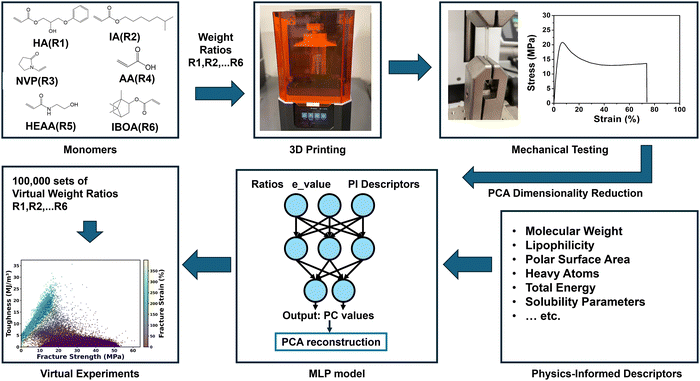
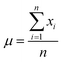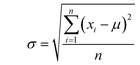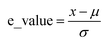Open Access Article
Open Access ArticlePhysics-informed machine learning enabled virtual experimentation for 3D printed thermoplastic†
Zhenru
Chen
a,
Yuchao
Wu
a,
Yunchao
Xie
b,
Kianoosh
Sattari
a and
Jian
Lin
 *a
*a
aDepartment of Mechanical and Aerospace Engineering, University of Missouri, Columbia, Missouri 65211, USA. E-mail: linjian@missouri.edu
bDepartment of Mechanical and Manufacturing Engineering, Miami University, Oxford, Ohio 45056, USA
First published on 2nd October 2024
Abstract
The performance of 3D printed thermoplastics largely depends on the ink formulation, which is composed of tremendous chemical space as an increased number of monomers, making it very difficult to identify an optimum one with desired properties. To tackle this challenge, we demonstrate a virtual experimentation platform that is enabled by a physics-informed machine learning algorithm. As a case study, the algorithm was trained based on a multilayer perceptron (MLP) model to predict the experimental stress–strain curves of the 3D printed thermoplastics given the ink compositions made of six monomers. To solve the issue of experimental data scarcity, we first reduced the dimensions of the curves to eight principal components (PCs), which serve as the outputs of the model. In addition, we incorporated the physics-informed descriptors into the input dataset. These two strategies afford the model with a prediction accuracy of R2 of 0.97 and an RMSE value of 1.01 for fracture strength, and an R2 of 0.95 and a RMSE of 0.40 for toughness. To perform virtual experimentation, the well-trained model was then utilized to predict 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 sets of the PCs from the randomly given 100
000 sets of the PCs from the randomly given 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ink formulations. The PC sets were then converted back to the corresponding stress–strain curves. To validate the prediction results, some of the virtual experiments were performed. The results showed a good match between the predicted and experimental curves. This methodology offers a general and efficient pathway to virtual experimentation for establishing the correlation between the complex input variables and the output performance metrics of new materials.
000 ink formulations. The PC sets were then converted back to the corresponding stress–strain curves. To validate the prediction results, some of the virtual experiments were performed. The results showed a good match between the predicted and experimental curves. This methodology offers a general and efficient pathway to virtual experimentation for establishing the correlation between the complex input variables and the output performance metrics of new materials.
New conceptsWe introduce a virtual experimentation platform that uses a physics-informed machine learning algorithm to predict the mechanical properties of 3D printed thermoplastics. By employing a multilayer perceptron (MLP) model, this innovative approach simulates the stress–strain behavior of materials based on their complex ink compositions. Our method effectively addresses the challenge of sparse experimental data by utilizing dimensionality reduction techniques and incorporating physics-informed descriptors into the model. This enables rapid prototyping and optimization of new material formulations while significantly reducing the need for extensive physical testing. The successful application of our model to a large dataset of ink formulations, validated against experimental results, showcases the platform's potential to transform material design in 3D printing. |
1. Introduction
Virtual experimentation represents a pivotal advancement in scientific research, enabling extensive pre-experimental screening that refines the scope of physical trials, thus saving resources on the most promising inquiries.1 Such preliminary simulations are especially critical in fields where experimental setups are costly and experimentations are labor intensive. A prime example is 3D printing, which offers rapid prototyping and manufacturing capabilities that have become indispensable across industries—from aerospace to healthcare—due to their ability to cost-effectively create objects with complex geometries.2,3 Despite these advantages, the development of new materials for 3D printing, especially thermoplastics, presents significant challenges. The mechanical properties of thermoplastics, crucial for their functional applications, depend heavily on ink formulations. The complex interactions and subsequent polymerization of various monomers in the inks profoundly impact their mechanical properties.4,5 The traditional experimental process, which involves the exploration of vast ink formulations to pinpoint the desired mechanical properties of 3D printed thermoplastics, requires extensive experimentation. This process becomes particularly laborious and time-consuming as the combinational chemical space dramatically increases. Virtual experimentation provides a significant advantage over traditional methods by allowing researchers to bypass the initial phases of testing, where intuition alone may not suffice to optimize experimental conditions.Virtual experimentation often relies on theoretical calculations or computational simulation techniques, such as density functional theory (DFT) and molecular dynamics (MD). These methods have been extensively applied in fields such as materials science6,7 and chemical engineering8 to predict the properties of materials at various scales. However, these approaches often face challenges in accurately scaling predictions to complex macroscopic phenomena. For instance, in the 3D printing processes, while molecular simulations are adept at modeling the intricate interactions between monomers,9 they struggle to extend these predictions to the overall mechanical properties of materials. This limitation suggests that alternative approaches, such as data-driven methods that bypass detailed microstructural modeling, may be necessary. Additionally, deriving practical characteristic curves, such as stress–strain (S–S) ones, poses another significant challenge because physics-based simulation results often rely on idealized material systems under conceptualized conditions, which may not accurately reflect real-world behaviors.
In contrast, data-driven algorithms, such as machine learning (ML), have recently emerged as a complementary approach,10 increasingly pervading the materials science in design,11,12 property prediction,13–15 synthesis planning,16–18 and automated data analysis.19–21 They are forming a new paradigm for virtual experimentation.1 For instance, in predicting material performance, the integration of vast datasets with advanced algorithms has enabled more precise and efficient predictions than ever before. By leveraging extensive data obtained from DFT simulations, ML algorithms can now be applied to predict the performance of composites22,23 and metamaterials24 with unprecedented accuracy and efficiency. To mitigate the data scarcity issue, physics-informed ML (PIML) by incorporating known physical laws into the ML training has been developed.5,25,26 This hybrid approach not only enhances prediction accuracy with a limited amount of data but also extends the capability of simulations to cover unexplored material systems. For example, our group incorporated the chemical and physical properties of metal salts and organic linkers as physics-informed descriptors to unravel complex synthesis parameters for accurately predicting the crystallization propensity of metal–organic nanocapsules.12 In our other work, we trained a scientific ML model that includes intermediate reaction variables obtained by simulations for predicting the reaction outcomes.27 Du and coworkers utilized six mechanistic variables that represent the physics of balling defects to train a ML model for predicting defects formed during the 3D printing processes.28 The use of PIML in virtual experimentation holds vast potential, particularly in refining the design and optimization processes in 3D printing, where understanding the detailed physical and chemical interactions crossing the multiple scales is often impractical. Despite the vast potential and recent research progress, in most literature reports that involve ML algorithms for property prediction, typically singular numerical features (e.g. strength and fractural strain) rather than a total performance profile were reported. In our recent work, we employed a multi-objective Bayesian optimization method to identify materials for 3D printing of thermoplastics that are both strong and tough, focusing specifically on optimizing these two singular numerical values.5 In contrast, the current study utilizes physics-informed machine learning (PIML) for predicting the whole stress–strain curves from which the complete mechanical performance of materials can be derived, thereby acting as a virtual experimentation platform.
