Beyond DAD: proposing a one-letter code for nucleobase-mediated molecular recognition†
Aiden J.
Ward
 and
Benjamin E.
Partridge
and
Benjamin E.
Partridge
 *
*
Department of Chemistry, University of Rochester, Rochester, NY 14627-0216, USA. E-mail: benjamin.partridge@rochester.edu
First published on 21st November 2024
Abstract
Nucleobase binding is a fundamental molecular recognition event central to modern biological and bioinspired supramolecular research. Underpinning this recognition is a deceptively simple hydrogen-bonding code, primarily based on the canonical nucleobases in DNA and RNA. Inspired by these biotic structures, chemists and biologists have designed abiotic hydrogen-bonding motifs that can interact with, augment, and reshape native molecular recognition, for applications ranging from genetic code expansion and nucleic acid recognition to supramolecular materials utilizing mono- and bifacial nucleobases. However, as the number of nucleobase-inspired motifs expands, the absence of a standard vocabulary to describe hydrogen bond (HB) patterns has led to a haphazard mixture of shorthand descriptors that are confusing and inconsistent. Alternative notations that specify individual HB sites (such as DAD for donor–acceptor–donor) are cumbersome for biological and supramolecular constructs that contain many such patterns. This situation creates a barrier to sharing and interpreting nucleobase-related research across sub-disciplines, hindering collaboration and innovation. In this perspective, we aim to initiate discourse on this issue by considering what would be needed to formulate a concise one-letter code for the HB patterns associated with synthetic nucleobases. We first summarize some of the issues caused by the current absence of a consistent naming scheme. Subsequently, we discuss some key considerations in designing a coherent naming system. Finally, we leverage chemical rationale and pedagogical mnemonic considerations to propose a succinct and intuitive one-letter code for supramolecular two- and three-HB motifs. We hope that this discussion will spark conversations within our interdisciplinary community, thereby facilitating collaboration and easing communication among researchers engaged in synthetic nucleobase design.
Introduction
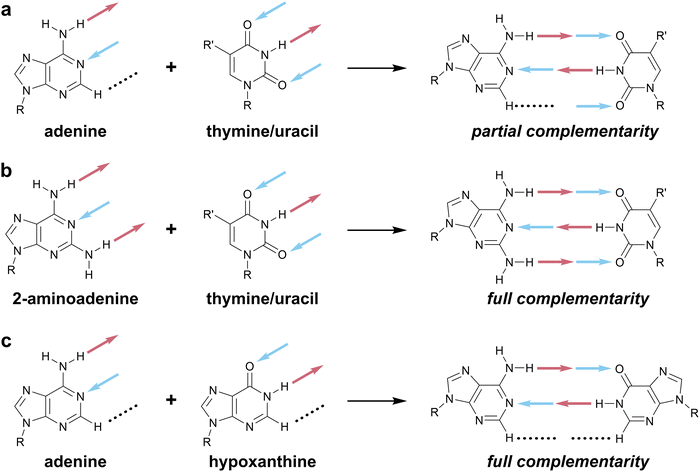
The molecular recognition of nucleobases via hydrogen bonds (HBs) is ubiquitous in living systems, enabling the efficient storage, communication, and transfer of genetic information within organisms and between generations. This information is primarily carried in just four patterns of HBs—those of guanine, cytosine, adenine, and thymine/uracil (Fig. 1a)—that ensure the fidelity of guanine–cytosine and adenine–thymine/uracil interactions via Watson–Crick–Franklin (WCF) base-pairing. Attracted by the efficiency, specificity, and tunability of nucleobase-mediated molecular recognition, chemists have repurposed DNA for materials design, both as a construction material capable of forming sophisticated architectures via DNA origami (Fig. 1b)1,2 and as a versatile and highly programmable interaction to organize nanoscale matter via colloidal crystal engineering (Fig. 1c).3,4 | ||
| Fig. 1 Natural nucleobases in biology and materials. (a) The canonical four nucleobases with hydrogen bonds on the WCF edge denoted by arrows. Color code: blue (HB acceptors) and maroon (HB donors). Black broken line denotes a null position not involved in WCF base-pairing. (b) Overview of DNA origami-engineered nanomaterials. Adapted from ref. 2. (c) Overview of colloidal crystal engineering with DNA. Reproduced from ref. 3. | ||
The versatility and potential impact of nucleobase-mediated molecular recognition across chemistry, biology, and materials science have spurred the design of new recognition motifs for different applications, including genetic code expansion, nucleic acid recognition, and supramolecular materials. Genetic code expansion has captivated chemists and biologists alike for decades,5–10 with recent examples such as hachimoji DNA successfully expanding the genetic alphabet from four to eight nucleobases (hence the name “hachimoji”, derived from the Japanese for “eight letters”) (Fig. 2a).11 These eight nucleobases rely on six unique HB patterns and the distinct geometries of purine and pyrimidine bases to encode a fully orthogonal four-base pair system that is transcribable in vitro using a modified T7 RNA polymerase.11 In the area of nucleic acid recognition, bifacial nucleobases, known also as Janus nucleobases after the Roman god of duality with two faces,12 have been designed to present two hydrogen-bonding arrays that each recognize a biotic nucleobase. Seminal work by Lehn and coworkers reported a Janus molecule capable of targeting the cytosine–uracil mismatch.13 More recently, the groups of Bong14,15 and Ly16,17 have utilized the bifacial HB motifs of melamine and Janus nucleobases, respectively, to bind to DNA and RNA double helices and form stable triplexes via double strand invasion (Fig. 2b). Triplex formation has also been explored by Rozners and coworkers via the formation of RNA bulges using modified peptide nucleic acids.18,19 For supramolecular materials, Lehn and coworkers were also instrumental in designing the G^C motif (Fig. 2c),20 the first synthetic bifacial nucleobase to assemble into hydrogen-bonded macrocycles, the formation of which was confirmed via XRD by Mascal et al.21,22 Subsequent in-depth studies by Fenniri and others explored these rosettes and their nanotubes as fibrous materials and mimics of ion channels.23–25 The synthesis and assembly of related motifs, including expanded G^C structures,26–28 A^T Janus nucleobases,29,30 and a G–C–T triple base-coded nucleobase,31 have also been reported.
