Open Access Article
Open Access ArticleDynamic satellite–parent liposome networks for quantitative microreactions†
Jia-Qi
Tian
a and
Nan-Nan
Deng
 *ab
*ab
aShanghai Jiao Tong University, School of Chemistry and Chemical Engineering, Shanghai 200240, China. E-mail: n.deng@sjtu.edu.cn
bShanghai Jiao Tong University Sichuan Research Institute, Chengdu, Sichuan 610213, China
First published on 6th November 2024
Abstract
The hierarchical assembly of liposomes into interconnected networks forms the basis for creating rudimentary artificial multicellular systems. Each vesicle performs specialized functions both temporally and spatially, replicating the complexity of living tissues. Controlling the size and number of liposomes in artificial multicellular systems and their dynamic interactions are necessary for quantitative bioprocesses but remain challenging. Here, we develop a satellite–parent liposome network—a central parent liposome surrounded by smaller satellite liposomes. This structure spontaneously forms during the dewetting transition of microfluidically prepared complex double emulsions. Intriguingly, the adhesion strength between the satellites and the parent liposome can be tuned using environmental stimuli. The varying numbers of satellite liposomes provide an excellent platform for studying quantitative microreactions. To illustrate, we first explore the differences in molecular affinity between parent and satellite liposomes to achieve directional molecular transfer against concentration gradients. Then, we mimic quantitative signal transfer by performing enzymatic reactions, supplying substrates from different numbers of satellites to the parent liposomes. After the reaction, the satellites can be separated from the parent liposome on demand upon osmotic stimuli. This work showcases an exceptional dynamic liposome network that will facilitate the mimicry of the complexity of multicellular systems in vitro.
Introduction
Modular compartmentalization of chemical and biological processes is one of the vital organizational features in living systems.1–3 Cells perform the functions of life in separate compartments that communicate,4 cooperate,5 and sense environmental changes,6 thereby generating emergent properties essential for life. Mimicking this biological compartmentalization at micrometre scales holds significant promise for technological development across myriad fields, including multistep micro-/nano-reactors, synergistic drug delivery systems, and synthetic cell networks.7–13Liposomes, synthetic compartments containing bilayer membranes, have been widely utilized as artificial cells due to their ability to encapsulate and protect active substances, and thus have been used as microcompartments to mimic fundamental aspects of natural cells,12–14 such as gene transcription,15,16 protein synthesis,17,18 molecular transport,19 oscillation,20,21 cell division22,23 and energy production and storage.17,24 Furthermore, the hierarchical assembly of liposomes into interconnected networks forms the basis for creating rudimentary artificial multicellular systems, in which each vesicle performs specialized functions both temporally and spatially, replicating the complexity of living tissues. Activities such as cascade enzymatic reactions,25–27 thermoresponsive volume changes,28,29 predatory behaviour,30 signal transmission31,32 and power generation33,34 have already been demonstrated. Molecular transfer between each microcompartment is essential in driving these collective behaviors. To facilitate transmembrane transport of molecules, membrane proteins like α-hemolysin are often inserted into the bilayer membranes, forming nanopores that allow free diffusion driven by concentration gradients of cargoes.35–38 However, this free diffusion typically results in a homogeneous distribution of the target molecules within the sub-microcompartments,36,37 leading to relatively low transport efficiency. Moreover, current research on tissue-like multicompartment systems has primarily focused on qualitatively demonstrating the proof-of-concept of coupled cascade reactions among their subunits. Controlling the size and number of liposomes in artificial multicellular systems and their dynamic interaction necessary for more complex and quantitative microreactions remains challenging.
