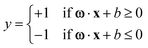Unveiling brain disorders using liquid biopsy and Raman spectroscopy
Jeewan C.
Ranasinghe
,
Ziyang
Wang
and
Shengxi
Huang

Department of Electrical and Computer Engineering, Rice University, Houston, TX 77005, USA. E-mail: shengxi.huang@rice.edu
First published on 29th May 2024
Abstract
Brain disorders, including neurodegenerative diseases (NDs) and traumatic brain injury (TBI), present significant challenges in early diagnosis and intervention. Conventional imaging modalities, while valuable, lack the molecular specificity necessary for precise disease characterization. Compared to the study of conventional brain tissues, liquid biopsy, which focuses on blood, tear, saliva, and cerebrospinal fluid (CSF), also unveils a myriad of underlying molecular processes, providing abundant predictive clinical information. In addition, liquid biopsy is minimally- to non-invasive, and highly repeatable, offering the potential for continuous monitoring. Raman spectroscopy (RS), with its ability to provide rich molecular information and cost-effectiveness, holds great potential for transformative advancements in early detection and understanding the biochemical changes associated with NDs and TBI. Recent developments in Raman enhancement technologies and advanced data analysis methods have enhanced the applicability of RS in probing the intricate molecular signatures within biological fluids, offering new insights into disease pathology. This review explores the growing role of RS as a promising and emerging tool for disease diagnosis in brain disorders, particularly through the analysis of liquid biopsy. It discusses the current landscape and future prospects of RS in the diagnosis of brain disorders, highlighting its potential as a non-invasive and molecularly specific diagnostic tool.
Introduction
Brain disorders encompass a wide spectrum of conditions, spanning from mild concussions to severe TBI and NDs such as Alzheimer's disease (AD) and Parkinson's disease (PD).1–4 The diagnostic complexities of these disorders arise from their intricate and varied biochemical manifestations, presenting critical challenges in accurate and timely diagnosis. Globally, the impact of these disorders on public health is substantial. They contribute significantly to the burden of diseases, affecting millions of lives and necessitating extensive and often lifelong care and management. Furthermore, NDs are a growing concern, especially with an aging global population. AD, for instance, affects millions worldwide, with projections indicating a significant rise in cases in the coming decades. According to the “Alzheimer's disease facts and figures” report by the Alzheimer's association, over 6 million Americans currently have AD, projected to reach nearly 13 million by 2050.5 It's the 6th leading cause of death among US adults and the 5th among those aged 65+.6,7 Treatment costs could surpass $500 billion annually by 2040, up from $215 billion in 2010.5 One in three seniors dies with AD or another dementia, exceeding breast and prostate cancer combined.5 By 2023, an estimated 6.7 million Americans aged 65 and older will have AD, projected to grow to 12.7 million by 2050.5 This highlights the urgent need for research, support, and resources to address this public health challenge. According to the World Health Organization, AD and other dementia affect 24 million of the one billion people worldwide affected by neurological disorders.8 This staggering prevalence underscores the urgent need for advanced and precise diagnostic approaches. AD is not only a medical stigma but also imposes a significant economic burden due to the high costs associated with diagnosis, medication, and caregiving. Early detection of AD can play a crucial role in alleviating this financial strain on the economy by reducing expenses related to medication and overall well-being of AD patients. The socioeconomic burden observed in AD is not unique; it extends to other brain disorders, including PD. Like AD, PD imposes significant costs on healthcare systems and society as a whole.9 The economic impact encompasses treatment expenses, lost productivity, and caregiving costs. Addressing the socioeconomic challenges posed by PD, alongside AD, requires comprehensive strategies and increased support for research, patient care, and public awareness initiatives. The multifaceted nature of brain disorders demands innovative methodologies to enable accurate and timely diagnoses, thus facilitating early intervention and improved patient outcomes. As such, the ongoing pursuit of sophisticated diagnostic tools and technologies remains pivotal in addressing these complex and pervasive health challenges.In the pursuit of more accurate and timely diagnostic tools for brain injuries, the integration of advanced spectroscopic techniques into clinical practice has garnered considerable attention. Traditional diagnostic methods such as computed tomography (CT) scans, magnetic resonance imaging (MRI), electroencephalogram (EEG) and neurological examination while fundamental, often lack the precision and timeliness required for effective clinical intervention.2 In addition, they are costly, time-consuming, and lack of providing molecular specific information. Liquid biopsy is an emerging field within early detection and diagnosis of diseases. The integration of liquid biopsy techniques and RS has emerged as a promising avenue in the quest to unveil the intricacies of brain disorders. The exploration of novel diagnostic modalities has spurred interest in RS, a non-invasive analytical technique that holds promise in unraveling the molecular intricacies present within human biofluids like blood, urine, and CSF.10,11 In the realm of medical diagnostics, liquid biopsy offers a minimally- to non-invasive means to probe the inner workings of the body by analyzing various biomarkers present in bodily fluids. When applied to the study of brain disorders, this approach allows for the detection and monitoring of specific molecules or markers associated with neurological conditions, providing valuable insights into disease pathology. RS, with its ability to provide unique molecular fingerprints and discern subtle biochemical alterations, holds immense potential in identifying biomarkers associated with brain injuries and NDs. This spectroscopic technique offers a novel means to study the intricate biochemical changes that occur within biofluids due to these disorders.12–14 By analyzing the vibrational patterns of molecules, RS enables researchers to detect and quantify specific biomolecular signatures indicative of disease pathology. Notably, its high sensitivity and specificity in detecting molecular variations make it a promising tool for early and precise diagnosis.
RS offers a valuable tool for the study of NDs related biomarkers using various biofluids, including blood, urine, saliva, tears, and CSF. Biomarkers are measurable indicators that can provide information about normal or pathological biological processes. For instance, RS provides molecular signatures of various biomolecules present in biofluids, including proteins, lipids, nucleic acids, and metabolites.15–18 In addition, RS is non-invasive and label-free, allowing for direct analysis of biofluids without the need for additional chemical labels or dyes. This preserves the natural composition of biofluids and minimizes sample preparation requirements. More importantly, RS is sensitive to changes in protein conformations. It can be used to characterize different protein conformations, including misfolded or aggregated states, which are relevant to NDs.19 Metabolic changes in biofluids, such as alterations in lipid profiles and metabolite concentrations, are associated with NDs. Moreover, those biofluids are easily accessible and minimally invasive for patients. RS can monitor these metabolic changes, offering insights into disease-related variations.20 With the emergence of machine learning techniques RS, combined with advanced statistical analysis methods, can differentiate between healthy and diseased states based on the spectral differences associated with biomarkers.21–25 This differentiation is crucial for early diagnosis and monitoring disease progression. Detecting specific biomarkers in biofluids can be crucial for early diagnosis, prognosis, and monitoring the progression of NDs. By analyzing these molecular signatures, researchers can identify specific biomarkers associated with NDs. The chemical composition of human body fluids is intricate, with intricate associations between different bands in the acquired spectra, as they originate from the same chemical bonds. Additionally, Raman spectra may be affected by noise, autofluorescence background, Raman shift drift, and contributions from external spectral noise sources.26,27 While improving detector sensitivity and utilizing higher-quality optical elements can enhance Raman signal quality and facilitate a more detailed analysis of body fluid component composition, these improvements come with added costs and experimental setup complexity. An alternative method for enhancing pathology detection is surface-enhanced Raman spectroscopy (SERS).
The primary objective of this comprehensive review is to thoroughly explore and highlight the expanding role of RS in decoding intricate biochemical signatures present within various biofluids, with a particular focus on its application in the detection and diagnosis of brain diseases. Moreover, it aims to meticulously survey the current landscape of its applications in neurological diagnostics, encompassing its use in identifying specific biomarkers, elucidating molecular alterations, and exploring the potential for clinical translation. This review further endeavors to critically examine the challenges encountered in the implementation of RS for neurological diagnostics, including considerations of standardization, reproducibility, and technological advancements necessary for widespread clinical adoption. Additionally, it seeks to present a comprehensive overview of the prospects and future directions, envisioning the transformative impact of RS as a pioneering tool in enhancing the diagnostic armamentarium for neurological disorders. We commence our discussion by exploring the theoretical foundations and working principles of RS. Subsequently, we delve into the practical considerations associated with conducting Raman spectroscopic investigations on biological fluids. Moving forward, we provide a concise overview of Raman spectra pertaining to biomolecules, emphasizing the distinctive contributions of different biomolecules across various regions of the Raman spectrum. Following this, we extensively examine the sensing of biomarkers in biofluids using both spontaneous and enhanced Raman spectroscopic techniques. Notably, our focus is on biofluids such as blood, tears, saliva, and CSF. In the conclusion part, we address the current challenges, opportunities, and future prospects inherent in RS investigations of biofluids. Through this thorough exploration of RS principles, applications, challenges, and future prospects, this review endeavors to underscore its pivotal and transformative role in revolutionizing the landscape of neurological diagnostics, advocating for its potential as an invaluable tool for clinicians and researchers alike.
Working principles of Raman spectroscopy
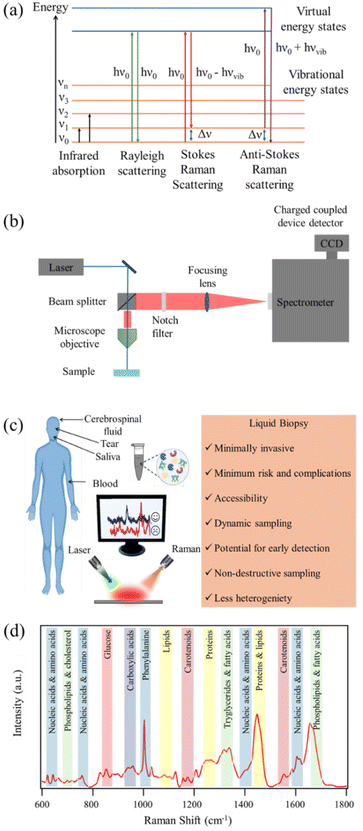
RS is a powerful analytical technique that provides detailed information about the chemical structure and phase of a substance. The working principle of RS relies upon the inelastic scattering of photons, a process known as Raman scattering. In the Raman effect, the scattered photons experience a change in energy due to interaction with molecular vibrations. The resulting Raman scattered light contains information about the vibrational modes within the sample. Raman spectra are characterized by bands corresponding to molecular vibrations, and the shifts in these bands provide insights into molecular structure, composition, and bonding. Stokes scattering involves lower energy photons, while anti-Stokes scattering involves higher energy photons. Schematic representation of the energy level diagram of Raman scattering process is depicted in Fig. 1(a). RS is non-destructive and can be applied to a variety of materials, making it a versatile tool in fields ranging from chemistry and physics to biology and materials science.The working principle of RS involves several key components as depicted in Fig. 1(b). A laser source emits monochromatic light, typically in the visible or near-infrared range, onto the sample. The scattered light is collected and passes through a monochromator to separate different wavelengths. Filtering out the Rayleigh line in a Raman spectrometer is essential to accurately measure Raman signals without interference from the excitation laser's Rayleigh scattering. To filter out the Rayleigh line in a Raman spectrometer, notch filters or edge pass filters are commonly employed. These filters are designed to block the Rayleigh scattered laser light while transmitting the Raman scattered light. Long pass edge filters are also used to transmit light from the sample that has been Raman scattered at lower energies, effectively reducing unwanted Rayleigh scattered signal laser light. The use of spectral filtering is pivotal for the successful acquisition of Raman spectra, as it ensures that only the Raman scattered light, contributing to chemical analysis, is directed to the detector. The dispersed light is then detected by a sensitive detector, typically a charge coupled device (CCD) detector, and the resulting spectrum reveals the Raman shifts corresponding to vibrational modes in the sample. The intensity and frequency of these shifts provide detailed information about the molecular structure and chemical composition. RS is employed in various modes, including conventional point measurements, confocal microscopy for spatial resolution, and even SERS for increased sensitivity. This technique has proven invaluable in identifying and characterizing materials at the molecular level, making it an indispensable tool in scientific research and industrial applications.
Spontaneous Raman signal is inherently weak. To overcome this limitation enhancement mechanisms are desired. SERS is a powerful analytical technique that enhances the Raman signals of molecules adsorbed on or near noble metal surfaces.28–31 The enhancement in signal intensity in SERS arises from two main mechanisms: electromagnetic enhancement and chemical enhancement. The electromagnetic mechanism involves the excitation of localized surface plasmon resonances (LSPRs) on the metal surface by incident light.32–35 This creates intense electromagnetic fields near the metal surface, enhancing the Raman scattering cross-section of nearby molecules. The chemical mechanism involves charge transfer between the molecule and the metal surface, further amplifying the Raman signals.36,37 In the electromagnetic mechanism, the enhancement is highly dependent on the shape, size, and composition of the nanostructured metal surfaces. Nanostructures such as silver and gold nanoparticles, nanorods, or nanostars are commonly employed to generate intense localized fields.33–35,38 The proximity of the analyte to these nanostructures results in significant signal enhancement. Chemical enhancement, on the other hand, involves the formation of charge transfer complexes between the adsorbed molecule and the metal surface. SERS has found wide applicability in investigating biological fluids due to its exceptional sensitivity. It enables the detection of low concentrations of biomolecules, making it invaluable for studying complex biological systems. In the realm of biological fluid analysis, SERS has been utilized for the identification and quantification of biomarkers associated with various diseases.39 Its capability to provide detailed molecular information makes SERS a promising tool for advancing our understanding of biochemical processes in biological fluids, contributing to both basic research and clinical diagnostics.
Experimental considerations
The analysis of biofluids using RS, both spontaneous and SERS, demands meticulous attention to various experimental parameters to ensure accurate and reliable results.21,24,25 Biofluid sample preparation is a critical initial step, involving careful handling and, at times, dilution to achieve optimal concentrations. Spectral acquisition involves exposing the sample to a laser source, where spontaneous Raman scattering, or SERS enhancement occurs. An illustrative diagram depicting the use of RS in analyzing liquid biopsy for the detection and diagnosis of brain disorders is shown in Fig. 1(c). Selection of an appropriate laser wavelength is crucial to avoid sample damage and fluorescence interference. In biofluid analysis, spectral correction becomes essential due to inherent variations in the background signals, arising from water and other biochemical constituents. Fluorescence background removal is particularly pertinent in spontaneous Raman and SERS, where the fluorescence emitted by certain biomolecules can overshadow Raman signals. Advanced signal processing techniques are employed to subtract background noise and enhance the specificity of the acquired spectra.Several experimental parameters impact the success of biofluid analysis, such as laser power, integration time, and the choice of substrate for SERS.40 Optimizing these parameters ensures the sensitivity and specificity required for detecting low-concentration biomarkers in complex biological matrices. Careful consideration of experimental conditions is paramount, as variations can significantly influence the obtained spectra. The selection of an appropriate laser wavelength is a crucial factor in RS, influencing the quality and sensitivity of the acquired spectra.41–43 The laser wavelength determines the energy of the incident photons and, consequently, the energy of the scattered photons. This choice impacts the efficiency of the Raman scattering process and the ability to excite specific molecular vibrations. For instance, resonant Raman scattering occurs when the laser wavelength closely matches an electronic transition of the molecule under investigation.44–47 Matching the resonance condition enhances the Raman signals significantly, making certain vibrations more pronounced and detectable. In biofluid analysis, avoiding fluorescence interference is crucial. Some biomolecules exhibit fluorescence when excited by certain wavelengths. Selecting a laser wavelength that minimizes fluorescence ensures that Raman signals are not obscured by background noise. Overall, successful biofluid analysis using RS, especially with the enhanced capabilities of SERS, relies on a holistic approach encompassing sample preparation, spectral acquisition, correction techniques, and meticulous control of experimental parameters.
Variants of Raman spectroscopy and microscopy techniques
RS presents a number of variants and imaging modalities that find application in protein-based analysis.48–50 These diverse approaches offer unique advantages, such as providing insights into protein structure, conformational changes, interactions, and dynamics with high sensitivity and spatial resolution. By understanding the mechanisms and underlying theory of Raman scattering, researchers can effectively harness these techniques to tackle a myriad of questions in protein science and biomedicine. Raman imaging techniques offer the ability to visualize and map the distribution of biomolecules within biological samples at the microscopic level.51,52 This can provide valuable insights into the spatial heterogeneity and localization of pathological features associated with brain disorders.Conventional (spontaneous) Raman spectroscopy
Conventional RS serves as a valuable tool for identifying and characterizing biomolecules such as proteins, lipids, and nucleic acids implicated in brain disorders.44,53,54 This is an inelastic scattering process where a molecule in the ground state interacts with an incident photon, and the resulting scattered photon has a different energy (frequency) from the incident photon. The energy difference corresponds to the vibrational energy levels of the molecule. Through the analysis of Raman spectra obtained from biofluids, researchers can discern spectral markers indicative of pathological changes, such as protein aggregation in NDs. This label-free approach offers a robust means of scrutinizing the molecular composition and structure of the brain, thereby unveiling crucial insights into neurological disorders. In addition, this technique has been instrumental in exploring changes in lipid and protein composition observed in AD, PD, and other brain disorders.54,55 By probing the vibrational modes of biomolecules, conventional RS provides molecular-specific information, thereby elucidating alterations in molecular composition, structure, and interactions associated with brain disorders.Resonance Raman spectroscopy (RRS), stimulated Raman spectroscopy (SRS), and coherent anti-stokes Raman spectroscopy (CARS)
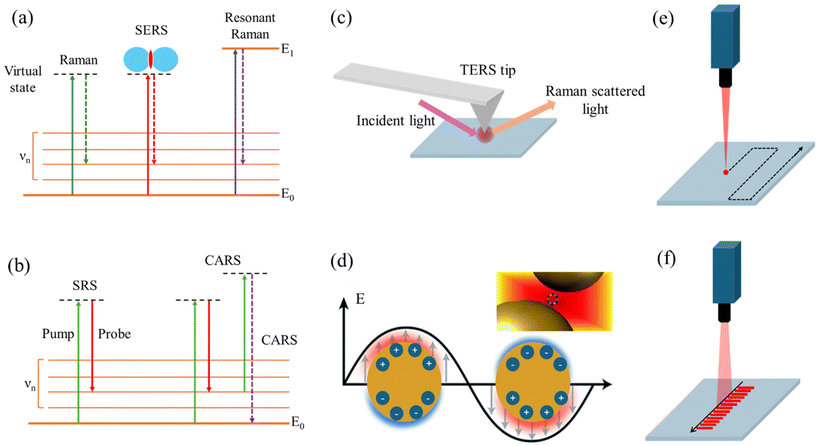
In these techniques, the involvement of two or more photons in the Raman scattering process leads to enhanced signals and unique spectroscopic capabilities compared to spontaneous Raman scattering. While RRS relies on resonance with electronic transitions, SRS and CARS involve the coherent interaction of multiple laser beams with the molecule, leading to stimulated or coherent Raman signals, respectively.RRS enables the selective detection and characterization of specific molecular species implicated in brain disorders. In RRS, the incident photon energy is close to an electronic transition energy of the molecule. This leads to an enhancement of the Raman signal, as the molecule is temporarily promoted to a virtual excited electronic state, increasing the polarizability and the Raman scattering cross-section (Fig. 2a). By tuning the laser wavelength to align with the absorption bands of target molecules, researchers can enhance Raman signals, achieving higher sensitivity and specificity for detecting disease-related biomolecules, such as amyloid-beta (Aβ) peptides in AD and other brain disorders.44,56 RRS enhances Raman signals by matching the excitation energy of incident photons to the electronic transition energy of target molecules, increasing the efficiency of Raman scattering and enabling the selective detection of disease-specific biomolecules. By selectively enhancing Raman signals from specific molecular vibrations through laser tuning, RRS provides detailed structural information about proteins and other key biomolecules involved in NDs and brain tumors, offering insights into changes in protein conformation, aggregation, and interactions, hallmarks of many brain disorders.1,57,58
SRS is a nonlinear RS technique used for imaging and analyzing biological samples, including biofluids relevant to diagnosing brain disorders. It offers several advantages, such as high sensitivity, rapid acquisition times, and the ability to perform label-free imaging and detect specific biomolecules.59,60 SRS relies on the simultaneous interaction of two laser beams with the sample: a pump beam and a Stokes beam (Fig. 2b). The frequency difference between these beams matches the vibrational frequency of a specific molecular bond in the sample, resulting in an amplification of the Raman signal known as stimulated Raman gain or loss. SRS can analyze biofluids like CSF, blood, or urine for specific biomarkers associated with brain disorders.59,61 For instance, in AD, SRS can detect and quantify Aβ peptides or tau proteins in CSF samples, known disease biomarkers. SRS microscopy provides high-resolution, chemically selective images of biofluids, revealing the spatial distribution of biomolecules of interest. This offers insights into the presence and localization of disease-related molecules or aggregates within the sample. As a label-free technique, SRS preserves the native state of the biofluid and its components, avoiding potential alterations or artifacts introduced by labeling procedures. With its rapid acquisition times and high sensitivity, SRS enables high-throughput screening of biofluid samples, facilitating analysis of large sample sets and potentially enabling early detection or monitoring of brain disorders.
