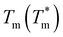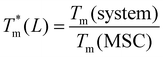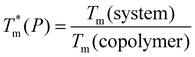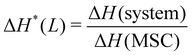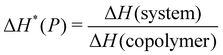Biophysical investigation of liposome systems decorated with bioconjugated copolymers in the presence of amantadine†
Rosendo
Pérez-Isidoro
 *a,
Alejandra Gabriela
Valdez-Lara
b,
Alma Jessica
Díaz-Salazar
c,
Stephanie
Hoeppener
*a,
Alejandra Gabriela
Valdez-Lara
b,
Alma Jessica
Díaz-Salazar
c,
Stephanie
Hoeppener
 de,
Carlos
Guerrero-Sánchez
de,
Carlos
Guerrero-Sánchez
 *de,
Patricia
Quintana-Owen
f,
Jesus Carlos
Ruiz-Suárez
*de,
Patricia
Quintana-Owen
f,
Jesus Carlos
Ruiz-Suárez
 b,
Ulrich S.
Schubert
b,
Ulrich S.
Schubert
 de,
Guadalupe
Ayora-Talavera
d,
Marco Antonio
De Jesús-Téllez
*a and
Enrique
Saldívar-Guerra
de,
Guadalupe
Ayora-Talavera
d,
Marco Antonio
De Jesús-Téllez
*a and
Enrique
Saldívar-Guerra
 *a
*a
aCentro de Investigación en Química Aplicada (CIQA), Enrique Reyna, 140, 25294 Saltillo, Coahuila, Mexico. E-mail: rosendo.perez@ciqa.edu.mx; marco.tellez@ciqa.edu.mx; enrique.saldivar@ciqa.edu.mx
bCINVESTAV-Monterrey, PIIT, 66600 Apodaca, Nuevo León, Mexico
cLaboratorio de Bio-fisicoquímica, Departamento de Fisicoquímica, Facultad de Química, Universidad Nacional Autónoma de México, 04510 México City, Mexico
dLaboratory of Organic and Macromolecular Chemistry (IOMC), Friedrich Schiller University Jena, Humboldtstrasse 10, 97743 Jena, Germany. E-mail: carlos.guerrero.sanchez@uni-jena.de
eJena Center for Soft Matter (JCSM), Friedrich-Schiller-University Jena, Philosophenweg 7, 07743 Jena, Germany
fDepartamento de Física Aplicada, CINVESTAV-IPN, Unidad Mérida, A.P. 73, Cordemex, 97310 Mérida, Yucatán, Mexico
gLaboratorio de Virología, Centro de Investigaciones Regionales Dr. Hideyo Noguchi, Universidad Autónoma de Yucatán, Mérida, Yucatán, Mexico
First published on 6th May 2024
Abstract
Liposome-based technologies derived from lipids and polymers (e.g., PEGylated liposomes) have been recognized because of their applications in nanomedicine. However, since such systems represent myriad challenges and may promote immune responses, investigation of new biomaterials is mandatory. Here, we report on a biophysical investigation of liposomes decorated with bioconjugated copolymers in the presence (or absence) of amantadine (an antiviral medication). First, copolymers of poly(N,N-dimethylacrylamide-co-fluoresceinacrylate-co-acrylic acid-N-succinimide ester)-block-poly(N-isopropylacrylamide) (PDMA-b-PNIPAM) containing a fluorescence label were biofunctionalized with short peptides that resemble the sequence of the loops 220 and 130 of the binding receptor of the hemagglutinin (HA) protein of the influenza A virus. Then, the bioconjugated copolymers were self-assembled along with liposomes composed of 1,2 dimyristoyl-sn-glycero-3-phosphocholine, sphingomyelin, and cholesterol (MSC). These biohybrid systems, with and without amantadine, were systematically characterized using differential scanning calorimetry (DSC), dynamic light scattering (DLS), and cryogenic transmission electron microscopy (cryoTEM). Finally, the systems were tested in an in vitro study to evaluate cytotoxicity and direct immunofluorescence in Madin Darbin Canine Kidney (MDCK) cells. The biohybrid systems displayed long-term stability, thermo-responsiveness, hydrophilic–hydrophobic features, and fluorescence properties and were presumable endowed with cell targeting properties intrinsically integrated into the amino acid sequences of the utilized peptides, which indeed turn them into promising nanodevices for biomedical applications.
1 Introduction
In recent years, an increasing number of contributions in the field of nanomedicine have focused on the usage of new biohybrid materials to formulate systems for improving drug administration.1–4 In this regard, it has been documented that combinations of biocompatible polymers with bioactive molecules can enable synergistic properties for biomedical applications.5–9 Fundamental knowledge related to biophysical properties and intermolecular interactions of cell membranes (or models) with other materials (e.g., smart polymers) is crucial for developing new drug delivery systems.4,10,11 Modern reversible deactivation radical polymerization (RDRP) techniques have enabled the synthesis of well-defined bioconjugated polymer materials functionalized with specific bioactive groups, such as lipids, carbohydrates, proteins, short active peptides, drugs, nucleic acids, etc., with exceptional properties for applications in drug delivery systems.6,8,12–15 From this perspective, a proper understanding of specific and non-specific interactions between drug delivery systems and cell membranes is essential for implementing new applications in nanomedicine.4,16 Taking the aforementioned information into account, one approach in the design and investigation of efficient drug delivery systems corresponds to the self-assembly of nanoplatforms that mimic relevant biological systems. Therefore, fundamental knowledge about interactions between the different components of the systems is crucial. For instance, in biohybrid systems composed of lipids and polymers and proposed as drug nanocarriers, it is very important to characterize their thermodynamic properties, which will provide valuable information about the intermolecular interactions between the components of the nanocarriers and their payloads.5–7,17 In this context, the investigation of biohybrid systems that combine lipids, polymers, and bioactive moieties has led to considerable progress in the development of new therapeutic applications.2,6,18,19 Furthermore, liposomes have been extensively used as model systems in the research of biomembranes and drug delivery approaches.20–22 For example, combinations of liposomes with synthetic (co)polymers enable the self-assembly of vesicular nanostructures via diverse (but demanding) formulation processes, including microfluidic manufacturing.22,23 However, lipid vesicles located at an aqueous interface are exposed to non-specific interactions, including electrostatic, hydrophobic, and steric interactions, which may lead to oxidation and hydrolysis.3,17,24 Such processes are related to relatively low stability of liposomes in aqueous media, their chemical degradation, subsequent aggregation or flocculation and, hence, a potential premature loss of encapsulated payloads.24–28 Therefore, it is thought that the limitations of liposome-based drug delivery systems could be properly addressed and overcome with a better understanding of the different physicochemical interactions between the different components of these systems. In addition, the utilization of biocompatible (co)polymeric materials with suitable bioconjugated moieties could also provide specific cell targeting features to liposome-based nanocarriers. Thus, these biohybrid platforms may also be used to encapsulate bioactive molecules or drugs of interest either inside the lumen, into the lipid bilayer or in the polymer envelope. In connection with this, synthetic polymers have been bioconjugated with peptides to improve the affinity of the respective nanocarrier for a specific cell or binding site.13,29 In a similar fashion, it is known that viral infections involve not only a specific binding of a virus to the surface of a cell but also a fusion of the cell membrane with the viral envelope.30,31 Therefore, viruses have evolved a capability to self-replicate by infecting in a very specific manner either specific cells, tissues, or host species; this ability is known as viral tropism.30,31 Hence, characteristics similar to those found in viral tropism embedded into specifically designed drug nanocarriers could enhance their ability to deliver their payloads (including antivirals) in more efficient and specific manners via a mechanism that resembles viral tropism. For instance, influenza A viruses (Orthomyxoviridae family) possess negative-sense single-stranded ribonucleic acid (RNA) molecules and a lipid membrane, which bears two kinds of proteins (i.e., neuraminidase (NA) and hemagglutinin (HA)) around its envelope.32,33 In turn, the HA protein can specifically recognize and penetrate host cells. In particular, HA can bind to the sialic receptors available on the surface of cell membranes. The receptor-binding site of HA is composed of the 190-helix, and the 130- and 220-loops at the most exposed and peripheral site of the protein.32 Following this natural approach, such an evolved mechanism could be implemented in currently available drug delivery systems to improve their cell targeting capabilities. Hence, the main objective in this contribution is to study the interaction of bioconjugate copolymers with a lipid membrane composed of DMPC, sphingomyelin, and cholesterol to generate new biohybrid nanovesicles for biomedical applications. We developed and characterized biohybrid systems from bioconjugated copolymers and liposome vesicles in an attempt to resemble cell-targeting properties like those exhibited by the HA protein of the influenza virus. To this end, we first carried out the design and synthesis of the bioconjugated copolymers. Thus, the utilized copolymer precursors poly(N,N-dimethylacrylamide-co-fluoresceinacrylate-co-acrylic acid-N-succinimide ester)-block-poly(N-isopropylacrylamide) (PDMA-b-PNIPAM) were synthesized by the reversible–addition–fragmentation chain-transfer (RAFT) polymerization technique; fluorescein moieties were also incorporated as labels along the polymer chains throughout the course of the polymerization reactions. Thereafter, the copolymers were functionalized with the peptide sequences Arg–Asp–Gln–Glu–Gly (L220) and Ala–Cys–Pro–His (L130) that resemble the sequences of the 220 and 130 loops, respectively, of the receptor-binding site of the HA of the influenza A virus (Scheme 1).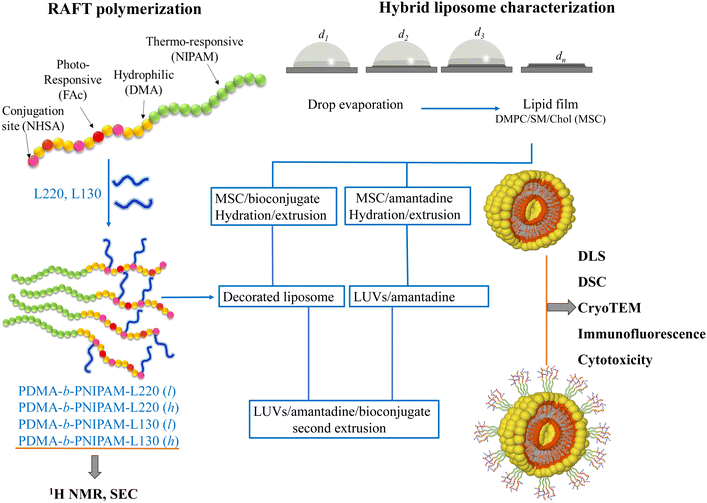 | ||
| Scheme 1 Schematic diagram of the preparation and investigation of liposome systems decorated with bioconjugated copolymers in the presence (or absence) of amantadine. | ||
