Open Access Article
Open Access ArticleA biomimetic nanofluidic tongue for highly selective and sensitive bitterness perception†
Yanan Lia,
Hua Zhenga,
Lei Xua,
Linfeng Chen *a,
Fan Xia
*a,
Fan Xia a and
Yanlin Song
a and
Yanlin Song b
b
aState Key Laboratory of Geomicrobiology and Environmental Changes, Engineering Research Center of Nano-Geomaterials of Ministry of Education, Faculty of Materials Science and Chemistry, China University of Geosciences, Wuhan 430074, China. E-mail: chenlinfeng@cug.edu.cn
bKey Laboratory of Green Printing, CAS Research/Education Center for Excellence in Molecular Sciences, Institute of Chemistry, Chinese Academy of Sciences (CAS), Beijing 100190, P. R. China
First published on 7th May 2025
Abstract
Precise detection of bitter compounds is crucial for enhancing life quality, ensuring food safety, and improving pharmaceutical efficacy. However, the structural diversity of bitter compounds often limits detection specificity and sensitivity. Inspired by the human taste perception mechanism, we have developed a biomimetic nanofluidic tongue based on bitter receptor-integrated nanochannels for the highly selective and sensitive perception of bitter molecules. This hybrid platform is constructed by leveraging the T2R38 receptor as a model protein to chemically modify the nanochannel surfaces, enabling a sensitive recognition of specific bitter molecules in a wide linear detection range from 10−4 M to 0.1 pM. An ultralow detection limit of 0.018 pM is achieved for structurally diverse bitter molecules, including allyl isothiocyanate, yohimbine hydrochloride, and limonin. The sensor remains stable at 4 °C for a week and is reusable. Additionally, it has also been successfully applied to detect bitter limonin in spoiled orange juice. This artificial bitter sensor holds significant potential in the food and beverage industries, pharmaceuticals, and humanoid robots, and provides valuable insights for the design of future bitter sensors.
Introduction
Bitterness, one of the five basic taste sensations (sour, sweet, bitter, salty, and umami), plays a crucial role in mammalian survival by enabling the detection and avoidance of potentially toxic substances.1–4 Bitter compounds are prevalent in everyday foods, beverages, and medications, significantly impacting taste and posing health risks when found in spoiled or stale food products.5 Detecting bitter substances is thus vital for ensuring food safety, enhancing flavor profiles, and improving pharmaceutical efficacy.Bitterness is caused by thousands of bitter compounds which have diverse chemical structures, including polyphenols, flavonoids, terpenes, amino acids, glucosinolates, etc.2,6 Traditional chemical techniques face significant challenges in detecting this vast array of bitter compounds due to their structural diversity. However, the human TAS2R family, comprising only 25 receptors, has the remarkable ability to perceive these bitter compounds.7 Sensory evaluation is thus a primary method for testing various tastes, which offers a direct taste experience.8 Despite its effectiveness, this method requires trained tasters in specialized, qualified laboratories and is subject to individual biases, preferences and sensory acuity.9–11 Moreover, it is not suitable for evaluating bitter compounds that are toxic or pose health risks. Electronic tongues,12–14 which employ an array of low-selectivity chemical sensors, are prevalent but face several limitations, including high sample usage, limited detection scope and insufficient selectivity.15–18 These limitations make it difficult to accurately identify bitter compounds in complex systems.19,20 Therefore, developing new bitter sensors with high selectivity and sensitivity to detect the structural diversity and complexity of bitter compounds remains a significant challenge.
The human taste system's remarkable ability to detect bitterness has inspired the creation of innovative bitter sensors. Bioelectronic sensors,21 leveraging bioactive components such as taste receptors,22 cells,23 and tissues,24 have gained increasing attention for their biomimetic taste detection capabilities and well-defined detection mechanism.25 Most bioactive components used as taste-sensitive elements are coupled with microelectrode arrays or immobilized on the surfaces of quartz crystal microbalances and field-effect transistor devices.26–28 Owing to the presence of bioactive components, such biosensors have shown considerable potential in taste detection. However, these sensors still face challenges that cannot be ignored, such as insufficient sensitivity, poor stability and complex manufacturing processes. These issues are critical factors that determine whether biosensors can be widely applied.