CARS, a nonlinear RS technique, is utilized for analyzing biological samples to diagnose brain disorders. In this process, three laser beams (pump, Stokes, and probe) interact with the sample in a four-wave mixing process.1,62 Similar to SRS, CARS relies on the frequency difference between the pump and Stokes beams matching the vibrational frequency of a specific molecular bond in the sample (Fig. 2b). However, in CARS, the probe beam interacts with the vibrationally excited molecules, generating a signal at a higher frequency than the incident beams, termed the anti-Stokes frequency. The coherent nature of the CARS signal allows for highly sensitive detection of biomolecules in biofluids, even at low concentrations. CARS microscopy, renowned for label-free, high-resolution imaging of brain tissue, exploits coherent excitation of molecular vibrations to generate anti-Stokes photons at specific vibrational frequencies. By scanning laser beams across brain tissue, CARS microscopy produces three-dimensional images with molecular contrast, enabling visualization of disease-related changes in tissue morphology and composition. Valuable insights into brain tumor identification are provided by CARS through detailed examination of lipid and biomolecule distribution within the brain. Its high sensitivity, speed, and three-dimensional vibrational imaging capabilities make it ideal for analyzing brain structures and monitoring changes associated with neurological conditions. By providing contrast for visualizing the distribution and dynamics of key biomolecules, CARS reveals pathological alterations in brain structure and function. Furthermore, CARS has been crucial in imaging lipid alterations in TBI models and protein aggregation in ND models.
Tip-enhanced Raman spectroscopy (TERS) and surface-enhanced Raman spectroscopy (SERS)
SERS and TERS are techniques that enhance the Raman scattering signal from molecules by exploiting the interaction between the molecules and metallic nanostructures or sharp metallic tips, respectively. Both SERS and TERS rely on the EM enhancement due to localized surface plasmons, but TERS benefits from the additional lightning rod effect and spatial confinement provided by the sharp metallic tip.TERS, a variant of RS, merges scanning probe microscopy (SPM) principles with SERS to achieve enhanced spatial resolution and Raman signal sensitivity. Although predominantly used for surface analysis and imaging of solid samples, TERS finds application in biofluid analysis for diagnosing brain disorders. In TERS, a metallic tip, often silver or gold, is positioned in close proximity (a few nanometers) to the sample surface, acting as a nanoscale antenna. This enhances the electromagnetic field via LSPR, amplifying Raman signals from nearby molecules and detecting weak signals with precision.63,64 The electromagnetic mechanism enhancement in TERS stems from the LSPR at the sharp metallic tip, which functions as a nanoantenna concentrating the electromagnetic field at its apex, thus creating a highly localized “hot spot” with a significantly enhanced field (Fig. 2c). Acting as a lightning rod, the sharp metallic tip in TERS further boosts the electric field at the tip apex. The field enhancement increases with the sharpness of the tip, with sharper tips yielding higher enhancements due to the inverse relationship with the tip's radius of curvature. In TERS, the Raman signal emanates from a highly confined region, typically within a few nanometers from the tip apex, facilitating high-resolution imaging and spectroscopy of nanoscale structures and individual molecules. TERS delves into biofilm formation on surfaces relevant to certain brain disorders like AD, where Aβ peptides’ aggregation leads to biofilm creation. It furnishes high-resolution chemical insights into biofilm composition and structure, aiding disease mechanism comprehension. While TERS is surface-focused, it extends to dried biofluid sample analysis, such as dried blood spots or CSF droplets.65–68 Here, the metallic tip approaches the dried sample closely, facilitating disease biomarker detection with high spatial resolution and sensitivity. TERS synergizes with other techniques like mass spectrometry or fluorescence microscopy to provide comprehensive biomarker composition and distribution information in biofluids. For instance, TERS identifies and locates specific molecular species, while mass spectrometry supplements with molecular masses and identities of detected compounds.
SERS is a technique that combines principles of RS with metallic nanostructure-induced signal enhancement. It holds promise for diagnosing brain disorders through biofluid analysis. Renowned for its high sensitivity, molecular specificity, and ability to detect minute biomarker concentrations, SERS capitalizes on the LSPR phenomenon. Herein, the collective oscillation of conduction electrons in metallic nanostructures, predominantly gold or silver, resonates with incident electromagnetic radiation, inducing a robust enhancement of the local electromagnetic field near the nanostructures.25,33–35 Consequently, Raman signals from molecules in proximity to or adsorbed on the metallic surface are amplified. SERS is adept at scrutinizing biofluids like CSF, blood, or urine for brain disorder-associated biomarkers.69–72 In AD, for instance, SERS discerns and quantifies Aβ peptides, tau proteins, and other pertinent molecules in CSF samples with remarkable sensitivity and specificity.73,74 A pivotal advantage of SERS lies in its capability to detect and analyze biomarkers at low concentrations, challenging to discern via conventional methods. Multiplexed detection of multiple biomarkers concurrently is feasible with SERS, accomplished by employing distinct SERS-active nanoparticles or substrates tailored with specific recognition elements (e.g., antibodies or aptamers) for diverse biomarkers.75 The distinct Raman signatures of these nanoparticles or substrates facilitate the identification and quantification of multiple biomarkers in a single analysis. SERS delves into protein structure, folding, and interactions, furnishing insights into the molecular mechanisms underpinning brain disorders. With potential for in situ biofluid analysis sans extensive sample preparation or labeling, SERS holds promise for real-time biomarker monitoring. Integration with complementary analytical techniques like mass spectrometry or fluorescence spectroscopy enriches our understanding of biomarker composition and structure in biofluids.
Various substrates commonly employed for SERS
The choice of the substrate material and its surface morphology plays a crucial role in achieving strong SERS enhancement. It's worth noting that the choice of substrate depends on factors such as the desired SERS enhancement, reproducibility, cost, and compatibility with the analyte and experimental conditions. Additionally, various substrate modifications, such as the addition of hot spots or incorporation of additional materials, can further enhance the SERS signals.Metallic nanoparticles
Gold and silver nanoparticles are the most widely used SERS substrates due to their strong plasmonic properties. They can be synthesized as colloidal solutions or deposited on solid supports. These nanoparticles can significantly enhance the Raman signal through electromagnetic and chemical enhancement mechanisms. Noble metal SERS substrates have limitations in practical applications due to disadvantages such as high cost, complex synthesis, and the need for large-scale production.Anisotropic plasmonic nanostructures
Gold or silver nanowires, nanoprisms, nanorods, and nanostars exhibit strong plasmonic properties, leading to highly enhanced SERS signals. Their anisotropic shapes allow for tuning of the plasmon resonance wavelength. Anisotropic plasmonic nanostructures may exhibit SERS blinking, which can affect the stability and reproducibility of the signal.76 In addition, the optical response of anisotropic nanostructures may vary, impacting the uniformity of Raman enhancement.Bimetallic nanoparticles
Bimetallic nanoparticles composed of two types of metal atoms, such as alloyed and core–shell configurations, have also been explored as SERS substrates. They offer tunable properties and enhanced SERS performance compared to monometallic nanoparticles.77,78 There are challenges associated with bimetallic nanoparticles, such as high cost, stability issues, and limitations in practicality based on comparison with other substrates.Metal nanoparticle aggregates
Aggregates of gold or silver nanoparticles can provide highly enhanced SERS signals compared to individual nanoparticles due to electromagnetic hot spots created between closely spaced nanoparticles.79,80 Aggregates may have limited reproducibility and stability, and the aggregation process can be challenging to control.Nanostructured metal films
Thin metal films (gold, silver, copper) deposited on nanostructured surfaces like silicon nanowires or nanoparticles provide high SERS enhancement due to the high density of hot spots.81,82 Metal films may suffer from surface roughness and non-uniformity, affecting SERS reproducibility. Film thickness and morphology need to be carefully controlled.Self-assembled metal nanoparticle arrays
Colloidal metal nanoparticles can be assembled into ordered arrays using techniques like Langmuir–Blodgett deposition, self-assembly, or lithographic patterning.83,84 Ordered arrays of gold or silver nanoparticles on solid substrates, such as silicon or glass, can provide reproducible and tunable SERS enhancement by controlling the nanoparticle size, shape, and spacing. These nanoparticle arrays exhibit strong plasmonic coupling and SERS activity. However, these substrates have limited control over nanoparticle organization and interparticle spacing. In addition, substrates have potential for defects and non-uniformities in the assembly.Metal–organic frameworks (MOFs)
MOFs loaded with metal nanoparticles or grown on metal surfaces can serve as highly sensitive SERS substrates due to their high porosity and large surface area.85,86 They provide chemical stability and tunable properties. On the other hand, MOF synthesis and modification can be complex, and achieving uniform nanoparticle dispersion within the MOF matrix may be challenging.Two-dimensional (2D) materials
2D materials like graphene, MoS2, WS2 combined with metal nanostructures have shown promising SERS performance due to their unique electronic and optical properties.24,87 For these types of substrates, control over morphology and defects may be challenging, and optimization of functionalization methods is required for maximum enhancement.Different scanning and imaging techniques
Raman hyperspectral imaging, an advanced analytical technique, merges Raman spectroscopy with optical microscopy, offering detailed chemical and structural data by capturing spatial and spectral information from samples.88–90 Also known as Raman chemical imaging or Raman mapping, it diverges from conventional RS by gathering spectra from various points across a sample's surface. This facilitates visualization and analysis of the sample's chemical components or molecular species distribution. The process entails scanning a focused laser beam over the sample while simultaneously collecting Raman spectra at each scan point. The resulting dataset comprises spectra corresponding to specific sample locations, enabling the generation of hyperspectral images displaying spatial distribution patterns. This technique holds promise for chemical analysis and materials characterization, providing comprehensive insights into sample composition and distribution. Its applications extend to biomedical research, offering valuable information for various analytical purposes. Point-scanning Raman imaging has been employed to study the distribution of lipids, proteins, and other biomolecules in brain tissue sections from patients with AD, PD, and other neurological conditions.91–93 In this approach, a tightly focused laser beam is raster-scanned across the sample, and the Raman spectrum is collected at each pixel position (Fig. 2e). The collected data are then processed to generate chemical maps and images based on the specific Raman signatures of the molecules present in the sample. This can reveal the formation of plaques, protein aggregates, or chemical changes in specific brain regions.94The line scanning technique in Raman microscopy is a method used to acquire Raman spectral data from a sample in a line-by-line fashion, rather than collecting data from a single point at a time (Fig. 2f).95,96 This technique offers several advantages, including faster data acquisition times, reduced photobleaching or photodamage to the sample, and the ability to generate high-resolution Raman images.97 In the line scanning technique, the laser beam is focused into a line shape instead of a diffraction-limited spot. This line-shaped illumination is achieved by using a cylindrical lens in the excitation path of the microscope. The sample is then raster-scanned in a direction perpendicular to the illumination line, and the Raman scattered light from the entire line is collected simultaneously by a detector, typically a CCD or an electron-multiplying CCD (EMCCD). Super-resolution Raman microscopy (SRRM) is an advanced imaging technique that surpasses the diffraction limit of conventional Raman microscopy, enabling the visualization of nanoscale features with enhanced spatial resolution.98 Unlike traditional Raman microscopy, which is limited by diffraction to a spatial resolution of approximately half the wavelength of the incident light, SRRM employs various strategies to achieve resolutions beyond this limit, often down to tens of nanometers or even single-digit nanometer scales.99,100 One common approach in SRRM is to utilize near-field Raman techniques, such as tip-enhanced Raman spectroscopy (TERS), which involves scanning a sharp metallic tip in close proximity to the sample surface.101 Another approach involves the use of super-resolution optical techniques, such as stimulated emission depletion (STED) microscopy or structured illumination microscopy (SIM), in combination with RS.102–105 These techniques exploit the principles of fluorescence microscopy to achieve super-resolution imaging, which can then be coupled with Raman scattering to provide chemically specific information about the sample.
Overall, the various RS techniques, each with their unique mechanisms and capabilities, have significantly contributed to unveiling the underlying mechanisms and characteristics of brain disorders. From the label-free visualization of pathological structures to the detailed biochemical characterization of relevant biomolecules, these Raman-based approaches have provided invaluable insights that can aid in the diagnosis, monitoring, and understanding of neurological conditions. Leveraging the molecular specificity and non-destructive nature of Raman techniques, researchers can gain unprecedented insights into the biochemical changes underlying brain disorders. This enables earlier diagnosis, monitoring of disease progression, and evaluation of therapeutic interventions. These techniques have the potential to aid in the early detection, diagnosis, and monitoring of neurological conditions, as well as contribute to the development of new therapeutic strategies by enhancing our understanding of the underlying molecular mechanisms.
Raman spectra of biomolecules
RS of biomolecules relies on the polarizability of chemical bonds, where distinct properties of chemical bonds result in varying light scattering. Complex biological samples consist of diverse chemical bonds, each inducing unique Raman shifts. The combination of Raman shift intensities forms the Raman spectrum of a sample. Biomolecules, integral to life processes, contribute to Raman spectra due to their specific chemical signatures. The Raman spectrum of biomolecules exhibits distinct characteristics, with unique peaks corresponding to different molecular components. These molecular signatures represent distinctive spectral patterns obtained through RS analysis of biofluids collected via liquid biopsy techniques. The Stokes Raman spectrum is categorized into four vibrational frequency regions.106,107 (i) The “low frequency (LF) region” below 300 cm−1 provides crucial information about biomolecular structural conformation and environmental conditions. For instance, chemical processes like ligand binding, enzymatic activity, electron transfer, or intermolecular interactions, induce fluctuations in these modes providing valuable information about the conformational state of the biomolecule in LF region.106,107 (ii) The “fingerprint region” (300–1800 cm−1) is essential for chemical identification, containing signals from proteins, nucleic acids, lipids, and carbohydrates. Specific vibrational bands, such as amide I and III, aid in determining protein secondary structure. In proteins, the Raman spectrum prominently features peaks associated with the side chain of aromatic amino acids and the peptide bond. Notably, amide I (1645–1680 cm−1, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching) and amide III (1225–1280 cm−1, C–N stretching coupled to N–H bending) vibrational bands are preferentially utilized for determining protein secondary structure.108–110 Nucleic acids contribute to the spectrum with signals from individual bases (600–800 cm−1, ring breathing) and the sugar-phosphate backbone. Strong Raman peaks in the region of 600 to 800 cm−1, attributed to ring breathing modes of various bases, facilitate the differentiation of nucleic acid bases.111–113 Nucleic acids exhibit distinctive bands providing information about their conformation and base pairing. Lipids contribute to the fingerprint region with details about hydrocarbon chains and C–C stretching. Lipids, crucial biomolecules with roles in energy storage and cellular functions, showcase distinctive Raman peaks. The long hydrocarbon chains contribute signals related to scissoring and twisting of CH2 and CH3 groups (1300 and 1400–1500 cm−1) and C–C stretching (1050–1200 cm−1).114 Additionally, lipid headgroups exhibit various structures, creating bands in the 710–890 cm−1 region. Carbohydrates create bands in this region, crucial for cellular recognition. Some common features are C–O–C stretching at 850 and 1125 cm−1 and CH3 rocking at 925 cm−1.115 Carbohydrates, essential compounds in living organisms, introduce characteristic bands in the fingerprint region. Raman spectra of human serum from a healthy donor highlighting the peak assignment for various metabolic groups and endogenous biomolecules are shown in Fig. 1(d) in the fingerprint region. (iii) The “silent region” (1800–2800 cm−1) excludes biomolecular contributions, suitable for specific tags in RS. This region is particularly valuable for incorporating specific molecular fingerprints or tags that are suitable for RS, without interference from the inherent signals of biological constituents.116,117 In this silent region, researchers often introduce tags or markers that have distinctive Raman signals, allowing for precise identification and tracking of specific molecules or functional groups. The absence of interference from the biomolecular background enhances the sensitivity and specificity of RS for detecting and characterizing these specific tags. (iv) The “high wavenumber region” (>2800 cm−1) is dominated by stretching vibrations of hydrogen bonds, ideal for studying lipids and long-chain hydrocarbons. The stretching vibrations of carbon–hydrogen (C–H) bonds are prominent in this region. Different types of C–H bonds, such as aliphatic and aromatic, exhibit characteristic peaks.114 These vibrations provide information about the types of chemical bonds and the environments in which they are present. The stretching vibrations of nitrogen–hydrogen (N–H) and oxygen–hydrogen (O–H) bonds also contribute to this region. These vibrations are significant in studying biomolecules like proteins and nucleic acids. The high wavenumber region is particularly informative for studying lipids. Stretching vibrations of C–H groups in the long hydrocarbon chains of lipids are observed, providing insights into lipid structure. Additionally, vibrations related to the C
O stretching) and amide III (1225–1280 cm−1, C–N stretching coupled to N–H bending) vibrational bands are preferentially utilized for determining protein secondary structure.108–110 Nucleic acids contribute to the spectrum with signals from individual bases (600–800 cm−1, ring breathing) and the sugar-phosphate backbone. Strong Raman peaks in the region of 600 to 800 cm−1, attributed to ring breathing modes of various bases, facilitate the differentiation of nucleic acid bases.111–113 Nucleic acids exhibit distinctive bands providing information about their conformation and base pairing. Lipids contribute to the fingerprint region with details about hydrocarbon chains and C–C stretching. Lipids, crucial biomolecules with roles in energy storage and cellular functions, showcase distinctive Raman peaks. The long hydrocarbon chains contribute signals related to scissoring and twisting of CH2 and CH3 groups (1300 and 1400–1500 cm−1) and C–C stretching (1050–1200 cm−1).114 Additionally, lipid headgroups exhibit various structures, creating bands in the 710–890 cm−1 region. Carbohydrates create bands in this region, crucial for cellular recognition. Some common features are C–O–C stretching at 850 and 1125 cm−1 and CH3 rocking at 925 cm−1.115 Carbohydrates, essential compounds in living organisms, introduce characteristic bands in the fingerprint region. Raman spectra of human serum from a healthy donor highlighting the peak assignment for various metabolic groups and endogenous biomolecules are shown in Fig. 1(d) in the fingerprint region. (iii) The “silent region” (1800–2800 cm−1) excludes biomolecular contributions, suitable for specific tags in RS. This region is particularly valuable for incorporating specific molecular fingerprints or tags that are suitable for RS, without interference from the inherent signals of biological constituents.116,117 In this silent region, researchers often introduce tags or markers that have distinctive Raman signals, allowing for precise identification and tracking of specific molecules or functional groups. The absence of interference from the biomolecular background enhances the sensitivity and specificity of RS for detecting and characterizing these specific tags. (iv) The “high wavenumber region” (>2800 cm−1) is dominated by stretching vibrations of hydrogen bonds, ideal for studying lipids and long-chain hydrocarbons. The stretching vibrations of carbon–hydrogen (C–H) bonds are prominent in this region. Different types of C–H bonds, such as aliphatic and aromatic, exhibit characteristic peaks.114 These vibrations provide information about the types of chemical bonds and the environments in which they are present. The stretching vibrations of nitrogen–hydrogen (N–H) and oxygen–hydrogen (O–H) bonds also contribute to this region. These vibrations are significant in studying biomolecules like proteins and nucleic acids. The high wavenumber region is particularly informative for studying lipids. Stretching vibrations of C–H groups in the long hydrocarbon chains of lipids are observed, providing insights into lipid structure. Additionally, vibrations related to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds in lipids are distinct from those in proteins. Researchers leverage the high wavenumber region for detailed molecular analysis, especially in the study of biomolecules like lipids, where the distinctive features of this region offer valuable information about their structure and interactions. Raman liquid biopsy molecular signatures hold significant promise for non-invasive diagnosis, prognosis, and personalized management of diseases, including neurological disorders. Overall, the Raman spectrum serves as a powerful tool for identifying and characterizing the molecular composition of proteins, lipids, nucleic acids, and carbohydrates in biomolecules.
O bonds in lipids are distinct from those in proteins. Researchers leverage the high wavenumber region for detailed molecular analysis, especially in the study of biomolecules like lipids, where the distinctive features of this region offer valuable information about their structure and interactions. Raman liquid biopsy molecular signatures hold significant promise for non-invasive diagnosis, prognosis, and personalized management of diseases, including neurological disorders. Overall, the Raman spectrum serves as a powerful tool for identifying and characterizing the molecular composition of proteins, lipids, nucleic acids, and carbohydrates in biomolecules.
Advanced chemometric methods applicable in Raman spectroscopy and microscopy
RS is a powerful analytical technique that can provide detailed chemical information about samples. However, the large amount of data generated can be challenging to analyze. This is where advanced chemometric methods step in, playing a pivotal role in Raman spectroscopic investigations. These methods are instrumental in gathering valuable information from complex spectral data, tackling issues like overlapping bands, spectral interferences, and data interpretation. To choose the appropriate method, the first step is to determine if each Raman spectrum responds to a known class of chemical property. If the Raman spectra are unlabeled, one can perform unsupervised methods to visualize and find clusters. If the label or response variable is known, one can choose supervised classification or regression methods. Linear models should first be tried since they are more accessible for interpretation and reveal essential Raman regions used in classification.Principal component analysis (PCA)
PCA stands out as a robust, unsupervised machine learning tool extensively employed in RS and microscopy for data analysis and interpretation. Its utility is important when grappling with extensive and intricate datasets, as it aids in pattern recognition, dimensionality reduction, and distilling key insights from the data. Within RS, PCA proves invaluable for delving into data, spotting outliers, and discerning spectral differences tied to distinct sample attributes or compositions.118 At its core, PCA transforms a set of potentially correlated variables into a fresh array of uncorrelated variables known as principal components (PCs).Before applying PCA, Raman spectral data typically undergo preprocessing steps like baseline correction, normalization, and noise filtering to ensure data quality and comparability. Following preprocessing, the Raman spectral data undergoes PCA calculations involving computing the covariance or correlation matrix and performing eigenvalue decomposition to derive principal components. These components are then ordered by associated eigenvalues, reflecting the variance explained by each. By selecting the initial few principal components that account for a substantial portion of the total variance (e.g., 90% or 95%), data dimensionality can be reduced while retaining pertinent information. The PCA scores, projections of the original data onto the new principal component space, are often visualized using scatter plots or other graphical representations, unveiling patterns or clusters challenging to discern in the original high-dimensional space. Analyzing PCA loadings, representing contributions of original variables (wavenumbers) to each principal component, allows the identification of spectral features or molecular signatures underlying observed data patterns or variations.