Herein, to tackle the challenge, we demonstrate a PIML for predicting full stress–strain curves of 3D printed thermoplastics, which serves as an efficient virtual experimentation platform for screening ink formulations that lead to thermoplastics with desired mechanical properties. To realize this goal, a total of 216 S–S curves were first collected from thermoplastics that were 3D printed using six monomers. Then, the dimensions of these S–S curves were reduced by principal component analysis (PCA) into eight principal components (PCs). After that, the compositions of the six monomers together with the physics-informed descriptors (including molecular weight, lipophilicity, Hbond donor/acceptor, rotatable bonds, polar surface area, heavy atoms, complexity, total energy and several solubility scores) serve as the inputs while the corresponding sets of PCs serve as the outputs to train a multiple layer perceptron (MLP) model. Given 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 sets of hypothesized ink compositions, the MLP can predict new PCs, which were then converted back to the corresponding S–S curves. Among them, six ink formulations featuring three different types of mechanical profiles were chosen for experimental validation. The obtained S–S curves from these experiments fell within the ranges predicted by the virtual experiments. A quantitative study shows that the model achieves prediction accuracy with a satisfactory R2 value of 0.97 and a root mean squared error (RMSE) of 1.01 for fracture strength, an R2 of 0.95 and an RMSE of 0.40 for toughness. These results affirm the success of the virtual experimentation for large scale screening, opening the way for designing new thermoplastics with desired properties.
000 sets of hypothesized ink compositions, the MLP can predict new PCs, which were then converted back to the corresponding S–S curves. Among them, six ink formulations featuring three different types of mechanical profiles were chosen for experimental validation. The obtained S–S curves from these experiments fell within the ranges predicted by the virtual experiments. A quantitative study shows that the model achieves prediction accuracy with a satisfactory R2 value of 0.97 and a root mean squared error (RMSE) of 1.01 for fracture strength, an R2 of 0.95 and an RMSE of 0.40 for toughness. These results affirm the success of the virtual experimentation for large scale screening, opening the way for designing new thermoplastics with desired properties.
2. Results and discussion
2.1. Workflow
Fig. 1 illustrates the workflow of developing a PIML based virtual experimentation platform for thermoplastic 3D printing. First, 2-hydroxy-3-phenoxypropyl acrylate (HA), iso-octyl acrylate (IA), N-vinylpyrrolidone (NVP), acrylic acid (AA), N-(2-hydroxyethyl) acrylamide (HEAA) and isobornyl acrylate (IBOA) were selected as the six monomers.5 This diverse selection was strategically chosen to demonstrate the robustness and adaptability of our machine learning model for a complex chemical space, showcasing the necessity and effectiveness of the proposed virtual experimentation workflow. Then, inks were prepared via mixing these six monomers in different weight ratios for printing using a liquid crystal display (LCD) printer. After that, the S–S curves of the resulting thermoplastics were collected using a tensile testing machine (Mark-10) according to American Society for Testing and Materials standards. The collected curves were preprocessed and reduced in dimensions by PCA detailed as follows. Following this, a multiple layer perceptron (MLP) model was trained by using the ink compositions together with the physics-informed descriptors as the input to predict these dimension-reduced representations. The culmination of this process employed an inverse PCA technique to reconstruct the S–S curves from the predicted PCs.2.2. Data collection and preprocessing
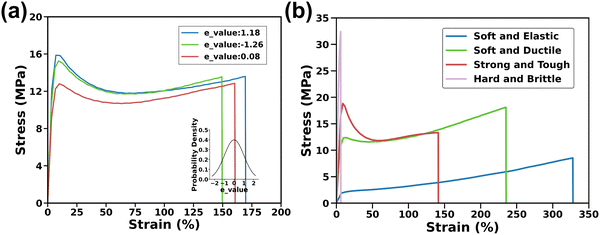
Experimental datasets were collected from 62 ink formulations, with each formulation represented by 2–4 individual S–S curves. 3D thermoplastics printed from the six monomers involve enormous chemical space. Training ML models with only the ratio of the six monomers to predict the high dimensional outputs could suffer from a serious overfitting issue. To overcome this issue, an additional thirteen physics-informed descriptors were chosen as the input parameters, which are the molecular weight, lipophilicity, h-bond donor, n-bond acceptor, rotatable bonds, polar surface area, heavy atoms, complexity, total energy, and solubility parameters.29–32 After normalization, these physics-informed descriptors were multiplied by the ratios of six monomers, leading to 78 cross-features.25 Details on these descriptors and more information about the methodology can be found in Supplementary Note S1 and Table S1 (ESI†).The S–S curves of the specimens with the same ink formulation underwent analysis to ensure the high quality of training data. As depicted in Fig. 2a, the three stress–strain curves of three specimens exhibit variation even though they were printed from the same ink formulation, indicating the unavoidable experimental uncertainty. If using the ink formulation and the corresponding S–S curves as the input and output for the model training, a ‘one-to-many’ prediction issue may arise, where each input corresponds to multiple outputs.33,34 It underscores the importance of using a model capable of adeptly handling such inherent data variability. To address this uncertainty, an e_value based on a normal distribution was introduced to encapsulate the inherent experimental variation. This e_value, analogous to the Z-score in a normal distribution, quantifies how many standard deviations that experimental data point deviates from the mean. Implementation of the e_value is elaborated in the Methods section. The e_value is combined with the ratios of the six monomers and the 78 cross-features to form a total of 85 features in the model.
Depending on different monomer ratios of the inks from which the samples were printed, these S–S curves represent four distinct soft/elastic, soft/tough, strong/tough, and hard/brittle samples, presenting the diversity of the training data, which imposes an additional challenge for the model training (Fig. 2b). The stress–strain curve for the soft/elastic sample shows a typical elastomer behavior, with minimal stress at the low strain and a constant stress level in elongation. The soft/tough and strong/tough samples begin with a steep initial slope, indicating stiffness, but as strain increases, the stress shows a continuous rise without peaking, reflecting substantial plastic deformation. Conversely, the stress in the hard/brittle sample's curve displays a linear increase followed by a sharp drop, characteristic of minimal plastic deformation before fracture. Due to significant variations in the length of data collected, preprocessing steps such as trimming, and interpolation were necessary to standardize the datasets for model training. Details of these preprocessing methods are provided in Methods Section 4.4.