Here, we report an interesting satellite–parent liposome network—a central parent liposome surrounded by smaller satellite liposomes. This structure spontaneously forms during the dewetting transition of microfluidically prepared complex double emulsions, where the inner cores can be precisely controlled. Intriguingly, the adhesion strength between the satellites and the parent liposome can be tuned using environmental stimuli, such as osmotic pressure. We demonstrate that the liposome clusters can function as chemical reaction networks, with different reactants loaded into separate compartments. The varying numbers of satellite liposomes provide an excellent platform for studying quantitative microreactions. To illustrate, we first explore the differences in molecular affinity between parent and satellite liposomes to achieve directional molecular transfer against concentration gradients. Then, we mimic quantitative signal transfer by performing enzymatic reactions, supplying substrates from different numbers of satellites to the parent liposomes. After the reactions are complete, the satellites can be separated from the parent liposome on demand using osmotic stimuli. This work showcases an exceptional dynamic liposome network, facilitating the mimicry of the complexity of multicellular systems in vitro.
Results and discussion
Microfluidic construction of satellite–parent liposome networks
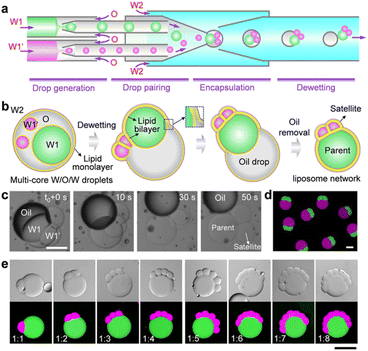
To construct satellite–parent liposome networks, we used a strategy of dewetting transition of water-in-oil-in-water (W/O/W).15 A two-stage microcapillary-based microfluidic device that consists of two droplet generators at the first stage was employed to prepare two different water-in-oil (W/O) droplets which were subsequently paired and encapsulated into W/O/W droplets at the second stage of the device, thereby forming complex W/O/W droplets with different inner droplets (W1 and W1′) (Fig. 1a and S1b†). The key to successful fabrication of satellite–parent liposomes is to control the size ratios of the W1/O to W1′/O droplets. The large one was identified as the parent, and the smaller ones were the satellites. The W1 and W1′ phases were aqueous solutions of 2% (w/v) PVA and 8% (w/v) PEG labelled with fluorescein isothiocyanate (FITC)-dextran and tetramethyl rhodamine (TRITC)-dextran, respectively. The O and W2 phases were respectively mixtures of chloroform and hexane (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 64, v/v) with 5 mg mL−1 DOPC, and aqueous solutions of 10% (w/v) PVA and 1.5% (w/v) Pluronic F-68. In the W/O/W templates, droplet interface bilayers (DIB) were first formed when the interior W1/O and W1′/O droplets were brought in contact. Then the DIB-connected interior droplet networks underwent a dewetting transition to form liposome networks via the spontaneous removal of the oil phases (Fig. 1b). The whole processes were driven by interfacial energy. By simply varying the flow rates, we prepared satellite–parent liposome networks with 1–8 and more satellite compartments (Fig. 1c and S2†). The key to successful fabrication of satellite–parent liposomes is to control the size ratios of the W1/O to W1′/O droplets. It requires a large W1/O droplet and several much smaller W1′/O droplets, and vice versa. When their sizes are comparable, they will form random liposome networks (Fig. S3†), instead of satellite–parent ones. Interestingly, the flow in the microchannels drags the “satellite” compartments into contact with each other (Fig. S4†). Furthermore, different types of satellite liposomes can also be created by adding more droplet generators at the first stage of the microfluidic device. For demonstration, satellite–parent liposome networks with two different types and numbers of satellites were successfully prepared (Fig. S5†). Similarly, when W1/O, W1′/O, W1′′/O droplets sizes are comparable, they may form liposome networks with various configurations but not the satellite–parent manner (Fig. S6†).
64, v/v) with 5 mg mL−1 DOPC, and aqueous solutions of 10% (w/v) PVA and 1.5% (w/v) Pluronic F-68. In the W/O/W templates, droplet interface bilayers (DIB) were first formed when the interior W1/O and W1′/O droplets were brought in contact. Then the DIB-connected interior droplet networks underwent a dewetting transition to form liposome networks via the spontaneous removal of the oil phases (Fig. 1b). The whole processes were driven by interfacial energy. By simply varying the flow rates, we prepared satellite–parent liposome networks with 1–8 and more satellite compartments (Fig. 1c and S2†). The key to successful fabrication of satellite–parent liposomes is to control the size ratios of the W1/O to W1′/O droplets. It requires a large W1/O droplet and several much smaller W1′/O droplets, and vice versa. When their sizes are comparable, they will form random liposome networks (Fig. S3†), instead of satellite–parent ones. Interestingly, the flow in the microchannels drags the “satellite” compartments into contact with each other (Fig. S4†). Furthermore, different types of satellite liposomes can also be created by adding more droplet generators at the first stage of the microfluidic device. For demonstration, satellite–parent liposome networks with two different types and numbers of satellites were successfully prepared (Fig. S5†). Similarly, when W1/O, W1′/O, W1′′/O droplets sizes are comparable, they may form liposome networks with various configurations but not the satellite–parent manner (Fig. S6†).
Dynamic morphologies in satellite–parent liposome networks
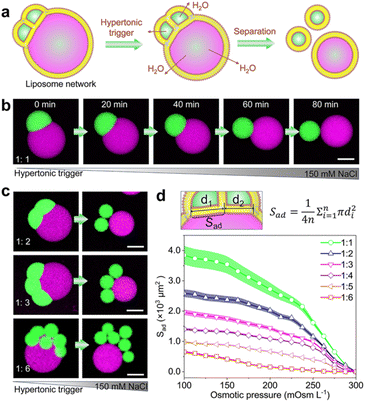
Membrane dynamics and remodelling are the key features of biological membranes and dominate diverse cellular processes, such as cell division,39 cell movements,40 and cargo transport.41 Next, we show that the satellite–parent liposome networks readily exhibit morphological dynamics of adhesion in response to environmental changes. As Fig. 2a shows, we used an osmotic shock to induce the separation of the satellites and the parent liposomes. When a hypertonic trigger (150 mM NaCl) was added into the satellite–parent liposome networks, the contact area between subunits was reduced gradually until complete separation (Fig. 2b). We attribute this dynamic process to the increase of surface-to-volume ratios of the sub-compartments due to the loss of internal water through the semipermeable lipid bilayers. The satellite–parent liposome networks deform and separate to minimize the energy associated with the line tension at the phase boundary.42 Similarly, for the networks with more satellite liposomes, hypertonic osmosis resulted in the complete separation of the liposomal sub-compartments (Fig. 2c, S7 and S8†). The adhesion areas (Sad) between the single satellite liposomes and the parent liposomes were calculated according to the morphological changes (Fig. 2d), representing the variations of the areas for mass transfer between the subunits. The adhesion area (Sad) decreases over time until satellite liposomes are completely separated from the parent liposomes and dropped to 0 (Fig. 2d).Directional and quantitative transport of molecular cargoes
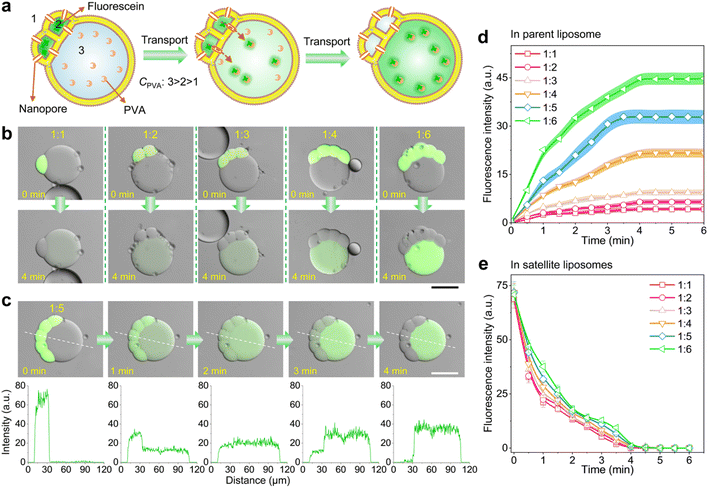
The resultant satellite–parent liposomes and their membrane dynamics provide exceptional models for quantitative transmembrane transport of molecular cargoes. To demonstrate, we here used a membrane peptide melittin to create nanopores in the bilayer membranes of the liposome networks, allowing for molecular transfer, and explored the differences of molecular binding affinity between cargoes and the different interiors of the satellite–parent liposomes to achieve the directional transport. Binding affinity describes the strength of the binding between a single biomolecule (such as protein or DNA) and its ligand/binding partner (such as drug or inhibitor), and is influenced by non-covalent intermolecular interactions like hydrogen bonding, electrostatic interactions, hydrophobic forces and van der Waals forces between the two molecules.43,44 We chose fluorescein as a model molecule, as it is impermeable to the lipid bilayer, and has a higher binding affinity with PVA than that with PEG.19 As Fig. 3a shows, we encapsulated fluorescein (0.5 μM L−1) and melittin monomers (2 μM) into the satellite compartments, and meanwhile created a concentration gradient of PVA as C3 > C2 > C1 across the bilayer membranes. After incubation, we observed that the mass transfer of fluorescein from the satellite compartments to the parent liposomes over time (Fig. 3b, c and S10†). The whole transport process took about 4 min. The fluorescence intensity of the parent compartments increased colligatively with the number of satellite compartments (Fig. 3d). The more satellites, the higher fluorescent signals in the parents. Strikingly, the speed and efficiency of the molecular binding affinity-directed transport is much higher than that of free diffusion without PVA gradients (Fig. S11†). After equilibrium was reached, the residual fluorescein molecules in the satellites were undetectable under a confocal microscope (Fig. 3e), indicating that the transport even occurs against the concentration gradients. No leakage of fluorescein to the outer environment was detected (Fig. 3c), though the nanopores exist in the whole membranes of the satellites. Additionally, binding affinity-mediated directional transport mechanisms are widely applicable and have great potential to enhance the design and functionality of synthetic tissues. By utilizing the differential binding affinities of biomolecules such as protein–protein interactions, DNA binding, and ligand-receptor dynamics proteins, one can create highly specific and directional transport systems that replicate the efficiency and precision seen in natural systems.Satellite–parent liposomes for quantitative micro-reactions
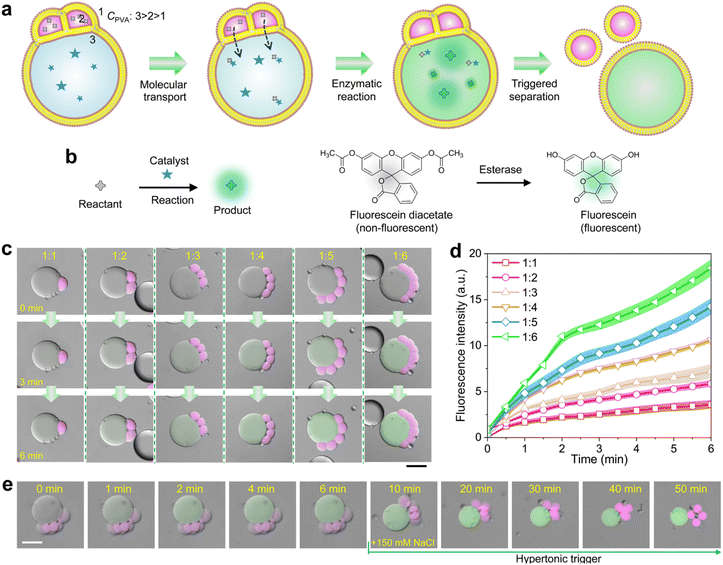
We finally demonstrate the satellite–parent liposome networks are promising platforms for quantitatively performing micro-reactions. We encapsulated 5% (w/v) PVA and enzymes (esterase, 0.1 units per mL) into the parent compartments, and 3% (w/v) PVA and the substrates (fluorescein diacetate) into the satellite, respectively. The PVA concentration in the outer phase was 1% (w/v) which is lower than that in all the liposomes.In this case, the substrates in satellite compartments were successfully transported into the parent liposomes, inducing the enzymatic degradation of the substrates to generate fluorescein in the parents (Fig. 4a–d and S12†). As Fig. 4c shows, the fluorescence intensity of fluorescein inside the parent compartments increases colligatively when the number of satellite compartments increase from 1 to 6 (Fig. 4d). After the reaction was completed, the detachment between the satellite compartments from the parent was performed by applying a hypertonic trigger, finally forming independent liposomes (Fig. 4e). Notably, by incorporating magnetic Fe3O4 nanoparticles into the satellite compartments, we can effectively collect either parent or satellite liposomes after the reaction and separation process. The satellite liposomes can be directed to the target site using a magnet, while the parent liposomes remain stationary (Fig. S13 and S14†). This method allows for the selective collection of different products post-reaction.