In Raman microscopy, PCA can be particularly useful for analyzing hyperspectral Raman images, where each pixel contains a full Raman spectrum. PCA can help to identify and separate different chemical components or molecular species present in the sample based on their unique Raman spectral signatures.119 Consequently, it facilitates the creation of chemical maps or component distribution images, offering valuable insights into the sample's spatial distribution and heterogeneity. Fig. 3(a) shows an example of PCA analysis of SERS spectra measured in AD mice serum with different disease stages. By applying PCA, the SERS spectra are projected into lower dimensions. The PCA scores visualize the spectra and reveal the separation of different disease stages. The PCA loadings further produce more information on the SERS peaks that distinguish the AD disease stages in the mice serum.
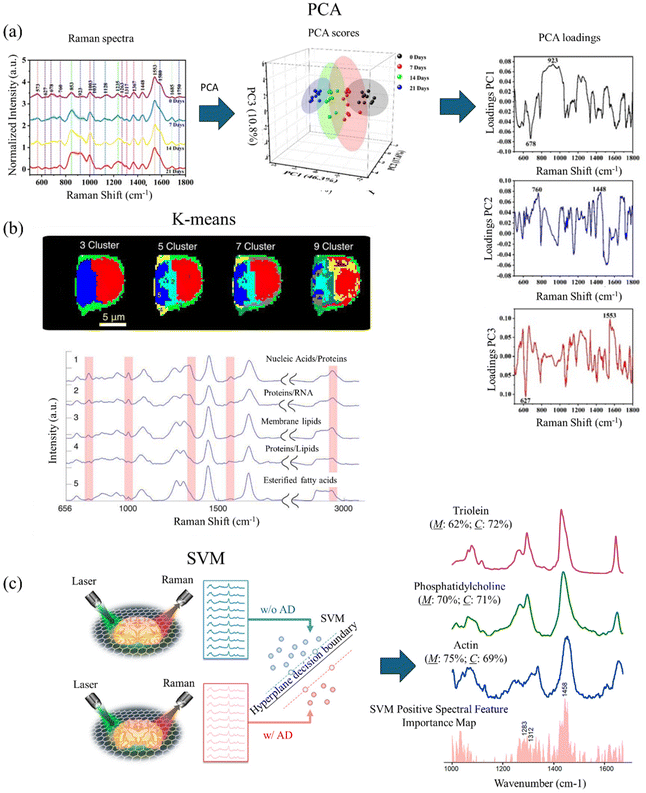 | ||
| Fig. 3 (a) Using PCA to analyze SERS spectra measured from AD mice serum at different stages. PCA score plot for SERS measured spectra of AD mice serum at different stages. PCA loading plot of first three principal components. Adapted with permission from ref. 119. Copyright 2023 Elsevier. (b) K-means cluster-based Raman mapping of lymphocyte cells with different numbers of clusters. Mean spectra of clusters with the number of clusters equal to 5. Adapted with permission from ref. 121. Copyright 2013 SAGE Publications. (c) Using SVM to classify Raman spectra measured from mice brain samples with and without AD. Linear SVM trained, which classifies the brain samples with and without AD using a hyperplane decision boundary. The spectral feature importance obtained from SVM and Raman spectra of potential biomarkers of AD. Adapted with permission from ref. 23. Copyright 2022 American Chemical Society. | ||
K-means clustering
K-means clustering represents an unsupervised machine learning method extensively employed in RS and microscopy for data analysis and pattern recognition. This partitioning clustering algorithm seeks to divide the data into K distinct clusters predicated on their likeness or divergence. In RS and microscopy, K-means clustering finds application in grouping or segmenting Raman spectra or sample points according to their spectral resemblances. This enables the delineation of discrete chemical constituents, materials, or regions within a given sample.120The application of K-means clustering in RS and microscopy typically involves several steps. First, an essential aspect of K-means clustering is determining the appropriate number of clusters (K) to partition the data into. This can be achieved through various techniques, including the elbow method, silhouette analysis, or leveraging domain knowledge. The K-means algorithm necessitates initial centroids or cluster centers to commence the iterative process. Each data point (Raman spectrum or sample) is then assigned to the nearest cluster centroid based on a chosen distance measure, typically Euclidean distance. Following the assignment of all data points to clusters, the algorithm iteratively recalculates the centroids of each cluster by computing the mean of all data points within that cluster. Upon convergence, the final clusters are obtained and can be analyzed and interpreted based on the spectral characteristics of the Raman spectra within each cluster. This analysis offers valuable insights into the chemical composition, molecular structures, or spatial distributions of different components or regions within the sample. Fig. 3(b) shows an example of using K-means to determine the biochemical composition of lymphocyte cells. By coloring the clusters on Raman mapping, K-means reveals different components of the lymphocyte cell.121 Leveraging domain knowledge, Schie et al. select the number of clusters equal to 5, plot the means (centroids) of each cluster, and assign components to the clusters. In the means of clusters, the highlighting areas show significant Raman peaks to determine the biochemical composition.
Partial least squares (PLS) regression
PLS regression stands as a powerful supervised chemometric technique extensively utilized in RS and microscopy for quantitative analysis and calibration model development. Its efficacy shines notably in situations involving collinear and high-dimensional data, a common occurrence in RS where the number of variables (wavenumbers or Raman shifts) can be extensive. The underlying principle of PLS regression revolves around establishing a linear relationship between the Raman spectral data (predictor variables, X) and a collection of reference measurements or properties (response variables, y).122 This is achieved by projecting both the predictor and response variables onto a new set of latent variables or components, adept at capturing the maximum covariance between X and y.| X = ZVT + E |
| y = Zb + e |
The initial step involves organizing the Raman spectral data (X) and the corresponding reference measurements or properties (y) into matrices. The n × p matrix X comprises n Raman spectra as rows, with each spectrum having p wavenumbers or Raman shifts, X = [x1,…,xp]. Meanwhile, the n × 1 matrix y contains the reference measurements or property values for each spectrum. Prior to constructing the PLS model, it is advisable to partition the data into calibration and validation sets, which ensures proper model validation and mitigates the risk of overfitting. The PLS algorithm decomposes the X matrix into n × k scores matrix Z (projections onto the k latent variables) and k × p loading matrix V (transformation of the original variables to the latent variables). Then, it decomposes the y matrix into Z and k × 1 PLS coefficient b (coefficient for predicting response). The scores capture the systematic variation in the data, while the loadings unveil the significance of each variable (wavenumber or Raman shift) in the model. The E and e are residuals, which change depending on the selection of k. Determining the optimal number of latent variables (components) k to incorporate in the PLS model involves evaluating the model's performance through cross-validation or other validation techniques. Insufficient components may lead to underfitting, whereas excessive components can result in overfitting and diminished predictive capability. Once developed, the PLS model undergoes validation using the independent validation set to gauge its predictive efficacy. Various metrics like root mean squared error (RMSE), coefficient of determination (R2), or bias are employed to assess the model's performance and robustness. If the PLS model demonstrates satisfactory performance, it can be utilized for the quantitative analysis of new Raman spectral data. Additionally, insights into relevant spectral features or molecular signatures contributing to the prediction of the response variable can be gleaned from the model loadings.
Linear discriminant analysis (LDA)
LDA, a supervised chemometric technique, is employed for classification and pattern recognition in RS and microscopy. Its objective is to identify the optimal linear combination of variables (Raman shifts or wavenumbers) that maximizes the distinction between predefined classes or groups while minimizing within-class variance. In the realm of RS and microscopy, LDA serves various purposes, including sample classification, identification of spectral markers or discriminating features linked to different classes, and the development of diagnostic models based on Raman spectral data.122The Raman spectral data is structured into a matrix, with each row representing a spectrum. Alongside, class labels or group assignments for each sample or spectrum are required. To streamline computational efficiency while retaining essential information, LDA can be preceded by dimensionality reduction techniques like PCA. The LDA algorithm then identifies the linear combination of variables (Raman shifts or wavenumbers) maximizing the ratio of between-class variance to within-class variance. This combination, termed the discriminant function, ensures optimal separation between predefined classes or groups. To evaluate the model, cross-validation or an independent test set is employed to gauge its classification accuracy and robustness using metrics like the confusion matrix, sensitivity, specificity, and overall classification rate. Following satisfactory performance, the LDA model is utilized to classify new Raman spectral data into predefined classes or groups. Furthermore, analysis of discriminant function coefficients or loadings aids in identifying the most discriminating Raman shifts or spectral features associated with each class, offering insights into molecular or chemical differences between groups.
In Raman microscopy, LDA proves especially valuable for scrutinizing hyperspectral Raman images, where every pixel includes a complete Raman spectrum. By incorporating spatial information with spectral data, LDA models are crafted to categorize diverse regions or constituents within the sample, leveraging their unique Raman spectral signatures. Consequently, this facilitates the creation of classification maps or discernment of spatial patterns linked to distinct chemical or molecular species.
Support vector machines (SVMs)
SVMs stand out as robust supervised machine learning algorithms widely employed in RS and microscopy for both classification and regression tasks. SVMs excel when confronted with high-dimensional datasets, such as Raman spectra, and proficiently manage non-linear relationships between the input data and target variables. Within the realm of RS and microscopy, SVMs find application in various tasks, including sample classification, identification of spectral markers or discriminating features linked to distinct classes, and the formulation of quantitative models for predicting properties or concentrations based on Raman spectral data.10,23The application of SVMs in RS and microscopy typically involves several steps. Firstly, the Raman spectral data is structured into a matrix X, with each row representing a sample or spectrum x. Additionally, class labels or target variables (for classification or regression tasks) y must be assigned to each sample or spectrum. To optimize computational efficiency and interpretability, feature selection techniques (e.g., variable importance in projection, recursive feature elimination) or dimensionality reduction methods (e.g., principal component analysis) can be employed on the Raman spectral data. The SVM algorithm then constructs a hyperplane or decision boundary in a high-dimensional space ω·x + b = 0, maximizing the margin between different classes or groups. The prediction is defined as the following, where +1 and −1 are two classes.
By transforming the input data into a higher-dimensional feature space using kernel functions (e.g., polynomial, radial basis function), SVM is able to handle non-linear relationships. To optimize the SVM model's performance, hyperparameters such as the kernel function, the regularization parameter (C), and the kernel parameters (e.g., gamma for the RBF kernel) are tuned using cross-validation or grid search techniques. The developed SVM model is evaluated using an independent test set or cross-validation to assess its performance. For classification tasks, metrics such as accuracy, precision, recall, and F1-score are employed, while for regression tasks, metrics like mean squared error (MSE), coefficient of determination (R2), and root mean squared error (RMSE) are utilized. If the SVM model achieves satisfactory performance, it can be deployed to classify new Raman spectral data into predefined classes or predict target variables based on the Raman spectral input. Furthermore, techniques like permutation importance or weight vector (ω) analysis for linear SVM can be utilized to identify the most discriminative or relevant Raman shifts or spectral features contributing to the model's predictions.
In Raman microscopy, SVMs are particularly valuable for analyzing hyperspectral Raman images, where every pixel encompasses a complete Raman spectrum. By integrating spatial information with spectral data, SVM models can be crafted to classify various regions or components within the sample, relying on their distinctive Raman spectral signatures. This capability facilitates the creation of classification maps or the anticipation of spatial distributions of chemical components or properties within the sample. Fig. 3(c) shows an example of using linear SVM to diagnose AD mice brain slices and identify potential AD biomarkers. The Raman spectra measured with and without AD are fed into the linear SVM model. After training, the linear SVM constructs a hyperplane decision boundary. By extracting the coefficient, Wang et al. plot spectra feature importance maps, identify essential Raman peaks that are correlated to AD, and reveal new potential biomarkers.23
Liquid biopsy investigation for brain disorders through spontaneous and enhanced Raman spectroscopic techniques
Blood-based investigations
Liquid biopsy of AD biomarkers, such as Aβ and tau proteins, in blood samples holds great promise for cost-effective, widely accessible, easily-administered, and minimally-invasive detection and follow-up of AD. Blood is a valuable reservoir of potential biomarkers essential for disease screening, risk assessment, detection, and prognosis. It contains free-floating erythrocytes, leukocytes, platelets, and a nutrient-rich plasma fluid, making it easier to collect samples compared to solid tissue biopsies.123 This enables the regular monitoring of a patient's health status over time. Blood-based biomarkers are of particular interest due to their minimally-invasive nature and ease of sample collection.124,125 Research has focused on identifying proteins such as tau, Aβ, microRNA, and other molecules associated with NDs in blood. The spectra obtained through Raman analysis of blood exhibit multicollinearity, coupled with the presence of autofluorescence background and various types of noise.26,126 Selecting an appropriate method for processing experimental data from blood spectra is imperative to derive statistically reliable insights into a pathological process within the body. Previous studies have investigated multivariate analysis methods of blood Raman spectra classification which is useful in detecting AD.26 Typically, diagnosis of AD through RS involves utilizing spectrochemical analysis of blood. This analytical approach aims to identify distinct chemical patterns or biomarkers in the blood that may distinguish AD from other conditions. By examining the unique spectrochemical profiles, researchers and clinicians seek to improve the accuracy and specificity of AD diagnosis, paving the way for more effective and targeted interventions. Spectrochemical analysis in blood holds promise as a minimally-invasive and potentially reliable method for discriminating AD from other disorders.Early studies used platelets from animal models of NDs to distinguish healthy and diseased cases. For instance, Chen et al. achieved over 90% accuracy of distinguishing AD, PD, and vascular dementia.127 In a subsequent work by the same research group extended this work to detect AD by Raman spectra of rat's platelets. Recent developments in machine learning enabled new opportunities for accurate diagnosis of AD. For instance, in a recent work by Lin et al. reported laser tweezers RS combined with machine learning for diagnosis of AD.128 By utilizing AD platelets from transgenic rats at different ages, the researchers were able to differentiate between normal and diseases platelets of 3-, 6-, and 12-month AD samples (Fig. 4a and b). Raman spectra of diseased samples and healthy controls showed unique features. Firstly, the intensity ratio of 1127 cm−1 peak to 1001 cm−1 peak increases significantly with the AD severity. This was attributed to changes of amyloid precursor protein metabolism of AD and normal platelets. Another feature is the intensity ratio of 1654 cm−1 peak to 1600 cm−1 peak decreases with the AD severity. This was explained based on the α helix structure and amyloid precursor protein changes of AD platelets. With the help of machine learning techniques, the researchers achieved accuracy of 91%, 68% and 97% for distinguishing normal and diseased samples of 3-, 6- and 12-month. Wang et al. developed classification models for early and advanced AD as well as the control group using fewer features.129 They applied mechanisms for noise reduction to enhance the accuracy of the models. Raman spectra of platelets were acquired from animal models, encompassing early and advanced stages of AD transgenic mice, non-transgenic controls, and PD mice (Fig. 4c–f). Here they adopted an adaptive classification method based on the Gaussian process for classification. The study conducted by Ralbovsky et al. introduced a statistical algorithm design to differentiate between two groups of rats: those subjected to a standard diet and those exposed to a high-fat diet leading to the pre-AD state.130 The algorithm was constructed and trained using a calibration dataset, and its diagnostic capabilities were assessed through external validation with new, unseen data. The application of partial least squares discriminant analysis achieved an 89% sensitivity and specificity at the donor level during cross-validation. External validation further confirmed the accuracy of the algorithm, achieving a 100% rate of correctly predicting the class of a donor. Notably, genetic algorithm analysis tentatively identified proteins and lipids as influential factors in discriminating between the two classes of blood serum donors.
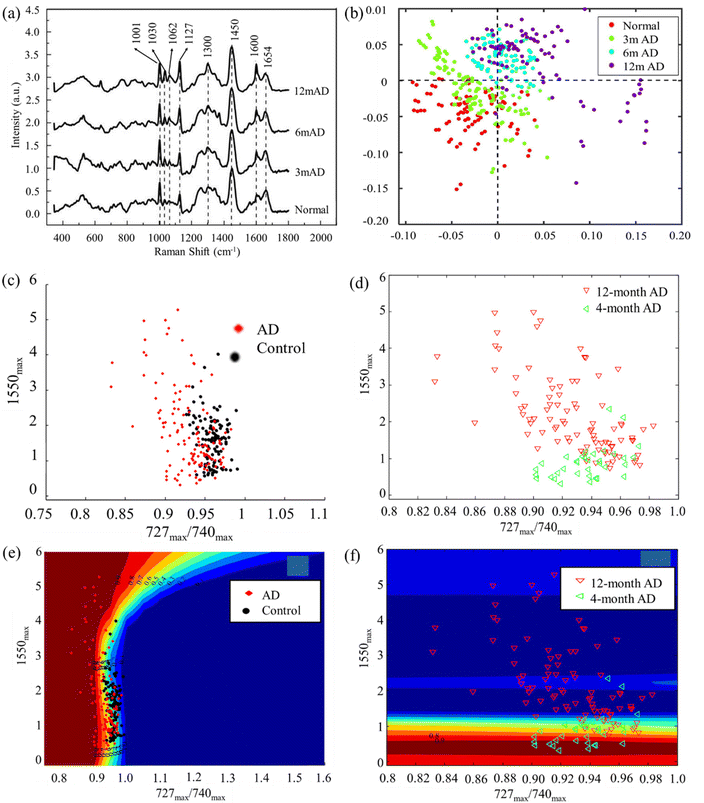 | ||
| Fig. 4 (a) Raman spectra of platelets at different stages of AD from different 3xTg-AD transgenic rats. (b) The discriminant scores plot result of partial least square discriminant analysis (PLS-DA) algorithm related to 3xTg-AD transgenic rats. Adapted with permission from ref. 128. Copyright 2022 Elsevier. (c and e) The visual display of first step and (d and f) second step of Gaussian process (GP) classification of spectral data from 4 month AD and 12 month AD platelets, and the control data based on two features. Adapted with permission from ref. 129. Copyright 2014 IOP Science. | ||
Carmona et al. investigated diagnostic potential of RS for prion disease.131,132 This was done through Raman analysis of prion protein in blood cell membranes from naturally affected scrapie sheep. Membrane fractions obtained from the blood of 150 healthy sheep and 31 sheep infected with scrapie were examined to detect the presence of β-sheet structure in the amide I region (1670 cm−1), indicating the conversion of cellular prion protein (PrPC) to proteinase-resistant prion protein (PrPSC). The diagnosis of scrapie-infected sheep achieved a 100% accuracy rate, as confirmed by postmortem analysis. Moreover, this research showcased the potential of RS in monitoring disease progression, as evidenced by continuous increases in β-sheet intensity observed one month after the onset of sickness. Alvarez-Puebla et al. conducted a study on the rapid direct detection of prions in serum and blood utilizing the SERS effect of gold nanorods.133 They developed a SERS substrate based on supercrystals of gold nanorods and performed scrambled prion detection. The primary chemical structure of both PrPC and PrPSC prions is very similar, and their respective vibrational patterns were detected. Bands located at 762 cm−1, assigned to the interaction of Au–S–C, and a triplet in PrPC (1390, 1416, and 1446 cm−1) which becomes a singlet (1448 cm−1) in the scrambled version, were observed. The SERS spectrum of PrPSC:PrPC in serum showed dominant bands corresponding to the C–N stretching (1118 cm−1), phenylalanine (1003 and 1033 cm−1), tryptophan (1011 and 1560 cm−1), tyrosine (845 cm−1), and cystine (720 cm−1). Even upon sequential dilution of the prion mixture (1% of PrPSC in 99% of PrPC) in serum, the characteristic prion bands remained clearly recognizable down to concentrations as low as 10−10 M.
Rickard et al. presented a novel sensing strategy by integrating SERS with an optofluidic device for the rapid and label-free detection of biomarkers associated with TBI in biofluids, achieving a picomolar limit of detection (LOD).134 The authors fabricated a SERS-active substrate using electrohydrodynamically produced submicrometer pillars, which were then incorporated into an optofluidic chip. A plasmon-active nanometric gold layer was applied to coat the pillars, creating the SERS-active platform. Utilizing this device, the researchers successfully detected N-acetylaspartate, a potential biomarker released from the central nervous system post-TBI, directly from finger-prick blood samples (Fig. 5a–d). This study demonstrates the suitability of the proposed strategy for constructing SERS devices, offering the potential for on-site and real-time target detection. Notably, the authors engineered a miniaturized Raman system for multiplexed and high-throughput analysis of biomarkers. Harris and colleagues employed RS-based detection to characterize a panel of 18 TBI-indicative biomarkers, encompassing both raw (human, animal, and synthetically derived) samples and their aqueous solutions.135 Colorimetric paper lateral flow strips (PLFS) could have drawbacks their low sensitivity and susceptibility to interference from complex sample matrices like blood. To address these issues, a new PLFS has been developed, incorporating SERS for signal transduction.136 Here gold nano-pyramid array chip was integrated into the detection zone of the PLFS for SERS enhancement. The design includes a hierarchical three-dimensional nanostructure creating “hot spots” to amplify SERS signals, resulting in high sensitivity. This PLFS demonstrates a low LOD of 5.0 pg mL−1 for the TBI biomarker S-100β in blood plasma (Fig. 5e and f). It has been successfully applied for rapid S-100β measurement in clinical samples from TBI patients in emergency departments. The availability of this PLFS for blood testing is transformative in TBI patient management, and its adaptability suggests potential for rapid detection of various human diseases by measuring low levels of protein blood biomarkers in complex human fluids.
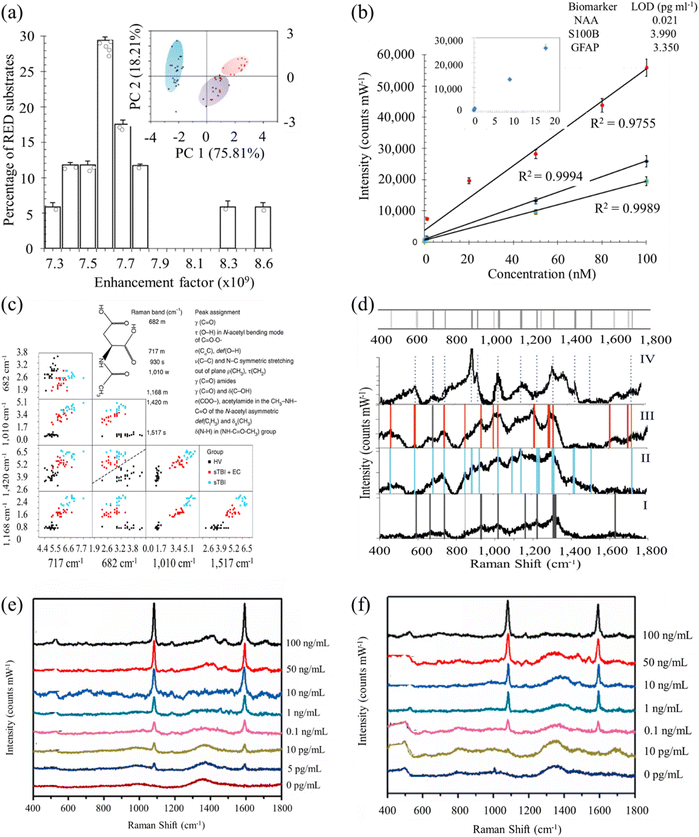 | ||
| Fig. 5 (a) Histogram of the measured electromagnetic enhancement factors of the SERS substrate. Inset: principal component score plots of PC1 and PC2 show the relationship between the multiplex spectra of the three single biomarkers. The blue cluster is N-acetylasparate (NAA) spectra (n = 23), the purple cluster represents S100B (n = 18) and the red is glial-fibrillary acidic protein (n = 13). (b) Calibration curves of SERS spectra acquired with an excitation laser of 785 nm. Inset: representative NAA levels as a function of SERS intensity for the dilution series and the calculated LOD values for each biomarker (inset table). (c) Classification matrices of the feature selection of subset of relevant features, used to establish the important peaks and their correlations reveals decision boundaries of multilayer perception with distribution of the selected peaks with clear separation at each subset between the STBI and the healthy volunteer patients. Inset: the NAA molecular structure and the major assignments of major SERS peaks of NAA on RED substrate. σ, stretching vibration; δ, bending vibration; δs, symmetric bending vibration; ρ, rocking, in-plane bending; γ, wagging; ν, breathing; τ, twisting. Raman intensity: s, strong; m, medium; w, weak. (d) Average SERS spectrum (n = 5) of healthy volunteers (i; bottom panel) excited at 785 nm are compared to the SERS spectrum of STBI only (ii), STBI + EC (iii) and to the fingerprint spectrum of NAA (iv; top panel) with the representative significant peaks highlighted with vertical grey (i), blue (ii), red (iii) and dotted (iv) lines, accordingly, highlighting the correspondence or the absence of the NAA peaks with some vibrational frequencies of the bands being unchanged in SERS spectra whereas several are red-shifted or not evident in the healthy volunteer spectrum. Inset: barcode derived from SERS spectra shown in e for severe traumatic brain injury (STBI) diagnostics. Adapted with permission from ref. 134. Copyright 2020 Nature Publishing Group. (e) SERS spectra of PLFS acquired from the buffer solution containing various concentrations of S-100β. (f) SERS spectra of PLFS taken at various concentrations of S-100β. Adapted with permission from ref. 136. Copyright 2021 Elsevier. | ||
Serum and plasma from clinical samples have been employed in distinguishing NDs from healthy controls. Carmona et al. utilized plasma Raman spectral data to categorize cases of mild, moderate, and severe AD.137,138 Their findings indicate an elevated ratio between spectral bands at 758 and 744 cm−1 in the blood plasma of patients with both mild and severe AD. Specifically, the 758 cm−1 band corresponds to the tryptophan side chain indole-ring breathing mode, while the 744 cm−1 band likely corresponds to mitochondrial cytochrome c from platelets, demonstrating a potential biomarker for disease severity. More recent research has revealed that the blood concentration levels of p-tau at threonine 181 (p-tau181) could aid in the diagnosis and differentiation of AD from other NDs.139 Habartová et al. reported a protocol focus on blood-based analyses for AD using chiroptical spectroscopy, including Raman optical activity (ROA) and electronic circular dichroism.140 This approach was supplemented with conventional vibrational spectroscopy (infrared, Raman) and metabolomics (high-performance liquid chromatography with high-resolution mass detection). The combination of these techniques allows for the identification of spectral patterns associated with AD and variations in metabolite levels (Fig. 6a and b). Through linear discriminant analysis, the spectral data can differentiate between AD patients and control subjects. For instance, average Raman spectra of the blood plasma of patients with AD showed notably lower intensities of the carotenoid spectral band. This was attributed to their utilization for protection against oxidative stress and disruptions in lipid metabolism. No differences were observed for the amide I band but intensity decrease was detected for amide III region which is due to AD-induced structural change in proteins/peptides. The less invasive nature of this approach highlighted its strong potential for identifying disease-related changes in essential plasmatic biomolecules and metabolites. Recently, it has been revealed that hollow gold nanospheres in conjunction with the redox molecules 4-mercaptobenzoic acid and Nile blue A, exhibit the potential to enhance the speed and accuracy of visually detecting neuron-specific enolase (NSE) and S100-β protein, both associated with brain damage, using the SERS technique.141 This study determined the lowest concentration for NSE and S100-β protein, with a linear range spanning from 0.2 to 22 ng mL−1, to be 0.1 and 0.06 ng mL−1, respectively. In a different report, Davies and coworkers presented utility of RS as a neuromonitoring tool in TBI.142 In this work, the researchers presented clinical potential of RS in diagnosis of brain disorders (Fig. 6c).
 | ||
| Fig. 6 (a) Average Raman spectra of the blood plasma of patients with AD (red; n = 35) and without AD (black; n = 29). (b) Average Raman optical activity spectra of the blood plasma of patients with AD (red; n = 35) and control group (black; n = 29). Adapted with permission from ref. 140. Copyright 2019 Elsevier. (c) Illustration of a simplified RS system and its potential application in clinical environments. Adapted with permission from ref. 142. Copyright 2022 MDPI. (d) FDTD simulations at different settings showing the electromagnetic field distributions of the energized nanoarray on glass (i) and Au substrate (ii–iv). The distance between the two nanorod cylinders was 150 nm (i–iii) and 50 nm (iv). (e) SERS spectra of AD plasma samples before and after isolation treatment. (f) Difference spectra obtained from subtracting the AD sample spectra from the healthy control sample spectra. Adapted with permission from ref. 143. Copyright 2021 Elsevier. | ||
Hao et al. proposed a diagnostic system that incorporates acoustofluidic principles and employed a multimodal approach for rapid detection of AD biomarkers from human plasma.143 In this work, a surface acoustic wave-based separation device has been designed to enhance signal-to-noise ratio (SNR) by isolating and purifying AD biomarkers. By utilizing ZnO nanorods and Ag nanoparticles, the developed system enabled label-free detections through SERS and electrochemical immunosensors (Fig. 6d). Raman spectral analysis identified key biochemical differences in human blood plasma for healthy individuals and AD group (Fig. 6e and f). AD patients were characterized by higher concentration of xanthine, uric acid, and ascorbic acid based on difference Raman spectrum. Xanthine and uric acid primarily serve as purine metabolites within the salvage pathway in the brain. The system demonstrates high sensitivity and specificity in label-free detections of clinical plasma samples from both AD patients and healthy controls. The efficient integration of these techniques presents promising solutions for the early diagnosis of AD.
In another work, a device utilizing Au nanostar@Raman reporter@silica sandwich nanoparticles as SERS probes, was developed ensuring high sensitivity for the detection of NSE, a TBI protein biomarker.144 The device achieved a LOD of 0.86 ng mL−1 in diluted blood plasma samples. Due to the merits of SERS this device exhibits superior sensitivity and a lower LOD in blood plasma-containing samples. Furthermore, the SERS-PLFS demonstrated successful NSE level measurement in clinical blood plasma samples from deidentified TBI patients. This study highlights the potential of SERS-PLFS for point-of-care screening of TBI patients. Yang et al. reported sandwich immunoassay using silver nanogap shells (AgNGSs) functionalized with Aβ antibody as SERS nanoprobes.145 These nanoprobes demonstrated sensitive, selective, and multiplexed detection of Aβ1–40 and Aβ1–42 peptides in blood. Through precise control of AgNGSs formation as plasmonic hot spots on silica nanoparticles at a one-nanometer resolution, the assay achieved a remarkable LOD of 0.25 pg mL−1 for Aβ1–40 and 0.33 pg mL−1 for Aβ1–42. Notably, these LODs were one order of magnitude lower than those of the ELISA test. Phung et al. presented SERS detection of dopamine levels in human blood plasms.146 By Ag-plated AuNPs deposited on ITO glass SERS substrate was developed and achieved lowest detection limit of ∼10−11 M for dopamine. NSE in blood plasma was detected using lateral flow glass-hemostix (FGH) in conjunction with Au nanocage as SERS substrates, achieving a LOD of 0.74 ng mL−1.147
In a work by Bedoni and coworkers, the analysis of serum using RS was enhanced by introducing nanostructures to induce a SERS signal, resulting in more detailed and intense Raman spectra.71 Standardization of all analyzed parameters was conducted to ensure the best repeatability and minimize variables. Following the methodological optimization, the established parameters were applied to analyze serum samples from 10 AD and 11 healthy control subjects. The obtained results, utilizing an innovative nanotechnology-based biosensor, were correlated with MRI findings in evaluating AD patients. This correlation provides a robust foundation for further exploration of the biosensor's applicability in monitoring AD progression and rehabilitation treatments. Overall, SERS technology contributes to the development of sensitive and noninvasive methods for detecting biomarkers with low concentrations. This advancement is conducive to early diagnosis and can potentially delay the progression of AD and other NDs.
Ryzhikova et al. employed an artificial neural network for classifying spectroscopic data, conducting learning and subsequent validation on subsets of the measured data.148 The neural network demonstrated a capability to distinguish between AD, other forms of dementia, and healthy controls with specificity and sensitivity exceeding 95%. However, the algorithm couldn't pinpoint the specific regions in the Raman spectra crucial for this differentiation. To address this, a genetic algorithm was employed, facilitating the identification of specific spectral regions deemed most significant for discriminating the measured spectra. Carotenoids, renowned for their antioxidant properties, are present in various biofluids and could potentially serve as a diagnostic tool for RS-based biopsies. Studies have shown that the levels of these carotenoids in biofluids, such as blood serum and plasma, may serve as biomarkers for NDs like AD and PD. Resonant Raman serves as the primary mechanism for amplification in the case of carotenoids, leading to the augmentation of three specific Raman bands positioned approximately at 1005, 1155, and 1520 cm−1. Kralova et al. conducted a comparative analysis of data derived from blood plasma samples collected from individuals with AD and healthy elderly controls.44 Four distinct techniques/experimental setups—RS with excitations at 532 and 785 nm, ROA, and SERS were employed for this purpose. The study revealed the impact of experimental design on the Raman spectra of blood plasma. Each of the four experimental setups resulted in distinctive spectral signatures. Conventional RS (excitations at 785 and 532 nm) and ROA primarily exhibited sensitivity to the protein fraction of blood plasma. However, due to resonance enhancement, Raman and ROA spectra obtained with 532 nm excitation also featured intense bands associated with carotenoids. ROA, utilizing circularly polarized light, produced spectral bands, especially in amide I and extended amide III, providing valuable information about the spatial structure of biomolecules. This suggests the potential of ROA to offer insights into disease-induced changes in the structure of blood plasma components. SERS spectroscopy, with its signal enhancement near plasmonic nanoparticles, particularly influenced low-molecular-weight metabolites. The SERS spectra of the low-molecular-weight blood plasma fraction and the whole blood plasma primarily exhibited bands related to uric acid, hypoxanthine, and ergothioneine. Overall, the study demonstrated that each RS technique provides distinct information about biomolecules in blood plasma or their conformation, offering diverse perspectives on the underlying biochemical processes associated with the disease.
Paraskevaidi and colleagues employed RS on blood plasma along with machine learning techniques to effectively distinguish AD from both healthy individuals and patients with dementia with Lewy bodies.149 Given the shared symptoms and clinical characteristics between these two diseases, the potential for misdiagnosis exists. The study involved the analysis of 56 samples categorized into four groups, including early and severe stages of AD, dementia with Lewy bodies, and healthy controls. Spectral data were processed using cross-validated PCA-LDA and a support vector machine algorithm. Six distinct statistical models were developed to compare all groups. Multiple binary algorithms were developed using support vector machines (SVM) to discern between healthy controls, early-stage AD donors, late-stage AD donors, and donors with dementia with Lewy bodies. The classification algorithms demonstrated an average sensitivity of 81.3% and specificity of 85.7%, underscoring the importance of pairing an optimal biological sample with an appropriate statistical analysis method for accurate analysis. Significant spectral bands crucial for discrimination were identified, such as amide I (∼1650 cm−1) and amide II (∼1530 cm−1). The latter exhibited an up-shift in the spectra of both early and severe stages of AD, attributed to an increase in tau protein or NFL in plasma. Additionally, a lower intensity band at ∼1432 cm−1 was observed, suggested to be due to decreased lipid levels resulting from oxidative stress-induced damage to phospholipid membranes. Moreover, the level of phenylalanine increased in dementia with Lewy bodies compared to healthy controls. In another study, Sharma et al. utilized RS to identify internal variances in erythrocytes of PD patients.150 α-Synuclein is closely associated with PD and other related conditions known as synucleinopathies. However, no discernible erythrocytic behavioral changes (eryptosis) or variations in hemoglobin were observed due to presence of α-synuclein. An increased level of plasmin–antiplasmin complexes was noted in the plasma of PD patients, suggesting activation of the fibrinolytic system. Huefner et al. employed RS to distinguish Huntington's disease (HD) patients from healthy controls using serum samples.151 Their study revealed notable alterations in the spectra associated with the progression of the disease. Additionally, the researchers identified differences corresponding to genotype and gender when analyzing serum samples from individuals with HD and those without the condition (Fig. 7a–f). The use of RS in this context showcases its potential as a valuable tool for probing molecular changes associated with HD and offers insights into potential biomarkers for diagnostic purposes. Schipper and colleagues employed a combination of RS and near-infrared spectroscopy (NIRS) to differentiate between blood samples of PD patients and a control group by analyzing various spectroscopic properties linked to oxidative stress.152 Patients with PD exhibited a reduction in bands associated with hydrocarbons (2990 cm−1), coupled with an increase in bands related to amines (3200 cm−1) and alcohols (3300 cm−1). These changes are likely indicative of oxidative stress. By utilizing these identified bands as biomarkers, the researchers established a discrimination model with 75% sensitivity and specificity.
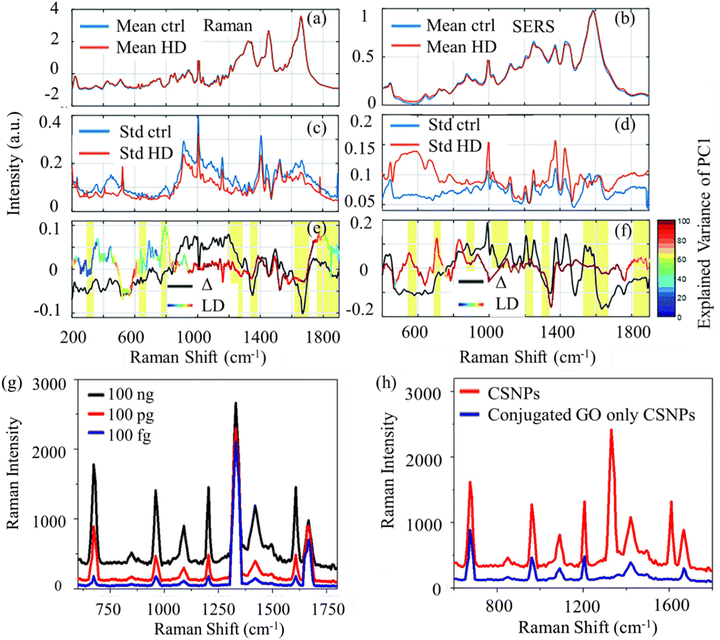 | ||
| Fig. 7 (a) Average Raman spectrum and (b) SERS spectra of serum from healthy control subjects (blue lines) and HD patients (red line) and standard deviations (c) and (d), respectively. (e) The different spectra of the averages for Raman spectrum (black line). (f) The different spectra of the averages for SERS (black line). LD loadings are shown by colored lines and yellow marked regions indicate important peaks. Adapted with permission from ref. 151. Copyright 2020 The Royal Society of Chemistry. (g) Concentration dependent SERS spectra from tau protein conjugated nanoplatform after magnetic separation of the nanocomposite. (h) SERS enhancement of Raman signal from 50 ng tau protein in the presence of CSNPs and from 500 ng tau protein in the presence of CSNPs attached graphene oxide hybrid. Adapted with permission from ref. 153. Copyright 2015 American Chemical Society. | ||
Two-dimensional (2D) materials have garnered significant attention for their unique properties and diverse applications, including their potential role in the field of biomarker sensing for NDs. These materials, such as graphene and transition metal dichalcogenides, offer high surface area, excellent conductivity, and biocompatibility. Their unique electronic and optical properties make them promising candidates for developing highly sensitive biosensors. Demeritte et al. discovered that the modification of graphene oxide with magnetic nanoparticles enables the early detection of AD by identifying Aβ42 peptide and total-tau (t-tau) proteins.153,154 In this study, researchers employed magnetic core and plasmonic shell nanoparticles, which were conjugated with graphene oxide (GO) and further modified with anti-Aβ42 antibodies and anti-tau antibodies. This innovative design serves as a robust 3D SERS platform specifically tailored for the detection of Aβ and tau in whole blood samples. The platform demonstrated the capability to detect target AD biomarkers at remarkably low concentrations, detecting Aβ42 peptide as low as 500 fg mL−1 and t-tau protein as low as 100 fg mL−1 showcasing its high sensitivity in detecting minute concentrations of the target biomarker in complex biological samples. This multifunctional nanoplatform was utilized to selectively collect more than 98% of AD biomarkers from whole blood samples. In their continued research, researchers expanded their approach by developing iron–gold core–shell nanoparticles (CSNPs) affixed to shell hybrid graphene oxide.153 This advancement aimed at identifying Aβ and tau from whole blood samples, resulting in remarkable detection limits of 100 fg mL−1 for Aβ and 0.15 ng mL−1 for tau (Fig. 7g and h). Notably, the nanoplatform exhibited the capability to differentiate between Aβ and tau biomarkers and human serum albumin, a highly abundant protein in the CSF. This underscores the specificity and versatility of the developed nanoplatform for effective biomarker discrimination in complex biological samples.
Furthermore, Yu and colleagues devised a SERS-based immunoassay employing specific antibodies tethered to Fe3O4@GOs and silver probes labeled with 4-mercaptobenzoic acid (4-MBA).74 The resulting SERS spectrum of 4-MBA signifies the capture of the target protein, showcasing a detection limit in clinical serum samples that can reach the femtomolar level. The established SERS-based immunoassay proficiently probed Aβ1–42 and phosphorylated-tau (p-tau)-181 in human serum samples, positioning it as a promising approach for the early detection of AD. This innovation opens up novel avenues for detecting clinical biomarkers with increased sensitivity. In the context of NDs, the use of 2D materials in biomarker sensing could revolutionize diagnostic approaches by providing rapid and accurate detection of specific biomolecules associated with conditions like AD or PD. The enhanced sensitivity and specificity offered by these materials hold great potential for advancing early disease diagnosis and monitoring. Collectively, these findings suggest that blood-based liquid biopsy investigations are a promising avenue for early diagnosis, disease prevention, and biomarker detection for a range of brain disorders.
