. Lee, Nanomaterials, 2022, 12, 264, DOI:10.3390/nano12020264.
- Y. J. Hwang, K. K. Lee, J. W. Kim, K. H. Chung, S. J. Kim, W. S. Yun and C. S. Lee, Biomolecules, 2021, 11, 841, DOI:10.3390/biom11060841.
- T. Lee, G. H. Kim, S. M. Kim, K. Hong, Y. Kim, C. Park, H. Sohn and J. Min, Colloids Surf., B, 2019, 182, 110341, DOI:10.1016/j.colsurfb.2019.06.070.
- C. Song, Y. Liu, X. Jiang, J. Zhang, C. Dong, J. Li and L. Wang, Talanta, 2019, 205, 120137, DOI:10.1016/j.talanta.2019.120137.
- M. Xiao, K. Xie, X. Dong, L. Wang, C. Huang, F. Xu, W. Xiao, M. Jin, B. Huang and Y. Tang, Anal. Chim. Acta, 2019, 1053, 139–147 CrossRef CAS PubMed.
- Z. Su, G. Zhao and W. Dou, Anal. Methods, 2021, 13, 2313–2319 RSC.
- R. Chi, P. Y. Lin, Y. S. Jhuo, F. Y. Cheng and J. a. A. Ho, Talanta, 2024, 267, 125159, DOI:10.1016/j.talanta.2023.125159.
- S. Lu, F. Li, Q. Chen, J. Wu, J. Duan, X. Lei, Y. Zhang, D. Zhao, Z. Bu and H. Yin, Cell Discovery, 2020, 6, 18, DOI:10.1038/s41421-020-0151-5.
- C. Aira, A. Monedero, S. Hernández-Antón, J. Martínez-Cano, A. Camuñas, N. Casado, R. Nieto, C. Gallardo, M. García-Durán, P. Rueda and A. Fresco-Taboada, Pathogens, 2023, 12, 811, DOI:10.3390/pathogens12060811.
- H. Lee, S. Lee, C. Park, M. Yeom, J. W. Lim, T. T. H. Vu, E. Kim, D. Song and S. Haam, Small, 2023, 19, 2207117, DOI:10.1002/smll.202207117.
- M. N. A. Uda, C. M. Hasfalina, A. A. Samsuzana, U. Hashim, S. A. B. Ariffin, I. Zamri, W. Nur Sabrina, B. B. Siti Noraini, S. Faridah, M. Mazidah and S. C. B. Gopinath, AIP Conference Proceedings, American Institute of Physics Inc., 2017, 1808 Search PubMed.
- R. M. Thangavelu, N. Kadirvel, P. Balasubramaniam and R. Viswanathan, Sci. Rep., 2022, 12, 4144, DOI:10.1038/s41598-022-07950-6.
- Z. Liu, X. Xia, C. Yang and L. Wang, RSC Adv., 2015, 5, 100891–100897 RSC.
- C. Zhang, M. Guo, J. Dong, L. Liu, X. Zhou and J. Wu, Viruses, 2023, 15, 1607, DOI:10.3390/v15071607.
- R. Lei, R. Kuang, X. Peng, Z. Jiao, Z. Zhao, H. Cong, Z. Fan and Y. Zhang, Front. Plant Sci., 2023, 14, 1088544, DOI:10.3389/fpls.2023.1088544.
- M. A. Tahir, S. Z. Bajwa, S. Mansoor, R. W. Briddon, W. S. Khan, B. E. Scheffler and I. Amin, J. Hazard. Mater., 2018, 346, 27–35 CrossRef CAS PubMed.
- R. Lavanya and V. Arun, Sci. Rep., 2021, 11, 14203, DOI:10.1038/s41598-021-93615-9.
- T. M. Wang and J. T. Yang, Sci. Rep., 2019, 9, 15146, DOI:10.1038/s41598-019-51650-7.
- S. C. Razo, V. G. Panferov, I. V. Safenkova, Y. A. Varitsev, A. V. Zherdev and B. B. Dzantiev, Anal. Chim. Acta, 2018, 1007, 50–60 CrossRef CAS PubMed.
- N. Verma, B. S. Tiwari and A. Pandya, ACS Agric. Sci. Technol., 2021, 1, 558–565 CrossRef CAS.
- S. Majumder and S. Johari, J. Virol. Methods, 2018, 255, 23–28 CrossRef CAS PubMed.
- M. Khater, A. de la Escosura-Muñiz, D. Quesada-González and A. Merkoçi, Anal. Chim. Acta, 2019, 1046, 123–131 CrossRef CAS PubMed.
- J. Qin, Z. Yin, D. Shen, H. Chen, X. Chen, X. Cui and X. Chen, Phytopathol. Res., 2021, 3, 3, DOI:10.1186/s42483-021-00080-3.
- M. Arvin, Encyclopedia of Food Safety, Elsevier, 2024, vol. 2, pp. 218–226 Search PubMed.
- J. Wang, Z. Gao, Z. R. Yang, K. Liu and H. Zhang, BMC Infect. Dis., 2023, 23, 595, DOI:10.1186/s12879-023-08519-y.
- M. A. Zaczek-Moczydlowska, A. Beizaei, M. Dillon and K. Campbell, Trends Food Sci. Technol., 2021, 114, 684–695, DOI:10.1016/j.tifs.2021.06.027.
- US CDC, https://www.cdc.gov/hepatitis/hev/hevfaq.html.
