Specific and efficient knockdown of intracellular miRNA using partially neutralized phosphate-methylated DNA oligonucleic acid-loaded mesoporous silica nanoparticles†
Yi-Jung
Sung
 a,
Wei-Ting
Cai
a,
Yi-Ping
Chen
b,
Hardy Wai-Hong
Chan
c,
Cong-Kai
Lin
d,
Po-Hsiang
Wang
a,
Wei-Ting
Cai
a,
Yi-Ping
Chen
b,
Hardy Wai-Hong
Chan
c,
Cong-Kai
Lin
d,
Po-Hsiang
Wang
 *ae and
Wen-Yih
Chen
*a
*ae and
Wen-Yih
Chen
*a
aDepartment of Chemical and Materials Engineering, National Central University, Taoyuan 320, Taiwan
bGraduate Institute of Nanomedicine and Medical Engineering, Taipei Medical University, Taipei 110, Taiwan
cHelios Bioelectronics Inc., Hsinchu 302, Taiwan
dGraduate Institute of Biomedical Materials and Tissue Engineering, College of Biomedical Engineering, Taipei Medical University, Taipei 110, Taiwan
eGraduate Institute of Environmental Engineering, National Central University, Taoyuan 320, Taiwan
First published on 4th June 2024
Abstract
Antisense oligonucleotides (ASOs) are molecules used to regulate RNA expression by targeting specific RNA sequences. One specific type of ASO, known as neutralized DNA (nDNA), contains site-specific methyl phosphotriester (MPTE) linkages on the phosphate backbone, changing the negatively charged DNA phosphodiester into a neutralized MPTE with designed locations. While nDNA has previously been employed as a sensitive nucleotide sequencing probe for the PCR, the potential of nDNA in intracellular RNA regulation and gene therapy remains underexplored. Our study aims to evaluate the regulatory capacity of nDNA as an ASO probe in cellular gene expression. We demonstrated that by tuning MPTE locations, partially and intermediately methylated nDNA loaded onto mesoporous silica nanoparticles (MSNs) can effectively knock down the intracellular miRNA, subsequently resulting in downstream mRNA regulation in colorectal cancer cell HCT116. Additionally, the nDNA ASO-loaded MSNs exhibit superior efficacy in reducing miR-21 levels over 72 hours compared to the efficacy of canonical DNA ASO-loaded MSNs. The reduction in the miR-21 level subsequently resulted in the enhanced mRNA levels of tumour-suppressing genes PTEN and PDCD4. Our findings underscore the potential of nDNA in gene therapies, especially in cancer treatment via a fine-tuned methylation location.
1. Introduction
Colorectal cancer is the second leading cause of cancer-related death worldwide, largely attributed to challenges in early diagnosis and its high rate of metastasis.1,2 Conventional treatments, such as chemotherapy, are non-specific and usually accompanied by serious side effects. Therefore, there is a pressing need for more efficient therapies without these adverse effects, including treatments based on oligonucleotide derivatives.3 It is important to note that most cancers arise from genetic mutations. Emerging evidence suggests that the abnormal expression of specific proteins in cancer cells significantly impacts tumour growth.4–6 Therefore, regulating messenger RNA (mRNA) levels, which in turn control the downstream protein expression, offers a promising strategy for anti-tumour therapies with potentially fewer serious side effects.Antisense oligonucleotides (ASOs) are short DNA or RNA strands between 18–30 nucleotides long that target specific RNA sequences to modulate mRNA expression and translation.7 By binding to microRNA (miRNA, small RNA molecules that usually suppress mRNA expression through the RNA-induced silencing complex (RISC) to degrade mRNA), ASOs can regulate specific mRNA translation and protein expression.8 Their therapeutic action can also be achieved by directly binding to target mRNA, known as steric blockage, or triggering RNase H-dependent RNA degradation to decrease mRNA and protein levels.7 While several ASOs have been approved for diseases linked to the abnormal production of certain proteins, none have been approved for cancer, likely due to delivery challenges.9,10 Other issues with ASOs include weak binding to their target RNA and susceptibility to enzymatic breakdown due to poor bio-stability.
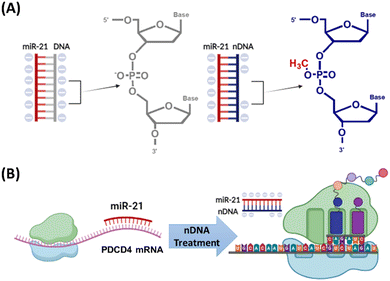
One common strategy to increase RNA affinity is through the chemical modification of ASO probes. For example, the phosphorothioate (PS) backbone involves replacing one of the non-bridging oxygen atoms in the phosphate backbone with a sulfur atom.11 While PS-modified ASOs perform in both the cytoplasm and nucleus, their therapeutic effectiveness can be influenced by their localization to various subcellular regions.12 Another strategy uses locked nucleic acid (LNA). The LNA modification connects the 2′-oxygen and 4′-carbon of the ribose through a methylene linkage, thereby locking the ribose in the 3′-endo conformation.11 This improves binding affinity to target RNA and enhances nuclease resistance. Beyond these strategies, we have introduced a novel approach. We developed a DNA analogue (Fig. 1A) containing site-specific methyl phosphotriester (MPTE) linkages on its backbone (methylating the phosphate groups of the phosphodiester bonds).13–16 This is termed as neutralized DNA (nDNA). The nDNA probes reduce the negative charge on the phosphate backbone through MPTE modification (Fig. 1A), thereby decreasing the electrostatic repulsion between nucleotides of the probe/RNA duplex, which results in enhanced hybridization affinity and duplex stability.13–16 Our several studies have demonstrated that nDNA probes exhibit higher hybridization affinity than canonical DNA probes when used as sensitive sequencing probes and polymerase chain reaction (PCR) primers.13–16 We can also synthesize an nDNA probe with designed MPTE frequency and location. From our latest study, we showed that nDNA with partial MPTE modification exhibited a heightened in situ hybridization (ISH) affinity to miR-21 (a microRNA frequently overexpressed in various cancers) compared to fully and highly MPTE-modified nDNA due to the reduced steric hindrance of nDNA.17 The enhanced hybridization affinity between the nDNA probe and target RNA also provides resistance to enzymatic degradation without compromising binding specificity. Thus, partially MPTE-modified nDNA also showed DNAase resistance compared to fully and highly MPTE-modified nDNA; the ISH affinity of the nDNA probe was comparable to that of the LNA probe. However, the potential of nDNA for using in antisense therapy at the cellular level remains unexplored.
