Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A multi-valent polymyxin-based fluorescent probe for the detection of Gram-negative infections†
Richa
Sharma‡
 a,
Maria
Rodriguez-Rios§
a,
Maria
Rodriguez-Rios§
 a,
James
Crossland
a,
Maulida
Septiyana
ab,
Alicia
Megia-Fernandez¶
a,
James
Crossland
a,
Maulida
Septiyana
ab,
Alicia
Megia-Fernandez¶
 a,
Maxime
Klausen
a,
Maxime
Klausen
 *ac and
Mark
Bradley
*ac and
Mark
Bradley
 *d
*d
aSchool of Chemistry, University of Edinburgh, David Brewster Road, Edinburgh EH9 3FJ, UK
bDepartment of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Mataram, Mataram, Indonesia
cChimie ParisTech, PSL University, CNRS, Institute of Chemistry for Life and Health Sciences, Laboratory for Inorganic Chemical Biology, 75005 Paris, France. E-mail: maxime.klausen@chimieparistech.psl.eu
dPrecision Healthcare University Research Institute, Queen Mary University of London, Empire House, London, E1 1HH, UK. E-mail: m.bradley@qmul.ac.uk
First published on 17th December 2024
Abstract
A multi-branched fluorogenic probe for the rapid and specific detection of Gram-negative bacteria is reported. Three Gram-negative-targeting azido-modified polymyxins were clicked onto a trivalent scaffold functionalised with the environmental green-emitting fluorophore 7-nitrobenz-2-oxa-1,3-diazole. The probe allowed wash-free detection of target bacteria with increased sensitivity and lower limits of detection compared to monovalent probes.
Introduction
Bacterial infections (from both antibiotic-resistant and susceptible species) remain a major cause of death globally, with an estimated 13.7 million infection-related deaths in 2019 (of which approx. 10 million were from sepsis).1 The most predominant organisms (in order of deaths caused, excluding TB) are Staphylococcus aureus, Escherichia coli, Streptococcus pneumoniae, Klebsiella pneumoniae, Pseudomonas aeruginosa and Acinetobacter baumannii, four of which are Gram-negative and are responsible for many life-threatening infections, such as sepsis, wound infections, urinary tract infections and pneumonia.2–4Unfortunately, the accurate and rapid identification of infections remains a challenge as current diagnostic techniques often rely on invasive biopsies and microbial culture, which causes a delay between patient sampling and diagnoses.5 Consequently, pre-emptive treatments are commonly employed, which typically leads to overuse of broad-spectrum antibiotics with serious adverse side-effects (e.g. deafness with aminoglycosides,6 nephrotoxicity with glycopeptides,7etc.). This approach also promotes the utilisation of “last resort” highly potent antibiotics thereby driving the antimicrobial resistance problem.8,9
The development of tools and methodologies for the rapid and accurate identification of pathogenic agents is crucial to prevent this “butterfly effect”, and therefore allow early bedside decision of tailored treatments. One approach to achieve this goal is to create targeted reporters that can label pathogens and be visualised in real time, for example by the attachment of a fluorophore to a targeting ligand with specific affinity for a class of pathogens,10–13 generating highly pathogen-specific imaging probes with high potential for clinical diagnosis.14–17
Antibiotics specific to classes of pathogens (i.e. targeted to a specific Gram status) have become popular targeting ligands for the design of fluorescent probes to detect bacterial infections,18–21 with broad applications in disease diagnostics, resistance mechanisms studies and drug susceptibility assessment.22–26 Probes targeting Gram-negative bacteria27–30 specifically can be designed using polymyxins, a class of naturally occurring cationic cyclic lipopeptides. These compounds selectively bind to the outer membrane of Gram-negative bacteria via interactions with lipid A, a key component of the lipopolysaccharide (LPS).19,31,32 The long aliphatic chain of unmodified polymyxins is able to insert into the membrane giving enhanced (entropically driven) binding. This chain can be removed to develop