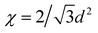Photo- and electropatterning of hydrogel-encapsulated living cell arrays
Dirk R.
Albrecht
a,
Valerie Liu
Tsang
a,
Robert L.
Sah
a and
Sangeeta N.
Bhatia
*ab
aDepartment of Bioengineering, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0412, USA. E-mail: sbhatia@ucsd.edu; Fax: 858-822-4203; Tel: 858-822-3142
bDepartment of Medicine, University of California San Diego, La Jolla, CA 92093, USA
First published on 24th November 2004
Abstract
Living cells have the potential to serve as sensors, naturally integrating the response to stimuli to generate predictions about cell fate (e.g., differentiation, migration, proliferation, apoptosis). Miniaturized arrays of living cells further offer the capability to interrogate many cells in parallel and thereby enable high-throughput and/or combinatorial assays. However, the interface between living cells and synthetic chip platforms is a critical one wherein the cellular phenotype must be preserved to generate useful signals. While some cell types retain tissue-specific features on a flat (2-D) surface, it has become increasingly apparent that a 3-D physical environment will be required for others. In this paper, we present two independent methods for creating living cell arrays that are encapsulated within a poly(ethylene glycol)-based hydrogel to create a local 3-D microenvironment. First, ‘photopatterning’ selectively crosslinks hydrogel microstructures containing living cells with ∼100 µm feature size. Second, ‘electropatterning’ utilizes dielectrophoretic forces to position cells within a prepolymer solution prior to crosslinking, forming cell patterns with micron resolution. We further combine these methods to obtain hierarchical control of cell positioning over length scales ranging from microns to centimeters. This level of microenvironmental control should enable the fabrication of next-generation cellular microarrays in which robust 3-D cultures of cells are presented with appropriate physical and chemical cues and, consequently, report on cellular responses that resemble in vivo behavior.
Introduction
Living cells offer unique advantages as the sensing element of biological assays compared with existing strategies based on biomolecules (e.g., antibodies, nucleic acids, proteins). In particular, the integrated, functional response of a mammalian cell can report on the activity, mechanism of action, and ultimate consequences of exposure to exogenous agents and stimuli, all of which may be difficult to predict with biomolecular detection assays.1 Such cell- and tissue-based sensors have numerous applications in drug development (e.g., toxicity screening),2 medical diagnostics,3 and detection of known and unknown pathogens.4Miniaturized platforms for cell-based sensors are expected to provide the same economies of scale, increased sample throughput at reduced cost, that have revolutionized the study of genomics with nucleic acid and protein microarrays5,6 and accelerated histological and molecular profiling via microarrays of nonvital cells7 or tissues.8 Current tools for micropatterning arrays of living cells rely on selective adhesion of cells onto flat two-dimensional (2-D) substrates;9 in this manner, researchers have shown that cell-cell interactions,10,11 cell shape,12 and extracellular matrix13 can regulate cell fate and tissue specific functions. However, these micropatterns often deteriorate over time14 and cells require several hours to adhere and spread over the substrate thereby adopting a flattened morphology. While two-dimensional culture can provide an adequate representation of cellular behavior in many instances, other cell types respond favorably to a 3-D physical microenvironment by alterations in cell shape, gene expression, and resemblance to in vivo responses.15–20 Thus, the development of cell microarray platforms that retain cells in 3-D gel culture, rather than on rigid 2-D substrates, represents an important step toward cell-based assays that accurately predict in vivo behavior.
Hydrogels are increasingly popular biomaterials for 3-D cell culture because their high water content and mechanical properties resemble those of tissues in the body.21 Additionally, many hydrogels can be formed in the presence of cells by photocrosslinking. Poly(ethylene glycol) (PEG)-based hydrogels are of particular interest because of their biocompatibility, hydrophilicity, and ability to be customized by varying chain length or chemically adding biological molecules.22 These materials have been used to immobilize various types of cells that can attach, proliferate, and produce matrix within millimeter-scale hydrogels.23,24 Hydrogel microstructures containing living cells were recently demonstrated by selective photocrosslinking,25,26 but these methods are limited to ∼100 µm feature size and are unable to define the cell-cell interactions that potentially modulate cell behavior. Thus, high resolution patterning methods capable of controlling cell organization within the hydrogel environment would enable reproducible control over the cellular microenvironment and could be beneficial for the maintenance of cell functions.
Electropatterning systems based on dielectrophoretic (DEP) forces are able to localize single cells within a fluid volume to micron-scale resolution, providing precise control over cell shape, organization, and interactions.27,28 Such electrokinetic systems move living cells rapidly toward positions dictated by the local electric field strength established via micropatterned surface electrodes.29–31 DEP force advantageously operates on all cells in parallel, in contrast to serial methods such as optical tweezers that move cells one by one or in small groups.32–34 Furthermore, careful selection of electrical properties has allowed efficient cell manipulation without compromising cell viability or apparent behavior.35–37 Thus, DEP electropatterning is well suited for adaptation to the photosensitive PEG-based hydrogel system.