 | ||
| Fig. 2 Non-natural nucleobases in biology and materials. (a) Four nucleobases that operate orthogonally to canonical ACGT bases in hachimoji DNA. Adapted from ref. 11. (b) MD simulation of a DNA–PNA–DNA triplex in which the PNA is modified with Janus nucleobases. Adapted from ref. 17. (c) The G^C motif.20 (d) Synthetic rosette nanotube porin mimic assembled from fused G^C bases functionalized with peripheral porphyrin units. Adapted from ref. 25. | ||
Though the development of a rich library of new hydrogen-bonding motifs has been motivated by the transdisciplinary appeal of nucleobase-inspired recognition,32 new structures have typically been designed and named by separate research communities. These names may have been selected with varying (or absent) rationales to maintain consistency within a specific research group or community, irrespective of usage beyond that community. Such inconsistency stymies collaboration across disciplines. The best alternative at present is to utilize descriptive three-letter codes based on the arrangement of HB donors and acceptors (e.g. ADD for guanine, cf.Fig. 1a). While this is suitable for single base pairs, these descriptions quickly become cumbersome for multiple, connected HB motifs such as DNA and RNA triplexes or extended supramolecular materials, both enabled by bifacial nucleobases.17,33 This inefficiency can be analogized to the inconvenience of referring to amino acids by three-letter abbreviations rather than one-letter codes, which becomes infeasible even for small peptide fragments like Aβ16–22.34
As nucleobase-inspired molecular recognition continues to broaden its relevance in biology and materials science, a succinct, common vocabulary for HB motifs will become ever more necessary to facilitate the community's ability to communicate, compare, and collaborate. Indeed, standard vocabularies play a crucial role in chemical and biological research.35 Three of many examples include the standard one-letter codes for amino acids,36 the Leontis–Westhof classification of RNA base-pairing patterns,37 and IUPAC's nucleic acid sequencing notation that not only denotes individual bases (such as G and C) but also all possible combinations thereof (for example, H denotes any base except G).38,39 However, a common shorthand to describe the arrangement of HB sites in nucleobases and related supramolecular building blocks simply does not exist.
In this perspective, we seek to initiate discussions to address this need by reflecting on some of the considerations needed to formulate a standard vocabulary for nucleobase-inspired HB motifs. Specifically, we consider all possible arrangements of three HB positions (acceptors, donors, and null sites) originating from a single molecular edge, comparable to a WCF or Hoogsteen edge; the result is a collection of 16 unique two- and three-HB patterns (Scheme 1). We begin with a brief discussion of the current state of the field, highlighting some of the consequences of uncoordinated naming and missed opportunities that arise therefrom. Subsequently, we outline some considerations for, and inherent contradictions of, designing a cohesive and intuitive naming system. Finally, we conclude by proposing a standard vocabulary and exemplifying its utility. Rather than being a complete solution to this nomenclature challenge, we instead intend that this perspective act as a starting point, to spark discussion around how best to leverage the strengths of the entire community of researchers interested in nucleobase-mediated molecular recognition, for the benefit of all.