non-cytotoxic imaging probes while keeping the cyclic peptide structure, essential for specific membrane recognition, thus enabling the development of targeted imaging probes for Gram-negative bacteria. In previous work, “switch-on” fluorescent probes, designed by derivatization of the antibiotic polymyxin B (PMB) with green- or red-emitting solvato-fluorogenic dyes, were used to detect Gram-negative pathogens with good specificity.33,34 The use of solvato-fluorogenic dyes, whose fluorescence is switched-on specifically in lipophilic environments (such as bacterial cell membranes), allowed wash-free labelling of pathogens with minimal background signal. This property brings key advantages to optical in vivo diagnostics, for example, the wash-free detection of Gram-negative infections in the distal airways of human lungs.33
Screening methods for infections often require high sensitivity, with low limits of detection (LoD). In addition, the synthesis of the probe should be straightforward, and scalable, to promote clinical translation and application. To address these challenges, we hereby developed a tri-branched Gram-negative specific imaging agent based on a scaffold bearing three PMB-derived targeting units and conjugated to one small, green-emitting, solvato-fluorogenic 7-nitrobenz-2-oxa-1,3-diazole (NBD) dye.35 Tri-branched fluorogenic probes have previously been used to enhance signal amplification in the detection of proteases and promote in vivo probe stability.36–38 Tripling the number of PMB ligands per fluorescent unit was expected to improve targeting efficiency and lower the LoD thanks to the greater number of binding domains. The ligand used was the polymyxin B cycloheptapeptide (PMB7) scaffold which proved to be readily synthetically accessible, and available en masse compared to the polymyxin B nonapeptide (PMB9) used in previous work.33 The properties of the resulting tri-branched probe NBD-Tris (PMB7) were then compared with the truncated linear (mono-branched) analogue NBD-PMB7 to study the effect of the branching/amplification strategy, and with the reported NBD-PMB9 probe (Fig. S1, ESI†) to study the influence of the dipeptide truncation on the bacterial detection assays. A synergistic enhancement of the fluorescent labelling was observed using the tri-branched scaffold compared to the mono-branched analogues, leading to more sensitive detection of Gram-negative bacteria.
Results and discussion
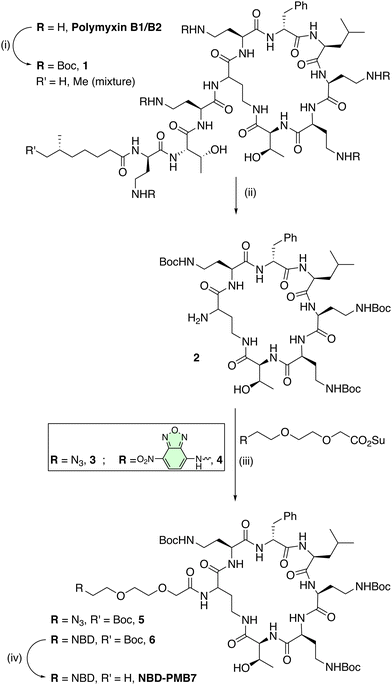
The design of the probe was based on conjugation of the cyclic polymyxin lipid A binding domain (PMB7) to the fluorophore NBD, as both mono- and tri-branched entities, preserving the three diaminobutyric acid (Dab) residues within the cyclic peptide for bacterial binding. Thus, polymyxin B sulphate was fully Boc-protected with Boc-anhydride, followed by enzymatic cleavage with Savinase, a protease that removed both the fatty acyl tail and the exocyclic tripeptide (Scheme 1). This afforded the truncated (Boc)3PMB7 moiety (compound 2) on gram scale, in excellent yield with minimal purification needed, with Savinase cleavage providing a unique attachment point for subsequent chemistry (Scheme 1). This strategy essentially reversed the protection and cleavage steps39 that have previously been reported for the generation of (Boc)nPMB building blocks, avoiding the time-consuming and tedious purification needed for the separation of mixtures of Boc-protected (Boc)3,4,5PMB9 and the various positional isomers.In order to verify that the absence of the three terminal amino-acid residues and the fatty acid tail in the PMB cycloheptapeptide did not unduly affect its binding to bacteria or selectivity, the mono-branched analogue NBD-PMB7 was prepared and compared to the established probe NBD-PMB9.33 The probe NBD-PMB7 was synthesized by amide coupling of the tris-Boc-protected cyclopeptide 2 with the NBD derivative 4, pre-functionalised with a diethylene glycol (EG)2 spacer, to give intermediate 6. Boc deprotection with trifluoroacetic acid (TFA) afforded the heptapeptide probe NBD-PMB7.
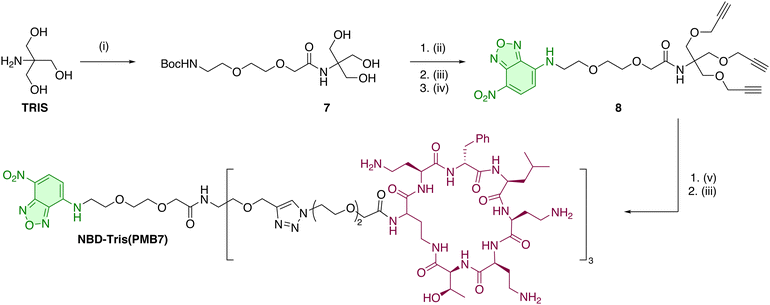
The synthesis of the tri-branched probe from the PMB cycloheptapeptide intermediate 2 was performed using a click-based strategy,40 utilising the (EG)2 spacer 3, containing an azide on one end, and an NHS ester on the other. This was synthesized from Cl-(EG)2-OH by oxidation to the carboxylic acid and nucleophilic substitution with sodium azide, followed by conversion to the active ester with EDC·HCl and NHS (see ESI†). The active ester was reacted with 2 in presence of DIPEA to yield the azide module 5 (Scheme 1). The tri-branched scaffold was based on a tris(hydroxymethyl)aminomethane (TRIS) scaffold that was modified with three propargyl groups to enable CuAAC. Thus, the amine group of TRIS was functionalised with the commercially available ethylene glycol linker BocNH-(EG)2-CO2H, with amide bond formation on this hindered amine position efficiently promoted by the coupling agent EEDQ under microwave irradiation, cleanly affording 7 (Scheme 2). The hydroxyl groups were alkylated with propargyl bromide under microwave heating to yield the corresponding tris-alkyne, which was then deprotected using TFA (see ESI†). The resulting crude amine was used in an SNAr reaction with NBD-Cl to give the NBD-functionalised fluorescent tri-branched scaffold 8 in a 4-step sequence with only a single final purification needed (34% yield over 4 steps). Finally, CuAAC between the tris-alkyne platform and three equivalents of 5 was performed using the CuI/THPTA catalytic system under microwave irradiation. Completion of the reaction was monitored using LC-MS to maximise the formation of the tri-functionalised compound over the partially (mono- or bi-) functionalised scaffold. The final bacterial-targeting probe NBD-Tris(PMB7) was then purified by preparative RP-HPLC (see ESI†). The copper-catalysed azide–alkyne cycloaddition (CuAAC) provided a clean, efficient, and high yielding reaction, even when constructing multi-branched scaffolds with biologically complex ligands.
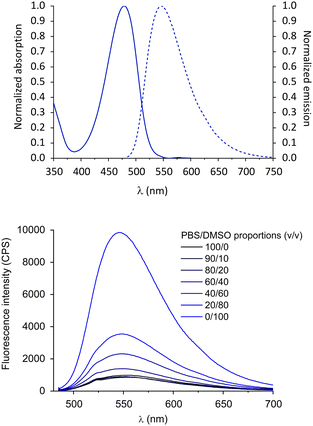
The optical properties of the tri-branched probe NBD-Tris(PMB7) were initially investigated to evidence their emissive character in lipophilic media. The compound showed a similar behaviour to other known NBD derivatives (ESI,† Fig. S1) and to the monomeric probe NBD-PMB9,33 with an intense absorption band at 464 nm, and broad green emission (λmax = 545 nm) (Fig. 1, top). The large band tailing beyond 600 nm and its large Stokes’ shift are characteristic of the NBD fluorophore.
We explored the sensitivity of the probe to the solvation environment by increasing the DMSO content of aqueous solutions of the probe. DMSO was selected to generate different lipophilic environments, whilst maintaining good probe solubility. The absorption and emission wavelengths were practically unaffected by the polarity and proticity changes (Fig. S2, ESI†), however, in accordance with the well-known solvato-fluorogenic character of NBD (i.e. known sensitivity to the polarity, viscosity, and hydrogen bonding capacity of their environment), higher DMSO contents led to a significant increase in fluorescence (Fig. 1, bottom). Indeed, a progressive 11-fold brighter fluorescence signal was observed for NBD-Tris(PMB7) in DMSO compared to in PBS. This strong sensitivity of the probe to its microenvironment is ideal to allow labelling of pathogen membranes without the need to wash excess probe from the sample. Interestingly, this 11-fold switch-on effect was much superior to that for the mono-branched NBD-PMB9 conjugate (≈3.5-fold),33 which could be instrumental in lowering the LoD. Interestingly, the probe was less fluorogenic than the model compound NBD-(EG)2-CO2H for which a 17-fold emission increase was observed in 90% DMSO (Fig. S4, ESI†). This indicates that the micro-environment generated on one hand by the solvent molecules and on the other by the spacer and binding ligands, has a strong influence on the relaxation pathways of the NBD dye. Hydrogen bonding with the nitro-group of the dye has been suggested as a predominant phenomenon affecting the properties of NBD and is likely to play a role in this brightness difference. The bulky and slightly hydrophobic tri-branched scaffold of the probe may also protect from direct interaction with water, leading to improved brightness compared to the monovalent analogue.
The labelling ability of NBD-PMB7 was evaluated on E. coli and compared with NBD-PMB9 to investigate the effect of the two amino acid truncations on probe binding. Labelling with the NBD probes (0.1–100 μM, without washing) was performed, and samples were imaged on a Leica SP5 confocal spinning disk microscope (λex = 480 nm, GFP filter settings, λem = 540 nm) (Fig. 2 and Fig. S5, ESI†). The truncated heptapeptide probe NBD-PMB7 showed slightly improved labelling compared to the nonapeptide NBD-PMB9 at all concentrations, this is consistent with the original reports on NBD-PMB9 in which the use of longer linkers led to a loss of affinity.33 The removal of the exocyclic dipeptide in NBD-PMB9 did not have a detrimental effect on the fluorescent labelling efficiency of Gram-negative bacteria when comparing the monomeric nonapeptide and heptapeptide (NBD-PMB7) probes.
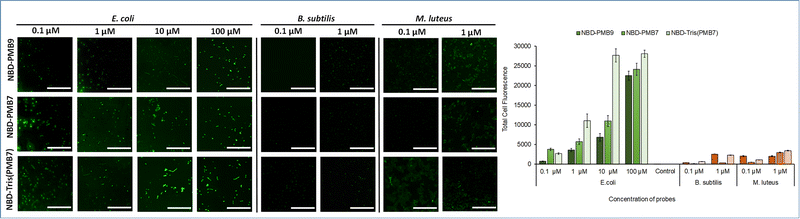 | ||
| Fig. 2 Left Panel: Fluorescent labelling of E. coli (Gram-negative, target) at increasing concentrations of probes (0.1 to 100 μM of NBD-PMB9, NBD-PMB7 and NBD-Tris(PMB7)) and B. subtilis and M. luteus (both Gram-positive, non-target) at 0.1 and 1 μM of NBD-PMB9, NBD-PMB7 and NBD-Tris(PMB7) (λex = 480 nm, GFP filter settings, λem = 540 nm). Scale bar is 100 μm. Bacterial concentration: 5 × 108 cfu mL−1. Brightfield images are presented in Fig. S5 (ESI†). Right panel: Quantification of the fluorescence of the imaged bacteria as obtained using Fiji (ImageJ, NIH). For each concentration, columns in the chart represent from left to right NBD-PMB9, NBD-PMB7 and NBD-Tris(PMB7) respectively. Total bacterial fluorescence was calculated by subtracting the mean of the background (10 background feature intensities of the same size as bacterial cells) from the mean of the fluorescent bacteria (20 fluorescent labelled bacteria). Error bars represent the standard error calculated using the errors of fluorescent and background features. | ||
The tri-branched probe contains two additional binding moieties compared to NBD-PMB7 and NBD-PMB9, but the same number of dye moieties. When both monomeric probes were compared with the tri-branched probe NBD-Tris(PMB7), at low concentrations (0.1 μM), no significant differences in labelling between the three probes were observed. However, at 1 μM NBD-Tris(PMB7), a 3-fold higher intensity was observed for the tri-branched probe over NBD-PMB9, and a 2-fold enhancement over NBD-PMB7 (which is consistent with the improved fluorogenicity of monomeric PMB7vs. PMB9). Binding saturation was observed at 100 μM for all probes.
The monovalent probes required three times the concentration to achieve fluorescence saturation, as 33 μM NBD-Tris(PMB7) showed equal brightness to 100 μM NBD-PMB7 (Fig. S6, ESI†). This proved that even with considerably reduced relative dye content per peptide, strong labelling of bacteria could be achieved thanks to the branching scaffold and click synthesis allowing an increased number of binding ligands. The improved signal may also be related to the improved solvato-fluorogenic character of NBD-Tris(PMB7), allowing a stronger switch-on effect.
Conclusions
A tri-branched fluorogenic probe with three bacterial-binding ligands was synthesized using a savinase-mediated cleavage of the non-cyclic components off the antibiotic polymyxin B, which provided access to the polymyxin core in high yields and purities, followed by a CuAAC reaction with a tri-branched-alkyne scaffold.The tri-branched probe NBD-Tris(PMB7) proved to be an efficient environmentally sensitive probe with significant brightness increases in lipophilic media compared to the original monovalent probe NBD-PMB9. One possible mechanism for this relies on the sensitivity of NBD to excited state hydrogen bonding. As such, in NBD-PMB9, the fluorophore is more likely to be exposed to unfavourable interactions with the peptide and with water molecules, which may limit the fluorescence increase. In contrast, with two diethylene glycol spacers attached via the TRIS platform, the NBD moiety in NBD-Tris(PMB7) may be more sterically shielded from unfavourable interactions, therefore leading to a stronger brightness increase. This is a further benefit of the tri-branched structure in our probe design, and allowed wash-free, selective fluorescent labelling of Gram-negative bacteria with low levels of cross-labelling. In bacterial assays, the tri-branched probe also showed higher sensitivity compared to the monovalent analogues based on PMB9 and the shorter PMB7 targeting ligands. This allowed up to 4-times more sensitive detection of Gram-negative bacteria, promoted synergistically by the presence of three PMB binding units and the greater solvato-fluorogenic properties of the NBD unit.
This new tri-branched tool could open the way to more precise optical detection of Gram-negative infections. Future iterations of the tri-branched probes could lead to the incorporation of three fluorophores to increase signal amplification, and the synthesis of multi-modal probes with additional diagnostic or therapeutic functionality. This also warrants further investigation of tri-branched fluorescent probes on in vivo infection models and clinical samples.
Author contributions
Conceptualization, M. K., M. R.-R., R. S., M. B.; synthesis M. K., M. R.-R., J. C., M. S., A. M.-F, photophysical characterization M. K.; biological validation, R.S.; writing – original draft preparation, M. K., R. S., M. R.-R, J. C.; writing – review and editing, M. K., R. S., M. R.-R., J. C., A. M.-F., M. B. All authors have read and agreed to the published version of the manuscript.This research was funded by Engineering and Physical Sciences Research Council (EPSRC, United Kingdom) (grant number EP/R005257/1 and EP/R018669/1). We thank the Royal Society (RS, United Kingdom) and the Science and Engineering Research Board (SERB, India) (grant number NIF/R1/192688) for a Newton International Fellowship. The authors thank Stuart Duncan (University of Edinburgh) for assistance with the synthesis of the probes.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare no conflict of interest.References
- K. S. Ikuta, L. R. Swetschinski, G. Robles Aguilar, F. Sharara, T. Mestrovic, A. P. Gray, N. Davis Weaver and E. E. Wool, et al. , Lancet, 2022, 400, 2221–2248 CrossRef PubMed.
- J. O'Neill, The Rev on AMR, H. M Government/Wellcome Trust, 2016.
- R. Laxminarayan, P. Matsoso, S. Pant, C. Brower, J.-A. Røttingen, K. Klugman and S. Davies, Lancet, 2016, 387, 168–175 CrossRef.
- M. S. Mulani, E. E. Kamble, S. N. Kumkar, M. S. Tawre and K. R. Pardesi, Front. Microbiol., 2019, 10, 539 CrossRef.
- P. Nauclér, A. Huttner, C. H. van Werkhoven, M. Singer, P. Tattevin, S. Einav and T. Tängdén, Clin. Microbiol. Infect., 2021, 27, 175–181 CrossRef.
- R. E. Brummett and R. B. Morrison, Arch. Otolaryngol., Head Neck Surg., 1990, 116, 406–410 CrossRef CAS.
- R. E. Campbell, C. H. Chen and C. L. Edelstein, Kidney Int. Rep., 2023, 8, 2211–2225 CrossRef PubMed.
- M. Exner, S. Bhattacharya, B. Christiansen, J. Gebel, P. Goroncy-Bermes, P. Hartemann, P. Heeg, C. Ilschner, A. Kramer, E. Larson, W. Merkens, M. Mielke, P. Oltmanns, B. Ross, M. Rotter, R. M. Schmithausen, H. G. Sonntag and M. Trautmann, GMS Hyg. Infect. Control, 2017, 12, Doc05 Search PubMed.
- U. Theuretzbacher, Curr. Opin. Microbiol., 2017, 39, 106–112 CrossRef CAS.
- J. A. Kim, D. J. Wales and G.-Z. Yang, Prog. Biomed. Eng., 2020, 2, 042001 CrossRef.
- H. M. Schouw, L. A. Huisman, Y. F. Janssen, R. H. J. A. Slart, R. J. H. Borra, A. T. M. Willemsen, A. H. Brouwers, J. M. van Dijl, R. A. Dierckx, G. M. van Dam, W. Szymanski, H. H. Boersma and S. Kruijff, Eur. J. Nucl. Med. Mol. Imaging, 2021, 48, 4272–4292 CrossRef CAS.
- P. Giacomo, R. Sheryl, K. Susanne and R. Thomas, J. Nucl. Med., 2020, 61, 1419 CrossRef PubMed.
- H. Kobayashi, M. Ogawa, R. Alford, P. L. Choyke and Y. Urano, Chem. Rev., 2010, 110, 2620–2640 CrossRef CAS.
- A. A. Ordonez, M. A. Sellmyer, G. Gowrishankar, C. A. Ruiz-Bedoya, E. W. Tucker, C. J. Palestro, D. A. Hammoud and S. K. Jain, Sci. Transl. Med., 2019, 11, eaax8251 CrossRef PubMed.
- M. M. Welling, A. W. Hensbergen, A. Bunschoten, A. H. Velders, H. Scheper, W. K. Smits, M. Roestenberg and F. W. B. van Leeuwen, Clin. Transl. Imaging, 2019, 7, 125–138 CrossRef.
- Z. Wang and B. Xing, Chem. Commun., 2022, 58, 155–170 RSC.
- Y. Huang, W. Chen, J. Chung, J. Yin and J. Yoon, Chem. Soc. Rev., 2021, 50, 7725–7744 RSC.
- P. Pristovšek and J. Kidrič, J. Med. Chem., 1999, 42, 4604–4613 CrossRef.
- T. Velkov, P. E. Thompson, R. L. Nation and J. Li, J. Med. Chem., 2010, 53, 1898–1916 CrossRef CAS PubMed.
- Z. Z. Deris, J. D. Swarbrick, K. D. Roberts, M. A. K. Azad, J. Akter, A. S. Horne, R. L. Nation, K. L. Rogers, P. E. Thompson, T. Velkov and J. Li, Bioconjug. Chem., 2014, 25, 750–760 CrossRef CAS PubMed.
- B. Yun, K. D. Roberts, P. E. Thompson, R. L. Nation, T. Velkov and J. Li, Sensors, 2017, 17, 2598 CrossRef.
- L. Miao, W. Liu, Q. Qiao, X. Li and Z. Xu, J. Pharm. Anal., 2020, 10, 444–451 CrossRef PubMed.
- M. R. L. Stone, M. S. Butler, W. Phetsang, M. A. Cooper and M. A. T. Blaskovich, Trends Biotechnol., 2018, 36, 523–536 CrossRef CAS.
- B. Mills, M. Bradley and K. Dhaliwal, Clin. Transl. Imaging, 2016, 4, 163–174 CrossRef.
- H.-Y. Kwon, X. Liu, E. G. Choi, J. Y. Lee, S.-Y. Choi, J.-Y. Kim, L. Wang, S.-J. Park, B. Kim, Y.-A. Lee, J.-J. Kim, N. Y. Kang and Y.-T. Chang, Angew. Chem., Int. Ed., 2019, 58, 8426–8431 CrossRef CAS.
- H. Chen, C. Liu, D. Chen, K. Madrid, S. Peng, X. Dong, M. Zhang and Y. Gu, Mol. Pharm., 2015, 12, 2505–2516 CrossRef CAS.
- C. Anaya, N. Church and J. P. Lewis, Proteomics, 2007, 7, 215–219 CrossRef CAS.
- F. Ishikawa, S. Konno, K. Takashima, H. Kakeya and G. Tanabe, Org. Biomol. Chem., 2021, 19, 8906–8911 RSC.
- W. Wang, Y. Wang, L. Lin, Y. Song and C. J. Yang, Anal. Bioanal. Chem., 2019, 411, 4017–4023 CrossRef CAS.
- M. Zheng, M. Zheng, S. Epstein, A. P. Harnagel, H. Kim and T. J. Lupoli, ACS Chem. Biol., 2021, 16, 1841–1865 CrossRef CAS.
- T. Velkov, K. D. Roberts, R. L. Nation, P. E. Thompson and J. Li, Future Microbiol., 2013, 8, 711–724 CrossRef CAS PubMed.
- Z. Yu, W. Qin, J. Lin, S. Fang and J. Qiu, BioMed. Res. Int., 2015, 679109 Search PubMed.
- A. R. Akram, S. V. Chankeshwara, E. Scholefield, T. Aslam, N. McDonald, A. Megia-Fernandez, A. Marshall, B. Mills, N. Avlonitis, T. H. Craven, A. M. Smyth, D. S. Collie, C. Gray, N. Hirani, A. T. Hill, J. R. Govan, T. Walsh, C. Haslett, M. Bradley and K. Dhaliwal, Sci. Transl. Med., 2018, 10, eaal0033 CrossRef PubMed.
- A. Megia-Fernandez, M. Klausen, B. Mills, G. E. Brown, H. McEwan, N. Finlayson, K. Dhaliwal and M. Bradley, Chemosensors, 2021, 9, 117 CrossRef CAS.
- C. Jiang, H. Huang, X. Kang, L. Yang, Z. Xi, H. Sun, M. D. Pluth and L. Yi, Chem. Soc. Rev., 2021, 50, 7436–7495 RSC.
- J. M. Ellard, T. Zollitsch, W. J. Cummins, A. L. Hamilton and M. Bradley, Angew. Chem., Int. Ed., 2002, 41, 3233–3236 CrossRef.
- A. R. Akram, N. Avlonitis, E. Scholefield, M. Vendrell, N. McDonald, T. Aslam, T. H. Craven, C. Gray, D. S. Collie, A. J. Fisher, P. A. Corris, T. Walsh, C. Haslett, M. Bradley and K. Dhaliwal, Sci. Rep., 2019, 9, 8422 CrossRef PubMed.
- M. Rodriguez-Rios, G. Rinaldi, A. Megia-Fernandez, A. Lilienkampf, C. T. Robb, A. G. Rossi and M. Bradley, Chem. Commun., 2023, 59, 11660–11663 RSC.
- J. M. Balkovec, D. C. Bensen, T. Blizzard, A. Borchardt, T. P. Brady, Z. Chen, Q.-Q. Thuy Do, W. Jiang, T. Lam, J. B. Locke, A. Noncovich and L. W. Tari, US Pat., WO2018006063A1, 2018 Search PubMed.
- G. Franc and A. K. Kakkar, Chem. Soc. Rev., 2010, 39, 1536–1544 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb01786b |
| ‡ Present address: School of Chemistry, Food and Pharmacy, University of Reading, Pepper Ln, Reading RG6 6DX, UK. |
| § Present address: Centre of Targeted Protein Degradation, University of Dundee, 1 James Lindsay Place, Dundee, DD1 5JJ, UK. |
| ¶ Present address: Department of Organic Chemistry, Faculty of Sciences, University of Granada, 18071 Granada, Spain. |
| This journal is © The Royal Society of Chemistry 2025 |