Saliva-based investigations
Saliva is readily accessible and has been investigated for potential biomarkers associated with NDs. Due to its non-invasive collection method, minimal or no pre-processing requirements before analysis, and possessing a molecular composition similar to blood, this intricate biofluid has garnered growing scientific interest.155,156 Saliva has shown its success as a biofluid for disease diagnosis and biomarker sensing of different diseases.157,158 Saliva reflects the systemic changes occurring in the body. Saliva-based RS liquid biopsy investigations for brain disorders represent a burgeoning field with transformative potential in non-invasive diagnostics. By probing molecular signatures present in saliva samples, this approach offers a convenient and accessible means of detecting biomarkers associated with various brain disorders, including AD and PD. With advancements in RS techniques and data analysis algorithms, researchers can uncover subtle molecular changes indicative of disease pathology, enabling early detection and intervention. Saliva-based liquid biopsy holds promise for facilitating population-wide screening initiatives, enabling scalable and cost-effective disease monitoring strategies. Moreover, the non-invasive nature of saliva sampling enhances patient compliance and reduces procedural barriers, making it an attractive option for routine diagnostic assessments and longitudinal monitoring of brain disorders. As NDs often involve systemic alterations, analyzing saliva provides a holistic view of the overall health status and specific molecular changes associated with these conditions. Saliva can be collected repeatedly over time, allowing for longitudinal studies and real-time monitoring of disease progression. This is especially beneficial for assessing the dynamic nature of NDs. Moreover, saliva contains a diverse range of molecules, including proteins, nucleic acids, and metabolites. Changes in the composition of these molecules can serve as potential biomarkers for NDs, and RS can effectively analyze these alterations.Saliva contains detectable levels of t-tau, p-tau, Aβ, and α-synuclein proteins. Initial investigations indicate promising potential for the use of saliva in diagnostic applications.159 Several pilot studies have demonstrated that the presence of salivary Aβ42 is detectable and elevated in AD, while Aβ40 levels remain unchanged.160–163 RS also demonstrated its utility in the field.156,164 For instance, Carlomagno et al. investigated the efficacy of RS in distinguishing various NDs and gaining insights into their pathogenesis by analyzing saliva samples from 10 healthy controls, 19 patients with amyotrophic lateral sclerosis (ALS), 10 with PD, and 10 with AD.165 SERS combined with PCA-LDA proved successful in revealing significant differences between groups, particularly distinguishing ALS from healthy controls, where RS peaks corresponding to phosphatidylinositol, phospholipids, nucleic acids, glycogen, and glucose played a differentiating role (Fig. 8b and c). Their findings suggested the involvement of carbohydrate metabolism, protein aggregation and misfolding, damage to membrane lipids, and alterations in DNA/RNA in ALS. The authors also highlighted the significant role of membrane phospholipids in distinguishing between ALS, AD, and PD, where peaks associated with phosphatidylinositol (500 and 576 cm−1) exhibited higher intensity in ALS samples, indicating increased activity of the phosphatidylinositol 3-kinase enzyme (Fig. 8d and e). The Lednev group employed RS in conjunction with machine learning to diagnose early AD by identifying potential biomarkers.166 This project utilized Raman hyper-spectroscopy and focused on saliva samples obtained from both a normal individual and someone with AD and mild cognitive impairment. The results indicated that Raman hyper-spectroscopic analysis of saliva holds promise as an effective diagnostic method for early-stage AD.
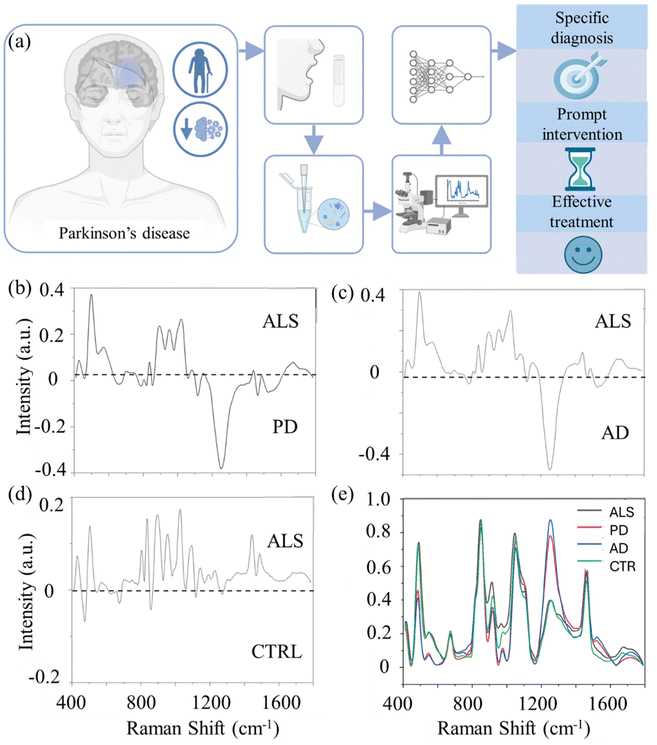 | ||
| Fig. 8 (a) Schematic representation of saliva preparation RS measurements, and machine learning data analysis. Adapted with permission from ref. 158. Copyright 2021 Frontiers. (b) PD average signal, (c) AD average signal and (d) CTRL average signal. (e) Overlapped average spectra of the experimental groups. Adapted with permission from ref. 165. Copyright 2020 Springer Nature. | ||
Raman analysis was employed to capture the comprehensive signal from the saliva of 23 PD patients, along with relevant pathological and healthy control subjects. Utilizing both machine and deep learning approaches, the acquired spectra were processed. Leveraging a Raman database, a classification model was developed, demonstrating the capability to accurately distinguish each spectrum into the correct group with accuracy, specificity, and sensitivity exceeding 97% for individual spectrum attribution. Additionally, each patient was correctly assigned with a discriminatory power exceeding 90% representing innovative non-invasive procedures for early detection of the diseases, with potential use in the future clinical applications. Furthermore, the extracted data exhibited significant correlations with clinical data currently employed for PD diagnosis and monitoring. Altuntas and Buyukserin examined artificial saliva containing various candidate biomarkers for AD.167 They integrated a SERS substrate for the detection of Aβ in artificial saliva, successfully achieving a LOD of 0.5 pg mL−1 for Aβ. Initially, polymeric films featuring surfaces with multibranched nanopillars (MNS) were crafted through the drop-casting of polycarbonate solutions onto anodized aluminum oxide molds, characterized by hierarchically branched pores. Following the extraction from the nanoporous molds, a 20 nm gold coating was applied to enable these MNS substrates to detect sub-picomolar concentrations of thioflavin-T, a SERS-active dye commonly employed in clinical settings for diagnosing the presence of amyloid plaques. Incorporating RS into the analysis of saliva adds a layer of specificity and sensitivity, enabling the identification of molecular changes associated with NDs. This non-invasive and patient-friendly approach holds great promise for advancing early diagnosis, understanding disease mechanisms, and monitoring treatment responses. Collectively, these findings suggest that saliva-based liquid biopsy investigations are a promising avenue for early diagnosis, disease prevention, and biomarker detection for a range of brain disorders.
Tear-based investigations
Human tears are a valuable reservoir of information reflecting the health status of the eyes and overall bodily functions. The richness of tear composition arises from the diverse array of salts and organic components present, encompassing proteins, lipids, metabolites, nucleic acids, and electrolytes.168–172 Changes in the concentrations of these constituents can serve as indicators of various pathologies, eye-related disorders, and inflammatory processes. Tear-based RS liquid biopsy investigations for brain disorders represent a novel and non-invasive approach to detecting biomarkers associated with neurological conditions. By analyzing molecular signatures present in tears, this method offers a convenient and accessible means of early detection and monitoring of brain disorders such as AD and PD. Tear-based sampling is relatively simple and can be performed without specialized equipment, making it suitable for widespread screening and remote monitoring. Tears have gained attention for their potential role in reflecting changes related to NDs. For instance, AD patients exhibit a noteworthy increase in tear production and tear protein concentrations compared to healthy individuals.173 Alterations in tear composition are also evident in AD patients, characterized by an elevated concentration of dermcidin and reduced levels of lysozyme-C, lactotransferrin, prolactin, lipocalin-1, extracellular glycoprotein lacritin, among others.174 Moreover, there is a significant rise in t-tau and Aβ42 levels in the tears of AD patients compared to controls.175 Another study highlights elevated levels of microRNA-200b-5p in the tear fluids of AD patients relative to controls.174 A noticeable reduction in tear volume is well-correlated with the progression and severity of PD. This decline in tear secretion in PD individuals is likely attributed to autonomic dysfunction.176,177 Additionally, elevated levels of soluble α-synuclein are observed in tear fluids of PD patients when compared to age-matched healthier individuals.178–180 Furthermore, PD subjects exhibit higher levels of TNF-α in tears than healthy subjects, suggesting TNF-α as a potential diagnostic biomarker for PD.181Some studies have explored proteins and other molecules in tears as potential biomarkers for conditions like AD with the assistance of RS. In a study by Ami et al., the researchers employed a combination of FTIR and RS to analyze tear samples, which contain proteins associated with ALS, from both healthy individuals and ALS patients.182 Utilizing a variety of machine learning methods, including multivariate analysis, PLS-DA, neural networks, and extreme gradient boosting, the authors successfully characterized the Raman spectra of tears from ALS patients with a specificity and sensitivity of 100% (Fig. 9a–d). The investigation revealed that phenylalanine bands exhibited significantly lower intensity in ALS patient samples compared to the healthy cohort, suggesting a rewiring of amino acid metabolism in ALS (Fig. 9c). Peaks associated with protein β-sheet structures at ∼1670 cm−1 and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of lipids at 1770 cm−1 had higher intensity in ALS patients, indicating potential alterations in protein conformation linked to the evidence of the role of protein misfolding and aggregation in NDs.
O stretching of lipids at 1770 cm−1 had higher intensity in ALS patients, indicating potential alterations in protein conformation linked to the evidence of the role of protein misfolding and aggregation in NDs.
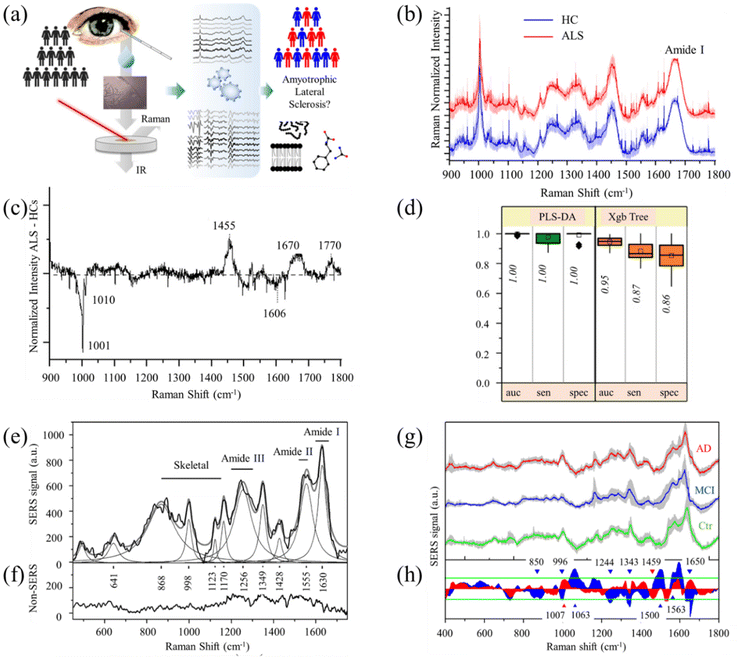 | ||
| Fig. 9 (a) Schematic representation of tear sample collection from ALS patients for Raman data acquisition and analysis. (b) Overall performances of PLS-DA and xgbTree methods in the 900–1800 cm−1 spectral range. (c) Comparison of the mean Raman spectra obtained by considering all the measured tears from ALS patients and healthy controls. The shadowed area refers to the standard deviation of the data. (d) Spectrally resolved differential average Raman spectra of the two investigated groups. Adapted with permission from ref. 182. Copyright 2021 American Chemical Society. (e) SERS spectrum of tears from a healthy subject. (f) Spectrum of tear obtained by conventional RS in similar acquisition conditions. (g) Averaged SERS spectra of tear samples from healthy subjects (Ctr-green line), mild cognitive disease-affected subjects (MCI-blue line), and AD-affected subjects (AD-red line). The gray areas represent the standard deviation of the signal intensities within the considered data. (h) Signal differences concerning the control data of AD (red area) and MCI (blue area) spectrum. The green lines indicate the signal dispersion range (0.68 of the standard deviation). The statistically significant signal differences (p-value <0.05 in the one-way ANOVA statistics) are indicated by blue (MCI) and red (AD) marks, respectively. Adapted with permission from ref. 72. Copyright 2020 SPIE. | ||
Cennamo et al. introduced a diagnostic approach utilizing SERS of tears for the detection of NDs, encompassing various forms of dementia and AD.72 The study enlisted 18 AD patients, 8 individuals with mild cognitive impairment, and 6 control volunteers. SERS measurements of tear fluids from individuals with AD and healthy subjects revealed changes in the acquired spectra, indicating conformational alterations in tear proteins. Spectral variations were observed in classes associated with lactoferrin and lysozyme protein components (Fig. 9e–h). Additionally, the quantitative assessment of changes related to pathological conditions was achieved by identifying the I1342/I1243 ratio, attributed to the C–H deformation and amide III β-sheet. Overall, these findings suggest that delving into tear-based liquid biopsy techniques shows potential for early diagnosis, disease prevention, and biomarker detection related to a variety of brain disorders.
Cerebrospinal fluid (CSF)-based investigations
CSF-based RS liquid biopsy investigations for brain disorders represent a promising avenue for minimally-invasive diagnosis and monitoring of neurological conditions. By analyzing molecular signatures present in the CSF, this approach offers insights into the biochemical changes associated with brain disorders such as AD and PD. CSF is in direct contact with the central nervous system and is considered a rich source of biomarkers for NDs. CSF sampling allows for direct access to the central nervous system and provides a rich source of biomarkers reflective of brain health and pathology. RS enables the identification and quantification of specific molecules in the CSF, facilitating early detection, disease monitoring, and personalized treatment strategies. Commonly studied CSF biomarkers include tau, Aβ, and p-tau.183 Yu and colleagues developed a SERS biosensing platform for the simultaneous detection of Aβ1–42 oligomers and tau protein.184 This platform utilized different Raman dye-coded polyA aptamer-AuNPs (PAapt-AuNPs) conjugates. The conjugation of PAapt-AuNPs occurs through the self-assembly of polyA block nucleotides and nonfluorescent Raman dyes on the surface of AuNPs. Facilitating attachment to the AuNPs surface, the polyA block plays a crucial role, while protein recognition is executed by polyA block nucleotides that consist of oligonucleotides. Upon introducing a protein biomarker into the reaction system, the polyA block nucleotide separates from the AuNP surface. This separation is facilitated by the hybridization of the target protein with its specific aptamer. Consequently, aggregates of AuNPs form, creating enhanced electromagnetic hotspots within the gaps between nanoparticles and yielding a significantly stronger SERS signal. The researchers achieved LOD of 4.2 × 10−4 pM for tau protein and LOD of 3.7 × 10−2 nM Aβ1–42 oligomer (Fig. 10c and d). The effectiveness of this strategy was demonstrated by successfully detecting tau protein and Aβ1–42 oligomers in artificial CSF samples, yielding satisfactory results. Lhiyani et al. introduced a RS method employing machine learning for the in vivo detection of Aβ and tau in human CSF.185 Their findings highlight the significance of the proteins’ concentration in the CSF as indicative of AD condition.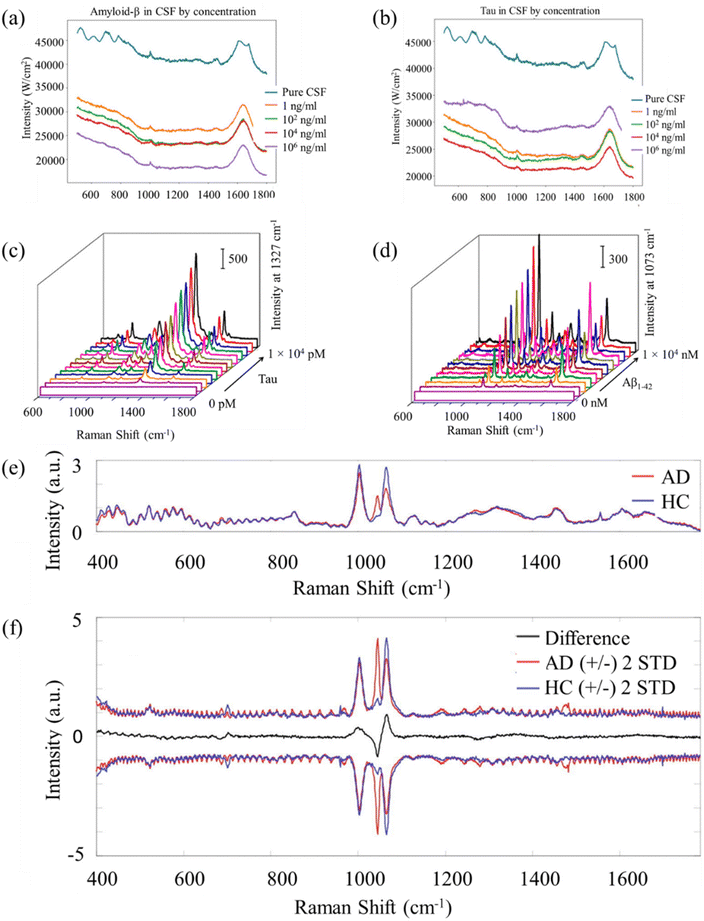 | ||
| Fig. 10 (a) Raman signature for samples of pure CSF and CSF with different volumes of Aβ scheme. (b) Raman signature for samples of pure CSF and CSF with different volumes of tau scheme. Adapted with permission from ref. 185. Copyright 2023 Optica Publishing Group. (c) SERS signals in responses to tau protein of varying concentrations. (d) SERS signals in response to Aβ1–42 oligomers of varying concentrations. Adapted with permission from ref. 184. Copyright 2019 American Chemical Society. (e) Mean Raman spectra of CSF from AD (red line) and healthy control (blue line) cohorts. (f) Difference spectrum (black line) and spectral variations around the mean (±2 standard deviations). Adapted with permission from ref. 187. Copyright 2020 Elsevier. | ||
Chou and colleagues introduced a nanofluidic biosensor utilizing SERS for the detection of conformational states of the Aβ peptide.186 The biosensor comprises a fluidic channel with a specific design, featuring a shallow region in the middle that induces capillary flow within the microchannel. This capillary flow is instrumental in creating SERS active sites by trapping large gold nanoparticles (AuNPs) at the entrance to the nanochannel. Simultaneously, the capillary flow enhances the concentration of target molecules, including Aβ, by transporting them through the interstices of the nanoparticle clusters. The device demonstrated effectiveness in distinguishing Aβ from confounding proteins commonly present in CSF. Ryzhikova and team achieved AD diagnoses by analyzing CSF using RS, attaining 84% sensitivity and specificity.187 In this study, the researchers presented a novel diagnostic approach for AD utilizing CSF through near-infrared (NIR) RS combined with machine learning analysis (Fig. 10e and f). NIR Raman spectra were obtained from CSF samples collected from 21 patients diagnosed with AD and 16 healthy control (HC) subjects. Artificial neural networks and support vector machine discriminant analysis (SVM-DA) were employed for differentiation, resulting in classification models demonstrating a high discriminative power. This suggests the method holds significant potential for effective AD diagnostics. Taken together, these results indicate that exploring CSF-based liquid biopsy techniques holds promise for early diagnosis, preventing diseases, and detecting biomarkers associated with various brain disorders.
In addition, extracellular vesicles also emerged as a liquid biopsy for neurological disorders expanding the utility of liquid biopsies.188 We would like to note that researchers have utilized diseased cells or tissues in order to diagnose different NDs.23,189 For instance, in HD, RS has previously been used to study fibroblast from patients.190–192 Wang et al. introduced graphene-assisted RS for rapid biomarker sensing of AD.23 Applying a single layer of graphene to brain slices significantly enhanced the SNR in Raman measurements taken from brain tissue. This improvement led to an increase in the accuracy of classification from 77% to 98%. In-depth discussion of these methods is beyond the scope of this article.
León-Bejarano and colleagues utilized SERS to analyze α-synuclein levels in skin biopsies from PD patients.189 Their study revealed distinct alterations in the Raman bands of the protein, with shifts observed from 1655, 1664, and 1680 cm−1 to 1650, 1670, and 1687 cm−1 when comparing control subjects to those with PD. These spectral changes were attributed to protein aggregation, suggesting the potential of SERS in detecting aggregated α-synuclein in skin samples. This non-invasive approach offers a promising avenue for disease detection, minimizing the need for invasive diagnostic procedures. This expansion of the types of biopsies for Raman investigation in the context of brain disorders represents a significant advancement in diagnostic techniques. This expansion opens up new possibilities for early diagnosis, disease monitoring, and understanding the underlying molecular mechanisms of brain disorders. Additionally, it offers the potential for personalized medicine approaches tailored to individual patients based on their specific biomarker profiles. Overall, the diversification of biopsy sources for Raman investigation holds great promise for improving the management and treatment of brain disorders.