Conclusions
Dynamic satellite–parent liposome networks represent a versatile and efficient platform for quantitative bioprocesses. Their ability to maintain stable yet dynamic interactions enhance the precision and efficiency of biochemical processes. We demonstrated the directional transport of molecular cargoes from the satellites to their parent compartments by exploring molecular affinity. To show the potential of constructing synthetic cells, we performed life-like predator–prey interactions in the satellite–parent liposome networks by accomplishing enzymatic reactions. The satellite–parent liposome networks can be expanded to encompass many other multi-step chemical reaction pathways and more advanced life-like systems. Additionally, changes in the morphology of liposome networks, such as the detachment among liposomes, may provide insights into the remodelling of lipid bilayers during (synthetic) cell division. Compared to lipid-stabilized droplet networks,45–47 our liposome networks are oil-free and more biocompatible to physiological environments, making them particularly useful for the exploration of quantitative bioprocesses, construction of life-like functional microsystems, and application in real-time biosensing.Experimental
Microfluidics
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–23
000–23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1, 87–88% hydrolyzed, Sigma-Aldrich), a mixture of chloroform (Chl) and hexane (Hex)(36
000 g mol−1, 87–88% hydrolyzed, Sigma-Aldrich), a mixture of chloroform (Chl) and hexane (Hex)(36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 64, v/v) containing 5 mg mL−1 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Avanti Polar Lipids) as well as 10% (w/v) PVA with 1.5% (w/v) Pluronic F-68 (Sigma-Aldrich) were respectively utilized as inner water phase (W1), middle oil phase (O) and outer water phase (W2). 0.03% (w/v) Fluorescein isothiocyanate-dextran (FITC-Dextran, MW 40
64, v/v) containing 5 mg mL−1 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Avanti Polar Lipids) as well as 10% (w/v) PVA with 1.5% (w/v) Pluronic F-68 (Sigma-Aldrich) were respectively utilized as inner water phase (W1), middle oil phase (O) and outer water phase (W2). 0.03% (w/v) Fluorescein isothiocyanate-dextran (FITC-Dextran, MW 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1, Sigma-Aldrich) and 0.03% (w/v) tetramethyl rhodamine-dextran (TRITC-Dextran, MW 40
000 g mol−1, Sigma-Aldrich) and 0.03% (w/v) tetramethyl rhodamine-dextran (TRITC-Dextran, MW 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1, Sigma-Aldrich) were added in the W1 and W1′, respectively.
000 g mol−1, Sigma-Aldrich) were added in the W1 and W1′, respectively.