In this context, PDMA-b-PNIPAM block copolymers have recently been regarded as promising candidates for the design of drug delivery systems due to their thermo-responsiveness, ability to stabilize cubosomes, and affinity to lipid membranes.34 Beyond the field of lipid membranes, it has been established that sphingomyelin and cholesterol enable the formation of certain lipid domains that are important for cell communication.35–39 Hence, we hypothesize that these lipid domains, enriched with cholesterol and sphingolipids, could favorably interact with the bioconjugated copolymers and lead to a more effective binding of the latter onto the corresponding lipid membranes. This capability has been previously observed in other biological systems where lipid domains can function as protein-anchoring platforms,35,36,38 and along these lines, we have recently reported important features derived from non-specific lipid–protein interactions of a lipid model composed of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), sphingomyelin (SM), and cholesterol (chol) under the influence of lysozyme and ovalbumin.40 Thereafter, we combined the peptide-functionalized block-copolymers with large unilamellar vesicles (LUVs) made of DMPC/SM/chol (herein denoted as MSC) to obtain biohybrid liposomes decorated with such bioconjugated copolymers, where the MSC lipid mixture was employed as a platform for anchoring the bioconjugated copolymers. Then, biophysical properties of such biohybrid liposomes decorated with bioconjugated copolymers were thoroughly evaluated by means of differential scanning calorimetry (DSC), dynamic light scattering (DLS), cryogenic transmission electron microscopy (cryoTEM), cytotoxicity assays, and fluorescence microscopy (Scheme 1). Additionally, thermal properties and long-term size stability of these bioconjugated copolymers/lipid systems were investigated in the presence of amantadine (AMT), an antiviral/antiparkinsonian compound. The obtained results suggest that combinations of synthetic bioconjugated copolymers with lipid vesicles enable the access to self-assembled biohybrid nanovesicles of long-term stability, with the capability to encapsulate hydrophobic bioactive compounds and with potential features to mimic viral tropism mechanisms. This kind of copolymer/lipid biohybrid platform could be implemented for the development of nanodevices for biomedical applications, for example, innovative drug delivery systems.
2 Materials and methods
2.1 Reagents
1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC, P-0888, 99%), 4-(2-hydroxyethyl)-piperazine-1-ethanesulfonic acid (HEPES, H3375 ≥ 99.5%), sodium chloride (NaCl, S9888 ≥ 99.0%), cholesterol (chol, C8667 ≥ 99%), and amantadine hydrochloride (A1260, ≥ 98%) were purchased from Sigma-Aldrich, USA. N-Octadecanoyl-D-erythro-sphingosylphosphoryl choline (sphingomyelin, SM brain porcine, 860062P) was obtained from Avanti Polar Lipids, Inc., USA (chemical structures for lipids are depicted in Fig. S5, ESI†). It is important to comment that the SM reagent is a natural mixture of sphingolipids where the main component corresponds to the structure depicted in Fig. S5 (ESI†) (SM). Deionized water (18.2 MΩ cm) was obtained from a Millipore system (Simplicity, F1CA58004A, France). Four bioconjugated block copolymers, PDMA-b-PNIPAM-L220(l) (19.4 kDa), PDMA-b-PNIPAM-L220(h) (30.5 kDa), PDMA-b-PNIPAM-L130(l) (18.5 kDa), and PDMA-b-PNIPAM-L130(h) (29.6 kDa) were synthesized via reversible–addition fragmentation chain-transfer (RAFT) polymerization followed by a grafting-through process for bioconjugation with different peptide sequences.13,41 Note that the acronym PDMA, as utilized in the nomenclature of this contribution, refers, for simplicity and in general, to the hydrophilic block of the bioconjugated block copolymers, which are composed of DMA, fluorescein acrylate (FAc) and acrylic acid-N-succinimide ester (NHSA) comonomers, where the DMA content is at least 88 mol%. Bioconjugated block copolymers were purified by dialysis and characterized by means of proton nuclear magnetic resonance (1H NMR, Bruker Avance 300 MHz spectrometer) and size-exclusion chromatography (SEC, Agilent 1200 series). More details related to the synthesis and characterization of the investigated peptide-functionalized block-copolymers can be found in the ESI.†2.2 Method to obtain lipid films of DMPC/SM/chol for preparing liposomes
Lipid films, free of organic solvents, were prepared from a suspension of DMPC/SM/chol (MSC, 80/10/10 mol%) utilizing the small-volume evaporation methodology.42–44 To this end, lipids (powders) were carefully weighed and deposited in a volumetric flask. The lipid powder was hydrated with deionized water and stirred at 1000 rpm for 3 h at 37 °C to homogenize the lipid mixture. Thereafter, the suspension was brought to a volume of 50 mL at 25 °C. Next, the lipid suspension was subjected to an evaporation process via the small-volume evaporation methodology. Briefly, a small volume (50 μL) of the lipid suspension was deposited onto different spots of a porcelain plate (containing 12 sample depressions) and dried at 45 °C in a dry block heater (Ohaus, HB1DG, USA). The suspensions were completely dried, and subsequently, additional amounts of suspension were deposited onto the film that resulted from a previous drying step. Next, the lipid films were recovered, hydrated with deionized water, and brought to a volume of 50 mL in a volumetric flask. This new suspension was divided into 50 subsamples (1.0 mL each), which were deposited in microtubes. Each of these 50 subsamples was subjected to a second round of drying steps onto a polypropylene surface using an amount of 25 μL for each step. Final DMPC/SM/chol (MSC) films were hermetically sealed and stored at −20 °C prior to usage.2.3 Preparation of MSC LUVs/bioconjugated PDMA-b-PNIPAM block copolymers self-assemblies
An MSC film and bioconjugated copolymer (powder) were combined and hydrated with a HEPES/NaCl (10 mM/145 mM) buffer solution (pH 7.4) at 37 °C. To obtain multilamellar vesicles (MLVs), the sample was homogenized for 1 h at 1000 rpm at 37 °C. After that, the MLVs suspension was extruded in a mini-extruder (610020, Avanti Polar Lipids, Inc., USA) using polycarbonate membranes (800309, nucleopore track-etched membrane, Whatman Inc., USA) with a pore size of 100 nm to obtain large unilamellar vesicles (LUVs of ca. ∼100 nm). To obtain a homogeneous size distribution of LUVs, the MLV samples were extruded 21 times across the polycarbonate membrane pores at ∼37 °C. For DSC analysis, the utilized concentration of MSC and bioconjugated block copolymer was 1.5 and 0.14 mM, respectively. The concentration of MSC represents the total concentration of the lipid mixture, considering a relative average molar mass where the contribution of DMPC, SM, and chol is 80, 10, and 10 mol%, respectively. We used this ratio of DMPC, SM, and chol owing to its importance in the formation of lipid domains in the field of biophysics of membranes, where this combination plays a crucial role as an anchoring platform for hosting several macromolecules. Similar to a previous contribution from our group,44 a total lipid concentration of 1.5 mM was selected (considering the molar mass of DMPC for the whole lipid formulation as this substance is the major component (80 mol%) of the investigated lipid mixture). Under these considerations, the utilized bioconjugated block copolymer/MSC molar ratio was ca. ∼0.09. Similar to our previous contribution,44 this molar ratio was selected under the assumption that the proposed multifunctional bioconjugated block copolymers act as compatibilizers/stabilizers between the different components of the investigated biohybrid systems/aqueous media. Hence, with the lipid mixture being the major component of the biohybrid formulation, it seems reasonable to assume that a “stabilizer” of the system might be present in a lower proportion (up to a molar ratio of ca. 0.1 in our current investigations).442.4 Preparation of MSC LUVs/amantadine (AMT)/bioconjugated PDMA-b-PNIPAM block copolymers self-assemblies
To favor the incorporation of AMT in the lipid bilayer, the previous method was slightly modified. Thus, AMT was weighed in a vial containing an MSC film and hydrated with HEPES/NaCl (10 mM/145 mM) buffer solution (pH 7.4) at 37 °C. The MSC/AMT LUVs system was obtained in a similar way to that described in the previous subsection. After that, to incorporate the PDMA-b-PNIPAM block copolymer bioconjugates onto the MSC/AMT liposomes, the powder of the copolymer bioconjugates was dispersed with the MSC/AMT LUVs solution previously prepared and homogenized at 1000 rpm at 40 °C for 1 h. Then, the samples were subjected to a second cycle of extrusion in a similar fashion described before for the preparation of LUVs. This second method was preferred because we expect that it facilitates the incorporation of AMT into the lipid membrane without the influence of the utilized bioconjugated copolymers. The concentration used for DSC analysis was 1.5 mM, 10 mM and 0.14 mM for MSC, AMT and bioconjugated copolymer, respectively. The concentration ratio for AMT/MSC was equal to 6.6 (r = 10 mM/1.5 mM).2.5 Differential scanning calorimetry
Thermodynamic properties of the copolymer-decorated liposomes were estimated using differential scanning calorimetry experiments. For this purpose, the reversible thermotropic gel–fluid transition of the MSC model and the thermal response of bioconjugated copolymers were both separately and jointly evaluated using a power compensation microcalorimeter NanoDSC (Microcalorimeter NanoDSC, TA Instruments, USA). This equipment utilizes capillary cells to achieve high sensitivity within a volume of 300 μL. From the recorded raw calorimetric profiles of the investigated systems, we obtained their molar heat capacity (Cp) value at constant pressure, as a function of temperature (T). The sample was equilibrated at room temperature (25 °C) for 15 min and degassed by applying low pressure (635 mmHg in a degassing station, TA Instruments, USA) for 15 min at 25 °C before the corresponding calorimetric analysis. Each sample was scanned in heating mode. After 600 s of equilibrium time, the profile was recorded at a heating rate of 1.0 °C min−1 and a cell pressure of 3 atm. Cp values were obtained from the corresponding calorimetric profile after subtracting physical (buffer–buffer trace) and chemical (a spline function joining the pre- and post-regions of the phase transition) baselines. To recover the MSC and the bioconjugated copolymer contributions in the DSC traces, raw data below and above 40 °C were normalized to MSC and the corresponding bioconjugated copolymer weights, respectively. The temperature where Cp is maximum (Tm, phase transition temperature), the enthalpy change (ΔH) and the width of the transition (ΔT1/2) at half peak height were obtained from the recorded DSC traces. ΔH was calculated by obtaining the integral value of the molar heat capacity from the initial (Ti) and final (Tf) temperatures of the phase transition, i.e.,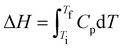 . Such thermodynamic parameters were estimated using homemade Python scripts. For the DSC analysis, each thermogram and the related parameters represent an average of at least six scans, considering three scans from each replicate.