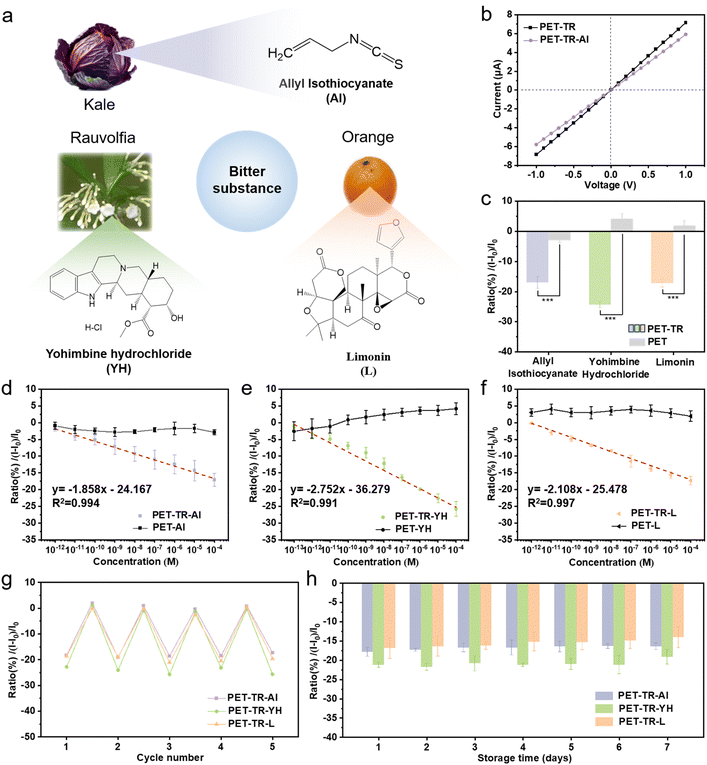
As shown in Fig. 1a, selective bitterness perception in humans involves two key processes: (1) selective recognition of bitter molecules by bitter receptors, and (2) biological channel-mediated electrical signal generation. The binding of bitter molecules to a receptor initiates intracellular signaling, activating biological channels and allowing the influx of Na+, which subsequently generates a potential signal.29–32 Finally, the taste is perceived as this electrical signal is transmitted to the brain. Drawing inspiration from human taste sensation, we propose a novel nanofluidic tongue that integrates solid-state nanochannels with bitter taste receptors for highly selective and sensitive detection of bitter compounds (Fig. 1b). Solid-state nanochannels have attracted considerable attention for their excellent mechanical properties,33,34 adjustable geometries,35–38 outstanding biocompatibility,39 and favorable electrochemical properties.40–46 Most importantly, these nanochannels can confine target analytes within extremely small volumes, thereby greatly enhancing sensitivity to ionic current perturbations and offering an ideal platform for molecular sensing.47–60 As a proof-of-concept, T2R38, one of the 25 known bitter taste receptors,61,62 was selected as a bitter probe to chemically functionalize the surfaces of nanoporous poly(ethylene terephthalate) (PET). Due to the advantages of nanochannel technology, this functionalization enabled the construction of a bitter sensor, PET-TR, which can detect specific bitter molecules with high sensitivity. Upon exposure to these molecules, PET-TR translates the recognition event into detectable changes in ionic current signals. The biomimetic sensor demonstrates high sensitivity for bitter compounds across a wide concentration range. Moreover, it maintains good stability, retaining activity for over 7 days when stored at 4 °C and can be reused multiple times without significant loss of performance. Importantly, we successfully applied PET-TR to detect limonin, a bitter compound, in spoiled orange juice, highlighting the sensor's potential in food quality monitoring and safety in real-world scenarios.
Experimental
Materials
The polyethylene terephthalate (PET) track etched membrane was produced by IT4IP S.A. in Belgium. The protein T2R38 was purchased from Shanghai Wu hao Biotechnology Co., Ltd. The bovine serum protein (BSA) was purchased from Guangzhou Saiguo Biotech Co., Ltd (China). 1-(3-Dimethylaminopropyl)-3-ethyl-carbodiimide hydrochloride (EDC), N-hydroxy succinimide (NHS, ≥98.0%), sodium chloride (NaCl, ≥99.0%), monosodium glutamate (MSG), quinine, allyl isothiocyanate (AI), and yohimbine hydrochloride (YH) were obtained from Shanghai Aladdin Biochemical Technology Co., Ltd (China). Phosphate buffered saline (PBS, pH 7.4) and sucrose (≥99%) were supplied by Shanghai Macklin Biochemical Technology Co., Ltd (China). Guanidine hydrochloride (GuHCl) was sourced from Biofroxx in Germany. Citric acid was procured from Sinopharm Chemical Reagent Co., Ltd (China). Limonin (L) was obtained from the Energy Chemical brand of Sarn Chemical Technology Co., Ltd (Shanghai). Vitamin C was purchased from ChengDu Chron Chemicals Co., Ltd (China).Method
Solutions of allyl isothiocyanate (AI), yohimbine hydrochloride (YH), and limonin (L) at concentrations ranging from 10−4 M to 10−12 M in PBS buffer solution (10−3 M, pH 7.4) were prepared. 1 mL of a 10−4 M solution of each bitter compound was introduced into the chamber, replacing the initial PBS buffer solution. The electrochemical measurement was repeated as described above, yielding a new I–V curve and recording the transmembrane ion current at 1 V, denoted as I1. These tests were conducted at least three times to calculate the average current values. The differences in current before and after the introduction of bitter substances, (I1 − I0)/I0 × 100%, were calculated for the 10−4 M solutions of AL, YH, and L. Subsequently, the concentration of bitter substances was varied to further investigate the transmembrane ion current. The current change ratios for different concentrations of each bitter substance were analyzed to determine the sensor's sensitivity and specificity in detecting bitter compounds. In addition, a 10−4 M solution of limonin was prepared in PBS (pH 7.4), and high-performance liquid chromatography (HPLC) analyses were performed at 3 hours, Day 1, and Day 3 to evaluate the stability of limonin in PBS.
To assess the stability, the same procedure was applied to calculate current change ratios for 10−4 M solutions of AI, YH, and L against the PBS buffer. After each measurement, the PET-TR membrane was gently washed and stored at 4 °C to minimize degradation. The current change ratios were monitored over a period of 7 days to determine whether there were any significant fluctuations, thus providing insights into the sensor's long-term stability. In addition, another batch of PET-TR membranes was stored at room temperature after each measurement, and their current change ratios over the same 7 days were also monitored to assess the sensor's stability at room temperature.
