The biosynthesis of bisorbicillinoids: evidence for a biosynthetic route from bisorbicillinol to bisorbibutenolide and bisorbicillinolide
Naoki Abe*, Kazunobu Yamamoto, Tadaharu Arakawa and Akira Hirota
School of Food and Nutritional Sciences, University of Shizuoka, 52-1 Yada, Shizuoka, 422-8526, Japan.. E-mail: abe@fns1.u-shizuoka-ken.ac.jp; Tel: +81-54-264-5555; Fax: +81-54-264-5099
First published on 11th December 2000
Abstract
Biosynthetic incorporation of labelled sodium acetates into bisorbicillinol in Trichoderma sp. USF-2690 suggests that bisorbicillinol is derived from 12 acetate units. Successful bioconversion of the labelled bisorbicillinols to bisorbibutenolides (bislongiquinolides) and bisorbicillinolides using the washed mycelium of the strain suggests that there are biosynthetic routes from bisorbicillinol to bisorbibutenolide and from bisorbicillinol to bisorbicillinolide.
The ‘Bisorbicillinoids’, comprised of dimeric sorbicillin-related natural products, have recently been described by Nicolaou et al.1 Several compounds in the group, possessing complex structures, exhibit interesting biological activities, e.g., inhibition of the production of TNF-α by macrophages and monocytes,2 inhibition of β-1,6-glucan biosynthesis,3 and scavenging of the 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical.4–7 The biosynthesis of bisorbicillinoids is of interest to many investigators5,8–10 and, recently, biomimetic total synthesis has been investigated.11–13 In particular, the biosynthetic pathway of bisorbibutenolide5 (bislongiquinolide, 3)14 has been proposed to involve several distinct routes.5,10 We recently reported that Trichoderma sp. USF-2690 produced seven bisorbicillinoids, including bisorbicillinol 1, bisorbibutenolide 2, and bisorbicillinolide 3,4,5 with presumed biogenetic routes from 1 to 2 and from 1 to 3 through a branch point as a common intermediate anion (Scheme 1).5 The routes were postulated based on the finding that 13C-labeled sodium acetate was incorporated into 1via a polyketide pathway prior to incorporation into 2 and 3. We wished to establish the biosynthetic relationship between 1, 2, and 3, and report herein the result of a preliminary labelling experiment that provides evidence of the biosynthetic routes from 1 to 2 and from 1 to 3.
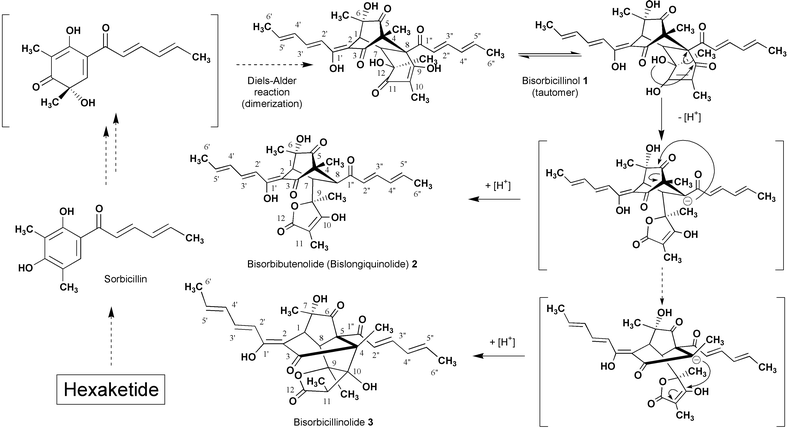 | ||
| Scheme 1 Biosynthetic pathway from bisorbicillinol 1 to bisorbibutenolide (bislongiquinolide 2) and bisorbicillinolide 3. | ||
Trichoderma sp. USF-2690 was first cultivated on a reciprocal shaker at 30 °C for 2 d.† The washed mycelium from the fermentation broth was then suspended in five 0.5-liter flasks each holding 150 mL of the medium (pH 7) containing 0.1% sodium [1-13C]acetate or 0.1% sodium [2-13C]acetate and 0.5% polypeptone, and again fermented with reciprocal shaking at 30 °C for 2 d. The filtered broth (1 L) was extracted with ethyl acetate (0.5 L × 2) at pH 3. The organic extract, concentrated in vacuo, was applied to a Sephadex LH-20 column and eluted with methanol. The fraction including 1 was rechromatographed using medium-pressure liquid chromatography (MPLC) under the following conditions: support, YMC-ODS-AQ 120-S50; solvent, acetonitrile–H2O (1∶1, containing 0.1% trifluoroacetic acid); detection, UV at 370 nm. Two types of labelled bisorbicillinol 1 were obtained. The experiment using sodium [1-13C] acetate gave 1 ([1-13C]-1) with 10 13C-enriched carbons (C-1, C-3, C-5, C-7, C-1′, C-3′, C-5′, C-1″, C-3″, and C-5″) in the 13C-NMR spectrum. The expected incorporations at C-9 and C-11 were not observed. We previously reported that bisorbicillinol 1 had a keto–enol tautomation between C-9 and C-11 in solution, and the 13C signals of C-9 and C-11 did not appear in the 13C-NMR spectrum.4 Therefore, the lack of peaks at C-9 and C-11 did not necessarily mean that the 13C atoms from sodium [1-13C]acetate were not incorporated at C-9 and C-11. On the other hand, the feeding experiment using sodium [2-13C]acetate enhanced the peak strength of 12 carbons (C-2, C-4, C-6, C-8, C-10, C-12, C-2′, C-4′, C-6′, C-2″, C-4″, and C-6″) in the 13C-NMR spectrum of 1 ([2-13C]-1). These results suggested that 1 was formed by the dimerization of six acetate units, which combined in the head-to-tail manner typical of fatty acids and polyketides, to give linkages between C-1 and C-7 and between C-4 and C-8 according to the Diels–Alder reaction.
The incorporation study employing each labelled bisorbicillinol 1 as a precursor of bioconversion was accomplished in the following manner. The fungus, inoculated in 0.5 L flasks containing 150 mL of the medium (pH 7) composed of 2.0% glucose and 0.5% polypeptone, was preincubated on a reciprocal shaker at 30 °C for 9 d. Mycelia were washed with sterilized water and then the washed mycelium was inoculated in 0.5 L flasks containing 150 mL of sterilized water with 10 mg of non-labelled and 5 mg of 13C-labelled bisorbicillinol 1. The cultures for isolation of bisorbibutenolide 2 and bisorbicillinolide 3 were incubated for 72 and 10 h, respectively. The filtrate obtained from each broth was adjusted to pH 3 and extracted with ethyl acetate. LH-20 column chromatography and repetitive MPLC yielded 2 and 3, respectively.
The 13C-NMR spectrum of bisorbibutenolide 2 obtained from the broth fed with [1-13C]-1 had 12 13C-enriched peaks at C-1, C-3, C-5, C-7, C-10, C-12, C-1′, C-3′, C-5′, C-1″, C-3″, and C-5″. This definitive result established the existence of a biological synthetic route from 1 to 2, as we had proposed previously.5 In addition, the enrichment of C-10 and C-12 of 2 meant that the two invisible carbons in the 13C-NMR spectrum of 1 were enriched by sodium [1-13C]acetate. Furthermore, the consecutive feeding study of [2-13C]-1 confirmed the validity of bioconversion from 1 to 2 in the fungal strain. This finding was consistent with the expectation based on a typical polyketide pathway that the 13C–enrichments at C-2, C-4, C-6, C-8, C-9, C-11, C-2′, C-4′, C-6′, C-2″, C-4″, and C-6″ were observed in the 13C-NMR spectrum of 2.
Proof of biosynthesis of bisorbicillinolide 3via bisorbicillinol 1 as a precursor, was then performed in the same manner. The bisorbicillinolide 3 obtained from the broth fed with [1-13C]-1 gave the characteristic 13C-NMR spectrum of 3, which had 12 peaks enhanced at C-1, C-3, C-6, C-8, C-10, C-12, C-1′, C-3′, C-5′, C-1″, C-3″, and C-5″. The continued experiment using [2-13C]-1 led to the expected 3, into which excess 13C-atoms were incorporated at C-2, C-4, C-5, C-7, C-9, C-11, C-2′, C-4′, C-6′, C-2″, C-4″, and C-6″. The results of a feeding study using [1-13C]-1 and [2-13C]-1 ascertained that bisorbicillinolide 3 was biosynthesized from bisorbicillinol 1 as a precursor.
To examine the possibility that the biosynthesis from bisorbicillinol 1 to bisorbibutenolide 2 occurred via bisorbicillinolide 3, or from 1 to 3via2, time course studies on bioconversion of products in the washed mycelium were performed using high performance liquid chromatography analysis. Non-labeled bisorbibutenolide 2 or bisorbicillinolide 3 was added to the washed mycelium prepared above, and the culture broth was incubated at 30 °C for 72 h. No other products were detected in either experiment.
These observations from the 13C-feeding studies indicate that there are biosynthetic routes from 1 to 2 and from 1 to 3 in Trichoderma sp. USF-2690. The supporting experiments might rule out the possibility of biosynthesis from 1 to 2via3 or from 1 to 3via2 and the reverse biosynthesis from 2 or 3 to 1. These results suggest the presence of the first anion as a branch point and the second anion following the C–C bond cleavage. Further incorporation studies will clarify the detailed biosynthetic mechanisms through the two precursor anions.
Acknowledgements
This work is supported in part by a grant-in-aid for Scientific Research (C) (No. 12660100) to A. H. from the Ministry of Education, Science, Sports and Culture of Japan.Notes and references
- K. C. Nicolaou, R. Jautelat, G. Vassilikogiannakis, P. S. Baran and K. B. Simonsen, Chem. Eur. J., 1999, 5, 3651 CrossRef CAS.
- G. A. Warr, J. A. Veitch, A. W. Walsh, G. A. Hesler, D. M. Pirnik, J. E. Lett, P-F. M. Lin, I. A. Medina, K. D. McBrien, S. Forenza and J. M. Clark and K. S. Lam, J. Antibiot., 1996, 49, 234 Search PubMed.
- M. Kontani, Y. Sakagami and S. Marumo, Tetrahedron Lett., 1994, 35, 2577 CrossRef CAS.
- N. Abe, T. Murata and A. Hirota, Biosci. Biotechnol. Biochem., 1998, 62, 661 Search PubMed.
- N. Abe, T. Murata and A. Hirota, Biosci. Biotechnol. Biochem., 1998, 62, 2120 Search PubMed.
- N. Abe, T. Murata, K. Yamamoto and A. Hirota, Tetrahedron Lett., 1999, 40, 5203 CrossRef CAS.
- N. Abe, K. Yamamoto and A. Hirota, Biosci. Biotechnol. Biochem., 2000, 64, 620 Search PubMed.
- L. S. Trifonov, H. Hilpert, P. Floersheim, A. S. Dreiding, D. M. Rast, R. Skrivanova and L. Hoesch, Tetrahedron, 1986, 42, 3157 CrossRef CAS.
- O. Shirota, V. Pathak, C. F. Hossain, S. Sekita, K. Takataori and M. Satake, J. Chem. Soc., Perkin Trans. 1, 1997, 2961 RSC.
- S. Sperry, G. J. Samuels and P. Crews, J. Org. Chem., 1998, 63, 10011 CrossRef CAS.
- D. Barnes-Seeman and E. J. Corey, Org. Lett., 1999, 1, 1503 CrossRef CAS.
- K. C. Nicolaou, K. B. Simonsen, G. Vassilikogiannakis, P.S. Baran, V. P. Vidali, E. N. Pitsinos and E. A. Couladouros, Angew. Chem., Int. Ed., 1999, 38, 3555 CrossRef CAS.
- K. C. Nicolaou, G. Vassilikogiannakis, K. B. Simonsen, P. S. Baran, Y.-L. Zhong, V. P. Vidali, E. N. Pitsinos and E. A. Couladouros, J. Am. Chem. Soc., 2000, 122, 3071 CrossRef CAS.
- R. Andrade, W. A. Ayer and L. S. Trifonov, Aust. J. Chem., 1997, 50, 255 CrossRef CAS.
Footnote |
| † The fermentation broth was pooled from ten 0.5 L flasks each containing 150 mL of the following medium: 2.0% glucose, 0.05% polypeptone, 0.2% yeast extract, 0.1% KH2PO4, 0.1% MgSO4•7H2O, and 0.1% trace salt mixture at pH 7. The filtered mycelial cake was washed with 2 L of sterilized water. |
| This journal is © The Royal Society of Chemistry 2001 |