The consequences of inconsistent naming
As an example of the current state of nomenclature in this area, we consider the co-assembly of melamine and cyanuric acid (Scheme 2a), extensively studied by Whitesides and coworkers in a series of papers in the early 1990s.33,40,41 Each edge of melamine features three HB sites, in the order donor–acceptor–donor (abbreviated here in the format DAD). Upon mixing with cyanuric acid, which presents the fully complementary acceptor–donor–acceptor (ADA) HB pattern, extended two-dimensional (2D) melamine–cyanuric acid (M–CA) structures are formed (Scheme 2, X = NH).40 However, cyanuric acid is not unique as an ADA motif: barbiturates contain the same HB pattern and also co-assemble with melamine into 2D M–BA structures (Scheme 2, X = CR2).42 The ADA-capable motif is most commonly found in the nucleobases thymine and uracil, which have three HB sites despite forming only two HBs in their WCF base pairs with adenine (Fig. 1a). Indeed, melamine–thymine/uracil co-assembly has been studied in the context of nucleic acid recognition14,15 and utilized for DNA materials.43 Describing this recognition using an explicit listing of HB sites is unwieldy: a cyanuric acid–melamine–cyanuric acid triplex would be styled as ADA^ADA·DAD^DAD·ADA^ADA, which is sufficiently cumbersome so as to be effectively useless. Moreover, the multiple names used to describe these motifs—M–CA,40 BA–Mel,42 M–U,15 and T–MA–T43—obfuscate the HB-mediated molecular recognition event that is common to all of these systems. | ||
| Scheme 2 Extended two-dimensional HB arrays. Melamine (black) co-assembles with cyanuric acid (blue, X = NH) or barbituric acid (blue, X = CH2) to form periodic 2D assemblies. | ||
Considerations for a cohesive naming system
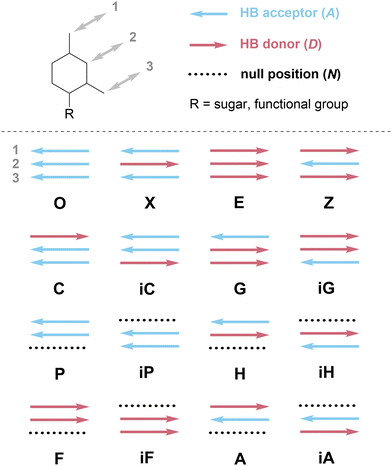
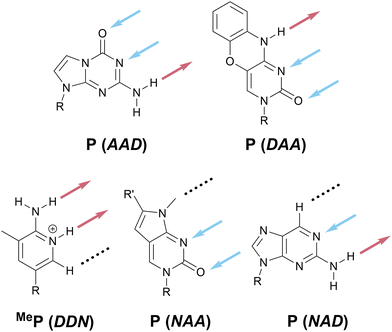
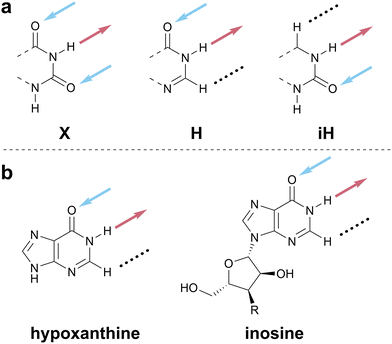
The absence of a cohesive and intuitive shorthand reflects the difficulty of assigning nomenclature that is simultaneously satisfactory to multiple research communities, each with different norms, priorities, and preferences.57Scheme 1 presents a potential naming system that describes all possible 2- and 3-HB patterns based on a nucleobase-like scaffold (that is, a six-membered planar heterocycle). In this section, we outline the key factors that we considered in assigning these one-letter codes to specific HB patterns; note that one-letter codes that form part of our proposed vocabulary are styled in bold. We recognize that the list of factors we considered is likely non-exhaustive. Instead, we intend that this scheme serve as a starting point for discussion in the broader research community and iteration towards a universally acceptable solution.There are several ways to resolve the A–T problem. One option would be to assign the HB patterns involved in the biological A–T base pair (that is, DAN and ADN, respectively) with the one-letter codes A and T. In this case, A and T remain complementary but a thymine nucleobase would no longer properly be described as having a T HB pattern; this is clearly unsatisfactory. An alternative would be to associate A and T with the HB patterns presented by adenine (DAN) and thymine (ADA), which would maintain the link to these specific molecules but would result in A and T no longer being complementary; this feels counterintuitive and would introduce a clear contradiction with those working with DNA and RNA. Our favored approach avoids the use of T (and, by extension, U) entirely as a descriptor of HB patterns, reserving these letters for the nucleobases themselves, and instead denotes the ADA pattern with X, derived from the xanthine moiety to which thymine is related. Analogously, the complement of A, ADN, is denoted H for the hypoxanthine motif that exhibits this pattern (Scheme 5c). Though this choice means that the complement of A is not T, but H, we believe this solution to the A–T problem strikes an appropriate balance between the interests of chemists, biologists, and materials scientists, because it removes a potential source of ambiguity without seeking to redefine a fundamental biological abbreviation (that is, the meaning of the one-letter code, T).
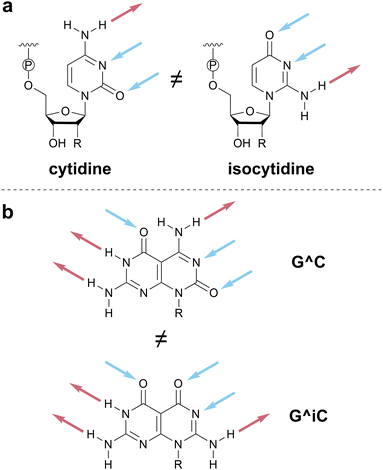
We invoke the term iso as a directionality descriptor, consistent with literature precedent for the names of isoguanine and isocytosine.59 Therefore, the pattern for cytosine (DAA) is denoted C and its rotated analog, AAD, is denoted isoC or iC for short. In assigning the iso designation, the HB pattern should be read from top to bottom when the point of covalent attachment (e.g., to a sugar–phosphate backbone or functional group) is positioned at the bottom of the HB motif (Scheme 6 and top of Scheme 1). Although the use of a modifier strays from a strictly one-letter code, we favored using a modifier to denote the relationship between inverted HB patterns, to minimize the number of unique one-letter codes that would need to be memorized. The modifier ‘i’ was chosen because alternative modifiers (e.g., asterisk (*), prime (′)) are otherwise defined in nucleic acid notation.