Mechanism analysis
In addition, to exclude the possibility of structural changes in the nanochannels, BSA was employed as a non-specific protein to modify the surface of the PET nanochannels (PET-BSA), and the current change ratios in the presence of 10−4 M solutions of AI, YH, and L were collected.
Additionally, T2R38 (10 μL) was applied to a silicon wafer and allowed to react with AI, YH, and L solutions (10 μL, 10−4 M) for 2 h. AFM was employed to characterize any size changes in the T2R38 receptor after ligand interaction.
To verify the reliability of the bitter sensor, a 6 × 10−5 M (C0) solution of limonin was prepared using a PBS buffer solution (10−3 M, pH 7.4). The biomimetic bitter sensor was used to detect the current of the prepared limonin solution, and a linear fitting current change diagram was applied to determine the concentration Ca.
High-performance liquid chromatography (HPLC) was employed to measure the peak areas of five different concentrations of limonin solutions (0.002 mM, 0.01 mM, 0.02 mM, 0.1 mM, 0.2 mM) at 210 nm, and a linear fitting was performed. The peak area of the limonin solution prepared for sensor testing was measured, and the concentration Cb was derived from the linear fitting peak area variation diagram. The Ca and Cb obtained by both the bitter sensor and HPLC methods were compared with C0 to validate the sensor's accuracy.
Fresh orange juice (FOJ) was allowed to deteriorate naturally by storage in air for 3 days to prepare deteriorated orange juice (DOJ). Subsequently, the PET-TR membrane was placed between two chambers, and 1 mL of each sample-FOJ, DOJ, vinegar, cola, coffee, monosodium glutamate (MSG) and NaCl salt solution was injected into the chambers for testing. It should be noted that FOJ, DOJ, vinegar and cola samples were diluted 500-fold with PBS butter (pH 7.4), while the coffee, MSG and NaCl salt solution samples were prepared directly at a concentration of 0.047 g L−1 in PBS butter (pH 7.4). All diluted samples were tested under the same conditions as previous experiments, and the transmembrane ion current at a voltage of 1 V was recorded. The tests aimed to observe whether there is significant difference in the ratio of current change between DOJ and other foods. To eliminate the potential interference from other components in orange juice, artificial orange juice samples containing water (adjusted to a final volume of 100 mL), sucrose (6 g), citric acid (0.65 g), and vitamin C (0.05 g) were specially prepared for the test. Additionally, the sample with the addition of limonin (1.2 mg) was used for the comparison.
Results and discussion
Construction of the biomimetic bitter sensor PET-TR
To construct the biomimetic bitter sensor PET-TR, PET nanochannel membranes with an average pore diameter of 28.85 nm were selected as the substrates (Fig. 2a and S1†). The details of the T2R38 sequence can be found in Fig. S2.† The protein structure of T2R38 (UniProt: P59533), obtained from the PDB file, has dimensions of 8.803 nm, 4.940 nm, and 7.092 nm along the X, Y, and Z axes, respectively (Fig. S2b†). T2R38 was immobilized on the PET nanochannel surfaces through an amidation reaction between the amine residues in T2R38 and the carboxylic groups abundantly present on the PET membrane surfaces (Fig. S3†). After T2R38 modification, the PET-TR membrane surface exhibited granular deposits, and the average pore diameter decreased to approximately 18.47 nm, as compared to the unmodified PET membrane (Fig. 2b and S1†). Simultaneously, the water contact angle (CA) of the membrane decreased from 73.1° to 58.3° (Fig. 2c), a change attributed to the increased surface roughness post-modification, which was consistent with the atomic force microscopy (AFM) analysis (Fig. S4†). Energy-dispersive X-ray spectroscopy (EDS) and X-ray photoelectron spectroscopy (XPS) were used to further characterize the functionalization, confirming the presence of sulfur, a key element in the cysteine (C) and methionine (M) residues of T2R38 (Fig. 2d–f, Table S1 and S2†). In addition, the surface of PET-TR became more negative, as indicated by kelvin probe force microscopy (KPFM) (Fig. 2g). Fourier transform infrared spectroscopy (FTIR) analysis showed two new peaks in the 1600–1500 cm−1 range for PET-TR, which correspond to the amine groups of T2R38 (Fig. S5†).Bitterness detection performance of the biomimetic bitter sensor
The detection of the bitter sensor PET-TR for bitter substances is briefly illustrated in Fig. 1b. The PET membranes were fixed between two home-made electrochemical cells, each filled with a PBS buffer solution (10−3 M, pH 7.4). Two electrodes were separately inserted into the cells, and a voltage sweep from −1 V to +1 V was applied. The resulting ionic current was monitored, and the plotted ionic current–voltage (I–V) curves under different conditions were analyzed. More experimental details can be found in the Experimental section of the ESI and are illustrated in Fig. S6.† The I–V curve showed a decrease in current after T2R38 modification (Fig. 2h), primarily due to the reduced diameter of the nanochannels, a finding that aligns with the statistical analysis of the diameter changes.Allyl isothiocyanate (AI), a typical bitter ligand for T2R38,62 is a natural compound found in cruciferous vegetables such as kale (Fig. 3a), and was selected for the detection. It was found that in the presence of 10−4 M AI, there was a noticeable decrease in the ionic current (Fig. 3b). To quantitatively correlate the ionic current change with the AI concentration, the current at 1 V for PET-TR before (I0) and after (I1) the addition of bitter ligand AI was further analyzed. The change in current was quantified using the formula (I1 − I0)/I0 × 100%. About 17.11% of current reduction for AI was determined (Fig. 3c). Another two bitter ligands, i.e., yohimbine hydrochloride (YH), and limonin (L),62 which are commonly found in Rauvolfia genus plants and in citrus fruits (e.g., oranges) respectively (Fig. 3a), were also selected for the test. At a concentration of 10−4 M, a reduction in current was observed for both YH and L, with current change ratios of 24.45% and 17.30%, respectively. Limonin remained stable in the PBS buffer within the test period (Fig. 3c, S7 and S8†). Moreover, within the tested concentration range, the current change ratios for these three bitter substances exhibited a linear correlation (Fig. 3d–f). As the concentration of the bitter substances increased, the current gradually decreased (Fig. S9†). The limit of detection (LOD) for AI, YH, and L using the PET-TR membrane was calculated to be 0.262 pM, 0.018 pM, and 0.231 pM, respectively, using the formula LOD = 3 × (SD/|m|), where SD represents the standard deviation of the blank solution, and m is the slope of the calibration curve. In contrast, the control PET membrane without bitter receptor modification, exhibited no obvious and consistent current changes in the presence of these three bitter substances (Fig. 3d–f and S10†). These findings underscore the biomimetic bitter sensor's capacity for highly sensitive detection of AI, YH, and L. A detailed comparison between the nanofluidic bitter sensor and other existing techniques is presented in Table S3.† Collectively, the new bitter sensor demonstrates superior performance, characterized by high selectivity and sensitivity, an ultralow LOD, a broad linear range, minimal sample requirement, and a shorter response time.
To evaluate the reusability of the biomimetic taste sensor, PET-TR membranes were alternatively applied to test PBS solutions with and without bitter substances (10−4 M). As shown in Fig. 3g, the artificial bitter sensors maintained consistent detection performance across five consecutive cycles, with no significant deterioration in sensitivity observed. Furthermore, the stability of the biomimetic bitter sensor was assessed over an extended period. The PET-TR sensors stored at 4 °C and room temperature were used for bitterness detection over a period of 7 days, respectively. As shown in Fig. 3h, the biomimetic bitter sensors stored at 4 °C retained a substantial portion of their initial response currents for AI, YH, and L, at 91.81%, 90.24%, and 83.15%, respectively. In contrast, when stored at room temperature the sensors gradually lost their activity, retaining only 13.79%, 9.86% and 8.74% of their initial response currents for AI, YH, and L, respectively, and basically lost their performance in identifying bitterness (Fig. S11†). Essentially, the sensors stored at room temperature lost their ability to identify bitterness after 7 days. These results emphasize the repeatability and good stability of the PET-TR sensor when stored at 4 °C, highlighting its potential for practical applications in environments where controlled storage conditions can be maintained.
Bitterness detection mechanism analysis
The biomimetic bitter sensor PET-TR for the detection of bitter molecules is attributed to the specific binding of T2R38 to the bitter ligands, resulting in the decrease of effective nanochannel size and the reduction of ionic current (Fig. 4a). To verify the pivotal role of T2R38 in bitter substance detection, PET-TR membranes were subjected to a denaturation process by immersing in a guanidine hydrochloride (GuHCl) solution (5 M) overnight. GuHCl is a potent denaturant that could denature T2R38 by disrupting its intramolecular hydrogen bonds and enhancing hydrophobic interactions.63,64 Post-denaturation, the PET-TR membranes were used to measure the response to 10−4 M solutions of AI, YH, and L, as shown in Fig. 4b. The structure alteration of T2R38 after the denaturation treatment rendered the protein incapable of binding to bitter molecules, resulting in a significant reduction in current change ratios when compared to untreated samples (Fig. 4b). In addition, PET membranes modified with non-specific protein BSA (PET–BSA) were also prepared and measured (Fig. S12†). No significant current change was observed in the presence of 10−4 M solutions of AI, YH, and L, indicating that the PET–BSA did not exhibit recognition of bitter substances. These results underscore the indispensable nature of the bitter taste protein in the detection of bitter substances, thereby validating the sensor's reliance on the integrity of the T2R38 protein for accurate sensing.We further examined the PET-TR by SEM after interaction with bitter ligands (Fig. S13†). As shown in Fig. 4c, the average diameter of the PET-TR reduced to about 15.38 nm, which supports our hypothesis that the ionic current decrease is due to the reduction of the nanochannels. Additionally, we employed AFM to analyze the size change of the bitter receptor after binding with the bitter ligands. It was observed that the particle size significantly increased for T2R38-AI, T2R38-YH and T2R38-L complexes. Concurrently, the surface roughness (Rq) increased from 0.477 nm for the control to 0.678 nm for T2R38-AI, 0.813 nm for T2R38-YH and 0.708 nm for T2R38-L, respectively (Fig. 4d).
To gain a deep understanding of the interaction mechanism between T2R38 and the bitter ligands, we conducted molecular docking studies of T2R38. The T2R38 protein structure was retrieved from the AlphaFold protein structure database (UniProt: P59533). Using PyMOL software, molecular docking was performed to obtain the complex structures (Fig. 4e). In the T2R38-AI complex, AI forms a hydrogen bond with the amino acid residue Serine-63 (SER-63). YH engages with T2R38 residues Threonine-155 (THR-155) and Serine-151 (SER-151), while L forms three hydrogen bonds with T2R38 residues Serine-51 (SER-51), Aspartic acid-54 (ASP-54) and Serine-248 (SER-248). The molecular affinity of all three bitter molecules for T2R38 is favorable (Table S4†). The molecular docking results provide a clear understanding of the binding interaction between T2R38 and the three bitter compounds, elucidating the molecular basis for the sensor's detection capabilities. However, it should be noted that the interaction between bitter molecules and the bitter receptor (i.e., molecular affinity) and the ionic current change ratio (reflecting protein structure change) may not be strictly linear. For instance, although AI exhibits a weaker binding affinity to the receptor compared to YH and L, the LOD values for AI and L are similar. This suggests that other factors, such as the conformational changes induced by binding or the specific interactions within the receptor, may also play a significant role in the detection mechanism. Further studies are warranted to explore this relationship more thoroughly.
Application demonstration of the biomimetic bitter sensor
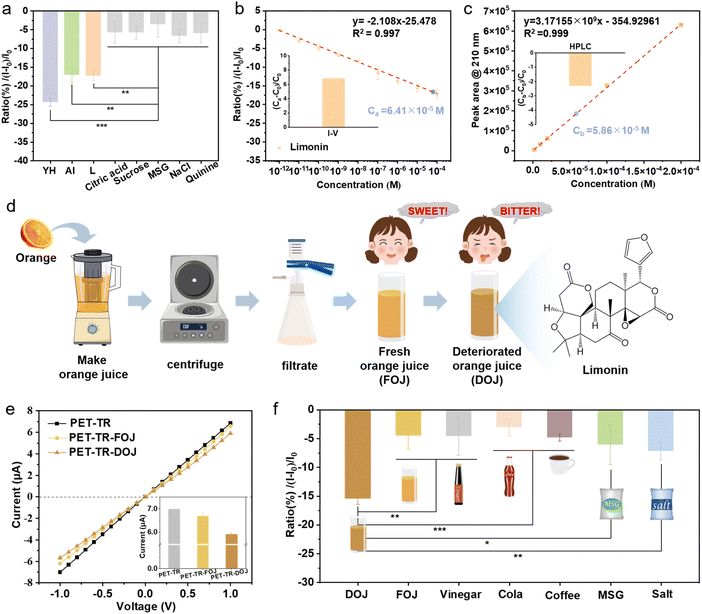
To evaluate the capability of the biomimetic taste sensor for selectively detecting bitter taste, 10−4 M of three bitter substances (AI, YH, and L) and the other taste molecules, including sour citric acid, sweet sucrose, umami monosodium glutamate (MSG), and salty sodium chloride (NaCl) were measured. As shown in Fig. 5a, the current change ratios for citric acid, sucrose, MSG and NaCl are significantly lower than those for AI, YH, and L, with ratios of −5.76%, −5.79%, −3.53%, and −6.66%, respectively. In addition, we also tested another typical bitter molecule quinine, which could not be recognized by T2R38. The ratio of current change is about −5.98%, further confirming the selective responsibility of PET-TR.To demonstrate the reliability of the biomimetic taste sensor for detecting bitter substances, comparative measurements of a certain concentration of L (C0 = 6.00 × 10−5 M) by using both high performance liquid chromatography (HPLC) and our bitter sensor were performed. The concentration of the as-prepared L was determined to be Ca = 6.41 × 10−5 M by the bitter sensor platform while the result was about 5.86 × 10−5 M by the HPLC method (Fig. 5b, c and S14†). The deviations of the two methods were about 6.83% and −2.30%, respectively, indicating that the biomimetic bitter sensor has relatively high accuracy and reliability for detecting bitter molecules.
We further demonstrated the potential of the biomimetic bitter sensor in the detection of real samples, where the presence of bitter compounds can be indicative of food spoilage and pose health risks. Bitterness detection is important for ensuring food safety and maintaining product quality. As a proof-of-concept, we selected orange juice for the demonstration. Freshly squeezed orange juice is rich in vitamin C and flavonoids, which can enhance the immune system, counteract free radical damage, and promote digestion. However, the acidic pH of the juice gradually induces the conversion of limonoate A-ring lactone into the bitter limonin, a process further accelerated by limonin D-ring lactone hydrolase (Fig. S15†),65,66 resulting in a slight bitterness to the juice. Extended storage under acidic conditions and in open air increases the production of bitter limonin (i.e., deterioration), thus intensifying the bitterness and compromising the juice's quality.
The preparation of fresh orange juice (FOJ) and a deteriorated sample (DOJ) after 3 days of storage in air is briefly shown in Fig. 5d. Our bitter sensor found that both FOJ and DOJ showed a decrease in ionic current, but the DOJ sample exhibited a much larger current change ratio, indicating a higher concentration of L (Fig. 5e). The other major components did not interfere with the test, confirmed by the results with artificial orange juice samples (Fig. S16†). As a comparison, real samples collected from table edible vinegar, cola, coffee, MSG, and salt solutions were also tested (Fig. 5f). These results indicate that the PET-TR bitter sensor holds promise for the detection of bitter compounds in various real sample, demonstrating its potential for broader application in food quality assessment and safety monitoring.
Conclusions
Inspired by biological taste systems to selectively sense bitter compounds, we have proposed and developed an innovative bitter sensor platform that integrates a biological bitter receptor with solid-state nanochannels. As a proof-of-concept, bitter receptor protein T2R38 was functionalized on the surface of PET nanochannels to give the artificial bitter sensor PET-TR. We have successfully demonstrated that this bitter sensor is capable of selectively detecting typical bitter ligands, including AI, YH, and L. The sensor offers wide linear detection ranges with ultra-sensitive detection limits of 0.262 pM for AI, 0.018 pM for YH, and 0.231 pM for L, respectively. This novel bitter sensor exhibits good reusability and could work efficiently for a week when stored properly. We also effectively applied the bitter sensor for L detection in a real sample, such as orange juice. The results highlight the practical utility of the biomimetic bitter sensors in quality monitoring of food and beverages, and in ensuring food safety. The potential applications of these biomimetic bitter sensors extend beyond quality control to include the food industry, pharmaceuticals, and even humanoid robots. This work not only contributes to the advancement of taste sensing technologies but also paves the way for the design of new bitter sensors with broad applications in various industries.It is worth noting that while we are actively improving our bitter taste sensor system to ensure its compatibility with commercial electronic devices, such as mobile phones and chips, the sensor still faces significant challenges in large-scale production and industrialization. Addressing the limitations will be a central focus of our future research efforts.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Conceptualization: L. C., methodology: Y. L. and L. C., investigation: Y. L., H. Z. and L. X., visualization: Y. L., H. Z. and L. X., supervision: L. C. and F. X., writing – original draft: Y. L., writing – review & editing: L. C., F. X., and Y. S.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (2021YFA1200403), the National Natural Science Foundation of China (22275171, 22090050 and U24A20502), Guangdong Basic and Applied Basic Research Foundation (2024A1515012749), and the Natural Science Foundation of Shenzhen (JCYJ20220530162406014).References
- D. A. Yarmolinsky, C. S. Zuker and N. J. P. Ryba, Cell, 2009, 139, 234–244 CrossRef CAS PubMed.
- X. Chu, W. Zhu, X. Li, E. Su and J. Wang, Food Res. Int., 2024, 183, 114234 CrossRef.
- J. Chandrashekar, K. L. Mueller, M. A. Hoon, E. Adler, L. Feng, W. Guo, C. S. Zuker and N. J. Ryba, Cell, 2000, 100, 703–711 CrossRef CAS.
- J. Chandrashekar, M. A. Hoon, N. J. Ryba and C. S. Zuker, Nature, 2006, 444, 288–294 CrossRef CAS PubMed.
- A. A. Bachmanov and G. K. Beauchamp, Annu. Rev. Nutr., 2007, 27, 389–414 CrossRef CAS PubMed.
- G. Vinci and M. L. Antonelli, Food Control, 2002, 13, 519–524 CrossRef CAS.
- W. Xu, L. Wu, S. Liu, X. Liu, X. Cao, C. Zhou, J. Zhang, Y. Fu, Y. Guo, Y. Wu and Q. Tan, Science, 2022, 377, 1298–1304 CrossRef CAS.
- M. P. Sáenz-Navajas, E. Campo, J. M. Avizcuri, D. Valentin, P. Fernández-Zurbano and V. Ferreira, Anal. Chim. Acta, 2012, 732, 64–72 CrossRef.
- T. Xia, S. Su, L. Wang, Z. Tang, J. Huo and H. Song, Food Chem., 2024, 457, 140150 CrossRef CAS.
- S. Scharbert and T. Hofmann, J. Agric. Food Chem., 2005, 53, 5377–5384 CrossRef CAS PubMed.
- M. Yu, T. Li, A. Razaa, L. Wang, H. Song, Y. Zhang, L. Li and Y. Hua, Food Res. Int., 2020, 129, 108880 CrossRef CAS PubMed.
- M. Ghasemi-Varnamkhasti, C. Apetrei, J. Lozano and A. Anyogu, Trends Food Sci. Technol., 2018, 80, 71–92 CrossRef CAS.
- C. W. Lee, S. E. Jun, S. J. Kim, T. H. Lee, S. A. Lee, J. W. Yang, J. H. Cho, S. Choi, C. J. Kim, S. Y. Kim and H. W. Jang, InfoMat, 2023, 5, e12427 CrossRef CAS.
- Y. Hou, M. Genua, D. Tada Batista, R. Calemczuk, A. Buhot, P. Fornarelli, J. Koubachi, D. Bonnaffé, E. Saesen, C. Laguri and H. Lortat-Jacob, Angew. Chem., Int. Ed., 2012, 51, 10394–10398 CrossRef CAS PubMed.
- Z. Zhang and B. Yan, Adv. Funct. Mater., 2024, 34, 2316195 CrossRef CAS.
- P. Ciosek-Skibińska, K. Cal, D. Zakowiecki and J. Lenik, Materials, 2024, 17, 5016 CrossRef.
- Z. Zhao, F. Song, S. Kimura, T. Onodera, T. Uchida and K. Toko, Microchem. J., 2024, 200, 110248 CrossRef CAS.
- X. Zhang, H. Wu, X. Yu, H. Luo, H. Yang, X. Li, Z. Li, L. Tang and Z. Wang, Molecules, 2018, 23, 3362 CrossRef.
- X. Wei, C. Qin, C. Gu, C. He, Q. Yuan, M. Liu, L. Zhuang, H. Wan and P. Wang, Biosens. Bioelectron., 2019, 145, 111673 CrossRef CAS.
- M. Podrazka, E. Baczynska, M. Kundys, P. S. Jelen and E. W. Nery, Biosensors, 2018, 8, 3 CrossRef PubMed.
- E. Tønning, S. Sapelnikova, J. Christensen, C. Carlsson, M. Winther-Nielsen, E. Dock, R. Solna, P. Skladal, L. Nørgaard, T. Ruzgas and J. Emnéus, Biosens. Bioelectron., 2005, 21, 608–617 CrossRef PubMed.
- H. S. Song, H. J. Jin, S. R. Ahn, D. Kim, S. H. Lee, U. K. Kim, C. T. Simons, S. Hong and T. H Park, ACS Nano, 2014, 8, 9781–9789 CrossRef CAS.
- Q. Liu, C. Wu, H. Cai, N. Hu, J. Zhou and P. Wang, Chem. Rev., 2014, 114, 6423–6461 CrossRef CAS.
- Q. Liu, D. Zhang, F. Zhang, Y. Zhao, K. J. Hsia and P. Wang, Sens. Actuators, B, 2013, 176, 497–504 CrossRef CAS.
- T. Wasilewski, W. Kamysz and J. Gębicki, J. Bioelectr., 2020, 150, 111923 CrossRef CAS.
- L. Du, W. Chen, Y. Tian, P. Zhu, C. Wu and P. A. Wang, Sensor. Actuator. B Chem., 2020, 312, 127949 CrossRef CAS.
- J. Wu, C. Chen, C. Qin, Y. Li, N. Jiang, Q. Yuan, Y. Duan, M. Liu, X. Wei, Y. Yu, L. Zhuang and P. Wang, Adv. Sci., 2023, 10, 2206101 CrossRef CAS PubMed.
- J. Y. Hwang, K. H. Kim, S. E. Seo, Y. Nam, S. Jwa, I. Yang, T. H. Park, O. S. Kwon and S. H. Lee, Adv. Funct. Mater., 2023, 33, 2304997 CrossRef CAS.
- C. A. Pérez, L. Huang, M. Rong, J. A. Kozak, A. K. Preuss, H. Zhang, M. Max and R. F. Margolskee, Nat. Neurosci., 2002, 5, 1169–1176 CrossRef.
- Z. Zhang, Z. Zhao, R. Margolskee and E. Liman, J. Neurosci., 2007, 27, 5777–5786 CrossRef CAS.
- Y. Huang, Y. Maruyama, G. Dvoryanchikov, E. Pereira, N. Chaudhari and S. D. Roper, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 6436–6441 CrossRef CAS PubMed.
- V. Lyall, R. I. Alam, D. Q. Phan, G. L. Ereso, T. H. Phan, S. A. Malik, M. H. Montrose, S. Chu, G. L. Heck, G. M. Feldman and J. A. DeSimone, J. Cell. Physiol., 2001, 281, C1005–C1013 CrossRef CAS PubMed.
- K. Chuah, Y. Wu, S. R. Vivekchand, K. Gaus, P. J. Reece, A. P. Micolich and J. J. Gooding, Nat. Commun., 2019, 10, 2109 CrossRef.
- C. Yuan, X. Wu, R. Gao, X. Han, Y. Liu, Y. Long and Y. Cui, J. Am. Chem. Soc., 2019, 141, 20187–20197 CrossRef CAS PubMed.
- Z. Zhu, D. Wang, Y. Tian and L. Jiang, J. Am. Chem. Soc., 2019, 141, 8658–8669 CrossRef CAS.
- H. Zhang, Y. Tian and L. Jiang, Nano today, 2016, 11, 61–81 CrossRef CAS.
- Z. Han, K. Wang, H. Min, J. Xu, W. Shi and P. Cheng, Angew. Chem., Int. Ed., 2022, 134, e202204066 CrossRef.
- K. Xiao, L. Chen, Z. Zhang, G. Xie, P. Li, X. Kong, L. Wen and L. Jiang, Angew. Chem., Int. Ed., 2017, 56, 8168–8172 CrossRef CAS.
- D. K. Ban, T. Bodily, A. G. Karkisaval, Y. Dong and S. Natani, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2206521119 CrossRef CAS.
- P. Gao, L. Hu, N. Liu, Z. Yang, L. Lou, T. Zhai, H. Li and F. Xia, Adv. Mater., 2015, 28, 460–465 CrossRef.
- E. T. Acar, S. F. Buchsbaum, C. Combs, F. Fornasiero and Z. S. Siwy, Sci. Adv., 2019, 5, eaav2568 CrossRef CAS PubMed.
- J. Pan, W. Xu, Y. Zhang, Y. Ke, J. Dong, W. Li, L. Wang, B. Wang, B. Meng, Q. Zhou and F. Xia, Nano Energy, 2024, 24, 110412 CrossRef.
- X. Yu, C. Li, J. Chang, Y. Wang, W. Xia, J. Suo, X. Guan, V. Valtchev, Y. Yan, S. Qiu and Q. Fang, Angew. Chem., Int. Ed., 2022, 134, e202200820 CrossRef.
- Y. Zhang, H. Wang, J. Wang, L. Li, H. Sun and C. Wang, Chem.–Asian J., 2023, 18, e202300876 CrossRef CAS PubMed.
- H. Wang, Y. Zhang, J. Wang, H. Saijilahu, H. Sun, H. Yang, X. Xia and C. Wang, Adv. Funct. Mater., 2025, 35, 2412477 CrossRef CAS.
- J. Wang, C. Zhu, J. Tan, J. Xu and C. Wang, J. Am. Chem. Soc., 2025, 147, 12821–12832 CrossRef CAS PubMed.
- Y. Qiao, J. Hu, Y. Hu, C. Duan, W. Jiang, Q. Ma, Y. Hong, W. Huang, F. Xia and X. Lou, Angew. Chem., Int. Ed., 2023, 62, e202309671 CrossRef CAS.
- J. R. Burns, A. Seifert, N. Fertig and S. A. Howorka, Nat. Nanotechnol., 2016, 11, 152–156 CrossRef CAS.
- Y. Ying, Y. Li, J. Mei, R. Gao, Y. Hu, Y. Long and H. Tian, Nat. Commun., 2018, 9, 3657 CrossRef PubMed.
- B. Liang, H. Wang, X. Shi, B. Shen, X. He, Z. A. Ghazi, N. A. Khan, H. Sin, A. M. Khattak, L. Li and Z. Tang, Nat. Chem., 2018, 10, 961–967 CrossRef CAS.
- X. Hou, Y. Hu, A. Grinthal, M. Khan and J. Aizenberg, Nature, 2015, 519, 70–73 CrossRef CAS PubMed.
- M. Lozada-Hidalgo, S. Hu, O. Marshall, A. Mishchenko, A. N. Grigorenko, R. A. Dryfe, B. Radha, I. V. Grigorieva and A. K. Geim, Science, 2016, 351, 68–70 CrossRef CAS.
- J. Lu, H. Zhang, J. Hou, X. Li, X. Hu, Y. Hu, C. D. Easton, Q. Li, C. Sun, A. W. Thornton and M. R. Hill, Nat. Mater., 2020, 7, 767–774 CrossRef.
- A. Siria, P. Poncharal, A. L. Biance, R. Fulcrand, X. Blase, S. T. Purcell and L. Bocquet, Nature, 2013, 494, 455–458 CrossRef CAS PubMed.
- J. Feng, M. Graf, K. Liu, D. Ovchinnikov, D. Dumcenco, M. Heiranian, V. Nandigana, N. R. Aluru, A. Kis and A. Radenovic, Nature, 2016, 536, 197–200 CrossRef CAS PubMed.
- J. Gao, W. Guo, D. Feng, H. Wang, D. Zhao and L. Jiang, J. Am. Chem. Soc., 2014, 136, 12265–12272 CrossRef CAS.
- W. Zhang, Y. Tu, H. Liu, R. Liu, X. Zhang, L. Jiang, Y. Huang and F. Xia, Angew. Chem., Int. Ed., 2024, 63, e202316434 CrossRef CAS PubMed.
- Z. Shang, D. Ding, Z. Deng, J. Zhao, M. Yang, Y. Xiao, W. Chu, S. Xu, Z. Zhang, X. Yi and M. Lin, Angew. Chem., Int. Ed., 2024, e202417280 Search PubMed.
- X. Zhang, L. Chen, K. H. Lim, S. Gonuguntla, K. W. Lim, D. Pranantyo, W. P. Yong, W. J. T. Yam, Z. Low, W. J. Teo and H. P. Nien, Adv. Mater., 2019, 31, 1804540 CrossRef.
- Z. Chu, W. Zhang, Q. You, X. Yao, T. Liu, G. Liu, G. Zhang, X. Gu, Z. Ma and W. Jin, Angew. Chem., Int. Ed., 2020, 132, 18860–18867 CrossRef.
- S. Wooding, H. Gunn, P. Ramos, S. Thalmann, C. Xing and W. Meyerhof, Chem. Senses, 2010, 35, 685–692 CrossRef CAS PubMed.
- W. Meyerhof, C. Batram, C. Kuhn, A. Brockhoff, E. Chudoba, B. Bufe, G. Appendino and M. Behrens, Chem. Senses, 2010, 35, 157–170 CrossRef CAS PubMed.
- V. V. Acharya and P. Chaudhuri, Int. J. Pharm. Sci. Rev. Res., 2021, 69, 19–24 Search PubMed.
- M. Z. Chen, N. S. Moily, J. L. Bridgford, R. J. Wood, M. Radwan, T. A. Smith, Z. Song, B. Z. Tang, L. Tilley, X. Xu and G. E. Reid, Nat. Commun., 2017, 8, 1–11 CrossRef.
- M. Kita, Y. Hirata, T. Moriguchi, T. Endo-Inagaki, R. Matsumoto, S. Hasegawa, C. G. Suhayda and M. Omura, FEBS Lett., 2000, 469, 173–178 CrossRef CAS.
- M. R. Karim and F. Hashinaga, Process Biochem., 2002, 38, 809–814 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ta02127h |
| This journal is © The Royal Society of Chemistry 2025 |