The objective of this paper is to demonstrate the fabrication of living cell arrays encapsulated within 3-D hydrogels by two methods: photopatterning and DEP electropatterning. Furthermore, we show that these methods can be combined into a versatile system capable of defining hydrogel and cell micropatterns over length scales ranging from microns to centimeters. This capability should enable the fabrication of next-generation microarrays of cells that are presented with appropriate 3-D physical and chemical cues and, consequently, report on cellular responses that resemble in vivo behavior.
Experimental
Microfabrication
To establish regions of high and low electric field strength for cell localization during DEP electropatterning, reusable planar microelectrode arrays were fabricated using a single-layer photolithographic process. A thin layer of electrically insulative photoresist was micropatterned upon a conductive bus plane such that all electrodes (i.e., holes within the resist layer) were energized using a single external connection. Aluminosilicate glass slides coated with indium tin oxide (ITO), a transparent conductor (Delta Technologies, Stillwater, MN), were cleaned in sequential solvent washes, exposed to oxygen plasma (Technics 500 II Asher, 100W) for 5 min, and baked at 110 °C to promote resist adhesion. The negative, epoxy-based photoresist, SU-8 2 (Microchem, Newton, MA), was statically dispensed and ramped to a final spin speed of 2500 RPM for 20 sec. The photoresist was soft baked for 12 min at 95 °C to evaporate the solvent, cooled, and exposed on a contact mask aligner (Kasper 2001, 2 mW cm−2, 365 nm) for 25 s through a patterned photomask. Emulsion masks were designed with CorelDraw and commercially printed at 8000 dpi (CAD/Art Services, Poway, CA). The slides were then baked for 75 min at 65 °C, developed in Nano XP, and cured at 185 °C for 60 min to complete crosslinking and to enhance the physical stability of the 1.5 µm thick film. The array was then soaked in distilled water overnight to leach out any residual species from the patterning process and ensure the biocompatibility of the SU-8 film.Patterning apparatus
A closed, transparent, parallel-plate flow chamber was designed to permit external UV light exposure while preventing oxygen quenching of the hydrogel polymerization reaction. A thin silicone rubber gasket (100 µm or 250 µm thick) was sandwiched between the electrode microarray slide and another ITO-coated slide outfitted with fluidic connections to enclose an 8 × 20 mm chamber area (Fig. 1, Step 1). The chamber components were assembled and sterilized with 70% ethanol for 15 min followed by several buffer washes. To discourage cell adhesion to the walls, surfaces were treated with Pluronic F108 (1% in H2O; Spectrum Quality Products, Gardena, CA), a triblock copolymer previously shown to resist protein adsorption and decrease cell adhesion.38 Also, the adhesion of hydrogel microstructures to internal chamber surfaces was either prevented by exposure to vapors of tridecafluoro-1,1,2,2-tetrahydrooctyl 1-trichlorosilane (United Chemical Technologies, Bristol, PA), or promoted with 3-(trimethoxysilyl) propyl methacrylate (Sigma) to provide free surface-bound reactive groups that bind with the polymer during UV exposure.25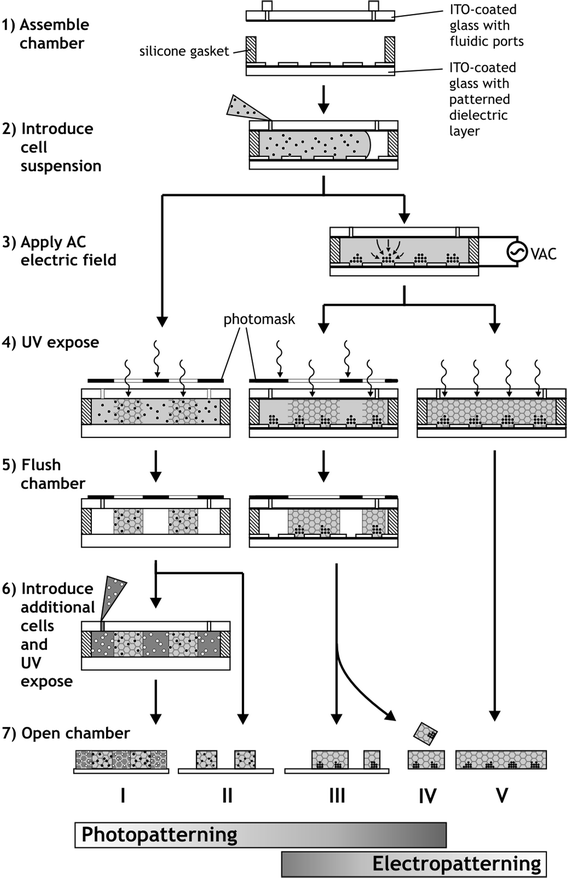 | ||
| Fig. 1 Process flow for the formation of hydrogel microstructures containing living cells by hydrogel photopatterning, cell electropatterning, or a combination of both techniques. Classified by Protocol number, resulting hydrogels are either photopatterned (I, II, III, IV) or unpatterned (V), contain cells randomly encapsulated (I, II) or in clusters (III, IV, V), are linked to a substrate (I, II, III) or free-floating (IV, V), and may contain multiple cell types (e.g., I). | ||
Cell culture