Further observation shows that the numerical range of the strain axis varies considerably, even though both the strain and stress axes consist of 50 data points each in the standardized data format. Given the limited datasets and a 100-dimension output, a concern known as the ‘curse of dimensionality’ arises, a phenomenon where the volume of the space increases so fast that the available data becomes sparse.35 This sparsity is problematic as it can severely impact the performance of machine learning models by making it difficult to extract meaningful patterns without overfitting. Given the limited datasets and the high-dimensional output, dimension reduction becomes essential to mitigate these issues. Previous studies adopted a manual extraction strategy to identify five feature points, i.e., linear limit, maximum yielding, strain softening end, steady flow limit, and fracture points.24,33 In our research, however, the S–S curves in our dataset are more diverse, making the manual extraction of these critical points either cumbersome or inconsistent. To address those concerns, PCA, a powerful dimension reduction technique, was employed.36 PCA is an unsupervised method that does not require predefined criteria for extracting information. It simplifies the dataset by transforming it into a new coordinate system, where the most significant features are summarized in the principal components (PCs). This process not only makes the data more manageable for the ML model but also preserves essential information, thereby facilitating accurate predictions. Instead of directly predicting the whole S–S curves, our model predicts the PC values, which can be then converted back to the S–S curves.
2.3. PCA on stress–strain curves
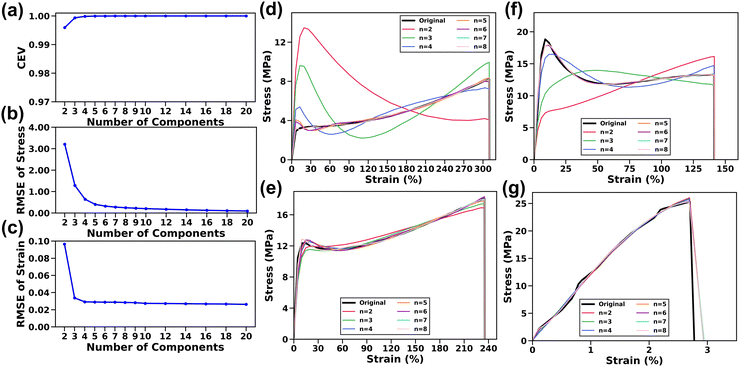
The impact of the number of principal components (denoted as n) on the capacity of the ML model to encapsulate data variance was initially investigated, with a focus on the explained variance which refers to the cumulative proportion of the dataset variance explained with the increase of n. As shown in Fig. 3a, the cumulative explained variance (CEV) increases sharply as n reaches 5, beyond which there is negligible change, indicating the efficacy of PCA in capturing key information from the S–S curves (see Supplementary Note S2 for details, ESI†). This trend is also evident when using the PCs to reconstruct the S–S curves (Fig. 3b and c). The RMSE37 was chosen to determine the difference between the reconstructed and original values of both stress and strain axes (Supplementary Note S3, ESI†). Specifically, the strain RMSE decreases to ∼0.02% when n reaches 4, while the stress RMSE remains nearly unchanged (∼0.03 MPa) at n of 7. Furthermore, the impact of n on the accuracy of the reconstructed S–S curves was also investigated visually across the collected datasets. Fig. 3d–g show a few examples, illustrating typical representative S–S curves as discussed in Fig. 2b. It is found that samples show good agreement between the original and reconstructed curves when n reaches 6. Based on these observations, to encapsulate more subtle variations, the n value is set to 8 for the subsequent analysis.2.4. Interpretability of PCA
After exploring the influence of n on the reconstructed S–S curves, we thoroughly examined the interpretability of each PC during the reconstruction process. By analyzing how the PC values influence the S–S curves, we demonstrate how the PCs reflect essential features of the S–S curves. To do it, each PC is varied by ±100%, ±50%, ±20%, and ±5%, while keeping other PCs the same. As shown in Fig. 4a and Fig. S1 (ESI†), an increase in PC1 prompts a shift of the S–S curves towards larger strains, while increase in PC2 results in a decrease in the slope of the plastic deformation region. It is determined that PC1 has the most pronounced effect on the variations of the S–S curves. An increase in PC3 leads to a decrease in the slope of the post-yield hardening region, whereas an increase in PC4 results in a decrease in the slopes of the plastic deformation region while an increase in the post-yield hardening region. Furthermore, the fractural strain remains constant regardless of the changes in PC2, PC3, and PC4. While the influence of PC5 to PC8 is less significant to be directly interpreted for analyzing the core material properties, these components still contribute to a certain degree of detail in the curves, such as minor fluctuations or inflection points in certain regions of the curves. For a brittle sample (Fig. S2, ESI†), close observation reveals that an increase in PC1 leads to a shift of the curve toward smaller strain, while an increase in PC2 results in an increase in the slope of the elastic deformation, fracture strength and fracture strain. There are no obvious changes in the S–S curves with the changes in PCs from PC3 to PC8. To further explore the hidden information, the relationship between PCs and mechanical properties was analyzed (Fig. S3, ESI†). Clearly, PC1 exhibits a linear relationship with the fractural strain. PC2 is proportional to toughness. PC3 is positively and negatively correlated with the fracture strength and the slope in the strain-hardening area, respectively. The observations are well aligned with the fundamental mechanical characteristics observed in the S–S curves (Fig. 4b).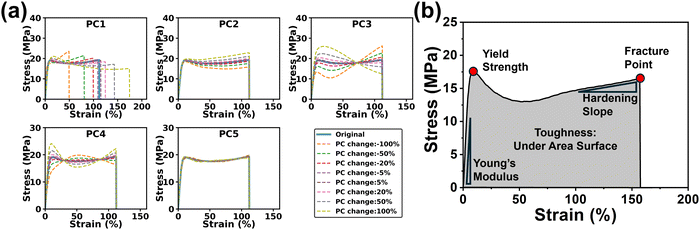 | ||
| Fig. 4 (a) Changes of each PC (PC1 to PC5) vs. change of the reconstructed S–S curves of a strong/tough sample. (b) A typical S–S curve with labeled characteristic points. | ||
2.5. Machine learning model
After establishing the input and output datasets, it is about to train an MLP model. The model takes 85 distinct and cross-features as the inputs to predict the outputs of the eight PCs. Given the relatively small data size, a combined approach of dropout and L1 regularization was employed to prevent overfitting. Dropout operates by randomly deactivating a subset of neurons during the training process, which is beneficial for reducing the model's dependency on specific features.38 Meanwhile, the L1 regularization introduces a penalty to the loss function proportional to the absolute magnitude of the feature coefficients.39 It prioritizes more influential features by pushing the coefficients of less significant ones towards zero. Both the dropout and L1 regularization work in concert to enhance the model's capacity to be generalized effectively. Furthermore, the model is designed to favor the utilization of beneficial physics-informed descriptors, while reducing reliance on those with less impact. This selective approach ensures that the model not only stays accurate but also remains relevant and grounded in the practical aspects of domain science. The mean squared error between the eight predicted and true PCs is chosen as the loss function since it can effectively reflect the hierarchy of significance by preserving the original difference among the PCs.Out of the 62 ink formulations, 50 (representing 180 S–S curves) were chosen as the training datasets, while the remaining 12 (representing 36 S–S curves) were the testing datasets. Here, the test set comprises a balanced combination of materials consisting of 7 elastic ones and 5 brittle ones. Details on the model's intricacies, computation specifics, and information about the hardware and software utilized in this study are comprehensively documented in Supplementary Note S4 (ESI†).