. Such thermodynamic parameters were estimated using homemade Python scripts. For the DSC analysis, each thermogram and the related parameters represent an average of at least six scans, considering three scans from each replicate.
2.6 Dynamic light scattering
Dynamic light scattering (DLS) was used to monitor the hydrodynamic diameter of the investigated biohybrid systems. Each measurement was carried out using 50 μL of each sample housed in disposable cuvettes ZEN0040. The instrument used was a Zsizer Nano ZSP (Malvern Instruments, USA). The laser wavelength and the detector angle location were 633 nm and 173°, respectively. The intensity fluctuations were registered followed by their analysis using the Stokes–Einstein equation R = KBT/6πηD, where R, KB, T, η and D correspond to the hydrodynamic radius, the Boltzmann constant, the temperature, the dynamic viscosity, and the diffusion coefficient, respectively. Samples were measured at 25 °C, in duplicate with at least five repetitions each. For DLS measurements, the sample prepared for DSC was diluted to 0.5% with a buffer solution of HEPES/NaCl (10 mM/145 mM) and subjected to scattering analysis.2.7 Cryogenic transmission electron microscopy (cryoTEM)
LUV solutions for cryoTEM investigations were obtained by direct dispersion into a buffer of HEPES/NaCl (10 mM/145 mM) at pH 7.4 according to the concentrations summarized in Table S3 (ESI†). The measurements were performed on an FEI Tecnai G2 20 platform with a LaB6 filament at 200 kV acceleration voltage. Samples were prepared on quantifoil grids (R2/2) which were treated with Ar plasma prior to use for hydrophilization and cleaning. 8.5 μL of the dispersions (Table S3, ESI†) was applied onto the grids utilizing a FEI Vitrobot Mark IV system (offset: −5 mm, blotting time: 1 s). After blotting, the samples were immediately plunged into liquid ethane to achieve vitrification. Samples were transferred to a Gatan cryo stage and subsequently into a Gatan cryo holder (Gatan 626) and were transferred into the microscope by always maintaining a temperature below −168 °C during the whole transfer and measurement process after vitrification. Images were acquired using a Mega View (OSIS, Olympus Soft Imaging Systems) or an Eagle 4k CCD camera. Due to the amount of effort required for the cryoTEM measurements and the large number of samples, only selected samples were imaged using this technique, where the selection was based on the DLS results.2.8 In vitro cytotoxicity of copolymer-bioconjugates decorated liposomes
Cytotoxic evaluations were performed using Madin Darbin Canine Kidney (MDCK) cells. Briefly, MDCK cells were seeded in 96 well plates at a cell density of 1 × 105 cells per well. After cells reached confluency, they were washed 2× with a phosphate-buffered saline (PBS) solution (pH 7.2) and incubated with the corresponding copolymer-bioconjugate system at six different concentrations (from 100 to 3.25 μg mL−1) diluted in Dulbecco's Modified Eagles Media (D-MEM) in quadruplicate. After 72 h of incubation time, the cells were fixed and stained with 0.4% crystal violet in methanol for 30 min, washed under running tap water and let to dry. To determine cytotoxicity, plates were analyzed at a wavelength of 490 nm in an automated UV-vis plate reader Victor 3×. The cytotoxic response was fitted to the Hill equation,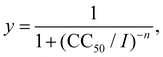 where y is the output cytotoxic response, I is the input concentration, CC50 is the cytotoxic concentration and n is the Hill coefficient. Fitting was carried out using the non-linear least-squares minimization and curve-fitting for Python (LMFIT) methods.45
where y is the output cytotoxic response, I is the input concentration, CC50 is the cytotoxic concentration and n is the Hill coefficient. Fitting was carried out using the non-linear least-squares minimization and curve-fitting for Python (LMFIT) methods.45
2.9 Direct immunofluorescence of liposome/copolymer-bioconjugate systems
MDCK cells were seeded in circular 22 mm coverslips. After the cells reached 60% confluency, they were washed 2× with the PBS solution and incubated with the corresponding liposome/copolymer-bioconjugate system in D-MEM at a concentration of 50 μM. Cells and the biohybrid system were incubated for 48 h at 37 °C under an atmosphere containing 5% CO2, thereafter, the inoculum was removed and the cells were washed 1× with PBS solution, fixed in cold acetone and incubated at room temperature with 10 μL of 0.2 mM Hoechst stain solution for 5 min. Cells were washed 1× with PBS solution and mounted into a Vectashield mounting medium (Vector Laboratories, Inc). Cells were directly observed using a fluorescence microscope (DM4000 B LED; Leica, Germany) equipped with a CCD camera. Images were taken with a filter L5, at λ 550 nm for green fluorescence and a filter A at λ 350 nm.3 Results and discussion
3.1 Physicochemical characterization of DMPC/SM/chol (MSC) LUVs and peptide-block copolymer bioconjugates dispersed in an aqueous buffer at pH 7.4
Liposome-based nanocarriers are established platforms in nanomedicine. Nonetheless, stability issues have been reported during long storage periods owing to aggregation and flocculation effects that substantially impact the vesicle size over time.10,25,46 Hence, knowledge about the size stability of liposome-containing nanocarriers is an important aspect of biomedical formulations derived from these compounds.22 Thus, the first analysis of the biohybrid systems investigated herein utilized the DLS technique to determine the hydrodynamic size distribution of biohybrid LUVs based on the MSC (DMPC/SM/chol, 80/10/10 mol%) lipid system and of the four PDMA-b-PNIPAM block copolymer bioconjugates: PDMA-b-PNIPAM-L220(l), PDMA-b-PNIPAM-L220(h), PDMA-b-PNIPAM-L130(l), and PDMA-b-PNIPAM-L130(h) (all materials dispersed in a buffer solution of HEPES/NaCl (10 mM/145 mM) at a pH value of 7.4). The MSC mixture served as a lipid membrane platform for the subsequent self-assembly of the bioconjugated copolymers that formed the investigated biohybrid nanocarrier systems described in the next section. The bioconjugated copolymers contain peculiar functionalities provided by their comonomer units such as thermo-responsiveness in the PNIPAM block, hydrophilicity ascribed to the PDMA segment, fluorescence due to the presence of the FAc units, and cell-targeting properties related to the amino acid sequences. The latter derived from the 220-loop (L220, Arg–Asp–Gln–Glu–Gly) and the 130-loop (L130, Ala–Cys–Pro–His) of the receptor binding site of the hemagglutinin protein of the influenza virus.32 Furthermore, the hydrophobic end group of the bioconjugated copolymers ascribed to the n-dodecyl (C12H25) moiety of the RAFT agent utilized during their synthesis might also influence the self-assembly properties of these materials as discussed elsewhere.47 This could also promote hydrophobic interactions and activate the incorporation of these materials into the lipid membranes. Fig. 1(a)–(c) displays the hydrodynamic size distributions for reference MSC LUVs (i.e., free of bioconjugated copolymers) and bioconjugated block copolymers (i.e., free of MSC liposomes) dispersed in a buffer solution. The hydrodynamic size distributions are shown in terms of intensity and volume with an attempt to visualize subpopulations of different sizes and the relative proportion of components of multiple sizes contained in the samples. As it can be observed for LUVs of pure MSC, both distributions in terms of volume and intensity are similar to each other indicating the presence of only one population with a size centered at 134 nm. The hydrodynamic diameter of the four block copolymers bioconjugated with peptides (i.e., free of liposomes) displayed interesting changes after extrusion (i.e., samples were extruded across a polycarbonate membrane with a pore size of 100 nm). PDMA-b-PNIPAM-L220(l) and PDMA-b-PNIPAM-L130(l) (bioconjugates featuring a low molar mass, 19.4 and 18.5 kDa, respectively) displayed more than one size distribution when they were evaluated in terms of intensity, being the subpopulation centered around 10 nm the most abundant as indicated in the size estimation as a function of volume. In contrast, for the case of copolymer bioconjugates with high molar mass (PDMA-b-PNIPAM-L220(h) and PDMA-b-PNIPAM-L130(h), 30.5 and 29.6 kDa, respectively), they only exhibited one size distribution featuring sizes >100 nm. However, it is important to consider that the DLS technique applied to polydisperse and non-spherical shape systems, like polymers, can potentially cause some drawbacks for the particle size interpretation.48,49 Thus, our results indicate that these bioconjugated copolymers tend to form aggregates in aqueous buffer solution (pH 7.4).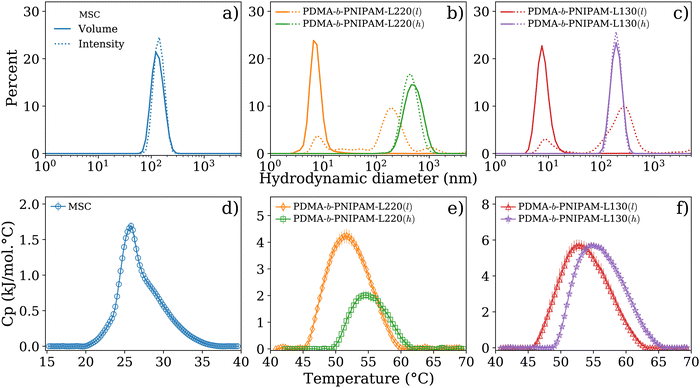 | ||
| Fig. 1 Hydrodynamic diameter distributions estimated from DLS measurements (a)–(c), and experimental calorimetric profiles obtained via DSC (d)–(f) for LUVs of DMPC/SM/chol (MSC, 80/10/10 mol%) and peptide–copolymer bioconjugates measured in HEPES/NaCl (10 mM/145 mM) buffer at a pH value 7.4. LUVs and block-copolymer bioconjugated with peptides were extruded across polycarbonate membranes with a pore size of 100 nm. PDMA-b-PNIPAM block copolymer is bioconjugated with the sequence L220 (Arg–Asp–Gln–Glu–Gly) and the sequence L130 (Ala–Cys–Pro–His). These amino acid sequences are related to the 220 and 130 loops of the receptor binding site of the hemagglutinin protein of the influenza virus.32 Peptide–copolymer bioconjugates with low and high molar mass are represented by l and h, respectively. Solid and dotted lines in DLS curves indicate the size distributions in terms of volume and intensity, respectively. The concentrations used for DSC analysis were 1.5 mM and 0.14 mM for MSC and bioconjugated copolymers, respectively. Whereas for DLS, the samples were diluted to 0.5% with the buffer solution as described in the Materials and methods section. | ||
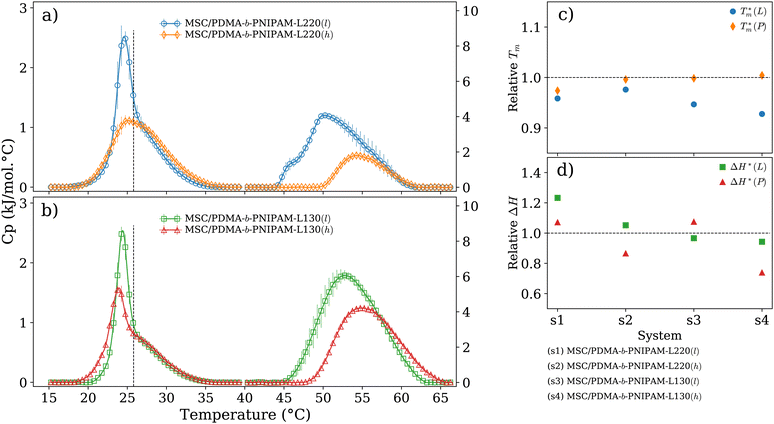
It is known that phospholipids hydrate in aqueous media to form self-assembled structures with different lamellar and non-lamellar phases, which is a property inherent to cell membranes and key in nanomedicine. Due to the amphiphilic nature of phospholipids, the polar regions (“head”) of these compounds favorably interact with water molecules when they self-assemble as lipid bilayers, maintaining a permanent electrostatic repulsion to each other, which is compensated by van der Waals forces existing between the hydrophobic regions of the lipids (“tail”). These interactions minimize the free energy of the system leading to the formation of a membrane morphology, which effectively expels water molecules from the lipid region. Lipid membranes exhibit a thermotropic behavior that can be examined using DSC and used to investigate effects that membrane-active molecules can inflict on them. To characterize the thermotropic behavior of the MSC system, we measured the calorimetric profile of the corresponding LUVs dispersed in a buffer solution of HEPES/NaCl at pH = 7.4 and in the absence of bioconjugated copolymers. Similarly, we characterized the thermotropic behavior of peptide-bioconjugated block copolymers in the absence of liposomes provided that these materials contain two thermo-responsive segments, i.e., the PNIPAM block and the peptide sequences. Fig. 1d reveals that LUVs, derived from the MSC lipids mixture, at pH 7.4 and under ionic strength (145 mM NaCl) have an expanded thermal transition ranging from 20 to 36 °C with a Cp max around 25.8 °C and a calorimetric enthalpy of ca. 8.7 kJ mol−1 (see Table 1). However, the observed endotherm was lower in cooperativity with a ΔT1/2 value of ca. 4 °C and revealed an asymmetrical profile. This suggests the formation of domains, where sphingomyelin and cholesterol might be heterogeneously distributed within the DMPC bilayer, indicating a thermal and dynamic reorganization process.35,50 Although sphingomyelin and cholesterol can migrate from different domains with specific Tm contributions, consecutive heating scans (repeated at least three times for each sample) demonstrated that the phase transition for the MSC system is reversible and proceeds close to equilibrium. Importantly, it has been observed that sphingolipid/cholesterol domains can diffuse across the plasma membrane as small units.51 Based on these observations, the prepared MSC membrane probably contains domains with different sphingomyelin and cholesterol ratios, which agrees with other experimental and theoretical investigations of similar membrane models where the formation of lipid domains induced by such compounds has been reported.35,50 This property, next to the capability of anchoring macromolecules such as proteins, might be two main factors for driving the observed assembly of the lipid/copolymer biohybrid systems herein investigated. In addition, the DSC thermograms in Fig. 1(e) and (f) confirm that, owing to the thermo-responsive PNIPAM block, the peptide-bioconjugated block copolymers have a thermotropic response with a broad phase transition ranging from ∼45 to 65 °C and a high ΔT1/2 value (see Table 1), which is also affected by the presence of the PDMA segment, the fluorescein units, and the corresponding peptide sequence. Similar effects have been also reported for lysozyme-bioconjugated PNIPAM-b-PDMA block copolymers.52
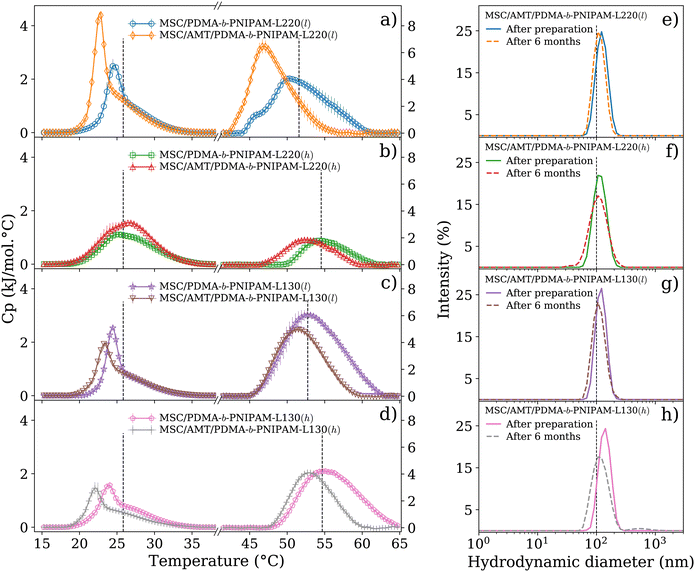
| System | T m (°C) | ΔHcal (kJ mol−1) | ΔT1/2 (°C) |
|---|---|---|---|
| (l), low molar mass (considering 19.4 and 18.5 kDa for PDMA-b-PNIPAM-L220 and PDMA-b-PNIPAM-L130, respectively). (h), high molar mass (considering 30.5 and 29.6 kDa for PDMA-b-PNIPAM-L220 and PDMA-b-PNIPAM-L130, respectively). L, thermodynamic parameters estimated in terms of the gel–fluid transition of MSC. P, thermodynamic parameters estimated in terms of the “coil–globule” transition of peptide-bioconjugated-copolymers. | |||
| MSC | 25.8 ± 0.1 | 8.66 ± 0.24 | 4.1 ± 0.3 |
| PDMA-b-PNIPAM-L220(l) | 51.6 ± 0.1 | 34.03 ± 2.24 | 8.2 ± 0.1 |
| PDMA-b-PNIPAM-L220(h) | 54.5 ± 0.1 | 13.97 ± 1.30 | 7.0 ± 0.1 |
| PDMA-b-PNIPAM-L130(l) | 52.7 ± 0.4 | 52.03 ± 3.52 | 9.0 ± 0.3 |
| PDMA-b-PNIPAM-L130(h) | 54.7 ± 0.1 | 52.17 ± 0.50 | 9.3 ± 0.1 |
| MSC/PDMA-b-PNIPAM-L220(l) L | 24.8 ± 0.3 | 10.68 ± 0.82 | 2.5 ± 0.3 |
| MSC/PDMA-b-PNIPAM-L220(l) P | 50.2 ± 0.3 | 36.46 ± 3.98 | 8.6 ± 1.1 |
| MSC/PDMA-b-PNIPAM-L220(h) L | 25.2 ± 0.7 | 9.11 ± 0.33 | 7.4 ± 0.4 |
| MSC/PDMA-b-PNIPAM-L220(h) P | 54.3 ± 0.4 | 12.09 ± 1.54 | 6.6 ± 0.6 |
| MSC/PDMA-b-PNIPAM-L130(l) L | 24.5 ± 0.1 | 8.37 ± 0.24 | 1.9 ± 0.1 |
| MSC/PDMA-b-PNIPAM-L130(l) P | 52.7 ± 0.7 | 55.91 ± 4.22 | 9.1 ± 0.5 |
| MSC/PDMA-b-PNIPAM-L130(h) L | 24.0 ± 0.1 | 8.17 ± 0.17 | 3.6 ± 0.4 |
| MSC/PDMA-b-PNIPAM-L130(h) P | 54.9 ± 0.2 | 38.52 ± 2.18 | 9.0 ± 0.2 |
| MSC/AMT | 23.7 ± 0.2 | 10.33 ± 0.61 | 2.8 ± 0.4 |
| MSC/AMT/PDMA-b-PNIPAM-L220(l) L | 22.7 ± 0.1 | 15.45 ± 0.83 | 1.9 ± 0.1 |
| MSC/AMT/PDMA-b-PNIPAM-L220(l) P | 46.9 ± 0.1 | 40.53 ± 2.11 | 6.0 ± 0.8 |
| MSC/AMT/PDMA-b-PNIPAM-L220(h) L | 26.3 ± 0.6 | 12.79 ± 0.63 | 7.6 ± 0.3 |
| MSC/AMT/PDMA-b-PNIPAM-L220(h) P | 52.5 ± 0.2 | 13.54 ± 1.30 | 7.4 ± 0.2 |
| MSC/AMT/PDMA-b-PNIPAM-L130(l) L | 23.4 ± 0.1 | 9.00 ± 0.17 | 2.8 ± 0.1 |
| MSC/AMT/PDMA-b-PNIPAM-L130(l) P | 51.4 ± 0.2 | 37.22 ± 2.52 | 7.4 ± 0.2 |
| MSC/AMT/PDMA-b-PNIPAM-L130(h) L | 22.1 ± 0.2 | 7.04 ± 0.95 | 2.6 ± 0.2 |
| MSC/AMT/PDMA-b-PNIPAM-L130(h) P | 52.9 ± 0.2 | 26.98 ± 2.32 | 6.5 ± 0.2 |
The endothermic phase transition observed for the peptide-bioconjugated block copolymers occurs reversibly and under equilibrium as suggested by repeated DSC measurements. PDMA-b-PNIPAM-L220(l) and PDMA-b-PNIPAM-L130(l) revealed a phase transition temperature of ca. 52 °C, which is slightly lower than that one observed for PDMA-b-PNIPAM-L220(h) and PDMA-b-PNIPAM-L130(h), Tm ∼ 54 °C (see Table 1). In this regard, it has been reported that the phase transition of PNIPAM occurs in a relatively broad temperature range from 30 to 40 °C.53 Moreover, the ΔH value of the investigated peptide-bioconjugated copolymer ranges from 13 to 52 kJ mol−1 (see Table 1). It is worth emphasizing that the thermo-responsiveness of PNIPAM discussed in polymer research often refers to a phase transition where this polymer in aqueous media at a specific concentration goes from a soluble to a non-soluble state upon heating, which is manifested like a change in the turbidity of the solution featuring the so-called lower critical solution temperature (LCST).54 The dehydration process that polymers undergo showing an LCST behavior implies a loss of hydrogen bonds between the polymer chains and water molecules, which involves an entropically driven endothermic process.54,55 This process exhibits hydrated coiled polymer chains/dehydrated globular polymer conformations, which are often referred to as “coil–globule” polymer transitions.54 In our investigation, Tm refers to the temperature value where Cp shows a maximum value in the DSC thermogram of the corresponding peptide-bioconjugated copolymer and does not only engulf the dehydration process of the PNIPAM block but also indicates that this phase transition is influenced by the PDMA segment, fluorescein units and the corresponding peptide sequence. This latter component may also feature a thermo-responsive behavior due to its composition of amino acids.
3.2 Thermal response and size stability of biohybrid liposomes decorated with bioconjugated copolymers
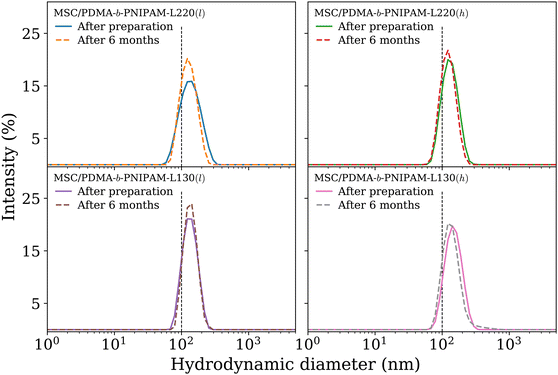
To improve the performance of liposome-based drug delivery systems, we hypothesize that combinations of phosphocholine, sphingomyelin, and cholesterol with block-copolymer bioconjugates could enhance not only the stability of liposome formulations derived thereof but also increase their specificity. In this section, we investigate the thermal effects induced by the presence of peptide-bioconjugated copolymers in MSC LUVs dispersed in a HEPES/NaCl buffer via DSC measurements. The corresponding endotherms and thermal parameters were analyzed considering both contributions derived from the gel–fluid phase transition of the phospholipid vesicles and the “coil–globule” polymer transition ascribed to the respective peptide-bioconjugate copolymer. The thermal responses of these biohybrid systems (Fig. 2) were compared with those discussed in the previous section, where MSC liposomes and the peptide-bioconjugated copolymers were individually evaluated. In general, it appears that the presence of bioconjugated block copolymers shifts Tm to a lower temperature (∼1–2 °C) and produces enthalpic changes (Fig. 2c and d), suggesting that the block copolymers are able to interact with the lipid bilayer. However, for the case of the MSC/PDMA-b-PNIPAM-L220(h) system, the shape of the calorimetric profile in the region of the lipid transition (20–36 °C) presents an attenuated-broadened peak. This result could indicate a different level of copolymer/bilayer interactions, where some moieties of PDMA-b-PNIPAM-L220(h) may be incorporated either totally or partially in the polar region of the surface of the lipid membrane. The effect on Tm is more clearly observed in Fig. 2c for the MSC/bioconjugated systems where the relative Tm is below 1.0. This relative value represents the ratio of the estimated DSC parameter (Tm or ΔH) of the MSC/bioconjugated systems divided by the thermal parameter of either pure MSC or the respective pure bioconjugated copolymer. The system with the highest effect on Tm was MSC/PDMA-b-PNIPAM-L130(h). However, this system did not reveal any significant change in the transition of the “coil–globule” copolymer counterpart. Regarding the observed ΔH changes, note that PDMA-b-PNIPAM block copolymers functionalized with peptide L220 (Arg–Asp–Gln–Glu–Gly) yielded a higher calorimetric ΔH value than that observed for the transition of the lipid counterpart. In contrast, those copolymers functionalized with peptide L130 (Ala–Cys–Pro–His) displayed a lower value for this thermodynamic parameter. Indeed, these changes in ΔH values suggest that copolymer moieties are adsorbed onto the lipid membrane at the polar regions of the bilayer. Considering that the L220 or L130 peptide functionality represents the main difference between the four investigated bioconjugated copolymers, and being the nature of these amino acid sequences polar, it is reasonable to assume that these functionalities play an important role in the interaction between the copolymers and the polar region of the surface of the MSC lipid bilayer. Furthermore, n-dodecyl (C12H25) (i.e., hydrophobic moiety) attached to the RAFT agent could favor the anchoring of the bioconjugated copolymers onto the lipid membrane. In summary, these results suggest that these bioconjugated copolymers are preferably located at the surface of the lipid bilayer.To evaluate the size stability of the MSC/bioconjugated systems over time, we carried out DLS experiments. First, MSC/bioconjugated samples were measured after preparation, and then they were maintained in storage in a refrigerator (4 °C) for a long period (months). The size of these systems was monitored at different times over the storage. Fig. 3 displays the hydrodynamic diameter distributions for the MSC/bioconjugated systems. It is worth noting that the size of these biohybrid liposomes remains stable for at least six months.
These findings might be useful to enhance the performance of formulations derived from these proposed systems in biomedical applications. As such, these nanocarrier systems are thermoresponsive and highly stable over time and contain features to potentially improve cell-specificity. Due to the nature of the lipid and peptide moieties used to self-assemble these biohybrid vesicles, we believe that these systems possess biomimetic and biocompatible capabilities, which make them attractive candidates for potential medical applications. Furthermore, the size distributions in terms of both intensity and volume corroborate the presence of only one size population in the samples. This fact suggests that the bioconjugated block copolymers preferably interact with the lipid membrane and do not produce bulk aggregates as observed for the individual bioconjugated copolymers (i.e., in the absence of liposomes).
3.3 MSC/bioconjugated bilayer in interaction with amantadine
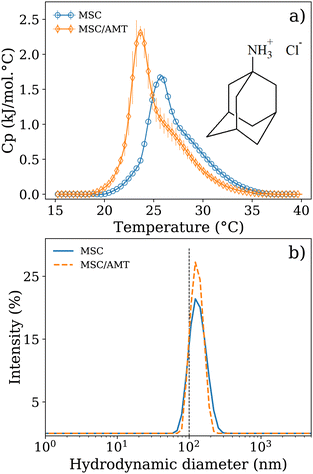
In the above section, we presented biophysical properties of MSC/bioconjugated copolymer systems, which revealed that the bioconjugated copolymers preferably interact with the surface of the lipid bilayer enabling the successful formation of self-assembled biohybrid LUVs. These biohybrid vesicles revealed a high stability in size for a considerable period of time and possess interesting properties for potential biological applications. These observations next to the intrinsic cell-targeting features of the utilized bioconjugated copolymers suggest that the proposed systems might be potentially good candidates for smart drug delivery carriers. For this reason, we have formulated MSC/bioconjugated copolymer nanovesicles in the presence of the antiviral/antiparkinsonian compound amantadine (AMT). AMT is a tricyclic amine (see chemical structure in Fig. 4) with lipophilic properties and exerts its antiviral activity by blocking the function of the M2 proton channel in a virus, which prevents virus replication and virus membrane fusion.56–58 On the other hand, the antiparkinsonian effect of AMT is related to the modulation of the dopamine system and the inhibition of the N-methyl-D-aspartic acid (NMDA) receptor.59 However, interesting results have also been reported for AMT in interaction with lipid membranes.57,58 Hence, we herein report our first investigations of interactions between AMT and a lipid DMPC/SM/chol membrane with improved properties via the incorporation of the proposed bioconjugated block copolymers that may mimic a viral tropism mechanism.To evaluate whether AMT interacts with an MSC lipid membrane, a suspension of MSC LUVs in the presence of AMT was prepared in a HEPES/NaCl buffer solution at a pH value of 7.4, whose thermal response was evaluated via DSC. The AMT/MSC ratio was equal to 6.6, (i.e., r = AMT (10 mM)/MSC (1.5 mM)). Fig. 4a reveals that AMT is incorporated into the MSC bilayer as suggested by the differences between the displayed calorimetric profiles, where Tm shifts to a lower temperature in the presence of AMT. The calorimetric enthalpy also increased for the MSC LUVs containing AMT (Table 1). In this respect, a shifting of Tm toward lower temperatures together with an increment in ΔH indicates that AMT can interact with the lipid molecules of the MSC membrane via electrostatic (at the “head”) and hydrophobic interactions (at the “tail”). The preferential location of AMT at the hydrophilic/hydrophobic lipid interface has been described by Conggang Li and coworkers60 using NMR and molecular dynamic simulation. They reported that AMT is located parallel to the bilayer normal, where its amine group aligns toward the head of a DMPC bilayer. Furthermore, as expected, DLS experiments in Fig. 4b also show that MSC and MSC/AMT LUVs have a very similar size distribution centered around 100 nm. We also evaluate some additional biophysical properties of the MSC/AMT LUVs with improved characteristics via the incorporation of peptide-bioconjugated PDMA-b-PNIPAM block copolymers.
As expected, endotherms of MSC/bioconjugated LUVs containing AMT suggest that AMT is encapsulated into the hybrid vesicles. Fig. 5(a)–(d) compare the calorimetric profiles of the four MSC/bioconjugated systems, which show the effect of adding AMT in the gel–fluid MSC transition as well as in the “coil–globule” bioconjugated block copolymer transition. These results indicate that the system with the lower impact is the MSC/AMT/PDMA-b-PNIPAM-L220(h), suggesting that this system may be less effective in preventing a prompt release of AMT. As observed, AMT tends to shift Tm of the gel–fluid phase transition to lower values, which is an indication that at least a fraction of the drug is incorporated into the lipid bilayer. Complementarily, when the effect of the presence of AMT in the MSC/bioconjugated systems is evaluated considering the “coil–globule” polymer transition, we can also observe that AMT shifts Tm to lower values, which suggests that AMT also incorporates into the bioconjugated copolymer domains. We may speculate that the excess of AMT that was not incorporated into the lipid membrane may be encapsulated into the copolymer envelope as indicated by the endotherms at the right-hand side of the DSC thermograms of Fig. 5. Thus, the MSC and the peptide-bioconjugated block copolymers might synergistically cooperate to make AMT encapsulation more efficient. Furthermore, we monitored the stability of the MSC/AMT/bioconjugated systems via DLS measurements of the size distribution of freshly prepared samples and after six months of being stored at 4 °C. Fig. 5(e)–(h) demonstrate that the MSC/AMT/bioconjugated formulations revealed a “long-term” stability (i.e., up to 6 months), which suggests their potential use for drug delivery applications.
3.4 Morphological characterization, in vitro cytotoxicity, and direct immunofluorescence for MSC and MSC/AMT liposomes decorated with bioconjugated copolymers
In addition, cryoTEM investigations revealed important morphological aspects of the biohybrid liposomes of MSC (Fig. S6, ESI†) and the MSC/AMT systems decorated with the bioconjugated copolymers (Fig. 6). Micrographs of the biohybrid liposomes confirm the formation of vesicles with a size of ca. 100 nm in agreement with the previously discussed DLS results. The micrographs also revealed the presence of vesicles of a smaller size in the samples containing PDMA-b-PNIPAM-L220(h) as compared to the other investigated systems. Moreover, the cryoTEM images corresponding to the biohybrid systems show the coexistence of vesicles with a wider peripheral thickness than those observed for pure MSC and MSC/AMT samples (some examples are pointed out by red arrows in Fig. S6 and Fig. 6, ESI†). In addition, the coexistence of multilamellar vesicles along with single vesicles can be mainly observed for systems containing the block copolymer of lower molar mass and in the absence of AMT (Fig. S6, ESI†). This suggests that the presence of AMT may also have an influence on the morphology of the investigated biohybrid systems.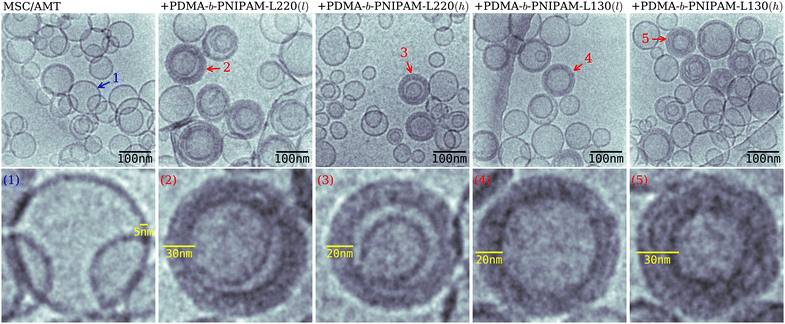 | ||
| Fig. 6 Representative cryoTEM micrographs of MSC/AMT liposomes decorated with bioconjugated block copolymers. | ||
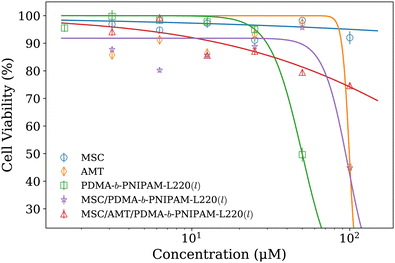
Furthermore, the results of performed cytotoxicity assays showed that pure PDMA-b-PNIPAM-L220(l) was the most cytotoxic case of the investigated systems (CC50 = 50 μM) followed by AMT (CC50 = 99 μM). In contrast, bare MSC liposomes were not cytotoxic even at the highest tested concentration of 100 μM (Fig. 7). However, the hybrid system MSC/PDMA-b-PNIPAM-L220(l) shows an attenuated cytotoxic effect (CC50 = 94 μM). And, the MSC/AMT/PDMA-b-PNIPAM-L220(l) system reveals a lower cytotoxicity with an estimated CC50 value of 569 μM. Balestri and coworkers61 recently reported a similar effect in PDMA-b-PNIPAM stabilized cubosomes, where the higher lipophilicity of the stabilizer plays a preponderant role in the internalization and cytotoxicity of this kind of nanovectors proposed as vehicles to deliver anticancer drugs. This finding further supports the potential applications of these biohybrid systems as drug delivery platforms.
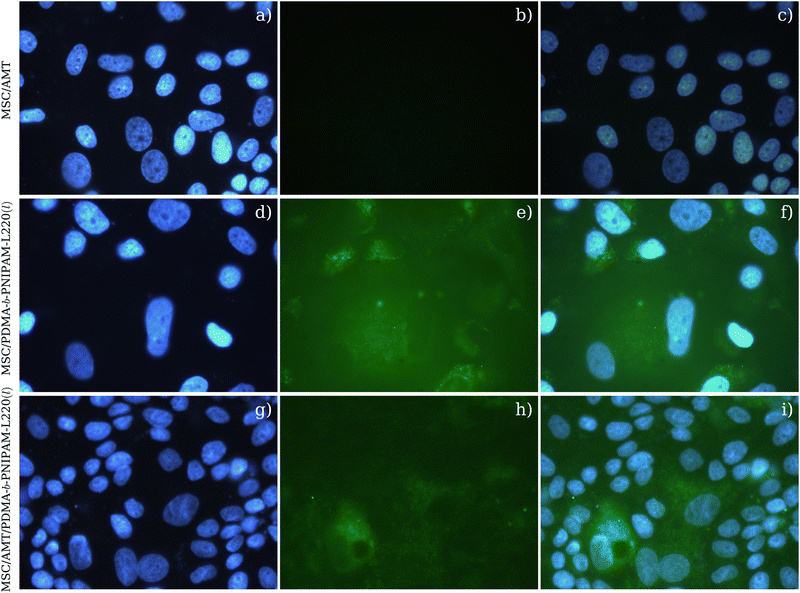
To evaluate if the biohybrid systems can be absorbed in the plasma membrane and reach the cell cytoplasm, we carried out immunofluorescence assays in MDCK cells. Once CC50 concentrations were determined, direct immunofluorescence assays were performed for the MSC/AMT, MSC/PDMA-b-PNIPAM-L220(l), and MSC/AMT/PDMA-b-PNIPAM-L220(l) systems at a concentration of 50 μM. As expected, the results showed that the MSC/AMT system does not exhibit bright fluorescence; in contrast, in the MSC/PDMA-b-PNIPAM-L220(l) and MSC/AMT/PDMA-b-PNIPAM-L220(l) systems, fluorescence is observed as small green dots distributed throughout the cell cytoplasm, in some cells, the accumulation of fluorescent particles closest to the nucleus is visible (Fig. 8f and i). These results confirm that liposome systems decorated with bioconjugated copolymers enhance preponderant features for biomedical applications.
From this perspective, several studies have well reviewed the relevance of progress toward the engineering of smart materials. For example, Meyer and coworkers6 presented an interesting review related to the advantages of biohybrid materials resulting from the combination of biomolecules such as proteins and lipids with synthetic polymeric arrays. Chen and coworkers8 summarized how polymer bioconjugates and their combination with biological molecules represent superior materials for the development of new technologies for biomedical applications. Also, Moulahoum et al.17 recognized that one of the main problems of nanocarriers based on nanovesicles such as liposomes and polymersomes is the passive way that they deliver their content, which leads to prompt degradation and random tissue interactions. They discuss that liposomes and polymersomes bioconjugated with antibodies potentially provide a high level of specificity. Furthermore, Fang-Yi and coworkers62 demonstrated that biohybrid systems composed of polymers and liposomes can be engineered with targeting and pH responses to deliver antibiotics. It has been reported that the ion traffic in hybrid lipid/polymer vesicles can be modulated, which is a relevant point for considering the development of technologies for drug delivery.63 Interestingly, Pippa et al.64 reported that biohybrid lipid/polymer systems composed of phospholipids (DPPC and HSPC) and poly(oligoethylene glycol acrylate)-b-poly(lauryl acrylate) have important physicochemical, biological, and low cytotoxicity properties to be suitable for nanocarriers. Such systems were demonstrated to be stable for weeks in aqueous dispersion media. This group also reported that phosphatidylcholines such as DPPC, DMPC, and DSPC can lead to the formation of polymer–lipid nanostructures when interacting with poly(2-(dimethylamino)ethyl methacrylate)-b-poly(lauryl methacrylate), where Tm of the phospholipid determines the final morphology and physicochemical properties of the hybrid systems.65
4 Conclusions
The investigation of biohybrid materials to improve the controlled release of drugs has gained extensive attention. The purpose of the present work was to design, assemble, and investigate the properties of innovative platforms for biomedical applications. First, the synthesis of block copolymer-bioconjugates with differences in composition, molar mass, and specific features was carried out by reversible–addition–fragmentation chain-transfer polymerization, followed by their characterization using proton nuclear magnetic resonance and size-exclusion chromatography. Second, a systematic physical characterization of liposome systems decorated with bioconjugated copolymers was performed via DSC, DLS and cryoTEM. Finally, such biohybrid systems were subjected to an in vitro study to evaluate cytotoxicity and direct immunofluorescence. Such copolymer/lipid hybrid platforms reveal important biophysical and in vitro properties that could be of practical relevance for improving drug delivery systems mimicking viral tropism as observed in the influenza A virus via its corresponding HA protein. These nanovesicles possess excellent biocompatibility and stability over time (at least for six months) and have potential target capabilities, which are preponderant features for the designing of effective drug nanocarrier systems for encapsulating hydrophobic molecules such as antiviral amantadine. In future work, we look forward to extending this investigation to characterize encapsulation efficiency and kinetics of drug delivery.Author contributions
Conceptualization: RPI, CGS, and ESG; synthesis and analysis of polymers: MAJT and CGS; methodology and data analysis: AGVL, AJDS, MAJT, GAT, SH, and RPI; original draft preparation: RPI, AGVL, CGS, GAT, and MAJT; review and editing: AGVL, MAJT, AJDS, CGS, JCRS, USS, PQO, RPI, GAT, and ESG; resources: RPI, CGS, USS, JCRS, PQO, GAT, and ESG. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
RPI and MAJT acknowledge the Consejo Nacional de Humanidades, Ciencias y Tecnologias (CONAHCYT, Mexico) for the corresponding postdoctoral grants through the programme Estancias Posdoctorales por México (project numbers 3969865 and 76219). AGVL thanks CONAHCYT for the financial support provided via a PhD scholarship. We thank CIQA-Unidad Monterrey for providing laboratory infrastructure. This work was also supported by the Deutsche Forschungsgemeinschaft (DFG, Germany) and the Collaborative Research Center PolyTarget (SFB 1278 – grant number 316213987, projects A01, B02 and Z01). Transmission electron microscopy facilities of the Jena Center of Soft Matter (JCSM) were established with a grant from the DFG and the European Funds for Regional Development (EFRE).References
- C. Alvarez-Lorenzo and A. Concheiro, Chem. Commun., 2014, 50, 7743–7765 RSC
.
- T. Ahmed, F. C. F. Liu, B. Lu, H. Lip, E. Park, I. Alradwan, J. F. Liu, C. He, A. Zetrini, T. Zhang, A. Ghavaminejad, A. M. Rauth, J. T. Henderson and X. Y. Wu, Mol. Pharmaceutics, 2022, 19, 1722–1765 CrossRef CAS PubMed
.
- M. Kumar, A. Jha, K. Bharti, G. Parmar and B. Mishra, Nanomedicine, 2022, 17, 913–934 CrossRef CAS PubMed
.
-
K. Karsauliya, S. P. Singh and M. Sharma, Smart Nanodevices for Point-of-Care Applications, CRC Press, 2022, pp. 157–168 Search PubMed
.
- T. Róg, M. Girych and A. Bunker, Pharmaceuticals, 2021, 14, 1062 CrossRef PubMed
.
- C. E. Meyer, S. L. Abram, I. Craciun and C. G. Palivan, Phys. Chem. Chem. Phys., 2020, 22, 11197–11218 RSC
.
- V. D. Leo, F. Milano, A. Agostiano and L. Catucci, Polymers, 2021, 13, 1027 CrossRef PubMed
.
- C. Chen, D. Yuen, W. Ng and T. Weil, Prog. Polym. Sci., 2020, 105, 101241 CrossRef CAS
.
- A. Krywko-cendrowska, S. Di Leone, M. Bina, S. Yorulmaz-Avsar, C. G. Palivan and W. Meier, Polymers, 2020, 12, 1003 CrossRef CAS PubMed
.
- D. L. Gbian and A. Omri, Biomedicines, 2022, 10, 2137 CrossRef CAS PubMed
.
- S. Shah, P. Famta, D. Bagasariya, K. Charankumar, E. Amulya, D. Kumar Khatri, R. Singh Raghuvanshi, S. Bala Singh and S. Srivastava, Int. J. Pharm., 2022, 625, 122101 CrossRef CAS PubMed
.
- A. A. Smith, K. Zuwala, M. B. Kryger, B. M. Wohl, C. Guerrero-Sanchez, M. Tolstrup, A. Postma and A. N. Zelikin, Chem. Sci., 2015, 6, 264–269 RSC
.
- C. Chen, F. Richter, C. Guerrero-Sanchez, A. Traeger, U. S. Schubert, A. Feng and S. H. Thang, ACS Macro Lett., 2020, 9, 260–265 CrossRef CAS PubMed
.
- M. A. De Jesús-Téllez, S. De la Rosa-García, I. Medrano-Galindo, I. Rosales-Peñafiel, S. Gómez-Cornelio, C. Guerrero-Sanchez, U. S. Schubert and P. Quintana-Owen, React. Funct. Polym., 2021, 163, 104887 CrossRef
.
- T. M. Hinton, A. Challagulla, C. R. Stewart, C. Guerrero-Sanchez, F. A. Grusche, S. Shi, A. G. Bean, P. Monaghan, P. A. Gunatillake, S. H. Thang and M. L. Tizard, Nanomedicine, 2014, 9, 1141–1154 CrossRef CAS PubMed
.
- V. Adibnia, M. Mirbagheri, S. Salimi, G. De Crescenzo and X. Banquy, Curr. Opin. Colloid Interface Sci., 2020, 47, 70–83 CrossRef CAS
.
- H. Moulahoum, F. Ghorbanizamani, F. Zihnioglu and S. Timur, Bioconjugate Chem., 2021, 32, 1491–1502 CrossRef CAS PubMed
.
- Y. Gao, K. Yang, A. N. Shelling and Z. Wu, Encyclopedia, 2021, 1, 773–780 CrossRef
.
- Z. R. Sia, X. He, A. Zhang, J. C. Ang, S. Shao, A. Seffouh, W.-C. Huang, M. R. D’Agostino, A. Teimouri Dereshgi, S. Suryaprakash, J. Ortega, H. Andersen, M. S. Miller, B. A. Davidson and J. F. Lovell, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2025759118 CrossRef CAS PubMed
.
- M. Schulz and W. H. Binder, Macromol. Rapid Commun., 2015, 36, 2031–2041 CrossRef CAS PubMed
.
- J. F. Le Meins, C. Schatz, S. Lecommandoux and O. Sandre, Mater. Today, 2013, 16, 397–402 CrossRef CAS
.
- D. Guimarães, A. Cavaco-Paulo and E. Nogueira, Int. J. Pharm., 2021, 601, 120571 CrossRef PubMed
.
- S. Shah, V. Dhawan, R. Holm, M. S. Nagarsenker and Y. Perrie, Adv. Drug Delivery Rev., 2020, 154–155, 102–122 CrossRef CAS PubMed
.
- T. M. Allen and P. R. Cullis, Adv. Drug Delivery Rev., 2013, 65, 36–48 CrossRef CAS PubMed
.
- T. Hernández-Caselles, J. Villalaín and J. C. Gómez-Fernández, J. Pharm. Pharmacol., 1990, 42, 397–400 CrossRef PubMed
.
- A. Akbarzadeh, R. Rezaei-Sadabady, S. Davaran, S. W. Joo, N. Zarghami, Y. Hanifehpour, M. Samiei, M. Kouhi and K. Nejati-Koshki, Nanoscale Res. Lett., 2013, 8, 102 CrossRef PubMed
.
- A. Jash, A. Ubeyitogullari and S. S. Rizvi, J. Mater. Chem. B, 2021, 9, 4773–4792 RSC
.
- M. Xiaomei and Z. Zhensheng, Int. J. Pharm., 2006, 318, 55–61 CrossRef PubMed
.
- C. Chen, F. Richter, J. Zhang, C. Guerrero-Sanchez, A. Traeger, U. S. Schubert, A. Feng and S. H. Thang, Eur. Polym. J., 2021, 160, 110777 CrossRef CAS
.
- K. Morizono and I. S. Y. Chen, Curr. Opin. Virol., 2011, 1, 13–18 CrossRef CAS PubMed
.
- G. Mcfadden, M. R. Mohamed, M. M. Rahman and E. Bartee, Nat. Rev. Immunol., 2009, 9, 645–655 CrossRef CAS PubMed
.
- N. Sriwilaijaroen and Y. Suzuki, Proc. Jpn. Acad., Ser. B, 2012, 88, 226–249 CrossRef CAS PubMed
.
- G. Ayora-Talavera, J. Recept., Ligand Channel Res., 2018, 10, 1–11 CrossRef
.
- A. Balestri, B. Lonetti, S. Harrisson, B. Farias-Mancilla, J. Zhang, H. Amenitsch, U. S. Schubert, C. Guerrero-Sanchez, C. Montis and D. Berti, Colloids Surf., B, 2022, 220, 112884 CrossRef CAS PubMed
.
- T. Róg and I. Vattulainen, Chem. Phys. Lipids, 2014, 184, 82–104 CrossRef PubMed
.
- E. Sezgin, I. Levental, S. Mayor and C. Eggeling, Nat. Rev. Mol. Cell Biol., 2017, 18, 361–374 CrossRef CAS PubMed
.
- A. K. Dunker, I. Silman, V. N. Uversky and J. L. Sussman, Curr. Opin. Struct. Biol., 2008, 18, 756–764 CrossRef CAS PubMed
.
- K. Simons and E. Ikonen, Nature, 1997, 387, 569–572 CrossRef CAS PubMed
.
- A. Keyvanloo, M. Shaghaghi, M. J. Zuckermann and J. L. Thewalt, Biophys. J., 2018, 114, 1344–1356 CrossRef CAS PubMed
.
- R. Pérez-Isidoro, A. J. Díaz-Salazar and M. Costas, J. Therm. Anal. Calorim., 2024, 149, 1219–1229 CrossRef
.
- W. A. Braunecker and K. Matyjaszewski, Progress Polym. Sci., 2007, 32, 93–146 CrossRef CAS
.
- E. Oropeza-Guzman and J. C. Ruiz-Suárez, Langmuir, 2018, 34, 6869–6873 CrossRef CAS PubMed
.
- J. González-Gutiérrez, R. Pérez-Isidoro, M. I. Pérez-Camacho and J. C. Ruiz-Suárez, Colloids Surf., B, 2017, 155, 215–222 CrossRef PubMed
.
- R. Pérez-Isidoro, F. J. Guevara-Pantoja, C. Ventura-Hunter, C. Guerrero-Sánchez, J. C. Ruiz-Suárez, U. S. Schubert and E. Saldívar-Guerra, Biochim. Biophys. Acta, Gen. Subj., 2023, 1867, 130287 CrossRef PubMed
.
- M. Newville, T. Stensitzki, D. B. Allen, M. Rawlik, A. Ingargiola and A. Nelson, Astrophysics Source Code Library, 2016, ascl-1606 Search PubMed
.
- M. Guimarães Sá Correia, M. L. Briuglia, F. Niosi and D. A. Lamprou, Int. J. Pharm., 2017, 516, 91–99 CrossRef PubMed
.
- M. L. Ohnsorg, J. M. Ting, S. D. Jones, S. Jung, F. S. Bates and T. M. Reineke, Polym. Chem., 2019, 10, 3469–3479 RSC
.
- K. Fischer and M. Schmidt, Biomaterials, 2016, 98, 79–91 CrossRef CAS PubMed
.
- P. M. Carvalho, M. R. Felício, N. C. Santos, S. Gonçalves and M. M. Domingues, Front. Chem., 2018, 6, 1–17 CrossRef PubMed
.
- D. A. Mannock, R. N. Lewis and R. N. McElhaney, Biophys. J., 2006, 91, 3327–3340 CrossRef CAS PubMed
.
- A. Pralle, P. Keller, E.-L. Florin, K. Simons and J. H. Hörber, J. Cell Biol., 2000, 148, 997–1008 CrossRef CAS PubMed
.
- H. Li, M. Li, X. Yu, A. P. Bapat and B. S. Sumerlin, Polym. Chem., 2011, 2, 1531–1535 RSC
.
- A. Halperin, M. Kröger and F. M. Winnik, Angew. Chem., Int. Ed., 2015, 54, 15342–15367 CrossRef CAS PubMed
.
- Q. Zhang, C. Weber, U. S. Schubert and R. Hoogenboom, Mater. Horiz., 2017, 4, 109–116 RSC
.
- M. Sponchioni, U. Capasso Palmiero and D. Moscatelli, Mater. Sci. Eng., C, 2019, 102, 589–605 CrossRef CAS PubMed
.
- A. J. Hay, A. J. Wolstenholme, J. J. Skehel and M. H. Smith, EMBO J., 1985, 4, 3021–3024 CrossRef CAS PubMed
.
- M. Suwalsky, M. Jemiola-rzeminska, M. Altamirano, F. Villena, N. Dukes and K. Strzalka, Biophys. Chem., 2015, 202, 13–20 CrossRef CAS PubMed
.
- S. D. Cady, T. V. Mishanina and M. Hong, J. Mol. Biol., 2009, 385, 1127–1141 CrossRef CAS PubMed
.
- T. Müller, W. Kuhn and J. D. Möhr, Expert Opin. Pharmacother., 2019, 20, 1181–1187 CrossRef PubMed
.
- C. Li, M. Yi, J. Hu, H.-X. Zhou and T. A. Cross, Biophys. J., 2008, 94, 1295–1302 CrossRef CAS PubMed
.
- A. Balestri, L. Gibot, H. Amenitisch, L. Cervelli, C. Montis, B. Lonetti and D. Berti, Colloids Surf., B, 2023, 231, 113532 CrossRef CAS PubMed
.
- F.-Y. Su, J. Chen, H.-N. Son, A. M. Kelly, A. J. Convertine, T. E. West, S. J. Skerrett, D. M. Ratner and P. S. Stayton, Biomater. Sci., 2018, 6, 1976–1985 RSC
.
- W. F. Paxton, P. T. McAninch, K. E. Achyuthan, S. H. R. Shin and H. L. Monteith, Colloids Surf., B, 2017, 159, 268–276 CrossRef CAS PubMed
.
- N. Pippa, D. Stellas, A. Skandalis, S. Pispas, C. Demetzos, M. Libera, A. Marcinkowski and B. Trzebicka, Eur. J. Pharm. Biopharm., 2016, 107, 295–309 CrossRef CAS PubMed
.
- M. Chountoulesi, D. R. Perinelli, A. Forys, H. Katifelis, D. Selianitis, V. Chrysostomou, N. Lagopati, G. Bonacucina, B. Trzebicka and M. Gazouli,
et al.
, J. Drug Delivery Sci. Technol., 2022, 77, 103830 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis and characterization of bioconjugated copolymers. CryoTEM images of biohybrid liposomes decorated with bioconjugated copolymers. See DOI: https://doi.org/10.1039/d4tb00171k |
| This journal is © The Royal Society of Chemistry 2024 |