Table 1 summarizes the most important Raman signatures and biomarkers to identify different brain disorders using liquid biopsy. The primary constraints associated with all the mentioned molecules and biomarkers stem from their overlap with biomarkers found in other comparable conditions (such as PD and AD) and the methodologies utilized for their characterization. One of the key challenges in the field of biomarkers is the issue of specificity. Many biomolecules and biomarkers can be present in more than one condition, making it difficult to differentiate between them. This overlap can lead to difficulties in accurately identifying and characterizing biomarkers for specific conditions, such as PD and AD. The intricate similarities in molecular signatures across these NDs necessitate precise characterization techniques to distinguish specific biomarkers unique to each condition. In addressing these constraints, ongoing research and technological advancements aim to enhance the specificity of biomarkers, develop more refined characterization methodologies, and explore innovative approaches such as multi-modal biomarker strategies to improve accuracy and reliability in disease diagnosis and monitoring. Overall, the challenges associated with biomolecules and biomarkers underscore the need for continued innovation and collaboration within the scientific community to overcome these constraints and advance the field of biomarker research for improved disease detection, treatment, and management.
| Brain disorder | Most important Raman signatures, biomolecules, and biomarkers | |||
|---|---|---|---|---|
| Blood | Tear | Saliva | CSF | |
| AD | Aβ143,145,184 | Urea182 | Aβ166,167 | Aβ140,187 |
| Tau143,184 | Phenylalanine182 | Tau166 | Tau140,187 | |
| Exosomes143 | Amide I182 | Lactoferrin159 | Phenylalanine186 | |
| Carotenoids140 | α-Helices182 | AChE159 | Tyrosine186 | |
| Tryptophan140 | β-Sheet182 | α-Synuclein159 | Histidine186,187 | |
| Alanine140 | Lipids182 | Acetylcholinesterase159 | Amide III (β-sheet)186 | |
| Tyrosine140 | Dermcidin172 | Amide III (α-helix)186 | ||
| Phenylalanine140 | Lysozyme-C lactotransferrin172 | Glycine187 | ||
| β-Components140 | Prolactin172 | Proline187 | ||
| Intensity ratio of indole and tryptophan137 | Lipocalin-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 172 |
Arginine187 | ||
| Histidine153 | Extracellular glycoprotein lacritin172 | Valine187 | ||
| Proteinic side chain structure127 | Tau172 | |||
| Amide I127 | Aβ172 | |||
| Intensity ratio of 1127 to 1001 cm−1 (protein, lipids to phenylalanine)128 | microRNA-200b-5p172 | |||
| Intensity ratio of 1654 to 1600 cm−1 (ref. 128) (α-helix structure to amyloid precursor protein) | Tryptophan72 | |||
| Ascorbic acid71 | Tyrosine72 | |||
| Hypoxantine71 | Lactoferrin72 | |||
| Uric acid71 | Lysozyme72 | |||
| Lipocalin72 | ||||
| Albumin72 | ||||
| PD | α-Synuclein150 | CCL-2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 172 |
α-Synuclein150 | α-Synuclein150 |
| Plasmin-antiplasmin150 | DJ-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 172 |
Phosphatidylinositol158 | Autosomal enzymes158 | |
| Extracellular vesicles193 | Proteins from S100 superfamily172 | Ester of cholesterol158 | Amyloid species158 | |
| Autosomal enzymes158 | Peroxiredoxin-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 172 |
Tryptophan158 | Micro-RNA158 | |
| Amide I193 | Annexin-X5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 172 |
Cytosine158 | Cytokines expression patterns158 | |
| Lipids193 | Glutathione-stransferase-A1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 172 |
Guanine158 | ||
| Carbohydrates193 | Apolipoprotein superfamily-ApoD172 | Phenylalanine158 | ||
| Porphyrin193 | ApoA4 and ApoA1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 172 |
Phospholipids158 | ||
| Pyrimidine193 | TNF-α172 | Glucose/glycogen158 | ||
| Carotenoids193 | α-Synuclein172 | Lipids158 | ||
| Saccharide and disaccharide193 | Amide I158 | |||
| Dopamine146 | Tyrosine158 | |||
| Nucleic acids158 | ||||
| Amide III158 | ||||
| Mucins158 | ||||
| HD | Cholesterol190 | Mutant huntingtin protein194 | Huntingtin protein195 | Neurogranin and TREM2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 196 196 |
| β-Sheet190 | Amylase195 | T-tau197 | ||
| Phospholipids190 | Cortisol195 | mHTT197 | ||
| Phenylalanine190 | C-reactive protein195 | Neurofilament light chain (NfL)198 | ||
| Proteins190 | Uric acid195 | |||
| DNA190 | ||||
| Mutant huntingtin protein151 | ||||
| Neurofilament light chain151 | ||||
| Proline151 | ||||
| β-Sheet protein151 | ||||
| Uric acid151 | ||||
| Adenine151 | ||||
| Amide III151 | ||||
| Amide I151 | ||||
| TBI | S-100β134,136,141,147 | GFAP199 | Micro-RNA200 | S-100β136,141,147 |
| NSE136,141,144,147 | Tau199 | Salivary extracellular vesicles200 | NSE136,141,144,147 | |
| Myelin basic protein (MBP)136 | NfL199 | NAA134 | ||
| Glial fibrillary acidic protein (GFAP)134,136,147 | MBP134 | |||
| NAA134 | ||||
| Cleaved tau134 | ||||
| T-tau134 | ||||
| P-tau134 | ||||
| Ubiquitin C-terminal hydrolase134 | ||||
Aβ1–42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 134 134 |
||||
| αII-Spectrin breakdown products134 | ||||
| Gamma-enolase134 | ||||
Selectivity and sensitivity of Raman spectroscopy with other analytical techniques routinely employed in biomedical applications
RS offers distinct advantages in terms of selectivity and sensitivity compared to other analytical techniques commonly used in biomedical applications. One notable advantage is its high molecular specificity, allowing for the identification and characterization of individual chemical bonds and functional groups within complex biological samples.201–203 This molecular specificity arises from the unique fingerprint-like Raman spectra produced by different biomolecules, enabling the differentiation of closely related species and the detection of subtle structural variations. In contrast, techniques such as infrared spectroscopy may struggle with spectral overlap and lack the ability to distinguish between similar molecular species. Furthermore, RS is inherently non-destructive and non-invasive, making it particularly well-suited for analyzing delicate biological samples without the need for extensive sample preparation or labeling.28,204 This preserves the integrity of the sample and allows for repeated measurements over time, facilitating longitudinal studies and minimizing experimental artifacts. In contrast, techniques such as mass spectrometry often require sample ionization or derivatization, which can alter the native state of biomolecules and introduce artifacts into the analysis.In terms of sensitivity, RS offers advantages over traditional fluorescence spectroscopy, especially in complex biological matrices with high autofluorescence background.201,205 Combining RS with techniques like SERS, SRS, and CARS can significantly enhance the sensitivity and provide advantages over traditional techniques like fluorescence spectroscopy.206 The extraordinary signal enhancement allows the detection of extremely low concentrations of analytes, down to single-molecule levels. Raman signals arise from inelastic scattering processes and are therefore independent of excitation wavelength, allowing for excitation at longer wavelengths where autofluorescence is minimized. This enables the detection of low concentrations of analytes within highly autofluorescent samples, enhancing sensitivity and enabling the analysis of a wide range of biomolecules. Overall, Raman spectroscopy's combination of molecular specificity, non-destructiveness, and sensitivity makes it a valuable tool for a wide range of biomedical applications, including disease diagnosis, drug discovery, and tissue engineering. While other analytical techniques may offer complementary advantages in certain scenarios, RS remains a powerful and versatile tool for understanding the molecular basis of health and disease.
CARS, TERS, and SRRM spectroscopy have been explored in various biomedical applications, including the detection and diagnosis of NDs. CARS has shown promise in distinguishing Aβ, a key biomarker for AD, through label-free vibrational imaging.207,208 Cunha et al. applied a multimodal imaging approach, including spontaneous Raman, CARS, SRS, and Second harmonic generation, to examine the core and halo of Aβ plaques in the hippocampus and cortex of brain tissues from an AD mouse model.207 Additionally, Tabatabaei et al. conducted TERS on neuronal spines exposed to Aβ treatment, revealing insights into Aβ accumulations at the surface of spines.65 Super-resolution vibrational imaging using expansion SRS microscopy has been utilized to track nanoscale features of protein synthesis in protein aggregates using metabolic labeling of small metabolites.209 Aβ proteins forming the plaques show distinctive Raman spectral features. While these techniques have shown promise in imaging biological samples such as tissues with high spatial resolution and chemical specificity, their application to liquid biopsy for the diagnosis of brain disorders has been limited due to several factors. These include the low concentration of biomarkers in blood, the complexity of blood composition, and the need for highly sensitive detection methods.
Practical challenges associated with Raman techniques and transitioning into a quantitative technique
Raman spectroscopic techniques offer great potential for biomedical applications due to their ability to provide detailed molecular information and chemical fingerprints of biological samples. However, there are several practical challenges associated with these techniques that need to be addressed to enable their widespread adoption and transition into quantitative techniques in biomedical applications.Reproducibility challenges
Variations in sample preparation protocols, such as handling, drying, and substrate or nanoparticle immobilization, can introduce inconsistencies in acquired Raman or SERS spectra, impacting reproducibility. Furthermore, differences in instrumentation, including laser sources, optics, detectors, and calibration procedures, can contribute to spectral measurement variability. Environmental factors such as temperature, humidity, and ambient light conditions may also influence Raman or SERS signals, adding to the variability. To address reproducibility challenges, standardized sample preparation protocols, instrument calibration procedures, and stringent environmental controls are essential.107,210 Additionally, employing internal standards, spectral normalization techniques, and robust data analysis methods can help mitigate variations.Quantification challenges
The intensity of Raman or SERS signals can vary significantly due to factors like sample concentration, substrate or nanoparticle properties, and molecular orientations, posing challenges for quantitative analysis. Biological samples, such as biofluids or tissues, present complex matrices that can interfere with these signals, further complicating quantitative analysis. Moreover, biological samples are inherently complex, with numerous molecular species having overlapping Raman spectra, making it difficult to interpret signals and identify specific biomarkers or molecular signatures associated with disease states. Unlike other analytical techniques, there is a lack of well-established and widely accepted calibration standards for Raman or SERS techniques in biomedical applications. To address these quantification challenges, several approaches can be adopted. Firstly, robust calibration models can be developed using multivariate data analysis techniques such as PLS regression or artificial neural networks (ANNs) to accommodate signal variations and matrix effects. Additionally, employing internal standards or spiked samples with known concentrations can establish quantitative relationships between signal intensity and analyte concentration. Exploring advanced SERS substrates or nanoparticles with controlled and reproducible enhancement factors can improve signal reproducibility and eliminate quantification challenges.28,201 Lastly, combining Raman or SERS techniques with other analytical methods such as chromatography or mass spectrometry can provide complementary quantitative information, enhancing the overall analytical capabilities.Sample complexity and interference
Biological samples often exhibit intrinsic fluorescence, which can interfere with Raman signals, complicating data acquisition and interpretation. Moreover, biological samples are frequently heterogeneous, with varying chemical compositions and distributions, further complicating the interpretation of Raman spectra. Background signals from substrates, solvents, or other sample components can also affect Raman spectra, necessitating careful background subtraction and data processing. To overcome these challenges, various strategies can be employed. Advanced data processing techniques such as baseline correction, background subtraction, and multivariate curve resolution can be implemented to isolate the desired Raman signals from interfering signals.205,211 Exploring alternative excitation wavelengths or resonance Raman techniques to minimize autofluorescence interference is also crucial. Additionally, developing sample pretreatment protocols such as extraction, purification, or separation techniques can simplify complex biological matrices, reducing interference and improving the quality of Raman measurements.Time and spatial resolution
RS faces numerous challenges in clinical applications, particularly concerning time and spatial resolution. Time resolution limitations are notably evident in achieving acceptable SNR for practical use. Longer exposure times and optimized data acquisition strategies are necessary to maximize Raman signal and minimize noise for improved SNR. Spatial resolution challenges significantly affect the technique's ability to capture Raman spectra from deeper tissue layers accurately. Ensuring high spatial resolution requires precise sample focus and alignment of optics to optimize the Raman signal, critical for reliable clinical diagnostics and guidance. However, maintaining a balance between high spatial resolution and SNR is essential for obtaining dependable results. Conventional Raman microscopy techniques often suffer from limited spatial resolution, hampering the accurate characterization of cellular or subcellular features. To address this, advanced microscopy techniques such as confocal Raman microscopy, TERS, or SRS microscopy can be employed to achieve sub-micron spatial resolution.28,67,98,101,103 These methods utilize confocal optics, plasmonic enhancement, or nonlinear optical processes to enhance spatial resolution and enable detailed imaging of biological samples at the cellular or subcellular level.Technological advancements have been developed to tackle challenges related to time and spatial resolution in RS for clinical applications. These include enhanced spectral resolution, confocality, and the use of shorter wavelength lasers and high numerical aperture (NA) immersion objectives. Optimizing exposure time and utilizing multiple exposures are fundamental practices to improve Raman spectra quality. Additionally, advancements aim to enhance the anti-interference capacity of RS for faster data acquisition and analysis, facilitating timely medical treatment. Spatial resolution enhancements are achieved through high NA immersion objectives and confocal RS implementation. Furthermore, advancements in super-resolution imaging restoration techniques and structured illumination methods have contributed to improving spatial resolution, particularly for clinical applications.98–100,105 These advancements enable detailed spectral information retrieval related to specific molecular structures and substances, aiding in precise disease diagnosis such as cancer, infections, and neurodegenerative conditions. Integration of RS with other imaging modalities, like optical coherence tomography (OCT) or fluorescence microscopy, can provide complementary information and enhance spatial resolution in clinical settings.212–214 Combining RS with high-resolution imaging techniques enables clinicians to obtain comprehensive molecular and structural information from biological samples, facilitating accurate disease diagnosis and monitoring.
Data interpretation and classification
RS holds significant promise for clinical applications, yet it encounters challenges in accurately interpreting and classifying acquired data. A primary hurdle is the inherently weak Raman scattering signal, necessitating extended acquisition times to achieve an acceptable SNR for practical use. This limitation in signal strength hampers rapid spectral imaging, limiting the technique's applicability in real-time clinical diagnostics and monitoring. Additionally, the complexity of Raman spectra from biological samples presents a substantial challenge. The broad peaks in these spectra represent a mix of molecules and neighboring molecular structures, complicating the identification of specific molecular fingerprints. This complexity impedes the accurate interpretation of data for clinical applications.To address the challenges in interpreting and classifying RS data for clinical use, various strategies and technological advancements have been explored. These include the use of genetic algorithms to optimize preprocessing strategies and classification models, such as support SVM. These approaches aim to enhance the accuracy and reliability of Raman spectral analysis for clinical diagnostics. Furthermore, advancements in machine learning techniques have been investigated to facilitate precise classification methods for clinical diagnoses using RS. For instance, there is recognition of Raman spectroscopy's potential to aid clinical decision-making, such as in classifying oncological samples, prompting the application of machine learning approaches to address the complexities of Raman spectra interpretation.215,216 Additionally, efforts have been directed towards developing effective data processing techniques, including advanced spectral preprocessing and statistical analysis methods, to extract pure Raman signals from in vivo or ex vivo tissue samples.
Instrumentation and cost
Raman/SERS techniques often require specialized and expensive instrumentation, such as high-quality lasers, spectrometers, and detectors, which can limit their widespread adoption in biomedical settings. Moreover, Raman/SERS instrumentation demands regular maintenance, calibration, and expertise to operate and interpret the data, posing challenges in resource-limited settings. To address these instrumentation and cost challenges, several approaches can be explored. Firstly, it is crucial to develop cost-effective and portable Raman/SERS instrumentation tailored for specific biomedical applications, leveraging advances in miniaturization and integrated photonics. Additionally, exploring the use of handheld or fiber-optic Raman/SERS probes for in vivo or point-of-care applications can broaden accessibility.46,217–219 Establishing dedicated Raman/SERS facilities or core facilities in research institutions or hospitals can centralize expertise and resources, facilitating broader access to these techniques. Moreover, collaborating with instrument manufacturers to develop user-friendly and cost-effective solutions for biomedical applications can help overcome barriers related to instrumentation and expertise.By addressing these practical challenges through a combination of technological advancements, robust data analysis methods, standardized protocols, and interdisciplinary collaborations among physicists, chemists, biologists, and clinicians, Raman and SERS techniques can be transformed into quantitative and reliable analytical tools for various biomedical applications, such as disease diagnosis, therapeutic monitoring, and biomarker discovery. Ongoing advancements in instrumentation, data analysis algorithms, and sample preparation methodologies will continue to drive the progress of Raman-based biomedical research and facilitate its translation into clinical practice.
Conclusion and future prospects
Researchers continue to explore and validate potential biomarkers with the goal of improving early detection and understanding the underlying mechanisms of diseases such as AD, PD, and others. The future of Raman-based liquid biopsy detection systems for NDs and brain disorders holds significant promise in revolutionizing diagnostic and monitoring approaches.220,221 Early detection is crucial for initiating interventions before significant neurological damage occurs. Continued efforts to identify novel biomarkers associated with NDs in biofluids may lead to the discovery of early and predictive markers. Research is focused on integrating multiple biomarkers from various biofluids to create comprehensive panels.107 Leveraging the sensitivity and specificity of RS, liquid biopsy techniques offer a minimally- to non-invasive and highly accurate means of detecting molecular biomarkers associated with various NDs, including AD and PD. With advancements in technology and methodologies, Raman-based liquid biopsy platforms are poised to provide real-time, point-of-care diagnostics, enabling early disease detection and intervention. Multimodal approaches combining genomics, proteomics, metabolomics, and imaging techniques may offer a more holistic understanding of NDs.46 The concept of “liquid biopsy” for NDs may gain traction, allowing for minimally invasive diagnostic procedures. Biofluids, such as blood and CSF, could serve as liquid biopsy sources, providing valuable information about disease status. The synergy between liquid biopsy and RS holds great promise for advancing our understanding of brain disorders. This integrated approach not only provides a less-invasive means to access crucial diagnostic information but also offers molecular insights into the intricate biochemical processes underlying neurological conditions. As ongoing research endeavors seek to refine and validate these techniques, the potential impact on early detection, disease monitoring, and treatment assessment for brain disorders becomes increasingly apparent, paving the way for more effective and personalized healthcare strategies. Ongoing advancements in analytical techniques, including RS, mass spectrometry, and next-generation sequencing, may enhance the sensitivity and specificity of biomarker detection. Integration of these technologies could contribute to more accurate and reliable diagnostic tools. Development of point-of-care and wearable diagnostic devices may facilitate on-the-spot biomarker analysis. These devices could enable continuous monitoring, early detection of changes, and timely intervention. The use of big data analytics and AI algorithms may help process complex datasets generated from biofluid analyses.222,223 Machine learning approaches could assist in identifying subtle patterns and correlations in biomarker profiles for improved diagnostic accuracy. Collaborative efforts between academia, industry, and healthcare institutions are likely to accelerate biomarker discovery and validation. Large-scale studies involving diverse patient populations may provide a more comprehensive understanding of the heterogeneity of NDs. Biomarkers from biofluids may be crucial for patient stratification in clinical trials, helping to identify suitable participants and monitor treatment responses. This could enhance the efficiency of drug development and facilitate the discovery of disease-modifying therapies. Successful validation of biofluid-based biomarkers may lead to regulatory approvals and their incorporation into routine clinical practice. Widespread adoption of validated biomarkers could significantly impact the early diagnosis and management of NDs.In summary, the future of using biofluids for NDs diagnosis holds great potential for early detection, personalized treatment approaches, and advancements in our understanding of these complex conditions. Ongoing interdisciplinary research and technological innovations are likely to shape the landscape of NDs diagnostics in the coming years. Raman-based liquid biopsy detection systems are expected to play a pivotal role in personalized medicine, guiding treatment strategies, monitoring disease progression, and ultimately improving patient outcomes in the realm of NDs and brain disorders.
Author contributions
J. C. R. and Z. W. drafted the manuscript. S. H. supervised the project. All authors have commented on and edited the manuscript. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We acknowledge the funding support from National Institutes of Health under grant no. 1R01AG077016-02, and National Science Foundation under grants ECCS-2246564 and ECCS-1943895.References
- T. D. Payne, A. S. Moody, A. L. Wood, P. A. Pimiento, J. C. Elliott and B. Sharma, Analyst, 2020, 145, 3461–3480 RSC.
- K. Hrubešová, M. Fousková, L. Habartová, Z. Fišar, R. Jirák, J. Raboch and V. Setnička, Clin. Biochem., 2019, 72, 39–51 CrossRef PubMed.
- M. Paraskevaidi, P. L. Martin-Hirsch and F. L. Martin, ACS Chem. Neurosci., 2018, 9, 446–461 CrossRef CAS PubMed.
- G. G. Klamminger, K. B. Frauenknecht, M. Mittelbronn and F. B. K. Borgmann, Free Neuropathol., 2022, 3, 19 Search PubMed.
- Alzheimer's disease facts and figures, https://www.alz.org/media/Documents/alzheimers-facts-and-figures.pdf, (accessed 03/20/2024).
- Burden of neurological conditions: level by country, https://www.paho.org/en/enlace/burden-neurological-conditions, (accessed 03/20/2024).
- Alzheimer's disease facts and statistics, https://www.alzinfo.org/understand-alzheimers/alzheimers-disease-facts-and-statistics/, (accessed 03/20/2024).
- Neurological disorders affect millions globally: WHO report, https://www.who.int/news/item/27-02-2007-neurological-disorders-affect-millions-globally-who-report, (accessed 03/20/2024).
- Statistics get informed about Parkinson's disease with these key numbers, https://www.parkinson.org/understanding-parkinsons/statistics, (accessed 03/20/2024).