Based on the test set, the performance of the MLP model in predicting the eight PCs is presented in Table 1. While the specific PC values lack direct physical meanings, the R2 values in comparison of the predicted PCs and respectively true PCs reveal the model's accuracy. The R2 values were notably high for the first three principal components (0.97, 0.76, and 0.77 for PC1, PC2, and PC3, respectively) and gradually declined for the remaining five PCs. This trend is expected, i.e., the importance of PCs slightly decreases as the number of PCs increases. This trend also holds true for other evaluation metrics including RMSE, MAE, and MSE, indicating that the MLP model prioritizes the key PCs. RMSE exhibits an opposite trend, starting at 5.40% for PC1 and 10.94% for PC2, and then gradually increasing to 25.18% for PC8. It also underlines the model's ability to concentrate on the most impactful PCs for balancing the accuracy by prevention of overfitting. This inherent characteristic originates from the L1 regularization and dropout to ensure a robust fit for the most significant features.
| PC values | R 2 | RMSE | MAE | MSE | Max | Min | Range | RMSE/range (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.97 | 78.84 | 44.48 | 6215.58 | 1263.16 | −197.45 | 1460.61 | 5.40 |
| 2 | 0.76 | 9.79 | 7.58 | 95.82 | 65.84 | −23.65 | 89.49 | 10.94 |
| 3 | 0.77 | 6.47 | 5.32 | 41.86 | 31.38 | −26.13 | 57.51 | 11.25 |
| 4 | 0.58 | 4.63 | 3.82 | 21.43 | 16.21 | −12.54 | 28.76 | 16.10 |
| 5 | 0.29 | 2.17 | 1.78 | 4.69 | 3.4 | −6.33 | 9.72 | 22.32 |
| 6 | 0.21 | 1.21 | 0.97 | 1.47 | 2.84 | −2.96 | 5.79 | 20.99 |
| 7 | 0.41 | 1.25 | 0.9 | 1.55 | 6.96 | −2.71 | 9.67 | 12.93 |
| 8 | 0.19 | 0.69 | 0.53 | 0.48 | 1.13 | −1.61 | 2.74 | 25.18 |
2.6. Evaluating stress–strain curves
The results indicate the high accuracy of the MLP model in predicting the eight PCs. We then evaluated how well the reconstructed S–S curves from these predicted PCs agree with the true ones. It is impractical to evaluate the reconstruction performance by directly calculating the difference between the reconstructed and true values at each point of the S–S curves. This is because the complexities of material behaviors and testing conditions lead to huge variations of the S–S curves. To mitigate this issue, two critical mechanical performance matrices, i.e., fracture strength and toughness, which can be derived from the S–S curves, were deployed for evaluation. As shown in Table 2, the R2 values are relatively high for fracture strength (0.97) and toughness (0.95), while RMSE and MAE of the fracture strength are 1.01 and 0.82 MPa and for toughness they are 0.40 and 0.31 MJ m−3. After considering their ranges, the RMSE of the fracture strength and toughness are relatively low, i.e., ∼4% for the fracture strength and ∼6% for the toughness. These results indicate the model's robust ability to account for a significant portion of the observed data variance.| Metric | R 2 | RMSE | MAE | Max | Min | Range | RMSE/range (%) |
|---|---|---|---|---|---|---|---|
| Fracture strength | 0.97 | 1.01 | 0.82 | 39.29 | 11.76 | 27.53 | 4.43 |
| Toughness | 0.95 | 0.40 | 0.31 | 10.48 | 4.03 | 6.45 | 5.90 |
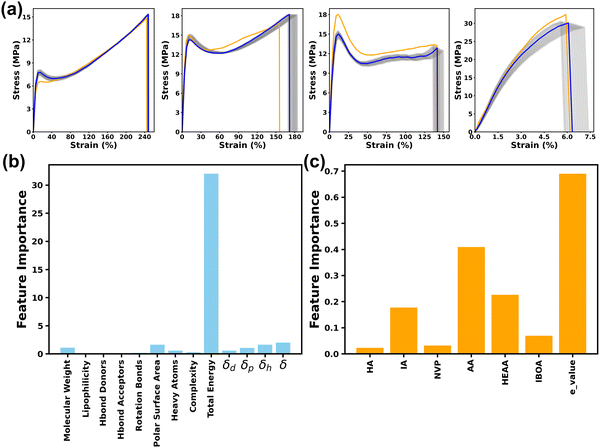
To visually evaluate the model prediction performance, the true and predicted S–S curves (reconstructed from the predicted PCs by the MLP) of the four samples from the test set with various fracture strengths and ductility are shown in Fig. 5a. Additionally, all 36 S–S curves from the test set are provided in Fig. S4 (ESI†). The yellow lines correspond to the original S–S curves, while the blue lines represent the reconstructed S–S curves with the corresponding e_values. To effectively adapt to the variations originating from the experimental and testing conditions, the e_values varying from −2 to 2 were incorporated to reconstruct multiple S–S curves (grey lines). The grey range encompasses 95% of probability the cases according to the Z-score definition in a normal distribution. It is found that these reconstructed S–S curves all fall within the grey areas. Their shapes and trends are matched well with the ground truth S–S curves. These results affirm the high effectiveness of the combination of the MLP model and PCA technique in predicting the S–S curves.
