DNA tetrahedral nanocages as a promising nanocarrier for dopamine delivery in neurological disorders†
Ramesh
Singh‡§
 a,
Krupa
Kansara‡
a,
Pankaj
Yadav‡
a,
Sandip
Mandal
a,
Krupa
Kansara‡
a,
Pankaj
Yadav‡
a,
Sandip
Mandal
 b,
Ritu
Varshney¶
b,
Ritu
Varshney¶
 a,
Sharad
Gupta
a,
Sharad
Gupta
 a,
Ashutosh
Kumar
a,
Ashutosh
Kumar
 c,
Prabal K.
Maiti
c,
Prabal K.
Maiti
 b and
Dhiraj
Bhatia
b and
Dhiraj
Bhatia
 *a
*a
aDepartment of Biological Sciences and Engineering, Indian Institute of Technology, Gandhinagar, Gujrat, India. E-mail: dhiraj.bhatia@iitgn.ac.in
bDepartment of Physics, Indian Institute of Science, Bangalore, India
cBiological and Life Sciences, School of Arts and Sciences, Ahmedabad University, India
First published on 25th July 2024
Abstract
Dopamine is a neurotransmitter in the central nervous system that is essential for many bodily and mental processes, and a lack of it can cause Parkinson's disease. DNA tetrahedral (TD) nanocages are promising in bio-nanotechnology, especially as a nanocarrier. TD is highly programmable, biocompatible, and capable of cell differentiation and proliferation. It also has tissue and blood–brain barrier permeability, making it a powerful tool that could overcome potential barriers in treating neurological disorders. In this study, we used DNA TD as a carrier for dopamine to cells and zebrafish embryos. We investigated the mechanism of complexation between TD and dopamine hydrochloride using gel electrophoresis, fluorescence and circular dichroism (CD) spectroscopy, atomic force microscopy (AFM), and molecular dynamic (MD) simulation tools. Further, we demonstrate that these dopamine-loaded DNA TD nanostructures enhanced cellular uptake and differentiation ability in SH-SY5Y neuroblastoma cells. Furthermore, we extended the study to zebrafish embryos as a model organism to examine survival and uptake. The research provides valuable insights into the complexation mechanism and cellular uptake of dopamine-loaded DNA tetrahedral nanostructures, paving the way for further advancements in nanomedicine for Parkinson's disease and other neurological disorders.
Introduction
Neurotransmitters are endogenous chemical messengers that play a crucial role in the complex network of neurons in the body. They facilitate neurons to communicate and transmit signals from neurons to targeted cells.1,2 An imbalance in the physiological concentration of specific neurotransmitters causes neurological disorders such as Parkinson's disease (PD) and Alzheimer's disease. Dopamine is a critical neurotransmitter in the central nervous system that is essential for many bodily and mental processes.2–4 Dopamine is involved in a delicate balance between other neurotransmitters to facilitate coordination between millions of nerves and muscle cell movement. A decrease in dopamine levels can cause Parkinson's disease.3,5 Currently, L-DOPA (a dopamine precursor) is mainly used to treat Parkinson's disease at an initial stage, but at the advanced level, with the loss of dopa decarboxylase enzyme, this drug ultimately loses its therapeutic effect.6,7 Therefore, researchers are exploring dopamine-loaded nanoparticles to treat Parkinson's disease, as direct dopamine delivery is required but cannot cross the blood–brain barrier on its own.8,9 Nanomaterials such as gold nanoparticles, liposomes, quantum dots, and other polymeric drug delivery systems have been used for loading and delivering dopamine.7–9 However, concerns have arisen regarding the neurotoxicity triggered by these nanoparticles. Many nano-delivery methods suffer from neuroglial toxicity, undesired interactions, intra-brain accumulation, and reactive oxidative stress.10DNA nanocages are an advanced nanocarrier category with excellent inherent biocompatibility and non-cytotoxicity. The Watson–Crick base pairing principle makes these structures highly programmable, allowing for precise control of shape, size, and versatile functionality.11–14 Recent research has demonstrated the wide use of DNA nanostructures as drug delivery systems for brain disease because of their ability to pass the blood–brain barrier (BBB).15–20 These properties indicate that DNA nanocages can potentially be used as drug-delivery systems for neurodegenerative diseases and be more suitable than conventional nanoscale drug carriers.12,15–17 DNA tetrahedral (TD) nanocages formed by self-assembling four synthetic ssDNA oligonucleotides exhibit remarkable mechanical stiffness, nontoxicity, and resistance to nuclease degradation.21,22 DNA tetrahedron provides enough space for small molecules as a nanocarrier and is a promising candidate in drug delivery and bioimaging platforms.23,24 It has been utilized as a nanocarrier for therapeutic drugs/small molecules15 such as doxorubicin,25 methylene blue,26 curcumin,27 actinomycin D,28 ampicillin,29 vitamin B12 (VB12),30 and nucleic acid drug CpG.31,32 The DNA TD nanocages showed excellent cellular uptake rate among other shaped nanocages.21,33
Recent research has demonstrated the wide use of DNA tetrahedrons as drug-delivery vehicles to treat brain tumors and neurological disorders.15,18,24,34 DNA TD with and without Angio-peptide-2 penetrated the BBB and were used in brain tumor imaging agents.24 D Y Tam et al. demonstrated that DNA nanostructure with and without BBB-penetrating peptide can cross the BBB in brain cancer treatment.35 They reported that ligand modification is not essential for DNA TD nanocages; it could still cross the BBB by endocytosis in both in vitro and in vivo models.35 W Cui et al. deliver vitamin B12-loaded DNA TD across BBB, which inhibits LRRK2 protein kinase and restores autophagy for treating Parkinson's disease.30 In addition to the excellent drug delivery ability,23,36,37 DNA TD has various neurotherapeutic efficiencies, such as neuroprotective and neurotherapeutic effects,38 neural stem cell proliferation, and neuronal differentiation ability.39 Many physical, chemical, and computational studies have reported that dopamine can bind to a double helical linear DNA molecule.40–42 Therefore, based on the above literature reports, the properties of DNA TD nanocages as blood–brain barrier crossing nanoparticles15,35 and their excellent drug delivery efficacy23 and neurotherapeutic efficiencies,15,38 we hypothesize that DNA TD can be a viable delivery system for effective delivery of dopamine. The dopamine-loaded DNA TD can be potential therapeutics in treating Parkinson's disease.15 As the protonated dopamine is an active form of the neuronal dopamine transporter, we used dopamine hydrochloride to make the TD-dopamine complex.43 The protonated form of dopamine can also enhance DNA binding through electrostatic interaction with a negatively charged phosphate backbone and intrudes into the dsDNA.
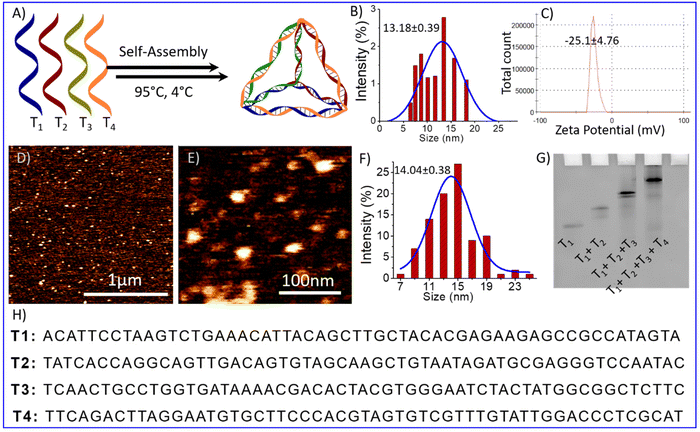
The self-assembled DNA tetrahedrons (TD) were synthesized using previously reported protocols.21,22 The synthesis involved mixing four complementary ss-oligonucleotides of 55 bases, T1, T2, T3, and T4, in equimolar concentration with 2 mM magnesium chloride. Thermal annealing was performed to self-assemble nucleotide into tetrahedron. It is performed by following these steps: starting temperature of 95 °C, decreasing the temperature by 5 °C, holding for 15 minutes, and repeating the process until reaching a temperature of 4 °C (Fig. 1A). The self-assembly of these four oligos into TDs was confirmed by the electrophoretic mobility shift assay, which showed the retardation in the migration of the self-assembled TDs (T1 + T2 + T3 + T4) compared to other lower combinations or single oligo (Fig. 1G). The particle size distribution of the self-assembled structures was observed using dynamic light scattering (DLS), which revealed an average hydrodynamic diameter of self-assembled structures is 13.18 ± 0.39 nm (Fig. 1B), similar to the estimated size. Then, atomic force microscopy (AFM) was used to perform a topographic analysis of nanostructures. AFM imaging showed the formation of homogenously distributed self-assembled tetrahedral DNA nanostructures (Fig. 1D and E). The histogram of particle size measurement of AFM images and subsequent Gaussian curve fit revealed that the average nanostructure size is 14.04 ± 0.38 nm (Fig. 1F). All the above characterization results obtained agree well with each other and confirm the formation of tetrahedral nanostructures.
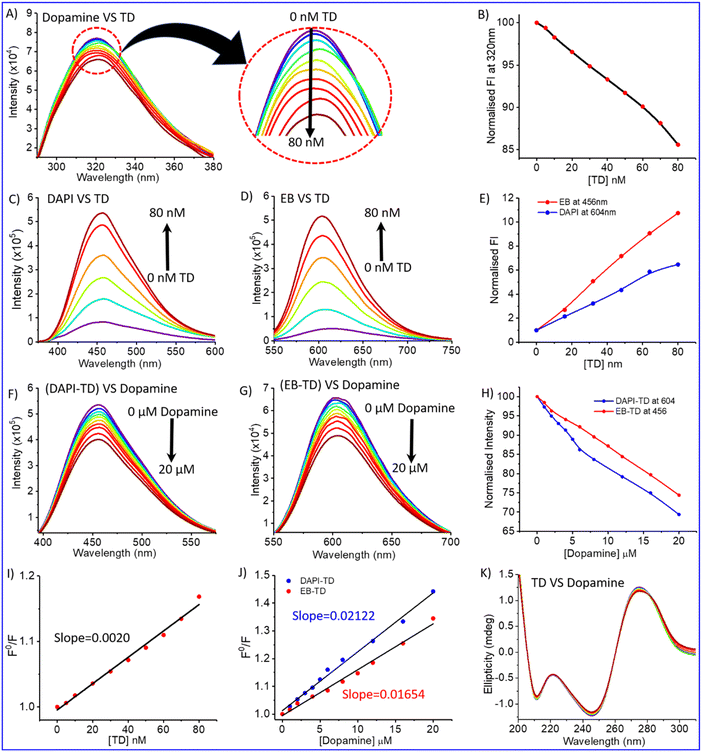
DNA molecules can bind to various ligands or small molecules through different binding modes, including electrostatic interactions, major or minor groove space binding, and intercalation into the base pairs.44 Fluorescence spectroscopy is a technique used to study binding interactions without altering or destroying them.44,45 To investigate the binding interaction of dopamine hydrochloride with tetrahedral DNA nanostructure, a fluorescence titration experiment of dopamine hydrochloride (20 μM) with the gradual addition of DNA tetrahedron (up to 80 nM) was performed. Dopamine exhibits intrinsic fluorescence emission at around 320 nm on excitation at 280 nm wavelength (Fig. 2A). Fluorescence spectra showed a quenching in fluorescence intensity on the gradual increase of DNA-TDs (Fig. 2B). The quenching in fluorescence intensity indicates the interaction between dopamine and TD. The Stern–Volmer constant, KSV (=2.01 × 106 M−1), has been determined from the slope of the plot of F0/F versus [TD] (Fig. 2I).44,46 The high value of the quenching constant indicates the quenching is static due to the complex formation between DNA-TD and dopamine.47
Previous research proved that dopamine binds DNA in multi-mode binding interactions via electrostatic interaction and major or minor groove binding.42 To get more clear information about the binding mode between dopamine chloride and tetrahedral DNA nanostructures, fluorescence quenching experiments were carried out by displacement of DNA binding dyes DAPI and ethidium bromide (EB) from DAPI-TD/EB-TD complex.48,49 DAPI is known to bind DNA at the groove region; however, ethidium bromide is an intercalating dye.48,50Fig. 2C and D represent the fluoresce spectra of DAPI and ethidium bromide dyes bound with DNA tetrahedron. Only dye (DAPI/EB) has a minimum fluorescence intensity, but the addition of TD forms a complex with the dye, which increases in intensity (Fig. 2C–E). A titration of DAPI-TD and EB-TD with the gradual addition of dopamine decreases the fluorescence intensity of the DAPI-TD complex at 604 nm wavelength (Fig. 2F) and the EB-TD complex at 456 nm (Fig. 2G). This quenching of fluorescence indicates the displacement of dye (DAPI/EB) by dopamine and the binding with DNA with their respective sites (Fig. 2H). The displacement of DAPI suggests that dopamine binds the minor groove side of ds-DNA edges of tetrahedron nanostructure, and displacement of EB indicates dopamine as an intercalator. The Stern Volmer plot represents that the quenching constant for the displacement of DAPI (K = 2.122 × 104 M−1) is much greater than that of the displacement of EB (1.645 × 104 M−1) (Fig. 2J). That means protonated dopamine binds the groove sides more effectively than stacked regions. The quenching constant obtained for dye displacement for DAPI and EB by dopamine is smaller than that of dopamine vs. TD titration. That means dopamine also interacts with DNA TD through another mode. It may be electrostatic interaction between a negatively charged phosphate backbone and positively charged dopamine.