Swiss 3T3 murine fibroblasts (American Type Culture Collection, Manassas, VA) were cultured in 150 cm2 flasks (Fisher, Springfield, NJ) and passaged in preconfluency no more than 15 times. The cells were maintained in Dulbecco's Modified Eagle Medium (DMEM; Gibco, Grand Island, NY) supplemented with 10% bovine calf serum, 100 μg mL−1 penicillin, and 100 μg mL−1 streptomycin and incubated in 5% CO2 at 37 °C. Prior to patterning experiments, cells were released from tissue culture flasks into suspension. Alternatively, freshly isolated primary rat hepatocytes38 were used for some experiments. Some cells were fluorescently labeled with chloromethylfluorescein diacetate (CMFDA; C-2925, Molecular Probes, Eugene, OR) or chloromethylbenzoylaminotetramethyl rhodamine (CMTMR; C-2927, Molecular Probes) for identification at (ex/em) 492/517 and 514/565 nm wavelengths, respectively.Hydrogel chemistry
Poly(ethylene glycol) diacrylate (PEGDA) (3.4 kDa; Nektar Hydrogels, Huntsville, AL) was obtained in lyophilized form and dissolved in either phosphate-buffered saline (pH 7.4) or a custom electropatterning buffer (see below). The photoinitiator 2-hydroxy-1-[4-(hydroxyethoxy)phenyl]-2-methyl 1-propanone (Irgacure 2959; Ciba, Tarrytown, NY) was dissolved in 70% ethanol in water or 1-vinyl-2-pyrrolidinone (Sigma, St. Louis, MO) and added to the prepolymer solution. Upon UV exposure, the photosensitive PEGDA solution crosslinks covalently via radical chemistry.25,39Photopatterning
A prepolymer solution containing 20% w/v PEGDA, 20 × 106 cells mL−1 and 0.1% w/v Irgacure 2959 photoinitiator in PBS was injected into the chamber (Fig. 1, Step 2). A photomask was secured to the chamber top with emulsion side facing the glass. Exposed regions of hydrogel were crosslinked by UV illumination (365 nm filter and collimating lens, EXFO Lite, Mississauga, ON) at 64 mW cm−2 for 100 s (Fig. 1, Step 4). The remaining uncrosslinked prepolymer solution and cells were then flushed from the chamber with PBS (Fig. 1, Step 5). To add additional cell types, the next cell/prepolymer solution was injected into the chamber and flowed around already crosslinked hydrogel microstructures (Fig. 1, Step 6). Further exposure to UV light through different masks formed additional cell-laden hydrogel domains. This procedure is illustrated as Protocols I and II in Fig. 1.Electropatterning
Cells prepared for DEP electropatterning were washed twice in a HEPES-buffered glucose solution (HBG; 306 mOsm, pH 7.35). The low ionic strength of this custom electropatterning buffer (conductivity: 21.4 mS m−1) is required for rapid DEP cell patterning towards regions of high electric field strength.29,30 Immediately prior to patterning, cells were added to form a prepolymer suspension with final concentrations of 20% w/v PEGDA, 6–25 × 106 cells mL−1, and 0.1% w/v Irgacure 2959 in HBG.The flow chamber with a 100 µm gasket was flushed with buffer and filled with ∼20 µL PEGDA cell suspension (Fig. 1, Step 2). A sinusoidal AC signal (3 MHz, 3.0 Vrms) was then applied between the conductive top slide and the bottom microelectrode array via a function generator (Agilent 33120A, Palo Alto, CA) and monitored by an oscilloscope (Tektronix TDS2014, Beaverton, OR) connected in parallel. The resulting AC electric field is strongest near the electrodes at the microarray surface, and DEP-induced cell motion toward these locations was observed by microscopy (Fig. 1, Step 3). Following cell localization, the chamber was exposed to UV light to crosslink the entire hydrogel (Fig. 1, Step 4), depicted by Protocol V in Fig. 1. Because the chamber is transparent on both top and bottom (Fig. 2A), illumination was possible either from the top with an external UV lamp, or from below with the microscope's mercury arc lamp (Fig. 2B), filtered to emit 340–380 nm wavelengths at 25 mW cm−2.