2.7. Feature importance
Importance of the physics-informed descriptors was explored via a comparative study training the MLP model using only the ratios of six monomers and the e_value without PI inputs. As shown in Table S2 (ESI†), the MLP model attained the highest R2 value for PC1, while delivering much lower R2 values for PC3 and PC4. This indicates that the model cannot effectively capture the underlying characteristics of the training datasets if only using PC3 and PC4. Furthermore, the presence of negative R2 values for PC2, PC5, PC6, PC7 and PC8 reveals that the predictive accuracy of the MLP model is even worse than the prediction results using the average of all sampling data. This underscores a substantial limitation in the MLP model without the physics-informed descriptors. This phenomenon was also found in the predicted S–S curves (Table S3 and Fig. S5, ESI†). The R2 values for both true stress (0.52) and toughness (0.38) are lower than those of the MLP model trained with included physics-informed descriptors. As shown in Fig. S5 (ESI†), nearly all the predicted S–S curves exhibited huge variations, revealing the poor prediction capability of the model without the physics-informed descriptors. These results indicate that the incorporation of physics-informed descriptors not only increases the predictive accuracy but also aid in accurately capturing the nuances of the S–S curves.The significance of these physics-informed descriptors was further quantified. An integrated gradients (IG) method was applied to investigate the interpretability of the MLP model.40 The IG method works by examining how change in the gradients of each feature influences the output. Specifically, for each PI descriptor, we calculated its interaction feature importance with each of the six monomers. To synthesize this information and provide a clearer understanding of the overall impact of each PI, we averaged the importance scores across these six monomers for every individual PI. The feature importance scores for 13 physics-informed descriptors, the ratio of six monomers, and e_values were shown in Fig. 5b and c. Detailed methodologies regarding this process are elaborated in the Methods section. As shown in Fig. 5b, the total energy is the primary dominant descriptor among these physics-informed descriptors, which well agrees with expertise and domain knowledge. It is reported that total energy plays a crucial role in determining the structural cohesion, arrangement, and consequent mechanical properties of polymeric materials.41 Other physics-informed descriptors such as solubility, molecular weight, polar surface area, and the number of heavy atoms exhibit relatively lower importance. This suggests that the model effectively leverages these classical features to capture complementary information related to chain entanglement, intermolecular forces, and steric effects, which are known to influence polymer performance.42,43 The remaining descriptors, including complexity, lipophilicity, Hbond donor, Hbond acceptor, and rotatable bonds, exhibit comparatively lower feature importance scores. These descriptors primarily pertain to molecular size, hydrophobicity, and conformational flexibility. The direct impact of these descriptors on intermolecular interactions and electronic structures, which play pivotal roles in determining the mechanical properties of polymers, may be relatively limited.
As shown in Fig. 5c, the e_value, used to account for the experimental uncertainty, was notably discernible. This highlights the model's capability to establish a predictive range based on e_value rather than a simple one-to-one prediction. The feature importance scores for the six monomers follow the order of AA > HEAA > IA > IBOA > NVP > HA. Monomers like AA and HEAA are noteworthy for their propensity to form hydrogen bonds, significantly impacting the intermolecular interactions of the 3D-printed thermoplastics.44 The presence of IA can be attributed to its function as a softer segment than HA, contributing significantly to the flexibility and toughness of 3D printed thermoplastics, despite the potential of HA to form hydrogen bonds.5 These feature importance scores well agree with the empirical understanding of the experiments, thus reinforcing the significance and practical applicability of these descriptors in the MLP model. This method underscores the effectiveness of combining data-driven machine learning with domain-specific expertise, paving the way to more sophisticated and accurate predictive models in materials science.
2.8. Virtual experimentation for screening new ink formulation candidates
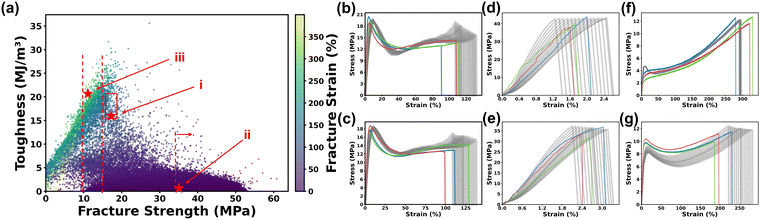
We expect that the developed MLP can be used as a surrogate model to virtually explore the combination space to accelerate the ink formulation to make the thermoplastics that show desired S–S curves. First of all, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 virtual ink formulations were randomly generated using the Dirichlet distribution method since it ensures a uniform distribution of each monomer.45 This approach guarantees an equitable representation of all possible monomer ratios, providing a balanced and comprehensive exploration of the design space. Details on generating virtual ink formulations are provided in Method. After that, a pre-trained random forest model that we previously demonstrated was employed to predict the printability of these ink formulations.5 Only the printable ink formulations were fed into the MLP model to predict the corresponding eight PCs. It is noteworthy that the prediction of these ink formulations took only 1 minute, highlighting the exceptional speed and efficiency of the virtual screening. Then, the S–S curves were reconstructed from the predicted PCs. Then, the fracture strength, maximum strain and toughness were extracted from these reconstructed S–S curves and plotted in Fig. 6a. It was observed that most datapoints were clustered in the region associated with lower toughness, possibly because out of six monomers, four of them are harder monomers including NVP, HA, HEAA and IBOA. If they are dominant in the ratio combinations, they considerably favor the formation of brittle thermoplastics with low toughness.
000 virtual ink formulations were randomly generated using the Dirichlet distribution method since it ensures a uniform distribution of each monomer.45 This approach guarantees an equitable representation of all possible monomer ratios, providing a balanced and comprehensive exploration of the design space. Details on generating virtual ink formulations are provided in Method. After that, a pre-trained random forest model that we previously demonstrated was employed to predict the printability of these ink formulations.5 Only the printable ink formulations were fed into the MLP model to predict the corresponding eight PCs. It is noteworthy that the prediction of these ink formulations took only 1 minute, highlighting the exceptional speed and efficiency of the virtual screening. Then, the S–S curves were reconstructed from the predicted PCs. Then, the fracture strength, maximum strain and toughness were extracted from these reconstructed S–S curves and plotted in Fig. 6a. It was observed that most datapoints were clustered in the region associated with lower toughness, possibly because out of six monomers, four of them are harder monomers including NVP, HA, HEAA and IBOA. If they are dominant in the ratio combinations, they considerably favor the formation of brittle thermoplastics with low toughness.
Following the virtual screening guided by the MLP model, new experiments were conducted to validate the prediction results. We chose these experiments with an aim of identifying the ink formulations leading to three types of thermoplastics (strong/tough, strong/brittle, and soft/elastic). For each type, two ink formulations were randomly selected to print three specimens. Fig. 6b–g show the profiles and trends of the predicted S–S curves by the MLP model.
The first one showing the strong/tough S–S curve has a fracture strength in the range of 15–20 MPa and a toughness in the range of 15–20 MJ m−3. As a result, a total of 143 ink formulations were screened, from which two ink formulations with HA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IA
IA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NVP
NVP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AA
AA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HEAA
HEAA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IBOA weight ratios 0.16
IBOA weight ratios 0.16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.39
0.39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.25
0.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.13
0.13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.02
0.02![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05 (Fig. 6b) and 0.34
0.05 (Fig. 6b) and 0.34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.32
0.32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.21
0.21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.09
0.09![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.02
0.02![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.02 (Fig. 6c) were randomly selected for experiments. As depicted in Fig. 6b and c the resulting S–S curves from these two selections conform to the trend predicted by the MLP model, in which both cases exhibited an instance of premature fracture. Moreover, to further support our mechanical testing data and elucidate the failure mechanisms, we conducted microstructural analysis of the fractured surfaces of the sample shown in Fig. 6b. For this ductile sample, a digital microscope reveals a ductile fracture surface (Fig. S6a, ESI†), which indicates a great degree of plastic deformation before fracture. The second type is the strong/brittle one with a fracture strength exceeding 35 MPa and a fracture strain of 2–5%, resulting in >10
0.02 (Fig. 6c) were randomly selected for experiments. As depicted in Fig. 6b and c the resulting S–S curves from these two selections conform to the trend predicted by the MLP model, in which both cases exhibited an instance of premature fracture. Moreover, to further support our mechanical testing data and elucidate the failure mechanisms, we conducted microstructural analysis of the fractured surfaces of the sample shown in Fig. 6b. For this ductile sample, a digital microscope reveals a ductile fracture surface (Fig. S6a, ESI†), which indicates a great degree of plastic deformation before fracture. The second type is the strong/brittle one with a fracture strength exceeding 35 MPa and a fracture strain of 2–5%, resulting in >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ink formulations. This is because lots of formulations in the virtual experiments show hard and brittle behaviors due to dominant compositions of NVP, HA HEAA or IBOA in the formula. The experimental S–S curves of the six specimens from the selected two ink formulations with HA
000 ink formulations. This is because lots of formulations in the virtual experiments show hard and brittle behaviors due to dominant compositions of NVP, HA HEAA or IBOA in the formula. The experimental S–S curves of the six specimens from the selected two ink formulations with HA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IA
IA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NVP
NVP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AA
AA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HEAA
HEAA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IBOA weight ratios 0.16
IBOA weight ratios 0.16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.18
0.18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05
0.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.42
0.42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.18
0.18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.01 and 0.26
0.01 and 0.26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.29
0.29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05
0.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.29
0.29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.03
0.03![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.08 are within the predicted range (Fig. 6d and e). For a brittle sample represented in Fig. 6d, the fracture surface is notably smoother (Fig. S6b, ESI†), indicating a different brittle fracture mechanism. These microstructural observations further validate our experimental results and provide deeper insights into the different fracture behaviors. The third type is the soft/elastic one. The ink formulations with a predicted fractural strain of >250% and a fracture strength in the range of 10–15 MPa were screened, resulting in 148 formulations. The selected two ink formulations with HA
0.08 are within the predicted range (Fig. 6d and e). For a brittle sample represented in Fig. 6d, the fracture surface is notably smoother (Fig. S6b, ESI†), indicating a different brittle fracture mechanism. These microstructural observations further validate our experimental results and provide deeper insights into the different fracture behaviors. The third type is the soft/elastic one. The ink formulations with a predicted fractural strain of >250% and a fracture strength in the range of 10–15 MPa were screened, resulting in 148 formulations. The selected two ink formulations with HA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IA
IA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NVP
NVP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AA
AA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HEAA
HEAA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IBOA weight ratios of 0.4
IBOA weight ratios of 0.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.28
0.28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.01
0.01![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.0
0.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.09
0.09![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.22 and 0.35
0.22 and 0.35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.38
0.38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.02
0.02![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.18
0.18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.07
0.07![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.0 led to the soft/elastic thermoplastics. Their S–S curves are shown in Fig. 6f and g. We can see that the predicted S–S profiles agree well with the experimental ones despite the little discrepancy in their fractural strains. They are out of the range of the predicted uncertainty range. These experimental validation results show that the developed MLP for virtual experiments is reliable and rapid because the prediction of 100
0.0 led to the soft/elastic thermoplastics. Their S–S curves are shown in Fig. 6f and g. We can see that the predicted S–S profiles agree well with the experimental ones despite the little discrepancy in their fractural strains. They are out of the range of the predicted uncertainty range. These experimental validation results show that the developed MLP for virtual experiments is reliable and rapid because the prediction of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ink formulations is within one minute. This rapid and efficient virtual experimentation process can significantly facilitate the exploration of design space for identification of ink formulations that lead to materials with desired properties, thus accelerating the development of new materials.
000 ink formulations is within one minute. This rapid and efficient virtual experimentation process can significantly facilitate the exploration of design space for identification of ink formulations that lead to materials with desired properties, thus accelerating the development of new materials.
3. Conclusions
In this study, a PIML model was developed for virtual experimentation to accelerate the discovery of 3D printed thermoplastics. The collected 216 S–S curves from 62 ink formulations were dimensionally reduced into eight PCs. Meanwhile, 13 physics-informed descriptors were included using domain knowledge to increase the robustness and generalization of the model. The developed physics informed MLP model achieved superior R2 and RMSE values when predicting the values of the eight PCs. The reconstructed S–S curves from the predicted PCs matched well with the true ones. Feature importance analysis confirmed the importance of physics-informed descriptors, showing that the total energy is the most important one. After mapping the mechanical properties of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ink formulations by the MLP model, six representative ink formulations that are expected to lead to three different types of thermoplastics were chosen. Validation experiments demonstrated a strong agreement between the predicted and experimental S–S curves. The methodologies and workflow can be readily extended to other materials for predicting other performance curves such as Raman and electrochemistry curves. This underscores the versatility and potential of this approach in a range of materials science and chemical research scenarios, offering a robust framework for expedited and accurate material and chemical analyses.
000 ink formulations by the MLP model, six representative ink formulations that are expected to lead to three different types of thermoplastics were chosen. Validation experiments demonstrated a strong agreement between the predicted and experimental S–S curves. The methodologies and workflow can be readily extended to other materials for predicting other performance curves such as Raman and electrochemistry curves. This underscores the versatility and potential of this approach in a range of materials science and chemical research scenarios, offering a robust framework for expedited and accurate material and chemical analyses.
4. Materials and methods
4.1. Materials
2-Hydroxy-3-phenoxypropyl acrylate (HA), isooctyl acrylate (IA, >90%), and acrylic acid (AA, 98%) were purchased from Sigma Aldrich (St. Louis, MO, US). Diphenyl(2,4,6-trimethylbenzoyl) phosphine oxide (TPO, >97%), isobornyl acrylate (IBOA, >90%), N-vinylpyrrolidone (NVP, >99%), and N-(2-hydroxyethyl) acrylamide (HEAA, >98%) were purchased from Fisher Scientific (Pittsburgh, PA, US).4.2. 3D printing and mechanical testing
In this study, the LCD 3D printing process was executed using a resin mixture comprising six monomers: HA, IA, AA, IBOA, NVP, and HEAA with carefully measured weight ratios. Each monomer's ratio in the mixture can vary continuously from 0 to 1. For the sake of experimental precision, the ratios have two decimal places. The total sum of the ratios for all monomers equals 1. To make the mixture, a photoinitiator, diphenyl(2,4,6-trimethylbenzoyl) phosphine oxide (TPO), was added at a concentration of 2 wt%. The mixture was then subjected to magnetic stirring for one minute to ensure thorough and uniform mixing. The resulting homogenized resin was used in an Anycubic Photon Mono 4K printer, operating at a 405 nm irradiation wavelength. The printing parameters included a power density of about 5 mW cm−2, a layer thickness of 50 μm, and an exposure time of 15 seconds per layer. Following the printing process, the samples were further cured under 405-nm UV light for 60 seconds. For the mechanical assessment of the 3D-printed samples, tensile testing was carried out using a Mark-10 universal testing machine at a loading rate of 50 mm min−1. To ensure a comprehensive statistical analysis, a minimum of 5 samples were printed and tested for each monomer ratio.4.3. S–S curve collection
326 S–S curves were collected from 62 distinct formulations, each of which was subjected to 5–7 independent mechanical tensile tests. To ensure reliability and quality, the S–S curves with significant errors such as measurement inconsistencies, premature breakage, or excessive mechanical testing noise were excluded. Consequently, a refined dataset comprising 216 S–S curves was obtained, with each thermoplastic represented by 2–4 individual curves. To demonstrate the diversity and balance of the dataset, when considering a maximum strain of 10% as the threshold, the data showed a distribution where approximately half of the materials displayed brittle properties (106 samples), while the other half exhibited higher ductility (80 samples).4.4. Data processing of S–S curves
The preliminary cleaning of the raw data from the tensile testing machine involves trimming the initial segments of each S–S curve to eliminate any measurements taken before the machine commenced operation by standardizing the starting points to a baseline of zero stress and zero strain (0,0). Then, a critical aspect of the preprocessing involves identifying the point of failure within each sample's S–S curve. By pinpointing and marking the exact location of sample failure on each curve, the final data point represents the moment of fracture by capturing the complete mechanical profile of each specimen. The last step in the data preprocessing routine is to apply an interpolation technique to standardize the data representation. Each S–S curve is interpolated to consist of 50 data points uniformly distributed in the x-axis (strain).4.5. Experimental uncertainty
In this study, the e_value is calculated based on normal distribution to capture the inherent uncertainties in the S–S data at the fracture point. This refinement involves analyzing the final strain values at fracture for each dataset as illustrated in Fig. 2a. By aggregating these values, a comprehensive picture of the strain behavior at fracture across various samples was obtained. To encapsulate the variability in the fracture strains of the materials, first, their means are calculated, providing a reference for the average material behavior under stress. Then the standard deviation is computed to quantify the dispersion among these values, a crucial step in highlighting the heterogeneity in material responses. This approach normalizes each fracture strain relative to this mean, adjusting for variance. This process results in the e_values, the standard deviations indicating the deviation of each sample's fracture point from the average, Mathematically, this normalization is expressed as:Where, μ is the mean of the fracture strain for all samples, n is the number of samples (S–S curves) and xi is the fracture strain value for each sample.
where, σ is the standard deviation, (xi − μ) represents the deviation of each sample's fracture strain value.
4.6. Uncovering features' importance
Due to the inherent complexity and ‘black box’ characteristic of the MLP model, we utilized the integrated gradients (IG) method for the interpretability study.40 This approach is particularly adept at illuminating the contribution of each input feature to the model's output. It works by calculating the gradient of the model's prediction with respect to each input feature. It then integrates these gradients along a path from a baseline input (a zero vector) to the actual input. This process effectively captures the importance of each feature in the model's prediction, highlighting both linear and non-linear relationships within the model. To do that, the analysis was expanded to include the entire dataset (both training and testing datasets) to ensure a comprehensive assessment of the feature importance. The IG method, applied to each data point, calculated the significance of every feature in relation to the model's predictions, thereby providing a quantitative measure of each feature's contribution. This process involved aggregating importance scores across all samples to derive an average importance for each feature. Additionally, focused analysis was conducted on cross-features: where Physics-Informed descriptors interact with monomer ratios. For each PI descriptor, the average importance across all its interactions was calculated, allowing for an assessment of the overall influence of each PI descriptor on the model's predictions.4.7. Virtual experiment ratio generation details
In the generation of random experiment formulations within our study, we employed the Dirichlet distribution. This distribution is commonly utilized for generating random proportions under specific constraints, like that the sum of the monomer ratios equals 1, making it particularly suitable for simulating a diverse range of monomer mixtures.45 Additionally, an important characteristic of the Dirichlet distribution is its uniformity and symmetry, when the parameters of the distribution, known as ‘alpha’, are all set equal to 1. This equal setting means that each component of the distribution has an equal chance of being sampled, leading to an evenly spread of probabilities across all ratios. For each generated combination, the first five ratios were rounded to two decimal places. The sixth ratio in each combination was then determined by subtracting the sum of these first five rounded ratios from one.Author contributions
Z. C. designed and implemented data preprocessing, model training and testing, and data analysis. Y. W. conducted all the experiments and analyzed the model interpretability aspects. K. S. used DFT calculations for physics-informed descriptor data collection. Y. X. contributed to the feature-important analysis of the model. J. L. conceived the idea, managed the research progress, and provided regular guidance. Z. C., Y. W. and Y. X. drafted the first manuscript, which was thoroughly revised by J. L. All authors commented and agreed on the final version of the manuscript.Data availability
The code supporting the findings of this study is available on GitHub at https://github.com/linresearchgroup/VirtualEXP_3Dprinting. The repository also includes original experimental datasets used in this study.Conflicts of interest
The authors declare no competing interests.Acknowledgements
J. L. thanks the financial support from the National Science Foundation (award number: 2154428) and the U.S. Army Corps of Engineers, ERDC (grant number: W912HZ-21-2-0050).References
- R. Gómez-Bombarelli, et al., Design of efficient molecular organic light-emitting diodes by a high-throughput virtual screening and experimental approach, Nat. Mater., 2016, 15, 1120–1127 CrossRef PubMed.
- P. K. Penumakala, J. Santo and A. Thomas, A critical review on the fused deposition modeling of thermoplastic polymer composites, Composites, Part B, 2020, 201, 108336 CrossRef CAS.
- P. Awasthi and S. S. Banerjee, Fused deposition modeling of thermoplastic elastomeric materials: Challenges and opportunities, Addit. Manuf., 2021, 46, 102177 CAS.
- Y. Wu, et al., A photocured Bio-based shape memory thermoplastics for reversible wet adhesion, Chem. Eng. J., 2023, 470, 144226 CrossRef CAS PubMed.
- K. Sattari, et al., Physics-constrained multi-objective bayesian optimization to accelerate 3d printing of thermoplastics, Addit. Manuf., 2024, 86, 104204 CAS.
- B. Cox and Q. Yang, In Quest of Virtual Tests for Structural Composites, Science, 2006, 314, 1102–1107, DOI:10.1126/science.1131624.
- B. Zhang, Z. Yang, X. Sun and Z. Tang, A virtual experimental approach to estimate composite mechanical properties: Modeling with an explicit finite element method, Comput. Mater. Sci., 2010, 49, 645–651 CrossRef CAS.
- N. Zhang, B. Lu, W. Wang and J. Li, Virtual experimentation through 3D full-loop simulation of a circulating fluidized bed, Particuology, 2008, 6, 529–539, DOI:10.1016/j.partic.2008.07.013.
- Y. L. Xue, J. Huang, C. H. Lau, B. Cao and P. Li, Tailoring the molecular structure of crosslinked polymers for pervaporation desalination, Nat. Commun., 2020, 11, 1461 CrossRef CAS PubMed.
- Y. Xie, K. Sattari, C. Zhang and J. Lin, Toward autonomous laboratories: Convergence of artificial intelligence and experimental automation, Prog. Mater. Sci., 2023, 132, 101043 CrossRef.
- P. Raccuglia, et al., Machine-learning-assisted materials discovery using failed experiments, Nature, 2016, 533, 73–76, DOI:10.1038/nature17439.
- Y. Xie, et al., Machine learning assisted synthesis of metal–organic nanocapsules, J. Am. Chem. Soc., 2019, 142, 1475–1481 CrossRef PubMed.
- Y. Dong, et al., Bandgap prediction by deep learning in configurationally hybridized graphene and boron nitride, npj Comput. Mater., 2019, 5, 26, DOI:10.1038/s41524-019-0165-4.
- Z. Rao, et al., Machine learning–enabled high-entropy alloy discovery, Science, 2022, 378, 78–85, DOI:10.1126/science.abo4940.
- B. A. Koscher, et al., Autonomous, multiproperty-driven molecular discovery: From predictions to measurements and back, Science, 2023, 382, eadi1407, DOI:10.1126/science.adi1407.
- B. Mikulak-Klucznik, et al., Computational planning of the synthesis of complex natural products, Nature, 2020, 588, 83–88, DOI:10.1038/s41586-020-2855-y.
- N. I. Rinehart, et al., A machine-learning tool to predict substrate-adaptive conditions for Pd-catalyzed C–N couplings, Science, 2023, 381, 965–972, DOI:10.1126/science.adg2114.
- C. W. Coley, et al., A robotic platform for flow synthesis of organic compounds informed by AI planning, Science, 2019, 365, eaax1566, DOI:10.1126/science.aax1566.
- J.-P. Correa-Baena, et al., Accelerating materials development via automation, machine learning, and high-performance computing, Joule, 2018, 2, 1410–1420 CrossRef CAS.
- Z. Ren, et al., Machine learning–aided real-time detection of keyhole pore generation in laser powder bed fusion, Science, 2023, 379, 89–94, DOI:10.1126/science.add4667.
- Z. Chen, et al., An interpretable and transferrable vision transformer model for rapid materials spectra classification, Digital Discovery, 2024, 3, 369–380, 10.1039/D3DD00198A.
- C. Yang, Y. Kim, S. Ryu and G. X. Gu, Prediction of composite microstructure stress–strain curves using convolutional neural networks, Mater. Des., 2020, 189, 108509 CrossRef.
- M.-L. Tsai, C.-W. Huang and S.-W. Chang, Theory-inspired machine learning for stress–strain curve prediction of short fiber-reinforced composites with unseen design space, Extreme Mech. Lett., 2023, 65, 102097 CrossRef.
- C. S. Ha, et al., Rapid inverse design of metamaterials based on prescribed mechanical behavior through machine learning, Nat. Commun., 2023, 14, 5765 CrossRef CAS PubMed.
- G. E. Karniadakis, et al., Physics-informed machine learning, Nat. Rev. Phys., 2021, 3, 422–440 CrossRef.
- K. Sattari, et al., A scientific machine learning framework to understand flash graphene synthesis, Digital Discovery, 2023, 2, 1209–1218 RSC.
- K. Sattari, et al., De novo molecule design towards biased properties via a deep generative framework and iterative transfer learning, Digital Discovery, 2024, 3, 410–421 RSC.
- Y. Du, T. Mukherjee and T. DebRoy, Physics-informed machine learning and mechanistic modeling of additive manufacturing to reduce defects, Appl. Mater. Today, 2021, 24, 101123 CrossRef.
- K. C. Chin, J. Cui, R. M. O’Dea, T. H. Epps III and A. J. Boydston, Vat 3D printing of bioderivable photoresins–toward sustainable and robust thermoplastic parts, ACS Sustainable Chem. Eng., 2023, 11, 1867–1874 CrossRef CAS.
- S. Kim, et al. , Nucleic Acids Res., 2023, 51, D1373–D1380 CrossRef PubMed.
- S. H. Bertz, The first general index of molecular complexity, J. Am. Chem. Soc., 1981, 103, 3599–3601 CrossRef CAS.
- N. M. O'Boyle, et al., Open Babel: An open chemical toolbox, J. Cheminf., 2011, 3, 1–14 Search PubMed.
- H. Liu, F.-Y. Wu, G.-J. Zhong and Z.-M. Li, Predicting the complex stress-strain curves of polymeric solids by classification-embedded dual neural network, Mater. Des., 2023, 227, 111773 CrossRef CAS.
- A. Kościuszko, D. Marciniak and D. Sykutera, Post-processing time dependence of shrinkage and mechanical properties of injection-molded polypropylene, Materials, 2020, 14, 22 CrossRef PubMed.
- T. Poggio, H. Mhaskar, L. Rosasco, B. Miranda and Q. Liao, Why and when can deep-but not shallow-networks avoid the curse of dimensionality: a review, Int. J. Autom. Comput., 2017, 14, 503–519 CrossRef.
- S. Wold, K. Esbensen and P. Geladi, Principal component analysis, Chemom. Intell. Lab. Syst., 1987, 2, 37–52 CrossRef CAS.
- T. Chai and R. R. Draxler, Root mean square error (RMSE) or mean absolute error (MAE)? –Arguments against avoiding RMSE in the literature, Geosci. Model Dev., 2014, 7, 1247–1250 CrossRef.
- P. Baldi and P. J. Sadowski, Understanding dropout, Advances in neural information processing systems, 2013, vol. 26 Search PubMed.
- R. Tibshirani, Regression shrinkage and selection via the lasso, J. R. Stat. Soc., Ser. B, Methodol., 1996, 58, 267–288 CrossRef.
- Z. Qi, S. Khorram and F. Li, CVPR Workshops, 2019, vol. 2, pp. 1–4 Search PubMed.
- Y. Wu, et al., Photocuring three-dimensional printing of thermoplastic polymers enabled by hydrogen bonds, ACS Appl. Mater. Interfaces, 2021, 13, 22946–22954, DOI:10.1021/acsami.1c02513.
- J. A. Pugar, C. M. Childs, C. Huang, K. W. Haider and N. R. Washburn, Elucidating the physicochemical basis of the glass transition temperature in linear polyurethane elastomers with machine learning, J. Phys. Chem. B, 2020, 124, 9722–9733 CrossRef CAS PubMed.
- D. W. T. N. Van Krevelen, Properties of polymers, Cohesive Properties and Solubility, Elsevier, 4th edn, ch. 7, 2009, pp. 189–227 Search PubMed.
- Y. Wu, et al., H-bonds and metal-ligand coordination-enabled manufacture of palm oil-based thermoplastic elastomers by photocuring 3D printing, Addit. Manuf., 2021, 47, 102268, DOI:10.1016/j.addma.2021.102268.
- A. H. Briggs, A. Ades and M. J. Price, Probabilistic sensitivity analysis for decision trees with multiple branches: use of the Dirichlet distribution in a Bayesian framework, Med. Decis. Making, 2003, 23, 341–350 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4mh01022a |
| This journal is © The Royal Society of Chemistry 2024 |