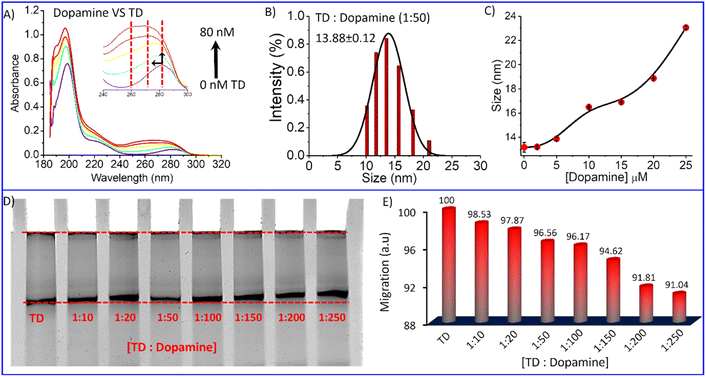
Circular dichroism (CD) is a spectroscopy tool that is used to investigate conformational changes in bio-molecules such as proteins, peptides, and DNA.51–53 It is an absorption spectroscopy method based on the differential absorption of left and right circularly polarized light. Therefore, we used it to investigate TD's conformational change in complexation with dopamine. CD spectra of tetrahedral DNA nanostructures in the UV region from 190 to 300 nm showed characteristic bands of double-strand DNA, including a positive band at 275 nm and two negative bands at 245 nm and 211 nm (Fig. 2K).51 The CD spectra remain almost similar on the addition of increasing concentrations of dopamine (up to 50 μM) into the DNA tetrahedron solution (200 nM). The lack of change in the CD spectra indicates that the complexation of dopamine with TD does not destroy the helical confirmation or complementary base pairing. The fluorescence and CD spectral analysis suggest multimode binding of dopamine with TD into groove sites, intercalation, and electrostatic interaction. Further, on UV-Vis titration, spectra of dopamine with TD showed that on increasing TD, the absorption maximum for dopamine shifted towards a higher wavelength with an increase in intensity (Fig. 3A). These results indicated the complexation between dopamine and DNA TD.54
Polyacrylamide gel electrophoresis is a widely used technique in biochemistry to analyze ligand interaction with proteins/DNA molecules. We performed an electrophoretic titration at constant Tetrahedral DNA against increasing amounts of dopamine hydrochloride to get more insight into the complexation between DNA-TD and dopamine hydrochloride. Separate solutions of a mixture of tetrahedron with 10 eq., 20 eq., 50 eq., 100 eq., 200 eq., and 250 eq. of dopamine hydrochloride, respectively, incubated for about 30 minutes and then performed electrophoresis experiment. Ethidium bromide staining visualized that the nanostructure migration distance decreases with increasing dopamine hydrochloride concentrations in the presence of dopamine (Fig. 3D). Quantifying migration distance by these nanostructures to bare tetrahedron (100%) showed the retention of migration on an increasing amount of dopamine (Fig. 3E). In the presence of 250 equivalent of dopamine, it is reduced to around 91% compared to TD alone. The retention in migration distance indicates an increase in the mass of the nanoparticles, which means dopamine gets attached to the DNA tetrahedron, and a complex has been formed. Further, the DLS analysis of the TD-dopamine complex in different ratios also proved an increase in the hydrodynamic size of tetrahedral nanostructures (Fig. S1†). The hydrodynamic size of TD in the presence of 10 eq., 20 eq., 50 eq., 100 eq., and 200 eq. dopamine hydrochloride 13.21 nm, 13.88 nm, 16.5 nm, 16.91 nm, 18.87 nm, 23.06 nm were obtained, respectively (Fig. 3C). The size increase also supports the formation of a TD-dopamine complex or successful full loading of dopamine on TD.
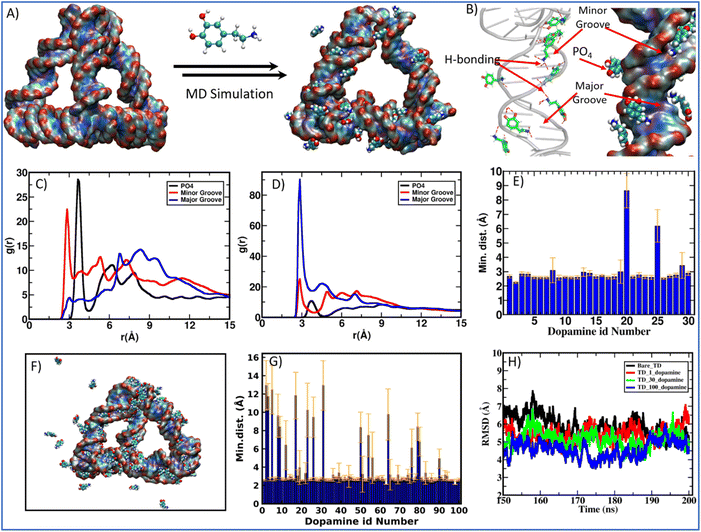
Molecular dynamics (MD) simulation is a sophisticated computational method that can be utilized to gain insights into the connections between the structure and function of macromolecules. By studying the time-dependent behavior of microscopic systems, MD simulations can provide valuable information about the binding affinities of ligands to DNA and specific ways in which ligands bind to DNA.55,56 After observing the complexation of DNA and dopamine hydrochloride using biophysical and chemical techniques, we performed the MD simulations of DNA tetrahedron for 200 ns in the presence of protonated dopamine at physiological conditions. The initial structure of the TD was built using the polygen, and the dopamine structure corresponding to the PubChem CID: 681, was extracted from the PubChem database.57 Then, the geometry optimization of the dopamine molecule was carried out using the B3LYP functional and a 6-311G* basis set with the Gaussian 09 program. The built structures were then solvated in a cubic box with a buffer length of 15 Å using the TIP3P water model58 with xLEAP module of AMBER20.59 MD simulations are carried out for three systems: first TD in the presence of only one dopamine, second in the presence of 30 dopamine and 100 dopamine molecules (Fig. 4A, F and Fig. S2†). The simulation of TD with single dopamine reveals that the dopamine molecule binds to the minor groove site of DNA-TD and remains there throughout the 200 ns long simulation. Our simulations with more dopamine (30 and 100) suggest that dopamine can bind both at the minor and major groove of TD and at the phosphate backbone of DNA-TD (Fig. 4B). The histogram of the minimum distance of dopamine from DNA-TD shows that 28 out of 30 dopamine molecules bind the TD (Fig. 4E). Further, the radial distribution function (RDF) of the hydroxyl group and amine group of dopamine and the DNA-TD suggests that protonated dopamine interacts with minor grooves through hydroxyl and NH3+ groups via hydrogen bonding; in major grooves, it binds mainly through NH3+ groups (Fig. 4C and D). In the case of 100 dopamine molecules, we also found the multi-mode binding of dopamine molecules over the DNA tetrahedron. The histogram of the minimum distance of dopamine from tetrahedral nanostructures shows that 82 molecules bond to the DNA tetrahedron, and 18 are far from it (Fig. 4F and G).
The instantaneous snapshot of the TD-dopamine mixture at the end of 200 ns long MD simulation showed that the helical structure of ds-DNA is not affected by binding the dopamine molecules, as confirmed by our spectroscopic observation. For closer insight into the effect of dopamine on TD structure, we calculate the root mean squire deviation (RMSD) for the entire simulation period. RMSD as a function of simulation time showed that in the presence of 100 protonated dopamine molecules, RMSD is slightly lower than that of bare TD alone (Fig. 4H). So, in the presence of dopamine molecules, the DNA TD structure is slightly more stable compared to bare TD. Otherwise, the DNA-TD structure maintains its canonical form both in the absence and presence of dopamine molecules. We also calculate the degree of hydrogen bonding (base pairing) responsible for the self-assembly of oligos into the tetrahedral nanostructure. The number of hydrogen bonds for bare TD is 246.8 ± 1.2, while in the presence of 100 dopamine, it increases to 253.7 ± 2.1. The increase in hydrogen bonding further supports the stabilization of DNA TD in the presence of dopamine molecules. In the groove region of TD, dopamine forms a multimodal hydrogen bond with base pairs. This suggests that there may be an interaction between the dopamine benzene ring and the nucleobases (A, T, G, and C). Since the DNA bases (cytosine and thymine) and the dopamine benzene ring face each other in both hydrogen bonding scenarios, there is a chance of π–π interaction between them. To better understand how dopamine interacts with TD, we have looked at the different types of hydrogen bonding between dopamine and bases in the grooves of TD (Fig S3†). Furthermore, we found that dopamine primarily interacts with thymine (T) of TD in the minor and major grooves. Strong π–π interaction and groove binding are indicated by the most prominent hydrogen bonding between the minor groove's thymine and dopamine benzene ring (Fig. S10†). The hydrogen bond between Watson–Crick base pairs is strengthened by this interaction, stabilizing TD without altering the molecule's structure or conformation. An aromatic planner molecule is inserted between stacked bases in TD during intercalation, which disrupts the base-pair stacking pattern; therefore, this groove binding interaction is not quite an intercalation interaction.