 | ||
| Fig. 2 Photograph and diagram of the patterning apparatus. (A) A photomask placed beneath demonstrates transparency of the flow chamber. (B) The apparatus is loaded onto the microscope stage for visualizing cell patterning and for UV exposure by either the microscope or an external source. | ||
Combined photo- and electropatterning
The transparent DEP electropatterning apparatus is amenable to photopatterning, allowing for further microstructure complexity. Cell electropatterning proceeded as in the previous section, except that UV illumination through a photomask selectively crosslinked hydrogel structures only the exposed regions (Fig. 1, Steps 4). Subsequently, uncrosslinked polymer was flushed with buffer and, in some cases, additional prepolymer/cell suspensions were introduced into the chamber and further UV exposed through additional masks (Fig. 1, Steps 5–6). This process is described by Protocols III and IV in Fig. 1.Microscopy and quantitation
Crosslinked hydrogel microstructures, either free or attached to the glass substrate, were removed from the apparatus, transferred into Petri dishes (Fig. 1, Step 7), and observed in epifluorescence and Hoffman modulation contrast using a Nikon TE300 inverted microscope and digital camera (Photometrics CoolSnap HQ; Roper Scientific, Tucson, AZ). To characterize cell cluster size, some electropatterned hydrogels were treated sequentially with 4% paraformaldehyde and 0.1% Triton-X to fix and permeabilize cells, followed by 4 µg mL−1 ethidium homodimer-1 (Molecular Probes) for 30 min to fluorescently stain cell nuclei. Gels were then mounted on slides and viewed by epifluorescence or laser scanning confocal microscopy (Bio-Rad MRC-1024UV, Hercules, CA). The number of cells per cluster was quantified with a semi-automated cell counting program (MetaMorph, Universal Imaging, Westchester, PA) using an intensity-based algorithm and verified in several microscope fields by manual counts.Results and discussion
Photopatterned cell arrays
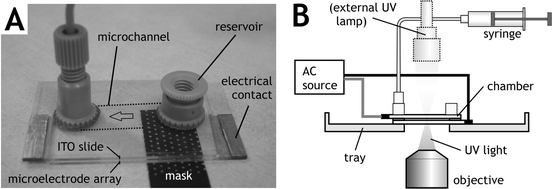
Hydrogel microstructures encapsulating a random distribution of rat hepatocytes were photopatterned in an array format with two distinct cellular constituents (Fig. 3). One array contained circular 500 µm diameter cell-laden islands surrounded by a field of another cell type, both embedded within a uniform 250 µm thick PEGDA hydrogel (Fig. 3A). This microstructure was formed by sequential crosslinking steps: photopatterning the first cell type (green), rinsing away the uncrosslinked polymer and cells, flooding the chamber with the second cell type (red), and crosslinking again by overall exposure to UV light (Protocol I). Each island of ∼50 nL volume contained approximately 1000 homogeneously distributed cells. This type of array would be useful when cells of primary interest require proximity of a secondary supportive cell type to maintain their normal cellular function, such as non-parenchymal cell stabilization of hepatocyte function in co-culture.11 | ||
| Fig. 3 Examples of photopatterned hydrogel microstructures containing living cells. (A) Red and green-labeled cells encapsulated within distinct 500 µm diameter domains of a 250 µm thick hydrogel layer (Protocol I), viewed in brightfield montage and epifluorescence. (B) Cells encapsulated within an array of micropatterned hydrogel islands (Protocol II). Repeated photopatterning through an incrementally shifted mask forms a multiphase microstructure array containing many cell types (C). | ||
A second multiphase example contained individual cell-laden hydrogel islands (Fig. 3B). This array was formed as in the preceding example, by first photopatterning one cell type (red), rinsing away the uncrosslinked polymer and cells, flowing in the second cell type (green), and photopatterning and rinsing again (Protocol II). For the second UV exposure step, the photomask was shifted, such that the circular green cell islands were separated from the red cell islands by a 200 µm gap. While a two-step process is shown, the sequence could be repeated as many times as desired, for example, to create a regular hexagonal array of islands containing four distinct cell types (Fig. 3C).
Because hydrogel microstructure dimensions are defined in the x–y-plane by the printed photomask features, noncircular islands of arbitrary shape may be formed. Furthermore, hydrogel thickness in the z-direction is easily adjustable by the silicone spacer thickness, although structures less than 50 µm tall may impose significant shear forces on large cells during chamber loading. For some applictions, smaller diameter features may be desired. However, the minimum feature size of photopatterned hydrogels is on the order of 100 µm when limited to cytocompatible gelling conditions, due to feature widening from hydrogel swelling, light scattering, or radical diffusion.25,26 Because this length scale is significantly greater than cell dimensions, it is currently not possible to define cell-cell interactions solely by photopatterning. The high-resolution electrokinetic cell patterning method addresses this limitation.