While recent advancements have enhanced ASO hybridization efficiency and stability, delivering ASO probes to tumour tissues still has significant challenges, mainly due to various physiological barriers (e.g. enzyme degradation and interstitial fluid pressure).18 Therefore, the importance of carrier systems cannot be overstated. These carriers not only protect ASOs from nucleases in the bloodstream but also offer a way to overcome delivery barriers by tweaking properties like size, charge, and hydrophilicity.18 Among the various carriers developed in the past decade, mesoporous silica nanoparticles (MSNs) continue to garner much recent attention for their biocompatibility, high loading capacity, tuneable physicochemical properties, and site-specific functionalization.19,20 In particular, the unique pore channel and the large surface area of MSNs provide an excellent platform for multifunctional cargo loading and modification (onto particle pores and surfaces) compared to other nanocarriers. Typically, MSNs are functionalized with polyethylene glycol (PEG) to bolster their resistance to unintended biological interactions via creating a hydration layer around MSNs, termed anti-fouling.21 To elevate their loading and transfection efficiency, a cationic component like polyethyleneimine (PEI) is often introduced.21,22 PEI facilitates stronger interactions with negatively charged nucleotides and enhances cellular uptake, contributing to the interactions with the anionic phospholipid headgroups of the cell membrane. Furthermore, while PEI-coated MSNs are internalized through endocytosis, PEI induces osmotic swelling in endosomes, leading to endosome rupture and subsequently releasing the cargo into the cytoplasm, a phenomenon known as the proton sponge effect.22 We recently developed an MSN variant functionalized with both PEI and PEG (MSN-PEG/PEI).23 This design harnesses the stability benefits of PEG and the cellular uptake and endosomal escape properties of PEI, effectively addressing cytosolic barriers. Subsequently, MSN-PEG/PEI refers to MSNs. Given these attributes, we sought to perfect a strategy employing MSNs to transport loaded nDNA to target RNA in colorectal cancer cells.
In this study, we successfully developed the first nDNA-loaded MSN system tailored for the cytosolic transport of nDNA ASO probes, achieving effective miRNA silencing via a fine-tuned MPTE location on the ASO backbone, which resulted in an elevated downstream mRNA level (Fig. 1B). Our results revealed that partial MPTE-modification in the middle position of nDNA efficiently suppressed miR-21 levels in colorectal cancer cell HCT116, consequently altering downstream mRNA expression. Specifically, miR-21 regulates the expression of various genes at the post-transcriptional level such as tumour suppressor programmed cell death 4 (PDCD4) and phosphatase and tensin homolog (PTEN). Successful knockdown of miR-21 by nDNA ASO probes resulted in enhanced PDCD4 and PTEN mRNA levels in HCT116 and indirectly led to tumour growth suppression. Through a well-tuned MPTE location, the nDNA-loaded MSN system effectively regulated miRNA and mRNA expression in cancer cells. This foundational work paves the way for future in vivo studies, suggesting a promising avenue for next-generation oligonucleotide therapeutics in oncology.
2. Results and discussion
2.1 Inhibition of miR-21 RT by N4-mid nDNA in synthetic human plasma
When DNA-based ASO probes bind to miR-21 strands, they can interfere with the reverse transcription (RT) of miR-21. The efficiency of this inhibition can be measured using quantitative RT-PCR (RT-qPCR). Specifically, a higher number of cycle thresholds (Ct) indicate a stronger binding affinity of duplex and stronger RT inhibition by the probe, therefore resulting in a lower miR-21 amount. In our previous study, we confirmed that the partially methylated N4 nDNA, featuring four MPTE modifications at the 5′ end, exhibited enhanced ISH with the miR-21 strand and increased enzyme resistance compared to fully or highly methylated nDNA and canonical DNA.17 In this study, we further investigate the inhibitory potential of specific MPTE locations within the N4 nDNA, including those at the 5′ end (N4-5′ nDNA), 3′ end (N4-3′ nDNA), and middle positions (N4-mid nDNA), within synthetic human plasma (Fig. 2). According to the RT-qPCR results, all ASO probes exhibited significantly more miR-21 suppression than both nontreatment (NT) and scramble groups (Fig. 2B) suggesting successful ISH with the miR-21 strand using ASO probes. Due to the strong duplex binding affinity and enzyme resistance, all nDNA probes showed significantly reduced miR-21 levels than the DNA probe even in the presence of human plasma (Fig. 2B). The greater suppression of N4-5′ nDNA compared to that of N4-3′ nDNA (Fig. 2B) was likely attributed to the cellular machinery that typically degrades RNA in a 5′ to 3′ direction.24,25 Thus, modifications at the 5′ end might be more effective than those at the 3′ end. Among these probes, N4-mid nDNA displayed the highest miR-21 suppression, significantly surpassing those of N4-5′ nDNA, N4-3′ nDNA, and canonical DNA (Fig. 2B) indicating that N4-mid nDNA possesses the strongest hybridization affinity than N4-5′ nDNA, N4-3′ nDNA, and DNA. Computer simulations reveal that a duplex structure is more easily unzipped from middle locations than terminal positions.26 This suggests that stabilizing the middle of the duplex with MPTE modifications might more effectively sustain nDNA–RNA binding compared to modifications at both ends. Therefore, the enhanced miR-21 inhibition via N4-mid nDNA over N4-5′ and N4-3′ nDNA (Fig. 2B) possibly resulted from both the difficulty in duplex unwinding from middle locations on the duplex and general enhanced duplex stability via partial MPTE modification. These results indicate that methylation at different positions resulted in varying levels of RT suppression, attributed to differences in hybridization affinity. The N4-mid nDNA probe exhibited the most efficient inhibition of miR-21 RT, consequently making the N4-mid nDNA the preferred option for subsequent assessments.2.2 MSN characterization
