Open Access Article
Open Access ArticleThriving in extremes: harnessing the potential of pH-resilient algal strains for enhanced productivity and stability
Neha Arora *ab,
Shweta Tripathic,
George P. Philippidisbd and
Shashi Kumarc
*ab,
Shweta Tripathic,
George P. Philippidisbd and
Shashi Kumarc
aDepartment of Biology, Skidmore College, Saratoga Springs, NY 12866, USA. E-mail: nehaarora@skidmore.edu; nehaarora@usf.edu
bPatel College of Global Sustainability, University of South Florida, Tampa, 33620, USA
cInternational Centre for Genetic Engineering and Biotechnology, New Delhi, 110067, India
dDepartment of Integrative Biology, University of South Florida, Tampa, 33620, USA
First published on 18th February 2025
Abstract
Algal biomass can play a multifaceted role in advancing the sustainable developmental goals (SDGs) as a means of carbon sequestration and waste mitigation. Outdoor algal cultivation, typically conducted in open raceway ponds, while a cost-effective approach for biofuel and bioproduct production, suffers from several challenges, including weather variability, contamination, nutrient mixing, and challenges in harvesting and dewatering. Notably, large-scale cultivation of neutrophilic algae grown at pH 7 necessitates pH stabilization measures due to fluctuations induced by CO2 uptake, nutrient concentration, photosynthesis, and competing microbial activity, resulting in significant operating costs. The exploitation of pH-resilient algae encompassing acidophilic, acid-tolerant, alkaliphilic, and alkali-tolerant strains can maximize growth and productivity across a wide range of pH from acidic to alkaline. As a result, the repertoire of water sources used for cultivation can be expanded to include wastewater treatment and industrial effluents, reducing use of scarce freshwater and dependence on costly pH regulation measures. Extremophilic strains possess the intrinsic capacity to withstand pH fluctuations that limit invaders, hence minimizing culture crashes. In the present review we highlight the unique adaptations of pH-resilient algal strains that can strengthen the resilience of large-scale algal cultivations and overcome the challenges of outdoor operations. We delve into the pH adaptation mechanism of extremophilic algae and their applicability in diverse fields of bioremediation, carbon capture, and bioproduct manufacture. Recent strides in strain improvement for enhancing the metabolic prowess of pH-resilient algae have been discussed, emphasizing their critical role towards shaping the future of a sustainable bioeconomy.
Environmental significancePhotosynthetic algae can provide sustainable solutions across various sectors, including food security, health, clean energy, water treatment, and environmental conservation. However, large scale outdoor cultivation of neutrophilic algae in open raceway ponds offen suffers from low biomass productivity due to fluctuations in temperature, light, pH, and salinity. Moreover, the outdoor cultivation systems are more prone to contamination by invaders leading to frequent culture crashes. In this regard, pH-resilient algae capable of surviving in extreme pH are often more tolerant to other abiotic and biotic stressors resulting in higher productivity. Thanks to their unique metabolism, these algae can be exploited for several applications such as wastewater remediation, CO2 sequestration, biofuels and generation of value-added compounds. The present review highlights the potential of these pH-resilient algae for driving the transition to a sustainable algae-based economy. |
1. Introduction
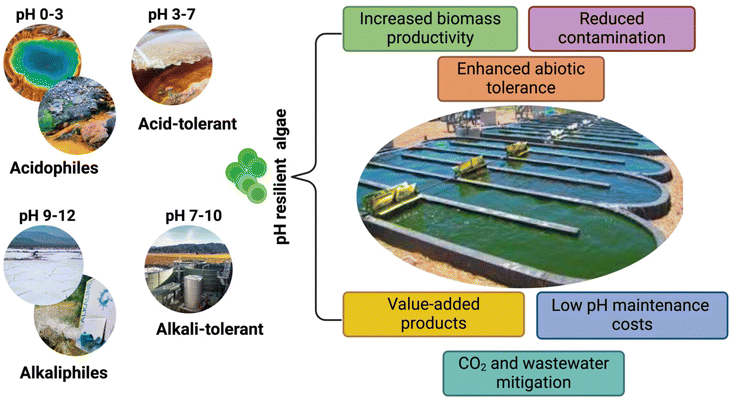
Photosynthetic algae are a promising enabler to fulfilling the 5 Fs of the sustainable developmental goals (SDGs), namely food, fiber, fuel, feed, and fertilizer.1 Flourishing across a range of habitats in freshwater, marine ecosystems, and extreme environments, like hot springs, polar ice, soda lakes, acidic mines, and alkaline wastewaters, is testament to their impressive robustness and evolutionary adjustment. In particular, extremophilic algae play a vital role as primary food producers in harsh environments forming the basis of the food web and supporting diverse microbial communities and nutrient recycling. The value of algal extremophiles lies in the diversity of their metabolic make-up that helps them survive in harsh surroundings. Among the extremophiles, algae capable of tolerating either acidic (pH 0–5) or alkaline conditions (pH 9–12) harbor immense potential to address several global concerns related to wastewater mitigation, CO2 capture, and production of renewable fuels and biomaterials. They have the upper hand over their neutrophile counterparts particularly in outdoor large-scale cultivation, which traditionally suffer from low productivity due to a higher incidence of contamination by invaders and the constant need to stabilize pH near neutrality via costly chemicals and infrastructure. The key advantages of cultivating pH resilient algae outdoors for biotechnological applications is due to their ability to maintain optimum growth even under fluctuating pH and CO2, ensuring higher productivity (Fig. 1). Moreover, algae cultivated at low pH (0–3) or high pH (9–14) grow in essence free of undesired neutrophilic microorganisms, such as bacteria, yeast, and grazers, thus reducing expensive culture crashes (Fig. 1).2 Moreover, these pH-resilient algae exhibit tolerance towards other abiotic stressors, such as fluctuations in temperature, light, and salinity, making them a preferrable choice for low-cost sustainable algal biomass production.Algae that can thrive in pH between 0–3, but grow poorly at neutral pH are termed acidophilic, while species able to grow at pH as low as 4, but can also grow at neutral pH are called acid-tolerant.3 Extreme acidic conditions (pH 0–3) are typical in natural volcanic waters and fumaroles, which are rich in elemental sulfur and sulfides along with high concentrations of CO2 and H2S.4 The inhabitants of such environments are very limited to a few archaebacterial, fungal, and algal species. Several mesophilic acidophilic algae, like Euglena spp., Chlorella spp., Chlamydomonas acidophila, Ulothrix zonata and Klebsormidium fluitans, have been reported in such acidic environments.4 Notably, the unicellular red alga Galdieria sulphuraria, isolated from geothermal acidic springs of the Yellowstone National Park in the USA, is reported to grow at pH 0.5–4.0 and temperature 42–45 °C.5 Due to its unique metabolic flexibility, this red alga has been extensively researched for wastewater mitigation and value-added products.6 On the other hand, anthropogenic low-pH wastewaters originating from coal and metal mines rich is sulfuric acid and ferric iron may consist of both acidophilic and acid-tolerant algae.7 Algal strains, such as Spirulina sp., Chlorella, Scenedesmus, Cladophora, Oscillatoria, Anabaena, and Phaeodactylum tricornutum, have been reported to thrive in acid mine wastewaters in addition to exhibiting excellent remediation potential for these recalcitrant effluents.8
Likewise, algae that exhibit good growth at pH values between 10 and 11, but cannot grow at neutral pH, are known as alkaliphilic, while alkali-tolerant algae can grow at pH values higher than 9, but can grow equally well at neutral pH.9 Natural highly alkaline environments, such as soda lakes and alkaline springs and soils, provide unique habitats for several extremophiles. In particular, soda lakes have been reported as one of the most productive ecosystems despite their extreme alkaline pH and salinity due to the increased light penetration and high nitrogen, phosphorous, and bicarbonate content.10,11 Such an environment is perfect for harboring several alkaliphilic algal species, including Spirulina sp., Chlorella spp., Dunaliella spp. Chlamydomonas spp., and diatoms, which exhibit increased tolerance to both pH and salinity.9 Moreover, large industrial processes, including textile, paper, chemical, and agricultural run offs, have created man-made alkaline wastewaters that also harbor alkaliphilic and alkali-tolerant algae, which provide a practical means for remediating these toxic wastewaters.
The biotechnological applications of these pH-resilient algae expand across wastewater treatment, carbon sequestration, and production of a spectrum of renewable commodities. In particular, to achieve high biomass production in algal cultivation systems, pure CO2 gas is typically sparged into the cultures at an estimated cost of $1.47–$7.33 per kg in addition to infrastructure costs.12 Instead, the use of CO2 emissions in industrial flue gases for cultivating algal biomass can help reduce costs and also assist in achieving carbon neutrality. While common neutrophilic algae growing at pH 6–7 often suffer from media acidification over time and are inhibited by flue gases having >2% CO2, acidophilic/acid-tolerant algae offer the advantage of CO2 tolerance by limiting their carbon concentration mechanism (CCM), while enhancing cellular energy (ATP) production, elevating proton pumping, and remodeling the membranal fatty acid constituents to maintain neutral cytosolic pH.13 On the other hand, at alkaline pH > 10, bicarbonate is the main carbon source available for algal uptake and has been proposed as one of the best alternatives to CO2 sparging.12 Moreover, conversion of captured CO2 to bicarbonate/carbonate facilitates its transport via pipelines operating under normal pressure.14 As opposed to neutrophilic algae, alkaliphilic and alkali-tolerant algal strains are reported to tolerate high levels of bicarbonate in the media, which significantly enhances their growth. Two well-known algae, Spirulina sp., and Dunaliella sp., are already commercially exploited for high protein (as animal feed or functional food ingredient) and β-carotene content (nutraceutical), respectively, thanks to their high-pH tolerance resulting in highly productive and stable outdoor operations.12 Such CO2- and carbonate-induced metabolic remodeling and carbon rerouting in low- and high-pH resilient algae, respectively, can be leveraged towards production of commercially important renewable materials, like fatty acids for biofuels or nutraceuticals and carotenoids for nutraceuticals. Furthermore, it has been reported that 1.238 billion gallons of water and 564 million kg of nitrogen are required to produce 1 billion gallons of biodiesel from microalgal biomass.15 Thus, integrating treatment of acidic/alkaline and heavy metal containing wastewaters from a variety of industrial activities with biodiesel could turn major environmental liabilities into productive assets.
In this review, we provide a critical review of the adaptive features and evolved mechanisms of pH-resilient microalgae including cyanobacteria, highlight their metabolic flexibility, and identify putative genetic targets for pursuing higher algal productivity in the future. Based on this context, we examine promising biotechnological applications of these strains to overcome the challenges of scalability, cost effectiveness, and sustainability that are key impediments to the future of the algae industry.