Osmotic pressure-induced separation of satellite–parent liposome networks
The freshly prepared satellite–parent liposome networks were collected in semi-enclosed silicone isolation chambers (diameter 9 mm, height 0.12 mm, SecureSealTM) covered with a glass coverside. To induce separation of liposome networks, we added 150 mM NaCl to the sample at a rate of 5 μL h−1 for a total volume of 7 μL. The entire process was captured by a CLSM (FV3000, Olympus).Transport of molecular cargoes between subcompartments of satellite–parent liposome networks
To achieve transport of fluorescent molecules from the satellite compartments (W1′) to the parent compartments (W1), the W1′, W1, O, and W2 phases were used as follows: W1′: an aqueous solution with 3% (w/v) PVA, 7% (w/v) PEG, 0.5 μM L−1 fluorescein (Sigma-Aldrich) and 2 μM melittin (Sigma-Aldrich); W1: an aqueous solution with 5% (w/v) PVA and 5% (w/v) PEG; O: a mixture of chloroform and hexane (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 64, v/v) containing 5 mg mL−1 DOPC; W2: an aqueous solution with 1% (w/v) PVA, 9% (w/v) PEG and 1.5% (w/v) Pluronic®F-68. The freshly prepared liposome networks were collected in a closed silicone isolation chamber (diameter 9 mm, height 0.12 mm, SecureSeal) covered with a glass coverslide, and then were captured at intervals of 0.5 min under a CLSM (FV3000, Olympus). Images were analyzed by ImageJ.
64, v/v) containing 5 mg mL−1 DOPC; W2: an aqueous solution with 1% (w/v) PVA, 9% (w/v) PEG and 1.5% (w/v) Pluronic®F-68. The freshly prepared liposome networks were collected in a closed silicone isolation chamber (diameter 9 mm, height 0.12 mm, SecureSeal) covered with a glass coverslide, and then were captured at intervals of 0.5 min under a CLSM (FV3000, Olympus). Images were analyzed by ImageJ.
Micro-reaction stoichiometry in satellite–parent liposome networks
W1′: an aqueous solution with 3% (w/v) PVA, 7% (w/v) PEG, fluorescein diacetate (10 μg mL−1, Sigma-Aldrich) in PBS buffer (pH 7.0). W1: an aqueous solution with 5% (w/v) PVA, 5% (w/v) PEG and 0.1 units per mL esterase (Shyuanye) in PBS buffer (pH 7.0). O: a mixture of chloroform and hexane (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 64, v/v) containing 5 mg mL−1 DOPC. W2: an aqueous solution with 1% (w/v) PVA, 9% (w/v) PEG and 1.5% (w/v) Pluronic®F-68 in PBS buffer (pH 7.0). The samples were collected and observed in the same manner as the transport of molecular cargoes between subcompartments of satellite–parent liposome networks. After the reaction was complete, we slowly added 150 mM NaCl into the samples to induce the satellite compartments to detach from the parent compartment. The processes were captured at intervals of 2 min under a CLSM (FV3000, Olympus).
64, v/v) containing 5 mg mL−1 DOPC. W2: an aqueous solution with 1% (w/v) PVA, 9% (w/v) PEG and 1.5% (w/v) Pluronic®F-68 in PBS buffer (pH 7.0). The samples were collected and observed in the same manner as the transport of molecular cargoes between subcompartments of satellite–parent liposome networks. After the reaction was complete, we slowly added 150 mM NaCl into the samples to induce the satellite compartments to detach from the parent compartment. The processes were captured at intervals of 2 min under a CLSM (FV3000, Olympus).
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
N. N. D. supervised the research. N. N. D. conceived the research and designed the experiments. J. Q. T. performed the experiments. J. Q. T. and N. N. D. wrote the paper. All authors approved the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 22278264), the Natural Science Foundation of Shanghai (Grant No. 22ZR1429000), the Interdisciplinary Program of Shanghai Jiao Tong University (project No. YG2022QN097), and Sichuan Emei talent plan (No. 2017).References
- J. W. Szostak, D. P. Bartel and P. L. Luisi, Nature, 2001, 409, 387–390 CrossRef CAS PubMed.
- C. Xu, N. Martin, M. Li and S. Mann, Nature, 2022, 609, 1029–1037 CrossRef CAS.
- N. A. Yewdall, A. F. Mason and J. C. Van Hest, Interface Focus, 2018, 8, 20180023 CrossRef.
- D. T. Hughes and V. Sperandio, Nat. Rev. Microbiol., 2008, 6, 111–120 CrossRef CAS PubMed.
- M. Archetti and K. J. Pienta, Nat. Rev. Cancer, 2019, 19, 110–117 CrossRef CAS PubMed.
- H. Du, J. M. Bartleson, S. Butenko, V. Alonso, W. F. Liu, D. A. Winer and M. J. Butte, Nat. Rev. Immunol., 2023, 23, 174–188 CrossRef CAS.
- P. A. Beales and T. K. Vanderlick, Adv. Colloid Interface Sci., 2014, 207, 290–305 CrossRef CAS PubMed.
- I. Ivanov, S. K. Smoukov, E. Nourafkan, K. Landfester and P. Schwille, Confl. Models. Orig. Life, 2023, 303–326 Search PubMed.
- W. Mu, Z. Ji, M. Zhou, J. Wu, Y. Lin and Y. Qiao, Sci. Adv., 2021, 7, eabf9000 CrossRef CAS.
- E. Rideau, R. Dimova, P. Schwille, F. R. Wurm and K. Landfester, Chem. Soc. Rev., 2018, 47, 8572–8610 RSC.
- N.-N. Deng, M. A. Vibhute, L. Zheng, H. Zhao, M. Yelleswarapu and W. T. S. Huck, J. Am. Chem. Soc., 2018, 140, 7399–7402 CrossRef CAS.
- N.-N. Deng, M. Yelleswarapu, L. Zheng and W. T. S. Huck, J. Am. Chem. Soc., 2017, 139, 587–590 CrossRef CAS.
- N.-N. Deng and W. T. S. Huck, Angew. Chem., Int. Ed., 2017, 56, 9736–9740 CrossRef CAS.
- Z.-H. Luo, C. Chen, Q.-H. Zhao and N.-N. Deng, Sci. Adv., 2024, 10, eadj4047 CrossRef CAS.
- N.-N. Deng, M. Yelleswarapu and W. T. Huck, J. Am. Chem. Soc., 2016, 138, 7584–7591 CrossRef CAS PubMed.
- J. M. Smith, D. Hartmann and M. J. Booth, Nat. Chem. Biol., 2023, 19, 1138–1146 CrossRef CAS PubMed.
- S. Berhanu, T. Ueda and Y. Kuruma, Nat. Commun., 2019, 10, 1325 CrossRef PubMed.
- T. Furusato, F. Horie, H. T. Matsubayashi, K. Amikura, Y. Kuruma and T. Ueda, ACS Synth. Biol., 2018, 7, 953–961 CrossRef CAS PubMed.
- J.-Q. Tian, M.-Y. Chang, C. Chen, Z.-H. Luo, W. T. Huck and N.-N. Deng, Nat. Chem. Eng., 2024, 1, 450–461 CrossRef.
- T. Litschel, B. Ramm, R. Maas, M. Heymann and P. Schwille, Angew. Chem., Int. Ed., 2018, 57, 16286–16290 CrossRef CAS PubMed.
- E. Godino, J. N. López, D. Foschepoth, C. Cleij, A. Doerr, C. F. Castellà and C. Danelon, Nat. Commun., 2019, 10, 4969 CrossRef.
- M.-Y. Chang, H. Ariyama, W. T. Huck and N.-N. Deng, Chem. Soc. Rev., 2023, 52, 3307–3325 RSC.
- N. De Franceschi, R. Barth, S. Meindlhumer, A. Fragasso and C. Dekker, Nat. Nanotechnol., 2024, 19, 70–76 CrossRef CAS PubMed.
- J. Zhao and X. Han, Nat. Commun., 2024, 15, 4956 CrossRef CAS PubMed.
- Y. Elani, R. V. Law and O. Ces, Nat. Commun., 2014, 5, 5305 CrossRef CAS PubMed.
- X. Wang, L. Tian, H. Du, M. Li, W. Mu, B. W. Drinkwater, X. Han and S. Mann, Chem. Sci., 2019, 10, 9446–9453 RSC.
- A. Galanti, R. O. Moreno-Tortolero, R. Azad, S. Cross, S. Davis and P. Gobbo, Adv. Mater., 2021, 33, 2100340 CrossRef CAS PubMed.
- P. Gobbo, A. J. Patil, M. Li, R. Harniman, W. H. Briscoe and S. Mann, Nat. Mater., 2018, 17, 1145–1153 CrossRef CAS.
- K. Ramsay, J. Levy, P. Gobbo and K. S. Elvira, Lab Chip, 2021, 21, 4574–4585 RSC.
- Y. Qiao, M. Li, D. Qiu and S. Mann, Angew. Chem., 2019, 131, 17922–17927 CrossRef.
- C. E. Hoskin, V. R. Schild, J. Vinals and H. Bayley, Nat. Chem., 2022, 14, 650–657 CrossRef CAS PubMed.
- C. Qi, X. Ma, J. Zhong, J. Fang, Y. Huang, X. Deng, T. Kong and Z. Liu, ACS Nano, 2023, 17, 16787–16797 CrossRef CAS PubMed.
- X. Zhang, C. Li, F. Liu, W. Mu, Y. Ren, B. Yang and X. Han, Nat. Commun., 2022, 13, 2148 CrossRef CAS PubMed.
- Y. Zhang, J. Riexinger, X. Yang, E. Mikhailova, Y. Jin, L. Zhou and H. Bayley, Nature, 2023, 620, 1001–1006 CrossRef CAS PubMed.
- M. J. Booth, V. R. Schild, A. D. Graham, S. N. Olof and H. Bayley, Sci. Adv., 2016, 2, e1600056 CrossRef.
- L. Song, M. R. Hobaugh, C. Shustak, S. Cheley, H. Bayley and J. E. Gouaux, Science, 1996, 274, 1859–1865 CrossRef CAS PubMed.
- A. Dupin and F. C. Simmel, Nat. Chem., 2019, 11, 32–39 CrossRef CAS PubMed.
- B. C. Buddingh’, J. Elzinga and J. C. M. van Hest, Nat. Commun., 2020, 11, 1652 CrossRef PubMed.
- J. G. Carlton, H. Jones and U. S. Eggert, Nat. Rev. Mol. Cell Biol., 2020, 21, 151–166 CrossRef CAS PubMed.
- S. SenGupta, C. A. Parent and J. E. Bear, Nat. Rev. Mol. Cell Biol., 2021, 22, 529–547 CrossRef CAS PubMed.
- W. O. Hancock, Nat. Rev. Mol. Cell Biol., 2014, 15, 615–628 CrossRef CAS.
- F. Jülicher and R. Lipowsky, Phys. Rev. E., 1996, 53, 2670 CrossRef PubMed.
- J. Nas, Phil. J. Health. Res. Dev., 2020, 24, 9–19 Search PubMed.
- X. Du, Y. Li, Y.-L. Xia, S.-M. Ai, J. Liang, P. Sang, X.-L. Ji and S.-Q. Liu, Int. J. Mol. Sci., 2016, 17, 144 CrossRef PubMed.
- Y. Elani, X. C. I. Solvas, J. B. Edel, R. V. Law and O. Ces, Chem. Commun., 2016, 52, 5961–5964 RSC.
- D. K. Baxani, A. J. Morgan, W. D. Jamieson, C. J. Allender, D. A. Barrow and O. K. Castell, Angew. Chem., Int. Ed., 2016, 55, 14240–14245 CrossRef CAS PubMed.
- G. Villar, A. D. Graham and H. Bayley, Science, 2013, 340, 48–52 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc04925j |
| This journal is © The Royal Society of Chemistry 2024 |