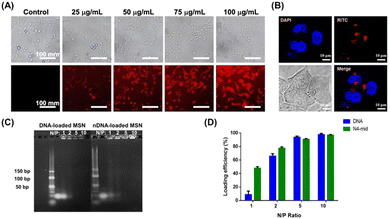
MSNs were initially functionalized with PEG to enhance their anti-fouling properties and with PEI to facilitate nucleotide adsorption and endosomal escape. The successful modification was confirmed by thermogravimetric analysis (TGA) (Fig. S1, ESI†). Subsequently, MSNs refer to MSN-PEG/PEI unless otherwise indicated. To investigate the potential uptake of MSNs by colorectal tumour cells, we labelled MSNs with fluorescent rhodamine B isothiocyanate (RITC). The RITC-labelled MSNs were then administered to the cells, and the cellular uptake within 2.5 hours was analysed through fluorescence microscopy and confocal laser scanning microscopy (CLSM). Intracellular fluorescence was only observed in cells treated with RITC-labelled MSNs indicating that MSNs can be internalized into cells within 2.5 hours (Fig. 3A and B). While PEI, which was modified onto MSNs, might be cytotoxic in certain situations,27 we confirmed that no significant cytotoxicity was observed upon MSN internalization for up to 72 hours (Fig. S2, ESI†). MSN-loaded nDNA (N4-mid nDNA) was then prepared by directly mixing negatively charged ASO probes and positively charged MSNs at various N/P ratios (the ratio of amine groups to nucleic acid phosphate groups) to optimize the amount of nDNA charged onto the MSNs (Fig. 3C and D). The amount of nucleic acid charged onto MSNs increased with the N/P ratio, primarily due to the higher concentration of positively charged PEI in the system. At an N/P ratio below 2, the amount of nDNA charged onto MSNs was higher than that of DNA. Variations in the electrostatic interaction with PEI possibly influenced these differences between the nDNA and DNA loading. Comparatively, the nDNA probe carries a lower negative charge, allowing more nDNA probes to be adsorbed by the positively charged PEI. At an N/P ratio of 5, nDNA and DNA loading onto MSNs exceeded 95% (Fig. 3C and D). This indicates that nDNA and DNA probes can be efficiently loaded onto MSNs under these conditions. Consequently, an N/P ratio of 5 was selected for all subsequent experiments.The hydrodynamic size of the MSNs and probe-loaded MSNs in water was approximately 40 nm (Table 1 and Fig. S3, ESI†). This size range, larger than 5 nm, effectively prevents renal filtration,18 yet remains small enough (less than 100 nm) to penetrate tumour cells through the enhanced permeability and retention (EPR) effect.18 In addition, the ζ-potential of the MSNs and probe-loaded MSNs in water was measured to be around +20 mV (Table 1 and Fig. S3, ESI†). As mentioned above, the slight positive charge is expected to enhance cellular uptake by interacting with the negatively charged lipid heads in cell membranes and facilitate endosomal escape by the proton sponge effect. The similarity in both the hydrodynamic size and ζ-potential between the MSNs and probe-loaded MSNs is likely attributed to the adsorption of most anionic ASO probes within the cationic PEI-modified mesoporous structure of MSNs. We subsequently examined serum stability using probe-loaded MSNs (Fig. S4, ESI†). The slightly increased particle size observed was attributed to the positively charged MSNs, which might induce protein adsorption in the presence of serum (Fig. S4A, ESI†). The slightly increased polydisperse index (PDI) resulted from the presence of both MSNs and serum (Fig. S4B, ESI†). There were no significant changes in the particle size, and the PDI was observed over 24 hours (Fig. S4, ESI†). Additionally, both nDNA- and DNA-loaded MSNs exhibited less than 10% release of ASO probes for 24 hours in the presence of serum (Fig. S5, ESI†). The results indicate that probe-loaded MSNs possessed serum stability.
| Size (nm) | PDI | ζ-potential (mV) | |
|---|---|---|---|
| MSNs | 38 ± 0.2 | 0.220 ± 0.001 | 23 ± 0.5 |
| DNA-loaded MSNs | 40 ± 0.2 | 0.190 ± 0.019 | 19 ± 0.5 |
| nDNA-loaded MSNs | 42 ± 0.2 | 0.271 ± 0.009 | 23 ± 0.5 |
2.3 Knockdown of miR-21 in HCT116 by nDNA-loaded MSNs
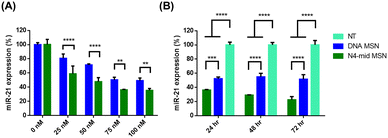
Given the efficient binding affinity of the N4-mid nDNA probe to miR-21 and the ability to induce miR-21 RT inhibition in synthetic human plasma medium (Fig. 2), and considering that the probe-loading, particle size, and ζ-potential of MSNs were suitable for probe delivery to cancer cells (Table 1 and Fig. 3C, D), our next objective was to evaluate the potential of the N4-mid nDNA probe to suppress miR-21 expression in cancer cells HCT116. At 0 nM ASO concentration, miR-21 expression remained unchanged (Fig. 4A). In both the N4-mid nDNA-loaded and DNA-loaded MSN-treated groups, miR-21 expression decreased, and the reduction level increased with the ASO concentration (Fig. 4A). Successful MSN internalization (Fig. 3A) without causing cytotoxicity (Fig. S2, ESI†) enabled the ASO probe targeting miR-21 to reduce miR-21 levels once the ASO probes were released into the cytosol. Remarkably, the N4-mid nDNA-treated group exhibited significant suppression of miR-21 expression compared to the DNA-treated group at all concentrations (Fig. 4A). The suppression level showed no differences at a probe concentration of 75 nM or higher in the given cell density. Therefore, cells were treated with a 75 nM probe concentration in subsequent assessments. Next, we investigate the knockdown efficiency during a 72-h incubation (cell viability started to decrease after a 72-h incubation (Fig. S6, ESI†) due to the limited area of the well-plate). Both N4-mid nDNA-loaded and DNA-loaded MSN treatments showed significant miR-21 repression over the NT group from 24 to 72-h incubation (Fig. 4B). Notably, the N4-mid nDNA probe demonstrated a significantly greater repression level than the DNA probe during 72-h incubation (Fig. 4B). These results suggest that the reduced negative charge on the phosphate backbone of nDNA enhances the affinity of the nDNA/miR-21 duplex by mitigating electrostatic repulsion between nDNA and the miR-21 duplex in comparison with the canonical DNA/miR-21 duplex. Consequently, this leads to the suppression of miR-21 levels in HCT116. Since miRNA is known to participate in the degradation of target mRNA through RISC, we subsequently investigated whether the reduction in miR-21 by N4-mid nDNA would increase downstream mRNA expression.2.4 PDCD4 and PTEN mRNA upregulation in HCT116 via miR-21 knockdown
miR-21 has been reported to regulate the expression of numerous target mRNAs, including PDCD4 and PTEN.28 PDCD4 is involved in apoptosis regulation and acts as a tumour suppressor, while PTEN is a tumour suppressor gene that regulates cell growth and death. Therefore, we further evaluated the mRNA expression of PDCD4 and PTEN using N4-mid nDNA-loaded MSNs in 72-h treatment. According to the RT-qPCR results, both N4-mid nDNA-loaded MSN treatment and DNA-loaded MSN treatment showed significantly higher PDCD4 and PTEN mRNA levels than the NT group (Fig. 5A and B). Notably, the N4-mid nDNA group exhibited significantly greater PDCD4 and PTEN mRNA levels than the DNA group (Fig. 5A and B). These results indicate that by knocking down miR-21 expression with the N4-mid nDNA probe, we successfully induced an increase in PDCD4 and PTEN mRNA expression, preventing miR-21 from targeting PDCD4 and PTEN mRNA to cause mRNA degradation. Since PDCD4 and PTEN are tumour suppressors, we observed whether N4-mid nDNA-loaded MSNs could indirectly lead to the suppression of cancer cell growth. According to the cell viability assay, N4-mid nDNA-loaded MSN treatment significantly suppressed cell growth compared to DNA-loaded MSN treatment and NT group during 72-h incubation (Fig. S7, ESI†). In contrast, the DNA group showed no significant difference compared to the NT group (Fig. S7, ESI†). The results suggested that enhanced PDCD4 and PTEN mRNA levels via N4-mid nDNA treatment indirectly suppressed cancer cell growth. Overall, we successfully utilized the N4-mid nDNA probe to target intracellular miRNA and induce miRNA knockdown, and downstream mRNA regulation.2.5 Discussion
nDNA synthesis confers flexibility in frequencies and locations of MPTE modification. The modification location influences the binding affinity of nDNA to the miR-21 duplex. The data in this study confirmed that nDNA with partial MPTE modification at the middle position effectively inhibited miR-21 reverse transcription when nDNA and miR-21 shared similar nucleotide numbers. Our results highlight the promising potential of nDNA as an ASO probe in gene cancer therapy. The regulation of gene expression involves a complex, multi-step process. The complexity extends from transcription to translation regulation. Among the genes regulated, miR-21 plays a significant role in controlling tumour suppressors like PDCD4 and PTEN, which were specifically studied in this research. Notably, these genes can also be influenced by various other miRNAs. For instance, miR-25 has been identified as targeting PTEN mRNA in certain cancer types,29,30 while miR-182 has been reported to target PDCD4 mRNA.31,32 However, in this study, relying on a single nDNA ASO probe solely to regulate one miR-21 is far from an effective cancer cell treatment due to the complex and dynamic interactions between miRNA and gene. Promising prospects lie in future improvements for this system. For example, from an ASO perspective, delivering multiple nDNA probes to knockdown several miRNAs simultaneously could present a more robust and precise strategy to enhance anti-tumour capabilities by elevating mRNA levels of tumour suppressors. Additionally, directly targeting mRNA using MPTE-modified oligonucleotide may be a potential method to silence overexpressed mRNAs such as polo-like kinase 1 (PLK1) in cancer cells.33,34PEG and PEI are commonly employed as surface modification agents for nanoparticles. However, some studies have reported that PEG-modified nanoparticles may induce immune responses. The permanent positive charge of PEI may lead to protein adsorption during circulation. Therefore, exploring alternative surface modification materials for MSNs is crucial to enhance the delivery efficacy. For instance, pH-sensitive ethylenediamine-based polyzwitterions exhibit a surface charge switch from neutral to positive under tumourous and endosomal pH conditions.35,36 This property enhances tumour accumulation by facilitating improved cellular uptake and endosomal escape.35,36 Similarly, the tertiary amine-based polymers convert to a positive charge within tumour microenvironments and exhibit comparable hydrophilicity to PEG.37 Additionally, pH-labile materials facilitate drug release within slightly acidic tumour microenvironments.38 Improvements in surface modification for MSNs hold the potential to advance the delivery system's efficiency and tumour-targeting capabilities in the future.
3. Conclusions
In this study, we developed a novel ASO-MSN complex using nDNA with site-specific MPTE modifications on its backbone to improve its targeting affinity and stability by reducing negative charge repulsion in the duplex. The binding affinity of the duplex was affected by fine-tuning the MPTE locations. We successfully knocked down cytosolic miR-21 of HCT116 through the N4-mid nDNA-loaded MSNs with sustained knockdown efficacy for up to 72 hours. Additionally, miR-21 silencing with the nDNA probes led to enhanced downstream mRNA expression of PDCD4 and PTEN, resulting in suppressed tumour growth compared to that with the canonical DNA probes. These findings lay the foundation for future in vivo investigations and signify a promising strategy for ASO therapeutics in cancer treatment.4. Experimental section
4.1 Materials
Chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless specified otherwise. McCoy's 5A medium, fetal bovine serum (FBS), penicillin, phosphate-buffered saline (PBS), trypsin–EDTA, and human plasma-like medium (HPLM) were purchased from Invitrogen (Waltham, MA, USA). Bovine serum albumin (BSA) was obtained from Merck Millipore (Burlington, MA, USA).4.2 Cells
Human colorectal cancer cells (HCT116) were provided by Professor Li-Jen Su from the Institute of Systems Biology and Bioinformatics, National Central University (Taoyuan, Taiwan). Cells were cultured in McCoy's 5A medium with 10% FBS and 1% penicillin (w/v) and were maintained at 37 °C in an incubator (5% CO2, 95% humidified environment).4.3 nDNA probes
nDNA and DNA oligonucleotide primers were purchased from Helios Biotech, Inc. (Taipei, Taiwan). An antisense DNA oligonucleotide against miR-21 with the sequence 5′-TCA ACA TCA GTC TGA TAA GCTA-3′ was modified through site-specific inter-nucleoside methyl phosphotriester (MPTE) linkages in the designated positions (Table S1, ESI†). MPTE modification was carried out according to procedures developed in our previous research.174.4 RNA hybridization
The hybridization of nDNA with miR-21 strand and DNA with miR-21 strand was performed following established protocols described previously.17 Briefly, the 10 μL hybridization reaction contained 1 μL miR-21 (1 nM) (mdbio, Taipei, Taiwan), one microliter of DNA or nDNA probe (40 nM), eight microliters of 1× HPLM, and was incubated at 37 °C for 30 min.4.5 RT-qPCR
To stabilize miR-21 (template sequence: 5′-UAG CUU AUC AGA CUG AUG UUG A-3′), poly(A) tails were added to both using E. coli poly(A) polymerase (New England Biolabs (NEB), Ipswitch, Massachusetts, USA) according to the manufacturer's instructions. miRNA containing poly(A) (1 nM) was then converted into cDNA using 2× Fast Premix (10 μL). RT primer sequence was described in our previous research.17 RT primer (2 μL), and HiScript I Reverse Transcriptase (1 μL) were used according to the manufacturer's instructions. RT was conducted at 42 °C for 30 min, followed by heating to 85 °C to inactivate reverse transcriptase. A two-step RT-qPCR was performed by first preparing a mixture (10 μL) containing 1 μL cDNA mixed with 1 μL forward/reverse primers (mdbio) (forward: 5′-TCA GTA GCT TAT CAG ACT GAT G-3′; reverse: 5′-CGT CCA GTT TTT TTT TTT TTT TCA AC-3′), RealQ Plus 2× Master Mix Green (5 μL) (Ampliqon, Odense, Denmark), and UitraPure distilled water (2 μL) (Invitrogen) followed by analysis on a StepOne™ Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) as described previously.17 Ct values were analyzed through StepOne™ software v2.3. The PCR amplification cycles consisted of an initial heat activation for 15 minutes at 95 °C, followed by denaturation for 15 seconds at 95 °C and annealing for 1 minute at 60 °C, repeated for 40 cycles.4.6 Synthesis and characterization of MSNs
MSNs functionalized with PEI and PEG (MSN-PEG/PEI) were synthesized following our previously published procedure.23 Briefly, an aqueous ammonia solution was prepared to dissolve hexadecyltrimethylammonium bromide (CTAB) as a surfactant. 3-Aminopropyltrimethoxysilane (APTMS)-conjugated RITC was added, followed immediately by the dropwise addition of tetraethyl orthosilicate (TEOS) diluted in 99.5% ethanol. PEI-silane and PEG-silane diluted in ethanol were introduced for surface modification. The nanoparticles were aged and subjected to hydrothermal treatment followed by the removal of surfactants. Subsequently, MSN refers to MSN-PEG/PEI. TGA was performed to confirm the successful modification (Fig. S1, ESI†). Hydrodynamic size, PDI, and ζ-potential of MSNs were determined using a Zetasizer Nano ZS90 (Malvern Panalytical, Worcestershire, UK).4.7 Preparation of nDNA-loaded MSNs
N4-5′, N4-mid, and N4-3′ nDNA probes (75 nM) and MSNs were mixed at different N/P ratios and dissolved in ultrapure water (Thermo Scientific, Waltham, MA, USA) to a total volume of 6 μL followed by a one-hour incubation at room temperature to form an nDNA/MSN complex. Control DNA-loaded MSN complexes were prepared following the same method using a canonical ASO sequence (Table S1, ESI†). Hydrodynamic size, PDI, and ζ-potential of nDNA–MSN were determined using a Zetasizer Nano ZS90 (Malvern Panalytical).4.8 Agarose gel electrophoresis and loading efficiency of nDNA or DNA onto MSNs
Agarose gel electrophoresis was performed in 4% (w/v) agarose gels containing 1× TAE buffer (Bioman Scientific, New Taipei, Taiwan), also used as the running buffer. Each sample (97.6 μg mL−1) (probe-loaded MSNs and probes) was mixed with 1.2 μL 6× orange DNA loading dye (Thermo Scientific), and 6 μL was loaded onto the gel, followed by gel running at 120 V for 30 min at room temperature. DNA and nDNA were stained using 1× SYBRTM Gold nucleic acid gel stain (Invitrogen) at 60 rpm for 30 min. Agarose gel electrophoresis results were analyzed through ImageJ software (1.52a) (National Institutes of Health, Bethesda, Maryland, USA). The amount of nucleic acid charged onto the MSNs was calculated by comparing the fluorescence of the band representing probe-loaded MSNs (which represents the amount of nucleic acid) to the band representing the probe.4.9 Serum stability of the probe-loaded MSNs
Serum stability was performed by incubating probe-loaded MSNs in McCoy’ 5A medium containing 10% FBS at 37 °C at designated intervals. Hydrodynamic size and PDI were measured at different time points using a Zetasizer Nano ZS90 (Malvern Panalytical).4.10 Release test
Suspended nDNA-loaded MSNs (43.94 μg mL−1; 100 μL) and DNA-loaded MSNs (43.94 μg mL−1; 100 μL) were placed into separate dialysis membranes (Slide-A-Lyzer™ MINI Dialysis Devices, 10k MWCO, Thermo Scientific), each containing 1.5 mL of McCoy’ 5A medium with 10% FBS. These membranes were then maintained at 37 °C using a water bath. At designated intervals, a 20 μL sample was extracted from each membrane to measure the DNA concentration. After sampling, 20 μL of fresh medium was added to replenish the system. The DNA concentration over time was assessed using a BioDrop μLite+ (BioDrop Ltd, Cambridge, UK).4.11 Cellular uptake
RITC-conjugated MSNs were synthesized as previously reported.23 HCT116 cells were seeded into a 6-well plate (Thermo Scientific) at a density of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ×
× ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105 cells per well. After 24-h incubation in an incubator (37 °C, 5% CO2, 95% humidified environment), cells were washed with 1× PBS and the culture medium (McCoy's 5A medium containing 10% FBS and 1% penicillin) was replaced with 2.5 mL fresh medium containing various concentrations of RITC-conjugated MSNs, followed by a 2.5 h treatment. Subsequently, the fluorescence of the cells was measured using a fluorescence microscope (10× objective) (IM-3FL4, OPTIKA, Ponteranica, Italy). HCT116 cells were seeded into a 6-well plate at a density of 2
105 cells per well. After 24-h incubation in an incubator (37 °C, 5% CO2, 95% humidified environment), cells were washed with 1× PBS and the culture medium (McCoy's 5A medium containing 10% FBS and 1% penicillin) was replaced with 2.5 mL fresh medium containing various concentrations of RITC-conjugated MSNs, followed by a 2.5 h treatment. Subsequently, the fluorescence of the cells was measured using a fluorescence microscope (10× objective) (IM-3FL4, OPTIKA, Ponteranica, Italy). HCT116 cells were seeded into a 6-well plate at a density of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ×
× ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105 cells per dish. After 24-h incubation in an incubator (37 °C, 5% CO2, 95% humidified environment), cells were washed with 1× PBS and the culture medium (McCoy's 5A medium containing 10% FBS and 1% penicillin) was replaced with 2.5 mL fresh medium containing 100 μg mL−1 of RITC-conjugated MSNs, followed by a 2.5 h treatment. Subsequently, cells were washed with 1× PBS and stained with DAPI (4′,6-diamidino-2-phenylindole, SouthernBiotech, Birmingham, AL, USA) according to the manufacturer's instructions. The fluorescence of the cells was observed using a CLSM (Leica STELLARIS 8 Confocal Spectral Scanning System with Multi-Band Spectrophotometer, Wetzlar, Germany).
105 cells per dish. After 24-h incubation in an incubator (37 °C, 5% CO2, 95% humidified environment), cells were washed with 1× PBS and the culture medium (McCoy's 5A medium containing 10% FBS and 1% penicillin) was replaced with 2.5 mL fresh medium containing 100 μg mL−1 of RITC-conjugated MSNs, followed by a 2.5 h treatment. Subsequently, cells were washed with 1× PBS and stained with DAPI (4′,6-diamidino-2-phenylindole, SouthernBiotech, Birmingham, AL, USA) according to the manufacturer's instructions. The fluorescence of the cells was observed using a CLSM (Leica STELLARIS 8 Confocal Spectral Scanning System with Multi-Band Spectrophotometer, Wetzlar, Germany).
4.12 Cell viability assay
HCT116 cells were seeded into 96-well microplates (Corning, Corning, NY, USA) at a density of 7 × 103 cells per well with culture medium containing 10% FBS and 1% penicillin. After 24-h incubation at 37 °C, cells were treated with a fresh 0.1 mL medium containing various concentrations of MSNs or ASO probe-loaded MSNs followed by further incubation for 24 to 72 h. Cell viability was evaluated using Cell Counting Kit-8 (Abcam, Cambridge, UK) according to the manufacturer's instructions. The living cell density was estimated in a 96-well plate by measuring λ450nm using a SpectraMax iD3 microplate reader (Molecular Devices, Downingtown, PA, USA).4.13 RNA extraction from HCT116
HCT116 cells were seeded into a 6-well plate (Thermo Scientific) at a density of 2 × 105 cells per well with culture medium containing 10% FBS and 1% penicillin. After 24-h incubation in an incubator (37 °C, 5% CO2, 95% humidified environment), cells were washed with 1× PBS and the culture medium (McCoy's 5A medium containing 10% FBS and 1% penicillin) was replaced with 2.5 mL fresh medium containing various concentrations of ASO, followed by incubation at designated intervals. After incubation (37 °C, 5% CO2, 95% humidified environment), the culture medium was removed and cells were washed twice with 1× PBS. Cells were lysed and RNAs were extracted using the miRNeasy Micro Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions.4.14 miR-21 knockdown assessment
HCT116 cells were seeded into a 6-well plate at a density of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ×
× ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105 cells per well followed by 24-h incubation (37 °C, 5% CO2, 95% humidified environment). After cells were washed twice using 1× PBS, cells were treated with fresh medium containing probes or probe-loaded MSNs (25 to 100 nM). A group of non-treatment was prepared as a negative control. After 24 to 72-h incubation (37 °C, 5% CO2, 95% humidified environment), cells were lysed and RNAs were extracted using the miRNeasy Micro Kit (Qiagen). After RNAs were extracted from cells, RT-qPCR was performed as described above. The cDNA reaction mixtures were prepared with appropriate primer pairs and RealQ Plus 2× Master Mix Green (5 μL) (Ampliqon). miR-21 levels were calculated using the comparative Ct method with U6 as the endogenous housekeeping gene. Gene expression was normalized to the value for nontreated cells. The following primer pairs were used: miR-21: forward: 5′-TCA GTA GCT TAT CAG ACT GAT G-3′; reverse: 5′-CGT CCA GTT TTT TTT TTT TTT TCA AC-3′; U6: 5′-CTCGCTTCGGCAGCACAT-3′ (forward); 5′-TTTGCGTGTCATCCTTGCG-3′ (reverse).
105 cells per well followed by 24-h incubation (37 °C, 5% CO2, 95% humidified environment). After cells were washed twice using 1× PBS, cells were treated with fresh medium containing probes or probe-loaded MSNs (25 to 100 nM). A group of non-treatment was prepared as a negative control. After 24 to 72-h incubation (37 °C, 5% CO2, 95% humidified environment), cells were lysed and RNAs were extracted using the miRNeasy Micro Kit (Qiagen). After RNAs were extracted from cells, RT-qPCR was performed as described above. The cDNA reaction mixtures were prepared with appropriate primer pairs and RealQ Plus 2× Master Mix Green (5 μL) (Ampliqon). miR-21 levels were calculated using the comparative Ct method with U6 as the endogenous housekeeping gene. Gene expression was normalized to the value for nontreated cells. The following primer pairs were used: miR-21: forward: 5′-TCA GTA GCT TAT CAG ACT GAT G-3′; reverse: 5′-CGT CCA GTT TTT TTT TTT TTT TCA AC-3′; U6: 5′-CTCGCTTCGGCAGCACAT-3′ (forward); 5′-TTTGCGTGTCATCCTTGCG-3′ (reverse).
4.15 mRNA expression evaluation
HCT116 cells were seeded into a 6-well plate at a density of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ×
× ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 105 cells per well followed by 24-h incubation (37 °C, 5% CO2, 95% humidified environment). After cells were washed twice using 1× PBS, cells were treated with fresh medium containing probes or probe-loaded MSNs (75 nM). A group of non-treatment was prepared as a negative control. After 72-h incubation (37 °C, 5% CO2, 95% humidified environment), cells were lysed and RNAs were extracted using the miRNeasy Micro Kit (Qiagen). After RNAs were extracted from cells, RT-qPCR was performed as described above. The cDNA reaction mixtures were prepared with appropriate primer pairs and RealQ Plus 2× Master Mix Green (5 μL) (Ampliqon). mRNA levels were calculated using the comparative Ct method with GAPDH as the endogenous housekeeping gene. Gene expression was normalized to the value for nontreated cells. The following primer pairs were used: PTEN mRNA: 5′-TGAGT TCCCT CAGCC GTTAC CT-3′ (forward); 5′-GAGGT TTCCT CTGGT CCTGG TA-3′ (reverse); PDCD4 mRNA: 5′-ATGAG ACTGT GGTTC TGCCC-3′ (forward); 5′-TCCCT TAACA TCTCC GCGAC-3′ (reverse); GAPDH: 5′-ACGGA TTTGG TCGTA TTGGG-3′ (forward); 5′-CGCTC CTGGA AGATG GTGAT-3′ (reverse).
105 cells per well followed by 24-h incubation (37 °C, 5% CO2, 95% humidified environment). After cells were washed twice using 1× PBS, cells were treated with fresh medium containing probes or probe-loaded MSNs (75 nM). A group of non-treatment was prepared as a negative control. After 72-h incubation (37 °C, 5% CO2, 95% humidified environment), cells were lysed and RNAs were extracted using the miRNeasy Micro Kit (Qiagen). After RNAs were extracted from cells, RT-qPCR was performed as described above. The cDNA reaction mixtures were prepared with appropriate primer pairs and RealQ Plus 2× Master Mix Green (5 μL) (Ampliqon). mRNA levels were calculated using the comparative Ct method with GAPDH as the endogenous housekeeping gene. Gene expression was normalized to the value for nontreated cells. The following primer pairs were used: PTEN mRNA: 5′-TGAGT TCCCT CAGCC GTTAC CT-3′ (forward); 5′-GAGGT TTCCT CTGGT CCTGG TA-3′ (reverse); PDCD4 mRNA: 5′-ATGAG ACTGT GGTTC TGCCC-3′ (forward); 5′-TCCCT TAACA TCTCC GCGAC-3′ (reverse); GAPDH: 5′-ACGGA TTTGG TCGTA TTGGG-3′ (forward); 5′-CGCTC CTGGA AGATG GTGAT-3′ (reverse).
4.16 Statistical analysis
Statistical analysis was conducted using ANOVA with Tukey's multiple comparison tests. A p-value <0.05 was considered statistically significant.Author contributions
Y.-J. S. conceptualized this research. W.-T. C. performed the experiments. All authors participated in experimental design and data analysis. P.-H. W. and W.-Y. C. supervised the whole project and conceived the concepts of this study.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Professor Li-Jen Su, National Central University for providing cell line HCT116. This study is supported by the Ministry of Science and Technology of Taiwan (MOST). W.-Y. C. is supported by MOST (MOST 110-2222-E-008-002-MY2; MOST 108-2221-E-008-056-MY3). Y.-P. C. is supported by MOST (MOST 110-2113-M-038-003; MOST 111-2124-M-038-001).Notes and references
- B. A. Sullivan, M. Noujaim and J. Roper, Gastrointest. Endosc. Clin., 2022, 32, 177–194 CrossRef PubMed.
- F. Baidoun, K. Elshiwy, Y. Elkeraie, Z. Merjaneh, G. Khoudari, M. T. Sarmini, M. Gad, M. Al-Husseini and A. Saad, Curr. Drug Targets, 2021, 22, 998–1009 CrossRef CAS PubMed.
- P. Raguraman, A. A. Balachandran, S. Chen, S. D. Diermeier and R. N. Veedu, Cancers, 2021, 13, 5555 CrossRef CAS PubMed.
- D. Kaloni, S. T. Diepstraten, A. Strasser and G. L. Kelly, Apoptosis, 2023, 28, 20–38 CrossRef CAS PubMed.
- S. Iliaki, R. Beyaert and I. S. Afonina, Biochem. Pharmacol., 2021, 193, 114747 CrossRef CAS PubMed.
- U. Degirmenci, M. Wang and J. Hu, Cells, 2020, 9, 198 CrossRef CAS PubMed.
- L. Zhu, J. Luo and K. Ren, J. Mater. Chem. B, 2023, 11, 261–279 RSC.
- G. M. Traber and A.-M. Yu, J. Pharmacol. Exp. Ther., 2023, 384, 133–154 CrossRef CAS PubMed.
- T. C. Roberts, R. Langer and M. J. A. Wood, Nat. Rev. Drug Discovery, 2020, 19, 673–694 CrossRef CAS PubMed.
- H. Xiong, R. N. Veedu and S. D. Diermeier, Int. J. Mol. Sci., 2021, 22, 3295 CrossRef CAS PubMed.
- G. Clavé, M. Reverte, J.-J. Vasseur and M. Smietana, RSC Chem. Biol., 2021, 2, 94–150 RSC.
- S. T. Crooke, S. Wang, T. A. Vickers, W. Shen and X.-H. Liang, Nat. Biotechnol., 2017, 35, 230–237 CrossRef CAS PubMed.
- T.-L. Li, M.-W. Wu, W.-C. Lin, C.-H. Lai, Y.-H. Chang, L.-J. Su and W.-Y. Chen, Anal. Bioanal. Chem., 2019, 411, 3871–3880 CrossRef CAS PubMed.
- C.-J. Huang, Z.-E. Lin, Y.-S. Yang, H. W.-H. Chan and W.-Y. Chen, Biosens. Bioelectron., 2018, 99, 170–175 CrossRef CAS PubMed.
- T. Kuo, M. Wu, W. Lin, D. Matulis, Y. Yang, S. Li and W. Chen, J. Taiwan Inst. Chem. Eng., 2020, 110, 1–7 CrossRef CAS.
- S. Chou, Y. Lai, X. Zhuo, W. Chen and S. Li, J. Taiwan Inst. Chem. Eng., 2022, 137, 104297 CrossRef CAS.
- P. Wang, T. Jia, C. Chang, B. Tan, Y. Chi and W. Chen, GEN Biotechnol., 2022, 1, 402–403 CrossRef.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed.
- R. Kumar, K. Mondal, P. K. Panda, A. Kaushik, R. Abolhassani, R. Ahuja, H.-G. Rubahn and Y. K. Mishra, J. Mater. Chem. B, 2020, 8, 8992–9027 RSC.
- M. Vallet-Regí, F. Schüth, D. Lozano, M. Colilla and M. Manzano, Chem. Soc. Rev., 2022, 51, 5365–5451 RSC.
- N.-T. Chen, S.-H. Cheng, J. S. Souris, C.-T. Chen, C.-Y. Mou and L.-W. Lo, J. Mater. Chem. B, 2013, 1, 3128–3135 RSC.
- Z. Chen, Z. Lv, Y. Sun, Z. Chi and G. Qing, J. Mater. Chem. B, 2020, 8, 2951–2973 RSC.
- J. Dembélé, J.-H. Liao, T.-P. Liu and Y.-P. Chen, ACS Appl. Mater. Interfaces, 2023, 15, 432–451 CrossRef PubMed.
- M. L. Salvador, L. Suay and U. Klein, Nucleic Acids Res., 2011, 39, 6213–6222 CrossRef CAS PubMed.
- G. Stoecklin, T. Mayo and P. Anderson, EMBO Rep., 2006, 7, 72–77 CrossRef CAS PubMed.
- S. A. Harris, Z. A. Sands and C. A. Laughton, Biophys. J., 2005, 88, 1684–1691 CrossRef CAS PubMed.
- V. Kafil and Y. Omidi, Bioimpacts, 2011, 1, 23–30 CAS.
- L. Buscaglia and Y. Li, Chin. J. Cancer, 2011, 30, 371–380 CrossRef PubMed.
- Y. Yao, F. Sun and M. Lei, Biosci. Rep., 2018, 38, BSR20171511 CrossRef CAS PubMed.
- J.-Z. Ning, C.-M. Chu, Y. Du and L. Zuo, J. Cancer, 2021, 12, 6706–6714 CrossRef CAS PubMed.
- J. Hu, Z. Wang, J. Wang, Y. Jian, J. Dai, X. Wang and W. Xiong, OncoTargets Ther., 2020, 13, 9159–9167 CrossRef CAS PubMed.
- Y.-Q. Wang, R.-D. Guo, R.-M. Guo, W. Sheng and L.-R. Yin, J. Cell. Biochem., 2013, 114, 1464–1473 CrossRef CAS PubMed.
- Z. Liu, Q. Sun and X. Wang, Transl. Oncol., 2017, 10, 22–32 CrossRef PubMed.
- E. Montaudon, J. Nikitorowicz-Buniak, L. Sourd and L. Morisset, et al. , Nat. Commun., 2020, 11, 4053 CrossRef CAS PubMed.
- X. Shen, A. Dirisala, M. Toyoda, Y. Xiao, H. Guo, Y. Honda, T. Nomoto, H. Takemoto, Y. Miura and N. Nishiyama, J. Controlled Release, 2023, 360, 928–939 CrossRef CAS PubMed.
- Y.-J. Sung, H. Guo, A. Ghasemizadeh, X. Shen, W. Chintrakulchai, M. Kobayashi, M. Toyoda, K. Ogi, J. Michinishi, T. Ohtake, M. Matsui, Y. Honda, T. Nomoto, H. Takemoto, Y. Miura and N. Nishiyama, Cancer Sci., 2022, 113, 4339–4349 CrossRef CAS PubMed.
- Z. Li, Y. Yang, H. Wei, X. Shan, X. Wang, M. Ou, Q. Liu, N. Gao, H. Chen, L. Mei and X. Zeng, J. Controlled Release, 2021, 338, 719–730 CrossRef CAS PubMed.
- Q. Li, Z. Shi, M. Ou, Z. Li, M. Luo, M. Wu, X. Dong, L. Lu, F. Lv, F. Zhang and L. Mei, J. Controlled Release, 2022, 352, 450–458 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb00509k |
| This journal is © The Royal Society of Chemistry 2024 |