2. Mechanism of pH-tolerance in acidophiles and alkaliphiles
Extreme pH environments, acidic or alkaline, present a particular challenge for all microorganisms, including algae, as survival requires the development of specialized mechanisms for maintaining structural integrity, bioenergetics, and intracellular pH homeostasis. A key metabolic characteristic of algae that survive or thrive in extreme pH (acidic or alkaline) is tight regulation of their H+ fluxes. Cytosolic pH regulation involves H+ buffering and metabolic H+ consumption and production in synergy with transmembrane H+ transport and compartmentalization.16 Algae utilize a plasma membrane P-type H+ ATPase and vacuolar V-type ATPases and pyrophosphatases (V-PPases) for pH regulation. It is important to note that for both acidophilic and alkaliphilic algae, cytosolic pH needs to be operated close to neutral pH for ATP synthesis to take place, which involves the coupling of H+ translocation by ATP synthetase via the plasma membrane (in cyanobacteria) or via the mitochondria and the thylakoid membrane (in eukaryotic algae).2.1. pH-tolerance in acidophilic and acid-tolerant algal strains
For ATP synthesis to occur in algae, the external pH should be more acidic than the internal one, resulting in formation of electrochemical gradient and inflow of H+ via ATP synthetase.9 Although in acidic conditions the external pH is indeed higher, acidophiles exhibit a low conductance of H+ through the plasma membrane along with high export capacity to maintain pH homeostasis.17 By combined virtue of low conductance and high export, neutral cytosolic pH in acidophiles is maintained and they tend to hold 104-fold higher proton gradient across their membrane than neutrophilic algae (Fig. 2).17 In addition to plasma membrane bound H+-ATPases (PMA) as the primary regulator of the intracellular pH, additional symporters of H+ and carbonic acid, such as a K+–H+ symport, has also been reported in Dunaliella acidophila.3 The enormous proton motive force utilized by these symporters and the coupled uptake of both K+/protonated acids and H+ ions help to maintain a positive membrane potential, which is essential for the survival of acidophiles in a low potassium environment.3 Similarly, uptake of ammonium (NH4+) and nitrite sources from the acidic environment is regulated through the proton motive force. Despite the downhill gradient of NH4+/nitrites across the membrane of acidophiles, the positive membrane potential of acidophiles provides an additional driving force for uptake of these species.3 Notably, the capacity of acidophiles to consume NH4+/nitrites as nitrogen source make them different from neutrophiles, expanding their biotechnological application to utilize toxic wastewaters from various industries.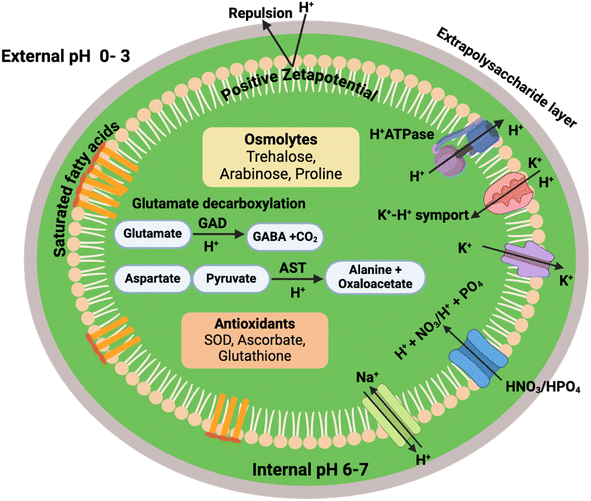 | ||
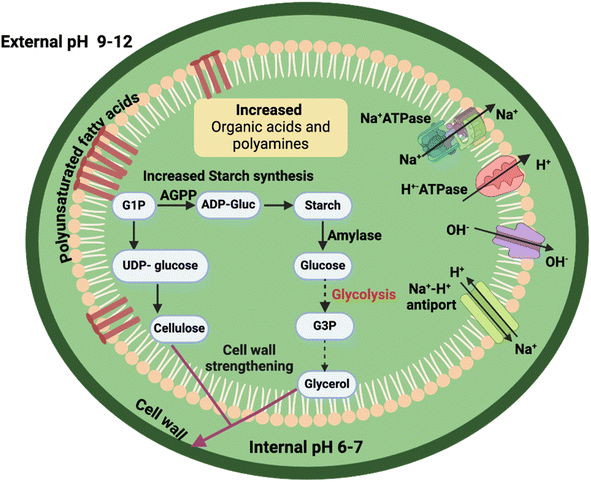
| Fig. 2 Mechanisms of pH adaptation in acidophilic algae. AST: aspartate aminotransferase, GABA: gamma-aminobutyric acid, GAD: glutamic acid decarboxylase, SOD: superoxide dismutase. | ||
Another strategy of pH tolerant algae for surviving in extreme pH is via modulation of their plasma membrane. Several acid-tolerant microalgal species have been reported to undergo significant phenotypic changes as an adaptive response in addition to prominent changes in the membranal composition. These changes generally include increase in cell size or developing an additional mucilaginous layer of extracellular polysaccharides on their surface.18,19 The acid-tolerant species Desmodesmus sp. MAS1 and Heterochlorella sp. MAS3, when cultivated in acidic mine drainage water, exhibited enlarged cell size and enhanced lipid content as adaptive response to the low pH.18 Notably, the majority of acidophilic algal strains, including C. acidophila and Dunaliella acidophila, exhibit a positive net surface charge and membrane potential, which aids in the repulsion of H+ to prevent excessive proton flux inside the cell.20 In general, the plasma membrane of the acid-tolerant/acidophilic algae is found to be rich in saturated fatty acids, bipolar tetraether lipids, and proteins, all of which aid in rigidity and reduced permeability of H+ ions, which could be true for algae as well.21 A recent study on the acid-tolerant alga Graesiella revealed a decrease in unsaturated fatty acids and increase in saturated fatty acids on long term acid exposure.22 Further, significant enrichment in lipid fractions has been also reported in acid-tolerant algae such as Graesiella sp. MA1, Heterochlorella sp. MAS3, Desmodesmus sp. MAS1, Chlamydomonas acidophila and Scenedesmus sp., isolated from acid mine drainages.19,22–25 However, detailed compositional analysis of Graesiella sp. MA1 revealed the higher content of glycerophospholipids, sphingolipids, and highly saturated fatty acids in the membrane to prevent influx of proton from the acidic environment.22 Detailed compositional analysis of acidophilic and acid-tolerant algal membranes is an unexplored subject that needs further attention to unravel the intrinsic differences among neutrophilic and pH-resilient strains.
In addition, synthesis of osmo-protectant and compatible solutes along with enrichment in stress-related proteins in response to extreme pH has also been reported in these algae. For instance, detailed physiological and metabolic profiling of the acid-tolerant Graesiella sp. MA1 over 81 days of cultivation highlighted the existence of an adaptation phase for 7 days before growth resumed.22 The initial slow growth rate lasted while the external pH increased from 3.5 to 5 with a gradual rise in biomass concentration to ∼3.5 g L−1. To maintain a neutral cytosolic pH, this acid-tolerant strain undergoes a significant loss in photosynthetic activity as well as total protein content, highlighting the metabolic re-routing of energy to overcome the proton gradient rather than growth. In addition, elevated levels of antioxidant components, such as ascorbate, malondialdehyde, superoxide dismutase, and glutathione, were also observed during the adaptation phase ranging from day 7–18, suggesting a metabolic adaptability under acidic stress. However, during the subsequent growth phase, more prominent and elevated concentrations of protein were recorded in comparison to carbohydrates, an unusual phenomenon, which is not reported in the cases of other abiotic stressors. Notably, metabolic profiling during day 18, day 60, and day 81 identified an enrichment in acid-responsive amino acids, such as aspartate, glutamate, lysine, arginine, histidine, and proline, along with osmotic sugars, including trehalose, cellobiose, xylobiose, and arabinose, to regulate the metabolic functions of Graesiella sp.22 In acid-tolerant strains, the amino acids play a crucial role in maintaining the neutral intracellular pH via the process of decarboxylation, energy production, and neutralization. Among the amino acids, arginine and glutamate help to neutralize the excess intracellular proton levels through production of CO2 via glutamate decarboxylation (Fig. 2).26,27 In parallel, conversion of aspartic acid to alanine utilizes intracellular H+ ions, while the combined action of histidine and lysine along with proline maintain a strong buffering action to prevent osmotic imbalance and thus proton influx under acidic conditions.28,29
A unique feature of the acidophilic alga Chlamydomonas eustigma, highlighted by a comparative genomics and transcriptomics study with its neutrophilic counterpart C. reinhardtii, showed a higher basal level of genes encoding PMA and heat shock proteins Hsp70 and Hsp60.17 Similarly, a proteomic study of acidophilic Chlamydomonas sp. in natural acidic metal-rich water indicated significant downregulation of photosynthesis-related enzymes, including ribulose-1,5-biphosphate carboxylase, whereas upregulated expression of phytochrome B, phosphoribulokinase, phosphoglycerate kinase, Hsp70, and Hsp90, as well as other stress-related enzymes.30 Expression of PMA and Hsps (Hsp70, Hsp60, and small Hsps) is considered a constitutive expression across acidophiles, including C. eustigma, C. acidophila, and Dunaliella acidophila.17,31,32 More specifically, a higher basal level of PMA genes in acidophiles is responsible for maintaining a high proton pump activity and intracellular pH of 6.5 even in external pH of 2.33 On the other hand, Hsps are known for maintaining protein structure by preventing irreversible aggregation and mediating folding, reassembly, and maintenance of denatured proteins in a transient-folding competent state under stress conditions to allow the cells to carry out essential metabolic activity in acidic conditions.34 The higher basal level of Hsps in acidophiles suggests the constant dealing of these cells with the acidic environment. Examining the expression of Hsp60, Hsp70, and small Hsps under a wide pH range of 1.5 to 7 revealed the unique expression of small Hsps to regulate the internal acidification in cells exposed to pH 4.21 On the other hand, elevated accumulation of Hsp60 and Hsp70 was found to be related to morphological and membranal changes reflecting the hyper/hypotonic conditions that C. acidophila undergoes during the exposure to low pH.30,31 Furthermore, acidophilic algae are also distinguished by a complete loss of their fermentative pathway, including key enzymes such as lactate dehydrogenase, pyruvate formate lyase (PFL), acetate kinase, and phosphate acyltransferase.34 The absence of genes encoding these enzymes prevents production of organic acids and averts further acidification of the cytosol to maintain a strong buffering capacity for neutral cytosolic pH until low external pH conditions exist.17
2.2. pH-tolerance in alkaliphiles and alkali-tolerant algae
Unlike, acidophilic and acid-tolerant algae, the reverse ΔpH in alkaliphiles could result in decreased electrochemical gradient leading to reduced growth. Surprisingly, alkaliphilic algae in soda lakes are known to have a higher productivity compared to strains from freshwater rivers and lakes. To understand how alkaliphiles counteract the high pH levels, it's important to look closely at the composition of their ecosystems. For example, soda lakes are characterized by high Na+ in the form of NaCl and Na2CO3, which is utilized by the algae to drive Na+/H+ antiporters across the plasma membrane (Fig. 3). This is the reason why well-known alkaliphiles, such as A. platensis NIES-39 and D. salina NIES-2257, cannot survive in a media without Na+.9 However, excess Na+ in the cytoplasm may disrupt the cellular homeostasis, so it is actively expelled outside the cell by alkaliphilic and alkali-tolerant algae. Exactly how this is done is still unknown, but the presence of Na+-ATPases in the cyanobacterium Synechococcus R2 and the marine alga Tetraselmis virdis indicates their role in active expulsion of Na+ from the algal cells.9,35 In addition, proton pumps (H+-ATPases) actively pump H+ out to counteract the influx of OH− ions in addition to supporting the active Na+ efflux from the cells (Fig. 3). Another unique feature of alkaliphile algae is the formation of massive starch grains and glycerol under alkali conditions. Glycerol is synthesized through catalytic conversion of starch in presence of glycerophosphate dehydrogenase, resulting in strengthened cell wall, providing resistance to alkaline conditions.36,37 On the other hand, high cellulose content in the cell wall of Chlorella JB17 has been reported to increase the cell wall thickness by 2–3 folds, that might partly support the alkali resistance in the cells.38 Similarly, a study in an alkaliphilic microalgae Chlorella sp. BLD showed re-routing of carbon from proteins and starch reservoir towards the synthesis of organic acids and lipids under alkaline condition.39In the case of alkaliphile microorganisms, such as bacteria and yeasts, high amounts of polyunsaturated fatty acids and acidic phospholipids have been reported.40 Presence of polyunsaturated fatty acids ensures proper membrane fluidity and flexibility in addition to preventing H+ leakage across the plasma membrane, while an increase in acidic phospholipids may contribute to the stability of the plasma membrane in alkaline pH.40,41 Although studies of alkaline pH algae are significantly fewer than for their acidic pH counterparts, investigations of alkaliphilic and alkali-tolerant algae have shown an increase in carbonic anhydrase activity in response to alkaline pH.42,43 Intracellular carbonic anhydrase in photosynthetic microorganisms participates in the CO2-concentration mechanism by catalyzing rapid bicarbonate conversion to CO2 in the direct vicinity of Rubisco, thus increasing CO2 uptake.44 At higher pH, increased carbonic anhydrase levels have been reported in several cyanobacteria and eukaryotic algae to increase the dissolved inorganic carbon (DIC) uptake. In the alkaliphilic alga Chlorella sp. BLD, an upregulation of Rubisco, light harvesting complex, and photosynthesis genes was reported under alkaline conditions revealing an adaptation mechanism to high pH conditions.39 Notably, an increase in organic acids and polyamines was reported verifying their role in pH stability and ion balance buffering their cytoplasm (Fig. 3). Overexpression of acyl-CoA-binding protein 1 (ACBP1) isolated from alkaliphilic Chlorella sp. JB6 in Arabidopsis resulted in increased resistance to high salinity, heavy metals (Pb and Cd), and low temperature stresses.45 The ACBP1 based on its structural similarly was hypothesized to transport multiple phosphocholine (PCs) associated with phospholipid metabolism, hence increasing the stability plasma membrane under stress. Likewise, overexpression of a novel bZIP transcription factor ChbZIP1 isolated from the alkaliphilic Chlorella sp. BLD in Arabidopsis resulted in increased alkali resistance due to an upregulation in oxygen detoxification pathway.46 Indeed, these two genes are promising targets for future genetic engineering of neutrophilic algae to improve their tolerance to high pH.
3. Application of acidophilic and acid-tolerant algae in sustainable biomass production
In the course of evolution, algae inhabiting extreme pH environments undergo adaptive rearrangements across their genetic profile to thrive in such unfavorable conditions.33 The phenomenon of metabolic flexibility in these strains holds potential for an array of biotechnological applications, including mitigation of recalcitrant pollutants like heavy metals, CO2 sequestration, and production of renewable materials, such as fuels, antioxidants, and cosmetics (Table 1). An acidic environment with pH ranging from 1 to 3 largely enhances the availability, as well as toxicity, of several heavy metals such as chromium, cadmium, copper, due to their increased solubility.47 In this context, several acidophilic algal strains, such as C. acidophila, D. acidophila, Coccomyxa subellipsoidea, Cyanidioschyzon merolae, G. sulphuraria, and C. eustigma, have gained remarkable interest due to their inherent potential to tolerate higher concentration of toxic heavy metals in comparison to neutrophilic microalgae.17,48–50 For instance, high arsenic tolerance of C. esutigma was directly related to the higher number of genes responsible for arsenic transformation and detoxification compared to neutrophilic C. reinhardtii.17 It is believed that the genes ACR3 and ArsB have been acquired by C. subellipsoidea and G. sulphuraria via horizontal gene transfer, imparting these acidophilic algae with approximately 10-fold higher arsenic tolerance.17,51 Similarly, tolerance of copper and cadmium by C. acidophila, and D. acidophila with the help of transcriptomics studies showed that even at the concentration of 500 μM, the cells did not exhibit any inhibition in the photosynthetic and growth activity.49,50 In fact, these extremophiles harbor constitutive expression of genes related to antioxidants and overproduction of proton-ATPases as an adaptive phenomenon, that not only enhances their resistance to H+ but also an passive hindrance to positively charged cations like Cd2+, and Cu2+.49,50| Algae | pH | pH threshold | Cultivation condition | Biomass density (g per L per day) | Bioproduct | Reference |
|---|---|---|---|---|---|---|
| a — not reported; BBM, Bold's basal media; BG-11, Blue green media. | ||||||
| Scenedesmus parvus | 3.00 | 3.0–9.0 | Flask culture with 15% CO2 concentration | 0.078 | Carbohydrate: 15.47 mg per L per day | 59 |
| outdoor using 15L fabricated PBR | 0.1036 | Carbohydrate: 44.35 mg per L per day | ||||
| Indoor using 15L fabricated PBR | 0.094 | Carbohydrate: 30.33 mg per L per day | ||||
| Tetratostichococcus sp. P1 | 5 | 3.00–8.00 | Flask culture with BG 11 and 1% CO2 | — | Fatty acid: 102.88 μg mg−1 | 56 |
| Coccomyxa sp. (strain onubensis) | 2.5 | — | 5L culture bottles with 5% CO2 in N-starved media | 0.28 | Lipid: 0.35 g g−1 | 60 |
| Lutein: 8 mg g−1 | ||||||
| β-carotene: 1.3 mg g−1 | ||||||
| Coccomyxa sp. (strain onubensis) | 4 | 2.5–9.0 | Flask cultures with 5% CO2 | 0.22 | — | 112 |
| Chlamydomonas acidophila LAFIC-004 | 3.6 | — | — | — | Oil: 54.63% | 113 |
| Coccomyxa onubensis | 2.5 | 2.5–3.0 | Indoor plastic bag 400 L with 2.5% CO2 | 0.09 | Lutein: 9.7 mg g−1 | 64 |
| Outdoor vertical tubular PBR 800 L with 2.5% CO2 | 0.14 | Lutein: 10.0 mg g−1 | ||||
| C. onubensis | — | 400 L transparent plastic bags with 5% CO2 | — | Protein: 44.60% | 62 | |
| Coccomyxa acidophila | — | Flask culture with urea and 5% CO2 | 0.25 | 3.55 mg g−1 | 58 | |
| Galdieria phlegrea | — | Flask cultures with wastewater | 0.065 | Lipid: 20% | 54 | |
| Chlamydomonas acidophila | — | 1 L batch reactor with 5% CO2 | 0.48 | Lutein: 5.18 mg g−1 | 114 | |
| Chlamydomonas acidophila LAFIC-004 | 7.5 | 2.5–8.0 | Flask culture with wastewater and 1% CO2 | 0.08 | — | 115 |
| Galdieria sulphuraria 074 G | 2.0 | — | Flask cultures with natural acidic water | 0.58 | Phycocyanin: 107.4 μg mg−1 | 116 |
| Coccomyxa sp | 3.0 | 3.0–7.0 | Flask cultures | 0.005 | Antioxidants: 48.0 (Trolox equivalent) micromole per g biomass | 117 |
| Pseudochlorella sp. YKT1 | 3.0–5.0 | 0.29 | Lipids: 30% | 118 | ||
| Euglena gracilis Z | 3.0 | 3.0–8.0 | 0.023 | Paramylon: 58.3% | 119 | |
| Euglena sp | 5 | 3.0–8.0 | 0.03 | Succinate 183 mg per L | 120 | |
Acid-tolerant algal strains gain their unique tolerance either as a natural adaptation or via adaptive lab evolution employed to custom-tailor strains with desired phenotypic characteristics. For instance, two non-acidophilic algae, Desmodesmus sp. MAS1 and Heterochlorella sp. MAS3, gradually adapted to acidic conditions (pH 3.5) also exhibited excellent removal capacity of Cd (20 ppm) along with an increase in intracellular lipids.52,53 These two adapted acid-tolerant algae were also able to efficiently remediate alkaline winery waste water (pH > 10.50).42 Both acid-tolerant strains outperformed the wild-type (non-adapted strains) resulting in 45–52%, 54–58%, and 65–75% reduction in total organic carbon, total nitrogen and total phosphate, respectively. Further, several studies have highlighted the feasibility of utilizing acid-tolerant/acidophilic algae to remediate wastewater and utilize CO2 with concomitant production of valuable compounds, like fatty acids, lutein (nutraceutical), and carbohydrates (for production of bioethanol, bioplastics, or nutraceuticals) (Table 1). Another acidophilic alga, Galdieria phlegrea, showed remarkable efficacy to mitigate ∼50% of the ammonium and ∼25% of the phosphate content in raw municipal wastewater, while augmenting to ∼22% their lipid content and to 94 mg L−1 their phycocyanin content (a food additive).54 The application of this acidophile alga to effluents rich in ammoniacal content provides a significant advantage over neutrophiles and makes possible the valorization of raw effluent in a circular economy model. Furthermore, employing these acidophiles for reclamation of abandoned mining sites and acidic soil is yet another advantage. Inoculation of acid-tolerant algal strains in acidic soil for 90 days resulted in an increase in pH of the soil from 4.8 to 5.6 due to the release of exopolysaccharides by algae.55 Along with algal crust development, the soil quality and fertility were significantly improved thanks to a carbon content enhancement of 57%, higher indole acetic acid (IAA) content, and increased dehydrogenase activity in soil. The unique capacity of acid-tolerant species for passive uptake of CO2 may be a practical means of increasing soil pH, where needed, in addition to the symbiotic interaction between algae and soil microbes accelerated through heterotrophic respiration.55 Interestingly, it is believed that the increase in soil IAA content represents enhanced signaling and communication between algae and microbes in the soil microbiome. In light of the positive impact acid-tolerant algae have on soil reclamation, artificial biofilms containing acid-tolerant/acidophilic algae and non-acidophilic bacterial communities have been proposed as a feasible approach for remediation of acidic mine drainages and mining sites.53
The novel acid-tolerant alga Tetratostichococcus sp. P1, isolated from peatland, was found suitable for utilizing tropical peat wastewater (pH 4.5) to produce biomass rich in fatty acids for cost-effective production of biodiesel.56 The alga was capable of achieving a specific growth rate of 0.22 per day with a maximum proportion of C20:0 (∼24%) as a chief fatty acid in biomass under supplementation of peat wastewater with air. However, cultivation with peat wastewater on 1% CO2 significantly remodeled the fatty acid composition to ∼23% C18:0, ∼27% C18:3, and ∼34% C16:0, which is suitable for biofuel production. Similarly, a positive influence of CO2 supplementation (2.5%) on the growth rate Elliptochloris sp. was observed (∼80% higher than phototrophic growth with air only), while saturated fatty acid production increased to ∼30% of the dry cell weight under mixotrophic condition with vegetal glycerin (5 mM) as organic carbon source.57 In addition, cultivation of this acid-tolerant strain in pH 2.5 reduced the risk of contaminant microbes suggesting promising applicability of this strain to outdoor cultivation. Similarly, mixotrophic cultivation of C. acidophila in urea and 5% CO2 significantly enhanced its biomass productivity (250 mg per L per day) and lutein content (3.5 mg g−1).58 Urea, a low-cost source of carbon and nitrogen, is viewed as cost effective nutrient source for cultivation of algae. When urea use is combined with 5% CO2 supplementation, the slower growth rate of acidophilic strains is improved, while still preventing contamination of outdoor cultivation.58
A study of the acidophilic strain Scenedesmus parvus isolated from mining sites established the feasibility of outdoor cultivation with better biomass and carbohydrate productivity in comparison to indoor cultivation systems.59 Outdoor cultivation of this alga in Bolds basal media (BBM) at pH 3 and supplementation with 15% CO2 resulted in a final biomass concentration of 0.9 g L−1. Outdoor cultivation conditions at higher temperature (∼34 °C) and light intensities (∼20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 648 lux) significantly improved the overall carbohydrate productivity by 33% compared to indoor controlled conditions.59 On a similar note, various acidophilic algae have been explored for sustainable production of high-value products, like lutein and carotenoids, while sequestering CO2. For instance, C. onubensis, capable of surviving at pH 2.5, has been reported to synthesize high levels of lutein.60,61 Nutrient deprivation studies in this strain also revealed the presence of phosphorus and sulphur reservoirs that aided the stable survival of C. onubensis for more than 15 days at a specific growth rate of 0.14–0.16 per day, which was comparable to growth under controlled conditions (0.19 per day). Such an ability to maintain photosynthetic activity even under phosphorus and sulphur deprivation conditions probably indicates an adaptive evolution in acidophilic strains to survive high heavy metal toxicity in acidic mines. Generally, high solubility of heavy metals at low pH enhances algal uptake through membrane transporters of essential ions. However, nitrogen starvation still affected the growth of C. onubesis cells, as reflected in the increase in content of fatty acids (C18:3), lutein (∼20%), and β-carotene (∼14%) in the initial span of 2–3 days, suggesting enhanced carbon fixation in the short term.60 Furthermore, augmentation of polyunsaturated fatty acids (PUFA) also makes these acidophiles a rich source of antioxidants. For instance, a study highlighted the nutritional application of acidophilic C. onubensis biomass in laboratory rats for monitoring their growth and health parameters. This biomass, consisting of protein (44%), carbohydrate (24%), fiber (16%), and lipids (5.4%) with more than 65% of them being PUFA, reduced the cholesterol and glyceride levels in rats at an inclusion rate of 6.5%.62 In addition to such antioxidant production, C. onubensis algal extract prepared in non-polar solvent (hexane/chloroform) demonstrated strong antimicrobial activity against both Gram positive/negative and pathological Candida albicans.63 Fatty acid profiling of the extract revealed the prominence of C16:0, C18:3, C18:2, and C18:1 that individually or in combination with glycerides are known to have antimicrobial properties. Indeed, the antimicrobial activity reported against the Gram-positive Staphylococcus aureus (zone of 13.3 mm) was the highest recorded for this algal extract. Moreover, the efficacy of the crude algal extract against Gram negative Salmonella enterica, Escherichia coli, and Proteus mirabilis was found to be equivalent to the commercial antibiotic amoxycillin.63 These studies suggest the feasibility of large-scale cultivation of acidophilic strains and utilization of the generated biomass in functional foods, nutraceuticals, animal feed, and antimicrobial formulations.