A dopamine loading and release experiment have for the quantification of dopamine loading capacity of TD with different amount of dopamine. A 80 nM DNA TD were incubated with 0.4, 0.8, and 1.6 μM dopamine, resulting in TD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dopamine ratios of 1
dopamine ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, 1
50, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, and 1
100, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200. After overnight incubation, samples were centrifuged to separate bound and unbound dopamine. Absorbance measurements at 280 nm were taken for unbound dopamine and compared to control samples. In case of 1
200. After overnight incubation, samples were centrifuged to separate bound and unbound dopamine. Absorbance measurements at 280 nm were taken for unbound dopamine and compared to control samples. In case of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (TD
50 (TD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dopamine) 89% of dopamine get binded with TD that is 44.5 nM dopamine per nM of TD. In case of 1
dopamine) 89% of dopamine get binded with TD that is 44.5 nM dopamine per nM of TD. In case of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (TD
100 (TD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dopamine) 72% binding efficiency were obtained that means per nM of TD can load around 72 nM of dopamine. These result quite similar to MD simulations results. Further increase of dopamine that in case of 1
dopamine) 72% binding efficiency were obtained that means per nM of TD can load around 72 nM of dopamine. These result quite similar to MD simulations results. Further increase of dopamine that in case of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 (TD
200 (TD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dopamine) 61% dopamine can binds the TD (Fig. S4†).
dopamine) 61% dopamine can binds the TD (Fig. S4†).
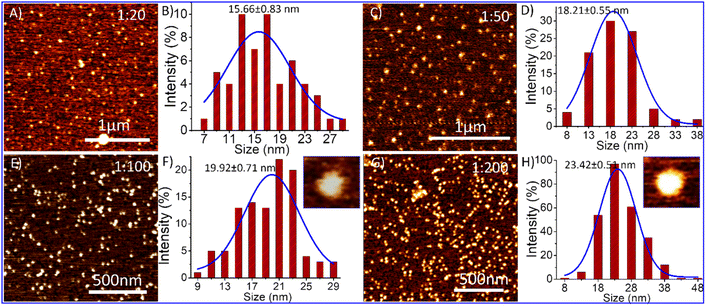
Considering the above-mentioned analysis, we investigate TD's morphological change in the presence of dopamine using atomic force microscopy (AFM). We added dopamine hydrochloride in the solution of DNA-TD in different ratios separately and incubated them for 2 h before AFM imaging. AFM imaging of the TD-dopamine complex revealed that the diameter size of nanostructures increases from 13 nm to 23 nm with increasing dopamine concentration (Fig. 5). The AFM image of DNA tetrahedral nanostructures in the presence of 20 eq. of dopamine showed a slight increase in size; however, no significant change was observed in morphology (Fig. 5A). A particle size distribution was done with the help of ImageJ followed by Gaussian fitting in origin pro, revealing the diameter range from 7 to 29 nm with an average size of 15.66 ± 0.83 nm (Fig. 5B). In the presence of 50 eq., dopamine also showed similar nanoparticles of the size ranging from 8–38 nm with an average diameter of 18.21 ± 0.55 nm (Fig. 5C and D). The morphology of the TD-dopamine complex, similar to DNA-TD nanostructures, suggests that most of the dopamine molecules bind with the ds-DNA edge of the tetrahedron without disturbing the original conformation of TD. These AFM results are in agreement with the results obtained from spectroscopy and simulation analysis. With further increases in dopamine concentrations, TD-dopamine morphology seems to transform into spherical or distorted sphere-like structures (Fig. 5E). The average particle size of the TD-dopamine complex (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) was around 19.92 ± 0.71 nm (Fig. 5F). In the case of the TD-dopamine complex in a 1
100) was around 19.92 ± 0.71 nm (Fig. 5F). In the case of the TD-dopamine complex in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 ratio, the atomic force microscopy image revealed the formation of homogeneously distributed sphere-like structures (Fig. 5G). The particle size distribution of the AFM image revealed an increase in the average size of the nanostructures, which is 23.17 ± 0.82 nm (Fig. 5H). Our MD simulation study showed that few dopamine molecules remain unbound in the case of 100 dopamine molecules during the 200 ns long dynamics. More prolonged incubation of dopamine with DNA-TD may allow them to deposit onto the negative backbone of TD. The TD nanoparticles get coated with dopamine molecules on increased dopamine concentration, which is also shown in the AFM images.
200 ratio, the atomic force microscopy image revealed the formation of homogeneously distributed sphere-like structures (Fig. 5G). The particle size distribution of the AFM image revealed an increase in the average size of the nanostructures, which is 23.17 ± 0.82 nm (Fig. 5H). Our MD simulation study showed that few dopamine molecules remain unbound in the case of 100 dopamine molecules during the 200 ns long dynamics. More prolonged incubation of dopamine with DNA-TD may allow them to deposit onto the negative backbone of TD. The TD nanoparticles get coated with dopamine molecules on increased dopamine concentration, which is also shown in the AFM images.
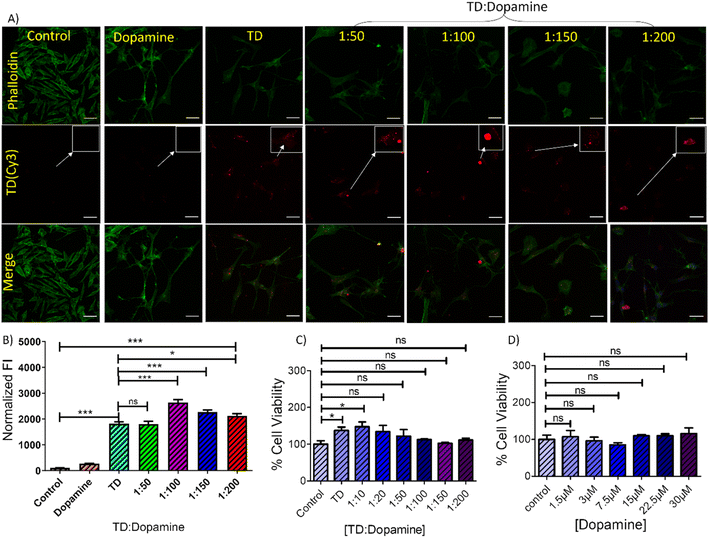
The study of DNA TD-dopamine complexation from spectroscopy, microscopy, and computational simulation has proved that DNA-tetrahedron can load a high amount of dopamine and be used as a dopamine carrier. DNA and dopamine are crucial components in the human body and possess inherent biocompatibility. However, a cell viability experiment using the MTT assay was conducted to assess the cytotoxicity of TD-dopamine complex nanoparticles on SH-SY5Y cells, and the results showed no significant decrease in cellular viability (Fig. 6C and D). Human SH-SY5Y cells have been extensively used to study in vitro neurodegenerative illnesses and as an in vitro model for neuroscience and neurotoxicity investigations.60,61 The SH-SY5Y human neuroblastoma cell line, either undifferentiated or developed into neuron-like cells, is one of the most often used cell lines in neuroscience.60,61 Cellular internalization of the nanostructure is a primary step to work as a delivery system; therefore, we did the cellular uptake study of TD-dopamine nanostructures on both differentiated and undifferentiated SH-SY5Y neuroblastoma cells. In the cellular uptake study, one of the four oligonucleotides (T4) was labeled with cyanine-3 (cy3) dye at 5′ ends for tracking purposes. Our previous report found that Cy3-label DNA tetrahedron (150 nM) showed significant uptake in SH-SY5Y cells. Therefore, we used 150 nM DNA tetrahedron alone and different amounts of dopamine hydrochloride, 50 eq., 100 eq., 150 eq., and 200 eq., for cell treatment (Fig. 6A). The SHSY5Y cells were incubated with all these combinations for 15 min at 37 °C and subjected the samples to laser scanning confocal microscopy after nucleus staining with Hoechst. Negligible signals were observed in the red channel for untreated cells and cells treated with dopamine (30 μM, 200 eq. of [TD]) on confocal imaging (Fig. 6A). The confocal image of the cells treated with TD and TD-dopamine complexes clearly showed the presence of Cy3 labeled TD. A quantification analysis of Cy3 signal intensity of confocal images reveals that the dopamine-loaded DNA-TD nanostructures are significantly uptaken by cells. In the case of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (TD-dopamine) combination, maximum cellular uptake is found (Fig. 6B). Similar trends were found in the cellular uptake study in nondifferentiated SHSY5Y cells (Fig. S5†).
100 (TD-dopamine) combination, maximum cellular uptake is found (Fig. 6B). Similar trends were found in the cellular uptake study in nondifferentiated SHSY5Y cells (Fig. S5†).
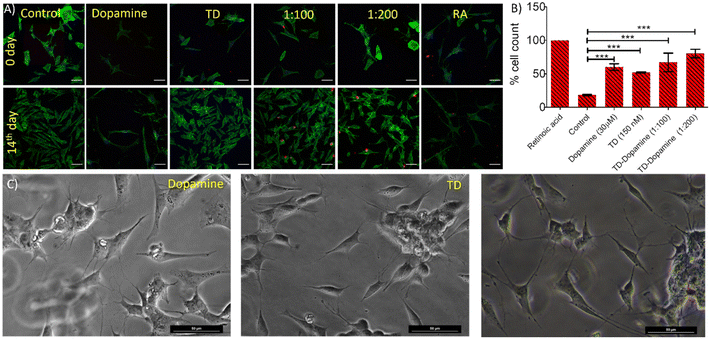
The ability to differentiate SH-SY5Y neuroblastoma cells into a more mature, neuron-like phenotype through manipulating the culture medium has provided numerous benefits in neuroscience research.60,61 A progressive death of dopaminergic neurons (dopamine-synthesizing neurons) causes a reduction of dopamine levels in the brain striatum. The differentiation of dopaminergic cells can boost the treatment of Parkinson's disease.5,62 Therefore, cell differentiation also plays an essential role in treating neurological diseases. DNA tetrahedral nanostructures are reported to promote neuronal NE-4C stem cell differentiation and proliferation.39 Therefore, an experiment was performed with DNA-TD, dopamine, and different combinations of TD-dopamine complex to check the ability to differentiate SH-SY5Y cells. The differentiation experiment was done by following a well-established protocol.60,63,64 Around 105 cells per well were seeded on a 10 mm glass coverslip in a four-well plate 24 h before the experiment. On the first day, cells were treated with DNA-TD, dopamine TD-dopamine complex at different differentiation media (DM)-1 composition. Untreated cells cultured only in DM-1 were used as a control, and retinoic acid was used as a standard differentiator. The same treatment was repeated on day 3 and day 5. On day 7 and day 9, the same treatment was done in DM-2, and on day 11 was treated in DM-3.
The growth and morphological changes due to differentiation were regularly monitored using an optical microscope (Fig. S6–S12†). The confocal image showed morphological differences between undifferentiated and differentiated SH-SY5Y cells (Fig. 7A). The undifferentiated cells have fewer neuritic projections with a linear type morphology; however, the differentiated cells showed multiple neuritic arms with larger area inflated morphology. The confocal images revealed that DNA-TD alone and combined with dopamine can differentiate the SH-SY5Y cells. The treatment with free dopamine also showed differentiation of SH-SY5Y cells. Quantification of these differentiated cells showed that retinoic acid, a standard differentiating agent, differentiates all the cells into neurons (Fig. 7B). However, the cells treated with TD, dopamine, and TD-dopamine combinations also showed significant differentiation compared to the control. Optical microscopy images also found some of the differentiated cells are like typical neurons (Fig. 7C). These cellular studies proved that DNA-TD can serve as a nanocarrier as well as a differentiating agent. These results, combined with the efficiency of TD nanostructures to cross the blood–brain barrier, suggest that dopamine-loaded DNA-TD can be an excellent nano-delivery system for Parkinson's disease treatment. This approach can potentially improve dopamine delivery to the brain and enhance its therapeutic efficacy.
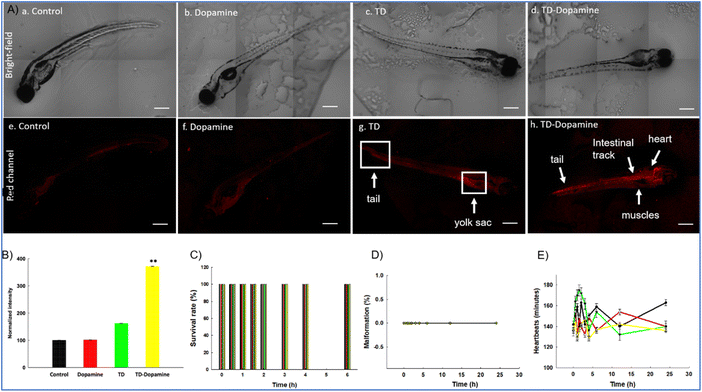
Zebrafish larvae are a commonly used model organism in research due to their transparency, high fecundity, and suitable brain dimensions. They are used in various studies, such as toxicity testing, behavioural assays, circuit modelling, and uptake studies.65,66 The zebrafish genome shares 70% of our genetic makeup, making it a superior in vivo model for studying how different biological substances are absorbed.67 Our previous study showed that TD did not cause any alterations in the survival rate and hatching success of developing zebrafish embryos.68 Study also demonstrated a significant uptake of DNA-TD compared to other geometries of DNA nanocages.68,69 At this time, to assess the biocompatible nature of TD and TD-dopamine complex on developing zebrafish larvae, the 8 dpf zebrafish larvae were exposed to TD (300 nM) and TD-dopamine (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100). The survival rate, malformation, and heart rate were assessed for the period of 6 and 24 hours, respectively. We observed that TD and TD-dopamine exhibited a biocompatible nature toward the developing zebrafish larvae post 6 hours of treatments (Fig. 8C). The survival rate for TD and TD-dopamine from zero to 6 hours was the same as for controls. Dopamine also exhibited a biocompatible nature towards developing zebrafish larvae until 6 hours of exposure. No malformation was observed in larvae until 24 hours of exposure to TD, dopamine, and TD-dopamine (1
100). The survival rate, malformation, and heart rate were assessed for the period of 6 and 24 hours, respectively. We observed that TD and TD-dopamine exhibited a biocompatible nature toward the developing zebrafish larvae post 6 hours of treatments (Fig. 8C). The survival rate for TD and TD-dopamine from zero to 6 hours was the same as for controls. Dopamine also exhibited a biocompatible nature towards developing zebrafish larvae until 6 hours of exposure. No malformation was observed in larvae until 24 hours of exposure to TD, dopamine, and TD-dopamine (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) (Fig. 8D). The heart rate per minute of developing larvae was 150, 142, 154, and 138 for control, TD, dopamine, and TD-dopamine (Fig. 8E). However, the initial heart rates in larvae exposed by alone dopamine were 175 per minute till 3 hours of exposure, and the highly active behaviour of larvae was also observed. The results depict that the TD and TD-dopamine exerted no toxic effect on the survival and heart rate of the zebrafish larvae compared to the control.
100) (Fig. 8D). The heart rate per minute of developing larvae was 150, 142, 154, and 138 for control, TD, dopamine, and TD-dopamine (Fig. 8E). However, the initial heart rates in larvae exposed by alone dopamine were 175 per minute till 3 hours of exposure, and the highly active behaviour of larvae was also observed. The results depict that the TD and TD-dopamine exerted no toxic effect on the survival and heart rate of the zebrafish larvae compared to the control.
The uptake capacity of any nanocarrier is crucial for its potential biomedical applications. Further, we proceed to analyze and quantify the uptake potential of TD-dopamine in zebrafish larvae. The larvae were exposed to TD (300 nM) and TD-dopamine complex (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) for 4 hours of treatment (Fig. 8A). TD and TD-dopamine (1
100) for 4 hours of treatment (Fig. 8A). TD and TD-dopamine (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) uptake was quantified using a laser-scanning confocal microscope. One of the four oligonucleotides (T4) was labelled with cyanine-5 (Cy5) dye at their 5′ ends for tracking purposes. The signal for TD-dopamine (1
100) uptake was quantified using a laser-scanning confocal microscope. One of the four oligonucleotides (T4) was labelled with cyanine-5 (Cy5) dye at their 5′ ends for tracking purposes. The signal for TD-dopamine (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) was significantly higher than alone TD in larvae post 4 h treatment (Fig. 8A). Additionally, the precise location of the targeted organelles in vivo depends on the physiological mechanism of intracellular transport and the unique characteristics of the subcellular structures. The usefulness of DNA nanostructures for imaging organelles for diagnostic applications has been demonstrated by precise targeting at the organelle level using fluorescently tagged DNA nanodevices; the same can be examined for therapeutic applications. Understanding the rules of appropriate transmission between organelles, cells, and tissues may help develop DNA nanodevices as diagnostic and therapeutic tools. Moreover, our uptake studies indicated that dopamine-modified TD enhanced the uptake and internalization in zebrafish larvae. The confocal image of larvae clearly shows the uptake and internalization of the TD-dopamine nanostructures. Quantification analysis of Cy5 signal intensity of TD-dopamine exerted the significant uptake with 100 eq. Dopamine compared to TD alone (Fig. 8B).
100) was significantly higher than alone TD in larvae post 4 h treatment (Fig. 8A). Additionally, the precise location of the targeted organelles in vivo depends on the physiological mechanism of intracellular transport and the unique characteristics of the subcellular structures. The usefulness of DNA nanostructures for imaging organelles for diagnostic applications has been demonstrated by precise targeting at the organelle level using fluorescently tagged DNA nanodevices; the same can be examined for therapeutic applications. Understanding the rules of appropriate transmission between organelles, cells, and tissues may help develop DNA nanodevices as diagnostic and therapeutic tools. Moreover, our uptake studies indicated that dopamine-modified TD enhanced the uptake and internalization in zebrafish larvae. The confocal image of larvae clearly shows the uptake and internalization of the TD-dopamine nanostructures. Quantification analysis of Cy5 signal intensity of TD-dopamine exerted the significant uptake with 100 eq. Dopamine compared to TD alone (Fig. 8B).
Conclusion
In this work, we studied the complexation mechanism of DNA tetrahedron nanocage and dopamine hydrochloride. The results showed that dopamine binds with the tetrahedral nanostructure through hydrogen bonding and electrostatic interaction. Through fluorescence spectroscopy analysis, we confirmed the binding of dopamine with the tetrahedral nanostructure. The dye displacement experiments and CD spectral observations showed that dopamine primarily binds to the groove region of DNA-TD without disrupting the tetrahedron's original helical structures of edges. Molecular dynamic simulations further supported the binding of protonated dopamine in both the major and minor groove regions, along with the negatively charged phosphate backbone. A combination of hydrogen bonding and electrostatic interactions facilitated this binding. Simulation results indicated that out of 100 dopamine molecules, approximately 82 molecules bound to TD within a 200 ns simulation. The binding of dopamine increased the degree of hydrogen bonding between DNA base pairs, thereby enhancing the stability of TD. Further, the dynamic light scattering analysis reveals an increase in the hydrodynamic diameter of nanostructures on increasing dopamine. Electrophoretic migration gel analysis and AFM imagining further supported the increased size. The rise in nanostructure size may be the deposition of dopamine molecules on the DNA backbone of TD through electrostatic interactions. They were subjected to biological application after successfully loading dopamine onto the TD. The dopamine-loaded TD nanostructures demonstrated significant cellular internalization with differentiated and nondifferentiated SH-SY5Y neuroblastoma cells. Further, it was found that DNA-TD and TD-dopamine complexes promote the differentiation of SH-SY5Y neuroblastoma cells. Additionally, we demonstrated the bio-compatibility and uptake of TD-dopamine complex in vivo model zebrafish larvae. The result showed a significant uptake of TD and TD-dopamine complex in zebrafish larvae, and no significant changes were obtained regarding survival rate, malformations, and heartbeats. These findings imply that dopamine-loaded DNA-TD can be a highly effective nano-delivery system for treating Parkinson's disease, along with high biocompatibility, non-cytotoxicity, and the effectiveness of TD nanostructures in crossing the blood–brain barrier. This strategy may enhance dopamine's therapeutic efficacy by improving its transport to the brain.Live subject statement
All zebrafish animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Ahmedabad University and approved by the Animal Ethics Committee of Ahmedabad University.Data availability
All the data and associated raw data from the manuscript is available upon reasonable request from authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
RS thanks, Gujcost-DST and IITGN-MoE, GoI, for the Postdoctoral Fellowship. KK and RV thanks SERB, and GoI, for the National Postdoctoral Fellowship. SM thanks CSIR, India for research fellowship. PY thanks MOE and IIT Gandhinagar for Director's Fellowship. PKM thanks SERB, India for funding. DB thanks SERB, and GoI for the Core research grant, MoES for STARS grant, IITGN for the start-up grant, Gujcost-DST, and GSBTM for research grants. SG thanks SERB and GSBTM for research grants. We thank CIF ITTGN for the instrumental facilities.References
- S. E. Hyman, Curr. Biol., 2005, 15, R154–R158 CrossRef CAS PubMed
.
- R. I. Teleanu, A.-G. Niculescu, E. Roza, O. Vladâcenco, A. M. Grumezescu and D. M. Teleanu, Int. J. Mol. Sci., 2022, 23, 5954 CrossRef CAS PubMed
.
- H. Juárez Olguín, D. Calderón Guzmán, E. Hernández García and G. Barragán Mejía, Oxid. Med. Cell. Longevity, 2016, 2016, 1–13 CrossRef PubMed
.
- D. L. Robinson, A. Hermans, A. T. Seipel and R. M. Wightman, Chem. Rev., 2008, 108, 2554–2584 CrossRef CAS PubMed
.
- F. N. Emamzadeh and A. Surguchov, Front. Neurosci., 2018, 12 DOI:10.3389/fnins.2018.00612
.
- D. Salat and E. Tolosa, J. Parkinson's Dis., 2013, 3, 255–269 CAS
.
- R. Pahuja, K. Seth, A. Shukla, R. K. Shukla, P. Bhatnagar, L. K. S. Chauhan, P. N. Saxena, J. Arun, B. P. Chaudhari, D. K. Patel, S. P. Singh, R. Shukla, V. K. Khanna, P. Kumar, R. K. Chaturvedi and K. C. Gupta, ACS Nano, 2015, 9, 4850–4871 CrossRef CAS PubMed
.
- J. Baskin, J. E. Jeon and S. J. G. Lewis, J. Neurol., 2021, 268, 1981–1994 CrossRef PubMed
.
- V. Monge-Fuentes, A. Biolchi Mayer, M. R. Lima, L. R. Geraldes, L. N. Zanotto, K. G. Moreira, O. P. Martins, H. L. Piva, M. S. S. Felipe, A. C. Amaral, A. L. Bocca, A. C. Tedesco and M. R. Mortari, Sci. Rep., 2021, 11, 15185 CrossRef CAS PubMed
.
- D. Teleanu, C. Chircov, A. Grumezescu and R. Teleanu, Nanomaterials, 2019, 9, 96 CrossRef PubMed
.
- W. Ma, Y. Zhan, Y. Zhang, C. Mao, X. Xie and Y. Lin, Signal Transduction Targeted Ther., 2021, 6, 351 CrossRef CAS PubMed
.
- Q. Hu, H. Li, L. Wang, H. Gu and C. Fan, Chem. Rev., 2019, 119, 6459–6506 CrossRef CAS PubMed
.
- A. R. Gada, P. Vaswani, R. Singh and D. Bhatia, ChemBioChem, 2023, 24, e202200634 CrossRef CAS PubMed
.
- D. Menon, R. Singh, K. B. Joshi, S. Gupta and D. Bhatia, ChemBioChem, 2023, 24, e202200580 CrossRef CAS PubMed
.
- X. Chen, Y. Xie, Z. Liu and Y. Lin, Front. Bioeng. Biotechnol., 2021, 9, 782237 CrossRef PubMed
.
- X. Chen, D. Liu, Y. Wu, H. Yao, Q. Xia and Y. Yang, ChemBioChem, 2022, 23, e202200459 CrossRef CAS PubMed
.
- P. Hivare, C. Panda, S. Gupta and D. Bhatia, ACS Chem. Neurosci., 2021, 12, 363–377 CrossRef CAS PubMed
.
- J. Yan, X. Zhan, Z. Zhang, K. Chen, M. Wang, Y. Sun, B. He and Y. Liang, J. Nanobiotechnol., 2021, 19, 412 CrossRef CAS PubMed
.
- F. Xiao, L. Lin, Z. Chao, C. Shao, Z. Chen, Z. Wei, J. Lu, Y. Huang, L. Li, Q. Liu, Y. Liang and L. Tian, Angew. Chem., Int. Ed., 2020, 59, 9702–9710 CrossRef CAS PubMed
.
- S. Shi, W. Fu, S. Lin, T. Tian, S. Li, X. Shao, Y. Zhang, T. Zhang, Z. Tang, Y. Zhou, Y. Lin and X. Cai, Nanomedicine, 2019, 21, 102061 CrossRef CAS PubMed
.
- A. Rajwar, S. R. Shetty, P. Vaswani, V. Morya, A. Barai, S. Sen, M. Sonawane and D. Bhatia, ACS Nano, 2022, 16, 10496–10508 CrossRef CAS PubMed
.
- N. M. Rao, Chem. Phys. Lipids, 2010, 163, 245–252 CrossRef CAS PubMed
.
- Y. Hu, Z. Chen, H. Zhang, M. Li, Z. Hou, X. Luo and X. Xue, Drug Delivery, 2017, 24, 1295–1301 CrossRef CAS PubMed
.
- T. Tian, J. Li, C. Xie, Y. Sun, H. Lei, X. Liu, J. Xia, J. Shi, L. Wang, W. Lu and C. Fan, ACS Appl. Mater. Interfaces, 2018, 10, 3414–3420 CrossRef CAS PubMed
.
- K.-R. Kim, D.-R. Kim, T. Lee, J. Y. Yhee, B.-S. Kim, I. C. Kwon and D.-R. Ahn, Chem. Commun., 2013, 49, 2010 RSC
.
- N. Chen, S. Qin, X. Yang, Q. Wang, J. Huang and K. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 26552–26558 CrossRef CAS PubMed
.
- M. Zhang, X. Zhang, T. Tian, Q. Zhang, Y. Wen, J. Zhu, D. Xiao, W. Cui and Y. Lin, Bioact. Mater., 2022, 8, 368–380 CAS
.
- M. I. Setyawati, R. V. Kutty, C. Y. Tay, X. Yuan, J. Xie and D. T. Leong, ACS Appl. Mater. Interfaces, 2014, 6, 21822–21831 CrossRef CAS PubMed
.
- Y. Sun, S. Li, Y. Zhang, Q. Li, X. Xie, D. Zhao, T. Tian, S. Shi, L. Meng and Y. Lin, ACS Appl. Mater. Interfaces, 2020, 12, 36957–36966 CrossRef CAS PubMed
.
- W. Cui, X. Yang, X. Chen, D. Xiao, J. Zhu, M. Zhang, X. Qin, X. Ma and Y. Lin, Adv. Funct. Mater., 2017, 27, 1603524 CrossRef PubMed
.
- H. Lee, A. K. R. Lytton-Jean, Y. Chen, K. T. Love, A. I. Park, E. D. Karagiannis, A. Sehgal, W. Querbes, C. S. Zurenko, M. Jayaraman, C. G. Peng, K. Charisse, A. Borodovsky, M. Manoharan, J. S. Donahoe, J. Truelove, M. Nahrendorf, R. Langer and D. G. Anderson, Nat. Nanotechnol., 2012, 7, 389–393 CrossRef CAS PubMed
.