- Y. Wang, L. Fang, Y. Wang and Z. Xiong, Adv. Sci., 2023, 2300668 Search PubMed.
- M. J. Baker, S. R. Hussain, L. Lovergne, V. Untereiner, C. Hughes, R. A. Lukaszewski, G. Thiéfin and G. D. Sockalingum, Chem. Soc. Rev., 2016, 45, 1803–1818 RSC.
- D. K. Medipally, A. Maguire, J. Bryant, J. Armstrong, M. Dunne, M. Finn, F. M. Lyng and A. D. Meade, Analyst, 2017, 142, 1216–1226 RSC.
- N. M. Ralbovsky, P. Dey, A. Galfano, B. K. Dey and I. K. Lednev, Sci. Rep., 2020, 10, 11734 CrossRef CAS PubMed.
- S. F. El-Mashtoly and K. Gerwert, Anal. Chem., 2021, 94, 120–142 CrossRef PubMed.
- D. Cialla-May, C. Krafft, P. Rösch, T. Deckert-Gaudig, T. Frosch, I. J. Jahn, S. Pahlow, C. Stiebing, T. Meyer-Zedler and T. Bocklitz, Anal. Chem., 2021, 94, 86–119 CrossRef PubMed.
- K. C. Doty and I. K. Lednev, ACS Cent. Sci., 2018, 4, 862–867 CrossRef CAS PubMed.
- E. Al-Hetlani, L. Halámková, M. O. Amin and I. K. Lednev, J. Biophotonics, 2020, 13, e201960123 CrossRef PubMed.
- A. N. Masterson, S. Hati, G. Ren, T. Liyanage, N. E. Manicke, J. V. Goodpaster and R. Sardar, Anal. Chem., 2021, 93, 2578–2588 CrossRef CAS PubMed.
- N. M. Ralbovsky and I. K. Lednev, Chem. Soc. Rev., 2020, 49, 7428–7453 RSC.
- A. Subaihi, L. Almanqur, H. Muhamadali, N. AlMasoud, D. I. Ellis, D. K. Trivedi, K. A. Hollywood, Y. Xu and R. Goodacre, Anal. Chem., 2016, 88, 10884–10892 CrossRef CAS PubMed.
- J. C. Ranasinghe, A. Jain, W. Wu, K. Zhang, Z. Wang and S. Huang, J. Mater. Res., 2022, 37, 1689–1713 CrossRef CAS PubMed.
- J. C. Ranasinghe, Z. Wang and S. Huang, Biosensors, 2022, 13, 27 CrossRef PubMed.
- Z. Wang, J. Ye, K. Zhang, L. Ding, T. Granzier-Nakajima, J. C. Ranasinghe, Y. Xue, S. Sharma, I. Biase and M. Terrones, ACS Nano, 2022, 16, 6426–6436 CrossRef CAS PubMed.
- K. Zhang, A. Jain, W. Wu, J. Ranasinghe, Z. Wang and S. Huang, in Novel Optical Materials, World Scientific, 2024, pp. 1–30 Search PubMed.
- K. Zhang, Z. Wang, H. Liu, N. Perea-López, J. C. Ranasinghe, G. Bepete, A. M. Minns, R. M. Rossi, S. E. Lindner and S. X. Huang, ACS Photonics, 2022, 9, 2963–2972 CrossRef CAS PubMed.
- L. A. Bratchenko, I. A. Bratchenko, A. A. Lykina, M. V. Komarova, D. N. Artemyev, O. O. Myakinin, A. A. Moryatov, I. L. Davydkin, S. V. Kozlov and V. P. Zakharov, J. Raman Spectrosc., 2020, 51, 279–292 CrossRef CAS.
- Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2007, 42, 493–541 CrossRef CAS.
- C. Zong, M. Xu, L.-J. Xu, T. Wei, X. Ma, X.-S. Zheng, R. Hu and B. Ren, Chem. Rev., 2018, 118, 4946–4980 CrossRef CAS PubMed.
- P. Polykretis, M. Banchelli, C. D'Andrea, M. de Angelis and P. Matteini, Front. Biosci.-Scholar, 2022, 14, 22 CrossRef.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. Bell, A. Boisen and A. G. Brolo, ACS Nano, 2019, 14, 28–117 CrossRef PubMed.
- J. Pérez-Juste, I. Pastoriza-Santos, L. M. Liz-Marzán and P. Mulvaney, Coord. Chem. Rev., 2005, 249, 1870–1901 CrossRef.
- S.-Y. Ding, E.-M. You, Z.-Q. Tian and M. Moskovits, Chem. Soc. Rev., 2017, 46, 4042–4076 RSC.
- A. S. Dikkumbura, P. Hamal, M. Chen, D. A. Babayode, J. C. Ranasinghe, K. Lopata and L. H. Haber, J. Phys. Chem. C, 2021, 125, 25615–25623 CrossRef CAS PubMed.
- R. A. Khoury, J. C. Ranasinghe, A. S. Dikkumbura, P. Hamal, R. R. Kumal, T. E. Karam, H. T. Smith and L. H. Haber, J. Phys. Chem. C, 2018, 122, 24400–24406 CrossRef CAS.
- J. C. Ranasinghe, A. S. Dikkumbura, P. Hamal, M. Chen, R. A. Khoury, H. T. Smith, K. Lopata and L. H. Haber, J. Chem. Phys., 2019, 151, 224701 CrossRef PubMed.
- Q. Ding, J. Wang, X. Chen, H. Liu, Q. Li, Y. Wang and S. Yang, Nano Lett., 2020, 20, 7304–7312 CrossRef CAS PubMed.
- S. Huang, R. Pandey, I. Barman, J. Kong and M. Dresselhaus, ACS Photonics, 2018, 5, 2978–2982 CrossRef CAS.
- I. Zare, M. T. Yaraki, G. Speranza, A. H. Najafabadi, A. Shourangiz-Haghighi, A. B. Nik, B. B. Manshian, C. Saraiva, S. J. Soenen and M. J. Kogan, Chem. Soc. Rev., 2022, 51, 2601–2680 RSC.
- S. Aitekenov, A. Sultangaziyev, P. Abdirova, L. Yussupova, A. Gaipov, Z. Utegulov and R. Bukasov, Crit. Rev. Anal. Chem., 2023, 53, 1561–1590 CrossRef CAS PubMed.
- J. Plou, P. S. Valera, I. Garcia, C. D. de Albuquerque, A. Carracedo and L. M. Liz-Marzan, ACS Photonics, 2022, 9, 333–350 CrossRef CAS PubMed.
- W. Premasiri, J. Lee and L. Ziegler, J. Phys. Chem. B, 2012, 116, 9376–9386 CrossRef CAS PubMed.
- D. Tuschel, Spectroscopy, 2016, 31, 14–23 Search PubMed.
- D. A. Long , New York, 1977, 1.
- K. Kralova, M. Kral, O. Vrtelka and V. Setnicka, Spectrochim. Acta, Part A, 2024, 304, 123392 CrossRef CAS PubMed.
- C. Krafft, G. Steiner, C. Beleites and R. Salzer, J. Biophotonics, 2009, 2, 13–28 CrossRef CAS PubMed.
- D. DePaoli, É. Lemoine, K. Ember, M. Parent, M. Prud'homme, L. Cantin, K. Petrecca, F. Leblond and D. C. Côté, J. Biomed. Opt., 2020, 25, 050901–050901 Search PubMed.
- G. Ramachandran, E. A. Milan-Garces, J. B. Udgaonkar and M. Puranik, Biochemistry, 2014, 53, 6550–6565 CrossRef CAS PubMed.
- Y. Yang, S. Wu, Y. Chen and H. Ju, Chem. Sci., 2023, 14, 12869–12882 RSC.
- J. Wang, L. Cong, W. Shi, W. Xu and S. Xu, Anal. Chem., 2023, 95, 11019–11027 CrossRef CAS PubMed.
- W. Mo, J. Wen, J. Huang, Y. Yang, M. Zhou, S. Ni, W. Le, L. Wei, D. Qi and S. Wang, J. Appl. Spectrosc., 2023, 89, 1203–1211 CrossRef CAS PubMed.
- R. Krishna and I. Colak, Anal. Lett., 2023, 56, 576–617 CrossRef CAS.
- S. Siddhanta, A. N. Kuzmin, A. Pliss, A. S. Baev, S. K. Khare, P. K. Chowdhury, A. K. Ganguli and P. N. Prasad, Adv. Opt. Photonics, 2023, 15, 318–384 CrossRef.
- E. Lopez, J. Etxebarria-Elezgarai, M. García-Sebastián, M. Altuna, M. Ecay-Torres, A. Estanga, M. Tainta, C. López, P. Martínez-Lage and J. M. Amigo, Int. J. Mol. Sci., 2024, 25, 4737 CrossRef CAS PubMed.
- K. Mamatkulov, S. Zavatski, Y. Arynbek, H. A. Esawii, A. Burko, H. Bandarenka and G. Arzumanyan, J. Biomol. Struct. Dyn., 2024, 1–14 CrossRef PubMed.
- S. Prasad, P. S. Nayak and P. D’Silva, Mater. Adv., 2024, 5, 2388–2399 RSC.
- C. Chen, J. Qi, Y. Li, D. Li, L. Wu, R. Li, Q. Chen and N. Sun, Front. Neurosci., 2024, 18, 1301107 CrossRef.
- H. Ren, Y. Zhang, S. Guo, N. Lin, L. Deng, T. Yue and F. Huang, Phys. Chem. Chem. Phys., 2017, 19, 31103–31112 RSC.
- B. Tian, C. Cheng, T. Yue, N. Lin and H. Ren, Chem. Phys., 2018, 513, 1–6 CrossRef CAS.
- M. Ji, M. Arbel, L. Zhang, C. W. Freudiger, S. S. Hou, D. Lin, X. Yang, B. J. Bacskai and X. S. Xie, Sci. Adv., 2018, 4, eaat7715 CrossRef CAS PubMed.
- V. Schweikhard, A. Baral, V. Krishnamachari, W. C. Hay and M. Fuhrmann, bioRxiv, 2019, preprint, 789248, DOI:10.1101/789248.
- B. Lochocki, B. D. Boon, S. R. Verheul, L. Zada, J. J. Hoozemans, F. Ariese and J. F. de Boer, Commun. Biol., 2021, 4, 474 CrossRef CAS PubMed.
- M. Ji, Biomed. Photonic Technol., 2023, 117–136 CAS.
- Z. Zhang, S. Sheng, R. Wang and M. Sun, Anal. Chem., 2016, 88, 9328–9346 CrossRef CAS.
- S. Bonhommeau and S. Lecomte, ChemPhysChem, 2018, 19, 8–18 CrossRef CAS PubMed.
- M. Tabatabaei, F. A. Caetano, F. Pashee, S. S. Ferguson and F. Lagugné-Labarthet, Analyst, 2017, 142, 4415–4421 RSC.
- S. Bonhommeau, D. Talaga, J. Hunel, C. Cullin and S. Lecomte, Angew. Chem., 2017, 129, 1797–1800 CrossRef.
- B. Zikic, A. Bremner, D. Talaga, S. Lecomte and S. Bonhommeau, Chem. Phys. Lett., 2021, 768, 138400 CrossRef CAS.
- D. Talaga, W. Smeralda, L. Lescos, J. Hunel, N. a. Lepejova-Caudy, C. Cullin, S. Bonhommeau and S. Lecomte, Angew. Chem., 2018, 130, 15964–15968 CrossRef.
- B. Yang, S. Jin, Y. Wang, H. Bao, J. Sun, L. Chen, H. Chung and Y. M. Jung, Appl. Spectrosc. Rev., 2019, 54, 856–872 CrossRef CAS.
- E. Ryzhikova, N. M. Ralbovsky, L. Halámková, D. Celmins, P. Malone, E. Molho, J. Quinn, E. A. Zimmerman and I. K. Lednev, Appl. Sci., 2019, 9, 3256 CrossRef CAS.
- C. Carlomagno, M. Cabinio, S. Picciolini, A. Gualerzi, F. Baglio and M. Bedoni, J. Biophotonics, 2020, 13, e201960033 CrossRef PubMed.
- G. Cennamo, D. Montorio, V. B. Morra, C. Criscuolo, R. Lanzillo, E. Salvatore, C. Camerlingo, M. Lisitskiy, I. Delfino and M. Portaccio, J. Biomed. Opt., 2020, 25, 087002–087002 CAS.
- S. J. Yang, J. U. Lee, M. J. Jeon and S. J. Sim, Anal. Chim. Acta, 2022, 1195, 339445 CrossRef CAS PubMed.
- D. Yu, Q. Yin, J. Wang, J. Yang, Z. Chen, Z. Gao, Q. Huang and S. Li, Int. J. Nanomed., 2021, 1901–1911 CrossRef CAS.
- Z. Zhang, R. Guan, J. Li and Y. Sun, Chemosensors, 2023, 11, 110 CrossRef CAS.
- B. Wrzosek, Y. Kitahama and Y. Ozaki, J. Phys. Chem. C, 2020, 124, 20328–20339 CrossRef CAS.
- M. Fan, F.-J. Lai, H.-L. Chou, W.-T. Lu, B.-J. Hwang and A. G. Brolo, Chem. Sci., 2013, 4, 509–515 RSC.
- Y. Cui, B. Ren, J.-L. Yao, R.-A. Gu and Z.-Q. Tian, J. Phys. Chem. B, 2006, 110, 4002–4006 CrossRef CAS PubMed.
- A. M. Schwartzberg, C. D. Grant, A. Wolcott, C. E. Talley, T. R. Huser, R. Bogomolni and J. Z. Zhang, J. Phys. Chem. B, 2004, 108, 19191–19197 CrossRef CAS.
- S. Corni and J. Tomasi, J. Chem. Phys., 2002, 116, 1156–1164 CrossRef CAS.
- H. Ko, S. Singamaneni and V. V. Tsukruk, Small, 2008, 4, 1576–1599 CrossRef CAS PubMed.
- E. Fazio, F. Neri, C. D'Andrea, P. Ossi, N. Santo and S. Trusso, J. Raman Spectrosc., 2011, 42, 1298–1304 CrossRef CAS.
- M. Tahghighi, I. Mannelli, D. Janner and J. Ignés-Mullol, Appl. Surf. Sci., 2018, 447, 416–422 CrossRef CAS.
- M. Swierczewski and T. Burgi, Langmuir, 2023, 39, 2135–2151 CrossRef CAS PubMed.
- L. E. Kreno, N. G. Greeneltch, O. K. Farha, J. T. Hupp and R. P. Van Duyne, Analyst, 2014, 139, 4073–4080 RSC.
- K. Sugikawa, Y. Furukawa and K. Sada, Chem. Mater., 2011, 23, 3132–3134 CrossRef CAS.
- W. Wu, J. C. Ranasinghe, A. Chatterjee and S. Huang, Mater. Chem. Phys., 2024, 129281 CrossRef CAS.
- Y. Wu, Z. Xu, W. Yang, Z. Ning and H. Dong, Front. Bioeng. Biotechnol., 2022, 10, 906728 CrossRef PubMed.
- Y. Wang, N. Reder, S. Kang, A. Glaser and J. Liu, Nanotheranostics, 2017, 1, 369–388 CrossRef PubMed.
- H. Mitsutake, R. J. Poppi and M. C. Breitkreitz, J. Braz. Chem. Soc., 2019, 30, 2243–2258 CAS.
- B. M. Davis, A. J. Hemphill, D. Cebeci Maltaş, M. A. Zipper, P. Wang and D. Ben-Amotz, Anal. Chem., 2011, 83, 5086–5092 CrossRef CAS PubMed.
- R. Michael, A. Lenferink, G. F. Vrensen, E. Gelpi, R. I. Barraquer and C. Otto, Sci. Rep., 2017, 7, 15603 CrossRef PubMed.
- E. Lipiec, J. Kaderli, J. Kobierski, R. Riek, K. Skirlińska-Nosek, K. Sofińska, M. Szymoński and R. Zenobi, Angew. Chem., Int. Ed., 2021, 60, 4545–4550 CrossRef CAS PubMed.
- D. a. Mrđenović, B. F. Combes, R. Ni, R. Zenobi and N. Kumar, ACS Chem. Neurosci., 2023, 15, 78–85 CrossRef PubMed.
- Y. Yu, Y. Dai, X. Wang, K. Chu and Z. J. Smith, Analyst, 2023, 148, 2809–2817 RSC.
- H. He, M. Xu, C. Zong, P. Zheng, L. Luo, L. Wang and B. Ren, Anal. Chem., 2019, 91, 7070–7077 CrossRef CAS PubMed.
- S. Schlücker, M. D. Schaeberle, S. W. Huffman and I. W. Levin, Anal. Chem., 2003, 75, 4312–4318 CrossRef PubMed.
- H. Xiong, N. Qian, Y. Miao, Z. Zhao, C. Chen and W. Min, Light: Sci. Appl., 2021, 10, 87 CrossRef CAS PubMed.
- K. Fujita, Label-Free Super-Resolution Microscopy, 2019, pp. 195–211 Search PubMed.
- C. D. L. De Albuquerque and Z. D. Schultz, Anal. Chem., 2020, 92, 9389–9398 CrossRef CAS PubMed.
- T. Deckert-Gaudig and V. Deckert, Phys. Chem. Chem. Phys., 2010, 12, 12040–12049 RSC.
- H. Blom and J. Widengren, Chem. Rev., 2017, 117, 7377–7427 CrossRef CAS PubMed.
- B. R. Rankin, R. R. Kellner and S. W. Hell, Opt. Lett., 2008, 33, 2491–2493 CrossRef PubMed.
- H. Lee, H. Yoo, G. Moon, K.-A. Toh, K. Mochizuki, K. Fujita and D. Kim, J. Chem. Phys., 2021, 155, 144202 CrossRef CAS PubMed.
- R. Heintzmann and T. Huser, Chem. Rev., 2017, 117, 13890–13908 CrossRef CAS PubMed.
- O. Terrones, J. Olazar-Intxausti, I. Anso, M. Lorizate, J. A. Nieto-Garai and F.-X. Contreras, Int. J. Mol. Sci., 2023, 24, 2384 CrossRef PubMed.
- N. Kuhar, S. Sil, T. Verma and S. Umapathy, RSC Adv., 2018, 8, 25888–25908 RSC.
- J. Lippert, D. Tyminski and P. Desmeules, J. Am. Chem. Soc., 1976, 98, 7075–7080 CrossRef CAS PubMed.
- R. W. Williams, in Methods in enzymology, Elsevier, 1986, vol. 130, pp. 311–331 Search PubMed.
- N. C. Maiti, M. M. Apetri, M. G. Zagorski, P. R. Carey and V. E. Anderson, J. Am. Chem. Soc., 2004, 126, 2399–2408 CrossRef CAS PubMed.
- J. Benevides and G. Thomas Jr, Nucleic Acids Res., 1983, 11, 5747–5761 CrossRef CAS PubMed.
- J. De Gelder, K. De Gussem, P. Vandenabeele and L. Moens, J. Raman Spectrosc., 2007, 38, 1133–1147 CrossRef CAS.
- J. Duguid, V. A. Bloomfield, J. Benevides and G. Thomas, Biophys. J., 1993, 65, 1916–1928 CrossRef CAS PubMed.
- K. Czamara, K. Majzner, M. Z. Pacia, K. Kochan, A. Kaczor and M. Baranska, J. Raman Spectrosc., 2015, 46, 4–20 CrossRef CAS.
- E. Wiercigroch, E. Szafraniec, K. Czamara, M. Z. Pacia, K. Majzner, K. Kochan, A. Kaczor, M. Baranska and K. Malek, Spectrochim. Acta, Part A, 2017, 185, 317–335 CrossRef CAS PubMed.
- M. Noestheden, Q. Hu, L.-L. Tay, A. M. Tonary, A. Stolow, R. MacKenzie, J. Tanha and J. P. Pezacki, Bioorg. Chem., 2007, 35, 284–293 CrossRef CAS PubMed.
- H. Yamakoshi, K. Dodo, M. Okada, J. Ando, A. Palonpon, K. Fujita, S. Kawata and M. Sodeoka, J. Am. Chem. Soc., 2011, 133, 6102–6105 CrossRef CAS PubMed.
- Y. Qi, E. X. Chen, D. Hu, Y. Yang, Z. Wu, M. Zheng, M. A. Sadi, Y. Jiang, K. Zhang and Z. Chen, Food Front., 2024, 5, 392–419 CrossRef CAS.
- W. Li, G. Chen, M. Chen, K. Shen, C. Wu, W. Shen and F. Zhang, Spectrochim. Acta, Part A, 2023, 302, 123088 CrossRef CAS PubMed.
- Y. Qi, D. Hu, Y. Jiang, Z. Wu, M. Zheng, E. X. Chen, Y. Liang, M. A. Sadi, K. Zhang and Y. P. Chen, Adv. Opt. Mater., 2023, 11, 2203104 CrossRef CAS.
- I. W. Schie and T. Huser, Appl. Spectrosc., 2013, 67, 813–828 CrossRef CAS PubMed.
- X. Bi, L. Lin, Z. Chen and J. Ye, Small Methods, 2024, 8, 2301243 CrossRef PubMed.
- C. Ciobanu, K. J. Ember, B. J. Nyíri, S. Rajan, V. Chauhan, F. Leblond and S. Murugkar, IEEE Instrumentation & Measurement Magazine, 2022, 25, 62–68.