648 lux) significantly improved the overall carbohydrate productivity by 33% compared to indoor controlled conditions.59 On a similar note, various acidophilic algae have been explored for sustainable production of high-value products, like lutein and carotenoids, while sequestering CO2. For instance, C. onubensis, capable of surviving at pH 2.5, has been reported to synthesize high levels of lutein.60,61 Nutrient deprivation studies in this strain also revealed the presence of phosphorus and sulphur reservoirs that aided the stable survival of C. onubensis for more than 15 days at a specific growth rate of 0.14–0.16 per day, which was comparable to growth under controlled conditions (0.19 per day). Such an ability to maintain photosynthetic activity even under phosphorus and sulphur deprivation conditions probably indicates an adaptive evolution in acidophilic strains to survive high heavy metal toxicity in acidic mines. Generally, high solubility of heavy metals at low pH enhances algal uptake through membrane transporters of essential ions. However, nitrogen starvation still affected the growth of C. onubesis cells, as reflected in the increase in content of fatty acids (C18:3), lutein (∼20%), and β-carotene (∼14%) in the initial span of 2–3 days, suggesting enhanced carbon fixation in the short term.60 Furthermore, augmentation of polyunsaturated fatty acids (PUFA) also makes these acidophiles a rich source of antioxidants. For instance, a study highlighted the nutritional application of acidophilic C. onubensis biomass in laboratory rats for monitoring their growth and health parameters. This biomass, consisting of protein (44%), carbohydrate (24%), fiber (16%), and lipids (5.4%) with more than 65% of them being PUFA, reduced the cholesterol and glyceride levels in rats at an inclusion rate of 6.5%.62 In addition to such antioxidant production, C. onubensis algal extract prepared in non-polar solvent (hexane/chloroform) demonstrated strong antimicrobial activity against both Gram positive/negative and pathological Candida albicans.63 Fatty acid profiling of the extract revealed the prominence of C16:0, C18:3, C18:2, and C18:1 that individually or in combination with glycerides are known to have antimicrobial properties. Indeed, the antimicrobial activity reported against the Gram-positive Staphylococcus aureus (zone of 13.3 mm) was the highest recorded for this algal extract. Moreover, the efficacy of the crude algal extract against Gram negative Salmonella enterica, Escherichia coli, and Proteus mirabilis was found to be equivalent to the commercial antibiotic amoxycillin.63 These studies suggest the feasibility of large-scale cultivation of acidophilic strains and utilization of the generated biomass in functional foods, nutraceuticals, animal feed, and antimicrobial formulations.
In a larger scale study of 800 L, outdoor cultivation of Coccomyxa onubensis was performed in a tubular closed photobioreactor (PBR) for production of lutein.64 Nitrogen, phosphorous, and potassium fertilizer media at pH of 2.5–3 with 2.5% CO2 supply were used to reduce cultivation cost for the acidophilic C. onubensis, which achieved a stable biomass productivity of 140 mg per L per day and lutein content up to 10 mg g−1 of biomass over a period of 30 days. Despite the slow growth rate, C. onubensis in outdoor large scale achieved a higher biomass productivity than indoor cultivation in plastic bags (400L) due to higher light intensity and improved solubility of CO2 in form of bicarbonates at low pH 2.5.64 In general, at pH value less than 3, CO2 is predominantly present as gaseous CO2 rather than its dissolved form, bicarbonate, resulting in rapid loss of CO2 to the ambient environment. Therefore, cultivation of acidophiles for CO2 capture/sequestration in closed tubular PBR is a preferable choice over large scale open raceways. Although the capital cost for establishing PBRs is higher than for open ponds, their lower risk of contamination, higher CO2 solubility, and higher biomass, fatty acids, and carotenoid productivity offer a sustainable and economically feasible model for production of specialty biochemicals.
Although there are currently no studies assessing the techno-economic feasibility of utilizing acidophile and acid-tolerant algae for bioremediation, biofuel, and bioproduct, existing studies on neutrophilic algae could provide crucial insights into the overall economic feasibility. For instance, a recent study utilizing Scenedesmus acuminatus for copper and zinc removal from wastewater reported an economic assessment of daily 1000 tons diesel production through Na4SiO4 transesterification with the lowest biodiesel selling price of US$ 147.36 per barrel for economic viability, provided the cost of algal biomass should not exceed US$ 462 per ton.65 The authors reported the algal business was viable, with a return on high investment (16.4%) and a payback period of 5 years. Another study established treatment of dairy wastewater plant with a bioreactor unit (1.68 * 104 m3) using Scenedesmus sp. SDEC-13 to yield an annual biomass of 1.44 × 106 kg along with capture of 2.58 × 106 kg CO2 per year.66 Moreover, after considering all the operational costs, the cost of wastewater treatment was reported to reach 0.01–0.02 $ per m3. Similarly, a V-shaped pond was reported to be cost-effective for large-scale production of Ascochloris sp. biomass cultivated in dairy wastewater.67 This techno-economic study suggested that algal biomass production in a high-volume V-shaped pond (volume of 3 m3 and area of 4 m2) was feasible, provided treatment of >1 megaliters per day of dairy wastewater over 20 years. This approach showed the high commercial feasibility of an algae-based treatment plant with an annual production of ∼500 tons algal biomass at the cost of US$ 0.48 per kg and 24 × 107 liters of treated water, while having an internal rate of return of 118% and 1.9 year payback time.67 Another techno-economic study focused on integrated biorefinery showcased the comparative study of Coelastrella striolata to produce biogas, biocrude, and fertilizer, biogas and biocrude, and only biocrude, while utilizing wastewater, flue gases, and excess energy generated from palm oil mills.68 The cultivation of Coelastrella sp. in an open pond of 1.2 ha area and hydrothermal liquefaction for downstream processing showed commercial feasibility of fertilizer and biogas over a period of 20 years. Moreover, the system allows daily production of 68.43 kg of biocrude and 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 470.70 kg of biogas that could be sold at US$ 2.0 per L and US$ 0.24 per kg, respectively, with an additional daily output of 261 kg of fertilizer at a price of US$ 0.05 per kg. The study revealed that both the routes showed a profitability index of 2.5%, an internal rate of return of 25%, and a payback period of 3.7 years, while producing 188 tons of annual biomass.68 On a similar note, utilization of palm oil mill wastewater was reported to reduce the overall cost of Arthrospira platensis biomass production from 87.45 € per kg to less than 50 € per kg.69 Further, technoeconomic assessment (TEA) using Nannochloropsis sp. demonstrated that utilizing fertilizer-based media for cultivation in a 10 h area installed with tubular PBR can reduce the biomass production cost to 36.21 € per kg dry weight.70 Indeed, the biomass production cost varies for different algal species and most importantly, it largely depends on the cultivation mode, type of media/wastewater utilized, electricity, location and distance of the plant, and downstream processing technologies.
470.70 kg of biogas that could be sold at US$ 2.0 per L and US$ 0.24 per kg, respectively, with an additional daily output of 261 kg of fertilizer at a price of US$ 0.05 per kg. The study revealed that both the routes showed a profitability index of 2.5%, an internal rate of return of 25%, and a payback period of 3.7 years, while producing 188 tons of annual biomass.68 On a similar note, utilization of palm oil mill wastewater was reported to reduce the overall cost of Arthrospira platensis biomass production from 87.45 € per kg to less than 50 € per kg.69 Further, technoeconomic assessment (TEA) using Nannochloropsis sp. demonstrated that utilizing fertilizer-based media for cultivation in a 10 h area installed with tubular PBR can reduce the biomass production cost to 36.21 € per kg dry weight.70 Indeed, the biomass production cost varies for different algal species and most importantly, it largely depends on the cultivation mode, type of media/wastewater utilized, electricity, location and distance of the plant, and downstream processing technologies.
However, to utilize acidophile and acid-tolerant algae for bioremediating acidic or low-pH effluents, several cultivation and techno-economic barriers need to be overcome before large-scale deployment. For instance, to cultivate acidophilic algae in acidic wastewater, specialized reactors (open or closed) and control sensors need to be built to withstand the corrosion from the acidic conditions and toxic metals and pollutants.71 Moreover, additional energy input may be required to maintain CO2 aeration and temperature control in PBRs, which could lead to increased operational costs. Additionally, depending on the wastewater composition, the efficiency of algae to remove heavy metals and degrade pollutants might vary between batches, which could involve multiple cultivation runs or additional treatment technologies to reach acceptable remediation levels, reducing the overall cost-effectiveness of the process. Another challenge is harvesting the heavy metal rich algal biomass which requires careful processing involving costly chemicals, drying, and extraction techniques to avoid secondary environmental contamination.
4. Application of alkaliphilic and alkali-tolerant algae in sustainable biomass production
Several industrial effluents originating from textile, paper, mining, pharmaceutical, food processing and agricultural run-offs generally have alkaline pH due to the presence of calcium hydroxide or sodium hydroxide. If these wastewaters are not properly neutralized and treated, they can seep into aquatic bodies, including freshwater aquifers. High pH levels (>9.0) can significantly impact biodiversity and degrade water quality and ecosystem health. Alkaliphilic and alkali-tolerant algae can thrive in high pH environments, constituting a source of food for primary producers in natural environments, such as soda lakes, alkaline soils, and hot springs. These high pH-tolerant algae also play a crucial role in sequestering carbon and recycling waste nitrogen and phosphorous from alkaline wastewaters. Thanks to their unique metabolic adaptations and robustness they are also being considered as promising candidates for sustainable large-scale cultivation using non-potable water, such as alkaline wastewaters (Table 2). Notably, the culture pH rises steadily as the algae grow due to the conversion of CO2 to bicarbonate (HCO3−) during photosynthesis, thus resulting in the accumulation of OH− ions with the pH fluctuation being greater in outdoor cultivation systems due to constant changes in temperature and photosynthesis rates.72 Moreover, at neutral pH, the diffusion and solubility of atmospheric CO2 in the cultivation media is low, so to increase CO2 uptake by algae, operators need to sparge pure or enriched gas into the culture.73 However, this significantly increases the cost of algal production. It is important to note that between pH 6.3 and 10.3, HCO3− is the dominant carbon species in water, which shifts to carbonate (CO3−) at pH >10.3. Thus, it will be beneficial to cultivate alkali-tolerant algae that can adapt and switch their metabolism based on the alterations in the culture pH resulting in higher biomass productivity. Importantly, alkaline cultures have been reported to be less infested by grazers and other undesirable microorganisms.73 For instance, Arthrospira (Spirulina)platensis, one of the most widely studied and used alkaliphilic algae for large scale cultivations, is able to tolerate high levels of both pH and salinity.74,75 This cyanobacterium possesses ATP-dependent sodium pumps that extrude excess sodium out of the cell allowing it to thrive in hypersaline lakes.74 Comparison of three alkali-halophilic microalgae, A. platensis NIES-39, D. salina NIES-2257, and Euhalothece sp. Z-M001 (cyanobacterium) revealed that latter grows optimally at pH 10 with a maximum specific growth rate of 1.67 per day and carbon assimilation of 0.422 g C per L per day thanks to its high pH stability.75 Oleaginous alkaliphilic Chlorella sp. ALP2 isolated from Soda Lake in the U.S. State of Washington showed an ability to grow at pH ranging from 7.0–9.0 at a bicarbonate concentration of 202 mM attaining maximum biomass concentration of 0.98 g L−1 in a two-stage heterotrophic cultivation that was started as heterotrophic at neutral pH and was switched to phototrophic at pH 9.0.76 Alkaline pH was reported to assist in auto-flocculation, resulting in 64.1% harvesting efficiency that represents yet another advantage of using alkaliphilic and alkali-tolerant algae for large scale outdoor cultivation. Twenty algae species belonging to Chlorococum sp. Chlamydomonas sp., Scenedesmus sp., Chlorella sp., Tribonema sp., Stigeoclonium sp. and Navicula sp. isolated from saline-alkali soil in China were characterized for their salt and bicarbonate tolerance.77 Among them, Chlorella sp., and Nannochloris sp. showed extreme salt (600 mM) and bicarbonate (300 mM) tolerance in addition to accumulating higher starch content (Table 2). Another study reported that alkaliphile Chlorella sorokiniana str. SLA-04 successfully grew at pH 10 in outdoor raceway ponds during autumn/winter cultivation using both phototrophic and mixotrophic modes without any noticeable contamination by invaders.73 These results indicate that alkaliphiles can be the preferred algae for mixotrophic cultivation, which typically is more prone to contamination compared to autotrophic cultivation. Likewise, Chlorella sp. BLD strain isolated from a soda lake in China exhibited pH tolerance from 4–12 and attained a maximum biomass concentration (0.97 g L−1) and photosynthetic activity at pH 10.39 The authors reported enhanced light harvesting, CO2 fixation, and carbon flow towards lipid biosynthesis, revealing a well-orchestrated metabolism of the alga under alkaline pH. A native diatom, Nitzschia plea (Bacillariophyceae) isolated from a soda lake in China was reported to accumulate high EPA (Eicosapentanoic Acid, C20:5) at pH 9, signifying yet another valuable application of these pH tolerant algae.78 These studies indicate that soda lakes and soils are promising isolation sites for bioprospecting novel haloalkaliphilic algal strains, since these lakes are not only characterized by high pH, but also high salinity and sometimes high temperature. Such lakes and soils represent stable pH environments, where large amounts of carbonates result in pH >11.5.11| Algae | Optimal pH | pH threshold | Cultivation conditions | Biomass density (g per L per day) | Bioproduct | Reference |
|---|---|---|---|---|---|---|
| a — not reported; EPA, eicosapentaenoic acid; PBR, photobioreactor. | ||||||
| Chlorella sp. (ALP2) | 10 | 7–10 | Two stage cultivation with 1 L fermenter (heterotrophic) and 40 L open tank PBR | 0.02 | Lipid: 39.78% | 76 |
| Chlorella sp. (JB6) | — | Flask cultures with 100 mM bicarbonate | 0.1 | — | 121 | |
| Nannochloris sp. (JB 17) | — | Flask cultures with 400 mM bicarbonate | 0.12 | — | ||
| Trebouxiophyte (adapted to high bicarbonate) | 9.61 | 8.5–10.0 | Floating outdoor PBR with 300 mmol L−1 bicarbonate | 0.80 | Protein: 50.80% | 81 |
| Chlorella sp. AT 1 (mutant strain) | 10 | 6.0–11.0 | Semi-continuous in PBR with intermittent 10% CO2 | 0.73 | Lipid:15.9% | 72 |
| Chlorella sp. BLD | 4.0–12.0 | Flask culture | 0.1 | Lipid: 31.86% | 39 | |
| Micractinium sp. (MA, SG, and TM) | >10 | 8.0–12.0 | Flask cultures with biogas derived CO2 | — | Lutein: 7.3 mg per g per dry cells | 79 |
| Monoraphidium dybowskii LB50 | 8.48–9.04 | — | Outdoor PBR with 20 g per L per NaCl | 0.081 | Lipid: 30.87% | 122 |
| Euhalothece sp. Z-M001 | 10 | 8.5–10.5 | Flask cultures | 0.84 | — | 75 |
| Chlorella sorokiniana str. SLA-04 | — | Outdoor raceway ponds | 0.05 | — | 73 | |
| — | Outdoor raceway ponds under mixotrophy (4 g per L per glucose) | 0.18 | Lipid: 36.7% | |||
| Chlorella vulgaris LC8 | 9.1 | — | Flask cultures | 0.02 | Lipid: 42.1 | 123 |
| C. vulgaris | 9.6–9.8 | 9.0–12.0 | PBR | 0.03 | Lipid: 38% | 124 |
| Nitzschia plea | 9 | 8.5–11.0 | Flask cultures | 0.30 | EPA: 15% | 78 |
As noted above, pH plays a vital role in determining the solubility of CO2 in the culture media. At higher pH >8.3, dissolved CO2 rapidly changes to HCO3− or CO3−, eventually reaching stable equilibrium that results in higher CO2 sequestration.79 Moreover, when sufficient buffering capacity is maintained, CO2 absorption increases.80 Thus, alkaline conditions allow for a higher absorption of CO2 and pH stability resulting in a higher carbon efficiency uptake (CEU) and biomass production. Not surprisingly, aquatic photosynthesis rates are higher in alkaline waters.73 Moreover, at higher pH more CO2 can be captured from emission sources into algal culture media, enhancing carbon sequestration as a means of combating climate change. For instance, the alkalihalophilic alga Trebouxiophyte sp. that was adapted to high bicarbonate tolerance, when cultivated in an outdoor pilot PBR, attained maximum areal productivity of 10.1 g per m2 per day in media supplemented with 300 mmol L−1 bicarbonate.81 The alga showed a higher CEU of 46.01%, when the media was supplemented with bicarbonate as compared to the usual 2% CO2 (10.28% CEU) indicating bicarbonate was a better source of carbon than CO2 or air, while also maintaining the high pH levels via bicarbonate buffering. Likewise, an alkali-tolerant mutant of Chlorella sp. AT1 capable of tolerating pH 11 showed a 5-fold higher CEU, when the culture was supplemented with intermittent 10% CO2 (30 min at 3 h intervals) compared to continuous 10% CO2 supply.72
Another potential use of high pH-tolerant algae is for biogas upgrading, which can easily grow in biogas plants operated at alkaline conditions in addition to minimizing biological contamination by other undesirable microorganisms. Biogas typically consists of methane (40–60%) and CO2 (30–40%), and traces of other compounds, such as hydrogen sulfide.82 However, to use biogas as a fuel, CO2 must be removed to increase its specific heat by upgrading the methane content to >90%. Algae, as a photosynthetic microorganism, can sequester CO2 from biogas providing a sustainable alternative as compared to use of costly chemical catalysts or membranes. For instance, alkaliphilic algae belonging to the genus Micractinium isolated from an alkaline river (pH 7.6–9.7) in Japan exhibited pH tolerance (pH 8–12) in 10% CO2.79 In addition, the strain grew well in biogas-derived CO2 reducing CO2 in the biogas to an undetectable level, thereby accomplishing successful upgrading. Notably, the strains showed high yields of antioxidant carotenoids, such as lutein, making them viable sources of nutraceuticals (Table 2). Another study reported an algal community isolated from Texcoco Soda Lake in Mexico capable of growing in alkaline conditions and adapted to high CO2 in the presence of H2S utilized 550–1000 mg CO2 per day from synthetic biogas.83 The authors identified Picochlorum sp. and Scenedesmus sp. as the prominent genera in the consortia.
Although to date limited studies are available on the application of alkaliphiles and alkali-tolerant algae, the published data clearly suggest that outdoor cultivation of these algae under high pH significantly improves CO2 absorption rates, resulting in high biomass productivity, minimizing culture crashes, and reducing the need for sparging additional CO2 into the media. In fact, CO2 has been estimated to account for over 50% of the raw material cost in algal cultivation.84 Typically, CO2 is compressed to 150 atm pressure for transport through the pipeline to an algal cultivation plant.14 However, sparging air or CO2 enriched gas directly into the media is considered impractical due to the high cost associated with the gas compression, which can vary depending on the size of the reactor, along with significant rates of evaporation in large-scale open raceway ponds.85 In this regard, flue gas sparging could potentially reduce the cost of CO2 delivery, provided the flue gas plant is located near the algae cultivation site, as increasing the distance would significantly increase the cost of transportation.84 Comparing the supply of costs of CO2 from raw flue gas and purified CO2 extracted from flue gas (14%) with a 100 km distance between the gas source and cultivation site resulted in US$ 57.20 per ton for compression, drying, and transportation for the raw flue gas while US$ 40.50 per ton for extracted CO2 due to reduced volume.86 Moreover, a recent study reported that the cost of carbon capture and utilization could be considerably reduced by 52% by utilizing algal strains such as acidophiles that show more tolerance and high carbon efficiency to CO2 compared to strains with low CO2 tolerance.87 In addition, to avoid compression and pumping of gas, particularly for alkaliphile and alkali-tolerant algae, the addition of sodium bicarbonate to the medium is also an alternative that significantly reduces the cost of transport for 100 km to US$ 0.05–0.06 per m by canal and US$ 0.0104–0.125 per m3 by water tunnel.88 However, this option may not be economically viable since the cost of sodium bicarbonate (US$ 380 per ton) is higher than purified CO2 (US$ 3–55 per ton).84 Another emerging alternative is CO2-loaded solvents, such as carbonates (a cheaper option than sodium carbonate) or amine-based solutions (methylethanolamine or triethanolamine), aimed at improving the efficiency of CO2 capture and avoiding energy and losses associated with pumping the gas.89 Indeed, further research is necessary to investigate the potential applications of these algae in biofuel production, bioremediation, carbon sequestration, and the synthesis of animal feed (protein) and nutraceuticals.
5. pH-resilient algae for CO2 fixation
The inherent flexibility of acidophilic and alkalophilic algae to survive under extreme pH conditions makes them promising candidates for efficient CO2 fixation. However, depending on the acidic and alkaline media, the carbon fixation pathways alter, affecting the CO2 fixation and bioproduct yields. For instance, low pH results in reduced dissolved inorganic carbon, which might limit the CO2 fixation rates. For this reason, acidophilic and acid-tolerant algae have evolved to have high affinity Rubisco and carbonic anhydrase to efficiently capture CO2.90 In addition, acidophilic alga such as G. sulphuraria rely on the utilization of both inorganic and organic carbon sources for its growth.91 Moreover, acidophilic algae divert their metabolism to synthesize acid-stable bioactive compounds, including phycobiliproteins and organic acids such as glutathione, for maintaining hemostasis.91 For instance, genetically engineered C. reinhardtii generated by overexpressing H+ pump to acquire acid resistance resulted in enhanced CO2 and tolerance to pH 5.5.92 The mixotrophic cultivation of the modified strain in 20% CO2 exhibited a 3-fold increase in biomass with a carbon fixation rate of 0.11 gCO2 per L per day in 7 days (Table 3).92 This study also evidenced the feasibility of the engineered strain C. reinhardtii to be utilized for outdoor cultivation in a 10 L bubble column PBR under exposure to real coal fire gas enriched with 13% CO2, 20 ppm NOX, 32 ppm SOX, and ∼80% N2. The authors reported an average biomass and a carbon fixation rate of 92 mg per L per day and 162 mg per L per day respectively (Table 3). Similarly, Chlamydomonas acidophila LAFIC-004 was assessed for its CO2 sequestration capacity by providing 5% to 20% CO2 with secondary effluent.93 The alga showed higher growth in 5–10% CO2 as compared to control, attaining biomass productivity of 0.06 g per L per day with a carbon fixation rate of 0.092–0.094 g-CO2 per L per day (Table 3). Besides CO2 removal, there was a significant rise in total PUFA content in C. acidophila biomass grown with CO2, highlighting the adaptive response of these cells to acclimatize to the stressful environment.93 Another study comparing the CO2 fixation rate of three different acidophilic algal strains revealed that Coccomyxa sp. CPCC508 showed a higher CO2 fixation rate of 0.43 g per L per day equivalent to 4.3 g per CO2 per g biomass, being 2–3 fold higher than those captured by E. garcilis and E. mutabilis.10 The study also highlighted that Coccomyxa sp. and E. garcilis were able to grow in a wide pH range of 3–7.5, whereas growth of E. mutabilis growth was limited to pH 3.10 Similarly, Desmosdesmus sp. SZ1 was able to survive in a pH range of 3.5–9, attaining a maximum biomass and carbon fixation rate of 3.5 g L−1 and 563 mg per L per day at pH 5.94 In fact, even at low pH of 3.5, this acid-tolerant strain exhibited a similar carbon fixation rate of 497 mg per L per day while producing 3.1 g L−1 biomass under supply of 10% CO2 showing a phenomenal carbon sequestration capacity under acidic conditions (Table 3).94| Algae | Nature | pH | Input CO2 mode and quantity | CO2 fixation rate | Biomass productivity | Reference |
|---|---|---|---|---|---|---|
| a — not reported; PBR, photobioreactor. | ||||||
| Chlorella sp. AT1 | Alkali-tolerant | 11 | 10% CO2 intermittent feeding 30 min at 3 h intervals for 21 days | 1.329 g per L per day | 0.726 g per L per day | 72 |
| Cyanobacterium sp. PNNL-SSL1 | Alkaliphile | Simulated raceway pond atmospheric CO2 (direct air capture) | 21.6 g CO2 per m2 per day | 15.2 g per m2 per day | 96 | |
| Thalassiosira pseudonana | 9 | 2.5 L glass vessel bioreactors | 0.6 g CO2 per g biomass | — | 10 | |
| Phaeodactylum tricornutum | 0.8 g CO2 per g biomass | — | 10 | |||
| Chlamydomonas acidophila LAFIC-004 | Acidophile | 7.5 | Flask culture with wastewater and 5 and 10% CO2 | 0.092–0.094 g per L per day | 0.06 g per L per day | 93 |
| Coccomyxa sp.CPCC508 | 3 | 2.5 L glass vessel bioreactors purged with 2.3% CO2 | 4.3 g per CO2 per g biomass | — | 10 | |
| Euglena mutabilis | 1.2 g per CO2 per g biomass | — | ||||
| E. gracilis | Acid-tolerant | 2.4 g per CO2 per g biomass | — | |||
| C. reinhardtii | 5.5 | 10 L (working volume) bubble column PBR purged with coal combustion flue gas having ∼13% CO2, 20 ppm NOX, and 32 ppm SOX | 162.2 mg per CO2 per L per day | 0.09 g per L per day | 92 | |
| Desmodesmus sp. SZ-1 | 3.5 | PBR with 10% CO2 | 497.6 mg per CO2 per L day | 0.3 g per L per day | 94 | |
In contrast, in high pH environments, an increase in the bicarbonate availability enhances the CO2 solubility in the growth media, resulting in the accumulation of lipids and proteins in the algae. Recent studies with alkali-resistant algae have reported the addition of both bicarbonate and CO2 to boost the carbon capture and biomass production cultivated in PBRs and open cultivation systems (Table 3). For instance, a cyanobacterial consortium was used for CO2 capture and its conversion to biomass in tubular PBR with media pH 11.2.95 In media with pH 11.2, the cyanobacterial consortium attained a total biomass productivity of 15 g per m2 per day during the fourth cycle of cultivation with a CO2 fixation rate of 4.6 g carbon per m2 per day.95 Similarly, a novel alkaliphilic cyanobacteria, Cyanobacterium sp. PNNL-SSL1 was reported to achieve an average productivity of 15.2 g per m2 per day by capturing CO2 at the rate of 21.6 g CO2 per m2 per day.96 Further, comparing the CO2 capture efficiency of six different acidophiles and alkaliphiles reported that under similar operational conditions, alkaliphiles (Thalassiosira pseudonana, Phaeodactylum tricornutum, and Chlamydomonas sp.) captured 50 to 65% of CO2 from air compared to only 38% removed by acidophiles (Coccomyxa sp., E. mutabilis, and E. gracilis).10 Indeed, both categories have their own limitations and merits and therefore can be implemented as per requirement. Especially for capturing concentrated flue gases containing SOx, NOx, and heavy metals, algal species tolerant to both high CO2 concentration and low pH are imperative.
6. Synthetic biology advancements
Recent advancements in genetic engineering of neutrophilic algal strains with acid/alkali-tolerance genes through advanced molecular biology approaches like RNA interference (RNAi) and clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 have enabled the fine-tuning of the metabolism of algae with enhanced performance.97 For instance, an alkali-tolerant strain of Chlamydomonas sp. was developed by transforming a cell wall synthesis gene named JbKOBITO 1 that encodes glycosyl transferase-like protein from Chlorella JB17.38 The gene was evidenced to provide tolerance to alkali medium in the presence of 300 mM sodium bicarbonate by enhancing 10.5% cellulose content in the cell wall and remodeling the sugar metabolome for an exclusive upsurge of trehalose and rhamnose (40-fold higher), compared to the wild-type strain.38 Moreover, the model algal strain, C. reinhardtii, was engineered to express the Proton-ATPase (PMA) to augment the CO2 tolerance to 20%.98 On a similar note, acid tolerance in P. tricornutum was developed by overexpressing transmembrane transporter genes, PtCPA (cation/proton antiporter) and PtSCL4 (HCO3− transporter).99 Further, to harness the full capacity of these extremophiles, these pH-tolerant strains have been genetically engineered to for generating high value metabolites. For instance, silencing of the genes encoding 3-ketoacyl-CoA thiolase (KAT 1/2) in E. gracilis, an acidophilic strain, resulted in an increased production of wax ester with a short chain.100 The gene KAT is known to catalyze the condensation reaction during fatty acid synthesis in the mitochondria. The knock-out of six different isozymes (EgKAT 1–6) in E. gracilis revealed the crucial role of KAT1/2 in the truncation of the C28 fatty acid acyl chain, resulting in modulated wax ester composition with 70% and 41% higher ratios of <C26 carbon in comparison to the wild-type.100 Moreover, silencing these genes did not cause any changes in the alga's composition under aerobic conditions but significantly modified the composition of the wax ester under hypoxia, improving the cold flow property of derived biodiesel.100 Further to achieve a stable compositional modification of wax ester in the Euglena strain, the authors demonstrated CRISPR-Cas9-based genome editing by creating multiple knock out strains of KAT 1/2 and acyl-CoA dehydrogenase (ACD).101 Similarly, CRISPR-based genome editing has also been performed to create modified Euglena strains with improved cell harvesting efficiency.102 This study adopted Cas9 ribonucleoprotein (RNP)-based genome editing for targeted deletion of Bardet–Biedl syndrome genes (BBS). The gene BBS 7/8 is responsible for the formation of full flagellar structure essential for the motility of E. gracilis, and after mutation resulted in a 32–38% reduced time for settling in comparison to wild-type cells.102 A recent study engineered a novel alkali-, salinity-, and temperature-resistant alga Chlamydomonas pacifica, through the combined action of mutagenesis, evolutionary adaptation, and overexpression of soybean-derived Dof and phosphoglucomutase (PGM1) resulting in an increase in lipid (28%) and starch (27%) content.103 The engineered Dof- and PGM1 strains attained a biomass productivity similar to the evolved C. pacifica strain (3.90 g per m2 per day) in 80 L pilot scale plant.103 Further, nitrogen limited conditions was used for cultivating both evolved and Dof-evolved strains, resulting in the production of 3–12% biodiesel on a dry weight basis. With a purity of 80%, the relative abundance of 42–48% was observed for the C16:0 fatty acid chain. Notably, thermoplastic polyurethane (TPU, A2141) with a molecular weight of 91 kDa was synthesized using diacids, linear diols, and diisocyanates derived from C. pacifica biomass. The algae-derived TPU, as a coating material increased the flexural rigidity of cotton canvas by 12-fold in comparison to the uncoated one.103 Overall, the study suggested that the combined action of biotechnological interventions and strategic engineering approaches can help to alleviate the burden of the huge cost associated with a single product. However, the commercial feasibility of utilizing these engineered strains at large scale further requires detailed techno-economical and life cycle assessment to establish a tangible route for sustained productivity and recovery of sustainable bioproducts. Indeed, to facilitate genetic improvements in algal strains for enhancing pH-tolerance, whole genome sequencing of these extremophiles is warranted.7. Challenges and future recommendations
Despite the immense potential of pH-resilient algae for bioenergy, bioproduct production, bioremediation, carbon sequestration, challenges persist, particularly strain selection, scaling up, performance optimization, and downstream processing in outdoor cultivation systems. For instance, the exact mechanism underlying pH-resilience in these algae is still not completely understood necessitating whole-genome sequencing in conjunction with other omics studies (transcriptomic, proteomic, metabolomics, and fluxomic) to decipher the key genes and metabolic pathways involved in maintaining cellular homeostasis under extreme pH conditions. A deeper understanding of the genetic makeup of these algae will aid in strain selection and custom-genetic engineering approaches. Indeed, bioprospecting novel pH-resilient strains, particularly wastewaters with wide pH variability, would be instrumental in establishing effective and stable treatment processes along with maximizing productivity. Furthermore, these wastewaters exhibit vast genetic diversity, and algal strains isolated from these environments could unravel novel biochemical pathways that could be harnessed for biotechnological applications. However, in natural environments, pH-resilient algal strains often co-exist with bacterial species, necessitating several rounds of antibiotic treatments to isolate axenic cultures, particularly for high-value metabolites, which could be challenging and time-consuming.Indeed, several parameters, such as cultivation, harvesting, drying, and metabolite extraction, determine the cost of algal biomass.104 Other parameters that influence overall production costs include operational and maintenance expenditures such as nutrients, CO2 sparging, energy, and water.104 However, one of the major barriers to scaling up the production of pH-resilient algae from lab to industrial level is the construction and installation of growth reactors (open raceway or PBR) as well as system modifications to avoid corrosion from low/high pH media, increasing the capital cost. Moreover, cultivating pH-resilient algae in open raceway ponds (ORPs) would be a more economical option due to the significant reduction in contamination by other invading organisms. A TEA conducted on Nannochloropsis sp. reported an overall biomass production cost of US$ 500 per ton in ORPs while US$ 9560 per ton in PBRs.105 Likewise, TEA of G. sulphuraria (acidophilic algae) cultivated in corn-stover-derived anaerobic digestate, and CO2-fed showed that the minimum biomass selling price was US $921 per dry metric ton in covered ponds (assuming productivity of 0.8 kg per m3 per day) compared to US$ 2869 per dry metric ton in PBRs (assuming productivity of 1.57 kg per m3 per day).106 The high cost of biomass production in PBRs was attributed to the cost of hydrolysate sugars and power required to operate the PBRs, accounting for 85% of the total cost. The authors reported a minimum renewable diesel selling price (MDSP) of US$ 8.24 and US$ 3.32 per dm per GE (gasoline equivalent) for the PBR and covered pond, respectively. To attain economic parity with conventional fuels (US$ 0.79 per dm per GE), using covered ponds, the authors proposed diverting 30% of algal biomass towards high value products such as phycocyanin, which sells for US 500–$100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per kg, depending on its purity. Furthermore, the life cycle assessment revealed a global warming potential of 39 and 9.1 gCO2-eq. per MJ on a well-to-wheels basis and a net energy ratio of 2.21 and 0.25 MJ/MJ for the PBRs and covered pond systems, respectively.106 Nonetheless, ORP cultivation systems require large tracts of land closer in specific locations/regions to maximize biomass productivity, which may not be feasible, particularly in densely populated areas posing an economic and logistic challenge. A recent economic feasibility analysis of microalgae-based biodiesel production using two marine algae species, Nannochloropsis oceanica and Dunaliella tertiolecta evaluated 12 international locations, including North and South America, Europe, the Middle East, India, Asia, and Oceania, to identify important conditions for scaling up using 500 ha ORPs.107 The authors identified growth, conversion through hydrothermal liquefaction, and harvesting/centrifugation as the largest cost components, ranging between US$ 128–245 million. Importantly, 10 out of these 12 locations, except for Turkey and the Netherlands, achieved an MSDP under US$ 6.99 per gal and nine under US$ 6.04 per gal bringing them closer to the non-renewable diesel benchmark of 3.02 per gal. Notably, adopting pH-resilient algae that could grow over the full 12 month period has the potential to boost productivity reducing the harvesting costs.107 Indeed, a trade-off between pH-resilience and desirable traits (growth rate, metabolic efficiency, and bioproduct yield) needs to be carefully balanced to attain the targeted productivity. For example, cultivating extremophile algae such as Arthrospira or Dunaliella resulted in a reduction in biomass production costs from € 14.5 per kg to € 9.46 per kg when biomass productivity improved to 26 g per m2 per day for high value bioproducts such as food additives and PUFAs.108 It is also important to restrict the pH-resilient algae from escaping the ORPs and invading local ecosystems to avoid loss of genetic diversity and alter or disrupt local biodiversity.109
000 per kg, depending on its purity. Furthermore, the life cycle assessment revealed a global warming potential of 39 and 9.1 gCO2-eq. per MJ on a well-to-wheels basis and a net energy ratio of 2.21 and 0.25 MJ/MJ for the PBRs and covered pond systems, respectively.106 Nonetheless, ORP cultivation systems require large tracts of land closer in specific locations/regions to maximize biomass productivity, which may not be feasible, particularly in densely populated areas posing an economic and logistic challenge. A recent economic feasibility analysis of microalgae-based biodiesel production using two marine algae species, Nannochloropsis oceanica and Dunaliella tertiolecta evaluated 12 international locations, including North and South America, Europe, the Middle East, India, Asia, and Oceania, to identify important conditions for scaling up using 500 ha ORPs.107 The authors identified growth, conversion through hydrothermal liquefaction, and harvesting/centrifugation as the largest cost components, ranging between US$ 128–245 million. Importantly, 10 out of these 12 locations, except for Turkey and the Netherlands, achieved an MSDP under US$ 6.99 per gal and nine under US$ 6.04 per gal bringing them closer to the non-renewable diesel benchmark of 3.02 per gal. Notably, adopting pH-resilient algae that could grow over the full 12 month period has the potential to boost productivity reducing the harvesting costs.107 Indeed, a trade-off between pH-resilience and desirable traits (growth rate, metabolic efficiency, and bioproduct yield) needs to be carefully balanced to attain the targeted productivity. For example, cultivating extremophile algae such as Arthrospira or Dunaliella resulted in a reduction in biomass production costs from € 14.5 per kg to € 9.46 per kg when biomass productivity improved to 26 g per m2 per day for high value bioproducts such as food additives and PUFAs.108 It is also important to restrict the pH-resilient algae from escaping the ORPs and invading local ecosystems to avoid loss of genetic diversity and alter or disrupt local biodiversity.109
Another challenge is the high cost associated with the downstream processing, which must be reduced to make microalgae biomass a preferred feedstock for biofuels and bioproducts. Typically, for large-scale algal cultivation systems, flocculation or filtration are the preferred choices owing to low energy requirements compared to centrifugation.110 However, due to the extreme pH conditions, it might be challenging to aggregate cells using typical flocculants, or pH-adjustment would be needed before harvesting. Moreover, the cell walls of some of the pH-resilient algae might contain complex polysaccharides, resulting in thicker and more robust cell walls, resisting lysis, thereby making the extraction of metabolites difficult with standardized cell breakage techniques such as mechanical disruption, or enzymatic or chemical lysis developed for neutrophilic algae. Thus, future efforts are needed to develop flocculants, such as electrocoagulants that work across a range of pH conditions, along with optimization of cell disruption technologies, such as high-pressure homogenization, that could disrupt the cell wall, increasing the metabolite yields without significantly increasing the cost. Notably, using artificial intelligence (AI) to digitize and automate microalgae culture and downstream processing has the potential to boost the efficiency and feasibility of microalgal biotechnology.109 For example, AI has been used to describe and predict the quantity and quality of high value bioproducts, as well as to monitor algal development in real time to enhance productivity (a thorough review is published elsewhere).109
Another potential application that could be explored is using pH-resilient algae for microbial fuel cells (MFCs). Algae-assisted cathodes offer several advantages compared to bacterial species, including high power generation, reducing aeration requirements, and biomass production, which could be utilized towards biofuel production.15 Compared to neutrophilic algae, utilization of pH-resilient algae in MFCs can allow the generation of electricity under fluctuating pH conditions, offering greater stability. The use of agricultural run-offs, industrial effluents, or mining wastewater for an integrated MFC systems using pH-resilient algae offers more flexibility, reducing the operational complexities such as maintaining external pH, enabling constant performance over longer periods. Finally, to make the large-scale cultivation of pH-resilient algae environmentally sustainable and economically feasible, a biorefinery approach is needed.111 A well-integrated algae biorefinery could be a cost-effective model for manufacturing biofuels and bioproducts such as carbohydrates, proteins, pigments, lipids, vitamins, and omega-fatty acids.
8. Conclusion
The use of pH-resilient algae offers a promising avenue for sustainable outdoor cultivation thanks to their ability to not only thrive in extreme pH conditions, but simultaneously exhibit high tolerance to other abiotic stresses, such as light, salinity, temperature, and heavy metals. In addition, these extremophilic algae offer a distinct advantage to commercial outdoor cultivation by preventing contamination by invaders and other neutrophilic microorganisms without the need for costly pH adjustments. Moreover, thanks to their unique metabolic adaptation to high concentrations of CO2 (for acidophilic and acid-tolerant) or bicarbonate (for alkaliphilic and alkali-tolerant), they are ideal candidates for sequestering CO2 from industrial flue gases in addition to enhancing the overall efficiency of the cultivation system. However, optimization of several parameters including cultivation, productivity, and down-stream processing are needed for scalability and economic feasibility. The present review highlights the unique pH adaptation mechanism of such extremophiles, along with their diverse biotechnological potential. It also acknowledges the challenges and underscores the importance of TEA and life-cycle analysis in determining the cost-effectiveness, environmental impact, and overall competitiveness of algae-based solutions, paving the way for their widespread use in sustainable biotechnology and industrial applications.Data availability
The data used in the present review consists of previously published studies and publicly available datasets. All relevant data sources and references are cited within the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
Figures were made using https://Biorender.com.References
- E. M. Ammar, N. Arora and G. P. Philippidis, Energies, 2020, 13, 6427 CrossRef CAS.
- F. Abiusi, E. Trompetter, A. Pollio, R. H. Wijffels and M. Janssen, Front. Microbiol., 2022, 13, 820907 CrossRef PubMed.
- H. Gimmler, Algal Adapt. Environ. Stresses, 2001, 259–290 CrossRef CAS PubMed.
- D. B. Johnson, FEMS Microbiol. Ecol., 1998, 27, 307–317 CrossRef CAS.
- W. Gross and C. Schnarrenberger, Plant Cell Physiol., 1995, 36, 633–638 CAS.
- G. Graziani, S. Schiavo, A. Nicolai and S. Buono, Food Funct., 2013, 144–152 RSC.
- S. Hedrich and A. Schippers, Curr. Issues Mol. Biol., 2021, 40, 25–48 CrossRef PubMed.
- J. K. Bwapwa, A. T. Jaiyeola and R. Chetty, S. Afr. J. Chem. Eng., 2017, 24, 62–70 Search PubMed.
- H. Gimmler and B. Degenhard, in Algal Adaptation to Environmental Stresses: Physiological, Biochemical and Molecular Mechanisms, Springer, 2001, pp. 291–321 Search PubMed.
- J. Piiparinen, D. Barth, N. T. Eriksen, S. Teir, K. Spilling and M. G. Wiebe, Algal Res., 2018, 32, 321–328 CrossRef.
- B. E. Jones, W. D. Grant, A. W. Duckworth and G. G. Owenson, Extremophiles, 1998, 2, 191–200 CrossRef CAS PubMed.
- J. Zhu, B. Guo, F. Qie, X. Li, X. Zhao, J. Rong and B. Zong, J. Energy Chem., 2022, 73, 13–25 CrossRef CAS.
- A. S. I. Khozin-goldberg, Biotechnol. Lett., 2013, 35, 1745–1752 CrossRef PubMed.
- Z. Chi, J. V. O. Fallon and S. Chen, Trends Biotechnol., 2011, 29, 537–541 CrossRef CAS PubMed.
- A. Azarpour, S. Zendehboudi, O. Mohammadzadeh and A. Reza, Energy Convers. Manage., 2022, 267, 115757 CrossRef CAS.
- A. R. Taylor, C. Brownlee and G. L. Wheeler, Trends Plant Sci., 2012, 17, 675–684 CrossRef CAS PubMed.
- S. Hirooka, Y. Hirose, Y. Kanesaki, S. Higuchi, T. Fujiwara, R. Onuma, A. Era, R. Ohbayashi, A. Uzuka, H. Nozaki, H. Yoshikawa and S. Y. Miyagishima, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E8304–E8313 CrossRef CAS PubMed.
- S. Abinandan, I. A. Perera, S. R. Subashchandrabose, K. Venkateswarlu, N. Cole and M. Megharaj, FEMS Microbiol. Ecol., 2020, 96, 1–12 CrossRef PubMed.
- S. Abinandan, K. Venkateswarlu and M. Megharaj, Curr. Res. Microb. Sci., 2021, 2, 100081 CAS.
- H. Gimmler, B. Treffny, M. Kowalski and U. Zimmermann, J. Plant Physiol., 1991, 138, 708–716 CrossRef CAS.
- W. Gross, Hydrobiologia, 2000, 433, 31–37 CrossRef CAS.
- A. Liu, L. Zhang, A. Zhou, F. Yang, Z. Yue and J. Wang, Environ. Sci. Pollut. Res., 2023, 30, 97209–97218 CrossRef CAS PubMed.
- H. Tatsuzawa, E. Takizawa, M. Wada and Y. Yamamoto, J. Phycol., 1996, 32, 598–601 CrossRef CAS.
- J. Poerschmann, E. Spijkerman and U. Langer, Microb. Ecol., 2004, 48, 78–89 CrossRef CAS PubMed.
- J. K. Eibl, J. D. Corcoran, G. N. A. Senhorinho, K. Zhang, N. S. Hosseini, J. Marsden, C. A. Laamanen, J. A. Scott and G. M. Ross, AMB Express, 2014, 4, 1–8 CrossRef CAS PubMed.
- M. Margalef-Català, I. Araque, A. Bordons, C. Reguant and J. Bautista-Gallego, Front. Microbiol., 2016, 7, 1554 Search PubMed.
- H. Zhao, L. Liu, S. Peng, L. Yuan, H. Li and H. Wang, Front. Microbiol., 2019, 10, 1393 CrossRef PubMed.
- H.-H. Wu, Y.-N. Zou, M. M. Rahman, Q.-D. Ni and Q.-S. Wu, Sci. Rep., 2017, 7, 42389 CrossRef CAS PubMed.
- S. Hayat, Q. Hayat, M. N. Alyemeni, A. S. Wani, J. Pichtel and A. Ahmad, Plant Signaling Behav., 2012, 7, 1456–1466 CrossRef CAS PubMed.
- C. Cid, L. Garcia-Descalzo, V. Casado-Lafuente, R. Amils and A. Aguilera, Proteomics, 2010, 10, 2026–2036 CrossRef CAS PubMed.
- A. Gerloff-Elias, D. Barua, A. Mölich and E. Spijkerman, FEMS Microbiol. Ecol., 2006, 56, 345–354 CrossRef CAS PubMed.
- I. Sekler, H.-U. Glaser and U. Pick, J. Membr. Biol., 1991, 121, 51–57 CrossRef CAS PubMed.
- M. A. Messerli, L. A. Amaral-Zettler, E. Zettler, S. K. Jung, P. J. S. Smith and M. L. Sogin, J. Exp. Biol., 2005, 208, 2569–2579 CrossRef CAS PubMed.
- S. Hirooka, Y. Hirose, Y. Kanesaki, S. Higuchi, T. Fujiwara, R. Onuma, A. Era, R. Ohbayashi, A. Uzuka, H. Nozaki, H. Yoshikawa and S. Y. Miyagishima, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E8304–E8313 CrossRef CAS PubMed.
- K. Wiangnon, W. Raksajit and A. Incharoensakdi, FEMS Microbiol. Lett., 2007, 270, 139–145 CrossRef CAS PubMed.
- A. Goyal, Plant Physiol. Biochem., 2007, 45, 696–704 CrossRef CAS PubMed.
- C. Liu, J. Liu, S. Hu, X. Wang, X. Wang and Q. Guan, PeerJ, 2019, 7, e7189 CrossRef PubMed.
- J. Qiu, J. Zhang, H. Zhao, C. Wu, C. Jin, X. Hu, J. Li, X. Cao, S. Liu and X. Jin, Front. Microbiol., 2023, 14, 1285796 CrossRef PubMed.
- D. Qu and X. Miao, Chemosphere, 2021, 265, 129046 CrossRef CAS PubMed.
- G. Mamo, Alkaliphiles Biotechnol., 2020, pp. 85–133 Search PubMed.
- K. Enomoto and N. Koyama, Curr. Microbiol., 1999, 39, 270–273 CrossRef CAS PubMed.
- K. Praveen, S. Abinandan, K. Venkateswarlu and M. Megharaj, ACS ES&T Eng., 2023, 4, 455–465 Search PubMed.
- E. V. Kupriyanova, M. A. Sinetova, S. M. Cho, Y. I. Park, A. G. Markelova and N. A. Pronina, Plants, 2013, 60, 465–471 CAS.
- T. Li, C. E. Sharp, M. Ataeian, M. Strous and D. De Beer, Front. Microbiol., 2018, 9, 2490 CrossRef PubMed.
- K. Qiao, M. Wang, T. Takano and S. Liu, Front. Plant Sci., 2018, 9, 1772 CrossRef PubMed.
- D. Qu, P.-L. Show and X. Miao, Int. J. Mol. Sci., 2021, 22, 2387 CrossRef CAS PubMed.
- A. Król, K. Mizerna and M. Bożym, J. Hazard. Mater., 2020, 384, 121502 CrossRef PubMed.
- F. Puente-sánchez, S. Díaz, V. Penacho, A. Aguilera and S. Olsson, Aquat. Toxicol., 2018, 200, 62–72 CrossRef PubMed.
- S. Olsson, F. Puente-Sánchez, M. J. Gómez and A. Aguilera, Extremophiles, 2015, 19, 657–672 CrossRef CAS PubMed.
- F. Puente-sánchez, S. Olsson and A. Aguilera, Microb. Ecol., 2016, 595–607 CrossRef PubMed.
- G. Schönknecht, W.-H. Chen, C. M. Ternes, G. G. Barbier, R. P. Shrestha, M. Stanke, A. Bräutigam, B. J. Baker, J. F. Banfield and R. M. Garavito, Science, 2013, 339, 1207–1210 CrossRef PubMed.
- S. Abinandan, S. R. Subashchandrabose and K. Venkateswarlu, Bioresour. Technol., 2019, 281, 469–473 CrossRef CAS PubMed.
- S. Abinandan, S. R. Subashchandrabose and N. Cole, Bioresour. Technol., 2019, 271, 316–324 CrossRef CAS PubMed.
- M. R. di Cicco, M. Palmieri, S. Altieri, C. Ciniglia and C. Lubritto, Int. J. Environ. Res. Public Health, 2021, 18, 2291 CrossRef CAS PubMed.
- S. Shanthakumar, S. Abinandan, K. Venkateswarlu, S. R. Subashchandrabose and M. Megharaj, Land Degrad. Dev., 2021, 32, 3157–3166 CrossRef.
- E. Sahabudin, J. Lee, R. Asada, E. A. Marsid, N. Yusof, N. S. Ahmad Sabri, H. Susanti, M. A. Muhammad Yuzir, F. N. Md Akhir and N. Othman, J. Appl. Phycol., 2022, 34, 1881–1892 CrossRef CAS.
- M. Robles, C. Ostojic, M. C. Ruiz-Domínguez, M. Cuaresma, C. Gonzalo, V. Obregón, J. L. Fuentes, A. Bartolomé and C. Vílchez, J. Appl. Phycol., 2024, 36, 2489–2502 CrossRef CAS.
- C. Casal, M. Cuaresma, J. M. Vega and C. Vilchez, Mar. Drugs, 2010, 9, 29–42 CrossRef PubMed.
- K. M. Tan, M. A. Kassim, Z. J. Ng and J. Lalung, in IOP Conference Series: Materials Science and Engineering, IOP Publishing, 2020, vol. 716, pp. 12011 Search PubMed.
- M. C. Ruiz-Domínguez, I. Vaquero, V. Obregón, B. de la Morena, C. Vílchez and J. M. Vega, J. Appl. Phycol., 2015, 27, 1099–1108 CrossRef.
- I. Vaquero, M. C. Ruiz-Domínguez, M. Márquez and C. Vílchez, Process Biochem., 2012, 47, 694–700 CrossRef CAS.
- F. Navarro, E. Forján, M. Vázquez, Z. Montero, E. Bermejo, M. Á. Castaño, A. Toimil, E. Chagüaceda, M. Á. García-Sevillano and M. Sánchez, Food Nutr. Res., 2016, 60, 30472 CrossRef PubMed.
- F. Navarro, E. Forján, M. Vázquez, A. Toimil, Z. Montero, M. del C. Ruiz-Domínguez, I. Garbayo, M. Á. Castaño, C. Vílchez and J. M. Vega, Phycol. Res., 2017, 65, 38–43 CrossRef CAS.
- J.-L. Fuentes, Z. Montero, M. Cuaresma, M.-C. Ruiz-Domínguez, B. Mogedas, I. G. Nores, M. González del Valle and C. Vílchez, Processes, 2020, 8, 324 CrossRef CAS.
- S. M. Hamed, H. I. El Shimi, J. R. van Dijk, A. I. Osman, S. M. Korany and H. AbdElgawad, J. Environ. Chem. Eng., 2022, 10, 108804 CrossRef CAS.
- M. Ma, Z. Yu, L. Jiang, Q. Hou, Z. Xie, M. Liu, S. Yu and H. Pei, J. Cleaner Prod., 2023, 390, 136105 CrossRef CAS.
- A. K. Kumar, S. Sharma, G. Dixit, E. Shah and A. Patel, Renewable Energy, 2020, 145, 1620–1632 CrossRef.
- K. Iwan, A. Kurniawan, H. Susanti, D. Saka and B. Mandra, J. Cleaner Prod., 2024, 477, 143857 CrossRef.
- M. Moglie, G. Biancini and L. Cioccolanti, Int. J. Life Cycle Assess., 2024, 29, 1000–1020 CrossRef CAS.
- B. Vázquez-Romero, J. A. Perales, H. Pereira, M. Barbosa and J. Ruiz, Sci. Total Environ., 2022, 837, 155742 CrossRef PubMed.
- V. Masindi, S. Foteinis, P. Renforth, J. Ndiritu, J. P. Maree and M. Tekere, Ecol. Eng., 2022, 183, 106740 CrossRef.
- C. Kuo, T. Lin, Y. Yang, W. Zhang, J. Lai and H. Wu, Bioresour. Technol., 2017, 244, 243–251 CrossRef CAS PubMed.
- A. Vadlamani, S. Viamajala, B. Pendyala and S. Varanasi, ACS Sustain. Chem. Eng., 2017, 5, 7284–7294 CrossRef CAS.
- S. Berry, Y. V Bolychevtseva, M. Rögner and N. V Karapetyan, Photosynth. Res., 2003, 78, 67–76 CrossRef CAS PubMed.
- M. Kishi and T. Toda, J. Appl. Phycol., 2018, 30, 401–410 CrossRef CAS.
- P. Wensel, G. Helms, B. Hiscox, W. C. Davis, H. Kirchhoff, M. Bule, L. Yu and S. Chen, Algal Res., 2014, 4, 2–11 CrossRef.
- Q. H. Shen, J. W. Jiang, L. P. Chen, L. H. Cheng, X. H. Xu and H. L. Chen, Bioresour. Technol., 2015, 190, 257–263 CrossRef CAS PubMed.
- D. Zhang, S. Wen, X. Wu and W. Cong, Bioprocess Biosyst. Eng., 2018, 41, 831–839 CrossRef CAS PubMed.
- Y. Kikuchi, D. Kanai, K. Sugiyama and K. Fujii, Fermentation, 2024, 10, 134 CrossRef CAS.
- K. A. Canon-Rubio, C. E. Sharp, J. Bergerson, M. Strous and H. De la Hoz Siegler, Appl. Microbiol. Biotechnol., 2016, 100, 1611–1622 CrossRef CAS PubMed.
- C. Zhu, Y. Xi, X. Zhai, J. Wang, F. Kong and Z. Chi, J. Cleaner Prod., 2021, 283, 124648 CrossRef CAS.
- C. I. Granada-Moreno, A. Aburto-Medina, D. de Los Cobos Vasconcelos and A. González-Sánchez, J. Appl. Microbiol., 2017, 123, 903–915 CrossRef CAS PubMed.
- L. Moreno-Garcia, K. Adjallé, S. Barnabé and G. S. V. Raghavan, Renew. Sustain. Energy Rev., 2017, 76, 493–506 CrossRef.
- Q. Zheng, X. Xu, G. J. O. Martin and S. E. Kentish, Chin. J. Chem. Eng., 2018, 26, 2219–2228 CrossRef CAS.
- N. M. Langley, S. T. L. Harrison and R. P. Van Hille, Biochem. Eng. J., 2012, 68, 70–75 CrossRef CAS.
- K. L. Kadam, Energy Convers. Manage., 1997, 38, S505–S510 CrossRef CAS.
- H. Leflay, J. Pandhal and S. Brown, J. CO2 Util., 2021, 52, 101657 CrossRef CAS.
- Y. Zhou and R. S. J. Tol, Water Resour. Res., 2005, 41, W03003 CrossRef.
- J. D. Noel, W. J. Koros, B. A. McCool and R. R. Chance, Ind. Eng. Chem. Res., 2012, 51, 4673–4681 CrossRef CAS.
- C. Gerotto, A. Norici and M. Giordano, Front. Energy Res., 2020, 8, 213 CrossRef.
- B. Retta, M. Iovinella and C. Ciniglia, Plants, 2024, 13, 1786 CrossRef CAS PubMed.
- H. Il Choi, S.-W. Hwang, J. Kim, B. Park, E. Jin, I.-G. Choi and S. J. Sim, Nat. Commun., 2021, 12, 6049 CrossRef PubMed.
- F. de Farias Neves, L. Hoinaski, L. R. Rörig, R. B. Derner and H. de Melo Lisboa, Environ. Technol., 2019, 40, 3308–3317 CrossRef PubMed.
- Y. Wang, A. Liu, C. Amanze, N. C. Ontita and W. Zeng, Bioresour. Technol., 2024, 414, 131572 CrossRef CAS PubMed.
- M. Ataeian, Y. Liu, K. A. Canon-Rubio, M. Nightingale, M. Strous and A. Vadlamani, Biotechnol. Bioeng., 2019, 116, 1604–1611 CrossRef CAS PubMed.
- S. Gao, K. Pittman, S. Edmundson, M. Huesemann, M. Greer, W. Louie, P. Chen, D. Nobles, J. Benemann and B. Crowe, J. CO2 Util., 2023, 69, 102399 CrossRef CAS.
- H. Alishah Aratboni, N. Rafiei, R. Garcia-Granados, A. Alemzadeh and J. R. Morones-Ramírez, Microb. Cell Fact., 2019, 18, 1–17 CrossRef CAS PubMed.
- J. Kim and B. Park, Nat. Commun., 2021, 1–15 Search PubMed.
- Y. Su, J. Chen, J. Hu, C. Qian, J. Ma, S. Brynjolfsson and W. Fu, iScience, 2024, 27, 110482 CrossRef CAS PubMed.
- M. Nakazawa, H. Andoh, K. Koyama, Y. Watanabe, T. Nakai, M. Ueda, T. Sakamoto, H. Inui, Y. Nakano and K. Miyatake, Lipids, 2015, 50, 483–492 CrossRef CAS PubMed.
- S. Nagamine, R. Oishi, M. Ueda, T. Sakamoto and M. Nakazawa, Bioresour. Technol., 2024, 410, 131255 CrossRef CAS PubMed.
- M. Ishikawa, T. Nomura, S. Tamaki, K. Ozasa, T. Suzuki, K. Toyooka, K. Hirota, K. Yamada, K. Suzuki and K. Mochida, Plant Biotechnol. J., 2022, 20, 2042 CrossRef PubMed.
- A. Gupta, J. V. Dutra Molino, K. M. J. Wnuk-Fink, A. Bruckbauer, M. Tessman, K. Kang, C. J. Diaz, B. Saucedo, A. Malik and M. D. Burkart, ACS ES&T Eng. Search PubMed.
- B. O. Abo, E. A. Odey, M. Bakayoko and L. Kalakodio, Rev. Environ. Health, 2019, 34, 91–99 CrossRef CAS PubMed.
- S. Banerjee and S. Ramaswamy, Bioresour. Technol. Rep., 2019, 8, 100328 CrossRef.
- M. D. Somers, P. Chen, J. Clippinger, J. R. Cruce, R. Davis, P. J. Lammers and J. C. Quinn, Algal Res., 2021, 59, 102419 CrossRef.
- J. Roles, J. Yarnold, K. Hussey and B. Hankamer, Biotechnol. Biofuels, 2021, 14, 133 CrossRef CAS PubMed.
- B. Llamas, M. C. Suárez-Rodríguez, C. V González-López, P. Mora and F. G. Acién, Algal Res., 2021, 57, 102339 CrossRef.
- A. S. Alzahmi, S. Daakour, D. Nelson, D. Al-Khairy, J.-C. Twizere and K. Salehi-Ashtiani, Front. Sustain. Food Syst., 2024, 8, 1331251 CrossRef.
- S. Khanra, M. Mondal, G. Halder, O. N. Tiwari, K. Gayen and T. K. Bhowmick, Food Bioprod. Process., 2018, 110, 60–84 CrossRef CAS.
- R. Araújo, F. Vazquez Calderón, J. Sánchez Lopez, I. C. Azevedo, A. Bruhn, S. Fluch, M. Garcia Tasende, F. Ghaderiardakani, T. Ilmjarv and M. Laurans, Front. Mar. Sci., 2021, 7, 626389 CrossRef.
- J. L. Fuentes, V. A. R. Huss, Z. Montero, R. Torronteras, M. Cuaresma, I. Garbayo and C. Vílchez, J. Appl. Phycol., 2016, 28, 3269–3279 CrossRef.
- L. S. Souza, C. Simioni, Z. L. Bouzon, R. De Cassia, S. Schneider, P. Gressler, M. C. Miotto, M. J. Rossi and L. R. Rorig, Protoplasma, 2017, 254, 1385–1398 CrossRef CAS PubMed.
- M. Cuaresma, C. Casal, E. Forján and C. Vílchez, J. Ind. Microbiol. Biotechnol., 2011, 38, 167–177 CrossRef CAS PubMed.
- F. de Farias Neves, R. B. Dernerb, L. R. Rorigc, H. de Melo and J. Algal, Biomass Utln., 2017, 8, 125–130 Search PubMed.
- S. Hirooka and S. Miyagishima, Front. Microbiol., 2016, 7, 2022 Search PubMed.
- M. R. Gauthier, G. N. A. Senhorinho, N. Basiliko, S. Desjardins and J. A. Scott, Ind. Biotechnol., 2022, 18, 168–175 CrossRef CAS.
- S. Hirooka, S. Higuchi, A. Uzuka, H. Nozaki and S. Miyagishima, PLoS One, 2014, 9, 3–9 CrossRef PubMed.
- S. Kim, R. Wirasnita, D. Lee, J. Yu and T. Lee, Appl. Sci., 2021, 11, 8182 CrossRef CAS.
- K. Yoshioka, K. Suzuki and T. Osanai, Algal Res., 2020, 51, 102084 CrossRef.
- K. Qiao, T. Takano and S. Liu, Algal Res., 2015, 9, 245–253 CrossRef.
- H. Yang, Q. He, J. Rong, L. Xia and C. Hu, Bioresour. Technol., 2014, 172, 131–137 CrossRef CAS PubMed.
- R. Selvarajan, T. Felföldi, T. Tauber, E. Sanniyasi, T. Sibanda and M. Tekere, Energies, 2015, 8, 7502–7521 CrossRef.
- R. Chowdhury, P. L. Keen and W. Tao, Bioresour. Technol. Rep., 2019, 6, 229–236 CrossRef.
| This journal is © The Royal Society of Chemistry 2025 |