- J. Li, H. Pei, B. Zhu, L. Liang, M. Wei, Y. He, N. Chen, D. Li, Q. Huang and C. Fan, ACS Nano, 2011, 5, 8783–8789 CrossRef CAS PubMed
.
- R. Duangrat, A. Udomprasert and T. Kangsamaksin, Cancer Sci., 2020, 111, 3164–3173 CrossRef CAS PubMed
.
- Y. Chen, X. Chen, B. Zhang, Y. Zhang, S. Li, Z. Liu, Y. Gao, Y. Zhao, L. Yan, Y. Li, T. Tian and Y. Lin, Signal Transduction Targeted Ther., 2024, 9, 28 CrossRef CAS PubMed
.
- D. Y. Tam, J. W.-T. Ho, M. S. Chan, C. H. Lau, T. J. H. Chang, H. M. Leung, L. S. Liu, F. Wang, L. L. H. Chan, C. Tin and P. K. Lo, ACS Appl. Mater. Interfaces, 2020, 12, 28928–28940 CrossRef CAS PubMed
.
- Y. Li, Z. Cai, W. Ma, L. Bai, E. Luo and Y. Lin, Bone Res., 2024, 12, 14 CrossRef CAS PubMed
.
- H. Wang, J. Gong, W. Chen, Q. Sun, T. Zhang, Y. Lin and X. Cai, Nano Today, 2024, 56, 102252 CrossRef CAS
.
- W. Cui, Y. Zhan, X. Shao, W. Fu, D. Xiao, J. Zhu, X. Qin, T. Zhang, M. Zhang, Y. Zhou and Y. Lin, ACS Appl. Mater. Interfaces, 2019, 11, 32787–32797 CrossRef CAS PubMed
.
- W. Ma, X. Shao, D. Zhao, Q. Li, M. Liu, T. Zhou, X. Xie, C. Mao, Y. Zhang and Y. Lin, ACS Appl. Mater. Interfaces, 2018, 10, 7892–7900 CrossRef CAS PubMed
.
- K. Skúpa, M. Melicherčík and J. Urban, J. Mol. Model, 2015, 21, 241 CrossRef PubMed
.
- J. Liu, Z.-H. Wang, G.-A. Luo, Q.-W. Li and H.-W. Sun, Anal. Sci., 2002, 18, 751–755 CrossRef CAS PubMed
.
- S. Sarkar, A. Chowdhury and P. C. Singh, J. Phys. Chem. B, 2019, 123, 10700–10708 CrossRef CAS PubMed
.
- J. L. Berfield, L. C. Wang and M. E. A. Reith, J. Biol. Chem., 1999, 274, 4876–4882 CrossRef CAS PubMed
.
- M. Sirajuddin, S. Ali and A. Badshah, J. Photochem. Photobiol., B, 2013, 124, 1–19 CrossRef CAS PubMed
.
- M. Khorasani-Motlagh, M. Noroozifar, A. Moodi and S. Niroomand, J. Photochem. Photobiol., B, 2013, 127, 192–201 CrossRef CAS PubMed
.
- R. Singh, N. K. Mishra, P. Gupta and K. B. Joshi, Chem.– Asian J., 2020, 15, 531–539 CrossRef CAS PubMed
.
- R. Singh, N. Kumar Mishra, V. Kumar, V. Vinayak and K. Ballabh Joshi, ChemBioChem, 2018, 19, 1630–1637 CrossRef CAS PubMed
.
- L. A. Reis and M. S. Rocha, Biopolymers, 2017, 107, e23015 CrossRef PubMed
.
- E. F. Healy, J. Chem. Educ., 2007, 84, 1304 CrossRef CAS
.
- P. O. Vardevanyan, A. P. Antonyan, M. A. Parsadanyan, H. G. Davtyan and A. T. Karapetyan, Exp. Mol. Med., 2003, 35, 527–533 CrossRef CAS PubMed
.
- V. I. Dodero, Front. Biosci., 2011, 16, 61 CrossRef PubMed
.
- V. Kumar, R. Singh and K. B. Joshi, New J. Chem., 2018, 42, 3452–3458 RSC
.
- R. Singh, V. Suryavashi, V. Vinayak and K. B. Joshi, ChemistrySelect, 2019, 4, 5810–5816 CrossRef CAS
.
- J. Liu, Q. Li, Y. Yu and X. Fang, Anal. Sci., 2003, 19, 1099–1102 CrossRef CAS PubMed
.
- V. Maingi, M. V. S. Kumar and P. K. Maiti, J. Phys. Chem. B, 2012, 116, 4370–4376 CrossRef CAS PubMed
.
- A. K. Sahoo, B. Bagchi and P. K. Maiti, J. Chem. Phys., 2019, 151, 164902 CrossRef PubMed
.
- C. Alves, F. Iacovelli, M. Falconi, F. Cardamone, B. Morozzo, C. L. Della Rocca, P. de Oliveira and A. Desideri, J. Chem. Inf. Model., 2016, 56, 941–949 CrossRef CAS PubMed
.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS
.
- R. Salomon-Ferrer, D. A. Case and R. C. Walker, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2013, 3, 198–210 CAS
.
- J. Kovalevich and D. Langford, Methods Mol. Biol., 2013, 1078, 9–21 CrossRef CAS PubMed
.
- H. Xicoy, B. Wieringa and G. J. M. Martens, Mol. Neurodegener., 2017, 12, 10 CrossRef PubMed
.
- P. P. Michel, E. C. Hirsch and S. Hunot, Neuron, 2016, 90, 675–691 CrossRef CAS PubMed
.
- M. M. Shipley, C. A. Mangold and M. L. Szpara, J. Vis. Exp., 2016, 108, 53193 Search PubMed
.
- P. Hivare, A. Gangrade, G. Swarup, K. Bhavsar, A. Singh, R. Gupta, P. Thareja, S. Gupta and D. Bhatia, Nanoscale, 2022, 14, 8611–8620 RSC
.
- S. Cassar, I. Adatto, J. L. Freeman, J. T. Gamse, I. Iturria, C. Lawrence, A. Muriana, R. T. Peterson, S. Van Cruchten and L. I. Zon, Chem. Res. Toxicol., 2020, 33, 95–118 Search PubMed
.
- R. Basnet, D. Zizioli, S. Taweedet, D. Finazzi and M. Memo, Biomedicines, 2019, 7, 23 Search PubMed
.
- K. Mhalhel, M. Sicari, L. Pansera, J. Chen, M. Levanti, N. Diotel, S. Rastegar, A. Germanà and G. Montalbano, Cells, 2023, 12, 252 Search PubMed
.
- K. Kansara, R. Singh, P. Yadav, A. Mansuri, A. Kumar and D. Bhatia, ACS Appl. Nano Mater., 2023, 6, 13443–13452 Search PubMed
.
- K. Kansara, A. Mansuri, A. Rajwar, P. Vaswani, R. Singh, A. Kumar and D. Bhatia, Nanoscale Adv., 2023, 5, 2558–2564 RSC
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr00612g |
| ‡ Authors contributed equally. |
| § Present address: Department of Bioengineering, College of Engineering and Computing, George Mason University, Fairfax, Virginia, United States. |
| ¶ Present address: P. D. Patel Institute of Applied Sciences, Charotar University of Science and Technology, CHARUSAT Campus, Changa, Anand - 388421, India. |
| This journal is © The Royal Society of Chemistry 2024 |