Electropatterned cell arrays
Large-scale arrays of single cells or cell clusters (up to ∼100 cells each) were embedded within thin hydrogel slabs using the DEP electropatterning method. These examples utilized a 15 × 20 mm array of hexagonally-packed electrodes with 25 µm diameter and 75 or 100 µm spacing. The sinusoidal potential applied across the electropatterning chamber, from the bottom microelectrode array to the top conductive plane, generated local regions of high electric field strength at the surface of each electrode. Resulting DEP forces moved cells rapidly toward the electrodes and formed an array of cell clusters within the prepolymer solution in 90–150 s. The entire chamber was then exposed to UV light to initiate crosslinking (Protocol V). The pattern of arrayed, encapsulated cells was preserved during careful opening of the chamber and transfer of the hydrogel to a Petri dish (Fig. 4). Very few cells remained on the electrode array slide, indicating low cell adhesion to the chamber surface and excellent pattern retention within the gel. In Fig. 4A, a free-floating hydrogel slab containing electropatterned fibroblasts in a uniform cluster array with 100 µm spacing is shown folded over to indicate the 100 µm thickness. The area density of clusters in a hexagonal array iswhere d is array spacing, such that hydrogels with d = 100 or 75 µm contain 115 or 205 clusters mm−2, and nearly 12 000 or 21 000 total clusters per >100 mm2 gel, respectively. At higher magnification, individual cluster organization is visible (Fig. 4B). Cluster size is controlled via the cell suspension density, ρ, with cell number per cluster defined by N = ρh/χ, where h is chamber height. Average cluster size in Fig. 4 was 4.8 ± 1.9 (SD) cells per cluster, with 83% of clusters containing 3–7 cells each (Fig. 4C). The size histogram matches a Poisson distribution due to the stochastic nature of electropatterning, in which cells at random initial locations move independently towards the nearest regularly-spaced electrode.
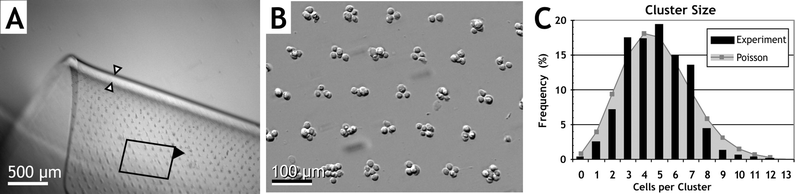 | ||
| Fig. 4 DEP electropatterned hydrogel formed via Protocol V. (A) Low-magnification view of fibroblast clusters arrayed 100 µm apart within a 100 µm thick PEGDA hydrogel (arrows). The gel is free-floating and folded, showing the embedded cluster array in the upper portion; lower portion is out of the focal plane. (B) Magnified view of the array in panel A as indicated. (C) Histogram of cell cluster size for a similar construct with a density of 115 clusters mm−2 and average of 4.8 ± 1.9 (SD) cells per cluster. Poisson distribution is computed using average cluster size. | ||
We have demonstrated that electropatterning localizes cells into clusters of controlled size within a 3-D hydrogel, at a resolution capable of defining cell-cell contacts and communication. The planar flow chamber geometry allows straightforward design of cell patterns that correspond directly to the surface electrode pattern. In this example, electrode spacing and chamber height (gel thickness) were selected based on theoretical models indicating that DEP electropatterning would occur most rapidly when these parameters are approximately equal.40 Because patterning slows exponentially as feature spacing increases,28,40 the maximum spacing (e.g., between adjacent clusters of an array) is limited to a few hundred µm in practice. Thus, the operating length scales of photopatterning (∼100 µm) and electropatterning (between one and hundreds of micrometres) do not overlap appreciably, and a combination of both methods would greatly expand overall resolution capabilities.
Combined photo- and electropatterned cell arrays
Several types of two-phase hierarchical cell arrays were constructed in which multiple circular hydrogel microstructures each contained a high-resolution array of embedded cell clusters (Fig. 5). In the first example (Fig. 5A–D), cells were electropatterned into clusters 75 µm apart and then photopatterned into circular hydrogel islands via UV exposure through a mask, as described by Protocol III. Next, a cell-free prepolymer solution was introduced into the chamber, thereby flushing out uncrosslinked cells and polymer. Overall UV exposure then formed a single 8 × 10 mm rectangular hydrogel slab containing the complex cell array. At higher magnification (Fig. 5C,D), encapsulated cells are seen centered over the visible electrodes. Each 450 µm diameter array element contained approximately 35 cell clusters with an average of ∼6 cells per cluster.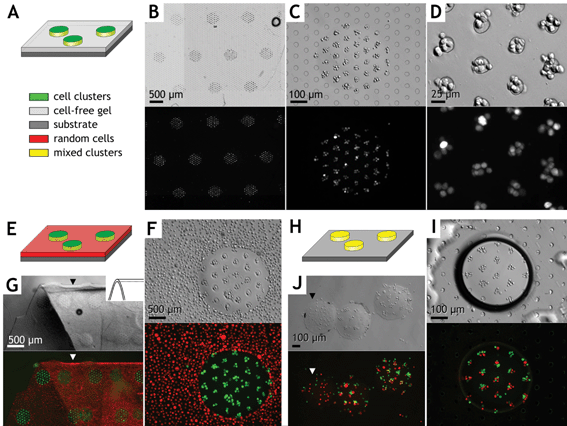 | ||
| Fig. 5 Examples of combined photo- and electropatterned hydrogel microstructures. Micrographs are shown in Hoffman modulation contrast above and identical fields in epifluorescence below. Example 1 (A–D): Green-labeled cell clusters were arrayed 75 µm apart within 450 µm diameter domains of a 100 µm thick rectangular hydrogel slab (Protocol III). Low (B), medium (C), and high (D) magnification images of a single construct are shown. The microelectrode array is visible in panels B, C, and I (circles). Example 2 (E–G): Hydrogel microstructure array with electropatterned green-labeled cells surrounded by a field of randomly distributed red-labeled cells (i.e., without electropatterning). The hydrogel is shown released from the chamber and folded (G) to demonstrate cross sectional thickness (arrows) and retention of complex cell patterns within the flexible hydrogel. Example 3 (H–J): A mixed population of green and red labeled cells formed heterogeneous clusters 100 µm apart and within individual 400 µm diameter hydrogel microstructures and surrounded by air (I, dark boundary in Hoffman modulation contrast). (J) Free cylindrical hydrogel “pucks” containing the heterogeneous cluster arrays after release from the glass substrate (Protocol IV). Note one “puck” (J, arrow) contains unclustered cells created via Protocol II from the same mixed cell population. | ||
The second example represents a co-culture array with two cellular constituents (Fig. 5E–G). A cluster array of cells labeled with a green fluorophore was first formed similar to the previous example. Next, uncrosslinked prepolymer was flushed from the chamber with buffer and replaced with a second cell/prepolymer solution containing red-labeled cells. In this example, the red cells were encapsulated in a random configuration by not energizing the electrodes. Each cell type was spatially confined within the hydrogel, indicating effective washing between crosslinking events. The hydrogel slab was then carefully removed from the chamber using sterile forceps and retained all cell patterns when floating freely (Fig. 5G).
A third example demonstrates the fabrication of mixed cell clusters containing multiple cell types (Fig. 5H–J). A prepolymer solution with equal proportions of red- and green-labeled cells was patterned into clusters 100 µm apart within 400 µm diameter hydrogel microstructures, as described by Protocol III. Uncrosslinked monomers were flushed with air, resulting in individual hydrated gel islands (Fig. 5I). Some islands were then removed from the glass substrate (Protocol IV), resulting in free cylindrical hydrogel “pucks” containing an array of heterogeneous clusters (Fig. 5J). Such prefabricated cellular microstructures would be useful as elements in nonpositional format multiplexed assays or incorporated into lab-on-a-chip devices when their formation in situ is not feasible.
The combined photo- and electropatterning system offers numerous advantages over current cell micropatterning techniques. Because they do not rely on adhesion events that are relatively slow (hours), these methods are significantly faster (minutes) and equally effective for both adherent and nonadherent cell types. These methods are independent and therefore retain full functionality when combined. In particular, photopatterning allows for the multiplexing of different cell types and/or hydrogel formulations, as has proven useful for high-throughput 2-D cell-matrix screening,41 while electropatterning provides micrometre-scale resolution for defining cell–cell interactions.
Cell viability assays
The transparent hydrogel microstructures are amenable to imaging and detection via fluorescence or colorimetric cell assays, even when the hydrogel slab remains on a transparent chamber slide. The viability of cultured fibroblasts and primary hepatocytes following photo- and electropatterning was assessed with CellTracker dyes (Fig. 5B–D, lower panels). In this assay, cell viability is indicated by the fluorescent product of cytoplasmic esterase activity and the retention of these fluorophores within the cytosol (Figs. 3 and 5) that would otherwise diffuse away if the cell membrane was compromised. Additionally, fibroblast viability was quantified after 1–2 h for some microstructures (Live/Dead kit, Molecular Probes) and found to be consistent among electropatterned hydrogels (95 ± 1%), photocrosslinked but unpatterned hydrogels (93 ± 1%), and also the initial prepolymer/cell solution (97% by Trypan Blue exclusion). Thus, cell damage was not observed during hydrogel photopatterning or cell electropatterning. The UV and electric field exposure parameters utilized here were previously determined to be safe for various mammalian cell types;25,27,28 nonetheless, both are potentially harmful for living cells and merit discussion. UV-induced cytotoxicity is primarily due to free radical damage rather than direct radiation effects.25 Sensitivity to UV exposure intensity, wavelength, and duration, and photoinitiator type and concentration is highly dependent on cell type. While minimization of these parameters reduces cytotoxicity, photopatterning performance may also be compromised. Previously, hydrogel pattern resolution was shown to decrease appreciably with low intensity and extended exposure time, possibly due to the diffusion of radicals within the solution.25 Thus, the UV exposure parameters used in this paper represent a balance between high resolution hydrogel microstructures and good cell viability.The electric field required for DEP localization is also potentially dangerous to living cells, due primarily to induced transmembrane voltages and secondarily to current-induced Joule heating of the prepolymer.37 In our system, high frequency AC excitation minimizes the induced membrane potential, and the low conductivity buffer and microscale electrode dimensions eliminate the temperature hazard. To reduce electric field exposure time, we patterned cells rapidly but safely by applying the greatest voltage that demonstrated no discernible effect on endothelial cell morphology and proliferation rate,27 two metrics that are broadly indicative of cell health. Other mammalian, yeast, and bacterial cells have also been manipulated via DEP in aqueous media,31 and we have electropatterned primary or cultured fibroblasts, epithelial cells, chondrocytes, and hepatocytes from human, bovine, and murine sources in hydrogels.28
For long-term cultures, photo- and electropatterned hydrogel microstructures fabricated aseptically remained free of contamination for 7 weeks (data not shown). Furthermore, pattern fidelity of 3-D encapsulated cells was preserved up to this time point, unlike most selective adhesion methods.14
Hydrogel properties
In addition to interactions with neighboring cells, the cell microenvironment is defined by contacts with extracellular matrix, soluble factors, and physical forces.42 In these preliminary studies, the PEG-based hydrogel was biologically inert and nondegradable. The 20% w/v solution of 3400 MW PEGDA produced a relatively firm hydrogel (360 kPa) with an estimated mesh size of 6 nm.43 While this network allowed quick diffusion of small molecules, such as cell nutrients and fluorescent assay substrates, antibody-based detection methods may not be compatible with this hydrogel formulation due to diffusion limitations (IgG hydrodynamic radius ∼7–8 nm). However, the hydrogel transport and mechanical properties can be tailored by the molecular weight and polymer fraction of the PEG monomers to alter pore size, diffusion characteristics, and hydrogel stiffness.22,43,44 Finally, photo- and electropatterning are compatible with recent techniques to present a well-defined chemical environment to encapsulated cells. For example, acrylated biomolecules become covalently incorporated into the PEG hydrogel when mixed into the prepolymer to specify cell–matrix interactions, provide cell anchorage sites, or allow hydrolytic or enzyme-specific hydrogel degradation.23,24,45Conclusion
Effective cell-based assays are required to be stabile, sensitive, and responsive to stimuli in a useful manner. The patterning methods described herein advance toward this goal by incorporating into a cell array platform a hydrated 3-D cellular microenvironment that is critical for the viability and phenotypic stability of many cell types. We have demonstrated the formation of various PEG hydrogel microstructures containing living cells organized in specific cell micropatterns or randomly dispersed, and illustrated their compatibility with a fluorescence-based assay. Hydrogel photopatterning allows the formation of multiple domains that may contain many cell types or hydrogel formulations but does not specify microscale cell organization, whereas DEP electropatterning provides high resolution cell localization to control cell-cell interactions within a single domain. These methods were then combined into a rapid and versatile system with both multiplexed and high resolution cell pattern capabilities, a wide operating range from microns to centimeters, and compatibility with both adherent and nonadherent cell types. Along with emerging hydrogel biomaterials with defined chemistry and physical properties, these tools for cell organization within a 3-D microenvironment may improve the sensitivity and performance of future cell-based sensors.Acknowledgements
Funding provided by the Whitaker Foundation (DRA, VLT), American Association of University Women (VLT), NSF CAREER (SNB), the David and Lucille Packard Foundation, NIH NIDDK, and NASA.References
- J. J. Pancrazio, J. P. Whelan, D. A. Borkholder, W. Ma and D. A. Stenger, Ann. Biomed. Eng., 1999, 27(6), 697–711 CrossRef CAS.
- K. Durick and P. Negulescu, Biosens. Bioelectron., 2001, 16(7–8), 587–92 CrossRef CAS.
- A. S. Rudolph and J. Reasor, Biosens. Bioelectron., 2001, 16(7–8), 429–31 CrossRef CAS.
- D. A. Stenger, G. W. Gross, E. W. Keefer, K. M. Shaffer, J. D. Andreadis, W. Ma and J. J. Pancrazio, Trends Biotechnol., 2001, 19(8), 304–9 CrossRef CAS.
- A. C. Pease, D. Solas, E. J. Sullivan, M. T. Cronin, C. P. Holmes and S. P. Fodor, Proc. Natl. Acad. Sci. USA, 1994, 91(11), 5022–6.
- R. Z. Wu, S. N. Bailey and D. M. Sabatini, Trends Cell Biol., 2002, 12(10), 485–8 CrossRef CAS.
- K. Oode, T. Furuya, K. Harada, S. Kawauchi, K. Yamamoto, T. Hirano and K. Sasaki, Am. J. Pathol., 2000, 157(3), 723–8 Search PubMed.
- O. P. Kallioniemi, U. Wagner, J. Kononen and G. Sauter, Hum. Mol. Genet., 2001, 10(7), 657–62 CAS.
- K. Bhadriraju and C. S. Chen, Drug Discovery Today, 2002, 7(11), 612–20 CrossRef CAS.
- C. M. Nelson and C. S. Chen, FEBS Lett., 2002, 514(2–3), 238–42 CrossRef CAS.
- S. N. Bhatia, U. J. Balis, M. L. Yarmush and M. Toner, FASEB J., 1999, 13(14), 1883–900 Search PubMed.
- C. S. Chen, M. Mrksich, S. Huang, G. M. Whitesides and D. E. Ingber, Science, 1997, 276(5317), 1425–1428 CrossRef CAS.
- P. Clark, S. Britland and P. Connolly, J Cell Sci., 1993, 105(Pt 1), 203–12 Search PubMed.
- C. M. Nelson, S. Raghavan, J. L. Tan and C. S. Chen, Langmuir, 2003, 19(5), 1493–1499 CrossRef CAS.
- A. Abbott, Nature, 2003, 424(6951), 870–2 CrossRef CAS.
- W. Mueller-Klieser, Am. J. Physiol., 1997, 273(4 Pt 1), C1109–23 Search PubMed.
- S. Levenberg, N. F. Huang, E. Lavik, A. B. Rogers, J. Itskovitz-Eldor and R. Langer, Proc. Natl. Acad. Sci. USA, 2003, 100(22), 12741–6 CrossRef CAS.
- P. D. Benya and J. D. Shaffer, Cell, 1982, 30(1), 215–24 CrossRef CAS.
- E. Cukierman, R. Pankov and K. M. Yamada, Curr. Opin. Cell Biol., 2002, 14(5), 633–9 CrossRef CAS.
- J. P. Stegemann and R. M. Nerem, Exp. Cell Res., 2003, 283(2), 146–55 CrossRef CAS.
- K. Y. Lee and D. J. Mooney, Chem. Rev., 2001, 101(7), 1869–79 CrossRef.
- N. A. Peppas, P. Bures, W. Leobandung and H. Ichikawa, Eur. J. Pharm Biopharm, 2000, 50(1), 27–46 CrossRef CAS.
- A. S. Gobin and J. L. West, FASEB J., 2002, 16(7), 751–3 Search PubMed.
- D. L. Hern and J. A. Hubbell, J. Biomed. Mater. Res., 1998, 39(2), 266–76 CrossRef CAS.
- V. A. Liu and S. N. Bhatia, Biomed. Microdev., 2002, 4(4), 257–266 Search PubMed.
- W. G. Koh, A. Revzin and M. V. Pishko, Langmuir, 2002, 18(7), 2459–62 CrossRef CAS.
- D. S. Gray, J. L. Tan, J. Voldman and C. S. Chen, Biosens. Bioelectron., 2003 Search PubMed In press.
- D. R. Albrecht, R. L. Sah and S. N. Bhatia, IEEE Proceedings of the 24th Annual International Conference of the Engineering in Medicine and Biology Society, 2002, vol. 2, p. 1708–1709 Search PubMed.
- H. A. Pohl, Dielectrophoretic Cell Patterning within Tissue Engineering Scaffolds, Cambridge University Press, Cambridge, 1978, p. 579 Search PubMed.
- T. B. Jones, Electromechanics of Particles, Cambridge University Press, Cambridge, 1995, p. 265 Search PubMed.
- R. Pethig, Crit. Rev. Biotechnol., 1996, 16(4), 331–348 Search PubMed.
- A. Ashkin, Proc. Natl. Acad. Sci. USA, 1997, 94(10), 4853–60 CrossRef CAS.
- M. Ozkan, T. Pisanic, J. Scheel, C. Barlow, S. Esener and S. N. Bhatia, Langmuir, 2003, 19(5), 1532–1538 CrossRef CAS.
- R. A. Flynn, A. L. Birkbeck, M. Gross, M. Ozkan, B. Shao, M. M. Wang and S. C. Esener, Sens. Actuators B, 2002, 87(2), 239–243 CrossRef.
- A. Docoslis, N. Kalogerakis and L. A. Behie, Cytotechnology, 1999, 30(1–3), 133–142 CrossRef.
- S. Archer, T. T. Li, A. T. Evans, S. T. Britland and H. Morgan, Biochem. Biophys. Res. Commun., 1999, 257(3), 687–98 CrossRef CAS.
- H. Glasser and G. Fuhr, Bioelectrochem. Bioenerg., 1998, 47(2), 301–310 CrossRef CAS.
- V. A. Liu, W. E. Jastromb and S. N. Bhatia, J. Biomed. Mater. Res., 2002, 60(1), 126–34 CrossRef CAS.
- M. B. Mellott, K. Searcy and M. V. Pishko, Biomaterials, 2001, 22(9), 929–41 CrossRef CAS.
- D. R. Albrecht, R. L. Sah and S. N. Bhatia, Biophys. J., 2004, 87(4), 2131–47 CrossRef CAS.
- D. G. Anderson, S. Levenberg and R. Langer, Nat. Biotechnol., 2004, 22(7), 863–6 CrossRef CAS.
- B. Palsson and S. Bhatia, Tissue Engineering, Pearson Prentice Hall, Upper Saddle River, NJ, 2004 Search PubMed.
- S. J. Bryant and K. S. Anseth, J. Biomed. Mater. Res., 2002, 59(1), 63–72 CrossRef CAS.
- G. M. Cruise, D. S. Scharp and J. A. Hubbell, Biomaterials, 1998, 19(14), 1287–94 CrossRef CAS.
- B. K. Mann, A. S. Gobin, A. T. Tsai, R. H. Schmedlen and J. L. West, Biomaterials, 2001, 22(22), 3045–51 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2005 |