A proposed naming scheme for HB motifs
Scheme 1 at the start of this perspective outlines our proposed one-letter codes for the 16 unique two- and three-HB patterns of relevance to planar nucleobase-inspired HB motifs (a complete set of all 27 possible combinations of HB acceptors, HB donors, and null sites is shown in Scheme S2, ESI†). In this section, we introduce each one-letter code and briefly provide specific rationale for its choice.Hypoxanthine is found naturally in tRNA as the nucleobase component of the nucleoside inosine (Scheme 8b); this is a rare example where the names of the nucleobase and nucleoside do not share a common root. We have opted to use hypoxanthine as the source of the name of the ADN pattern for two reasons: first, the hypoxanthine motif is more relevant for both biological and materials applications than the full inosine nucleoside; second, using H avoids confusion between upper-case I (for inosine) and lower-case i (used here as an abbreviation for iso).
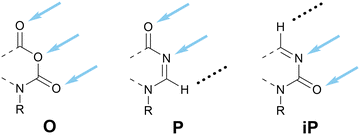
In the previous subsection, we assigned consecutive letters to DDD (E) and its analog without a HB site in the 3-position, DDN (F). Analogously, given that AAA is denoted O, we propose that its analog without a HB site in the 3-position, AAN, be denoted P (Scheme 11). P is non-centrosymmetric and therefore the final unnamed pattern, NDD, is denoted iP.
A standard vocabulary in practice
The one-letter code proposed in Scheme 1 and discussed in the previous section is succinct and unambiguous. The naming of these 16 HB patterns naturally leads to eight base pairs that are fully complementary to each other: G-C, iG-iC, A-H, iA-iH, F-P, iF-iP, X-Z, and O-E. These base pairs concisely convey information about the nature and number of HBs involved in each base-pairing interaction.The value of one-letter codes is especially evident when discussing supramolecular systems that invoke more than one HB molecular recognition event. Consider, for example, the cyanuric acid–melamine–cyanuric acid triplex discussed earlier (Scheme 2). Using our proposed vocabulary, the HB pattern of melamine is described as Z^Z, where the caret (^) is used in the bifacial nucleobase community to denote the HB patterns on each face of the molecule (see, for example, G^C in Fig. 2c). Similarly, the HB pattern of cyanuric acid is described as X^X. Hence the triplex noted above could be concisely represented as X^X·Z^Z·X^X, rather than the ADA^ADA·DAD^DAD·ADA^ADA description required by bond-by-bond donor–acceptor notation.
It is important to clarify that the proposed naming scheme is not intended to supersede efforts to name HB-capable molecules with trivial or more convenient names. We recognize, for example, that referring to melamine only as Z^Z would be non-sensical, as valuable chemical information about the molecular structure that presents the Z^Z HB pattern would be lost. Instead, our vocabulary is meant to augment the current literature and provide a means of communicating HB patterns, rather than molecular structures, clearly and unambiguously. As an example, consider the Janus-AT (JAT) bifacial nucleobase shown in Scheme 12.29 Under our proposed standard vocabulary, the JAT motif would be denoted by X^Z. While this may be less practical for the goals of the original authors’ work, including this formalized name makes the connection between the JAT motif and other HB motifs, such as the cyanuric acid (X^X) and melamine (Z^Z) system, much more readily apparent. Our proposed naming scheme maps readily onto reported bifacial nucleobases (Scheme S3, ESI†). Therefore, we encourage researchers to include this formal description of their hydrogen bonding motifs in their future work, to facilitate collaboration and the exchange of ideas within our community.
 | ||
| Scheme 12 A Janus-AT (JAT) bifacial nucleobase reported by Asadi et al.29 | ||
Summary and outlook
In summary, we have identified a need for a consistent naming scheme for nucleobase-like HB patterns and proposed a standard vocabulary to address this need. As far as possible, literature precedent, chemical rationale, and consistency with DNA have been considered while designating one-letter codes. Otherwise, mnemonics based on shape or structural relationships have been employed, specifically to assist with memorization and encourage uptake of our proposed alphabet by researchers.We recognize that a vocabulary is only useful if it is adopted broadly and meets consensus in its intended community of use. Therefore, this perspective is intended as a starting point, both to solicit feedback and to spur conversations among researchers in our field, to consider and debate how best we can communicate and share our ideas and findings. In this way, we hope that we can contribute to maximizing our collective progress towards our respective goals, whether in the biological, chemical, or materials context, applied or fundamental. With this in mind, we encourage feedback and commentary, to help shape a common vocabulary useful to all.
Author contributions
A. J. W.: conceptualization, funding acquisition, investigation, visualization, writing – review & editing; B. E. P.: conceptualization, funding acquisition, supervision, visualization, writing – original draft, review & editing.Data availability
No primary research results, software or code have been included and no new data were generated or analyzed as part of this perspective.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the Air Force Office of Scientific Research (AFOSR) through the Young Investigator Program (Award FA9550-24-1-0104). A. J. W. acknowledges the American Chemical Society (ACS) Division of Organic Chemistry for financial support through a Summer Undergraduate Research Fellowship (SURF). The authors thank Parbhat Kumar and Alejandro Lazaro (University of Rochester) for scientific discussions.References
- Y. Tian, J. R. Lhermitte, L. Bai, T. Vo, H. L. Xin, H. Li, R. Li, M. Fukuto, K. G. Yager, J. S. Kahn, Y. Xiong, B. Minevich, S. K. Kumar and O. Gang, Ordered three-dimensional nanomaterials using DNA-prescribed and valence-controlled material voxels, Nat. Mater., 2020, 19, 789–796 CrossRef CAS.
- P. Zhan, A. Peil, Q. Jiang, D. Wang, S. Mousavi, Q. Xiong, Q. Shen, Y. Shang, B. Ding, C. Lin, Y. Ke and N. Liu, Recent Advances in DNA Origami-Engineered Nanomaterials and Applications, Chem. Rev., 2023, 123, 3976–4050 CrossRef CAS PubMed.
- C. R. Laramy, M. N. O’Brien and C. A. Mirkin, Crystal engineering with DNA, Nat. Rev. Mater., 2019, 4, 201–224 CrossRef CAS.
- D. Samanta, W. Zhou, S. B. Ebrahimi, S. H. Petrosko and C. A. Mirkin, Programmable Matter: The Nanoparticle Atom and DNA Bond, Adv. Mater., 2022, 34, 2107875 CrossRef CAS PubMed.
- D. A. Malyshev, K. Dhami, T. Lavergne, T. Chen, N. Dai, J. M. Foster, I. R. Corrêa and F. E. Romesberg, A semi-synthetic organism with an expanded genetic alphabet, Nature, 2014, 509, 385–388 CrossRef CAS PubMed.
- J. W. Chin, Expanding and reprogramming the genetic code, Nature, 2017, 550, 53–60 CrossRef CAS PubMed.
- M. A. Shandell, Z. Tan and V. W. Cornish, Genetic Code Expansion: A Brief History and Perspective, Biochemistry, 2021, 60, 3455–3469 CrossRef CAS.
- M. Kimoto and I. Hirao, Genetic alphabet expansion technology by creating unnatural base pairs, Chem. Soc. Rev., 2020, 49, 7602–7626 RSC.
- T. Yoshida, K. Morihiro, Y. Naito, A. Mikami, Y. Kasahara, T. Inoue and S. Obika, Identification of nucleobase chemical modifications that reduce the hepatotoxicity of gapmer antisense oligonucleotides, Nucleic Acids Res., 2022, 50, 7224–7234 CrossRef CAS PubMed.
- A. Berdis, Nucleobase-modified nucleosides and nucleotides: Applications in biochemistry, synthetic biology, and drug discovery, Front. Chem., 2022, 10, 1051525 CrossRef CAS PubMed.
- S. Hoshika, N. A. Leal, M.-J. Kim, M.-S. Kim, N. B. Karalkar, H.-J. Kim, A. M. Bates, N. E. Watkins, H. A. SantaLucia, A. J. Meyer, S. DasGupta, J. A. Piccirilli, A. D. Ellington, J. SantaLucia, M. M. Georgiadis and S. A. Benner, Hachimoji DNA and RNA: A genetic system with eight building blocks, Science, 2019, 363, 884–887 CrossRef CAS.
- S. J. Cristol and D. C. Lewis, Bridged Polycyclic Compounds. XLV. Synthesis and Some Properties of 5,5a,6,11,11a,12-Hexahydro-5,12:6,11-di-o-benzenonaphthacene (Janusene), J. Am. Chem. Soc., 1967, 89, 1476–1483 CrossRef CAS.
- N. Branda, G. Kurz and J.-M. Lehn, JANUS WEDGES: a new approach towards nucleobase-pair recognition, Chem. Commun., 1996, 2443 RSC.
- Y. Zeng, Y. Pratumyot, X. Piao and D. Bong, Discrete Assembly of Synthetic Peptide–DNA Triplex Structures from Polyvalent Melamine–Thymine Bifacial Recognition, J. Am. Chem. Soc., 2012, 134, 832–835 CrossRef CAS PubMed.
- S. Rundell, O. Munyaradzi and D. Bong, Enhanced Triplex Hybridization of DNA and RNA via Syndiotactic Side Chain Presentation in Minimal bPNAs, Biochemistry, 2022, 61, 85–91 CrossRef CAS PubMed.
- S. A. Thadke, J. D. R. Perera, V. M. Hridya, K. Bhatt, A. Y. Shaikh, W.-C. Hsieh, M. Chen, C. Gayathri, R. R. Gil, G. S. Rule, A. Mukherjee, C. A. Thornton and D. H. Ly, Design of Bivalent Nucleic Acid Ligands for Recognition of RNA-Repeated Expansion Associated with Huntington's Disease, Biochemistry, 2018, 57, 2094–2108 CrossRef CAS PubMed.
- S. A. Thadke, V. M. Hridya, J. D. R. Perera, R. R. Gil, A. Mukherjee and D. H. Ly, Shape selective bifacial recognition of double helical DNA, Commun. Chem., 2018, 79 CrossRef PubMed.
- T. Endoh, N. Brodyagin, D. Hnedzko, N. Sugimoto and E. Rozners, Triple-Helical Binding of Peptide Nucleic Acid Inhibits Maturation of Endogenous MicroRNA-197, ACS Chem. Biol., 2021, 16, 1147–1151 CrossRef CAS.
- C. A. Ryan, M. M. Rahman, V. Kumar and E. Rozners, Triplex-Forming Peptide Nucleic Acid Controls Dynamic Conformations of RNA Bulges, J. Am. Chem. Soc., 2023, 145, 10497–10504 CrossRef CAS PubMed.
- A. Marsh, M. Silvestri and J.-M. Lehn, Self-complementary hydrogen bonding heterocycles designed for the enforced self-assembly into supramolecular macrocycles, Chem. Commun., 1996, 1527 RSC.
- M. Mascal, N. M. Hext, R. Warmuth, M. H. Moore and J. P. Turkenburg, Programming a Hydrogen-Bonding Code for the Specific Generation of a Supermacrocycle, Angew. Chem., Int. Ed. Engl., 1996, 35, 2204–2206 CrossRef CAS.
- M. Mascal, N. M. Hext, R. Warmuth, J. R. Arnall-Culliford, M. H. Moore and J. P. Turkenburg, The G−C DNA Base Hybrid: Synthesis, Self-Organization and Structural Analysis, J. Org. Chem., 1999, 64, 8479–8484 CrossRef CAS.
- R. L. Beingessner, Y. Fan and H. Fenniri, Molecular and supramolecular chemistry of rosette nanotubes, RSC Adv., 2016, 6, 75820–75838 RSC.
- H. Fenniri, G. Tikhomirov, D. H. Brouwer, S. Bouatra, M. El Bakkari, Z. Yan, J.-Y. Cho and T. Yamazaki, High Field Solid-State NMR Spectroscopy Investigation of 15N-Labeled Rosette Nanotubes: Hydrogen Bond Network and Channel-Bound Water, J. Am. Chem. Soc., 2016, 138, 6115–6118 CrossRef CAS.
- P. Tripathi, L. Shuai, H. Joshi, H. Yamazaki, W. H. Fowle, A. Aksimentiev, H. Fenniri and M. Wanunu, Rosette Nanotube Porins as Ion Selective Transporters and Single-Molecule Sensors, J. Am. Chem. Soc., 2020, 142, 1680–1685 CrossRef CAS PubMed.
- A. Asadi, B. O. Patrick and D. M. Perrin, G^C Quartet—A DNA-Inspired Janus-GC Heterocycle: Synthesis, Structural Analysis, and Self-Organization, J. Am. Chem. Soc., 2008, 130, 12860–12861 CrossRef CAS PubMed.
- G. Borzsonyi, R. S. Johnson, A. J. Myles, J.-Y. Cho, T. Yamazaki, R. L. Beingessner, A. Kovalenko and H. Fenniri, Rosette nanotubes with 1.4 nm inner diameter from a tricyclic variant of the Lehn–Mascal G ∧ C base, Chem. Commun., 2010, 46, 6527 RSC.
- G. Borzsonyi, A. Alsbaiee, R. L. Beingessner and H. Fenniri, Synthesis of a Tetracyclic G ∧ C Scaffold for the Assembly of Rosette Nanotubes with 1.7 nm Inner Diameter, J. Org. Chem., 2010, 75, 7233–7239 CrossRef CAS PubMed.
- A. Asadi, B. O. Patrick and D. M. Perrin, Janus-AT Bases: Synthesis, Self-Assembly, and Solid State Structures, J. Org. Chem., 2007, 72, 466–475 CrossRef CAS PubMed.
- D. Shin and Y. Tor, Bifacial Nucleoside as a Surrogate for Both T and A in Duplex DNA, J. Am. Chem. Soc., 2011, 133, 6926–6929 CrossRef CAS PubMed.
- M. A. Wagh, R. Maity, R. J. Bhosale, D. Semwal, S. Tothadi, R. Vaidhyanathan and G. J. Sanjayan, Three in One: Triple G-C-T Base-Coded Brahma Nucleobase Amino Acid: Synthesis, Peptide Formation, and Structural Features, J. Org. Chem., 2021, 86, 15689–15694 CrossRef CAS PubMed.
- M. López-Tena, S.-K. Chen and N. Winssinger, Supernatural: Artificial Nucleobases and Backbones to Program Hybridization-Based Assemblies and Circuits, Bioconjugate Chem., 2023, 34, 111–123 CrossRef.
- J. A. Zerkowski, C. T. Seto and G. M. Whitesides, Solid-state structures of rosette and crinkled tape motifs derived from the cyanuric acid melamine lattice, J. Am. Chem. Soc., 1992, 114, 5473–5475 CrossRef CAS.
- E. R. Piedmont, E. E. Christensen, T. D. Krauss and B. E. Partridge, Amphiphilic dendrons as supramolecular holdase chaperones, RSC Chem. Biol., 2023, 4, 754–759 RSC.
- N. B. Leontis, R. B. Altman, H. M. Berman, S. E. Brenner, J. W. Brown, D. R. Engelke, S. C. Harvey, S. R. Holbrook, F. Jossinet, S. E. Lewis, F. Major, D. H. Mathews, J. S. Richardson, J. R. Williamson and E. Westhof, The RNA Ontology Consortium: An open invitation to the RNA community, RNA, 2006, 12, 533–541 Search PubMed.
- M. Saffran, Amino acid names and parlor games: from trivial names to a one-letter code, amino acid names have strained students’ memories. Is a more rational nomenclature possible?, Biochem. Educ., 1998, 26, 116–118 CrossRef CAS.
- N. B. Leontis and E. Westhof, Geometric nomenclature and classification of RNA base pairs, RNA, 2001, 7, 499–512 CrossRef CAS.
- A. Cornish-Bowden, Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984, Nucleic Acids Res., 1985, 13, 3021–3030 CrossRef CAS.
- Nomenclature committee of the International Union of Biochemistry (NC-IUB). Nomenclature for incompletely specified bases in nucleic acid sequences. Recommendations 1984, J. Biol. Chem., 1986, 261, 13–17 Search PubMed.
- C. T. Seto and G. M. Whitesides, Self-assembly based on the cyanuric acid-melamine lattice, J. Am. Chem. Soc., 1990, 112, 6409–6411 CrossRef CAS.
- J. A. Zerkowski and G. M. Whitesides, Steric Control of secondary, solid-state architecture in 1:1 complexes of melamines and barbiturates that crystallize as crinkled tapes, J. Am. Chem. Soc., 1994, 116, 4298–4304 CrossRef CAS.
- T. P. Roche, P. J. Nedumpurath, S. C. Karunakaran, G. B. Schuster and N. V. Hud, One-Pot Formation of Pairing Proto-RNA Nucleotides and Their Supramolecular Assemblies, Life, 2023, 13, 2200 CrossRef CAS PubMed.
- Q. Li, J. Zhao, L. Liu, S. Jonchhe, F. J. Rizzuto, S. Mandal, H. He, S. Wei, H. F. Sleiman, H. Mao and C. Mao, A poly(thymine)–melamine duplex for the assembly of DNA nanomaterials, Nat. Mater., 2020, 19, 1012–1018 CrossRef CAS.
- G. E. Plum, Y. W. Park, S. F. Singleton, P. B. Dervan and K. J. Breslauer, Thermodynamic characterization of the stability and the melting behavior of a DNA triplex: a spectroscopic and calorimetric study, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 9436–9440 CrossRef CAS.
- A. Ono, P. O. P. Ts’o and L. S. Kan, Triplex formation of an oligonucleotide containing 2’-O-methylpseudoisocytidine with a DNA duplex at neutral pH, J. Org. Chem., 1992, 57, 3225–3230 CrossRef CAS.
- M. Egholm, L. Christensen, K. L. Deuholm, O. Buchardt, J. Coull and P. E. Nielsen, Efficient pH-independent sequence-specific DNA binding by pseudoisocytosine-containing bis-PNA, Nucleic Acids Res., 1995, 23, 217–222 CrossRef CAS PubMed.
- A. B. Eldrup, B. B. Nielsen, G. Haaima, H. Rasmussen, J. S. Kastrup, C. Christensen and P. E. Nielsen, 1,8-Naphthyridin-2(1H)-ones − Novel Bicyclic and Tricyclic Analogues of Thymine in Peptide Nucleic Acids (PNAs), Eur. J. Org. Chem., 2001, 1781–1790 CrossRef CAS.
- G. Devi, Z. Yuan, Y. Lu, Y. Zhao and G. Chen, Incorporation of thio-pseudoisocytosine into triplex-forming peptide nucleic acids for enhanced recognition of RNA duplexes, Nucleic Acids Res., 2014, 42, 4008–4018 CrossRef CAS.
- Q. Ma, D. Lee, Y. Q. Tan, G. Wong and Z. Gao, Synthetic genetic polymers: advances and applications, Polym. Chem., 2016, 7, 5199–5216 RSC.
- C. R. Geyer, T. R. Battersby and S. A. Benner, Nucleobase Pairing in Expanded Watson-Crick-like Genetic Information Systems, Structure, 2003, 11, 1485–1498 CrossRef CAS.
- S. Hildbrand and C. Leumann, Enhancing DNA Triple Helix Stability at Neutral pH by the Use of Oligonucleotides Containing a More Basic Deoxycytidine Analog, Angew. Chem., Int. Ed. Engl., 1996, 35, 1968–1970 CrossRef CAS.
- D. A. Rusling, Four base recognition by triplex-forming oligonucleotides at physiological pH, Nucleic Acids Res., 2005, 33, 3025–3032 CrossRef CAS PubMed.
- R. T. Ranasinghe, D. A. Rusling, V. E. C. Powers, K. R. Fox and T. Brown, Recognition of CG inversions in DNA triple helices by methylated 3H-pyrrolo[2,3-d]pyrimidin-2(7H)-one nucleoside analogues, Chem. Commun., 2005, 2555 RSC.
- P. Gupta, T. Zengeya and E. Rozners, Triple helical recognition of pyrimidine inversions in polypurine tracts of RNA by nucleobase-modified PNA, Chem. Commun., 2011, 47, 11125 RSC.
- S. Hildbrand, A. Blaser, S. P. Parel and C. J. Leumann, 5-Substituted 2-Aminopyridine C -Nucleosides as Protonated Cytidine Equivalents: Increasing Efficiency and Selectivity in DNA Triple-Helix Formation, J. Am. Chem. Soc., 1997, 119, 5499–5511 CrossRef CAS.
- J.-A. Ortega, J. R. Blas, M. Orozco, A. Grandas, E. Pedroso and J. Robles, Binding Affinities of Oligonucleotides and PNAs Containing Phenoxazine and G-Clamp Cytosine Analogues Are Unusually Sequence-Dependent, Org. Lett., 2007, 9, 4503–4506 CrossRef CAS PubMed.
- Editorial, Catalyzing collaboration, Nat. Chem. Biol., 2007, 3, 239 CrossRef.
- S. Kuchin, Covering All the Bases in Genetics: Simple Shorthands and Diagrams for Teaching Base Pairing to Biology Undergraduates, J. Microbiol. Biol. Educ., 2011, 12, 64–66 CrossRef.
- C. Roberts, R. Bandaru and C. Switzer, Theoretical and Experimental Study of Isoguanine and Isocytosine: Base Pairing in an Expanded Genetic System, J. Am. Chem. Soc., 1997, 119, 4640–4649 CrossRef CAS.
- B. A. Schweitzer and E. T. Kool, Hydrophobic, Non-Hydrogen-Bonding Bases and Base Pairs in DNA, J. Am. Chem. Soc., 1995, 117, 1863–1872 CrossRef CAS.
- F. Li, P. S. Pallan, M. A. Maier, K. G. Rajeev, S. L. Mathieu, C. Kreutz, Y. Fan, J. Sanghvi, R. Micura, E. Rozners, M. Manoharan and M. Egli, Crystal structure, stability and in vitro RNAi activity of oligoribonucleotides containing the ribo-difluorotoluyl nucleotide: insights into substrate requirements by the human RISC Ago2 enzyme, Nucleic Acids Res., 2007, 35, 6424–6438 CrossRef CAS PubMed.
- V. Kumar and E. Rozners, Fluorobenzene Nucleobase Analogues for Triplex-Forming Peptide Nucleic Acids, ChemBioChem, 2022, 23, e202100560 CrossRef CAS PubMed.
- A. D. Boese, Density Functional Theory and Hydrogen Bonds: Are We There Yet?, ChemPhysChem, 2015, 16, 978–985 CrossRef CAS.
- H. Kwon, A. Kumar, M. Del Ben and B. M. Wong, Electron/Hole Mobilities of Periodic DNA and Nucleobase Structures from Large-Scale DFT Calculations, J. Phys. Chem. B, 2023, 127, 5755–5763 CrossRef CAS PubMed.
- P. Mazumdar, A. Kashyap and D. Choudhury, Investigation of hydrogen bonding in small nucleobases using DFT, AIM, NCI and NBO technique, Comput. Theor. Chem., 2023, 1226, 114188 CrossRef CAS.
- I. V. Kutyavin, R. L. Rhinehart, E. A. Lukhtanov, V. V. Gorn, R. B. Meyer and H. B. Gamper, Oligonucleotides Containing 2-Aminoadenine and 2-Thiothymine Act as Selectively Binding Complementary Agents, Biochemistry, 1996, 35, 11170–11176 CrossRef CAS PubMed.
- K. G. Devine and S. Jheeta, De Novo Nucleic Acids: A Review of Synthetic Alternatives to DNA and RNA That Could Act as Bio-Information Storage Molecules, Life, 2020, 10, 346 CrossRef CAS PubMed.
- G. Haaima, Increased DNA binding and sequence discrimination of PNA oligomers containing 2,6-diaminopurine, Nucleic Acids Res., 1997, 25, 4639–4643 CrossRef CAS PubMed.
- J. Lohse, O. Dahl and P. E. Nielsen, Double duplex invasion by peptide nucleic acid: A general principle for sequence-specific targeting of double-stranded DNA, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11804–11808 CrossRef CAS PubMed.
- Y. Zhou, X. Xu, Y. Wei, Y. Cheng, Y. Guo, I. Khudyakov, F. Liu, P. He, Z. Song, Z. Li, Y. Gao, E. L. Ang, H. Zhao, Y. Zhang and S. Zhao, A widespread pathway for substitution of adenine by diaminopurine in phage genomes, Science, 2021, 372, 512–516 CrossRef CAS.
- D. Sleiman, P. S. Garcia, M. Lagune, J. Loc’h, A. Haouz, N. Taib, P. Röthlisberger, S. Gribaldo, P. Marlière and P. A. Kaminski, A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes, Science, 2021, 372, 516–520 CrossRef CAS.
- V. Pezo, F. Jaziri, P.-Y. Bourguignon, D. Louis, D. Jacobs-Sera, J. Rozenski, S. Pochet, P. Herdewijn, G. F. Hatfull, P.-A. Kaminski and P. Marliere, Noncanonical DNA polymerization by aminoadenine-based siphoviruses, Science, 2021, 372, 520–524 CrossRef CAS PubMed.
- T. Zengeya, P. Gupta and E. Rozners, Triple-Helical Recognition of RNA Using 2-Aminopyridine-Modified PNA at Physiologically Relevant Conditions, Angew. Chem., Int. Ed., 2012, 51, 12593–12596 CrossRef CAS PubMed.
- I. Kumpina, N. Brodyagin, J. A. MacKay, S. D. Kennedy, M. Katkevics and E. Rozners, Synthesis and RNA-Binding Properties of Extended Nucleobases for Triplex-Forming Peptide Nucleic Acids, J. Org. Chem., 2019, 84, 13276–13298 CrossRef CAS.
- P. Champagne, J. Desroches and J.-F. Paquin, Organic Fluorine as a Hydrogen-Bond Acceptor: Recent Examples and Applications, Synthesis, 2014, 306–322 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb01999g |
| This journal is © The Royal Society of Chemistry 2025 |