- A. N. Olaimat, A. O. Taybeh, A. Al-Nabulsi, M. Al-Holy, M. M. Hatmal, J. Alzyoud, I. Aolymat, M. H. Abughoush, H. Shahbaz, A. Alzyoud, T. Osaili, M. Ayyash, K. M. Coombs and R. Holley, Life, 2024, 14, 190 CrossRef CAS PubMed.
- S. You, T. Liu, M. Zhang, X. Zhao, Y. Dong, B. Wu, Y. Wang, J. Li, X. Wei and B. Shi, Nat. Food, 2021, 2, 802–808 CrossRef PubMed.
- W. A. Gebreyes, D. Jackwood, C. J. B. de Oliveira, C.-W. Lee, A. E. Hoet and S. Thakur, Microbiol. Spectrum, 2020, 8(2), 1–21, DOI:10.1128/microbiolspec.ame-0011-2019.
- J. S. M. Peiris, M. D. de Jong and Y. Guan, Clin. Microbiol. Rev., 2007, 20(2), 243–267, DOI:10.1128/CMR.00037-06.
- W. W. Hsiao, G. Fadhilah, C. C. Lee, R. Endo, Y. J. Lin, S. Angela, C. C. Ku, H. C. Chang and W. H. Chiang, Talanta, 2023, 265, 124892, DOI:10.1016/j.talanta.2023.124892.
- S. Yang, C. Miao, W. Liu, G. Zhang, J. Shao and H. Chang, Front. Microbiol., 2023, 14, 1043129, DOI:10.3389/fmicb.2023.1043129.
- T. Nguyen-Thi, L. Pham-Thi-Ngoc, Q. Nguyen-Ngoc, S. Dang-Xuan, H. S. Lee, H. Nguyen-Viet, P. Padungtod, T. Nguyen-Thu, T. Nguyen-Thi, T. Tran-Cong and K. M. Rich, Front. Vet. Sci., 2021, 8, 686038, DOI:10.3389/fvets.2021.686038.
- P. Babaie, A. Saadati and M. Hasanzadeh, J. Biomed. Mater. Res., 2021, 109, 548–571, DOI:10.1002/jbm.b.34723.
- E. R. Duke, B. D. Williamson, B. Borate, J. L. Golob, C. Wychera, T. Stevens-Ayers, M. L. Huang, N. Cossrow, H. Wan, T. C. Mast, M. A. Marks, M. E. Flowers, K. R. Jerome, L. Corey, P. B. Gilbert, J. T. Schiffer and M. Boeckh, J. Clin. Invest., 2021, 131, e133960, DOI:10.1172/JCI133960.
- H. Xi, H. Jiang, M. Juhas and Y. Zhang, ACS Omega, 2021, 6, 25846–25859, DOI:10.1021/acsomega.1c04024.
- S. Y. Ow, L. Sutarlie, S. W. Y. Lim, N. A. B. M. Salleh, Y. Tanaka, C. K. I. Tan and X. Su, TrAC, Trends Anal. Chem., 2024, 173, 117630, DOI:10.1016/j.trac.2024.117630.
- J. A. Lowther, A. Bosch, S. Butot, J. Ollivier, D. Mäde, S. A. Rutjes, G. Hardouin, B. Lombard, P. in't Veld and A. Leclercq, Int. J. Food Microbiol., 2019, 288, 82–90 CrossRef CAS PubMed.
- D. Pissuwan, C. Gazzana, S. Mongkolsuk and M. B. Cortie, Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol., 2020, 12, e1584, DOI:10.1002/wnan.1584.
- Statista, Number of mobile phone users worldwide from 2015–2020, https://www.statista.com/statistics/274774/forecast-of-mobile-phone-users-worldwide/.
- M. Xiao, F. Tian, X. Liu, Q. Zhou, J. Pan, Z. Luo, M. Yang and C. Yi, Adv. Sci., 2022, 9, 2105904, DOI:10.1002/advs.202105904.
- H. Khan, Z. Jan, I. Ullah, A. Alwabli, F. Alharbi, S. Habib, M. Islam, B. J. Shin, M. Y. Lee and J. K. Koo, Nanotechnol. Rev., 2024, 13, 20240056, DOI:10.1515/ntrev-2024-0056.
- K. Zhang, J. Wang, T. Liu, Y. Luo, X. J. Loh and X. Chen, Adv. Healthcare Mater., 2021, 10, 2100734, DOI:10.1002/adhm.202100734.
- S. Ramalingam, C. M. Collier and A. Singh, Biosensors, 2021, 11, 29, DOI:10.3390/bios11020029.
- F. Cui, Y. Yue, Y. Zhang, Z. Zhang and H. S. Zhou, ACS Sens., 2020, 5, 3346–3364, DOI:10.1021/acssensors.0c01424.
- A. M. Zeid, I. M. Mostafa, B. Lou and G. Xu, Lab Chip, 2023, 23, 4160–4172, 10.1039/d3lc00674c.
- Z. Zhou, D. Tian, Y. Yang, H. Cui, Y. Li, S. Ren, T. Han and Z. Gao, Curr. Res. Food Sci., 2024, 8, 100679, DOI:10.1016/j.crfs.2024.100679.
- Y. X. Leong, E. X. Tan, S. X. Leong, C. S. L. Koh, L. B. T. Nguyen, J. R. T. Chen, K. Xia and X. Y. Ling, ACS Nano, 2022, 16, 13279–13293, DOI:10.1021/acsnano.2c05731.
| This journal is © The Royal Society of Chemistry 2025 |