- M. Huber, K. V. Kepesidis, L. Voronina, M. Božić, M. Trubetskov, N. Harbeck, F. Krausz and M. Žigman, Nat. Commun., 2021, 12, 1511 CrossRef CAS PubMed.
- J. Ollesch, S. L. Drees, H. M. Heise, T. Behrens, T. Brüning and K. Gerwert, Analyst, 2013, 138, 4092–4102 RSC.
- R. F. Dos Santos, M. Paraskevaidi, D. M. Mann, D. Allsop, M. C. Santos, C. L. Morais and K. M. Lima, Sci. Rep., 2022, 12, 16199 CrossRef CAS.
- P. Chen, Q. Tian, S. Baek, X. Shang, A. Park, Z. Liu, X. Yao, J. Wang, X. Wang and Y. Cheng, Laser Phys. Lett., 2011, 8, 547–552 CrossRef CAS.
- M. Lin, H. Ou, P. Zhang, Y. Meng, S. Wang, J. Chang, A. Shen and J. Hu, Spectrochim. Acta, Part A, 2022, 280, 121542 CrossRef CAS PubMed.
- L. Wang, X. Du, Z. Du, Y. Yang, P. Chen, Q. Tian, X. Shang, Z. Liu, X. Yao and J. Wang, Laser Phys., 2014, 24, 085702 CrossRef.
- N. M. Ralbovsky, G. S. Fitzgerald, E. C. McNay and I. K. Lednev, Spectrochim. Acta, Part A, 2021, 254, 119603 CrossRef CAS.
- P. Carmona, E. Monleón, M. Monzón, J. J. Badiola and J. Monreal, Chem. Biol., 2004, 11, 759–764 CrossRef CAS.
- P. Carmona, M. Monzon, E. Monleon, J. J. Badiola and J. Monreal, J. Gen. Virol., 2005, 86, 3425–3431 CrossRef CAS PubMed.
- R. A. Alvarez-Puebla, A. Agarwal, P. Manna, B. P. Khanal, P. Aldeanueva-Potel, E. Carbó-Argibay, N. Pazos-Pérez, L. Vigderman, E. R. Zubarev and N. A. Kotov, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 8157–8161 CrossRef CAS PubMed.
- J. J. Rickard, V. Di-Pietro, D. J. Smith, D. J. Davies, A. Belli and P. G. Oppenheimer, Nat. Biomed. Eng., 2020, 4, 610–623 CrossRef CAS.
- G. Harris, C. A. Stickland, M. Lim and P. Goldberg Oppenheimer, Cells, 2023, 12, 2589 CrossRef CAS PubMed.
- X. Gao, J. Boryczka, P. Zheng, S. Kasani, F. Yang, E. B. Engler-Chiurazzi, J. W. Simpkins, J. G. Wigginton and N. Wu, Biosens. Bioelectron., 2021, 177, 112967 CrossRef CAS PubMed.
- P. Carmona, M. Molina, M. Calero, F. Bermejo-Pareja, P. Martínez-Martín and A. Toledano, J. Alzheimer's Dis., 2013, 34, 911–920 CAS.
- P. Carmona, M. Molina, E. López-Tobar and A. Toledano, Anal. Bioanal. Chem., 2015, 407, 7747–7756 CrossRef CAS PubMed.
- T. K. Karikari, T. A. Pascoal, N. J. Ashton, S. Janelidze, A. L. Benedet, J. L. Rodriguez, M. Chamoun, M. Savard, M. S. Kang and J. Therriault, Lancet Neurol., 2020, 19, 422–433 CrossRef CAS PubMed.
- L. Habartová, K. Hrubešová, K. Syslová, J. Vondroušová, Z. Fišar, R. Jirák, J. Raboch and V. Setnička, Clin. Biochem., 2019, 72, 58–63 CrossRef PubMed.
- Y. Wang, P. Zhao, L. Mao, Y. Hou and D. Li, RSC Adv., 2018, 8, 3143–3150 RSC.
- A. R. Stevens, C. A. Stickland, G. Harris, Z. Ahmed, P. Goldberg Oppenheimer, A. Belli and D. J. Davies, Cells, 2022, 11, 1227 CrossRef CAS PubMed.
- N. Hao, Z. Wang, P. Liu, R. Becker, S. Yang, K. Yang, Z. Pei, P. Zhang, J. Xia and L. Shen, Biosens. Bioelectron., 2022, 196, 113730 CrossRef CAS PubMed.
- X. Gao, P. Zheng, S. Kasani, S. Wu, F. Yang, S. Lewis, S. Nayeem, E. B. Engler-Chiurazzi, J. G. Wigginton and J. W. Simpkins, Anal. Chem., 2017, 89, 10104–10110 CrossRef CAS PubMed.
- J. K. Yang, I. J. Hwang, M. G. Cha, H. I. Kim, D. Yim, D. H. Jeong, Y. S. Lee and J. H. Kim, Small, 2019, 15, 1900613 CrossRef PubMed.
- V.-D. Phung, W.-S. Jung, T.-A. Nguyen, J.-H. Kim and S.-W. Lee, Nanoscale, 2018, 10, 22493–22503 RSC.
- D. Li, M. Yang, H. Li, L. Mao, Y. Wang and B. Sun, New J. Chem., 2019, 43, 5925–5931 RSC.
- E. Ryzhikova, O. Kazakov, L. Halamkova, D. Celmins, P. Malone, E. Molho, E. A. Zimmerman and I. K. Lednev, J. Biophotonics, 2015, 8, 584–596 CrossRef PubMed.
- M. Paraskevaidi, C. L. Morais, D. E. Halliwell, D. M. Mann, D. Allsop, P. L. Martin-Hirsch and F. L. Martin, ACS Chem. Neurosci., 2018, 9, 2786–2794 CrossRef CAS PubMed.
- A. Sharma, J. Müller, K. Schuetze, V. Rolfes, R. Bissinger, N. Rosero, A. Ahmad, B. S. Franklin, B. Zur and H. Fröhlich, Biology, 2021, 10, 716 CrossRef CAS PubMed.
- A. Huefner, W.-L. Kuan, S. L. Mason, S. Mahajan and R. A. Barker, Chem. Sci., 2020, 11, 525–533 RSC.
- H. M. Schipper, C. S. Kwok, S. M. Rosendahl, D. Bandilla, O. Maes, C. Melmed, D. Rabinovitch and D. H. Burns, Biomark. Med., 2008, 2, 229–238 CrossRef CAS PubMed.
- T. Demeritte, B. P. Viraka Nellore, R. Kanchanapally, S. S. Sinha, A. Pramanik, S. R. Chavva and P. C. Ray, ACS Appl. Mater. Interfaces, 2015, 7, 13693–13700 CrossRef CAS PubMed.
- S. S. Sinha, S. Jones, A. Pramanik and P. C. Ray, Acc. Chem. Res., 2016, 49, 2725–2735 CrossRef CAS PubMed.
- E. Buchan, L. Kelleher, M. Clancy, J. J. S. Rickard and P. G. Oppenheimer, Anal. Chim. Acta, 2021, 1185, 339074 CrossRef CAS PubMed.
- E. Buchan, M. Hardy, P. d. C. Gomes, L. Kelleher, H. O. M. Chu and P. G. Oppenheimer, Appl. Spectrosc. Rev., 2022, 1–38 Search PubMed.
- S. Derruau, J. Robinet, V. Untereiner, O. Piot, G. D. Sockalingum and S. Lorimier, Molecules, 2020, 25, 4142 CrossRef CAS PubMed.
- C. Carlomagno, D. Bertazioli, A. Gualerzi, S. Picciolini, M. Andrico, F. Rodà, M. Meloni, P. I. Banfi, N. Ticozzi and V. Silani, Front. Neurosci., 2021, 15, 704963 CrossRef PubMed.
- N. J. Ashton, M. Ide, H. Zetterberg and K. Blennow, Neurol. Ther., 2019, 8, 83–94 CrossRef PubMed.
- M. Lee, J.-P. Guo, K. Kennedy, E. G. McGeer and P. L. McGeer, J. Alzheimer's Dis., 2017, 55, 1175–1182 CAS.
- F. Bermejo-Pareja, D. Antequera, T. Vargas, J. A. Molina and E. Carro, BMC Neurol., 2010, 10, 1–7 CrossRef.
- C.-B. Kim, Y. Y. Choi, W. K. Song and K.-B. Song, J. Biomed. Opt., 2014, 19, 051205–051205 CrossRef PubMed.
- M. N. Sabbagh, J. Shi, M. Lee, L. Arnold, Y. Al-Hasan, J. Heim and P. McGeer, BMC Neurol., 2018, 18, 1–4 CrossRef.
- M. Hardy, L. Kelleher, P. de Carvalho Gomes, E. Buchan, H. O. M. Chu and P. Goldberg Oppenheimer, Appl. Spectrosc. Rev., 2022, 57, 177–233 CrossRef.
- C. Carlomagno, P. Banfi, A. Gualerzi, S. Picciolini, E. Volpato, M. Meloni, A. Lax, E. Colombo, N. Ticozzi and F. Verde, Sci. Rep., 2020, 10, 10175 CrossRef CAS PubMed.
- N. M. Ralbovsky, L. Halámková, K. Wall, C. Anderson-Hanley and I. K. Lednev, J. Alzheimer's Dis., 2019, 71, 1351–1359 CAS.
- S. Altuntas and F. Buyukserin, J. Raman Spectrosc., 2018, 49, 1247–1256 CrossRef CAS.
- C. Camerlingo, M. Lisitskiy, M. Lepore, M. Portaccio, D. Montorio, S. D. Prete and G. Cennamo, Sensors, 2019, 19, 1177 CrossRef CAS PubMed.
- S. S. Adigal, A. Rizvi, N. V. Rayaroth, R. V. John, A. Barik, S. Bhandari, S. D. George, J. Lukose, V. B. Kartha and S. Chidangil, Expert Rev. Mol. Diagn., 2021, 21, 767–787 CrossRef CAS PubMed.
- A. Capaccio, A. Sasso and G. Rusciano, Sensors, 2019, 19, 3392 CrossRef CAS PubMed.
- S. Alotaibi, M. Markoulli, J. Ozkan and E. Papas, Clin. Exp. Optom., 2022, 105, 166–176 CrossRef PubMed.
- S. K. Nandi, D. Singh, J. Upadhay, N. Gupta, N. Dhiman, S. K. Mittal and N. Mahindroo, Int. J. Biol. Macromol., 2021, 193, 838–846 CrossRef CAS PubMed.
- G. Kalló, M. Emri, Z. Varga, B. Ujhelyi, J. Tőzsér, A. Csutak and É. Csősz, PLoS One, 2016, 11, e0158000 CrossRef PubMed.
- A. Kenny, E. M. Jiménez-Mateos, M. A. Zea-Sevilla, A. Rábano, P. Gili-Manzanaro, J. H. Prehn, D. C. Henshall, J. Ávila, T. Engel and F. Hernandez, Sci. Rep., 2019, 9, 15437 CrossRef PubMed.
- M. Gijs, R. M. Nuijts, I. Ramakers, F. Verhey and C. A. Webers, Invest. Ophthalmol. Visual Sci., 2019, 60, 1744–1744 Search PubMed.
- M. Boerger, S. Funke, A. Leha, A.-E. Roser, A.-K. Wuestemann, F. Maass, M. Bähr, F. Grus and P. Lingor, Parkinsonism Relat. Drd., 2019, 63, 3–9 CrossRef PubMed.
- O. Y. Kwon, S. H. Kim, J. H. Kim, M. H. Kim and M. K. Ko, J. Korean Med. Sci., 1994, 9, 239–242 CrossRef CAS PubMed.
- M. C. Edman, S. R. Janga, S. S. Kakan, C. T. Okamoto, D. Freire, D. Feigenbaum, M. Lew and S. F. Hamm-Alvarez, Biomarkers Med., 2020, 14, 151–163 CrossRef CAS PubMed.
- S. F. Hamm-Alvarez, S. R. Janga, M. C. Edman, D. Feigenbaum, D. Freire, W. J. Mack, C. T. Okamoto and M. F. Lew, Biomarkers Med., 2019, 13, 1447–1457 CrossRef CAS PubMed.
- F. Maass, S. Rikker, V. Dambeck, C. Warth, L. Tatenhorst, I. Csoti, M. Schmitz, I. Zerr, A. Leha and M. Bähr, Sci. Rep., 2020, 10, 8507 CrossRef CAS PubMed.
- S. S. Çomoğlu, H. Güven, M. Acar, G. Öztürk and B. Koçer, Neurosci. Lett., 2013, 553, 63–67 CrossRef PubMed.
- D. Ami, A. Duse, P. Mereghetti, F. Cozza, F. Ambrosio, E. Ponzini, R. Grandori, C. Lunetta, S. Tavazzi and F. Pezzoli, Anal. Chem., 2021, 93, 16995–17002 CrossRef CAS PubMed.
- Y. Song, H. Bian, T. Zeng, T. Lin, Y. Liu, S. Deng, J. Liao, Z. Mao and S. Chen, Curr. Nanosci., 2023, 19, 338–349 CrossRef CAS.
- X. Zhang, S. Liu, X. Song, H. Wang, J. Wang, Y. Wang, J. Huang and J. Yu, ACS Sens., 2019, 4, 2140–2149 CrossRef CAS PubMed.
- N. Lhiyani, A. Sanjeev, A. Mor, Y. Beiderman, J. Garcia and Z. Zalevsky, Opt. Continuum, 2023, 2, 847–855 CrossRef CAS.
- I.-H. Chou, M. Benford, H. T. Beier, G. L. Coté, M. Wang, N. Jing, J. Kameoka and T. A. Good, Nano Lett., 2008, 8, 1729–1735 CrossRef PubMed.
- E. Ryzhikova, N. M. Ralbovsky, V. Sikirzhytski, O. Kazakov, L. Halamkova, J. Quinn, E. A. Zimmerman and I. K. Lednev, Spectrochim. Acta, Part A, 2021, 248, 119188 CrossRef CAS PubMed.
- A. Kumar, M. A. Nader and G. Deep, Pharmacol. Rev., 2024, 76, 199–227 CrossRef PubMed.
- F. León-Bejarano, M. O. Méndez, A. Alba, I. Rodríguez-Leyva, F. J. González, M. d. C. Rodríguez-Aranda, E. Guevara, R. A. Guirado-López and M. G. Ramírez-Elías, Appl. Spectrosc., 2022, 76, 1317–1328 CrossRef PubMed.
- D. Tsikritsis, A. Elfick and A. Downes, Spectrosc. Lett., 2016, 49, 535–541 CrossRef CAS.
- M. Muratore, Anal. Chim. Acta, 2013, 793, 1–10 CrossRef CAS PubMed.
- Z. Shahnavaz, M. J. Shiddiky, K. Saremi, G. Qiao and N.-Y. Kim, in Analytical Techniques for Biomedical Nanotechnology, IOP Publishing, Bristol, UK, 2023, pp. 5-1-5-93 Search PubMed.
- A. Gualerzi, S. Picciolini, C. Carlomagno, F. Terenzi, S. Ramat, S. Sorbi and M. Bedoni, Nanomedicine, 2019, 22, 102097 CrossRef CAS PubMed.
- M. Gijs, N. Jorna, N. Datson, C. Beekman, C. Dansokho, A. Weiss, D. E. Linden and M. Oosterloo, J. Mov. Disord., 2024, 17, 181–188 CrossRef PubMed.
- J. Corey-Bloom, A. M. Aikin, S. Park, A. Haque, A. S. Nathan, D. A. Granger, S. W. Granger and E. A. Thomas, Neurotherapeutics, 2018, 15, 241–242 Search PubMed.
- L. M. Byrne, F. Rodrigues, E. Johnson, E. De Vita, K. Blennow, R. Scahill, H. Zetterberg, A. Heslegrave and E. Wild, Sci. Rep., 2018, 8, 4260 CrossRef PubMed.
- F. Pio and B. R. Leavitt, in Biomarkers for Huntington's Disease: Improving Clinical Outcomes, Springer, 2023, pp. 19–44 Search PubMed.
- M. K. Bondulich, J. Phillips, M. Cañibano-Pico, I. M. Nita, L. M. Byrne, E. J. Wild and G. P. Bates, Brain Commun., 2024, 6, fcae030 CrossRef PubMed.
- U. M. Deshetty and P. Periyasamy, J. Clin. Med., 2023, 12, 3923 CrossRef CAS PubMed.
- S. D. Hicks, C. Onks, R. Y. Kim, K. J. Zhen, J. Loeffert, A. C. Loeffert, R. P. Olympia, G. Fedorchak, S. DeVita and Z. Gagnon, J. Sport Health Sci., 2023, 12, 369–378 CrossRef PubMed.
- D. Cialla-May, X.-S. Zheng, K. Weber and J. Popp, Chem. Soc. Rev., 2017, 46, 3945–3961 RSC.
- H. Abramczyk and B. Brozek-Pluska, Chem. Rev., 2013, 113, 5766–5781 CrossRef CAS PubMed.
- I. Pence and A. Mahadevan-Jansen, Chem. Soc. Rev., 2016, 45, 1958–1979 RSC.
- K. Kong, C. Kendall, N. Stone and I. Notingher, Adv. Drug Delivery Rev., 2015, 89, 121–134 CrossRef CAS PubMed.
- K. J. Ember, M. A. Hoeve, S. L. McAughtrie, M. S. Bergholt, B. J. Dwyer, M. M. Stevens, K. Faulds, S. J. Forbes and C. J. Campbell, npj Regener. Med., 2017, 2, 12 CrossRef PubMed.
- L. Yang, P. Li, H. Liu, X. Tang and J. Liu, Chem. Soc. Rev., 2015, 44, 2837–2848 RSC.
- R. Cunha, L. Lafeta, E. A. Fonseca, A. Barbosa, M. A. Romano-Silva, R. Vieira, A. Jorio and L. M. Malard, Analyst, 2021, 146, 2945–2954 RSC.
- S. Li, Z. Luo, R. Zhang, H. Xu, T. Zhou, L. Liu and J. Qu, Biosensors, 2021, 11, 365 CrossRef CAS PubMed.
- L. Shi, A. Klimas, B. Gallagher, Z. Cheng, F. Fu, P. Wijesekara, Y. Miao, X. Ren, Y. Zhao and W. Min, Adv. Sci., 2022, 9, 2200315 CrossRef PubMed.
- L. F. d. C. e Silva and M. S. Nogueira, Analyst, 2018, 143, 6037–6048 RSC.
- S. Tanwar, S. K. Paidi, R. Prasad, R. Pandey and I. Barman, Spectrochim. Acta, Part A, 2021, 260, 119957 CrossRef CAS PubMed.
- I. W. Schie, F. Placzek, F. Knorr, E. Cordero, L. M. Wurster, G. G. Hermann, K. Mogensen, T. Hasselager, W. Drexler and J. Popp, Sci. Rep., 2021, 11, 9951 CrossRef CAS PubMed.
- S. Fitzgerald, J. Akhtar, E. Schartner, H. Ebendorff-Heidepriem, A. Mahadevan-Jansen and J. Li, J. Biophotonics, 2023, 16, e202200231 CrossRef CAS PubMed.
- N. Wang, X. Wang, T. Yan, H. Xie, L. Wang, F. Ren, D. Chen, D. Zhang, Q. Zeng and S. Zhu, Sci. Adv., 2023, 9, eadf3504 CrossRef CAS PubMed.
- F. Conti, M. D'Acunto, C. Caudai, S. Colantonio, R. Gaeta, D. Moroni and M. A. Pascali, Sci. Rep., 2023, 13, 7282 CrossRef CAS PubMed.
- L. Huang, H. Sun, L. Sun, K. Shi, Y. Chen, X. Ren, Y. Ge, D. Jiang, X. Liu and W. Knoll, Nat. Commun., 2023, 14, 48 CrossRef CAS PubMed.
- C. Banbury, G. Harris, M. Clancy, R. J. Blanch, J. J. S. Rickard and P. Goldberg Oppenheimer, Sci. Adv., 2023, 9, eadg5431 CrossRef CAS PubMed.
- I. A. Bratchenko, L. A. Bratchenko, A. A. Moryatov, Y. A. Khristoforova, D. N. Artemyev, O. O. Myakinin, A. E. Orlov, S. V. Kozlov and V. P. Zakharov, Exp. Dermatol., 2021, 30, 652–663 CrossRef CAS PubMed.
- M. Jermyn, K. Mok, J. Mercier, J. Desroches, J. Pichette, K. Saint-Arnaud, L. Bernstein, M.-C. Guiot, K. Petrecca and F. Leblond, Sci. Transl. Med., 2015, 7, 274ra19 CAS.
- B. T. Murti, A. D. Putri, Y.-J. Huang, S.-M. Wei, C.-W. Peng and P.-K. Yang, RSC Adv., 2021, 11, 20403–20422 RSC.
- J. M. Cameron, C. Rinaldi, S. H. Rutherford, A. Sala, A. G. Theakstone and M. J. Baker, Appl. Spectrosc., 2022, 76, 393–415 CrossRef CAS PubMed.
- D. Kavungal, P. Magalhães, S. T. Kumar, R. Kolla, H. A. Lashuel and H. Altug, Sci. Adv., 2023, 9, eadg9644 CrossRef CAS PubMed.
- M. Paraskevaidi, C. L. Morais, K. M. Lima, J. S. Snowden, J. A. Saxon, A. M. Richardson, M. Jones, D. M. Mann, D. Allsop and P. L. Martin-Hirsch, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E7929–E7938 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |