Mitochondrial membrane potential-independent near-infrared fluorescent probes for viscosity-exclusive imaging†
Xiu
Pan‡
ab,
Yu
Zhao‡
b,
Jia-Li
Wang
b,
Shun
Feng
b,
Xiao-Qi
Yu
 c and
Ming-Yu
Wu
c and
Ming-Yu
Wu
 *a
*a
aSchool of Biomedical Engineering, Sichuan University, Chengdu, 610065, China. E-mail: wumy1050hx@swjtu.edu.cn
bSchool of Life Science and Engineering, Southwest Jiaotong University, Chengdu 610031, P. R. China
cAsymmetric Synthesis and Chiral Technology Key Laboratory of Sichuan Province, Department of Chemistry, Xihua University, Chengdu 610039, P. R. China
First published on 30th October 2024
Abstract
Elucidating the intrinsic relationship between disease and mitochondrial viscosity is crucial for early diagnosis. However, current mitochondrial viscosity fluorescent probes are highly dependent on mitochondrial membrane potential (MMP) and are sensitive to other mitochondrial microenvironment parameters. To address these issues, a mitochondria-targeting MMP-independent and viscosity exclusive near-infrared (NIR) fluorescent probe, ACR-DMA, was developed. ACR-DMA consists of thiophene acetonitrile as the skeleton and viscosity-sensitive unit, a pyridinium cation for the mitochondria-targeting group, and a benzyl bromide subunit for mitochondrial immobilization. It is very sensitive to viscosity and shows significant “turn-on” fluorescence behavior at 710 nm with a more than 150-fold fluorescence intensity increase. Furthermore, ACR-DMA can be firmly immobilized in mitochondria and can monitor viscosity changes induced by nystain, monensin, and lipopolysaccharide. Additionally, it was successfully used to visualize mitochondrial viscosity changes resulting from tumors, inflammation, and drug-induced acute kidney injury, revealing the relationship between viscosity and disease both in vitro and in vivo. ACR-DMA is expected to be a promising candidate for diagnosing mitochondrial viscosity-related diseases.
Introduction
Mitochondria are vital organelles that serve as energy factories. They regulate cellular energy metabolism, signal transduction, cellular differentiation, and apoptosis.1–4 Mitochondrial microenvironmental parametrs, such as MMP, pH, polarity, viscosity, etc., are crucial for maintaining mitochondrial homeostasis and function. Among these, viscosity plays a key role in controlling cellular processes such as the transport of substances and signals, protein aggregation, and interactions between different biomolecules.5–8 Mitochondrial viscosity is closely related to mitochondrial respiration states and metabolism.9,10 Changes in mitochondrial viscosity may affect the organization of the mitochondrial network and the diffusion rate of metabolites.11,12 Abnormal changes in mitochondrial viscosity are closely associated with various diseases, including atherosclerosis, diabetes, neurodegenerative diseases, inflammation, and even malignant tumors.13–16 Therefore, developing effective tools to monitor mitochondrial viscosity variations for elucidating the pathogenesis and early diagnosis of related diseases is urgent.Traditional viscosity measurement methods, such as capillary viscometry, falling ball viscometry and rotational viscometry, are only applicable to macroscopic fluids and difficult to be used in complex organisms.17,18 In contrast, fluorescence imaging is an ideal technology for complex organisms, offering high sensitivity, non-invasiveness, and excellent spatiotemporal resolution. It has been widely adopted in the fields of life and medical sciences.19–21 In recent years, numerous mitochondria-targeted viscosity probes based on molecular rotors have been reported.22–27 These probes exhibit weak fluorescence emission in low viscosity environments, where energy is released through intramolecular rotation, but show viscosity-activated “turn-on” fluorescence in high viscosity environments, where intramolecular rotation is restricted and energy is released by emitting fluorescence.28–31 However, most reported mitochondrial probes rely on negative MMP, using lipophilic cationic groups to accumulate in the mitochondria via electrostatic interactions.12,32,33 When mitochondrial metabolic dysfunction occurs, it is often accompanied by changes in MMP. Upon MMP collapse, these MMP-dependent fluorescent probes leak out from the mitochondria, potentially leading to inaccurate localization or unreliable signals.12,34,35 To address this issue, Feng and Tang enhanced the affinity between the probes and mitochondria by introducing a C12 chain to embed in the phospholipid bilayer of the mitochondrial membrane for long-time tracking of mitochondria and real-time monitoring mitochondrial viscosity in mitophagy.36,37 Besides, Dong and Tang et al. designed mitochondria-targeted MMP-independent viscosity-sensitive fluorescent probes for monitoring mitochondrial viscosity in living cells or mice.38,39 A benzyl chloride moiety was decorated on the fluorophore which can immobilize on mitochondria through nucleophilic substitution with a sulfhydryl group on mitochondrial proteins. However, the complex microenvironmental parameters of mitochondria, such as pH, polarity, reactive oxygen species (ROS), reactive sulfur species (RSS), etc., are important factors that interfere the viscosity detection. All these reported viscosity probes were molecular rotors based on the twisted intramolecular charge transfer (TICT) mechanism. Probes with TICT characteristics are easily affected by polarity, showing strong fluorescence intensity with blue-shifted emission in low-polarity environments. Besides, the abundant ROS and RSS in mitochondria can break the double bond. These factors may lead to false signals and erroneous results.32 Therefore, the development of viscosity-exclusive mitochondrial targeted MMP-independent fluorescent probes is highly desirable but remains a challenging.
Herein, a novel mitochondrial targeted viscosity specific probe, ACR-DMA, was developed which can be firmly immobilized in mitochondria for tracking of mitochondrial viscosity changes in vitro and in vivo (Scheme 1). ACR-DMA emitted fluorescence in the NIR region (710 nm) with a 181 nm Stokes shift. It not only demonstrated excellent mitochondrial immobilization with an MMP-independent property but also exhibited high selectivity in responding to viscosity while remaining insensitive to polarity, pH, ROS, and RSS. These characteristics will be beneficial for accurately detecting viscosity in complicated biological environments. Consequently, ACR-DMA can be successfully used for detecting viscosity changes of mitochondria in living HeLa cells induced by nystain (Nys), monensin (Mon) and lipopolysaccharide (LPS). Furthermore, it could discern tumor tissues and image inflammation, and drug-induced acute kidney injuries in vivo. ACR-DMA is not only expected to be a powerful tool for in vivo viscosity imaging, but also has the potential to be applied in the diagnosis of other viscosity abnormalities, which is of great significance for exploring their clinical application.
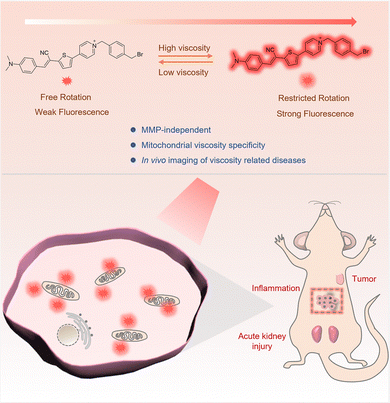 | ||
| Scheme 1 Illustration of the structure, viscosity response mechanism, and imaging in live mice of the mitochondria-immobilized NIR fluorescent probe ACR-DMA. | ||
Results and discussion
Design and synthesis of the probe ACR-DMA
Thiophene acrylonitrile, as an electronic system, is widely used to construct push–pull electronic systems to develop fluorescent probes and optical materials with NIR absorption/emission through an intramolecular charge transfer mechanism. Among these, the single bond connecting acrylonitrile and thiophene is viscosity responsive as a rotor structure.40,41 Therefore, we used thiophene acrylonitrile as the backbone and introduced N,N-dimethylaniline as a strong electron donor and pyridinium as an electron-acceptor to construct a NIR probe. At the same time, the pyridinium cation is expected to be mutually attracted to the negative inner membrane potential of mitochondria to contribute to mitochondria-targeting ability. And then we introduced a benzyl bromide group on the pyridinium salt in anticipation of the probe being immobilized in the mitochondria by nucleophilic substitution with mitochondrial sulfhydryl group-containing proteins or polypeptides, which are unaffected by MMP. Based on these, a viscosity-responsive MMP-independent NIR fluorescence probe ACR-DMA was successfully designed and synthesized (Scheme S1, ESI†) and was characterized by 1H NMR, 13C NMR, and HRMS.Optical sensing properties of ACR-DMA to viscosity
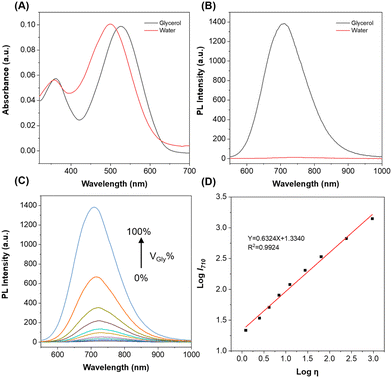
First, the photophysical properties of ACR-DMA were investigated. As shown in Fig. 1A and B, the maximum absorption peak of ACR-DMA was at 498 nm with a molar absorbance (ε) of 2.017 × 104 L mol−1 cm−1 in water, while almost no fluorescence emission was found. However, in glycerol, the absorption peak shifted to 528 nm (ε = 1.976 × 104 L mol−1 cm−1) with strong fluorescence at 710 nm. We then investigated the viscosity response ability in water with different glycerol contents. As the glycerol ratio gradually increased from 0% to 99.9%, the fluorescence intensity of ACR-DMA at 710 nm significantly enhanced by nearly 150-fold, which was more sensitive than the most reported viscosimetry probes (Fig. 1C).42–46 As shown in Table S1 (ESI†), the fluorescence quantum yield of ACR-DMA in water was only 0.20%, while in glycerol the fluorescence quantum yield was significantly increased to 25.24%, which was due to the restriction of intramolecular rotation. According to the Förster–Hoffmann equation, there was a good linear relationship between log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) I710 and log
I710 and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) η (R2 = 0.9924, slope = 0.6324) over a wide viscosity range of 1.22–945 cP (Fig. 1D and Table S2, ESI†), suggesting that ACR-DMA can sensitively detect viscosity.47
η (R2 = 0.9924, slope = 0.6324) over a wide viscosity range of 1.22–945 cP (Fig. 1D and Table S2, ESI†), suggesting that ACR-DMA can sensitively detect viscosity.47
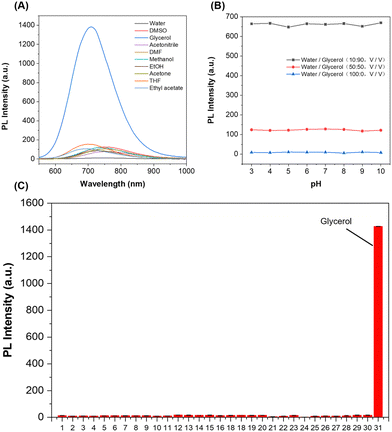
High selectivity toward viscosity is crucial for the biological application in a complex cellular microenvironment. The solvent polarity response was investigated in different solvents with various polarities, including tetrahydrofuran, ethyl acetate, acetonitrile, acetone, ethanol, methanol, dimethyl sulfoxide, N,N-dimethylformamide, phosphate-buffered saline, and glycerol. As shown in Fig. 2A and Fig. S1 (ESI†), the absorption and emission spectra of ACR-DMA hardly changed in solutions with different polarities except glycerol. The quantum yield in glycerol was far higher than those in other solvents, indicating that ACR-DMA was insensitive to polarity. Meanwhile, the effect of pH on ACR-DMA was further studied. In solvent systems containing different glycerol ratios (0%, 50% and 90%), the fluorescence intensity of ACR-DMA remained constant under different pH conditions ranging from pH 3–10 (Fig. 2B), demonstrating that ACR-DMA was stable in different pHs. The selectivity of ACR-DMA for viscosity was subsequently studied (Fig. 2C and Fig. S2, ESI†). Whenever metal ions (Na+, K+, Mg2+, Ca2+, Fe2+, Fe3+, Cu2+, Ni2+, and Zn2+), anions (Cl−, CO32−, HCO3−, CH3COO−, C2O42−, SO42−, and NO2−), amino acids (Gly, Glu, Trp), RSS (GSH, SO32−, S2O32−, HSO3−, and HS−), and ROS (ONOO−, 1O2, ˙OH, ClO−, and H2O2) were present, they can hardly change the fluorescence. These results indicate that ACR-DMA is highly exclusively responsive to viscosity with minimal interference from polarity, pH, and other biologically relevant species, and is promising for viscosity detection in complex biological systems.
Monitoring mitochondrial viscosity with ACR-DMA in living cells
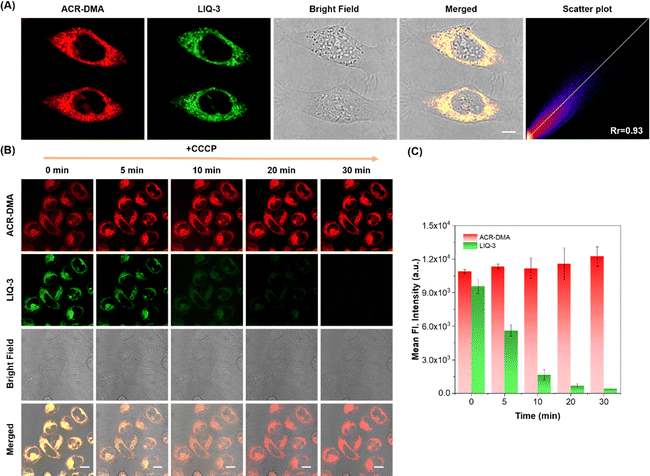
Benefiting from the viscosity specificity, we explored the ability of ACR-DMA to sense viscosity changes at the cellular level. The cytotoxicity of ACR-DMA was tested with the CCK-8 assay. More than 95% cells survived even after incubating with 30 μM ACR-DMA for 12 h (Fig. S3, ESI†), indicating good biocompatibility of the ACR-DMA. Colocalization experiments were performed in HeLa cells using the reported mitochondria targeting fluorescent probe LIQ-3, lysosome targeting probe Lyso-Tracker and lipid droplet targeting probe BODIPY.48 As shown in Fig. 3A and Fig. S4 (ESI†), the red fluorescence signal of ACR-DMA overlapped well with the green fluorescence signal of LIQ-3 with the Pearson correlation coefficient (Rr) as high as 0.93. However, the Rr values of ACR-DMA and lysosome or lipid droplets were only 0.50 or 0.32, respectively, suggesting that ACR-DMA can selectively accumulate in mitochondria.As mentioned above, conventional mitochondrial probes highly rely on electrostatic attraction, which can easily leak out from mitochondria once MMP decreases or disappears, resulting in inaccurate and even false information. To assess the immobilizing ability of ACR-DMA in mitochondria, we disrupted MMP using carbonyl cyanide m-chlorophenyl hydrazone (CCCP, mitochondrial membrane potential uncoupling reagent).49 HeLa cells were co-stained with ACR-DMA and LIQ-3 and then treated with CCCP. As shown in Fig. 3B and C, with a prolonged incubation time with CCCP, the red fluorescence intensity of ACR-DMA in HeLa cells remained stable without obvious leakage. In contrast, LIQ-3 was highly sensitive to MMP and its green fluorescence gradually disappeared. These results indicate that ACR-DMA is MMP-independent, which facilitates real-time monitoring of mitochondrial microenvironmental changes.
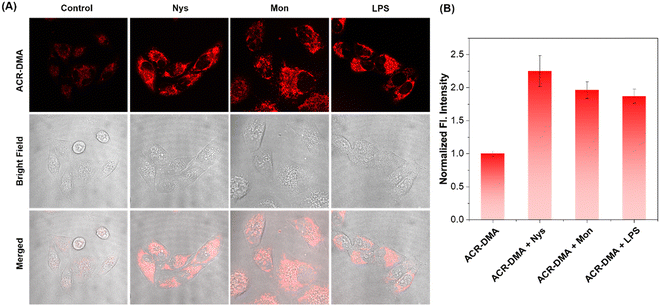
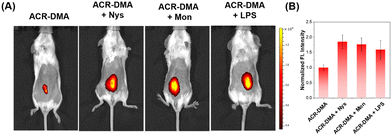
Nys and Mon are ionophores, which can disrupt intracellular ionic homeostasis and induce structural changes or swelling of mitochondria, thereby leading to mitochondrial dysfunction accompanied by mitochondrial viscosity changes. LPS is a proinflammatory agent. LPS attacks inevitably cause oxidative stress and mitochondrial dysfunction. Spectroscopy experiments show that these drugs did not affect the fluorescence of ACR-DMA as shown in Fig. S5 (ESI†). Nys, Mon and LPS were used to stimulate HeLa cells. As shown in Fig. 4A, very weak fluorescence could be observed in the cells after incubation with ACR-DMA only. In contrast, the red fluorescence signal of ACR-DMA in cells significantly enhanced after treating with Nys, Mon, or LPS, respectively, with a mean fluorescence intensity increase of 2.25-fold, 1.96-fold, and 1.87-fold (Fig. 4B). ACR-DMA can act as a powerful tool for sensitive detection of intracellular mitochondrial viscosity.
Fluorescence imaging of tumor tissue with ACR-DMA
The changes in extracellular secretions and extracellular matrix components caused by the unique proliferation of cancer cells affect the viscosity fluctuations in a tumor microenvironment. Therefore, viscosity is a potential biomarker of a tumor microenvironment.16 We constructed a tumor mouse model, and then obtained the tumor tissues and other primary organs. As shown in Fig. 5, after incubated with the ACR-DMA for 2 h, hardly fluorescence was observed in major organs such as heart, liver, spleen, lungs and kidneys, while strong fluorescence was recorded in tumors, indicating that ACR-DMA can diagnose cancer by imaging mitochondrial viscosity changes. Therefore, ACR-DMA showed great potential for the diagnosis of other diseases associated with viscosity abnormalities.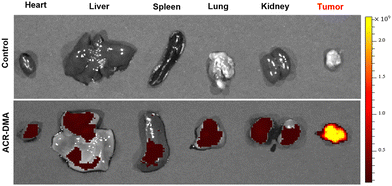 | ||
| Fig. 5 Fluorescence imaging of viscosity with ACR-DMA (20 μM) in tumors and organs (heart, liver, spleen, lung and kidneys), respectively. ACR-DMA: λex = 500 nm, λem = 740 nm. | ||
Fluorescence imaging of viscosity changes of inflammation in vivo
Motivated by the results of the above experiments, the fluorescence imaging of mitochondrial viscosity changes was conducted in vivo. Firstly, we injected Nys, Mon or LPS subcutaneously into the mice, respectively, to induce the viscosity change. Subsequently, in vivo viscosity imaging was performed using ACR-DMA. As expected, the subcutaneous fluorescence intensity of the drug-stimulated mice significantly enhanced 1.85-fold, 1.76-fold, and 1.59-fold, respectively, compared with that of mice treated with PBS (Fig. 6A and B), which coincided with the cellular viscosity imaging results.Fluorescence imaging of viscosity changes of kidneys in an acute kidney injury
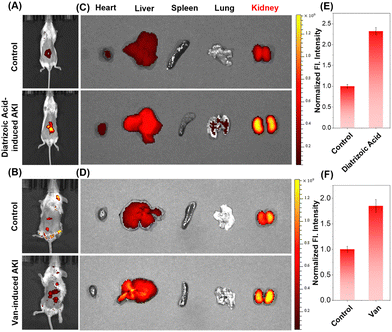
The kidney is one of the most important organ in the body's life activities. Studies showed that many factors could induce acute kidney injury (AKI), such as sepsis, low blood pressure, organ failure, kidney stones, and overuse of medications by patients. During AKI, overexpression of inflammatory factors in the renal organ may lead to deranged viscosity levels.22,50 Diatrizoic acid and vancomycin were used to construct an AKI mice model in vivo, respectively. The normal renal tissues stained by hematoxylin and eosin (H&E) staining had intact peritubular membranes and intact tubular structures. In contrast, the AKI group's renal tissues showed cellular swelling, tissue hyperplasia, and increased neutrophils, indicating that the construction of the model was successful (Fig. S6, ESI†).Then, imaging was performed under the same imaging conditions. As shown in Fig. 7A and B, the fluorescence from mice treated with PBS was very weak, while the fluorescence signals from mice in the AKI groups were significantly enhanced. Subsequently, the mice were dissected to obtain the organs. Strong fluorescent signals were observed in the kidney areas of mice treated with diatrizoic acid or vancomycin, compared to the tissues from PBS-treated mice (Fig. 7C–F). ACR-DMA is successfully used to image viscosity changes of AKI both in vivo and ex vivo, and is expected to be a powerful tool for the diagnosis of AKI and is important for exploring its pathogenesis and clinical research.
Conclusions
In summary, we rationally designed a mitochondrial targeted MMP-independent viscosity sensitive fluorescent probe ACR-DMA. ACR-DMA possesses NIR luminescence characteristics and large Stokes shift, and is exclusively responsive to viscosity and insensitive to other microenvironment parameters, such as polarity, pH, ROS, etc. ACR-DMA demonstrated excellent mitochondrial targeting and immobilization abilities, facilitating long-term tracking of mitochondria. Benefiting from the excellent viscosity specifity, it was successfully used for real-time monitoring of viscosity changes during drug-induced mitochondrial disorders. More importantly, this powerful tool permits visualization of mitochondrial viscosity changes in tumor tissues and inflammatory mice, as well as drug-induced acute kidney injury in mice. Therefore, we believe that ACR-DMA may serve as a promising imaging tool for real-time tracking of the mitochondrial disorder and is expected to be used in diagnosis of diseases associated with viscosity abnormalities.Author contributions
Xiu Pan: writing – original draft, visualization, and investigation. Yu Zhao: writing – original draft, formal analysis, and methodology. Jia-Li Wang: investigation and visualization. Shun Feng: conceptualization and supervision. Xiao-Qi Yu: conceptualization and supervision. Ming-Yu Wu: conceptualization, funding acquisition, and writing – review and editing.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors M.-Y. W., and S. F. declare that they have filed a Chinese patent application (2023107146198) containing the synthesis of ACR-DMA. Other authors declare no conflict of interest.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (22177094 and 22174117) and the Fundamental Research Funds for the Central Universities (YJ202419).Notes and references
- D. R. Green and J. C. Reed, Science, 1998, 281, 1309–1312 CrossRef CAS PubMed.
- B. Spinelli and M. C. Haigis, Nat. Cell Biol., 2018, 20, 745–754 CrossRef PubMed.
- S. Javadov, A. V. Kozlov and A. K. S. Camara, Cell, 2020, 9, 1177 CrossRef PubMed.
- H. Huang, C. Dong, M. Chang, L. Ding, L. Chen, W. Feng and Y. Chen, Exploration, 2021, 1, 50–60 CrossRef PubMed.
- J. Shawn Goodwin, K. R. Drake, C. L. Remmert and A. K. Kenworthy, Biophys. J., 2005, 89, 1398–1410 CrossRef PubMed.
- D. Su, C. L. Teoh, L. Wang, X. Liu and Y.-T. Chang, Chem. Soc. Rev., 2017, 46, 4833–4844 RSC.
- H. Li, C. Xin, G. Zhang, X. Han, W. Qin, C.-W. Zhang, C. Yu, S. Jing, L. Li and W. Huang, J. Mater. Chem. B, 2019, 7, 4243–4251 RSC.
- X. Liu, K. Li, L. Shi, H. Zhang, Y.-H. Liu, H.-Y. Wang, N. Wang and X.-Q. Yu, Chem. Commun., 2021, 57, 2265–2268 RSC.
- L. Ellgaard and A. Helenius, Nat. Rev. Mol. Cell Biol., 2003, 4, 181–191 CrossRef CAS PubMed.
- Y. Zhang, Z. Li, W. Hu and Z. Liu, Anal. Chem., 2019, 91, 10302–10309 CrossRef CAS PubMed.
- P. Mecocci, M. F. Beal, R. Cecchetti, M. C. Polidori, A. Cherubini, F. Chionne, L. Avellini, G. Romano and U. Senin, Mol. Chem. Neuropathol., 1997, 31, 53–64 CrossRef CAS PubMed.
- Y. Ma, Y. Zhao, R. Guo, L. Zhu and W. Lin, J. Mater. Chem. B, 2018, 6, 6212–6216 RSC.
- A. P. West, G. S. Shadel and S. Ghosh, Nat. Rev. Immunol., 2011, 11, 389–402 CrossRef CAS PubMed.
- D. C. Wallace, Nat. Rev. Cancer, 2012, 12, 685–698 CrossRef CAS PubMed.
- Y. Guo, H. Leng, Q. Chen, J. Su, W.-J. Shi, C. Xia, L. Zhang and J. Yan, Sens. Actuators, B, 2022, 372, 132648 CrossRef CAS.
- J. Yin, X. Kong and W. Lin, Anal. Chem., 2021, 93, 2072–2081 CrossRef CAS PubMed.
- X. Yang, D. Zhang, Y. Ye and Y. Zhao, Coord. Chem. Rev., 2022, 453, 214336 CrossRef CAS.
- C. Ma, W. Sun, L. Xu, Y. Qian, J. Dai, G. Zhong, Y. Hou, J. Liu and B. Shen, J. Mater. Chem. B, 2020, 8, 9642–9651 RSC.
- S. Li, P. Wang, W. Feng, Y. Xiang, K. Dou and Z. Liu, Chem. Commun., 2020, 56, 1050–1053 RSC.
- Y. Liao, R. Wang, S. Wang, Y. Xie, H. Chen, R. Huang, L. Shao, Q. Zhu and Y. Liu, ACS Appl. Mater. Interfaces, 2021, 13, 54783–54793 CrossRef CAS.
- Y. Ma, J. Niu, X. Liang, L. Wang, Y. Zhang, H. Lv, T. Wang, J. Wang, X. Zhang, S. Xu, Q. Zhu, Z. Jiang and W. Lin, Chem. Eng. J., 2023, 464, 142767 CrossRef CAS.
- Y. Tang, J. Peng, Q. Zhang, S. Song and W. Lin, Talanta, 2022, 249, 123647 CrossRef CAS PubMed.
- H. Song, W. Zhang, Y. Zhang, C. Yin and F. Huo, Chem. Eng. J., 2022, 445, 136448 CrossRef CAS.
- H. Li, W. Shi, X. Li, Y. Hu, Y. Fang and H. Ma, J. Am. Chem. Soc., 2019, 141, 18301–18307 CrossRef CAS PubMed.
- Y. Wu, C. Yin, W. Zhang, Y. Zhang and F. Huo, Anal. Chem., 2022, 94, 5069–5074 CrossRef CAS PubMed.
- S.-Y. Wang, J.-R. Liu, Z.-H. Ju, D.-H. Tian, Z.-H. Chai, Y. Zhang, F. Dai, S. Zhang and B. Zhou, Sens. Actuators, B, 2023, 381, 133470 CrossRef CAS.
- L.-L. Li, K. Li, M.-Y. Li, L. Shi, Y.-H. Liu, H. Zhang, S.-L. Pan, N. Wang, Q. Zhou and X.-Q. Yu, Anal. Chem., 2018, 90, 5873–5878 CrossRef CAS PubMed.
- S.-C. Lee, C.-L. Lee, J. Heo, C.-U. Jeong, G.-H. Lee, S. Kim, W. Yoon, H. Yun, S. O. Park, S. K. Kwak, S.-H. Park and O.-P. Kwon, Chem. – Eur. J., 2018, 24, 2888–2897 CrossRef CAS.
- L. Wang, Y. Xiao, W. Tian and L. Deng, J. Am. Chem. Soc., 2013, 135, 2903–2906 CrossRef CAS PubMed.
- L. Fan, Q. Zan, X. Wang, X. Yu, S. Wang, Y. Zhang, Q. Yang, W. Lu, S. Shuang and C. Dong, Chem. Eng. J., 2022, 449, 137762 CrossRef CAS.
- L. Fan, Q. Yang, Q. Zan, K. Zhao, W. Lu, X. Wang, Y. Wang, S. Shuang and C. Dong, Anal. Chem., 2023, 95, 5780–5787 CrossRef CAS PubMed.
- T. Gu, S. Mo, Y. Mu, X. Huang and L. Hu, Sens. Actuators, B, 2020, 309, 127731 CrossRef CAS.
- M. Ren, B. Deng, K. Zhou, X. Kong, J.-Y. Wang and W. Lin, Anal. Chem., 2017, 89, 552–555 CrossRef CAS PubMed.
- N. Zhao, S. Chen, Y. Hong and B. Z. Tang, Chem. Commun., 2015, 51, 13599–13602 RSC.
- M.-Y. Li, K. Li, Y.-H. Liu, H. Zhang, K.-K. Yu, X. Liu and X.-Q. Yu, Anal. Chem., 2020, 92, 3262–3269 CrossRef CAS PubMed.
- J. Hong, X. Guan, Y. Chen, X. Tan, S. Zhang and G. Feng, Anal. Chem., 2023, 95, 5687–5694 CrossRef CAS PubMed.
- X. Wu, R. Zhang, Y. Li, Y. Gai, T. Feng, J. Kou, F. Kong, L. Li and B. Tang, Anal. Chem., 2023, 95, 7611–7619 CrossRef CAS PubMed.
- X. Wang, L. Fan, S. Wang, Y. Zhang, F. Li, Q. Zan, W. Lu, S. Shuang and C. Dong, Anal. Chem., 2021, 93, 3241–3249 CrossRef CAS PubMed.
- X. Wu, G. Fu, Y. Li, S. Li, Q. Zhao, F. Kong, L. Li and B. Tang, Anal. Chem., 2023, 95, 3544–3549 CrossRef CAS PubMed.
- P. Cole, J. S. Mandel and J. J. Collins, Regul. Toxicol. Pharmacol., 2008, 52, 342–351 CrossRef CAS PubMed.
- R. K. Grasselli and F. Trifirò, Top. Catal., 2016, 59, 1651–1658 CrossRef CAS.
- P. Ning, P.-Y. Dong, Q. Geng, L. Bai, Y. Q. Ding, X.-H. Tian, R. Shao, L. Li and X.-M. Meng, J. Mater. Chem. B, 2017, 5, 2743–2749 RSC.
- W. H. Du, Y. Gu, X. Zhou, Z.-L. Wang and S.-F. Wang, Analyst, 2024, 149, 789–799 RSC.
- L.-X. Dai, M.-G. Ren, Z.-H. Li, L. Wang and W.-Y. Lin, Anal. Methods, 2019, 11, 4561–4565 RSC.
- Z.-X. Peng, D. Zhang, H. Yang, Z. Zhou, F.-Y. Wang, Z. Wang, J. Ren and E.-F. Wang, Analyst, 2024, 149, 3356–3362 RSC.
- X. Dai, B.-L. Dong, M.-G. Ren and W.-Y. Lin, J. Mater. Chem. B, 2018, 6, 381–385 RSC.
- J. Liu, W. Zhang, C. Zhou, M. Li, X. Wang, W. Zhang, Z. Liu, L. Wu, T. D. James, P. Li and B. Tang, J. Am. Chem. Soc., 2022, 144, 13586–13599 CrossRef CAS PubMed.
- L. Liu, Q. Zou, J.-K. Leung, J.-L. Wang, C. Kam, S. Chen, S. Feng and M.-Y. Wu, Chem. Commun., 2019, 55, 14681–14684 RSC.
- X. Li, M. Tian, G. Zhang, R. Zhang, R. Feng, L. Guo, X. Yu, N. Zhao and X. He, Anal. Chem., 2017, 89, 3335–3344 CrossRef CAS PubMed.
- Y. Ding, R. Zhong, R. Jiang, X. Yang, L. He, L. Yuan and D. Cheng, ACS Sens., 2023, 8, 914–922 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Spectral experiments, cytotoxicity assay, H&E staining analysis, NMR spectra and HRMS. See DOI: https://doi.org/10.1039/d4tb01785d |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |